Abstract
Transmission electron microscopy has become a valuable tool to investigate tissues of COVID‐19 patients because it allows visualisation of SARS‐CoV‐2, but the ‘virus‐like particles’ described in several organs have been highly contested. Because most electron microscopists in pathology are not accustomed to analysing viral particles and subcellular structures, our review aims to discuss the ultrastructural changes associated with SARS‐CoV‐2 infection and COVID‐19 with respect to pathology, virology and electron microscopy. Using micrographs from infected cell cultures and autopsy tissues, we show how coronavirus replication affects ultrastructure and put the morphological findings in the context of viral replication, which induces extensive remodelling of the intracellular membrane systems. Virions assemble by budding into the endoplasmic reticulum–Golgi intermediate complex and are characterised by electron‐dense dots of cross‐sections of the nucleocapsid inside the viral particles. Physiological mimickers such as multivesicular bodies or coated vesicles serve as perfect decoys. Compared to other in‐situ techniques, transmission electron microscopy is the only method to visualise assembled virions in tissues, and will be required to prove SARS‐CoV‐2 replication outside the respiratory tract. In practice, documenting in tissues the characteristic features seen in infected cell cultures seems to be much more difficult than anticipated. In our view, the hunt for coronavirus by transmission electron microscopy is still on.
Keywords: coronavirus, virus replication, ultrastructure, electron microscopy

Abbreviations
- ACE2
angiotensin converting enzyme 2
- CM
convoluted membranes
- CMS
cubic membrane structures
- COPI
COPII, coat protein complex I/II
- CoV
coronavirus
- COVID‐19
coronavirus disease 2019
- DMV
double membrane vesicle
- EM
electron microscopy
- ER
endoplasmic reticulum
- ERGIC
endoplasmic reticulum Golgi intermediate complex
- ILV
intraluminal vesicle
- LVCV
large virion containing vacuole
- MERS
middle‐east respiratory syndrome
- MVB
multivesicular body
- QNAT
quantitative nucleic acid amplification technique
- RMW
replication membranous web
- RTC
replication transcription complex
- SARS
severe acute respiratory syndrome
- TEM
transmission electron microscopy
Introduction
SARS‐CoV‐2 currently dominates all headlines as a highly contagious pandemic with considerable mortality. 1 , 2 , 3 , 4 While hygiene precautions and lockdown measures have changed our personal and professional daily lives during the last months, many investigators are eager to understand the biological basis of the contagiousness of SARS‐CoV‐2 and its pathogenesis leading to respiratory and multi‐organ failure. Autopsy studies have established the morphological changes associated with COVID‐19 and have tried to visualise the virus in tissues. 5 , 6 , 7 , 8 , 9 Transmission electron microscopy (TEM) seems to be a logical tool to look for SARS‐CoV‐2 infection, but some of the published results are highly contested (kidney, 8 , 22 endothelium, 8 , 9 , 23 , 24 , 25 , 26 , 27 , 28 intestine, 8 liver 29 , 30 , 31 , 32 and placenta 33 , 34 , 35 , 36 , 37 ). Because most of us are not virologists or electron microscopists dedicated to the study of viral diseases, this review aims at combining multidisciplinary expertise of SARS‐CoV‐2 pathology, virology and electron microscopy.
Pathologists are good at detecting some viral infections – at least in identifying unusual inclusions on a haematoxylin and eosin (H&E)‐stained slide. However, unless there is a knowledge of virus morphology (what they look like) and morphogenesis (how and where in the cell they are assembled), it is difficult to identify them. Depending on the virus, we use immunohistochemistry targeting viral proteins or in‐situ hybridisation to highlight their DNA or RNA. Molecular pathology techniques allow us to test for viruses in tissues when in situ techniques are not yielding results. All these techniques have been applied successfully in the context of SARS‐CoV‐2 (Figure 1; 5 , 38 , 39 ). It is important to note that any of these tests require an a priori notion of what is present; otherwise, it is difficult to choose the right reagent (e.g. if a herpesvirus is suspected and an anti‐herpesvirus antibody is used, but the infection is caused by an adenovirus, then the test is negative and the diagnosis is no closer to being made).
Figure 1.
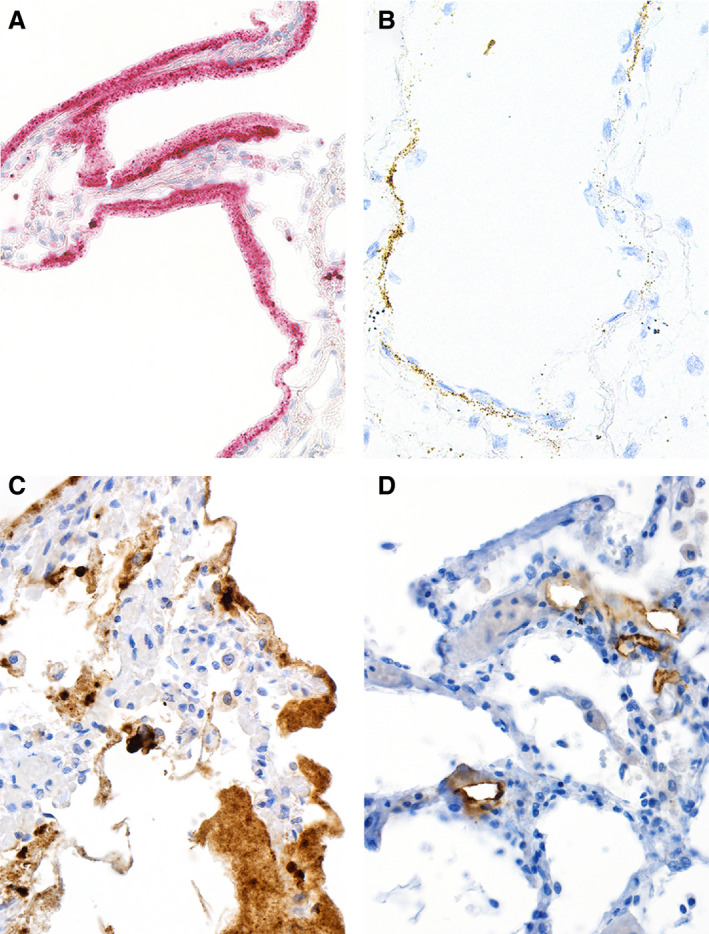
In‐situ detection of severe acute respiratory syndrome (SARS)‐CoV‐2 RNA and proteins. In‐situ imaging of SARS‐CoV‐2 RNA and proteins reflects the disproportionate production of various virus RNA and antigen compounds. A, In‐situ hybridisation (ISH) applying the 845701 RNAscope probe – V‐nCoV2019‐S‐sense duplexed with the 859151 RNAscope probe – V‐nCoV2019‐orf1ab‐sense from Advanced Cell Diagnostics (Hayward, CA, USA) yields abundant SARS‐CoV‐2 RNA within the hyaline membranes of an affected lung. This labelling corresponds to viral RNA, but not to complete virions. B, ISH using the 845701 RNAscope probe – V‐nCoV2019‐S‐sense highlights SARS‐CoV‐2 RNA within alveolar walls, most probably attributable to viral RNA within endothelial cells. C, Immunohistochemistry (IHC) for SARS‐CoV‐2 nucleocapsid antigens with the polyclonal rabbit anti‐nucleocapsid antibody from SinoBiological (Wayne, PA, USA) labels excessive amounts of viral protein present within the hyaline membranes of an affected lung, but does not correspond to intact virions. D, IHC staining for SARS‐CoV‐2 S protein with the clone 007 rabbit anti‐spike antibody from SinoBiological is confined to alveolar vessels. C and D were performed on autopsy cases from our published cohort 5 by Mattia Bugatti in the laboratory of Fabio Facchetti.
Virus detection by TEM is rarely deployed in the routine setting (outside the viral EM diagnostic laboratories that examine tissues and fluids) because it is expensive, time‐consuming, covers only minute portions of the tissue and is not available in most laboratories. Nephropathology is one of a few areas in pathology that routinely performs TEM. Therefore, it is not surprising that renal pathologists were among the first to search for SARS‐CoV‐2 infection in the kidneys by TEM. 10 , 11 , 12 , 40 , 41 Most changes they routinely look for in kidney biopsies are visible at magnifications of between × 1000 and × 10 000 (which can be considered as ‘low power’ in the context of TEM). Thus, the magnifications required to identify viral structures are a long way from their normal ‘comfort zone’. As a consequence, ‘hunting coronavirus by electron microscopy’ takes us to subcellular structures that we usually do not study because they are not important in the context of the standard pathology diagnostic work‐up. Complicating matters further, ultrastructural preservation is limited in autopsy samples by delayed fixation which may obscure subtle changes of the intracellular membrane systems associated with replication of enveloped viruses.
To more clearly understand the ultrastructural morphology of SARS‐CoV‐2 infection and COVID‐19, we will first briefly discuss the pathogenesis of COVID‐19 and coronavirus replication in general and then examine the TEM findings in more detail.
Pathogenesis of COVID‐19 and target organs
Transmission of community‐acquired respiratory viruses including SARS‐CoV‐2 usually results from close‐range contacts through respiratory droplets or aerosols making contact with mucus membranes of the upper respiratory tract. 42 , 43 , 44 , 45 SARS‐CoV‐2 infection of host cells involves specific binding of the viral spike glycoprotein S (S protein) to its receptor angiotensin‐converting enzyme 2 (ACE2). The S protein is then cleaved by host cell proteases to allow for conformational changes mediating fusion between the viral envelope and the host cell membrane. The viral RNA is uncoated and released into the cytoplasm. Host cells are ciliated epithelial cells of the upper and lower airways and Type II pneumocytes. 46 , 47 , 48 , 49 , 50 The level of innate immune response appears to increase as the infection progresses to the lower respiratory tract and the patient increasingly shows symptoms. 51 This gradual progression may explain the range of the clinical manifestations from asymptomatic to severe disease.
In the alveoli, SARS‐CoV‐2 infects Type II pneumocytes as well as Type I pneumocytes via local cell‐to‐cell transmission according to preclinical primate models. 52 SARS‐CoV‐2 infection impairs production of surfactant and fluid resorption leading to increased transmural microcapillary pressure and microvascular leakage, finally resulting in adult respiratory distress syndrome clinically and diffuse alveolar damage histologically. 2 , 58 While direct infection of the cells in the upper and lower respiratory tract drives these initial phases, the innate immune response is mostly responsible for the hyperinflammatory phase (‘cytokine storm’) characteristic of severe COVID‐19. 51 , 59 , 60
Some evidence suggests that SARS‐CoV‐2 infection may not be restricted to the respiratory tract, but can spread to other organs. Viral RNA has been detected by quantitative nucleic acid amplification technique (QNAT) initially in the blood of severely ill patients, in faeces, and rarely in urine samples. 61 , 62 , 63 , 64 , 65 , 66 , 67 It can also be amplified from multiple tissues obtained at autopsy, including heart, liver, kidney, intestine, skin and brain. 5 , 38 , 39 , 68 In‐situ hybridisation and immunohistochemistry studies support the idea of viral spread throughout the body. 8 , 12 , 33 , 38 , 69 , 70 , 71 However, except for lower respiratory fluids such as sputum or bronchoalveolar lavage, 61 , 64 , 72 , 73 , 74 and very rarely faeces, 65 other viral RNA‐containing samples have not been reported to allow for a productive infection in cell cultures. 64 , 75 , 76 These data suggest limited sensitivity of current culture assays and/or low infectiousness. The time‐point of viral dissemination is unclear but – from a virological viewpoint – would require significant local replication and access to blood, blood cells and release at new and distant sites. A clearer knowledge of these events may help to predict the clinical symptoms and their relevance to the disease course.
Currently, our knowledge of the morphological changes associated with COVID‐19 infection is mainly based on autopsy tissue obtained from severely affected individuals in the pulmonary or hyperinflammatory phase, making it difficult to differentiate between changes driven by local viral replication, changes due to the systemic inflammatory response and repair, or possibly therapy effects. Indisputable detection of SARS‐CoV‐2 by TEM would confirm viral replication outside the upper respiratory tract and lungs and firmly establish a role of direct viral infection to some of the organs mentioned above.
Coronavirus replication
Interpretation of TEM findings in tissues of COVID‐19 patients benefits from a good understanding of coronavirus replication in cells. 77 , 78 , 79 Like all members of this family, SARS‐CoV‐2 virions are enveloped infectious particles with a diameter of 60–140 nm. 80 Their genome of approximately 30 000 nucleotides consists of a single positive‐sense RNA strand which is associated with the nucleocapsid protein (N). The envelope consists of host cell‐derived lipids with three structural viral proteins: the membrane glycoprotein M, the envelope protein E and the trimeric spike glycoprotein S.
Similar to other positive‐sense single‐strand RNA viruses, coronavirus genomes are directly translated on ribosomes in the cytoplasm of cells giving rise to large non‐structural polyproteins that are co‐ and post‐translationally cleaved. The resulting replicase complex, including the viral RNA polymerase and smaller proteins, builds the cytoplasmic replication complex in association with membranes of the endoplasmic reticulum (ER) and the Golgi complex. This has been studied in detail in cell cultures infected with different strains of coronaviruses, including SARS‐CoV and Middle East respiratory syndrome (MERS)‐CoV. 81 , 82 , 83 , 84 , 85 , 86 , 87 Recent data show that SARS‐CoV‐2 replication induces very similar ultrastructural changes. 88
SARS‐CoV‐2 infection and replication can be arbitrarily divided into three phases (Figure 2), which may occur simultaneously.
Figure 2.
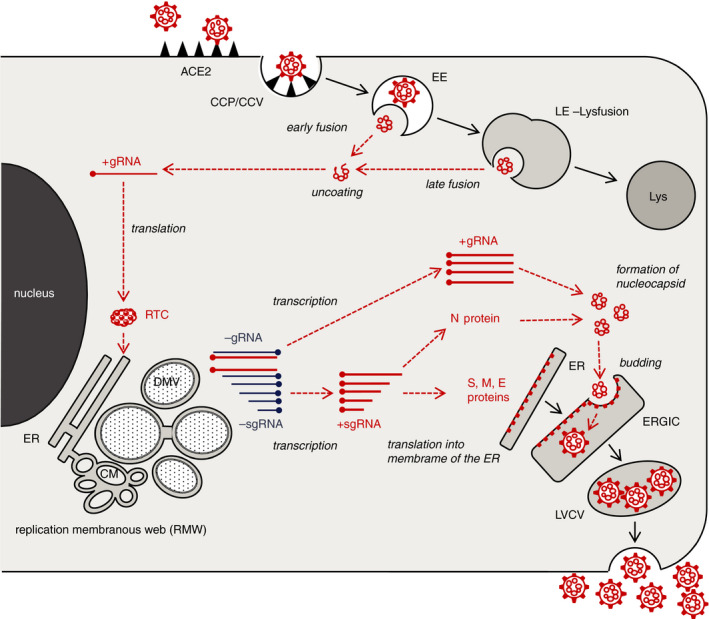
Cartoon of SARS‐CoV‐2 replication in cells. Severe acute respiratory syndrome (SARS)‐CoV‐2 infection and replication can be arbitrarily divided into three phases. (1) After docking to its receptor, the virus is internalised. Proteolytic cleavage of the S protein results in the fusion of the viral envelope with the endosomal membrane, followed by release and uncoating of the viral RNA (upper right). (2) The genomic viral RNA (+gRNA) is translated by ribosomes producing the replication–transcription complex. This initiates an extensive remodelling of intracellular membranes forming the replication membranous web where genomic and subgenomic viral RNA (+sgRNA) are generated (left). (3) The +sgRNA coding for the structural envelope proteins of the virus is directly translated into the membranes of the ER. The nucleocapsid protein assembles with the +gRNA to form the nucleocapsid. Its invagination into the ERGIC forms new virions residing in vesicles, which are subsequently released (lower right). Viral structures are shown in red, cell components in black. For clarity, ribosomes are not depicted. CCP, clathrin‐coated pit; CCV, clathrin‐coated vesicle; EE, early endosome; LE, late endosome; Lys, lysosome; RTC, replication–transcription complex; ER, endoplasmic reticulum; DMV, double membrane vesicle; CM, convoluted membranes; ERGIC, endoplasmic reticulum Golgi intermediate complex.
After binding to its receptor ACE2, SARS‐CoV‐2 is shuttled into the endosomal pathway, probably by clathrin‐coated vesicles. 89 Depending on the presence of furin, a serine protease, cleavage of the S protein triggers early fusion of the viral and the endosomal membranes and causes the release of viral genomes into the cytoplasm. 46 , 47 Cleavage may also occur by other proteases after fusion of the late endosome with a lysosome. 90
Ribosomes recognise the positive‐sense genomic RNA strand (+gRNA) as mRNA and translate the viral proteins making up the replication–transcription complex (RTC) at the ER. This initiates an extensive remodelling of intracellular membranes forming a three‐dimensional structure, referred to as the ‘replication membranous web’ (RMW). 83 , 84 , 91 , 92 Virus replication within the RMW has several advantages: all factors necessary are concentrated in close proximity to each other, making the process very efficient and, additionally, the RMW may hide viral RNA from innate immune sensors within the cytoplasm. Morphologically, the RMW is a fascinating and confusing structure containing multiple interconnected vesicles with single or double membranes (termed ‘double‐membrane vesicles’ and ‘convoluted membranes’). It has to be assumed that the RMW is a dynamic structure with multiple fission and scission events which, unfortunately, cannot be detected by TEM. The double‐membrane vesicles (DMV) are the main site of viral RNA replication via the minus‐sense strand and double‐strand intermediates. 93 , 94 , 95 Newly transcribed +gRNA and positive‐sense subgenomic RNA strands (+sgRNA), necessary for the production of SARS‐CoV‐2 structural proteins, are the results of this replication process.
In the final phase, new virions assemble. While +sgRNA of the M, S and E proteins are directly translated into the membrane of the ER, the N protein assembles with the +gRNA in the cytoplasm to form the nucleocapsid. The latter invaginates the membrane containing the M, S and E proteins in the endoplasmic reticulum‐Golgi intermediate compartment (ERGIC) forming new virions residing in vesicles. 77 After fusion with the cell plasma membrane, the virions are released into the extracellular space.
Electron microscopic findings in infected cell cultures
Coronaviruses including SARS‐CoV‐2 and the morphological changes associated with replication can be visualised by TEM in infected cell lines (Figure 3A–G) 81 , 82 , 83 , 84 , 85 , 87 , 88 or organoids. 96 , 97 Non‐infected cultures serve as controls (Figure 3H–K). For comparison with tissues from COVID‐19 patients, understanding the morphology of the assembled viruses and the replication membranous web is most important.
Figure 3.
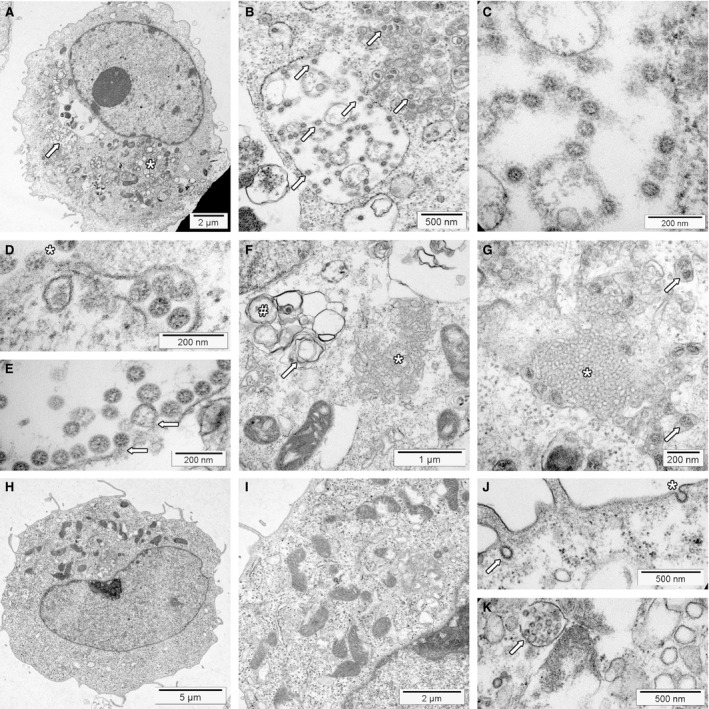
Transmission electron microscopy of severe acute respiratory syndrome (SARS)‐CoV‐2 infected and non‐infected Vero cells. Cell blocks were prepared from SARS‐CoV‐2 infected and non‐infected Vero cells (see Supporting information for details), processed for transmission electron microscopy according to standard procedures and investigated using an FEI Morgagni 268D transmission electron microscope (TEM). Unmarked and uncropped high‐resolution images of all panels are provided in the Supporting information. A, SARS‐CoV‐2‐infected Vero cell at low power. The replication organelle is visible below the nucleus (*). The cell contains a large virion‐containing vacuole (LVCV; arrow) shown at higher magnification in B. B, At higher magnification, a LVCV as well as smaller vesicles contain multiple assembled virions (some marked by arrows). C, High magnification depicts SARS‐CoV‐2 virions inside the LVCV. Within virions, the nucleocapsid is visible as small electron‐dense dots. Depending on the cross‐sectional plane, the centre can be electron lucent. The ‘corona’ formed by the S protein is not visible using standard staining protocols. D, High magnification of a vesicle containing virions docked to the cell surface (*) for virus release; vacuole just under the plasma membrane contains virions to be exocytosed. E, High magnification of LVCV showing virus budding (arrows) and assembled virions. F, Area with convoluted membranes (*) next to mitochondria and double membrane vesicles (#) with signs of deterioration (myelin figure, arrow). Nucleus is shown in the upper left corner under the figure label (F). G, Area showing a cubic membrane structures with membranes arranged in an ordered fashion (*). The surrounding cytoplasm contains several vesicles containing virions (arrows). H, Non‐infected Vero cell. The cytoplasm contains mitochondria, rough endoplasmic reticulum and few vesicles shown at higher magnification in panel I. I, Higher magnification of a non‐infected Vero cell. The intracellular membranes are not prominent. J, Surface of a non‐infected Vero cell showing a coated pit at the cell surface (*) and a coated vesicle (arrow). K, Deteriorated non‐infected cell with a multivesicular body (MVB; arrow) with intraluminal vesicles inside, mimicking an LVCV. Note that the intraluminal vesicles do not show the dots of the nucleocapsid. The structure next to the MVB is a lysosome.
The virus with its circular shape is formed by budding of the nucleocapsid into the membranes containing the structural proteins (Figure 3C). Thus, assembled SARS‐CoV‐2 virions reside within vacuoles and cannot be seen freely in the cytoplasm (Figure 3B,D,E). 82 Cross‐sections of viruses measure 60–140 nm, 80 and show the membrane of the envelope with the helical electron‐dense nucleocapsid inside as several small granules of approximately 12 nm. Frequently, the centre of the viral cross‐section is electron lucent. Unless a negative staining procedure for viewing viruses in fluids or tannic acid staining of tissue for thin sections is used, the S proteins forming the ‘corona’ are not readily discernible. 98
Visualisation of the RMW suggests present or past viral replication (Figure 3A,F). Infected cells contain abundant membrane‐limited vesicles, which may give them a vacuolated appearance by light microscopy. It is often impossible to precisely identify a vesicle in routine cross‐sections, especially in samples with poor preservation of the membranes (e.g. autopsy tissue with delay in fixation).
DMV mark the site of initial viral RNA replication early after infection (Figure 3F), being the most prominent structure of the RMW. 82 , 83 , 84 , 93 , 94 , 95 Probably originating from the ER, they are round or oval, of variable size (usually 200–300 nm), and frequently arranged in clusters. A characteristic double‐membrane associated with the RTC limits the DMV from the cytoplasm. The complex biogenesis of DMV formation is not completely understood, but non‐structural viral proteins are critical for this process. 92 , 93
Convoluted membranes (CM) are less common structures that develop early after infection (Figure 3F). 83 , 84 They associate with the DMV and the ER, forming a disorganised reticular structure measuring 200–600 nm with a single limiting membrane. CM may be involved in the formation of the DMV. 83 Another hypothesis suggests that these structures may serve as a storage for, or are generated by, an excess of non‐structural proteins not incorporated into the DMV. 84
Although rare, cubic membrane structures (CMS) are the most eye‐catching membrane rearrangement in coronavirus‐infected cells because the membranes are highly organised (Figure 3G). 84 In cell cultures, they emerge late after infection. CMS consist of highly curved membranes arranged in a recurrent (three‐dimensional) pattern. Their function is unknown; one hypothesis proposes that they are formed from membranes with an excess of S protein. 82 , 84 , 99 CMS are not specific for coronavirus infection. 100 For example, CMS resulting from chloroquine therapy may be seen in kidney biopsies of patients with lupus nephritis (termed ‘curvilinear bodies’ in this context). 101
Late after infection, cell cultures frequently contain large virion‐containing vacuoles (LVCV, Figure 3B). 81 , 82 , 83 These are large circular vesicles belonging to the secretory pathway containing multiple cross‐sections of assembled virus and evidence of additional virus budding. Marker studies suggest that LVCV are ERGIC/Golgi‐derived cisternae. 84
TEM findings in COVID‐19 tissues
Based on the cell culture findings outlined above, we expect to find the same SARS‐CoV‐2 morphology and distribution in vesicles of autopsy and biopsy tissues of COVID‐19 patients. Published reports have focused on the detection of assembled virus and mostly neglected the membrane rearrange‐ments. If shown and reported, many of the depicted cells contain abundant membrane structures (Figure 4A). 5 , 102 , 103 In our material, some of the vesicles have double membranes and areas with convoluted membranes, and cubic membrane structures are also present (Figure 4B–D). While this suggests at least past virus replication and supports the idea of viral replication outside the respiratory tract, this cannot be substantiated further in the absence of complete virions.
Figure 4.
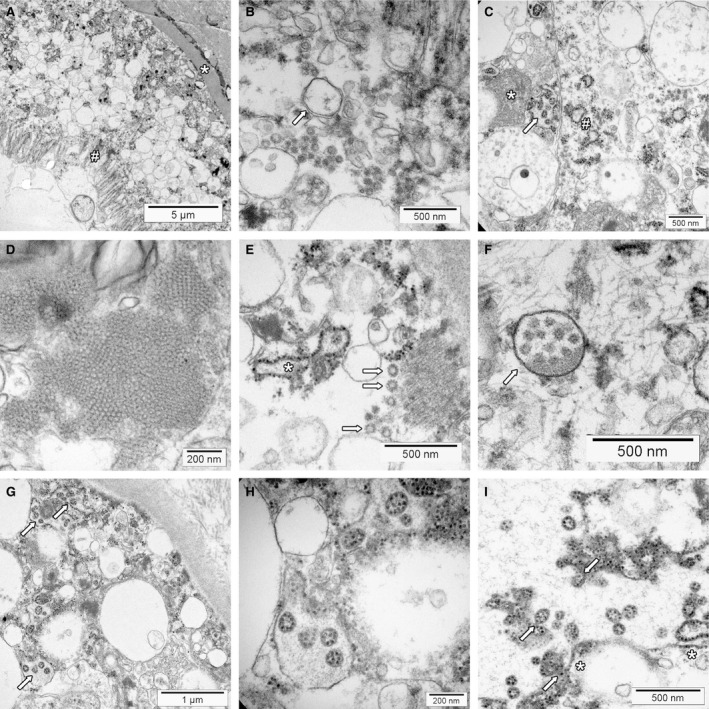
Transmission electron microscopy of COVID‐19 autopsy tissue. Membrane alterations and mimickers of severe acute respiratory syndrome (SARS)‐CoV‐2. Tissues were collected during autopsies and fixed in 4% formalin for at least 72 h. Small tissue blocks were cut and transferred to 3% glutaraldehyde. Processing for transmission electron microscopy was performed according to standard procedures. The tissues were investigated with FEI Morgagni 268D transmission electron microscopy (TEM). Unmarked and uncropped high‐resolution images of all panels are provided in the Supporting information. A, Overview of kidney proximal tubular epithelial cells on top of basement membrane (*). The interstitium is seen in the upper right corner, the brush border at the apical surface (#). The cytoplasm contains multiple membrane‐limited vesicles. B, Higher magnification reveals that some of the vesicles have double membranes (arrows) suggesting the presence of double membrane vesicles. C, This area of a kidney tubular epithelial cell shows convoluted membranes (*) and dilated rough endoplasmic reticulum (#). Some of the larger vacuoles contain vesicles with prominent electron‐dense dots that have the same size as the ribosomes of the endoplasmic reticulum (arrow, ‘outside‐in’ ribosomes). D, Cubic membrane structures within a pneumocyte of the lung. E, Coated vesicles (arrows) are seen in the basal part of this kidney proximal tubular epithelial cell. The tubular basement membrane is sectioned in the upper right corner. The rough endoplasmic reticulum (*) is dilated. F, A multivesicular body within a podocyte of a glomerulus (arrow). Note that the intraluminal vesicles do not show electron‐dense dots. G, Vesicles with ‘outside‐in’ ribosomes (arrows) mimicking SARS‐CoV‐2 virions in a proximal tubule of the kidney. Some of the surrounding vesicles show ribosomes at the outside, which have the same size as the electron‐dense dots on the inside. The tubular basement membrane and collagen fibrils of the interstitium are visible in the upper right corner. H, Higher magnification of vesicles with ‘outside‐in’ ribosomes. The surrounding membrane has no ribosomes attached. I, Part of a larger vesicle with some ribosomes attached on the cytoplasmic side of the membrane (*). The vesicles with ‘outside‐in’ ribosomes seem to be generated by invagination of the membrane (arrows).
There are few reports on upper airway and lung tissue demonstrating assembled SARS‐CoV‐2 virions that morphologically replicate the cell culture findings. 58 , 80 Others, including some of us, have reported on ‘virus‐like particles’ by TEM not completely matching the cell culture findings in a variety of organs, including lung, 5 , 8 , 26 , 56 , 102 , 104 kidney, 5 , 8 , 10 , 11 , 12 , 23 , 104 liver, 29 heart, 104 intestine, 8 , 103 skin 71 and placenta. 33 , 36 , 69 , 70 Colleagues have rightfully raised their concerns that these particles are not consistent with SARS‐CoV‐2, 14 , 15 , 16 , 17 , 18 , 34 but depict other subcellular structures, making identification of SARS‐CoV‐2 by TEM much more challenging than expected.
Morphological mimickers of SARS‐CoV‐2
Physiological structures including coated vesicles, multivesicular bodies and cross‐sections of the rough ER are morphological lookalikes of genuine coronaviruses. 105
Coated vesicles (CV) are single membrane‐bound vesicles of variable size (typically 50–150 nm) characterised by ‘spiny adornments on their limiting membrane’ (Ghadially 106 ) (Figure 4E). They are involved in endocytosis and membrane trafficking (reviewed in Robinson 107 ).
In clathrin‐coated vesicles, the best‐studied example, the CV bud off from so‐called coated pits on the cell surface during micropinocytosis. Clathrin and other quantitatively minor proteins provide a three‐dimensional structural lattice, which is readily seen in electron micrographs. Morphologically identical structures with coats provided by the main proteins, COPI or COPII, are involved in transport processes of the trans‐Golgi network.
Internalisation of SARS‐CoV‐2, after binding to its receptor ACE2, involves this mechanism. 46 , 47 While CV may transport viral proteins, as shown for vesicular stomatitis virus, 108 and may be used for replication of poliovirus 109 and, further, have a similar size to that of coronavirus, they are not the assembled virus itself. However, although the projections appear as a perfect ‘corona’ in cross‐sections, CV lack the nucleocapsid present inside coronavirus cross‐sections, and they are located within the cytoplasm and not within vacuoles.
Multivesicular bodies (MVB) are other structures of the endosomal pathway visible by TEM (Figure 4F) (reviewed in Huotari and Helenius 110 ). They consist of a round or oval vacuole (250–1000 nm) with a limiting membrane, and depending on the density of the matrix, a light or dense background. The larger vacuoles contain multiple smaller membrane‐confined intraluminal vesicles (ILV) with a diameter of 50–100 nm. MVB are the prototypical structures of late endosomes. The ILV form by membrane invagination and fission. The process is used to sort and target membrane‐associated proteins for lysosomal degradation, which happens after fusion of a MVB with a lysosome. A divergent pathway for MVB is the release of the ILV as exosomes after fusion with the cell membrane. Interestingly, the molecular machinery involved in these processes is also used for the budding of enveloped viruses such as HIV (reviewed in Ahmed et al. 111 ).
MVB are the perfect ‘decoy’ for electron microscopists searching for viral particles. Some of us have been misled by them in our COVID‐19 autopsy series 5 because the ILV have a similar size to SARS‐CoV‐2 and are located within vesicles. The key difference is that ILVs do not show the electron‐dense granules of the nucleocapsid.
Pathologists regularly performing TEM are familiar with the ER. Two types of this largest closed and interconnected membrane structure in eukaryotic cells are recognised: the smooth ER, the site of lipid synthesis and metabolism, and the rough ER, where ribosomes attached to the outside of the membrane translate proteins for membranes, organelles and secretion. 112 Dilatation of the ER occurs in the context of cell stress of various aetiologies. In COVID‐19 tissues the ER is frequently swollen, and the accumulation of membrane structures further adds to the confusing ultrastructure (Figure 4C). A cross‐section through rough ER can easily be mistaken for a ‘virus‐like particle’, but these are located within the cytoplasm and not in vesicles and lack the nucleocapsid structures inside.
In kidneys from COVID‐19 autopsies, we encountered a peculiar subcellular structure closely mimicking SARS‐CoV‐2 but probably related to the ER (Figure 4C,G–I). 5 , 13 Larger vesicles with a smooth outside membrane contained several round to oval small vesicles with prominent electron‐dense granules on the inside. These granules were bigger than SARS‐CoV‐2 nucleocapsid seen in our infected cell cultures and had the same size as the ribosomes visible in areas containing rough ER (ribosomes: 20–21 nm (range = 17–23 nm) versus nucleocapsid: 12 nm (range = 9–16 nm). These vesicles with ‘outside‐in’ ribosomes are possibly derived from the rough ER by membrane invaginations, as suggested in some of the TEM pictures (Figure 4I). Because the particles inside are larger than nucleocapsid cross‐sections, we believe that they probably do not represent assembled virions. However, it is theoretically possible that the particles may consist of deteriorated and swollen nucleocapsid due to autolysis, approaching the appearance of ribosomes.
Summary and conclusions
Because of SARS, MERS and the importance of coronavirus infections for agriculture, the ultrastructural changes induced by infection and replication of CoV have been investigated and described in great detail prior to the current pandemic. In infected cell cultures, SARS‐CoV‐2 behaves in a similar manner, and we expect to see the same morphology in tissues of COVID‐19 patients.
The published in‐vivo data are less convincing. Coated vesicles, multivesicular bodies and swollen rough endoplasmic reticulum are important mimics of assembled virions, all of which lack the electron‐dense dots of the nucleocapsid inside the particles. High magnification (~×90 000) is required to clearly identify them. Autolysis of autopsy tissue further adds to the difficulty of identifying viral particles by TEM.
Despite evidence suggesting viral replication outside the respiratory tract, unassailable TEM evidence is still missing in tissues from COVID‐19 patients. Possible explanations include timing (too late: the virus has already been cleared by the immune system) and sensitivity (too insensitive: there are too few infected cells or too few virions, such that detection by TEM becomes very unlikely; the low viral loads found by quantitative nucleic acid amplification techniques (QNAT) in tissues other than lung would support this argument). We think that the observed extensive intracellular membrane remodelling could be a result of direct infection, but this is difficult to prove in the absence of newly formed viral particles. Ideally, TEM morphology will be backed by other in‐situ techniques in the same case.
Another important concept to keep in mind is that viral components (RNA and proteins) are not produced in balanced amounts (as suggested in Figure 1 and Massoth et al. 113 ). Therefore, surplus viral RNA and proteins may be encountered at the site of infection, in the circulation and at distant sites. Detection of viral RNA and proteins does not necessarily reflect the presence of intact and infectious particles. It is also conceivable that pathology in non‐respiratory organs could be the result of distant viral disease, due to transport of viral components, and not a direct result of infection. Therefore, TEM investigation is essential to verify assembled virions in SARS‐CoV‐2 infection and COVID‐19.
In our view, the hunt for coronavirus by TEM in tissues from COVID‐19 patients beyond the upper airways and lungs is still open.
Conflicts of interest
The authors declare no conflicts of interest.
Author contribution
HH had the idea, designed the concept and Figures 2, 3, 4 of the review, and together with TM and AT drafted the manuscript. AT created Figure 1. MCH prepared the samples and together with HH performed the TEM investigation. JH initiated the SARS‐CoV‐2 infected and non‐infected Vero cell culture provided by RG and HHH. RG and HHH provided virology and SEM electron microscopy expertise to the text and figures. HH, AT, RG, HHH, and SEM participated in a detailed analysis and discussion of all figures during a video conference. All authors revised the text and figures. HH was responsible for the final editing.
Supporting information
Data S1. Supplementary material.
Acknowledgements
This study was supported by the Botnar Research Centre for Child Health.
Hopfer H, Herzig MC, Gosert R, Menter T, Hench J, Tzankov A, Hirsch HH & Miller SE. (2021) Histopathology 78, 358–370. 10.1111/his.14264 Hunting coronavirus by transmission electron microscopy – a guide to SARS‐CoV‐2‐associated ultrastructural pathology in COVID‐19 tissues
References
- 1. Li Q, Guan X, Wu P et al. Early transmission dynamics in Wuhan, China, of novel coronavirus‐infected pneumonia. N. Engl. J. Med. 2020; 382; 1199–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Huang C, Wang Y, Li X et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan. China. Lancet 2020; 395; 497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Battegay M, Kuehl R, Tschudin‐Sutter S et al. 2019‐novel Coronavirus (2019‐nCoV): estimating the case fatality rate – a word of caution. Swiss Med. Wkly 2020; 150; w20203. [DOI] [PubMed] [Google Scholar]
- 4. Leuzinger K, Roloff T, Gosert R et al. Epidemiology of SARS‐CoV‐2 emergence amidst community‐acquired respiratory viruses. J. Infect. Dis. 2020; 222; 1270–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Menter T, Haslbauer JD, Nienhold R et al. Postmortem examination of COVID‐19 patients reveals diffuse alveolar damage with severe capillary congestion and variegated findings in lungs and other organs suggesting vascular dysfunction. Histopathology 2020; 77; 198–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wichmann D, Sperhake J‐P, Lütgehetmann M et al. Autopsy findings and venous thromboembolism in patients with COVID‐19. Ann. Intern. Med. 2020; 173; 268–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lax SF, Skok K, Zechner P et al. Pulmonary arterial thrombosis in COVID‐19 with fatal outcome: results from a prospective, single‐center, clinicopathologic case series. Ann. Intern. Med. 2020; 173; 350–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bradley BT, Maioli H, Johnston R et al. Histopathology and ultrastructural findings of fatal COVID‐19 infections in Washington State: a case series. Lancet 2020; 396; 320–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bösmüller H, Traxler S, Bitzer M et al. The evolution of pulmonary pathology in fatal COVID‐19 disease: an autopsy study with clinical correlation. Virchows Arch. 2020; 477; 349–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kissling S, Rotman S, Gerber C et al. Collapsing glomerulopathy in a COVID‐19 patient. Kidney Int. 2020; 98; 228–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Farkash EA, Wilson AM, Jentzen JM. Ultrastructural evidence for direct renal infection with SARS‐CoV‐2. J. Am. Soc. Nephrol. 2020; 31; 1683–1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Su H, Yang M, Wan C et al. Renal histopathological analysis of 26 postmortem findings of patients with COVID‐19 in China. Kidney Int. 2020; 98; 219–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Werion A, Belkhir L, Perrot M et al. SARS‐CoV‐2 causes a specific dysfunction of the kidney proximal tubule. Kidney Int. 2020;. 10.1016/j.kint.2020.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Miller SE, Brealey JK. Visualization of putative coronavirus in kidney. Kidney Int. 2020; 98; 231–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Calomeni E, Satoskar A, Ayoub I et al. Multivesicular bodies mimicking SARS‐CoV‐2 in patients without COVID‐19. Kidney Int. 2020; 98; 233–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Roufosse C, Curtis E, Moran L et al. Electron microscopic investigations in COVID‐19: not all crowns are coronas. Kidney Int. 2020; 98; 505–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Smith KD, Akilesh S, Alpers CE et al. Am I a coronavirus? Kidney Int. 2020; 98; 506–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Goldsmith CS, Miller SE. Caution in identifying coronaviruses by electron microscopy. J. Am. Soc. Nephrol. 2020; 31; 2223–2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kissling S, Rotman S, Fakhouri F. The authors reply. Kidney Int. 2020; 98; 232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Su H, Gao D, Yang H‐C et al. The authors reply. Kidney Int. 2020; 98; 232–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Farkash EA, Wilson AM, Jentzen JM. Authors’ reply. J. Am. Soc. Nephrol. 2020; 31; 2225–2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Santoriello D, Khairallah P, Bomback AS et al. Postmortem kidney pathology findings in patients with COVID‐19. J. Am. Soc. Nephrol. 2020; 31; 2158–2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Varga Z, Flammer AJ, Steiger P et al. Endothelial cell infection and endotheliitis in COVID‐19. Lancet 2020; 395; 1417–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Goldsmith CS, Miller SE, Martines RB et al. Electron microscopy of SARS‐CoV‐2: a challenging task. Lancet 2020; 395; e99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Varga Z, Flammer AJ, Steiger P et al. Electron microscopy of SARS‐CoV‐2: a challenging task – authors’ reply. Lancet 2020; 395; e100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ackermann M, Verleden SE, Kuehnel M et al. Pulmonary vascular endothelialitis, thrombosis, and angiogenesis in Covid‐19. N. Engl. J. Med. 2020; 383; 120–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Scholkmann F, Nicholls J. Pulmonary vascular pathology in Covid‐19. N. Engl. J. Med. 2020; 383; 887–888. [DOI] [PubMed] [Google Scholar]
- 28. Ackermann M, Mentzer SJ, Jonigk D. Pulmonary vascular pathology in Covid‐19. Reply. N. Engl. J. Med. 2020; 383; 888–889. [DOI] [PubMed] [Google Scholar]
- 29. Wang Y, Liu S, Liu H et al. SARS‐CoV‐2 infection of the liver directly contributes to hepatic impairment in patients with COVID‐19. J. Hepatol. 2020; 73; 807–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bangash MN, Patel JM, Parekh D et al. SARS‐CoV‐2: is the liver merely a bystander to severe disease? J. Hepatol. 2020; 73; 995–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Philips CA, Ahamed R, Augustine P. SARS‐CoV‐2 related liver impairment – perception may not be the reality. J. Hepatol. 2020; 73; 991–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wang Y, Lu F, Zhao J. Reply to: Correspondence relating to ‘SARS‐CoV‐2 infection of the liver directly contributes to hepatic impairment in patients with COVID‐19’. J. Hepatol. 2020; 73; 996–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Algarroba GN, Rekawek P, Vahanian SA et al. Visualization of SARS‐CoV‐2 virus invading the human placenta using electron microscopy. Am. J. Obstet. Gynecol. 2020; 223; 275–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kniss DA. Alternative interpretation to the findings reported in visualization of severe acute respiratory syndrome coronavirus 2 invading the human placenta using electron microscopy. Am. J. Obstet. Gynecol. 2020; 223; 785–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Algarroba GN, Rekawek P, Vahanian SA et al. Reply. Am. J. Obstet. Gynecol. 2020; 223; 786–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Facchetti F, Bugatti M, Drera E et al. SARS‐CoV2 vertical transmission with adverse effects on the newborn revealed through integrated immunohistochemical, electron microscopy and molecular analyses of placenta. EBioMed. 2020; 59; 102951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Algarroba GN, Hanna NN, Rekawek P et al. Confirmatory evidence of visualization of SARS‐CoV‐2 virus invading the human placenta using electron microscopy. Am. J. Obstet. Gynecol. 2020; 10.1016/j.ajog.2020.08.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Puelles VG, Lütgehetmann M, Lindenmeyer MT et al. Multiorgan and renal tropism of SARS‐CoV‐2. N. Engl. J. Med. 2020; 383; 590–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Skok K, Stelzl E, Trauner M et al. Post‐mortem viral dynamics and tropism in COVID‐19 patients in correlation with organ damage. Virchows Arch. 2020. 10.1007/s00428-020-02903-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Larsen CP, Bourne TD, Wilson JD et al. Collapsing glomerulopathy in a patient with coronavirus disease 2019 (COVID‐19). Kidney Int. Rep. 2020; 5; 935–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Peleg Y, Kudose S, D’Agati V et al. Acute kidney injury due to collapsing glomerulopathy following COVID‐19 Infection. Kidney Int. Rep. 2020; 5; 940–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Centers for Disease Control and Prevention . Coronavirus disease 2019 (COVID‐19) – transmission: how COVID‐19 spreads. Available at: https://www.cdc.gov/coronavirus/2019‐ncov/faq.html#Spread (accessed 9 September 2020).
- 43. van Doremalen N, Bushmaker T, Morris DH et al. Aerosol and surface stability of SARS‐CoV‐2 as compared with SARS‐CoV‐1. N. Engl. J. Med. 2020; 382; 1564–1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Prather KA, Wang CC, Schooley RT. Reducing transmission of SARS‐CoV‐2. Science 2020; 368; 1422–1424. [DOI] [PubMed] [Google Scholar]
- 45. Ison MG, Hirsch HH. Community‐acquired respiratory viruses in transplant patients: diversity, impact, unmet clinical needs. Clin. Microbiol. Rev. 2019; 32; e00042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hoffmann M, Kleine‐Weber H, Schroeder S et al. SARS‐CoV‐2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 2020; 181(271–280); e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ou X, Liu Y, Lei X et al. Characterization of spike glycoprotein of SARS‐CoV‐2 on virus entry and its immune cross‐reactivity with SARS‐CoV. Nat. Commun. 2020; 11; 1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sungnak W, Huang N, Bécavin C et al. SARS‐CoV‐2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat. Med. 2020; 26; 681–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hou YJ, Okuda K, Edwards CE et al. SARS‐CoV‐2 reverse genetics reveals a variable infection gradient in the respiratory tract. Cell 2020; 182(429–446); e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hui KPY, Cheung M‐C, Perera RAPM et al. Tropism, replication competence, and innate immune responses of the coronavirus SARS‐CoV‐2 in human respiratory tract and conjunctiva: an analysis in ex‐vivo and in‐vitro cultures. Lancet Respir. Med. 2020; 8; 687–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Mason RJ. Thoughts on the alveolar phase of COVID‐19. Am. J. Physiol. Lung Cell. Mol. Physiol. 2020; 319; L115–L120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Rockx B, Kuiken T, Herfst S et al. Comparative pathogenesis of COVID‐19, MERS, and SARS in a nonhuman primate model. Science 2020; 368; 1012–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wang D, Hu B, Hu C et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus–infected pneumonia in Wuhan. China. JAMA 2020; 323; 1061–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Matthay MA, Zemans RL, Zimmerman GA et al. Acute respiratory distress syndrome. Nat. Rev. Dis. Primers 2019; 5; 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Tian S, Hu W, Niu L et al. Pulmonary pathology of early‐phase 2019 novel coronavirus (COVID‐19) pneumonia in two patients with lung cancer. J. Thorac. Oncol. 2020; 15; 700–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Fox SE, Akmatbekov A, Harbert JL et al. Pulmonary and cardiac pathology in African American patients with COVID‐19: an autopsy series from New Orleans. Lancet Respir. Med. 2020; 8; 681–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Konopka KE, Nguyen T, Jentzen JM et al. Diffuse alveolar damage (DAD) from coronavirus disease 2019 infection is morphologically indistinguishable from other causes of DAD. Histopathology 2020; 77; 570–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Martines RB, Ritter JM, Matkovic E et al. Pathology and pathogenesis of SARS‐CoV‐2 associated with fatal coronavirus disease. United States. Emerg. Infect. Dis. 2020; 26; 2005–2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Mangalmurti N, Hunter CA. Cytokine storms: understanding COVID‐19. Immunity 2020; 53; 19–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Polidoro RB, Hagan RS, de Santis SR et al. Overview: systemic inflammatory response derived from lung injury caused by SARS‐CoV‐2 infection explains severe outcomes in COVID‐19. Front. Immunol. 2020; 11; 1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Wang W, Xu Y, Gao R et al. Detection of SARS‐CoV‐2 in different types of clinical specimens. JAMA 2020; 323; 1843–1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wu Y, Guo C, Tang L et al. Prolonged presence of SARS‐CoV‐2 viral RNA in faecal samples. Lancet Gastroenterol. Hepatol. 2020; 5; 434–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Lescure F‐X, Bouadma L, Nguyen D et al. Clinical and virological data of the first cases of COVID‐19 in Europe: a case series. Lancet Infect. Dis. 2020; 20; 697–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Wölfel R, Corman VM, Guggemos W et al. Virological assessment of hospitalized patients with COVID‐2019. Nature 2020; 581; 465–469. [DOI] [PubMed] [Google Scholar]
- 65. Xiao F, Sun J, Xu Y et al. Infectious SARS‐CoV‐2 in feces of patient with severe COVID‐19. Emerg. Infect. Dis. 2020; 26; 1920–1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Young BE, Ong SWX, Kalimuddin S et al. Epidemiologic features and clinical course of patients infected with SARS‐CoV‐2 in Singapore. JAMA 2020; 323; 1488–1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Zheng S, Fan J, Yu F et al. Viral load dynamics and disease severity in patients infected with SARS‐CoV‐2 in Zhejiang province, China, January–March 2020: retrospective cohort study. BMJ 2020; 369; m1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Jamiolkowski D, Mühleisen B, Müller S et al. SARS‐CoV‐2 PCR testing of skin for COVID‐19 diagnostics: a case report. Lancet 2020; 396; 598–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Sisman J, Jaleel MA, Moreno W et al. Intrauterine transmission of SARS‐CoV‐2 infection in a preterm infant. Pediatr. Infect. Dis. J. 2020; 39; e265–e267. [DOI] [PubMed] [Google Scholar]
- 70. Hosier H, Farhadian SF, Morotti RA et al. SARS‐CoV‐2 infection of the placenta. J. Clin. Invest. 2020; 130; 4947–4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Colmenero I, Santonja C, Alonso‐Riaño M et al. SARS‐CoV‐2 endothelial infection causes COVID‐19 chilblains: histopathological, immunohistochemical and ultrastructural study of 7 paediatric cases. Br. J. Dermatol. 2020; 10.1111/bjd.19327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Perera RAPM, Tso E, Tsang OTY et al. SARS‐CoV‐2 Virus culture and subgenomic RNA for respiratory specimens from patients with mild coronavirus disease. Emerg. Infect. Dis. 2020;26; 2701–2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. COVID‐19 Investigation Team . Clinical and virologic characteristics of the first 12 patients with coronavirus disease (COVID‐19) in the United States. Nat. Med. 2019; 2020(26); 861–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Huang C‐G, Lee K‐M, Hsiao M‐J et al. Culture‐based virus isolation to evaluate potential infectivity of clinical specimens tested for COVID‐19. J. Clin. Microbiol. 2020; 58; e01068‐20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Kim J‐M, Kim HM, Lee EJ et al. Detection and isolation of SARS‐CoV‐2 in serum, urine, and stool specimens of COVID‐19 patients from the Republic of Korea. Osong Public Health Res. Perspect. 2020; 11; 112–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Li X, Chan JF‐W, Li KK‐W et al. Detection of SARS‐CoV‐2 in conjunctival secretions from patients without ocular symptoms. Infection 2020;. 10.1007/s15010-020-01524-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Baker SC. Coronaviruses: molecular biology. In Mahy BWJ, Van Regenmortel MHV eds. Encyclopedia of virology. 3rd ed. Oxford: Academic Press, 2008; 554–562. [Google Scholar]
- 78. Perlman S, Netland J. Coronaviruses post‐SARS: update on replication and pathogenesis. Nat. Rev. Microbiol. 2009; 7; 439–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Fehr AR, Perlman S. Coronaviruses: an overview of their replication and pathogenesis. In Maier HJ, Bickerton E, Britton P eds. Coronaviruses: methods and protocols. methods in molecular biology. New York, NY: Springer, 2015; 1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Zhu N, Zhang D, Wang W et al. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020; 382; 727–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Ng M‐L, Tan S‐H, See E‐E et al. Proliferative growth of SARS coronavirus in Vero E6 cells. J. Gen. Virol. 2003; 84; 3291–3303. [DOI] [PubMed] [Google Scholar]
- 82. Goldsmith CS, Tatti KM, Ksiazek TG et al. Ultrastructural characterization of SARS coronavirus. Emerg. Infect. Dis. 2004; 10; 320–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Knoops K, Kikkert M, van den Worm SHE et al. SARS‐coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLOS Biol. 2008; 6; e226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Ulasli M, Verheije MH, de Haan CAM et al. Qualitative and quantitative ultrastructural analysis of the membrane rearrangements induced by coronavirus. Cell. Microbiol. 2010; 12; 844–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Lednicky JA, Waltzek TB, McGeehan E et al. Isolation and genetic characterization of human coronavirus NL63 in primary human renal proximal tubular epithelial cells obtained from a commercial supplier, and confirmation of its replication in two different types of human primary kidney cells. Virol. J. 2013; 10; 213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. de Wilde AH, Raj VS, Oudshoorn D et al. MERS‐coronavirus replication induces severe in vitro cytopathology and is strongly inhibited by cyclosporin A or interferon‐α treatment. J. Gen. Virol. 2013; 94; 1749–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Doyle N, Hawes PC, Simpson J et al. The porcine deltacoronavirus replication organelle comprises double‐membrane vesicles and zippered endoplasmic reticulum with double‐membrane spherules. Viruses 2019; 11; 1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Ogando NS, Dalebout TJ, Zevenhoven‐Dobbe JC et al. SARS‐coronavirus‐2 replication in Vero E6 cells: replication kinetics, rapid adaptation and cytopathology. J. Gen. Virol. 2020; 101; 925–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Inoue Y, Tanaka N, Tanaka Y et al. Clathrin‐dependent entry of severe acute respiratory syndrome coronavirus into target cells expressing ACE2 with the cytoplasmic tail deleted. J. Virol. 2007; 81; 8722–8729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Jaimes JA, Millet JK, Whittaker GR. Proteolytic cleavage of the SARS‐CoV‐2 spike protein and the role of the novel S1/S2 site. iScience 2020; 2; 101212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. van den Worm SHE, Knoops K, Zevenhoven‐Dobbe JC et al. Development and RNA‐synthesizing activity of coronavirus replication structures in the absence of protein synthesis. J. Virol. 2011; 8; 5669–5673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Harak C, Lohmann V. Ultrastructure of the replication sites of positive‐strand RNA viruses. Virology 2015; 479–480; 418–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Wolff G, Melia CE, Snijder EJ et al. Double‐membrane vesicles as platforms for viral replication. Trends Microbiol. 2020; 10.1016/j.tim.2020.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Gosert R, Kanjanahaluethai A, Egger D et al. RNA replication of mouse hepatitis virus takes place at double‐membrane vesicles. J. Virol. 2002; 76; 3697–3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Snijder EJ, Limpens RWAL, de Wilde AH et al. A unifying structural and functional model of the coronavirus replication organelle: tracking down RNA synthesis. PLOS Biol. 2020; 18; e3000715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Monteil V, Kwon H, Prado P et al. Inhibition of SARS‐CoV‐2 infections in engineered human tissues using clinical‐grade soluble human ACE2. Cell 2020; 181(905–913); e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Mallapaty S. Mini organs reveal how the coronavirus ravages the body. Nature 2020; 58; 15–16. [DOI] [PubMed] [Google Scholar]
- 98. Park WB, Kwon NJ, Choi SJ et al. Virus isolation from the first patient with SARS‐CoV‐2 in Korea. J. Korean Med. Sci. 2020; 35; e84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Almsherqi ZA, McLachlan CS, Mossop P et al. Direct template matching reveals a host subcellular membrane gyroid cubic structure that is associated with SARS virus. Redox Rep. 2005; 10; 167–171. [DOI] [PubMed] [Google Scholar]
- 100. Almsherqi ZA, Kohlwein SD, Deng Y. Cubic membranes: a legend beyond the Flatland of cell membrane organization. J. Cell Biol. 2006; 173; 839–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Müller‐Höcker J, Schmid H, Weiss M et al. Chloroquine‐induced phospholipidosis of the kidney mimicking Fabry’s disease: case report and review of the literature. Hum. Pathol 2003; 34; 285–289. [DOI] [PubMed] [Google Scholar]
- 102. Yao X‐H, He Z‐C, Li T‐Y et al. Pathological evidence for residual SARS‐CoV‐2 in pulmonary tissues of a ready‐for‐discharge patient. Cell Res. 2020; 30; 541–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Qian Q, Fan L, Liu W et al. Direct evidence of active SARS‐CoV‐2 replication in the intestine. Clin. Infect. Dis. 2020; 10.1093/cid/ciaa925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Pesaresi M, Pirani F, Tagliabracci A et al. SARS‐CoV‐2 identification in lungs, heart and kidney specimens by transmission and scanning electron microscopy. Eur. Rev. Med. Pharmacol. Sci. 2020; 24; 5186–5188. [DOI] [PubMed] [Google Scholar]
- 105. Neil D, Moran L, Horsfield C et al. Ultrastructure of cell trafficking pathways and coronavirus: how to recognise the wolf amongst the sheep. J. Pathol. 2020; 10.1002/path.5547. [DOI] [PubMed] [Google Scholar]
- 106. Ghadially F ed. Endocytotic vesicles and vacuoles. Ultrastructural pathology of the cell and matrix, vol. 2. 3rd ed. London: Butterworth: 1988; 1134–1139. [Google Scholar]
- 107. Robinson MS. Forty years of clathrin‐coated vesicles. Traffic 2015; 16; 1210–1238. [DOI] [PubMed] [Google Scholar]
- 108. Rothman JE, Bursztyn‐Pettegrew H, Fine RE. Transport of the membrane glycoprotein of vesicular stomatitis virus to the cell surface in two stages by clathrin‐coated vesicles. J. Cell Biol. 1980; 86; 162–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Rust RC, Landmann L, Gosert R et al. Cellular COPII proteins are involved in production of the vesicles that form the poliovirus replication complex. J. Virol. 2001; 75; 9808–9818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Huotari J, Helenius A. Endosome maturation. EMBO J. 2011; 30; 3481–3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Ahmed I, Akram Z, Iqbal HMN et al. The regulation of Endosomal sorting complex required for transport and accessory proteins in multivesicular body sorting and enveloped viral budding – an overview. Int. J. Biol. Macromol. 2019; 127; 1–11. [DOI] [PubMed] [Google Scholar]
- 112. Lodish H, Baltimore D, Berk A et al. Cell organization, subcellular structure, and cell division. In Lodish M, Berk A, Zipursky SL, Matsudaira P, Baltimore D, Darnell J eds. Molecular Cell Biology. 3rd ed. New York, Oxford: Scientific American Books, 1995; 141–188. [Google Scholar]
- 113. Massoth LR, Desai N, Szabolcs A et al. Comparison of RNA in situ hybridization and immunohistochemistry techniques for the detection and localization of SARS‐CoV‐2 in human tissues. Am. J. Surg. Pathol. 2020; 10.1097/PAS.0000000000001563. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1. Supplementary material.


