A pollen-specific protein activates callose synthases by sequestering an RNA binding protein away from the mRNA encoding a transcriptional repressor, leading to the callose synthesis and deposition required for male gametogenesis and pollen germination.
Abstract
Spatiotemporally regulated callose deposition is an essential, genetically programmed phenomenon that promotes pollen development and functionality. Severe male infertility is associated with deficient callose biosynthesis, highlighting the significance of intact callose deposition in male gametogenesis. The molecular mechanism that regulates the crucial role of callose in production of functional male gametophytes remains completely unexplored. Here, we provide evidence that the gradual upregulation of a previously uncharacterized cotton (Gossypium hirsutum) pollen-specific SKS-like protein (PSP231), specifically at the post pollen-mitosis stage, activates callose biosynthesis to promote pollen maturation. Aberrant PSP231 expression levels caused by either silencing or overexpression resulted in late pollen developmental abnormalities and male infertility phenotypes in a dose-dependent manner, highlighting the importance of fine-tuned PSP231 expression. Mechanistic analyses revealed that PSP231 plays a central role in triggering and fine-tuning the callose synthesis and deposition required for pollen development. Specifically, PSP231 protein sequesters the cellular pool of RNA-binding protein GhRBPL1 to destabilize GhWRKY15 mRNAs, turning off GhWRKY15-mediated transcriptional repression of GhCalS4/GhCalS8 and thus activating callose biosynthesis in pollen. This study showed that PSP231 is a key molecular switch that activates the molecular circuit controlling callose deposition toward pollen maturation and functionality and thereby safeguards agricultural crops against male infertility.
Male gametogenesis within the anthers of flowering plants involves a series of spatiotemporally highly orchestrated complex molecular events, leading to the production of functional male gametophytes (pollen). During male gametogenesis, the microspore first undergoes an asymmetric cell division called pollen mitosis I (PMI). Then, as one of the products of PMI, the larger vegetative cell engulfs the small germ cell in its cytoplasm. This asymmetric division enables the establishment of differential cell fates for the germ cell and the vegetative cell. After PMI, the large vegetative cell exits the cell cycle at the G1 phase, while the smaller germ cell continues through a further cell division, PMII, to produce twin sperm cells (Sprunck and Gross-Hardt, 2011; Russell and Jones, 2015). In most plant species that shed pollen at the bicellular state, including tobacco (Nicotiana tabacum) and cotton (Gossypium hirsutum), PMII takes place inside the growing pollen tube. For other plant species, such as Arabidopsis (Arabidopsis thaliana), that shed tricellular pollen, PMII takes place inside the pollen grain before anthesis (Borg et al., 2009). Following their release from the anther, pollen grains land on receptive stigmas and germinate to produce tip-growing pollen tubes that grow through stigmatic tissue and into the transmitting tract of the style (Dresselhaus et al., 2016). In this process, cell walls of pollen tubes play crucial regulatory and structural roles. Distinct from other plant cells, a pollen tube wall does not contain the conventional secondary cell wall layer, but is composed of two characteristic layers, an inner layer mainly consisting of pectin surrounded by an outer layer formed by deposition of callose molecules (Chebli et al., 2012).
Callose deposition is well known to be transient yet indispensable for various highly specialized developmental and cellular processes, especially pollen maturation and cell division plate formation. Callose typically consists of a linear 1,3-β-glucan polymer that participates multiple times as a transitory component during pollen development in plants (Chen and Kim 2009). During microsporogenesis, callose deposition occurs in the walls of microsporocytes and microspores from meiosis to the tetrad stage. This callose wall is transient and quickly degraded to release individual microspores. Previous reports highlight that callose deposition is vital for pollen development. In Arabidopsis, the callose synthase CalS5 knockout mutant lacks normal callose walls at the tetrad stage, impacting the exine pattern of the microspore and resulting in male sterility (Dong et al., 2005). Both rice (Oryza sativa) callose synthase GSL5 and Arabidopsis CalS5 are expressed in meiocytes, tetrads, and microspores, with the highest levels of expression in late meiosis and newly released microspores. Loss of function in these plants results in abnormal tetrads (Dong et al., 2005; Shi et al., 2015). After PMI, callose forms a prominent but transient cell wall that separates the vegetative cell and the germ cell (Park and Twell, 2001). The gsl8 and gsl10 mutants harbor aborted pollen grains that fail to enter mitosis, suggesting that callose synthesis is crucial for pollen cell mitosis (Töller et al., 2008). During pollen tube elongation within the style, callose becomes the major component of the pollen tube wall and the characteristic plugs that form along the length of the tube (Li et al., 1997; Ferguson et al., 1998). Specifically, the callose wall is continuously built around the pollen tube behind its growing tip, mainly providing structural support. On the other hand, callose plugs are generated at regular intervals along the growing pollen tube to keep the twin sperm cells together, with cytoplasmic components concentrated near the tip of the growing pollen tube, preparing the sperm cells for fertilization (Feijó et al., 2004).
Although the SKS (SKU5-similar) family is structurally related to multiple copper oxidases, none of the conserved His residues used to bind copper are observed in the SKS proteins (Sedbrook et al., 2002; Jacobs and Roe, 2005). Also, our understanding of the SKS family members, especially in relation to pollen development, remains limited and largely phenotypical. In Brassica napus, a SKS-like gene, Bp10, was reported to be specifically expressed in developing pollen (Albani et al., 1992). Furthermore, the tobacco SKS-like gene NTP303 displays pollen-specific expression patterns, with its highest levels occurring during pollen maturation and tube growth (Weterings et al., 1992, 1995), while it encodes a glycosylated protein that localizes at the vegetative plasma membranes (Capková et al., 1997; Wittink et al., 2000). In NTP303-silenced plants, the pollen tubes display a slower growth rate and arrest in the style before reaching the ovary (de Groot et al., 2004). However, the molecular mechanisms by which these SKS-like proteins promote pollen development remain unknown. Here, we report comprehensive in vitro and in vivo functional and mechanistic studies on a cotton pollen-specific SKS-like protein, PSP231. Mechanistically, PSP231 promotes male gametogenesis and pollen germination by activating a previously dormant molecular pathway controlling callose biosynthesis. This study not only provides evidence on the identity of genetic factors but also describes the mechanism responsible for switching on and off the multiple transient callose deposition events indispensable for pollen maturation and germination.
RESULTS
PSP231 Encodes a Glycosylated Protein with a High Level of Expression in Late Maturing Pollen
To investigate the molecular mechanisms driving cotton pollen development, we constructed a cotton flower complementary DNA (cDNA) library (Wang and Li 2009) aimed at identifying molecules specifically present during the generation of functional male gametophytes for functional and mechanistic studies. Within the cotton flower cDNA library, the highly abundant presence of the cDNA clone-231, encoding a previously uncharacterized protein, caught our interest. Thus, we isolated the full-length clone-231 gene, including its promoter fragment. Bioinformatic analysis suggested that clone-231 was likely related to the plant SKS-like protein family. The clone-231 transcripts were most abundantly present in late developing anthers, but were undetectable in other tissues (Fig. 1A; Supplemental Fig. S1M).
Figure 1.
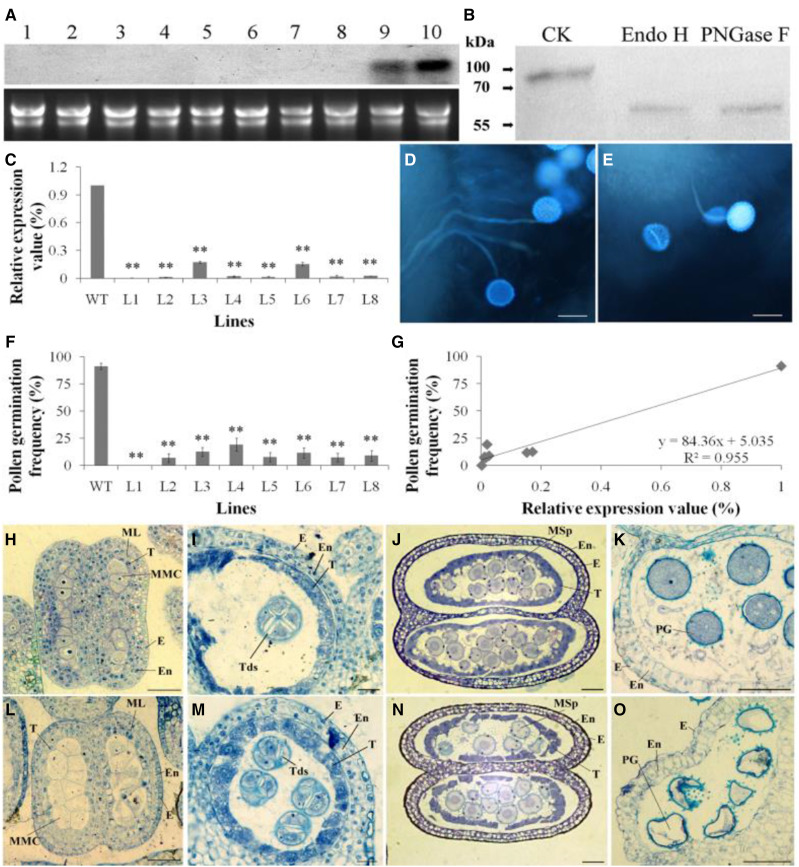
PSP231 deficiency impairs pollen development and germination. A, RNA gel blot analysis of PSP231 expression (top) in cotton cotyledons (lane 1), leaves (lane 2), roots (lane 3), hypocotyls (lane 4), petals (lane 5), fibers (lane 6), ovules (lane 7), 15-d-old anthers (lane 8), 20-d-old anthers (land 9), and 25-d-old anthers (lane 10), and loading of RNA samples (bottom). B, Assay of PSP231 glycosylation. Cotton anther proteins were treated with buffer as control (CK), Endo H, and PNGase F, respectively. Protein gel blot was performed to determine the migration of PSP231 using PSP231 antibody. C, RT-qPCR analysis of PSP231 expression in anthers of RNAi cotton lines (L1–L8). Expression level of PSP231 in the wild type (WT) is set to 1. Data are means ± sd for three replicates. Dunnett's t test demonstrated that there were very significant differences (**P < 0.01) in gene expression level between the PSP231 RNAi plants and the wild type. D and E, In situ pollen germination of wild-type (D) and PSP231 RNAi cotton (E). Scale bars = 100 μm. F, Percentage of in vivo pollen germination of wild-type and PSP231 RNAi cotton. More than 200 pollen grains for each PSP231 RNAi line or the wild type were examined for in vivo pollen germination (see “Materials and Methods”). Data are means ± sd of three replicates. Dunnett's t test demonstrated that there was a very significant difference in the percentage of germinated pollen grains between the PSP231 RNAi plants and the wild type (**P < 0.01). G, Correlation between pollen germination and relative expression value of PSP231 for the RNAi plants. H to O, Comparative analysis of the histological features of anther development in 9-d-old (H), 11-d-old (I), 12-d-old (J), and 20-d-old (K) wild-type anther and 9-d-old (L), 11-d-old (M), 12-d-old (N), and 20-d-old PSP231 RNAi anther (O). Images show cross sections through a locule of the anther. Scale bars = 25 μm (H, I, J, L, M, and N) and 50 μm (K and O). E, Epidermis; En, endothecium; ML, middle layer; MMC, microspore mother cell; MSp, microspore; T, tapetum; Tds, tetrad; PG, pollen grain; Fb, fibrous bands.
Given the high specificity and stepwise upregulation of its expression during cotton anther maturation, we hypothesized that clone-231 expression is specifically turned on only in certain cell types within the anther during a developmental stage for a specific functional purpose. Notably, in situ hybridization analysis revealed that clone-231 transcripts accumulated at the highest level specifically in maturing pollen within 25-d-old anthers. In contrast, no or very weak signals were detected in other cell types inside these 25-d-old anther tissues, or in younger anthers, compared with negative controls (Supplemental Fig. S1, A–F).
In order to confirm our in situ hybridization results at the protein level, we generated an antibody against clone-231 protein. The expected Mr of clone-231 protein as deduced from its sequence is about 61 kD. However, clone-231 proteins migrated substantially more slowly than expected on a SDS-PAGE gel, with an apparent Mr of ∼100 kD. Mass spectrometric analysis demonstrated that this 100-kD protein is indeed the clone-231 protein (Supplemental Fig. S2), suggesting that it probably has large posttranslational modifications such as glycosylation. Notably, western blot analysis revealed that after treatment with endoglycosidase H (Endo H) or peptide N-glycosidase F (PNGase F), clone-231 proteins from 25-d-old cotton anthers migrated faster than those from the untreated control (Fig. 1B), indicating that clone-231 is a glycosylated protein. Additionally, immunofluorescence analysis revealed that the clone-231 protein was localized in the pollen wall and cytoplasm (Supplemental Fig. S3). Subsequently, we determined the clone-231 protein expression profile in situ via immunohistochemistry. As expected, staining of tissue sections from anthers at different developmental stages (with normal rabbit serum as a negative control) generated no detectable signal (Supplemental Fig. S1, G–I). Immunohistochemistry analysis with the anti-clone-231 antibody on anthers of different developmental stages resulted in robust positive signals specifically in maturing pollen grains of 25-d-old cotton anthers, although no detectable signal was observed in other cell types within these tissues, nor in any cell types of the younger anthers (Supplemental Fig. S1, J–L). In addition, histochemical assay of GUS activity confirmed that the clone-231 promoter is pollen specific (Supplemental Fig. S1, N–U). Therefore, the protein encoded by clone-231 was named Pollen-Specific Protein 231 (PSP231).
Collectively, these results indicate that PSP231 expression is tightly spatiotemporally regulated in cotton. The highly specific accumulation of PSP231 proteins in maturing pollen suggests that its presence at the late developmental stage of anthers likely promotes cellular activities specifically needed for pollen maturation.
PSP231-Deficiency Impairs Pollen Development and Germination
To investigate the functional significance of the accumulation of PSP231 proteins specifically in maturing pollen, we employed RNA interference (RNAi) to knock down PSP231 expression stably in cotton plants. More than 120 plants of eight independent lines of PSP231 RNAi cotton were regenerated and transferred into soil to grow to maturation. PSP231 expression was dramatically repressed in 25-d-old anthers of the PSP231 RNAi plants (Fig. 1C). In accordance with the absence of PSP231 expression prior to anther maturation, phenotypic analysis of the PSP231 RNAi plants indicated that they grew normally relative to the wild type throughout the vegetative stage. However, these PSP231-silenced cotton plants exhibited a near-complete male-sterile phenotype once they reached the reproductive stage. Thus, we performed a more detailed analysis on three independent lines (L3, L5, and L8) of PSP231 RNAi cotton. Given the specific upregulation of PSP231 expression in maturing pollen, we examined the capability of PSP231-silenced pollen to germinate in vivo. Mature styles of wild-type cotton plants were pollinated with the same amount of pollen grains collected from either PSP231-silenced plants or wild-type controls. After 4 h, the pollinated styles were harvested and stained with aniline blue to examine pollen germination. As expected, almost all wild-type pollen grains (91.3% ± 3.0%) successfully germinated on the stigma, and pollen tubes grew normally within styles (Fig. 1, D and F). In striking contrast, only 0% to 19.1% ± 5.9% of pollen grains from the PSP231-silenced cotton lines could germinate on stigmas under the same conditions (Fig. 1, E and F). Regression analysis indicated that pollen germination frequency was correlated with expression level of PSP231 in anthers (Fig. 1G), demonstrating that upregulation of PSP231 expression specifically during late anther maturation is essential for production of fertile pollen.
To delineate the cause of PSP231-deficiency-associated male infertility at the cellular level, we histomorphologically analyzed all tissue architecture and cell types from PSP231-silenced and wild-type cotton during the key stages of anther maturation. Consistent with the undetectable expression of PSP231 in early developing anthers, there was no substantial difference in microsporogenesis between PSP231-silenced cotton plants and the wild type. Both wild-type (Fig. 1, H–J) and PSP231-silenced plants (Fig. 1, L–N) formed normal pollen mother cells, which underwent meiosis to produce microspores. As the anther further matured to the imminent stage of pollen release, pollen grains from wild-type plants exhibited a healthy round shape and a strong positivity for toluidine blue, with uniform blue staining inside the cytoplasm surrounded by a dark-blue staining of the cell wall (Fig. 1K). In striking contrast, almost all pollen grains from PSP231-silenced plants displayed an irregular shape and an almost complete absence of toluidine blue staining inside the cytoplasm, with light-blue-stained cell walls. Also, withered pollen grains were frequently observed in these mutant plants (Fig. 1O). The above data collectively demonstrate that PSP231 deficiency impairs pollen development of cotton, leading to the formation of aberrant, unhealthy pollen and the observed male infertility phenotype associated with PSP231 silencing.
PSP231 Overexpression Undermines Pollen Viability and Functionality in an Expression-Dependent Manner
Given the negative impact of PSP231 deficiency on pollen development and the stepwise upregulation of PSP231 expression highly specific to the final stage of anther maturation, we hypothesized that the expression level of PSP231 needs to be tightly regulated and fine-tuned to an optimal level such that either loss or gain of function of PSP231 can impair male gametogenesis. Since tobacco plants are routinely employed to examine the effects of gene overexpression, we generated knockin models of the PSP231 gene under the control of its own promoter in tobacco. The PSP231 overexpression in 20-d-old anthers of PSP231 knockin tobacco plants was confirmed by reverse transcription quantitative PCR (RT-qPCR) analysis (Fig. 2A). To address whether the role of PSP231 during pollen maturation is expression dependent, we employed three independent lines of PSP231 knock-in tobacco with mildly (L2), moderately (L6), and substantially (L3) higher expression levels of PSP231 compared with the wild type for further detailed analyses. Consistent with the undetectable expression of PSP231 during early stages of anther development, we did not observe any phenotypic difference between the transgenic plants and the wild type during the vegetative and generative developmental stages. However, as the anther matured, we observed a drastic decrease in the number of pollen grains contained in anther locules from the L3 transgenic line (with the highest expression level of PSP231) compared with wild-type controls. In contrast to the wild-type anthers, which contained an abundance of uniformly rounded, healthy-looking pollen grains (Fig. 2B), anther locules from the L3 transgenic line harbored substantially fewer pollen grains, and these frequently displayed a much smaller, irregularly shaped, unhealthy appearance (Fig. 2C), suggesting that the abnormally high level of PSP231 adversely affects pollen development.
Figure 2.
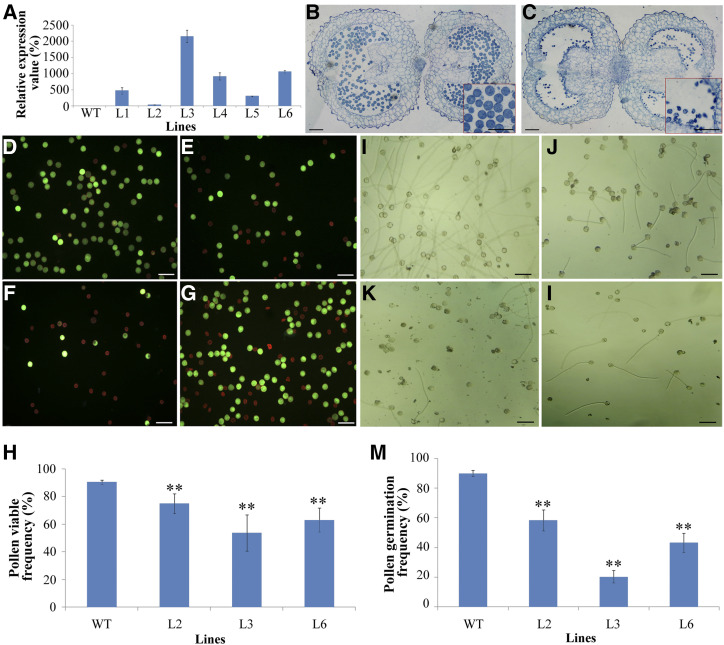
PSP231 overexpression dose-dependently undermines pollen viability and functional ability. A, RT-qPCR analysis of PSP231 expression in anthers of T3-generation wild type and transgenic lines (L1–L6) in tobacco. B and C, Semithin sections of wild-type (WT; B) and transgenic (C) mature anthers. Insets (red boxes) show amplified segments of the images. D to G, Assay of pollen viability in PSP231 transgenic tobacco (see “Materials and Methods”) for pollen grains from the wild type (D) and three PSP231 transgenic lines (L2 [E], L3 [F], and L6 [G]). Viable pollen grains display green fluorescence, whereas unviable pollen grains are red. H, Statistical analysis of the percentage of viable pollen in different transgenic plants and the wild type. I to M, Assay of in vitro pollen germination of the wild type and PSP231 transgenic lines for germinating pollen grains from the wild type (I) and three PSP231 transgenic lines (L2 [J], L3 [K], and L6 [L]). M, Statistical analysis of pollen germination rate (%) for different transgenic plants and the wild type. For H and M, data are means ± sd for three replicates. Dunnett's t test demonstrated that there was significant difference (*P < 0.05) or very significant difference (**P < 0.01) in percentage of viable pollen and pollen germination rate between the PSP231 transgenic plants and the wild type. Scale bars = 100 μm.
To distinguish live from dead pollen grains inside anthers, we double-stained mature pollen grains from the wild type and transgenic lines with fluorescein diacetate (FDA) and propidium iodide (PI), which stain live cells green and dead cells red, respectively. We performed a quantitative analysis of pollen viability on the wild type and transgenic lines L2, L6, and L3; see “Materials and Methods”). As expected, 90.4% ± 1.7% of wild-type pollen grains were viable and stained green in color. In striking contrast, the L2, L6, and L3 PSP231 knockin lines had 75.0% ± 1.7%, 63.0% ± 8.7%, and 53.8% ± 10.1% viable pollen grains, respectively (Fig. 2, D–H). Collectively, these data demonstrate that abnormally high expression levels of PSP231 undermine pollen productivity and viability in an expression-dependent manner.
Since the reduced pollen cell viability caused by excessive amounts of PSP231 is likely a gradual process, we further hypothesized that the pollen grains in the transgenic lines are also functionally impaired in a PSP231 expression dependent manner. We examined the capability of pollen grains from the transgenic lines and the wild type to germinate in vitro. As expected, 89.9% ± 2.0% of wild-type pollen grains could germinate, and their pollen tubes grew rapidly (Fig. 2, I and M). Interestingly, the germination capability of pollen grains from the transgenic lines decreased significantly compared with that of the wild type, while the severity of the PSP231 overexpression-associated functional defect correlated positively with the expression levels of PSP231. Specifically, only about 58.4% ± 7.1%, 43.3% ± 6.4%, and 20.3% ± 4.2% of pollen grains from the L2, L6, and L3 transgenic lines, respectively, were capable of germination (Fig. 2, J–M). Notably, for the germinated transgenic pollen grains, the elongated pollen tubes were markedly shorter than those of wild-type controls, suggesting their unhealthy cellular state and abnormal functionality.
Collectively, PSP231 overexpression undermines pollen development, viability, and functionality. Given that both abnormal upregulation and downregulation of PSP231 significantly impair pollen development, this gene represents an important player during male gametogenesis. Notably, its expression is not only tightly regulated to be turned on specifically during the late stage of anther maturation, but also needs to be fine-tuned to an optimal level for male gametogenesis.
PSP231 Overexpression Triggers Abnormal Mitotic Cell Death
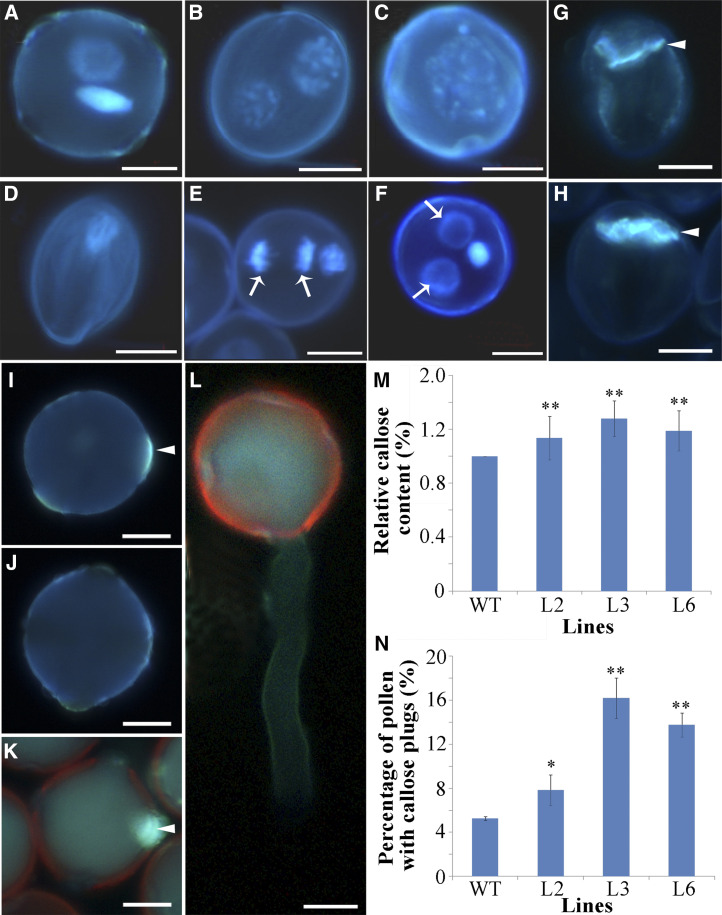
To gain insight into the molecular mechanism driving PSP231-overexpression-induced pollen defects during male gametogenesis, we carefully examined pollen at different developmental stages in the transgenic plants by staining with 4′,6-diamino-2-phenylindole (a nucleus-specific dye), using wild-type plants as controls. Microscopic observations revealed no substantial difference between transgenic plants and the wild type in the early stage of pollen development. Like the wild type, the transgenic plants formed normal microspores at the uninucleate stage. After microspore division, bicellular mature pollen grains were observed in the wild type (Fig. 3A). Conversely, a range of abnormal phenotypes of the transgenic plants were observed during pollen development. In the transgenic plants, pollen grains developed into defective male gametophytes with two vegetative-like nuclei nearly identical in size (Fig. 3B), one giant nucleus arising from nuclear fusion (Fig. 3C), an aborted nucleus (Fig. 3D), abnormal cell division (Fig. 3E), or two vegetative cells and one germ cell (Fig. 3F). Pollen with aborted nuclei comprised the majority of the aberrant cellular phenotypes at the mature pollen stage in the transgenic plants (Fig. 3D). These results demonstrate that overexpression of PSP231 hindered normal cell division during male gametogenesis of the transgenic plants.
Figure 3.
Overexpression of PSP231 results in aberrant hyper-callose deposition impairing pollen cell division. A to F, Pollen grains of T3-generation transgenic tobacco lines L2, L3, and L6 and the wild type were stained by 4',6-diamidino-2-phenylindole (a nucleus-specific dye that emits stronger fluorescence when bound to DNA). Shown are mature and bicellular pollen of the wild type (control; A), two equal nuclei in transgenic pollen (B), fusion nuclei in transgenic pollen (C), aborted transgenic pollen (D), transgenic pollen with abnormal cell division (E), and two vegetative cell nuclei in transgenic pollen (F). Arrows indicate extra vegetative cell nuclei. G and H, Decolorized aniline blue staining showing the callose wall between the vegetative cell and the germ cell in pollen from wild-type (G) and PSP231 transgenic tobacco (H). I and J, Aniline blue-stained mature pollen from PSP231 transgenic tobacco (I) and the wild type (J). K and L, Aniline blue-stained pollen germinated in vitro from PSP231 transgenic tobacco lines (K) and the wild type (L). M, Statistical analysis of the relative callose content in dividing pollen cells. Relative callose content in the wild type (WT) is set to 1. N, Statistical analysis of the percentage (%) of pollen grains with callose plugs in germ pores in the transgenic lines and wild-type tobacco. For M and N, data are means ± sd for three replicates. Dunnett's t test demonstrated that there was significant difference (*P < 0.05) or very significant difference (**P < 0.01) between the PSP231 transgenic plants and the wild type. Arrowheads in G, H, I, and K indicate callose deposition. Scale bars = 10 μm (A–H) and 2 μm (I–L).
To obtain further insight into the molecular mechanism of PSP231-overexpression-associated cell division defects, we overexpressed PSP231 in fission yeast (Schizosaccharomyces pombe) cells, and examined cell division status in the presence or absence of induction of exogenous PSP231 expression. As expected, no difference was observed for yeast cells harboring the empty vector under noninductive or inductive culture conditions, and ∼20% of cells in both conditions were at the late division phase (Supplemental Fig. S4, G–I), characterized by the formation of a visible callose-rich cell division plate. Interestingly, for the yeast cells harboring the inducible PSP231-overexpression construct, >50% of the cells cultured under the inductive condition for PSP231 overexpression had the obvious presence of cell division plates, indicating an ∼2.5-fold increase compared with the same cells cultured under the noninductive condition (Supplemental Fig. S4, E, F, and I). The above data corroborate the PSP231 transgenic pollen data on the remarkable impact of PSP231 overexpression on cell division (Fig. 3, A–F).
To determine PSPS231-overexpression-associated cell division defects, growth of the above yeast cells was analyzed on solid medium in both the induction and noninduction conditions. As expected, both yeast cells harboring the empty vector and those harboring the PSP231 overexpression construct showed similar growth rates on noninduction solid medium (Supplemental Fig. S4A). However, when they grew on solid medium with induced exogenous PSP231 expression, cells harboring the PSP231 overexpression construct displayed an obvious growth retardation phenotype compared with controls (Supplemental Fig. S4B). To determine the cause of this PSP231-overexpression-induced growth defect, the viability of these yeast cells was examined by FDA-PI, which stains dead cells red. As expected, there was no significant difference in cell viability between the PSP231 transformants and controls cultured in noninduction medium. Notably, whereas the control cells in induction medium displayed ∼98% viability and healthy growth, the viability of the PSP231-transformed lines in induction medium significantly decreased to be only 70.2% to 80.8% (Supplemental Fig. S4, J–N). Consistently, under induction conditions, the cumulative cell growth curves of the PSP231 transformants were significantly lower than those of controls (Supplemental Fig. S4, C and D). Collectively, these results suggest that overexpression of PSP231 abnormally traps dividing cells at the late division phase with their cell division plates unable to be efficiently dissolved, consequently resulting in abnormal mitotic cell death that probably caused the diminished pollen production in PSP231 knockin plants.
Aberrant Hyperproduction and Deposition of Callose Causes PSP231-Overexpression-Associated Cell Death
Given the association of GhPSPS231 overexpression with the observed failure of dissolution of cell division plates and the accompanying increase in cell death, we hypothesized that PSP231 overexpression impacts the components at the cell division plate or septum, a shared characteristic structure of both developing pollen and dividing fission yeast cells. To gain a deeper understanding of their trapped status with the aberrant cell division plates, the above yeast cells were examined by optical microscopy and transmission electron microscopy. As shown in Supplemental Figure S5, A and C, control cells present a clear septum structure. However, septa of PSP231-overexpressing yeast cells are significantly thicker than those of controls (Supplemental Fig. S5, B, D, and J). Given that the primary septum formed in cell division mainly consists of callose, we used the callose-specific dye aniline blue to examine whether PSP231-overexpressing cells suffered abnormal callose formation and deposition (see “Materials and Methods”). In contrast to the normal callose septa in control cells at the late division phase (Supplemental Fig. S5E), >50% of the PSP231-overexpressing cells displayed obviously aberrant callose deposition phenotypes, including cells with multiple callose septa (Supplemental Fig. S5F), excessive callose deposition in the septum (Supplemental Fig. S5G), mislocalized callose deposition (Supplemental Fig. S5H), and highly elongated cells containing multiple aberrant deposits of callose-rich materials (Supplemental Fig. S5I).
Similar to S. pombe septa, the plant cell plate is mainly composed of callose (Chen and Kim, 2009). Given that callose is also one of the most abundant components in pollen tubes and has been shown to participate in pollen development of multiple plant species, we hypothesized that PSP231 overexpression in plants leads to abnormal callose deposition that hinders further development of pollen. Unfortunately, an in vitro experimental system for cotton pollen germination has not been successfully developed, although our lab has also attempted many strategies. To circumvent this technological limitation, we employed the well-established in vitro experimental model system for tobacco pollen germination for this study. Consistent with the data from the yeast model system, in vitro tobacco pollen germination analysis demonstrated that the callose wall between the vegetative cell and the generative cell in the PSP231-overexpressing pollen was much thicker than that in wild-type pollen (Fig. 3, G and H). The callose content in the PSP231-overexpressing pollen was statistically significantly greater than that in wild-type pollen (Fig. 3M). As expected, only weak staining of callose was detected in wild-type mature pollen (Fig. 3J) and pollen tubes (Fig. 3L), consistent with the concept of a tightly regulated, fine-tuned callose level in these cells. In contrast, an abnormally high amount of callose was deposited in germinal apertures of the mature pollen grains of PSP231-overexpressing transgenic tobacco (Fig. 3, I and K). Interestingly, the atypically germinated PSP231 transgenic pollen grains contained statistically significantly more callose plugs in germ pores compared to wild-type pollen grains (Fig. 3N). Collectively, the above results corroborate that PSP231 positively contributes to callose formation and deposition.
PSP231 Dose-Dependently Activates Callose Biosynthesis to Govern Pollen Maturation
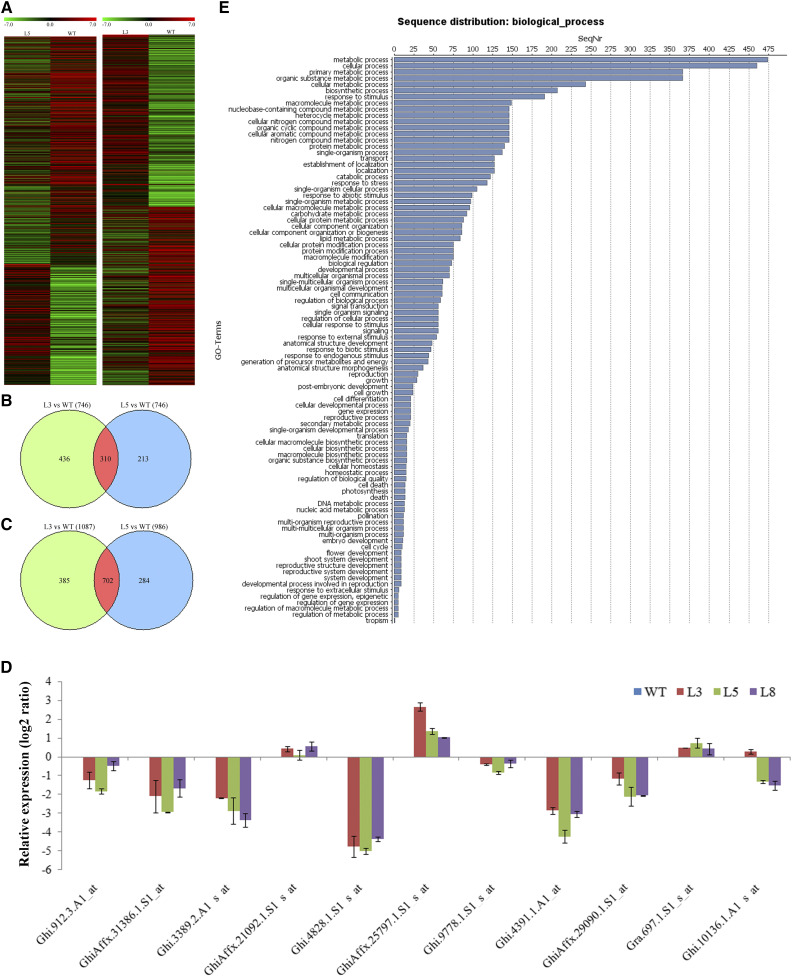
To further characterize the overall effects of PSP231 loss on cotton pollen development on a global molecular scale, we performed transcriptome analysis on anthers of the PSP231 RNAi knockdown cotton and the wild type at the same developmental stage (see “Materials and Methods”). Consistent with the drastic male sterile phenotype of PSP231-silenced cotton plants, large numbers of probesets reported that genes were induced and repressed in anthers of the PSP231-silenced cotton lines compared with those in the wild type, demonstrating that PSP231 loss has profound molecular consequences for cotton anther tissues (Fig. 4A). Specifically, compared with wild-type controls, a total of 310 upregulated and 702 downregulated genes were identified in the PSP231 RNAi cotton lines (Fig. 4, B and C; Supplemental Dataset S1). To validate the microarray data, the expressions of 10 genes (including upregulated, unchanged, and downregulated genes identified by microarray analysis) in the RNAi anthers were further analyzed by RT-qPCR. As shown in Figure 4D, the RT-qPCR results were consistent with the microarray data. Transcriptional changes in genes involved in metabolic and cellular processes represented the major proportions, suggesting that PSP231 loss directly and indirectly impacts many components of these biological processes (Fig. 4E).
Figure 4.
Microarray analysis of the genes differentially expressed in anthers of PSP231 RNAi cotton. A, Heatmap displays of ratios of the differentially expressed genes in PSP231 RNAi cotton lines L5 (left) and L3 (right) to those in the wild type (WT). B and C, Numbers of upregulated (B) and downregulated (C) genes in PSP231 RNAi lines L3 and L5 compared with the wild type. D, RT-qPCR analysis of expression of isolated genes in 25-d-old anthers of the PSP231 RNAi cotton lines. The histogram is drawn based on the log2 scale of the ratio of gene expression levels in the PSP231 RNAi anthers to those in the wild type. Probe identification of genes in the microarray analysis is shown on the x axis. Data are means ± sd for three replicates. E, Substantially changed genes in two PSP231 RNAi lines were subcategorized by biological process. SeqNr, Sequence number; GO, gene ontology.
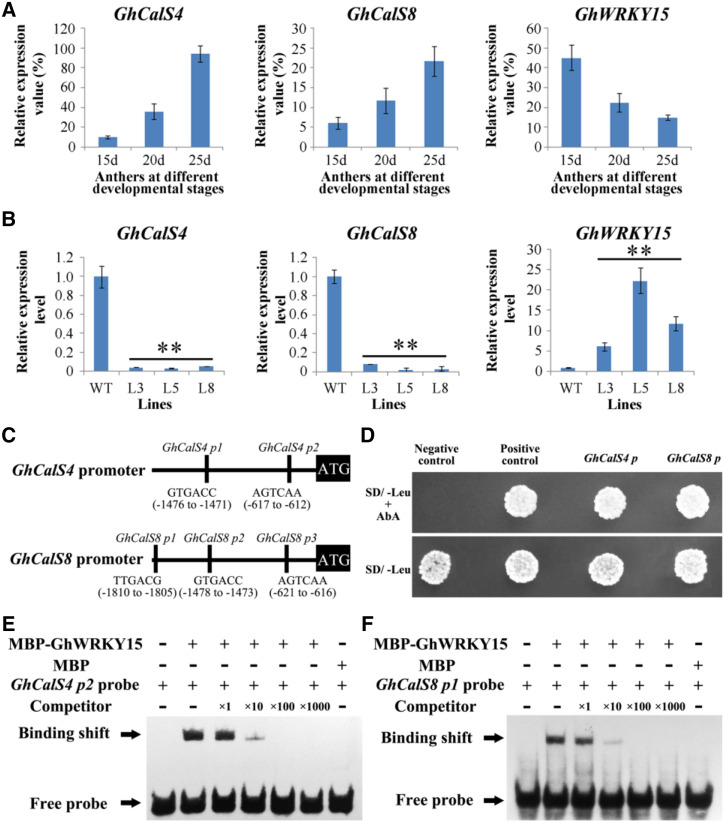
Given that PSP231 overexpression leads to increased aberrant callose formation and deposition (Fig. 3), we analyzed the above microarray datasets for the impact of PSP231 loss on callose-related genes. Interestingly, two anther-specific callose synthase genes (GhCalS4 and GhCalS8) were significantly downregulated in anthers of the PSP231 RNAi cotton compared with age-matched wild-type controls. RT-qPCR analysis revealed a specific and stepwise upregulation of GhCalS4 and GhCalS8 expression levels in wild-type developing anthers, reminiscent of the expression profile of GhPSPS231 (Fig. 5A; Supplemental Fig. S6). Consistently, measurements at 25 d showed that PSP231 RNAi cotton anthers suffered an ∼19- to 29-fold decrease in GhCalS4 expression level and an ∼12- to 37-fold decrease in the GhCalS8 expression level compared with wild-type controls (Fig. 5B). Collectively, the above data from both gain- and loss-of-function studies of PSP231 consistently demonstrated a strong positive effect of PSP231 expression on pollen-specific callose biosynthesis.
Figure 5.
GhWRKY15 transcriptionally represses GhCalS4 and GhCalS8 during anther development. A, RT-qPCR analysis of expression of CalS4, CalS8, and WRKY15 in wild-type cotton anthers. Total RNA isolated from anthers of 15-, 20-, and 25-d-old plants and relative values of CalS4, CalS8, and WRKY15 expression in cotton are shown as a percentage of GhUBI1 (EU604080) expression activity. Data are means ± sd for three replicates. B, RT-qPCR analysis of expression of CalS4, CalS8, and WRKY15 in 25-d-old anthers of PSP231 RNAi cotton lines L3, L5, and L8. Expression level of CalS4, CalS8, and WRKY15 in the wild type (WT) is set to 1. Data are means ± sd for three replicates. Dunnett's t test demonstrated that there was very significant difference (**P < 0.01) in gene expression level between the PSP231 RNAi plants and the wild type. C, Schematic diagram of the GhCalS4 and GhCalS8 promoters showing relative positions of the W-box elements (p1–p3). D, Yeast one-hybrid assay showed interaction of GhWRKY15 with the GhCalS4 and GhCalS8 promoter fragments. Transformants were grown on SD/-Leu nutritional selection medium with 600 ng mL−1 AbA (top), and SD/-Leu medium served as the control (bottom; see “Materials and Methods”). E and F, EMSA of GhWRKY15 binding to the W-box elements of the GhCalS4 (E) and GhCalS8 (F) promoters. MBP-GhWRKY15 protein was incubated with a biotin-labeled probe, using 1×, 10×, 100×, and 1,000× unlabeled probe as competitor. Biotin-labeled probes incubated with MBP protein or without any protein served as the negative control.
Since PSP231 has no predicted DNA binding domain, we hypothesized that PSP231 acts as a molecular switch for transcriptional activators or repressors of pollen-specific callose synthase genes. We further analyzed the above microarray datasets, and found that GhWRKY15, a member of the WRKY family of transcriptional repressors, was significantly upregulated in PSP231 RNAi cotton compared with wild-type controls. RT-qPCR analysis revealed a specific and stepwise downregulation of GhWRKY15 expression levels in wild-type developing anthers (Fig. 5A; Supplemental Fig. S6), inversely mimicking the specific stepwise upregulation of PSP231 expression levels during the same developmental time window. Furthermore, GhWRKY15 expression levels increased ∼7- to 22-fold in 25-d-old PSP231 RNAi cotton anthers compared with age-matched wild-type controls (Fig. 5B), indicating that PSP231 negatively regulates GhWRKY15 expression either directly or indirectly.
Interestingly, bioinformatics analysis revealed the existence of two and three putative W-boxes in the promoter regions of GhCalS4 and GhCalS8, respectively (Fig. 5C). To determine whether GhWRKY15 can directly repress pollen-specific callose synthase expression, we performed a yeast one-hybrid assay to test the interaction of GhWRKY15 protein and GhCalS4/GhCalS8 promoters in cells. As expected, the control yeast cells containing empty pGADT7 vector and GhCalS4/GhCalS8 promoter fragments failed to grow in the selective synthetic dropout (SD) medium containing aureobacidin A (AbA). In striking contrast, yeast cells with GhWRKY15 and GhCalS4/GhCalS8 promoter fragments grew well in the selective SD medium containing AbA (Fig. 5D), indicating that GhWRKY15 directly binds to GhCalS4/GhCalS8 promoters. To further determine whether GhWRKY15 binds to GhCalS4/GhCalS8 promoters through the W-box elements, we employed the electrophoretic mobility shift assay (EMSA) with purified GhWRKY15 proteins and W-box-containing fragments of GhCalS4/GhCalS8 promoters to test this hypothesis. As shown in Figure 5, E and F, incubation of the biotin-labeled GhCalS4p2 or GhCalS8p1 probe with purified GhWRKY15 protein produced strong signals of band shifts, demonstrating their direct binding relationship. Notably, upon addition of increasing concentrations of unlabeled probes as competitors, the intensities of the shifted bands were gradually reduced to undetectable levels comparable to negative control levels, indicating the specificity of this direct intermolecular interaction between GhWRKY15 proteins and the W-box regions of GhCalS4 and GhCalS8 promoters. On the other hand, EMSA results revealed that GhWRKY15 could not bind to GhCalS4p1 and GhCalS8p2/p3, indicating that these regions may not be GhWRKY15 binding sites in GhCalS4 and GhCalS8 promoters. Collectively, the above data shed light on a plausible mechanistic explanation for the observed PSP231-mediated activation of callose biosynthesis molecular machinery: PSP231 negatively regulates GhWRKY15 expression levels to relieve the GhWRKY15-mediated transcriptional repressive effect on GhCalS4 and GhCalS8.
PSP231 Interacts with GhRBPL1 to Turn Off GhWRKY15 Repression of Callose Biosynthesis
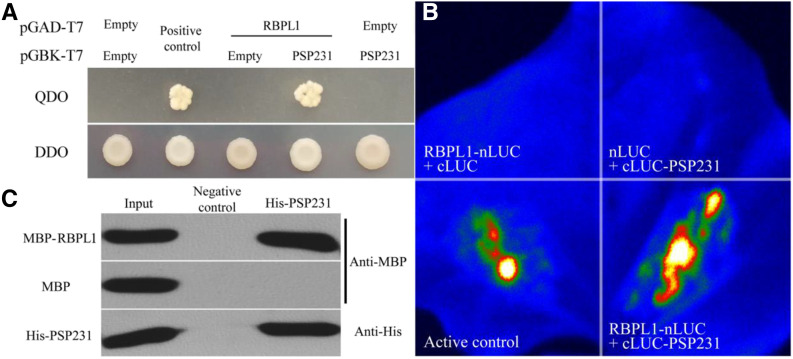
To investigate the molecular mechanism by which PSP231 controls GhWRKY15 expression, we analyzed PSP231-associated direct protein-protein interactions in wild-type cotton anthers by screening a yeast two-hybrid library of cotton anther cDNAs (see “Materials and Methods”). Interestingly, this screening revealed cotton RNA-binding protein-like protein (designated GhRBPL1) as the top candidate binding partner of PSP231. Yeast two-hybrid assay confirmed the direct interaction between PSP231 and GhRBPL1, while a binding mapping analysis revealed that amino acid residues 158 to 390 of PSP231 (PSP231∆2) are key to mediation of the intermolecular binding event with GhRBPL1 (Fig. 6A; Supplemental Fig. S7). To further examine their interaction in vivo, we employed the bimolecular fluorescence complementation (BiFC) system to visualize whether PSP231 can directly interact with GhRBPL1 in leaves of Nicotiana benthamiana. As shown in Figure 6B, coexpression of cLUC-PSP231 with GhRBPL1-nLUC in tobacco leaf cells produced strong luciferase (LUC) fluorescence signals, whereas the negative controls (cLUC-PSP231+nLUC and GhRBPL1-nLUC+cLUC) did not show any detectable signal. Moreover, the in vitro pull-down assay confirmed the protein-protein complex formation between purified maltose-binding protein (MBP)-GhRBPL1 proteins and His-PSP231 proteins (Fig. 6C). Collectively, the above in vitro and in vivo data corroborated that PSP231, via residues 158 to 390, directly interacts with the RNA-binding protein GhRBPL1 to form a protein-protein complex, and implies a potential link between GhRBPL1 and PSP231-controlled GhWRKY15 mRNA levels.
Figure 6.
PSP231 directly interacts with GhRBPL1 to form a complex. A, Yeast two-hybrid assay of the interaction between PSP231 and GhRBPL1. Yeast transformants containing pGBKT7-PSP231 or pGADT7-GhRBPL1 were cultivated on quadruple- (QDO) or double-dropout (DDO) nutritional selection medium. Negative controls were pGBKT7 and pGADT7 empty vectors, and positive controls were P53 and SV40 large T antigen. B, BiFC assay of the interaction between PSP231 and GhRBPL1. PSP231-LUCc and GhRBPL1-LUCn were coexpressed in leaves of N. benthamiana, using nLUC and cLUC as negative controls (see “Materials and Methods”). C, In vitro pull-down assay of the interaction between PSP231 and GhRBPL1. MBP-GhRBPL1 fusion protein was pulled down with His-PSP231 fusion protein and detected by immunoblotting with anti-MBP antibody, using MBP protein as a control. As a negative control, no His-PSP231 protein was added in the experiment.
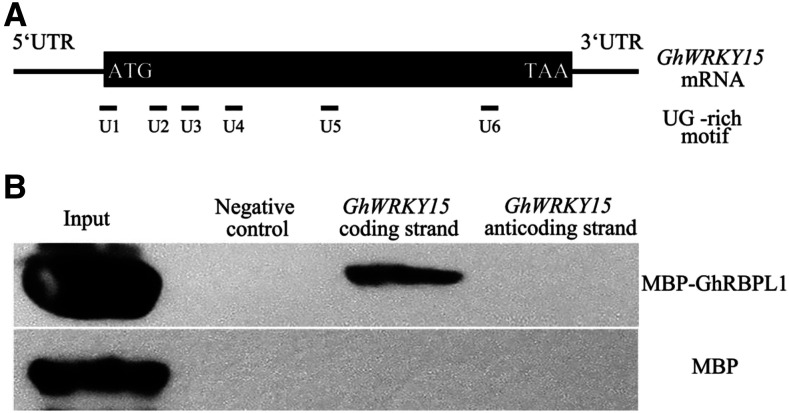
In Arabidopsis, the UG-rich motif is identified as a Gly-rich RNA-binding protein (GRP)-binding motif (Bailey et al., 2009). Notably, sequence analysis revealed that GhWRKY15 mRNAs harbor six putative UG-rich motifs (Fig. 7A). To investigate whether GhRBPL1 directly binds to GhWRKY15 mRNA, we performed the in vitro RNA protein pull-down assay. Specifically, purified MBP-tagged GhRBPL1 proteins were mixed with magnetic bead-bound, biotin-labeled GhWRKY15 mRNA coding strand or anticoding strand, and then subjected to magnetic pull-down of RNA-protein complexes. As expected, MBP tag proteins alone were not pulled-down by the GhWRKY15 mRNA coding strand (Fig. 7B). Remarkably, the GhWRKY15 mRNA coding strand, but not its anticoding strand, directly interacted with GhRBPL1 protein, suggesting that GhRBPL1 protein can specifically recognize and bind with GhWRKY15 mRNA to influence its stability and expression levels.
Figure 7.
GhRBPL1 specifically recognizes GhWRKY15 mRNA to form a stable RNA-protein complex. A, Schematic diagram of the positions of putative GhRBPL1 binding motifs (UG-rich motifs U1–U6) in the GhWRKY15 mRNA sequence. B, RNA pull-down assay of the interaction between GhRBPL1 and GhWRKY15 mRNA. MBP-GhRBPL1 fusion protein was pulled down with GhWRKY15 mRNA and detected by immunoblotting with anti-MBP antibody, using MBP protein as a control. As a negative control, no mRNA of GhWRKY15 was added in the experiment.
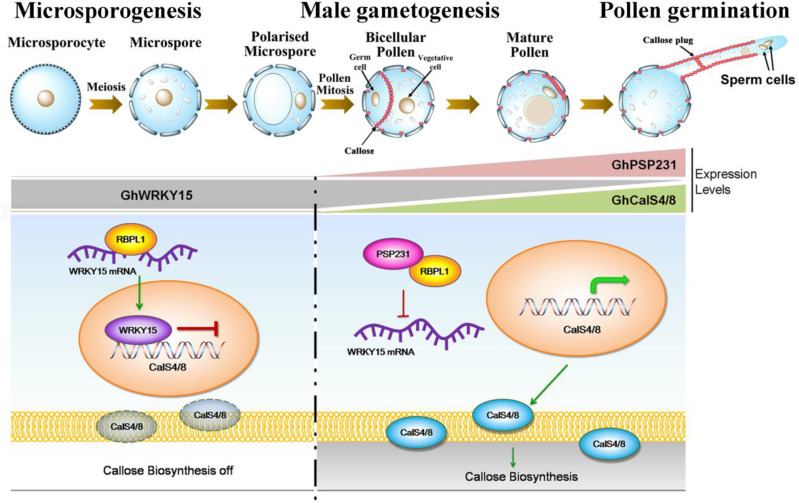
Collectively, the above in vitro and in vivo results comprehensively unveiled the key role of the previously uncharacterized gene PSP231 in triggering callose deposition during late pollen development, as well as the molecular mechanisms involved. Mechanistically, PSP231 activates a molecular circuit through its specific, gradual upregulation of expression at the postpollen mitosis stage to gradually sequester and deplete the pool of the RNA-binding protein GhRBPL1, which was previously available to bind with GhWRKY15 mRNAs. This PSP231-spearheaded series of intermolecular interactions may impair the stability of GhWRKY15 mRNAs and thereby gradually decrease cellular levels of GhWRKY15 proteins, turning off GhWRKY15 repression of the expression of the pollen-specific callose synthetase genes GhCalS4 and GhCalS8 as developing pollen matures further. Overall, this study identified PSP231 as a pollen-specific molecular switch capable of dose-dependently adjusting and fine-tuning levels of callose biosynthesis and deposition required for the production of functional male gametophytes.
DISCUSSION
PSP231 Is a Regulatory Factor for Asymmetric Cell Division in Late Pollen Development
PSP231 belongs to the SKS-like family and has not been characterized prior to this study. So far, only a few plant SKS-like genes (including Bp10, NTP303 and AtSKS11/12/13/14) have been reported to be pollen specific, but their functions and mechanisms are still elusive and largely unexplored (Albani et al., 1992; Wittink et al., 2000; Becker et al., 2003; Honys and Twell 2003). Interestingly, our study reveals that PSP231 expression is highly specific to male gametophytes of cotton at the late developmental stage of their maturation process, highlighting a recurring theme of SKS-like family member involvement in pollen development.
Notably, overexpression of PSP231 under its own promoter resulted in the generation of defective pollen grains unable to undergo normal pollen cell division, suggesting that abnormally high levels of PSP231 expression negatively impact vegetative/germ cell differentiation during male gametogenesis. Consistently, previous studies also support the concept that the ability of plants to produce functional pollen grains diminishes when the asymmetric microspore cell division process is defective during male gametogenesis (Schmidt et al., 2015). For instance, in Arabidopsis, GEM1 deficiency impairs asymmetric microspore cell division and germ cell fate establishment, leading to pollen abortion (Park et al., 1998; Twell et al., 2002). The Arabidopsis gem2 mutant shows repositioning of the pollen cell division plate and a failure of cytokinesis, resulting in abnormal symmetrical divisions and altered pollen cell fate. Both daughter cells in gem2 mutant pollen display vegetative cell fate, and consequently the gem2 mutant is male sterile (Park et al., 2004). Similarly, the scp mutant is unable to produce male germ cells, because its microspore shows delayed cell mitosis and aberrant orientation (Oh et al., 2010a). Also, LBD proteins have been shown to modulate asymmetric cell division in pollen (Kim et al., 2015, 2016). Furthermore, downregulation of TMBP200 in tobacco leads to defective cell division associated with altered spindle position and orientation, producing binucleate and uninucleate cells arising from nuclear fusion or aborted pollen (Oh et al., 2010b). Overexpression of TMS in Arabidopsis hinders pollen development, leading to defective PMI cell plate formation and failure of intine deposition (Li et al., 2019). Interestingly, our results demonstrate that overexpression of PSP231 in tobacco impairs normal pollen cell mitosis during male gametogenesis (Fig. 3). Moreover, both PSP231 RNAi cotton and PSP231-overexpressing transgenic tobacco plants suffer drastically reduced pollen viability and aberrant pollen germination, suggesting that a fine-tuned level of PSP231 plays an important role in male gametogenesis through regulating asymmetric microspore cell division.
PSP231 Dose-Dependently Activates Callose Biosynthesis by Sequestering RBPL1 from WRKY15 mRNAs
Both pollen and fission yeast cells utilize callose as a key component of their transient cell division plates or septa. The discovery of inefficient septum dissolution and excessive aberrant callose deposition in PSP231-overexpressing fission yeast cells have led us to hypothesize that this protein functionally associates with cellular machineries responsible for callose formation and deposition in pollen development. Callose is one of the most dynamic components in the plant cell wall and plays an important role during pollen development. The study of rice Ugp1-silenced plants links disrupted callose deposition to a complete pollen collapse phenotype (Chen et al., 2007). Notably, B. napus can’t form normal pollen grains if the callose deposited in their locules is not degraded on time after the tetrad stage (Zhou et al., 2012), highlighting the fact that transient callose participation in pollen development must be very tightly regulated. Our results on gene expression profiling have revealed that PSP231 is transcriptionally activated in pollen specifically at the PMI stage, implying that it may participate in modulating callose formation and deposition in pollen cells at the gametogenesis stage. Notably, overexpression of PSP231 in tobacco leads to aberrantly excessive and often mislocalized callose deposition that probably is the cause of the observed failure of pollen mitosis and unviable pollen, as the aberrant excessive callose wall persists and hinders the timely release of generative cells into vegetative cells.
GRPs in plants perform multiple functions in RNA processing, including mRNA transport (Kim et al., 2008), mRNA splicing (Daras et al., 2019), primary microRNA processing (Köster et al., 2014), and regulation of mRNA abundance (Foley et al., 2017). By means of RNA affinity chromatography analysis, the highly conserved GRPs were found to bind to a UG-rich motif in the mRNA sequence to regulate RNA processing (Bailey et al., 2009; Xiao et al., 2015; Foley et al., 2017). In this study, we find that six UG-rich motifs exist in the GhWRKY15 mRNA sequence, and GhRBPL1 directly binds to GhWRKY15 mRNA (Fig. 6), suggesting that GhRBPL1 probably affects GhWRKY15 mRNA stability in late developing pollen. On the other hand, RBPs can be coordinated by their interaction partners. For example, Arabidopsis JAC1 was found to bind to GRP7 to influence the nucleocytoplasmic distribution of GRP7 (Xiao et al., 2015). In this study, we propose a plausible mechanistic explanation for PSP231-expression-associated callose synthesis and provide mechanistic evidence for the existence of a molecular circuit controlling the transient yet indispensable callose formation and deposition during pollen development. The physical interaction between PSP231 and GhRBPL1 may hinder GhRBPL1 binding to the mRNAs of GhWRKY15, thereby impairing its mRNA stability. Consequently, this diminished cellular level of GhWRKY15 mRNAs would obviously lead to a diminished nuclear pool of GhWRKY15 proteins, thereby activating callose synthesis and deposition. Consistently, PSP231 deficiency in cotton leads to transcriptional activation of the canonical transcriptional repressor GhWRKY15, which can directly interact with the promoters of cotton pollen-specific callose synthase genes (GhCalS4 and GhCalS8), leading to their transcriptional repression. Therefore, PSP231-deficient cotton suffers defective male gametogenesis and pollen abortion because it is unable to activate the required callose biosynthesis molecular machinery. Moreover, given the well-known role of the callose wall as an indispensable component of pollen tubes, our analyses of pollen germination capability for both PSP231-overexpressing transgenic tobacco and PSP231 RNAi cotton further demonstrate the varied transient yet important participation of PSP231 fine-tuned callose deposition in pollen germination, suggesting the importance of this integral PSP231-triggered regulatory mechanism of callose biosynthesis for pollen germination and pollen tube growth. Additionally, tobacco NTP303 protein, a homolog of PSP231, is also localized to callose plugs of pollen tubes, and NTP303-silenced pollen tubes cannot grow through the style (Capková et al., 1997; Wittink et al., 2000). Similarly, an in vivo pollen germination test revealed the incapability of PSP231-silenced pollen to grow normally in the style of cotton, probably because there are not enough callose plugs formed to sustain pollen tube elongation.
Collectively, this study not only identifies a previously uncharacterized protein, PSP231, as an important participant with high specificity in cotton male gametogenesis, but also establishes the existence of a PSP231-initiated regulatory mechanism/pathway for activating pollen-specific callose biosynthesis machinery to drive pollen maturation in cotton (Fig. 8). During microsporogenesis, in the absence of PSP231 expression, GhRBPL1 can interact with GhWRKY15 mRNA to stabilize these mRNA molecules that can be translated into an abundance of GhWRKY15 proteins. Then, these GhWRKY15 transcriptional repressors can directly bind to GhCalS4 and GhCalS8 promoters to suppress their expression and thus turn off the callose biosynthesis machinery. The subsequent pollen cell mitosis developmentally triggers the progressive transcriptional activation of PSP231 expression. The activated PSP231 directly interacts with GhRBPL1 to inhibit GhRBPL1 binding to GhWRKY15 mRNA, thereby leading to diminished GhWRKY15 mRNA stability and GhWRKY15 protein production. Thus, the GhWRKY15-mediated transcriptional repressive effect on the pollen-specific callose biosynthesis machinery involving GhCalS4 and GhCalS8 can be relieved, so that callose biosynthesis and deposition required for male gametogenesis and pollen germination can be activated and fine-tuned in a PSP231-expression-dependent manner. This proposed mechanism establishes PSP231 as the key molecular switch to activate a previously unknown pathway to fine-tune callose biosynthesis for several previously known pollen-specific developmental events, including generation of the callose-rich transient cell division plate between the germ cell and the vegetative cell in bicellular pollen, formation of concentrated callose surrounding the germination pore in mature pollen, and generation of callose plugs inside the pollen tube and concentrated callose along the germinated pollen tube wall (except in the tip region). Notably, both loss and gain of function of PSP231 impair male gametogenesis and pollen germination capabilities in a dose-dependent manner. Collectively, this study highlights the importance of the integrity of this PSP231-spearheaded regulatory signaling pathway of callose biosynthesis for normal pollen development and functionality and suggests that PSP231 operates as a molecular switch for manipulating pollen fertility of crops with agricultural value.
Figure 8.
Schematic model depicting the proposed molecular mechanism of PSP231-mediated activation of a molecular circuit that controls callose biosynthesis required for late pollen development and pollen germination.
MATERIALS AND METHODS
Plant Growth Conditions and Transformation
Cotton (Gossypium hirsutum ‘Coker 312’) seeds were surface-sterilized with 70% ethanol for 1 min and then 10% hydrogen peroxide for 2 h, followed by washing with sterile water. Subsequently, the seeds were germinated on one-half strength Murashige and Skoog medium (pH 5.8) under a 16 h light/8 h dark cycle at 28°C for 5 to 6 d. Hypocotyls from these seedlings were used as explants for genetic transformation as described previously (Li et al., 2002).
Tobacco (Nicotiana tabacum) seeds were germinated on Murashige and Skoog medium under a 16 h light/8 h dark cycle at 25°C. Leaves of 4- to 6-week-old tobacco plants were used as explants. Tobacco transformation was performed by the Agrobacterium tumefaciens-mediated leaf slice method. Transgenic plants of both cotton and tobacco were transplanted into soil in the greenhouse or field to grow to maturation.
RNA Gel Blot Analysis
Total RNA was isolated from roots, cotyledons, hypocotyls, leaves, anthers, petals, ovules, and fibers of cotton plants, and 20-μg RNA samples of the different cotton tissues were separated on 1.2% (w/v) agarose-formaldehyde gels and transferred onto a Hybond-N nylon membrane (GE Healthcare) by capillary blotting. A PCR fragment (250 bp) of the 3′-untranslated region of PSP231 was used as a gene-specific probe (primers are PSP231-l and PSP231-R in Supplemental Table S1) for northern hybridization as described previously (Li et al., 2002).
RT-qPCR Analysis
Total RNA isolated from different cotton tissues was reverse-transcribed to cDNAs with Moloney murine leukemia virus reverse transcriptase (Promega) according to the manufacturer’s instructions. RT-qPCR analysis was performed to evaluate the expression levels of the target genes in cotton tissues, using SYBR Green real-time PCR master mix (Toyobo) in a DNA detection system (Applied Biosystems) according to a method described earlier (Li et al., 2005). A cotton polyubiquitin gene (GhUBI1, EU604080) was used as a standard control for normalization. All reactions were performed in triplicate, and three independent biological replicates were performed. Mean values of normalized target gene expression levels from three biological experiments were calculated to determine all target gene expression levels relative to GhUBI1. All the primers were listed in Supplemental Table S1.
Anti-PSP231 Antiserum Preparation and Western Blot Analysis
PSP231 antigen production was achieved using a His-tagged protein expression system (Qiagen). The partial coding region of PSP231 was amplified using Pfu DNA polymerase and primers (PSP231-exL and PSP231-exR; Supplemental Table S1) and cloned into pET-28a vector. This plasmid was transferred into the bacterial host BL21 and protein synthesis was induced with 0.5 mm isopropylthio-β-galactoside for 4 h at 37°C. The purified protein was used to immunize rabbit. The polyclonal antibodies against PSP231 were tested on purified recombinant proteins by western blotting.
Extraction of total proteins from cotton tissues was performed according to the method described by Yao et al. (2006). Briefly, 2 g (fresh weight) of cotton tissues were ground to a fine powder in liquid nitrogen with 10% (w/v) SiO2 and 10% (percent of sample weight) polyvinylpyrrolidone, and were then washed with 15 mL ice-cold acetone. After being lyophilized for 30 min, the powders were resuspended in 5 mL of extraction buffer (50 mm Tris-HCl [pH8.65], 2% [w/v] SDS, 30% [w/v] Suc, and 2% [v/v] β-mercaptoethanol) and an equal volume of Tris-saturated phenol (pH 7.9). The samples were centrifuged for 10 min at 12,000g. The phenolic phase was collected and precipitated with five volumes of 0.1 m ammonium acetate in methanol at –20°C for 30 min. After centrifugation, the pellet was washed three times with ice-cold 80% acetone, then lyophilized and stored at –80°C until use.
Proteins were dissolved in a protein elution buffer (7 m urea, 2 m thiourea, and 4% [w/v] CHAPS) and quantified using the Bradford protein assay kit (Bio-Rad). Then, 50 μg of total protein extracts was loaded per lane, separated by 10% (w/v) SDS-PAGE, and electroblotted using semidry transfer to polyvinylidene difluoride membranes. The membranes were then blocked in 5% (w/v) nonfat dry milk in Tris-buffered saline (50 mm Tris-HCl and 150 mm NaCl [pH 7.5]) containing 0.1% (v/v) Tween 20 (TBST) for 1 h followed by incubation with the anti-PSP231 antibody (1:10,000 dilution) in TBST with 5% (w/v) nonfat dry milk for 1 h at room temperature. Subsequently, the membranes were washed three times with TBST and then incubated with horseradish peroxidase-conjugated goat anti-rabbit IgG secondary antibody (CWBIO). Signals were visualized with a diaminobenzidine kit (CWBIO).
Assay of PSP231 Glycosylation
Endo H can cleave high Man and some hybrid types of N-linked carbohydrates on proteins, while PNGase F is capable of removing almost all types of N-linked carbohydrates regardless of type (Sedbrook et al., 2002). Total proteins isolated from 25-d-old anthers of cotton were dissolved in the denaturing buffer, and incubated with Endo H and PNGase F, respectively, according to the manufacturer’s instruction manual (New England Biolabs). Then, the treated or untreated proteins were separated by 10% (w/v) SDS-PAGE, and detected by western blot analysis using anti-PSP231 antibody. Beta-actin was used as a loading control.
Construction of PSP231 Overexpression and RNAi Recombinant Plasmids
For the PSP231 yeast (Schizosaccharomyces pombe) overexpression construct, the coding region of PSP231 was cloned into the pREP-5N vector (primers are PSP231-YL and PSP231-YR). For the PSP231 plant overexpression construct, the promoter of PSP231 was first cloned into the PBI101 vector to create the PBI101-PSP231p construct (Primers are PSP231-P1 and PSP231-P2). Then, the coding sequence of PSP231 was cloned into the PBI101-PSP231p plasmid, replacing the GUS gene. For the PSP231 RNAi construct, a 250bp fragment of PSP231 was cloned into pBluescript SK vector to create an inverted repeat transgene and then cloned into the PBI121 vector (primers are PSP231-L1, PSP231-R1, PSP231-L2, and PSP231-R2). All primers used are listed in Supplemental Table S1.
Pollen Viability Assay
Pollen viability was assayed by double staining with FDA and PI as described previously (Mandaokar and Browse, 2009). The stained pollen grains were observed under a Nikon Digital Sight DS-5Mc microscope equipped with a UV light and CCD camera. Quantitative analysis of pollen viability on wild-type controls and transgenic lines was performed by examining at least 30 independent flowers per genotype. More than 1,000 pollen grains per flower were analyzed, with green-colored pollen grains counted as alive and red-colored pollen grains counted as dead. The data were input into SPSS software, and Dunnett's t test was performed for statistical analysis.
Floral Sectioning
Whole flowers were collected and fixed in formaldehyde-acetic acid solution overnight, embedded into SPI-PON 812 resin, and sliced into semithin (∼3-μm) sections by a Leica microtome (http://www.leica.com). The samples were stained with 0.25% toluidine blue and photographed under Nikon Digital Sight DS-5Mc microscope equipped with a CCD camera.
In Vitro and In Vivo Pollen Germination
For in vitro pollen germination, mature pollen grains of tobacco were collected and immediately transferred into germination medium consisting of 5% (w/v) Suc, 0.03% (w/v) Amicase (casein acid-lydrolysate), 12.5% (w/v) polyethylene glycol 6000, 15 mm MES-KOH, 1 mm CaCl2, 1 mm KCl, 0.8 mm MgSO4, 1.6 mm H3PO3, and 30 μm CuSO4 at 25°C for 2 h in the dark. At least nine independent anthers per genotype, with >500 pollen grains in each sample, were analyzed for in vitro pollen germination.
For in vivo pollen germination, cotton pollen grains were collected and pollinated to wild-type pistils. Six hours later, the pistils were cut down and fixed in ethanol: acetic acid (3:1) for 24 h at room temperature, followed by washing in 1 m phosphate buffer (pH 7.0) and softening for 20 h in 1 m NaOH. Afterward, the pistils were stained with 0.01% (w/v) decolorized aniline blue solution for 2 to 4 h. After being stained, the pistils were placed in glycerol for 1 h, then transferred to a microscope slide, squashed under a coverslip, and observed under a fluorescence microscope.
Yeast One-Hybrid Assay
GhCalS4 and GhCalS8 promoter fragments were integrated into the pAbAi vector separately. The constructs were transferred into yeast strain Y1HGold using the PEG/LiAc method described in the user manual. Prior to testing for interactions between GhWRKY15 and the DNA fragment, a self-activation test was conducted for the DNA sequence bait to determine concentration of AbA (600 ng mL−1) in this experiment. Then, the coding region of GhWRKY15 was cloned into pGADT7 vector and transformed into the yeast strain Y1HGold with bait vector. Interaction between GhWRKY15 and the GhCalS4 or GhCalS8 promoter was tested by SD/-Leu with 600 ng mL−1 AbA. The empty pGADT7 vector was transferred into Y1HGold with bait vector as a negative control. The Y1HGold yeast strain containing p53-pAbAi and p53-pGADT7 was used as the positive control. All primers used are listed in Supplemental Table S1.
EMSA
The coding sequence of GhWRKY15 was cloned into pMal-c2 vector. Then, MBP-GhWRKY15 fusion protein was expressed in Escherichia coli strain BL21 (DE3) and subsequently purified through an amylose resin column. About 40 bp of synthetic biotin-labeled DNA fragments containing the W-box were used as the detection probe (sequences shown in Supplemental Table S1). The same unlabeled fragments were used as the competitor probe. EMSA was performed in accordance with previously reported methods (Li et al., 2018). Typically, 10 nmol protein and 20 pmol probe were used in each binding reaction. The samples were separated by 6% native polyacrylamide gel and transferred to a nylon membrane. Then the labeled probe signals were detected using the Light Shift Chemiluminescent EMSA kit (Thermo Fisher Scientific).
Construction of the Cotton Anther cDNA Yeast Two-Hybrid Library and the Yeast Two-Hybrid Assay
The yeast two-hybrid library of cotton anther cDNAs was constructed using the Clontech BD Matchmaker Library Construction Kit according to the manufacture's instruction (BD Biosciences Clontech). Then, fragments of the PSP231 coding sequence (Supplemental Fig. S6A) were fused to the GAL4 DNA binding domain in bait vector pGBK-T7 and transformed into yeast strain Y187. The transformants were used for mating to the yeast two-hybrid library of cotton anther cDNAs. After mating, the diploid cells were selected on QDO medium (SD/-Trp/-Leu/-His/-Ade) at 30°C. Yeast plasmid DNA was isolated from the positive clones for sequencing.
The PSP231∆2 was fused in frame to the GAL4-BD coding sequence in the pGBKT7 vector and then transferred into yeast strain Y187. The coding sequence of GhRBPL1 was fused in frame to the GAL4-AD coding sequence in the pGADT7 vector and then transferred into yeast strain AH109. After mating reactions between the two haploid strains, the diploid strains were selected on the SD/-Leu/-Trp medium, and positive interactions were detected on a selective medium (SD/-Leu/-Trp/-His/-Ade; Zhang et al., 2010). All primers used are listed in Supplemental Table S1.
BiFC Assay
The BiFC assay was performed according to the method described previously (Gou et al., 2011). The PSP231 coding sequence was fused in frame to the N terminus of LUC in the JW771 vector to generate the JW771-PSP231-nLUC construct, and the coding sequence of GhRBPL1 was fused in frame to the C terminus of LUC in the JW772 vector to produce the JW772-cLUC-GhRBPL1 construct. The constructs were transferred individually into A. tumefaciens strain GV3101. The A. tumefaciens cells containing cLUC-GhRBPL1 or PSP231-nLUC were suspended in tobacco-transformed buffer (10 mm MgCl2, 10 mm MES [pH5.7], and 150 μm acetosyringone) and injected into leaves of Nicotiana benthamiana, using cLUC and nLUC as negative controls. After coexpression of cLUC-GhRBPL1 and PSP231-nLUC in tobacco leaves for 72 h, the luciferase (LUC) fluorescence signals were detected by a low-light cooled 16-bit CCD imaging apparatus (CHEMIPROHT 1300B/LND, Roper Scientific). All primers used are listed in Supplemental Table S1.
Pull-Down Assay
The PSP231-His proteins were expressed in mammalian cells at AtaGenix and purified with anti-His-tag mAb-Magnetic Beads (MBL International). The immobilized PSP231-His proteins were incubated with 3 μg MBP-GhRBPL1 proteins at 4°C overnight in phosphate-buffered saline, using MBP as control. Then, the protein samples were washed with phosphate-buffered saline three times and boiled in SDS loading buffer for 10 min. After separation by SDS-PAGE gel, the proteins were detected by immunoblotting analysis using anti-MBP antibody and anti-His antibody (CWBIO).
RNA Pull-Down Assay
The coding and anticoding strands of GhWRKY15 mRNA were obtained by in vitro transcription with T7 and SP6 RNA polymerase, respectively, and subsequently labeled with biotin using the Pierce RNA 3′-End Desthiobiotinylation Kit (Thermo Fisher Scientific). The RNA pull-down assay was performed according to the Pierce Magnetic RNA-Protein Pull-Down Kit user manual (Thermo). Briefly, 100 pmol labeled RNA was captured with streptavidin magnetic beads in RNA Capture buffer (20 mm Tris-Cl [pH 7.5], 1 m NaCl, and 1 mm EDTA). After washing in 20 mm Tris-Cl (pH 7.5), the magnetic beads were incubated with the purified MBP-GhRBPL1 or MBP (control) proteins in protein-RNA binding buffer (20 mm Tris-Cl [pH 7.5], 50 mm NaCl, 2 mm MgCl2, and 0.1% [v/v] Tween20) and then washed in an equal volume of wash buffer (20 mm Tris-Cl [pH 7.5], 10 mm NaCl, and 0.1% [v/v] Tween20). Finally, the protein samples were eluted in Biotin Elution buffer, separated on a 10% (w/v) SDS-PAGE gel, and detected by immunoblotting analysis using MBP antibody (CWBIO).
Microarray Analysis
Total RNA from 25 d-old anthers of wild-type and PSP231 RNAi cotton plants was prepared for microarray analysis. Labeling and hybridization were performed at the Shanghai Biochip Company according to the protocols in the Affymetrix GeneChip 3′ IVT Express Kit User Manual. Three biological replicates processed for each PSP231 RNAi cotton line and the wild type, with overall correlation coefficient values of >0.95, were further used for the final data analysis. Analysis to determine whether a gene is differentially expressed was based on fold change (FC ≥ 3 or FC ≤ −3) and P-value (P ≤ 0.01). To predict gene function and calculate the functional category distribution frequency, Blast2 GO software was employed. After that, maps from the Kyoto Encyclopedia of Genes and Genomes were used to investigate the pathways in which differentially expressed genes are involved.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers KF270893 (Gh_A13G1836; PSP231), Gh_A01G1273 (GhWRKY15), Gh_D10G1068 (GhCalS4), Gh_A10G1388 (GhCalS8), and Gh_A12G0257 (GhRBPL1).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Analysis of PSP231 expression in cotton.
Supplemental Figure S2. Mass spectrometric analysis of PSP231 protein from cotton anther.
Supplemental Figure S3. Subcellular localization of PSP231 in germinating pollen.
Supplemental Figure S4. Overexpression of PSP231 in fission yeast negatively affects yeast cell growth, viability, and cell division.
Supplemental Figure S5. Aberrant callose deposition of PSP231-overexpressing yeast cells.
Supplemental Figure S6. RT-qPCR analysis of of CalS4, CalS8, and WRKY15 expression in cotton tissues.
Supplemental Figure S7. Yeast two-hybrid assay mapping potential interactions between PSP231Δ1/2/3/4/5 and GhRBPL1.
Supplemental Table S1. Primers used in the experiments.
Supplemental Dataset S1. Summary of genes differentially expressed in anthers of PSP231 RNAi cotton versus the wild type.
DIVE Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Footnotes
This work was supported by the National Natural Sciences Foundation of China (grant nos. 31070281 and 31500987).
References
- Albani D, Sardana R, Robert LS, Altosaar I, Arnison PG, Fabijanski SF(1992) A Brassica napus gene family which shows sequence similarity to ascorbate oxidase is expressed in developing pollen. Molecular characterization and analysis of promoter activity in transgenic tobacco plants. Plant J 2: 331–342 [PubMed] [Google Scholar]
- Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS(2009) MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res 37: W202–W208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker JD, Boavida LC, Carneiro J, Haury M, Feijó JA(2003) Transcriptional profiling of Arabidopsis tissues reveals the unique characteristics of the pollen transcriptome. Plant Physiol 133: 713–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borg M, Brownfield L, Twell D(2009) Male gametophyte development: A molecular perspective. J Exp Bot 60: 1465–1478 [DOI] [PubMed] [Google Scholar]
- Capková V, Fidlerová A, van Amstel T, Croes AF, Mata C, Schrauwen JA, Wullems GJ, Tupý J(1997) Role of N-glycosylation of 66 and 69 kDa glycoproteins in wall formation during pollen tube growth in vitro. Eur J Cell Biol 72: 282–285 [PubMed] [Google Scholar]
- Chebli Y, Kaneda M, Zerzour R, Geitmann A(2012) The cell wall of the Arabidopsis pollen tube—spatial distribution, recycling, and network formation of polysaccharides. Plant Physiol 160: 1940–1955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Zhao X, Shao Z, Wei Z, Wang Y, Zhu L, Zhao J, Sun M, He R, He G(2007) Rice UDP-glucose pyrophosphorylase1 is essential for pollen callose deposition and its cosuppression results in a new type of thermosensitive genic male sterility. Plant Cell 19: 847–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen XY, Kim JY(2009) Callose synthesis in higher plants. Plant Signal Behav 4: 489–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daras G, Rigas S, Alatzas A, Samiotaki M, Chatzopoulos D, Tsitsekian D, Papadaki V, Templalexis D, Banilas G, Athanasiadou AM, et al. (2019) LEFKOTHEA regulates nuclear and chloroplast mRNA splicing in plants. Dev Cell 50: 767–779 [DOI] [PubMed] [Google Scholar]
- de Groot P, Weterings K, de Been M, Wittink F, Hulzink R, Custers J, van Herpen M, Wullems G(2004) Silencing of the pollen-specific gene NTP303 and its family members in tobacco affects in vivo pollen tube growth and results in male sterile plants. Plant Mol Biol 55: 715–726 [DOI] [PubMed] [Google Scholar]
- Dong X, Hong Z, Sivaramakrishnan M, Mahfouz M, Verma DPS(2005) Callose synthase (CalS5) is required for exine formation during microgametogenesis and for pollen viability in Arabidopsis. Plant J 42: 315–328 [DOI] [PubMed] [Google Scholar]
- Dresselhaus T, Sprunck S, Wessel GM(2016) Fertilization mechanisms in flowering plants. Curr Biol 26: R125–R139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feijó JA, Costa SS, Prado AM, Becker JD, Certal AC(2004) Signalling by tips. Curr Opin Plant Biol 7: 589–598 [DOI] [PubMed] [Google Scholar]
- Ferguson C, Teeri TT, Siika-aho M, Read SM, Bacic A(1998) Location of cellulose and callose in pollen tubes and grains of Nicotiana tabacum. Planta 206: 452–460 [Google Scholar]
- Foley SW, Gosai SJ, Wang D, Selamoglu N, Sollitti AC, Köster T, Steffen A, Lyons E, Daldal F, Garcia BA, et al. (2017) A global view of RNA-protein interactions identifies post-transcriptional regulators of root hair cell fate. Dev Cell 41: 204–220.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gou JY, Felippes FF, Liu CJ, Weigel D, Wang JW(2011) Negative regulation of anthocyanin biosynthesis in Arabidopsis by a miR156-targeted SPL transcription factor. Plant Cell 23: 1512–1522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honys D, Twell D(2003) Comparative analysis of the Arabidopsis pollen transcriptome. Plant Physiol 132: 640–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs J, Roe JL(2005) SKS6, a multicopper oxidase-like gene, participates in cotyledon vascular patterning during Arabidopsis thaliana development. Planta 222: 652–666 [DOI] [PubMed] [Google Scholar]
- Kim JS, Jung HJ, Lee HJ, Kim KA, Goh CH, Woo Y, Oh SH, Han YS, Kang H(2008) Glycine-rich RNA-binding protein 7 affects abiotic stress responses by regulating stomata opening and closing in Arabidopsis thaliana. Plant J 55: 455–466 [DOI] [PubMed] [Google Scholar]
- Kim MJ, Kim M, Lee MR, Park SK, Kim J(2015) LATERAL ORGAN BOUNDARIES DOMAIN (LBD)10 interacts with SIDECAR POLLEN/LBD27 to control pollen development in Arabidopsis. Plant J 81: 794–809 [DOI] [PubMed] [Google Scholar]
- Kim M, Kim MJ, Pandey S, Kim J(2016) Expression and protein interaction analyses reveal combinatorial interactions of LBD transcription factors during Arabidopsis pollen development. Plant Cell Physiol 57: 2291–2299 [DOI] [PubMed] [Google Scholar]
- Köster T, Meyer K, Weinholdt C, Smith LM, Lummer M, Speth C, Grosse I, Weigel D, Staiger D(2014) Regulation of pri-miRNA processing by the hnRNP-like protein AtGRP7 in Arabidopsis. Nucleic Acids Res 42: 9925–9936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Li Y, Liu F, Tan X, Rui Q, Tong Y, Qiao L, Gao R, Li G, Shi R, et al. (2019) Overexpressed Tomosyn binds syntaxins and blocks secretion during pollen development. Plant Physiol 181: 1114–1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Bacic A, Read SM(1997) Activation of pollen tube callose synthase by detergents. Evidence for different mechanisms of action. Plant Physiol 114: 1255–1265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XB, Cai L, Cheng NH, Liu JW(2002) Molecular characterization of the cotton GhTUB1 gene that is preferentially expressed in fiber. Plant Physiol 130: 666–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XB, Fan XP, Wang XL, Cai L, Yang WC(2005) The cotton ACTIN1 gene is functionally expressed in fibers and participates in fiber elongation. Plant Cell 17: 859–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Wang NN, Wang Y, Liu D, Gao Y, Li L, Li XB(2018) The cotton XLIM protein (GhXLIM6) is required for fiber development via maintaining dynamic F-actin cytoskeleton and modulating cellulose biosynthesis. Plant J 96: 1269–1282 [DOI] [PubMed] [Google Scholar]
- Mandaokar A, Browse J(2009) MYB108 acts together with MYB24 to regulate jasmonate-mediated stamen maturation in Arabidopsis. Plant Physiol 149: 851–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh SA, Park KS, Twell D, Park SK(2010a) The SIDECAR POLLEN gene encodes a microspore-specific LOB/AS2 domain protein required for the correct timing and orientation of asymmetric cell division. Plant J 64: 839–850 [DOI] [PubMed] [Google Scholar]
- Oh SA, Pal MD, Park SK, Johnson JA, Twell D(2010b) The tobacco MAP215/Dis1-family protein TMBP200 is required for the functional organization of microtubule arrays during male germline establishment. J Exp Bot 61: 969–981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SK, Howden R, Twell D(1998) The Arabidopsis thaliana gametophytic mutation gemini pollen1 disrupts microspore polarity, division asymmetry and pollen cell fate. Development 125: 3789–3799 [DOI] [PubMed] [Google Scholar]
- Park SK, Rahman D, Oh SA, Twell D(2004) gemini pollen 2, a male and female gametophytic cytokinesis defective mutation. Sex Plant Reprod 17: 63–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SK, Twell D(2001) Novel patterns of ectopic cell plate growth and lipid body distribution in the Arabidopsis gemini pollen1 mutant. Plant Physiol 126: 899–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell SD, Jones DS(2015) The male germline of angiosperms: Repertoire of an inconspicuous but important cell lineage. Front Plant Sci 6: 173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt A, Schmid MW, Grossniklaus U(2015) Plant germline formation: Common concepts and developmental flexibility in sexual and asexual reproduction. Development 142: 229–241 [DOI] [PubMed] [Google Scholar]
- Sedbrook JC, Carroll KL, Hung KF, Masson PH, Somerville CR(2002) The Arabidopsis SKU5 gene encodes an extracellular glycosyl phosphatidylinositol-anchored glycoprotein involved in directional root growth. Plant Cell 14: 1635–1648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X, Sun X, Zhang Z, Feng D, Zhang Q, Han L, Wu J, Lu T(2015) GLUCAN SYNTHASE-LIKE 5 (GSL5) plays an essential role in male fertility by regulating callose metabolism during microsporogenesis in rice. Plant Cell Physiol 56: 497–509 [DOI] [PubMed] [Google Scholar]
- Sprunck S, Gross-Hardt R(2011) Nuclear behavior, cell polarity, and cell specification in the female gametophyte. Sex Plant Reprod 24: 123–136 [DOI] [PubMed] [Google Scholar]
- Töller A, Brownfield L, Neu C, Twell D, Schulze-Lefert P(2008) Dual function of Arabidopsis glucan synthase-like genes GSL8 and GSL10 in male gametophyte development and plant growth. Plant J 54: 911–923 [DOI] [PubMed] [Google Scholar]
- Twell D, Park SK, Hawkins TJ, Schubert D, Schmidt R, Smertenko A, Hussey PJ(2002) MOR1/GEM1 has an essential role in the plant-specific cytokinetic phragmoplast. Nat Cell Biol 4: 711–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XL, Li XB(2009) The GhACS1 gene encodes an acyl-CoA synthetase which is essential for normal microsporogenesis in early anther development of cotton. Plant J 57: 473–486 [DOI] [PubMed] [Google Scholar]
- Weterings K, Reijnen W, van Aarssen R, Kortstee A, Spijkers J, van Herpen M, Schrauwen J, Wullems G(1992) Characterization of a pollen-specific cDNA clone from Nicotiana tabacum expressed during microgametogenesis and germination. Plant Mol Biol 18: 1101–1111 [DOI] [PubMed] [Google Scholar]
- Weterings K, Reijnen W, Wijn G, van de Heuvel K, Appeldoorn N, de Kort G, van Herpen M, Schrauwen J, Wullems G(1995) Molecular characterization of the pollen-specific genome clone Ntpg303 and in situ localization of expression. Sex Plant Reprod 8: 11–17 [Google Scholar]
- Wittink FRA, Knuiman B, Derksen J, Capkova V, Twell D, Schrauwen JAM, Wullems GJ(2000) The pollen-specific gene NTP303 encodes a 69-kDa glycoprotein associated with the vegetative membranes and the cell wall. Sex Plant Reprod 12: 276–284 [Google Scholar]
- Xiao J, Li C, Xu S, Xing L, Xu Y, Chong K(2015) Jacalin-lectin like1 regulates the nuclear accumulation of glycine-rich RNA-binding protein7, influencing the RNA processing of FLOWERING LOCUS C antisense transcripts and flowering time in Arabidopsis. Plant Physiol 169: 2102–2117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y, Yang YW, Liu JY(2006) An efficient protein preparation for proteomic analysis of developing cotton fibers by 2-DE. Electrophoresis 27: 4559–4569 [DOI] [PubMed] [Google Scholar]
- Zhang ZT, Zhou Y, Li Y, Shao SQ, Li BY, Shi HY, Li XB(2010) Interactome analysis of the six cotton 14-3-3s that are preferentially expressed in fibres and involved in cell elongation. J Exp Bot 61: 3331–3344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Dun X, Xia S, Shi D, Qin M, Yi B, Wen J, Shen J, Ma C, Tu J, et al. (2012) BnMs3 is required for tapetal differentiation and degradation, microspore separation, and pollen-wall biosynthesis in Brassica napus. J Exp Bot 63: 2041–2058 [DOI] [PMC free article] [PubMed] [Google Scholar]