Abstract
Context
Approximately 70% of women report experiencing vasomotor symptoms (VMS, hot flashes and/or night sweats). The etiology of VMS is not clearly understood but may include genetic factors.
Evidence Acquisition
We searched PubMed and Embase in accordance with the Preferred Reporting Items for Systematic Reviews and Meta-Analyses guidance. We included studies on associations between genetic variation and VMS. We excluded studies focused on medication interventions or prevention or treatment of breast cancer.
Evidence Synthesis
Of 202 unique citations, 18 citations met the inclusion criteria. Study sample sizes ranged from 51 to 17 695. Eleven of the 18 studies had fewer than 500 participants; 2 studies had 1000 or more. Overall, statistically significant associations with VMS were found for variants in 14 of the 26 genes assessed in candidate gene studies. The cytochrome P450 family 1 subfamily A member 1 (CYP1B1) gene was the focus of the largest number (n = 7) of studies, but strength and statistical significance of associations of CYP1B1 variants with VMS were inconsistent. A genome-wide association study reported statistically significant associations between 14 single-nucleotide variants in the tachykinin receptor 3 gene and VMS. Heterogeneity across trials regarding VMS measurement methods and effect measures precluded quantitative meta-analysis; there were few studies of each specific genetic variant.
Conclusions
Genetic variants are associated with VMS. The associations are not limited to variations in sex-steroid metabolism genes. However, studies were few and future studies are needed to confirm and extend these findings.
Keywords: hot flashes, night sweats, menopause, vasomotor, gene, genome-wide association study
Approximately 70% of midlife women experience vasomotor symptoms ([VMS], hot flashes and/or night sweats) (1). VMS are typically experienced as episodes of heat accompanied by sweating and flushing, particularly about the head, neck, chest, and upper back. For many women, VMS persist for more than a decade (2).
Despite the high prevalence of VMS, the physiology underlying VMS is not fully understood. The cytochrome P450 enzymes are involved in estrogen biosynthesis and metabolism, and there are known genetic variants in the genes encoding those enzymes (3). Sex-steroid hormone levels are associated with VMS reporting, yet their association with VMS is modest (4), and other mechanisms are known to be important to the physiology of VMS. Thermoregulatory mechanisms appear to have a role in VMS, whereby heat dissipation results from a narrowing of the thermoneutral zone, the zone in which core body temperature is maintained without triggering thermoregulatory homeostatic mechanisms (sweating or shivering) (5). Administration of estradiol to postmenopausal women with VMS reduces VMS and widens the thermoneutral zone (6). Other systems, including the sympathetic and parasympathetic systems, have been implicated in the etiology of VMS (7). Animal studies and recent clinical trials suggest that the neurokinin B pathway may play a role in the etiology of VMS (8-10).
In the United States, the prevalence, persistence, and severity of VMS is highest among African American women, lowest among Asian women, and intermediate among Hispanic and non-Hispanic white women (1). Therefore, in addition to the physiologic systems mentioned previously, the racial/ethnic patterns suggest the possibility of genetic variation as one potential mechanism involved in VMS etiology. The goal of this systematic review was to investigate the association between genetic variation and VMS in women. We hypothesized that specific genetic variants would be associated with VMS, and that these genetic variants are involved in sex-steroid metabolism pathways as well as other physiologic pathways.
Materials and Methods
This systematic review was conducted in accordance with PRISMA (Preferred Reporting Items for Systematic Reviews and Meta-Analysis) guidance (11). We addressed the following key question: What is the association between genetic variation and VMS in women? The study protocol was submitted to the PROSPERO international prospective register of systematic reviews (https://www.crd.york.ac.uk/prospero/) April 2, 2020 (identification number 17800).
We included studies on associations between genetic variation (candidate gene studies or genome-wide association studies [GWAS]) and vasomotor symptoms in women. We did not apply any restrictions on participant age or menopausal status. We excluded studies focused on medication interventions (e.g. hormone therapy) or prevention or treatment of cancer, editorials, and review articles.
We searched PubMed (1966-present) and Embase (1947-present) April 17, 2020, using the search terms in Table 1.
Table 1.
Search terms for literature searchers (April 17, 2020)
| Search terms for PubMed | No. of citations retrieved |
|---|---|
| Candidate gene AND “menopausal symptoms” | 2 |
| Candidate gene AND hot flashes | 4 |
| Candidate gene AND hot flushes | 5 |
| Candidate gene AND vasomotor | 11 |
| Genetic variation AND “menopausal symptoms” | 42 |
| Genetic variation AND hot flashes | 49 |
| Genetic variation AND hot flushes | 54 |
| Genome wide association study AND hot flashes | 1 |
| Genome wide association study AND hot flushes | 1 |
| Genome wide association study AND menopausal symptoms | 69 |
| Genome wide association study AND vasomotor | 3 |
| GWAS AND “menopausal symptoms” | 1 |
| GWAS AND hot flashes | 1 |
| GWAS AND hot flushes | 1 |
| GWAS AND vasomotor | 3 |
| Total number of citations retrieved | 247 |
| After exclusion of duplicate citations | 170 |
| Search terms for Embase, limited to human studies | No. of citations retrieved |
| “Candidate gene” AND “hot flashes” | 3 |
| “Candidate gene” AND “hot flushes” | 0 |
| “Candidate gene” AND “menopausal symptoms” | 1 |
| “Candidate gene” AND vasomotor | 6 |
| “Genetic variation” AND “hot flashes” | 22 |
| “Genetic variation” AND “hot flushes” | 3 |
| “Genetic variation” AND “menopausal symptoms” | 5 |
| “Genetic variation” AND vasomotor | 10 |
| “Genome wide association study” AND “hot flashes” | 4 |
| “Genome wide association study” AND “hot flushes” | 0 |
| “Genome wide association study” AND “menopausal symptoms” | 0 |
| “Genome wide association study” AND vasomotor | 7 |
| GWAS AND “hot flashes” | 4 |
| GWAS AND “hot flushes” | 1 |
| GWAS AND “menopausal symptoms” | 1 |
| GWAS AND vasomotor | 4 |
| Total number of citations retrieved | 71 |
| After exclusion of duplicate citations | 44 |
| After exclusion of citations also found on PubMed | 44 – 12 = 32 |
Abbreviation: GWAS, genome-wide association study.
Reference lists of all relevant citations were manually reviewed for additional relevant citations.
Retrieved citations were independently screened in duplicate (by authors C.J.C. and A.L.D.) for inclusion. Any disagreements regarding inclusion were resolved by discussion between the 2 screeners.
Study quality was rated using the QUIPS (Quality in Prognostic Factor Studies) tool (12, 13). Results of retrieved studies were not quantitatively summarized across studies (eg, by risk ratio, difference in means) because there were too few studies to allow meta-analysis. Therefore, results of each study are described individually. In extracting results, we retained the original term used to describe VMS (“hot flashes,” “hot flushes,” “night sweats,” “vasomotor symptoms”) from each study.
We made the a priori decision to summarize results of candidate gene studies separately from those of GWAS.
Results
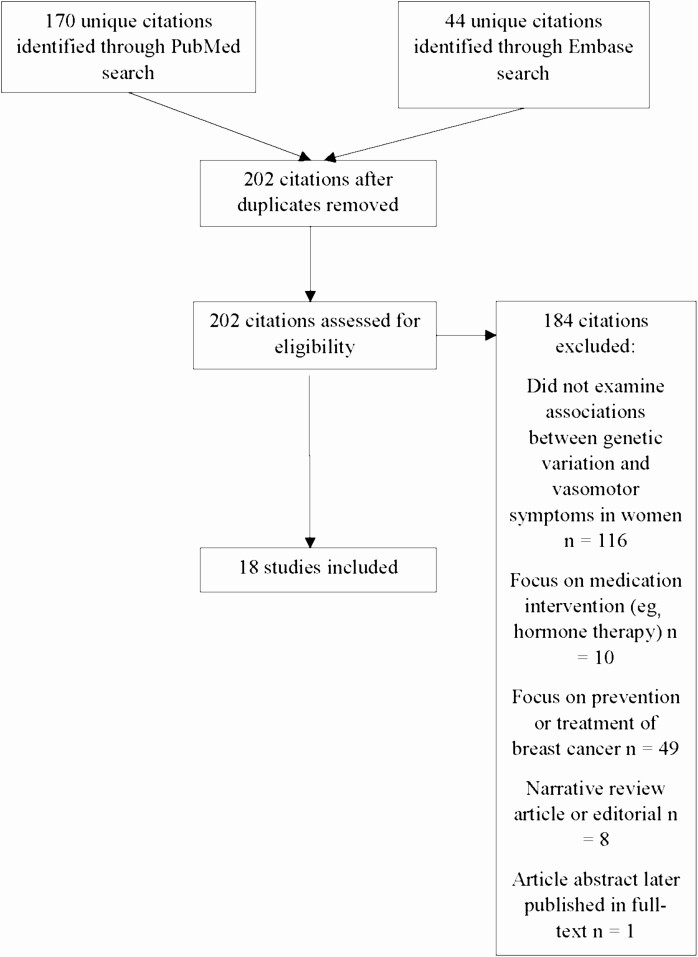
We identified 170 citations in the PubMed search and 44 citations through the Embase search, resulting in 202 citations after exclusion of duplicates (Fig. 1). Of these 202 citations, 116 were excluded because they did not examine associations between genetic variation and VMS, 10 were excluded because they focused on medication interventions (eg, hormone therapy), 49 were excluded because they focused on prevention or treatment of cancer, 8 were excluded because they were reviews or editorials, and 1 citation was excluded because it was an abstract of a study that was subsequently published in full-text form. No additional relevant citations were found from manual review of references lists of the included studies. Therefore, 18 studies met inclusion criteria (Table 2 lists PubMed citations and Table 3 lists Embase citations) (3, 14-30). Of these 18 studies, some studies were performed using data from the same study population (2 from the Penn Ovarian Aging Study (3, 20), 4 from the Baltimore Midlife Health Study (14, 17, 27, 28), 2 from the Seattle Midlife Women’s Study (15, 16), and 2 studies from Slovakia (21, 29).
Figure 1.
Article flow.
Table 2.
Citations retrieved in PubMed searches
| First author and year | Inclusion or reason for exclusion | Citation |
|---|---|---|
| 1.Abubakar 2014 | BC | (31) |
| 2.Aguilar-Zavala 2012 | INCLUDE | (30) |
| 3.Atkinson 2015 | EO | (32) |
| 4.Bacon 2019 | EO | (33) |
| 5.Bahl 2015 | EO | (34) |
| 6.Baker 2012 | EO | (35) |
| 7.Barber 2013 | EO | (36) |
| 8.Barrdahl 2014 | BC | (37) |
| 9.Bechlioulis 2012 | MED | (38) |
| 10.Birrer 2018 | EO | (39) |
| 11.Bonanni 2006 | BC | (40) |
| 12.Bonfa 2015 | EO | (41) |
| 13.Brand 2015 | EO | (42) |
| 14.Brauch and Jordan 2014 | BC | (43) |
| 15.Brauch and Murdter 2009 | BC | (44) |
| 16.Brooks 2012 | EO | (45) |
| 17.Butts 2014 | EO | (46) |
| 18.Butts 2012 | INCLUDE | (20) |
| 19.Campa 2011 | EO | (47) |
| 20.Campa 2010 | EO | (48) |
| 21.Campfield 2011 | EO | (49) |
| 22.Candrakova 2018 | INCLUDE | (21) |
| 23.Cao 2019 | EO | (50) |
| 24.Chan 2015 | EO | (51) |
| 25.Chang 2018 | MED | (52) |
| 26.Chapman 2011 | BC | (53) |
| 27.Chiang 2012 | EO | (54) |
| 28.Cho 2018 | EO | (55) |
| 29.Crandall 2017 | INCLUDE | (22) |
| 30.Crandall 2006 | INCLUDE | (23) |
| 31.D’Alonzo 2018 | BC | (56) |
| 32.Dagan 2009 | BC | (57) |
| 33.Day 2018 | EO | (58) |
| 34.De Rooij 2010 | EO | (59) |
| 35.Dezentje 2014 | BC | (60) |
| 36.Didriksen 2013 | EO | (61) |
| 37.Domchek 2007 | BC | (62) |
| 38.Domchek 2006 | BC | (63) |
| 39.Dorjgochoo 2012 | EO | (64) |
| 40.Edwards 2019 | EO | (65) |
| 41.Elands 2017 | EO | (66) |
| 42.Essemine 2011 | EO | (67) |
| 43.Federici 2016 | EO | (68) |
| 44.Fehringer 2010 | EO | (69) |
| 45.Fernandez-Navarro 2013 | EO | (70) |
| 46.Finch 2012 | BC | (71) |
| 47.Finch and Narod 2011 | BC | (72) |
| 48.Finch, Metcalf 2011 | BC | (73) |
| 49.Fox, Heard-Costa 2007 | EO | (74) |
| 50.Fujita 2014 | EO | (75) |
| 51.Gabriel 2009 | EO | (76) |
| 52.Gadduci 2010 | BC | (77) |
| 53.Gao 2016 | BC | (78) |
| 54.Garber 2005 | EO | (79) |
| 55.Goetz 2005 | BC | (80) |
| 56.Goto 2018 | EO | (81) |
| 57.Greenwood 2011 | EO | (82) |
| 58.Guidozzi 2016 | BC | (83) |
| 59.Guo 2016 | EO | (84) |
| 60.Hall 2019 | EO | (85) |
| 61.Harge 2009 | EO | (86) |
| 62.Harvey 2015 | EO | (87) |
| 63.He 2010 | EO | (88) |
| 64.He 2013 | EO | (89) |
| 65.Hein 2013 | BC | (90) |
| 66.Henry 2009 | BC | (91) |
| 67.Higgins 2011 | BC | (92) |
| 68.Higgins 2010 | BC | (93) |
| 69.Hoeijmakers 2012 | EO | (94) |
| 70.Ingle 2010 | BC | (95) |
| 71.Intharuksa 2020 | EO | (96) |
| 72.Irarrazaval 2011 | BC | (97) |
| 73.Ishiguro 2019 | BC | (98) |
| 74.Jansen 2018 | BC | (99) |
| 75.Jin 2015 | EO | (100) |
| 76.Johansson 2016 | BC | (101) |
| 77.Jung, Mancuso 2019 PLoS One | EO | (102) |
| 78.Jung Mancuro 2019 Cancer Prev Res (Phila) | EO | (103) |
| 79.Jung 2018 | EO | (104) |
| 80.Jung 2017 | EO | (105) |
| 81.Justenhoven 2012 | BC | (106) |
| 82.Kawase 2009 | EO | (107) |
| 83.Kim 2016 | REV | (108) |
| 84.Koller 2010 | EO | (109) |
| 85.Komm 2001 | REV | (110) |
| 86.Lapid 2014 | EO | (111) |
| 87.Lee 2016 | EO | (112) |
| 88.Leyland-Jones 2015 | BC | (113) |
| 89.Li 2011 | EO | (114) |
| 90.Li 2019 | REV | (115) |
| 91.Lim 2012 | EO | (116) |
| 92.Lintermans 2016 | BC | (117) |
| 93.Luptakova Sivakova 2012 Menopause | EO | (29) |
| 94.Luptakova Sivakova 2012 Anthropol Anz | INCLUDE | (118) |
| 95.Mai 2017 | EO | (119) |
| 96.Malacara 2004 | INCLUDE | (25) |
| 97.Mallin 2008 | EO | (120) |
| 98.Mariapun 2016 | EO | (121) |
| 99.Markus 1998 | EO | (122) |
| 100.Massad-Costa 2008 | INCLUDE | (26) |
| 101.Matchkov 2010 | EO | (123) |
| 102.Mirhaegue 2005 | EO | (124) |
| 103.Mizutani 2002 | EO | (125) |
| 104.Montasser 2015 | INCLUDE | (27) |
| 105.Moriwaki 2017 | EO | (126) |
| 106.Moyer 2018 | MED | (127) |
| 107.Moyer 2016 | MED | (128) |
| 108.Mendez 2013 | EO | (129) |
| 109.Nachtigall 2011 | EO | (130) |
| 110.Niederhofer 2007 | REV | (131) |
| 111.Nogueira 2011 | MED | (132) |
| 112.O’Brien 2016 | BC | (133) |
| 113.Ochs-Balcom 2018 | EO | (134) |
| 114.Ohshima 2011 | EO | (135) |
| 115.Passarelli 2011 | EO | (136) |
| 116.Pausova 2009 | EO | (137) |
| 117.Petherick 2012 | EO | (138) |
| 118.Pezaro 2012 | EO | (139) |
| 119.Pilling 2017 | EO | (140) |
| 120.Prentice 2009 | BC | (141) |
| 121.Pru 2014 | EO | (142) |
| 122.Pulit 2017 | EO | (143) |
| 123.Qin 2012 | EO | (144) |
| 124.Qin 2013 | BC | (145) |
| 125.Rask-Anderson 2017 | EO | (146) |
| 126.Rebbeck 2010 | INCLUDE | (3) |
| 127.Regan 2012 | BC | (147) |
| 128.Reid 2018 | BC | (148) |
| 129.Rojas-Roldan 2014 | EO | (149) |
| 130.Rudolph 2015 | EO | (150) |
| 131.Rudolph 2013 | BC | (151) |
| 132.Saskova | BC | (152) |
| 133.Schilling 2007 | INCLUDE | (28) |
| 134.Schneider 2009 | INCLUDE | (19) |
| 135.Schneider 2003 | REV | (153) |
| 136.Schogor 2014 | EO | (154) |
| 137.Sestak 2012 | BC | (155) |
| 138.Shang 2013 | EO | (156) |
| 139.Shigehiro 2016 | EO | (157) |
| 140.Singh 2011 | BC | (158) |
| 141.Somekawa 1998 | MED | (159) |
| 142.Takeo 2005 | INCLUDE | (18) |
| 143.Tamimi 2008 | EO | (160) |
| 144.Tenenbaum-Rakover 2015 | EO | (161) |
| 145.Thomin 2014 | BC | (162) |
| 146.Thompson 2016 | EO | (163) |
| 147.Toth 2011 | EO | (164) |
| 148.Ushiroyama 2001 | EO | (165) |
| 149.Varghese 2012 | EO | (166) |
| 150.Velez Edwards 2013 | EO | (167) |
| 151.Visvanathan 2005 | INCLUDE | (17) |
| 152.Warran Endersen 2014 | EO | (168) |
| 153.Warren Andersen 2013 | EO | (169) |
| 154.Wise 2012 | EO | (170) |
| 155.Woad 2006 | EO | (171) |
| 156.Woods 2018 | INCLUDE | (16) |
| 157.Woods 2006 | INCLUDE | (15) |
| 158.Woyka 2014 | EO | (172) |
| 159.Yan 2009 | EO | (173) |
| 160.Yang 2004 | EO | (174) |
| 161.Younis 2012 | EO | (175) |
| 162.Zaffanello 2011 | EO | (176) |
| 163.Zeller 2008 | EO | (177) |
| 164.Zembutsu 2017 | BC | (178) |
| 165.Zhang 2014 | EO | (179) |
| 166.Zhang 2013 | EO | (180) |
| 167.Zhao 2011 | EO | (181) |
| 168.Zingue 2016 | EO | (182) |
| 169.Ziv-Gal 2012 | INCLUDE | (14) |
| 170.Zig-Gal 2010 | REV | (183) |
Abbreviations: EO, excluded outcome (did not focus on association between genetic variation and hot flashes or vasomotor symptoms); MED, medication intervention trial (eg, hormone therapy); BC, breast cancer prevention or treatment trial; REV, review or editorial (narrative reviews were excluded; systematic reviews published more than 10 years ago were excluded).
Table 3.
Citations retrieved in Embase searches
| First author and year | Inclusion or reason for exclusion | Citation | Duplicate found in PubMed |
|---|---|---|---|
| 1.Brown 2016 | EO | (184) | |
| 2.Butts 2012 | INCLUDE | (185) | X |
| 3.Cerril 2007 | BC | (186) | |
| 4.Chae 2006 | EO | (187) | |
| 5.Chollet 2017 | EO | (188) | |
| 6.Crandall 2015 | INCLUDE but published as full-text in reference (22) | (189) | |
| 7.Crandall 2017 | INCLUDE | (22) | X |
| 8.Depypere 2017 | MED | (190) | |
| 9.Dern 1947 | EO | (191) | |
| 10.Fraser 2017 | MED | (192) | |
| 11.Fujita 2007 | EO | (193) | |
| 12.Hartmaier 2009 | BC | (194) | |
| 13.Hayes 2017 | MED | (195) | |
| 14.He 2013 | EO | (89) | X |
| 15.Hertz 2017 | BC | (196) | |
| 16.Houtsma 2013 | BC | (197) | |
| 17.Ingle 2013 | BC | (198) | |
| 18.Jansen 2018 | BC | (99) | X |
| 19.Kapoor 2019 | INCLUDE | (24) | |
| 20.Kim 2016 | REV | (108) | X |
| 21.Lorenz 2010 | REV | (199) | |
| 22.Lu 2009 | EO | (200) | |
| 23.Luo 2019 | EO | (201) | |
| 24.Markus 1998 | EO | (122) | X |
| 25.Meirhaeghe 2005 | EO | (124) | |
| 26.Minoretti 2006 | EO | (202) | |
| 27.Mizutani 2002 | EO | (125) | X |
| 28.Moyer 2016 | MED | (128) | X |
| 29.Moyer 2018 | MED | (127) | X |
| 30.Moyer 2014 | MED | (203) | |
| 31.Murphy 2004 | EO | (204) | |
| 32.O’Sullivan 2018 | REV | (205) | |
| 33.Prague 2017 | EO | (8) | |
| 34.Rumianowski 2012 | EO | (206) | |
| 35.Waage 2018 | EO | (207) | |
| 36.Welzen 2015 | EO | (208) | |
| 37.Weng 2013 | EO | (209) | |
| 38.Woad 2006 | EO | (171) | X |
| 39.Wolff 1973 | EO | (210) | |
| 40.Yiannakopoulou 2012 | BC | (211) | |
| 41.Zaffanello 2011 | EO | (176) | X |
| 42.Zembutsu 2015 | BC | (212) | |
| 43.Zembutsu 2016 | BC | (213) | |
| 44.Zembutsu 2017 | BC | (178) | X |
Abbreviations: EO, excluded outcome (ie, did not focus on association between genetic variation and hot flashes or vasomotor symptoms); MED, medication intervention trial (eg, hormone therapy); BC, breast cancer prevention or treatment trial; REV, review or editorial (narrative reviews were excluded; systematic reviews published more than 10 years ago were excluded).
Study characteristics (“PICOS” characteristics, including participants, interventions, comparisons, outcomes, study design) of the 18 studies that met inclusion criteria are displayed in Table 4. Sixteen studies were cross-sectional (14-22, 24-28) and 2 studies were longitudinal (3, 23). Sample sizes ranged from 51 (18) to 17 695 participants (22). Eleven of the 16 studies had fewer than 500 participants. Twelve studies examined data from women in the United States (3, 14-17, 19, 20, 22-24, 27, 28); 6 examined data from women from other countries (18, 21, 25, 26, 29, 30). Although we excluded studies that were focused on women with cancer, we did include one study in which 20% of women were taking breast cancer hormonal therapy, and 15% were taking medications that could decrease hot flashes (eg, selective serotonin reuptake inhibitors, clonidine, and menopausal hormone therapy) (19).
Table 4.
Characteristics of studies that met inclusion criteria
| First author and publication year | No., race/ethnicity, location | Age | Study design | Citation | ||
|---|---|---|---|---|---|---|
| Range, y | Mean, y | Cross-sectional | Longitudinal | |||
| Aguilar-Zavala 2012 | 290 postmenopausal women from 3 cities in Mexico, race/ethnicity not specified | Unstated | 54 | X | (30) | |
| Butts 2012 | 157 EA; 139 AA in Philadelphia County (Penn Ovarian Aging Study) | 35-47 late reproductive age at enrollment, VMS assessed 11 y later | EA 51 (median); AA 51 (median) | X | (20) | |
| Candrakova 2018 | 367 women Western and Central Slovakia, race/ethnicity not specified | 40-60 | Premenopausal 45, perimenopausal 49, postmenopausal 54 | X | (21) | |
| Crandall 2017 | 17 695 postmenopausal women at 40 US clinical centers; EA 8185; AA 6732, Hispanic 2778 | 50-79 | 64 | X | (22) | |
| Crandall 2006 | 1467 premenopausal (55%) and perimenopausal (45%) women; 359 AA, 791 Caucasian, 151 Chinese, 166 Japanese women in Pittsburgh, PA; Boston, MA; Detroit, MI; Chicago, IL; Los Angeles, CA; Oakland, CA; and Jersey City, NJ | 42-52 | 46 | X | (23) | |
| Kapoor 2019 | 140 perimenopausal and postmenopausal women in the Women’s Health Clinic at Mayo Clinic in Rochester, MN, race/ethnicity and location of participants not stated | Unstated | Unstated | X | (24) | |
| Luptakova 2012 | 399 premenopausal, perimenopausal, or postmenopausal women from Western and Central Slovakia, race/ethnicity not specified | 39-60 | 53 postmenopausal, 46 premenopausal, and perimenopausal | X | (29) | |
| Malacara 2004 | 177 postmenopausal women living in Mexico; race and locations of participants not stated | Not stated | 53 (PvuII group), 53-54 (XbaI group)a | X | (25) | |
| Massad-Costa 2008 | 93 postmenopausal women, race and locations of participants not stated | Not stated | 53 | X | (26) | |
| Montasser 2015 | 788 EA, 206 AA premenopausal and perimenopausal Baltimore-area women | 45-54 | 48 | X | (27) | |
| Rebbeck 2010 | 436 premenopausal women (206 AA, 207 EA) from Penn Ovarian Aging Study, Philadelphia County | 35-47 | Unstated | X | (3) | |
| Schilling 2007 | 639 women, 532 white, 94 black in Baltimore | 45-54 | Unstated (413 aged 45-59, 226 aged 50-54) | X | (28) | |
| Schneider 2009 | 441 premenopausal and 533 postmenopausal Caucasian women, single site, in “Friends for Life” project, area of Indianapolis, IN, women with (520) and without (715) breast cancer, 20% taking breast cancer hormonal therapy, 15% taking medications that could decrease hot flashes (eg, selective serotonin reuptake inhibitors, clonidine, menopausal hormone therapy) | Unstated | Unstated | X | (19) | |
| Takeo 2005 | 51 postmenopausal women in Japan, single-site study, location and race/ethnicity of participants not specified | Unstated | Mean age 56 in group with extremely short (≤ 17) and 1 long (≥ 22 repeats) C-A allele; mean age 54 in group with 2 short (18-21) C-A repeat alleles; mean age 55 in group with 1 short and 1 long C-A repeat allele; mean age 56 in group with 2 long C-A repeat alleles | X | (18) | |
| Visvanathan 2005 | 354 women with hot flushes, 258 women without hot flushes in Midlife Health Study, Baltimore residents. 82% black and 18% AA among women with hot flushes; 87% white, 11% AA among women with hot flushes | 45-54 | 49 (women with hot flushes), 48y (women without hot flushes) | X | (17) | |
| Woods 2018 | 140 women, 3% AA, 9% Asian/Pacific Islander, 88% Caucasian, 1% Hispanic or mixed race/ethnicity, Seattle Midlife Women’s Health Study, locations of participants not further described | 35-55 | 41 | X | (16) | |
| Woods 2006 | 104 women (4% AA, 5% Asian American, 0% Hispanic, 89% white non-Hispanic), Seattle Midlife Women’s Health Study, locations of participants not further described | Unstated | 53 | X | (15) | |
| Ziv-Gal 2012 | 639 women (413 aged 45-49; 226 aged 50-54), 532 Caucasian, 94 AA, 11 other race), Baltimore city and surrounding counties | 45-54 | Unstated | X | (14) | |
Abbreviations: AA, African American; CA, California; C-A, cytosine-adenine; EA, European American; IL, Illinois; IN, Indiana; MA, Massachusetts; MI, Michigan; MN, Minnesota; PA, Pennsylvania; VMS, vasomotor symptoms.
a PvuII and XbaI are genetic variants in the estrogen receptor gene.
Measurement methods for VMS (any vs none, frequency, severity, and years of duration) in each study are described in Table 5. Some studies described hot flashes or hot flushes without night sweats, some described hot flashes and night sweats, some did not provide detailed information regarding how VMS were ascertained (21). Similarly, the time horizon over which VMS were assessed varied: within the past month, within the past 2 weeks, current vs not current, ever vs never, and sometimes time horizons over which VMS were ascertained were not described (24, 25, 28). Eight studies analyzed VMS frequency and 10 studies analyzed VMS severity.
Table 5.
Methods of assessment of vasomotor symptoms
| First author and publication year | VMS any vs none | VMS frequency | VMS severity | Additional comments | Citation |
|---|---|---|---|---|---|
| Aguilar-Zavala 2012 | NA | NA | Severity of current hot flashes (slight = 1 to severe = 3) | (30) | |
| Butts 2012 | Hot flashes within past month assessed at 1 visit | Hot flashes within past mo. | Hot flashes 0 (none)-4 (severe) | (20) | |
| Candrakova 2018 | “Vasomotor symptoms,” time horizon and specific definition of “vasomotor” not described | NA | NA | Kaczmarek 2007 questionnaire (Poland) | (21) |
| Crandall 2017 | Hot flashes and/or night sweats, ever vs never, assessed at baseline | NA | NA | (22) | |
| Crandall 2006 | NA | ≥ 6 d vs < 6 d of any VMS (hot flashes, cold sweats, night sweats) in past 2 wks, assessed annually for 7 visits (repeated-measures analysis) | NA | (23) | |
| Kapoor 2019, meeting abstract | NA | NA | VMS severity specified in “Methods” in abstract, further details not stated, but “Results” specifies “hot flash severity,” not VMS severity | Menopause Rating Scale questionnaire | (24) |
| Luptakova 2012 | NA | NA | Hot flashes and night sweats separately scored 1 (weakly affected) to 7 (extremely affected) | Scale developed Kaczmarck et al | (29) |
| Malacara 2004 | Hot flashes yes vs no, time horizon not specified | NA | NA | (25) | |
| Massad-Costa 2008 | NA | NA | Hot flushes scored mild = 4, moderate = 8, severe = 12, time frame for hot flushes not stated | Kupperman Menopause Index | (26) |
| Montasser 2015 | Hot flashes ever/never | Hot flashes daily, weekly, or monthly; duration of mos/y of hot flashes | NA | (27) | |
| Rebbeck 2010 | Hot flashes in past month, assessed at 9-mo intervals (11-y follow-up) | Hot flash frequency in past month, assessed at 9-mo intervals (11-y follow-up) | Hot flashes severity in past month (0 = none to 4 = severe), assessed at 9-mo intervals (11-y follow-up) | (3) | |
| Schilling 2007 | “Menopausal symptoms” by survey, no further details stated, time window not stated | “Menopausal symptoms” by survey, frequency assessed including “at least weekly,” no further details stated, beginning of time window not stated but hot flashes for ≥ 1 y assessed | “Menopausal symptoms” by survey, severity categories of moderate or severe, no further details stated, time window not stated | (28) | |
| Schneider 2009 | Ever or never experienced hot flashes, currently experiencing hot flashes in past 2 wks, yes or no | NA | NA | (19) | |
| Takeo 2005 | Hot flushes yes/no currently | Hot flush index reflecting frequency and severity; daily frequency at time of maximal symptoms (5 levels) | 5-point scale of hot flush severity (1 = none to 5 = debilitating) | (18) | |
| Visvanathan 2005 | Hot flushes ever vs never, hot flushes in past 30 d | Number of hot flushes in past 30 d, frequency of hot flushes, duration of hot flushes | Severity of hot flushes | (17) | |
| Woods 2018 | NA | NA | Hot flashes from 3-d symptoms diary regarding hot flashes over past 24 h, scored from 0 = not present to 4 = extreme), 3-d rating averaged together | (16) | |
| Woods 2006 | NA | NA | Hot flashes and cold sweats from 3-d symptoms diary regarding symptoms over past 24 h, scored from 0 = not present to 4 = extreme), 3-d rating averaged together | (15) | |
| Ziv-Gal 2012 | Ever experienced hot flashes | Hot flash frequency daily, weekly, or monthly (time window not specified); hot flash duration (no. of mos/y) | Hot flash severity mild, moderate, or severe (time window not specified) | (14) |
Abbreviations: NA, not applicable to the citation; VMS, vasomotor symptoms.
Study quality ratings are displayed in Table 6. Risk of bias regarding study participation was high in 2 studies, bias related to the prognostic factor (genotype) was low in all studies. However, risk of bias regarding the outcome measurement (VMS) was high in 2 studies and risk of bias related to study confounding and/or statistical analysis was high in 5 studies.
Table 6.
Study quality ratings summary (Quality in Prognostic Factor Studies tool)
| Component rated | Risk of bias |
|---|---|
| Aguilar-Zavala, 2012 | |
| Study participation | Moderate—women were invited via announcements in public places in 3 cities. Unclear how sample compares to demographics of population. |
| Study attrition | Not applicable—cross sectional |
| Prognostic factor | Low—all used serology with good sensitivity and/or specificity |
| Outcome measurement | Moderate—self-reported via severity scale form 0-3, no reliability or validity data or reference cited |
| Study confounding | High—did not adjust for important confounders, studies co-variates that seem less relevant |
| Statistical analysis and presentation | High—simple mean severity scores between genetic groups, no significant differences |
| Penn Ovarian Aging Study: Butts, 2012; Rebbeck, 2010 | |
| Study participation | Low—random-digit dialing and stratified enrollment to achieve representation by race |
| Study attrition | Low—cross-sectional analysis of longitudinal study at 1 time point |
| Prognostic factor | Low—serology with good validity |
| Outcome measurement | Low—used validated symptom list |
| Study confounding | Low—logistic regression adjusted for co-variates |
| Statistical analysis and presentation | Moderate—not sure how Latinas were classified; all participants classified as “European American” or “African American” |
| Two studies from a Slovakian cohort: Candrakova, 2018; Luptakova 2012 | |
| Study participation | Moderate—letters sent to prospective women; unclear if representative sample |
| Study attrition | Not applicable—cross-sectional |
| Prognostic factor | Low—JetQuick Tissue DNA test |
| Outcome measurement | Low—validity of questionnaire published 2007 |
| Study confounding | High—reports binary associates with each covariate separately |
| Statistical analysis and presentation | High—includes unvalidated measures (“Do you feel obese?” and “Are you satisfied with your life?” etc) |
| Crandall, 2006 | |
| Study participation | Low—community-based sample, > 3300 women, sampling for ethnic and/or racial representation |
| Study attrition | Low |
| Prognostic factor | Low—validated serology test |
| Outcome measurement | Moderate—No. of d had symptoms in past 2 wks |
| Study confounding | Low—adjusted for important covariates |
| Statistical analysis and presentation | Low |
| Crandall, 2017 | |
| Study participation | Low—per Women’s Health Initiative documentation. Large representative sample |
| Study attrition | Unclear |
| Prognostic factor | Low—serology validated |
| Outcome measurement | Moderate—“ever experienced VMS”; no measure of frequency or severity |
| Study confounding | Low—adjusted for important covariates |
| Statistical analysis and presentation | Low |
| Kapoor, 2019 | |
| Study participation | Unclear—Mayo Clinic Right study |
| Study attrition | Not applicable—cross-sectional study |
| Prognostic factor | Low—serology |
| Outcome measurement | Low—Menopausal Rating Scale has been validated |
| Study confounding | High—reports only adjustment for hormone therapy |
| Statistical analysis and presentation | High—states multivariate analysis is forthcoming |
| Malacara, 2004 | |
| Study participation | High—“volunteers recruited by house visit”; unclear if representative sample |
| Study attrition | Not applicable—cross-sectional study |
| Prognostic factor | Low—sensitivity and specificity of serology reported |
| Outcome measurement | High—self-report by face-to-face interview; no validation reported |
| Study confounding | Moderate—some potential confounders not analyzed |
| Statistical analysis and presentation | High—inappropriate stepwise regression models do not adjust for all potential confounders simultaneously |
| Massad-Costa, 2008 | |
| Study participation | High—no information on how sample recruited or how representative, demographics not described |
| Study attrition | Not applicable—cross-sectional study |
| Prognostic factor | Low—serology validated |
| Outcome measurement | High—Kupperman Menopause Index developed in 1950s has been widely critiqued |
| Study confounding | High—no adjustment for potential confounders |
| Statistical analysis and presentation | High—simple percentages |
| Baltimore Midlife Health Study: Ziv-Gal 2012; Visvanathan, 2005; Montasser, 2015; Schilling, 2007 | |
| Study participation | Moderate—recruitment used letters; no mention of response rate or comparison of responders to nonresponders |
| Study attrition | Not applicable—cross-sectional study |
| Prognostic factor | Low—validated serology methods |
| Outcome measurement | Low—severity, frequency, and duration quantified |
| Study confounding | Low—adjusted for important potential confounders |
| Statistical analysis and presentation | Low |
| Schneider, 2009 | |
| Study participation | Low—representative population based sample. (However, only data for white women analyzed). |
| Study attrition | Not applicable—cross-sectional study |
| Prognostic factor | Low—serology validated |
| Outcome measurement | Moderate—ever or currently experiencing hot flashes |
| Study confounding | Low—multivariate analyses adjusted for potential confounders |
| Statistical analysis and presentation | Low—no flaws |
| Takeo, 2005 | |
| Study participation | Moderate—recruitment from outpatient records; unclear refusal rate, unclear if representative |
| Study attrition | Not applicable—cross-sectional study |
| Prognostic factor | Low—serology |
| Outcome measurement | Low—assessed frequency and severity using validated measures |
| Study confounding | Moderate—reported data for each genetic “group” but did not adjust for in regression analyses |
| Statistical analysis and presentation | Low |
| Seattle Midlife Women’s Study: Woods, 2006; Woods, 2018 | |
| Study participation | Low—recruited by complete ascertainment of entire multiethnic neighborhoods |
| Study attrition | Low—cross-sectional analysis |
| Prognostic factor | Low—serology |
| Outcome measurement | Low—prospective collection: 3-d diary of frequency, severity |
| Study confounding | Moderate—some potential confounders not adjusted for |
| Statistical analysis and presentation | Low |
Abbreviation: VMS, vasomotor symptoms.
Detailed results of the candidate gene studies are provided in Table 7. Candidate gene studies are studies that preselect specific genetic variants based on a priori hypotheses, for example, because the genes are involved in estrogen biosynthesis. Candidate gene studies evaluated single-nucleotide polymorphisms (SNPs) in the following genes: aryl hydrocarbon receptor (14), aryl-hydrocarbon receptor repressor (14), aryl hydrocarbon receptor nuclear translocator (14), catechol-O-methyltransferase (3, 20, 28), cytochrome P450 family 1 subfamily A member 1 (15, 17, 23), cytochrome P450 family 1 subfamily A member 2 (3, 20, 24), cytochrome P450 family 1 subfamily B member 1 (3, 14, 15, 17, 20, 21, 23, 28, 29), cytochrome P450 family 2 subfamily C member 9 (24), cytochrome P450 family 3 subfamily A member 4 (3, 20, 24), cytochrome P450 family 17 subfamily A member 1 (15, 17, 26), cytochrome P450 family 19 subfamily A member 1 (3, 15, 28), nitric oxide synthase 3 (19), estrogen receptor 1 (15, 23, 25, 30), estrogen receptor 2 (18), 3-β-hydroxysteroid dehydrogenase (28), hydroxysteroid 17-β dehydrogenase 1 (16, 23), hypoxia inducible factor 1 subunit α (19), neuropilin 1 (19), neuropilin 2 (19), uridine diphosphate glucuronosyltransferase family 1 member A1 (24), serotonin transporter gene (27, 30), sulfotransterase family 1A member 1 (3), sulfotransferase family 1E member 1 (3), vascular endothelial growth factor A (19), fms-related receptor tyrosine kinase 1 (19), and kinase insert domain receptor (19). The results of GWAS (one published study met inclusion criteria, reference [22]) are displayed in Table 8. In that study, 14 SNPs were statistically significantly associated with HF (ever vs never) at a P value threshold of less than 5 × 10–8, and all of those were located in the tachykinin receptor 3 (TACR3) gene (22).
Table 7.
Associations between genetic variants and vasomotor symptoms in candidate gene studiesa
| First author and publication year | SNPs examinedb | Results | Citation |
|---|---|---|---|
| Aguilar-Zavala 2012 | ERα PvuII, rs number not stated | Hot flash severity score mean +/– SD 0.78+/– 0.52 for PP genotype group; 1.21 +/– 0.94 for Pp genotype group; 1.18 +/– 1.04 for pp genotype group, with P = .14 across genotype groups | (30) |
| ERα XbaI, rs number not stated | Hot flash severity score mean +/– SD 1.08 +/– 0.90 for XX genotype group, 1.21 +/– 0.95 for Xx genotype groups, and 1.12 +/– 0.97 for xx genotype group, with P = .68 across genotype groups | ||
| Serotonin transporter promoter region variant (5-HTTLPR short vs L allele), rs number not provided | Hot flash severity score mean +/– SD 0.95 +/– 0.95 for SS genotype group; 1.22 +/– 0.93 for SL genotype group; 1.26 +/– 1.02 for LL genotype group, with P = .68 across genotype groups | ||
| Butts 2012 | Catechol-O-methyltransferase (COMT) rs4680 (COMTVal158Met) | EA smokers with +/+ genotype had aOR 6.15 (95% CI 1.32-28.78) (ref EA nonsmokers with +/+ genotype); EA smokers with +/– genotype had aOR of 0.72 (0.22-2.35) (ref EA nonsmokers with +/– genotype); EA smokers with –/– genotype had aOR of 1.68 (0.26-10.70) (ref EA nonsmokers with –/– genotype) for any HF within past mo. + designates variant allele | (20) |
| EA smokers with +/+ genotype had aOR 4.35 (0.95-19.93) (ref EA nonsmokers with +/+ genotype); EA smokers with +/– genotype had aOR of 1.68 (0.48-5.81) (ref EA nonsmokers with +/– genotype); EA smokers with –/– genotype had aOR of 0.63 (95% CI 0.06-6.71) (ref EA nonsmokers with –/– genotype) for moderate and severe HF within past mo. + designates variant allele | |||
| AA smokers with +/+ genotype had insufficient data for estimate; AA smokers with +/– genotype had aOR of 2·89 (0.74-11.25) (ref AA nonsmokers with +/– genotype); AA smokers with –/– genotype had aOR of 0.95 (0.29-3.08) (ref AA nonsmokers with –/– genotype) for any HF within past mo. + designates variant allele | |||
| AA smokers with +/+ genotype had aOR 1.11 (95% CI 0.1-12.17) (ref AA nonsmokers with +/+ genotype); AA smokers with +/– genotype had aOR of 1.78 (0.52-0.61) (ref AA nonsmokers with +/– genotype); AA smokers with –/– genotype had aOR of 1.99 (0.62-6.46) (ref AA nonsmokers with –/– genotype) for moderate and severe HF within past mo. + designates variant allele | |||
| CYP3A4 rs2740574 (CYP3A4*1B) | EA smokers with +/+ genotype had insufficient data for estimate; EA smokers with +/– genotype had aOR of 2·63 (0.15-44.97) (ref EA nonsmokers with +/– genotype); EA smokers with –/– genotype had aOR of 1.49 (0.65-3.43) (ref EA nonsmokers with –/– genotype) for any HF within past mo. + designates variant allele | ||
| EA smokers with +/+ genotype had insufficient data for estimate; EA smokers with +/– genotype had aOR of 2.65 (0.11-66.8) (ref EA nonsmokers with +/– genotype); EA smokers with –/– genotype had aOR of 1.87 (95% CI, 0.78-4.46) (ref EA nonsmokers with –/– genotype) for moderate and severe HF within past mo. + designates variant allele | |||
| AA smokers with +/+ genotype had aOR 3.35 (95% CI 0.86-13.11) (ref AA nonsmokers with +/+ genotype); AA smokers with +/– genotype had aOR of 1.83 (0.48-7.02) (ref AA nonsmokers with +/– genotype); AA smokers with –/– genotype had aOR of 0.64 (0.09-4.34) (ref AA nonsmokers with –/– genotype) for any HF within past month. + designates variant allele | |||
| AA smokers with +/+ genotype had aOR 2.51 (95% CI 0.74-8.55) (ref AA nonsmokers with +/+ genotype); AA smokers with +/– genotype had aOR of 2.14 (0.62-7.34) (ref AA nonsmokers with +/– genotype); AA smokers with –/– genotype had aOR of 0.76 (0.11-5.06) (ref AA nonsmokers with –/– genotype) for moderate and severe HF within past mo. + designates variant allele | |||
| Cytochrome P450 family 1 subfamily A member 2 (CYP1A2) rs762551 (CYP1A2*1F) | EA smokers with +/+ genotype had aOR 1.43 (95% CI 0.46-4.44) (ref EA nonsmokers with +/+ genotype) and EA smokers with +/– genotype had aOR 1.47 (0.42-5.17) (ref EA nonsmokers with +/– genotype) and EA smokers with –/– genotype had aOR 2.79 (0.27-28.98) (ref EA nonsmokers with –/– genotype) for any hot flashes within past mo. + designates variant allele | ||
| EA smokers with +/+ genotype had aOR of 1.07 (0.31-3.73) (ref EA nonsmokers with +/+ genotype); EA smokers with +/– genotype had aOR of 2.91 (95% CI 0.84-10.04) (ref EA nonsmokers with +/– genotype); EA smokers with –/– genotype had aOR of 11.1 (95% CI, 0.65-189.29) (ref EA nonsmokers with –/– genotype) for moderate and severe HF within past mo. + designates variant allele | |||
| AA smokers with +/+ genotype had aOR 0.88 (95% CI 0.35-3.15) (ref AA nonsmokers with +/+ genotype); AA smokers with +/– genotype had aOR of 6.16 (1.11-33.91) (ref AA nonsmokers with +/– genotype); AA smokers with –/– genotype had aOR of 1.13 (0.13-9.97) (ref AA nonsmokers with –/– genotype) for any HF within past mo. + designates variant allele | |||
| AA smokers with +/+ genotype had aOR 1.78 (95% CI 0.5-6.39) (ref AA nonsmokers with +/+ genotype); AA smokers with +/– genotype had aOR of 2.67 (0.82-8.70) (ref AA nonsmokers with +/– genotype); AA smokers with –/– genotype had aOR of 0.64 (0.07-6.19) (ref AA nonsmokers with –/– genotype) for moderate and severe HF within past mo. + designates variant allele | |||
| Cytochrome P450 family 1 subfamily B member 1 (CYP1B1) rs1056836 (CYP1B1*3, Leu432Val) | EA smokers with +/+ genotype had insufficient data for analysis; EA smokers with +/– genotype had aOR 1.51 (0.5-4.59) (ref EA nonsmokers with +/– genotype) and EA smokers with –/– genotype had aOR 0.49 (0.11-2.10) (ref EA nonsmokers with –/– genotype) for any hot flashes within past mo. + designates variant allele | ||
| EA smokers with +/+ genotype had aOR 20.6 (95% CI 1.64-257.93) (ref EA nonsmokers with +/+ genotype) and EA smokers with +/– genotype had aOR of 1.22 (0.37-4.05) (ref EA nonsmokers with +/– genotype) and EA smokers with –/– genotype had aOR 1.58 (0.37-6.79) (ref EA nonsmokers with –/– genotype) for moderate and severe HF within past mo. + designates variant allele | |||
| AA smokers insufficient data re any HF within past mo | |||
| AA smokers insufficient data re moderate and severe HF within past mo | |||
| Cytochrome P450 family 1 subfamily B member 1 (CYP1B1) rs1800440 (CYP1B1*4, Asn452Ser) | EA smokers with +/+ genotype had insufficient data; EA smokers with +/– genotype had aOR 0.52 (0.11-2.46) (ref EA nonsmokers with +/– genotype) and EA smokers with –/– genotype had aOR 2.46 (0.96-6.30) (ref EA nonsmokers with –/– genotype) for any hot flashes within past mo. + designates variant allele | ||
| EA smokers with +/+ genotype had insufficient data; EA smokers with +/– genotype had aOR 0.71 (0.12-4.32) (ref EA nonsmokers with +/– genotype); EA smokers with –/– genotype had aOR 2.63 (1.02-6.78) (ref EA nonsmokers with –/– genotype) for moderate and severe HF. + designates variant allele | |||
| AA smokers with +/+ genotype had insufficient data; AA smokers with +/– genotype had aOR 2.35 (0.12-46.80) (ref AA nonsmokers with +/– genotype) and AA smokers with –/– genotype had aOR 1.71 (0.67-4.36) (ref AA nonsmokers with –/– genotype) for any hot flashes within past month. + designates variant allele | |||
| AA smokers with +/+ genotype had insufficient data; AA smokers with +/– genotype had aOR 2.6 (0.13-51.81) (ref AA nonsmokers with +/– genotype); AA smokers with –/– genotype had aOR 1.86 (0.81-4.26) (ref AA nonsmokers with –/– genotype) for moderate and severe HF. + designates variant allele | |||
| Candrakova 2018 | Cytochrome P450 family 1 subfamily B member 1 (CYP1B1) Arg48Gly rs10012 | In postmenopausal women, those with Gly/Gly genotype had unadjOR 20.98 (95% CI 3.28-134.02) and those with Arg/Gly genotype had unadjOR 0.70 (0.267-1.848) (ref Arg/Arg genotype) for VMS yes/no; race/ethnicity not stated but Slovakian women. Time frame for VMS not stated | (21) |
| In premenopausal women, no statistically significant association for VMS yes/no; race/ethnicity not stated but Slovakian women. Time frame for VMS not stated. Effect estimate not stated | |||
| Cytochrome P450 family 1 subfamily B member 1 (CYP1B1) Ala119Ser rs1056827 | No statistically significant association in premenopausal women for VMS yes/no; race/ethnicity not stated but Slovakian women. Time frame for VMS not stated. Effect estimate not stated | ||
| No statistically significant association in postmenopausal women for VMS yes/no; race/ethnicity not stated but Slovakian women. Time frame for VMS not stated. Effect estimate not stated. | |||
| Cytochrome P450 family 1 subfamily B member 1 (CYP1B1 Leu432Val) rs1056836 | No statistically significant association in premenopausal women for VMS yes/no; race/ethnicity not stated but Slovakian women. Time frame for VMS not stated. Effect estimate not stated | ||
| No statistically significant association in postmenopausal women for VMS yes/no; race/ethnicity not stated but Slovakian women. Time frame for VMS not stated. Effect estimate not stated | |||
| cytochrome P450 family 1 subfamily B member 1 (CYP1B1) Asn453Ser rs1800440 | No significant association in premenopausal women for VMS yes/no; race/ethnicity not stated but Slovakian women. Time frame for VMS not stated | ||
| No significant association in postmenopausal women for VMS yes/no; race/ethnicity not stated but Slovakian women. Time frame for VMS not stated. Effect estimate not stated | |||
| Crandall 2006 | Cytochrome P450 family 1 subfamily B member 1 (CYP1B1) rs1056836 (CYP1056836, CYP1B1*3, Leu432Val, 4326C>G, C1294G) | AA aOR 0.87 (0.44-1.70) for GG vs CC genotype; aOR·0.56 (0.28-1.12) for GC genotype vs CC genotype for VMS ≥ 6 d in past 2 wks | (23) |
| SNP not in Hardy-Weinberg equilibrium in other racial/ethnic groups | |||
| Estrogen receptor 1 (ESR1) rs2234693 (ESRA418, PvuII RFLP) | SNP not in Hardy-Weinberg equilibrium in any racial/ethnic group | ||
| Estrogen receptor 1 (ESR)1 rs9340799 (ESRA464, XbaI RFLP) | SNP not in Hardy-Weinberg equilibrium in any racial/ethnic group | ||
| Hydroxysteroid 17-beta dehydrogenase 1 (17HSD) rs2830 (HSD17B2830) | In Caucasian womenc, women with AG genotype had aOR 0.66 (0.47-0.90) and women with GG genotype had aOR 0.81 (0·56-1.18) for VMS ≥ 6 d in past 2 wks (ref Caucasian women with AA genotype) | ||
| SNP not in Hardy-Weinberg equilibrium in any racial/ethnic group | |||
| Hydroxysteroid 17-beta dehydrogenase 1 (17HSD) rs592389 (HSD592389) | In Caucasian women, women with TG genotype had aOR 0.65 (0.47-0.90) and women with TT genotype had aOR 0.81 (0.56-1.17) (ref Caucasian women with GG genotype) VMS ≥ 6 d in past 2 wks | ||
| SNP was not in Hardy-Weinberg equilibrium in any racial/ethnic group | |||
| Hydroxysteroid 17-beta dehydrogenase 1 (17HSD) rs615942 (HSD615942) | In Caucasian women, women with TG genotype had aOR 0.64 (0.46-0.88) and women with GG genotype had aOR 0.77 (0.53-1.11 (ref Caucasian women with TT genotype) VMS ≥ 6 d in past 2 wks | ||
| SNP not in Hardy-Weinberg equilibrium in any racial/ethnic group | |||
| Cytochrome P450 family 1 subfamily A member 1 (CYP1A1) rs2606345 (CYP2606345,-1806) | In Chinese women, women with AC genotype had aOR 0.24 (0.08-0.72) (ref Chinese women with CC genotype, no women had AA genotype) VMS ≥ 6 d in past 2 wks | ||
| SNP not in Hardy-Weinberg equilibrium in any racial/ethnic group | |||
| Kapoor 2019 | “Enzymes or transporters involved in estrogen metabolism”; meeting abstract not published in full text at time of this systematic review, so unclear what other SNPs were examined | (24) | |
| Cytochrome P450 family 1 subfamily A member 2 (CYP1A2) rs number not stated | “Decreased activity was associated with decreased HF severity P = .08.” Effect measure not specified (eg, OR, RR). Association no longer statistically significant after adjustment for hormone therapy use | ||
| Cytochrome P450 family 2 subfamily C member 9 (CYP2C9) rs number not stated | “Decreased activity was associated with decreased HF severity P = .04.” Effect measure not specified (eg, OR, RR). Association no longer statistically significant after adjustment for hormone therapy use | ||
| Cytochrome P450 family 3 subfamily A member 4 (CYP3A4) rs number not stated | “Decreased activity was associated with decreased HF severity P = .01.” Effect measure not specified (eg, OR, RR) | ||
| Uridine diphosphate (UDP) glucuronosyltransferase family 1 member A1 (UGT1A1) rs number not stated | “Decreased activity was associated with increased HF severity P = .01.” Effect measure not specified (eg, OR, RR) | ||
| Luptakova 2012 | Cytochrome P450 family 1 subfamily B member 1 (CYP1B1, Leu432Val), rs number not provided | Premenopausal women: prevalence of HF 29% for Leu/Leu genotype, 42% for Leu/Val genotype, 29% for Val/Val genotype. Time frame not specified. P = .4 | (29) |
| Premenopausal women: prevalence of night sweats 33% for Leu/Leu genotype, 42% for Leu/Val genotype, 24% for Val/Val genotype. Time frame not specified. P = .2 | |||
| Perimenopausal or postmenopausal women: prevalence of HF 38% for Leu/Leu genotype, 40% for Leu/Val genotype, 21% for Val/Val genotype. Time frame not specified. P = .7 | |||
| Perimenopausal or postmenopausal women: prevalence of night sweats 32% for Leu/Leu genotype, 46% for Leu/Val genotype, 22% for Val/Val genotype. Time frame not specified. P = .3 | |||
| Malacara 2004 | Estrogen receptor 1 (ESRαXbaI, rs number not stated but author communicated rs number rs9340799 | Prevalence of HF 68% in XX genotype group, 66% in Xx genotype group, and 66% in xx genotype group, all unadjusted estimates. P = .98 for comparison across genotypes. Race/ethnicity not stated. Study performed in Mexico. Time frame for HF not stated. Uppercase letter designates absence and lowercase letter designates presence of restriction sites. | (25) |
| Estrogen receptor 1 (ESRαPvuII, rs number not stated but author has communicated rs number rs2234693 | Prevalence of hot flashes 72% in pp genotype group, 57% in Pp genotype group, and 81% in PP genotype group, all unadjusted estimates. P = .048 for comparison between Pp and pp group; P = .03 across all 3 genotype groups. P no longer significant in stepwise multiple regression (details not provided). Race/ethnicity not stated. Study performed in Mexico. Time frame for HF not stated. Uppercase letter designates absence and lowercase letter designates presence of restriction sites. | ||
| Massad-Costa 2008 | CYP17 MspAI (single base pair change C→T in the 5’ UTR, which creates a recognition site for MspAI restriction enzyme, rs number not stated | No statistically significant association with HF severity. For A1/A1 genotype group, prevalence of HF (from Kupperman Menopause Index) was 25% for mild symptoms, 32% for moderate symptoms, and 19% for severe symptoms. For A1/A2 genotype group, prevalence of HF (from Kupperman Menopause Index) was 61% for mild symptoms, 44% for moderate symptoms, and 63% for severe symptoms. For A2/A2 genotype group, prevalence of HF (from Kupperman Menopause Index) was 14% for mild symptoms, 24% for moderate symptoms, and 19% for severe symptoms. Race/ethnicity and time frame for HF severity not stated. Wild allele A1 is C allele; variant allele A2 is T allele. P = .58 across all 3 genotype groups | (26) |
| Montasser 2015 | Serotonin transporter gene rs11080121 | EA P = .003 any vs never HF, effect was aOR but magnitude of aOR not stated | (27) |
| AA P = .4211 any vs never HF, effect was aOR but magnitude of aOR not stated | |||
| Serotonin transporter gene rs140700 | EA P = .78 any vs never HF, effect was aOR but magnitude of aOR not stated | ||
| AA P = 1 any vs never HF, effect was aOR but magnitude of aOR not stated | |||
| Serotonin transporter gene rs8076005 | EA P = .84 any vs never HF, effect was aOR but magnitude of aOR not stated | ||
| AA P = .28 any vs never hot flashes, effect was aOR but magnitude of aOR not stated | |||
| Serotonin transporter gene rs2066713 | EA P = .0009 any vs never HF, effect was aOR but magnitude of aOR not stated | ||
| AA P = .27 any vs never HF, effect was aOR but magnitude of aOR not stated | |||
| Serotonin transporter gene rs4251417 | EA P = .35 any vs never HF, effect was aOR but magnitude of aOR not stated | ||
| AA P = 1 any vs never hot flashes, effect was aOR but magnitude of aOR not stated | |||
| Serotonin transporter gene rs16965628 | EA P = .62 any vs never HF, effect was aOR but magnitude of aOR not stated | ||
| AA P = .36 any vs never HF, effect was aOR but magnitude of aOR not stated | |||
| Serotonin transporter gene rs4404067 | EA P = .10 any vs never HF, effect was aOR but magnitude of aOR not stated | ||
| AA P = .37 any vs never hot flashes, effect was aOR but magnitude of aOR not stated | |||
| Serotonin transporter gene rs4238784 | EA P = .28 any vs never HF, effect was aOR but magnitude of aOR not stated | ||
| AA P = .17 any vs never HF, effect was aOR but magnitude of aOR not stated | |||
| Serotonin transporter gene rs747107 | EA P = .89 any vs never HF, effect was aOR but magnitude of aOR not stated | ||
| AA P = .27 any vs never HF, effect was aOR but magnitude of aOR not stated | |||
| Serotonin transporter gene rs13333066 | EA P = 1 any vs never HF, effect was aOR but magnitude of aOR not stated | ||
| AA P = .69 any vs never HF, effect was aOR but magnitude of aOR not stated | |||
| Serotonin transporter gene rs187715 | EA P = .62 any vs never HF, effect was aOR but magnitude of aOR not stated | ||
| AA P = .41 any vs never HF, effect was aOR but magnitude of aOR not stated | |||
| Serotonin transporter gene rs36026 | EA P = .20 any vs never HF, effect was aOR but magnitude of aOR not stated | ||
| AA P = .05 any vs never HF, effect was aOR but magnitude of aOR not stated | |||
| Serotonin transporter gene rs36024 | EA P = .48 any vs never HF, effect was aOR but magnitude of aOR not stated | ||
| AA P = 1 any vs never HF, effect was aOR but magnitude of aOR not stated | |||
| Serotonin transporter gene rs36021 | EA P = .22 any vs never HF, effect was aOR but magnitude of aOR not stated | ||
| AA P = .04 any vs never HF, effect was aOR but magnitude of aOR not stated | |||
| Serotonin transporter gene rs3785151 | EA P = 1 any vs never HF, effect was aOR but magnitude of aOR not stated | ||
| AA P = .28 any vs never HF, effect was aOR but magnitude of aOR not stated | |||
| Serotonin transporter gene rs36020 | EA P = .30 any vs never hot flashes, effect was aOR but magnitude of aOR not stated | ||
| AA P = .17 any vs never hot flashes, effect was aOR but magnitude of aOR not stated | |||
| Serotonin transporter gene rs16955591 | EA P = 1 any vs never hot flashes, effect was aOR but magnitude of aOR not stated | ||
| AA P = .43 any vs never hot flashes, effect was aOR but magnitude of aOR not stated | |||
| Serotonin transporter gene rs3785152 | EA P = .80 any vs never hot flashes, effect was aOR but magnitude of aOR not stated | ||
| AA P = .42 any vs never hot flashes, effect was aOR but magnitude of aOR not stated | |||
| Serotonin transporter gene rs40147 | EA P = .40 any vs never hot flashes, effect was aOR but magnitude of aOR not stated | ||
| AA P = .008 any vs never hot flashes, effect was aOR but magnitude of aOR not stated | |||
| Serotonin transporter gene rs1814270 | EA P = .41 any vs never hot flashes, effect was aOR but magnitude of aOR not stated | ||
| AA P = .70 any vs never hot flashes, effect was aOR but magnitude of aOR not stated | |||
| Serotonin transporter gene rs5564 | EA P = .33 any vs never hot flashes, effect was aOR but magnitude of aOR not stated | ||
| AA P = .25 any vs never HF, effect was aOR but magnitude of aOR not stated | |||
| Serotonin transporter gene rs5568 | EA P = .58 any vs never HF, effect was aOR but magnitude of aOR not stated | ||
| AA P = .53 any vs never HF, effect was aOR but magnitude of aOR not stated | |||
| Serotonin transporter gene rs1566652 | EA P = .32 any vs never HF, effect was aOR but magnitude of aOR not stated | ||
| AA P = 1 any vs never HF, effect was aOR but magnitude of aOR not stated | |||
| Serotonin transporter gene rs5569 | EA P = .16 any vs never HF, effect was aOR but magnitude of aOR not stated | ||
| AA P = .29 any vs never HF, effect was aOR but magnitude of aOR not stated | |||
| Serotonin transporter gene rs2242447 | EA P = .75 any vs never HF, effect was aOR but magnitude of aOR not stated | ||
| AA P = .65 any vs never hot flashes, effect was aOR but magnitude of aOR not stated | |||
| Serotonin transporter gene rs424605 | EA P = .71 any vs never HF, effect was aOR but magnitude of aOR not stated | ||
| AA P = .86 any vs never hot flashes, effect was aOR but magnitude of aOR not stated | |||
| Serotonin transporter gene rs16955708 | EA P = .84 any vs never HF, effect was aOR but magnitude of aOR not stated | ||
| AA P = .51 any vs never HF, effect was aOR but magnitude of aOR not stated | |||
| Serotonin transporter gene rs12596924 | EA P = .50 any vs never HF, effect was aOR but magnitude of aOR not stated | ||
| AA P = .84 any vs never HF, effect was aOR but magnitude of aOR not stated | |||
| Serotonin transporter gene rs258099 | EA P = .79 any vs never HF, effect was aOR but magnitude of aOR not stated | ||
| AA P = .74 any vs never HF, effect was aOR but magnitude of aOR not stated | |||
| Rebbeck 2010 | Catechol-O-methyltransferase (COMT) Val158Met rs4680 | AA Met/Met vs any Val aOR 1.29 (0.67-2.48) for HF severity (moderate or severe vs none or mild) | (3) |
| EA Met/Met vs any Val aOR 0.82 (0.49-1.35) for HF severity (moderate or severe vs none or mild) | |||
| Cytochrome P450 family 19 subfamily A member 1 (CYP19) Arg264Cys rs700519 | AA 264 Arg/Arg vs any 264Cys aOR 0.68 (0.43-1.07) for HF severity (moderate or severe vs none or mild) | ||
| EA 264 Arg/Arg vs any 264Cys aOR 0.86 (0.41-1.80) for HF severity (moderate or severe vs none or mild) | |||
| Cytochrome P450 family 1 subfamily A member 2 (CYP1A2 *1F) rs762551 | AA any *1F vs *1/*1 aOR 1·04 (0.64-1.69) for HF severity (moderate or severe vs none or mild) | ||
| EA any *1F vs *1/*1 aOR 0.96 (0.48-1.92) for HF severity (moderate or severe vs none or mild) | |||
| Cytochrome P450 family 1 subfamily B member 1 (CYP1B1*3, Leu432Val) rs1056836 | AA aOR 0.62 (0.40-0.95) for any *3 vs *1/*1 associated with HF severity (moderate or severe vs none or mild) | ||
| EA any *3 vs *1/*1 aOR 0.94 (0.55-1.59) for HF severity (moderate or severe vs none or mild) | |||
| Cytochrome P450 family 1 subfamily B member 1 (CYP1B1*4, Asn452Ser) rs1800440 | AA any *4 vs *1/*1 aOR 0.66 (0.33-1.30) for HF severity (moderate or severe vs none or mild) | ||
| EA any *4 vs *1/* aOR 11.20 (0.76-1.89) for HF severity (moderate or severe vs none or mild) | |||
| Cytochrome P450 family 3 subfamily A member 4 (CYP3A4 *1B) rs2740574 | AA any *1B vs *1/*1 aOR 1.10 (0.63-1.94) for HF severity (moderate or severe vs none or mild) | ||
| EA any *1B vs *1/*1 aOR 0.87 (0.39-1.94) for HF severity (moderate or severe vs none or mild) | |||
| Sulfotransferase family 1A member 1 (SULT1A1*2, Arg213His) rs9282861 | AA any *2 vs *1/*1 aOR 0.77 (0.48-1.24) for HF severity (moderate or severe vs none or mild) | ||
| EA any *2 vs *1/*1 aOR 0.75 (0.49-1.14) for HF severity (moderate or severe vs none or mild) | |||
| Sulfotransferase family 1A member 1 (SULTA1*3, Met223Val) rs1801030 | AA any *3 vs *1/*1 aOR 0.95 (0.59-1.50) for HF severity (moderate or severe vs none or mild) | ||
| EA any *3 vs *1/*1 genotype had aOR 2.08 (1.64-2.63) associated with hot flash severity (moderate or severe vs none or mild) | |||
| Sulfotransferase family 1E member 1 (SULT1E1, 5’UTR promoter variant -64G→A) rs3736599 | AA any A vs G/G aOR 1.39 (0.86-2.23) for HF severity (moderate or severe vs none or mild) | ||
| EA any A vs G/G aOR 1.43 (0.77-2.63) for HF severity (moderate or severe vs none or mild) | |||
| Sulfotransferase family 1E member 1 (SULT1E1, A220G) rs3786599 | AA any C vs T/T aOR 1.01 (0.49-2.10) for HF severity (moderate or severe vs none or mild) | ||
| EA any C vs T/T aOR 0.86 (0.57-1.31) for HF severity (moderate or severe vs none or mild) | |||
| Schilling 2007 | Hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 1 (3βHSD) rs number not provided | No statistically significant association between +/– or –/– (ref +/+) and any HF (time window not specified), adjusted for race. RR 1.18 (0.96, 1.46) + designates WT allele | (28) |
| No statistically significant association between +/– or –/– (ref +/+) and moderate or severe HF (time window not specified); adjusted for race. RR 1.38 (1.00, 1.90) + designates WT allele | |||
| No statistically significant association between +/– or –/– (ref +/+) and at least weekly HF (time window not specified); adjusted for race. RR 1.29 (0.91, 1.83) + designates WT allele | |||
| No statistically significant association between +/– or –/– (ref +/+) and HF for ≥ 1 y, adjusted for race. RR 1.36 (0.99, 1.86) + designates WT allele | |||
| Cytochrome P450 family 19 subfamily A member 1 (CYP19) rs number not provided | No statistically significant association between +/– or –/– (ref +/+) genotype and any HF (time window not specified), adjusted for race. RR 1.00 (0.86, 1.17) + designates WT allele | ||
| No statistically significant association between +/– or –/– (ref +/+) genotype and moderate or severe HF (time window not specified), adjusted for race. RR 1.03 (0.83, 1.28) + designates WT allele | |||
| No statistically significant association between +/– or –/– (ref +/+) genotype and at least weekly hot flashes (time window not specified), adjusted for race. RR 1.10 (0.85, 1.43) + designates WT allele | |||
| No statistically significant association between +/– or –/– (ref +/+) genotype and hot flashes for ≥ 1 y, adjusted for race. RR 0.99 (0.81, 1.22) + designates WT allele | |||
| Catechol-O-methyltransferase (COMT) rs number not provided | No statistically significant association between +/– or –/– (ref +/+) genotype and any HF (time window not specified), adjusted for race. RR 0.98 (0.84, 1.14) + designates WT allele | ||
| No statistically significant association between +/– or –/– (ref +/+) genotype and moderate or severe HF (time window not specified), adjusted for race. RR 1.06 (0.84, 1.34) + designates WT allele | |||
| No statistically significant association between +/– or –/– (ref +/+) genotype and at least weekly HF (time window not specified), adjusted for race. RR 0.94 (0.73, 1.21) + designates WT allele | |||
| No statistically significant association between +/– or –/– (ref +/+) genotype and HF for ≥ 1 y, adjusted for race. RR 1.00 (0.81, 1.25) + designates WT allele | |||
| Cytochrome P450 family 1 subfamily B member 1 (CYP1B1) rs number not provided | No statistically significant association between +/– or –/– (ref +/+) genotype and any HF (time window not specified); adjusted for race. RR 1.18 (1.00, 1.40) + designates WT allele | ||
| RR 1.33 (1.04-1.71) for +/– or –/– (ref +/+) moderate or severe hot flashes (time window not specified), adjusted for race. + designates WT allele | |||
| RR 1.37 (1.02-1.84) for +/– or –/– (ref +/+) at least weekly HF (time window not specified); adjusted for race. + designates WT allele | |||
| RR 1.33 (1.05-1.69) for +/– or –/– (ref +/+) HF for ≥ 1 y (time window not specified), adjusted for race. + designates WT allele | |||
| Hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 1 (3βHSD) and cytochrome P450 family 1 subfamily B member 1 (CYP1B1) rs number not provided | RR 1.23 (0.75, 2.01) for +/+ and +/− or −/−, or +/− or −/− and +/+ (ref +/+, +/+) for any HF (time window not specified); adjusted for race. + designates WT allele | ||
| RR 1.22 (0.60, 2.45) for +/+ and +/− or −/−, or +/− or −/− and +/+. (ref +/+, +/+) for moderate or severe HF (time window not specified); adjusted for race. + designates WT allele | |||
| RR 1.50 (0.61, 3.67) for +/+ and +/− or −/−, or +/− or −/− and +/+ (ref +/+, +/+) for at least weekly HF (time window not specified); adjusted for race. + designates WT allele | |||
| RR 2.24 (0.79, 6.35) for +/+ and +/− or −/−, or +/− or −/− and +/+ (ref +/+, +/+) for HF for ≥1 year-adjusted for race. + designates wild-type allele | |||
| RR 1.42 (0.88, 2.30) for +/− or −/−, or +/− or −/− and +/+ (ref +/+, +/+) for any HF (time window not specified); adjusted for race. + designates WT allele | |||
| RR 1.63 (0.82, 3.21) for +/− or −/−, or +/− or −/− and +/+ (ref +/+, +/+) for moderate or severe HF (time window not specified), adjusted for race. + designates WT allele | |||
| RR 1.91 (0.80, 4.60) for +/− or −/−, or +/− or −/− and +/+ (ref +/+, +/+) for at least weekly HF (time window not specified), adjusted for race. + designates WT allele | |||
| RR 2.77 (0.99, 7.80) for +/− or −/−, or +/− or −/− and +/+ (ref +/+, +/+) for HF for ≥ 1 y, adjusted for race. + designates WT allele | |||
| Schneider 2009 | Nitric oxide synthase 3 (eNOS-786 T/C, T minor allele) rs number not provided | Premenopausal women TT or CT vs CC genotype OR 8.89 (1.20-65.5) for current HF but association not statistically significant after adjustment for covariates (no OR presented) | (19) |
| Postmenopausal women no statistically significant associated with current HF. Effect estimate not provided in published paper | |||
| Hypoxia inducible factor 1 subunit alpha (HIFα 1744 C/T, T minor allele) rs number not provided | Postmenopausal women TT or CT vs CC genotype aOR 1.27 (1.01-1.59) for current HF after adjustment for covariates | ||
| Premenopausal women no statistically significant association with current HF. Effect estimate not provided in published paper | |||
| Hypoxia inducible factor 1 subunit alpha (HIFα 1762 A/G,A minor allele) rs number not provided | Postmenopausal women no statistically significant association with current HF. Effect estimate not provided in published paper | ||
| Premenopausal women no statistically significant association with current HF. Effect estimate not provided in published paper | |||
| Vascular endothelial growth factor A (VEGF-2578 C/A, A minor allele) rs number not provided | Postmenopausal women no statistically significant association with current HF. Effect estimate not provided in published paper | ||
| Premenopausal women no statistically significant association with current HF. Effect estimate not provided in published paper | |||
| Vascular endothelial growth factor A (VEGF-634 G/C, C minor allele) rs number not provided | Postmenopausal women no statistically significant association with current HF. Effect estimate not provided in published paper | ||
| Premenopausal women no statistically significant association with current HF. Effect estimate not provided in published paper | |||
| Vascular endothelial growth factor A (VEGF-1154 G/C, A minor allele) rs number not provided | Postmenopausal women no statistically significant association with current HF. Effect estimate not provided in published paper | ||
| Premenopausal women no statistically significant association with current HF. Effect estimate not provided in published paper | |||
| Vascular endothelial growth factor A (VEGF 936 C/T, T minor allele) rs number not provided | Postmenopausal women no statistically significant association with current HF. Effect estimate not provided in published paper | ||
| Premenopausal women no statistically significant association with current HF. Effect estimate not provided in published paper | |||
| Nitric oxide synthase 3 (eNOS 894 G/T, T minor allele) rs number not provided | Postmenopausal women no statistically significant association with current HF. Effect estimate not provided in published paper | ||
| Premenopausal women no statistically significant association with current HF. Effect estimate not provided in published paper | |||
| Fms-related receptor tyrosine kinase 1 (VEGFR1-962 C/T, T minor allele) rs number not provided | Postmenopausal women no statistically significant association with current HF. Effect estimate not provided in published paper | ||
| Premenopausal women no statistically significant association with current HF. Effect estimate not provided in published paper | |||
| Kinase insert domain receptor (VEGFR2 889 A/G, A minor allele) rs number not provided | Postmenopausal women no statistically significant association with current HF. Effect estimate not provided in published paper | ||
| Premenopausal women no statistically significant association with current HF. Effect estimate not provided in published paper | |||
| Kinase insert domain receptor (VEGFR2 1416 A/T, T minor allele) rs number not provided | Postmenopausal women no statistically significant association with current HF. Effect estimate not provided in published paper | ||
| Premenopausal women no statistically significant association with current HF. Effect estimate not provided in published paper | |||
| Neuropilin 1 (NRP1 1683 C/G, G minor allele) rs number not provided | Postmenopausal women no statistically significant association with current HF. Effect estimate not provided in published paper | ||
| Premenopausal women no statistically significant association with current HF. Effect estimate not provided in published paper. | |||
| Neuropilin 1 (NRP1 2197 A/G, A minor allele) rs number not provided | Postmenopausal women no statistically significant association with current HF. Effect estimate not provided in published paper | ||
| Premenopausal women no statistically significant association with current HF. Effect estimate not provided in published paper | |||
| Neuropilin 2 (NRP2 368 A/G, G minor allele) rs number not provided | Postmenopausal women no statistically significant association with current HF. Effect estimate not provided in published paper | ||
| Premenopausal women no statistically significant association with current HF. Effect estimate not provided in published paper | |||
| Takeo 2005 | Estrogen receptor 2 (ERβ cytosine-adenine dinucleotide repeat length in intron 5), no rs number provided | In postmenopausal women, unadj OR 7.0 (1.25-39.15) (P = .025) for 2 short (18-21 repeats) alleles and 5.6 (0.97-32.2) (P = .046) for 2 long (≥ 22 repeats) alleles for current HF (ref 1 short and 1 long allele of cytosine-adenine repeat length. OR could not be calculated for group with 1 extremely short (≤ 17 repeats) allele and 1 long (≥22 repeats) allele because there were no asymptomatic women in that group | (18) |
| Visvanathan 2005 | Cytochrome P450 family 17 subfamily A member 1 (CYPc17α MspA1 polymorphisms resulting from single base pair change from T to C in 5’ untranslated region), rs number not provided | No statistically significant association between any HF and +/– or –/– genotype (ref +/+). RR 0.99 (0.86-1.14). No significant difference across menopausal status strata so menopausal status groups combined. + designates WT allele | (17) |
| No statistically significant association between moderate or severe HF and +/– or –/– genotype group (ref +/+). RR 0.96 (0.79-1.17). No significant difference across menopausal status strata so menopausal status groups combined. + designates WT allele | |||
| No statistically significant association between at least weekly hot flashes and +/– or –/– genotype (ref +/+). RR 1.01 (0.79-1.28). No significant difference across menopausal status strata so menopausal status groups combined. + designates WT allele | |||
| No statistically significant association between HF for > 1 y and +/– or –/– genotype (ref +/+). RR 1.02 (0.84-1.25). No significant difference across menopausal status strata so menopausal status groups combined. + designates WT allele | |||
| Cytochrome P450 family 1 subfamily B member 1 (CYP1B1) single base pair substitution of leucine for valine at codon 432, rs number not provided | No statistically significant association between any HF and +/– or –/– genotype (ref +/+). RR 1.16 (0.98-1.37). No significant difference across menopausal status strata so menopausal status groups combined. + designates WT allele | ||
| RR 1.33 (1.03-1.70) for moderate or severe HF in +/– or –/– genotype group (ref +/+). No significant difference across menopausal status strata so menopausal status groups combined. + designates WT allele | |||
| No statistically significant association between at least weekly HF and +/- or -/- genotype (reference +/+) RR 1.32 (0.99-1.77). No significant difference across menopausal status strata so menopausal status groups were combined. + designates WT allele | |||
| No statistically significant association between HF for > 1 y and +/– or –/– genotype (ref +/+). RR 1.28 (1.00-1.63). No significant difference across menopausal status strata so menopausal status groups combined. + designates WT allele | |||
| Cytochrome P450 family 1 subfamily A member 1 (CYP1A1), rs number not provided | No statistically significant association between any HF and +/– or –/– genotype (ref +/+). aRR 1.03 (0.88-1.21). No significant difference across menopausal status strata so menopausal status groups combined. + designates WT allele | ||
| aRR 1.15 (0.94-1.42) for moderate or severe HF if +/– or –/– genotype group (ref +/+). No significant difference across menopausal status strata so menopausal status groups were combined. + designates WT allele | |||
| No statistically significant association between at least weekly HF and +/– or –/– genotype (reference +/+). RR 1.11 (0.86-1.44). No significant difference across menopausal status strata so menopausal status groups combined. + designates WT allele | |||
| No statistically significant association between HF for > 1 y and +/– or –/– genotype (ref +/+). RR 1.05 (0.84-1.31). No significant difference across menopausal status strata so menopausal status groups combined. + designates WT allele | |||
| Cytochrome P450 family 1 subfamily B member 1 (CYP1B1 single base pair substitution of leucine for valine at codon 432, rs number not provided) or cytochrome P450 family 1 subfamily A member 1 (CYP1A1, rs number not provided) | No statistically significant association between any HF and +/+ and +/– or –/– genotype. aRR 1.11 (0.92-1.34) (ref +/+,+/+). No significant difference across menopausal status strata so menopausal status groups combined. + designates WT allele | ||
| No statistically significant association between moderate or severe HF and +/+ and +/– or –/– genotype group. aRR 1.24 (0.92-1.66) (ref +/+, +/+). No significant difference across menopausal status strata so menopausal status groups combined. + designates WT allele | |||
| aRR 1.17 (0.85-1.62) for at least weekly hot flashes if +/+ and +/– or –/– genotype (ref +/+, +/+). No significant difference across menopausal status strata so menopausal status groups combined. + designates WT allele | |||
| No statistically significant association between hot flushes for > 1 y and +/+ and +/– or –/– genotype. aRR 1.27 (0.95-1.69) (reference +/+,+/+). No significant difference across menopausal status strata so menopausal status groups were combined. + designates WT allele | |||
| aRR 1.20 (0.96-1.51) for +/-, -- and +/-,-/- genotype (ref +/+,+/+). No significant difference across menopausal status strata so menopausal status groups were combined. + designates WT allele | |||
| aRR 1.53 (1.11-2.12) for +/–, –/– and +/–, –/– genotype (ref +/+,+/+). No significant difference across menopausal status strata so menopausal status groups combined. + designates WT allele | |||
| aRR 1.47 (1.02-2.13) for at least weekly hot flashes if +/–, –/–, and +/–, –/– genotype (ref +/+, +/+) No significant difference across menopausal status strata so menopausal status groups combined. + designates WT allele | |||
| aRR 1.37 (0.97-1.92) for +/–, –/–, and +/–, –/– genotype (ref +/+, +/+). No significant difference across menopausal status strata so menopausal status groups combined. + designates WT allele | |||
| Woods 2018 | Hydroxysteroid 17-β dehydrogenase 1 (HSDB1) rs615942 (T > G) | Variant TT vs GG associated with aOR 6.84 (2.81-6.66) for high-severity HF symptoms in past 24 h (vs low-severity). No significant association between race/ethnicity and genotype so race/ethnicity not included in models | (16) |
| Hydroxysteroid 17-β dehydrogenase 1 (HSDB1) rs2830 | AA vs T/T genotype associated with aOR 0.553 (0.21-1.44) for high-severity HF in past 24 h. No significant association between race/ethnicity and genotype so race/ethnicity not included in models | ||
| Hydroxysteroid 17-β dehydrogenase 1 (HSDB1) rs592389 (G > T) | Variant GG vs TT associated with aOR 10.00 (4.84-22.31) for high-severity HF symptoms in past 24 hours. No significant association between race/ethnicity and genotype so race/ethnicity not included in models | ||
| Cytochrome P450 family 19 subfamily A member 1 (CYP19 TTTA(n)), rs2389 | 7-repeat genotype associated with aOR 1.35 (0.14-3.31) for high-severity HF in past 24 h. 11-repeat genotype associated with aOR 1.04 (0.47-2.29) for high-severity HF in past 24 h. No significant association between race/ethnicity and genotype so race/ethnicity not included in models | ||
| Woods 2006 | Cytochrome P450 family 1 subfamily A member 1 (CYP1A1m2, rs number not stated) | No statistically significant association between genotype and VMS, expressed either as any VMS, HF, sweats, or % d with HF in past 24 h. Details of effect estimates not provided | (15) |
| Cytochrome P450 family 1 subfamily B member 1 (CYP1B1*2, rs number not stated) | No statistically significant association between genotype and VMS, expressed either as any VMS, HF, sweats, or % d with hot flashes in past 24 h. Details of effect estimates not provided | ||
| Cytochrome P450 family 1 subfamily B member 1 (CYP1B1*3, rs number not stated) | No statistically significant association between genotype and VMS, expressed either as any VMS, hot flashes, sweats, or % d with hot flashes in past 24 h. Details of effect estimates not provided | ||
| Cytochrome P450 family 17 subfamily A member 1 (CYP17 5’UTR, rs number not stated) | No statistically significant association between genotype and VMS, expressed either as any VMS, HF, sweats, or % d with hot flashes in past 24 h. Details of effect estimates not provided | ||
| Cytochrome P450 family 19 subfamily A member 1 (CYP19) 3’UTR, rs number not stated | No statistically significant association between genotype and VMS, expressed either as any VMS, HF, sweats, or % d with hot flashes in past 24 h. Details of effect estimates not provided | ||
| Cytochrome P450 family 19 subfamily A member 1 (CYP1911r, rs number not stated) | Middle menopausal transition group severity of VMS (HF or cold sweats) (adjusted mean, SD) in past 24 h higher with 2 variant alleles (0.35, 0.22) than with 1 variant allele (0.13, 0.21) or 0 alleles (0.16, 0.27). Kruskal-Wallis test P = .01 | ||
| Middle menopausal transition group severity of HF symptoms (adjusted mean, SD) in past 24 h was higher with 2 variant alleles (0.58, 0.38) than with 1 variant allele (0.21, 0.36) or 0 alleles (0.22, 0.37). Kruskal-Wallis test P = .007 | |||
| Middle menopausal transition group severity of sweats (adjusted mean, SD) in past 24 h higher with 2 variant alleles (0.11, 0.12) than with 1 variant allele (0.05, 0.16) or 0 alleles (0.10, 0.24). Kruskal-Wallis test P = .08 | |||
| Middle menopausal transition group % of days with HF in past 24 h higher with 2 variant alleles (0.35, 0.23) than with 1 variant allele (0.14, 0.23) or 0 alleles (0.12, 0.23). Kruskal-Wallis test P = .006 | |||
| Late menopausal transition group severity of VMS (HF or cold sweats) (adjusted mean, SD) in past 24 h higher with 2 variant alleles (0.90 0.45) than with 1 variant allele (0.23, 0.21) or 0 alleles (0.53, 0.57). Kruskal-Wallis test P = .002 | |||
| Late menopausal transition group severity of HF symptoms (adjusted mean, SD) in past 24 h higher with 2 variant alleles (1.40, 0.60) than with 1 variant allele (0.36, 0.33) or 0 alleles (0.87, 0.92). Kruskal-Wallis test P = .001 | |||
| Late menopausal transition group genotype sweats severity in past 24 h adjusted mean (SD) were 0.37 (0.40) with 2 variant alleles, 0.12 (0.24) with 1 variant allele, and 0.19 (0.40) with no variant alleles. Kruskal-Wallis test P = .117 | |||
| Late menopausal transition group % of days with HF in past 24 h higher with 2 variant alleles (0.71, 0.21) than with 1 variant allele (0.24, 0.25) or 0 alleles (0.35, 0.32). Kruskal-Wallis test P = .002 | |||
| Postmenopausal group severity of VMS (HF or cold sweats) in past 24 h (adjusted mean, SD) higher with 2 variant alleles (1.06, 0.66) than with 1 variant allele (0.40, 0.32) or 0 alleles (0.67, 0.44). Kruskal-Wallis test P = .06 | |||
| Postmenopausal group severity of HF symptoms (adjusted mean, SD) in past 24 h higher with 2 variant alleles (1.60, 0.94) than with 1 variant allele (0.72, 0.54) or 0 alleles (1.23, 0.74). Kruskal-Wallis test P = .02 | |||
| Postmenopausal group severity of sweats (adjusted mean, SD) in past 24 h higher with 2 variant alleles (0.51, 0.59) than with 1 variant allele (0.08, 0.18) or 0 alleles (0.10, 0.35). Kruskal-Wallis test P = .02 | |||
| Postmenopausal group % of days with hot flashes in past 24 h (adjusted mean, SD) higher with 2 variant alleles (0.75, 0.28) than with 1 variant allele (0.50, 0.29) or 0 alleles (0.75, 0.25). Kruskal-Wallis test P = .01 | |||
| Estrogen receptor 1 (ESR1XbaI, rs number not stated) | No statistically significant association between genotype and VMS, expressed either as any VMS, HF, sweats, or % d with HF in past 24 h. Details of effect estimates not provided | ||
| Estrogen receptor 1 (ESR1PvuII, rs number not stated) | No statistically significant association between genotype and VMS, expressed either as any VSM, HF, sweats, or % days with HF in past 24 h. Details of effect estimates not provided | ||
| Ziv-Gal 2012 | Aryl hydrocarbon receptor (AHR) rs2066853 | unadjOR 2.67 (1.13-6.33) for –/– (AA) and 1.16 (0.81-1.67) for +/– (GA) genotype ever experiencing HF (ref +/+ ie, GG) never experienced HF. aOR 2.44 (0.99-6.01) for –/– (AA) and 1.08 (0.72-1.60) for +/– (GA) genotype ever experiencing HF (ref +/+ ie, GG who never experienced HF) | (14) |
| aOR 2.21 (0.82-5.96) for –/– (AA) genotype and aOR 1.14 (0.73-1.76) for +/– (GA) genotype for moderate or severe HF (ref GG, ie, +/+ who never experienced HF) | |||
| aOR 2.37 (0.90-6.21) for –/– (AA) and aOR 0.95 (0.61-1.49) for +/–(GA) genotype for HF ≥ 1 y (ref GG, ie, +/+ who never experienced HF) | |||
| aOR 2.66 (0.70-0.11) for –/– (AA) and aOR 1.46 (0.81-2.64) for +/–(GA) genotype for daily HF (ref GG, ie, +/+ who never experienced HF) | |||
| Aryl-hydrocarbon receptor repressor (AHRR) rs2292596 | aOR 0.84 (0.36-1.98) for –/– (GG) genotype and aOR 1.03 (0.73-1.46) for +/– (CG) genotype ever experiencing hot flashes (ref CC, ie, +/+ who never experienced hot flashes) | ||
| aOR 1.30 (0.53-3.18) for –/– (GG) genotype and aOR 1.22 (0.83-1.79) for +/– (CG) genotype for moderate or severe HF (ref CC, ie, +/+ who never experienced HF) | |||
| aOR 0.60 (0.22-1.66) for –/– (GG) and aOR 0.97 (0.66-1.42) for +/–(CG) genotype for HF ≥ 1 y (ref CC, ie, +/+ who never experienced HF) | |||
| aOR 0.59 (0.12-3.02) for –/– (GG) and aOR 1.12 (0.66-1.90) for +/–(CG) genotype for daily HF (ref CC, ie, +/+ who never experienced HF) | |||
| Aryl hydrocarbon receptor nuclear translocator (ARNT) rs2228099 | aOR 0.83 (0.51-1.35) for –/– (CC) genotype and aOR 0.86 (0.59-1.24) for +/– (GC) genotype ever experiencing HF (ref GG, ie, +/+ who never experienced HF) | ||
| aOR 0.81 (0.47-1.39) for –/– (CC) genotype and aOR 0.77 (0.51-1.16) for +/– (GC) genotype for moderate or severe HF (ref GG, ie, +/+ who never experienced HF) | |||
| aOR 0.83 (0.48-1.43) for –/– (CC) and aOR 0.87 (0.58-1.31) for +/– (GC) genotype for HF ≥ 1 y (ref GG, ie, +/+ who never experienced HF) | |||
| aOR 0.79 (0.36-1.70) for –/– (CC) and aOR 0.82 (0.45-1.47) for +/–(GC) genotype for daily hot flashes (ref. GG, ie, +/+ who never experienced hot flashes) | |||
| Cytochrome P450 family 1 subfamily B member 1 (CYP1B1) rs1800440 | aOR 2.49 (0.95-6.48) for –/– (GG) genotype and aOR 1.07 (0.74-1.55) +/– (AG) genotype ever experiencing HF (ref AA, ie, +/+ who never experienced HF) | ||
| aOR 2.62 (0.94-7.31) for –/– (GG) genotype and aOR 1.04 (0.68-1.58) for +/– (AG) genotype for moderate or severe HF (ref AA, ie, +/+ who never experienced HF) | |||
| aOR 3.05 (1.12-8.25) for –/– (GG) and aOR 1.10 (0.72-1.67) for +/–(AG) genotype for HF ≥ 1 y (ref AA ie, +/+ who never experienced HF) | |||
| aOR 1.60 (0.36–7.20) for –/– (GG) and aOR 1.14 (0.63–2.05) for +/– (AG) genotype for daily hot flashes (ref AA i.e. +/+ who never experienced hot flashes) | |||
| Aryl hydrocarbon receptor (AHR) rs 2066853 and aryl hydrocarbon receptor nuclear translocator (AHRR) rs2292596 | aOR 1.12 (0.67-1.86) for +/–, –/– and +/–, –/– –/– genotype and aOR 1.29 (0.88-1.88) for +/+ and +/– or –/– genotype ever experiencing HF (ref +/+ and +/+ who never experienced HF) (for AHR + denotes G allele; for AHRR, + denotes C allele) | ||
| aOR 1.44 (0.81-2.55) for +/–, –/– and +/–, –/– –/– genotype and aOR 1.38 (0.89-2.13) for +/+ and +/– or –/– genotype for moderate or severe HF (ref +/+ and +/+ who never experienced HF) (for AHR + denotes G allele; for AHRR, + denotes C allele) | |||
| aOR 0.99 (0.55–1.74) for +/–, –/– and +/–, –/– –/– genotype and aOR 0.98 (0.64–1.49) for +/+ and +/– or –/– genotype for HF ≥1 y (ref +/+ and +/+ who never experienced hot flashes) (for AHR + denotes G allele; for AHRR, + denotes C allele) | |||
| aOR 1.61 (0.74-3.50) for +/–, –/–, and +/–, –/– –/– genotype and aOR 1.24 (0.66-2.31) for +/+ and +/– or –/– genotype for daily HF (ref +/+ and +/+ who never experienced HF) (for AHR + denotes G allele; for AHRR, + denotes C allele) | |||
| Aryl hydrocarbon receptor (AHR) rs 2066853 and aryl hydrocarbon receptor nuclear translocator (ARNT) rs2228099 | aOR 0.99 (0.60-1.65) for +/–, –/–, and +/–, –/– –/– genotype and aOR 0.92 (0.61-1.38) for +/+ and +/– or –/– genotype ever experiencing HF (ref +/+ and +/+ who never experienced HF) (for AHR, + denotes G allele; for ARNT, + denotes G allele) | ||
| aOR 0.94 (0.53-1.64) for +/–, –/– and +/–, –/– –/– genotype and aOR 0.79 (0.51-1.24) for +/+ and +/– or –/– genotype for moderate or severe HF (ref +/+ and +/+ who never experienced HF) (for AHR, + denotes G allele; for ARNT, + denotes G allele) | |||
| aOR 0.90 (0.51-1.60) for +/–, –/–, and +/–, –/– –/– genotype and aOR 0.93 (0.59-1.46) for +/+ and +/– or –/– genotype for HF ≥ 1 y (ref +/+ and +/+ who never experienced hot flashes) (for AHR, + denotes G allele; for ARNT, + denotes G allele) | |||
| aOR 1.24 (0.54-2.83) for +/–, –/–, and +/–, –/– –/– genotype and aOR 1.11 (0.56-2.20) for +/+ and +/– or –/– genotype for daily HF (ref. +/+ and +/+ who never experienced hot flashes) (for AHR, + denotes G allele; for ARNT, + denotes G allele) | |||
| Aryl hydrocarbon receptor (AHR) rs 2066853 and cytochrome P450 family 1 subfamily B member 1 (CYP1B1) rs1800440 | aOR 1.87 (0.95-3.70) for +/–, –/– and +/–, –/– –/– genotype and aOR 1.00 (0.72-1.41) for +/+ and +/– or –/– genotype ever experiencing HF (ref +/+ and +/+ who never experienced HF) (for AHR + denotes G allele; for CYP1B1, + denotes A allele) | ||
| aOR 1.72 (0.82–3.64) for +/–, –/– and +/–, –/– –/– genotype and aOR 1.06 (0.72–1.55) for +/+ and +/– or –/– genotype for moderate or severe hot flashes (ref +/+ and +/+ who never experienced hot flashes) (for AHR + denotes G allele; for CYP1B1, + denotes A allele) | |||
| aOR 1.71 (0.81–3.62) for +/–, –/– and +/–, –/– –/– genotype and aOR 1.01 (0.69–1.48) for +/+ and +/– or –/– genotype for HF ≥ 1 y (ref +/+ and +/+ who never experienced HF) (for AHR + denotes G allele; for CYP1B1, + denotes A allele) | |||
| aOR 2.11 (0.75-5.95) for +/–, –/– and +/–, –/– –/– genotype and aOR 1.28 (0.74-2.22) for +/+ and +/– or –/– genotype for daily hot flashes (ref +/+ and +/+ who never experienced hot flashes) (for AHR + denotes G allele; for CYP1B1, + denotes A allele) | |||
| Cytochrome P450 family 1 subfamily B member 1 (CYP1B1) rs 1800440 and CYP1B1 rs1056836 | aOR 1.54 (0.97-2.42) for +/–, –/– and +/–, –/– –/– genotype and aOR 1.51 (1.01-2.31) for +/+ and +/– or –/– genotype ever experiencing HF (ref +/+ and +/+ who never experienced HF) (for rs 1800440 + denotes A allele; for rs1056836, + denotes C allele) | ||
| aOR 1.68 (1·00-2.82) for +/–, –/– and +/–, –/– –/– genotype and aOR 1.81 (1.13-2.89) for +/+ and +/– or –/– genotype for moderate or severe HF (ref +/+ and +/+ who never experienced HF) (for rs 1800440 + denotes A allele; for rs1056836, + denotes C allele) | |||
| aOR 1.77 (1.06-2.96) for +/–, –/–, and +/–, –/– –/– genotype and aOR 1.64 (1.02-2.62) for +/+ and +/– or –/– genotype for HF ≥ 1 y (ref +/+ and +/+ who never experienced HF) (for rs 1800440 + denotes A allele; for rs1056836, + denotes C allele) | |||
| aOR 1.74 (0.82-3.69) for +/–, –/–, and +/–, –/– –/– genotype and aOR 1.84 (0.93-3.65) for +/+ and +/– or –/– genotype for daily HF (ref +/+ and +/+ who never experienced HF) (for rs 1800440 + denotes A allele; for rs1056836, + denotes C allele) |
Abbreviations: AA, African Americans; aOR, adjusted odds ratio; aRR, adjusted risk ratio; EA, European Americans; GWAS, genome-wide association study; HF, hot flashes; ref, reference; SNP, single nucleotide polymorphism; unadjOR, unadjusted OR; VMS, vasomotor symptoms; WT, wild-type.
Table 8.
Associations between genetic variants and vasomotor symptoms in genome-wide association studies
| RefsnpID | Chromosome:position | GARNET study (EA) | SHARe-AA study | SHARe-HA study | WHIMS+ study | P | Meta-analysis P | First author (citation) | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| P | OR (SE) | OR (SE) | P | OR (SE) | P | OR (SE) | |||||
| rs74827081 | 4:104556732 | 1.65 (0.18) | 7.88e-6 | 1.83 (0.34) | 1.25e-3 | 1.64 (0.24) | 7.46e-4 | 1.54 (0.15) | 6.41e-6 | 4.77e -15 |
Crandall 2017 (22) |
| rs74589515 | 4:104584258 | 1.60 (0.18) | 2.71e-5 | 1.80 (0.32) | 1.11e-3 | 1.61 (0.23) | 9.62e-4 | 1.57 (0.15) | 2.92e-6 | 7.11e -15 |
Crandall 2017 (22) |
| rs79246187 | 4:104580809 | 1.61 (0.18) | 2.1e-5 | 1.51 (0.23) | 7.46e-3 | 1.60 (0.23) | 1.02e-3 | 1.57 (0.15) | 2.63e-6 | 2.64e -14 |
Crandall 2017 (22) |
| rs112390256 | 4:104575473 | 1.64 (0.18) | 9.46e-6 | 1.51 (0.24) | 8.92e-3 | 1.61 (0.23) | 9.87e-4 | 1.54 (0.15) | 4.97e-6 | 2.81e -14 |
Crandall 2017 (22) |
| rs75544266 | 4:104584997 | 1.60 (0.18) | 2.84e-5 | 1.51 (0.23) | 6.55e-3 | 1.60 (0.23) | 9.42e-4 | 1.57 (0.15) | 2.89e-6 | 3.15e -14 |
Crandall 2017 (22) |
| rs78154848 | 4:104562840 | 1.65 (0.18) | 8.16e-6 | 1.46 (0.24) | 1.94e-2 | 1.62 (0.24) | 8.61e-4 | 1.54 (0.15) | 6.05e-6 | 5.65e -14 |
Crandall 2017 (22) |
| rs76643670 | 4:104562842 | 1.65 (0.18) | 8.16e-6 | 1.46 (0.24) | 1.94e-2 | 1.62 (0.24) | 8.59e-4 | 1.54 (0.15) | 6.05e-6 | 5.65e -14 |
Crandall 2017 (22) |
| rs78289784 | 4:104580155 | 1.61 (0.18) | 2.12e-5 | 1.46 (0.23) | 1.51e-2 | 1.60 (0.23) | 9.92e-4 | 1.56 (0.15) | 3.28e-6 | 6.55e -14 |
Crandall 2017 (22) |
| rs77322567 | 4:104569676 | 1.64 (0.18) | 8.62e-6 | 1.23 (0.13) | 5.12e-2 | 1.55 (0.22) | 2.24e-3 | 1.53 (0.15) | 6.52e-6 | 2.17e -12 |
Crandall 2017 (22) |
| rs78141901 | 4:104593977 | 1.42 (0.16) | 2.67e-3 | 1.80 (0.37) | 3.88e-3 | 1.35 (0.23) | 7.62e-2 | 1.55 (0.16) | 1.31e-5 | 7.27e -10 |
Crandall 2017 (22) |
| rs78844131 | 4:104600029 | 1.41 (0.16) | 2.85e-3 | 1.49 (0.26) | 2.08e-2 | 1.30 (0.22) | 1.17e-1 | 1.56 (0.16) | 1.14e-5 | 3.34e -09 |
Crandall 2017 (22) |
| rs79852843 | 4:104628587 | 1.44 (0.17) | 1.68e-3 | 1.49 (0.29) | 4.45e-2 | 1.22 (0.20) | 2.22e-1 | 1.58 (0.16) | 5.08e-6 | 4.38e -09 |
Crandall 2017 (22) |
| rs80328778 | 4:104612447 | 1.42 (0.16) | 2.15e-3 | 1.46 (0.27) | 3.67e-2 | 1.24 (0.21) | 1.95e-1 | 1.57 (0.16) | 7.06e-6 | 4.97e -09 |
Crandall 2017 (22) |
| rs112623956 | 4:104623714 | 1.42 (0.16) | 2.33e-3 | 1.49 (0.27) | 2.83e-2 | 1.20 (0.20) | 2.67e-1 | 1.57 (0.16) | 6.19e-6 | 6.19e -09 |
Crandall 2017 (22) |
Odds ratio (OR) is expressed as allelic OR (SE). Reference group: never had vasomotor symptoms. Adjusted for bilateral oophorectomy, age, smoking, alcohol intake, physical activity, population structure, and body mass index. Refsnp identification numbers were obtained from dbSNP Build 144. Chr:Pos denotes chromosome assignment and position of single nucleotide polymorphism (SNP) according to Build 37.
Abbreviations: AA: African American; GARNET, Genome-Wide Association Studies of Treatment Response in Randomized Clinical Trials; HA, Hispanic American; EA, non-Hispanic European ancestry; SHARe, SNP Health Association Resource cohort; WHIMS+: WHI Memory Study cohort.
a Parentheses contain 95% CI if available unless otherwise stated.
b rs numbers were not mentioned in some publications. Single-nucleotide abbreviations are retained as they appear in the publications. Gene names corresponding to abbreviations are taken from the National Center for Biotechnology Information gene database at https://www.ncbi.nlm.nih.gov/gene/.
c In this study, Caucasian race was a choice of racial/ethnic category on the baseline questionnaire.
To aid in interpretation of study results, we display the functions of the proteins encoded by each of the examined genetic variants (Table 9) and the locations and functional consequences of each genetic variant in the included studies (Table 10).
Table 9.
Functions of proteins examined in included studies
| Gene abbreviation | Gene name (per NCBI Gene database) and function of encoded protein (link to Kyoto Encyclopedia of Genes and Genomes [KEGG] website if available)a | Gene databaseb |
|---|---|---|
| AHR | Aryl hydrocarbon receptor | https://www.ncbi.nlm.nih.gov/gene/196 |
| Regulates xenobiotic-metabolizing enzymes such as cytochrome P450. Involved in Th17 cell differentiation and Cushing syndrome https://www.genome.jp/dbget-bin/www_bget?hsa:196 Also see reference (14). |
||
| Also known as RP85; bHLHe76 | ||
| AHRR | Aryl-hydrocarbon receptor repressor | https://www.ncbi.nlm.nih.gov/gene/57491 |
| Participates in aryl hydrocarbon receptor signaling cascade; involved in regulation of cell growth and differentiation. Is a feedback modulator by repressing aryl hydrocarbon receptor-dependent gene expression. KEGG webpage https://www.genome.jp/dbget-bin/www_bget?hsa:57491 Blocks activation of aryl hydrocarbon receptor signaling pathway (14). |
||
| Also known as AHH; AHHR; bHLHe77 | ||
| ARNT | Aryl hydrocarbon receptor nuclear translocator | https://www.ncbi.nlm.nih.gov/gene/405 |
| Binds to ligand-bound aryl hydrocarbon receptor and aids in movement of this complex to the nucleus, where it promotes expression of genes involved in xenobiotic metabolism. Is a co-factor for transcriptional regulation by hypoxia-inducible factor 1. Implicated in Cushing syndrome and carcinogenesis https://www.genome.jp/dbget-bin/www_bget?hsa:405 Also see reference (14). |
||
| Also known as HIF1B; TANGO; bHLHe2; HIF1BETA; HIF-1β; HIF1-β; HIF-1-β | ||
| COMT | Catechol-O-methyltransferase | https://www.ncbi.nlm.nih.gov/gene/1312 |
| Converts 2-hydroxyestrone to 2-methoxyestrone; converts 2-hydroxy-estradiol-17β to 2-methoxy-estradiol-17β (2.1.16) https://www.genome.jp/dbget-bin/www_bget?hsa:1312 | ||
| Also known as HEL-S-98n | ||
| CYP1A1 | cytochrome P450 family 1 subfamily A member 1 | https://www.ncbi.nlm.nih.gov/gene/1543 |
| Converts dehydroepiandrosterone to 16αhydroxyandrost-4ene-3,17-dione; converts estradiol-17β to estriol; converts estrone to 2-hydroxyestrone. (1.14.14.1) https://www.genome.jp/dbget-bin/www_bget?hsa:1543 | ||
| Also known as AHH; AHRR; CP11; CYP1; CYPIA1; P1-450; P450-C; P450DX | ||
| CYP1A2 | Cytochrome P450 family 1 subfamily A member 2 | https://www.ncbi.nlm.nih.gov/gene/1544 |
| Converts dehydroepiandronsterone to 16α-hydroxyandrost-4-ene-3,17-dione. (1.14.14.1) https://www.genome.jp/dbget-bin/www_bget?hsa:1544 Also see references (20) and (3) |
||
| Also known as CP12; CYPIA2; P3-450; P450(PA) | ||
| CYP1B1 | Cytochrome P450 family 1 subfamily B member 1 | https://www.ncbi.nlm.nih.gov/gene/1545 |
| Converts estradiol-17β to 2-hydroxyestradiol-17β; (1.14.14.1) https://www.genome.jp/dbget-bin/www_bget?hsa:1545 Also see references (3, 20, 21, 23) |
||
| Also known as CP1B; ASGD6; GLC3A; CYPIB1; P4501B1 | ||
| CYP2C9 | Cytochrome P450 family 2 subfamily C member 9 | https://www.ncbi.nlm.nih.gov/gene/1559 |
| This gene encodes a member of the cytochrome P450 superfamily of enzymes. The cytochrome P450 proteins are monooxygenases which catalyze many reactions involved in drug metabolism and synthesis of cholesterol, steroids and other lipids. KEGG website https://www.genome.jp/dbget-bin/www_bget?hsa:1559 |
||
| Also known as CPC9; CYP2C; CYP2C10; CYPIIC9; P450IIC9 | ||
| CYP3A4 | Cytochrome P450 family 3 subfamily A member 4 | https://www.ncbi.nlm.nih.gov/gene/1576 |
| Converts estrone to 16α-hydroxyestrone; converts dehydroepiandrosterone to 16α-hydroxyandrost-4-ene-3,17-dione; (1.14.13.32; 1.14.14.55, 1.14.14.56, 1.14.14.57, 1.14.14.73, 1.14.13.-) https://www.genome.jp/dbget-bin/www_bget?hsa:1576 Also see references (3, 20) |
||
| Also known as HLP; CP33; CP34; CYP3A; NF-25; CYP3A3; P450C3; CYPIIIA3; CYPIIIA4; P450PCN1 | ||
| CYP17A1 | Cytochrome P450 family 17 subfamily A member 1 | https://www.ncbi.nlm.nih.gov/gene/1586 |
| Converts 20α-hydroxy-cholesterol to 17α,20α-dihydroxy-cholesterol; converts; converts 17α-hydroxy-pregnenolone to dehydroepiandrosterone; converts pregnenolone to 17α-hydroxy-pregnenolone; converts progesterone to 17α-hydroxy-progesterone; converts 17α-hydroxy-progesterone to androst-4-ene-3,17-dione; converts 21-deoxycortisol to 11β-hydroxy-progesterone (1.14.14.32, 1.14.14.19)) https://www.genome.jp/dbget-bin/www_bget?hsa:1586 |
||
| Also known as CPT7; CYP17; S17AH; P450C17 | ||
| CYP19A1 | Cytochrome P450 family 19 subfamily A member 1 | https://www.ncbi.nlm.nih.gov/gene/1588 |
| Converts testosterone to 19-hydroxy-testosterone; converts 19-hydroxy-testosterone to 19-oxotestosterone; converts 19-oxotestosterone to estradiol-17β; converts androst-4-ene-3,17-dione to 19-hydroxyandrost-4-ene-3,17-dione; converts 19-hydroxyandrost-4-ene-3,17-dione to 19-oxoandrost-4-ene-3,17-dione; converts 19-oxoandrost-4-ene-3,17-dione to estrone (1.14.14.14) https://www.genome.jp/dbget-bin/www_bget?hsa:1588 |
||
| Also known as ARO; ARO1; CPV1; CYAR; CYP19; CYPXIX; P-450AROM | ||
| ESR1 | Estrogen receptor 1 | https://www.ncbi.nlm.nih.gov/gene/2099 |
| This gene encodes an estrogen receptor, a ligand-activated transcription factor composed of several domains important for hormone binding, DNA binding, and activation of transcription. The protein localizes to the nucleus where it may form a homodimer or a heterodimer with estrogen receptor 2. Estrogen and its receptors are essential for sexual development and reproductive function, but also play a role in other tissues such as bone. Receptor to which estradiol binds, ultimately activating cascade that leads to vascular smooth muscle relaxation, influence on gene transcription involving regulation of cell cycle, apoptosis, and cell adhesion molecules https://www.genome.jp/kegg-bin/show_pathway?hsa04915 + 2099 |
||
| Also known as ER; ESR; Era; ESRA; ESTRR; NR3A1 | ||
| ESR2 | Estrogen receptor 2 | https://www.ncbi.nlm.nih.gov/gene/2100 |
| This gene encodes a member of the family of estrogen receptors and superfamily of nuclear receptor transcription factors. The gene product contains an N-terminal DNA binding domain and C-terminal ligand binding domain and is localized to the nucleus, cytoplasm, and mitochondria. On binding to 17β-estradiol or related ligands, the encoded protein forms homodimers or heterodimers that interact with specific DNA sequences to activate transcription. Some isoforms dominantly inhibit the activity of other estrogen receptor family members. Receptor to which estradiol binds, ultimately activating cascade that leads to vascular smooth muscle relaxation, influence on gene transcription involving regulation of cell cycle, apoptosis, and cell adhesion molecules https://www.genome.jp/kegg-bin/show_pathway?hsa04915 + 2099 |
||
| Also known as Erb; ESRB; ODG8; ESTRB; NR3A2; ER-BETA; ESR-BETA | ||
| FLT1 | Fms-related receptor tyrosine kinase 1 | https://www.ncbi.nlm.nih.gov/gene/2321 |
| This gene encodes a member of the vascular endothelial growth factor receptor (VEGFR) family. VEGFR family members are receptor tyrosine kinases (RTKs) which contain an extracellular ligand-binding region with 7 immunoglobulin (Ig)-like domains, a transmembrane segment, and a tyrosine kinase (TK) domain within the cytoplasmic domain. This protein binds to VEGFR-A, VEGFR-B and placental growth factor and plays an important role in angiogenesis and vasculogenesis. KEGG pathway at https://www.genome.jp/dbget-bin/www_bget?hsa:2321 Also see reference (19) |
||
| Also known as FLT; FLT-1; VEGFR1; VEGFR-1 | ||
| HIFA | Hypoxia inducible factor 1 subunit α | https://www.ncbi.nlm.nih.gov/gene/3091 |
| This gene encodes the α subunit of transcription factor hypoxia-inducible factor-1 (HIF-1), which is a heterodimer composed of an α and a β subunit. HIF-1 functions as a master regulator of cellular and systemic homeostatic response to hypoxia by activating transcription of many genes, including those involved in energy metabolism, angiogenesis, apoptosis, and other genes whose protein products increase oxygen delivery or facilitate metabolic adaptation to hypoxia. Link to KEGG pathway: https://www.genome.jp/kegg-bin/show_pathway?hsa04066 + 3091 Also see reference (19) |
||
| Also known as HIF1; MOP1; PASD8; HIF-1A; bHLHe78; HIF-1α; HIF1-ALPHA; HIF-1-α | ||
| HSD3B1 | Hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 1 | https://www.ncbi.nlm.nih.gov/gene/3283 |
| The protein encoded by this gene is an enzyme that catalyzes the oxidative conversion of delta-5-3-beta-hydroxysteroid precursors into delta-4-ketosteroids, which leads to the production of all classes of steroid hormones. The encoded protein also catalyzes the interconversion of 3-β-hydroxy- and 3-keto-5-α-androstane steroids. Link to KEGG pathway: https://www.genome.jp/kegg-bin/show_pathway?map00140 Converts 21-hydroxy-pregnenolone to 11-deoxy-corticosterone, interconverts pregnenolone and protesterone, interconverts 17α-hydroxy-progesterone and 17α-hydroxy-pregnenolone, converts 17α,21-dihydroxyo-pregnenolone to 11-deoxycortisol, converts 11β,17α,21-trihydroxy-pregnenolone to cortisol, converts dehydroepiandronaterone to androst-4-ene-3,17-dione, converts 3β,17β-dihydroxy-androst-5-ene to testosterone |
||
| Also known as HSD3B; HSDB3; HSDB3A; SDR11E1; 3BETAHSD | ||
| HSD17B1 | Hydroxysteroid 17-β dehydrogenase 1 | https://www.ncbi.nlm.nih.gov/gene/3292 |
| Converts 4-androsten-11beta-ol-3,17-dione to 11β-hydroxytestosterone; Interconverts estrone to estradiol-17β; interconverts 16-α-hydroxyestrone and estriol (1.1.62) https://www.genome.jp/dbget-bin/www_bget?hsa:3292 |
||
| Also known as E2DH; HSD17; EDHB17; EDH17B2; SDR28C1; 17-beta-HSD; 20-alpha-HSD | ||
| KDR | Kinase insert domain receptor | https://www.ncbi.nlm.nih.gov/gene/3791 |
| Vascular endothelial growth factor (VEGF) is a major growth factor for endothelial cells. This gene encodes 1 of the 2 receptors of the VEGF. This receptor, known as kinase insert domain receptor, is a type III receptor tyrosine kinase. It functions as the main mediator of VEGF-induced endothelial proliferation, survival, migration, tubular morphogenesis, and sprouting. KEGG pathway at https://www.genome.jp/kegg-bin/show_pathway?hsa04370 + 3791 Also see reference (19) |
||
| Also known as FLK1; CD309; VEGFR; VEGFR2 | ||
| NRP1 | Neuropilin 1 | https://www.ncbi.nlm.nih.gov/gene/8829 |
| Neuropilins bind many ligands and various types of co-receptors; they affect cell survival, migration, and attraction. Some of the ligands and co-receptors bound by neuropilins are vascular endothelial growth factor (VEGF) and semaphorin family members. | ||
| Also known as NP1; NRP; BDCA4; CD304; VEGF165R | ||
| NRP2 | Neuropilin 2 | https://www.ncbi.nlm.nih.gov/gene/8828 |
| The encoded transmembrane protein binds to SEMA3C protein (sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C} and SEMA3F protein (sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F}, and interacts with vascular endothelial growth factor (VEGF). | ||
| Also known as NP2; NPN2; PRO2714; VEGF165R2 | ||
| NOS3 | Nitric oxide synthase 3 | https://www.ncbi.nlm.nih.gov/gene/4846 |
| Involved in estrogen signaling pathway https://www.genome.jp/dbget-bin/www_bget?hsa:4846; after estradiol binds to estrogen receptor, converts Akt (protein kinase B) to nitric oxide https://www.genome.jp/kegg-bin/show_pathway?hsa04915 + 4846 Also see reference (19) |
||
| Also known as eNOS; ECNOS | ||
| Serotonin transporter gene see SLC6A4 below | Solute carrier family 6 member 4 | https://www.ncbi.nlm.nih.gov/gene/6532 |
| SLC6A4 | Solute carrier family 6 member 4 | https://www.ncbi.nlm.nih.gov/gene/6532 |
| This gene encodes an integral membrane protein that transports the neurotransmitter serotonin from synaptic spaces into presynaptic neurons. The encoded protein terminates the action of serotonin and recycles it in a sodium-dependent manner. This protein is a target of psychomotor stimulants, such as amphetamines and cocaine, and is a member of the sodium:neurotransmitter symporter family. KEGG pathway: https://www.genome.jp/dbget-bin/www_bget?hsa:6532 Also see reference (27) |
||
| Also known as HTT; 5HTT; OCD1; SERT; 5-HTT; SERT1; hSERT; 5-HTTLPR | ||
| SULT1A1 | Sulfotransferase family 1A member 1 | https://www.ncbi.nlm.nih.gov/gene/6817 |
| Sulfotransferase enzymes catalyze the sulfate conjugation of many hormones, neurotransmitters, drugs, and xenobiotic compounds. KEGG pathway page https://www.genome.jp/dbget-bin/www_bget?hsa:6817 Interconverts 4-hydroxyestrdaiol and 4-hydroxy-estrdaiol-sulfate; interconverts estradiol and estradiol sulfate; converts 2-hydroxyestrdaiol to 2-hydroxy-estrdaiol sulfate https://www.wikipathways.org/index.php/Pathway:WP697 |
||
| Also known as PST; STP; STP1; P-PST; ST1A1; ST1A3; TSPST1; HAST1/HAST2 | ||
| SULT1E1 | Sulfotransferase family 1E member 1 | https://www.ncbi.nlm.nih.gov/gene/6783 |
| Converts estrone to estrone 3-sulfate (2.8.2.4) https://www.genome.jp/dbget-bin/www_bget?hsa:6783 |
||
| Also known as EST; STE; EST-1; ST1E1 | ||
| TACR3 | Tachykinin receptor 3 | https://www.ncbi.nlm.nih.gov/gene/6870 |
| This gene encodes the receptor for the tachykinin neurokinin 3, also referred to as neurokinin B. Neurokinin B is a neuropeptide and mutations in the gene for neurokinin B are associated with normosmic hypogonadotropic hypogonadism. https://www.ncbi.nlm.nih.gov/gene/6866 | ||
| Also known as NK3; NKR; HH11; NK3R; NK-3R; TAC3R; TAC3RL | ||
| UGT1A1 | Uridine diphosphate (UDP) glucuronosyltransferase family 1 member A1 | https://www.ncbi.nlm.nih.gov/gene/54658 |
| This gene encodes a UDP-glucuronosyltransferase, an enzyme of the glucuronidation pathway that transforms small lipophilic molecules, such as steroids, bilirubin, hormones, and drugs, into water-soluble, excretable metabolites. converts estradiol-17β to estradiol-17β-3-glucuronide; converts estrone to estrone glucuronide; 2-methoxyestrone to 2-methoxyestrone-3-glucuronide; converts estriol to 16-glucuronide-estriol; converts etiocholan-3α-ol-17-one to etiocholan-3α-ol-17-one 3-glucuronide; converts androsterone to androsterone-glucuronide; converts 2-methoxy-estradiol-17β to 2-methoxy-estradiol-17β-3-glucuronide; converts testosterone to testosterone glucuronide (2.4.1.117) https://www.genome.jp/kegg-bin/show_pathway?hsa00140 + 10720 |
||
| Also known as GNT1; UGT1; UDPGT; UGT1A; HUG-BR1; BILIQTL1; UDPGT 1-1 | ||
| VEGFA | Vascular endothelial growth factor A | https://www.ncbi.nlm.nih.gov/gene/7422 |
| This gene is a member of the PDGF/VEGF growth factor family. It encodes a heparin-binding protein, which exists as a disulfide-linked homodimer. This growth factor induces proliferation and migration of vascular endothelial cells, and is essential both for physiological and pathological angiogenesis. KEGG signaling pathway: https://www.genome.jp/kegg-bin/show_pathway?hsa04370 + 7422 Also see reference (19) |
||
| Also known as VPF; VEGF; MVCD1 | ||
| VEGFR1 see FLT1 above | Fms-related receptor tyrosine kinase 1 | https://www.ncbi.nlm.nih.gov/gene/2321 |
| VEGFR2 see KDR above | kinase insert domain receptor | https://www.ncbi.nlm.nih.gov/gene/3791 |
ahttps://www.genome.jp/kegg-bin/show_pathway?ec00140 + 2.8.2.4. If not listed in KEGG, then alternate reference for function of enzyme is cited.
bNCBI gene database at https://www.ncbi.nlm.nih.gov/gene
Table 10.
Characteristics of genetic variants examined in included studies
| Gene name | dbSNP rs number (alternate name) | Position | Alleles | Consequence | Amino acid or amino acid change | Uniform Resource Locator for dbSNP websitea |
|---|---|---|---|---|---|---|
| AHR | rs2066853 | chr7:17339486 | G>A | Missense variant | Arg > Lys | https://www.ncbi.nlm.nih.gov/snp/rs2066853 |
| AHRR | rs2292596 | chr5:422840 | C>G/C>T | Missense variant | Pro > Ala or Pro > Ser | https://www.ncbi.nlm.nih.gov/snp/rs2292596 |
| ARNT | rs2228099 | chr1:150836413 | C>G | Synonymous variant | Val>Val | https://www.ncbi.nlm.nih.gov/snp/rs2228099 |
| COMT | rs4680 (Val158Met) | chr22:19963748 | G>A | Missense variant | Val>Met | https://www.ncbi.nlm.nih.gov/snp/rs4680 |
| CYP1A1 | rs2606345 (CYP2606345,-1806) | chr15:74724835 | C>A | Intron variant | NA | https://www.ncbi.nlm.nih.gov/snp/rs2606345 |
| CYP1A2 | rs762551 (CYP1A2*1F) | chr15:74749576 | C>A | Intron variant | NA | https://www.ncbi.nlm.nih.gov/snp/rs762551 |
| CYP1B1 | rs10012 (Arg48Gly) | chr2:38075247 | G>C | Missense variant | Arg > Gly | https://www.ncbi.nlm.nih.gov/snp/rs10012 |
| rs1056827 (Ala119Ser) | chr2:38075034 | C>A | Missense variant | Ala > Ser | https://www.ncbi.nlm.nih.gov/snp/rs1056827 | |
| rs1056836 (CYP1B1*3, Leu432Val, 4326C>G, C1294G) | chr2:38071060 | G>C | Missense variant | Leu > Val | https://www.ncbi.nlm.nih.gov/snp/rs1056836 | |
| rs1800440 (CYP1B1*4, Asn452Ser, Asn453Ser) | chr2:38070996 | T>C/T>G | Missense variant | Asn > Ser or Asn > Thr | https://www.ncbi.nlm.nih.gov/snp/rs1800440 | |
| CYP3A4 | rs2740574 (CYP3A4*1B) | chr7:99784473 | C>T | 2KB upstream variant | NA | https://www.ncbi.nlm.nih.gov/snp/rs2740574 |
| CYP19A1 (CYP19) | rs700519 (Arg264Cys) | chr15:51215771 | G>A | Missense variant | Arg > Cys | https://www.ncbi.nlm.nih.gov/snp/rs700519 |
| rs2389 (TTTA[n]) | chr3:104780426 | T>A/T>C | Intron variant | NA | https://www.ncbi.nlm.nih.gov/snp/rs2389 | |
| ESR1 | rs2234693 (ESRA418, PvuII) | chr6:151842200 | T>C | Intron variant | NA | https://www.ncbi.nlm.nih.gov/snp/rs2234693 |
| rs9340799 (XbaI, ESRA 464) | chr6:151842246 | A>G | Intron variant | NA | https://www.ncbi.nlm.nih.gov/snp/rs9340799 | |
| HSD17B1 | rs2830 (HSD17B2830) | chr17:42552545 | G>A | 2KB upstream variant | NA | https://www.ncbi.nlm.nih.gov/snp/rs2830 |
| rs592389 (HSD592389) | chr17:42555426 | A>C | 500b downstream variant | NA | https://www.ncbi.nlm.nih.gov/snp/rs592389 | |
| rs615942b (HSD615942) | chr17:42562786 | C>A | Missense variant | Ser > Tyr | https://www.ncbi.nlm.nih.gov/snp/rs615942 | |
| SLC6A4 (serotonin transporter gene) | rs11080121 | chr17:30201824 | T>C | Intron variant | NA | https://www.ncbi.nlm.nih.gov/snp/rs11080121 |
| rs140700 | chr17:30216371 | C>A/C>G/C>T | Intron variant | NA | https://www.ncbi.nlm.nih.gov/snp/rs140700 | |
| rs8076005 | chr17:30220192 | G>A | Intron variant | NA | https://www.ncbi.nlm.nih.gov/snp/rs8076005 | |
| rs2066713 | chr17:30224647 | G>A | Intron variant | NA | https://www.ncbi.nlm.nih.gov/snp/rs2066713 | |
| rs4251417 | chr17:30224840 | C>T | Intron variant | NA | https://www.ncbi.nlm.nih.gov/snp/rs4251417 | |
| rs16965628 | chr17:30228407 | G>C | Intron variant | N/A | https://www.ncbi.nlm.nih.gov/snp/rs16965628 | |
| rs4404067c | chr16:55649770 | G>A | None | https://www.ncbi.nlm.nih.gov/snp/rs4404067 | ||
| rs4238784d | chr16:55653153 | A>G | None | https://www.ncbi.nlm.nih.gov/snp/rs4238784 | ||
| rs747107 | chr16:55661809 | T>A/T>C | Intron variant | NA | https://www.ncbi.nlm.nih.gov/snp/rs747107 | |
| rs13333066 | chr16:55669136 | T>A/T>C | Intron variant | NA | https://www.ncbi.nlm.nih.gov/snp/rs13333066 | |
| rs187715 | chr16:55670130 | T>C | Intron variant | NA | https://www.ncbi.nlm.nih.gov/snp/rs187715 | |
| rs36026 | chr16:55670883 | C>A | Intron variant | NA | https://www.ncbi.nlm.nih.gov/snp/rs36026 | |
| rs36024 | chr16:55672479 | A>G | Intron variant | NA | https://www.ncbi.nlm.nih.gov/snp/rs36024 | |
| rs36021 | chr16:55678038 | T>A | Intron variant | NA | https://www.ncbi.nlm.nih.gov/snp/rs36021 | |
| rs3785151 | chr16:55678607 | G>C | Intron variant | NA | https://www.ncbi.nlm.nih.gov/snp/rs3785151 | |
| rs36020 | chr16:55679176 | C>T | Intron variant | NA | https://www.ncbi.nlm.nih.gov/snp/rs36020 | |
| rs16955591 | chr16:55679874 | C>T | Intron variant | NA | https://www.ncbi.nlm.nih.gov/snp/rs16955591 | |
| rs3785152 | chr16:55682638 | T>C | Intron variant | NA | https://www.ncbi.nlm.nih.gov/snp/rs3785152 | |
| rs40147 | chr16:55682928 | G>A/G>C | Intron variant | NA | https://www.ncbi.nlm.nih.gov/snp/rs40147 | |
| rs1814270 | chr16:55683165 | T>C | Intron variant | NA | https://www.ncbi.nlm.nih.gov/snp/rs1814270 | |
| rs5564 | chr16:55692063 | A>G | Intron variant | NA | https://www.ncbi.nlm.nih.gov/snp/rs5564 | |
| rs5568 | chr16:55696212 | A>C/A>G | Intron variant | NA | https://www.ncbi.nlm.nih.gov/snp/rs5568 | |
| rs1566652 | chr16:55697663 | G>T | Intron variant | NA | https://www.ncbi.nlm.nih.gov/snp/rs1566652 | |
| rs5569 | chr16:55697923 | G>A/G>C | Missense variant | Gly > Arg | https://www.ncbi.nlm.nih.gov/snp/rs5569 | |
| rs2242447 | chr16:55702000 | C>T | Intron variant | NA | https://www.ncbi.nlm.nih.gov/snp/rs2242447 | |
| rs424605e | chr7:138324023 | A>G | None | https://www.ncbi.nlm.nih.gov/snp/rs424605 | ||
| rs16955708f | chr16:55707544 | T>C | None | https://www.ncbi.nlm.nih.gov/snp/rs16955708 | ||
| rs12596924g | chr16:55710735 | T>G | None | https://www.ncbi.nlm.nih.gov/snp/rs12596924 | ||
| rs258099 | chr16:55713265 | C>G/C>T | None | https://www.ncbi.nlm.nih.gov/snp/rs258099 | ||
| SULT1A1 | rs9282861 (SULT1A1*2 Arg213His)—has merged into rs1042028h | chr16:28606193 | C>T | Missense variant | Arg > His | https://www.ncbi.nlm.nih.gov/snp/rs1042028 |
| rs1801030 (SULTA1*3 Met223Val) | chr16:28606164 | C>G/C>T | Missense variant | Val > Leu or Val > Met | https://www.ncbi.nlm.nih.gov/snp/rs1801030 | |
| SULT1E1 | rs3736599 (5’UTR promoter variant –64G→A) | chr4:69860103 | C>T | 5 prime UTR variant | NA | https://www.ncbi.nlm.nih.gov/snp/rs3736599 |
| rs3786599 (A220G)i | chr19:16618799 | C>T | Intron variant | NA | https://www.ncbi.nlm.nih.gov/snp/rs3786599 | |
| VEGFR1 see FLT1 above | ||||||
| VEGFR2 see KDR above |
Abbreviations: chr, chromosome; NA, not applicable to gene.
a The homepage for NCBI dbSNP is https://www.ncbi.nlm.nih.gov/snp/, version released July 9, 2019
b In dbSNP, this single-nucleotide variant is listed as being in the gene for Coenzyme A synthase.
c dbSNP does not list this single nucleotide variant as being located in the serotonin transporter gene.
d dbSNP does not list this single nucleotide variant as being located in the serotonin transporter gene.
e dbSNP does not list this single nucleotide variant as being located in the serotonin transporter gene.
f dbSNP does not list this single nucleotide variant as being located in the serotonin transporter gene.
g dbSNP does not list this single nucleotide variant as being located in the serotonin transporter gene.
i dbSNP lists this single nucleotide variant as being located in the mediator complex subunit 26.
Guidelines for reporting genetic association studies recommend that studies clearly define genetic exposures (genetic variants) using a widely used nomenclature system (214). The typical nomenclature system is the dbSNP Reference SNP cluster identification numbers (rs numbers), accessible in a public domain database maintained by the National Center for Biotechnology Information (https://www.ncbi.nlm.nih.gov/snp/). For a given genetic variant, the RefSNP rs number is the stable accession, regardless of differences in genomic assemblies. Of the 18 citations that met the inclusion criteria, 8 studies included rs numbers of the examined genetic variants in the published reports (3, 14, 16, 20-23, 27) and rs numbers were obtained from communication with the author of one further study (Montasser 2015, [25]). Those 9 studies for which rs numbers were provided are summarized separately in Table 11.
Table 11.
Summary table of findings classified by rs numbera
| Rs No. and gene name | Studies that reported statistically significant associations with vasomotor symptoms | Studies that reported absence of statistically significant associations with vasomotor symptoms | ||
|---|---|---|---|---|
| No. of studies | Rs No., first author, reference No. | No. of studies | Rs No., first author, reference No. | |
| Aryl hydrocarbon receptor (AHR) | 1 | rs2066853 Ziv-Gal 2012 (14) | – | – |
| Aryl-hydrocarbon receptor repressor (AHRR) | – | – | 1 | rs2292596 Ziv-Gal 2012 (14) |
| Aryl hydrocarbon receptor nuclear translocator (ARNT) | – | – | 1 | rs2228099 Ziv-Gal 2012 (14) |
| Catechol-O-methyltransferase (COMT) | 1 | rs4680 Butts 2012 (20)b | 1 | rs4680 Rebbeck 2010 (3) |
| Cytochrome P450 family 1 subfamily A member 1 (CYP1A1) | 1 | rs2606345 Crandall 2006 (23) | – | – |
| Cytochrome P450 family 1 subfamily A member 2 (CYP1A2) | 1 | rs762551 Butts 2012 (20)c | 1 | rs762551 Rebbeck 2010 (3) |
| Cytochrome P450 family 1 subfamily B member 1 (CYP1B1) | 4 | rs1056836, rs1800440, Butts 2012 (20)d; rs10012 Candrakova 2018 (21); rs1056836 Rebbeck 2010 (3); rs1800440 Ziv-Gal 2012 (14) | 3 | rs1056827, rs1056836, rs1800440 Candrakova 2018 (21); rs1056836 Crandall 2006 (23); rs1800440 Rebbeck 2010 (3) |
| Cytochrome P450 family 3 subfamily A member 4 (CYP3A4) | – | – | 2 | rs2740574 Butts 2012 (185)e; rs2740574 Rebbeck 2010 (3) |
| Cytochrome P450 family 19 subfamily A member 1 (CYP19A1) | – | – | 2 | rs700519 Rebbeck 2010 (3); rs2389 Woods 2018 (16) |
| Estrogen receptor 1 (ESR1) | 1 | rs2234693 Malacara 2004 (25) | 1 | rs9340799 Malacara 2004 (25) |
| Hydroxysteroid 17-β dehydrogenase 1 (HSD17B1) | 2 | rs2830, rs592389, rs615942 Crandall 2006; rs615942, rs592389 Woods 2018 (16) | 1 | rs2830 Woods 2018 (16) |
| Solute carrier family 6 member 4 (SLC6A4) (serotonin transporter gene) | 1 | rs11080121; rs2066713; rs40147 Montasser 2015 (27) | 1 | rs140700, rs8076005, rs4251417, rs16965628, rs4404067, rs4238784, rs747107, rs13333066, rs187715, rs36026, rs36024, rs36021, rs3785151, rs36020, rs16955591, rs3785152, rs1814270, rs5564, rs5568, rs1566652, rs5569, rs2242447, rs424605, rs16955708, rs12596924, rs258099 Montasser 2015 (27) |
| Sulfotransferase family 1A member 1 (SULT1A1) | 1 | rs1801030 Rebbeck 2010 (3) | 1 | rs9282861 Rebbeck 2010 (3) |
| sulfotransferase family 1E member 1 (SULT1E1) | – | – | 1 | rs3736599, rs3786599 Rebbeck 2010 (3) |
| Tachykinin receptor 3 | 1 | rs74827081, rs74589515, rs79246187, rs112390256, rs75544266, rs78154848, rs76643670, rs78289784, rs77322567, rs78141901, rs78844131, rs79852843, rs80328778, rs112623956 Crandall 2017 (22) | – | – |
a – indicates no studies fulfill the criteria
b Studies focused on differences between smokers and nonsmokers with a given genotype
c Studies focused on differences between smokers and nonsmokers with a given genotype
d Studies focused on differences between smokers and nonsmokers with a given genotype
e Studies focused on differences between smokers and nonsmokers with a given genotype
In candidate gene studies, significant associations were found between SNPs in several genes and VMS. These genes include the aryl hydrocarbon receptor, aryl hydrocarbon receptor repressor, aryl hydrocarbon receptor nuclear translocator, catechol-O-methyltransferase, cytochrome P450 (CYP) family 1 subfamily A member 1, CYP family 1 subfamily A member 2, CYP450 family 1 subfamily B member 1 (CYP1B1), CYP3 subfamily A member 4, CYP19 subfamily A member 1, estrogen receptor 1, hydroxysteroid 17-β dehydrogenase 1, solute carrier family 6 member 4 (also known as serotonin transporter), sulfotransferase family 1A member 1, and sulfotransferase family 1E genes.
There were few studies of each specific genetic variant in relation to VMS, which precluded quantitative meta-analysis.
The gene that was the focus of the largest number of studies was CYP1B1; it was the focus of 7 studies. Most SNPs were the focus of only a single study. Results of studies of the same genetic variant sometimes conflicted with each other, and sometimes the studies focused on whether magnitudes of associations between CYP1B1 variants and VMS were different in smokers and nonsmokers, rather than associations between the genetic variants and VMS per se (20). One study found that CYP1B1 rs1800440 was significantly associated with VMS, with 3-fold higher odds of having hot flashes for 1 or more years among women with the GG genotype compared with women who had the AA genotype (adjusted odds ratio [aOR] 3.05, 95% CI, 1.12-8.25) after adjustment for covariates (14). However, the same variant was not significantly associated with VMS in 2 other studies; 1 of the 2 studies reported an aOR of 0.66 with a 95% CI of 0.33 to 1.30 for the any 4* variant allele (vs reference genotype *1/*1) (3), and the other study did not report the effect size and 95% CI (21). Similarly, rs1056836 was associated with VMS in one study (3); African American women with the rs1056836 variant (at least one allele, Leu432Val) had approximately 40% lower odds of moderate or severe hot flashes (aOR 0.62, 95% CI, 0.40-0.95), but other studies found no association between rs1056836 and VMS; 1 of the 2 studies found an aOR of 0.87 (95% CI, 0.44-1.70) for the GG genotype (vs CC genotype referent) among African American women (23) and the other did not mention the effect size and 95% CI (21).
Among CYP1A2 studies, results were conflicting regarding rs 762551 (3, 20). In the 2 studies of hydroxysteroid 17-β dehydrogenase 1, results were conflicting for rs2830 (16, 23).
In a sensitivity analyses, for studies that did not provide SNP rs numbers, we attempted to obtain rs numbers corresponding to the specific locations of SNPs described (Table 12). Compared with the main results, the sensitivity analysis revealed additional evidence of an absence of associations between several SNPs in CYP1B1, CYP1A1, and estrogen receptor genes and VMS, but added to evidence suggesting associations between the CYP1B1 rs1056836 SNP and VMS. The sensitivity analysis also revealed 2 genes, CYP17A1 and estrogen receptor 2, that were not included in the studies described in Table 10. Specifically, 1 study found a significant association between the estrogen receptor 2 cytosine-adenine dinucleotide repeat length in intron 5 and VMS; conversely, 2 studies reported that there were no significant associations between the CYP17A1 variant (rs743572) and VMS. Finally, several SNPs in the hypoxia inducible factor 1 subunit α, kinase insert domain receptor, nitric oxide synthase, and vascular endothelial growth factor A genes were included in the sensitivity analysis, each in only a single study, and only one reporting a statistically significant association between the SNP (rs11549465) and VMS (aOR 1.27, 95% CI, 1.01-1.59).
Table 12.
Summary table of findings classified by rs number from sensitivity analysisa
| Rs No. and gene name | No. of studies that reported statistically significant associations | First author, reference No. | No. of studies that reported absence of statistically significant associations | First author, reference No. |
|---|---|---|---|---|
| Aryl hydrocarbon receptor (AHR) | 1 | rs2066853 Ziv-Gal 2012 (14) | - | - |
| Aryl-hydrocarbon receptor repressor (AHRR) | – | – | 1 | rs2292596 Ziv-Gal 2012 (14) |
| Aryl hydrocarbon receptor nuclear translocator (ARNT) | – | – | 1 | rs2228099 Ziv-Gal 2012 (14) |
| Catechol-O-methyltransferase (COMT) | 1 | rs4680 Butts 2012 (20)b | 1 | rs4680 Rebbeck 2010 (3) |
| Cytochrome P450 family 1 subfamily A member 1 (CYP1A1) | 1 | rs2606345 Crandall 2006 (23) | 1 | rs1048943 c (CYP1A1m2, missense variant) Woods 2006 (15) |
| Cytochrome P450 family 1 subfamily A member 2 (CYP1A2) | 1 | rs762551 Butts 2012 (20)d | 1 | rs762551 Rebbeck 2010 (3) |
| Cytochrome P450 family 1 subfamily B member 1 (CYP1B1) | 4 | rs1056836, rs1800440, Butts 2012 (20)e; rs10012 Candrakova 2018 (21); rs1056836 Rebbeck 2010 (3) rs1800440 Ziv-Gal 2012 (14); rs1056836f (Leu->Val at codon 432) Visvanathan 2005 (17) | 4 | rs1056827; rs1056836; rs1800440 Candrakova 2018 (21); rs1056836 Crandall 2006 (23); rs1800440 Rebbeck 2010 (3), rs1056827g (CYP1B1*2, missense variant); rs1056836h (CYP1B1*3, missense variant) Woods 2006 (15); rs1056836i (Leu432Val) Luptakova 2012 (29) |
| Cytochrome P450 family 3 subfamily A member 4 (CYP3A4) | – | – | 2 | rs2740574 Butts 2012 (185)j; rs2740574 Rebbeck 2010 (3) |
| Cytochrome P450 family 17 subfamily A member 1 (CYP17A1) | 2 | rs743572 k (MspA1 5’ untranslated region variant) Massad-Costa 2008 (26); rs743572l (MspA1 5’ untranslated region variant) Visvanathan 2005 (17) | ||
| Cytochrome P450 family 19 subfamily A member 1 (CYP19A1) | – | - | 2 | rs700519 Rebbeck 2010 (3); rs2389 Woods 2018 (16) |
| Estrogen receptor 1 (ESR1) | 1 | rs2234693 Malacara 2004 (25) | 2 | rs9340799 Malacara 2004 (25), rs 9340799m (ESRXbaI, intron variant); rs2234693n (ESR1PvuII, intron variant) Woods 2006 (15), rs9340799 (ESRXbaI) Aguilar Zavala 2012 (30), rs2234693p (ESR1PvuII, intron variant) Aguilar-Zavala 2012 (30) |
| Estrogen receptor 2 (ESR2) | 1 | Cytosine-adenine dinucleotide repeat length in intron 5 Takeo 2005 (18) | ||
| Hydroxysteroid 17-β dehydrogenase 1 (HSD17B1) | 2 | rs2830; rs592389; rs615942 Crandall 2006; rs615942; rs592389 Woods 2018 (16) | 1 | rs2830 Woods 2018 (16) |
| Hypoxia inducible factor 1 subunit α (HIFA) | rs11549465 (HIFα 1744 C/T) q Schneider 2009 (19) | rs11549467 (HIFα 1762 A/G) r Schneider 2009 (19) | ||
| Kinase insert domain receptor (KDR) | rs2305948 (VEGFR2 889 A/G) s , rs1870377 (VEGFR2 1416 A/T) t Schneider 2009 (19) | |||
| Nitric oxide synthase 3 (eNOS) | rs2070744 (eNOS-786 T/C) u , rs1799983 (eNOS 894 G/T) v Schneider 2009 (19) | |||
| Solute carrier family 6 member 4 (SLC6A4) (serotonin transporter gene) | 1 | rs11080121; rs2066713; rs40147 Montasser 2015 (27) | 1 | rs140700; rs8076005; rs4251417; rs16965628; rs4404067; rs4238784; rs747107; rs13333066; rs187715; rs36026; rs36024; rs36021; rs3785151; rs36020; rs16955591; rs3785152; rs1814270; rs5564; rs5568; rs1566652; rs5569; rs2242447; rs424605; rs16955708; rs12596924; rs258099; Montasser 2015 (27) |
| Sulfotransferase family 1A member 1 (SULT1A1) | 1 | rs1801030 Rebbeck 2010 (3) | 1 | rs9282861 Rebbeck 2010 (3) |
| sulfotransferase family 1E member 1 (SULT1E1) | – | – | 1 | rs3736599; rs3786599 Rebbeck 2010 (3) |
| Tachykinin receptor 3 | 1 | rs74827081; rs74589515; rs79246187; rs112390256; rs75544266; rs78154848; rs76643670; rs78289784; rs77322567; rs78141901; rs78844131; rs79852843; rs80328778; rs112623956 Crandall 2017 (22) | – | – |
| Vascular endothelial growth factor A (VEGFA) | rs699947 (VEGF-2578 C/A)x, rs2010963 (VEGF-634 G/C)y, rs1570360 (VEGF-1154 G/C)z, rs3025039 (VEGF 936 C/T)aa Schneider 2009 (19) |
a – indicates that no studies fulfill the criteria; studies in boldface indicate additional results revealed by the sensitivity analysis.
b Studies were focused on differences between smokers and nonsmokers with a given genotype
c Reference for corresponding rs number is Naif et al 2019 PMID 29707532; dbsnp link https://www.ncbi.nlm.nih.gov/snp/rs1048943
d Studies were focused on differences between smokers and nonsmokers with a given genotype
e Studies were focused on differences between smokers and nonsmokers with a given genotype
f Reference for corresponding rs number is Candrakova 2018 PMID: 29285838; dbSNP link https://www.ncbi.nlm.nih.gov/snp/rs1056836
g Reference for corresponding rs number is Reding et al 2009 PMID 19383894; dbSNP link https://www.ncbi.nlm.nih.gov/snp/rs1056827
h Reference for corresponding rs number is Martinez-Ramirez 2015 PMID 26123186; dbSNP link https://www.ncbi.nlm.nih.gov/snp/rs1056836
i Reference for corresponding rs number is Butts 2012 PMID 22466345; dbSNP link https://www.ncbi.nlm.nih.gov/snp/rs1056836
j Studies were focused on differences between smokers and nonsmokers with a given genotype
k Reference for corresponding rs number is Mao et al 2010 PMID 20033766; dbSNP link https://www.ncbi.nlm.nih.gov/snp/rs743572
l Reference for corresponding rs number is Mao et al 2010 PMID 20033766; dbSNP link https://www.ncbi.nlm.nih.gov/snp/rs743572
m Reference for corresponding rs number is Hayes et al 2010 PMID 20827267; dbSNP link https://www.ncbi.nlm.nih.gov/snp/rs9340799
n Reference for corresponding rs number is Hayes et al 2010 PMID 20827267; dbsnp link https://www.ncbi.nlm.nih.gov/snp/rs2234693
o Reference for corresponding rs number is Hayes et al 2010 PMID 20827267; dbSNP link https://www.ncbi.nlm.nih.gov/snp/rs9340799
p Reference for corresponding rs number is Hayes et al 2010 PMID 20827267; dbSNP link https://www.ncbi.nlm.nih.gov/snp/rs2234693
q Reference for corresponding rs number is Heino et al 2008 PMID 18980686; dbSNP link https://www.ncbi.nlm.nih.gov/snp/rs11549465
r Reference for corresponding rs number is Heino et al 2008 PMID 18980686; dbSNP link https://www.ncbi.nlm.nih.gov/snp/rs11549467
s Reference for corresponding rs number is Lopes-Aguiar et al 2017 PMID 28665417; dbSNP link https://www.ncbi.nlm.nih.gov/snp/rs2305948
t Reference for corresponding rs number is Lakkireddy et al 2016 PMID 26476544; dbSNP link https://www.ncbi.nlm.nih.gov/snp/rs1870377
u Reference for corresponding rs number is Loo et al 2012 PMID 22594584; dbSNP link: https://www.ncbi.nlm.nih.gov/snp/rs2070744
v Reference for corresponding rs number is Loo et al 2012 PMID 22594584; dbSNP link: https://www.ncbi.nlm.nih.gov/snp/rs1799983
x Reference for corresponding rs number is Schneider et al 2008 PMID 17891484; dbSNP link: https://www.ncbi.nlm.nih.gov/snp/rs699947
y Reference for corresponding rs number is Schneider et al 2008 PMID 17891484; dbSNP link: https://www.ncbi.nlm.nih.gov/snp/rs2010963
z Reference for corresponding rs number is Schneider et al 2008 PMID 17891484; dbSNP link: https://www.ncbi.nlm.nih.gov/snp/rs1570360
aa Reference for corresponding rs number is Schneider et al 2008 PMID 17891484; dbSNP link: https://www.ncbi.nlm.nih.gov/snp/rs3025039
Studies varied regarding whether results of analyses were adjusted for covariates and which specific covariates were selected (Table 13). Some studies did not mention adjustment for covariates (18, 26), one was not adjusted but was stratified by menopausal stage (15), and one (a meeting abstract) did not mention which effect measure was used (eg, OR, risk ratio) (24). Other studies included covariates in their analyses (3, 14, 16, 17, 19-21, 23, 25, 27-30, 189). Covariates commonly included were age, body mass index (BMI), menopausal status, race, and smoking. It is possible that differences in results across various studies are partly attributable to differences in covariates selected for inclusion. Studies often did not adjust analyses to account for multiple statistical comparisons; not correcting for multiple statistical comparisons increases the chance of committing a Type I error, i.e. rejecting the null hypothesis (of no association between genotype and VMS) when such an association truly does not exist.
Table 13.
Covariates included in the studies
| Study first author (citation) | Covariates | Adjustment for multiple statistical comparisons described |
|---|---|---|
| Aguilar-Zavala (30) | Age, y since menopause, pregnancies, scores for dominance, submission, circulating levels of follicle-stimulating hormone and estradiol | Yes (Bonferroni) |
| Butts 2012 (20) | BMI, alcohol consumption, MT stage, age, anxiety, stratified by race | No |
| Candrakova 2018 (21) | Age, BMI, physical activity, education, smoking | No |
| Crandall 2006 (23) | Baseline premenstrual symptoms, MT status, serum sex hormone-binding globulin level, serum estradiol level, serum follicle-stimulating hormone level, d of menstrual cycle on which phlebotomy occurred, symptoms sensitivity, passive smoking, active smoking, anxiety, age, depression symptoms, education, BMI, stratified by race | No |
| Crandall 2017 (22) | Age, bilateral oophorectomy, age, smoking, alcohol intake, physical activity, population structure, BMI, education, income, menopausal estrogen therapy use, stratified by race/ethnicity | No |
| Kapoor 2019 (24) | None mentioned; effect measure not specified (eg, odds ratio, risk ratio) | Stringent P threshold prespecified (5 X 10–8) |
| Luptakova 2012 (29) | Age, BMI, waist-to-hip ratio, stratified by premenopausal/perimenopausal and postmenopausal groups | No |
| Malacara 2004 (25) | Age, education, parity, y since menopause, BMI, serum estrone level, serum estradiol level | No |
| Massad-Costa 2008 (26) | None mentioned | No |
| Montasser 2015 (27) | Race, age, MT status, smoking, BMI | Yes (false discovery rate) |
| Rebbeck 2010 (3) | Smoking, BMI, race/ethnicity, MT stage | No |
| Schilling 2007 (28) | Age, race, smoking, BMI, d since last menstrual period | No |
| Schneider 2009 (19) | Age, MT stage, use of selective serotonin reuptake inhibitor, menopausal hormone therapy, clonidine, tamoxifen, or aromatase inhibitor | No |
| Takeo 2005 (18) | None mentioned | No |
| Visvanathan 2005 (17) | BMI, smoking, alcohol use, time since last menstrual period, age, race, prior menopausal hormone therapy use, prior oral contraceptive use | No |
| Woods 2018 (16) | Age, education, BMI, smoking, race/ethnicity | No |
| Woods 2006 (15) | Stratified by MT stage | No |
| Ziv-Gal 2012 (14 | Race, BMI, smoking, age | No |
Abbreviations: BMI, body mass index; MT, menopausal transition.
Discussion
To our knowledge, this is the first systematic review to focus on associations between genetic variants and VMS. Our results highlight that there are few studies of the genetics of VMS. Several studies have reported associations between genetic variants and VMS. Statistically significant associations were found in 14 of the 26 genes that were evaluated across studies. The gene that was evaluated in the largest number of studies was CYP1B1, a gene for an enzyme involved in estradiol metabolism, which was evaluated in 7 studies. CYP1B1 rs1056836 was associated with lower odds, and CYP1B1 rs1800440 was associated with higher odds, of VMS in one study each, but other studies reported that these variants were not significantly associated with VMS. However, effect measures and the means of assessing VMS varied across studies, making comparison of strength of associations difficult. Moreover, for most genetic variants, results were not replicated in more than one study. One GWAS study met inclusion criteria; in that study, the 14 SNPs with the highest P value (< 5 × 10–8) were all in the tachykinin receptor 3 gene, which is the receptor for neurokinin B (NKB).
Genetic variation in sex-steroid metabolism genes, such as CYP1B1, might be expected. CYP1B1 converts 17β-estradiol to 2-hydroextradiol-17β, thus altering the balance of the major female sex hormone estradiol. Fluctuations in estradiol levels that occur during the menopausal transition are implicated in the etiology of VMS. GWAS are agnostic, that is, they do not involve an a priori knowledge of biological mechanisms. In this way, they may hold advantages of candidate gene studies in the examination of outcomes for which pathophysiology is incompletely understand, such as VMS. Only one GWAS met inclusion criteria, and it found associations between 14 different genetic variants in the tachykinin receptor 3 gene and VMS. Tachykinin receptor 3 is the receptor for NKB. Notably, NKB gene expression increases after menopause; postmenopausal women have hypertrophy of neurons expressing NKB relative to premenopausal women (215). Also, in monkeys and rats, oophorectomy induces increases in NKB gene expression that can be reversed by estradiol therapy (215, 216). Moreover, recent animal and human studies highlight the probable role of the NKB pathway in the genesis of VMS (10). Drugs affecting the neurokinin receptor pathway (neurokinin 3 receptor antagonists) effectively treat VMS in randomized controlled trials (8, 9). and many clinical trials of these agents are ongoing.
Inconsistent results across studies can be puzzling. One example is found in the study by Rebbeck and colleagues (3) in which the rs1056836 variant (at least one allele, Leu432Val) was associated with a 40% lower odds of moderate or severe hot flashes among African American women, but the association of this variant with VMS was not statistically significant among European Americans (OR = 0.94 95% CI, = 0.55-1.89). The reason may not be due to any difference in genetic etiology of VMS between African American and European American women. Instead, the apparent lack of replication of associations from study to study, or even across ethnic groups within the same study, can be due to a number of statistical and population historical reasons. The sample sizes can be different, resulting in lack of statistical power to demonstrate associations that exist. Although the sample sizes for African American and European American women are nearly the same in the study of Rebbeck and colleagues, the allele frequencies at marker rs1056836 are not. In this example, allele frequencies for African American women are more favorable to detecting a true signal than the allele frequencies in the European American women. Of course, the result for the African American women could be a false-positive association, a common occurrence in candidate gene studies (217, 218). Both the allele frequencies and the prevalence and severity of VMS could be associated with degree of African ancestry and hence the observed association between the marker and VMS is a result of confounding by ancestry (219). Finally, the marker may not be a causal variant, but rather may be in linkage disequilibrium (LD) with a causal variant. In this event, the strength of LD is an important determinant of the power to detect an association (217). African Americans have longer haplotype blocks because of their population history of recent African and European admixture, so the LD could be stronger, thus allowing association to be detected in the African American women but not in the European American women (220).
Genetic variants may influence risk of VMS differentially among smokers and nonsmokers. This possibility was examined in one study (20). European American smokers with the +/+ genotype (where + denotes the variant allele) for CYP1B1 rs1056836 had 20-fold higher odds of moderate and severe hot flashes within the past month, compared with nonsmokers after adjustment for other variables. The adjusted OR was 20.6 (95% CI, 1.64-257.93), where the reference was European American nonsmokers with the +/+ genotype.
The included studies had several potential limitations. Several studies had small sample sizes, many specific SNPs were examined in only 1 or 2 studies each (precluding quantitative meta-analysis), and there was substantial heterogeneity across studies regarding how information regarding VMS was ascertained. Also, genetic variants associated with VMS may vary in specific clinical settings, such as primary ovarian insufficiency, early menopause, surgical menopause, and chemotherapy-induced menopause. The heterogeneity of the participants of the included studies precluded us from separately examining each of these clinically important subgroups.
In conclusion, several genetic variants were significantly associated with VMS, but the small number of candidate gene and GWAS studies, small numbers of participants in most studies, and conflicting results regarding the same genetic variants highlight the need for future research.
Acknowledgments
Financial Support: This work was supported by National Institutes of Health/National Human Genome Research Institute (NIH/NHGRI) (HG009120, Integrative Approaches for mapping the Genetic Risk of Complex Traits, principal investigator, B. Pasaniuc, to J.S.).
Author Contributions: Conceptualization, screening of citations, study design, interpretation of results, and manuscript drafting: C.J.C; screening of citations, critical revision of manuscript: A.L.D.; rating of quality of studies, critical revision of manuscript: M.M.; critical revision of manuscript: R.C.T; and interpretation of results and critical revision of manuscript: J.S.
Glossary
Abbreviations
- BMI
body mass index
- GWAS
genome-wide association study
- LD
linkage disequilibrium
- SNP
single-nucleotide polymorphism
- VMS
vasomotor symptoms
Additional Information
Disclosure Summary: C.J.C., M.M., A.L.D., and J.S. have nothing to disclose. RCT is a member of the advisory board and/or has consulted for Astellas, Virtue Health, Pfizer, and Procter and Gamble.
Data Availability
Data sharing is not applicable to this article because no data sets were generated or analyzed during the present study.
References
- 1. Gold EB, Colvin A, Avis N, et al. Longitudinal analysis of the association between vasomotor symptoms and race/ethnicity across the menopausal transition: Study of Women’s Health Across the Nation. Am J Public Health. 2006;96(7):1226-1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Avis NE, Crawford SL, Greendale G, et al. ; Study of Women’s Health Across the Nation . Duration of menopausal vasomotor symptoms over the menopause transition. JAMA Intern Med. 2015;175(4):531-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rebbeck TR, Su HI, Sammel MD, et al. Effect of hormone metabolism genotypes on steroid hormone levels and menopausal symptoms in a prospective population-based cohort of women experiencing the menopausal transition. Menopause. 2010;17(5):1026-1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Randolph JF Jr, Sowers M, Bondarenko I, et al. The relationship of longitudinal change in reproductive hormones and vasomotor symptoms during the menopausal transition. J Clin Endocrinol Metab. 2005;90(11):6106-6112. [DOI] [PubMed] [Google Scholar]
- 5. Freedman RR, Krell W. Reduced thermoregulatory null zone in postmenopausal women with hot flashes. Am J Obstet Gynecol. 1999;181(1):66-70. [DOI] [PubMed] [Google Scholar]
- 6. Freedman RR, Blacker CM. Estrogen raises the sweating threshold in postmenopausal women with hot flashes. Fertil Steril. 2002;77(3):487-490. [DOI] [PubMed] [Google Scholar]
- 7. Thurston RC. Vasomotor symptoms: natural history, physiology, and links with cardiovascular health. Climacteric. 2018;21(2):96-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Prague JK, Dhillo WS. Neurokinin 3 receptor antagonism—the magic bullet for hot flushes? Climacteric. 2017;20(6):505-509. [DOI] [PubMed] [Google Scholar]
- 9. Depypere H, Timmerman D, Donders G, et al. Treatment of menopausal vasomotor symptoms with fezolinetant, a neurokinin 3 receptor antagonist: a phase 2a trial. J Clin Endocrinol Metab. 2019;104(12):5893-5905. [DOI] [PubMed] [Google Scholar]
- 10. Rance NE, Dacks PA, Mittelman-Smith MA, Romanovsky AA, Krajewski-Hall SJ. Modulation of body temperature and LH secretion by hypothalamic KNDy (kisspeptin, neurokinin B and dynorphin) neurons: a novel hypothesis on the mechanism of hot flushes. Front Neuroendocrinol. 2013;34(3): 211-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Moher D, Liberati A, Tetzlaff J, Altman DG, Group P. Preferred Reporting Items for Systematic Reviews and Meta-Analyses: the PRISMA statement. Ann Intern Med. 2009;151:264-269, W264. [DOI] [PubMed] [Google Scholar]
- 12. Riley RD, Moons KGM, Snell KIE, et al. A guide to systematic review and meta-analysis of prognostic factor studies. BMJ. 2019;364:k4597. [DOI] [PubMed] [Google Scholar]
- 13. Hayden JA, van der Windt DA, Cartwright JL, Côté P, Bombardier C. Assessing bias in studies of prognostic factors. Ann Intern Med. 2013;158(4):280-286. [DOI] [PubMed] [Google Scholar]
- 14. Ziv-Gal A, Gallicchio L, Miller SR, Zacur HA, Flaws JA. Genetic polymorphisms in the aryl hydrocarbon receptor signaling pathway as potential risk factors of menopausal hot flashes. Am J Obstet Gynecol. 2012;207:202.e9-202.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Woods NF, Mitchell ES, Tao Y, Viernes HM, Stapleton PL, Farin FM. Polymorphisms in the estrogen synthesis and metabolism pathways and symptoms during the menopausal transition: observations from the Seattle Midlife Women’s Health Study. Menopause. 2006;13(6):902-910. [DOI] [PubMed] [Google Scholar]
- 16. Woods NF, Cray LA, Mitchell ES, Farrin F, Herting J. Polymorphisms in estrogen synthesis genes and symptom clusters during the menopausal transition and early postmenopause: observations from the Seattle Midlife Women’s Health Study. Biol Res Nurs. 2018;20(2):153-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Visvanathan K, Gallicchio L, Schilling C, et al. Cytochrome gene polymorphisms, serum estrogens, and hot flushes in midlife women. Obstet Gynecol. 2005;106(6):1372-1381. [DOI] [PubMed] [Google Scholar]
- 18. Takeo C, Negishi E, Nakajima A, et al. Association of cytosine-adenine repeat polymorphism of the estrogen receptor-beta gene with menopausal symptoms. Gend Med. 2005;2(2):96-105. [DOI] [PubMed] [Google Scholar]
- 19. Schneider BP, Radovich M, Flockhart DA, et al. Exploratory study evaluating the association of polymorphisms of angiogenesis genes with hot flashes. Breast Cancer Res Treat. 2009;116(3):543-549. [DOI] [PubMed] [Google Scholar]
- 20. Butts SF, Freeman EW, Sammel MD, Queen K, Lin H, Rebbeck TR. Joint effects of smoking and gene variants involved in sex steroid metabolism on hot flashes in late reproductive-age women. J Clin Endocrinol Metab. 2012;97(6):E1032-E1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Candráková Čerňanová V, Danková Z, Vorobeľová L, Cvíčelová M, Siváková D. Vasomotor, urogenital, psychological, and somatic symptoms in association with CYP1B1 polymorphisms in Slovak women of different menopausal status. Am J Hum Biol. 2018;30(3):e23094. [DOI] [PubMed] [Google Scholar]
- 22. Crandall CJ, Manson JE, Hohensee C, et al. Association of genetic variation in the tachykinin receptor 3 locus with hot flashes and night sweats in the Women’s Health Initiative Study. Menopause. 2017;24(3):252-261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Crandall CJ, Crawford SL, Gold EB. Vasomotor symptom prevalence is associated with polymorphisms in sex steroid-metabolizing enzymes and receptors. Am J Med. 2006;119(9 Suppl 1):S52-S60. [DOI] [PubMed] [Google Scholar]
- 24. Kapoor E, Faubion S, Miller VM, et al. Do genetic variations in estrogen transportation and metabolism affect the severity of menopausal symptoms? Menopause. 2019;26:1456. [Google Scholar]
- 25. Malacara JM, Pérez-Luque EL, Martínez-Garza S, Sánchez-Marín FJ. The relationship of estrogen receptor-alpha polymorphism with symptoms and other characteristics in post-menopausal women. Maturitas. 2004;49(2):163-169. [DOI] [PubMed] [Google Scholar]
- 26. Massad-Costa AM, Nogueira-de-Souza NC, de Carvalho CV, et al. CYP17 polymorphism and hot flushes in postmenopausal women. Climacteric. 2008;11(5):404-408. [DOI] [PubMed] [Google Scholar]
- 27. Montasser ME, Ziv-Gal A, Brown JP, Flaws JA, Merchenthaler I. A potentially functional variant in the serotonin transporter gene is associated with premenopausal and perimenopausal hot flashes. Menopause. 2015;22(1):108-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Schilling C, Gallicchio L, Miller SR, Langenberg P, Zacur H, Flaws JA. Genetic polymorphisms, hormone levels, and hot flashes in midlife women. Maturitas. 2007;57(2):120-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Luptáková L, Sivaková D, Srámeková D, Cvíčelová M. The association of cytochrome P450 1B1 Leu432Val polymorphism with biological markers of health and menopausal symptoms in Slovak midlife women. Menopause. 2012;19(2):216-224. [DOI] [PubMed] [Google Scholar]
- 30. Aguilar-Zavala H, Pérez-Luque EL, Luna-Martínez F, et al. Symptoms at postmenopause: genetic and psychosocial factors. Menopause. 2012;19(10):1140-1145. [DOI] [PubMed] [Google Scholar]
- 31. Abubakar MB, Wei K, Gan SH. The influence of genetic polymorphisms on the efficacy and side effects of anastrozole in postmenopausal breast cancer patients. Pharmacogenet Genomics. 2014;24(12):575-581. [DOI] [PubMed] [Google Scholar]
- 32. Atkinson EJ, Eckel-Passow JE, Wang A, et al. The association of copy number variation and percent mammographic density. BMC Res Notes. 2015;8:297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bacon ER, Mishra A, Wang Y, Desai MK, Yin F, Brinton RD. Neuroendocrine aging precedes perimenopause and is regulated by DNA methylation. Neurobiol Aging. 2019;74:213-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bahl A, Pöllänen E, Ismail K, et al. Hormone replacement therapy associated white blood cell DNA methylation and gene expression are associated with within-pair differences of body adiposity and bone mass. Twin Res Hum Genet. 2015;18(6):647-661. [DOI] [PubMed] [Google Scholar]
- 35. Baker DA, Stevenson DW, Little DP. DNA barcode identification of black cohosh herbal dietary supplements. J AOAC Int. 2012;95(4):1023-1034. [DOI] [PubMed] [Google Scholar]
- 36. Barber TM, Franks S. Genetics of polycystic ovary syndrome. Front Horm Res. 2013;40:28-39. [DOI] [PubMed] [Google Scholar]
- 37. Barrdahl M, Canzian F, Joshi AD, et al. Post-GWAS gene-environment interplay in breast cancer: results from the Breast and Prostate Cancer Cohort Consortium and a meta-analysis on 79,000 women. Hum Mol Genet. 2014;23(19):5260-5270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bechlioulis A, Naka KK, Kalantaridou SN, et al. Short-term hormone therapy improves sCD40L and endothelial function in early menopausal women: potential role of estrogen receptor polymorphisms. Maturitas. 2012;71(4):389-395. [DOI] [PubMed] [Google Scholar]
- 39. Birrer N, Chinchilla C, Del Carmen M, Dizon DS. Is hormone replacement therapy safe in women with a BRCA mutation? A systematic review of the contemporary literature. Am J Clin Oncol. 2018;41(3):313-315. [DOI] [PubMed] [Google Scholar]
- 40. Bonanni B, Macis D, Maisonneuve P, et al. Polymorphism in the CYP2D6 tamoxifen-metabolizing gene influences clinical effect but not hot flashes: data from the Italian Tamoxifen Trial. J Clin Oncol. 2006;24:3708-3709; author reply 3709. [DOI] [PubMed] [Google Scholar]
- 41. Bonfá AC, Seguro LP, Caparbo V, Bonfá E, Pereira RM. RANKL and OPG gene polymorphisms: associations with vertebral fractures and bone mineral density in premenopausal systemic lupus erythematosus. Osteoporos Int. 2015;26(5):1563-1571. [DOI] [PubMed] [Google Scholar]
- 42. Brand JS, Li J, Humphreys K, et al. Identification of two novel mammographic density loci at 6Q25.1. Breast Cancer Res. 2015;17:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Brauch H, Jordan VC. Targeting of tamoxifen to enhance antitumour action for the treatment and prevention of breast cancer: the ‘personalised’ approach? Eur J Cancer. 2009;45(13):2274-2283. [DOI] [PubMed] [Google Scholar]
- 44. Brauch H, Mürdter TE, Eichelbaum M, Schwab M. Pharmacogenomics of tamoxifen therapy. Clin Chem. 2009;55(10):1770-1782. [DOI] [PubMed] [Google Scholar]
- 45. Brooks JD, Bernstein L, Teraoka SN, et al. ; WECARE Study Collaborative Group . Variation in genes related to obesity, weight, and weight change and risk of contralateral breast cancer in the WECARE Study population. Cancer Epidemiol Biomarkers Prev. 2012;21(12):2261-2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Butts SF, Sammel MD, Greer C, Rebbeck TR, Boorman DW, Freeman EW. Cigarettes, genetic background, and menopausal timing: the presence of single nucleotide polymorphisms in cytochrome P450 genes is associated with increased risk of natural menopause in European-American smokers. Menopause. 2014;21(7):694-701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Campa D, Kaaks R, Le Marchand L, et al. Interactions between genetic variants and breast cancer risk factors in the Breast and Prostate Cancer Cohort Consortium. J Natl Cancer Inst. 2011;103(16):1252-1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Campa D, Hüsing A, McKay JD, et al. The INSIG2 rs7566605 polymorphism is not associated with body mass index and breast cancer risk. BMC Cancer. 2010;10:563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Campfield Bonadies D, Moyer A, Matloff ET. What I wish I’d known before surgery: BRCA carriers’ perspectives after bilateral salipingo-oophorectomy. Fam Cancer. 2011;10(1):79-85. [DOI] [PubMed] [Google Scholar]
- 50. Cao T, Jiang Y, Wang Z, et al. H19 lncRNA identified as a master regulator of genes that drive uterine leiomyomas. Oncogene. 2019;38(27):5356-5366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Chan KH, Chacko SA, Song Y, et al. Genetic variations in magnesium-related ion channels may affect diabetes risk among African American and Hispanic American women. J Nutr. 2015;145(3):418-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Chang BY, Kim DS, Kim HS, Kim SY. Evaluation of estrogenic potential by herbal formula, HPC 03 for in vitro and in vivo. Reproduction. 2018;155(2):105-115. [DOI] [PubMed] [Google Scholar]
- 53. Chapman JS, Powell CB, McLennan J, et al. Surveillance of survivors: follow-up after risk-reducing salpingo-oophorectomy in BRCA 1/2 mutation carriers. Gynecol Oncol. 2011;122(2):339-343. [DOI] [PubMed] [Google Scholar]
- 54. Chiang CW, Liu CT, Lettre G, et al. ; Genetic Investigation of ANthropometric Traits Consortium . Ultraconserved elements in the human genome: association and transmission analyses of highly constrained single-nucleotide polymorphisms. Genetics. 2012;192(1):253-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Cho HG, Ransohoff KJ, Yang L, et al. Melanoma risk prediction using a multilocus genetic risk score in the Women’s Health Initiative cohort. J Am Acad Dermatol. 2018;79:36-41.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. D’Alonzo M, Piva E, Pecchio S, et al. Satisfaction and impact on quality of life of clinical and instrumental surveillance and prophylactic surgery in BRCA-mutation carriers. Clin Breast Cancer. 2018;18(6):e1361-e1366. [DOI] [PubMed] [Google Scholar]
- 57. Dagan E, Shochat T. Quality of life in asymptomatic BRCA1/2 mutation carriers. Prev Med. 2009;48(2):193-196. [DOI] [PubMed] [Google Scholar]
- 58. Day F, Karaderi T, Jones MR, et al. ; 23andMe Research Team . Large-scale genome-wide meta-analysis of polycystic ovary syndrome suggests shared genetic architecture for different diagnosis criteria. PloS Genet. 2018;14(12):e1007813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. de Rooij AM, Gosso MF, Alsina-Sanchis E, Marinus J, van Hilten JJ, van den Maagdenberg AM. No mutations in the voltage-gated NaV1.7 sodium channel alpha1 subunit gene SCN9A in familial complex regional pain syndrome. Eur J Neurol. 2010;17(6):808-814. [DOI] [PubMed] [Google Scholar]
- 60. Dezentjé VO, Gelderblom H, Van Schaik RHN, et al. CYP2D6 genotype in relation to hot flashes as tamoxifen side effect in a Dutch cohort of the Tamoxifen Exemestane Adjuvant Multinational (TEAM) trial. Breast Cancer Res Treat. 2014;143(1):171-179. [DOI] [PubMed] [Google Scholar]
- 61. Didriksen A, Grimnes G, Hutchinson MS, et al. The serum 25-hydroxyvitamin D response to vitamin D supplementation is related to genetic factors, BMI, and baseline levels. Eur J Endocrinol. 2013;169(5):559-567. [DOI] [PubMed] [Google Scholar]
- 62. Domchek SM, Rebbeck TR. Prophylactic oophorectomy in women at increased cancer risk. Curr Opin Obstet Gynecol. 2007;19(1):27-30. [DOI] [PubMed] [Google Scholar]
- 63. Domchek SM, Stopfer JE, Rebbeck TR. Bilateral risk-reducing oophorectomy in BRCA1 and BRCA2 mutation carriers. J Natl Compr Canc Netw. 2006;4(2):177-182. [DOI] [PubMed] [Google Scholar]
- 64. Dorjgochoo T, Shi J, Gao YT, et al. Genetic variants in vitamin D metabolism-related genes and body mass index: analysis of genome-wide scan data of approximately 7000 Chinese women. Int J Obes (Lond). 2012;36(9):1252-1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Edwards TL, Giri A, Hellwege JN, et al. A trans-ethnic genome-wide association study of uterine fibroids. Front Genet. 2019;10:511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Elands RJ, Simons CC, Riemenschneider M, et al. A systematic SNP selection approach to identify mechanisms underlying disease aetiology: linking height to post-menopausal breast and colorectal cancer risk. Sci Rep. 2017;7:41034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Essemine J, Govindachary S, Ammar S, Bouzid S, Carpentier R. Abolition of photosystem I cyclic electron flow in Arabidopsis thaliana following thermal-stress. Plant Physiol Biochem. 2011;49(3):235-243. [DOI] [PubMed] [Google Scholar]
- 68. Federici LM, Roth SD, Krier C, et al. Anxiogenic CO2 stimulus elicits exacerbated hot flash-like responses in a rat menopause model and hot flashes in postmenopausal women. Menopause. 2016;23(11):1257-1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Fehringer G, Ozcelik H, Knight JA, et al. Family-based association study of IGF1 microsatellites and height, weight, and body mass index. J Hum Genet. 2010;55(4):255-258. [DOI] [PubMed] [Google Scholar]
- 70. Fernandez-Navarro P, Pita G, Santamariña C, et al. Association analysis between breast cancer genetic variants and mammographic density in a large population-based study (Determinants of Density in Mammographies in Spain) identifies susceptibility loci in TOX3 gene. Eur J Cancer. 2013;49(2):474-481. [DOI] [PubMed] [Google Scholar]
- 71. Finch A, Evans G, Narod SA. BRCA carriers, prophylactic salpingo-oophorectomy and menopause: clinical management considerations and recommendations. Womens Health (Lond). 2012;8(5):543-555. [DOI] [PubMed] [Google Scholar]
- 72. Finch A, Narod SA. Quality of life and health status after prophylactic salpingo-oophorectomy in women who carry a BRCA mutation: a review. Maturitas. 2011;70(3):261-265. [DOI] [PubMed] [Google Scholar]
- 73. Finch A, Metcalfe KA, Chiang JK, et al. The impact of prophylactic salpingo-oophorectomy on menopausal symptoms and sexual function in women who carry a BRCA mutation. Gynecol Oncol. 2011;121(1):163-168. [DOI] [PubMed] [Google Scholar]
- 74. Fox CS, Heard-Costa N, Cupples LA, Dupuis J, Vasan RS, Atwood LD. Genome-wide association to body mass index and waist circumference: the Framingham Heart Study 100K project. BMC Med Genet. 2007;8(Suppl 1):S18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Fujita K, Roforth MM, Demaray S, et al. Effects of estrogen on bone mRNA levels of sclerostin and other genes relevant to bone metabolism in postmenopausal women. J Clin Endocrinol Metab. 2014;99(1):E81-E88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Gabriel CA, Tigges-Cardwell J, Stopfer J, Erlichman J, Nathanson K, Domchek SM. Use of total abdominal hysterectomy and hormone replacement therapy in BRCA1 and BRCA2 mutation carriers undergoing risk-reducing salpingo-oophorectomy. Fam Cancer. 2009;8(1):23-28. [DOI] [PubMed] [Google Scholar]
- 77. Gadducci A, Biglia N, Cosio S, Sismondi P, Genazzani AR. Gynaecologic challenging issues in the management of BRCA mutation carriers: oral contraceptives, prophylactic salpingo-oophorectomy and hormone replacement therapy. Gynecol Endocrinol. 2010;26(8):568-577. [DOI] [PubMed] [Google Scholar]
- 78. Gao C, Patel CJ, Michailidou K, et al. ; the Colorectal Transdisciplinary Study (CORECT); Discovery, Biology and Risk of Inherited Variants in Breast Cancer (DRIVE); Elucidating Loci Involved in Prostate Cancer Susceptibility (ELLIPSE); Follow-up of Ovarian Cancer Genetic Association and Interaction Studies (FOCI); and Transdisciplinary Research in Cancer of the Lung (TRICL) . Mendelian randomization study of adiposity-related traits and risk of breast, ovarian, prostate, lung and colorectal cancer. Int J Epidemiol. 2016;45(3):896-908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Garber K. Tamoxifen pharmacogenetics moves closer to reality. J Natl Cancer Inst. 2005;97(6):412-413. [DOI] [PubMed] [Google Scholar]
- 80. Goetz MP, Rae JM, Suman VJ, et al. Pharmacogenetics of tamoxifen biotransformation is associated with clinical outcomes of efficacy and hot flashes. J Clin Oncol. 2005;23(36):9312-9318. [DOI] [PubMed] [Google Scholar]
- 81. Goto A, Chen BH, Chan KK, et al. ; MEta-analysis of type 2 DIabetes in African Americans (MEDIA) Consortium . Genetic variants in sex hormone pathways and the risk of type 2 diabetes among African American, Hispanic American, and European American postmenopausal women in the US. J Diabetes. 2018;10(6):524-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Greenwood CM, Paterson AD, Linton L, et al. A genome-wide linkage study of mammographic density, a risk factor for breast cancer. Breast Cancer Res. 2011;13(6):R132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Guidozzi F. Hormone therapy after prophylactic risk-reducing bilateral salpingo-oophorectomy in women who have BRCA gene mutation. Climacteric. 2016;19(5):419-422. [DOI] [PubMed] [Google Scholar]
- 84. Guo Y, Warren Andersen S, Shu XO, et al. Genetically predicted body mass index and breast cancer risk: mendelian randomization analyses of data from 145,000 women of European descent. PloS Med. 2016;13(8):e1002105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Hall E, Finch A, Jacobson M, et al. Effects of bilateral salpingo-oophorectomy on menopausal symptoms and sexual functioning among women with a BRCA1 or BRCA2 mutation. Gynecol Oncol. 2019;152:145-150. [DOI] [PubMed] [Google Scholar]
- 86. Hartge P. Genetics of reproductive lifespan. Nat Genet. 2009;41(6):637-638. [DOI] [PubMed] [Google Scholar]
- 87. Harvey RE, Coffman KE, Miller VM. Women-specific factors to consider in risk, diagnosis and treatment of cardiovascular disease. Womens Health (Lond). 2015;11(2):239-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. He C, Kraft P, Chasman DI, et al. A large-scale candidate gene association study of age at menarche and age at natural menopause. Hum Genet. 2010;128:515-527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. He X, Yu C, Zhao P, et al. The genetics of Henoch-Schönlein purpura: a systematic review and meta-analysis. Rheumatol Int. 2013;33(6):1387-1395. [DOI] [PubMed] [Google Scholar]
- 90. Hein R, Flesch-Janys D, Dahmen N, et al. ; GENICA Network . A genome-wide association study to identify genetic susceptibility loci that modify ductal and lobular postmenopausal breast cancer risk associated with menopausal hormone therapy use: a two-stage design with replication. Breast Cancer Res Treat. 2013;138(2):529-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Henry NL, Rae JM, Li L, et al. ; Consortium on Breast Cancer Pharmacogenomics Investigators . Association between CYP2D6 genotype and tamoxifen-induced hot flashes in a prospective cohort. Breast Cancer Res Treat. 2009;117(3):571-575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Higgins MJ, Stearns V. Pharmacogenetics of endocrine therapy for breast cancer. Annu Rev Med. 2011;62:281-293. [DOI] [PubMed] [Google Scholar]
- 93. Higgins MJ, Stearns V. CYP2D6 polymorphisms and tamoxifen metabolism: clinical relevance. Curr Oncol Rep. 2010;12(1):7-15. [DOI] [PubMed] [Google Scholar]
- 94. Hoeijmakers JG, Han C, Merkies IS, et al. Small nerve fibres, small hands and small feet: a new syndrome of pain, dysautonomia and acromesomelia in a kindred with a novel NaV1.7 mutation. Brain. 2012;135(Pt 2):345-358. [DOI] [PubMed] [Google Scholar]
- 95. Ingle JN, Schaid DJ, Goss PE, et al. Genome-wide associations and functional genomic studies of musculoskeletal adverse events in women receiving aromatase inhibitors. J Clin Oncol. 2010;28(31):4674-4682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Intharuksa A, Kitamura M, Peerakam N, et al. Evaluation of white Kwao Krua (Pueraria candollei Grah. ex Benth.) products sold in Thailand by molecular, chemical, and microscopic analyses. J Nat Med. 2020;74(1):106-118. [DOI] [PubMed] [Google Scholar]
- 97. Irarrazaval OM. Antagonism of tamoxifen and antidepressants among women with breast cancer. Rev Med Chil. 2011;139:89-99. [PubMed] [Google Scholar]
- 98. Ishiguro H, Ohno S, Yamamoto Y, et al. Pharmacogenomic-pharmacokinetic study of selective estrogen-receptor modulators with intra-patient dose escalation in breast cancer. Breast Cancer. 2019;26(5):535-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Jansen LE, Teft WA, Rose RV, Lizotte DJ, Kim RB. CYP2D6 genotype and endoxifen plasma concentration do not predict hot flash severity during tamoxifen therapy. Breast Cancer Res Treat. 2018;171(3):701-708. [DOI] [PubMed] [Google Scholar]
- 100. Jin HS, Kim J, Park S, et al. Association of the I264T variant in the sulfide quinone reductase-like (SQRDL) gene with osteoporosis in Korean postmenopausal women. PloS One. 2015;10(8):e0135285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Johansson H, Gray KP, Pagani O, et al. ; the TEXT principal investigators . Impact of CYP19A1 and ESR1 variants on early-onset side effects during combined endocrine therapy in the TEXT trial. Breast Cancer Res. 2016;18(1):110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Jung SY, Mancuso N, Papp J, Sobel E, Zhang ZF. Post genome-wide gene-environment interaction study: The effect of genetically driven insulin resistance on breast cancer risk using Mendelian randomization. PloS One. 2019;14(6):e0218917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Jung SY, Mancuso N, Yu H, Papp J, Sobel E, Zhang ZF. Genome-wide meta-analysis of gene-environmental interaction for insulin resistance phenotypes and breast cancer risk in postmenopausal women. Cancer Prev Res (Phila). 2019;12(1):31-42. [DOI] [PubMed] [Google Scholar]
- 104. Jung SY, Papp JC, Sobel EM, Zhang ZF. Genetic variants in metabolic signaling pathways and their interaction with lifestyle factors on breast cancer risk: a random survival forest analysis. Cancer Prev Res (Phila). 2018;11(1):44-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Jung SY, Sobel EM, Papp JC, Zhang ZF. Effect of genetic variants and traits related to glucose metabolism and their interaction with obesity on breast and colorectal cancer risk among postmenopausal women. BMC Cancer. 2017;17(1):290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Justenhoven C, Obazee O, Brauch H. The pharmacogenomics of sex hormone metabolism: breast cancer risk in menopausal hormone therapy. Pharmacogenomics. 2012;13(6):659-675. [DOI] [PubMed] [Google Scholar]
- 107. Kawase T, Matsuo K, Suzuki T, et al. FGFR2 intronic polymorphisms interact with reproductive risk factors of breast cancer: results of a case control study in Japan. Int J Cancer. 2009;125(8):1946-1952. [DOI] [PubMed] [Google Scholar]
- 108. Kim SK, Massett MP. Genetic regulation of endothelial vasomotor function. Front Physiol. 2016;7:571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Koller DL, Ichikawa S, Lai D, et al. Genome-wide association study of bone mineral density in premenopausal European-American women and replication in African-American women. J Clin Endocrinol Metab. 2010;95(4):1802-1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Komm BS, Lyttle CR. Developing a SERM: stringent preclinical selection criteria leading to an acceptable candidate (WAY-140424) for clinical evaluation. Ann N Y Acad Sci. 2001;949:317-326. [DOI] [PubMed] [Google Scholar]
- 111. Lapid K, Lim A, Clegg DJ, Zeve D, Graff JM. Oestrogen signalling in white adipose progenitor cells inhibits differentiation into brown adipose and smooth muscle cells. Nat Commun. 2014;5:5196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Lee AW, Bomkamp A, Bandera EV, et al. ; Ovarian Cancer Association Consortium . A splicing variant of TERT identified by GWAS interacts with menopausal estrogen therapy in risk of ovarian cancer. Int J Cancer. 2016;139(12):2646-2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Leyland-Jones B, Gray KP, Abramovitz M, et al. ; BIG 1-98 Collaborative Group . ESR1 and ESR2 polymorphisms in the BIG 1-98 trial comparing adjuvant letrozole versus tamoxifen or their sequence for early breast cancer. Breast Cancer Res Treat. 2015;154(3):543-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Li G, Xiang YB, Courtney R, et al. Association of a single nucleotide polymorphism at 6q25.1,rs2046210, with endometrial cancer risk among Chinese women. Chin J Cancer. 2011;30(2):138-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Li M, Hung A, Li H, Yang AWH. A classic herbal formula guizhi fuling wan for menopausal hot flushes: from experimental findings to clinical applications. Biomedicines. 2019;7(3):60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Lim U, Ernst T, Wilkens LR, et al. Susceptibility variants for waist size in relation to abdominal, visceral, and hepatic adiposity in postmenopausal women. J Acad Nutr Diet. 2012;112(7):1048-1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Lintermans A, Van Asten K, Jongen L, et al. Genetic variant in the osteoprotegerin gene is associated with aromatase inhibitor-related musculoskeletal toxicity in breast cancer patients. Eur J Cancer. 2016;56:31-36. [DOI] [PubMed] [Google Scholar]
- 118. Luptáková L, Sivtáková D, Cernanová V, Cvicelová M. Menopausal complaints in Slovak midlife women and the impact of CYP1B1 polymorphism on their incidence. Anthropol Anz. 2012;69(4):399-415. [PubMed] [Google Scholar]
- 119. Mai PL, Piedmonte M, Han PK, et al. Factors associated with deciding between risk-reducing salpingo-oophorectomy and ovarian cancer screening among high-risk women enrolled in GOG-0199: an NRG Oncology/Gynecologic Oncology Group study. Gynecol Oncol. 2017;145(1):122-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Mallin EA, Pavlicek W. 55-year-old woman with menopausal symptoms and a family history of breast cancer. Mayo Clin Proc. 2008;83(4):485-488. [DOI] [PubMed] [Google Scholar]
- 121. Mariapun S, Ho WK, Kang PC, et al. Variants in 6q25.1 are associated with mammographic density in Malaysian Chinese women. Cancer Epidemiol Biomarkers Prev. 2016;25(2):327-333. [DOI] [PubMed] [Google Scholar]
- 122. Markus HS, Ruigrok Y, Ali N, Powell JF. Endothelial nitric oxide synthase exon 7 polymorphism, ischemic cerebrovascular disease, and carotid atheroma. Stroke. 1998;29(9):1908-1911. [DOI] [PubMed] [Google Scholar]
- 123. Matchkov VV. Mechanisms of cellular synchronization in the vascular wall. Mechanisms of vasomotion. Dan Med Bull. 2010;57(10):B4191. [PubMed] [Google Scholar]
- 124. Meirhaeghe A, Cottel D, Amouyel P, Dallongeville J. Lack of association between certain candidate gene polymorphisms and the metabolic syndrome. Mol Genet Metab. 2005;86(1-2):293-299. [DOI] [PubMed] [Google Scholar]
- 125. Mizutani K, Sugimoto K, Okuda T, et al. Kynureninase is a novel candidate gene for hypertension in spontaneously hypertensive rats. Hypertens Res. 2002;25(1):135-140. [DOI] [PubMed] [Google Scholar]
- 126. Moriwaki M, Moore B, Mosbruger T, et al. POLR2C mutations are associated with primary ovarian insufficiency in women. J Endocr Soc. 2017;1(3):162-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Moyer AM, de Andrade M, Faubion SS, et al. SLCO1B1 genetic variation and hormone therapy in menopausal women. Menopause. 2018;25(8):877-882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Moyer AM, de Andrade M, Weinshilboum RM, Miller VM. Influence of SULT1A1 genetic variation on age at menopause, estrogen levels, and response to hormone therapy in recently postmenopausal white women. Menopause. 2016;23(8):863-869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Méndez JP, Rojano-Mejía D, Coral-Vázquez RM, et al. Impact of genetic variants of IL-6, IL6R, LRP5, ESR1 and SP7 genes on bone mineral density in postmenopausal Mexican-Mestizo women with obesity. Gene. 2013;528(2):216-220. [DOI] [PubMed] [Google Scholar]
- 130. Nachtigall MJ, Jessel RH, Flaumenhaft R, et al. The selective estrogen receptor modulator DT56a (Femarelle) does not affect platelet reactivity in normal or thrombophilic postmenopausal women. Menopause. 2011;18(3):285-288. [DOI] [PubMed] [Google Scholar]
- 131. Niederhofer H. Re: Polymorphisms in the estrogen synthesis and metabolism pathways and symptoms during the menopausal transition: observations from the Seattle Midlife Women’s Health Study. Menopause. 2007;14(6):1069; author reply 1069. [DOI] [PubMed] [Google Scholar]
- 132. Nogueira RC Jr, Costa AM, Silva ID, et al. Influence of the CYP17 polymorphism on vasomotor symptoms in postmenopausal women: a pilot study. Climacteric. 2011;14(5):537-543. [DOI] [PubMed] [Google Scholar]
- 133. O’Brien KM, Shi M, Sandler DP, et al. A family-based, genome-wide association study of young-onset breast cancer: inherited variants and maternally mediated effects. Eur J Hum Genet. 2016;24:1316-1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Ochs-Balcom HM, Preus L, Nie J, et al. Physical activity modifies genetic susceptibility to obesity in postmenopausal women. Menopause. 2018;25(10):1131-1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Ohshima S, Hatori Y, Honma S, Terasawa K, Saitoh Y, Kobayashi D. Analysis of body constitution of fifty-two patients with Stevens-Johnson syndrome (SJS) using Kampo Medical Questionnaires: prediction of SJS based on body constitution using decision tree. Yakugaku Zasshi. 2011;131(5):745-756. [DOI] [PubMed] [Google Scholar]
- 136. Passarelli MN, Coghill AE, Hutter CM, et al. Common colorectal cancer risk variants in SMAD7 are associated with survival among prediagnostic nonsteroidal anti-inflammatory drug users: a population-based study of postmenopausal women. Genes Chromosomes Cancer. 2011;50(11):875-886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Pausova Z, Syme C, Abrahamowicz M, et al. A common variant of the FTO gene is associated with not only increased adiposity but also elevated blood pressure in French Canadians. Circ Cardiovasc Genet. 2009;2(3):260-269. [DOI] [PubMed] [Google Scholar]
- 138. Petherick A. Environment and genetics: making sense of the noise. Nature. 2012;485(7400):S64-S65. [DOI] [PubMed] [Google Scholar]
- 139. Pezaro C, James P, McKinley J, Shanahan M, Young MA, Mitchell G. The consequences of risk reducing salpingo-oophorectomy: the case for a coordinated approach to long-term follow up post surgical menopause. Fam Cancer. 2012;11(3):403-410. [DOI] [PubMed] [Google Scholar]
- 140. Pilling LC, Atkins JL, Duff MO, et al. Red blood cell distribution width: genetic evidence for aging pathways in 116,666 volunteers. PloS One. 2017;12(9):e0185083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Prentice RL, Huang Y, Hinds DA, et al. Variation in the FGFR2 gene and the effects of postmenopausal hormone therapy on invasive breast cancer. Cancer Epidemiol Biomarkers Prev. 2009;18(11):3079-3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Pru JK. Genetic predisposition to ovotoxic effects of smoking may hasten time to menopause. Menopause. 2014;21(7):685-686. [DOI] [PubMed] [Google Scholar]
- 143. Pulit SL, Karaderi T, Lindgren CM. Sexual dimorphisms in genetic loci linked to body fat distribution. Biosci Rep. 2017;37(1):BSR20160184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Qin Y, Sun M, You L, et al. ESR1, HK3 and BRSK1 gene variants are associated with both age at natural menopause and premature ovarian failure. Orphanet J Rare Dis. 2012;7:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Qin Z, Xue J, He Y, et al. Potentially functional polymorphisms in ATG10 are associated with risk of breast cancer in a Chinese population. Gene. 2013;527(2):491-495. [DOI] [PubMed] [Google Scholar]
- 146. Rask-Andersen M, Karlsson T, Ek WE, Johansson Å. Gene-environment interaction study for BMI reveals interactions between genetic factors and physical activity, alcohol consumption and socioeconomic status. PloS Genet. 2017;13(9):e1006977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Regan MM, Leyland-Jones B, Bouzyk M, et al. ; Breast International Group (BIG) 1-98 Collaborative Group . CYP2D6 genotype and tamoxifen response in postmenopausal women with endocrine-responsive breast cancer: the Breast International Group 1-98 Trial. J Natl Cancer Inst. 2012;104(6):441-451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Reid RL. Hormone therapy in breast cancer survivors and those at high risk for breast cancer. Clin Obstet Gynecol. 2018;61(3):480-487. [DOI] [PubMed] [Google Scholar]
- 149. Rojas-Roldan L, Wilkins T. Fatigue, arthralgia, amenorrhea—Dx? J Fam Pract. 2014;63(6):305-308. [PubMed] [Google Scholar]
- 150. Rudolph A, Fasching PA, Behrens S, et al. A comprehensive evaluation of interaction between genetic variants and use of menopausal hormone therapy on mammographic density. Breast Cancer Res. 2015;17:110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Rudolph A, Hein R, Lindström S, et al. ; GENICA Network; kConFab Investigators; AOCS Management Group; Breast Cancer Association Consortium . Genetic modifiers of menopausal hormone replacement therapy and breast cancer risk: a genome-wide interaction study. Endocr Relat Cancer. 2013;20(6):875-887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Šašková P, Pavlišta D, Dostálek L. Satisfaction and overall quality of life in BRCA positive women after prophylactic surgeries [article in Czech]. Rozhl Chir. 2017;96(8):328-333. [PubMed] [Google Scholar]
- 153. Schneider H. Hormone replacement therapy, better aging, molecular genetic diagnostics and the image of the gynaecologist. Gynakol Geburtshilfliche Rundsch. 2003;43(2):75-78. [DOI] [PubMed] [Google Scholar]
- 154. Schogor AL, Huws SA, Santos GT, et al. Ruminal Prevotella spp. may play an important role in the conversion of plant lignans into human health beneficial antioxidants. PloS One. 2014;9(4):e87949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Sestak I, Kealy R, Nikoloff M, et al. Relationships between CYP2D6 phenotype, breast cancer and hot flushes in women at high risk of breast cancer receiving prophylactic tamoxifen: results from the IBIS-I trial. Br J Cancer. 2012;107(2):230-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Shang M, Lin L, Cui H. Association of genetic polymorphisms of RANK, RANKL and OPG with bone mineral density in Chinese peri- and postmenopausal women. Clin Biochem. 2013;46(15):1493-1501. [DOI] [PubMed] [Google Scholar]
- 157. Shigehiro M, Kita M, Takeuchi S, Ashihara Y, Arai M, Okamura H. Study on the psychosocial aspects of risk-reducing salpingo-oophorectomy (RRSO) in BRCA1/2 mutation carriers in Japan: a preliminary report. Jpn J Clin Oncol. 2016;46(3):254-259. [DOI] [PubMed] [Google Scholar]
- 158. Singh MS, Francis PA, Michael M. Tamoxifen, cytochrome P450 genes and breast cancer clinical outcomes. Breast. 2011;20(2):111-118. [DOI] [PubMed] [Google Scholar]
- 159. Somekawa Y, Wakabayashi A. Relationship between apolipoprotein E polymorphism, menopausal symptoms, and serum lipids during hormone replacement therapy. Eur J Obstet Gynecol Reprod Biol. 1998;79(2):185-191. [DOI] [PubMed] [Google Scholar]
- 160. Tamimi RM, Cox D, Kraft P, Colditz GA, Hankinson SE, Hunter DJ. Breast cancer susceptibility loci and mammographic density. Breast Cancer Res. 2008;10(4):R66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Tenenbaum-Rakover Y, Weinberg-Shukron A, Renbaum P, et al. Minichromosome maintenance complex component 8 (MCM8) gene mutations result in primary gonadal failure. J Med Genet. 2015;52(6):391-399. [DOI] [PubMed] [Google Scholar]
- 162. Thomin A, Friszer S, Fajac A, Daraï É, Chabbert-Buffet N. Hormonal prevention of breast cancer. Ann Endocrinol (Paris). 2014;75(3):148-155. [DOI] [PubMed] [Google Scholar]
- 163. Thompson DJ, O’Mara TA, Glubb DM, et al. ; Australian National Endometrial Cancer Study Group (ANECS); National Study of Endometrial Cancer Genetics Group (NSECG); for RENDOCAS; AOCS Group . CYP19A1 fine-mapping and Mendelian randomization: estradiol is causal for endometrial cancer. Endocr Relat Cancer. 2016;23(2):77-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Tóth SZ, Nagy V, Puthur JT, Kovács L, Garab G. The physiological role of ascorbate as photosystem II electron donor: protection against photoinactivation in heat-stressed leaves. Plant Physiol. 2011;156(1):382-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Ushiroyama T, Heishi M, Ikeda A, Ueki M. Does the estrogen receptor gene polymorphism relate to undefined menopausal symptoms? Res Commun Mol Pathol Pharmacol. 2001;109(1-2):3-14. [PubMed] [Google Scholar]
- 166. Varghese JS, Thompson DJ, Michailidou K, et al. ; MODE Consortium . Mammographic breast density and breast cancer: evidence of a shared genetic basis. Cancer Res. 2012;72(6):1478-1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Velez Edwards DR, Naj AC, Monda K, et al. Gene-environment interactions and obesity traits among postmenopausal African-American and Hispanic women in the Women’s Health Initiative SHARe Study. Hum Genet. 2013;132(3):323-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Warren Andersen S, Trentham-Dietz A, Gangnon RE, et al. Breast cancer susceptibility loci in association with age at menarche, age at natural menopause and the reproductive lifespan. Cancer Epidemiol. 2014;38(1):62-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Warren Andersen S, Trentham-Dietz A, Gangnon RE, et al. The associations between a polygenic score, reproductive and menstrual risk factors and breast cancer risk. Breast Cancer Res Treat. 2013;140(2):427-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Wise LA, Ruiz-Narvaez EA, Palmer JR, et al. African ancestry and genetic risk for uterine leiomyomata. Am J Epidemiol. 2012;176(12):1159-1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Woad KJ, Watkins WJ, Prendergast D, Shelling AN. The genetic basis of premature ovarian failure. Aust N Z J Obstet Gynaecol. 2006;46(3):242-244. [DOI] [PubMed] [Google Scholar]
- 172. Woyka J. Practice observed. Post Reprod Health. 2014;20(4):151-153. [DOI] [PubMed] [Google Scholar]
- 173. Yan H, Liu YJ, Zhou Q, Xiao P, Recker RR, Deng HW. Comparison of whole genome linkage scans in premenopausal and postmenopausal women: no bone-loss-specific QTLs were implicated. Osteoporos Int. 2009;20(5):771-777. [DOI] [PubMed] [Google Scholar]
- 174. Yang Y, Wang Y, Li S, et al. Mutations in SCN9A, encoding a sodium channel alpha subunit, in patients with primary erythermalgia. J Med Genet. 2004;41(3):171-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Younis JS. Ovarian aging and implications for fertility female health. Minerva Endocrinol. 2012;37(1):41-57. [PubMed] [Google Scholar]
- 176. Zaffanello M, Tardivo S, Cataldi L, Fanos V, Biban P, Malerba G. Genetic susceptibility to renal scar formation after urinary tract infection: a systematic review and meta-analysis of candidate gene polymorphisms. Pediatr Nephrol. 2011;26(7):1017-1029. [DOI] [PubMed] [Google Scholar]
- 177. Zeller J, Poulsen KT, Sutton JE, et al. CGRP function-blocking antibodies inhibit neurogenic vasodilatation without affecting heart rate or arterial blood pressure in the rat. Br J Pharmacol. 2008;155(7):1093-1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Zembutsu H, Nakamura S, Akashi-Tanaka S, et al. Significant effect of polymorphisms in CYP2D6 on response to tamoxifen therapy for breast cancer: a prospective multicenter study. Clin Cancer Res. 2017;23(8):2019-2026. [DOI] [PubMed] [Google Scholar]
- 179. Zhang HJ, Li JW, Liu YJ, et al. Quantitative trait loci analysis of individual and total isoflavone contents in soybean seeds. J Genet. 2014;93(2):331-338. [DOI] [PubMed] [Google Scholar]
- 180. Zhang L, Spencer KL, Voruganti VS, et al. Association of functional polymorphism rs2231142 (Q141K) in the ABCG2 gene with serum uric acid and gout in 4 US populations: the PAGE Study. Am J Epidemiol. 2013;177(9):923-932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Zhao L, Cui B, Liu JM, et al. Interactions of osteoporosis candidate genes for age at menarche, age at natural menopause, and maximal height in Han Chinese women. Menopause. 2011;18(9):1018-1025. [DOI] [PubMed] [Google Scholar]
- 182. Zingue S, Tchoumtchoua J, Ntsa DM, et al. Estrogenic and cytotoxic potentials of compounds isolated from Millettia macrophylla Benth (Fabaceae): towards a better understanding of its underlying mechanisms. BMC Complement Altern Med. 2016;16(1):421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Ziv-Gal A, Flaws JA. Factors that may influence the experience of hot flushes by healthy middle-aged women. J Womens Health (Larchmt). 2010;19(10):1905-1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Brown JT, Abdel-Rahman SM, van Haandel L, Gaedigk A, Lin YS, Leeder JS. Single dose, CYP2D6 genotype-stratified pharmacokinetic study of atomoxetine in children with ADHD. Clin Pharmacol Ther. 2016;99(6):642-650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Butts SF, Freeman EW, Sammel MD, Queen K, Lin H, Rebbeck TR. Joint effects of smoking and gene variants involved in sex steroid metabolism on hot flashes in late reproductive-age women. J Clin Endocrinol Metab. 2012;97(6):E1032-E1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Cerril E, Falcone A, Innocenti F. Cancer pharmacogenomics: germline DNA, tumor DNA, or both? Curr Pharmacogenomics. 2007;5:87-101. [Google Scholar]
- 187. Chae SC, Park YR, Li CS, et al. Analysis of the variations in IL-28RA gene and their association with allergic rhinitis. Exp Mol Med. 2006;38(3):302-309. [DOI] [PubMed] [Google Scholar]
- 188. Chollet C, Placier S, Chatziantoniou C, et al. Genetically increased angiotensin 1-converting enzyme and peripheral and renal vascular reactivity in mice. Hypertension. 2017;70(Suppl 1):AP412. [DOI] [PubMed] [Google Scholar]
- 189. Crandall CJ, Hohensee C, Horvath S, et al. Genome-wide association study of hot flashes in the women’s health initiative study. Menopause. 2015;22:1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. Depypere H, Timmerman D, Donders G, et al. Clinical evaluation of the NK3 receptor antagonist fezolinetant (a.k.a. ESN364) for the treatment of menopausal hot flashes. Maturitas. 2017;103:89-90. [Google Scholar]
- 191. Dern RJ, Fenn WO. The effect of varying pulmonary pressure on the arterial pressures in man and anesthetized cats. J Clin Invest. 1947;26(3):460-467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Fraser GL, Depypere H, Timmerman D, et al. Clinical evaluation of the NK3 receptor antagonist fezolinetant (a.k.a. ESN364) for the treatment of menopausal hot flashes. Endocr Rev. 2017;38. [Google Scholar]
- 193. Fujita K, Sasaki Y. Pharmacogenomics in drug-metabolizing enzymes catalyzing anticancer drugs for personalized cancer chemotherapy. Curr Drug Metab. 2007;8(6):554-562. [DOI] [PubMed] [Google Scholar]
- 194. Hartmaier RJ, Skaar TC, Rae J, Wang J, Li L, Oesterreich S. Impact of SNPs in ERα corepressor SMRT on breast cancer risk and response to endocrine therapy. Cancer Res. 2009;69(2 Suppl):Abstract 1043. [Google Scholar]
- 195. Hayes MG, Lin FTJ, Diamond MP, et al. Genetic associations with outcomes after letrozole or clomiphene citrate treatment in women of European ancestry with PCOS. Endocr Rev. 2017;38. [Google Scholar]
- 196. Hertz DL, Henry NL, Rae JM. Germline genetic predictors of aromatase inhibitor concentrations, estrogen suppression and drug efficacy and toxicity in breast cancer patients. Pharmacogenomics. 2017;18(5):481-499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197. Houtsma D, Fontein D, van der Straaten T, et al. Genetic variation in CYP19A1 and occurrence of adverse events in exemestane treatment with early breast cancer patients in the Dutch TEAM trial. J Clin Oncol. 2013;31(15 Suppl):e22152. [Google Scholar]
- 198. Ingle J. Postmenopausal women with hormone receptor-positive breast cancer: balancing benefit and toxicity from aromatase inhibitors. Breast. 2013;22(22 Suppl):S180-–S183.. [DOI] [PubMed] [Google Scholar]
- 199. Lorenz M, Prause S, Eidens M, et al. Translation of CYP2D6 human genetic variation into medical practice: lessons learned and the way forward. Curr Pharmacogenomics Person Med. 2010;8:306-319. [Google Scholar]
- 200. Lu JC, Coca SG, Patel UD, Cantley L, Parikh CR; Translational Research Investigating Biomarkers and Endpoints for Acute Kidney Injury (TRIBE-AKI) Consortium . Searching for genes that matter in acute kidney injury: a systematic review. Clin J Am Soc Nephrol. 2009;4(6):1020-1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201. Luo S, Ruan X, Wang Y, et al. The first family group of α1-AT-P in the world with repeated hematomas: 10-year follow-up. Climacteric. 2019;22(5):527-530. [DOI] [PubMed] [Google Scholar]
- 202. Minoretti P, Falcone C, Aldeghi A, et al. A novel Val734Ile variant in the ABCC9 gene associated with myocardial infarction. Clin Chim Acta. 2006;370(1-2):124-128. [DOI] [PubMed] [Google Scholar]
- 203. Moyer A, Miller V, De Andrade M. Impact of genetic variation in SULT1A1 and in mode of estrogen administration on serum estrogen levels in newly menopausal caucasian women. J Women’s Health. 2014;23:13-14. [Google Scholar]
- 204. Murphy DL, Lerner A, Rudnick G, Lesch KP. Serotonin transporter: gene, genetic disorders, and pharmacogenetics. Mol Interv. 2004;4(2):109-123. [DOI] [PubMed] [Google Scholar]
- 205. O’Sullivan CC, Loprinzi CL, Haddad TC. Updates in the Evaluation and Management of Breast Cancer. Mayo Clin Proc. 2018;93(6):794-807. [DOI] [PubMed] [Google Scholar]
- 206. Rumianowski B, Brodowska A, Karakiewicz B, Grochans E, Ryterska K, Laszczyńska M. Environmental factors influencing age at natural menopause in women. Prz Menopauzalny. 2012;16:412-416. [Google Scholar]
- 207. Waage J, Standl M, Curtin JA, et al. ; 23andMe Research Team; AAGC collaborators . Genome-wide association and HLA fine-mapping studies identify risk loci and genetic pathways underlying allergic rhinitis. Nat Genet. 2018;50(8):1072-1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208. Welzen MEB, Dezentjé VO, Van Schaik RHN, et al. The effect of tamoxifen dose increment in patients with impaired CYP2D6 activity. Ther Drug Monit. 2015;37:501-507. [DOI] [PubMed] [Google Scholar]
- 209. Weng L, Ziliak D, Im HK, et al. ; Consortium on Breast Cancer Pharmacogenomics (COBRA) . Genome-wide discovery of genetic variants affecting tamoxifen sensitivity and their clinical and functional validation. Ann Oncol. 2013;24(7):1867-1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210. Wolff PH. Vasomotor sensitivity to alcohol in diverse Mongoloid populations. Am J Hum Genet. 1973;25(2):193-199. [PMC free article] [PubMed] [Google Scholar]
- 211. Yiannakopoulou EC. Pharmacogenomics of breast cancer targeted therapy: focus on recent patents. Recent Pat DNA Gene Seq. 2012;6(1):33-46. [DOI] [PubMed] [Google Scholar]
- 212. Zembutsu H. Pharmacogenomics toward personalized tamoxifen therapy for breast cancer. Pharmacogenomics. 2015;16(3):287-296. [DOI] [PubMed] [Google Scholar]
- 213. Zembutsu H, Nakamura S, Akashi-Tanaka S, et al. Association between CYP2D6 genotype and response to tamoxifen in a prospective multicenter study in Japan. Cancer Res. 2016;76(14 Suppl):Abstract 2031. [Google Scholar]
- 214. Little J, Higgins JP, Ioannidis JP, et al. STrengthening the REporting of Genetic Association studies (STREGA): an extension of the STROBE Statement. Ann Intern Med. 2009;150(3):206-215. [DOI] [PubMed] [Google Scholar]
- 215. Rance NE, Krajewski SJ, Smith MA, Cholanian M, Dacks PA. Neurokinin B and the hypothalamic regulation of reproduction. Brain Res. 2010;1364:116-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216. Rance NE, Bruce TR. Neurokinin B gene expression is increased in the arcuate nucleus of ovariectomized rats. Neuroendocrinology. 1994;60(4):337-345. [DOI] [PubMed] [Google Scholar]
- 217. Marigorta UM, Rodríguez JA, Gibson G, Navarro A. Replicability and prediction: lessons and challenges from GWAS. Trends Genet. 2018;34(7):504-517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218. Ioannidis JP. Why most published research findings are false. PloS Med. 2005;2(8):e124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219. Hellwege JN, Keaton JM, Giri A, Gao X, Velez Edwards DR, Edwards TL. Population stratification in genetic association studies. Curr Protoc Hum Genet. 2017;95:12221-212223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220. Pritchard JK, Przeworski M. Linkage disequilibrium in humans: models and data. Am J Hum Genet. 2001;69(1):1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article because no data sets were generated or analyzed during the present study.