Abstract
Recent studies have shown that cancer stem cells (CSCs) are involved in the occurrence and development of hepatocellular carcinoma (HCC). However, potential mechanisms for this have not yet been elucidated. We constructed a model based on the Progenitor Cell Biology Consortium database to generate stemness indices. We then utilized RNA-seq data and clinical information from the Cancer Genome Atlas (CGA) and International Cancer Genome Consortium (ICGC) for model predictions and verification. An mRNA gene expression-based stemness index (mRNAsi) and a DNA methylation-based stemness index (mDNAsi) were both calculated through one-class logistic regression. By applying univariate Cox regression analysis, we found that the mRNAsi and the mDNAsi correlated significantly with overall survival. Functional prediction analyses were used to characterize implicated genes and their degree of involvement as network hubs through protein-protein interaction analysis, and Spearman’s rank correlation coefficient test was used to assess the relationship between hub genes and indices for stemness. The mRNAsi values for CGA and ICGC carcinoma samples correlated significantly with negative clinical characteristics and overall survival, whereas gene and protein-protein interaction analyses revealed that SNAP25, KPT19, GABBR1, and EPCAM were negatively associated with clinical mDNAsi scores. Collectively, the data suggest that our new stemness model based on related genes may predict patient prognoses.
Keywords: Hepatocellular carcinoma, cancer stem cell, mRNAsi, mDNAsi, prognosis
Introduction
Hepatocellular carcinoma (HCC) is the sixth most common cancer, and the third most frequent cause of cancer death [1,2]. Although surgical resection is the most promising treatment during early stages, the 5-year survival rate for HCC is less than 20%, mainly due to limited treatment efficacy for advanced HCC [3-6]. Due to the absence of clinical symptoms during early stages, most patients with HCC are diagnosed at advanced stages, with poor prognoses [4,7]. As the pathogenesis of HCC is unclear, it is essential to explore the mechanisms of HCC development and progression so that early diagnosis and targeted treatments can be achieved.
The theory of cancer stem cells (CSCs) indicated to researchers that malignant solid tumors contained a highly heterogeneous population of CSCs [8,9]. CSCs represent the small number of cancer cells that can self-replicate as stem cells and support both tumor growth and tumor characteristics, just as normal stem cells can renew and maintain organs and tissues [10,11]. Growing evidence suggests that CSCs play crucial roles in tumorigenesis, metastasis, recurrence, and resistance to both radiation and chemotherapy [12-15]. However, how CSCs maintain malignant tumors, including potential molecular mechanisms and signaling pathways involved, is not clear.
Recent studies have shown the potential for CSCs to be targets of cancer therapy [16,17]. Yang et al. [18] found that HGF/c-Met promoted the enrichment of renal CSCs, which could be a mechanism for metastasis and recurrence to leverage as a new target for treating renal cell carcinoma. Additional evidence has suggested that clarifying the source of CSCs may offer novel methods for individualized cancer treatment [19], but for HCC, little is known about the source, expression patterns, and molecular mechanisms involved.
In this study, we constructed a model based on stem-cell sequencing data from the Progenitor Cell Biology Consortium (PCBC) to predict stem cell-associated molecular-feature (stemness) indices for HCC samples. We then utilized a Cancer Genome Atlas (CGA) cohort to make predictions and validated our findings through an independent dataset, analyzing relationships between stemness indices, clinical characteristics, and overall patient survival. Furthermore, we evaluated genes based on stemness and performed protein-protein interaction (PPI) analyses to identify potential hub genes in signaling networks, which could provide CSC-specific targets for the treatment of HCC.
Methods
Data processing
The following databases were used: the Cancer Genome Atlas (CGA, https://cancergenome.nih.gov/) database, International Cancer Genome Consortium (ICGC, https://icgc.org/) database, Progenitor Cell Biology Consortium (PCBC, https://progenitorcells.org/frontpage) database, and the STRING (https://string-db.org/) database. Expression data (syn2701943) for pluripotent stem-cell samples, including embryonic stem cells (ESCs) and induced pluripotent stem cells (iPSCs), were downloaded from the PCBC using the R package synapse (v0.6.61). RNA-Seq data and clinical follow-up information were downloaded concurrently for 424 samples in the CGA, and 203 samples in the ICGC database, the latter used as a validation set. Clinical follow-up information included gender, age, pathology grading, Tumor Node Metastasis (TNM) staging, hepatitis B virus (HBV) and hepatitis C virus (HCV) infection status, immune-cell score, and iCluster typing. Data were processed as previously described [20-22].
Calculation of a stemness index based on the one-class logistic regression (OCLR) method
To predict and calculate stemness indices, ESC and iPSC expression data from the PCBC database were evaluated using the OCLR method, and this method was performed as previously described [23]. There were 78 total ESC and iPSC samples with 12,998 mRNA expression profiles of genes per sample. For DNA methylation-based stemness index (mDNAsi) calculations, 99 samples with 219 stem-cell probe signatures in the PCBC database were divided into two groups, of which 44 samples were stem-cell samples, and 55 samples were non-stem-cell samples. Expression profiles and methylation data were centralized by the average value for each sample. Both the mRNA gene expression-based stemness index (mRNAsi), and the mDNAsi weight vector for each gene were then calculated by the OCLR method using the R package gel net (v1.2.1).
For the expression profiles and methylation data from the CGA and ICGC HCC samples, we calculated Spearman’s rank correlation coefficient for each sample gene with the model’s gene weight vector. We then performed a linear conversion to the Spearman’s rank correlation coefficient obtained from the sample. Conversion methods were used to identify a Spearman’s rank correlation coefficient for each sample, minus a minimum, which was then divided by the maximum value. The values obtained became the mRNAsi and mDNAsi for each sample, with a distribution range of stemness indices between zero and one.
Analysis and screening of differentially expressed genes
We used the R package DESeq2 (v1.24.0) to perform a differential analysis of the RNA-seq data from mRNAsi-high/mDNAsi-high and mRNAsi-low/mDNAsi-low samples and then determined differentially expressed genes (DEGs) between the these two samples [23,24]. These DEGs were then filtered according to a threshold false discovery rate (FDR) < 0.05 and |log2FC| > 1 [22].
Enrichment analysis of gene function and pathways
A Kyoto Encyclopedia of Genes and Genomes (KEGG) (https://www.kegg.jp/ or https://www.genome.jp/kegg/) pathway-enrichment analysis was used to determine networks for metabolic, signaling, and other molecular interactions [25]. Gene Ontology (GO) functional enrichment analysis is an international standard classification system for gene function, including Biological Process (BP), Cellular Components (CC), and Molecular Function (MF) [26]. Through these categories, gene functions were defined and described accordingly [27]. To examine possible functions related to the stemness indices of DEGs, we performed KEGG-pathway analyses and GO functional enrichment analyses with mRNAsi and mDNAsi through the R package cluster Profiler (v3.14.0). The significance level was defined as FDR < 0.05.
PPI network analysis
The STRING database was used to analyze the PPI network of DEGs, and the degree algorithm in the cytoHubba plugin of Cityscape software was applied to identify hub genes. PPI network analysis is commonly used to elucidate molecular bases of diseases, and provides detailed knowledge about protein interactions which can be used to determine underlying factors for prevention, diagnosis, and treatment of disease [28]. This PPI analysis also provides information about the genes and proteins shared in a disease and describes their interactions.
Statistics
A t-test was used for continuous variables. Statistical analyses were performed using IBM SPSS 24.0 software. All statistical tests were two-sided, and the significance level was defined as P < 0.05. Spearman’s rank correlation coefficients were used to analyze correlations between two variables. Kaplan-Meier survival curves were generated, after stratification of the data, and compared using a log-rank test.
Results
Relationships between indices for mRNA expression- and DNA methylation-based stemness and clinical characteristics
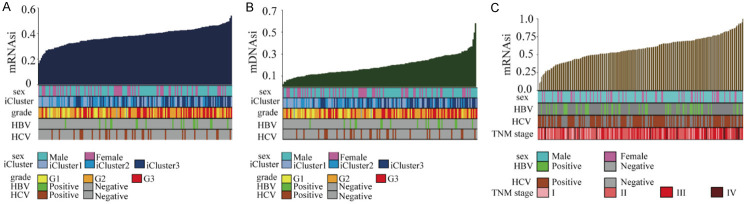
We first investigated relationships between stem-cell index distributions and clinical characteristics by ranking HCC samples according to their mRNAsi and mDNAsi values, and then tested whether any clinical features correlated with low- or high-stemness values. Clinical features for tumor samples included gender, age, pathology staging, HBV or HCV infection status, immune-cell score, and iCluster typing. mRNAsi and mDNAsi exponential distributions for the CGA HCC samples are shown in Figure 1A, 1B, respectively. The mRNAsi exponential distributions for the ICGC HCC samples are shown in Figure 1C.
Figure 1.
Exponential mRNAsi and mDNAsi distributions based on clinical characteristics of HCC samples. A. The mRNAsi distribution based on the clinical characteristics of the CGA HCC samples. B. The mDNAsi distribution based on the clinical characteristics of the CGA HCC samples. C. The mRNAsi distribution based on the clinical characteristics of the ICGC HCC samples. HCC: hepatocellular carcinoma; mRNAsi: mRNA gene expression-based stemness index; mDNAsi: DNA methylation-based stemness index; CGA: Cancer Genome Atlas; ICGC: International Cancer Genome Consortium.
We then performed linear-regression testing for the above characteristics. As shown in Table 1, the mRNAsi values for the CGA HCC samples correlated significantly with pathology grading, HBV infection, immune-cell score, and iCluster typing. mDNAsi values were significantly associated with immune-cell score and iCluster typing. Analysis of the ICGC HCC validation cohort dataset indicated that mRNAsi values were significantly associated with TNM stage (Table 2).
Table 1.
Correlations between CGA HCC sample mRNAsi and mDNAsi values and clinical features and molecular typing
| Percent/Average | p (mRNAsi) | p (mDNAsi) | |
|---|---|---|---|
| Sex | Male: 65.6% | 0.80 | 0.12 |
| Female: 34.4% | |||
| Age | 60.13 | 0.47 | 0.23 |
| G1 | |||
| Grade | G2 | 3.71E-05 | 0.67 |
| G3 | |||
| HBV | 23.00% | 3.04E-04 | 0.47 |
| HCV | 18.6% | 0.76 | 0.15 |
| Immune score | 166.12 | 1.2E-03 | 0.018 |
| iCluster1 | |||
| iCluster | iCluster2 | 1.29E-07 | 5.64E-31 |
| iCluster3 |
Table 2.
Correlations between ICGC HCC sample mRNAsi values and clinical features and molecular typing
| Percent/Average | p (mRNAsi) | |
|---|---|---|
| Sex | Male: 75.4% | 0.12 |
| Female: 24.6% | ||
| Age | 67 | 0.2 |
| I: 16.3% | ||
| TNM stage | II: 47.3% | 9.09E-03 |
| III: 29.1% | ||
| IV: 7.4% | ||
| HBV | 23.00% | 0.81 |
| HCV | 18.6% | 0.64 |
| Immune score | 166.12 | 0.97 |
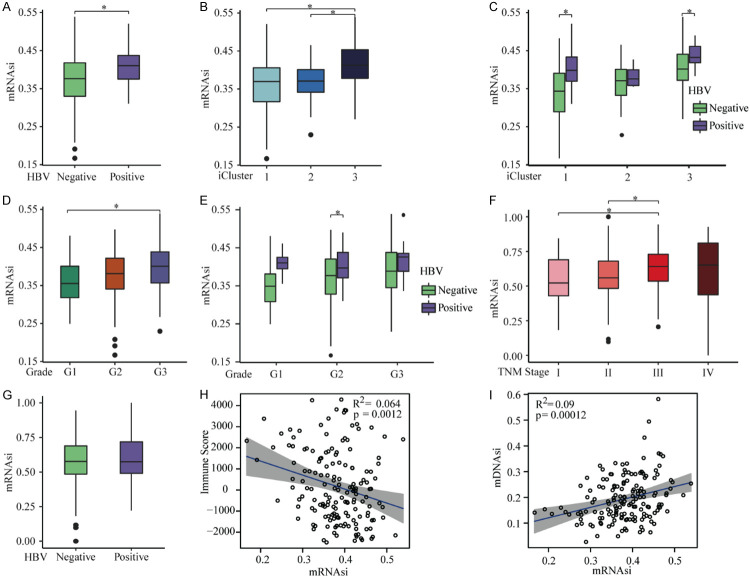
For the CGA HCC samples, mRNAsi scores were significantly higher in HBV-infected samples compared to uninfected samples (Figure 2A), and in iCluster3 samples compared to iCluster1 and iCluster2 samples (Figure 2B, 2C). In addition, samples from HCC patients with late pathology grading showed higher mRNAsi scoring (Figure 2D, 2E). iCluster typing also significantly correlated with mDNAsi values. mDNAsi scores were lowest in iCluster1 samples and highest in iCluster3 samples, while HBV infection and pathology grading had no obvious correlations with mDNAsi scores (Figure S1A-E). In the ICGC HCC samples, TNM stage was significantly associated with mRNAsi values in an increasing manner for the first three stages, but no relationship between HBV infection and mRNAsi values was found (Figure 2F, 2G). Immune scores were negatively correlated with both mRNAsi and mDNAsi scores in the CGA HCC samples (Figures 2H, S1F). Furthermore, there was a positive correlation between the mRNAsi and mDNAsi scores in the CGA HCC samples (R = 0.30, Figure 2I). Taken together, higher stemness-index values in HCC samples correlated with worse clinical characteristics, demonstrating that stemness indices may characterize tumor progression.
Figure 2.
Relationships between the mRNAsi values and clinical characteristics. A. mRNAsi scores were higher in samples infected with HBV in the CGA HCC samples. B. mRNAsi scores were higher in iCluster3 compared to scores in iCluster1 and iCluster2 for all CGA HCC samples. C. mRNAsi scores were highest in iCluster3, and lowest in iCluster1, for CGA HCC samples without HBV infection. D. mRNAsi scores were highest for grade 3, and lowest for grade 1, for all CGA HCC samples. E. mRNAsi scores were highest for grade 3, and lowest for grade 1, for CGA HCC samples without HBV infection. F. For ICGC HCC samples, mRNAsi values increased with TNM staging only for the first three staging levels. G. There was no significant correlation between mRNAsi values and HBV infection for ICGC HCC samples. H. Immune scores were inversely related to mRNAsi scores in the CGA HCC samples. I. mRNAsi and mDNAsi values were positively correlated for CGA HCC samples. *P < 0.05. mRNAsi: mRNA gene expression-based stemness index; mDNAsi: DNA methylation-based stemness index; HBV: hepatitis B virus; CGA: Cancer Genome Atlas. ICGC: International Cancer Genome Consortium.
Correlations between HCC stemness indices and overall survival
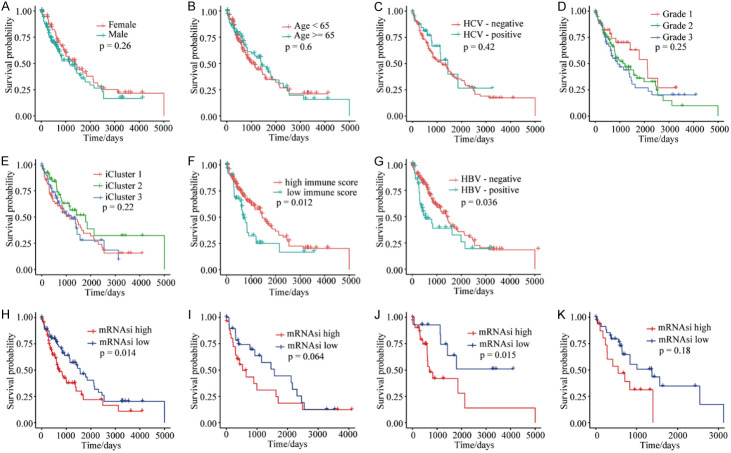
To better understand the relationship between stemness-index values and prognosis, we first compared overall survival with clinical features such as gender, age, pathology grading, iCluster typing, immune-cell scores, and HBV or HCV infection status, among the HCC samples. The results demonstrated that gender, age, HCV infection, pathology grade, and iCluster typing were not significantly correlated with overall survival (Figure 3A-E). Significant survival differences were identified in these samples based on HBV infection and immune-cell scoring. The high immune-score group demonstrated higher survival compared to the low immune-score group (Figure 3F), and patient survival for HBV-infected samples was lower than for non-infected samples (Figure 3G). Importantly, overall survival for the mRNAsi-low group was higher compared to the mRNAsi-high group (Figure 3H), and survival for the mDNAsi-high group was higher compared to the mDNAsi-low group (Figure S2A). Samples grouped by mRNAsi and mDNAsi values were then assessed according to iCluster typing (Table 3). Only iCluster2 samples grouped according to mRNAsi values showed a significant survival difference (Figures 3I-K and S2B-D). However, this evaluation was complicated by the small sample sizes and short survival times of the three groups.
Figure 3.
Kaplan-Meier survival analyses of the CGA HCC samples. A. Survival probabilities for sex-grouped samples showed no significant differences. B. Survival probabilities for age-grouped samples showed no significant differences. C. Survival probabilities for samples grouped by HCV infection showed no significant differences. D. Survival probabilities for samples grouped by pathology grading showed no significant differences. E. Survival probabilities for iCluster-grouped samples showed no significant differences. F. The high immune-score group showed higher survival probabilities than the low immune-score group. G. The survival probabilities for HBV-infected samples was lower than for HBV-negative samples. H. The survival probabilities for the mRNAsi-low group were higher than for the mRNAsi-high group for all CGA HCC samples. I. The survival probabilities for the mRNAsi-low group and the mRNAsi-high group in iCluster1 were not significantly different. J. The survival probabilities for the mRNAsi-low group were higher than those for the mRNAsi-high group in iCluster2. K. The survival probabilities for the mRNAsi-low group and the mRNAsi-high group in iCluster3 were not significantly different. *P < 0.05. mRNAsi: mRNA gene expression-based stemness index; CGA: Cancer Genome Atlas; HCV: hepatitis C virus.
Table 3.
Hazard ratio (HR) analysis and optimal thresholds for the CGA stem-cell index grouping
| iCluster | mRNAsi | |||||
|
| ||||||
| HR | Cut p | |||||
|
|
|
|||||
| HR | Lower | Upper | p | Cut | p (Cut) | |
|
| ||||||
| C1 | 1.80 | 0.96 | 3.40 | 6.77E-02 | 0.37 | 0.23 |
| C2 | 3.61 | 1.20 | 10.84 | 2.21E-02 | 0.35 | 0.08 |
| C3 | 1.60 | 0.79 | 3.23 | 0.19 | 0.43 | 0.77 |
| All | 1.68 | 1.11 | 2.53 | 0.0145 | 0.38 | 0.08 |
|
| ||||||
| iCluster | mDNAsi | |||||
|
| ||||||
| HR | Cut p | |||||
|
|
|
|||||
| HR | Lower | Upper | p | Cut | p (Cut) | |
|
| ||||||
| C1 | 0.69 | 0.37 | 1.31 | 0.26 | 0.12 | 0.87 |
| C2 | 0.52 | 0.20 | 1.34 | 1.75E-01 | 0.20 | 0.67 |
| C3 | 1.85 | 0.91 | 3.76 | 0.09 | 0.30 | 0.48 |
| All | 0.62 | 0.39 | 0.98 | 4.04E-02 | 0.12 | 0.51 |
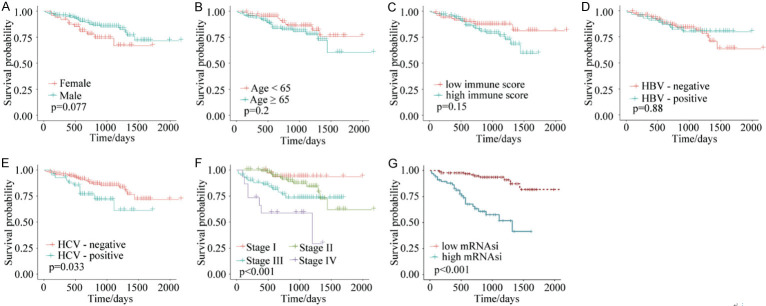
We next used the above analysis on the ICGC HCC dataset, and validated that there were no correlations between survival and gender, age, immune-cell scoring, and HBV infection (Figure 4A-D). Overall survival was lower in the presence of HCV infection (Figure 4E) and was lower with increasing TNM stage (Figure 4F). For the stemness indices, overall survival of the mRNAsi-low group was longer than for the mRNAsi-high group (Figure 4G). Collectively, these data suggest that higher mRNAsi values correlate with poorer survival, which may offer new criteria for determining prognoses in patients with HCC.
Figure 4.
Kaplan-Meier survival analyses of ICGC HCC samples. A. Survival probabilities for samples grouped by gender showed no significant differences. B. Survival probabilities for age-grouped samples showed no significant differences. C. Survival probabilities for samples grouped by immune-cell scores showed no significant differences. D. Survival probabilities for samples grouped by HBV infection showed no significant differences. E. The survival probabilities for HCV-infected samples were lower than for HCV-negative samples. F. Survival probabilities were lower with increasing TNM staging. G. The survival probabilities for the mRNAsi-low group were higher than those for the mRNAsi-high group. *P < 0.05. mRNAsi: mRNA gene expression-based stemness index. ICGC: International Cancer Genome Consortium. TNM: Tumor Node Metastasis.
Screening for stem cell-related genes in HCC samples
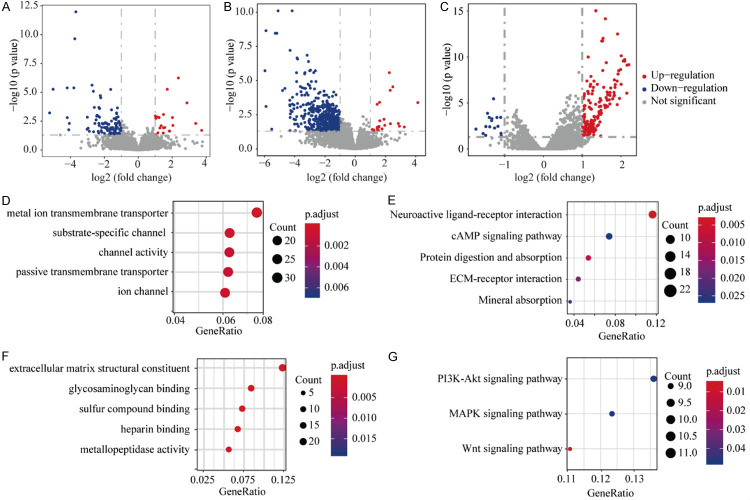
Based on the above results, we further analyzed differentially expressed genes (DEGs) in the CGA HCC cohort. After threshold-filtering, 136 DEGs were identified, with 110 genes downregulated and 26 genes upregulated in the mRNAsi-high group compared to the mRNAsi-low group (Figure 5A). In the mDNAsi set, 569 DEGs were identified, with 546 genes downregulated and 23 genes upregulated in the mDNAsi-high group compared to mDNAsi-low group (Figure 5B). Common DEGs between the mRNAsi and mDNAsi sets were APOBEC3C, C1orf116, GABBR2, and TFCP2L1. In the filtered ICGC cohort, 190 DEGs were observed, with 16 genes downregulated and 174 genes upregulated in the mRNAsi-high group compared with the mRNAsi-low group (Figure 5C). The only DEG in common between the CGA and ICGC cohorts was upregulated ALDH1A3. These differences in gene expression reflect index-value differences (high versus low) among HCC samples.
Figure 5.
Volcano plots, and GO and KEGG annotation maps, for differentially expressed genes (DEGs). A. 110 genes were downregulated, and 26 genes were upregulated, in the mRNAsi-high group compared to the mRNAsi-low group for CGA HCC samples. B. 546 genes were downregulated, and 23 genes were upregulated, in the mDNAsi-high group compared to the mDNAsi-low group for CGA HCC samples. C. 16 genes were downregulated, and 174 genes were upregulated in the mDNAsi-high group compared to the mRNAsi-low group for ICGC HCC samples. D. The GO annotation map for the top 5 (out of 569) mDNAsi DEGs in the CGA HCC samples. E. The KEGG annotation map for the top 5 mDNAsi DEGs in the CGA HCC samples. F. The GO annotation map for the top 5 (out of 569) mRNAsi DEGs in the ICGC HCC samples. G. The KEGG annotation map for the top 3 mRNAsi DEGs in the ICGC HCC samples. *FDR < 0.05. GO: Gene Ontology; KEGG: Kyoto Encyclopedia of Genes and Genomes; DEGs: differentially expressed genes.
Functional analyses of differentially expressed genes
To explore the possible roles of these DEGs in the CGA HCC samples for tumorigenesis, metastasis, recurrence, and drug resistance, we performed Gene Ontology (GO) functional analyses for the 136 mRNAsi DEGs that showed no significant enrichments (FDR < 0.05). GO functional annotations for the 569 DEGs identified using mDNAsi values indicated that the “metal ion transmembrane transporter activity” pathway, and “monovalent inorganic action transmembrane transporter activity” were significantly enriched (Figure 5D). KEGG-pathway enrichment analysis of mRNAsi DEGs found no significantly enriched pathways (FDR < 0.05), while KEGG-pathway enrichment analysis of mDNAsi DEGs demonstrated that the “cAMP signaling pathway” was significantly enriched (Figure 5E). GO functional annotations of the 190 mRNAsi-grouped DEGs in the ICGC HCC samples showed significant enrichment for “extracellular matrix structural constituents” and “glycosaminoglycan binding” (Figure 5F). KEGG-pathway enrichment analysis of mRNAsi DEGs in the ICGC HCC samples found three significantly enriched pathways, including the PI3K-AKT signaling pathway (Figure 5G). Taken together, we found that the DEGs from high- and low-stemness indices were enriched for pathways related to tumor formation, metastasis and recurrence. These enrichments should aid in our understanding of tumorigenesis mechanisms and may guide the development of targeted therapies for HCC.
PPI and hub-gene analyses of differentially expressed genes
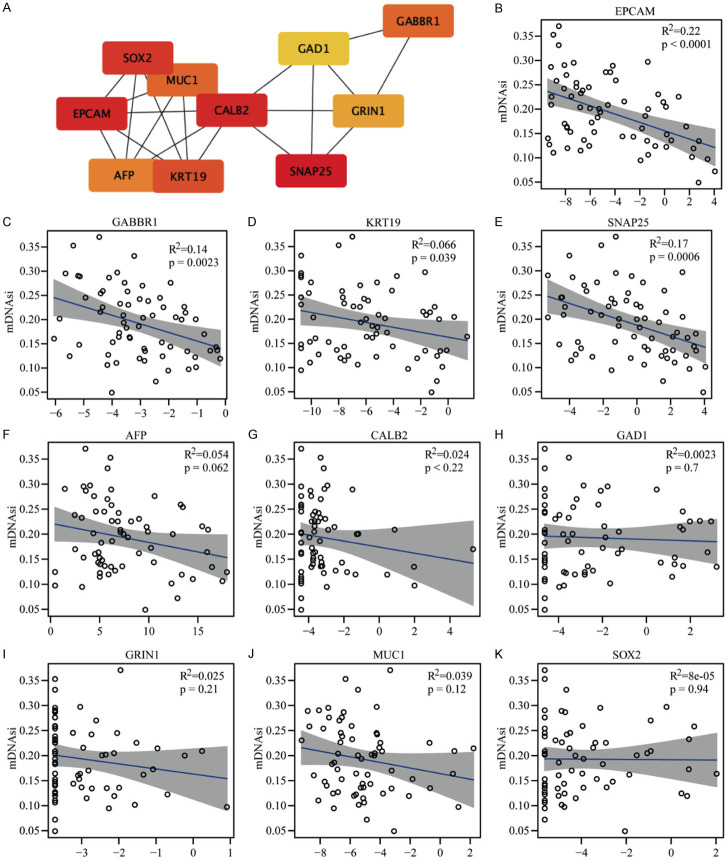
To identify hub genes related to these stemness-index values, PPI analyses were performed based on DEGs. Ten genes (SNAP25, EPCAM, CALB2, SOX2, KRT19, GABBR1, MUC1, AFP, GRIN1, and GAD1) were identified as hub genes among mDNAsi DEGs (Figure 6A). Correlation analyses between these hub genes and mDNAsi values were performed, and significant negative correlations were found be-tween SNAP25, KPT19, GABBR1, and EPCAM and these index values (Figure 6B-E) while six other hub genes had no prominent connections (Figure 6F-K). These findings suggested that tumor stemness-index values could be utilized as new prognosticators for patients with HCC. In addition, SNAP25, KPT19, GABBR1, and EPCAM could be key genes involved in HCC tumorigenesis, metastasis and recurrence.
Figure 6.
PPI analyses and correlations between hub genes and mDNAsi scores. A. Protein interactions between the 10 hub genes; darker colors indicate higher scores that represent more network involvement and significance. B. EPCAM had a significant negative correlation with mDNAsi scores. C. GABBR1 has a significant negative correlation with mDNAsi scores. D. KRT19 had a significant negative correlation with mDNAsi scores. E. SNAP25 has a significant negative correlation with mDNAsi scores. F. AFP was not significantly correlated with mDNAsi scores. G. CALB2 was not significantly correlated with mDNAsi scores. H. GAD1 was not significantly correlated with mDNAsi scores. I. GRIN1 was not significantly correlated with mDNAsi scores. J. MUC1 was not significantly correlated with mDNAsi scores. K. SOX2 was not significantly correlated with mDNAsi scores. *P < 0.05. mDNAsi: DNA methylation-based stemness index; PPI: protein-protein interaction.
Discussion
In this study, we found that stemness indices were related to pathology grading, HBV infection, immune-cell score, iCluster typing, and TNM staging. Consistent with our HCC findings, several other studies have demonstrated that stemness-index values were associated with pathological characteristics in many cancers. For example, Pan et al. [24] observed that mRNAsi values in bladder cancer increased as tumor stage increased, with T3 staging having the most stem-cell characteristics. Lower mRNAsi scores also had better overall survival and treatment outcomes. Another recent study reported that the expression of CSC marker OCT4 was correlated with poor differentiation, tumor size, and N stage in patients with rectal cancer, and concluded that OCT4 could be an independent prognostic biomarker [29]. These results suggest that stemness-index scores could play an essential role in defining tumor progression.
An increasing number of studies have also linked stemness indices with patient prognosis. In our study, we constructed a novel model to predict overall survival in HCC patients, and observed a relationship between HCC stemness-index values, clinical characteristics, and survival. Our results showed that patients with worse clinical phenotypes usually possessed higher stemness-index scores. Furthermore, patients with higher mRNAsi scores tended to have lower overall survival. However, we observed the opposite result in mDNAsi-grouped samples which may have been due to small sample size. Consistent with our findings, previous studies have shown that increased expression of CSC marker CD133 confers a poor prognosis for invasive breast cancer [30], and Qin et al. [31] reported that an mRNAsi score was an independent prognostic factor in lung squamous-cell carcinoma. Additionally, Zhao et al. [32] reported that TBX21 was both a patient prognosis predictor and a CSC maintenance driver via the TBX21-IL-4 pathway in lung adenocarcinoma, suggesting that TBX21 may serve as a novel predictive biomarker and therapeutic target. Lastly, Lian et al. determined that a prognostic signature based on mRNAsi values may predict sonic-hedgehog medulloblastoma prognosis and such a signature could be a potential biomarker for informing treatment-options in clinical practice [23]. These findings demonstrate that stemness-index scores may be reliable predictors for HCC prognoses.
Our investigation also examined DEGs between stemness indices and their possible functions. The functional pathways for “metal ion transmembrane transporter activity”, “monovalent inorganic action transmembrane transporter activity”, “cAMP signaling pathway”, “extracellular matrix structural constituent”, “glycosaminoglycan binding”, and “PI3K/AKT signaling pathway” were significantly enriched. Previously, Roberto et al. found that synergistic inhibition of HCC and liver CSC proliferation could occur by targeting RAS/RAF/MAPK and WNT/β-Catenin pathways [33], and microRNA-28-5p was reported to regulate liver CSC expansion via the IGF-1 pathway [34]. Moreover, Si et al. identified the miR219/E-cadherin axis as a potential therapeutic target against liver CSCs and as a predictor for sorafenib treatment in HCC patients [35]. Based on these results, stem cell-related DEGs correlated with a variety of tumorigenesis-related pathways, offering mechanistic insight into how to target CSC maintenance in patients with HCC.
Finally, we performed PPI analyses to identify HCC hub genes based on the identified DEGs. We found that SNAP25, KPT19, GABBR1, and EPCAM were significantly correlated with mDNAsi towards negative outcomes. Similar to our results, prior work showed that SENP1 activity sustained CSCs in hypoxic HCC [36]. Li et al. observed that activated STAT3 played a pivotal role in maintaining stemness in HCC CSCs [37], and Ritu et al. found that miR-26b-5p imparted metastatic properties and helped maintain Ep+ CSCs via HSPA8. Thus, miR-26b-5p and HSPA8 may serve as molecular targets for selectively eliminating the Ep+ CSC population in human HCC [38]. Collectively, these findings further support the idea that identifying hub genes related to CSCs in HCC could inform about its pathogenesis and indicate novel targets for HCC therapy.
The stem-cell index results of this study may advance the development of objective diagnostic tools for quantitating HCC stemness, prognosticating judgments, and treatment-response predictions. This provides new directions for clinical treatment, especially for developing personalized treatment protocols. Further research is needed to explore stemness index-related genes and their roles in tumor proliferation and metastasis to advance this promising area.
Acknowledgements
We thank the patients and investigators who participated in TCGA, ICGC and PCBC for providing data. And this study was supported by the National Natural Science Foundation of China (U1904164 and 81702927), and National S&T Major Project of China (2018ZX10301201-008).
Consent for participation from all patients was obtained through the Cancer Genome Atlas Project, the International Cancer Genome Consortium and the Progenitor Cell Biology Consortium.
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Wang D, Hu X, Xiao L, Long G, Yao L, Wang Z, Zhou L. Prognostic nutritional index and systemic immune-inflammation index predict the prognosis of patients with HCC. J Gastrointest Surg. 2020 doi: 10.1007/s11605-019-04492-7. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang X, Shi J, Chen X, Jiang Y, Zhao H. Efficacy of cabozantinib and nivolumab in treating hepatocellular carcinoma with RET amplification, high tumor mutational burden, and PD-L1 expression. Oncologist. 2020;25:470–474. doi: 10.1634/theoncologist.2019-0563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li X, Li C, Zhang L, Wu M, Cao K, Jiang F, Chen D, Li N, Li W. The significance of exosomes in the development and treatment of hepatocellular carcinoma. Mol Cancer. 2020;19:1. doi: 10.1186/s12943-019-1085-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jiao J, González Á, Stevenson HL, Gagea M, Sugimoto H, Kalluri R, Beretta L. Depletion of S100A4+ stromal cells does not prevent HCC development but reduces the stem cell-like phenotype of the tumors. Exp Mol Med. 2018;50:e422. doi: 10.1038/emm.2017.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ding J, Xu K, Sun S, Qian C, Yin S, Xie H, Zhou L, Zheng S, Zhang W. SOCS1 blocks G1-S transition in hepatocellular carcinoma by reducing the stability of the CyclinD1/CDK4 complex in the nucleus. Aging (Albany NY) 2020;12:3962–3975. doi: 10.18632/aging.102865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ding J, Liu X, Tang B, Bai X, Wang Y, Li S, Li J, Liu M, Wang X. Inflammation-induced LINC00665 increases the malignancy through activating PKR/NF-κB pathway in hepatocellular carcinoma. Parasite. 2020;27:47. [Google Scholar]
- 7.Li C, Wang X, Song Q. MicroRNA 885-5p inhibits hepatocellular carcinoma metastasis by repressing AEG1. Onco Targets Ther. 2020;13:981–988. doi: 10.2147/OTT.S228576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marcu LG. Cancer stem cells as therapeutic targets of pancreatic cancer. Fundam Clin Pharmacol. 2020;34:200–201. doi: 10.1111/fcp.12536. [DOI] [PubMed] [Google Scholar]
- 9.Liu D, Hong Y, Li Y, Hu C, Yip TC, Yu WK, Zhu Y, Fong CC, Wang W, Au SK, Wang S, Yang M. Targeted destruction of cancer stem cells using multifunctional magnetic nanoparticles that enable combined hyperthermia and chemotherapy. Theranostics. 2020;10:1181–1196. doi: 10.7150/thno.38989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang L, Lian J, Chen X, Qin G, Zheng Y, Zhang Y. WASH overexpression enhances cancer stem cell properties and correlates with poor prognosis of esophageal carcinoma. Cancer Sci. 2017;108:2358–2365. doi: 10.1111/cas.13400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hajighasemlou S, Pakzad S, Ai J, Muhammadnejad S, Mirmoghtadaei M, Hosseinzadeh F, Gharibzadeh S, Kamali A, Ahmadi A, Verdi J. Characterization and validation of hepatocellular carcinoma (HCC) xenograft tumor as a suitable liver cancer model for preclinical mesenchymal stem cell studies. Asian Pac J Cancer Prev. 2018;19:1627–1631. doi: 10.22034/APJCP.2018.19.6.1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abad E, Graifer D, Lyakhovich A. DNA damage response and resistance of cancer stem cells. Cancer Lett. 2020;474:106–117. doi: 10.1016/j.canlet.2020.01.008. [DOI] [PubMed] [Google Scholar]
- 13.Wang S, Cai L, Zhang F, Shang X, Xiao R, Zhou H. Inhibition of EZH2 attenuates sorafenib resistance by targeting NOTCH1 Activation-dependent liver cancer stem cells via NOTCH1-related microRNAs in hepatocellular carcinoma. Transl Oncol. 2020;13:100741. doi: 10.1016/j.tranon.2020.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Versini A, Colombeau L, Hienzsch A, Gaillet C, Retailleau P, Debieu S, Müller S, Cañeque T, Rodriguez R. Salinomycin derivatives kill breast cancer stem cells via lysosomal iron targeting. Chemistry. 2020;26:7416–7424. doi: 10.1002/chem.202000335. [DOI] [PubMed] [Google Scholar]
- 15.Liang W, Lin Z, Du C, Qiu D, Zhang Q. mRNA modification orchestrates cancer stem cell fate decisions. Mol Cancer. 2020;19:38. doi: 10.1186/s12943-020-01166-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guo Y, Li B, Yan X, Shen X, Ma J, Liu S, Zhang D. Bisphenol A and polychlorinated biphenyls enhance the cancer stem cell properties of human ovarian cancer cells by activating the WNT signaling pathway. Chemosphere. 2019;246:125775. doi: 10.1016/j.chemosphere.2019.125775. [DOI] [PubMed] [Google Scholar]
- 17.Kalogirou C, Ellinger J, Kristiansen G, Hatzichristodoulou G, Kübler H, Kneitz B, Busch J, Fendler A. Inhibiting WNT and NOTCH in renal cancer stem cells and the implications for human patients. Transl Androl Urol. 2020;11:929. doi: 10.21037/tau.2020.03.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang J, Yang L, Li S, Hu N. HGF/c-Met promote renal carcinoma cancer stem cells enrichment through upregulation of Cir-CCDC66. Technol Cancer Res Treat. 2020;19:1533033819901114. doi: 10.1177/1533033819901114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yen CH, Lai CC, Shia TH, Chen M, Yu HC, Liu YP, Chang FR. Gynura divaricata attenuates tumor growth and tumor relapse after cisplatin therapy in HCC xenograft model through suppression of cancer stem cell growth and Wnt/β-catenin signalling. J Ethnopharmacol. 2018;213:366–375. doi: 10.1016/j.jep.2017.07.019. [DOI] [PubMed] [Google Scholar]
- 20.He Y, Dang Q, Li J, Zhang Q, Yu X, Xue M, Guo W. Prediction of hepatocellular carcinoma prognosis based on expression of an immune-related gene set. Aging (Albany NY) 2020;12:965–977. doi: 10.18632/aging.102669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He Y, Xue C, Yu Y, Chen J, Chen X, Ren F, Ren Z, Cui G, Sun R. CD44 is overexpressed and correlated with tumor progression in gallbladder cancer. Cancer Manag Res. 2018;10:3857–3865. doi: 10.2147/CMAR.S175681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pei J, Wang Y, Li Y. Identification of key genes controlling breast cancer stem cell characteristics via stemness indices analysis. J Transl Med. 2020;18:74. doi: 10.1186/s12967-020-02260-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lian H, Han YP, Zhang YC, Zhao Y, Yan S, Li QF, Wang BC, Wang JJ, Meng W, Yang J, Wang QH, Mao WW, Ma J. Integrative analysis of gene expression and DNA methylation through one-class logistic regression machine learning identifies stemness features in medulloblastoma. Mol Oncol. 2019;13:2227–2245. doi: 10.1002/1878-0261.12557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pan S, Zhan Y, Chen X, Wu B, Liu B. Identification of biomarkers for controlling cancer stem cell characteristics in bladder cancer by network analysis of transcriptome data stemness Indices. Front Oncol. 2019;9:613. doi: 10.3389/fonc.2019.00613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kanehisa M, Sato Y. KEGG Mapper for inferring cellular functions from protein sequences. Protein Sci. 2020;29:28–35. doi: 10.1002/pro.3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou X, Zhen X, Liu Y, Cui Z, Yue Z, Xu A, Han J. Identification of key modules, hub genes, and noncoding RNAs in chronic rhinosinusitis with nasal polyps by weighted gene coexpression network analysis. Biomed Res Int. 2020;2020:6140728. doi: 10.1155/2020/6140728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bhushan R, Rani A, Ali A, Singh VK, Dubey PK. Bioinformatics enrichment analysis of genes and pathways related to maternal type 1 diabetes associated with adverse fetal outcomes. J Diabetes Complications. 2020;34:107556. doi: 10.1016/j.jdiacomp.2020.107556. [DOI] [PubMed] [Google Scholar]
- 28.Silverbush D, Sharan R. A systematic approach to orient the human protein-protein interaction network. Nat Commun. 2019;10:3015. doi: 10.1038/s41467-019-10887-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.You L, Guo X, Huang Y. Correlation of cancer stem-cell markers OCT4, SOX2, and NANOG with clinicopathological features and prognosis in operative patients with rectal cancer. Yonsei Med J. 2018;59:35–42. doi: 10.3349/ymj.2018.59.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Joseph C, Arshad M, Kurozomi S, Althobiti M, Miligy IM, Al-Izzi S, Toss MS, Goh FQ, Johnston SJ, Martin SG, Ellis IO, Mongan NP, Green AR, Rakha EA. Overexpression of the cancer stem cell marker CD133 confers a poor prognosis in invasive breast cancer. Breast Cancer Res Treat. 2019;174:387–399. doi: 10.1007/s10549-018-05085-9. [DOI] [PubMed] [Google Scholar]
- 31.Qin S, Long X, Zhao Q, Zhao W. Co-expression network analysis identified genes associated with cancer stem cell characteristics in lung squamous cell carcinoma. Cancer Invest. 2020;38:13–22. doi: 10.1080/07357907.2019.1697281. [DOI] [PubMed] [Google Scholar]
- 32.Zhao S, Shen W, Yu J, Wang L. TBX21 predicts prognosis of patients and drives cancer stem cell maintenance via the TBX21-IL-4 pathway in lung adenocarcinoma. Stem Cell Res Ther. 2018;9:89. doi: 10.1186/s13287-018-0820-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Galuppo R, Maynard E, Shah M, Daily MF, Chen C, Spear BT, Gedaly R. Synergistic inhibition of HCC and liver cancer stem cell proliferation by targeting RAS/RAF/MAPK and WNT/β-catenin pathways. Anticancer Res. 2014;34:1709–1713. [PMC free article] [PubMed] [Google Scholar]
- 34.Feng MX, Zhang JX, Wan P, Qiu BJ, Gu LH, Zhang JJ, Xia Q. MicroRNA-28-5p regulates liver cancer stem cell expansion via IGF-1 pathway. Hepatobiliary Pancreat Dis Int. 2019;2019:8734362. doi: 10.1155/2019/8734362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Si A, Wang L, Miao K, Zhang R, Ji H, Lei Z, Cheng Z, Fang X, Hao B. miR-219 regulates liver cancer stem cell expansion via E-cadherin pathway. Cell Cycle. 2019;18:3550–3561. doi: 10.1080/15384101.2019.1691762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Conigliaro A, Tripodi M, Parola M. SENP1 activity sustains cancer stem cell in hypoxic HCC. Gut. 2017;66:2051–2052. doi: 10.1136/gutjnl-2017-313946. [DOI] [PubMed] [Google Scholar]
- 37.Li W, Zhang Q, Chen K, Sima Z, Liu J, Yu Q, Liu J. 2-Ethoxystypandrone, a novel small-molecule STAT3 signaling inhibitor from Polygonum cuspidatum, inhibits cell growth and induces apoptosis of HCC cells and HCC cancer stem cells. BMC Complement Altern Med. 2019;19:38. doi: 10.1186/s12906-019-2440-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Khosla R, Hemati H, Rastogi A, Ramakrishna G, Sarin SK, Trehanpati N. miR-26b-5p helps in EpCAM+cancer stem cells maintenance via HSC71/HSPA8 and augments malignant features in HCC. Liver Int. 2019;39:1692–1703. doi: 10.1111/liv.14188. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.