Abstract
Wastewater systems are recognized pathways for the spread of antibiotic resistant bacteria, but relatively little is known about the microbial ecology of the sewer environment. Sewer biofilm colonization by antibiotic resistance gene (ARG) carrying bacteria may impact interpretations of sewage epidemiology data, water quality during sewer overflows, and hazard to utility workers. The objectives of this research were to evaluate the (1) microbiome of real and simulated sewer biofilms and their potential to accumulate ARGs and (2) susceptibility of simulated sewer biofilms to bleach disinfection. First, biofilm samples were collected from sewer municipal systems. Next, an annular biofilm reactor was used to simulate the sewer environment while controlling the pipe material (concrete vs. PVC). The reactor was operated either as fed semi-batch with sewer sediment and synthetic wastewater (Sed-SB) or fed with a continuous flow of raw sewage (WW-CF). The abundance of ARGs, human fecal marker HF183, and 16S rRNA gene copies in these biofilm samples was measured with qPCR. Amplicon sequencing was performed to compare the prokaryotic diversity between samples. Finally, the susceptibility of reactor biofilm to a 4.6% bleach disnfection protocol was evaluated using viability qPCR and amplicon sequencing. Field and WW-CF biofilms contained the most ARG copies and the microbial community compositions varied between the different biofilm samples (field, Sed-SB, and WW-CF). Pipe material did not affect the abundance of ARGs in the reactor samples. However, log removal following bleach treatment suggested that the biofilm grown on PVC surface was primarily dislodged from the surface by the bleach treatment whereas more bacteria were lysed within the biofilm that remained on the concrete surface. Viable bacteria carrying ARGs were observed following 10 minutes of treatment. This study showed that sewer biofilms can accumulate bacteria carrying ARGs and that while bleach can reduce sewer biofilm density, the protocol tested here will not completely remove the biofilms.
Graphical abstract
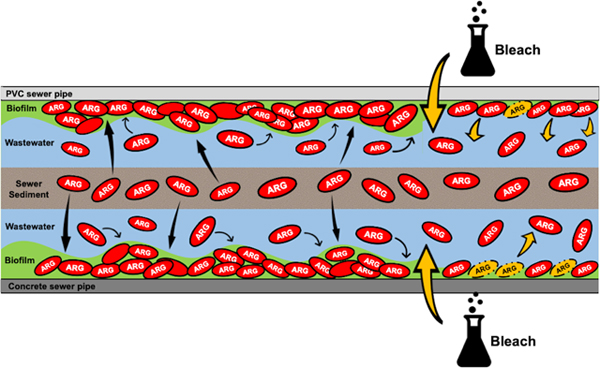
Introduction
Recent microbial disease outbreaks remind us of the need for continued study of the fate of microbial agents in sewage collection systems. In the 2019 Antibiotic Resistance Treats Report,1 the US CDC recognized wastewater systems as an environmental source of pathogenic antibiotic resistant bacteria. Better understanding of the sewer microbiome can provide insight into (1) protecting public health in the case of sewer overflows, (2) understanding utility worker exposure hazards when working in the sewer environment,2 (3) improving our interpretation of wastewater based epidemiology data,3 and (4) evaluating decontamination techniques. Sewers contain multiple matrices: the waste or stormwater being conveyed, sediments that may accumulate at joints or discontinuities, and biofilms. Bacterial concentration in sewer biofilm is on the scale of 108 and 1011 cells per gram of biofilm dry weight.4 The most studied biofilms in sewers are likely those containing the microbes responsible for pipe corrosion.5 But, there is growing interest in studying the microbial communities and antibiotic resistome of sewer sediment and biofilms in addition to the much better studied sewage matrix .6–11
Sewer pipes are made of several materials depending on the pipe size and date of installation often including concrete, clay, steel, or plastic. The pipe material is of potential interest given that it affects the microbial attachment and detachment processes. Generally, an increase in surface roughness and hydrophobicity results in an increase in microbial colonization.12–13 This has been confirmed in drinking water distribution systems14 but to our knowledge, no study has assessed the effect of pipe material in sewer biofilm development.
Of particular interest is whether sewer biofilms can support the proliferation of antibiotic resistance genes (ARGs) and the selection for ARBs. The high load of nutrient in sewers11 make these systems suitable environments for microbial growth and gene exchange. Selective pressure is present in sewers given that the concentrations of antibiotics in sewage are often above the predicted no-effect level for selection. However, fluoroquinolones in a simulated hospital sewer study were not shown to select for resistance in sewer sediments,15 likely because this antibiotic sorbed to sewer sediments and was not bioavailable. This fate is not the expected for all antibiotics which have a range of Kow’s and Koc’s. e.g.,16 With respect to what is known about the sewer biofilm resistome, a recent field analysis of a municipal sewer system in Spain detected more than 20 species of potential human pathogens and the presence of 8 ARGs in sewer biofilms, the latter with an abundance of up to one gene copy per 10 copies of 16S rRNA gene.17
Limited data is available on the impact of disinfectants on sewer biofilms besides what is known about stopping microbial corrosion processes or on sink drain U-bends.18–20 However, there is a wealth of knowledge from other systems that indicate that bacterial communities growing as biofilms have been proven to be 100 to 1,000 times more resistant to disinfecting and antimicrobial agents such as alkaline hypochlorite (1000 mg/L; pH 11), chlorosulfamate (1000 mg/L; pH 5.5), and chlorhexidine gluconate (4 % w/v), triclosan (1 % w/v), and benzalkonium chloride (1 % w/v) than planktonic cells.21–23 Disinfection of a sewer line may be desirable prior to sewer maintenance particularly in the event of a disease outbreak or bioterrorism resulting in elevated loads of of a high-risk pathogn in a sewer line. Therefore, there is a desire to learn more about potential methods for sewer biofilm disinfection.
To address the identified gaps in knowledge, a simulated sewer experiment was conducted and the results were compared to sewer biofilm samples from the field. The objectives of this research were to evaluate (1) the microbiome of real and simulated sewer biofilms and their potential to accumulate ARGs and (2) susceptibility of simulated sewer biofilms to bleach disinfection. Field sewer biofilm samples were analyzed to compare the relative abundance of selected ARGs and microbial diversity. Next, an annular biofilm reactor was used to simulate the sewer environment. The reactor study was designed to provide insight into the relative contributions of sewer sediment and wastewater to ARG concentrations and microbial diversity in sewer biofilms as a function of inoculum matrix and pipe material. Next, the effect of bleach on sewer biofilms was measured using qPCR for total and propidium monoazide treated samples for viable cell 16S rRNA gene copies, fecal indicator marker, select ARGs, and microbial diversity. Overall, the results obtained by this research indicate the potential for sewer biofilm to accumulate ARGs and provide insight into the mechanisms (detachment vs. cell membrane disruption) following standardized disinfection.
Methodology
Field Sampling
To study the abundance of ARGs and the microbial community composition of sewer biofilm, biofilm grab samples were collected from two different separate sanitary sewer systems in the eastern United States. To provide anonymity to the collaborating utilities, samples were labeled as “System 1” and “System 2” (more information is provided in Supplemental Table S1A). Six samples were collected from System 1 at least one week apart in fall of 2016 (9/7/16, 9/12/16, 9/21/16) and summer 2017 (6/29/17, 7/5/17 and 7/14/17). Given that access to several sites in System 1 was possible, these samples were each collected at a different location. Five samples were collected from the same location in System 2, due to limitations in access, two during winter 2016–2017 (12/19/16, 1/9/17) and three during summer 2017 (8/22/17, 8/28/17 and 9/19/17). These sampling dates coincided with a larger sampling campaign of five sewer systems for both wastewater and sewer sediment, Eramo et al. 2020.24 Sampling was performed by scraping a 19 cm2 portion of the biofilm from the sewer pipe using a sterile cotton swab. The swab was stored in a 2 mL O-ring tube and transported in a cooler with ice to the lab for processing.
Annular Biofilm Reactor
The sewer pipe environment was simulated using an annular biofilm reactor (Fig. 1A) to understand if pipe material and/or the inoculum source matrix (sewage vs. sewer sediments) and reactor operation has an effect on the prevalence of ARGs and the microbial community structure in sewer biofilms. The reactor consisted of an outer poly(methyl methacrylate) cylindrical tank of 3.5 L (inner diameter 17.1 cm, outer diameter 17.78 cm, 30.48 cm long) and an inner cylinder with slots for coupons of different pipe material (BST 1106 Standard Inner Cylinder for 1320 Biofilm Annular Reactor, BioSurface Technology Corporation, Bozeman, MT), similar to the one used by Sun et al. 2014.25 The inner cylinder was continuously spun at a rotational speed of 150 rpm, generating a flow velocity of 1.1 m/s which creates the shear stress consistent with the turbulent flow expected inside a sewer pipe. Coupons of polyvinylchloride (PVC) and concrete material (Bio Surface Technology Corporation, Bozeman, MT) were inserted into the inner cylinder and served as the surface for the biofilm growth.
fig. 1:
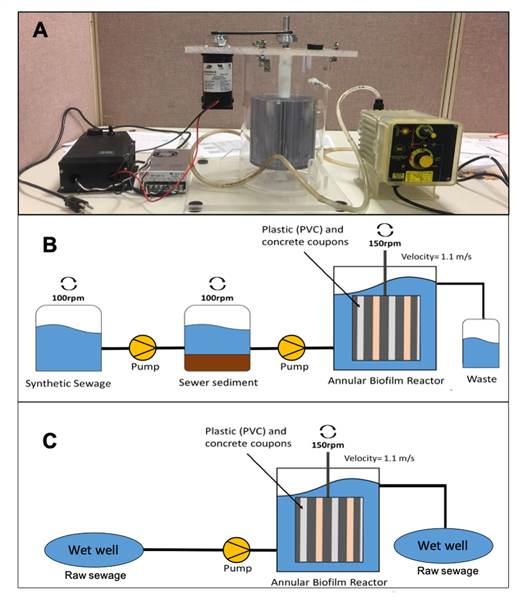
Annular biofilm reactor used to simulate sewer pipe conditions. (a) picture of the biofilm reactor system connected to the measuring pump. (b) Sed-SB biofilm reactor setup and (c) WW-CF biofilm reactor setup.
The reactor was operated for two experiments with either (1) semi-continuous batch feeding of simulated wastewater inoculated with sewer sediment (Sed-SB, Fig. 1B) or (2) continuous feeding with raw sewage (WW-CF, Fig. 1C). For the experiments with simulated wastewater and actual sewer sediment, a 1000X solution of synthetic sewage was prepared as described by Hiraishi et al. 1998.26 The solution was filter sterilized (0.45 μm, Cellulose Ester Membrane, Millipore Sigma) and stored at 4 °C until use. Sewer sediment samples for feeding the reactor were collected from two different sewer system wet wells containing raw sewage located at a separate sanitary and combined sewer system (System 1 and System A respectively, Table S1A). In System 1, sediment samples were collected directly from a wet well while in System A, sediment samples were obtained from a sewer sediment pile outside of the sewer pipe. The sediment samples were collected in 6 sterile 1 L polypropylene Nalgene® bottles per location using a metal shovel that was disinfected with 95% ethanol. Samples were then stored in a cooler with ice and transported to the lab. To prepare the reactor, a mixture of sediment from both locations [1 Kg (wet weight) of a 50:50 (weight:weight) mixtures] was added to an 2 L acrylic jar (Phipps & Bird,® Richmond, VA). Another of these acrylic jars served as the feed container for the synthetic sewage (1X) to feed the outer tank of the reactor (total volume of 3.5 L). The synthetic sewage was pumped to the sediment jar at a flow rate of 30 mL/min using a metering pump (Iwaki® model EX-A 15 VC T, Iwaki America, Holliston, MA). Both sewage and sediment tanks were stirred with paddles (Phipps & Bird® Stirrer model 7790–400, Richmond, VA) at 100 rpm during the pumping. After the sewage and sediment were mixed in the sediment tank, the liquid portion was pumped (30 mL/min) into the reactor until the water level reached the upper “waste” outlet (Fig. 1B), using a metering pump (LMI B721–91S TP, Ivyland, PA). The fluid was exchanged from the reactor every two days by draining the simulated sewage from the reactor and immediately refilling via the procedure described above.
To determine the contribution of ARGs and microbes from wastewater in a continuous flow (WW-CF) reactor, the experiment described above was repeated with the same reactor (Fig. 1C) that instead was continuously fed with untreated sewage. The reactor was located inside the cabin of a wastewater pump station. Using a 7.62 m long, 1 cm diameter PVC clear vinyl tube, raw sewage was continuously pumped for 28 days from the wet well to the reactor with the same metering pump as above (LMI B721–91S TP, Ivyland, PA ). When the water level reached the upper “waste” outlet of the reactor, the wastewater was discarded back to the wet well through a rubber tube by gravity.
Reactor biofilm sampling
Biofilm samples from each pipe material were collected at day 1, 4, 8 and 28 for Sed-SB, (Fig. 1B), and at day 7, 14, 21 and 28 for WW-CF (Fig. 1C). Differences in sampling days between reactor types was due to accessibility to the pump station cabin. Sampling consisted of turning off motor controlling the inner rotating cylinder to allow for removal of the desired coupon using sterile tweezers. For each sampling event a unique PVC and concrete coupon was gently rinsed with sterile ddH2O to remove planktonic cells. Then, a sterile swab was used to scrape the biofilm from half of the coupon (10 cm2). The other half of the coupon was sampled the same way using a new swab to serve an experimental replicate. Each swab was then stored in an individual plastic 2 mL O-ring tube at −20ºC. For Sed-SB, the reactor was run twice with identical conditions and one coupon per experimental condition was sampled in each of the two runs to have a total of four samples per experimental condition. For WW-CF, the reactor was run once but two coupons were sampled for each experimental condition for a total of four samples.
Wastewater and sewer sediment analysis
Wastewater samples were collected from inside the reactor while the cylinder was rotating using a sterile pipette. For Sed-SB, wastewater samples were collected before the fluid exchange on days 2 and 16. For WW-CF, wastewater samples from inside the reactor were collected on days 7, 21 and 28 for a total of three samples. Total suspended solids (TSS), chemical oxygen demand (COD), pH and conductivity were measured on preserved wastewater samples. TSS was measured according to Environmental Sciences Section (ESS) Method 340.2. COD was analyzed using Hach COD vials (20–1500 mg/L range) and a DR2700 spectrophotometer (Hach, Loveland, CO) according to Hach Method 8000. pH and conductivity were measured using a calibrated multimeter (Orion Star A329, Thermo Scientific). In addition, pH and conductivity were measured in the sewer sediment samples used to feed reactor Sed-SB. To do this, 10 g of sediment were collected from the sediment tank before the experiment started (Fig. 1B) and mixed with 50 mL of ddH20. pH and conductivity measurements were taken using a calibrated multimeter (Orion Star A329, Thermo Scientific).
Disinfection experiment
The susceptibility/resistance of the biofilm to bleach treatment (a suggested standardized method for cleaning sewage spills) was assessed. After a stationary phase in the biofilm development was observed based on 16S rRNA gene copy qPCR data during each experiment, the reactor was treated with a final concentration of 4.6% bleach. This concentration was chosen based on the Vermont Department of Environmental Conservation Standard Procedures for Cleaning Up Domestic Wastewater Spills protocol.27 Biofilm samples from concrete and PVC coupons were collected before the bleach treatment (Time 0) and after 1 and 10 minutes of exposure to the treatment. Samples were collected with a sterile swab as previously described and placed in a 15 mL falcon tube containing 10 mL of phosphate buffered saline (PBS). Immediately after each swab was placed inside the PBS tube, the oxidation reaction of bleach was quenched with 150 μL of a 1% catalase solution and a 2% sodium thiosulfate solution (experimentally validated through spectrophotometry to reduce free chlorine concentration in the samples to 0 ppm, as via EPA protocol 334.028).
After quenching, biofilm samples were dislodged from the swabs (described below) and either treated with propidium monoazide (PMA) to facilitate viability PCR or not treated to facilitate PCR for total DNA analysis. PMA is a dye that binds to free dsDNA and intracellular dsDNA after penetration into cells with damaged/impaired membranes, inhibiting PCR amplification. To dislodge cells, swabs suspended in PBS were sonicated for 5 min at 500 watt, 20 kHz, 20% Amp (Qsonica sonicators, model Q500 with a 20kHz converter, model CL-334 and a 12.7 mm probe) following the protocol described by Branck et al. 2017.29 Sonication has been demonstrated to be one of the most effective methods for dislodging biofilm cells for collection and downstream analyses including cultivation.29–31 The tubes with the dislodged cells were then centrifuged at 6000×g for 10 min at 4 °C. The supernatant was discarded and the cell pellets from were resuspended in 1 mL of PBS and divided in two tubes (500 μL of cells solution each). For each sample, one of the tubes (500 μL cell solution) was stored at −20 ºC and analyzed for the total DNA per volume in the sample. In the other tube (500 μL cell solution), 1.25 μL of a solution of 20 mM of PMA was added following the protocol described in Eramo et al. 2017 and Eramo et al. 2019.32–33 to analyze the genes from “viable” cells (defined here as cells with sufficiently intact membranes to prevent PMA penetration) only. Then, to activate the crosslinking of PMA to extracellular DNA and DNA from cells with compromised membranes, all tubes were exposed to light using a PMA-Lite™ LED Photolysis Device (Biotium, Fremont, CA) for 15 minutes. Tubes were then stored at −20 ºC until analysis. A positive control consisting of a swab sample of Escherichia coli (strain TOP10) culture in logarithmic growth phase was used to determine if the sonication and centrifugation affects the membrane integrity of the samples and therefore biases the viable cell measurements. To do this, the E. coli culture swab was suspended in PBS, sonicated, centrifuged, and resuspended in PBS, as described above. The dislodged cells were divided for analysis of either total DNA or the viability treatment, as described above. Sonication and centrifugation did not impact the “viable” E. coli measurements (Supplemental Information).
Biomolecular analyses
DNA was extracted from preserved swabs or pelleted cells using the FastDNA® Spin Kit for Soil (MP Biomedicals, Solon, OH, USA), following the procedure in the instructional manual. DNA was resuspended in 100 μL DES buffer until use. DNA extracts were diluted 1:25 – 1:100 to reduce inhibition and qPCR was performed to quantify 16S rRNA gene34 copies, human-specific HF183 Bacteriodes 16S rRNA genetic marker,35 and multiple ARGs. 16S rRNA was chosen as a surrogate for total bacterial abundance. HF183 was chosen given that water quality regulations are often still based upon fecal indicator organisms and there is data comparing observation of this marker to fecal indicator data for a variety of environmental matrices. The targeted ARGs encode resistance for sulfonamide (sul136), beta-lactam (blaTEM37), tetracycline [ tet(G),38 tet(O),39 tet(W),39], macrolide ermF40 and beta-lactam including the carbapenem family (NDM-141). Further discussion of the choice to study these ARGs is included in the Supplemental Information.
qPCR reaction mixtures for the analysis of sul1, blaTEM, tet(G) tet(O), tet(W), ermF, HF183 and the 16S rRNA gene consisted of 5 μL of SsoFast Eva Green® SuperMix (BioRad, Hercules, CA), 0.4 μM of the forward and reverse primers, 2.4 μL of molecular biology grade water, and 1 μL of diluted DNA extract from the samples. For NDM-1, the qPCR reaction recipe consisted of 5 μL of SsoAdvanced Universal Probes SuperMix (BioRad, Hercules, CA), 0.22 μM of the forward and reverse primers, 0.07 μM of the probe, 1 μL of molecular biology grade water, and 1 μL of diluted DNA extract from the samples. All qPCR reactions were performed in a Real Time Thermocycler (BioRad CFX96 Touch, Hercules, CA). Thermocycler conditions and primers/probes sequences for each gene are summarized in Table S2. All samples were analyzed in triplicate in order to have technical replicates. A standard curve of seven points (102–108 gene copies) and a no-template control (NTC) were run in triplicate in each qPCR 96-well plate. The average standard curve R2 and efficiency of each gene are specified in Table S2. qPCR results are presented as gene copies/cm2 based on the area swabbed during sampling. The limit of quantitation (LOQ) was determined based on the lowest qPCR standard (102, for all genes). The LOQ was 3.40 Log10 gene copies/cm2 for the field samples and biofilm growth experiment in the reactor and 3.08 log10 gene copies/cm2 for the disinfection experiment. Melt curve analysis was performed for all SYBR green qPCR reactions. The amplicon length of selected qPCR products from all genes was verified using agarose gel electrophoresis. To allow for comparison of the qPCR results between matrixes (biofilm and the feed wastewater), copies of each studied gene were normalized to 16S rRNA gene copies. Disinfection efficiency was measured as log removal, by comparing the reduction in total gene copies and gene copies from viable cells with exposure times of 0 to 1 minute and 0 to 10 minutes.
To study the prokaryotic diversity in the biofilm samples, amplicon sequencing (Illumina MiSeq, 300-bp, paired-end) was performed on selected samples targeting the V3–V4 region of the 16S rRNA gene at a commercial laboratory (MrDNA, Shallowater, TX). Sequencing was performed on all of the field biofilm samples (N=10 samples total). For the biofilm reactor experiments (Sed-SB and WW-CF), replicate samples from day 28 from each reactor type and pipe material were sequenced (N=8). To understand the impact of disinfection, two samples collected after 10 min of treatment (one of total DNA and one of DNA from viable cells), for each reactor type (Sed-SB and WW-CF) and each pipe material (concrete and PVC), were sequenced (N=8).
Sequencing results were processed through Qiime2–2019.442 following the “Atacama soil microbiome” tutorial in https://qiime2.org and a previously published pipeline43 in order to generate operational taxonomic unit (OTU) tables. Because the number of sequences in samples treated with bleach were low relative to the untreated (samples collected at time 0 of disinfection which is the same as day 28 of biofilm development) and field samples, the sequences were randomly subsampled (rarefied) at two different sequence sampling depths. To study changes in the prokaryotic community composition after the bleach treatment 500 sequences per sample were used, given this was the maximum common number of sequences per sample observed (Fig. S1A). (Note, the sequencing depth corresponds only to viable-cell samples post-disinfection and plateau of the rarefaction curves for these samples indicates the sequencing depth can reasonably represent the post-disinfection sample richness.) To compare differences in biofilm prokaryotic communities from different locations, pipe materials and source of microbes 3500 sequences per sample were used (Fig. S1B). The non-disinfected reactor samples resulted in 3519 to 7198 sequences per sample while the field biofilm samples yielded higher number of sequences (>29,000 sequences/sample). To corroborate if rarefaction to a given sequence depth (3500 and 500) affects the alpha diversity analysis results, an OTU table without subsampling was generated and analyzed. Sequences are available at NCBI database (https://www.ncbi.nlm.nih.gov) under Accession Numbers SAMN14464860 and SAMN10356326-SAMN10356393.
Statistical and microbial diversity analysis
All statistical analyses were performed in R (http://www.r-project.org).44 To compare the abundance of 16S rRNA genes, fecal indicator marker, and ARGs between field samples and reactor sample types (pipe material and inoculum/reactor operation), a Kruskal–Wallis test with a post hoc pairwise t-test with a Bonferroni correction for multiple comparisons was performed. To compare the log-removal of total and viable-cell gene copies for a reactor sample type, a Wilcoxon Rank Sum test was performed for non-parametric data and a Student’s t-test for parametric data. Normality of the data was determined through a Shapiro-Wilk test.
To analyze the prokaryotic diversity in each sample, alpha diversity tests (richness, evenness, Shannon, Simpson, inverse Simpson and Fisher alpha diversity indices) were calculated using the Vegan: Community Ecology Package, version 2.5–6.45 All alpha diversity indices were determined for both subsampled sequences (3500 and 500 sequences, as previously explained) and for the same sequences but without subsampling (rarefaction). This was done to compare and determine if rarefaction causes changes in diversity indices that can affect the interpretation of the results (as discussed in Supplemental Information). Differences in alpha-diversity indices were determined with a Kruskal–Wallis test followed by a post hoc pairwise t-test with a Bonferroni correction. For beta diversity analysis, a Bray–Curtis similarity matrix was calculated at the family level for both subsampled OTU depths (N= 3500 or 500 sequences). Then, non-metric multidimensional scaling (nMDS) analysis was performed to the Log10 plus one transformed OTU values using a Bray Curtis distance, two dimension and 20 random starts (Vegan package)45 for both subsamples (sequencing depths). nMDS plots were generated using the ggplot2 package in R.46–47 To test for significant differences in beta-diversity between samples as a function of sample type (field vs. reactor), pipe material, inoculum/reactor operation, and treatment a Permutational multivariate analysis of variance (PERMANOVA) was done.45 A post hoc pairwise PERMANOVA with a Bonferroni p adjustment was performed by comparing the dissimilarity matrix for the samples between factors (inoculum/reactor operation and pipe material) using the “pairwiseAdonis” package version 0.3.48 A heatmap was created44 for each subsampled sequencing depth (N= 3500 and 500) at the family level.
The linear discriminant analysis (LDA) effect size (LEfSe) method as described by Segata et al. 201149 was used to identify significant prokaryotic biomarkers in the biofilm samples (Supplemental Information).
Results
Field Sewer Biofilm samples
Two sewer systems were sampled during winter and summer to understand the presence of ARGs, overall microbial community structure, and to allow for comparison to the laboratory biofilm reactor. All the studied ARGs except NDM-1, were detected above the LOQ in field biofilm samples (Fig. S2). Human fecal indicator marker HF183 was detected above the LOQ in the field samples from System 1 but not in System 2 (Fig. S2). All ARGs except ermF had similar relative abundances (ARG gene copies/16S rRNA gene copies) in the two field system biofilms (p > 0.176, Kruskal–Wallis test).
Biofilm ARGs as a factor of pipe material and inoculum/reactor operation
To understand if sewer pipe material and/or inoculum (sediment vs. wastewater)/feed are associated with elevated ARGs, two experiments were performed: Experiment 1 with sewer sediment fed semi-batch (Sed-SB) and Experiment 2 with continuous feed of wastewater (WW-CF). No significant differences were observed for the qPCR results between PVC or cement pipe material in paired samples from either of the experiments (all p > 0.32, Kruskal–Wallis test). Based on quantification of the 16S rRNA gene, concentration of gene copies reached a plateau by the first sampling event (24 hours for SS-SB and 7 days for WW-CF) regardless of pipe material and inoculum source/reactor feed (Fig. 2). These results were consistent for sul1 and blaTEM measurements in the Sed-SB biofilm, where no significant increase in abundance was detected after 24 hours. (Fig. 2). In these biofilms, only sul1 and blaTEM were consistently detected above the LOQ in qPCR analysis. The other five ARGs and HF183 were not detected or below LOQ by qPCR, however, their presence was confirmed by gel electrophoresis.
fig. 2.
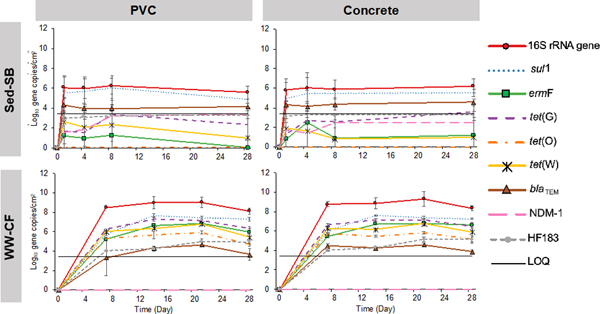
concentration of 16s rrna gene, args and human fecal indicator gene hf183 throughout 28 days of biofilm development in both reactor setups (sed-sb and ww-cf) and both pipe materials (pvc and concrete). error bars represent the standard deviation of the mean (n=4). loq is based on the qpcr lowest value of the standard curve.
In the Sed-SB biofilms, all targeted ARGs except NDM-1 were above the LOQ(Fig. 2). In the WW-CF biofilms, sul1 continued to increase in abundance until day 14 on both pipe materials (p < 1.0×10−6, Kruskal–Wallis test). The other six ARGs studied and HF183 did not show any significant increase in abundance after day 7 (Fig. 2, p> 0.078, Kruskal–Wallis test). NDM-1 was not detected in any of the WW-CF or field biofilm samples. Comparing the biofilms from the different inoculum/feed conditions, the genes quantified in this study were more abundant in the WW-CF biofilms than in Sed-SB biofilms except for blaTEM which was similar in both reactor types and NDM-1 that was undetected in WW-CF biofilms.
To help explain differences observed in the biofilms in the two experiments, qPCR was performed on both source wastewaters used to feed the reactors. The Sed-SB wastewater had higher 16S rRNA gene copies per volume than the wastewater feeding WW-CF (p = 0.0023, Wilcoxon Rank Sum test, Fig. S3).
All the of the other genes (targeted ARGs and HF183) except NDM-1 were present in the feed of both reactors setups (Sed-SB and WW-CF, Fig. S4). NDM-1 was observed in the wastewater used to feed Sed-SB but not detected in the wastewater for WW-CF. Next, the relative abundance of the ARGs in the feed wastewater was compared to the relative abundance of these genes in the biofilms (ARG copies/16S rRNA gene copies). In Sed-SB wastewater (Fig. S4A), sul1 had higher relative abundance (sul1 copies/16S rRNA gene copies) than the other ARGs (p < 0.042, Kruskal–Wallis test), which had similarly relative abundance to one another (p > 0.077, Kruskal–Wallis test). There was no difference in the relative abundance of sul1 between the Sed-SB wastewater and the Sed-SB biofilms (p > 0.32, Wilcoxon Rank Sum test ), but more blaTEM was detected in the Sed-SB biofilm than in the wastewater. In the WW-CF reactor (Fig. S4B), all ARGs except for NDM-1 (below LOQ) had similar relative abundance in the wastewater matrix (p > 0.074, Kruskal–Wallis test). ermF, tet(G), tet(O), and tet(W) were similarly abundant between both matrices (WW-CF wastewater and biofilm). However, sul1 was more abundant in the biofilm than in wastewater (p < 0.008, Kruskal–Wallis test) and blaTEM was more abundant in the wastewater than in the biofilm (p < 0.029, Kruskal–Wallis test).
COD, TSS and conductivity were significantly higher in wastewater from Sed-SB than WW-CF (all p > 0.0062, Wilcoxon Rank Sum test, Table S1B). pH measurements were between 6.6 and 7.5. The sewer sediment samples used to feed Sed-SB reactor had conductivity between 1242 and 59.33 μS/cm and pH between 5.72 and 7.88 (Table S1C).
Comparison of qPCR data from field and simulated sewer biofilm:
The biofilm developed in simulated sewer reactors were compared to the field sewer pipe biofilms to understand how representative the laboratory results were of field conditions. The 16S rRNA gene abundance per area swabbed was similar for the field biofilm samples and biofilms samples from the WW-CF collected at day 28 (p=1, Kruskal–Wallis test). The Sed-SB biofilm had significantly lower 16S rRNA gene copies than the field or WW-CF samples (p < 0.0018, Kruskal–Wallis test), regardless of the pipe material (Fig. S5). Human fecal indicator HF183 was detected and quantifiable in WW-CF biofilm on both pipe materials but not in Sed-SB biofilms. In the field biofilm samples, HF183 was only detected above LOQ in the biofilm of System 1.
To compare the ARG profiles between the field and experimental biofilm samples from Day 28, qPCR data were compared and a non-metric multidimensional scaling (nMDS) analysis was performed on 16S rRNA normalized ARG qPCR data (Fig 3). All ARGs except NDM-1 were above LOQ in biofilms from the field and WW-CF. The ARG profiles observed in biofilm from the field, Sed-SB, and WW-CF samples were significantly distant from one and other (p= 0.003, PERMANOVA, Fig. 3A). ARG profiles did not vary by pipe material during a given experiment or between the field sampling sites (System 1 and 2; p > 0.87, PERMANOVA). Seasonal differences were not observed in the field biofilm samples (data not shown).
figure 3.
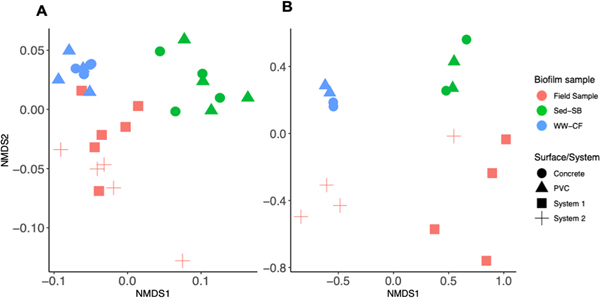
non-metric multidimensional scaling (nmds) showing the (a) profile of args (stress = 0.088) and (b) prokaryote community structure (stress = 0.041) as a factor of biofilm sample type (colors) and surface or system where the biofilm sample was collected (shapes).
Prokaryotic Diversity Analysis
An nMDS was performed on the microbial community structures observed in the field and experimental biofilm samples. Similar conclusions were obtained from nMDS analysis performed on the microbial community composition at the family level (Fig. 3B) to that observed for the ARG profiles: significant differences were observed between the three different biofilm sample types (Field, Sed-SB, WW-CF; p < 0.038, PERMANOVA) and no differences were observed between pipe material (PVC and concrete) from the experiments or between the two field systems sampled (System 1 and System 2, p > 0.053). Analysis of the microbiome showed that each biofilm sample type (Field, Sed-SB, WW-CF) was dominated by different family taxa. The families observed included those containing potential human pathogens, obligate anerobic hydrogenotrophs known to cause corrosion, and nitrogen oxidizing bacteria (Fig. 4 and S6).
fig. 4.
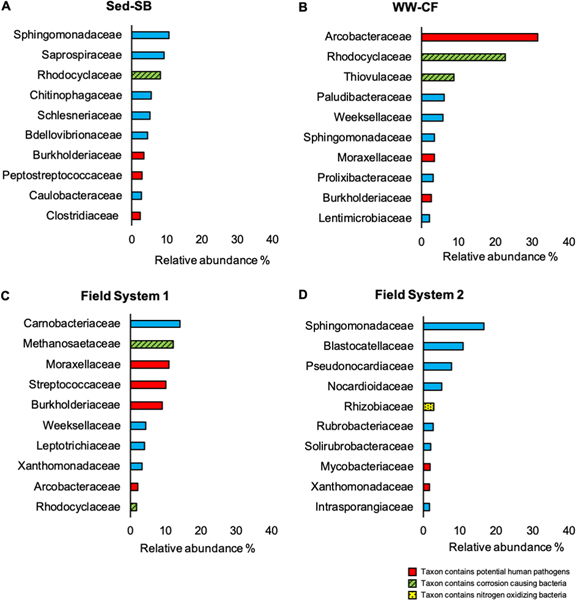
bar plots representing the relative abundance of the 10 most dominant family taxa in each biofilm sample type: (a) sed-sb (b) cf-ww (c) field system 1 and (d) field system 2. taxa containing relevant potential human pathogens, corrosion causing bacteria and nitrogen oxidizers are color coded.
To compare the prokaryotic diversity at the family level between biofilm samples the Shannon-Wiener diversity Simpson, inverse Simpson, and Fisher indices of diversity along with richness and evenness were calculated (Supplemental Information, Table S3 and Table S4A). Results of the Shannon diversity index at a depth of 3500 sequences showed that field biofilm samples had similar diversity compared to the biofilm communities grown in both experiments (p > 0.072).
Biofilm disinfection
Disinfection of the reactor biofilms was performed to determine the persistence of sewer biofilms following a standardized sewage spill treatment protocol. After 28 days of biofilm growth, the reactors were treated with a final concentration 4.6% of bleach. At treatment time 0 (day 28), viable cell and total 16S rRNA gene copies, HF183 copies, and almost all ARGs were more abundant in the WW-CF biofilms compared to the Sed-SB biofilms regardless of pipe material (Fig. S7). After the bleach treatment, viable cell 16S rRNA was detected above LOQ via PMA-qPCR in both reactor biofilms (Sed-SB and WW-CF) after 10 minutes of exposure to the disinfectant (Fig. S7).
Log-removals for total and viable-cell gene copies were calculated and compared to provide insight into the mechanism of biofilm loss during disinfection. For Sed-SB biofilms on the PVC surface, the log-removal of total 16S rRNA gene copies was greater than the log-removal of viable cell 16S rRNA gene copies after both 1 and 10 minutes of exposure (p = 0.018 and 0.023 respectively, Kruskal–Wallis test; Fig. 5). Likewise, for the WW-CF biofilm, greater log-removal of total 16S rRNA gene copies compared to viable cell 16S rRNA gene copies was observed after 1 minute on the PVC surface (p = 0.042, Kruskal–Wallis test; Fig. 5). No differences between the log removal of viable-cell and total 16S rRNA gene copies were observed for biofilm grown on the concrete surface in either experiment (p > 0.074, Kruskal–Wallis test; Fig. 4).
fig. 5.
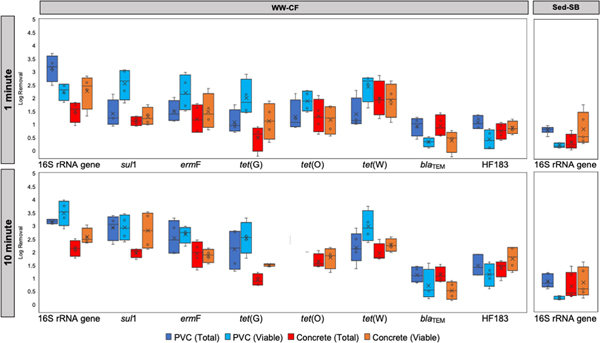
boxplots comparing the biofilm log removal of total and viable cells carrying 16s rrna gene, args and human fecal indicator hf183 in both pipe materials (pvc and concrete) and reactor setups (sed-sb and ww-cf) after 1 and 10 minutes of exposure to 4.6% of bleach. for ww-cf biofilms n =4 and for sed-sb n =5. the genes that are not shown in sed-sb were not detected or below loq after 1 minute of exposure to the treatment.
After 1 minute of exposure to the bleach treatment, the concentration of total ARG copies and viable-cell carrying ARGs in the Sed-SB biofilm were below the LOQ (Fig. S7A). For the WW-CF biofilms, viable cells carrying all the studied ARGs (except for NDM-1 and tet(O) on PVC), remained detectable above LOQ even after 10 minutes of exposure (Supplemental Fig. S7B). Log removal of total ARGs and viable cells carrying ARGs varied gene-to-gene in the WW-CF biofilms (Fig. 5): for some genes (i.e., sul1 and tet(G) on concrete, p < 0.049, Wilcoxon Rank Sum test) viable-cell log removal was greater than total log removals, for blaTEM on concrete total log removals was greater than viable-cell log removal (p = 0.029, Wilcoxon Rank Sum test), and for the rest of the genes in both surfaces there was no difference (p > 0.17, Wilcoxon Rank Sum test).
Effect of disinfection on the sewer biofilm prokaryotic community
The effect of bleach on the prokaryotic biofilm community was analyzed via nMDS (Fig. 6). Before the treatment, the WW-CF biofilms were clustered together regardless of pipe material (Fig. 6). Similarly, the Sed-SB biofilms on both PVC and concrete, clustered together, but segregated apart from the WW-CF biofilm cluster. After the bleach treatment, the viable- and total cell biofilm community did not cluster as a function of source of inoculum/feed and pipe material (Fig. 6 and Fig. S9). None of the samples clustered by any factor (source of inoculum/reactor operation, pipe material, or viability) after the bleach treatment.
fig. 6.
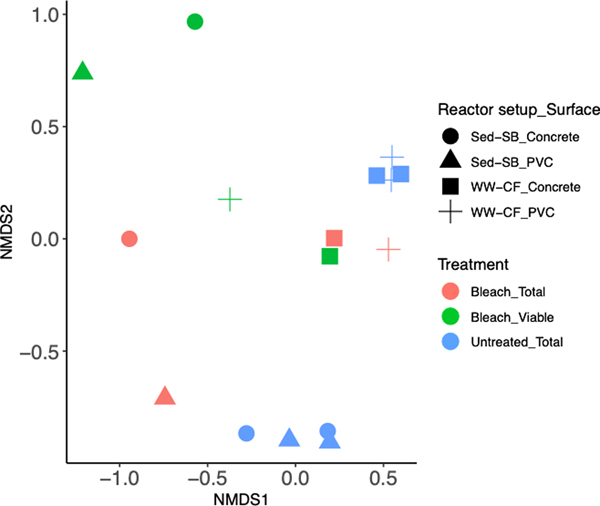
non-metric multidimensional scaling (nmds) showing the prokaryote community structure (stress = 0.089) as a factor of treatment and cell viability (colors), and biofilm type and surface (shapes).
The Shannon alpha diversity index, richness, and evenness were calculated to understand the impact of bleach treatment on the prokaryotic diversity in the simulated sewer biofilms. Disinfection did not significantly change the Shannon diversity measurement, richness or evenness from the WW-CF biofilm: indices were similar among all treated and untreated biofilm samples (Table S5 and Table S4B). Disinfection with bleach for 10 minutes resulted in a significant decrease in the Shannon index (p=0.024, Wilcoxon Rank Sum test) and in the community richness (p= 0.00011, Wilcoxon Rank Sum test) for the Sed-SB biofilms (Table S5 and Table S4B). There was no difference in evenness between these samples.
Discussion
Antibiotic resistance in sewer biofilms
An annular biofilm reactor simulating the conditions in a sewer pipe was used to study the biofilm development and accumulation of ARGs as a function of pipe material (PVC and concrete) and source of inoculum/reactor operation (Sed-SB or WW-CF). Concentrations of 16S rRNA genes per area, human fecal indicator gene HF183, and ARGs were similar for biofilms from both PVC and concrete within a given experiment, despite the differences in surface roughness for the materials studied (0.0015mm for PVC and 0.3–3.0mm for concrete). Surface topography and material chemistry have been proven to affect bacterial attachment to surfaces in other systems including water distribution systems.13–14, 50–54 Higher surface roughness at the nanometer scale was previously shown to result in higher adhesion force and early Streptococcus mutans attachment to zirconia (ZrO2), a material used for the production of hard ceramics and prostodontics, however it did not affect the further bacterial accumulation in the biofilm.51 Other reports suggests that in drinking water distribution systems, pipe material has ultimately little impact on long term cell accumulation in the biofilm 13, 55 In all the reactor experiments, the maximum load of the genes analyzed except sul1 was achieved before the first sampling event, one day for Sed-SB and seven days for WW-CF (Fig. 2). This suggests biofilm colonization and maximum load of cells is achieved in less than 7 days for both newly installed concrete and PVC pipes. HF183 concentrations in the biofilm followed a similar pattern (no changes over 28 days of operation) indicating this human fecal indicator marker had similar behavior to ARGcarrying cells. Some Bacteroides species from the HF183 cluster can carry multiple ARGs including tetracycline and erythromycin resistance genes,56 which could explain the similar behavior.
The annular biofilm reactor was operated with different inocula/reactor operations. For Sed-SB the reactor was operated with a semi-batch feed with sewer sediment as the major source of microbes carried by synthetic sewage. For WW-CF, wastewater was the only source of microbes and the reactor was continuously fed. The WW-CF biofilms accumulated significantly more 16S rRNA gene copies and ARGs than the Sed-SB biofilms despite the Sed-SB wastewater having more 16S rRNA copies per volume and higher COD than WW-CF feed water. This along with the fact that TSS was higher in Sed-SB wastewater may suggest that bacteria from the sewer sediment remained attached to sediment particles in wastewater rather than attaching to pipe biofilms. Our measurements were for bulk wastewater 16S rRNA gene copies and did not differentiate genes that were attached to suspended particles. Another possibility could be that there was more 16S rRNA from non-viable cells in the Sed-SB wastewater samples.
Relative abundances of the targeted ARGs in the feed wastewaters were compared to the relative abundances of these genes (ARG copies/16S rRNA gene copies) in the biofilms. All target ARGs were detected in the feed wastewater of the Sed-SB reactor but only sul1 and blaTEM were detected above LOQ in the biofilm. Sed-SB reactor biofilms had higher blaTEM relative abundances than the feed wastewater indicating potential enrichment of the gene within the biofilm community or that community members carrying this gene were more likely to attach to the coupon biofilms. In the case of WW-CF, sul1 had higher relative abundance in the biofilm compared to the feed wastewater, again indicating potential enrichment or likelihood of the host cells to attach to the biofilm. Thus, the ARGs observed to accumulate in the biofilms from the two reactor types were different. This observation may be explained by differences in community structure (i.e., which bacteria attach) which will be discussed further below.
The 16S rRNA gene concentration (gene copies/area) in the sewer biofilm samples from the field were similar to the concentrations detected in the WW-CF biofilms from day seven to day 28 (Fig. 2 and Fig. S5). Thus, to capture the kinetics of the simulated sewer biofilm colonization one should sample more frequently during the first week of reactor operation. Earlier sampling was possible for the Sed-SB reactor and results indicated that less than 1 day may be needed to capture 16S rRNA gene copy kinetics. Previous laboratory-scale studies in continuously fed sewer biofilm reactors captured biofilm growth kinetics by measuring biofilm thickness between zero and 25 days of operation.57–58 Differences in the feed wastewater (here simulated and real municipal wastewater from the US compared to either wastewater57 or a mix of wastewater and seawater China58) and reactor operation (i.e., one referenced study recirculated the feed wastewater57 as compared to fresh wastewater being used here), and reactor type (i.e. annular biofilm reactor used here and elsewhere57 compared to a plug flow reactor58) may explain the apparent sampling period needed to capture simulated sewer biofilm growth kinetics.
The sulfonamide resistance gene sul1 was consistently the ARG with the highest relative abundance (ARG copies/16S rRNA gene copies) in all the studied biofilm samples (field and experimental biofilms Fig. S2). These results are consistent with the findings by Auguet et al. 2017,17 who reported that sulfonamide resistance genes were the most abundant ARG in sewer biofilm samples collected from sewer pump stations located in Palamos, Spain. Sulfonamide resistance genes have been consistently the most abundant ARG reported in all sewer systems matrices (raw sewage, combined sewer overflow, WWTP effluent, sediment and biofilms).17, 32–33, 59–61 This gene has been suggested as a marker ARG for qPCR in different matrices given that it is widely observed and that continues in this less well studied matrix.
Of particular interest is the prevalence of NDM-1, which confers resistance to carbapenem antibiotics and was discovered in 2008 in India and further became epidemic in the same country and in the Middle East.62 In recent years this gene has been detected in bacterial isolates from patients in hospitals in the US.63 However, to our knowledge, the detection of NDM-1 in environmental samples in the US is rarely reported. NDM-1 has been mainly detected in Enterobacteria but with more frequency in the species Klebsiella pneumoniae, a human pathogen.62–64 NDM-1 was below LOQ in all reactor and field biofilm samples, however it was detected by gel electrophoresis in biofilm samples from the Sed-SB experiment. NDM-1 was not observed in the WW-CF or field biofilm samples. Moreover, NDM-1 was detected in the feed in wastewater for Sed-SB but not in that for WW-CF. This is consistent with the findings in Eramo et al. (in review) who reported NDM-1 was detected in higher relative abundance in sewer sediment samples than in paired wastewater influent field samples. The presence of this gene in sewer sediment and biofilms may indicate these matrices serve as a niche for bacteria with the gene or indicate that it is observed transiently in WW.
Structure of microbial communities
Amplicon sequencing and ARGs profile of Sed-SB and WW-CF biofilm samples (Fig. 3) showed similar clustering: samples cluster by biofilm sample type (field, Sed-SB, or WW-CF) with statistically significant different distance between each other. For the field biofilm microbiome and ARG profile (Fig. 3) there was no difference between sample locations, consistent with the findings of Su et al. 20179 who reported no geographical variation in the microbiome and antibiotic resistome for WWTP influent detected across 17 cities in China. Interestingly, the biofilms from the field (System 1 and 2) and WW-CF achieved similar 16S rRNA gene copies per area (Fig. S5). One of the field sampling sites (System 2) was also used to feed the reactor for the WW-CF experiment (Table S1A), but the biofilm microbial community structures were dissimilar between the two (Field system 2 and WW-CF). Therefore, temporal differences in when the reactor was fed (fall 2018) compared to when the field biofilm samples were collected (winter 2016/ 2017) may explain the biofilm community differences. Another possibility is that biofilm age may explain the difference between the WW-CF and field sample. A previous laboratory scale study65 showed that in early stages of sewer biofilm development sulfate reducing bacteria were the dominant (highest relative abundance) hydrogenotrophic prokaryotes and then methanogenic archaea outcompete them and became dominant in the mature biofilm. nMDS analyses of the microbiome and ARG profiles showed no significant distance between both pipe material (PVC and concrete) in the two experiments (Sed-SB and WW-CF). Again, supporting the observation that pipe material (PVC or concrete) does not affect biofilm development and accumulation of ARGs.
Multiple studies have found that the genus Arcobacter is consistently one of the organisms that dominates wastewater influent microbial communities.10–11, 66–68 This is consistent with our observation that the taxonomical family Arcobacteraceae was the most dominant in the WW-CF biofilm composing the 31% of the prokaryotic relative abundance (Fig. S4). Arcobacter has been detected in sewer sediment and biofilms via 16S rRNA gene amplicon sequencing11, 17, 69 but in comparison, it is relatively more abundant as planktonic cells in wastewater.11 In the biofilm samples from the field, the abundance of the family Arcobacteraceae was 2% or less while in the Sed-SB biofilm it was less than 1%. This may suggest that because Arcobacteraceae is highly abundant in wastewater, it is an early colonizer of sewer biofilms. However, the low abundance of this taxon in the System 2 biofilms (same system used to feed WW-CW) suggests that in mature sewer biofilms they are outcompeted. This observation was consistent with previous reports of field samples of sewer biofilms and wastewater from the same location in Spain where the abundance of Epsilonproteobacteria (class affiliated to Arcobacteraceae) was 37% in wastewater, but 8% in the biofilm.17
Sed-SB biofilm on both PVC and concrete was marked by sulfate-reducing bacteria belonging to multiple families (i.e., Desulfovibrionales and Desulfobacterales, Fig. S10, LEfSe analysis described in Supplemental Information). These bacteria are known to cause MIC that is the major cause of deterioration in concrete sewer systems.70 According to Auguet et al. 201565 these strict anaerobic hydrogenotrophic bacteria are early colonizers in sewer biofilms before they are displaced by methanogenic archaea. This is consistent with the results from the presumably mature field biofilm samples from System 1, where the family Methanosaetaseae is the second most dominant prokaryote of the community (12%).
In all three biofilm sample types (Sed-SB, WW-CF and Field samples), taxa containing human pathogens were detected as biomarkers (Supplemental Information and Fig. S10). Family taxon containing pathogenic species were detected in the 10 most abundant organisms of each biofilm sample type including Arcobacteraceae, Clostridiaceae, Streptococcaceae and Mycobacteraceae (Fig. S4). Taxonomic analysis of the species level (level 7) showed the presence of sequences with 100% similarities to the human pathogens Streptococcus anginosus and Streptoccocus salivarius. Pathogenic species from all of these family taxa were detected in field sewer biofilm samples in the Spanish study.17 These observations could indicate a potential hazard for utility workers and may have implications for interpreting sewage epidemiology data if pathogens were found to indeed be multiplying or attenuated in sewer biofilm.
Effect of disinfection on cell viability and structure of microbial communities
A standardized bleach treatment for disinfecting following sewage spills was applied to the biofilm reactors andMedian log removals for total genes varied between 0.88 and 3.16 for the WW-CF reactor and 0.39 and 0.76 in the Sed-SB reactor after 10 minutes of exposure to the disinfectant. Greater log removal of total 16S rRNA gene copies than viable cell 16S rRNA gene copies were observed in biofilms grown on PVC (Fig. 5). This result suggests that dislodging of cells from the PVC biofilm is the predominant mechanism of loss from the biofilm following bleach disinfection. In contrast, no significant differences between viable and total 16S rRNA gene copies per area were observed on the concrete surface. This may be explained by the fact that PVC has a smoother surface topology compared to concrete. The detachment rate of cells from biofilms resulting from physical disturbances was previously shown to be greater for smoother surfaces than rougher surfaces.71 Likewise, Karin et al. 201772 tested disinfection methods on different surfaces (nitrile, stainless steel and heavy-duty tarp) inoculated with E. coli and reported that the cells were more challenging to disinfect on the heavy-duty tarp, the roughest of the three tested surfaces.
Observation of viable-cell gene copies following 10 minutes of disinfection indicated that further treatment would be needed to disrupt the membranes of cells carrying some or all of the target genes in sewer biofilms from the two experiments. Viable-cell 16S rRNA gene copies were above LOQ following disinfection for both reactors and HF183 for the WW-CF biofilms (Fig S7). A higher abundance of viable cells carrying ARGs were detected following disinfection in the WW-CF biofilms than the Sed-SB biofilms, the latter of which were visibly thinner and had lower 16S rRNA gene copies per area. Biofilm thickness has been demonstrated to increase antimicrobial resistance (including disinfectants) in biofilm because it reduces the contact of disinfectant with the cells deeper within the biofilm.73 A study by Cochran et al. 200074 showed that thicker biofilms with more cell density were more resistant to H2O2, an oxidant disinfectant like bleach, than thinner biofilms with less cell abundance.
nMDS analysis of the microbial community composition after disinfection showed no consistent shift in the community profile (Fig. 6), demonstrating that chlorine did not select for a specific microbial community structure in these sewer biofilms. In fact, chlorine generally is considered a nonselective disinfectant that reacts with multiple cellular components thus affecting cellular metabolism.75–76 Alpha diversity measurements from amplicon sequencing of the WW-CF biofilm with and without PMA treatment showed that even though the disinfectant caused loss of both total genes and viable-cell genes based on 16S rRNA gene qPCR, the microbial diversity did not change (Table S4 and Table S5). In contrast, in the Sed-SB experiment the richness and diversity significantly decreased after the disinfectant treatment. Because the biofilm community from the Sed-SB experiment was more diverse but had less initial cell density before the treatment, cells and DNA from whole taxa were more likely to be completely detached from the biofilm by the disinfectant, thus causing a decrease in richness and diversity. In the case of WW-CF, before the treatment biofilm had less prokaryotic diversity but higher cell density. In other words, the biofilm was dominated by a greater number of cells of a smaller pool of taxa. Thus, detachment of cells by the disinfectant does not change the richness and relative diversity because the few but very abundant taxa are less likely to be completely lost from the biofilm.
Conclusions
Field and simulated sewer biofilm samples provided insight into the abundance of ARGs, microbial community composition, and susceptibility to disinfection of sewer. ARGs and bacterial taxa containing pathogens and corrosion-causing bacteria were observed in both field and simulated sewer biofilms. Pipe material (PVC and concrete) did not cause a significant effect in sewer biofilm 16S rRNA gene copies per area, accumulation of ARGs, or prokaryotic community composition. However, the inoculum/reactor operation in the simulated sewer experiments resulted in a different prokaryote community and ARG profiles in the sewer biofilms. The WW-CF reactor resulted in a less diverse prokaryote community but with higher 16S rRNA concentrations and ARG relative abundances than the Sed-SB. Bleach based standardized sewage spills treatment resulted in median log removals of total 16S rRNA gene copies of up to 0.76 to 3.16. Thus, the treatment did not completely detach or kill cells carrying ARGs in sewer biofilms. Total cell dislodging from the biofilm by the disinfectant was the major mechanism of biofilm loss from the PVC coupons. No differences between log removal for cell dislodging and membrane disruption in the biofilm was detected for the biofilm on the concrete coupons. Overall, these results indicate that sewer biofilms are reservoirs of bacteria carrying ARGs that can survive standardized sewage disinfection treatments.
Supplementary Material
Water Impact Statement.
Sewer biofilms may retard or allow for growth of antibiotic resistant microbes that could skew interpretation of sewage epidemiology data and present a hazard to utility workers or to the public if dislodged during sewer overflows. Results are presented for in-situ and reactor sewer biofilms including the potential for removal or loss of cell viability by standard disinfection.
Acknowledgements
Thanks our utility partners who wish to remain anonymous. Funding for this research was provided by the National Science Foundation 1510461, the Rutgers Aresty Research program, and NIH Bridges to the Doctorate (R25GM058389).
References
- 1.US Centers for Disease Control and Prevention (CDC), Antibiotic resistance threats in the Univved States. Department of Health, Ed. www.cdc.gov, 2019. [Google Scholar]
- 2.LeChevallier MW; Mansfield TJ; Gibson JM, Protecting wastewater workers from disease risks: Personal protective equipment guidelines. Water Environ Res 2019. [DOI] [PubMed] [Google Scholar]
- 3.Fahrenfeld N; Bisceglia KJ, Emerging investigators series: sewer surveillance for monitoring antibiotic use and prevalence of antibiotic resistance: urban sewer epidemiology. Environ Sci Water Res Technol 2016, 2 (5), 788–799. [Google Scholar]
- 4.Li W; Zheng T; Ma Y; Liu J, Current status and future prospects of sewer biofilms: Their structure, influencing factors, and substance transformations. Sci Total Environ 2019, 695, 133815. [DOI] [PubMed] [Google Scholar]
- 5.Reynolds JH; Barrett MH, A review of the effects of sewer leackage on groundwater quality. Water Environ J2003, 17 (1), 34–39. [Google Scholar]
- 6.Buelow E; Bayjanov JR; Majoor E; Willems RJ; Bonten MJ; Schmitt H; van Schaik W, Limited influence of hospital wastewater on the microbiome and resistome of wastewater in a community sewerage system. FEMS Microbiol Ecol 2018, 94 (7). [DOI] [PubMed] [Google Scholar]
- 7.Buelow E; Rico A; Gaschet M; Lourenco J; Kennedy SP; Wiest L; Ploy MC; Dagot C, Hospital discharges in urban sanitation systems: Long-term monitoring of wastewater resistome and microbiota in relationship to their eco-exposome. Water Res X 2020, 7, 100045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ng C; Tay M; Tan B; Le TH; Haller L; Chen H; Koh TH; Barkham TMS; Gin KY, Characterization of metagenomes in urban aquatic compartments reveals high prevalence of clinically relevant antibiotic resistance genes in wastewaters. Front Microbiol 2017, 8, 2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Su JQ; An XL; Li B; Chen QL; Gillings MR; Chen H; Zhang T; Zhu YG, Metagenomics of urban sewage identifies an extensively shared antibiotic resistome in China. Microbiome 2017, 5 (1), 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Newton RJ; McLellan SL; Dila DK; Vineis JH; Morrison HG; Eren AM; Sogin ML, Sewage reflects the microbiomes of human populations. mBio 2015, 6 (2), e02574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McLellan SL; Roguet A, The unexpected habitat in sewer pipes for the propagation of microbial communities and their imprint on urban waters. Curr Opin Biotechnol 2019, 57, 34–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Donlan RM, Biofilms: microbial life on surfaces. Emerg Infect Dis 2002, 8 (9), 881–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu Z; Wu C; Zhong D; Yuan Y; Shan L; Zhang J, Effects of pipe materials on chlorine-resistant biofilm formation under long-term high chlorine level. Appl Biochem Biotechnol 2014, 173 (6), 1564–78. [DOI] [PubMed] [Google Scholar]
- 14.Cowle MW; Webster G; Babatunde AO; Bockelmann-Evans BN; Weightman AJ, Impact of flow hydrodynamics and pipe material properties on biofilm development within drinking water systems. Environ Technol 2019, 1–13. [DOI] [PubMed] [Google Scholar]
- 15.Jarnheimer PA; Ottoson J; Lindberg R; Stenstrom TA; Johansson M; Tysklind M; Winner MM; Olsen B, Fluoroquinolone antibiotics in a hospital sewage line; occurrence, distribution and impact on bacterial resistance. Scan J Infect Dis 2004, 36 (10), 752–5. [DOI] [PubMed] [Google Scholar]
- 16.Wunder DB; Bosscher VA; Cok RC; Hozalski RM, Sorption of antibiotics to biofilm. Water Research 2011, 45 (6), 2270–80. [DOI] [PubMed] [Google Scholar]
- 17.Auguet O; Pijuan M; Borrego CM; Rodriguez-Mozaz S; Triado-Margarit X; Giustina SVD; Gutierrez O, Sewers as potential reservoirs of antibiotic resistance. Sci Total Environ 2017, 605–606, 1047–1054. [DOI] [PubMed] [Google Scholar]
- 18.Swan JS; Deasy EC; Boyle MA; Russell RJ; O’Donnell MJ; Coleman DC, Elimination of biofilm and microbial contamination reservoirs in hospital washbasin U-bends by automated cleaning and disinfection with electrochemically activated solutions. J Hosp Infect 2016, 94 (2), 169–74. [DOI] [PubMed] [Google Scholar]
- 19.Deasy EC; Moloney EM; Boyle MA; Swan JS; Geoghegan DA; Brennan GI; Fleming TE; O’Donnell MJ; Coleman DC, Minimizing microbial contamination risk simultaneously from multiple hospital washbasins by automated cleaning and disinfection of U-bends with electrochemically activated solutions. J Hosp Infect 2018, 100 (3), e98–e104. [DOI] [PubMed] [Google Scholar]
- 20.Wang H; Hu C; Hu X; Yang M; Qu J, Effects of disinfectant and biofilm on the corrosion of cast iron pipes in a reclaimed water distribution system. Water Res 2012, 46 (4), 1070–8. [DOI] [PubMed] [Google Scholar]
- 21.Gander S, Bacterial biofilms: resistance to antimicrobial agents. J Antimicrob Chemother 1996, 37 (6), 1047–50. [DOI] [PubMed] [Google Scholar]
- 22.Smith K; Hunter IS, Efficacy of common hospital biocides with biofilms of multi-drug resistant clinical isolates. J Med Microbiol 2008, 57 (Pt 8), 966–73. [DOI] [PubMed] [Google Scholar]
- 23.Stewart PS; Rayner J; Roe F; Rees WM, Biofilm penetration and disinfection efficacy of alkaline hypochlorite and chlorosulfamates. J Appl Microbiol 2001, 91 (3), 525–32. [DOI] [PubMed] [Google Scholar]
- 24.Eramo A, Morales Medina WR, Fahrenfeld NL Factors associated with elevated levels of antibiotic resistance genes in sewer sediments and wastewater. Environ Sci Water Res Technol 2020, 6: 1697–1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun J; Hu S; Sharma KR; Ni BJ; Yuan Z, Stratified microbial structure and activity in sulfide- and methane-producing anaerobic sewer biofilms. Appl Environ Microbiol 2014, 80 (22), 7042–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hiraishi A; Ueda Y; Ishihara J, Quinone profiling of bacterial communities in natural and synthetic sewage activated sludge for enhanced phosphate removal. 1998, 64 (3), 992–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vermont Department of Environmental Conservation, Vermont Statutes Specific to Spills and releases. dec.vermont.gov, 2017.
- 28.US Environmental Protection Agency, Determination of residual chlorine in drinking water. epa.gov/safewater, 2009.
- 29.Branck TA; Hurley MJ; Prata GN; Crivello CA; Marek PJ, Efficacy of a sonicating swab for removal and capture of Listeria monocytogenes in biofilms on stainless steel. Appl Environ Microbiol 2017, 83 (11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ahnrud GP; Mendoza AJ; Hurley MJ; Marek PJ, Efficacy of a sonicating swab for removal and capture of mcroorganisms from experimental and natural contaminated surfaces. Appl Environ Microbiol 2018, 84 (9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bjerkan G; Witso E; Bergh K, Sonication is superior to scraping for retrieval of bacteria in biofilm on titanium and steel surfaces in vitro. Acta Orthop 2009, 80 (2), 245–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eramo A; Morales Medina WR; Fahrenfeld NL, Viability-based quantification of antibiotic resistance genes and human fecal markers in wastewater effluent and receiving waters. Sci Total Environ 2019, 656, 495–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eramo A; Medina WM; Fahrenfeld NL, Peracetic acid disinfection kinetics for combined sewer overflows: indicator organisms, antibiotic resistance genes, and microbial community. Environ Sci Water Res Technol 2017, 3 (6), 1061–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Muyzer G; de Waal EC; Uitterlinden AG, Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol 1993, 59 (3), 695–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seurinck S; Defoirdt T; Verstraete W; Siciliano SD, Detection and quantification of the human-specific HF183 Bacteroides 16S rRNA genetic marker with real-time PCR for assessment of human faecal pollution in freshwater. Environ Microbiol 2005, 7 (2), 249–59. [DOI] [PubMed] [Google Scholar]
- 36.Pei R; Kim SC; Carlson KH; Pruden A, Effect of river landscape on the sediment concentrations of antibiotics and corresponding antibiotic resistance genes (ARG). Water Res 2006, 40 (12), 2427–35. [DOI] [PubMed] [Google Scholar]
- 37.Narciso-da-Rocha C; Varela AR; Schwartz T; Nunes OC; Manaia CM, blaTEM and vanA as indicator genes of antibiotic resistance contamination in a hospital–urban wastewater treatment plant system. J Global Antimicrob Resist 2014, 2 (4), 309–315. [DOI] [PubMed] [Google Scholar]
- 38.Aminov RI; Chee-Sanford JC; Garrigues N; Teferedegne B; Krapac IJ; White BA; Mackie RI, Development, validation, and application of PCR primers for detection of tetracycline efflux genes of gram-negative bacteria. Appl Environ Microbiol 2002, 68 (4), 1786–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aminov RI; Garrigues-Jeanjean N; Mackie RI, Molecular ecology of tetracycline resistance: development and validation of primers for detection of tetracycline resistance genes encoding ribosomal protection proteins. Appl Environ Microbiol 2001, 67 (1), 22–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen J; Yu Z; Michel FC Jr.; Wittum T; Morrison M, Development and application of real-time PCR assays for quantification of erm genes conferring resistance to macrolides-lincosamides-streptogramin B in livestock manure and manure management systems. Appl Environ Microbiol 2007, 73 (14), 4407–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Diene SM; Bruder N; Raoult D; Rolain JM, Real-time PCR assay allows detection of the New Delhi metallo-beta-lactamase (NDM-1)-encoding gene in France. Int J Antimicrob Agents 2011, 37 (6), 544–6. [DOI] [PubMed] [Google Scholar]
- 42.Bolyen E; Rideout JR; Dillon MR; Bokulich NA; Abnet CC; Al-Ghalith GA; Alexander H; Alm EJ; Arumugam M; Asnicar F; Bai Y; Bisanz JE; Bittinger K; Brejnrod A; Brislawn CJ; Brown CT; Callahan BJ; Caraballo-Rodríguez AM; Chase J; Cope EK; Da Silva R; Diener C; Dorrestein PC; Douglas GM; Durall DM; Duvallet C; Edwardson CF; Ernst M; Estaki M; Fouquier J; Gauglitz JM; Gibbons SM; Gibson DL; Gonzalez A; Gorlick K; Guo J; Hillmann B; Holmes S; Holste H; Huttenhower C; Huttley GA; Janssen S; Jarmusch AK; Jiang L; Kaehler BD; Kang KB; Keefe CR; Keim P; Kelley ST; Knights D; Koester I; Kosciolek T; Kreps J; Langille MGI; Lee J; Ley R; Liu Y-X; Loftfield E; Lozupone C; Maher M; Marotz C; Martin BD; McDonald D; McIver LJ; Melnik AV; Metcalf JL; Morgan SC; Morton JT; Naimey AT; Navas-Molina JA; Nothias LF; Orchanian SB; Pearson T; Peoples SL; Petras D; Preuss ML; Pruesse E; Rasmussen LB; Rivers A; Robeson MS; Rosenthal P; Segata N; Shaffer M; Shiffer A; Sinha R; Song SJ; Spear JR; Swafford AD; Thompson LR; Torres PJ; Trinh P; Tripathi A; Turnbaugh PJ; Ul-Hasan S; van der Hooft JJJ; Vargas F; Vázquez-Baeza Y; Vogtmann E; von Hippel M; Walters W; Wan Y; Wang M; Warren J; Weber KC; Williamson CHD; Willis AD; Xu ZZ; Zaneveld JR; Zhang Y; Zhu Q; Knight R; Caporaso JG, Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nature Biotechnology 2019, 37 (8), 852–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Payne RB; Ghosh U; May HD; Marshall CW; Sowers KR, A pilot-scale field study: in situ treatment of PCB-impacted sediments with bioamended activated carbon. Environ Sci Technol 2019, 53 (5), 2626–2634. [DOI] [PubMed] [Google Scholar]
- 44.R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria, 2018. [Google Scholar]
- 45.Jari Oksanen, F. G. B., Friendly Michael, Kindt Roeland, Legendre Pierre, Dan McGlinn, O’Hara PRM,RB, Simpson Gavin L, Solymos Peter, H M. Henry.; Stevens E. S. a. H. W. Vegan: Community Ecology Package, R package version 2.5–6. https://CRAN.R-project.org/package=vegan, 2019.
- 46.Wickham H ggplot2: Elegant Graphics for Data Analysis, Springer-Verlag; NewYork: 2016. [Google Scholar]
- 47.Xing L; Yu H; Qi J; Jiang P; Sun B; Cui J; Ou C; Chang W; Hu Q, ErmF and ereD are responsible for erythromycin resistance in Riemerella anatipestifer. PLoS One 2015, 10 (6), e0131078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Arbizu PM pairwiseAdonis: Pairwise multilevel comparison using adonis, R packege version 0.3; 2019. [Google Scholar]
- 49.Segata N; Izard J; Waldron L; Gevers D; Miropolsky L; Garrett WS; Huttenhower C, Metagenomic biomarker discovery and explanation. Genome Biol 2011, 12 (6), R60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Borel N; Polkinghorne A; Pospischil A, A review on Chlamydial diseases in animals: Still a challenge for pathologists? Vet Pathol 2018, 55 (3), 374–390. [DOI] [PubMed] [Google Scholar]
- 51.Yu P; Wang C; Zhou J; Jiang L; Xue J; Li W, Influence of surface properties on adhesion forces and attachment of Streptococcus mutans to zirconia in vitro. Biomed Res Int 2016, 2016, 8901253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Werb M; Falcon Garcia C; Bach NC; Grumbein S; Sieber SA; Opitz M; Lieleg O, Surface topology affects wetting behavior of Bacillus subtilis biofilms. NPJ Biofilms Microbiomes 2017, 3, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lagree K; Mon HH; Mitchell AP; Ducker WA, Impact of surface topography on biofilm formation by Candida albicans. PLoS One 2018, 13 (6), e0197925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mohamed Azab El-Liethy BH, Effect of corroded and non corroded pipe materials on biofilm formation in water distribution systems. World Appl Sci J 2016, 34 (2), 214. [Google Scholar]
- 55.Bachmann RT, R. G. J. E., Biofouling: an historic and contemporary review of its causes, consequences and control in drinking water distribution systems. Biofilms 2005, 2 (3), 197–227. [Google Scholar]
- 56.Veloo ACM; Baas WH; Haan FJ; Coco J; Rossen JW, Prevalence of antimicrobial resistance genes in Bacteroides spp. and Prevotella spp. Dutch clinical isolates. Clin Microbiol Infect 2019, 25 (9), 1156 e9–1156 e13. [DOI] [PubMed] [Google Scholar]
- 57.Hainan Ai JX, Wei Huang, Qiang He, Bingjie Ni and Yinliang Wang, Mechanism and kinetics of biofilm growth process influenced by shear stress in sewers. Water Sci Technol 2015, 73 (7), 1572–1582. [DOI] [PubMed] [Google Scholar]
- 58.Zan F; Liang Z; Jiang F; Dai J; Chen G, Effects of food waste addition on biofilm formation and sulfide production in a gravity sewer. Water Res 2019, 157, 74–82. [DOI] [PubMed] [Google Scholar]
- 59.Laht M; Karkman A; Voolaid V; Ritz C; Tenson T; Virta M; Kisand V, Abundances of tetracycline, sulphonamide and beta-lactam antibiotic resistance genes in conventional wastewater treatment plants (WWTPs) with different waste load. PLoS One 2014, 9 (8), e103705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Luprano ML; De Sanctis M; Del Moro G; Di Iaconi C; Lopez A; Levantesi C, Antibiotic resistance genes fate and removal by a technological treatment solution for water reuse in agriculture. Sci Total Environ 2016, 571, 809–18. [DOI] [PubMed] [Google Scholar]
- 61.Xu J; Xu Y; Wang H; Guo C; Qiu H; He Y; Zhang Y; Li X; Meng W, Occurrence of antibiotics and antibiotic resistance genes in a sewage treatment plant and its effluent-receiving river. Chemosphere 2015, 119, 1379–1385. [DOI] [PubMed] [Google Scholar]
- 62.Pesesky MW, T H Wallace M, Wang B, Andleeb S, Burnham CD, KPC and NDM-1 genes in related Enterobacteriaceae strains and plasmids from Pakistan and the United States. Emerg Infect Dis 2015, 21 (6), 1034–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rasheed JK; Kitchel B; Zhu W; Anderson KF; Clark NC; Ferraro MJ; Savard P; Humphries RM; Kallen AJ; Limbago BM, New Delhi metallo-beta-lactamase-producing Enterobacteriaceae, United States. Emerg Infect Dis 2013, 19 (6), 870–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Norsigian CJ; Attia H; Szubin R; Yassin AS; Palsson BO; Aziz RK; Monk JM, Comparative genome-scale metabolic modeling of metallo-beta-lactamase-producing multidrug-resistant Klebsiella pneumoniae clinical isolates. Front Cell Infect Microbiol 2019, 9, 161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Auguet O; Pijuan M; Batista J; Borrego CM; Gutierrez O, Changes in microbial biofilm communities during colonization of sewer systems. Appl Environ Microbiol 2015, 81 (20), 7271–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.McLellan SL; Huse SM; Mueller-Spitz SR; Andreishcheva EN; Sogin ML, Diversity and population structure of sewage-derived microorganisms in wastewater treatment plant influent. Environ Microbiol 2010, 12 (2), 378–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Millar JA; Raghavan R, Accumulation and expression of multiple antibiotic resistance genes in Arcobacter cryaerophilus that thrives in sewage. PeerJ 2017, 5, e3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fisher JC; Levican A; Figueras MJ; McLellan SL, Population dynamics and ecology of Arcobacter in sewage. Front Microbiol 2014, 5, 525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liu Y; Dong Q; Shi H, Distribution and population structure characteristics of microorganisms in urban sewage system. Appl Microbiol Biotechnol 2015, 99 (18), 7723–34. [DOI] [PubMed] [Google Scholar]
- 70.Vinayak Kaushal MN, Love J Qualitative investigation of microbially induced corrosion of concrete in sanitary sewer pipe and manholes. In Pipelines 2018: Condition Assessment, Construction, and Rehabilitation, 2018. [Google Scholar]
- 71.Shen Y; Monroy GL; Derlon N; Janjaroen D; Huang C; Morgenroth E; Boppart SA; Ashbolt NJ; Liu WT; Nguyen TH, Role of biofilm roughness and hydrodynamic conditions in Legionella pneumophila adhesion to and detachment from simulated drinking water biofilms. Environ Sci Technol 2015, 49 (7), 4274–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gallandat K, Wolfe MK, Lantagne D. Surface cleaning and disinfection: Efficacy assessment of four chlorine types using Escherichia coli and the Ebola surrogate Phi6. Environ Sci Technol2017, 51 (8), 4624–4631. [DOI] [PubMed] [Google Scholar]
- 73.Mah TF; O’Toole GA, Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol 2001, 9 (1), 34–9. [DOI] [PubMed] [Google Scholar]
- 74.Cochran WL; McFeters GA; Stewart PS, Reduced susceptibility of thin Pseudomonas aeruginosa biofilms to hydrogen peroxide and monochloramine. J Appl Microbiol 2000, 88 (1), 22–30. [DOI] [PubMed] [Google Scholar]
- 75.Virto R; Manas P; Alvarez I; Condon S; Raso J, Membrane damage and microbial inactivation by chlorine in the absence and presence of a chlorine-demanding substrate. Appl Environ Microbiol 2005, 71 (9), 5022–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chii Shang ERB, Differentiation and quantification of free chlorine and inorganic chloramines in aqueous solution by MIMS. Environ Sci Technol 1999, 33, (13), 2218–2223. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


