Abstract
Viruses such as human cytomegalovirus (HCMV), human papillomavirus (HPV), Epstein–Barr virus (EBV), human immunodeficiency virus (HIV), and coronavirus (severe acute respiratory syndrome coronavirus 2 [SARS‐CoV‐2]) represent a great burden to human health worldwide. FDA‐approved anti‐parasite drug ivermectin is also an antibacterial, antiviral, and anticancer agent, which offers more potentiality to improve global public health, and it can effectively inhibit the replication of SARS‐CoV‐2 in vitro. This study sought to identify ivermectin‐related virus infection pathway alterations in human ovarian cancer cells. Stable isotope labeling by amino acids in cell culture (SILAC) quantitative proteomics was used to analyze human ovarian cancer cells TOV‐21G treated with and without ivermectin (20 μmol/L) for 24 h, which identified 4447 ivermectin‐related proteins in ovarian cancer cells. Pathway network analysis revealed four statistically significant antiviral pathways, including HCMV, HPV, EBV, and HIV1 infection pathways. Interestingly, compared with the reported 284 SARS‐CoV‐2/COVID‐19‐related genes from GencLip3, we identified 52 SARS‐CoV‐2/COVID‐19‐related protein alterations when treated with and without ivermectin. Protein–protein network (PPI) was constructed based on the interactions between 284 SARS‐CoV‐2/COVID‐19‐related genes and between 52 SARS‐CoV‐2/COVID‐19‐related proteins regulated by ivermectin. Molecular complex detection analysis of PPI network identified three hub modules, including cytokines and growth factor family, MAP kinase and G‐protein family, and HLA class proteins. Gene Ontology analysis revealed 10 statistically significant cellular components, 13 molecular functions, and 11 biological processes. These findings demonstrate the broad‐spectrum antiviral property of ivermectin benefiting for COVID‐19 treatment in the context of predictive, preventive, and personalized medicine in virus‐related diseases.
Keywords: ivermectin, quantitative proteomics, SARS‐CoV‐2/COVID‐19, stable isotope labeling by amino acids in cell culture, virus‐related pathways
-
1.
This study sought to identify ivermectin‐related virus infection pathway alterations in human cells.
-
2.
Quantitative proteomics revealed that ivermectin‐related proteins are involved in four statistically significant antiviral pathways, including human cytomegalovirus (HCMV), human papillomavirus (HPV), Epstein–Barr virus (EBV), human immunodeficiency virus 1 (HIV1), and COVID‐19 infection pathways.
-
3.
We identified 52 severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2)/COVID‐19‐related protein alterations when treated with and without ivermectin, and these proteins were involved in cytokines and growth factor family, MAP kinase and G‐protein family, and HLA class proteins.
-
4.
These findings demonstrate the broad‐spectrum antiviral property of ivermectin benefiting for COVID‐19 treatment in the context of predictive, preventive, and personalized medicine in virus‐related diseases.
1. INTRODUCTION
Ōmura discovered a unique and extraordinary microorganism that could produce ivermectin in 1973 (Burg et al., 1979). Ivermectin was subsequently commercialized because it showed great safety and effectivity in human health. The current status of ivermectin was continuing to surprise and excite scientists (Laing, Gillan, & Devaney, 2017). It was originally intended to be a broad‐spectrum antiparasitic agent, and treat onchocerciasis, strongyloidiasis, lymphatic filariasis, and scabies in veterinary and human medicine (Chabala et al., 1980). There was an outstanding advantage of ivermectin that no confirmed or increased drug resistance appears in parasites, even in those human populations who have been receiving ivermectin as a monotherapy for more than 30 years (van Wyk & Malan, 1988). In terms of mechanism, the primary target of ivermectin is glutamate‐gated chloride channels (Abdeltawab et al., 2020). However, it was increasingly believed that ivermectin was closely related to the immune defense mechanism and acted like immunomodulatory agents to help suppress the parasite's ability to evade the host's immune (Schaller et al., 2017). Today, the new use of ivermectin made it become a relatively unknown drug. Drug repurposing and repositioning has been shown to control a completely new range of diseases (Ashour, 2019). For example, orbital myiasis, trichinosis, malaria, leishmaniasis, African trypanosomiasis, asthma, epilepsy, neurological disease, antiviral (e.g., human immunodeficiency virus [HIV], dengue, encephalitis; Yang et al., 2020), antibacterial (tuberculosis and Buruli ulcer; Csóka et al., 2018), anticancer (breast cancer, leukemia, glioblastoma, cervical cancer, gastric cancer, ovarian cancer, colon cancer, melanoma, and lung cancer; Crump, 2017). Multifaceted ‘wonder’ ivermectin may become an even more exceptional drug in the future. An international patent ‘Use of ivermectin and derivatives thereof’ caught people's increasing attention to ivermectin those years. Ivermectin would be developed to use for metabolic‐related diseases (diabetes, hypercholesterolemia, insulin resistance, obesity, hypertriglyceridemia, and hyperglycemia), Famesoid X receptor‐mediated diseases (atherosclerosis, nonalcohol fatty liver disease, cholestasia, and gallstones), inflammation, and cancer (Crump, 2017).
Viruses such as HIV, human cytomegalovirus (HCMV), Epstein–Barr virus (EBV), human papillomavirus (HPV), and novel severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) represent a great burden for human health worldwide. For example, there were almost 37 million people infected with HIV‐1 in the whole world, and nearly 1 million patients died of human immunodeficiency virus infection and acquired immune deficiency syndrome (AIDS)‐related disease each year (Huynh & Gulick, 2020). HIV damaged the immune system and majorly killed CD4 cells to make patients vulnerable to various illnesses, including pneumonia, cytomegalovirus, cryptococcal meningitis, tuberculosis, cryptosporidiosis, oral thrush, toxoplasmosis, and cancer (Kaposi's sarcoma and lymphoma; (Nash & Robertson, 2019). HCMV is a β‐herpesvirus that closely has a prevalence of 55%–100% within the human population. HCMV is one of the most common infection in all live births (1%–2.5%) in the Western world (Buxmann, Hamprecht, Meyer‐Wittkopf, & Friese, 2017). HCMV intrauterine infection could lead to congenital abnormalities, including visual impairment, low birth weight, hearing loss, varying degrees of mental retardation, hepatosplenomegaly, and microcephaly (Zavattoni et al., 2014). HCMV acquired different mechanisms to evade the human immune response (Britt, 2008). For example, the HCMV prevented NK cell activity by virus UL16 and UL142 proteins. HCMV has acquired a viral homolog of IL‐10 to suppress anti‐cytomegalovirus immunity (Holder & Grant, 2019). HCMV also downregulated the expression of major histocompatibility complex to prevent the antigen processing and presentation by virus US11, US2, and US3 proteins (Britt, 2008). HCMV has also evolved proteins (UL36 and UL37) to prevent apoptosis of infected cells, which promoted HCMV dissemination within the host (Andoniou & Degli‐Esposti, 2006). EBV was a member of the herpesvirus family that could cause mononucleosis. Though lots of people were asymptomatic infections, but potential links between EBV and other lymphoproliferative diseases (nonmalignant, premalignant, and malignant diseases) were widely studied (Rezk, Zhao, & Weiss, 2018), such as Burkitt lymphoma, Hodgkin's lymphoma, hemophagocytic lymphohistiocytosis, gastric cancer, central nervous system lymphomas, acute cerebellar ataxia (Nussinovitch, Prais, Volovitz, Shapiro, & Amir, 2003), nasopharyngeal carcinoma, and hairy leukoplakia (Marques‐Piubelli et al., 2020). EBV can infect different kinds of cells, but viral tropism is preferred to B cells and epithelial cells. B cell membrane fusion was mediated by the three‐part glycoprotein complexes of gHgL gp42; although epithelial cell membrane fusion was mediated by the two‐part complexes of gHgL (Shannon‐Lowe, Rowe, 2014). About 90% of HPV were asymptomatic infections, but HPV infection would lead to either warts or precancerous lesions. The infected sites by HPV, especially the subtype HPV16 and HPV18, showed high risk of cancer, including cervix, vagina, vulva, mouth, penis, throat, and anus (Athanasiou et al., 2020). HPV was believed to cause cancers in nonintegrated episomes and integrating into DNA. Some of the HPV genes (E6 and E7), acted as oncogenes to promote malignant transformation (Hoppe‐Seyler, Bossler, Braun, Herrmann, & Hoppe‐Seyler, 2018). E6 protein bound to p53 protein and resulted in the inactivation of p53 (Almeida, Queiroz, Sousa, & Sousa, 2019). E7 acted as the transforming protein and competed between retinoblastoma protein (pRb) for binding to transcription factor E2F, which pushed the cell cycle forward (Almeida et al., 2019). SARS‐CoV‐2 lead to the outbreak of coronavirus disease 2019 (COVID‐19) and rapidly grew into a global pandemic. Scientists set out to develop a treatment for COVID‐19, but no anti‐SARS‐CoV‐2 drug or vaccine has been approved to solve the serious challenge (H. Li et al., 2020). In the whole world, more than 7 million people have infected SARS‐CoV‐2, including more than 400,000 deaths at the national level (Lai, Shih, Ko, Tang, & Hsueh, 2020). Further studies to develop the safest and most effective ways to combat viral infections were urgent. Ivermectin has been demonstrated to limit infection by a number of viruses with potential broad‐spectrum activity (Yang et al., 2020). For example, ivermectin has been reported anti‐HIV‐1 reliant on importin α/β nuclear import (Wagstaff, Sivakumaran, Heaton, Harrich, & Jans, 2012). Ivermectin could reduce MAPK pathway activation through the inhibition of PAK‐1 activity. The high content screening also identified ivermectin as a promising drug against EBV‐positive and EBV‐negative nasopharyngeal carcinoma cells (Gallardo, Mariamé, Gence, & Tilkin‐Mariamé, 2018). Herpes genitalis and infections, which are caused by HPV in males, might have an effective treatment choice for oral ivermectin, but it has not been officially approved until now (Buechner, 2002). More importantly, ivermectin was reported as an inhibitor of the SARS‐CoV‐2, which was able to produce an effect ~5000‐fold reduction in viral RNA with a single addition to cells infected with SARS‐CoV‐2 (Caly, Druce, Catton, Jans, & Wagstaff, 2020).
Viruses remain one of the least well‐understood pathogens. The lack of knowledge about mechanisms and host–parasite interactions limited success in developing vaccines. It is facing challenges on several fronts, including limitations in availability, high cost of production, high mutation probability, and high prevalence of resistance. Ivermectin, as an antiparasitic, anticancer, antibacterial, and antiviral agent, provided more potentiality to improve global public health. The present study used stable isotope labeling by amino acids in cell culture (SILAC) quantitative proteomics analysis to reveal ivermectin‐related proteomics profiling and molecular network alterations. We focused our attention on the virus‐related pathways, such as HCMV infection, HPV infection, EBV infection, and HIV1 infection. More interestingly, a large number of identified proteins were reported to be related to SARS‐CoV‐2/COVID‐19. These results indicated that ivermectin might be a broad‐spectrum antiviral drug. SILAC quantitative proteomics proved the molecular mechanisms of ivermectin in virus‐related pathways. Furthermore, protein–protein interaction (PPI)‐based hub modules for SARS‐CoV‐2‐related proteins discovered a key molecule in COVID‐19 disease in the context of predictive, preventive, and personalized medicine (PPPM) in COVID‐19.
2. MATERIALS AND METHODS
2.1. SILAC‐treated cells
Human ovarian cell line (TOV‐21G; Keibai Academy of Science, Nanjing, China) was cultured with two different SILAC reagents (Thermo Fisher Scientific) (One was RPMI 1640 medium without l‐lysine [K] and l‐Arginine [R] supplemented with 100 mg/L l‐lysine‐2HCl and 100 mg/L l‐arginine‐HCl [“light” labeling reagent = L] and 10% fetal bovine serum [FBS; Gibco], and another was RPMI 1640 medium without l‐lysine [K] and l‐arginine [R] supplemented with 100 mg/L l‐lysine‐2HCl[13C6,15N2] and 100 mg/L l‐arginine‐HCl[13C6,15N4] [“heavy” labeling reagent = H; [13C6,15N2] means 8 mass units increased in residue K, [13C6,15N4] means 10 mass units increased in residue R] and 10% FBS), and maintained with 5% CO2 and 37°C and medium renewal every 2 days. A total of 10 passages were treated with SILAC reagents with 12C14N (light = L) and 13C15N (heavy = H)‐labeled amino acids to ensure complete incorporation of stable isotope into the cultured cells.
2.2. Ivermectin treatment of SILAC‐labeled cells
Our previous study found that when TOV‐21G cells were treated with ivermectin (0–60 μM) for 24 h, the IC50 was 22.54 μM for ivermectin, and also 20 μM ivermectin (it was less than IC50 22.54 μM) significantly suppressed cell proliferation and migration of TOV‐21G, and maintained TOV‐21G cells in good shape (N. Li & Zhan, 2020). Thus, TOV‐21G cells cultured in the H‐ and L‐stable isotope‐labeled media were treated with 20 μM ivermectin in dimethyl sulfoxide (DMSO) or with the same amount of the DMSO as control, for 24 h. Ivermectin‐treated TOV‐21G cells were centrifuged (800g), washed with PBS (×3), and then suspended (30 min, 4°C) in protein isolation buffer [7 M urea, 2 mM thiourea, 4% CHAPS (3‐[(3‐cholamidopropyl)‐dimethylammonio]‐1‐propane), 100 mM dithiothreitol (DTT), and 2% ampholyte] with a vortex (×5). The extracted protein solution was centrifuged (13,000g, 20 min, 4°C). The supernatants were the extracted protein samples whose protein concentrations were examined with 2‐D quant kit.
2.3. SILAC‐labeling efficiency analysis
The extracted protein samples were ultrasonicated and centrifuged (14,000g, 25°C, 40 min). The H‐ and L‐stable isotope‐labeled proteins were equally mixed (1:1), separated with 12.5% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS‐PAGE; 20 μg/lane; constant current 14 mA, 90 min), and stained with Coomassie brilliant blue. SDS‐PAGE‐separated proteins were subjected to reduction, alkylation, digestion with trypsin, and identification with mass spectrometry (MS). The efficiency of SILAC‐isotope incorporation into proteins was estimated with Rappsilber's method (Rappsilber, Ishihama, & Mann, 2003). For this study, the SILAC labeling efficiency was up to 97%.
2.4. Protein digestion and LC‐fractionation
The extracted protein samples were treated with a final concentration of 100 mM DTT, boiled (water bath; 5 min), transferred to a 10 kD ultrafiltration centrifuge tube with 200 μl of 8 M urea in 0.1 M Tris–HCl, pH 8.5, and centrifuged (14,000g, 15 min; ×2). The protein samples in ultrafiltration centrifuge tube were treated (dark room, 30 min, room temperature) with 100 μl solution of 0.05 M iodoacetamide, 8 M urea, and 0.1 M Tris–HCl, pH 8.5), followed by centrifugation (14,000g, 15 min). The iodoacetamide‐treated protein sample was treated with 100 μl of 8 M urea in 0.1 M Tris–HCl, pH 8.5, and centrifuged (14,000g, 15 min; ×3), followed by treatment with 100 μl of 25 mM NH4HCO3 solution, and centrifugation (14,000g, 15 min; ×3). The treated protein samples were mixed (shaking with 600 rpm, 1 min) with 40 μl of 2 μg trypsin in 40 μl 100 mM NH4HCO3, shaked, stayed (37°C, 16–18 h), and transferred into a new collection tube for centrifugation (14,000g, 15 min), followed by mixing with 40 μl of 25 mM NH4HCO3, and centrifugation (14,000g, 15 min) to collect the filtrate as the tryptic peptide mixture. The peptide content was quantified (OD280). Liquid chromatography (LC) was used to fractionate the tryptic peptide mixture into 15 peptide fractions for reverse LC‐tandem mass spectrometry (LC‐MS/MS) analysis.
2.5. LC–MS/MS
Each peptide fraction was subjected to LC–MS/MS analysis for 60 min on an Easy nLC (Proxeon Biosystems, now Thermo Fisher Scientific) coupled with Q Exactive mass spectrometer (Thermo Fisher Scientific). The obtained MS/MS spectra data were used to identify and quantify proteins with MaxQuant software against the protein database. The intensities of the light and heavy isotopes were used to determine the protein differentially expressed levels between TOV‐21G cells treated with (heavy labeling = H) and without (light labeling = L) ivermectin.
2.6. Bioinformatics and statistical analysis
Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis was performed with clusterProfiler (https://bioconductor.org/packages/release/bioc/html/clusterProfiler.html; Yu, Wang, Han, & He, 2012) to find the signaling pathway based on the identified protein (p < .05, and adjusted p < .05). The reported SARS‐CoV‐2‐related genes were searched by GenCLiP3 (http://ci.smu.edu.cn/genclip3/; Wang et al., 2019). Cytoscape ClueGO (Bindea et al., 2009) was used to reveal the biological processes (BPs), molecular functions (MFs), and cellular components (CCs) enriched from identified proteins (two‐sided hypergeometric test, adjusted p < .05 corrected with Benjamini–Hochberg). The reported SARS‐CoV‐2‐related genes were analyzed by STRING 10.0 (http://string-db.org/cgi/input.pl; Szklarczyk et al., 2015) with the confidence of parameter (co‐expression score > 0.700) for PPI network construction. Then, the entire PPIs were analyzed with the molecular complex detection (MCODE; Bader & Hogue, 2003) using Cytoscape software (version 3.2.1; National Resource for Network Biology) to obtain hub modules (score > 6). The statistical significance was set as p < .05. A Benjamini–Hochberg was used to adjust p value for the probability of the association between the proteins in the pathway.
3. RESULTS
3.1. SILAC quantitative proteomics analysis revealed a broad‐spectrum antiviral property of ivermectin
A total of 4447 ivermectin‐related proteins were identified in ovarian cancer cells treated with and without ivermectin with SILAC‐based quantitative proteomics (Table S1). Further KEGG pathway analysis revealed four virus‐related pathways (Table 1), including HCMV (Figure S1A), HPV (Figure S1b), EBV (Figure S1C), and HIV1 (Figure S1d) infection pathways, and eight bacteria‐related pathways (Table 1), including bacterial invasion of epithelial cells, vibrio cholerae infection, epithelial cell signaling in Helicobacter pylori infection, pathogenic Escherichia coli infection, shigellosis, salmonella infection, legionellosis, and yersinia infection (Figure S2). Those enriched pathways indicated that ivermectin was an antibacterial and antiviral agent, and provided clues to relevant mechanisms of ivermectin.
Table 1.
Statistically significant bacteria‐ and virus‐related pathways identified with ivermectin‐related proteins by KEGG pathway enrichment analysis
| Number | Pathway ID | Pathway name | Ratio of matched versus total genes | p value | Adjusted p value | q value | Gene ID of ivermectin‐related proteins |
|---|---|---|---|---|---|---|---|
| 1 | hsa05100 | Bacterial invasion of epithelial cells | 44/2091 | 5.95E − 10 | 1.46E − 08 | 9.92E − 09 | CLTB, CTNNB1, SEPTIN3, DNM1, FN1, CTNNA1, RAC1, CBL, ACTG1, CDC42, CRKL, RHOA, ARPC3, ARPC5, SEPTIN2, ARPC2, ARPC5L, ELMO2, SEPTIN11, SEPTIN9, SHC1, CD2AP, DNM2, PIK3R2, SRC, CLTC, ARPC1B, CRK, CTTN, PTK2, ACTB, PXN, ITGB1, MET, WASF1, SEPTIN8, WASF2, CAV1, CLTA, WASL, ARHGEF26, ITGA5, DOCK1, PIK3R1 |
| 2 | hsa05110 | Vibrio cholerae infection | 28/2091 | 6.22E − 06 | 5.69E − 05 | 3.85E − 05 | KDELR2, ATP6V0D1, ATP6V0A1, ATP6V1B2, ATP6V1C1, ATP6V1A, ATP6V1H, ACTG1, PRKCA, PRKACA, PRKACB, SLC12A2, ATP6V1E1, ARF1, ATP6V0C, TJP2, PLCG1, TJP1, ACTB, GNAS, SEC61G, ATP6V1G1, ATP6V1F, ERO1A, SEC61B, TCIRG1, SEC61A1, KDELR1 |
| 3 | hsa05120 | Epithelial cell signaling in Helicobacter pylori infection | 33/2091 | 1.07E − 04 | 7.31E − 04 | 4.96E − 04 | PAK1, F11R, ATP6V0D1, MAPK8, CHUK, IKBKG, ATP6V0A1, ATP6V1B2, RAC1, ATP6V1C1, ATP6V1A, ATP6V1H, ADAM10, LYN, CDC42, MAPK14, GIT1, ATP6V1E1, MAP2K4, RELA, ATP6V0C, CSK, SRC, PLCG1, TJP1, NFKB1, MET, PTPN11, EGFR, ATP6V1G1, CASP3, ATP6V1F, TCIRG1 |
| 4 | hsa05130 | Pathogenic Escherichia coli infection | 90/2091 | 6.13E − 09 | 1.31E − 07 | 8.86E − 08 | IRAK1, MYD88, MYH9, PAK1, ROCK1, TUBB1, TUBB2B, WIPF2, ARHGEF12, TMBIM6, BAIAP2, TUBB4A, CYFIP2, SEC24A, TUBA4A, MAPK8, CHUK, IKBKG, SLC9A3R1, TUBA1B, TUBB2A, MAPK1, RAC1, ABI1, ACTG1, LYN, CYFIP1, CDC42, FADD, BRK1, CYCS, SEC24C, MAPK14, PAK2, RHOA, ARPC3, ARPC5, ARHGEF2, TUBA1C, TUBB, NCL, RAB1A, ARPC2, ARPC5L, CASP8, MYO1B, RELA, ROCK2, ARF1, BAIAP2L1, TUBA1A, ABCF2, ARF6, SEC24B, TUBB3, MYH10, MYO1C, MYO1E, TJP1, ARPC1B, CTTN, SEC24D, TMED10, ACTB, EZR, NCKAP1, NFKB1, TUBAL3, TRAF2, ARHGEF1, RIPK1, TUBB8, ITGB1, MYH3, RPS3, CLDN1, PTPN11, WASF1, WASF2, CASP3, BAX, CASP7, CLDN11, WASL, TUBB6, MYO6, IL18, GNA13, TRADD, NCK1 |
| 5 | hsa05131 | Shigellosis | 98/2091 | 2.10E‐07 | 2.68E − 06 | 1.82E − 06 | BCL10, IRF3, MYD88, ROCK1, RPS6KB1, RPS6KB2, SEPTIN3, HKDC1, MAPK8, CHUK, PFN2, CBX3, IKBKG, MAPK1, RAC1, PRKCD, ACTG1, UBE2D3, CDC42, CRKL, PFN3, RBX1, TBK1, CYCS, GSK3B, MYL12A, AKT1, MAPK14, RHOA, RNF31, ARPC3, ARPC5, PPID, UBE2V2, ARHGEF2, CAST, SEPTIN2, ARPC2, ARPC5L, CUL1, ELMO2, FNBP1L, GLMN, PFN1, RELA, ROCK2, RPS6KA5, UBA52, VDAC1, ARF1, MTOR, SEPTIN11, SEPTIN9, ACTN1, PIK3R2, RPTOR, SKP1, SRC, TLN1, PLCG1, ACTN4, AKT1S1, ARPC1B, CRK, CTTN, PTK2, ACTB, NFKB1, PIK3C3, PXN, TRAF2, HK1, RIPK1, U2AF1L5, ITGB1, PLCB3, SQSTM1, DIAPH1, PLCD1, WASF1, CAPN1, EGFR, SEPTIN8, WASF2, RPS27A, BAX, WASL, ITPR3, CD44, ATG5, CAPN2, CAPNS1, IL18, ITGA5, DOCK1, TLN2, TRADD, PIK3R1 |
| 6 | hsa05132 | Salmonella infection | 106/2091 | 7.10E − 14 | 2.84E − 12 | 1.92E − 12 | ACBD3, CTNNB1, FBXO22, IRAK1, MAP2K1, MYD88, MYH9, MYL6, PAK1, DYNLL2, CYFIP2, MAPK8, CHUK, PFN2, TXN, IKBKG, MAPK1, RAC1, DYNC1I2, EXOC2, ABI1, ACTG1, CYFIP1, MAP2K2, CDC42, FADD, PFN3, BRK1, CYCS, MYL12A, AKT1, CSE1L, MAPK14, PKN1, RHOA, VPS39, ARPC3, ARPC5, KPNA1, KPNA4, RAB5B, ARPC2, ARPC5L, CASP8, DYNC1LI1, MAP2K4, PFN1, RELA, ROCK2, ARF1, FHOD1, KPNA3, AHNAK, ARF6, DNM2, FLNB, HSP90AB1, SKP1, VPS11, DYNC1H1, FLNA, MYH10, VPS18, ARPC1B, RAB9A, ACTB, EXOC7, MAP2K3, NCKAP1, NFKB1, PIK3C3, RAB5C, DYNLT1, TRAF2, DYNLRB2, RIPK1, MYH3, RPS3, DYNC1LI2, EXOC5, SNX9, KLC2, RHOB, RRAS, AHNAK2, RAB7A, CASP3, EXOC4, BAX, CASP7, FYCO1, RAB5A, S100A10, DYNLT3, WASL, GSDMD, MYO6, ARHGEF26, RALA, STX10, IL18, FLNC, VPS16, M6PR, VPS33A, TRADD |
| 7 | hsa05134 | Legionellosis | 26/2091 | 1.05E − 03 | 5.89E − 03 | 3.99E − 03 | APAF1, HSPA1L, MYD88, EEF1A2, HSPA8, CYCS, HBS1L, BCL2L13, RAB1A, CASP8, RELA, SAR1B, ARF1, EEF1A1, EEF1G, HSPA1B, RAB1B, HSPD1, NFKB1, SEC22B, SAR1A, CASP3, CASP7, HSPA6, NFKB2, IL18 |
| 8 | hsa05135 | Yersinia infection | 54/2091 | 4.64E − 06 | 4.58E − 05 | 3.10E − 05 | IRAK1, IRF3, MAP2K1, MYD88, ROCK1, WIPF2, FN1, ARHGEF12, BAIAP2, RAC2, ARHGEF28, MAPK8, CHUK, IKBKG, MAPK1, RAC1, ACTG1, MAP2K2, CDC42, CRKL, TBK1, GSK3B, AKT1, MAPK14, PKN1, RHOA, ACTR3, MAP2K4, RELA, ROCK2, RPS6KA3, ARF6, PIK3R2, SRC, PLCG1, ACTR2, CRK, PTK2, ACTB, MAP2K3, NFKB1, PXN, TRAF2, ARHGEF1, ITGB1, PKN2, VAV2, WASF2, WASL, IL18, ITGA5, RPS6KA1, DOCK1, PIK3R1 |
| 9 | hsa05163 | Human cytomegalovirus infection | 85/2091 | 5.57E − 05 | 4.15E − 04 | 2.81E − 04 | CTNNB1, GNAO1, GNG2, HLA‐B, HLA‐C, IRF3, MAP2K1, PRKCB, ROCK1, RPS6KB1, RPS6KB2, PPP3R1, ARHGEF12, PPP3CA, PPP3CB, TSC2, RAC2, CHUK, IKBKG, HLA‐A, MAPK1, RAC1, GNB1, GNB2, CREB1, PRKCA, MAP2K2, STAT3, CCND1, CRKL, FADD, TBK1, CYCS, GSK3B, PRKACA, AKT1, MAPK14, RHOA, B2M, GNG12, PRKACB, GRB2, CALR, CASP8, RELA, ROCK2, GNG5, MTOR, CDKN2A, PIK3R2, SRC, GNAI2, ITGAV, CRK, GNAQ, PTK2, RHEB, GNAI1, NFKB1, CDK6, GNAS, PXN, TRAF2, ARHGEF1, RIPK1, NRAS, GNAI3, PLCB3, EGFR, BID, CASP3, BAX, GNA11, PDIA3, CALM3, ITPR3, RB1, TAP1, GNB4, GNA13, TAP2, TAPBP, TRADD, EIF4EBP1, PIK3R1 |
| 10 | hsa05165 | Human papillomavirus infection | 107/2091 | 4.68E − 03 | 2.03E − 02 | 1.37E − 02 | COL6A1, CTNNB1, HLA‐B, HLA‐C, IRF3, MAP2K1, RPS6KB1, RPS6KB2, SPP1, TNR, FN1, THBS1, COMP, RELN, TSC2, ATP6V0D1, CHUK, DLG3, IKBKG, PPP2R5C, PRKCI, HLA‐A, ITGA1, SLC9A3R1, ATP6V0A1, ATP6V1B2, MAPK1, STAT2, ATP6V1C1, PPP2R1B, ATP6V1A, ATP6V1H, CCNA2, COL1A2, CREB1, SCRIB, MAP2K2, CCND1, CDC42, EP300, FADD, HDAC2, PPP2CB, TBK1, CHD4, GSK3B, LLGL1, PRKACA, STAT1, AKT1, PPP2R5E, PPP2R5D, PRKACB, DLG1, GRB2, TUBG1, LAMC1, PPP2R1A, PPP2R2A, ATP6V1E1, CASP8, EIF2AK2, PSMC1, RELA, ATP6V0C, MTOR, CDK2, PIK3R2, ITGAV, HDAC1, LAMA4, LAMB1, PTK2, RHEB, LAMC2, NFKB1, PARD6B, CDK6, GNAS, LLGL2, PXN, ISG15, ITGB4, PARD3, ITGB1, NRAS, COL4A1, UBR4, EGFR, ATP6V1G1, CASP3, BAX, BCAP31, ATP6V1F, ITGB5, COL4A2, ITGA2, RB1, ITGA3, ITGA5, ITGA6, UBE3A, TCIRG1, TRADD, EIF4EBP1, PIK3R1, PKM |
| 11 | hsa05169 | Epstein–Barr virus infection | 79/2091 | 2.03E − 05 | 1.63E − 04 | 1.10E − 04 | APAF1, HLA‐B, HLA‐C, IRAK1, IRF3, MYD88, MAPK8, CHUK, IKBKG, HLA‐A, NCOR2, RAC1, STAT2, CCNA2, LYN, STAT3, CCND1, FADD, HDAC2, TBK1, CYCS, STAT1, AKT1, MAPK14, B2M, PSMD4, USP7, CALR, CASP8, EIF2AK2, MAP2K4, PSMC1, RELA, SAP30, ADRM1, CDK2, PIK3R2, PSMC2, PSMC4, PSMC5, SNW1, HDAC1, PSMC3, PSMC6, PSMD1, MAP2K3, NFKB1, PSMD11, CDK6, PSMD3, TRAF2, ISG15, PSMD12, PSMD14, PSMD2, RIPK1, PSMD13, SIN3A, BID, CASP3, PSMD8, BAX, PDIA3, PSMD7, PSMD6, DDX58, MAVS, NEDD4, NFKBIE, RB1, CD44, TAP1, NFKB2, DDB2, TAP2, TAPBP, ICAM1, TRADD, PIK3R1 |
| 12 | hsa05170 | Human immunodeficiency virus 1 infection | 91/2091 | 4.59E − 08 | 7.00E − 07 | 4.74E − 07 | GNAO1, GNG2, HLA‐B, HLA‐C, IRAK1, IRF3, MAP2K1, MYD88, PAK1, PRKCB, RPS6KB1, RPS6KB2, PPP3R1, PPP3CA, PPP3CB, RAC2, MAPK8, CHUK, SAMHD1, IKBKG, HLA‐A, MAPK1, RAC1, CCNB1, CUL5, GNB1, GNB2, AP1B1, APOBEC3C, PRKCA, RNF7, MAP2K2, PAK4, CRKL, FADD, RBX1, TBK1, CYCS, AKT1, CFL1, ELOC, MAPK14, PAK2, B2M, GNG12, CDK1, CFL2, DCAF1, CALR, CASP8, CUL1, RELA, GNG5, MTOR, AP1M1, PIK3R2, SKP1, GNAI2, PLCG1, AP1S1, CRK, CUL4B, GNAQ, PTK2, DDB1, GNAI1, MAP2K3, NFKB1, PXN, TRAF2, APOBEC3B, CUL4A, RIPK1, NRAS, ELOB, GNAI3, AP1G1, BID, CASP3, BAX, GNA11, PDIA3, CALM3, AP1G2, ITPR3, TAP1, GNB4, TAP2, TAPBP, TRADD, PIK3R1 |
| 13 | hsa05203 | Viral carcinogenesis | 78/2091 | 6.98E − 05 | 4.97E − 04 | 3.37E − 04 | GTF2H3, HDAC3, HLA‐B, HLA‐C, IRF3, MAD1L1, VAC14, GTF2E2, ATP6V0D1, IKBKG, GTF2A2, HLA‐A, MAPK1, RAC1, GTF2E1, CCNA2, CREB1, LYN, SCRIB, STAT3, YWHAG, CCND1, CDC42, EP300, HDAC2, YWHAH, CHD4, GSN, PRKACA, DNAJA3, MRPS18B, RHOA, YWHAZ, GTF2B, PRKACB, RANBP1, CDK1, DLG1, GRB2, HNRNPK, YWHAQ, SCIN, USP7, VDAC3, CASP8, EIF2AK2, PSMC1, RELA, DDX3X, ACTN1, CDK2, CDKN2A, PIK3R2, SRC, SND1, SNW1, ACTN4, GTF2H4, HDAC1, HDAC7, DDB1, NFKB1, CDK6, PXN, TRAF2, NRAS, SP100, UBR4, CASP3, BAX, HDAC6, GTF2A1, RB1, NFKB2, UBE3A, TRADD, PIK3R1, PKM |
3.2. SARS‐CoV‐2‐related proteins regulated by ivermectin
The reported SARS‐CoV‐2‐related genes were searched by GenCLiP3, and 284 SARS‐CoV‐2‐related genes were summarized (Table S2). Furthermore, 52 SARS‐CoV‐2‐related proteins were regulated by ivermectin (Table 2), including AKT1, ALB, ANPEP, APOE, APP, ATG5, B2M, BSG, CASP3, CAV1, CDC42, COL4A2, COPB2, CTSB, EEF1A2, EGFR, G6PD, HLA‐A, HMGB1, HSPA4, IDH2, IFITM3, IL18, ITCH, ITPA, KPNA2, KPNB1, LIMS1, MAPK1, MAPK14, MAPK8, MB, MGMT, MTOR, NFKB1, PAK1, PARP1, PML, PPP1CA, PRKAA1, RB1, SARS, SARS2, SLTM, STAT1, TGFB1, TIMM8A, TRPV1, UBL5, ZC3HAV1, HMOX1, and IL1F10. Those ivermectin‐regulated SARS‐CoV‐2‐related proteins included 50 proteins with the decreased abundance in ivermectin‐treated TOV‐21G group (SILAC: H) compared with normal TOV‐21G group (SILAC: L) with the abundance ratio (H/L) < 1 and two (HMOX1 and IL1F10) proteins with the increased abundance in ivermectin‐treated TOV‐21G group (SILAC: H) compared with normal TOV‐21G group (SILAC: L), with an increased abundance ratio (H/L) > 1, identified with SILAC quantitative proteomics.
Table 2.
SARS‐CoV‐2/COVID‐19‐related proteins identified in ovarian cancer cells treated with (SILAC: H) and without (SILAC: L) ivermectin
| Protein ID | Gene name | Protein name | Peptides | Unique peptides | Sequence coverage (%) | Mol. weight (kDa) | Sequence length | Score | q value | Intensity H | Intensity L | Ratio H/L |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| B0LPE5 | AKT1 | Nonspecific serine/threonine protein kinase | 4 | 4 | 10.2 | 55.6 | 480 | 6.8 | 0.00E + 00 | 17,030,000 | 58,303,000 | 0.42 |
| A0A0C4DGB6 | ALB | Serum albumin | 8 | 0 | 11.1 | 66.5 | 585 | 4.0 | 0.00E + 00 | 0 | 36,434,000 | – |
| A0A024RC61 | ANPEP | Aminopeptidase | 23 | 23 | 27.1 | 109.5 | 967 | 185.9 | 0.00E + 00 | 5,262,400,000 | 6,701,300,000 | 0.77 |
| A0A0S2Z3D5 | APOE | Apolipoprotein E isoform 1 (Fragment) | 3 | 3 | 12.3 | 36.2 | 317 | 8.0 | 0.00E + 00 | 18,205,000 | 47,756,000 | 0.62 |
| E9PG40 | APP | Amyloid‐β precursor protein | 6 | 6 | 11.8 | 80.8 | 714 | 11.3 | 0.00E + 00 | 57,717,000 | 129,820,000 | 0.55 |
| A0A2R8Y718 | ATG5 | Autophagy protein 5 | 3 | 3 | 15.0 | 27.7 | 233 | 3.8 | 0.00E + 00 | 32,604,000 | 46,003,000 | 0.68 |
| B2MG | B2M | β‐2‐Microglobulin | 3 | 3 | 34.4 | 13.9 | 122 | 31.5 | 0.00E + 00 | 530,490,000 | 1,221,900,000 | 0.43 |
| BASI | BSG | Basigin | 8 | 5 | 37.8 | 28.4 | 262 | 82.0 | 0.00E + 00 | 2095,000,000 | 4,575,700,000 | 0.41 |
| CASP3 | CASP3 | Caspase‐3 | 5 | 5 | 17.7 | 31.6 | 277 | 8.9 | 0.00E + 00 | 133,130,000 | 284,300,000 | 0.58 |
| Q2TNI1 | CAV1 | Caveolin | 4 | 1 | 25.3 | 20.5 | 178 | 29.4 | 0.00E + 00 | 310,770,000 | 997,490,000 | 0.58 |
| A0A024RAE4 | CDC42 | Cell division cycle 42 (GTP binding protein, 25 kDa), isoform CRAa | 5 | 2 | 31.4 | 21.3 | 191 | 14.7 | 0.00E + 00 | 1,085,900,000 | 3,109,100,000 | 0.40 |
| CO4A2 | COL4A2 | Collagen α‐2(IV) chain | 3 | 3 | 2.5 | 167.6 | 1712 | 20.5 | 0.00E + 00 | 85,865,000 | 171,350,000 | 0.64 |
| COPB2 | COPB2 | Coatomer subunit β | 25 | 25 | 38.6 | 102.5 | 906 | 168.3 | 0.00E + 00 | 2,000,400,000 | 4,163,000,000 | 0.58 |
| B4DMY4 | CTSB | cDNA FLJ59133, highly similar to Cathepsin B | 6 | 6 | 33.9 | 26.9 | 245 | 33.9 | 0.00E + 00 | 445,650,000 | 1,172,100,000 | 0.50 |
| A0A2U3TZH3 | EEF1A2 | Elongation factor 1‐α | 13 | 3 | 36.3 | 54.3 | 496 | 43.1 | 0.00E + 00 | 223,680,000 | 767,560,000 | 0.30 |
| E7BSV0 | EGFR | Receptor protein‐tyrosine kinase | 10 | 10 | 10.7 | 125.8 | 1136 | 33.5 | 0.00E + 00 | 152,970,000 | 203,050,000 | 0.57 |
| Q0PHS4 | G6PD | Glucose‐6‐phosphate dehydrogenase (Fragment) | 4 | 2 | 58.3 | 7.2 | 60 | 10.7 | 0.00E + 00 | 181,720,000 | 418,590,000 | 0.39 |
| D9UB05 | HLA‐A | MHC class I antigen | 11 | 0 | 35.9 | 40.9 | 365 | 4.0 | 0.00E + 00 | 6,615,900 | 14,287,000 | 0.34 |
| Q5T7C4 | HMGB1 | High mobility group protein B1 | 11 | 7 | 54.4 | 18.3 | 158 | 97.6 | 0.00E+00 | 2,916,000,000 | ########### | 0.40 |
| Q6FH11 | HMOX1 | Heme oxygenase | 8 | 8 | 37.2 | 32.8 | 288 | 80.0 | 0.00E + 00 | 2,721,500,000 | 1,934,900,000 | 1.37 |
| HSP74 | HSPA4 | Heat shock 70 kDa protein 4 | 31 | 29 | 47.1 | 94.3 | 840 | 323.3 | 0.00E + 00 | 4,012,100,000 | 9,034,800,000 | 0.43 |
| IDHP | IDH2 | Isocitrate dehydrogenase [NADP], mitochondrial | 18 | 17 | 48.0 | 50.9 | 452 | 174.3 | 0.00E + 00 | 1,281,200,000 | 2,994,300,000 | 0.46 |
| IFM3 | IFITM3 | Interferon‐induced transmembrane protein 3 | 3 | 2 | 48.9 | 14.6 | 133 | 38.0 | 0.00E + 00 | 55,750,000 | 542,360,000 | 0.13 |
| A0A024R3E0 | IL18 | Interleukin‐18 | 2 | 2 | 9.0 | 21.9 | 189 | 2.6 | 1.26E − 03 | 154,860,000 | 261,690,000 | 0.69 |
| IL1FA | IL1F10 | Interleukin‐1 family member 10 | 1 | 1 | 5.3 | 16.9 | 152 | ‐2.0 | 1.00E + 00 | 31,451,000 | 27,990,000 | 1.13 |
| Q59ER4 | ITCH | Itchy homolog E3 ubiquitin protein ligase variant (Fragment) | 9 | 9 | 20.2 | 71.4 | 605 | 22.9 | 0.00E + 00 | 36,320,000 | 155,710,000 | 0.52 |
| A0A0S2Z3W7 | ITPA | Nucleotide diphosphatase (Fragment) | 4 | 4 | 25.3 | 21.4 | 194 | 7.5 | 0.00E + 00 | 115,320,000 | 316,960,000 | 0.36 |
| Q7Z726 | KPNA2 | Importin subunit α | 16 | 16 | 37.2 | 57.9 | 529 | 239.8 | 0.00E + 00 | 1,355,200,000 | 5,127,000,000 | 0.28 |
| IMB1 | KPNB1 | Importin subunit β1 | 28 | 28 | 40.6 | 97.2 | 876 | 323.3 | 0.00E + 00 | 4,067,400,000 | 8,484,500,000 | 0.49 |
| LIMS1 | LIMS1 | LIM and senescent cell antigen‐like‐containing domain protein 1 | 8 | 8 | 31.7 | 37.3 | 325 | 23.4 | 0.00E + 00 | 206,310,000 | 633,960,000 | 0.41 |
| Q1HBJ4 | MAPK1 | Mitogen‐activated protein kinase | 16 | 12 | 52.2 | 41.4 | 360 | 126.6 | 0.00E + 00 | 608,480,000 | 2,451,900,000 | 0.35 |
| A0A024RD15 | MAPK14 | Mitogen‐activated protein kinase | 5 | 5 | 16.1 | 41.3 | 360 | 16.3 | 0.00E + 00 | 36,701,000 | 134,140,000 | 0.42 |
| B5BUB8 | MAPK8 | Mitogen‐activated protein kinase (Fragment) | 2 | 2 | 7.6 | 44.3 | 384 | 4.0 | 0.00E + 00 | 10,294,000 | 54,886,000 | 0.31 |
| U6FM64 | MB | Myoglobin (Fragment) | 1 | 1 | 45.7 | 3.9 | 35 | 3.8 | 0.00E + 00 | 0 | 39,839,000 | – |
| B4DEE8 | MGMT | Methylated‐DNA‐‐protein‐cysteine methyltransferase | 4 | 4 | 21.8 | 25.1 | 238 | 36.0 | 0.00E + 00 | 26,168,000 | 181,230,000 | 0.43 |
| MTOR | MTOR | Serine/threonine‐protein kinase mTOR | 9 | 9 | 4.7 | 288.9 | 2549 | 13.7 | 0.00E + 00 | 52,206,000 | 144,780,000 | 0.47 |
| A0A494C157 | NFKB1 | Nuclear factor NF‐κB p105 subunit (fragment) | 3 | 3 | 14.6 | 27.6 | 253 | 17.0 | 0.00E + 00 | 27,964,000 | 159,750,000 | 0.51 |
| E9PM17 | PAK1 | Serine/threonine‐protein kinase PAK1 | 5 | 1 | 16.0 | 50.9 | 455 | 2.4 | 1.98E − 03 | 0 | 19,986,000 | – |
| A0A024R3T8 | PARP1 | Poly [ADP‐ribose] polymerase | 26 | 26 | 32.3 | 111.1 | 993 | 123.8 | 0.00E + 00 | 813,590,000 | 1,981,300,000 | 0.53 |
| PML | PML | Protein PML | 3 | 3 | 4.2 | 97.6 | 882 | 8.4 | 0.00E + 00 | 7,258,200 | 30,941,000 | 0.43 |
| PP1A | PPP1CA | Serine/threonine‐protein phosphatase PP1‐α catalytic subunit | 17 | 5 | 52.4 | 37.5 | 330 | 245.9 | 0.00E + 00 | 3,461,200,000 | 7,539,100,000 | 0.47 |
| AAPK1 | PRKAA1 | 5‐AMP‐activated protein kinase catalytic subunit α1 | 5 | 5 | 10.2 | 64.0 | 559 | 10.7 | 0.00E + 00 | 63,860,000 | 150,730,000 | 0.64 |
| A0A3B3IS71 | RB1 | Retinoblastoma‐associated protein | 6 | 6 | 8.5 | 104.2 | 907 | 14.6 | 0.00E + 00 | 8,727,900 | 98,240,000 | 0.64 |
| Q0VGA5 | SARS | SARS protein | 12 | 12 | 30.3 | 58.4 | 511 | 149.3 | 0.00E + 00 | 1,216,000,000 | 2,647,300,000 | 0.53 |
| M0QWZ7 | SARS2 | Serine—tRNA ligase, mitochondrial | 4 | 4 | 12.2 | 58.2 | 518 | 35.8 | 0.00E + 00 | 77,202,000 | 153,790,000 | 0.44 |
| H0YLW7 | SLTM | SAFB‐like transcription modulator | 1 | 1 | 30.6 | 6.6 | 62 | 23.4 | 0.00E + 00 | 0 | 0 | / |
| STAT1 | STAT1 | Signal transducer and activator of transcription 1‐α/β | 18 | 18 | 32.8 | 87.3 | 750 | 112.0 | 0.00E + 00 | 617,100,000 | 1,708,900,000 | 0.41 |
| A0A499FJK2 | TGFB1 | Transforming growth factor β | 4 | 4 | 11.8 | 44.3 | 390 | 5.7 | 0.00E + 00 | 36,764,000 | 91,605,000 | 0.53 |
| TIM8A | TIMM8A | Mitochondrial import inner membrane translocase subunit Tim8A | 2 | 2 | 22.7 | 11.0 | 97 | 6.2 | 0.00E + 00 | 38,301,000 | 81,190,000 | 0.35 |
| B3KP31 | TRPV1 | cDNA FLJ31047 fis, clone HSYRA2000424, very similar to carbohydrate kinase‐like protein | 5 | 5 | 13.8 | 51.5 | 478 | 12.6 | 0.00E + 00 | 29,446,000 | 40,309,000 | 0.60 |
| A0A024R7B0 | UBL5 | Testicular tissue protein Li 217 | 1 | 1 | 12.3 | 8.5 | 73 | 2.5 | 1.50E − 03 | 33,400,000 | 156,360,000 | 0.20 |
| ZCCHV | ZC3HAV1 | Zinc finger CCCH‐type antiviral protein 1 | 13 | 13 | 23.8 | 101.4 | 902 | 50.6 | 0.00E + 00 | 159,350,000 | 339,740,000 | 0.62 |
Note: – means the protein expressed in L group but not in H group Protein ID.
The protein‐protein interacted molecules with a coexpression score more than 0.7 were selected to construct PPI network of 284 SARS‐CoV‐2‐related genes (Figure 1a and Table S3). The entire PPI network was further analyzed using MCODE, which found three modules (module 1 score = 18, module 2 score = 11, and module 3 score = 7; Figure 1b–d). Those hub molecules assisted in improving the understanding of the key molecular mechanisms underlying SARS‐CoV‐2. The hub module 1 was mainly involved in cytokines and growth factor family, including 20 hub molecules IL1B, IL1A, IL18, CCL2, CCL3, IL13, CCL4, CXCL8, CCL5, TNF, IL10, IL7, IL5, CSF2, CSF3, TLR2, IFNG, CXCL10, IL4, and IL17A. The hub module 2 was mainly involved in MAP kinase family and G‐protein family, including 23 hub molecules MAPK14, MAPK3, VEGFA, AGT, AGTR2, MAPK1, IL6R, STAT1, MAPK8, P2RY12, JUN, KNG1, JAK1, EGFR, JAK2, CRP, PTGS2, HMOX1, C3, IL6, TLR4, IL2, and SAA1. The hub module 3 was mainly involved in HLA class proteins, including 7 hub molecules HLA‐A, HLA‐DRB1, B2M, NCAM1, PML, HLA‐DPB1, and HLA‐E. Furthermore, 52 SARS‐CoV‐2‐related proteins that were regulated by ivermectin were also used to construct a PPI network (Figure 1e). Some important molecules were well‐known to play crucial roles in proliferation and growth (MAPK1, MAPK14, MAPK8, EGFR, and PAK1), autophagy (AKT1, MTOR, and ATG5), and inflammation (IL18, IL1F10, STAT1, TGFB1, and NFKB1). It should be concerned that those ivermectin‐regulated SARS‐CoV‐2‐related proteins located in the center of PPI network.
Figure 1.
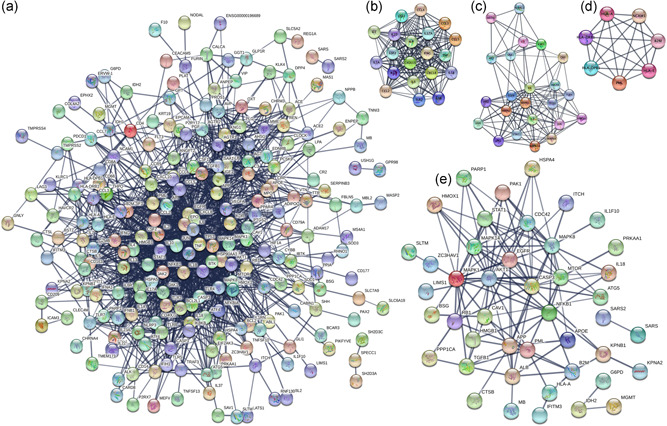
Construction of the PPI network. (a) Construction of the PPI network of 284 SARS‐CoV‐2‐related genes with a co‐expression score more than 0.7. (b–d) MCODE analysis of the entire PPI network identified three modules (module 1 score = 18, module 2 score = 11, and module 3 score = 7). (e) Construction of the PPI network of 52 ivermectin‐regulated SARS‐CoV‐2‐related proteins with co‐expression score more than 0.7. PPI, protein–protein interaction; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2
3.3. Cellular process changes of 52 SARS‐CoV‐2‐related proteins regulated by ivermectin
Gene Ontology (GO) enrichment analysis of SARS‐CoV‐2‐related proteins regulated by ivermectin obtained many cellular process changes (Figure 2 and Table S4), including 11 statistically significant BPs, 10 cellular components (CCs), and 13 MFs. BP analysis showed that many SARS‐COV‐2‐related proteins were enriched in protein import, regulation of reactive oxygen species metabolic process, muscle cell proliferation, positive regulation of binding, interaction with the host, cellular response to lipopolysaccharide, cellular response to a metal ion, positive regulation of intracellular protein transport, myeloid cell homeostasis, homeostasis of a number of cells, and erythrocyte homeostasis (Figure 2a). CC analysis showed that many SARS‐CoV‐2‐related proteins were enriched in caveola, early endosome membrane, specific granule lumen, platelet α granule, platelet α granule lumen, ficolin‐1‐rich granule, ficolin‐1‐rich granule lumen, endosome lumen, COPII‐coated ER to Golgi transport vesicle, and phagocytic vesicle membrane (Figure 2b). MF analysis showed that many SARS‐CoV‐2‐related proteins were enriched in phosphoprotein binding, negative regulation of protein binding, lipid kinase activity, regulation of lipid kinase activity, positive regulation of binding, positive regulation of protein binding, regulation of DNA binding, positive regulation of DNA binding, regulation of oxidoreductase activity, oxidoreductase activity, regulation of monooxygenase activity, nitric‐oxide synthase activity, and regulation of nitric‐oxide synthase activity (Figure 2c).
Figure 2.
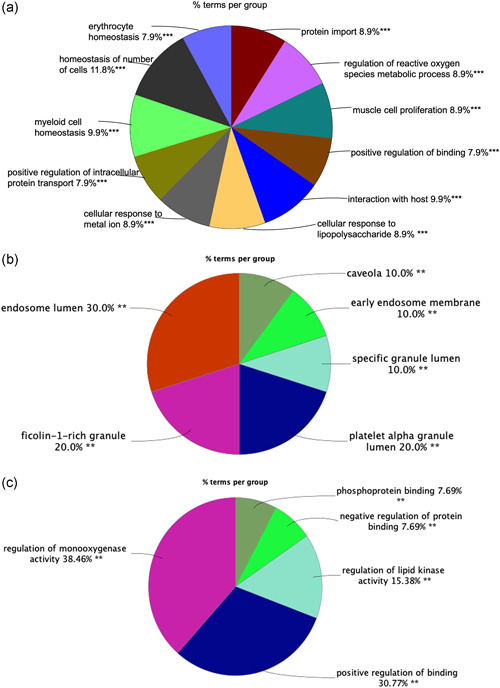
Functional and pathway enrichment analysis. (a) The biological process enrichment analysis of 52 ivermectin‐regulated SARS‐CoV‐2‐related proteins. (b) The cellular component enrichment analysis of 52 ivermectin‐regulated SARS‐CoV‐2‐related proteins. (c) The molecular function enrichment analysis of 52 ivermectin‐regulated SARS‐CoV‐2‐related proteins. Only gene sets with adjusted p value < .05 corrected with the Benjamini–Hochberg procedure were considered significant. The less p value and more significant enrichment were shown with the greater node size. The same color indicated the same function group. Among the groups, we chose a representative of the most significant term and lag highlighted. SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2
3.4. The overlap of 52 ivermectin‐regulated SARS‐CoV‐2‐related proteins among virus‐related pathways
The overlap of ivermectin‐regulated SARS‐CoV‐2‐related proteins on virus‐related pathways was constructed by Venn diagrams (Figure 3a and Table S5), and four ivermectin‐regulated SARS‐CoV‐2‐related proteins were identified among those five groups (EBV, HCMV, HIV, HPV, and SARS‐COV‐2), including HLA‐A, AKT1, NFKB1, and CASP3. SILAC quantitative proteomics analysis revealed a broad‐spectrum antiviral property of ivermectin, so further study of the overlap of ivermectin‐regulated SARS‐CoV‐2‐related proteins among virus‐related pathways might be important. Because of the importance of SARS‐COV‐2 currently, the specially SARS‐CoV‐2‐related proteins were also specifically mentioned, including ZC3HAV1, ITPA, ALB, COPB2, IL1F10, KPNB1, SLTM, HMOX1, CTSB, IDH2, LIMS1, G6PD, UBL5, TGFB1, PML, IFITM3, CAV1, SARS, ITCH, MGMT, ATG5, HSPA4, SARS2, KPNA2, PRKAA1, ANPEP, APP, MB, BSG, TRPV1, IL18, TIMM8A, PPP1CA, HMGB1, APOE, PARP1, and EEF1A2.
Figure 3.
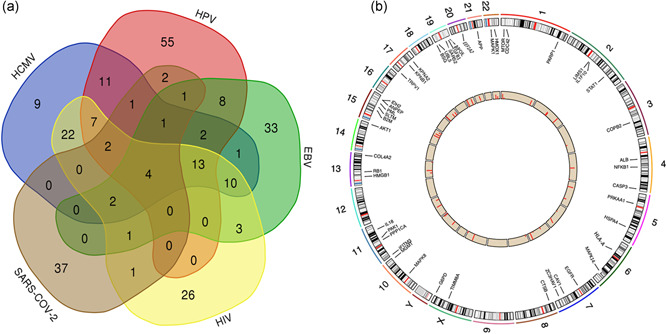
The overlapping analysis of ivermectin‐regulated SARS‐CoV‐2‐related proteins among virus‐related pathways and their chromosomal locations. (a) The overlap of ivermectin‐regulated SARS‐CoV‐2‐related proteins among virus‐related pathways was constructed by Venn diagrams. (b) The chromosomal locations corresponding with protein expression of 52 SARS‐CoV‐2‐related proteins that were regulated by ivermectin. EBV, Epstein–Barr virus; HCMV, human cytomegalovirus; HPV, human papillomavirus; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2
The chromosomal locations corresponding with protein expression of SARS‐CoV‐2‐related proteins that were regulated by ivermectin were plotted. Four ivermectin‐regulated SARS‐CoV‐2‐related proteins identified among those five groups (EBV, HCMV, HIV, HPV, and SARS‐COV‐2) were localized in different chromosomes, including HLA‐A in chromosome 6, AKT1 in chromosome 14, and NFKB1 and CASP3 in chromosome 4 (Figure 3b and Table S6).
4. DISCUSSION
Ivermectin, as an antiparasitic drug for a long time, was proved very safe in highly developed animals because the major mechanism was targeting the chloride‐dependent channels of both glutamate and γ‐aminobutyric acid that interrupts neurotransmission in invertebrates (lower developed animals). In human, a blood–brain barrier exists, which can well protect the central nervous system (Develoux, 2004). Ivermectin rarely provoked drug resistance, and most of the side effects were related to the release of antigen, not ivermectin itself (Boussinesq, 2005). The good tolerance of ivermectin was even shown in children or infants. A total of 170 infants and children (weight < 15 kg) were treated with oral ivermectin, and only seven subjects were reported mild adverse events but not very serious (Levy et al., 2020). When evaluated the existing evidence for serious events (stillbirths, spontaneous abortions, neonatal death, and congenital anomalies) after ivermectin exposure in pregnant women, 893 women with pregnancy did not report low birth weight, neonatal deaths, preterm births, or maternal morbidity, which indicated that high safety of ivermectin, but it was still insufficient evidence to conclude the certain safety of ivermectin during pregnancy (Nicolas et al., 2020). The study of pharmacokinetics for the antiparasitic drug ivermectin provided some reference values, which would be helpful for ivermectin used in other diseases. Subjects (n = 68) were treated with higher or more frequent doses than currently approved for human use (the highest FDA‐approved ivermectin dose of 200 μg/kg). The results showed that ivermectin was generally well‐tolerated even at 10 times the highest FDA‐approved dose (2000 μg/kg), and rarely appeared associated with CNS toxicity. Additionally, the mean area under the curve ratios were 1.24 and 1.40 for the 30 and 60 mg doses, respectively, which indicated that the accumulation of ivermectin was minimal (Guzzo et al., 2002). The great number of patients treated with ivermectin showed that it was a safe and well‐tolerated drug. It made ivermectin more likely to turn to great value in clinical application.
Ivermectin appeared to be a basis for the future development of antiviral agents, and many studies have been reported as a broad antiviral activity of ivermectin. For example, ivermectin caused the reduced synthesis of Chikungunya virus RNA, as well as downregulation of viral protein expression, to affect viral infectious cycle (Varghese et al., 2016). Ivermectin has nuclear transport inhibitory properties and was proved to be a broad‐spectrum inhibitor of importin α/β nuclear import through a high‐throughput screen. Further, ivermectin was able to inhibit the replication of HIV‐1 and dengue virus (Wagstaff et al., 2012). One study also demonstrated that ivermectin treatment inhibits pseudorabies virus infection by disrupting viral DNA synthesis and progeny virus production in a dose‐dependent manner. In this process, the nuclear localization of UL42 was also affected by ivermectin via targeting the nuclear localization signal pathways (Lv et al., 2018). In the present study, KEGG pathway analysis showed four virus‐related pathways, including HCMV, HPV, EBV, and HIV1 infection pathways. Those four ivermectin‐regulated virus‐related pathways totally contained 362 proteins. Many of these proteins were closely associated with the outcomes of virus infection. NFKB is a transcription regulator that is activated by various intra‐ and extracellular stimuli such as ultraviolet irradiation, oxidant‐free radicals, cytokines, and bacterial or viral products. Herpes simplex virus ICP0 protein, a viral E3 ubiquitin ligase, significantly suppressed tumor necrosis factor‐α (TNF‐α)‐mediated nuclear factor‐κB (NF‐κB) activation by binding with the p65 and p50 subunits of NF‐κB, which may contribute to pathogenesis and immune evasion of herpes simplex virus (J. Zhang, Wang, Wang, & Zheng, 2013). DDX58 encoded a protein containing RNA helicase‐DEAD box protein motifs and a caspase recruitment domain. It is involved in viral regulation of immune response and double‐stranded RNA recognition. Additionally, DDX58 mediated the transcriptional induction of other host‐derived genes and type I interferons, which lead to immunopathology alteration (Rehwinkel & Gack, 2020). EIF2AK2 encoded a serine/threonine protein kinase that is activated by autophosphorylation after binding to dsRNA. The activated form of the encoded protein can phosphorylate translation initiation factor EIF2S1, which, in turn, inhibits protein synthesis. EIF2AK2, as one of Type I interferon‐stimulated genes, showed important biological and immunological functions. In viral infections, EIF2AK2 inhibited or promoted viral replication (Wei et al., 2020). In terms of the HCMV infection pathway, a total of 85 ivermectin‐related proteins have been identified. Some of them have been reported mediated by the ivermectin in previous studies. For example, ivermectin induced apoptosis of epithelial cells through loss of mitochondrial calcium ion overload, mitochondrial membrane potential, and reactive oxygen species generation. As a mechanistic approach, ivermectin regulated cell signaling pathways, including AKT, PI3K, and MAPK pathways (Lee et al., 2019). Ivermectin also regulated cell cycle arrest at the G1 phase via downregulation of CCND1 and CDK4 to inhibit cell growth (Diao et al., 2019). In terms of HPV infection pathway, a total of 107 ivermectin‐related proteins have been identified. Some of the identified and related proteins have been reported mediated by the ivermectin in previous studies. For example, ivermectin induced apoptosis by the downregulation of BCL‐2 expression, and upregulation of BAX expression, cleaved poly [ADP‐ribose] polymerase, and CASP3 activity (Deng, Xu, Long, & Xie, 2018). Ivermectin reduced the transcription of P‐glycoprotein by bounding with the extracellular domain of the EGFR to inhibit the activation of EGFR and its downstream signaling, not by directly inhibiting P‐glycoprotein activity (Jiang, Wang, Sun, & Wu, 2019). In terms of EBV infection pathway, a total of 79 ivermectin‐related proteins have been identified. Some of them have been reported mediated by ivermectin in previous studies. For example, ivermectin was proved to inhibit nitric oxide synthase and cyclooxygenase‐2 enzymes by inhibiting phosphorylation of mitogen‐activated protein kinases (MAPK8) after stimulated cells with LPS (X. Zhang et al., 2009). Ivermectin could be from an antiparasitic agent to a repositioned antibacterial, antiviral, and anticancer drug because ivermectin interacts with multitargeted, including certain epigenetic deregulator SIN3A (Juarez, Schcolnik‐Cabrera, & Dueñas‐Gonzalez, 2018). In terms of HIV1 infection pathway, a total of 91 ivermectin‐related proteins have been identified. Some of them have been reported mediated by ivermectin in previous studies. For example, ivermectin‐induced autophagy was associated with decreased P21‐activated kinase 1 (PAK1) expression via the ubiquitination‐mediated degradation pathway (Dou et al., 2016).
Due to the outbreak and pandemic of SARS‐CoV‐2, the whole world is concerned about this public health emergency. Epidemiological studies showed that SARS‐CoV‐2 had a quick transmission, and it estimated that each infection might result in 1.4 to 3.9 new infections when no preventive measures are taken (Benvenuto et al., 2020). The virus primarily spreads through close contact or respiratory droplets. Many researchers proved that SARS‐CoV‐2 could bind to the receptor angiotensin‐converting enzyme 2 (ACE2) to enter human cells (Letko, Marzi, & Munster, 2020). Ivermectin, an FDA‐approved antiparasitic drug, was reported many times in recent studies as an inhibitor of the SARS‐CoV‐2 (Caly et al., 2020). Ivermectin mediated viral import by inhibiting the importin (IMPα/β1) and creating the acidic environment (Caly et al., 2020). Caly et al. reported a 5000‐fold reduction between the ivermectin treatment group (5 μM ivermectin) and the control group in SARS‐CoV‐2 RNA levels. The IC50 of ivermectin for the SARS‐CoV‐2 was calculated at approximately 2.5 μM. According to previous pharmacokinetic studies in healthy volunteers, it suggested that single doses up to 120 mg of ivermectin proved to be safe and well‐tolerated (Chaccour, Hammann, Ramón‐García, & Rabinovich, 2020). In recent study, quantitative translatomics and SILAC‐based proteomics identified the signaling pathway profile of the cellular responses to SARS‐CoV‐2 infection in human colon epithelial carcinoma cell line, including glycolysis, translation, splicing, proteostasis, and nucleotide synthesis (Bojkova et al., 2020). In this study, SILAC was used to analyze the human ovarian cancer cell line TOV‐21G. After 10 passages, TOV‐21G cells were treated by 20 μmol/L ivermectin for 24 h. Interestingly, compared with reported SARS‐CoV‐2/COVID‐19‐related genes from GencLip3 (n = 284), we identified 52 SARS‐CoV‐2/COVID‐19‐related protein alterations when treated with and without ivermectin. For example, CD147 (BSG)‐encoded protein was also a member of the immunoglobulin superfamily, and the reported possible direct viral invasion of progenitor/stem cells was via CD147 (BSG; Ulrich & Pillat, 2020). RB1 was a negative regulator of the cell cycle and was the first tumor suppressor gene found. Structural homology with SARS‐CoV‐1 indicated that SARS‐CoV‐2 might directly impair pRb. Considering preeminent inflammatory response and strong oxidative stress by SARS‐CoV‐2, whether SARS‐CoV‐2 would be associated with high carcinogenic risk should be watched for long periods (Alpalhão, Ferreira, & Filipe, 2020). Expression of elevated levels of pro‐inflammatory cytokines was closely related to the acute lung injury and pathogenesis in SARS‐CoV‐infected patients, including IL‐1β, MCP‐1, IL‐6, TNF‐α, and TGF‐β1 (He et al., 2006). Our data also identified that ivermectin‐regulated key interleukins in SARS‐CoV‐2‐induced cytokine storm, such as TNFB1, IL18, and IL1F10. Ivermectin seemed to potentially act against novel coronavirus infection. We provided mechanisms of ivermectin used in the treatment of SARS‐CoV‐2 infection.
5. CONCLUSION
This study, to best of our knowledge, was the first to provide ivermectin‐regulated virus‐related pathways by SILAC quantitative proteomics analysis, which revealed a broad‐spectrum antiviral property of ivermectin. More exciting thing was that the identified ivermectin‐regulated proteins included some reported SARS‐CoV‐2‐related proteins, and it could assist in exploiting potential ivermectin‐related biomarkers and the novel mechanisms in the treatment of SARS‐CoV‐2 infection. The combination of ivermectin with other drugs might result in more favorable prognoses for patients with COVID‐19. For example, one study hypothesized that the combination of hydroxychloroquine and ivermectin might show a consequential and synergistic action for treatment of COVID‐19 (Patrì & Fabbrocini, 2020). We anticipate our results to guide efforts to understand the molecular mechanisms underlying ivermectin used for the treatment of SARS‐CoV‐2 infection. Furthermore, our findings provide insight into the development of ivermectin as an option for the treatment of COVID‐19 in the context of PPPM research and practice.
CONFLICT OF INTERESTS
The authors have declared that no competing interests exist.
AUTHOR CONTRIBUTIONS
Na Li performed SILAC cell experiments, analyzed the data, prepared figures and tables, and drafted the manuscript. Lingfeng Zhao participated in bioinformatics analysis. Xianquan Zhan conceived the concept, guided experiments and data analysis, supervised results, wrote and critically revised the manuscript, and was responsible for the financial supports and corresponding works. All authors approved the final manuscript.
Supporting information
Supporting information.
Supporting information.
Supporting information.
Supporting information.
Supporting information.
Supporting information.
Supporting information.
Supporting information.
ACKNOWLEDGMENTS
This study was supported by the Shandong First Medical University Talent Introduction Funds (to X.Z.), and the Hunan Provincial Hundred Talent Plan (to X.Z.).
Li N, Zhao L, Zhan X. Quantitative proteomics reveals a broad‐spectrum antiviral property of ivermectin, benefiting for COVID‐19 treatment. J Cell Physiol. 2021;236:2959–2975. 10.1002/jcp.30055
REFERENCES
- Abdeltawab, M. S. A. , Rifaie, S. A. , Shoeib, E. Y. , El‐Latif, H. A. A. , Badawi, M. , Salama, W. H. , & El‐Aal, A. A. A. (2020). Insights into the impact of Ivermectin on some protein aspects linked to Culex pipiens digestion and immunity. Parasitology Research, 119(1), 55–62. [DOI] [PubMed] [Google Scholar]
- Almeida, A. M. , Queiroz, J. A. , Sousa, F. , & Sousa, Â. (2019). Cervical cancer and HPV infection: Ongoing therapeutic research to counteract the action of E6 and E7 oncoproteins. Drug Discovery Today, 24(10), 2044–2057. [DOI] [PubMed] [Google Scholar]
- Alpalhão, M. , Ferreira, J. A. , & Filipe, P. (2020). Persistent SARS‐CoV‐2 infection and the risk for cancer. Medical Hypotheses, 143, 109882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andoniou, C. E. , & Degli‐Esposti, M. A. (2006). Insights into the mechanisms of CMV‐mediated interference with cellular apoptosis. Immunology and Cell Biology, 84(1), 99–106. [DOI] [PubMed] [Google Scholar]
- Ashour, D. S. (2019). Ivermectin: From theory to clinical application. International Journal of Antimicrobial Agents, 54(2), 134–142. [DOI] [PubMed] [Google Scholar]
- Athanasiou, A. , Bowden, S. , Paraskevaidi, M. , Fotopoulou, C. , Martin‐Hirsch, P. , Paraskevaidis, E. , & Kyrgiou, M. (2020). HPV vaccination and cancer prevention. Best Practice & Research Clinical Obstetrics & Gynaecology, 65, 109–124. [DOI] [PubMed] [Google Scholar]
- Bader, G. D. , & Hogue, C. W. (2003). An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics, 4, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bindea, G. , Mlecnik, B. , Hackl, H. , Charoentong, P. , Tosolini, M. , Kirilovsky, A. , … Galon, & J. (2009). ClueGO: A cytoscape plug‐in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics, 25(8), 1091–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benvenuto, D. , Giovanetti, M. , Ciccozzi, A. , Spoto, S. , Angeletti, S. , & Ciccozzi, M. (2020). The 2019‐new coronavirus epidemic: Evidence for virus evolution. Journal of Medical Virology, 92(4), 455–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bojkova, D. , Klann, K. , Koch, B. , Widera, M. , Krause, D. , Ciesek, S. , … Münch, C. (2020). Proteomics of SARS‐CoV‐2‐infected host cells reveals therapy targets. Nature, 583(7816), 469–472. [DOI] [PubMed] [Google Scholar]
- Boussinesq, M. (2005). [Ivermectin]. Medecine tropicale, 65(1), 69–79. [PubMed] [Google Scholar]
- Britt, W. (2008). Manifestations of human cytomegalovirus infection: Proposed mechanisms of acute and chronic disease. Current Topics in Microbiology and Immunology, 325, 417–470. [DOI] [PubMed] [Google Scholar]
- Buechner, S. A. (2002). Common skin disorders of the penis. BJU International, 90(5), 498–506. [DOI] [PubMed] [Google Scholar]
- Burg, R. W. , Miller, B. M. , Baker, E. E. , Birnbaum, J. , Currie, S. A. , Hartman, R. , … Omura, S. (1979). Avermectins, new family of potent anthelmintic agents: Producing organism and fermentation. Antimicrobial Agents Chemother, 15(3), 361–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buxmann, H. , Hamprecht, K. , Meyer‐Wittkopf, M. , & Friese, K. (2017). Primary human cytomegalovirus (HCMV) infection in pregnancy. Deutsches Ärzteblatt International, 114(4), 45–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caly, L. , Druce, J. D. , Catton, M. G. , Jans, D. A. , & Wagstaff, K. M. (2020). The FDA‐approved drug ivermectin inhibits the replication of SARS‐CoV‐2 in vitro. Antiviral Research, 178, 104787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabala, J. C. , Mrozik, H. , Tolman, R. L. , Eskola, P. , Lusi, A. , Peterson, L. H. , … Ostlind, D. A. (1980). Ivermectin, a new broad‐spectrum antiparasitic agent. Journal of Medicinal Chemistry, 23(10), 1134–1136. [DOI] [PubMed] [Google Scholar]
- Chaccour, C. , Hammann, F. , Ramón‐García, S. , & Rabinovich, N. R. (2020). Ivermectin and COVID‐19: Keeping rigor in times of urgency. American Journal of Tropical Medicine and Hygiene, 102(6), 1156–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crump, A. (2017). Ivermectin: Enigmatic multifaceted 'wonder' drug continues to surprise and exceed expectations. Journal of Antibiotics, 70(5), 495–505. [DOI] [PubMed] [Google Scholar]
- Csóka, B. , Németh, Z. H. , Szabó, I. , Davies, D. L. , Varga, Z. V. , Pálóczi, J. , … Haskó, G. (2018). Macrophage P2X4 receptors augment bacterial killing and protect against sepsis. JCI Insight, 3, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng, F. , Xu, Q. , Long, J. , & Xie, H. (2018). Suppressing ROS‐TFE3‐dependent autophagy enhances ivermectin‐induced apoptosis in human melanoma cells. Journal of Cellular Biochemistry, 120, 1702–1715. 10.1002/jcb.27490 [DOI] [PubMed] [Google Scholar]
- Develoux, M. (2004). [Ivermectin]. Annales de Dermatologie et de Vénéréologie, 131(6‐7 Pt 1), 561–570. [DOI] [PubMed] [Google Scholar]
- Diao, H. , Cheng, N. , Zhao, Y. , Xu, H. , Dong, H. , Thamm, D. H. , … Lin, D. (2019). Ivermectin inhibits canine mammary tumor growth by regulating cell cycle progression and WNT signaling. BMC Veterinary Research, 15(1), 276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou, Q. , Chen, H. N. , Wang, K. , Yuan, K. , Lei, Y. , Li, K. , … Huang, C. (2016). Ivermectin induces cytostatic autophagy by blocking the PAK1/Akt axis in breast cancer. Cancer Research, 76(15), 4457–4469. [DOI] [PubMed] [Google Scholar]
- Gallardo, F. , Mariamé, B. , Gence, R. , & Tilkin‐Mariamé, A. F. (2018). Macrocyclic lactones inhibit nasopharyngeal carcinoma cells proliferation through PAK1 inhibition and reduce in vivo tumor growth. Drug Design, Development and Therapy, 12, 2805–2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzzo, C. A. , Furtek, C. I. , Porras, A. G. , Chen, C. , Tipping, R. , Clineschmidt, C. M. , … Lasseter, K. C. (2002). Safety, tolerability, and pharmacokinetics of escalating high doses of ivermectin in healthy adult subjects. Journal of Clinical Pharmacology, 42(10), 1122–1133. [DOI] [PubMed] [Google Scholar]
- He, L. , Ding, Y. , Zhang, Q. , Che, X. , He, Y. , Shen, H. , … Jiang, S. (2006). Expression of elevated levels of pro‐inflammatory cytokines in SARS‐CoV‐infected ACE2+ cells in SARS patients: Relation to the acute lung injury and pathogenesis of SARS. Journal of Pathology, 210(3), 288–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holder, K. A. , & Grant, M. D. (2019). Human cytomegalovirus IL‐10 augments NK cell cytotoxicity. Journal of Leukocyte Biology, 106(2), 447–454. [DOI] [PubMed] [Google Scholar]
- Hoppe‐Seyler, K. , Bossler, F. , Braun, J. A. , Herrmann, A. L. , & Hoppe‐Seyler, F. (2018). The HPV E6/E7 Oncogenes: Key factors for viral carcinogenesis and therapeutic targets. Trends in Microbiology, 26(2), 158–168. [DOI] [PubMed] [Google Scholar]
- Huynh, K. , & Gulick, P. G. (2020). HIV prevention. Treasure Island, FL: StatPearls Publishing. PMID: 29261888. [PubMed] [Google Scholar]
- Jiang, L. , Wang, P. , Sun, Y. J. , & Wu, Y. J. (2019). Ivermectin reverses the drug resistance in cancer cells through EGFR/ERK/Akt/NF‐κB pathway. Journal of Experimental and Clinical Cancer Research, 38(1), 265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juarez, M. , Schcolnik‐Cabrera, A. , & Dueñas‐Gonzalez, A. (2018). The multitargeted drug ivermectin: From an antiparasitic agent to a repositioned cancer drug. American Journal of Cancer Research, 8(2), 317–331. [PMC free article] [PubMed] [Google Scholar]
- Lai, C. C. , Shih, T. P. , Ko, W. C. , Tang, H. J. , & Hsueh, P. R. (2020). Severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) and coronavirus disease‐2019 (COVID‐19): The epidemic and the challenges. International Journal of Antimicrobial Agents, 55(3), 105924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laing, R. , Gillan, V. , & Devaney, E. (2017). Ivermectin—Old drug, new tricks? Trends Parasitol, 33(6), 463–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J. Y. , Lim, W. , Ham, J. , Kim, J. , You, S. , & Song, G. (2019). Ivermectin induces apoptosis of porcine trophectoderm and uterine luminal epithelial cells through loss of mitochondrial membrane potential, mitochondrial calcium ion overload, and reactive oxygen species generation. Pesticide Biochemistry and Physiology, 159, 144–153. [DOI] [PubMed] [Google Scholar]
- Letko, M. , Marzi, A. , & Munster, V. (2020). Functional assessment of cell entry and receptor usage for SARS‐CoV‐2 and other lineage B betacoronaviruses. Nature Microbiology, 5(4), 562–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy, M. , Martin, L. , Bursztejn, A. C. , Chiaverini, C. , Miquel, J. , Mahé, E. , … Boralevi, F. (2020). Ivermectin safety in infants and children under 15 kg treated for scabies: A multicentric observational study. British Journal of Dermatology, 182(4), 1003–1006. [DOI] [PubMed] [Google Scholar]
- Li, H. , Zhou, Y. , Zhang, M. , Wang, H. , Zhao, Q. , & Liu, J. (2020). Updated Approaches against SARS‐CoV‐2. Antimicrob Agents Chemother, 64, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, N. , & Zhan, X. (2020). Anti‐parasite drug ivermectin can suppress ovarian cancer by regulating lncRNA‐EIF4A3‐mRNA axes. EPMA Journal, 11, 289–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv, C. , Liu, W. , Wang, B. , Dang, R. , Qiu, L. , Ren, J. , … Wang, X. (2018). Ivermectin inhibits DNA polymerase UL42 of pseudorabies virus entrance into the nucleus and proliferation of the virus in vitro and vivo. Antiviral Research, 159, 55–62. [DOI] [PubMed] [Google Scholar]
- Marques‐Piubelli, M. L. , Salas, Y. I. , Pachas, C. , Becker‐Hecker, R. , Vega, F. , & Miranda, R. N. (2020). Epstein–Barr virus‐associated B‐cell lymphoproliferative disorders and lymphomas: A review. Pathology, 52(1), 40–52. [DOI] [PubMed] [Google Scholar]
- Nash, D. , & Robertson, M. (2019). How to evolve the response to the global HIV epidemic with new metrics and targets based on pre‐treatment CD4 counts. Current HIV/AIDS Reports, 16(4), 304–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas, P. , Maia, M. F. , Bassat, Q. , Kobylinski, K. C. , Monteiro, W. , Rabinovich, N. R. , … Chaccour, C. (2020). Safety of oral ivermectin during pregnancy: A systematic review and meta‐analysis. Lancet Global Health, 8(1), e92–e100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussinovitch, M. , Prais, D. , Volovitz, B. , Shapiro, R. , & Amir, J. (2003). Post‐infectious acute cerebellar ataxia in children. Clinical Pediatrics, 42(7), 581–584. [DOI] [PubMed] [Google Scholar]
- Patrì, A. , & Fabbrocini, G. (2020). Hydroxychloroquine and ivermectin: A synergistic combination for COVID‐19 chemoprophylaxis and treatment? Journal of the American Academy of Dermatology, 82(6), e221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappsilber, J. , Ishihama, Y. , & Mann, M. (2003). Stop and go extraction tips for matrix‐assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Analytical Chemistry, 75(3), 663–670. [DOI] [PubMed] [Google Scholar]
- Rehwinkel, J. , & Gack, M. U. (2020). RIG‐I‐like receptors: Their regulation and roles in RNA sensing. Nature Reviews Immunology, 20, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezk, S. A. , Zhao, X. , & Weiss, L. M. (2018). Epstein–Barr virus (EBV)‐associated lymphoid proliferations, a 2018 update. Human Pathology, 79, 18–41. [DOI] [PubMed] [Google Scholar]
- Schaller, M. , Gonser, L. , Belge, K. , Braunsdorf, C. , Nordin, R. , Scheu, A. , & Borelli, C. (2017). Dual anti‐inflammatory and anti‐parasitic action of topical ivermectin 1% in papulopustular rosacea. Journal of the European Academy of Dermatology and Venereology, 31(11), 1907–1911. [DOI] [PubMed] [Google Scholar]
- Shannon‐Lowe, C. , & Rowe, M. (2014). Epstein Barr virus entry; kissing and conjugation. Current Opinion in Virology, 4, 78–84. [DOI] [PubMed] [Google Scholar]
- Szklarczyk, D. , Franceschini, A. , Wyder, S. , Forslund, K. , Heller, D. , Huerta‐Cepas, J. , … von Mering, C. (2015). STRING v10: Protein–protein interaction networks, integrated over the tree of life. Nucleic Acids Research, 43, D447–D452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich, H. , & Pillat, M. M. (2020). CD147 as a target for COVID‐19 treatment: Suggested effects of azithromycin and stem cell engagement. Stem Cell Reviews and Reports, 16(3), 434–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wyk, J. A. , & Malan, F. S. (1988). Resistance of field strains of Haemonchus contortus to ivermectin, closantel, rafoxanide and the benzimidazoles in South Africa. Veterinary Record, 123(9), 226–228. [DOI] [PubMed] [Google Scholar]
- Varghese, F. S. , Kaukinen, P. , Gläsker, S. , Bespalov, M. , Hanski, L. , Wennerberg, K. , … Ahola, T. (2016). Discovery of berberine, abamectin and ivermectin as antivirals against chikungunya and other alphaviruses. Antiviral Research, 126, 117–124. [DOI] [PubMed] [Google Scholar]
- Wagstaff, K. M. , Sivakumaran, H. , Heaton, S. M. , Harrich, D. , & Jans, D. A. (2012). Ivermectin is a specific inhibitor of importin α/β‐mediated nuclear import able to inhibit replication of HIV‐1 and dengue virus. Biochemical Journal, 443(3), 851–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J. H. , Zhao, L. F. , Wang, H. F. , Wen, Y. T. , Jiang, K. K. , Mao, X. M. , … Huang, Z. X. (2019). GenCLiP 3: Mining human genes' functions and regulatory networks from PubMed based on co‐occurrences and natural language processing. Bioinformatics, btz807. [DOI] [PubMed] [Google Scholar]
- Wei, J. , Zang, S. , Li, C. , Zhang, X. , Gao, P. , & Qin, Q. (2020). Grouper PKR activation inhibits red‐spotted grouper nervous necrosis virus (RGNNV) replication in infected cells. Developmental and Comparative Immunology, 111, 103744. [DOI] [PubMed] [Google Scholar]
- Yang, S. N. Y. , Atkinson, S. C. , Wang, C. , Lee, A. , Bogoyevitch, M. A. , Borg, N. A. , & Jans, D. A. (2020). The broad spectrum antiviral ivermectin targets the host nuclear transport importin α/β1 heterodimer. Antiviral Research, 177, 104760. [DOI] [PubMed] [Google Scholar]
- Yu, G. , Wang, L. , Han, Y. , & He, Q. (2012). ClusterProfiler: An R package for comparing biological themes among gene clusters. OMICS, 16(5), 284–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavattoni, M. , Furione, M. , Arossa, A. , Iasci, A. , Spinillo, A. , Lombardi, G. , … Baldanti, F. (2014). Diagnosis and counseling of fetal and neonatal HCMV infection. Early Human Development, 90(Suppl. 1), S29–S31. [DOI] [PubMed] [Google Scholar]
- Zhang, J. , Wang, K. , Wang, S. , & Zheng, C. (2013). Herpes simplex virus 1 E3 ubiquitin ligase ICP0 protein inhibits tumor necrosis factor alpha‐induced NF‐κB activation by interacting with p65/RelA and p50/NF‐κB1. Journal of Virology, 87(23), 12935–12948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X. , Song, Y. , Xiong, H. , Ci, X. , Li, H. , Yu, L. , … Deng, X. (2009). Inhibitory effects of ivermectin on nitric oxide and prostaglandin E2 production in LPS‐stimulated RAW 264.7 macrophages. International Immunopharmacology, 9(3), 354–359. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information.
Supporting information.
Supporting information.
Supporting information.
Supporting information.
Supporting information.
Supporting information.
Supporting information.


