Abstract
The genus Pseudogymnoascus encompasses soil psychrophilic fungi living also in caves. Some are opportunistic pathogens; nevertheless, they do not cause outbreaks. Pseudogymnoascus destructans is the causative agent of the white-nose syndrome, which is decimating cave-hibernating bats. We used comparative eco-physiology to contrast the enzymatic potential and conidial resilience of P. destructans with that of phylogenetically diverse cave fungi, including Pseudogymnoascus spp., dermatophytes and outdoor saprotrophs. Enzymatic potential was assessed by Biolog MicroArray and by growth on labelled substrates and conidial viability was detected by flow cytometry. Pseudogymnoascus destructans was specific by extensive losses of metabolic variability and by ability of lipid degradation. We suppose that lipases are important enzymes allowing fungal hyphae to digest and invade the skin. Pseudogymnoascus destructans prefers nitrogenous substrates occurring in bat skin and lipids. Additionally, P. destructans alkalizes growth medium, which points to another possible virulence mechanism. Temperature above 30 °C substantially decreases conidial viability of cave fungi including P. destructans. Nevertheless, survival of P. destructans conidia prolongs by the temperature regime simulating beginning of the flight season, what suggests that conidia could persist on the body surface of bats and contribute to disease spreading during bats active season.
Subject terms: Microbial ecology, Pathogens
Introduction
Pseudogymnoascus destructans (Pseudeurotiaceae, Ascomycota)1 is a fungus that infects the skin of hibernating bats and causes the disease called the white-nose syndrome (WNS)2–4. This fungus is native to Eurasian bat hibernacula, where it behaves as a not too virulent cutaneous pathogen. It causes sporadic mortality and health deterioration, but not apparent changes in bat populations5,6. This fact, together with the high genetic diversity of the European P. destructans populations, suggests its long co-evolution with its hosts6–10. A single P. destructans clone was introduced from Europe or Asia to the United States, where it encountered naïve hosts and has been causing massive mortality observed since 200611,12. Despite of its clonal growth P. destructans manifests some fenotypical divergence13,14.
Pseudogymnoascus destructans infects naked parts of the bat skin, such as the muzzle, ears and flight membranes, where it forms cup-like epidermal erosions and ulcerations. Unlike dermatophytes15, P. destructans invades no only the epidermis, but grows into deeper parts of the skin, the dermis2,15–17. The ability to make a lesion is the same between P. destructans strains in the United States and Europe18. The lethal progress described as WNS is thus a result of complex interactions of variables, which are under debate19–22. Despite availability of extensive studies on P. destructans biology, the decided answer about its virulence and biology still missing and further research is desired to get closer. P. destructans shares some virulence factors with dermatophytes23,24, such as subtilisin-like proteases degrading collagen25, but lacks others, for example keratinase26. Comparative studies16,24 have identified siderophore secretion as virulence factor19,27, which is similar to fungi forming deeper mycotic infections28, where siderophores play an important role in the pathogenesis. Pseudogymnoascus destructans, in contrast to non-virulent Pseudogymnoascus species, is also unique in its overproduction of riboflavin, the role of which in virulence needs to be assessed further19. The WNS transcriptome revealed over fifty genes that are upregulated during the infection process on the side of the fungus29–31. They include numerous secreted proteases (including destructins and a homolog of the Aspergillus fumigatus major allergen Aspf2), heat shock proteins, ureases, metal-binding siderophores, enzymes responsible for fatty acid utilization and protein kinases.
WNS is spread by bat-to-bat transmission during hibernation32 and by infections from environmental pool of the cave sediments33,34. Non-pathogenic Pseudogymnoascus species are common soil fungi also living outside of hibernacula1. To the contrary, alternative habitats for P. destructans are so far unknown. It is unclear which physiological constraints restrict this fungus to the cold and humid habitats of caves. P. destructans is a psychrophilic fungus growing at temperatures between 3 and 19 °C, with an optimal range of 12–15 °C. Its growth stops at 10 °C and above 20 °C35–37. The optimal relative humidity for the growth of P. destructans is around 80%, which is the moisture level found in bats’ hibernacula and their body surface during hibernation15. Its psychrophilic nature enables bats to clear any visible infection during summer, when ambient temperatures exceed the upper limit for P. destructans growth. Nevertheless, we have only sparse information about its conidial resilience13,38,39. It therefore remains unknown whether P. destructans conidia can disseminate on the body surface of bats during the active flight season to new hibernacula and participate in the spreading of the disease.
The goal of our study was to compare phenotype of P. destructans with that of (1) other Pseudogymnoascus species to find traits specific to P. destructans. We propose that these traits are potentially under selective pressure and are possible virulence factors, (2) cave fungi to identify common traits related to growth in cave habitat and thus not directly linked with P. destructans pathogenesis and (3) dermatophytes which are pathogens and thus shared traits could be important in P. destructans virulence. Our study combines knowledge of prior studies on virulence factors of P. destructans and compares phenotype of P. destructans with phenotype of ecologically related species, which enables to estimate selective pressure leading to pathogenicity of P. destructans.
Results
Biolog analysis
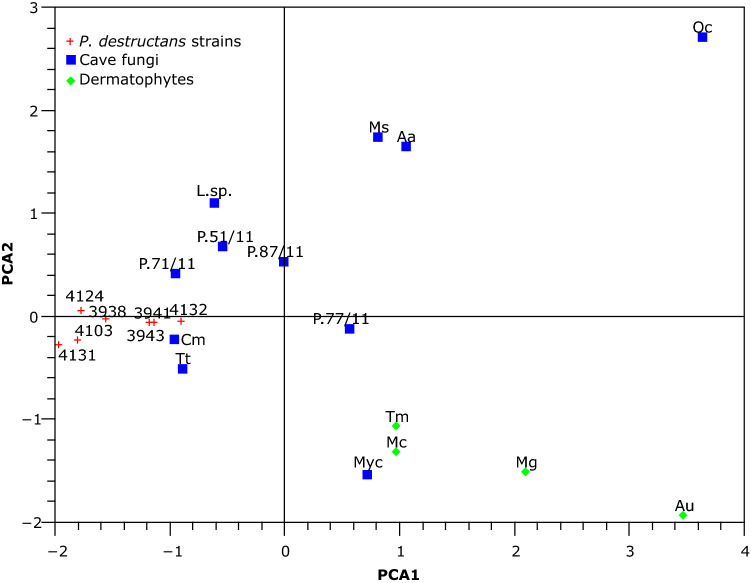
Analyzed ecological groups formed well-defined clusters on PCA analysis (Fig. 1), only a cave inhabiting fungus Myotisia cremea (Onygenales), which lives on the bats guano, clustered with dermatophytes. Based on one-way NPMANOVA analysis, P. destructans significantly differed in growth potential from dermatophytes on each of the Biolog MicroPlates tested (p < 0.05). Pseudogymnoascus destructans was also distinct from cave fungi (p < 0.05) except growth on sulfur and phosphorus sources (p = 0.75). The main differences between these groups resided in the limitation of P. destructans to only a few nitrogen substrates and its increased growth potential on nutrient supplements (Supplementary Dataset online). Pseudogymnoascus destructans was significantly distinct from Pseudogymnoascus species on carbon and nutrient supplements (p < 0.05). Pseudogymnoascus species were similar to cave fungi on all microplates (p > 0.7). When all microplates were integrated into one analysis (Fig. 1), P. destructans was significantly separated from cave fungi, Pseudogymnoascus spp. and dermatophytes (p < 0.02), and dermatophytes differed from cave fungi (p = 0.01). Pseudogymnoascus spp. were significantly different from P. destructans (p < 0.02) and were similar to cave fungi (p = 1).
Figure 1.
PCA analysis showing dissimilarities in nutrient metabolism between P. destructans, other cave fungi and dermatophytes on Biolog MicroPlates. The plotted PCA axes describe 52.3% of the variability in the data. Tt Trichophyton terestre, Cm Chrysosporium merdarium, L.sp. Leotiomycetes sp. CCF6130, P.71/11 Pseudogymnoascus sp. 2 AK71/11, P.77/11 P. sp. 2 AK77/11, P.87/11 P. sp. 4 AK87/11, P.51/11 P.sp. 1 AK51/11, Ms Metapochonia suchlasporia, Aa Aspergillus askiburgiensis, Oc Oidiodendron cerealis, Myc Myotisia cremea CCF5407, Tm T. mentagrophytes, Au Arthroderma uncinatum, Mc Microsporum canis, Mg M. gypseum.
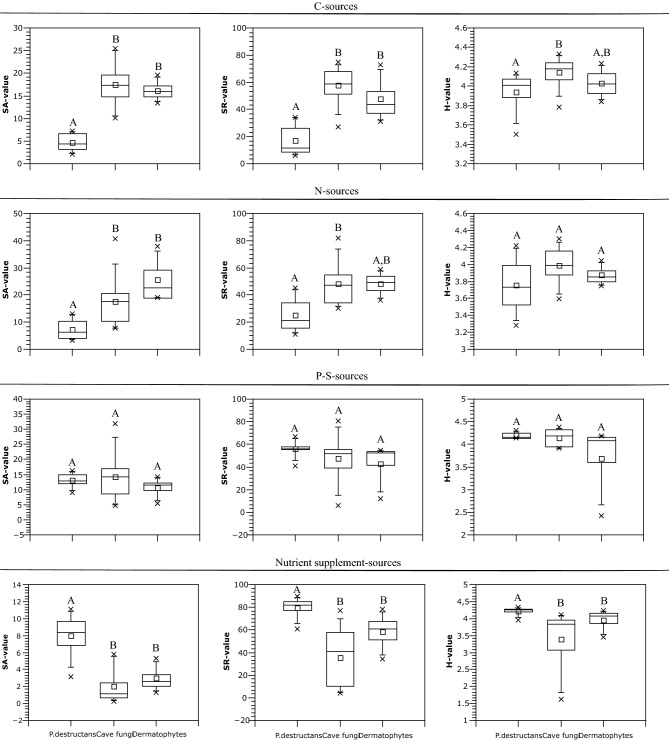
Pseudogymnoascus destructans was remarkable for its relatively low SA and SR but high H values on microplates containing carbon and nitrogen sources (Fig. 2). This means that even if P. destructans growth is supported by only a few carbon and nitrogen sources, it also preserves a weak growth capability on most of them. The most growth-supporting carbon and nitrogen substrates for P. destructans were the lipid Tween 80, glycogen, and urea, allantoin, uric acid and ammonia (Table 1, Supplementary Table S1 online). Cave fungi and dermatophytes also favoured these nitrogen substrates. P. destructans grew well on nutrient supplements, especially on amino acids. All groups had similar growth potential on phosphorus and sulphur substrates. Cave fungi displayed versatility in carbon utilization (highest value of SA, SR and H). Dermatophytes were specific in their preference for nitrogenous carbon sources (mostly amino acids and N-acetyl-D-glucosamine).
Figure 2.
Growth characteristics on Biolog MicroPlates described by SA, SR and H values. Different letters indicate significant differences between groups and same letters indicate similarity based on Kruskall-Wallis statistics supplemented with Mann–Whitney pairwise comparison and Bonferroni correction.
Table 1.
Top ten utilized substrates by P. destructans on each Biolog MicroPlates, nutrient sources in parentheses.
| FF (C) | PM3 (N) | PM4 (S + P) | PM5 (nutrient supplements) |
|---|---|---|---|
| Tween 80 | Gly-Gln | Cytidine-2-monophosphate | L-lysine |
| D-mannose | Urea | O-phosphoryl-ethanolamine | L-leucine |
| Gentibiose | Guanosine | O-phospho-L-serine | L-valine |
| D-trehalose | Gly-Asn | 6-phospho-gluonic acid | Orotic acid |
| D-fructose | Gly-Glu | Guanosine-2-monophosphate | L-isoleucine + L-valine |
| α-D-glucose | Allantoin | 2-deoxy-D-glucose 6-phosphate | D,L-α,δ-diamino-pimelic acid |
| Glycogen | Ala-Gly | D-3-phospho-glyceric acid | D-alanine |
| L-glutamic acid | L-glutamine | Carbamyl phosphate | L-tyrosine |
| Turanose | Ammonia | Uridine-5-monophosphate | L-cysteine |
| D-psicose | Uric acid | Guanosine-5-monophosphate | Trans-4-hydroxy L-proline |
Semi-quantitative extracellular enzymes activities
We compared the production of extracellular enzymes cleaving connective tissue between P. destructans and other Pseudogymnoascus species. We detected the production of lipases, elastases and collagenases. Pseudogymnoascus destructans was specific in the production of lipases and lack of elastases. There was no difference in the production of collagenases (Table 2).
Table 2.
Semi-quantitative extracellular enzymes activities indicate the presence of lipases and lack of elastases in Pseudogymnoascus destructans compared to P. spp.
| Species | Strain | Lipases (olive oil) | Elastases pH 5.5 | Elastases pH 7 | Elastases pH 8.5 | Colagenases pH 5.5 | Colagenases pH7 | Colagenases pH 8 |
|---|---|---|---|---|---|---|---|---|
| P. destructans | 20,631-21 T | + | − | − | − | + | + | + |
| CCF3941 | + | − | − | − | + | + | + | |
| CCF3943 | + | − | − | − | + | + | + | |
| CCF4103 | + | − | − | − | + | + | + | |
| CCF4987 | + | − | − | − | + | + | + | |
| CCF4986 | + | − | − | − | + | + | + | |
| P. sp. 1 | CCF5025 | − | − | − | − | + | + | + |
| P. sp. 2 | CCF5030 | − | − | + + | + | + | + | + |
| P. sp. 2 | CCF5027 | − | − | + + | − | + | + | + |
| P. sp. 2 | CCF5029 | − | − | + + + | + + + | + | + | + |
| P. sp. 3 | CCF5026 | − | − | + + | + + | + | + | + |
− Activity absent, + / + + / + + + detected activity from weak to strong.
Medium alkalization by urea degradation
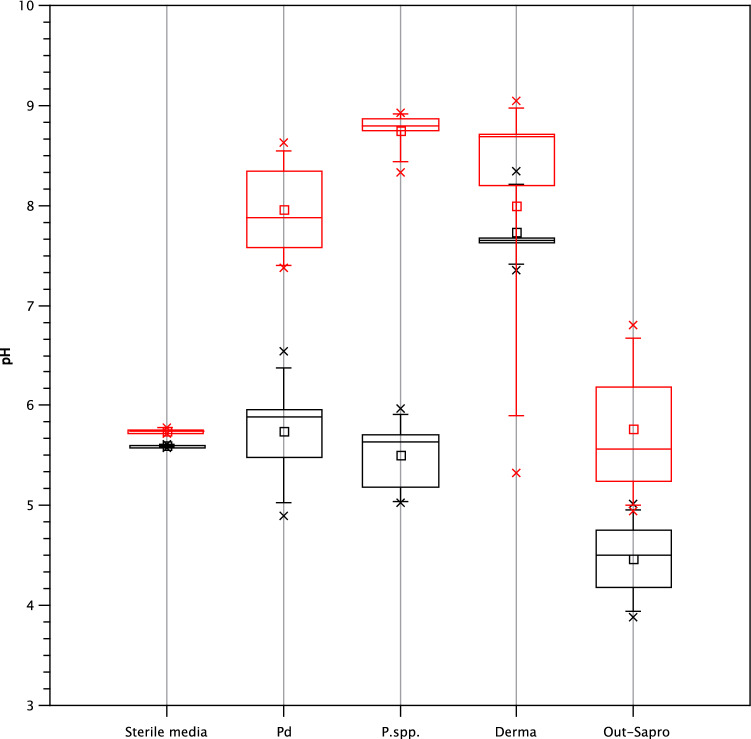
Pseudogymnoascus species including P. destructans significantly increase pH in Sabouraud medium supplemented with urea (permutation test, p = 0.0001) (Fig. 3, Supplementary Table S2 online). Dermatophytes increased the pH of both media equally (p = 0.8), except for Arthroderma uncinatum which only alkalized medium without the presence of urea. Outdoor saprotrophs alkalized none of media analysed (p = 0.29). We detected a slight pH decrease (< 0.2) in both of sterile media over the course of the experiment. This means that the detected changes in the pH of inoculated media were the result of fungal growth.
Figure 3.
Increase of growth medium pH caused by urea metabolism. The Sabouraud medium is plotted in black and the Sabouraud medium supplemented with urea is plotted in red. Pd Pseudogymnoascus destructans, P.spp. Pseudogymnoascus species, Derma dermatophytes, Out-Sapro outdoor saprotrophs.
Conidial viability under stressful conditions
We evaluated conidial viability under stressful conditions over a period of 42 days for nine P. destructans strains, three non-pathogenic Pseudogymnoascus species and two air-borne Aspergillus species (Fig. 4, Supplementary Table S3 online).
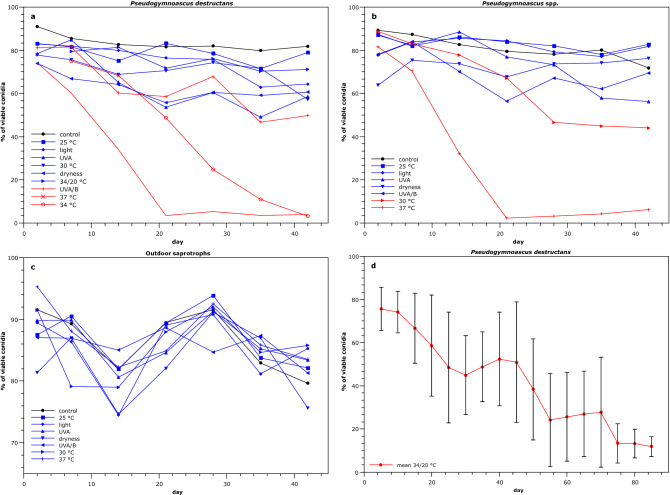
Figure 4.
Test of conidial viability under stressful conditions. A-C: Conidial viability affected by variable stress factors for 42 days. D: prolonged effect of periodic temperature switches from 34 to 20 °C (12 h period, 85 days) on conidial viability of P. destructans. Mean values with standard deviations for seven strains are presented. Stress factors with a minor effect are plotted in blue. Stress factors causing a decrease in conidial viability under 50% are marked red.
All Pseudogymnoascus species, including P. destructans, were sensitive to temperatures higher than 30 °C. At the end of the incubation at 30 and 37 °C, conidial viability of Pseudogymnoascus spp. dropped to 44.1% and 6.3%, respectively. P. destructans conidia exhibited greater resilience to incubation in 30 °C, final viability of its conidia was 57.2%. Nevertheless, its conidial viability was less than 4% at temperatures between 34 and 37 °C. P. destructans was also mildly sensitive to UVA/B radiation, with 49.8% of viable conidia. Our next experiment traced the conidial viability of P. destructans strains over a 12-h period of repeated temperature switches from 20 to 34 °C. The changing of temperature markedly enhanced their conidial viability to 64.5%. Even after 85 days of exposure, their conidial viability remained between 6 and 17%, with a mean value of 11.1%. The Aspergillus species were resistant to all of the stressful conditions. Sensitivity to the dry conditions was similar across the tested species.
Discussion
Our aims were (1) to ascertain the enzymatic potential and ability of environmental alkalization of P. destructans, to compare it with a phylogenetically diverse set of cave fungi (including non-pathogenic Pseudogymnoascus spp.), dermatophytes and outdoor saprotrophs, and (2) to characterize the viability of P. destructans conidia under stressful conditions. This enabled us to trace potential virulence factors of P. destructans and to propose the physiological constraints of P. destructans that limit its distribution to bat-inhabited caves. Information about conidial stress tolerance is necessary for a complex evaluation of the dissemination potential of P. destructans and the spreading of the white-nose syndrome. We found that P. destructans has extensively reduced metabolic potential compared to species tested, including other Pseudogymnoascus species. P. destructans produces lipases, which may be important for its pathogenicity. Conidia of P. destructans are more resilient than mycelium to harsh environmental conditions. Thus, it could mean that conidia contribute to disease spreading after bat departures from caves after winter.
The enzymatic profile of P. destructans was more similar to saprotrophic cave fungi than to dermatophytes. The similarity of P. destructans with cave fungi found in this study was expectable and points to its descent from the saprotrophic Pseudogymnoascus species. P. destructans is generally isolated directly from bats, but rarely also from sediments of abandoned caves33 where it grows on cellulose and protein/lipid substrates40,41. Our Biolog analyses indicate that P. destructans is able to use a broad range of carbon, nitrogen, phosphorus and sulfur sources, but its growth on most such sources is weak. This suggests that P. destructans could survive with low fitness on a variety of substrates. Nevertheless, faster-growing cave fungi probably outcompete P. destructans on these non-preferred substrates.
Despite the supposed saprophytic origin of P. destructans, our results from enzymatic and total Biolog assimilation profiles clearly separate P. destructans from cave fungi, including Pseudogymnoascus spp. Compared to cave fungi, P. destructans strains display intensive enzymatic curtailment on Biolog MicroPlates. We propose that it could be result of P. destructans long-term specialization to a specific niche with limited set of available nutrients, which led to subsequent selective loses of metabolic pathways. This supports previous evidence that the metabolism of P. destructans is restricted compared to other Pseudogymnoascus species38,42–44 and confirms the long co-evolution between this pathogen and its hosts.
Both dermatophytes and P. destructans grow on mammal skin, and thus have similar nutrient sources available for growth. Our sampling unable us to draw decided conclusions; however, it seems that they have different nutritive strategy. While dermatophytes prefer nitrogenous substrates, P. destructans is rather specialized on lipid substrates. P. destructans even differs from analyzed cave fungi, including non-pathogenic Pseudogymnoascus spp., that prefer saccharides as carbon sources. These results correspond with prior findings of lipid degradation by P. destructans40,45. We propose that P. destructans preference of lipids is one of virulence factors, which enables P. destructans to colonize bat tissues. Lipases enable fungi to damage epidermal and epithelial tissue. They also influence fungal growth, adherence and dissemination inside the host46. Our comparative eco-physiological approach proposes outstanding role of lipid and fatty acid degradation in the virulence of P. destructans, as already noted by Donaldson et al.29 based on transcriptomic data. Given the facts that P. destructans lacks keratinolytic activity and that phospholipids are a major component of all cell membranes, lipases are probably the most important enzymes allowing fungal hyphae to digest and invade the integumentary structures of infected skin, forming cup-like erosions diagnostic of the white-nose syndrome.
Similarly to cave fungi including Pseudogymnoascus spp., P. destructans prefers urea, allantoin, ammonia and uric acid as nitrogen sources. These substances are products of the metabolic breakdown of nitrogenous compounds. Beside occurrence in sediments, these substances are also contained in urine and sweat. In bat skin, they are typical components of sweat glands, where hyphal growth of P. destructans was described2 and thus where are easily accessible for its nutrition. Beside nitrogen metabolism, urease has a versatile functions in plants, animals and microorganisms, as is reviewed in47. It participates in fungal pathogenesis48,49, for example by production of ammonia which cause environmental alkalization50 and subsequent spontaneous creation of reactive oxygen species51 and tissue damage. We detected an increase in pH caused by the degradation of urea during cultivation of P. destructans. Nevertheless, we were not successful in direct pH measurements in bat necrosis, thus further experiments are needed to examine the role of pH manipulation in virulence of P. destructans.
Similarly to other Pseudogymnoascus species, P. destructans produces collagenases that erode connective tissues and enable fungi to penetrate the skin of bats. Collagenase production is well known in P. destructans25,52, but from our data we propose that it is probably a preadaptation present in the whole genus, not an adaptation primarily responsible for the behavior of P. destructans. This corresponds with the fact that P. destructans has an equal level of protease expression (including that of destructins) in agar culture and on the wings of bats29. Similarly, subtilisin-like serine proteases, whose role in virulence is not yet clear, are present both in P. destructans and P. pannorum25. Our results presume that P. destructans lacks the ability to cleave other constitutive proteins in the skin, elastin and keratin, which may suggests that the pathogen does not use enzymatic skin degradation primarily for nutrient acquisition. Thus, we propose that other nutrient sources like lipids, urea and various nitrogenous sources found in sweat glands are targets for assimilation by P. destructans.
Preceding studies have shown that temperatures above 20 °C37 and relative humidity lower than 70%53 impedes the mycelial growth of P. destructans. Our viability tests revealed that conidia of P. destructans are more resilient to higher temperatures and lower humidity than its mycelium. Recent study39 has proposed that conidia of P. destructans could germinate even after exposition to elevated temperatures. Our simulation of temperature changes on the surface of a bat’s body after its departure from a cave indicates even more prolonged conidial viability of P. destructans at higher temperatures. In vitro conditions are far away from that in nature, where more factors are combined and could have cumulative negative effect on viability of conidia. Thus, we cannot foresee how long conidia of P. destructans stay viable on the bat body surface after its departure from cave. However, can state that conidia of P. destructans are much more resilient to harsh environmental conditions than its mycelium. As conidia are the primary dissemination agent of fungi, we wonder if they might be able to survive on the surface of the bat body reaching the next hibernation site.
Conclusion
Analyzed cave fungi, despite being phylogenetically unrelated, represent a metabolically well-defined group. Among them, Pseudogymnoascus destructans evinces specific physiological profile. We suggest that it is a result of its strong specialization to life on bat skin and pathogenicity. Pseudogymnoascus destructans is specific by loss of redundant metabolic variability and production of lipases. We propose that lipases are probably the most important enzymes allowing fungal hyphae to digest and invade the integumentary structures of infected skin. The future investigations of virulence could test our hypothesis by specific knock-out of these genes. Pseudogymnoascus destructans conidia show great viability potential under various stressful condition, which offers possibility of their inactive persistence on the body surface of bats during their active season. These resting conidia are maybe able to germinate when bats reach their hibernation sites, where environmental conditions enable the growth of P. destructans. This could mean that once infected bats might serve as a reservoir and vector for the spread of the disease to new hibernacula.
Methods
Fungal material and cultivation
The type strain from the USA and nine European strains of P. destructans strains covering different haplotypes and mating types identified according to Zukal et al.6 were used. Representatives of typical non-pathogenic cave-inhabiting fungi (8 genera from four Ascomycota orders, 16 strains) and dermatophytes (2 genera, 5 strains) or saprotrophic fungi (2 genera, 3 strains) living outside of caves were chosen for comparison (Tables 3, 4). Relatively low representation of dermatophyte and outside living saprotrophic strains prevents general conclusion about their physiology. Nevertheless, it was not attempt of our study. Inclusion of these species served us for suggestions of potential selective pressures that could play a role in formation of P. destructans physiology. Fungal isolates were identified by ITS rDNA barcoding using the methods of Kolařík et al.54, and their sequences were deposited in the EMBL sequence database. Cultures were deposited in the Culture Collection of Fungi (abbreviations CCF or AK) at the Department of Botany, Faculty of Science, Charles University in Prague. Pseudogymnoascus species including P. destructans were grown in Petri dish containing glucose yeast extract agar (GYEA, 20 g glucose, 5 g yeast extract, 15 g agar, 1 l distilled water) at 10 °C unless stated otherwise. Cave fungi Trichophyton terrestre AK44/09, Leotiomycetes sp. CCF6130 (88/11) and Chrysosporium merdarium CCF6131 (AK91/11) were grown on 4° malt agar (MA) at 10 °C. Other fungi were cultivated on 4° MA at 25 °C.
Table 3.
Fungal strains used in this study.
| Group | Fungus | Source | Sequence accession number | Reference |
|---|---|---|---|---|
|
Pseudogymnoascus destructans (Leotiomycetes) |
CCF3938 | CZE, Solenice, Myotis myotis, 2010 | HM584954 | 66 |
| CCF3941 | CZE, Bohemian Karst, Malá Amerika mine, Myotis myotis, 2010 | HM584956 | 66 | |
| CCF3943 | CZE, Stříbro, Myotis myotis, 2010 | HM584957 | 66 | |
| CCF4103 | CZE, Herlíkovice, Krkonoše Mts., Pleurotus auritus, 2011 | LN852366 | 19 | |
| CCF4124 | CZE, Horní Albeřice, Krkonoše, Myotis myotis, 2011 | KJ938421 | 9 | |
| CCF4131 | CZE, Vyškov u Chodové Plané, Myotis myotis, 2011 | KJ938420 | 9 | |
| CCF4132 | CZE, Pernink, Myotis myotis, 2011 | nd | 9 | |
| CCF4987 | CZE, Kašperské Hory, Myotis myotis, 2014 | LN871252 | 6 | |
| CCF4986 | Rusia, Ural mts., cave Smolinskaya, Myotis dasycneme, 2014 | LN852359 | 6 | |
| 20631-21T | USA, Williams Hotel, NY, Myotis lucifugus, 2008 | EU884921 | 35 | |
| Saprotrophic cave fungi | Pseudogymnoascus sp. 1 CCF50251 | CZE, Bohemian Karst, Alkazar tunnel, bat excrement, 2009 | LN852360 | 19 |
| P. sp. 2 CCF50302 | CZE, Moravia, Myotis myotis, 2012 | LN852361 | 19 | |
| P. sp. 2 CCF50272 | CZE, cave sediment, Moravia, Javoříčske Caves, 2012 | LN852363 | 19 | |
| P. sp. 2 CCF50292 | CZE, Moravia, Javoříčske caves, cave sediment, 2012 | LN852364 | 19 | |
| P. sp. 3 CCF50263 | CZE, Moravian Karst, Sloupsko-Šošůvké Cave, Rhinolophus hipposideros, 2013 | LN852365 | 19 | |
| P. sp. 1 AK51/114 | CZE, Herlíkovice, Krkonoše, Eptesicus nilssonii | LN714595 | 67 | |
| P. sp. 2 AK71/115 | CZE, Velká Amerika mine, sediment, 2011 | Submitted to EMBL | This study | |
| P. sp. 4 AK87/116 | CZE, Bohemian Karst, Koněpruské jeskyně caves, sediment, 2011 | Submitted to EMBL | This study | |
| P. sp. 2 AK 77/117 | CZE, Bohemian Karst, Velká Amerika mine, sediment, 2011 | Submitted to EMBL | This study | |
| Trichophyton terrestre AK44/09 (Onygenales) | CZE, Bohemian Karst, Alkazar tunnel, excrement, 2009 | LN714614 | 67 | |
| Myotisia cremea CCF5407 (Onygenales) | CZE, Bohemian Karst, Malá Amerika mine, bat excrement, 2009 | LT627243 | 68 | |
| Metapochonia suchlasporia CCF6128 (Clavicipitaceae) | CZE, Bohemian Karst, Velká Amerika mine, sediment, 2011 | Submitted to EMBL | This study | |
| Leotiomycetes sp. CCF6130 (= AK88/11)8 | CZE, Bohemian Karst, Velká Amerika mine, sediment, 2011 | Submitted to EMBL | This study | |
| Chrysosporium merdarium CCF6131 (AK 91/11) (Leotiomycetes) | CZE, Karlštejn castle, sediment in the castle well, 2011 | Submitted to EMBL | This study | |
| Oidiodendron cerealis CCF3491 (Leotiomycetes) | CZE, Bedřichov, tunnel wall, 2004 | ng | This study | |
| Aspergillus askiburgiensis CCF4085 (Eurotiales) | CZE, Bohemian Karst, Malá Amerika mine, WNS positive Myotis myotis, 2010 | LN873940 | 69 | |
| Saprotroph from the outside of underground spaces | Aspergillus luchuensis CCF3984 (Eurotiales) | CZE, Praha, tea bag (Yerba maté ), 2010 | FR727131 | V. Hubka, unpublished |
| A. flavus CCF3154 (Eurotiales) | Brno ČR, black papper, 1999 | ng | ||
| Penicillium oxalicum 2315T (Eurotiales) | USA, soil, Connecticut, 1914 | HE651152 | 70 | |
| Dermatophyte | Microsporum canis CCF3443 (Onygenales) | CZE, Ostrava, human skin, 2003 | ng | |
| Microsporum gypseum CCF3100 (Onygenales) | CZE, Šumperk, human skin, 1998 | ng | ||
| Arthroderma uncinatum CCF2907 (Onygenales) | CZE, Horní Počaply ČR, soil with industrial ash deposits, 1994 | ng | ||
| Trichophyton mentagrophytes CCF3954 (Onygenales) | CZE, Pardubice, human skin, 2009 | ng | ||
| Trichophyton interdigitale CCF4473 (Onygenales) | CZE, tinea corporis, human skin, 2012 | LN736306 | 71 |
CZE Czech Republic.
1ITS rDNA identical with JX270356. “Clade L” sensu1.
2ITS rDNA identical with JX845296. “Clade B” sensu1.
3ITS rDNA identical with JX270432. “Clade J” sensu1.
4ITS rDNA 99% (466/467 bp) similarity with JX270356. “Clade L” sensu1.
5ITS rDNA 99% (570/571 bp) similarity with JX270614. “Clade B” sensu1.
6ITS rDNA 99% (884/894 bp) similarity with JX270621. Phylogenetic position outside of the clades delimited by1.
7ITS rDNA 99% (893/896 bp) similarity with JX270443. “Clade B” sensu1.
8Best BlastN hits (90% for ITS rDNA) are various Botrytis species (e.g. Botrytis cinerea strain CBS 261.71, MH860108).
Table 4.
List of analysed species and used methods.
| Group | Fungal strain | Biolog | Extracellular enzymes | pH test | Conidial viability test |
|---|---|---|---|---|---|
| Pseudogymnoascus destructans | 20631-21T | − | + | + | − |
| CCF3938 | + | − | − | + | |
| CCF3941 | + | + | + | + | |
| CCF3943 | + | + | + | + | |
| CCF4103 | + | + | + | + | |
| CCF4124 | + | − | − | + | |
| CCF4131 | + | − | − | + | |
| CCF4132 | + | − | − | + | |
| CCF4986 | − | + | + | − | |
| CCF4987 | − | + | + | − | |
| Cave fungi | Aspergillus askiburgiensis CCF4085 | + | − | − | − |
| Chrysosporium merdarium CCF6131 | + | − | − | − | |
| Leotiomycetes sp. CCF6130 | + | − | − | − | |
| Metapochonia suchlasporia CCF6128 | + | − | − | − | |
| Myotisia cremea CCF5407 | + | − | − | − | |
| Oidiodendron cerealis CCF3491 | + | − | − | − | |
| Pseudogymnoascus sp. 1 AK51/11 | + | − | + | + | |
| P. sp. 1 CCF5025 | − | + | + | − | |
| P. sp. 2 CCF5027 | − | + | + | − | |
| P. sp. 2 CCF5029 | − | + | + | − | |
| P. sp. 2 CCF5030 | − | + | + | − | |
| P. sp. 2 AK71/11 | + | − | − | + | |
| P. sp. 2 AK 77/11 | + | − | − | + | |
| P. sp. 3 CCF5026 | − | + | + | − | |
| P. sp. 4 AK87/11 | + | − | − | − | |
| Trichophyton terrestre AK44/09 | + | − | − | − | |
| Saprotroph | Aspergillus flavus CCF3154 | − | − | + | + |
| A. luchuensis CCF3984 | − | − | + | + | |
| Penicillium oxalicum 2315 T | − | − | + | − | |
| Dermatophyte | Arthroderma uncinatum CCF2907 | + | − | + | − |
| Microsporum canis CCF3443 | + | − | + | − | |
| M. gypseum CCF3100 | + | − | + | − | |
| T. interdigitale CCF4473 | − | − | + | − | |
| T. mentagrophytes CCF3954 | + | − | + | − |
+/− Analyses done/not done.
Biolog analysis
Biolog MicroPlate for Filamentous Fungi (FF) and Biolog Phenotype Micro-Arrays (PM) (Biolog, Inc., Hayward, CA) were used to evaluate the assimilation profiles of carbon (FF), nitrogen (PM3B), phosphorus, sulphur (PM4A) and nutrient supplements (PM5) according to the manufacturer’s instructions. Fungal conidia from grown cultures were transferred into the inoculating fluid (0.25% Phytagel, 0.03% Tween 40) by rolling a swab across sporulating areas to get the final transmittance of 75 ± 2% or 62 ± 2% for FF and PM, respectively. Inoculated plates were incubated at convenient temperatures as is described in “Fungal material and cultivation” section, and absorbance at 750 nm was used to measure mycelial growth. Two technical replicates per strain were prepared for the FF plates and one replicate was prepared for the PM plates. The isolates analysed differed in their growth rate. For this reason, the last readings before reaching a growth plateau of substrate utilization were used for analysis.
The absorbance of the negative control was subtracted from all substrates within one plate and negative values were assigned a value of zero55. Some substrates were omitted from the analysis due to their low solubility (FF—B1, B3, G2, H10; PM3B—C1, G1; PM4A—A3, B9). Functional diversity was evaluated based on substrate activity (SA), substrate richness (SR)56 and the Shannon index (H)57. In brief, SA was defined as the sum of the optical densities of all substrates on one plate greater than zero. SR is defined as the number of substrates on a microtitre plate that exhibit an optical density greater than the threshold. The optical density of 0.1 was fixed as the threshold for FF and PM3B plates, and the optical density of 0.01 was fixed for PM4A and PM5 plates58.
Extracellular enzyme detection
Lipolytic activities were detected using the rhodamine B assay described by59. Rhodamine B plates were made using the Czapek substrate as the basal medium with the addition of rhodamine B (0.0001% w/v) and olive oil (1%) as a lipid substrate. Inoculated plates were incubated at 4 °C for three weeks, as this temperature is similar to that in bat hibernacula. Fungal colonies with lipolytic activity showed an orange fluorescent halo under UV light.
For collagenase assays, strains were grown as described by60 for two weeks at 4 °C. An adaptation of the double-layer plate assay was employed for the detection of enzymatic activity61. Briefly, Petri dishes were filled with a bottom layer containing 15 ml of 0.7% (w/v) agarose in 50 mM citric acid/sodium citrate buffer (pH 5.5) and 50 mM phosphate buffer (pH 7 and pH 8), and then overlaid with 5 ml of the same media with the addition of 2% w/v collagen. The plates were then inoculated with 10 μl drops from the cultures of the strains in the Czapek medium supplemented with collagen. Undigested collagen was detected by Coomassie brilliant blue G staining60. An identical procedure, in which the collagen was replaced by elastin, was followed to test the production of elastases. Elastin hydrolysis was detected based on the observation of halos62.
Medium alkalization by urea degradation
The ability to alkalize the growth medium by the degradation of urea was investigated by comparison of pH changes during cultivation on a liquid Sabouraud medium (40 g glucose, 10 g pepton, 1 l distilled water) versus a liquid Sabouraud medium supplemented with 50 mM of filter sterilised urea. Erlenmeyer flasks containing 150 ml of the medium were inoculated by fungal strains and pH was measured by a digital pH meter until the maximum value was reached. Non-inoculated sterile media were used as a control. Penicillium oxalicum CCF2315 served as a negative control because of its known acidification ability. Pseudogymnoascus species were cultivated at 15 °C for up to 5 weeks, dermatophytes and outdoor saprotrophs were cultivated at 25 °C for up to 3 weeks.
Conidial survival under stressful conditions
We compared the conidial viability of P. destructans under stressful conditions with that of several non-pathogenic Pseudogymnoascus species living in caves and two air-borne Aspergillus saprotrophic species. The fungi were grown under their optimal growth conditions to attain sufficient conidial production. Petri dishes (one replication for each set of conditions) with well-established fungal cultures were exposed for up to six weeks to ten sets of physical conditions: in the dark at 25 °C, 30 °C, 34 °C and 37 °C; at 25 °C with white light, UVA, UVA plus UVB radiation, and in dryness (dark). UVA and UVB radiation, and white light was administered by a fluorescent tube lamp UV 11 W/G23-DZ (11 W, 350–410 nm), a fluorescent tube lamp Repti Glo 10.0 (20 W, T8, 10% UVB and 33% UVA) and a white light bulb (Philips, LUX BL, 11 W, 230–240 V, ~ 50/60 Hz, 2700 K), respectively. The influence of dryness was tested on conidia obtained by rolling a swab across a well-established culture. Swabs with conidia were then placed into glass jars containing silica gel and sealed with Parafilm to keep humidity below 20%. Conidial viability was evaluated weekly. The next experiment traced conidial viability of P. destructans strains during a 12 h period of repeated temperature switches from 20 to 34 °C for 85 days in the dark, by which we simulated temperature changes during the day and night on the body surface of bats after their departure from a cave, i.e. temperatures of homeothermy and daily torpor during the bats’ active season63.
Evaluation of conidial viability by flow cytometry
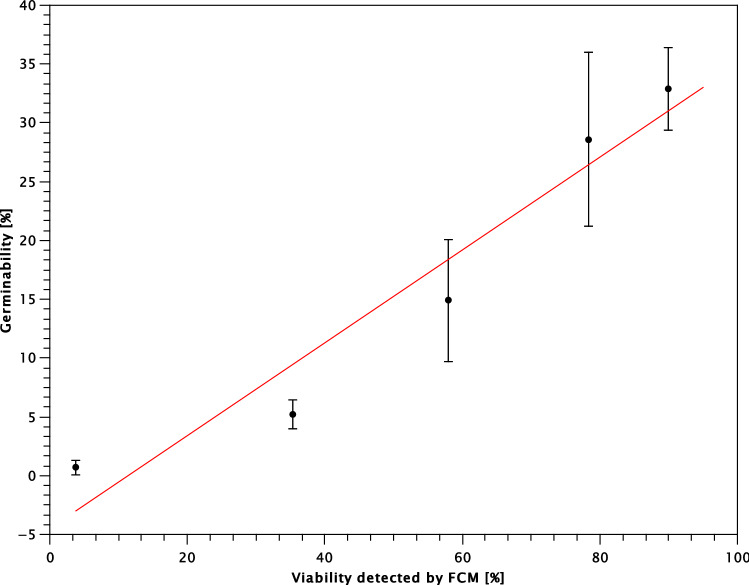
Conidia from stressed cultures were collected using a cotton swab into a 10 × phosphate-buffered saline solution (PBS, 1.370 M NaCl, 27 mM KCl, 0.1 M Na2PO4, 18 mM KH2PO4), pH 7.4, and their concentration was adjusted to 5–7 × 106 conidia/ml. Propidium iodide (PI) was added to attain the final concentration of 2 μg/ml. Samples were incubated with PI for 30 min at room temperature. Three technical replicates per sample were measured using an LSRII (Becton Dickinson, New Jersey, USA) flow cytometer equipped with FACSdiva 6 Software at the Service Centre for Cytometry and Microscopy at the Institute of Microbiology of the Czech Academy of Sciences. Excitation was performed using a Melles Griot 85-YCA-025 laser (23 mW) with a wavelength of 561 nm. Fluorescence was collected through a 590 LP filter and a 610/20 BP filter. Data were analysed in FlowJo 7.6.1 (Tree Star, Inc., Ashland, USA). We verified the precision of this method by testing the viability test of conidia (1) obtained from a non-stressed culture grown under convenient conditions, (2) conidia fixed with 70% ethanol, and (3) a mixture of non-stressed and fixed conidia in various proportions. The mean percentages of viable conidia from untreated and fixed cultures were 90% and 3.8%, respectively. The detected percentages of viable conidia corresponded to the predefined proportions of non-stressed and fixed conidia. We therefore consider flow cytometry with PI staining a suitable method for testing conidial viability. As the viability does not mean germinability, we also cultivated these samples on GYEA for 14 days. Each sample was cultivated at three different conidial concentrations in three replications. Linear Regression Analysis revealed strong correlation between the mean proportions of germinating conidia and proportions of viable conidia detected by FCM (Fig. 5), R2 = 9.3. The proportion of germinated conidia was lower than proportion of viable conidia detected by FCM, only 36% of non-stressed conidia are able to germinate. Thus, P. destructans conidia have relatively low natural germinability or they need specific cultivation conditions for germination. The germination process is a complex result of environmental and physiological factors64. Thus, we cannot foresee the germination efficiency of stressed conidia in nature. However, observed correlation between conidial in vitro germination and conidial viability tests enables insight into approximate germination potential of conidia. Data analysis and gating strategy is presented in Supplementary Figure S1 online.
Figure 5.
Strong correlation between proportion of germinated conidia on GYEA and proportion of viable conidia detected by FCM. Mean values with standard deviations of proportions of germinated conidia are presented. Linear Regression curve in marked in red, R2 = 9.3.
Statistics
Biolog data were visualized using PCA (Principal Component Analysis) in PAST65. The statistical significance of the type of ecology was evaluated by one-way NPMANOVA with Bonferroni-corrected p-values using Bray–Curtis distance and 9,999 permutations. Differences in SA, SR and H between ecological groups were evaluated using Kruskal–Wallis statistics supplemented with Mann–Whitney pairwise comparison and Bonferroni correction. Significance of medium alkalization by urea degradation was computed by permutation test using 9999 permutations.
Supplementary information
Acknowledgements
The study was supported by the Czech Science Foundation Grant no. 17-20286S.
Author contributions
The present study was part of the interdisciplinary project co-ordinated by N.M. and J.P. The study was outlined by M.K. with the help of N.M., J.P and A.K. and supervised by M.K, A.K. and T.V. K.H. performed majority of the experiments with the contribution of P.G.F. The data were analysed and interpreted by T.V., K.H. P.G.F and M.K. The manuscript draft was written by T.V. and revised by all other authors.
Data availability
All primary data are presented in Supplementary material.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-73619-7.
References
- 1.Minnis AM, Lindner DL. Phylogenetic evaluation of geomyces and allies reveals no close relatives of Pseudogymnoascus destructans, comb. Nov., in bat hibernacula of eastern North America. Fungal Biol. 2013;117:638–649. doi: 10.1016/j.funbio.2013.07.001. [DOI] [PubMed] [Google Scholar]
- 2.Blehert DS, et al. Bat white-nose syndrome: An emerging fungal pathogen? Science. 2009;323:227. doi: 10.1126/science.1163874. [DOI] [PubMed] [Google Scholar]
- 3.Chaturvedi V, Chaturvedi S. Editorial: What is in a Name? A proposal to use geomycosis instead of white nose syndrome (WNS) to describe bat infection caused by geomyces destructans. Mycopathologia. 2011;171:231–233. doi: 10.1007/s11046-010-9385-3. [DOI] [PubMed] [Google Scholar]
- 4.Frick WF, Puechmaille SJ, Willis CK. Bats in the Anthropocene: Conservation of Bats in a Changing World. Berlin: Springer; 2016. pp. 245–262. [Google Scholar]
- 5.Bandouchova H, et al. Alterations in the health of hibernating bats under pathogen pressure. Sci. Rep. 2018;8:6067. doi: 10.1038/s41598-018-24461-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zukal J, et al. White-nose syndrome without borders: Pseudogymnoascus destructans infection tolerated in Europe and Palearctic Asia but not in North America. Sci. Rep. 2016;6:19829. doi: 10.1038/srep19829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Drees KP, et al. Phylogenetics of a fungal invasion: Origins and widespread dispersal of white-nose syndrome. mBio. 2017;8:11941–11917. doi: 10.1128/mBio.01941-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leopardi S, Blake D, Puechmaille SJ. White-nose syndrome fungus introduced from Europe to North America. Curr. Biol. 2015;25:217–219. doi: 10.1016/j.cub.2015.01.047. [DOI] [PubMed] [Google Scholar]
- 9.Palmer JM, et al. Molecular characterization of a heterothallic mating system in Pseudogymnoascus destructans, the fungus causing white-nose syndrome of bats. G3 Genes Genomes Genet. 2014;4:1755–1763. doi: 10.1534/g3.114.012641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Trivedi JN. Population genomics and mutational history of the invasive, epidemic clone of Pseudogymnoascus destructans, causal agent of White-nose Syndrome in bats. Toronto: University of Toronto; 2017. [Google Scholar]
- 11.Rajkumar SS, et al. Clonal genotype of Geomyces destructans among bats with white nose syndrome, New York, USA. Emerg. Infect. Dis. 2011;17:1273–1276. doi: 10.3201/eid1707.102056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lorch JM, et al. First detection of bat white-nose syndrome in western North America. mSphere. 2016;1:e00148–e1116. doi: 10.1128/mSphere.00148-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Forsythe A, Giglio V, Asa J, Xu J. Phenotypic divergence along geographic gradients reveals potential for rapid adaptation of the White-nose syndrome pathogen, Pseudogymnoascus destructans, North America. Appl. Environ. Microbiol. 2018;84:e00863–e1818. doi: 10.1128/aem.00863-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khankhet J, et al. Clonal expansion of the Pseudogymnoascus destructans genotype in North America is accompanied by significant variation in phenotypic expression. PLoS ONE. 2014;9:e104684. doi: 10.1371/journal.pone.0104684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cryan PM, Meteyer CU, Boyles JG, Blehert DS. Wing pathology of white-nose syndrome in bats suggests life-threatening disruption of physiology. BMC Biol. 2010;8:1–8. doi: 10.1186/1741-7007-8-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meteyer CU, et al. Histopathologic criteria to confirm white-nose syndrome in bats. J. Vet. Diagn. Invest. 2009;21:411–414. doi: 10.1177/104063870902100401. [DOI] [PubMed] [Google Scholar]
- 17.Pikula J, et al. White-nose syndrome pathology grading in nearctic and palearctic bats. PLoS ONE. 2017;12:e0180435. doi: 10.1371/journal.pone.0180435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Warnecke L, et al. Inoculation of bats with European Geomyces destructans supports the novel pathogen hypothesis for the origin of white-nose syndrome. Proc. Natl. Acad. Sci. 2012;109:6999–7003. doi: 10.1073/pnas.1200374109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Flieger M, et al. Vitamin B2 as a virulence factor in Pseudogymnoascus destructans skin infection. Sci. Rep. 2016;6:33200. doi: 10.1038/srep33200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hayman DTS, Pulliam JRC, Marshall JC, Cryan PM, Webb CT. Environment, host, and fungal traits predict continental-scale white-nose syndrome in bats. Sci. Adv. 2016;2:e1500831. doi: 10.1126/sciadv.1500831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Warnecke, L. et al. Pathophysiology of white-nose syndrome in bats: A mechanistic model linking wing damage to mortality. Vol. 9 (2013). [DOI] [PMC free article] [PubMed]
- 22.Wibbelt G. in Emerging and Epizootic Fungal Infections in Animals. Berlin: Springer; 2018. pp. 289–307. [Google Scholar]
- 23.Achterman RR, White TC. Dermatophyte virulence factors: Identifying and analyzing genes that may contribute to chronic or acute skin infections. Int. J. Microbiol. 2011;20:12. doi: 10.1155/2012/358305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chinnapun D. Virulence factors involved in pathogenicity of dermatophytes. Walailak J. Sci. Technol. (WJST) 2015;12:573–580. [Google Scholar]
- 25.Pannkuk EL, Risch TS, Savary BJ. Isolation and identification of an extracellular subtilisin-like serine protease secreted by the bat pathogen Pseudogymnoascus destructans. PLoS ONE. 2015;10:e0120508. doi: 10.1371/journal.pone.0120508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Raudabaugh DB, Miller AN. Nutritional capability of and substrate suitability for Pseudogymnoascus destructans, the causal agent of bat white-nose syndrome. PLoS ONE. 2013;8:e78300. doi: 10.1371/journal.pone.0078300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mascuch SJ, et al. Direct detection of fungal siderophores on bats with white-nose syndrome via fluorescence microscopy-guided ambient ionization mass spectrometry. PLoS ONE. 2015;10:e0119668. doi: 10.1371/journal.pone.0119668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Burik JAH, Magee PT. Aspects of fungal pathogenesis in humans. Annu. Rev. Microbiol. 2001;55:743–772. doi: 10.1146/annurev.micro.55.1.743. [DOI] [PubMed] [Google Scholar]
- 29.Donaldson ME, et al. Growth medium and incubation temperature alter the Pseudogymnoascus destructans transcriptome: Implications in identifying virulence factors. Mycologia. 2018;110:300–315. doi: 10.1080/00275514.2018.1438223. [DOI] [PubMed] [Google Scholar]
- 30.Field KA, et al. The white-nose syndrome transcriptome: activation of anti-fungal host responses in wing tissue of hibernating little brown Myotis. PLoS Pathog. 2015;11:e1005168. doi: 10.1371/journal.ppat.1005168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reeder SM, et al. Pseudogymnoascus destructans transcriptome changes during white-nose syndrome infections. Virulence. 2017;8:1695–1707. doi: 10.1080/21505594.2017.1342910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lorch JM, et al. Experimental infection of bats with Geomyces destructans causes white-nose syndrome. Nature. 2011;480:376. doi: 10.1038/nature10590. [DOI] [PubMed] [Google Scholar]
- 33.Lorch JM, et al. A culture-based survey of fungi in soil from bat hibernacula in the eastern United States and its implications for detection of Geomyces destructans, the causal agent of bat white-nose syndrome. Mycologia. 2012;105:237–252. doi: 10.3852/12-207. [DOI] [PubMed] [Google Scholar]
- 34.Meyer AD, Stevens DF, Blackwood JC. Predicting bat colony survival under controls targeting multiple transmission routes of white-nose syndrome. J. Theor. Biol. 2016;409:60–69. doi: 10.1016/j.jtbi.2016.08.033. [DOI] [PubMed] [Google Scholar]
- 35.Gargas A, Trest M, Christensen M, Volk TJ, Blehert D. Geomyces destructans sp. nov. associated with bat white-nose syndrome. Mycotaxon. 2009;108:147–154. doi: 10.5248/108.147. [DOI] [Google Scholar]
- 36.Chaturvedi V, et al. Morphological and molecular characterizations of psychrophilic fungus Geomyces destructans from New York bats with white nose syndrome (WNS) PLoS ONE. 2010;5:e10783. doi: 10.1371/journal.pone.0010783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Verant ML, Boyles JG, Waldrep W, Jr, Wibbelt G, Blehert DS. Temperature-dependent growth of Geomyces destructans, the fungus that causes bat white-nose syndrome. PLoS ONE. 2012;7:e46280. doi: 10.1371/journal.pone.0046280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Palmer JM, Drees KP, Foster JT, Lindner DL. Extreme sensitivity to ultraviolet light in the fungal pathogen causing white-nose syndrome of bats. Nat. Commun. 2018;9:35. doi: 10.1038/s41467-017-02441-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Campbell LJ, Walsh DP, Blehert DS, Lorch JM. Long-term survival of Pseudogymnoascus destructans at elevated temperatures. J. Wildlife Dis. 2020;56:278–287. doi: 10.7589/2019-04-106. [DOI] [PubMed] [Google Scholar]
- 40.Reynolds HT, Barton HA. Comparison of the white-nose syndrome agent Pseudogymnoascus destructans to cave-dwelling relatives suggests reduced saprotrophic enzyme activity. PLoS ONE. 2014;9:e86437. doi: 10.1371/journal.pone.0086437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smyth C, Schlesinger S, Overton B, Butchkoski C. The alternative host hypothesis and potential virulence genes in Geomyces destructans. Bat Res. News. 2013;54:17–24. [Google Scholar]
- 42.Chaturvedi V, DeFiglio H, Chaturvedi S. Phenotype profiling of white-nose syndrome pathogen Pseudogymnoascus destructans and closely-related Pseudogymnoascus pannorum reveals metabolic differences underlying fungal lifestyles. F1000Research. 2018;7:2. doi: 10.12688/f1000research.15067.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vanderwolf KJ, Malloch D, McAlpine DF, Forbes GJ. A world review of fungi, yeasts, and slime molds in caves. Int. J. Speleol. 2013;42:9. doi: 10.5038/1827-806X.42.1.9. [DOI] [Google Scholar]
- 44.Wilson MB, Held BW, Freiborg AH, Blanchette RA, Salomon CE. Resource capture and competitive ability of non-pathogenic Pseudogymnoascus spp. and P. destructans, the cause of white-nose syndrome in bats. PLoS ONE. 2017;12:e0178968. doi: 10.1371/journal.pone.0178968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gabriel KT, Neville JJ, Pierce GE, Cornelison CT. Lipolytic activity and the utilization of fatty acids associated with bat sebum by Pseudogymnoascus destructans. Mycopathologia. 2019;184:625–636. doi: 10.1007/s11046-019-00381-4. [DOI] [PubMed] [Google Scholar]
- 46.Park M, Do E, Jung WH. Lipolytic enzymes involved in the virulence of human pathogenic fungi. Mycobiology. 2013;41:67–72. doi: 10.5941/myco.2013.41.2.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Carlini CR, Ligabue-Braun R. Ureases as multifunctional toxic proteins: A review. Toxicon. 2016;110:90–109. doi: 10.1016/j.toxicon.2015.11.020. [DOI] [PubMed] [Google Scholar]
- 48.Cox GM, Mukherjee J, Cole GT, Casadevall A, Perfect JR. Urease as a virulence factor in experimental cryptococcosis. Infect. Immun. 2000;68:443–448. doi: 10.1128/IAI.68.2.443-448.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vylkova S, Lorenz MC. Modulation of phagosomal pH by Candida albicans promotes hyphal morphogenesis and requires Stp2p, a regulator of amino acid transport. PLoS Pathog. 2014;10:e1003995. doi: 10.1371/journal.ppat.1003995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vylkova S. Environmental pH modulation by pathogenic fungi as a strategy to conquer the host. PLoS Pathog. 2017;13:e1006149. doi: 10.1371/journal.ppat.1006149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shawcross DL, et al. Ammonia impairs neutrophil phagocytic function in liver disease. Hepatology. 2008;48:1202–1212. doi: 10.1002/hep.22474. [DOI] [PubMed] [Google Scholar]
- 52.O’Donoghue AJ, et al. Destructin-1 is a collagen-degrading endopeptidase secreted by Pseudogymnoascus destructans, the causative agent of white-nose syndrome. PNAS. 2015;112:7478–7483. doi: 10.1073/pnas.1507082112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Marroquin CM, Lavine JO, Windstam ST. Effect of humidity on development of Pseudogymnoascus destructans, the causal agent of bat white-nose syndrome. Northeastern Nat. 2017;24:54–64. doi: 10.1656/045.024.0105. [DOI] [Google Scholar]
- 54.Kolařík M, et al. Geosmithia associated with bark beetles and woodborers in the western USA: Taxonomic diversity and vector specificity. Mycologia. 2017;109:185–199. doi: 10.1080/00275514.2017.1303861. [DOI] [PubMed] [Google Scholar]
- 55.Garland JL. Analytical approaches to the characterization of samples of microbial communities using patterns of potential C source utilization. Soil Biol. Biochem. 1996;28:213–221. doi: 10.1016/0038-0717(95)00112-3. [DOI] [Google Scholar]
- 56.Dobranic JK, Zak JC. A microtiter plate procedure for evaluating fungal functional diversity. Mycologia. 1999;91:756–765. doi: 10.1080/00275514.1999.12061081. [DOI] [PubMed] [Google Scholar]
- 57.Harch BD, Correll RL, Meech W, Kirkby CA, Pankhurst CE. Using the Gini coefficient with BIOLOG substrate utilisation data to provide an alternative quantitative measure for comparing bacterial soil communities. J. Microbiol. Methods. 1997;30:91–101. doi: 10.1016/s0167-7012(97)00048-1. [DOI] [Google Scholar]
- 58.Sobek EA, Zak JC. The Soil FungiLog procedure: Method and analytical approaches toward understanding fungal functional diversity. Mycologia. 2003;95:590–602. doi: 10.1080/15572536.2004.11833063. [DOI] [PubMed] [Google Scholar]
- 59.Kouker G, Jaeger K-E. Specific and sensitive plate assay for bacterial lipases. Appl. Environ. Microbiol. 1987;53:211–213. doi: 10.1128/AEM.53.1.211-213.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lupan DM, Nziramasanga P. Collagenolytic activity of Coccidioides immitis. Infect. Immun. 1986;51:360–361. doi: 10.1128/IAI.51.1.360-361.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Saleh-Rastin N, Petersen M, Coleman S, Hubbell D. The rhizosphere and plant growth. Berlin: Springer; 1991. pp. 188–188. [Google Scholar]
- 62.NziramasangaM P, Lupan D. Elastase activity of Coccidioides immitis. J. Med. Microbiol. 1985;19:109–114. doi: 10.1099/00222615-19-1-109. [DOI] [PubMed] [Google Scholar]
- 63.Dietz M, Kalko EK. Seasonal changes in daily torpor patterns of free-ranging female and male Daubenton’s bats (Myotis daubentonii) J. Comp. Physiol. B. 2006;176:223–231. doi: 10.1007/s00360-005-0043-x. [DOI] [PubMed] [Google Scholar]
- 64.Sephton-Clark PCS, Voelz K. In: Advances in applied microbiology. Sima S, Geoffrey MG, editors. New York: Academic Press; 2018. pp. 117–157. [DOI] [PubMed] [Google Scholar]
- 65.Hammer O, Harper DAT, Ryan PD. PAST: Paleontological statistics software package for education and data analysis. Palaeontol. Electron. 2001;4:1–9. [Google Scholar]
- 66.Martínková N, et al. Increasing incidence of Geomyces destructans fungus in bats from the Czech Republic and Slovakia. PLoS ONE. 2010;5:e13853. doi: 10.1371/journal.pone.0013853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Větrovský T, Kolařík M, Žifčáková L, Zelenka T, Baldrian P. The rpb2 gene represents a viable alternative molecular marker for the analysis of environmental fungal communities. Mol. Ecol. Resour. 2016;16:388–401. doi: 10.1111/1755-0998.12456. [DOI] [PubMed] [Google Scholar]
- 68.Crous P, et al. Fungal planet description sheets: 558–624. Persoonia. 2017;38:240. doi: 10.3767/003158517X698941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hubka V, et al. A reappraisal of Aspergillus section Nidulantes with descriptions of two new sterigmatocystin-producing species. Plant Syst. Evol. 2016;302:1267–1299. doi: 10.1007/s00606-016-1331-5. [DOI] [Google Scholar]
- 70.Kubátová A, Hujslová M, Frisvad JC, Chudíčková M, Kolařík M. Taxonomic revision of the biotechnologically important species Penicillium oxalicum with the description of two new species from acidic and saline soils. Mycol. Progr. 2019;18:215–228. doi: 10.1007/s11557-018-1420-7. [DOI] [Google Scholar]
- 71.Gabrielová A, et al. The oomycete Pythium oligandrum can suppress and kill the causative agents of dermatophytoses. Mycopathologia. 2018;183:751–764. doi: 10.1007/s11046-018-0277-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All primary data are presented in Supplementary material.