Abstract
Background & objectives:
SARS-CoV-2 (Severe acute respiratory syndrome coronavirus-2) is evolving with the progression of the pandemic. This study was aimed to investigate the diversity and evolution of the coronavirus SARS-CoV-2 with progression of the pandemic over time and to identify similarities and differences of viral diversity and evolution across geographical regions (countries).
Methods:
Publicly available data on type definitions based on whole-genome sequences of the SARS-CoV-2 sampled during December and March 2020 from 3636 infected patients spread over 55 countries were collected. Phylodynamic analyses were performed and the temporal and spatial evolution of the virus was examined.
Results:
It was found that (i) temporal variation in frequencies of types of the coronavirus was significant; ancestral viruses of type O were replaced by evolved viruses belonging to type A2a; (ii) spatial variation was not significant; with the spread of SARS-CoV-2, the dominant virus was the A2a type virus in every geographical region; (iii) within a geographical region, there was significant micro-level variation in the frequencies of the different viral types, and (iv) the evolved coronavirus of type A2a swept rapidly across all continents.
Interpretation & conclusions:
SARS-CoV-2 belonging to the A2a type possesses a non-synomymous variant (D614G) that possibly eases the entry of the virus into the lung cells of the host. This may be the reason why the A2a type has an advantage to infect and survive and as a result has rapidly swept all geographical regions. Therefore, large-scale sequencing of coronavirus genomes and, as required, of host genomes should be undertaken in India to identify regional and ethnic variation in viral composition and its interaction with host genomes. Further, careful collection of clinical and immunological data of the host can provide deep learning in relation to infection and transmission of the types of coronavirus genomes.
Keywords: Host genome interaction, phylogeny, RNA sequence, SARS-CoV-2, viral type coronavirus
Coronaviruses have emerged as major human respiratory pathogens. Before the emergence of SARS-CoV-2, six other coronaviruses were known to infect humans. All of those cause clinical symptoms. Two of these, SARS-CoV and MERS-CoV, caused severe disease and often death as was observed in the epidemics of 2003 and 2012, respectively1. The remaining four (HKU1, NL63, OC43 and 229E) cause mild respiratory distress. Coronaviruses are positive-sense, single-stranded (+ss) RNA viruses. The RNA genome of SARS-CoV-2 has about 30,000 nucleotides, encoding for 29 proteins1. The structural proteins include the spike (S), the envelope (E), the membrane (M) and the nucleocapsid (N) proteins. Three coronaviruses have crossed species barriers from bat to civet cat (SARS-CoV) or camel (MERS-CoV) or pangolin (SARS-CoV-2), before crossing to human. The causes or mechanisms of species barrier crossing are not completely known. Based on the fact that the sequence identity of eight SARS-CoV-2 whole genomes sampled from China immediately after the outbreak in Wuhan exceeds 99.98 per cent1, it may be inferred that SARS-CoV-2 emerged in humans very recently. Further, the SARS-CoV-2 strains were less genetically similar to SARS-CoV (about 79%) and MERS-CoV (about 50%)1. Based on the extent of sequence identity, it has been inferred that SARS-CoV-2 has descended from SARS-CoV1.
SARS-CoV-2 is extremely contagious. However, the case fatality rate of SARS-CoV-2 (2-3%)2 is much lower compared to the SARS-CoV (11%)3 or MERS-CoV (34%)4. One reason why SARS-CoV-2 is so successful in infecting humans is because of its ability to use human angiotensin converting enzyme 2 (ACE2)1 as a receptor and enter the target cells in the human lung. The spike (S) protein mediates receptor binding and membrane fusion5. The spike protein of coronaviruses has two functional domains – S1, responsible for receptor binding, and S2 domain, responsible for cell membrane fusion6. Five key residues in the receptor-binding domain enable efficient binding of SARS-CoV-2 to human ACE2; these are Asn439, Asn501, Gln493, Gly485 and Phe4861. Another mutation, A23403G, located in the gene encoding the spike glycoprotein results in an amino acid change (D614G) from aspartic acid to glycine. Although the effect of the D614G mutation is unclear, this mutation is located in the S1-S2 junction near the furin recognition site (R667) for the cleavage of S protein that is required for the entry of the virion into the host cell7.
Currently, a large number of sequences of SARS-CoV-2 sampled from infected individuals from various geographical regions (after the infection was first reported from Wuhan, China, in December 2019) – are publicly available (https://www.gisaid.org/). The evolution of SARS-CoV-2, in relation to coronaviruses found in bats, pangolins and other animals, has been studied on the basis of 103 sequences that were available from a limited geographical region in January 20208. This study identified that SARS-CoV-2 has evolved into two major types8. A more recent study9 has identified three major types based on 160 sequences that were collected before March 3, 2020. Both of these studies have failed to identify the major features of temporal evolution of SARS-CoV-2 because of small sample sizes and inclusion of sequences of samples that were essentially collected before March 2020. The geographical spread of SARS-CoV-2 was extremely rapid after/ during March 2020.
A much larger data set on SARS-CoV-2 sequences is now available, from isolates that have been sampled throughout the period of spread of this infection and from multiple geographical regions. We undertook an analysis of genomic sequences of SARS-CoV-2 with the following objectives: (i) to investigate the diversity and evolution of SARS-CoV-2 with progression of the pandemic over time; (ii) to investigate similarities and differences of viral diversity and evolution, along with transmission, across geographical regions (countries); and (iii) to formulate relevant questions relating the evolution of this virus in India with clinical and immunological outcomes.
Material & Methods
The data dump was downloaded from www.nextstrain.org (https://nextstrain.org/ncov/) on April 6, 2020. The data contained information on 3639 nCov2019 viral strains. The developers of this portal use SARS-CoV-2 sequences deposited to the Global Initiative on Sharing All Influenza Data (GISAID; https://www.gisaid.org/), carry out quality checks and use a highly stringent analysis pipeline comprising a bioinformatics workflow manager, Augur, and a data visualization front-end web framework, Auspice, to uniformly process all quality passed sequences. The multiple sequence alignment and site numbering for amino acids uses the first viral genome sequence named ncov2019-Wuhan-hu-1/2019 (Genbank accession no: MN908947) as reference. The viral type assignment is rooted and based on the early samples from Wuhan, People's Republic of China. The data dump from the Nextstrain portal contains information on various parameters such as viral strain name, viral sample collection data, sampled from the country and State level information as available from the submitter, viral type information, age, GISAID accession number and sequence submission date. Three sequences – two collected from non-human species (canine and panther) and one collected from a human in April 2020, were excluded (we excluded the sample collected from a human since we attempted to analyze data by month from December 2019 through March 2020). Therefore, our analysis was based on 3636 nCov2019 viral samples. Type defining marker mutations (mostly amino acid changes) were obtained from the Nextstrain github repository (https://github.com/nextstrain/ncov). To draw global inferences, specific sets of analysis were carried out on the pool of all 3636 sequences. To draw more focused inferences, some sets of analysis were performed on data from nine countries (China, Italy, USA, United Kingdom, Spain, Iceland, Australia, Brazil and Congo) from where sequence data were available in large numbers. To understand contrasting patterns of viral transmission, State-level data from four specific countries (USA, United Kingdom, Spain and Canada) were used. Standard Unix tools and data visualization packages were used to partition data over time, countries and to define types. Sample collection date was used for all temporal analyses. For many pathogens, in particular RNA viruses, the timescale on which evolutionary processes and epidemiological processes (within-host diversity and transmission) occur is essentially the same. Therefore, pathogen evolutionary inferences from genetic sequences must simultaneously consider host dynamics and pathogen genetics; this is called 'phylodynamics analysis'10. Phylodynamic analyses were performed using TreeTime11, as implemented in the Nextstrain pipeline12.
To formally test for selection, we computed Tajima's D13.
Results
Figure 1A presents the evolutionary relationships among the 3636 RNA sequences of SARS-CoV-2, combining phylogenetic and transmission information. The tree is radially displayed in concentric circles, with the date of sequence data deposition during the period marked on each concentric circle. There are various types with differing numbers of sequences in these types; the types are colour coded in Figure 1A. The defining mutations of each type are provided in Table I. The earliest sequences emanating from the innermost concentric circle form a distinct type – type O – which is the ancestral type. Sequences of type O were collected from patients initially infected in Wuhan, People's Republic of China. The remaining types are all derived ones. Only two sequences were contributed from India during the period under consideration (December 2019 to March 2020) in this study; both sequences belong to the O type. In addition to the ancestral type (O), there were 10 derived types. The order in which the derived types have evolved, as determined by the data on sequence diversity and date of viral sample collection, is provided in Table I. Five types (O, B, B1, A1a and A2a) have high frequencies (Table I). It is noteworthy that 51 per cent of the viral sequences belonged to a single derived type A2a (Table I and Fig. 1A). There was considerable sequence variation across isolates along the entire length of the genome of SARS-CoV-2 (Fig. 1B top panel), many of which were non-synonymous (Fig. 1B middle panel). The non-synonymous D614G mutation in the spike protein occurred at a high frequency. This is the defining mutation of type A2a.
Fig. 1.
(A) Radially displayed phylogenetic tree of 3636 RNA sequences of SARS-CoV-2. The various types (O, A2, B, etc.) are colour coded. (B) Top and middle panels depict variations at the nucleotide and amino acid levels, respectively, along the RNA sequence of SARS-CoV-2. For each variant, the entropy value [δ=- plog2p–(1- p)log2 (1- p), where p is the variant allele frequency] is provided on the Y-axis for ease of display. The bottom panel provides a description of the structure of the genome of the virus. (Source: https://nextstrain.org).
Table I.
Numbers of SARS-CoV-2 sequences belonging to a specific phylogenetic type
| Phylogenetic type | Type order | Defining mutation(s) for type | Numbers of viral sequence belonging to type (total number of sequences used=3636) |
|---|---|---|---|
| O | 1 | Ancestral type | 582 |
| B | 2 | ORF8 - L84S | 191 |
| B1 | 3 | ORF8 - L84S, nt - C18060T | 505 |
| B2 | 4 | ORF8 - L84S, nt - C29095T | 20 |
| B4 | 5 | ORF8 - L84S, N - S202N | 24 |
| A3 | 6 | ORF1a - V378I, ORF1a - L3606F | 87 |
| A6 | 7 | nt - T514C | 53 |
| A7 | 8 | ORF1a - A3220V | 4 |
| A1a | 9 | ORF3a - G251V, ORF1a - L3606F | 321 |
| A2 | 10 | S - D614G | 1 |
| A2a | 11 | S - D614G, ORF1b - P314L | 1848 |
nt, nucleotide. A total of 11 distinct mutations define 10 derived types
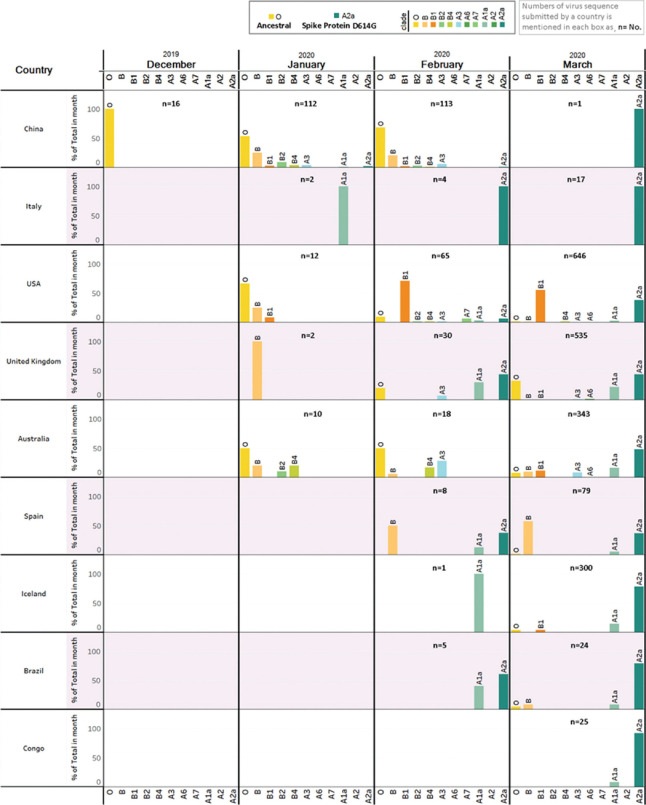
The temporal changes of the five most frequent types of SARS-CoV-2 were studied as it spread geographically. This was done by calculating proportions of sequences belonging to the five types in each of the four months under consideration in this study. The results are presented in Figure 2. In each country, except China, temporal variation of frequencies of virus types was notable. The essential feature was that initially after the pandemic struck, the vast majority of viruses were of the ancestral Chinese (Wuhan) type (Type O). This is more clearly seen from (Fig. 3) that pertains to nine countries most affected by SARS-CoV-2. In each country, diversity of the virus type initially increased and then decreased. The ancestral virus was replaced by viruses that belonged to the evolved type A2a, in each of the most affected countries (Fig. 3) and also globally (Fig. 2). In China, the virus does not seem to have evolved; the ancestral virus of type O has remained the dominant type, although the diversity of viral type has increased over time. Sequence diversity in Italy remained low over time, with A2a being the dominant type. The pattern in USA was interesting as sequence diversity decreased, frequency of ancestral type O diminished remarkably, and the A2a type seemed to be replacing the B1 type. Results based on weekly submissions were similar (Data not shown).
Fig. 2.
Temporal (monthly) change in frequencies of SARS-CoV-2 belonging to the five major types as the virus spread globally (Within each type, the intensity of the colour of each circle is directly proportional to the number of sequences belonging to the type). Source: https://nextstrain.org/ncov/.
Fig. 3.
Temporal (monthly) change in frequencies of five major types of SARS-CoV-2 in five countries in which the prevalence of infection has been high. Source: https://nextstrain.org/ncov/.
Within each of the four countries with high prevalence of infection, there was considerable variation in frequencies of viruses that belonged to the various types (Fig. 4). In the USA, the States of Washington and New York showed contrasting patterns of modal viral types. In Washington, type B1 was the modal (83% of viruses belong to this type), while in New York, the modal type was A2a (81%). This was possibly because of differences in patterns of travel contact with China and Europe. Others have also noted this feature and made similar speculations14.
Fig. 4.
Contrasting frequencies of viral types in different geographical regions within the same country. Source: https://nextstrain.org/ncov/.
It was observed that there was significant temporal, but not spatial variation in frequencies of the different types of SARS-CoV-2; ancestral viruses of type O was replaced by evolved viruses belonging to type A2a. The value of Tajima's D was −2.7. This signifies an excess of low-frequency variants among the coronaviruses in the A2a type and indicates a rapid expansion in its population size and positive selection15,16.
New data submissions from India: Even though only two SARS-CoV-2 sequences were submitted from India until April 6, 2020, there were 33 new submissions to GISAID. The total number of sequences deposited from India was 35 on April 22, 2020. An analysis of 21 sequences, with special reference to Indians returning from abroad, has recently been published17. The 35 viral sequences belonged to four types: the ancestral type O (n=5; 14.3%) and derived types A2a (n=16; 45.7%), A3 (n=13; 37.1%) and B (n=1; 2.9%). Interestingly, two types – A3 and A2a – predominated. As seen from Table II, all persons infected with type A3 coronavirus had travel history to Iran, while most persons with type A2a had no known travel history to countries outside of India.
Table II.
Number of SARS-CoV-2 collected from infected Indians belonging to various types by date of collection and exposure history
| Type | Exposure history | 27 January | 31 January | 02 March | 03 March | 10 March | 12 March | 17 Mar | 05 April | 06 April | 08 April | 10 April | 14 April | Total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| O | China | 1 | 1 | |||||||||||
| Unknown | 2 | 2 | 4 | |||||||||||
| B | China | 1 | 1 | |||||||||||
| A3 | Iran | 8 | 5 | 13 | ||||||||||
| A2a | Europe | 5 | 5 | |||||||||||
| Iran | 1 | 1 | ||||||||||||
| Unknown | 1 | 2 | 2 | 2 | 2 | 1 | 10 | |||||||
| Total | 1 | 1 | 1 | 7 | 9 | 5 | 2 | 2 | 2 | 1 | 2 | 2 | 35 |
Source: https://nextstrain.org/ncov/asia
Discussion
This rapid spread of SARS-CoV-2 to perhaps all countries across the globe is facilitated by the ability of the coronavirus to bind to the human ACE2 receptor that enabled it to enter the alveoli. As the coronavirus spread over the geographical space, it has also evolved. Many mutations that arose throughout the genome of the coronavirus rose to high frequency, among which D614G was notable. These mutations have given rise to clusters of similar sequences that have resulted in the formation of 11 types, of which one is ancestral (O type) that arose in China. The three types (A, B and C) defined by Forster et al9 on the basis of a small number of sequences are broad and some of these types have been split into finer subtypes. The B type defined by C28144T (ORF8: L>S) and T8782C comprises the collection of B, B1, B2 and B4 types of this study9. The A type defined by T29095C is a mutation that is possessed by all sequences of type B2 of this study9. The C type defined by G26144T (ORF3a:G251>V) is the A1a type of this study9. Forster et al9 have not reported the A2a type because A2a evolved and primarily spread widely during March 2020; the data analyzed by Forster et al9 did not include many sequences generated from samples collected in March 2020.
In all countries, initially the ancestral type was the most frequent, possibly because of return of travellers from China but was replaced by the A2a type that is characterized by the D614G non-synonymous mutation located in the S1-S2 junction near the furin recognition site (R667) for the cleavage of S protein required for the entry of the virion into the host cell. Thus, there was a temporal decline in diversity of SARS-CoV-2 types in every geographical region. This selective sweep was consistent with a selective/transmission advantage of the A2a type. It is not clear whether the derived allele producing glycine directly provides a selective/transmission advantage for the entry of the virion or whether the polymorphic locus (Orf1b:P314L; Fig. 1B) with which it is in linkage disequilibrium, provides advantage for entry. Functional studies are required to settle this issue. An earlier genetic analysis of human SARS-CoV has revealed that the spike protein is subjected to a very strong positive selection pressure during transmission and that amino acid residues within the RBD of the S protein is potentially important for progression and tropism18. Further, this study also showed that two-amino acid substitutions (N479K/T487S) in the RBD of SARS-CoV had strong impact on the potential of the coronavirus to infect human cells expressing ACE218.
In India, we need to sequence a large number of viral genomes, relate the type and other genomic features of SARS-CoV-2 with clinical features of the infected persons and, as required, sequence the host genomes to understand the nature and extent of host-virus interaction. In countries with high prevalence of SARS-CoV-2 infection, there were regional differences in frequencies of different types of the coronavirus. It is not clear whether these regional differences are because of differences in patterns of travel of residents or visitors, or whether these are because of differences in ethnic composition. There is a need to investigate regional differences within India in respect of viral genomic diversity and frequencies of virus types. This will inform the relationship of coronavirus type with host ethnicity, perhaps mediated through differences in frequencies of variants in genes of the immune system among ethnic groups in India. Large-scale sequencing of SARS-CoV-2 is essential because it is likely that this virus also mutates rapidly as the influenza virus. Rapid mutations, for certain types of the influenza virus, have significantly reduced sensitivities with many commercial reverse transcription-PCR tests19. RNA sequencing of coronavirus isolates can provide early indication. Further, co-infection with other respiratory viruses is also a possibility for COVID-19, since the presentation of SARS-CoV-2 infection varies from asymptomatic to fatal. Sequencing can identify co-infection more easily than any other test, especially when the co-infecting partner pathogen of SARS-CoV-2 is unknown.
Acknowledgment
Authors thank all those who submitted the coronavirus sequence data to the GISAID database and database managers, developers and scientists engaged with GISAID (www.gisaid.org) and Nextstrain (www.nextstrain.org) for making these data publicly available in a user-friendly format. Authors acknowledge the help rendered by OpenStreetMap® by making their data available in the public domain via Open Data Commons Open Database License (ODbL) by the OpenStreetMap Foundation (OSMF). These data and map were used to create (Fig. 2) of this paper. We are also grateful to Ms. Chitrarpita Das for computational help and to Drs Analabha Basu and Souvik Mukherjee for their valuable comments during the early phase of this work.
Footnotes
Financial support & sponsorship: None.
Conflicts of Interest: None.
References
- 1.Lu R, Zhao X, Li J, Niu P, Yang B, Wu H, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. Lancet. 2020;395:565–74. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Verity R, Okell LC, Dorigatti I, Winskill P, Whittaker C, Imai N, et al. Estimates of the severity of coronavirus disease 2019: A model-based analysis. Lancet Infect Dis. 2020 doi: 10.1016/S1473-3099(20)30243-7. pii: S1473-3099(20)30243-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.World Health Organization. Update 49 - SARS case fatality ratio, incubation period. WHO; 2003. [accessed on April 9, 2020]. Available from: https://wwwwhoint/csr/sars/archive/2003_05_07a/en/ [Google Scholar]
- 4.World Health Organization. Middle East respiratory syndrome coronavirus (MERS-CoV) WHO; 2020. [accessed on April 9, 2020]. Available from: https://wwwwhoint/emergencies/mers-cov/en/">https://wwwwhoint/emergencies/mers-cov/en/ [Google Scholar]
- 5.Li F. Structure, function, and evolution of coronavirus spike proteins. Annu Rev Virol. 2016;3:237–61. doi: 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.He Y, Zhou Y, Liu S, Kou Z, Li W, Farzan M, et al. Receptor-binding domain of SARS-CoV spike protein induces highly potent neutralizing antibodies: Implication for developing subunit vaccine. Biochem Biophys Res Commun. 2004;324:773–81. doi: 10.1016/j.bbrc.2004.09.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Follis KE, York J, Nunberg JH. Furin cleavage of the SARS coronavirus spike glycoprotein enhances cell-cell fusion but does not affect virion entry. Virology. 2006;350:358–69. doi: 10.1016/j.virol.2006.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tang X, Wu C, Li X, Song Y, Yao X, Wu X, et al. On the origin and continuing evolution of SARS-CoV-2. Natl Sci Rev. 2020 doi: 10.1093/nsr/nwaa036. doi: 101093/nsr/nwaa036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Forster P, Forster L, Renfrew C, Forster M. Phylogenetic network analysis of SARS-CoV-2 genomes. Proc Natl Acad Sci USA. 2020 doi: 10.1073/pnas.2004999117. doi: 101073/pnas2004999117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grenfell BT, Pybus OG, Gog JR, Wood JL, Daly JM, Mumford JA, et al. Unifying the epidemiological and evolutionary dynamics of pathogens. Science. 2004;303:327–32. doi: 10.1126/science.1090727. [DOI] [PubMed] [Google Scholar]
- 11.Sagulenko P, Puller V, Neher RA. TreeTime: Maximum-likelihood phylodynamic analysis. Virus Evol. 2018;4:vex042. doi: 10.1093/ve/vex042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hadfield J, Megill C, Bell SM, Huddleston J, Potter B, Callender C, et al. Nextstrain: Real-time tracking of pathogen evolution. Bioinformatics. 2018;34:4121–3. doi: 10.1093/bioinformatics/bty407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–95. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coronavirus has mutated to become far deadlier in Europe than the milder strain that made its way to the US west coast, Chinese study claims. [accessed on April 21, 2020]. Available from: https://wwwdailymailcouk/health/article-8237849/Coronavirus-mutated-Strains-evolved-far-deadlier-spread-Europe-New-Yorkhtmlito= email_share_article-top,">https://wwwdailymailcouk/health/article-8237849/Coronavirus-mutated-Strains-evolved-far-deadlier-spread-Europe-New-Yorkhtmlito= email_share_article-top .
- 15.Krietman M. Methods to detect selection in populations with applications to the human. Annu Rev Genomics Hum Genet. 2000;1:539–59. doi: 10.1146/annurev.genom.1.1.539. [DOI] [PubMed] [Google Scholar]
- 16.Biswas S, Akey JM. Genomic insights into positive selection. Trends Genet. 2006;22:437–46. doi: 10.1016/j.tig.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 17.Potdar V, Cherian SS, Deshpande GR, Ullas PT, Yadav PD, Choudhary ML, et al. Genomic analysis of SARS-CoV-2 strains among Indians returning from Italy, Iran & China, & Italian tourists in India. Indian J Med Res. 2020;151:255–60. doi: 10.4103/ijmr.IJMR_1058_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qu XX, Hao P, Song XJ, Jiang SM, Liu YX, Wang PG, et al. Identification of two critical amino acid residues of the severe acute respiratory syndrome coronavirus spike protein for its variation in zoonotic tropism transition via a double substitution strategy. J Biol Chem. 2005;280:29588–95. doi: 10.1074/jbc.M500662200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stellrecht KA. The drift in molecular testing for influenza: Mutations affecting assay performance. J Clin Microbiol. 2018:56. doi: 10.1128/JCM.01531-17. pii: e01531-17. [DOI] [PMC free article] [PubMed] [Google Scholar]