FcRn and CD32a form a ternary complex under acidic conditions and act together in determining responses to IgG immune complexes. The human autoimmune disease–associated CD32a-H131 variant more avidly forms this ternary complex, leading to greater FcRn-dependent immune responses to IgG.
Abstract
IgG immune complexes (ICs) promote autoimmunity through binding fragment crystallizable (Fc) γ-receptors (FcγRs). Of these, the highly prevalent FcγRIIa (CD32a) histidine (H)-131 variant (CD32aH) is strongly linked to human autoimmune diseases through unclear mechanisms. We show that, relative to the CD32a arginine (R)-131 (CD32aR) variant, CD32aH more avidly bound human (h) IgG1 IC and formed a ternary complex with the neonatal Fc receptor (FcRn) under acidic conditions. In primary human and mouse cells, both CD32a variants required FcRn to induce innate and adaptive immune responses to hIgG1 ICs, which were augmented in the setting of CD32aH. Conversely, FcRn induced responses to IgG IC independently of classical FcγR, but optimal responses required FcRn and FcγR. Finally, FcRn blockade decreased inflammation in a rheumatoid arthritis model without reducing circulating autoantibody levels, providing support for FcRn’s direct role in IgG IC-associated inflammation. Thus, CD32a and FcRn coregulate IgG IC-mediated immunity in a manner favoring the CD32aH variant, providing a novel mechanism for its disease association.
Introduction
Immune responses to IgG immune complexes (ICs) result from interactions between the fragment crystallizable (Fc) regions of IgG antibodies and diverse classical and atypical γ-receptors (FcγRs) primarily on hematopoietic cells (Pincetic et al., 2014; Bournazos et al., 2017). Many FcγRs are characterized by nonsynonymous single-nucleotide polymorphisms (SNPs), some of which affect the binding of FcγR variants to specific IgG subclasses (Bruhns et al., 2009; Shashidharamurthy et al., 2009). Of these, CD32a is particularly important, as it is widely expressed on hematopoietic cells and is characterized by the highly prevalent R131H SNP (rs1801274; Bournazos et al., 2017; Lehrnbecher et al., 1999). The H131 (CD32aH) is a high-affinity variant that is associated with several human autoimmune diseases, such as inflammatory bowel disease and rheumatoid arthritis (Shashidharamurthy et al., 2009; Zhang et al., 2016; Li et al., 2014), in contrast to its lower-affinity R131 counterpart (CD32aR), which is linked to an increased risk of infectious disease complications (Endeman et al., 2009; Bredius et al., 1994; Li et al., 2014). Critically, the residue 131 affected by this CD32a SNP is located on the CD32a-IgG Fc binding interface, where the R131H substitution alters CD32a binding to different isoforms of human IgGs (hIgGs) and the ICs they form (Shashidharamurthy et al., 2009; Bruhns et al., 2009). Specifically, one study found an ∼1.5- to 2-fold greater binding of ICs formed from hIgG1 and hIgG3 to CD32aH relative to CD32aR (Shashidharamurthy et al., 2009). In addition, CD32aR notably exhibits little binding to hIgG2 ICs (Shashidharamurthy et al., 2009; Parren et al., 1992; Salmon et al., 1992; Sanders et al., 1995). However, little is known about the functional consequences of these differences in CD32a variant binding or the precise intracellular mechanisms underlying their associations with increased risk of specific human diseases.
While CD32a variants disparately bind IgG ICs on the cell surface and direct their internalization into acidic intracellular compartments (Pincetic et al., 2014; Bournazos et al., 2017), their function in acidic conditions has not been examined. This is important, because the H131 residue (pKa ∼6.0) of CD32aH is prone to protonation in the pH 4.5–6.5 range observed in IgG IC-containing intracellular compartments (Baker et al., 2011; Haynes et al., 2016). Furthermore, intracellular endosomal compartments also contain FcRn, which binds monomeric IgGs and IgG ICs under mildly acidic conditions (pH <6.5) and at a site on IgG Fc that is distinct from FcγRs (Kim et al., 1999; Martin et al., 2001; Wines et al., 2000). FcRn is a nonpolymorphic atypical FcγR that is expressed in parenchymal and hematopoietic cells of mammals throughout life as a heterodimer of a MHC class I (MHCI)–related α-chain noncovalently associated with β-2-microglobulin (Pyzik et al., 2019; Simister and Mostov, 1989; Martin and Björkman, 1999). The pH-dependent binding of FcRn to IgG Fc occurs through salt bridge formation between several acidic residues of FcRn and critical histidine (H) residues on IgG Fc (Kim et al., 1999; Martin et al., 2001), whose pKa also renders them prone to protonation in the pH range observed in endosomes (pH <6.5), but not at neutral pH (Baker et al., 2011; Haynes et al., 2016). Functionally, FcRn is primarily recognized as an IgG salvage receptor, protecting IgG and IgG ICs from degradation and extending their circulating half-lives via pH-dependent binding in acidic endosomes and release under neutral pH cell surface conditions (Qiao et al., 2008; Blumberg et al., 2019; Roopenian et al., 2003; Borvak et al., 1998; Prabhat et al., 2007; Dickinson et al., 1999). Moreover, FcRn has also been shown to regulate a variety of functions in hematopoietic cells previously considered to be primarily determined by the classical FcγRs, such as phagocytosis (Vidarsson et al., 2006), innate cytokine production (Baker et al., 2013), antigen presentation (Qiao et al., 2008), and cross-presentation (Baker et al., 2011). However, the mechanisms whereby hematopoietic cells integrate CD32a- and FcRn-mediated processes in responding to IgG ICs is unclear. Because FcRn and CD32a both interact with IgG ICs under acidic conditions and regulate a variety of cellular responses to IgG ICs, we hypothesized that FcRn and CD32a functions are intimately coordinated in a CD32a allele-specific manner. We therefore investigated the mode of FcRn and CD32a interactions and the role of FcRn in CD32a-mediated immune processes. We assessed the effects of these interactions on innate and adaptive immune responses to IgG ICs in vitro in murine and human cells in the context of the CD32a variants and in vivo using the IC-mediated K/BxN model of rheumatoid arthritis.
Results
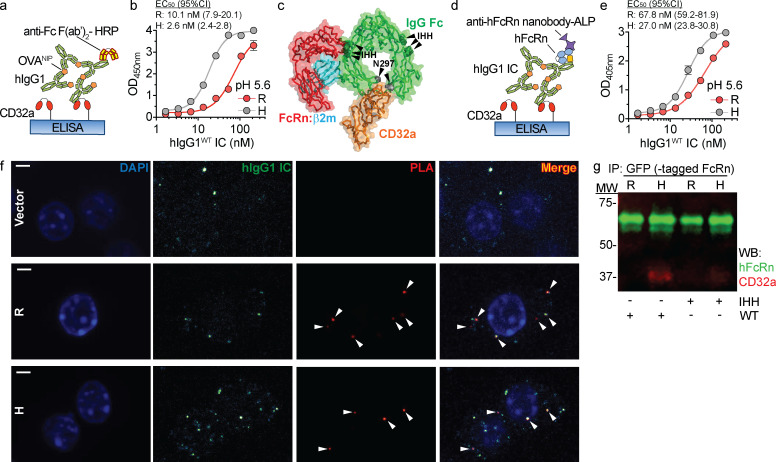
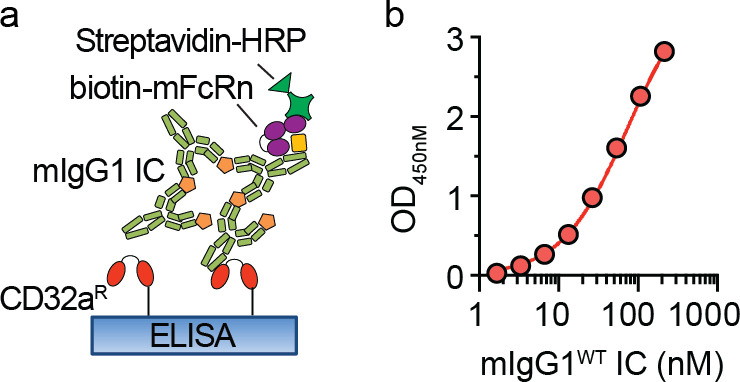
We first assessed whether acidic conditions affected IgG IC binding to CD32a variants using an ELISA. As the FcRn-bearing compartments containing IgG ICs attain pH ∼5.6 (Baker et al., 2011), CD32aR and CD32aH variants were exposed at pH 5.6 to increasing concentrations of model hIgG1 ICs (Fig. 1 a), which were prepared from 4-hydroxy-3-iodo-5-nitrophenylacetyl (NIP)–conjugated OVA (OVANIP) and a monoclonal anti-NIP hIgG1 antibody (hIgG1WT). Under these conditions, CD32aH demonstrated significantly increased avidity for hIgG1WT IC relative to CD32aR (Fig. 1 b).
Figure 1.
CD32aH exhibits increased bridging with IgG IC and FcRn under acidic conditions. (a) Schematic representation of ELISA of hIgG1 ICs binding to CD32a variants. Neutravidin-immobilized C-terminal biotinylated CD32aR or CD32aH variants were exposed to titrated concentrations of hIgG1 ICs at pH 5.6. hIgG1WT and all derived mutants were monoclonal mouse/human chimeric IgG1, composed of hIgG1 heavy chain associated with murine λ light chain, both with NIP specificity. Bound ICs were detected with anti-Fc HRP-conjugated F(ab′)2 fragments. (b) Log of hIgG1WT IC concentration (1.67–214.3 nM) versus OD mean values ± SEM of triplicate technical replicates fitted with nonlinear regression curves are shown (solid line; R2 = 0.99; EC50 compared by extra sum-of-squares F test; P < 0.0001), representative of three independent experiments. (c) FcRn–hIgG1 IC–CD32a ternary complex structural model based on the superposition of the FcRn–hIgG1 Fc (PDB 4N0U) and CD32aR–hIgG1 Fc (PDB 3RY6) crystal structures with root mean square deviation of 1.378 Å. The binding sites on IgG Fc (green) for FcRn (red) and CD32a (orange) are between ∼39 and 53 Å apart on the ipsilateral and contralateral Fc heavy chain, respectively. The hIgG1 Fc residues critical for binding to FcRn (IHH; green spheres) and CD32a (N297; gray spheres) are indicated by black arrowheads. (d) Schematic representation of ELISA setup used to detect hIgG1 IC bridging between FcRn and CD32a. CD32aR or CD32aH variants were immobilized and incubated with hIgG1WT ICs as described in panel a, followed by incubation with soluble hFcRn. hFcRn was detected with anti-hFcRn-ALP–conjugated nanobody. (e) Log of hIgG1 IC concentration (1.67–214.3 nM) versus OD mean values ± SEM of triplicate technical replicates fitted with nonlinear regression curves are shown (solid line; R2 = 0.99; EC50 compared by extra sum-of-squares F test; P < 0.0001), representative of three independent experiments. (f) Confocal microscopic images of PLA performed between CD32a and FcRn, using PLA probes targeting the cytoplasmic tails of CD32a and mFcRn, respectively, in CD32aR (R)–, CD32aH (H)–, or vector control (Vector) plasmid–transfected RAW 264.7 cells treated with fluorescent hIgG1WT ICs. Amplification of adequately proximate PLA probe oligonucleotides that enabled hybridization of fluorescent complementary oligos were visualized at 63× magnification under glycerol immersion and are indicated by white arrowheads. Representative images are shown of nuclei (blue), hIgG1 ICs (green), and PLA signals (red) and merged images (red and yellow). Scale bars = 3 µm. (g) Representative multiplex immunoblot showing coimmunoprecipitation of CD32aR or CD32aH variants (red) with hFcRn (green) after treatment with hIgG1WT or hIgG1IHH ICs. Data are representative of two (f and g) or three (b and e) independent experiments. H, CD32aH; MW, molecular weight; R, CD32aR; Vector, control vector; IP, immunoprecipitation; WB, Western blot.
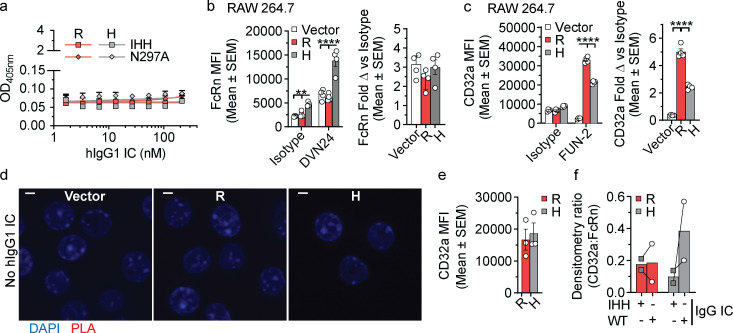
IgG contains two potential FcRn binding sites on each IgG at critical amino acids (I253, H310, and H435; IHH) contained within the CH2:CH3 domain interface of IgG Fc (Kim et al., 1999; Martin et al., 2001). These FcRn binding sites on IgG Fc are distinct from those associated with CD32a binding (Wines et al., 2000). Superimposition of FcRn:hIgG1 Fc and CD32aR:hIgG1 Fc crystal structures further suggested that a single hIgG1 Fc could accommodate simultaneous CD32a and FcRn engagement and formation of a ternary complex without steric clashing, as the FcRn and FcγR binding sites were separated by >39–53 Å (Fig. 1 c; Oganesyan et al., 2014; Ramsland et al., 2011). Alternatively, taking into consideration the tethering of the receptors to the cell membrane, it is also conceivable that distinct IgG Fc contained within ICs could also serve as a platform for ternary complex formation. We therefore next tested for simultaneous IgG IC binding by the CD32a variants and FcRn under acidic conditions by ELISA (Fig. 1 d). Consistent with formation of a ternary complex, hFcRn bound to hIgG1WT ICs captured by both CD32a variants, but in a manner that favored the CD32aH variant (Fig. 1 e). This ternary complex formation was abrogated if the applied ICs were composed of mutant hIgG1s that were specifically unable to bind FcRn (hIgG1IHH) or classical FcγRs (hIgG1N297A), including the CD32a variants (Table S1 and Fig. S1 a; Medesan et al., 1997; Kim et al., 1999; Tao and Morrison, 1989). We next used a proximity ligation assay (PLA) to investigate CD32a–IgG IC–FcRn ternary complex formation in an intracellular environment. To do so, mouse RAW 264.7 macrophage cells expressing endogenous mouse FcRn (mFcRn), which binds hIgG1 at acidic pH with comparable affinity to human FcRn (hFcRn; Neuber et al., 2014), were transfected with either the CD32aR or CD32aH variant (Fig. S1, b and c). PLA probes targeting CD32a and mFcRn yielded a fluorescence signal with both CD32a variants but only when both hIgG1 ICs and CD32a were present (Fig. 1 f and Fig. S1 d). We next used coimmunoprecipitation to biochemically identify the CD32a–IgG IC–FcRn ternary complex in the intracellular milieu. For this, we used human embryonic kidney cells (HEK 293) cells transfected with either CD32a variant and GFP-conjugated hFcRn (HEK 293hFcRn-GFP), which does not impact its hIgG1 binding or transcytosis functions, as a tractable tag for immunoprecipitation (Fig. S1 e; Christianson et al., 2012). Upon treatment of these cells with hIgG1WT ICs, but not hIgG1IHH ICs, more CD32aH relative to CD32aR was coimmunoprecipitated with hFcRn (Fig. 1 g and Fig. S1 f). Together, these data confirm ternary complex formation both under acidic conditions and in the intracellular environment when IgG IC can engage both CD32a and FcRn.
Figure S1.
Ternary complex (FcRn–hIgG1 IC–CD32a) formation requires hIgG1WT ICs. (a) Log of hIgG1IHH or hIgG1N297A IC concentration (1.67–214.3 nM) versus OD values, with nonlinear regression curves (solid line; R2 = 0.99). This ELISA was performed in triplicate, concomitantly with and using a setup identical to the experiment reported in Fig. 1, d and e, except for the use of hIgG1IHH and hIgG1N297A mutant ICs, which display specifically abrogated binding to FcRn or classical FcγR (CD32a in this instance), respectively. (b and c) FcRn (b) or CD32a (c) expression by RAW 264.7 cells stably transfected with CD32aR (R)-, CD32aH (H)-, or vector control (Vector) plasmids. To detect CD32a, FUN-2 clone was used. To detect mFcRn, mAb DVN24 was employed. Bar graphs display average mean fluorescence intensity (MFI) ± SEM of four technical replicates. Data are shown without (left) and with (right) normalization to isotype IgG2a control antibody staining. (d) Confocal microscopic merged images of PLA control experiments in CD32a variant– or control vector–transfected RAW 264.7 cells as in Fig. 1 f, but without hIgG1 IC treatment. Representative images are shown of nuclei (blue). Note the absence of red PLA signals in the absence of IgG ICs, in contrast to Fig. 1 f. Scale bars = 3 µm. (e) CD32a expression on transfected HEK 293GFP-hFcRn cells. Bar graphs display average MFI ± SEM of three technical replicates. (f) Cumulative CD32a/FcRn densitometry ratio of multiplex immunoblot shown in Fig. 1 g. Bar graphs show mean of two independent experiments; no statistical comparison was performed. Data are representative of two (d–f), or three (a–c) independent experiments. H, CD32aH; R = CD32aR; Vector, control vector. Statistical comparisons were performed via unpaired t test for two comparisons (e) or two-way ANOVA for three or more comparisons (a–c) followed by the two-stage linear step-up procedure of Benjamin, Krieger, and Yekutieli with FDR <0.05 (a–c). **, P < 0.01; ****, P < 0.0001.
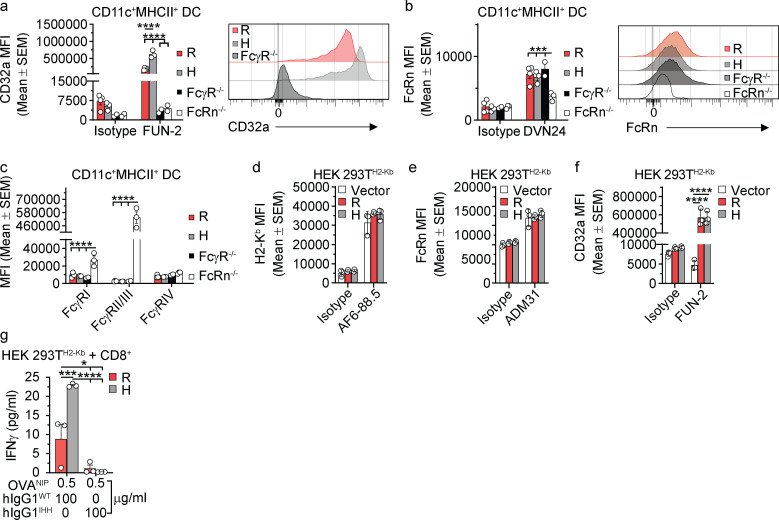
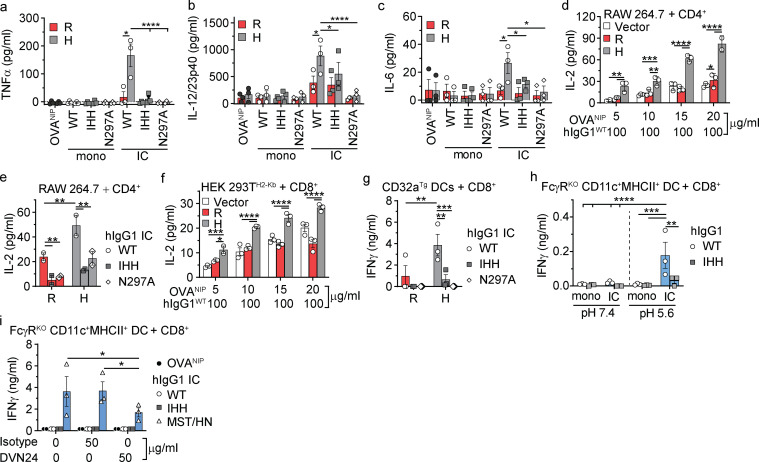
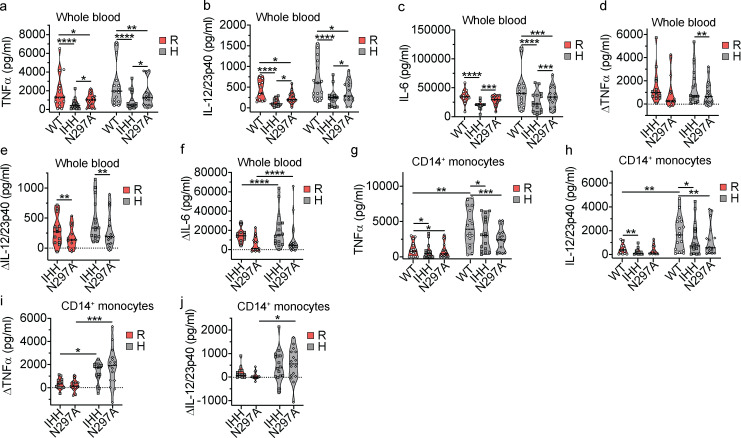
We next sought to determine whether formation of the CD32a–IgG IC–FcRn ternary complex was necessary for cellular responses to hIgG1 ICs. We first assessed innate immune responses in primary cells obtained from C57BL/6 mice transgenic (Tg) for either the CD32aR or CD32aH variant (hereafter “CD32aTg”; Fig. S2 a). To isolate any observed functional effects in these primary cells to CD32a and FcRn, these CD32aTg mice also lacked all classical FcγRs but maintained mFcRn expression (Fcgr2b−/−/Fcer1g−/−/Fcgrt+/+; Table S2; Fig. S2, b and c). Therefore, we isolated CD11c+MHCII+ dendritic cells (DCs) from spleens of CD32aR-Tg or CD32aH-Tg mice and observed increased secretion of TNF-α, IL-12/23p40, and IL-6 upon stimulation with hIgG1WT ICs relative to that observed with hIgG1IHH ICs and hIgG1N297A ICs, which are specifically disabled in FcRn or CD32a binding, respectively (Fig. 2, a–c; Medesan et al., 1997; Tao and Morrison, 1989; Kim et al., 1999). These data indicate that innate cytokine production in response to IgG ICs in APCs is both FcRn and CD32a dependent, consistent with the function of a CD32a–IgG IC–FcRn ternary complex.
Figure S2.
Increased CD32aH-associated presentation and cross-presentation is codependent on FcRn and CD32a. (a–c) CD32a (a), mFcRn (b), and classical FcγR (c) surface expression by primary splenic CD11c+MCHII+ DCs from CD32aR-Tg (R; red), CD32aH-Tg (H; gray), FcγR−/− (black), and mFcRn−/− (white) mice (n = 3). Bar graphs representing the average MFI ± SEM, and representative histograms (right for a and b) are shown. The Fcgrt−/− mice (n = 3) served as negative and positive controls for mFcRn and classical FcγR staining, respectively. (d–f) H2-Kb (d), hFcRn (e), and CD32a (f) surface expression on CD32aR- or CD32aH- or vector control plasmid–transfected HEK 293TH2-Kb cells. For panels d–f, bar graphs display average MFI ± SEM in triplicate. (g) IFN-γ production by CD8+ OT-I T cells after 48 h of co-culture with HEK 293TH2-Kb cells expressing CD32aR or CD32aH and loaded with hIgG1WT ICs or hIgG1IHH ICs. Data are representative of two or three (a–g) independent experiments with individual points representing triplicate technical replicates. H, CD32aH; R, CD32aR; Vector, control vector. All data were analyzed by two-way ANOVA followed by the two-stage linear step-up procedure of Benjamin, Krieger, and Yekutieli with FDR controlled at <0.05. *, P < 0.05; ***, P < 0.001; ****, P < 0.0001.
Figure 2.
FcRn regulates CD32a-induced responses to IgG IC. (a–c) Absolute TNF-α (a), IL-12/23p40 (b), and IL-6 (c) production by primary splenic CD11c+MHCII+ DCs from CD32aR-Tg (R) or CD32aH-Tg (H) mice after 24 h of exposure to antigen (OVANIP) alone (1 µg/ml) or 100 µg/ml anti-NIP hIgG1WT (WT), hIgG1IHH (IHH), or hIgG1N297A (N297A) in monomeric (mono) or OVANIP-IC form. Black-filled circles, OVANIP; white-filled circles, monomeric or hIgG1WT ICs; gray-filled squares, monomeric or hIgG1IHH ICs; white-filled diamonds, monomeric or hIgG1N297A ICs. (d and e) IL-2 production by MHCII-restricted, OVA-specific CD4+ T cells after 24 h of co-culture with CD32aR- or CD32aH- or vector control plasmid–transfected RAW 264.7 cells that were treated with hIgG1WT ICs prepared with increasing concentrations of OVANIP (d) or treated with hIgG1 ICs composed of hIgG1WT, hIgG1IHH, or hIgG1N297A mutants and 10 µg/ml OVANIP (e). (f) IL-2 production by OVA-specific CD8+ OT-I T cells after 48 h of co-culture with CD32aR- or CD32aH- or vector control plasmid–transfected HEK 293H2-Kb cells loaded with OVANIP-containing hIgG1WT ICs as in Fig. 1 d. (g) IFN-γ production by CD8+ OT-I T cells after 48 h of co-culture with primary CD11c+MHCII+ CD32aR-Tg or CD32aH-Tg DCs loaded with hIgG1WT ICs, hIgG1IHH ICs, or hIgG1N297A ICs. (h and i) IFN-γ production by CD8+ OT-I T cells co-cultured for 48 h with CD11c+MHCII+ FcγRKO DCs loaded with hIgG1WT ICs or hIgG1IHH ICs at pH 7.4 or pH 5.6 (h) or pretreated with an isotype IgG2a control antibody or anti-m/hFcRn mAb DVN24 for 30 min before treatment with OVANIP only, or hIgG1WT, hIgG1IHH, or hIgG1MST/HN ICs (white-filled triangles) at pH 7.4 (i). All data represent arithmetic mean ± SEM of duplicate or triplicate technical replicates from three independent experiments (a–c), or are representative of three independent experiments (d–i), with triplicate technical replicates shown. All data were analyzed by two-way ANOVA followed by the two-stage linear step-up procedure of Benjamin, Krieger, and Yekutieli with FDR controlled at <0.05. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
As FcRn and classical FcγR are required for efficient antigen presentation and cross-presentation (Bonnerot et al., 1998; Qiao et al., 2008; Regnault et al., 1999; Baker et al., 2011), we examined the role of the CD32a–IgG IC–FcRn tripartite complex in these pathways. To do so, we used the antigen presentation machinery of the RAW 264.7 cells (MHC haplotype H-2d) described above. CD32aR or CD32aH variant- or control plasmid-transfected RAW 264.7 cells were loaded with hIgG1WT ICs at neutral pH and compared for their ability to stimulate co-cultured CD4+ T cells from DO11.10 mice Tg for a H2-Ad–restricted, OVA-specific T cell receptor (Qiao et al., 2008). Despite higher expression of CD32aR (Fig. S1 b), the CD32aH variant induced significantly more IL-2 production by CD4+ T cells over a range of hIgG1WT IC concentrations (Fig. 2 d), which was significantly decreased in the context of hIgG1IHH ICs or hIgG1N297A ICs (Fig. 2 e). We next examined cross-presentation in cells endogenously expressing hFcRn but lacking any classical FcγRs. For this, we turned to a previously described HEK 293T cell line stably expressing the murine MHCI molecule H2-Kb (HEK 293TH2-Kb; Faure et al., 2009; Giodini et al., 2009), which we transfected with either CD32a variant (Fig. S2, d–f). CD32aH-expressing HEK 293TH2-Kb cells treated with hIgG1WT ICs, but not hIgG1IHH ICs, stimulated greater IL-2 (Fig. 2 f) or IFN-γ (Fig. S2 g) production by primary H2-Kb–restricted, OVA-specific CD8+ OT-I T cells compared with CD32aR-expressing or vector-transfected HEK 293TH2-Kb cells (Hogquist et al., 1994; Faure et al., 2009). Similarly, treatment of primary CD11c+MHCII+ DCs from CD32aR-Tg and CD32aH-Tg mice (Fig. S2, a–c) with hIgG1WT ICs stimulated IFN-γ secretion by CD8+ OT-I T cells, which was lost if the hIgG1 ICs were composed of hIgG1IHH or hIgG1N297A (Fig. 2 g). Thus, antigen presentation and cross-presentation depend on both CD32a and FcRn, with the CD32aH variant inducing greater FcRn-dependent adaptive immune responses relative to the CD32aR variant.
We next sought to more precisely delineate the role of FcRn relative to FcγRs in eliciting these adaptive immune responses. DCs and monocytes uniquely express high levels of FcRn on the cell surface (Fig. S2 b; Blumberg et al., 2019; Zhu et al., 2001), which allowed us to examine whether FcRn could mediate cross-presentation independently of FcγR under conditions that enable cell surface FcRn–IgG interactions (e.g., acidic pH; Kim et al., 1999; Medesan et al., 1997; Dickinson et al., 1999). To do so, we incubated hIgG1WT ICs with DCs derived from C57BL/6 mice lacking all classical FcγRs but maintaining mFcRn expression (Fcgr2b−/−Fcer1g−/−Fcgrt+/+, hereafter "FcγRKO"; Table S2 and Fig. S2, a–c). When FcγRKO DCs were loaded with hIgG1WT at pH 5.6, but not at neutral pH or with hIgG1IHH ICs, co-cultured CD8+ OT-I T cells were induced to secrete IFN-γ (Fig. 2 h), though at lower levels than when CD32a is present (Fig. 2 g). Next, we performed cross-presentation experiments with hIgG1 ICs composed of the well-characterized hIgG1MST/HN mutant that exhibits augmented FcRn binding at both neutral (equilibrium dissociation constant [KD] = 7.4 nM at pH 7.2) and acidic (KD = 1.2 nM at pH 6) pH relative to hIgG1WT and therefore can bind FcRn on the cell surface (Table S1; Vaccaro et al., 2005; Grevys et al., 2015). When FcγRKO DCs were loaded with hIgG1MST/HN ICs at pH 7.4, we observed robust IFNγ production by co-cultured CD8+ OT-I T cells (Fig. 2 i). Pretreatment with a monoclonal mouse h/mFcRn-blocking monoclonal antibody, DVN24 (Christianson et al., 2012), significantly diminished cross-presentation of hIgG1MST/HN IC-delivered antigen to CD8+ OT-I T cells (Fig. 2 i). Thus, IgG IC-driven innate and adaptive immune responses require FcRn and can occur in the absence of classical FcγRs, but maximal responses result when both FcRn and FcγRs are engaged.
Previous studies have evaluated IgG IC interactions with CD32a variants (Shashidharamurthy et al., 2009; Zhu et al., 2001), yet no studies have directly assayed the contribution of FcRn to CD32a variant–associated immune responses in human cells. We therefore extended our investigations to leukocytes from healthy human volunteers homozygous for CD32aR (FCGR2AG/G) or CD32aH (FCGR2AA/A). Whole-blood samples from these donors were stimulated in vitro with hIgG1WT ICs, and the production of proinflammatory cytokines (TNF-α, IL-12/23p40, and IL-6) was assessed (Blumberg et al., 2019). Innate cytokine secretion from these cells incubated with soluble OVANIP or monomeric hIgG was negligible (data not shown). While TNF-α, IL-12/23p40, and IL-6 levels trended higher in CD32aH than CD32aR cells upon treatment with hIgG1WT ICs (Fig. 3, a–c), both the CD32aR and CD32aH cells exhibited significantly decreased cytokine production in response to the hIgG1IHH ICs and hIgG1N297A ICs, indicative of FcRn and classical FcγR dependence, respectively. Furthermore, we compared the differences in the FcRn-dependent production of TNF-α, IL-12/23p40, and IL-6 (Fig. 3, d–f) upon treatment with these hIgG1 mutants relative to hIgG1WT ICs. Notably, we observed that the reduction of IL-6 as a consequence of disabling hIgG1 interactions with FcRn (hIgG1IHH ICs) or FcγR (hIgG1N297A ICs) was significantly more prominent in cells from CD32aH individuals in vitro, although absolute IL-6 production levels were not different between CD32aH- and CD32aR-expressing cells (Fig. 3, c and f). A similar trend was noted for IL-12/23p40 (Fig. 3, b and e). This suggests not only that are human leukocyte innate immune responses to hIgG1 ICs dependent upon FcRn and classical FcγRs but also that individuals who are homozygous for CD32aH may be more affected by a loss of FcRn function compared with CD32aR-homozygous individuals.
Figure 3.
CD32aH expression confers higher FcRn-dependent innate immune responses to IgG1 IC in human leukocytes. (a–c) Absolute TNF-α (a), IL-12/23p40 (b), and IL-6 (c) cytokine production by human whole blood collected from healthy volunteer human subjects, homozygous for FCGR2AG/G (CD32aR = R; n = 13) or FCGR2AA/A (CD32aH = H; n = 16), after 24 h of stimulation with hIgG1WT ICs, hIgG1IHH ICs, or hIgG1N297A ICs. (d–f) Relative TNF-α (d), IL-12/23p40 (e), and IL-6 (f) cytokine production calculated as differences (Δ) between hIgG1WT IC- and either hIgG1IHH IC- or hIgG1N297A IC-treated whole blood, respectively. (g and h) Absolute TNF-α (g) and IL-12/23p40 (h) production by CD14+ monocytes isolated from whole blood from the same healthy human donors and treated as in panels a–c. (i and j) Relative TNF-α (i) and IL-12/23p40 (j) production differences (Δ) between hIgG1WT IC- and either hIgG1IHH IC- or hIgG1N297A IC-treated CD14+ monocytes, respectively, are shown for panels g and h. Individual points in panels a–j represent the mean of two technical replicates of cellular responses to hIgG1 IC stimulation for each individual donor on one occasion, and groups of values for each genotype and treatment condition are presented as violin plots, with dashes indicating group arithmetic means. White filled circles, hIgG1WT ICs; gray filled squares, hIgG1IHH ICs; white filled diamonds, hIgG1N297A ICs. Statistical analysis of absolute cytokine production (a–c, g, and h) in response to hIgG1WT ICs between R and H was performed by unpaired two-tailed Mann–Whitney test and by matched Friedman test for comparison of hIgG1WT ICs, hIgG1IHH ICs, and hIgG1N297A ICs within each genotype. The relative (Δ) cytokine production for panels d–f and i were compared by two-way ANOVA of loge-transformed values. All multiple comparison tests (a–i) underwent post-hoc analysis by the two-stage linear step-up procedure of Benjamin, Krieger, and Yekutieli with FDR controlled at <0.05. Change in CD14+ monocyte production of IL-12/23p40 (j) were non-Gaussian when transformed and therefore were compared by unpaired Mann–Whitney testing. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
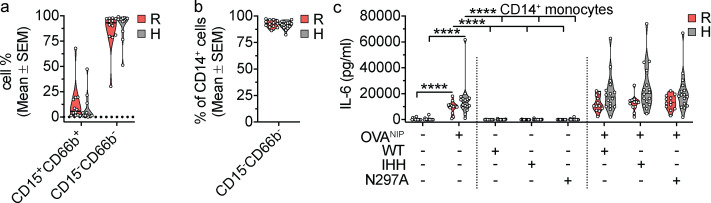
Given the heterogenous nature of FcRn- and FcγR-expressing cells in the peripheral blood, we next focused our attention on peripheral blood monocytes that express CD32a and FcRn and secrete innate cytokines when stimulated by IgG ICs (Pincetic et al., 2014; Bournazos et al., 2017; Blumberg et al., 2019; Zhu et al., 2001). We isolated CD14+ monocytes (Fig. S3, a and b) from the blood of the same cohort of healthy volunteers described above and assessed their responses to hIgG1 ICs. Strikingly, CD14+ monocytes from the CD32aH donors produced significantly more TNF-α and IL-12/23p40 in response to hIgG1WT ICs compared with CD14+ monocytes from CD32aR donors (Fig. 3, g and h). The IC-dependent IL-6 responses by CD14+ monocytes could not be assessed due to the high nonspecific responses observed during treatment with soluble OVANIP only (Fig. S3 c). Furthermore, in response to the hIgG1IHH and hIgG1N297A ICs, monocytes displayed significant reductions in TNF-α and IL-12/23p40 levels compared with the IgG1WT ICs irrespective of CD32aR or CD32aH status (Fig. 3, g and h). Still, the reductions in TNF-α and, to a lesser extent, IL-12/23p40 production from hIgG1IHH and hIgG1N297A mutant ICs were most prominent in cells from CD32aH-homozygous donors (Fig. 3, i and j). As observed with whole-blood samples, these effects were observed in the absence of overall differences in the responses of CD32aR and CD32aH donors to hIgG1N297A ICs (Fig. 3, g–j). Together, these data confirm that the CD32aH variant induces augmented inflammatory innate immune responses to hIgG1 ICs in human leukocytes compared with the CD32aR variant. Further, these results suggest that immune cells expressing CD32aH exhibit greater dependence on FcRn interactions, relative to CD32aR-expressing cells, which is consistent with increased formation of a ternary complex by the CD32aH variant.
Figure S3.
Assessment of CD14+ monocyte phenotype and responses to OVANIP. (a) Frequency of live CD15+CD66b+ (granulocytes) and CD15−CD66b− (nongranulocytes) from healthy human volunteers homozygous for CD32aR or CD32aH, separated from heparinized whole blood by Mono-Poly density gradient, and enriched for CD14+ cells by immunomagnetic cell separation. (b) Frequency of live CD14+ cells (monocytes) within the CD15−CD66b− cell fraction. (c) Absolute IL-6 cytokine production by human CD14+ monocytes untreated or upon stimulation with OVANIP alone, monomeric hIgG1 controls, or OVANIP-containing hIgG1WT ICs, hIgG1IHH ICs, or hIgG1N297A ICs. Individual points represent the mean of two technical replicates of cellular responses to hIgG1 IC stimulation for each individual donor on one occasion, and groups of values for each genotype and treatment condition are presented as violin plots, with dashes indicating group arithmetic means of individual mean values. All IL-6 levels resulting from treatment with OVANIP and IgG ICs were significantly different at P< 0.0001, from all monomeric IgG control conditions (hIgG1WT, hIgG1IHH, or hIgG1N297A alone); significance (*) symbols indicating this were omitted to minimize clutter. All data (a–c) were analyzed by two-way ANOVA of loge-transformed mean values of duplicate technical replicates, followed by the two-stage linear step-up procedure of Benjamin, Krieger, and Yekutieli with FDR controlled at <0.05. ****, P < 0.0001.
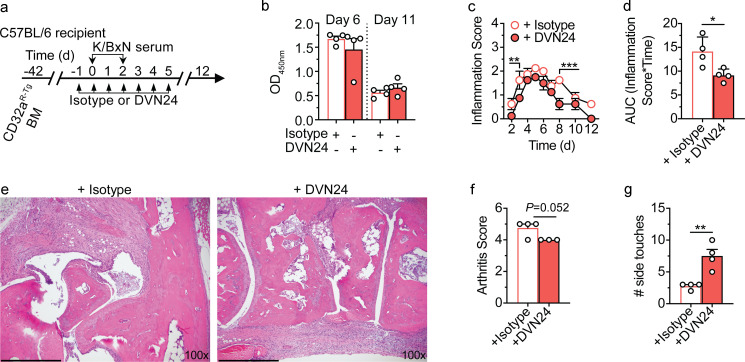
We finally sought to determine whether FcRn can regulate IgG IC-mediated inflammation in an FcγR-dependent disease model. To do so, we examined the K/BxN serum transfer model of rheumatoid arthritis (Matsumoto et al., 1999; Monach et al., 2008). As the K/BxN model is primarily mediated by murine IgG1 autoantibodies which avidly bind CD32aR, but not CD32aH (Matsumoto et al., 1999; Monach et al., 2008; Mancardi et al., 2011; Clark et al., 1991; Tate et al., 1992), we focused on the CD32aR-Tg mouse model (Table S2). We first verified that mIgG1 ICs can form a ternary complex with mFcRn and CD32aR at acidic pH (Fig. S4, a and b). To investigate the importance of FcRn in this ternary complex in vivo, we adoptively transferred bone marrow from CD32aR-Tg mice lacking other classical FcγRs (Fig. S2, a–c) into irradiated WT C57BL/6 mice, followed by K/BxN serum injection in the presence of isotype IgG2a control treatment or FcRn blockade with the DVN24 antibody by the schedule described in Fig. 4 a. As FcRn blockade has been shown to decrease circulating IgG levels in mice and humans (Blumberg et al., 2019), we designed an anti-FcRn DVN24 antibody treatment regimen that did not affect circulating autoantibody levels (Fig. 4 b) in order to more precisely investigate FcRn’s role in regulating CD32a-mediated cellular immune responses to IgG ICs in vivo. Despite the lack of associated decreases in autoantibody levels, DVN24 antibody treatment significantly decreased inflammatory disease burden as assessed by clinical inflammatory scoring (Fig. 4, c and d), histopathologic arthritic inflammation (Fig. 4, e and f) and animal mobility (Fig. 4 g). These data demonstrate that inflammation derived from CD32aR-expressing bone marrow cells is dependent upon FcRn.
Figure S4.
CD32aR forms ternary complexes with mFcRn and mIgG1 IC. (a) Schematic representation of bridging ELISA with mIgG1 ICs, mFcRn, and CD32aR. His-tagged CD32aR was directly immobilized to the ELISA plate followed by titration of mIgG1 IC concentrations at pH 5.6, the binding of which was detected by addition of biotinylated mFcRn prebound to streptavidin-HRP. (b) Log of mIgG1 IC concentrations (1.67–214.3 nM) versus OD (mean ± SEM of triplicate technical replicates), with nonlinear regression fit shown (solid line) with R2 = 0.99. Data are representative of three independent experiments.
Figure 4.
FcRn blockade can ameliorate IC-mediated arthritis without increasing IgG clearance. (a) Schematic representation of K/BxN model of rheumatoid arthritis in CD32aR-Tg bone marrow chimeric mice. 6-wk-old male C57BL/6 mice were lethally irradiated and injected with sex-matched CD32aR-Tg bone marrow (BM) cells. 6 wk later, BM chimeric CD32aR-Tg mice were treated with 0.2 mg isotype IgG2a antibody or DVN24 daily (n = 4/group) for 5 d and administered K/BxN serum twice to induce arthritis. The mice were then monitored for 12 d and evaluated for disease progression and severity. (b) ELISA measurements (mean ± SEM of triplicate technical replicates) of total anti-GPI mIgG levels in serum for each mouse on days 6 and 11 following the initial K/BxN serum transfer. (c–g) Cumulative arthritis inflammation scores (c), displayed as area under the inflammation–time curve (AUC; d; mean ± SEM of individual mouse AUC) and ankle joint histopathology (e; day 12 representative images; scale bars = 400 µm), with blinded histopathologic scoring of inflammation (f; mean ± SEM of least three consecutive sections) for individual animals, and mobility measured on day 7 (g) as the number of cylinder side touches of individual mice in 1 min. Individual data points represent individual animals. To minimize clutter, mean ± SEM are shown in the inflammation score panel (c). Data shown are representative of two independent experiments. Analysis was by unpaired t test (d, f, and g) or two-way ANOVA (b and c), with the two-stage linear step-up procedure of Benjamin, Krieger, and Yekutieli with FDR <0.05, as appropriate. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Discussion
In summary, we show that CD32a and FcRn directly cooperate through formation of a ternary complex on an IgG Fc scaffold under acidic conditions, as occurs in intracellular organelles (Baker et al., 2011). This ternary complex formation could impact a wide range of FcγR-mediated processes, including IgG IC induction of thrombosis, which has been previously recognized as a FcγRIIa-mediated process that is regulated by and dependent upon FcRn (Cines et al., 2020). Furthermore, we demonstrate that FcRn can independently execute adaptive cellular immune responses to IgG ICs when FcRn can engage IgG on the cell surface, but that the presence of an FcγR coreceptor for IgG ICs significantly augments these responses. Our studies thus predict that enhanced FcRn cooperation with classical FcγR will amplify their responses to IgG ICs. In line with this, passive immunization in nonhuman primates using anti-HIV IgG antibodies engineered to possess stronger FcRn binding demonstrated increased passive immune protection from HIV that depends on CD8+ T cells (Nishimura et al., 2017; Gaudinski et al., 2018). Thus, while engineered IgG with enhanced FcRn binding exhibit increased half-life, which probably improves their effectiveness (Pyzik et al., 2019), these engineered IgG antibodies will likely also induce greater FcRn-dependent cellular immune responses than would be expected of otherwise identical IgG antibodies with “normal” affinity for FcRn. Similarly, augmentation of IgG IC interactions with FcRn enhances antitumor CD8+ T cell responses (Baker et al., 2013).
Our studies also highlight the importance of acidic environments in shaping FcRn- and classical FcγR-dependent immune responses to IgG ICs. For FcγRs specifically, this is consistent with a study which found that hIgG1 IC binding to nongenotyped human neutrophils increases as extracellular pH declines (López et al., 1999). Our data are also consistent with published crystal structures of hIgG1 Fc complexed with CD32aH and CD32aR, respectively, which show distinct homodimers under neutral and acidic conditions that are predicted to affect both interactions with IgG Fc and conceivably formation of a ternary complex with FcRn (Ramsland et al., 2011), as illustrated here. Such pH-dependent synergy between CD32a and FcRn may also evolve from events at the cell surface, given our demonstration that surface FcRn can mediate cross-presentation of hIgG1WT ICs independent of classical FcγR when APCs encounter ICs under acidic conditions. This suggests that engagement of FcRn on the cell surface in a ternary complex with classical FcγRs could also amplify cellular immune responses to IgG ICs. Therefore, both intracellular and extracellular acidic environments, such as found in endosomes (Baker et al., 2011) or disease-associated tissue acidosis (e.g., neoplasia and inflammation; Riemann et al., 2016; Lindner and Raghavan, 2009), respectively, may affect FcRn-dependent immune responses that are regulated by classical FcγRs.
Further, we have found that FcRn-dependent innate and adaptive immune responses to IgG ICs are more actively promoted in the setting of CD32aH relative to CD32aR. This occurred through the increased propensity of CD32aH to associate with hIgG1 ICs and foster recruitment of FcRn to form a ternary complex under acidic conditions. Thus, our data also highlight the importance of allele-specific differences in CD32a’s handling of hIgG1 ICs and its functional interactions with FcRn, thereby delineating a potential mechanism underlying CD32aH contributions to human autoimmune disease (Carcao et al., 2003; Zhang et al., 2016; Li et al., 2014; Lehrnbecher et al., 1999; Dijstelbloem et al., 2000).
The functional demonstration of CD32a–hIgG1 IC–FcRn ternary complex formation may further implicate FcRn in classical FcγR-mediated immune responses more broadly. As observed with CD32a, FcRn may similarly cooperate with other classical FcγRs such as the polymorphic CD16a and CD16b, which are associated with human disease and expressed together with FcRn in polymorphonuclear leukocytes (Pincetic et al., 2014; Wu et al., 1997; Morgan et al., 2000; Lee et al., 2015; Li et al., 2014). This suggests that the monomorphic FcRn acts as a pH-dependent coreceptor of the highly polymorphic classical FcγR system and in an allele-specific manner as shown here for CD32a. These studies have implications for current clinical trials of anti-FcRn and anti-CD32a therapies (Chen et al., 2019; Kiessling et al., 2017; Ulrichts et al., 2018; Ling et al., 2019; Nixon et al., 2015; Blumberg et al., 2019), as they suggest that the therapeutic efficacy of these agents may derive in part from their effects on FcRn and classical FcγR interactions associated with cellular immunity independent of circulating IgG levels.
Materials and methods
Human ethics
All experiments involving human volunteers were performed consistent with ethical guidelines and with the approval of the Clinical Research Ethics Board at the University of British Columbia (H13-03524 and H14-00622). All volunteer subjects provided written, informed consent for blood collection for DNA isolation and genotyping, immune cell isolation, and functional characterization, as described previously (Kozicky et al., 2018).
Animals and cell lines
Animal experiments were approved by the institutional care and use committees of Harvard Medical School and Brigham and Women’s Hospital (Boston, MA). Mice (Table S2) were housed in specific pathogen–free facilities. WT C57BL/6, C57BL/6-Tg(TcraTcrb)1100Mjb/J (OT-I mice), and C.Cg-Tg(DO11.10)10Dlo/J mice were from The Jackson Laboratory. Fcgrt−/−, Fcgr2b−/−/Fcer1g−/− (FcγRKO), and CD32aR-Tg (FCGR2AR-Tg/Fcgr2b−/−/Fcer1g−/−) mice have been previously described (Roopenian et al., 2003; McKenzie et al., 1999; Smith et al., 2012), and the latter two strains were generously provided by Dr. Jeffery Ravetch (The Rockefeller University, New York, NY), who also provided the CD32aH-Tg mice, the generation of which is described below.
HEK 293T cells stably expressing the mouse MHC I molecule H2-Kb (HEK 293TH2-Kb) were kindly provided by Dr. Peter Cresswell (Yale University, Hartford, CT; Giodini et al., 2009). RAW 264.7 cell lines were from American Type Culture Collection (#TIB-71). The GM-CSF–secreting B16-F10 melanoma cell line was previously described and was a kind gift from Dr. Glenn Dranoff (Dana Farber Cancer Institute; currently at Novartis, Boston, MA; Mach et al., 2000). Primary APCs and T cells were grown in RPMI 1640 medium (Corning; #10-040) with 10% fetal bovine serum (Life Technologies; #26400044), 1% sodium pyruvate (Lonza; #13-115E), 1% antibiotics (penicillin-streptomycin; Thermo Fisher Scientific; #15070063), 1% nonessential amino acids (Thermo Fisher Scientific; #11140076), and 8.6 µM β-mercaptoethanol (Sigma-Aldrich; #M6250; hereafter “cRPMI”) at 37°C, 5% CO2. Human CD14+ monocytes were maintained in RPMI with 10% fetal bovine serum. B16-F10, HEK 293GFP-hFcRn, and HEK 293TH2-Kb, and all derived cell lines were grown in complete DMEM (Corning; #10-017) in an environment and with additives as for cRPMI plus Hepes 1% (Corning; #25-060) but without β-mercaptoethanol (hereafter “cDMEM”). Details of cloning and associated primers, as well as methods of transfection and transduction of CD32a variants into these cell types, are outlined below. All unique reagents and mouse strains generated for or described in this article will be readily available upon request, with applicable restrictions as outlined in associated material transfer agreements.
Proteins and reagents
NIP hapten-conjugated OVA (OVANIP) was from Biosearch Technologies (N-5041-10), with 11 NIP per OVA. The mAbs DVN24 and ADM31 were produced as described previously (Christianson et al., 2012). Isotype IgG2a control was from BioXCell (clone c1.18.4, #BE0085). Staining for flow cytometry was done using the following antibodies against mouse tissues: CD64 (clone x54-5/7.2; BioLegend; #139307), CD16 (clone 93; BioLegend; #101302), CD16.2 (clone 9E9; BioLegend; #149517), and 2Kb (clone AF6-88.5; BioLegend; #116511). Antibodies for staining hFcγR for flow cytometry were specific for CD64 (clone 10.1; BioLegend; #305005), CD32a (clone FUN-2; BioLegend; #303208), CD16 (clone 3G8; BioLegend; #302010) and other cell markers, including CD15 (clone SSEA-1; BioLegend; #301924), CD66b (clone G10F5; BioLegend; #305112), and CD14 (clone M5E2; BioLegend; #301842). Staining for m/hFcRn or hFcRn was performed using mAbs DVN24 or ADM31, respectively, conjugated to a fluorophore by custom conjugation service (BioLegend). Flow cytometry isotype controls included mIgG2a,κ (clone MOPC-173; BioLegend #400240), mIgG2b,κ (clone 27–35; BioLegend; #402204), mIgG1,κ (clone MOPC-21; BioLegend; #400150). All cell acquisition for flow cytometry experiments were performed on MACSQuant (Miltenyi Biotec) or CytoFLEX flow cytometers (Beckman Coulter) and data were analyzed using FlowJo software (TreeStar), gating on whole cells→single cells→live cells.
Generation of CD32aH-Tg mice
CD32aH-Tg mice were generated by recombineering in EL350 cells (gift from Neal Copeland, National Cancer Institute, Frederick, MD) as described previously (Lee et al., 2001; Kotzamanis and Huxley, 2004). Homology arms were amplified by PCR, (including 16 kb upstream of Exon 1 and 6 kb downstream of Exon 7 of the FCGR2AH gene), subcloned into a pBeloBAC vector (gift from Clare Huxley, Imperial College, London, UK; Kotzamanis and Huxley, 2004) and electroporated into EL350 cells (CTD-2514J12 positive; Invitrogen; #96012) capturing ∼37 kb genomic DNA containing the FCGR2A locus, as described previously (Lee et al., 2001). The presence of rs1801274_His allele of FCGR2A was confirmed by DNA sequencing. The resulting captured construct was linearized using Not1 restriction enzyme (New England Biolabs; #R0189S) and microinjected into the pronuclei of fertilized oocytes from C57BL/6 mice. Tg FCGR2AH+/− founders were mated with C57BL/6 mice and maintained on this background. The CD32aH-Tg strain of mice expressing the FCGR2AH transgene as the only FcγR was created by crossing FCGR2AH mice with FcγRKO mice, producing FCGR2AH/Fcgr2b−/−/Fcer1g−/−, or CD32aH-Tg mice.
Generation of CD32a variant–expressing cell lines
Full-length FCGR2AH cDNA was purchased from Origene (#SC112914). The CD32aR variant was generated via site-directed mutagenesis using the QuickChange II site-directed mutagenesis kit (Agilent Genomics; 200523) and the following overlapping primer pairs: forward primer, 5′-ATCCCAGAAATTCTCCCGTTTGGATCCCACCTTCT-3′; reverse primer, 5′-AGAAGGTGGGATCCAAACGGGAGAATTTCTGGGAT-3′. The cDNAs encoding for CD32aR and CD32aH were subcloned into a pcDNA3.1 vector and sequences verified by DNA sequencing. HEK 293TH2-Kb and RAW 264.7 cells were transfected with pcDNA3.1-CD32aR, pcDNA3.1-CD32aH or empty pcDNA3.1 vector using the Lipofectamine 2000 reagent (Life Technologies) or by electroporation using the Amaxa Cell line nucleofector Kit V (Lonza; #VACA-1003), respectively, and maintained under constant selection with 0.2 mg/ml hygromycin B (Hygromycin B Gold; Invivogen; #ant-hg-1). CD32aR- or CD32aH-transfected HEK 293TH2-Kb and RAW 264.7 were processed by FACS (BD FACSAria II SORP). Transient transfection with pcDNA3.1 vectors was performed by separately mixing Opti-MEM (Thermo Fisher Scientific; #31985070) with DNA (40 µg/ml) and polyethylenimine (PEI; 65 µg/ml) MAX (Polysciences; #24765-1), adding DNA-Opti-MEM to the PEI-Opti-MEM for 30 min at room temperature before incubation with cells, and addition of growth medium, which was changed after 24 h and maintained for 72 h before transfection analysis and experimentation.
Production of recombinant human and mouse FcRn, and anti-NIP IgG mutants
His-tagged soluble mouse and human forms of FcRn proteins were produced using an insect cell–based system, as described previously (Firan et al., 2001; Popov et al., 1996), and site-specifically biotinylated FcRn variants were purchased from Immunitrack (#ITF07 or ITF02). Anti-NIP IgG mutants, as summarized in Table S1, were mouse/human chimeric mAbs and detailed previously (Grevys et al., 2015). hIgG1WT and derived mutants were produced by transient transfection of a vector containing the heavy chain gene from hIgG1 cotransfected with a vector encoding the mouse λ light chain, both with specificity for NIP, or from a stably transfected J558L cell line, as previously described (Grevys et al., 2015; Qiao et al., 2008). The IgG antibodies were purified either by affinity chromatography with anti-mouse λ L chain CaptureSelect resin (Thermo Fisher Scientific; #194323005), anti-hIgG-CH1 CaptureSelect column (Thermo Fisher Scientific; #494320001), or a column coupled with NIP followed by size exclusion chromatography using Superdex 200 10/300 column (GE Healthcare; #17517501).
ELISA
ELISA was used to assess IgG IC binding to CD32a variants. Site-specific biotinylated monomeric human CD32aR and CD32aH (Sino Biological; #10374-H27H1-B-50 and 10374-H27H-B-50) were captured overnight on neutravidin-coated ELISA plates, blocked for 1 h at room temperature with 5% skim milk in PBS, washed with a pH 5.6 buffer (164 mM KH2PO4, 13 mM Na2HPO4 7H2OM NaCl, and 0.05% Tween [PBS-T], pH 5.6), and assayed at pH 5.6. Serial dilutions of ICs containing the anti-NIP IgGs (1.67–214.3 nM) in complex with OVANIP (Biosearch Technologies; #N-5041-100) at a ratio of 2:1 were added to the wells, incubated for 1 h, and washed at pH 5.6. The amount of IgG bound by CD32a variants was measured at pH 5.6 using peroxidase-conjugated goat anti-IgG Fc F(ab′)2 (Jackson ImmunoResearch; #109-036-008) diluted 1:1,000.
To assess IgG IC bridging of CD32a and FcRn, CD32aH and CD32aR were captured (4 µg/ml) on ELISA plates and incubated with titrated amounts of IgG ICs (1.7–214 nM) as described above. For hIgG ICs, biotinylated CD32a variants were captured on neutravidin-coated ELISA plates as above. For mIgG1 ICs, unconjugated CD32a variants (Sino Biological; #10374-H08H1-100 and 10374-H08H-100) were coated directly on ELISA plates overnight. Plates were then washed and blocked as above. His-tagged hFcRn or biotinylated mFcRn (10 µg/ml; Immunitrack; #ITF07) was added and incubated for 1 h at pH 5.6 with 2 µg/ml alkaline phosphatase (ALP)–conjugated anti-hFcRn nanobody for hIgG1 IC (Nb218-H4, which binds hFcRn at acidic pH and does not interfere with IgG binding; Andersen et al., 2013) or Streptavidin-HRP for mIgG1 ICs. After washing with PBS-T, pH 5.6, bound receptor was detected by adding 100 µl p-nitrophenyl phosphate (Thermo Fisher Scientific; #37620), or 3,3′,5,5′-tetramethylbenzidine substrate (Sera Care; #5120) for ALP and HRP, respectively. Protein concentrations were measured using a NanoDrop 2000c spectrophotometer (Thermo Fisher Scientific). ELISA plates were analyzed using a VERSAmax microplate reader (Molecular Devices).
Structural models
Superimposition of the hFcRn-hIgG1 Fc crystal structure (PDB accession number 4N0U; Oganesyan et al., 2014) and hIgG1 Fc-CD32aR complex (PDB accession number 3RY6; Ramsland et al., 2011) with root mean square deviation of 1.378 Å was performed using PyMol (DeLano Scientific), yielding an hFcRn-hIgGFc-CD32aR ternary complex structural model.
IgG IC formation
For OVANIP antigen presentation and cross-presentation studies, innate immune stimulation, and ELISA experiments, anti-NIP IgG ICs were pre-formed in buffer or serum-free RPMI (for primary APCs) or DMEM (all others) at 37°C for 1 h, mixing every 15 min, and using 100 µg/ml of recombinant anti-NIP hIgG1 mutants and 0.5–20 µg/ml OVANIP, as specified.
PLA
RAW 264.7 cells were seeded onto sterile glass coverslips (12 mm, #1.0) coated with 0.1 mM poly-L-lysine, at 5 × 105 cells/ml and incubated overnight at 37°C, 5% CO2. ICs were formed in serum-free DMEM by complexing 0.1 mg/ml hIgG1 (hIgG1κ; Sigma-Aldrich; #15154) with 0.05 mg/ml Dylight 594–conjugated goat F(ab′)2 anti-human F(ab′)2 IgG (Jackson ImmunoResearch; #109-516-006) for 60 min at 37°C. Cells were washed with serum-free DMEM, treated with IC-containing DMEM for 15 min at 37°C and 5% CO2, and then washed with ice-cold PBS and fixed with 3% paraformaldehyde. Permeabilization and blocking was performed with PBS containing 0.1% saponin wt/vol, 1% BSA, and 5% goat serum for 1 h at room temperature. Primary antibody incubation occurred at 4°C overnight using antibodies specific for the cytoplasmic tails of CD32a (5 µg/ml mouse mAb clone 11B6; EMD Millipore; #mabf841) and rat/mFcRn (8 µg/ml rabbit polyclonal antibody made in-house). Goat anti-mouse and goat anti-rabbit secondary antibody PLA probes conjugated to complementary oligonucleotides (Sigma-Aldrich; #DUO92011 and #DUO92013, respectively) were then applied. Ligation of PLA probes in adequate proximity to form a closed, circular DNA template containing repeating sequences was performed followed by DNA amplification. After amplification, FarRed-labeled detection oligo probes (Sigma-Aldrich; #DUO92013) complementary to the repeated sequences were applied per instructions supplied by the manufacturer, except that the PLA probe incubation step was shortened from 60 to 30 min. Coverslips were mounted in antifade mounting medium containing DAPI (Invitrogen; #P36935). Images were acquired at 63× magnification under glycerin immersion using an inverted DMi6000 microscope (Leica) equipped with a CSU-X1 Yokogawa spinning disk, ZYLA SL150 sCMOS camera (Andor), with image analysis and overlay performed with ImageJ version 2.0.0-rc-68/1.52h, with Fiji plugins (https://imagej.net/Fiji/Downloads).
Coimmunoprecipitation
As FcγRs, including CD32a, and FcRn inherently contain binding sites for hIgG1 Fc and bind avidly to IgG from multiple species, we sought a system that would enable selective, Fc-free immunoprecipitation of the ternary complex. We therefore turned to previously published HEK 293hFcRn-GFP cells expressing full-length hFcRn with N-terminal GFP tag (Christianson et al., 2012), which could be immunoprecipitated using magnetic beads coated with anti-GFP high-affinity nanobodies, which lack a Fc region (Rothbauer et al., 2008) and therefore avoid CD32a binding to the immunoprecipitating agent per se. Accordingly, HEK 293hFcRn-GFP cells were transiently transfected using PEI-MAX transfection reagent and pcDNA3.1 vectors expressing either CD32aR of CD32aH (Genscript; #OHu27189D; CD32aR-containing vector was custom prepared by GenScript, introducing a A500G base substitution using OHu27189D as template). 3 d after transfection, CD32a transfection was assessed by flow cytometry and cells were treated for 15 min with hIgGWT or hIgGIHH ICs formed as described above for PLA but using an unconjugated anti–hIgG-Fc F(ab′)2 (Jackson ImmunoResearch; #109-006-006) and anti-NIP hIgG1WT or hIgG1IHH previously conjugated to a disulfide-containing, UV-activated, 13.5-Å, N-hydroxysuccinimide diazirine cross-linker (Thermo Fisher Scientific; #26169) per the manufacturer’s instructions. After IgG IC treatment, cells were washed with ice-cold PBS, exposed to direct UV light (380 nm) on ice for 30 min, and then lysed with ice-cold pH 8.0 CHAPS (3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate hydrate) lysis buffer containing 0.5% (wt/vol) CHAPS, 5% (vol/vol) glycerol, 150 mM NaCl, 2 mM CaCl2, 25 mM Tris-HCl, pH 8.0, and HALT Protease and Phosphatase Inhibitor (Thermo Fisher Scientific; #78442). Insoluble material was removed by centrifugation, protein content was quantified by bicinchoninic acid assay (Thermo Fisher Scientific; kit #23227), and immunoprecipitation of hFcRnGFP from lysate containing 0.5 mg protein was performed using GFP-Trap Magnetic Agarose (ChromoTek; #grma-20) or empty control agarose (data not shown; ChromoTek; #bmab-20) per the manufacturer’s protocol, washing with pH 7.4 wash buffer (0.01 M Tris-HCl, 0.15 M NaCl, and 0.5 mM EDTA). Anti-GFP magnetic agarose-bound proteins were eluted with Laemmli buffer containing β-mercaptoethanol at 95°C for 10 min. The eluate was resolved on 4–20% Tris-Glycine SDS-PAGE gel (Thermo Fisher Scientific; #XP04205BOX), followed by wet transfer to a nitrocellulose membrane. After membrane blocking with Odyssey Blocking Buffer (TBS; LI-COR; #927-50100) for 1 h at room temperature, immunoblotting was performed for hFcRn (Sigma-Aldrich; #HPA012122) and CD32a (clone 11B6) at 1:4,000 and 1:1,000 dilutions, respectively, in Odyssey Blocking Buffer 0.1% Tween 20 overnight at 4°C. Multiplex detection of hFcRn and CD32a was by a IRDye 800CW- or IRDye 680RD-conjugated antibodies (LI-COR; #926-32213 and #926-68072, respectively). Near-infrared–based imaging and densitometry quantification was performed using a LI-COR Odyssey Fc Imaging System running Image Studio version 5.6 (LI-COR).
Innate and adaptive APC stimulation with IgG ICs
APCs (RAW 264.7 cells, HEK 293TH2-Kb cells, or primary DCs; 5 × 104 cells/well) stably expressing CD32aR, CD32aH, or pcDNA3.1 control vector were incubated in 96-well plates in serum-free RMPI containing IgG ICs prepared as above at indicated concentrations for 3 h at 37°C. After washing, cells were co-cultured with 105 OVA-restricted T cells in cRPMI per well using CD8+ T cells for cross-presentation and CD4+ T cells for presentation experiments. Specifically, CD8+ OT-I T cells from mice Tg for a T cell receptor recognizing OVA257–264 peptide in the context of MHCI H-2b (expressed by CD11c+MHCII+ DCs and HEK 293TH2-Kb) or CD4+ T from DO11.10 mice Tg for a T cell receptor recognizing OVA323–339 peptide in the context of the MHCII H-2d (expressed by RAW 264.7) were purified from spleens and peripheral LNs by magnetically activated cell sorting using CD8α+ T cell Isolation kit (Miltenyi Biotec; #130-104-075) or CD4+ T cell Isolation kit (Miltenyi Biotec; #130-104-454), respectively. The cells were co-cultured with IC-stimulated APCs in cRPMI and supernatant collected at 24 or 48 h. The levels of IL-2 and/or IFN-γ in the co-culture supernatant were quantified by ELISA according to the manufacturer’s instructions (BD Biosciences; #555148 or 555138, respectively). For primary cell studies, CD11c+MHCII+ DCs were purified rom spleen by magnetically activated cell sorting in two steps, first using negative selection (CD19 MicroBeads; Miltenyi; #130-052-201) followed by positive selection (CD11c MicroBeads UltraPure; Miltenyi; #130-108-338), from CD32aH-Tg, CD32aR-Tg, or FcγRKO mice that had been inoculated subcutaneously with 5 × 106 GM-CSF–secreting B16-F10 melanoma cells 2 wk before spleen harvest, as described previously (Baker et al., 2013; Mach et al., 2000). For FcRn inhibition experiments, DCs were pretreated for 30 min with the indicated concentrations of the isotype IgG2a control or DVN24 before IC exposure.
For innate immune response assessment, murine splenic CD11c+MHCII+ DCs plated at 5 × 104 cells per well in 96-well plates were treated (or untreated) for 24 h with 100 µg/ml anti-NIP hIgG1 mutants in monomeric or IC form with or without OVANIP (1 µg/ml), formed as above. Mouse TNF-α, IL-12/23p40, and IL-6 levels in clarified supernatant were quantified by ELISA kits according to the manufacturer’s instructions (BD Biosciences; #555165, 558534, or 555240, respectively).
For studies of primary human APC, DNA was isolated from blood collected from volunteer human blood donors and genotyped for FCGR2A SNP rs1801274 by a Taqman assay (Thermo Fisher Scientific; #C_9077561_20) according to the manufacturer’s instructions as described (Kozicky et al., 2018). Blood for functional studies was collected in heparinized vials and either plated immediately or separated by density gradient using Mono-Poly medium (MP Biomedicals; #1698049). CD14+ cells were isolated from the upper Mono-Poly fraction by magnetic separation using EasySep Human CD14 Positive Selection Kit II (StemCell; #17858), according to the manufacturer’s instructions. Both whole-blood and CD14+ monocytes were stimulated as described above for murine cells (Blumberg et al., 2019), and TNF-α, IL-12/23p40, and IL-6 were quantified in clarified supernatant using ELISA kits according to the manufacturer’s instructions (BD Biosciences; #555171, 555212, or 555220, respectively).
K/BxN rheumatoid arthritis model
Rheumatoid arthritis was induced and assessed as described previously (Monach et al., 2008) in bone marrow chimeric mice prepared by adoptive transfer of bone marrow from sex-matched CD32aR-Tg donor mice to irradiated WT C57BL/6 recipient mice. mAb isotype IgG2a control or DVN24 (0.2 mg in 0.2 ml) was administered i.p. daily beginning on day −1 through day 5 from K/BxN sera injection (Fig. 4 a). Of note, the DVN24 dose employed in these experiments reflects its ∼17-fold lower affinity for mFcRn versus hFcRn (Sand et al., 2014). Therefore, these experiments required a proportionately higher DVN24 dose relative to that expected to be efficacious in the setting of hFcRn. After 1 wk (day 7), mobility was assessed by the cylinder test or the number side touches per mouse per minute in a 1-liter glass beaker (Brooks and Dunnett, 2009). Histopathological scoring of rear ankle joint inflammation and bone erosion on day 12 was by a blinded pathologist (J.N. Glickman) as described previously (Pettit et al., 2001). Escherichia coli expressing recombinant glucose-6-phosphate isomerase (GPI) were kindly provided by Dr. Christopher Benoist (Harvard Medical School, Boston, MA), and GPI production, purification, and quantification of circulating anti-GPI IgG by ELISA was performed as previously described (Monach et al., 2008).
Statistical analysis
Prism for Mac OS X version 8.2.1 (GraphPad Software) was used for statistical analysis. Analysis of ELISA binding curves was by nonlinear regression using variable slope (agonist) versus response equation for least squares fitting, with EC50 confidence intervals calculated using the likelihood ratio asymmetric method, goodness of fit assessed by R2, and extra sum-of-squares F test to detect differences between the EC50 values associated with the resulting best-fit curves. Nonnormal data were processed by loge transformation and resulting data evaluated for a normal distribution by inspection of homoscedasticity and QQ plots. Comparisons of two unpaired groups was made by a two-tailed Student’s t test for normal data or Mann–Whitney test for nonnormal data. For three or more groups with two parameters, two-way ANOVA was used as appropriate for data with a normal distribution, or a matched Friedman test was employed for nonnormally distributed data. Analysis of responses to multiple stimuli of a single cell type or of cells from a single donor were analyzed using paired tests. Post-hoc analysis to correct for multiple comparisons and detect differences between groups was by the two-stage linear step-up procedure of Benjamin, Krieger, and Yekutieli with a false discovery rate (FDR) < 0.05. A two-sided probability (P) of α error < 0.05 defined significance.
Online supplemental material
Fig. S1 (related to Fig. 1) shows control ELISA and PLA experiments, as well as flow cytometry assessments of transfection efficiency. Fig. S2 (related to Fig. 2) shows FcRn and CD32a transgene expression measured by flow cytometry and cellular activation data supportive of data in Fig. 2. Fig. S3 (related to Fig. 3) shows flow cytometric assessments of primary human cell preparation purity. Fig. S4 (related to Fig. 4) shows ELISA of ternary complex formation with CD32a, mFcRn, and mIgG1. Table S1 shows mutations defining anti-NIP hIgG1 mutants, with a summary of qualitative, relative pH-dependent binding strengths to FcRn and low-affinity FcγRs. Table S2 shows a summary of Tg mouse strains utilized in these studies.
Supplementary Material
shows mutations defining anti-NIP hIgG1 mutants, with a summary of qualitative, relative pH-dependent binding strengths to FcRn and low-affinity FcγRs.
shows a summary of Tg mouse strains utilized in these studies.
Acknowledgments
The authors thank Victoria M. Thiele, Garrett Hauck, Thomas Hanley, Arianna Degruttola, Samantha Torquato, Mario Sablon, Victoria G. Aveson, and Anh Do for excellent technical assistance and Drs. Nitesh Shashikanth and Jerrold Turner for expert assistance with and access to confocal microscopy equipment. The authors also thank Jennifer Cusick for managerial assistance and Dr. Amanjot Riar for technical and managerial assistance. Dr. Jeffrey Ravetch (The Rockefeller University, New York, NY) generated and very generously shared the CD32aR-Tg and CD32aH-Tg mice. H2-Kb–expressing HEK 293T cells were a generous gift of Dr. Peter Cresswell (Yale University, Harford, CT). E. coli expressing recombinant GPI were kindly provided by Dr. Christopher Benoist (Harvard Medical School, Boston, MA).
This work was funded by Deutsche Forschungsgemeinschaft (RA 2040/1-1; T. Rath), the Canadian Institutes of Health Research (K. Baker, M. Pyzik, and L.K. Kozicky) and Canadian Institutes of Health Research/Canadian Blood Services project grant (CIHR2016-LS) to L.M. Sly, the North American Society for Pediatric Gastroenterology, Hepatology and Nutrition Foundation Fellow to Faculty Transition Award NF-17-100 (J.J. Hubbard), the Crohn’s and Colitis Foundation Research Fellowship Award 511426 (J.J. Hubbard), the National Institutes of Health grants DK53056, DK053162, DK088199, DK044319 (R.S. Blumberg), and 1F32AI131511 (J.J. Hubbard), and the Harvard Digestive Diseases Center National Institutes of Health grant P30DK034854 (R.S. Blumberg). J.J. Hubbard was supported by the National Institutes of Health T32 grant DK007477 through the Division of Gastroenterology and Nutrition at Boston Children’s Hospital. I. Sandlie and J.T. Andersen were supported in part by the Research Council of Norway through its Centre of Excellence funding scheme (project 179573), the Research Council of Norway (grants 230526, 179573, and 287927), and South-Eastern Norway Regional Health Authority (grant 40018). K.M.K. Sand was supported by the University of Oslo, The U.S.A.-Norway Fulbright Foundation for Educational Exchange, and University of Oslo Life Science.
Author contributions: Conceptualization: J.J. Hubbard, M. Pyzik, T. Rath, L.K. Kozicky, K.M.K. Sand, J.T. Andersen, L.M. Sly, D.C. Roopenian, K. Baker, R.S. Blumberg. Methodology: J.J. Hubbard, M. Pyzik, T. Rath, L.K. Kozicky, K.M.K. Sand, A.K. Gandhi, A. Grevys, S. Foss, S.C. Menzies, J.T. Andersen, J.N. Glickman, I. Sandlie, L.M. Sly, K. Baker, and R.S. Blumberg. Validation: J.J. Hubbard, M. Pyzik, T. Rath, K.M.K. Sand, L.K. Kozicky, S.C. Menzies, J.T. Andersen, K. Baker, and R.S. Blumberg. Formal analysis: J.J. Hubbard, M. Pyzik, T. Rath, L.K. Kozicky, K.M.K. Sand, J.T. Andersen, and J.N. Glickman. Investigation: J.J. Hubbard, M. Pyzik, L.K. Kozicky, K.M.K. Sand, T. Rath, J.T. Andersen, K. Baker, S. Foss, and A. Grevys. Resources: J.T. Andersen, L.M. Sly, I. Sandlie, D.C. Roopenian, and R.S. Blumberg. Writing (original draft): J.J. Hubbard, M. Pyzik, L.K. Kozicky, K.M.K. Sand, L.M. Sly, K. Baker, and R.S. Blumberg. Writing (review and editing): J.J. Hubbard, M. Pyzik, T. Rath, L.K. Kozicky, K.M.K. Sand, J.T. Andersen, A.K. Gandhi, A. Grevys, E. Fiebiger, D.C. Roopenian, I. Sandlie, L.M. Sly, K. Baker, R.S. Blumberg. Visualization: J.J. Hubbard, M. Pyzik, L.K. Kozicky, K.S.M., T. Rath, J.T. Andersen, A.K. Gandhi, A. Grevys, I. Sandlie, K. Baker, and R.S. Blumberg. Supervision, L.M. Sly, I. Sandlie, K. Baker, and R.S. Blumberg. Project administration, K. Baker and R.S. Blumberg. Funding acquisition, R.S. Blumberg (primarily) but also L.M. Sly, J.J. Hubbard, M. Pyzik, T. Rath, K.M.K. Sand, J.T. Andersen, L.K. Kozicky, and K. Baker.
References
- Andersen J.T., Gonzalez-Pajuelo M., Foss S., Landsverk O.J.B., Pinto D., Szyroki A., de Haard H.J., Saunders M., Vanlandschoot P., and Sandlie I.. 2013. Selection of nanobodies that target human neonatal Fc receptor. Sci. Rep. 3:1118 10.1038/srep01118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker K., Qiao S.W., Kuo T.T., Aveson V.G., Platzer B., Andersen J.T., Sandlie I., Chen Z., de Haar C., Lencer W.I., et al. . 2011. Neonatal Fc receptor for IgG (FcRn) regulates cross-presentation of IgG immune complexes by CD8-CD11b+ dendritic cells. Proc. Natl. Acad. Sci. USA. 108:9927–9932. 10.1073/pnas.1019037108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker K., Rath T., Flak M.B., Arthur J.C., Chen Z., Glickman J.N., Zlobec I., Karamitopoulou E., Stachler M.D., Odze R.D., et al. . 2013. Neonatal Fc receptor expression in dendritic cells mediates protective immunity against colorectal cancer. Immunity. 39:1095–1107. 10.1016/j.immuni.2013.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumberg L.J., Humphries J.E., Jones S.D., Pearce L.B., Holgate R., Hearn A., Cheung J., Mahmood A., Del Tito B., Graydon J.S., et al. . 2019. Blocking FcRn in humans reduces circulating IgG levels and inhibits IgG immune complex-mediated immune responses. Sci. Adv. 5:x9586 10.1126/sciadv.aax9586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnerot C., Briken V., Brachet V., Lankar D., Cassard S., Jabri B., and Amigorena S.. 1998. syk protein tyrosine kinase regulates Fc receptor gamma-chain-mediated transport to lysosomes. EMBO J. 17:4606–4616. 10.1093/emboj/17.16.4606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borvak J., Richardson J., Medesan C., Antohe F., Radu C., Simionescu M., Ghetie V., and Ward E.S.. 1998. Functional expression of the MHC class I-related receptor, FcRn, in endothelial cells of mice. Int. Immunol. 10:1289–1298. 10.1093/intimm/10.9.1289 [DOI] [PubMed] [Google Scholar]
- Bournazos S., Wang T.T., Dahan R., Maamary J., and Ravetch J.V.. 2017. Signaling by Antibodies: Recent Progress. Annu. Rev. Immunol. 35:285–311. 10.1146/annurev-immunol-051116-052433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bredius R.G.M., Derkx B.H.F., Fijen C.A.P., de Wit T.P.M., de Haas M., Weening R.S., van de Winkel J.G.J., and Out T.A.. 1994. Fc gamma receptor IIa (CD32) polymorphism in fulminant meningococcal septic shock in children. J. Infect. Dis. 170:848–853. 10.1093/infdis/170.4.848 [DOI] [PubMed] [Google Scholar]
- Brooks S.P., and Dunnett S.B.. 2009. Tests to assess motor phenotype in mice: a user’s guide. Nat. Rev. Neurosci. 10:519–529. 10.1038/nrn2652 [DOI] [PubMed] [Google Scholar]
- Bruhns P., Iannascoli B., England P., Mancardi D.A., Fernandez N., Jorieux S., and Daëron M.. 2009. Specificity and affinity of human Fcgamma receptors and their polymorphic variants for human IgG subclasses. Blood. 113:3716–3725. 10.1182/blood-2008-09-179754 [DOI] [PubMed] [Google Scholar]
- Carcao M.D., Blanchette V.S., Wakefield C.D., Stephens D., Ellis J., Matheson K., and Denomme G.A.. 2003. Fcgamma receptor IIa and IIIa polymorphisms in childhood immune thrombocytopenic purpura. Br. J. Haematol. 120:135–141. 10.1046/j.1365-2141.2003.04033.x [DOI] [PubMed] [Google Scholar]
- Chen B., Vousden K.A., Naiman B., Turman S., Sun H., Wang S., Vinall L.M.K., Kemp B.P., Kasturiangan S., Rees D.G., et al. . 2019. Humanised effector-null FcγRIIA antibody inhibits immune complex-mediated proinflammatory responses. Ann. Rheum. Dis. 78:228–237. 10.1136/annrheumdis-2018-213523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson G.J., Sun V.Z., Akilesh S., Pesavento E., Proetzel G., and Roopenian D.C.. 2012. Monoclonal antibodies directed against human FcRn and their applications. MAbs. 4:208–216. 10.4161/mabs.4.2.19397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cines D.B., Zaitsev S., Rauova L., Rux A.H., Stepanova V., Krishnaswamy S., Sarkar A., Kowalska M.A., Zhao G., Mast A.E., et al. . 2020. FcRn augments induction of tissue factor activity by IgG-containing immune complexes. Blood. 135:2085–2093. 10.1182/blood.2019001133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark M.R., Stuart S.G., Kimberly R.P., Ory P.A., and Goldstein I.M.. 1991. A single amino acid distinguishes the high-responder from the low-responder form of Fc receptor II on human monocytes. Eur. J. Immunol. 21:1911–1916. 10.1002/eji.1830210820 [DOI] [PubMed] [Google Scholar]
- Dickinson B.L., Badizadegan K., Wu Z., Ahouse J.C., Zhu X., Simister N.E., Blumberg R.S., and Lencer W.I.. 1999. Bidirectional FcRn-dependent IgG transport in a polarized human intestinal epithelial cell line. J. Clin. Invest. 104:903–911. 10.1172/JCI6968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dijstelbloem H.M., Bijl M., Fijnheer R., Scheepers R.H.M., Oost W.W., Jansen M.D., Sluiter W.J., Limburg P.C., Derksen R.H.W.M., van de Winkel J.G.J., et al. . 2000. Fcgamma receptor polymorphisms in systemic lupus erythematosus: association with disease and in vivo clearance of immune complexes. Arthritis Rheum. 43:2793–2800. [DOI] [PubMed] [Google Scholar]
- Endeman H., Cornips M.C.A., Grutters J.C., van den Bosch J.M., Ruven H.J.T., van Velzen-Blad H., Rijkers G.T., and Biesma D.H.. 2009. The Fcgamma receptor IIA-R/R131 genotype is associated with severe sepsis in community-acquired pneumonia. Clin. Vaccine Immunol. 16:1087–1090. 10.1128/CVI.00037-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faure F., Mantegazza A., Sadaka C., Sedlik C., Jotereau F., and Amigorena S.. 2009. Long-lasting cross-presentation of tumor antigen in human DC. Eur. J. Immunol. 39:380–390. 10.1002/eji.200838669 [DOI] [PubMed] [Google Scholar]
- Firan M., Bawdon R., Radu C., Ober R.J., Eaken D., Antohe F., Ghetie V., and Ward E.S.. 2001. The MHC class I-related receptor, FcRn, plays an essential role in the maternofetal transfer of gamma-globulin in humans. Int. Immunol. 13:993–1002. 10.1093/intimm/13.8.993 [DOI] [PubMed] [Google Scholar]
- Gaudinski M.R., Coates E.E., Houser K.V., Chen G.L., Yamshchikov G., Saunders J.G., Holman L.A., Gordon I., Plummer S., Hendel C.S., et al. ; VRC 606 Study Team . 2018. Safety and pharmacokinetics of the Fc-modified HIV-1 human monoclonal antibody VRC01LS: A Phase 1 open-label clinical trial in healthy adults. PLoS Med. 15 e1002493 10.1371/journal.pmed.1002493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giodini A., Rahner C., and Cresswell P.. 2009. Receptor-mediated phagocytosis elicits cross-presentation in nonprofessional antigen-presenting cells. Proc. Natl. Acad. Sci. USA. 106:3324–3329. 10.1073/pnas.0813305106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grevys A., Bern M., Foss S., Bratlie D.B., Moen A., Gunnarsen K.S., Aase A., Michaelsen T.E., Sandlie I., and Andersen J.T.. 2015. Fc Engineering of Human IgG1 for Altered Binding to the Neonatal Fc Receptor Affects Fc Effector Functions. J. Immunol. 194:5497–5508. 10.4049/jimmunol.1401218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes W., Lide D., and Bruno T., editors. 2016. CRC Handbook of Chemistry and Physics. Ninety seventh edition CRC Press, Boca Raton, Florida: 10.1201/9781315380476 [DOI] [Google Scholar]
- Hogquist K.A., Jameson S.C., Heath W.R., Howard J.L., Bevan M.J., and Carbone F.R.. 1994. T cell receptor antagonist peptides induce positive selection. Cell. 76:17–27. 10.1016/0092-8674(94)90169-4 [DOI] [PubMed] [Google Scholar]
- Kiessling P., Lledo-Garcia R., Watanabe S., Langdon G., Tran D., Bari M., Christodoulou L., Jones E., Price G., Smith B., et al. . 2017. The FcRn inhibitor rozanolixizumab reduces human serum IgG concentration: A randomized phase 1 study. Sci. Transl. Med. 9 eaan1208 10.1126/scitranslmed.aan1208 [DOI] [PubMed] [Google Scholar]
- Kim J.-K., Firan M., Radu C.G., Kim C.-H., Ghetie V., and Ward E.S.. 1999. Mapping the site on human IgG for binding of the MHC class I-related receptor, FcRn. Eur. J. Immunol. 29:2819–2825. [DOI] [PubMed] [Google Scholar]
- Kotzamanis G., and Huxley C.. 2004. Recombining overlapping BACs into a single larger BAC. BMC Biotechnol. 4:1–10. 10.1186/1472-6750-4-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozicky L.K., Menzies S.C., Zhao Z.Y., Vira T., Harnden K., Safari K., Del Bel K.L., Turvey S.E., and Sly L.M.. 2018. IVIg and LPS Co-stimulation Induces IL-10 Production by Human Monocytes, Which Is Compromised by an FcγRIIA Disease-Associated Gene Variant. Front. Immunol. 9:2676 10.3389/fimmu.2018.02676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E.-C., Yu D., Martinez de Velasco J., Tessarollo L., Swing D.A., Court D.L., Jenkins N.A., and Copeland N.G.. 2001. A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics. 73:56–65. 10.1006/geno.2000.6451 [DOI] [PubMed] [Google Scholar]
- Lee Y.H., Bae S.C., and Song G.G.. 2015. FCGR2A, FCGR3A, FCGR3B polymorphisms and susceptibility to rheumatoid arthritis: a meta-analysis. Clin. Exp. Rheumatol. 33:647–654. [PubMed] [Google Scholar]
- Lehrnbecher T., Foster C.B., Zhu S., Leitman S.F., Goldin L.R., Huppi K., and Chanock S.J.. 1999. Variant genotypes of the low-affinity Fcgamma receptors in two control populations and a review of low-affinity Fcgamma receptor polymorphisms in control and disease populations. Blood. 94:4220–4232. 10.1182/blood.V94.12.4220 [DOI] [PubMed] [Google Scholar]
- Li X., Gibson A.W., and Kimberly R.P.. 2014. Human FcR polymorphism and disease. Curr. Top. Microbiol. Immunol. 382:275–302. 10.1007/978-3-319-07911-0_13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindner D., and Raghavan D.. 2009. Intra-tumoural extra-cellular pH: a useful parameter of response to chemotherapy in syngeneic tumour lines. Br. J. Cancer. 100:1287–1291. 10.1038/sj.bjc.6605022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling L.E., Hillson J.L., Tiessen R.G., Bosje T., van Iersel M.P., Nix D.J., Markowitz L., Cilfone N.A., Duffner J., Streisand J.B., et al. . 2019. M281, an Anti-FcRn Antibody: Pharmacodynamics, Pharmacokinetics, and Safety Across the Full Range of IgG Reduction in a First-in-Human Study. Clin. Pharmacol. Ther. 105:1031–1039. 10.1002/cpt.1276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López D.H., Trevani A.S., Salamone G., Andonegui G., Raiden S., Giordano M., and Geffner J.R.. 1999. Acidic pH increases the avidity of FcgammaR for immune complexes. Immunology. 98:450–455. 10.1046/j.1365-2567.1999.00884.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mach N., Gillessen S., Wilson S.B., Sheehan C., Mihm M., and Dranoff G.. 2000. Differences in dendritic cells stimulated in vivo by tumors engineered to secrete granulocyte-macrophage colony-stimulating factor or Flt3-ligand. Cancer Res. 60:3239–3246. [PubMed] [Google Scholar]
- Mancardi D.A., Jönsson F., Iannascoli B., Khun H., Van Rooijen N., Huerre M., Daëron M., and Bruhns P.. 2011. Cutting Edge: The murine high-affinity IgG receptor FcγRIV is sufficient for autoantibody-induced arthritis. J. Immunol. 186:1899–1903. 10.4049/jimmunol.1003642 [DOI] [PubMed] [Google Scholar]
- Martin W.L., and Björkman P.J.. 1999. Characterization of the 2:1 complex between the class I MHC-related Fc receptor and its Fc ligand in solution. Biochemistry. 38:12639–12647. 10.1021/bi9913505 [DOI] [PubMed] [Google Scholar]
- Martin W.L., West A.P. Jr., Gan L., and Björkman P.J.. 2001. Crystal structure at 2.8 A of an FcRn/heterodimeric Fc complex: mechanism of pH-dependent binding. Mol. Cell. 7:867–877. 10.1016/S1097-2765(01)00230-1 [DOI] [PubMed] [Google Scholar]
- Matsumoto I., Staub A., Benoist C., and Mathis D.. 1999. Arthritis provoked by linked T and B cell recognition of a glycolytic enzyme. Science. 286:1732–1735. 10.1126/science.286.5445.1732 [DOI] [PubMed] [Google Scholar]
- McKenzie S.E., Taylor S.M., Malladi P., Yuhan H., Cassel D.L., Chien P., Schwartz E., Schreiber A.D., Surrey S., and Reilly M.P.. 1999. The role of the human Fc receptor Fc gamma RIIA in the immune clearance of platelets: a transgenic mouse model. J. Immunol. 162:4311–4318. [PubMed] [Google Scholar]
- Medesan C., Matesoi D., Radu C., Ghetie V., and Ward E.S.. 1997. Delineation of the amino acid residues involved in transcytosis and catabolism of mouse IgG1. J. Immunol. 158:2211–2217. [PubMed] [Google Scholar]
- Monach P.A., Mathis D., and Benoist C.. 2008. The K/BxN arthritis model. Curr. Protoc. Immunol. Chapter 15:22 10.1002/0471142735.im1522s81 [DOI] [PubMed] [Google Scholar]
- Morgan A.W., Griffiths B., Ponchel F., Montague B.M., Ali M., Gardner P.P., Gooi H.C., Situnayake R.D., Markham A.F., Emery P., et al. . 2000. Fcgamma receptor type IIIA is associated with rheumatoid arthritis in two distinct ethnic groups. Arthritis Rheum. 43:2328–2334. [DOI] [PubMed] [Google Scholar]
- Neuber T., Frese K., Jaehrling J., Jäger S., Daubert D., Felderer K., Linnemann M., Höhne A., Kaden S., Kölln J., et al. . 2014. Characterization and screening of IgG binding to the neonatal Fc receptor. MAbs. 6:928–942. 10.4161/mabs.28744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura Y., Gautam R., Chun T.-W., Sadjadpour R., Foulds K.E., Shingai M., Klein F., Gazumyan A., Golijanin J., Donaldson M., et al. . 2017. Early antibody therapy can induce long-lasting immunity to SHIV. Nature. 543:559–563. 10.1038/nature21435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nixon A.E., Chen J., Sexton D.J., Muruganandam A., Bitonti A.J., Dumont J., Viswanathan M., Martik D., Wassaf D., Mezo A., et al. . 2015. Fully human monoclonal antibody inhibitors of the neonatal fc receptor reduce circulating IgG in non-human primates. Front. Immunol. 6:176 10.3389/fimmu.2015.00176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oganesyan V., Damschroder M.M., Cook K.E., Li Q., Gao C., Wu H., and Dall’Acqua W.F.. 2014. Structural insights into neonatal Fc receptor-based recycling mechanisms. J. Biol. Chem. 289:7812–7824. 10.1074/jbc.M113.537563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parren P.W., Warmerdam P.A., Boeije L.C., Arts J., Westerdaal N.A., Vlug A., Capel P.J., Aarden L.A., and van de Winkel J.G.. 1992. On the interaction of IgG subclasses with the low affinity Fc gamma RIIa (CD32) on human monocytes, neutrophils, and platelets. Analysis of a functional polymorphism to human IgG2. J. Clin. Invest. 90:1537–1546. 10.1172/JCI116022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettit A.R., Ji H., von Stechow D., Müller R., Goldring S.R., Choi Y., Benoist C., and Gravallese E.M.. 2001. TRANCE/RANKL knockout mice are protected from bone erosion in a serum transfer model of arthritis. Am. J. Pathol. 159:1689–1699. 10.1016/S0002-9440(10)63016-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pincetic A., Bournazos S., DiLillo D.J., Maamary J., Wang T.T., Dahan R., Fiebiger B.-M., and Ravetch J.V.. 2014. Type I and type II Fc receptors regulate innate and adaptive immunity. Nat. Immunol. 15:707–716. 10.1038/ni.2939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popov S., Hubbard J.G., Kim J., Ober B., Ghetie V., and Ward E.S.. 1996. The stoichiometry and affinity of the interaction of murine Fc fragments with the MHC class I-related receptor, FcRn. Mol. Immunol. 33:521–530. 10.1016/0161-5890(96)00004-1 [DOI] [PubMed] [Google Scholar]
- Prabhat P., Gan Z., Chao J., Ram S., Vaccaro C., Gibbons S., Ober R.J., and Ward E.S.. 2007. Elucidation of intracellular recycling pathways leading to exocytosis of the Fc receptor, FcRn, by using multifocal plane microscopy. Proc. Natl. Acad. Sci. USA. 104:5889–5894. 10.1073/pnas.0700337104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyzik M., Sand K.M.K., Hubbard J.J., Andersen J.T., Sandlie I., and Blumberg R.S.. 2019. The Neonatal Fc Receptor (FcRn): A Misnomer? Front. Immunol. 10:1540 10.3389/fimmu.2019.01540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao S.-W.W., Kobayashi K., Johansen F.-E.E., Sollid L.M., Andersen J.T., Milford E., Roopenian D.C., Lencer W.I., and Blumberg R.S.. 2008. Dependence of antibody-mediated presentation of antigen on FcRn. Proc. Natl. Acad. Sci. USA. 105:9337–9342. 10.1073/pnas.0801717105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsland P.A., Farrugia W., Bradford T.M., Sardjono C.T., Esparon S., Trist H.M., Powell M.S., Tan P.S., Cendron A.C., Wines B.D., et al. . 2011. Structural basis for Fc gammaRIIa recognition of human IgG and formation of inflammatory signaling complexes. J. Immunol. 187:3208–3217. 10.4049/jimmunol.1101467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regnault A., Lankar D., Lacabanne V., Rodriguez A., Théry C., Rescigno M., Saito T., Verbeek S., Bonnerot C., Ricciardi-Castagnoli P., et al. . 1999. Fcgamma receptor-mediated induction of dendritic cell maturation and major histocompatibility complex class I-restricted antigen presentation after immune complex internalization. J. Exp. Med. 189:371–380. 10.1084/jem.189.2.371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riemann A., Wußling H., Loppnow H., Fu H., Reime S., and Thews O.. 2016. Acidosis differently modulates the inflammatory program in monocytes and macrophages. Biochim. Biophys. Acta. 1862:72–81. 10.1016/j.bbadis.2015.10.017 [DOI] [PubMed] [Google Scholar]
- Roopenian D.C., Christianson G.J., Sproule T.J., Brown A.C., Akilesh S., Jung N., Petkova S., Avanessian L., Choi E.Y., Shaffer D.J., et al. . 2003. The MHC class I-like IgG receptor controls perinatal IgG transport, IgG homeostasis, and fate of IgG-Fc-coupled drugs. J. Immunol. 170:3528–3533. 10.4049/jimmunol.170.7.3528 [DOI] [PubMed] [Google Scholar]
- Rothbauer U., Zolghadr K., Muyldermans S., Schepers A., Cardoso M.C., and Leonhardt H.. 2008. A versatile nanotrap for biochemical and functional studies with fluorescent fusion proteins. Mol. Cell. Proteomics. 7:282–289. 10.1074/mcp.M700342-MCP200 [DOI] [PubMed] [Google Scholar]
- Salmon J.E., Edberg J.C., Brogle N.L., and Kimberly R.P.. 1992. Allelic polymorphisms of human Fc gamma receptor IIA and Fc gamma receptor IIIB. Independent mechanisms for differences in human phagocyte function. J. Clin. Invest. 89:1274–1281. 10.1172/JCI115712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sand K.M.K., Dalhus B., Christianson G.J., Bern M., Foss S., Cameron J., Sleep D., Bjørås M., Roopenian D.C., Sandlie I., et al. . 2014. Dissection of the neonatal Fc receptor (FcRn)-albumin interface using mutagenesis and anti-FcRn albumin-blocking antibodies. J. Biol. Chem. 289:17228–17239. 10.1074/jbc.M113.522565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders L.A., Feldman R.G., Voorhorst-Ogink M.M., de Haas M., Rijkers G.T., Capel P.J., Zegers B.J., and van de Winkel J.G.. 1995. Human immunoglobulin G (IgG) Fc receptor IIA (CD32) polymorphism and IgG2-mediated bacterial phagocytosis by neutrophils. Infect. Immun. 63:73–81. 10.1128/IAI.63.1.73-81.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shashidharamurthy R., Zhang F., Amano A., Kamat A., Panchanathan R., Ezekwudo D., Zhu C., and Selvaraj P.. 2009. Dynamics of the interaction of human IgG subtype immune complexes with cells expressing R and H allelic forms of a low-affinity Fc gamma receptor CD32A. J. Immunol. 183:8216–8224. 10.4049/jimmunol.0902550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simister N.E., and Mostov K.E.. 1989. An Fc receptor structurally related to MHC class I antigens. Nature. 337:184–187. 10.1038/337184a0 [DOI] [PubMed] [Google Scholar]
- Smith P., DiLillo D.J., Bournazos S., Li F., and Ravetch J.V.. 2012. Mouse model recapitulating human Fcγ receptor structural and functional diversity. Proc. Natl. Acad. Sci. USA. 109:6181–6186. 10.1073/pnas.1203954109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao M.H., and Morrison S.L.. 1989. Studies of aglycosylated chimeric mouse-human IgG. Role of carbohydrate in the structure and effector functions mediated by the human IgG constant region. J. Immunol. 143:2595–2601. [PubMed] [Google Scholar]
- Tate B.J., Witort E., McKenzie I.F., and Hogarth P.M.. 1992. Expression of the high responder/non-responder human Fc gamma RII. Analysis by PCR and transfection into FcR-COS cells. Immunol. Cell Biol. 70:79–87. 10.1038/icb.1992.12 [DOI] [PubMed] [Google Scholar]
- Ulrichts P., Guglietta A., Dreier T., van Bragt T., Hanssens V., Hofman E., Vankerckhoven B., Verheesen P., Ongenae N., Lykhopiy V., et al. . 2018. Neonatal Fc receptor antagonist efgartigimod safely and sustainably reduces IgGs in humans. J. Clin. Invest. 128:4372–4386. 10.1172/JCI97911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaccaro C., Zhou J., Ober R.J., and Ward E.S.. 2005. Engineering the Fc region of immunoglobulin G to modulate in vivo antibody levels. Nat. Biotechnol. 23:1283–1288. 10.1038/nbt1143 [DOI] [PubMed] [Google Scholar]
- Vidarsson G., Stemerding A.M., Stapleton N.M., Spliethoff S.E., Janssen H., Rebers F.E., de Haas M., and van de Winkel J.G.. 2006. FcRn: an IgG receptor on phagocytes with a novel role in phagocytosis. Blood. 108:3573–3579. 10.1182/blood-2006-05-024539 [DOI] [PubMed] [Google Scholar]
- Wines B.D., Powell M.S., Parren P.W., Barnes N., and Hogarth P.M.. 2000. The IgG Fc contains distinct Fc receptor (FcR) binding sites: the leukocyte receptors Fc gamma RI and Fc gamma RIIa bind to a region in the Fc distinct from that recognized by neonatal FcR and protein A. J. Immunol. 164:5313–5318. 10.4049/jimmunol.164.10.5313 [DOI] [PubMed] [Google Scholar]
- Wu J., Edberg J.C., Redecha P.B., Bansal V., Guyre P.M., Coleman K., Salmon J.E., and Kimberly R.P.. 1997. A novel polymorphism of FcgammaRIIIa (CD16) alters receptor function and predisposes to autoimmune disease. J. Clin. Invest. 100:1059–1070. 10.1172/JCI119616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Wang W., Zhang H., Wei L., and Guo S.. 2016. Association of FCGR2A rs1801274 polymorphism with susceptibility to autoimmune diseases: A meta-analysis. Oncotarget. 7:39436–39443. 10.18632/oncotarget.9831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X., Meng G., Dickinson B.L., Li X., Mizoguchi E., Miao L., Wang Y., Robert C., Wu B., Smith P.D., et al. . 2001. MHC class I-related neonatal Fc receptor for IgG is functionally expressed in monocytes, intestinal macrophages, and dendritic cells. J. Immunol. 166:3266–3276. 10.4049/jimmunol.166.5.3266 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
shows mutations defining anti-NIP hIgG1 mutants, with a summary of qualitative, relative pH-dependent binding strengths to FcRn and low-affinity FcγRs.
shows a summary of Tg mouse strains utilized in these studies.