Abstract
Chlamydia trachomatis is an obligate intracellular bacterium that causes the most common sexually transmitted bacterial diseases in the world. With a biphasic developmental cycle, the bacteria utilize a type III secretion system (T3SS) to invade host cells as infectious elemental bodies, which then differentiate into actively dividing reticulate bodies. The regulation of the developmental cycle and the T3SS are linked by the bi-functional protein, specific Chlamydia chaperone 4 (Scc4). Scc4 is a class I T3SS chaperone forming a heterodimer with specific Chlamydia chaperone 1 (Scc1) to chaperone the essential virulence effector, Chlamydia outer membrane protein N (CopN). Scc4 also functions as a transcription factor by binding to the RNA polymerase holoenzyme between the flap region of the β subunit and region 4 of σ66. In order to investigate the mechanism behind Scc4’s dual functions and target its protein-protein interactions as a route for drug development, the structure and dynamics of Scc4 are being pursued. In the course of this effort, we assigned 89.2% of the backbone and sidechain 1H, 15N, and 13C resonances of full-length Scc4. The assigned chemical shifts were used to predict the secondary structure and dynamic properties. The type and order of Scc4’s determined secondary structure are consistent with the X-ray crystal structures of other bacterial T3SS chaperones.
Keywords: Chlamydia trachomatis, Scc4, type III secretion system, transcription regulation, RNAP binding protein, chaperone
Biological context
Scc41 was identified in Chlamydia trachomatis as a type III secretion system (T3SS) chaperone (Spaeth et al. 2009) and a transcription factor that directly binds the β subunit2 and the primary sigma factor, σ66,3 of the RNA polymerase holoenzyme (Rao et al. 2009). As a T3SS chaperone, Scc4 functions as an unusual heterodimer with Scc14 to chaperone CopN5 (Shen et al. 2015), an essential virulence factor that interferes with the host’s microtubule network (Huang et al. 2008). The chaperone function of Scc4 occurs primarily in elemental bodies between the late stage of development and initiation of infection (when CopN is secreted into the host cell). The activity of Scc4 as a transcription factor occurs in reticulate bodies during the middle stage of development, when σ66-dependent transcription of housekeeping genes are active (Gao et al. 2020). The stage-specific activity and multiple protein-protein interactions of Scc4 offer several pathways to approach drug development. As part of that effort, the high-resolution NMR structure of Scc4 is being pursued, and the assignments and secondary structure presented herein are the first results of these efforts.
Sequence analysis and structural homology modeling of Scc4 strongly support its T3SS chaperone function, but there is no evidence of homologous transcription factors. Sequence homology among the class I T3SS chaperones is low (typically 5 – 15%), but structural homologs can be identified with high confidence using remote homology detection with the Phyre2 protein fold recognition server (Kelley et al. 2015). The best structural templates for Scc4 are all X-ray structures of homodimer, Gram-negative bacterial class I T3SS chaperones (Shen et al. 2015).6 The only structure to date of a heterodimer T3SS chaperone is from the X-ray crystal structure of the Yersinia pestis complex of the chaperones, SycN and YscB, and the effector, YopN (PDB id 1XKP) (Schubot et al. 2005). No NMR structures in the Protein Data Bank (Berman et al. 2000) or chemical shift assignments in the Biological Magnetic Resonance Data Bank (Ulrich et al. 2008) are available for any class I T3SS chaperones to date. The absence of sequence or structural homologs of Scc4 to transcription factors provides the possibility of a new fold for this function.
Here, we present 89.2% of the backbone and sidechain 1H, 15N, and 13C NMR resonance assignments of Scc4, the chemical shift-based secondary structure, and the random coil index order parameter predictions. We also discuss the relevance of the unassigned residues, which map to a distinct region on Scc4’s structural homology model.
Methods and experiments
Sample preparation
Full length Scc4 (residues 1–133) was expressed and purified according to the methods previously reported (Ukwaththage et al. 2019). Briefly, Scc4 and Scc1-His6 were co-expressed from the vectors, pACYCScc1-His6 and pET28Scc4 (Shen et al. 2015), in E. coli BL21-Gold (DE3) cells using minimal medium supplemented with 2 gL−1 15N-ammonium chloride and 4 gL−1 13C-glucose. The Scc4:Scc1-His6 complex from the cleared lysate was immobilized on Ni2+-charged immobilized metal affinity chromatography resin, and Scc4 was dissociated from the complex and eluted with 0.5% sarkosyl in 20 mM tris(hydroxymethyl)aminomethane hydrochloride, 300 mM NaCl, and 5% glycerol at pH 8.0 buffer. The sarkosyl was removed from the Scc4 sample by diluting the sample 5-fold with running buffer (50 mM sodium phosphate, pH 7.3) and purifying with size exclusion chromatography (Bio-Rad P-4 polyacrylamide beads). The Scc4 sample was concentrated using a 10 kDa molecular weight cutoff centrifugal filter to 0.5 mM concentration. The purity of the Scc4 samples was analyzed by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE), and the protein concentration was determined using the Bradford protein assay following the manufacturer’s protocol.
NMR experiments
The NMR samples consisted of 300 μL of 0.5 mM Scc4 in 50 mM sodium phosphate (pH 7.3) with 5 μM sodium 3-(trimethylsilyl)-1-propanesulfonate and 10% D2O, for chemical shift referencing and locking, respectively, in 5 mm D2O-magnetic susceptibility matched tubes. The spectra were collected at the National Magnetic Resonance Facility at Madison on Bruker Avance III and Varian VNMRS spectrometers operating at 900, 800, and 600 MHz (1H) and equipped with cryogenically-cooled probes. The temperature of the sample was regulated to 298 K for all experiments. The stability of the protein was tested by comparing 2D 1H-15N HSQC spectra before and after each 3D experiment. In order to assign the backbone resonances, spectra of Scc4 were acquired using 2D 1H,15N-HSQC, 3D CBCA(CO)NH, 3D HNCACB, 3D HNCO, 3D HNCA, and 3D HN(CO)CA experiments. Additional 2D and 3D spectra were collected to assign both aliphatic and aromatic sidechain chemical shifts. The experiments recorded to assign the aliphatic sidechains are 2D constant-time 1H,13C-HSQC, 3D C(CO)NH, 3D HBHA(CO)NH, 3D H(CCO)NH, and 3D HC(C)H-TOCSY. The aromatic sidechains were assigned by recording aromatic 2D constant-time 1H,13C-HSQC, aromatic Cβ 2D constant-time 13C-HSQC (edited to observe only Cβ groups of aromatic residues), 2D (HB)CB(CGCD)HD, 2D (HB)CB(CGCDCE)HE, and aromatic 3D HC(C)H-TOCSY spectra (Table 1). All spectra, except for the aliphatic and aromatic 3D HC(C)H-TOCSY spectra, were recorded using non-uniform sampling (NUS) with sampling rates ranging between 30 and 40%. In addition, 3D HNCA and HN(CO)CA spectra were recorded using band-selective excitation short-transient sequences (BEST-type experiments) with a recovery delay of 0.3 s between transients (Lescop et al. 2007). All spectra were processed using the NMRPipe software package (Delaglio et al. 1995). 3D spectra recorded with NUS were reconstructed using the SMILE algorithm available in NMRPipe (Ying et al. 2017). The spectral analysis was done using the NMRFAM-SPARKY software package (Lee et al. 2015). These are the first Scc4 backbone and sidechain assignments; hence all assignments are de novo. The initial peaks were picked using the APES automation plugin (Shin et al. 2008) and verified manually. Cα, Cβ, and C peaks were manually identified and verified using strip plots from CBCA(CO)NH, HNCACB, HNCA, HNCOCA, and HNCO spectra. Sidechain 1H and 13C resonances were identified with the aliphatic and aromatic sidechain experiments starting from the assigned backbone resonances and taking advantage of the “transfer and simulate assignments” extension in NMRFAM-SPARKY. The secondary structure predictions and the residue specific random coil index (RCI-S2) were determined using the backbone assignments with TALOS-N (Shen and Bax 2013) and PECAN (Eghbalnia et al. 2005) web servers.
Table 1.
Experimental details of 2D and 3D spectra collected for backbone and sidechain assignments.
| Experiment | No. of scans | Spectral window (ppm) | Complex points | Offset (ppm) |
|---|---|---|---|---|
| 15N -HSQC& | 8 | 16.67 × 26.318 | 1024 × 128 | 4.773 × 117.88 |
| CBCA(CO)NH* | 32 | 16.67 × 57.009 × 26.318 | 1024 × 52 × 42 | 4.773 × 43.75 × 117.88 |
| HNCACB* | 16 | 15.44 × 63.141 × 32.256 | 1024 × 66 × 42 | 4.771 × 43.718 × 117.666 |
| HNCO* | 32 | 16.285 × 13.924 × 29.619 | 1024 × 48 × 48 | 4.773 × 175.945 × 117.692 |
| HN(CO)CA* | 52 | 16.67 × 27.841 × 26.318 | 800 × 64 × 42 | 4.773 × 55.8 × 117.88 |
| HNCA* | 52 | 16.67 × 27.841 × 26.318 | 800 × 64 × 42 | 4.773 × 55.8 × 117.88 |
| 1H-13C-HSQC-CT$ | 16 | 20.04 × 72.918 | 1024 × 256 | 4.797 × 42.964 |
| C(CO)NH* | 64 | 20.04 × 76.233 × 26.318 | 1024 × 64 × 32 | 4.797 × 42.964 × 117.904 |
| HBHA(CO)NH# | 32 | 20.04 × 6.0 × 26.318 | 1024 × 60 × 40 | 4.797 × 4.797 × 117.904 |
| H(CCO)NH# | 64 | 20.04 × 8.335 × 26.318 | 1024 × 64 × 32 | 4.797 × 4.797 × 117.904 |
| HC(C)H-TOCSY@ | 8 | 20.04 × 8.335 × 72.925 | 1024 × 64 × 64 | 4.797 × 4.797 × 42.964 |
| Aromatic 1H, 13CHSQC-CT$ | 32 | 16.669 × 29.167 | 1024 × 70 | 4.797 × 125.569 |
| Aromatic Cβ-13CHSQC-CT$ | 32 | 16.669 × 18.561 | 1024 × 124 | 4.797 × 35.186 |
| (HB)CB(CGCD)HD$ | 512 | 16.669 × 19.887 | 1024 × 23 | 4.797 × 35.186 |
| (HB)CB(CGCDCE)HE$ | 512 | 16.669 × 19.887 | 1024 × 23 | 4.797 × 35.186 |
| Aromatic HCCHTOCSY@ | 16 | 20.04 × 3.334 × 29.165 | 1024 × 64 × 48 | 4.773 × 4.773 × 125.552 |
(1H × 15N);
(1H × 13C × 15N);
(1H × 13C);
(1H × 1H × 15N);
(1H × 1H × 13C)
Assignment and data deposition
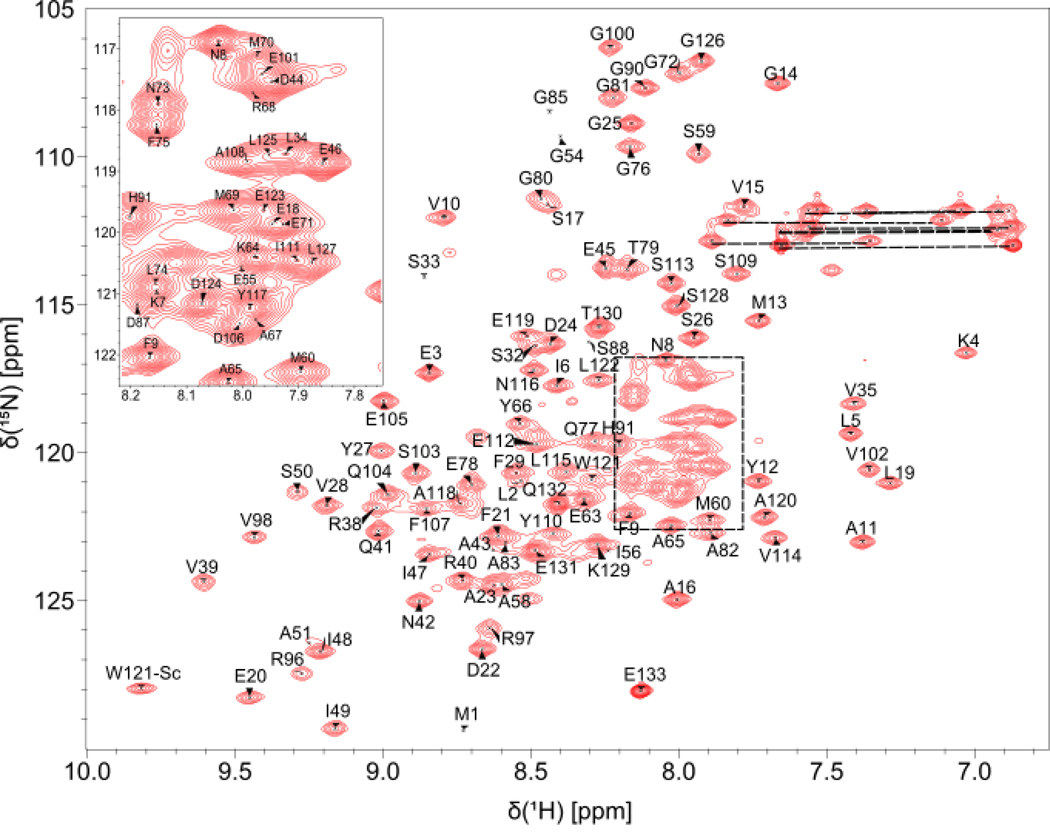
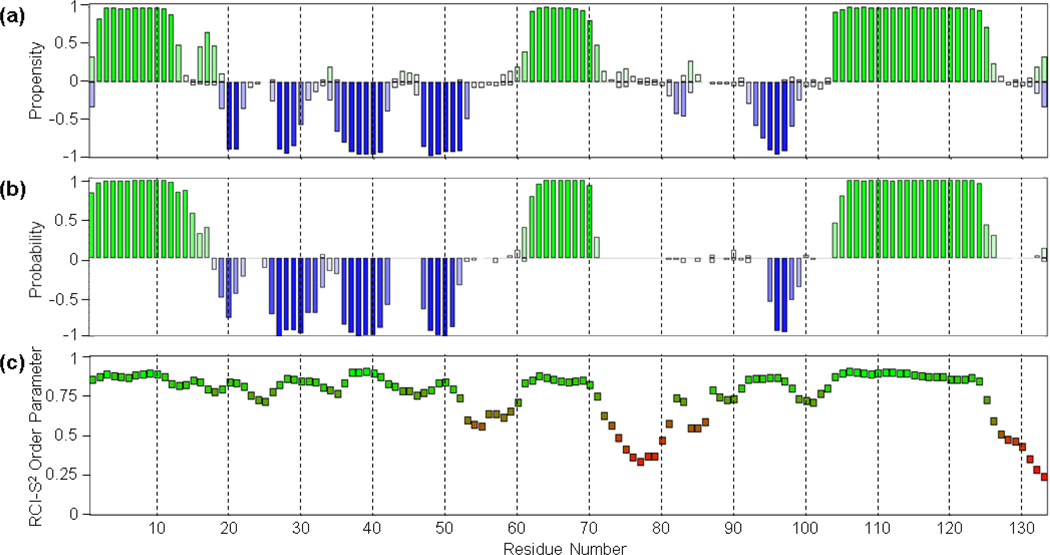
The Scc4 backbone and side chain assignments of 1H, 15N, and 13C resonances were deposited in the Biological Magnetic Resonance Data Bank (http://www.bmrb.wisc.edu/) with the accession number 28101. The assigned backbone amide (15N-1HN) peaks are shown in the 2D 1H,15N HSQC spectrum of Scc4 in Fig. 1. Scc4 consists of 133 amino acids including 3 proline residues (P30, P57 and P99). Hence, from 130 residues, the 15N-1HN chemical shifts of 116 residues (89.2%) were assigned in the 1H,15N HSQC spectrum. The 15N-1HN peaks of some residues (namely S33, A43, A51, G54, E55, I56, G85, D87, and S88) have lower intensity compared to the majority of the peaks; these residues were assigned using the sidechain spectra, including the 3D HC(C)H-TOCSY experiment. In addition, some of the residues exhibit multiple peaks in the 1H,15N HSQC spectrum, indicating the presence of multiple conformations in slow exchange for these residues (Online Resource 1, Supplementary Fig. 1 and 2). The major peaks with higher intensity were assigned to determine the structure of the most abundant conformational state. The minor peaks, which could be identified, include E3’, K4’, I6’, F9’, N42’, I47’, N73’, E78’, L127’ (labeled with an apostrophe to differentiate from the major peak). The cis/trans isomerization of 3 proline residues (P30, P57 and P99) was investigated using the PROMEGA (Proline Omega angle prediction from sequence and chemical shifts) server (Shen and Bax 2010). All three prolines in the major conformation of the protein were found to be in the trans configuration. On the other hand, we cannot rule out that the minor conformation may include one or more of the prolines in the cis configuration. Unassigned residues (14) include I31, R36, M37, F52, L53, D61, I62, L84, L86, D89, and A92-V95. Unassigned peaks (12) in the 1H,15N HSQC are shown in Supplementary Fig. 1 (Online Resource 1) and were not assigned due to weak or ambiguous Cα and Cβ resonances (Online Resource 1, Supplementary Figs. 3 and 4). The assignment completeness of the Cα, Cβ, and C resonances was calculated using the completeness counter in NMRFAM-SPARKY: 130 of 133 Cα (97.7%), 118 of 122 Cβ (96.7%), and 115 of 133 C (86.5%). The secondary structure predictions of Scc4 with TALOS-N and PECAN servers were obtained using the chemical shift assignments. According to both predictions shown in Fig. 2a (TALOS-N) and 2b (PECAN), with the RSI-S2 values from TALOS-N shown in Fig. 2c, Scc4 contains 3 α-helices and 5 β-strands. The RCI-S2 values calculated using TALOS-N (Fig. 2c) indicate that residues 1–52, 60–71, 83–84, 88–125 are ordered (RCI-S2 > 0.7) and residues 53–59, 72–82, 85–87, and 126–133 are disordered (RCI-S2 < 0.7).
Fig. 1.
The 1H,15N HSQC spectrum of Scc4 with assigned resonances. The dashed lines indicate the side chain 1H,15N peaks.
Fig. 2.
Secondary structure predictions of Scc4 by (a) TALOS-N and (b) PECAN, and (c) the random coil index order parameters (RCI-S2) determined using TALOS-N. (a,b) Green and blue bars indicate the propensity/probability of a residue to adopt alpha helical or beta strand secondary structures, respectively. (c) The green to red indicates the determined protein flexibility from ordered to flexible.
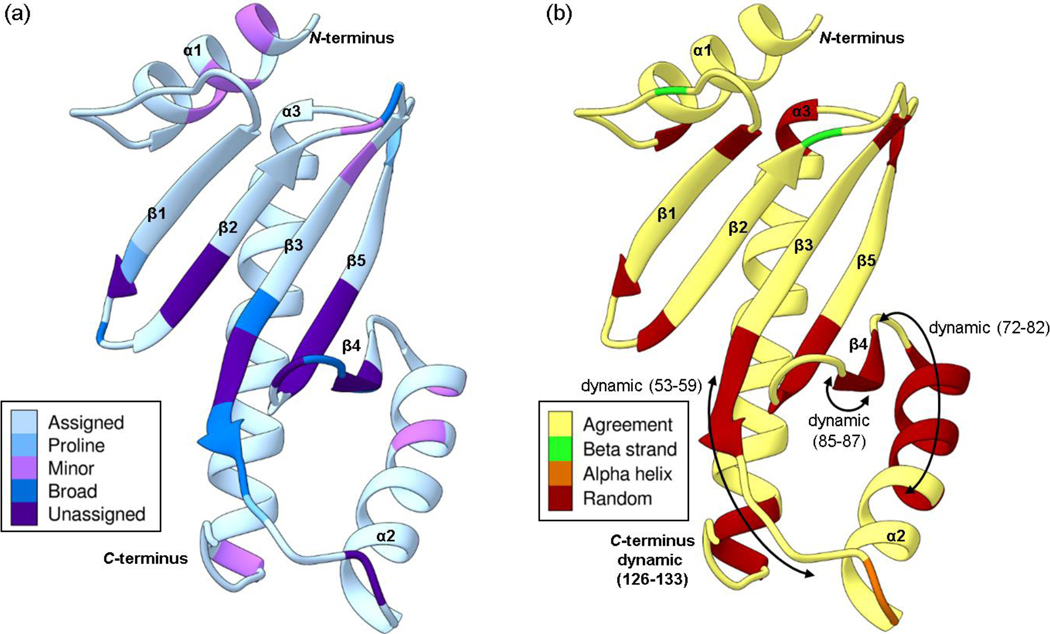
Mapping the results onto the homology model of Scc4 provides insight into the dynamic behavior of Scc4. We hypothesize that this dynamic behavior plays a role in Scc4’s dual functions and the mechanism of switching between functions. Unlike most class I T3SS chaperones, Scc4 does not function as a homodimer to bind CopN, but rather binds Scc1 to form a heterodimer. The Scc4:Scc1 interaction has very high affinity, while Scc4 alone forms various aggregation states that depend on the buffer conditions (Shen et al. 2015). In Fig. 3a, the unassigned residues are shaded purple and the assigned residues with broad peaks in the 1H,15N-HSQC are shaded in dark blue. These residues fall predominantly across the beta sheet and include each strand, with most of the residues converging at one end of the β3-β5-β4 structure, indicating a dynamic region across the beta sheet. Resonances that exhibited minor peaks (residues shaded in pink in Fig. 3a) do not cluster well with the exception of the residues in the α1 helix, which could result from the slow exchange of the N-terminal helix between two conformations. The proximity of the proline residues (shaded in medium blue) makes it unlikely that cis/trans isomerization is involved in this chemical exchange. The determined secondary structure from the NMR data (Fig. 2a,b) differ in some regions from the predicted secondary structure from Phyre2 (Fig. 3b). The differences are indicated in Fig. 3b and include an overall decrease in secondary structure, especially within the unassigned β3-β5-β4 region and the α2 helix. The RCI-S2 parameters in this region are consistent with low values (RCI-S2 < 0.7) for residues 53–59 (β3), 85–87 (β4), and 72–82 (α2) as shown in Fig. 3b with double sided arrows. The order of the NMR-determined secondary structural elements is consistent with the topology of class I T3SS chaperones (Page and Parsot 2002), with the exception of β4, which is determined to be random coil (PECAN) with a low beta strand propensity (TALOS-N) and dynamic residues (85–87). For the α2 helix, the NMR data predicts that residues 72–82 are random coil (TALOS-N and PECAN) and dynamic (RCI-S2 < 0.7) while the Phyre2 model predicts this region to be alpha helical. Considering that the Phyre2 model is based on the X-ray crystal structures of homodimer class I T3SS chaperones and the homodimer interface is β4-α2, the NMR data likely describe the correct state of free Scc4, and the Scc4 Phyre2 model likely resembles the structure of Scc4 in complex with Scc1. Further investigation into the structures of Scc4 alone and in complex with its T3SS and RNAP partners are necessary to confirm these speculations.
Fig. 3.
The Phyre2 structural homology model of Scc4 with the (a) assignment status and (b) secondary structure comparison. (a) The assigned residues are shaded in light blue, the prolines in medium blue, and the unassigned residues are in purple. Residues that show minor peaks in the 1H,15N-HSQC spectrum are shaded in pink, and residues with broad resonances are shaded in dark blue. (b) The secondary structure of the Phyre2 homology model is shown as a ribbon drawing. Residues shaded in yellow have NMR-determined secondary structure in agreement with the homology model. Residues with NMR-determined secondary structures that are different from the model are shaded in green for beta strands, orange for alpha helices, and red for random coil. The arrows indicate residues that are dynamic with RCI-S2 values < 0.7. The color schemes are based on palettes for color blindness and interpretability in grayscale (Krzywinski 2020).
Supplementary Material
Acknowledgments
This work was supported by funding from the National Institutes of Health grant R15GM109413 (Megan A. Macnaughtan). Thilini O. Ukwaththage thanks the Louisiana State University Department of Chemistry for providing support through teaching assistantships. This study made use of the National Magnetic Resonance Facility at Madison, which is supported by NIH grant P41GM103399 (NIGMS), old number: P41RR002301. Equipment was purchased with funds from the University of Wisconsin-Madison, the NIH P41GM103399, S10RR02781, S10RR08438, S10RR023438, S10RR025062, S10RR029220), the NSF (DMB-8415048, OIA-9977486, BIR-9214394), and the USDA. The authors thank Dr. Woonghee Lee for software assistance with NMRFAM-SPARKY and Dr. Ronnie Frederick for assistance in concentrating the NMR samples prior to data acquisition.
Funding
This work was supported by funding from the National Institutes of Health grant R15GM109413 (MAM). TOU thanks the Louisiana State University Department of Chemistry for providing support through teaching assistantships. This study made use of the National Magnetic Resonance Facility at Madison, which is supported by NIH grant P41GM103399 (NIGMS), old number: P41RR002301. Equipment was purchased with funds from the University of Wisconsin-Madison, the NIH P41GM103399, S10RR02781, S10RR08438, S10RR023438, S10RR025062, S10RR029220), the NSF (DMB-8415048, OIA-9977486, BIR-9214394), and the USDA.
Footnotes
Ethics approval
Not applicable
Consent to participate
Not applicable
Consent for publication
Not applicable
Availability of data and material (data transparency)
The raw NMR data and assignments are available through the Biological Magnetic Resonance Data Bank (http://www.bmrb.wisc.edu/), the accession number 28101.
Code availability
Not applicable
Electronic supplementary material
Online Resource 1 containing Supplementary Fig. 1–4.
Ethical standards
The experiments comply with the current laws of the United States, the country in which they were performed.
specific Chlamydia chaperone 4, formerly CT663, UniprotKB O84670.
RpoB, UniprotKB P0CE09.
σ66 or RpoD, UniprotKB P18333.
specific Chlamydia chaperone 1 or CT088, UniprotKB O84090.
low calcium response E or Chlamydia outer membrane protein N (CopN) or CT089, UniprotKB O84091.
Escherichia coli CesT (PDB id 1K3E chain A, Uniprot P58233), Yersinia pestis SycH (PDB id 1TTW chain A, Uniprot Q7BTX0), Salmonella enterica SicP (PDB id 1JYO chain A, Uniprot P0CL16), Pseudomonas syringae Shc (PDB id 4G6T chain A, Uniprot Q87UE6), and Salmonella enterica STM2138 (PDB id 3EPU chain B, Uniprot Q8ZNP3).
Conflicts of interest
The authors declare that they have no conflict of interest.
Publisher's Disclaimer: This Author Accepted Manuscript is a PDF file of an unedited peer-reviewed manuscript that has been accepted for publication but has not been copyedited or corrected. The official version of record that is published in the journal is kept up to date and so may therefore differ from this version.
References
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE (2000) The Protein Data Bank. Nucleic Acids Res 28:235–242. 10.1093/nar/28.1.235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A (1995) NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J Biomol NMR 6:277–293. 10.1007/bf00197809 [DOI] [PubMed] [Google Scholar]
- Eghbalnia HR, Wang L, Bahrami A, Assadi A, Markley JL (2005) Protein energetic conformational analysis from NMR chemical shifts (PECAN) and its use in determining secondary structural elements. J Biomol NMR 32:71–81. 10.1007/s10858-005-5705-1 [DOI] [PubMed] [Google Scholar]
- Gao L, Cong Y, Plano GV, Rao X, Gisclair LN, Schesser Bartra S, Macnaughtan M, Shen L (2020) Context-dependent action of Scc4 reinforces control of the type III secretion system. J Bacteriol 10.1128/JB.00132-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Lesser CF, Lory S (2008) The essential role of the CopN protein in Chlamydia pneumoniae intracellular growth. Nature 456:112–115. 10.1038/nature07355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJE (2015) The Phyre2 web portal for protein modeling, prediction and analysis. Nature Protocols 10:845–858. 10.1038/nprot.2015.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski M (2020) Designing for color blindness: color choices and transformations for deuteranopia and other afflictions. http://mkweb.bcgsc.ca/colorblind/index.mhtml. [Google Scholar]
- Lee W, Tonelli M, Markley JL (2015) NMRFAM-SPARKY: Enhanced software for biomolecular NMR spectroscopy. Bioinformatics 31:1325–1327. 10.1093/bioinformatics/btu830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescop E, Schanda P, Brutscher B (2007) A set of BEST triple-resonance experiments for time-optimized protein resonance assignment. J Magn Reson 187:163–169. 10.1016/j.jmr.2007.04.002 [DOI] [PubMed] [Google Scholar]
- Page AL, Parsot C (2002) Chaperones of the type III secretion pathway: Jacks of all trades. Mol Microbiol 46:1–11. 10.1046/j.1365-2958.2002.03138.x [DOI] [PubMed] [Google Scholar]
- Rao X, Deighan P, Hua Z, Hu X, Wang J, Luo M, Wang J, Liang Y, Zhong G, Hochschild A, Shen L (2009) A regulator from Chlamydia trachomatis modulates the activity of RNA polymerase through direct interaction with the beta subunit and the primary sigma subunit. Genes Dev 23:1818–1829. 10.1101/gad.1784009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubot FD, Jackson MW, Penrose KJ, Cherry S, Tropea JE, Plano GV, Waugh DS (2005) Three-dimensional structure of a macromolecular assembly that regulates type III secretion in Yersinia pestis. J Mol Biol 346:1147–1161. 10.1016/j.jmb.2004.12.036 [DOI] [PubMed] [Google Scholar]
- Shen L, Macnaughtan MA, Frohlich KM, Cong YG, Goodwin OY, Chou CW, LeCour L, Krup K, Luo M, Worthylake DK (2015) Multipart chaperone-effector recognition in the type III secretion system of Chlamydia trachomatis. J Biol Chem 290:28141–28155. 10.1074/jbc.M115.670232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Bax A (2010) Prediction of Xaa-Pro peptide bond conformation from sequence and chemical shifts. J Biomol NMR 46:199–204. 10.1007/s10858-009-9395-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Bax A (2013) Protein backbone and sidechain torsion angles predicted from NMR chemical shifts using artificial neural networks. J Biomol NMR 56:227–241. 10.1007/s10858-013-9741-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J, Lee W, Lee W (2008) Structural proteomics by NMR spectroscopy. Expert Rev Proteomics 5:589–601. 10.1586/14789450.5.4.589 [DOI] [PubMed] [Google Scholar]
- Spaeth KE, Chen Y-S, Valdivia RH (2009) The Chlamydia type III secretion system C-ring engages a chaperone-effector protein complex. PLOS Pathog 5:1–11. 10.1371/journal.ppat.1000579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ukwaththage TO, Goodwin OY, Songok AC, Tafaro AM, Shen L, Macnaughtan MA (2019) Purification of tag-free Chlamydia trachomatis Scc4 for structural studies using sarkosyl-assisted on-column complex dissociation. Biochemistry 58:4284–4292. 10.1021/acs.biochem.9b00665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich EL, Akutsu H, Doreleijers JF, Harano Y, Ioannidis YE, Lin J, Livny M, Mading S, Maziuk D, Miller Z, Nakatani E, Schulte CF, Tolmie DE, Kent Wenger R, Yao H, Markley JL (2008) BioMagResBank. Nucleic Acids Res 36:D402–D408. 10.1093/nar/gkm957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying J, Delaglio F, Torchia DA, Bax A (2017) Sparse multidimensional iterative lineshape-enhanced (SMILE) reconstruction of both non-uniformly sampled and conventional NMR data. J Biomol NMR 68:101–118. 10.1007/s10858-016-0072-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.