Abstract
Abstract
Recently in China, a novel coronavirus outbreak took place which caused pneumonia-like symptoms. This coronavirus belongs to the family of SARS and MERS and causes respiratory system disease known as COVID-19. At present we use polymerase chain reaction (PCR) based molecular biology methods for the detection of coronavirus. Other than these PCR based methods, some improved methods also exist such as microarray-based techniques, Real time-quantitative PCR, CRISPR-Cas13 based tools but almost all of the available methods have advantages and disadvantages. There are many limitations associated with this method and hence there is a need for a fast, more sensitive, and specific diagnostic tool which can detect a greater number of samples in less time. Here we have summarised currently available nucleic acid-based diagnostic methods for the detection of coronavirus and the need for developing a better technique for a fast and sensitive detection of coronavirus infections.
Graphic abstract
Keywords: Coronavirus, COVID-19, qRT-PCR, Microarray, Amplification, Nucleic acid detection
Introduction
Coronaviruses, a member of the coronaviridae virus family causes a simple cough cold to much more severe and complicated disease such as Severe Acute Respiratory Syndrome (SARS-CoV), Middle East Respiratory Syndrome (MERS-CoV), and Coronavirus Disease 2019 (COVID-19). The genetic material of SARS-CoV-2 is a positive-sense single-stranded RNA of 26-32 kb enclosed in an envelope [1]. Till nowfour different genera of coronavirus have been discovered including alpha, beta, gamma, and delta. A total of six human infecting coronaviruses have been discovered which belongs to alpha (HCoV-229E and HCoV-NL63) and beta (MERS-CoV, SARS-CoV, HCoV-OC43, and HCoV-HKU1) genera [2, 3]. Out of these six coronaviruses, MERS-CoV, and SARS-CoV resulted in pandemic [4].
Recently, in December 2019, a novel coronavirus (SARS-CoV-2) emerged in Wuhan city of China and then subsequently spread throughout the world with reported cases in 150 countries. Early reports suggested the onset of a potential coronavirus outbreak which now has been named as COVID-19 by WHO on 11th February 2020 [5]. As of 18th September 2020, the total confirmed cases of COVID-19 is 3,02,39,914 and a total of 948,382 death (https://www.worldometers.info/coronavirus/). The immunity against viral infections remains a challenge and never-ending task despite huge efforts from the researchers and scientists. WHO has declared the COVID-19 as pandemic on 11th March 2020. The laboratory diagnosis of coronavirus (SARS-CoV-2) depends on the epidemiological history of patients, signs, and symptoms, and some specific laboratory tests such as nucleic acid-based detection, CT scanning, some immunological identification technologies of immunoglobulin M/G, ELISA and blood cultures. The sign and symptoms in the case of COVID-19 are very atypical (includes, high fever, cough, dyspnea, pneumonia-like symptoms, and respiratory problems) and hence the role of the laboratory-based diagnostic methodologies is significant.
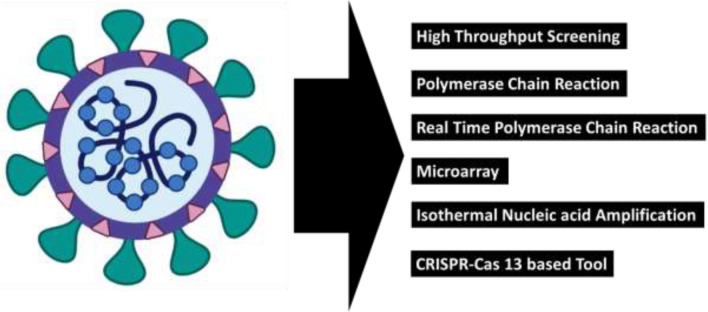
In the current epidemic time, the detection and identification of SARS-CoV2 RNA genome are one of the useful tools in the diagnosis, which is very helpful for the management of infection source as well as to help patients for better recovery from the illness. Along with the growth of molecular biology and genetic engineering techniques, the nucleic acid-based identification and detection methodologies is a revolution especially for virus detection (Fig. 1). One of the qualitative and quantitative SARS-CoV2 detection method is Enzyme-Linked Immunosorbent Assay (ELISA) based method where IgM or IgGantibody produced against the spikes protein of SARS-CoV2 is measured. This is having high sensitivity and throughput. The important sample used during ELISA is blood, plasma or serum from the suspect and introduced into the microtiter multi well plate coated with viral spikes proteins. After incubation secondary antibodies labelled with enzymes is used and signal generated is measured. The assay format can be adapted for different detection modalities, including colorimetric, fluorescent, and electrochemical methods. Among all the nucleic acid-based methods, the polymerase chain reaction (PCR) based methodologies are very much accurate, having high specificity and sensitivity, and very fast. Dues to these reasons the PCR is a monetary standard method for virus detection. Along with PCR based methods, several non-PCR based methodologies also exist such as loop-mediated isothermal amplification (LAMP) and nucleic acid sequence-based amplification. These are isothermal nucleic acid amplification methods developed for the spotting and identification of COVID-19 RNA genetic material. In this manuscript, coronavirus detection methodologies are reviewed for the benefit of the scientific community in fast and accurate detection of viral RNA.
Fig. 1.
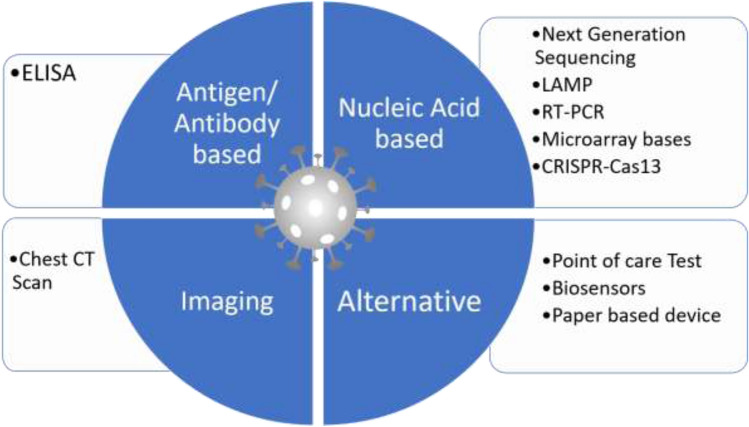
Commonly available test for the detection of SARS-CoV2
Nucleic acid detection-based technology
Two commonly preferred nucleic acid-based detection methodology is available for the detection of coronavirus COVID-19 are high throughput genome sequencing and PCR based method, real-time quantitative polymerase chain reaction (RT-qPCR) [6].
High throughput sequencing
As the cost of high throughput sequencing of genome is very high and it also depends on the availability of sequencer and hence its applicability in the clinical diagnostic methodologies is very limited and restricted. The dependency on the RT-qPCR based methodology is very high and it is one of the best methods to detect coronavirus with huge sensitivity.
PCR based methods
Due to the limited application of genome sequencing-based methodologies, the significance of PCR based method is very high. Among PCR based methods, RT-qPCR is a commonly used method which is a very easy, effective, and straightforward method for the identification and detection of pathogenic coronavirus from the respiratory swabs and blood samples [7]. PCR is a molecular technique for the amplification of the copies number of gene fragments through the thermostable DNA polymerase reactions. In the PCR, first the double helix DNA is separated into two single-strand DNA in the denaturing stage and then a sequence-specific primer is ligated to both the strands in annealing steps and finally both the primer is elongated by a thermostable DNA polymerase using single-stranded DNA as the template in extending stage. These cycles are repeated multiple times to get multiple copies of DNA from a single copy of DNA. This method increases the sample DNA to get a sufficient quantity of genetic material for laboratory analysis. For COVID-19 detection this sequence-specific method is highly utilized [8, 9]. The genome of SARS-CoV2 is single-stranded RNA which is first converted into complementary DNA (cDNA) through the reverse transcriptase-based process, cDNA amplified through PCR followed by and quantification and detection through specific methods. Conventional detection methods for the PCR products are visualization on an agarose gel and DNA sequencing [10, 11]. Time-consuming, sequencing cost, and instrument dependency this detection method is not preferred for the clinical samples.
Currently, real-time reverse transcriptase polymerase chain reaction (RT-PCR) is the most commonly used and preferred method for the coronavirus due to its sensitivity, specificity, and simple quantitative methods [12, 13]. The RT-PCR is used for the detection of almost all the coronavirus including SARS-CoV2 [7, 14, 15]. Even so, the RT-PCR is continuously improving to increase its specificity and sensitivity. Due to the enhanced time consumption in the sample preparation, sensitivity towards contaminations, issues in handling and analysis, an improved TaqMan based RT-PCR method has been introduced by van Elden et al., for the routine diagnosis of HCoV [16]. Also, to increase the sensitivity, Yip and their team structured an ongoing RT-qPCR examine for SARS-CoV by using 2 TaqMan tests, instead of 1 probe [17]. This straightforward alteration utilizing double TaqMan tests for evaluation has huge applications in regions in which ultra-sensitivity is fundamentally required, with the SARS-CoV identification limit of 1 copy RNA per cycle.
In clinical identification, the absence of protected and stable outer positive controls (OPC) could turn into a difficult issue in the identification of SARS-CoV-2 and a lot of consideration has been engaged to address this issue. However, these issues can be very well avoided in the case of real-time RT-PCR where OPC is a significant element. Yu et al. have developed an RT-PCR based assay in which the defensively covered RNA was utilized as OPC to identify the SARS-CoV-2, with a discovery cut-off value of 10 duplicates/μL [18]. The fast mutating behavior of coronaviruses increases the requirement for a sensitive and specific method for the detection and recognition of genetic variants of coronaviruses. Hence, to ameliorate the ability to detect and identify coronavirus correctly with reduced risk of false-negative results (which occur due to genetic variability in the sequences), scientists have developed multiplex real-time RT-PCR protocol with high sensitivity for the multitarget detection of CoV. Hadjinicolaou et al. built up a constant RT-PCR measure utilizing befuddle tolerant sub-atomic signals to recognize disease-causing and non-disease causing strains [19]. The test joined four reference points, focusing on four qualities not withstanding an inside positive control. It was approved utilizing clinical examples, which showed target recognition capacity and explicitness with the identification breaking point of 5 duplicates for each response.
As soon as the outbreak of COVID-19 occurs in China in December 2019, several industries launched the qRT-PCR based diagnostic kit for the CoV. The Chinese Centre for Disease Control and Prevention (China CDC) has commended to use the sequence-specific probes as well as primers in the “ORF1ab” and “N-gene region of SARS-CoV-2” through the qRT-PCR process. Theo separate gene locus (ORF1ab and N) on the SARS-CoV-2 gene has been detected by one-step RT-qPCR reported by Bustin and Nolan [20]. In this described method, the negative control was affirmed as negative and the test sample of two patients having COVID-19 was affirmed as positive. These RT-qPCR based methods have shown high sensitivity and specificity for SARS and MERS group of coronavirus [21].
Five patients showed a negative SARS-CoV2 result through RT-qPCR but showed positive results through a chest CT scan. These patients were tested again by taking a swab sample and RT-qPCR has been performed and confirmed as infected with SARS-CoV-2 [22]. Based on the protocol used for the RT-qPCR and the total number of samples collected, this shows a sensitivity of 50–79% for the detection of SARS-CoV [23]. And hence there is the scope of improvement in the detection rate of RT-qPCR for SARS-CoV-2 disease. Furthermore, RT-qPCR has drawbacks which include the biological hazards, cumbersome nucleic detection operations, and long sitting tight time for results.
Microarrays based methodologies
Microarray technology is a rapid and high throughput molecular biology detection tool which is very capable of quantifying thousands of gene transcript from the provided test cell or tissue at one moment. The microarray has a huge number of gene fragments (in picomoles) of already known sequences arrayed in a known sequence (called as probes) of rows and columns on a glass microscopic slide. In this method of coronavirus detection, the coronavirus RNA genome is first converted to a specific probe labeled cDNA by the reverse transcription process. These labeled cDNA will be charged into each well of the microarray plate containing prefixed oligonucleotides on solid phase and allowed to hybridize. Free unbound DNA is then washed out in a series of washing steps. The hybridized sequence is detected and due to the transcendency, the microarray-based methods are highly used protocol for the detection of coronavirus [24]. A group of researched lead by Shi et al. has designed and developed a microarray of 60 mer oligonucleotides as per the sequence of phosphatidylinositol kinase-related protein kinase (TOR2) and succeeded in the detection of coronavirus of SARS family from an isolated clinical sample. They created a thirty 60mer TOR2 sequence-specific oligo which covers the whole genome of the first submitted sequence of coronavirus strain [25]. But after the consideration of high and fast mutation rates in SARS-CoV, another group of scientists leads by Guo have designed a microarray for the detection of 24 single nucleotide polymorphism in the spike gene with 100% accuracy [26]. Due to the sudden outbreak of SARS-CoV2, it will be of great significance to design a diagnostic assay tool to detect a wide range of coronavirus and it may be placed at the Point of Care (POC) center. Accordance with this, Luna and their team have developed a cost-effective, nonfluorescent based low-density oligonucleotide assay tool for the detection of complete coronavirus genome with the same sensitivity as to real-time PCR. The limit of detection was 15.7 copies per reaction [27]. And also, Hardick et al. have also developed a microarray-based diagnostic tool, called Mobile Analysis Platform, with an acceptable detection limit and shown good performance in the detection [28].
Isothermal nucleic acid-based amplification
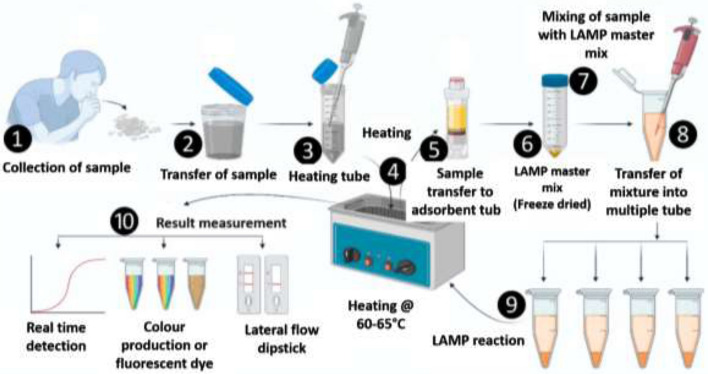
Isothermal amplification of nucleic acid is a tool for the rapid and efficient accumulation of nucleic acid at a specific constant temperature. Isothermal amplification is an alternative to the PCR technique which is used for various biosensing targets like nucleic acids (DNA and RNA), proteins and peptides, and ions. The amplicons of these isothermal amplification techniques have been used for the generation of various nucleic acid-based nanomaterials for their application in the field of biomaterials, biosensors, and biomedicines [29]. The association of the isothermal nucleic acid amplification and the microsystem on any portable device increases its utility in the on-site nucleic acid-based diagnostic assays and also delivers high sensitivity (Fig. 2).
Fig. 2.
The steps of the LAMP
Regular loop-mediated isothermal amplification (LAMP) based methods
“Loop-mediated isothermal amplification (LAMP)” is a low-cost method for the amplification of DNA in a single tube and mainly done for the detection of specific disease [30]. In this case, the target DNA sequence is amplified at a specific temperature of 60–65 °C by using 2–3 sets of primers and a DNA polymerase of large DNA strand displacement activity as well as DNA replication activity. Commonly four different primers are used to amplify 6-different regions of the target DNA sequences and it induces sensitivity of the process. Along with the four specific primers, a pair of loop primer is also used to increase the speed of the process [31]. The amount of amplified DNA produced by LAMP is high compared to the conventional PCR process. The amplified product can be detected and evaluated through a photometric reaction where turbidity caused due to magnesium pyrophosphate precipitate or fluorescence dye is measured [32]. LAMP is having advantages over other DNA amplifying methods due to its simplicity, cost efficiency, and toughness. It can be used for both screening purposes as well as for the point of care by diagnostic technicians. As the process is isothermal and takes place at a specific temperature which removes the requirement of costly thermocyclers. LAMP is already used for the diagnosis of a large number of different diseases such as malaria [33], tuberculosis [34], sleeping sickness [35] etc. This might be one of the cost-effective diagnostic tests for the coronavirus diagnosis [36]. There are many LAMP-based detection methods that have been designed and developed for the detection of coronavirus.
LAMP-based SARS assay has been developed by Poon and his team and also demonstrated its feasibility in the diagnosis of SARS-CoV [37]. ORF1b region of SARS-CoV was amplified through a LAMP based amplification reaction by using 6-primer. The amplified product was assayed through gel electrophoresis with detection rate and sensitivity of LAMP-based amplification similar to the conventional PCR based amplification. Another group of scientists leads by Pyrc developed a LAMP-based amplification method for the detection of HCoV-NL63 on agarose gel electrophoresis with desirable sensitivity and specificity [38].
A valuable RT-LAMP test for the conclusion and epidemiologic reconnaissance of human MERS-CoV was created by Shirato et al. [39], which is fit for identifying as not many as 3.4 duplicates of MERS-CoV RNA and is exceptionally explicit, with no cross-response with other respiratory infections. Thai et al. [40] built an one-advance single-tube continuous quantitative RT-LAMP test checked by ongoing estimation of turbidity in a photometer for the early and fast analysis of SARS-CoV. When compared with clinical samples the detection was reported to be 100-fold more sensitive than that of conventional polymerase chain reaction-based methods with a detection limit of 0.01 plaque-forming units (PFU).
Sequence-specific loop-mediated isothermal amplification (LAMP) methods
Turbidity generation due to the production of pyrophosphates at the time of polymerizing reaction or intercalation of fluorescence dyes inside the dsDNA amplicons possibility that the signal may come from primer dimerization or non-primer interactions [41]. This issue can be resolved by the use of a sequence-specific supervising of LAMP and some other temperature specific DNA amplification methods which produce true signal that can be differentiated from nonspecific signals. Shirato and his team had modified and improved the RT-LAMP detection tool by adding quenching probes (QProbe) to detect, and supervise true signal and at the same time it has a similar performance as the normal standard RT-PCR assays for the diagnosis of MERS-CoV [42]. A more developed nucleic acid visualization tool has been reported by combining RT-LAMP and a vertical flow visualization strip (RT-LAMP-VF) for the detection and diagnosis of MERS-CoV [3]. The optimum temperature for the LAMP is near to 65 °C which is a limiting factor for its applicability. To overcome this, Cai et al., has developed a modified version of LAMP by using phosphorothioated primers (PS-LAMP), which increases the efficient hairpin creation and elongation at the terminus of concatemers and it works at much low temperature than the optimum temperature of LAMP [43]. A better specificity and sensitivity was observed with the performance of PS-LAMP at 40 °C when comparable with the conventional LAMP at 65 °C.
CRISPR based newly developed methods
CRISPR (clustered regularly interspaced short palindromic repeats)-Cas is a genome-editing tool which allows scientist to add, remove or modify the genome at a desired specific site in the DNA. Recently, RNA targeting CRISPR-Cas13 has been reported for the fast and portable nucleic acid detection process [44, 45]. A group of scientists leads by Zhang reported the use of Cas13 to detect and destroy different mammalian viruses’ single-stranded RNA genome [46]. This group has created a specific platform-specific high-sensitivity enzymatic reporter unlocking (SHERLOCK) by combing two different tools, isothermal preamplification with Cas13 for the detection and digestion of ssRNA or ssDNA [47]. This has already been used for the detection of dengue and Zika virus ssRNA genome and also in the liquid biopsy sample of some patients. Their recent protocol for the detection and diagnosis of COVID-19 is published on a website (https://broad.io/sherlockprotocol) with the title, “A protocol for detection of COVID-19 using CRISPR diagnostics”. This online protocol may provide some reference points to the researchers for the rapid, quantitative detection of COVID-19.
CT scan and other diagnostic methodology
Although the RT-qPCR is a sensitive and specific tool for the detection of coronavirus, but its false-positive rate could not be ignored. Hence, some clinicians have suggested that CT scanning can be one of the compulsory auxiliary diagnostic tools due to its sensitivity. Combination of the repetition of RT-qPCR along with the chest CT scanning should be performed in the patients with negative RT-PCR but symptoms of SARS-CoV2. The high-resolution CT scanning of the chest (HRCT) is necessary for the preliminary diagnosis of the severity of COVID-19 in the patients infected with SARS-CoV2 [48]. There are many pieces of literature already available for the use of CT scanning in the case of COVID-19 [49, 50]. A common visualization of CT scanning shows the bilateral pulmonary parenchymal ground-glass and consolidative pulmonary opacities, sometimes with a rounded morphology and a peripheral lung distribution. Lung affected in the patients with SARS-CoV and MERS-CoV and the CT scanning of the chest reveals that the abnormalities progress along with the ground-glass opacities in the lung. A similar type of results is also seen in the case of SARS-CoV2 infections [51, 52]. These findings suggest that chest CT scanning can be a great help in the diagnosis of COVID-19 in the area with high prevalence. One of the major abnormalities of CT scanning is that it cannot differentiate between pneumonia caused due to other viruses and hysteresis of abnormal CT imaging.
Summary and future prospects
Presently the detection and diagnosis of SARS-CoV-2 mainly work through the viral ssRNA genome detection of coronavirus. In the diagnosis of a disease, the selection of a diagnostic method plays a very significant role. All the methods and tools discussed have their advantages and disadvantages (Table 1). A very high specific and sensitive method for the detection of the virus is through PCR methods, but its drawback is it needs a sophisticated instrument, costly reagents, well-established laboratory, and trained analysts. LAMP is another ultrasensitive amplification method for a nucleic acid that can detect a very small amount of DNA or RNA in less than an hour time but its drawback is the requirement of high temperature. In the case of microarray, its cost limits its application commonly in the detection of SARS-CoV-2. Hence there is still a requirement of cost-efficient COVID-19 detection method which can be used very easily in a cost-efficient and less time along with high sensitivity and specificity.
Table 1.
Comparison of various detection techniques for SARS-CoV2
| Detection methods | Detecting material | Advantages | Disadvantages |
|---|---|---|---|
| A. Nucleic acid detection-based technology | |||
| High throughput sequencing | Nucleic acid |
• Precise and sensitive • Not subject to cross-hybridization, and hence high accuracy • Larger dynamic range (> 105) |
• High cost • Require sequencer |
| PCR based methods (RT-PCR; RT-qPCR) | Viral RNA/mRNA |
• Detect virus directly • Highly accurate and sensitive • RT-qPCR is gold standard (96–100% specificity) • Time reuired: 2–4 h |
• High cost • False positive result possible |
| B. Microarrays based methodologies | |||
| Microarray |
• Relatively low cost • Well defined protocol and SOP |
• Small dynamic range (102) • Relies on hybridization which is non specific |
|
| C. Isothermal nucleic acid-based amplification | |||
| Regular loop-mediated isothermal amplification-based methods | DNA/RNA |
• High amount of DNA produced compared to PCR • Simple, Low cost • No requirement of thermocyclers • 99% specificity, Time required 15–60 min |
• Detect total DNA amplification in a reaction and thus limited to detection in a single target |
| Sequence-specific loop-mediated isothermal amplification methods | DNA/RNA |
• High amount of DNA produced compared to PCR • Simple, Low cost • High sensitive • No requirement of thermocyclers |
• Detect total DNA amplification in a reaction and thus limited to detection in a single target |
| D. CRISPR based methods | |||
| CRISPR based technology | ssDNA/ssRNA | • Rapid and quantitative detection of SARS-CoV2 | • Off target effect and imprecise effect |
| E. Antigen–antibody based methods | |||
| Rapid antigen test (RAT) | Nucleocapsid protein as antigen |
• Sensitive and specific • Easy handling • No requirement of any sophisticated instruments • Rapid detection efficacy • Cost-effective |
• Can be detected only after 7–9 days of infection • Antigenic variations make it difficult to generate similar antibodies |
| F. CT scan and other diagnostic methodology | |||
| CT scan | NA | • Detect the severity of the COVID-19 |
• Not specific but sensitive • Not a confirmatory test • Can be auxillary test |
Compliance with ethical standards
Conflict of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper. The authors declare that there is no conflict of interest.
Research involving human participants and/or animals
The work submitted under this manuscript doesn’t include any human participation or any kind of animal studies.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Su S, Wong G, Shi W, Liu J, Lai AC, Zhou J, et al. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. 2016;24:490–502. doi: 10.1016/j.tim.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Perlman S, Netland J. Coronaviruses post-SARS: update on replication and pathogenesis. Nat Rev Microbiol. 2009;7:439–450. doi: 10.1038/nrmicro2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang P, Wang H, Cao Z, Jin H, Chi H, Zhao J, et al. A rapid and specific assay for the detection of MERS-CoV. Front Microbiol. 2018;9:1101. doi: 10.3389/fmicb.2018.01101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jiang S, Xia S, Ying T, Lu L. A novel coronavirus (2019-nCoV) causing pneumonia-associated respiratory syndrome. Cell Mol Immunol. 2020;17:554. doi: 10.1038/s41423-020-0372-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhao S, Lin Q, Ran J, Musa SS, Yang G, Wang W, et al. Preliminary estimation of the basic reproduction number of novel coronavirus (2019-nCoV) in China, from 2019 to 2020: a data-driven analysis in the early phase of the outbreak. Int J Infect Dis. 2020;92:214–217. doi: 10.1016/j.ijid.2020.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou P, Yang X-L, Wang X-G, Hu B, Zhang L, Zhang W, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Corman VM, Landt O, Kaiser M, Molenkamp R, Meijer A, Chu DK, et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance. 2020;25:2000045. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Balboni A, Gallina L, Palladini A, Prosperi S, Battilani M. A real-time PCR assay for bat SARS-like coronavirus detection and its application to Italian greater horseshoe bat faecal sample surveys. Sci World J. 2012 doi: 10.1100/2012/989514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Uhlenhaut C, Cohen JI, Pavletic S, Illei G, Gea-Banacloche JC, Abu-Asab M, et al. Use of a novel virus detection assay to identify coronavirus HKU1 in the lungs of a hematopoietic stem cell transplant recipient with fatal pneumonia. Transpl Infect Dis. 2012;14:79–85. doi: 10.1111/j.1399-3062.2011.00657.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Adachi D, Johnson G, Draker R, Ayers M, Mazzulli T, Talbot P, et al. Comprehensive detection and identification of human coronaviruses, including the SARS-associated coronavirus, with a single RT-PCR assay. J Virol Methods. 2004;122:29–36. doi: 10.1016/j.jviromet.2004.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Setianingsih TY, Wiyatno A, Hartono TS, Hindawati E, Dewantari AK, Myint KS, et al. Detection of multiple viral sequences in the respiratory tract samples of suspected Middle East respiratory syndrome coronavirus patients in Jakarta, Indonesia 2015–2016. Int J Infect Dis. 2019;86:102–107. doi: 10.1016/j.ijid.2019.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wan Z, Zhang Y, He Z, Liu J, Lan K, Hu Y, et al. A melting curve-based multiplex RT-qPCR assay for simultaneous detection of four human coronaviruses. Int J Mol Sci. 2016;17:1880. doi: 10.3390/ijms17111880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Noh JY, Yoon S-W, Kim D-J, Lee M-S, Kim J-H, Na W, et al. Simultaneous detection of severe acute respiratory syndrome, Middle East respiratory syndrome, and related bat coronaviruses by real-time reverse transcription PCR. Arch Virol. 2017;162:1617–1623. doi: 10.1007/s00705-017-3281-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Corman V, Eckerle I, Bleicker T, Zaki A, Landt O, Eschbach-Bludau M, et al. Detection of a novel human coronavirus by real-time reverse-transcription polymerase chain reaction. Eurosurveillance. 2012;17:20285. doi: 10.2807/ese.17.39.20285-en. [DOI] [PubMed] [Google Scholar]
- 15.Lu X, Whitaker B, Sakthivel SKK, Kamili S, Rose LE, Lowe L, et al. Real-time reverse transcription-PCR assay panel for Middle East respiratory syndrome coronavirus. J Clin Microbiol. 2014;52:67–75. doi: 10.1128/JCM.02533-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Elden LJ, Anton MAM, van Alphen F, Hendriksen KA, Hoepelman AI, van Kraaij MG, et al. Frequent detection of human coronaviruses in clinical specimens from patients with respiratory tract infection by use of a novel real-time reverse-transcriptase polymerase chain reaction. J Infect Dis. 2004;189:652–657. doi: 10.1086/381207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yip SP, To SST, Leung PH, Cheung TS, Cheng PK, Lim WW. Use of dual TaqMan probes to increase the sensitivity of 1-step quantitative reverse transcription-PCR: application to the detection of SARS coronavirus. Clin Chem. 2005;51:1885–1888. doi: 10.1373/clinchem.2005.054106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu X-F, Pan J-C, Ye R, Xiang H-Q, Kou Y, Huang Z-C. Preparation of armored RNA as a control for multiplex real-time reverse transcription-PCR detection of influenza virus and severe acute respiratory syndrome coronavirus. J Clin Microbiol. 2008;46:837–841. doi: 10.1128/JCM.01904-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hadjinicolaou AV, Farcas GA, Demetriou VL, Mazzulli T, Poutanen SM, Willey BM, et al. Development of a molecular-beacon-based multi-allelic real-time RT-PCR assay for the detection of human coronavirus causing severe acute respiratory syndrome (SARS-CoV): a general methodology for detecting rapidly mutating viruses. Arch Virol. 2011;156:671–680. doi: 10.1007/s00705-010-0906-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bustin SA, Nolan T. RT-qPCR Testing of SARS-CoV-2: A Primer. Int J Mol Sci. 2020;21(8):3004. doi: 10.3390/ijms21083004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Woo PC, Lau SK, Wong BH, Tsoi H-w, Fung AM, Kao RY, et al. Differential sensitivities of severe acute respiratory syndrome (SARS) coronavirus spike polypeptide enzyme-linked immunosorbent assay (ELISA) and SARS coronavirus nucleocapsid protein ELISA for serodiagnosis of SARS coronavirus pneumonia. J Clin Microbiol. 2005;43:3054–3058. doi: 10.1128/JCM.43.7.3054-3058.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xie X, Zhong Z, Zhao W, Zheng C, Wang F, Liu J. Chest CT for typical 2019-nCoV pneumonia: relationship to negative RT-PCR testing. Radiology. 2020 doi: 10.1148/radiol.2020200343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yam W, Chan K, Poon L, Guan Y, Yuen K, Seto W, et al. Evaluation of reverse transcription-PCR assays for rapid diagnosis of severe acute respiratory syndrome associated with a novel coronavirus. J Clin Microbiol. 2003;41:4521–4524. doi: 10.1128/JCM.41.10.4521-4524.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen Q, Li J, Deng Z, Xiong W, Wang Q, Hu Y-q. Comprehensive detection and identification of seven animal coronaviruses and human respiratory coronavirus 229E with a microarray hybridization assay. Intervirology. 2010;53:95–104. doi: 10.1159/000264199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shi R, Ma W, Wu Q, Zhang B, Song Y, Guo Q, et al. Design and application of 60mer oligonucleotide microarray in SARS coronavirus detection. Chin Sci Bull. 2003;48:1165–1169. doi: 10.1007/BF03183928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guo X, Geng P, Wang Q, Cao B, Liu B. Development of a single nucleotide polymorphism DNA microarray for the detection and genotyping of the SARS coronavirus. J Microbiol Biotechnol. 2014;24:1445–1454. doi: 10.4014/jmb.1404.04024. [DOI] [PubMed] [Google Scholar]
- 27.de Souza Luna LK, Heiser V, Regamey N, Panning M, Drexler JF, Mulangu S, et al. Generic detection of coronaviruses and differentiation at the prototype strain level by reverse transcription-PCR and nonfluorescent low-density microarray. J Clin Microbiol. 2007;45:1049–1052. doi: 10.1128/JCM.02426-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hardick J, Metzgar D, Risen L, Myers C, Balansay M, Malcom T, et al. Initial performance evaluation of a spotted array Mobile Analysis Platform (MAP) for the detection of influenza A/B, RSV, and MERS coronavirus. Diagn Microbiol Infect Dis. 2018;91:245–247. doi: 10.1016/j.diagmicrobio.2018.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao Y, Chen F, Li Q, Wang L, Fan C. Isothermal amplification of nucleic acids. Chem Rev. 2015;115:12491–12545. doi: 10.1021/acs.chemrev.5b00428. [DOI] [PubMed] [Google Scholar]
- 30.Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N, et al. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28:E63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nagamine K, Hase T, Notomi T. Accelerated reaction by loop-mediated isothermal amplification using loop primers. Mol Cell Probes. 2002;16:223–229. doi: 10.1006/mcpr.2002.0415. [DOI] [PubMed] [Google Scholar]
- 32.Mori Y, Nagamine K, Tomita N, Notomi T. Detection of loop-mediated isothermal amplification reaction by turbidity derived from magnesium pyrophosphate formation. Biochem Biophys Res Commun. 2001;289:150–154. doi: 10.1006/bbrc.2001.5921. [DOI] [PubMed] [Google Scholar]
- 33.Ponaka C, Curioso C, Patel D, Elagin S, Slepnev V, Lucchi NW et al (2015) Detection of plasmodium parasites with loop mediated isothermal amplification (LAMP) using simple sample preparation methods (poster ID25). In: Association for molecular pathology (AMP) 2015 Annual Meeting Austin, Tx, USA
- 34.Geojith G, Dhanasekaran S, Chandran SP, Kenneth J. Efficacy of loop mediated isothermal amplification (LAMP) assay for the laboratory identification of Mycobacterium tuberculosis isolates in a resource limited setting. J Microbiol Methods. 2011;84:71–73. doi: 10.1016/j.mimet.2010.10.015. [DOI] [PubMed] [Google Scholar]
- 35.Njiru Z, Mikosza A, Matovu E, Enyaru J, Ouma J, Kibona S, et al. African trypanosomiasis: sensitive and rapid detection of the sub-genus Trypanozoon by loop-mediated isothermal amplification (LAMP) of parasite DNA. Int J Parasitol. 2008;38:589–599. doi: 10.1016/j.ijpara.2007.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Enosawa M, Kageyama S, Sawai K, Watanabe K, Notomi T, Onoe S, et al. Use of loop-mediated isothermal amplification of the IS900 sequence for rapid detection of cultured Mycobacterium avium subsp. paratuberculosis. J Clin Microbiol. 2003;41:4359–4365. doi: 10.1128/JCM.41.9.4359-4365.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Poon LL, Leung CS, Tashiro M, Chan KH, Wong BW, Yuen KY, et al. Rapid detection of the severe acute respiratory syndrome (SARS) coronavirus by a loop-mediated isothermal amplification assay. Clin Chem. 2004;50:1050–1052. doi: 10.1373/clinchem.2004.032011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pyrc K, Milewska A, Potempa J. Development of loop-mediated isothermal amplification assay for detection of human coronavirus-NL63. J Virol Methods. 2011;175:133–136. doi: 10.1016/j.jviromet.2011.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shirato K, Yano T, Senba S, Akachi S, Kobayashi T, Nishinaka T, et al. Detection of Middle East respiratory syndrome coronavirus using reverse transcription loop-mediated isothermal amplification (RT-LAMP) Virol J. 2014;11:139. doi: 10.1186/1743-422X-11-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thai HTC, Le MQ, Vuong CD, Parida M, Minekawa H, Notomi T, et al. Development and evaluation of a novel loop-mediated isothermal amplification method for rapid detection of severe acute respiratory syndrome coronavirus. J Clin Microbiol. 2004;42:1956–1961. doi: 10.1128/JCM.42.5.1956-1961.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Njiru ZK. Loop-mediated isothermal amplification technology: towards point of care diagnostics. PLOS Negl Trop Dis. 2012;6:e1572. doi: 10.1371/journal.pntd.0001572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shirato K, Semba S, El-Kafrawy SA, Hassan AM, Tolah AM, Takayama I, et al. Development of fluorescent reverse transcription loop-mediated isothermal amplification (RT-LAMP) using quenching probes for the detection of the Middle East respiratory syndrome coronavirus. J Virol Methods. 2018;258:41–48. doi: 10.1016/j.jviromet.2018.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cai S, Jung C, Bhadra S, Ellington AD. Phosphorothioated primers lead to loop-mediated isothermal amplification at low temperatures. Anal Chem. 2018;90:8290–8294. doi: 10.1021/acs.analchem.8b02062. [DOI] [PubMed] [Google Scholar]
- 44.Wright AV, Nuñez JK, Doudna JA. Biology and applications of CRISPR systems: harnessing nature’s toolbox for genome engineering. Cell. 2016;164:29–44. doi: 10.1016/j.cell.2015.12.035. [DOI] [PubMed] [Google Scholar]
- 45.Gootenberg JS, Abudayyeh OO, Lee JW, Essletzbichler P, Dy AJ, Joung J, et al. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science. 2017;356:438–442. doi: 10.1126/science.aam9321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Freije CA, Myhrvold C, Boehm CK, Lin AE, Welch NL, Carter A, et al. Programmable inhibition and detection of RNA viruses using Cas13. Mol Cell. 2019;76(826–37):e11. doi: 10.1016/j.molcel.2019.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gootenberg JS, Abudayyeh OO, Kellner MJ, Joung J, Collins JJ, Zhang F. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science. 2018;360:439–444. doi: 10.1126/science.aaq0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pan Y, Guan H, Zhou S, Wang Y, Li Q, Zhu T, et al. Initial CT findings and temporal changes in patients with the novel coronavirus pneumonia (2019-nCoV): a study of 63 patients in Wuhan, China. Eur Radiol. 2020 doi: 10.1007/s00330-020-06731-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shi H, Han X, Zheng C. Evolution of CT manifestations in a patient recovered from 2019 novel coronavirus (2019-nCoV) pneumonia in Wuhan, China. Radiology. 2020;295:20. doi: 10.1148/radiol.2020200269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chung M, Bernheim A, Mei X, Zhang N, Huang M, Zeng X, et al. CT imaging features of 2019 novel coronavirus (2019-nCoV) Radiology. 2020;295:202–207. doi: 10.1148/radiol.2020200230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ooi GC, Khong PL, Müller NL, Yiu WC, Zhou LJ, Ho JCM, et al. Severe acute respiratory syndrome: temporal lung changes at thin-section CT in 30 patients. Radiology. 2004;230:836–844. doi: 10.1148/radiol.2303030853. [DOI] [PubMed] [Google Scholar]
- 52.Ajlan AM, Ahyad RA, Jamjoom LG, Alharthy A, Madani TA. Middle East Respiratory Syndrome Coronavirus (MERS-CoV) infection: chest CT findings. Am J Roentgenol. 2014;203:782–787. doi: 10.2214/AJR.14.13021. [DOI] [PubMed] [Google Scholar]