Abstract
Significance: Nuclear factor erythroid 2 (NFE2)-related factor 2 (NFE2L2, or NRF2) is a transcription factor predominantly affecting the expression of antioxidant genes. NRF2 plays a significant role in the control of redox balance, which is crucial in cancer cells. NRF2 activation regulates numerous cancer hallmarks, including metabolism, cancer stem cell characteristics, tumor aggressiveness, invasion, and metastasis formation. We review the molecular characteristics of the NRF2 pathway and discuss its interactions with the cancer hallmarks previously listed.
Recent Advances: The noncanonical activation of NRF2 was recently discovered, and members of this pathway are involved in carcinogenesis. Further, cancer-related changes (e.g., metabolic flexibility) that support cancer progression were found to be redox- and NRF2 dependent.
Critical Issues: NRF2 undergoes Janus-faced behavior in cancers. The pro- or antineoplastic effects of NRF2 are context dependent and essentially based on the specific molecular characteristics of the cancer in question. Therefore, systematic investigation of NRF2 signaling is necessary to clarify its role in cancer etiology. The biggest challenge in the NRF2 field is to determine which cancers can be targeted for better clinical outcomes. Further, large-scale genomic and transcriptomic studies are missing to correlate the clinical outcome with the activity of the NRF2 system.
Future Directions: To exploit NRF2 in a clinical setting in the future, the druggable members of the NRF2 pathway should be identified. In addition, it will be important to study how the modulation of the NRF2 system interferes with cytostatic drugs and their combinations.
Keywords: NRF2, cancer, breast cancer, cancer metabolism, reactive species
Introduction
The redox balance in cells is tightly regulated to meet physiological needs by balancing the abundance of pro-oxidants and reductants. In cancer, the redox balance is commonly dysregulated. However, whether the redox imbalance induces or inhibits cancer formation and cell growth is rather controversial, as the pro- or anticancer effects vary between different cancers and may change as a function of the cancer type (48, 49, 73–75, 80, 95, 180, 183, 218, 225, 227, 295). Nuclear factor erythroid 2 (NFE2)-related factor 2 (NFE2L2, commonly referred to as NRF2) is a transcription factor that plays a crucial role in maintaining the expression of antioxidant genes. Therefore, NRF2 is vital in maintaining the cellular redox balance.
NRF2 is present in model organisms such as Caenorhabditis elegans and Drosophila melanogaster, which have an antioxidant system similar to mammals, suggesting that the NRF2 pathway represents an evolutionary conserved cytoprotective program (71). However, NRF2 signaling spans beyond redox signaling toward metabolic pathways and the induction of collective cellular changes exemplified by cancer metabolism. In this review, we set out to provide a critical focus on NRF2 activity in cancer. We discuss the aberrant activation of NRF2 and its critical cellular targets.
The Molecular Determinants of NRF2 Signaling
The molecular structure of NRF2
NRF2 is a basic leucine zipper domain (bZIP) transcription factor and contains a Cap'N'Collar structure. NRF2 comprises 605 amino acids and has a calculated molecular weight of 68 kDa. However, recent studies claim that NRF2 may have a higher apparent molecular weight (up to ∼130 kDa) (155, 292). In our hands, the actual molecular weight of NRF2 and the proportions between the different molecular weight isoforms vary between experimental models. Thus, the use of validated antibodies is crucial in investigating NRF2.
The NRF2 transcription factor contains seven conserved NRF2-ECH homology (Neh) domains, known as Neh1-Neh7 (Fig. 1A). The Neh1 domain allows the binding of NRF2 to the antioxidant response sequences (electrophilic responsive/antioxidant-response elements [EpRE/ARE]; 5′-RTGABNNNGCR-3′). The Neh2 domain, the major regulatory domain located in the N-terminal region of NRF2, is essential for the Kelch-like ECH-associated protein 1 (KEAP1)-dependent degradation of NRF2 (see later). The Neh2 domain possesses ETGE and DLG motifs, which are responsible for direct interaction with the negative regulator, KEAP1 (191, 192, 287). The Neh3, Neh4, and Neh5 domains are transactivation domains. Neh3 is located in the C-terminal region of NRF2 and binds to the chromo-ATPase/helicase DNA binding protein family member CHD6, which functions as an NRF2 transcriptional coactivator (204). Neh4 and Neh5 are responsible for NRF2-mediated ARE transactivation by binding to a transcriptional coactivator CREB-binding protein (130). The Neh6 domain is a serine-rich region that regulates NRF2 stability. The redox-insensitive Neh6 domain consists of two motifs (DSGIS and DSAPGS) that are responsible for the binding of β-transducin repeat-containing protein (β-TrCP). Phosphorylation of the DSGIS motif by glycogen synthase kinase 3 (GSK3) enhances the ability of β-TrCP to ubiquitinate NRF2 (37, 223). The Neh7 domain was shown to interact with the retinoic acid receptor X receptor α (RAR α), which represses NRF2 target gene expression (297).
FIG. 1.
Schematic representation of the domain structure of NRF2 and KEAP1 proteins. (A) NRF2 comprises seven domains (Neh1–7). The Neh1 is responsible for DNA binding and dimerization with small Maf proteins. The Neh2 domain contains DLG and ETGE motifs that interact with KEAP1. The Neh3, Neh4, and Neh5 domains are transactivation domains; the Neh6 domain regulates NRF2 stability, and the Neh7 domain is responsible for RXRα binding. (B) KEAP1 contains five domains: the amino terminal NTR, CTR, and three major domains, BTB, IVR, and Kelch/DGR domains. The BTB domain associates with Cul3 and mediates KEAP1 dimerization. The IVR domain contains cysteine residues and connects the BTB and Kelch/DGR domains. The Kelch/DGR domain maintains the interaction between KEAP1 and NRF2. BTB, Broad complex/Tramtrack/bric-a-brac; CTR, carboxy-terminal; DGR, double glycine repeats; IVR, intervening region; KEAP1, Kelch-like ECH-associated protein 1; Neh, NRF2-ECH homology; NFE2, nuclear factor erythroid 2; NRF2, NFE2-related factor 2; NTR, N-terminal region; RXRα, retinoid X receptor α. Color images are available online.
The repressor protein of NRF2, KEAP1; the canonical activation of the KEAP1-NRF2 pathway
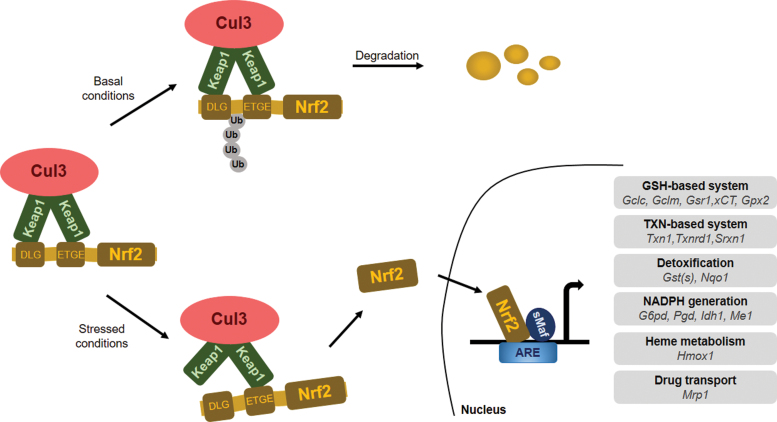
The main function of NRF2 is to protect the cells against exogenous and endogenous insults elicited by oxidative stress, which impairs cell function by damaging intracellular lipids, proteins, nucleic acids, and carbohydrates (45). KEAP1 is a repressor of NRF2 located mainly in the cytoplasm, which acts as a substrate adaptor for Cul3-containing E3 ubiquitin ligase (108, 202, 280). E3 ubiquitin ligase is responsible for the ubiquitination of NRF2 followed by its degradation in the proteasomes, resulting in low cellular NRF2 levels under unstressed conditions (41, 145, 332).
KEAP1 contains five domains, including the amino-terminal N-terminal region, the Broad complex/Tramtrack/Bric-a-brac (BTB) domain, the intervening region (IVR), the six Kelch/double glycine repeats (DGR), and the carboxy-terminal (CTR) domain (Fig. 1B). The BTB domain interacts with Cul3 and is required for KEAP1 dimerization (347). The IVR contains several cysteine residues and regulates the activity of KEAP1 (207). The Kelch/DGR domain maintains the interaction between NRF2 and KEAP1 (191). The DGR region recognizes ETGE and DLG motifs in the Neh2 domain of NRF2 protein.
Under normal conditions, NRF2 interacts with KEAP1 through the Neh2 domains (DLG and ETGE motifs), which drive NRF2 to the Cul-3-RBx1-E3 (Cullin-3-ring box-1 E3) ubiquitin ligase complex for degradation (108, 202, 280). KEAP1 is a cysteine-rich protein; in humans, it contains 27 cysteine residues that can be modified by different oxidant and electrophilic compounds (54, 190). The highly reactive cysteine residues, Cys151, Cys171, Cys273, and Cys288, are located in the BTB-IVR domains of KEAP1 protein. Cysteines are susceptible to oxidation and modification (e.g., alkylation or Michael addition) of the cysteine thiols of KEAP1 and NRF2 during oxidative stress is a key step in NRF2 activation (184). Cys151 of KEAP1 was found to facilitate NRF2 activity, whereas Cys288 was shown to diminish NRF2 activity (94, 321). Modification of redox-sensitive cysteines (Cys119, Cys235) of NRF2 prevents KEAP1 recognition and binding (93). Modification of those residues alters the conformation of KEAP1, causing detachment of the DLG motif (low-affinity binding site) from the KEAP1-NRF2 complex. However, NRF2 remains in association with KEAP1 through the ETGE motif (high-affinity binding site), according to the hinge and latch model by Tong et al. (287, 288). This steric change, when the DLG motif of KEAP1 releases NRF2, prevents the degradation of NRF2.
During cellular stress, Sekhar and co-workers (251) suggested that the binding between KEAP1 and Cul3 can also be disrupted, leading to NRF2 escape from proteasomal degradation. NRF2 can also be degraded and eliminated by KEAP1-independent mechanisms with the help of β-TrCP, which can recognize and bind to phosphorylated NRF2 and trigger its ubiquitination and degradation (37, 223, 224). A third degradation procedure has been reported in cirrhotic liver, where endoplasmic reticulum stress triggers E3 ubiquitin ligase (namely, HRD1) interaction with NRF2 domains, promoting NRF2 elimination (313). Subsequently, free NRF2 translocates to the nucleus where it dimerizes with transactivation partners [for instance, Small Maf protein family (293) or Jun (289, 328)] and binds to the promoter region of EpRE/ARE-response genes, inducing their transcription (101, 219). After NRF2 exerts its effect in the nucleus, it is phosphorylated by tyrosine kinases that promote NRF2 nuclear export and cytosolic degradation (110).
Role of NRF2 in antioxidant defense
There are more than 200 NRF2-dependent target genes identified in humans (186, 346). A more recent study suggests that there might be more than 1000 genes regulated by NRF2 (186). The majority of NRF2-dependent target genes encode for detoxifying enzymes to overcome oxidative damage of DNA, proteins, and lipids, for example, genes involved in the synthesis of glutathione, antioxidant proteins, drug-metabolizing enzymes, transporters, and numerous other stress response proteins [for review, see Hayes and Dinkova-Kostova (92) and Suzuki et al. (277)] (Fig. 2).
FIG. 2.
The classical KEAP1/NRF2 signaling pathway. Under basal conditions, KEAP1 binds to NRF2 and links NRF2 to the KEAP1-Cul3-E3 ubiquitin ligase complex, resulting in ubiquitination and degradation of NRF2. In response to stress, KEAP1-NRF2 binding is disrupted, NRF2 is stabilized, and free NRF2 translocates to the nucleus, where it heterodimerizes with the small Maf proteins, binds to AREs, and induces the transcription of its target genes. ARE, antioxidant response element; Cul3, cullin 3; Gclc, glutamate-cysteine ligase catalytic subunit; Gclm, glutamate-cysteine ligase modifier subunit; Gsr1, glutathione reductase 1; GST, glutathione S-transferase; Gpx2, glutathione peroxidase 2; G6pd, glucose-6-phosphate dehydrogenase; Hmox1, heme oxygenase 1; Idh1, isocitrate dehydrogenase 1; Me1, malic enzyme 1; Mrp1, multidrug resistance-associated protein 1; NRF2 or Nfe2l2, nuclear factor E2-related factor 2; Nqo1, NAD(P)H quinone dehydrogenase 1; Pgd, 6-phosphogluconate dehydrogenase; Srxn1, sulfiredoxin 1; Txn, thioredoxin; Txnrd1, thioredoxin reductase 1. Color images are available online.
Oxidative, nitrosative, and electrophilic stress promote NRF2 stabilization and activity. NRF2 maintains the appropriate intracellular reduced glutathione (GSH)/oxidized glutathione (GSSG) ratio by maintaining glutathione synthesis and glutathione reduction. This is achieved through controlling the expression of glutamate-cysteine ligase catalytic (GCLC) and modifier (GCLM) subunits of the glutamate-cysteine ligase (GCL) complex, which collectively catalyze the rate-limiting step in GSH synthesis (99). NRF2 also regulates the expression of glutathione reductase 1 (Gsr1) (282). Further, NRF2 regulates the expression of the SLC7A11 (subunit of the cystine/glutamate antiporter system Xc−), which imports cystine into cells in exchange for glutamate, and hence, regulates cysteine and glutamate availability in cells (163). Finally, NRF2 regulates the pentose-phosphate shunt and through that NADPH availability that is needed for reducing GSSG (4, 35, 92, 159, 334).
Besides the regulation of glutathione levels, NRF2 controls the expression of a broad set of enzymes that play a role in the detoxification of H2O2, peroxide radicals, and oxidized thiols, including glutathione peroxidase 2 (Gpx2) (88), thioredoxin 1 (Txn1), thioredoxin reductase 1 (Txnrd1), sulfiredoxin 1 (Srxn1) (2, 90), and glutathione S-transferases (282). Production of NADPH, which is used as a cofactor by many antioxidant systems in redox reactions, is also under the control of NRF2. The expression of glucose-6-phosphate dehydrogenase (G6pd), 6-phosphogluconate dehydrogenase (Pgd), isocitrate dehydrogenase 1 (Idh1), and malic enzyme 1 (Me1) is NRF2 dependent. Another prominent cytoprotective enzyme regulated by NRF2 is heme oxygenase 1 (HMOX1), the enzyme catalyzing the breakdown of heme molecules (7) and catalase, which reduces H2O2 to water and oxygen.
Natural and synthetic inductors and inhibitors of NRF2 signaling
Other small-molecule inhibitors and activators are vital in fine-tuning NRF2 activity (184). In cancer, certain intermediates of cell metabolism can modulate the NRF2-KEAP1 system by introducing post-translational modifications into NRF2 or KEAP1 that impact NRF2 function or stability. For instance, the mutation of fumarate hydratase in papillary renal cell carcinoma leads to fumarate accumulation, which induces KEAP1 succination and NRF2 stabilization (209). The inhibition of glycolysis and concomitant build-up of glycolytic intermediates in several cell lines promote NRF2 activity through methylglyoxal-induced (elimination product of dihydroxyacetone phosphate and glyceraldehyde-3-phosphate) formation of a covalent bond between arginines and cysteines (so-called MICA modification) that leads to the dimerization of KEAP1 (23). In glucose-free conditions, a decrease in KEAP1 O-glycosylation with β-N-acetylglucosamine leads to NRF2 accumulation in breast cancer cells (30). Finally, glycation destabilizes NRF2 via increased proteasomal degradation in hepatocellular carcinoma (HCC) (245).
Several artificial NRF2 activators function by disrupting KEAP1-NRF2 interaction, including tert-butylhydroquinone (166) or RA839 (310). Removing KEAP1 from the KEAP1-NRF2 complex leads to stabilization and activation of NRF2. The inhibition of the proteasome (e.g., by MG132) can prevent the degradation of NRF2 (149). A food additive and pro-carcinogen, butylated hydroxyanisole, can induce NRF2 (179) and NAD(P)H quinone dehydrogenase 1 (NQO1), which boosts the expression of 8-Oxoguanine glycosylase (OGG1) (268). Increased OGG1 prevents the oxidative catabolism of 4-hydroxyestradiol (4-OHE2) to the catechol estrogen quinone metabolites, which readily reacts with DNA to produce depurinated adducts. Superphysiological concentrations of vitamin C can also decrease the nuclear/cytosolic NRF2 ratio, indicating NRF2 activation (200).
A nucleoside analog antimetabolite, Cordycepin, downregulates NRF2 signaling and, consequently, increases the cytoplasmic levels of oxygen-centered reactive species (ROS). Cordycepin was employed as a radiosensitizer in breast cancer in a preclinical study (56). Although this study did not elucidate a mechanism of action, Cordycepin may induce caveolin-1 (Cav-1) expression (119). Cav-1 can suppress the NRF2-elicited effects by reducing manganese-dependent superoxide dismutase (MnSOD) expression, an important downstream target of the NRF2 pathway (86). In agreement with this theory, epidemiologic nested case–control studies show that Cav-1 and NRF2/MnSOD are inversely expressed in cases of invasive ductal carcinoma, where low Cav-1 and high NRF2 and MnSOD expression are associated with lower 5-year survival rates and the poorest prognosis (86).
The n-6 and n-3 polyunsaturated fatty acids (PUFAs) serve as substrates for cyclooxygenases that synthesize PUFA-derived hormones. One of the final products of the cyclooxygenase-2 (COX-2) pathway, 15-deoxy-prostaglandin J2 (15d-PGJ2), is switched on by inflammatory stimuli (258). In breast cancer cells, NRF2 translocation to the nucleus is induced by 15d-PGJ2, resulting in enhanced interaction between NRF2 with ARE (161) to drive the expression of a variety of phase 2 detoxifying and antioxidant enzymes (131). Nuclear accumulation and the transactivation of NRF2 appears to be dependent on the presence of the characteristic cyclopentenone moiety in 15d-PGJ2 (161). The electrophilic α,β-unsaturated carbonyl group, constituting the cyclopentenone ring of 15d-PGJ2, is anticipated to bind covalently to specific cysteine residues of the KEAP1 molecule, resulting in the liberation and nuclear translocation of NRF2 (59).
There is a firm link between the etiology of several cancers and bacterial dysbiosis (65, 67, 76, 77, 98, 118, 150, 196). In breast cancer, bacterial metabolites (estrogen metabolites, short chain fatty acids, lithocholic acid [LCA], and cadaverine) were shown to play key roles in regulating the behavior of cancer cells by influencing breast cancer stemness, anticancer immunity, cancer cell migration, metastasis formation, epithelial-to-mesenchymal transitions, and cancer cell metabolism (109, 149, 150, 194–196). Apparently, the capacity of the microbiome to synthesize these metabolites is reduced in breast cancer as compared with controls (76, 150, 196). The secondary bile acid, LCA was shown to exert cytostatic properties on breast cancer cells. Further, bacterial LCA production is largely reduced in the early stages of breast cancer (196). LCA reduces NRF2 expression, and the consequent reduction in antioxidant defenses exerts cytostatic properties in breast cancer. This mechanism is ineffective in nontransformed cells (149).
Hormones regulating NRF2 signaling
Hormones, such as gonadotropins and sex steroids (follicle-stimulating hormone, luteinizing hormone, estrogen), can promote NRF2 signaling through the induction of ROS [measured by dichlorofluorescein (DCF)—please note that DCF autooxidation may produce reactive species and, hence, provide a false positive result (124, 236)] in ovarian cancer (167). Follicle-stimulating hormone can further activate hypoxia-inducible factor 1 (HIF1) and vascular endothelial growth factor (VEGF) to promote tumor angiogenesis (339). Hormone regulation of NRF2 was found in ovarian cancer and may also play a major role in the development of other hormone-related cancers such as prostate and breast cancer (122).
Noncanonical NRF2 activation
Noncanonical NRF2 activation is based on the presence of proteins that activate NRF2 transcription by inducing the disassembly of the NRF2-KEAP1 complex. Certain ETGE motif-containing proteins can compete with KEAP1 for dimerization with NRF2, to prevent NRF2 degradation. Proteins that activate NRF2 include Wilms tumor gene on the X chromosome (WTX, AMER1) (25), partner and localizer of BRCA2 (PALB2) (181), dipeptidyl peptidase 3 (DPP3) (89), Cyclin-dependent kinase 20 (CDK20) (300), sequestosome 1 (p62) (107), p21 (31), and additional proteins identified in (89). The spectrum of competing proteins might represent a divergence of oxidative stress-related and -unrelated cellular signals, resulting in NRF2-induced cytoprotection.
Although several noncanonical activators of NRF2 were described, only a few have been studied in the context of cancer etiology, including kidney (25), breast (178), and lung (89) cancers, and osteosarcoma (181). DPP3 contains an ETGE motif, which binds to the Kelch domain of KEAP1 and activates NRF2 (89) in breast cancer cells in response to ROS. Overexpression of DPP3 in MCF7 cells causes accumulation of NRF2 and promotes resistance to H2O2 and diquat (178). Hast et al. (89) performed TCGA-based data mining analysis in tumor gene expression datasets to assess DPP3 expression in relation to NFE2L2 and KEAP1. DPP3 expression and copy number were increased in NFE2L2 wild-type and KEAP1 mutant tumors as compared with NFE2L2 mutant or KEAP1 wild-type tumors. This implicates that DPP3 enhances NRF2 activity under conditions when KEAP1 activity is attenuated by hypomorphic mutations. Consequently, the function of NRF2 is amplified even though hypomorphic KEAP1 activity is still able to suppress NRF2. On the other hand, NRF2 mutations often lead to hyperactivation of NRF2 and eliminate the need for additional amplifying factors. Further, this phenomenon might apply to other ETGE-containing proteins, too.
More recent research (32) identified family with sequence similarity 129, member B (FAM129B) as another positive regulator of NRF2 signaling, active in several cancer cell lines. FAM129B contains DLG and ETGE sequences, and FAM129B expression correlates with poor clinical outcome in breast and lung cancers (32). Similarly, PALB2 competes effectively with KEAP1 for NRF2 binding and the overexpression of PALB2 induces NRF2 levels and transcription via interaction with the nuclear NRF2-KEAP1 complex (181). The depletion of PALB2 in vitro impairs NRF2 function.
Another example of noncanonical activation of NRF2 is p21, a cyclin-dependent kinase inhibitor. In the cell model of colorectal cancer, p21 can recognize and directly bind to the DLG/ETGE motif of NRF2 to prevent NRF2 degradation. Hence, the expression of antioxidant genes, such as HMOX1 an NQO1 (31), in response to H2O2 treatment is enhanced. NRF2, in turn, can elevate p21 expression by binding to the promoter region of p21, as demonstrated in a study focused on lung carcinoma with deficient p53 (111).
During the initiation of the autophagy cascade, p62 binds to KEAP1 and initiates NRF2 transactivation of target genes. As an autophagy substrate, p62 can also interfere with NRF2-KEAP1 interaction through binding to the DLG/ETGE motif and activating NRF2 (107). The role of p62 in NRF2 transactivation was confirmed in hepatic adenoma (107), HCC (261), breast and ovarian cancers (233, 315), and in glioma (340). In contrast, in normal hepatocytes, NRF2 is degraded under the same conditions (317). The expression of p62 and oligomerization in the cytosol is also a negative prognostic factor in HCC recurrence (317). In breast cancer, p62-dependent NRF2 activation induces the proportions of CD44-positive cancer stem cells (CSCs) that are associated with an aggressive phenotype with enhanced tumor growth and elevated therapeutic resistance (239).
Role of NRF2 in Cancer
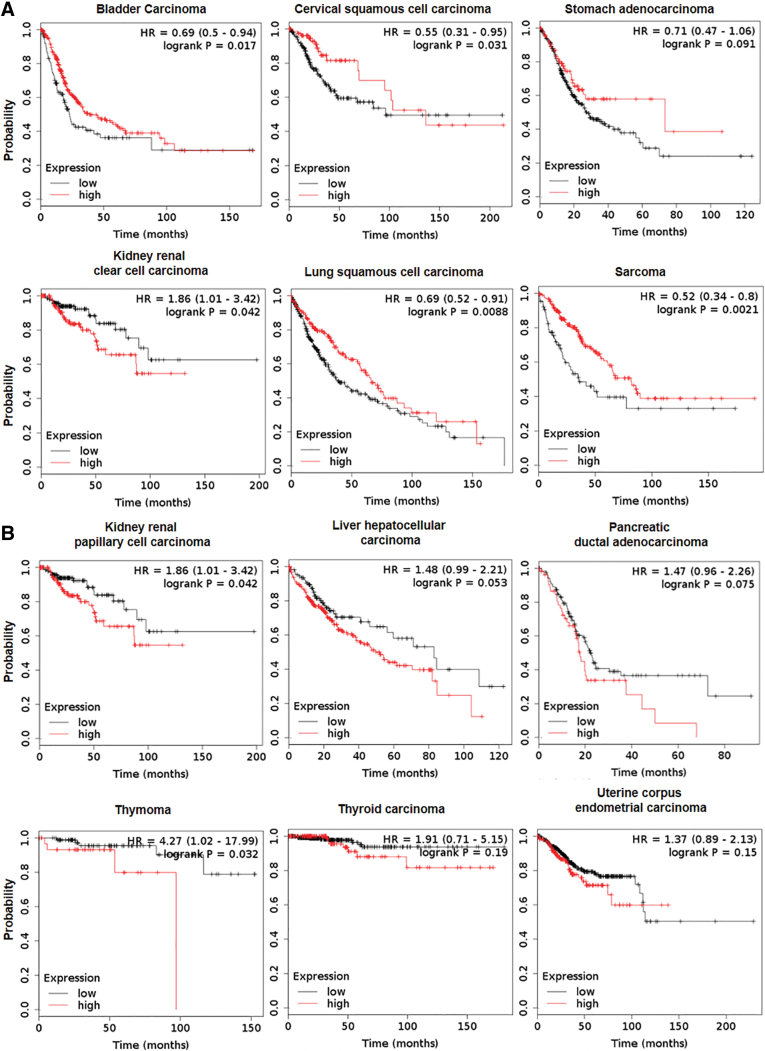
Cancers and cancer cell behavior can be characterized by cancer hallmarks (85), which contribute to the ability to sustain proliferation and to evade cell death. NRF2, as a strategic cytoprotective factor, positively regulates several cancer hallmarks, including the promotion of metastasis, evasion of cell death, and metabolic changes. In normal cells, activation of NRF2 complies with the period of acute detoxification; whereas in cancer cells, a newly described phenomenon called “NRF2 addiction” occurs (144). NRF2 addiction refers to sustained NRF2 signaling due to genetic, epigenetic, or general oncogenic signaling. There is growing experimental evidence that NRF2 activation enhances cancer growth and increases therapy resistance (14, 177) due to the expression of phase 1 and phase 2 drug-metabolizing enzymes (92) and multidrug-resistance-associated transporters (Mrps) (185), which support metastasis formation (100, 246), cancer growth (52, 232), and poor survival (115, 178) (Fig. 3 and Table 1). It is interesting to note that, despite the large number of examples where NRF2 activation is associated with negative outcomes, NRF2 overexpression can support longer survival in certain cancers (58, 75, 80, 95, 128, 156, 172, 180, 277) (Fig. 4). In this article, we will review the molecular prerequisites of aberrant NRF2 signaling in cancer.
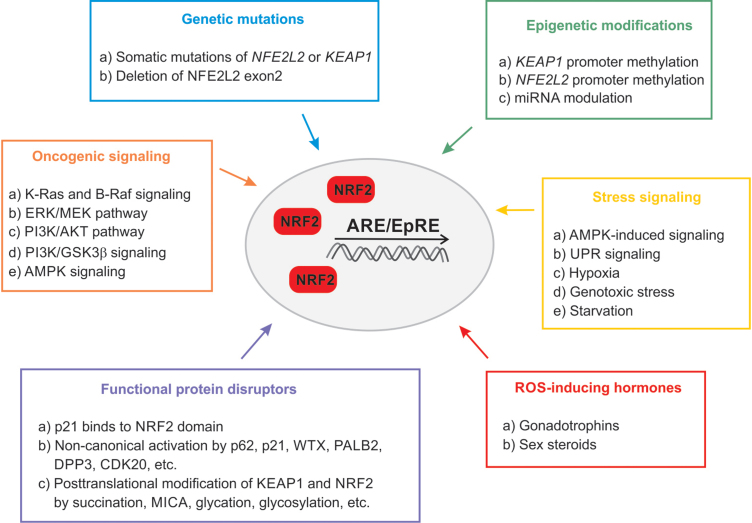
FIG. 3.
Mechanisms involved in the aberrant activation of KEAP1/NRF2 pathway in cancer. Color images are available online.
Table 1.
Aberrant Activation of NRF2 in Cancer and NRF2-Addicted Cancer Types
| Tissue or organ | Cancer/cell type | Mechanism of action | Reference |
|---|---|---|---|
| Nerve system | Malignant glioma | Hypermethylation of KEAP1 promoter | (201) |
| Neuroblastoma | miR-7 inhibits KEAP1 expression | (123) | |
| miR-144 inhibits NRF2 and antioxidant gene expression | (341) | ||
| Glioma | p62 disrupts KEAP1-NRF2 complex | (340) | |
| Endoplasmatic reticulum stress induces NRF2 | (27) | ||
| Bone | Osteosarcoma | PALB2 disrupts KEAP1-NRF2 complex | (181) |
| Breast | Breast carcinoma | NRF2 promotes cell survival | (178) |
| LOF mutation of KEAP1 | (203, 271) | ||
| Hypermethylation of KEAP1 promoter | (16) | ||
| miR-28 inhibits NRF2 expression | (324) | ||
| miR-93a supports malignant transformation | (269) | ||
| miR153 supports malignant transformation | (296) | ||
| PI3K/AKT influence NRF2 signaling | (239) | ||
| AMPK induces NRF2 target gene expression and modulate autophagy | (294) | ||
| miR-200a targets KEAP1 and induces NRF2 | (58) | ||
| Hypermethylation of KEAP1 promoter | (84) | ||
| Overexpression of p62 facilitate tumorigenesis | (233) | ||
| Colon | Colorectal carcinoma | Hypomethylation of NRF2 promoter | (125) |
| PI3K/AKT influence NRF2 signaling | (175) | ||
| LOF mutation of KEAP1 | (271) | ||
| Head and neck | Head and neck cancer | Cul3 copy number loss | (187) |
| Esophageal carcinoma | Cul3 copy number loss | (170) | |
| miR-144 directly suppresses NRF2 | (320) | ||
| Deletion of NRF2 exon 2 | (142) | ||
| Genetic mutation of NRF2 affect DLG/ETGE motif | (142, 257) | ||
| Hematologic malignancies | AML | Stress signaling induces NF-κB signaling | (238) |
| Endoplasmatic reticulum stress induces NRF2 signaling | (8) | ||
| Deletion of NRF2 exon 2 | (78) | ||
| miR-144-3p suppresses NRF2 activation | (274) | ||
| Lung | NSCLC | Hypermethylation of KEAP1 promoter | (301) |
| Lung cancer | miR-155 promotes malignant transformation | (28) | |
| p21 disrupts KEAP1-NRF2 connection | (111) | ||
| DPP3 disrupts KEAP1-NRF2 connection | (89) | ||
| CDK20 disrupts KEAP1-NRF2 connection | (300) | ||
| K-RAS promotes ERK/MEK pathway | (279) | ||
| PI3K/AKT influences NRF2 signaling | (197) | ||
| Liver | Hepatocellular carcinoma | Genetic mutation of NRF2 affects DLG/ETGE motif | (81) |
| miR-144 directly suppresses NRF2 | (344) | ||
| p62 disrupts KEAP1-NRF2 complex | (261) | ||
| AICAR induces NRF2 activity | (264) | ||
| p62 disrupts KEAP1-NRF2 complex | (107) | ||
| Skin | Squamous cell carcinoma | Deletion of NRF2 exon 2 | (142) |
| Pancreas | Pancreas adenocarcinoma | Hypermethylation of KEAP1 promoter through URHF1 | (3, 331, 337) |
| Prostate | Prostate cancer | Hypermethylation of NRF2 promoter | (105, 135) |
| Kidney | Papillary renal cell carcinoma | Hypermethylation of KEAP1 promoter | (61) |
| Genetic mutation of NRF2 affects DLG/ETGE motif FH disrupts KEAP1-NRF2 connection |
(208, 209) | ||
| Wilms tumor | K-RAS promotes ERK/MEK pathway | (13) | |
| Cell model of renal cell carcinoma | miR-148b represses NRF2 and enhances ROS | (220) | |
| Clear renal cell carcinoma | WTX disrupts KEAP1-NRF2 connection | (25) | |
| Ovary | Ovarian cancer | Cul3 copy number loss | (188) |
| Hormone-induced NRF2 overexpression | (144, 339) | ||
| p62 disrupts KEAP1-NRF2 complex | (315) | ||
| Endoplasmatic reticulum stress induce FGFR4/GSK3β/NRF2 axis | (167) |
AKT, protein kinase B; AML, acute myeloid leukemia; AMPK, AMP-activated protein kinase; BRCA2, breast cancer type 2 susceptibility protein; CDK20, cyclin-dependent kinase 20; DPP3, dipeptidyl peptidase 3; FH, fumarate hydratase; GSK3β, glycogen synthase kinase 3β; LOF, loss-of-function; MEK, mitogen-activated protein kinase kinase; NSCLC, nonsmall-cell lung carcinoma; p62, sequestosome 1; PALB2, partner and localizer of BRCA2; PI3K, phosphoinositide 3-kinases; ROS, oxygen-centered reactive species; WTX, Wilms tumor gene on the X chromosome.
FIG. 4.
Tumor NRF2 expression affects survival. Patient survival data were retrieved from the pan-cancer RNAseq segment of the kmplot.com database (154). (A) Tumors where high expression of NRF2 prolongs survival. (B) Tumors where high expression of NRF2 shortens survival. The y axis represents the probability for survival, whereas the x axis represents time in months. Numerical values for the Kaplan-Mayer survival curves can be found in Table 3. The red line depicts the high expression quartile of the population, while the black line depicts the low expression quartile. Color images are available online.
Genetic mutations in KEAP1 and NRF2
The occurrence of somatic mutations in KEAP1, NRF2, or Cul3 is the most well-known mechanism of sustained NRF2 activation in cancer (193). According to public genomic databases, KEAP1 and NFE2L2 seem to be the most affected genes in lung cancer, followed by esophageal and endometrial cancer, as summarized (134). Studies assessing statistics of genetic alterations in lung cancer (226) demonstrate frequent loss-of-function (LOF) mutations in KEAP1 (213) and loss-of-heterozygosity at the genetic locus of KEAP1 (266). KEAP1 missense or nonsense mutations have been also identified in malignant melanoma (198), thyroid cancer (44), HCC (38), endometrial carcinoma (311), and breast (203, 271) and ovarian cancers (148). In summary, mutations of KEAP1 that confer loss of function or hypomorphic function induce NRF2 activity.
Alleles of NRF2 that lead to its activation have been described as well. The genetic mutation of NFE2L2 affecting the DLG/ETGE motif of the Neh2 domain was shown in a lung cancer cell line (259), renal cell carcinoma (208), HCC (81), skin cancer (142), and esophageal carcinoma (257). A unique genetic mutation, deletion of the NFE2L2 exon 2, was found to stabilize NRF2 and enhance its activity in nonsmall-cell lung carcinoma (NSCLC) and head-neck cancer (78). Moreover, copy number loss of Cul3 or inactivating mutation of RBx1, which controls NRF2 degradation, was shown in esophageal (170) and ovarian cancer (188), as well as in thyroid (187) and renal cell carcinoma (208).
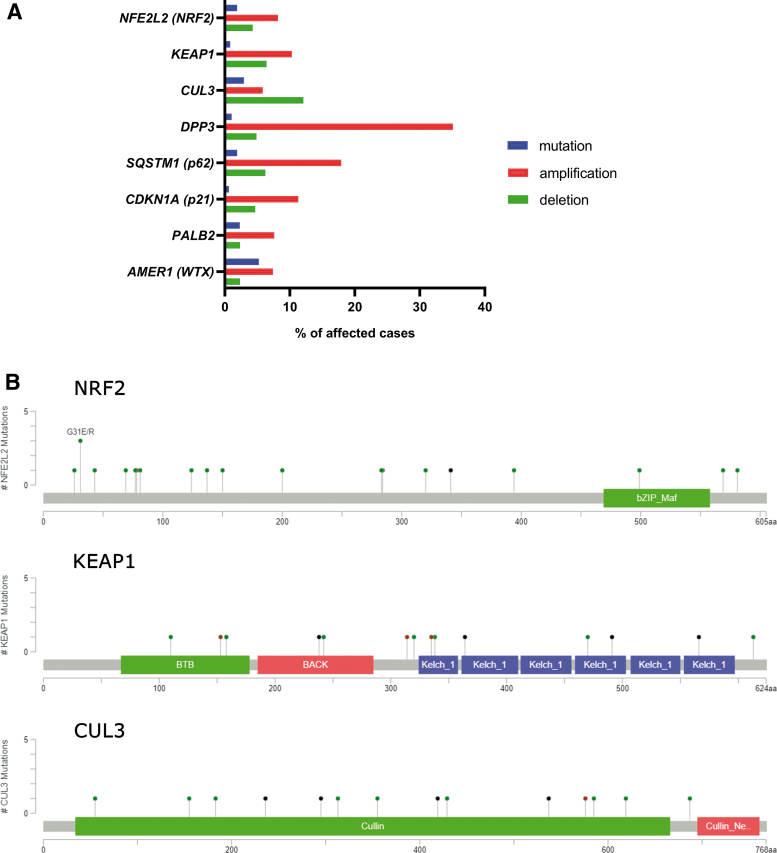
A recent study identified two single nucleotide polymorphisms of KEAP1 associated with shorter relapse-free survival in breast carcinoma (87). A study by Kovács et al. (149) showed that NRF2 overexpression and the downregulation of downstream pro-oxidant genes negatively correlates with patient survival in nontriple negative breast cancer. Our search in the TCGA-BRCA dataset revealed prominent amplification of DPP3 and p62 genes, which suggests amplification of NRF2 activity in breast cancer by noncanonical mechanisms (Fig. 5).
FIG. 5.
Genetic aberrations of NRF2 signaling pathway members in breast carcinoma. (A) Chart depicting percentages of mutations and copy number variations (amplification and deletion, respectively) of NRF2, KEAP1, CUL3, and several genes involved in noncanonical activation of NRF2 as discussed in the Strategic role of p62 in NRF2 signaling: autophagy and metabolic rewiring section. The data are based on the TCGA portal search (data set TCGA BRCA, Ductal and Lobular Neoplasms, containing 513 cases). (B) Mutation maps (lollipop plots) depicting positions of the mutations in NRF2 (NFE2L2), KEAP1, and CUL3 proteins. The data are based on a cBIOPORTAL search, combining several studies containing data on breast carcinoma samples. Color images are available online.
Epigenetic modifications of KEAP1 and NRF2
Epigenetic modification in KEAP1 or NFE2L2 promoter regions can also contribute to NRF2 stabilization in many cancers (122). Both hypo- and hypermethylation of the NFE2L2 promoter have been reported. Hypomethylation, due to the reduced expression of EZH2 methyltransferase, is associated with NRF2 overexpression in colon cancer (125). Hypermethylation of the NFE2L2 promoter is associated with prostate cancer (135) and leads to reduced NRF2 activity, a rare example among malignant diseases. Bromodomain protein 4 (BRD4) was implicated in the transcriptional regulation of KEAP1. Thus, BRD4 might represent a strong modulator of the NRF2/KEAP1 pathway in prostate cancer (105).
Hypermethylation of the KEAP1 promoter (CpG islands) is catalyzed by methyl-CpG-binding domain protein 1 and ubiquitin-like, containing PHD and RING finger domains-1 (UHRF1), leading to reduced KEAP1 protein levels and augmented expression of NRF2-dependent genes (3, 331). KEAP1 hypermethylation is associated with lung (301), colorectal (84), breast (16), prostate (337), thyroid (187), and renal (61) cancers, as well as malignant glioma (201).
The regulatory role of microRNAs in NRF2 signaling
NRF2 and KEAP1 levels can be modulated at the post-transcriptional level through altering mRNA splicing or microRNA (miRNA) levels. In the breast cancer cell line, MCF-7, miR-28 inhibits NRF2 action without the contribution of KEAP1 (324). The miRNAs, miR-507, miR-634, miR450a, miR129–5p miR-340, miR-146b, and miR144, were reported to directly suppress NRF2 activation in different cancer types, including acute myeloid leukemia (AML) (274), breast cancer (104), and esophageal cancer (320). In HCC, miR-144 suppressed NRF2 levels (344); whereas in neuroblastoma, the same miRNA induced ROS-induced apoptosis (341). Recent studies demonstrate that miRNAs promote migration and support malignant transformation in different neoplasias. For example, miR-155 promotes lung cancer (28) and miR93a and miR153 drive the malignant transformation of the breast (269, 296). Last but not least, NRF2 can influence and modulate the expression of numerous miRNAs in tumors, creating a complex feedback-regulatory system in humans (253).
Several miRNAs were shown to modulate KEAP1, NRF2, and target gene translation or transcription. The miRNAs, miR-432-3p and miR-200a, impair KEAP1 translation in esophageal and breast carcinoma (6, 58); whereas miR-7 and miR-148b increase the expression of NRF2-dependent antioxidant genes (HMOX1 and GCLM) in neuroblastoma (123) or elevate ROS production due to the repression of HIF1 and NRF2 expression (220), respectively.
Oncogenic signaling
Although NRF2 activity promotes cancer progression by various mechanisms, as previously discussed, we should emphasize that NRF2 accumulation can only contribute to malignant proliferation in the presence of active oncogenic signaling and, therefore, we refer to its role as the oncogenic driver. Keap1-deficient mice develop cancer only in the presence of oncogenic mutations, such as those in Kras/Hras or Tp53 (113, 234). The contribution of NRF2 to carcinogenesis differs between cancer types. For example, the Kras:Tp53:Keap1 triple mutation is associated with an aggressive phenotype in lung carcinoma in ∼30% of cases, whereas in the pancreas this combination of mutations does not initiate cancer but leads to fibrosis (83).
Recent evidence suggests that cancer-associated signaling pathways can influence the activity of NRF2 by increasing its mRNA level rather than improving its stability. In this respect, K-Ras and B-Raf signaling pathways can enhance NRF2 mRNA levels to promote ROS elimination (52). K-Ras promotes NRF2 transcription through the extracellular signal–regulated kinase (ERK)/mitogen-activated protein kinase kinase (MEK) pathway in NSCLC (279) and renal carcinoma (13). The importance of Ras signaling is highlighted by the findings that H-Ras-activated HO-1 overexpression is abolished by ERK inhibitors or NFE2L2 knockdown (13). In K-Ras-associated malignancies, the elevated expression of antioxidant genes (HMOX1 and NQO1) is associated with an aggressive phenotype, whereas the elimination of NRF2 disrupts malignant progression and restores sensitivity to cancer therapy (254). NRF2 signaling also actively improves cancer survival by maintaining independence of growth factors in K-Ras-associated malignancies; in pancreatic ductal adenocarcinoma (PDAC), NRF2 signaling is upregulated, resulting in improved cancer survival via maintaining phosphorylated levels of EGFR and ERBB2, and by maintaining intact mRNA translation efficiency (33). Interestingly, NFE2L2 knockdown in normal organoids was tolerated, whereas NRF2 knockdown was detrimental in tumor-derived organoids.
The nuclear accumulation of NRF2 increases when different proliferative pathways are active (263, 278). Phosphoinositide 3-kinases (PI3K)/protein kinase B (AKT) activation is associated with elevated NRF2 mRNA levels and nuclear accumulation of NRF2, leading to increased cell proliferation, survival, and metabolic reprogramming (197). In breast cancer, PI3K/AKT activation, in response to estradiol, is accompanied by elevated expression of NRF2 and, consequently, increased expression of a large set of antioxidant genes (79). As mentioned earlier, GSK3 can phosphorylate NRF2 and induce its degradation by the Cult1-βTrCP complex. When the PI3K-AKT pathway is active, AKT phosphorylates and inactivates GSK3, leading to inactivation of Cult1-βTrCP-driven NRF2 degradation in cancer cells (37). Further, in Keap1-deficient mice, PI3K-AKT activation leads to NRF2 nuclear accumulation and NRF2-driven cell proliferation of hepatocytes (263).
NRF2 in unfolded protein response
Accumulation of unfolded proteins in the lumen of endoplasmic reticulum, termed endoplasmic reticulum stress, activates the unfolded protein response (UPR) (235). Endoplasmic reticulum stress and UPR are frequently found in tumor cells due to nutrient deficiency, hypoxia, and exposure to therapeutic agents. The association of UPR with the KEAP1/NRF2 pathway has been investigated in both cancer cells and normal cells. Starvation activates NRF2 through AMP-activated protein kinase (AMPK) and through protein kinase R-like endoplasmic reticulum kinase (PERK), which is located in the endoplasmic reticulum membrane and induced during UPR (40–42). NRF2 is phosphorylated by PERK, which leads to the dissociation of the NRF2/KEAP1 complex and increased expression of ARE genes to promote survival via reduced ROS levels (42).
In cancer, the PERK-NRF2 pathway drives endoplasmic reticulum stress-resistance and multidrug resistance as demonstrated in colorectal cancer (243), multiple myeloma (276), and AML cells (8). Further, in oral (tongue) squamous carcinoma cells, heat shock 70 kDa protein 5 (GRP78) and PERK-dependent activation of NRF2 induced Warburg-type metabolism and maintained cancer-initiating cells independently of ROS production (27). Moreover, fibroblast growth factor 19, which is overexpressed in HCC, can promote the cytoprotective FGFR4/GSK3β/NRF2 axis to protect against endoplasmic reticulum stress (281).
NRF2 regulating specific aspects of cancer metabolism
Cancer metabolism is usually perceived in the context of maintaining high energy standards for growing cells and includes enhanced catabolic pathways, enhanced biosynthetic pathways, and maintenance of redox homeostasis associated with high substrate turnover. The primary function of NRF2 is still the regulation of oxidative damage. However, since a number of NRF2 targets are rate-limiting enzymes in the respective pathways, the function of NRF2 logically encompasses metabolic rewiring also. Indeed, several metabolic pathways in NRF2-overexpressing cancer cells have been described as elevated, including the pentose-phosphate pathway (PPP), and nucleotide-, glutathione- (234), and serine synthesis in a lung cancer (51) model. In this section, we draw attention to certain metabolic pathways that tend to be affected by NRF2 signaling as a primary or secondary function, and we evaluate the role of NRF2 in cell survival based on metabolic rewiring (Fig. 6).
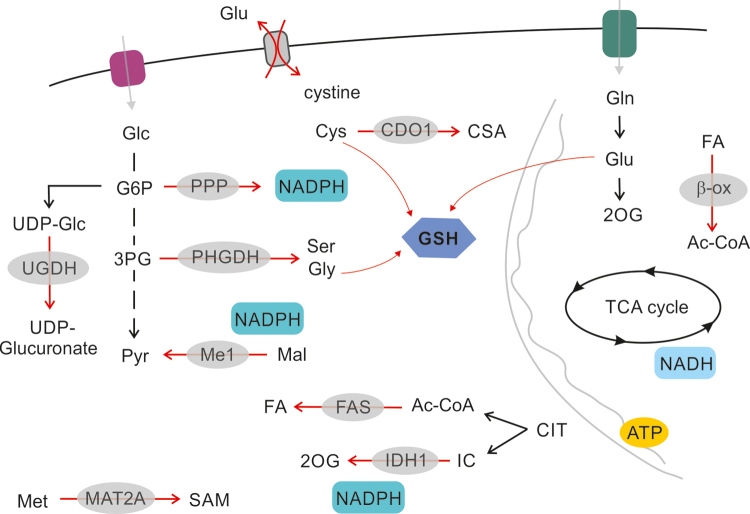
FIG. 6.
The role of NRF2 in regulation of cancer cell metabolism. NRF2, activated through canonical or noncanonical pathways, regulates carbon utilization. Glycolytic flux at the level of G6P is driven toward UDP-Glc and glucoronate synthesis, as well as toward the PPP cycle, to support the synthesis of ribose and NADPH. Glycolytic flux supports Ser and Gly synthesis by upregulating PHGDH. Glutathione synthesis is supported by cystine import and glycine synthesis. Cystine import is connected to glutamate export by Xc− transporter. Fatty acid synthesis and fatty acid oxidation (β-ox) are both regulated by NRF2. NRF2 also regulates synthesis of polyamines by regulation of MAT2A expression. Pathways upregulated by NRF2 signaling are designated by red arrows. G6P, glucose-6-phosphate; Gly, glycine; PPP, pentose-phosphate pathway; Ser, serine; UDP-Glc, UDP-glucose. Color images are available online.
Strategic role of p62 in NRF2 signaling: autophagy and metabolic rewiring
A considerable body of evidence points toward a link between NRF2 and autophagy, which represents a conserved strategy to overcome starvation and nutrient deprivation (162, 329) by recycling cellular macromolecules and organelles for essential metabolic substrates (222). During autophagy, p62 functions as a marker of proteins and organelles that are targeted for degradation by the autophagosome. The NRF2 signaling pathway is activated by p62 through the noncanonical mechanism connecting NRF2 to a spectrum of conditions related to starvation.
In HCC, Parkin-induced autophagy promotes p62 phosphorylation on Ser349 by a mechanistic (or mammalian) target of rapamycin complex 1 (mTORC1), leading to a concomitant rise in NRF2 protein expression (106). In fact, p62 binds to the STGE motif of KEAP1 with low affinity, which increases when p62 is phosphorylated. Hence, phosphorylation of p62 effectively induces NRF2 expression during autophagy (146). The p62-dependent NRF2 activation also occurs in a carfilzomib- (proteasome inhibitor) resistant model of multiple myeloma (228).
Other reports contradict the induction of NRF2 in autophagy by p62. A Crispr-based screen focused on finding novel negative regulators of NRF2 signaling (133) also identified a number of autophagy-related genes in human kidney HK2 cells. However, subsequent functional analyses demonstrated that changes in the phosphorylation of p-p62 do not necessarily induce NRF2 in HK2 cells, indicating that the autophagy-NRF2 loop can proceed in a p62-independent manner. In addition, the p62-NRF2 axis is not activated in a TCS2-deficient kidney cancer model, where upregulated levels of p62 do not activate NRF2 (153). Thus, NRF2 function in autophagy requires tissue- and context-specific evaluation. On the other hand, in Atg7-deficient mice, NRF2 activation remains p62 dependent and enhances cellular growth of HCC in 3D cultures (107).
The p62-NRF2 axis also interacts with signaling pathways through other pathways besides mTORC1. In HCC, H2O2 and hypoxia-induced ROS accumulation and NRF2 activation are p62 dependent (317). In this case, p62 is activated by phosphorylation at S28 by ketohexokinase-A (KHK-A), a splice isoform of phosphofructokinase that possesses protein kinase activity and KHK-A activation depends on AMPK activation (317). These changes collectively lead to NRF2-mediated gene expression and contribute to the development of HCC in xenograft animals (317). AMPK, a well-known energy sensor, can also influence NRF2 activity in breast cancer directly, as AMPK phosphorylates NRF2 and facilitates its nuclear accumulation (120). In MCF-7 and T47D breast cancer cells, AMPK activity, induced by glucose deprivation during nutrient starvation, is necessary for the expression of NRF2-driven genes (294). In addition, in breast cancer, a positive feedback loop between p62 and NRF2 has been demonstrated. The S349 residue of p62 can be phosphorylated by class III PtdIns3 lipid kinase (VPS34), whereas VPS34 induces NRF2-dependent expression of p62 and induces the noncanonical activation of NRF2 (117). Moreover, VPS34 promotes PKCδ-dependent phosphorylation of p62, also on S349, that not only strengthens the interaction between NRF2 and p62 in response to caspase inhibitor treatment but also activates downstream signaling pathways leading to tumor formation, including the MEK/ERK pathway (117).
In terms of generic carbon rewiring for energy metabolism, HCC expressing pS349–p62, reprogram both glucose and glutamine metabolism, marked by the induction of the glucuronate pathway and glutamine-derived glutathione synthesis in an NRF2-dependent manner (242). The resulting increase in glutathione levels is vital to support cancer growth and chemoresistance. Activation of NRF2 is also critical for the survival of breast cancer cells in glucose-deprived conditions, as NRF2 induction protected cells from autophagy and oxidative stress (294). Under glucose-free conditions, NRF2 enhances cellular survival in a p62-independent fashion (248). The authors suggest that the mechanism is initiated by ATP deficiency followed by increases in the levels of NQO1 (248). In accordance with this suggestion, reduced ATP induces NRF2 activity, causing an alteration in redox balance, as recently described in lung carcinoma cells (248).
To summarize, a substantial body of evidence supports the notion that NRF2 can be upregulated by p62, which connects NRF2 signaling to various means of signal transduction, such as AMPK and mTORC1. Consequently, NRF2 induction supports growth in a spectrum of cancer types.
NRF2 regulated ferroptosis and metabolic rewiring through SLC7A11 expression
Ferroptosis is a form of programmed cell death that is dependent on iron and independent of caspases, which is induced specifically by oxidative stress (55). In principle, inhibition of SLC7A11, depletion of intracellular cysteine and glutathione, and lipid peroxidation suppress GPX4 activity and initiate ferroptosis. NRF2 regulates the expression of genes involved in the prevention of ferroptosis, including iron metabolism, NADPH synthesis, glutathione reduction, and nonessential amino acid interconversion. The SLC7A11 gene codes for xCT protein, a subunit of a membrane-localized cystine-glutamate antiporter (Xc−), which imports a cysteine dimer (cystine) in exchange for glutamate. SLC7A11 is a direct target of NRF2, activating transcription factor 4 (ATF4) (286), and ATF3 (298) and is also a prominent target of NRF2 signaling and ferroptosis in a number of malignancies.
Consistent evidence demonstrates that the NRF2 transcriptional response, involving the expression of SLC7A11 and xCT, is protective against ferroptosis. This implies that NRF2-dependent transcription programs lead to escape from ferroptosis and, in turn, support cell growth. xCT is induced by H2O2 and NRF2 activation in MCF7 breast cancer cells, with a concomitant increase in GSH synthesis (82). Also, in KrasG12D, TP53−/− lung adenocarcinoma cells, the expression of the KEAP1 LOF mutant induces SLC7A11, which boosts GSH and reduces ROS levels (247). Moreover, the repression of xCT impairs colony formation in Ras-transformed cell models (169). As a result, xCT induction due to NRF2 signaling confers a refractory phenotype in response to the ferroptosis-inducing agents, erastin or RSL3, in glioma cell models (62). Further, NRF2 expression coincides with worse clinical outcome in glioma patients. Recent evidence also indicates that depletion of ovarian tumor family member deubiquitinase (OTUB1) (174) or the inactivation of breast cancer type 1 susceptibility protein (BRCA1) associated protein 1 (BAP1) tumor suppressor in breast cancer (338) leads to induction of xCT in an NRF2/hypoxia-inducible factor 1α (HIF1α)-dependent fashion, which protects cancer cells from ferroptosis.
It should be noted that a high level of NRF2 accumulation, which is achieved by the functional impairment of KEAP1 or NRF2 hyperactivation, combined with sustained oncogenic signaling, allows NRF2 to modulate metabolism under pathological conditions. NRF2 redirects glucose and glutamine into anabolic pathways in metabolic reprogramming (197). In addition, xCT alters cellular glucose and glutamine utilization to promote cell survival (Fig. 7). The tendency of cells to prefer glucose and glycolysis in response to xCT expression level was demonstrated in several cell models (114). The knockdown of SLC7A11 rescues viability in glucose-free conditions. Inhibition of xCT in nonsmall lung cancer cells improves survival in glucose-free medium, when bioenergetics rely primarily on mitochondrial OXPHOS through glutamine oxidation (114). Increased xCT expression sensitizes breast cancer cells to glucose, but not glutamine withdrawal (262). Thus, the export glutamate via the Xc− system due to the expression of SLC7A11 maintains glucose addiction for survival.
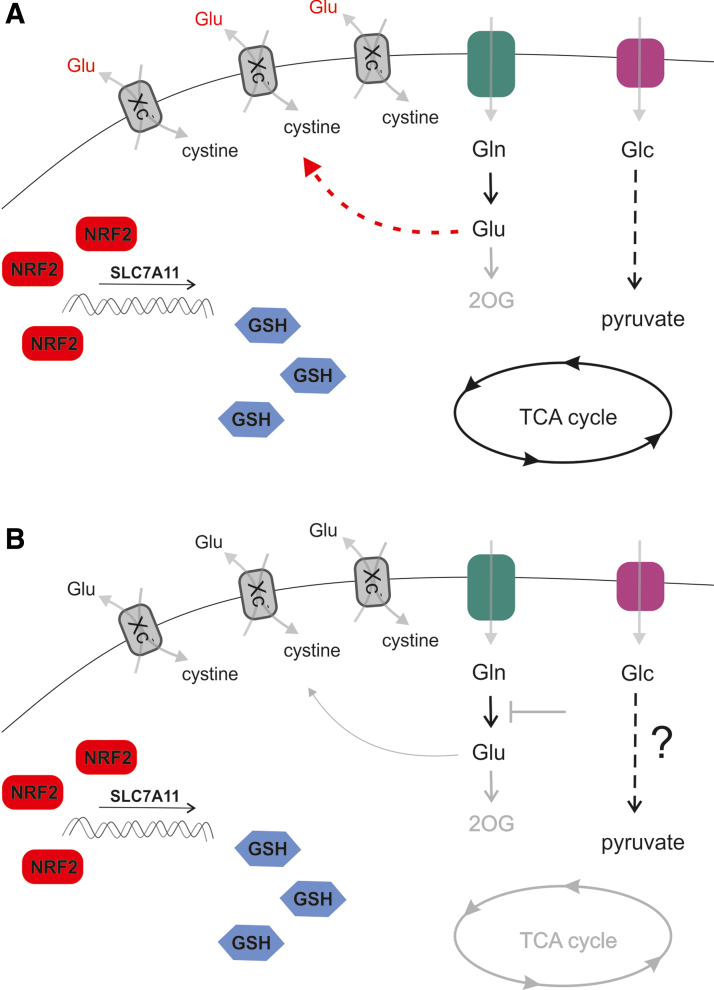
FIG. 7.
NRF2 regulates metabolism by expression of SLC7A11. SLC7A11 is a prominent target of NRF2 signaling in cancer cells. Upregulation of NRF2 results in expression of Xc−, an antiporter importing cystine, which results in enhanced antioxidant defense by GSH synthesis. Glutamate is exported in exchange for cystine, which compromises glutaminolytic flux. A subset of cancer cells are able to support the TCA cycle (A), whereas another subset of cancer cells are dependent on glutaminolysis and 2OG anaplerosis, because they are unable to supply the TCA cycle from glycolysis (B). Glutamine auxotrophs are, therefore, susceptible to glutaminolysis inhibitors, such as CB839, when NRF2 signaling is upregulated. GSH, reduced glutathione; TCA, tricarboxylic acid cycle. Color images are available online.
Comparing low- and high-expressing SLC7A11 cell lines revealed that OXPHOS-related gene expression negatively correlates with the expression level of SLC7A11 in breast cancer. In turn, xCT expression improved GSH levels and ROS homeostasis, while diminishing the ability of breast cancer cells to utilize glutamine instead of glucose (262).
A study that grouped breast cancer cell lines based on the ability to proliferate in glutamine-free conditions revealed that cells overexpressing SLC7A11 are sensitive to glutamine withdrawal, as glutamate is excessively exported (284). This offers the possibility to target both Xc− and glutamine import in glutamine auxotrophs (284). Another subset of malignancies is unable to upregulate glycolysis to compensate for loss of glutamine as a carbon source (Fig. 7). In accordance with these data, studies using KRASG12V, TP53−/− cell models of lung adenocarcinoma expressing an LOF form of KEAP1, demonstrate the vulnerability of xCT overexpressing cells to glutamine withdrawal. These cell models also have an increased sensitivity to glutaminase 1 inhibitor CB-839 (247), which can be overcome by pretreatment with erastin or knockdown of SLC7A11. In this case, active export of glutamate limits mitochondrial anaplerosis, which, in turn, sensitizes the cells to CB-839 treatment. Thus, a combination of KI696, a compound inhibiting KEAP1-NRF2 interaction, with CB-839 is another plausible strategy to target NRF2 overexpressing cancers. In support of this therapeutic approach, another line of evidence also documents enhanced glutamine dependence due to KEAP1-NRF2 in the KRAS/LKB1/KEAP1 transformed lung cancer (72). Further, a recent study (157) also demonstrated that the NRF2-mediated induction of the Xc− system limits intracellular glutamate content that, consequently, leads to an external dependency on nonessential amino acids (serine, glycine, and asparagine) in cancer cells, rendering cells metabolically inflexible. The antioxidant capacity of cancer cells, indeed, shows an inverse correlation with nonessential amino acids synthesizing capacity. As cells are reliant on nonessential amino acids, a diet that avoids these amino acids can limit cancer cell proliferation.
A recent publication (126) suggested that the NRF2-mediated accumulation of cysteine in nonsmall cell lung cancer may induce metabolic liability. The mechanism of metabolic liability is that cystein in superphysiological concentrations becomes a substrate for cysteine dioxygenase 1 (CDO1), creating cysteine sulfinic acid that apparently hampers cellular proliferation. From a broader perspective, this metabolic inflexibility can be exploited in combating nonsmall cell lung cancer.
NRF2 interference with hypoxia signaling and neovascularization
Hypoxia, a common feature of the tumor microenvironment, leads to the stabilization of HIF1α, which modulates cellular metabolism as an adaptive response (252). HIF1α expression initiates transcriptional programs that lead to survival and overcome hypoxia by inducing angiogenesis, glycolysis, and a decrease in pyruvate oxidation in mitochondria. NRF2 activation supports glycolysis by promoting the stabilization of transcription factor BTB and CNC homology 1 (Bach1) that supports the activity of hexokinase (168, 309). This effect of NRF2 is mediated by induction of HMOX1, which catabolizes heme, the main inhibitor of Bach1. Both NRF2 and HIF1α are critical factors for sensing O2 and are dependent on each other when responding to oxygen tension changes or ROS production. Of note, HIF1α is probably a target of NRF2; an ARE was identified in the HIF1α promoter (152). Strikingly, NRF2 silencing suppresses HIF1α accumulation in breast cancer cells under hypoxic conditions. Thus, HIF1α-mediated metabolic adaptations, including the activation of glycolysis and PPP, are inhibited. Inhibition of metabolic adaptations eventually impairs the viability of NRF2-silenced cancer cells in a hypoxic environment (159). Further evidence suggests that NRF2 is induced in hypoxia due to elevated ROS (250) and the subsequent induction of GCLC and GSH synthesis. Hence, chemoresistance is promoted (11). A recent in vitro study showed that both NRF2 and HIF1α are upregulated in MCF-7 and MDA-MB-231 breast cancer cells, compared with MCF-10A cells (a benign breast epithelial cell line). These results suggest that NRF2 and HIF1α simultaneously contribute to cell proliferation and tumor formation in breast cancer (333). Further, NRF2 can promote glycolysis in breast cancer cells in cooperation with HIF1α (333).
In addition to the well-known angiogenic role of HIF1α to overcome hypoxia, the NRF2 pathway also contributes to neovascularization in cancer models. The induction of NRF2 in endothelial cells promotes cell motility and angiogenesis (102). In addition, the expression of platelet derived growth factor and VEGF, strategic angiogenesis inductors, are regulated by HIF1α and NRF2, as demonstrated in a prostate cancer model (323). Moreover, loss of NRF2 inhibited HIF1α accumulation and VEGF induction, resulting in reduced angiogenesis in colorectal carcinoma cells (140); this effect was mediated by the induction of HIF1α hydroxylation in hypoxia.
NRF2 in the regulation of PPP flux
PPP is upregulated in many types of tumors, including breast cancer where PPP promotes proliferation, differentiation, and survival. Breast cancer cells either overexpress NRF2 or cells, in which KEAP1 is silenced, or upregulate G6pd (318, 334). This observation appears to be generalizable; the activities of G6pd and transketolase (TKT), key PPP enzymes, are commonly increased in cancer cells (318). Oxidative PPP converts G6P, a glycolytic intermediate, into ribulose-5-phosphate and generates NADPH, which is used for GSH regeneration, detoxification, and biosynthesis of lipids. NRF2 directly activates five genes involved in PPP and NADPH production pathways, including G6pd, PGD, TKT, transaldolase 1, and ME1, through binding to AREs (306). The G6pd target for NRF2 is a critical enzyme in the PPP, catalyzing the rate-limiting step that produces NADPH (24).
A recent report showed that NRF2 contributes to the metastatic ability of basal type breast cancer cells through the G6pd/HIF-1α/Notch1 signaling axis. NRF2-dependent G6pd/HIF-1α activation elicits Notch1 signaling by upregulating Jagged1 and Hes1, and, thereby, promoting epithelial-mesenchymal transition (EMT) in breast cancer cells (334). In addition, a clinical study showed that PPP-related proteins are differentially expressed according to the molecular subtype of breast cancer; in particular, G6pd and 6-phosphogluconolactone are highly expressed in HER2-type breast cancer (35). Thus, understanding the role of this pathway in breast cancers, including the effects of targeting this pathway, is needed to clarify the clinical implications of PPP in breast cancers. Benito et al. (19) demonstrated critical rewiring of glucose flux due to the NRF2-induced G6pd and TKT expression; the changes in glucose flux included increased ribose and NADPH synthesis, which are associated with poor outcomes in breast cancer patients. However, in contradiction to previous studies, the NRF2 level was downregulated in all types of breast cancers in the TCGA dataset, suggesting a possible role of NRF2 as a tumor suppressor in breast cancer (35). It is also possible that the result of the RNA sequencing may not reflect the actual activity of NRF2 in cells due to post-translational regulation by ubiquitination and degradation (35).
Methionine adenosyltransferase 2A
Polyamines, such as putrescine, spermine, and spermidine, have unknown biological function and accumulate in cancer cells, resulting in increased proliferation. Upregulated enzymes of polyamine pathways have been implicated in several cancer types (26). Methionine adenosyltransferase (MAT) is an essential cellular enzyme that is ubiquitously expressed in mammalian cells; MAT catalyzes the formation of S-adenosylmethionine, the principal biological methyl donor and the ultimate source of the propylamine moiety used in polyamine biosynthesis (189). Altered expression of methionine adenosyltransferase 2A (MAT2A), an isoform of MAT, in tamoxifen-resistant (TAMR) breast cancer and the role of MAT2A in tumor growth and chemoresistance have been described in the context of NRF2 regulation. MAT2A expression is upregulated in TAMR-MCF-7 cells compared with control MCF-7 cells. Moreover, MAT2A expression, associated with altered NRF2 expression levels, is more frequent in TAM-resistant human breast cancer tissues than in TAM-responsive cases (216). This genetic correlation between MAT and NRF2 has been partially clarified; the promoter of MAT2A contains several potential binding sites for transcription factors, including c-Myb, NRF2, NF-κB, and AP-1 (322), suggesting that NRF2 could be vital in regulating the expression of MAT2A. MAT2A immunoreactivity is significantly higher in TAMR cases of human breast cancer compared with TAM-sensitive cases, and it is associated with the overexpression of NRF2 (215). However, whether this association is direct or acts via another intermediate pathway that links MAT2A and NRF2 is unclear. NF-κB also regulates MAT2A gene transcription, suggesting an underlying mechanism of enhanced NF-κB activity related to miR-146b downregulation, which is associated with NRF2 expression, in TAMR breast cancer (104). Therefore, decreased miR-146b and MAT2A induction could be a new phenotype contributing to the growth of breast cancer cells and tamoxifen resistance in breast cancer cells.
NRF2 in the regulation of stemness, EMT, and metastasis
EMT is a trans-differentiation process that enables cell migration. EMT constitutes the initial step of metastatic progression. During EMT, typical markers of the mesenchymal phenotype, such as Vimentin, N-cadherin, Snail, Slug, Twist, and ZEB1, are increased, whereas epithelial markers, such as cell adhesion markers (E-cadherin, MUC-1, and laminin) are lost. Mesenchymal cells also merge phenotypically with circulating tumor cells (CTCs), which are considered a negative prognostic marker of cancer (326). Isolated CTCs usually form clusters, which is a feature of the mesenchymal phenotype (327). In addition, CTCs express the stem cell marker, CD133 (9). On the other hand, mesenchymal–epithelial transition (MET) is the reverse process of EMT by which cells initiate in situ metastases.
Although research in EMT advanced in recent years, there is persistent controversy regarding the existence of EMT/MET phenotype in CSCs, which are also implicated in tumor maintenance and metastasis. EMT mediators can lead to increments of the CSC phenotype (249). CSC are defined by specific expression of cell surface markers (CD24low, CD44high), the expression of aldehyde-dehydrogenase 1 (ALDH1), and low proliferation rate (50). The typical experimental in vitro models, leading to CSCs enrichment, are “spheroids” cultivated from cell cultures. These spheroids are also used for xenograft studies, due to the improved engraftment compared with conventional 2D cell cultures (129). The overlap of CSC issues with the EMT phenomenon is further underlined by the existence of defined epithelial-like and mesenchymal-like CSCs classes (273). Moreover, a hybrid epithelial/mesenchymal (E/M) state of cells has been described recently as the mixed phenotype preceding full mesenchymal state (151). Nevertheless, studying EMT and CSCs in vivo is rather challenging, given that the phenomenon is typically difficult to capture in real-time, due to the rapid exchange of the markers associated with EMT/MET once the transition is completed.
Information regarding the involvement of NRF2 in EMT, MET, and CSC phenotypes is rather ambiguous, due to the variety of experimental models. NRF2 induction most likely promotes spheroid formation and the CSC phenotype. A study by Luo et al. (180) demonstrated that transition from CD24low CD44high CSCs (mesenchymal-like stem cells) into ALDH+ breast cancer CSCs (epithelial-like stem cells) is accompanied by the NRF2-dependent gene expression signature in response to metabolic perturbation such as 2-deoxyglucose treatment (180). Inhibition of NRF2 and the antioxidant system (Txns and GSH) inhibits sphere formation and cancer growth in vivo (180). In breast cancer cells, the expression of CD44 coincides with higher NRF2 expression and markers of CSC-like cancer cells (239, 240). The induction of NRF2 in CD44+ cells is dependent on the noncanonical activation of NRF2 through p62, and NRF2 induction facilitates spheroid formation and induces chemoresistance to 5-fluorouracil and doxorubicin (239, 240).
Formation of spheroids and xenografts is suppressed after NRF2 silencing in CD44high cells in the same studies (239, 240). Moreover, Kipp et al. (143) used a remarkable fluorescence-based in vitro model of monitoring NRF2 expression to demonstrate that NRF2 is clearly induced during spheroid formation. NRF2 induction by the Txnrd1 inhibitor, auranofin, promotes spheroid growth, evidenced by proliferation and differentiation markers, rather than the quiescent (CSC-like) phenotype. In line with this, Kovács et al. (149) showed that the 4-hydroxynonenal content in breast cancer tissue negatively correlated with the proliferation of cancer cells. On the contrary, another point of view was provided in a study by Bocci (21) showing that the knockdown of NRF2 induces mesenchymal markers and the transition from hybrid E/M to the mesenchymal phenotype. NRF2 expression in TCGA datasets also corresponds with the expression of E/M markers (high ZEB1, high miR-200) and elevated NRF2 expression correlates with a worse prognosis of breast cancer (21).
Another aspect of CSCs is cell migration and invasion, which can also be supported by NRF2 signaling. In cell models of breast cancer, NRF2 overexpression or KEAP1 knockdown promotes cell migration (assessed by wound healing assay) and invasion (transwell invasion assay) through G6pd and possibly Notch1 induction (334). Moreover, the CD24−CD44+ mesenchymal subpopulation of several human breast cancer cell lines is characterized by upregulated TrkB-AKT signaling that suppresses KEAP1 expression (138). In agreement with that, NRF2 accumulation in the nucleus can be prevented by the presence of E-cadherin, which is present in epithelial cells, but not in mesenchymal cells (141). The mechanism of this process requires sequestration of the NRF2/KEAP1 complex in the proximity of the plasma membrane and involves the interaction of NRF2 with β-catenin (141).
Of note, NRF2 is involved in the crosstalk between associated cell species and tumor bulk cells in HCC and PDAC (63). Lactate produced and secreted by cancer cells into the environment and elevated ROS levels induce NRF2 in tumor-associated macrophages, converting them to the M2 (CD163+) polarized phenotype. NRF2 in M2 macrophages was essential for the induction of migration and expression of EMT markers in tumor bulk cells via VEGF signaling, which also induces NRF2 levels.
The role of NRF2 in DNA repair
As previously mentioned, the induction of NQO1 by NRF2 prevents DNA damage (267, 270). NRF2 is responsible for DNA protection at multiple levels and for different kinds of DNA damage inducers, such as irradiation or chemical exposure. Reduction of NRF2 levels sensitizes cells to irradiation, a process involving the decreased ability to repair DNA via homologous recombination, by inhibiting RAD51 levels (112). The study by Jayakumar (112) also indicates additional targets containing ARE sequences that are active in DNA repair, including possible targets of NRF2. NRF2 also regulates expression of ATM and ATR after cysplatin exposure in an ovarian cancer model via stress-induced kinases specific to the DNA damage response (267). In addition, synthetic tripertenoids activate NRF2 and protect cells from radiation damage (60, 139). In this case, the role of NRF is controversial (285) in terms of cancer progression, since NRF2 might prevent DNA damage and cancer progression.
The role of NRF2 in aging and lifespan regulation
In Drosophila, NRF2 expression extends the lifespan and induces tolerance to paraquat treatment (285). A straightforward study (164) investigated NRF2 signaling on naked mole rats and stratified NRF2 expression according to the maximum species lifespan potential (MSLP). The study revealed that NRF2 activity and downstream targets exhibit positive correlation with MSLP, due to the diminished activity or expression of canonical and noncanonical negative regulators. In addition, a spectrum of NRF2 target genes and proteins were recently identified in a high-throughput study; these NRF targets were differentially regulated in the long-lived naked mole rat and the short-lived guinea pig, its close relative (96). Nevertheless, there are contradicting reports concerning the role of NRF2 in longevity. For example, in Nqo1-deficient mice, the lifespan is improved under caloric restriction, but not under ad libitum conditions (53). Taken together, the lifespan-prolonging effect of NRF2 has not been conclusively demonstrated.
Future Prospects
NRF2 is a vital component of the cellular machinery that conserves cellular redox balance through the expression of antioxidant proteins. Reactive oxygen and nitrogen species have a dual nature in carcinogenesis. On one hand, enhanced reactive species production increases the risk for neoplastic diseases by increasing the mutation rate and, hence, increasing the chance of transformation (158). On the other hand, in cancer cells, reactive species negatively regulate cancer cell survival (149, 158). It is also of note that although low NRF2 expression is associated with senescence, the induction of NRF2 expression is apparently associated with cancer cell survival (5, 46, 68, 149, 158, 290, 343). This dual nature of free radicals confers an interesting, ambiguous nature to NRF2 in cancer biology. The pro- or antineoplastic effects of NRF2 are context dependent; thus, the effects of NRF2 depend on the nature and molecular characteristics of the cancer in question. Large-scale studies to carefully evaluate cancer subtypes, where NRF2 signaling can be targeted, should be a focus for the future.
NRF2 promotes several enabling cancer hallmarks, such as metabolism, CSC characteristics, tumor aggressiveness, invasion, and metastasis formation. The role of NRF2 in regulating cancer cell stemness and EMT seems to be a central feature in defining the role of NRF2 in regulating metastasis and progression among the neoplastic processes. The level of NRF2 activity is apparently a switch in metabolism and ferroptosis that enables cancer cells to mobilize nutrients to support growth, enable metabolic flexibility, and avoid cell death. The bulk of cancer cells are sensitive to chemotherapy and are rather reliant on Warburg metabolism, whereas stem cells are metabolically flexible and resistant to chemotherapy (66, 214). Therefore, from a translational point of view, fine-tuning NRF2 activity may help in reprogramming CSCs to become more sensitive to chemotherapy (126). Understanding the complex metabolic rearrangements behind NRF2 activation will be crucial in understanding the potential interference of ROS signaling and metabolic rewiring in cancer and other pathologies.
Immune destruction is a major pathway to eliminate cancer cells, involving the production of reactive species. It remains an open question of whether NRF2 is involved in protecting against immune destruction. Logically, the NRF2-driven antioxidant response is probably protective against immune cell-derived free radicals and, therefore, may be pro-neoplastic. Further, NRF2 activity modulates the function of antigen-presenting dendritic cells and T cells (165, 330), implying a modality to improve anticancer immune response.
There are only a few papers on the interactions between the NRF2 pathway and chemotherapy regimens (12) (Table 2); understanding these interactions can help identify the best therapeutic window for NRF2 activators or inhibitors. Most likely, the picture will be very complex, as some chemotherapeutic agents produce superoxide [e.g., doxorubicin (10, 211, 212)], which interferes with the NRF2 system. Further, the scavenging of free radicals produced by chemotherapeutic drugs is controversial (176). The modulation of NRF2 tone will likely impact the sensitivity of tumors to chemotherapy.
Table 2.
Interactions Between the NRF2-KEAP1 System and Cytostatic Drugs
| Drug | Effect | Cancer | Reference |
|---|---|---|---|
| Platinium compounds | |||
| Cysplatin | NRF2 overexpression confers resistance to cisplatin | Cervix cancer | (303) |
| Lung cancer | (20, 137, 265, 283, 299) | ||
| Ovarian cancer | (15, 34, 275) | ||
| Laryngeal cancer | (345) | ||
| Bladder cancer | (43, 91) | ||
| Head and neck tumors | (230) | ||
| NRF2 overexpression protects against Cysplatin nephrotoxicity | N/A | (302) | |
| NRF2 overexpression protects against Cysplatin ototoxicity | N/A | (64) | |
| Oxaliplatin | High NRF2 expression leads to resistance to Cysplatin | Colon cancer | (217, 305) |
| NRF2 knockout protects against Oxaliplatin-induced peripheral neuropathy | N/A | (325) | |
| Carboplatin | NRF2 upregulation protects against Carboplatin treatment | Epithelial ovarian cancer | (147) |
| Nucleotide analogs | |||
| Cordycepin | Cordycepin induces NRF2 that protects against radiation ulcer | N/A | (307) |
| Cordycepin induces NRF2 | Breast cancer | (56) | |
| Cytarabine | NRF2 overexpression confers Cytarabine resistance | Myelosdysplasy | (171) |
| 5-fluorouracil | Vitamin D in combination with 5-fluorouracil protects against hepatocellular carcinoma | Hepatocellular carcinoma | (1, 255) |
| NRF2 overexpression leads to 5-fluorouracil resistance | Colorectal cancer | (125, 173) | |
| NRF2 downregulation or Keap1 loss-of-function enhances the effect of 5-fluorouracil | Cholangiocarcinoma | (244, 256) | |
| NRF2 overexpression leads to 5-fluorouracil resistance | Gastric cancer | (103) | |
| NRF2 activation supports cell proliferation under 5-fluorouracil treatment | Pancreatic cancer | (172) | |
| NRF2 induction protects against 5-fluorouracil-induced myelosuppression | N/A | (206) | |
| Gemcitabine | Reduction of NRF2 sensitizes pancreatic cancer cells to gemcitabine | Pancreatic cancer | (57, 121, 316) |
| Gemcitabine effects are enhanced by the downregulation of NRF2 | Cholangiocarcinoma | (244) | |
| Monoclonal antibodies | |||
| Bevacizumab | NRF2 overexpression confers resistance to VEGF inhibition | Glioblastoma | (319) |
| Vincalkaloids | |||
| Vinorelbine | NRF2 overexpression confers Vinorelbine resistance | Lung cancer | (314) |
| Vincristine | NRF2 protects against Vincristine-induced neuropathic pain | N/A | (342) |
| Anthracycline drugs | |||
| Doxorubicin | Doxorubicine decreases NRF2 expression | N/A | (205) |
| NRF2 activation confers doxorubicin resistance | Ovarian cancer | (260) | |
| Tyrosine kinase inhibitors | |||
| Lapatinib | NRF2 overexpression leads to resistance against lapatinib | Ovarian cancer | (127) |
| Lapatinib inhibits NRF2 expression | N/A | (210) | |
| Erlotinib | NRF2 overexpression leads to resistance against erlotinib | Ovarian cancer | (127) |
| Vorinostat in combination with Gefitinib or Erlotinib suppresses NRF2 expression | Nonsmall-cell lung cancer | (160) | |
| Gefitinib | Vorinostat in combination with Gefitinib or Erlotinib suppresses NRF2 expression | Nonsmall-cell lung cancer | (160) |
| Topoisomerase inhibitors | |||
| Camptotechin | Camptotechin suppresses NRF2 expression | Hepatocellular carcinoma | (29) |
| Alkylating agents | |||
| Cyclophosphamide | NRF2 activation counteracts Cyclophosphamide-induced myelosuppression | N/A | (221) |
| Themozolomide | NRF2 overexpression confers resistance to Themozolomide | Glioma | (229) |
| Themozolomide and radiation therapy induces NRF2 expression | Glioblastoma | (39) | |
| Proteasome inhibitors | |||
| Bortezomib | NRF2 overexpression confers protection against Bortezomib treatment | Neuroblastoma | (69, 70) |
| Bortezomib induces NRF2 expression | Multiple myeloma | (17) | |
| NRF2 overexpression protects against Bortezomib treatment | Mantle cell lymphoma | (308) | |
| Nuclear receptor modulators | |||
| Tamoxifen | NRF2 overexpression contributes to Tamoxifen resistance | Breast cancer | (18, 215) |
| Retinoic acid | Retinoic acid suppresses NRF2 | Breast cancer | (304) |
| Other cytostatic drugs | |||
| Vorinostat | Vorinostat in combination with Gefitinib or Erlotinib suppresses NRF2 expression | Nonsmall-cell lung cancer | (160) |
| Arsenic trioxide | Arsenic trioxide induces NRF2 | Acute promyelocytic leukemia | (199) |
| Radiotherapy | |||
| NRF2 overexpression protects against radiotherapy | Ovarian cancer | (291) | |
| NRF2 overexpression protects against radiotherapy | Esophageal squamous cancer | (132) | |
| Gain-of-function NRF2 mutation confers malignant potential | Esophageal squamous cancer | (257) | |
| NRF2 activation supports cell proliferation under radiotherapy | Pancreatic cancer | (172) | |
| Studies with no drug defined | |||
| Not defined | NRF2 overexpression confers resistance to chemotherapy | Hepatobiliary cancer | (136) |
| NRF2 overexpression confers resistance to chemoradiotherapy | Esophageal squamous cell cancer | (336) | |
| NRF2 inhibition confers sensitivity to chemotherapy | Cholangiocarcinoma | (272) | |
| NRF2 expression shows negative correlation with neoadjuvant chemotherapy success | Breast cancer | (231) | |
| NRF2 activation protects against chemotherapy | Colorectal cancer | (241) | |
| NRF2 downregulation increases the efficacy of chemotherapy | Cervical cancer | (182) | |
| NRF2 protects against chemotherapy | Leukemia | (237) | |
| High NRF2 expression confers chemoresistance | Endometrial cancer | (116) | |
| NRF2 downregulation is vital for chemotherapy success | Chronic lymphocytic leukemia | (312) | |
| NRF2 expression is associated with chemotherapy resistance | Ovarian cancer | (97) | |
N/A, not applicable; VEGF, vascular endothelial growth factor.
Table 3.
Values for the Survival Curve on Figure 4
| Number at risk (months) | |||||
|---|---|---|---|---|---|
| Bladder carcinoma | |||||
| HR = 0.69 | |||||
| p = 0.021 | |||||
| 0 | 50 | 100 | 150 | ||
| Low expression | 115 | 16 | 3 | 2 | |
| High expression | 289 | 50 | 9 | 1 | |
| Cervical | |||||
| Cervical squamous cell carcinoma | |||||
| HR = 0.53 | |||||
| p = 0.25 | |||||
| 0 | 50 | 100 | 150 | 200 | |
| Low expression | 214 | 37 | 9 | 4 | 1 |
| High expression | 90 | 24 | 11 | 3 | 1 |
| Stomach adenocarcinoma | |||||
| HR = 0.66 | |||||
| p = 0.05 | |||||
| 0 | 20 | 40 | 60 | 80 | |
| Low expression | 277 | 96 | 27 | 10 | 3 |
| High expression | 94 | 38 | 8 | 5 | 1 |
| Kidney renal clear cell carcinoma | |||||
| HR = 0.48 | |||||
| p = 1.6 × 10–6 | |||||
| 0 | 50 | 100 | 150 | ||
| Low expression | 159 | 46 | 6 | 0 | |
| High expression | 371 | 160 | 34 | 1 | |
| Lung squamous cell carcinoma | |||||
| HR = 0.68 | |||||
| p = 0.0087 | |||||
| 0 | 50 | 100 | 150 | ||
| Low expression | 309 | 52 | 16 | 2 | |
| High expression | 186 | 53 | 11 | 3 | |
| Sarcoma | |||||
| HR = 0.52 | |||||
| p = 0.0021 | |||||
| 0 | 50 | 100 | 150 | ||
| Low expression | 64 | 14 | 4 | 2 | |
| High expression | 195 | 59 | 12 | 3 | |
| Kidney renal papillary cell carcinoma | |||||
| HR = 1.73 | |||||
| p = 0.07 | |||||
| 0 | 50 | 100 | 150 | 200 | |
| Low expression | 155 | 38 | 9 | 1 | 0 |
| High expression | 132 | 35 | 3 | 0 | 0 |
| Liver hepatocellular carcinoma | |||||
| HR = 1.48 | |||||
| p = 0.053 | |||||
| 0 | 20 | 40 | 60 | 80 | |
| Low expression | 114 | 54 | 27 | 17 | 8 |
| High expression | 256 | 128 | 57 | 25 | 11 |
| Pancreatic ductal adenocarcinoma | |||||
| HR = 1.56 | |||||
| p = 0.45 | |||||
| 0 | 20 | 40 | 60 | 80 | |
| Low expression | 127 | 46 | 15 | 8 | 1 |
| High expression | 50 | 12 | 2 | 0 | 0 |
| Thymoma | |||||
| HR = 4.27 | |||||
| p = 0.032 | |||||
| 0 | 50 | 100 | 150 | ||
| Low expression | 87 | 40 | 11 | 2 | |
| High expression | 31 | 10 | 0 | 0 | |
| Thyroid carcinoma | |||||
| HR = 1.93 | |||||
| p = 0.19 | |||||
| 0 | 50 | 100 | 150 | ||
| Low expression | 365 | 90 | 22 | 3 | |
| High expression | 137 | 38 | 12 | 4 | |
| Uterine corpus endometrial carcinoma | |||||
| HR = 1.37 | |||||
| p = 0.16 | |||||
| 0 | 50 | 100 | 150 | 200 | |
| Low expression | 377 | 115 | 17 | 3 | 1 |
| High expression | 165 | 32 | 3 | 0 | 0 |
HR, hazard ratio.
The exploitation of the NRF2 pathway clearly holds promise in the clinical setting. Nevertheless, substantial studies are needed to sharply pinpoint the diseases, patient subgroups, and therapeutic schemes where NR2-directed therapies will be beneficial.
Acknowledgment
The authors are grateful to Dr. Karen Uray (University of Debrecen, Department of Medical Chemistry) for the critical revision of the text.
Abbreviations Used
- β-TrCP
β-transducin repeat-containing protein
- 4-OHE2
4-Hydroxyestradiol
- 15d-PGJ2
15-deoxy-prostaglandin J2
- AKT
protein kinase B
- ALDH1
aldehyde-dehydrogenase 1
- AMER1
Wilms tumor gene on the X chromosome
- AML
acute myeloid leukemia
- AMPK
AMP-activated protein kinase
- ARE
antioxidant response element
- ATF4
activating transcription factor 4
- Bach1
BTB and CNC homology 1
- BAP1
BRCA1 associated protein 1
- BRCA1
breast cancer type 1 susceptibility protein
- BRCA2
breast cancer type 2 susceptibility protein
- BRD4
bromodomain protein 4
- BTB
Broad complex/Tramtrack/Bric-a-brac
- bZIP
basic leucine zipper domain
- Cav-1
caveolin-1
- CDK20
cyclin-dependent kinase 20
- CDO1
cysteine dioxygenase 1
- CHD6
chromodomain helicase DNA binding protein 6
- COX-2
cyclooxygenase-2
- CSC
cancer stem cell
- CTCs
circulating tumor cells
- CTR
carboxy-terminal
- Cul3
cullin 3
- DGR
double glycine repeats
- DPP3
dipeptidyl peptidase 3
- E/M
epithelial/mesenchymal
- EMT
epithelial-mesenchymal transition
- EpRE
electrophilic response element
- ERK
extracellular signal–regulated kinase
- FAM129B
family with sequence similarity 129, member B
- FH
fumarate hydratase
- G6pd
glucose-6-phosphate dehydrogenase
- GCL
glutamate-cysteine ligase
- GCLC
glutamate-cysteine ligase catalytic
- GCLM
glutamate-cysteine ligase modifier
- Gpx2
glutathione peroxidase 2
- GRP78
heat shock 70 kDa protein 5
- GSH
reduced glutathione
- GSK3
glycogen synthase kinase 3
- GSK3β
glycogen synthase kinase 3β
- Gsr1
glutathione reductase 1
- GSSG
oxidized glutathione
- GST
glutathione S-transferase
- HCC
hepatocellular carcinoma
- HER2
human epidermal growth receptor 2, HER2/neu, cErbb2
- HIF1α
hypoxia-inducible factor 1α
- HMOX1
heme oxygenase (decycling) 1
- Idh1
isocitrate dehydrogenase 1
- IVR
intervening region
- KEAP1
Kelch-like ECH-associated protein 1
- KHK-A
ketohexokinase-A
- LCA
lithocholic acid
- LOF
loss-of-function
- MAT
methionine adenosyltransferase
- MAT2A
methionine adenosyltransferase 2A
- Me1
malic enzyme 1
- MEK
mitogen activated protein kinase kinase
- MET
mesenchymal–epithelial transition
- MICA
not defined as abbreviation
- miRNA
microRNA
- MnSOD
manganese-dependent superoxide dismutase
- Mrps
multidrug-resistance-associated transporters
- Mrp1
multidrug resistance-associated protein 1
- MSLP
maximum species lifespan potential
- mTORC1
mechanistic (or mammalian) target of rapamycin complex 1
- N/A
not applicable
- NFE2
nuclear factor erythroid 2
- Neh
NRF2-ECH homology
- NQO1
NAD(P)H quinone dehydrogenase 1
- NRF2
NFE2-related factor 2 (Nfe2l2, commonly called NRF2)
- NSCLC
nonsmall-cell lung carcinoma
- OGG1
8-Oxoguanine glycosylase
- OTUB1
ovarian tumor family member deubiquitinase
- p62
sequestosome 1
- PALB2
partner and localizer of BRCA2
- PDAC
pancreatic ductal adenocarcinoma
- PERK
protein kinase R-like endoplasmic reticulum kinase
- Pgd
6-phosphogluconate dehydrogenase
- PI3K
phosphoinositide 3-kinases
- PPP
pentose-phosphate pathway
- PUFA
polyunsaturated fatty acid
- RARα
retinoic acid receptor X receptor α
- ROS
oxygen-centered reactive species
- RXRα
retinoid X receptor α
- SLC7A11
solute carrier family 7 member 11
- Srxn1
sulfiredoxin 1
- TAMR
tamoxifen-resistant
- TCA
tricarboxylic acid cycle
- TKT
transketolase
- TP53
gene encoding for tumor protein P53
- Txn1
thioredoxin 1
- Txnrd1
thioredoxin reductase 1
- UHRF1
ubiquitin-like, containing PHD and RING finger domains-1
- UPR
unfolded protein response
- VEGF
vascular endothelial growth factor
- WTX
Wilms tumor gene on the X chromosome
- xCT
protein encoded by SLC7A11
Funding Information
This work was supported by the NKFIH (K123975, PD124110, FK128387, GINOP-2.3.2-15-2016-00006) to P.B., the Hungarian Academy of Sciences (NKM-26/2019) to P.B., the Czech Health Research Council project number NV19-01-00101 to K.S., and institutional funding of IPHYS CAS number RVO67985823. E.M. was supported by the Bolyai fellowship from the Hungarian Academy of Sciences. The research was financed by the Higher Education Institutional Excellence Programme (NKFIH-1150-6/2019) of the Ministry of Innovation and Technology in Hungary, within the framework of the Biotechnology thematic programme of the University of Debrecen. E.M. is supported by the ÚNKP-19-4-DE-79 New National Excellence Program of the Ministry of Human Capacities.
References
- 1. A RE, El-Mesery M, El-Karef A, and Eissa LA. Vitamin D potentiates anti-tumor activity of 5-fluorouracil via modulating caspase-3 and TGF-beta1 expression in hepatocellular carcinoma-induced in rats. Can J Physiol Pharmacol 96: 1218–1225, 2018 [DOI] [PubMed] [Google Scholar]
- 2. Abbas K, Breton J, Planson AG, Bouton C, Bignon J, Seguin C, Riquier S, Toledano MB, and Drapier JC. Nitric oxide activates an Nrf2/sulfiredoxin antioxidant pathway in macrophages. Free Radic Biol Med 51: 107–114, 2011 [DOI] [PubMed] [Google Scholar]
- 3. Abu-Alainin W, Gana T, Liloglou T, Olayanju A, Barrera LN, Ferguson R, Campbell F, Andrews T, Goldring C, Kitteringham N, Park BK, Nedjadi T, Schmid MC, Slupsky JR, Greenhalf W, Neoptolemos JP, and Costello E. UHRF1 regulation of the Keap1-Nrf2 pathway in pancreatic cancer contributes to oncogenesis. J Pathol 238: 423–433, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ahmad F, Dixit D, Sharma V, Kumar A, Joshi SD, Sarkar C, and Sen E. Nrf2-driven TERT regulates pentose phosphate pathway in glioblastoma. Cell Death Dis 7: e2213, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ahn B, Pharaoh G, Premkumar P, Huseman K, Ranjit R, Kinter M, Szweda L, Kiss T, Fulop G, Tarantini S, Csiszar A, Ungvari Z, and Van Remmen H. Nrf2 deficiency exacerbates age-related contractile dysfunction and loss of skeletal muscle mass. Redox Biol 17: 47–58, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Akdemir B, Nakajima Y, Inazawa J, and Inoue J. miR-432 induces NRF2 stabilization by directly targeting KEAP1. Mol Cancer Res 15: 1570–1578, 2017 [DOI] [PubMed] [Google Scholar]
- 7. Alam J, Stewart D, Touchard C, Boinapally S, Choi AM, and Cook JL. Nrf2, a Cap'n'Collar transcription factor, regulates induction of the heme oxygenase-1 gene. J Biol Chem 274: 26071–26078, 1999 [DOI] [PubMed] [Google Scholar]
- 8. Ali D, Mohammad DK, Mujahed H, Jonson-Videsater K, Nore B, Paul C, and Lehmann S. Anti-leukaemic effects induced by APR-246 are dependent on induction of oxidative stress and the NFE2L2/HMOX1 axis that can be targeted by PI3K and mTOR inhibitors in acute myeloid leukaemia cells. Br J Haematol 174: 117–126, 2016 [DOI] [PubMed] [Google Scholar]
- 9. Armstrong AJ, Marengo MS, Oltean S, Kemeny G, Bitting RL, Turnbull JD, Herold CI, Marcom PK, George DJ, and Garcia-Blanco MA. Circulating tumor cells from patients with advanced prostate and breast cancer display both epithelial and mesenchymal markers. Mol Cancer Res 9: 997–1007, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bai P, Mabley JG, Liaudet L, Virag L, Szabo C, and Pacher P. Matrix metalloproteinase activation is an early event in doxorubicin-induced cardiotoxicity. Oncol Rep 11: 505–508, 2004 [PubMed] [Google Scholar]
- 11. Bai X, Chen Y, Hou X, Huang M, and Jin J. Emerging role of NRF2 in chemoresistance by regulating drug-metabolizing enzymes and efflux transporters. Drug Metab Rev 48: 541–567, 2016 [DOI] [PubMed] [Google Scholar]
- 12. Bala A and Panditharadyula SS. Role of nuclear factor erythroid 2-related factor 2 (NRF-2) mediated antioxidant response on the synergistic antitumor effect of L-arginine and 5-fluro uracil (5FU) in breast adenocarcinoma. Curr Pharm Des 25: 1643–1652, 2019 [DOI] [PubMed] [Google Scholar]
- 13. Banerjee P, Basu A, Datta D, Gasser M, Waaga-Gasser AM, and Pal S. The heme oxygenase-1 protein is overexpressed in human renal cancer cells following activation of the Ras-Raf-ERK pathway and mediates anti-apoptotic signal. J Biol Chem 286: 33580–33590, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bao L, Wu J, Dodson M, Rojo de la Vega EM, Ning Y, Zhang Z, Yao M, Zhang DD, Xu C, and Yi X. ABCF2, an Nrf2 target gene, contributes to cisplatin resistance in ovarian cancer cells. Mol Carcinog 56: 1543–1553, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bao LJ, Jaramillo MC, Zhang ZB, Zheng YX, Yao M, Zhang DD, and Yi XF. Nrf2 induces cisplatin resistance through activation of autophagy in ovarian carcinoma. Int J Clin Exp Pathol 7: 1502–1513, 2014 [PMC free article] [PubMed] [Google Scholar]
- 16. Barbano R, Muscarella LA, Pasculli B, Valori VM, Fontana A, Coco M, la Torre A, Balsamo T, Poeta ML, Marangi GF, Maiello E, Castelvetere M, Pellegrini F, Murgo R, Fazio VM, and Parrella P. Aberrant Keap1 methylation in breast cancer and association with clinicopathological features. Epigenetics 8: 105–112, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Barrera LN, Rushworth SA, Bowles KM, and MacEwan DJ. Bortezomib induces heme oxygenase-1 expression in multiple myeloma. Cell Cycle 11: 2248–2252, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bekele RT, Venkatraman G, Liu RZ, Tang X, Mi S, Benesch MG, Mackey JR, Godbout R, Curtis JM, McMullen TP, and Brindley DN. Oxidative stress contributes to the tamoxifen-induced killing of breast cancer cells: implications for tamoxifen therapy and resistance. Sci Rep 6: 21164, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Benito A, Polat IH, Noe V, Ciudad CJ, Marin S, and Cascante M. Glucose-6-phosphate dehydrogenase and transketolase modulate breast cancer cell metabolic reprogramming and correlate with poor patient outcome. Oncotarget 8: 106693–106706, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bialk P, Wang Y, Banas K, and Kmiec EB. Functional gene knockout of NRF2 increases chemosensitivity of human lung cancer A549 cells in vitro and in a xenograft mouse model. Mol Ther Oncolytics 11: 75–89, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bocci F, Tripathi SC, Vilchez Mercedes SA, George JT, Casabar JP, Kin Wong P, Hanash SM, Levine H, Onuchic JN, and Kumar Jolly M. NRF2 activates a partial epithelial-mesenchymal transition and is maximally present in a hybrid epithelial/mesenchymal phenotype. Integr Biol 11: 251–263, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. This reference has been deleted
- 23. Bollong MJ, Lee G, Coukos JS, Yun H, Zambaldo C, Chang JW, Chin EN, Ahmad I, Chatterjee AK, Lairson LL, Schultz PG, and Moellering RE. A metabolite-derived protein modification integrates glycolysis with KEAP1-NRF2 signalling. Nature 562: 600–604, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Brewer AC, Mustafi SB, Murray TVA, Rajasekaran NS, and Benjamin IJ. Reductive stress linked to small HSPs, G6PD, and Nrf2 pathways in heart disease. Antioxid Redox Signal 18: 1114–1127, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Camp ND, James RG, Dawson DW, Yan F, Davison JM, Houck SA, Tang X, Zheng N, Major MB, and Moon RT. Wilms tumor gene on X chromosome (WTX) inhibits degradation of NRF2 protein through competitive binding to KEAP1 protein. J Biol Chem 287: 6539–6550, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Casero RA, Murray Stewart T, and Pegg AE. Polyamine metabolism and cancer: treatments, challenges and opportunities. Nat Rev Cancer 18: 681–695, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chang CW, Chen YS, Tsay YG, Han CL, Chen YJ, Yang CC, Hung KF, Lin CH, Huang TY, Kao SY, Lee TC, and Lo JF. ROS-independent ER stress-mediated NRF2 activation promotes warburg effect to maintain stemness-associated properties of cancer-initiating cells. Cell Death Dis 9: 194, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chen C, Jiang X, Gu S, and Zhang Z. MicroRNA-155 regulates arsenite-induced malignant transformation by targeting Nrf2-mediated oxidative damage in human bronchial epithelial cells. Toxicol Lett 278: 38–47, 2017 [DOI] [PubMed] [Google Scholar]
- 29. Chen F, Wang H, Zhu J, Zhao R, Xue P, Zhang Q, Bud Nelson M, Qu W, Feng B, and Pi J. Camptothecin suppresses NRF2-ARE activity and sensitises hepatocellular carcinoma cells to anticancer drugs. Br J Cancer 117: 1495–1506, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chen PH, Smith TJ, Wu J, Siesser PF, Bisnett BJ, Khan F, Hogue M, Soderblom E, Tang F, Marks JR, Major MB, Swarts BM, Boyce M, and Chi JT. Glycosylation of KEAP1 links nutrient sensing to redox stress signaling. EMBO J 36: 2233–2250, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chen W, Sun Z, Wang XJ, Jiang T, Huang Z, Fang D, and Zhang DD. Direct interaction between Nrf2 and p21(Cip1/WAF1) upregulates the Nrf2-mediated antioxidant response. Mol Cell 34: 663–673, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cheng K-C, Lin R-J, Cheng J-Y, Wang S-H, Yu J-C, Wu J-C, Liang Y-J, Hsu H-M, Yu J, and Yu AL. FAM129B, an antioxidative protein, reduces chemosensitivity by competing with Nrf2 for Keap1 binding. EBioMedicine 45: 25–38, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chio IIC, Jafarnejad SM, Ponz-Sarvise M, Park Y, Rivera K, Palm W, Wilson J, Sangar V, Hao Y, Ohlund D, Wright K, Filippini D, Lee EJ, Da Silva B, Schoepfer C, Wilkinson JE, Buscaglia JM, DeNicola GM, Tiriac H, Hammell M, Crawford HC, Schmidt EE, Thompson CB, Pappin DJ, Sonenberg N, and Tuveson DA. NRF2 promotes tumor maintenance by modulating mRNA translation in pancreatic cancer. Cell 166: 963–976, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cho JM, Manandhar S, Lee HR, Park HM, and Kwak MK. Role of the Nrf2-antioxidant system in cytotoxicity mediated by anticancer cisplatin: implication to cancer cell resistance. Cancer Lett 260: 96–108, 2008 [DOI] [PubMed] [Google Scholar]
- 35. Choi J, Kim E-S, and Koo JS. Expression of pentose phosphate pathway-related proteins in breast cancer. Dis Markers 2018: 9369358, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. This reference has been deleted
- 37. Chowdhry S, Zhang Y, McMahon M, Sutherland C, Cuadrado A, and Hayes JD. Nrf2 is controlled by two distinct beta-TrCP recognition motifs in its Neh6 domain, one of which can be modulated by GSK-3 activity. Oncogene 32: 3765–3781, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cleary SP, Jeck WR, Zhao X, Chen K, Selitsky SR, Savich GL, Tan TX, Wu MC, Getz G, Lawrence MS, Parker JS, Li J, Powers S, Kim H, Fischer S, Guindi M, Ghanekar A, and Chiang DY. Identification of driver genes in hepatocellular carcinoma by exome sequencing. Hepatology 58: 1693–1702, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cong ZX, Wang HD, Zhou Y, Wang JW, Pan H, Zhang DD, Zhang L, and Zhu L. Temozolomide and irradiation combined treatment-induced Nrf2 activation increases chemoradiation sensitivity in human glioblastoma cells. J Neurooncol 116: 41–48, 2014 [DOI] [PubMed] [Google Scholar]
- 40. Cullinan SB and Diehl JA. PERK-dependent activation of Nrf2 contributes to redox homeostasis and cell survival following endoplasmic reticulum stress. J Biol Chem 279: 20108–20117, 2004 [DOI] [PubMed] [Google Scholar]
- 41. Cullinan SB, Gordan JD, Jin J, Harper JW, and Diehl JA. The Keap1-BTB protein is an adaptor that bridges Nrf2 to a Cul3-based E3 ligase: oxidative stress sensing by a Cul3-Keap1 ligase. Mol Cell Biol 24: 8477–8486, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Cullinan SB, Zhang D, Hannink M, Arvisais E, Kaufman RJ, and Diehl JA. Nrf2 is a direct PERK substrate and effector of PERK-dependent cell survival. Mol Cell Biol 23: 7198–7209, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Daga M, Pizzimenti S, Dianzani C, Cucci MA, Cavalli R, Grattarola M, Ferrara B, Scariot V, Trotta F, and Barrera G. Ailanthone inhibits cell growth and migration of cisplatin resistant bladder cancer cells through down-regulation of Nrf2, YAP, and c-Myc expression. Phytomedicine 56: 156–164, 2019 [DOI] [PubMed] [Google Scholar]
- 44. Danilovic DLS, de Mello ES, Frazzato EST, Wakamatsu A, de Lima Jorge AA, Hoff AO, and Marui S. Oncogenic mutations in KEAP1 disturbing inhibitory Nrf2-Keap1 interaction: activation of antioxidative pathway in papillary thyroid carcinoma. Head Neck 40: 1271–1278, 2018 [DOI] [PubMed] [Google Scholar]
- 45. Das J, Sarkar A, and Sil PC. Hexavalent chromium induces apoptosis in human liver (HepG2) cells via redox imbalance. Toxicol Rep 2: 600–608, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Davies KJA and Forman HJ. Does Bach1 & c-Myc dependent redox dysregulation of Nrf2 & adaptive homeostasis decrease cancer risk in ageing? Free Radic Biol Med 134: 708–714, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. This reference has been deleted
- 48. Davila A, Liu L, Chellappa K, Redpath P, Nakamaru-Ogiso E, Paolella LM, Zhang Z, Migaud ME, Rabinowitz JD, and Baur JA. Nicotinamide adenine dinucleotide is transported into mammalian mitochondria. Elife 7: 33246, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Davila-Gonzalez D, Choi DS, Rosato RR, Granados-Principal SM, Kuhn JG, Li WF, Qian W, Chen W, Kozielski AJ, Wong H, Dave B, and Chang JC. Pharmacological inhibition of NOS activates ASK1/JNK pathway augmenting docetaxel-mediated apoptosis in triple-negative breast cancer. Clin Cancer Res 24: 1152–1162, 2018 [DOI] [PubMed] [Google Scholar]
- 50. De Francesco EM, Sotgia F, and Lisanti MP. Cancer stem cells (CSCs): metabolic strategies for their identification and eradication. Biochem J 475: 1611–1634, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. DeNicola GM, Chen PH, Mullarky E, Sudderth JA, Hu Z, Wu D, Tang H, Xie Y, Asara JM, Huffman KE, Wistuba II, Minna JD, DeBerardinis RJ, and Cantley LC. NRF2 regulates serine biosynthesis in non-small cell lung cancer. Nat Genet 47: 1475–1481, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. DeNicola GM, Karreth FA, Humpton TJ, Gopinathan A, Wei C, Frese K, Mangal D, Yu KH, Yeo CJ, Calhoun ES, Scrimieri F, Winter JM, Hruban RH, Iacobuzio-Donahue C, Kern SE, Blair IA, and Tuveson DA. Oncogene-induced Nrf2 transcription promotes ROS detoxification and tumorigenesis. Nature 475: 106–109, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Diaz-Ruiz A, Di Francesco A, Carboneau BA, Levan SR, Pearson KJ, Price NL, Ward TM, Bernier M, de Cabo R, and Mercken EM. Benefits of caloric restriction in longevity and chemical-induced tumorigenesis are transmitted independent of NQO1. J Gerontol A Biol Sci Med Sci 74: 155–162, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Dinkova-Kostova AT, Holtzclaw WD, Cole RN, Itoh K, Wakabayashi N, Katoh Y, Yamamoto M, and Talalay P. Direct evidence that sulfhydryl groups of Keap1 are the sensors regulating induction of phase 2 enzymes that protect against carcinogens and oxidants. Proc Natl Acad Sci U S A 99: 11908–11913, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Dodson M, Castro-Portuguez R, and Zhang DD. NRF2 plays a critical role in mitigating lipid peroxidation and ferroptosis. Redox Biol 11: 101107, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Dong J, Li Y, Xiao H, Luo D, Zhang S, Zhu C, Jiang M, Cui M, Lu L, and Fan S. Cordycepin sensitizes breast cancer cells toward irradiation through elevating ROS production involving Nrf2. Toxicol Appl Pharmacol 364: 12–21, 2019 [DOI] [PubMed] [Google Scholar]
- 57. Duong HQ, Yi YW, Kang HJ, Hong YB, Tang W, Wang A, Seong YS, and Bae I. Inhibition of NRF2 by PIK-75 augments sensitivity of pancreatic cancer cells to gemcitabine. Int J Oncol 44: 959–969, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Eades G, Yang M, Yao Y, Zhang Y, and Zhou Q. miR-200a regulates Nrf2 activation by targeting Keap1 mRNA in breast cancer cells. J Biol Chem 286: 40725–40733, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Eggler AL, Liu G, Pezzuto JM, van Breemen RB, and Mesecar AD. Modifying specific cysteines of the electrophile-sensing human Keap1 protein is insufficient to disrupt binding to the Nrf2 domain Neh2. Proc Natl Acad Sci U S A 102: 10070–10075, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Eskiocak U, Kim SB, Roig AI, Kitten E, Batten K, Cornelius C, Zou YS, Wright WE, and Shay JW. CDDO-Me protects against space radiation-induced transformation of human colon epithelial cells. Radiat Res 174: 27–36, 2010 [DOI] [PubMed] [Google Scholar]
- 61. Fabrizio FP, Costantini M, Copetti M, la Torre A, Sparaneo A, Fontana A, Poeta L, Gallucci M, Sentinelli S, Graziano P, Parente P, Pompeo V, De Salvo L, Simone G, Papalia R, Picardo F, Balsamo T, Flammia GP, Trombetta D, Pantalone A, Kok K, Paranita F, Muscarella LA, and Fazio VM. Keap1/Nrf2 pathway in kidney cancer: frequent methylation of KEAP1 gene promoter in clear renal cell carcinoma. Oncotarget 8: 11187–11198, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Fan Z, Wirth AK, Chen D, Wruck CJ, Rauh M, Buchfelder M, and Savaskan N. Nrf2-Keap1 pathway promotes cell proliferation and diminishes ferroptosis. Oncogenesis 6: e371, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Feng R, Morine Y, Ikemoto T, Imura S, Iwahashi S, Saito Y, and Shimada M. Nrf2 activation drive macrophages polarization and cancer cell epithelial-mesenchymal transition during interaction. Cell Commun Signal 16: 54, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Fetoni AR, Paciello F, Mezzogori D, Rolesi R, Eramo SL, Paludetti G, and Troiani D. Molecular targets for anticancer redox chemotherapy and cisplatin-induced ototoxicity: the role of curcumin on pSTAT3 and Nrf-2 signalling. Br J Cancer 113: 1434–1444, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Flores R, Shi J, Fuhrman B, Xu X, Veenstra TD, Gail MH, Gajer P, Ravel J, and Goedert JJ. Fecal microbial determinants of fecal and systemic estrogens and estrogen metabolites: a cross-sectional study. J Transl Med 10: 253, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Fodor T, Szanto M, Abdul-Rahman O, Nagy L, Der A, Kiss B, and Bai P. Combined treatment of MCF-7 cells with AICAR and methotrexate, arrests cell cycle and reverses Warburg metabolism through AMP-activated protein kinase (AMPK) and FOXO1. PLoS One 11: e0150232, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Fuhrman BJ, Feigelson HS, Flores R, Gail MH, Xu X, Ravel J, and Goedert JJ. Associations of the fecal microbiome with urinary estrogens and estrogen metabolites in postmenopausal women. J Clin Endocrinol Metab 99: 4632–4640, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Fulop GA, Kiss T, Tarantini S, Balasubramanian P, Yabluchanskiy A, Farkas E, Bari F, Ungvari Z, and Csiszar A. Nrf2 deficiency in aged mice exacerbates cellular senescence promoting cerebrovascular inflammation. Geroscience 40: 513–521, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Furfaro AL, Piras S, Domenicotti C, Fenoglio D, De Luigi A, Salmona M, Moretta L, Marinari UM, Pronzato MA, Traverso N, and Nitti M. Role of Nrf2, HO-1 and GSH in neuroblastoma cell resistance to bortezomib. PLoS One 11: e0152465, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Furfaro AL, Piras S, Passalacqua M, Domenicotti C, Parodi A, Fenoglio D, Pronzato MA, Marinari UM, Moretta L, Traverso N, and Nitti M. HO-1 up-regulation: a key point in high-risk neuroblastoma resistance to bortezomib. Biochim Biophys Acta 1842: 613–622, 2014 [DOI] [PubMed] [Google Scholar]
- 71. Fuse Y and Kobayashi M. Conservation of the Keap1-Nrf2 system: an evolutionary journey through stressful space and time. Molecules 22: pii: , 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Galan-Cobo A, Sitthideatphaiboon P, Qu X, Poteete A, Pisegna MA, Tong P, Chen P-H, Boroughs LK, Rodriguez MLM, Zhang W, Parlati F, Wang J, Gandhi V, Skoulidis F, DeBerardinis RJ, Minna JD, and Heymach JV. LKB1 and KEAP1/NRF2 pathways cooperatively promote metabolic reprogramming with enhanced glutamine dependence in KRAS-mutant lung adenocarcinoma. Cancer Res 79: 3251–3267, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Garrido P, Shalaby A, Walsh EM, Keane N, Webber M, Keane MM, Sullivan FJ, Kerin MJ, Callagy G, Ryan AE, and Glynn SA. Impact of inducible nitric oxide synthase (iNOS) expression on triple negative breast cancer outcome and activation of EGFR and ERK signaling pathways. Oncotarget 8: 80568–80588, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Gill JG, Piskounova E, and Morrison SJ. Cancer, oxidative stress, and metastasis. Cold Spring Harb Symp Quant Biol 81: 163–175, 2016 [DOI] [PubMed] [Google Scholar]
- 75. Girotti AW. Upregulation of nitric oxide in tumor cells as a negative adaptation to photodynamic therapy. Lasers Surg Med 50: 590–598, 2018 [DOI] [PubMed] [Google Scholar]
- 76. Goedert JJ, Hua X, Bielecka A, Okayasu I, Milne GL, Jones GS, Fujiwara M, Sinha R, Wan Y, Xu X, Ravel J, Shi J, Palm NW, and Feigelson HS. Postmenopausal breast cancer and oestrogen associations with the IgA-coated and IgA-noncoated faecal microbiota. Br J Cancer 23: 435, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Goedert JJ, Jones G, Hua X, Xu X, Yu G, Flores R, Falk RT, Gail MH, Shi J, Ravel J, and Feigelson HS. Investigation of the association between the fecal microbiota and breast cancer in postmenopausal women: a population-based case-control pilot study. J Natl Cancer Inst 107: pii: , 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Goldstein LD, Lee J, Gnad F, Klijn C, Schaub A, Reeder J, Daemen A, Bakalarski CE, Holcomb T, Shames DS, Hartmaier RJ, Chmielecki J, Seshagiri S, Gentleman R, and Stokoe D. Recurrent loss of NFE2L2 Exon 2 is a mechanism for Nrf2 pathway activation in human cancers. Cell Rep 16: 2605–2617, 2016 [DOI] [PubMed] [Google Scholar]
- 79. Gorrini C, Gang BP, Bassi C, Wakeham A, Baniasadi SP, Hao Z, Li WY, Cescon DW, Li YT, Molyneux S, Penrod N, Lupien M, Schmidt EE, Stambolic V, Gauthier ML, and Mak TW. Estrogen controls the survival of BRCA1-deficient cells via a PI3K-NRF2-regulated pathway. Proc Natl Acad Sci U S A 111: 4472–4477, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Gorrini C, Harris IS, and Mak TW. Modulation of oxidative stress as an anticancer strategy. Nat Rev Drug Discov 12: 931–947, 2013 [DOI] [PubMed] [Google Scholar]
- 81. Guichard C, Amaddeo G, Imbeaud S, Ladeiro Y, Pelletier L, Maad IB, Calderaro J, Bioulac-Sage P, Letexier M, Degos F, Clement B, Balabaud C, Chevet E, Laurent A, Couchy G, Letouze E, Calvo F, and Zucman-Rossi J. Integrated analysis of somatic mutations and focal copy-number changes identifies key genes and pathways in hepatocellular carcinoma. Nat Genet 44: 694–698, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Habib E, Linher-Melville K, Lin H-X, and Singh G. Expression of xCT and activity of system xc− are regulated by NRF2 in human breast cancer cells in response to oxidative stress. Redox Biol 5: 33, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Hamada S, Shimosegawa T, Taguchi K, Nabeshima T, Yamamoto M, and Masamune A. Simultaneous K-ras activation and Keap1 deletion cause atrophy of pancreatic parenchyma. Am J Physiol Gastrointest Liver Physiol 314: G65–G74, 2018 [DOI] [PubMed] [Google Scholar]
- 84. Hanada N, Takahata T, Zhou Q, Ye X, Sun R, Itoh J, Ishiguro A, Kijima H, Mimura J, Itoh K, Fukuda S, and Saijo Y. Methylation of the KEAP1 gene promoter region in human colorectal cancer. BMC Cancer 12: 66, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Hanahan D and Weinberg RA. Hallmarks of cancer: the next generation. Cell 144: 646–674, 2011 [DOI] [PubMed] [Google Scholar]
- 86. Hart PC, Ratti BA, Mao M, Ansenberger-Fricano K, Shajahan-Haq AN, Tyner AL, Minshall RD, and Bonini MG. Caveolin-1 regulates cancer cell metabolism via scavenging Nrf2 and suppressing MnSOD-driven glycolysis. Oncotarget 7: 308–322, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Hartikainen JM, Tengstrom M, Winqvist R, Jukkola-Vuorinen A, Pylkas K, Kosma VM, Soini Y, and Mannermaa A. KEAP1 genetic polymorphisms associate with breast cancer risk and survival outcomes. Clin Cancer Res 21: 1591–1601, 2015 [DOI] [PubMed] [Google Scholar]
- 88. Harvey CJ, Thimmulappa RK, Singh A, Blake DJ, Ling G, Wakabayashi N, Fujii J, Myers A, and Biswal S. Nrf2-regulated glutathione recycling independent of biosynthesis is critical for cell survival during oxidative stress. Free Radic Biol Med 46: 443–453, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Hast BE, Goldfarb D, Mulvaney KM, Hast MA, Siesser PF, Yan F, Hayes DN, and Major MB. Proteomic analysis of ubiquitin ligase KEAP1 reveals associated proteins that inhibit NRF2 ubiquitination. Cancer Res 73: 2199–2210, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Hawkes HJ, Karlenius TC, and Tonissen KF. Regulation of the human thioredoxin gene promoter and its key substrates: a study of functional and putative regulatory elements. Biochim Biophys Acta 1840: 303–314, 2014 [DOI] [PubMed] [Google Scholar]
- 91. Hayden A, Douglas J, Sommerlad M, Andrews L, Gould K, Hussain S, Thomas GJ, Packham G, and Crabb SJ. The Nrf2 transcription factor contributes to resistance to cisplatin in bladder cancer. Urol Oncol 32: 806–814, 2014 [DOI] [PubMed] [Google Scholar]
- 92. Hayes JD and Dinkova-Kostova AT. The Nrf2 regulatory network provides an interface between redox and intermediary metabolism. Trends Biochem Sci 39: 199–218, 2014 [DOI] [PubMed] [Google Scholar]
- 93. He X and Ma Q. NRF2 cysteine residues are critical for oxidant/electrophile-sensing, Kelch-like ECH-associated protein-1-dependent ubiquitination-proteasomal degradation, and transcription activation. Mol Pharmacol 76: 1265–1278, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. He X and Ma Q. Critical cysteine residues of Kelch-like ECH-associated protein 1 in arsenic sensing and suppression of nuclear factor erythroid 2-related factor 2. J Pharmacol Exp Ther 332: 66–75, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Hegedus C, Kovacs K, Polgar Z, Regdon Z, Szabo E, Robaszkiewicz A, Forman HJ, Martner A, and Virag L. Redox control of cancer cell destruction. Redox Biol 16: 59–74, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Heinze I, Bens M, Calzia E, Holtze S, Dakhovnik O, Sahm A, Kirkpatrick JM, Szafranski K, Romanov N, Sama SN, Holzer K, Singer S, Ermolaeva M, Platzer M, Hildebrandt T, and Ori A. Species comparison of liver proteomes reveals links to naked mole-rat longevity and human aging. BMC Biol 16: 82, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Helleman J, Smid M, Jansen MP, van der Burg ME, and Berns EM. Pathway analysis of gene lists associated with platinum-based chemotherapy resistance in ovarian cancer: the big picture. Gynecol Oncol 117: 170–176, 2010 [DOI] [PubMed] [Google Scholar]
- 98. Helmink BA, Khan MAW, Hermann A, Gopalakrishnan V, and Wargo JA. The microbiome, cancer, and cancer therapy. Nat Med 25: 377–388, 2019 [DOI] [PubMed] [Google Scholar]
- 99. Higgins LG, Kelleher MO, Eggleston IM, Itoh K, Yamamoto M, and Hayes JD. Transcription factor Nrf2 mediates an adaptive response to sulforaphane that protects fibroblasts in vitro against the cytotoxic effects of electrophiles, peroxides and redox-cycling agents. Toxicol Appl Pharmacol 237: 267–280, 2009 [DOI] [PubMed] [Google Scholar]
- 100. Hiramoto K, Satoh H, Suzuki T, Moriguchi T, Pi J, Shimosegawa T, and Yamamoto M. Myeloid lineage-specific deletion of antioxidant system enhances tumor metastasis. Cancer Prev Res (Phila) 7: 835–844, 2014 [DOI] [PubMed] [Google Scholar]
- 101. Hirotsu Y, Katsuoka F, Funayama R, Nagashima T, Nishida Y, Nakayama K, Engel JD, and Yamamoto M. Nrf2-MafG heterodimers contribute globally to antioxidant and metabolic networks. Nucleic Acids Res 40: 10228–10239, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Hojo T, Maishi N, Towfik AM, Akiyama K, Ohga N, Shindoh M, Hida Y, Minowa K, Fujisawa T, and Hida K. ROS enhance angiogenic properties via regulation of NRF2 in tumor endothelial cells. Oncotarget 8: 45484–45495, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Hu XF, Yao J, Gao SG, Wang XS, Peng XQ, Yang YT, and Feng XS. Nrf2 overexpression predicts prognosis and 5-FU resistance in gastric cancer. Asian Pac J Cancer Prev 14: 5231–5235, 2013 [DOI] [PubMed] [Google Scholar]
- 104. Hurst DR, Edmonds MD, Scott GK, Benz CC, Vaidya KS, and Welch DR. Breast cancer metastasis suppressor 1 up-regulates miR-146, which suppresses breast cancer metastasis. Cancer Res 69: 1279–1283, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Hussong M, Borno ST, Kerick M, Wunderlich A, Franz A, Sultmann H, Timmermann B, Lehrach H, Hirsch-Kauffmann M, and Schweiger MR. The bromodomain protein BRD4 regulates the KEAP1/NRF2-dependent oxidative stress response. Cell Death Dis 5: e1195, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Ichimura Y, Waguri S, Sou Y-S, Kageyama S, Hasegawa J, Ishimura R, Saito T, Yang Y, Kouno T, Fukutomi T, Hoshii T, Hirao A, Takagi K, Mizushima T, Motohashi H, Lee M-S, Yoshimori T, Tanaka K, Yamamoto M, and Komatsu M. Phosphorylation of p62 activates the Keap1-Nrf2 pathway during selective autophagy. Mol Cell 51: 618–631, 2013 [DOI] [PubMed] [Google Scholar]
- 107. Inami Y, Waguri S, Sakamoto A, Kouno T, Nakada K, Hino O, Watanabe S, Ando J, Iwadate M, Yamamoto M, Lee MS, Tanaka K, and Komatsu M. Persistent activation of Nrf2 through p62 in hepatocellular carcinoma cells. J Cell Biol 193: 275–284, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Itoh K, Wakabayashi N, Katoh Y, Ishii T, Igarashi K, Engel JD, and Yamamoto M. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev 13: 76–86, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Ivan J, Major E, Sipos A, Kovacs K, Horvath D, Tamas I, Bay P, Dombradi V, and Lontay B. The short-chain fatty acid propionate inhibits adipogenic differentiation of human chorion-derived mesenchymal stem cells through the free fatty acid receptor 2. Stem Cells Dev 26: 1724–1733, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Jain AK and Jaiswal AK. Phosphorylation of tyrosine 568 controls nuclear export of Nrf2. J Biol Chem 292: 2051, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Jana S, Patra K, Jana J, Mandal DP, and Bhattacharjee S. Nrf-2 transcriptionally activates P21(Cip/WAF1) and promotes A549 cell survival against oxidative stress induced by H2O2. Chem Biol Interact 285: 59–68, 2018 [DOI] [PubMed] [Google Scholar]
- 112. Jayakumar S, Pal D, and Sandur SK. Nrf2 facilitates repair of radiation induced DNA damage through homologous recombination repair pathway in a ROS independent manner in cancer cells. Mutat Res 779: 33–45, 2015 [DOI] [PubMed] [Google Scholar]
- 113. Jeong Y, Hoang NT, Lovejoy A, Stehr H, Newman AM, Gentles AJ, Kong W, Truong D, Martin S, Chaudhuri A, Heiser D, Zhou L, Say C, Carter JN, Hiniker SM, Loo BW Jr., West RB, Beachy P, Alizadeh AA, and Diehn M.. Role of KEAP1/NRF2 and TP53 mutations in lung squamous cell carcinoma development and radiation resistance. Cancer Discov 7: 86–101, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Ji X, Qian J, Rahman SMJ, Siska PJ, Zou Y, Harris BK, Hoeksema MD, Trenary IA, Heidi C, Eisenberg R, Rathmell JC, Young JD, and Massion PP. xCT (SLC7A11)-mediated metabolic reprogramming promotes non-small cell lung cancer progression. Oncogene 37: 5007–5019, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Jia Y, Chen J, Zhu H, Jia ZH, and Cui MH. Aberrantly elevated redox sensing factor Nrf2 promotes cancer stem cell survival via enhanced transcriptional regulation of ABCG2 and Bcl-2/Bmi-1 genes. Oncol Rep 34: 2296–2304, 2015 [DOI] [PubMed] [Google Scholar]
- 116. Jiang T, Chen N, Zhao F, Wang XJ, Kong B, Zheng W, and Zhang DD. High levels of Nrf2 determine chemoresistance in type II endometrial cancer. Cancer Res 70: 5486–5496, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Jiang X, Bao Y, Liu H, Kou X, Zhang Z, Sun F, Qian Z, Lin Z, Li X, Liu X, Jiang L, and Yang Y. VPS34 stimulation of p62 phosphorylation for cancer progression. Oncogene 36: 6850–6862, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Jones GS, Spencer Feigelson H, Falk RT, Hua X, Ravel J, Yu G, Flores R, Gail MH, Shi J, Xu X, and Goedert JJ. Mammographic breast density and its association with urinary estrogens and the fecal microbiota in postmenopausal women. PLoS One 14: e0216114, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Joo JC, Hwang JH, Jo E, Kim YR, Kim DJ, Lee KB, Park SJ, and Jang IS. Cordycepin induces apoptosis by caveolin-1-mediated JNK regulation of Foxo3a in human lung adenocarcinoma. Oncotarget 8: 12211–12224, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Joo MS, Kim WD, Lee KY, Kim JH, Koo JH, and Kim SG. AMPK facilitates nuclear accumulation of Nrf2 by phosphorylating at serine 550. Mol Cell Biol 36: 1931–1942, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Ju HQ, Gocho T, Aguilar M, Wu M, Zhuang ZN, Fu J, Yanaga K, Huang P, and Chiao PJ. Mechanisms of overcoming intrinsic resistance to gemcitabine in pancreatic ductal adenocarcinoma through the redox modulation. Mol Cancer Ther 14: 788–798, 2015 [DOI] [PubMed] [Google Scholar]
- 122. Jung BJ, Yoo HS, Shin S, Park YJ, and Jeon SM. Dysregulation of NRF2 in cancer: from molecular mechanisms to therapeutic opportunities. Biomol Ther (Seoul) 26: 57–68, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Kabaria S, Choi DC, Chaudhuri AD, Jain MR, Li H, and Junn E. MicroRNA-7 activates Nrf2 pathway by targeting Keap1 expression. Free Radic Biol Med 89: 548–556, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Kalyanaraman B, Darley-Usmar V, Davies KJ, Dennery PA, Forman HJ, Grisham MB, Mann GE, Moore K, Roberts LJ 2nd, and Ischiropoulos H. Measuring reactive oxygen and nitrogen species with fluorescent probes: challenges and limitations. Free Radic Biol Med 52: 1–6, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Kang KA, Piao MJ, Kim KC, Kang HK, Chang WY, Park IC, Keum YS, Surh YJ, and Hyun JW. Epigenetic modification of Nrf2 in 5-fluorouracil-resistant colon cancer cells: involvement of TET-dependent DNA demethylation. Cell Death Dis 5: e1183, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Kang YP, Torrente L, Falzone A, Elkins CM, Liu M, Asara JM, Dibble CC, and DeNicola GM. Cysteine dioxygenase 1 is a metabolic liability for non-small cell lung cancer. Elife 8: 45572, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Kankia IH, Khalil HS, Langdon SP, Moult PR, Bown JL, and Deeni YY. NRF2 Regulates HER1 signaling pathway to modulate the sensitivity of ovarian cancer cells to lapatinib and erlotinib. Oxid Med Cell Longev 2017: 1864578, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Kansanen E, Kuosmanen SM, Leinonen H, and Levonen AL. The Keap1-Nrf2 pathway: mechanisms of activation and dysregulation in cancer. Redox Biol 1: 45–49, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Kapalczynska M, Kolenda T, Przybyla W, Zajaczkowska M, Teresiak A, Filas V, Ibbs M, Blizniak R, Luczewski L, and Lamperska K. 2D and 3D cell cultures—a comparison of different types of cancer cell cultures. Arch Med Sci 14: 910–919, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Katoh Y, Itoh K, Yoshida E, Miyagishi M, Fukamizu A, and Yamamoto M. Two domains of Nrf2 cooperatively bind CBP, a CREB binding protein, and synergistically activate transcription. Genes Cells 6: 857–868, 2001 [DOI] [PubMed] [Google Scholar]
- 131. Kawamoto Y, Nakamura Y, Naito Y, Torii Y, Kumagai T, Osawa T, Ohigashi H, Satoh K, Imagawa M, and Uchida K. Cyclopentenone prostaglandins as potential inducers of phase II detoxification enzymes. J Biol Chem 275: 11291–11299, 2000 [DOI] [PubMed] [Google Scholar]
- 132. Kawasaki Y, Okumura H, Uchikado Y, Kita Y, Sasaki K, Owaki T, Ishigami S, and Natsugoe S. Nrf2 is useful for predicting the effect of chemoradiation therapy on esophageal squamous cell carcinoma. Ann Surg Oncol 21: 2347–2352, 2014 [DOI] [PubMed] [Google Scholar]
- 133. Kerins MJ, Liu P, Tian W, Mannheim W, Zhang DD, and Ooi A. Genome-wide CRISPR screen reveals autophagy disruption as the convergence mechanism that regulates the NRF2 transcription factor. Mol Cell Biol 39: pii: , 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Kerins MJ and Ooi A. A catalogue of somatic NRF2 gain-of-function mutations in cancer. Sci Rep 8: 12846, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Khor TO, Fuentes F, Shu L, Paredes-Gonzalez X, Yang AY, Liu Y, Smiraglia DJ, Yegnasubramanian S, Nelson WG, and Kong AN. Epigenetic DNA methylation of antioxidative stress regulator NRF2 in human prostate cancer. Cancer Prev Res (Phila) 7: 1186–1197, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Kiesslich T, Mayr C, and Neureiter D. NRF2: the key to tumor- and patient-dependent chemosensitivity in biliary tract cancer? EBioMedicine 49: 9–10, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Kim HR, Kim S, Kim EJ, Park JH, Yang SH, Jeong ET, Park C, Youn MJ, So HS, and Park R. Suppression of Nrf2-driven heme oxygenase-1 enhances the chemosensitivity of lung cancer A549 cells toward cisplatin. Lung Cancer 60: 47–56, 2008 [DOI] [PubMed] [Google Scholar]
- 138. Kim MS, Lee WS, and Jin W. TrkB promotes breast cancer metastasis via suppression of Runx3 and Keap1 expression. Mol Cells 39: 258–265, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Kim SB, Pandita RK, Eskiocak U, Ly P, Kaisani A, Kumar R, Cornelius C, Wright WE, Pandita TK, and Shay JW. Targeting of Nrf2 induces DNA damage signaling and protects colonic epithelial cells from ionizing radiation. Proc Natl Acad Sci U S A 109: E2949–E2955, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Kim TH, Hur EG, Kang SJ, Kim JA, Thapa D, Lee YM, Ku SK, Jung Y, and Kwak MK. NRF2 blockade suppresses colon tumor angiogenesis by inhibiting hypoxia-induced activation of HIF-1alpha. Cancer Res 71: 2260–2275, 2011 [DOI] [PubMed] [Google Scholar]
- 141. Kim WD, Kim YW, Cho IJ, Lee CH, and Kim SG. E-cadherin inhibits nuclear accumulation of Nrf2: implications for chemoresistance of cancer cells. J Cell Sci 125: 1284–1295, 2012 [DOI] [PubMed] [Google Scholar]
- 142. Kim YR, Oh JE, Kim MS, Kang MR, Park SW, Han JY, Eom HS, Yoo NJ, and Lee SH. Oncogenic NRF2 mutations in squamous cell carcinomas of oesophagus and skin. J Pathol 220: 446–451, 2010 [DOI] [PubMed] [Google Scholar]
- 143. Kipp AP, Deubel S, Arnér ESJ, and Johansson K. Time- and cell-resolved dynamics of redox-sensitive Nrf2, HIF and NF-κB activities in 3D spheroids enriched for cancer stem cells. Redox Biol 12: 403–409, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Kitamura H and Motohashi H. NRF2 addiction in cancer cells. Cancer Sci 109: 900–911, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Kobayashi A, Kang MI, Okawa H, Ohtsuji M, Zenke Y, Chiba T, Igarashi K, and Yamamoto M. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol Cell Biol 24: 7130–7139, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Komatsu M, Kurokawa H, Waguri S, Taguchi K, Kobayashi A, Ichimura Y, Sou YS, Ueno I, Sakamoto A, Tong KI, Kim M, Nishito Y, Iemura S, Natsume T, Ueno T, Kominami E, Motohashi H, Tanaka K, and Yamamoto M. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat Cell Biol 12: 213–223, 2010 [DOI] [PubMed] [Google Scholar]
- 147. Konstantinopoulos PA, Fountzilas E, Pillay K, Zerbini LF, Libermann TA, Cannistra SA, and Spentzos D. Carboplatin-induced gene expression changes in vitro are prognostic of survival in epithelial ovarian cancer. BMC Med Genomics 1: 59, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Konstantinopoulos PA, Spentzos D, Fountzilas E, Francoeur N, Sanisetty S, Grammatikos AP, Hecht JL, and Cannistra SA. Keap1 mutations and Nrf2 pathway activation in epithelial ovarian cancer. Cancer Res 71: 5081–5089, 2011 [DOI] [PubMed] [Google Scholar]
- 149. Kovács P, Csonka T, Kovács T, Sári Z, Ujlaki G, Sipos A, Karányi Z, Szeőcs D, Hegedűs C, Uray K, Jankó L, Kiss M, Kiss B, Laoui D, Virág L, Méhes G, Bai P, and Mikó E. Lithocholic acid, a metabolite of the microbiome, increases oxidative stress in breast cancer. Cancers (Basel) 11: pii: , 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Kovács T, Mikó E, Vida A, Sebő É, Toth J, Csonka T, Boratkó A, Ujlaki G, Lente G, Kovács P, Tóth D, Árkosy P, Kiss B, Méhes G, Goedert JJ, and Bai P. Cadaverine, a metabolite of the microbiome, reduces breast cancer aggressiveness through trace amino acid receptors. Sci Rep 9: 1300, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Kröger C, Afeyan A, Mraz J, Eaton EN, Reinhardt F, Khodor YL, Thiru P, Bierie B, Ye X, Burge CB, and Weinberg RA. Acquisition of a hybrid E/M state is essential for tumorigenicity of basal breast cancer cells. Proc Natl Acad Sci U S A 116: 7353–7362, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Lacher SE, Levings DC, Freeman S, and Slattery M. Identification of a functional antioxidant response element at the HIF1A locus. Redox Biol 19: 401–411, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Lam HC, Baglini CV, Lope AL, Parkhitko AA, Liu H-J, Alesi N, Malinowska IA, Ebrahimi-Fakhari D, Saffari A, Yu JJ, Pereira A, Khabibullin D, Ogorek B, Nijmeh J, Kavanagh T, Handen A, Chan SY, Asara JM, Oldham WM, Diaz-Meco MT, Moscat J, Sahin M, Priolo C, and Henske EP. p62/SQSTM1 Cooperates with hyperactive mTORC1 to regulate glutathione production, maintain mitochondrial integrity, and promote tumorigenesis. Cancer Res 77: 3255–3267, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Lanczky A, Nagy A, Bottai G, Munkacsy G, Szabo A, Santarpia L, and Gyorffy B. miRpower: a web-tool to validate survival-associated miRNAs utilizing expression data from 2178 breast cancer patients. Breast Cancer Res Treat 160: 439–446, 2016 [DOI] [PubMed] [Google Scholar]
- 155. Lau A, Tian W, Whitman SA, and Zhang DD. The predicted molecular weight of Nrf2: it is what it is not. Antioxid Redox Signal 18: 91–93, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Lau A, Villeneuve NF, Sun Z, Wong PK, and Zhang DD. Dual roles of Nrf2 in cancer. Pharmacol Res 58: 262–270, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. LeBoeuf SE, Wu WL, Karakousi TR, Karadal B, Jackson SR, Davidson SM, Wong KK, Koralov SB, Sayin VI, and Papagiannakopoulos T. Activation of oxidative stress response in cancer generates a druggable dependency on exogenous non-essential amino acids. Cell Metab 31: 339–350.e4, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Lee JD, Cai Q, Shu XO, and Nechuta SJ. The role of biomarkers of oxidative stress in breast cancer risk and prognosis: a systematic review of the epidemiologic literature. J Womens Health (Larchmt) 26: 467–482, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Lee S, Hallis SP, Jung K-A, Ryu D, and Kwak M-K. Impairment of HIF-1α-mediated metabolic adaption by NRF2-silencing in breast cancer cells. Redox Biol 24: 101210, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Leone A, Roca MS, Ciardiello C, Terranova-Barberio M, Vitagliano C, Ciliberto G, Mancini R, Di Gennaro E, Bruzzese F, and Budillon A. Vorinostat synergizes with EGFR inhibitors in NSCLC cells by increasing ROS via up-regulation of the major mitochondrial porin VDAC1 and modulation of the c-Myc-NRF2-KEAP1 pathway. Free Radic Biol Med 89: 287–299, 2015 [DOI] [PubMed] [Google Scholar]
- 161. Levonen A-L, Landar A, Ramachandran A, Ceaser EK, Dickinson DA, Zanoni G, Morrow JD, and Darley-Usmar VM. Cellular mechanisms of redox cell signalling: role of cysteine modification in controlling antioxidant defences in response to electrophilic lipid oxidation products. Biochem J 378: 373–382, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Levy JMM, Towers CG, and Thorburn A. Targeting autophagy in cancer. Nat Rev Cancer 17: 528–542, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Lewerenz J, Hewett SJ, Huang Y, Lambros M, Gout PW, Kalivas PW, Massie A, Smolders I, Methner A, Pergande M, Smith SB, Ganapathy V, and Maher P. The cystine/glutamate antiporter system x(c)(-) in health and disease: from molecular mechanisms to novel therapeutic opportunities. Antioxid Redox Signal 18: 522–555, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Lewis KN, Wason E, Edrey YH, Kristan DM, Nevo E, and Buffenstein R. Regulation of Nrf2 signaling and longevity in naturally long-lived rodents. Proc Natl Acad Sci U S A 112: 3722–3727, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Li N, Wang M, Barajas B, Sioutas C, Williams MA, and Nel AE. Nrf2 deficiency in dendritic cells enhances the adjuvant effect of ambient ultrafine particles on allergic sensitization. J Innate Immun 5: 543–554, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Li W and Kong AN. Molecular mechanisms of Nrf2-mediated antioxidant response. Mol Carcinog 48: 91–104, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Liao H, Zhou Q, Zhang Z, Wang Q, Sun Y, Yi X, and Feng Y. NRF2 is overexpressed in ovarian epithelial carcinoma and is regulated by gonadotrophin and sex-steroid hormones. Oncol Rep 27: 1918–1924, 2012 [DOI] [PubMed] [Google Scholar]
- 168. Lignitto L, LeBoeuf SE, Homer H, Jiang S, Askenazi M, Karakousi TR, Pass HI, Bhutkar AJ, Tsirigos A, Ueberheide B, Sayin VI, Papagiannakopoulos T, and Pagano M. Nrf2 activation promotes lung cancer metastasis by inhibiting the degradation of Bach1. Cell 178: 316–329.e18, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Lim JKM, Delaidelli A, Minaker SW, Zhang H-F, Colovic M, Yang H, Negri GL, von Karstedt S, Lockwood WW, Schaffer P, Leprivier G, and Sorensen PH. Cystine/glutamate antiporter xCT (SLC7A11) facilitates oncogenic RAS transformation by preserving intracellular redox balance. Proc Natl Acad Sci U S A 116: 9433–9442, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Lin DC, Dinh HQ, Xie JJ, Mayakonda A, Silva TC, Jiang YY, Ding LW, He JZ, Xu XE, Hao JJ, Wang MR, Li C, Xu LY, Li EM, Berman BP, and Phillip Koeffler H. Identification of distinct mutational patterns and new driver genes in oesophageal squamous cell carcinomas and adenocarcinomas. Gut 67: 1769–1779, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Lin P, Ren Y, Yan X, Luo Y, Zhang H, Kesarwani M, Bu J, Zhan D, Zhou Y, Tang Y, Zhu S, Xu W, Zhou X, Mei C, Ma L, Ye L, Hu C, Azam M, Ding W, Jin J, Huang G, and Tong H. The high NRF2 expression confers chemotherapy resistance partly through up-regulated DUSP1 in myelodysplastic syndromes. Haematologica 104: 485–496, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Lister A, Nedjadi T, Kitteringham NR, Campbell F, Costello E, Lloyd B, Copple IM, Williams S, Owen A, Neoptolemos JP, Goldring CE, and Park BK. Nrf2 is overexpressed in pancreatic cancer: implications for cell proliferation and therapy. Mol Cancer 10: 37, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Liu C, Zhao Y, Wang J, Yang Y, Zhang Y, Qu X, Peng S, Yao Z, Zhao S, He B, Mi Q, Zhu Y, Liu X, Zou J, Zhang X, and Du Q. FoxO3 reverses 5-fluorouracil resistance in human colorectal cancer cells by inhibiting the Nrf2/TR1 signaling pathway. Cancer Lett 470: 29–42, 2020 [DOI] [PubMed] [Google Scholar]
- 174. Liu T, Jiang L, Tavana O, and Gu W. The deubiquitylase OTUB1 mediates ferroptosis via stabilization of SLC7A11. Cancer Res 79: 1913–1924, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Liu X, Abe-Kanoh N, Liu Y, Zhu B, Munemasa S, Nakamura T, Murata Y, and Nakamura Y. Inhibition of phosphatidylinositide 3-kinase impairs the benzyl isothiocyanate-induced accumulation of autophagic molecules and Nrf2 in human colon cancer cells. Biosci Biotechnol Biochem 81: 2212–2215, 2017 [DOI] [PubMed] [Google Scholar]
- 176. Lodi M, Priksz D, Fulop GA, Bodi B, Gyongyosi A, Nagy L, Kovacs A, Kertesz AB, Kocsis J, Edes I, Csanadi Z, Czuriga I, Kisvarday Z, Juhasz B, Lekli I, Bai P, Toth A, Papp Z, and Czuriga D. Advantages of prophylactic versus conventionally scheduled heart failure therapy in an experimental model of doxorubicin-induced cardiomyopathy. J Transl Med 17: 229, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Lu BC, Li J, Yu WF, Zhang GZ, Wang HM, and Ma HM. Elevated expression of Nrf2 mediates multidrug resistance in CD133(+) head and neck squamous cell carcinoma stem cells. Oncol Lett 12: 4333–4338, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Lu K, Alcivar AL, Ma J, Foo TK, Zywea S, Mahdi A, Huo Y, Kensler TW, Gatza ML, and Xia B. NRF2 induction supporting breast cancer cell survival is enabled by oxidative stress-induced DPP3-KEAP1 interaction. Cancer Res 77: 2881–2892, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Luo L, Chen Y, Wu D, Shou J, Wang S, Ye J, Tang X, and Wang XJ. Butylated hydroxyanisole induces distinct expression patterns of Nrf2 and detoxification enzymes in the liver and small intestine of C57BL/6 mice. Toxicol Appl Pharmacol 288: 339–348, 2015 [DOI] [PubMed] [Google Scholar]
- 180. Luo M, Shang L, Brooks MD, Jiagge E, Zhu Y, Buschhaus JM, Conley S, Fath MA, Davis A, Gheordunescu E, Wang Y, Harouaka R, Lozier A, Triner D, McDermott S, Merajver SD, Luker GD, Spitz DR, and Wicha MS. Targeting breast cancer stem cell state equilibrium through modulation of redox signaling. Cell Metab 28: 69–86, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Ma J, Cai H, Wu T, Sobhian B, Huo Y, Alcivar A, Mehta M, Cheung KL, Ganesan S, Kong AN, Zhang DD, and Xia B. PALB2 interacts with KEAP1 to promote NRF2 nuclear accumulation and function. Mol Cell Biol 32: 1506–1517, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Ma X, Zhang J, Liu S, Huang Y, Chen B, and Wang D. Nrf2 knockdown by shRNA inhibits tumor growth and increases efficacy of chemotherapy in cervical cancer. Cancer Chemother Pharmacol 69: 485–494, 2012 [DOI] [PubMed] [Google Scholar]
- 183. Mabley JG, Pacher P, Bai P, Wallace R, Goonesekera S, Virag L, Southan GJ, and Szabo C. Suppression of intestinal polyposis in Apcmin/+ mice by targeting the nitric oxide or poly(ADP-ribose) pathways. Mutat Res 548: 107–116, 2004 [DOI] [PubMed] [Google Scholar]
- 184. Magesh S, Chen Y, and Hu L. Small molecule modulators of Keap1-Nrf2-ARE pathway as potential preventive and therapeutic agents. Med Res Rev 32: 687–726, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Maher JM, Dieter MZ, Aleksunes LM, Slitt AL, Guo G, Tanaka Y, Scheffer GL, Chan JY, Manautou JE, Chen Y, Dalton TP, Yamamoto M, and Klaassen CD. Oxidative and electrophilic stress induces multidrug resistance-associated protein transporters via the nuclear factor-E2-related factor-2 transcriptional pathway. Hepatology 46: 1597–1610, 2007 [DOI] [PubMed] [Google Scholar]
- 186. Malhotra D, Portales-Casamar E, Singh A, Srivastava S, Arenillas D, Happel C, Shyr C, Wakabayashi N, Kensler TW, Wasserman WW, and Biswal S. Global mapping of binding sites for Nrf2 identifies novel targets in cell survival response through ChIP-Seq profiling and network analysis. Nucleic Acids Res 38: 5718–5734, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Martinez VD, Vucic EA, Pikor LA, Thu KL, Hubaux R, and Lam WL. Frequent concerted genetic mechanisms disrupt multiple components of the NRF2 inhibitor KEAP1/CUL3/RBX1 E3-ubiquitin ligase complex in thyroid cancer. Mol Cancer 12: 124, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Martinez VD, Vucic EA, Thu KL, Pikor LA, Hubaux R, and Lam WL. Unique pattern of component gene disruption in the NRF2 inhibitor KEAP1/CUL3/RBX1 E3-ubiquitin ligase complex in serous ovarian cancer. Biomed Res Int 2014: 159459, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Mato JM and Lu SC. Role of S-adenosyl-L-methionine in liver health and injury. Hepatology 45: 1306–1312, 2007 [DOI] [PubMed] [Google Scholar]
- 190. McMahon M, Lamont DJ, Beattie KA, and Hayes JD. Keap1 perceives stress via three sensors for the endogenous signaling molecules nitric oxide, zinc, and alkenals. Proc Natl Acad Sci U S A 107: 18838–18843, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191. McMahon M, Thomas N, Itoh K, Yamamoto M, and Hayes JD. Redox-regulated turnover of Nrf2 is determined by at least two separate protein domains, the redox-sensitive Neh2 degron and the redox-insensitive Neh6 degron. J Biol Chem 279: 31556–31567, 2004 [DOI] [PubMed] [Google Scholar]
- 192. McMahon M, Thomas N, Itoh K, Yamamoto M, and Hayes JD. Dimerization of substrate adaptors can facilitate cullin-mediated ubiquitylation of proteins by a “tethering” mechanism: a two-site interaction model for the Nrf2-Keap1 complex. J Biol Chem 281: 24756–24768, 2006 [DOI] [PubMed] [Google Scholar]
- 193. Menegon S, Columbano A, and Giordano S. The dual roles of NRF2 in cancer. Trends Mol Med 22: 578–593, 2016 [DOI] [PubMed] [Google Scholar]
- 194. Miko E, Kovacs T, Sebo E, Toth J, Csonka T, Ujlaki G, Sipos A, Szabo J, Mehes G, and Bai P. Microbiome-microbial metabolome-cancer cell interactions in breast cancer-familiar, but unexplored. Cells 8: E293, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195. Miko E, Vida A, and Bai P. Translational aspects of the microbiome-to be exploited. Cell Biol Toxicol 32: 153–156, 2016 [DOI] [PubMed] [Google Scholar]
- 196. Miko E, Vida A, Kovacs T, Ujlaki G, Trencsenyi G, Marton J, Sari Z, Kovacs P, Boratko A, Hujber Z, Csonka T, Antal-Szalmas P, Watanabe M, Gombos I, Csoka B, Kiss B, Vigh L, Szabo J, Mehes G, Sebestyen A, Goedert JJ, and Bai P. Lithocholic acid, a bacterial metabolite reduces breast cancer cell proliferation and aggressiveness. Biochim Biophys Acta 1859: 958–974, 2018 [DOI] [PubMed] [Google Scholar]
- 197. Mitsuishi Y, Taguchi K, Kawatani Y, Shibata T, Nukiwa T, Aburatani H, Yamamoto M, and Motohashi H. Nrf2 redirects glucose and glutamine into anabolic pathways in metabolic reprogramming. Cancer Cell 22: 66–79, 2012 [DOI] [PubMed] [Google Scholar]
- 198. Miura S, Shibazaki M, Kasai S, Yasuhira S, Watanabe A, Inoue T, Kageshita Y, Tsunoda K, Takahashi K, Akasaka T, Masuda T, and Maesawa C. A somatic mutation of the KEAP1 gene in malignant melanoma is involved in aberrant NRF2 activation and an increase in intrinsic drug resistance. J Invest Dermatol 134: 553–556, 2014 [DOI] [PubMed] [Google Scholar]
- 199. Morales AA, Gutman D, Cejas PJ, Lee KP, and Boise LH. Reactive oxygen species are not required for an arsenic trioxide-induced antioxidant response or apoptosis. J Biol Chem 284: 12886–12895, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200. Mostafavi-Pour Z, Ramezani F, Keshavarzi F, and Samadi N. The role of quercetin and vitamin C in Nrf2-dependent oxidative stress production in breast cancer cells. Oncol Lett 13: 1965–1973, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 201. Muscarella LA, Barbano R, D'Angelo V, Copetti M, Coco M, Balsamo T, la Torre A, Notarangelo A, Troiano M, Parisi S, Icolaro N, Catapano D, Valori VM, Pellegrini F, Merla G, Carella M, Fazio VM, and Parrella P. Regulation of KEAP1 expression by promoter methylation in malignant gliomas and association with patient's outcome. Epigenetics 6: 317–325, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202. Nguyen T, Sherratt PJ, Huang HC, Yang CS, and Pickett CB. Increased protein stability as a mechanism that enhances Nrf2-mediated transcriptional activation of the antioxidant response element. Degradation of Nrf2 by the 26 S proteasome. J Biol Chem 278: 4536–4541, 2003 [DOI] [PubMed] [Google Scholar]
- 203. Nioi P and Nguyen T. A mutation of Keap1 found in breast cancer impairs its ability to repress Nrf2 activity. Biochem Biophys Res Commun 362: 816–821, 2007 [DOI] [PubMed] [Google Scholar]
- 204. Nioi P, Nguyen T, Sherratt PJ, and Pickett CB. The carboxy-terminal Neh3 domain of Nrf2 is required for transcriptional activation. Mol Cell Biol 25: 10895–10906, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205. Niringiyumukiza JD, Cai H, Chen L, Li Y, Wang L, Zhang M, Xu X, and Xiang W. Protective properties of glycogen synthase kinase-3 inhibition against doxorubicin-induced oxidative damage to mouse ovarian reserve. Biomed Pharmacother 116: 108963, 2019 [DOI] [PubMed] [Google Scholar]
- 206. Numazawa S, Sugihara K, Miyake S, Tomiyama H, Hida A, Hatsuno M, Yamamoto M, and Yoshida T. Possible involvement of oxidative stress in 5-fluorouracil-mediated myelosuppression in mice. Basic Clin Pharmacol Toxicol 108: 40–45, 2011 [DOI] [PubMed] [Google Scholar]
- 207. Ogura T, Tong KI, Mio K, Maruyama Y, Kurokawa H, Sato C, and Yamamoto M. Keap1 is a forked-stem dimer structure with two large spheres enclosing the intervening, double glycine repeat, and C-terminal domains. Proc Natl Acad Sci U S A 107: 2842–2847, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208. Ooi A, Dykema K, Ansari A, Petillo D, Snider J, Kahnoski R, Anema J, Craig D, Carpten J, Teh BT, and Furge KA. CUL3 and NRF2 mutations confer an NRF2 activation phenotype in a sporadic form of papillary renal cell carcinoma. Cancer Res 73: 2044–2051, 2013 [DOI] [PubMed] [Google Scholar]
- 209. Ooi A, Wong JC, Petillo D, Roossien D, Perrier-Trudova V, Whitten D, Min BW, Tan MH, Zhang Z, Yang XJ, Zhou M, Gardie B, Molinie V, Richard S, Tan PH, Teh BT, and Furge KA. An antioxidant response phenotype shared between hereditary and sporadic type 2 papillary renal cell carcinoma. Cancer Cell 20: 511–523, 2011 [DOI] [PubMed] [Google Scholar]
- 210. Ozgen M, Koca SS, Karatas A, Dagli AF, Erman F, Gundogdu B, Sahin K, and Isik A. Lapatinib ameliorates experimental arthritis in rats. Inflammation 38: 252–259, 2015 [DOI] [PubMed] [Google Scholar]
- 211. Pacher P, Liaudet L, Bai P, Mabley JG, Kaminski PM, Virag L, Deb A, Szabo E, Ungvari Z, Wolin MS, Groves JT, and Szabo C. Potent metalloporphyrin peroxynitrite decomposition catalyst protects against the development of doxorubicin-induced cardiac dysfunction. Circulation 107: 896–904, 2003 [DOI] [PubMed] [Google Scholar]
- 212. Pacher P, Liaudet L, Bai P, Virag L, Mabley JG, Hasko G, and Szabo C. Activation of poly(ADP-ribose) polymerase contributes to development of doxorubicin-induced heart failure. J Pharmacol Exp Ther 300: 862–867, 2002 [DOI] [PubMed] [Google Scholar]
- 213. Padmanabhan B, Tong KI, Ohta T, Nakamura Y, Scharlock M, Ohtsuji M, Kang MI, Kobayashi A, Yokoyama S, and Yamamoto M. Structural basis for defects of Keap1 activity provoked by its point mutations in lung cancer. Mol Cell 21: 689–700, 2006 [DOI] [PubMed] [Google Scholar]
- 214. Peiris-Pages M, Martinez-Outschoorn UE, Pestell RG, Sotgia F, and Lisanti MP. Cancer stem cell metabolism. Breast Cancer Res 18: 55, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215. Phuong NT, Kim SK, Im JH, Yang JW, Choi MC, Lim SC, Lee KY, Kim YM, Yoon JH, and Kang KW. Induction of methionine adenosyltransferase 2A in tamoxifen-resistant breast cancer cells. Oncotarget 7: 13902–13916, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216. Phuong NTT, Kim SK, Lim SC, Kim HS, Kim TH, Lee KY, Ahn S-G, Yoon J-H, and Kang KW. Role of PTEN promoter methylation in tamoxifen-resistant breast cancer cells. Breast Cancer Res Treat 130: 73–83, 2011 [DOI] [PubMed] [Google Scholar]
- 217. Pirpour Tazehkand A, Akbarzadeh M, Velaie K, Sadeghi MR, and Samadi N. The role of Her2-Nrf2 axis in induction of oxaliplatin resistance in colon cancer cells. Biomed Pharmacother 103: 755–766, 2018 [DOI] [PubMed] [Google Scholar]
- 218. Pons DG, Nadal-Serrano M, Torrens-Mas M, Valle A, Oliver J, and Roca P. UCP2 inhibition sensitizes breast cancer cells to therapeutic agents by increasing oxidative stress. Free Radic Biol Med 86: 67–77, 2015 [DOI] [PubMed] [Google Scholar]
- 219. Primiano T, Sutter TR, and Kensler TW. Antioxidant-inducible genes. Adv Pharmacol 38: 293–328, 1997 [DOI] [PubMed] [Google Scholar]
- 220. Qu J, Zhang L, Li L, and Su Y. miR-148b functions as a tumor suppressor by targeting endoplasmic reticulum metallo protease 1 in human endometrial cancer cells. Oncol Res 27: 81–88, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 221. Que L, He L, Yu C, Yin W, Ma L, Cao B, and Yu S. Activation of Nrf2-ARE signaling mitigates cyclophosphamide-induced myelosuppression. Toxicol Lett 262: 17–26, 2016 [DOI] [PubMed] [Google Scholar]
- 222. Rabinowitz JD and White E. Autophagy and metabolism. Science 330: 1344–1348, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223. Rada P, Rojo AI, Chowdhry S, McMahon M, Hayes JD, and Cuadrado A. SCF/{beta}-TrCP promotes glycogen synthase kinase 3-dependent degradation of the Nrf2 transcription factor in a Keap1-independent manner. Mol Cell Biol 31: 1121–1133, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224. Rada P, Rojo AI, Evrard-Todeschi N, Innamorato NG, Cotte A, Jaworski T, Tobon-Velasco JC, Devijver H, Garcia-Mayoral MF, Van Leuven F, Hayes JD, Bertho G, and Cuadrado A. Structural and functional characterization of Nrf2 degradation by the glycogen synthase kinase 3/beta-TrCP axis. Mol Cell Biol 32: 3486–3499, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225. Ranganathan S, Krishnan A, and Sivasithambaram ND. Significance of twist and iNOS expression in human breast carcinoma. Mol Cell Biochem 412: 41–47, 2016 [DOI] [PubMed] [Google Scholar]
- 226. Rekhtman N, Pietanza MC, Hellmann MD, Naidoo J, Arora A, Won H, Halpenny DF, Wang H, Tian SK, Litvak AM, Paik PK, Drilon AE, Socci N, Poirier JT, Shen R, Berger MF, Moreira AL, Travis WD, Rudin CM, and Ladanyi M. Next-generation sequencing of pulmonary large cell neuroendocrine carcinoma reveals small cell carcinoma-like and non-small cell carcinoma-like subsets. Clin Cancer Res 22: 3618–3629, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227. Reuter S, Gupta SC, Chaturvedi MM, and Aggarwal BB. Oxidative stress, inflammation, and cancer: how are they linked? Free Radic Biol Med 49: 1603–1616, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228. Riz I, Hawley TS, Marsal JW, Hawley RG, Riz I, Hawley TS, Marsal JW, and Hawley RG. Noncanonical SQSTM1/p62-Nrf2 pathway activation mediates proteasome inhibitor resistance in multiple myeloma cells via redox, metabolic and translational reprogramming. Oncotarget 7: 66360–66385, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229. Rocha CR, Kajitani GS, Quinet A, Fortunato RS, and Menck CF. NRF2 and glutathione are key resistance mediators to temozolomide in glioma and melanoma cells. Oncotarget 7: 48081–48092, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230. Roh JL, Kim EH, Jang H, and Shin D. Nrf2 inhibition reverses the resistance of cisplatin-resistant head and neck cancer cells to artesunate-induced ferroptosis. Redox Biol 11: 254–262, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231. Roininen N, Haapasaari KM, and Karihtala P. The role of redox-regulating enzymes in inoperable breast cancers treated with neoadjuvant chemotherapy. Oxid Med Cell Longev 2017: 2908039, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232. Rojo AI, Rada P, Mendiola M, Ortega-Molina A, Wojdyla K, Rogowska-Wrzesinska A, Hardisson D, Serrano M, and Cuadrado A. The PTEN/NRF2 axis promotes human carcinogenesis. Antioxid Redox Signal 21: 2498–2514, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233. Rolland P, Madjd Z, Durrant L, Ellis IO, Layfield R, and Spendlove I. The ubiquitin-binding protein p62 is expressed in breast cancers showing features of aggressive disease. Endocr Relat Cancer 14: 73–80, 2007 [DOI] [PubMed] [Google Scholar]
- 234. Romero R, Sayin VI, Davidson SM, Bauer MR, Singh SX, LeBoeuf SE, Karakousi TR, Ellis DC, Bhutkar A, Sanchez-Rivera FJ, Subbaraj L, Martinez B, Bronson RT, Prigge JR, Schmidt EE, Thomas CJ, Goparaju C, Davies A, Dolgalev I, Heguy A, Allaj V, Poirier JT, Moreira AL, Rudin CM, Pass HI, Vander Heiden MG, Jacks T, and Papagiannakopoulos T. Keap1 loss promotes Kras-driven lung cancer and results in dependence on glutaminolysis. Nat Med 23: 1362–1368, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235. Ron D and Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol 8: 519–529, 2007 [DOI] [PubMed] [Google Scholar]
- 236. Rota C, Chignell CF, and Mason RP. Evidence for free radical formation during the oxidation of 2′-7′-dichlorofluorescin to the fluorescent dye 2′-7′-dichlorofluorescein by horseradish peroxidase: possible implications for oxidative stress measurements. Free Radic Biol Med 27: 873–881, 1999 [DOI] [PubMed] [Google Scholar]
- 237. Rushworth SA and Macewan DJ. The role of nrf2 and cytoprotection in regulating chemotherapy resistance of human leukemia cells. Cancers (Basel) 3: 1605–1621, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238. Rushworth SA, Zaitseva L, Murray MY, Shah NM, Bowles KM, and MacEwan DJ. The high Nrf2 expression in human acute myeloid leukemia is driven by NF-kappaB and underlies its chemo-resistance. Blood 120: 5188–5198, 2012 [DOI] [PubMed] [Google Scholar]
- 239. Ryoo IG, Choi BH, Ku SK, and Kwak MK. High CD44 expression mediates p62-associated NFE2L2/NRF2 activation in breast cancer stem cell-like cells: implications for cancer stem cell resistance. Redox Biol 17: 246–258, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240. Ryoo IG, Choi BH, and Kwak MK. Activation of NRF2 by p62 and proteasome reduction in sphere-forming breast carcinoma cells. Oncotarget 6: 8167–8184, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241. Sadeghi MR, Jeddi F, Soozangar N, Somi MH, and Samadi N. The role of Nrf2-Keap1 axis in colorectal cancer, progression, and chemoresistance. Tumour Biol 39: 1010428317705510, 2017 [DOI] [PubMed] [Google Scholar]
- 242. Saito T, Ichimura Y, Taguchi K, Suzuki T, Mizushima T, Takagi K, Hirose Y, Nagahashi M, Iso T, Fukutomi T, Ohishi M, Endo K, Uemura T, Nishito Y, Okuda S, Obata M, Kouno T, Imamura R, Tada Y, Obata R, Yasuda D, Takahashi K, Fujimura T, Pi J, Lee M-S, Ueno T, Ohe T, Mashino T, Wakai T, Kojima H, Okabe T, Nagano T, Motohashi H, Waguri S, Soga T, Yamamoto M, Tanaka K, and Komatsu M. p62/Sqstm1 promotes malignancy of HCV-positive hepatocellular carcinoma through Nrf2-dependent metabolic reprogramming. Nat Commun 7: 12030, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243. Salaroglio IC, Panada E, Moiso E, Buondonno I, Provero P, Rubinstein M, Kopecka J, and Riganti C. PERK induces resistance to cell death elicited by endoplasmic reticulum stress and chemotherapy. Mol Cancer 16: 91, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244. Samatiwat P, Prawan A, Senggunprai L, and Kukongviriyapan V. Repression of Nrf2 enhances antitumor effect of 5-fluorouracil and gemcitabine on cholangiocarcinoma cells. Naunyn Schmiedebergs Arch Pharmacol 388: 601–612, 2015 [DOI] [PubMed] [Google Scholar]
- 245. Sanghvi VR, Leibold J, Mina M, Mohan P, Berishaj M, Li Z, Miele MM, Lailler N, Zhao C, de Stanchina E, Viale A, Akkari L, Lowe SW, Ciriello G, Hendrickson RC, and Wendel HG. The oncogenic action of NRF2 depends on de-glycation by fructosamine-3-kinase. Cell 178: 807–819, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246. Satoh H, Moriguchi T, Taguchi K, Takai J, Maher JM, Suzuki T, Winnard PT Jr., Raman V, Ebina M, Nukiwa T, and Yamamoto M.. Nrf2-deficiency creates a responsive microenvironment for metastasis to the lung. Carcinogenesis 31: 1833–1843, 2010 [DOI] [PubMed] [Google Scholar]
- 247. Sayin VI, LeBoeuf SE, Singh SX, Davidson SM, Biancur D, Guzelhan BS, Alvarez SW, Wu WL, Karakousi TR, Zavitsanou AM, Ubriaco J, Muir A, Karagiannis D, Morris PJ, Thomas CJ, Possemato R, Vander Heiden MG, and Papagiannakopoulos T. Activation of the NRF2 antioxidant program generates an imbalance in central carbon metabolism in cancer. eLife 6: pii: , 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248. Schafer ZT, Grassian AR, Song L, Jiang Z, Gerhart-Hines Z, Irie HY, Gao S, Puigserver P, and Brugge JS. Antioxidant and oncogene rescue of metabolic defects caused by loss of matrix attachment. Nature 461: 109–113, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249. Schatten H. Cell and Molecular Biology of Breast Cancer New York, NY: Humana Press, 2013 [Google Scholar]
- 250. Schwab LP, Peacock DL, Majumdar D, Ingels JF, Jensen LC, Smith KD, Cushing RC, and Seagroves TN. Hypoxia-inducible factor 1α promotes primary tumor growth and tumor-initiating cell activity in breast cancer. Breast Cancer Res 14: R6, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251. Sekhar KR, Rachakonda G, and Freeman ML. Cysteine-based regulation of the CUL3 adaptor protein Keap1. Toxicol Appl Pharmacol 244: 21–26, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252. Semenza GL. Regulation of mammalian O 2 homeostasis by hypoxia-inducible factor 1. Annu Rev Cell Dev Biol 15: 551–578, 1999 [DOI] [PubMed] [Google Scholar]
- 253. Shah NM, Rushworth SA, Murray MY, Bowles KM, and MacEwan DJ. Understanding the role of NRF2-regulated miRNAs in human malignancies. Oncotarget 4: 1130–1142, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254. Shao J, Glorieux C, Liao J, Chen P, Lu W, Liang Z, Wen S, Hu Y, and Huang P. Impact of Nrf2 on tumour growth and drug sensitivity in oncogenic K-ras-transformed cells in vitro and in vivo. Free Radic Res 52: 661–671, 2018 [DOI] [PubMed] [Google Scholar]
- 255. Shi L, Wu L, Chen Z, Yang J, Chen X, Yu F, Zheng F, and Lin X. MiR-141 Activates Nrf2-dependent antioxidant pathway via down-regulating the expression of Keap1 conferring the resistance of hepatocellular carcinoma cells to 5-fluorouracil. Cell Physiol Biochem 35: 2333–2348, 2015 [DOI] [PubMed] [Google Scholar]
- 256. Shibata T, Kokubu A, Gotoh M, Ojima H, Ohta T, Yamamoto M, and Hirohashi S. Genetic alteration of Keap1 confers constitutive Nrf2 activation and resistance to chemotherapy in gallbladder cancer. Gastroenterology 135: 1358–1368, 2008 [DOI] [PubMed] [Google Scholar]
- 257. Shibata T, Kokubu A, Saito S, Narisawa-Saito M, Sasaki H, Aoyagi K, Yoshimatsu Y, Tachimori Y, Kushima R, Kiyono T, and Yamamoto M. NRF2 mutation confers malignant potential and resistance to chemoradiation therapy in advanced esophageal squamous cancer. Neoplasia 13: 864–873, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258. Shibata T, Kondo M, Osawa T, Shibata N, Kobayashi M, and Uchida K. 15-Deoxy-Δ 12,14 -prostaglandin J2. J Biol Chem 277: 10459–10466, 2002 [DOI] [PubMed] [Google Scholar]
- 259. Shibata T, Ohta T, Tong KI, Kokubu A, Odogawa R, Tsuta K, Asamura H, Yamamoto M, and Hirohashi S. Cancer related mutations in NRF2 impair its recognition by Keap1-Cul3 E3 ligase and promote malignancy. Proc Natl Acad Sci U S A 105: 13568–13573, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260. Shim GS, Manandhar S, Shin DH, Kim TH, and Kwak MK. Acquisition of doxorubicin resistance in ovarian carcinoma cells accompanies activation of the NRF2 pathway. Free Radic Biol Med 47: 1619–1631, 2009 [DOI] [PubMed] [Google Scholar]
- 261. Shimizu T, Inoue K, Hachiya H, Shibuya N, Aoki T, and Kubota K. Accumulation of phosphorylated p62 is associated with NF-E2-related factor 2 activation in hepatocellular carcinoma. J Hepatobiliary Pancreat Sci 23: 467–471, 2016 [DOI] [PubMed] [Google Scholar]
- 262. Shin CS, Mishra P, Watrous JD, Carelli V, D'Aurelio M, Jain M, and Chan DC. The glutamate/cystine xCT antiporter antagonizes glutamine metabolism and reduces nutrient flexibility. Nat Commun 8: 15074, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263. Shirasaki K, Taguchi K, Unno M, Motohashi H, and Yamamoto M. NF-E2-related factor 2 promotes compensatory liver hypertrophy after portal vein branch ligation in mice. Hepatology 59: 2371–2382, 2014 [DOI] [PubMed] [Google Scholar]
- 264. Sid B, Glorieux C, Valenzuela M, Rommelaere G, Najimi M, Dejeans N, Renard P, Verrax J, and Calderon PB. AICAR induces Nrf2 activation by an AMPK-independent mechanism in hepatocarcinoma cells. Biochem Pharmacol 91: 168–180, 2014 [DOI] [PubMed] [Google Scholar]
- 265. Silva MM, Rocha CRR, Kinker GS, Pelegrini AL, and Menck CFM. The balance between NRF2/GSH antioxidant mediated pathway and DNA repair modulates cisplatin resistance in lung cancer cells. Sci Rep 9: 17639, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266. Singh A, Misra V, Thimmulappa RK, Lee H, Ames S, Hoque MO, Herman JG, Baylin SB, Sidransky D, Gabrielson E, Brock MV, and Biswal S. Dysfunctional KEAP1-NRF2 interaction in non-small-cell lung cancer. PLoS Med 3: e420, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267. Singh B, Bhat NK, and Bhat HK. Induction of NAD(P)H-quinone oxidoreductase 1 by antioxidants in female ACI rats is associated with decrease in oxidative DNA damage and inhibition of estrogen-induced breast cancer. Carcinogenesis 33: 156–163, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268. Singh B, Chatterjee A, Ronghe AM, Bhat NK, and Bhat HK. Antioxidant-mediated up-regulation of OGG1 via NRF2 induction is associated with inhibition of oxidative DNA damage in estrogen-induced breast cancer. BMC Cancer 13: 253, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269. Singh B, Ronghe AM, Chatterjee A, Bhat NK, and Bhat HK. MicroRNA-93 regulates NRF2 expression and is associated with breast carcinogenesis. Carcinogenesis 34: 1165–1172, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270. Singh B, Shoulson R, Chatterjee A, Ronghe A, Bhat NK, Dim DC, and Bhat HK. Resveratrol inhibits estrogen-induced breast carcinogenesis through induction of NRF2-mediated protective pathways. Carcinogenesis 35: 1872–1880, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271. Sjoblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber TD, Mandelker D, Leary RJ, Ptak J, Silliman N, Szabo S, Buckhaults P, Farrell C, Meeh P, Markowitz SD, Willis J, Dawson D, Willson JK, Gazdar AF, Hartigan J, Wu L, Liu C, Parmigiani G, Park BH, Bachman KE, Papadopoulos N, Vogelstein B, Kinzler KW, and Velculescu VE. The consensus coding sequences of human breast and colorectal cancers. Science 314: 268–274, 2006 [DOI] [PubMed] [Google Scholar]
- 272. Sompakdee V, Prawan A, Senggunprai L, Kukongviriyapan U, Samathiwat P, Wandee J, and Kukongviriyapan V. Suppression of Nrf2 confers chemosensitizing effect through enhanced oxidant-mediated mitochondrial dysfunction. Biomed Pharmacother 101: 627–634, 2018 [DOI] [PubMed] [Google Scholar]
- 273. Sulaiman A, McGarry S, Han X, Liu S, and Wang L. CSCs in breast cancer-one size does not fit all: therapeutic advances in targeting heterogeneous epithelial and mesenchymal CSCs. Cancers (Basel) 11: pii: , 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274. Sun X, Liu D, Xue Y, and Hu X. Enforced miR-144-3p expression as a non-invasive biomarker for the acute myeloid leukemia patients mainly by targeting NRF2. Clin Lab 63: 679–687, 2017 [DOI] [PubMed] [Google Scholar]
- 275. Sun X, Wang S, Gai J, Guan J, Li J, Li Y, Zhao J, Zhao C, Fu L, and Li Q. SIRT5 promotes cisplatin resistance in ovarian cancer by suppressing DNA damage in a ROS-dependent manner via regulation of the Nrf2/HO-1 pathway. Front Oncol 9: 754, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276. Sun Y, Abdul Aziz A, Bowles K, and Rushworth S. High NRF2 expression controls endoplasmic reticulum stress induced apoptosis in multiple myeloma. Cancer Lett 412: 37–45, 2018 [DOI] [PubMed] [Google Scholar]
- 277. Suzuki T, Motohashi H, and Yamamoto M. Toward clinical application of the Keap1-Nrf2 pathway. Trends Pharmacol Sci 34: 340–346, 2013 [DOI] [PubMed] [Google Scholar]
- 278. Taguchi K, Hirano I, Itoh T, Tanaka M, Miyajima A, Suzuki A, Motohashi H, and Yamamoto M. Nrf2 enhances cholangiocyte expansion in Pten-deficient livers. Mol Cell Biol 34: 900–913, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279. Tao S, Wang S, Moghaddam SJ, Ooi A, Chapman E, Wong PK, and Zhang DD. Oncogenic KRAS confers chemoresistance by upregulating NRF2. Cancer Res 74: 7430–7441, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280. Tebay LE, Robertson H, Durant ST, Vitale SR, Penning TM, Dinkova-Kostova AT, and Hayes JD. Mechanisms of activation of the transcription factor Nrf2 by redox stressors, nutrient cues, and energy status and the pathways through which it attenuates degenerative disease. Free Radic Biol Med 88: 108–146, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281. Teng Y, Zhao H, Gao L, Zhang W, Shull AY, and Shay C. FGF19 protects hepatocellular carcinoma cells against endoplasmic reticulum stress via activation of FGFR4-GSK3beta-Nrf2 signaling. Cancer Res 77: 6215–6225, 2017 [DOI] [PubMed] [Google Scholar]
- 282. Thimmulappa RK, Mai KH, Srisuma S, Kensler TW, Yamamoto M, and Biswal S. Identification of Nrf2-regulated genes induced by the chemopreventive agent sulforaphane by oligonucleotide microarray. Cancer Res 62: 5196–5203, 2002 [PubMed] [Google Scholar]
- 283. Tian Y, Wu K, Liu Q, Han N, Zhang L, Chu Q, and Chen Y. Modification of platinum sensitivity by KEAP1/NRF2 signals in non-small cell lung cancer. J Hematol Oncol 9: 83, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284. Timmerman LA, Holton T, Yuneva M, Louie RJ, Padro M, Daemen A, Hu M, Chan DA, Ethier SP, van ‘t Veer LJ, Polyak K, McCormick F, and Gray JW. Glutamine sensitivity analysis identifies the xCT antiporter as a common triple-negative breast tumor therapeutic target. Cancer Cell 24: 450–465, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285. To C, Ringelberg CS, Royce DB, Williams CR, Risingsong R, Sporn MB, and Liby KT. Dimethyl fumarate and the oleanane triterpenoids, CDDO-imidazolide and CDDO-methyl ester, both activate the Nrf2 pathway but have opposite effects in the A/J model of lung carcinogenesis. Carcinogenesis 36: 769–781, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286. Tonelli C, Chio IIC, and Tuveson DA. Transcriptional regulation by Nrf2. Antioxid Redox Signal 29: 1727–1745, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287. Tong KI, Kobayashi A, Katsuoka F, and Yamamoto M. Two-site substrate recognition model for the Keap1-Nrf2 system: a hinge and latch mechanism. Biol Chem 387: 1311–1320, 2006 [DOI] [PubMed] [Google Scholar]
- 288. Tong KI, Padmanabhan B, Kobayashi A, Shang C, Hirotsu Y, Yokoyama S, and Yamamoto M. Different electrostatic potentials define ETGE and DLG motifs as hinge and latch in oxidative stress response. Mol Cell Biol 27: 7511–7521, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289. Tsuji Y. JunD activates transcription of the human ferritin H gene through an antioxidant response element during oxidative stress. Oncogene 24: 7567–7578, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290. Ungvari Z, Tarantini S, Nyul-Toth A, Kiss T, Yabluchanskiy A, Csipo T, Balasubramanian P, Lipecz A, Benyo Z, and Csiszar A. Nrf2 dysfunction and impaired cellular resilience to oxidative stressors in the aged vasculature: from increased cellular senescence to the pathogenesis of age-related vascular diseases. Geroscience 41: 727–738, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291. van der Wijst MG, Huisman C, Mposhi A, Roelfes G, and Rots MG. Targeting Nrf2 in healthy and malignant ovarian epithelial cells: protection versus promotion. Mol Oncol 9: 1259–1273, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292. Venkatraman G, Benesch MG, Tang X, Dewald J, McMullen TP, and Brindley DN. Lysophosphatidate signaling stabilizes Nrf2 and increases the expression of genes involved in drug resistance and oxidative stress responses: implications for cancer treatment. FASEB J 29: 772–785, 2015 [DOI] [PubMed] [Google Scholar]
- 293. Venugopal R and Jaiswal AK. Nrf2 and Nrf1 in association with Jun proteins regulate antioxidant response element-mediated expression and coordinated induction of genes encoding detoxifying enzymes. Oncogene 17: 3145–3156, 1998 [DOI] [PubMed] [Google Scholar]
- 294. Walker A, Singh A, Tully E, Woo J, Le A, Nguyen T, Biswal S, Sharma D, and Gabrielson E. Nrf2 signaling and autophagy are complementary in protecting breast cancer cells during glucose deprivation. Free Radic Biol Med 120: 407–413, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295. Walsh EM, Keane MM, Wink DA, Callagy G, and Glynn SA. Review of triple negative breast cancer and the impact of inducible nitric oxide synthase on tumor biology and patient outcomes. Crit Rev Oncog 21: 333–351, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296. Wang B, Teng Y, and Liu Q. MicroRNA-153 regulates NRF2 expression and is associated with breast carcinogenesis. Clin Lab 62: 39–47, 2016 [DOI] [PubMed] [Google Scholar]
- 297. Wang H, Liu K, Geng M, Gao P, Wu X, Hai Y, Li Y, Li Y, Luo L, Hayes JD, Wang XJ, and Tang X. RXRalpha inhibits the NRF2-ARE signaling pathway through a direct interaction with the Neh7 domain of NRF2. Cancer Res 73: 3097–3108, 2013 [DOI] [PubMed] [Google Scholar]
- 298. Wang L, Liu Y, Du T, Yang H, Lei L, Guo M, Ding HF, Zhang J, Wang H, Chen X, and Yan C. ATF3 promotes erastin-induced ferroptosis by suppressing system Xc. Cell Death Differ 27: 662–675, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299. Wang N, Song L, Xu Y, Zhang L, Wu Y, Guo J, Ji W, Li L, Zhao J, Zhang X, and Zhan L. Loss of scribble confers cisplatin resistance during NSCLC chemotherapy via Nox2/ROS and Nrf2/PD-L1 signaling. EBioMedicine 47: 65–77, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300. Wang Q, Ma J, Lu Y, Zhang S, Huang J, Chen J, Bei JX, Yang K, Wu G, Huang K, Chen J, and Xu S. CDK20 interacts with KEAP1 to activate NRF2 and promotes radiochemoresistance in lung cancer cells. Oncogene 36: 5321–5330, 2017 [DOI] [PubMed] [Google Scholar]
- 301. Wang R, An J, Ji F, Jiao H, Sun H, and Zhou D. Hypermethylation of the Keap1 gene in human lung cancer cell lines and lung cancer tissues. Biochem Biophys Res Commun 373: 151–154, 2008 [DOI] [PubMed] [Google Scholar]
- 302. Wang SW, Xu Y, Weng YY, Fan XY, Bai YF, Zheng XY, Lou LJ, and Zhang F. Astilbin ameliorates cisplatin-induced nephrotoxicity through reducing oxidative stress and inflammation. Food Chem Toxicol 114: 227–236, 2018 [DOI] [PubMed] [Google Scholar]
- 303. Wang T, Liu Q, and Duan L. MBNL1 regulates resistance of HeLa cells to cisplatin via Nrf2. Biochem Biophys Res Commun 522: 763–769, 2020 [DOI] [PubMed] [Google Scholar]
- 304. Wang XJ, Hayes JD, Henderson CJ, and Wolf CR. Identification of retinoic acid as an inhibitor of transcription factor Nrf2 through activation of retinoic acid receptor alpha. Proc Natl Acad Sci U S A 104: 19589–19594, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305. Wang XJ, Li Y, Luo L, Wang H, Chi Z, Xin A, Li X, Wu J, and Tang X. Oxaliplatin activates the Keap1/Nrf2 antioxidant system conferring protection against the cytotoxicity of anticancer drugs. Free Radic Biol Med 70: 68–77, 2014 [DOI] [PubMed] [Google Scholar]
- 306. Wang YY, Chen J, Liu XM, Zhao R, and Zhe H. Nrf2-mediated metabolic reprogramming in cancer. Oxid Med Cell Longev 2018: 9304091, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307. Wang Z, Chen Z, Jiang Z, Luo P, Liu L, Huang Y, Wang H, Wang Y, Long L, Tan X, Liu D, Jin T, Wang Y, Wang Y, Liao F, Zhang C, Chen L, Gan Y, Liu Y, Yang F, Huang C, Miao H, Chen J, Cheng T, Fu X, and Shi C. Cordycepin prevents radiation ulcer by inhibiting cell senescence via NRF2 and AMPK in rodents. Nat Commun 10: 2538, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308. Weniger MA, Rizzatti EG, Perez-Galan P, Liu D, Wang Q, Munson PJ, Raghavachari N, White T, Tweito MM, Dunleavy K, Ye Y, Wilson WH, and Wiestner A. Treatment-induced oxidative stress and cellular antioxidant capacity determine response to bortezomib in mantle cell lymphoma. Clin Cancer Res 17: 5101–5112, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309. Wiel C, Le Gal K, Ibrahim MX, Jahangir CA, Kashif M, Yao H, Ziegler DV, Xu X, Ghosh T, Mondal T, Kanduri C, Lindahl P, Sayin VI, and Bergo MO. BACH1 stabilization by antioxidants stimulates lung cancer metastasis. Cell 178: 330–345, 2019 [DOI] [PubMed] [Google Scholar]
- 310. Winkel AF, Engel CK, Margerie D, Kannt A, Szillat H, Glombik H, Kallus C, Ruf S, Gussregen S, Riedel J, Herling AW, von Knethen A, Weigert A, Brune B, and Schmoll D. Characterization of RA839, a noncovalent small molecule binder to Keap1 and selective activator of Nrf2 signaling. J Biol Chem 290: 28446–28455, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311. Wong TF, Yoshinaga K, Monma Y, Ito K, Niikura H, Nagase S, Yamamoto M, and Yaegashi N. Association of keap1 and nrf2 genetic mutations and polymorphisms with endometrioid endometrial adenocarcinoma survival. Int J Gynecol Cancer 21: 1428–1435, 2011 [DOI] [PubMed] [Google Scholar]
- 312. Wu RP, Hayashi T, Cottam HB, Jin G, Yao S, Wu CC, Rosenbach MD, Corr M, Schwab RB, and Carson DA. Nrf2 responses and the therapeutic selectivity of electrophilic compounds in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A 107: 7479–7484, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313. Wu T, Zhao F, Gao B, Tan C, Yagishita N, Nakajima T, Wong PK, Chapman E, Fang D, and Zhang DD. Hrd1 suppresses Nrf2-mediated cellular protection during liver cirrhosis. Genes Dev 28: 708–722, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314. Wu YW, Lin CF, Lin YS, Su WC, and Chiu WH. Autophagy regulates vinorelbine sensitivity due to continued Keap1-mediated ROS generation in lung adenocarcinoma cells. Cell Death Discov 4: 33, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315. Xia M, Yu H, Gu S, Xu Y, Su J, Li H, Kang J, and Cui M. p62/SQSTM1 is involved in cisplatin resistance in human ovarian cancer cells via the Keap1-Nrf2-ARE system. Int J Oncol 45: 2341–2348, 2014 [DOI] [PubMed] [Google Scholar]
- 316. Xiang Y, Ye W, Huang C, Yu D, Chen H, Deng T, Zhang F, Lou B, Zhang J, Shi K, Chen B, and Zhou M. Brusatol enhances the chemotherapy efficacy of gemcitabine in pancreatic cancer via the Nrf2 signalling pathway. Oxid Med Cell Longev 2018: 2360427, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317. Xu D, Li X, Shao F, Lv G, Lv H, Lee J-H, Qian X, Wang Z, Xia Y, Du L, Zheng Y, Wang H, Lyu J, and Lu Z. The protein kinase activity of fructokinase A specifies the antioxidant responses of tumor cells by phosphorylating p62. Sci Adv 5: eaav4570, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318. Xu X, zur Hausen A, Coy JF, and Löchelt M. Transketolase-like protein 1 (TKTL1) is required for rapid cell growth and full viability of human tumor cells. Int J Cancer 124: 1330–1337, 2009 [DOI] [PubMed] [Google Scholar]
- 319. Yamaguchi Y, Kamai T, Higashi S, Murakami S, Arai K, Shirataki H, and Yoshida KI. Nrf2 gene mutation and single nucleotide polymorphism rs6721961 of the Nrf2 promoter region in renal cell cancer. BMC Cancer 19: 1137, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320. Yamamoto S, Inoue J, Kawano T, Kozaki K, Omura K, and Inazawa J. The impact of miRNA-based molecular diagnostics and treatment of NRF2-stabilized tumors. Mol Cancer Res 12: 58–68, 2014 [DOI] [PubMed] [Google Scholar]
- 321. Yamamoto T, Suzuki T, Kobayashi A, Wakabayashi J, Maher J, Motohashi H, and Yamamoto M. Physiological significance of reactive cysteine residues of Keap1 in determining Nrf2 activity. Mol Cell Biol 28: 2758–2770, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322. Yang H, Huang Z-Z, Zeng Z, Chen C, Selby RR, and Lu SC. Role of promoter methylation in increased methionine adenosyltransferase 2A expression in human liver cancer. Am J Physiol Gastrointest Liver Physiol 280: G184–G190, 2001 [DOI] [PubMed] [Google Scholar]
- 323. Yang KQ, Liu Y, Huang QH, Mo N, Zhang QY, Meng QG, and Cheng JW. Bone marrow-derived mesenchymal stem cells induced by inflammatory cytokines produce angiogenetic factors and promote prostate cancer growth. BMC Cancer 17: 878, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324. Yang M, Yao Y, Eades G, Zhang Y, and Zhou Q. MiR-28 regulates Nrf2 expression through a Keap1-independent mechanism. Breast Cancer Res Treat 129: 983–991, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325. Yang Y, Luo L, Cai X, Fang Y, Wang J, Chen G, Yang J, Zhou Q, Sun X, Cheng X, Yan H, Lu W, Hu C, and Cao P. Nrf2 inhibits oxaliplatin-induced peripheral neuropathy via protection of mitochondrial function. Free Radic Biol Med 120: 13–24, 2018 [DOI] [PubMed] [Google Scholar]
- 326. Yoneda K, Tanaka F, Kondo N, Hashimoto M, Takuwa T, Matsumoto S, Okumura Y, Tsubota N, Sato A, Tsujimura T, Kuribayashi K, Tabata C, Nakano T, and Hasegawa S. Circulating tumor cell (CTC) as a prognostic marker in primary lung cancer. J Clin Oncol 33: e22041, 2015 [Google Scholar]
- 327. Yu M, Bardia A, Wittner BS, Stott SL, Smas ME, Ting DT, Isakoff SJ, Ciciliano JC, Wells MN, Shah AM, Concannon KF, Donaldson MC, Sequist LV, Brachtel E, Sgroi D, Baselga J, Ramaswamy S, Toner M, Haber DA, and Maheswaran S. Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science 339: 580–584, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328. Yuan X, Xu C, Pan Z, Keum YS, Kim JH, Shen G, Yu S, Oo KT, Ma J, and Kong AN. Butylated hydroxyanisole regulates ARE-mediated gene expression via Nrf2 coupled with ERK and JNK signaling pathway in HepG2 cells. Mol Carcinog 45: 841–850, 2006 [DOI] [PubMed] [Google Scholar]
- 329. Yun CW and Lee SH. The roles of autophagy in cancer. Int J Mol Sci 19: pii: , 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330. Zagorski JW, Turley AE, Freeborn RA, VanDenBerg KR, Dover HE, Kardell BR, Liby KT, and Rockwell CE. Differential effects of the Nrf2 activators tBHQ and CDDO-Im on the early events of T cell activation. Biochem Pharmacol 147: 67–76, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331. Zhang B, Xu J, Li C, Shi S, Ji S, Xu W, Liu J, Jin K, Liang D, Liang C, Liu L, Liu C, Qin Y, and Yu X. MBD1 is an epigenetic regulator of KEAP1 in pancreatic cancer. Curr Mol Med 16: 404–411, 2016 [DOI] [PubMed] [Google Scholar]
- 332. Zhang DD, Lo SC, Cross JV, Templeton DJ, and Hannink M. Keap1 is a redox-regulated substrate adaptor protein for a Cul3-dependent ubiquitin ligase complex. Mol Cell Biol 24: 10941–10953, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333. Zhang HS, Du GY, Zhang ZG, Zhou Z, Sun HL, Yu XY, Shi YT, Xiong DN, Li H, and Huang YH. NRF2 facilitates breast cancer cell growth via HIF1a-mediated metabolic reprogramming. Int J Biochem Cell Biol 95: 85–92, 2018 [DOI] [PubMed] [Google Scholar]
- 334. Zhang HS, Zhang ZG, Du GY, Sun HL, Liu HY, Zhou Z, Gou XM, Wu XH, Yu XY, and Huang YH. Nrf2 promotes breast cancer cell migration via up-regulation of G6PD/HIF-1alpha/Notch1 axis. J Cell Mol Med 23: 3451–3463, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335. This reference has been deleted
- 336. Zhang J, Jiao Q, Kong L, Yu J, Fang A, Li M, and Yu J. Nrf2 and Keap1 abnormalities in esophageal squamous cell carcinoma and association with the effect of chemoradiotherapy. Thorac Cancer 9: 726–735, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337. Zhang P, Singh A, Yegnasubramanian S, Esopi D, Kombairaju P, Bodas M, Wu H, Bova SG, and Biswal S. Loss of Kelch-like ECH-associated protein 1 function in prostate cancer cells causes chemoresistance and radioresistance and promotes tumor growth. Mol Cancer Ther 9: 336–346, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338. Zhang Y, Shi J, Liu X, Feng L, Gong Z, Koppula P, Sirohi K, Li X, Wei Y, Lee H, Zhuang L, Chen G, Xiao Z-D, Hung M-C, Chen J, Huang P, Li W, and Gan B. BAP1 links metabolic regulation of ferroptosis to tumour suppression. Nat Cell Biol 20: 1181–1192, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339. Zhang Z, Wang Q, Ma J, Yi X, Zhu Y, Xi X, Feng Y, and Jin Z. Reactive oxygen species regulate FSH-induced expression of vascular endothelial growth factor via Nrf2 and HIF1alpha signaling in human epithelial ovarian cancer. Oncol Rep 29: 1429–1434, 2013 [DOI] [PubMed] [Google Scholar]
- 340. Zhao M, Xu H, Zhang B, Hong B, Yan W, and Zhang J. Impact of nuclear factor erythroid-derived 2-like 2 and p62/sequestosome expression on prognosis of patients with gliomas. Hum Pathol 46: 843–849, 2015 [DOI] [PubMed] [Google Scholar]
- 341. Zhou C, Zhao L, Zheng J, Wang K, Deng H, Liu P, Chen L, and Mu H. MicroRNA-144 modulates oxidative stress tolerance in SH-SY5Y cells by regulating nuclear factor erythroid 2-related factor 2-glutathione axis. Neurosci Lett 655: 21–27, 2017 [DOI] [PubMed] [Google Scholar]
- 342. Zhou L, Ao L, Yan Y, Li C, Li W, Ye A, Liu J, Hu Y, Fang W, and Li Y. Levo-corydalmine attenuates vincristine-induced neuropathic pain in mice by upregulating the Nrf2/HO-1/CO pathway to inhibit Connexin 43 expression. Neurotherapeutics 17: 340–355, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343. Zhou L, Zhang H, Davies KJA, and Forman HJ. Aging-related decline in the induction of Nrf2-regulated antioxidant genes in human bronchial epithelial cells. Redox Biol 14:35–40, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344. Zhou S, Ye W, Zhang Y, Yu D, Shao Q, Liang J, and Zhang M. miR-144 reverses chemoresistance of hepatocellular carcinoma cell lines by targeting Nrf2-dependent antioxidant pathway. Am J Transl Res 8: 2992–3002, 2016 [PMC free article] [PubMed] [Google Scholar]
- 345. Zhou Z, Xu J, Bao X, Shi J, Liu B, Chen Y, and Li J. Nuclear Nrf2 activity in laryngeal carcinoma is regulated by SENP3 after cisplatin-induced reactive oxygen species stress. J Cancer 10: 3427–3434, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346. Zhu M and Fahl WE. Functional characterization of transcription regulators that interact with the electrophile response element. Biochem Biophys Res Commun 289: 212–219, 2001 [DOI] [PubMed] [Google Scholar]
- 347. Zipper LM and Mulcahy RT. The Keap1 BTB/POZ dimerization function is required to sequester Nrf2 in cytoplasm. J Biol Chem 277: 36544–36552, 2002 [DOI] [PubMed] [Google Scholar]