Abstract
Dysregulation of the human microbiome has been linked to various disease states, which has galvanized the efforts to modulate human health through microbiomes. Currently, human microbiome research is going through several phases to identify the constituent components of the microbiome, associate microbiome changes with physiological and pathological states, understand causative relationships, and finally translate this knowledge into therapeutics and diagnostics. The convergence of microfluidic technologies with molecular and cell profiling, microbiology, and tissue engineering can potentially be applied to these different phases of microbiome research to overcome the existing challenges faced by conventional approaches. The goal of this paper is to discuss and highlight the opportunities of applying different microfluidic technologies to specific areas of microbiome research as well as unique challenges that microfluidics must overcome when working with microbiome-relevant biological materials, e.g., micro-organisms, host tissues, and fluids. We will discuss the applicability of integrated microfluidic systems for processing biological samples for genomic sequencing analyses. For functional analysis of the microbiota, we will cover state-of-the-art microfluidic devices for microbiota cultivation and functional measurements. Finally, we highlight the use of organs-on-chips to model various microbiome–host tissue interactions. We envision that microfluidic technologies may hold great promise in advancing the knowledge on the interplay between microbiome and human health, as well as its eventual translation into microbiome-based diagnostics and therapeutics.
I. INTRODUCTION
The human microbiome refers to the collection of micro-organisms that live symbiotically in the human body (defined as the “microbiota”), their genetic material, and the surrounding environmental habitat.1 It is now appreciated that the microbiome plays an important role in human health and diseases. Various disease states have been linked to dysregulation of the gut microbiota, including neurodegenerative, cardiovascular, and metabolic diseases.2 The composition of the gut microbiome can also affect responses to therapies, most notably in cancer treatment. The gut microbiota can partake in drug metabolism and, therefore, result in unintended adverse drug reactions. For instance, severe diarrhea happens due to reactivation of the inactive metabolite of irinotecan, a first line chemotherapeutic drug for colorectal cancer, back into its active form.3 In immunotherapy, antibiotic induced alteration of the gut microbiota can contribute to checkpoint inhibitor resistance.4 Therefore, the endeavor to modulate human health through microbiomes has become the next frontier of biomedical research.
As with other fields of biological research, human microbiome research is going through several phases to (1) identify the constituent components, (2) associate the components with different physiological or pathological states, (3) understand the functional and causative relationships between the microbiome and human host, and (4) translate that knowledge into applications such as therapies or diagnostics.5 The advent of -omics technologies, such as next-generation sequencing (NGS), has been instrumental in catapulting the ability to profile microbial lineages present in humans. This has led to the establishment of various microbial censuses in the human body, such as the Human Microbiome Project (HMP) led by the National Institutes of Health (NIH) in the USA6 and the MetaHIT, led by a European consortium.7 At present, a majority of microbiome research has been focusing on identifying associations between the microbiome and diseases, host factors (e.g., age, ethnicity, diet), or a wider environment. However, associations identified through metagenome wide association studies (MWAS) are purely correlative and do not shed insights into causal links and the involvement of any indirect confounding factors. Hence, perturbation studies conducted through loss or gain-of-function experiments in experimental models, such as fecal transplantation in germ-free mice,8,9 are important to gain mechanistic understanding into the causal relationships between a particular microbiome signature and a physiological state. The practical translation of microbiome research into new therapies or diagnostics is still at a nascent stage. The recent years we have seen an emergence of a plethora of start-ups attempting to monetize microbiome research. These can range from either providing services to sequence an individual's microbiome or developing therapies via fecal transplantation or engineered probiotics strains.10–12 However, many of these commercial offerings have yet to qualify as bona fide companion diagnostics or therapeutics since this would require the ability to reliably predict drug responses based on the microbiome signatures (for companion diagnostics) or deliver targeted modulation of an individual's microbiome with predictable efficacy and side-effects (for therapeutics). Realization of these goals will need to be built on a more fundamental understanding of how microbes modulate mammalian cellular functions to cause diseases. Therefore, the field has been advocating for a push beyond associative studies to perturbation-based study designs to gain a functional understanding of the human microbiome.5,13
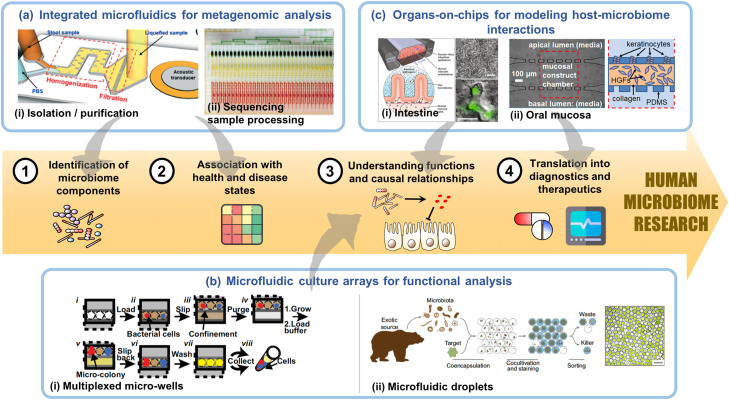
Over the past few decades, microfluidic technologies have made a significant impact in the translation of fundamental cell and molecular biology knowledge into diverse technological platforms for cell analyses, molecular diagnostics, and in vitro tissue models, popularly referred to as organs-on-chips. Similarly, microfluidic technologies can add value to the workflow in different aspects of microbiome research as mentioned above. Here, we will be discussing the potential of applying different microfluidic technologies to specific areas of microbiome research as well as unique challenges that microfluidics must overcome when working with microbiome-relevant biological materials, e.g., micro-organisms, host tissues, and fluids (Fig. 1).
FIG. 1.
Microfluidic technologies can value add to the different phases of human microbiome research. (a) Integrated microfluidics devices can potentially streamline (i) isolation and purification of genetic materials from clinical specimens such as stool14 and saliva with (ii) sequencing sample preparation to overcome current limitations with sample quantity and throughput during metagenomic analysis, which are important for the identification of microbiome components and metagenome wide association studies (MWAS).15 (b) Multiplexable microfluidic culture arrays enable culture and functional measurements of various microbes in the microbiota, which will facilitate mechanistic understanding of causal relationships. The two major classes of culture arrays are (i) micro-well systems as exemplified by the SlipChip16 and (ii) microfluidic droplets, which can co-encapsulate microbes with different functional reporters.17 (c) Organs-on-chips are physiological human tissue analogs, such as (i) intestinal epithelium18–20 and (ii) oral mucosa,21 which serve as an animal-alternative experimental system to functionally validate mechanisms of host–microbiome interactions. They can also potentially be used to evaluate the efficacies to therapeutics aimed at modulating the microbiome. (a-i) Reproduced with permission from Zhao et al., Lab Chip 19, 941 (2019). Copyright 2019 Royal Society of Chemistry. (a-ii) Reproduced with permission from Kim et al., Nat. Commun. 8, 13919 (2017). Copyright 2017 Author(s), licensed under a Creative Commons Attribution (CC BY) license. (b-i) Reproduced with permission from Ma et al., Proc. Natl. Acad. Sci. U.S.A. 111, 9768 (2014). (b-ii) Reproduced with permission from Terekhov et al., Proc. Natl. Acad. Sci. U.S.A. 115, 9551 (2018). Copyright 2018 National Academy of Sciences. (c-i) Reproduced with permission from Bein et al., Cell. Mol. Gastroenterol. Hepatol. 5, 659 (2018). Copyright 2018 Author(s), licensed under a Creative Commons Attribution (CC BY) license. (c-ii) Reproduced with permission from Rahimi et al., Biomicrofluidics 12, 054106 (2018). Copyright 2018 AIP Publishing LLC.
II. BIOLOGICAL SAMPLE PROCESSING FOR MICROBIOME SEQUENCING-BASED ANALYSES
The workflow for microbiome profiling typically involves (1) isolation and purification of genetic material from clinical specimens collected from donors; (2) Polymerase chain reaction (PCR) amplification and tagging to create an amplicon library from individual samples; and (3) next-generation sequencing.22 This is often accomplished with the aid of various commercial kits optimized for each step. Each step comprises of complex experimental logistics and is often labor intensive and time-consuming, which pose a limiting step for sample throughput. Even with the use of commercial liquid handling systems, there is a need for large quantity sample input since the different steps are discrete.15
Microfluidic lab-on-a-chip systems can potentially integrate and automate DNA purification together with sample preparation for DNA sequencing. This will circumvent the existing limitations with low input sample quantity and low throughput. Although microfluidic devices for DNA analysis have been extensively developed as evidenced by a large body of literature on this topic,23–25 it is important to note that they are often meant to be point-of-care diagnostics for detecting a single microbial pathogen or molecular biomarker. In the attempt to determine microbiome signatures as prognostication biomarkers, the extraction and purification processes of microbial DNA must be effective for a diverse microbial community without any bias. This often requires a combination of physical, chemical, and enzymatic lysis approaches to effectively extract DNA from both gram-positive and gram-negative bacteria.26 In addition, the choice of different sequencing-based analyses may require a different process workflow. Amplicon-based sequencing typically involves tagging and amplification of taxonomy-specific amplicons, e.g., bacterial 16S rRNA gene before sequencing the amplified PCR products, while metagenomic analysis uses the whole-genome shotgun (WGS) approach to fragment and sequence the entire DNA of a microbiome sample instead of just the 16S rRNA gene fragments or other target amplicons alone.27 As compared to 16S rRNA sequencing, WGS is more comprehensive; however, it requires extensive computational resources (and is therefore more expensive) to tease out which reads originate from which microbe (i.e., bacteria, virus, fungi). However, from an engineering perspective of designing a microfluidic system that can integrate the workflow processes, WGS is more simplistic, as it does not involve a PCR amplification step, which would necessitate the incorporation of heating and cooling components to perform thermal cycling.
To date, numerous compartmentalized microfluidic systems have been developed. These devices have achieved varying degrees of integration between microbial isolation, DNA purification, and sample preparation for sequencing-based analyses. The technical challenges in the process integration of microbial DNA isolation and purification from clinical specimens are not trivial, particularly for fecal samples, which are important for microbiome research because majority of microbes reside in the intestinal tract. Although fecal samples contain ample amount of microbial materials, they are one of the most challenging samples to process because they are complex in their composition, comprising of microbes, fibers, undigested food materials, nucleases, and human cells.22 This necessitates pre-treatment steps, such as homogenization and filtration to remove particulates before cell lysis, and DNA extraction steps can occur. Mechanical homogenization and filtration can be implemented by incorporating external magnetic stirrers and filters into the microfluidic device.28 Alternatively, acoustofluidics has been used for homogenization while micro-pillars act as filters to remove solid particles.14 Cell lysis and DNA capture from samples are often accomplished with the use of heat, chemical lysis buffers in conjunction with magnetic particles that can selectively bind to DNA and be moved to a different locality within the microfluidic device with an external magnet.15,29,30 To date, microfluidic systems that have demonstrated DNA isolation and purification from human fecal samples have either relied on off-chip genomic analysis14,28 or have only attained integration with reverse transcription polymerase polymerase chain reaction (RT-PCR) detection of single bacteria strains, such as Helicobactor pylori.29
As compared to fecal samples, other clinical specimens, such as urine, sweat, saliva, virginal discharge, and buccal swaps, are less complex and heterogeneous in their composition. However, their microbial DNA content may be more diluted. To overcome this limitation, microfluidic technologies specifically developed to isolate and concentrate low abundance genetic materials, e.g., cell free DNA, have been adopted.31,32 They can generally be categorized into those that incorporate silica membranes or nanoparticles to perform solid phase isolation or those that rely on liquid phase separation by using electrophoresis (EP) or dielectrophoresis (DEP) forces to enrich negatively charged DNA molecules.32 Microbial DNA harvested from these clinical specimens will be relevant for tissue-specific microbiome profiling. At present, the microfluidic technologies that can isolate microbes as well as extract and purify microbial DNA for pathogen detection from environmental (e.g., soil, water), blood, saliva samples are relatively more mature than microfluidic devices that can process stool samples and have been previously reviewed.33,34 Therefore, researchers have been able to build on existing know-how on extracting microbial genetic materials from these clinical samples and further integrate downstream genomic sequencing sample preparation procedures into a single device. For instance, Shi et al. used size-fractionation microfluidics to enrich microbes from clinical sputum samples, which was then coupled to a microfluidic droplet generator to perform droplet-based whole-genome shotgun (WGS) sequencing for profiling the airway metagenomics.35 In another study, a high throughput microfluidic multiplex array with integrated “Quake” valves was used to integrate key steps in microbial DNA isolation and WGS sample preparation, including cell concentration, lysis, fragmentation, adaptor tagging, fragment purification, and size selection.15 This integrated system has been used to perform sequencing on Mycobacterium tuberculosis from clinical bronchial lavage or sputum samples to determine antibiotics resistance related signatures. Looking forward, the key challenge for the microfluidic community is to improve existing microfluidic DNA analyses as well as pathogen capture and purification technologies to cope with a diverse microbial population in more complex clinical specimens.
III. MICROBIOTA CULTIVATION AND FUNCTIONAL ANALYSES
Metagenomic analyses of the human microbiome can profile and measure relative changes in microbiota composition. However, to understand causal links to human normal and pathophysiology and eventually translate into therapeutic or diagnostic applications, it is important to determine the functions of constituent microbial population. This would require the ability to cultivate different microbial species and measure their functions, such as carbon source utilization and antibiotics resistance. Traditional microbial cultivation techniques are tailored for large-scale production of a small number of bacterial strains that are of commercial interest in the food and biotechnology industries. Besides being large scale, traditional microbiology culture conditions often cannot support a vast majority of micro-organisms that exist in the human body because they do not recapitulate the native microenvironment, where effects such as cross-feeding and microbial–host interactions are present.16 Therefore, many bacteria identified by various microbiome census to be most relevant to human health are considered “unculturable.” In addition, the accompanying microbial functional testing kits and assays (e.g., bioMérieux API® test strips) tend to require large quantities of microbes, which is not amenable to microbiome research. Therefore, a different repertoire of microbiological culture and assay technologies needs to be specifically tailored for microbiome research.
A technical hurdle that culture and assay technologies must overcome stems from the shift in having large quantities of a few bacterial strains in traditional microbiology research to minute quantities of a massive number of bacterial species in microbiome research. Microfluidics offers an extremely attractive platform such that large numbers of discrete, nano/micro-scale culture chambers can be created to partition a mixed microbial population into single cells for subsequent expansion and functional assessment. A comprehensive review on the various microfluidic technologies that have the potential capability to perform single microbial cell compartmentalization and analyses have been previously presented.36 Here, we will be focusing on two major classes of microfluidic devices, which have been successfully translated to enable the isolation, culture, and functional analyses of microbial communities in the microbiome. They include (1) single phase multiplexed microfluidic systems, which use elastomeric micro-valves or physical micro-wells to compartmentalize the cell-containing aqueous phase; and (2) droplet microfluidics, which has the aqueous phase broken up into droplets that are suspended in a continuous immiscible oil phase.
Single phase microfluidic devices with varying design architectures have been extensively used for studying microbial physiology and ecology, including chemotaxis, motility, adhesion, biofilm formation, and quorum sensing. These studies have been previously discussed in a number of reviews.37,38 Here, we will be specifically highlighting designs that enable high throughput compartmentalization and cultivation of previously “unculturable” microbes that constitute the environmental or human microbiome. Typically, physical microwells are fabricated out of polydimethylsiloxane (PDMS) or plastics to partition an environmental or clinical sample containing a mixed microbial population. In one embodiment, an isolation chip (iChip) was fabricated by machining an array of 1 mm diameter through-holes in a hydrophobic plastic (Delrin) sheet. When the iChip is dipped and removed into a pre-cured agarose solution containing microbes, surface tension would spontaneously partition the microbes in each through-hole as an agar plug at single-cell resolution.39 Ma's group designed a more sophisticated microwell array known as the SlipChip to enable simultaneous culture and genetic characterization of the same gut bacteria identified by the Human Microbiome Project.16 The SlipChip comprises of two conjoined plates, each containing a microwell array that is juxtaposed to each other. Slippage between the two plates can merge or split the microwells for culture and analysis.40 In both instances, 16S rRNA sequencing was used to identify the microbial population partitioned into each microwell.16,39 The throughput achievable for single phase microwell arrays is typically between 102 and 103 discrete wells per device, which is limited by the scale at which the physical microwells or microvalves can be fabricated.
The inherent characteristics of droplet microfluidics allow for the formation of massive number of uniformly sized (∼10 000 Hz and a coefficient of variation of droplet diameter of less than 2%) “micro-reactors,”41 which are ideal for encapsulating discrete microbial strains present in the microbiome and analyzing their functions. The working principles underpinning the generation and manipulation of microfluidic droplets have been previously reviewed41 and will not be further discussed here. Instead, we will highlight specific applications of droplet microfluidics for culturing and functional analysis of microbiome-derived microbes. For instance, a microfluidic streak plate was developed by partitioning a microbial population into microfluidic droplets, followed by dispensing each droplet onto an agar plate for subsequent culture and functional analyses, including the growth rate and biofilm formation by imaging-based morphological assessment.42 This technology has been applied to isolate and culture live microbes from the termite gut microbiome.43 Microfluidic droplets can also be formed from hydrogels, such as agarose, to provide more conducive microenvironments to support the long-term culture of fastidious microbes.44 Besides encapsulating microbes into single droplets for enrichment and culture, single-cell functional assays can be implemented by co-encapsulating the microbes with functional reporters. For example, Terekhov's group has employed an ultrahigh throughput microfluidic droplet platform to co-encapsulate the microbiota obtained from saliva with a GFP-labeled Staphylococcus aureus (S. aureus) target to functionally screen for microbes that has antimicrobial activity against S. aureus.17 Droplets containing microbes that can selectively kill S. aureus would be identified by a loss of fluorescence intensity via optical microscopy.
The ability to perform biological assays on microbes compartmentalized within microwells or droplets is crucial to determine the identity and functionality of constituent microbial communities of the microbiome. The different design and working principles of single phase multiplexed microwell and droplet microfluidic devices imply that there may be pros and cons associated with each class of devices to perform such analyses. Since both classes of microfluidic devices are usually fabricated from transparent plastics or elastomer (e.g., PDMS), they are amenable to light and fluorescence imaging, which is often used to enumerate and track the morphology of the microbes to indicate for survival and proliferative capacity.16,39,40,42 Expanded bacterial colonies are then typically collected for off-chip 16S rRNA sequencing to identify the expanded microbial population within each compartment.16,39 In such an analysis workflow, integrated microwell systems may have the advantage of being able to sustain longer term culture and facilitate cell retrieval because it is relatively straightforward to execute these processes. In comparison, microfluidic droplets, which are stabilized in an aqueous-oil two phase milieu, are usually not amenable to medium changes. Cell retrieval would entail using detergent washes to break up droplets, which can result in cell losses.
Future integration of multiplexed microbial culture systems with single-cell DNA or RNA sequencing platforms will offer a means to functionally screen how environmental perturbations (e.g., dietary supplements, antibiotics, and probiotics) affect the microbiome composition and functions. At present, microfluidics-enabled microbial single-cell genomic and transcriptomic profiling have already been demonstrated as reviewed by Liu and Walther-Antonio.36 Microbial single-cell genomic sequencing has been implemented in both droplet45,46 and integrated microwell47 platforms. In comparison, the implementation of microbial single-cell RNA sequencing to determine transcriptome profiles is technically more challenging due to the presence of a tough cell wall and a lack of polyadenylated mRNA that can be easily captured.48 To date, microbial single-cell transcriptomic sequencing has only been shown on an integrated micro-well system.49 As new sequencing reagents and probes are being developed to address the challenges of working with microbial RNA,50 we anticipate that this will be an area of opportunity for microfluidic technologists. To eventually perform single-cell genomic and transcriptomic sequencing on individual microbial cells cultured in a microwell or droplet, multiple reagents, such as cell lysis buffers and barcoded beads, will need to be loaded sequentially into each compartment. For single phase micro-well systems, multi-step fluid manipulation of individual wells will require the incorporation of integrated micro-valve arrays, such as those used in integrated microfluidic systems described above for genetic profiling,15 in order to create parallelized, addressable microwell arrays, which would increase the overall complexity of the device design and operation. The miniaturized and massively parallelizable nature of droplet microfluidics is well-suited to cope with this unique challenge. The technologies for generating and manipulating microfluidic droplets are relatively well-developed due to the popularity of single-cell analysis in mammalian cell biology and have been extensively reviewed.51,52 With the development of more droplet-compatible assays that are specially tailored for evaluating microbial functions, droplet microfluidics will likely play an increasingly important role in human microbiome research.
IV. MODELING OF HOST–MICROBIOME INTERACTIONS
Until date, most microbiome studies are conducted in animal models. An earlier review has discussed on the huge contributions of animal models toward understanding host–microbiome interactions.53 However, from the financial and temporal standpoints, such a method is costly and time-consuming. Besides, to a certain extent, animal models used in human microbiome research are physiologically not representative since there is intrinsic variability in microbiota composition.54 As such, the number of animals required to allow meaningful statistical comparisons across groups will be higher than what researchers generally have used for purely immune-based analyses. Conventional static cell culture systems established in labs prove that co-culturing epithelial cells with bacteria in the same medium to study their interactions is exceedingly difficult. The reason being the bacterial cells are freely suspended and may not associate with the epithelial cells. Furthermore, overgrowing of bacterial cells in the nutrient-rich and favorable temperatures of epithelial cell culture conditions is a common sight. This resulted in rapid nutrient depletion and dramatic changes in pH, thus rendering the medium unsuitable for epithelial cell growth.55 Advancements in biomaterials and microfabrication has given rise to a plethora of interesting and unique in vitro physiological platforms, better known as “organs-on-chips.” These engineered microfluidic-based cell culture chips allow for co-culture of multiple cell types and continual perfusion of nutrients and removal of wastes. These promising “organs-on-chips” provide a wide spectrum of opportunities to revolutionize host–microbe research (Table I) and personalized medicine. A range of new opportunities are provided for bridging gaps in studying complex host–microbe interactions (HMIs). In this section, we will highlight some host–microbe studies that these organs-on-chips have aided and the advantages and disadvantages of the various systems for recapitulating the biology and physiology of host–microbe interactions.
TABLE I.
Summary of reported microfluidic-based host–microbe models developed for different applications.
| Target | Microfluidic device | Major achievements | Pathological mode | Reference |
|---|---|---|---|---|
| Lung | Compartmentalized multi-layered microfluidic device. Sandwiched between the upper and lower chambers is a semi-permeable membrane coated with ECM | E. coli co-cultured with lung epithelium | Lung inflammation caused by bacteria | 56 |
| Commensal bacteria co-cultured with intestinal epithelial cells | … | 19 | ||
| Gut peristalsis, inflammatory cells and gut microbiome independently contribute to health and barrier property of gut epithelial layer | Intestinal inflammation and bacteria overgrowth | 20 | ||
| Gut | Oxygen sensors were integrated into compartmentalized microfluidic device | Co-culture of aerobic and anaerobic bacteria with gut epithelial under anaerobic conditions Complex microbiome isolated from fresh fetal stool samples inoculated in gut chips maintained high bacteria richness without compromising epithelial barrier | … | 57 |
| Compartmentalized multi-layered microfluidic device where bacteria were cultured in a separate chamber on semi-permeable membrane coated with mucin | A community of aerobic and anaerobic commensal bacteria in close contact with gut epithelial cells Transcriptional, metabolic and immunological responses of Caco-2 cells were presented | … | 58 and 59 | |
| Oral | Three-channel microfluidic device where central channel contained gingival fibroblast-laden collagen hydrogel. This was separated from two side channels by micropillars | Keratinocytes were cultured in one side channel while the other channel was to introduce toxic dental materials or Streptococcus mutants | S. mutans disrupt oral epithelial property | 21 |
| Cancer | Microfluidic device comprised of three channels, where chemotactic microchannels separate cell culture chamber from central bacterial channel | Preference of S. typhimurium toward liver cancer cells (HepG2) as compared to healthy liver cells Preference of E. coli toward lung cancer cells as compared to healthy lung cells | Bacterial mediated cancer therapy | 60 and 61 |
A. Lung microbiome
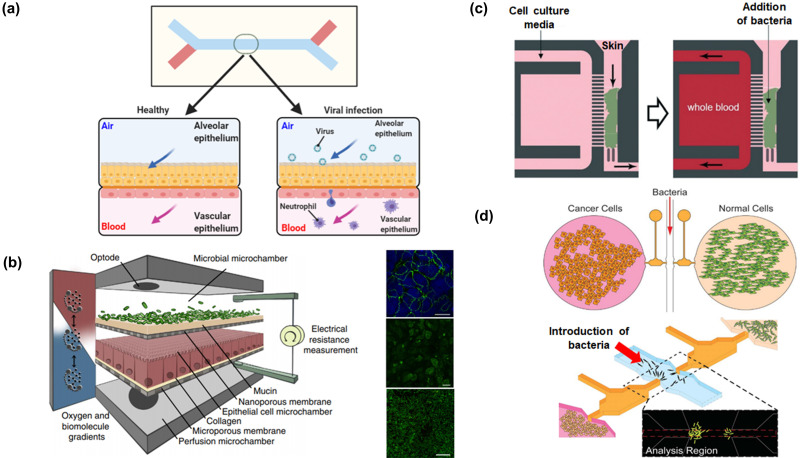
One of the earliest organ-on-chip models is a “breathing” lung-on-chip, in which the alveolar–capillary interface was recapitulated by using two channels separated by a porous extracellular matrix (ECM)-coated membrane.56 Human lung alveolar epithelial cells lined the upper channel, which will eventually be air-filled. In the lower microvascular channel, the semi-permeable membrane was lined with lung microvascular endothelial cells. To mimic inflammation, fluorescently labeled Escherichia coli (E. coli) were added to the alveolar channel, and this activated the endothelium. Increased expressions of surface ICAM-1 and recruitment of human neutrophils (that were perfused through the vascular channel) was observed. Furthermore, the recruited immune cells underwent diapedesis and migrated through both cell layers to the upper chamber, where they engulfed the living E. coli. The findings from the lung-on-chip system provided a means for direct visualization and quantitative analysis of diverse biological processes of the intact lung organ in ways that is not possible in traditional cell culture or animal models. Nevertheless, there are still differences between the lung-on-chip device and in vivo alveolar–capillary barrier, such as barrier thickness, cellular composition, lack of alveolar macrophages, and changes in air pressure and flow. Furthermore, responses of native alveolar epithelial cells might not have been fully reproduced by the transformed lung cells. A few years later, the group improved the system by using primary human airway epithelial cells isolated from healthy individuals or people with chronic obstructive pulmonary disease62 [Fig. 2(a)]. Definitely, these lung-on-chip devices may be used in conjunction with the existing rapid cell-based screening assays to study human disease pathogenesis and expedite drug repurposing in biothreat crises caused by pandemic viruses such as SARS-CoV-2.63 Not only will this allow a better understanding of viral entry into cells, viral replication, host cytokine production, and recruitment of circulating immune cells in response to infection by a virus, experimenting with potential antiviral therapeutics as well as effects of existing and novel therapeutics becomes a possibility. Certainly, lungs-on-chips can bring about these exciting avenues and capabilities; however, these microengineered lungs-on-chips are still in their nascent stages. Better knowledge of lung microbiome and its metabolic fallout (local and systemic) are still required before better physiologically relevant lung microbiome models can be developed.
FIG. 2.
Microfluidic-based in vitro models for mimicking host–microbe interactions. (a) Multi-layered microchip with semi-permeable membrane coated with ECM for investigating viral infection in a lung-on-chip.63 (b) A four-layer microfluidic device with integrated TEER electrodes for studying gut–microbiome59 interactions. Reproduced with permission from Shah et al., Nat. Commun. 7, 11535 (2016). Copyright 2016 Author(s), licensed under a Creative Commons Attribution (CC BY) license. (c) Compartmentalized microfluidic device with migrating channels for analyzing neutrophil migration when skin columns were inoculated with Staphylococcus aureus.64 Reproduced with permission from Kim et al., Lab Chip 19(18), 3094–3103 (2019). Copyright 2019 Royal Society of Chemistry.121 (d) Chemotatic channels in microfluidic device for allowing biochemical factors to diffuse into collagen gel and for observing bacterial preference for cancer cells as compared to healthy cells.61 Reproduced with permission from Hong et al., Lab Chip 13(15), 3033–3040. Copyright 2013 Royal Society of Chemistry.
B. Gut microbiome
Of the microbial mass in a human, 99% is within the gastrointestinal tract, and it exerts both local and long-distance effects. It has been hypothesized that the gastrointestinal microbiome has the greatest effect on the overall health and metabolic status as compared to other microbiomes. Until date, gastrointestinal microbiome is the most and best-investigated microbiome and serves as a model for understanding host–microbiota interactions and diseases. 99% of our gut microbes are anaerobic.65 Specialized bioreactor models, such as the mucosal-simulator of human intestinal microbial ecosystem (M-SHIME), was developed earlier to sustain the growth of luminal and mucosal gut micro-organisms in vitro.66,67 The authors reported seeing different communities of bacteria at different regions of the M-SHIME; for instance, Akkermansia muciniphila, a mucin degrader, is virtually absent in the ascending compartments but abundantly present in the transverse and distal compartments. Although this model has given us a better insight that microbial colonization and community development upon inoculation is possible in a well-controlled multi-compartment gastrointestinal in vitro model and preference of different bacterial communities in different regions of the intestinal compartments, living human intestinal epithelium is absent in M-SHIME. Furthermore, the M-SHIME system is complex and huge.
To allow better insights into host–microbe interactions and develop a more physiological relevant model, co-culturing gut microbiome with intestinal epithelial cells in organs-on-chips is an area that has garnered lots of activities and attention. Initially, these “mini-guts” are formed by culturing Caco-2 cells (a cell line derived from human colorectal adenocarcinoma) on an extracellular matrix (ECM)-coated porous membrane sandwiched between microfluidic channels and subjected to continuous fluidic flow68–74 and/or peristaltic-like movements.19,57,75,76 Marzorati et al. developed a two-compartment device [host–microbe interaction (HMI)] module, where the upper chamber simulated the luminal side of the gut and the lower chamber containing gut epithelial cells simulated the host.58 A semi-permeable membrane with a layer of mucus (200 μm in thickness, facing the upper chamber) separated the chambers. Anaerobic conditions that are representative of the gastrointestinal human–microbe interface were simulated in the system. This device allowed co-culture of intestinal cells with complex microbial communities under microaerophilic conditions for up to 48 h. The authors showed that the short chain fatty acids produced by commensal bacteria (bifidobacterial) helped reduce the pro-inflammatory marker IL-8 in Caco-2 cells. Furthermore, under relevant shear stress (3 dyn cm−2), bacteria could adhere on the mucosal layer. Shah et al. later reported on a better-designed microbiota-gut-chip platform that permitted the co-culture of human and anaerobes59 [Fig. 2(b)]. The reported device composed of a modular stacked assembly of three elastomeric gaskets with a distinct spiral-shaped microchannel, sandwiched between two polycarbonate blocks. Semi-permeable membranes were affixed to elastomeric gaskets that demarcated three channels of 400 ml each where the top microchannel was for culturing bacteria under anaerobic conditions, middle microchannel for culturing Caco-2 under aerobic conditions, and the bottom channel functioned as a perfusion chamber. By co-culturing human epithelial cells with obligate anaerobes, Bacteroides caccae and Lactobacillus rhamnosus GG (LGG) resulted in transcriptional responses, which was distinct from that of a co-culture solely comprising LGG. A closer look at the microfluidic device, however, raises the question on how reliable the reported model was, since there was no direct contact of microbes with human epithelial cells. The microbes were cultured in a chamber that was separated from the human epithelial cells by a porous membrane coated with mucin. Although both systems reported by Marzorati et al. and Shah et al. have shed more insights into host–microbe interactions, both systems lacked direct contact of microbial communities with the epithelial layer and immune cells. Studies have shown that direct contact of microbial communities with the epithelial layer and immune cells may affect the morphology and functionality of the intestinal epithelium and vice versa.77
To enable human intestinal epithelium, capillary endothelium, immune cells, and commensal microbes to multiply, coexist, and interact, while experiencing physiologically relevant flow and peristalsis-like motions in vitro, a two-compartment gut-chip was microengineered.78 Over a period of five days, the Caco-2 cells formed three-dimensional villi-like structures and secreted mucus. These mini-guts were predominately used for studying the physiology of host cells under various treatment conditions, including co-culture with specific micro-organisms. When these mini-guts were colonized with gut dwelling microbes, epithelial cells were inflamed and the intestinal barrier was compromised.19,20 However, the commensal bacteria used in these studies were cultured under aerobic conditions. Recently, the team had reported on how establishing a hypoxia gradient across the engineered tissue–tissue (endothelium–epithelium) interface by using primary human intestinal epithelial allowed for stable co-culture of highly complex communities of anaerobic and aerobic human commensal gut bacteria for up to five days.57 The authors reported that controlling oxygen tension in the gut chips affected the abundance of various bacteria. For instance, mucus degrading, obligate anaerobe genus Akkermansia was found in greater abundance in the anaerobic gut chips, which contain human intestinal epithelial cells that secrete mucus, as compared to cultures maintained under similar anaerobic conditions, which were artificially supplemented with mucin. Complex microbiome isolated from fresh fetal stool samples collected from four different infants (stool samples contained more than 200 different bacteria) cultured in the device did not compromise the epithelial barrier, and bacteria richness was maintained. These gut chips have demonstrated their capability to discriminate between healthy and injury responses of human intestinal epithelium in the presence of commensal and pathogenic bacteria. Furthermore, they offer an avenue for studying the direct intercellular interactions under controlled conditions in vitro. Clearly, these microfluidic-based intestines-on-chips offer an attractive physiologically relevant in vitro model for understanding how different commensal microbes may contribute to the numerous gastrointestinal diseases as well as defining the relationship between the availability of each genus of microbe and the distinct co-cultured intestinal epithelium. Moreover, these gut chips can be used for analyzing fluctuations of microbial communities, their functions, and locations within the chip. Such as when the intestinal epithelium is exposed to hormonal or antibiotics stimulations, what would be the outcome on intestinal pathophysiology. Certainly, in the future, it is possible to develop patient-, disease-, and location- (different regions of the intestine)specific host–microbiome co-culture models in these gut chips in advancing toward personalized medicine.
Oxygen tension in the gut plays an important role in maintaining the spatial distribution and metabolism of gut flora.79 Numerous attempts were done to recapitulate the intestinal microenvironment, such as bubbling nitrogen gas in the culture medium before introducing medium to cells57,59 or using an oxygen scavenger.80 One group modified Transwell systems by integrating microfluidics technology. This modified system allows for generating different oxygen tension environments. One such study reported on developing a new Transwell compatible system whereby oxygen was perfused through the basal compartment while the apical compartment was kept anaerobic by placing the entire system in a hypoxic chamber.81 In another study, a self-sustaining oxygen gradient platform was developed by incorporating a polycarbonate plug in a Transwell insert to study the interaction of the crypt base and obligate anaerobes.82 Other strategies use 3D culture models like sacrificial micromolding and microfluidic tissue culture arrays83 to mimic the intestinal–bacterial environment. The basic limitation of these approaches is the lack of perfusion across the cells. Studies have shown that perfusion is important for cell function and maintenance of tight junctions.20
C. Oral microbiome
A diverse community of microbiome (including bacteria, fungi, archaea, viruses, and protozoa) resides in the human oral mucosa.84 Human oral mucosal health is partially determined by cell and tissue responses to potentially toxic compounds from dental materials and pathogenic bacteria. As exemplified by a three-channel microfluidic device, the central channel contained a gingival fibroblast-laden collagen hydrogel separated from two side channels [where one channel contained keratinocytes and the other was to introduce hydroxylethyl methacrylate (HEMA) or an oral bacterium, Streptococcus mutans (S. mutans)] by micropillars.21 Exposure to HEMA lowered mucosal cell viability, while exposure to S. mutans disrupted the epithelial barrier property. This study has shown that oral mucosal remodeling and responses are dependent on the epithelial and subepithelial layer interactions with bacteria and biomaterials. However, the downside of the study was that all investigations were carried out in static conditions. Studies have shown that morphology of keratinocyte layer and cell turnover in the oral epithelium are affected by saliva or gingival crevicular fluid.85 Furthermore, in the oral cavity, researchers will need to bear in mind that saliva plays a key role influencing the oral microbiome. Therefore, when designing oral host–microbe studies in microfluidic devices, reagents that are similar to saliva components86 will be ideal for mimicking the oral environment. Additionally, the mucosa-on-a-chip was cultured in normoxic conditions and colonized with oxygen-tolerant S. mutans, and this best represents bacterial association with tissue at the gingival margins.87 However, to represent periodontitis in the gingival sulcus, anaerobic conditions will need to be implemented with the oral mucosa chip. Furthermore, to our knowledge, even for static cultures, better oral mucosa-on-chip systems can be developed. Perhaps, in the near future, some can consider including Langerhans’s cells or endothelial or leveraging on some biocompatible hydrogels to develop complex 3D models for better characterization of mouth-on-chip systems.
D. Skin microbiome
Skin is an essential and complex barrier in the human body. It has passive functions such as preventing dehydration and maintaining a gradient of gas exchanges and concentrations. It even regulates the composition of the microbiome in conjunction with skin resident dendritic cells. Over the last decade, skin tissue engineering has advanced greatly. To mimic the complexity of skin models, microfluidic-based devices have been leveraged to develop in vitro skin models that include resident dendritic cells,88 vascular channels, and non-contracting fibrin-based dermal matrices for testing skin responses toward drugs,89 chemicals,88,90 and UV treatment.88 Currently, these skin-on-chip models have not fully captured the cellular and structural characteristics of human skin. This could probably be one of the reasons why limited studies have been reported on studying host and skin microbiota in microfluidic devices. Furthermore, when working with skin-derived micro-organisms, one must always bear in mind that the physiology of bacteria is highly affected when skin milieu is switched to conventional culture media. Hence, instead of culturing skin cells using cell lines before introducing bacteria to the cultures, one group had explored on harvesting human microscopic skin tissue columns collected by micro-biopsy to study neutrophil responses toward Staphylococcus aureus (S. aureus) skin infections.64 Samples of human microscopic skin tissue columns were integrated into one channel of a compartmentalized microfluidic device where it comprised of two main channels separated by an array of chemotaxis microchannels. The other channel contained a drop of whole blood (the source of neutrophils) [Fig. 2(c)]. The number of migrating neutrophils was dependent on the concentration of bacteria that the skin column was exposed to. When the infected samples were treated with penicillin and loaded into a microfluidic device, neutrophils were not seen migrating toward the skin column. Testing various antibiotics at various doses with the reported skin-on-chip may allow one to evaluate the efficiency of antibiotic treatment by monitoring the change in the immune responses. Although the reported skin-on-chip may have the capability to carry out such evaluations to help investigate and compare various treatments on human tissue samples before applying the same treatment to the patients, it would have been advantageous if the bacteria was apical side of the skin under an air interface to better mimic the pathophysiological model of the microbiome on skin. Nevertheless, a better understanding of bacteria such as S. aureus on skin inflammation may help advance the development of antimicrobial treatments such as eczema creams.
E. Cancer microbiome
Cancer remains one of the most common causes of death worldwide.91 Significant attention has been bestowed on microbiome and its influence toward cancer development and cancer therapy,91 especially the area of bacteria-assisted cancer therapy.92–96 For instance, tumors and metastases less than 6–8 mm in diameter are often not detected by positron emission tomography (PET), magnetic resonance imaging (MRI), and computed tomography (CT). Researchers have turned to commensal or engineered bacteria to target tumors and release biomarkers for detection. One group had shown that using a microfluidic device with cultured 3D tumor masses, a biomarker, ZsGreen, in engineered Salmonella (Salmonella has a preference accumulate and replicate in tumors97) was released when Salmonella colonized the tumor masses. This allowed for tumor masses detection that are 2600-fold smaller than the current limits of tomography.98 Such studies would not have been possible to carry out in animal models. Potentially, the exciting results may serve as a non-invasive technique to detect and monitor treatment to tumor masses that are undetectable by current techniques. Moreover, engineering and manipulating geometrical factors of microfluidic channels allow for analyzing cancer-targeting mechanisms of bacteria. One of the earliest studies conducted was to study the interactions between E. coli (wild type (BW25113) and enterohemorrhagic strains (O157:H7) with HeLa cells in a microfluidic device that was controlled by pressure driven microfluidic valves.55 By means of pneumatic control, physical separation was maintained between bacterial and epithelial cells. Once the valve was activated, time-resolved infiltration of E. coli into HeLa cells was measured while preventing premature epithelial cell death, which occurred after 6 h. This study demonstrated that bacteria displayed a spatial bias to colonization and provided insights into how E. coli O157:H7 may outcompete commensal bacteria (BW25113) in the gastrointestinal tract, even though HeLa cells were not of gastrointestinal origin. In another study, a microfluidic device was developed to gain insights into bacterial proliferation in a 3D in vitro tumor model.99 Findings from the study suggested that bacterial proliferation is dependent on the amount of interactions of invasin (a class of proteins associated with the penetration of pathogens into host cells100) with the β1 integrins on the tumor cells. Additionally, it was revealed that tumor cells have the affinity of secreting inhibitors to reduce bacteria proliferation. This information may be useful for engineering bacteria-mediated gene therapy and control of bacterial population dynamics within the host tissues. In another instance, a microengineered device containing four independent cell culture chambers and a central bacteria channel was constructed.61 The presence of the collagen barrier allowed for physical isolation of bacteria from the cell culture chambers. This microfluidic configuration allowed for comparisons of fluorescence intensities in the analyzing region (regions between two cell culture chambers), and bacterial chemotaxis against different cell types can be quantitatively determined [Fig. 2(d)]. The results showed that Salmonella typhimurium has a preference for cancer hepatocytes, HepG2, as compared to healthy liver cells. A similar study was reported on E. coli preference toward lung cancer (NCI-H460) as compared to healthy lung cells.60 Additionally, by secretome analysis and validation experiments, chemotaxis of E. coli in targeting lung cancer was regulated by clusterin (CLU). Co-cultures of bacteria with tumor cells in microfluidic devices allow for a better understanding of bacterial preferences, and this can provide better insights into the pathophysiology of cancer or development of bacterial-based therapeutic means, which are not possible in animal models. However, major downsides of the above reported studies did not reveal how bacteria will perform in media that resembles that of blood nor did they include immune cells such as neutrophils and T-cells. Since bacteria are “foreign” components, there is a possibility, immune cells may destroy the bacteria before they can reach the tumor sites.
Studies have shown that cancer development and progression at a variety of body sites (e.g., stomach, colon, liver, lung, and skin) is dependent on the microbiome community, and microbial abundance may vary at locations within the organs. Such differences might explain for cancer occurrences in particular locations within an organ; for example, there are higher chances of contracting cancer in the large intestine—where microbial densities are much higher as compared to small intestines.101 Perhaps, one of the best examples of bacteria causing cancer is the link of Helicobacter pylori with gastric adenocarcinoma. Gastritis, a precursor of cancer, can be induced by H. pylori as demonstrated by Dr. Barry Marshall infecting himself with H. pylori to illustrate that H. pylori is an etiologic agent of gastritis and gastric ulcers. There may be many other cancers out there that may emanate from microbiome dysbiosis, and such discoveries are waiting for researchers to unveil them. These are opportunities that microbiome-cancer-on-chips can be leveraged to better understand the mechanisms behind specific bacteria undertake to cause certain cancers in humans. This is more beneficial than carrying out such studies in animal models, since animals have different microbiome and biology makeup as compared to humans.53 Having such knowledge is incredibly powerful as it will help advance the discovery of new therapies for cancer.
F. Current hurdles and future steps of organs-on-chips for host–microbe interactions
1. Multi-organ-chips for mimicking systemic microbiome–host interactions
The appearance of organs-on-chips has provided a means to better understand the importance of microbiome and their roles in modulating hosts’ health. It is important to note that studying host–microbe interactions in organs-on-chips devices is still in its emerging phase. Therefore, we must keep in mind their current limitations and challenges. At present, much attention is paid to gut–epithelial and gut–microbiome interactions. This is partly due to the knowledge that the human gut houses the largest communities of microbiota, and a large variety could be acquired from processing fecal samples. The gut microbiota can not only influence the local host tissue that they reside in but also act on other distant organs either directly via microbial metabolites (e.g., short chain fatty acids) or indirectly through modulation of immune-stimulatory and endocrine factors.102 Numerous reports have shown that dysbiosis of gut microbiome may cause metabolic disorders such as obesity and diabetes103,104 and as well as mental illness.105,106 For instance, specific strains of microbes (e.g., Bacteroidetes, E. coli Nissle) have been singled out to study the interactions of gut microbiome with distant organs (e.g., brain, liver) in relation to gut microbiota–brain axis or gut microbiota–liver axis. Organs-on-chips can be extended to encompass multi-organ compartments that are fluidically connected to emulate the systemic circulation between different organs. These microfluidic multi-organ perfusion culture systems are often popularly termed “human/body-on-a-chip.”107–109 Thus, multi-organ perfusion systems can be used to construct systems that allow for the study of interactions between the microbiome with more than one type of host cells. For example, gut microbial organisms are involved in the development of numerous cardio-metabolic phenotypes (i.e., gut microbes participate in a meta-organismal pathway that contributes to the development of atherosclerosis). Multi-organ-chips can provide the means to allow these three different organ-chips (i.e., gut, liver, and endothelial) to simulate the outcome. Moreover, host–microbe studies conducted with organ-chips110 are not only restricted to the gut microbiome but also can be explored with other microbiomes, such as oral microbiome as causative agents for endocarditis111 and brain112 and liver abscess. Undoubtedly, numerous host–microbe interactions remain yet to be explored by researchers (e.g., ocular epithelial–microbe, nasal epithelial–microbe, stomach epithelial–microbe, and vagina epithelial–microbe interactions). This could be partly due to the challenges of not having sufficient knowledge on the different strains of bacteria that have adverse or beneficial effects on the above-mentioned organs. Additionally, the technical challenges with co-culturing microbes with mammalian cells or tissues of interest are not trivial owing to disparate in vitro culture conditions for these two cell types. Some have explored using bacteria isolated from clinical samples co-culture with primary human cells.57 This is certainly an advancement toward personalized medicine; however, much more can be accomplished.
2. Oxygen control in microbial–mammalian cell coculture systems
A major challenge of co-culturing gut microbiota and mammalian cells stems from the different oxygen levels that they must be maintained in. A large proportion of gut microbes are obligate anaerobes and need to be maintained in oxygen-free environments while mammalian cells are often cultured under 21% oxygen. If aerobic and anaerobic conditions are required for host–microbe studies, the microchip material must be capable of keeping both conditions for long durations and/or the external system must be well designed and developed to support such conditions. Additionally, such host–microbe studies require direct or indirect interactions of bacteria with mammalian cells, one major challenge faced is the increased risk of bacterial contamination on the mammalian cell cultures since antibiotics (e.g., penicillin–streptomycin) cannot be present in culture media. Another common challenge that many faced is overgrowth of bacteria leading to cytopathic effects on the mammalian cells.18,58 To resolve such issues, peristalsis actions19,20 or membranes59,66,67 and/or gels21 can be integrated in microchips to prevent bacterial migration from one chamber to another. Additionally, some reports have shown that co-culturing of different bacterial communities may lead to colonization competition in the microenvironment.79 Therefore, one must pay attention when deciding on the strains of bacteria used during co-culture, duration of culture, amount and type of nutrients, aerobic or anaerobic conditions, etc., when carrying out host–microbe studies in microfluidic devices. Furthermore, the conditions in microfluidic devices are generated artificially and often only incorporate one to two cell types that are representative of an organ. Immune, endothelial, and endocrinal cells that can support and modulate organ functions are often not represented in organs-on-chips models. Therefore, the biomarkers identified during co-cultures with microbes in vitro may not fully reflect the in vivo equivalent.
Conducting host–microbe studies with organs-on-chips110 are definitely more beneficial than animal studies since human cells and human commensal bacteria are used in the models. This allow for more relevant pathophysiological observations and outcome. Certainly, more strains of microbiomes will need to be identified for reliable host–microbe studies to be conducted. Although researchers are culturing isolated bacteria from stool samples for microbial studies, they may never be able to identify all the thousands of different types of bacteria that are available in the human body. For instance, the human intestine contains hundreds of different types of bacteria, where many are obligate anaerobes that are incapable of multiplying nor surviving in the atmospheric environment. Therefore, existing in vitro models are not capable of analyzing the direct interactions among complex communities of anaerobic and aerobic microbiota with human intestinal mucosa, which is crucial for understanding gut health and diseases.
3. Hydrogels for creating localized microenvironments in microbial–mammalian cell coculture systems
Tissue- and organ-specific functions can be further improved when eukaryotic cells are cultured on ECM-coated membrane or encapsulated in hydrogels. Likewise, for prokaryotic cells, integrating ECM and synthetic-polymer gels used in 3D culture systems into microfluidic channels provides an opportunity to create localized complex tissue microenvironments that more optimally maintain the microbial and mammalian cell populations. Thus, compartmentalization of different cell types using biomaterials such as hydrogels allows different cell types to become accustomed in the micro-engineered environment without compromising their viability. For instance, some bacteria are not capable of growing unless they are grown close to other bacteria in the same environment like in the case of Haemophilus influenzae dependency on Staphylococcus aureus.113 Since bacteria are encapsulated within ECM and nutrient-rich droplets, localized stable partitioning of bacteria and epithelial cell layer can be established, as opposed to having bacteria throughout the media as found in traditional bulk phase culture.114 This is especially important when studying host–pathogen interactions since increased association between the bacteria and the epithelial cell layer is a requirement. Furthermore, encapsulating bacteria communities makes it possible to study and evaluate the dose-dependent effects of virulent strains on pathogenesis within the organ-chips. Studies revealed that cross-feeding of metabolites between commensal bacteria could serve as a mechanism to preserve certain species diversities. This is achievable by limiting some bacterium from dominating and eliminating other species.115,116 Droplet microfluidics could be leveraged to encapsulate bacteria in microdroplets.117 This approach allows symbiotic microbial communities to be brought together for co-culture and to better understand their synergistic interactions. For instance, a model system using auxotrophic laboratory strains in which a synthetic pair of symbiotic Escherichia coli (E. coli) mutants were diluted into 1 nl microdroplets. Growth occurred only when both variants were present in the same drop.118 However, bacteria require different conditions from human cells in order to grow.
Co-culturing microbes with host mammalian cells is a challenging task. The behavior of microbial communities in vivo differs substantially from their behavior and growth in monoclonal cultures in laboratories. One approach is to culture bacterium on biomimetic materials such as incorporating mucus (e.g., Mucin2) onto a substrate59 or encapsulating the bacterium in hydrogel with mucin. Incorporation of biomimetic compounds has the potential to facilitate viable multispecies culture systems and increase the number of culturable species114 and understand the localization preference of various bacteria at the various sites on the host tissue.119 A study by Kim et al. revealed that 3D printed scaffolds with and without mucin coating (MUC17) co-incubated with Salmonella typhimurium and Escherichia coli O157:H7 revealed that epithelial cells were protected against bacterial infection for the former and enhanced bacterial invasion was observed for the latter.120 Earlier, we have discussed that multi-organ interactions are possible with organs-on-chips. Advancement in bioinks and 3D bioprinting may also be another avenue for constructing modular units with 3D bioprinted hydrogel for studying multi-organ or multi-tissue interactions. Dinh et al. reported that hydrogel microparticles produced by droplet-based microfluidics incubated with mesenchymal stem cell were used as assembly blocks to form 3D microtissues.117 Cell laden hydrogels can be manipulated to form patterns before assembly and cell proliferation. Certainly, this technology can be leveraged in the future for constructing the villi-like structures of the intestines to culture different intestinal cells or alveolar epithelial region for co-culturing different bacterial communities. One major stumbling block in 3D bioprinting is the lack of appropriate bioinks. Surely, a suitable compatible bioink should mimic the mechanical behavior of tissues of interest and yet avoid their structural collapse after several layers have been printed or assembled. In this regard, biomaterials resembling the tissue ECM generated from a variety of natural polymers (such as agarose or collagen) or synthetic polymers [such as poly(ethyleneglycol) or poly(vinylalcohol)] hold promise.
V. CONCLUDING REMARKS AND OUTLOOK
In this paper, we have highlighted various opportunities and challenges of using microfluidic technologies in human microbiome research. By using microengineered microbiome-on-chips to conduct metagenomics, culture arrays for functional analysis, and study on host–microbe interactions, we can have better insights into inter-kingdom communication and signaling. A major advantage of microfluidic-based devices for microbial studies is the possibility of scaling up to achieve high throughput measurements while minimizing sample materials and costs. Potentially, this will reap benefits for technological advancements in systems and synthetic biology, biomaterials, and traditional biotechnology. This may inadvertently bring us closer to initiating personalized medicine and microbial-based therapeutics. However, it is important to keep in mind their current limitations and challenges.
Understandably, the volume of microchannels and/or microchambers in microfluidic devices is small (e.g., in microliters or lesser range). Therefore, collecting samples on microfluidic devices may interfere with their operations. This will result in changes in concentrations of various microbial metabolites. Perhaps, suitable microsensors may need to be integrated onto the microdevices to provide in-line monitoring and measurements. Most crucially, in conducting host–microbe interactions studies, as the number of organ-chips increases, complexities in the functionality becomes more apparent. This may also increase the risk of artifactual and non-translatable issues in the data.
In the upcoming decade, we foresee that advancement of microfluidic technologies discussed in this article will result in the emergence of hybrid systems (e.g., bioprinted organ–host microchips, integration of sample preparation with microbial microdevices). These systems may significantly contribute to appearances of many other sophisticated microfluidic-based sample preparation and analysis devices, organ–microbiome models, and even patient-derived host–microbe models.
AUTHORS’ CONTRIBUTIONS
All authors contributed to the discussion of the contents and reviewed and edited the manuscript before submission.
ACKNOWLEDGMENTS
We acknowledge support from the Australian Research Council Future Fellowship (No. FT180100157) awarded to Y.C. Toh; the Singapore-MIT Alliance for Research & Technology (SMART) (No. ING-000534 BIO); and the Institute for Health Innovation and Technology (No. R-722-000-004-731).
DATA AVAILABILITY
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- 1.Marchesi J. R. and Ravel J., Microbiome 3, 31 (2015). 10.1186/s40168-015-0094-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Flint H. J., Scott K. P., Louis P., and Duncan S. H., Nat. Rev. Gastroenterol. Hepatol. 9, 577 (2012). 10.1038/nrgastro.2012.156 [DOI] [PubMed] [Google Scholar]
- 3.Guthrie L., Gupta S., Daily J., and Kelly L., NPJ Biofilms Microbiomes 3, 1 (2017). 10.1038/s41522-017-0034-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Routy B., Le Chatelier E., Derosa L., Duong C. P. M., Alou M. T., Daillère R., Fluckiger A., Messaoudene M., Rauber C., Roberti M. P., Fidelle M., Flament C., Poirier-Colame V., Opolon P., Klein C., Iribarren K., Mondragón L., Jacquelot N., Qu B., Ferrere G., Clémenson C., Mezquita L., Masip J. R., Naltet C., Brosseau S., Kaderbhai C., Richard C., Rizvi H., Levenez F., Galleron N., Quinquis B., Pons N., Ryffel B., Minard-Colin V., Gonin P., Soria J. C., Deutsch E., Loriot Y., Ghiringhelli F., Zalcman G., Goldwasser F., Escudier B., Hellmann M. D., Eggermont A., Raoult D., Albiges L., Kroemer G., and Zitvogel L., Science 359, 91 (2018). 10.1126/science.aan3706 [DOI] [PubMed] [Google Scholar]
- 5.Schmidt T. S. B., Raes J., and Bork P., Cell 172, 1198 (2018). 10.1016/j.cell.2018.02.044 [DOI] [PubMed] [Google Scholar]
- 6.Huttenhower C., Gevers D., Knight R., Abubucker S., Badger J. H., Chinwalla A. T., Creasy H. H., Earl A. M., Fitzgerald M. G., Fulton R. S., Giglio M. G., Hallsworth-Pepin K., Lobos E. A., Madupu R., Magrini V., Martin J. C., Mitreva M., Muzny D. M., Sodergren E. J., Versalovic J., Wollam A. M., Worley K. C., Wortman J. R., Young S. K., Zeng Q., Aagaard K. M., Abolude O. O., Allen-Vercoe E., Alm E. J., Alvarado L., Andersen G. L., Anderson S., Appelbaum E., Arachchi H. M., Armitage G., Arze C. A., Ayvaz T., Baker C. C., Begg L., Belachew T., Bhonagiri V., Bihan M., Blaser M. J., Bloom T., Bonazzi V., Paul Brooks J., Buck G. A., Buhay C. J., Busam D. A., Campbell J. L., Canon S. R., Cantarel B. L., Chain P. S. G., Chen I. M. A., Chen L., Chhibba S., Chu K., Ciulla D. M., Clemente J. C., Clifton S. W., Conlan S., Crabtree J., Cutting M. A., Davidovics N. J., Davis C. C., Desantis T. Z., Deal C., Delehaunty K. D., Dewhirst F. E., Deych E., Ding Y., Dooling D. J., Dugan S. P., Michael Dunne W., Scott Durkin A., Edgar R. C., Erlich R. L., Farmer C. N., Farrell R. M., Faust K., Feldgarden M., Felix V. M., Fisher S., Fodor A. A., Forney L. J., Foster L., Di Francesco V., Friedman J., Friedrich D. C., Fronick C. C., Fulton L. L., Gao H., Garcia N., Giannoukos G., Giblin C., Giovanni M. Y., Goldberg J. M., Goll J., Gonzalez A., Griggs A., Gujja S., Kinder Haake S., Haas B. J., Hamilton H. A., Harris E. L., Hepburn T. A., Herter B., Hoffmann D. E., Holder M. E., Howarth C., Huang K. H., Huse S. M., Izard J., Jansson J. K., Jiang H., Jordan C., Joshi V., Katancik J. A., Keitel W. A., Kelley S. T., Kells C., King N. B., Knights D., Kong H. H., Koren O., Koren S., Kota K. C., Kovar C. L., Kyrpides N. C., La Rosa P. S., Lee S. L., Lemon K. P., Lennon N., Lewis C. M., Lewis L., Ley R. E., Li K., Liolios K., Liu B., Liu Y., Lo C. C., Lozupone C. A., Dwayne Lunsford R., Madden T., Mahurkar A. A., Mannon P. J., Mardis E. R., Markowitz V. M., Mavromatis K., McCorrison J. M., McDonald D., McEwen J., McGuire A. L., McInnes P., Mehta T., Mihindukulasuriya K. A., Miller J. R., Minx P. J., Newsham I., Nusbaum C., Oglaughlin M., Orvis J., Pagani I., Palaniappan K., Patel S. M., Pearson M., Peterson J., Podar M., Pohl C., Pollard K. S., Pop M., Priest M. E., Proctor L. M., Qin X., Raes J., Ravel J., Reid J. G., Rho M., Rhodes R., Riehle K. P., Rivera M. C., Rodriguez-Mueller B., Rogers Y. H., Ross M. C., Russ C., Sanka R. K., Sankar P., Fah Sathirapongsasuti J., Schloss J. A., Schloss P. D., Schmidt T. M., Scholz M., Schriml L., Schubert A. M., Segata N., Segre J. A., Shannon W. D., Sharp R. R., Sharpton T. J., Shenoy N., Sheth N. U., Simone G. A., Singh I., Smillie C. S., Sobel J. D., Sommer D. D., Spicer P., Sutton G. G., Sykes S. M., Tabbaa D. G., Thiagarajan M., Tomlinson C. M., Torralba M., Treangen T. J., Truty R. M., Vishnivetskaya T. A., Walker J., Wang L., Wang Z., Ward D. V., Warren W., Watson M. A., Wellington C., Wetterstrand K. A., White J. R., Wilczek-Boney K., Wu Y., Wylie K. M., Wylie T., Yandava C., Ye L., Ye Y., Yooseph S., Youmans B. P., Zhang L., Zhou Y., Zhu Y., Zoloth L., Zucker J. D., Birren B. W., Gibbs R. A., Highlander S. K., Methé B. A., Nelson K. E., Petrosino J. F., Weinstock G. M., Wilson R. K., and White O., Nature 486, 207 (2012). 10.1038/nature11234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qin J., Li R., Raes J., Arumugam M., Burgdorf K. S., Manichanh C., Nielsen T., Pons N., Levenez F., Yamada T., Mende D. R., Li J., Xu J., Li S., Li D., Cao J., Wang B., Liang H., Zheng H., Xie Y., Tap J., Lepage P., Bertalan M., Batto J. M., Hansen T., Le Paslier D., Linneberg A., Nielsen H. B., Pelletier E., Renault P., Sicheritz-Ponten T., Turner K., Zhu H., Yu C., Li S., Jian M., Zhou Y., Li Y., Zhang X., Li S., Qin N., Yang H., Wang J., Brunak S., Doré J., Guarner F., Kristiansen K., Pedersen O., Parkhill J., Weissenbach J., Bork P., Ehrlich S. D., Wang J., Antolin M., Artiguenave F., Blottiere H., Borruel N., Bruls T., Casellas F., Chervaux C., Cultrone A., Delorme C., Denariaz G., Dervyn R., Forte M., Friss C., Van De Guchte M., Guedon E., Haimet F., Jamet A., Juste C., Kaci G., Kleerebezem M., Knol J., Kristensen M., Layec S., Le Roux K., Leclerc M., Maguin E., Melo Minardi R., Oozeer R., Rescigno M., Sanchez N., Tims S., Torrejon T., Varela E., De Vos W., Winogradsky Y., and Zoetendal E., Nature 464, 59 (2010). 10.1038/nature08821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fritz J. V., Desai M. S., Shah P., Schneider J. G., and Wilmes P., Microbiome 1, 14 (2013). 10.1186/2049-2618-1-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chevrette M. G., Bratburd J. R., Currie C. R., and Stubbendieck R. M., mSystems 4, e00175 (2019). 10.1128/mSystems.00175-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gershoni K. and Eberhard A., See https://techcrunch.com/2016/08/03/how-the-microbiome-will-lead-a-revolution-in-the-consumerization-of-personalized-medicine-and-diet/ for Techcrunch (2016).
- 11.Yule A., See https://www.alacrita.com/blog/monetizing-the-microbiome for information on efforts in commercializing microbiome products (2019).
- 12.Taroncher-Oldenburg G., Jones S., Blaser M., Bonneau R., Christey P., Clemente J. C., Elinav E., Ghedin E., Huttenhower C., Kelly D., Kyle D., Littman D., Maiti A., Maue A., Olle B., Segal L., Vlieg J. E. T. V. H., and Wang J., Nat. Biotechnol. 36, 1037 (2018). 10.1038/nbt.4287 [DOI] [PubMed] [Google Scholar]
- 13.Proctor L., Nature 569, 623 (2019). 10.1038/d41586-019-01654-0 [DOI] [PubMed] [Google Scholar]
- 14.Zhao S., He W., Ma Z., Liu P., Huang P.-H., Bachman H., Wang L., Yang S., Tian Z., Wang Z., Gu Y., Xie Z., and Huang T. J., Lab Chip 19, 941 (2019). 10.1039/C8LC01310A [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim S., De Jonghe J., Kulesa A. B., Feldman D., Vatanen T., Bhattacharyya R. P., Berdy B., Gomez J., Nolan J., Epstein S., and Blainey P. C., Nat. Commun. 8, 13919 (2017). 10.1038/ncomms13919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ma L., Kim J., Hatzenpichler R., Karymov M. A., Hubert N., Hanan I. M., Chang E. B., and Ismagilov R. F., Proc. Natl. Acad. Sci. U.S.A. 111, 9768 (2014). 10.1073/pnas.1404753111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Terekhov S. S., Smirnov I. V., Malakhova M. V., Samoilov A. E., Manolov A. I., Nazarov A. S., Danilov D. V., Dubiley S. A., Osterman I. A., Rubtsova M. P., Kostryukova E. S., Ziganshin R. H., Kornienko M. A., Vanyushkina A. A., Bukato O. N., Ilina E. N., Severinov K. V., Gabibov A. G., and Altman S., Proc. Natl. Acad. Sci. U.S.A. 115, 9551 (2018). 10.1073/pnas.1811250115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bein A., Shin W., Jalili-Firoozinezhad S., Park M. H., Sontheimer-Phelps A., Tovaglieri A., Chalkiadaki A., Kim H. J., and Ingber D. E., Cell. Mol. Gastroenterol. Hepatol. 5, 659 (2018). 10.1016/j.jcmgh.2017.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim H. J., Huh D., Hamilton G., and Ingber D. E., Lab Chip 12, 2165 (2012). 10.1039/c2lc40074j [DOI] [PubMed] [Google Scholar]
- 20.Kim H. J., Li H., Collins J. J., and Ingber D. E., Proc. Natl. Acad. Sci. U.S.A. 113, E7 (2015). 10.1073/pnas.1522193112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rahimi C., Rahimi B., Padova D., Rooholghodos S. A., Bienek D. R., Luo X., Kaufman G., and Raub C. B., Biomicrofluidics 12, 054106 (2018). 10.1063/1.5048938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kumar R., Eipers P., Little R. B., Crowley M., Crossman D. K., Lefkowitz E. J., and Morrow C. D., Curr. Protoc. Hum. Genet. 82, 18.8.1 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bruijns B., Van Asten A., Tiggelaar R., and Gardeniers H., Biosensors 6, 41 (2016). 10.3390/bios6030041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reinholt S. J. and Baeumner A. J., Angew. Chem. Int. Ed. 53, 13988 (2014). 10.1002/anie.201309580 [DOI] [PubMed] [Google Scholar]
- 25.Ritzi-Lehnert M., Expert Rev. Mol. Diagn. 12, 189 (2014). 10.1586/erm.11.98 [DOI] [PubMed] [Google Scholar]
- 26.Bag S., Saha B., Mehta O., Anbumani D., Kumar N., Hansen T., Arumugam M., Vestergaard H., and Pedersen O., Sci. Rep. 6, 26775 (2016). 10.1038/srep26775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fricker A. M., Podlesny D., and Fricke W. F., J. Adv. Res. 19, 105 (2019). 10.1016/j.jare.2019.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kang J. S., Park C., Lee J., Namkung J., Hwang S. Y., and Kim Y. S., Biochip J. 11, 76 (2017). 10.1007/s13206-016-1205-5 [DOI] [Google Scholar]
- 29.Mosley O., Melling L., Tarn M. D., Kemp C., Esfahani M. M. N., Pamme N., and Shaw K. J., Lab Chip 16, 2108 (2016). 10.1039/C6LC00228E [DOI] [PubMed] [Google Scholar]
- 30.Kang J. H., Krause S., Tobin H., Mammoto A., Kanapathipillai M., and Ingber D. E., Lab Chip 12, 2175 (2012). 10.1039/c2lc40072c [DOI] [PubMed] [Google Scholar]
- 31.Hyun K.-A., Gwak H., Lee J., Kwak B., and Jung H.-I., Micromachines 9, 340 (2018). 10.3390/mi9070340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu Z., Qiao Y., and Tu J., Micromachines 10, 672 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Foudeh A. M., Didar T. F., Veres T., and Tabrizian M., Lab Chip 12, 3249 (2012). 10.1039/c2lc40630f [DOI] [PubMed] [Google Scholar]
- 34.Zhang D., Bi H., Liu B., and Qiao L., Anal. Chem. 90, 5512 (2018). 10.1021/acs.analchem.8b00399 [DOI] [PubMed] [Google Scholar]
- 35.Shi X., Shao C., Luo C., Chu Y., Wang J., Meng Q., Yu J., Gao Z., and Kang Y., mSystems 4, e00198 (2019). 10.1128/mSystems.00198-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu Y. and Walther-Antonio M., Biomicrofluidics 11, 061501 (2017). 10.1063/1.5002681 [DOI] [Google Scholar]
- 37.Hol F. J. H. and Dekker C., Science 346, 1251821 (2014). 10.1126/science.1251821 [DOI] [PubMed] [Google Scholar]
- 38.Rusconi R., Garren M., and Stocker R., Annu. Rev. Biophys. 43, 65 (2014). 10.1146/annurev-biophys-051013-022916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nichols D., Cahoon N., Trakhtenberg E. M., Pham L., Mehta A., Belanger A., Kanigan T., Lewis K., and Epstein S. S., Appl. Environ. Microbiol. 76, 2445 (2010). 10.1128/AEM.01754-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ma L., Datta S. S., Karymov M. A., Pan Q., Begolo S., and Ismagilov R. F., Integr. Biol. 6, 796 (2014). 10.1039/C4IB00109E [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kaminski T. S., Scheler O., and Garstecki P., Lab Chip 16, 2168 (2016). 10.1039/C6LC00367B [DOI] [PubMed] [Google Scholar]
- 42.Jiang C.-Y., Dong L., Zhao J.-K., Hu X., Shen C., Qiao Y., Zhang X., Wang Y., Ismagilov R. F., Liu S.-J., and Du W., Appl. Environ. Microbiol. 82, 2210 (2016). 10.1128/AEM.03588-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou N., Sun Y.-T., Chen D., Du W., Yang H., and Liu S.-J., MicrobiologyOpen 8, e00654 (2019). 10.1002/mbo3.654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Eun Y., Utada A. S., Copeland M. F., Takeuchi S., and Weibel D. B., ACS Chem. Biol. 6, 260 (2011). 10.1021/cb100336p [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chijiiwa R., Hosokawa M., Kogawa M., Nishikawa Y., Ide K., Sakanashi C., Takahashi K., and Takeyama H., Microbiome 8, 5 (2020). 10.1186/s40168-019-0779-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Leung K., Zahn H., Leaver T., Konwar K. M., Hanson N. W., Pagé A. P., Lo C. C., Chain P. S., Hallam S. J., and Hansen C. L., Proc. Natl. Acad. Sci. U.S.A. 109, 7665 (2012). 10.1073/pnas.1106752109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Landry Z. C., Giovanonni S. J., Quake S. R., and Blainey P. C., Methods Enzymol. 531, 61 (2013). 10.1016/B978-0-12-407863-5.00004-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mazutis L., Gilbert J., Ung W. L., Weitz D. A., Griffiths A. D., and Heyman J. A., Nat. Protoc. 8, 870 (2013). 10.1038/nprot.2013.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu Y., Jeraldo P., Jang J. S., Eckloff B., Jen J., and Walther-Antonio M., Anal. Chem. 91, 8036 (2019). 10.1021/acs.analchem.8b04773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Blattman S. B., Jiang W., Oikonomou P., and Tavazoie S., Nat. Microbiol. 5, 1192 (2020). 10.1038/s41564-020-0729-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rakszewska A., Tel J., Chokkalingam V., and Huck W. T. S., NPG Asia Mater. 6, 1 (2014). 10.1038/am.2014.86 [DOI] [Google Scholar]
- 52.Matuła K., Rivello F., and Huck W. T. S., Adv. Biosyst. 4, 1900188 (2020). 10.1002/adbi.201900188 [DOI] [PubMed] [Google Scholar]
- 53.Kostic A. D., Howitt M. R., and Garrett W. S., Genes Dev. 27, 701 (2013). 10.1101/gad.212522.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nguyen T. L. A., Vieira-Silva S., Liston A., and Raes J., Dis. Model. Mech. 8, 1 (2015). 10.1242/dmm.017400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim J., Hegde M., and Jayaraman A., Lab Chip 10, 43 (2010). 10.1039/B911367C [DOI] [PubMed] [Google Scholar]
- 56.Huh D., Matthews B. D., Mammoto A., Montoya-Zavala M., Hsin H. Y., and Ingber D. E., Science 328, 1662 (2010). 10.1126/science.1188302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jalili-Firoozinezhad S., Gazzaniga F. S., Calamari E. L., Camacho D. M., Fadel C. W., Bein A., Swenor B., Nestor B., Cronce M. J., Tovaglieri A., Levy O., Gregory K. E., Breault D. T., Cabral J. M. S., Kasper D. L., Novak R., and Ingber D. E., Nat. Biomed. Eng. 3, 520 (2019). 10.1038/s41551-019-0397-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Marzorati M., Vanhoecke B., De Ryck T., Sadaghian Sadabad M., Pinheiro I., Possemiers S., Van Den Abbeele P., Derycke L., Bracke M., Pieters J., Hennebel T., Harmsen H. J., Verstraete W., and Van De Wiele T., BMC Microbiol. 14, 133 (2014). 10.1186/1471-2180-14-133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shah P., Fritz J. V., Glaab E., Desai M. S., Greenhalgh K., Frachet A., Niegowska M., Estes M., Jager C., Seguin-Devaux C., Zenhausern F., and Wilmes P., Nat. Commun. 7, 11535 (2016). 10.1038/ncomms11535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Song J., Zhang Y., Zhang C., Du X., Guo Z., Kuang Y., Wang Y., Wu P., Zou K., Zou L., Lv J., and Wang Q., Sci. Rep. 8, 1 (2018). 10.1038/s41598-017-17765-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hong J. W., Song S., and Shin J. H., Lab Chip 13, 3033 (2013). 10.1039/c3lc50163a [DOI] [PubMed] [Google Scholar]
- 62.Benam K. H., Villenave R., Lucchesi C., Varone A., Hubeau C., Lee H. H., Alves S. E., Salmon M., Ferrante T. C., Weaver J. C., Bahinski A., Hamilton G. A., and Ingber D. E., Nat. Methods 13, 151 (2016). 10.1038/nmeth.3697 [DOI] [PubMed] [Google Scholar]
- 63.Si L., Bai H., Rodas M., Cao W., Oh C. Y., Jiang A., Nurani A., Zhu D. Y., Goyal G., Gilpin S. E., Prantil-Baun R., and Ingber D. E., “Human organs-on-chips as tools for repurposing approved drugs as potential influenza and COVID19 therapeutics in viral pandemics,” bioRxiv.
- 64.Kim J. J., Ellett F., Thomas C. N., Jalali F., Anderson R. R., Irimia D., and Raff A. B., Lab Chip 19, 3094 (2019). 10.1039/C9LC00399A [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vedantam G. and Hecht D. W., Curr. Opin. Microbiol. 6, 457 (2003). 10.1016/j.mib.2003.09.006 [DOI] [PubMed] [Google Scholar]
- 66.Van den Abbeele P., Grootaert C., Marzorati M., Possemiers S., Verstraete W., Gérard P., Rabot S., Bruneau A., Aidy Ei S., Derrien M., Zoetendal E., Kleerebezem M., Smidt H., and Van De Wiele T., Appl. Environ. Microbiol. 76, 5237 (2010). 10.1128/AEM.00759-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Van Den Abbeele P., Belzer C., Goossens M., Kleerebezem M., De Vos W. M., Thas O., De Weirdt R., Kerckhof F. M., and Van De Wiele T., ISME J. 7, 949 (2013). 10.1038/ismej.2012.158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zahavi E. E., Ionescu A., Gluska S., Gradus T., Ben-Yaakov K., and Perlson E., J. Cell Sci. 128, 1241 (2015). 10.1242/jcs.167544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yu Z. T. F., Kamei K., Takahashi H., Shu C. J., Wang X., He G. W., Silverman R., Radu C. G., Witte O. N., Lee K.-B., and Tseng H.-R., Biomed. Microdevices 11, 547 (2009). 10.1007/s10544-008-9260-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yu L. C.-H., Wang J.-T., Wei S.-C., and Ni Y.-H., World J. Gastrointest. Pathophysiol. 3, 27 (2012). 10.4291/wjgp.v3.i1.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Younossi Z. M., Koenig A. B., Abdelatif D., Fazel Y., Henry L., and Wymer M., Hepatology 64, 73 (2016). 10.1002/hep.28431 [DOI] [PubMed] [Google Scholar]
- 72.Young M. E., Carroad P. A., and Bell R. L., Biotechnol. Bioeng. 22, 947 (1980). 10.1002/bit.260220504 [DOI] [Google Scholar]
- 73.Du H., Zha G., Gao L., Wang H., Li X., Shen Z., and Zhu W., Polym. Chem. 5, 4002 (2014). 10.1039/C4PY00030G [DOI] [Google Scholar]
- 74.Tan H.-Y., Trier S., Rahbek U. L., Dufva M., Kutter J. P., and Andresen T. L., PLoS One 13, e0197101 (2018). 10.1371/journal.pone.0197101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shin W. and Kim H. J., Proc. Natl. Acad. Sci. U.S.A. 115, E10539 (2018). 10.1073/pnas.1810819115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shin W., Hinojosa C. D., Ingber D. E., and Kim H. J., IScience 15, 391 (2019). 10.1016/j.isci.2019.04.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Okumura R. and Takeda K., Exp. Mol. Med. 49, e338 (2017). 10.1038/emm.2017.20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kim H. J. and Ingber D. E., Integr. Biol. 5, 1130 (2013). 10.1039/c3ib40126j [DOI] [PubMed] [Google Scholar]
- 79.Ohland C. L. and Jobin C., Cell. Mol. Gastroenterol. Hepatol. 1, 28 (2015). 10.1016/j.jcmgh.2014.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Skolimowski M., Nielsen M. W., Emnéus J., Molin S., Taboryski R., Sternberg C., Dufva M., and Geschke O., Lab Chip 10, 2162 (2010). 10.1039/c003558k [DOI] [PubMed] [Google Scholar]
- 81.Fofanova T. Y., Stewart C. J., Auchtung J. M., Wilson R. L., Britton R. A., Grande-Allen K. J., Estes M. K., and Petrosino J. F., “A novel human enteroid-anaerobe co-culture system to study microbial-host interaction under physiological hypoxia,” bioRxiv.
- 82.Kim R., Attayek P. J., Wang Y., Furtado K. L., Tamayo R., Sims C. E., and Allbritton N. L., Biofabrication 12, 015006 (2020). 10.1088/1758-5090/ab446e [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Trietsch S. J., Naumovska E., Kurek D., Setyawati M. C., Vormann M. K., Wilschut K. J., Lanz H. L., Nicolas A., Ng C. P., Joore J., Kustermann S., Roth A., Hankemeier T., Moisan A., and Vulto P., Nat. Commun. 8, 262 (2017). 10.1038/s41467-017-00259-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wade W. G., Pharmacol. Res. 69, 137 (2013). 10.1016/j.phrs.2012.11.006 [DOI] [PubMed] [Google Scholar]
- 85.Jones K. B. and Klein O. D., Int. J. Oral Sci. 5, 121 (2013). 10.1038/ijos.2013.46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Seo S. H., Han I., Lee H. S., Choi J. J., Choi E. H., Kim K. N., Park G., and Kim K. M., Sci. Rep. 7, 1 (2017). 10.1038/s41598-016-0028-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Law V., Seow W. K., and Townsend G., Aust. Dent. J. 52, 93 (2007). 10.1111/j.1834-7819.2007.tb00471.x [DOI] [PubMed] [Google Scholar]
- 88.Ramadan Q. and Ting F. C. W., Lab Chip 16, 1899 (2016). 10.1039/C6LC00229C [DOI] [PubMed] [Google Scholar]
- 89.Wufuer M., Lee G. H., Hur W., Jeon B., Kim B. J., Choi T. H., and Lee S. H., Sci. Rep. 6, 1 (2016). 10.1038/srep37471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Abaci H. E., Gledhill K., Guo Z., Christiano A. M., and Shuler M. L., Lab Chip 15, 882 (2015). 10.1039/C4LC00999A [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bhatt A. P., Redinbo M. R., and Bultman S. J., CA Cancer J. Clin. 67, 326 (2017). 10.3322/caac.21398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhou S., Gravekamp C., Bermudes D., and Liu K., Nat. Rev. Cancer 18, 727 (2018). 10.1038/s41568-018-0070-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhang Z., Tang H., Chen P., Xie H., and Tao Y., Signal Transduct. Target. Ther. 4, 41 (2019). 10.1038/s41392-019-0074-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gopalakrishnan V., Helmink B. A., Spencer C. N., Reuben A., and Wargo J. A., Cancer Cell 33, 570 (2018). 10.1016/j.ccell.2018.03.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Fessler J., Matson V., and Gajewski T. F., J. Immunother. Cancer 7, 108 (2019). 10.1186/s40425-019-0574-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Charbonneau M. R., Isabella V. M., Li N., and Kurtz C. B., Nat. Commun. 11, 1738 (2020). 10.1038/s41467-020-15508-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ganai S., Arenas R. B., Sauer J. P., Bentley B., and Forbes N. S., Cancer Gene Ther. 18, 457 (2011). 10.1038/cgt.2011.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Panteli J. T., Forkus B. A., Van Dessel N., and Forbes N. S., Integr. Biol. 7, 423 (2015). 10.1039/c5ib00047e [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Elliott N., Lee T., You L., and Yuan F., Integr. Biol. 3, 696 (2011). 10.1039/c0ib00137f [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Isberg R. R., Voorhis D. L., and Falkow S., Cell 50, 769 (1987). 10.1016/0092-8674(87)90335-7 [DOI] [PubMed] [Google Scholar]
- 101.O’Hara A. M. and Shanahan F., EMBO Rep. 7, 688 (2006). 10.1038/sj.embor.7400731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Smith P. A., Nature 526, 312 (2015). 10.1038/526312a [DOI] [PubMed] [Google Scholar]
- 103.Tilg H., Cani P. D., and Mayer E. A., Gut 65, 2035 (2016). 10.1136/gutjnl-2016-312729 [DOI] [PubMed] [Google Scholar]
- 104.Schnabl B. and Brenner D. A., Gastroenterology 146, 1513 (2014). 10.1053/j.gastro.2014.01.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Parker A., Fonseca S., and Carding S. R., Gut Microbes 11, 135 (2019). 10.1080/19490976.2019.1638722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Fung T. C., Olson C. A., and Hsiao E. Y., Nat. Neurosci. 20, 145 (2017). 10.1038/nn.4476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sung J. H., Wang Y. I., Narasimhan Sriram N., Jackson M., Long C., Hickman J. J., and Shuler M. L., Anal. Chem. 91, 330 (2019). 10.1021/acs.analchem.8b05293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Edington C. D., Chen W. L. K., Geishecker E., Kassis T., Soenksen L. R., Bhushan B. M., Freake D., Kirschner J., Maass C., Tsamandouras N., Valdez J., Cook C. D., Parent T., Snyder S., Yu J., Suter E., Shockley M., Velazquez J., Velazquez J. J., Stockdale L., Papps J. P., Lee I., Vann N., Gamboa M., Labarge M. E., Zhong Z., Wang X., Boyer L. A., Lauffenburger D. A., Carrier R. L., Communal C., Tannenbaum S. R., Stokes C. L., Hughes D. J., Rohatgi G., Trumper D. L., Cirit M., and Griffith L. G., Sci. Rep. 8, 1 (2018). 10.1038/s41598-018-22749-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ong L. J. Y., Ching T., Chong L. H., Arora S., Li H., Hashimoto M., Dasgupta R., Yuen P. K., and Toh Y. C., Lab Chip 19, 2178 (2019). 10.1039/C9LC00160C [DOI] [PubMed] [Google Scholar]
- 110.Raimondi M. T., Albani D., and Giordano C., Trends Mol. Med. 25, 737 (2019). 10.1016/j.molmed.2019.07.006 [DOI] [PubMed] [Google Scholar]
- 111.Lockhart P. B., Brennan M. T., Thornhill M., Michalowicz B. S., Noll J., Bahrani-Mougeot F. K., and Sasser H. C., J. Am. Dent. Assoc. 140, 1238 (2009). 10.14219/jada.archive.2009.0046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Li X., Tronstad L., and Olsen I., Endod. Dent. Traumatol. 15, 95 (1999). 10.1111/j.1600-9657.1999.tb00763.x [DOI] [PubMed] [Google Scholar]
- 113.Davis D. J., J. Infect. Dis. 29, 178 (1921), see http://www.jstor.org/stable/30082202. [Google Scholar]
- 114.Dwidar M., Leung B. M., Yaguchi T., Takayama S., and Mitchell R. J., PLoS One 8, 1 (2013). 10.1371/journal.pone.0067165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Embree M., Liu J. K., Al-Bassam M. M., and Zengler K., Proc. Natl. Acad. Sci. U.S.A. 112, 15450 (2015). 10.1073/pnas.1506034112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Reese A. T. and Dunn R. R., MBio 9, e01294 (2018). 10.1128/mBio.01294-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Dinh N.-D., Luo R., Christine M. T. A., Lin W. N., Shih W.-C., Goh J. C.-H., and Chen C.-H., Small 13, 1700684 (2017). 10.1002/smll.201700684 [DOI] [PubMed] [Google Scholar]
- 118.Park J., Kerner A., Burns M. A., and Lin X. N., PLoS One 6, e17019 (2011). 10.1371/journal.pone.0017019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Derrien M., van Passel M. W. J., van de Bovenkamp J. H. B., Schipper R. G., de Vos W. M., and Dekker J., Gut Microbes 1, 254 (2010). 10.4161/gmic.1.4.12778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kim S. H., Chi M., Yi B., Kim S. H., Oh S., Kim Y., Park S., and Sung J. H., Integr. Biol. 6, 1122 (2014). 10.1039/c4ib00157e [DOI] [PubMed] [Google Scholar]
- 121.Xing S., Jiang J., and Pan T., Lab Chip 13(10), 1937–1947 (2013). 10.1039/C3LC41255E [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.