The application of artificial intelligence (AI) in medicine is an old idea1–3, but methods for this in the past involved programming computers with patterns or rules ascertained from human experts, which resulted in deterministic, rules-based systems. The study of AI in medicine has grown tremendously in the past few years due to increasingly available datasets from medical practice, including clinical images, genetics, and electronic health records, as well as the maturity of methods that use data to teach computers4–6. The use of data labeled by clinical experts to train machine, probabilistic, and statistical models is called ‘supervised machine learning’. Successful uses of these new machine-learning approaches include targeted real-time early-warning systems for adverse events7, the detection of diabetic retinopathy8, the classification of pathology and other images, the prediction of the near-term future state of patients with rheumatoid arthritis9, patient discharge disposition10, and more.
These newer machine-learning methods have clear advantages, including higher levels of performance, adaptability to more complex inputs (such as images), and scalability, over older rules-based systems. However, older rules-based systems had one clear advantage: by definition, the methodologies implemented in the programming code were more interpretable by medical professionals, as these actually came from experts. Newer methodologies have the danger of becoming more complex and less interpretable, even when sophisticated interpretation technique are used11. Indeed, the potential lack of method interpretability has been called out as an area of worry12. Unclear documentation on training and test-cohort selection, development methodology, and how systems were validated has added to the confusion. This is particularly important as more of these models make their way into clinical testing and into medical products and services, with many of these already bring approved by the US Food and Drug Administration in the past few years. More calls for transparency of the ‘explainability’, and probably the interpretability, of machine-learning models can be expected, as these models in other fields have shown serious shortcomings when researchers have attempted to generalize across populations.
As the field progresses, an increasing number of machine-learning models are being tested in interventional clinical trials, and new reporting guidelines have now been proposed for clinical-trial protocols and trial reports involving AI as an intervention13,14. However, there is still a need for guidelines that better inform readers and users about the machine-learning models themselves, especially about how they were developed and tested in retrospective studies.
In the past, ‘minimum information’ guidelines have substantially improved the downstream utility, transparency, and interpretability of data deposited in repositories and reported in publications across many other research domains, including data on randomized control trials15, RNA (gene) expression, diagnostic accuracy15, observational studies16, and meta-analyses. Here we propose the first steps toward a minimum set of documentation to bring similar levels of transparency and utility to the application of AI in medicine: minimum information about clinical artificial intelligence modeling (MI-CLAIM). With this work, we are targeting medical-algorithm designers, repository managers, manuscript writers and readers, journal editors, and model users.
General principles of the MI-CLAIM design
Sharing of raw clinical data is often neither possible, due to institutional patient-privacy policies, nor advisable without such safeguards in place. Furthermore, the same methods that are enabling new analyses of clinical data may also enable the re-identification of patients in sometimes unpredictable ways17. In any case, validation of the exact results is generally of less interest than whether or not the results are validated in a new cohort of patients. Therefore, MI-CLAIM has two purposes: first, to enable a direct assessment of clinical impact, including fairness and bias; and second, to allow rapid replication of the technical design process of any legitimate clinical AI study.
The six parts of the MI-cLAIM
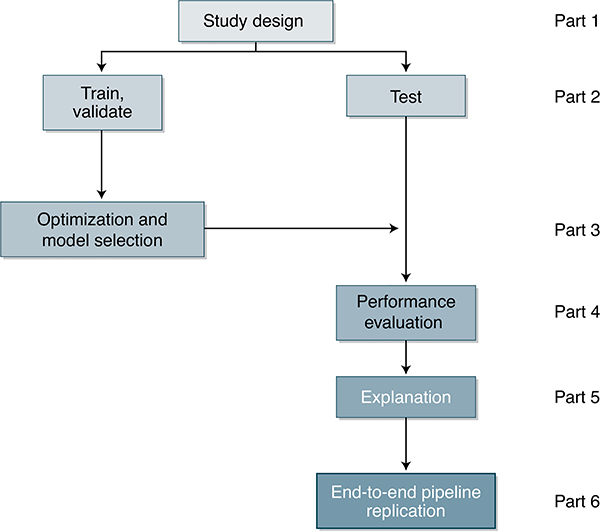
There are six parts to the MI-CLAIM process (Fig. 1). These are outlined below.
Fig. 1 |.
A schematic representation of the six components of a clinical AI study.
Part 1: study design
This section describes the study as a whole. It can be broken down into four subsections: (a) clinical setting, (b) performance measures, (c) population composition, and (d) current baselines to measure performance against.
The clinical problem and the workflow by which a successful model would be employed should be described. Formulating the exact question the algorithm is supposed to answer, as well as how this fits into specific clinical decision-making, is critical, to assess both accuracy and bias. A recent example of this is how the choice of predicting future healthcare costs as a proxy for healthcare needs induced large-scale racial bias in a population health-management algorithm18.
The performance measurements that were used to evaluate the results and how those measurements translate into successes and failures in the clinical setting should be described in detail. Part 4 (below) provides greater detail on performance measurements.
Details should be provided that explain how representative the setting and cohorts are of real-world settings for the clinical question at hand. Additionally, whether it is important that performance is comparable among certain subgroups of the cohorts (e.g., people with diabetes who also have hypertension, or all people with type 2 diabetes) should be stated.
What the clinical baseline abilities are, such as current standard models or methodologies employed in the clinical setting that can act as a proxy for the standard of care in order to gauge how useful the new model is above and beyond current standards, should be included.
Part 2: separation of data into partitions for model training and model testing
Models are said to be overfit if they have learned very specific patterns within the noise of the training data that do not reflect predictive patterns in the real-world cohorts that the model will be applied to. Information leakage occurs when information that would not be available to a model under real-world circumstances is used to train the model. Examples of this include leaking information from the future into the past in time-series modeling, and leaking information from the test set into the training set, such as the same person having one record in the training set and another in the testing set. These two factors are the strongest drivers of poor generalization out of sample and misrepresentations of model efficacy. Documentation detailing the steps that were taken to prevent overfitting and information leakage is critically important for understanding the impact and implications of any medical AI study.
There are two main methods for testing for algorithm generalization: internal and external. Multiple different data conventions exist for internal training and testing, such as cross-validation, two-way splits (training, test), and three-way splits (training, validation, and test). Testing with an external cohort from an independent clinic or hospital system is the highest level of validation.
Within this document, we specifically define the test cohort as a group of cases, set aside at the beginning of the study, against which the final selected model or algorithm is evaluated a single time. We refer to all other cases as members of the training cohort, used for model training, optimization, and selection, which allows researchers to determine the best approach for using the training data for their specific study.
Clarity on how samples were partitioned into separate groups for training and testing at the beginning of the study is essential. Ideally, members of the test cohort will reflect the target clinical population, including the distribution of the clinical outcomes of interest. Methods used to create representative test populations, such as stratified sampling and reporting a comparison of statistics that describe the distribution of variables and outcomes within training and testing populations, should be clearly documented. Documentation must include how any information from the test set was excluded from all activities before the final performance validation. For example, that information should not be considered during feature normalization, model selection, or hyperparameter determination. Cross-validation is not a replacement for a separate test cohort and should not be described as such.
Part 3: optimization and final model selection
With a test set established, the training cohort can now safely be used to estimate (a) the best format of data, (b) the type of model to be used, and (c) the optimal model hyperparameters. This section should begin with data provenance, clearly specifying where the data (in the most raw form) originated, how the data were cleaned and formatted, and, if relevant, what data were additionally available but not used. Transformations (such as de-identification, feature engineering, normalization, and encoding) that were done to the data prior input of the data into the model or algorithm should be described.
The type of models that were evaluated and the process used for selecting the top performing combination of model type, hyperparameters, and data formatting should be clearly described. An example statement is provided in Box 1: the process of preparing the baseline standard-of-care models should be described in equivalent detail, and should include information about the availability or existing use of baseline methods and data.
Box 1 |. Example model selection and optimization statement from Part 3.
Samples were randomly divided into three partitions: training, validation, and test (60:20:20). Fivefold cross-validation (stratified on age, sex, and the outcome variable) on the validation cohort was used to evaluate the results of a grid search on the training cohort comparing number of input features, number of variables to consider at each split, number of splits, and number of trees for random forest models. No other model types were considered. The top performing approach was selected on the basis of median AUC on the validation cohort. The code for implementing the process for feature engineering and model selection is in the ‘Code availability’ section.
Part 4: performance evaluation
Model performance should be reported at two levels. First, how does the model itself perform (F scores, Dice coefficient, or area under the curve (AUC))? Second, how do the model predictions translate into the most relevant clinical performance metrics (sensitivity, specificity, positive predictive value, negative predictive value, numbers needed to treat, and AUC)? This section will include a typical results table with the performance of the baseline and new models tested, along with appropriate statistics for significance. If any important subgroups of patients were identified a priori in Part 1, the performance of the baseline and model in each of those subgroups should also be provided in an identically formatted table.
Part 5: model examination
The provision of some intuition as to how complex models are behaving is useful for many clinical problems and typically serves many purposes. First, it may provide a ‘sanity check’ that the model reached its accuracy by focusing on relevant inputs and not by focusing on unanticipated artifacts of the data. Second, it can uncover biases that model users should be aware of. These biases could relate to adequate representation of clinical and social subgroup samples during training or anticipated points of failure. Third, it provides an understanding of how the model will behave as shifts are seen in the underlying inputs19. Fourth, there are many potential tasks that clinical AI models might be applied to that no human is definitively capable of performing well20. In these cases, it may be useful to harness what the model has learned to generate testable hypotheses to move those fields of science forward.
The appropriate type of examination to perform is dependent upon the type of data being used in the study and the type of model being employed. Clinical AI models accept two broad categories of input data: structured and unstructured. Raw features for structured data can be explicitly defined and understood by researchers; examples include medication names, diagnoses, procedures, laboratory values, and demographic variables. Unstructured data can be loosely defined as the absence of explicitly definable raw features. The most common examples of unstructured data types in the clinical setting include images, whose raw features are individual pixels; natural language, whose raw features are characters; and time series, whose raw features are points in the series.
Visual explanations such as saliency maps21 and their equivalents can be obtained through the use of various methods22. Case-level coefficients, or their equivalents that provide direction and magnitude of effect, can be generated for structured data by multiple methods (such as MAgEC, SHAP). Sensitivity analysis for classification models should include a description of the features of the top five cases in which the model was most confident and correct, most confident and incorrect, and least confident. The same philosophy can be applied to regression models through examination of the cases in which the model had the largest error above the true answer, the largest error below the true answer, and the smallest error. For unsupervised models, domain experts can compare learned representations to known archetypes for fidelity.
The results of model examination must always be considered in the context of the model’s performance. This means that the results of the examination of a model with excellent performance metrics for a particular clinical task should be considered more relevant than the resulting examination of a lower-performing model for the same task. Furthermore, no single examination or interpretation methodology is perfect. For this reason, we require that a minimum of two of the techniques noted above be included as a part of every manuscript. Examination is necessary; however, reporting authors should be free to select and justify any examination approach that addresses the underlying clinical task for which the model or algorithm was designed.
Part 6: reproducible pipeline
Ideally, the code for the complete model-building pipeline should be provided, as well-documented scripts or notebooks, including the exact computer environment requirements, so that an independent researcher can run the pipeline end to end without any modifications to the code being necessary. Preferably, the entire pipeline, beginning with a few examples of properly formatted raw input data and ending with performance evaluation, should be shared via a single container with all of the appropriate versions of necessary dependencies (e.g., through the use of containers and virtualization tools such as Docker). Code for building and running the model should output as many intermediate results as possible so that independent researchers can identify points of divergence when they attempt to reproduce it.
The goal here is not for an independent researcher to replicate the exact results but instead for them to replicate the exact process by which the results were generated, providing that second researcher with everything necessary to rapidly validate the results in their own cohorts. This enables the new researcher to determine whether the results are validated in their own clinical settings and also facilitates the transfer of pipelines from one clinical task to another, which rapidly speeds up prototyping and helps the entire field to develop best practices.
Real-world circumstances may prevent the complete sharing of code in certain situations, such as for results published by commercial entities that view their code base as a proprietary trade secret. Accordingly, we propose a tiered system of transparency. Tier 1 represents complete open sharing of all the software code and scripts. Tier 2 would allow a trusted neutral third party to evaluate the code for accuracy and fairness and provide a report that details the results to accompany the manuscript. For tier 3, authors could release a virtual computing machine or container running the code as an executable (binary) to enable external researchers to test model results against new data without anything about the underlying model itself being revealed. Tier 4 represents no sharing of the underlying model or codebase. We expect model repositories and journals to pick whichever tier thresholds they want to adopt, on the basis of their own needs and standards.
Discussion
Our goal is to develop a documentation standard that can serve clinical scientists, data scientists, and the clinicians of the future who will be using these tools. To that end, a checklist is provided as a part of MI-CLAIM that should be included along with each clinical AI model or manuscript (Table 1). Additionally, we hope that this description will stimulate discussion of the proposed MI-CLAIM standards, and we encourage the clinical community, as well as the AI community, to provide us with their views on how this standard can be improved. For this purpose, a public Github repository has been set up to coincide with the release of this Comment that will allow the community to comment on existing sections and suggest additions.
Table 1 |.
The MI-CLAIM checklist
| Before paper submission | |||
| Study design (Part 1) | Completed: page number | Notes if not completed | |
| The clinical problem in which the model will be employed is clearly detailed in the paper. | ☐ | ||
| The research question is clearly stated. | ☐ | ||
| The characteristics of the cohorts (training and test sets) are detailed in the text. | ☐ | ||
| The cohorts (training and test sets) are shown to be representative of real-world clinical settings. | ☐ | ||
| The state-of-the-art solution used as a baseline for comparison has been identified and detailed. | ☐ | ||
| Data and optimization (Parts 2, 3) | Completed: page number | Notes if not completed | |
| The origin of the data is described and the original format is detailed in the paper. | ☐ | ||
| Transformations of the data before it is applied to the proposed model are described. | ☐ | ||
| The independence between training and test sets has been proven in the paper. | ☐ | ||
| Details on the models that were evaluated and the code developed to select the best model are provided. | ☐ | ||
| Is the input data type structured or unstructured? | ☐ Structured | ☐ Unstructured | |
| Model performance (Part 4) | Completed: page number | Notes if not completed | |
| The primary metric selected to evaluate algorithm performance (e.g., AUC, F-score, etc.), including the justification for selection, has been clearly stated. | ☐ | ||
| The primary metric selected to evaluate the clinical utility of the model (e.g., PPV, NNT, etc.), including the justification for selection, has been clearly stated. | ☐ | ||
| The performance comparison between baseline and proposed model is presented with the appropriate statistical significance. | ☐ | ||
| Model examination (Part 5) | Completed: page number | Notes if not completed | |
| Examination technique 1a | ☐ | ||
| Examination technique 2a | ☐ | ||
| A discussion of the relevance of the examination results with respect to model/algorithm performance is presented. | ☐ | ||
| A discussion of the feasibility and significance of model interpretability at the case level if examination methods are uninterpretable is presented. | ☐ | ||
| A discussion of the reliability and robustness of the model as the underlying data distribution shifts is included. | ☐ | ||
| Reproducibility (Part 6): choose appropriate tier of transparency | Notes | ||
| Tier 1: complete sharing of the code | ☐ | ||
| Tier 2: allow a third party to evaluate the code for accuracy/fairness; share the results of this evaluation | ☐ | ||
| Tier 3: release of a virtual machine (binary) for running the code on new data without sharing its details | ☐ | ||
| Tier 4: no sharing | ☐ | ||
PPV, positive predictive value; NNT, numbers needed to treat.
Common examination approaches based on study type: for studies involving exclusively structured data, coefficients and sensitivity analysis are often appropriate; for studies involving unstructured data in the domains of image analysis or natural language processing, saliency maps (or equivalents) and sensitivity analyses are often appropriate.
Footnotes
Competing interests
I.S.K. is on the scientific advisory boards of Pulse Data and Medaware, both companies involved in predictive analytics. S.S. is a founder of, and holds equity in, Bayesian Health. The results of the study discussed in this publication could affect the value of Bayesian Health. This arrangement has been reviewed and approved by Johns Hopkins University in accordance with its conflict-of-interest policies. S.S. is a member of the scientific advisory board for PatientPing. B.K.B.-J. is a cofounder of Salutary, Inc. B.N. is employed by Anthem. A.J.B. is a cofounder of and consultant to Personalis and NuMedii; consultant to Samsung, Mango Tree Corporation and, in the recent past, 10x Genomics, Helix, Pathway Genomics and Verinata (Illumina); has served on paid advisory panels or boards for Geisinger Health, Regenstrief Institute, Gerson Lehman Group, AlphaSights, Covance, Novartis, Genentech, Merck and Roche; is a shareholder in Personalis and NuMedii; is a minor shareholder in Apple, Facebook, Google, Microsoft, 10x Genomics, Amazon, Biogen, Illumina, Snap, Nuna Health, Royalty Pharma, Sanofi, AstraZeneca, Assay Depot, Vet24seven, Regeneron, Moderna and Sutro, many of which use AI and predictive modeling, and several other non-health-related companies and mutual funds; and has received honoraria and travel reimbursement for invited talks from Genentech, Takeda, Varian, Roche, Pfizer, Merck, Lilly, Mars, Siemens, Optum, Abbott, Celgene, AstraZeneca, AbbVie, Johnson & Johnson, Westat and many academic institutions, state or national agencies, medical or disease specific foundations and associations, and health systems. A.J.B. receives royalty payments through Stanford University for several patents and other disclosures licensed to NuMedii and Personalis. A.J.B. has research funded by the NIH, Northrup Grumman (as the prime on an NIH contract), Genentech, Johnson & Johnson, FDA, US Department of Defense, Robert Wood Johnson Foundation, Leon Lowenstein Foundation, Intervalien Foundation, Priscilla Chan and Mark Zuckerberg, Barbara and Gerson Bakar Foundation and, in the recent past, the March of Dimes, Juvenile Diabetes Research Foundation, California Governor’s Office of Planning and Research, California Institute for Regenerative Medicine, L’Oreal and Progenity.
Code availability
A public Github repository (https://github.com/beaunorgeot/MI-CLAIM) has been set up to coincide with the release of this manuscript, which will allow the community to comment on existing sections and suggest additions.
Here we present the mI-CLAIm checklist, a tool intended to improve transparent reporting of AI algorithms in medicine.
References
- 1.Schwartz WB N. Engl. J. Med. 283, 1257–1264 (1970). [DOI] [PubMed] [Google Scholar]
- 2.Shortliffe EH, Axline SG, Buchanan BG, Merigan TC & Cohen SN Comput. Biomed. Res. 6, 544–560 (1973). [DOI] [PubMed] [Google Scholar]
- 3.Shortliffe EH et al. Comput. Biomed. Res. 8, 303–320 (1975). [DOI] [PubMed] [Google Scholar]
- 4.Ching T et al. J. R. Soc. Interface 15, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Esteva A et al. Nat. Med. 25, 24–29 (2019). [DOI] [PubMed] [Google Scholar]
- 6.Zou J et al. Nat. Genet. 51, 12–18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Henry KE, Hager DN, Pronovost PJ & Saria S Sci. Transl. Med 7, 299ra122 (2015). [DOI] [PubMed] [Google Scholar]
- 8.Gulshan V et al. J. Am. Med. Assoc. 316, 2402–2410 (2016). [Google Scholar]
- 9.Norgeot B et al. JAMA Netw. Open 2, e190606 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rajkomar A et al. NPJ Digit Med. 1, 18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lipton ZC Queue 16, 31–57 (2018). [Google Scholar]
- 12.Topol EJ Deep Medicine: How Artificial Intelligence Can Make Healthcare Human Again 1st edn. (Basic Books, 2019). [Google Scholar]
- 13.Rivera SC et al. Nat. Med 10.1038/s41591-020-1037-7 (2020). [DOI]
- 14.Liu X et al. Nat. Med 10.1038/s41591-020-1034-x (2020). [DOI]
- 15.Moher D et al. Br. Med. J. 340, c869 (2010).20332511 [Google Scholar]
- 16.von Elm E et al. Ann. Intern. Med. 147, 573–577 (2007). [DOI] [PubMed] [Google Scholar]
- 17.Schwarz CG et al. N. Engl. J. Med. 381, 1684–1686 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Obermeyer Z, Powers B, Vogeli C & Mullainathan S Science 366, 447–453 (2019). [DOI] [PubMed] [Google Scholar]
- 19.Subbaswamy A & Saria S Biostatistics 21, 345–352 (2020). [DOI] [PubMed] [Google Scholar]
- 20.Poplin R et al. Nat Biomed Eng. 2, 158–164 (2018). [DOI] [PubMed] [Google Scholar]
- 21.Pan J, McGuinness K, Sayrol E, O’Connor N & Giro-i-Nieto X arXiv https://ui.adsabs.harvard.edu/abs/2016arXiv160300845P (2016).
- 22.Lundberg S & Lee S-I arXiv https://ui.adsabs.harvard.edu/abs/2017arXiv170507874L (2017).