Abstract
Altered expression and function of the transcription factor cyclic AMP response-binding protein (CREB) has been identified to play an important role in cancer and is associated with the overall survival and therapy response of tumor patients. This review focuses on the expression and activation of CREB under physiologic conditions and in tumors of distinct origin as well as the underlying mechanisms of CREB regulation by diverse stimuli and inhibitors. In addition, the clinical relevance of CREB is summarized, including its use as a prognostic and/or predictive marker as well as a therapeutic target.
Electronic supplementary material
The online version of this article (10.1007/s00018-020-03525-8) contains supplementary material, which is available to authorized users.
Keywords: Transcription factor, CREB, Carcinogenesis, Prognosis, Clinical outcome
Major characteristics of CREB
Cyclic AMP (cAMP)-response element-binding protein 1 (CREB) is a 43 kDa stimulus-induced transcription factor (TF). It can bind to the cAMP response element (CRE) sequence TGACGTCA or the conserved half CRE TGACG and was first identified in the somatostatin gene promoter [1]. Genome-wide screening for CREB-binding sites suggested that more than 4000 genes might be controlled by CREB, postulating CREB as a general transcriptional activator [2].
Regarding its structure, CREB is made up of different domains with distinct functions. While the DNA binding and dimerization of CREB is mediated by a basic leucine zipper (bZIP) domain, CREB has nine serine residues in the kinase inducible domain (KID) that can be phosphorylated and activated by different kinases. Activated CREB can recruit coactivators, such as CREB-binding protein (CBP). The interaction between CREB and CBP is mediated via the interacting domain of CBP, named KIX. The CREB/CBP complex recruits the transcription machinery at the gene promoter to initiate CREB-dependent gene transcription [3]. The CREB complex upregulates the methylation of histones H3 and H4, which is essential for the initiation of the transcriptional machinery [4]. CREB activity is regulated by the phosphorylation of amino acid (aa) residues, which are mainly localized in the KID region, thereby influencing the dimerization of CREB and its binding to the CRE sequence [5]. Phosphorylation of CREB at the Ser133 residue frequently occurs, whereas phosphorylation at other serine tyrosine and threonine residues of CREB is observed at a lower frequency [5]. Interestingly, the different phosphorylation patterns of CREB are correlated with distinct cellular functions (Table 1) and can exert opposite effects: CREBSer111 and CREBSer121 inhibit transcription, while CREBSer129 and CREBSer133 induce transcription.
Table 1.
Distinct functions of the phosphorylation sites in CREB
| Serine residue | Molecular association | Induction of activity | Inhibition of activity | Cell growth | Cell mobility | Inhibition of apoptosis | Cell differentiation | Induction of transcription | Inhibition of transcription |
|---|---|---|---|---|---|---|---|---|---|
| Ser108 | X | ||||||||
| Ser111 | X | X | |||||||
| Ser114 | X | ||||||||
| Ser117 | |||||||||
| Ser121 | X | X | |||||||
| Ser129 | X | X | |||||||
| Ser133 | X | X | X | X | X | X |
In the following chapters, the knowledge about CREB expression, activation and clinical relevance in tumors of distinct origin and modulators of CREB that could be used as therapeutics for the treatment of diverse cancers are summarized.
Function of CREB as a mediator of carcinogenesis: a general dogma
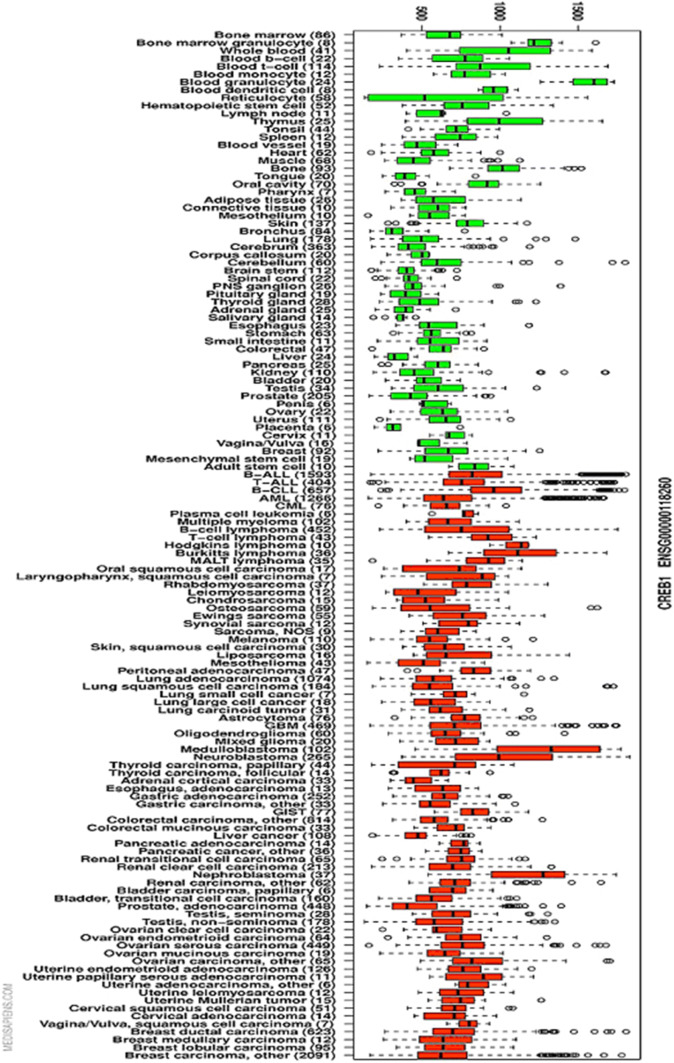
Under physiological conditions, CREB is expressed in all nucleated cells. Its expression is essential for major cellular functions, as CREB knockout mice exhibit embryonal and neuronal deficits and have a reduced lifespan [6–8]. CREB is often overexpressed in hematopoietic and solid tumors compared with control tissues, which has led to the identification of CREB-associated cancers (Fig. 1). These include acute lymphoblastic leukemia (ALL), acute myeloid leukemia (AML), Hodgkin’s lymphoma, chronic lymphatic leukemia (CLL), melanoma, hepatocellular, renal cell, ovarian, prostate, lung, gastric, esophageal, pancreatic and breast carcinoma, and brain tumors [9–11] (Supplementary Table 1).
Fig. 1.
CREB expression in cancer patients. The in silico transcriptomics database (http://ist.medisapiens.com/) was employed for CREB expression in cancer and normal tissues (tissue boxplot). Green represents healthy tissue, while red represents tumor tissues
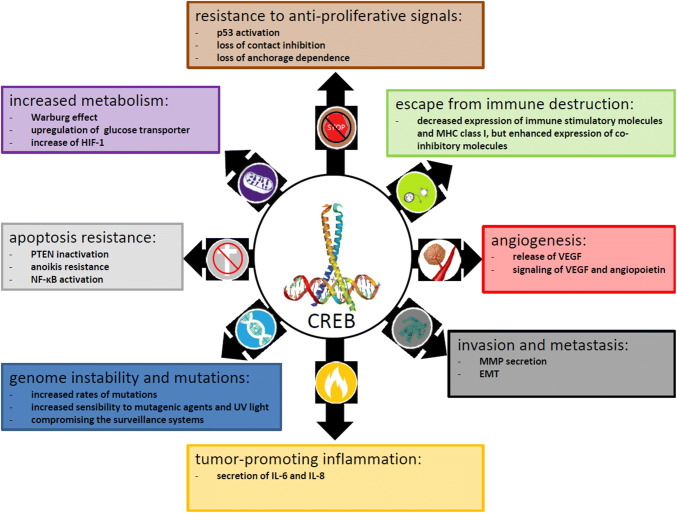
In these malignancies, overexpression of CREB is associated with aberrant signal transduction caused by the deregulated expression of downstream genes that control the hallmarks of cancer, such as proliferation, apoptosis, angiogenesis, metastasis, immune surveillance, and metabolism, and the generation of tumor stem cells, which lead to the initiation and progression of tumors (Fig. 2). These different CREB activities result in increased tumor growth, resistance to antiproliferative signals, decreased apoptosis, enhanced angiogenesis, increased metabolism, and reduced immunogenicity [11–18].
Fig. 2.
Link of the hallmarks of cancer with CREB expression and activation
Opposing the clinical relevance of CREB in different cancers and its association with therapeutic resistance
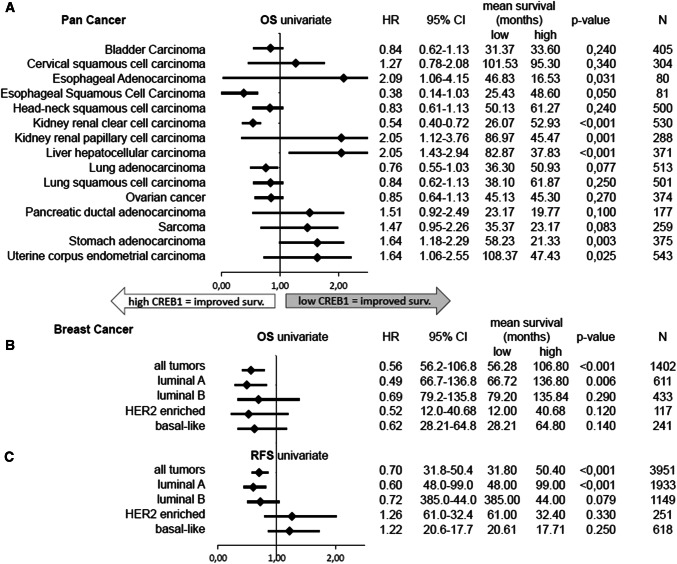
In addition to the role of CREB expression and activity in different tumor entities, CREB protein levels are often correlated with clinical parameters. These include tumor grading and staging, metastasis formation, increased recurrence rates, and worse prognosis of tumor patients [19–23]. Using the KMplot mRNA gene chip and RNA-seq analysis (https://kmplot.com/analysis/), a link between CREB mRNA expression and the overall survival (OS) of patients with different tumors and tumor subtypes was reported and is summarized in Fig. 3 [24]. For example, ALL and AML patients with enhanced expression and phosphorylation of CREB at Ser133 had a decreased OS and a higher risk of tumor relapse [19, 25]. Similar data were obtained for hepatocellular carcinoma (HR 2.05, CI 1.43-2.94, p < 0.01), esophageal adenocarcinoma (HR 2.09, 95% CI 1.06–4.15, p = 0.031), and stomach adenocarcinoma (HR 1.64, 95% CI 1.18–2.29, p = 0.003), in which low CREB expression was associated with reduced OS (Fig. 3a). In contrast, other tumor types benefit from high CREB expression, such as clear cell renal cell carcinoma (ccRCC) (HR 0.38, 95% CI 0.14–1.03, p < 0.001), lung adenocarcinoma (HR 0.76, 95% CI 0.55–1.03, p = 0.077), esophageal squamous cell carcinoma (HR 0.38, 95% CI 0.14–1.03, p = 0.05), and breast cancer (BC) (HR 0.56, 95% CI 0.41–0.79, p < 0.001).
Fig. 3.
Log-rank test of continuous CREB1 expression as a prognostic marker for recurrence-free survival and overall survival. a Hazard ratio (HR) of overall survival from different cancer entities (pan cancer). b HR of overall survival from breast cancer and intrinsic subtypes. c HR of recurrence-free survival from breast cancer and intrinsic subtypes
Regarding BC, overexpression of CREB in all intrinsic BC subtypes has been associated with improved survival of patients (Fig. 3b). In contrast, patients with HER-2/neu-positive or basal-like BC expressing high CREB levels had worse recurrence-free survival (RFS), while luminal-type A BC had an even higher RFS with enhanced CREB expression (Fig. 3c). Since CREB is often overexpressed in different tumor types, but is associated with different outcomes, the quantity of CREB (expression levels of mRNA and protein) might be less important than the quality of CREB (posttranslational modifications and dimerization). Therefore, the dual role of CREB in different tumor entities must be addressed in additional studies to obtain further insights into the underlying mechanisms of CREB regulation and function.
In addition, there is limited information about the role of CREB in therapy resistance. In BC, downregulation of CREB was associated with altered BRCA1 expression and increased expression of aromatase, a key enzyme in estrogen biosynthesis. The latter is transcriptionally regulated by CREB and associated with the development of resistance to tamoxifen [26]. Furthermore, CREB phosphorylation is involved in the development of tumor resistance to inhibitors of the MEK–ERK and PI3K/AKT pathways [26, 27]. In contrast, resistance against MAPK inhibition in BC is induced by reactivation of CREB, which is linked to an altered histone acetylation pattern [27]. In-depth characterization of the mechanisms involved in CREB-mediated therapy resistance is mandatory and relevant for improved therapeutic decision-making in tumor patients.
Regulation of CREB activity by influencing its phosphorylation
There exists evidence that CREB activity is tightly regulated and could be either upregulated or inhibited by diverse factors. Until now, a large number of modulators known to induce CREB phosphorylation have been described in tumor cell lines and tissues of distinct origin, which include growth factors, steroid and peptide hormones, cytokines, stress, lipids, calcium and nitric oxide signaling, various viral, bacterial, and plant components, chemotherapeutics, and others (Supplementary Table 2). These stimuli exert their activities by phosphorylation of distinct CREB residues, but mainly of CREBSer133, thereby affecting different signal transduction pathways, such as ERK1/2, MAPK, PI3K/AKT, CaMK, PKC, and PKA, which are known to be activated in different tumor entities.
Furthermore, high-throughput screening with compound libraries identified 1800 additional substances that were able to enhance CREB-mediated gene transcription [28]. However, most of these substances have not yet been tested in vitro and in vivo in (tumor) cell models.
In addition to stimulators of CREB activity, inhibitors of CREB phosphorylation at Ser133 have been identified, which might have therapeutic potential. To date, no information exists about substances negatively interfering with other CREB phosphorylation residues, as shown for the c-MET inhibitor SU11274 targeting the MET pathway [29] and for serotonin [30]. Other signal transduction inhibitors were tested in various murine and human cell systems regarding their effect on CREB expression and phosphorylation, such as H89, lapatinib, LY294002, PD98059, Ro31-8220, trametinib, the COX-2 inhibitor NS398, and the EP4 inhibitor AH23848 [31, 32]. However, these signal transduction inhibitors were not helpful for functional analysis of CREB, since they do not specifically block the phosphorylation of CREB or influence the activity of other signaling molecules. More specifically, the phosphatase-mediated dephosphorylation of CREB leads to reduced CREB activity. Most phosphatases that inactive CREB, such as protein phosphatase 2A (PP2A) [33], protein phosphatase 1 (PP1) [34], or the nuclear form of PTEN phosphatase [35], are localized to the nucleus. These phosphatases can be targeted by inhibitors, such as okadaic acid blocking both PP2A and PP1 [36]. Their deregulation is associated with altered CREB activity: under hypoxic conditions, these phosphatases are inactive, while CREB is activated and hyperphosphorylated under oxygen limitations, a condition that often occurs in the tumor microenvironment (TME).
Other regulators of CREB expression and/or activity
Mutations in the CREB gene
Structural alterations of CREB have been reported in tumor cell lines and lesions of distinct origin, but their frequency is highly varied. These alterations are diverse and include amplification, homozygous deletions, missense, and in-frame and truncated mutations and fusions, as summarized in Supplementary Tables 3 and 4. Mutations of the CREB gene are most common in neuroendocrine prostate cancer and cervical carcinoma. Missense mutations or truncated mutations have been found, but are not associated with the function of CREB. Furthermore, amplification of the CREB gene has been reported in many tumors. The structural abnormalities of CREB are often accompanied by alterations of other genes known to be involved in tumorigenicity.
A number of studies described gene rearrangements of the Ewing’s sarcoma (EWS) gene with CREB1 in various rare diseases, such as clear cell sarcoma (CCS), CCS associated with the gastrointestinal tract, and angiomatoid fibrous histiocytoma [37–40]. EWS/CREB fusion in CCS associated with the gastrointestinal tract resulted in the loss of KID and was accompanied by melanin pigmentation of these tumors [37]. This is caused by low expression levels of genes involved in melanogenesis, such as MITF and TYR, representing an indicator of deregulated CREB activity. In addition, gene fusions were reported for the CREB family members ATF1 and CREM, particularly in tumors of young children [39], in rare cases of angiomyeloid fibrous histiocytoma [41] and in myxoid neoplasms [40].
Regulation by epigenetic modification, such as methylation and histone modification
There exists only limited information on the epigenetic control of CREB. By employing TF arrays, CREB was identified among 42 TFs to interact with the DNA methyltransferases DNMT3A and DNMT3B [42]. In addition, there is an epigenetic modification switch mediated by the CRE element. After methylation of the central CpG, binding of CREB and related TFs to CpMetG is not possible [43, 44], while binding of the TF C/EBPα is promoted [45], resulting in the control of tissue-specific gene expression [46]. A well-studied example is the CRE site in the BRCA1 gene [47]. CREB is considered a positive regulator of BRCA1, since the methylation of CpG islands significantly reduces its expression. A similar relationship has been reported for MMP-13 [48], which is only transcribed upon demethylation. Nuclear magnetic resonance studies suggest that methylation affects the flexibility of DNA, thereby reducing the ability of TFs to bind to DNA [49].
microRNAs
Posttranscriptional regulation is of crucial importance for the control of gene expression and is mainly mediated by the interaction of RNA-binding proteins (RBPs) and/or microRNAs (miRNAs) with the 3′-untranslated region (3′-UTR) of the respective gene [50–53]. Discordant CREB mRNA and protein expression has been found in some tumor cells, suggesting the regulation of CREB at the posttranscriptional level. Furthermore, it is noteworthy that the annotated ~ 9.000-nucleotide-long 3′-UTR of CREB (ENST00000432329.2) is well above average (~ 800 nucleotides) [54] and gives rise to extensive regulation via this region. In silico analysis and CLIP data revealed a number of well-characterized RBPs potentially binding to CREB, e.g., FUS/TLS (Fused in Sarcoma/Translated in Sarcoma) and RBM10 (RNA-binding protein motif 10) [55, 56]. However, to the best of our knowledge, there is no proven interaction between RBPs and CREB, illustrating the need for further research. In contrast, a number of CREB-regulating and CREB-regulated miRNAs have been recently described in tumor cell lines and in tumors of distinct origin, which are summarized in Table 2 [57].
Table 2.
Characterization of CREB-regulating (RC) miRNAs or CREB-regulated (CR) miRNAs in human tumors and tissues or cell lines
| Name | Cell line/tumor | CR, RC miRNAs |
References |
|---|---|---|---|
| miR-181b | Gastric cancer | RC | [132] |
| miR-34b | AML | RC | [60] |
| miR-200b | Astrocytoma | RC | [133] |
| miR-181a | PC12 (pheochromocytoma) | RC | [134] |
| miR-9 | Glioblastoma | RC, CR | [61] |
| miR-433-3p | Glioblastoma | RC | [135] |
| miR-372 | Liver cancer | RC | [136] |
| miR-1271 | Prostate cancer | RC | [137] |
| miR-760 | Colorectal cancer | RC | [138] |
| miR-23a | Glioma | CR | [139] |
| miR-27b | HepaRG liver cells | CR, RC | [223] |
In leukemia, the CREB protein is overexpressed, which is associated with a poor outcome in these patients [58, 59]. Pigazzi and coworkers demonstrated that miR-34b is involved in the oncogenesis of various tumors and is a major regulator of CREB expression. A direct interaction of this miRNA with the 3′-UTR of CREB was described. In AML, the miR-34b/-34c promoter is hypermethylated and provides a mechanism for the low miR-34b expression in this disease [60].
However, particularly in the context of a general TF such as CREB, it is obvious that miRNA-dependent deregulation is more than a one-to-one relationship. For example, a regulatory mechanism was reported for miR-9 and CREB, whereby CREB promotes the transcriptional expression of miR-9, and in turn, miR-9 directly targets the 3′-UTR of CREB. The balance between these two players is supposed to coordinate the migration and proliferation potential of glioma cells, which may help cells adapt rapidly to environmental changes [61]. Furthermore, miR-27b targeted CREB, demonstrating a positive correlation between CREB and miR-27b in gastric cancer, suggests a bidirectional CREB–miR-27b interaction. This hypothesis is supported by the presence of several CREB-binding sites in the putative promoter of miR-27b [62]. Thus, a better understanding of the CREB–miRNA regulatory networks may open new perspectives for novel therapeutic targets in human malignancies.
Posttranslational modifications with the exception of phosphorylation
It is generally accepted that posttranslational modifications (PTMs), such as acetylation, phosphorylation, glycosylation, SUMOylation, and ubiquitination, often occur (Supplementary Fig. 1) and are altered during physiologic and pathophysiologic cellular processes. Furthermore, these PTMs were also found for CREB and were associated either with increased or decreased CREB activity, which was mediated by distinct mechanisms, as summarized in Table 3. Several PTMs of CREB can affect the progression of cancer and have been recently extensively reviewed [63].
Table 3.
Different PTMs of CREB and their functional relevance
| Modification | aa residue in CREB | CREB activity | Mechanism | Species | References |
|---|---|---|---|---|---|
| Acetylation | K136 | Increased | Recruitment of CBP/p300 | M, 3T3-L | [140] |
| K136 | Increased | Deacetylation by SirT1 | H, HEK293T | [141] | |
| K91, 94, 136 | Decreaseda | Acetylation by CBP/p300 | R, F9; Mo, COS-7 | [142] | |
| n/a | Increased | HDAC9 regulating CREB mRNA | H, HuH7 | [143] | |
| K136 | Increased | CREB acetylation increased by low glucose | M, hippocampal cells | [144] | |
| Ubiquitination | K48-linkedb | Decreased | TRAF3 increasing ubiquitination | M, B cells | [145] |
| n/a | Decreased | MTUS1 deubiquitinating CREB | H, THP-1 cells | [146] | |
| K48-linkedc | Decreased | Hypoxia-mediated ubiquitination | M, NIH3T3 cells | [147] | |
| n/a | Decreased | H2O2-induced ubiquitination | D, in vivo | [148] | |
| n/ac | Decreased | PDGF-stimulated phosphorylation of S103/S107 | R, pulmonary artery | [149] | |
| n/a | Decreased | Hypoxia-mediated loss of PP1 activity | H, CaCo-2 cells | [150] | |
| n/a | Decreased | Hypoxia-mediated ubiquitination | H, HeLa; BT, T84 | [151] | |
| SUMOylation | K271, K290 | Increased | PIAS1-induced modification with SUMO-1 | H, HEK293T | [152] |
| K285, K304 | Increased | Hypoxia mediated by SUMO-1 | H, HeLa; BT, T84 | [151] | |
| K285, K304 | Increased | Hypoxia mediated by SUMO-1,2,3 | M, NIH3T3 cells | [147] | |
| O glycosylation | S40, T228 | Decreased | Elevated CRTC/TORC interaction | R, neuronal cells | [153] |
| n/a | Decreased | Nuclear import under high glucose | H, HuH7 | [154] | |
| T256, S260 | Decreased | Disrupted interaction with TAFII130 | R, brain | [155] | |
| n/a | Decreased | Iron-induced decreased levels of O-GlcNAcylated | M, 3T3-L | [156] | |
| phosphorylation (not in KID) | S270/S271 | Decreased | DNA damage | H, HeLa; H, HEK293T | [157, 158] |
| S271 | Increased | Genotoxic stress | H, SH-SY5Y; H K562 | [158] |
Species: M mouse, H human, R rat, D dog, BT cow, Mo monkey; n/a not analyzed
aTriple mutants only; in single mutants, no changes were observed; enhanced CREB-mediated gene expression, when inhibition of histone deacetylase activity by trichostatin A
bPolyubiquitinated chain, CREB aa not assigned
cPolyubiquitinated chain and monoubiquitination, presumably CREB-K330 or K339
Dimer formation of CREB (homodimers and heterodimers)
The dimer formation of CREB has been controversially discussed. For example, CREB dimerization with ATF1 was described in HeLa cells, but these heterodimers had a lower stability and CRE binding activity than the CREB homodimers [64]. Furthermore, the CREB:ATF1 heterodimers were predominantly found in undifferentiated cells, while homodimer formation was mainly detected in differentiated cells [65, 66]. Regarding jun/fos, CREB:fos heterodimers exist, but their formation is ineffective [67]. In contrast, Muchardt and coauthors reported that neither jun nor fos form heterodimers with CREB, suggesting cell-specific control of this process [68]. In line with these data, no ATF1:jun or ATF1:fos heterodimers could be detected, but heterodimer formation between ATF4 and jun/fos occurred [69]. However, dimer formation of CREB with other bZip TFs has not yet been analyzed in detail in different tumor entities.
Localization-dependent activity of CREB
Under physiological conditions, CREB is localized in the nucleus, while under pathophysiological conditions, e.g., in a hypoxic microenvironment, CREB is shuttled to the mitochondrial matrix [57], where it binds to the mitochondrial CRE sequence. This process results in the control of mitochondrial gene transcription [70], which can be blocked by H89. These data suggest a localization-dependent activity of CREB. Chalovich and coauthors demonstrated that the equilibrium between nuclear and cytoplasmic CREB can be triggered to the site of cytoplasmic localization by 6-hydroxydopamine (and therefore enhancing the levels of mitochondrial CREB) [71, 72]. While Cammarota and coworkers localized phosphorylated CREB in the mitochondria [73], the antibody reacts with an epitope of mitochondrial pyruvate dehydrogenase, suggesting a non-CREB-specific signal [74]. In more recent studies, different CREB-specific antibodies directed against different epitopes of the non-phosphorylated form, gel shift assays [57, 72, 75] or 35S-methionine-labeled CREB have been applied, demonstrating that CREB could be localized in mitochondria under certain conditions [70]. In addition, irradiation can increase the amount of CREBSer131 in the nucleus, which might represent a resistance mechanism of prostate cancer cells [21]. Furthermore, the quantity and activity of the CREB protein in the nucleus can be increased by high glucose levels [76], which are often associated with enhanced tumor cell metabolism, calcium influx [77], or thrombin [78].
Experimental modulation of CREB expression and/or activity
Molecular approaches by CREB silencing
In addition to chemical compounds, diverse experimental approaches, e.g., shCREB, siCREB, double negative (DN) CREB, and CRISPR/CAS, have been used to downregulate or inhibit CREB expression. Although CREB protein expression can be transiently repressed by siRNA binding to CREB1 mRNA [79, 80], long-term experiments exceeding 96 h were not possible. Therefore, shRNA constructs against CREB1 have been commonly used for analyses of the long-term effects of CREB [32, 81]. The specificity of these constructs was proven by monitoring the expression of CREB-related ATF1 and CREM. The implementation of a dominant negative construct as well as reconstitution of CREB knockdown is necessary to rule out unspecific effects. Different dominant negative forms of CREB to block its expression or activity were developed, including a construct named A-CREB, in which the bZIP domain was replaced with an acid amphipathic sequence [82]. This construct mimics the polarity of the CRE sequence and can form a heterodimeric complex with CREB, resulting in decreased CREB binding to the CRE sequence. Another dominant negative form of CREB is the overexpression of a mutated CREB protein, which contains a KID with a replaced amino acid. Furthermore, CREBSer133 has been mutated to CREBAla133, which prevents CREB phosphorylation at this position [83, 228]. A similar approach has been employed for the inhibition of phosphorylation at other serine residues [84]. Furthermore, the DNA-binding domain has also been mutated [83], while Aucoin and coworkers (2004) used double-negative forms of CREB to efficiently block the invasion potential of melanoma cells [226]. Dominant CREB repressors were successfully used both in vitro and in vivo [85], resulting in increased oxidative stress in a transgenic mouse model. In this context, it is noteworthy that silencing or deleting CREB by, e.g., CRISPR/Cas-9, has not been successfully established [86], since CREB is critical for the survival of cells. CREB knockout is lethal in mice, as CREB knockout causes deficits in embryonal development [6, 87]. To circumvent cell death mediated by CREB knockout, the generation of inducible constructs is suggested. Interestingly, the CREB-mediated transduction of cAMP signaling and CREB function in vivo could be partially compensated by CREM [7].
Chemical compounds
Small molecule inhibitors
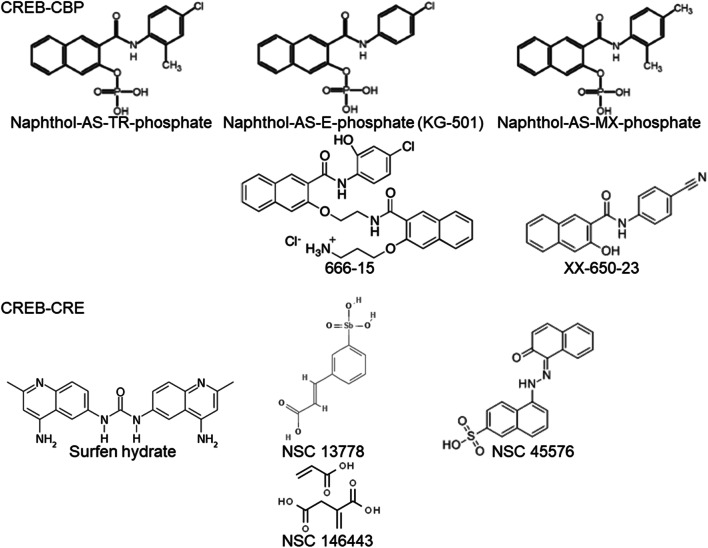
Two different strategies are currently used to block CREB activity with high specificity using chemicals/inhibitors. Based on nuclear magnetic resonance (NMR) analysis demonstrating the binding of CREB KID to CBP KIX [88–90], the interaction between CREB and the coactivator CBP was targeted using CREB-CBP inhibitors, such as different naphthol derivatives [91] (Fig. 4). Furthermore, the binding of CREB to the CRE-DNA element can be blocked with substances binding to the DNA major groove (positively loaded substances) or directly to the bZIP of the TF (negatively loaded substances). A live imaging system using a bioluminescence-based detector system for the analysis of the interaction of KID and KIX was developed by Ishimoto and coworkers [92], which enables screening for CREB inhibitors, e.g., in herbal extracts [93].
Fig. 4.
Chemical structure of CREB-specific small molecule inhibitors. NSC 146443 is a 1:1 mixture of 2-methylenesuccinic acid and acrylic acid and can form a polymer (Pubchem CID: 161509)
CREB-CBP inhibitors targeting the KID-KIX interaction
Various CREB-CBP inhibitors exhibit distinct activities, which are summarized in Supplementary Table 5. The CREB-CBP inhibitor naphthol-AS-E-phosphate (KG-501) was identified by Best and coworkers [94] based on molecular modeling for chemicals binding to the pocket of the KID domain, and was the first inhibitor used for this blocking mechanism in cell culture [95]. KG-501 is specific for CREB, as it blocks the interaction between KID and KIX only at the CREB-specific binding site of KIX, named the c-myb site [96], where the nonphosphorylated proteins c-myb, p53, and BRCA1 can bind [97, 98]. The other binding site of KIX (MLL), where MLL, c-jun, and HIV-1 TAT can bind, is blocked by pamoic acid (KG-122). However, CREB Ser133 had the highest affinity for the c-myb site (Kd CREB − CBP = 700 nM; Kd c-myb − CBP = 15 µM, Kd p53 − CBP > 90 µM) [99]. The unphosphorylated form as a physiological inhibitor was analyzed in different tumor cells using an FRET-based test system. In ALL cell lines, this inhibitor caused increased apoptosis [25], while it blocked CREB phosphorylation induced by curculigoside A and diminished tube formation [100]. In HER-2/neu-overexpressing cell lines, KG-501 decreased migration and anchorage-independent growth without influencing CREB expression and phosphorylation [32]. Due to the relatively low potency of this inhibitor (Ki ~ 90 µM) and its reduced solubility, different structural analogs have been synthesized in recent years [91, 101–103], such as naphthol-MX-phosphate and naphthol-AS-TR-phosphate. However, MX-phosphate is less efficient than KG-501 (IC50 9.7 µM vs 6.9 µM), while AS-TR-phosphate is more potent, as decreased anchorage-independent growth and cyclin expression were detected at lower concentrations (IC50 3.7 µM) [104].
The 3-(3-aminopropoxy)-N-[2-[[3-[[(4-chloro-2-hydroxyphenyl)amino]carbonyl]-2-naphthalenyl]-oxy]ethyl]-2-naphthalenecarboxamide hydrochloride inhibitor (666-15) is an improved, highly efficient CREB-CBP inhibitor (IC50 ~ 80 nM) [94, 167] that weakly affects NF-κB activity by blocking the CBP–NF–κB interaction (IC50 5290 nM). In vivo experimental murine studies of 666-15 revealed its quick bioavailability; no effects were found on kidney and heart functions [105], and it was, therefore, well tolerated in the mouse model. The synthesis of this chemical was first described by Xie et al. [183], followed by the synthesis of different regioisomers [106]. Some modifications for higher aqueous solubility were recently introduced to the backbone of this inhibitor, and results showed that 666-15 had a higher IC50 but inhibitor combinations conferred an additional effect [107]. In a murine xenograph model, 666-15 suppressed the tumor growth [183].
The N-(4-cyanophenyl)-3-hydroxy-2-naphthamide inhibitor (XX-650-23) was synthesized by Li et al. and Xie et al. [101, 103]. XX-650-23 blocks the interaction of CREB and CBP in AML cells expressing high CREB levels, leading to cell cycle arrest and apoptosis by activating caspase-3 activity and decreasing the expression of the antiapoptotic CREB-regulated BCL-2 protein [108]. XX-650-23 is more efficient than KG-501 (IC50 ~ 3 µM in a luciferase detection system). AML cells with higher CREB protein expression, such as HL-60 cells, had an IC50 < 1000 nM, while the CREB low-expressing MOLM-13 cells had an IC50 > 2000 nM. Thus, a specific inhibitory potential seems to be possible for patients with higher CREB levels. Indeed, patients with primary AML or relapsed AML showed higher CREB expression than healthy individuals, and treatment of bone marrow with 2 µM XX-650-23 for 48 h increased the number of dead cells in AML samples but not in normal bone marrow cells [108]. Niclosamide, a molluscicide, had similar effects to XX-650-23 on CREB activity and viability of AML cell lines, but lacks the naphthalene ring [109]. As shown in a recent study further modifications of the XX-650-23 compound lead to better physiological stability and improve the potency [230]. N-(4-Chlorophenyl)-3-hydroxy-2-naphthamide is a cell permeable naphthamide compound that directly binds to the KIX of CBP with an IC50 < 3 µM. It blocks firefly luciferase activity (IC50 ~ 1 µM) but not Renilla luciferase activity.
CREB-CRE inhibitors targeting the interaction of CREB and DNA
Different CREB-CRE inhibitors have also been developed, but are currently less frequently used than KID-KIX inhibitors. These include Surfen (Surfen hydrate, Alias: NSC 12155; CAS-No: 3811-56-1), which is commonly used as a disinfection agent in wound healing solutions or as a depot in combination with insulin, but was withdrawn due to strong allergic reactions. Surfen is an antagonist for heparan sulfate [110], and its potential CREB-CRE blocking mechanism was described by Rishi and coworkers (2005), who reported that Surfen has a higher specificity for CREB than for C/EBPβ (EC50 0.6 µM vs. 2.5 µM) [112]. Surfen at lower concentrations has been shown to block the binding of CREB to a CRE oligonucleotide, accompanied by reduced proliferation of BC cell lines [111].
Stibavirin (Alias: NSC 13778; CAS-No: not registered) is an arylstibonic acid that was proven to bind the basic leucine zipper of CREB but not to DNA [112]. It is, therefore, a specific inhibitor for CREB but also for fos/junD (EC50 13.9 vs. 2.5) [113] and binds to CD4+ T cells [114]. Furthermore, NSC 13778 blocks the binding of TFE3 type 1/2 to the specific promoter element [115], while its derivative P6981 had a stronger effect on CREB inhibition [116]. However, neither substance is commercially available.
The inhibitor 5-[(2-hydroxy-1-naphthalenyl)azo]-2-naphthalenesulfonic acid (Alias: NSC 45576; CAS-No: 68133-05-1) has been proposed as a therapeutic agent for AML, because it decreases the proliferation of AML cell lines. Furthermore, NSC45576 influences cAMP/PKA signaling by reducing the activity of PKA holoenzymes [117, 118].
CREB, ATF2, and c-jun: “It stays in the family”
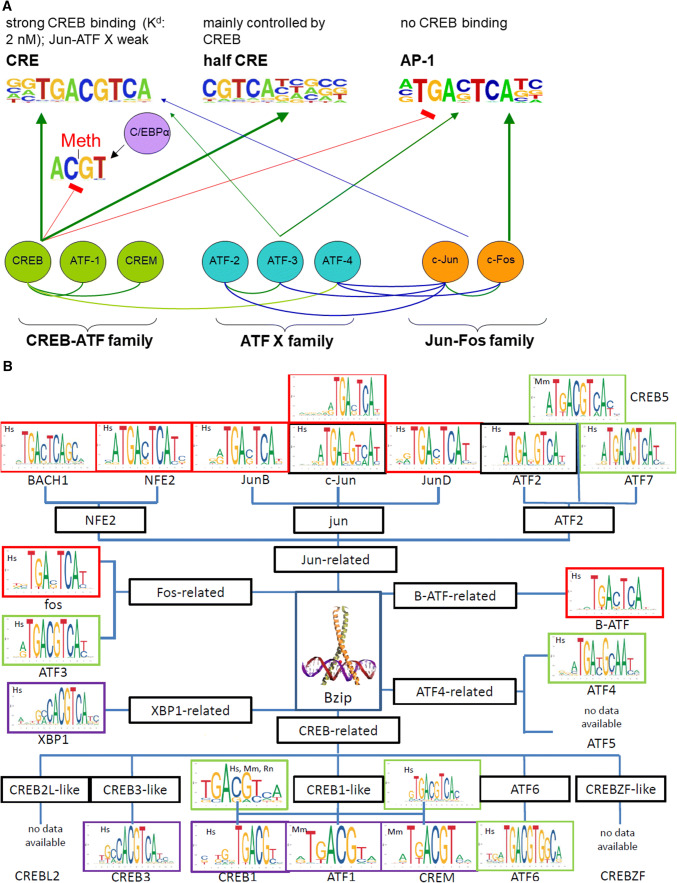
CREB is a member of the bZIP TF family consisting of approximately 20 ATF/CREB family members [119, 120]. Since many TFs can bind to CREB-binding elements, the analysis of whether CREB can be replaced by other TFs is crucial for targeted therapies. Studies have revealed that knocking down CREB expression or activity significantly decreases the transcription of many CRE-regulated genes, such as bcl-2 [18], suggesting that CREB is the major regulator of these genes. Furthermore, genes with a nonpalindromic CRE regulator element, e.g., a half CRE element such as TCAGC, are often downregulated in CREB deficiency and sometimes more efficient than full CRE genes. This could be explained by a stronger induction of CREB at a half CRE sequence than at a full CRE sequence [68] due to a higher competition to members of the CREB-ATF family at the complete CRE site. Interestingly, the activity of the full CRE sequence is higher than that of half CRE sequences in the absence of CREB [68]. Therefore, it is likely that ATF1 can partially compensate for the loss of CREB activity, which is limited due to the lower stability of ATF1 and CREM homo- and heterodimers [3].
Other bZIP TFs, such as ATF2 or ATF3, which cannot form heterodimers with CREB or ATF1 [119], can also bind to the CRE element. They can form heterodimers with jun and fos, and allow binding to the CRE element but with a lesser affinity than CREB. Experiments performed by Hai and Curran [69] revealed that jun/fos heterodimers with ATF2/3 can bind to CRE but not to half CRE. Furthermore, jun and fos heterodimers can bind with higher affinity to AP-1 and full CRE sequences compared to half CRE sites [225]. Therefore, CREB competes with the heterodimers jun-fos/ATF2/3 at CRE but not at half CRE, as summarized in Fig. 5a, b.
Fig. 5.
Model for heterodimerization and the interaction of CREB and other bZIP TFs with DNA elements. a Shown are three bZIP TF families (CREB-ATF, ATF X, and jun-fos) with representative examples. Possible heterodimerization processes are displayed by curved lines: dark green lines represent intrafamily heterodimerization, dark blue lines represent heterodimerization between ATF X and the jun-fos family, and bright green lines represents (rare) heterodimerization between the CREB-ATF and ATF X family. The arrows show the binding of homo- and heterodimers at CRE, half CRE, and AP1 sequences; the line thickness represents the binding affinity, and the line color represents the specificity of the complexes. Jun and fos can bind only as heterodimers with ATF X on a CRE element (blue arrow). CREB-ATF cannot interact with a CpG-methylated CRE or AP1 sequence (red arrows), but C/EBPα can bind to a methylated CRE (black arrow). The conservation of binding sequences was taken from the HOMER database (http://homer.ucsd.edu/homer/motif/motifDatabase.html). b The conservation sequence of the DNA-binding motifs for different bZIP TFs was taken from the JASPAR2020 database (http://jaspar.genereg.net/). The bZIP TFs were classified according to the entrance into the TFClass library (http://tfclass.bioinf.med.uni-goettingen.de/). Common bZIP TFs are presented. The colored boxes around the sequences are as follows: green = typical 8 bp full CRE sequence with a central conserved CG (TGACGTCG), violet = half site CRE (TGACG), red = typical 7 bp AP-1 site without a central CG (TGACTCA or TGAGTCA), and black = 8 bp binding site without a conserved central CG. The species are abbreviated as follows: Hs = Homo sapiens, Mm = Mus musculus, and Rn = Rattus norvegicus. The central bZIP motif is the CREB1 leucine zipper domain binding to the CRE-DNA and was taken from the PyMOL data bank PDB1DH3
Furthermore, CREB prefers the central CpG dinucleotide of CRE/half CRE, which explains the lack of CREB binding to the CRE-similar AP1 sequence [121]. A methylated CpG dinucleotide is a binding site for C/EBPα but not for CREB [46]. Genes with a CRE element and a TATA box could show different regulation than CRE genes without a TATA box. Binding of CREB to the 8 bp CRE sequence and the variable half CREs depends on different mechanisms [122]. Exclusively, CREB can bind to two different half CRE motifs with a dissociation constant that is comparable to that of the full CRE sequence [68, 119, 123].
In addition to ATF1 and CREM, the cAMP response transcriptional coactivators (CRTCs), comprising the three members CRTC1, CRTC2, and CRTC3, represent an additional family of CREB coactivators with similar modular structures. CRTCs are evolutionarily highly conserved and sequestered in the cytoplasm [124]. They have been shown to regulate transcriptional activation and pre-mRNA splicing via distinct functional domains [125]. CRTCs upregulate the activity of CREB by association with residues in the bZIP domain. However, CRTCs not only regulate CREB-dependent target genes but also CREB-independent transcriptional responses. The aberrant activation of CRTCs in tumors is linked with oncogenic activities, such as migration, invasion, and metastasis formation, representing all hallmarks of cancer [126, 127]. This is also strengthened by the fact that mutations in CRTCs have been shown to be key drivers in the development and progression of cancer [128].
Conclusion: CREB as a prognostic biomarker or therapeutic target?
Based on the central role of CREB in the initiation, maintenance, and progression of many cancer types (Supplementary Fig. 2), CREB is considered a prognostic biomarker and an excellent therapeutic target structure for tumors. This claim is supported by expression analyses of the early, inducible cAMP repressor (ICER), an inhibitor of CREB, which is downregulated in BM cells of AML patients [60]. An advantage of using CREB as a target structure is its ability to regulate different signal transduction pathways, which are often aberrantly activated in tumors. However, it is noteworthy that high CREB expression in some tumor types is associated with better patient outcomes. To date, the underlying mechanisms of these opposing effects are not well understood and require further investigation.
Several strategies are currently used to inhibit CREB function in tumor cells: (1) Initial studies focused on dominant negative CREB mutants (KCREB) to block CREB transcription. KCREB cannot bind to CRE sequences but forms heterodimers with wild-type CREB. Overexpression of KCREB in metastatic tumor cells decreases the metastatic potential in vitro and in vivo [129]. (2) CREB decoy oligonucleotides that efficiently inhibit CREB-mediated gene transcription and therefore negatively influence tumor growth have been developed [130]. (3) CREB expression is silenced by RNA interference, which not only modulates cell viability and growth properties but also enhances apoptosis. shRNA-mediated silencing of CREB expression is coupled with diminished growth of tumor cells, increased apoptosis, cell cycle arrest in the G0/G1 phase and suppression of anchorage-independent growth [32, 61, 131].
Since these proof-of-principle studies have revealed therapeutic effects, alternative strategies to inhibit CREB-mediated gene transcription with small molecule inhibitors have been developed. For example, kinase inhibitors can prevent phosphorylation and, therefore, inhibit the activation of CREB. In addition, chemical inhibitors can block the interaction of CREB-CRE or CREB-CBP [91, 94, 101, 103]. Naphthol-AS-E-phosphate (KG-501) reversibly and dose-dependently disrupts the interaction between the KID domain of CREB and the KIX domain of CBP but not forskolin-stimulated phosphorylation at Ser133. Micromolar concentrations of KG-501 can modulate the cAMP-dependent expression of CREB target genes without off-target inhibition. Another strategy is the modulation of CREB-regulating miRNAs [22]. Since CREB has many oncogenic properties and participates in the induction of resistance mechanisms, it is a promising target for the treatment of many tumor types; nevertheless, for tumor types in which high levels of CREB expression are associated with better outcomes, this approach may not be suitable.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
Open Access funding provided by Projekt DEAL. We are grateful to Maria Heise for excellent secretarial help.
Abbreviations
- ALL
Acute lymphatic leukemia
- AML
Acute myeloid leukemia
- ATF-1
Activating transcription factor 1
- BC
Breast cancer
- bZIP
Basic leucine zipper
- CaMK
Calcium-activated calmodulin kinase
- CBP
CREB-binding protein
- CLL
Chronic lymphatic leukemia
- CRE
cAMP response element
- CREB
cAMP response element-binding protein
- CREM
cAMP response element modulator
- CRTC
cAMP response transcriptional co-activator
- DNMT
DNA methyltransferase
- ERK
Extracellular signal-regulated kinase
- EWS
Ewing’s sarcoma
- HR
Hazard ratio
- KID
Kinase-inducible domain
- KIX
KID-interacting domain
- MAPK
Mitogen-activated protein kinase
- MFS
Metastasis-free survival
- miRNA
microRNA
- OS
Overall survival
- PI3K
Phosphatidylinositol 3-kinase
- PK
Protein kinase
- PP1
Protein phosphatase 1
- PP2A
Protein phosphatase 2A
- PTM
Post-translational modification
- RBP
RNA-binding protein
- RCC
Renal cell cancer
- RFS
Recurrence-free survival
- TCGA
The Cancer Genome Atlas
- TF
Transcription factor
- TME
Tumor microenvironment
- TNBC
Triple negative breast cancer
- UTR
Untranslated region
Author contributions
MF and NH searched for miRNAs connected with CREB expression and wrote the chapter “MicroRNAs”; PJ analyzed the survival of patients with different CREB statuses; PJ and CD wrote the chapter “Clinical relevance of CREB”; AS recorded the regulation of CREB by phosphorylation and other approaches, possible modulation strategies for CREB, and the CREB-associated hallmarks of cancer; JB and AS performed the cBioPortal analysis; AS and BS generated the concept of the review, structured and corrected the different chapters provided by the coauthors, and wrote the remaining parts of the manuscript. All authors reviewed the final version of this manuscript.
Availability of data and material
All data are available upon request from the corresponding author.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest for any of the authors.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Montminy MR, Bilezikjian LM. Binding of a nuclear protein to the cyclic-AMP response element of the somatostatin gene. Nature. 1987;328(6126):175–178. doi: 10.1038/328175a0. [DOI] [PubMed] [Google Scholar]
- 2.Zhang X, et al. Genome-wide analysis of cAMP-response element binding protein occupancy, phosphorylation, and target gene activation in human tissues. Proc Natl Acad Sci USA. 2005;102(12):4459–4464. doi: 10.1073/pnas.0501076102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shaywitz AJ, Greenberg ME. CREB: a stimulus-induced transcription factor activated by a diverse array of extracellular signals. Annu Rev Biochem. 1999;68:821–861. doi: 10.1146/annurev.biochem.68.1.821. [DOI] [PubMed] [Google Scholar]
- 4.Sen N, Snyder SH. Neurotrophin-mediated degradation of histone methyltransferase by S-nitrosylation cascade regulates neuronal differentiation. Proc Natl Acad Sci USA. 2011;108(50):20178–20183. doi: 10.1073/pnas.1117820108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johannessen M, Delghandi MP, Moens U. What turns CREB on? Cell Signal. 2004;16(11):1211–1227. doi: 10.1016/j.cellsig.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 6.Bleckmann SC, et al. Activating transcription factor 1 and CREB are important for cell survival during early mouse development. Mol Cell Biol. 2002;22(6):1919–1925. doi: 10.1128/MCB.22.6.1919-1925.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hummler E, et al. Targeted mutation of the CREB gene: compensation within the CREB/ATF family of transcription factors. Proc Natl Acad Sci USA. 1994;91(12):5647–5651. doi: 10.1073/pnas.91.12.5647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mantamadiotis T, et al. Disruption of CREB function in brain leads to neurodegeneration. Nat Genet. 2002;31(1):47–54. doi: 10.1038/ng882. [DOI] [PubMed] [Google Scholar]
- 9.Chen P, et al. Targeting the overexpressed CREB inhibits esophageal squamous cell carcinoma cell growth. Oncol Rep. 2018;39(3):1369–1377. doi: 10.3892/or.2017.6167. [DOI] [PubMed] [Google Scholar]
- 10.Siu YT, Jin DY. CREB–a real culprit in oncogenesis. FEBS J. 2007;274(13):3224–3232. doi: 10.1111/j.1742-4658.2007.05884.x. [DOI] [PubMed] [Google Scholar]
- 11.Xia Y, et al. Targeting CREB pathway suppresses small cell lung cancer. Mol Cancer Res. 2018;16(5):825–832. doi: 10.1158/1541-7786.MCR-17-0576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang S, et al. The transcription factor creb is involved in sorafenib-inhibited renal cancer cell proliferation. Migration and invasion. Acta Pharm. 2018;68(4):497–506. doi: 10.2478/acph-2018-0033. [DOI] [PubMed] [Google Scholar]
- 13.Son J, et al. cAMP-response-element-binding protein positively regulates breast cancer metastasis and subsequent bone destruction. Biochem Biophys Res Commun. 2010;398(2):309–314. doi: 10.1016/j.bbrc.2010.06.087. [DOI] [PubMed] [Google Scholar]
- 14.Suarez CD, Deng X, Hu CD. Targeting CREB inhibits radiation-induced neuroendocrine differentiation and increases radiation-induced cell death in prostate cancer cells. Am J Cancer Res. 2014;4(6):850–861. [PMC free article] [PubMed] [Google Scholar]
- 15.Wang X, et al. Decrease of phosphorylated proto-oncogene CREB at Ser 133 site inhibits growth and metastatic activity of renal cell cancer. Expert Opin Ther Targ. 2015;19(7):985–995. doi: 10.1517/14728222.2015.1053208. [DOI] [PubMed] [Google Scholar]
- 16.Wang X, et al. Cyclic AMP responsive element-binding protein induces metastatic renal cell carcinoma by mediating the expression of matrix metallopeptidase-2/9 and proteins associated with epithelial-mesenchymal transition. Mol Med Rep. 2017;15(6):4191–4198. doi: 10.3892/mmr.2017.6519. [DOI] [PubMed] [Google Scholar]
- 17.Zhang ZX, et al. CREB promotes laryngeal cancer cell migration via MYCT1/NAT10 axis. Onco Targ Ther. 2018;11:1323–1331. doi: 10.2147/OTT.S156582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zheng KB, et al. Knockdown of CERB expression inhibits proliferation and migration of glioma cells line U251. Bratisl Lek Listy. 2019;120(4):309–315. doi: 10.4149/BLL_2019_049. [DOI] [PubMed] [Google Scholar]
- 19.Cheng JC, et al. Potential role of CREB as a prognostic marker in acute myeloid leukemia. Future Oncol. 2007;3(4):475–480. doi: 10.2217/14796694.3.4.475. [DOI] [PubMed] [Google Scholar]
- 20.Chhabra A, et al. Expression of transcription factor CREB1 in human breast cancer and its correlation with prognosis. Oncol Rep. 2007;18(4):953–958. [PubMed] [Google Scholar]
- 21.Deng X, et al. Ionizing radiation induces prostate cancer neuroendocrine differentiation through interplay of CREB and ATF2: implications for disease progression. Cancer Res. 2008;68(23):9663–9670. doi: 10.1158/0008-5472.CAN-08-2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sakamoto KM, Frank DA. CREB in the pathophysiology of cancer: implications for targeting transcription factors for cancer therapy. Clin Cancer Res. 2009;15(8):2583–2587. doi: 10.1158/1078-0432.CCR-08-1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seo HS, et al. Cyclic AMP response element-binding protein overexpression: a feature associated with negative prognosis in never smokers with non-small cell lung cancer. Cancer Res. 2008;68(15):6065–6073. doi: 10.1158/0008-5472.CAN-07-5376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gyorffy B, et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat. 2010;123(3):725–731. doi: 10.1007/s10549-009-0674-9. [DOI] [PubMed] [Google Scholar]
- 25.van der Sligte NE, et al. Essential role for cyclic-AMP responsive element binding protein 1 (CREB) in the survival of acute lymphoblastic leukemia. Oncotarget. 2015;6(17):14970–14981. doi: 10.18632/oncotarget.3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Phuong NT, et al. Aromatase induction in tamoxifen-resistant breast cancer: role of phosphoinositide 3-kinase-dependent CREB activation. Cancer Lett. 2014;351(1):91–99. doi: 10.1016/j.canlet.2014.05.003. [DOI] [PubMed] [Google Scholar]
- 27.Johannessen CM, et al. A melanocyte lineage program confers resistance to MAP kinase pathway inhibition. Nature. 2013;504(7478):138–142. doi: 10.1038/nature12688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xia M, et al. Identification of compounds that potentiate CREB signaling as possible enhancers of long-term memory. Proc Natl Acad Sci USA. 2009;106(7):2412–2417. doi: 10.1073/pnas.0813020106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Coudriet GM, et al. Hepatocyte growth factor modulates interleukin-6 production in bone marrow derived macrophages: implications for inflammatory mediated diseases. PLoS One. 2010;5(11):e15384. doi: 10.1371/journal.pone.0015384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klavdianou K, et al. Decreased serotonin levels and serotonin-mediated osteoblastic inhibitory signaling in patients with ankylosing spondylitis. J Bone Miner Res. 2016;31(3):630–639. doi: 10.1002/jbmr.2724. [DOI] [PubMed] [Google Scholar]
- 31.Sirianni R, et al. Inhibition of cyclooxygenase-2 down-regulates aromatase activity and decreases proliferation of Leydig tumor cells. J Biol Chem. 2009;284(42):28905–28916. doi: 10.1074/jbc.M109.041020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Steven A, et al. HER-2/neu mediates oncogenic transformation via altered CREB expression and function. Mol Cancer Res. 2013;11(11):1462–1477. doi: 10.1158/1541-7786.MCR-13-0125. [DOI] [PubMed] [Google Scholar]
- 33.Wadzinski BE, et al. Nuclear protein phosphatase 2A dephosphorylates protein kinase A-phosphorylated CREB and regulates CREB transcriptional stimulation. Mol Cell Biol. 1993;13(5):2822–2834. doi: 10.1128/mcb.13.5.2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Genoux D, et al. Protein phosphatase 1 is a molecular constraint on learning and memory. Nature. 2002;418(6901):970–975. doi: 10.1038/nature00928. [DOI] [PubMed] [Google Scholar]
- 35.Gu T, et al. CREB is a novel nuclear target of PTEN phosphatase. Cancer Res. 2011;71(8):2821–2825. doi: 10.1158/0008-5472.CAN-10-3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Choe ES, et al. The protein phosphatase 1/2A inhibitor okadaic acid increases CREB and Elk-1 phosphorylation and c-fos expression in the rat striatum in vivo. J Neurochem. 2004;89(2):383–390. doi: 10.1111/j.1471-4159.2003.02334.x. [DOI] [PubMed] [Google Scholar]
- 37.Antonescu CR, et al. EWS-CREB1: a recurrent variant fusion in clear cell sarcoma–association with gastrointestinal location and absence of melanocytic differentiation. Clin Cancer Res. 2006;12(18):5356–5362. doi: 10.1158/1078-0432.CCR-05-2811. [DOI] [PubMed] [Google Scholar]
- 38.Segawa K, et al. Detection of specific gene rearrangements by fluorescence in situ hybridization in 16 cases of clear cell sarcoma of soft tissue and 6 cases of clear cell sarcoma-like gastrointestinal tumor. Diagn Pathol. 2018;13(1):73. doi: 10.1186/s13000-018-0752-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Konstantinidis A, et al. Intracranial angiomatoid fibrous histiocytoma with EWSR1-CREB family fusions: a report of 2 pediatric cases. World Neurosurg. 2019;126:113–119. doi: 10.1016/j.wneu.2019.02.107. [DOI] [PubMed] [Google Scholar]
- 40.Kao YC, et al. EWSR1 fusions with CREB family transcription factors define a novel myxoid mesenchymal tumor with predilection for intracranial location. Am J Surg Pathol. 2017;41(4):482–490. doi: 10.1097/PAS.0000000000000788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rekhi B, et al. Angiomatoid fibrous histiocytoma: clinicopathological spectrum of five cases, including EWSR1-CREB1 positive result in a single case. Indian J Pathol Microbiol. 2016;59(2):148–152. doi: 10.4103/0377-4929.182023. [DOI] [PubMed] [Google Scholar]
- 42.Hervouet E, Vallette FM, Cartron PF. Dnmt3/transcription factor interactions as crucial players in targeted DNA methylation. Epigenetics. 2009;4(7):487–499. doi: 10.4161/epi.4.7.9883. [DOI] [PubMed] [Google Scholar]
- 43.Iguchi-Ariga SM, Schaffner W. CpG methylation of the cAMP-responsive enhancer/promoter sequence TGACGTCA abolishes specific factor binding as well as transcriptional activation. Genes Dev. 1989;3(5):612–619. doi: 10.1101/gad.3.5.612. [DOI] [PubMed] [Google Scholar]
- 44.Mancini DN, et al. Site-specific DNA methylation in the neurofibromatosis (NF1) promoter interferes with binding of CREB and SP1 transcription factors. Oncogene. 1999;18(28):4108–4119. doi: 10.1038/sj.onc.1202764. [DOI] [PubMed] [Google Scholar]
- 45.Bakker O, Parker MG. CAAT/enhancer binding protein is able to bind to ATF/CRE elements. Nucleic Acids Res. 1991;19(6):1213–1217. doi: 10.1093/nar/19.6.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rishi V, et al. CpG methylation of half-CRE sequences creates C/EBPalpha binding sites that activate some tissue-specific genes. Proc Natl Acad Sci USA. 2010;107(47):20311–20316. doi: 10.1073/pnas.1008688107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.DiNardo DN, et al. Functional analysis of CpG methylation in the BRCA1 promoter region. Oncogene. 2001;20(38):5331–5340. doi: 10.1038/sj.onc.1204697. [DOI] [PubMed] [Google Scholar]
- 48.Bui C, et al. cAMP response element-binding (CREB) recruitment following a specific CpG demethylation leads to the elevated expression of the matrix metalloproteinase 13 in human articular chondrocytes and osteoarthritis. FASEB J. 2012;26(7):3000–3011. doi: 10.1096/fj.12-206367. [DOI] [PubMed] [Google Scholar]
- 49.Derreumaux S, et al. Impact of CpG methylation on structure, dynamics and solvation of cAMP DNA responsive element. Nucleic Acids Res. 2001;29(11):2314–2326. doi: 10.1093/nar/29.11.2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Martin KC, Ephrussi A. mRNA localization: gene expression in the spatial dimension. Cell. 2009;136(4):719–730. doi: 10.1016/j.cell.2009.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Moore MJ, Proudfoot NJ. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell. 2009;136(4):688–700. doi: 10.1016/j.cell.2009.02.001. [DOI] [PubMed] [Google Scholar]
- 52.Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136(4):731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mignone F, et al. Untranslated regions of mRNAs. Genome Biol. 2002;3(3):REVIEWS0004. doi: 10.1186/gb-2002-3-3-reviews0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dweep H, et al. miRWalk–database: prediction of possible miRNA binding sites by “walking” the genes of three genomes. J Biomed Inform. 2011;44(5):839–847. doi: 10.1016/j.jbi.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 56.Dweep H, Gretz N. miRWalk20: a comprehensive atlas of microRNA-target interactions. Nat Methods. 2015;12(8):697. doi: 10.1038/nmeth.3485. [DOI] [PubMed] [Google Scholar]
- 57.Steven A, Seliger B. Control of CREB expression in tumors: from molecular mechanisms and signal transduction pathways to therapeutic target. Oncotarget. 2016;7(23):35454–35465. doi: 10.18632/oncotarget.7721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shankar DB, et al. The role of CREB as a proto-oncogene in hematopoiesis and in acute myeloid leukemia. Cancer Cell. 2005;7(4):351–362. doi: 10.1016/j.ccr.2005.02.018. [DOI] [PubMed] [Google Scholar]
- 59.Cheng JC, et al. CREB is a critical regulator of normal hematopoiesis and leukemogenesis. Blood. 2008;111(3):1182–1192. doi: 10.1182/blood-2007-04-083600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pigazzi M, et al. miR-34b targets cyclic AMP-responsive element binding protein in acute myeloid leukemia. Cancer Res. 2009;69(6):2471–2478. doi: 10.1158/0008-5472.CAN-08-3404. [DOI] [PubMed] [Google Scholar]
- 61.Tan X, et al. The CREB-miR-9 negative feedback minicircuitry coordinates the migration and proliferation of glioma cells. PLoS One. 2012;7(11):e49570. doi: 10.1371/journal.pone.0049570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang YW, et al. High expression of cAMP-responsive element-binding protein 1 (CREB1) is associated with metastasis, tumor stage and poor outcome in gastric cancer. Oncotarget. 2015;6(12):10646–10657. doi: 10.18632/oncotarget.3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wu Z, Huang R, Yuan L. Crosstalk of intracellular post-translational modifications in cancer. Arch Biochem Biophys. 2019;676:108138. doi: 10.1016/j.abb.2019.108138. [DOI] [PubMed] [Google Scholar]
- 64.Johannessen M, et al. Synergistic activation of CREB-mediated transcription by forskolin and phorbol ester requires PKC and depends on the glutamine-rich Q2 transactivation domain. Cell Signal. 2004;16(10):1187–1199. doi: 10.1016/j.cellsig.2004.03.009. [DOI] [PubMed] [Google Scholar]
- 65.Hurst HC, Totty NF, Jones NC. Identification and functional characterisation of the cellular activating transcription factor 43 (ATF-43) protein. Nucleic Acids Res. 1991;19(17):4601–4609. doi: 10.1093/nar/19.17.4601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ellis MJ, Hurst HC, Goodbourn S. A novel cyclic AMP response element-binding protein-1 (CREB-1) splice product may down-regulate CREB-1 activity. J Mol Endocrinol. 1995;14(2):191–198. doi: 10.1677/jme.0.0140191. [DOI] [PubMed] [Google Scholar]
- 67.Benbrook DM, Jones NC. Heterodimer formation between CREB and JUN proteins. Oncogene. 1990;5(3):295–302. [PubMed] [Google Scholar]
- 68.Muchardt C, et al. CREB regulation of cellular cyclic AMP-responsive and adenovirus early promoters. J Virol. 1990;64(9):4296–4305. doi: 10.1128/jvi.64.9.4296-4305.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hai T, Curran T. Cross-family dimerization of transcription factors Fos/Jun and ATF/CREB alters DNA binding specificity. Proc Natl Acad Sci USA. 1991;88(9):3720–3724. doi: 10.1073/pnas.88.9.3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.De Rasmo D, et al. cAMP response element-binding protein (CREB) is imported into mitochondria and promotes protein synthesis. FEBS J. 2009;276(16):4325–4333. doi: 10.1111/j.1742-4658.2009.07133.x. [DOI] [PubMed] [Google Scholar]
- 71.Chalovich EM, et al. Functional repression of cAMP response element in 6-hydroxydopamine-treated neuronal cells. J Biol Chem. 2006;281(26):17870–17881. doi: 10.1074/jbc.M602632200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lee J, et al. Mitochondrial cyclic AMP response element-binding protein (CREB) mediates mitochondrial gene expression and neuronal survival. J Biol Chem. 2005;280(49):40398–40401. doi: 10.1074/jbc.C500140200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cammarota M, et al. Cyclic AMP-responsive element binding protein in brain mitochondria. J Neurochem. 1999;72(6):2272–2277. doi: 10.1046/j.1471-4159.1999.0722272.x. [DOI] [PubMed] [Google Scholar]
- 74.Platenik J, et al. Apparent presence of Ser133-phosphorylated cyclic AMP response element binding protein (pCREB) in brain mitochondria is due to cross-reactivity of pCREB antibodies with pyruvate dehydrogenase. J Neurochem. 2005;95(5):1446–1460. doi: 10.1111/j.1471-4159.2005.03471.x. [DOI] [PubMed] [Google Scholar]
- 75.Ryu H, et al. Antioxidants modulate mitochondrial PKA and increase CREB binding to D-loop DNA of the mitochondrial genome in neurons. Proc Natl Acad Sci USA. 2005;102(39):13915–13920. doi: 10.1073/pnas.0502878102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Habib SL, et al. Novel mechanism of transcriptional regulation of cell matrix protein through CREB. Cell Cycle. 2015;14(16):2598–2608. doi: 10.1080/15384101.2015.1064204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hongpaisan J, Winters CA, Andrews SB. Calcium-dependent mitochondrial superoxide modulates nuclear CREB phosphorylation in hippocampal neurons. Mol Cell Neurosci. 2003;24(4):1103–1115. doi: 10.1016/j.mcn.2003.09.003. [DOI] [PubMed] [Google Scholar]
- 78.Liu Y, et al. CaMKIIdelta-dependent inhibition of cAMP-response element-binding protein activity in vascular smooth muscle. J Biol Chem. 2013;288(47):33519–33529. doi: 10.1074/jbc.M113.490870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kim SJ, et al. Glucose-dependent insulinotropic polypeptide-mediated up-regulation of beta-cell antiapoptotic Bcl-2 gene expression is coordinated by cyclic AMP (cAMP) response element binding protein (CREB) and cAMP-responsive CREB coactivator 2. Mol Cell Biol. 2008;28(5):1644–1656. doi: 10.1128/MCB.00325-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Satpathy M, et al. Tissue transglutaminase regulates matrix metalloproteinase-2 in ovarian cancer by modulating cAMP-response element-binding protein activity. J Biol Chem. 2009;284(23):15390–15399. doi: 10.1074/jbc.M808331200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dobroff AS, et al. Silencing cAMP-response element-binding protein (CREB) identifies CYR61 as a tumor suppressor gene in melanoma. J Biol Chem. 2009;284(38):26194–26206. doi: 10.1074/jbc.M109.019836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ahn S, et al. A dominant-negative inhibitor of CREB reveals that it is a general mediator of stimulus-dependent transcription of c-fos. Mol Cell Biol. 1998;18(2):967–977. doi: 10.1128/mcb.18.2.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Amorino GP, et al. Dominant-negative cAMP-responsive element-binding protein inhibits proliferating cell nuclear antigen and DNA repair, leading to increased cellular radiosensitivity. J Biol Chem. 2003;278(32):29394–29399. doi: 10.1074/jbc.M304012200. [DOI] [PubMed] [Google Scholar]
- 84.Barton K, et al. Defective thymocyte proliferation and IL-2 production in transgenic mice expressing a dominant-negative form of CREB. Nature. 1996;379(6560):81–85. doi: 10.1038/379081a0. [DOI] [PubMed] [Google Scholar]
- 85.Lee B, et al. The CREB/CRE transcriptional pathway: protection against oxidative stress-mediated neuronal cell death. J Neurochem. 2009;108(5):1251–1265. doi: 10.1111/j.1471-4159.2008.05864.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jinek M, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337(6096):816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lonze BE, et al. Apoptosis, axonal growth defects, and degeneration of peripheral neurons in mice lacking CREB. Neuron. 2002;34(3):371–385. doi: 10.1016/s0896-6273(02)00686-4. [DOI] [PubMed] [Google Scholar]
- 88.Radhakrishnan I, et al. Structural analyses of CREB-CBP transcriptional activator-coactivator complexes by NMR spectroscopy: implications for mapping the boundaries of structural domains. J Mol Biol. 1999;287(5):859–865. doi: 10.1006/jmbi.1999.2658. [DOI] [PubMed] [Google Scholar]
- 89.Radhakrishnan I, et al. Conformational preferences in the Ser133-phosphorylated and non-phosphorylated forms of the kinase inducible transactivation domain of CREB. FEBS Lett. 1998;430(3):317–322. doi: 10.1016/s0014-5793(98)00680-2. [DOI] [PubMed] [Google Scholar]
- 90.Gee CT, Koleski EJ, Pomerantz WC. Fragment screening and druggability assessment for the CBP/p300 KIX domain through protein-observed 19F NMR spectroscopy. Angew Chem Int Ed Engl. 2015;54(12):3735–3739. doi: 10.1002/anie.201411658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Jiang M, et al. Design, synthesis, and biological evaluation of conformationally constrained analogues of naphthol AS-E as inhibitors of CREB-mediated gene transcription. J Med Chem. 2012;55(8):4020–4024. doi: 10.1021/jm300043c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ishimoto T, et al. Measuring CREB activation using bioluminescent probes that detect KID-KIX interaction in living cells. Bioconjug Chem. 2012;23(5):923–932. doi: 10.1021/bc200491j. [DOI] [PubMed] [Google Scholar]
- 93.Mano H, et al. Discovery of novel adenylyl cyclase inhibitor by cell-based screening. Biol Pharm Bull. 2014;37(10):1689–1693. doi: 10.1248/bpb.b14-00283. [DOI] [PubMed] [Google Scholar]
- 94.Best JL, et al. Identification of small-molecule antagonists that inhibit an activator: coactivator interaction. Proc Natl Acad Sci USA. 2004;101(51):17622–17627. doi: 10.1073/pnas.0406374101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sun H, et al. Cyclic AMP-responsive element binding protein- and nuclear factor-kappaB-regulated CXC chemokine gene expression in lung carcinogenesis. Cancer Prev Res (Phila) 2008;1(5):316–328. doi: 10.1158/1940-6207.CAPR-07-0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.De Guzman RN, et al. Structural basis for cooperative transcription factor binding to the CBP coactivator. J Mol Biol. 2006;355(5):1005–1013. doi: 10.1016/j.jmb.2005.09.059. [DOI] [PubMed] [Google Scholar]
- 97.Thakur AS, et al. Synthesis and oral hypoglycemic effect of novel thiazine containing trisubstituted benzenesulfonylurea derivatives. Saudi Pharm J. 2015;23(5):475–482. doi: 10.1016/j.jsps.2014.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Uttarkar S, et al. Naphthol AS-E phosphate inhibits the activity of the transcription factor Myb by blocking the interaction with the KIX domain of the coactivator p300. Mol Cancer Ther. 2015;14(6):1276–1285. doi: 10.1158/1535-7163.MCT-14-0662. [DOI] [PubMed] [Google Scholar]
- 99.Wang F, Marshall CB, Ikura M. Transcriptional/epigenetic regulator CBP/p300 in tumorigenesis: structural and functional versatility in target recognition. Cell Mol Life Sci. 2013;70(21):3989–4008. doi: 10.1007/s00018-012-1254-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kang Z, et al. Curculigoside A induces angiogenesis through VCAM-1/Egr-3/CREB/VEGF signaling pathway. Neuroscience. 2014;267:232–240. doi: 10.1016/j.neuroscience.2014.02.050. [DOI] [PubMed] [Google Scholar]
- 101.Li BX, Yamanaka K, Xiao X. Structure-activity relationship studies of naphthol AS-E and its derivatives as anticancer agents by inhibiting CREB-mediated gene transcription. Bioorg Med Chem. 2012;20(23):6811–6820. doi: 10.1016/j.bmc.2012.09.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Li BX, et al. Novel type of prodrug activation through a long-range O, N-acyl transfer: a case of water-soluble CREB inhibitor. ACS Med Chem Lett. 2014;5(10):1104–1109. doi: 10.1021/ml500330n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Xie F, Li BX, Xiao X. Synthesis and evaluation of an O-aminated naphthol AS-E as a prodrug of CREB-mediated gene transcription inhibition. Lett Org Chem. 2013;10(5):380–384. doi: 10.2174/1570178611310050014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lee JW, et al. A novel small-molecule inhibitor targeting CREB-CBP complex possesses anti-cancer effects along with cell cycle regulation, autophagy suppression and endoplasmic reticulum stress. PLoS One. 2015;10(4):e0122628. doi: 10.1371/journal.pone.0122628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Li BX, et al. Systemic Inhibition of CREB is Well-tolerated in vivo. Sci Rep. 2016;6:34513. doi: 10.1038/srep34513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Xie F, Li BX, Xiao X. Design, synthesis and biological evaluation of regioisomers of 666-15 as inhibitors of CREB-mediated gene transcription. Bioorg Med Chem Lett. 2017;27(4):994–998. doi: 10.1016/j.bmcl.2016.12.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Xie F, et al. Discovery of a synergistic inhibitor of cAMP-response element binding protein (CREB)-mediated gene transcription with 666-15. J Med Chem. 2019;62(24):11423–11429. doi: 10.1021/acs.jmedchem.9b01207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Mitton B, et al. Small molecule inhibition of cAMP response element binding protein in human acute myeloid leukemia cells. Leukemia. 2016;30(12):2302–2311. doi: 10.1038/leu.2016.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Chae HD, et al. Niclosamide suppresses acute myeloid leukemia cell proliferation through inhibition of CREB-dependent signaling pathways. Oncotarget. 2018;9(4):4301–4317. doi: 10.18632/oncotarget.23794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Schuksz M, et al. Surfen, a small molecule antagonist of heparan sulfate. Proc Natl Acad Sci USA. 2008;105(35):13075–13080. doi: 10.1073/pnas.0805862105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Steven A, et al. Linking CREB function with altered metabolism in murine fibroblast-based model cell lines. Oncotarget. 2017;8(57):97439–97463. doi: 10.18632/oncotarget.22135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Rishi V, et al. A high-throughput fluorescence-anisotropy screen that identifies small molecule inhibitors of the DNA binding of B-ZIP transcription factors. Anal Biochem. 2005;340(2):259–271. doi: 10.1016/j.ab.2005.02.012. [DOI] [PubMed] [Google Scholar]
- 113.Rishi V, et al. 12 Arylstibonic acids that inhibit the DNA binding of five B-ZIP dimers. J Struct Biol. 2010;170(2):216–225. doi: 10.1016/j.jsb.2010.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Yang QE, et al. Discovery of small-molecule human immunodeficiency virus type 1 entry inhibitors that target the gp120-binding domain of CD4. J Virol. 2005;79(10):6122–6133. doi: 10.1128/JVI.79.10.6122-6133.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Krosky PM, et al. NSC13778 disrupts interactions between transcription factors TFE3, ASPL-TFE3 type 1, and ASPL-TFE3 type 2 and cognate DNA. Proc Amer Assoc Cancer Res. 2006;47:4622. [Google Scholar]
- 116.Zhao J, et al. P6981, an arylstibonic acid, is a novel low nanomolar inhibitor of cAMP response element-binding protein binding to DNA. Mol Pharmacol. 2012;82(5):814–823. doi: 10.1124/mol.112.080820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Tsalkova T, Mei FC, Cheng X. A fluorescence-based high-throughput assay for the discovery of exchange protein directly activated by cyclic AMP (EPAC) antagonists. PLoS One. 2012;7(1):e30441. doi: 10.1371/journal.pone.0030441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Mizuta K, et al. The dopamine D1 receptor is expressed and facilitates relaxation in airway smooth muscle. Respir Res. 2013;14:89. doi: 10.1186/1465-9921-14-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Vlahopoulos SA, et al. The role of ATF-2 in oncogenesis. BioEssays. 2008;30(4):314–327. doi: 10.1002/bies.20734. [DOI] [PubMed] [Google Scholar]
- 120.Li G, Jiang Q, Xu K. CREB family: a significant role in liver fibrosis. Biochimie. 2019;163:94–100. doi: 10.1016/j.biochi.2019.05.014. [DOI] [PubMed] [Google Scholar]
- 121.Smith B, et al. Evolution of motif variants and positional bias of the cyclic-AMP response element. BMC Evol Biol. 2007;7(Suppl 1):S15. doi: 10.1186/1471-2148-7-S1-S15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Craig JC, et al. Consensus and variant cAMP-regulated enhancers have distinct CREB-binding properties. J Biol Chem. 2001;276(15):11719–11728. doi: 10.1074/jbc.M010263200. [DOI] [PubMed] [Google Scholar]
- 123.Wu X, et al. cAMP response element-binding protein monomers cooperatively assemble to form dimers on DNA. J Biol Chem. 1998;273(33):20820–20827. doi: 10.1074/jbc.273.33.20820. [DOI] [PubMed] [Google Scholar]
- 124.Iourgenko V, et al. Identification of a family of cAMP response element-binding protein coactivators by genome-scale functional analysis in mammalian cells. Proc Natl Acad Sci USA. 2003;100(21):12147–12152. doi: 10.1073/pnas.1932773100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Amelio AL, Caputi M, Conkright MD. Bipartite functions of the CREB co-activators selectively direct alternative splicing or transcriptional activation. EMBO J. 2009;28(18):2733–2747. doi: 10.1038/emboj.2009.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Gu Y, et al. Altered LKB1/CREB-regulated transcription co-activator (CRTC) signaling axis promotes esophageal cancer cell migration and invasion. Oncogene. 2012;31(4):469–479. doi: 10.1038/onc.2011.247. [DOI] [PubMed] [Google Scholar]
- 127.Ma R, et al. LINC01585 functions as a regulator of gene expression by the CAMP/CREB signaling pathway in breast cancer. Gene. 2019;684:139–148. doi: 10.1016/j.gene.2018.10.063. [DOI] [PubMed] [Google Scholar]
- 128.Tasoulas J, et al. Adaptive transcriptional responses by CRTC coactivators in cancer. Trends Cancer. 2019;5(2):111–127. doi: 10.1016/j.trecan.2018.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Linnerth NM, et al. IGF-II induces CREB phosphorylation and cell survival in human lung cancer cells. Oncogene. 2005;24(49):7310–7319. doi: 10.1038/sj.onc.1208882. [DOI] [PubMed] [Google Scholar]
- 130.Alper O, et al. Apoptosis, growth arrest and suppression of invasiveness by CRE-decoy oligonucleotide in ovarian cancer cells: protein kinase A downregulation and cytoplasmic export of CRE-binding proteins. Mol Cell Biochem. 2001;218(1–2):55–63. doi: 10.1023/a:1007205205131. [DOI] [PubMed] [Google Scholar]
- 131.Steven A, et al. Colorectal carcinogenesis: connecting K-RAS-induced transformation and CREB activity in vitro and in vivo. Mol Cancer Res. 2015;13(8):1248–1262. doi: 10.1158/1541-7786.MCR-14-0590. [DOI] [PubMed] [Google Scholar]
- 132.Chen L, et al. MicroRNA-181b targets cAMP responsive element binding protein 1 in gastric adenocarcinomas. IUBMB Life. 2012;64(7):628–635. doi: 10.1002/iub.1030. [DOI] [PubMed] [Google Scholar]
- 133.Zhang JQ, et al. Prognostic value of coexistence of abnormal expression of micro-RNA-200b and cyclic adenosine monophosphate-responsive element-binding protein 1 in human astrocytoma. Hum Pathol. 2014;45(10):2154–2161. doi: 10.1016/j.humpath.2014.01.025. [DOI] [PubMed] [Google Scholar]
- 134.Liu Y, et al. microRNA-181a is involved in insulin-like growth factor-1-mediated regulation of the transcription factor CREB1. J Neurochem. 2013;126(6):771–780. doi: 10.1111/jnc.12370. [DOI] [PubMed] [Google Scholar]
- 135.Sun S, et al. MiR-433-3p suppresses cell growth and enhances chemosensitivity by targeting CREB in human glioma. Oncotarget. 2017;8(3):5057–5068. doi: 10.18632/oncotarget.13789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Wang J, et al. CREB up-regulates long non-coding RNA, HULC expression through interaction with microRNA-372 in liver cancer. Nucleic Acids Res. 2010;38(16):5366–5383. doi: 10.1093/nar/gkq285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Wang M, et al. MiR-1271 inhibits cell growth in prostate cancer by targeting ERG. Pathol Oncol Res. 2018;24(2):385–391. doi: 10.1007/s12253-017-0254-y. [DOI] [PubMed] [Google Scholar]
- 138.Xian D, Zhao Y. LncRNA KCNQ1OT1 enhanced the methotrexate resistance of colorectal cancer cells by regulating miR-760/PPP1R1B via the cAMP signalling pathway. J Cell Mol Med. 2019;23(6):3808–3823. doi: 10.1111/jcmm.14071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Tan X, et al. cAMP response element-binding protein promotes gliomagenesis by modulating the expression of oncogenic microRNA-23a. Proc Natl Acad Sci USA. 2012;109(39):15805–15810. doi: 10.1073/pnas.1207787109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Paz JC, et al. Combinatorial regulation of a signal-dependent activator by phosphorylation and acetylation. Proc Natl Acad Sci USA. 2014;111(48):17116–17121. doi: 10.1073/pnas.1420389111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Qiang L, et al. Proatherogenic abnormalities of lipid metabolism in SirT1 transgenic mice are mediated through Creb deacetylation. Cell Metab. 2011;14(6):758–767. doi: 10.1016/j.cmet.2011.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Lu Q, et al. Acetylation of cAMP-responsive element-binding protein (CREB) by CREB-binding protein enhances CREB-dependent transcription. J Biol Chem. 2003;278(18):15727–15734. doi: 10.1074/jbc.M300546200. [DOI] [PubMed] [Google Scholar]
- 143.Chen J, et al. Role of HDAC9-FoxO1 axis in the transcriptional program associated with hepatic gluconeogenesis. Sci Rep. 2017;7(1):6102. doi: 10.1038/s41598-017-06328-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Fusco S, et al. A CREB-Sirt1-Hes1 circuitry mediates neural stem cell response to glucose availability. Cell Rep. 2016;14(5):1195–1205. doi: 10.1016/j.celrep.2015.12.092. [DOI] [PubMed] [Google Scholar]
- 145.Mambetsariev N, et al. Nuclear TRAF3 is a negative regulator of CREB in B cells. Proc Natl Acad Sci USA. 2016;113(4):1032–1037. doi: 10.1073/pnas.1514586113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Wang Y, et al. MTUS1 silencing promotes E-selectin production through p38 MAPK-dependent CREB ubiquitination in endothelial cells. J Mol Cell Cardiol. 2016;101:1–10. doi: 10.1016/j.yjmcc.2016.10.012. [DOI] [PubMed] [Google Scholar]
- 147.Steven A, et al. Hypoxia-mediated alterations and their role in the HER-2/neuregulated CREB status and localization. Oncotarget. 2016;7(32):52061–52084. doi: 10.18632/oncotarget.10474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Ozgen N, et al. Determinants of CREB degradation and KChIP2 gene transcription in cardiac memory. Heart Rhythm. 2010;7(7):964–970. doi: 10.1016/j.hrthm.2010.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Garat CV, et al. Platelet-derived growth factor BB induces nuclear export and proteasomal degradation of CREB via phosphatidylinositol 3-kinase/Akt signaling in pulmonary artery smooth muscle cells. Mol Cell Biol. 2006;26(13):4934–4948. doi: 10.1128/MCB.02477-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Taylor CT, et al. Phosphorylation-dependent targeting of cAMP response element binding protein to the ubiquitin/proteasome pathway in hypoxia. Proc Natl Acad Sci USA. 2000;97(22):12091–12096. doi: 10.1073/pnas.220211797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Comerford KM, et al. Small ubiquitin-related modifier-1 modification mediates resolution of CREB-dependent responses to hypoxia. Proc Natl Acad Sci USA. 2003;100(3):986–991. doi: 10.1073/pnas.0337412100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Chen YC, et al. CREB SUMOylation by the E3 ligase PIAS1 enhances spatial memory. J Neurosci. 2014;34(29):9574–9589. doi: 10.1523/JNEUROSCI.4302-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Rexach JE, et al. Dynamic O-GlcNAc modification regulates CREB-mediated gene expression and memory formation. Nat Chem Biol. 2012;8(3):253–261. doi: 10.1038/nchembio.770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Azuma Y, et al. Protein O-N-acetylglucosaminylation modulates promoter activities of cyclic AMP response element and activator protein 1 and enhances E-selectin expression on HuH-7 human hepatoma cells. Biol Pharm Bull. 2007;30(12):2284–2289. doi: 10.1248/bpb.30.2284. [DOI] [PubMed] [Google Scholar]
- 155.Lamarre-Vincent N, Hsieh-Wilson LC. Dynamic glycosylation of the transcription factor CREB: a potential role in gene regulation. J Am Chem Soc. 2003;125(22):6612–6613. doi: 10.1021/ja028200t. [DOI] [PubMed] [Google Scholar]
- 156.Gao Y, et al. Iron down-regulates leptin by suppressing protein O-GlcNAc modification in adipocytes, resulting in decreased levels of O-glycosylated CREB. J Biol Chem. 2019;294(14):5487–5495. doi: 10.1074/jbc.RA118.005183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Trinh AT, et al. Cyclin-dependent kinase 1-dependent phosphorylation of cAMP response element-binding protein decreases chromatin occupancy. J Biol Chem. 2013;288(33):23765–23775. doi: 10.1074/jbc.M113.464057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Sakamoto K, et al. Regulation of genotoxic stress response by homeodomain-interacting protein kinase 2 through phosphorylation of cyclic AMP response element-binding protein at serine 271. Mol Biol Cell. 2010;21(16):2966–2974. doi: 10.1091/mbc.E10-01-0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Shabestari RM, et al. CREB knockdown inhibits growth and induces apoptosis in human pre-B acute lymphoblastic leukemia cells through inhibition of prosurvival signals. Biomed Pharmacother. 2017;87:274–279. doi: 10.1016/j.biopha.2016.12.070. [DOI] [PubMed] [Google Scholar]
- 160.Chen S, et al. Polydatin down-regulates the phosphorylation level of Creb and induces apoptosis in human breast cancer cell. PLoS One. 2017;12(5):e0176501. doi: 10.1371/journal.pone.0176501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Fujii K, et al. Carnosine activates the CREB pathway in Caco-2 cells. Cytotechnology. 2017;69(3):523–527. doi: 10.1007/s10616-017-0089-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Kesh K, et al. Association of MMP7-181A–>G Promoter Polymorphism with Gastric Cancer Risk: influence of nicotine in differential allele-specific transcription via increased phosphorylation of cAMP-response element-binding protein (CREB) J Biol Chem. 2015;290(23):14391–14406. doi: 10.1074/jbc.M114.630129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Shen J, Li M, Min L. HSPB8 promotes cancer cell growth by activating the ERKCREB pathway and is indicative of a poor prognosis in gastric cancer patients. Oncol Rep. 2018;39(6):2978–2986. doi: 10.3892/or.2018.6376. [DOI] [PubMed] [Google Scholar]
- 164.Barresi V, et al. p-CREB expression in human gliomas: potential use in the differential diagnosis between astrocytoma and oligodendroglioma. Hum Pathol. 2015;46(2):231–238. doi: 10.1016/j.humpath.2014.10.011. [DOI] [PubMed] [Google Scholar]
- 165.Kovach SJ, et al. Role of cyclic-AMP responsive element binding (CREB) proteins in cell proliferation in a rat model of hepatocellular carcinoma. J Cell Physiol. 2006;206(2):411–419. doi: 10.1002/jcp.20474. [DOI] [PubMed] [Google Scholar]
- 166.Shneor D, et al. Stable knockdown of CREB, HIF-1 and HIF-2 by replication-competent retroviruses abrogates the responses to hypoxia in hepatocellular carcinoma. Cancer Gene Ther. 2017;24(2):64–74. doi: 10.1038/cgt.2016.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Yu L, et al. Cyclic adenosine monophosphate-responsive element-binding protein activation predicts an unfavorable prognosis in patients with hepatocellular carcinoma. Onco Targets Ther. 2014;7:873–879. doi: 10.2147/OTT.S63594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Lu F, et al. Downregulation of CREB promotes cell proliferation by mediating G1/S phase transition in hodgkin lymphoma. Oncol Res. 2016;24(3):171–179. doi: 10.3727/096504016X14634208142987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Zhuang H, et al. Cyclic AMP responsive element-binding protein promotes renal cell carcinoma proliferation probably via the expression of spindle and kinetochore-associated protein 2. Oncotarget. 2016;7(13):16325–16337. doi: 10.18632/oncotarget.7017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Cho EC, Mitton B, Sakamoto KM. CREB and leukemogenesis. Crit Rev Oncog. 2011;16(1–2):37–46. doi: 10.1615/critrevoncog.v16.i1-2.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Crans-Vargas HN, et al. Expression of cyclic adenosine monophosphate response-element binding protein in acute leukemia. Blood. 2002;99(7):2617–2619. doi: 10.1182/blood.v99.7.2617. [DOI] [PubMed] [Google Scholar]
- 172.Pigazzi M, et al. MicroRNA-34b promoter hypermethylation induces CREB overexpression and contributes to myeloid transformation. Haematologica. 2013;98(4):602–610. doi: 10.3324/haematol.2012.070664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Illiano M, et al. Adiponectin down-regulates CREB and inhibits proliferation of A549 lung cancer cells. Pulm Pharmacol Ther. 2017;45:114–120. doi: 10.1016/j.pupt.2017.05.009. [DOI] [PubMed] [Google Scholar]
- 174.Barresi V, et al. p-CREB expression in human meningiomas: correlation with angiogenesis and recurrence risk. J Neurooncol. 2015;122(1):87–95. doi: 10.1007/s11060-014-1706-9. [DOI] [PubMed] [Google Scholar]
- 175.Liu W, et al. Protein signature for non-small cell lung cancer prognosis. Am J Cancer Res. 2014;4(3):256–269. [PMC free article] [PubMed] [Google Scholar]
- 176.de Alexandre RB, et al. Phosphodiesterase sequence variants may predispose to prostate cancer. Endocr Relat Cancer. 2015;22(4):519–530. doi: 10.1530/ERC-15-0134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Linnerth NM, et al. cAMP response element-binding protein is expressed at high levels in human ovarian adenocarcinoma and regulates ovarian tumor cell proliferation. Int J Gynecol Cancer. 2008;18(6):1248–1257. doi: 10.1111/j.1525-1438.2007.01177.x. [DOI] [PubMed] [Google Scholar]
- 178.Braeuer RR, et al. Transcriptional control of melanoma metastasis: the importance of the tumor microenvironment. Semin Cancer Biol. 2011;21(2):83–88. doi: 10.1016/j.semcancer.2010.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Jean D, Bar-Eli M. Regulation of tumor growth and metastasis of human melanoma by the CREB transcription factor family. Mol Cell Biochem. 2000;212(1–2):19–28. [PubMed] [Google Scholar]
- 180.Melnikova VO, et al. CREB inhibits AP-2alpha expression to regulate the malignant phenotype of melanoma. PLoS One. 2010;5(8):e12452. doi: 10.1371/journal.pone.0012452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Guan H, et al. Transmembrane protease serine 4 promotes thyroid cancer proliferation via CREB phosphorylation. Thyroid. 2015;25(1):85–94. doi: 10.1089/thy.2014.0155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Terazawa S, et al. Withaferin A abolishes the stem cell factor-stimulated pigmentation of human epidermal equivalents by interrupting the auto-phosphorylation of c-KIT in human melanocytes. Arch Dermatol Res. 2015;307(1):73–88. doi: 10.1007/s00403-014-1518-y. [DOI] [PubMed] [Google Scholar]
- 183.Xie F, et al. Identification of a potent inhibitor of CREB-mediated gene transcription with efficacious in vivo anticancer activity. J Med Chem. 2015;58(12):5075–5087. doi: 10.1021/acs.jmedchem.5b00468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Somlo G, et al. A phase I/II prospective, single arm trial of gefitinib, trastuzumab, and docetaxel in patients with stage IV HER-2 positive metastatic breast cancer. Breast Cancer Res Treat. 2012;131(3):899–906. doi: 10.1007/s10549-011-1850-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Buzzi N, et al. MAP kinases in proliferating human colon cancer Caco-2 cells. Mol Cell Biochem. 2009;328(1–2):201–208. doi: 10.1007/s11010-009-0090-9. [DOI] [PubMed] [Google Scholar]
- 186.Girgert R, Emons G, Grundker C. Inhibition of GPR30 by estriol prevents growth stimulation of triple-negative breast cancer cells by 17beta-estradiol. BMC Cancer. 2014;14:935. doi: 10.1186/1471-2407-14-935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Moeenrezakhanlou A, et al. 1alpha,25-dihydroxycholecalciferol activates binding of CREB to a CRE site in the CD14 promoter and drives promoter activity in a phosphatidylinositol-3 kinase-dependent manner. J Leukoc Biol. 2007;81(5):1311–1321. doi: 10.1189/jlb.1106681. [DOI] [PubMed] [Google Scholar]
- 188.Evans AN, et al. Regulation of hypothalamic corticotropin-releasing hormone transcription by elevated glucocorticoids. Mol Endocrinol. 2013;27(11):1796–1807. doi: 10.1210/me.2013-1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Kang Y, et al. Adrenergic stimulation of DUSP1 impairs chemotherapy response in ovarian cancer. Clin Cancer Res. 2016;22(7):1713–1724. doi: 10.1158/1078-0432.CCR-15-1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Kikuchi D, Tanimoto K, Nakayama K. CREB is activated by ER stress and modulates the unfolded protein response by regulating the expression of IRE1alpha and PERK. Biochem Biophys Res Commun. 2016;469(2):243–250. doi: 10.1016/j.bbrc.2015.11.113. [DOI] [PubMed] [Google Scholar]
- 191.Barlow CA, et al. Protein kinase A-mediated CREB phosphorylation is an oxidant-induced survival pathway in alveolar type II cells. Apoptosis. 2008;13(5):681–692. doi: 10.1007/s10495-008-0203-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Shi Y, et al. Direct regulation of CREB transcriptional activity by ATM in response to genotoxic stress. Proc Natl Acad Sci USA. 2004;101(16):5898–5903. doi: 10.1073/pnas.0307718101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Meyuhas R, et al. A Key role for cyclic AMP-responsive element binding protein in hypoxia-mediated activation of the angiogenesis factor CCN1 (CYR61) in Tumor cells. Mol Cancer Res. 2008;6(9):1397–1409. doi: 10.1158/1541-7786.MCR-07-2086. [DOI] [PubMed] [Google Scholar]
- 194.Horibe I, et al. Induction of melanogenesis by 4′-O-methylated flavonoids in B16F10 melanoma cells. J Nat Med. 2013;67(4):705–710. doi: 10.1007/s11418-012-0727-y. [DOI] [PubMed] [Google Scholar]
- 195.Zhou WJ, et al. Angelica sinensis polysaccharides promotes apoptosis in human breast cancer cells via CREB-regulated caspase-3 activation. Biochem Biophys Res Commun. 2015;467(3):562–569. doi: 10.1016/j.bbrc.2015.09.145. [DOI] [PubMed] [Google Scholar]
- 196.Zhang C, et al. Sulforaphene induces apoptosis and inhibits the invasion of esophageal cancer cells through MSK2/CREB/Bcl-2 and cadherin pathway in vivo and in vitro. Cancer Cell Int. 2019;19:342. doi: 10.1186/s12935-019-1061-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Fernandez-Araujo A, et al. Key role of phosphodiesterase 4A (PDE4A) in autophagy triggered by yessotoxin. Toxicology. 2015;329:60–72. doi: 10.1016/j.tox.2015.01.004. [DOI] [PubMed] [Google Scholar]
- 198.Saha B, et al. Activation of the Mitf promoter by lipid-stimulated activation of p38-stress signalling to CREB. Pigment Cell Res. 2006;19(6):595–605. doi: 10.1111/j.1600-0749.2006.00348.x. [DOI] [PubMed] [Google Scholar]
- 199.Shanware NP, et al. Conserved and distinct modes of CREB/ATF transcription factor regulation by PP2A/B56gamma and genotoxic stress. PLoS One. 2010;5(8):e12173. doi: 10.1371/journal.pone.0012173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Mizutani N, et al. Increased SPHK2 transcription of human colon cancer cells in serum-depleted culture: the involvement of CREB transcription factor. J Cell Biochem. 2015;116(10):2227–2238. doi: 10.1002/jcb.25173. [DOI] [PubMed] [Google Scholar]
- 201.Okoshi R, et al. Transcriptional regulation of tumor suppressor p53 by cAMP-responsive element-binding protein/AMP-activated protein kinase complex in response to glucose deprivation. Genes Cells. 2009;14(12):1429–1440. doi: 10.1111/j.1365-2443.2009.01359.x. [DOI] [PubMed] [Google Scholar]
- 202.Fiol CJ, et al. A secondary phosphorylation of CREB341 at Ser129 is required for the cAMP-mediated control of gene expression. A role for glycogen synthase kinase-3 in the control of gene expression. J Biol Chem. 1994;269(51):32187–32193. [PubMed] [Google Scholar]
- 203.Shanware NP, et al. Coregulated ataxia telangiectasia-mutated and casein kinase sites modulate cAMP-response element-binding protein-coactivator interactions in response to DNA damage. J Biol Chem. 2007;282(9):6283–6291. doi: 10.1074/jbc.M610674200. [DOI] [PubMed] [Google Scholar]
- 204.Bachelor MA, et al. Inhibition of p38 mitogen-activated protein kinase and phosphatidylinositol 3-kinase decreases UVB-induced activator protein-1 and cyclooxygenase-2 in a SKH-1 hairless mouse model. Mol Cancer Res. 2005;3(2):90–99. doi: 10.1158/1541-7786.MCR-04-0065. [DOI] [PubMed] [Google Scholar]
- 205.Kutcher LW, et al. Nuclear CaMKII inhibits neuronal differentiation of PC12 cells without affecting MAPK or CREB activation. Am J Physiol Cell Physiol. 2003;284(6):C1334–C1345. doi: 10.1152/ajpcell.00510.2002. [DOI] [PubMed] [Google Scholar]
- 206.Vaziri-Gohar A, Houston KD. GPER1-mediated IGFBP-1 induction modulates IGF-1-dependent signaling in tamoxifen-treated breast cancer cells. Mol Cell Endocrinol. 2016;422:160–171. doi: 10.1016/j.mce.2015.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Sayan M, et al. Extracellular signal-regulated kinase 5 and cyclic AMP response element binding protein are novel pathways inhibited by vandetanib (ZD6474) and doxorubicin in mesotheliomas. Am J Respir Cell Mol Biol. 2014;51(5):595–603. doi: 10.1165/rcmb.2013-0373TR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Langner E, et al. Quinaldic acid inhibits proliferation of colon cancer ht-29 cells in vitro: effects on signaling pathways. Eur J Pharmacol. 2015;757:21–27. doi: 10.1016/j.ejphar.2015.02.053. [DOI] [PubMed] [Google Scholar]
- 209.Jiang M, et al. Retinoic acid induces caspase-8 transcription via phospho-CREB and increases apoptotic responses to death stimuli in neuroblastoma cells. Biochim Biophys Acta. 2008;1783(6):1055–1067. doi: 10.1016/j.bbamcr.2008.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Robertson AG, et al. Comprehensive molecular characterization of muscle-invasive bladder cancer. Cell. 2017;171(3):540–556. doi: 10.1016/j.cell.2017.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Cancer Genome Atlas Research N et al (2017) Integrated genomic characterization of oesophageal carcinoma. Nature 541(7636):169–175 [DOI] [PMC free article] [PubMed]
- 212.Cancer Genome Atlas Research N (2014) Comprehensive molecular profiling of lung adenocarcinoma. Nature 511(7511):543–550 [DOI] [PMC free article] [PubMed]
- 213.Abida W, et al. Genomic correlates of clinical outcome in advanced prostate cancer. Proc Natl Acad Sci USA. 2019;116(23):11428–11436. doi: 10.1073/pnas.1902651116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.de Sam Lazaro S, et al. p16 staining of cervical biopsies may decrease the frequency of unnecessary loop electrosurgical excision procedures. J Low Genit Tract Dis. 2016;20(3):201–206. doi: 10.1097/LGT.0000000000000189. [DOI] [PubMed] [Google Scholar]
- 215.Campbell JD, et al. Distinct patterns of somatic genome alterations in lung adenocarcinomas and squamous cell carcinomas. Nat Genet. 2016;48(6):607–616. doi: 10.1038/ng.3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Armenia J, et al. The long tail of oncogenic drivers in prostate cancer. Nat Genet. 2018;50(5):645–651. doi: 10.1038/s41588-018-0078-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Kubota K, et al. The traditional japanese herbal medicine hachimijiogan elicits neurite outgrowth effects in PC12 cells and improves cognitive in AD model rats via phosphorylation of CREB. Front Pharmacol. 2017;8:850. doi: 10.3389/fphar.2017.00850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Muller-Langle A, et al. NMDA receptor-mediated signaling pathways enhance radiation resistance, survival and migration in glioblastoma cells-a potential target for adjuvant radiotherapy. Cancers (Basel) 2019;11(4):503. doi: 10.3390/cancers11040503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Xie Y, et al. Functional cyclic AMP response element in the breast cancer resistance protein (BCRP/ABCG2) promoter modulates epidermal growth factor receptor pathway- or androgen withdrawal-mediated BCRP/ABCG2 transcription in human cancer cells. Biochim Biophys Acta. 2015;1849(3):317–327. doi: 10.1016/j.bbagrm.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Rodriguez-Ubreva J, et al. Prostaglandin E2 leads to the acquisition of DNMT3A-dependent tolerogenic functions in human myeloid-derived suppressor cells. Cell Rep. 2017;21(1):154–167. doi: 10.1016/j.celrep.2017.09.018. [DOI] [PubMed] [Google Scholar]
- 221.Srinivasan S, et al. Tobacco carcinogen-induced production of GM-CSF activates CREB to promote pancreatic cancer. Cancer Res. 2018;78(21):6146–6158. doi: 10.1158/0008-5472.CAN-18-0579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Li BX, Xiao X. Discovery of a small-molecule inhibitor of the KIX-KID interaction. ChemBioChem. 2009;10(17):2721–2724. doi: 10.1002/cbic.200900552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Hsu SH, Chen YJ. CREB-regulated miR-27b is linked to hepatic insulin resistance by targeting insulin/Akt signaling. FASEB J. 2019 doi: 10.1096/fasebj.2019.33.1_supplement.lb2. [DOI] [Google Scholar]
- 224.Resende C, Regalo G, Durães C, Pinto MT, Wen X, Figueiredo C, Carneiro F, Machado JC. Interleukin-1B signalling leads to increased survival of gastric carcinoma cells through a CREB-C/EBPβ-associated mechanism. Gastric Cancer. 2016;19(1):74–84. doi: 10.1007/s10120-014-0448-x. [DOI] [PubMed] [Google Scholar]
- 225.Glick Y, Orenstein Y, Chen D, Avrahami D, Zor T, Shamir R, Gerber D. Integrated microfluidic approach for quantitative high-throughput measurements of transcription factor binding affinities. Nucleic Acids Res. 2016;44(6):e51. doi: 10.1093/nar/gkv1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Aucoin R, Reiland J, Roy M, Marchetti D. Dominant-negative CREB inhibits heparanase functionality and melanoma cell invasion. J Cell Biochem. 2004;93(2):215–223. doi: 10.1002/jcb.20231. [DOI] [PubMed] [Google Scholar]
- 227.Chen S, Yin W, Bi K, Lu B. MicroRNA-497 attenuates cerebral infarction in patients via the TLR4 and CREB signaling pathways. Int J Mol Med. 2018;42(1):547–556. doi: 10.3892/ijmm.2018.3611. [DOI] [PubMed] [Google Scholar]
- 228.Fernandes ND, Sun Y, Price BD. Activation of the kinase activity of ATM by retinoic acid is required for CREB-dependent differentiation of neuroblastoma cells. J Biol Chem. 2007;282(22):16577–16584. doi: 10.1074/jbc.M609628200. [DOI] [PubMed] [Google Scholar]
- 229.Zomerman WW, et al. Essential role for cyclic-AMP responsive element binding protein 1 (CREB1) phosphorylation in the survival of medulloblastoma patients [abstract] Cancer Res. 2017;77((13 Suppl)):Abstract nr 5826. [Google Scholar]
- 230.Chae H-D, Cox N, Capolicchio S, Lee JW, Horikoshi N, Kam S, Ng AA, Edwards J, Butler T-L, Chan J, Lee Y, Potter G, Capece MC, Liu CW, Wakatsuki S, Smith M, Sakamoto KM. SAR optimization studies on modified salicylamides as a potential treatment for acute myeloid leukemia through inhibition of the CREB pathway. Bioorg Med Chem Lett. 2019;29(16):2307–2315. doi: 10.1016/j.bmcl.2019.06.023. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available upon request from the corresponding author.