Abstract
Time is of the essence in evaluating potential drugs and biologics for the treatment and prevention of COVID‐19. There are currently 876 randomized clinical trials (phase 2 and 3) of treatments for COVID‐19 registered on clinicaltrials.gov. Covariate adjustment is a statistical analysis method with potential to improve precision and reduce the required sample size for a substantial number of these trials. Though covariate adjustment is recommended by the U.S. Food and Drug Administration and the European Medicines Agency, it is underutilized, especially for the types of outcomes (binary, ordinal, and time‐to‐event) that are common in COVID‐19 trials. To demonstrate the potential value added by covariate adjustment in this context, we simulated two‐arm, randomized trials comparing a hypothetical COVID‐19 treatment versus standard of care, where the primary outcome is binary, ordinal, or time‐to‐event. Our simulated distributions are derived from two sources: longitudinal data on over 500 patients hospitalized at Weill Cornell Medicine New York Presbyterian Hospital and a Centers for Disease Control and Prevention preliminary description of 2449 cases. In simulated trials with sample sizes ranging from 100 to 1000 participants, we found substantial precision gains from using covariate adjustment–equivalent to 4–18% reductions in the required sample size to achieve a desired power. This was the case for a variety of estimands (targets of inference). From these simulations, we conclude that covariate adjustment is a low‐risk, high‐reward approach to streamlining COVID‐19 treatment trials. We provide an R package and practical recommendations for implementation.
Keywords: covariate adjustment, COVID‐19, ordinal outcomes, randomized trial, survival analysis
1. INTRODUCTION
This paper builds on our report (Benkeser et al., 2020) written in response to a request by the U.S. Food and Drug Administration (FDA) for statistical analysis recommendations for COVID‐19 treatment trials. We aim to help inform the choice of estimand (ie, target of inference) and analysis method to be used in COVID‐19 treatment trials. To this end, we describe treatment effect estimands for binary, ordinal, and time‐to‐event outcomes. Importantly, the interpretability of these estimands does not rely on correct specification of models. For binary outcomes, we consider the risk difference, relative risk, and odds ratio. For ordinal outcomes, we consider the difference in means, the Mann‐Whitney (rank‐based) estimand, and the average of the cumulative log odds ratios over levels of the outcome. For time‐to‐event outcomes, we consider the difference in restricted mean survival times, the difference in survival probabilities, and the ratio of survival probabilities.
For each estimand, we give a corresponding covariate‐adjusted estimator that (1) leverages information from baseline variables and (2) is robust to model misspecification. We introduce a new covariate adjustment method for ordinal outcomes, but use existing methods for binary and time‐to‐event outcomes. By incorporating baseline variable information, covariate‐adjusted estimators often enjoy smaller variance compared to estimators that ignore this information, thereby resulting in reductions in the required sample size to achieve a desired power.
To evaluate the performance of covariate‐adjusted estimators, we simulated two‐arm, randomized trials comparing a hypothetical COVID‐19 treatment versus standard of care. Our simulated distributions are derived from two sources: longitudinal data on over 500 patients hospitalized at Weill Cornell Medicine New York Presbyterian Hospital prior to March 28, 2020 and a preliminary description of 2449 cases reported to the Centers for Disease Control and Prevention (CDC) from February 12 to March 16, 2020. We focused on hospitalized, COVID‐19 positive patients and specified distributions for binary, ordinal, and time‐to‐event outcomes based on information collected on intensive care unit (ICU) admission, intubation with ventilation, and death. We conducted simulations using all three estimands when the outcome is ordinal, but only evaluated the risk difference when the outcome is binary and the restricted mean survival time and risk difference when the outcome is time to event.
After our aforementioned report (which contains some of our simulation results for ordinal and time‐to‐event outcomes), the FDA released a guidance for industry on COVID‐19 treatment and prevention trials (FDA, 2020). The guidance contains the following statement, which is similar to our key recommendation regarding covariate adjustment: “To improve the precision of treatment effect estimation and inference, sponsors should consider adjusting for prespecified prognostic baseline covariates (eg, age, baseline severity, comorbidities) in the primary efficacy analysis and should propose methods of covariate adjustment.”
There is already an extensive literature on the theory and practice of covariate adjustment, for example, Yang and Tsiatis (2001), Tsiatis et al. (2008), Zhang et al. (2008), Moore and van der Laan (2009a), Austin et al. (2010), Zhang and Gilbert (2010), and Jiang et al. (2019). However, covariate adjustment is underutilized, particularly for trials with a binary, ordinal, or time‐to‐event outcome. Since many COVID‐19 treatment trials focus on these types of outcomes, our goal is to demonstrate the potential benefits of covariate adjustment in these contexts. Recent examples of COVID‐19 treatment trials include a trial of dexamethasone with 28‐day mortality as the primary outcome (The RECOVERY Collaborative Group, 2020) and three trials of remdesivir with the following primary outcomes: clinical status on day 14 using a 7‐point ordinal scale (Goldman et al., 2020), time to clinical improvement (Wang et al., 2020), and time to death (Beigel et al., 2020).
The remainder of this paper is organized as follows. A brief background on covariate adjustment in randomized trials is provided in Section 2. Section 3 describes estimands and estimation strategies when the outcome is binary, ordinal, or time‐to‐event. Section 4 describes the methods underlying the simulation study, and Section 5 presents the simulation study results. Section 6 presents our recommendations for COVID‐19 treatment trials. A brief discussion is given in Section 7.
2. BACKGROUND ON COVARIATE ADJUSTMENT IN RANDOMIZED TRIALS
The ICH E9 Guidance on Statistical Methods for Analyzing Clinical Trials (FDA and EMA, 1998) states that “Pretrial deliberations should identify those covariates and factors expected to have an important influence on the primary variable(s), and should consider how to account for these in the analysis to improve precision and to compensate for any lack of balance between treatment groups.” The term “covariates” refers to baseline variables. Adjusting for prespecified, prognostic baseline variables (ie, variables that are correlated with the outcome) is called covariate adjustment. The primary goal of covariate adjustment is to improve precision in estimating the marginal treatment effect (Tsiatis et al., 2008). Examples of such marginal treatment effects, called estimands, are given in Section 3.
Though there appears to be a general agreement among regulators (EMA, 2015; FDA, 2019) that when the outcome is continuous, analysis of covariance (ANCOVA) may be used to appropriately adjust for baseline variables, there is a dearth of specific guidance for ordinal and time‐to‐event outcomes, which are of keen interest in COVID‐19 treatment trials. Even for binary outcomes, for which one possible adjustment method (Ge et al., 2011) was cited in the recent FDA COVID‐19 guidance (FDA, 2020), there has not been any study showing how much precision gain is to be expected by using covariate adjusted, rather than unadjusted, methods in the context of COVID‐19 treatment trials. In this work, we evaluate the performance of covariate‐adjusted estimators (hence, simply adjusted estimators) for binary, ordinal, and time‐to‐event outcomes. We explain the intuition for how covariate adjustment can lead to precision gains in Appendix A of the Supporting Information.
3. ESTIMANDS AND ANALYSIS METHODS
Throughout, we assume that treatment is assigned independently of baseline variables. All estimands are intention‐to‐treat in that they are contrasts between outcome distributions under assignment to treatment and under assignment to control. We let A denote study arm assignment, taking on the value 0 for a control group and 1 for a treatment group. We let Y denote the outcome of interest and denote a vector of baseline covariates.
We assume that participant data vectors are independent, identically distributed (i.i.d.) draws from an unknown, superpopulation distribution; this assumption is commonly made in statistical analyses of randomized trials. Our goal is to draw inferences about the superpopulation distribution.
3.1. Binary outcomes
We consider three estimands, though our simulation studies only involve the first. All probabilities below are marginal (as opposed to conditional on baseline variables). The outcome is coded as “good” (1) or “bad” (0). In what follows we let denote a random treatment‐outcome pair.
Estimand 1: Risk Difference. Difference between probability of bad outcome comparing treatment to control arms, that is, .
Estimand 2: Relative Risk. Ratio of probability of bad outcome comparing treatment to control arms, that is, .
Estimand 3: Odds Ratio. Ratio of odds of bad outcome, comparing treatment to control arms, that is, , where for each .
Estimators of each estimand 1–3 above can be constructed from estimators of the probability of a bad outcome for each study arm; for example, the risk difference can be estimated by the difference between the arm‐specific estimators. The unadjusted estimator of is the sample proportion of bad outcomes among patients assigned to arm . A covariate‐adjusted estimator of this quantity can be based on the standardization approach of Ge et al. (2011), as indicated in the FDA COVID‐19 guidance (FDA, 2020). This estimator is identical to that of Moore and van der Laan (2009a) and for the risk difference it is a special case of estimators from Scharfstein et al. (1999). First, a logistic regression model is fit for the probability of bad outcome given study arm and baseline variables. Next, for each participant (from both arms), a predicted probability of bad outcome is obtained under each possible arm assignment by plugging in the participant's baseline variables and setting arm assignment and , respectively, in the logistic regression model fit. Lastly, the covariate‐adjusted estimator of is the sample mean over all participants (pooling across arms) of the predicted probability of bad outcome setting .
3.2. Ordinal outcomes
We consider three estimands when the outcome is ordinal, with levels . Without loss of generality, we assume that higher values of the ordinal outcome are preferable. In what follows, we let and denote independent treatment–outcome pairs.
- Estimand 1: Difference in means (DIM). For a prespecified, real‐valued transformation of an outcome, the estimand is defined as
In most settings, this transformation will be monotone increasing so that larger values of the ordinal outcome will result in larger, and therefore preferable, transformed outcomes. Transformations could incorporate, for example, utilities assigned to each level, as has been done in some stroke trials (Chaisinanunkul et al., 2015; Nogueira et al., 2018). - Estimand 2: Mann‐Whitney (MW) estimand. This estimand reports the probability that a random individual assigned to treatment will have a better outcome than a random individual assigned to control, with ties broken at random. The estimand is defined as
- Estimand 3: Log‐odds ratio (LOR). We consider a nonparametric extension of the LOR (Díaz et al., 2016) defined as the average of the cumulative log odds ratios over levels 1 to of the outcome, namely
In the case that the distribution of the outcome given study arm is accurately described by a proportional odds model of the outcome against treatment (McCullagh, 1980), this estimand is equal to the coefficient associated with treatment.
All three estimands are smooth summaries of the treatment‐specific cumulative distribution functions (CDFs) of the ordinal outcome. The CDF for arm evaluated at is denoted by , and the corresponding probability mass function is denoted by . The estimands can be equivalently expressed in terms of the CDFs as follows:
To estimate these quantities, it suffices to estimate the arm‐specific CDFs and then to evaluate the summaries; such estimators are called plug‐in estimators.
The unadjusted estimator of the CDF in each arm is the empirical distribution in that arm. The resulting plug‐in estimator for the DIM is the difference between arms of sample means of the transformed outcomes. Also, the resulting plug‐in estimator (denoted M) for the MW estimand is closely related to the usual Mann‐Whitney U‐statistic , where n 0 and n 1 are the total sample sizes in the two study arms.
Model‐robust, covariate‐adjusted estimators are available for estimation of the MW estimand, for example, Vermeulen et al. (2015), and for the LOR estimand, for example, Díaz et al. (2016). We use a slightly different approach as described below. It is an area of future research to compare the performance of our method to the those from related works.
Our covariate‐adjusted estimator of the CDF in each arm, presented in Appendix B of the Supporting Information, leverages prognostic information in baseline variables. It uses working models, that is, models that are fit in the process of computing the estimator but which we do not assume to be correctly specified. Specifically, the adjusted estimator of the CDF for each study arm is based on the following arm‐specific, proportional odds working model for the cumulative probability of the outcome given the baseline variables: , for each with parameters and β ; the model for the other study arm is the same but with a separate set of parameters. Each model is fit using data from the corresponding study arm, yielding two working covariate‐conditional CDFs (one per arm). For each arm, the estimated marginal CDF is then obtained by averaging the corresponding conditional CDF across the empirical distribution of baseline covariates pooled across the two study arms. The above methods are implemented in an accompanying R package, drord.
The validity (ie, consistency and asymptotic normality) of the adjusted CDF estimator given above in no way relies on correct specification of the aforementioned working model. This property also holds for the estimators of Vermeulen et al. (2015) and Díaz et al. (2016).
3.3. Time‐to‐event outcomes
We consider three treatment effect estimands in the time‐to‐event setting, all of which are interpretable under violations of a proportional hazards assumption. To define these estimands, we let T be a time‐to‐event outcome, C be a right‐censoring time, A be a treatment indicator, and be a collection of baseline covariates.
- Estimand 2: Survival probability difference (also called risk difference, RD). Difference between arm‐specific probabilities of survival to a specified time , that is,
- Estimand 3: Relative risk (RR). Ratio of the arm‐specific probabilities of survival to a specified time , that is,
Analogous to the ordinal outcome case, estimators of these parameters can be constructed from estimators of the survival functions for each arm. One approach to constructing adjusted estimators, used here, involves discretizing time and then: (i) estimating the time‐specific hazard conditional on baseline variables, (ii) transforming to survival probabilities using the product‐limit formula, and (iii) marginalizing using the estimated covariate distribution (pooled across arms). The adjusted approach as implemented here (and elsewhere—see references below) has two key benefits relative to unadjusted alternatives such as using the unadjusted Kaplan‐Meier estimator (Kaplan and Meier, 1958). First, the adjusted estimator's consistency depends on an assumption of censoring being independent of the outcome given study arm and baseline covariates (), rather than an assumption of censoring in each arm being independent of the outcome marginally (). The former may be a more plausible assumption. Second, in large samples and under regularity conditions, the adjusted estimator is at least as precise as the unadjusted estimator in the case that censoring is completely at random, that is, that in each arm , .
Covariate‐adjusted estimators for time‐to‐event outcomes include, for example, Chen and Tsiatis (2001), Rubin and van der Laan (2008b), Moore and van der Laan (2009b), Lu and Tsiatis (2011), Stitelman et al. (2011), Brooks et al. (2013), Parast et al. (2014), Zhang (2014), and Benkeser et al. (2018, 2019). Díaz et al. (2019) compare the properties of some of these estimators. We used the covariate‐adjusted estimator of the RMST (specifically, the targeted minimum loss‐based estimator of the RMST) from Díaz et al. (2019) implemented in the R package survtmlerct. Time was discretized at the day level. Similar covariate‐adjusted estimators for the RD and RR are also available (Moore and van der Laan, 2009b; Benkeser et al., 2018, 2019). Both Díaz et al. (2019) and Benkeser et al. (2018) provide approaches that can be used to develop Wald‐type confidence intervals and corresponding tests of the null hypothesis of no treatment effect.
4. SIMULATION METHODS
4.1. Data‐generating distributions
In each setting below, we simulated trials with 1:1 randomization to the two arms and total enrollment of n = 100, 200, 500, and 1000. In each case, 1000 trials were simulated. In each simulated trial, the n participant data vectors are i.i.d. draws from a population data‐generating distribution that depends on the setting.
Data for simulated control‐arm participants were simulated based on real data, while data for simulated treatment‐arm participants were simulated by modifying the outcome distribution observed in the real data to achieve a desired level of treatment effect (details below). In all simulation settings, covariates are approximately, equally prognostic across arms and there is no treatment effect heterogeneity.
4.1.1. Binary outcomes
The data‐generating distributions are the same as for ordinal outcomes (below), except that we dichotomized the outcome into “bad” (death or survival with ICU admission) and “good” (survival without ICU admission).
4.1.2. Ordinal outcomes
We generated data based on CDC COVID‐19 Response Team (2020), which reported outcomes for individuals with COVID‐19. We focus on hospitalized patients. (See Appendix C of the Supporting Information for additional results pertaining to the non‐hospitalized population.) The ordinal outcome has three levels: death (level 1), survival with ICU admission (level 2), and survival without ICU admission (level 3). The following age categories define the single baseline variable (which is used for adjustment): 0‐19, 20‐44, 45‐54, 55‐64, 65‐74, 75‐84, and ≥85. In CDC COVID‐19 Response Team (2020), lower and upper estimates were reported for each age group‐specific outcome probability; we used the average of these within each age group to define our data‐generating distributions. For the hospitalized COVID‐19 positive population, the resulting outcome probabilities for each age group are listed in Table 1.
TABLE 1.
Hospitalized, COVID‐19 positive population: age and conditional outcome distributions based on data from CDC COVID‐19 Response Team (2020) that we use for defining the control arm distribution in the ordinal outcome simulation studies. “ICU” represents ICU admission
| Age | P (age) | P (death | age) | P (ICU and survived | age) | P (no ICU and survived | age) |
|---|---|---|---|---|
| 0–19 | 0.004 | 0.000 | 0.000 | 1.000 |
| 20‐44 | 0.189 | 0.009 | 0.177 | 0.815 |
| 45‐54 | 0.162 | 0.026 | 0.319 | 0.655 |
| 55‐64 | 0.165 | 0.079 | 0.314 | 0.607 |
| 65‐74 | 0.225 | 0.105 | 0.373 | 0.521 |
| 75‐84 | 0.143 | 0.166 | 0.465 | 0.369 |
| ≥ 85 | 0.112 | 0.371 | 0.347 | 0.281 |
We separately considered two types of treatment effects in our data‐generating distributions: no treatment effect and an effective treatment. For the former, we randomly sampled n age‐outcome pairs according to the distribution in Table 1 and then independently assigned study arm with probability 1/2 for each arm.
For the latter case (effective treatment), we randomly generated control arm participants as in the previous paragraph and randomly generated treatment arm participants by modifying the values in Table 1 to achieve a desired level of true treatment effect. Specifically, the probabilities P(ICU admission and survived | age) in column 4 were proportionally reduced, while P(no ICU admission and survived | age) were increased by an equal amount. The probabilities of death given age in column 3 were not changed. This modified table corresponds to a scenario where the treatment has no effect on the probability of death but decreases the odds of ICU admission among those who survive by the same relative amount in each age category.
The aforementioned relative reduction (and the resulting treatment effect) was separately selected for each sample size n = 100, 200, 500, 1000. For the DIM estimand at each sample size, we selected treatment effect sizes such that a t‐test using the unadjusted estimator would achieve roughly 50% and 80% power, respectively, to reject the null hypothesis of no treatment effect. For sample size , we instead set this relative reduction to achieve roughly 30% and 40% power, respectively, because there did not exist a relative reduction that achieved 80% power at this sample size. The same data‐generating distributions used for the DIM estimand were also used for the MW and LOR estimands.
In our simulations, we used the adjusted estimator described in Section 3.2, where age is coded using the categories in Table 1. Specifically, these age categories are included as the main terms in the linear parts of the proportional odds working models.
For simplicity, for binary and ordinal outcomes we simulated trials with no missing data. However, the methods we used can adjust for missing outcomes. (See Appendix B of the Supporting Information.)
4.1.3. Time‐to‐event outcomes
In this simulation, the outcome is time from hospitalization to the first of intubation or death, and the predictive variables used are sex, age, whether the patient required supplemental oxygen at Emergency Department (ED) presentation, dyspnea, hypertension, and the presence of bilateral infiltrates on the chest x‐ray. We focus on RMST 14 days after hospitalization, and the RD of remaining intubation‐free and alive 7 days after hospitalization.
Our data generation distribution is based on a database of over 500 patients hospitalized at Weill Cornell Medicine New York Presbyterian Hospital prior to March 28, 2020. Outcome information was known for all patients through at least day 14. Patient data were resampled with replacement to generate 1000 datasets, for each of the sizes n = 100, 200, 500, and 1000. For each dataset, a hypothetical treatment variable was drawn from a Bernoulli distribution with probability 0.5 independently of all other variables. Positive treatment effects were simulated by adding an independent random draw from a χ2 distribution to each participant's outcome in the treatment arm; we used χ2 distributions with two and four degrees of freedom, respectively, to generate two different effect sizes. These data‐generating distributions correspond to a difference in RMST of 0.507 and 1.004 at 14 days, and an RD of 3.5% and 8.8% at 7 days, respectively. Five percent of the patients were selected at random to be censored, and their censoring time was drawn from a uniform distribution on .
We compare the performance of the unadjusted, Kaplan‐Meier‐based estimator to the covariate‐adjusted estimator. These estimators are defined in Sections 4 and 6 of Díaz et al. (2019), respectively, and implemented in the R package survtmlerct. Wald‐type confidence intervals and corresponding tests of the null hypothesis of no effect are reported.
4.2. Performance criteria
We compare the type I error and power of tests of the null hypothesis H 0 of no treatment effect based on unadjusted and adjusted estimators, both within and across estimands. For each estimand, we also compare the bias, variance, and mean squared error of the unadjusted and the adjusted estimators.
We approximate the relative efficiency of the unadjusted relative to the adjusted estimator by the ratio of the mean squared error of the latter to the mean squared error of the former. In all of our simulation studies, this is similar to the corresponding ratio of variances, since the bias squared was always much smaller than the variance. One minus this relative efficiency is approximately the proportion reduction in sample size needed for a covariate‐adjusted estimator to achieve the same power as the unadjusted estimator (van der Vaart, 1998, pp. 110–111).
5. SIMULATION RESULTS
For binary and ordinal outcomes, we present results that use the nonparametric BCa bootstrap (Efron and Tibshirani, 1994) for confidence intervals and hypothesis tests. We used 1000 replicates for each BCa bootstrap confidence interval. While we recommend 10 000 replicates in practice, the associated computational time was too demanding for our simulation study. Nonetheless, we expect similar or slightly better performance with an increased number of bootstrap samples. Results that use closed‐form, Wald‐based inference methods are presented in Appendix C of the Supporting Information.
For time‐to‐event outcomes, we used Wald‐based confidence intervals since these made the computations faster compared to the BCa bootstrap method.
5.1. Binary outcomes
Table 2 compares the performance of the unadjusted and adjusted estimators when “bad outcome” is defined as death or survival with ICU admission, and the estimand is the risk difference. The relative efficiency of the unadjusted method relative to the adjusted method varied from 0.92 to 0.86. This is roughly equivalent to needing 8–14% smaller sample size when using the adjusted estimator compared to the unadjusted estimator, to achieve the same power.
TABLE 2.
Results for the binary outcome and risk difference (RD) estimand in the hospitalized population
| n | Estimator type | Effect | P(reject H 0) | MSE | Bias | Variance | Rel. Eff. |
|---|---|---|---|---|---|---|---|
| 100 | Unadjusted | 0.000 | 0.030 | 0.995 | 0.022 | 0.996 | 1.000 |
| 100 | Adjusted | 0.000 | 0.052 | 0.900 | 0.023 | 0.900 | 0.904 |
| 100 | Unadjusted | −0.161 | 0.307 | 0.877 | 0.011 | 0.878 | 1.000 |
| 100 | Adjusted | −0.161 | 0.420 | 0.791 | 0.009 | 0.792 | 0.902 |
| 100 | Unadjusted | −0.201 | 0.463 | 0.829 | 0.025 | 0.829 | 1.000 |
| 100 | Adjusted | −0.201 | 0.607 | 0.755 | 0.023 | 0.755 | 0.911 |
| 200 | Unadjusted | 0.000 | 0.038 | 1.006 | −0.024 | 1.007 | 1.000 |
| 200 | Adjusted | 0.000 | 0.049 | 0.907 | −0.030 | 0.906 | 0.901 |
| 200 | Unadjusted | −0.147 | 0.527 | 0.917 | 0.002 | 0.918 | 1.000 |
| 200 | Adjusted | −0.147 | 0.633 | 0.801 | −0.009 | 0.802 | 0.873 |
| 200 | Unadjusted | −0.201 | 0.821 | 0.864 | 0.010 | 0.865 | 1.000 |
| 200 | Adjusted | −0.201 | 0.895 | 0.749 | −0.001 | 0.750 | 0.867 |
| 500 | Unadjusted | 0.000 | 0.036 | 1.038 | 0.020 | 1.039 | 1.000 |
| 500 | Adjusted | 0.000 | 0.043 | 0.897 | 0.024 | 0.898 | 0.864 |
| 500 | Unadjusted | −0.093 | 0.542 | 0.994 | −0.017 | 0.995 | 1.000 |
| 500 | Adjusted | −0.093 | 0.611 | 0.863 | −0.012 | 0.863 | 0.868 |
| 500 | Unadjusted | −0.126 | 0.798 | 0.979 | −0.013 | 0.980 | 1.000 |
| 500 | Adjusted | −0.126 | 0.862 | 0.850 | −0.007 | 0.851 | 0.868 |
| 1000 | Unadjusted | 0.000 | 0.033 | 0.932 | 0.012 | 0.933 | 1.000 |
| 1000 | Adjusted | 0.000 | 0.038 | 0.829 | 0.019 | 0.829 | 0.889 |
| 1000 | Unadjusted | −0.058 | 0.440 | 0.932 | 0.014 | 0.933 | 1.000 |
| 1000 | Adjusted | −0.058 | 0.507 | 0.857 | 0.021 | 0.857 | 0.919 |
| 1000 | Unadjusted | −0.091 | 0.837 | 0.898 | 0.012 | 0.899 | 1.000 |
| 1000 | Adjusted | −0.091 | 0.892 | 0.817 | 0.020 | 0.818 | 0.910 |
BCa bootstrap is used for confidence intervals and hypothesis testing. “Effect” denotes the true estimand value; “MSE” denotes mean squared error; “Rel. Eff.” denotes relative efficiency, which we approximate as the ratio of the MSE of the estimator under consideration to the MSE of the unadjusted estimator. In each block of six rows, the first two rows involve no treatment effect and the last four rows involve a benefit from treatment. MSE and variance are scaled by n; bias is scaled by .
Type I error of the covariate‐adjusted method was comparable to that of the unadjusted method. The covariate‐adjusted method achieved higher power across all settings. Absolute gains in power varied from 5% to 14%.
5.2. Ordinal outcomes
Tables 3, 4, and 5 display results for the DIM, MW, and LOR estimands, respectively. The relative efficiency of the unadjusted methods relative to adjusted methods varied from 0.94 to 0.85 for the DIM, 0.94 to 0.85 for the MW estimand, and 0.93 to 0.85 for the LOR. This is roughly equivalent to needing 6‐15% (DIM), 6‐15% (MW), and 7–15% (LOR) smaller sample sizes, respectively, when using the adjusted estimator compared to the unadjusted estimator, to achieve the same power.
TABLE 3.
Results for the ordinal outcome and DIM estimand in the hospitalized population
| n | Estimator type | Effect | P(reject H 0) | MSE | Bias | Variance | Rel. Eff. |
|---|---|---|---|---|---|---|---|
| 100 | Unadjusted | 0.000 | 0.059 | 1.853 | −0.038 | 1.854 | 1.000 |
| 100 | Adjusted | 0.000 | 0.054 | 1.640 | −0.046 | 1.639 | 0.885 |
| 100 | Unadjusted | 0.190 | 0.287 | 1.757 | −0.036 | 1.758 | 1.000 |
| 100 | Adjusted | 0.190 | 0.296 | 1.606 | −0.038 | 1.606 | 0.914 |
| 100 | Unadjusted | 0.244 | 0.419 | 1.645 | −0.035 | 1.646 | 1.000 |
| 100 | Adjusted | 0.244 | 0.449 | 1.543 | −0.025 | 1.544 | 0.938 |
| 200 | Unadjusted | 0.000 | 0.048 | 1.848 | 0.023 | 1.850 | 1.000 |
| 200 | Adjusted | 0.000 | 0.054 | 1.640 | 0.033 | 1.641 | 0.888 |
| 200 | Unadjusted | 0.195 | 0.531 | 1.838 | −0.022 | 1.839 | 1.000 |
| 200 | Adjusted | 0.195 | 0.587 | 1.623 | −0.004 | 1.624 | 0.883 |
| 200 | Unadjusted | 0.252 | 0.763 | 1.798 | 0.019 | 1.800 | 1.000 |
| 200 | Adjusted | 0.252 | 0.811 | 1.565 | 0.060 | 1.563 | 0.870 |
| 500 | Unadjusted | 0.000 | 0.056 | 1.898 | −0.061 | 1.896 | 1.000 |
| 500 | Adjusted | 0.000 | 0.042 | 1.604 | −0.066 | 1.601 | 0.845 |
| 500 | Unadjusted | 0.126 | 0.533 | 2.013 | −0.025 | 2.014 | 1.000 |
| 500 | Adjusted | 0.126 | 0.581 | 1.786 | −0.036 | 1.786 | 0.887 |
| 500 | Unadjusted | 0.171 | 0.781 | 1.986 | −0.022 | 1.987 | 1.000 |
| 500 | Adjusted | 0.171 | 0.820 | 1.788 | −0.022 | 1.789 | 0.900 |
| 1000 | Unadjusted | 0.000 | 0.050 | 1.852 | −0.005 | 1.854 | 1.000 |
| 1000 | Adjusted | 0.000 | 0.044 | 1.661 | −0.013 | 1.663 | 0.897 |
| 1000 | Unadjusted | 0.089 | 0.558 | 1.842 | −0.006 | 1.844 | 1.000 |
| 1000 | Adjusted | 0.089 | 0.586 | 1.662 | −0.021 | 1.664 | 0.903 |
| 1000 | Unadjusted | 0.126 | 0.839 | 1.819 | 0.003 | 1.821 | 1.000 |
| 1000 | Adjusted | 0.126 | 0.881 | 1.658 | −0.006 | 1.660 | 0.911 |
BCa bootstrap is used for confidence intervals and hypothesis testing. “Effect” denotes the true estimand value; “MSE” denotes mean squared error; “Rel. Eff.” denotes relative efficiency, which we approximate as the ratio of the MSE of the estimator under consideration to the MSE of the unadjusted estimator. In each block of six rows, the first two rows involve no treatment effect and the last four rows involve a benefit from treatment. MSE and variance are scaled by n; bias is scaled by .
TABLE 4.
Results for ordinal outcome and MW estimand in the hospitalized population
| n | Estimator type | Effect | P(reject H 0) | MSE | Bias | Variance | Rel. Eff. |
|---|---|---|---|---|---|---|---|
| 100 | Unadjusted | 0.500 | 0.054 | 0.263 | −0.014 | 0.263 | 1.000 |
| 100 | Adjusted | 0.500 | 0.050 | 0.234 | −0.017 | 0.234 | 0.890 |
| 100 | Unadjusted | 0.585 | 0.389 | 0.226 | −0.010 | 0.226 | 1.000 |
| 100 | Adjusted | 0.585 | 0.431 | 0.208 | −0.011 | 0.208 | 0.919 |
| 100 | Unadjusted | 0.609 | 0.625 | 0.205 | −0.009 | 0.205 | 1.000 |
| 100 | Adjusted | 0.609 | 0.670 | 0.194 | −0.006 | 0.194 | 0.944 |
| 200 | Unadjusted | 0.500 | 0.053 | 0.264 | 0.010 | 0.264 | 1.000 |
| 200 | Adjusted | 0.500 | 0.056 | 0.236 | 0.014 | 0.236 | 0.895 |
| 200 | Unadjusted | 0.587 | 0.720 | 0.232 | −0.006 | 0.232 | 1.000 |
| 200 | Adjusted | 0.587 | 0.776 | 0.205 | −0.001 | 0.206 | 0.886 |
| 200 | Unadjusted | 0.612 | 0.924 | 0.217 | 0.008 | 0.217 | 1.000 |
| 200 | Adjusted | 0.612 | 0.953 | 0.190 | 0.021 | 0.190 | 0.879 |
| 500 | Unadjusted | 0.500 | 0.063 | 0.271 | −0.018 | 0.271 | 1.000 |
| 500 | Adjusted | 0.500 | 0.044 | 0.230 | −0.020 | 0.230 | 0.848 |
| 500 | Unadjusted | 0.556 | 0.710 | 0.262 | −0.001 | 0.262 | 1.000 |
| 500 | Adjusted | 0.556 | 0.749 | 0.231 | −0.008 | 0.231 | 0.882 |
| 500 | Unadjusted | 0.576 | 0.935 | 0.249 | 0.000 | 0.250 | 1.000 |
| 500 | Adjusted | 0.576 | 0.958 | 0.224 | −0.002 | 0.224 | 0.897 |
| 1000 | Unadjusted | 0.500 | 0.039 | 0.255 | −0.004 | 0.255 | 1.000 |
| 1000 | Adjusted | 0.500 | 0.040 | 0.227 | −0.007 | 0.228 | 0.894 |
| 1000 | Unadjusted | 0.540 | 0.722 | 0.243 | −0.005 | 0.243 | 1.000 |
| 1000 | Adjusted | 0.540 | 0.745 | 0.220 | −0.013 | 0.220 | 0.906 |
| 1000 | Unadjusted | 0.556 | 0.956 | 0.234 | −0.003 | 0.234 | 1.000 |
| 1000 | Adjusted | 0.556 | 0.970 | 0.214 | −0.009 | 0.214 | 0.915 |
BCa bootstrap is used for confidence intervals and hypothesis testing. “Effect” denotes the true estimand value; “MSE” denotes mean squared error; “Rel. Eff.” denotes relative efficiency, which we approximate as the ratio of the MSE of the estimator under consideration to the MSE of the unadjusted estimator. In each block of six rows, the first two rows involve no treatment effect and the last four rows involve a benefit from treatment. MSE and variance are scaled by n; bias is scaled by .
TABLE 5.
Results for the ordinal outcome and LOR estimand in the hospitalized population
| n | Estimator type | Effect | P(reject H 0) | MSE | Bias | Variance | Rel. Eff. |
|---|---|---|---|---|---|---|---|
| 100 | Unadjusted | 0.000 | 0.063 | 23.243 | 0.149 | 23.244 | 1.000 |
| 100 | Adjusted | 0.000 | 0.060 | 20.663 | 0.178 | 20.652 | 0.889 |
| 100 | Unadjusted | −0.432 | 0.108 | 25.783 | 0.015 | 25.809 | 1.000 |
| 100 | Adjusted | −0.432 | 0.120 | 23.461 | 0.065 | 23.480 | 0.910 |
| 100 | Unadjusted | −0.593 | 0.163 | 28.183 | −0.063 | 28.207 | 1.000 |
| 100 | Adjusted | −0.593 | 0.183 | 26.138 | −0.039 | 26.163 | 0.927 |
| 200 | Unadjusted | 0.000 | 0.037 | 20.717 | −0.032 | 20.736 | 1.000 |
| 200 | Adjusted | 0.000 | 0.031 | 18.285 | −0.064 | 18.300 | 0.883 |
| 200 | Unadjusted | −0.447 | 0.229 | 24.220 | −0.008 | 24.244 | 1.000 |
| 200 | Adjusted | −0.447 | 0.239 | 21.329 | −0.008 | 21.351 | 0.881 |
| 200 | Unadjusted | −0.619 | 0.383 | 26.778 | −0.231 | 26.751 | 1.000 |
| 200 | Adjusted | −0.619 | 0.436 | 23.233 | −0.277 | 23.180 | 0.868 |
| 500 | Unadjusted | 0.000 | 0.048 | 20.373 | 0.269 | 20.321 | 1.000 |
| 500 | Adjusted | 0.000 | 0.039 | 17.249 | 0.284 | 17.186 | 0.847 |
| 500 | Unadjusted | −0.272 | 0.252 | 23.800 | 0.134 | 23.806 | 1.000 |
| 500 | Adjusted | −0.272 | 0.277 | 21.157 | 0.209 | 21.134 | 0.889 |
| 500 | Unadjusted | −0.383 | 0.442 | 24.797 | 0.099 | 24.812 | 1.000 |
| 500 | Adjusted | −0.383 | 0.473 | 22.250 | 0.170 | 22.244 | 0.897 |
| 1000 | Unadjusted | 0.000 | 0.055 | 20.669 | −0.020 | 20.690 | 1.000 |
| 1000 | Adjusted | 0.000 | 0.048 | 18.547 | 0.001 | 18.566 | 0.897 |
| 1000 | Unadjusted | −0.189 | 0.243 | 21.127 | −0.028 | 21.147 | 1.000 |
| 1000 | Adjusted | −0.189 | 0.267 | 18.864 | 0.055 | 18.880 | 0.893 |
| 1000 | Unadjusted | −0.272 | 0.464 | 21.606 | −0.071 | 21.623 | 1.000 |
| 1000 | Adjusted | −0.272 | 0.504 | 19.444 | 0.017 | 19.464 | 0.900 |
BCa bootstrap is used for confidence intervals and hypothesis testing. “Effect” denotes the true estimand value; “MSE” denotes mean squared error; “Rel. Eff.” denotes relative efficiency, which we approximate as the ratio of the MSE of the estimator under consideration to the MSE of the unadjusted estimator. In each block of six rows, the first two rows involve no treatment effect and the last four rows involve a benefit from treatment. MSE and variance are scaled by n; bias is scaled by .
Type I error control of the covariate‐adjusted methods was comparable to that of the unadjusted methods. The covariate‐adjusted methods achieved higher power across all settings. Absolute gains in power varied from 1% to 6% for the DIM, 1% to 6% for the MW estimand, and 1% to 5% for the LOR.
5.3. Time‐to‐event outcomes
Table 6 displays the results for RMST estimators, where the baseline variables adjusted for include age and sex along with the four other variables described in Section 4.1.3. First consider the no treatment effect case. At sample sizes , the relative efficiencies were 0.96, 0.89, 0.83, 0.82, respectively; this is roughly equivalent to needing 4%, 11%, 17%, 18% smaller sample size to achieve the same power as using the unadjusted estimator, respectively. The results were similar for the positive treatment effect cases.
TABLE 6.
Results for difference in RMST at 14 days estimand in hospitalized population, when the adjusted estimator uses all six baseline variables from Section 4.1.3
| Sample size | Estimator type | Effect | P(reject H 0) | MSE | Bias | Variance | Rel. Eff. |
|---|---|---|---|---|---|---|---|
| 100 | Unadjusted | 0.000 | 0.011 | 76.296 | 0.014 | 76.304 | 1.000 |
| 100 | Adjusted | 0.000 | 0.035 | 73.480 | 0.013 | 73.488 | 0.963 |
| 100 | Unadjusted | 0.507 | 0.025 | 67.882 | −0.938 | 67.008 | 1.000 |
| 100 | Adjusted | 0.507 | 0.063 | 62.857 | −0.668 | 62.418 | 0.926 |
| 100 | Unadjusted | 1.004 | 0.087 | 53.738 | −2.030 | 49.622 | 1.000 |
| 100 | Adjusted | 1.004 | 0.154 | 50.988 | −1.804 | 47.738 | 0.949 |
| 200 | Unadjusted | 0.000 | 0.044 | 95.651 | −0.131 | 95.644 | 1.000 |
| 200 | Adjusted | 0.000 | 0.055 | 85.260 | −0.176 | 85.238 | 0.891 |
| 200 | Unadjusted | 0.507 | 0.108 | 80.512 | −0.332 | 80.410 | 1.000 |
| 200 | Adjusted | 0.507 | 0.131 | 71.970 | −0.187 | 71.943 | 0.894 |
| 200 | Unadjusted | 1.004 | 0.330 | 62.739 | −1.014 | 61.718 | 1.000 |
| 200 | Adjusted | 1.004 | 0.399 | 56.397 | −0.770 | 55.810 | 0.899 |
| 500 | Unadjusted | 0.000 | 0.051 | 100.299 | −0.042 | 100.307 | 1.000 |
| 500 | Adjusted | 0.000 | 0.054 | 83.466 | −0.008 | 83.474 | 0.832 |
| 500 | Unadjusted | 0.507 | 0.226 | 87.159 | 0.085 | 87.160 | 1.000 |
| 500 | Adjusted | 0.507 | 0.274 | 71.673 | 0.155 | 71.656 | 0.822 |
| 500 | Unadjusted | 1.004 | 0.735 | 72.850 | −0.032 | 72.856 | 1.000 |
| 500 | Adjusted | 1.004 | 0.816 | 62.236 | 0.150 | 62.220 | 0.854 |
| 1000 | Unadjusted | 0.000 | 0.052 | 99.702 | 0.113 | 99.700 | 1.000 |
| 1000 | Adjusted | 0.000 | 0.053 | 81.859 | 0.144 | 81.846 | 0.821 |
| 1000 | Unadjusted | 0.507 | 0.411 | 87.420 | 0.282 | 87.349 | 1.000 |
| 1000 | Adjusted | 0.507 | 0.492 | 71.611 | 0.329 | 71.510 | 0.819 |
| 1000 | Unadjusted | 1.004 | 0.958 | 76.466 | 0.282 | 76.394 | 1.000 |
| 1000 | Adjusted | 1.004 | 0.980 | 63.461 | 0.360 | 63.339 | 0.830 |
Confidence intervals and hypothesis tests are Wald‐based. “Effect” denotes the true estimand value; “MSE” denotes mean squared error; “Rel. Eff.” denotes relative efficiency, which we approximate as the ratio of the MSE of the estimator under consideration to the MSE of the unadjusted estimator. In each block of four rows, the first two rows involve no treatment effect and the last two rows involve a benefit from treatment. MSE and variance are scaled by n; bias is scaled by .
Type I error control of the covariate‐adjusted method was comparable to that of the unadjusted method. The covariate‐adjusted methods achieved higher power across all settings, with absolute gains varying from 2% to 8%.
To evaluate the importance of adjusting for multiple baseline variables, we also evaluated an adjusted RMST estimator that only adjusts for age and sex; see Appendix C of the Supporting Information. The gains of the covariate‐adjusted methods relative to the unadjusted methods were small, with absolute gains in power of approximately 0‐1% and relative efficiency ranging from 0.96 to 1.00. These results suggest that there can be a meaningful benefit from adjusting for prognostic covariates beyond just age and sex.
We also considered the RD estimand; see Appendix C of the Supporting Information. The results (when adjusting for age and sex along with the four other variables described in Section 4.1.3) are qualitatively similar, except with slightly smaller precision gains, to those for the RMST in Table 6.
The type I error in Table 6 for the unadjusted estimator at is 1.1%, much smaller than the nominal level. We conjecture this is due to the Wald‐type asymptotic inference procedure being a poor approximation at this sample size. This is illustrated by the fact that the scaled variance at is much smaller than the scaled variance at . Similar comments apply to some of the results in Appendix C of the Supporting Information.
6. RECOMMENDATIONS FOR TARGET OF INFERENCE AND PRIMARY EFFICACY ANALYSIS
Recommendations below that do not reference related work or our simulation results are based on the authors' experience.
-
(1)
Estimand when the outcome is ordinal. If a utility function can be agreed upon to transform the outcome to a score with a clinically meaningful scale, then we recommend using the difference between the transformed means in the treatment and control arms. Otherwise, we recommend using the unweighted difference between means or the MW estimand. We recommend against estimating LOR, since clinical interpretation requires considerable nuance (Díaz et al., 2016) and the corresponding estimators (even unadjusted ones) can be unstable at small sample sizes (Appendix C of the Supporting Information).
-
(2)
Covariate adjustment. Based on our simulations, we recommend adjustment for prognostic baseline variables to improve precision and power. In the context of COVID‐19 trials, we expect improvements to be substantial since there are already several known prognostic baseline variables, for example, age and comorbidities. We did not consider it here, but one may consider using an algorithm for variable selection from a prespecified list of candidate variables; see, for example, Tsiatis et al. (2008), Rubin and van der Laan (2008a), Moore et al. (2011), Bloniarz et al. (2016), Wager et al. (2016), and Tian et al. (2019). The entire statistical procedure should be prespecified (FDA and EMA, 1998).
-
(3)
Confidence intervals and hypothesis testing. Based on our simulations for binary and ordinal outcomes, hypothesis tests and confidence intervals had improved performance when using the bootstrap (BCa method) compared to using Wald statistics. (For time‐to‐event outcomes, only Wald statistics were used due to computational limitations in implementing the bootstrap in simulations.) We recommend that the nonparametric bootstrap (BCa method) be used with 10 000 replicates for constructing a confidence interval. The entire estimation procedure, including any model fitting, should be repeated in each replicate dataset. Hypothesis tests can be conducted either by inverting the confidence interval or by permutation methods—the latter may be especially useful in smaller sample size trials in order to achieve the desired Type I error rate. Vermeulen et al. (2015) present such a permutation‐based test for the MW estimand based on a different covariate‐adjusted estimator than presented here.
-
(4)
Information monitoring. We summarize our recommendations below and give more detailed recommendations in Appendix E of the Supporting Information. First, consider trials without interim analyses. Information monitoring can be used to determine how long the trial will continue. Before starting the trial, one computes the information level required to achieve the desired power at a fixed alternative. Then during the trial, the accrued information (defined as the reciprocal of the estimator's variance) is monitored and the trial is continued until the required information level is surpassed. In this way, covariate adjustment can lead to faster trials even when the treatment effect is zero (ie, when the null hypothesis is true); this may be more ethical in settings where it is desirable to stop as early as possible to avoid unnecessary exposure to side effects.
Next, consider trials with interim analyses. For the estimands and adjusted estimators that we considered for continuous, binary, or ordinal outcomes, one can directly apply the group sequential, information‐based designs of Scharfstein et al. (1997); Jennison and Turnbull (1997, 1999). This can be done as long as data from pipeline participants, that is, participants who enrolled but have not been in the study long enough to have their primary outcomes measured, are not used when conducting interim analyses. This is because the key property needed to apply the aforementioned group sequential designs, called the independent‐increments property, is only guaranteed to hold if pipeline participant information is not used. There are methods for modifying the estimators through orthogonalization so that the independent increments property holds even when using pipeline participant information (and similarly when the outcome is time‐to‐event), but this was not simulated in our paper and is an area of future research.
-
(5)
Plotting the CDF and the probability mass function when the outcome is ordinal. Regardless of which treatment effect definition is used in the primary efficacy analysis, we recommend that the covariate‐adjusted estimate of the probability mass function (PMF) and/or CDF of the primary outcome be plotted for each study arm when the outcome is ordinal. Pointwise and simultaneous confidence intervals should be displayed (where the latter account for multiple comparisons). This is analogous to plotting Kaplan‐Meier curves for time‐to‐event outcomes, which can help in interpreting the trial results. For example, Figure 1 shows covariate‐adjusted estimates of the CDF and PMF for a dataset from our simulation study. From the plots, it is evident that the effect of the treatment on the ordinal outcome is primarily through preventing ICU admission, with no impact on probability of death.
-
(6)
Missing covariates. We do not recommend adjusting for baseline covariates that are expected to have high levels of missing data. For the situation with low levels of missing data, it is simplest to singly impute missing values based only on data from those baseline covariates that were observed. To ensure that treatment assignment is independent of all baseline covariates (including imputed ones), no treatment or outcome information should be used in this imputation. For performing inference based on the bootstrap, the bootstrap sample should be drawn first, then missing covariates should be imputed.
-
(7)
Missing ordinal outcomes. We recommend handling missing ordinal outcomes using methods that are robust to model misspecification, such as the one described in Appendix B of the Supporting Information. Compared to a complete‐case analysis, these approaches weaken the assumption of missingness from missing completely at random to missing at random. Nevertheless, these methods are still subject to bias in the presence of unmeasured factors that influence the study outcome and missingness probability. Therefore, trials should seek to minimize the likelihood of missing outcomes and employ relevant sensitivity analyses to address robustness of studying findings to assumptions about missing data (National Research Council, 2010); this applies to all outcome types.
-
(8)
Loss to follow up with time‐to‐event outcomes. We recommend accounting for loss‐to‐follow‐up using methods that are robust to model misspecification such as those described in Benkeser et al. (2018) and Díaz et al. (2019). These methods rely on a potentially more plausible condition on the censoring distribution than do unadjusted methods, as discussed in Section 3.3. The covariate‐adjusted estimator that we used for the restricted mean survival time in the time‐to‐event setting is robust to misspecification of one of its working models (as long as the other is correctly specified) under censoring being independent of the outcome given baseline variables and arm assignment.
FIGURE 1.
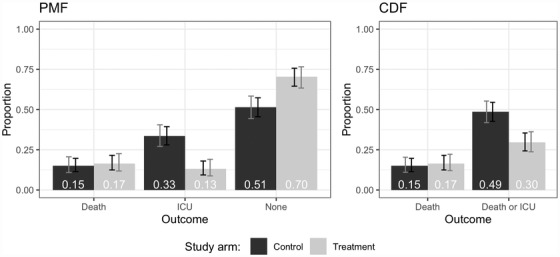
Example figures illustrating covariate‐adjusted estimates of the PMF and CDF by study arm with pointwise (black) and simultaneous (gray) confidence intervals. “ICU” represents survival and ICU admission; “None” represents survival and no ICU admission
7. DISCUSSION
In our simulated data‐generating distributions, the correlations between baseline variables and the outcome were similar for each arm. Because we designed our data‐generating distributions to mimic the correlations between baseline variables and outcomes from observational study data, these may be reasonable approximations to the control arm (ie, standard of care) of a trial involving the same population. If the treatment arm in such a trial has similar correlations between baseline variables and the outcome, then the precision gains in such a trial may be similar to those in our simulations. However, if the treatment arm in such a trial has smaller correlations between baseline variables and the outcome, then the precision gains in such a trial may be smaller than those in our simulations.
Adjusting for baseline variables beyond just age and sex led to substantial improvements in precision in our simulations involving time‐to‐event outcomes, as described in Section 5.3. For the other outcome types, that is, binary and ordinal, our data‐generating distributions only had one baseline variable, age; this is all that was available in the CDC data, so we were not able to investigate the value added by adjusting for more variables.
The described methods for binary and ordinal outcomes can be adapted to handle the case where stratified randomization on a subset of the measured baseline covariates is used. Specifically, one can apply the general method of Wang et al. (2019), which gives a formula for consistently estimating the asymptotic variance of covariate‐adjusted estimators under stratified randomization. This method can be applied to any M‐estimator and therefore applies to the estimators that we considered for binary and ordinal outcomes. To the best of our knowledge, for time‐to‐event outcomes it is an open problem to prove consistency and asymptotic normality for the TMLE estimators considered here under stratified randomization; we conjecture that the approach of Wang et al. (2019) can be extended to do this.
Treatment effect heterogeneity refers to differences in treatment effects among groups defined by baseline variables. Variance reductions due to covariate adjustment can occur both in the presence or absence of treatment effect heterogeneity (Qian et al., 2016). For example, when the data‐generating distributions are identical under treatment and control (in which case there is no treatment effect and no treatment effect heterogeneity), there can still be substantial variance reductions due to covariate adjustment if baseline variables are correlated with the outcome. This was observed in each of Tables 2– 6 for rows corresponding to no treatment effect.
Vermeulen et al. (2015) derived an adjusted estimator that is directly targeted at maximizing precision for the MW estimand, and similarly Díaz et al. (2016) derived an adjusted estimator that is directly targeted at the LOR estimand. In contrast, our adjusted estimators for ordinal outcomes target the entire treatment‐specific CDFs. A potential benefit of targeting the entire CDFs is that these estimates can be plugged in to estimate any smooth contrast of the treatment‐specific distributions, including, but not limited to, the MW estimand or the LOR estimand. It is an open research question to compare the statistical properties of our method to those above.
Supporting information
Web Appendices referenced in Sections 2–6 are available with this paper at the Biometrics website on Wiley Online Library. Appendix A gives intuition for how covariate adjustment can lead to precision gains in randomized trials. Appendix B defines our covariate‐adjusted estimators for ordinal outcomes. Appendix C presents additional simulation studies, including data‐generating distributions for non‐hospitalized COVID‐19 patients. Appendix D describes the availability of code on Github that reproduces our simulation experiments and that implements the estimators and confidence intervals. Appendix E gives our recommendations for information monitoring in trials that use covariate adjustment.
ACKNOWLEDGMENTS
David Benkeser, Iván Díaz, and Alex Luedtke are co‐first authors and contributed equally to this manuscript. AL was supported by the National Institutes of Health under award number DP2‐LM013340. MR was supported by the Johns Hopkins Center of Excellence in Regulatory Science and Innovation, which is funded by the Food and Drug Administration (FDA) of the U.S. Department of Health and Human Services (HHS) as part of a financial assistance award (U01FD005942). The contents are those of the author(s) and do not necessarily represent the official views of, nor an endorsement, by any of the aforementioned organizations, the FDA/HHS, nor the U.S. Government.
Benkeser D, Díaz I, Luedtke A, Segal J, Scharfstein D, Rosenblum M. Improving precision and power in randomized trials for COVID‐19 treatments using covariate adjustment, for binary, ordinal, and time‐to‐event outcomes. Biometrics. 2021;77:1467–1481. 10.1111/biom.13377
Contributor Information
David Benkeser, Email: benkeser@emory.edu.
Iván Díaz, Email: ild2005@med.cornell.edu.
Alex Luedtke, Email: aluedtke@uw.edu.
Jodi Segal, Email: jsegal@jhmi.edu.
Daniel Scharfstein, Email: daniel.scharfstein@hsc.utah.edu.
Michael Rosenblum, Email: mrosen@jhu.edu.
DATA AVAILABILITY STATEMENT
The data and code needed to reproduce the simulations for ordinal and binary outcomes are available on GitHub at https://github.com/mrosenblum/COVID‐19‐RCT‐STAT‐TOOLS. These data were derived from the following resource available in the public domain: (CDC COVID‐19 Response Team, 2020). The code for the survival simulations is also included in that repository. However, because the simulation is based on private data from Weill Cornell Medicine (research data not shared), the results of the survival simulation reported in the manuscript are not reproducible based on the available code. We provide a simulated dataset (not based on real data) with the same structure of the real dataset and that can be used to run the simulation code.
REFERENCES
- Austin, P.C. , Manca, A. , Zwarenstein, M. , Juurlink, D.N. and Stanbrook, M.B. (2010) A substantial and confusing variation exists in handling of baseline covariates in randomized controlled trials: a review of trials published in leading medical journals. Journal of Clinical Epidemiology, 63, 142–153. [DOI] [PubMed] [Google Scholar]
- Beigel, J.H. , Tomashek, K.M. , Dodd, L.E. , Mehta, A.K. , Zingman, B.S. , Kalil, A.C et al. (2020) Remdesivir for the treatment of COVID‐19 – preliminary report. New England Journal of Medicine. 10.1056/NEJMoa2007764. [DOI] [PubMed] [Google Scholar]
- Benkeser, D. , Carone, M. and Gilbert, P.B. (2018) Improved estimation of the cumulative incidence of rare outcomes. Statistics in Medicine, 37, 280–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benkeser, D. , Diaz, I. , Luedtke, A. , Segal, J. , Scharfstein, D. and Rosenblum, M. (2020) Improving precision and power in randomized trials for COVID‐19 treatments using covariate adjustment, for ordinal or time‐to‐event outcomes. Available at: https://www.medrxiv.org/content/10.1101/2020.04.19.20069922v1?versioned=true. Accessed April 23, 2020.
- Benkeser, D. , Gilbert, P.B. and Carone, M. (2019) Estimating and testing vaccine sieve effects using machine learning. Journal of the American Statistical Association, 114, 1038–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloniarz, A. , Liu, H. , Zhang, C. , Sekhon, J. and Yu, B. (2016) Lasso adjustments of treatment effect estimates in randomized experiments. Proceedings of the National Academy of Sciences of the United States of America, 113, 7383–7390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks, J.C. , van der Laan, M.J. , Singer, D.E. and Go, A.S. (2013) Targeted minimum loss‐based estimation of causal effects in right‐censored survival data with time‐dependent covariates: warfarin, stroke, and death in atrial fibrillation. Journal of Causal Inference, 1, 235–254. [Google Scholar]
- CDC COVID‐19 Response Team (2020) Severe Outcomes among patients with coronavirus disease 2019 (COVID‐19) — United States, February 12–March 16, 2020. Available at: https://www.cdc.gov/mmwr/volumes/69/wr/mm6912e2.htm. MMWR. Morbidity and Mortality Weekly Report, 69, 343–346. Accessed March 30, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaisinanunkul, N. , Adeoye, O. , Lewis, R.J. , Grotta, J.C. , Broderick, J. , Jovin, T.G et al. (2015) Adopting a patient‐centered approach to primary outcome analysis of acute stroke trials using a utility‐weighted modified Rankin scale. Stroke, 46, 2238–2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, P.‐Y. and Tsiatis, A.A. (2001) Causal inference on the difference of the restricted mean lifetime between two groups. Biometrics, 57, 1030–1038. [DOI] [PubMed] [Google Scholar]
- Díaz, I. , Colantuoni, E. , Hanley, D.F. and Rosenblum, M. (2019) Improved precision in the analysis of randomized trials with survival outcomes, without assuming proportional hazards. Lifetime Data Analysis, 25, 439–468. [DOI] [PubMed] [Google Scholar]
- Díaz, I. , Colantuoni, E. and Rosenblum, M. (2016) Enhanced precision in the analysis of randomized trials with ordinal outcomes. Biometrics, 72, 422–431. [DOI] [PubMed] [Google Scholar]
- Efron, B. and Tibshirani, R.J. (1994) An Introduction to the Bootstrap. Boca Raton, FL: CRC Press. [Google Scholar]
- EMA (2015) Guideline on adjustment for baseline covariates in clinical trials. Reference number EMA/CHMP/295050/2013. Committee for Medicinal Products for Human Use. Available at: https://www.ema.europa.eu/documents/scientific-guideline/guideline-adjustment-baseline-covariates-clinical-trials_en.pdf. Accessed March 30, 2020.
- FDA (2019) Adjusting for covariates in randomized clinical trials for drugs and biologics with continuous outcomes. Draft guidance for industry. Available at: https://www.fda.gov/media/123801/download. Accessed March 30, 2020.
- FDA (2020) COVID‐19: Developing drugs and biological products for treatment or prevention. Guidance for industry. Available at: https://www.fda.gov/media/137926/download. Accessed June 1, 2020.
- FDA and EMA (1998) E9 statistical principles for clinical trials. U.S. Food and Drug Administration: CDER/CBER. European Medicines Agency: CPMP/ICH/363/96.
- Ge, M. , Durham, L.K. , Meyer, R.D. , Xie, W. and Thomas, N. (2011) Covariate‐adjusted difference in proportions from clinical trials using logistic regression and weighted risk differences. Drug Information Journal, 45, 481–493. [Google Scholar]
- Goldman, J.D. , Lye, D.C. , Hui, D.S. , Marks, K.M. , Bruno, R. , Montejano, R et al. (2020) Remdesivir for 5 or 10 days in patients with severe covid‐19. New England Journal of Medicine. 10.1056/NEJMoa2015301. Accessed August 1, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennison, C. and Turnbull, B.W. (1997) Group‐sequential analysis incorporating covariate information. Journal of the American Statistical Association, 92, 1330–1341. [Google Scholar]
- Jennison, C. and Turnbull, B.W. (1999) Group Sequential Methods with Applications to Clinical Trials. Boca Raton, FL: CRC Press. [Google Scholar]
- Jiang, F. , Tian, L. , Fu, H. , Hasegawa, T. and Wei, L.J. (2019) Robust alternatives to ANCOVA for estimating the treatment effect via a randomized comparative study. Journal of the American Statistical Association, 114, 1854–1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan, E.L. and Meier, P. (1958) Nonparametric estimation from incomplete observations. Journal of the American Statistical Association, 53, 457–481. [Google Scholar]
- Lu, X. and Tsiatis, A.A. (2011) Semiparametric estimation of treatment effect with time‐lagged response in the presence of informative censoring. Lifetime Data Analysis, 17, 566–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullagh, P. (1980) Regression models for ordinal data. Journal of the Royal Statistical Society: Series B (Methodological), 42, 109–127. [Google Scholar]
- Moore, K.L. , Neugebauer, R. , Valappil, T. and van der Laan, M.J. (2011) Robust extraction of covariate information to improve estimation efficiency in randomized trials. Statistics in Medicine, 30, 2389–2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, K.L. and van der Laan, M.J. (2009a) Covariate adjustment in randomized trials with binary outcomes: targeted maximum likelihood estimation. Statistics in Medicine, 28, 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, K.L. and van der Laan, M.J. (2009b) Increasing power in randomized trials with right censored outcomes through covariate adjustment. Journal of Biopharmaceutical Statistics, 19, 1099–1131. PMID: 20183467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Research Council (2010) The Prevention and Treatment of Missing Data in Clinical Trials. Washington, DC: The National Academies Press. [PubMed] [Google Scholar]
- Nogueira, R.G. , Jadhav, A.P. , Haussen, D.C. , Bonafe, A. , Budzik, R.F. , Bhuva, P et al. (2018) Thrombectomy 6 to 24 hours after stroke with a mismatch between deficit and infarct. New England Journal of Medicine, 378, 11–21. PMID: 29129157. [DOI] [PubMed] [Google Scholar]
- Parast, L. , Tian, L. and Cai, T. (2014) Landmark estimation of survival and treatment effect in a randomized clinical trial. Journal of the American Statistical Association, 109, 384–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian, T. , Rosenblum, M. and Qiu, H. (2016) Improving power in group sequential, randomized trials by adjusting for prognostic baseline variables and short‐term outcomes. Johns Hopkins University, Department of Biostatistics Working Papers. Working Paper 285. Available at: https://biostats.bepress.com/jhubiostat/paper285. Accessed August 1, 2020.
- Royston, P. and Parmar, M.K. (2011) The use of restricted mean survival time to estimate the treatment effect in randomized clinical trials when the proportional hazards assumption is in doubt. Statistics in Medicine, 30, 2409–2421. [DOI] [PubMed] [Google Scholar]
- Rubin, D. and van der Laan, M. (2008a) Covariate adjustment for the intention‐to‐treat parameter with empirical efficiency maximization. U.C. Berkeley Division of Biostatistics Working Paper Series. Available at: https://biostats.bepress.com/ucbbiostat/paper229. Accessed August 1, 2020.
- Rubin, D.B. and van der Laan, M.J. (2008b) Empirical efficiency maximization: improved locally efficient covariate adjustment in randomized experiments and survival analysis. The International Journal of Biostatistics, 4, 5. [PMC free article] [PubMed] [Google Scholar]
- Scharfstein, D.O. , Rotnitzky, A. and Robins, J.M. (1999) Adjusting for nonignorable drop‐out using semiparametric nonresponse models. Journal of the American Statistical Association, 94, 1096–1120. [Google Scholar]
- Scharfstein, D.O. , Tsiatis, A.A. and Robins, J.M. (1997) Semiparametric efficiency and its implication on the design and analysis of group‐sequential studies. Journal of the American Statistical Association, 92, 1342–1350. [Google Scholar]
- Stitelman, O.M. , De Gruttola, V. and van der Laan, M.J. (2011) A general implementation of TMLE for longitudinal data applied to causal inference in survival analysis. The International Journal of Biostatistics, 8, 26. [DOI] [PubMed] [Google Scholar]
- The RECOVERY Collaborative Group (2020) Dexamethasone in Hospitalized Patients with Covid‐19 – Preliminary Report. New England Journal of Medicine. 10.1056/NEJMoa2021436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, L. , Jiang, F. , Hasegawa, T. , Uno, H. , Pfeffer, M. and Wei, L. (2019) Moving beyond the conventional stratified analysis to estimate an overall treatment efficacy with the data from a comparative randomized clinical study. Statistics in Medicine, 38, 917–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsiatis, A.A. , Davidian, M. , Zhang, M. and Lu, X. (2008) Covariate adjustment for two‐sample treatment comparisons in randomized clinical trials: a principled yet flexible approach. Statistics in Medicine, 27, 4658–4677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Vaart, A.W. (1998) Asymptotic Statistics. Cambridge, UK: Cambridge University Press. [Google Scholar]
- Vermeulen, K. , Thas, O. and Vansteelandt, S. (2015) Increasing the power of the Mann‐Whitney test in randomized experiments through flexible covariate adjustment. Statistics in Medicine, 34, 1012–1030. [DOI] [PubMed] [Google Scholar]
- Wager, S. , Du, W. , Taylor, J. and Tibshirani, R. (2016) High‐dimensional regression adjustments in randomized experiments. Proceedings of the National Academy of Sciences of the United States of America, 113, 12673–12678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, B. , Susukida, R. , Mojtabai, R. , Amin‐Esmaeili, M. and Rosenblum, M. (2019) Model‐robust inference for clinical trials that improve precision by stratified randomization and covariate adjustment [Preprint]. arXiv. Available at: https://arxiv.org/abs/1910.13954v2.
- Wang, Y. , Zhang, D. , Du, G. , Du, R. , Zhao, J. , Jin, Y et al. (2020) Remdesivir in adults with severe COVID‐19: a randomised, double‐blind, placebo‐controlled, multicentre trial. The Lancet, 395, 1569–1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, L. and Tsiatis, A.A. (2001) Efficiency study of estimators for a treatment effect in a pretest–posttest trial. The American Statistician, 55, 314–321. [Google Scholar]
- Zhang, M. (2014) Robust methods to improve efficiency and reduce bias in estimating survival curves in randomized clinical trials. Lifetime Data Analysis, 21, 119–137. [DOI] [PubMed] [Google Scholar]
- Zhang, M. and Gilbert, P.B. (2010) Increasing the efficiency of prevention trials by incorporating baseline covariates. Statistical Communications in Infectious Diseases, 2, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, M. , Tsiatis, A.A. and Davidian, M. (2008) Improving efficiency of inferences in randomized clinical trials using auxiliary covariates. Biometrics, 64, 707–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Web Appendices referenced in Sections 2–6 are available with this paper at the Biometrics website on Wiley Online Library. Appendix A gives intuition for how covariate adjustment can lead to precision gains in randomized trials. Appendix B defines our covariate‐adjusted estimators for ordinal outcomes. Appendix C presents additional simulation studies, including data‐generating distributions for non‐hospitalized COVID‐19 patients. Appendix D describes the availability of code on Github that reproduces our simulation experiments and that implements the estimators and confidence intervals. Appendix E gives our recommendations for information monitoring in trials that use covariate adjustment.
Data Availability Statement
The data and code needed to reproduce the simulations for ordinal and binary outcomes are available on GitHub at https://github.com/mrosenblum/COVID‐19‐RCT‐STAT‐TOOLS. These data were derived from the following resource available in the public domain: (CDC COVID‐19 Response Team, 2020). The code for the survival simulations is also included in that repository. However, because the simulation is based on private data from Weill Cornell Medicine (research data not shared), the results of the survival simulation reported in the manuscript are not reproducible based on the available code. We provide a simulated dataset (not based on real data) with the same structure of the real dataset and that can be used to run the simulation code.


