Abstract
Background:
Intraflagellar transport is a motor-driven trafficking system that is required for the formation of cilia. Intraflagellar transport protein 20 (IFT20) is a master regulator for the control of spermatogenesis and male fertility in mice. However, the mechanism of how IFT20 regulates spermatogenesis is unknown.
Results:
Spermatogenesis associated 1 (SPATA1) was identified to be a major potential binding partner of IFT20 by a yeast two-hybrid screening. The interaction between SPATA1 and IFT20 was examined by direct yeast two-hybrid, colocalization, and co-immunoprecipitation assays. SPATA1 is highly abundant in the mouse testis, and is also expressed in the heart and kidney. During the first wave of spermatogenesis, SPATA1 is detectable at postnatal day 24 and its expression is increased at day 30 and 35. Immunofluorescence staining of mouse testis sections and epididymal sperm demonstrated that SPATA1 is localized mainly in the acrosome of developing spermatids but not in epididymal sperm. IFT20 is also present in the acrosome area of round spermatids. In conditional Ift20 knockout mice, testicular expression level and acrosomal localization of SPATA1 are not changed.
Conclusions:
SPATA1 is an IFT20 binding protein and may provide a docking site for IFT20 complex binding to the acrosome area.
Keywords: acrosome, intraflagellar transport 20, spermiogenesis
1 |. INTRODUCTION
Intraflagellar transport (IFT) was originally found in the green algae Chlamydomonas reinhardtii and is an evolutionarily conserved motor-driven trafficking system that moves IFT particles including cargo proteins and signaling molecules along the microtubular axoneme of motile and primary cilia.1–3 IFT machinery is required for maintaining the precise balance between ciliary/flagellar assembly and disassembly.3 The IFT particles are transported to the assembly site at the axonemal tip of cilia/flagella in a kinesin-2-dependent manner (anterograde trafficking), and the movement of turnover products from the tip back to the cell body is catalyzed by cytoplasmic dynein-2 (retrograde trafficking).4–7 The IFT particle isolated from flagella of C reinhardtii shows that it is composed of at least 22 subunits that are divided into two complexes, IFT-A containing 6 subunits and IFT-B comprising 16 subunits.3,4 Even though originally discovered in Chlamydomonas, IFT orthologues are present in mammals. Mutations disrupting IFT subunits have been linked to a range of human diseases such as nephronophthisis, Bardet-Biedl syndrome, asphyxiating thoracic dystrophy, and Mainzer-Saldino syndrome.8–12
IFT20 was originally reported to be a subunit of the IFT-B complex in Chlamydomonas.4 The role of mouse orthologue IFT20 has been studied in vitro and in vivo. It has been shown that IFT20 is located in the Golgi complex of mouse IMCD3 cells and its cellular localization is dependent on GMAP210.13–15 It can move between the Golgi complex and the basal body of the cilium, as well as along the cilium in mammalian cells, suggesting that it plays a role in anterograde transport of IFT particles in cilia.13,14 To study the role of IFT20 in vivo, tissue-specific Ift20 knockout mice have been generated. Knockout of Ift20 in mouse kidney causes cystic kidney disease.16 IFT20 is also indispensable for rhodopsin and opsin transport and development and maintenance of photoreceptor outer segment.17 Disruption of Ift20 in the cochlea results in shorter cochlear ducts and mis-oriented hair cell stereociliary bundles.18 Besides these ciliary cells, IFT20 has been found to be expressed in nonciliated T cells and involved in immune synapse formation in T lymphocytes. Knockdown of Ift20 by RNA interference disrupts the polarized recycling of the T-cell receptor/CD3 complex to the immune synapse.19 Deletion of Ift20 in the early stage of T-cell differentiation suppresses the development of collagen-induced arthritis via controlling T-cell development.20 These results provide novel insights into the trafficking-related function of the IFT system beyond ciliogenesis.
Of all mammalian cells, sperm have the longest motile cilia. In male germ cells, IFT20 has been reported to be present in the Golgi complex of late spermatocytes and round spermatids, and in the manchette and basal body of elongating spermatids.21 In our previous study, IFT20 was identified to express in male germ cells, and it is essential for male fertility and spermiogenesis in mice, and one of its major function is to transport cargo proteins for sperm flagella formation.22 To elucidate the mechanism of IFT20, particularly in spermatogenesis, a yeast-two hybrid screen was conducted, and a number of potential binding partners were identified. One of them was spermatogenesis-associated protein 1 (SPATA1). We thus further characterized the gene. SPATA1 protein was found to be present in the acrosome during spermiogenesis, and its expression level and acrosomal localization were independent of IFT20. These findings suggest that SPATA1 may play a role upstream of IFT20 action in spermiogenesis.
2 |. RESULTS
2.1 |. Identification of IFT20 binding partners
To gain insights into the mechanism of IFT20 action in spermatogenesis, we performed a yeast two-hybrid screen using IFT20 as a bait under stringent conditions (quadruple-dropout medium). Potential binding partners were identified after sequencing the plasmid DNA isolated from yeast clones. The IFT20 binding candidates are listed in Table S1. IFT81 is the most frequently identified binding partner. In this study, we characterized the second most frequently identified binding partner spermatogenesis-associated protein 1 (SPATA1).
2.2 |. IFT20 interacts with SPATA1 in vitro
To confirm the interaction between IFT20 and SPATA1, a direct yeast two-hybrid assay was carried out by cotransformation of yeast cells AH109 with IFT20/pGBKT7 and SPATA1/pGADT7. Like the positive control, the yeast transformed with IFT20/pGBKT7 and SPATA1/pGADT7 plasmids grew on selective media, indicating that SPATA1 is capable of interacting with IFT20 (Figure 1A). The interaction between SPATA1 and IFT20 was further examined in transfected mammalian cells. When CHO cells were transfected with SPATA1 expression constructs, the protein was distributed as cytoplasmic vesicles in the cells (Figure 1B, upper panel). Similarly, when expressed alone, IFT20 protein was present as vesicles in the cytoplasm of CHO cells (Figure 1B, upper panel). When IFT20 and SPATA1 were co-expressed, they were partially co-localized and formed clusters in the cells (Figure 1B, lower panel). In addition, co-immunoprecipitation assay validated the interaction between SPATA1 and IFT20. When lysates of transfected cells containing IFT20/Flag and SPATA1/GFP expression constructs, the Flag antibody pulled down both Flag-tagged IFT20 and SPATA1. Immunoprecipitation with mouse IgG was used as a negative control (Figure 1C).
FIGURE 1.

IFT20 interacts with SPATA1. A, Direct yeast two-hybrid assay to examine the interaction between IFT20 and SPATA1. Yeast AH109 cells were transformed the indicated plasmids and grown on nonselective (SD/–Leu/–Trp) and selective media (SD/-His/–Leu/–Trp), respectively. Pair of pGADT7-T/pGBKT7-p53 was used as a positive control. B, Co-localization of IFT20 and SPATA1 in CHO cells. (Upper panel) CHO cells were transfected with SPATA1/pcDNA3 or IFT20/pEGFP-N2 plasmids, respectively. SPATA1 or IFT20 protein was present as cytoplasmic vesicle in the cells (lower panel). CHO cells were transfected with both SPATA1/pcDNA3 and IFT20/pEGFP-N2. When both proteins were co-expressed, they were partially co-localized and formed clusters in the cells. DNA was stained with DAPI (blue). C, Co-immunoprecipitation of IFT20 with SPATA1. COS-1 cells were co-transfected with IFT20/Flag and SPATA1/GFP. The cell lysate was immunoprecipitated with anti-Flag antibody and then subjected to Western blot analysis with anti-Flag and anti-GFP antibodies
2.3 |. Spata1 is highly expressed in mouse testis
The expression profile of Spata1 in different mouse tissues was investigated at the transcriptional and protein levels. High level of Spata1 transcripts was detected in the kidney and testis. Moderate expression of Spata1 was observed in the brain and lung, and extremely low level of the Spata1 transcript was detected in the heart (Figure 2A). To further study the function of SPATA1, N-terminus of SPATA1 was purified to boost rabbits to generate a specific polyclonal antibody. To examine specificity of the antibody, Western blotting was conducted using cell lysates extracted from COS-1 cells transfected with a SPATA1/pEGFP-N2 plasmid and a control pEGFP-N2 plasmid. When an anti-GFP antibody was used, the 26 kDa free GFP protein was detected in the cell lysates from pEGFP-N2 plasmid transfection and a protein with 78 kDa that could correspond to the SPATA1/GFP fusion protein was detected in the cell lysates from SPATA1/pEGFP-N2 plasmid transfection (Figure 2B-a). When the specific anti-SPATA1 antibody was used, only the 78 kDa protein was detected (Figure 2B-b).
FIGURE 2.
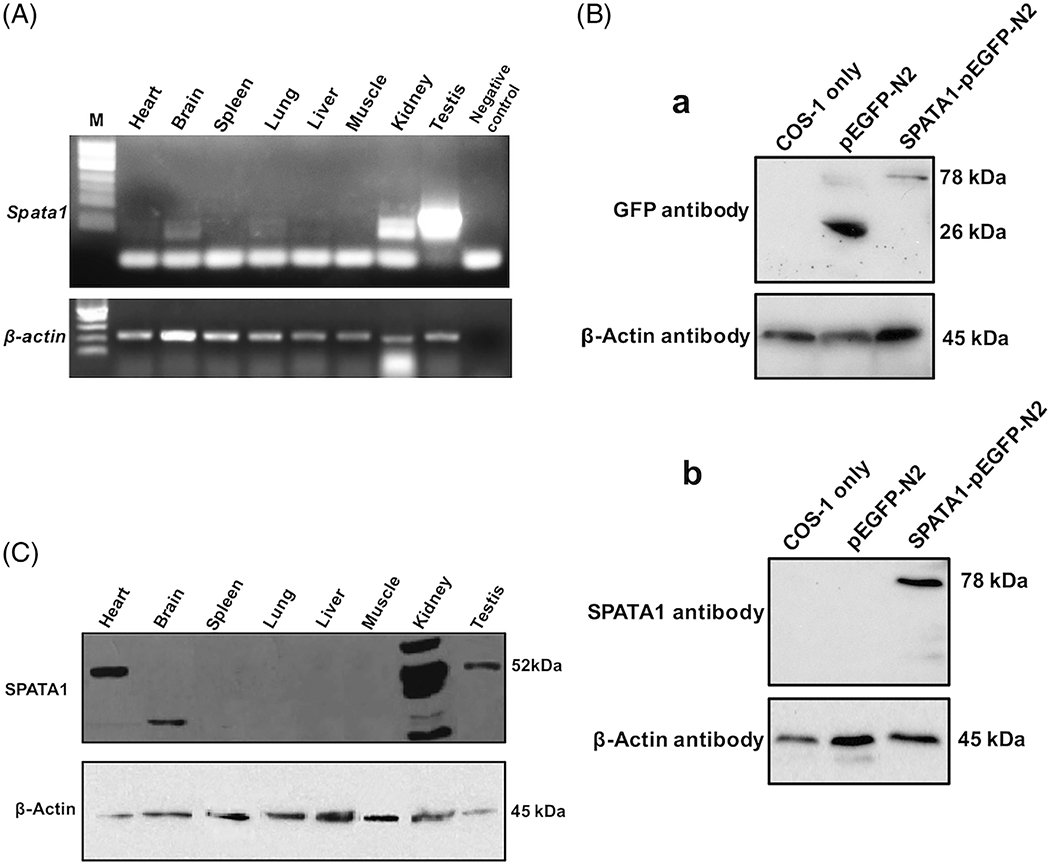
Tissue distribution of Spata1 mRNA and protein. A, Total RNA was extracted from indicated mouse tissues and the distribution of Spata1 transcripts was analyzed by RT-PCR. Notice that Spata1 mRNA was highly expressed in the testis and kidney, and it was also expressed in other tissues at a low level. B, Generation of a specific anti-SPATA1 polyclonal antibody. COS-1 cells were transfected with pEGFP-N2 and SPATA1/pEGFP-N2 plasmids respectively, and the protein lysates were subjected to Western blotting with an anti-GFP antibody (a) or an anti-SPATA1 antibody (b). C, Expression profile of the SPATA1 protein was examined in the indicated tissues by Western blotting. The antibody recognized the predicted 52 kDa SPATA1 in the heart and testis; a smaller protein in the brain and multiple proteins in the kidney, presumably representing other SPATA1 isoforms
The expression pattern of SPATA1 protein was analyzed by Western blot using the anti-SPATA1 antibody. A single protein band at approximately 52 kDa, which corresponds to the predicted molecular weight of SPATA1, was observed in heart and testis. Besides the 52 kDa protein, the antibody also recognized multiple proteins with different sizes in the kidney, and a smaller protein in the brain (Figure 2C).
2.4 |. SPATA1 expression is upregulated during spermiogenesis and is localized in the acrosome, where IFT20 localizes in round spermatids, but not in epididymal sperm
To determine the expression pattern of SPATA1 during the first wave of spermatogenesis, Western blotting was performed using mouse testes obtained at different times after birth. Expression of SPATA1 is developmentally regulated. SPATA1 was initially detected at postnatal day 24, at a time that corresponds to round spermatids, and considerably increased at day 30 and 35 after birth (Figure 3A). The elevated expression of SPATA1 in the late stage of spermatogenesis suggests that SPATA1 is involved in spermiogenesis. Given that both IFT20 and SPATA1 antibodies were made from rabbits, it is challenging to conduct co-immunostaining to determine if the two proteins interact in vivo. Thus, we performed co-immunoprecipitation using mouse testis extracts. When the anti-SPATA1 antibody was used to pull-down the testis extract, IFT20 was co-pulled down (Figure 3B).
FIGURE 3.

Expression and localization of SPATA1 in male germ cells of wild-type mice. A, Testicular expression of SPATA1 during the first wave of spermatogenesis analyzed by Western blotting. Notice that SPATA1 was detected from day 24 after birth, and the expression level was higher at day after day 30. B, IFT20 and SPATA1 were present in the same complex in mouse testis. Testicular extract was pulled down with an anti-SPATA1 antibody and Western blotting was conducted using anti-IFT20 antibody. IFT20 was pulled down by the anti-SPATA1 antibody. C, Examination of SPATA1 localization in isolated male germ cells by immunofluorescence staining. SPATA1 (red signals, white arrows) was co-localized with an acrosomal marker, Sp56 (green signals) in the acrosomal region of spermatids. D, Immunofluorescence analysis of SPATA1 protein in early stages of testicular germ cells. The protein is localized in the acrosome of germ cells from stages I to III. E, Representative image of localization of IFT20 and peanut-lectin in a round spermatid. The isolated male germ cells were double-stained with an anti-IFT20 antibody and fluorescence labeled peanut-lectin. Notice that IFT20 was partially co-localized with peanut-lectin (white arrow). Most IFT20 signal was on the top of lectin
To examine the localization of SPATA1 during spermatogenesis, we performed immunofluorescence (IF) staining on isolated testicular germ cells. SPATA1 signals were observed in the acrosomal region of spermatids as shown by its colocalization with Sp56, an acrosomal marker (Figure 3C). In addition, IF staining on the testicular sections from control mice showed that SPATA1 is also located in the acrosome region of germ cells at early stages (Figure 3D). We tried to rule out the possibility for SPATA1 in the Golgi bodies. However, the antibodies (TGN38, Golgin-97, GM130 and Golgin-84) specifically against the Golgi bodies did not work in the present study.
Given that SPATA1 is localized in the acrosome, we next examined if IFT20 is also present in this organelle. Isolated testicular germ cells were double-stained with a rabbit polyclonal IFT20 antibody and peanut-lectin. In round spermatids, some IFT20 signal partially overlapped with peanut-lectin, indicating that some IFT20 is also present in the acrosome. However, most IFT20 signal seems to be on top of the lectin signal (Figure 3E).
To determine if SPATA1 is expressed in mature sperm, Western blotting and IF were carried out using epididymal sperm collected from wild-type adult mice. As shown in Figure 4A, a protein band with molecular weight of 52 kDa was detected in the testis but not in sperm. Consistently, SPATA1 was not detected in the acrosome of epididymal sperm although it was present in the acrosomal region of spermatids (Figure 4B).
FIGURE 4.
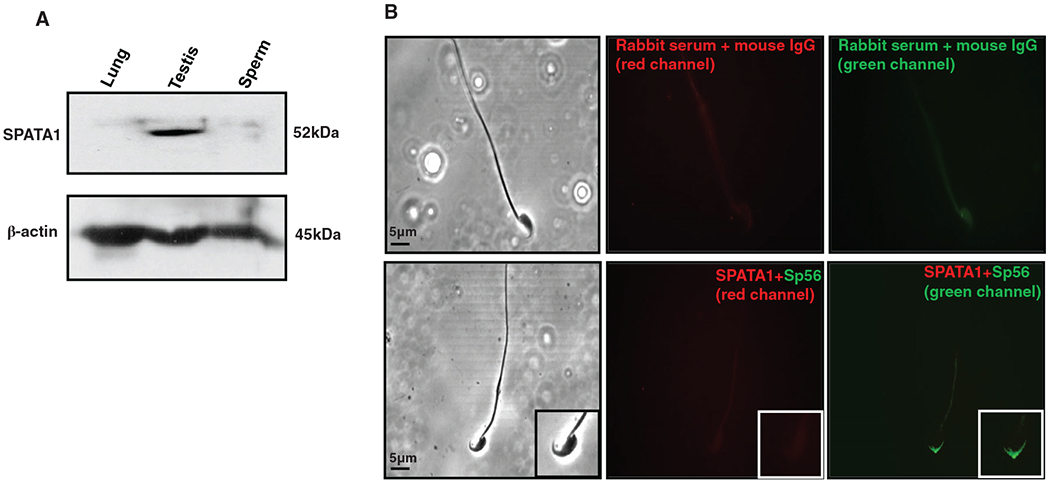
Expression and localization of SPATA1 in epididymal sperm. A, Protein samples from lung, testis and epididymal sperm were immunoblotted with the anti-SPATA1 antibody. SPATA1 was only expressed in the testis but not in the lung and epididymal sperm. B, Immunofluorescence analysis of SPATA1 protein expression in epididymal sperm. Sperm were double-stained with the anti-SPATA1 and anti-Sp56 antibodies. As a negative control, sperm were reacted with preimmune serum (upper panel). SPATA1 was not detected in epididymal sperm (lower panel)
2.5 |. Expression levels and localization of SPATA1 were not changed in the Ift20 knockout mice
To explore functional relationship between IFT20 and SPATA1, protein levels of SPATA1 in the Ift20 knockout mice and the control mice were first examined by Western blotting. Knockout of Ift20 had little effect on the testicular expression of SPATA1 (Figure 5A). We next compared the localization of SPATA1 in testis sections between the control and Ift20 mutant mice at different stages of spermatogenesis. In the control mice, specific signals were present on the nuclear surface of round spermatids at stage IV (Figure 5B-a). During germ cell development, SPATA1 was detected in the acrosome of spermatids and expended over the anterior surface of the nuclear head at stage IX-X (Figure 5B-b). At stage XII, SPATA1 was still lining the acrosome (Figure 5B-c). In the Ift20 knockout mice, localization of SPATA1 was similar to that observed in the control mice before stage XII and the protein was still present in the acrosome. As disruption of Ift20 impairs spermatogenesis in mice,22 step 12 spermatid nuclei of the Ift20 mutant mice did not elongate properly and some SPATA1 proteins seemed to be scattered rather than lining the acrosome (Figure 5B-c). As a negative control, no specific signal was observed when a preimmune rabbit serum was used (Figure 5B-d).
FIGURE 5.
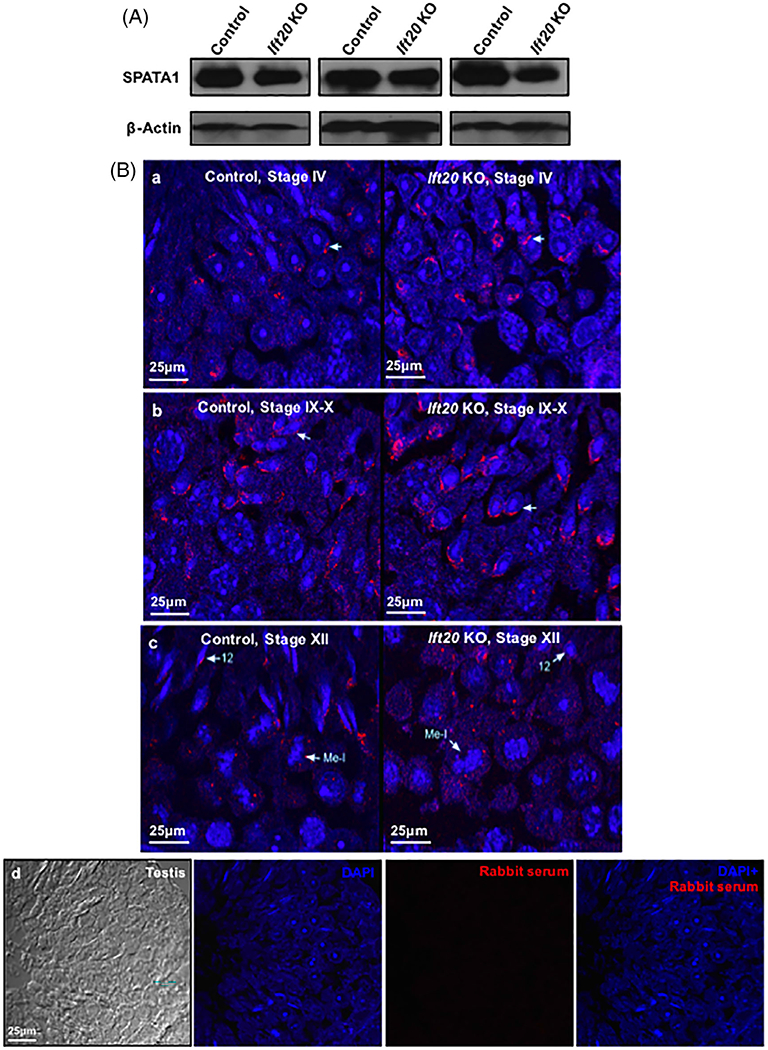
Examination of SPATA1 expression and localization in the conditional Ift20 knockout mice. A, Representative Western blot results of testicular SPATA1 expression in the control and Ift20 knockout mice. β-actin was used as the internal control. There was no significantly difference in SPATA1 expression between the control and Ift20 knockout mice. B, Examination of SPATA1 localization in the testicular section of control and Ift20 knockout mice at different developmental stages by immunofluorescence. In the control mice, SPATA1 was concentrated on top of the nuclear membrane (white arrow) at stage IV (a), and covered the whole acrosome of spermatids at stages IX to X (b). The protein was still lining the acrosome at stage XII (c). In the conditional Ift20 knockout mice, SPATA1 had a similar localization as observed in the control mice before stage XII. At step 12, spermatid nuclei of Ift20 KO mice did not elongate properly, and SPATA1 was scattered rather than lining the acrosome (c). As a control, testis sections were reacted with preimmune serum (d)
3 |. DISCUSSION
It has been previously shown that IFT20 is required for the development of germ cells and male fertility in mice.22 The present study demonstrates the potential function of SPATA1 in spermatogenesis. A yeast two-hybrid screen was performed using the full-length mouse IFT20 protein as a bait and found a number of putative binding partners. Interestingly, IFT81 was identified to be the strongest binding partner as evaluated by the frequency appeared. IFT81 is another component of the IFT-B complex.23 It is not surprising for these two proteins to interact as IFT components usually form a complex with other IFT proteins.23,24 It is highly possible that IFT20 directly associates with IFT81 during spermatogenesis. Floxed Ift81 mice were generated in our laboratory, and the role of IFT81 in male fertility and spermatogenesis is currently under investigation. IFT20 was identified to be a binding partner of sperm flagellar 2 (SPEF2) in a yeast two-hybrid screen when SPEF2 was used as a bait.21 The protein was not identified in our screen when IFT20 was used as a bait. It is possible that IFT20 is a major binding partner of SPEF2 and is easily selected under less stringent conditions (triple-dropout medium) using SPEF2 as a bait. However, in our screen, SPEF2 may not be the major binding partner and it is more difficult to be selected under stringent conditions (quadruple-dropout medium) using IFT20 as a bait. Similarly, GMAP210, another IFT20 binding partner,13,14 was not identified in the screen. Our recently investigation demonstrated that GMAP210 is essential for acrosome biogenesis and to determine IFT20 localization in the Golgi bodies and acrosome area.15 In the present study, we decided to characterize SPATA1 as this protein was identified to be another major binding partner of IFT20.
Spata1 mRNA is widely expressed in multiple mouse tissues, being the most abundant in the testis. To further investigate this gene, we generated a specific antibody. The protein is expressed as predicted size in the heart and testis. However, a smaller protein was detected in the brain, and multiple proteins, including the protein of the same size detected in the testis, were found in the kidney. These proteins may represent the translated proteins from other Spata1 transcripts that were reported in the Ensembl database (ENSMUST00000093951, ENSMUST00000200488, and ENSMUST00000195949). Even though the Spata1 mRNA expression in the heart was lower than that in the testis, the protein level of SPATA1 was higher. This might be due to higher translation efficiency in the heart, or lower translation efficiency in the testis. Spata1 transcripts were also detected in the lung, but no protein was detected by Western blot analysis. It might also be due to lower translation efficiency of Spata1 mRNA in the lung.
The dynamic expression pattern of SPATA1 protein during the first wave of spermatogenesis strongly suggests a role in spermiogenesis, and this notion is supported by its localization. SPATA1 was found to be localized in the acrosome, a unique membranous organelle formed only in male germ cells.25,26 Given that SPATA1 is present in the testicular germ cells rather than the epididymal sperm, it is likely that SPATA1 only has a role in the developing germ cells, and suggests that SPATA1 might be involved in biosynthesis of the acrosome of male germ cells.
It has been previously shown that IFT20 is present in the Golgi complex of late spermatocytes and round spermatids.21 This is consistent with its expression pattern during the first wave of spermatogenesis.22 IFT20 was detected from day 16 after birth, which was earlier than SPATA1 detection. IFT20 is present in the Golgi bodies in spermatocytes.21 However, we do not have evidence showing that SPATA1 is localized in the Golgi bodies in the present study, suggesting that the Golgi bodies may not be the major settlement of SPATA1. SPATA1 may represent other resources instead of Golgi bodies for acrosome biogenesis.27,28
In CHO cells, both SPATA1 and IFT20 were present as cytoplasmic vesicles; but when co-expressed, two proteins co-localized as large cytoplasmic clusters. Moreover, co-immunoprecipitation assay showed an interaction between SPATA1 and IFT20 in vitro and in vivo. IFT20 has been reported to be present in the Golgi complex of late spermatocytes and round spermatids, and in the manchette and basal body of elongating spermatids.21 The fact that SPATA1 localization in the acrosome prompted us to re-examine IFT20 localization in male germ cells using peanut-lectin, an acrosomal marker. The result confirmed our hypothesis that some IFT20 is also present in the acrosome area, and this localization is also supported by our recent studies on GMAP210 and COPS5, the latter is another IFT20 binding partner also identified in the same yeast two-hybrid screen.15,29 Thus, the acrosome area might be the major location for the two proteins, IFT20 and SPATA1 to interact. Interestingly, even though some IFT20 signal overlapped with the lectin, most IFT20 seemed to be on the top of lectin (Figure 3E). Given that SPATA1 signal is almost identical to the acrosomal marker, we can image that a small proportion of IFT20 binds to SPATA1 in the acrosome, but most IFT20 should be outside SPATA1. Thus, SPATA1 seems to provide the docking site of IFT20 complex binding to the acrosome area, and the model is now displayed as Figure 6. This hypothesis will be further examined when SPATA1 deficient mice are available. Our original speculation was that the acrosomal localization of SPATA1 is dependent on IFT20 in germ cells. However, our data showed that testicular SPATA1 expression remained at similar levels in the control and Ift20 knockout mice, and it was still present in the acrosome of Ift20-null spermatids before stage XII. It seems that IFT20 is not required for SPATA1 transport. At stage XII, the scattered localization of SPATA1 in the germ cells of Ift20 mutant mice might be due to secondary effects of impaired spermiogenesis. It has been reported that IFT20 is involved in the movement from the Golgi complex to the base of the cilium.31 Thus, IFT20 has a longer expression period during spermatogenesis than SPATA1. SPATA1 provides a docking site in the acrosome so that the IFT20 complex can attach on the acrosome surface in order to carry cargo proteins for sperm flagella formation. The function seems to be different from GMAP210 and COPS5. GMAP210 determines IFT20 localization in the Golgi bodies and is essential for IFT20-containing vesicles trafficking to the acrosome for acrosome biogenesis.15 COPS5 was initially identified as c-Jun activation domain-binding protein-1 (Jab1),32 a c-Jun coactivator, and subsequently discovered to be the fifth component of the constitutive photomorphogenic-9 signalosome (COP9, CSN), which phosphorylates target proteins, leading to their ubiquitination and degradation by the 26S proteasome.33 COPS5 is essential for acrosome formation in male germ cells.29 Testicular IFT20 level was significantly reduced in germ cell-specific Cops5 knockout mice, and the protein was no longer seen in the spermatid acrosome. However, the protein was present as small vesicles near one side of the nucleus, probably due to the fact that acrosome was not developed normally, and IFT20 was not able to fuse here. COPS5 might also modulate IFT20 level through Ubiquitin-Proteasome System.
FIGURE 6.
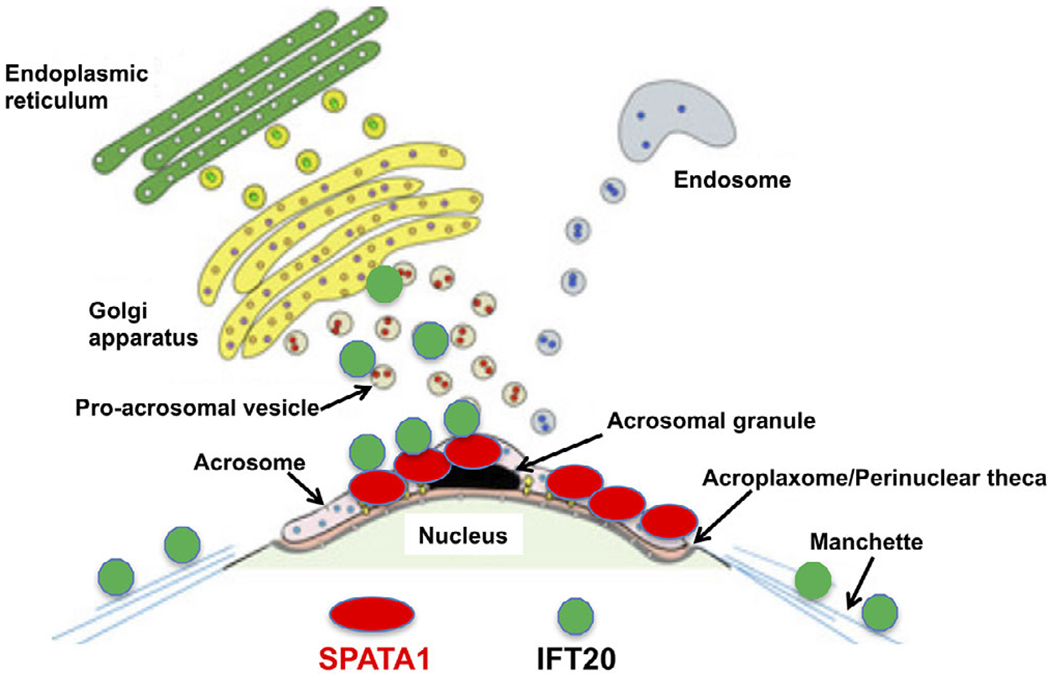
Working model of SPATA1/IFT20 interaction in male germ cells. The cartoon was drawn based on a model for acrosome biogenesis published previously.30 Two major systems, Golgi body vesicles and endosomal vesicles, contribute to acrosome formation.27 IFT20 is present in the Golgi bodies, acrosome area, and manchette. SPATA1 is detected in the acrosome, where it provides a docking site for IFT20
A number of proteins localized in the acrosome have been reported to be important for sperm head formation and fertility.30,34–37 Human SPATA1 is a strong candidate for influencing the shape of the sperm head.38 SPATA1 has also been shown to be associated with pregnancy for stallions.39,40 Given that IFT20 has not been shown to be involved in acrosome formation or function,22 it seems that IFT20 and SPATA1 have separate functions during spermatogenesis. We believe that SPATA1 may function as a key player for sperm head formation and is essential for mouse fertility. Our present studies provide a foundation for further examining the role of SPATA1 in the male germ cell development.
4 |. EXPERIMENTAL PROCEDURES
4.1 |. Ethics statement
All rodent work was approved by Virginia Commonwealth University’s Institutional Animal Care & Use Committee (protocol AD10000167) and Wayne State University (license number: IACUC-18-02-0534) in concordance with all federal and local regulations regarding the use of nonprimate vertebrates in scientific research.
4.2 |. Mice used in the study
Conditional Ift20 knockout mice were generated in our laboratory and described in the previous publication.22 Ift20flox/+; Stra8-Cre mice were used as the controls.
4.3 |. Yeast two-hybrid experiments
The full-length of mouse IFT20 coding sequence was amplified using the following primer set: IFT20forward: 5′-CATATGGCCAAGGACATCTTGGGC-3′; and IFT20reverse: 5′-GGATCCTCATTTCTGAAAAATAAATTGG-3′. After sequencing, the cDNA was cloned into pGBKT7, and the resulting plasmid was used to screen a Mate & Plate Library-Universal mouse normalized cDNA library (Clontech) according to the manufacturer’s instructions. A stringent protocol was used, and the yeast was grown on plates with quadruple-dropout medium lacking four amino acids (-Ade-His-Leu-Trp) (Clontech). The potential binding partners were sequenced using BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems) with a T7 sequencing primer, and the genes were identified by BLAST search using the NCBI service.
To detect interaction between mouse SPATA1 and IFT20, full-length Spata1 cDNA was amplified using the following primers: SPATA1forward: 5′-GAATTCGACAACATGTCGCTCAATCCAAGT-3′, and SPATA1reverse: 5′-CTCGAGCCTAGGTTGGCACACTTCCAGAAAA-3′. After sequencing, the cDNA was cloned into pGADT7 vector. SPATA1/pGADT7 and IFT20/pGBKT7 were cotransformed into the AH109 host strain using the Matchmaker Two-Hybrid System 3 (Clontech) according to the manufacturer’s instructions. Two plasmids containing simian virus (SV) 40 large T antigen (LgT) in pGADT7 and p53 in pGBKT7 were cotransformed into AH109 and used a positive control. The AH109 transformants harboring both SPATA1/pGADT7 and IFT20/pGBKT7 were streaked out in complete drop-out medium (SCM) lacking tryptophan, leucine, and histidine to test for histidine prototrophy. SCM lacking tryptophan and leucine was also used for a negative control.
4.4 |. Generation of a specific anti-SPATA1 polyclonal antibody
The coding sequence of the N-terminal portion from Spata1 was PCR-amplified from a Spata1 cDNA clone. The two primers used were: 5′-GAATTCGACAACATGTCGCTCAAT.
CCAAGT-3′ (forward) and 5′-CTCGAGCTCACAACTTGGAATTTCCAAAGTG-3′ (reverse). The PCR product was inserted into EcoRI/XhoI restriction sites of the pET28a expression vector (Novagen, Madison, Wisconsin). Notably, His6 tags at both the N and C termini were contained in the resulting fusion protein. The construct was transformed into Escherichia coli strain BL21 (DE3) cells, and the fusion protein was induced and subsequently purified as reported previously.41 The purified recombinant protein was used to generate polyclonal antisera in rabbits by a commercial organization (Antibody Research Center, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences, China).
4.5 |. Primary antibodies
Expression of the indicated proteins was examined using the following specific antibodies: SPATA1 antibody (1:1,000 dilution for Western blot, 1:200 dilution of IF); IFT20 antibody (1:1,000 dilution, generated by Gregory J. Pazour’s Laboratory at University of Massachusetts, Amherst, MA); FLAG, GFP and β-actin antibodies (1:1000 dilution, Cell Signaling Technology, Danvers, MA); Sp56 antibody (1:500 dilution, QED Bioscience, San Diego, CA); mouse IgG was used at the same concentration as a negative control (Cell Signaling Technology); TGN38(C-15) antibody (1:200 dilution, Santa Cruz Biotechnology, Dallas, TX); Golgin-97 antibody (1.0 μg/mL, ThermoFisher Scientific, Rockford, Illinois); GM130 (H-7) antibody (1:100, Santa Cruz Biotechnology); Golgin-84 (D-5) antibody (1:200 dilution, Santa Cruz Biotechnology); peanut-lectin antibody (20 μg/mL, ThermoFisher Scientific).
4.6 |. Western blot analysis
Tissue samples were homogenized in RIPA buffer (Thermo Scientific) with protease inhibitors (Complete mini, Roche Diagnostics, Indianapolis, IN) using an Ultra Turrax homogenizer and centrifuged at 13,000 rpm for 10 minutes at 4°C. Protein concentrations were quantified using DC protein assay kit (Bio-Rad, Hercules, California). Equal amount of protein (~50 μg per lane) was heated to 95°C for 10 minutes in 4xSDS sample buffer. Samples were resolved on a 10% sodium dodecyl sulfate-polyacrylamide gels and electro-transferred onto polyvinylidene difluoride membranes (Millipore, Billerica, Massachusetts). The membranes were then blocked in a Tris-buffered saline (TBST) solution containing 5% nonfat dry milk powder and 0.1% Tween 20 (TBST), and incubated with indicated primary antibodies at 4°C for 16 hours. After washing with TBST three times, the PVDF membrane was immersed in horseradish peroxidaselabeled goat anti-rabbit IgG with dilution of 1:2,000 at room temperature for 2 hours. Following by washing with TBST, the bound antibodies were detected with Super Signal Chemiluminescent Substrate (Pierce, Rockford, Illinois).
4.7 |. Reverse transcription-polymerase chain reaction (RT-PCR)
Total RNA was extracted from the indicated tissues (testis, brain, liver, kidney, heart, skeletal muscle, spleen, and lung) using TRIzol reagent (Invitrogen, Grand Island, New York), and RT was performed using the first-stand cDNA synthesis kit from Fermentas (Beijing, China). Briefly, the same amount of total RNA (1 μg) from each tissue was pretreated with DNase I (Promega, Madison, Wisconsin) and reverse transcribed with Moloney murine leukemia virus reverse transcriptase in the presence of random primers. One microliter of cDNA from each tissue was used as a template for PCR. A specific region of the Spata1 transcript was amplified by PCR with primers 5′-GAATTCGACAACATGTCGCTCAATCCAAGT-3′ (forward) and 5′-CCTCGTAATACTCCTTGGGT-3′ (reverse) using DreamTaq Green PCR master mix (Thermo Scientific, Beijing, China) and the following PCR amplification conditions: 35 cycles of 95°C for 30 seconds, 52°C for 30 seconds, and 72° C for 1 minute. β-actin was served as a control and amplified under the same conditions using the primers 5′-CTGTCCCTGTATGCCTCTG-3′ (forward) and 5′-ATGTCACGCACGATTTCC-3′ (reverse).
4.8 |. Cell culture and transient transfection
Chinese hamster ovary (CHO) and COS-1 were cultured with DMEM (with 10% fetal bovine serum) at 37°C. The cells were transfected with indicated plasmids using Fugene6 transfection reagent (Promega). After 48 hours transfection, the cells were processed either for immunofluorescence staining (CHO) or Western blot analysis (COS-1 cells).
4.9 |. Mammalian expression constructs
Mouse Spata1 coding region was amplified by PCR using testis cDNA as a template and the following primers: 5′-GAATTCGACAACATGTCGCTCAATCCAAGT-3′ (forward) and 5′-GGATCCCGGTTGGCACACTTCCAGAAAACAT-3′ (reverse). The correct PCR product was ligated into pEGFP-N2 vector (Clontech, California). To make SPATA1/pcDNA3 construct, the Spata1 coding region was amplified using the following primers: 5′-GAATTCGACAACATGTCGCTCAATCCAAGT-3′ (forward), and 5′-CTCGAGCCTAGGTTGGCACACTTCCAGAAAA-3′ (reverse). The correct PCR product was cloned into pcDNA3 vector (Invitrogen, New York). A plasmid used to express IFT20/Flag fusion protein in mammalian cells was kindly provided by Dr. Gregory J. Pazour at University of Massachusetts Medical School.
4.10 |. Co-immunoprecipitation assays
Protein expression plasmids IFT20/Flag and SPATA1/pEGFP-N2 were co-transfected into COS-1 cells and co-immunoprecipitation assays were performed with lysates from transfected mammalian cells following previously described methods.42 Briefly, the supernatant of cell lysates was precleaned with protein A beads at 4°C for 30 minutes and the precleared lysate was then incubated with the indicated antibodies or preimmune serum as a negative control at 4°C for 2 hours. The mixture was then incubated with protein A beads at 4°C overnight. The beads were washed with immunoprecipitation buffer (50 mM NaCl, 50 mM Tris-HCl, pH 8.0, 5 mM EDTA, 1% Triton X-100, 1 mM PMSF, proteinase inhibitor) three times. The beads were resuspended in 2× SDS sample buffer (100 mM Tris-HCl, pH 6.8, 4% SDS, 20% glycerol, 10% β-mercaptoethanol, 0.004% bromophenol blue) and boiled at 100° C for 10 minutes. The samples were centrifuged at 3,000 g for 30 seconds and the supernatant was then subjected to Western blot analysis with the indicated antibodies.
For tissue lysates, testicular extracts were prepared from adult wild-type mice using an immunoprecipitation buffer (150 mM NaCl/50 mM Tris-HCl, pH 8.0/5 mM EDTA/1% Triton X-100/1 mM PMSF/proteinase inhibitor mixture). Before the IP experiment, the extracts were precleared with protein A beads at 4°C for 30 minutes. After centrifuging at 12,000 rpm for 5 minutes, the beads were discarded and the supernatants were used for IP experiment. For each IP experiment, 1 mg of testicular protein was incubated with 1 μg of anti-SPATA1 antibody (rabbit IgG) or normal rabbit IgG (as a negative control) at 4°C for 2 hours, and protein A beads were added with a further incubation period at 4°C overnight. The beads were washed with immunoprecipitation buffer four times, and loading buffer was then added to the beads, which were then boiled at 100° C for 10 minutes; the samples were then processed for Western blotting with an anti-IFT20 antibody.
4.11 |. Preparation of testicular cells, epididymal sperm, and IF analysis
Enzyme-dissociated testicular cells were prepared as previously described with modification.43 Briefly, testes from adult mice were dissected in 5 mL DMEM containing 0.5 mg/mL collagenase IV and 1.0 mg/mL DNase I (Sigma-Aldrich, St. Louis, Missouri), and was then incubated at 32° C for 30 minutes with agitation. The released testicular cells were collected by centrifugation at 1,000 rpm for 5 minutes at 4° C. Spermatozoa from dissected cauda epididymis were directly released into PBS at 37° C for 5 minutes. After washing with PBS, the testicular cells and sperm were fixed by immersion in 5 mL of PBS containing 4% paraformaldehyde (PFA) and 0.1 M sucrose at room temperature for 10 minutes. The mixed testicular cells and sperm were washed with PBS three times and then resuspended in 2 mL of PBS. 50 μL of cell suspension was loaded to the slide and allowed to air-dry. For IF analysis, the cells were permeabilized with 0.1% Triton X-100 at 37°C for 5 minutes. The cells were washed with PBS, followed by blocking with 10% goat serum in PBS at room temperature for 1 hour. The cells were washed with PBS three times and incubated with indicated primary antibodies at 4°C overnight. For double-straining, the cells were incubated with a mixture of two primary antibodies. Then the cells were washed with PBS three times and incubated at room temperature with Cy3-conjugated anti-rabbit IgG secondary antibody (1:5,000; Jackson ImmunoResearch Laboratories, West Grove, Pennsylvania) or Alexa 488-conjugated anti-mouse IgG secondary antibody (1:3,000; Jackson ImmunoResearch Laboratories) for 1 hour. The slides were washed with PBS and mounted in VectaMount with DAPI (H1200, Vector Laboratories, Burlingame, California) and sealed with a cover slip. Images of testicular cells were captured by confocal laser-scanning microscopy (Zeiss LSM 700). Sperm images were taken by confocal microscopy (Leica CTR7000 HS) and processed using the MetaMorph software.
4.12 |. IF analysis of testis sections
Testis sections in IF analysis were prepared as previously described.44 Testes from adult control and conditional Ift20 knockout mice were fixed with 4% PFA in 0.1 M PBS (pH 7.4) by rocking at 4°C overnight, embedded in paraffin wax (Tisseu-Tek) and frozen at −80° C. Five micrometer sections of paraffin-embedded tissue were dewaxed in 100% xylene and rehydrated through a series of decreasing concentrations of ethanol (100%, 90%, 70%) and deionized water. Antigen retrieval was performed by immersing slides in prewarmed (95° C) Dako Target Retrieval Solution (S1699, Agilent) for 20 minutes. The slides were cooled down on the bench for 30 minutes and immersed in distilled water for 5 minutes. The tissue sections were then permeabilized in 0.1% Triton X-100 in PBS at 37°C for 5 minutes. The sections were blocked with 10% goat serum in PBS for 1 hour, followed by incubation with the preimmune serum or anti-SPATA1 antibody at 4°C overnight. The slides were washed with PBS and incubated with Cy3-conjugated anti-rabbit IgG secondary antibody (1:5,000; Jackson ImmunoResearch Laboratories) at room temperature for 1 hour. The slides were washed with PBS three times, and mounted using VectaMount with DAPI, and sealed with a cover slip. Images were captured by confocal laser-scanning microscopy (Zeiss LSM 700).
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Scott C. Henderson and Judy C. Williamson for their assistance with the confocal microscopy in Microscopy Core Facility of Virginia Commonwealth University.
Funding information
National Institutes of Health, Grant/Award Numbers: HD076257, HD090306; National Natural Science Foundation of China, Grant/Award Number: 81671514; Natural Science Foundation of Hubei Province, Grant/Award Number: 2018CFA040; Wayne State University Research Fund; Wayne State University Start up fund
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
SUPPORTING INFORMATION
Additional supporting information may be found online in the Supporting Information section at the end of this article.
REFERENCES
- 1.Kozminski KG, Johnson KA, Forscher P, Rosenbaum JL. A motility in the eukaryotic flagellum unrelated to flagellar beating. Proc Natl Acad Sci USA. 1993;90:5519–5523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mourao A, Christensen ST, Lorentzen E. The intraflagellar transport machinery in ciliary signaling. Curr Opin Struct Biol. 2016;41:98–108. [DOI] [PubMed] [Google Scholar]
- 3.Prevo B, Scholey JM, Peterman EJG. Intraflagellar transport: mechanisms of motor action, cooperation, and cargo delivery. FEBS J. 2017;284:2905–2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cole DG, Diener DR, Himelblau AL, Beech PL, Fuster JC, Rosenbaum JL. Chlamydomonas kinesin-II-dependent intraflagellar transport (IFT): IFT particles contain proteins required for ciliary assembly in Caenorhabditis elegans sensory neurons. J Cell Biol. 1998;141:993–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pazour GJ, Dickert BL, Witman GB. The DHC1b (DHC2) isoform of cytoplasmic dynein is required for flagellar assembly. J Cell Biol. 1999;144:473–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Porter ME, Bower R, Knott JA, Byrd P, Dentler W. Cytoplasmic dynein heavy chain 1b is required for flagellar assembly in Chlamydomonas. Mol Biol Cell. 1999;10:693–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scholey JM. Kinesin-2: a family of heterotrimeric and homodimeric motors with diverse intracellular transport functions. Annu Rev Cell Dev Biol. 2013;29:443–469. [DOI] [PubMed] [Google Scholar]
- 8.Aldahmesh MA, Li Y, Alhashem A, et al. IFT27, encoding a small GTPase component of IFT particles, is mutated in a consanguineous family with Bardet-Biedl syndrome. Hum Mol Genet. 2014;23:3307–3315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beales PL, Bland E, Tobin JL, et al. IFT80, which encodes a conserved intraflagellar transport protein, is mutated in Jeune asphyxiating thoracic dystrophy. Nat Genet. 2007;39:727–729. [DOI] [PubMed] [Google Scholar]
- 10.Halbritter J, Bizet AA, Schmidts M, et al. Defects in the IFT-B component IFT172 cause Jeune and Mainzer-Saldino syndromes in humans. Am J Hum Genet. 2013;93:915–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Perrault I, Halbritter J, Porath JD, et al. IFT81, encoding an IFT-B core protein, as a very rare cause of a ciliopathy phenotype. J Med Genet. 2015;52:657–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schaefer E, Stoetzel C, Scheidecker S, et al. Identification of a novel mutation confirms the implication of IFT172 (BBS20) in Bardet-Biedl syndrome. J Hum Genet. 2016;61:447–450. [DOI] [PubMed] [Google Scholar]
- 13.Follit JA, San Agustin JT, Xu F, et al. The Golgin GMAP210/TRIP11 anchors IFT20 to the Golgi complex. PLoS Genet. 2008;4:e1000315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Follit JA, Tuft RA, Fogarty KE, Pazour GJ. The intraflagellar transport protein IFT20 is associated with the Golgi complex and is required for cilia assembly. Mol Biol Cell. 2006;17:3781–3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Z, Shi Y, Ma S, et al. Abnormal fertility, acrosome formation, IFT20 expression and localization in the conditional Gmap210 knockout mice. Am J Physiol Cell Physiol. 2019. 10.1152/ajpcell.00517.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jonassen JA, San Agustin J, Follit JA, Pazour GJ. Deletion of IFT20 in the mouse kidney causes misorientation of the mitotic spindle and cystic kidney disease. J Cell Biol. 2008;183:377–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Keady BT, Le YZ, Pazour GJ. IFT20 is required for opsin trafficking and photoreceptor outer segment development. Mol Biol Cell. 2011;22:921–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.May-Simera HL, Petralia RS, Montcouquiol M, et al. Ciliary proteins Bbs8 and Ift20 promote planar cell polarity in the cochlea. Development. 2015;142:555–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Finetti F, Paccani SR, Riparbelli MG, et al. Intraflagellar transport is required for polarized recycling of the TCR/CD3 complex to the immune synapse. Nat Cell Biol. 2009;11:1332–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yuan X, Garrett-Sinha LA, Sarkar D, Yang S. Deletion of IFT20 in early stage T lymphocyte differentiation inhibits the development of collagen-induced arthritis. Bone Res. 2014;2:14038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sironen A, Hansen J, Thomsen B, et al. Expression of SPEF2 during mouse spermatogenesis and identification of IFT20 as an interacting protein. Biol Reprod. 2010;82:580–590. [DOI] [PubMed] [Google Scholar]
- 22.Zhang Z, Li W, Zhang Y, et al. Intraflagellar transport protein IFT20 is essential for male fertility and spermiogenesis in mice. Mol Biol Cell. 2016;27:3705–3716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lucker BF, Behal RH, Qin H, et al. Characterization of the intraflagellar transport complex B core: direct interaction of the IFT81 and IFT74/72 subunits. J Biol Chem. 2005;280:27688–27696. [DOI] [PubMed] [Google Scholar]
- 24.Behal RH, Miller MS, Qin H, Lucker BF, Jones A, Cole DG. Subunit interactions and organization of the Chlamydomonas reinhardtii intraflagellar transport complex A proteins. J Biol Chem. 2012;287:11689–11703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Berruti G, Paiardi C. Acrosome biogenesis: revisiting old questions to yield new insights. Spermatogenesis. 2011;1:95–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hermo L, Pelletier RM, Cyr DG, Smith CE. Surfing the wave, cycle, life history, and genes/proteins expressed by testicular germ cells. Part 2: changes in spermatid organelles associated with development of spermatozoa. Microsc Res Tech. 2010;73: 279–319. [DOI] [PubMed] [Google Scholar]
- 27.Berruti G, Paiardi C. USP8/UBPy-regulated sorting and the development of sperm acrosome: the recruitment of MET. Reproduction. 2015;149:633–644. [DOI] [PubMed] [Google Scholar]
- 28.Escalier D, Gallo JM, Albert M, et al. Human acrosome biogenesis: immunodetection of proacrosin in primary spermatocytes and of its partitioning pattern during meiosis. Development. 1991;113:779–788. [DOI] [PubMed] [Google Scholar]
- 29.Huang Q, Liu H, Zeng J, et al. COP9 signalosome complex subunit 5, an IFT20 binding partner, is essential to maintain male germ cell survival and acrosome biogenesis. Biol Reprod. 2019;pii: ioz154 10.1093/biolre/ioz154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fujihara Y, Satouh Y, Inoue N, Isotani A, Ikawa M, Okabe M. SPACA1-deficient male mice are infertile with abnormally shaped sperm heads reminiscent of globozoospermia. Development. 2012;139:3583–3589. [DOI] [PubMed] [Google Scholar]
- 31.Crouse JA, Lopes VS, Sanagustin JT, Keady BT, Williams DS, Pazour GJ. Distinct functions for IFT140 and IFT20 in opsin transport. Cytoskeleton. 2014;71:302–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Claret F-X, Hibi M, Dhut S, Toda T, Karin M. A new group of conserved coactivators that increase the specificity of AP-1 transcription factors. Nature. 1996;383:453–457. [DOI] [PubMed] [Google Scholar]
- 33.Cope GA, Suh GS, Aravind L, et al. Role of predicted metalloprotease motif of Jab1/Csn5 in cleavage of Nedd8 from Cul1. Science. 2002;298:608–611. [DOI] [PubMed] [Google Scholar]
- 34.Geyer CB, Inselman AL, Sunman JA, Bornstein S, Handel MA, Eddy EM. A missense mutation in the Capza3 gene and disruption of F-actin organization in spermatids of repro32 infertile male mice. Dev Biol. 2009;330:142–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kherraf ZE, Christou-Kent M, Karaouzene T, et al. SPINK2 deficiency causes infertility by inducing sperm defects in heterozygotes and azoospermia in homozygotes. EMBO Mol Med. 2017;9:1132–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xiao N, Kam C, Shen C, et al. PICK1 deficiency causes male infertility in mice by disrupting acrosome formation. J Clin Invest. 2009;119:802–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yao R, Ito C, Natsume Y, et al. Lack of acrosome formation in mice lacking a Golgi protein, GOPC. Proc Natl Acad Sci U S A. 2002;99:11211–11216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.L’Hote D, Serres C, Laissue P, et al. Centimorgan-range one-step mapping of fertility traits using interspecific recombinant congenic mice. Genetics. 2007;176:1907–1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Giesecke K, Hamann H, Stock KF, Woehlke A, Sieme H, Distl O. Evaluation of SPATA1-associated markers for stallion fertility. Anim Genet. 2009;40:359–365. [DOI] [PubMed] [Google Scholar]
- 40.Giesecke K, Sieme H, Distl O. Infertility and candidate gene markers for fertility in stallions: a review. Vet J. 2010;185:265–271. [DOI] [PubMed] [Google Scholar]
- 41.Zhang Z, Jones BH, Tang W, et al. Dissecting the axoneme interactome: the mammalian orthologue of Chlamydomonas PF6 interacts with sperm-associated antigen 6, the mammalian orthologue of Chlamydomonas PF16. Mol Cell Proteomics. 2005;4:914–923. [DOI] [PubMed] [Google Scholar]
- 42.Zhang Z, Shen X, Gude DR, et al. MEIG1 is essential for spermiogenesis in mice. Proc Natl Acad Sci U S A. 2009;106:17055–17060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Steiner R, Ever L, Don J. MEIG1 localizes to the nucleus and binds to meiotic chromosomes of spermatocytes as they initiate meiosis. Dev Biol. 1999;216:635–645. [DOI] [PubMed] [Google Scholar]
- 44.Robertson D, Savage K, Reis-Filho JS, Isacke CM. Multiple immunofluorescence labelling of formalin-fixed paraffin-embedded (FFPE) tissue. BMC Cell Biol. 2008;9:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


