Abstract
We developed a reverse line blot (RLB) hybridization-, and rolling circle amplification (RCA)-based assays for the identification of Trichoporon species and evaluated them with 48 isolates that had been previously recognized as belonging to eight species (Trichosporon asahii, T. cutaneum, T. dermatis, T. domesticum, T. inkin, T. japonicum, T. jirovecii, and T. laibachii). Results were compared to those obtained with DNA sequencing of three rRNA gene loci, i.e., the internal transcribed spacer (ITS) region, D1/D2 domain of the 28S rRNA gene and intergenic spacer 1 (IGS1) region. Using species-specific, or group-specific probes targeted at the ITS region and the D1/D2 domain, the RLB assay permitted accurate species identification of all 48 isolates with 100% specificity. Species-specific RLB probes correctly assigned 45/48 (94%) of the isolates (six species) with the exception of T. dermatis and T. japonicum isolates which were not targeted by the assay. Identification of T. dermatis relied on a positive hybridization result with the group-specific probe hybridizing with T. dermatis and T. jirovecii and the absence of a signal with the T. jirovecii-specific probe. T. japonicum strains were first assigned to the T. asahii-T. japonicum group by hybridization with the two species group-specific probe and then as T. japonicum by the absence of signal with a T. asahii-specific probe. Twelve species-specific RCA probes targeting the eight species studied detected templates of all 48 Trichosporon isolates and an artificial template of T. asteroides, all with good specificity. Both RLB and RCA are potential alternatives to DNA sequencing for the identification of Trichosporon species. The RLB approach is suited for the batched simultaneous analysis of large numbers of isolates, while RCA is more appropriate for the immediate study of single isolates. Comparative costs are US$7 and US$2 per assay for the RLB and RCA methods, respectively.
Keywords: Trichosporon, reverse line blot hybridization assay, rolling circle amplification
Introduction
Trichosporon species are important emerging fungal pathogens associated with high mortality (42–80%). Typically, infection occurs in severely immunocompromised patients, including those receiving chemotherapy for underlying hematological malignancies and organ transplantations [1–4].
Species identification of Trichosporon clinical isolates is important to help to define species-specific clinical associations and geographic variation in their epidemiology. In addition, certain Trichosporon species may be resistant or less susceptible to antifungal drugs such as echinocandin and/or amphotericin B [1,4–7]. Since the introduction of molecular identification techniques for fungi, the nomenclature within this genus has been significantly revised. While Trichosporon beigelii was considered to be the most important etiologic agent [8], at least 16 species have now been reported to be associated with human disease [1,5,6,9–11]. DNA sequencing-based methods have primarily been required to delineate the taxonomy and epidemiology of Trichosporon species [1,6,9,12,13]. We previously characterized 48 Trichosporon strains from China belonging to eight species (Trichosporon asahii, T. cutaneum, T. dermatis, T. domesticum, T. inkin, T. japonicum, T. jirovecii and T. laibachii) by sequencing three fungal rRNA gene loci, i.e., the internal transcribed spacer (ITS), D1/D2 domain of the 28S rRNA gene, and intergenic spacer 1 (IGS1), regions. The study of nucleotide polymorphisms within the ITS and D1/D2 regions have especially proved to be sensitive techniques in species identification [12].
However, DNA sequencing may be slow (turnaround time of 2–3 days), expensive, and not easily adaptable to use in the routine clinical laboratory. Two PCR-based approaches, the PCR-reverse line blot (RLB) and rolling cycle amplification (RCA) methods, have been shown to be accurate and practical alternatives to DNA sequencing in the identification/detection of medically important fungal species [14–20]. RCA methodology is particularly well suited for the sensitive detection of single nucleotide polymorphisms (SNPs) and can discriminate between genetically closely-related species, and within species [14,20]. In the present study, by using species-, and group-specific probes targeted against members of the genus Trichosporon, we designed and evaluated a (i) RLB-, and (ii) RCA-based assay for species identification of the same 48 strains previously-characterized by combined ITS/D1-D2 sequencing [12].
Materials and methods
Trichosporon strains and DNA extraction
The identification of the 48 Trichosporon isolates (Supplementary Table 1, available online at http://informahealthcare.com/doi/abs/10.3109/13693786.2012.723223) employed in this study had been previously determined by combined ITS and D1-D2 sequencing [12]. DNA extraction was performed as described before [12] and stored at −20°C prior to use.
Primers for PCR, RLB and RCA
For all isolates, primer pairs ITS1/ITS4 and F63/R635 [12] were used to amplify the ITS region and D1/D2 domain, respectively. For PCR in preparation for the RLB assay, 5′-end biotin-labelled primers, ITS1b/R635b, (Table 1) were employed. For the RCA assay, all four primers (ITS1/ITS4 and F63/R635) were non-labeled. Two additional primers (RCA primer 1 and RCA primer 2; Table 1) were specifically designed to bind the linker region of the padlock probes [14,21]. All primers were synthesized at Beijing AuGCT Biotechnology Co. Ltd. (Beijing, P. R. China).
Table 1.
Primers, probes and artificial template used in the present study.
| Oligonucleotide | Specificity | Target gene | GenBank accession no. | Sequence (5′–3′)* |
|---|---|---|---|---|
| Primers | ||||
| ITS1/ITS1b† | Panfungal | 18S rRNA gene | HM802135 | 4 TCCGTAGGTGAACCTGCG 21 |
| ITS4 | Panfungal | 28S rRNA gene | HM802135 | 534 TCCTCCGCTTATTGATATGC 515 |
| F63 | Panfungal | 28S rRNA gene | EU882103 | 1 GCATATCAATAAGCGGAGGAAAAG 24 |
| R635/R635b† | Panfungal | 28S rRNA gene | EU882103 | 640 GGTCCGTGTTTCAAGACG 623 |
| RCA primer 1‡ | – | Padlock probe linker region | – | ATGGGCACCGAAGAAGCA |
| RCA primer 2‡ | – | Padlock probe linker region | – | CGCGCAGACACGATA |
| RLB probes† | ||||
| 12AP | Panfungal | ITS region | AF455524 | 211 CCAAGAGATCCGTTGTTGAAAG 190 |
| 23SP | Panfungal | ITS region | AF455524 | 271 GTGAATCATCGARTCTTTGAACG 293 |
| Tri-ITS-1 | T. asahii, T. domesticum and T. japonicum | ITS region | AF444473 | 31 GTGATTGCCTTAATTGGCTTATAAC 55 |
| Tri-ITS-2 | T. asahii, T. inkin and T. japonicum | ITS region | AF444473 | 93 ACGCAAGTCGAGTATTTTTACAAAC 117 |
| Tri-ITS-3 | T. asahii and T. japonicum | ITS region | AJ864867 | 10 GTGATTGCCTTTATAGGCTTATAAC 34 |
| Tri-ITS-4 | T. cutaneum | ITS region | AF444325 | 89 TTCGGTCAATTGATTTTACAAAC 111 |
| Tri-ITS-5 | T. dermatis and T. jirovecii | ITS region | AF444437 | 88 CTCCGGTCAATTACTTTACAAAC 110 |
| Tri-ITS-6 | T. jirovecii | ITS region | AF444437 | 373 GAGTTAGCGTGTTTAACTTGTCGAT 397 |
| Tri-ITS-7 | T. dermatis and T. jirovecii | ITS region | AF444437 | 385 TTAACTTGTCGATCTGGCGTA 405 |
| Tri-ITS-8 | T. domesticum | ITS region | AF444422 | 85 TTGAATCTTCGGATTCGATTTTATACAAA 113 |
| Tri-ITS-9 | T. domesticum and T. laibachii | ITS region | AF444422 | 377 AAAGAGTTAGCAAGTTGAACTATTGCTAT 405 |
| Tri-ITS-10 | T. inkin | ITS region | AF444420 | 31 GTGATTGCCTTTACAGGCTTAACTA 55 |
| Tri-ITS-11 | T. laibachii | ITS region | AF444421 | 84 TTGAATCTCTGATTCAATTTTACAAAC 110 |
| Tri-ITS-12 | T. laibachii | ITS region | AF444421 | 31 GTGATTGCCATCTTGGCTTAAAC 53 |
| Tri-D1D2-1 | T. cutaneum, T. dermatis and T. jirovecii | D1/D2 domain | AF105398 | 181 GCTTGATACGACGACCAGTGCTCT 204 |
| Tri-D1D2-2 | T. cutaneum, T. dermatis, T. domesticum and T. jirovecii | D1/D2 domain | AF105398 | 405 ATTCAGCTGGTTCTTCCAGTCTACT 429 |
| Tri-D1D2-3 | T. dermatis and T. domesticum | D1/D2 domain | EU559351 | 413 GATTCAGCTAGTTCTTCTAGTCTACTTCC 441 |
| Tri-D1D2-4 | T. domesticum | D1/D2 domain | AF189874 | 181 ACTTGACACAACAATCAGTGCTCT 204 |
| Tri-D1D2-5 | T. asahii | D1/D2 domain | AF337949 | 535 GGCCGGCCTTCGGGCACGTT 554 |
| Tri-D1D2-6 | T. asahii, T. inkin and T. japonicum | D1/D2 domain | AF308657 | 407 TCAGCCAGTTCTGCTGGTCTACT 429 |
| Tri-D1D2-7 | T. domesticum, T. inkin and T. laibachii | D1/D2 domain | AJ749822 | 566 GGCCGGGGTTCGCCCACGTT 585 |
| Tri-D1D2-8 | T. laibachii | D1/D2 domain | AJ507664 | 157 ACTTTACACAATCATCAGTGCTCT 180 |
| Tri-D1D2-9 | T. laibachii | D1/D2 domain | AJ507664 | 381 ATTCAGCCGGTCTTCGGTGTACT 403 |
| RCA padlock probes‡ | ||||
| Tri-jap | T. japonicum | ITS region | AF444473 | 51 TAAGCCAATTAAGGCAATCACTAATG 26 –linker-74 ACAGGTGTAAGTGGATATAGTTA 52 |
| Tri-cut | T. cutaneum | ITS region | AF444325 | 354 ACTGGCAGCGCCCAA 340 –linker-366 GCGAGCCAGGCT 355 |
| Tri-der-a | T. dermatis | ITS region | EU559351 | 441 GGAAGTAGACTAGAAGAACTAGCTGAATCC 412
–linker-457 TGTTGACCCGTTCAAT 442 |
| Tri-dom-a | T. domesticum | ITS region | AF444454 | 393 AACTTGCTAACTCTTTTAAGAGGAGCC 367
–linker-411 GCCAGATAGCAATAGTTC 394 |
| Tri-asa-a | T. asahii | D1/D2 domain | AF337949 | 540 CCGGCCATAAAGGCGA 525 –linker-552 CGTGCCCGAAGG 541 |
| Oligonucleotide | Specificity | Target gene | GenBank accession no. | Sequence (5′–3′)* |
| Tri-asa-b | T. asahii | D1/D2 domain | AF337949 | 547 CCGAAGGCCGGCCA 534 –linker-563 CTAAGCTCGAACGTGC 548 |
| Tri-der-b | T. dermatis | D1/D2 domain | EU559351 | 494 TACATTCCTACTATCTTTATCCACCGG 468
–linker-505 CCCGGGGAGC 495 |
| Tri-dom-b | T. domesticum | D1/D2 domain | AF189874 | 510 TAGGCTATAACACTCCCGAGGGA 488 –linker-528 ACCCAGTGTATGTGAACC 511 |
| Tri-ink | T. inkin | D1/D2 domain | AJ749822 | 519 CAGGCTATAACACTTCCGGAGAAG 496
–linker-537 ACCCAGTGTATGTGACAG 520 |
| Tri-jir-a | T. jirovecii | D1/D2 domain | AF105398 | 130 TCCAGCACGGAAAACACG 113 –linker-148 CAAGGGACTTAGATACGA 131 |
| Tri-jir-b | T. jirovecii | D1/D2 domain | AF105398 | 214 CGTGTATCACAGAGCACTGGTC 193 –linker-233 AACAACTCGACTCGTAGAA 215 |
| Tri-lai | T. laibachii | D1/D2 domain | EU559352 | 431 ACCGAAGACCGGCTGAAT 414 –linker-450 ACCCGTTTAAAGGAAGTAC 432 |
| Tri-ast§ | T. asteroides | D1/D2 domain | AF075513 | 490 GAAGTCACATTCCTACTACCTTTATCCAC 462
–linker-507 GCTATAACACTTCCGGG 491 |
| Artificial template§ | ||||
| T. asteroides-AT§ | T. asteroides | D1/D2 domain | AF075513 | 457 GTCCGGTGGATAAAGGTAGTAGGAATGT GACTTCCCCGGAAGTGTTATAGCCTATT 512 |
Abbreviations: D1/D2 domain, D1/D2 domain of 28S rRNA gene; ITS region, internal transcribed spacer region; RLB, reverse line blot; RCA, rolling cycle amplification. *Numbers represent the numbered base positions where the oligonucleotide sequences start or finish (commencing at point 1 of the corresponding gene GenBank sequence). †Primers ITS1b and R635b were based on previously published ITS1 and R635, respectively, with 5′ end biotin labeled for PCR in preparation for RLB. All RLB probes were 5′-hexylamine modified. ‡All RCA padlock probes were with 5′-end phosphorylation. 5′ and 3′ binding arms of the probes derived from reference GenBank sequence and their positions are showed. These are joined by the 64 bp linker region including (i) binding site of RCA primer 1 to the padlock probe (reverse compliment sequence of RCA primer 1), generating a long single-stranded DNA; (ii) binding site of RCA primer 2 (with the same sequence of RCA primer 2), primer binds to nascent single-stranded DNAs as their binding sites become available; and (iii) the non-specific linker region. §An additional probe Tri-ast was designed to identify T. asteroides, which has high sequence similarity with T. japonicum studied (1 bp difference each within the ITS region and D1/D2 domain, respectively). As no T. asteroides isolates were available for study, an artificial template ‘T. asteroides-AT’ was used along with the 48 Trichosporon isolates to test the sensitivity and specificity of the probe Tri-ast.
Oligonucleotide probes and PCR for RLB
Oligonucleotide probes for the RLB assay were designed to achieve maximum specificity according to published protocols [22]. Six species- and eight group-specific probes were designed to target the ITS region, including two panfungal oligonucleotide probes, i.e., 12AP and 23SP (Table 1) [18]. In addition, four species- and five group-specific probes were directed at nucleotide polymorphisms in the D1/D2 domain of Trichosporon spp. Thus, a total of 23 probes were employed in the RLB assay (Table 1), all of which were 5′-hexylamine modified and synthesized by Beijing AuGCT Biotechnology Co. Ltd.
In preparation for the RLB assay, each PCR reaction (total volume 30 μl) contained: 5 μl template DNA, 10 × PCR buffer (Qiagen, Doncaster, Australia); 0.2 mM each dNTP (Roche Diagnostics, Mannheim, Germany), 1.5 U HotStar Taq polymerase (Qiagen); 0.5 μM each of forward primer and reverse primers and biology grade water (Eppendorf, North Ryde, Australia). Amplification was performed in a Mastercycler gradient thermocycler (Eppendorf, GmbH, Germany), under cycling conditions previously described [12] and consisted of 10 min at 95°C for initial activation, 30 cycles of 30 sec at 95°C for denaturing, 30 sec at 50°C for annealing, and 30 sec at 72°C for extension, followed by 7-min final extension at 72°C. After amplification, equal volumes of ITS and D1/D2 PCR- amplified products (20 μl each) were added to 160 μl 2 × SSPE (Amresco) in 0.1% SDS (Sigma), then denatured at 100°C for 10 min, and cooled immediately on ice. The RLB procedure was then performed as previously described [18,22].
Design of padlock probes and RCA
Initially 14 species-specific RCA padlock probes to detect and identify Trichosporon species were designed using established methods to target nucleotide polymorphisms (including single nucleotide polymorphisms [SNPs]) within the ITS region and D1/D2 domain (see Table 1) [14,21,23]. However, preliminary experiments showed that the ‘T. cutaneum-specific’ and ‘T. inkin-specific’ ITS-targeted probes failed to detect the species. These two probes were not further used in the study and hence a total of 12 species-specific probes were employed (Table 1). Briefly, the probes were 91–113 bp in length, consisting of (i) a 5′-binding arm region, (ii) a 64-bp linker region, and (iii) a 3′-binding arm region (Table 1). The 5′- and 3′-binding arms of the probes are complementary to the 5′ and 3′ termini of the target sequence in reverse. To optimize the binding to target DNA, probes were designed with a minimum of secondary structure and with a Tm of the 5′-end probe binding arm greater than the temperature used for probe ligation (65 ± 1°C). To increase the specificity, the 3′-end binding arm was designed to have a Tm (51–56°C) below the ligation temperature. Careful attention was paid to the linker region for each point mutation-specific probe to (i) minimize similarity to those mutations closely located to the mutation of interest and (ii) to allow primer binding during RCA and amplification of the probe-specific signal. Binding sites of RCA primers 1 and 2 were designed within 64 nucleotides linker region as previously described [14,21,23].
In addition, a T. asteroides-specific probe (Tri-ast) was designed, targeting a 55-bp artificial template of the species (T. asteroides-AT) derived from Tri-ast target region of T. asteroides sequences AF075513 accessed from the GenBank database (Table 1). An artificial template for this species was constructed because we were unable to obtain T. asteroides isolates for study. All padlock probes were synthesized by Beijing AuGCT Biotechnology Co. Ltd.
For PCR in preparation for RCA analysis, the ITS regions and D1/D2 domains of the test organisms were amplified as described for preparation for the RLB assay using the primer pairs ITS1/R635. Procedures for ligation of the padlock probe, exonucleolysis and hyperbranched RCA reactions were performed as previously described [14,21,23].
Results
Species identification by RLB
The panfungal primers ITS1b/ITS4 and F63/R635b amplified the ITS region and D1/D2 domain, respectively, of all 48 study isolates. All species-, and group-specific, RLB probes were 100% specific for their target species or species group. The ITS-targeted panfungal probes 12AP and 23SP (Table 1) hybridized with amplified DNA of all isolates (Fig. 1).
Fig. 1.
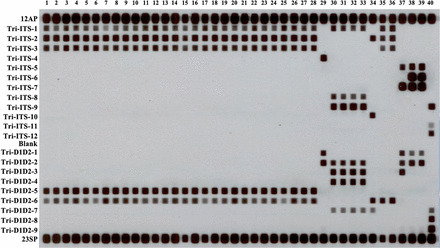
Reverse line blot assay results of eight Trichosporon species studied. The left column shows the probe list (see Table 1). Lanes 1–28 (T. asahii), PUMCHBY12-PUMCHBY19, PUMCH30401-PUMCH30410, PUMCH5Z6443, PUMCH5Z6527, PUMCH6W5203, PUMCH6Z10766, PUMCH6Z2782, PUMCH6Z6579, PUMCH6Z8369, PUMCH6Z9690, PUMCH7R7615, PUMCH7Z102; Lane 29 (T. cutaneum), PUMCHBY28; Lanes 30–33 (T. domesticum), PUMCHBY20-PUMCHBY22, PUMCHBY24; Lane 34 (T. inkin), PUMCHBY23; Lanes 35–36 (T. japonicum), PUMCHBY11, PUMCHBY27; Lane 37 (T. dermatis), PUMCHBY24; Lanes 38–39 (T. jirovecii), PUMCH6Z2374 PUMCH6Z10950; Lane 40 (T. laibachii), PUMCHMC31.
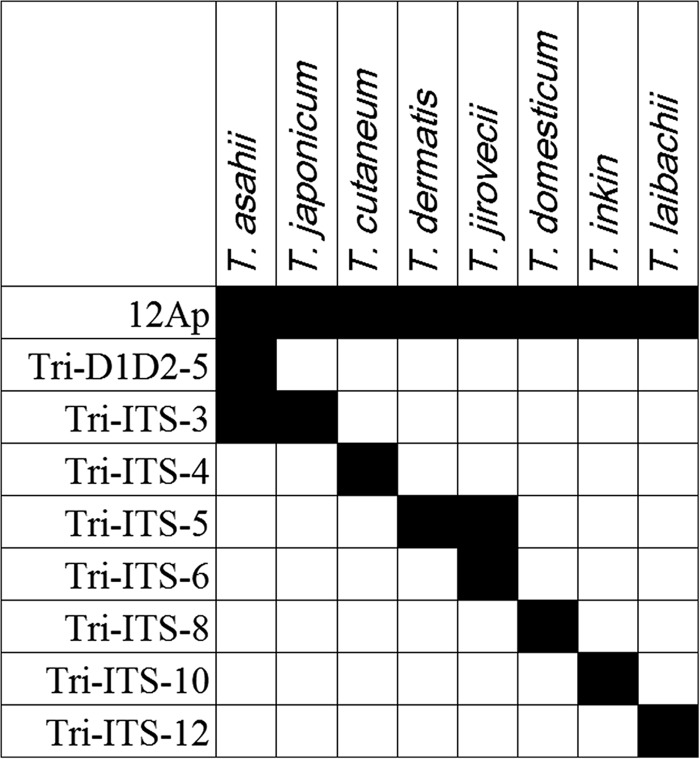
Species-specific RLB probes correctly identified 45 of 48 (94%) Trichosporon isolates as compared with DNA sequencing [12] (Table 2). The exceptions were the non-target species, i.e., T. dermatis and T. japonicum, and as a result T. dermatis PUMCHBY24 and two T. japonicum isolates were not assigned to species. Since we were unable to design ‘T. dermatis’- or ‘T. japonicum’-specific RLB probes, species identification of ‘T. dermatis’ relied on obtaining a positive hybridization result with the group-specific probe Tri-ITS-5 (specifically hybridizing with T. dermatis and T. jirovecii; Table 1) and the subsequent absence of a positive signal using the T. jirovecii-specific probe, Tri-ITS-6 (Figs. 1 and 2). Similarly, T. japonicum strains PUMCHBY11 and strain PUMCHBY27 were first assigned to the T. asahii-T. japonicum group by hybridization with probe Tri-ITS-3 (T. asahii and T. japonicum group-specific) and then as T. japonicum by absence of a probe signal with the T. asahii-specific probe, Tri-D1D2-5 (Figs. 1 and 2).
Table 2.
Comparison of species ID by three-locus sequencing, RLB and RCA amongst eight Trichosporon species studied.
| Strain ID by three-locus sequencing* | No. of isolates (IGS1 genotype*) |
Strain ID by RLB | Strain ID by RCA |
| Trichosporon asahii | 36 (4) | T. asahii | T. asahii |
| Trichosporon cutaneum | 1 (1) | T. cutaneum | T. cutaneum |
| Trichosporon dermatis | 1 (1) | T. dermatis† | T. dermatis |
| Trichosporon domesticum | 4 (1) | T. domesticum | T. domesticum |
| Trichosporon inkin | 1 (1) | T. inkin | T. inkin |
| Trichosporon japonicum | 2 (2) | T. japonicum† | T. japonicum |
| Trichosporon jirovecii | 2 (2) | T. jirovecii | T. jirovecii |
| Trichosporon laibachii | 1 (1) | T. laibachii | T. laibachii |
Abbreviations: ID, identification; IGS1, intergenic spacer 1 region; RLB, reverse line blot hybridization assay; RCA, rolling circle amplification. *Genotypes within species were further assigned based on IGS1 sequences as previously described [12]. †Identification of T. dermatis relied on obtaining a positive hybridization result with the group-specific probe which hybridizes with T. dermatis and T. jirovecii and subsequent absence of a RLB signal using a T. jirovecii-specific probe. Similarly, T. japonicum strains were first assigned to the T. asahii-T. japonicum group by hybridization with a combined T. asahii and T. japonicum group-specific probe and then as T. japonicum by absence of a probe signal with a T. asahii-specific probe (Figs. 1 and 2).
Fig. 2.
The in silico results obtained by reverse line blot (RLB) assay for identification of Trichosporon species studied. RLB probes proposed for the species identification of eight medically important Trichosporon species. The black squares represent positive signals on the RLB membrane. The left column shows the probe list (see Table 1) and the top row shows the species identity.
For the eight Trichosporon species tested in this study, we proposed a set of nine RLB probes (eight species- specific probes and one panfungal probe as an internal ‘positive’ control) for species identification (Fig. 2).
Species identification by RCA
The accumulation of double-stranded DNA was detected by staining with Sybr Green I. RCA signals obtained for the target species are shown as exponential increases in fluorescence (Fig. 3). The 12 RCA probes correctly detected their respective DNA templates with 100% specificity and were 100% concordant with DNA sequencing (Table 1 and Supplementary Table 1, available online http://informahealthcare.com/doi/abs/10.3109/13693786.2012.723223). Probe signals were obtained only with matched template-probe mixtures, with no RCA signal observed when templates from isolates of non-targeted species were present. In contrast to results from the RLB assay, T. dermatis and T. japonicum were identified to species level by species-specific RCA probes (Fig. 3).
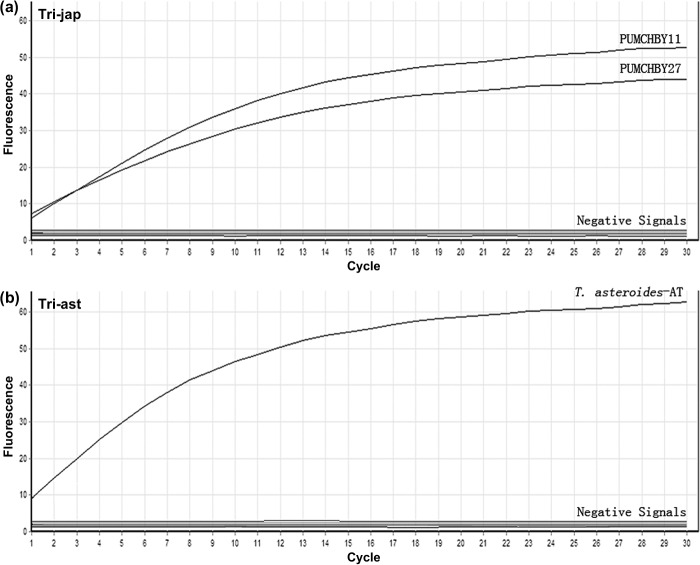
Fig. 3.
Rolling circle amplification (RCA) results of isolates PUMCHBY11, PUMCHBY27 and of the artificial template T. asteroides-AT. RCA results monitored by the RotorGene 6000 real-time PCR machine (Corbett research). The experiment was conducted using the T. japonicum-specific RCA probe Tri-jap in (a) and T. asteroides-specific probe Tri-ast in (b) tested on all 48 Trichosporon study isolates and the artificial template T. asteroides-AT. The accumulation of double-stranded DNA was detected by staining with Sybr Green I. RCA signals indicating the species identified are shown as exponential increases in fluorescence. Ligation-mediated RCA with matched templates [DNA from T. japonicum isolates PUMCHBY11, PUMCHBY27 in (a) and artificial template T. asteroides-AT in (b)] produced ‘positive signals’. Other templates showed an absence of signal (labeled as ‘Negative Signals’).
In experiments using the artificial template T. asteroides- AT and probe Tri-ast, RCA analysis yielded a signal but produced no signal with all of the 48 Trichosporon isolates studied as illustrated by the negative signals in Fig. 3b.
Discussion
In this study, we evaluated two non-DNA sequencing-based approaches, the RLB and RCA methods, for the identification of 48 Trichosporon isolates representing eight clinically-relevant species in China.
Although the RLB assay correctly assigned all 48 Trichosporon isolates to species, with 100% specificity, species-specific RLB probes only correctly assigned 94% of isolates belonging to six species. Of note, we were not able to design ‘T. japonicum-specific’ and ‘T. dermatis-specific’ RLB probes. Instead, the identification of these two species required the use of group-specific probes – Tri-ITS-5 (T. dermatis and T. jirovecii group-specific) and Tri-ITS-3 (T. asahii and T. japonicum group-specific), followed by the absence of a positive signal using the T. jirovecii-specific probe (Tri-ITS-6) and T. asahii-specific probe (Tri-D1D2-5), respectively. The inability to successfully identify T. dermatis or T. japonicum with species-specific RLB probe is unexplained. Approximately 2% sequence dissimilarity was observed between the ITS and D1/D2 domain sequences of T. dermatis and T. jirovecii. Yet some of these polymorphic sites contained only a SNP and thus may have been unsuitable as RLB-based probe hybridization sites [22]. In addition, preliminary experiments showed cross-hybridization of the ‘T. dermatis- specific’ probe with other Trichosporon species (including the most common species T. asahii, data not shown). Sequence similarity within the ITS region, as well as in the D1/D2 domain between T. asahii and T. japonicum was also high with only 4-bp differences observed. Only the T. asahii-specific probe, but not the ‘T. japonicum-specific’ probe, was successfully designed.
As an alternative approach, RCA has been established as being highly sensitive for detecting SNPs in fungi [14,19,20]. In the present study, RCA assay not only successfully identified all 48 Trichosporon isolates to species, but also correctly identified T. dermatis and T. japonicum and distinguished these species from T. jirovecii and T. asahii, respectively. Although species outside of those tested were not available for analysis in the present study, the return of a positive RCA signal using the artificial template T. asteroides-AT and the T. asteroides-specific probe, Tri-ast, was a proof of the concept that such probe-template combinations may be used with good results to establish working molecular models of probe-based RCA assays [14,23].
In the present study, the RLB assay had a turnaround time of ˜1.5 days [17,18,22] and the RCA method, ˜0.5 days [19–21] (the time needed for the isolation and preliminary identification of yeast isolates was not included). Both approaches were less time-consuming than DNA sequencing methodology which requires ˜2–3 days for a definitive result. DNA sequencing following PCR has been used successfully to detect and identify fungal pathogens directly from clinical samples [18,24]. The PCR/RLB hybridization approach has also been reported to accurately detect pathogenic fungi in clinical specimens [16,18], yet the ability of either RLB or RCA assay to detect Trichosporon species from clinical specimens has not been studied.
Test costs are likely to influence decisions as to their implementation in clinical laboratories. The set-up costs of the RCA assay are relatively high (US$300 per probe), but a typical commercial batch of probes provides sufficient material for up to 5000 assays. Thus, running costs of RCA are no more than US$2 per assay (for consumables) [19,21]. The costs of the RLB method are approximately US$7 per isolate (for consumables), compared to US$14–35 for DNA sequencing [17]. Hence, either RLB or RCA may be employed as a potential alternative tool to DNA sequencing for the identification of Trichosporon species in the clinical laboratory. The RLB approach is suited for the simultaneous analysis of a large number of isolates (up to 43) whereas RCA assays can be used to identify isolates, either as the need arises or in batches. RCA is better suited to detect SNPs or polymorphisms which may differ between species by only a few bp. In the present study, although only 23 RLB probes and 12 RCA padlock probes were utilized for identification of eight Trichosporon species, incorporating more probes will extend the target range of species. This flexibility allows laboratories to customize the assays to meet their specific requirements according to local epidemiology. RLB and/or RCA assays have been successfully applied in the identification of various fungal pathogens including Aspergillus, Candida, Cryptococcus and Fusarium [15–20]. The RLB assay may also be adapted for intra-species genotyping by designing and incorporating ‘genotype-specific’ probes into the blot as has been demonstrated for Fusarium spp. [17].
Limitations of the present study include the relative small number of species tested since only isolates representing eight species were available. Other pathogenic Trichosporon species such as T. dohaense, T. faecale and T. mycotoxinivorans [1,6,10] may not able to be identified to species level using the probes in this study. T. mycotoxinivorans has been reported as an emerging pathogen in cystic fibrosis [10]. Hence assays that target fungal pathogens should take into consideration the context in which they will be applied.
In conclusion, two simple and practical molecular assays, RLB and RCA were evaluated and both assays obtained accurate results for species identification of Trichosporon fungi. The methods can be extended to detect other Trichosporon species not included in the study.
Supplementary material available online
Acknowledgments
Declaration of interest: The authors report no conflicts of interest. The authors alone are responsible for the content and the writing of the paper.
This study was supported by (i) National Science Research Megaproject against Major Infectious Diseases in China 2009ZX10004-102; and (ii) The Ministry of Health of the People's Republic of China, public service sectors funds 200802026.
Contributor Information
Sharon C-A. Chen, Email: sharon.chen@swahs.health.nsw.gov.au.
Ying-Chun Xu, Email: xycpumch@yahoo.com.cn.
References
- 1.Chagas-Neto TC, Chaves GM, Melo AS, Colombo AL. Bloodstream infections due to Trichosporon spp.: species distribution, Trichosporon asahii genotypes determined on the basis of ribosomal DNA intergenic spacer 1 sequencing, and antifungal susceptibility testing. J Clin Microbiol. 2009;47:1074–1081. doi: 10.1128/JCM.01614-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Girmenia C, Pagano L, Martino B, et al. Invasive infections caused by Trichosporon species and Geotrichum capitatum in patients with hematological malignancies: a retrospective multicenter study from Italy and review of the literature. J Clin Microbiol. 2005;43:1818–1828. doi: 10.1128/JCM.43.4.1818-1828.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Matsue K, Uryu H, Koseki M, Asada N, Takeuchi M. Breakthrough trichosporonosis in patients with hematologic malignancies receiving micafungin. Clin Infect Dis. 2006;42:753–757. doi: 10.1086/500323. [DOI] [PubMed] [Google Scholar]
- 4.Araujo Ribeiro M, Alastruey-Izquierdo A, Gomez-Lopez A, Rodriguez-Tudela JL, Cuenca-Estrella M. Molecular identification and susceptibility testing of Trichosporon isolates from a Brazilian hospital. Rev Iberoam Micol. 2008;25:221–225. [PubMed] [Google Scholar]
- 5.Rodriguez-Tudela JL, Diaz-Guerra TM, Mellado E, et al. Susceptibility patterns and molecular identification of Trichosporon species. Antimicrob Agents Chemother. 2005;49:4026–4034. doi: 10.1128/AAC.49.10.4026-4034.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Taj-Aldeen SJ, Al-Ansari N, El Shafei S, et al. Molecular identification and susceptibility of Trichosporon species isolated from clinical specimens in Qatar: isolation of Trichosporon dohaense Taj-Aldeen, Meis & Boekhout sp. nov. J Clin Microbiol. 2009;47:1791–1799. doi: 10.1128/JCM.02222-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paphitou NI, Ostrosky-Zeichner L, Paetznick VL, et al. In vitro antifungal susceptibilities of Trichosporon species. Antimicrob Agents Chemother. 2002;46:1144–1146. doi: 10.1128/AAC.46.4.1144-1146.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gueho E, Smith MT, de Hoog GS, et al. Contributions to a revision of the genus Trichosporon. Antonie Van Leeuwenhoek. 1992;61:289–316. doi: 10.1007/BF00713938. [DOI] [PubMed] [Google Scholar]
- 9.Rodriguez-Tudela JL, Gomez-Lopez A, Alastruey-Izquierdo A, et al. Genotype distribution of clinical isolates of Trichosporon asahii based on sequencing of intergenic spacer 1. Diagn Microbiol Infect Dis. 2007;58:435–440. doi: 10.1016/j.diagmicrobio.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 10.Hickey PW, Sutton DA, Fothergill AW, et al. Trichosporon mycotoxinivorans, a novel respiratory pathogen in patients with cystic fibrosis. J Clin Microbiol. 2009;47:3091–3097. doi: 10.1128/JCM.00460-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tambe SA, Dhurat SR, Kumar CA, et al. Two cases of scalp white piedra caused by Trichosporon ovoides. Indian J Dermatol Venereol Leprol. 2009;75:293–295. doi: 10.4103/0378-6323.51256. [DOI] [PubMed] [Google Scholar]
- 12.Guo LN, Xiao M, Kong F, et al. Three-locus identification, genotyping, and antifungal susceptibilities of medically important Trichosporon species from China. J Clin Microbiol. 2011;49:3805–3811. doi: 10.1128/JCM.00937-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sugita T, Ikeda R, Nishikawa A. Analysis of Trichosporon isolates obtained from the houses of patients with summer-type hypersensitivity pneumonitis. J Clin Microbiol. 2004;42:5467–5471. doi: 10.1128/JCM.42.12.5467-5471.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kong F, Tong Z, Chen X, et al. Rapid identification and differentiation of Trichophyton species, based on sequence polymorphisms of the ribosomal internal transcribed spacer regions, by rolling-circle amplification. J Clin Microbiol. 2008;46:1192–1199. doi: 10.1128/JCM.02235-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu Q, van den Ende AH, de Hoog GS, et al. Reverse line blot hybridisation screening of Pseudallescheria/Scedosporium species in patients with cystic fibrosis. Mycoses. 2011;54((Suppl. 3)):5–11. doi: 10.1111/j.1439-0507.2011.02108.x. [DOI] [PubMed] [Google Scholar]
- 16.Playford EG, Kong F, Sun Y, et al. Simultaneous detection and identification of Candida, Aspergillus, and Cryptococcus species by reverse line blot hybridization. J Clin Microbiol. 2006;44:876–880. doi: 10.1128/JCM.44.3.876-880.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang H, Xiao M, Kong F, et al. Accurate and practical identification of 20 Fusarium species by seven-locus sequence analysis and reverse line blot hybridization, and an in vitro antifungal susceptibility study. J Clin Microbiol. 2011;49:1890–1898. doi: 10.1128/JCM.02415-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zeng X, Kong F, Halliday C, et al. Reverse line blot hybridization assay for identification of medically important fungi from culture and clinical specimens. J Clin Microbiol. 2007;45:2872–2880. doi: 10.1128/JCM.00687-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou X, Kong F, Sorrell TC, et al. Practical method for detection and identification of Candida, Aspergillus, and Scedosporium spp. by use of rolling-circle amplification. J Clin Microbiol. 2008;46:2423–2427. doi: 10.1128/JCM.00420-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaocharoen S, Wang B, Tsui KM, et al. Hyperbranched rolling circle amplification as a rapid and sensitive method for species identification within the Cryptococcus species complex. Electrophoresis. 2008;29:3183–3191. doi: 10.1002/elps.200700903. [DOI] [PubMed] [Google Scholar]
- 21.Wang H, Kong F, Sorrell TC, et al. Rapid detection of ERG11 gene mutations in clinical Candida albicans isolates with reduced susceptibility to fluconazole by rolling circle amplification and DNA sequencing. BMC Microbiol. 2009;9:167. doi: 10.1186/1471-2180-9-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kong F, Gilbert GL. Multiplex PCR-based reverse line blot hybridization assay (mPCR/RLB) – a practical epidemiological and diagnostic tool. Nat Protoc. 2006;1:2668–2680. doi: 10.1038/nprot.2006.404. [DOI] [PubMed] [Google Scholar]
- 23.Wang B, Potter SJ, Lin Y, et al. Rapid and sensitive detection of severe acute respiratory syndrome coronavirus by rolling circle amplification. J Clin Microbiol. 2005;43:2339–2344. doi: 10.1128/JCM.43.5.2339-2344.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lau A, Chen S, Sorrell T, et al. Development and clinical application of a panfungal PCR assay to detect and identify fungal DNA in tissue specimens. J Clin Microbiol. 2007;45:380–385. doi: 10.1128/JCM.01862-06. [DOI] [PMC free article] [PubMed] [Google Scholar]