Abstract
Tissue engineering aims to capture the structural and functional aspects of diverse tissue types in vitro. However, most approaches are limited in their ability to produce complex 3D geometries that are essential for tissue function. Tissues, such as the vasculature or chambers of the heart, often possess curved surfaces and hollow lumens that are difficult to recapitulate given their anisotropic architecture. Cell-sheet engineering techniques using thermoresponsive substrates provide a means to stack individual layers of cells with spatial control to create dense, scaffold-free tissues. In this study, we developed a novel method to fabricate complex 3D structures by layering multiple sheets of aligned cells onto flexible scaffolds and casting them into hollow tubular geometries using custom molds and gelatin hydrogels. To enable the fabrication of 3D tissues, we adapted our previously developed thermoresponsive nanopatterned cell-sheet technology by applying it to flexible substrates that could be folded as a form of tissue origami. We demonstrated the versatile nature of this platform by casting aligned sheets of smooth and cardiac muscle cells circumferentially around the surfaces of gelatin hydrogel tubes with hollow lumens. Additionally, we patterned skeletal muscle in the same fashion to recapitulate the 3D curvature that is observed in the muscles of the trunk. The circumferential cell patterning in each case was maintained after one week in culture and even encouraged organized skeletal myotube formation. Additionally, with the application of electrical field stimulation, skeletal myotubes began to assemble functional sarcomeres that could contract. Cardiac tubes could spontaneously contract and be paced for up to one month. Our flexible cell-sheet engineering approach provides an adaptable method to recapitulate more complex 3D geometries with tissue specific customization through the addition of different cell types, mold shapes, and hydrogels. By enabling the fabrication of scaled biomimetic models of human tissues, this approach could potentially be used to investigate tissue structure-function relationships, development, and maturation in the dish.
Keywords: Cell-sheet engineering, 3D tissue engineering, thermoresponsive polymer, nanofabrication
Introduction
Tissues throughout the body possess complex three-dimensional (3D) structures with many degrees of organization and function. For example, the vasculature, like many other tissues, is organized by stratification of several layers of different cell types that perform complementary functions to modulate blood pressure and tissue perfusion [1,2]. The endothelial cells in the lining of the blood vessel’s lumen are oriented parallel to the direction of blood flow, whereas the surrounding smooth muscle cells that encircle the endothelium are aligned perpendicularly. Similar patterns of differential organization are observed in the helical fiber organization of the myocardium in the heart and in the radial fan patterns seen in the trapezius and pectoral muscles of the trunk. The function of each of these tissues is highly dependent upon their structure and 3D geometry, and when their organization is compromised by disease it can be detrimental or potentially fatal [3-5].
To study tissue function and their associated diseases, advancements have been made in tissue engineering to recapitulate tissue micro- and macroenvironments in vitro. For example, cell-dense cardiac tissue patches made from induced pluripotent stem cell-derived cardiomyocytes (iPSC-CMs) can mimic action potential conduction velocities close to those of adult cardiac tissues [6-8]; vascular grafts have been made from cell-deposited matrix and then decellularized before implantation [9-11]; and bioprinting with cellularized-inks (or bioinks) has enabled fabrication of intricate 3D tissue-specific structures with corresponding function [12-15]. A challenge facing each of these approaches is that tissues often have complex 3D geometries, including curved surfaces and hollow lumens. Such structures have been difficult to recreate in vitro due to limitations of available fabrication techniques. Specifically, there are few fabrication approaches that allow for production of curved 3D geometries while also having control over spatial organization at the cell-layer level. The ability to recapitulate these structures would impart function that better mimics native tissues and organs.
To address this need, our group has previously established a nanofabrication technique to pattern sheets of organized cells and stack them to create multi-layered tissue patches using a novel gel-casting technique in conjunction with thermoresponsive substrates [16,17]. In the present study, we sought to improve upon this technology by introducing flexible substrates and custom molds to enable the fabrication of organized 3D tissue structures. We found that all three cell types could be patterned to form an intact monolayer with a uniform orientation in the direction of the nanotopography. Each monolayer was lifted from the surface through temperature-mediated release provided by the thermoresponsive poly(N-isopropylacrylamide) (pNIPAM) functional layer. Multiple organized monolayers were stacked onto a single flexible film and were folded into a cylindrical shape, as a form of tissue origami, where the organized cell layers could be casted into free-standing a 3D tubular tissue. We demonstrated the diverse application of this technology by fabricating tubular tissues with curved surfaces from three muscle types: smooth, skeletal, and cardiac muscle. This approach enabled patterning of all three cell types in 3D multilayered tissues with circumferential alignment that was maintained for two weeks in culture. Additionally, with application of electrical field stimulation, skeletal myotubes assembled functional sarcomeres that could contract, and cardiac tubes could be paced for over one month. This flexible patterned film technology can readily be adapted to fabricate tissues with other complex geometries by changing the shape of the flexible film and custom mold, producing better tissues for the study of development and disease.
Materials and methods
1.1. Fabrication of flexible thermoresponsive nanofabricated substrates (fTNFS)
To fabricate flexible films with nanotopographical cues and thermoresponsive properties, capillary force lithography was utilized as described in our previously established protocol [16-18]. Briefly, nanopatterned films were fabricated using 100 μL of a polymer curable by ultraviolet light (UV), polyurethane acrylate (PUA, Norland Optical Adhesive #76) mixed with either 1% or 20% (w/w) glycidyl methacrylate (GMA). The UV-curable polymer was sandwiched and spread between a 23 μm-thick flexible poly-ethylene terephthalate (PET) film and a PUA master mold with parallel ridges and grooves that were 800 nm in width and 600 nm in depth (Figure 1A). The mold and film construct were exposed to high intensity 365 nm wavelength UV light for 1 minute to polymerize the PUA-GMA solution. After initial polymerization of the sandwiched polymer layer, the flexible film and adhered nanopatterned polymer layer were carefully removed from the master mold using forceps (Figure 1B). The newly constructed nanopatterned film was placed under low intensity 365 nm UV light for 24 hours to ensure complete polymerization of the PUA-PGMA polymer. To provide thermoresponsive functionality, nanopatterned substrates were then dip-coated with an amine-terminated poly(N-isopropylacrylamide) (pNIPAM) solution (13 μM in H2O, Mn = 2500 Sigma-Aldrich) for 24 hours on a tabletop rocker (55 rpm, room temperature). After 24 hours, excess pNIPAM was removed from the flexible thermoresponsive nanofabricated substrates (fTNFS) through three 5-minute washes with deionized water and allowed to dry overnight. The films were cut into rectangular sheets (1.25 cm x 1.5 cm) using a die cutter. The fTNFS were dipped in 70% ethanol and exposed to 294 nm UV light for one hour in a biosafety cabinet for sterilization prior to use.
Figure 1. Fabrication of anisotropic multilayered tissues using flexible thermoresponsive nanofabricated stubstrates (fTNFS) and nanopatterned cell sheet engineering.
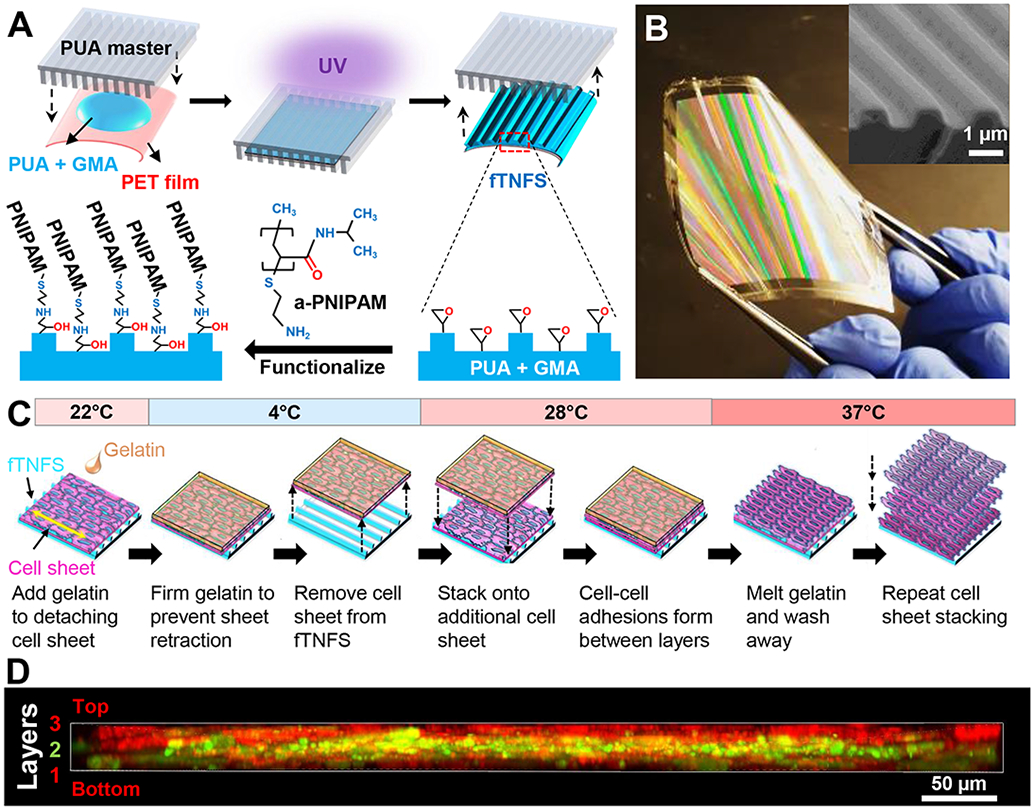
A. Fabrication of flexible nanopatterned substrates (fTNFS) using capillary force lithography and subsequent thermoresponsive functionalization with amine-terminated PNIPAM (a-PNIPAM).
B. Image of flexible-TNFS after curing and a-PNIPAM functionalization. Rainbow coloring is caused by the nanotopography deflecting light. (Inset) Scanning electron micrograph of fTNFS surface demonstrating high fidelity fabrication of the ridge-groove nanotopography.
C. Schematic of gel casting and stacking of organized cell monolayers from flexible TNFS.
D. Z-stack cross-sectional image of smooth muscle cell tri-layer tissue stack 24 hours after stacking. Top and bottom sheets were membrane-dyed red (Cell Tracker Red) and middle sheet was membrane-dyed green (Cell Tracker Green) before stacking.
In order to restrict cell-seeding to the fTNFS surface and minimize cell waste, two fTNFS were temporarily affixed into the bottoms of custom polydimethylsiloxane (PDMS, Sylgard 181) rectangular wells (13.5 mm x 30 mm) using porcine gelatin (7.5% w/v, Sigma) crosslinked with transglutaminase (MooGloo™ TI-TG, Modernist Pantry) as an adhesive. Flexible TNFS were incubated with fetal bovine serum (FBS, Sigma) overnight at 4°C before cell seeding to deposit a thin protein layer to promote cell adhesion to the surface.
1.2. Scanning electron microscopy of fTNFS
Poly-NIPAM-functionalized fTNFS were sputter-coated with Au/Pd alloy prior to imaging using scanning electron microscopy (Sirion XL30, FEI, OR, USA). Images were taken with an acceleration voltage of 5 kV and spot size of 2.
1.3. Cell culture and cell-seeding of fTNFS
Mouse aortic smooth muscle cells (SMCs) were generously provided by Dr. Marta Scatena’s group [19]. Mouse SMCs were cultured on tissue-culture treated plastic dishes with Dulbecco’s Modified Eagle’s Medium (DMEM, Gibco) supplemented with 1% penicillin-streptomycin (p/s, Sigma), 10% FBS. Cells were passaged at 80% confluency during expansion and only passages 30 and below were used to minimize confounding effects of cell senescence on tissue fabrication. Cells were split and seeded onto fTNFS at a density of 175,000 cells/cm2 in 1 mL of medium and allowed to adhere overnight at 37°C and 5% CO2. Seeded cells were cultured for 5-7 days before cell sheet stacking and tissue fabrication to allow a highly confluent monolayer of cells to form. Observation of cell growth was conducted using a bright-field microscope (Nikon TS100).
C2C12 mouse muscle myoblasts (C2C12s; ATCC) were cultured under the same conditions as the SMCs as described above and seeded at 175,000 cells/cm2 onto fTNFS. However, seeded cells were cultured for 2-3 days before cell sheet stacking and tissue fabrication as C2C12 cells were found to proliferate at a faster rate than SMCs. Three to four days after tissue fabrication, tissue constructs were cultured in a low-serum containing medium (DMEM, 2% horse serum (HS), 1% p/s) to promote fusion and differentiation of myoblasts into myotubes. To promote further functional and structural maturation of myotubes, chronic broad-field electrical stimulation was applied (1 Hz, 3V, 24 ms; IonOptix C-Pace) after 3-4 days of culturing tissues with low-serum differentiating medium and once myotube formation was observed over the entire tissue surface area.
Cardiomyocytes (CMs) and endocardial-like endothelial cells (ECs) were differentiated from human induced pluripotent stem cells (hiPSCs, UC 3-4) derived from patient urine samples [20]. Endothelial cells were included to promote cell sheet formation as monolayers of pure cardiomyocytes were found to clump during tissue fabrication (Supplemental Figure 1). Established monolayer-based differentiation protocols were used that modulate Wnt-signaling pathways with small molecules to specify cardiac mesoderm lineages [21,22]. In brief, UC 3-4 hiPSC colonies were maintained on Matrigel (1:60, Corning) coated tissue-culture plates in mTeSR medium until 80% confluency. Colonies were then replated into a monolayer at 250,000 cells/cm2 (high density) or 100,000 cells/cm2 (low density) for directed differentiation of cardiomyocytes (CMs) or endothelial cells (ECs), respectively [22,23]. To drive differentiation toward the cardiomyocyte lineage, high density monolayers were exposed to CHIR-99021 (Fischer Technologies) in Roswell Park Memorial Institute 1640 (RPMI) medium with B27 without insulin (Gibco) on day 0 (induction) to activate the Wnt signaling pathway and specify mesoderm gene expression. High-density monolayers were exposed to the Wnt-inhibitor IWP4 (Stemgent) on day 3 to further specify cardiac mesoderm and were then cultured in RPMI-B27 medium with insulin from day 7 and onward. Beating monolayers of cardiomyocytes were observed as early as day 9. Cardiac-differentiated populations were then subjected to a lactate-rich glucose-poor selection medium at day 14 for 3 days to enrich the cardiomyocyte population [24]. Cells were harvested on day 17 and fixed in 4% paraformaldehyde as a single-cell solution and prepared for flow cytometry to determine cardiomyocyte purity. Cells were stained with a mouse-anti-cardiac troponin T (cTnT) antibody (1:100, Thermo-Scientific) and counterstained with a goat-anti-mouse Alexa Fluor 488-counjugated antibody (1:200, Invitrogen). Cell populations used for this study were at least 95% cTnT-positive when analyzed by flow cytometry (Supplemental Figure 2).
Similarly, to drive differentiation towards the cardiac endothelial lineage, low density monolayers were exposed to activin-A (R&D Systems) and Matrigel in RPMI with B27 on day 0 (induction). On day 1 post induction, low-density monolayers were exposed to bone morphogenic protein-4 (BMP-4; R&D Systems) and CHIR-99021 in RPMI-B27 medium. To specify the endothelial lineage, low-density monolayers were switched into StemPro-34 medium, containing vascular endothelial growth factor (VEGF; PeproTech), BMP-4, basic fibroblast growth factor (bFGF; R&D Systems), ascorbic acid, and monothioglycerol. On day 5, the low-density monolayers were re-plated at 13k/cm2 into gelatin-coated tissue culture plates and expanded in Endothelial Growth Medium-2 (EGM-2, Lonza) supplemented with VEGF, bFGF, and CHIR-99021 until day 11. Live cells were stained with a mouse-anti-CD31 antibody pre-conjugated with an Alexa Fluor 488 fluorophore (R&D Systems) for 1 hour on ice and flow cytometry was performed immediately. All EC populations used in this study were at least 90% CD31-positive when analyzed by live-cell flow cytometry on day 11 (Supplemental Figure 3). ECs were then seeded with cardiomyocytes immediately or cryopreserved for later use.
To limit possible confounding factors associated with age variation in the CM or EC population, CMs were used within 17-30 days post-induction and ECs used between 12-15 days post-induction for tissue fabrication. Purified CMs and ECs were seeded onto fTNFS such that the final proportion of CMs and ECs was 88% and 12% (~7:1 CMs:ECs) of the total cell number, respectively. This CM:EC ratio was optimized during preliminary experiments to yield highly aligned and confluent cell sheets that could withstand our previously published cell-sheet detachment and stacking process [16,17]. Cardiac cell sheets were cultured for 10-14 days in cardiac growth medium (75% RPMI-B27 + insulin, 25% EGM-2, 10% FBS, 1% p/s) before cell sheet stacking and tissue fabrication to allow for the formation of highly dense and confluent monolayers.
1.4. Cell sheet stacking
Cell sheets that form dense monolayers through cell-cell connection and ECM deposition exhibit a tendency to clump or fold inward on themselves once detached from cell culture surfaces. To prevent this, we previously developed a gel casting method for stacking aligned cell sheets that conferred layer-by-layer control over tissue architecture (Figure 1C) [16,17]. Briefly, patterned cell sheets were detached from the fTNFS by lowering the culture temperature to 22°C (room temperature), below the lower critical solution temperature point of pNIPAM (32°C), where the hydrophobicity of pNIPAM abruptly and dramatically switches to a hydrophilic hydrogel. This hydrogel polymer then swells and dislodges intact cell sheets [25,26]. After incubation for 30 minutes (for C2C12 and cardiac sheets) or 1 hour (SMC sheets) at room temperature, and just before complete detachment, cell sheets were cast in a 7.5% w/v gelatin solution at 4°C for 30 minutes to maintain the anisotropic organization of the cell sheet and prevent sheet retraction. The gel-casted cell sheets were moved into a 28°C incubator for one hour to further promote cell detachment from the fTNFS without melting the gelatin that maintains the cellular alignment. The gel-casted sheets were then incubated at 4°C for 15 minutes to allow the gelatin to further solidify for better handleability. The gel-casted cell sheet was then removed from the fTNFS with forceps and stacked on top of another cell sheet with parallel cellular orientation to produce an aligned, bi-layered laminar tissue. Once stacking was complete, the gelatin was completely dissolved 37°C and the construct washed with warm (37°C) medium to ensure the remaining tissue structure constitutes a scaffold-free, bi-layered cell sheet construct on top of an fTNFS. This process was then repeated to add a third and final cell layer with parallel orientation. To visualize maintenance of three discrete cell layers, each cell sheet was labeled prior to stacking by incubating with either a red or green cell dye (2 μM CellTracker CMFDA Green or 2 μM CellTracker Red CMTPX, Invitrogen) for 30 minutes. Z-stacks were then taken with a confocal microscope (Nikon A1R, 10x objective) to visualize the alternately layered red, green, then red cell sheets (Figure 1D).
1.5. Flexible TNFS manipulation and 3D tissue casting
After stacking three-layered tissues as described above (Figure 1C), tri-layered tissue constructs were then cast into a 3D tubular geometry using polystyrene cylindrical molds and custom 3D-printed casting implements (Figure 2 & 3A-D). The tri-layered tissue and fTNFS were first incubated in room temperature phosphate buffered saline (PBS, Gibco) for 30 minutes to promote the basement layer to detach from the fTNFS. Simultaneously, the polystyrene center mandrel (Figure 3A.i.), cylindrical mold (Figure 3A.ii.), and 3D-printed end cap (Figure 3A.iii.) were coated with a 1% bovine serum albumin in PBS (BSA, Sigma) solution to prevent the final tissue construct from sticking upon removal from the mold. The fTNFS with cell sheets were manipulated with forceps into a cylindrical shape with the cell layers facing inward and inserted into the cylindrical mold (Figure 2 & 3A.ii.). The end cap (Figure 3A.iii.) was then placed on the end of the cylindrical mold and the center mandrel (Figure 3A.i.) was inserted into the assembly through the hole in the end cap. The final casting assembly (Figure 3B) ensured that the lumen created in the tissue construct was straight and the resulting tissue walls were of uniform thickness on all sides. The remaining negative space within the casting tube was then filled with 200 μL of warmed gelatin and transglutaminase crosslinker (10% TG in PBS, MooGloo™ TI-TG; 10% porcine gelatin w/v in DMEM, Sigma) and allowed to crosslink at 28°C for 1 hour; the final concentration of crosslinked gelatin was 5%. After the gelatin was crosslinked, the molded tissue constructs were incubated at 4°C for 30 minutes to allow the basement layer of cells connected to the fTNFS to detach. The fTNFS and tubular tissue with cells was then carefully removed from the casting assembly with forceps and the fTNFS was unwrapped from the tubular tissue. The final tubular tissue was attached to a custom 3D printed tissue housing (Figure 3C) to prevent tissue damage and placed into a culture well with medium.
Figure 2. Schematic of tubular tissue casting process using multilayered cell-sheet stacks with fTNFS and cylindrical molds.
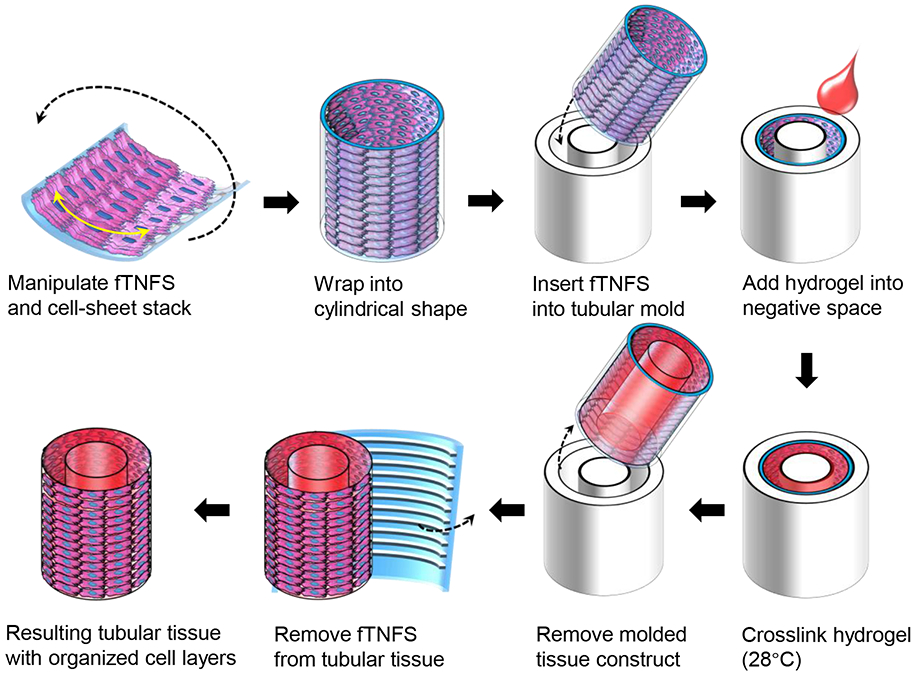
Figure 3. Fabrication of 3D tubular tissues with circumferential cellular alignment.
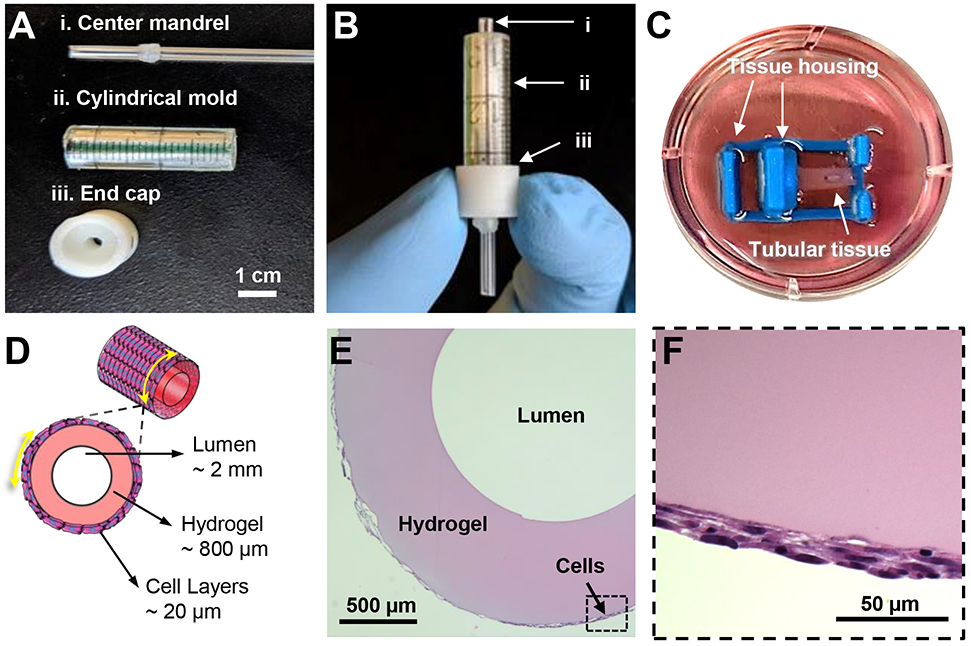
A. Image of tissue casting implements. (i) mandrel that is inserted through the 3D-printed end cap (iii) into the cylindrical mold (ii) to create a hollow lumen through the center of the resulting tubular tissue.
B. Casting mold pieces in B assembled as during tissue casting. The fTNFS and cell layers are manipulated into the cylindrical mold (ii), the cap end (iii) is fastened over one end of the cylindrical mold (ii), and the mandrel (i) is inserted through the bottom of the end cap (iii), and the hydrogel is pipetted into the open end of the mold (ii) to cast the cell sheets around a hydrogel tube.
C. Image of resulting tubular tissue attached to a custom 3D-printed housing in a culture well after removal from the casting mold.
D. Cross-sectional schematic of expected tissue dimensions and structure. Thickness of cell layers is dependent on cell type and number of layers.
E. Histological cross-section of SMC tube stained with hematoxylin and eosin showing a hollow central lumen encircled by a gelatin hydrogel layer and cell layers.
F. Boxed inset of E demonstrating three layers of SMCs with elongated nuclei along the curvature of the tube’s outer edge.
1.6. Tissue histology and dimensional measurements
To visualize cellular orientation and quantify tissue dimensions after 7 days in culture, SMC tubular tissues were fixed in 4% paraformaldehyde (PFA in PBS) for 30 minutes at room temperature and washed with PBS. Tissues were then dehydrated by serial washes in ethanol (50%, 60%, and 70%) for 20 minutes before embedding in paraffin blocks for sectioning. Tubular tissues were cut using a cryostat (Leica CM 1950) to make 4 μm cross-sections along the short access of the tube allowing for visualization of the center lumen, tissue wall thickness, and cell layer thickness (Figure 3E-F). Sections were stained with hematoxylin and eosin to visualize extracellular matrix and cytoplasm (pink) and DNA (blue). ImageJ (National Institutes of Health) measurement tools were used to measure tissue wall thickness, cell layer thickness, lumen diameter, and outer diameter across 30 sections. Measurements of each dimension were averaged and standard deviation was calculated (GraphPad, Prism).
1.7. Immunostaining and confocal imaging
Tubular tissues were cultured for 7 days after casting and fixed in 4% paraformaldehyde (PFA in PBS) for 30 minutes at room temperature and washed with PBS. To visualize intercellular proteins, tissues were permeabilized in 0.2% Triton-X 100, 0.5% BSA, and 5% goat serum in PBS at room temperature for 1 hour and transferred into a blocking solution of 5% goat serum with 0.5% BSA in PBS for 2 hours to prevent non-specific antibody binding. Primary antibodies (mouse-anti-smooth muscle α-actin (1:200, SMα-actin, Abcam), mouse-anti-myosin heavy chain (1:50, MYH, A4.1025 Developmental Studies Hybridoma Bank, The University of Iowa, Department of Biology; deposited by the Baxter Lab for Stem Cell Biology at Stanford University), rabbit-anti-titin (1:300, Myomedix)) were diluted in a staining solution of 1.5% goat serum in PBS and incubated with the respective tissues overnight at 4°C. Excess primary antibodies were washed away through three 5-minute washes with PBS before corresponding secondary antibodies (1:400, Alexa 488, 594, or 647, Invitrogen) and conjugated phalloidin (1:200, F-actin, 488 or 647, Invitrogen) were applied in 0.5% BSA in PBS for 2 hours at room temperature. Excess secondary antibodies were washed away through three 5-minute washes with PBS before a nuclear counterstain (DAPI, Invitrogen) was applied.
Given the relatively large 3D geometry of the tubular tissues, custom mounting chambers were developed by placing a square 3 mm-thick PDMS frame around the tissue and sandwiching them between two rectangular cover-glasses (0.17 mm thickness, Fisher Scientific). The tissues were stored in anti-fade mounting medium (VECTASHIELD, Vector Laboratories) within the PDMS mounting chambers. The rounded surfaces of the tubular tissues were slightly flattened to visualize their cellular layers with a confocal microscope, but the overall curvature of the tissue was maintained. Confocal z-stacks were taken of tubular tissues using either a Nikon A1R and or a Yokogawa W1 spinning-disk confocal microscope, and 10x-dry, 20x-dry, or 40x oil-immersion objectives.
1.8. Cellular orientation analysis
To quantify cellular orientation in 3D tubular tissues, confocal images of cytoskeletal filamentous actin (F-actin) for SMC, C2C12, and cardiac tubes were taken of three different areas using a 40x oil-immersion objective. These images were analyzed using a modified MATLAB script (MathWorks) that utilizes pixel gradient analysis to determine the distribution of orientation angles within an image [16,27]. Briefly, a Gaussian low pass filter and Sobel horizontal edge-emphasize filter are applied (as predefined by the MATLAB Image Analysis Toolbox) to create a 2D convolution. The Sobel filter is then transposed to extract horizontal and vertical edges and then used to calculate the gradient magnitude of each pixel within the image. The images were then thresholded to define the edges of single cells and calculate their orientation angles relative to the x-axis at 0°. These orientation angles were then binned and plotted as histograms to represent the overall cell alignment of the 3D tissue (Figure 5G-I).
Figure 5. Patterned cellular orientation in tubular tissues is maintained after 7 days in culture.
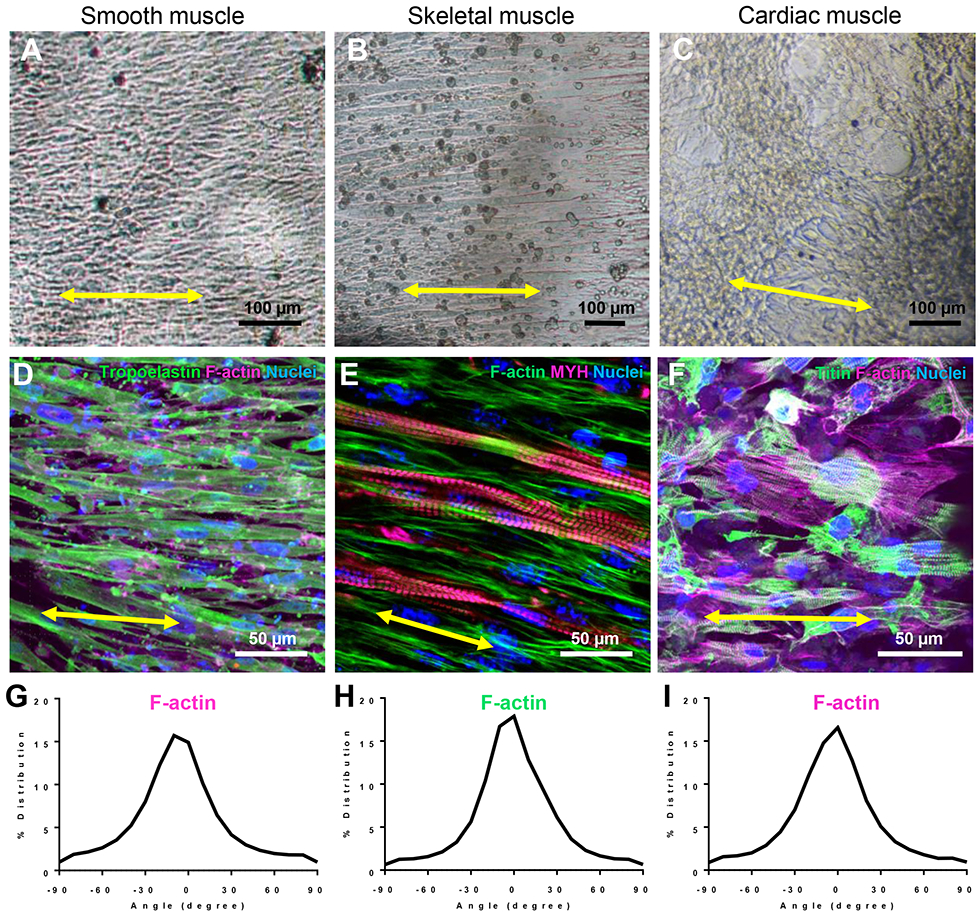
A-C. Brightfield images of SMC (A), skeletal (B), and cardiac (C) tubes after 7,14, or 7 days in culture, respectively.
D-F. Confocal images of each tubular tissues imaged in A-C, respectively. Each tissue was immunostained for cytoskeletal and or contractile proteins as listed and color-coded in the upper right corner of each panel.
G-I. Quantitative analysis of filamentous-actin (F-actin) cytoskeletal alignment of cells in each tissue type.
Results and Discussion
1.9. Flexible TNFS fabrication and cell sheet stacking
To develop a tissue engineering platform that would enable fabrication of 3D tissue geometries with control over local and global cellular patterning, we sought to adapt our established capillary force lithography techniques [16-18,28] and amine-terminated poly(N-isopropylacrylamide) (pNIPAM)-mediated surface chemistry to produce flexible thermoresponsive nanofabricated substrates (fTNFS). Thermoresponsive functionalization was included to mediate the release of organized cell sheets from the nanopatterned surfaces without the use of digestive enzymes, such as trypsins, which are required to detach cells from traditional culture surfaces. Flexible films were chosen in this study to enable the fabrication of 3D tissues with curved surfaces by their capacity to be folded into a cylindrical shape. Large area flexible films (5 cm x 5 cm) were patterned using a stiff polyurethane (PUA, 19.8 MPa) master mold with 800 nm ridges and groves and 600 nm depth (Figure 1A,B). When examined by scanning electron microscopy, fTNFS were found to have high pattern fidelity even after functionalization with a pNIPAM layer and bending with forceps (Figure 1B, inset).
The speed and ease of cell-sheet detachment were optimized for each cell type by modulating the percentage of bound pNIPAM groups through increasing or decreasing the amount of glycidyl methacrylate (GMA) that is incorporated into the PUA layer during fTNFS fabrication[16,17]. We found that smooth muscle cell (SMC) sheets required more pNIPAM-mediated release from the fTNFS using our gel-casting method (Figure 1C) and 20% GMA was therefore blended into the PUA layer of the scaffolds. In contrast, skeletal myoblast (C2C12s) and cardiac sheets exhibited a tendency to spontaneously detach from fTNFS with higher GMA concentrations, and required much lower (1% GMA) levels of pNIPAM-mediated release. These differences in detachment may be due to the spontaneously contractile behavior of cardiomyocytes and the migration and fusion of muscle myoblasts during differentiation into myotubes. After optimization of GMA content, all cell sheet types could be detached and stacked to form multilayered tissues with maintenance of three discrete cell layers (Figure 1D).
1.10. Fabrication of 3D smooth muscle tissues
To demonstrate how the flexible nature of fTNFS allows for fabrication of 3D tissues with control over global cellular orientation, we first aimed to model a simplified tubular structure to mimic the geometry of vascular structures. In blood vessels, vascular tone and blood flow are regulated by SMC contraction and relaxation. Smooth muscle cells make up the medial layer of blood vessels, the tunica media, and are organized in a circumferential pattern [1,29]. To recapitulate architecture of the tunica media, we patterned the fTNFS such that the nanogrooves and ridges were parallel to the long axis of the rectangular scaffold. To form a cylinder with circumferentially layered SMC-sheets, the fTNFS were then rolled along the short axis with the cell layers on the inside of the lumen (Figure 2). This cylinder was then inserted into a cylindrical mold with a capped end and center mandrel (Figure 3A,B). The void space between the mandrel and the SMC-sheet cylinder was filled with a crosslinking gelatin hydrogel to provide a structured tubular shape of the final tissue. Finally, the SMC-sheet cylinder and crosslinked hydrogel were removed from the mold followed by the unwrapping of the fTNFS. The resulting tubular tissue possessed a hollow lumen (diameter = 2.0 +/− 0.9 mm) surrounded by hydrogel walls (thickness ~ 800 μm) (Figure 3C,D). The cell layers were wrapped around the outer edge of the hydrogel tube (cell layer thickness = 17 +/− 3 μm) and were not encased by the hydrogel during the casting process (Figure 3E). The tissue was then gently manipulated with forceps onto a custom tissue housing for culture and visualization with an inverted microscope (Figure 3C). After 7 days in culture, SMC tubes were cross-sectioned and histologically stained. Three distinct cell layers were maintained around the outer edge of the hydrogel walls with the center lumen still intact (Figure 3F). Furthermore, the cell bodies and their nuclei had maintained circumferential alignment and elongation along the hydrogel’s edge after several days in culture.
1.11. Fabrication of 3D cardiac and skeletal muscle tissues
The muscle structures throughout the body have multiple stratified layers of organized cells and varying curved 3D geometries. For example, limb muscles have a spindled shape with tapered ends, while trunk muscles, such as the transvers abdominis and oblique muscles, are curved around the side of the body. Furthermore, cardiovascular and digestive organs possess hollow lumens with layers of organized muscle, such as the stomach and intestines and the chambers of the heart. There have been several approaches to modeling these tissue organizations in vitro, such as seeding engineered scaffolds[30-32], 3D bioprinting[13,33], cell sheet layering[26,34-37], and tissue casting[38-41]. However, few of these approaches can recapitulate the anisotropic layering of organized cell-sheets that ultimately gives rise to tissue functionality. To address this limitation, we sought to apply our fTNFS technology and tissue casting process to fabricating organized multilayered skeletal and cardiac muscle tubes with curved surfaces.
The fTNFS were seeded with either mouse skeletal muscle myoblasts (C2C12 cells) or hiPSC-derived cardiomyocytes and endothelial cells to form organized monolayers. Endothelial cells were included in cardiac monolayers as a stromal cell component which improves the integrity of formed and detached cell sheets through a combination of increased intercellular coupling and additional ECM deposition. In contrast, monolayers of cardiomyocytes alone did not maintain a contiguous cell-sheet during detachment, but rather individual cells pulled away from one another, resulting in the detachment of small clusters (Supplemental Figure 1). Skeletal and cardiac monolayers were detached and stacked to create multilayered constructs using our gel-casting process and subsequently cast into tubular geometries as described above. Upon removal from the casting mold and unwrapping of the fTNFS, skeletal and cardiac tubes were found to have global cell coverage on the curved outer edges of the tissues and possessed hollow lumens, similar to the SMC tubes (Figure 4). Cardiac tubes began coordinated, spontaneous contractions after 1-2 days in culture, demonstrating that the cell-cell connections had been maintained within the cardiac sheets after the casting process.
Figure 4. Fabrication of patterned 3D tubular tissues with three muscle cell types.
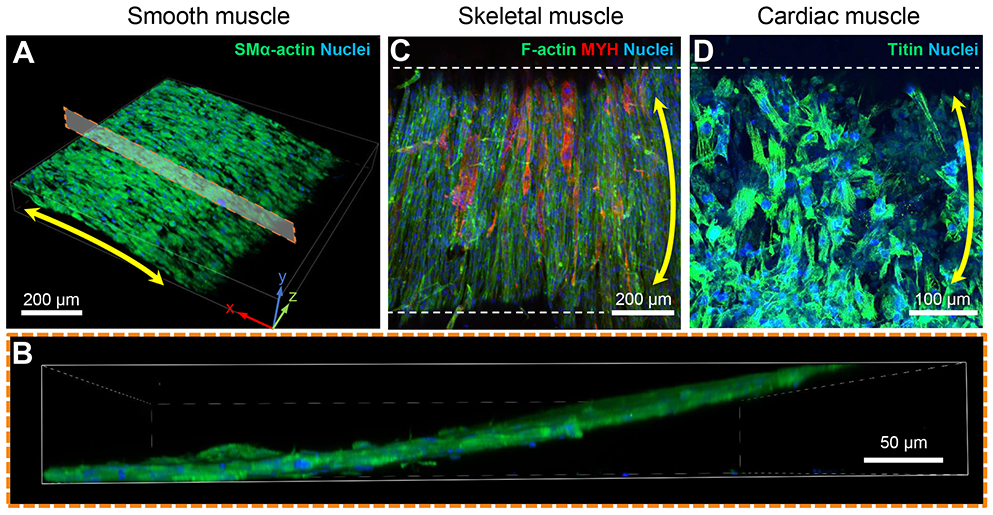
A. 3D rendered image of a confocal z-stack of a SMC tube. Image was rotated to show the curvature of the tubular tissue’s outer surface.
B. Boxed inset of A. Cross-sectional view of confocal z-stack demonstrates cell layers are attached to the outer edge of the hydrogel wall.
C. Maximum intensity projection of a confocal z-stack taken of a tubular tissue circumferentially patterned with mouse muscle myoblasts (C2C12s) and cultured in differentiation medium to promote fusion of myoblasts into elongated myotubes (MYH, all isoforms).
D. Maximum intensity projection of a confocal z-stack taken of a tubular tissue circumferentially patterned with iPSC-derived cardiomyocytes.
The double-headed yellow arrows denote global circumferential cellular alignment perpendicular to the tube’s long axis. Dashed white lines outline the edges of the tubular constructs in C and D.
In the case of skeletal muscle tubes, tissues were cultured in a serum-rich (20% FBS) medium for 3-4 days after fabrication to promote additional cell growth before switching into a serum-poor (2% HS) medium. Once is low serum conditions, myoblasts began to fuse into multinucleated myotubes that elongated circumferentially around the tube’s curved surface (Figure 4C). This result suggests that pre-patterning individual myoblast cell sheets before incorporation into 3D tissues is sufficient to provide robust organizational cues from within the cell-sheet’s structure and does not require sustained external cues to generate aligned myotubes. We hypothesize that the deposited ECM during cell-sheet formation was also organized and provided robust directional cues that promote consistent cellular alignment after casting into a 3D tissue.
We have investigated this hypothesis in a previous study where sheets of aligned C2C12 myoblasts were transferred onto another sheet with either parallel or orthogonal alignment [17]. We found that the alignment of the deposited ECM within each sheet while cultured on fTNFS was maintained after stacking and promoted the formation of parallel or orthogonally organized myotubes within each layer, respectively. Furthermore, in our previous study, sheets of myoblasts stacked in parallel alignment were found to have longer myotubes and higher fusion indices compared to sheets stacked in an orthogonal orientation [17]. Our previous results [17] taken together with those described in this study demonstrate the significant influence that the ECM has on tissue development and structure.
1.12. Cellular organization is maintained in 3D tubular tissues
To investigate if circumferential patterning of cellular alignment was maintained over longer culture periods, smooth, skeletal, and cardiac muscle tubes were cultured for 7 or 14 days. Each engineered tissue was stained for filamentous actin (F-actin) and its organization was quantified using alignment analysis MATLAB scripts as described previously (Figure 5) [16,27]. Smooth, skeletal, and cardiac muscle tubes demonstrated similar levels of circumferential cellular alignment around the tubes’ surfaces (Figure 5D-I).
Skeletal muscle tubes showed formation of elongated circumferential myotubes after 3-4 days in culture with medium containing low-serum, which promotes fusion and differentiation of myoblasts [33,42,43]. We observed that myosin heavy chain (MYH, all isoforms) was expressed throughout fused myotubes at earlier timepoints (Figure 4C). However, with application of broad-field electrical stimulation (1 Hz, 10V, 24 ms pulses) no myotube contraction was observed suggesting contractile proteins had not yet been assembled into functional sarcomeres. To promote the formation of functional sarcomeres, we applied chronic broad field stimulation at lower voltages (1 Hz, 3V, 24 ms) 3-5 days after myotube fusion was apparent in differentiation medium conditions, as shown by others [44,45]. As early as 2-3 days after application of chronic electrical stimulation, myotube twitching followed by robust contraction was visualized in sync with the 1 Hz stimulation pacing and halted in the absence of an electrical pulse (Supplemental video 1). After 9 days of chronic electrical stimulation, registered sarcomeres were easily detectable within myotubes when visualized with immunocytochemistry (Figure 5E). Other groups have also substantiated the role of electrical stimulation in the formation of functional skeletal myotubes in vitro and have demonstrated that sodium and calcium flux, through voltage-gated ion channels, may be required for Z- and A-band formation [44-47]. Together, these data support observations made in this study and suggest that incorporation of both internal and external developmental cues may be required in tissue engineering approaches to recapitulate in vivo-like environments and to promote functional maturation.
Cardiac tubes were also subjected to chronic stimulation pulses (1 Hz, 3V, 8 ms) for up to 37 days in culture. After 37 days in stimulated culture, cross-sectional videos of cardiac tubes contracting under broad-field electrical stimulation showed that the hydrogel walls could be deformed during contraction (Supplemental Video 2). This result demonstrated that patterning and layering aligned cardiomyocytes onto curved three-dimensional tissues was possible and that their contractile function was maintained in long term culture. In future applications of this technology, long term electrical stimulation protocols with increasingly challenging pacing frequencies could be applied to promote maturation of cardiac tubes as shown by other groups [6,48,49]. It would be interesting to explore if pre-patterning of cardiomyocyte architecture within 3D ventricular models would enhance or accelerate maturation when combined with electrical and or mechanical conditioning. Additionally, this technique provides a novel approach for recapitulating more complex myocardial architectures. For example, in the myocardium of the left ventricle, every four to five layers of cardiomyocytes (or myolaminae) are aligned in a single plane but the alignment direction of each myolamina shifts by approximately 10°. This allows the myocardium to encompass a helical fiber architecture with a 180° range of orientations and efficiently maximize its ejection fraction of blood from the ventricles with each contraction of the heart [50-52]. By patterning and stacking individual sheets of cardiomyocytes, this approach could be used to model microenvironments that cardiomyocytes experience at cleavage planes during contraction. Furthermore, if wrapped into a 3D ventricle shape, this platform could be used to study how varied cardiac tissue organizations contribute to ventricle-level function.
2. Conclusions
In this study, we developed a novel method for patterning and layering individual cell sheets and casting them into 3D tubular geometries with curved surfaces. We used fabricated custom molds to cast tubular tissues inspired by the vasculature and the curved tissue structures of the heart ventricles and skeletal muscles in the body’s trunk. We found that pre-patterning individual cell sheets promoted cellular alignment in 3D tissues for several weeks after tissue casting. In addition to providing tissue-level alignment cues, we provided broad-field electrical stimulation for skeletal tubes and found that electrical conditioning was required to promote contractile function. These results suggest that a combination of internal and external conditioning cues may be required to further mature tissues fabricated using fTNFS-enabled cell-sheet casting.
Given the versatile nature of our fTNFS platform, this approach be adapted to fabricate uniquely shaped flexible films and tissue-specific shaped molds for even more complex tissue architectures, such as the conical ventricles of the heart. In this study, we created tissues with thicknesses of 3 – 4 cell layers.
However, we could also generate thicker tissues that surpass the limits of nutrient and oxygen diffusion and prevent tissue necrosis, by incorporating vascular networks or proangiogenic factors [53]. Providing vascular networks could enable long-term culture of thicker tissues for maturation studies. Additionally, this system could be further adapted by incorporating biochemically tunable hydrogels (e.g. fibrin, photo-crosslinking gels, decellularized-ECM, etc.) for tissue specific customization and or presentation of embedded signaling factors for developmental and maturation studies. Flexible TNFS could enable the fabrication of more advanced engineered tissues that could be used to investigate complex structure-function relationships, development, and maturation in the dish.
Supplementary Material
Acknowledgments
We would like to thank the core facilities and staff of the Institute for Stem Cell and Regenerative Medicine (ISCRM), particularly the Tom and Sue Ellison Stem Cell Core and the Lynn and Mike Garvey Imaging Core. We thank Dr. Jonathan Tsui for extensive discussion and editing of this work. Dr. Eunpyo Choi was instrumental in the creation of graphics for this publication as well. This work was supported by the National Institutes of Health: R01HL146436, UG3EB028094, R01NS094388, R01HL94388 (to D.H.K.), TR002317 (to A.S.T.S.), and 1F31HL145809-01A1 (to N.P.W.). The Human Frontier Science Program (RGP0038/2018 to D.H.K). The Jaconette L. Tietze Young Scientist Award (to A.S.T.S.). Lastly, we are grateful for the institutional funding sources that supported this work: the Washington State funded ISCRM Fellows Program and the generous support from the Gree Real Estate company through the ISCRM Gree Scholars Program.
Footnotes
Competing Interests
D-H.K. is a scientific founder and equity holder of NanoSurface Biomedical Inc. A.J. is an employee of NanoSurface Biomedical. A.S.T.S. is a scientific advisor to NanoSurface Biomedical, a company that cells patterned cultureware, and has uncompensated stock in the business. C.E.M is an employee and equity holder in Sana Biotechnology.
Data Availability
The raw data required to reproduce these findings are available to download from https://data.mendeley.com/datasets/nkk5mdywwg/draft?a=b5adeee7-c75c-447b-8418-fb999cd1f39b.
References
- [1].Patel A, Fine B, Sandig M, Mequanint K, Elastin biosynthesis: The missing link in tissue-engineered blood vessels, Cardiovasc. Res 71 (2006) 40–49. 10.1016/j.cardiores.2006.02.021. [DOI] [PubMed] [Google Scholar]
- [2].Roostalu U, Wong JK, Arterial smooth muscle dynamics in development and repair, Dev. Biol 435 (2018) 109–121. 10.1016/j.ydbio.2018.01.018. [DOI] [PubMed] [Google Scholar]
- [3].Domenga V, Fardoux P, Lacombe P, Monet M, Maciazek J, Krebs LT, Klonjkowski B, Berrou E, Mericskay M, Li Z, Tournier-Lasserve E, Gridley T, Joutel A, Notch3 is required for arterial identity and maturation of vascular smooth muscle cells., Genes Dev. 18 (2004) 2730–5. 10.1101/gad.308904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Phillips HM, Rhee HJ, Murdoch JN, Hildreth V, Peat JD, Anderson RH, Copp AJ, Chaudhry B, Henderson DJ, Disruption of Planar Cell Polarity Signaling Results in Congenital Heart Defects and Cardiomyopathy Attributable to Early Cardiomyocyte Disorganization, Circ. Res 101 (2007) 137–145. 10.1161/CIRCRESAHA.106.142406. [DOI] [PubMed] [Google Scholar]
- [5].Weinstein BM, Stemple DL, Driever W, Fishman MC, gridlock, a localized heritable vascular patterning defect in the zebrafish, Nat. Med 1 (1995) 1143–1147. 10.1038/nm1195-1143. [DOI] [PubMed] [Google Scholar]
- [6].Ronaldson-Bouchard K, Ma SP, Yeager K, Chen T, Song L, Sirabella D, Morikawa K, Teles D, Yazawa M, Vunjak-Novakovic G, Advanced maturation of human cardiac tissue grown from pluripotent stem cells, Nature. 556 (2018) 239–243. 10.1038/s41586-018-0016-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Liau B, Christoforou N, Leong KW, Bursac N, Pluripotent stem cell-derived cardiac tissue patch with advanced structure and function, Biomaterials. 32 (2011) 9180–9187. 10.1016/j.biomaterials.2011.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Nunes SS, Miklas JW, Liu J, Aschar-Sobbi R, Xiao Y, Zhang B, Jiang J, Massé S, Gagliardi M, Hsieh A, Thavandiran N, Laflamme MA, Nanthakumar K, Gross GJ, Backx PH, Keller G, Radisic M, Biowire: a platform for maturation of human pluripotent stem cell–derived cardiomyocytes, Nat. Methods 10 (2013) 781–787. 10.1038/nmeth.2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Syedain ZH, Meier LA, Lahti MT, Johnson SL, Tranquillo RT, Implantation of completely biological engineered grafts following decellularization into the sheep femoral artery., Tissue Eng. Part A 20 (2014) 1726–34. 10.1089/ten.TEA.2013.0550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Quint C, Arief M, Muto A, Dardik A, Niklason LE, Allogeneic human tissue-engineered blood vessel, J. Vase. Surg 55 (2012) 790–798. 10.1016/j.jvs.2011.07.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Dahl SLM, Kypson AP, Lawson JH, Blum JL, Strader JT, Li Y, Manson RJ, Tente WE, DiBernardo L, Hensley MT, Carter R, Williams TP, Prichard HL, Dey MS, Begelman KG, Niklason LE, Readily available tissue-engineered vascular grafts., Sci. Transl. Med 3 (2011) 68ra9 10.1126/scitranslmed.3001426. [DOI] [PubMed] [Google Scholar]
- [12].Skardal A, Zhang J, Prestwich GD, Bioprinting vessel-like constructs using hyaluronan hydrogels crosslinked with tetrahedral polyethylene glycol tetracrylates, Biomaterials. 31 (2010) 6173–6181. 10.1016/J.BIOMATERIALS.2010.04.045. [DOI] [PubMed] [Google Scholar]
- [13].Hinton TJ, Jallerat Q, Palchesko RN, Park JH, Grodzicki MS, Shue H-J, Ramadan MH, Hudson AR, Feinberg AW, Three-dimensional printing of complex biological structures by freeform reversible embedding of suspended hydrogels, Sci. Adv 1 (2015) e1500758 10.1126/sciadv.1500758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Lee A, Hudson AR, Shiwarski DJ, Tashman JW, Hinton TJ, Yerneni S, Bliley JM, Campbell PG, Feinberg AW, 3D bioprinting of collagen to rebuild components of the human heart, Science (80-. ). 365 (2019) 482–487. 10.1126/science.aav9051. [DOI] [PubMed] [Google Scholar]
- [15].Miri AK, Khalilpour A, Cecen B, Maharjan S, Shin SR, Khademhosseini A, Multiscale bioprinting of vascularized models, Biomaterials. 198 (2019) 204–216. 10.1016/j.biomaterials.2018.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Jiao A, Trosper NE, Yang HS, Kim J, Tsui JH, Frankel SD, Murry CE, Kim D-H, Thermoresponsive nanofabricated substratum for the engineering of three-dimensional tissues with layer-by-layer architectural control., ACS Nano. 8 (2014) 4430–9. 10.1021/nn4063962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Jiao A, Moerk CT, Penland N, Perla M, Kim J, Smith AST, Murry C, Kim D-H, Regulation of skeletal myotube formation and alignment by nanotopographically controlled cell-secreted extracellular matrix, J. Biomed. Mater. Res. Part A (2018). 10.1002/jbm.a.36351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Penland N, Choi E, Perla M, Park J, Kim D-H, Facile fabrication of tissue-engineered constructs using nanopatterned cell sheets and magnetic levitation, Nanotechnology. 28 (2017) 075103 10.1088/1361-6528/aa55e0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Callegari A, Coons ML, Ricks JL, Rosenfeld ME, Scatena M, Increased calcification in osteoprotegerin-deficient smooth muscle cells: Dependence on receptor activator of NF-kB ligand and interleukin 6., J. Vase. Res 51 (2014) 118–31. 10.1159/000358920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Guan X, Mack DL, Moreno CM, Strande JL, Mathieu J, Shi Y, Marked CD, Wang Z, Liu G, Lawlor MW, Moorefield EC, Jones TN, Fugate JA, Furth ME, Murry CE, Ruohola-Baker H, Zhang Y, Santana LF, Childers MK, Dystrophin-deficient cardiomyocytes derived from human urine: New biologic reagents for drug discovery, Stem Cell Res. 12 (2014) 467–480. 10.1016/j.scr.2013.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Lian X, Zhang J, Azarin SM, Zhu K, Hazeltine LB, Bao X, Hsiao C, Kamp TJ, Palecek SP, Directed cardiomyocyte differentiation from human pluripotent stem cells by modulating Wnt/β-catenin signaling under fully defined conditions, Nat. Protoc 8 (2012) 162–175. 10.1038/nprot.2012.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Palpant NJ, Pabon L, Roberts M, Hadland B, Jones D, Jones C, Moon RT, Ruzzo WL, Bernstein I, Zheng Y, Murry CE, Inhibition of -catenin signaling respecifies anterior-like endothelium into beating human cardiomyocytes, Development. 142 (2015) 3198–3209. 10.1242/dev.117010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Palpant NJ, Pabon L, Friedman CE, Roberts M, Hadland B, Zaunbrecher RJ, Bernstein I, Zheng Y, Murry CE, Generating high-purity cardiac and endothelial derivatives from patterned mesoderm using human pluripotent stem cells, Nat. Protoc 12 (2017) 15–31. 10.1038/nprot.2016.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Hemmi N, Tohyama S, Nakajima K, Kanazawa H, Suzuki T, Hattori F, Seki T, Kishino Y, Hirano A, Okada M, Tabei R, Ohno R, Fujita C, Haruna T, Yuasa S, Sano M, Fujita J, Fukuda K, A Massive Suspension Culture System With Metabolic Purification for Human Pluripotent Stem Cell-Derived Cardiomyocytes, Stem Cells Transl. Med. 3 (2014) 1473–1483. 10.5966/sctm.2014-0072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Nagase K, Yamato M, Kanazawa H, Okano T, Poly( N -isopropylacrylamide)-based thermoresponsive surfaces provide new types of biomedical applications, Biomaterials. 153 (2018) 27–48. 10.1016/j.biomaterials.2017.10.026. [DOI] [PubMed] [Google Scholar]
- [26].Takahashi H, Okano T, Thermally-triggered fabrication of cell sheets for tissue engineering and regenerative medicine, Adv. Drug Deliv. Rev (2019). 10.1016/j.addr.2019.01.004. [DOI] [PubMed] [Google Scholar]
- [27].Cho H, Jönsson H, Campbell K, Melke P, Williams JW, Jedynak B, Stevens AM, Groisman A, Levchenko A, Self-organization in High-Density Bacterial Colonies: Efficient Crowd Control, PLoS Biol. 5 (2007) e302 10.1371/journal.pbio.0050302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Kim D-H, Seo C-H, Han K, Kwon KW, Levchenko A, Suh K-Y, Guided Cell Migration on Microtextured Substrates with Variable Local Density and Anisotropy., Adv. Funct. Mater 19 (2009) 1579–1586. 10.1002/adfm.200990041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Owens GK, Kumar MS, Wamhoff BR, Molecular Regulation of Vascular Smooth Muscle Cell Differentiation in Development and Disease, Physiol. Rev 84 (2004) 767–801. 10.1152/physrev.00041.2003. [DOI] [PubMed] [Google Scholar]
- [30].Chen Y, Lin Y, Davis KM, Wang Q, Rnjak-Kovacina J, Li C, Isberg RR, Kumamoto CA, Mecsas J, Kaplan DL, Robust bioengineered 3D functional human intestinal epithelium, Sci. Rep 5 (2015) 13708 10.1038/srep13708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].MacQueen LA, Sheehy SP, Chantre CO, Zimmerman JF, Pasqualini FS, Liu X, Goss JA, Campbell PH, Gonzalez GM, Park S-J, Capulli AK, Ferrier JP, Kosar TF, Mahadevan L, Pu WT, Parker KK, A tissue-engineered scale model of the heart ventricle, Nat. Biomed. Eng (2018) 1 10.1038/s41551-018-0271-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Finkbeiner SR, Freeman JJ, Wieck MM, El-Nachef W, Altheim CH, Tsai YH, Huang S, Dyal R, White ES, Grikscheit TC, Teitelbaum DH, Spence JR, Generation of tissue-engineered small intestine using embryonic stem cell-derived human intestinal organoids, Biol. Open 4 (2015) 1462–1472. 10.1242/bio.013235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Merceron TK, Burt M, Seol Y-J, Kang H-W, Lee SJ, Yoo JJ, Atala A, A 3D bioprinted complex structure for engineering the muscle–tendon unit, Biofabrication. 7 (2015) 035003 10.1088/1758-5090/7/3/035003. [DOI] [PubMed] [Google Scholar]
- [34].Sasaki D, Matsuura K, Seta H, Haraguchi Y, Okano T, Shimizu T, Contractile force measurement of human induced pluripotent stem cell-derived cardiac cell sheet-tissue, PLoS One. 13 (2018) e0198026 10.1371/journal.pone.0198026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Sakaguchi K, Shimizu T, Okano T, Construction of three-dimensional vascularized cardiac tissue with cell sheet engineering, J. Control. Release 205 (2015) 83–88. 10.1016/JJCONREL.2014.12.016. [DOI] [PubMed] [Google Scholar]
- [36].Juthani N, Howell C, Ledoux H, Sotiri I, Kelso S, Kovalenko Y, Tajik A, Vu TL, Lin JJ, Sutton A, Aizenberg J, Infused polymers for cell sheet release, Sci. Rep (2016). 10.1038/srep26109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Haraguchi Y, Shimizu T, Yamato M, Okano T, Scaffold-free tissue engineering using cell sheet technology, RSC Adv. (2012). 10.1039/c2ra00704e. [DOI] [Google Scholar]
- [38].Tiburcy M, Hudson JE, Balfanz P, Schlick S, Meyer T, Chang Liao M-L, Levent E, Raad F, Zeidler S, Wingender E, Riegler J, Wang M, Gold JD, Kehat I, Wettwer E, Ravens U, Dierickx P, van Laake LW, Goumans MJ, Khadjeh S, Toischer K, Hasenfuss G, Couture LA, Unger A, Linke WA, Araki T, Neel B, Keller G, Gepstein L, Wu JC, Zimmermann W-H, Defined Engineered Human Myocardium With Advanced Maturation for Applications in Heart Failure Modeling and Repair., Circulation. 135 (2017) 1832–1847. 10.1161/CIRCULATIONAHA.116.024145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Li RA, Keung W, Cashman TJ, Backeris PC, Johnson BV, Bardot ES, Wong AOT, Chan PKW, Chan CWY, Costa KD, Bioengineering an electro-mechanically functional miniature ventricular heart chamber from human pluripotent stem cells, Biomaterials. (2018). 10.1016/j.biomaterials.2018.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Maffioletti SM, Sarcar S, Henderson ABH, Mannhardt I, Pinton L, Moyle LA, Steele-Stallard H, Cappellari O, Wells KE, Ferrari G, Mitchell JS, Tyzack GE, Kotiadis VN, Khedr M, Ragazzi M, Wang W, Duchen MR, Patani R, Zammit PS, Wells DJ, Eschenhagen T, Tedesco FS, Three-Dimensional Human iPSC-Derived Artificial Skeletal Muscles Model Muscular Dystrophies and Enable Multilineage Tissue Engineering, Cell Rep. 23 (2018). 10.1016/j.celrep.2018.03.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Zhou W, Chen Y, Roh T, Lin Y, Ling S, Zhao S, Lin JD, Khalil N, Cairns DM, Manousiouthakis E, Tse M, Kaplan DL, Multifunctional Bioreactor System for Human Intestine Tissues, ACS Biomater. Sci. Eng 4 (2018) 231–239. 10.1021/acsbiomaterials.7b00794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Matsumoto T, Sasaki J.-l., Alsberg E, Egusa H, Yatani H, Sohmura T, Three-Dimensional Cell and Tissue Patterning in a Strained Fibrin Gel System, PLoS One. 2 (2007) e1211 10.1371/journal.pone.0001211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Xu B, Magli A, Anugrah Y, Koester SJ, Perlingeiro RCR, Shen W, Nanotopography-responsive myotube alignment and orientation as a sensitive phenotypic biomarker for Duchenne Muscular Dystrophy, Biomaterials. 183 (2018) 54–66. 10.1016/J.BIOMATERIALS.2018.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Fujita H, Nedachi T, Kanzaki M, Accelerated de novo sarcomere assembly by electric pulse stimulation in C2C12 myotubes, Exp. Cell Res 313 (2007) 1853–1865. 10.1016/j.yexcr.2007.03.002. [DOI] [PubMed] [Google Scholar]
- [45].De Deyne PG, Formation of sarcomeres in developing myotubes: role of mechanical stretch and contractile activation, Am. J. Physiol. Physiol 279 (2000) C1801–C1811. 10.1152/ajpcell.2000.279.6.C1801. [DOI] [PubMed] [Google Scholar]
- [46].Hara M, Tabata K, Suzuki T, Do M-KQ, Mizunoya W, Nakamura M, Nishimura S, Tabata S, Ikeuchi Y, Sunagawa K, Anderson JE, Allen RE, Tatsumi R, Calcium influx through a possible coupling of cation channels impacts skeletal muscle satellite cell activation in response to mechanical stretch, Am. J. Physiol. Physiol 302 (2012) C1741–C1750. 10.1152/ajpcell.00068.2012. [DOI] [PubMed] [Google Scholar]
- [47].Cooper ST, Maxwell AL, Kizana E, Ghoddusi M, Hardeman EC, Alexander IE, Allen DG, North KN, C2C12 Co-culture on a fibroblast substratum enables sustained survival of contractile, highly differentiated myotubes with peripheral nuclei and adult fast myosin expression, Cell Motil. Cytoskeleton 58 (2004) 200–211. 10.1002/cm.20010. [DOI] [PubMed] [Google Scholar]
- [48].Physiologic force-frequency response in engineered heart muscle by electromechanical stimulation, Biomaterials. 60 (2015) 82–91. 10.1016/J.BIOMATERIALS.2015.03.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Radisic M, Park H, Shing H, Consi T, Schoen FJ, Langer R, Freed LE, Vunjak-Novakovic G, Functional assembly of engineered myocardium by electrical stimulation of cardiac myocytes cultured on scaffolds., Proc. Natl. Acad. Sci. U. S. A 101 (2004) 18129–34. 10.1073/pnas.0407817101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Streeter DD, Bassett DL, An engineering analysis of myocardial fiber orientation in pig’s left ventricle in systole, Anat. Rec 155 (1966) 503–511. 10.1002/ar.1091550403. [DOI] [Google Scholar]
- [51].Pope AJ, Sands GB, Smaill BH, LeGrice IJ, Three-dimensional transmural organization of perimysial collagen in the heart, Am. J. Physiol. Circ. Physiol 295 (2008) H1243–H1252. 10.1152/ajpheart.00484.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Ae DJH, Anderson RH, The Development and Structure of the Ventricles in the Human Heart, (n.d.). 10.1007/s00246-009-9390-9. [DOI] [PubMed] [Google Scholar]
- [53].Sekine H, Shimizu T, Sakaguchi K, Dobashi I, Wada M, Yamato M, Kobayashi E, Umezu M, Okano T, In vitro fabrication of functional three-dimensional tissues with perfusable blood vessels, Nat. Commun 4 (2013). 10.1038/ncomms2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data required to reproduce these findings are available to download from https://data.mendeley.com/datasets/nkk5mdywwg/draft?a=b5adeee7-c75c-447b-8418-fb999cd1f39b.


