ABSTRACT
Cellular aging occurs as a cell loses its ability to maintain homeostasis. Aging cells eliminate damaged cellular compartments and other senescence factors via self-renewal. The mechanism that regulates cellular rejuvenation remains to be further elucidated. Using budding yeast gametogenesis as a model, we show here that the endosomal sorting complex required for transport (ESCRT) III regulates nuclear envelope organization. During gametogenesis, the nuclear pore complex (NPC) and other senescence factors are sequestered away from the prospore nuclei. We show that the LEM-domain protein Heh1 (Src1) facilitates the nuclear recruitment of ESCRT-III, which is required for meiotic NPC sequestration and nuclear envelope remodeling. Furthermore, ESCRT-III-mediated nuclear reorganization appears to be critical for gamete rejuvenation, as hindering this process curtails either directly or indirectly the replicative lifespan in gametes. Our findings demonstrate the importance of ESCRT-III in nuclear envelope remodeling and its potential role in eliminating senescence factors during gametogenesis.
KEYWORDS: ESCRT-III, lem-domain protein, nuclear pore complex, nuclear envelope, meiosis, replicative lifespan, cellular aging
Introduction
Cellular aging occurs as a cell’s ability to maintain homeostasis deteriorates, affecting both the chronological age, the cell can attain and its replicative lifespan. Replicative lifespan refers to the number of daughter cells a mother cell generates before death; dysregulation of replicative lifespan is characteristic of diseases of aging and cancer [1,2]. The resetting of replicative potential is best characterized in budding yeast, S. cerevisiae [3–5]. Vegetative yeast cells undergo asymmetric cell division, in which the mother cell retains aging factors such as old or damaged cellular compartments, protein aggregates, and extra chromosomal rDNA circles, allowing the daughter cell to inherit a full replicative lifespan [6–10]. Interestingly, self-renewal has also been observed during gametogenesis in budding yeast [11], in which nuclear divisions are symmetrical. However, the molecular mechanism regulating gametogenic self-renewal remains to be determined.
A key factor in mitigating cellular aging is proper nuclear trafficking between the cytoplasm and nucleoplasm [12]. The eukaryotic nucleus is enclosed by two membranes that separate these regions, forming a selective barrier. The outer nuclear membrane (ONM) is continuous with the endoplasmic reticulum (ER), whereas the inner nuclear membrane (INM) interacts with the nucleoplasm [13]. Embedded within the nuclear envelope are nuclear pore complexes (NPCs). NPCs form a ~ 50 MD protein channel that is composed of concentric inner, outer, and membrane ring assemblies that scaffold a central transport channel, cytosolic filaments, and nuclear basket [14,15]. These NPC domains are constructed from modular subcomplexes of proteins termed nucleoporins or nups [15,16]. NPCs are the primary method of nuclear trafficking, imposing a diffusion barrier to molecules larger than ~40 kD, while also providing binding sites for the rapid and selective transport of molecules containing nuclear import and export signals [17–19]. The semipermeable barrier provided by NPCs aids in maintaining nuclear integrity and nuclear envelope homeostasis. Loss of nuclear compartmentalization through NPC damage or assembly defects is associated with cellular aging [12,20,20,21].
Approximately 100 NPCs are formed during budding yeast mitosis [22]. NPC biogenesis likely occurs through an inside-out evagination of the INM followed by membrane fusion with the ONM [23], resulting in nuclear envelope holes containing NPC assembly intermediates that develop into mature NPCs [24,25]. Due to the crucial function of NPCs in maintaining cellular health, a surveillance mechanism is in place to ensure proper biogenesis [21]. Surveillance of NPC biogenesis is mediated by the LEM (Lap2-emerin-MAN1) family of integral INM proteins, Heh1 (also called Src1; for consistency, we use Heh1 hereafter) and Heh2, in addition to the endosomal sorting complex required for transport (ESCRT) III subunits Chm7, Snf7, and the AAA-ATPase Vps4 which function to destabilize and clear defective NPC assembly intermediates [21,25]. It has been proposed that Heh1 is required for the focal accumulation of Chm7 at the nuclear envelope in genetic backgrounds where NPC assembly is inhibited and ESCRT-III activation at the nuclear envelope is likely through a direct interaction between Snf7 and Chm7 in conjunction with Heh1 and Heh2 [24,25].
In the absence of the ESCRT-III surveillance mechanism, malformed NPCs in mitosis accumulate in a storage of improperly assembled nuclear pore complexes compartment, or SINC [21]. The SINC, containing misassembled or damaged NPCs, is not passed on to daughter cells but instead remains in the mother with other unwanted materials, resulting in a daughter cell free of senescence factors [21,26,27]. While there is sufficient evidence for ESCRT-III and its associated proteins function in NPC quality control during budding yeast mitosis, little is known about the regulation of NPCs and cellular aging in meiosis. In contrast to mitosis, meiosis consists of two rounds of chromosome segregation resulting in four gametes, also referred to as spores in budding yeast [28]. Unlike the asymmetric division in vegetative yeast cells, meiotic cells reset aging symmetrically such that all gametes have a renewed replicative lifespan regardless of the age of the mother cell [11,29]. Importantly, senescence factors originally present in the mother cell, such as protein aggregates and extrachromosomal rDNA circles, are no longer present in the newly formed gametes [11,29–31].
We show here that nucleoporins are selectively sequestered away from the newly formed gametes during meiosis II, and this process is regulated by the ESCRT-III complex. In the absence of ESCRT-III function, gametes exhibit a reduced replicative lifespan, suggesting that ESCRT-III-mediated nuclear envelope remodeling is crucial for gamete rejuvenation. Our work provides evidence for a potential ESCRT-III mediated meiotic mechanism for sequestering unwanted nuclear materials in a mother cell, resulting in the renewal of the newly formed gametes.
Materials and Methods
Yeast strains and plasmids used in this study
Yeast strains used in this study are isogenic to the SK1 background and are listed in Table 1. To tag the C-terminus of Hta1, Heh1, Vps4, Nup49, Pom34, and other nucleoporins with either GFP or mApple, we used a standard PCR-based homologous recombination method [32]. Similarly, we used this technique to tag Vps4 with V5 and an auxin-inducible degron (AID) in tandem to generate Vps4-V5-AID and to generate Vps4-GFP. These tagged alleles are the only functional copies of the corresponding genes in the yeast genome. A comparable PCR-based method was used to replace the HEH1, HEH2, CHM7, and SPO21 open reading frames with either a KanMX4 or Hygromycin-B cassette to generate gene deletions. Correct transformations were further confirmed by colony-based diagnostic PCR. Primers are listed in Table 2.
Table 1.
Yeast strains used in this study.
| Strain | Background | Genotype | Experiment |
|---|---|---|---|
| HY6229 | SK1 | MATa/MATα, his3∆200, leu2-k, ura3, lys2, ho::LYS2, POM34-GFP//his3∆200, leu2-k, ura3, lys2, ho::LYS2, POM34-GFP, HTA1-mApple | Figures 1b, 4e, 4f, 4g, 4i, and 6 |
| HY6279 | SK1 | MATa/MATα, his3∆200, leu2-k, ura3, lys2, ho::LYS2, POM34-GFP, NUP49-mApple//his3∆200, leu2-k, ura3, lys2, ho::LYS2, POM34-GFP, NUP49-mApple | Figure 1c |
| HY6200 | SK1 | MATa/MATα, his3∆200, leu2-k, ura3, lys2, ho::LYS2, HTA1-mApple//his3∆200, leu2-k, ura3, lys2, ho::LYS2, PDMC1-MPS3-NC | Figure 2a |
| HY6695 | SK1 | MATa/MATα, ho::LYS2, lys2, ura3, leu::hisG, his3::hisG, trp1::hisG, PGAL-NDT80::TRP1, ura3::PGPD1-GAL4.ER::URA3, PGAL1-GFP-Mps2//ho::LYS2, lys2, ura3, leu::hisG, his3::hisG, trp1::hisG, PGAL-NDT80::TRP1, ura3::PGPD1-GAL4.ER::URA3, PGAL1-GFP-Mps2, HTA1-mApple | Figures 2b and 2c |
| HY6586-2 | SK1 | MATa/MATα, his3∆200, leu2-k, ura3, lys2, ho::LYS2, heh1∆::KAN, POM34-GFP//his3∆200, leu2-k, ura3, lys2, ho::LYS2, heh1∆::KAN, POM34-GFP, HTA1-mApple | Figures 3b, 3d, 3e, 4e, and 6 |
| HY6356 | SK1 | MATa/MATα, his3∆200, leu2-k, ura3, lys2, ho::LYS2, heh2∆::KAN, POM34-GFP//his3∆200, leu2-k, ura3, lys2, ho::LYS2, heh2∆::KAN, POM34-GFP, HTA1-mApple | Figures 3c, 3d, and 3e |
| HY6389 | SK1 | MATa/MATα, his3∆200, leu2-k, ura3, lys2, ho::LYS2, heh1∆::KAN, heh2∆::KAN, POM34-GFP, HTA1-mApple//his3∆200, leu2-k, ura3, lys2, ho::LYS2, heh1∆::KAN, heh2∆::KAN, POM34-GFP, HTA1-mApple | Figures 3d and 3e |
| HY6541 | SK1 | MATa/MATα, his3∆200, leu2-k, ura3, lys2, ho::LYS2, chm7∆::HB, POM34-GFP//his3∆200, leu2-k, ura3, lys2, ho::LYS2, chm7∆::HB, POM34-GFP, HTA1-mApple | Figures 4a, 4e, 4f, 4i, and 6 |
| HY6585 | SK1 | MATa/MATα, his3∆200, leu2-k, ura3, lys2, ho::LYS2, heh1∆::KAN, chm7∆::HB, POM34-GFP, HTA1-mApple//his3∆200, leu2-k, ura3, lys2, ho::LYS2, heh1∆::KAN, chm7∆::HB, POM34-GFP, HTA1-mApple | Figures 4b, 4e, 4g, 4i, and 6 |
| HY6608 | SK1 | MATa/MATα, his3∆200, leu2-k, ura3, lys2, ho::LYS2, spo21∆::KAN, POM34-GFP//his3∆200, leu2-k, ura3, lys2, ho::LYS2, spo21∆::KAN, POM34-GFP, HTA1-mApple | Figures 4 C, 4E, 4 H, and 4I |
| HY6614 | SK1 | MATa/MATα, his3∆200, leu2-k, ura3, lys2, ho::LYS2, spo21∆::KAN, chm7∆::HB, POM34-GFP//his3∆200, leu2-k, ura3, lys2, ho::LYS2, spo21∆::KAN, chm7∆::HB, POM34-GFP, HTA1-mApple | Figures 4D, 4E, 4 H, and 4I |
| HY6724 | SK1 | MATa/MATα, his3Δ200, leu2-k, ura3, lys2, ho::LYS2, PDMC1-HEH1-mApple, VPS4-GFP::HIS5//his3Δ200, leu2-k, ura3, lys2, ho::LYS2, PDMC1-HEH1-mApple, VPS4-GFP::HIS5 | Figure 5A |
| HY6663 | SK1 | MATa/MATα, ho::LYS2, lys2, ura3, leu::hisG, his3::hisG, trp1::hisG, PGAL-NDT80::TRP1, ura3::PGPD1-GAL4.ER::URA3, PDMC1-TIR1::LEU2, VPS4-V5-AID::HIS5, POM34-GFP, HTA1-mApple//ho::LYS2, lys2, ura3, leu::hisG, his3::hisG, trp1::hisG, PGAL-NDT80::TRP1, ura3::PGPD1-GAL4.ER::URA3, PDMC1-TIR1::LEU2, VPS4-V5-AID::HIS5, POM34-GFP, HTA1-mApple |
Figures 5B, 5 C, 5D, 5 F, 5 G, 5 H, 5I and 6 |
| HY6644 | SK1 | MATa/MATα, ho::LYS2, lys2, ura3, leu::hisG, his3::hisG, trp1::hisG, PGAL-NDT80::TRP1, ura3::PGPD1-GAL4.ER::URA3, POM34-GFP, HTA1-mApple//ho::LYS2, lys2, ura3, leu::hisG, his3::hisG, trp1::hisG, PGAL-NDT80::TRP1, ura3::PGPD1-GAL4.ER::URA3, POM34-GFP, HTA1-mApple | Figures 5B, 5E and 5I |
| HY6705 | SK1 | MATa/MATα, ho::LYS2, lys2, ura3, leu::hisG, his3::hisG, trp1::hisG, PGAL-NDT80::TRP1, ura3::PGPD1-GAL4.ER::URA3, PDMC1-TIR1::LEU, VPS4-V5-AID, heh1∆::KAN, POM34-GFP, HTA1-mApple//ho::LYS2, lys2, ura3, leu::hisG, his3::hisG, trp1::hisG, PGAL-NDT80::TRP1, ura3::PGPD1-GAL4.ER::URA3, PDMC1-TIR1::LEU2, VPS4-V5-AID, heh1∆::KAN, POM34-GFP, HTA1-mApple | Figures 5B, 5 F, 5 G, 5 H and 5I |
Table 2.
Primers used in this study.
| Primer Name | Sequence |
|---|---|
| POM34-tagF | GCAAATATGCATATATGATGAACTCACAGTCCCCAAGGGGGAAAATAGCGGCCGCTCTAGAACTAGT |
| POM34-tagR | TATATAGCTATGGAAAGTATTAAATGTTTTTTTGCTGTTTTCCCCCTCGAGGTCGACGGTA |
| HTA1-tagF | GTTGCCAAAGAAGTCTGCCAAGGCTACCAAGGCTTCTCAAGAATTAGCGGCCGCTCTAGAACTAGTGG |
| HTA1-tagR | GCAGTTTAGTTCCTTCCGCCTTCTTTAAAATACCAGAACCGATCTCCCCCTCGAGGTCGACGGTATCG |
| NUP49-tagF | GTTACATCAAAAAACGAAAACACTGGCATCATTGAGCATAGCGGCCGCTCTAGAACTAGT |
| NUP49-tagR | ACTTGTTATACGCACTATATAAACTTTCAGGGCGATTTACCCCCCTCGAGGTCGACGGTA |
| HEH1-deletionF | TCACCCTGAACGGAAATCAA |
| HEH1-deletionR | TTTCTTTCCTCCATGTGTCG |
| HEH2-deletionF | TGACAAGCACTATCTTCCAAAGT |
| HEH2-deletionR | GTATGCGTAGGGGAAGGGAT |
| HEH1-tagF | TCGATGAAAAGGTTAAACCGCAGATCCCGCAGTTACGGAAAtCAGGGGCATGATGTGACT |
| HEH1-tagR | TTCCGCCAATGTTGTTGTCAGTGGGTGATAACTAGAGAAAGCTCGTTTTCGACACTGGAT |
| CHM7-deletionF | AGTGCAGCGTTAGTAGAGACAATAAGAGGAGTTTTAAATTCTTAAACAGGGGCATGATGTGACT |
| CHM7-deletionR | TGCACAGGTCCTTCATTTGTATTTATCTTCAGATTATTCAATCTCTTTAATAGCTCGTTTTCGACACTGGAT |
| SPO21-deletionF | TCTGGGTTCAAGAATTCCTCAGA |
| SPO21-deletionR | GTTTCTTCGGCAACCCTGTA |
| VPS4-tagF | CTTGCTGAAGCAAGAACAGTTCACTAGAGAGATTTTGGTCAAGAAGGTAACGCGGCCGCTCTAGAACTAGTGG |
| VPS4-tagR | TATTTTCATGTACACAAGAAATCTACATTAGCACGTTAATCAATTGACCCCTCGAGGTCGACGGTATCG |
Plasmids used in this study are included in Table 3. To sufficiently deplete Vps4-V5-AID, the plant F-box protein Tir1 is present in addition to auxin. We generated a PDMC1-TIR1 (pHG273) construct to effectively overexpress TIR1 in meiosis. Integration was achieved by linearizing pHG273 with StuI and integrating at the URA3 locus by yeast transformation. Uracil – positive colonies were confirmed by colony-based diagnostic PCR. To overproduce Mps2 in meiosis II, we used the GAL promoter to construct PGAL-GFP-MPS2 (pHG527) by placing the MPS2 open reading frame under the control of the GAL promoter. We linearized plasmid pHG527 with StuI and integrated it at the endogenous MPS2 locus by yeast transformation. Note that the endogenous MPS2 remains intact and functional. Leucine-positive colonies were confirmed by diagnostic PCR. Similarly, we generated a PDMC1-HEH1-mApple (pHG742) construct to produce Heh1-mApple specifically in meiosis. Integration was achieved by linearizing pHG742 with XbaI and integrating at the HEH1 locus. Note that the endogenous HEH1 remains intact and functional. Positive colonies were confirmed by diagnostic PCR.
Table 4.
Gene deletions used in genetic screen.
| Systematic name | Ploidy | Standard name |
|---|---|---|
| YML107C | MAT a | PML39 |
| YMR129W | MAT a | POM152 |
| YHR004 C | MAT a | NEM1 |
| YER027C | MAT a | GAL83 |
| YDR205W | MAT a | MSC2 |
| YBR150C | MAT a | TBS1 |
| YIL030C | MAT a | SSM4 |
| YAL009W | MAT a | SPO7 |
| YAR002W | MAT a | NUP60 |
| YAR027W | MAT a | UIP3 |
| YAR042W | MAT a | SWH1 |
| YAR044W | MAT a | SWH1 |
| YBL079W | MAT a | NUP170 |
| YBR097W | MAT a | VPS15 |
| YBR170C | MAT a | NPL4 |
| YDL019C | MAT a | OSH2 |
| YDL088C | MAT a | ASM4 |
| YDL089W | MAT a | NUR1 |
| YDL116W | MAT a | NUP84 |
| YDR073W | MAT a | SNF11 |
| YDR120C | MAT a | TRM1 |
| YDR159W | MAT a | SAC3 |
| YDR192 C | MAT a | NUP42 |
| YDR395W | MAT a | SXM1 |
| YDR410C | MAT a | STE14 |
| YDR458C | MAT a | HEH2 |
| YDR532 C | MAT a | KRE28 |
| YEL017W | MAT a | GTT3 |
| YGL016W | MAT a | KAP122 |
| YGL035C | MAT a | MIG1 |
| YGL086W | MAT a | MAD1 |
| YGL115W | MAT a | SNF4 |
| YGL241W | MAT a | KAP114 |
| YGR202 C | MAT a | PCT1 |
| YGR212W | MAT a | SLI1 |
| YHL020C | MAT a | OPI1 |
| YHR076W | MAT a | PTC7 |
| YHR134W | MAT a | WSS1 |
| YHR195W | MAT a | NVJ1 |
| YIL016W | MAT a | SNL1 |
| YIL149C | MAT a | MLP2 |
| YJL080C | MAT a | SCP160 |
| YJL079C | MAT a | PRY1 |
| YJL073W | MAT a | JEM1 |
| YJL030W | MAT a | MAD2 |
| YKL057C | MAT a | NUP120 |
| YKL068W | MAT a | NUP100 |
| YKR044W | MAT a | UIP5 |
| YLL023 C | MAT a | POM33 |
| YLR018C | MAT a | POM34 |
| YLR262 C | MAT a | YPT6 |
| YLR240W | MAT a | VPS34 |
| YLR265C | MAT a | NEJ1 |
| YLR335W | MAT a | NUP2 |
| YML034W | MAT a | SRC1 |
| YMR153W | MAT a | NUP53 |
| YMR255W | MAT a | GFD1 |
| YMR284W | MAT a | YKU70 |
| YNL199C | MAT a | GCR2 |
| YNL159C | MAT a | ASI2 |
| YNL008C | MAT a | ASI3 |
| YNL012W | MAT a | SPO1 |
| YOR112W | MAT a | CEX1 |
| YOR311 C | MAT a | DGK1 |
| YOL072W | MAT a | THP1 |
| YPL200W | MAT a | CSM4 |
| YPL192 C | MAT a | PRM3 |
| YPL186C | MAT a | UIP4 |
| YPL125W | MAT a | KAP120 |
| YLR064W | MAT a | PER33 |
| YBR273 C | MAT a | UBX7 |
| YCR045C | MAT a | RRT12 |
| YCR086W | MAT a | CSM1 |
| YKR082W | MAT a | NUP133 |
| YLR450W | MAT a | HMG2 |
| YER110C | MAT a | KAP123 |
| YER120W | MAT a | SCS2 |
| YER123W | MAT a | YCK3 |
| YMR065W | MAT a | KAR5 |
| YNR075W | MAT a | COS10 |
| YDR424C | MAT a | DYN2 |
| YML103 C | MAT a | NUP188 |
Table 3.
Plasmids used in this study.
| Plasmid Name | Description |
|---|---|
| pHG363 | PDMC1-GFP-MPS3-NC, LEU2 |
| pHG527 | PGAL1-GFP-MPS2, LEU2 |
| pHG496 | PGAL1-HEH1, LEU2 |
| pHG273 | PDMC1-TIR1, URA3 |
| pHG740 | PDMC1-TIR1, LEU2 |
| pHG742 | PDMC1-HEH1-mApple, LEU2 |
To determine nuclear envelope permeability during meiosis we utilized a nucleus-localized TetR-GFP fusion protein we previously reported [33].
The following alleles have been reported previously: PDMC1-GFP-MPS3-NC, PGPD1-GAL4.ER, and PGAL-NDT80 [34,35]. The gene mutations and tagged alleles were first generated in MATa and MATα haploid cells; homozygous diploids (zygotes) were obtained by mating the corresponding haploids.
Yeast culture methods
Yeast cells were grown in YPD (1% yeast extract, 2% peptone, and 2% dextrose) at 30°C. To induce meiosis, YPD cultures were diluted with YPA (1% yeast extract, 2% peptone, and 2% potassium acetate) to reach OD (optical density, λ = 600 nm) of 0.2 and incubated at 30°C for approximately 14 h to reach a final OD of ~1.6–1.8. Yeast cells were then washed once in water and resuspended in 2% potassium acetate to induce meiosis. Cells were allowed to grow for approximately 6 h before microscopy to ensure that the majority of cells had initiated meiosis II. The onset of meiosis II is referred to as time zero in our time-lapse microscopy experiments.
To induce the expression of the GAL promoter in meiosis, 2 μM β-estradiol (final concentration) was added to a portion of the culture medium 6 h after transferring to 2% potassium acetate. An equal volume of ethanol was added to the control strains. To initiate the degradation of AID tagged proteins, 2 mM 1-napthylene acetic acid (final concentration) was added to a portion of the culture medium 6 h after transferring to 2% potassium acetate. An equal volume of ethanol was added to the control strains. Cultures were shaken vigorously at 30°C and protein extracts collected at designated time points prior to time-lapse microscopy.
Time-lapse fluorescence microscopy
Prior to microscopy, yeast cells were prepared as described previously [33]. Briefly, an agarose pad with 2% potassium acetate was prepared on a concave slide, and a small aliquot of yeast cells were placed on the agarose, sealed with a coverslip, and scoped for the desired time duration. Time-lapse fluorescence microscopy was carried out on a DeltaVision imaging system (GE Healthcare Life Sciences) at 30°C. We used a 60x (NA = 1.40) objective lens on an inverted microscope (IX-71, Olympus). Microscopic images were acquired with a CoolSNAP HQ2 charge-coupled device camera (Photometrics). Pixel size was set at 0.10700 μm. Time intervals were set at 5 min, and optical sections at 12, each with 0.5 μm thickness. Ultra-high signal-to-background coated custom filter sets were used. For GFP, the excitation spectrum was at 470/40 nm, emission spectrum at 525/50 nm; for RFP, excitation was at 572/35 nm, and emission at 632/60 nm. To minimize photo toxicity to the cells and photo bleaching to fluorophores, we used neutral density filters to limit excitation light to 32% or less of the normal equipment output.
Microscopy data analysis
Acquired microscopy images were deconvolved using the SoftWorx package (GE Healthcare Life Sciences). Projected images are used for figure display. To determine the fluorescence intensity of Pom34-GFP at the midzone and around the meiotic nuclei, we defined a 16 × 16 pixel area covering the entirety of the cell and obtained the total GFP fluorescence intensity from single optical sections. We then defined a 10 × 10 pixel area covering a single nucleus based on the Hta1-mApple signal and obtained the total GFP fluorescence intensity from single optical sections. The net percent intensity was determined by subtracting the background from each nucleus and the total GFP signal of the cell. The mean percentage of GFP localized to the nuclei of at least 25 cells was analyzed in each strain. Mutant phenotypes were analyzed similarly, or based on observed Hta1-mApple signal for cells with more than four masses. We used one-way ANOVA tests to determine statistical significance.
Genetic screen to identify NPC interacting factors
From the yeast deletion collection (ATCC-GSA-5), a pooled MATa library containing deletions of individual open reading frames that encode known nuclear-envelope-associated proteins was used to screen the localization of Pom34-GFP. The library was crossed to a yeast strain containing Pom34-GFP and Hta1-mApple. After mating, meiosis was induced in diploids by resuspending in 2% potassium acetate. Resulting tetrads were dissected, genotyped, and corresponding homozygous zygotes were generated. Yeast cells were induced to undergo meiosis in 2% potassium acetate and visualized periodically for progression through meiosis II with a 100× objective lens (NA = 1.40) mounted on a motorized epifluorescence microscope (AxioImager, Zeiss).
Protein extraction and western blotting
Yeast aliquots were withdrawn at indicated times for protein extraction with the trichloroacetic acid (TCA) method as described previously [36]. Briefly, 4 mL of yeast cells were collected, resuspended in 2.5% ice cold TCA, and then incubated at 4°C for 10 min. Cell pellets were stored at −80°C, and proteins were extracted by bead beating with a mini bead-beater homogenizer for 90 seconds at 4°C. Pom34-GFP was detected by an anti-GFP mouse monoclonal antibody (1:10 K dilution, Thermo Fisher Scientific, cat#GF28R). Vps4-V5-AID was detected by an anti-V5 antibody (1:5 K, Thermo Fisher Scientific, cat#R960-25). The level of Pgk1 was probed by a Pgk1 antibody (Thermo Fisher Scientific, cat#PA5-28,612) to serve as a loading control. Horseradish peroxidase-conjugated secondary antibodies, goat anti-mouse (Bio-Rad, cat#1,706,516), were used to probe the proteins of interest by an enhanced chemiluminescence (ECL) kit (Bio-Rad, cat#1,705,060). The ChemiDoc MP Imaging System (Bio-Rad, cat#17,001,402) was used to detect the ECL-based western blot. To calculate the relative protein abundance at each time point, individual band intensities were measured using the IPLab Imaging Software in conjunction with an in-house GelAnalyzer script and exported to Microsoft Excel. Target protein band intensities were made relative to those of the loading control (Pgk1).
Replicative lifespan analysis
Lifespan analysis was carried out by micromanipulation as described previously [37] with the following modification. Cells were grown to saturation in YPD at 30°C. To induce meiosis, YPD cultures were diluted and washed once in water. Cultures were then resuspended in 2% potassium acetate and incubated for 16 h at 30°C to ensure the completion of meiosis. These cells were digested with 2% glusulase at 4°C for about 12 hr and plated on YPD plates where tetrads were identified and each gamete was isolated. After each cell division of the gamete, the daughter cell was removed and the number of generations was recorded. At least 15 tetrads, totaling 60 spores, were used for each experiment. Cells were grown at 30°C and stored overnight at 4°C. Every cell was followed until cell lysis was observed.
Results
Nucleoporins are sequestered during meiosis II in budding yeast
During our investigation of the meiotic nuclear envelope in budding yeast, we observed that Nup49, an FG-nucleoporin of the NPC central core [38], aggregated to the mid plane of a dividing anaphase II nucleus (Figure 1a and see below). By live-cell fluorescence microscopy, we found similarly that Pom34, a subunit of the transmembrane ring of the NPC [39], aggregated like Nup49 Figures 1b and 1c. Our findings therefore confirm that nucleoporins are sequestered during meiosis II, leading to the formation of the fifth nuclear component [30,31], which is also called GUNC for gametogenesis uninherited nuclear compartment [40].
Figure 1.

Sequestration of nucleoporins Pom34 and Nup49 to the GUNC during meiosis II. (a) Schematic representation of meiosis II in budding yeast. Note that nucleoporins (purple) are sequestrated to the GUNC and are largely excluded from the newly formed gamete nuclei. Blue dots represent the yeast spindle pole body; red lines, prospore membrane. (b) Time-lapse live-cell microscopy showing Pom34-GFP localization in meiosis II. Time 0 refers to the perceived start point of meiosis II based on nuclear morphology elucidated by the Hta1-mApple marker. Note that during anaphase II the majority of Pom34-GFP is aggregated to the midzone of the dividing nucleus. Projected images from 12 optical sections are shown. (c) Colocalization of Pom34-GFP and Nup49-mApple in meiosis II. Time-lapse live-cell microscopy was performed as in B. Note that Pom34 and Nup49 colocalize and are both largely excluded from the developing nuclei. Dashed lines show cell boundary.
To visualize Pom34 and Nup49, we generated alleles with their encoding proteins tagged C-terminally with either GFP or mApple Figures 1b and 1c. The expression of these alleles was under the control of their endogenous promoters, and they served as the only functional copies in the yeast genome. We used histone H2A, one of the two copies encoded by HTA1, fused to mApple to serve as a marker for the nucleus, allowing us to determine meiotic cell progression on the basis of nuclear morphology. By time-lapse live-cell fluorescence microscopy, we found that Pom34-GFP was sequestered to the GUNC during anaphase II and was largely excluded from the newly forming nuclei Figure 1b. We also found the fusion protein, TetR-GFP, which harbors a nuclear localization signal, was retained inside the nucleus throughout meiosis, with no discernable GFP signal present in the cytoplasm (Supplemental Figure 1). Therefore, sequestration of Pom34 to GUNC does not impair nuclear trafficking during meiosis.
To confirm that nucleoporin aggregation is not limited to certain NPC subcomplexes, we analyzed Pom34-GFP and Nup49-mApple together and observed their colocalization throughout meiosis, resulting in a change in positioning from along the nuclear periphery to the GUNC compartment Figure 1c. We also observed the aggregation of nucleoporins Nup170 and Pom152, as shown previously [31]. We therefore conclude that nucleoporins, including Nup49, Nup170, Pom34 and Pom152, are sequestered to GUNC and are largely excluded from the developing nuclei after anaphase II.
Sequestration to the GUNC compartment is selective to nucleoporins
To determine if non-NPC proteins at the nuclear envelope are sequestered to GUNC, we used the INM protein Mps3 and ONM protein Mps2 as representatives. We have shown previously that Mps3, also localized to the spindle pole body (SPB), is cleaved and degraded during yeast meiosis [35]. To stabilize Mps3, we used a meiosis-specific non-degradable MPS3 allele, PDMC1-GFP-MPS3-NC [35]. As expected, GFP-Mps3-NC was concentrated at the SPB but also localized along the nuclear periphery throughout meiosis Figure 2a. Notably, Mps3-NC did not appear to be confined to the GUNC compartment during anaphase II like the nucleoporins Figure 2a.
Figure 2.

Localization of Mps3-NC and Mps2 in meiosis II. (a) Localization of Mps3-NC in meiosis. Time-lapse live-cell microscopy showing the localization of INM protein Mps3 during meiosis II. Time 0 refers to the perceived start point of meiosis II based on nuclear morphology as in Figure 1b. Note that during anaphase II GFP-Mps3-NC remains on the nuclear periphery and also forms distinct puncta, representing the spindle pole bodies. (b and c) Localization of Mps2 in meiosis. Schematic diagram shown in B illustrates the method of Mps2 overproduction in meiosis. Yeast cells were arrested at prophase I due to the lack of Ndt80, a meiosis-specific transcription factor required for expression of mid and later meiotic genes. Addition of estradiol led to production of Ndt80 and Mps2. Time-lapse live-cell microscopy was performed approximately 2 h after estradiol addition. Note that during anaphase II GFP-Mps2 localizes to both the spindle pole bodies and the nuclear periphery. Projected images from 12 optical sections are shown.
Next, we determined the localization of Mps2 in budding yeast meiosis Figures 2b and 2c. Like Mps3, the ONM protein Mps2 is also concentrated at the SPB in addition to its localization to the nuclear periphery Figure 2c. Mps2-GFP is mostly detectable as puncta corresponding to the SPB during meiosis (our unpublished data). In order to examine Mps2 distribution along the nuclear periphery, we generated a conditional PGAL-GFP-MPS2 allele to effectively overexpress MPS2 in meiosis Figure 2b. We used the GAL4.ER PGAL-NDT80 constructs [34], henceforth called PGAL-NDT80, to arrest yeast cells at prophase I, and upon the addition of β-estradiol, release the yeast cells from prophase I arrest, therefore achieving highly synchronized yeast cultures undergoing meiosis II. In cells containing PGAL-GFP-MPS2 PGAL-NDT80, the addition of β-estradiol also led to the overproduction of Mps2 in meiosis II Figures 2b and 2c. We found that GFP-Mps2 localized to both the SPB and the nuclear periphery Figure 2c. Notably, Mps2 behaved similarly to Mps3, distributing along the periphery throughout the GUNC compartment and also along the nascent gamete nuclei, indicating that both inner and outer nuclear membranes are present at the GUNC.
In addition, we observed an even distribution of INM-localized Heh1 throughout the nuclear periphery and the GUNC (Supplemental Figure 2 and [31]. Taken together, our findings indicate that nucleoporins, but not any NE-associated proteins, are selectively sequestered to the GUNC compartment.
Heh1 and Heh2 play a key role in nucleoporin sequestration
To identify factors that regulate nucleoporin sequestration, we used a genetic approach. From the yeast deletion collection library (ATCC-GSA-5), we pooled together 83 deletions of nonessential yeast genes that encode nuclear-envelope-associated proteins and determined their effect on Pom34-GFP confinement during anaphase II (Figure 3a and Supplemental Table 4). From this genetic screen, we found that yeast cells without HEH1, which is known to function in NPC assembly and surveillance in budding yeast [21], failed to retain Pom34-GFP to the GUNC Figure 3b. In the absence of Heh1, Pom34-GFP still aggregated during early meiosis II, but was not confined to GUNC; instead it became visible at the newly forming nuclei prior to the completion of meiosis Figures 1b and 3b). HEH1 has a paralog, HEH2; we, therefore, examined the role of Heh2 in nucleoporin sequestration by analyzing Pom34-GFP in heh2Δ cells Figure 3c. On the basis of fluorescence intensity, we estimated the percentage of Pom34-GFP on the newly forming nuclei compared to the total detectable cellular Pom34-GFP Figure 3d. In heh1Δ cells the distribution of Pom34-GFP to the newly forming nuclei showed approximately 1.5-fold increase compared to the wild type Figure 3d. Importantly, the heh1Δ heh2Δ double mutant showed a 2-fold increase of Pom34-GFP distributed to the newly forming nuclei Figure 3d, demonstrating that Heh1 and Heh2 are partially redundant in constricting nucleoporins to GUNC. In addition, we found that removal of Heh2 caused another striking phenotype, with Pom34-GFP circling a single Hta1-mApple mass Figure 3c. The appearance of Pom34-GFP encapsulating a single Hta1-mApple mass occurred in about 68% of the heh2Δ cells Figure 3e. Together, our findings demonstrate that Heh1, and to a lesser degree Heh2, plays a role in confining nucleoporins to the GUNC during meiosis II.
Figure 3.

Heh1 and Heh2 regulate Pom34 sequestration in meiosis II. (a) Schematic diagram showing targeted genetic screen to identify genes that regulate Pom34 localization. Briefly, targeted gene deletions were crossed to a strain containing POM34-GFP and HTA1-mApple. Selective media allowed for the identification of heterozygous diploids, which were induced to undergo meiosis. Subsequent tetrads were dissected and genotyped, and zygotes for homozygous gene deletions were created and induced to undergo meiosis, followed by fluorescence microscopy. (b) Heh1 regulates Pom34-GFP sequestration. Time-lapse live-cell microscopy was performed as described in Figure 1b. Note that in the absence of Heh1, Pom34-GFP was no longer restricted to the GUNC. (c) Heh2 mediates Pom34-GFP distribution during meiosis II. Time-lapse microscopy was performed as described in Figure 1b. Note that in the absence of Heh2, Pom34-GFP encapsulated one of the Hta1 masses, in addition to localizing to the midzone. (d) Quantification of Pom34-GFP distribution to the meiotic nuclei 30 min into meiosis II in WT, heh1Δ, heh2Δ, and heh1Δheh2Δ cells. (e) Quantification of the percent of cells showing Pom34-GFP encapsulating a single Hta1-mApple mass in WT, heh1Δ, heh2Δ, and heh1Δheh2Δ yeast cells as shown in C. A minimum of 25 cells were analyzed for each strain over 3 biological replicates.
The ESCRT-III component Chm7 regulates nucleoporin sequestration
Heh1 is known to function together with the ESCRT-III component Chm7 in NPC assembly and surveillance in vegetative yeast cells [24,25]; we therefore hypothesized that Chm7 is required for nucleoporin sequestration in meiosis. Using time-lapse live-cell microscopy, we observed that in chm7Δ cells, the accumulation of Pom34-GFP in the GUNC took place during anaphase II, however sequestration from the developing nuclei was severely hindered, with over 50% of the Pom34-GFP associating with the nascent nuclei Figures 4a and 4e. These findings suggest that the ESCRT-III component Chm7 is necessary for maintaining GUNC.
Figure 4.

Chm7 regulates GUNC in budding yeast meiosis. (a) Representative images showing Pom34-GFP localization in a chm7Δ cell in meiosis. Time-lapse live-cell microscopy was performed as described in Figure 1b. Projected images from 12 optical sections are shown. (b) Representative images showing Pom34-GFP localization in a heh1Δchm7Δ double mutant cell. (c) Representative images showing Pom34-GFP localization in a spo21Δ cell. (d) Representative images showing Pom34-GFP localization in a spo21Δchm7Δ double mutant cell. (e) Quantification of Pom34-GFP distribution to the meiotic nuclei 30 min into meiosis II in WT, heh1Δ, chm7Δ, heh1Δchm7Δ, spo21Δ, and spo21Δchm7Δ cells. A minimum of 25 cells were analyzed for each strain over 3 biological replicates. One-way ANOVA analysis between spo21Δ and spo21Δ chm7Δ shows F ratio = 5.35601, p > 0.01; and between heh1Δ, chm7Δ, heh1Δchm7Δ F ratio = 8.2218, p < 0.01. (f-h) Pom34-GFP protein level in WT, chm7Δ, heh1Δchm7Δ, spo21Δ, and spo21Δchm7Δ cells. Cell aliquots were withdrawn at the indicated times after meiosis induction. Protein extracts were prepared for western blotting. An anti-GFP antibody was used to probe Pom34-GFP. The level of Pgk1 serves as a loading control. (i) Quantification of Pom34-GFP protein level in meiosis. Error bars represent the standard deviation from the mean of biological replicates (n = 3). Note that there is no discernable difference in Pom34-GFP protein levels between WT, chm7Δ, and heh1Δchm7Δ yeast cells, but deletion of SPO21 alters Pom34-GFP protein level.
To determine whether Chm7 and Heh1 act together in nucleoporin sequestration, we examined Pom34-GFP distribution in heh1Δ chm7Δ double mutant cells Figure 4b. We found that Pom34-GFP was not confined to the GUNC, with 73% of the total Pom34-GFP signal distributed along the developing prospore nuclei Figure 4e, demonstrating that the lack of Pom34-GFP sequestration in the heh1Δ chm7Δ double mutant is more severe than in either the chm7Δ or heh1Δ single mutant Figure 4e. Because Heh1 is thought to recruit Chm7 to the nuclear envelope [25], the observed synergistic effect of Chm7 and Heh1 on Pom34-GFP distribution is unexpected and raises the possibility that additional factors from ESCRT-III are required for sequestration of nucleoporins to GUNC (see below). Alternatively, there may also be redundancies within the GUNC formation pathway that are capable of confining nucleoporins.
Recent work has shown that Spo21, an outer plaque component of the SPB, is required for nucleoporin sequestration [31]. In the absence of Spo21, sequestration of nucleoporins to GUNC became defective Figure 4c and [31]. To determine the epistatic relationship between SPO21 and CHM7, we examined Pom34-GFP localization in spo21Δ chm7Δ double mutant cells Figure 4d. Removal of Spo21 and Chm7 simultaneously did not significantly change the distribution of Pom34-GFP along the nascent nuclei, indicating that Spo21 acts upstream of Chm7 Figure 4e. We note that removal of Heh1 or Chm7, individually or simultaneously, did not alter the protein level of Pom34 Figures 4f, 4g and 4i, demonstrating that Heh1 and Chm7 regulate nucleoporin localization, but not protein stability. In contrast, removal of Spo21 altered Pom34 protein level, and mature spores never formed (Figure 4h and our unpublished data). This is likely due to the requirement of Spo21 in meiotic cell progression and spore formation [41,42]. Taken together, our findings suggest that SPO21 acts epistatically to CHM7 to sequester nucleoporins to the GUNC.
The ESCRT-III-associated factor Vps4 regulates nucleoporin sequestration
Vps4, which associates with the ESCRT-III complex, has been shown to function with Chm7 during NPC surveillance in budding yeast [24]. We hypothesized that Vps4 is required for nucleoporin sequestration in meiosis. First, we determined Vps4 localization with a VPS4-GFP allele, which served as the only functional copy of VPS4 in the budding yeast genome Figure 5a. In meiosis II, Vps4 formed multiple foci inside the cell Figure 5a. Approximately one discernible Vps4 focus was localized to the GUNC and attached to each nucleus Figure 5a, demonstrating that Vps4 is associated with GUNC and the meiotic nuclei. Next, we generated a conditional vps4-AID allele to deplete Vps4 specifically after the induction of meiosis Figures 5b-5d, because deletion of VPS4 prevented budding yeast cells from undergoing meiosis (our unpublished data). With PGAL-NDT80, yeast cells were arrested at prophase I, and addition of 1-napthylacetic acid (NAA) led to the degradation of Vps4-AID Figures 5b-5d. Cells were then released from prophase I with the addition of β-estradiol to activate PGAL-NDT80 and allowed to proceed through meiosis II in the absence of Vps4. We found that depletion of Vps4 in meiosis resulted in defects in nucleoporin sequestration and Hta1-mApple partitioning in over 90% of the cells observed Figures 5e-5i. The most common mutant phenotype observed in cells depleted of Vps4 was similar to that of the chm7Δ and heh1Δ mutants, in which Pom34-GFP was no longer confined to the GUNC Figure 5i. On the basis of the Pom34-GFP fluorescence intensity, we characterized meiotic cells in four categories: type 1, wild type, with less than 50% of the total Pom34-GFP presenting on the nascent nuclei; type 2, with over 50% of the total Pom34-GFP presenting on the nascent nuclei; type 3, with Pom34-GFP encapsulating one of the four Hta1-mApple masses; and type 4, with Pom34-GFP encapsulating a fifth Hta1-mApple mass Figures 5e-I. As expected, wild-type and vps4-AID cells without NAA treatment showed mostly the type 1 phenotype, 81% and 74%, respectively, Figures 5e and 5i. Upon depletion of Vps4-AID with NAA, we observed Pom34-GFP dispersed along the nuclear periphery, type 2, in 46% of cells Figures 5f and 5i. The accumulation of Pom34-GFP on a single Hta1-mApple mass, a phenotype similar to that of heh1Δ and heh2Δ mutants (type 3), was also observed in 34% of Vps4-depleted cells Figures 5g and 5i. Finally, we observed the formation of a 5th Hta1-mApple mass encapsulated by Pom34-GFP (type 4) occurred in about 18% of Vps4-depleted cells Figures 5h and 5i. We found that the vps4-AID heh1Δ double mutant showed an increase in the frequency of type 2 cells compared to the single mutants, with 58% of vps4-AID heh1Δ cells lacking Pom34-GFP sequestration Figure 5i. However, the percentage of type 3 and type 4 cells appeared comparable in the double mutant Figure 5i. Together, our findings demonstrate that the ESCRT-III factor Vps4 is required for nucleoporin sequestration during meiosis II.
Figure 5.

Vps4 localization and its role in Pom34 sequestration. (a) Localization of Vps4-GFP (green) in meiosis II. Time-lapse fluorescence microscopy was performed as in Figure 1b. Images from 7 continuous optical sections (z = 0.5 μm) were shown. Arrows indicate the localization of Vps4-GFP to the GUNC compartment. Note that Heh1-mApple (red) demarcates GUNC and the four nuclei, which are numerically labeled. (b) Schematic diagram showing the experimental procedure to deplete Vps4 in meiosis II. Addition of 1-napthylacetic acid (NAA) initiates Vps4 degradation through AID. Addition of estradiol releases cells from prophase I arrest. Time-lapse live-cell microscopy was performed approximately 2 h after estradiol addition to ensure cells were in meiosis II. Addition of ethanol serves as a negative control. (c and d) Depletion of Vps4-V5-AID in meiosis. Cells were prepared as described in B, and cell aliquots were withdrawn at indicated times. Time zero refers to the point of NAA addition. Protein extracts were prepared for western blotting. An anti-V5 antibody was used to probe Vps4-V5-AID. The level of Pgk1 serves as a loading control. Quantification of Vps4 protein abundance is shown in panel D. (e-h) Vps4 regulates Pom34-GFP localization and chromosome segregation. Cells were prepared as described in B. Projected images from 12 optical sections are shown. Four categories based on nuclear morphology were classified: Type 1, Pom34-GFP sequestered to GUNC as shown in E; Type 2, Pom34-GFP is concentrated on newly forming nuclei as shown in F; Type 3, Pom34-GFP encapsulates one of the four Hta1-mApple masses (nuclei) as shown in G; Type 4, Pom34-GFP encapsulates the fifth Hta1-mApple mass as shown in H. (i) Quantification of Pom34-GFP localization in cells with WT, vps4-AID with EtOH, vps4-AID with NAA, vps4-AID heh1Δ with EtOH, and vps4-AID heh1Δ with NAA. A minimum of 50 cells were analyzed for each strain background.
ESCRT-III and its associated factors regulate gamete replicative lifespan
To determine whether ESCRT-III-mediated nuclear envelope remodeling regulates gamete rejuvenation, we determined the replicative lifespan of gametes derived from mutants that impaired GUNC formation. Wild-type gametes had a maximal replicative lifespan of about 32 generations, with an average of 22 Figure 6. In contrast, the replicative lifespan of gametes derived from heh1Δ and chm7Δ mutants reduced to 15 generations on average Figure 6, demonstrating the importance of Heh1 and Chm7 in restoring replicative potential. Importantly, gametes from the heh1Δ chm7Δ mutant reduced the maximal replicative lifespan by nearly half, with an average replicative lifespan of 10 generations Figure 6. Similarly, depletion of Vps4 via the vps4-AID allele, reduced the replicative lifespan of its gametes Figure 6. Of note, the vps4-AID allele permitted us to deplete Vps4 specifically in meiosis Figures 5b-5d, thus eliminating a potential deleterious effect the removal of mitotic Vps4 may exert on replicative lifespan during the vegetative growth of the gamete. Taken together, these findings suggest that Heh1 and ESCRT-III are critical for maintaining gamete replicative lifespan.
Figure 6.
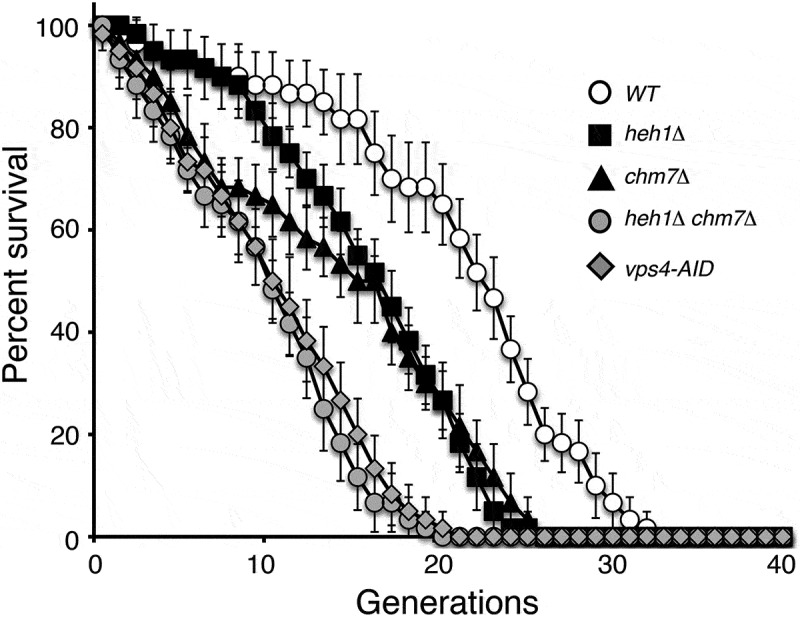
ESCRT-III and its associated factors regulate replicative lifespan in gametes. Replicative lifespan in wild-type and mutant gametes was determined by single-cell analysis using micromanipulation. Each cell analyzed was a gamete of the homozygous diploid originally analyzed, isolated at random from the population. The average life-span for WT gametes was 21.3, 17.3 for heh1Δ, 15.3 for chm7Δ, 9.9 for heh1Δchm7Δ, and 10.9 for vps4-AID. A total of 60 spores were assayed over 3 biological replicates for each strain. Error bars represent deviation from the mean.
Discussion
In this study, we confirm that nucleoporins are sequestered during meiosis II in budding yeast. Importantly, we show the LEM-domain protein Heh1 and the ESCRT-III complex act in concert to confine nucleoporins and regulate gamete rejuvenation. Our findings not only are consistent with a recent work showing the sequestration of NPCs and other age-induced damages away from the newly-formed gametes [30,31], but also provide a mechanistic understanding for the process of GUNC and its potential role in rejuvenation of gametes in budding yeast, which may have implications for understanding how self-renewal is achieved during animal gametogenesis.
How is GUNC formed and maintained in meiosis II?
We have shown that nucleoporins, but not any nuclear-envelope-associated proteins, are selectively sequestered to the GUNC. In their recent work, King et al. have shown that other age-induced factors, including extrachromosomal rDNA circles and heat shock proteins, also are aggregated in the GUNC [31], indicating that formation of the GUNC is a coordinated process involving the nuclear envelope and the nuclear plasma.
All nucleoporins we tested are sequestrated to the GUNC during anaphase II, suggesting that the NPC as a whole, not just individual nucleoporins, is sequestered. One exception is Nup2, the nucleoporin that binds to the cytoplasmic or nucleoplasmic surface of the NPC, that appears to not be confined at the GUNC. Instead, Nup2 is distributed evenly along the nuclear periphery and also potentially associated with the ER (our unpublished data and [31], which distribution is likely mediated by the ability of Nup2 to associate and dissociate with the NPC depending on the level of Ran-GTP present in the cell [43,44].
How then is GUNC formed? Previous work has shown that the SPB meiotic outer plaque component Spo21 is required for nucleoporin sequestration to GUNC [40]. In the absence of Spo21, nucleoporins largely spread out throughout the nuclear periphery without any evidence of aggregation. Spo21 and its associated outer plaque are uniquely modified on the cytoplasmic side of the SPB during meiosis to form a membrane-organizing center that is necessary for establishing the prospore membrane, which encircles the four gametes at the end of meiosis [28]. We speculate that newly formed prospore membrane, emanating from the SPB, actively ‘pushes’ NPCs to the GUNC Figure 7. Alternatively, rapid expansion of the nuclear membranes during anaphase II, which coincides with the formation of the prospore membrane, passively leaves the old NPCs behind in the GUNC. In this regard, the newly formed prospore membranes act as a selective filter, facilitating the constriction of old NPCs and hindering migration into the gamete nuclei. Our finding that ESCRT-III is required for sequestering nucleoporins to the GUNC lends support to this confinement model Figure 7. Furthermore, Spo21 appears to act epistatically to ESCRT-III (this study), indicating that ESCRT-III-mediated GUNC functions in the context of prospore membrane biogenesis.
Figure 7.

Model for ESCRT-III-mediated GUNC formation in budding yeast meiosis. Top, GUNC formation and maintenance in wild type. Note that NPCs are sequestered away from the newly formed gamete nuclei. Bottom, GUNC fails to form in the absence of Heh1/ESCRT-III function at the nuclear envelope. The GUNC compartment is shown in purple; Chm7/ESCRT-III in orange. Pink dots represent Vps4; red lines prospore membrane. NPC, nuclear pore complex; SPB, spindle pole body.
How does ESCRT-III constrict NPCs to GUNC?
We have shown that the ESCRT-III component Chm7 and the AAA-ATPase Vps4, a cofactor of ESCRT-III, are crucial for restricting nucleoporins to the GUNC. Functions of the highly conserved ESCRT machinery revolve around membrane fission reactions throughout the cell [45,46]. In addition, upon the completion of mitosis when the nuclear envelope reforms in animal cells, ESCRT-III/Vps4 facilitates sealing of nuclear membrane fenestrations [47–49]. We speculate that during late anaphase II, the LEM-domain protein Heh1 acts as a site-specific adaptor to recruit ESCRT-III, for example Chm7, to the sites between the expanding nuclei and the GUNC. Vps4 then likely binds to the ESCRT-III complex and constricts the nuclear envelope, resulting in the cleavage of the GUNC compartment from the newly formed nuclei Figure 7. This reasoning is consistent with the observation that HEH1, but not its paralog HEH2, is upregulated in late meiosis [50]. Furthermore, Vps4 forms foci at the junctions of the GUNC and the expanding prospore nuclear envelope (this study). In the absence of ESCRT-III function, NPCs would move freely from the GUNC without restriction to the expanding daughter nuclei Figure 7. This reasoning is analogous to nuclear division in budding yeast mitosis, in which the mother cell retains all of the detrimental morphological and physiological changes causing the mother cell to age [3,4]. By way of concerted actions from Heh1 and ESCRT-III, the GUNC thus functionally resembles the mother cell in mitosis and restricts the old or damaged NPCs and other nuclear senescence factors from segregating into the newly formed gametes Figure 7. Because Heh1 directs ESRCT-III during NPC biogenesis by exposure to the cytoplasm and subsequent binding of Chm7 [25], the activity of Heh1 may hold the key to where and how the GUNC constriction sites are determined. Because NPC sequestration is not completely abolished in the absence of ESCRT-III function, ESCRT-independent pathways could also be at work. Alternatively, NPCs may be passively left behind when the prospore nuclear membrane expands rapidly outward from the spindle pole body during late meiosis Figure 7. Without ESCRT-III, certain NPCs may fortuitously be trapped in the GUNC.
ESCRT-III-mediated GUNC regulates gamete rejuvenation
We show that ESCRT-III and its associated factors help maintain the replicative lifespan of gametes. Removal of meiotic ESCRT-III activity diminishes the replicative lifespan of gametes in budding yeast (this study). We speculate that one of the major functions of the GUNC is to restrict old or damaged NPCs from being incorporated into the newly formed gametes. NPCs are known to function as both barrier and gateway between the cytoplasm and nucleoplasm, ensuring cellular homeostasis between compartments, and the disruption of this barrier has been shown in several different contexts, including a variety of human diseases [51]. Alternatively, ESCRT-III-mediated NPC confinement may not be directly modulating aging but instead serve as a readout of cellular aging similar to other unwanted materials such as protein aggregates and extrachromosomal rDNA circles. In either case, sequestration of NPCs to the GUNC in budding yeast meiosis represents a means for nuclear quality control and provides a new paradigm for elucidating the mechanism of cellular aging.
We note that the nuclear envelope appears to be intact and its permeability remains normal in meiosis II when the GUNC occurs. Although budding yeast undergoes closed meiosis and mitosis, the GUNC process in meiosis functionally mimics the nuclear envelope breakdown seen in higher eukaryotes, which permits the removal of nuclear senescence factors. Indeed, in fission yeast at anaphase II, the nuclear envelope undergoes a ‘virtual’ breakdown, during which the permeability of the nuclear envelope is temporally increased [52,53]. In that regard, the GUNC process may represent a primordial version of nuclear envelope remodeling that precedes nuclear envelope breakdown. Future studies on the regulation of ESCRT-III-mediated GUNC formation and maintenance in budding yeast will provide further insight into the functionality of the GUNC and cellular rejuvenation.
Supplementary Material
Acknowledgments
We thank Robert Tomko and Yanchang Wang for insightful discussions. Jen Kennedy assisted with text editing, and Charles Badland with graphic design. This work was supported by the National Institute of General Medicine (GM117102) and the National Science Foundation (MCB1951313).
Funding Statement
This work was supported by the National Institute of General Medical Sciences [GM117102]; National Science Foundation, MCB1951313.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplemental data for this article can be accessed here.
References
- [1].Reya T, Morrison SJ, Clarke MF, et al. Stem cells, cancer, and cancer stem cells. Nature. 2001;414(6859):105–111. [DOI] [PubMed] [Google Scholar]
- [2].Sinclair D, Mills K, Guarente L.. Aging in saccharomyces cerevisiae. Annu Rev Microbiol. 1998;52(1):533–560. [DOI] [PubMed] [Google Scholar]
- [3].Kaeberlein M. Lessons on longevity from budding yeast. Nature. 2010;464(7288):513–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Longo VD, Shadel GS, Kaeberlein M, et al. Replicative and chronological aging in Saccharomyces cerevisiae. Cell Metab. 2012;16(1):18–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Mortimer RK, Johnston JR. Life span of individual yeast cells. Nature. 1959;183(4677):1751–1752. [DOI] [PubMed] [Google Scholar]
- [6].Aguilaniu H, Gustafsson L, Rigoulet M, et al. Asymmetric inheritance of oxidatively damaged proteins during cytokinesis. Science. 2003;299(5613):1751–1753. [DOI] [PubMed] [Google Scholar]
- [7].Egilmez NK, Jazwinski SM. Evidence for the involvement of a cytoplasmic factor in the aging of the yeast Saccharomyces cerevisiae. J Bacteriol. 1989;171(1):37–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Kennedy BK, Austriaco NR Jr., Guarente L. Daughter cells of Saccharomyces cerevisiae from old mothers display a reduced life span. J Cell Biol. 1994;127(6):1985–1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Sinclair DA, Guarente L. Extrachromosomal rDNA circles–a cause of aging in yeast. Cell. 1997;91(7):1033–1042. [DOI] [PubMed] [Google Scholar]
- [10].Spokoini R, Moldavski O, Nahmias Y, et al. Confinement to organelle-associated inclusion structures mediates asymmetric inheritance of aggregated protein in budding yeast. Cell Rep. 2012;2(4):738–747. [DOI] [PubMed] [Google Scholar]
- [11].Unal E, Kinde B, Amon A. Gametogenesis eliminates age-induced cellular damage and resets life span in yeast. Science. 2011;332(6037):1554–1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].D’Angelo MA, Raices M, Panowski SH, et al. Age-dependent deterioration of nuclear pore complexes causes a loss of nuclear integrity in postmitotic cells. Cell. 2009;136(2):284–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Ungricht R, Kutay U. Mechanisms and functions of nuclear envelope remodelling. Nat Rev Mol Cell Biol. 2017;18:229–245. [DOI] [PubMed] [Google Scholar]
- [14].Beck M, E. H. The nuclear pore complex: understanding its function through structural insight. Nat Rev Mol Cell Biol. 2017;18(2):73–89. [DOI] [PubMed] [Google Scholar]
- [15].Kim SJ, Fernandez-Martinez J, Nudelman I, et al. Integrative structure and functional anatomy of a nuclear pore complex. Nature. 2018;555(7697):475–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Teimer R, Kosinski J, von Appen A, et al. A short linear motif in scaffold Nup145C connects Y-complex with pre-assembled outer ring Nup82 complex. Nat Commun. 2017;8(1):1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Popken P, Ghavami A, Onck PR, et al. Size-dependent leak of soluble and membrane proteins through the yeast nuclear pore complex. Mol Biol Cell. 2015;26(7):1386–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Schmidt HB, Gorlich D. Transport selectivity of nuclear pores, phase separation, and membraneless organelles. Trends Biochem Sci. 2016;41(1):46–61. [DOI] [PubMed] [Google Scholar]
- [19].Timney BL, Raveh B, Mironska R, et al. Simple rules for passive diffusion through the nuclear pore complex. J Cell Biol. 2016;215(1):57–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Kelley JB, Datta S, Snow CJ, et al. The defective nuclear lamina in Hutchinson-gilford progeria syndrome disrupts the nucleocytoplasmic Ran gradient and inhibits nuclear localization of Ubc9. Mol Cell Biol. 2011;31(16):3378–3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Webster BM, Colombi P, Jager J, et al. Surveillance of nuclear pore complex assembly by ESCRT-III/Vps4. Cell. 2014;159(2):388–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Aitchison JD, Rout MP. The yeast nuclear pore complex and transport through it. Genetics. 2012;190(3):855–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Otsuka S, Bui KH, Schorb M, et al. Nuclear pore assembly proceeds by an inside-out extrusion of the nuclear envelope. Elife. 2016;5. doi:10.7554/eLife.19071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Thaller DJ, Allegretti M, Borah S, et al. An ESCRT-LEM protein surveillance system is poised to directly monitor the nuclear envelope and nuclear transport system. Elife. 2019;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Webster BM, Thaller DJ, Jager J, et al. Chm7 and Heh1 collaborate to link nuclear pore complex quality control with nuclear envelope sealing. Embo J. 2016;35(22):2447–2467. doi:10.7554/eLife.45284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Colombi P, Webster BM, Frohlich F, et al. The transmission of nuclear pore complexes to daughter cells requires a cytoplasmic pool of Nsp1. J Cell Biol. 2013;203(2):215–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Makio T, Lapetina DL, Wozniak RW. Inheritance of yeast nuclear pore complexes requires the Nsp1p subcomplex. J Cell Biol. 2013;203(2):187–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Neiman AM. Sporulation in the Budding Yeast Saccharomyces cerevisiae. Genetics. 2011;189(3):737–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Unal E, Amon A. Gamete formation resets the aging clock in yeast. Cold Spring Harb Symp Quant Biol. 2011;76(x):73–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Fuchs J, Loidl J. Behaviour of nucleolus organizing regions (NORs) and nucleoli during mitotic and meiotic divisions in budding yeast. Chromosome Res. 2004;12(5):427–438. [DOI] [PubMed] [Google Scholar]
- [31].King GA, Goodman JS, Schick JG, et al. Meiotic cellular rejuvenation is coupled to nuclear remodeling in budding yeast. Elife. 2019;8. doi: 10.7554/eLife.47156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Longtine MS, McKenzie A 3rd, Demarini DJ, et al. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14(10):953–961. [DOI] [PubMed] [Google Scholar]
- [33].Li P, Jin H, Yu HG. Condensin suppresses recombination and regulates double-strand break processing at the repetitive ribosomal DNA array to ensure proper chromosome segregation during meiosis in budding yeast. Mol Biol Cell. 2014;25(19):2934–2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Carlile TM, A. A. Meiosis I is established through division-specific translational control of a cyclin. Cell. 2008;133(2):280–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Li P, Jin H, Koch BA, et al. Cleavage of the SUN-domain protein Mps3 at its N-terminus regulates centrosome disjunction in budding yeast meiosis. PLoS Genet. 2017;13(6):e1006830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Jin H, Guacci V, Yu HG. Pds5 is required for homologue pairing and inhibits synapsis of sister chromatids during yeast meiosis. J Cell Biol. 2009;186(5):713–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Boselli M, J. R, E. U, et al. Effects of age on meiosis in budding yeast. Dev Cell. 2009;16(6):844–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Rout MP, Aitchison JD, Suprapto A, et al. The yeast nuclear pore complex: composition, architecture, and transport mechanism. J Cell Biol. 2000;148(4):635–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Miao M, Ryan KJ, Wente SR. The integral membrane protein Pom34p functionally links nucleoporin subcomplexes. Genetics. 2006;172(3):1441–1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].King GA, Unal E. The dynamic nuclear periphery as a facilitator of gamete health and rejuvenation. Curr Genet. 2020;66(3):487–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Knop M, Strasser K. Role of the spindle pole body of yeast in mediating assembly of the prospore membrane during meiosis. Embo J. 2000;19(14):3657–3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Neiman AM. Prospore membrane formation defines a developmentally regulated branch of the secretory pathway in yeast. J Cell Biol. 1998;140(1):29–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Denning D, Mykytka B, Allen NP, et al. The nucleoporin Nup60p functions as a Gsp1p-GTP-sensitive tether for Nup2p at the nuclear pore complex. J Cell Biol. 2001;154(5):937–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Dilworth DJ, Suprapto A, Padovan JC, et al. Nup2p dynamically associates with the distal regions of the yeast nuclear pore complex. J Cell Biol. 2001;153(7):1465–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].McCullough J, Frost A, Sundquist WI. Structures, functions, and dynamics of ESCRT-III/Vps4 membrane remodeling and fission complexes. Annu Rev Cell Dev Biol. 2018;34(1):85–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Stoten CL, Carlton JG. ESCRT-dependent control of membrane remodelling during cell division. Semin Cell Dev Biol. 2018;74:50–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Olmos Y, Hodgson L, Mantell J, et al. ESCRT-III controls nuclear envelope reformation. Nature. 2015;522(7555):236–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Vietri M, Schink KO, Campsteijn C, et al. Spastin and ESCRT-III coordinate mitotic spindle disassembly and nuclear envelope sealing. Nature. 2015;522(7555):231–235. [DOI] [PubMed] [Google Scholar]
- [49].von Appen A, LaJoie D, Johnson IE, et al. LEM2 phase separation promotes ESCRT-mediated nuclear envelope reformation. Nature. 2020;582(7810):115–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Chu S, DeRisi J, Eisen M, et al. The transcriptional program of sporulation in budding yeast. Science. 1998;282(5389):699–705. [DOI] [PubMed] [Google Scholar]
- [51].Hatch E, Hetzer M. Breaching the nuclear envelope in development and disease. J Cell Biol. 2014;205(2):133–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Arai K, Sato M, Tanaka K, et al. Nuclear compartmentalization is abolished during fission yeast meiosis. Curr Biol. 2010;20(21):1913–1918. [DOI] [PubMed] [Google Scholar]
- [53].Asakawa H, Kojidani T, Mori C, et al. Virtual breakdown of the nuclear envelope in fission yeast meiosis. Curr Biol. 2010;20:1919–1925. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


