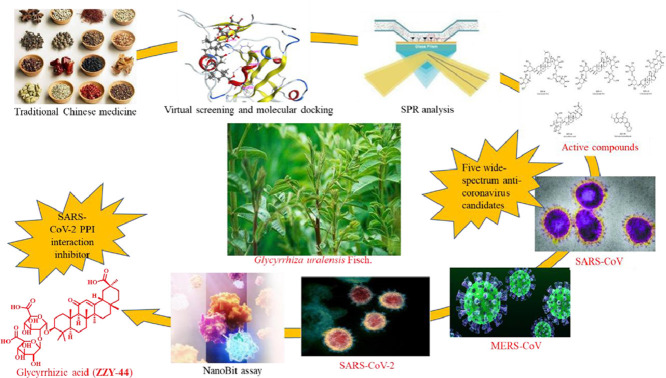
Abstract
Coronavirus causes a disease with high infectivity and pathogenicity, especially SARS in 2003, MERS in 2012, and COVID-2019 currently. The spike proteins of these coronaviruses are critical for host cell entry by receptors. Thus, searching for broad-spectrum anti-coronavirus candidates, such as spike protein inhibitors, is vital and desirable due to the mutations in the spike protein. In this study, a combination of computer-aided drug design and biological verification was used to discover active monomers from traditional Chinese medicine. Surface plasmon resonance (SPR) assays and NanoBit assays were used to verify the predicated compounds with their binding activities to spike proteins and inhibitory activities on the SARS-CoV-2 RBD/ACE2 interaction, respectively. Furthermore, an MTT assay was used to evaluate the cell toxicities of active compounds. As a result, glycyrrhizic acid (ZZY-44) was found to be the most efficient and nontoxic broad-spectrum anti-coronavirus molecule in vitro, especially, the significant effect on SARS-CoV-2, which provided a theoretical basis for the study of the pharmacodynamic material basis of traditional Chinese medicine against SARS-CoV-2 and offered a lead compound for further structural modification in order to obtain more effective candidate drugs against SARS-CoV-2.
Keywords: Traditional Chinese medicine (TCM), SARS-CoV-2, MERS-CoV, SARS-CoV, Spike protein
Abbreviations: ACE2, angiotensin-converting enzyme 2; CADD, computer-aided drug design; MASMC, mouse aorta smooth muscle cell; MERS, Middle East respiratory syndrome; PDB, protein data bank; RBD, receptor-binding domain; SARS, severe acute respiratory syndrome; SPR, surface plasmon resonance; TCM, traditional Chinese medicine; WHO, World Health Organization
Graphical abstract
Introduction
SARS-CoV-2 is a novel coronavirus that is also known as 2019-nCoV. The coronavirus disease 2019 caused by SARS-CoV-2 was named COVID-2019 by the WHO [Rubin et al., 2020; Wu et al., 2020]. Since the outbreak of the disease in late 2019, more than approximately 200 countries and regions have reported the disease to be prevalent. In addition, over 30 million people worldwide have been infected by the virus. It poses a great threat to people's lives and health. Additionally, it has had a huge impact on the global economy, forcing economic downturn and world economic losses [Ayittey et al., 2020]. So far, there are no effective measures to treat this infectious disease; thus, there is an urgent need to produce vaccines or antibody drugs to fight the disease or to screen new drugs for treating this extremely dangerous coronavirus disease.
Traditional Chinese medicine (TCM) is a large trove with a massive amount of bioactive ingredients and compounds that have cured a great quantity of diseases in China's 5000-year history [Normile, 2003; Chen and Xie, 1999]. The positive curative effect of TCM in treating COVID-19 has been widely recognized and drawn great attention from increasing numbers of countries and people [Wan et al., 2020a; Lai et al., 2020; Yang and Wang, 2020]. Recently, Zhang and colleagues reported over 30 clinically effective treatment schemes of TCM for treating COVID-19 [Zheng et al., 2020]. However, the precise anticoronavirus mechanisms of these medicines are still very unclear. Exploring active components of these TCM prescriptions and analyzing their molecular mechanisms are deep important for promoting acceptance of TCM and for developing TCM as an anticoronavirus therapy.
Spike proteins, also known as S proteins, are critical for coronavirus entry due to binding with different kinds of host receptors [Kirchdoerfer et al., 2016; Wan et al., 2020b; Li, 2016]. It is well-known that the ACE2 protein in the human body is targeted by the spike proteins of SARS-CoV and SARS-CoV-2, and the DPP4 protein is targeted by the S protein of MERS-CoV [Zhou et al., 2020; Moore et al., 2004; Raj et al., 2013]. Notably, receptor-binding domains (RBDs) in the spike protein fragment 1 (S1 subunit) of S proteins always play key roles by attacking human cells [Simmons et al., 2013; Weiss and Martin, 2005]. Due to their significant roles in several coronaviruses, they have become potential targets for drug design. Moreover, structural analysis of S proteins indicated that the mutations occurring within the RBD probably are the main reason for the easy spread of SARS-CoV-2 [Tian et al., 2020; Wrapp et al., 2020b]. Thus, searching for broad-spectrum anti-coronavirus candidates targeting S proteins holds promise for preventing coronavirus infections, especially those by the currently widespread SARS-CoV-2.
Here, we investigated the most frequent herbs within clinically effective TCM schemes and selected representative compounds for computer-aided drug design (CADD) targeting the S protein RBD of SARS-CoV-2. For the positive candidates, the surface plasmon resonance (SPR) technique was used to verify their binding activity to the S1 subunit of SARS-CoV-2. The binding activity to the MERS-CoV S1 subunit and SARS-CoV S1 subunit was also investigated. Furthermore, a NanoBit assay was used to evaluate the 5 active compounds verified by SPR for their activity disrupting the binding between the RBD and ACE2. Finally, we examined the cell toxicity of the active agents in mouse aorta smooth muscle cells (MASMCs) and human bronchial epithelial (16HBE) cells. These results identified that glycyrrhizic acid (ZZY-44) derived from Glycyrrhiza uralensis Fisch. (licorice) targeting S proteins of SARS-CoV-2, MERS-CoV and SARS-CoV, which also had good activity disrupting the interaction between the RBD of SARS-CoV-2 and ACE2. It might be a potential anti-coronavirus candidate.
Materials and methods
Compounds
All active ingredients of traditional Chinese medicine were purchased from commercial suppliers or were available in our laboratory compound library. They were used without any purification. Compound ID, compound name, chemical structure and the purity of each tested compound are shown in Supplementary Material Table S1.
Virtual screening
Small molecule optimization, protein preparation, binding-site definition and molecular docking were all performed in MOE software (version: 2019.0101). The site finder function of MOE 2019 was used to search and define binding sites. All compounds were tried to dock into spike proteins of SARS-CoV-2 (PDB ID = 6VSB) [Wrapp et al., 2020b], MERS-CoV (PDB ID = 5X5F) [Gui et al., 2017] and SARS-CoV (PDB ID = 5XLR) [Yuan et al., 2017]. The S1 subunits of the crystal structures were taken as docking targets, and RBDs were defined as binding sites. Triangle matcher of placement and rigid receptor of refinement were chosen as docking methods. The physicochemical properties and their scoring functions (a combination of S score, E refine score, hydrogen bonding and electrostatic interactions) were used to select the final pose.
Proteins
SARS-CoV-2 spike protein (S1 subunit, His tag) (catalog number: 40591-V08B1), MERS-CoV spike protein (S1 subunit, His tag) (catalog number: 40069-V08B1) and SARS-CoV spike protein (S1 subunit, His tag) (catalog number: 40150-V08B1) were purchased from Sino Biological Inc., China. Proteins were dissolved in an HBS-EP+ buffer for use.
Surface plasmon resonance (SPR) assay
The selected compounds from the virtual screening were verified with an SPR assay with a BIAcore T200 instrument ((BIAcore T200, GE Healthcare, Chicago, IL, USA) as described. S1 subunits of SARS-CoV-2, MERS-CoV and SARS-CoV were immobilized on a CM5 sensor chip, and a blank channel was employed as a negative control for each assay. A concentration of 100 µM selected compounds was used for the primary screening. For positive hits, compounds were serially diluted to different concentrations with the HBS-EP+ buffer and flowed through the chip. K D values were calculated with a steady affinity state model by BIAcore T200 analysis software, and the equations are shown below.
At a steady state, the net rate of complex formation was 0:
Thus,
R = Req (Req is the response level of the equilibrium state) yields:
ka/kd = K A yields:
With this equation, fitting Req and C could yield K A. K D is equal to the inverse of K A (K D = 1/K A). Additional details of the SPR assays are shown in Supplementary Material (Figs. S1−S3).
Construction of plasmids
NanoBiTⓇ PPI Vectors (CS1603B32, Promega, Madison, WI) were used for the cloning of SARS-CoV-2 S-RBD and ACE2 fusion proteins according to the manufacturer's instructions. SARS-CoV-2 S-RBD (S residues 319 to 591) or ACE2 (residues 19 to 615) was subcloned into the NanoBiT pBiT1.3-C [CMV/LgBiT/Hyg], pBiT1.3-N [CMV/LgBiT/Hyg], pBiT2.3-C [CMV/SmBiT/Blast], pBiT2.3-N [CMV/SmBiT/Blast] vectors using restriction enzymes to fuse the NanoBiT LgBiT or SmBiT subunit of NanoLuc luciferase to the C or N terminus of both proteins, respectively. The NanoBiT Negative Control Vector encoding the fusion protein (HaloTag-SmBit) was used as a negative control.
NanoBiT-based RBD-ACE2 interaction assays
A NanoBiT Protein:Protein Interaction (PPI) Assay (N2012, Promega, Madison, WI) was utilized to screen SARS-CoV-2 S-RBD/ACE2 PPI inhibitors. HEK293 cells were seeded into a 6-well cell culture plate and incubated in a 37 °C, 5% CO2 incubator for 16–24 h. Then, the SARS-CoV-2 S-RBD-LgBiT and SmBiT-ACE2 fusion plasmids were cotransfected into the HEK293 cells using FuGENE HD transfection reagent (Promega, Madison, WI). After 6 h, the HEK293 cells were reseeded into a 384-well plate for 16–24 h, and the compounds (final DMSO concentration of 1%) were added and incubated for 3 h. Finally, Nano-Glo live Cell Assay reagent was added, and luminescence was determined using an Envision plate reader (EnVision, Perkin Elmer, Waltham, MA, USA). To exclude false positives, the cytotoxicity of the compounds on the HEK293 cells was measured according to the same transfection conditions using a CellTiter-Glo Luminescent Cell Viability Assay (Promega, Madison, WI), and the inhibitory effects of the compounds on NanoLuc (HEK293/NanoLuc stable cells) were determined. The inhibitory effects (IC50) of the active compounds on the SARS-CoV-2 S-RBD/ACE2 interaction (NanoBiT inh%), NanoLuc luciferase (NanoLuc inh%) and the cell proliferation (Cytotox inh%, CC50) in HEK293 cells were analyzed.
Cell culture and MTT assays
Mouse aorta smooth muscle cells (MASMCs) were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin–streptomycin, human bronchial epithelial (16HBE) cells were cultured in Keratinocyte Medium (KM) supplemented with 1% keratinocyte growth supplement (KGS) and 1% penicillin–streptomycin. All the cells were cultured at 37 °C in a humidified 5% CO2 incubator.
For cell toxicity assays, the MTT method was adopted. Cells were seeded in a 96-well tissue culture plate, incubated for 12 h, and then treated with different concentrations of compounds. After 48 h of incubation, MTT solution (0.5 mg/ml) was added to the wells. The cells were then incubated for 4–5 h at 37 °C, and formazan crystals in viable cells were dissolved in 100 µl of DMSO. The solubilized formazan was spectrophotometrically quantified with a Tecan M1000Pro Multiscan Spectrum (Tecan) at 490 nm.
Statistical analysis
Sensorgram figures from the SPR assays were generated with the OriginPro 8.5 program. The raw data from the NanoBiT-based assays and MTT assays were analyzed using GraphPad Prism 6.0 software. All values are presented as the mean ± the standard (SD). The numbers of experimental replicates are shown in the figure legends.
Results
Frequency analysis of herbs
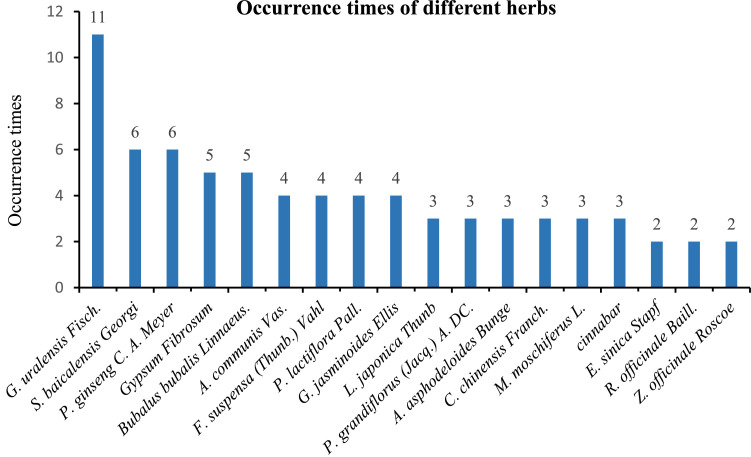
We first analyzed the frequency of different herbs used from approximately 30 TCM prescriptions that are effective for treating COVID-2019 in the clinic. Among them, 18 kinds of herbs were applied two or more times, as depicted in Fig. 1 . Overall, Glycyrrhiza uralensis Fisch. (licorice) was the most commonly used herb, appearing 11 times. This result may reveal that active ingredients from this herb are efficient. Several herbs such as Scutellaria baicalensis Georgi (baical skullcap root), Panax ginseng C. A. Meyer (ginseng), Gypsum Fibrosum (plaster), and Bubalus bubalis Linnaeus. (buffalo horn) were also utilized more frequently (five or six times). However, it was difficult to analyze the active ingredients from plaster and buffalo horn because their active compositions remain elusive. Other herbs occurred within 4 times, but some of them also attracted our attention due to their internal heat-clearing and detoxifying effects, including Lonicera japonica Thunb (honeysuckle flower), Platycodon grandiflorus (Jacq.) A. DC., (platycodon root) and Ephedra sinica Stapf (ephedra) [Yuan et al., 2002; Lee et al., 2017; Wang et al., 2004]. Active compounds from these 18 herbs were elected as virtual screening candidates in this study.
Fig. 1.
Occurrence times of herbs from clinically effective traditional Chinese medicine prescriptions.
Molecular docking
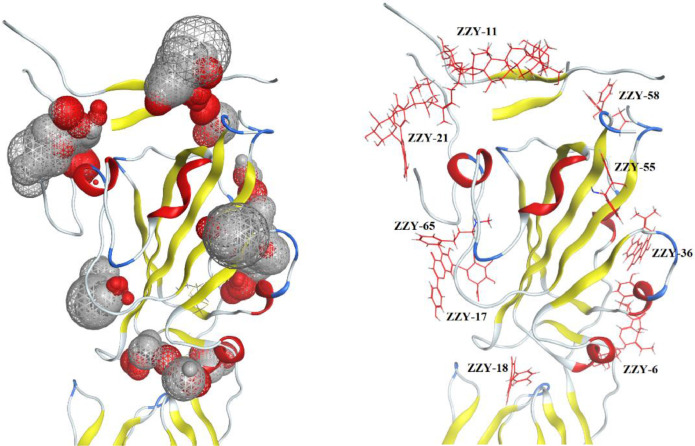
Our molecular docking strategy was utilized to identify approximately 500 active ingredients from the 18 most commonly used herbs for virtual screening, from which a large number of theoretically inactive compounds were filtered out. RBDs in coronaviruses are the most significant domains in the process of host entry receptor interactions [Simmons et al., 2013; Weiss and Martin, 2005]. The RBD of SARS-CoV-2 was selected as the important docking domain in this study. As a result, we obtained 66 compounds that could bind to the RBD of the S1 subunit of SARS-CoV-2. As shown in Table 1 , 28 of these compounds could bind to the interface of the complex of the RBD whereas ACE2, and other theoretically active compounds could dock into other pockets of the RBD (Fig. 2 ). Docking results are shown in the Supplementary Material.
Table 1.
Binding positions of each compound.
| Binding positions | Compounds (ZZY-) |
|---|---|
| Interface between RBD and ACE2 | 1–5, 8, 9, 11–13, 16, 22, 26–28, 30, 37, 38, 43, 44, 49, 51–54, 56, 59, 64 |
| Other pockets in RBD | 6, 14, 15, 17–21, 23–25, 29, 31–36, 40–42, 48, 55, 57, 58, 60–63, 65–73 |
Fig. 2.
Left: binding sites defined by MOE 2019; Right: compounds docked into different positions of RBD domain in S1 subunit.
Binding activity of TCM-derived components with SARS-CoV-2 S1 subunit
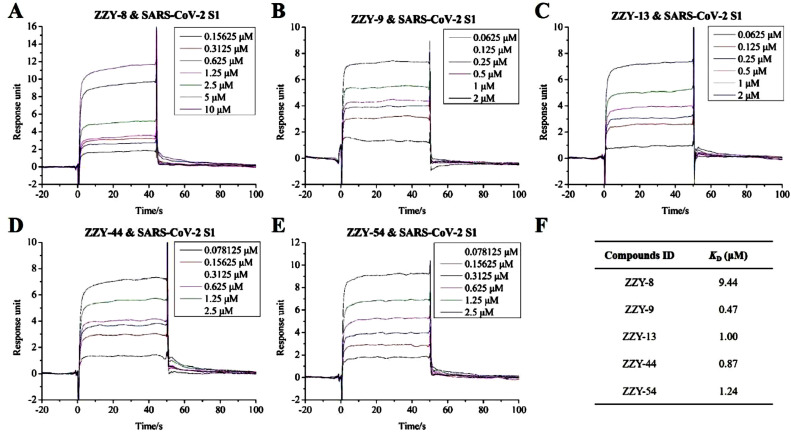
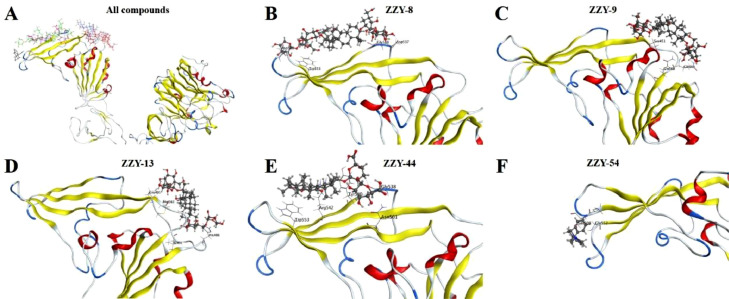
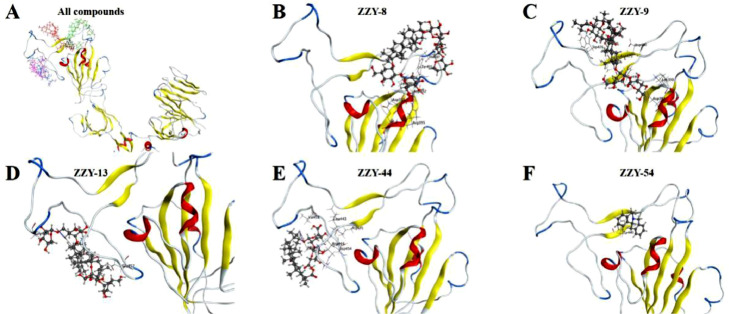
For the selected 66 TCM-derived components from the virtual screening, the SPR technique was used to detect their binding activities to the SARS-CoV-2 S1 subunit. Notably, 19 compounds were excluded from the detection, as they are insoluble in HBS-EP+ buffer. Among the other 44 compounds, ginsenoside Ra2 (ZZY-8), ginsenoside Rb1 (ZZY-9), ginsenoside Rb3 (ZZY-13), glycyrrhizic acid (ZZY-44) and berberine chloride (ZZY-54) showed high affinity for the SARS-CoV-2 S1 subunit. As shown in Fig. 3 , the interactions between these five compounds and the SARS-CoV-2 S1 subunit were concentration dependent. Fig. 3A–E indicate that the interactions reached a steady state rapidly upon injection, as well as rapid dissociation from the binding site. The K D values of the five compounds are shown in Fig. 3F: 9.44 μM for ginsenoside Ra2 (ZZY-8), 0.47 μM for ginsenoside Rb1 (ZZY-9), 1.00 μM for ginsenoside Rb3 (ZZY-13), 0.87 μM for glycyrrhizic acid (ZZY-44), and 1.24 μM for berberine chloride (ZZY-54). The simulated interaction modes of the different compounds and SARS-CoV-2 S1 subunit are shown in Fig. 4 . The five positive compounds all reacted with the binding interface of the RBD and ACE2 (Fig. 4A). These results showed that these five active compounds bind to SARS-CoV-2 S1 subunit with high affinity, which might have potential anti-SARS-CoV-2 activity by influencing the interaction between the S protein and human ACE2 protein.
Fig. 3.
SPR analysis of the interaction between TCM-derived components with S1 subunit of SARS-CoV-2. A to E exhibit kinetic behaviors of ginsenoside Ra2 (ZZY-8), ginsenoside Rb1 (ZZY-9), ginsenoside Rb3 (ZZY-13), glycyrrhizic acid (ZZY-44), berberine chloride (ZZY-54) and SARS-CoV-2 S1 subunit, respectively. F shown the KD value of the five TCM-derived components.
Fig. 4.
Molecular docking of active compounds with SARS-CoV-2 S1 subunit. A represents binding position of all the five active compounds within RBD. B to F exhibit interaction mode of ginsenoside Ra2 (ZZY-8), ginsenoside Rb1 (ZZY-9), ginsenoside Rb3 (ZZY-13), glycyrrhizic acid (ZZY-44), berberine chloride (ZZY-54) with RBD domain, respectively.
Broad-spectrum binding activity of TCM-derived components to the S1 subunits of coronaviruses
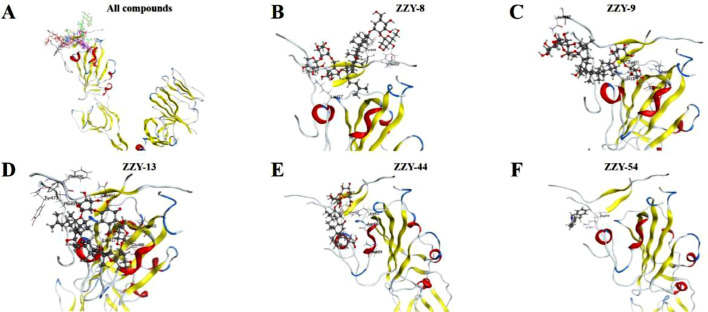
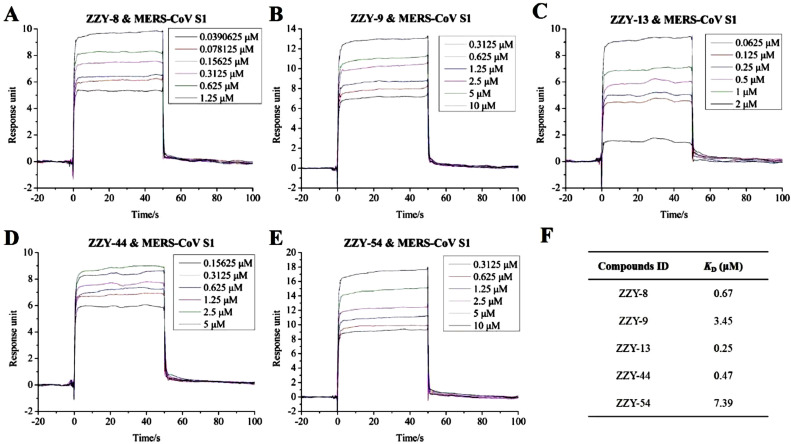
Spike proteins are conserved in different coronaviruses, including SARS-CoV-2, MERS-CoV and SARS-CoV [Kirchdoerfer et al., 2016; Wan et al., 2020b; Li, 2016]. Thus, we then investigated the binding activity of the five compounds to the S1 subunits of MERS-CoV and SARS-CoV with SPR assays. As shown in Fig. 5 , the interactions between the five compounds and the MERS-CoV S1 subunit were concentration-dependent. The K D values are shown in Fig. 5F: 0.67 μM for ginsenoside Ra2 (ZZY-8), 3.45 μM for ginsenoside Rb1 (ZZY-9), 0.25 μM for ginsenoside Rb3 (ZZY-13), 0.47 μM for glycyrrhizic acid (ZZY-44), and 7.39 μM for berberine chloride (ZZY-54). The simulated interaction modes of the different compounds and the MERS-CoV S1 subunit are expressed in Fig. 6 . The five positive compounds all reacted with the binding interface of RBD and its receptor DPP4 protein (Fig. 6A).
Fig. 5.
SPR analysis of the interaction between TCM-derived components with S1 subunit of MERS-CoV. A to E exhibiting kinetic behaviors of ginsenoside Ra2 (ZZY-8), ginsenoside Rb1 (ZZY-9), ginsenoside Rb3 (ZZY-13), glycyrrhizic acid (ZZY-44), berberine chloride (ZZY-54) and MERS-CoV S1 subunit, respectively. F shown the KD value of the five TCM-derived components.
Fig. 6.
Molecular docking of active compounds with MERS-CoV S1 subunit. A represents binding position of all the five active compounds within RBD. B to F exhibit interaction mode of ginsenoside Ra2 (ZZY-8), ginsenoside Rb1 (ZZY-9), ginsenoside Rb3 (ZZY-13), glycyrrhizic acid (ZZY-44), berberine chloride (ZZY-54) with RBD domain, respectively.
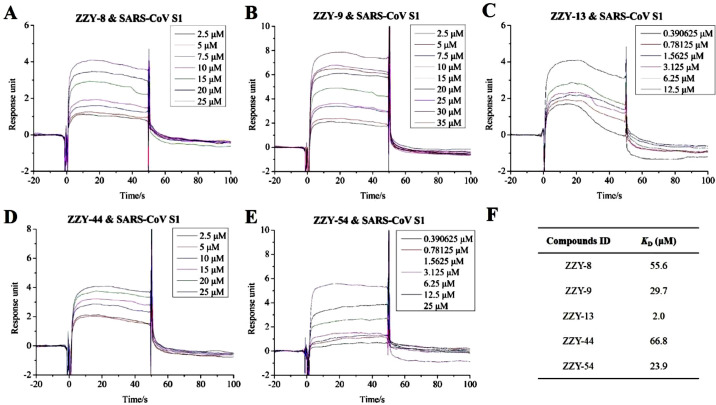
However, the binding activity of these five active compounds with the SARS-CoV S1 subunit was weak (Fig. 7 ). The K D values detected with the SPR assays were 55.6 μM for ginsenoside Ra2 (ZZY-8), 29.7 μM for ginsenoside Rb1 (ZZY-9), 2.0 μM for ginsenoside Rb3 (ZZY-13), 66.8 μM for glycyrrhizic acid (ZZY-44) and 23.9 μM for berberine chloride (ZZY-54). Molecular docking results (Fig. 8 ) also show different binding patterns: ginsenoside Ra2 (ZZY-8), ginsenoside Rb1 (ZZY-9) and berberine chloride (ZZY-54) interacted with the binding interface of the RBD and receptor ACE2, whereas ginsenoside Rb3 (ZZY-13) and glycyrrhizic acid (ZZY-44) bound to the outside of the interface. In conclusion, the selected five active compounds showed a high affinity for SARS-CoV-2 S1 and MERS-CoV S1 by interacting with their receptor-binding interface within the RBD, whereas reduced affinity was detected for the SARS-CoV S1 subunit due to their different interaction modes with the RBD.
Fig. 7.
SPR analysis of the interaction between TCM-derived components with S1 subunit of SARS-CoV. A to E exhibiting kinetic behaviors of ginsenoside Ra2 (ZZY-8), ginsenoside Rb1 (ZZY-9), ginsenoside Rb3 (ZZY-13), glycyrrhizic acid (ZZY-44), berberine chloride (ZZY-54) and SARS-CoV S1 subunit, respectively. F shown the KD value of the five TCM-derived components.
Fig. 8.
Molecular docking of active compounds with SARS-CoV S1 subunit. A represents binding position of all the five active compounds within RBD. B to F exhibit interaction mode of ginsenoside Ra2 (ZZY-8), ginsenoside Rb1 (ZZY-9), ginsenoside Rb3 (ZZY-13), glycyrrhizic acid (ZZY-44), berberine chloride (ZZY-54) with RBD domain, respectively.
Disruption effect of the active TCM-derived components on the SARS-CoV-2 S-RBD and ACE2 interaction
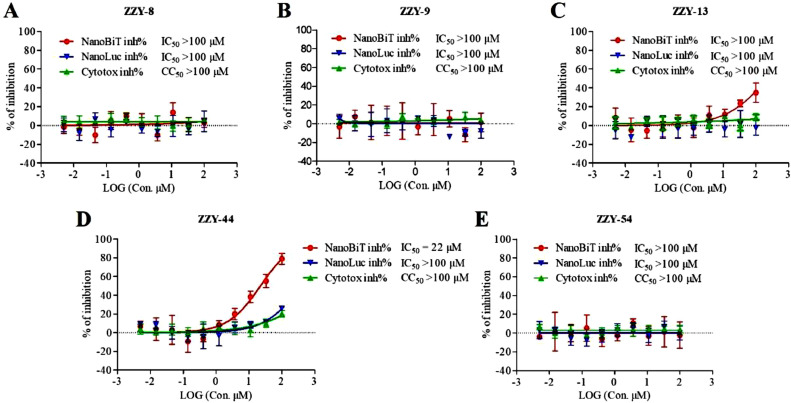
To investigate the disruption effect of the five active components on the S-RBD/ACE2 interaction, a NanoBiT-based SARS-CoV-2 S-RBD/ACE2 interaction was established. As shown in Fig. 9 , the inhibition rates for the SARS-CoV-2 S-RBD/ACE2 interaction (NanoBiT inh%), NanoLuc luciferase (NanoLuc inh%) and cell proliferation (Cytotox inh%) were analyzed for each compound. No obvious cell toxicity was observed for any of the five compounds in the transfected HEK293 cells, and the CC50 of cell toxicity exceeded 100 μM for all the compounds. Among the five compounds, ginsenoside Rb3 (ZZY-13) and glycyrrhizic acid (ZZY-44) could disrupt the SARS-CoV-2 S-RBD/ACE2 interaction in cells, and only glycyrrhizic acid (ZZY-44) disrupted the SARS-CoV-2 S-RBD/ACE2 interaction at low concentrations (IC50 = 22 μM, Fig. 9D). The detection of NanoLuc luciferase inhibition rates (NanoLuc inh%) of the five compounds showed that they are not false positive hits. Thus, glycyrrhizic acid (ZZY-44) could effectively inhibit the SARS-CoV-2 S-RBD/ACE2 interaction.
Fig. 9.
NanoBiT-based SARS-CoV-2 S-RBD-ACE2 interaction assays for active compounds: ginsenoside Ra2 (ZZY-8), ginsenoside Rb1 (ZZY-9), ginsenoside Rb3 (ZZY-13), glycyrrhizic acid (ZZY-44), berberine chloride (ZZY-54). NanoBiT inh% represents the inhibition rates of S-RBD/ACE2 interaction, NanoLuc inh% represents inhibition rates of NanoLuc luciferase, and Cytotox inh% represents inhibition rate to cell proliferation. n = 3.
Cell toxicity of the active TCM-derived components
Finally, we examined the cell toxicity to mouse aorta smooth muscle cells (MASMCs) and human bronchial epithelial (16HBE) cells of the five selected TCM-derived components. According to results shown in Table 2 , ginsenoside Ra2 (ZZY-8), ginsenoside Rb3 (ZZY-13) and glycyrrhizic acid (ZZY-44) have no cell toxicity to MASMCs even at high concentrations (100 μM). The CC50 of ginsenoside Rb1 (ZZY-9) was 13 μM, and the CC50 of berberine hydrochloride (ZZY-54) was 63 μM. Furthermore, the cell toxicity of the five TCM-derived components to 16HBE cells also showed that the CC50s of ginsenoside Ra2 (ZZY-8), ginsenoside Rb3 (ZZY-9), ginsenoside Rb3 (ZZY-13) and glycyrrhizic acid (ZZY-44) was over 100 μM, whereas the CC50 of berberine chloride (ZZY-54) was 13 μM. Thus, we demonstrated that ginsenoside Ra2 (ZZY-8), ginsenoside Rb3 (ZZY-13) and glycyrrhizic acid (ZZY-44) have low cell toxicities to normal human lung cells, and might be promising candidates for anti-coronavirus drugs.
Table 2.
Cell toxicity of TCM-derived components to MASMC cells and 16HBE cells.
| Compound ID\Cellsa | ZZY-8 | ZZY-9 | ZZY-13 | ZZY-44 | ZZY-54 |
|---|---|---|---|---|---|
| MASMC | > 100 | 13 | > 100 | > 100 | 63 |
| 16HBE | > 100 | > 100 | > 100 | > 100 | 13 |
The table shows the CC50 values of MASMC cells and 16HBE cells treat with the five TCM-derived components in cell toxicity assays. n = 3.
Discussion
Coronavirus disease 2019 is a high-mortality disease, with fever, cough and dyspnea as common symptoms [Sohrabi et al., 2020]. It is also characterized by high infectivity and pathogenicity [Chen, 2020]. In the 21st century, coronaviruses have caused several large epidemics, such as severe acute respiratory syndrome (SARS-CoV) in 2003, Middle East respiratory syndrome (MERS-CoV) in 2012, and COVID-2019 (SARS-CoV-2), which has been prevalent since 2019. Global economic turmoil was caused several times by the viruses, and limited of medical resources have unfortunately exist in many countries during these epidemics [Ayittey et al., 2020]. Thus, searching for anti-coronavirus candidates is currently an urgent task. Moreover, it is a long-term task to find new broad-spectrum anti-coronavirus drugs.
In recent studies, several small molecule inhibitors were reported to be effective against SARS-CoV-2, such as 11a (EC50 = 0.53 μM) and 11b (EC50 = 0.72 μM) by Liu and colleagues [Dai et al., 2020] and N3 (EC50 = 16.77 μM) by Rao's group [Jin et al., 2020]. These inhibitors were designed based on the X-ray crystal structures of the main proteases [Jin et al., 2020; Xue et al., 2007]. However, most of these groups focused their attention on inhibiting the Mpro enzyme. It is worth mentioning that focusing on proteases is just one way of looking for anti-coronavirus candidates. We noted that these viruses enter host cells through their spike proteins [Zhou et al., 2020; Moore et al., 2004; Raj et al., 2013]. In this study, we tested traditional Chinese medicine to target this common virus-entry pathway, which could provide an effective solution for this disastrous crisis [Wan et al., 2020a; Lai et al., 2020; Yang and Wang, 2020]. However, the active ingredients and molecular mechanisms still need to be clarified.
To solve these problems, we initially used molecular docking to judge whether ingredients from TCM could have interactions with the S protein of SARS-CoV-2. The result showed that 66 of the selected compounds could dock into the RBD of the S1 subunit. Following that, we analyzed the best docking pose of each molecule. We found that 28 of these candidates could bind to the interface between the RBD and ACE2. Other compounds also interacted with the RBD in the S1 subunit. We also found that very bulky compounds such as ginsenoside Ra2 (ZZY-8), ginsenoside Rb1 (ZZY-9), ginsenoside Rg1 (ZZY-11), ginsenoside Re (ZZY-12), ginsenoside Rb3 (ZZY-13), Astragaloside I (ZZY-43), and glycyrrhizic acid (ZZY-44), have higher docking scores and preferentially dock to the interface between the RBD and ACE2. Other smaller compounds, such as 4-(4-methoxyphenyl)-1-butanol (ZZY-14), umbelliferone (ZZY-33), ephedrine (ZZY-72), and pseudoephedrine (ZZY-73), preferentially dock to smaller pockets at the side face of the protein. However, according to the literature [Wrapp et al., 2020a], binding outside the interface of the RBD may also be active. Thus, biological evaluation was performed on these compounds under the same conditions. Notably, the bulky compounds exhibited better potential activities than the smaller ones. N-glucoside and O-glucoside substituted compounds showed better activities.
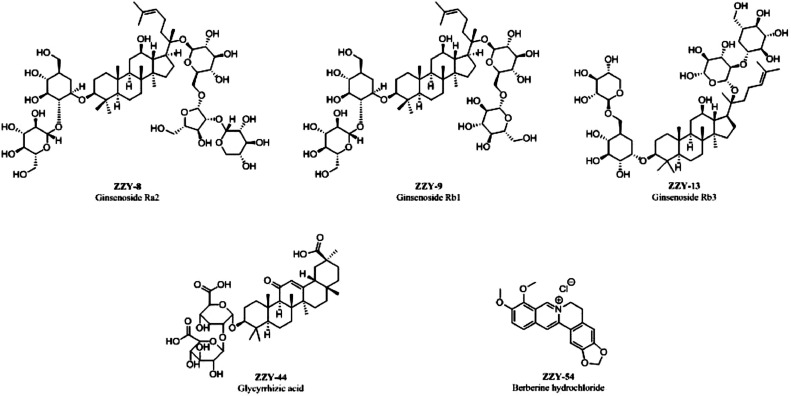
We further verified the above simulation results at a molecular level by SPR assay. Fortunately, we found that compounds ginsenoside Ra2 (ZZY-8), ginsenoside Rb1 (ZZY-9), ginsenoside Rb3 (ZZY-13), glycyrrhizic acid (ZZY-44), and berberine chloride (ZZY-54) (Fig. 10 ) performed with good activities in binding to the S1 subunit of SARS-CoV-2. Next, to investigate whether these compounds can be candidates for broad-spectrum anti-coronavirus activity, we performed further evaluation. The S1 subunits of MERS-CoV and SARS-CoV were used to verify the corresponding binding activity. The results showed that the five active compounds displayed similar binding activities with MERS-CoV S1 compared to those with the SARS-CoV-2 S1 subunit, and reduced affinity for SARS-CoV S1. The affinity variation of the five active compounds to different S1 subunits is not known. We speculate that the reduced binding activities of the five active compounds with the SARS-CoV S1 subunit might be due to mutations of key amino acids compared to those of SARS-CoV-2 S1, but additional evidence is needed.
Fig. 10.
Structures of active compounds.
The structure-activity relationships summarized from the virtual screening results were verified, as that the bulky compounds had better binding activity to the S1 subunit than other molecules. Interestingly, it was reported that the glycyrrhizic acid (ZZY-44) derivative is clinically effective due to its inhibitory activity to the ACE2 protein as well [Chen and Du, 2020]. This study might reveal another mechanism of glycyrrhizic acid (ZZY-44), specifically its binding activities to the S1 subunits of SARS-CoV-2, MERS-CoV, and SARS-CoV. All the results led to the conclusion that glycyrrhizic acid (ZZY-44) could probably be a multitarget inhibitor and broad-spectrum anti-coronavirus candidate.
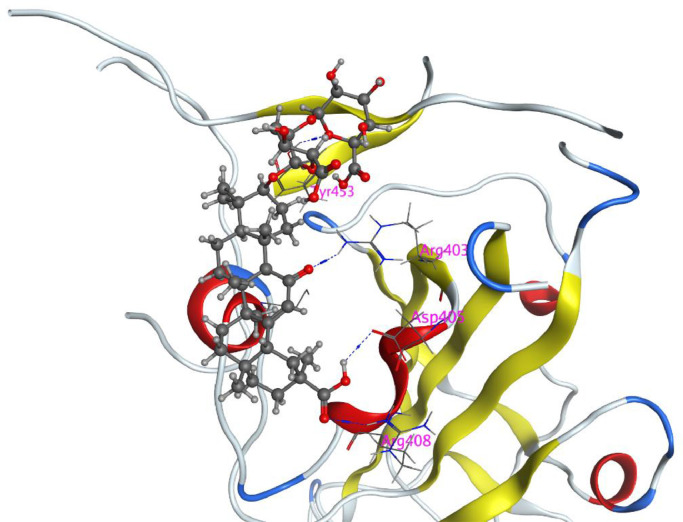
To explore the binding mode of the best active compound and spike protein of SARS-CoV-2, a molecular docking study was performed using glycyrrhizic acid (ZZY-44) and the S1 subunit of SARS-CoV-2. As shown in Fig. 11 (enlarged picture of Fig. 4E), the molecule was docked at the interface between the RBD and ACE2; thus, it probably contributed to the disruption of the interaction between the RBD and ACE2. The carboxyl in ring E had two strong hydrogen interactions with Asp405 and Arg408. The carbonyl in ring C had a strong hydrogen interaction with Arg403. Furthermore, the glycosyl had a strong hydrogen interaction with Tyr453. These interactions might verify that compound glycyrrhizic acid (ZZY-44) has potent activity against SARS-CoV-2.
Fig. 11.
Glycyrrhizic acid (ZZY-44) docked with RBD of SARS-CoV-2.
In addition, to further explore whether the 5 active compounds discovered by SPR could disrupt the interaction between the RBD of SARS-CoV-2 and ACE2, we performed a NanoBit assay to determine their activity. The results showed that compounds ginsenoside Rb3 (ZZY-13) and glycyrrhizic acid (ZZY-44) had certain activities in inhibiting the SARS-CoV-2 S-RBD/ACE2 interaction. Especially, glycyrrhizic acid (ZZY-44) exhibited better activity, as its IC50 was 22 μM. These results further revealed the mechanism of glycyrrhizic acid (ZZY-44) with its binding activities to S proteins and disruption activity to the RBD and ACE2 and probably proves that it is a potential clinically effective candidate.
Finally, the cytotoxicity of the five active compounds on mouse aorta smooth muscle cells (MASMCs) and human bronchial epithelial (16HBE) cells was evaluated. Among the five active compounds determined by SPR, ginsenoside Ra2 (ZZY-8), ginsenoside Rb3 (ZZY-13), and glycyrrhizic acid (ZZY-44) showed no toxicity even at high concentrations. In addition, given that glycyrrhizic acid (ZZY-44) performed the best in disrupting the interaction between the RBD of SARS-CoV-2 and ACE2, it might be predicted that the compound would be a highly effective and nontoxic broad-spectrum anti-coronavirus candidate for further exploration. While berberine chloride (ZZY-54) showed cell toxicity in both MASMC and 16HBE cells, and according to references, berberine chloride (ZZY-54) is a potential anticancer drug [Tan et al., 2011; Wang et al., 2014]. Ginsenoside Rb1 (ZZY-9) showed high cell toxicity to MASMCs, literatures indicated a derivative of ginsenoside Rb1 (ZZY-9), Compound K, with broad anticancer effect [Sharma and Lee, 2020]. Thus, ginsenoside Rb1 (ZZY-9) and berberine chloride (ZZY-54) might be promising in developing anticancer drugs.
Conclusions
In summary, we have discovered glycyrrhizic acid (ZZY-44) as a broad-spectrum anti-coronavirus candidate with low toxicity based on our screening of natural products. It or its derivatives might be used to inhibit SARS-CoV-2 and other novel and unknown coronaviruses in the future. In particular, it was also identified for its disruption activity on the RBD and ACE2 and could be considered as a multitarget inhibitor.
Author contributions
JG Zhang, GQ Lin, LL Chen and HZ Chen conceived and supervised the whole study. SP Yu performed molecular docking study. YY Zhu, JR Xu and MG Wang carried out the experiments and data analysis. SP Yu, YY Zhu and LL Chen drafted the manuscript. GT Yao, P Zhang and YF Zhao contributed to experimental consumables. JG Zhang, GQ Lin, LL Chen and HZ Chen revised the manuscript.
All data were generated in-house, and no paper mill was used. All authors agree to be accountable for all aspects of work ensuring integrity and accuracy.
Declaration of Competing Interest
The authors declare that they have no competing interests.
Acknowledgements
This work was supported by Scientific Research Project of Shanghai Municipal Health Commission on Traditional Chinese Medicine for Prevention and Treatment of COVID-19 (2020XGKY07; 2020XGKY09) and Emergency Scientific Research Program of Shanghai University of Traditional Chinese Medicine (2019YJ 06-01; 2019YJ 06-03) .
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.phymed.2020.153364.
Appendix. Supplementary materials
References
- Ayittey F.K., Ayittey M.K., Chiwero N.B., Kamasah J.S., Dzuvor C. Economic impacts of Wuhan 2019‐nCoV on China and the world. J. Med. Virol. 2020;92:473–475. doi: 10.1002/jmv.25706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J. Pathogenicity and transmissibility of 2019-nCoV—a quick overview and comparison with other emerging viruses. Microb. Infect. 2020;22:69–71. doi: 10.1016/j.micinf.2020.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Du Q. Preprints; https://www.preprints.org/manuscript/202001.0358/v1.: 2020. Potential Natural Compounds for Preventing SARS-CoV-2 (2019-nCoV) Infection. [Google Scholar]
- Chen P., Xie P. IOS Press; 1999. History and Development of Traditional Chinese Medicine. [Google Scholar]
- Dai W., Zhang B., Jiang X., Su H., Li J., Zhao Y., Xie X., Jin Z., Peng J., Liu F., Li C., Li Y., Bai F., Wang H., Cheng X., Cen X., Hu S., Yang X., Wang J., Liu X., Xiao G., Jiang H., Rao Z., Zhang L., Xu Y., Yang H., Liu H. Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease. Science. 2020 doi: 10.1126/science.abb4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gui M., Song W., Zhou H., Xu J., Chen S., Xiang Y., Wang X. Cryo-electron microscopy structures of the SARS-CoV spike glycoprotein reveal a prerequisite conformational state for receptor binding. Cell Res. 2017;27:119–129. doi: 10.1038/cr.2016.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y., Zhang B., Li X., Zhang L., Peng C., Duan Y., Yu J., Wang L., Yang K., Liu F., Jiang R., Yang X., You T., Liu X., Yang X., Bai F., Liu H., Liu X., Guddat L.W., Xu W., Xiao G., Qin C., Shi Z., Jiang H., Rao Z., Yang H. Structure of Mpro from COVID-19 virus and discovery of its inhibitors. Nature. 2020 doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- Kirchdoerfer R.N., Cottrell C.A., Wang N., Pallesen J., Yassine H.M., Turner H.L., Corbett K.S., Graham B.S., McLellan J.S., Ward A.B. Pre-fusion structure of a human coronavirus spike protein. Nature. 2016;531:118–121. doi: 10.1038/nature17200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai C., Shih T., Ko W., Tang H., Hsueh P. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and corona virus disease-2019 (COVID-19): the epidemic and the challenges. Int. J. Antimicrobe. Ag. 2020;55 doi: 10.1016/j.ijantimicag.2020.105924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y., Yeh S., Ruan X., Zhang H., Hsu S., Huang H., Hsieh C., Lin Y., Yeh T., Liu H., Gan D. Honeysuckle aqueous extract and induced let-7a suppress dengue virus type 2 replication and pathogenesis. J. Ethnopharmacol. 2017;198:109–121. doi: 10.1016/j.jep.2016.12.049. [DOI] [PubMed] [Google Scholar]
- Li F. Structure, function, and evolution of coronavirus spike proteins. Ann. Rev. Virol. 2016;3:237–261. doi: 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore M.J., Dorfman T., Li W., Won g S.K., Li Y., Kuhn J.H., Coderre J., Vasilieva N., Han Z., Greenough T.C., Farzan M., Choe H. Retroviruses pseudotyped with the severe acute respiratory syndrome coronavirus spike protein efficiently infect cells expressing angiotensin-converting enzyme 2. J. Virol. 2004;78:10628–10635. doi: 10.1128/JVI.78.19.10628-10635.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normile D. The new face of traditional Chinese medicine. Science. 2003;299:188–190. doi: 10.1126/science.299.5604.188. [DOI] [PubMed] [Google Scholar]
- Raj V.S., Mou H., Smits S.L., Dekkers D.H.W., Müller M.A., Dijkman R., Muth D., Demmers J.A.A., Zaki L., Fouchier R.A.M., Thiel V., Drosten C., Rottier P.J.M., Osterhaus A.D.M.E., Bosch B.J., Haagmans B.L. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature. 2013;495:251–254. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin E.J., Baden L.R., Morrissey S., Campion E.W. Medical Journals and the 2019-nCoV Outbreak. New Engl. J. Med. 2020;382:866. doi: 10.1056/NEJMe2001329. [DOI] [PubMed] [Google Scholar]
- Sharma A., Lee H. Ginsenoside compound k: insights into recent studies on pharmacokinetics and health-promoting activities. Biomolecules. 2020;10:1028. doi: 10.3390/biom10071028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons G., Zmora P., Gierer S., Heurich A., Pöhlmann S. Proteolytic activation of the SARS-coronavirus spike protein: cutting enzymes at the cutting edge of antiviral research. Antivir. Res. 2013;100:605–614. doi: 10.1016/j.antiviral.2013.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohrabi C., Alsafi Z., O'Neill N., Khan M., Kerwan A., Al-Jabir A., Iosifidis C., Aghad R. World Health Organization declares global emergency: a review of the 2019 novel coronavirus (COVID-19) Int. J. Surg. 2020;76:71–76. doi: 10.1016/j.ijsu.2020.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan W., Li C., Wang Y. Berberine hydrochloride: anticancer activity and nanoparticulate delivery system. Int. J. Nanomed. 2011;6:1773–1777. doi: 10.2147/IJN.S22683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, X., Li, C., Huang, A., Xia, S., Lu, S., Shi, Z., Lu, L., Jiang, S., Yang, Z., Wu, Y., Ying, T., 2020. Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus-specific human monoclonal antibody. bioRxiv 2020.01.28.923011. [DOI] [PMC free article] [PubMed]
- Wan S., Xiang Y., Fang W., Zheng Y., Li B., Hu Y., Lang C., Huang D., Sun Q., Xiong Y., Huang X., Lv J., Luo Y., Shen L., Yang H., Huang G., Yang R. Clinical features and treatment of COVID‐19 patients in northeast Chongqing. J. Med. Virol. 2020 doi: 10.1002/jmv.25783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y., Shang J., Graham R., Baric R.S., Li F. Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J. Virol. 2020;94:e00127. doi: 10.1128/JVI.00127-20. -20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Li H., Wang S., Liu R., Wu Z., Wang C., Wang Y., Chen M. Enhancing the antitumor activity of berberine hydrochloride by solid lipid nanoparticle encapsulation. AAPS PharmSciTech. 2014;15:834–844. doi: 10.1208/s12249-014-0112-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss S.R., Martin S.N. Coronavirus pathogenesis and the emerging pathogen severe acute respiratory syndrome coronavirus. Microbiol. Mol. Biol. Rev. 2005;69:635–664. doi: 10.1128/MMBR.69.4.635-664.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Levis G., Lee E., Levis W., Lee D., Kim B., Park S., Park E. Platycodon grandiflorum modulate the production of nitric oxide and secretion of TNF-α in activated RAW 264.7 cells. Int. Immunopharmacol. 2004;4:1039–1049. doi: 10.1016/j.intimp.2004.04.005. [DOI] [PubMed] [Google Scholar]
- Wrapp D., Vlieger D.D., Corbett K.S., Torres G.M., Wang N., Breedam W.V., Roose K., Schie L.V., COVID-19 Response Team VIB-CMB, Hoffmann M., Pöhlmann S., Graham B.S., Callewaert N., Schepens B., Saelens X., McLellan J.S. Structural basis for potent neutralization of betacoronaviruses by single-domain camelid antibodies. Cell. 2020 doi: 10.1016/j.cell.2020.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J.T., Leung K., Leung G.M. Nowcasting and forecasting the potential domestic and international spread of the 2019-nCoV outbreak originating in Wuhan, China: a modelling study. Lancet. 2020;395:689–697. doi: 10.1016/S0140-6736(20)30260-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue X., Yang H., Shen W., Zhao Q., Li J., Yang K., Chen C., Jin Y., Bartlam M., Rao Z. Production of authentic SARS-CoV M(pro) with enhanced activity: application as a novel tag-cleavage endopeptidase for protein overproduction. J. Mol. Biol. 2007;366:965–975. doi: 10.1016/j.jmb.2006.11.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P., Wang X. COVID-19: a new challenge for human beings. Cell. Mol. Immunol. 2020;17:555–557. doi: 10.1038/s41423-020-0407-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y., Cao D., Zhang Y., Ma J., Qi J., Wang Q., Lu G., Wu Y., Yan J., Shi Y., Zhang X., Gao G.F. Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains. Nature Commun. 2017;8:15092. doi: 10.1038/ncomms15092. -15092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan D., Sunouchi H., Sakurai T., Saito K., Kano Y. Pharmacological properties of traditional medicines (XXVII). Interaction between Ephedra Herb and Gypsum under hyperthermal conditions in rats. Bio. Pharm. Bull. 2002;25:872–874. doi: 10.1248/bpb.25.872. [DOI] [PubMed] [Google Scholar]
- Zheng W., Zhang J., Yang F., Wang Y., Liu Q., Zhang B. Comprehensive analysis of diagnosis and treatment schemes for prevention and treatment of novel coronavirus pneumonia by traditional Chinese medicine. J. Tradit. Chin. Med. 2020;61:277–280. [Google Scholar]
- Zhou P., Yang X., Wang X. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.