Summary
Non-small cell lung cancer (NSCLC) is the deadliest form of cancer worldwide, due in part to its proclivity to metastasize. Identifying novel drivers of invasion and metastasis holds therapeutic potential for the disease. We conducted a gain-of-function invasion screen, which identified two separate hits, IMPAD1 and KDELR2, as robust, independent drivers of lung cancer invasion and metastasis. Given that IMPAD1 and KDELR2 are known to be localized to the ER-Golgi pathway, we studied their common mechanism of driving in vitro invasion and in vivo metastasis and demonstrated that they enhance Golgi-mediated function and secretion. Therapeutically inhibiting matrix metalloproteases (MMPs) suppressed both IMPAD1- and KDELR2-mediated invasion. The hits from this unbiased screen and the mechanistic validation highlight Golgi function as one of the key cellular features altered during invasion and metastasis.
Introduction
Non-small cell lung cancer (NSCLC) is the leading cause of cancer-associated death in the United States accounting for approximately 25% of all cancer-related mortality (1). The predominant reason for this poor survival is the propensity of lung cancer cells to metastasize resulting in a 5-year survival of 4.5% (1).
To identify novel drivers of metastasis in NSCLC, we previously performed an in vivo gain-of-function screen using genetically engineered KRASLA1/+;p53R172HΔG/+ (KP) mice, and cell lines derived from their primary tumors and metastases with differential metastatic potential (2, 3). Cell lines expressing individual open-reading frames (ORFs) were previously pooled for a stringent, competitive in vivo positive selection resulting in enrichment of genes with a growth advantage. Furthermore, to identify drivers of invasion, here we have performed an in vitro screen utilizing the same candidate gene library from the in vivo screen. The invasive potential of the candidate genes was assessed by a high-throughput transwell invasion assay. We identified both known (MYC and SNAI2) and novel drivers of NSCLC invasion, including IMPAD1 and KDELR2.
Inositol Monophosphatase Domain Containing 1 (IMPAD1) is a Golgi-resident phosphatase that drives the sulfation of glycosaminoglycans (GAGs), which are then secreted from the Golgi and form the cartilaginous extracellular matrix (ECM) (4, 5). In humans, loss-of-function of the enzyme is associated with diseases involving bone deformities, and chondrodysplasia (6). In the context of cancer, IMPAD1 undergoes mRNA upregulation and is co-amplified with oncogenes like MYC (7, 8). However, nothing is known about the functional impact of IMPAD1 upregulation and its role in Golgi-secretion or cancer.
KDEL endoplasmic reticulum protein retention receptor 2 (KDELR2) is a transmembrane-domain protein belonging to the KDEL receptor (KDELR) family (9-11). KDELR2 localizes to the ER Golgi intermediary complex (ERGIC) and binds to ER-resident chaperone proteins with a KDEL ligand in the cis-Golgi and traffics them back to the ER via retrograde transport. In melanoma cells, KDELR activity promoted invadopodia formation and ECM degradation (12, 13). However, the function of KDELR2, specifically, in driving secretion from the Golgi into the ECM, and promoting lung cancer invasion and metastasis remains unstudied.
Here, we identified previously unknown functions of both, IMPAD1 and KDELR2 in enhancing Golgi-mediated secretion of matrix metalloproteases (MMPs) to drive cellular invasion and metastasis. Inhibiting MMPs by using Ilomastat successfully suppressed IMPAD1- or KDELR2-mediated lung cancer cell invasion.
Results
Novel screen identifies IMPAD1 and KDELR2 as drivers of lung cancer invasion and disease progression.
We have previously established model systems to study NSCLC metastasis in mice that recapitulate the disease phenotype of patients (14, 15). We used the KRASLA1/+;p53R172HΔG/+ (KP) mice to derive a panel of lung adenocarcinoma cell lines that were distinguished based on their invasive and metastatic abilities (16, 17). The KP cell line, 393P, is representative of the non-invasive and non-metastatic characteristics, whereas the 344SQ cell line is representative of the highly invasive and metastatic phenotype.
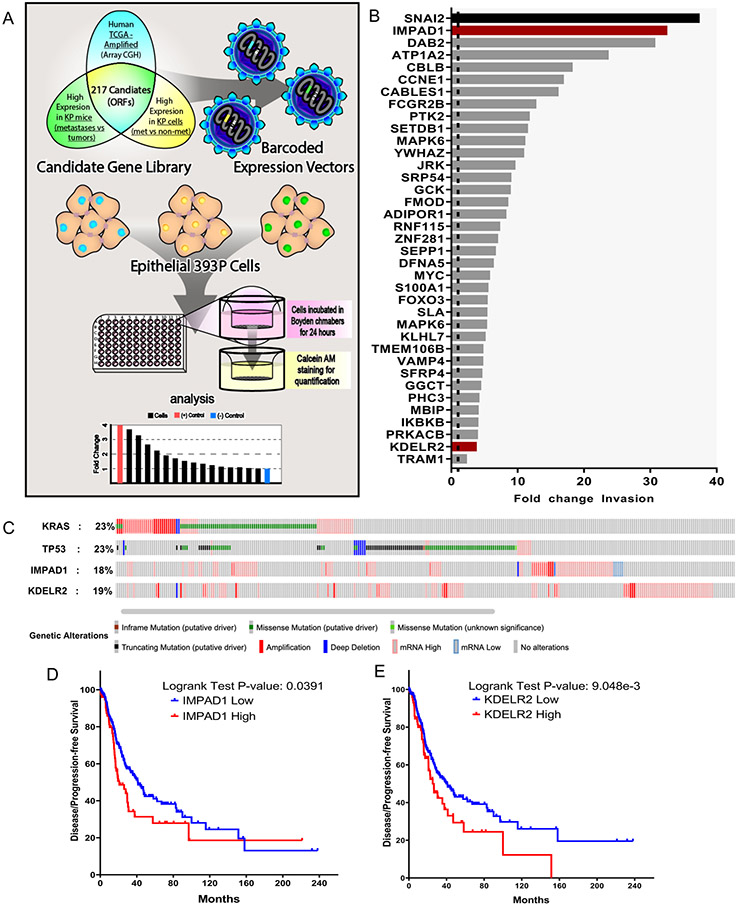
We published a gain-of-function (GOF) in vivo screen that identified drivers of metastasis in NSCLC. The 217 candidate genes for the screen were selected based on their increased expression in three independent datasets. First, in metastases compared to primary tumors in KP mice. Second, in syngeneic tumors formed by implantation of metastatic 344SQ cells compared to non-metastatic 393P cells; and third, in the human TCGA lung adenocarcinoma dataset with a 1.5-fold amplification in copy number (2, 3). The in vivo screen yielded only the top metastasis drivers after a competitive selection pressure. Therefore, to identify additional drivers of invasion, independent of this in vivo selective pressure, we performed an in vitro invasion screen in parallel by utilizing the same candidate genes (Fig. 1a). These cell lines were randomly grouped and were individually seeded in quadruplicates in 96-well invasion chambers. Cells that invaded were stained with calcein AM and the fluorescence quantified to determine their fold change invasion compared to mCherry within the respective groups. To minimize variation, we analyzed the results in two different ways. First, we identified hits with a fold-change invasion higher than 3X standard deviation (SD) of mCherry, across all the groups. To do this, we calculated fold-change invasion for each gene compared to mCherry from the respective cohorts. We then calculated the SD of fold-change values across genes from all groups. Finally, we identified the hits that had a fold-change invasion significantly higher than 3X SD of mCherry across all groups (Details in supplemental methods). The normalized invasion changes showed the drivers of lung cancer invasion (p-value<0.05) (Fig. 1b). Through this, we identified known oncogenes, such as SNAI2, CCNE1, and MYC, and many potential novel drivers, including IMPAD1 and KDELR2. IMPAD1 exhibited the highest invasive ability with a 32-fold increase in invasion, second to the positive control, SNAI2 (37-fold). Importantly, several of the hits were also identified as drivers of metastasis from the in vivo screen (TMEM106B and MBIP), demonstrating functional overlap between the two screening methodologies (3, 18).
Figure 1. Novel screen identifies IMPAD1 and KDELR2 as drivers of lung cancer invasion and disease progression.
a. Schematic representation of workflow for the in vitro screen. DNA barcoded candidate genes were overexpressed in non-invasive & non-metastatic 393P murine tumor cell line using lentiviral infection. Individual ORF-barcoded cell lines were randomly grouped and individually seeded in quadruplicates in 96-well Boyden chambers. Invaded cells were quantified and plotted as shown in representative histogram. b. Waterfall plot demonstrates 37 hits that showed a significant increase in fold change invasion across all cohorts when compared to mCherry. SNAI2 (in black) used as a positive control and mCherry as a negative control, which is denoted as a dotted line (fold change of 1). IMPAD1 and KDELR2, the hits that also showed a significant change in invasion upon further validation (See also Supplemental Fig. S2) with 3 different migration/invasion assays, are represented as red bars. c. IMPAD1 and KDELR2 amplification and mutation frequency in lung adenocarcinoma (LuAd) in relation to putative LuAd drivers, Kras and Tp53, as reported by TCGA. d-e. Kaplan–Meier survival analysis shows significantly poor outcome in disease-free survival of LuAd patients with increased expression of (d) IMPAD1, and (e) KDELR2. See also Supplemental Fig. S3.
Additionally, we determined the top hits from each cohort by comparing the fold-change invasion of the individual genes, that were higher than 3X SD of the respective mCherry control within the same group. This avoided statistical variation across mCherry samples from different cohorts and ascertained the top hits within the respective groups (Fig. S1, details in supplemental methods). These hits were functionally validated by 2D migration and invasion, and 3D invasion assays, where cells were plated in a collagen/Matrigel matrix, and organoids with invasive structures were quantified (Fig. S2A-E). IMPAD1 and KDELR2 were the only two hits that significantly increased invasion compared to mCherry across the three validation platforms (Fig. S2F).
Next, to determine the clinical relevance of IMPAD1 and KDELR2 expression in NSCLC patients, we mined the TCGA lung adenocarcinoma dataset containing 517 samples (provisional). 18% and 19% of the patients showed alterations in IMPAD1 and KDELR2, respectively (Fig. 1c) (https://www.cbioportal.org). 4% of patients showed an amplification in both IMPAD1 and KDELR2. Patients with IMPAD1 alterations had significantly worse disease-free survival compared to the patients with no alterations, with a median of 26.12 months for high expression and 41.23 months for low expression (log rank P = 0.0391) (Fig. 1d). Patients with alterations in KDELR2 with a median survival of 25.22 months also did significantly worse when compared to 41 months for patients with no alterations (log rank P = 9.048e-3) (Fig. 1e). We also found this negative correlation in other lung adenocarcinoma datasets (Fig. S3A, C) (19), as well as in the Human Protein Atlas dataset that uses protein expression instead of mRNA expression (Fig. S3B, D) (20, 21). Increased IMPAD1 and KDELR2 co-expression worsened disease-free survival (median for cases with alterations 25.33 months) similarly to only IMPAD1 or KDELR2 amplification, indicating that a single gene alteration is sufficient to impact survival (Fig. S3E). Hence, we proceeded to study IMPAD1 and KDELR2 as distinct drivers of NSCLC invasion and metastasis.
IMPAD1 or KDELR2 expression is sufficient to drive invasion and metastasis.
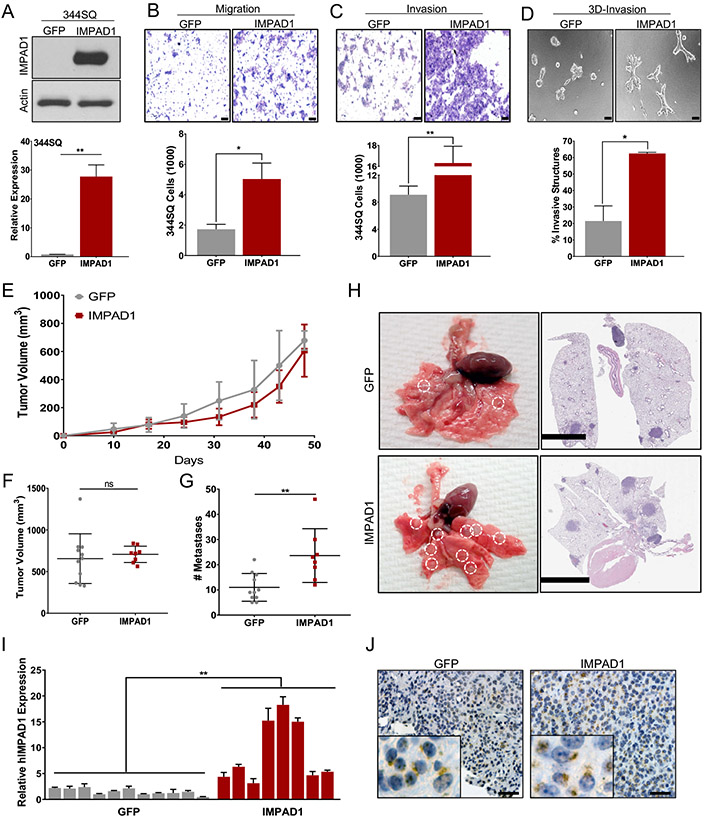
Using the KP murine cell line panel, we demonstrated that both IMPAD1 and KDELR2 are upregulated in the metastatic cells compared to the non-metastatic cells (Fig. S3F-I). To validate IMPAD1 as a driver of invasion and metastasis, it was overexpressed in murine 393P, 344SQ, and human HCC827 NSCLC cell lines either constitutively or in a doxycycline-inducible manner (Fig. 2a, Fig. S4A, G-J). Increased IMPAD1 expression made the cells more invasive in transwell assays, as well as enhanced their cellular motility in scratch assays (Fig. 2b-c, Fig. S4B-C, E, H-I, K). Additionally, using a collagen/Matrigel matrix, which recapitulates the ECM of the tumor, and quantifying the invasive spheroids formed, we demonstrated that IMPAD1 promotes invasion in 3D as well (Fig. 2d, Fig. S4F, L). Next, to study IMPAD1 as a driver of metastasis in vivo, 344SQ-GFP and -IMPAD1 constitutively overexpressing cells were subcutaneously implanted into WT 129/Sv syngeneic mice. The IMPAD1 cohort showed no difference in tumor volume compared to the control group (Fig. 2e-f). This effect was analogous to the phenotype observed in vitro where IMPAD1 expression had no significant consequence on cellular proliferation (Fig. S4N-O). However, IMPAD1 overexpression resulted in a significant increase in metastasis, determined by quantifying the macroscopic lung metastatic nodules and by the H&E-stained images (Fig. 2g-h). We confirmed that the primary tumors maintained IMPAD1 mRNA and protein overexpression (Fig. 2i-j Fig. S4P). Furthermore, we validated the phenotype with the 393P lung cancer cell line, which is innately non-metastatic (Fig. S4Q-V). We have previously demonstrated that alterations in EMT can drive tumor invasion and metastasis in the KP models (14-17). Therefore, we investigated whether IMPAD1 could alter the EMT status of the characteristically epithelial 393P cells by assessing for expression levels of EMT markers such as Zeb1, Vimentin, and E-cadherin (Fig. S4W-X). No significant change was observed upon IMPAD1 upregulation conferring that IMPAD1 could not alter the EMT status in cells.
Figure 2. IMPAD1 expression is sufficient to drive lung cancer invasion and metastasis.
a. RT-qPCR and western blot analysis for human IMPAD1 expression in 344SQ cells with stable overexpression compared to GFP control. b-c. 344SQ cells overexpressing IMPAD1 show a significant increase in (b) migration and (c) invasion compared to GFP controls (scale bar: 100uM). d. IMPAD1 overexpressing cells form significantly more invasive structures compared to GFP in 3D matrix comprising of 1.5 mg/ml Collagen in Matrigel by day 6 (scale bar: 100uM). e-f. Primary tumor growth for 344SQ GFP (n=11) and IMPAD1 (n=8) overexpressing cells implanted subcutaneously into syngeneic mice (e) over time, and (f) at time of euthanasia. g. IMPAD1 overexpressing cells form significantly more lung metastatic nodules compared to GFP control. h. Representative lungs and their respective H&E stained sections showing increased metastases in lungs from mice implanted with IMPAD1 overexpressing cells compared to control (scale bar 5mM). i-j. Analysis to confirm overexpression of IMPAD1 in SQ tumors by (i) RT-qPCR for RNA, and (j) immunohistochemistry for protein (scale bar: 20uM). See also Supplemental Fig. S4. Data are represented as mean ± SEM. Significance by Student’s T-test. P-value<0.05 - *; <0.002 - **
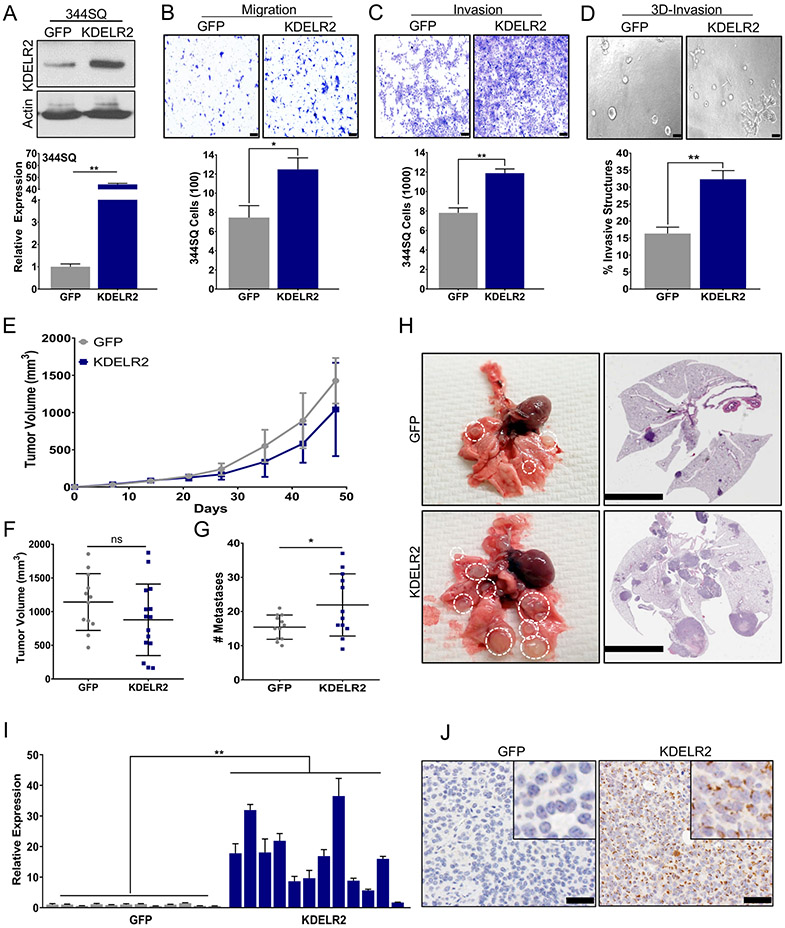
Like IMPAD1, we determined KDELR2 as a driver of invasion and metastasis upon doxycycline-dependent induction in 393P, 344SQ, and HCC827 cells (Fig 3a, Fig. S5B, G). Our data demonstrated that the cellular invasiveness and motility was dependent upon KDELR2 expression (Fig. 3b-d, Fig. S5A, C-F, H-I). To study the effect of KDELR2 in vivo we implanted 344SQ-GFP or -KDELR2 cells into mice with doxycycline induction. KDELR2 significantly increased lung cancer metastases in mice without altering primary tumor growth (Fig. 3e-h). This was once again recapitulative of the in vitro data where KDELR2 showed no significant effect on cellular proliferation (Fig. S5J-L). The metastatic phenotype was validated by gross lung images and the H&E-stained sections (Fig. 3H). We confirmed overexpression of KDELR2 in the primary tumors by RT-qPCR and IHC (Fig. 3i-j). We also determined no change in EMT status upon KDELR2 induction (Fig. S5M-N).
Figure 3. KDELR2 expression is sufficient to drive lung cancer invasion and metastasis.
a. RT-qPCR and western blot analysis for mouse KDELR2 expression in 344SQ cells with doxycycline-inducible overexpression compared to GFP control after 24-hour induction. b-c. 344SQ cells overexpressing Flag-tagged KDELR2 show a significant increase in (b) migration and (c) invasion compared to GFP after 24-hour induction (scale bar: 100uM). d. KDELR2 overexpressing cells pre-induced for 24 hours before seeding form significantly more invasive structures compared to GFP in 3D matrix comprising of 1.5 mg/ml Collagen in Matrigel by day 6 (scale bar: 100uM). e-f. Primary tumor growth for 344SQ GFP and KDELR2 inducible cells implanted subcutaneously in syngeneic mice (e) over time, and (f) at time of euthanasia. N=12. g. KDELR2 overexpressing cells form significantly more lung metastatic nodules compared to GFP control. h. Representative lungs and their respective H&E stained sections showing increased metastases in lungs from mice implanted with KDELR2 overexpressing cells compared to GFP control (scale bar: 5mM). i-j. Analysis to confirm overexpression of Flag-tagged KDELR2 in SQ tumors by (i) RT-qPCR for RNA, and (j) Flag IHC for protein (scale bar: 50uM). See also Supplemental Fig. S5. Data are represented as mean ± SEM. Significance by Student’s T-test. P-value<0.05 - *; <0.002 - **
Our in vitro and in vivo studies with IMPAD1- and KDELR2-overexpression indicate that upregulation of either genes is sufficient to independently drive lung cancer invasion and metastasis by an EMT-independent mechanism and without altering cellular proliferation.
IMPAD1 or KDELR2 expression is necessary for invasive and metastatic ability of lung cancer cells.
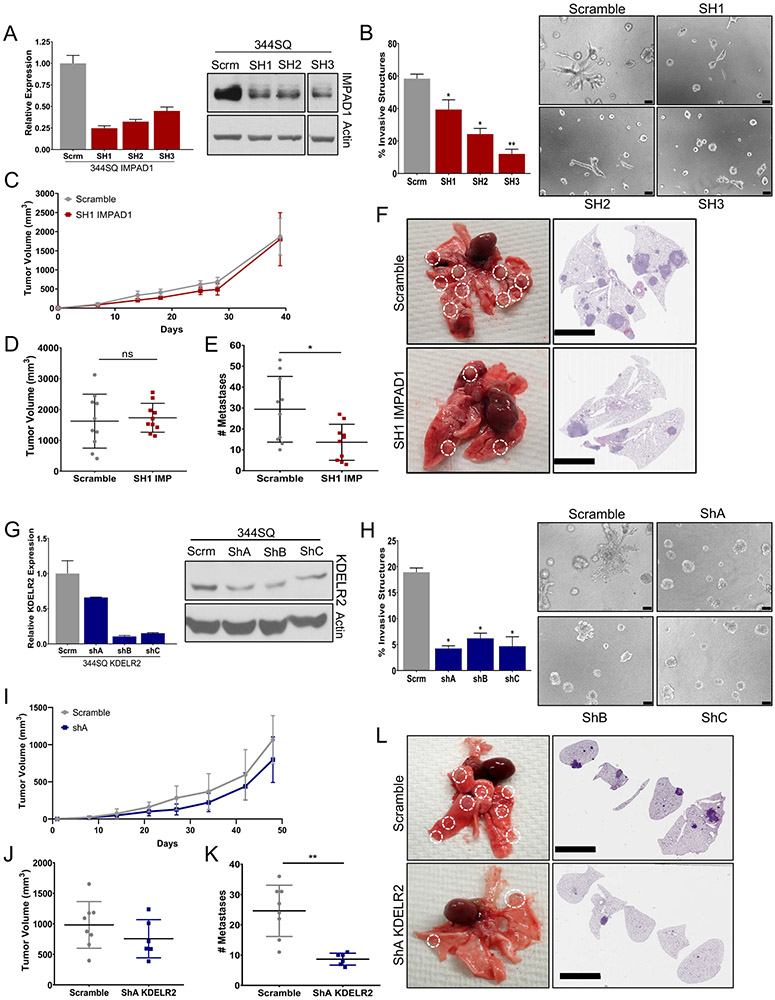
Given that IMPAD1 and KDELR2 are sufficient to drive invasion and metastasis, we next ascertained if they are also necessary for the same. Hence, we independently inhibited the genes by using three different shRNAs, in the metastatic 344SQ and 344P murine cells and confirmed their knockdown by RT-qPCR and western blotting (Fig. 4a, g. Fig. S6C, J). IMPAD1 or KDELR2 repression caused a significant decrease in the invasive abilities in 2D and 3D environments (Fig. 4b, h. Fig. S6A-B, D-F, I, K-L). Thus, we conclude that both IMPAD1 and KDELR2 are essential for tumor cell invasion. We also determined that IMPAD1 or KDELR2 repression does not alter the EMT status of the knockdown cells compared to the scramble controls (Fig. S6G-H, M-N).
Figure 4. IMPAD1 or KDELR2 expression is necessary for invasive and metastatic ability of lung cancer cells.
a. RT-qPCR and western blot analysis for mouse IMPAD1 expression in 344SQ cells with stable knockdown by shRNA compared to scramble control. b. IMPAD1 knockdown cells form significantly less invasive structures compared to scramble in 3D matrix comprising of 1.5 mg/ml Collagen in Matrigel by day 6 (scale bar: 100uM). See also Supplemental Fig. S6. c-d. Primary tumor growth for 344SQ scramble and SH1 IMPAD1 knockdown cells implanted subcutaneously into syngeneic mice (c) over time, and (d) at time of euthanasia. N=10. e. IMPAD1 knockdown cells form significantly less lung metastatic nodules compared to scramble control. f. Representative lungs and their respective H&E stained sections showing decreased metastases in lungs from mice implanted with SH1 IMPAD1 cells compared to control (scale bar 5mM). g. qPCR and western blot analysis for mouse KDELR2 expression in 344SQ cells with stable knockdown by shRNA compared to scramble control. h. KDELR2 knockdown cells form significantly less invasive structures compared to scramble in 3D matrix comprising of 1.5 mg/ml Collagen in Matrigel by day 6 (scale bar: 100uM). i-j. Primary tumor growth for 344SQ scramble and ShA KDELR2 knockdown cells implanted subcutaneously into syngeneic mice (i) over time, and (j) at time of euthanasia. N=8. k. KDELR2 knockdown cells form significantly less lung metastatic nodules compared to scramble control. l. Representative lungs and their respective H&E stained sections showing decreased metastases in lungs from mice implanted with ShA KDELR2 cells compared to GFP control (scale bar: 5mM). See also Supplemental Fig. S7. Data are represented as mean ± SEM. Significance by Student’s T-test. P-value<0.05 - *; <0.002 - **
To study the effect of IMPAD1 knockdown on metastasis, we implanted the 344SQ scramble control and knockdown cells into the flanks of the syngeneic WT 129/Sv mice and observed the tumor growth over 6 weeks. Similar to the proliferation assays in vitro (Fig. S7G), there was no difference in tumor volume between the scramble and the IMPAD1 suppression cohorts (Fig. 4c-d). Despite this, IMPAD1 knockdown significantly reduced the metastatic ability of the 344SQ cells, as confirmed by quantifying the macro-metastatic lung nodules and by the H&E-stained images (Fig. 4e-f). We validated the knockdown in the syngeneic tumors by mRNA and protein expression levels (Fig. S7A-B). Despite a significant repression of IMPAD1 expression that correlated with decreased invasiveness in vitro, there was no change in the metastases for the SH2-IMPAD1 and SH3-IMPAD1 tumor groups compared to control (Fig. S7C-E). Our data indicated that the tumors from these two cohorts did not retain IMPAD1 suppression over time in vivo, explaining the absence of a change in the metastatic phenotype (Fig. S7F). Thus, the knockdown data strongly suggests that IMPAD1 expression is required for NSCLC metastasis.
To investigate the requirement of KDELR2 in lung cancer metastasis, we similarly implanted the 344SQ scramble control and KDELR2 knockdown cells (shA, shB, and shC) into mice. Although the proliferation assays showed a significant reduction in cellular growth upon KDELR2 repression in vitro (Fig. S7N), there was no significant change in tumor volume (Fig. 4i-j Fig. S7J). This difference in proliferation could be due to cellular growth in vitro not recapitulating factors required for tumor growth in vivo. Additionally, we observed a decrease in metastasis upon KDELR2 knockdown (Fig. 4k-l Fig. S7K-L). We validated the knockdown in the primary tumors by RT-qPCR and IHC, which supported that cancer metastasis is dependent on KDELR2 expression (Fig. S7H-I, L-M).
IMPAD1 and KDELR2 independently regulate Golgi-mediated secretion of proteases such as MMPs to drive lung cancer invasion.
Although IMPAD1 and KDELR2 are amplified in lung adenocarcinoma, and literature suggests that they function in the Golgi and ER respectively, their role in Golgi-mediated secretion of proteins, as well as in lung cancer invasion and metastasis is still unknown (7, 8).
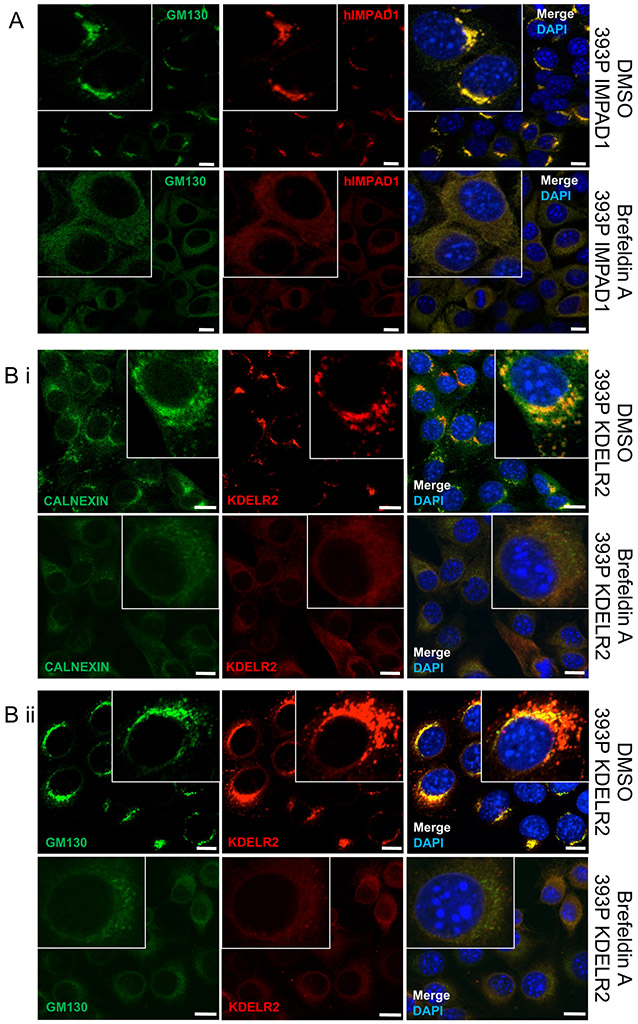
Hence to study this phenotype, we first confirmed the localization of IMPAD1 at the Golgi membrane in our murine and human models by fractionation, and co-IF assays with a Golgi marker, GM130 (Fig. 5a, Fig. S8A-D) (4, 22-24). Furthermore, Brefeldin A (BFA) treatment, which disrupts Golgi stacking (25), also abrogated IMPAD1 signal indicating that IMPAD1 localization to the Golgi is dependent on appropriate organelle formation. Next, in addition to showing that KDELR2 is a membrane-bound protein (Fig. S8E), we also validated its localization to the ER by using the ER marker, Calnexin (Fig. 5bi, Fig. S8Fi), and to the Golgi by using GM130 (Fig. 5bii, Fig. S8Fii) (26). Previous literature indicates that KDELR2 overexpression auto-activates the protein that then localizes to the ER (10-11), which is confirmed by our co-IF data upon KDELR2 induction. Furthermore, like IMPAD1, BFA-mediated disruption of KDELR2 signal validated that proper Golgi and ER assembly is necessary for accurate KDELR2 localization (Fig. 5bi, ii, Fig. S8Fi, ii). Here we demonstrated that IMPAD1 is Golgi-resident, whereas KDELR2 is at the ERGIC and transports between the Golgi and ER, and their localization is dependent upon appropriate Golgi formation. Therefore, we posited that IMPAD1 and KDELR2 are a part of the ER-Golgi pathway that drive the invasive phenotype by altering Golgi function and the secretory processes.
Figure 5. IMPAD1 and KDELR2 localize to the ER-Golgi pathway.
a. Co-immunofluorescence for human IMPAD1 (red) and Golgi marker, GM130 (green) in 393P cells with IMPAD1 overexpression. Nucleus was stained with Dapi. Cells were treated with DMSO (upper) or Brefeldin-A (BFA) at 1uM for 4 hours (lower). IMPAD1 localization to the Golgi is disrupted upon BFA treatment, which abrogates Golgi stacking. b-i. Co-immunofluorescence for FLAG-KDELR2 (red) and Calnexin (green) in 393P cells with mouse KDELR2 overexpression or empty vector control. Cells were treated with DMSO (upper) or Brefeldin-A (1uM 4 hours) (lower). b-ii. Co-immunofluorescence for FLAG-KDELR2 (red) and GM130 (green) in 393P cells with KDELR2 overexpression or empty vector control. Cells were treated with DMSO (upper) or Brefeldin-A (1uM 4 hours) (lower). Nucleus was stained with DAPI. Scale bar for all images:10uM. See also Supplemental Fig. S8.
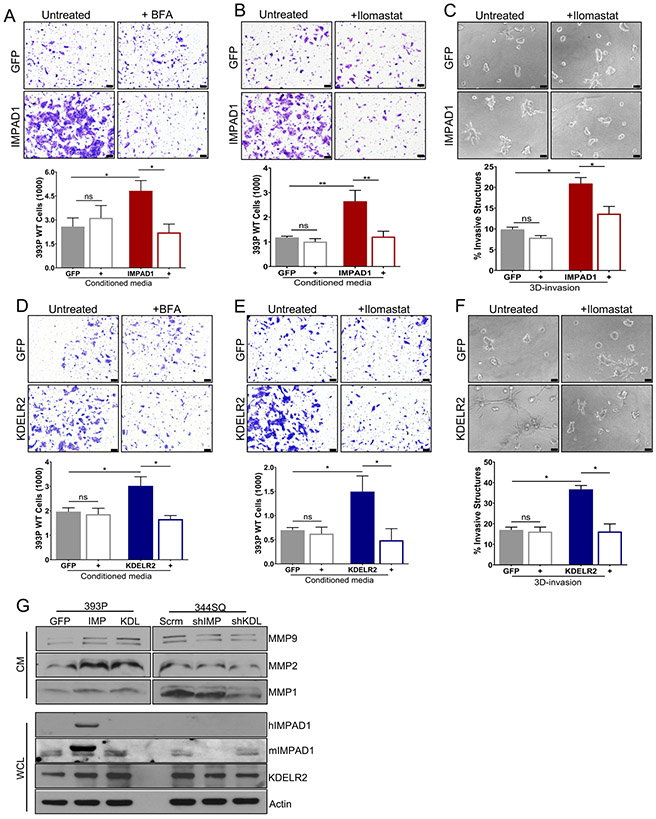
To test this hypothesis, we performed a secretome-mediated invasion assay by seeding non-invasive 393P parental cells in conditioned media (CM) collected from the GFP-, IMPAD1- or KDELR2-inducible cells with or without BFA treatment (Fig. S9A). Conditioned media from IMPAD1- or KDELR2- overexpressing cells significantly increased the invasiveness of the 393P cells, which was reversed upon BFA treatment. This indicated that IMPAD1 and KDELR2 independently enhance Golgi-regulated secretion to drive invasion. (Fig. 6a, d, Fig. S9B, G). To identify components of the secretome that assist IMPAD1- or KDELR2-mediated invasion, we investigated some of the known proteases secreted by the Golgi, such as matrix metalloproteases (MMPs) (27, 28). We performed a similar invasion assay but treated the conditioned media with a pan-MMP inhibitor, Ilomastat (Fig. S9C). Once again, conditioned media from IMPAD1-overexpressing cells and KDELR2-overexpressing cells was sufficient to promote invasion of the non-invasive 393P cells. This phenotype was suppressed upon Ilomastat treatment (Fig. 6b, e, Fig. S9D, H). To recapitulate this in 3D, we plated the IMPAD1- or KDELR2-overexpressing cells in the Matrigel/collagen matrix and treated them with DMSO or Ilomastat. Cells over expressing IMPAD1 or KDELR2 lost their invasive ability by days 3 (data not shown) and 5 (Fig. 6c, f, Fig. S9E-F, I-J) upon MMP inhibition. Furthermore, quantification of mRNA by RT-qPCR demonstrated that MMPs 1a, 2, and 9 were transcriptionally upregulated upon increased IMPAD1 and KDELR2 levels but showed no change upon gene repression (Fig. S9K-N). Western blots performed for MMPs1, 2, and 9 on conditioned media revealed that induction of either IMPAD1 or KDELR2 increased MMP secretion, which was abrogated upon knockdown of the two genes. IMPAD1 and KDELR2 expression was confirmed in the whole cell lysates. (Fig. 6g, Fig. S9O). Additionally, previous literature (12, 13) has shown KDELR2 as a regulator of Src signaling. Hence, we also analyzed the Src signaling pathway in the KDELR2- and IMPAD1-inducible cells. Increased Src phosphorylation was observed until 2 hours post KDELR2 induction, after which the pathway was inhibited, possibly by compensatory cellular mechanisms (Fig. S10A-B). However, the phenotypic changes observed in IMPAD1-overexpressing cells could not be attributed to changes in the Src pathway (Fig. S10C). Thus, we have established that two different hits from our invasion screen, IMPAD1 and KDELR2, similarly modulate the Golgi-mediated secretion of proteases such as MMPs into the ECM, to promote lung cancer migration and invasion.
Figure 6. IMPAD1 and KDELR2 independently induce Golgi-mediated secretion of proteases such as MMPs to drive lung cancer cell invasion.
a-b. Conditioned media from inducible IMPAD1 overexpressing cells significantly increase invasiveness of non-invasive 393P WT cells. This phenotype is reversed, when using conditioned media from inducible IMPAD1 overexpressing cells after treatment with (a) BFA, a Golgi secretion inhibitor (1uM 6 hours) or when conditioned media from IMPAD1 overexpressing cells was supplemented with (b) Ilomastat, an MMP inhibitor (1uM). c. Invasive structures formed by cells with inducible overexpression of IMPAD1 or GFP control, in matrix comprising of 1.5mg/ml collagen in Matrigel, upon inhibition of MMPs with Ilomastat (1uM) (day 5). d-e. Conditioned media from dox-inducible KDELR2 overexpressing cells is sufficient to promote invasiveness of non-invasive 393P WT cells. This phenotype is rescued, when using conditioned media from inducible KDELR2 overexpressing cells after treatment with (d) BFA (1uM 6 hours), or when conditioned media from KDELR2 overexpressing cells was supplemented with (e) Ilomastat (1uM). f. Invasive structures formed by cells with inducible overexpression of KDELR2 or GFP control, in matrix comprising of 1.5mg/ml collagen in Matrigel, upon inhibition of MMPs with Ilomastat (1uM) (day 5). Scale bar for all images:100uM. g. Western blot analysis of conditioned media (CM) collected from human IMPAD1 and mouse KDELR2 overexpressing and knockdown cells shows IMPAD1- and KDELR2-regulated secretion of MMPs 1, 2, and 9. Whole cell lysate (WCL) collected from the same cells confirms IMPAD1 and KDELR2 overexpression and knockdown. See also Supplemental Fig. S9. Data are represented as mean ± SEM. Significance by Student’s T-test. P-value<0.05 - *; <0.002 - **
Discussion
Metastasis still accounts for 90% of all cancer-related mortality and cancer cell invasion is an essential functional requirement for the disease (29, 30). Therefore, understanding regulators of these processes is important to therapeutically prevent lung cancer metastasis (31). In parallel to the previously published in vivo screen, we performed a gain-of-function in vitro invasion screen, where we identified several known and novel drivers of invasion (2, 3). Moreover, we further explored the two hits that promoted invasion in all the secondary validation assays, IMPAD1 and KDELR2, which were also demonstrated as drivers of metastasis in vivo. This was an intriguing revelation as neither of these genes were hits in the in vivo screen. This could be explained as the in vivo metastasis screen used a competitive positive selection to identify hits based on a growth advantage, but our data indicate that neither IMPAD1 nor KDELR2 can alter cellular proliferation or tumor growth, but robustly promote invasion and metastasis.
The TCGA dataset showed that IMPAD1 (18%) and KDELR2 (19%) overlap with and are altered to almost the same extent as putative drivers of lung adenocarcinoma, KRAS (23%) and TP53 (23%). Therefore, we used the syngeneic Kras and Tp53 mutant cell lines to study IMPAD1 and KDELR2 as drivers of NSCLC invasion and metastasis. Using the human non-invasive HCC827 cell line with an activating EGFR mutation, we demonstrated that the phenotypic effects of the two genes are independent of the species, mutational status, or baseline metastatic potential. Our study is the first to elucidate the individual roles of IMPAD1 and KDELR2 in driving tumorigenesis and metastasis.
IMPAD1 is a Golgi-resident protein (4, 5) and KDELR2 localizes to the ERGIC (9-11). Although KDELR2 has transient effects on the Src pathway in our models, given the function of the ER-Golgi pathway in regulating cellular secretion (28, 32), we establsihed that both IMPAD1 and KDELR2 drive invasion through enhanced Golgi-mediated secretion of proteases such as MMPs. (Fig. 7). In addition to MMPs, the Golgi also regulates the secretion of other proteins such as collagens, elastins, etc (28, 33, 34). Thus, there may be additional unexplored components in the secretome that can modulate ECM degradation and invasion upon IMPAD1 or KDELR2 overexpression. This is especially apparent in our 3D Matrigel/collagen invasion assays where we obtained only a partial suppression of IMPAD1-mediated invasive structures upon MMP inhibition (Fig. 6C). We also acknowledge that increased expression of IMPAD1 and KDELR2 may alter other aspects of the Golgi apparatus such as Golgi morphogenesis that can promote invasion (35, 36). However, this hypothesis needs further inquiry extending beyond the scope of this paper.
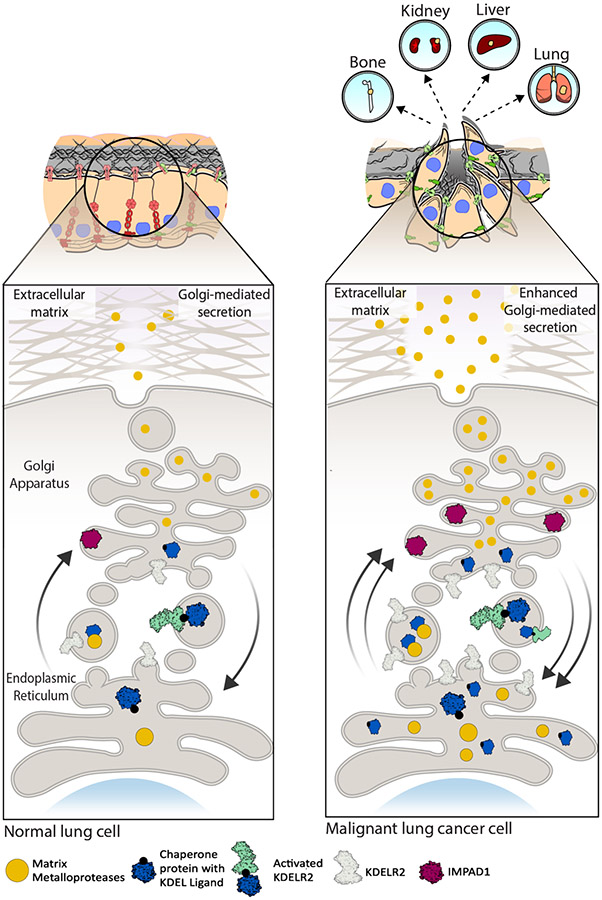
Figure 7. Working model for IMPAD1 and KDELR2 as part of the Golgi secretory cascade that regulate secretion of MMPs to drive NSCLC invasion and metastasis.
Cargo (secretory) proteins such as matrix metalloproteases (MMPs) in the ER bind to the chaperone proteins a with KDEL ligand. This leaves KDELR2 inactive during anterograde transport. Once in the Golgi, MMPs dissociate from the chaperone protein and are secreted into the ECM, whereas chaperone proteins bind to KDELR2 thus activating it during retrograde transport. IMPAD1 is localized to the Golgi and is involved in secretion of proteases like MMPs. Normal function of IMPAD1 and KDELR2 as part of the ER-Golgi secretion pathway in non-malignant lung cell lead to regular Golgi-mediated secretion. Increased IMPAD1 and KDELR2 expression in aggressive, metastatic lung cancer cells lead to hyperactive Golgi-mediated secretion of MMPs, thereby enhancing ECM degradation and promoting metastatic NSCLC.
Due to the stringent cut-offs applied, additional hits such as CBL-B, ADIPOR1, ATP1A2, and JRK were not explored. Casitas B-lineage lymphoma-b (CBL-B), an E3 ubiquitin ligase, drives degradation of TAM tyrosine kinase receptors, leading to repressed natural killer cells and increased metastasis (37). However, it has also been established as a metastasis repressor in gastric and breast cancers alluding to its unexplained opposing roles in cancer (38). Adiponectin Receptor 1 (ADIPOR1) activates the AMPK pathway and acts as a tumor and metastasis suppressor through AMPK signaling (39, 40). However, ADIPOR1 was a hit in our oncogene invasion screen indicating a hitherto unidentified dual functionality of both ADIPOR1 and AMPK as oncogenes and tumor suppressors. ATPase Na+/K+ Transporting Subunit Alpha 2 (ATP1A2) is part of the Na+/K+ -ATPases (NKP) family which are altered in and play a role in invasion of endocrine resistant breast cancer cells (41, 42). Jrk Helix-Turn-Helix (JRK) protein promotes hyper-activation of β-catenin, thereby driving cellular proliferation and tumorigenesis (43). Thus, our screen identified multiple genes which are potential metastasis drivers. Detailed mechanistic studies on these genes, which is beyond the scope of this paper, can help enhance our understanding of the complexities of metastasis and establish them as novel targets.
In this study, we identified two unique genes, IMPAD1 and KDELR2 which independently function in the ER-Golgi pathway altering Golgi-mediated secretion of MMPs to drive a pro-invasive and metastatic phenotype. Golgi secretory functions that alter the tumor ECM and promote cancer invasion and metastasis have been grossly under-studied as a potential target for the disease. Hence, our work has helped identify two new regulators of Golgi secretion that can be used to develop therapeutic strategies for lung cancer.
Materials and Methods
additional experimental details in supplemental methods. N=3 unless specified as it is the minimum requirement for performing a Student’s T-test. Data are represented as mean ± SEM. Significance by Student’s T-test. Variance is not equal so we use parametric t-test. P-value<0.05 - *; <0.002 - **
Candidate gene selection.
Genes upregulated in the metastases as compared to the primary tumors in the KRASLA1/+; p53R172HΔG/+ (KP) mice were overlapped with the genes upregulated in the tumors from the 344SQ metastatic cell line as compared to the tumors from the 393P non-metastatic cell line. These coinciding genes were then triangulated with human copy number amplifications established by TCGA. The 217 identified candidates were individually overexpressed in the 393P non-invasive cells as described before (2, 3).
Tissue culture and transfections.
For all tissue culture assays, cells were plated in 10% FBS + RPMI-1640 (complete media) in 5% CO2 at 37°C. Inducible cells were doxycycline-induced (2uM) for 24 hours before experiments unless specified. All transfections were done by using Lipofectamine 2000 (Life Technologies, Grand Island, NY). Cell lines were generated and authenticated in our lab and mycoplasma tested (14). HCC827 cells were procured from ATCC.
In vitro screen and data analysis.
15,000 cells/well were plated (n=4) in serum-free media in upper chamber of 96-well invasion plates (Corning BioCoatTM, #354167) and complete media was added to the basal chamber. Plates were incubated, washed, stained with Calcein AM, and the fluorescence read on a Victor II plate reader. Fluorescence reading output was statistically analyzed by two different methods as detailed in supplemental methods.
Validation of screen hits.
Top hits identified by method 2 that overlapped with those from method 1 were then functionally validated by 2D migration and invasion, and 3D invasion assays.
2D Migration and invasion assay.
Transwell migration (8uM inserts, Corning Star) and invasion (BD-Biosciences #354480) assays performed by using standard protocol (3, 44). 50,000 cells/well were plated in Boyden chamber with serum-free media.
3D invasion assay.
1,500 cells/well were seeded in a 1.5 mg/ml collagen/Matrigel matrix (BD-Biosciences #354230 and #354236) in 8-well chambered glass slides (Nunc, Naperville, IL) and incubated for 6-8 days as described in (4). Complete media with 2% matrigel was replenished every 2 days.
Plasmids and reagents.
Human IMPAD1 cDNA was gateway cloned into pLD6EF vector for constitutive overexpression and then sub-cloned into pTRIPZ vector for inducible expression (17). Mouse KDELR2 cDNA (ORIGENE #MR202320) was cloned into pTRIPZ vector. Mouse Impad1 and Kdelr2 shRNA constructs were from GE-Dharmacon and scramble control vectors from Thermo-Scientific. Primer and shRNA details in supplemental methods.
RT-qPCR and western blot.
Total RNA was isolated and RT-qPCR was performed with specific primers (supplemental methods) and SYBR® Green PCR Master Mix (Life technologies). L32 was used as the control. Western blot was performed with antibodies mentioned in supplemental methods.
Wound healing assay.
Induced cells were cultured to confluency in a 6-well plate. A scratch on the cell monolayer was made by a 200uL tip (45). Cells were washed, incubated with 1uM Mitomycin C for 4 hours, washed again, imaged at 0 hours, incubated with complete media for 24 hours, and imaged again at 24 hours. Percentage wound healing was calculated by comparing area migrated between 0 and 24 hours by experimental cells compared to control. N=3 images/well and 2 wells/sample.
In vivo metastasis experiments.
All experiments were reviewed and approved by the Institutional Animal Care and Use Committee at The University of Texas M.D. Anderson Cancer Center. Cells were implanted subcutaneously into the flank of syngeneic 129/sv male and female mice and tumor growth was monitored twice a week for 6-8 weeks. Once the tumors reached a volume of 1500 mm3, mice were euthanized with CO2 and metastatic nodules on the lung surface were manually quantified. Lung and tumor tissues were fixed in 10% Formalin and then proceeded for sectioning and H&E staining. Power analysis for sample size determination using 80% power and 5% two-sided type 1 error.
Immunohistochemistry
Formalin-fixed, paraffin-embedded sections were used for antigen retrieval. Sections were then incubated O/N at 4 °C with primary antibody and then with secondary antibody for 1 hour at RT. Immunohistochemistry was carried out using Streptavidin (Life technologies #SNN1004), Envision+ System (Dako), and HRP-DAB (Dako) colorimetric detection. Antibody details in supplemental methods.
Cell fractionation
Cell fractionation was performed using a kit as per the manufacturer’s protocol (Cell signaling technologies). Fractions were analyzed by western blotting.
Co-immunofluorescence for IMPAD1 and GM130.
Cells with or without 1uM Brefeldin-A treatment (4 hours at 37 °C) were grown on 8-well glass slides and fixed in 4% paraformaldehyde with 50uM CaCl2/PBS for 20 mins at 37 °C, and permeabilized in 0.1% Triton X-100/PBS for 10 mins at RT. Cells were then blocked with 5% goat serum/PBS for 1 hour at RT and then incubated with primary antibody for human IMPAD1 O/N at 4 °C. Cells were sequentially incubated with fluorophore-conjugated secondary antibodies. First, anti-mouse and then for GM130. Each of the secondary antibodies were incubated for 1 hour at RT and then counterstained with DAPI by mounting with VECTASHIELD Hard-Set Mounting Medium (Vector Laboratories, Burlingame, CA). Cells were imaged by confocal microscopy.
Co-immunofluorescence for KDELR2 and Calnexin, or GM130.
Cells were fixed in 4% paraformaldehyde in +Mg +Ca PBS (Corning #21-030-CV) for 10 mins at RT, and permeabilized with 0.1% Triton X-100/PBS for 5 mins at RT. Cells were then blocked with 2% goat serum+2% BSA/PBS for 1.5 hours at RT and incubated with primary antibody for FLAG for 1 hour at RT. Cells were sequentially incubated with fluorophore-conjugated secondary antibodies. First, anti-mouse and then for Calnexin or GM130. Each of the secondary antibodies were incubated for 1.5 hours at RT and then counterstained and imaged as mentioned above. Brefeldin-A treatment as mentioned above.
Secretome-mediated invasion assay for Golgi secretion.
Doxycycline-induced cells were plated in 60mm plates after 24-hour induction. Conditioned media (CM) was collected from confluent cells after 24 hours, where cells were treated with DMSO or 1uM BFA for the last 6 hours. 50,000 cells/well 393P WT parental cells were suspended in this conditioned media and plated in invasion Boyden chambers. RPMI with 20% FBS was added to the basal wells. Cells were incubated, stained, and quantified as mentioned in invasion assay. Western Blots were performed using CM.
Secretome-mediated invasion assay for MMP secretion.
Experiment done as mentioned above except cells were not treated with BFA. Instead, 1uM Ilomastat was added to the conditioned media in the Boyden chambers containing the 393P WT cells.
Supplementary Material
Acknowledgements
This work was supported by the DoD CDMRP Lung cancer research award W81XWH-12-16294 (D.L.G. and K.L.S.), NIH/NCI K08 CA151651, NIH R37CA214609, the MD Anderson Physician-Scientist Program and Rexanna’s Foundation for Fighting Lung Cancer (D.L.G.). D.L.G. is an R. Lee Clark Fellow of the University of Texas MD Anderson Cancer Center, supported by the Jeane F. Shelby Scholarship Fund. The work was also supported by the generous philanthropic contributions to The University of Texas MD Anderson Lung Cancer Moon Shots Program. We would like to thank the UTMDACC Department of Veterinary Medicine Facility.
Footnotes
Conflict of interest.
The authors declare no conflict of interest.
REFERENCES
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70(1):7–30. [DOI] [PubMed] [Google Scholar]
- 2.Grzeskowiak CL, Kundu ST, Mo X, Ivanov AA, Zagorodna O, Lu H, et al. In vivo screening identifies GATAD2B as a metastasis driver in KRAS-driven lung cancer. Nat Commun. 2018;9(1):2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kundu ST, Grzeskowiak CL, Fradette JJ, Gibson LA, Rodriguez LB, Creighton CJ, et al. TMEM106B drives lung cancer metastasis by inducing TFEB-dependent lysosome synthesis and secretion of cathepsins. Nat Commun. 2018;9(1):2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Frederick JP, Tafari AT, Wu SM, Megosh LC, Chiou ST, Irving RP, et al. A role for a lithium-inhibited Golgi nucleotidase in skeletal development and sulfation. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(33):11605–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sohaskey ML, Yu J, Diaz MA, Plaas AH, Harland RM. JAWS coordinates chondrogenesis and synovial joint positioning. Development. 2008;135(13):2215–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vissers LE, Lausch E, Unger S, Campos-Xavier AB, Gilissen C, Rossi A, et al. Chondrodysplasia and abnormal joint development associated with mutations in IMPAD1, encoding the Golgi-resident nucleotide phosphatase, gPAPP. Am J Hum Genet. 2011;88(5):608–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Parris TZ, Kovacs A, Hajizadeh S, Nemes S, Semaan M, Levin M, et al. Frequent MYC coamplification and DNA hypomethylation of multiple genes on 8q in 8p11-p12-amplified breast carcinomas. Oncogenesis. 2014;3:e95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Plasterer C, Tsaih SW, Lemke A, Schilling R, Dwinell M, Rau A, et al. Identification of a Rat Mammary Tumor Risk Locus That Is Syntenic with the Commonly Amplified 8q12.1 and 8q22.1 Regions in Human Breast Cancer Patients. G3-Genes Genom Genet. 2019;9(5):1739–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cancino J, Jung JE, Luini A. Regulation of Golgi signaling and trafficking by the KDEL receptor. Histochem Cell Biol. 2013;140(4):395–405. [DOI] [PubMed] [Google Scholar]
- 10.Capitani M, Sallese M. The KDEL receptor: New functions for an old protein. Febs Letters. 2009;583(23):3863–71. [DOI] [PubMed] [Google Scholar]
- 11.Wiersma VR, Michalak M, Abdullah TM, Bremer E, Eggleton P. Mechanisms of translocation of ER chaperones to the cell surface and immunomodulatory roles in cancer and autoimmunity. Front Oncol. 2015;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ruggiero C, Grossi M, Fragassi G, Di Campli A, Di Ilio C, Luini A, et al. The KDEL receptor signalling cascade targets focal adhesion kinase on focal adhesions and invadopodia. Oncotarget. 2018;9(12):10228–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ruggiero C, Fragassi G, Grossi M, Picciani B, Di Martino R, Capitani M, et al. A Golgi-based KDELR-dependent signalling pathway controls extracellular matrix degradation. Oncotarget. 2015;6(5):3375–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gibbons DL, Lin W, Creighton CJ, Rizvi ZH, Gregory PA, Goodall GJ, et al. Contextual extracellular cues promote tumor cell EMT and metastasis by regulating miR-200 family expression. Genes & development. 2009;23(18):2140–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gibbons DL, Lin W, Creighton CJ, Zheng S, Berel D, Yang Y, et al. Expression signatures of metastatic capacity in a genetic mouse model of lung adenocarcinoma. PLoS One. 2009;4(4):e5401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ahn YH, Gibbons DL, Chakravarti D, Creighton CJ, Rizvi ZH, Adams HP, et al. ZEB1 drives prometastatic actin cytoskeletal remodeling by downregulating miR-34a expression. The Journal of clinical investigation. 2012;122(9):3170–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kundu ST, Byers LA, Peng DH, Roybal JD, Diao L, Wang J, et al. The miR-200 family and the miR-183~96~182 cluster target Foxf2 to inhibit invasion and metastasis in lung cancers. Oncogene. 2016;35(2):173–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ochieng KO, Kundu ST, Rodriguez LB, Fradette JJ, Gibbons DL MBIP (MAP3K12 binding inhibitory protein) drives NSCLC metastasis by JNK-dependent activation of MMPs. Oncogene. 2020;In Revision. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gyorffy B, Surowiak P, Budczies J, Lanczky A. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS One. 2013;8(12):e82241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Uhlen M, Zhang C, Lee S, Sjostedt E, Fagerberg L, Bidkhori G, et al. A pathology atlas of the human cancer transcriptome. Science. 2017;357(6352). [DOI] [PubMed] [Google Scholar]
- 21.Uhlen M, Fagerberg L, Hallstrom BM, Lindskog C, Oksvold P, Mardinoglu A, et al. Tissue-based map of the human proteome. Science. 2015;347(6220). [DOI] [PubMed] [Google Scholar]
- 22.Halberg N, Sengelaub CA, Navrazhina K, Molina H, Uryu K, Tavazoie SF. PITPNC1 Recruits RAB1B to the Golgi Network to Drive Malignant Secretion. Cancer Cell. 2016;29(3):339–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tan XC, Banerjee P, Guo HF, Ireland S, Pankova D, Ahn YH, et al. Epithelial-to-mesenchymal transition drives a pro-metastatic Golgi compaction process through scaffolding protein PAQR11. J Clin Invest. 2017;127(1):117–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nakamura N, Rabouille C, Watson R, Nilsson T, Hui N, Slusarewicz P, et al. Characterization of a cis-Golgi matrix protein, GM130. The Journal of cell biology. 1995;131(6 Pt 2):1715–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bershadsky AD, Futerman AH. Disruption of the Golgi-Apparatus by Brefeldin-a Blocks Cell Polarization and Inhibits Directed Cell-Migration. Proceedings of the National Academy of Sciences of the United States of America. 1994;91(12):5686–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taylor A, Melton JV, Herrero LJ, Thaa B, Karo-Astover L, Gage PW, et al. Effects of an In-Frame Deletion of the 6k Gene Locus from the Genome of Ross River Virus. J Virol. 2016;90(8):4150–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sbai O, Ferhat L, Bernard A, Gueye Y, Ould-Yahoui A, Thiolloy S, et al. Vesicular trafficking and secretion of matrix metalloproteinases-2, −9 and tissue inhibitor of metalloproteinases-1 in neuronal cells. Mol Cell Neurosci. 2008;39(4):549–68. [DOI] [PubMed] [Google Scholar]
- 28.Viotti C ER to Golgi-Dependent Protein Secretion: The Conventional Pathway. Methods Mol Biol. 2016;1459:3–29. [DOI] [PubMed] [Google Scholar]
- 29.Friedl P, Locker J, Sahai E, Segall JE. Classifying collective cancer cell invasion. Nat Cell Biol. 2012;14(8):777–83. [DOI] [PubMed] [Google Scholar]
- 30.Lambert AW, Pattabiraman DR, Weinberg RA. Emerging Biological Principles of Metastasis. Cell. 2017;168(4):670–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Steeg PS, Theodorescu D. Metastasis: a therapeutic target for cancer. Nat Clin Pract Oncol. 2008;5(4):206–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brandizzi F, Barlowe C. Organization of the ER-Golgi interface for membrane traffic control. Nat Rev Mol Cell Biol. 2013;14(6):382–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Feizi A, Gatto F, Uhlen M, Nielsen J. Human protein secretory pathway genes are expressed in a tissue-specific pattern to match processing demands of the secretome. NPJ Syst Biol Appl. 2017;3:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tan XC, Banerjee P, Pham EA, Rutaganira FUN, Basu K, Bota-Rabassedas N, et al. PI4KIII beta is a therapeutic target in chromosome 1q-amplified lung adenocarcinoma. Sci Transl Med. 2020;12(527). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rodriguez-Boulan E, Macara IG. Organization and execution of the epithelial polarity programme. Nat Rev Mol Cell Bio. 2014;15(4):225–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Handler JS. Overview of epithelial polarity. Annu Rev Physiol. 1989;51:729–40. [DOI] [PubMed] [Google Scholar]
- 37.Paolino M, Choidas A, Wallner S, Pranjic B, Uribesalgo I, Loeser S, et al. The E3 ligase Cbl-b and TAM receptors regulate cancer metastasis via natural killer cells. Nature. 2014;507(7493):508–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu L, Zhang Y, Qu X, Che X, Guo T, Cai Y, et al. E3 Ubiquitin Ligase Cbl-b Prevents Tumor Metastasis by Maintaining the Epithelial Phenotype in Multiple Drug-Resistant Gastric and Breast Cancer Cells Neoplasia (New York, NY: ). 2017;19(4):374–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu X, Yan Q, Zhang Z, Du G, Wan X. Acrp30 inhibits leptin-induced metastasis by downregulating the JAK/STAT3 pathway via AMPK activation in aggressive SPEC-2 endometrial cancer cells. Oncology reports. 2012;27(5):1488–96. [DOI] [PubMed] [Google Scholar]
- 40.Tang CH, Lu ME. Adiponectin increases motility of human prostate cancer cells via adipoR, p38, AMPK, and NF-kappaB pathways. The Prostate. 2009;69(16):1781–9. [DOI] [PubMed] [Google Scholar]
- 41.Bogdanov A, Moiseenko F, Dubina M. Abnormal expression of ATP1A1 and ATP1A2 in breast cancer. F1000Research. 2017;6:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Khajah MA, Mathew PM, Luqmani YA. Na+/K+ ATPase activity promotes invasion of endocrine resistant breast cancer cells. PLoS One. 2018;13(3):e0193779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pangon L, Ng I, Giry-Laterriere M, Currey N, Morgan A, Benthani F, et al. JRK is a positive regulator of β-catenin transcriptional activity commonly overexpressed in colon, breast and ovarian cancer. Oncogene. 2016;35(22):2834–41. [DOI] [PubMed] [Google Scholar]
- 44.Justus CR, Leffler N, Ruiz-Echevarria M, Yang LV. In vitro cell migration and invasion assays. J Vis Exp. 2014(88). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Martinotti S, Ranzato E. Scratch Wound Healing Assay. Methods Mol Biol. 2019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.