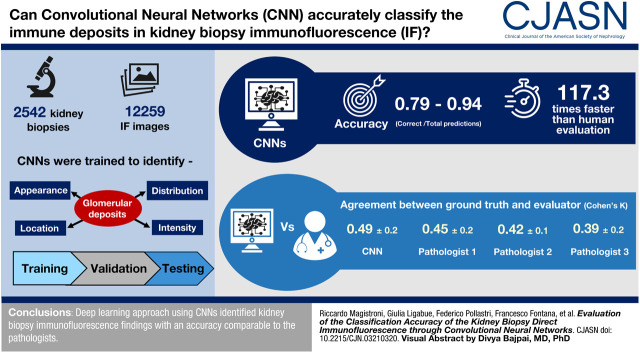
Visual Abstract
Keywords: renal pathology, renal biopsy, immunofluorescence, Convoluted Neural Network, artificial intelligence
Abstract
Background and objectives
Immunohistopathology is an essential technique in the diagnostic workflow of a kidney biopsy. Deep learning is an effective tool in the elaboration of medical imaging. We wanted to evaluate the role of a convolutional neural network as a support tool for kidney immunofluorescence reporting.
Design, setting, participants, & measurements
High-magnification (×400) immunofluorescence images of kidney biopsies performed from the year 2001 to 2018 were collected. The report, adopted at the Division of Nephrology of the AOU Policlinico di Modena, describes the specimen in terms of “appearance,” “distribution,” “location,” and “intensity” of the glomerular deposits identified with fluorescent antibodies against IgG, IgA, IgM, C1q and C3 complement fractions, fibrinogen, and κ- and λ-light chains. The report was used as ground truth for the training of the convolutional neural networks.
Results
In total, 12,259 immunofluorescence images of 2542 subjects undergoing kidney biopsy were collected. The test set analysis showed accuracy values between 0.79 (“irregular capillary wall” feature) and 0.94 (“fine granular” feature). The agreement test of the results obtained by the convolutional neural networks with respect to the ground truth showed similar values to three pathologists of our center. Convolutional neural networks were 117 times faster than human evaluators in analyzing 180 test images. A web platform, where it is possible to upload digitized images of immunofluorescence specimens, is available to evaluate the potential of our approach.
Conclusions
The data showed that the accuracy of convolutional neural networks is comparable with that of pathologists experienced in the field.
Introduction
The kidney histopathologic diagnosis involves a combined and complementary approach of different microscopic techniques: essentially, they consist of light microscopy, immunohistopathology, and electron microscopy. The kidney biopsy is frequently necessary to distinguish among diseases with similar clinical presentation. The evaluation of glomerular disease by light microscopy alone rarely allows a definitive diagnosis, for which the information from immunohistopathology and electron microscopy analyses is required. All of these elements must finally be integrated with the patient’s clinical history and laboratory findings to obtain the conclusive diagnosis. Immunofluorescence technique (or other immunohistopathologic approaches) is required to demonstrate deposits of Igs and complement components. Some specific kidney diseases, such as IgA nephropathy, antiglomerular basal membrane GN, and C3 glomerulopathy, can only be diagnosed by the result of immunofluorescence. The analysis of the immunofluorescence pattern of deposition in glomerular diseases is a time-consuming activity, and it relies on the availability of specific resources and requires an experienced operator for interpretation. In pathology, as in many other clinical disciplines, the ability of artificial intelligence algorithms to interpret medical images is taking on a dominant role. Some examples of the application of these technologies are also emerging in kidney pathology (1,2). An artificial neural network is an interconnected ensemble of simple processing elements with the ability of learning how to perform different tasks directly from data. Each element, or artificial neuron, receives input values from several preceding nodes, processes them, and computes the output in order to forward it to the next set of neurons. When this hierarchical architecture is formed by several layers, the strategy takes the name of deep learning. Each node has a set of weights used to multiply its inputs. These weights are the processing ability of a neural network. Convolutional neural networks present weights in the form of small filters, which learn to recognize specific characteristics regardless of their position within the input image. They are randomly initialized and then updated during the training process in order to optimize the final output. The learning strategy causes neural networks to adapt their filters in order to gain the ability to extract meaningful features directly from the set of images used during the training process. This is the reason why, unlike any other machine learning technique, deep learning algorithms require no hand-crafted feature. Given the aforementioned hierarchical architecture, automatically extracted features grow more complex as the depth of the network increases. However, very deep neural networks tend to learn to classify the training set with extremely high accuracy while failing to generalize their capabilities to samples that were not used during training. This phenomenon takes the name of overfitting (3) and causes networks to learn to recognize each training image instead of the semantic content within. Therefore, many efforts have been devoted to finding valid regularization techniques (4–6). Convolutional neural networks have been widely proven to outperform other strategies in countless computer vision tasks, such as semantic segmentation, object detection, object classification, and others (7–9). This kind of architecture is able to process images in an extremely effective and computationally efficient manner. In fact, convolutional neural networks have been extensively used in many medical imaging fields, such as skin lesion analysis (10,11) and retinopathy or pneumonia (12,13), growing into an extremely powerful tool to support specialists in clinical decision making. In this work, we aimed to exploit artificial neural networks to build an automated tool supporting the diagnostic process for the immunofluorescence classification of kidney biopsy.
Materials and Methods
Kidney Biopsy and Immunofluorescence Staining and Image Acquisition
The analysis of the immunofluorescence images used in this study was approved by the local ethical committee (“Comitato Etico dell’Area Vasta Emilia Nord,” Protocol #434/2019/OSS/AOUMO). The immunofluorescence images were collected in our center during the years 2001–2018; they were obtained from native or kidney allograft biopsies independently of the clinical indication for the biopsy collection. The only inclusion criterion for the study was the availability of images and of the pathology report. In the kidney biopsy procedure, two cores of kidney tissue were obtained in the cortical kidney area by a semiautomatic needle (16 or 18 gauge × 16 cm; Bard Max-Core). The core assigned to light microscopy was Bouin fixed and routinely processed according to conventional procedures. The second core, prepared for direct immunofluorescence, was quickly frozen in liquid nitrogen, sliced into 3-μm cryosections (Cryostat CM1950; Leica Microsystem), and stained with FITC-conjugated rabbit antisera directed against human Ig (IgG, IgA, IgM, and their fractions: λ- and κ-chains), fibrinogen, and complement components C1q and C3 (all antisera were from DakoCytomation). The slides were incubated with the primary antibodies at 20°C in the dark for 30 minutes and then washed two times for 15 minutes each in PBS (IgG and C3: 1:80; IgA, IgM, C1q, fibrinogen, κ, and λ: 1:40; each dilution with 1× PBS). Images were captured at ×400 apparent magnification with a fluorescence microscope (BX41 with U-RFL-T; Olympus) by a digital camera (XC30, Firmware version 4.0.2; Olympus) controlled by a dedicated software (CellB software; Olympus) and stored as gray scale uncompressed TIFF images with 16 bits per pixel. All of the images were captured with a standardized protocol and with a fixed time of exposure. The quality and sharpness of the available images can be checked online (https://nephronn.ing.unimore.it/public/) on a sample of 180 images (test set).
Ground Truth Training and Test Sets
The immunofluorescence report (Supplemental Table 1) adopted at our center describes the “appearance,” “distribution,” and “location” of immune deposits. Furthermore, the “intensity” of staining is described by a semiquantitative rank with discrete values from 0 to 3 (by 0.5 intervals).
A separate description is detailed for each anatomic compartment (glomeruli, tubules, interstitium, and vessels), but in this work, only the glomerular compartment was considered. The standard report includes the evaluation of Igs, fibrinogen, and complement deposition. The deposit “appearance” is categorized in the following predefined classes: granular (coarse granular or fine granular) and linear or pseudolinear (Supplemental Figure 1). The “distribution” of deposits is described as focal or diffuse and segmental or global (Supplemental Figure 2). With regard to “location,” the following predefined classes were annotated: mesangial or capillary wall (Supplemental Figure 3). When possible, capillary wall sublocation has been defined: continuous regular capillary wall (subepithelial), discontinuous regular capillary wall, and irregular capillary wall. All annotations underwent a quality control check, and when needed, they were corrected by an experienced kidney pathologist. Ground truth was provided by L.F. and M.L. (Pathologists 3 and 2, respectively). All of the pathologists reviewed the test set; F.F. (Pathologist 1) in particular did not contribute to the ground truth definition but participated to the test set evaluation. A detailed description of pathologist contribution is reported in the Acknowledgments.
Convolutional Neural Networks Design
For all of the features with the exception of “intensity,” the algorithm is required to assign an image to one of k categories. In order to do so, a neural network processes the input image and outputs k values, each of which is the likelihood that the input sample belongs to the kth class. One image can belong to only one class; therefore, the final prediction is the class with the highest score. A different strategy is used to predict the “intensity” of the deposits as a value ranging from 0 to 3 (with intervals of 0.5). For this task, a neural network is asked to output a probability for each of the possible intensity values (seven different classes). The final intensity prediction is obtained as
where n is the number of classes (n=7), p is the probability that the network assigns to kth class, and v is the value represented by kth class.
Convolutional neural networks designed for classification progressively reduce the image dimensions while increasing the number of feature maps obtained after each layer. ResNet-101 (14) exploits 101 convolutional layers to progressively transform input images composed of 512×512 pixels and the three channels (R, G, and B) in images with 16×16 pixels and 2048 feature maps. From each 16×16 feature map, a single value is obtained as the average of the 256 pixels, and finally, a nonconvolutional fully connected layer is trained to process the 2048 mean values of the channels of features and obtain the two output values, representing the final predicted probability for each class. Input images are given a square shape by adding black bands close to the borders when needed, and they are then resized to 512×512.
In order to help convolutional neural networks generalize their processing abilities and therefore avoid overfitting, two regularization techniques are applied during the training process: data augmentation (4) and dropout (5). To perform data augmentation, each image is randomly flipped and rotated before feeding it to the network because neither of these two transformations should change the content of the image and, therefore, the deposit characteristics. To put dropout into use, feature maps are randomly set to zero before the fully connected layer in order to boost the resilience of the model. Regularization techniques are excluded during inference, when the goal is to obtain the best possible prediction for a single novel image.
In this work, transfer learning is exploited by pretraining convolutional neural networks using ImageNet (15,16) and then fine-tuning them to correctly classify immunofluorescence images. The learning rate is set to 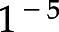 during the fine-tuning process, and a weighted crossentropy loss is used.
during the fine-tuning process, and a weighted crossentropy loss is used.
In order to improve the interpretability of the algorithm, the Grad-Cam method (17) is used to draw attention heat maps. This is done by first automatically analyzing how each convolutional filter affects the predictive score of each class and assigning a weight to each filter-class couple. Then, when a sample is fed to the network, the weights are used to highlight which pixels of the input image had a bigger effect on the prediction of the final class.
Assessment of Convolutional Neural Networks Performance
The convolutional neural networks classification performance in feature recognition of “appearance,” “distribution,” and “location” is assessed using four different metrics:
“Accuracy” is the ratio between the correct predictions and the total predictions;
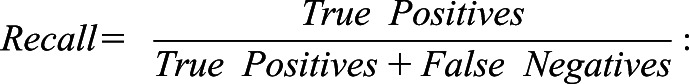 |
“Recall” has the same statistical meaning as “sensitivity”;
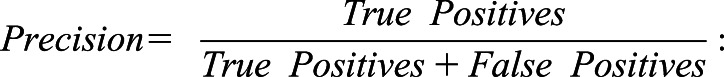 |
“Precision” has the same statistical meaning as “positive predictive value”; and
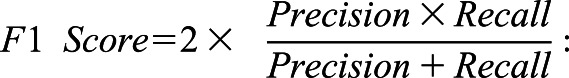 |
“F1 Score” has the same statistical meaning as “weighted average of precision and recall.”
Moreover, receiver operating characteristic curves are presented to display the relation between true positive rate and false positive rate and to compute the area under the curve. True positive rate is equal to the recall, whereas false positive rate is calculated according to the following equation:
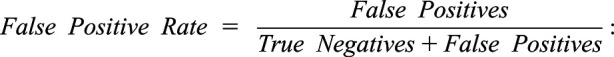 |
False Positive Rate has the same statistical meaning as “fallout.”
For the convolutional neural networks performance of the “intensity,” we represented the classification result in a confusion matrix. The confusion matrix is a specific type of data representation (18) that reports the classes of data provided by the algorithm on each row, while it represents the classes of observed data on each column (or vice versa). The name derives from the fact that it allows a simple visualization of mistakes that the system tends to make in the classification of objects.
Furthermore, we calculated mean absolute error (MAE) and mean squared error (MSE). Where f x ∈ X represent predicted values and y ∈ Y represent ground truth labels, MAE and MSE are defined as
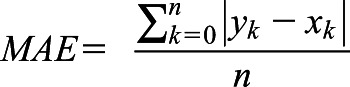 |
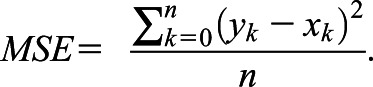 |
Agreement between evaluators, convolutional neural networks, and ground truth was calculated by the Cohen K (19). The average between features K presented in Table 1 is reported as mean ± SD. All of the statistical analyses were performed by the software package STATA/IC 11.2 for Windows (STATA Corp, College Station, TX).
Table 1.
Average agreement between evaluators
| Average | Ground Truth | Convolutional Neural Networks | Pathologist 1 | Pathologist 2 |
|---|---|---|---|---|
| Pathologist 3 | 0.39±0.18 | 0.43±0.22 | 0.56±0.15 | 0.49±0.17 |
| Pathologist 2 | 0.42±0.07 | 0.54±0.21 | 0.56±0.16 | |
| Pathologist 1 | 0.45±0.21 | 0.52±0.15 | ||
| Convolutional neural networks | 0.49±0.20 |
The average K of each feature ± SDs is reported (180 test images overall).
Results
The Dataset
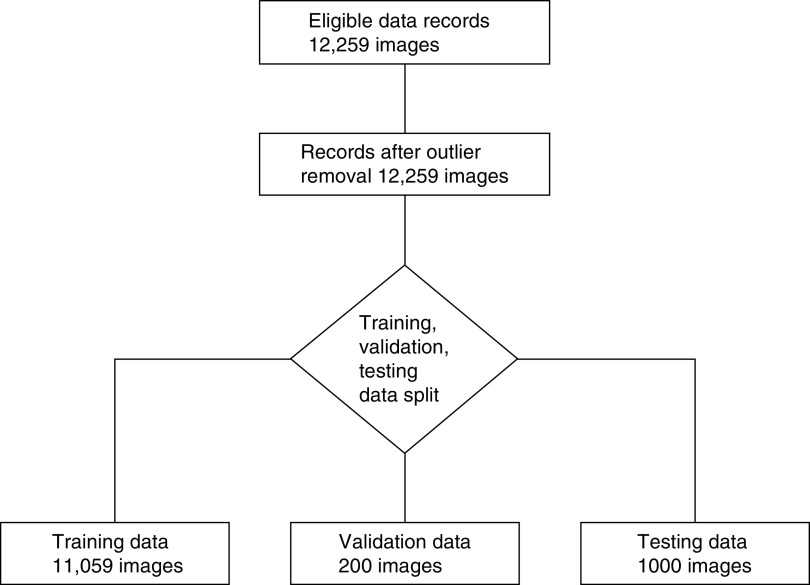
The database of immunofluorescence analysis collects the reports of 2542 consecutive subjects who underwent kidney biopsy between October 2001 and December 2018 in the Division of Nephrology and Dialysis of the AOU Policlinico di Modena, Italy. For each specimen, at least one image of a glomerulus for each antibody is available, but frequently, the images of more than one glomerulus are stored for the same antibody. The main clinical characteristics of the patients are reported in Table 2. The biopsy samples were obtained from native or transplant kidneys independently of the indication for the biopsy procedure. The only criterion for inclusion was the availability of images and of the pathology report; 2225 are reports of native kidneys, while 317 reports describe biopsies of transplanted kidneys. The first ten most frequent final diagnoses for the native kidneys are reported in Supplemental Table 1. Each report describes the result of the immunofluorescence analysis of the kidney specimen of the patient for the following deposits: IgG, IgA, IgM, C1q, C3, fibrinogen, and κ- and λ-light chains. The definition of the features collected in this study is described in Supplemental Figures 1–3. The distribution of the immunofluorescence features is reported in Supplemental Tables 3 and 4. According to the prevalent histologic diagnosis of IgA nephropathy, the most frequent “location” feature is mesangial, the most frequent “appearance” feature is coarse granular, and the most frequent “distribution” feature is global/diffuse. The image dataset collects 12,259 images. The partitioning of the images into a training set (11,059 images), a validation set (200 images), and a test set (1000 images) is shown in Figure 1. For each feature, different training, validation, and test sets were prepared. Given the fact that negative samples are over-represented for every feature, it is crucial that the three subsets have similar positive/negative samples ratio. In particular, a test set containing no positive samples would be futile. The presence (or absence) of the investigated feature is thus taken into account when splitting the dataset in order to force similar data distribution into every subset. Other patient metadata are ignored during the partitioning, and their distribution in the subsets is ruled by random distribution.
Table 2.
Main clinical characteristics of the patients
| Characteristic | Value |
|---|---|
| Subjects | 2542 |
| Men:women, % | 62:38 |
| Age, yr | 52 (38–66) |
| Creatinine, mg/dl | 1.6 (1.0–2.7) |
| eGFR, ml/min per 1.73 m2 | 45 (22–78) |
| CKD class, % | |
| 1 | 18 |
| 2 | 18 |
| 3a | 13 |
| 3b | 16 |
| 4 | 18 |
| 5 | 17 |
| Hb, g/dl | 12.0 (10.1–13.7) |
| Urine protein-to-creatinine ratio, g/g | 1.8 (0.5–4.3) |
| Hematuria, % | 56 |
| Systolic BP, mm Hg | 130 (120–142) |
| Diastolic BP, mm Hg | 80 (70–84) |
| Diabetes, % | 11 |
| Hypertension, % | 37 |
| Clinical presentation, % | |
| None | 11 |
| Urinary abnormalities | 44 |
| Nephrotic syndrome | 32 |
| Nephritic syndrome | 9 |
| Macrohematuria | 4 |
With the exception of the men-women ratio, all of the data are expressed as median (25°–75° percentile) or percentage. Hb, hemoglobin.
Figure 1.
The image dataset collects 12,259 images. The partitioning of the images into a training set (11,059 images), a validation set (200 images), and a test set (1000 images) is shown.
Performance of the Convolutional Neural Networks
We trained the neural networks to recognize the four different “locations”: mesangial, continuous regular capillary wall, discontinuous regular capillary wall, and irregular capillary wall. A general class named capillary wall obtained by merging all of the different kinds of capillary wall locations was added to the analysis. Regarding “location,” the pattern recognition tasks are completely independent classification problems because each image could present neither, none, or every combination of the investigated location characteristics. For each one of the tasks, a specific convolutional neural network is trained to classify a single image as either presenting or not presenting the pattern of interest. The same approach is taken to tackle the deposit “appearance” (linear/pseudolinear, coarse granular, and fine granular) and the deposit “distribution” (segmental and global). Because our dataset is composed of high-magnification images (×400) that do not allow a panoramic evaluation of the slice, the “location” features “diffuse” or “focal” have been left out from the analysis. The discontinuous regular capillary wall and the linear/pseudolinear features were not further assessed because of the low performance of the convolutional neural network on these parameters: the linear/pseudolinear and discontinuous regular capillary wall F1 scores were 0.26 and 0.09, respectively, much lower values than obtained by the other features (Table 3). We suspect that the bad performance of these features is mainly attributable to the poor representation in our dataset (linear/pseudolinear and discontinuous regular capillary: 88/335 and 232 features, respectively) (Supplemental Table 3). However, only the evaluation of a larger dataset with a greater representation of these features will allow us to verify this hypothesis.
Table 3.
Neural network performance in feature prediction
| Feature | Accuracy | Recall | Precision | F1 Score | Area under the Curve |
|---|---|---|---|---|---|
| Appearance | |||||
| Coarse granular | 0.84 | 0.61 | 0.34 | 0.44 | 0.85 |
| Fine granular | 0.94 | 0.76 | 0.43 | 0.47 | 0.83 |
| Distribution | |||||
| Segmental | 0.81 | 0.50 | 0.36 | 0.42 | 0.81 |
| Global | 0.82 | 0.74 | 0.87 | 0.79 | 0.89 |
| Location | |||||
| Mesangial | 0.84 | 0.78 | 0.71 | 0.74 | 0.89 |
| Capillary wall | 0.81 | 0.77 | 0.66 | 0.71 | 0.87 |
| Continuous regular capillary wall | 0.91 | 0.82 | 0.75 | 0.78 | 0.94 |
| Irregular capillary wall | 0.79 | 0.67 | 0.49 | 0.57 | 0.84 |
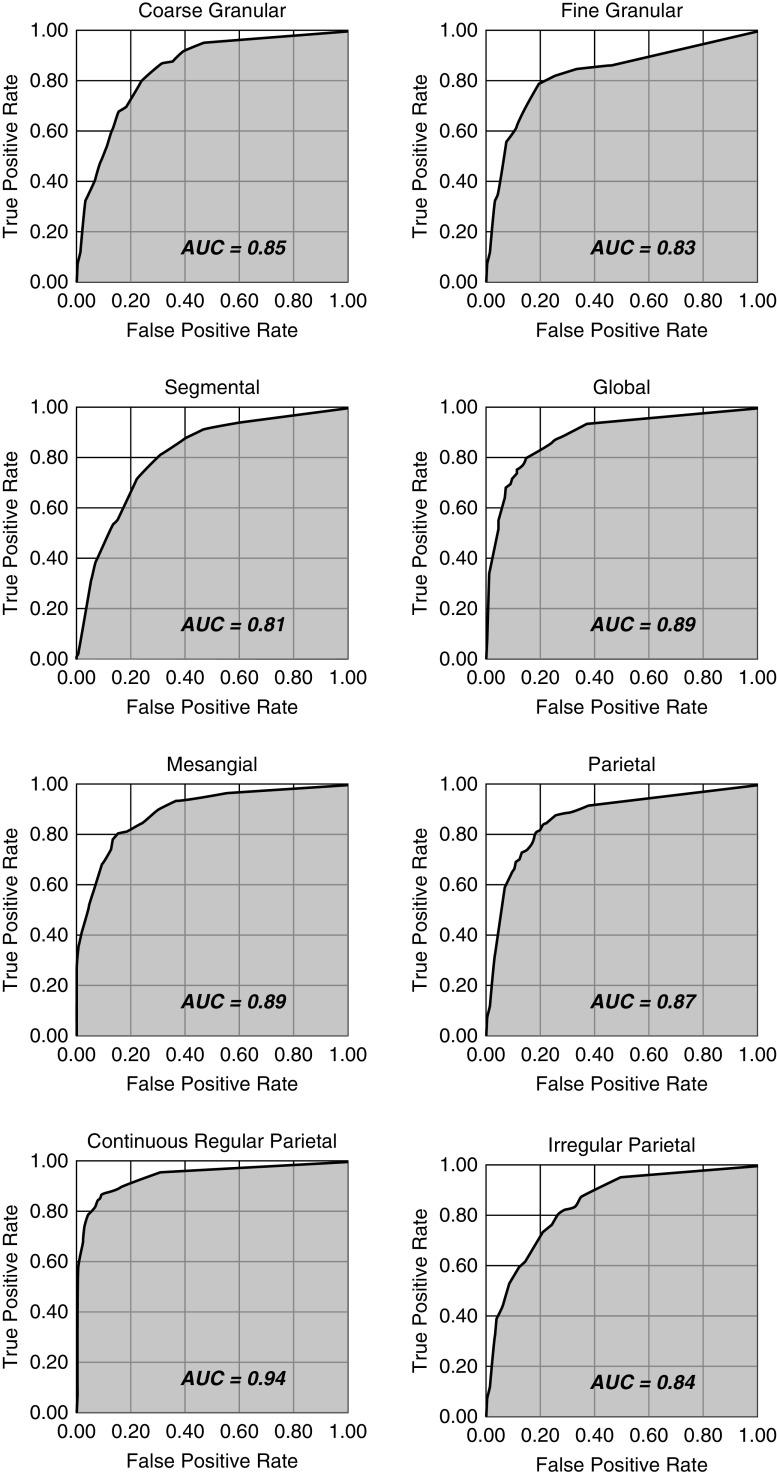
Table 3 represents the performance of the neural network to predict the immunofluorescence features. All of the features of “location,” “appearance,” and “distribution” obtained levels of accuracy around 0.8 and above. The highest performance was obtained by the “appearance” feature “fine granular” (0.94), followed by the “location” feature “continuous regular capillary wall” (0.91). The receiver operating characteristic curve of each feature is presented in Figure 2.
Figure 2.
Receiver operating characteristic curves of the convolutional neural networks performance of prediction of each feature. AUC, area under the curve.
Eight representative examples of the ground truth and automatic feature classification used in the test set are depicted in Figure 3. Furthermore, the figures show the results obtained through the Grad-Cam method. The heat maps indicate that neural networks correctly identify the glomerulus within an image and search for the pattern in the appropriate section of the image. The final decision to classify the images is notably influenced by the deposits on the glomerular structure.
Figure 3.
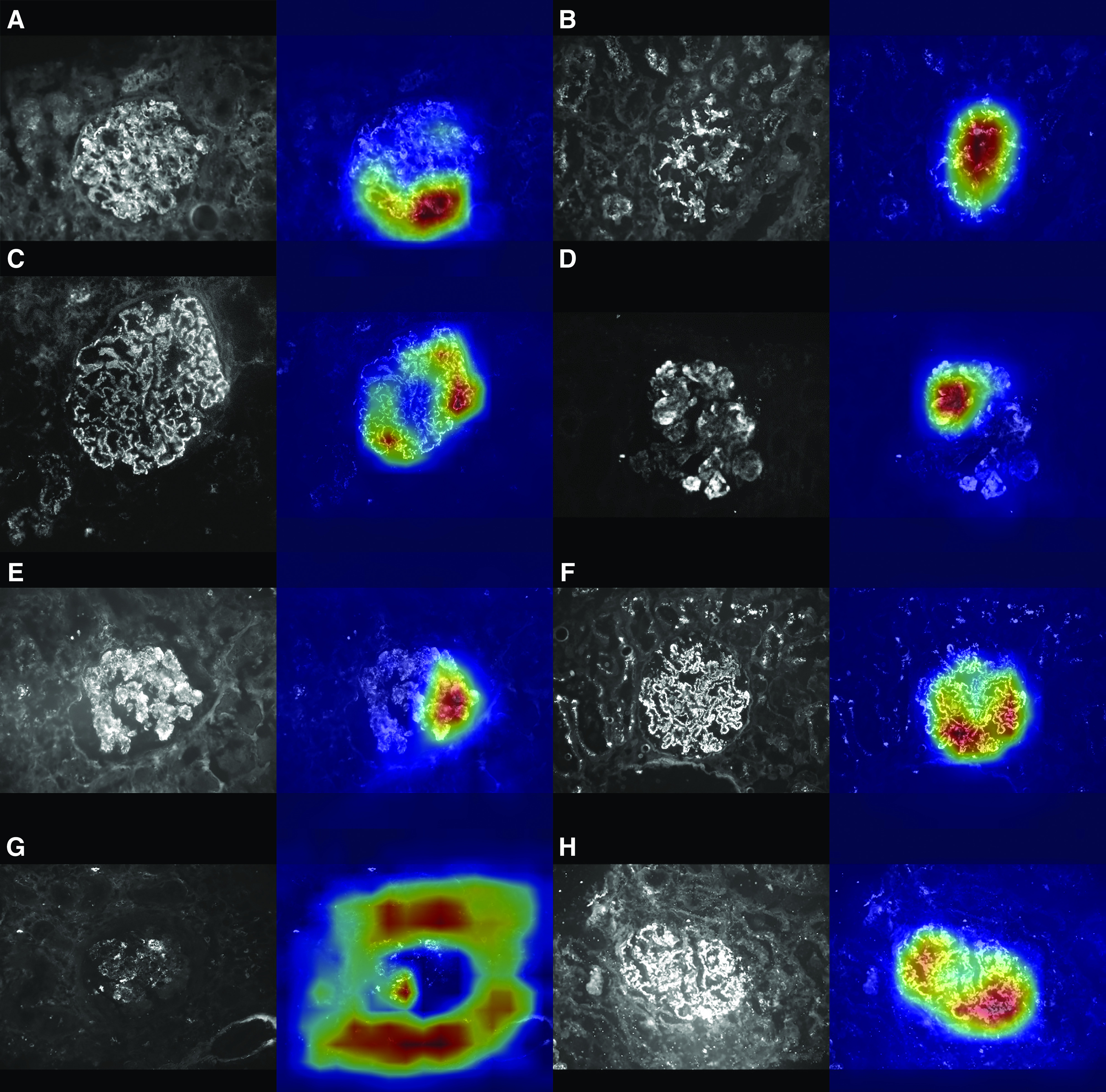
The Convolutional Neural Network classification is influenced by the deposits on the glomerular structure inside the image. Each panel of the figure represents the result of the feature-specific convolutional neural networks elaboration of a test image. In (A–H), left panels show the original test images, while right panels show their heat maps. In the heat map, the red area shows the sections of the image most involved in the classification process by the convolutional neural network. All of the presented images were correctly classified by the specific convolutional neural network: (A) parietal, (B) mesangial, (C) continuous regular capillary wall, (D) irregular capillary wall, (E) coarse granular, (F) fine granular, (G) segmental, (H) global.
For what concerns the “intensity,” we were able to predict the intensity level of images with a good level of approximation, as confirmed by the MAE of 0.398 and the MSE of 0.455, which are two of the most relevant metrics for regression tasks, such as intensity prediction (20). Table 4 presents the confusion matrix of the seven intensity classes, where the predicted class for each sample is the closest one to the real output value (which is a continuous number between zero and three).
Table 4.
Confusion matrix of the intensity classes
| Intensity | 0 | 0.5 | 1 | 1.5 | 2 | 2.5 | 3 |
|---|---|---|---|---|---|---|---|
| 0 | 0.82 | 0.11 | 0.04 | 0.00 | 0.02 | 0.00 | 0.01 |
| 0.5 | 0.31 | 0.29 | 0.22 | 0.05 | 0.07 | 0.02 | 0.05 |
| 1 | 0.16 | 0.16 | 0.28 | 0.06 | 0.24 | 0.00 | 0.10 |
| 1.5 | 0.16 | 0.10 | 0.20 | 0.04 | 0.20 | 0.02 | 0.28 |
| 2 | 0.03 | 0.03 | 0.10 | 0.10 | 0.34 | 0.00 | 0.39 |
| 2.5 | 0.06 | 0.03 | 0.07 | 0.01 | 0.25 | 0.03 | 0.55 |
| 3 | 0.00 | 0.00 | 0.02 | 0.00 | 0.06 | 0.00 | 0.92 |
The confusion matrix provides insight into how prediction is correctly distributed over the seven different classes of intensity. The ground truth labels are given vertically, and the predicted labels by the convolutional neural network are written on the horizontal axis.
We have preliminary data suggesting that the algorithm is robust enough to correctly recognize the glomerular pattern even in the presence of unusual tubular changes. We refer to some peculiar conditions with the presence of deposits along the tubular basement membrane as, for example, can occur in lupus nephritis or in light-chain deposition disease. We have included in Supplemental Figures 4 and 5 and Supplemental Tables 7 and 8 the figures, the ground truth, and the interpretation of the convolutional neural network of two cases. Nonetheless, given the small number of these cases, this statement will have to be confirmed in light of a broader series.
Agreement between Pathologists, Ground Truth, and Convolutional Neural Network
We compared the classifications of a set of 20 images for each feature (180 test images overall) between three different pathologists and convolutional neural networks with respect to ground truth (Supplemental Table 5). The human evaluators as well as the convolutional neural network were blinded about clinical information related to the images. The data suggest a fair to moderate agreement between pathologists and ground truth; convolutional neural networks had a performance with respect to ground truth comparable with human evaluators. We also calculated for the pairs between pathologist 1, 2, 3, convolutional neural network, and ground truth, an average Cohen K of all features (Table 1). Also in this case, the data suggest a moderate agreement between the different evaluators and, in particular, convolutional neural networks show comparable performance to human evaluators. The interagreement Cohen K of every feature is reported in Supplemental Table 6.
Time Analyses
The three pathologists spent 25, 23, and 18 minutes, respectively, in the analysis of the 180 test images. This corresponds to an average time of evaluation per image of 7.3±1.2 seconds. The execution time of the convolutional neural networks has been calculated on an Intel(R) Xeon(R) E5–2650 v2@2.60 GHz CPU with 32 cores and 126 GB of RAM running Ubuntu 18.04 LTS and using a Tesla K80 GPU with 12 GB of memory. The time for the complete analysis of the 180 test images by the convolutional neural networks was 11.2 seconds. This corresponds to 62.5 milliseconds per image, which is 117.3 times faster than the average human evaluator.
Discussion
In this study, we presented a deep learning approach for the automatic reporting of immunofluorescence specimens of kidney biopsy. The main finding of our work is that convolutional neural networks achieve an accurate definition of the main pathology features usually collected from this type of specimen: “appearance,” “distribution,” “location,” and “intensity” of the deposits. The comparison of agreement among three pathologists shows a moderate agreement, and, in this respect, convolutional neural networks show a similar performance to human evaluators. The interest in deep learning applied to nephropathology has grown significantly in recent years. The researchers adopted deep learning technology to achieve different goals: some focused on the automated identification of the main microscopic structures (glomeruli, tubules, vessels, etc.) (21–25), some authors worked on the segmentation of the identified structures (2), and others have used convolutional neural networks to obtain automated clinical classifications (1,2). We can consider our approach as part of the strategies for developing a tool for assisting in automated clinical classification. In this work, we did not deal with the problem of the preliminary extraction of glomerular structures, which was already addressed in many previous papers (21–25). Because our dataset was already made up of high-magnification images of individual glomeruli historically collected in the last 18 years of manual reporting of the kidney specimens, we focused our attention on the pure classification of these images. Immunohistopathologic analysis (in particular, the immunofluorescence approach) has never been previously addressed with deep learning techniques. We therefore focused on the ability to extract from these images the features normally collected by our pathologists during the examination. The immunofluorescence interpretation, like many other clinical imaging techniques, is a field in which the deep learning approach can offer a significant contribution. Indeed, any medical imaging technique involving human reporting is conditioned by a significant interpretative subjectivity. This poor reproducibility has been repeatedly documented in the old kidney histopathologic literature (26–29), and it has been recently confirmed in studies concerning the application of deep learning in kidney pathology (1,2). The indices of agreement between human pathologists vary according to the metrics used, the preparation of the specimen, the histologic parameters assessed, and the kidney disease considered; overall, reported agreement rate between human kidney pathologists is fair to moderate, with agreement ratio ranging from 0.3 to 0.6. The agreement among the pathologists of our center settles within the moderate level, according to Cohen K methodology (30). In this scenario, the convolutional neural network algorithms have ground truth agreement performances similar to the human operators. Our data suggest a high accuracy in identifying the analyzed features, which varies from 0.79 to 0.94. The feature in which the system performs best is the identification of the “fine granular appearance,” which, together with the “continuous regular capillary wall location” (the second-best identified feature), is often associated with the definition of the subepithelial deposits of membranous GN. With regard to these two patterns, the levels of agreements expressed between pathologists and ground truth have proven to be completely analogous to those between convolutional neural networks and ground truth. However, even if less outstanding, the agreement relating to the remaining features shows a convolutional neural networks performance comparable with that of the human evaluators. It can be assumed that the imperfect concordance between ground truth with respect to convolutional neural network and pathologists may be attributable to a different analytical setting (direct microscopic visualization of the specimen compared with captured images). However, we believe that this is a partial and not a substantial explanation of the phenomenon. In fact, our data suggest that, under the same analytical conditions, the pathologists show a rather modest agreement between them (Table 1), a well-known phenomenon against which an automated approach can bring improvements. Even for intensity, a critical parameter for the reporting, a substantial subjectivity is documented (data not shown). To the best of our knowledge, there are no literature data suggesting that a simple determination of the fluorescence intensity level could replace a subjective (or convolutional neural network) assessment. The classification of intensity still remains a semiquantitative evaluation because several preanalytical variables are not completely controllable and do not allow for a purely quantitative evaluation. These include the variability of fluorescence emission between commercial antibody batches, the decay of UV lamp fluorescence during its lifetime, and many other confounders that are not usually under control (laboratory temperature, solution batches, and manual skill of the technician, etc.).
The reproducibility of the evaluation, which in itself already has enormous value, is not the only advantage that deep learning can introduce in this field; the speed of interpretation, the amount of data that can be analyzed, and the relative economic savings all represent elements of great interest in the adoption of this technology. About this aspect, the speed of execution of the analyses is crucial: the pathologists spent, on average, 22 minutes in the assessment of 180 images. Convolutional neural networks processed the same images in 11.2 seconds with comparable accuracy. This in itself represents 117.3 times greater efficiency than the human evaluator. Nevertheless, compared with the evaluation of a specimen in a real scenario, it must be taken into account that our simulation does not consider the times of identification of the glomeruli and the image acquisition from the microscope because these were not goals of our study.
Our group has developed a web platform at https://nephronn.ing.unimore.it/, where it is possible to upload digitized images of glomeruli from immunofluorescence preparations at high magnification (×400). After the image is loaded, it is analyzed, and the prediction for each one of the nine features currently available in our algorithm is presented. Because of the current hardware constraints, the web application relies on shallower neural networks than the ones described in this paper, causing a classification accuracy drop of about 5%. This platform will allow the kidney pathology community to directly experience the potential of our approach. In particular, this service will allow the development of possible collaborations with other kidney pathology centers for sharing images and contributing to the development of a larger common immunofluorescence database. At the moment, in fact, this experience is on the basis of a relatively homogeneous dataset; indeed, in this study, all of the preparations were obtained from a single center that processed and digitized the images with a single protocol. It cannot be excluded that our algorithm may have lower performances when applied to images derived from other centers. Another limitation of our approach consists of the impossibility of recognizing with a significant accuracy the features that were less represented in our dataset (discontinuous regular capillary wall and the linear/pseudolinear) or are not captured by our standardized report (for example, the amorphous/globular-IgM/complement staining in segmental glomerulosclerosis). With collaborations already formalized with other laboratories, we plan to increase our dataset and incorporate these additional features into our algorithm in the near future. Another possible limitation of our approach could be identified in the double role of Pathologists 2 and 3 in defining the ground truth and in the analysis of the testing set. However, we do not believe that this negatively affects the result of our work. Indeed, the dual role of Pathologists 2 and 3 would have possibly inflated their K value of concordance with the ground truth to the detriment of Pathologist 1 and the convolutional neural network. This phenomenon, which, however, does not emerge from the data (Table 1), would eventually make the estimate of concordance K of the convolutional neural network more conservative.
We can speculate that our algorithm can be part of a larger computer-aided pathology diagnostic platform. This platform, in the context of a network of nephrologic centers engaged in kidney biopsy diagnostics, could provide for local scanning of immunofluorescence preparations (whole-slide imaging). The scanned image could be sent to the central processing center, which will apply a glomeruli identification algorithm (2,25) and the subsequent analysis of the individual glomeruli for the automatic definition of the characteristics of “appearance,” “distribution,” “location,” and “intensity.” An averaged summary of the results obtained by each glomerulus could therefore be organized in a report, which would be sent to the local center for validation and implementation in the flow chart of the biopsy diagnostic process.
In conclusion, we presented a deep learning approach capable of recognizing with significant accuracy the main features normally collected in the reporting of kidney immunofluorescence. The data showed an accuracy comparable with what is normally expressed by pathologists who are experts in the field. In the near future, the collaboration with other centers with experience in kidney pathology will allow us to complete our algorithm with features that are currently under-represented and will guarantee external validity of our analysis. We believe that the algorithm developed by our group can become a useful tool to support the reporting of kidney immunofluorescence.
Disclosures
All authors have nothing to disclose.
Funding
This work was supported by personal academic funds of R. Magistroni.
Supplementary Material
Acknowledgments
Dr. Riccardo Magistroni conceptualized the study; Dr. Costantino Grana and Dr. Riccardo Magistroni designed the study; Dr. Silvia Giovanella and Dr. Giulia Ligabue acquired immunofluorescence images; Dr. Federico Pollastri developed the convolutional neural networks; Dr. Francesco Fontana was responsible for the pathologic evaluation of the test set (Pathologist 1); Dr. Marco Leonelli was responsible for the pathologic definition of the ground truth and evaluation of the test set (Pathologist 2); Dr. Luciana Furci was responsible for the pathologic definition of the ground truth and evaluation of the test set (Pathologist 3); Dr. Riccardo Magistroni was responsible for data analysis; Dr. Federico Bolelli developed the web page; Dr. Giulia Ligabue, Dr. Riccardo Magistroni, and Dr. Federico Pollastri wrote the draft of the paper; and Dr. Gaetano Alfano, Dr. Federico Bolelli, Dr. Gianni Cappelli, Dr. Silvia Giovanella, Dr. Costantino Grana, Dr. Giulia Ligabue, Dr. Riccardo Magistroni, Dr. Federico Pollastri, and Dr. Francesca Testa revised the paper.
Footnotes
Published online ahead of print. Publication date available at www.cjasn.org.
See related editorial, “Artificial Intelligence: The Next Frontier in Kidney Biopsy Evaluation,” on pages 1389–1391.
Supplemental Material
This article contains the following supplemental material online at http://cjasn.asnjournals.org/lookup/suppl/doi:10.2215/CJN.03210320/-/DCSupplemental.
Supplemental Figure 1. Classification of the “appearance” of the deposits of the immunofluorescence specimen.
Supplemental Figure 2. Classification of the “distribution” of the deposits of the immunofluorescence specimen.
Supplemental Figure 3. Classification of the “location” of the deposits of the immunofluorescence specimen.
Supplemental Figure 4. IgG direct immunofluorescence of a lupus nephritis case.
Supplemental Figure 5. IgG direct immunofluorescence of light-chain deposition disease.
Supplemental Table 1. Immunofluorescence report.
Supplemental Table 2. Final diagnoses of the kidney biopsies.
Supplemental Table 3. Numeric distribution of the immunofluorescence features.
Supplemental Table 4. Percent distribution of the immunofluorescence features.
Supplemental Table 5. Ground truth agreement between pathologists and convolutional neural networks.
Supplemental Table 6. Interagreement Cohen K of each feature.
Supplemental Table 7. Comparison of the report of a case of lupus nephritis (Supplemental Figure 4) between ground truth and convolutional neural networks.
Supplemental Table 8. Ground truth agreement between pathologists and convolutional neural networks.
References
- 1.Ginley B, Lutnick B, Jen KY, Fogo AB, Jain S, Rosenberg A, Walavalkar V, Wilding G, Tomaszewski JE, Yacoub R, Rossi GM, Sarder P: Computational segmentation and classification of diabetic glomerulosclerosis. J Am Soc Nephrol 30: 1953–1967, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hermsen M, de Bel T, den Boer M, Steenbergen EJ, Kers J, Florquin S, Roelofs JJTH, Stegall MD, Alexander MP, Smith BH, Smeets B, Hilbrands LB, van der Laak JAWM: Deep learning-based histopathologic assessment of kidney tissue. J Am Soc Nephrol 30: 1968–1979, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hawkins DM: The problem of overfitting. J Chem Inf Comput Sci 44: 1–12, 2004. [DOI] [PubMed] [Google Scholar]
- 4.Pollastri F, Bolelli F, Paredes R, Grana C: Augmenting data with GANs to segment melanoma skin lesions. Multimedia Tools Appl 79: 15575–15592, 2020 [Google Scholar]
- 5.Srivastava N, Hinton G, Krizhevsky A, Sutskever I, Salakhutdinov R: Dropout: A simple way to prevent neural networks from overfitting. J Mach Learn Res 15: 1929–1958, 2014 [Google Scholar]
- 6.Ioffe S, Szegedy C: Batch normalization: Accelerating deep network training by reducing internal covariate shift. In: Proceedings of the 32nd International Conference on Machine Learning, edited by Francis B, David B, Proceedings of Machine Learning Research, PMLR, 2015, pp 448–456
- 7.Litjens G, Kooi T, Bejnordi BE, Setio AAA, Ciompi F, Ghafoorian M, van der Laak JAWM, van Ginneken B, Sánchez CI: A survey on deep learning in medical image analysis. Med Image Anal 42: 60–88, 2017. [DOI] [PubMed] [Google Scholar]
- 8.Canalini L, Pollastri F, Bolelli F, Cancilla M, Allegretti S, Grana C: Skin Lesion Segmentation Ensemble with Diverse Training Strategies, Cham, Switzerland, Springer International Publishing, 2019, pp 89–101 [Google Scholar]
- 9.Bolelli F, Baraldi L, Pollastri F, Grana C: A hierarchical quasi-recurrent approach to video captioning. Presented at the 2018 IEEE International Conference on Image Processing, Applications and Systems (IPAS), Sophia Antipolis, France, December 12–14, 2018 [Google Scholar]
- 10.Pollastri F, Bolelli F, Palacios RP, Grana C: Improving skin lesion segmentation with generative adversarial networks. Presented at the 2018 IEEE 31st International Symposium on Computer-Based Medical Systems (CBMS), Karlstad, Sweden, June 18–21, 2018 [Google Scholar]
- 11.Esteva A, Kuprel B, Novoa RA, Ko J, Swetter SM, Blau HM, Thrun S: Dermatologist-level classification of skin cancer with deep neural networks. Nature 542: 115–118, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gulshan V, Peng L, Coram M, Stumpe MC, Wu D, Narayanaswamy A, Venugopalan S, Widner K, Madams T, Cuadros J, Kim R, Raman R, Nelson PC, Mega JL, Webster DR: Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. JAMA 316: 2402–2410, 2016. [DOI] [PubMed] [Google Scholar]
- 13.Kermany DS, Goldbaum M, Cai W, Valentim CCS, Liang H, Baxter SL, McKeown A, Yang G, Wu X, Yan F, Dong J, Prasadha MK, Pei J, Ting MYL, Zhu J, Li C, Hewett S, Dong J, Ziyar I, Shi A, Zhang R, Zheng L, Hou R, Shi W, Fu X, Duan Y, Huu VAN, Wen C, Zhang ED, Zhang CL, Li O, Wang X, Singer MA, Sun X, Xu J, Tafreshi A, Lewis MA, Xia H, Zhang K: Identifying medical diagnoses and treatable diseases by image-based deep learning. Cell 172: 1122–1131.e9, 2018 [DOI] [PubMed] [Google Scholar]
- 14.He K, Zhang X, Ren S, Sun J: Deep residual learning for image recognition. Presented at the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, June 27–30, 2016 [Google Scholar]
- 15.Krizhevsky A, Sutskever I, Hinton GE: ImageNet classification with deep convolutional neural networks. Presented at the Advances in Neural Information Processing Systems, Lake Tahoe, NV, December 3–8, 2012
- 16.Deng J, Socher R, Fei-Fei L, Dong W, Li K, Li L-J: ImageNet: A large-scale hierarchical image database. Presented at the 2009 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Miami, FL, June 20–25, 2009 [Google Scholar]
- 17.Selvaraju RR, Cogswell M, Das A, Vedantam R, Parikh D, Batra D: Grad-CAM: Visual explanations from deep networks via gradient-based localization. Presented at the 2017 IEEE International Conference on Computer Vision (ICCV), Venice, Italy, October 22–29, 2017 [Google Scholar]
- 18.Tharwat A: Classification assessment methods [published online ahead of print August 21, 2018]. Appl Comput Info . Available at: 10.1016/j.aci.2018.08.003 [DOI] [Google Scholar]
- 19.Cohen J: A coefficient of agreement for nominal scales. Educ Psychol Meas 20: 37–46, 1960 [Google Scholar]
- 20.Xu Y, Pei J, Lai L: Deep learning based regression and multiclass models for acute oral toxicity prediction with automatic chemical feature extraction. J Chem Inf Model 57: 2672–2685, 2017. [DOI] [PubMed] [Google Scholar]
- 21.Bukowy JD, Dayton A, Cloutier D, Manis AD, Staruschenko A, Lombard JH, Solberg Woods LC, Beard DA, Cowley AW Jr.: Region-based convolutional neural nets for localization of glomeruli in trichrome-stained whole kidney sections. J Am Soc Nephrol 29: 2081–2088, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gadermayr M, Dombrowski AK, Klinkhammer BM, Boor P, Merhof D: CNN cascades for segmenting sparse objects in gigapixel whole slide images. Comput Med Imaging Graph 71: 40–48, 2019. [DOI] [PubMed] [Google Scholar]
- 23.Gallego J, Pedraza A, Lopez S, Steiner G, Gonzalez L, Laurinavicius A, Bueno G: Glomerulus classification and detection based on convolutional neural networks. J Imaging 4: 20, 2018 [Google Scholar]
- 24.Marsh JN, Matlock MK, Kudose S, Liu T-C, Stappenbeck TS, Gaut JP, Swamidass SJ: Deep learning global glomerulosclerosis in transplant kidney frozen sections. IEEE Trans Med Imaging 37: 2718–2728, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao K, Tang YJJ, Zhang T, Carvajal J, Smith DF, Wiliem A, Hobson P, Jennings A, Lovell BC: DGDI: A dataset for detecting glomeruli on renal direct immunofluorescence. Presented at 2018 Digital Image Computing: Techniques and Applications (DICTA), Canberra, Australia, December 10–13, 2018
- 26.Gamba G, Reyes E, Angeles A, Quintanilla L, Calva J, Peña JC: Observer agreement in the scoring of the activity and chronicity indexes of lupus nephritis. Nephron 57: 75–77, 1991. [DOI] [PubMed] [Google Scholar]
- 27.Grootscholten C, Bajema IM, Florquin S, Steenbergen EJ, Peutz-Kootstra CJ, Goldschmeding R, Bijl M, Hagen EC, van Houwelingen HC, Derksen RH, Berden JH: Interobserver agreement of scoring of histopathological characteristics and classification of lupus nephritis. Nephrol Dial Transplant 23: 223–230, 2008. [DOI] [PubMed] [Google Scholar]
- 28.Oni L, Beresford MW, Witte D, Chatzitolios A, Sebire N, Abulaban K, Shukla R, Ying J, Brunner HI: Inter-observer variability of the histological classification of lupus glomerulonephritis in children. Lupus 26: 1205–1211, 2017. [DOI] [PubMed] [Google Scholar]
- 29.Wernick RM, Smith DL, Houghton DC, Phillips DS, Booth JL, Runckel DN, Johnson DS, Brown KK, Gaboury CL: Reliability of histologic scoring for lupus nephritis: A community-based evaluation. Ann Intern Med 119: 805–811, 1993. [DOI] [PubMed] [Google Scholar]
- 30.Landis JR, Koch GG: The measurement of observer agreement for categorical data. Biometrics 33: 159–174, 1977. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.