Significance
Corticosteroids that are currently in clinical use bind to the glucocorticoid receptor (GR) to exert antiinflammation effects, yet are associated with undesirable side effects. Vamorolone is a recently developed drug for Duchenne muscular dystrophy; it decreases muscle inflammation and reduces side effects observed in other corticosteroid-based treatments. Our structure studies reveal a crucial GR-ligand hydrogen bond is not possible in vamorolone. An unprecedented allosteric intramolecular network derived from this hydrogen bond modulates GR dynamic motions and coregulator binding. Our work provides insights into how subtle modifications on a drug exploits structural and dynamic properties in the receptor to dissociate downstream side effects from therapeutic benefits.
Keywords: glucocorticoid receptor, dissociative agonist, allostery, molecular dynamics, nuclear receptor
Abstract
Duchenne muscular dystrophy is a genetic disorder that shows chronic and progressive damage to skeletal and cardiac muscle leading to premature death. Antiinflammatory corticosteroids targeting the glucocorticoid receptor (GR) are the current standard of care but drive adverse side effects such as deleterious bone loss. Through subtle modification to a steroidal backbone, a recently developed drug, vamorolone, appears to preserve beneficial efficacy but with significantly reduced side effects. We use combined structural, biophysical, and biochemical approaches to show that loss of a receptor-ligand hydrogen bond drives these remarkable therapeutic effects. Moreover, vamorolone uniformly weakens coactivator associations but not corepressor associations, implicating partial agonism as the main driver of its dissociative properties. Additionally, we identify a critical and evolutionarily conserved intramolecular network connecting the ligand to the coregulator binding surface. Interruption of this allosteric network by vamorolone selectively reduces GR-driven transactivation while leaving transrepression intact. Our results establish a mechanistic understanding of how vamorolone reduces side effects, guiding the future design of partial agonists as selective GR modulators with an improved therapeutic index.
Duchenne muscular dystrophy (DMD) is the most common and severe form of muscular dystrophy with an incidence rate of 1 in 5,000 boys (1). Symptoms begin early in childhood and manifest as weakness and degeneration of muscle tissues. The loss of muscle strength becomes most obvious in the pelvic area and gradually progresses to the upper limbs. Chronic muscle degeneration ultimately leads to cardiac and respiratory muscle weakness and wasting, with an average life expectancy below 30 y. DMD is an X-linked disorder that is characterized by mutations to the DMD gene, the largest gene in the human genome, and loss of the encoded dystrophin protein (2). The dystrophin protein provides structural support to muscle fiber plasma membranes and alleviates mechanical stress during muscle contraction by forming a complex to connect the intercellular cytoskeleton to extracellular matrix (3).
Activation of the proinflammatory nuclear factor-κB (NF-κB) pathway is observed in muscles of DMD patients from birth and is believed to cause chronic inflammation in muscle that contributes to disease onset and progression (4, 5). The current standard care for DMD is pharmacologic treatment with corticosteroids such as prednisone and deflazacort, which potently suppress NF-κB signaling. As a result of treatment, DMD patients’ muscle tissues exhibit reduced fibrosis and stabilized muscle strength while mouse DMD models showed improved muscle regeneration (6). However, like other corticosteroid treatments, DMD patients experience adverse side effects with long-term treatment. In particular, corticosteroid treatment frequently results in a decreased bone mineral density, resulting in an increased rate of osteoporosis and risk of vertebral and long bone fractures (7). Another noteworthy side effect of corticosteroid treatment is muscle weakness and atrophy caused by increased protein catabolism through atrogene pathways (8, 9). While different dosing regimens for prednisone and deflazacort have been tested to tip the balance toward greater efficacy with fewer side effects (10, 11), drugs able to separate (dissociate) efficacy from safety concerns are highly needed. Vamorolone (previously named as VBP15) was recently discovered to show properties of a dissociative steroidal drug, that decreased muscle inflammation and improved muscle strength in mouse models of DMD (12, 13). Moreover, open label initial clinical trials with vamorolone showed dose-responsive efficacy, while improving side effects associated with traditional corticosteroid treatments (14, 15).
The molecular target of corticosteroids is the glucocorticoid receptor (GR; NR3C1 gene), a member of the nuclear receptor superfamily (16). GR is a ligand-regulated transcription factor that plays key roles in inflammation, metabolism, and immunity (17). It has an N-terminal domain with an activation function-1 (AF-1) region that can interact with different coregulators for full transcriptional activity. GR also has a DNA binding domain that binds to specific glucocorticoid response elements (GREs) to transactivate or transrepress distinct target genes (18). Transrepression of proinflammatory genes occurs via distinct DNA sequences from transactivation (19). Current models suggest that transactivation drives many safety concerns of GR-mediated drug activity, whereas transrepression drives antiinflammatory efficacy. Serious safety concerns of corticosteroid drugs include transactivation of metabolic, catabolic, and apoptotic genes that lead to many side effects detracting from patient quality of life (20). The GR C terminus harbors a ligand binding domain (LBD) that recognizes both its endogenous ligand, cortisol, named hydrocortisone when used in medication, and synthetic ligands such as dexamethasone. The ligand binding pocket communicates allosterically with the activation function 2 (AF-2) region that is responsible for binding to coregulatory proteins. Coactivators with a conserved LXXLL motif or corepressors that contain conserved LXXX(I/L)XXX(I/L) motif (L, leucine; I, isoleucine; X, any amino acid) can be recruited to the AF-2 region which in turn controls chromatin remodeling and gene transcription. Helix 12 (also referred to as the activation function helix, AFH) in the AF-2 region is highly dynamic and adopts varying orientations through conformational change to facilitate the recognition of different coregulators (21, 22).
Vamorolone has shown high-affinity binding to the GR and related mineralocorticoid receptor (MR; NR3C2 gene) in cell-based assays, but lacks downstream gene transactivation activity (12, 13, 23). The molecular mechanism of these dissociative properties, and the observation that vamorolone-based DMD treatment appears to retain an efficacy while reducing side effects, is not understood at the molecular level. Here we leverage an integrated biochemical, structural, cellular, and computational approach to elucidate the vamorolone mechanism of action compared to active metabolites of two known corticosteroids used to treat DMD (prednisone and deflazacort). The reconstructed ancestral GR LBD (AncGR2 LBD), sharing 79% sequence identity and 96% similarity with the hGR LBD (SI Appendix, Fig. S1), has been successfully used in previous studies as it significantly improves protein expression and crystallization and reliably represents GR ligand binding, transcriptional responses, and allosteric regulation (24–27). Here, we present the high-resolution structures of AncGR2 LBD with three ligands. Structural comparisons reveal that an important hydrogen bond—typically observed in other GR-glucocorticoids interactions—is absent due to the unique chemical nature of vamorolone. Loss of this hydrogen bond does not significantly affect GR dimerization in cells but instead alters local protein dynamics at the AF-2 site to modulate coregulator binding. Molecular dynamic simulations and cellular assays highlight the key role of this hydrogen bond in allosterically controlling coregulators recognition, tipping the balance favorably between efficacy and side effects in GR-targeted treatment.
Results
All Three Drugs Bind Directly to AncGR2 with High Affinity.
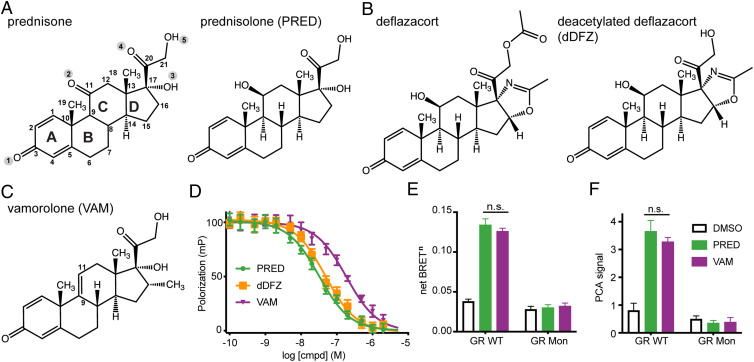
Prednisone and deflazacort administration results in increased strength and mobility in DMD patients, but also severe side effects (28, 29). Both drugs resemble the classical glucocorticoid structure with four rings labeled as A, B, C, and D from left to right (Fig. 1A). Prednisone is a prodrug, metabolized to its functional form, prednisolone, when the C11 oxo group is reduced to a hydroxyl group in the liver (Fig. 1A). Likewise, deflazacort is also biologically inert and is transformed into the active metabolite C21-desacetyl deflazacort. While prednisolone and C21-desacetyl deflazacort share ring structures A through C, C21-desacetyl deflazacort differs from prednisolone in ring D by replacing the C17 hydroxyl group with a 2-methyl-2-oxazoline ring (Fig. 1B). Ring D of vamorolone contains a methyl group (-CH3) at C16 compared to prednisolone and C21-desacetyl deflazacort (Fig. 1C). Most importantly, vamorolone has a double bond between C9 and C11 and lacks a C11 hydroxyl group.
Fig. 1.
Corticosteroids for DMD treatment have tight binding to AncGR2 LBD and induce similar GR dimerization effects. (A–C) Chemical structures of prednisolone (PRED), C21-desacetyl deflazacort (dDFZ), and vamorolone (VAM) used for DMD treatment. Key carbon and oxygen atom positions are highlighted. (D) All compounds bind to AncGR2 LBD directly with a nanomolar (nM) Ki, as measured by FP competition against fluorescein labeled dexamethasone (FAM-DEX). Error bars in D indicate SD from three replicates and from three independent experiments. (E and F) Effects of VAM on GR dimerization probed by BRETn and PCA assays with GR WT and Mon mutant upon treatment with 1,000 nM concentration of PRED and VAM. Bars indicate mean, and error bars represent SEM. Two-way ANOVA followed by Tukey’s post hoc test was performed. n.s., not significant.
Structural and biochemical studies of hGR LBD-drug complexes have been hampered by the challenge of obtaining soluble protein and require an extensive number of mutations in human receptor to facilitate crystallization (30, 31). We have overcome this problem by leveraging an ancestrally reconstructed GR variant, AncGR2 LBD. The AncGR2 LBD has increased protein expression, solubility, and improved crystallization. It also faithfully maintains ligand binding and allostery of hGR LBD and has been successfully leveraged for GR biochemistry and crystallization studies (25–27). Here, we used fluorescence-labeled dexamethasone, a strong agonist, to monitor the direct binding of these corticosteroids to the AncGR2 LBD in a fluorescence polarization (FP)-based competitive assay. All compounds have Ki values in the nanomolar (nM) range, representing tight binding (Fig. 1D). Prednisolone binds to AncGR2 LBD with Ki of 28 nM [25, 31] (95% confidence interval)—marginally tighter than C21-desacetyl deflazacort (Ki = 41 nM [36, 49]). Moreover, both have higher affinity for AncGR2 LBD than vamorolone (Ki = 156 nM [125, 194]).
Prednisolone and Vamorolone Induce Similar GR Dimerization Levels in Cells.
GR-agonist complexes homodimerize on canonical GR response elements to drive gene activation (16, 18). To determine if the reduced transactivation associated with vamorolone is caused by reduced GR-ligand dimerization, we compared its dimerization in cells treated with prednisolone or vamorolone by a NanoLuc luciferase-based bioluminescence resonance energy transfer (BRETn) assay and a protein-fragment complementation assay (NanoPCA) (32, 33). Both assays utilize NanoLuc, the smallest but the brightest luciferase to date, which has high sensitivity for detecting cellular protein-protein interactions including protein dimerization (32–34). Both vamorolone and prednisolone induce strong dimerization of WT GR and have no effects on GR Mon, a well-studied dimerization defective mutant (35) (Fig. 1 E and F). Unexpectedly, prednisolone and vamorolone result in a similar dimerization levels, suggesting that vamorolone prevents strong transactivation via an alternative mechanism.
High-Resolution Crystal Structures of AncGR2 LBD with Prednisolone, C21-Desacetyl Deflazacort and Vamorolone.
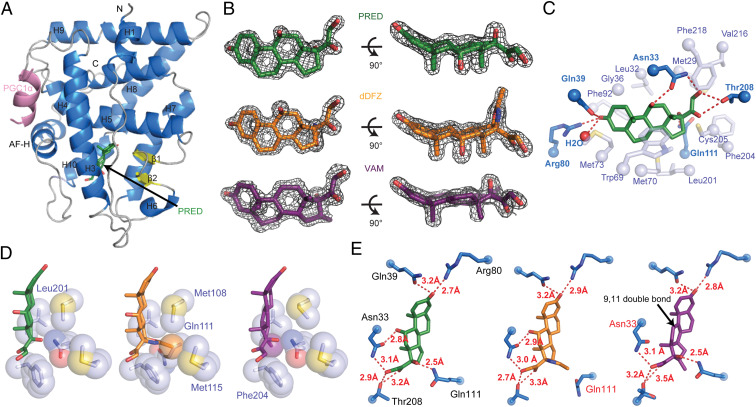
To gain insight into the molecular mechanism of vamorolone treatment, we determined the X-ray crystal structures of AncGR2 LBD complexed with prednisolone, C21-desacetyl deflazacort, or vamorolone, and to our knowledge, obtained the highest resolution structures of GR LBD (1.45 to 1.60 Å) to date (25–27, 30, 36, 37) (SI Appendix, Table S1). Each structure contains one protein molecule in the asymmetric unit. Overall, AncGR2 LBD contains 11 α-helices and four β-strands, is folded into a helical sandwich with three layers, and adopts the classical steroid receptor LBD structure (Fig. 2A). Both prednisolone- and C21-desacetyl deflazacort-bound AncGR2 LBD crystallized in complex with a peptide derived from peroxisome proliferator-activated gamma coactivator 1-α (PGC1α), whereas the AncGR2 LBD-vamorolone complex crystallized in the presence of a peptide derived from the atypical nuclear receptor small heterodimer partner (SHP). The coactivator peptides formed a short α-helix and bound at the activation function 2 (AF-2) surface comprised of helix 3, helix 4, and the AF-H (Fig. 2A). Unambiguous electron density shows all three ligands occupy the core of the AncGR2 LBD (Fig. 2B). Residues in the ligand binding pocket form extensive hydrophobic interactions and unique hydrogen bonds with all three ligands (Fig. 2 C–E). Gln39, Arg80, and a structurally conserved water molecule form a hydrogen bond network with carbonyl O1 on the A ring of PRED. Residue Asn33 makes two hydrogen bonds with both hydroxyls O2 and O5. Residue Gln111 hydrogen bonds with hydroxyl O3 on the D ring. Residue Thr208 hydrogen bonds with atoms O4 and O5 on the D ring (Fig. 2C). Residues Leu32, Met73, Phe-218, Val-216, etc. make extensive hydrophobic interactions with prednisolone (Fig. 2C). All five residues that H-bond with the ligands are identical between AncGR2 and hGR LBD. Residues involved in hydrophobic interactions are 83% identical with the remaining two residues showing a relative minor change in character (i.e., Ile vs. Val or Tyr vs. Phe) (SI Appendix, Fig. S1).
Fig. 2.
PRED, dDFZ, and VAM adopt similar orientations within the AncGR2 LBD but engage different H-bonding networks. (A) Overall structure of AncGR2 LBD with PRED (green) bound to PGC1α (light purple), with α-helices shown in light blue, β-strands in yellow, and loops in gray. (B) 2Fo-Fc omit electron density map (contoured to 2.0 σ) surrounding PRED (green), dDFZ (orange), and VAM (purple) in the ligand binding pocket. (C) Extensive hydrogen bonds (dark blue residues) and hydrophobic interactions (light blue residues) are formed between AncGR2 LBD-PRED. (D) Select hydrophobic interactions between AncGR2 LBD and different ligands. (E) Different hydrogen bond networks are formed between AncGR2 LBD and ligands with bond distances labeled.
Superimposition of the AncGR2 LBD C21-desacetyl deflazacort structure onto that of AncGR2 LBD-prednisolone shows close agreement, with a root mean square deviation (rmsd) of 0.12 Å out of 207 Cα atoms (SI Appendix, Fig. S2A). Closer inspection of two ligand binding sites shows that the oxazoline ring and its methyl group in the D ring of C21-desacetyl deflazacort make closer hydrophobic contacts with residues Met108, Glu111, and Met115 compared to the hydroxyl in the same position (C17) in prednisolone (Fig. 2D). However, this bulky oxazoline ring rotates the side chain of Gln111 away from its typical position, which would otherwise form a hydrogen bond with the hydroxyl group O3 as observed in both prednisolone- and vamorolone-bound AncGR2 LBD structures (Fig. 2E). The loss of this Gln111-mediated hydrogen bond, despite several newly formed hydrophobic interactions, may contribute to the weakened binding of C21-desacetyl deflazacort to AncGR2 LBD when compared to prednisolone.
Structural comparison between the AncGR2 LBD complexed with prednisolone and vamorolone shows relatively larger conformational variation than between AncGR2 LBD-prednisolone and C21-desacetyl deflazacort, as indicated by higher rmsd (0.3 Å out of 207 Cα atoms) with highest variation in the N- and C-terminal regions and the loops, such as the loop prior to AF-H (pre-AF-H loop) (SI Appendix, Fig. S2B). These differences are relatively minor, localized to the N terminus and the loop proceeding H3, and appear to be influenced by differential crystal packing in the AncGR2 LBD-vamorolone-SHP complex which crystallized in a space group different from the other two complexes. The hydrogen bond formed between Asn33 and O2, observed in the other two structures, is not possible with vamorolone (Fig. 2E). Asn33 directly bridges the ligand to the pre-AF-H loop via H-bonding with E217 (SI Appendix, Fig. S2B) (36), highlighting a critical role in stabilizing pre-AF-H loop and AF-H. Moreover, the double bond between C9 and C11 reorients the C ring, shifting C14 and O4, (1.2 Å and 0.8 Å, respectively) away from their positions as seen in the prednisolone-bound AncGR2 LBD structure (SI Appendix, Fig. S2B). Consequently, the side chain of residue Thr208 adopts an alternative configuration upon vamorolone binding. Weaker hydrogen bonds with both O4 and O5 are formed only with one configuration and with extended bond distances (2.9 Å and 3.2 Å in prednisolone-bound state versus 3.2 Å and 3.5 Å in the vamorolone-bound state) (Fig. 2E). Vamorolone has an additional methyl group on C16 and makes better contact with Phe-204 and Leu-201(Fig. 2D). However, these relatively weak hydrophobic interactions likely do not compensate for the loss of the C11-OH-Asn33 hydrogen bond.
Vamorolone Drives Weaker Association with Coactivators.
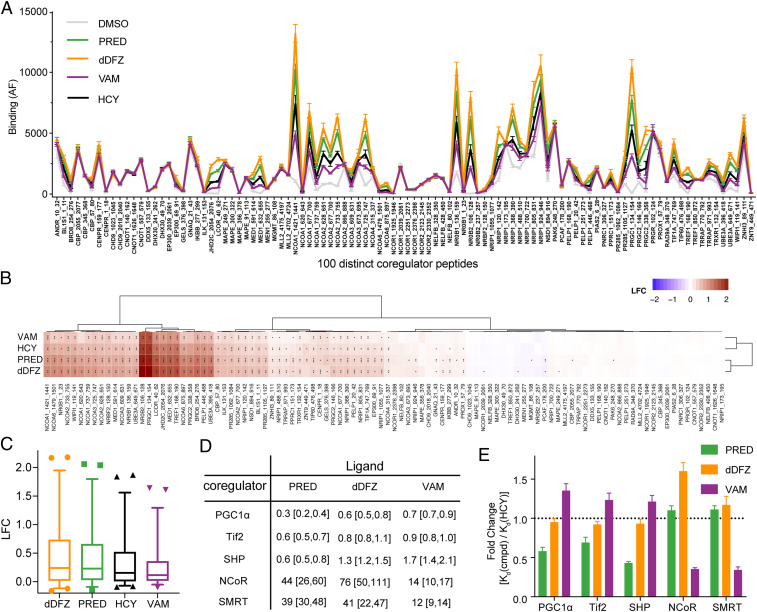
Ligand binding to GR drives recruitment of coregulators harboring the enzymatic functions required to remodel chromatin and facilitate transcriptional activity. We profiled 100 distinct coregulator peptides binding to GR in both apo and drug-bound forms using the microarray assay for real-time coregulator-nuclear receptor interaction (MARCoNI) assay. All four drugs (prednisolone, C21-desacetyl deflazacort, vamorolone, and hydrocortisone) confer significant coregulator peptide recruitment compared to apo hGR, and these coregulators contain NCOA, NRIP, and PGC1 family members (Fig. 3A). Unexpectedly, each drug drives an identical pattern of coregulator association varying only in the magnitude of coregulator peptide interaction. This result and the log-fold change modulation of binding compared to apo hGR suggest that the overall conformation adopted by the receptor is similar in all four complexes but relative population of GR in the active orientation differs in the solution state (Fig. 3B). Cluster analysis reveals that C21-desacetyl deflazacort and prednisolone promote enhanced coregulator binding relative to hydrocortisone or vamorolone, with vamorolone inducing the weakest binding. This pattern of coregulator recruitment in the presence of saturating ligand suggests that vamorolone may act as a partial agonist compared to C21-desacetyl deflazacort or prednisolone (Fig. 3C).
Fig. 3.
GR LBD-VAM complex has a distinct coregulator binding pattern. (A) Comparison of drug-induced coregulator peptide recruitment to hGR LBD by MARCoNI. (B) Hierarchical clustering of log-fold change (LFC) by comparing binding data of individual drug-bound and apo hGR LBD (dimethyl sulfoxide [DMSO] control) and colored continuously from blue to red. Statistically significant changes relative to DMSO control were identified by Student's t test, post hoc false discovery rate (FDR), *P < 0.05, **P < 0.01, or ***P < 0.001. (C) Box-and-whisker plot of LFC against DMSO control for hGR LBD in complex with PRED, dDFZ, VAM, and hydrocortisone (HCY). The line in the box is plotted at the median of all of the coregulator binding, whereas the whiskers are the 2.5 and 97.5 percentiles. (D) Summary of binding affinities for various coregulator peptides bound to AncGR2 LBD with different ligands are expressed as Kd (μM) with 95% confidence interval. (E) Fold change of binding to different coregulators compared to AncGR2 LBD with HCY as expressed by [Kd(cmpd)/Kd(HCY)] for each coregulator.
To quantitatively assess the binding strength between GR and representative FAM-labeled coregulator peptides in solution, we employed a FP assay using AncGR2 LBD in complex with different drugs. Overall, AncGR2 LBD bound to coactivators (Tif2, PGC1α, and SHP) more tightly than corepressors (NCoR and SMRT) (Fig. 3D and SI Appendix, Fig. S3). For the same coactivator, prednisolone confers tighter binding than C21-desacetyl deflazacort or vamorolone (Fig. 3D). Intriguingly, AncGR2 LBD-vamorolone binds to corepressors more tightly than AncGR2 LBD-prednisolone or C21-desacetyl deflazacort. For instance, Kd for AncGR2LBD-vamorolone binding to SMRT and NCoR was estimated to be <15 μM; this was approximately threefold more favorable than those for AncGR2 LBD-prednisolone and C21-desacetyl deflazacort binding to the same corepressors (Fig. 3D and SI Appendix, Fig. S3). Vamorolone is incapable of enhancing coactivator binding (fold change >1) but can bolster the corepressor binding (fold change <1) when compared with hydrocortisone, distinguishing from prednisolone and C21-desacetyl deflazacort (Fig. 3E). Together, this suggests that vamorolone is a partial agonist with weakened coactivator binding and slightly enhanced binding to representative corepressors compared to the other two drugs.
Vamorolone Stabilizes AncGR2 LBD Less than Other Drugs.
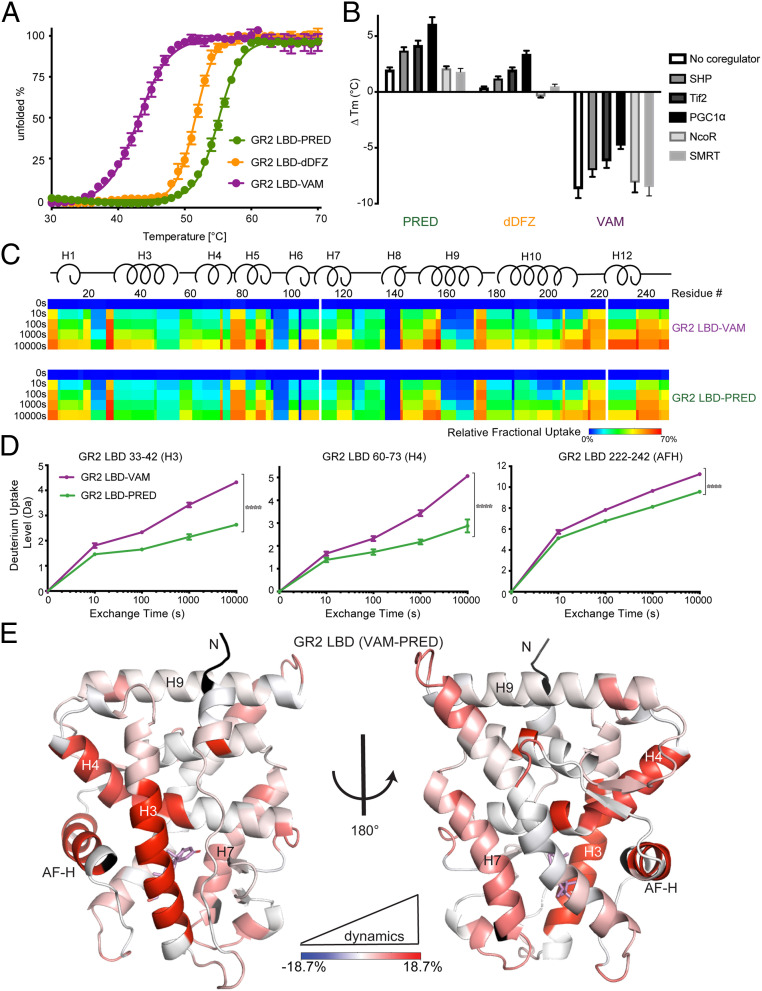
To determine whether weakened coactivator binding while enhanced corepressor association is due to a difference in protein conformational dynamics, we first used differential scanning fluorimetry (DSF)-based thermal denaturation to characterize the stability of GR-ligand/coregulator complexes. The AncGR2 LBD-vamorolone (melting temperature [Tm] = 43.7 °C) complex is significantly less stable than the prednisolone and C21-desacetyl deflazacort-containing complexes (Tm = 54.0 °C and 51.7 °C, respectively) and this holds true upon inclusion of coregulator peptides (Fig. 4 A and B). Among the coregulators tested, all three coactivators increased protein stability, whereas NCoR and SMRT had no significant effect in line with their weak binding (SI Appendix, Fig. S4 A–C). PGC1α shows the most stabilizing effect, followed by Tif2- and SHP-derived peptides which is consistent with the FP binding and MARCoNI data (SI Appendix, Fig. S4D). Structural analyses explain the weaker association of SHP. SHP and PGC1α interact similarly with the AF-2 region, creating several hydrophobic interactions and two primary charge clamps with Glu-224 and Lys-48, respectively (SI Appendix, Fig. S5 A and B). However, the Lys48 side chain -NH2 group fails to form the hydrogen bond with Ser27 (more than 4.0 Å away) that is observed in PGC1α, which likely explains the reduced binding affinity and protein stability (SI Appendix, Fig. S5C).
Fig. 4.
AncGR2 LBD with VAM has reduced stability and enhanced protein dynamics. (A) Thermal unfolding curves of AncGR2 LBD bound to PRED, dDFZ, and VAM. (B) Difference in thermostability (∆Tm) of GR2 LBD with different ligands in the presence of coregulators compared to GR2 LBD with HCY as expressed by ∆Tm = [Tm(ligand) − Tm(HCY)] for each coregulator. (C). Heat maps of deuterium uptake monitored by HDX-MS for AncGR2 LBD bound to PRED and VAM. Different time points of LBD incubation in D2O before measuring deuterium uptake are indicated on the Left. (D) Three representative HDX plots of peptic fragments from PRED- and VAM-bound AncGR2 LBD. (E) Differential deuterium uptakes are mapped on the structure of AncGR2 LBD in complex with VAM. Residues are colored in a continuous gradient from blue to red, with their intensity scaling to the difference in percentage of deuterium exchange [(VAM-bound) − (PRED-bound)]. Residues not covered by any peptides are shown in black.
Vamorolone Enhances the Local Conformational Dynamics of the Coregulator Binding Surface.
Local conformational dynamics, especially at the coregulator binding surface, play a critical role in nuclear receptor activation (38, 39). To determine how vamorolone affects GR’s local conformational dynamics in comparison to a strong agonist, we employed solution hydrogen deuterium exchange-mass spectrometry (HDX-MS). Differential HDX-MS can reveal conformational dynamics that are not readily observed in crystal structures; regions that are more susceptible to deuterium exchange indicate their relatively greater conformational flexibility.
Overall, 130 peptic fragments that cover 99.2% of the AncGR2 LBD sequence, with up to sevenfold redundancy, were sequenced and mapped for HDX analysis (SI Appendix, Fig. S6A). Comparison between vamorolone- and prednisolone-bound complexes reveal increased deuterium uptake of several regions in the vamorolone-bound complex, with three representative fragments including residues 33 to 42, 60 to 73, and 222 to 242, in line with the lower observed thermal stability (Fig. 4 C and D and SI Appendix, Fig. S6B). Mapping these residues onto the structure shows that the most prominent differences are in the AF-H, helix 3, and helix 4, all of which are adjacent to or part of the coregulator binding site, providing a mechanism to explain how vamorolone reduces coregulator association and downstream transcription (Fig. 4 D and E). This suggests that vamorolone selects for a more dynamic set of conformations at the AF-2 site than prednisolone which consequently affect coregulator binding and downstream transcription.
Dampened Allosteric Communication between Vamorolone and the AF-H.
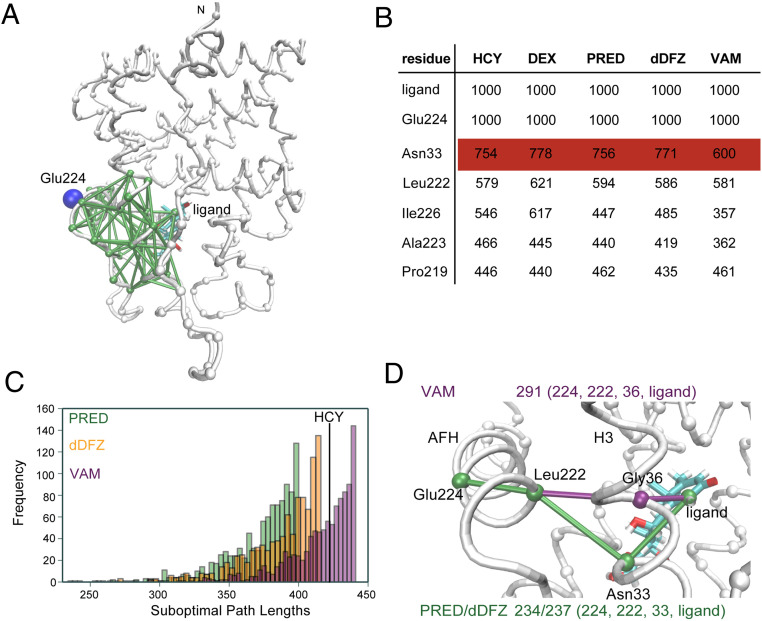
To understand how vamorolone allows for greater conformational dynamics of the AF-2, we conducted molecular dynamics simulations of different AncGR2 LBD complexes combined with dynamic network analyses. During the 1-μs simulation, all AncGR2 LBD molecules were characterized by stable global rmsd of <1.7 Å (SI Appendix, Fig. S7A). Root mean square fluctuations (RMSFs) of Ca atoms were calculated to measure the protein flexibility showing that the pre-AF-H loop, residues 217 to 225, are highly flexible in the AncGR2 LBD-vamorolone complex (SI Appendix, Fig. S7B).
Networks were constructed by selecting Cα atoms of the protein along with C2 atoms from the ligand as nodes. A pair of nodes was connected by their edges if they have satisfied a distance requirement (<4.5 Å) for at least 75% of the simulation time (Fig. 5A). Edge distance is inversely proportional to the pairwise correlations; thus, short distances indicate a strong correlation between two nodes (40). The ligand and Glu224, residing in the AF-H forming the primary charge clamp with coactivator, were selected as the nodes to study allosteric communication within the LBD (Fig. 5 A–C). Suboptimal path length analyses reveal that AncGR2 LBD-vamorolone has longer path lengths, and thus weaker ligand-AF-H allosteric communication, than the AncGR2 LBD-prednisolone, C21-desacetyl deflazacort or hydrocortisone complex (Fig. 5C). Likewise, the optimal path length in vamorolone-bound AncGR2 LBD is longer than the other two complexes. The most commonly utilized node along all pathways was residue Asn33 (hGR N564) in H3. It was used greater than 700 times over four different ligand-bound complexes simulations, including two previously reported GR-ligand bound structures (24), identifying Asn33 as the key node connecting the ligand to the AF-H. Asn33 engages in a C11-OH hydrogen bond with all ligands tested. Vamorolone that lacks a C11-OH preventing this critical H-bond thereby reducing the utilization of Asn33 in communication with the AF-H. In the vamorolone-bound complex, Gly36 takes on the nodal role (Fig. 5D). Together, these analyses indicate a critical role for Ans33 in relaying allosteric communication from the ligand to AF-H. Vamorolone selectively lacks this interaction weakening ligand-driven communication to and stabilization of the AF-H.
Fig. 5.
VAM binding reduces allosteric communication between AncGR2 ligand and AFH. (A) Suboptimal paths connecting nodes Glu224 (shown in blue) and ligand (shown in cyan) with edges (shown in green) displayed. The other nodes in the protein are shown in gray. (B) Top nodes used in the top 1,000 suboptimal pathway analyses in five different AncGR2-ligand complexes. (C) Histograms of top 1,000 suboptimal paths in AncGR2 with three ligands. (D) Optimal path length and nodes used in these paths in the AncGR2 with three ligands.
Vamorolone Uniquely Modulates the Asn33-Mediated Allosteric Network to Confer Dissociative Properties.
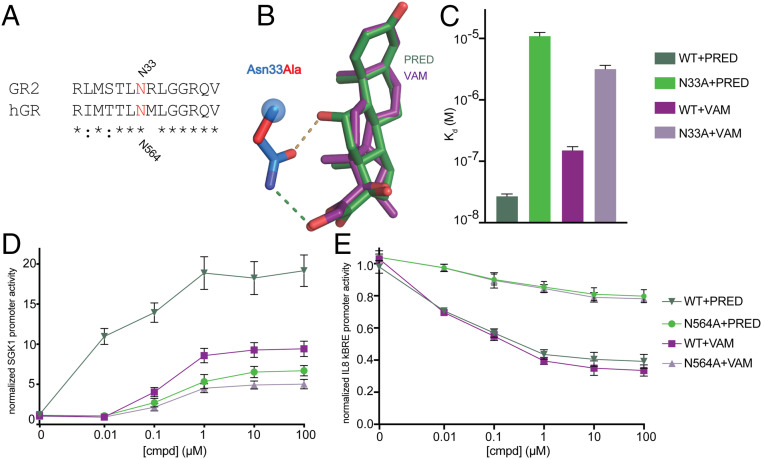
Asn-33 (N564 in hGR) is evolutionarily conserved (Fig. 6A) and is known to play a critical role in GR activation from classical GCs as mutation to alanine significantly reduces GC-induced transactivation (41). This residue makes two H-bond interactions with classical glucocorticoids, such as prednisolone, at the C11 and C21 oxygens (Fig. 6B). The role of this Asn in transrepression is unknown and we sought to determine if mutation of Asn to Ala would support transrepression or convert a strong agonist such as prednisolone to a weak or dissociative compound. N33A mutation reduced binding to both prednisolone and vamorolone compared to WT, with the larger effects on prednisolone given the loss of two H-bonds (Fig. 6C). Coregulator binding at saturating ligand was also tested via MARCoNI assays revealing that the N33A mutation diminishes the vast majority of binding in both prednisolone and vamorolone-bound AncGR2 LBD (SI Appendix, Fig. S8). Importantly, the hGR N564A mutation ablates both transactivation from a classical SGK promoter and transrepression from IL8 NF-κB promoter, indicating a crucial role for GR N564 in gene repression (Fig. 6 D and E). These results suggest that gene transrepression requires some level of ligand-mediated communication to AF-H. A complete ablation of this communication destroys activity but a partial reduction in communication can selectivity disrupt transactivation tipping the balance toward transrepression.
Fig. 6.
Residue N33 (in AncGR2) or N564 (in hGR) is crucial for ligand binding and gene transcription. (A) Sequence alignment between hGR and AncGR2 centered on N33 (AncGR2 numbering). (B) Hydrogen bonds formed between N33 and ligands (overlaid structures of PRED and VAM) with the N33A mutation modeled and shown in red. (C) Binding affinities of PRED and VAM interacting with AncGR2WT and N33A mutant that expressed as Kd (M) with 95% confidence interval. (D and E) Effects of N564A mutation on transactivation and transrepression using luciferase reporters containing SGK1 promoter (D) and κBRE promoter (E) upon treatment with PRED and VAM. Error bars in D and E represent SEM.
Discussion
GR is the most common drug target for chronic and dysregulated inflammation which is a hallmark of numerous diseases including asthma, arthritis, atherosclerosis, neurodegeneration, and muscular dystrophy (42). Chronic treatment of these diseases with GR-targeting corticosteroid drugs is highly effective, but extensive adverse side effects are seen, generally attributed to transactivation of GR target genes (43). Selective GR modulators minimizing undesired gene activation have been extremely challenging to produce (44). Vamorolone is a first-in-class dissociative steroid with antiinflammatory efficacy separated from safety concerns and represents a potential treatment for DMD (13, 45). Both nonclinical and open label phase II clinical studies show dose-responsive suppression of inflammation and improvement of muscle function that are typically associated with the prodrugs prednisone and deflazacort, the standard of care of DMD (14, 15). A debilitating safety concern of corticosteroids is their detrimental effect on bone (stunting of growth, osteopenia, increased bone breakage). Muscular dystrophy mouse models treated with vamorolone show no stunting of growth or osteopenia (13). Human DMD patients treated with vamorolone show normal bone formation biomarkers (e.g., osteocalcin), and no stunting of growth (45, 46).
To understand how vamorolone may improve GR-mediated DMD therapy, we compared the GR-vamorolone complex with other glucocorticoids that constitute standard of care using structural, biophysical, and biochemical approaches. Detailed structural and computational analysis reveals formation of the typical N33-C11-hydroxyl (O2) hydrogen bond is not possible with vamorolone. This hydrogen bond is generally formed with other GR ligands including prednisolone, C21-desacetyl deflazacort, dexamethasone, hydrocortisone, mometasone furoate, and triamcinolone acetonide (24, 25, 27), setting vamorolone apart from conventional corticosteroids. This missing hydrogen bond partly explains the weaker vamorolone binding affinity and reduced overall stability of the AncGR2-vamorolone complex (Figs. 1D and 4A). Differential HDX-MS revealed increased conformational dynamics of the AF-2 surface in the AncGR2 LBD-vamorolone versus AncGR2 LBD-prednisolone complex highlighting an important role for the N33- C11-hydroxyl (O2) hydrogen bond in transmitting information from the ligand binding pocket to the AF-H (Fig. 4D).
Ligand-dependent interaction between GR and various coregulators is the predominant molecular determinant of ligand-selective transcriptional differences in both potency and efficacy (47). Coregulator interaction screens have been viewed as a way to probe for ligand-specific conformations (48). The hierarchical clustering result of the MARCoNI profiling reveals that all glucocorticoids tested here drive a similar GR conformation (i.e., the “active” GR conformation). Vamorolone drives weaker interaction with coregulators without altering coregulator preference. This correlates well with the weak transactivation induced by vamorolone (Fig. 6D) (12, 13), suggesting that it achieves its dissociative properties by acting as a weak GR agonist through (+) GRE-mediated transcriptional pathways yet is still able to support transrepression. The enhanced AF-2 dynamics observed in HDX is in harmony with the increased ability of GR-vamorolone to sample multiple conformations to accommodate both coactivator and corepressor binding, compared to the more rigid AF-2 surface in the prednisolone-bound GR suitable for strong coactivator binding. Indeed, we found tighter association between GR-vamorolone and NCoR and SMRT than with other drugs (Fig. 3D), though corepressor peptide binding is still relatively weak in vitro. In cells, NCoR and SMRT can further recruit and activate histone deacetylases to repress gene activation by regulating chromatin condensation and accessibility (16, 49). Recruitment of histone deacetylases by GR corepressors to suppress activated inflammatory genes is the accepted mechanism of inhaled corticosteroids-based asthma treatment (50). Moreover, NCoR and HDAC3 are required for the suppression of the NF-κB target gene IL-6 (19). Vamorolone acts as a partial agonist but retains or even slightly increases transrepressive effects on inflammatory genes relative to other glucocorticoids, suggesting that selective destabilization of coactivator binding may favor corepressor interaction and gene repression.
AncGR2 residue N33 evolved following gene duplication of the estrogen-sensitive ancestral steroid receptor (AncSR) over 500 million years ago (51). This residue was then coopted into a critical ligand-sensing role in AncSR2 and remaining fixed in all 3-ketosteroid receptors (PR: N719; AR: N705; MR: N770; and GR: N564) (52). Indeed, this Asn-ligand interaction is integral for ligand-driven allosteric communication in all aforementioned receptor-ligand complexes (52) and mutation of this residue reduces activation of all 3-ketosteroid steroid receptors (41, 53–55). Using molecular dynamics simulation, we verified that the allosteric communication between the ligand and the AF-H routes through residue N33 in AncGR2 (N564 in hGR) (Fig. 5) and disruption of this pathway through either mutation or removal of the (C11)-OH reduces ligand and coregulator binding and gene transcription (Fig. 6). Total ablation of Asn33 interaction with the ligand through mutation reduces both transactivation and transrepression, suggesting that some level of Ans33-ligand interaction is required to suppress gene expression.
Chemical modifications on C11 through addition of large chemical groups intended to deform the ligand binding pocket near H3, is a strategy employed for a range of current steroidal and nonsteroidal compounds; however, this does not always yield predictable dissociative effects (56–58). By contrast, vamorolone contains a subtle modification to the corticosteroid backbone to weaken the communication network and enhance the conformational dynamics of the AF-2 region. This work suggests a strategy where weakening ligand-H3 interactions rather than increasing them may better guide the design future dissociative drugs targeting GR.
Vamorolone is also known to target and antagonize mineralocorticoid receptor (MR), the closest paralog of GR, for the treatment of cardiomyopathy found in late-stage DMD patients (23). Vamorolone lacks a C11 -OH group, a feature shared with other MR antagonists such as spironolactone and progesterone. MR contains an analogous residue N770 to GR2 N33 (and hGR N564). It is thus conceivable that vamorolone suppresses MR activation by affecting the ligand-N770-E955 (at pre-AF-H loop) H-bond network that is critical for activation as observed in the MR-progesterone structure (59). However, the precise mechanism of how vamorolone antagonizes MR and exerts different actions (partial agonist for GR vs. antagonist for MR) is still unclear. Future studies focusing on how vamorolone antagonizes MR signaling will be important for the development of next-generation dual receptor-targeting drugs with improved MR antagonism for DMD treatment.
Materials and Methods
Protein Expression and Purification.
All AncGR2 LBD constructs were expressed in Escherichia coli BL21 cells, induced in the presence of 50 μM drugs at 16 °C and purified as previously reported (24) as detailed in SI Appendix.
Crystallization and Structure Determination.
X-ray diffraction data of AncGR2 LBD-drug complexes were collected on the South East Regional Collaborative Access Team (SER-CAT) beamline 22-ID at the Advanced Photon Source (APS) at Argonne National Laboratory at 100 K and processed using HKL-2000 as detailed in SI Appendix.
Biochemical Experiments.
Ligand binding and coregulator binding assays, MARCoNI, DSF, and HDX-MS experiments were performed as detailed in SI Appendix.
Cellular Assays.
Reporter gene and GR dimerization assays were performed in HeLa and HEK293T cells, respectively, as detailed in SI Appendix.
Supplementary Material
Acknowledgments
These studies were supported by a W. M. Keck Foundation Medical Research Grant, ReveraGen Biopharma (Rockville, MD), and partially supported by the NIH (R01DK115213 to E.A.O.) X.L. was supported by an American Heart Association postdoctoral fellowship (17POST33660110). We thank the HDX-MS core in the Department of Pediatrics, Emory University School of Medicine for their technical assistance in data collection and analysis. We are also grateful to René Houtman from Precision Medicine Laboratory (The Netherlands) for his help with MARCoNI. X-ray data were collected at Southeast Regional Collaborative Access Team (SER-CAT) 22-ID beamline at the Advanced Photon Source, Argonne National Laboratory. Supporting institutions may be found at https://www.ser.aps.anl.gov/www.ser-cat.org/members.html. Use of the Advanced Photon source was supported by the Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract W-31-109-Eng-38.
Footnotes
Competing interest statement: The authors declare competing financial interests. J.M.D., K.N., and E.P.H. are employees of ReveraGen BioPharma, and hold stock. The other authors declare no competing interests.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2006890117/-/DCSupplemental.
Data Availability.
The atomic coordinates and structure factors have been deposited in the Protein Data Bank with the accession numbers 6W9K–6W9M for AncGR2 LBD prednisolone-PGC1α, AncGR2 LBD C21-desacetyl deflazacort-PGC1α, and AncGR2 LBD vamorolone-SHP complexes, respectively.
References
- 1.Yiu E. M., Kornberg A. J., Duchenne muscular dystrophy. J. Paediatr. Child Health 51, 759–764 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Hoffman E. P., Brown R. H. Jr., Kunkel L. M., Dystrophin: The protein product of the Duchenne muscular dystrophy locus. Cell 51, 919–928 (1987). [DOI] [PubMed] [Google Scholar]
- 3.Gao Q. Q., McNally E. M., The dystrophin complex: Structure, function, and implications for therapy. Compr. Physiol. 5, 1223–1239 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guiraud S. et al., The pathogenesis and therapy of muscular dystrophies. Annu. Rev. Genomics Hum. Genet. 16, 281–308 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Rosenberg A. S. et al., Immune-mediated pathology in Duchenne muscular dystrophy. Sci. Transl. Med. 7, 299rv4 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Manzur A. Y., Kuntzer T., Pike M., Swan A., Glucocorticoid corticosteroids for Duchenne muscular dystrophy. Cochrane Database Syst. Rev., CD003725 (2008). [DOI] [PubMed] [Google Scholar]
- 7.Singh A., Schaeffer E. K., Reilly C. W., Vertebral fractures in Duchenne muscular dystrophy patients managed with deflazacort. J. Pediatr. Orthop. 38, 320–324 (2018). [DOI] [PubMed] [Google Scholar]
- 8.Hoffman E. P. et al., Novel approaches to corticosteroid treatment in Duchenne muscular dystrophy. Phys. Med. Rehabil. Clin. N. Am. 23, 821–828 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoffman E. P., Nader G. A., Balancing muscle hypertrophy and atrophy. Nat. Med. 10, 584–585 (2004). [DOI] [PubMed] [Google Scholar]
- 10.Connolly A. M., Schierbecker J., Renna R., Florence J., High dose weekly oral prednisone improves strength in boys with Duchenne muscular dystrophy. Neuromuscul. Disord. 12, 917–925 (2002). [DOI] [PubMed] [Google Scholar]
- 11.Escolar D. M. et al., Randomized, blinded trial of weekend vs daily prednisone in Duchenne muscular dystrophy. Neurology 77, 444–452 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reeves E. K. M., Hoffman E. P., Nagaraju K., Damsker J. M., McCall J. M., VBP15: Preclinical characterization of a novel anti-inflammatory delta 9,11 steroid. Bioorg. Med. Chem. 21, 2241–2249 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heier C. R. et al., VBP15, a novel anti-inflammatory and membrane-stabilizer, improves muscular dystrophy without side effects. EMBO Mol. Med. 5, 1569–1585 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Conklin L. S. et al., Phase IIa trial in Duchenne muscular dystrophy shows vamorolone is a first-in-class dissociative steroidal anti-inflammatory drug. Pharmacol. Res. 136, 140–150 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoffman E. P. et al., Phase 1 trial of vamorolone, a first-in-class steroid, shows improvements in side effects via biomarkers bridged to clinical outcomes. Steroids 134, 43–52 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weikum E. R., Liu X., Ortlund E. A., The nuclear receptor superfamily: A structural perspective. Protein Sci. 27, 1876–1892 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kadmiel M., Cidlowski J. A., Glucocorticoid receptor signaling in health and disease. Trends Pharmacol. Sci. 34, 518–530 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weikum E. R., Knuesel M. T., Ortlund E. A., Yamamoto K. R., Glucocorticoid receptor control of transcription: Precision and plasticity via allostery. Nat. Rev. Mol. Cell Biol. 18, 159–174 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hudson W. H. et al., Cryptic glucocorticoid receptor-binding sites pervade genomic NF-κB response elements. Nat. Commun. 9, 1337 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vandewalle J., Luypaert A., De Bosscher K., Libert C., Therapeutic mechanisms of glucocorticoids. Trends Endocrinol. Metab. 29, 42–54 (2018). [DOI] [PubMed] [Google Scholar]
- 21.Kauppi B. et al., The three-dimensional structures of antagonistic and agonistic forms of the glucocorticoid receptor ligand-binding domain: RU-486 induces a transconformation that leads to active antagonism. J. Biol. Chem. 278, 22748–22754 (2003). [DOI] [PubMed] [Google Scholar]
- 22.Schoch G. A. et al., Molecular switch in the glucocorticoid receptor: Active and passive antagonist conformations. J. Mol. Biol. 395, 568–577 (2010). [DOI] [PubMed] [Google Scholar]
- 23.Heier C. R. et al., Vamorolone targets dual nuclear receptors to treat inflammation and dystrophic cardiomyopathy. Life Sci. Alliance 2, e201800186 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu X., Wang Y., Ortlund E. A., First high-resolution crystal structures of the glucocorticoid receptor ligand-binding domain-peroxisome proliferator-activated γ coactivator 1-α complex with endogenous and synthetic glucocorticoids. Mol. Pharmacol. 96, 408–417 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weikum E. R., Okafor C. D., D’Agostino E. H., Colucci J. K., Ortlund E. A., Structural analysis of the glucocorticoid receptor ligand-binding domain in complex with triamcinolone acetonide and a fragment of the atypical coregulator, small heterodimer partner. Mol. Pharmacol. 92, 12–21 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bridgham J. T., Ortlund E. A., Thornton J. W., An epistatic ratchet constrains the direction of glucocorticoid receptor evolution. Nature 461, 515–519 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kohn J. A., Deshpande K., Ortlund E. A., Deciphering modern glucocorticoid cross-pharmacology using ancestral corticosteroid receptors. J. Biol. Chem. 287, 16267–16275 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Campbell C., Jacob P., Deflazacort for the treatment of Duchenne dystrophy: A systematic review. BMC Neurol. 3, 7 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bello L. et al.; CINRG Investigators , Prednisone/prednisolone and deflazacort regimens in the CINRG Duchenne natural history study. Neurology 85, 1048–1055 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.He Y. et al., Structures and mechanism for the design of highly potent glucocorticoids. Cell Res. 24, 713–726 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Edman K. et al., Ligand binding mechanism in steroid receptors: From conserved plasticity to differential evolutionary constraints. Structure 23, 2280–2290 (2015). [DOI] [PubMed] [Google Scholar]
- 32.Mo X. L. et al., Enabling systematic interrogation of protein-protein interactions in live cells with a versatile ultra-high-throughput biosensor platform. J. Mol. Cell Biol. 8, 271–281 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mo X. et al., AKT1, LKB1, and YAP1 revealed as MYC interactors with NanoLuc-based protein-fragment complementation assay. Mol. Pharmacol. 91, 339–347 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hall M. P. et al., Engineered luciferase reporter from a deep sea shrimp utilizing a novel imidazopyrazinone substrate. ACS Chem. Biol. 7, 1848–1857 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Presman D. M. et al., Live cell imaging unveils multiple domain requirements for in vivo dimerization of the glucocorticoid receptor. PLoS Biol. 12, e1001813 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bledsoe R. K. et al., Crystal structure of the glucocorticoid receptor ligand binding domain reveals a novel mode of receptor dimerization and coactivator recognition. Cell 110, 93–105 (2002). [DOI] [PubMed] [Google Scholar]
- 37.Suino-Powell K. et al., Doubling the size of the glucocorticoid receptor ligand binding pocket by deacylcortivazol. Mol. Cell. Biol. 28, 1915–1923 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kojetin D. J., Burris T. P., Small molecule modulation of nuclear receptor conformational dynamics: Implications for function and drug discovery. Mol. Pharmacol. 83, 1–8 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hughes T. S. et al., Ligand and receptor dynamics contribute to the mechanism of graded PPARγ agonism. Structure 20, 139–150 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bowerman S., Wereszczynski J., Detecting allosteric networks using molecular dynamics simulation. Methods Enzymol. 578, 429–447 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dezitter X. et al., A structural explanation of the effects of dissociated glucocorticoids on glucocorticoid receptor transactivation. Mol. Pharmacol. 85, 226–236 (2014). [DOI] [PubMed] [Google Scholar]
- 42.Rhen T., Cidlowski J. A., Antiinflammatory action of glucocorticoids–New mechanisms for old drugs. N. Engl. J. Med. 353, 1711–1723 (2005). [DOI] [PubMed] [Google Scholar]
- 43.Schäcke H., Döcke W. D., Asadullah K., Mechanisms involved in the side effects of glucocorticoids. Pharmacol. Ther. 96, 23–43 (2002). [DOI] [PubMed] [Google Scholar]
- 44.Clark A. R., Belvisi M. G., Maps and legends: The quest for dissociated ligands of the glucocorticoid receptor. Pharmacol. Ther. 134, 54–67 (2012). [DOI] [PubMed] [Google Scholar]
- 45.Hoffman E. P. et al.; Cooperative International Neuromuscular Research Group , Vamorolone trial in Duchenne muscular dystrophy shows dose-related improvement of muscle function. Neurology 93, e1312–e1323 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smith E. C., et al. , Efficacy and safety of vamorolone in Duchenne muscular dystrophy: An 18-month interim analysis of a non-randomized open-label extension study. In press at PLoS Med. (2020). DOI: 10.1371/journal.pmed.1003222. [DOI] [PMC free article] [PubMed]
- 47.Ronacher K. et al., Ligand-selective transactivation and transrepression via the glucocorticoid receptor: Role of cofactor interaction. Mol. Cell. Endocrinol. 299, 219–231 (2009). [DOI] [PubMed] [Google Scholar]
- 48.Kazmin D. et al., Linking ligand-induced alterations in androgen receptor structure to differential gene expression: A first step in the rational design of selective androgen receptor modulators. Mol. Endocrinol. 20, 1201–1217 (2006). [DOI] [PubMed] [Google Scholar]
- 49.Watson P. J., Fairall L., Schwabe J. W., Nuclear hormone receptor co-repressors: Structure and function. Mol. Cell. Endocrinol. 348, 440–449 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Barnes P. J., Adcock I. M., Ito K., Histone acetylation and deacetylation: Importance in inflammatory lung diseases. Eur. Respir. J. 25, 552–563 (2005). [DOI] [PubMed] [Google Scholar]
- 51.Eick G. N., Thornton J. W., Evolution of steroid receptors from an estrogen-sensitive ancestral receptor. Mol. Cell. Endocrinol. 334, 31–38 (2011). [DOI] [PubMed] [Google Scholar]
- 52.Okafor C. D., Hercules D., Kell S. A., Ortlund E. A., Rewiring ancient residue interaction networks drove the evolution of specificity in steroid receptors. Structure 28, 196–205.e3 (2020). [DOI] [PubMed] [Google Scholar]
- 53.Petit-Topin I. et al., Met909 plays a key role in the activation of the progesterone receptor and also in the high potency of 13-ethyl progestins. Mol. Pharmacol. 75, 1317–1324 (2009). [DOI] [PubMed] [Google Scholar]
- 54.Poujol N. et al., Specific recognition of androgens by their nuclear receptor. A structure-function study. J. Biol. Chem. 275, 24022–24031 (2000). [DOI] [PubMed] [Google Scholar]
- 55.Rafestin-Oblin M. E. et al., 11beta-hydroxyprogesterone acts as a mineralocorticoid agonist in stimulating Na+ absorption in mammalian principal cortical collecting duct cells. Mol. Pharmacol. 62, 1306–1313 (2002). [DOI] [PubMed] [Google Scholar]
- 56.Safy M. et al., Efficacy and safety of selective glucocorticoid receptor modulators in comparison to glucocorticoids in arthritis, a systematic review. PLoS One 12, e0188810 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jin Z. et al., Synthesis of novel steroidal agonists, partial agonists, and antagonists for the glucocorticoid receptor. Bioorg. Med. Chem. Lett. 27, 347–353 (2017). [DOI] [PubMed] [Google Scholar]
- 58.Hu X. et al., The antagonists but not partial agonists of glucocorticoid receptor ligands show substantial side effect dissociation. Endocrinology 152, 3123–3134 (2011). [DOI] [PubMed] [Google Scholar]
- 59.Bledsoe R. K. et al., A ligand-mediated hydrogen bond network required for the activation of the mineralocorticoid receptor. J. Biol. Chem. 280, 31283–31293 (2005). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The atomic coordinates and structure factors have been deposited in the Protein Data Bank with the accession numbers 6W9K–6W9M for AncGR2 LBD prednisolone-PGC1α, AncGR2 LBD C21-desacetyl deflazacort-PGC1α, and AncGR2 LBD vamorolone-SHP complexes, respectively.