Abstract
Current biochemical studies of nitric oxide synthase enzymes (NOSs) are conducted in diluted solutions. However, the intracellular milieu where the proteins perform their biological functions is crowded with macromolecules. The crowding effect on the electron transfer kinetics of multidomain proteins is much less understood. Herein, we investigated the effect of macromolecular crowding on the FMN ‒ heme intraprotein interdomain electron transfer (IET), an obligatory step in NOS catalysis. A noticeable increase in the IET rate in the bi-domain oxygenase/FMN (oxyFMN) and the holoprotein of human inducible NOS (iNOS) was observed upon addition of Ficoll 70 in a non-saturatable manner. Additionally, the magnitude of IET enhancement for the holoenzyme is much higher than that that of the oxyFMN construct. Importantly, the enhancing extent is similar for the iNOS oxyFMN protein with added Ficoll 70 and Dextran 70, showing that specific interactions do not exist between the NOS protein and the crowder. The crowding effect is also evident under different ionic strength. Moreover, the population of the docked FMN-heme state is significantly increased upon adding Ficoll 70. The steady-state cytochrome c reduction by the holoenzyme is noticeably enhanced by the crowder, while the ferricyanide reduction is unchanged. The NO production activity of the iNOS holoenzyme is stimulated by Ficoll 70. The effect of macromolecular crowding on the kinetics can be rationalized based on excluded volume effect, with an entropic origin. The results indicate that the macromolecular crowding influences the NOS electron transfer through multiple pathways.
Keywords: nitric oxide synthase, electron transfer, excluded volume, macromolecular crowding, kinetics, laser flash photolysis
Graphical Abstract
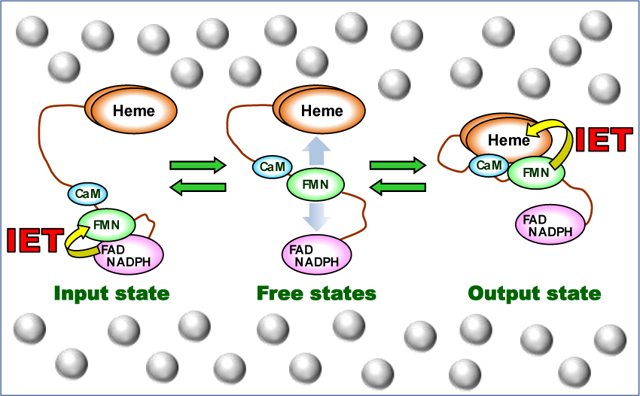
1. Introduction
Nitric oxide (NO) is a key signaling small molecule in the cardiovascular system and it exerts other functions in the neuron and immune systems.1, 2 NO synthases (NOSs) catalyze oxidation of l-Arginine (l-Arg) to NO. Eukaryotic NOS is a modular heme-flavoprotein. Each subunit of the homodimeric NOS has two domains: a C-terminal reductase domain with binding sites for NADPH (the electron source), FAD, and FMN, and an N-terminal heme-containing dimeric oxygenase domain (Figure 1A).3, 4 The l-Arg substrate and a 6R-5,6,7,8-tetrahydrobiopterin (H4B) cofactor bind near the catalytic heme center.5 The reductase and oxygenase (heme) domain are joined by a calmodulin (CaM)-binding linker. There are three mammalian NOS isoforms: neuronal, endothelial, and inducible NOS (nNOS, eNOS, and iNOS, respectively). In iNOS, CaM binds to the CaM-binding linker at basal Ca2+ concentration, whereas in eNOS and nNOS, the CaM binding requires an increase of calcium concentration.3, 6
Figure 1.
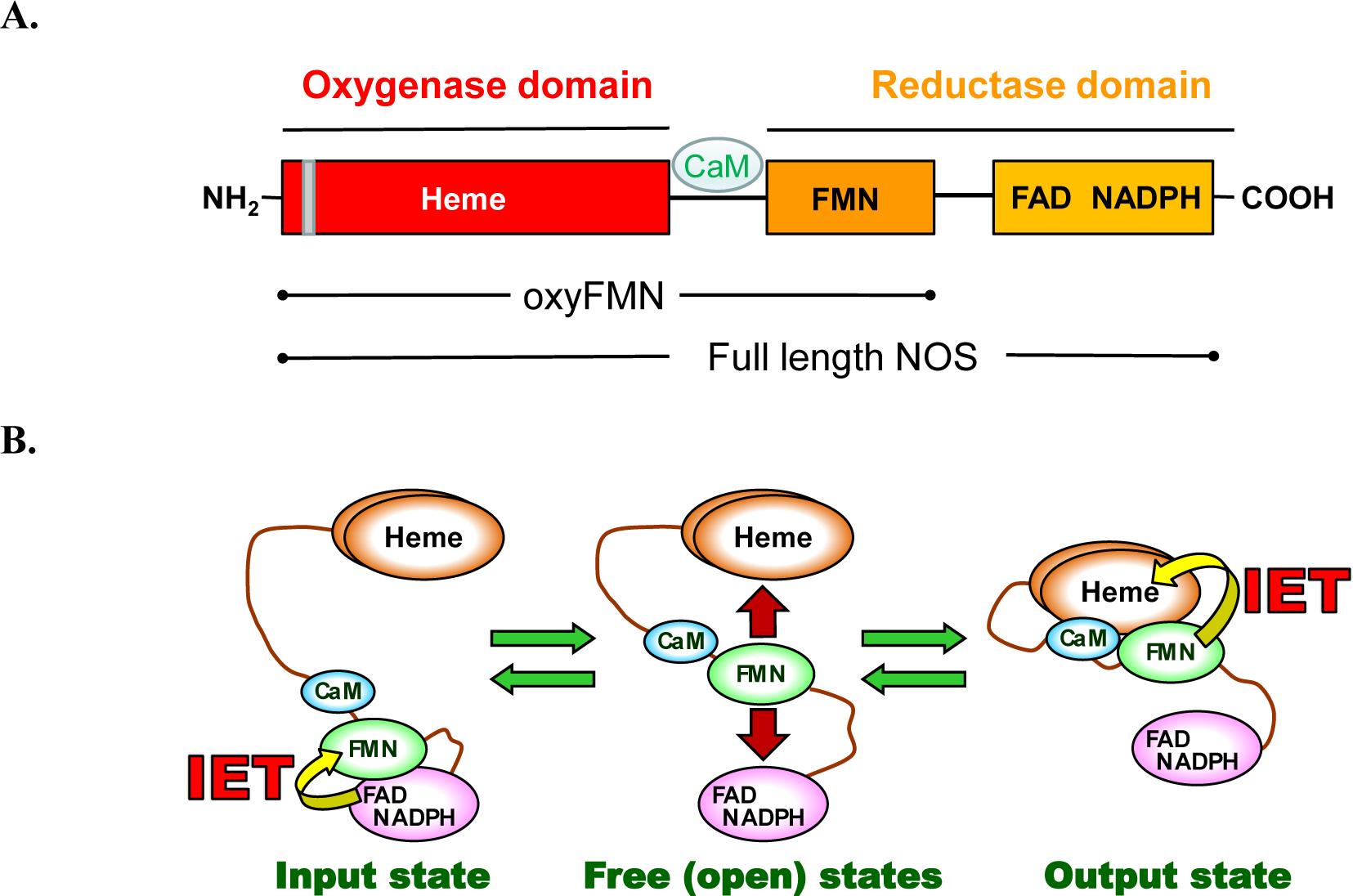
A. Domain organization of full-length and bi-domain oxyFMN iNOS proteins. The grey bar indicates the dimer interface in the heme-containing oxygenase domain. The oxyFMN and holo iNOS proteins used in this study are purified as dimers. B. Tether shuttle model for the NOS electron transport pathway. The FMN domain swings between the input and output states through the free/open states to transport the NADPH-derived electrons across the NOS domains.
The CaM-activated NO synthesis requires NOS to undergo significant conformational changes, in which the FMN domain moves back and forth between the “input” (electron-accepting) state and the “output” (electron-donating) state to deliver electrons across the enzyme (Figure 1B).7 Such tethered shuttle mechanism also functions in the ancestral electron transfer protein systems (e.g., diflavin oxidoreductase8), while the unique characteristic of the NOS protein is the contribution of CaM. CaM activates NOS by two mechanisms. First, CaM binding releases the FMN domain from the input state.9 The NO synthesis at the heme site further requires a CaM-dependent heme-FMN domain docking to form the output state.10 The CaM-dependent output state enables the FMN − heme interdomain electron transfer (IET),11 allowing O2 activation and subsequent NO synthesis at the heme site. A CO photolysis approach was developed by us for direct measurement of kinetics of the FMN – heme IET between the catalytically relevant heme Fe and FMN centers in the NOS proteins.12
The previous biochemical studies of NOS proteins including ours were conducted in dilute conditions in vitro. However, the intracellular milieu, in which the enzymes perform their biological functions in vivo, is packed with macromolecules such as polysaccharides and ribosomes. The macromolecules in the cytoplasm exist in the concentration range of 80 − 400 mg/ml and occupy 5 – 40 % of total aqua-based volume.13 A substantial fraction of the cellular space is physically occupied by the macromolecules and thus unavailable to other molecules, resulting in repulsive interactions and structural hurdles to the free movements of other molecules. The biochemical profiles of the proteins inside cells are thus different from those determined by in vitro experiments with dilute samples.14 In the cellular crowded conditions, nonspecific interactions can affect biomolecules’ behavior. For example, the effect of macromolecular crowding on protein behavior is substantial for those that experience conformational changes, and macromolecular crowding can considerably shift the equilibria between protein conformations and affect its function.15 It is thus interesting to study the effect of macromolecular crowding on structure and function of biomolecules16 including proteins17–19 and DNA.20 However, the crowding effect is much less understood in the context of kinetics facet of enzymatic reactions, and only few reports on effect of crowding on protein electron transfer are available in the literature, most of which are for inter-protein electron transfer complexes.21, 22 The intraprotein FMN − heme IET in NOS requires conformational cycling between open and docked conformational states (Figure 1B).23 The NOS’s large conformational changes inspire our examination of its sensitivity to macromolecular crowding. To determine the effect of crowding on the intraprotein FMN − heme IET kinetics in the NOS proteins, herein we utilized inert synthetic polymer crowding molecules, Ficoll 70 and Dextran 70, to imitate the crowding conditions. Ficoll 70 and Dextran 70 are common crowding agents used to mimic the crowded cellular environment because these neutral inert polysaccharides lack attractive interactions for proteins, and resemble more closely the proteins seen in a biological medium.24 Moreover, they are colorless and do not interfere with spectroscopic measurements of the electron transfer and enzymatic activities. Importantly, macromolecular crowding is dependent upon the hydrodynamic radius of the macromolecular crowder and the protein of interest, and the most operative conditions are those in which the macromolecular crowder and the protein are of similar size.25, 26 Ficoll 70 has comparable diameter as the NOS proteins: Ficoll 70 has a radius of ~ 5.5 nm,27 and the radius of NOS systems ranges between 4.2 and 8.6 nm.9, 28, 29 The radius of Dextran 70 is ~ 6.5 nm,30 which is also similar to the size of the NOS molecule.
In this work, we have determined and compared the macromolecular crowding effects on the FMN − heme IET rates of a truncated bi-domain oxygenase/FMN (oxyFMN) construct and the full-length protein of human iNOS (UniProtKB P35228). The oxyFMN construct comprises of only the oxygenase domain and the FMN domain, together with the CaM binding region (Figure 1A). This is done to preclude the FAD-FMN interdomain interactions and to promote the interdomain FMN-heme interactions.31 Biochemical results demonstrate that the oxyFMN construct is a valid model of the NOS output state.32–34 Indeed, the oxyFMN model35 can be fitted into the docked FMN/heme module without any alteration into the electron microscopy density of the holoprotein,36 showing that the docked interdomain FMN-heme complex structure is identical in the full-length NOS and truncated oxyFMN proteins. To determine whether specific interactions exist between the protein and the crowder, we also compared the IET rates with added Ficoll 70 and Dextran 70 that give the same solution viscosity. Steady-state cytochrome (cyt.) c reduction by the iNOS holoenzyme in the presence of the crowder was further measured to probe the electron transport through the entire reductase unit. The kinetics results, along with the fluorescence lifetime data, demonstrate that the macromolecular crowder enhances the FMN – heme IET by excluded volume effect, its origin being entropic.
2. Materials and Methods
2.1. Overexpression and purification of human iNOS oxyFMN and iNOS holoenzyme
The human iNOS oxyFMN or holoprotein plasmid was co-transfected with the CaM expression vector (p209) into E. coli BL21 (DE3) competent cells.37 Expression and purification of the human iNOS holoprotein were performed as described.38 The cloning and purification of the iNOS oxyFMN protein were conducted using the previous procedure.39
2.2. Laser flash photolysis
Laser flash photolysis experiments were conducted at 21 °C on an Edinburgh LP920 laser flash photolysis spectrometer, as previously described.40 Briefly, sample contained ~ 20 μM deazariboflavin (dRF) and 5 mM semicarbazide in a pH 7.6 buffer (40 mM bis-Tris propane, 400 mM NaCl, 1 mM Ca2+, 2 mM l-Arg, 20 μM H4Band 10% glycerol). The crowder (Ficoll 70 or Dextran 70) was added to the solution when necessary. The dRF solution without the protein in a cuvette was vigorously deaerated by a CO/Ar (v/v 1:3) mixed gas for ~ 90 minutes. The surface of introduced protein aliquot was then purged by the gas to remove traces of O2 before it was mixed into the dRF solution. The sample was illuminated for 3 – 4 minutes by white light to obtain the [Fe(II)−CO][FMNH•] form. The protein sample was subsequently flashed with 446 nm laser excitation to initiate the FMN ‒ heme IET, which can be monitored by the decrease of absorption of FMNH· and Fe(II) at 580 nm and 465 nm, respectively.41 The CO photolysis experiments were conducted at least twice for one sample. The averaged kinetic traces were analyzed by OriginPro 2019 (OriginLab, MA).
2.3. Fluorescence lifetime measurement
Time-resolved fluorescence intensity decays were recorded on a Mini-tau filter-based fluorescence lifetime spectrometer (Edinburgh, U.K.). The time-correlated single photon counting technique was used.42 The excitation at 450 nm was obtained using an EPL-450 pulsed diode laser (Edinburgh, U.K.) with a 2 MHz repetition rate. The emission was detected at 530 nm with a polarizer oriented at 54.7° with respect to the linearly polarized excitation light. This magic-angle placement is to avoid the effects of rotational diffusion on the intensity decays. The fluorescence lifetime measurements of human iNOS oxyFMN were performed at a final concentration of 2 μM in a pH 7.6 buffer (40 mM bis-Tris propane, 400 mM NaCl, 1 mM Ca2+, 2 mM l-Arg, 20 μM H4B, 1 mM dithiothreitol, 10% glycerol) with added crowder when needed. The instrument response was collected on a scattering Ludox solution.
The fluorescence intensity decays were analyzed by the fluorescence analysis software FAST (Edinburgh, U.K.) using a multiexponential model where the intensity is supposed to decay as the sum of individual single exponential decays. The goodness-of-fit can be judged by the value of χ2.
2.4. Steady-state NOS enzymatic activity assays
Steady-state rate of NO production by the human iNOS holoenzyme was determined in a pH 7.4 buffer (50 mM Tris, 100 mM NaCl, 5 μM H4B, and 200 μM CaCl2).43 The sample solution contained 100 μM l-Arg, 100 μM NADPH, and 8 μM oxyhemoglobin. As needed, the crowder was added into the assay buffer. Reaction was initiated by adding 20 nM final concentration of human iNOS holoprotein, and the rate of NO synthesis was measured at 401 nm, using an extinction coefficient of 60 mM−1 cm−1.43
Cyt. c reduction by the iNOS holoenzyme was determined in a pH 7.5 buffer (50 mM Tris, 100 mM NaCl, 200 μM CaCl2) with added crowder. Assay mixtures contained 1.5 nM human iNOS holoprotein and 40 μM horse heart cyt. c (Sigma-Aldrich, MO). The reduction reaction was started by adding 100 μM NADPH, and the rate of cyt. c reduction was measured at 550 nm, using an extinction coefficient of 21 mM−1 cm−1.43 Ferricyanide reduction assay was performed in the same manner except that the potassium ferricyanide concentration was 1 mM and the extinction coefficient is 1.02 mM−1 cm−1 at 420 nm.44 A semi-micro cuvette was used to give higher transmittance and better signal at 420 nm.
3. Results and Discussions
3.1. The FMN – heme IET kinetics of human iNOS oxyFMN with added macromolecular crowder
The FMN −heme IET kinetics of human iNOS oxyFMN were first measured by laser flash photolysis with added 0 − 14 % (w/v) Ficoll 70. The partial specific volume of Ficoll 70 is 0.69,45 which can be used to calculate its fractional volume occupancy at a given concentration.46 For example, 14% (w/v) Ficoll 70 gives a fractional volume occupancy of ~ 0.1, which is relevant to crowded intracellular environment with a volume occupancy of 0.05 − 0.4. Higher Ficoll 70 concentration impeded the IET kinetics measurements since the sample became too viscous to be well deaerated, a requirement for the laser flash photolysis experiments.23
A typical kinetic trace with added 14% Ficoll 70 is shown in Figure 2. The absorption at 580 nm of the partially reduced human iNOS oxyFMN decays rapidly below the pre-flash baseline with a rate constant of 401 ± 10 s−1 (red solid line, Figure 2). This depletion of FMNH· absorbance is attributable to the FMN – heme IET (Eq. 1, where FMNhq stands for FMN hydroquinone).38
| (1) |
Figure 2.
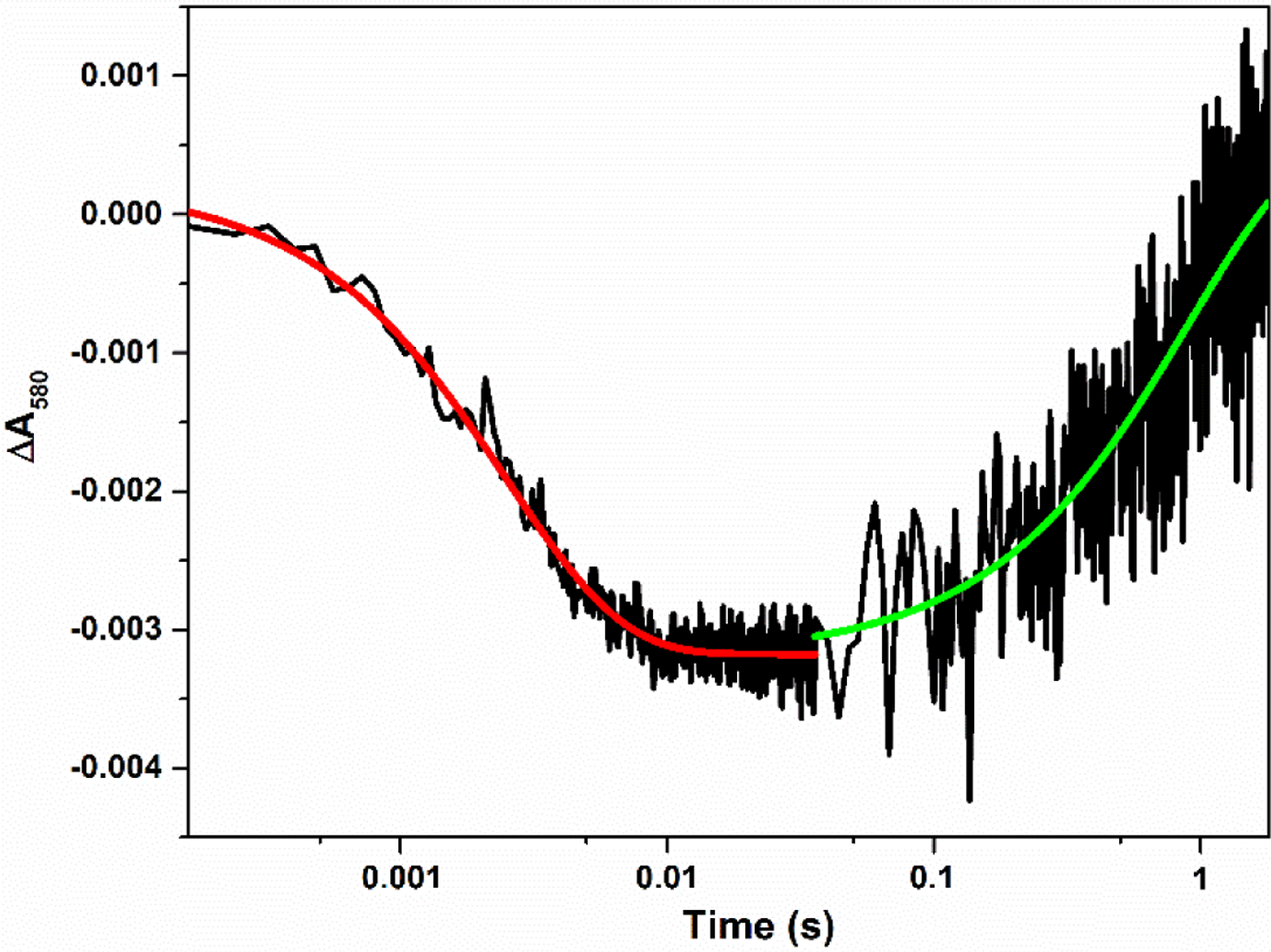
Transient absorbance at 580 nm obtained for the [Fe(II)–CO][FMNH•] form of human iNOS oxyFMN flashed by 446 nm laser; 14% (w/v) Ficoll 70 was present. The graph is a combined plot of two traces at 0 – 0.036 s and 0 – 1.8 s on a logarithmic time scale. Solid lines correspond to the best single-exponential fit to the data.
The decay is followed by a slow recovery toward baseline (see the green solid line in Figure 2), which is due to the CO re-binding to Fe(II).38 Importantly, the rate constant of the rapid decay, ket, is independent of the signal amplitude, i.e. concentration of the partially reduced NOS protein, affirming an intra-protein process. Moreover, the IET rate is independent of the signal amplitude at the other crowder concentrations, showing that the decay is due to an intramolecular process with added Ficoll 70 in the concentration range. The observed rate constants ket are listed in Table 1. The FMN−heme IET is noticeably enhanced by Ficoll 70 in a non-saturating manner (Figure 3). This behavior is a characteristic of crowding via volume exclusion.24
Table 1.
The FMN ‒ heme IET rate (s−1) of human iNOS oxyFMN and human iNOS holoenzyme with added Ficoll 70
| Ficoll 70 (w/v %) | iNOS oxyFMN | iNOS holoenzyme |
|---|---|---|
| 0 | 321 ± 8 | 34 ± 2 |
| 3 | 344 ± 13 | 42 ± 2 |
| 7 | 370 ± 11 | 48 ± 2 |
| 11 | 386 ± 12 | 54 ± 2 |
| 14 | 401 ± 10 | 60 ± 2 |
Figure 3.
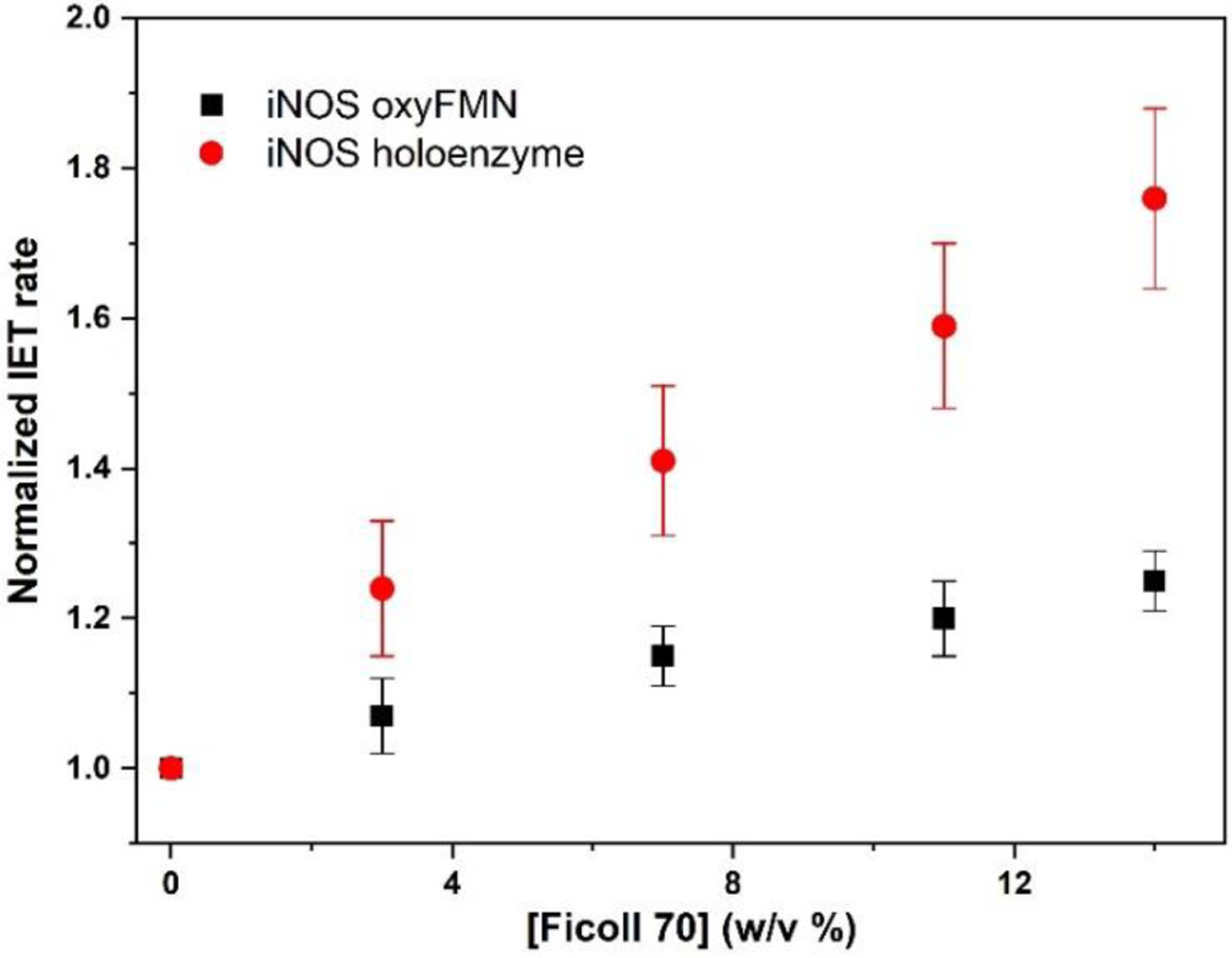
Plot of normalized values of the FMN – heme IET rates in human iNOS oxyFMN and iNOS holoenzyme with added Ficoll 70.
Mounting evidence demonstrates that conformational change limits the FMN − heme IET in NOS proteins, and the FMN domain explores the conformational space to dock productively onto the heme domain.36, 47 The volume occupied by Ficoll 70 is unavailable to the iNOS protein module because of the mutual impenetrability of the macromolecules. As a result, the macromolecular crowding agent can affect the FMN − heme IET process that depends on available volume. Upon increasing Ficoll 70 concentration, the number of means to place the FMN domain is gradually limited because the available volume is progressively constrained to the portion of space from which the FMN domain is not excluded. Consequently, randomness of the particle (i.e., the FMN domain) distribution in the solution is decreased, which leads to a reduction in the entropy of the more crowded solution. Restrictions on the conformational space available to the docking FMN domain and reduction of the randomness of the FMN domain motions augment the intraprotein electron transfer process. In other words, in response to adding of the crowding macromolecules, the NOS system will adapt to diminish the overall crowding by enhancing the docking of the FMN domain onto the heme domain, thereby reducing the total excluded volume.
Besides the excluded volume effect, the solution viscosity is inevitably increased by addition of Ficoll 70. For example, solution viscosity was increased to 2.7 cP with added 7% Ficoll 70. Our previous work showed that the FMN – heme IET rate value of an iNOS oxyFMN was decreased with added sucrose,39 the monomeric form of Ficoll 70. Addition of sucrose only increases the solution viscosity, but should not give macromolecular crowding effect because its hydrodynamic diameter in water (1.00 – 1.12 nm)48 is much smaller than the size of NOS protein noted in the Introduction section. We here observed that the IET rates of human iNOS oxyFMN were noticeably increased with added Ficoll 70 (Table 1), which brings forth both crowding and viscosity effects, while the rate was decreased by ~ 50 % when only viscosity effect is involved by adding sucrose (to give the same viscosity of 2.7 cP).39 This indicates that excluded volume has a more substantial effect on the FMN – heme IET kinetics than solution viscosity. The positive crowding effect of the Ficoll 70 obviously overrides the negative effect by solution viscosity, resulting in the observed increase in the IET rate (Figure 3).
In living cells, proteins are crowded by macromolecules in various shapes.49 It is thus necessary to examine the crowding effect using crowders with different shapes. This is also useful to determine whether specific interactions exist between the protein and the crowder. Ficoll 70 is compact and less flexible, while Dextran 70, a polymer of D-glucopyranose, is a linear polysaccharide and behaves as a quasirandom coil.27, 50–52 To determine whether the observed crowding effect depends on the specific crowing agent, the IET rate of human iNOS oxyFMN in the presence of Dextran 70 was measured under the concentration that gives the same solution viscosity as that of Ficoll 70 because of the abovementioned solution viscosity effect. The values of the IET rate constants obtained with added Dextran 70 (Table 2) are the same as that of Ficoll 70, within experimental error, for both the solution viscosity values (2.7 and 5.0 cP). These results with two distinctly different crowders (in shape and chemical properties) clearly demonstrate that the crowding effect on the FMN – heme IET rate is not dependent on the shape and other properties of the crowders and that specific interactions do not exist between the iNOS protein and the crowders. The crowding agent does not interact with the NOS protein under test, except via nonspecific steric repulsion (i.e., excluded volume effect).
Table 2.
The FMN ‒ heme IET rates of human iNOS oxyFMN protein with added Ficoll 70 or Dextran 70 under the same viscosity conditions
| [Crowder] (w/v) | η (cP) | ket (s−1) |
|---|---|---|
| 0% | 1.40 | 321 ± 8 |
| 7% Ficoll 70 | 2.7 | 370 ± 11 |
| 4.6% Dextran 70 | 2.7 | 351 ± 9 |
| 14% Ficoll 70 | 5.0 | 401 ± 10 |
| 7.5% Dextran 70 | 5.0 | 398 ± 9 |
3.2. The FMN – heme IET kinetics of human iNOS holoprotein as a function of crowding
We further measured the IET kinetics of iNOS holoprotein to determine whether the crowding effect is protein-specific, and to compare the effects on two iNOS proteins with different sizes. Adding Ficoll 70 increases the IET rate of the holoprotein in a non-saturatable manner (Figure 3), which is similar to oxyFMN. On the other hand, the magnitude of IET enhancement for the human iNOS holoenzyme (25 % − 76%) is much higher than that of the iNOS oxyFMN construct (10 % − 25 %) (Figure 3 and Table 1). Such difference can be rationalized by the fact that the excluded volume is substantially increased when the protein size is larger.53 We calculated the excluded volume of human iNOS oxyFMN and iNOS holoenzyme using the Voss Volume Voxelator54 and normalized the numbers to the volume of the first solvent shell.55 The human iNOS holoenzyme has a larger excluded volume than human iNOS oxyFMN (Figure S1). Note that Ficoll 70 has a radius of gyration of ~ 55 Å,27 at which the difference in the excluded volume for the two iNOS proteins is obvious. Therefore, the larger crowding effect on the iNOS holoenzyme is due to its larger excluded volume, compared to the bi-domain oxyFMN construct. Since the size of the iNOS holoenzyme molecule is larger than the iNOS oxyFMN, the available volume is substantially smaller for the holoenzyme, thereby facilitating the FMN domain to sample the conformational space in a more controlled manner.
It is of note that alike the NOS holoenzyme, the oxyFMN construct is purified as a homodimer,32 which is expected because both proteins contain the dimeric heme domain. In the NOS holoenzyme, the FMN domain docks onto the heme domain in the other subunit to enable the intersubunit FMN – heme IET.56 The oxyFMN construct is also a dimer and the FMN/heme interdomain alignment is identical in these two iNOS proteins, as shown in the recent literature.36 The dimeric nature of oxyFMN is important because macromolecular crowding affects dimeric protein to a different extent from its monomeric form: the excluded volume around a protein dimer is ~ twice the excluded volume of its monomer.57 The observed enhancing effect on the FMN − heme IET is thus not caused by the difference in the oligomeric state of the oxyFMN and full-length iNOSs. It is the obligatory conformational change that is the basis for the observed macromolecular crowding effect on the IET kinetics.
Our recent results showed that the IET rate can be affected by ionic strength and the optimal IET rate value was obtained at I = 200 mM.58 To determine whether ionic strength influences the IET rate under crowding conditions, the IET rates of human iNOS holoenzyme at I = 200 and 400 mM were measured in the presence of 14 % (w/v) Ficoll 70. Compared to I = 400 mM, the IET rate was increased at I = 200 mM with added Ficoll 70 (Table 3). On the other hand, the magnitude of the IET enhancement by Ficoll 70 for the human iNOS holoenzyme at I = 200 mM is 1.76, which is similar to that obtained at I = 400 mM (1.88). Although ionic strength still influences the IET kinetics under crowding conditions, there is no synergy between macromolecular crowding and ionic strength in affecting the FMN – heme IET process.
Table 3.
The FMN ‒ heme IET rate ket (s−1) of human iNOS holoenzyme with added Ficoll 70 under different ionic strength
| [Ficoll 70] (w/v %) | Ionic strength (mM) | ket (s−1) |
|---|---|---|
| 0 | 400 | 34 ± 2 |
| 0 | 200 | 40 ± 3 |
| 14 | 400 | 60 ± 2 |
| 14 | 200 | 75 ± 2 |
3.3. Fluorescence lifetimes of human iNOS oxyFMN as a function of crowding
Flavin fluorescence is a sensitive probe of conformational change in NOSs, and specific phases observed in the multi-exponential fluorescence lifetime decay can be assigned to certain NOS conformational states.59 To probe the effect of crowding on the protein conformations, the flavin fluorescence lifetime of human iNOS oxyFMN was measured. The raw time-resolved NOS flavin fluorescence intensity data (Figure 4) can be fitted with a multi-exponential function. The lifetime data for the iNOS oxyFMN protein with added 0 ‒ 14% (w/v) Ficoll 70 are listed in Table 4. The relative amplitude (A1) of the short fluorescence lifetime (τ1) state is significantly increased from 10 % to 31 % by adding the crowder, along with a decrease in the relative amplitude (A2) of the long fluorescence lifetime (τ2) state. This can also be clearly illustrated by a comparison of the raw decay for 0 % and 14 % Ficoll 70 (Figure 5). In the literature, the τ1 and τ2 states are assigned to the NOS output and free/open states, respectively.59 It is generally accepted that the crowder decreases the available volume of protein, resulting in the hard-core steric repulsions and a shift of the conformational equilibrium toward formation of a more compact protein state.60 The presence of crowder has been proposed to bias the compact state relative to more open state of other proteins.61–63 In the iNOS oxyFMN protein under volume-occupied conditions, excluded volume effect favors the formation of the more compact docked FMN/heme state over the free/open states, resulting in higher output state population, as observed from the fluorescence lifetime measurements. Shifting the conformational equilibrium may not be the only pathway in enhancing the IET, though.
Figure 4.
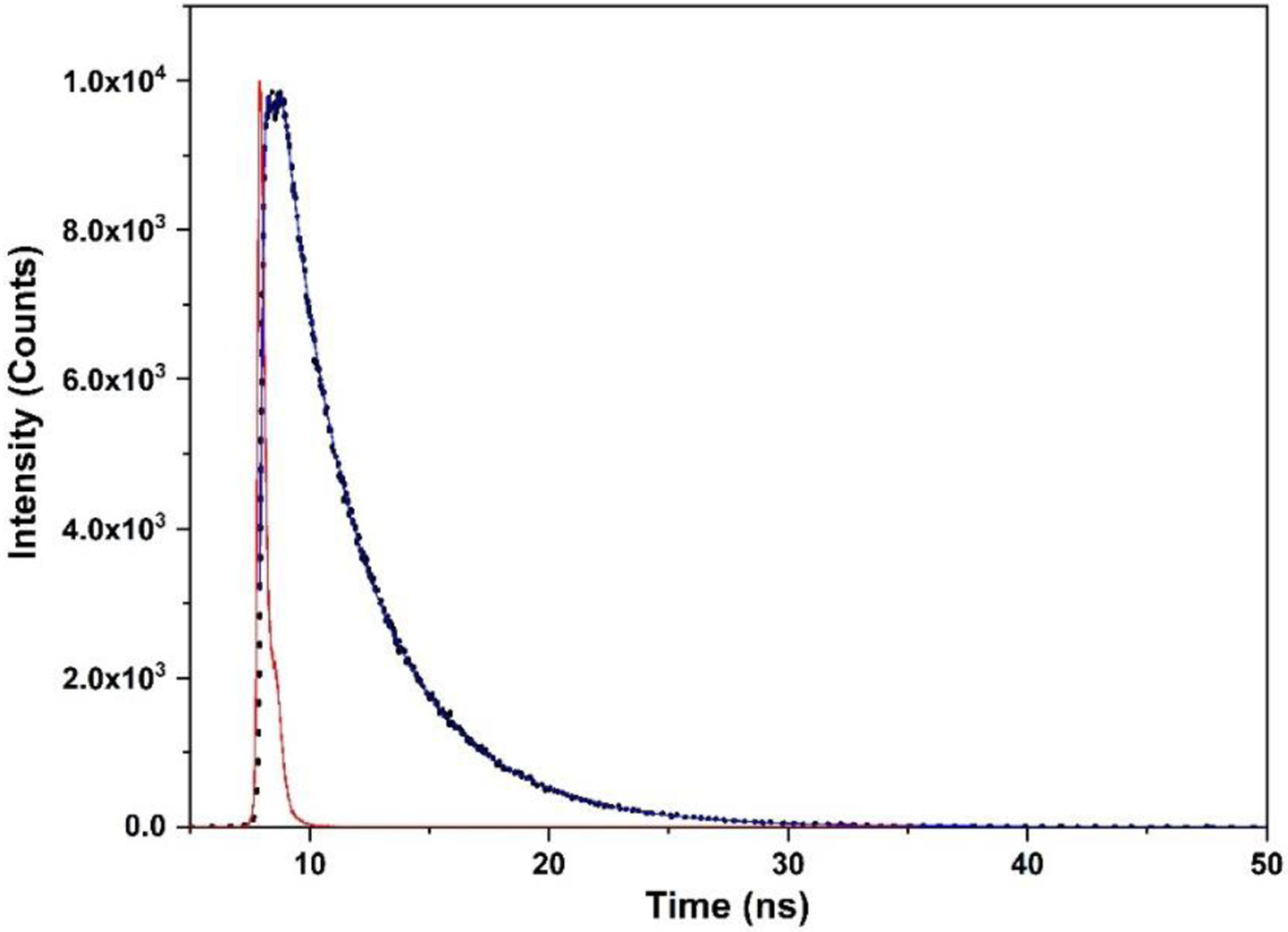
Plot of time-resolved fluorescence intensity decay of human iNOS oxyFMN with added14% (w/v) Ficoll 70. The fluorescence raw data are shown in dots, the multiexponential fit is a blue line, and the instrument response (IR) is shown in red.
Table 4.
Fluorescence lifetime data of human iNOS oxyFMN with added Ficoll 70
| Ficoll 70 (w/v %) | τ1 (ns) | A1 % | τ2 (ns) | A2 % | χ2 |
|---|---|---|---|---|---|
| 0 | 1.767 ± 0.031 | 9.30 ± 0.541 | 3.775 ± 0.0006 | 90.70 ± 0.542 | 1.188 |
| 3 | 1.613 ± 0.019 | 15.17 ± 0.231 | 3.860 ± 0.0005 | 84.83 ± 0.232 | 1.224 |
| 7 | 1.593 ± 0.013 | 22.29 ± 0.184 | 3.977 ± 0.0004 | 77.71 ± 0.184 | 1.240 |
| 11 | 1.500 ± 0.012 | 28.81 ± 0.141 | 4.054 ± 0.0004 | 71.19 ± 0.141 | 1.326 |
| 14 | 1.486 ± 0.011 | 31.43 ± 0.104 | 4.151 ± 0.0004 | 68.57 ± 0.106 | 1.314 |
Figure 5.
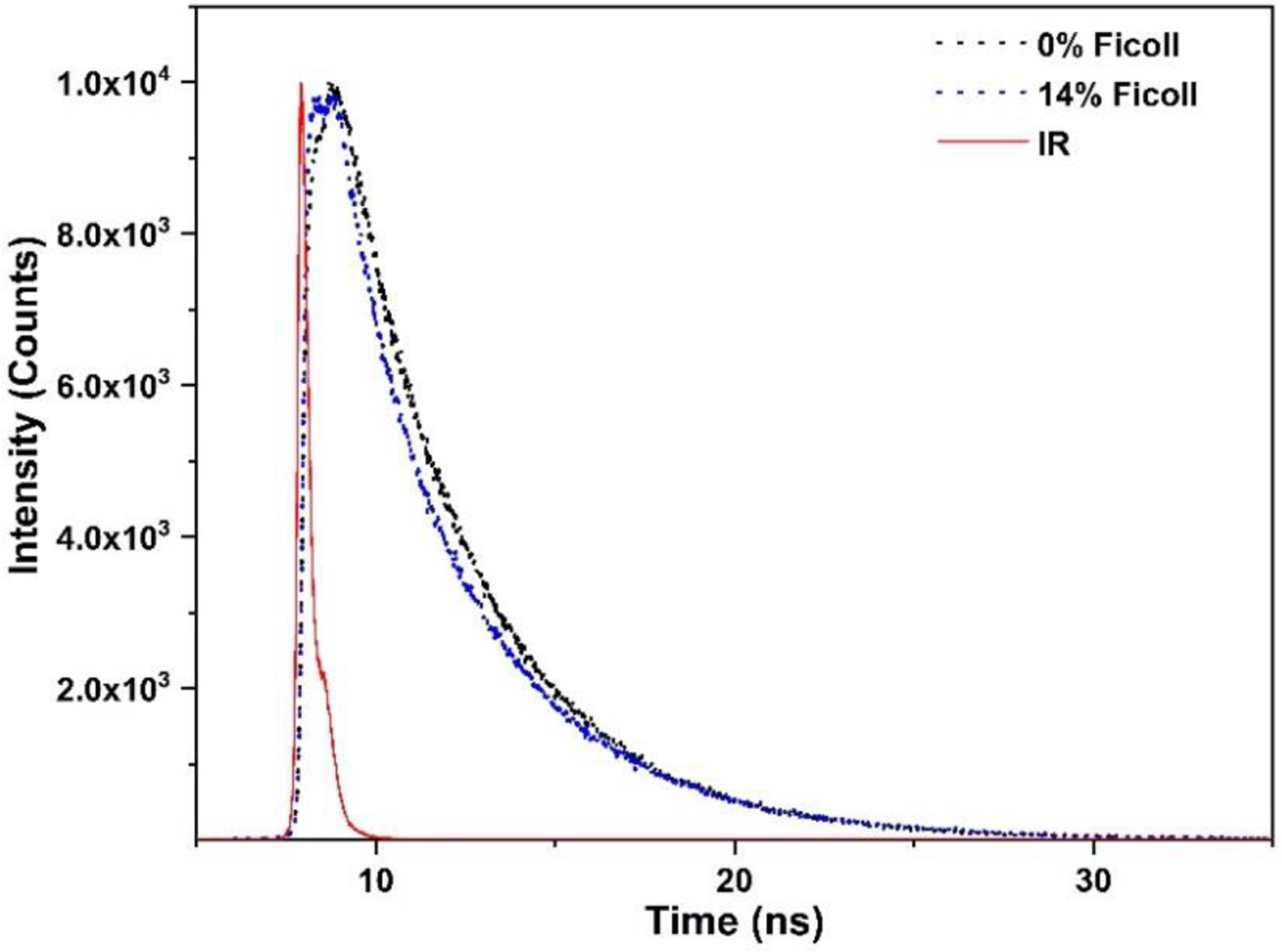
Comparison of raw fluorescence intensity decay of human iNOS oxyFMN without and with added 14 % (w/v) Ficoll 70; the instrument response is shown in red. Qualitatively, the decay with added 14 % Ficoll 70 possesses higher amplitude of fast decay τ1 species than that of 0 % Ficoll.
Note that τ1 and τ2 values with added Ficoll 70 do not correspond to those in the absence of Ficoll 70 (Table 4). The geometries of these conformational states may be altered in the crowded environment. The FMN domain docked at the heme domain is believed to go through short-range motions to productively dock onto the heme domain.58 This short-range exploration for the optimal docking position (i.e., conformational sampling) results in productive docking of the FMN domain with the heme domain. Quenching of the FMN fluorescence by the ferriheme in the docked state gives its short fluorescence lifetime τ1.59 Crowding narrows the available volume and restricts the degrees of motion of the FMN domain for the conformational sampling, and thus likely causes the decrease in τ1 through facilitating more productive FMN-heme docking with higher FMN fluorescence quenching. On the other hand, the increase in τ2 value indicates the increasing sampling from open state conformations, which will also facilitate the formation of the docked output state.
In the iNOS holoenzyme, the crowding similarly limits the range of motions of the tethered FMN domain (Figure 1B) and may shift the equilibrium towards both the input and output (docked) states of the NOS holoprotein, at the expense of the free/undocked states, in proportion to the decrease of the available volume. Consequently, the effective distance between the FAD/NADPH and heme domains (i.e., the FMN domain shuttling distance) will decrease, and it will take less time to transport the electron from the NADPH/FAD domain to the heme domain. If there is no NADPH/FAD domain (as in the oxyFMN construct), the range of the FMN domain motion is still decreasing. The rate of conformational rearrangements in both the iNOS proteins will therefore increase, which will lead to the observed increase in the bulk IET rate.64 It is of note that crowding does not necessarily stabilize the output state over the input state because both the docked states are more compact than the free/open states and both involve in conformational sampling of the proximal FMN domain at the heme or FAD domain. The conformational sampling in a more controlled manner may also contribute to the enhanced IET by the crowder. The detailed mechanism merits further investigations.
3.4. Steady-state cyt. c and ferricyanide reduction activities as a function of crowding
Steady-state cyt. c reduction is a measure of electron transport through the entire reductase unit of NOS because cyt. c is reduced exclusively by the FMN cofactor.65 We found that the cyt. c reduction rate was noticeably increased by addition of Ficoll 70 or Dextran 70 (Table 5), showing that the iNOS holoenzyme supports greater electron flux through its FMN domain to cyt. c in the presence of the crowder. These results are in line with the increase in the NO production activities by the crowder (see below). Moreover, the magnitudes of increase are similar for Ficoll 70 and Dextran 70 at the concentrations that give the same viscosity (Table 5). Therefore, the enhancing effect is independent of the chemical essence of the crowder and no specific interactions appear to exist between the crowder and the proteins (cyt. c and NOS). This has also been observed for the FMN − heme IET process (Table 2).
Table 5.
Cyt. c reduction rates of iNOS holoenzyme in the presence of crowder.a
| [Crowder] (w/v) | Cyt. c reduction (min−1) |
|---|---|
| 0 % | 1403 ± 49 |
| 3% Ficoll 70 | 1510 ± 38 |
| 7% Ficoll 70b | 1668 ± 70 |
| 11% Ficoll 70 | 1795 ± 108 |
| 14% Ficoll 70c | 1928 ± 130 |
| 4.6% Dextran 70b | 1680 ± 88 |
| 7.5% Dextran 70c | 1947 ± 122 |
Rates are the average of at least three assays. The final concentration of human iNOS holoenzyme is 1.5 nM.
The solution viscosity with the added 7 % Ficoll 70 or 4.6 % Dextran 70 is the same (2.7 cP).
The solution viscosity with the added 14 % Ficoll 70 or 7.5% Dextran 70 is the same (5.0 cP).
The reduction of cyt. c is a bimolecular process, which is different from the intramolecular FMN − heme IET where a tethered FMN domain is in motion. Conceivably, the bimolecular reaction in a crowded environment can be influenced by (i) the retardation of diffusion due to crowding, and (ii) a crowding-facilitated association of the two molecules (which is an entropic effect).25, 66 Since increased cyt. c reduction is observed here, it is more likely that crowding increases the reduction rate by favoring more effective association of cyt. c with the FMN domain. The cyt. c reduction results further support our model that the enhanced kinetics by macromolecular crowding is based on the excluded volume effect, with an entropic origin.
Cyt. c can receive electron from the FMN cofactor only when the FMN domain is dissociated from the reductase complex. This is because the FMN domain in the input state is shielded and inaccessible to its electron acceptor.9 Besides the abovementioned pathway in the biomolecular process, a greater degree of FMN deshielding under crowded environment may increase the steady-state cyt. c reduction activity. There is an inverse correlation between the FMN shielding degree and the steady-state cyt. c reduction rates for the nNOS reductase constructs, particularly in the absence of CaM.67 However, such a model where electron transfer to cyt. c is mainly controlled by conformational gating of the FMN domain of nNOS may not be applicable to iNOS, in which CaM is always bound and the autoinhibitory insert within the nNOS FMN domain does not exist in iNOS (the autoinhibitory insert in nNOS affects the conformational equilibrium of the reductase domain68). Even in the CaM-bound nNOS, other steps may become rate-limiting the steady-state cyt. c reduction under circumstance where a greater degree of FMN deshielding cannot further increase the steady-state activity beyond what is already achieved by CaM binding.69 Moreover, the crowding is reasonably proposed to favor the formation of the docked states including the input state at the expense of the deshielding/open states (see above). Therefore, the FMN deshielding is unlikely to participate in the observed enhancement of the steady-state cyt. c reduction in the presence of the crowder.
Because of its comparable size (relative to the NOS heme domain) and specific interaction with the FMN domain, cyt. c is a worthy model for NOS electron transfer (the nNOS reductase domain is homologous to P450 reductase that forms a specific inter-protein complex with cyt. c70). On the other hand, ferricyanide is small, and primarily receives the NADPH-derived electron from the FAD cofactor in NOS.71 Moreover, the FAD reduction by NADPH do not require a large-scale conformational change.9 Therefore, ferricyanide reduction is a good ‘negative’ control for studying the crowding effect on the NOS flavin-mediated electron transfer. Indeed, neither Ficoll 70 nor Dextran 70 changed the ferricyanide reduction activity of the holoenzyme, within the experimental error (Table 6). Note that the unchanged FAD reduction rate can still support the faster turnover in the presence of the crowder (see below). The FAD reduction by NADPH is insensitive to excluded volume, as expected.
Table 6.
Ferricyanide reduction rates of iNOS holoenzyme in the presence of crowder.a
| [Crowder] (w/v) | Ferricyanide reduction (min−1) |
|---|---|
| 0 % | 23486 ± 1321 |
| 3% Ficoll 70 | 24172 ± 394 |
| 7% Ficoll 70 | 22930 ± 754 |
| 11% Ficoll 70 | 23682 ± 1097 |
| 14% Ficoll 70 | 23921 ± 1364 |
| 4.6% Dextran 70 | 24270 ± 589 |
| 7.5% Dextran 70 | 23769 ± 983 |
Rates are the average of at least three assays. The final concentration of human iNOS holoenzyme is 1.5 nM.
During electron transport across the NOS domains, besides the IETs in the input state and output state and the related domain docking and conformational samplings,58 the FMN domain swings between the FAD and heme domains (Figure 1B). The conformational cycle involves (i) release of the FMN domain from the docked FMN/FAD state, (ii) passage of the free states to those near the heme domain, (iii) release of the FMN domain from its docked FMN/heme state after the FMN – heme IET, and (iv) passage of the free states to those near the FAD domain. In iNOS and CaM-bound eNOS and nNOS, the cyt. c reduction is order(s) of magnitude faster than the NOS heme reduction.35, 72 Release of the FMN (sub)domain from the rest of the reductase domain (step i) is thus not rate limiting delivery of the NADPH-derived electrons to the heme active site. The output state is transient, and the free/open states predominate in the conformational equilibrium.39, 73 These indicate that after the FMN − heme electron transfer, it is rather effortless for the FMN domain to dissociate from the heme domain (step iii) to return to its former input state to pick up a second electron from FAD. Salerno et al. proposed that the slowest step in NOS turnover is instead traverse of the FMN (sub)domain through a conformational bottleneck that ties the free states near the input state with other free states near the output state (steps ii and iv).59 The macromolecular crowding will very likely narrow the available space for the free states such that the passage of the conformational bottleneck will be accelerated, resulting in more efficient electron flow across the NOS domains.
It is interesting to investigate the structural and dynamic properties of NOSs in the presence of crowding agents. The FMN − heme IET studied here is an essential step in NOS catalysis because the electron delivery to the heme center is required for the O2 binding and activation and the subsequent substrate oxidation to NO.74 There are three IET steps in the process of NO production, in which the heme iron in redox states (i) l-Arg-Fe(III)-H4B, (ii) NOHA-Fe(III)-H4B+, and (iii) NOHA-Fe(III)-H4B, are reduced by FMNhq.75, 76 The heme reduction does not appear to directly involve the H4B cofactor,12 and bound l-Arg and NOHA both give high spin Fe(III) species. Therefore, the kinetics of step (i) studied here should be applicable to the other two steps. Emerging evidence suggests that the FMN − heme IET associated conformational change is in fact rate-limiting the NOS catalysis.77 There are three essential aspects of conformation control of NOSs: structural (constraints on the conformational space available to the docking modules), dynamic (the interconversions between the conformations), and energetic (docking energies and their effect on the lifetimes of the docked complexes). The crowding effect may affect more than one aspect of the conformational dynamics in NOS. It is unclear what are the main causes, and more dynamic and energetic results will be crucial to understand the crowding effect. Detailed spectroscopic, mass spectrometric and computational studies will provide much needed information on macromolecular crowding in regulation of this important family of multidomain redox enzymes.
3.5. NO production activity of human iNOS holoenzyme as a function of crowding
To determine whether macromolecular crowding affects the NOS function, we measured the NO production activities of the holoenzyme with added crowder (Table S1). The NO production rates of human iNOS holoenzyme are continuously increased upon adding Ficoll 70 (Figure 6). For example, the NO production rate was increased by 53% with added 14% (w/v) Ficoll 70. One would expect that the interaction of the small size l-Arg substrate with the NOS enzyme should not be changed much by the excluded volume. It is of note that, however, when the enzymatic catalysis involves conformational changes (which is the case for the NOS enzyme), macromolecular crowding can influence the enzymatic activity. For example, macromolecular crowding shifts the conformational equilibrium of a human translation initiation factors from an inactive open state towards a closed active conformation, resulting in enhanced ATPase activity of the multidomain protein.78 Besides the conformational equilibrium, it is tempting to speculate that the other effect of macromolecular crowding is increased rate of interconversion between the free/open and docked states, as discussed above. It is also of note that the interplay of rates of three NOS heme-related processes (NO dissociation from the Fe(III)–NO complex, oxidation of the Fe(II)–NO complex, and heme reduction) governs the steady-state NO production activity.79 Although it is unclear how exactly the NO production is enhanced by the macromolecular crowder, the enzymatic activity results clearly demonstrate that the crowding effect is functionally relevant. This work should inspire additional detailed studies of NOS enzymology in the presence of crowding reagent, which is closer to cellular environment than the common practice of testing NOSs in diluted solutions.
Figure 6.
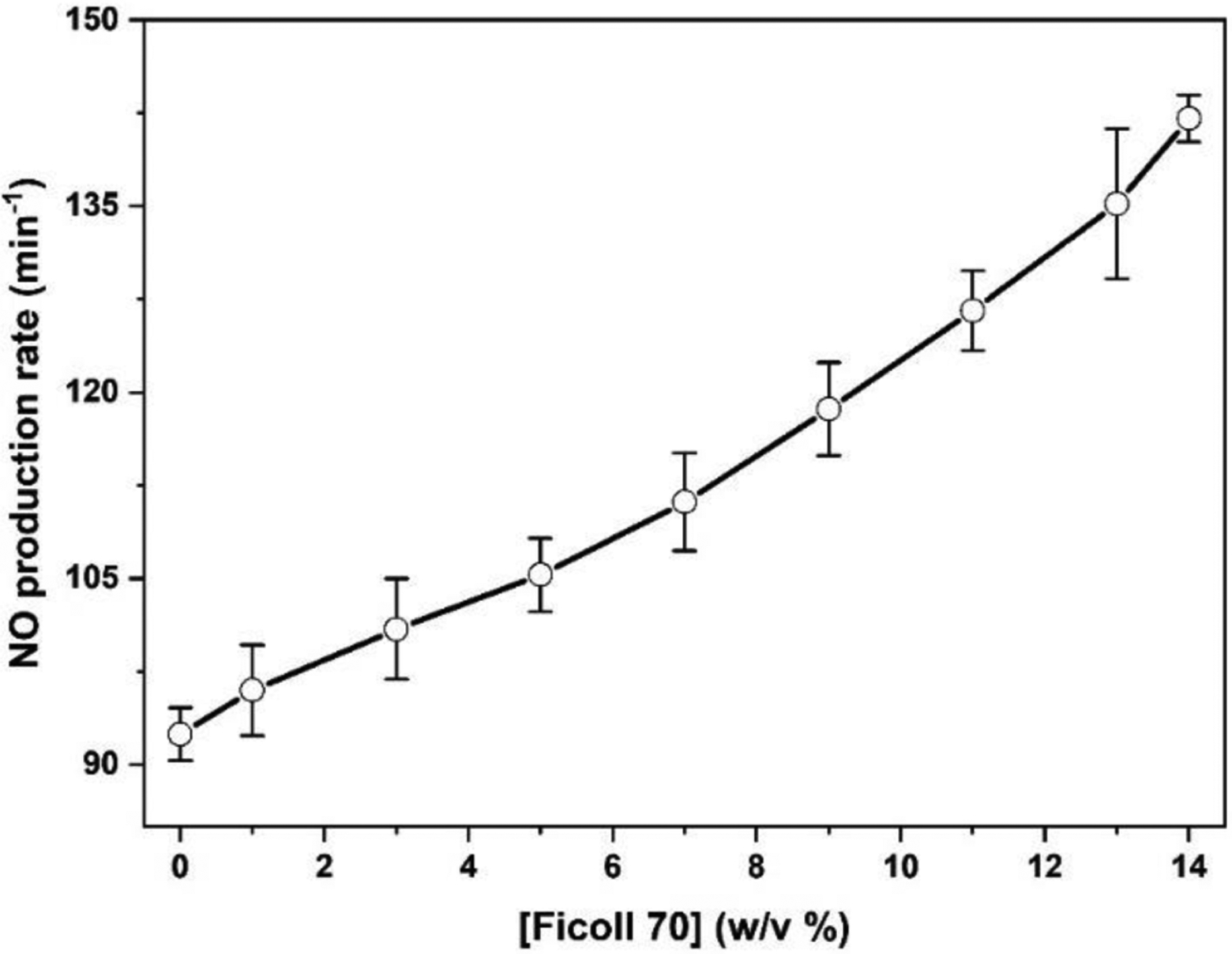
Plot of the NO production activity of human iNOS holoenzyme with added Ficoll 70.
In conclusion, a noticeable increase in the FMN – heme IET rate value of both human iNOS oxyFMN and holoenzyme proteins were observed upon addition of the macromolecular crowder. The effect is independent of the shape and chemical essence of the specific crowder. The magnitude of IET enhancement for the human holoenzyme is much higher than that of iNOS oxyFMN, which can be explained by the larger excluded volume of the holoenzyme. The electron transfer kinetics, fluorescence lifetime and steady-state enzymatic activity results together demonstrate that the FMN ‒ heme IET can be significantly enhanced by excluded volume effect, its origin being entropic. The macromolecular crowder narrows the available volume for the FMN domain motions, which include the long-range shuttling motion between the FAD and heme domains and the short-range sampling motions near these domains. As such, we propose that the macromolecular crowding affects the NOS electron transfer through multiple pathways such as (i) shifting the conformational equilibrium toward more compact docked states, (ii) facilitating conformational sampling near its electron transfer partner (the heme domain, the FAD domain, or cyt. c), and (iii) increasing the rates of conformational rearrangements. Such mechanism should be applicable to electron transfer in other multidomain redox proteins.
Supplementary Material
Acknowledgements
This work was supported by the National Institutes of Health (GM081811).
Abbreviations
- NOS
nitric oxide synthase
- iNOS
inducible NOS
- CaM
calmodulin
- oxyFMN
bi-domain oxygenase/FMN construct in which only the FMN and heme domains are present, along with the CaM binding region
- l-Arg
l-Arginine
- H4B, 6R-5,6,7,8
-tetrahydrobiopterin
- IET
interdomain electron transfer
- dRF
deazariboflavin
- FMNH•
FMN semiquinone
- cyt. c
cytochrome c
- FMNhq
FMN hydroquinone
Footnotes
Accession Codes.
Human iNOS P35228
Supporting Information. Plot of normalized excluded volumes for human iNOS oxyFMN and iNOS holoenzyme with different probe sizes; NO synthesis rates of human iNOS holoenzyme in the presence of Ficoll 70.
References
- 1.Schmidt HHHW, and Walter U (1994) NO at work, Cell 78, 919–925. [DOI] [PubMed] [Google Scholar]
- 2.Moncada S, and Higgs EA (2006) The discovery of nitric oxide and its role in vascular biology, Br J Pharmacol 147, S193–S201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roman LJ, Martásek P, and Masters BSS (2002) Intrinsic and Extrinsic Modulation of Nitric Oxide Synthase Activity, Chem. Rev 102, 1179–1190. [DOI] [PubMed] [Google Scholar]
- 4.Stuehr DJ, Santolini J, Wang Z-Q, Wei C-C, and Adak S (2004) Update on Mechanism and Catalytic Regulation in the NO Synthases, J. Biol. Chem 279, 36167–36170. [DOI] [PubMed] [Google Scholar]
- 5.Wei C-C, Crane BR, and Stuehr DJ (2003) Tetrahydrobiopterin Radical Enzymology, Chem. Rev 103, 2365–2384. [DOI] [PubMed] [Google Scholar]
- 6.Alderton WK, Cooper CE, and Knowles RG (2001) Nitric oxide synthases: Structure, function and inhibition, Biochem. J 357, 593–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feng CJ, Tollin G, Holliday MA, Thomas C, Salerno JC, Enemark JH, and Ghosh DK (2006) Intraprotein electron transfer in a two-domain construct of neuronal nitric oxide synthase: The output state in nitric oxide formation, Biochemistry 45, 6354–6362. [DOI] [PubMed] [Google Scholar]
- 8.Iyanagi T (2019) Molecular mechanism of metabolic NAD(P)H-dependent electron-transfer systems: The role of redox cofactors, Biochimica et Biophysica Acta (BBA) - Bioenergetics 1860, 233–258. [DOI] [PubMed] [Google Scholar]
- 9.Garcin ED, Bruns CM, Lloyd SJ, Hosfield DJ, Tiso M, Gachhui R, Stuehr DJ, Tainer JA, and Getzoff ED (2004) Structural basis for isozyme-specific regulation of electron transfer in nitric-oxide synthase, J. Biol. Chem 279, 37918–37927. [DOI] [PubMed] [Google Scholar]
- 10.Volkmann N, Martásek P, Roman LJ, Xu X-P, Page C, Swift M, Hanein D, and Masters BS (2014) Holoenzyme structures of endothelial nitric oxide synthase – An allosteric role for calmodulin in pivoting the FMN domain for electron transfer, J Struct Biol 188, 46–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feng CJ, Tollin G, Hazzard JT, Nahm NJ, Guillemette JG, Salerno JC, and Ghosh DK (2007) Direct measurement by laser flash photolysis of intraprotein electron transfer in a rat neuronal nitric oxide synthase, J. Am. Chem. Soc 129, 5621–5629. [DOI] [PubMed] [Google Scholar]
- 12.Feng C, Thomas C, Holliday MA, Tollin G, Salerno JC, Ghosh DK, and Enemark JH (2006) Direct Measurement by Laser Flash Photolysis of Intramolecular Electron Transfer in a Two-Domain Construct of Murine Inducible Nitric Oxide Synthase, J. Am. Chem. Soc 128, 3808–3811. [DOI] [PubMed] [Google Scholar]
- 13.Ellis RJ, and Minton AP (2003) Join the crowd, Nature 425, 27. [DOI] [PubMed] [Google Scholar]
- 14.Kuznetsova IM, Turoverov KK, and Uversky VN (2014) What Macromolecular Crowding Can Do to a Protein, 15, 23090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sasahara K, McPhie P, and Minton AP (2003) Effect of Dextran on Protein Stability and Conformation Attributed to Macromolecular Crowding, J. Mol. Biol 326, 1227–1237. [DOI] [PubMed] [Google Scholar]
- 16.Rivas G, and Minton AP (2016) Macromolecular Crowding In Vitro, In Vivo, and In Between, Trends Biochem. Sci 41, 970–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Y, Sarkar M, Smith AE, Krois AS, and Pielak GJ (2012) Macromolecular Crowding and Protein Stability, J. Am. Chem. Soc 134, 16614–16618. [DOI] [PubMed] [Google Scholar]
- 18.Kuzmak A, Carmali S, von Lieres E, Russell AJ, and Kondrat S (2019) Can enzyme proximity accelerate cascade reactions?, Scientific Reports 9, 455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guseman AJ, Perez Goncalves GM, Speer SL, Young GB, and Pielak GJ (2018) Protein shape modulates crowding effects, Proceedings of the National Academy of Sciences 115, 10965–10970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Takahashi S, Yamamoto J, Kitamura A, Kinjo M, and Sugimoto N (2019) Characterization of Intracellular Crowding Environments with Topology-Based DNA Quadruplex Sensors, Anal. Chem 91 2586–2590. [DOI] [PubMed] [Google Scholar]
- 21.Chen E, Christiansen A, Wang Q, Cheung MS, Kliger DS, and Wittung-Stafshede P (2012) Effects of Macromolecular Crowding on Burst Phase Kinetics of Cyt. c Folding, Biochemistry 51, 9836–9845. [DOI] [PubMed] [Google Scholar]
- 22.Hervas M, and Navarro JA (2011) Effect of crowding on the electron transfer process from plastocyanin and cyt. c6 to photosystem I: a comparative study from cyanobacteria to green algae, Photosynth Res 107, 279–286. [DOI] [PubMed] [Google Scholar]
- 23.Feng C (2012) Mechanism of nitric oxide synthase regulation: Electron transfer and interdomain interactions, Coord. Chem. Rev 256, 393–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou HX, Rivas G, and Minton AP (2008) Macromolecular crowding and confinement: biochemical, biophysical, and potential physiological consequences, Annu Rev Biophys 37, 375–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ellis RJ (2001) Macromolecular crowding: obvious but underappreciated, Trends Biochem. Sci 26, 597–604. [DOI] [PubMed] [Google Scholar]
- 26.Chen C, Loe F, Blocki A, Peng Y, and Raghunath M (2011) Applying macromolecular crowding to enhance extracellular matrix deposition and its remodeling in vitro for tissue engineering and cell-based therapies, Adv Drug Delivery Rev 63, 277–290. [DOI] [PubMed] [Google Scholar]
- 27.Wenner JR, and Bloomfield VA (1999) Crowding Effects on EcoRV Kinetics and Binding, Biophys. J 77, 3234–3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.List BM, Klosch B, Volker C, Gorren ACF, Sessa WC, Werner ER, Kukovetz WR, Schmidt K, and Mayer B (1997) Characterization of bovine endothelial nitric oxide synthase as a homodimer with down-regulated uncoupled NADPH oxidase activity: Tetrahydrobiopterin binding kinetics and role of haem in dimerization, Biochem. J 323, 159–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yokom AL, Morishima Y, Lau M, Su M, Glukhova A, Osawa Y, and Southworth DR (2014) Architecture of the nitric oxide synthase holoenzyme reveals large conformational changes and a calmodulin-driven release of the FMN domain, J. Biol. Chem 289, 16855–16865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Armstrong JK, Wenby RB, Meiselman HJ, and Fisher TC (2004) The hydrodynamic radii of macromolecules and their effect on red blood cell aggregation, Biophys J 87, 4259–4270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ghosh DK, Holliday MA, Thomas C, Weinberg JB, Smith SME, and Salerno JC (2006) Nitric-oxide synthase output state - Design and properties of nitric-oxide synthase oxygenase/FMN domain constructs, J. Biol. Chem 281, 14173–14183. [DOI] [PubMed] [Google Scholar]
- 32.Ghosh DK, Holliday MA, Thomas C, Weinberg JB, Smith SME, and Salerno JC (2006) Nitric-oxide Synthase Output State: DESIGN AND PROPERTIES OF NITRIC-OXIDE SYNTHASE OXYGENASE/FMN DOMAIN CONSTRUCTS, J. Biol. Chem 281, 14173–14183. [DOI] [PubMed] [Google Scholar]
- 33.Li H, Igarashi J, Jamal J, Yang W, and Poulos TL (2006) Structural studies of constitutive nitric oxide synthases with diatomic ligands bound, J. Biol. Inorg. Chem 11, 753–768. [DOI] [PubMed] [Google Scholar]
- 34.Ilagan RP, Tejero J, Aulak KS, Ray SS, Hemann C, Wang Z-Q, Gangoda M, Zweier JL, and Stuehr DJ (2009) Regulation of FMN Subdomain Interactions and Function in Neuronal Nitric Oxide Synthase, Biochemistry 48, 3864–3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smith BC, Underbakke ES, Kulp DW, Schief WR, and Marletta MA (2013) Nitric oxide synthase domain interfaces regulate electron transfer and calmodulin activation, Proc Natl Acad Sci U S A 110, E3577–E3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Campbell MG, Smith BC, Potter CS, Carragher B, and Marletta MA (2014) Molecular architecture of mammalian nitric oxide synthases, Proc Natl Acad Sci U S A 111, E3614–E3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Feng CJ, Dupont A, Nahm N, Spratt D, Hazzard JT, Weinberg J, Guillemette J, Tollin G, and Ghosh DK (2009) Intraprotein electron transfer in inducible nitric oxide synthase holoenzyme, J. Biol. Inorg. Chem 14, 133–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Feng C, Dupont AL, Nahm NJ, Spratt DE, Hazzard JT, Weinberg JB, Guillemette JG, Tollin G, and Ghosh DK (2009) Intraprotein Electron Transfer in Inducible Nitric Oxide Synthase Holoenzyme, J. Biol. Inorg. Chem 14, 133–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li W, Fan W, Elmore BO, and Feng C (2011) Effect of solution viscosity on intraprotein electron transfer between the FMN and heme domains in inducible nitric oxide synthase, FEBS Lett 585, 2622–2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Feng C, Tollin G, Hazzard JT, Nahm NJ, Guillemette JG, Salerno JC, and Ghosh DK (2007) Direct Measurement by Laser Flash Photolysis of Intraprotein Electron Transfer in a Rat Neuronal Nitric Oxide Synthase, J. Am. Chem. Soc 129, 5621–5629. [DOI] [PubMed] [Google Scholar]
- 41.Li W, Fan W, Chen L, Elmore B, Piazza M, Guillemette J, and Feng C (2012) Role of an isoform-specific serine residue in FMN–heme electron transfer in inducible nitric oxide synthase, J. Biol. Inorg. Chem 17, 675–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.(2006) Time-Domain Lifetime Measurements, In Principles of Fluorescence Spectroscopy (Lakowicz JR, Ed.), pp 97–155, Springer US, Boston, MA. [Google Scholar]
- 43.Panda SP, Li W, Venkatakrishnan P, Chen L, Astashkin AV, Masters BSS, Feng C, and Roman LJ (2013) Differential calmodulin-modulatory and electron transfer properties of neuronal nitric oxide synthase mu compared to the alpha variant, FEBS Lett. 587, 3973–3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roman LJ, and Masters BSS (2006) Electron transfer by neuronal nitric-oxide synthase is regulated by concerted interaction of calmodulin and two intrinsic regulatory elements J. Biol. Chem 281, 23111–23118. [DOI] [PubMed] [Google Scholar]
- 45.Christiansen A, Wang Q, Samiotakis A, Cheung MS, and Wittung-Stafshede P (2010) Factors Defining Effects of Macromolecular Crowding on Protein Stability: An in Vitro/in Silico Case Study Using Cyt. c, Biochemistry 49, 6519–6530. [DOI] [PubMed] [Google Scholar]
- 46.Minton AP (1983) The effect of volume occupancy upon the thermodynamic activity of proteins: some biochemical consequences, Mol. Cell. Biochem 55, 119–140. [DOI] [PubMed] [Google Scholar]
- 47.He Y, Haque MM, Stuehr DJ, and Lu HP (2015) Single-molecule spectroscopy reveals how calmodulin activates NO synthase by controlling its conformational fluctuation dynamics, Proc Natl Acad Sci U S A 112, 11835–11840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schultz SG, and Solomon AK (1961) Determination of the Effective Hydrodynamic Radii of Small Molecules by Viscometry, The Journal of General Physiology 44, 1189–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Goodsell DS (1991) Inside a living cell, Trends Biochem. Sci 16, 203–206. [DOI] [PubMed] [Google Scholar]
- 50.Luby-Phelps K, Castle PE, Taylor DL, and Lanni F (1987) Hindered diffusion of inert tracer particles in the cytoplasm of mouse 3T3 cells, Proc Natl Acad Sci U S A 84, 4910–4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rühlmann C, Thieme M, and Helmstedt M (2001) Interaction between dextran and human low density lipoproteins (LDL) observed using laser light scattering, Chem. Phys. Lipids 110, 173–181. [DOI] [PubMed] [Google Scholar]
- 52.Venturoli D, and Rippe B (2005) Ficoll and dextran vs. globular proteins as probes for testing glomerular permselectivity: effects of molecular size, shape, charge, and deformability, American Journal of Physiology-Renal Physiology 288, F605–613. [DOI] [PubMed] [Google Scholar]
- 53.Minton AP (2001) The influence of macromolecular crowding and macromolecular confinement on biochemical reactions in physiological media, J Biol Chem 276, 10577–10580. [DOI] [PubMed] [Google Scholar]
- 54.Voss NR, and Gerstein M (2010) 3V: cavity, channel and cleft volume calculator and extractor, Nucleic Acids Res. 38, W555–W562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Halpin JC, Huang B, Sun M, and Street TO (2016) Crowding Activates Heat Shock Protein 90, J. Biol. Chem 291, 6447–6455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Panda K, Ghosh S, and Stuehr DJ (2001) Calmodulin activates intersubunit electron transfer in the neuronal nitric-oxide synthase dimer, J. Biol. Chem 276, 23349–23356. [DOI] [PubMed] [Google Scholar]
- 57.Ralston GB (1990) Effects of “crowding” in protein solutions, J. Chem. Educ 67, 857. [Google Scholar]
- 58.Astashkin AV, Li J, Zheng H, Miao Y, and Feng C (2018) A docked state conformational dynamics model to explain the ionic strength dependence of FMN – heme electron transfer in nitric oxide synthase, J. Inorg. Biochem 184, 146–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ghosh DK, Ray K, Rogers AJ, Nahm NJ, and Salerno JC (2012) FMN fluorescence in inducible NOS constructs reveals a series of conformational states involved in the reductase catalytic cycle, FEBS J 279, 1306–1317. [DOI] [PubMed] [Google Scholar]
- 60.Shahid S, Hassan MI, Islam A, and Ahmad F (2017) Size-dependent studies of macromolecular crowding on the thermodynamic stability, structure and functional activity of proteins: in vitro and in silico approaches, Biochimica et Biophysica Acta (BBA) - General Subjects 1861, 178–197. [DOI] [PubMed] [Google Scholar]
- 61.Minton AP (1981) Excluded volume as a determinant of macromolecular structure and reactivity, Biopolymers 20, 2093–2120. [Google Scholar]
- 62.Minton AP (2000) Effect of a Concentrated “Inert” Macromolecular Cosolute on the Stability of a Globular Protein with Respect to Denaturation by Heat and by Chaotropes: A Statistical-Thermodynamic Model, Biophys. J 78, 101–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Minton AP (2005) Models for Excluded Volume Interaction between an Unfolded Protein and Rigid Macromolecular Cosolutes: Macromolecular Crowding and Protein Stability Revisited, Biophys. J 88, 971–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Astashkin AV, and Feng C (2015) Solving kinetic equations for the laser flash photolysis experiment on nitric oxide synthases: Effect of conformational dynamics on the interdomain electron transfer, J. Phys. Chem. A 119, 11066–11075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Matsuda H, and Iyanagi T (1999) Calmodulin activates intramolecular electron transfer between the two flavins of neuronal nitric oxide synthase flavin domain, Biochimica Et Biophysica Acta-General Subjects 1473, 345–355. [DOI] [PubMed] [Google Scholar]
- 66.Berezhkovskii AM, and Szabo A (2016) Theory of Crowding Effects on Bimolecular Reaction Rates, The Journal of Physical Chemistry B 120, 5998–6002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ilagan RP, Tiso M, Konas DW, Hemann C, Durra D, Hille R, and Stuehr DJ (2008) Differences in a conformational equilibrium distinguish catalysis by the endothelial and neuronal nitric-oxide synthase flavoproteins, J. Biol. Chem 283, 19603–19615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Guan Z-W, Haque MM, Wei C-C, Garcin ED, Getzoff ED, and Stuehr DJ (2010) Lys842 in neuronal nitric-oxide synthase enables the autoinhibitory insert to antagonize calmodulin binding, increase FMN shielding, and suppress interflavin electron transfer, J. Biol. Chem 285, 3064–3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tiso M, Konas DW, Panda K, Garcin ED, Sharma M, Getzoff ED, and Stuehr DJ (2005) C-terminal tail residue Arg(1400) enables NADPH to regulate electron transfer in neuronal nitric-oxide synthase, J. Biol. Chem 280, 39208–39219. [DOI] [PubMed] [Google Scholar]
- 70.Freeman SL, Martel A, Devos JM, Basran J, Raven EL, and Roberts GCK (2018) Solution structure of the cyt. P450 reductase–cyt. c complex determined by neutron scattering, J. Biol. Chem 293, 5210–5219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Roman LJ, McLain J, and Masters BSS (2003) Chimeric enzymes of cyt. P450 oxidoreductase and neuronal nitric-oxide synthase reductase domain reveal structural and functional differences, J. Biol. Chem 278, 25700–25707. [DOI] [PubMed] [Google Scholar]
- 72.Roman LJ, Martasek P, Miller RT, Harris DE, de la Garza MA, Shea TM, Kim JJP, and Masters BSS (2000) The C termini of constitutive nitric-oxide synthases control electron flow through the flavin and heme domains and affect modulation by calmodulin, J. Biol. Chem 275, 29225–29232. [DOI] [PubMed] [Google Scholar]
- 73.Ilagan RP, Tejero JS, Aulak KS, Ray SS, Hemann C, Wang Z-Q, Gangoda M, Zweier JL, and Stuehr DJ (2009) Regulation of FMN subdomain interactions and function in neuronal nitric oxide synthase, Biochemistry 48, 3864–3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Leferink NGH, Hay S, Rigby SEJ, and Scrutton NS (2015) Towards the free energy landscape for catalysis in mammalian nitric oxide synthases, FEBS J 282, 3016–3029. [DOI] [PubMed] [Google Scholar]
- 75.Woodward JJ, NejatyJahromy Y, Britt RD, and Marletta MA (2010) Pterin-centered radical as a mechanistic probe of the second step of nitric oxide synthase, J. Am. Chem. Soc 132, 5105–5113. [DOI] [PubMed] [Google Scholar]
- 76.Stoll S, NejatyJahromy Y, Woodward JJ, Ozarowski A, Marletta MA, and Britt RD (2010) Nitric oxide synthase stabilizes the tetrahydrobiopterin cofactor radical by controlling its protonation state, J. Am. Chem. Soc 132, 11812–11823. [DOI] [PubMed] [Google Scholar]
- 77.Arnett DC, Bailey SK, and Johnson CK (2018) Exploring the conformations of nitric oxide synthase with fluorescence, Frontiers in bioscience (Landmark edition; ) 23, 2133–2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Akabayov SR, Akabayov B, Richardson CC, and Wagner G (2013) Molecular Crowding Enhanced ATPase Activity of the RNA Helicase eIF4A Correlates with Compaction of Its Quaternary Structure and Association with eIF4G, J. Am. Chem. Soc 135, 10040–10047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Santolini J, Meade AL, and Stuehr DJ (2001) Differences in three kinetic parameters underpin the unique catalytic profiles of nitric-oxide synthases I, II, and III, J. Biol. Chem 276, 48887–48898. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


