Summary
With mounting interest in translating genome-wide association study (GWAS) hits from large meta-analyses (meta-GWAS) in diverse clinical settings, evaluating their generalizability in target populations is crucial. Here, we consider long-term survivors of childhood cancers from the St. Jude Lifetime Cohort Study, and we show the limited generalizability of 1,376 robust SNP associations reported in the general population across 12 complex anthropometric and cardiometabolic phenotypes (n = 2,231; observed-to-expected replication ratio = 0.70, p = 6.2 × 10−8). An examination of five comparable phenotypes in a second independent cohort of survivors from the Childhood Cancer Survivor Study corroborated the overall limited generalizability of meta-GWAS hits to survivors (n = 4,212; observed-to-expected replication ratio = 0.55, p = 5.6 × 10−15). Finally, in direct comparisons of survivor samples against independent equivalently powered general population samples from the UK Biobank, we consistently observed lower meta-GWAS hit replication rates and poorer polygenic risk score predictive performance in survivor samples for multiple phenotypes. As a possible explanation, we found that meta-GWAS hits were less likely to be replicated in survivors who had been exposed to cancer therapies that are associated with phenotype risk. Examination of complementary DNA methylation data in a subset of survivors revealed that treatment-related methylation patterns at genomic sites linked to meta-GWAS hits may disrupt established genetic signals in survivors.
Keywords: genome-wide association study, replication, polygenic risk score, cardiovascular disease, metabolic disease, childhood cancer survivor, chemotherapy, radiation therapy, DNA methylation, polygenic risk score generalizability
Introduction
Genetic associations reported in recent meta-analyses of genome-wide association studies (meta-GWAS) with large general population study samples (n > 10,000) of predominantly European ancestry have proven to be highly generalizable to other European cohorts.1 For example, an examination of genome-wide significant associations for 32 complex traits across five broad disease groups reported a median replication rate of 84% in a general population cohort with >13,000 individuals of European ancestry.2 As ever-larger meta-analyses continue to corroborate the generalizability of previous GWAS findings across European general population samples and discover novel susceptibility loci, polygenic risk scores (PRS)—typically weighted sums of an individual’s risk alleles at genome-wide significant SNPs identified in the literature—are increasingly viewed as viable genetic predictors of disease risk. PRS based on genome-wide significant SNPs have been shown to improve clinical prediction models for cardiovascular disease risk and have been used to support pharmaceutical interventions to target reductions in low-density lipoprotein (LDL) levels in high-risk individuals.3,4
However, the generalizability of robust genetic associations reported by these large-scale meta-GWAS (hereafter referred to as meta-GWAS hits) to specialized clinical populations has not been established. Given that the clinical utility of genetic risk prediction tools based on published meta-GWAS findings, e.g., PRS, depends on the extent to which these genetic associations are generalizable to target populations, it is imperative to evaluate the generalizability of established meta-GWAS hits in specialized clinical populations. Childhood cancer survivors are one such specialized clinical population that would greatly benefit from genetic predictors of disease risk. Today, approximately one in every 750 individuals is a survivor of childhood or adolescent cancer in the United States.5 This growing population of survivors differs markedly from the general population: studies have consistently shown that survivors are at greater risk for a wide range of serious health conditions earlier in life relative to general population or sibling controls, in part due to their exposures to treatments necessary to cure pediatric cancers;5, 6, 7, 8, 9 this includes greater risk for chronic cardiovascular and metabolic health conditions that are among the leading causes of morbidity and mortality among survivors.6,10, 11, 12, 13
Here, we report on the limited generalizability of 1,376 meta-GWAS hits (p < 5 × 10−8) identified from the literature for 12 anthropometric and cardiometabolic phenotypes to adult survivors of childhood cancer from the St. Jude Lifetime Cohort Study8 (SJLIFE; n = 2,231, European ancestry), a single-institution retrospective cohort study with longitudinal follow-up of survivors with clinically ascertained health outcomes. We evaluated the generalizability of meta-GWAS hits in a second cohort of survivors for five phenotypes available for comparison from the Childhood Cancer Survivor Study (CCSS; n = 4,212, European ancestry), a multi-center study with self-reported health conditions. We also compared meta-GWAS hit replication frequencies and corresponding PRS predictive performance for phenotypes that were evaluable in both SJLIFE and CCSS in equivalently powered independent general population samples. We found that depletions of replicated meta-GWAS hits for some phenotypes were exacerbated in survivor subgroups exposed to certain cancer treatments, particularly when treatments had larger contributions to phenotype variation. Lastly, we conducted ancillary analyses to explore the role of DNA methylation, an epigenetic alteration that is influenced by both inherited genetic variation and environmental factors.14 Among the 236 survivors in SJLIFE with both germline methylome and genotype data, we found that cancer treatments, particularly radiation therapy (RT), may obscure some robust meta-GWAS SNP associations in survivors.
Subjects and Methods
Compiling SNP Associations with Complex Traits and Diseases
We selected 12 complex traits and diseases that were: (A) related to cardiovascular and metabolic disease; (B) ascertained during SJLIFE study visits; and (C) examined in at least one recent (i.e., published after 01/01/2008) meta-GWAS with >10,000 participants of European ancestry. The 12 selected phenotypes included three anthropometric traits (height, body mass index [BMI], and waist-to-hip ratio [WHR]); two blood pressure traits (systolic [SBP] and diastolic [DBP]); four serum lipid traits (high-density lipoprotein levels [HDL], low-density lipoprotein levels [LDL], total cholesterol levels [TC], and triglycerides [TG]); and three cardiometabolic disease outcomes (coronary artery disease [CAD], obesity, and type 2 diabetes [T2D]). For each of the selected phenotypes, we searched all reports available in the NHGRI-EBI GWAS Catalog15 (accessed 11/20/2017). Using the following selection criteria, we retained any study that: (1) investigated single SNP associations with selected phenotypes; (2) included a replication analysis for novel findings; and (3) had discovery and/or replication sample size(s) with >10,000 participants of European ancestry (Figure S1). We reviewed each of the compiled studies to confirm the set of “index SNPs” for replication testing, i.e., published SNPs with genome-wide significant associations (p < 5 × 10−8), and their respective effect sizes, standard errors, p values, and effect alleles. To address the potential effects of “Winner’s curse” on replication power calculations, reported effect sizes and p values for each published SNP association were taken from the combined analysis of discovery and replication samples from the largest current reference meta-GWAS for the phenotype; if combined analysis results were not available, effect sizes were taken from the replication analysis. When necessary, we transformed effect sizes reported in different units across papers for comparability.
Description of Study Cohorts
This study was approved by the Institutional Review Boards at St. Jude Children’s Research Hospital (SJCRH; Memphis, Tennessee) and all participating study centers. All study participants provided informed consent. Brief descriptions of the two cohorts included in our study are provided below.
SJLIFE Cohort
Initiated in 2007, the St. Jude Lifetime Cohort Study16 (SJLIFE) is an ongoing retrospective cohort study dedicated to the longitudinal study of a wide-ranging set of health outcomes in survivors treated for pediatric cancer at SJCRH. The details of this study have been described previously.16 In brief, eligibility criteria include treatment for pediatric cancer at SJCRH and ≥5 years survival since diagnosis. Participants included in the current study were ≥18 years of age, had no history of allogeneic stem cell transplantation, participated in specimen biobanking, and had completed at least one study visit as of June 30, 2015.
SJCRH study visits include medical evaluations (with core laboratory and/or diagnostic studies), assessments of self-reported outcomes, and examinations of neurocognitive function and physical performance. Data for demographics, treatments (chemotherapeutic agent cumulative dosages, field and doses of RT, and surgical interventions), and primary cancer diagnosis were obtained from medical record review. Quantitative trait measurements were taken from the participant’s most recent SJLIFE study visit. Height and weight were measured using a stadiometer and an electronic scale (Scale-Tronix). Waist and hip circumferences were taken with a Gulick tape measure. BMI values were adjusted for amputation, and obesity was defined as BMI ≥ 30 kg/m2. Average SBP and DBP (mmHg) values taken with a calibrated sphygmomanometer after an initial 5-min rest were used for participants with at least two measurements. Fasting blood lipids (mg/dL), including HDL, calculated LDL, TC, and TG, were measured using an enzymatic spectrophotometric assay (Roche Diagnostics). CAD and diabetes mellitus were clinically assessed and graded according to the National Cancer Institute (NCI) Common Terminology Criteria for Adverse Events (CTCAE) v4.03 classification system.17 For CAD, use of medications to treat angina symptoms or evidence of abnormal cardiac enzymes, angina and ischemic heart disease, myocardial infarction, percutaneous transluminal coronary angioplasty (PTCA), or coronary artery bypass grafting (CABG) was used to define cases. Participants with symptomatic diabetes or use of oral medications or insulin to treat diabetes were treated as T2D cases given that >79% of cases in survivors can be classified as T2D.18 Resolved episodes occurring immediately after treatment or pregnancy were excluded.
CCSS Cohort
The Childhood Cancer Survivor Study19 (CCSS) is a retrospective cohort study of 5-year childhood cancer survivors with prospective follow-up. Descriptions for CCSS participant eligibility and study design have been published in detail elsewhere.20,21 CCSS participants included in this analysis were <21 years of age at primary cancer diagnosis between January 1, 1970 and December 31, 1986; received treatment for pediatric cancer at one of 26 participating study institutions in North America; responded to at least one CCSS questionnaire covering demographics, health conditions, health-related behaviors, and health care use; and provided a whole blood, saliva, or buccal sample for DNA sequencing.
All phenotypes assessed in CCSS (height, BMI, obesity, CAD, and T2D) were self-reported or reported by family proxies for survivors who could not complete surveys, were deceased, or were <18 years old. For CAD and T2D phenotypes, questionnaire responses related to these conditions, including relevant medication use, were graded using CTCAE v4.03. Information related to chemotherapy, radiotherapy, and surgery was abstracted from medical records. Participants with height values above or below ±4 standard deviations (SDs) of the sample mean or improbable BMI values (<10, >100 kg/m2) were excluded. All exclusion criteria, adjustment covariates, and case/phenotype definitions were consistent with those applied to the SJLIFE analysis.
Genotype Data
Our analysis was restricted to the common SNPs (≥1% effect allele frequency [EAF]) reported to have a genome-wide significant association (p < 5 × 10−8) with phenotypes in selected meta-GWAS. We also considered best common SNP proxies, or SNPs in high linkage disequilibrium (LD) with corresponding index SNPs in the European 1000 Genomes22 (1000G EUR) populations likely to fall in the same LD block.
SJLIFE Genotype Data
The SJLIFE genotype data used in this analysis were collected as a part of larger effort to sequence whole genomes of SJLIFE participants.23 Comprehensive details of DNA sample collection, extraction, sequencing, quality control, and variant mapping have been described previously.23,24 In brief, sequencing for 3,006 samples was completed at the HudsonAlpha Institute for Biotechnology Genomic Services Laboratory (Huntsville, Alabama) using the Illumina HiSeq X10 platform to yield 150 base pair paired-end reads with an average coverage per sample of 36.8×. Sequenced data were aligned to the GRCh38 human reference using BWA-ALN v0.7.12.25 Variant calls were processed with GATK v3.4.026 and BCFtools.27 PLINK v1.90b28 and VCFtools v0.1.1329 were used to perform additional quality control, applying the following sample exclusion criteria: excess missingness (≥5%), cryptic relatedness (pi-hat > 0.25), and excess heterozygosity (>3 SD). Variants with Hardy Weinberg Equilibrium (HWE) p < 1 × 10−10 and >10% missingness across samples were removed, leaving approximately 84.3 million autosomal single-nucleotide variants (SNVs) and small insertions and deletions (indels) in 2,986 samples. We then restricted our sample to the 2,364 participants that were identified as European (see Ancestry below).
CCSS Genotype Data
Details describing methods used to generate genotype data for the CCSS cohort can be found in previous papers.30,31 To summarize, DNA was extracted from whole blood, saliva, or buccal samples and genotyped at the Cancer Genomics Research Laboratory of the National Cancer Institute (Bethesda, Maryland) using the Illumina HumanOmni5Exome array. Genotyping Module v1.9 (Illumina GenomeStudio software v2011.1) was used to call genotypes. The following per-sample exclusion criteria were applied: ≥8% missingness, heterozygosity <0.11 or >0.16, X chromosome heterozygosity >5.0% for males or <20.0% for females, and identity-by-descent sharing >0.70. Genotypes were then imputed using Minimac332 and the Haplotype Reference Consortium r1.1 reference panel for the 5,739 samples meeting quality control thresholds. After we retained 4,513 survivors of European ancestry (see Ancestry below) with no overlap with SJLIFE, downstream analyses excluded SNPs with minor allele frequency <1% and missingness >5% and only considered SNPs with high imputation quality (r2 ≥ 0.8).
Ancestry
Procedures to identify the ancestry of SJLIFE and CCSS samples have been described elsewhere.24 In brief, PLINK v1.90b was used to perform an EIGENSTRAT-based Principal Component Analysis33 for each cohort by combining the cohort samples with samples from 1000G global reference populations. Cohort samples with principal component scores within 3 SD of the means of the first two principal components in the 1000G EUR populations were considered to be of European ancestry.
SJLIFE DNA Methylation Data
Whole blood DNA methylation was measured in 300 survivors in SJLIFE with a range of treatment histories through the use of the Infinium MethylationEPIC Array (Illumina) according to the manufacturer’s protocols. Genomic DNA (500 ng per sample; previously extracted for whole-genome sequencing) was treated with bisulfate using the Zymo EZ DNA Methylation Kit under the following thermos-cycling conditions: 16 cycles: 95°C for 30 s, 50°C for 1 h. Following bisulfite treatment, DNA samples were desulphonated, column purified, then eluted using 12 μl of elution buffer (Zymo Research). Bisulfite-converted DNA (4 μl) was then processed by following the Illumina Infinium Methylation Assay protocol, which includes hybridization to MethylationEPIC BeadChips, single-base extension assay, and staining and scanning using the Illumina HiScan system. The raw intensity data were exported from the Illumina Genome Studio Methylation Module as IDAT files for further downstream analysis.
Raw intensity data were processed with the “minfi” R package,34 including sample and probe quality controls, background correction, and normalization. Probes were mapped to the GRCh38 build to identify and remove cross-reactive and non-specific probes. We eliminated samples with a low call rate (<95% probes with a detection p value < 0.01) or sex discrepancies, along with probes located on sex chromosomes, with low detection rates (<95%), or with SNPs at CpG sites. A total of 689,742 high-quality probes were retained for 300 samples after preliminary quality control. Data from the BIOS Consortium35 (BIOS QTL) were used to identify significant (false discovery rate [FDR] < 0.05) cis-methylation quantitative trait loci (cis-meQTLs, ≤250 kb between SNP and CpG) linked to index SNPs; of the 15,481 probes in BIOS QTL contributing to significant cis-meQTLs with index SNPs, 11,458 probes were available after quality control.
SNP-Phenotype Association Testing and Replication Enrichment Analysis
We conducted association tests for index SNPs by using phenotype definitions, exclusion criteria, and adjustment covariates that were consistent with the compiled meta-GWAS (Table 1). Linear or logistic regression models were used for association testing using R v3.4.1. All association tests assumed an additive model of genetic inheritance. We used the first 10 principal components as covariates in all association analyses to account for population stratification. SNP-phenotype associations with p values <0.05 and the same direction of effect as the reference literature were considered to be successful replications. While we also evaluated replications under phenotype-specific Bonferroni-corrected p value thresholds, we regarded the p value threshold of 5% as the primary definition for replications.
Table 1.
Summary of Methodological Components for Each SNP-Phenotype Association Analysis in SJLIFE
| Phenotype | Phenotype Transformationa | Unit or Definitiona | GWAS Adjustment Covariatesa | Childhood Cancer Survivor Adjustment Covariatesb | Exclusionsa | Reference Meta-GWASc(PMID) |
|---|---|---|---|---|---|---|
| Anthropometric | ||||||
| Height | sex-standardized Z score | cm | age, ancestry | surgical procedures affecting spinal growth; scoliosis; hypothalamic-pituitary axis tumors; cranial or craniospinal radiation | genetic syndromes, health conditions affecting stature | 25282103, 20881960, 19570815, 19343178, 18391952, 18391951, 18391950 |
| Body mass index (BMI) | inverse normal transformation of residuals | kg/m2; BMI adjusted for amputation | age, age2, sex, ancestry | hypothalamic-pituitary axis tumors; cranial radiation; glucocorticoids | none | 25673413, 24064335, 23669352, 22982992, 20935630, 19079261 |
| Waist-to-hip ratio (WHR) | inverse normal transformation of sex-standardized residuals | ratio of waist and hip circumference (cm) | age, age2, BMI, ancestry | hypothalamic-pituitary axis tumors; cranial radiation; glucocorticoids | none | 28443625, 25673412, 20935629 |
| Blood Pressure | ||||||
| Systolic blood pressure (SBP) | +15 mmHg with use of blood pressure lowering medications | mmHg | age, age2, sex, BMI, ancestry | abdominal, pelvic radiation | prior myocardial infarction or heart failure | 28135244, 28739976, 26390057, 21909115, 19430483, 19430479 |
| Diastolic blood pressure (DBP) | +10 mmHg with use of blood pressure lowering medications | mmHg | age, age2, sex, BMI, ancestry | abdominal, pelvic radiation | prior myocardial infarction or heart failure | same as SBP |
| Blood Lipids | ||||||
| High-density lipoprotein (HDL) | inverse normal transformation of residuals | mg/dL | age, age2, sex, ancestry | hypothalamic-pituitary axis tumors; cranial radiation | use of lipid-lowering medications | 24097068, 19060906 |
| Low-density lipoprotein (LDL) | inverse normal transformation of residuals | mg/dL | age, age2, sex, ancestry | hypothalamic-pituitary axis tumors; cranial radiation | use of lipid-lowering medications | same as HDL |
| Total cholesterol (TC) | inverse normal transformation of residuals | mg/dL | age, age2, sex, ancestry | hypothalamic-pituitary axis tumors; cranial radiation | use of lipid-lowering medications | 24097068 |
| Triglycerides (TG) | inverse normal transformation of residuals | mg/dL | age, age2, sex, ancestry | hypothalamic-pituitary axis tumors; cranial radiation | use of lipid-lowering medications | same as HDL |
| Cardiometabolic Disease | ||||||
| Coronary artery disease (CAD) | none | cases: CTCAE grades ≥ 2 | age, sex, ancestry | BMI; smoking; cardiac-directed radiation; anthracyclines; platinums (cisplatin, carboplatin) | none | 28714975, 26950853, 26343387, 19198609 |
| Type 2 diabetes (T2D) | none | cases: CTCAE grades ≥ 2 | age, sex, BMI, ancestry | cranial radiation; abdominal radiation | none | 28869590, 28566273, 24509480, 20581827, 20418489, 19734900, 18372903 |
| Obesity | none | cases: BMI ≥ 30 kg/m2 | age, sex, ancestry | hypothalamic-pituitary axis tumors; cranial radiation; glucocorticoids | none | 23563607, 21708048 |
Abbreviations: genome-wide association study (GWAS); cm (centimeter); kg (kilogram); m (meter); mmHg (millimeter of mercury); CTCAE (Common Terminology Criteria for Adverse Events, modified v4.03).
Phenotype units and/or definitions and participant exclusion criteria from reference GWAS were reviewed and adapted when necessary for analysis in SJLIFE. GWAS covariates were defined by references.
Covariates specific to childhood cancer survivors, based on the childhood cancer survivorship research literature. Syndromes and health conditions affecting height include: Down syndrome; Turner syndrome; Neurofibromatosis, type 1; Russell-Silver syndrome; benign bone lesion and/or cysts; cartilage disorder; skeletal spine disorder.
Only includes reference GWAS from which summary statistics were compiled, where association statistics for each SNP-phenotype association were taken from the largest, most current study.
In SJLIFE, we also considered whether reported index SNPs were in high LD with potentially “causal” SNP candidates that would better capture the phenotype association at a given LD block. To this end, we tested all best SNP proxies for non-replicated SNP associations, where best proxies for an index SNP were defined as SNPs in strong LD with the index SNP in 1000G EUR (r2 > 0.8) within a 5-kb window of the index SNP (based on a median LD block size of ∼2.5 kb in 1000G EUR).36 Given that non-replication rates from clusters of high-LD SNPs without replication signals could inflate replication depletions, we also assessed replication rates for a pruned set of independent index SNPs (retaining the SNP with the highest EAF among SNPs in high LD or r2 > 0.8 within a 500-kb window in 1000G EUR), as well as a restricted set of SNPs from a single meta-GWAS with the largest sample size.
We used QUANTO v1.2.437 to estimate the power for replicating each reported SNP association in SJLIFE and CCSS. Power calculations assumed either a 5% or Bonferroni-corrected significance threshold, cohort sample sizes and case-control ratios, and an additive genetic model. Phenotype-specific power curves for our main analysis accounting for a range of effect allele frequencies and effect sizes are provided in Figures S2–S5. We used these power calculations to estimate the replication power for each SNP-phenotype association, assuming effect sizes in reference GWAS and the effect allele frequency observed in the survivor cohorts. We used the same procedure to also estimate replication power for each SNP-phenotype association in treatment-exposed and treatment-unexposed subsamples in SJLIFE, where treatment exposure was defined as any exposure to one or more curative agents for pediatric cancer previously associated with the specific phenotype (treatments listed in Table 1).
In order to evaluate whether the observed replication frequencies were greater or less than expected, we used a Poisson generalized estimating equations (GEE) regression approach with robust variance estimation.38 We estimated the expected number of replications for each phenotype based on the assumption that each SNP replication may be treated as a Bernoulli random variable with a replication probability equal to its estimated replication power, and under Le Cam’s theorem,39 the sum of independent Bernoulli variables that are not identically distributed approximately follows a Poisson distribution. The model assumed a log-link of the following form:
where and were observed replications and the expected replication probability, respectively. The exponentiated estimate served as the replication enrichment ratio (RER), or the ratio of observed to expected replication frequencies.
Although we computed phenotype-specific RERs in SJLIFE and CCSS separately, we also calculated combined cohort RERs by combining association test results for the evaluable phenotypes in both studies by using the fixed-effects inverse variance-weighted meta-analysis method implemented in METAL.40
Comparison of RERs and PRS Predictive Performance in Comparably Powered General Population Samples
We evaluated meta-GWAS hit replication rates and corresponding RER estimates in comparably powered general population samples for four distinct phenotypes (height, obesity, T2D, and CAD) that were evaluable in both SJLIFE and CCSS, using data from the UK Biobank (UKBB), an extensive genetic and phenotypic database with ∼500,000 individuals across the United Kingdom aged 40–69 years at recruitment.41 Specifically, we restricted analyses to 207,604 participants of White British genetic ancestry in the UKBB phase 2 genotype data release, because these participants had no overlap with SJLIFE or CCSS or with samples in compiled reference meta-GWAS. Determination of genetic ancestry and genotype imputation using the Haplotype Reference Consortium, UK10K, and 1000 Genomes panels for UKBB data were performed centrally.41 As previously reported,41 additional exclusions for excess heterozygosity, genotype missingness, sex discordance, putative sex chromosome aneuploidy, or withdrawal of informed consent were applied. Nearly all (∼99%) SNPs were available for comparison in the UKBB data, and all evaluated SNPs were of high imputation quality (INFO > 0.8). Phenotype ascertainment in the UKBB data consisted of: (A) measured height; (B) measured BMI to define obesity controls and cases (BMI ≥ 30 kg/m2); (C) self-reported diabetes to define T2D cases; and (D) algorithmically defined myocardial infarction to define CAD cases based on self-report or hospitalization or death records, including ICD-9 codes 410.x, 411.0, 411.1, 411.8, 412.x, or 429.79 or ICD-10 codes I21.x, I22.x, I23.x, I24.1, or I25.2.
For comparably powered UKBB samples, we took 100 random samples from the UKBB release 2 data for each phenotype that had: (1) equal sample sizes (and equal case-control ratios for disease phenotypes) to SJLIFE and CCSS cohorts; and (2) <1% absolute difference in the median EAF between GWAS-reported EAFs and sample EAFs. We then conducted power calculations and association testing using the same methods (e.g., phenotype transformations, statistical models) applied in the survivor cohorts (Table 1) for each “pseudo-survivor” sample drawn from the UKBB data. Median and interquartile range (IQR) statistics were examined for the UKBB pseudo-survivor sample RERs. To calculate phenotype-specific PRS in the UKBB samples and SJLIFE and CCSS cohorts that could be compared to sample RERs, we used summary statistics for genome-wide significant SNPs included in our primary analysis that were reported in the largest current study (n > 100K) among the reference meta-GWAS for height,42 obesity,43 T2D,44 and CAD45 to generate a 358-SNP polygenic risk score for height, a 61-SNP score for obesity, a 63-SNP score for T2D, and a 52-SNP score for CAD using PLINK v1.90b.28 To facilitate cross-study comparisons of PRS performance, we examined the mean change in the SD unit or log(odds ratio [OR]) per unit increase in PRS as a predictive performance metric.
Ancillary Analyses: Epigenetic and Functional Annotation Enrichments
We evaluated external epigenetic and functional annotations for index SNPs by using resources provided by the Roadmap Epigenomics Mapping Consortium46 (REMC), the Genotype-Tissue Expression Project47 (GTEx Analysis v7), Reactome,48 and BIOS QTL.35 For each of 127 cell types or cell lines, we compared the frequency of enhancer/promoter state overlap (from 15-state ChromHMM) in the set of SNPs with replicated associations (“replicated SNPs”) against the SNPs without replicated associations (“non-replicated SNPs”) in our SJLIFE main analysis with two-sided Fisher’s exact tests. Using GTEx, we counted the number of significant cis-expression quantitative trait loci (cis-eQTLs; SNPs within ±1 Mb of transcription start sites, FDR ≤ 0.05) for replicated SNPs and non-replicated SNPs and used a two-sided Fisher’s exact test to investigate enrichments in gene expressions among replicated SNPs for each of the 48 available cell or tissue types. Lastly, we compiled non-overlapping gene sets for replicated and non-replicated SNPs to conduct a biological pathway enrichment analysis with geneSCF v1.149 and Reactome gene pathway ontologies. Genes were mapped to SNPs based on co-location in gene bodies defined by RefSeq50 gene models. For each biological pathway, the number of genes in replicated and non-replicated SNP groups with a specific ontology was compared to the number of genes with the same ontology in all remaining genes in the genome. Top biological pathway enrichments were determined using FDR-adjusted p values from two-sided Fisher’s exact tests.
Cis-meQTLs and Treatment-Methylation Associations
DNA methylation at specific CpG sites has been linked to both GWAS-identified disease variants35 and many complex traits and diseases.51 As such, significant (FDR < 5%) BIOS QTL cis-meQTLs may reflect molecular mechanisms contributing to phenotypes. We validated significant (FDR < 5%) cis-meQTLs reported in BIOS QTL for compiled SNPs in SJLIFE participants with methylation and genotype data by testing associations between methylation M-values (log2-transformed ratio of the methylated to unmethylated probe intensities) at quality-controlled CpG sites and SNP genotypes assuming an additive inheritance model using linear regression, adjusting for sex, age, and genetic ancestry. Because additional analyses to evaluate potential confounding by inter-individual differences in blood cell composition revealed no significant differences in cell type distributions across samples, no adjustment covariates for blood cell composition were considered. Established cis-meQTLs (i.e., BIOS QTL with FDR < 5%) were considered validated in SJLIFE if associations had p < 0.05 and the same direction of allelic effect.
Recent studies have also shown that cancer therapies can induce persistent changes in DNA methylation in diverse cells and tissues.52, 53, 54, 55, 56, 57, 58 The set of BIOS QTL cis-meQTLs validated in SJLIFE survivors effectively nominates meta-GWAS SNPs and corresponding CpGs that would hypothetically be more likely to employ a DNA methylation mechanism to contribute to phenotypes in survivors, and as a consequence, identifies SNPs and corresponding CpGs that are plausible targets for modifying treatment-methylation effects. Using two-sided Fisher’s exact tests, we tested for enrichment of validated cis-meQTLs, first among non-replicated SNPs and then among groups of SNPs identified a priori as “treatment-sensitive” (not replicated in our main analysis, but replicated in samples without treatment exposures) and “treatment-insensitive” (replicated in treatment-unexposed and treatment-exposed samples).
After aligning the directionality of cis-meQTLs reported in BIOS QTL with GWAS-reported allelic effects on phenotypes for each SNP, we considered observations of treatment-methylation associations with directions of effect that were discordant with SJLIFE-validated cis-meQTL associations (p < 0.05) as potential indicators of disrupted cis-meQTL effects on phenotypes in survivors. We examined directionally discordant cis-meQTLs and treatment-methylation associations for CpGs linked to non-replicated SNPs (“non-replicated CpGs”) and replicated SNPs (“replicated CpGs”) for the cis-meQTLs we validated in SJLIFE. Among the eight treatment types we considered (cranial, chest, abdominal, and pelvic radiotherapies; anthracycline, corticosteroid, cisplatin, and carboplatin chemotherapies), we limited our analysis to seven treatment types where >5% of the experimental sample was exposed. To ascertain the direction of cis-meQTLs at CpGs with multiple associated SNPs without arbitrarily assigning a “best” cis-meQTL (i.e., smallest p value), we used simple majority voting classification to determine the direction of the cis-meQTL set for such CpGs. For each treatment type, treatment dose associations with M-values at CpGs contributing to SJLIFE-validated cis-meQTLs were tested with linear regression, adjusting for age and sex. We compared the discordance between directions of cis-meQTLs and treatment-methylation associations among replicated and non-replicated CpGs using a two-sided Fisher’s exact test. For additional details, see Supplemental Methods.
Results
Meta-GWAS Hits Show Limited Generalizability to Childhood Cancer Survivors
Using the National Human Genome Research Institute (NHGRI) and European Bioinformatics Institute (EBI) GWAS Catalog,15 we identified 149 GWAS for 12 anthropometric and cardiometabolic phenotypes, including height, BMI, WHR, blood pressure (SBP, DBP), blood lipid levels (HDL, LDL, TC, TG), obesity, CAD, and T2D. After reviewing the literature against criteria for relevance, ancestry, and study suitability, we compiled 1,415 genome-wide significant (p < 5 × 10−8) SNP-phenotype associations from 46 selected GWAS featuring meta-analyses with replication studies that included >10,000 participants of predominantly European ancestry (Figure S1). We limited our analysis to the 1,376 SNP-phenotype associations (97.2%) that could be directly tested using 1,231 quality-controlled SNPs measured in SJLIFE. Of these, 70.4% (969 SNP-phenotype associations) were from meta-analyses with n > 100,000.
Using the phenotype definitions, statistical models, and exclusion criteria described in reference GWAS (Table 1), we primarily aimed to replicate the 1,376 robust meta-GWAS hits in 2,231 adult long-term (≥5-year) survivors of childhood cancer of European ancestry in SJLIFE. Relevant descriptive statistics for the SJLIFE cohort are provided in Table 2. Most survivors had been exposed to at least one type of chemotherapeutic agent (85.3%) and over half (58.3%) had received RT. There was high correspondence between EAFs reported in the reference GWAS and SJLIFE, with a median absolute difference of 0.99% (IQR = 0.47%–1.71%).
Table 2.
Descriptive Statistics for Phenotypes, Treatments, and Demographic Variables in SJLIFE and CCSS
| Phenotypes/Variables | Unit | SJLIFE n | SJLIFE % or Median (IQR) | CCSS n | CCSS % or Median (IQR) |
|---|---|---|---|---|---|
| Demographic Variables | |||||
| Sex | |||||
| Male | % | 2,231 | 53.0% | 4,513 | 48.1% |
| Female | % | 2,231 | 47.0% | 4,513 | 51.9% |
| Age | years | 2,231 | 35.8 (13.3) | 4,513 | 40.9 (12.9) |
| Treatments (any exposure) | |||||
| Radiation, any type | % | 2,231 | 58.3% | 4,513 | 61.9% |
| Chemotherapeutic agent, any type | % | 2,231 | 85.3% | 4,513 | 73.9% |
| Cranial radiation | % | 2,199 | 31.0% | 4,227 | 30.9% |
| Cardiac-directed radiation | % | 2,199 | 22.9% | 4,224 | 26.7% |
| Abdominal radiation | % | 2,199 | 20.0% | 4,226 | 25.9% |
| Pelvic radiation | % | 2,199 | 17.5% | 4,226 | 20.5% |
| Anthracyclines | % | 2,231 | 57.9% | 4,290 | 35.8% |
| Glucocorticoids | % | 2,231 | 47.8% | 4,513 | 43.4% |
| Platinums (cisplatin, carboplatin) | % | 2,227 | 10.3% | 4,513 | 4.4% |
| Phenotypes | |||||
| Anthropometric | |||||
| Height | cm | 2,025 | 168.7 (14.6) | 4,212 | 168.0 (18.0) |
| Body mass index | kg/m2 | 2,229 | 27.6 (9.3) | 4,208 | 26.1 (7.3) |
| Waist-to-hip ratio | ratio | 2,204 | 0.9 (0.1) | – | – |
| Blood Pressure | |||||
| Systolic blood pressure | mmHg | 2,020 | 123.0 (17.7) | – | – |
| Diastolic blood pressure | mmHg | 2,020 | 75.5 (13.0) | – | – |
| Serum Lipids | |||||
| High-density lipoprotein | mg/dL | 1,984 | 49.0 (20.0) | – | – |
| Low-density lipoprotein | mg/dL | 1,964 | 107.0 (46.0) | – | – |
| Total cholesterol | mg/dL | 1,997 | 183.0 (50.0) | – | – |
| Triglycerides | mg/dL | 1,997 | 100.0 (80.0) | – | – |
| Cardiometabolic Disease | |||||
| Coronary artery disease | % cases | 2,079 | 4.7% | 4,036 | 4.1% |
| Obesity | % cases | 2,229 | 38.3% | 4,208 | 25.8% |
| Type 2 diabetes | % cases | 2,112 | 7.1% | 4,207 | 7.0% |
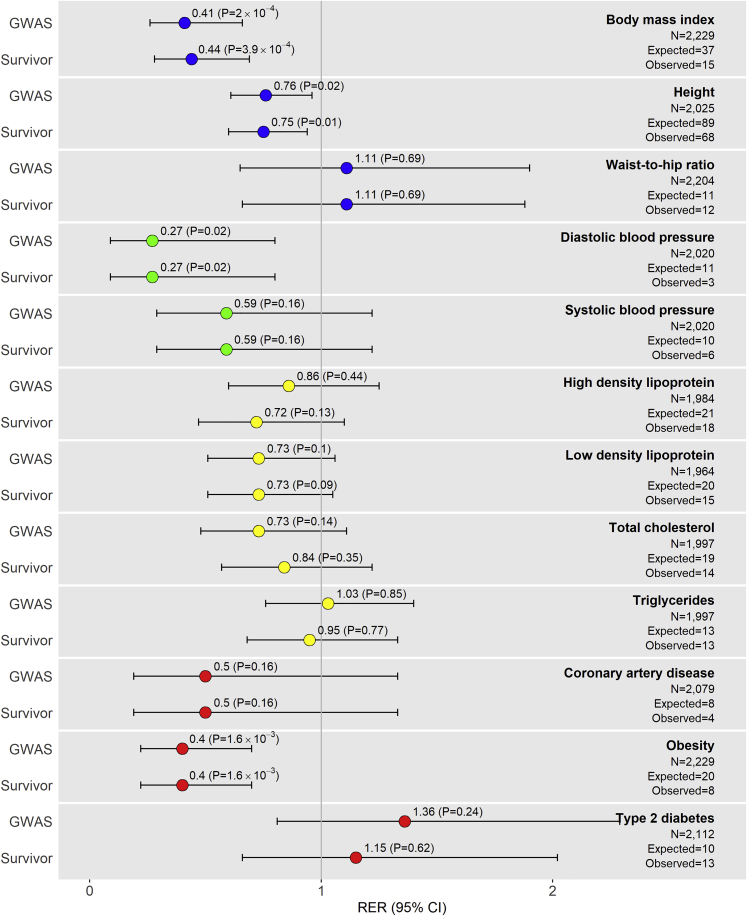
Of the 1,376 meta-GWAS hits, we expected to replicate ∼268 SNP-phenotype associations across all phenotypes based on power (replication was defined by association test p < 0.05, with same directions of effect in literature). We replicated 189 SNP-phenotype associations (replication rate = 13.7%) with models adhering to reference GWAS, and 185 SNP-phenotype associations (replication rate = 13.4%) after adjusting for additional covariates relevant to survivors (i.e., cancer treatment exposures, Table 1). All SJLIFE replication results are listed in Table S1. The RER (the ratio of observed-to-expected meta-GWAS hit replication frequencies) across all 12 phenotypes was 0.70 (95% confidence interval [CI]: 0.62–0.80, p = 6.2 × 10−8) when we used models adjusting for reference GWAS covariates only, indicating that the overall number of meta-GWAS hit replications observed in SJLIFE was significantly less than expected (Table S2). Significant replication depletion was also observed across all phenotypes when we used models adjusting for additional covariates relevant to survivors (RER = 0.69, 95% CI: 0.61–0.78, p = 1.2 × 10−8). While three phenotypes (WHR, T2D, TG) showed no evidence of replication depletion (RER > 1), the remaining nine phenotypes had either significant depletions of meta-GWAS hit replications (RER < 1 and p < 0.05 for height, BMI, DBP, and obesity) or suggestive evidence of replication depletions (RER < 1 and p < 0.2 for SBP, HDL, LDL, TC, and CAD) (Figure 1, Table S2).
Figure 1.
Plots of Replication Enrichment Ratios and Respective 95% Confidence Intervals by Phenotype in SJLIFE
Replication enrichment ratios (RERs) left of the vertical line corresponding to RER = 1 suggest meta-GWAS hit replication depletion, i.e., observations of fewer replications of meta-GWAS hits than expected. RERs considering adjustment covariates under two different models are presented for each phenotype: (1) covariates adhering to reference GWAS (“GWAS”) and (2) GWAS covariates along with covariates considered in childhood cancer survivor populations (“Survivor”). Phenotype RERs are color coded based on similarity: anthropometric (blue), blood pressure (green), lipid (yellow), and cardiometabolic disease (red). The observed numbers of replications included in the figure are under the “GWAS” model (Table 1). The expected numbers of replications are estimated based on the sum of the power to replicate each SNP-phenotype association assuming observed SNP effect allele frequencies, the cohort sample size, an additive genetic inheritance model, a = 0.05, and effect sizes in reference meta-GWAS.
Robustness of the Limited Meta-GWAS Hit Generalizability Finding in Survivors
We explored several alternative evaluation strategies. First, we examined an “extended” replication strategy, under the scenario in which all 1,187 non-replicated robust meta-GWAS hits are weak representatives for nearby causal variants but are in high LD with causal variants in the same LD block. We re-tested non-replicated meta-GWAS hits by using best SNP proxies for reported index SNPs, where best proxies were defined as SNPs in high LD with the index SNP (r2 > 0.8 in 1000G EUR) that were likely to fall in the same LD block.36 Although we re-tested 812 non-replicated SNP associations that each had at least one plausible proxy (median = three proxies per index SNP), only 12 additional meta-GWAS hits were replicated (overall RER = 0.75, 95% CI: 0.66–0.85, p = 4.1 × 10−6) (Table S3). In order to avoid bias in replication rate estimates, we also assessed replication rates for a set of independent SNP-phenotype associations by limiting the SNP set to those with the highest EAF in SJLIFE among clusters of SNPs in high LD (r2 > 0.8, 500-kb window in 1000G EUR) for each phenotype. The same nine phenotypes continued to show significant or suggestive replication depletion when we used the pruned SNP-phenotype associations (Table S4). We further restricted the set of evaluated SNP-phenotype associations to those reported from the single largest meta-analysis (all with n > 100K) for a given phenotype, and we continued to observe significantly fewer replications than expected (overall RER = 0.77, 95% CI: 0.66–0.90, p = 8.8 × 10−4) (Table S5). Finally, we examined replications of meta-GWAS hits under strict replication p value thresholds corrected for multiple testing. Although replication of ∼47 SNP-phenotype associations was expected under Bonferroni-corrected p value thresholds, only 25 SNP-phenotype associations were replicated when we used more stringent p value thresholds (Table S6).
Confirmation of the Limited Generalizability of Meta-GWAS Hits in a Second Independent Cohort of Survivors
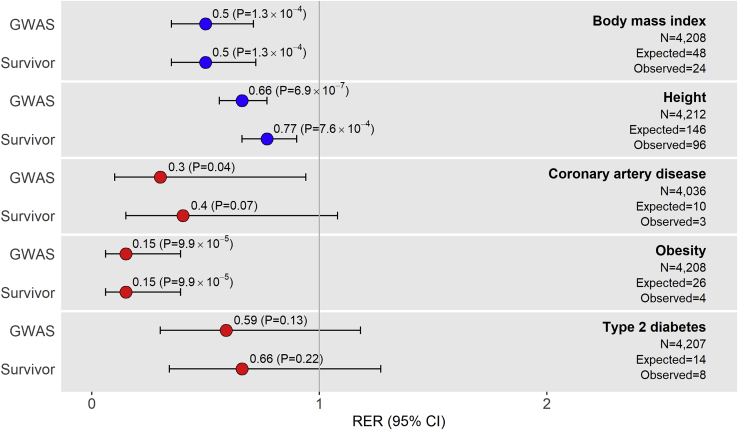
To assess our findings from SJLIFE in an independent cohort, we conducted a second analysis in survivors from CCSS. We examined five self-reported phenotypes available in CCSS that corresponded with our SJLIFE analysis (height, BMI, CAD, obesity, and T2D) in 4,513 survivors whose genotype data was available. Descriptive statistics for the CCSS study sample are provided in Table 2. Similar to those in the SJLIFE study, most CCSS survivors had been exposed to at least one type of chemotherapeutic agent (73.9%) or RT (61.9%). With power calculations for replication accounting for CCSS sample sizes and EAFs, we expected to replicate ∼244 meta-GWAS hits. A total of 135 SNP-phenotype associations were successfully replicated in CCSS survivors with complete genotype, phenotype, and covariate data (up to n = 4,212) when we used models adhering to reference GWAS. All five phenotypes showed significant (p < 0.05) or suggestive (p < 0.2) meta-GWAS hit replication depletions beyond what was expected (Figure 2, Table S2), for an overall RER of 0.55 (p = 5.6 × 10−15). RERs based on meta-analysis of SJLIFE and CCSS results revealed similar trends (Table S7).
Figure 2.
Plots of Replication Enrichment Ratios and Respective 95% Confidence Intervals by Phenotype in CCSS
Replication enrichment ratios (RERs) left of the vertical line corresponding to RER = 1 suggest meta-GWAS hit replication depletion, i.e., observations of fewer replications of meta-GWAS hits than expected. Phenotype RERs are color coded based on similarity: anthropometric (blue) and cardiometabolic disease (red). The observed numbers of replications included in the figure are under the “GWAS” model (Table 1).
Comparably Powered General Population Samples Show Larger Meta-GWAS RERs and Better PRS Performance Than Survivor Samples Show
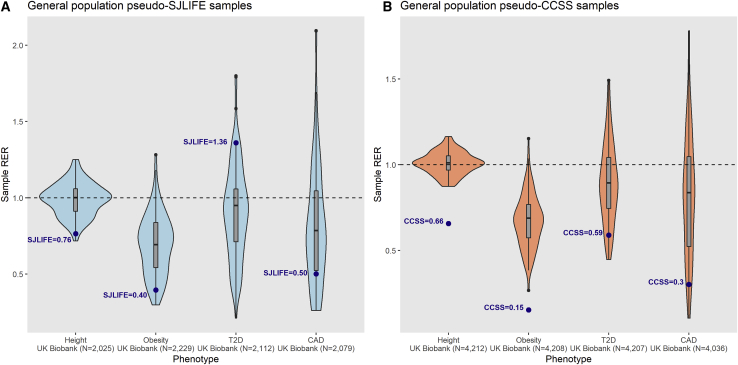
To directly compare meta-GWAS hit replication rates and RER estimates in independent general population samples against survivor cohort estimates, we used 100 random “pseudo-survivor” general population subsamples drawn from the UKBB that: (A) did not overlap with reference meta-GWAS, (B) had equal sample sizes and case-control ratios to those of SJLIFE and CCSS samples, and (C) were evaluated with identical models and phenotype transformations applied in survivor analyses for four distinct phenotypes (height, obesity, T2D, and CAD). Excluding T2D associations in pseudo-SJLIFE samples (where diabetes was clinically ascertained in SJLIFE versus self-reported in CCSS and UKBB), we found that the median replication frequency was ∼1.3–4.5-fold higher in the comparably powered UKBB samples (Table S8), and corresponding survivor cohort RERs fell below first-quartile UKBB RERs for all phenotypes (Figure 3). RER IQRs in UKBB samples overlapped RER = 1 (indicating observed replications equal to those expected) for all phenotypes except obesity.
Figure 3.
Comparison of Sample Replication Enrichment Ratios for Four Complex Phenotypes in Equivalently Powered General Population Samples
Using comparably powered samples drawn from the UK Biobank (UKBB) with no overlap with original reference GWAS, we tested associations among the same set of SNPs evaluated in the survivor cohort analyses for four phenotypes: height, obesity, type 2 diabetes (T2D), and coronary artery disease (CAD). Violin plots show distributions of phenotype-specific sample replication enrichment ratios (RERs) with power calculations based on the same set of reference GWAS summary statistics for 100 samplings of two general population sample types: “pseudo-SJLIFE” (A) and “pseudo-CCSS” (B), which observe the same samples sizes and case-control ratios analyzed in the respective survivor cohorts. Box and whisker plots for sample RER medians (vertical line inside box) and the lower and upper quartiles (bottom and top of box, respectively) are shown, with whiskers extending to highest and lowest estimates. The dashed horizontal line representing RER = 1 signifies that observed replications are equal to expected. Blue dots represent RER estimates observed in SJLIFE (A) and CCSS (B) cohorts.
We also evaluated the predictive performance of PRS (i.e., mean change in the SD unit or log[OR] per PRS unit increase) derived from the genome-wide significant SNPs assessed in our primary analysis in the same set of UKBB pseudo-survivor samples for height, obesity, T2D, and CAD. We observed that increases in RER estimates in UKBB samples, reflecting increasing numbers of meta-GWAS replications, were strongly associated with improved PRS performance for all phenotypes (p < 6.1 × 10−6) (Figure S6, Figure S7). Consistent with observations of smaller phenotype-specific RERs in survivor cohorts (excluding SJLIFE T2D), PRS performance was worse in survivor cohorts than in the UKBB samples for all phenotype comparisons (Figure S6, Figure S7). In particular, the median predictive performances of a 358-SNP PRS for height and 52-SNP PRS for CAD across UKBB pseudo-SJLIFE samples were ∼1.3-fold and ∼1.8-fold greater than in SJLIFE, respectively; in UKBB pseudo-CCSS samples, the median predictive performances of phenotype-specific PRS were ∼1.3 to ∼2.1-fold greater compared to those in CCSS.
The Generalizability of Meta-GWAS Hits in Survivors Differs by Phenotype-Relevant Functional and Regulatory Genomic Annotations
We speculated that meta-GWAS SNPs with replicated phenotype associations in survivors could have functional and/or epigenetic annotation enrichments that may distinguish them from SNPs with non-replicated associations. Using publicly available gene expression data from GTEx47 and REMC chromatin state annotations,46 we compared the set of 170 SNPs with at least one replicated association with the 12 phenotypes (“replicated SNPs”) against the set of 1,061 SNPs without any replicated associations (“non-replicated SNPs”) from our main analysis in SJLIFE. Similar proportions of replicated and non-replicated SNPs were mapped to RefSeq50 gene bodies (57.1% versus 58.7%, respectively; p = 0.74). However, replicated SNPs had greater odds of being a cis-eQTL SNP (FDR ≤ 0.05) in adipose and liver tissues than non-replicated SNPs had (nominal p < 0.05, Table S9). Top 15-state ChromHMM46 enhancer and promoter chromatin state annotation enrichments revealed that replicated SNPs also had greater odds of overlapping enhancer chromatin states in cell or tissue types related to the kidney, adipose, gut, and obesity-linked brain structures (nominal p < 0.05, Table S10). We also assessed top Reactome48 biological pathway enrichments for non-overlapping genes mapped to replicated and non-replicated SNPs against all other genes in the human genome (Figure S8). For the 79 genes that corresponded with the replicated SNPs, the lead biological pathway enrichments (FDR < 0.10) were more specific to cardiometabolic phenotypes (e.g., plasma lipoprotein metabolism is connected to serum lipid traits, elastic fiber assembly is related to arterial wall formation, and peroxisome proliferator-activated receptor alpha [PPAR-alpha]-mediated lipid metabolism is linked to metabolic phenotypes). In contrast, the vast majority of leading biological pathway enrichments (FDR < 0.10) for the 466 genes mapped to non-replicated SNPs were related to signal transduction.
Exposures to Treatments for Pediatric Cancer May Underlie the Limited Generalizability of Meta-GWAS Hits in Survivors
We assessed whether risk factors implicated in a range of late effects in long-term survivors, i.e., exposure to specific cancer treatments or age at cancer diagnosis (treatment), could “disrupt” robust genetic associations reported in the general population. We estimated RERs in SJLIFE survivor subgroups stratified either by treatment exposure (defined as any exposure to therapeutic agents for pediatric cancer associated with the phenotype of interest [all therapies described in Table 1]) or by age at diagnosis older or younger than the median (∼7 years). We hypothesized that if these factors contribute to phenotypic variation and distort meta-GWAS SNP-phenotype associations in survivors, the magnitude of replication would be greater in the survivor subgroup that was more similar to the general population (i.e., treatment-unexposed and older age at diagnosis).
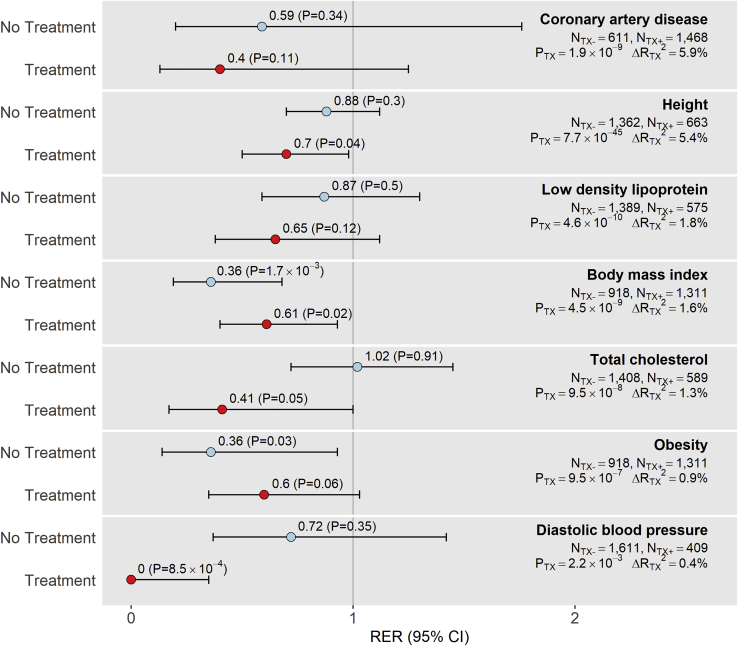
We found evidence of replication depletion in treatment-exposed survivor subgroups for seven phenotypes: the height, BMI, TC, and DBP phenotypes showed significant (p < 0.05) replication depletion, while CAD, LDL, and obesity phenotypes showed suggestive (p < 0.2) replication depletion. Among these seven phenotypes, CAD, height, LDL, TC, and DBP showed stronger evidence of replication depletion than expected in treatment-exposed subgroups compared to treatment-unexposed subgroups (Figure 4). For example, whereas replication power in treatment-unexposed subgroups for height, LDL, and TC was ∼32%–38% higher compared to replication power in treatment-exposed subgroups, replication frequencies were ∼85%–300% higher in the treatment-unexposed subgroups. Similarly, CAD meta-GWAS hit replications in the treatment-unexposed subgroup were equivalent to those in the treatment-exposed group despite having ∼45% lower power for replication. CAD, height, LDL, and TC also showed the greatest incremental changes in variance explained (change in adjusted R2 > 1%) when we compared clinical models with and without treatments and had the strongest treatment likelihood ratio test p values (p < 1 × 10−7). These results suggest that replication depletions in meta-GWAS hits are exacerbated in survivors when treatments have greater contributions to the phenotype risk. In comparison, we found the RERs stratified by median age at diagnosis to be similar, and replication frequencies were not affected to the same degree (Table S11).
Figure 4.
Plots of Phenotype-Specific Replication Enrichment Ratios and Respective 95% Confidence Intervals in SJLIFE Samples Unexposed or Exposed to Treatments
Treatments were defined as cancer treatments associated with phenotypes. Phenotypes with any evidence of replication depletion (RER < 1) in our main analysis that showed either significant (p < 0.05) or suggestive (p < 0.2) replication depletion in SJLIFE treatment-exposed samples are included in this figure. Sample sizes by exposure strata (NTX-, No Treatment; NTX+, Treatment) are provided, as well as likelihood ratio test p values representing treatment associations with phenotypes (PTX) and incremental changes in adjusted R2 (ΔRTX2) after removing treatment variables from clinical models. Phenotypes are ordered based on ΔRTX2 values, with larger ΔRTX2 values reflecting greater treatment influence on phenotype variation.
DNA Methylation as a Mechanism for Cancer Treatment Exposures to Limit the Generalizability of Meta-GWAS Hits in Survivors
Because BIOS QTL35 includes samples from the Lifelines Cohort Study (which recently reported a median meta-GWAS hit replication rate of 84% across 32 phenotypes2), we used BIOS QTL35 meQTL data as a reference resource for ancillary DNA methylation analyses. Whole blood cis-meQTLs from BIOS QTL for any of the 1,231 meta-GWAS SNPs of interest (FDR < 0.05) were regarded as established phenotype-variant-associated cis-meQTLs. Most of the meta-GWAS SNPs examined in our SJLIFE main analysis (87.5%, 1,077 SNPs) were mapped to at least one established cis-meQTL (Table S12).
First, we assessed whether established cis-meQTLs in the general population (BIOS QTL) could be generalized to childhood cancer survivors by using a subset of SJLIFE survivors in our main analysis with blood-derived methylome and genotype data (n = 236). We successfully validated 5,651 established cis-meQTLs for the meta-GWAS SNPs of interest (40.6%; 13,930 tested), where validation was defined by associations with p < 0.05 and the same directions of association as BIOS QTL (all aligned to be consistent with GWAS-reported effect alleles).
Non-replicated SNPs had greater odds of being SJLIFE-validated cis-meQTLs than replicated SNPs had (OR = 1.66, p = 0.02, Table S13). We therefore investigated whether meta-GWAS hit replications likely to be affected by childhood cancer treatments were also more likely to involve cis-meQTL mechanisms. Specifically, we compared 48 “treatment-sensitive” meta-GWAS SNPs that showed replicated associations only in the treatment-unexposed subgroup (i.e., meta-GWAS hit replications adversely affected by cancer treatments) and 66 “treatment-insensitive” meta-GWAS SNPs with robust replications (i.e., replicated in both treatment-unexposed and treatment-exposed subgroups). We found greater enrichment for SJLIFE-validated cis-meQTLs among treatment-sensitive SNPs (38/42, 90.5%) compared to treatment-insensitive SNPs (37/57, 64.9%; OR = 5.06, p = 4.1 × 10−3, Table S13); these results indicate that SNPs with phenotype association replications that were perturbed by treatment exposures in survivors were more likely to involve cis-meQTL mechanisms than were SNPs with robust replications.
Finally, we hypothesized that treatment-associated changes in the baseline CpG methylation associated with a meta-GWAS SNP could reduce the likelihood of its replication in survivors. We first split the 4,153 CpG sites linked to the 5,561 SJLIFE-validated cis-meQTLs into two mutually exclusive groups: 549 “replicated CpGs” linked to replicated meta-GWAS SNPs versus 3,604 “non-replicated CpGs” linked to non-replicated meta-GWAS SNPs. We then counted the frequency of discordance between cis-meQTL effects and the direction of methylation at the same CpG site associated with specific childhood cancer treatments. We examined different RT and chemotherapeutic exposures (Table S14). Non-replicated CpGs were enriched for directionally discordant cis-meQTL and treatment-methylation associations for multiple treatment types compared to the replicated CpGs (Table S15). The non-replicated CpGs showed the strongest enrichment for directionally discordant methylation associations for pelvic RT, with ∼54% of non-replicated CpGs bearing directionally discordant methylation associations in contrast to ∼29% of replicated CpGs (OR = 2.90, p = 8.7 × 10−4). The non-replicated CpGs were also significantly enriched for directionally discordant associations for chest RT (OR = 2.70, p = 5.3 × 10−4) and modestly enriched for abdominal RT (OR = 1.91, p = 0.06).
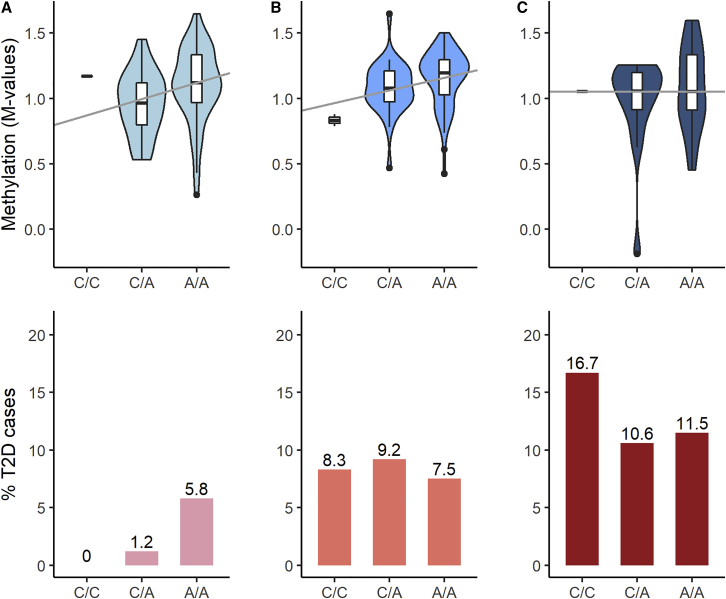
We illustrate these results by describing the failed replication of the T2D risk variant rs1552224 (chr11:72722053, GRCh38) in SJLIFE survivors as an example. Multiple meta-GWAS have linked the A allele of rs1552224 with increased T2D risk.44,59 However, this association was not replicated among survivors exposed to abdominal or pelvic RT, but it was replicated in survivors without these RT exposures (Table S16). Figure 5 demonstrates how abdominal and/or pelvic RT can obscure the replication of the rs1552224-T2D risk association in survivors by potentially disrupting cis-meQTL effects on T2D risk. The strongest cis-meQTL effect for rs1552224 was reported at cg04827223 in BIOS QTL (assessed allele = A, Z score = 34.8, p = 6.0 × 10−266) and was validated in SJLIFE (β = 0.12, p = 3.7 × 10−4). Figure 5 shows that increasing A allele dose for rs1552224 corresponds with increases in methylation at cg04827223 and T2D risk in survivors without exposures to abdominal or pelvic RT; this is consistent with the general population. But in survivors with increasing doses of abdominal or pelvic RT, increasing A allele dose for rs1552224 does not change methylation at cg04827223 or T2D risk, reflecting the inverse relationships between methylation levels at cg04827223 and pelvic (β = −4.0 × 10−6, p = 0.03) and abdominal RT (β = −3.4 × 10−6, p = 0.06) dose observed in SJLIFE.
Figure 5.
DNA Methylation Levels at cg04827223 and Percentage of T2D Cases by Genotype Classes for rs1552224 in SJLIFE Survivor Subgroups with Differential Exposures to Abdominal or Pelvic Radiation Therapy
No radiation therapy (RT) dose was defined as 0 Gy (column A), low-to-moderate RT dose was defined by >0 to <20 Gy (column B), and high dose was defined by ≥20 Gy (column C). The upper panels show the observed methylation level relationships with the SNP at the cg04827223 CpG site in the SJLIFE subset with methylome and genotype data (n = 236); boxes represent the median and interquartile range (IQR), with whiskers extending from the first or third quartile to 1.5 times the IQR. Methylation level trend by allele dose is shown with median regression lines. Genotype frequencies in this SJLIFE subset were as follows: 1.8% (C/C), 30.8% (C/A), and 67.4% (A/A). The lower panels show the percentage (%) of T2D cases by genotype in SJLIFE survivors in the main analysis (n = 2,112), with the following genotype frequencies: 1.9% (C/C), 26.9% (C/A), and 71.2% (A/A).
Discussion
There is growing interest in leveraging knowledge of established meta-GWAS hits though PRS in specialized clinical populations such as childhood cancer survivors.60 The suitability of translating this knowledge to such populations, however, depends on the generalizability of general population SNP associations to the clinical population of interest. We evaluated the generalizability of 1,376 SNP associations reported in 46 selected meta-GWAS for 12 anthropometric and cardiometabolic phenotypes in a large cohort of adult survivors of pediatric cancer in SJLIFE using genotypes from whole-genome sequencing and clinically ascertained phenotypes. Significantly fewer robust meta-GWAS hits than expected were replicated in SJLIFE survivors, with an observed-to-expected RER of 0.70 (p = 6.2 × 10−8) across all phenotypes. Replication depletion was also observed in a secondary analysis of five comparable phenotypes in an independent cohort of survivors from CCSS.
Decreased RER estimates in survivor cohorts may also reflect sampling variability and Winner’s Curse for some phenotypes (i.e., inflated power estimates due to overestimates in reported SNP-phenotype associations may artificially depress RERs). By evaluating comparably powered independent general population samples (UKBB) across multiple phenotypes (height, obesity, CAD, and T2D) with power calculated based on the same set of meta-GWAS summary statistics, we found: (1) survivor samples had smaller RERs than UKBB samples, and (2) increases in RER estimates correspond consistently to improved PRS predictive performance. It is noteworthy that height and CAD PRS in SJLIFE (where phenotype ascertainment was more similar to that in UKBB) and T2D PRS in CCSS (where phenotype ascertainment was more similar to that in UKBB) underperformed compared to UKBB samples with similar sample RERs. This was still the case even for height PRS, which had relatively strong associations with phenotypes in survivor cohorts (e.g., PRS p = 2.2 × 10−34 in SJLIFE; p = 7.5 × 10−65 in CCSS). These results suggest that vulnerable clinical populations like childhood cancer survivors may not see the same gains in genetic risk prediction conferred by PRS based on general population summary statistics relative to non-clinical populations for some phenotypes, particularly those for which clinical factors play a substantial role. This is not to say that PRS will universally have insufficient clinical utility in survivors. Instead, deliberate assessments of the predictive performance and validity of PRS based on population-based studies should be undertaken in specific clinical populations of interest in order to evaluate the extent to which such PRS are applicable and whether their clinical utility can be improved upon when they are not.
We discovered that, when cancer treatments had greater contributions to phenotype risk, greater replication depletions than expected were observed in treatment-exposed survivor subgroups. Recent studies have demonstrated that ionizing radiation can induce persistent dose-dependent changes in DNA methylation in cells or tissues targeted by radiation.52, 53, 54, 55, 56 Chemotherapies, e.g., cisplatin58 and carboplatin,57 have also been linked to differential methylation. Therefore, we assessed whether, among survivors, treatment-related DNA methylation could potentially “disrupt” robust SNP-phenotype relationships that are reported in the general population. We found that non-replicated SNPs were significantly enriched overall for SNPs with cis-meQTLs reported in BIOS QTL that were also validated in a subset of SJLIFE survivors. Furthermore, we discovered a ∼5-fold enrichment (p = 4.1 × 10−3) of validated cis-meQTL SNPs among SNPs with replications perturbed by cancer treatments in survivors compared to SNPs that were robustly replicated in survivors. Lastly, we observed enrichments of “disruptive” or directionally discordant methylation associations for chest (OR = 2.70, p = 5.3 × 10−4), pelvic (OR = 2.90, p = 8.7 × 10−4), and abdominal (OR = 1.91, p = 0.06) RT among CpGs linked to meta-GWAS SNPs that failed to replicate in survivors. Notably, chronic hematological toxicity has been well documented for RT to the chest, pelvic, and abdominal fields due to the volume of active bone marrow in these regions,61 which suggests that the DNA methylation patterns we see in the blood-derived methylome data are plausibly related to these RT exposures. Taken together, these results suggest that cancer treatments, particularly RT, may disrupt DNA methylation patterns at genomic sites linked to some disease- or trait-associated variants and interfere with their generalizability to survivors.
The main limitation of this analysis was the relatively small sample sizes of the survivor cohorts. Given the limited power to detect some SNP-phenotype replications (especially those with small effect sizes), we estimated the expected number of replications for comparison and replicated these results in a second survivor cohort with nearly double the sample size. Although we also provide comparisons of RER estimates in comparably powered general population samples, the RER comparisons for height should be considered cautiously due to potential sample-specific differences in residual variance after accounting for adjustment covariates. Interpretations of our analyses of cis-meQTLs and treatment associations with cross-sectional whole blood DNA methylation measurements also have several limitations. We were only able to evaluate DNA methylation associations in a small sample of survivors (n = 236), and this limited our ability to evaluate cis-meQTL effects in subsamples stratified based on treatment exposures, age at diagnosis (treatment), and other factors known to have profound effects on methylation (e.g., smoking62) or to conduct interaction analyses. Similar to other analyses of DNA methylation associations, we cannot ascertain the extent to which methylation levels at the selected CpGs associated with allelic variation at meta-GWAS SNPs truly contribute to phenotype variation. It is important to note that consideration of methylation associations with treatments that are discordant with cis-meQTL associations is a hypothetical indicator for disrupted cis-meQTL effects on phenotypes among survivors. Alternative mechanisms, e.g., cancer pathology or age at treatment, may also disrupt cis-meQTL effects on phenotypes. Examining associations between treatments and gene expression levels linked to these CpG sites would be a necessary first step in order to determine how treatment-related changes in DNA methylation disrupt SNP-phenotype associations.
In summary, we have shown that robust meta-GWAS SNP hits that were observed in general populations for a range of cardiometabolic phenotypes are only partially generalizable to childhood cancer survivor cohorts. Methodologies and applications that rely on established meta-GWAS hits from the general population to predict or clinically surveil some cardiometabolic outcomes or traits may have poorer performance in survivors than in the general population. A plausible explanation for the partial generalizability of robust meta-GWAS hits in survivors is that cancer treatment exposures obscure some genetic associations through epigenetic alterations such as DNA methylation. This analysis is among the first to provide evidence toward a hypothesis described in a recent review of the transferability of PRS across populations, specifically that the generalizability of PRS may also be limited in cohorts with differential environmental exposures.1 This phenomenon may also apply to other clinical populations.
Declaration of Interests
The authors declare no competing interests.
Acknowledgments
This work was funded by the National Cancer Institute (grant numbers U24 CA55727 to G.T.A., principal investigator, U01 CA195547 M.M.H. and L.L.R, principal investigators, CA21765 to C. Roberts, principal investigator, and R01 CA216354 to Y.Y. and J.Z., principal investigators), American Lebanese Syrian Associated Charities, and Alberta Machine Intelligence Institute. UK Biobank analyses were conducted via application 44891.
Published: September 17, 2020
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2020.08.014.
Web Resources
Database of Genotypes and Phenotypes (dbGaP), https://www.ncbi.nlm.nih.gov/gap/
St. Jude Cloud, https://www.stjude.cloud/
Data and Code Availability
The SJLIFE data used in this study may be accessed from the St. Jude Cloud under accession number SJC-DS-1002. The CCSS data used in this study may be accessed from dbGaP: phs001327.v1.p1.
Supplemental Data
SNP-Phenotype Association Test Results in the SJLIFE Cohort, Adjusted for Covariates Used in Reference Meta-GWAS
References
- 1.Martin A.R., Kanai M., Kamatani Y., Okada Y., Neale B.M., Daly M.J. Clinical use of current polygenic risk scores may exacerbate health disparities. Nat. Genet. 2019;51:584–591. doi: 10.1038/s41588-019-0379-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nolte I.M., van der Most P.J., Alizadeh B.Z., de Bakker P.I., Boezen H.M., Bruinenberg M., Franke L., van der Harst P., Navis G., Postma D.S. Missing heritability: is the gap closing? An analysis of 32 complex traits in the Lifelines Cohort Study. Eur. J. Hum. Genet. 2017;25:877–885. doi: 10.1038/ejhg.2017.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Natarajan P., Young R., Stitziel N.O., Padmanabhan S., Baber U., Mehran R., Sartori S., Fuster V., Reilly D.F., Butterworth A. Polygenic risk score identifies subgroup with higher burden of atherosclerosis and greater relative benefit from statin therapy in the primary prevention setting. Circulation. 2017;135:2091–2101. doi: 10.1161/CIRCULATIONAHA.116.024436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mega J.L., Stitziel N.O., Smith J.G., Chasman D.I., Caulfield M., Devlin J.J., Nordio F., Hyde C., Cannon C.P., Sacks F. Genetic risk, coronary heart disease events, and the clinical benefit of statin therapy: an analysis of primary and secondary prevention trials. Lancet. 2015;385:2264–2271. doi: 10.1016/S0140-6736(14)61730-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Robison L.L., Hudson M.M. Survivors of childhood and adolescent cancer: life-long risks and responsibilities. Nat. Rev. Cancer. 2014;14:61–70. doi: 10.1038/nrc3634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bhakta N., Liu Q., Ness K.K., Baassiri M., Eissa H., Yeo F., Chemaitilly W., Ehrhardt M.J., Bass J., Bishop M.W. The cumulative burden of surviving childhood cancer: an initial report from the St Jude Lifetime Cohort Study (SJLIFE) Lancet. 2017;390:2569–2582. doi: 10.1016/S0140-6736(17)31610-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Armstrong G.T., Kawashima T., Leisenring W., Stratton K., Stovall M., Hudson M.M., Sklar C.A., Robison L.L., Oeffinger K.C. Aging and risk of severe, disabling, life-threatening, and fatal events in the childhood cancer survivor study. J. Clin. Oncol. 2014;32:1218–1227. doi: 10.1200/JCO.2013.51.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hudson M.M., Ness K.K., Gurney J.G., Mulrooney D.A., Chemaitilly W., Krull K.R., Green D.M., Armstrong G.T., Nottage K.A., Jones K.E. Clinical ascertainment of health outcomes among adults treated for childhood cancer. JAMA. 2013;309:2371–2381. doi: 10.1001/jama.2013.6296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oeffinger K.C., Mertens A.C., Sklar C.A., Kawashima T., Hudson M.M., Meadows A.T., Friedman D.L., Marina N., Hobbie W., Kadan-Lottick N.S., Childhood Cancer Survivor Study Chronic health conditions in adult survivors of childhood cancer. N. Engl. J. Med. 2006;355:1572–1582. doi: 10.1056/NEJMsa060185. [DOI] [PubMed] [Google Scholar]
- 10.Bhakta N., Liu Q., Yeo F., Baassiri M., Ehrhardt M.J., Srivastava D.K., Metzger M.L., Krasin M.J., Ness K.K., Hudson M.M. Cumulative burden of cardiovascular morbidity in paediatric, adolescent, and young adult survivors of Hodgkin’s lymphoma: an analysis from the St Jude Lifetime Cohort Study. Lancet Oncol. 2016;17:1325–1334. doi: 10.1016/S1470-2045(16)30215-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nottage K.A., Ness K.K., Li C., Srivastava D., Robison L.L., Hudson M.M. Metabolic syndrome and cardiovascular risk among long-term survivors of acute lymphoblastic leukaemia - From the St. Jude Lifetime Cohort. Br. J. Haematol. 2014;165:364–374. doi: 10.1111/bjh.12754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mulrooney D.A., Yeazel M.W., Kawashima T., Mertens A.C., Mitby P., Stovall M., Donaldson S.S., Green D.M., Sklar C.A., Robison L.L., Leisenring W.M. Cardiac outcomes in a cohort of adult survivors of childhood and adolescent cancer: retrospective analysis of the Childhood Cancer Survivor Study cohort. BMJ. 2009;339:b4606. doi: 10.1136/bmj.b4606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mertens A.C., Liu Q., Neglia J.P., Wasilewski K., Leisenring W., Armstrong G.T., Robison L.L., Yasui Y. Cause-specific late mortality among 5-year survivors of childhood cancer: the Childhood Cancer Survivor Study. J. Natl. Cancer Inst. 2008;100:1368–1379. doi: 10.1093/jnci/djn310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jaenisch R., Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat. Genet. 2003;33(Suppl):245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 15.Welter D., MacArthur J., Morales J., Burdett T., Hall P., Junkins H., Klemm A., Flicek P., Manolio T., Hindorff L., Parkinson H. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014;42:D1001–D1006. doi: 10.1093/nar/gkt1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hudson M.M., Ness K.K., Nolan V.G., Armstrong G.T., Green D.M., Morris E.B., Spunt S.L., Metzger M.L., Krull K.R., Klosky J.L. Prospective medical assessment of adults surviving childhood cancer: study design, cohort characteristics, and feasibility of the St. Jude Lifetime Cohort study. Pediatr. Blood Cancer. 2011;56:825–836. doi: 10.1002/pbc.22875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hudson M.M., Ehrhardt M.J., Bhakta N., Baassiri M., Eissa H., Chemaitilly W., Green D.M., Mulrooney D.A., Armstrong G.T., Brinkman T.M. Approach for Classification and Severity Grading of Long-term and Late-Onset Health Events among Childhood Cancer Survivors in the St. Jude Lifetime Cohort. Cancer Epidemiol. Biomarkers Prev. 2017;26:666–674. doi: 10.1158/1055-9965.EPI-16-0812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meacham L.R., Sklar C.A., Li S., Liu Q., Gimpel N., Yasui Y., Whitton J.A., Stovall M., Robison L.L., Oeffinger K.C. Diabetes mellitus in long-term survivors of childhood cancer. Increased risk associated with radiation therapy: a report for the childhood cancer survivor study. Arch. Intern. Med. 2009;169:1381–1388. doi: 10.1001/archinternmed.2009.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Robison L.L., Armstrong G.T., Boice J.D., Chow E.J., Davies S.M., Donaldson S.S., Green D.M., Hammond S., Meadows A.T., Mertens A.C. The Childhood Cancer Survivor Study: a National Cancer Institute-supported resource for outcome and intervention research. J. Clin. Oncol. 2009;27:2308–2318. doi: 10.1200/JCO.2009.22.3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Robison L.L., Mertens A.C., Boice J.D., Breslow N.E., Donaldson S.S., Green D.M., Li F.P., Meadows A.T., Mulvihill J.J., Neglia J.P. Study design and cohort characteristics of the Childhood Cancer Survivor Study: a multi-institutional collaborative project. Med. Pediatr. Oncol. 2002;38:229–239. doi: 10.1002/mpo.1316. [DOI] [PubMed] [Google Scholar]
- 21.Leisenring W.M., Mertens A.C., Armstrong G.T., Stovall M.A., Neglia J.P., Lanctot J.Q., Boice J.D., Jr., Whitton J.A., Yasui Y. Pediatric cancer survivorship research: experience of the Childhood Cancer Survivor Study. J. Clin. Oncol. 2009;27:2319–2327. doi: 10.1200/JCO.2008.21.1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.1000 Genomes Project Consortium A global reference for human genetic variation. Nature. 2015;526:68. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Z., Wilson C.L., Easton J., Thrasher A., Mulder H., Liu Q., Hedges D.J., Wang S., Rusch M.C., Edmonson M.N. Genetic Risk for Subsequent Neoplasms Among Long-Term Survivors of Childhood Cancer. J. Clin. Oncol. 2018;36:2078–2087. doi: 10.1200/JCO.2018.77.8589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sapkota Y., Cheung Y.T., Moon W., Shelton K., Wilson C.L., Wang Z., Mulrooney D.A., Zhang J., Armstrong G.T., Hudson M.M. Whole-Genome Sequencing of Childhood Cancer Survivors Treated with Cranial Radiation Therapy Identifies 5p15.33 Locus for Stroke: A Report from the St. Jude Lifetime Cohort Study. Clin. Cancer Res. 2019;25:6700–6708. doi: 10.1158/1078-0432.CCR-19-1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., DePristo M.A. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., 1000 Genome Project Data Processing Subgroup The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J., Sham P.C. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Danecek P., Auton A., Abecasis G., Albers C.A., Banks E., DePristo M.A., Handsaker R.E., Lunter G., Marth G.T., Sherry S.T., 1000 Genomes Project Analysis Group The variant call format and VCFtools. Bioinformatics. 2011;27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morton L.M., Sampson J.N., Armstrong G.T., Chen T.-H., Hudson M.M., Karlins E., Dagnall C.L., Li S.A., Wilson C.L., Srivastava D.K. Genome-wide association study to identify susceptibility loci that modify radiation-related risk for breast cancer after childhood cancer. J. Natl. Cancer Inst. 2017;109:djx058. doi: 10.1093/jnci/djx058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sapkota Y., Turcotte L.M., Ehrhardt M.J., Howell R.M., Arnold M.A., Wilson C.L., Leisenring W., Wang Z., Sampson J., Dagnall C.L. Genome-Wide Association Study in Irradiated Childhood Cancer Survivors Identifies HTR2A for Subsequent Basal Cell Carcinoma. J. Invest. Dermatol. 2019;139:2042–2045.e8. doi: 10.1016/j.jid.2019.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Das S., Forer L., Schönherr S., Sidore C., Locke A.E., Kwong A., Vrieze S.I., Chew E.Y., Levy S., McGue M. Next-generation genotype imputation service and methods. Nat. Genet. 2016;48:1284–1287. doi: 10.1038/ng.3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Price A.L., Patterson N.J., Plenge R.M., Weinblatt M.E., Shadick N.A., Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 34.Aryee M.J., Jaffe A.E., Corrada-Bravo H., Ladd-Acosta C., Feinberg A.P., Hansen K.D., Irizarry R.A. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics. 2014;30:1363–1369. doi: 10.1093/bioinformatics/btu049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bonder M.J., Luijk R., Zhernakova D.V., Moed M., Deelen P., Vermaat M., van Iterson M., van Dijk F., van Galen M., Bot J., BIOS Consortium Disease variants alter transcription factor levels and methylation of their binding sites. Nat. Genet. 2017;49:131–138. doi: 10.1038/ng.3721. [DOI] [PubMed] [Google Scholar]
- 36.Whalen S., Pollard K.S. Most chromatin interactions are not in linkage disequilibrium. Genome Res. 2019;29:334–343. doi: 10.1101/gr.238022.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gauderman W.J. Sample size requirements for association studies of gene-gene interaction. Am. J. Epidemiol. 2002;155:478–484. doi: 10.1093/aje/155.5.478. [DOI] [PubMed] [Google Scholar]
- 38.Greenland S. Model-based estimation of relative risks and other epidemiologic measures in studies of common outcomes and in case-control studies. Am. J. Epidemiol. 2004;160:301–305. doi: 10.1093/aje/kwh221. [DOI] [PubMed] [Google Scholar]
- 39.Le Cam L. An approximation theorem for the Poisson binomial distribution. Pacific Journal of Mathematics. 1960;10:1181–1197. [Google Scholar]
- 40.Willer C.J., Li Y., Abecasis G.R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bycroft C., Freeman C., Petkova D., Band G., Elliott L.T., Sharp K., Motyer A., Vukcevic D., Delaneau O., O’Connell J. The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018;562:203–209. doi: 10.1038/s41586-018-0579-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wood A.R., Esko T., Yang J., Vedantam S., Pers T.H., Gustafsson S., Chu A.Y., Estrada K., Luan J., Kutalik Z., Electronic Medical Records and Genomics (eMEMERGEGE) Consortium. MIGen Consortium. PAGEGE Consortium. LifeLines Cohort Study Defining the role of common variation in the genomic and biological architecture of adult human height. Nat. Genet. 2014;46:1173–1186. doi: 10.1038/ng.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Berndt S.I., Gustafsson S., Mägi R., Ganna A., Wheeler E., Feitosa M.F., Justice A.E., Monda K.L., Croteau-Chonka D.C., Day F.R. Genome-wide meta-analysis identifies 11 new loci for anthropometric traits and provides insights into genetic architecture. Nat. Genet. 2013;45:501–512. doi: 10.1038/ng.2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhao W., Rasheed A., Tikkanen E., Lee J.-J., Butterworth A.S., Howson J.M.M., Assimes T.L., Chowdhury R., Orho-Melander M., Damrauer S., CHD Exome+ Consortium. EPIC-CVD Consortium. EPIC-Interact Consortium. Michigan Biobank Identification of new susceptibility loci for type 2 diabetes and shared etiological pathways with coronary heart disease. Nat. Genet. 2017;49:1450–1457. doi: 10.1038/ng.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nelson C.P., Goel A., Butterworth A.S., Kanoni S., Webb T.R., Marouli E., Zeng L., Ntalla I., Lai F.Y., Hopewell J.C., EPIC-CVD Consortium. CARDIoGRAMplusC4D. UK Biobank CardioMetabolic Consortium CHD working group Association analyses based on false discovery rate implicate new loci for coronary artery disease. Nat. Genet. 2017;49:1385–1391. doi: 10.1038/ng.3913. [DOI] [PubMed] [Google Scholar]
- 46.Kundaje A., Meuleman W., Ernst J., Bilenky M., Yen A., Heravi-Moussavi A., Kheradpour P., Zhang Z., Wang J., Ziller M.J., Roadmap Epigenomics Consortium Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.GTEx Consortium Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Croft D., Mundo A.F., Haw R., Milacic M., Weiser J., Wu G., Caudy M., Garapati P., Gillespie M., Kamdar M.R. The Reactome pathway knowledgebase. Nucleic Acids Res. 2014;42:D472–D477. doi: 10.1093/nar/gkt1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Subhash S., Kanduri C. GeneSCF: a real-time based functional enrichment tool with support for multiple organisms. BMC Bioinformatics. 2016;17:365. doi: 10.1186/s12859-016-1250-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.O’Leary N.A., Wright M.W., Brister J.R., Ciufo S., Haddad D., McVeigh R., Rajput B., Robbertse B., Smith-White B., Ako-Adjei D. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016;44(D1):D733–D745. doi: 10.1093/nar/gkv1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Portela A., Esteller M. Epigenetic modifications and human disease. Nat. Biotechnol. 2010;28:1057–1068. doi: 10.1038/nbt.1685. [DOI] [PubMed] [Google Scholar]
- 52.Reisz J.A., Bansal N., Qian J., Zhao W., Furdui C.M. Effects of ionizing radiation on biological molecules--mechanisms of damage and emerging methods of detection. Antioxid. Redox Signal. 2014;21:260–292. doi: 10.1089/ars.2013.5489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Antwih D.A., Gabbara K.M., Lancaster W.D., Ruden D.M., Zielske S.P. Radiation-induced epigenetic DNA methylation modification of radiation-response pathways. Epigenetics. 2013;8:839–848. doi: 10.4161/epi.25498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kuhmann C., Weichenhan D., Rehli M., Plass C., Schmezer P., Popanda O. DNA methylation changes in cells regrowing after fractioned ionizing radiation. Radiother. Oncol. 2011;101:116–121. doi: 10.1016/j.radonc.2011.05.048. [DOI] [PubMed] [Google Scholar]
- 55.Goetz W., Morgan M.N., Baulch J.E. The effect of radiation quality on genomic DNA methylation profiles in irradiated human cell lines. Radiat. Res. 2011;175:575–587. doi: 10.1667/RR2390.1. [DOI] [PubMed] [Google Scholar]
- 56.Pogribny I., Raiche J., Slovack M., Kovalchuk O. Dose-dependence, sex- and tissue-specificity, and persistence of radiation-induced genomic DNA methylation changes. Biochem. Biophys. Res. Commun. 2004;320:1253–1261. doi: 10.1016/j.bbrc.2004.06.081. [DOI] [PubMed] [Google Scholar]
- 57.Gifford G., Paul J., Vasey P.A., Kaye S.B., Brown R. The acquisition of hMLH1 methylation in plasma DNA after chemotherapy predicts poor survival for ovarian cancer patients. Clin. Cancer Res. 2004;10:4420–4426. doi: 10.1158/1078-0432.CCR-03-0732. [DOI] [PubMed] [Google Scholar]
- 58.Yu W., Jin C., Lou X., Han X., Li L., He Y., Zhang H., Ma K., Zhu J., Cheng L., Lin B. Global analysis of DNA methylation by Methyl-Capture sequencing reveals epigenetic control of cisplatin resistance in ovarian cancer cell. PLoS ONE. 2011;6 doi: 10.1371/journal.pone.0029450. e29450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Voight B.F., Scott L.J., Steinthorsdottir V., Morris A.P., Dina C., Welch R.P., Zeggini E., Huth C., Aulchenko Y.S., Thorleifsson G., MAGIC investigators. GIANT Consortium Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nat. Genet. 2010;42:579–589. doi: 10.1038/ng.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang Z., Liu Q., Wilson C.L., Easton J., Mulder H., Chang T.-C., Rusch M.C., Edmonson M.N., Rice S.V., Ehrhardt M.J. Polygenic determinants for subsequent breast cancer risk in survivors of childhood cancer: The St Jude Lifetime Cohort Study (SJLIFE) Clin. Cancer Res. 2018;24:6230–6235. doi: 10.1158/1078-0432.CCR-18-1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mauch P., Constine L., Greenberger J., Knospe W., Sullivan J., Liesveld J.L., Deeg H.J. Hematopoietic stem cell compartment: acute and late effects of radiation therapy and chemotherapy. Int. J. Radiat. Oncol. Biol. Phys. 1995;31:1319–1339. doi: 10.1016/0360-3016(94)00430-S. [DOI] [PubMed] [Google Scholar]
- 62.Joehanes R., Just A.C., Marioni R.E., Pilling L.C., Reynolds L.M., Mandaviya P.R., Guan W., Xu T., Elks C.E., Aslibekyan S. Epigenetic signatures of cigarette smoking. Circ Cardiovasc Genet. 2016;9:436–447. doi: 10.1161/CIRCGENETICS.116.001506. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SNP-Phenotype Association Test Results in the SJLIFE Cohort, Adjusted for Covariates Used in Reference Meta-GWAS
Data Availability Statement
The SJLIFE data used in this study may be accessed from the St. Jude Cloud under accession number SJC-DS-1002. The CCSS data used in this study may be accessed from dbGaP: phs001327.v1.p1.