Abstract
Background
Giardia lamblia is a pathogenic intestinal flagellate transmitted by the ingestion of contaminated water or food with the cyst stage of the parasite. Giardiasis can cause severe acute diarrhea and malabsorption or may persist as a chronic infection. Effective treatment and control measures depend on proper laboratory diagnosis using diagnostic methods with high sensitivity and specificity.
Objective
To compare the sensitivity and specificity of direct smear, Ritchie sedimentation technique, two brands of rapid chromatographic immunoassay test, and real-time polymerase chain reaction (PCR) for the detection of G. lamblia in clinical human fecal samples.
Materials and methods
Unpreserved 100 stool specimens were collected in clean plastic containers and labeled with the patient’s information and examined through light microscopy, immunochromatographic test (ICTs), and real-time PCR.
Results
Out of 100 fresh stool samples obtained from workers analyzed, real-time PCR targeting the SSU rRNA gene was able to detect Giardia deoxyribonucleic acid (DNA) in (42) samples followed by ImmunoCard STAT! (31) samples (Meridian Bioscience, Germany), direct smear (23) samples, CerTest (19) samples (Biotec, Zaragoza, Spain), and Ritchie technique (17) samples. Real-time PCR was the most sensitive for the diagnosis of G. lamblia in comparison to the other techniques.
Conclusions
All the techniques investigated were sensitive for the detection of G. lamblia in stool samples. Further studies are recommended using multiplex real-time PCR assay in order to increase the possibility of the presence or absence of the infection.
Keywords: giardia lamblia, diagnosis, parasites, real-time pcr, neglected diseases, saudi arabia, icts
Introduction
One of the most important causes of parasitic diseases is a flagellated protozoan called Giardia lamblia. The life cycle is simple and direct, including the motile trophozoite and nonmotile cyst stages while the ingestion of cysts in contaminated water or food (fecal-oral route) is the main mode of transmission [1]. The World Health Organization (WHO) reported that 748 million people drink unsafe water, 2.5 billion are without improved sanitation, more than 200 million cases are reported annually, and since 2004, Giardia has been included in the "neglected diseases initiative" [2-3]. Infection with Giardia in many people is asymptomatic while others suffer from acute watery diarrhea, abdominal cramps, nausea, epigastric pain, malabsorption, flatulence, and weight loss [4]. In some patients and in relation to several factors, the infection may sustain and become chronic [5]. Previous studies in Saudi Arabia reported high prevalence rates of intestinal parasitic diseases with a different rate of G. lamblia (2%-20%) among various populations [6-9].
The early accurate diagnosis of giardiasis is important for the successful treatment and prevention of diseases. The routine laboratory diagnosis is performed for the detection of trophozoites or cysts by microscopic examination of at least three stool samples collected independently [10-11]. This diagnosis depends upon the times of sampling, patient compliance, application of concentration methods, and the expertise of the technologists. Furthermore, in recent years, other methods, such as rapid immunochromatographic tests (ICTs) and molecular techniques, are used mainly for diagnostic or research proposes. The sensitivity and specificity of all diagnostic methods can be improved by including alternative diagnostic procedures that are more rapid and reliable [12]. Isolates of Giardia are classified into eight genotypes and the studies of these genes have indicated evidence of cryptic infection, suggesting the use of molecular diagnostics [13].
The aim of the present study was to compare the sensitivity and specificity of several diagnostic methods: direct smears, Ritchie sedimentation technique, two brands of ICTs, and real-time PCR for the detection of G. lamblia in clinical human fecal samples.
Materials and methods
Specimen collection
Stool specimens were collected in clean plastic containers from workers recently arrived in Jeddah, Saudi Arabia, during their screening visit to get a residence permit (officially known as Iqama).
Questionnaire form
The data of patients were collected by interviews in a standard questionnaire that was designed for this study. It included questions on demographic information (age, nationality, the period of residence in Saudi Arabia, and occupation) and some general information about intestinal parasitic infections.
Laboratory examination
Laboratory microscopic examination of feces was accomplished by using direct smears, the Ritchie technique, Para-Pak trichrome stain. Two brands of rapid chromatographic immunoassay tests were performed according to the manufacturer's instructions. ImmunoCard STAT! CGE (Meridian, Bioscience, Germany) and CerTest (Biotec, Zaragoza, Spain) are designed to identify G. lamblia, Cryptosporidium parvum, and Entamoeba histolytica. Real-time PCR was used to detect the small subunit ribosomal RNA gene (SSU rRNA) of G. lamblia.
Direct smears
About 2 mg for each stool sample was mixed with a drop of normal saline (0.9%) and iodine on a clean glass slide (75 X 26 mm). A cover glass (22 X 22 mm) was placed carefully and then examined by light microscopy using (10x) and (40x) objective lenses.
Ritchie (formal ether) sedimentation technique
About 2 gm of each stool sample was emulsified in 10 ml of 10% formal-saline. It was then filtered through two gauze layers fitted in a funnel and centrifuged for 5 min at 2000 rpm. The supernatant was discarded, and the sediment was re-suspended in 10 ml of formal saline (10%) and mixed very well. After that, 3 ml of diethyl ether was added, shaken vigorously for 10 sec, and then centrifuged for 5 min at 2000 rpm. Four layers were formed: the top layer of ether, the plug of the debris, 10% formal saline, and the sediment layer at the bottom of the tube. The top three layers were removed, and a drop of iodine was mixed with the sediment and then examined using (10x) and (40x) objective lenses [14].
Para-Pak trichrome stain
Stool samples were fixed in polyvinyl alcohol (PVA). Stool smears were prepared and stained by Wheatley trichrome stain according to the manufacturer's instructions, (Para-Pak® Trichrome Stain, Meridian, Bioscience, Germany). Smears were air-dried then stained with trichrome stain for 6-8 min, placed in acid ethanol for 5-10 sec, then two quick dips in (95%) ethanol. The slides were immersed two times in (95%) ethanol for 5 min, then in (96-100%) ethanol for 3 min, and finally placed in xylene for 3 min. Finally, each slide was mounted with a coverslip (22 X 40 mm) and examined using an oil immersion (100x) objective lens.
ImmunoCard STAT! CGE
For each specimen, 1 ml of the diluent was transferred into a 15 ml centrifuge tube. Thereafter, approximately 50 mg of each stool specimen was added and mixed gently, then centrifuged for 5 min at 3000 rpm. Next, 125 µl of each prepared specimen was placed into the sample port of the device and the results were interpreted after 10 min. A purple band appears at the control line (C), indicating that the test was completed successfully. The blue band at position 1 on the device frame is indicative of the presence of C. parvum antigens. In the presence of G. lamblia antigens, a red/pink color is observed at band 2. A green band appearing at position 3 is related to E. histolytica antigens.
CerTest
One-hundred twenty-five (125) mg was added into the diluent collection tube, mixed very well, and then shaken vigorously for seconds. Four drops of the prepared specimen were dispensed in each of the circular windows. The test was interpreted after 10 min with a green band-of-control line. The formation of a red band at the test line is indicative of the presence of the investigated parasite.
DNA isolation
Stool samples were subjected to DNA extraction using the QIAamp DNA Stool Mini Kit (Qiagen, Germany) protocol. Each DNA sample was eluted in a final volume of 100 μl and stored at -80°C until needed.
Real-time PCR amplification
The target gene of the real-time PCR assay is the SSU rRNA gene. DNA was amplified and detected for G. lamblia specifically. To select the specific Giardia sequence, alignments were made of 31 GenBank sequences that included 16 Giardia and 15 sequences of other parasites and bacteria (non-pathogenic). The Giardia-specific primers and probe set consisted of the forward primer Giardia F, the reverse primer Giardia R, and the Giardia specific double-labeled probe Giardia T (Humanizing Genomic Macrogen Clinical Lab, Korea) [15]: Giardia F (5՜-GACGGCTCAGGACAACGGTT-3՜; positions 80-99), Giardia R (5՜- TTGCCAGCGGTGTCCG-3՜; positions 142-127) and Giardia T (FAM-5՜-CCCGCGGCGGTCCCTGCTAG-3՜-TAMRA; positions 105-124).
Stock primers and probe
According to the manufacturer's instructions, to reach 100 pmol/µl, 250 µl of nuclease-free water was added for each primer and 280 µl was added for the probe.
Working primers and probe
Fifty µl from each stock primer was added to 50 µl nuclease-free water to prepare 50 pmol/µl. From this, 10 µl was mixed with 90 µl of nuclease-free water. Thirty µl from the stock probe was added to 70 µl nuclease-free water to prepare 30 pmol/µl. From this, 10 µl was mixed with 90 µl of nuclease-free water. All working primers and probes were stored at -20°C until needed.
Real-time PCR master mix preparation
The real-time PCR master mix was prepared according to QuantiTect® Probe PCR Kit instructions (Qiagen, Germany) in a 1.5 ml microcentrifuge tube, vortexed very well, and then briefly centrifuged at full speed. After that, the reaction mix was dispensed in a volume of 15.5 µl using a standard PCR plate containing 96 wells strips tubes. Then, 9.5 µl of the template DNA was added to the individual wells containing the reaction mix. The plate was then transferred into Applied Biosystems 7500 Fast cycler (Foster City, California), to start the cycling program.
Real-time PCR data analysis
Data analysis was performed with the Applied Biosystems 7500 Fast System by calculating the threshold cycle number (Ct value) that presented the positive amplification of gene in a real-time cycler number.
Statistical analysis
Categorical data were reported as frequency and percentage (%). Data were analyzed using SPSS (version 21, IBM Corp., Armonk, NY). A P-value of < 0.05 value was accepted as statistically significant (corresponding to 95% confidence intervals (CI)). Sensitivity, specificity, PPV, NPV, and accuracy were calculated to test the diagnostic yield.
Results
Out of 100 fecal samples analyzed, 48 were positive for G. lamblia, 18 samples for E. histolytica, nine samples for C. parvum, three samples for E. coli, three samples for hookworms, two samples for Trichuris trichiura, two samples for Enterobius vermicularis, and one sample for each of the following: Endolimax nana, Hymenolepis nana, and Taenia spp.
Table 1 summarizes the demographic characteristics of the study subjects. All subjects were male workers aged from 20 to 53 years (32.09 ± 8.598), mainly from Pakistan. Most of the stool samples were soft (49%) and loose (48%) while three patients produced watery stool.
Table 1. Demographic characteristics.
a: within the same nationality; b: within all infected cases (62)
| Country of origin | Frequencies | Infected cases | ||
| N | n | (%)a | (%)b | |
| Bangladesh | 7 | 3 | 42.86 | 4.84 |
| Egypt | 12 | 6 | 50 | 9.68 |
| India | 31 | 18 | 58.06 | 29.03 |
| Nepal | 2 | 2 | 100 | 3.23 |
| Pakistan | 36 | 27 | 75 | 43.55 |
| Philippines | 4 | 1 | 25 | 1.61 |
| Sri Lanka | 2 | 1 | 50 | 1.61 |
| Sudan | 4 | 3 | 75 | 4.84 |
| Yemen | 2 | 1 | 50 | 1.61 |
| Total | 100 | 62 | --- | 100 |
Out of the 100 samples screened for G. lamblia, real-time PCR was able to detect Giardia DNA in 42 samples followed by ImmunoCard STAT! CGE (31), direct smears (23), CerTest (19), and the Ritchie sedimentation technique (17).
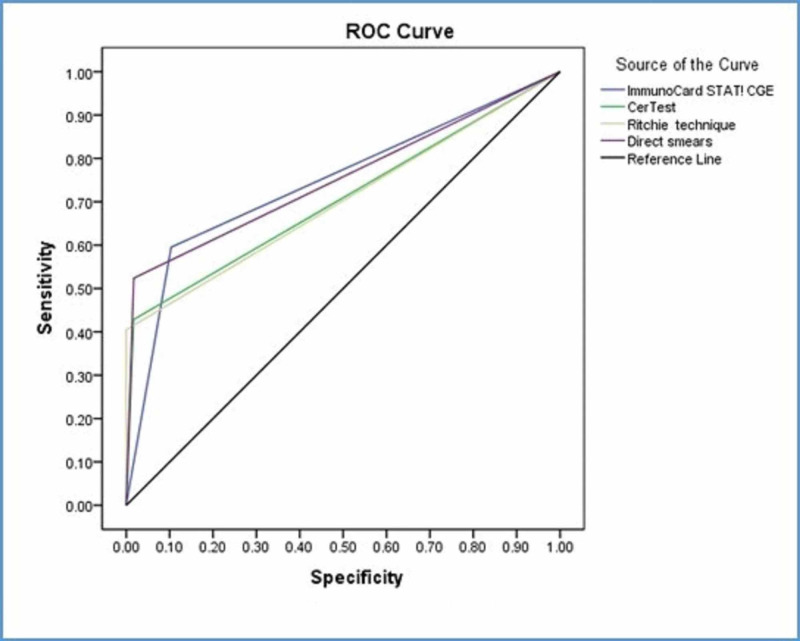
Considering real-time PCR as a nominated golden standard, ImmunoCard STAT! CGE revealed the highest sensitivity followed by direct smears (Table 2). ImmunoCard STAT! CGE was acceptable. The area under the curve is 0.746, Std. Error=0.053, p-value<0.0005 with 95% CI (0.643-0.849). CerTest was acceptable. The area under the curve is 0.706, Std. Error=0.056, p-value<0.0005 with 95% CI (0.596-0.815). The Ritchie technique was acceptable. The area under the curve is 0.702, Std. Error=0.056, p-value=0.001 with 95% CI (0.592-0.812). Direct smears were acceptable. The area under the curve is 0.753, Std. Error=0.053, p-value<0.0005 with 95% CI (0.649-0.857) (Figure 1).
Table 2. Sensitivity, specificity, and accuracy of diagnostic tools with real-time PCR.
N: Negative, P: Positive, TP: True-positive, FP: False-negative, TN: True-negative, FN: False-negative, PPV: Positive predictive value, NPV: Negative predictive value
ImmunoCard STAT!: Meridian Bioscience, Germany; CerTest: Biotec, Zaragoza, Spain
| Test | Real-time PCR | Sensitivity % | Specificity % | PPV % | NPV % | 95% CI | P-value | ||
| P | N | ||||||||
| Immuno Card STAT! CGE | P | TP25 | FP6 | (59.52) | (89.65) | (80.64) | (75.36) | (0.64-0.85) | P < 0.001 |
| N | FN17 | TN52 | |||||||
| CerTest | P | TP18 | FP1 | (42.86) | (98.27) | (94.74) | (70.37) | (0.60-0.82) | P < 0.001 |
| N | FN24 | TN57 | |||||||
| Ritchie | P | TP17 | FP0 | (40.47) | (100) | (100) | (69.88) | (0.60-0.81) | P < 0.001 |
| N | FN25 | TN58 | |||||||
| Direct smears | P | TP22 | FP1 | (52.38) | (98.27) | (95.65) | (74.02) | (0.65-0.86) | P < 0.001 |
| N | FN20 | TN57 | |||||||
Figure 1. Sensitivity, specificity, and accuracy of diagnosis tools with real-time PCR.
PCR: polymerase chain reaction
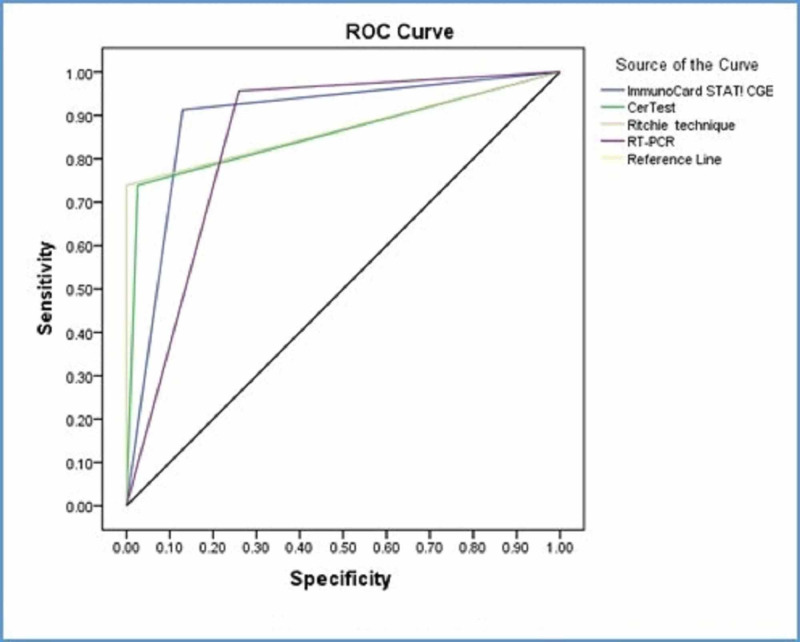
When direct smears assumed as a nominated golden standard, real-time PCR showed the highest sensitivity followed by ImmunoCard STAT! CGE and then the CerTest and Ritchie technique (Table 3). ImmunoCard STAT! CGE was excellent. The area under the curve is 0.892, Std. Error=0.041, p-value <0.0005 with 95% CI (0.812-0.972). CerTest was excellent. The area under the curve is 0.857, Std. Error=0.056, p-value<0.0005 with 95% CI (0.746-0.967). The Ritchie technique was excellent. The area under the curve is 0.870, Std. Error=0.056, p-value<0.0005 with 95% CI (0.760-0.979). Real-time PCR was excellent. The area under the curve is 0.848, Std. Error=0.041, p-value<0.0005 with 95% CI (0.768-0.929) (Figure 2).
Table 3. Sensitivity, specificity, and accuracy of diagnostic tools with direct smears.
N: Negative, P: Positive, TP: True-positive, FP: False-negative, TN: True-negative, FN: False-negative, PPV: Positive predictive value, NPV: Negative predictive value
ImmunoCard STAT!: Meridian Bioscience, Germany; CerTest: Biotec, Zaragoza, Spain
| Test | Direct smears | Sensitivity % | Specificity % | PPV % | NPV % | 95% CI | P value | ||
| P | N | ||||||||
| Immuno Card STAT! CGE | P | TP21 | FP10 | (91.3) | (87.01) | (67.74) | (97.10) | (0.81-0.98) | P < 0.001 |
| N | FN2 | TN67 | |||||||
| CerTest | P | TP17 | FP2 | (73.91) | (97.40) | (89.47) | (92.59) | (0.75-0.97) | P < 0.001 |
| N | FN6 | TN75 | |||||||
| Ritchie | P | TP17 | FP0 | (73.91) | (100) | (100) | (92.77) | (0.76-0.98) | P < 0.001 |
| N | FN6 | TN77 | |||||||
| Real-time PCR | P | TP22 | FP20 | (95.65) | (74.03) | (52.38) | (98.28) | (0.77-0.93) | P < 0.001 |
| N | FN1 | TN57 | |||||||
Figure 2. Sensitivity, specificity, and accuracy of diagnosis tools with direct smears.
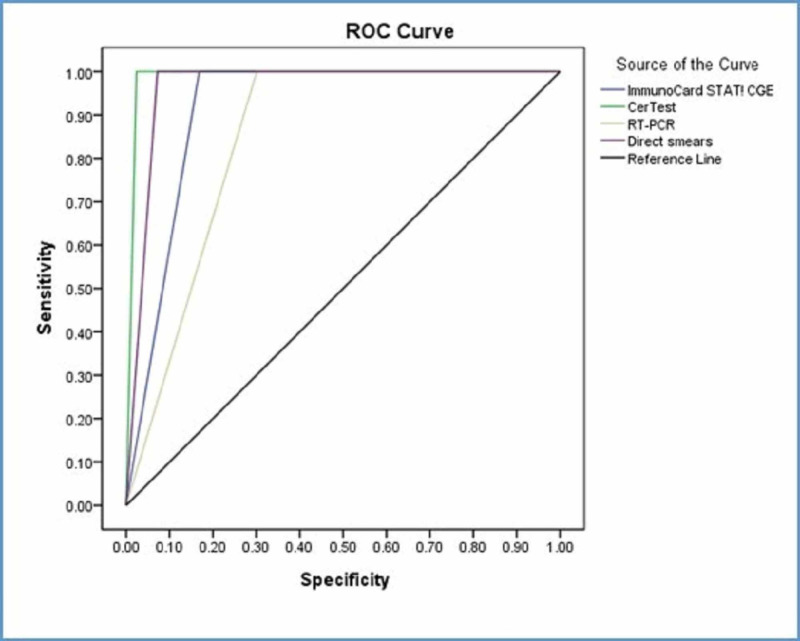
When the Ritchie technique considered as a nominated golden standard, all tests had (100%) sensitivity with the Ritchie technique (Table 4). ImmunoCard STAT! CGE was outstanding. The area under the curve is 0.916, Std. Error=0.027, p-value<0.0005 with 95% CI (0.862-0.969). CerTest was outstanding. The area under the curve is 0.988, Std. Error=0.010, p-value<0.0005 with 95% CI (0.969-1). Real-time PCR was excellent. The area under the curve is 0.849, Std. Error=0.038, p-value<0.0005 with 95% CI (0.776-0.923). Direct smears were outstanding. The area under the curve is 0.964, Std. Error=0.018, p-value<0.0005 with 95% CI (0.929-0.998) (Figure 3).
Table 4. Sensitivity, specificity, and accuracy of diagnostic tools with the Ritchie technique.
N: Negative, P: Positive, TP: True-positive, FP: False-negative, TN: True-negative, FN: False-negative, PPV: Positive predictive value, NPV: Negative predictive value
ImmunoCard STAT!: Meridian Bioscience, Germany; CerTest: Biotec, Zaragoza, Spain
| Test | Ritchie | Sensitivity % | Specificity % | PPV % | NPV % | 95% CI | P-value | ||
| P | N | ||||||||
| Immuno Card STAT! CGE | P | TP17 | FP14 | (100) | (83.13) | (54.84) | (100) | (0.86-0.97) | P < 0.001 |
| N | FN0 | TN69 | |||||||
| CerTest | P | TP17 | FP2 | (100) | (97.59) | (89.47) | (100) | (0.97-1.00) | P < 0.001 |
| N | FN0 | TN81 | |||||||
| Real-time PCR | P | TP17 | FP25 | (100) | (69.88) | (40.48) | (100) | (0.78-0.92) | P < 0.001 |
| N | FN0 | TN58 | |||||||
| Direct smears | P | TP17 | FP6 | (100) | (92.77) | (73.91) | (100) | (0.93-1.00) | P < 0.001 |
| N | FN0 | TN77 | |||||||
Figure 3. Sensitivity, specificity, and accuracy of diagnostic tools with the Ritchie technique.
Discussion
Intestinal parasitic infections are among the major public health problems in tropical and sub-tropical parts of the world, particularly in rural communities with poor sanitation and poor personal hygiene behaviors. The goal of our study was to compare the sensitivity and specificity of three diagnostic methods: light microscopy, ICTs, and real-time PCR for the detection of G. lamblia in clinical human fecal samples.
Light microscopy is still considered the reference standard access to the fecal parasitological laboratories but there is growing interest in alternative detection methods to overcome the current limitations of microscopic examination. We accomplished a microscopic examination by using direct smears, and the Ritchie technique. The Para-Pak trichrome stain was used to confirm the morphological features for trophozoites and cysts for protozoa parasites. Meanwhile, DNA detection by PCR is offering a better turnaround time and ICTs are commercially available and widely used in clinical laboratories, with limited capacities for diagnostic complexity [16].
Stool samples were collected from non-Saudi workers applying for a residence permit (Iqama). Non-Saudis represent around 30% of the population in Saudi Arabia [17]. All subjects submitted non-formed stool specimens ranging from soft to watery. Similar results were revealed by a study about the detection of gastrointestinal pathogens in migrant workers in Qatar [18].
Out of the 100 fecal samples analyzed for G. lamblia detection, 42 were found positive by real-time PCR, 31 by ImmunoCard STAT! CGE, 23 by direct smears, 19 by CerTest, and 17 by the Ritchie technique. In 2016, Ahmed and others [19] studied the prevalence of three recognized enteric protozoa; E. histolytica, G. lamblia, and Cryptosporidium spp. among children in Bangladesh. Regarding the variation in their study subject, the prevalence of giardiasis was (50.6%), (26.7%), and (3.3%) by using real-time PCR, ICTs, and direct microscopy respectively. The authors also observed that the positive samples by real-time PCR but negative by the other techniques showed higher Ct values due to the small number of parasites in those samples that fell below the other methods limit of detection. Our finding is in agreement with that observation. The variation in our results between microscopic examination using direct smears and the Ritchie technique is due to the ability of each technique for the detection of cyst and trophozoite. Direct smears detect both cyst and trophozoite stages while the Ritchie technique is applicable to the cyst stage only [10]. Using direct smears, we observed two cases with only trophozoites and one case with both trophozoites and cysts while none of the trophozoites was detected with the Ritchie technique.
A study conducted in Mozambique for the detection of intestinal parasites in fecal samples showed that the sensitivity of real-time PCR for the detection of each of the parasite species was higher than direct smears, Ritchie technique, Kato smear, Baermann, and coproculture [20]. The workers noticed that real-time PCR-positive samples, but negative during microscopy had higher Ct values (significantly lower DNA loads) than samples that were positive by both techniques. On the other hand, some samples were positive only by microscopy but negative by real-time PCR. Moreover, a recent study reported the detection of a low number of parasites by light microscopy but negative by PCR [21]. In our study, one sample was positive by routine microscopy and undetectable by real-time PCR; this was fairly in agreement with the above-mentioned studies. The microscopic examination and the DNA isolation usually use a small portion of feces so it might be in some instances that the parasite is observed by light microscopy but missed by PCR. This variation between techniques might be due to the dispersion of cysts within the subsampling due to the nonhomogeneous structure in some stool samples besides the reported non-uniform structure of the intestinal parasites' excretion in the stool [22].
Nominating real-time PCR as a gold standard, ImmunoCard STAT! CGE showed a higher sensitivity (59.5%) than both direct smears and the Ritchie technique (52.4% and 40.5%), respectively (Table 2). A study in Egypt revealed higher sensitivity for ICTs than microscopy when comparing the PCR technique detecting the 18S and tpi genes of giardiasis [23]. Rapid immunoassays kits provide sufficient sensitivities and specificities in direct detection, particularly when using a single stool sample [24].
Seven negative samples in real-time PCR were positive by other tests, therefore, they were suspected to be false-positive; six with ImmunoCard STAT! CGE and one with CerTest. The one giardiasis positive sample with CerTest could be due to cross-reactivity, as the specimen comprised other intestinal parasites, including E. histolytica. Interestingly, this sample was negative by direct smears as well. However, the suspected six false-positive samples in ImmunoCard STAT! CGE could be due to a cryptic giardiasis infection or the persistence of antigen shading after treatment rather than true cross-reactivity [25]. G. lamblia DNA can be found in feces after the parasite clearance and may occasionally produce false-positive PCR results. Moreover, real-time PCR and ICTs may generate false-negative results because of the intermittent patterns of cysts shedding and the number of stool samples required for analysis [26].
Using direct smears as the reference method, real-time PCR showed the highest sensitivity, followed by ImmunoCard STAT! CGE, then CerTest, and finally the Ritchie technique (95.7%, 91.3%, 73.9%, and 73.9%), respectively (Table 3). This finding contrasts with a study in Germany, as the two ICTs showed a sensitivity of 72.9% and 93.8%, respectively, while PCR showed a sensitivity of 85.4% [27]. This could be explained by the high concentration of DNase in the samples that inhibit the PCR diagnostics from this material.
Using the Ritchie technique as the reference method, the sensitivity for all the diagnostic tests has reached 100% while the specificities ranged from 69.9% to 97.6% (Table 4). Another study reported the low sensitivity of ImmunoCard STAT! CGE (65.3%) by using formol concentration as the golden standard for the detection of Giardia in fecal samples in Central Africa [28]. The low sensitivity of ICTs could be related to the non-specificity of the antibody-based method owing to cross-reactivity with other microorganisms. Meanwhile, a higher sensitivity of the ImmunoCard STAT! CGE (93.5%) was observed in a previous study [29]. A similar observation was reported after using saturated sodium nitrate flotation as the golden standard for the detection of G. lamblia [30], and the sensitivities of the PCR and the immunofluorescence assay were 97.3% and 91.9%, respectively, while the specificity of both was 100%.
Conclusions
In conclusion, with some variations, all the investigated techniques were applicable for the detection of G. lamblia in stool samples. The real-time PCR assay was revealed as the most accurate diagnosis of giardiasis. ICTs proved to be practical, easy to perform, time-saving, and beneficial to analyze a low number of samples. It can be a valuable diagnostic tool in case microscopic expertise is limited. Wet mount microscopy and the Ritchie technique are still important methods for the detection of gastrointestinal parasites when a skilled technologist tests the samples.
Acknowledgments
The authors would like to thank the Ministry of Health, Directorate of Health Affairs in Jeddah, for granting ethical approval and help in the collections of samples. This work was supported by funds from King Abdulaziz City for Science and Technology (37-1198). Thanks are also extended to the Faculty of Science/Department of Biology, the Faculty of Applied Medical Sciences/Department of Medical Laboratory Sciences, and finally to the Special Infectious Agents Unit, King Fahd Medical Research Center, KAU, for providing all the necessary facilities and support.
The content published in Cureus is the result of clinical experience and/or research by independent individuals or organizations. Cureus is not responsible for the scientific accuracy or reliability of data or conclusions published herein. All content published within Cureus is intended only for educational, research and reference purposes. Additionally, articles published within Cureus should not be deemed a suitable substitute for the advice of a qualified health care professional. Do not disregard or avoid professional medical advice due to content published within Cureus.
The authors have declared that no competing interests exist.
Human Ethics
Consent was obtained by all participants in this study. Saudi Ministry of Health, Directorate of Health Affairs in Jeddah issued approval A00366. Consent was obtained by all participants in this study. The Ethics and Research Committee of the Directorate of Health Affairs, Ministry of Health, Jeddah, Saudi Arabia, issued approval A00366
Animal Ethics
Animal subjects: All authors have confirmed that this study did not involve animal subjects or tissue.
References
- 1.Giardia and Vilém Dušan Lambl. Lipoldová M. PLoS Negl Trop Dis. 2014;8:0. doi: 10.1371/journal.pntd.0002686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Giardiasis: an update review on sensitivity and specificity of methods for laboratorial diagnosis. Soares R, Tasca T. J Microbiol Methods. 2016;129:98–102. doi: 10.1016/j.mimet.2016.08.017. [DOI] [PubMed] [Google Scholar]
- 3.Giardia and Cryptosporidium join the 'neglected diseases initiative'. Savioli L, Smith H, Thompson A. Trends Parasitol. 2006;22:203–208. doi: 10.1016/j.pt.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 4.Advances in understanding Giardia: determinants and mechanisms of chronic sequelae. Bartelt LA, Sartor RB. F1000Prime Rep. 2015;7:62. doi: 10.12703/P7-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Management of chronic Giardia infection. Escobedo AA, Hanevik K, Almirall P, Cimerman S, Alfonso M. Expert Rev Anti Infect Ther. 2014;12:1143–1157. doi: 10.1586/14787210.2014.942283. [DOI] [PubMed] [Google Scholar]
- 6.Prevalence of intestinal parasites among expatriate workers in Al-Khobar, Saudi Arabia. Abahussain N. https://pdfs.semanticscholar.org/bc13/96acb6b2140d3bc0a8cd00d467d0c450f590.pdf Middle East J Fam Med. 2005;3:17–21. [Google Scholar]
- 7.Distribution of intestinal parasites among food handlers in Jeddah, Saudi Arabia. Wakid MH. https://www.semanticscholar.org/paper/04fa56a073e2a1b545c7d45e68b6a698398c3e4d J Parasit Dis. 2006;30:146–152. [Google Scholar]
- 8.Intestinal Parasitic infection among food handlers in the Holy City of Makkah during Hajj season: 1428 Hegira (2007G) Wakid MH, Azhar E, Zafar TA. J King Saud Univ Sci. 2009;16:39–52. [Google Scholar]
- 9.The prevalence of intestinal parasitic infections among foreign workers in Madinah, Kingdom of Saudi Arabia. Imam NF, Abdulbaqi ZB, Fahad RA. Saudi J Med Med Sci. 2015;3:112–117. [Google Scholar]
- 10.Giardiasis in man: review and updates. Shalaby NM, Wakid MH. JKAU Med Sci. 2014;21:81–94. [Google Scholar]
- 11.Comparison of five diagnostic tests for Giardia duodenalis in fecal samples from young dogs. Uehlinger FD, Naqvi SA, Greenwood SJ, McClure JT, Conboy G, O’Handley R, Barkemae HW. Vet Parasitol. 2017;244:91–96. doi: 10.1016/j.vetpar.2017.07.030. [DOI] [PubMed] [Google Scholar]
- 12.A guide to aid the selection of diagnostic tests. Kosack CS, Page AL, Klatser PR. Bull World Health Organ. 2017;95:639–645. doi: 10.2471/BLT.16.187468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.A novel, multi-parallel, real-time polymerase chain reaction approach for eight gastrointestinal parasites provides improved diagnostic capabilities to resource-limited at-risk populations. Mejia R, Vicuña Y, Broncano N, et al. Am J Trop Med Hyg. 2013;88:1041–1047. doi: 10.4269/ajtmh.12-0726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Improvement of Ritchie technique by identifying the food that can be consumed pre-analysis. Wakid MH. https://kfmrc.kau.edu.sa/Files/0030238/Researches/49153_27274.pdf J Appl Sc Res. 2009;5:293–296. [Google Scholar]
- 15.Real-time PCR for the detection of Giardia lamblia. Verweij JJ, Schinkel J, Laeijendecker D, van Rooyen M A, van Lieshout L, Polderman AM. Mol Cell Probes. 2003;17:223–225. doi: 10.1016/s0890-8508(03)00057-4. [DOI] [PubMed] [Google Scholar]
- 16.Detection of intestinal protozoa in the clinical laboratory. Mchardy IH, Wu M, Shimizu-Cohen R, Couturier MR, Humphries RM. J Clin Microbiol. 2014;52:712–720. doi: 10.1128/JCM.02877-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saudi Arabia’s population statistics of 2019. [Aug;2019 ];https://www.globalmediainsight.com/blog/saudi-arabia-population-statistics/ 2019
- 18.Multiplex polymerase chain reaction for detection of gastrointestinal pathogens in migrant workers in Qatar. Humphrey JM, Ranbhise S, Ibrahim E, Al-Romaihi HE, Farag E, Abu-Raddad LJ, Glesby MJ. Am J Trop Med Hyg. 2016;95:1330–1337. doi: 10.4269/ajtmh.16-0464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Entamoeba histolytica, Giardia lamblia and Cryptosporidium spp. infection in children in an urban slum area of Bangladesh. Ahmed T, Khanum H, Uddin MS, Barua P, Arju T, Kabir M, Haque R. http://bioresearchcommunications.com/index.php/brc/article/view/69/63 Biores Commun. 2016;2:175–181. [Google Scholar]
- 20.Diagnosing polyparasitism in a high-prevalence setting in Beira, Mozambique: detection of intestinal parasites in fecal samples by microscopy and real-time PCR. Meurs L, Polderman AM, Melchers NVV, et al. PLoS Negl Trop Dis. 2017;11:5310. doi: 10.1371/journal.pntd.0005310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Application of a multiplex quantitative PCR to assess prevalence and intensity of intestinal parasite infections in a controlled clinical trial. Llewellyn S, Inpankaew T, Nery SV, et al. PLoS Negl Trop Dis. 2016;10:4380. doi: 10.1371/journal.pntd.0004380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Consistency of direct microscopic examination and ELISA in detection of Giardia in stool specimen among children. Torabi Z, Niksirat A, Mazloomzadeh S, Ahmadiafshar A. Asian Pac J Trop Dis. 2014;4:725–727. [Google Scholar]
- 23.Molecular detection of giardiasis among children at Cairo university pediatrics hospitals. Ghieth MA, Kotb MA, Abu-Sarea EY, El-Badry AA. J Parasit Dis. 2016;40:1470–1474. doi: 10.1007/s12639-015-0714-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Evaluation of an immunochromatographic dip strip test for simultaneous detection of Cryptosporidium spp, Giardia duodenalis, and Entamoeba histolytica antigens in human faecal samples. Goni P, Martin B, Villacampa M, García A, Seral C, Castillo FJ, Clavel A. Eur J Clin Microbiol Infect Dis. 2012;31:2077–2082. doi: 10.1007/s10096-012-1544-7. [DOI] [PubMed] [Google Scholar]
- 25.Stool diagnosis of giardiasis using a commercially available enzyme immunoassay to detect Giardia-specific antigen 65 (GSA 65) Rosoff JD, Sanders CA, Sonnad SS, et al. J Clin Microbiol. 1989;27:1997–2002. doi: 10.1128/jcm.27.9.1997-2002.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Comparison of microscopy, real-time PCR and a rapid immunoassay for the detection of in human stool specimens. Schuurman T, Lankamp P, Van Belkum A, Kooistra‐Smid M, Van Zwet A. Clin Microbiol Infect. 2007;13:1186–1191. doi: 10.1111/j.1469-0691.2007.01836.x. [DOI] [PubMed] [Google Scholar]
- 27.Detection of Giardia lamblia stool samples: a comparison of two enzyme-linked immunosorbent assays. Jelinek T, Neifer S. F1000Res. 2013;39:2–7. [Google Scholar]
- 28.Low sensitivity of the immunocardSTAT® Crypto/Giardia rapid assay test for the detection of Giardia and Cryptosporidium in fecal samples from children living in Libreville, Central Africa. Bouyou-Akotet MK, Owono-Medang M, Moussavou-Boussougou MN, Mamfoumbi MM, Mintsa-Nguema R, Mawili-Mboumba DP, Kombila M. J Parasit Dis. 2016;40:1179–1183. doi: 10.1007/s12639-015-0645-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Commercial assay for detection of Giardia lamblia and Cryptosporidium parvum antigens in human fecal specimens by rapid solid-phase qualitative immunochromatography. Garcia LS, Shimizu RY, Novak S, Carroll M, Chan F. J Clin Microbiol. 2003;41:209–212. doi: 10.1128/JCM.41.1.209-212.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Evaluation of the sensitivities of DNA extraction and PCR methods for detection of Giardia duodenalis in stool specimens. Nantavisai K, Mungthin M, Tan-ariya P, Rangsin R, Naaglor T, Leelayoova S. J Clin Microbiol. 2007;45:581–583. doi: 10.1128/JCM.01823-06. [DOI] [PMC free article] [PubMed] [Google Scholar]