Abstract
Cervical cancer is the fourth most common cancer in women worldwide. The current approaches still have limitations in predicting the therapy outcome of each individual because of cancer heterogeneity. The goal of this study was to establish a gene expression signature that could help when choosing the right therapeutic method for the treatment of advanced-stage cervical cancer. The 666 patients were collected from four independent datasets. The 70-gene expression signature was established using univariate Cox proportional hazard regression analysis. The 70-gene signature was significantly different between low- and high-risk groups in the training dataset (p = 4.24e−6) and in the combined three validation datasets (p = 4.37e−3). Treatment of advanced-stage cancer patients in the high-risk group with molecular-targeted therapy combined with chemoradiotherapy yielded a better survival rate than with only chemoradiotherapy (p = 0.0746). However, treatment of the patients in the low-risk group with the combined therapy resulted in significantly lower survival (p = 0.00283). Functional classification of 70 genes revealed involvement of the angiogenesis pathway, specifically phosphatidylinositol 3-kinase signaling (p = 0.040), extracellular matrix organization (p = 0.0452), and cell adhesion (p = 0.011). The 70-gene signature could predict the prognosis and indicate an optimal therapeutic modality in molecular-targeted therapy or chemotherapy for advanced-stage cervical cancer.
Keywords: cervical cancer, gene signature, survival, advanced stage, molecular-targeted therapy
Graphical Abstract
Patients with late-stage cervical cancer have significantly high mortality and resistance to anticancer drugs. We hypothesize that finding the patients with a sensitivity of anti-cancer drugs or molecular-targeted therapy can be applied to an effective cure against the high cost and low sensitivity to cervical cancer patients.
Introduction
Cervical cancer is the fourth most frequent cancer in women, and about 570,000 new cases were diagnosed in 2018, representing 6.6% of all female cancers.1 Early-stage cervical cancer rate increased because of the development of diagnosis technique and screening test. However, many patients are still diagnosed in the advanced stage with poor prognosis. Therefore, it is very important for cervical cancer patients to get a timely, accurate diagnosis and appropriate treatment to increase the survival rate. The National Comprehensive Cancer Network (NCCN) and the International Federation of Gynecology and Obstetrics (FIGO) suggested standard treatment guidelines, such as surgery, radiotherapy, and chemotherapy, for treatment of the patients since the time of diagnosis and according to the disease stage. Even though patients are treated based on these guidelines, each patient has a different prognosis because of the tumor heterogeneity. Therefore, proper stratification of patients, depending on their clinical conditions, is required. In this way, a more effective treatment could be provided to the patient.
Many clinical and pathological studies have descried a number of prognostic factors for cervical cancer, such as clinical stage, tumor histology, depth of invasion, tumor grade, size of primary tumor, lymph node involvement, parametrium involvement, and lymph-vascular space invasion.2, 3, 4, 5, 6, 7 A more successful result was obtained when the therapeutic methods, such as surgery, radiotherapy, and chemotherapy, were applied to patients with certain characteristics. NCCN recommends either surgery or radiotherapy for the treatment of patients with FIGO stage IA1–IIA1 called early stage. In contrast, the treatment of choice for advanced cancer with stages IIB–IVA consists of a combined therapy including external beam radiation therapy (EBRT), cisplatin-containing chemotherapy, and brachytherapy.8, 9, 10 In case of metastasis, targeted therapy and immunotherapy have been applied.11,12 Despite the development of many treatment options, the survival rate in advanced-stage cancer remains poor.
Recently, the gene expression profiles obtained from microarray, next generation sequencing (NGS), and subsequent oncogenic signaling pathway analysis have become useful for predicting prognosis of the disease and for discovering therapeutic targets in various cancers.13, 14, 15 In estrogen receptor-positive breast cancer, a 21-gene signature has been widely used to improve disease-free survival and to predict chemo-resistance to anthracyclines and a beneficial outcome with tamoxifen administration.16,17 In this regard, microarray data have been studied for diagnosis, prognosis, or prediction of therapeutic response in cervical cancer as well.18, 19, 20, 21 However, no favorable results have been obtained yet. Therefore, more reliable gene signatures are warranted to further achieve predictive accuracy of prognosis and effective treatments.
In this study, we established a novel prognostic gene signature to distinguish low- and high-risk patients. In addition, we assessed the association between the gene signatures and clinicopathological factors. Finally, we provided a patient profile that could receive the most benefit from molecular-targeted therapy.
Results
Stratification by Hierarchical Clustering
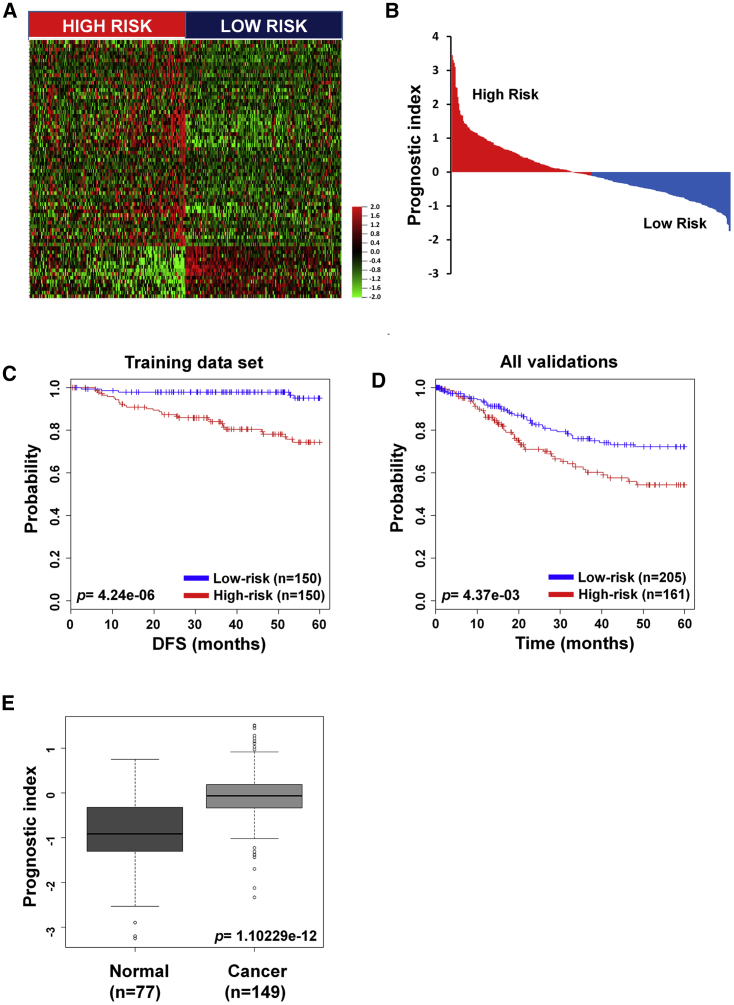
In order to identify a prognostic gene signature that distinguished low- and high-risk patients, we analyzed gene expression profiles based on survival data. Gene Expression Omnibus (GEO): GSE44001 was assigned as the training dataset; meanwhile, The Cancer Genome Atlas (TCGA) RNA sequencing (RNA-seq) and microarray data (GEO: GSE39001 and GSE52904) corresponded to validation datasets. Patients were classified into low- (n = 150) and high-risk (n = 150) groups by the expression pattern of 70 genes and their prognostic indexes (PIs) (Figures 1A and 1B). There was a significant survival difference between the low- and high-risk groups (p = 4.24e−6; Figure 1C). The 70-gene signature was applied to other datasets for validation, and a statistically significant difference of survival between the two groups was observed (p = 0.00437; Figure 1D). Healthy controls had lower PI than cervical cancer patients (p = 1.102e−12; Figure 1E). In both RNA-seq and microarray datasets, low-risk patients had a better survival rate than high-risk patients (p = 0.0311 and p = 0.0466, respectively; Figures S1A and S1B). The expression pattern of 70 genes was divided by low- and high-risk patients in both microarray and TCGA datasets (Figures S1C and S1D).
Figure 1.
Hierarchical Clustering and Classification by 70 Genes
(A) Heatmap of the median centered 70 genes’ expression profiles between high- and low-risk groups in the training dataset (red, relative high expression; green, relative low expression). (B) The relative prognostic index based on 70 genes’ expression of each patient. The weight of each gene was calculated by the Cox proportional hazard regression model. (C and D) The 70-gene signature of patients with disease-free survival (DFS) in the training dataset (C) and overall survival (OS) in combined validation datasets (D). Each group was classified by the 70-gene signature into low and high risk, and evaluated by Kaplan-Meier analysis. (E) Prognostic index value of the 70-gene signature in cervical cancer and healthy tissue. The p values were computed by the log rank test.
Validation of Predicted Gene Signature in Specific Tumor Stages
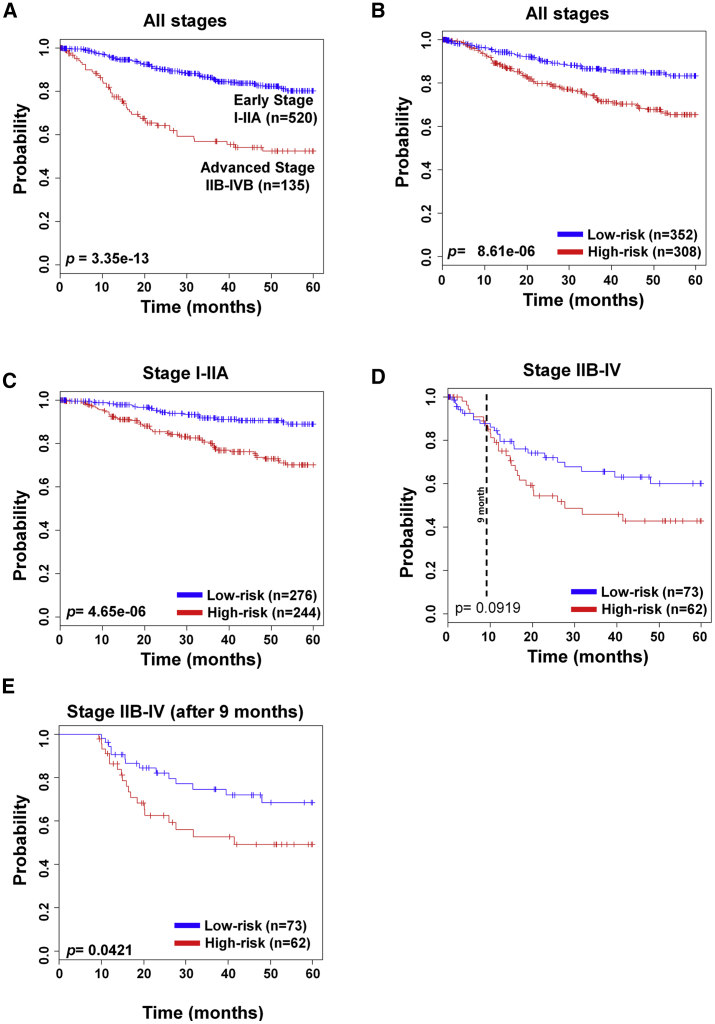
Next, cervical cancer patients were divided based on NCCN guidelines and FIGO staging system into two groups: early stage (stage I–IIA) and advanced stage (stage IIB–IV). Patients in the early-stage group can be cured by surgery and/or radiotherapy, whereas those with advanced-stage cancer require combined therapy, including radiation and chemotherapy.
As previously reported, the survival rate in our patient data was also different between the early and advanced stages (p = 3.35e−13; Figure 2A). Our 70-gene signature stratified patients’ survival in all stages (p = 8.61e−6; Figure 2B). Importantly, the low-risk group had a significantly higher survival rate than the high-risk group in the early stage (p = 4.65e−6; Figure 2C). However, advanced-stage patients did not show the statistical significance (p = 0.0919; Figure 2D), but patients showed significant survival difference between the two groups after 9 months (p = 0.0421; Figure 2E). To evaluate the prognostic accuracy of the 70-gene signature in relation to the cancer stage, we performed univariate and multivariate Cox proportional hazards regression analyses using all of the datasets. As shown in Table S1, stage was significantly associated with overall survival (OS) in both analyses (univariate: hazard ratio [HR], 3.547, 95% confidence interval [CI]: 2.466–5.103, p = 8.8e−12; multivariate: HR, 3.691, 95% CI: 2.564–5.313, p = 2.2e−12). The 70-gene signature was also significantly associated with OS according to univariate and multivariate analyses (univariate: HR, 2.265, 95% CI: 1.566–3.275, p = 1.4e−5; multivariate: HR, 2.369, 95% CI: 1.637–3.428, p = 4.8e−6).
Figure 2.
Validation of the 70-Gene Signature in Tumor Stage
(A) Patients were separated according to early- and advanced-stage cancer. (B–E) The 70-gene signature was applied to cancer patients of all stages (B), early stage (C), advanced stage (D), and advanced stage after 9 months (E). Each group was classified by the 70-gene signature into low and high risk, and evaluated by Kaplan-Meier analysis. The p values were computed by the log rank test.
Association of the 70-Gene Signature with Tumor Size and Age
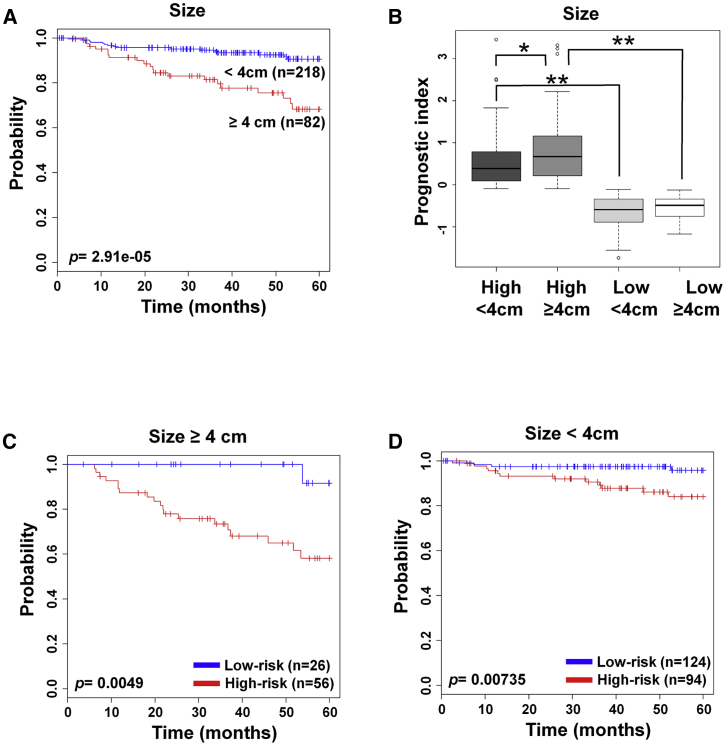
The tumor size is an important clinical diagnosis factor; the threshold size for our study was 4 cm in diameter. As reported, cervical cancer patients with tumor over 4 cm had a worse prognosis than under 4 cm (p = 2.91e−5; Figure 3A). The patients in the low-risk group had lower PI values than those of the high-risk group, even though they carried tumors of over 4 cm in size (Figure 3B). Regardless of tumor size, the high-risk group had poor survival rate in both groups: under and over 4 cm (p = 0.0049 and p = 0.00735, respectively; Figures 3C and 3D). Cox proportional hazard regression test was performed when analyzing the association with tumor size (univariate: HR, 3.703, 95% CI: 1.917–7.152, p = 9.7e−5; multivariate: HR, 2.724, 95% CI: 1.397–5.311, p = 0.00326) and 70-gene signature (univariate: HR, 6.769, 95% CI: 2.632–17.409, p = 7.3e−5; multivariate: HR, 5.491, 95% CI: 2.108–14.304, p = 0.00049) (Table S2).
Figure 3.
Survival Analysis by the 70-Gene Signature along with Tumor Size
(A) Patients were separated according to the tumor size: under 4 cm and over 4 cm in diameter. (B) The prognostic index of 70-gene signature in the cross-combination of risk group and tumor size. (C and D) The 70-gene signature was applied to patients with tumor sizes over 4 cm (C) and under 4 cm (D). Each group was classified by 70-gene signature into low and high risk, and evaluated by Kaplan-Meier analysis. The p values were computed by the log rank test. ∗p < 0.05, ∗∗p < 0.0001.
Patients younger than 65 years of age at the time of diagnosis of cervical cancers have higher chances of longer survival.22 The 70-gene signature further stratified patients according to age under 65 years into low- and high-risk groups (p = 0.00153; Figure S2A). In this age group, patients were stratified regardless of early or advanced stage (p = 0.0126 and p = 0.0653, respectively; Figures S2B and S2C). The Cox proportional hazard regression test was performed in advanced stage with under 65 years old (univariate: HR, 2.144, 95% CI: 1.346–3.416, p = 0.0013; multivariate: HR, 2.048, 95% CI: 1.284–3.265, p = 0.0033) and 70-gene signature (univariate: HR: 2.134, 95% CI: 1.328–3.432, p = 0.0017; multivariate: HR, 2.095, 95% CI: 1.303–3.368, p = 0.0023) (Table S3).
The Prediction of Therapeutic Effect by the 70-Gene Signature
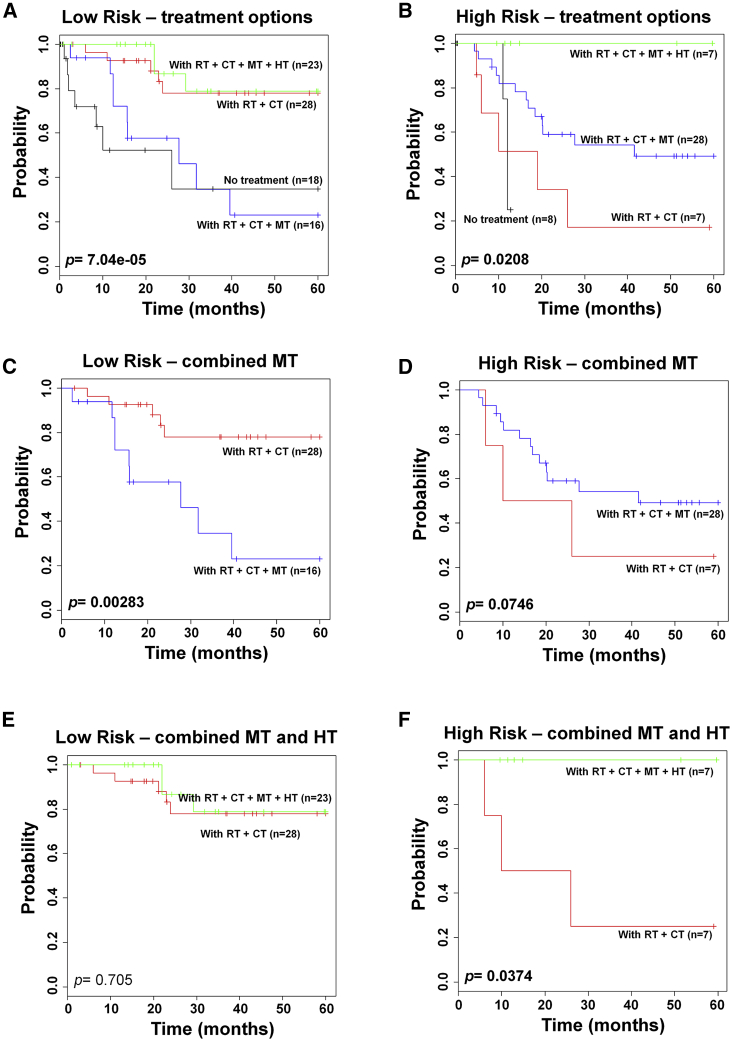
Next, we analyzed the treatment method, such as hysterectomy, radiotherapy, chemotherapy, and molecular-targeted therapy, that was applied to early-stage or late-stage cancer patients. Following the NCCN guidelines, the first-line treatment is hysterectomy and/or radiotherapy in early-stage patients. In early-stage patients, there was no difference in patient survival depending on the applied conventional therapy, such as hysterectomy and/or radiotherapy (Figure S3A). Chemotherapy did not affect the survival even in combination with conventional therapies (Figure S3B). However, chemoradiation therapy is the first-line therapy in advanced-stage patients. The chemoradiation therapy showed a significant difference between the low- and high-risk groups (p = 0.0341; Figure S4). Recently, molecular-targeted therapy was provided to increase survival of patients. Survival differences were observed among the various combination therapies in both low- and high-risk patients with advanced-stage cancer (p = 7.04e−5 and p = 0.0208, respectively; Figures 4A and 4B). However, molecular-targeted therapy resulted in the opposite effect between low- and high-risk patients (p = 0.00283 and p = 0.0746, respectively; Figures 4C and 4D). The combined molecular-targeted therapy, hysterectomy, and chemoradiation therapy affected the survival of patients in only the high-risk group (p = 0.705, and p = 0.0374, respectively; Figures 4E and 4F). In this study, the term hysterectomy included radical hysterectomy, simple hysterectomy, and radical trachelectomy.
Figure 4.
Survival Prediction of Therapeutic Effect by the 70-Gene Signature in Advanced-Stage Cervical Cancer
(A) The therapeutic advantage was evaluated by Kaplan-Meier analysis. Patients were separated according to the treatment options in low-risk. (B) The therapeutic advantage was evaluated by Kaplan-Meier analysis. Patients were separated according to the treatment options in high-risk. (C) Chemoradiation therapy with/without molecular-targeted therapy in low-risk. (D) Chemoradiation therapy with/without molecular-targeted therapy in high-risk. (E) Combined chemoradiation therapy, molecular-targeted therapy, with/without hysterectomy in low-risk. (F) Combined chemoradiation therapy, molecular-targeted therapy, with/without hysterectomy in high-risk. Each group was evaluated by Kaplan-Meier analysis. The p values were obtained from the χ2 test. CT, chemotherapy; HT, hysterectomy; MT, molecular-targeted therapy; RT, radiotherapy.
Classification Analysis Results of 70 Genes in Cervical Cancer
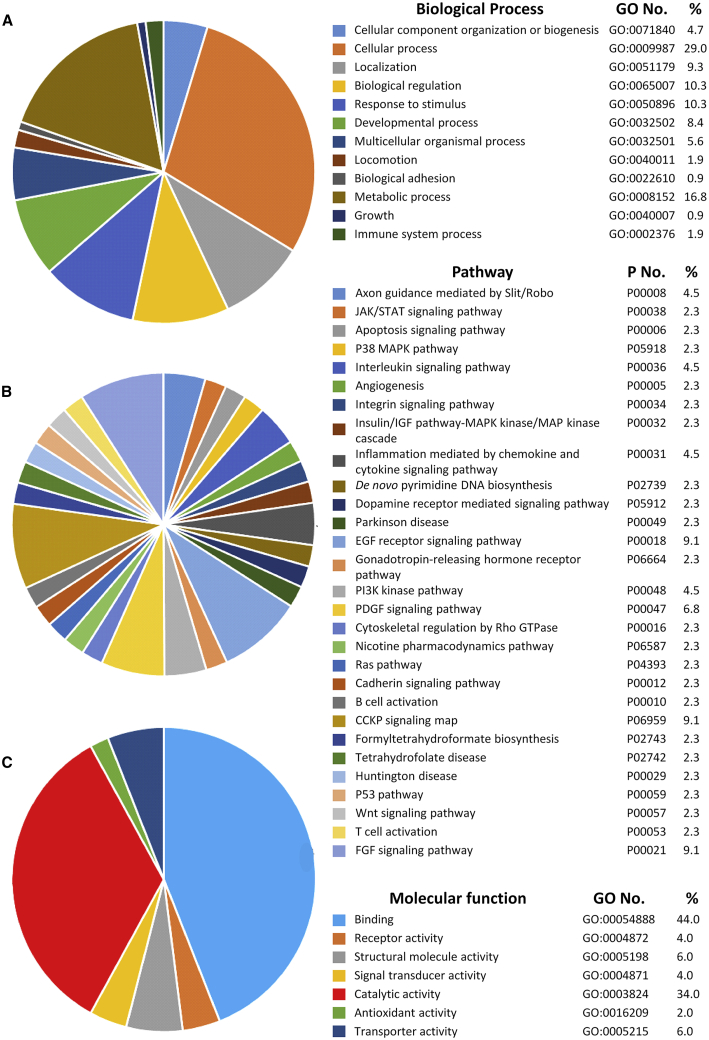
Gene Ontology enrichment analysis in DAVID (Database for Annotation, Visualization, and Integrated Discovery, National Institutes of Health, Bethesda, MD, USA) was used to identify the biological functions of the genes in the 70-gene signature (Table S4) and identified 31 significant terms (biological processes) (Table 1). These terms consist of 29% of cellular process, 16.8% metabolic process, 10.3% biological regulation, and 10.3% response to stimulus (Figure 5A). The 70 genes were analyzed to see the relationship with functional pathway using the PANTHER system. They were involved in various pathways (Figure 5B). The functions of 70 genes were related to binding (44.0%), catalytic activity (34.0%), and other processes (Figure 5C).
Table 1.
Gene Ontology Analysis
| Term | Name | Count | p Value | Genes |
|---|---|---|---|---|
| GO:0031623 | receptor internalization | 4 | 0.000698039 | GRB2, GHR, DNM2, BTN1A1 |
| GO:0040014 | regulation of multicellular organism growth | 3 | 0.004932049 | FGFR2, STAT5A, GHR |
| GO:0001764 | neuron migration | 4 | 0.008871214 | CXCL12, CDK5R2, BAX, NTRK2 |
| GO:0007155 | cell adhesion | 7 | 0.010592445 | CXCL12, CX3CL1, NCAN, PCDHB6, APLP1, HAPLN1, COL19A1 |
| GO:0032526 | response to retinoic acid | 3 | 0.011967854 | RBP4, NCOA1, SCAMP3 |
| GO:0006549 | isoleucine metabolic process | 2 | 0.01210023 | STAT5A, GHR |
| GO:0000255 | allantoin metabolic process | 2 | 0.01210023 | STAT5A, GHR |
| GO:0007595 | lactation | 3 | 0.012533686 | NCOA1, STAT5A, GHRHR |
| GO:0006573 | valine metabolic process | 2 | 0.016101492 | STAT5A, GHR |
| GO:0046449 | creatinine metabolic process | 2 | 0.016101492 | STAT5A, GHR |
| GO:0001501 | skeletal system development | 4 | 0.018107062 | ZBTB16, NCAN, HAPLN1, COL19A1 |
| GO:0006417 | regulation of translation | 3 | 0.018809178 | DDX25, EIF4E, TYMS |
| GO:0019530 | taurine metabolic process | 2 | 0.020086785 | STAT5A, GHR |
| GO:0048562 | embryonic organ morphogenesis | 2 | 0.020086785 | RBP4, FGFR2 |
| GO:0021987 | cerebral cortex development | 3 | 0.021620051 | NCOA1, BAX, NTRK2 |
| GO:0060041 | retina development in camera-type eye | 3 | 0.023086905 | BAX, SERPINF1, NTRK2 |
| GO:0018108 | peptidyl-tyrosine phosphorylation | 4 | 0.0241554 | BTC, FGFR2, STAT5A, NTRK2 |
| GO:0006101 | citrate metabolic process | 2 | 0.028009715 | STAT5A, GHR |
| GO:0051384 | response to glucocorticoid | 3 | 0.028530929 | TYMS, GHR, GHRHR |
| GO:0007568 | aging | 4 | 0.02932512 | GRB2, SERPINF1, TYMS, BTN1A1 |
| GO:0010518 | positive regulation of phospholipase activity | 2 | 0.031947477 | ARHGAP6, FGFR2 |
| GO:0060670 | branching involved in labyrinthine layer morphogenesis | 2 | 0.03586952 | GRB2, FGFR2 |
| GO:0060068 | vagina development | 2 | 0.03586952 | RBP4, BAX |
| GO:0030324 | lung development | 3 | 0.038002374 | RBP4, EIF4E, FGFR2 |
| GO:0006105 | succinate metabolic process | 2 | 0.039775906 | STAT5A, GHR |
| GO:0014066 | regulation of phosphatidylinositol 3-kinase signaling | 3 | 0.039837466 | GRB2, BTC, FGFR2 |
| GO:0006600 | creatine metabolic process | 2 | 0.043666696 | STAT5A, GHR |
| GO:0030198 | extracellular matrix organization | 4 | 0.045163581 | MFAP2, NCAN, HAPLN1, COL19A1 |
| GO:0006107 | oxaloacetate metabolic process | 2 | 0.047541952 | STAT5A, GHR |
| GO:0002031 | G-protein-coupled receptor internalization | 2 | 0.047541952 | DNM2, BTN1A1 |
| GO:0045778 | positive regulation of ossification | 2 | 0.047541952 | ZBTB16, BTN1A1 |
Figure 5.
The Functional Classification Analysis of 70 Genes in Cervical Cancer
PANTHER classification system was used to classify the 70 genes according to their functions. (A) Biological process analysis. (B) Functional pathway analysis. (C) Molecular function analysis of the proteins translated by 70 genes.
Discussion
The previous studies showed that the gene signatures can predict prognosis in cervical cancer patients. In locally advanced cervical cancer, it was suggested that ANXA2-NDRG1-STAT1 gene signature was a candidate for concurrent chemoradiation treatment.20 Fernandez-Retana et al.23 established a molecular signature comprising eight degradome-related genes that predicted which patients were at risk for developing distal metastasis among the locally advanced cervical cancer patients. The epigenetic gene regulation has also been extensively analyzed. The methylation pattern observed in promoters of nine genes constituted a potential biomarker for early detection and screening of cervical cancer.24 In the other studies, gene signatures of microRNAs showed correlation with early diagnosis, prognosis, treatment, occurrence, and cancer development.25, 26, 27, 28 The long non-coding RNAs also correlated with diagnosis, prognosis, recurrence, metastasis, and effective targeted therapy.29, 30, 31, 32
In clinic, FIGO stage system classifies cancer depending on tumor size, spread to a lymph node, and metastasis. Stages are further subdivided based on the lesion’s maximum diameter: stage IB3, diameter of ≥4 cm; and stage IIA2, diameter of ≥4 cm.33 Cervical cancer is diagnosed most frequently in middle-aged women between the ages of 35 and 44 years. It rarely develops in women younger than 20 years. The older women, over 65 years old, account for 10% of cervical cancer patients and are more likely to die of the disease because they are at advanced stage when diagnosed. Importantly, we showed that the low-risk patients stratified by our 70-gene signature had better survival even in the same tumor stage, size, and age, suggesting that cervical cancer patients were more accurately subclassified if the 70-gene signature was added to the conventional classification system.
Although the prediction of survival and prognosis depends on the FIGO stage, tumor size, and age, it is unknown why prognosis, survival, and drug response are different in the same stage. Radiotherapy and/or hysterectomy is the standard therapy for treatment of early-stage cancer, whereas chemoradiation therapy is generally used for advanced-stage patients. Generally, 80%–90% of patients in early stage are cured using surgery and/or radiotherapy, but it is still difficult to choose the right treatment method for women with advanced-stage cervical cancer. To increase the survival of the patients, chemotherapy and molecular-targeted therapy are currently recommended to early-stage and advanced-stage patients, respectively. In this study, we showed that the 70-gene signature increased accuracy for predicting prognosis of the patients. We observed that chemotherapy had no effect on survival in the early stage of cancer. In contrast, molecular-targeted therapy combined with chemoradiation therapy provided a survival benefit in high-risk patients (p = 0.0746), but had the opposite result in low-risk patients (p = 0.00283). Thus, the 70-gene signature could help to recommend molecular-targeted therapy to high-risk patients suffering from advanced-stage cancer.
Molecular-targeted therapy was approved in recurrent or metastatic cervical cancer.34 In clinical trials, molecular-targeted therapy increased OS and progression-free survival (PFS), but decreased HR. In a randomized controlled study, vascular endothelial growth factor (VEGF) increased median OS up to 4 months and decreased HR to 0.71.35 HER2 and epidermal growth factor receptor (EGFR) targeted therapies (clinical trial phase II, randomized controlled study) showed median OS (11 months), time to progression (4.27 months), and stable disease (44%).36 A single treatment of EGFR targeted agents showed minimal activity or no meaningful benefits.37, 38, 39, 40, 41 programed cell death (PD-1) and programmed cell death ligand 1 (PD-L1) inhibitors increased the response rate in recurrence and metastatic cervical cancer patients.42 In our study, 31 biological processes were identified from Gene Ontology term analysis of 70 genes. Analysis by the PANTHER classification system revealed the angiogenesis pathway as the major target of molecular-targeted therapy. Bevacizumab, an angiogenesis blocker, was associated with regulation of phosphatidylinositol 3-kinase signaling, extracellular matrix organization, and cell adhesion, which were identified in our Gene Ontology analysis (Table 1).
Our study had several strengths. First, we showed that the 70-gene signature was a good biomarker for better prognosis prediction of cervical cancer. Second, it allowed for stratification of the patients into two distinctive risk groups even when the patients were also classified in terms of stage, size, and age. Finally, it provided the guidance on how to select patients (high risk in the advanced stage) who could benefit from optimal molecular-targeted therapy. There are a few limitations in this study. Only a small number of patients were analyzed for outcome determination with each therapeutic modality, and more detailed analysis of each molecular-targeted therapy and chemotherapy drugs was not possible to perform due to limited information. The clinical stages used in this study corresponded to the previous FIGO version because the new staging system was announced in 2019.
In conclusion, the 70-gene signature could constitute a more accurate biomarker for further stratification of patients besides by stage, size, and age. It also provided guidance for identifying patients who could benefit from molecular-targeted therapy in advanced-stage cervical cancer.
Materials and Methods
Gene Expression Databases
Three gene expression datasets were obtained from the National Center for Biotechnology Information GEO database (http://www.ncbi.nlm.nih.gov/geo) and one NGS dataset was from TCGA database (https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga). Gene expression data from the GEO: GSE44001 was used as the training dataset. TCGA, GSE39001, and GSE52904 were used as validation datasets (Table S5). Table 2 showed clinicopathological and clinical information of patients in detail, which were provided by TCGA and GEO databases. Healthy controls data were collected from GEO: GSE39001 and GEO: GSE52904.
Table 2.
Clinical Characteristics of Patients in the Training and Validation Datasets with Cervical Cancer
| Variable | Training Set |
Validation Set |
||
|---|---|---|---|---|
| GEO: GSE44001 | TCGA | GEO: GSE39001 | GEO: GSE52094 | |
| Patients used, n | 300 | 290 | 21 | 55 |
| Median age (range), years | 46 (20–88) | 43 (32–67) | 50 (24–74) | |
| Tumor size | ||||
| <4 cm | 218 | |||
| ≥4 cm | 82 | |||
| FIGO stage (n) | ||||
| I | 258 | 155 | 14 | 27 |
| II | 42 | 66 | 7 | 8 |
| III | 0 | 42 | 0 | 16 |
| IV | 0 | 21 | 0 | 4 |
| DFS (range), months | 47.5 (0.43–104.13) | |||
| Follow-up (range), months | 61 (17–93) | 58 (1–86) | ||
| OS (range), months | 18.4 (0.03–214) | |||
| Hysterectomy | ||||
| Yes | 155 | 11 | 15 | |
| No | 9 | 10 | 40 | |
| Radiotherapy | ||||
| Yes | 175 | 17 | 41 | |
| No | 63 | 4 | 14 | |
| Chemotherapy | ||||
| Yes | 145 | 11 | 23 | |
| No | 144 | 10 | 32 | |
| Molecular-targeted therapy | ||||
| Yes | 84 | |||
| No | 44 | |||
DFS, disease-free survival; FIGO, International Federation of Gynecology and Obstetrics; OS, overall survival; TCGA, The Cancer Genome Atlas.
Development of the Prognostic Gene Expression Signature
The gene expression data in the GEO: GSE44001 dataset were used to develop the gene signature. At first, genes were filtered by more than 1.5-fold absolute value of log2 scale, which represented the same gene expression level. The filtering step leads to 26,156 probes that were selected among various probe sets. In the next step, the disease-free survival (DFS)-associated gene expression signature derived from the training dataset was identified by the univariate Cox proportional hazard regression (p < 0.001). Initially, 175 probes were found from the analysis. However, only 70 genes were common among the four datasets (one training and three validation datasets). Then the 70-gene signature was selected as a prognostic signature. To predict prognosis, we applied selected probes from the survival signature to the survival risk prediction analysis. The PI was computed by the formula:
where wi and xi were the weight and logged gene expression for the i-th gene, respectively.
The patients were divided into two groups based on a median PI of −0.107633. If the PIs were greater than −0.107633, patients were assigned to the high-risk group. In contrast, patients with PIs equivalent to or less than −0.107633 were assigned to the low-risk group. The cluster analysis was performed with Cluster 3.0. Initially, 175 probes were used for prediction model analysis, and only the 70 genes were common in all of the chip types (Figure S5).
Validation of the Prognostic Gene Expression Signature
The validation of the gene signature was accomplished on independent datasets. Gene expression data from validation datasets were individually adjusted by subtracting the median expression value across the samples. To integrate each validation dataset in order to construct the prediction models, we aligned the 70-gene set in each dataset.
To further refine this model and to sub-stratify the predicted outcomes, we used Compound Covariate Predictor (CCP) as a class prediction algorithm. Gene expression data in the training set were combined to form a classifier according to CCP. The robustness of the classifier was determined by the misclassification rate obtained during the leave-one-out cross-validation (LOOCV) in the training set. Kaplan-Meier (KM) survival analyses were performed after the patients were divided into two predicted subgroups, and chi-square (χ2) and log rank tests were used to evaluate the survival risk between the two predicted subgroups of patients. Univariate and multivariate Cox proportional hazard models were used to evaluate independent prognostic factors associated with survival, gene signature, stage, size, and age, as covariates.
Statistical Methods of Microarray Data
Heatmap was analyzed using BRB-Array Tools Version 4.3 (National Institutes of Health, MD, USA). All other statistical analyses were accomplished in the R language environment (R Foundation for Statistical Computing, Vienna, Austria) and Statistical Package for Social Sciences (SPSS) software (version 20; IBM, Armonk, NY, USA). Cluster analysis was performed with Cluster 3 and Tree View (Stanford University, CA, USA). Statistical significance was defined as p < 0.05.
Differential Protein and Gene Categorization and Network Modeling
DAVID was used to classify the genes. DAVID provides a comprehensive set of functional annotation and biological meaning of genes. PANTHER classification system (University of Southern California, CA, USA) was used to classify the proteins, and genes by the function. Protein functions were categorized according to the biological process, protein class, and molecular function through protein’s relationship analysis.
Author Contributions
N.N.Y.N., Y.H.J., and S.S.K. designed and performed the research, analyzed the data, and wrote the manuscript. T.G.C., J.K., and Y.S. assisted in bioinformatics and statistical analyses. M.H.J. and S.H.K. supervised the study and wrote the manuscript. I.K. and J.H. conceived the project and supervised the study. All authors revised and approved the manuscript. Y.H.J. and S.S.K. shared the corresponding authorship.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This study was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) grants funded by the Korean Government (NRF-2017R1A6A3A11028420 and NRF-2020R1I1A1A01065254 to Y.H.J.; NRF-2018R1A6A1A03025124 to S.S.K.).
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omto.2020.09.001.
Contributor Information
Sung Soo Kim, Email: sgskim@khu.ac.kr.
Yong Hwa Jo, Email: yonghwa.jo@gmail.com.
Supplemental Information
References
- 1.World Health Organization . 2018. Cervical cancer.http://www.who.int/health-topics/cervical-cancer [Google Scholar]
- 2.Aoki Y., Sasaki M., Watanabe M., Sato T., Tsuneki I., Aida H., Tanaka K. High-risk group in node-positive patients with stage IB, IIA, and IIB cervical carcinoma after radical hysterectomy and postoperative pelvic irradiation. Gynecol. Oncol. 2000;77:305–309. doi: 10.1006/gyno.2000.5788. [DOI] [PubMed] [Google Scholar]
- 3.Ayhan A., Al R.A., Baykal C., Demirtas E., Yüce K., Ayhan A. A comparison of prognoses of FIGO stage IB adenocarcinoma and squamous cell carcinoma. Int. J. Gynecol. Cancer. 2004;14:279–285. doi: 10.1111/j.1048-891X.2004.014211.x. [DOI] [PubMed] [Google Scholar]
- 4.Eifel P.J., Burke T.W., Morris M., Smith T.L. Adenocarcinoma as an independent risk factor for disease recurrence in patients with stage IB cervical carcinoma. Gynecol. Oncol. 1995;59:38–44. doi: 10.1006/gyno.1995.1265. [DOI] [PubMed] [Google Scholar]
- 5.Sevin B.U., Nadji M., Lampe B., Lu Y., Hilsenbeck S., Koechli O.R., Averette H.E. Prognostic factors of early stage cervical cancer treated by radical hysterectomy. Cancer. 1995;76(Suppl 10):1978–1986. doi: 10.1002/1097-0142(19951115)76:10+<1978::aid-cncr2820761313>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 6.Trattner M., Graf A.H., Lax S., Forstner R., Dandachi N., Haas J., Pickel H., Reich O., Staudach A., Winter R. Prognostic factors in surgically treated stage ib-iib cervical carcinomas with special emphasis on the importance of tumor volume. Gynecol. Oncol. 2001;82:11–16. doi: 10.1006/gyno.2001.6252. [DOI] [PubMed] [Google Scholar]
- 7.Kristensen G.B., Abeler V.M., Risberg B., Trop C., Bryne M. Tumor size, depth of invasion, and grading of the invasive tumor front are the main prognostic factors in early squamous cell cervical carcinoma. Gynecol. Oncol. 1999;74:245–251. doi: 10.1006/gyno.1999.5420. [DOI] [PubMed] [Google Scholar]
- 8.Monk B.J., Tewari K.S., Koh W.J. Multimodality therapy for locally advanced cervical carcinoma: state of the art and future directions. J. Clin. Oncol. 2007;25:2952–2965. doi: 10.1200/JCO.2007.10.8324. [DOI] [PubMed] [Google Scholar]
- 9.Gaffney D.K., Soisson A.P. Simple or complex: optimal therapy for cancer of the cervix. Gynecol. Oncol. 2010;119:401–403. doi: 10.1016/j.ygyno.2010.09.018. [DOI] [PubMed] [Google Scholar]
- 10.Abu-Rustum N.R., Yashar C.M., Bradley K., Campos S.M., Chon H.S., Chu C. NCCN; 2018. Cervical cancer (version 1. 2018)https://www.nccn.org/store/login/login.aspx?ReturnURL=https://www.nccn.org/professionals/physician_gls/pdf/cervical.pdf [Google Scholar]
- 11.Rotman J., Mom C.H., Jordanova E.S., de Gruijl T.D., Kenter G.G. ‘DURVIT’: a phase-I trial of single low-dose durvalumab (Medi4736) IntraTumourally injected in cervical cancer: safety, toxicity and effect on the primary tumour- and lymph node microenvironment. BMC Cancer. 2018;18:888. doi: 10.1186/s12885-018-4764-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Menderes G., Black J., Schwab C.L., Santin A.D. Immunotherapy and targeted therapy for cervical cancer: an update. Expert Rev. Anticancer Ther. 2016;16:83–98. doi: 10.1586/14737140.2016.1121108. [DOI] [PubMed] [Google Scholar]
- 13.Dang Y., Wang Y.C., Huang Q.J. Microarray and next-generation sequencing to analyse gastric cancer. Asian Pac. J. Cancer Prev. 2014;15:8033–8039. [PubMed] [Google Scholar]
- 14.Nguyen M.N., Choi T.G., Nguyen D.T., Kim J.H., Jo Y.H., Shahid M., Akter S., Aryal S.N., Yoo J.Y., Ahn Y.J. CRC-113 gene expression signature for predicting prognosis in patients with colorectal cancer. Oncotarget. 2015;6:31674–31692. doi: 10.18632/oncotarget.5183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shahid M., Choi T.G., Nguyen M.N., Matondo A., Jo Y.H., Yoo J.Y., Nguyen N.N., Yun H.R., Kim J., Akter S. An 8-gene signature for prediction of prognosis and chemoresponse in non-small cell lung cancer. Oncotarget. 2016;7:86561–86572. doi: 10.18632/oncotarget.13357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arranz E.E., Vara J.A., Gámez-Pozo A., Zamora P. Gene signatures in breast cancer: current and future uses. Transl. Oncol. 2012;5:398–403. doi: 10.1593/tlo.12244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Paik S., Shak S., Tang G., Kim C., Baker J., Cronin M., Watson D., Bryant J., Costontino J., Wolmark N. Expression of the 21 genes in the Recurrence Score assay and tamoxifen clinical benefit in the NSABP study B-14 of node negative, estrogen receptor positive breast cancer. J. Clin. Oncol. 2005;23(Suppl 16):510. [Google Scholar]
- 18.Kitahara O., Katagiri T., Tsunoda T., Harima Y., Nakamura Y. Classification of sensitivity or resistance of cervical cancers to ionizing radiation according to expression profiles of 62 genes selected by cDNA microarray analysis. Neoplasia. 2002;4:295–303. doi: 10.1038/sj.neo.7900251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harima Y., Ikeda K., Utsunomiya K., Shiga T., Komemushi A., Kojima H., Nomura M., Kamata M., Sawada S. Identification of genes associated with progression and metastasis of advanced cervical cancers after radiotherapy by cDNA microarray analysis. Int. J. Radiat. Oncol. Biol. Phys. 2009;75:1232–1239. doi: 10.1016/j.ijrobp.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 20.Buttarelli M., Babini G., Raspaglio G., Filippetti F., Battaglia A., Ciucci A., Ferrandina G., Petrillo M., Marino C., Mancuso M. A combined ANXA2-NDRG1-STAT1 gene signature predicts response to chemoradiotherapy in cervical cancer. J. Exp. Clin. Cancer Res. 2019;38:279. doi: 10.1186/s13046-019-1268-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xia L., Wang H., Cai S., Su X., Shen J., Meng Q., Chen Y., Li L., Yan J., Zhang C., Xu M. Integrated Analysis of a Competing Endogenous RNA Network Revealing a Prognostic Signature for Cervical Cancer. Front. Oncol. 2018;8:368. doi: 10.3389/fonc.2018.00368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yost S., Hoekstra A. Cervical cancer in women over 65: An analysis of screening. Gynecol. Oncol. Rep. 2018;25:48–51. doi: 10.1016/j.gore.2018.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fernandez-Retana J., Zamudio-Meza H., Rodriguez-Morales M., Pedroza-Torres A., Isla-Ortiz D., Herrera L., Jacobo-Herrera N., Peralta-Zaragoza O., López-Camarillo C., Morales-Gonzalez F. Gene signature based on degradome-related genes can predict distal metastasis in cervical cancer patients. Tumour Biol. 2017;39 doi: 10.1177/1010428317711895. 1010428317711895. [DOI] [PubMed] [Google Scholar]
- 24.Bhat S., Kabekkodu S.P., Varghese V.K., Chakrabarty S., Mallya S.P., Rotti H., Pandey D., Kushtagi P., Satyamoorthy K. Aberrant gene-specific DNA methylation signature analysis in cervical cancer. Tumour Biol. 2017;39 doi: 10.1177/1010428317694573. 1010428317694573. [DOI] [PubMed] [Google Scholar]
- 25.Sandhu S., Garzon R. Potential applications of microRNAs in cancer diagnosis, prognosis, and treatment. Semin. Oncol. 2011;38:781–787. doi: 10.1053/j.seminoncol.2011.08.007. [DOI] [PubMed] [Google Scholar]
- 26.Garzon R., Calin G.A., Croce C.M. MicroRNAs in Cancer. Annu. Rev. Med. 2009;60:167–179. doi: 10.1146/annurev.med.59.053006.104707. [DOI] [PubMed] [Google Scholar]
- 27.Liu S.S., Chan K.K.L., Chu D.K.H., Wei T.N., Lau L.S.K., Ngu S.F., Chu M.M.Y., Tse K.Y., Ip P.P.C., Ng E.K.O. Oncogenic microRNA signature for early diagnosis of cervical intraepithelial neoplasia and cancer. Mol. Oncol. 2018;12:2009–2022. doi: 10.1002/1878-0261.12383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun P., Shen Y., Gong J.M., Zhou L.L., Sheng J.H., Duan F.J. A New MicroRNA Expression Signature for Cervical Cancer. Int. J. Gynecol. Cancer. 2017;27:339–343. doi: 10.1097/IGC.0000000000000863. [DOI] [PubMed] [Google Scholar]
- 29.Mao Y., Dong L., Zheng Y., Dong J., Li X. Prediction of Recurrence in Cervical Cancer Using a Nine-lncRNA Signature. Front. Genet. 2019;10:284. doi: 10.3389/fgene.2019.00284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luo W., Wang M., Liu J., Cui X., Wang H. Identification of a six lncRNAs signature as novel diagnostic biomarkers for cervical cancer. J. Cell. Physiol. 2020;235:993–1000. doi: 10.1002/jcp.29015. [DOI] [PubMed] [Google Scholar]
- 31.Shen L., Yu H., Liu M., Wei D., Liu W., Li C., Chang Q. A ten-long non-coding RNA signature for predicting prognosis of patients with cervical cancer. OncoTargets Ther. 2018;11:6317–6326. doi: 10.2147/OTT.S175057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Taheri M., Ghafouri-Fard S. Long Non-Cod-ing RNA Signature in Cervical Cancer. Klin. Oncol. 2018;31:403–408. doi: 10.14735/amko2018403. [DOI] [PubMed] [Google Scholar]
- 33.Berek J.S., Matsuo K., Grubbs B.H., Gaffney D.K., Lee S.I., Kilcoyne A., Cheon G.J., Yoo C.W., Li L., Shao Y. Multidisciplinary perspectives on newly revised 2018 FIGO staging of cancer of the cervix uteri. J. Gynecol. Oncol. 2019;30:e40. doi: 10.3802/jgo.2019.30.e40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.FDA . 2019. FDA approval for bevacizumab.https://www.cancer.gov/about-cancer/treatment/drugs/bevacizumab [Google Scholar]
- 35.Tewari K.S., Sill M.W., Long H.J., 3rd, Penson R.T., Huang H., Ramondetta L.M., Landrum L.M., Oaknin A., Reid T.J., Leitao M.M. Improved survival with bevacizumab in advanced cervical cancer. N. Engl. J. Med. 2014;370:734–743. doi: 10.1056/NEJMoa1309748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Monk B.J., Mas Lopez L., Zarba J.J., Oaknin A., Tarpin C., Termrungruanglert W., Alber J.A., Ding J., Stutts M.W., Pandite L.N. Phase II, open-label study of pazopanib or lapatinib monotherapy compared with pazopanib plus lapatinib combination therapy in patients with advanced and recurrent cervical cancer. J. Clin. Oncol. 2010;28:3562–3569. doi: 10.1200/JCO.2009.26.9571. [DOI] [PubMed] [Google Scholar]
- 37.Schilder R.J., Sill M.W., Lee Y.C., Mannel R. A phase II trial of erlotinib in recurrent squamous cell carcinoma of the cervix: a Gynecologic Oncology Group Study. Int. J. Gynecol. Cancer. 2009;19:929–933. doi: 10.1111/IGC.0b013e3181a83467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goncalves A., Fabbro M., Lhommé C., Gladieff L., Extra J.M., Floquet A., Chaigneau L., Carrasco A.T., Viens P. A phase II trial to evaluate gefitinib as second- or third-line treatment in patients with recurring locoregionally advanced or metastatic cervical cancer. Gynecol. Oncol. 2008;108:42–46. doi: 10.1016/j.ygyno.2007.07.057. [DOI] [PubMed] [Google Scholar]
- 39.Farley J., Sill M.W., Birrer M., Walker J., Schilder R.J., Thigpen J.T., Coleman R.L., Miller B.E., Rose P.G., Lankes H.A. Phase II study of cisplatin plus cetuximab in advanced, recurrent, and previously treated cancers of the cervix and evaluation of epidermal growth factor receptor immunohistochemical expression: a Gynecologic Oncology Group study. Gynecol. Oncol. 2011;121:303–308. doi: 10.1016/j.ygyno.2011.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Santin A.D., Sill M.W., McMeekin D.S., Leitao M.M., Jr., Brown J., Sutton G.P., Van Le L., Griffin P., Boardman C.H. Phase II trial of cetuximab in the treatment of persistent or recurrent squamous or non-squamous cell carcinoma of the cervix: a Gynecologic Oncology Group study. Gynecol. Oncol. 2011;122:495–500. doi: 10.1016/j.ygyno.2011.05.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kurtz J.E., Hardy-Bessard A.C., Deslandres M., Lavau-Denes S., Largillier R., Roemer-Becuwe C., Weber B., Guillemet C., Paraiso D., Pujade-Lauraine E. Cetuximab, topotecan and cisplatin for the treatment of advanced cervical cancer: A phase II GINECO trial. Gynecol. Oncol. 2009;113:16–20. doi: 10.1016/j.ygyno.2008.12.040. [DOI] [PubMed] [Google Scholar]
- 42.Chung H.C., Schellens J.H.M., Delord J.-P., Perets R., Italiano A., Shapira-Frommer R., Manzuk L., Piha-Paul S.A., Wang J., Zeigenfuss S. Pembrolizumab treatment of advanced cervical cancer: Updated results from the phase 2 KEYNOTE-158 study. J. Clin. Oncol. 2018;36(Suppl 15):5522. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.