Multicolor imaging with SNAP-tagged tubulin and monitoring of PIN2 endocytosis with a chemical dye demonstrate that specific self-labeling of proteins can be used effectively in plants.
Abstract
Synthetic chemical fluorescent dyes promise to be useful for many applications in biology. Covalent, targeted labeling, such as with a SNAP-tag, uses synthetic dyes to label specific proteins in vivo for studying processes such as endocytosis or for imaging via super-resolution microscopy. Despite its potential, such chemical tagging has not been used effectively in plants. A major drawback has been the limited knowledge regarding cell wall and membrane permeability of the available synthetic dyes. Of 31 synthetic dyes tested here, 23 were taken up into BY-2 cells, while eight were not. This creates sets of dyes that can serve to measure endocytosis. Three of the dyes that were able to enter the cells, SNAP-tag ligands of diethylaminocoumarin, tetramethylrhodamine, and silicon-rhodamine 647, were used to SNAP-tag α-tubulin. Successful tagging was verified by live cell imaging and visualization of microtubule arrays in interphase and during mitosis in Arabidopsis (Arabidopsis thaliana) seedlings. Fluorescence activation-coupled protein labeling with DRBG-488 was used to observe PIN-FORMED2 (PIN2) endocytosis and delivery to the vacuole as well as preferential delivery of newly synthesized PIN2 to the actively forming cell plate during mitosis. Together, the data demonstrate that specific self-labeling of proteins can be used effectively in plants to study a wide variety of cellular and biological processes.
INTRODUCTION
Small-molecule fluorescent dyes have been widely used in biological research. However, challenges in efficient use were that they must be added exogenously and cannot be targeted to specific cellular compartments or cellular components, such as proteins. Fluorescent proteins as genetically encoded dyes enabled efficient labeling of proteins in live or fixed material. In 2003, Kai Johnsson’s lab developed a tool for covalent labeling of fusion proteins with small molecules in vivo, the O6-alkylguanine-DNA alkyltransferase (SNAP)-tag (Keppler et al., 2003). The SNAP-tag adds a site to a target protein for covalent fusion of a dye that has been modified with a benzylguanine moiety. Analogous tags such as Halo- and CLIP-tags were developed that enable simultaneous multicolor labeling of different proteins (Los et al., 2005; Gautier et al., 2008). A CLIP-tag in a protein reacts with O2-benzylcytosine, a cytosine conjugated to a dye, while the Halo-tag makes use of a mutated bacterial dehalogenase that creates stable bonds with any labeling chloroalkane ligand. Recently, technologies that make use of an engineered photoactive yellow protein, which can bind hydroxybenzylidene rhodamine derivatives, have been developed (Plamont et al., 2016) that present an alternative to tagging with synthetic dyes. Recently developed dyes include features such as ultra-resistance to photobleaching, allowing for super-resolution imaging with stimulated emission depletion (STED; Wang et al., 2017), fluorescence activation-coupled protein labeling (FAPL; Komatsu et al., 2011), and the ability to sense biochemical parameters such as pH or metabolites (Asanuma et al., 2014; Jiang et al., 2019).
Covalently tagging proteins with chemical dyes has advantages over using fluorescent proteins (FPs), including the following: access to a wide range of fluorescent dyes, which typically have higher quantum yield and photostability than FPs; initiation of fluorescence when the dye is conjugated to the target protein, providing temporal control (e.g., DRBG-488; Komatsu et al., 2011); and a smaller size relative to most FPs. Tagging with different dyes has been widely used in the animal field for in vivo labeling (Yang et al., 2015). Tagging with chemical fluorescent dyes has also been deployed for in vitro studies of the class II formin AtFH14 from Arabidopsis (Arabidopsis thaliana; Zhang et al., 2016). Surprisingly, only two reports have been published using SNAP-tagging in plants (Schiller et al., 2012; Steinebrunner et al., 2014). Both studies indicated that in vivo SNAP-tagging may not be feasible in plants. For example, an attempt to label apyrase (AtAPY1) found that labeling of fusion proteins in vivo with the cell-permeable fluorescent dyes tetramethylrhodamine-Star and BG-505 showed high background, and AtAPY1-SNAP–specific fluorescence could not be detected in Arabidopsis seedlings (Schiller et al., 2012). The dye entered the cell through the plasma membrane, but the free dye remained in the cytoplasm even after 14 h of washing. A second study reported an attempt to use SNAP-tagging for localizing the cytochrome c oxidase assembly factor HCC2 in mitochondria (Steinebrunner et al., 2014). The authors were unable to detect biotinylated HCC2-SNAP fusion proteins in mitochondria, possibly because the commercially available dyes did not efficiently pass through the cell wall or the plant membranes are impermeable or, although unlikely, the conditions in the respective cellular compartments interfered with the labeling reaction.
Given the large potential of small protein labeling in cell biology studies, we systematically tested the cell permeability of 31 different chemical dyes in cultured plant cells. The dyes were classified into three groups: dyes that enter the cytosol, dyes that enter in a pH-dependent fashion, and dyes that cannot be taken up by the cells. We subsequently used four different dyes to test SNAP-tagging of different cargo by tagging microtubules and the auxin transporter PIN2. We demonstrate self-labeling of microtubules by different dyes with different emission spectra in BY-2 cells and multicolor live cell imaging in Arabidopsis seedlings carrying genetically encoded fluorescent proteins. Localization and abundance of plasma membrane proteins such as transporters and receptors are dynamically controlled through endocytosis, recycling, and vacuolar degradation in addition to de novo synthesis (Luschnig and Vert, 2014; Yoshinari and Takano, 2017). Here, we show that PIN2 undergoes clathrin-mediated endocytosis and subsequent vacuolar sorting and that de novo–synthesized PIN2 protein is preferentially transported to the cell plate over the endocytosed/recycled PIN2 protein. The self-labeling of PIN2 shown here has advantages over alternative methods such as photoconversion of fluorescent proteins that is limited by the activation radius and artifacts that occur as a consequence of time-lapse imaging. Taken together, our data show that SNAP-tagging can be used for in vivo labeling in plants, thereby opening a wide range of applications to plant science studies.
RESULTS
Uptake of Synthetic Dyes in BY-2 Cells
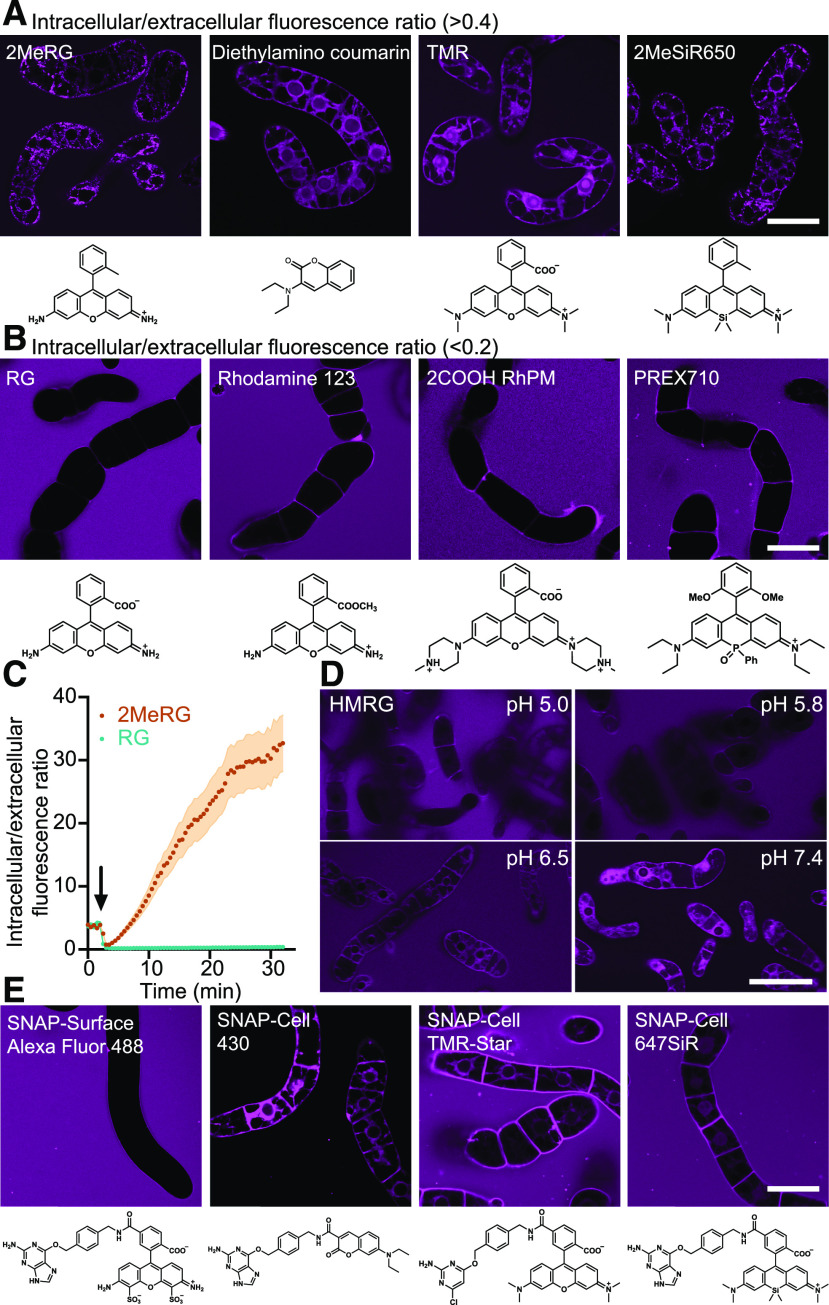
We collected 32 different fluorescent dyes from commercial resources and colleagues to add to those developed at Institute of Transformative Bio-Molecules (ITbM). The collection includes dyes that had been developed for SNAP-tagging in animal cells (Table 1) and potential SNAP-taggable dyes for measuring endocytosis of receptors; dyes for super-resolution microscopy; and dyes tested in plant cells such as rhodamine 123, rhodamine 6G, and fluorescein diacetate (Table 2; Vannini et al., 1988; Eisele et al., 2016; Jones et al., 2016). To evaluate which of these dyes, if any, can pass through the plant cell wall and which dyes can then enter the cell (through the cell membrane or another method), we systematically tested whether any of the 31 dyes could enter BY-2 cells by using confocal microscopy. DRBG-488, which is used for monitoring endocytosis of membrane proteins, was excluded since it carries a quenching group that is released for self-labeling of proteins through SNAP-tagging. As a proxy to uptake, we calculated the ratio between cytoplasmic and extracellular fluorescence in modified Linsmaier and Skoog (LS) medium at pH 5.8 (Katsuta et al., 1990). The dyes could be classified into two categories based on the relative fluorescence intensity inside versus outside the cell (Figure 1; Supplemental Figures 1 and 2). Twenty-three dyes were able to enter the cytoplasm of BY-2 cells within 1 min (Figure 1A), while 8 dyes did not show any substantial uptake over the short exposure period (Figure 1B; Supplemental Figures 1 and 2). Dyes such as rhodamine green (RG), rhodamine 123 (a dye that accumulates in mitochondria of animal cells and is used to monitor membrane potential), and PREX 710 (a long-wavelength dye for multicolor imaging) did not appear to be taken up to a measurable degree and thus at least after short incubation times do not seem suitable for plant cell biology. The dye 2COOH RhP-M (a pH-sensitive dye for membrane labeling) was also not taken up efficiently but could be suitable for measuring the pH during endocytosis in plant cells. We did not detect rhodamine 123 uptake into BY-2 cells, although the dye has previously been used for labeling mitochondria in leaves of Elodea and in suspension-cultured cells of Ranunculus (Vannini et al., 1988). Dyes that were not taken up by the BY-2 cells in our conditions may enter cells under other conditions or after longer exposure times.
Table 1. Photophysical Properties of Synthetic SNAP Dyes and Cellular Uptake in BY-2 Cells.
| Compound | Permeability | λabs (nm) | λem (nm) | φfl | Application | Reference, Catalog No. and Supplier |
|---|---|---|---|---|---|---|
| SNAP-Cell 430 | + | 421 | 444, 484 | Covalent labeling | S9109S, NEB | |
| SNAP-Surface Alexa Fluor 488 | - | 496 | 520 | covalent labeling | S9129S, NEB | |
| DRBG-488 | n.d.a | 496 | 520 | covalent labeling, pulse chase labeling | (Komatsu et al., 2011) | |
| SNAP-Cell TMR-Star | + | 554 | 580 | covalent labeling | S9105S, NEB | |
| SNAP-Cell 647SiR | + | 645 | 661 | covalent labeling | S9102S, NEB |
Plus (+) and minus (-) indicate intracellular/extracellular signal intensity at the 1-min time point >0.4 and <0.2, respectively. λabs, absorption maximum; λem, fluorescent emission maximum; φfl, fluorescence quantum yield. n.d., not determined. Blank cells indicate no data.
DRBG-488 is not fluorescent until reaction with a SNAP-tag.
Table 2. Photophysical Properties of Synthetic Dyes and Cellular Uptake in BY-2 Cells.
| Compound | Permeability | λabs (nm) | λem (nm) | φfl | Application | Reference, Catalog No. and Supplier |
|---|---|---|---|---|---|---|
| Diethylaminocoumarin | + | 387a | 467a | 0.05a | D0371, Tokyo Chemical Industry | |
| PhoxBright 430 | + | 426 | 542 | 0.66 | STED imaging | (Wang et al., 2017) |
| Fluorescein diacetate | + | 488 | 530 | esterase activity | F7378, Sigma-Aldrich | |
| Fluorescein | + | 494 | 521 | 0.7a | 065-00252, FUJIFILM Wako | |
| 2MeRG | + | 498 | 520 | (Piao et al., 2013) | ||
| RG (rhodamine 110) | - | 499 | 521 | CDX-R0016, AdipoGen Life Sciences | ||
| HMRG | + | 501 | 524 | 0.81 | pH sensitive | (Sakabe et al., 2013) |
| 2′,7′-Dichlorofluorescein | + | 504 | 529 | D0371, Tokyo Chemical Industry | ||
| Rhodamine 123 | - | 507 | 529 | mitochondria, membrane potential | 181-01701, FUJIFILM Wako | |
| HMDER | + | 525 | 543 | 0.14 | (Kamiya et al., 2011) | |
| Rhodamine 6G | + | 526 | 555 | 183-00622, FUJIFILM Wako | ||
| 2COOH RhP-M | - | 527a | 541b | 0.32a | pH sensor | (Asanuma et al., 2014), see Supplemental Methods |
| Rhodamine B | + | 527a | 570 | A5102, Tokyo Chemical Industry | ||
| HMDiMeR | + | 529 | 553 | 0.05 | photo-acoustic imaging | (Iwatate et al., 2016) |
| HMTMR | + | 552 | 576 | pH sensitive | (Kenmoku et al.,2007) | |
| Tetramethylrhodamine | + | 554 | 569a | 0.05a | See Supplemental Methods | |
| 2MeSiR600 | + | 593 | 613 | 0.38 | single molecule measurements | (Piao et al., 2013) |
| 2MeSiR620 h | + | 621 | 644 | 0.15 | (Hanaoka et al., 2018) | |
| HMJSiR | + | 637 | 662 | 0.20 | pH sensitive | (Iwatate et al., 2018) |
| 2MeSiR650 | + | 646 | 667 | 0.31 | (Umezawa et al., 2017) | |
| PREX 670 | + | 670 | 698 | 0.11 | STED imaging | (Grzybowski et al., 2017) |
| 2MeSiR700 | + | 691 | 720 | 0.13 | (Umezawa et al., 2017) | |
| BcPR 705 | + | 704 | 733 | 0.20 | SIM imaging | unpublished data |
| MePR 705 | - | 705 | 733 | 0.12 | unpublished data | |
| AzPR 710 | - | 708 | 734 | 0.14 | unpublished data | |
| PyPR 710 | - | 712 | 739 | 0.11 | unpublished data | |
| PREX 710 | - | 712 | 740 | 0.12 | mitochondrial dye | (Grzybowski et al., 2018) |
Plus (+) and minus (-) indicate intracellular/extracellular signal intensity at the 1-min time point >0.4 and <0.2, respectively. λabs, absorption maximum; λem, fluorescent emission maximum; φfl, fluorescence quantum yield. Blank cells indicate no data.
Measured in 0.5× MS buffer at pH 5.8.
Figure 1.
Representative Images of BY-2 Cells Exposed to Various Fluorescent Dyes under Various Conditions.
(A) Representative images of four dyes that entered BY-2 cells within 1 min. Permeability was calculated as the ratio of fluorescence intensity inside/outside the cell. An intracellular/extracellular fluorescence intensity ratio of >0.4 indicated uptake.
(B) Representative data for three dyes unable to enter BY-2 cells. Intracellular/extracellular fluorescence intensity ratios were <0.2.
(C) Time-lapse analysis of 2MeRG and RG accumulation, measured by intracellular/extracellular signal ratios. Plots and error bands represent mean (n = 6 cells) and se, respectively. Arrow indicates when dyes were added. Initial positive ratio is in absence of dye and due to autofluorescence.
(D) HMRG uptake at different pH values.
(E) Confocal images of BY-2 cells incubated with SNAP-Surface Alexa Fluor 488, SNAP-Cell 430, SNAP-Cell TMR-Star, and SNAP-Cell 647SiR for 1 min. Images were taken with same settings at different time points. Bars = 50 µm. Experiments were repeated independently three times with comparable results.
While our initial survey was performed at only the 1-min time point, we also tested for two compounds whether uptake was possible when using longer incubation periods. While RG did not show any detectable uptake even after 30 min, the methylated variant 2MeRG, which showed some accumulation after 1 min, displayed multiphasic uptake kinetics and accumulated 40-fold within 30 min (Figure 1C). All of the experiments mentioned above were performed at pH 5.8, a typical pH in the physiological range for the apoplasm. Since several dyes, for example, hydroxymethyl group–bearing O-rhodamines, alter their charge or change their structure in a pH-dependent manner, we tested whether uptake changed with pH. We found that hydroxymethyl rhodamine green (HMRG) showed increased accumulation at higher pH values (Figure 1D).
To test whether certain dyes may be suitable for protein self-labeling, four commercially available dyes frequently used for SNAP-tagging in animal cells were tested (Table 1). These SNAP-taggable dyes are derived from dyes tested here in their native form or a methylated form, by conjugation to a benzylguanine group. Surface Alexa Fluor 488, derived from Alexa Fluor 488, showed no accumulation in the cells. On the other hand, SNAP-Cell 430, a diethylaminocoumarin derivative; SNAP-Cell TMR-Star, a tetramethylrhodamine (TMR) derivative; and SNAP-Cell 647 silicon-rhodamine (SiR), a 2MeSiR650 derivative, did show low but detectable accumulation (Figure 1E). For these three dyes tested here, they could enter the BY-2 cells in their native or benzylguanine-conjugated forms (TMR and SNAP-Cell TMR-Star, diethylaminocoumarine and SNAP-Cell430, 2MeSiR650 and SNAP-Cell 647SiR). In summary, this screen identified permeable dyes suitable for SNAP-tagging of cytoplasmic processes and cell-impermeable dyes suitable for tagging membrane proteins at the cell surface.
SNAP-Tagging of α-Tubulin in BY-2 Cells
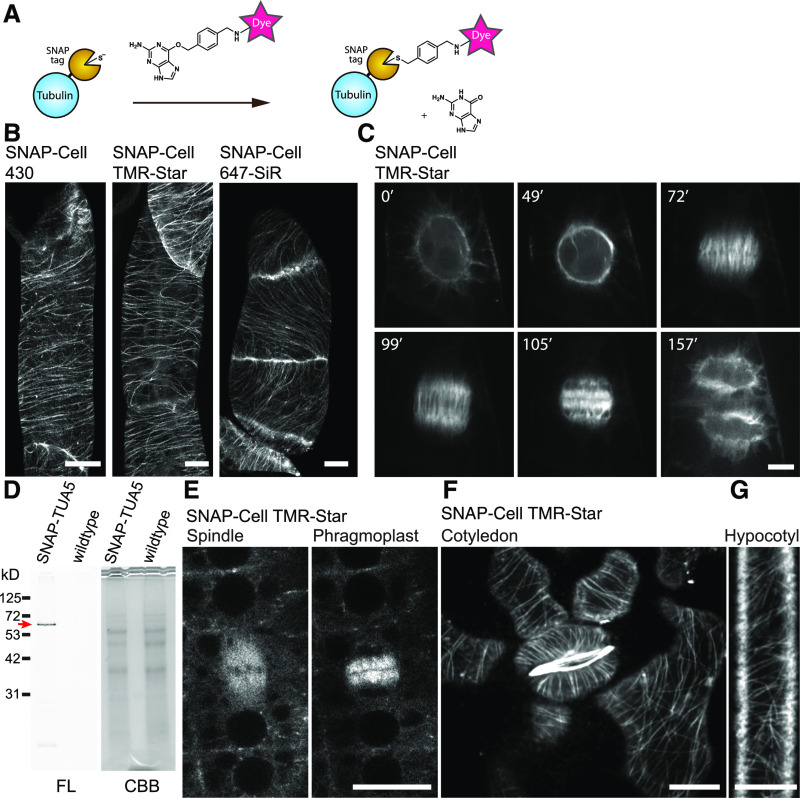
While the experiments with BY-2 cells may indicate that there is no fundamental issue with dye permeability, other factors could be responsible for reported failure to deploy SNAP-tagging in plant cells (Schiller et al., 2012; Steinebrunner et al., 2014). We therefore tested whether the commercially available dyes for self-labeling, namely, SNAP-Cell 430, SNAP-Cell TMR-Star, and SNAP-Cell 647SiR, would be suitable for labeling the cytoskeleton of BY-2 cells. When BY-2 cells stably transformed with the construct pUBQ10:SNAP-α-tubulin (TUA5) were incubated with any of the three dyes, efficient labeling of the microtubules was observed (Figure 2). In interphase cells, cortical microtubules were found to be highly ordered and oriented perpendicular to the growth axis (Figure 2B). In mitotic cells, microtubules first formed a preprophase band (49 min) and then generated the spindle during metaphase (72 min) and subsequently transitioned to a concentrated cylinder, the phragmoplast, during telophase (105 min; Figure 2C; Supplemental Movie 1; Supplemental Movie Legends). The observed microtubule structures were highly similar to those seen when using fluorescent protein-tagged TUA5 (Lindeboom et al., 2013). These results indicated that SNAP-tagging with these dyes is feasible in plants, at least in tobacco BY-2 cell cultures.
Figure 2.
SNAP-Tagging Enabled in Vivo Imaging of Tubulin in BY-2 Cells and Arabidopsis.
(A) Labeling mechanism of the SNAP-tag.
(B) Cortical microtubules in BY-2 cells expressing SNAP-TUA5 with SNAP dyes denoted above the images. Images show max intensity projection of confocal z-stack slices taken in 0.5-µm steps.
(C) Time-lapse imaging of mitotic microtubule dynamics via TMR-Star labeling of TUA5. Images were taken every 30 s, and elapsed time (minutes) is shown.
(D) Fourteen-day-old pUBQ10:SNAP-TUA5 and Columbia-0 (wild-type) seedlings were stained with 500 nM SNAP-Cell TMR-Star for 3 h and lysed. The crude protein extract was analyzed by SDS-PAGE. Left, Fluorescence (FL). Right, Coomassie Brilliant Blue (CBB) staining. SNAP-Cell TMR-Star selectively labeled the SNAP-TUA5 in protein extracts. A red arrow indicates the expected molecular weight of SNAP-TUA5.
(E) Confocal images of mitotic cells in the root epidermis of 4-d-old SNAP-TUA5 Arabidopsis seedlings stained with TMR-Star. Spindles and phragmoplasts were observed.
(F) Confocal image of cortical microtubules of pavement cells and stomata in the cotyledon of 4-d-old SNAP-TUA5 Arabidopsis seedlings stained with SNAP-Cell TMR-Star.
(G) Confocal image of cortical microtubules of etiolated hypocotyl epidermal cells stained with SNAP-Cell TMR-Star. Bars = 10 µm. Experiments were repeated independently three times with comparable results.
SNAP-Tagging of α-Tubulin in Arabidopsis Cells
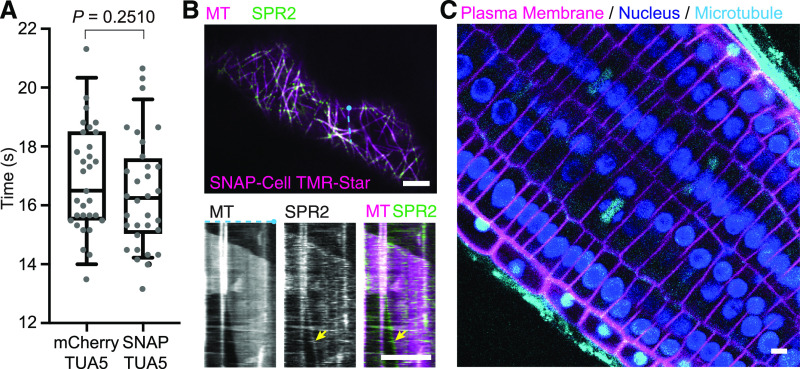
The experiments with BY-2 cells may indicate that tag labeling is feasible in plants; however, it is also conceivable that BY-2 cells, as single cells or cell files, were more accessible compared to cells in intact organs such as plant roots or that BY-2 cells might have particular features that allow passage of dyes through the cell wall and entry into the cytoplasm. We therefore tested whether the cytoskeleton could be labeled in a similar way in Arabidopsis plants stably transformed with pUBQ10:SNAP-TUA5, a label on α-tubulin 5 protein. SDS-PAGE demonstrated that SNAP-Cell TMR-Star selectively labeled SNAP-TUA5 in protein extracts from Arabidopsis seedlings (Figure 2D). In Arabidopsis roots incubated in a dye solution, cytoskeletal structures such as the spindle and phragmoplast were efficiently labeled with SNAP-Cell TMR-Star and SNAP-Cell 647SiR (Figure 2E; Supplemental Figure 3A). To image cotyledon and etiolated hypocotyls, 3- to 4-d-old seedlings were vacuum infiltrated with the SNAP dye in microcentrifuge tubes. Cortical microtubules in the cotyledon pavement cells and the etiolated hypocotyl were labeled with SNAP-Cell TMR Star and SNAP-Cell 647SiR (Figures 2F and 2G; Supplemental Figure 3B; Supplemental Movie 2; Supplemental Movie Legends). To investigate any potential toxicity of the SNAP-tag and the dyes, we compared mitotic cell divisions in the plants expressing SNAP-tag tubulin and incubated them with SNAP-Cell TMR-Star dye and with those carrying a FP-tagged microtubule marker, mCherry-TUA5 (Shaw et al., 2003). The duration from nuclear envelope breakdown to phragmoplast initiation in SNAP-TUA5 treated with 500 nM SNAP-Cell TMR-Star (mean = 16.41 min from 31 cells) was not significantly different from that in plants expressing FP-tagged tubulin (mean = 16.81 min from 30 cells; Figure 3A).
Figure 3.
SNAP-Tagging Did Not Have a Significant Impact on the Duration of Mitotic Cell Divisions and Enabled in Vivo Multicolor Imaging in Arabidopsis Seedlings.
(A) Comparison of the duration from nuclear envelope breakdown to the phragmoplast initiation between mitotic cells in mCherry-TUA5 plants (n = 31) and SNAP-TUA5 plants incubated in 500 nM SNAP-Cell TMR-Star (n = 32). There was no significant difference (P = 0.2510 by Mann-Whitney U test). In the box plots, the boxes represent the range from the 25th to 75th percentiles, the horizontal lines represent the median value, and the whiskers span from the 5th to 95th percentiles.
(B) Cotyledon epidermal cells of Arabidopsis coexpressing SPR2-GFP (SPR2) and pUBQ10:SNAP-TUA5 (MT). Four-day-old seedlings were incubated in 0.5× MS containing 500 nM SNAP-Cell TMR-Star. (Top) Representative confocal image of MTs and SPR2 in a pavement cell. (Bottom) Kymographs generated from the top panel (at the blue dotted line) showing SPR2 tracking the minus end of microtubule (yellow arrow).
(C) Root tip of Arabidopsis coexpressing p35S:YFP-LTI6b, p35:H2B-RFP, and pUBQ10:SNAP-TUA5. Three-day-old seedlings were incubated in 0.5× MS containing 500 nM SNAP-Cell 647SiR for 30 min. Bars = 5 µm. Experiments were repeated independently two times ( [A] and [B]) and three times (C)) with comparable results.
One of the major advantages of SNAP tagging is the availability of a wide range of dyes with specific spectral properties, which enables dual or multicolor in vivo imaging. The potential of dual-color imaging was tested here in plants coexpressing SNAP-TUA5 with the microtubule minus-end marker SPR2-GFP (Yao et al., 2003). SPR2-GFP accumulated at the minus ends of SNAP-Cell TMR-Star–labeled microtubules (Figure 3B; Supplemental Movie 3; Supplemental Movie Legends). The labeling of minus ends by SPR2-GFP is consistent with the previous observation of microtubules utilizing fluorescent proteins (Nakamura et al., 2018), further supporting the hypothesis that the SNAP dye does not negatively affect SPR2-GFP interaction with the minus end. Three-color live cell imaging was performed by crossing the SNAP-TUA5 line with plants coexpressing the plasma membrane marker yellow fluorescent protein (YFP)-LTI6b (Cutler et al., 2000) and the nuclear marker H2B-RFP (Federici et al., 2012). These cells were incubated with SNAP-Cell 647SiR, shown to enter BY-2 cells (Figure 3C), to label the SNAP-tagged TUA5 microtubules. In Arabidopsis root tips incubated for 30 min with SNAP-Cell 647SiR, microtubule arrays were detected, as well as YFP-labeled plasma membrane and red fluorescent protein (RFP)–marked nuclei (Figure 3C; Supplemental Movie 4; Supplemental Movie Legends). Thus, SNAP-tagging appears to be an efficient and simple-to-use tool for self-labeling abundant cytosolic proteins such as α-tubulin.
SNAP-Tagging of the Auxin Transporter PIN2
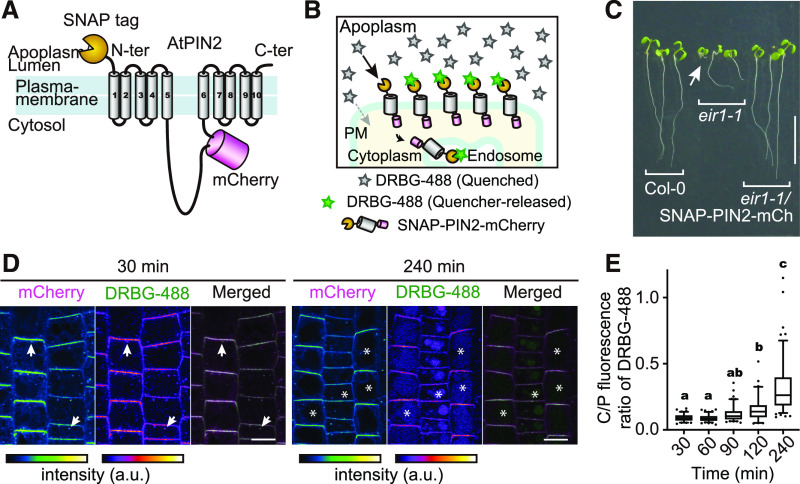
Probes that cannot pass through the cell membrane can be particularly suitable for studying endocytosis of membrane proteins, such as transmembrane receptors and transporters (Komatsu et al., 2011; Geiger et al., 2013; Roed et al., 2014). The cell membrane–impermeable probe DRBG-488 carries an intramolecular Förster resonance energy transfer quencher group that is released when reacting with a SNAP-tag. Thus, DRBG-488 only labels tagged proteins located at the cell surface and only becomes fluorescent after covalent linkage to a tagged protein of interest, providing high signal to background and proving useful for monitoring endocytosis (Komatsu et al., 2011). As membrane impermeability has been observed in animal cells, SNAP-TUA5 Arabidopsis seedlings were used to confirm that DRBG-488 was unable to enter plant cells (Supplemental Figure 4). To test whether DRBG-488 can be used to observe endocytosis of a plasma membrane transporter that is localized to polar ends of plant cells, we examined the SNAP-tagged auxin efflux carrier PIN2 (Adamowski and Friml, 2015). The SNAP-tag was fused to the extracellular N terminus of a PIN2 protein that also carried an mCherry tag in the central cytosolic loop, to be able to monitor the production of PIN2. PIN2 was expressed under its native promoter (Figures 4A and 4B).
Figure 4.
SNAP-Tagging of the PIN2 Auxin Transporter Enabled FAPL with DRBG-488.
(A) Topological model of SNAP-PIN2-mCherry fusion protein. SNAP-tag and mCherry are fused at the N terminus and cytosolic loop of PIN2, respectively.
(B) Conceptional illustration of labeling of SNAP-PIN2-mCherry by DRBG-488. DRBG-488 itself is impermeable to the cell membrane and is not fluorescent until reaction with a SNAP-tag. Once DRBG-488 binds to SNAP-tag, the quencher group is released and DRBG-488 becomes fluorescent. Since DRBG-488 reacts covalently with the SNAP-tag, it can be internalized with the chimeric SNAP-PIN2-mCherry.
(C) Phenotype of wild-type Columbia-0, eir1-1, and eir1-1 harboring pPIN2:SNAP-PIN2-mCherry grown for 7 d on solid media on vertical plates. Arrow indicates a root detached from the medium. Bar = 10 mm.
(D) Pulse-chase analysis of DRBG-488 labeling. Plants were incubated with 200 nM DRBG-488 for 30 min followed by washout and 30- or 240-min incubation in liquid medium. Arrows indicate signals in plasma membrane and intracellular punctate structures. Asterisks indicate vacuoles. Bars = 10 µm.
(E) Ratio of fluorescence in the C/P of DRBG-488-SNAP-PIN2–derived fluorescence in the root gradually increased over time. In the box plots, the boxes represent the range from the 25th to 75th percentiles, the horizontal lines represent the median value, and the dots indicate outliers. Different letters above the plots represent significant differences among means (P < 0.005 by one-way ANOVA with Tukey’s post hoc test; exact P-values are described in the Supplemental Data Set). n = 86, 104, 104, 62, and 118 cells from three roots. Experiments were repeated independently three times.
To validate that the double-tagged fusion SNAP-PIN2-mCherry did not negatively impact PIN2 activity in Arabidopsis, we complemented the pin2 mutant line (eir1-1) and investigated agravitropic root growth (Roman et al., 1995). SNAP-PIN2-mCherry fusions restored normal gravitropic growth of eir1-1 (Figure 4C), demonstrating that SNAP-PIN2-mCherry was functional. Moreover, consistent with previous reports (Roman et al., 1995), confocal microcopy using RFP emission showed that SNAP-PIN2-mCherry localized preferentially in apical and basal domains of the plasma membrane of the root epidermal and cortex cells, respectively (Supplemental Figures 5A and 5B). DRBG-488 effectively labeled SNAP-PIN2-mCherry within 30 min (Supplemental Figures 6A and 6B). Both RFP and green fluorescence derived from the unquenched DRBG-488 showed polar SNAP-PIN2-mCherry localization at the apical and basal membranes of epidermal and cortical cells, respectively (Supplemental Figure 7).
To monitor endocytic internalization of SNAP-PIN2-mCherry, a pulse-chase experiment using DRBG-488 was performed. Thirty minutes after washout of the dye, green (DRBG-488) and red fluorescence (mCherry) colocalized in puncta in the cytoplasm, presumably corresponding to endosomes (Figure 4D). Four hours after washout, DRBG-488-SNAP-PIN2–derived fluorescence was detected in vacuoles in addition to the punctate structures also seen earlier (Figure 4D). The ratio of fluorescence in the cytoplasm:plasma membrane (C/P) gradually increased over time (Figure 4E). These results indicated that DRBG-488–labeled SNAP-PIN2-mCherry was at the cell surface at first and then was endocytosed over approximately an hour, and finally was transported to the vacuole after ∼2 h. To corroborate that PIN2 is endocytosed, pharmacological inhibition of endocytosis by ES9-17, an inhibitor of the clathrin heavy chain that is required for clathrin-mediated endocytosis (Dejonghe et al., 2019), was performed. ES9-17 reduced DRBG-488–derived fluorescence in both endosomes and vacuoles (Figure 5A; Supplemental Figure 8A). The number of endosomes labeled by DRBG-488 within 60 min was reduced significantly from 3.0 (±0.2 se) in mock-inhibited roots to 1.6 (±0.1 se) in ES9-17–treated roots (Figure 5B). Similarly, the C/P ratio of DRBG-488 fluorescence was significantly decreased from 0.11 (±0.004 se) in mock-treated roots to 0.08 (±0.003 se) in ES9-17–treated roots (Figure 5C). The reduction in the number of endosomes and the C/P ratio by ES9-17 treatment was more substantial after 120 min of incubation with DRBG488 (Supplemental Figure 8B). The effect of ES9-17 on the C/P ratio of mCherry fluorescence was less compared to DRBG-488 (Supplemental Figure 9), due to continuous de novo synthesis of SNAP-PIN2-mCherry (mCherry positive and DRBG-488 negative) in the cytoplasm. Taken together, DRBG-488 only labels SNAP-PIN2-mCherry that is localized at the cell surface at the time of dye treatment, and the internalization of DRBG-488–labeled SNAP-PIN2-mCherry depends to a large extent on clathrin-mediated endocytosis.
Figure 5.
Evidence for Clathrin-Mediated Endocytosis of PIN2.
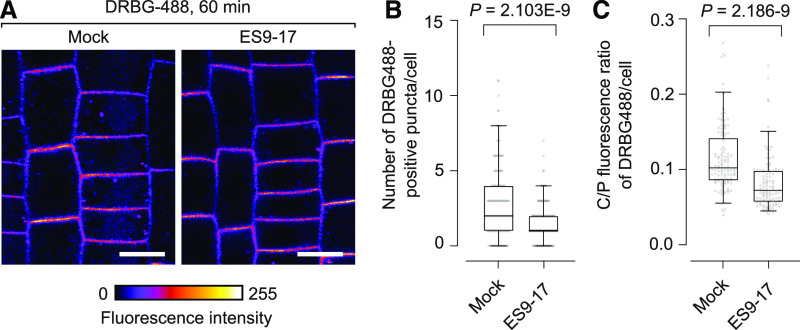
(A) DRBG-488 labeling of SNAP-PIN2-mCherry after DMSO (mock) or 30 µM ES9-17 treatment for 60 min. Bars = 10 µm.
(B) Number of DRBG-488–positive endosomes per cell in the presence or absence of ES9-17. Significance of difference was determined by unpaired two-tailed t test with Welch’s correction. n = 139 (DMSO) and 128 (ES9-17) cells from five roots.
(C) C/P fluorescence ratio of DRBG-488 in the presence or absence of ES9-17. Box plots are centered at the data median and mark from the 25th to the 75th percentile and whiskers span from 5th to 95th percentile. Dots indicate outliers. Significance of difference was determined by unpaired two-tailed t test with Welch’s correction. n = 117 (DMSO) and 115 (ES9-17) cells from five roots. Data that show comparable effects for 120-min treatments are shown in Supplemental Figure 7.
It had previously been reported that, after synthesis, PIN2 is preferentially transported to the cell plate during cytokinesis, followed by re-distribution of PIN2 in a polar manner (Mravec et al., 2011; Glanc et al., 2018). We thus hypothesized that the DRBG-488–free fraction of SNAP-PIN2-mCherry, which represents de novo–secreted PIN2, will accumulate at higher levels at the cell plate than the DRBG-488–labeled fraction, which is subject to endocytosis. We thus determined the ratio of mCherry:DRBG-488 fluorescence within different parts of the plasma membrane of dividing epidermal cells in the Arabidopsis root over time. After labeling with 200 nM DRBG-488 for 90 min, time-lapse imaging showed that while the ratio of DRBG-488- to mCherry-labeled SNAP-PIN2-mCherry was ∼1 in the apical part of the cells, newly formed cell plates had a ratio of DRBG-488– to mCherry-labeled SNAP-PIN2-mCherry of <0.1 (Figure 6). These data are consistent with the hypothesis that endocytosis of PIN2 from the apical domains does not contribute to a large extent to the PIN2 amount present at the cell plate and that the protein at the cell plate is mainly derived from de novo synthesis.
Figure 6.
Preferential Delivery of Newly Produced PIN2 to the Newly Forming Cell Plate.
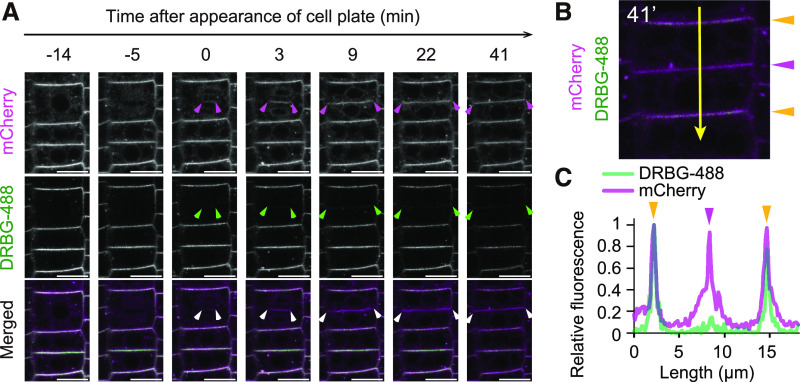
(A) Time-lapse imaging of PIN2 in a dividing cell. Arrowheads indicate edges of the cell plate and connection sites between newly formed plasma membrane and the lateral plasma membrane. Bars = 10 µm.
(B) Quantification of fluorescence intensities of DRBG-488 and mCherry along the line in the cell at 41 min after appearance of the cell plate. Orange and pink arrowheads indicate preexisting and newly formed plasma membrane, respectively.
(C) Scan of relative fluorescence of DRBG-488 and mCherry in images from (B), with length representing the position from the top of the image. Colored arrowheads on peaks correspond to the scan position in (B). Experiments were repeated independently three times.
These in planta experiments showed that the reaction of a membrane-impermeable, self-quenched dye with the SNAP-tag on the extracellular N terminus of a polytopic transporter in combination with the fusion of a fluorescent protein enabled the monitoring of endocytosis, intracellular trafficking, and delivery of newly synthesized protein to the cell plate. The use of the double tag allowed differentiation of the cellular pools of one protein.
DISCUSSION
Self-labeling of tagged fusion proteins in vivo has several advantages over the use of fusions with fluorescent proteins. For unknown reasons, it was widely thought that this extensive tool set, complete with many different tags and dyes, was not applicable to plant cells. Given the large potential of the technology, and the numerous advantages and wide range of possibilities offered by chemical dyes, we systemically reevaluated their utility in plants. A systematic screen of 31 dyes allowed us to classify the dyes into cell-permeable and cell-impermeable compounds. We subsequently demonstrated SNAP-tagging of microtubules in BY-2 cells using three structurally different dyes. We also showed multicolor labeling with SNAP-tagging in combination with FPs using a quenched dye that is unable to enter cells on its own to observe endocytosis of the SNAP-tagged auxin transporter PIN2.
Synthetic Dyes
The analysis of a total of 32 chemical dyes showed that some dyes are able to enter BY-2 cells within a minute; other dyes may enter more slowly, while for some no substantial uptake was observed. In addition, the permeability of the BY-2 cell membrane was pH dependent for certain dyes. For some dyes, uptake into roots of intact Arabidopsis plants was also observed, indicating that the survey in BY-2 provides a good way to determine whether such dyes can or cannot be taken up by plant cells. Notably, commercially available dyes used for SNAP-tagging in animal cells were also able to pass the cell wall and enter the plasma membrane of plant cells. In principle, dyes that enter the cells can be used for self-labeling of cytosolic and nuclear proteins and possibly proteins in subcompartments such as vacuoles, plastids, and mitochondria. Dyes that cannot enter the cells have the potential to be used to label proteins present at the cell surface, such as transporters and receptors. One caveat is that for covalent labeling of tagged proteins, the dyes have to be modified by conjugation to benzylguanine or an analogous reagent, which may affect membrane permeability. Thus, for dyes that have not been tested in their SNAP form, permeability must be determined. We could not detect permeability of DRBG-488, a dye that carries both a benzylguanine and a quencher that largely eliminates fluorescence of unreacted dye. DRBG-488 only becomes fluorescent once it is SNAP-tagged to a protein at the cell surface. DRBG-488 was able to label the auxin transporter PIN2 specifically at the cell surface. Taken together, the survey and the SNAP-tagging experiment show that a wide range of dyes with unique properties can in principle be used for self-labeling of plant proteins inside cells or at the cell surface.
For self-labeling of cytosolic proteins, SNAP-Cell 430, SNAP-Cell TMR-Star, and SNAP-Cell 647SiR were used to visualize microtubule arrays in BY-2 cells. By making use of the long wavelength excitation and emission maximum, SNAP-Cell 647SiR was also used to perform three-color live cell imaging in Arabidopsis seedlings. The near-infrared (NIR) fluorescent dyes (650 to 900 nm) are useful for fluorescence imaging of mammalian cells and tissues, which have high transparency and low autofluorescence in the NIR optical window (Weissleder and Ntziachristos, 2003). Although plants have phytochromes with absorption maxima at 660 and 730 nm (Li et al., 2011), the NIR dyes were shown here to be usable for self-labeling and for multicolor NIR fluorescence imaging in plant cells. In addition to SiR-based dyes, PREX 670 and BcPR 705 have long-wavelength emission spectra and pass through the plasma membrane, permitting simultaneous use with commonly used genetically integrated fluorescent protein probes. PB430 was also able to enter plant cells, and its high photostability and relatively long fluorescence lifetime (τ = 10.6 ns) make it suitable for STED microscopy imaging (Wang et al., 2017). Since it may be useful in plant cells, the synthesis of a SNAP-PB430 dye would be a useful tool for such applications as super-resolution imaging of cytosolic proteins. Dyes such as 2COOH RhP-M, which was unable to enter the plant cytoplasm, could be used as a pH sensor in endocytosed compartments.
Dynamic Analysis of Endocytosis and Intracellular Trafficking of PIN2 by SNAP-Tagging
FAPL with DRBG-488 has been successfully used to monitor endocytosis of the epidermal growth factor receptor in COS7 and Madin-Darby canine kidney animal cell cultures (Komatsu et al., 2011). In plants, endocytosis and intracellular trafficking also play important roles in the maintenance of membrane protein abundance and in subcellular localization (Luschnig and Vert, 2014). Here, we utilized the cell-impermeable fluorescent probe DRBG-488 that is de-quenched upon reaction with the SNAP-tag for labeling of SNAP-PIN2-mCherry at the cell surface. Our data indicate that PIN2 is internalized by clathrin-mediated endocytosis (Figure 4). This is consistent with the conclusion from previous studies (Dhonukshe et al., 2007; Kitakura et al., 2011; Adamowski et al., 2018; Glanc et al., 2018). These studies used photoconvertible fluorescent proteins, pharmacological experiments with brefeldin A (BFA), and genetic evidence. BFA is widely used to examine the endocytic rate in plant research. BFA inhibits a subset of the ADP ribosylation factor guanine nucleotide exchange factors, such as GNOM. GNOM, however, is localized to both the plasma membrane and the Golgi bodies, but not in the trans-Golgi network/early endosomes under normal conditions, and BFA induces dissociation of GNOM from both the Golgi apparatus and plasma membrane and accumulation in the trans-Golgi network/early endosomes (Naramoto et al., 2010, 2014). Consequently, BFA disturbs not only endocytosis of membrane proteins but also endosomal recycling (Naramoto et al., 2010, 2014). Furthermore, BFA alters the morphology of the Golgi apparatus and induces absorption of the Golgi cisternae into the endoplasmic reticulum in aerial tissues of Arabidopsis, implying that there are unknown molecular targets other than the ADP ribosylation factor guanine nucleotide exchange factors (Robinson et al., 2008). Thus, BFA treatment compromises GNOM-dependent endocytosis and natural intracellular trafficking, making it difficult to draw reliable conclusions on endocytosis. Photoconvertible fluorescent proteins (FPs) such as Dendra2 (Jásik et al., 2013; Glanc et al., 2018) are also widely used to monitor endocytosis of proteins. However, photoconvertible proteins potentially undergo undesirable photoconversion during time-lapse imaging, and photoconversion can be difficult in certain regions of interest. Moreover, photoconversion occurs over a radius of several micrometers in the cell, activating other proteins as well. Therefore, self-labeling thus has two major advantages for the study of cell surface proteins: it targets only proteins at the cell surface and permits high-resolution imaging that is otherwise limited by the undesirable photoconversion of FPs. One advantage of photoconvertible FPs is the ability to mark proteins specifically in a subset of the plasma membrane, which is not possible with a simple SNAP-tagging approach. Furthermore, we confirmed by pulse-chase observation of DRBG-488–labeled SNAP-PIN2-mCherry that newly formed PIN2 protein is transported to the cell plate rather than endocytosed/recycled PIN2 protein (Figure 5). This result is consistent with observations from a previous report in which a photoconvertible FP was instead used (Glanc et al., 2018). Notably, while cells in the root tip and elongation zone showed reliable and specific labeling of PIN2, cells in the calyptra, cells in older zones of the root, and dead cells also became fluorescent, even in the wild type (Supplemental Figure 6A), likely as a consequence of unspecific removal of the quencher from DRBG-488 in these cells. Although further investigation will be needed to understand potential limitations of SNAP technology, we demonstrate herein that SNAP-tagging provides a way for dynamic analysis of endocytosis and intracellular trafficking of functional membrane proteins such as PIN2.
Taken together, more than 30 chemical dyes were classified as cell-permeable or cell-impermeable compounds in BY-2 cultures. Furthermore, we showed that self-labeling of soluble proteins with chemical dyes that are taken up into plant cells is feasible in plants expressing a SNAP-tagged construct and by fluorescently tagging microtubules to observe cytoskeletal dynamics in vivo. Self-labeling techniques expand the options for multicolor labeling and enable the use of novel dyes with reporting functions, such as pH sensors, or compounds suitable for super-resolution microscopy, such as MitoPB Yellow, PhoxBright 430, or PREX 670 (Wang et al., 2017, 2019; Grzybowski et al., 2018). Self-labeling with chemical dyes via a SNAP-tag can be efficiently used to fluorescently tag membrane proteins at the cell surface, in particular for FAPL. Here, we also used DRBG-488, which cannot enter the plant cells, in Arabidopsis and to tag PIN2 at the cell surface and to monitor endocytosis. We demonstrated that SNAP-tagged PIN2 accumulates at newly forming cell plates to higher levels than mCherry-labeled PIN2, providing support for the hypothesis that newly synthesized PIN2 is delivered to the cell plate rather than being endocytosed.
METHODS
Sources and Synthesis of Dyes
Dyes were either obtained from commercial sources, the Yamaguchi lab (ITbM, Nagoya University), or the Urano lab (University of Tokyo). 6-[[4-(Aminomethyl) phenyl] methoxy]-9H-purin-2-amine was prepared according to a previous report (Keppler et al., 2004). TMR and 2COOH RhP-M were synthesized here (for full details, see Supplemental Methods).
DNA Constructs
The pUBQ10:SNAP-TUA5 binary vectors were generated using Gateway technology (Invitrogen) by following the Multisite Gateway Technology three-fragment vector construction kit protocol. UBQ10pro-p4p1r and t35S-p2rp3 constructs were kindly provided by Jelmer Lindeboom and David Ehrhardt (Carnegie Institution for Science, Stanford University). The SNAP-tag sequence was amplified from the pSNAPf vector (cat. no. N9183; New England Biolabs [NEB]) and inserted into the AscI and KpnI sites after the UBQ10 promoter in the UBQ10pro-p4p1r entry vector. The open reading frame of AtTUA5 (At5g19780) was cloned into pDONR/zeo. Three entry clones, UBQ10pro-p4p1r, TUA5-pDONR/zeo, and t35S-p2rp3, were transferred into the binary vector pHm43GW (Karimi et al., 2005) by LR Clonase II Plus enzyme (Invitrogen). The primers used in this study were shown in Supplemental Table 1. The plasmid map of the resulting vector, named pSTA5, is shown in Supplemental Figure 10A. For the construction of SNAP-PIN2-mCherry, the genomic sequence of AtPIN2 (At5g57090; –2784 bp from the start codon to +688 bp from the stop codon) was amplified from Arabidopsis (Arabidopsis thaliana) genomic DNA by PCR and integrated into the pGEM-T Easy vector (Promega). mCherry was introduced into the PIN2 construct by inverse PCR followed by an In-Fusion reaction (Clontech). The SNAP-tag with an N-terminal secretion signal peptide sequence (MKTNLFLFLIFSLLLSLSSAEF; the signal sequence likely improves import of the tag into the endoplasmic reticulum, since constructs lacking the signal sequence did not show mCherry fluorescence; Haseloff et al., 1997) was synthesized by IDT gBlocks Gene Fragments service (Supplemental Tables 2 and 3). The SNAP-tag was introduced into the PIN2-mCherry construct by inverse PCR and In-Fusion reaction. The resulting construct was digested by NotI and integrated into the pBIN40 binary vector by ligation (Miyashima et al., 2011). The resulting binary vector was introduced into Agrobacterium tumefaciens strain GV3101:MP90 and used to transform Arabidopsis and tobacco BY-2 cells. The plasmid map of the resulting vector, pAY230, is shown in Supplemental Figure 10B.
Plant Cell Cultures and SNAP-Tag Labeling with Fluorescent Dyes
BY-2 cells were cultured in modified LS medium (Katsuta et al., 1990) buffered with 0.05% (w/v) MES, pH 5.8. Cells were subcultured once a week by adding cell suspension (1.5 mL for the wild-type cells or 3 mL for SNAP-TUA5–expressing cells) into 80 mL of fresh medium. Cultures were maintained in the dark at 25°C with shaking (120 rpm). To generate transgenic BY-2 cell lines expressing SNAP-TUA5, Agrobacterium-mediated transformation was performed following published methods (http://2010.igem.org/Team:Nevada/BY-2_(NT1) Transformation_Protocol). Transformants were selected in LS medium containing 0.8% (w/v) agar, 40 µg mL−1 hygromycin, and 200 µg mL−1 carbenicillin. Resistant calli were transferred to new selection media and then cultured for 3 weeks before initiating a liquid culture in modified LS medium. To label SNAP-TUA5 with chemical dyes, 1- or 2-d-old SNAP-TUA5–expressing BY-2 cells were used. Subcultured cells (500 µL) were transferred into 1.5-mL tubes to replace the media with SNAP dye–containing medium. For SNAP-Cell TMR-Star (cat. no. S9105S; NEB) and SNAP-Cell 430 (cat. no. S9109S; NEB), solutions were prepared by diluting 1 mM dye stock solution in 100% DMSO to a final concentration of 500 nM. SNAP-Cell SiR647 (cat. no. S9102S; NEB) was prepared from 1 mM dye stock solution in DMSO to a final concentration of 1 µM, but to dissolve the dye into the media the final DMSO concentration was adjusted to 1% (v/v). Since covalent labeling of the SNAP-tag with O6-benzylguanine–conjugated dyes occurred within 5 to 10 min in vitro, and within 60 min in Escherichia coli (Keppler et al., 2003), incubation times between 5 and 60 min were tested, depending on the dye and experiments: 5 or 10 min for SNAP-Cell TMR-Star, 30 min for SNAP-Cell 430, and 60 min for SNAP-Cell 647SiR. After incubation with the SNAP dyes, cells were washed with 3% (w/v) Suc several times and then resuspended in fresh LS medium. For time-lapse imaging of microtubules, cells were attached to poly-l-lysine–coated glass-based dishes.
Plant Material and Treatments
For multicolor labeling, we generated Arabidopsis lines expressing p35S:H2B-RFP (nuclei; Federici et al., 2012), p35S:YFP-LTI6b (plasma membrane; Cutler et al., 2000), and pUBQ10:SNAP-TUA5 (this study) by crossing. Partially heterozygous F2 lines were used for imaging. Plants were grown on either 0.5× Murashige and Skoog (MS) media with 1% (w/v) Suc and 1% (w/v) agar or modified MGRL media (Takano et al., 2005) containing 1% (w/v) Suc and 1.5% (w/v) gellan gum (Wako Pure Chemicals) and 30 µM boric acid. Seeds were surface sterilized with 70% (v/v) ethanol and sown on media plates. To synchronize germination, plates were placed in the dark at 4°C for 2 d. Plants were grown vertically in a 16-h-light/8-h-dark cycle (40 μE m−2 s−1 of white light) for 4 d for imaging or for 7 d for growth analysis. For fluorometric analysis on SDS-PAGE, 14-d-old seedlings were incubated in 0.5× MS medium containing 500 nM SNAP-Cell TMR-Star for 3 h. Seedlings were homogenized and then lysed in 50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1% Triton X-100, 2 mM phenylmethylsulfonyl fluoride, and Complete Protease Inhibitor cocktail (Roche Diagnostics). The extracted proteins (2 µg) were mixed with 4× SDS sample loading buffer, separated by SDS-PAGE, and stained with Coomassie Brilliant Blue R 250. Fluorescent images were captured using a Typhoon FLA 9500 laser scanner (GE Healthcare). For microtubule imaging, whole seedlings were soaked in 0.5× MS medium containing 500 nM SNAP dye. For DRBG-488 staining, all experiments were performed in liquid culture. Seedlings were soaked in MGRL solution (modified MGRL lacking both Suc and gellan gum) containing 200 nM DRBG-488 under light (∼120 μE m−2 s−1). All reactions and analyses were performed at 23 to 25°C. For pulse-chase labeling by DRBG-488, plants were treated with 200 nM DRBG-488 for 30 min and then residual dye was washed out by adding fresh MGRL solution. Plants were subsequently incubated in MGRL in the light (∼120 μE m−2 s−1) for up to 210 min with recordings every 30 min.
Confocal Imaging
Fluorescence images were captured using a Leica Application Suite Advanced Fluorescence (LAS-AF) instrument with a TCS SP8 gated STED (gSTED) and a 20× objective (0.75 numerical aperture [NA] HC PL APO CS2; Leica), 63× oil-immersion objective (1.40 NA HC PL APO CS2; Leica), or a 100× oil-immersion objective (1.40 NA HC PL APO CS2; Leica). The light sources were a white-light laser, an argon laser, or a diode laser. The excitation wavelength was set according to the maximum wavelength of dye absorption: 458 nm for SNAP-Cell 430, 488 nm for DRBG-488, 552 nm for SNAP-Cell TMR-Star, 561 nm for mCherry, and 645 nm for SNAP-Cell SiR647. Detection with Leica HyD (GaAsP hybrid detection system) was set to a 50-nm range according to the maximum wavelength of dye fluorescence, 463 to 650 nm for SNAP-Cell 430, 500 to 550 nm for DRBG-488, 557 to 764 nm for SNAP-Cell TMR-Star, 570 to 650 nm for mCherry, and 648 to 749 nm for SNAP-Cell SiR647. Images were taken at 400 Hz, with a picture size of 1024 × 1024 pixels and 1.0 Airy unit. A time-gate pulse of 0.3 to 6.0 ns was used for detection of DRBG-488, SNAP-Cell TMR-Star, and SNAP-Cell SiR647 to reduce background signals. All dyes were prepared in DMSO (Wako Pure Chemicals) as a stock solution. The concentration of dyes was calculated and adjusted based on their molar absorption coefficient and the absorbance read with a spectrophotometer (UV-1800; Shimadzu).
For screening of synthetic dyes using BY-2 cells (Figure 1; Supplemental Figure 1), 100 μL of cells in LS media at pH 5.8 was loaded into eight-well chamber slides (Iwaki), and 1 min later 100 μL of 2 µM dye solution was added. After 1 min, images were taken using a LAS-AF instrument with a TCS SP8 gSTED and a 20× objective (0.75 NA HC PL APO CS2; Leica). The excitation wavelength was set according to the maximum wavelength of dye absorption. The emission wavelength, a 50-nm range of wavelength according to the maximum wavelength of the dye fluorescence, was detected with a Leica HyD (GaAsP hybrid detection system). To change the pH of the assay, 500 μL of BY-2 cells in LS media was left to settle for 5 min and then the supernatant was removed and replaced with 500 μL of buffer with a different pH. This procedure was repeated three times before cells were imaged as described above. For time-course analysis of staining BY-2 cells with 2MeRG and RG, 100 μL of cells in LS media at pH 5.8 was loaded into eight-well chamber slide (Iwaki), and fluorescent images were taken every 30 s using a LAS-AF instrument with a TCS SP8 gSTED and a 20× objective (0.75 NA HC PL APO CS2; Leica). Between 120 and 150 s, 100 μL of 2 µM dye solution was added into the well. The excitation wavelength was set to 498 and 499 nm for 2MeRG and RG, respectively, and fluorescence was detected with a Leica HyD (GaAsP hybrid detection system) set to a 50-nm range of 510 to 560 nm. For time-lapse imaging of SNAP dye–labeled microtubules in BY-2 cells and Arabidopsis seedlings, an Eclipse Ti2 inverted microscope (Nikon) equipped with a spinning disk confocal unit CSU-10 (Yokogawa) and a sCMOS camera PRIME 95B (Photometrics) was used. SNAP-Cell TMR-Star and mCherry were excited with a 561-nm laser, and emission signals were collected with a 605/64-nm filter (Semrock). GFP was excited with a 488-nm laser, and emission signals were collected with a 525/50-nm filter (Semrock). Images were taken with a 60× oil-immersion lens (CFI Plan Apo VC 1.40 NA; Nikon) or a 100× oil-immersion lens (CFI Plan Apo Lambda 1.45 NA; Nikon).
Quantitative Analysis of Cell Divisions
The duration of cell divisions was measured using Fiji and ImageJ software (version 2.0.0-rc-69/1.52p; Sterinebrunner et al., 2014). The box plots were generated and statistical analyses were performed with Prism8 (GraphPad Software).
Quantitative Analyses of PIN2 Dynamics
PIN2 dynamics were analyzed in epidermal cells of the meristematic and in transition zones in primary roots. Images were quantified using Fiji/ImageJ software. The number of endosomes was counted using the ImageJ macro SiCE spot detector with manual correction (Bayle et al., 2017). Manual correction involved ignoring puncta that overlapped with the plasma membrane and by manually assigning puncta that localized too close to the plasma membrane to be detected automatically. Finally, the number of puncta per cell was counted. For calculation of the C/P fluorescence ratio, the mean fluorescence value in the apical plasma membrane domain detected by a segmented line tool with 3-pixel thickness was defined as fluorescence in the plasma membrane, and the mean fluorescence value in the entire cytoplasm selected by a polygonal selection tool was assigned to the cytoplasm. Scatterplots were generated and statistical analyses were performed with Prism8.
Pharmacological Inhibition of Endocytosis
As described, all DRBG-488–labeling experiments were performed in liquid culture. ES9-17 (cat. no. BE17203; Carbosynth) was dissolved in DMSO at a concentration of 50 mM and stored in a glass vial at –30°C. Plants were incubated with 30 µM ES9-17 or an equivalent volume of DMSO (mock) diluted in MGRL solution for 30 min followed by incubation in MGRL solution containing 200 nM DRBG-488 and either 30 µM ES9-17 or DMSO for 60 or 120 min with supplemental light next to the microscope (∼120 μE m−2 s−1) until imaging was started (∼10 μE m−2 s−1 light level at the microscope stage without supplemental lighting). This light intensity was chosen since mCherry fluorescence in the vacuole is apparently pH dependent and thus lower intensity is observed at higher light intensities (Tamura et al., 2003).
Imaging of Cytokinesis in Root Epidermal Cells of Arabidopsis
To label most of the SNAP-PIN2-mCherry existing in the cells, plants were treated with 200 nM DRBG-488 for 90 min. Plants were then transferred into glass-bottomed chambers and overlaid with an MGRL soft-gel medium pad (MGRL medium containing 0.8% [w/v] gellan gum without Suc). Root epidermal cells were observed by inverted confocal microscopy. During the time-course experiment under the microscope, plants were exposed only to low light conditions, not taking the laser irradiation into account during imaging (∼1 μE m−2 s−1).
Accession Numbers
Sequence data from this article can be found in The Arabidopsis Information Resource under the following accession numbers: TUA5 (At5g19780); PIN2 (At5g57090); LTI6b (At3g05890); H2B (At3g45980); UBQ10 (At4g05320).
Supplemental Data
Supplemental Figure 1. Confocal images of BY-2 cells incubated with various dyes.
Supplemental Figure 2. Structures for dyes shown in Supplemental Figure 1.
Supplemental Figure 3. SNAP-tag enabled in vivo imaging of microtubules in Arabidopsis.
Supplemental Figure 4. DRBG-488 does not label cytosolic SNAP-TUA5.
Supplemental Figure 5. SNAP-tagging of the auxin transporter PIN2.
Supplemental Figure 6. SNAP-tag-dependent labeling of SNAP-PIN2 by DRBG-488.
Supplemental Figure 7. DRBG-488-labeling of SNAP-PIN2-mCherry in epidermis and cortex.
Supplemental Figure 8. Evidence for clathrin-mediated endocytosis of PIN2.
Supplemental Figure 9. Effect of ES9-17 on SNAP-PIN2-mCherry.
Supplemental Figure 10. Plasmids used in this study.
Supplemental Table 1. Primers used in this study.
Supplemental Table 2. Synthetic DNA sequence of SNAP.
Supplemental Table 3. Protein sequence of SNAP with secretion signal peptide sequence.
Supplemental Data Set. Statistical analysis data.
Supplemental Movie 1. Time-lapsed imaging of mitotic cell division in BY-2 expressing pUBQ10:SNAP-TUA5 stained with SNAP-Cell TMR-Star.
Supplemental Movie 2. Time-lapsed imaging of cortical microtubules in a pUBQ10:SNAP-TUA5 dark-grown hypocotyl epidermal cell stained with SNAP-Cell TMR-Star.
Supplemental Movie 3. Time series showing dynamic localization of SPR2-GFP and treated SNAP-TUA5 stained with SNAP-Cell TMR-Star in pavement cell.
Supplemental Movie 4. Time-lapsed imaging of Arabidopsis root expressing YFP-LTi6b, H2B-RFP, and SNAP-TUA5.
Acknowledgments
We thank Shunsuke Oishi (ITbM, Nagoya University) for HPLC assistance and Yoshikatsu Sato (ITbM Live Imaging Center, Nagoya University) for microscope use. We are grateful to Miyo Morita (National Institute of Basic Biology) for providing eir1-1 seeds; David W. Ehrhardt for YFP-LTI6b and H2B-RFP seeds; and Keitaro Umezawa (Tokyo Metropolitan Institute of Gerontology), Wen Piao, Kenjiro Hanaoka, Yasuteru Urano (University of Tokyo), and Chenguang Wang (ITbM, Nagoya University) for providing compounds. This research was supported by the Human Frontier Science Program (to M.N.), the Japan Society for the Promotion of Science (grant 18KK0195 to M.N. and A.Y.), and the Alexander von Humboldt Foundation (to W.B.F.). ITbM is supported by World Premier International Research Center Initiative (WPI), Japan.
AUTHOR CONTRIBUTIONS
R.J.I., M.N., and W.B.F. conceived the projects. R.J.I., A.Y., N.Y., and M.N. performed the experiments. R.J.I., H.O., M.G., M.K., T.K., and M.T. synthesized, prepared, and analyzed fluorescent dyes. M.N., W.B.F., R.J.I., A.K., and N.Y. analyzed the data and wrote the article. All authors discussed the results and commented on the article.
References
- Adamowski, M., Friml, J. (2015). PIN-dependent auxin transport: Action, regulation, and evolution. Plant Cell 27: 20–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamowski, M., Narasimhan, M., Kania, U., Glanc, M., De Jaeger, G., Friml, J. (2018). A functional study of AUXILIN-LIKE1 and 2, two putative clathrin uncoating factors in Arabidopsis. Plant Cell 30: 700–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asanuma, D., Takaoka, Y., Namiki, S., Takikawa, K., Kamiya, M., Nagano, T., Urano, Y., Hirose, K. (2014). Acidic-pH-activatable fluorescence probes for visualizing exocytosis dynamics. Angew. Chem. Int. Ed. Engl. 53: 6085–6089. [DOI] [PubMed] [Google Scholar]
- Bayle, V., Platre, M.P., Jaillais, Y. (2017). Automatic quantification of the number of Intracellular compartments in Arabidopsis thaliana root cells. Biol. Protoc. 7: e2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler, S.R., Ehrhardt, D.W., Griffitts, J.S., Somerville, C.R. (2000). Random GFP:cDNA fusions enable visualization of subcellular structures in cells of Arabidopsis at a high frequency. Proc. Natl. Acad. Sci. USA 97: 3718–3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejonghe, W., et al. (2019). Disruption of endocytosis through chemical inhibition of clathrin heavy chain function. Nat. Chem. Biol. 15: 641–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhonukshe, P., Aniento, F., Hwang, I., Robinson, D.G., Mravec, J., Stierhof, Y.-D., Friml, J. (2007). Clathrin-mediated constitutive endocytosis of PIN auxin efflux carriers in Arabidopsis. Curr. Biol. 17: 520–527. [DOI] [PubMed] [Google Scholar]
- Eisele, J.F., Fäßler, F., Bürgel, P.F., Chaban, C. (2016). A rapid and simple method for microscopy-based stomata analyses. PLoS One 11: e0164576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Federici, F., Dupuy, L., Laplaze, L., Heisler, M., Haseloff, J. (2012). Integrated genetic and computation methods for in planta cytometry. Nat. Methods 9: 483–485. [DOI] [PubMed] [Google Scholar]
- Gautier, A., Juillerat, A., Heinis, C., Corrêa, I.R., Jr., Kindermann, M., Beaufils, F., Johnsson, K. (2008). An engineered protein tag for multiprotein labeling in living cells. Chem. Biol. 15: 128–136. [DOI] [PubMed] [Google Scholar]
- Geiger, R., Luisoni, S., Johnsson, K., Greber, U.F., Helenius, A. (2013). Investigating endocytic pathways to the endoplasmic reticulum and to the cytosol using SNAP-trap. Traffic 14: 36–46. [DOI] [PubMed] [Google Scholar]
- Glanc, M., Fendrych, M., Friml, J. (2018). Mechanistic framework for cell-intrinsic re-establishment of PIN2 polarity after cell division. Nat. Plants 4: 1082–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grzybowski, M., Taki, M., Senda, K., Sato, Y., Ariyoshi, T., Okada, Y., Kawakami, R., Imamura, T., Yamaguchi, S. (2018). A highly photostable near-infrared labeling agent based on a phospha-rhodamine for long-term and deep imaging. Angew. Chem. Int. Ed. Engl. 57: 10137–10141. [DOI] [PubMed] [Google Scholar]
- Grzybowski, M., Taki, M., Yamaguchi, S. (2017). Selective conversion of P=O-bridged rhodamines into P=O-rhodols: Solvatochromic near-infrared fluorophores. Chemistry 23: 13028–13032. [DOI] [PubMed] [Google Scholar]
- Hanaoka, K., Kagami, Y., Piao, W., Myochin, T., Numasawa, K., Kuriki, Y., Ikeno, T., Ueno, T., Komatsu, T., Terai, T., Nagano, T., Urano, Y. (2018). Synthesis of unsymmetrical Si-rhodamine fluorophores and application to a far-red to near-infrared fluorescence probe for hypoxia. Chem. Commun. (Camb.) 54: 6939–6942. [DOI] [PubMed] [Google Scholar]
- Haseloff, J., Siemering, K.R., Prasher, D.C., Hodge, S. (1997). Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Proc. Natl. Acad. Sci. USA 94: 2122–2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwatate, R.J., Kamiya, M., Umezawa, K., Kashima, H., Nakadate, M., Kojima, R., Urano, Y. (2018). Silicon rhodamine-based near-infrared fluorescent probe for γ-glutamyltransferase. Bioconjug. Chem. 29: 241–244. [DOI] [PubMed] [Google Scholar]
- Iwatate, R.J., Kamiya, M., Urano, Y. (2016). Asymmetric rhodamine-based fluorescent probe for multicolour in vivo imaging. Chemistry 22: 1696–1703. [DOI] [PubMed] [Google Scholar]
- Jásik, J., Boggetti, B., Baluška, F., Volkmann, D., Gensch, T., Rutten, T., Altmann, T., Schmelzer, E. (2013). PIN2 turnover in Arabidopsis root epidermal cells explored by the photoconvertible protein Dendra2. PLoS One 8: e61403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, X., et al. (2019). Quantitative real-time imaging of glutathione with subcellular resolution. Antioxid. Redox Signal. 30: 1900–1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, K., Kim, D.W., Park, J.S., Khang, C.H. (2016). Live-cell fluorescence imaging to investigate the dynamics of plant cell death during infection by the rice blast fungus Magnaporthe oryzae. BMC Plant Biol. 16: 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiya, M., Asanuma, D., Kuranaga, E., Takeishi, A., Sakabe, M., Miura, M., Nagano, T., Urano, Y. (2011). β-Galactosidase fluorescence probe with improved cellular accumulation based on a spirocyclized rhodol scaffold. J. Am. Chem. Soc. 133: 12960–12963. [DOI] [PubMed] [Google Scholar]
- Karimi, M., De Meyer, B., Hilson, P. (2005). Modular cloning in plant cells. Trends Plant Sci. 10: 103–105. [DOI] [PubMed] [Google Scholar]
- Katsuta, J., Hashiguchi, Y., Shibaoka, H. (1990). The role of the cytoskeleton in positioning of the nucleus in premitotic tobacco BY-2 cells. J. Cell Sci. 95: 413–422. [Google Scholar]
- Keppler, A., Gendreizig, S., Gronemeyer, T., Pick, H., Vogel, H., Johnsson, K. (2003). A general method for the covalent labeling of fusion proteins with small molecules in vivo. Nat. Biotechnol. 21: 86–89. [DOI] [PubMed] [Google Scholar]
- Keppler, A., Pick, H., Arrivoli, C., Vogel, H., Johnsson, K. (2004). Labeling of fusion proteins with synthetic fluorophores in live cells. Proc. Natl. Acad. Sci. USA 101: 9955–9959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitakura, S., Vanneste, S., Robert, S., Löfke, C., Teichmann, T., Tanaka, H., Friml, J. (2011). Clathrin mediates endocytosis and polar distribution of PIN auxin transporters in Arabidopsis. Plant Cell 23: 1920–1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu, T., Johnsson, K., Okuno, H., Bito, H., Inoue, T., Nagano, T., Urano, Y. (2011). Real-time measurements of protein dynamics using fluorescence activation-coupled protein labeling method. J. Am. Chem. Soc. 133: 6745–6751. [DOI] [PubMed] [Google Scholar]
- Li, J., Li, G., Wang, H., Wang Deng, X. (2011). Phytochrome signaling mechanisms. Arabidopsis Book 9: e0148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindeboom, J.J., Lioutas, A., Deinum, E.E., Tindemans, S.H., Ehrhardt, D.W., Emons, A.M.C., Vos, J.W., Mulder, B.M. (2013). Cortical microtubule arrays are initiated from a nonrandom prepattern driven by atypical microtubule initiation. Plant Physiol. 161: 1189–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Los, G.V., et al. (2005). HaloTagTM interchangeable labeling technology for cell imaging, protein capture and immobilization. Promega Cell Notes 11: 2–6. [Google Scholar]
- Luschnig, C., Vert, G. (2014). The dynamics of plant plasma membrane proteins: PINs and beyond. Development 141: 2924–2938. [DOI] [PubMed] [Google Scholar]
- Miyashima, S., Koi, S., Hashimoto, T., Nakajima, K. (2011). Non-cell-autonomous microRNA165 acts in a dose-dependent manner to regulate multiple differentiation status in the Arabidopsis root. Development 138: 2303–2313. [DOI] [PubMed] [Google Scholar]
- Mravec, J., et al. (2011). Cell plate restricted association of DRP1A and PIN proteins is required for cell polarity establishment in Arabidopsis. Curr. Biol. 21: 1055–1060. [DOI] [PubMed] [Google Scholar]
- Naramoto, S., Kleine-Vehn, J., Robert, S., Fujimoto, M., Dainobu, T., Paciorek, T., Ueda, T., Nakano, A., Van Montagu, M.C.E., Fukuda, H., Friml, J. (2010). ADP-ribosylation factor machinery mediates endocytosis in plant cells. Proc. Natl. Acad. Sci. USA 107: 21890–21895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naramoto, S., Otegui, M.S., Kutsuna, N., de Rycke, R., Dainobu, T., Karampelias, M., Fujimoto, M., Feraru, E., Miki, D., Fukuda, H., Nakano, A., Friml, J. (2014). Insights into the localization and function of the membrane trafficking regulator GNOM ARF-GEF at the Golgi apparatus in Arabidopsis. Plant Cell 26: 3062–3076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piao, W., et al. (2013). Development of azo-based fluorescent probes to detect different levels of hypoxia. Angew. Chem. Int. Ed. Engl. 52: 13028–13032. [DOI] [PubMed] [Google Scholar]
- Plamont, M.-A., et al. (2016). Small fluorescence-activating and absorption-shifting tag for tunable protein imaging in vivo. Proc. Natl. Acad. Sci. USA 113: 497–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, D.G., Langhans, M., Saint-Jore-Dupas, C., Hawes, C. (2008). BFA effects are tissue and not just plant specific. Trends Plant Sci. 13: 405–408. [DOI] [PubMed] [Google Scholar]
- Roed, S.N., et al. (2014). Real-time trafficking and signaling of the glucagon-like peptide-1 receptor. Mol. Cell. Endocrinol. 382: 938–949. [DOI] [PubMed] [Google Scholar]
- Roman, G., Lubarsky, B., Kieber, J.J., Rothenberg, M., Ecker, J.R. (1995). Genetic analysis of ethylene signal transduction in Arabidopsis thaliana: Five novel mutant loci integrated into a stress response pathway. Genetics 139: 1393–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakabe, M., Asanuma, D., Kamiya, M., Iwatate, R.J., Hanaoka, K., Terai, T., Nagano, T., Urano, Y. (2013). Rational design of highly sensitive fluorescence probes for protease and glycosidase based on precisely controlled spirocyclization. J. Am. Chem. Soc. 135: 409–414. [DOI] [PubMed] [Google Scholar]
- Schiller, M., Massalski, C., Kurth, T., Steinebrunner, I. (2012). The Arabidopsis apyrase AtAPY1 is localized in the Golgi instead of the extracellular space. BMC Plant Biol. 12: 123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinebrunner, I., Gey, U., Andres, M., Garcia, L., Gonzalez, D.H. (2014). Divergent functions of the Arabidopsis mitochondrial SCO proteins: HCC1 is essential for COX activity while HCC2 is involved in the UV-B stress response. Front. Plant Sci. 5: 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano, J., Miwa, K., Yuan, L., von Wirén, N., Fujiwara, T. (2005). Endocytosis and degradation of BOR1, a boron transporter of Arabidopsis thaliana, regulated by boron availability. Proc. Natl. Acad. Sci. USA 102: 12276–12281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura, K., Shimada, T., Ono, E., Tanaka, Y., Nagatani, A., Higashi, S.I., Watanabe, M., Nishimura, M., Hara-Nishimura, I. (2003). Why green fluorescent fusion proteins have not been observed in the vacuoles of higher plants. Plant J. 35: 545–555. [DOI] [PubMed] [Google Scholar]
- Umezawa, K., Yoshida, M., Kamiya, M., Yamasoba, T., Urano, Y. (2017). Rational design of reversible fluorescent probes for live-cell imaging and quantification of fast glutathione dynamics. Nat. Chem. 9: 279–286. [DOI] [PubMed] [Google Scholar]
- Vannini, G.L., Pancaldi, S., Poli, F., Fasulo, M.P. (1988). Rhodamine 123 as a vital stain for mitochondria of plant cells. Plant Cell Environ. 11: 123–127. [Google Scholar]
- Wang, C., Taki, M., Sato, Y., Fukazawa, A., Higashiyama, T., Yamaguchi, S. (2017). Super-photostable phosphole-based dye for multiple-acquisition stimulated emission depletion imaging. J. Am. Chem. Soc. 139: 10374–10381. [DOI] [PubMed] [Google Scholar]
- Wang, C., Taki, M., Sato, Y., Tamura, Y., Yaginuma, H., Okada, Y., Yamaguchi, S. (2019). A photostable fluorescent marker for the superresolution live imaging of the dynamic structure of the mitochondrial cristae. Proc. Natl. Acad. Sci. USA 116: 15817–15822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissleder, R., Ntziachristos, V. (2003). Shedding light onto live molecular targets. Nat. Med. 9: 123–128. [DOI] [PubMed] [Google Scholar]
- Woodroofe, C.C., Lim, M.H., Bu, W., Luppard, S.J. (2005). Synthesis of isomerically pure carboxylate- and sulfonate-substituted xanthene fluorophores. Tetrahedron 12: 3097–3105. [Google Scholar]
- Yang, G., de Castro Reis, F., Sundukova, M., Pimpinella, S., Asaro, A., Castaldi, L., Batti, L., Bilbao, D., Reymond, L., Johnsson, K., Heppenstall, P.A. (2015). Genetic targeting of chemical indicators in vivo. Nat. Methods 12: 137–139. [DOI] [PubMed] [Google Scholar]
- Yoshinari, A., Takano, J. (2017). Insights into the mechanisms underlying boron homeostasis in plants. Front. Plant Sci. 8: 1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, S., Liu, C., Wang, J., Ren, Z., Staiger, C.J., Ren, H. (2016). A processive Arabidopsis formin modulates actin filament dynamics in association with profilin. Mol. Plant 9: 900–910. [DOI] [PubMed] [Google Scholar]