In this work, Slomka et al. have evolved the naturally competent B. subtilis in the lab, in the presence or absence of foreign genomic DNA. They examine the effects of foreign DNA acquisition on the growth.....
Keywords: horizontal gene transfer, natural transformation, experimental evolution, Bacillus subtilis
Abstract
Tracing evolutionary processes that lead to fixation of genomic variation in wild bacterial populations is a prime challenge in molecular evolution. In particular, the relative contribution of horizontal gene transfer (HGT) vs. de novo mutations during adaptation to a new environment is poorly understood. To gain a better understanding of the dynamics of HGT and its effect on adaptation, we subjected several populations of competent Bacillus subtilis to a serial dilution evolution on a high-salt-containing medium, either with or without foreign DNA from diverse pre-adapted or naturally salt tolerant species. Following 504 generations of evolution, all populations improved growth yield on the medium. Sequencing of evolved populations revealed extensive acquisition of foreign DNA from close Bacillus donors but not from more remote donors. HGT occurred in bursts, whereby a single bacterial cell appears to have acquired dozens of fragments at once. In the largest burst, close to 2% of the genome has been replaced by HGT. Acquired segments tend to be clustered in integration hotspots. Other than HGT, genomes also acquired spontaneous mutations. Many of these mutations occurred within, and seem to alter, the sequence of flagellar proteins. Finally, we show that, while some HGT fragments could be neutral, others are adaptive and accelerate evolution.
BACTERIAL evolution is driven not only by mutations but also by horizontal gene transfer (HGT), denoting the integration of external DNA into the genome via various mechanisms. HGT can be particularly evolutionarily beneficial in times of stress, as it facilitates the introduction of multiple genomic variations at once, at existing loci, or by introduction of novel genes (Ochman et al. 2000). The adaptive potential brought about by HGT comes with a phenotypic cost and possible negative implications, depending on the donor and recipient genetic compatibility, such as disruption of regulatory circuits and cytotoxic effects of transferred proteins (Baltrus 2013). The evidence for the high abundance of different HGT mechanisms and their footprints on bacterial genomes has accumulated rapidly in recent years thanks to dedicated homology comparison methods applied to entire genomes (Koonin et al. 2001; Ravenhall et al. 2015). Using such bioinformatic methods, it was possible to quantify the large extent of transfer events in extant bacterial genomes (Kloesges et al. 2011; Popa et al. 2017; González-Torres et al. 2019), and characterize the genetic traits most affected by this process and their chromosomal organization in hotspots (Oliveira et al. 2017). The clear abundance of transfer events in bacterial genomes could suggest an adaptive value of HGT (Wiedenbeck and Cohan 2011) despite its cost, but it does not exclude the possibility that at least some HGT events have neutral effects in evolution. Informatics analysis of HGT is best at providing a snapshot of an evolutionary process in nature, while tracing dynamic trajectories of HGT events is more challenging. Though some previous works have been able to utilize bioinformatics analyses of extant genomes for a better understanding of bacterial ecology and evolution (Hehemann et al. 2016; Arevalo et al. 2019), it is difficult to determine with such methods what is the contribution of HGT, relative to point mutations, in adaptation. A complementary approach for the investigation of evolutionary dynamics is experimental evolution (Barrick and Lenski 2013; Kaminski et al. 2019).
Two recent studies have employed experimental evolution to explore bacterial HGT in the context of evolution, by following the adaptation of Escherichia coli cells subjected to multiple rounds of conjugation with different E. coli strains. In one of the studies, HGT-based acquisition of foreign DNA did not provide a fitness advantage [first described in Souza et al. (1997), later analyzed in Maddamsetti and Lenski (2018)], whereas, in the other study, in E. coli too, gene acquisition was found to improve the adaptation of the evolving population in one out of two conditions tested (Chu et al. 2018). The scarcity of such studies, their inconsistent outcomes, the fact that they were limited to conjugation only as a mechanism of HGT, and their focusing on E. coli only, all call for broadening the scope of investigation of HGT in experimental evolution.
Here, we widen experimental evolutionary exploration to a main mode of HGT mechanism, that of natural transformation (Johnston et al. 2014). Since conjugation and natural transformation are fundamentally different, DNA acquisition via each mechanism could give rise to different genomic and evolutionary signatures. The average size of transferred fragments through natural transformation ranges between 1 and 10 kb (Fornili and Fox 1977; Lin et al. 2009), though larger transferred fragments have been detected in previous works (Coupat et al. 2008). On the other hand, the size of transferred plasmids through conjugation is typically an order of magnitude larger, with the median size of conjugative plasmids being 181 kb (Smillie et al. 2010; Derbyshire and Gray 2014). Previous works that sought to characterize the genomic signature of natural transformation, applied in vitro transformation of various naturally competent bacterial species with foreign DNA (Kulick et al. 2008; Lin et al. 2009; Mell et al. 2011, 2014; Croucher et al. 2012; Bubendorfer et al. 2016). These studies revealed that acquisition of foreign DNA fragments occurs in bursts of many fragments by the same cell, and that donor segments tend to be clustered along the genome. Yet, these experiments examined the immediate time period after the transformation, and, hence, they could not determine for how long the horizontally transferred DNA segments were retained in the genome. Other works focused on the effect of phylogenetic distance between the donor and recipient strains on the frequency of recombination (Majewski and Cohan 1999; Carrasco et al. 2016). They show a decline in the rate of in vitro recombination with increasing phylogenetic distance between donors and recipient; however, it is possible that during prolonged evolutionary adaptation, deviations from this trend might occur, e.g., due to selective advantage of more distant donor fragments, and rare events of integration that could occur and propagate.
We have started to close this gap by putting forward a serial dilution evolution experiment which aims to assess and characterize: (1) the contribution of HGT from adapted donors to the recipient’s adaptation process under abiotic stress (2) the dynamics of foreign DNA acquisition and its propagation in evolving populations compared to mutations, and (3) the dependence of the above on the phylogenetic distance between the donor and recipient genomes. For that purpose, we have chosen the highly studied, nonpathogenic model organism for natural transformation Bacillus subtilis.
We conducted this evolution experiment by evolving a competent B. subtilis strain that has the ability to uptake naked DNA from its environment under high salinity stress, either with or without providing foreign genomic DNA from alternative potential donors. The motivation for the choice of the stress applied, high salt concentration [Luria broth (LB) with 0.8 M NaCl], was the lack of pre-existing specific adaptation of our ancestral strain to high salinity on one hand, and the abundance of diverse naturally adapted strains that could serve as DNA donors on the other hand. Another consideration was the ease of applying this environmental stress in a prolonged evolutionary experiment. In addition, such general stress provides several possible adaptive solutions that can be acquired (Steil et al. 2003; López et al. 2006; Kohlstedt et al. 2014). The evolution was carried out in four separate evolution regimes, such that the source of DNA added in each regime varied based on phylogeny. While one experiment, that served as a control, was provided with no foreign DNA, the other three were supplemented with genomic DNA from salt-adapted species with different phylogenetic distance to the evolving strain.
By following the evolving populations over >500 generations, at both the molecular and phenotypic levels, we have identified basic evolutionary principles and properties of HGT-mediated adaptation. Most notably, foreign DNA acquisition, although highly restricted by phylogenetic distance, introduced a large amount of physically clustered DNA segments that introduced ample genetic variation to the evolving genomes. These HGT events were also accompanied by spontaneous point mutations. Further competition assays show that some of the repeatedly acquired HGT fragments have facilitated the increase of the populations’ fitness.
Materials and Methods
Strains and media
All experimental evolution experiments were performed using an erythromycin-resistant competent B. subtilis 168 strain in which the comK gene was placed under control of a xylose-induced promoter (Genotype: his met srfA-lacZ [tet] amyE::xylR Pxyl-comK [ery]. comQ comX comP replaced by Bacillus mojavensis RO-H-1 homologs). See Tortosa et al. (2001) for strain details. A total of 11 strains were used as DNA donors in the experimental evolution experiments, list of these strains can be found in Supplemental Material, File S1. Evolution and growth experiments were done in LB medium containing 0.8 M NaCl (10 g/liter tryptone, 5 g/liter yeast extract, and 46.7 g/liter NaCl). Competition experiments were done on either LB medium (10 g/liter tryptone, 5 g/liter yeast extract, and 10 g/liter NaCl) or LB medium containing 0.8 M NaCl.
Calculating the genetic distance between the various donors and the B. subtilis strain 168 recipient
Calculation of the pairwise average nucleotide identity (ANI) between the genome of the recipient strain, B. subtilis 168, and the genomes of each of the organisms used as DNA donors in the experimental evolution experiment was performed with the JSpeciesWS tool (Richter et al. 2016) using MUMmer. Briefly, each genome was split into 1 kb fragments and pairs of donor and recipient genomes were compared using MUMmer (ANIm) (Kurtz et al. 2004). The ANIm values obtained for the Bacilli donors were: RS-D-2: 97.99% for alignment of 91% of the genome; RO-FF-1: 98.47% for alignment of 90.91%; RO-E-2: 92.86% for alignment of 87.11% of the genome; RO-H-1: 87.66% for alignment of 81.79% of the genome. The ANIm values obtained for the bacteria donors were: Halobacillus halophilus: 87.9% for alignment of 1.29% of the genome; Halomonas elongate: 81.8% for alignment of 0.48% of the genome; Aliivibrio fischeri: 82.09% for alignment of 0.53% of the genome. The ANIm values for the Archaea donors were all 0% for alignment of 0% of the genome.
Constructing a phylogenetic tree
Sequence similarities among the studied genomes were calculated using JSpeciesWS tool (Richter et al. 2016) using MUMmer (ANIm). The distance values (taken as 1-similarity) were then used by hclust function of R, using the “average” method, to plot the hierarchical clustering tree.
Evolution of other-Bacillus donors to high salt
Each of the two B. subtilis strains (RO-FF-1 and RS-D-2) as well as Bacillus spizizenii strain RO-E-2 and B. mojavensis strain RO-H-1, was grown in 1.2 ml of LB medium containing 0.8 M NaCl. Cultures were grown at 30° under shaking conditions, and diluted every 24 hr by a factor of 1:120 into fresh medium. This procedure was repeated daily for 72 days. After 72 days, genomic DNA was extracted from each evolved population and used as a DNA source in the main evolution experiment.
Genomic DNA extraction
DNA was extracted from all bacterial strains and species added as donor DNA in the evolution experiment using the Wizard Genomic DNA Purification Kit (cat# A1620; Promega) according to the manufacturer’s guidelines. The protocol was adjusted for purification of large volumes as follows: cultures were grown in 10 ml medium, with the volumes of all reagents and buffers increased by a factor of 10. Incubation time at 80° was increased to 15 min. DNA was extracted from all archaeal species added as donor DNA in the evolution experiment according to the protocol specified in the Halohandbook (Nuttall et al. 2008).
Evolution experiments with and without foreign DNA
All experimental evolution experiments were carried out by serial dilution. Cells were grown in 1.2 ml LB medium containing 0.8 M NaCl and 5 µg/ml erythromycin (cat# E5389; Sigma) at 30° under shaking conditions, and diluted every 24 hr by a factor of 1:120 into fresh medium. This dilution rate amount to roughly seven generations a day according to optical density (OD) growth measurements at the beginning of the experiment (a generation was determined as a duplication in the OD value throughout the growth measurement). This procedure was repeated daily for 72 days (total of 504 generations). For evolution experiments in the presence of foreign DNA, the medium was supplemented with ∼2 μg of foreign DNA mixture dissolved in double-distilled water (DDW; ∼50 μl) which was added daily (see Figure 1 and File S1 for donor details). All mixtures were composed of equal amounts of the various DNA sources. Each experiment was done in three independent repeats (a total of 12 evolutionary lines). Every 42 generations, cell samples were frozen in 30% glycerol and kept at −80°.
Figure 1.
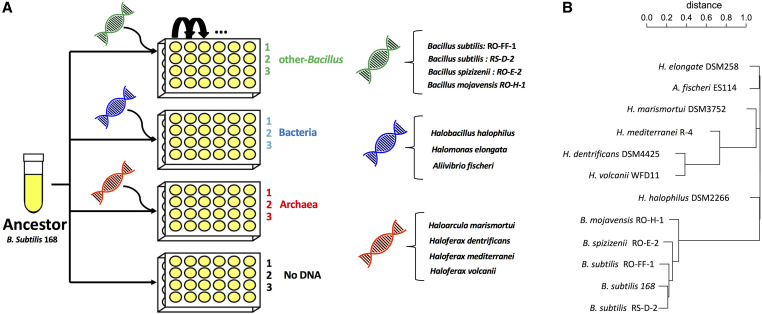
Experimental evolution setup and donor phylogeny. (A) The experimental design consisted of four evolution experiments in which the same ancestral competent Bacillus subtilis 168 strain was evolved with or without different sources of genomic DNA in the growth medium. In the other-Bacillus experiment (green), a mixture of genomic DNA from four closely related Bacillus donors was added (B. subtilis strains: RO-FF-1, RS-D-2, Bacillus spizizenii strain RO-E-2, and Bacillus mojavensis strain RO-H-1). In the Bacteria experiment (blue), the added mixture of DNA originated from more distant bacterial donors—members of a different family (Halobacillus halophilus) or phylum (Halomonas elongate and Aliivibrio fischeri). In the Archaea experiment (red), a mixture of DNA from extremely distant donors, belonging to a different kingdom, was used (Haloarcula marismortui, Haloferax dentrificans, Haloferax mediterranei, and Haloferax volcanii). In the No-DNA experiment, cells were evolved without the addition of genomic DNA into the growth medium. Each experiment was done as three independent biological repeats. Cells were evolved in high salt LB medium (0.8 M NaCl) by a daily serial dilution (1:120) procedure (see Materials and Methods for details). (B) A phylogenetic tree of all the donor strains as well as the recipient strain, calculated based on ANIm between all the genomes. Note that due to arbitrary left-right ordering of branches in the dendrogram drawing, some phylogenetically remote species appear adjacent on this tree.
Liquid growth measurements
Samples taken from frozen glycerol stocks were inoculated into 3 ml of LB medium and grown for 48 hr at 30°. Cells were diluted 1:100 into LB medium containing 0.8 M NaCl. Qualitative growth comparisons were performed using 96-well plates in which strains were divided on the plate in a checkerboard manner to cancel out positional effects. Cultures were grown at 30° under shaking conditions using a robotic system (Hamilton); OD600 measurements (using infinite 500 Tecan) were taken during growth at 60 min intervals for ∼24 hr. For each strain, a growth curve was obtained by averaging over 21 wells. Each plate of evolved populations included all three evolution populations from the same evolutionary regime (i.e., either other-Bacillus, Bacteria, Archaea or No-DNA), as well as the ancestor to account for minor differences between plates. Growth parameters were extracted from the growth curves using the curveball algorithm (Ram et al. 2019).
Whole-genome DNA sequencing
Whole genome sequencing was performed on the following DNA samples: The Ancestor (single colony), all 12 evolved populations at generation 504, the 3 other-Bacillus populations at 12 additional time points throughout the evolution time course, and 12 random single colonies of the other-Bacillus population three at generation 504. DNA sequencing was performed as previously described (Blecher-Gonen et al. 2013) with some modifications. For the detailed description of the modifications see File S1.
Assembly of donor genomes
For the four other-Bacillus donors with no sequenced genome (RO-FF-1, RS-D-2, RO-E-2, and RO-H-1) whole-genome DNA sequencing was done as described above. The reads obtained were used for genome assembly using SPAdes (v3.6.2) with the “–careful” option (Bankevich et al. 2012). The resulting contigs were filtered by length >200 and coverage >10.
Bioinformatics pipeline for identification of foreign DNA fragments and mutations
In order to define HGT-acquired regions and identify point mutations and insertions and deletions (indels) in each evolved population, we carried out two analyses in parallel. The first analysis included variant calling against the reference genome of B. subtilis 168. The second analysis was based on mapping the reads from each evolved population to a reference sequence, containing all the relevant donors of each evolutionary regime and the reference genome of B. subtilis 168. The final classification of genomic events was based on the integration of the results obtained by the two methods. The two analyses and integration process as well as the accession numbers of all the donor genomes are detailed in File S1.
Analyzing genomic proximity between HGT fragments and between HGT fragments and mutations
In order to test whether HGT events detected in the evolved populations are clustered around the chromosome, a randomization-based statistical test was carried out in which the proportion of pairwise distances between fragments that were shorter than a set threshold was compared to the proportion of such distances when the locations are randomized. A downstream analysis included the randomization of the clone identities to which each fragment belonged. A similar analysis was carried out to test the clustering between HGT events and de novo mutations. See Figure S5 for an illustration of the method and File S1 for a full description of the analysis.
Competition-based fitness measurement
A 5.6 kb fragment comprising the overlapping region of the foreign DNA fragment identified both in other-Bacillus population two and three and partly in population one was transformed to B. subtilis 168 competent cells. All resulting colonies (∼4100 colonies) were scraped and pooled together. This pool served as generation zero of the competition experiment. The competition was carried out by serial dilution with both standard LB medium (three replicates) and 0.8 M NaCl LB (six replicates) for 70 generations. To calculate the fitness of a foreign-DNA-containing cell, samples taken at generations 0, 42, and 70 of the competition experiment were sequenced and the fraction of donor variants in each time point was determined. For full description of the assay see File S1.
Performing GO enrichment analysis
For gene enrichment assays, the gene ontology server was used (Ashburner et al. 2000; The Gene Ontology Consortium 2019) (10.5281/zenodo.3873405 Released 2020-06-01). The PANTHER overrepresentation test (Mi et al. 2019) was performed with the B. subtilis reference list. A Fisher’s exact test was performed with a false discovery rate correction. For genes affected by de novo mutations, the test was performed on the GO cellular component complete data set. For the gene enrichment test on the foreign DNA fragments acquired, both GO molecular function and GO biological process data sets were used.
Determining the percentage of competent cells in the population
Cells from samples at different time points of the evolution experiment were diluted as described in Evolution experiment with and without foreign DNA, and were supplemented with genomic DNA extracted from B. subtilis strain 168 containing two genes conferring resistance to one of two antibiotics, Phleomycin or Chloramphenicol (kindly provided by Avidgdor Eldar, Tel Aviv University). After overnight growth, cells were plated on the appropriate selective plates as well as nonselective plates, and the percent of competent cells was estimated from the ratio between the number of colonies in each plate. For full description of the method, see File S1.
Data availability
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article. The genome assembly of the donor Bacillus strains has been deposited at DDBJ/ENA/GenBank, under the BioProject accession number PRJNA650388. See File S1 for the individual accession numbers. The populations’ raw sequencing data supporting the findings of this study have been deposited at the SRA database under the following SRA accession: PRJNA655387. The supplemental tables and figures have been deposited via the GSA figshare portal. File S1 contains the supplemental Materials and Methods. File S2 contains all the supplemental figures (Figures S1 – S6), Table S1 contains statistics derived from the growth assays performed on the evolution regimes, Table S2 contains a full list of genetic variations detected in the other-Bacillus evolutionary regime in the different time points, Table S3 contains a full list of all de novo mutations detected in all evolution lines in the last time point, Table S4 contains a list of HGT fragments and mutations detected in single colonies sequenced in the last time point. Supplemental material available at figshare: https://doi.org/10.25386/genetics.12800933.
Results
Experimental set-up of evolution through HGT
Our experimental evolution assay was performed on a competent B. subtilis 168 strain in the presence or absence of genomic DNA from other microorganisms. The particular strain used in our evolution experiment is the B. subtilis 168 laboratory strain, genetically modified to include an additional copy of the competence master regulator comK under a leaky xylose promoter (see Materials and Methods for full description of the strain), enabling constitutive competence and transformability (Hahn et al. 1996; Tortosa et al. 2001).
We carried out four parallel serial dilution experiments, in three of which we added foreign DNA into the growth media of the evolving cells daily, thus allowing these cells to utilize both mutations and foreign DNA as a means to generate variation, and one experiment in which cells evolved without foreign DNA added to their medium. The donors added to each of the three experiments represent species of increasing phylogenetic distance from B. subtilis (Figure 1). In the first experiment, the foreign DNA originated from two B. subtilis strains, as well as from the closely related B. spizizenii and B. mojavensis species (spanning an average nucleotide identity (ANI) of 87–99% to the recipient cells for 83–90% of the genomes, see Materials and Methods for sequence similarity calculation). Since we used high salt concentration as a selection pressure, a condition to which the Bacillus class are not adapted, we grew the donors to the same condition for 500 generations before their DNA was extracted and given to the recipient B. subtilis (hereinafter other-Bacillus). Although natural transformation possessed by B. subtilis was shown not to allow integration of DNA from distant species due to its dependence on homologous recombination (Carrasco et al. 2016), we initiated two other explorative evolutionary regimes in order to allow the detection of potential events of HGT from phylogenetically very remote species that are well adapted to salt. In one regime, we used genomic DNA from two halophilic bacteria and the marine halotolerant Aliivibrio fischeri (hereinafter Bacteria). In the other regime, we added DNA from Archaea belonging to two halophilic genera: Haloferax and Haloarcula (hereinafter Archaea). The phylogenetic relations between all donors, as well as the recipient B. subtilis strain 168 are shown in Figure 1B. In a fourth, control experiment, no foreign DNA was added to the growth medium of the evolving cells (hereinafter No-DNA). Each experiment was repeated in triplicate, amounting altogether to 12 independent evolution lines, all originating from the same ancestral B. subtilis strain. All evolution experiments were carried out for 504 generations via daily serial dilution.
Growth of evolved lines was manifested in maximal OD improvement, which was highest for populations that evolved with “other-Bacillus” donor DNA
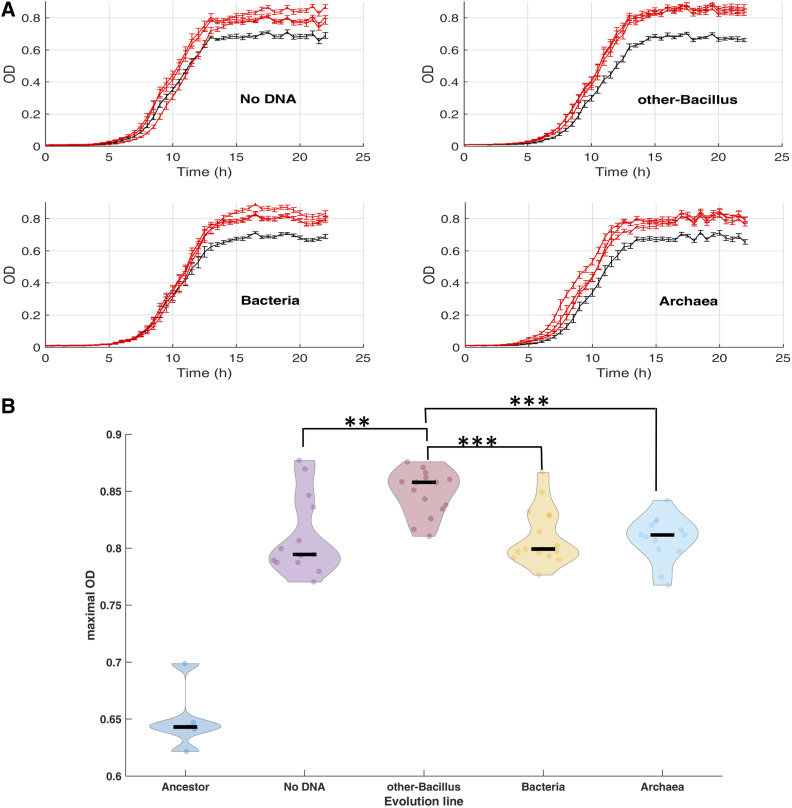
Following 504 generations of evolution, the extent of adaptation of each of the 12 evolving populations was determined by the measurement of their OD under the same conditions applied during the evolution stage. We conducted several independent growth assays in which we compared the OD of the same ancestral strain to each of the 12 evolutionary populations. Each three lines evolving in the presence of the same DNA donor (or without DNA) and ancestor were grown in the same plate (see Materials and Methods). Figure 2A shows one such experiment. By extracting and comparing the growth parameters of the evolved lines with that of the ancestor, we found that OD improvement in all 12 lines was manifested mainly by a 20–30% higher yield (quantified as the maximal OD) at the end of the growth cycle, when the culture enters the stationary phase (Figure 2A). The B. subtilis populations with the statistically significant higher yield values were those that grew in the presence of other-Bacillus donor DNA (Figure 2B shows the extracted yield values across all growth assays replicates). While the average mean maximal OD of the three lines of that evolutionary regime was 0.85 (with 0.02 SD), the yield of the other three evolutionary regimes was significantly lower and had an identical mean of 0.81 (see Figure S1 and Table S1 for the growth parameters statistics summary). The change in yield was the only growth parameter out of three examined that showed statistically significant improvement in the evolved lines compared to the ancestor. The two other parameters of the growth curve, the lag time and maximal growth rate, were not found to increase significantly during the evolution of the different populations (Figure S2, A and B, respectively, and Table S1). These results are consistent with either neutral or adaptive contributions of horizontally transferred fragments.
Figure 2.
Growth measurements of the evolved lines. (A) Growth measurements of each of the evolution lines (red) compared to the ancestor (gray). Following 504 generations of evolution, the growth of each population, as well as the ancestor, on high salinity media was measured. Each subplot presents the change in optical density (OD) as a function of time. For simplicity, only one growth experiment out of five biological replicates is shown. (B) Maximal OD values (y axis) were extracted from several growth experiments using the curveball algorithm (Ram et al. 2019). The violin plots present the distribution of curveball-derived maximal OD values for the ancestor as well as each of the four experiment types (grouping together all three evolutionary lines, each measured in three to five independent growth experiments). Black lines indicate median values (median, mean, and SD values can be found in Table S1). Statistically significant pairwise comparisons (rank sum tests) between other-Bacillus populations and each of the three additional populations is shown. **P-value < 0.01 ***P-value < 0.001. Comparisons between all other pairs of evolutionary populations were found to be not significant.
Foreign DNA was acquired extensively and exclusively from closely related Bacillus donors
Sequencing the genome of all 12 populations at the end of the evolutionary process and comparing the number of mismatches and indels detected (at a frequency of 10% of the population, or above) revealed a striking difference between the four experiments. While in the Bacteria, Archaea and No-DNA populations, a handful of substitutions were detected (ranging from 2 to 18 per population), those in their other-Bacillus counterparts was in the order of hundreds to thousands of nucleotides in each evolving line (Table 1). This observation strongly suggests that B. subtilis integrated donor Bacillus DNA segments.
Table 1. All 12 populations acquired mutations while foreign DNA acquisition is restricted to those evolved in the presence of closely related other-Bacillus donors.
| DNA donor | Biological repetition | # of mismatches | # of insertions | # of deletions | # of foreign DNA fragments | # of mismatches not in foreign fragments | # of insertions not in foreign fragments | # of deletions not in foreign fragments |
|---|---|---|---|---|---|---|---|---|
| No DNA | 1 | 9 | 0 | 0 | 0 | 9 | 0 | 0 |
| 2 | 2 | 0 | 0 | 0 | 2 | 0 | 0 | |
| 3 | 4 | 0 | 1 | 0 | 4 | 0 | 1 | |
| other-Bacillus | 1 | 246 | 3 | 1 | 1 | 0 | 0 | 1 |
| 2 | 103 | 0 | 1 | 1 | 2 | 0 | 0 | |
| 3 | 5585 | 39 | 44 | 36 | 12 | 0 | 1 | |
| Bacteria | 1 | 2 | 1 | 0 | 0 | 2 | 1 | 0 |
| 2 | 6 | 0 | 2 | 0 | 6 | 0 | 2 | |
| 3 | 10 | 1 | 0 | 0 | 10 | 1 | 0 | |
| Archaea | 1 | 6 | 0 | 0 | 0 | 6 | 0 | 0 |
| 2 | 17 | 1 | 0 | 0 | 17 | 1 | 0 | |
| 3 | 7 | 1 | 1 | 0 | 7 | 1 | 1 |
Deep sequencing results of all 12 evolved populations (indicated in the DNA donor and biological repetition columns) and the ancestor following 504 generations of evolution. The number of single-nucleotide mismatches and small insertions and deletions detected in each population (compared to the ancestor) is shown on the third to fifth columns respectively. The number of foreign DNA fragments detected in each population is shown in the sixth column. The seventh to ninth columns depict the number of single-nucleotide mismatches and small insertions and deletions that cannot be ascribed to a foreign DNA fragment, thus likely to result from mutations (for details on bioinformatic analysis see Material and Methods).
In order to identify such fragments and the potential donors, we developed a bioinformatics pipeline (see Materials and Methods) that can detect events of foreign DNA integration based on deep sequencing results. It should be noted that this analysis likely underestimated the actual extent and length of the transferred regions, since our ability to detect transfer events is limited to sites with sufficient mismatches between donor and recipient genomes. It is entirely possible that donor segments were transferred and replaced identical or near-identical counterparts, leaving no detectable mark on the genome.
Our bioinformatics results indicated that the three other-Bacillus populations acquired extensive foreign genetic material, all of which came from three out of the four Bacilli donors: B. subtilis strains RO-FF-1, RS-D-2, and B. spizizenii strain RO-E-2, which exhibit a respective average nucleotide identity with the recipient B. subtilis strain 168 of 98.47% (over 90.91% of the genome), 97.99% (for alignment of 91% of the genome), and 92.86% (for alignment of 87.11% of the genome). The fourth Bacillus donor belonging to a more distant species (See Figure 1B): B. mojavensis RO-H-1 strain, which showed 87.6% genetic similarity (for alignment of 81% of the genome), was not found to contribute any DNA, indicating that this extent of genetic similarity was no longer sufficient for successful transformation or propagation of fragments in this setting. This result is consistent with a previous study showing sexual isolation between the RO-H-1 strain and our recipient strain (Cohan et al. 1991). Among Bacillus donors that did contribute DNA (B. subtilis and B. spizizenii strains), we did not see a correlation between phylogenetic distance and the number or length of foreign DNA fragments contributed. The most phylogenetically remote donor out of the three (see Figure 1B)—the RO-E-2 donor—contributed 12 fragments overall, while the closer donor (based on percent identity)—RO-FF1—contributed only four fragments. The RS-D-2 donor contributed the highest number of fragments (14), but actually introduced fewer positions of variability overall than RO-E-2 donor (all the information can be derived from Table S2). The natural competence acquisition mechanism, which is strongly dependent on homologous recombination, facilitates the integration of foreign DNA, provided that the donor fragment resides within homologous ends (Majewski and Cohan 1999; de Vries and Wackernagel 2002). The efficiency of this process was shown to decrease log-linearly with increased phylogenetic distance (Carrasco et al. 2016), as well as the size of the integrated fragments (Zawadzki and Cohan 1995). This dependency explains the lack of contribution of DNA from phylogenetically remote donors. However, the fact that we did not see a correlation among the contributing Bacillus, between phylogenetic distance, and number of horizontally acquired fragments, can be due to the relatively small number of events detected in our experiment, or can be a result of differential selection operating on retention of HGT events that may have occurred in our experiment. It is also possible that short transformation events, which did not introduce any substitutions, went undetected, and, hence, a bias was created, as these events are more likely to come from donors with higher sequence similarity. As to be expected from the homology requirement of DNA integration, we determined that all HGT fragments detected in our experiment were integrated at the homologous genomic loci, with no ectopic integration observed. The size of the other-Bacillus integrated fragments ranged from a few-hundred bases to almost 20 kb, with a mean, median, and SD of 5.66, 4.32, and 5.2, respectively (Figure S3), this size range is in agreement with previous reports of natural transformation (Kulick et al. 2008; Mell et al. 2014). Consistent with the low sequence similarity, no foreign DNA fragments were detected in any of the Bacteria, Archaea, and No-DNA populations (Table 1). As such, most of the subsequent analyses on HGT fragments, described below, were performed on the said three other-Bacillus populations.
HGT events introduced mainly synonymous substitutions, whereas mutations were mostly nonsynonymous
Analysis of the set of nucleotide mismatches, and indels observed in the other-Bacillus populations at the final time point of the evolution experiment revealed that some of the mismatches cannot be ascribed to any of the acquired foreign DNA fragments. Thus, these genetic alterations were likely the outcome of spontaneous mutations. Table 1 specifies the number of point mutations and small insertion and deletions detected in each of the 12 evolved populations. In total, we identified 87 mutations that reached a frequency of at least 10% at generation 504 (77 point mutations, four insertions and six deletions, from now on will be considered together in our analysis). Out of these, 83% were located within coding regions, with the majority of them nonsynonymous substitutions or indels that likely affected the protein sequence (Table 2). Such a high rate of nonsynonymous mutations stands in sharp contrast to the nature of the genetic variations acquired by foreign DNA fragments, as the latter were dominated by synonymous substitutions (Table 2). Furthermore, some genes were repeatedly affected by mutations in the various populations. For example, mutations in the coding region of the arginine utilization transcriptional regulator -rocR gene appeared in 7 out of the 12 populations (in some cases, two different mutations were located within this gene in a single population). In the fliI coding region, coding for a flagellar-specific ATPase, the same mutation appeared in seven populations (see Table S3 for the full list of mutations and indels).
Table 2. Nature of genetic variation identified at generation 504 in all 12 populations.
| “No-DNA,” “Bacteria,” “Archaea” | “other-Bacillus” | ||
|---|---|---|---|
| Mutations | Mutations | HGT | |
| # of mismatches and indels | 71 | 16 | 6,006 |
| % of mismatches and indels in CDS | 82% | 87% | 92% |
| % of nonsynonymous substitutions or indels | 96% | 64% | 20% |
| % of substitutions in flagella CDS | 28% | 36% | 0% |
| % of flagella mutations that are nonsynonymous | 100% | 100% | — |
Table showing the summed number of mismatches and indels identified in the three other-Bacillus populations and that of the nine other populations (No-DNA, Bacteria, and Archaea) at generation 504. In the other-Bacillus populations, HGT-acquired fragments and mutations are considered separately. The percentage of mismatches and indels located in coding regions is calculated out of the total number of mismatches and indels observed. Out of those, the percentage of mismatches resulting in nonsynonymous substitution and indels (that are likely to change the protein sequence) is shown. Also presented is the percentage of mismatches and indels affecting the coding sequence of flagellar proteins and the percentage of nonsynonymous changes for this group of mismatches and indels.
Experimental evidence for positive selection on a chosen HGT fragment
To characterize the dynamics of the different genomic alterations that occurred within the three other-Bacillus populations, and to follow changes in population structures, we deep-sequenced the genome of the evolving other-Bacillus populations at multiple time points during the evolution. We then applied our HGT detection and characterization pipeline and identified all the HGT fragments, as well as mutations, acquired by the three other-Bacillus populations, along their respective evolutionary time course. Given the entire time course, we could merge proximal fragments that appeared as separate HGT events provided that they resided in close proximity on the genome (up to 1 kb), that they showed similar dynamics of change in population frequency throughout the evolutionary time course, and that they originated from the same donor. Using this refined analysis procedure, and employing a cut-off that admits only fragments that reached a frequency of at least 10% in the population at one time point at least, we detected four foreign DNA fragments in population 1, a single fragment in population 2, and 25 fragments in population 3 (Table S2). We further examined the genomic location of both mutations and the HGT fragments inserted in all three other-Bacillus populations (Figure 3).
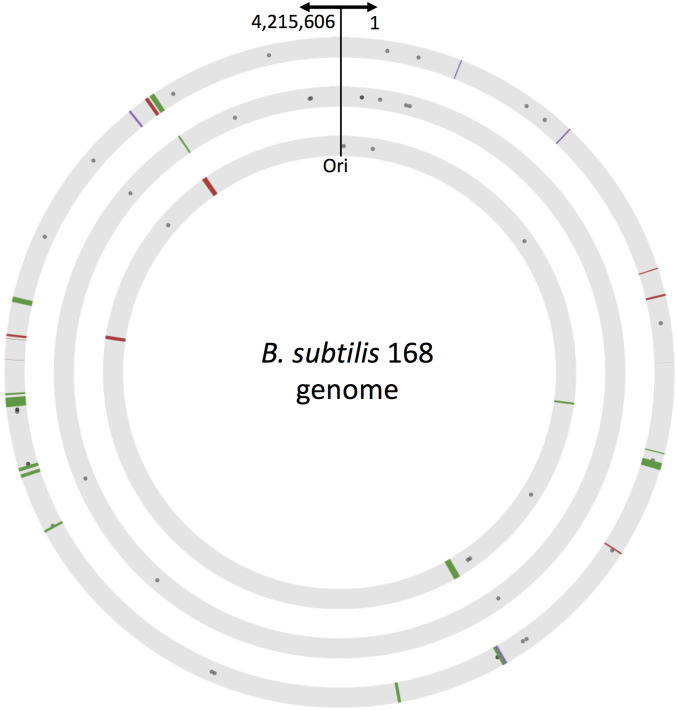
Figure 3.
Genomic location and distance distribution of mutations and HGT fragments. Graphic representation of the genomic location of mutations and HGT fragments identified in other-Bacillus populations 1 (inner circle), 2 (middle circle), and 3 (outer circle). Circles represent the genome of the recipient B. subtilis 168 (keeping the same scale); arrows indicate the location of the origin of replication and coordinates. The locations of all genomic alterations identified in each of the three other-Bacillus populations are marked to scale. Gray dots mark mutations, lines mark HGT-acquired fragments (red: B. spizizenii strain RO-E-2, green: B. subtilis strain RS-D-2 and purple: B. subtilis strain RO-FF-1). The width of each line is proportional to the size of the inserted fragment.
Interestingly, the one fragment identified in population 2, reached a high frequency at the final time point, and overlapped in location with fragments identified in populations 1 and 3 (Figure 3 and Table S2). The high frequency reached by this fragment in population 2, and the reoccurrence of this fragment in all populations, strongly suggests an adaptive value to this donor region. Given the large bottleneck employed in our experiment, of ∼106 cells, and the fact that the same fragment (or parts of it) reached considerable frequencies in all three independent populations, it is unlikely that the fragment’s rise in frequency is due to genetic drift, but rather due to some adaptive value given by the fragment itself, or by its linkage to another genetic event.
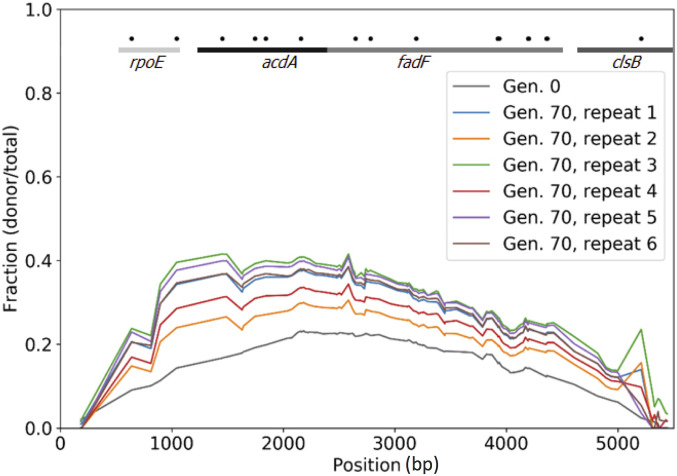
The straightforward approach to determine the adaptive value of a specific genetic change observed in an evolved population is to reintroduce it into the ancestral background and examine its effect on fitness in the absence of other genetic alterations. We used this approach to evaluate this ∼5 kb foreign DNA fragment that was integrated independently in other-Bacillus populations 2 and 3, and, in population 1 a shorter segment of this fragment was inserted. This fragment contained 115 nucleotide mismatches compared to the homologous region in the recipient B. subtilis 168 strain. We amplified this fragment by PCR from the donor strain, and transformed it into the ancestral strain, thus breaking the linkage between this fragment and all other genetic alterations occurred during the evolution. We pooled hundreds of transformed cells and subjected them to population-level sequencing (Figure 4, “Gen. 0”), which revealed that the fragment was integrated with different efficiencies across the region (including cells that did not received any foreign DNA). We competed the pooled cells for 70 generations either under high salinity conditions identical to those used in the main evolution experiment (six independent replicates) or under the standard salt concentration (three independent replicates). We then deep sequenced the 5 kb region among the competing cells and followed the change in frequency of the different mismatches in the population over competition time. We found that, at the beginning of the competition, the frequency of the different mismatches along the fragment ranged between 10 and 20%, a baseline value that simply represents the efficiency of the transformation (Figure 4, “Gen. 0”). As can be seen in Figure 4, the cells harboring the horizontally acquired fragment increased in frequency as evident by the increase of the frequency of the donor variants (Figure 4 “Gen. 70”) in all six replicates. This result is a clear indication that this fragment indeed conferred a fitness advantage. In order to assess statistical confidence in the observation, we have calculated a 95% confidence interval (CI) for the difference of proportions of donor reads between the ancestor and that competition replicate that showed the smallest increase in frequency out of the six replicates (replicate 2). We repeated the CI calculation at each SNP. The CI for all SNPs with a sufficient coverage (a condition met by 92% of the SNPs) was well above zero, indicating that, with 95% confidence, the donor proportions in the two populations are different. Note that the other five replicates of the competition experiment yielded even higher increase in donor variants, providing an even stronger indication that the increase in the frequency of cells that acquired the HGT fragment is due to an adaptive benefit rather than drift. An advantage of this type of competition is that it can be used to pinpoint regions along the fragment that are most likely to contain site(s) that contribute positively to fitness. We indeed identified such a segment, a region of ∼1 kb at the 5′ end of the 5 kb fragment, which overlapped two genes, rpoE [a nonessential subunit of RNA polymerase (Weiss and Shaw 2015)] and acdA [acyl-CoA dehydrogenase gene involved in fatty acid β-oxidation (Matsuoka et al. 2007)]. Interestingly, when competing the pool in LB with standard salt concentration (0.17 M), we observed an even higher increase in donor fragment frequency during the competition, with some subregions of the fragment reaching a frequency of 60% (Figure S4). These findings indicate that the donor DNA fragment provided a general fitness advantage under our experimental setting, rather than a specific adaptation to the high salt concentration.
Figure 4.
Acquisition of an HGT fragment provides a fitness advantage under similar conditions to those used for the experimental evolution experiment. Results of six competition assay replicates (70 generations) of a library of transformed B. subtilis 168 competent cells, containing different or no regions of a 5.6 kb DNA fragment from B. subtilis strain RS-D-2. The graph portrays the frequency of each mismatch along the fragment before and after the competition as calculated based on deep sequencing. The frequency of each mismatch at generation 0 (immediately after transformation) and in generation 70 is presented on the y-axis. The mismatch location along the fragment is presented on the x-axis. The lines and gene names at the top of the figure denote the genes located in the corresponding regions in the B. subtilis 168 genome. Black dots denote mismatches that result in nonsynonymous substitutions.
Reoccurring mutations in the flagellum suggest parallel evolution across all regimes
While the above analysis clearly shows that the acquisition of foreign DNA provided fitness advantage under our experimental settings, it does not exclude the possibility that some of the adaptation that occurred in the other-Bacillus populations is due to putative point mutations that may have occurred concomitantly with, but separately from, the HGT. Under such a scenario, some HGT fragments were neutral and rose in frequency simply as “hitchhikers.” Our above-noted finding that all 12 populations—those that acquired DNA through HGT and those that did not—increased their maximal yield compared to the ancestor (although the latter exhibited smaller increase) lends support to this alternative notion. Interestingly, exploring the identity of genes affected by mutations in all 12 populations revealed that ∼30% of the point mutations affected genes coding for flagellar proteins and that all of them resulted in a nonsynonymous substitution (Table S3 and Table 2). A gene ontology (GO) enrichment analysis performed on the genes affected by de novo mutations in all evolution lines indeed revealed an enrichment for the “Flagellum basal body cellular component” (FDR P value = 1.34*E−2), see Materials and Methods for details. Interestingly, none of the foreign DNA fragments integrated included flagellar genes (Table 2), ruling out the possibility that such genes were recombination hotspots. Furthermore, the detected mutations affected many fllagelar genes belonging in some cases to separate operons located in distant genomic locations. Thus, these mutations are unlikely to have occurred due to mutational hotspots. The reoccurrence of mutations in flagellar genes suggests the possibility that mutating the flagella is likely to be beneficial under our experimental setting. In agreement with that, mutations in flagellar proteins were detected in the past in various experimental evolution experiments in B. subtilis (Brown et al. 2011; Waters et al. 2015), as well as in E. coli and Salmonella enterica (Knöppel et al. 2018). These observations suggest a direct fitness advantage to some of the point mutations, sustaining two alternative possibilities of adaptive and neutral effect of some of the HGT fragments.
Foreign DNA integration events occurred in bursts, allowing simultaneous integration and propagation of several fragments
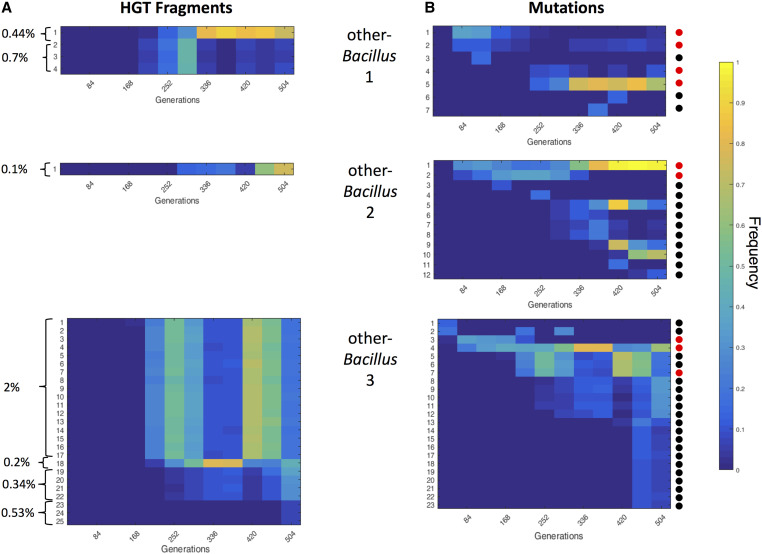
The frequency of each observed HGT fragment in the three other-Bacillus populations, at different time points throughout the time-course of the evolution is shown in Figure 5A. We defined “frequency group” as a set of HGT fragments that show a similar pattern of population frequency across the time course. We found in population 1, HGT fragments belonging to two frequency groups, one consisting of one fragment (replacing 0.44% of the genome) and the other consisting of three different fragments (replacing together 0.7% of the genome). In population 2, only one fragment was observed (replacing 0.1% of the genome), whereas, in population 3, the detected fragments correspond to four different frequency groups, the largest of which comprising 17 different foreign DNA fragments (replacing together 2% of the genome). This observation suggests that the 25 DNA fragments from population 3 resided in four competing clones. In support of this conclusion, sequencing the genomes of 12 single colonies randomly picked from population 3 at generation 504 revealed that each typically contained exclusively all the DNA fragments belonging to a single frequency group, but none of those belonging to any of the other frequency groups (except for two fragments that were either shared between two frequency groups or appeared in a different frequency group, see Table S4). The most parsimonious explanation for shared frequencies between fragments, given the population size and the daily dilution, is that the fragments that belong to the same frequency group were acquired horizontally by the same single cell, and were then spread in the population as one cohort of fragments (defined below as a clone). Such bursts thus resulted in the massive acquisition of genetic material in the receiving cells, which, in the extreme case (Figure 5A, population other-Bacillus 3, upper clone), resulted in the exchange of ∼86,000 nucleotides, corresponding to ∼2% of the genetic material of the cell. We conducted a GO enrichment analysis on fragments co-integrated in the same clone, to investigate the possibility that cotransformed fragments share similar functional properties; however, we found no statistical enrichment in any of the clones (no GO Biological Process or Molecular Function was found to be enriched, see Materials and Methods for the test description).
Figure 5.
Both HGT fragments and mutations show dynamic changes in frequency over time. The frequency of each HGT fragment (A) and mismatch (or indel) nucleotides (B), at different generations along the evolutionary process in other-Bacillus populations 1–3 (shown from top to bottom). The color bar indicates the frequency of each genomic event in the population. (A) Each fragment is labeled by a number and is presented as a separate line. Fragments are ordered based on time of detection; brackets mark fragments with a similar frequency pattern (i.e., belonging to the same clone). Only the fragments that reached a frequency of at least 10% in at least one of the generations examined are presented. The number to the left of each bracket indicates the overall percentage of genetic material replaced by all the fragments belonging to the same clone (frequency group). (B) Mismatches and indels are numbered and ordered based on time of detection. Only mismatches and indels that reached a frequency of at least 10% in at least one of the generations examined are presented. Colored dots to the right indicate whether the respective genetic alteration affects a gene coding for a flagellar protein (red) or is not located in the coding region of a flagellar protein (black).
As in the case of the HGT fragments, time-course sequencing of the three other-Bacillus populations, which identified all mutations and indels in each linage (i.e., frequency 10% or higher in at least one time point) (Figure 5B), revealed that many of the mutations could be divided into frequency groups, suggesting that they resided within the same clone. Comparing the HGT and mutation population frequency dynamics revealed that some of the mutations and indels clones largely resembled the HGT clones in dynamics, suggesting that both mutations and foreign DNA resided within the same genetic background. This suggestion was further supported by sequencing the genome of single colonies (Table S4). We also found that, typically, the alterations that could be detected earlier during the evolutionary time course were mutations and indels, while HGT fragments could be detected only later during the evolutionary process. Furthermore, the change in frequency of mutations and HGT fragments over time further attests for a fitness advantage of some of these genetic changes.
HGT fragments tended to occur in genomic hotspots
Following the genomic location of both mutations and HGT fragments in the other-Bacillus populations (Figure 3), and, given the partition of the populations to competing clones (Figure 5), we could further assign each of the genomic events that occurred in the three populations to a clone. Some clones were comprised of mutation only, others of HGT events only, and yet others comprised a combination of both types of events. We turned to investigate whether HGT fragments and point mutations were clustered next to one another around the chromosome, and, if so, whether clustering around the chromosome was a result of events that took place in the same clone (and hence, probably, integrated in the same original cell) or were they resulted from repeated independent integration events. We computed pairwise distances between HGT fragments, and assessed their tendency to be shorter relative to a null expectation, which was derived from a randomization test, using three different distance thresholds (see Materials and Methods and Figure S5 for schematic representation of the pipeline). This analysis yielded significant P-values for all three distance thresholds assessed (1, 0.5, and 0.25% of half genome size corresponding to ∼20, ∼10, and 5 kb, respectively), suggesting that the HGT fragments were clustered in physical proximity around the genome. We decided to continue with the threshold of 0.25% (5 kb), as it minimized the P-value (P- value <0.001), and hence better represented the characteristic clustering distance between HGT fragments. To investigate the temporal dimension (i.e., tendency of HGT events to co-occur simultaneously within the same clone), we randomly shuffled the clone identities of the HGT fragments 1000 times and asked whether the proportion of short distances (<5 kb) between HGT fragments within the same clone are significantly higher than when clone identities are randomized. The P-value obtained was not significant (P-value = 0.753), and, hence, the observed clustering cannot be attributed to events that took place simultaneously. Taken together, our findings suggest that the tendency of fragments to integrate at proximal locations was not dependent on their co-occurrence in the same cell, but rather indicates the existence of genomic regions that serve as recombination “hotspots,” i.e., regions with a higher tendency to integrate DNA.
We did a parallel pairwise distance computation, this time between HGT fragments and point mutations, which revealed a tendency for short distances between events occurring within the same clone (Figure S6A). Such a tendency is not observed when computing the distances between HGT and mutations that occurred in different clones (Figure S6B). In order to quantify this observation, and evaluate its statistical significance, we followed the same procedure as described above (using the same 0.25% threshold). We found that, similar to the case of HGT events, HGT fragments and mutations also clustered on the chromosome (P-value < 0.001). However, unlike the case of HGT–HGT distances, mutations–HGT distances are short when considering events that happened simultaneously at the same clone (P-value < 0.001, compared to a null model obtained by randomizing the clone identity of mutations). The observed close proximity between mutations and HGT fragments that occurred within the same cell (i.e., simultaneously in time) may suggests that acquisition of foreign DNA is an event that exerts a mutagenic effect in the genomic vicinity (at least up to 5 kb). However, although highly statistically significant, the clustering within the same clone between HGT fragments and mutations was based on a few data points. In addition, we cannot exclude the possibility that, in some cases, these apparent substitutions in fact result from very small HGT events.
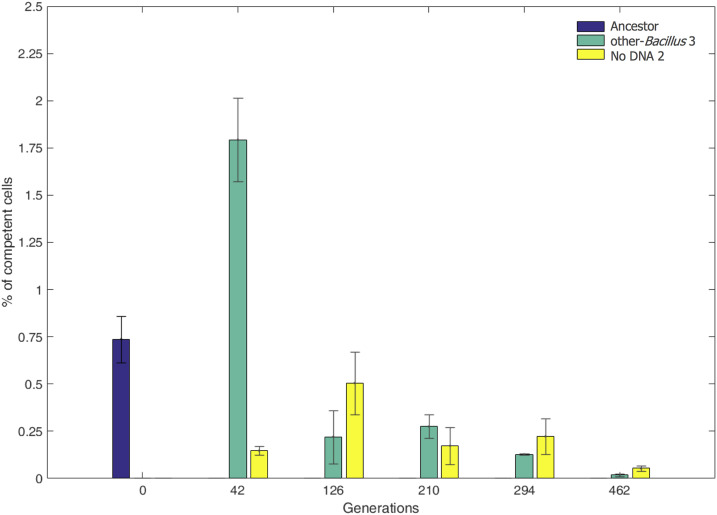
The competence of evolved populations decreased throughout the evolution experiment
While evolution through HGT depends on competence, the level of competence (e.g., percentage of cells that are competent) itself could evolve and change too. We thus estimated the percentage of competence in our system at different time points during the evolution. We did so by measuring the competence of the ancestor strain, other-Bacillus population 3, and No-DNA population 2. Competence was measured by transforming each population with genomic DNA of a B. subtilis 168 strain containing an antibiotic-resistance gene, followed by plating the transformed cells on plates with the corresponding drug. Using this analysis, we estimated the competent cells in the ancestor population to be ∼0.75% (see Materials and Methods for competence level calculation). When performing the competence assay on the evolved populations obtained from different generations during the evolution, we observed a decline of up to 50-fold in competence levels (Figure 6). The decline in competence was observed both for the No-DNA regime and for the other-Bacillus regime, implying the decline is not dependent on acquisition of HGT fragments or presence of genomic DNA in the medium.
Figure 6.
Decline in competence level during evolution. The percent of competent cells (y-axis) in evolved other-Bacillus population 3 (green) and evolved No-DNA population 2 (yellow) is shown at different time points (x-axis) throughout the evolution experiment, as well as in the ancestral population (blue). Presented are averaged values of three biological replicates; error bars correspond to the SD from the mean.
Discussion
In this work, we assessed the potential of HGT of donor DNA from various phylogenetic sources, its characteristics, and relative contribution compared to mutations to the adaptation process of B. subtilis. At the phenotypic level, we observed an 7% higher maximal OD for the three populations that acquired foreign DNA fragments than those that did not (Figure 2B). We further demonstrated on one re-occurring fragment that its acquisition was adaptive, suggesting that HGT can indeed accelerate evolution. On the other hand, the acquisition of foreign DNA did not give rise to detectable improvement of growth rate, suggesting potential neutral effects. The recipient B. subtilis accepted DNA only from other Bacillus donors, as can be expected given the homology constraints of B. subtilis natural transformation (Carrasco et al. 2016), and the previously reported general positive correlation between genetic similarity and HGT frequency (Popa et al. 2017).
Fragment acquisition appears to occur in bursts, in which multiple segments are acquired by the same cell. As can be seen in Figure 5, integrated donor fragments propagate in cohorts of similar frequencies, suggesting that they most likely were co-integrated into the same recipient genome. While we cannot rule out the possibility that the similarity in frequency dynamics is due to transfer events of individual fragments that are separated in time, such a scenario is less parsimonious as it requires sequential acquisition of multiple fragments (in one case, 17 different fragments) within consecutive descendant cells.
The HGT cohort sizes range between 1 fragment per genome to 17 fragments, and in the extreme case they appear to have replaced ∼2% of the recipient genome. This highly variable nature of integration was previously reported in transformation assays of other naturally competent bacteria: S. pneumonia and H. influenza (Mell et al. 2011; Croucher et al. 2012; Bubendorfer et al. 2016), likely reflecting a shared property of naturally competent bacteria. However, it is important to note that, while results from previous studies reflect mainly the acquisition phase of HGT, i.e., immediately following transformation, our results show for the first time the evolutionary fate of co-integrated fragments, which persist and propagate over time. From our three repeats and the large variability between them, we learn that even massive foreign DNA integration events, such as the one that occurred in one of the clones in population 3 of the other-Bacillus evolutionary regime (Figure 5A), can be sustained evolutionarily over time. An increased number of repeats would have helped to assess whether selection influences the abundance of massive acquisitions in populations, i.e., whether their frequency in the population during adaptation is different from, or similar to, their frequency upon transformation. The number of fragments (as well as mutations) identified was larger for population 3 compared to populations 1 and 2. It is possible that population 3 acquired a change that altered its competence characteristics, yet we did not find any mutations in competence related genes. Alternatively, it is possible that in all three populations a wide distribution of number of HGT events occurred at first, yet only in population 3 did a clone with many events prevail.
Analyzing the dynamics of clone frequency changes (as defined in Figure 5), we could determine for each clone the earliest generation in which it could be detected. The time-course dynamics revealed that clones that tend to appear first during evolution contain mutations only, whereas clones with HGT fragments (either alone or in combination with mutations) are detected only at later stages along the adaptation process. Our results further indicate that some clones converged, at least partially, to the same genotypes. In other-Bacillus line 2 (Figure 5), for example, the first mutation to appear eventually reached almost 100% of the population, with the HGT fragment appearing later and reaching 60% frequency by the end of the evolution experiment. Since the combined frequency of both events is higher than 100%, one must assume that both events converged into the same cell at some point. The most parsimonious explanation of the observed dynamics is that a clone containing a beneficial mutation started to accumulate in the population, and, at some point, one of the cells with this mutation incorporated the HGT fragment. Since that cell is probably only one of many cells that contain the founding mutation, a likely explanation for the observed rise in frequency of the HGT fragment is that it conferred an additional fitness advantage on top of that conferred by the point mutation. Indeed, our competition analysis revealed that, when this fragment is reinserted into the ancestor strain, it confers a fitness benefit (Figure 4). Additional examples of such dynamics can be observed in our data.
Interestingly, in line 3, competing clones fluctuated in frequency, with neither clone being extinct or fixating over the entire population. These dynamics might suggest that the different clones, even if adaptive, prevent each other’s fixation by competition in a process known as clonal interference. Another exciting possibility, other than competition, is that the different emerging clones form different ecotypes, i.e., ecologically distinct subpopulations that do not compete over the same resources, and therefore would not cause each other’s extinction. A previous work has demonstrated that such events of new ecotype emergence can be quite frequent in a rich medium (Koeppel et al. 2013) and could provide a hypothetical explanation to the lack of selective sweep observed in population 3.
Another intriguing finding is that HGT fragments were often integrated in closer proximity than expected by random (Figure 3). Clustering of HGT events along the chromosome could be attributed to three separate mechanisms: (1) Proximal integration of donor fragments originating from the same strand invasion or separate adjacent ones (Kulick et al. 2008; Lin et al. 2009). This process could result in multiple fragments inserted in close proximity within the same cell. Since our statistical analysis revealed that the clustering observed in our case is independent of the temporal co-occurrence of fragments, this mechanism cannot fully account for the observed results. (2) The emergence of recombination hotspots due to sequence features, e.g., high-sequence similarity between donor and recipient in the hotspot regions. Although this feature can theoretically account for the emergence of hotspots, previous analyses in various competent bacteria found no correlation between high local sequence similarity and a higher tendency of integration (Mell et al. 2011, 2014; Croucher et al. 2012; Bubendorfer et al. 2016). (3) While the two previous mechanisms evoke no selection argument, it is possible that the observed HGT hotspots are shaped, at least partly, by an adaptive benefit conferred by these fragments. Indeed, both bioinformatics (Oliveira et al. 2017) and experimental (Croucher et al. 2011; Chewapreecha et al. 2014) works demonstrate the role played by selection in generating recombination hotspots. It is therefore plausible that the clustering of HGT fragments observed in our populations results, at least partly, from selection. Our competition assay might serve as supporting evidence for such a claim, since the fragment we found to be beneficial was integrated (at least partly) in all three other-Bacillus lines.
Under our experimental design, both HGT-based DNA acquisition and point mutations can contribute to the generation of genetic variations. Yet the two processes differ in both the extent and type of variation introduced. Considering a point mutation rate of 10−9 (Drake et al. 1998) per base per generation, a genome size of ∼4 × 106, and the population size and dilution rate used in our experiment that results in ∼109 cell divisions per day, one would expect the generation of ∼4 × 106 nucleotide substitutions in the population per day of evolution through point mutations. The extent of genetic variation inserted per day by HGT can be also estimated. Considering a median fragment size of 4.3 kb, a sequence divergence of 1–7% between donor and recipient, a population reaching 109 cells every day, and a competence level of ∼0.75% of cells at the beginning of the evolution, we estimate that 3.2 × 108–2.2 × 109 nucleotides are altered daily in an evolving population via HGT. Thus, the amount of genetic variation inserted by HGT is two to three orders of magnitude higher than that introduced through point mutations. Yet, we observed up to 50-fold decline in competence during the evolution (Figure 6). Therefore, the relative contribution of HGT compared to mutations in generating genetic variation is reduced as evolution progresses. Furthermore, the majority of the non-neutral mutations in genomes tend to be deleterious (Eyre-Walker and Keightley 2007). In contrast, we suggest, mutations acquired by an organism through HGT likely have a lower proportion of deleterious mutations, as at least some of these would have been eliminated from the genome of the donor species. Thus, mutations introduced into a genome through HGT might show an improve prevalence of beneficial mutations. Further, one can speculate that the adaptive value of integrated fragments may depend on the genetic similarity between the donor and recipient and thus HGT from closely related donors is less likely to introduce deleterious mutations. Therefore, even if HGT introduces higher genetic variability than spontaneous mutations, the fitness burden could potentially be lower than that introduced by mutations.
The other very predominant difference between the effect of HGT and mutations in adaptation is that, whereas mutations introduce variability in a sequential manner, HGT can introduce many mutations at once, in genomic proximity. This difference can be particularly important when the genomic regions undergoing change manifest genetic interactions with one another. HGT allows the concerted acquisition of multiple epistatic regions at once into the genome. As such, HGT can serve as a means to cross fitness valleys, especially in cases of positive epistasis interactions. However, the opposite could also be that the introduction of many positions of variability at once, could hamper the effect of positions that, if introduced alone, would be more beneficial (negative epistasis), in that case favoring mutations over HGT. Our experiment began to uncover epistatic interactions, positive and negative, between regions of segments that were cotransferred by HGT (data not shown). Future research will allow to further explore the evolutionary utility of HGT in rugged fitness landscape that often feature complex epistasis.
Our analysis revealed an over-representation of mutations affecting genes coding for flagella in particular in the type III secretion system genes involved in the early steps of generating flagella (Mukherjee and Kearns 2014). Most of the flagellar mutations were nonsynonymous, strongly indicating that mutating the flagella is likely to be beneficial. In addition to its role in enabling motility, maintaining an active flagellum is essential for the activation of the competence system by affecting the phosphorylation status of DegU, a master regulator of competence in B. subtilis (Diethmaier et al. 2017; Hölscher et al. 2018). Thus, two types of driving forces may account for the fixation of flagellar mutations in our experiment: reducing the large cellular cost of generating and maintaining active flagella under conditions in which motility is not needed, and reducing the cost originating from switching into a competence state (Haijema et al. 2001). The link between flagella activity and competence may account for the decrease in competence levels observed during the adaptation process in our settings (Figure 6). Other than the pleiotropic effect of flagellar mutations on competence, it is possible that deleterious DNA acquisition events brought competent cells to extinction. A final thought-provoking possibility for the loss of competence is that, under a constant environment, as time progresses, competent cells are selected against, as those already exhausted the available solutions but still bear the cost of maintaining the system. In other settings not examined in this study, such as under frequently changing environmental conditions, acquisitions of foreign DNA fragments could remain beneficial for longer, and the adaptive potential might outweigh the cost. The decrease in competence could then be viewed in the context of the well-known trade-off between exploration vs. exploitation (Alba and Dorronsoro 2005). In the beginning of the evolutionary process, competence can be used to increase genetic variation, thus exploring the genotypic space. This exploration, however, comes with a cost of reduced growth and with a risk of introducing deleterious mutations or genetic incompatibilities. Therefore, as evolution progresses and beneficial genotypes are found, reducing competence level and “exploiting” the solutions obtained may be preferred.
Acknowledgments
We thank Uri Gophna, Avigdor Eldar and Neta Altman from Tel-Aviv University and Ilana Kolodkin-Gal from the Weizmann Institute of science from providing strains, materials and helpful advice. We thank Tsviya Olender for bioinformatics assistance. We thank Shlomit Gilad from the Crown Institute for Genomics at the INCPM for performing deep sequencing. We thank the Minerva foundation for granting the Minerva Center for Live Emulation of Genome Evolution. YP is incumbent of the Ben May Professorial Chair.
Footnotes
Supplemental material available at figshare: https://doi.org/10.25386/genetics.12800933.
Communicating editor: D. Weinreich
Literature Cited
- Alba E., and Dorronsoro B., 2005. The exploration/exploitation tradeoff in dynamic cellular genetic algorithms. IEEE Trans. Evol. Comput. 9: 126–142. 10.1109/TEVC.2005.843751 [DOI] [Google Scholar]
- Arevalo P., VanInsberghe D., Elsherbini J., Gore J., and Polz M. F., 2019. A reverse ecology approach based on a biological definition of microbial populations. Cell 178: 820–834.e14. 10.1016/j.cell.2019.06.033 [DOI] [PubMed] [Google Scholar]
- Ashburner M., Ball C. A., Blake J. A., Botstein D., Butler H. et al. , 2000. Gene Ontology: tool for the unification of biology. Nat. Genet. 25: 25–29. 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltrus D. A., 2013. Exploring the costs of horizontal gene transfer. Trends Ecol. Evol. 28: 489–495. 10.1016/j.tree.2013.04.002 [DOI] [PubMed] [Google Scholar]
- Bankevich A., Nurk S., Antipov D., Gurevich A. A., Dvorkin M. et al. , 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19: 455–477. 10.1089/cmb.2012.0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrick J. E., Lenski R. E.. 2013. Genome dynamics during experimental evolution. Nat Rev Genet. 14: 827–39. 10.1038/nrg3564. http://www.ncbi.nlm.nih.gov/pubmed/24166031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blecher-Gonen R., Barnett-Itzhaki Z., Jaitin D., Amann-Zalcenstein D., Lara-Astiaso D. et al. , 2013. High-throughput chromatin immunoprecipitation for genome-wide mapping of in vivo protein-DNA interactions and epigenomic states. Nat. Protoc. 8: 539–554. 10.1038/nprot.2013.023 [DOI] [PubMed] [Google Scholar]
- Brown C. T., Fishwick L. K., Chokshi B. M., Cuff M. A., Jackson J. M. et al. , 2011. Whole-genome sequencing and phenotypic analysis of Bacillus subtilis mutants following evolution under conditions of relaxed selection for sporulation. Appl. Environ. Microbiol. 77: 6867–6877. 10.1128/AEM.05272-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bubendorfer S., Krebes J., Yang I., Hage E., Schulz T. F. et al. , 2016. Genome-wide analysis of chromosomal import patterns after natural transformation of Helicobacter pylori. Nat. Commun. 7: 11995 10.1038/ncomms11995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrasco B., Serrano E., Sánchez H., Wyman C., and Alonso J. C., 2016. Chromosomal transformation in Bacillus subtilis is a non-polar recombination reaction. Nucleic Acids Res. 44: 2754–2768. 10.1093/nar/gkv1546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chewapreecha C., Harris S. R., Croucher N. J., Turner C., Marttinen P. et al. , 2014. Dense genomic sampling identifies highways of pneumococcal recombination. Nat. Genet. 46: 305–309. 10.1038/ng.2895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu H. Y., Sprouffske K., and Wagner A., 2018. Assessing the benefits of horizontal gene transfer by laboratory evolution and genome sequencing. BMC Evol. Biol. 18: 54 10.1186/s12862-018-1164-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohan F. M., Roberts M. S., and King E. C., 1991. The potential for genetic exchange by transformation within a natural population of Bacillus subtilis. Evolution 45: 1393–1421. 10.1111/j.1558-5646.1991.tb02644.x [DOI] [PubMed] [Google Scholar]
- Coupat B., Chaumeille-Dole F., Fall S., Prior P., Simonet P. et al. , 2008. Natural transformation in the Ralstonia solanacearum species complex: number and size of DNA that can be transferred. FEMS Microbiol. Ecol. 66: 14–24. 10.1111/j.1574-6941.2008.00552.x [DOI] [PubMed] [Google Scholar]
- Croucher N. J., Harris S. R., Fraser C., Quail M. A., Burton J., et al. . 2011. Rapid pneumococcal evolution in response to clinical interventions. Science 331: 430–434. 10.1126/science.1198545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croucher N. J., Harris S. R., Barquist L., Parkhill J., and Bentley S. D., 2012. A high-resolution view of genome-wide pneumococcal transformation. PLoS Pathog. 8: e1002745 10.1371/journal.ppat.1002745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derbyshire K. M., and Gray T. A., 2014. Distributive conjugal transfer: new insights into horizontal gene transfer and genetic exchange in mycobacteria. Microbiol. Spectr. 2: 04 10.1128/microbiolspec.MGM2-0022-2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries J., and Wackernagel W., 2002. Integration of foreign DNA during natural transformation of Acinetobacter sp. by homology-facilitated illegitimate recombination. Proc. Natl. Acad. Sci. USA 99: 2094–2099. 10.1073/pnas.042263399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diethmaier C., Chawla R., Canzoneri A., Kearns D. B., Lele P. P. et al. , 2017. Viscous drag on the flagellum activates Bacillus subtilis entry into the K-state. Mol. Microbiol. 106: 367–380. 10.1111/mmi.13770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake J., Charlesworth B., Charlesworth D. F., and Crow J., 1998. Rates of spontaneous mutation. Genetics 148: 1667–1686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyre-Walker A., and Keightley P. D., 2007. The distribution of fitness effects of new mutations. Nat. Rev. Genet. 8: 610–618. 10.1038/nrg2146 [DOI] [PubMed] [Google Scholar]
- Fornili S. L., and Fox M. S., 1977. Electron microscope visualization of the products of Bacillus subtilis transformation. J. Mol. Biol. 113: 181–191. 10.1016/0022-2836(77)90048-1 [DOI] [PubMed] [Google Scholar]
- González-Torres P., Rodríguez-Mateos F., Antón J., and Gabaldón T., 2019. Impact of homologous recombination on the evolution of prokaryotic core genomes. MBio 10: e02494–18 10.1128/mBio.02494-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn J., Luttinger A., and Dubnau D., 1996. Regulatory inputs for the synthesis of ComK, the competence transcription factor of Bacillus subtilis. Mol. Microbiol. 21: 763–775. 10.1046/j.1365-2958.1996.371407.x [DOI] [PubMed] [Google Scholar]
- Haijema B. J., Hahn J., Haynes J., and Dubnau D., 2001. A ComGA-dependent checkpoint limits growth during the escape from competence. Mol. Microbiol. 40: 52–64. 10.1046/j.1365-2958.2001.02363.x [DOI] [PubMed] [Google Scholar]
- Hehemann J.-H., Arevalo P., Datta M. S., Yu X., Corzett C. H. et al. , 2016. Adaptive radiation by waves of gene transfer leads to fine-scale resource partitioning in marine microbes. Nat. Commun. 7: 12860 10.1038/ncomms12860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hölscher T., Schiklang T., Dragoš A., Dietel A.-K., Kost C. et al. , 2018. Impaired competence in flagellar mutants of Bacillus subtilis is connected to the regulatory network governed by DegU. Environ. Microbiol. Rep. 10: 23–32. 10.1111/1758-2229.12601 [DOI] [PubMed] [Google Scholar]
- Johnston C., Martin B., Fichant G., Polard P., and Claverys J. P., 2014. Bacterial transformation: distribution, shared mechanisms and divergent control. Nat Rev Microbiol. 12: 181–196. 10.1038/nrmicro3199 [DOI] [PubMed] [Google Scholar]
- Kaminski Strauss S., Schirman D., Jona G., Brooks A. N., Kunjapur A. M. et al. , 2019. Evolthon: A community endeavor to evolve lab evolution. PLoS Biol. 17: e3000182. 10.1371/journal.pbio.3000182. http://www.ncbi.nlm.nih.gov/pubmed/30925180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloesges T., Popa O., Martin W., and Dagan T., 2011. Networks of gene sharing among 329 proteobacterial genomes reveal differences in lateral gene transfer frequency at different phylogenetic depths. Mol. Biol. Evol. 28: 1057–1074. 10.1093/molbev/msq297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knöppel A., Knopp M., Albrecht L. M., Lundin E., Lustig U. et al. , 2018. Genetic adaptation to growth under laboratory conditions in Escherichia coli and Salmonella enterica. Front. Microbiol. 9: 756 10.3389/fmicb.2018.00756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koeppel A. F., Wertheim J. O., Barone L., Gentile N., Krizanc D. et al. , 2013. Speedy speciation in a bacterial microcosm: new species can arise as frequently as adaptations within a species. ISME J. 7: 1080–1091. 10.1038/ismej.2013.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohlstedt M., Sappa P. K., Meyer H., Maaß S., Zaprasis A. et al. , 2014. Adaptation of Bacillus subtilis carbon core metabolism to simultaneous nutrient limitation and osmotic challenge: a multi-omics perspective. Environ. Microbiol. 16: 1898–1917. 10.1111/1462-2920.12438 [DOI] [PubMed] [Google Scholar]
- Koonin E. V., Makarova K. S., and Aravind L., 2001. Horizontal gene transfer in prokaryotes: quantification and classification. Annu. Rev. Microbiol. 55: 709–742. 10.1146/annurev.micro.55.1.709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulick S., Moccia C., Didelot X., Falush D., Kraft C. et al. , 2008. Mosaic DNA imports with interspersions of recipient sequence after natural transformation of Helicobacter pylori. PLoS One 3: e3797 10.1371/journal.pone.0003797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz S., Phillippy A., Delcher A. L., Smoot M., Shumway M. et al. , 2004. Versatile and open software for comparing large genomes. Genome Biol. 5: R12 10.1186/gb-2004-5-2-r12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin E. A., Zhang X. S., Levine S. M., Gill S. R., Falush D. et al. , 2009. Natural transformation of Helicobacter pylori involves the integration of short DNA fragments interrupted by gaps of variable size. PLoS Pathog. 5: e1000337 10.1371/journal.ppat.1000337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López C. S., Alice A. F., Heras H., Rivas E. A., and Sánchez-Rivas C., 2006. Role of anionic phospholipids in the adaptation of Bacillus subtilis to high salinity. Microbiology 152: 605–616. 10.1099/mic.0.28345-0 [DOI] [PubMed] [Google Scholar]
- Maddamsetti R., and Lenski R. E., 2018. Analysis of bacterial genomes from an evolution experiment with horizontal gene transfer shows that recombination can sometimes overwhelm selection. PLoS Genet. 14: e1007199 10.1371/journal.pgen.1007199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majewski J., and Cohan F. M., 1999. DNA sequence similarity requirements for interspecific recombination in Bacillus. Genetics 153: 1525–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka H., Hirooka K., and Fujita Y., 2007. Organization and function of the YsiA regulon of Bacillus subtilis involved in fatty acid degradation. J. Biol. Chem. 282: 5180–5194. 10.1074/jbc.M606831200 [DOI] [PubMed] [Google Scholar]
- Mell J. C., Shumilina S., Hall I. M., and Redfield R. J., 2011. Transformation of natural genetic variation into Haemophilus influenzae genomes. PLoS Pathog. 7: e1002151 10.1371/journal.ppat.1002151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mell J. C., Lee J. Y., Firme M., Sinha S., and Redfie R. J., 2014. Extensive cotransformation of natural variation into chromosomes of naturally competent Haemophilus influenzae. G3 (Bethesda) 4: 717–731. 10.1534/g3.113.009597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H., Muruganujan A., Ebert D., Huang X., and Thomas P. D., 2019. PANTHER version 14: more genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 47: D419–D426. 10.1093/nar/gky1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S., and Kearns D. B., 2014. The structure and regulation of flagella in Bacillus subtilis. Annu Rev Genet. 48: 319–40. 10.1146/annurev-genet-120213-092406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuttall, S., C. Bath, M. Pfeiffer, F. Santos, J. Eichler et al., 2008 The Halohandbook. Media.(March):1–144. Available at: http://scholar.google.com/scholar?hl=en&btnG=Search&q=intitle:The+Halohandbook#0.
- Ochman H., Lawrence J. G., Groisman E. A., 2000. Lateral gene transfer and the nature of bacterial innovation. Nature. 405: 299–304. 10.1038/35012500. http://www.ncbi.nlm.nih.gov/pubmed/10830951 [DOI] [PubMed] [Google Scholar]
- Oliveira, P. H., M. Touchon, J. Cury, and E. P. C. Rocha. 2017 The chromosomal organization of horizontal gene transfer in bacteria. Nat Commun. 8: 841 (erratum: Nat Commun 11: 1155). 10.1038/s41467-017-00808-w 10.1038/s41467-017-00808-w [DOI] [PMC free article] [PubMed]
- Popa O., Landan G., and Dagan T., 2017. Phylogenomic networks reveal limited phylogenetic range of lateral gene transfer by transduction. ISME J. 11: 543–554. 10.1038/ismej.2016.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ram Y., Dellus-Gur E., Bibi M., Karkare K., Obolski U. et al. , 2019. Predicting microbial growth in a mixed culture from growth curve data. Proc. Natl. Acad. Sci. USA 116: 14698–14707 [corrigenda: Proc. Natl. Acad. Sci. USA 117: 13848 (2020)]. 10.1073/pnas.1902217116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravenhall M., Škunca N., Lassalle F., and Dessimoz C., 2015. Inferring horizontal gene transfer. PLOS Comput. Biol. 11: e1004095 10.1371/journal.pcbi.1004095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter M., Rosselló-Móra R., Oliver Glöckner F., and Peplies J., 2016. JSpeciesWS: a web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 32: 929–931. 10.1093/bioinformatics/btv681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smillie C., Garcillán-Barcia M. P., Francia M. V., Rocha E. P. C., and de la Cruz F., 2010. Mobility of plasmids. Microbiol. Mol. Biol. Rev. 74: 434–452. 10.1128/MMBR.00020-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souza V., Turner P. E., and Lenski R. E., 1997. Long-term experimental evolution in Escherichia coli. V. Effects of recombination with immigrant genotypes on the rate of bacterial evolution. J. Evol. Biol. 10: 743–769. 10.1007/s000360050052 [DOI] [Google Scholar]
- Steil L., Hoffmann T., Budde I., Völker U., and Bremer E., 2003. Genome-wide transcriptional profiling analysis of adaptation of Bacillus subtilis to high salinity. J. Bacteriol. 185: 6358–6370. 10.1128/JB.185.21.6358-6370.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Gene Ontology Consortium , 2019. The gene ontology resource: 20 years and still GOing strong. Nucleic Acids Res. 47: D330–D338. 10.1093/nar/gky1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tortosa P., Logsdon L., and Dubnau D., 2001. Specificity and genetic polymorphism of the Bacillus competence quorum-. Sens. Syst. 183: 451–460. 10.1128/JB.183.2.451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters S. M., Zeigler D. R., and Nicholson W. L., 2015. Experimental evolution of enhanced growth by Bacillus subtilis at low atmospheric pressure: genomic changes revealed by whole-genome sequencing. Appl. Environ. Microbiol. 81: 7525–7532. 10.1128/AEM.01690-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss A., and Shaw L. N., 2015. Small things considered: the small accessory subunits of RNA polymerase in Gram-positive bacteria. FEMS Microbiol. Rev. 39: 541–554. 10.1093/femsre/fuv005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedenbeck J., and Cohan F. M., 2011. Origins of bacterial diversity through horizontal genetic transfer and adaptation to new ecological niches. FEMS Microbiol. Rev. 35: 957–976. 10.1111/j.1574-6976.2011.00292.x [DOI] [PubMed] [Google Scholar]
- Zawadzki P., and Cohan F. M., 1995. The size and continuity of DNA segments integrated in Bacillus transformation. Genetics 141: 1231–1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article. The genome assembly of the donor Bacillus strains has been deposited at DDBJ/ENA/GenBank, under the BioProject accession number PRJNA650388. See File S1 for the individual accession numbers. The populations’ raw sequencing data supporting the findings of this study have been deposited at the SRA database under the following SRA accession: PRJNA655387. The supplemental tables and figures have been deposited via the GSA figshare portal. File S1 contains the supplemental Materials and Methods. File S2 contains all the supplemental figures (Figures S1 – S6), Table S1 contains statistics derived from the growth assays performed on the evolution regimes, Table S2 contains a full list of genetic variations detected in the other-Bacillus evolutionary regime in the different time points, Table S3 contains a full list of all de novo mutations detected in all evolution lines in the last time point, Table S4 contains a list of HGT fragments and mutations detected in single colonies sequenced in the last time point. Supplemental material available at figshare: https://doi.org/10.25386/genetics.12800933.