ABSTRACT
Teleost zebrafish and neonatal mammalian hearts exhibit the remarkable capacity to regenerate through dedifferentiation and proliferation of pre-existing cardiomyocytes (CMs). Although many mitogenic signals that stimulate zebrafish heart regeneration have been identified, transcriptional programs that restrain injury-induced CM renewal are incompletely understood. Here, we report that mutations in gridlock (grl; also known as hey2), encoding a Hairy-related basic helix-loop-helix transcriptional repressor, enhance CM proliferation and reduce fibrosis following damage. In contrast, myocardial grl induction blunts CM dedifferentiation and regenerative responses to heart injury. RNA sequencing analyses uncover Smyd2 lysine methyltransferase (KMT) as a key transcriptional target repressed by Grl. Reduction in Grl protein levels triggered by injury induces smyd2 expression at the wound myocardium, enhancing CM proliferation. We show that Smyd2 functions as a methyltransferase and modulates the Stat3 methylation and phosphorylation activity. Inhibition of the KMT activity of Smyd2 reduces phosphorylated Stat3 at cardiac wounds, suppressing the elevated CM proliferation in injured grl mutant hearts. Our findings establish an injury-specific transcriptional repression program in governing CM renewal during heart regeneration, providing a potential strategy whereby silencing Grl repression at local regions might empower regeneration capacity to the injured mammalian heart.
KEY WORDS: Grl, Hey2, Heart regeneration, Cardiomyocyte proliferation, Lysine methyltransferase, Zebrafish
Highlighted Article: Novel mechanisms of the Grl-Smyd2 network govern vertebrate CM renewal and heart regeneration, which might be relevant in developing strategies for regeneration interventions in humans.
INTRODUCTION
The human heart has limited capacity to regenerate new cardiac muscle after myocardial injury. Instead, impaired myocardium is replaced by fibrotic scar, eventually leading to heart failure (Tzahor and Poss, 2017; Xin et al., 2013). By contrast, zebrafish heart possesses natural regeneration capacity by dedifferentiation and proliferation of pre-existing cardiomyocytes (CMs) after injury (Jopling et al., 2010; Kikuchi et al., 2010; Li et al., 2015). Hearts of mice can regenerate if injured in the few days after birth, which coincides with the transient capacity for CM proliferation (Porrello et al., 2011). The limited regenerative potential in the adult mammalian heart can be ascribed to cell-intrinsic barriers or molecular blocks that prevent CMs from entering proliferation and completing cytokinesis after damage (González-Rosa et al., 2018; Li et al., 2015; Tzahor and Poss, 2017). Various mitogenic factors and signaling pathways have been identified as initiating and achieving heart regeneration (Fang et al., 2013; Han et al., 2019; Harrison et al., 2019; Kikuchi et al., 2011; Marin-Juez et al., 2016; Mohamed et al., 2018; Monroe et al., 2019; Wu et al., 2016; Zhao et al., 2019). However, transcriptional programs and epigenetic signaling that serve as roadblocks to limit injury-induced CM renewal are incompletely understood.
Gridlock (Grl; also known as Hey2) is a basic helix-loop-helix (bHLH) transcriptional repressor belonging to the Hesr (Hairy/Enhancer-of-split related) family (also known as Hey or Hrt family) containing Hey1, Grl and HeyL, which play important functions via diverse mechanisms involving vascular, myocardial, endocardial and neuronal tissues during development (Fischer et al., 2002; Nakagawa et al., 1999; Sakamoto et al., 2003; Zhong et al., 2000). Hey proteins mediate transcriptional repression by directly binding an E-box DNA motif, and can repress GATA-mediated transcription via physically interacting with GATA factors (Heisig et al., 2012; Jia et al., 2007; Kathiriya et al., 2004). We and others previously reported that Grl and Hey1 regulate arterial-venous differentiation in zebrafish and mice (Fischer et al., 2004; Zhong et al., 2001). Grl also contributes to zebrafish heart development by limiting expansion of cardiac progenitor cells (CPC) and embryonic CMs (Gibb et al., 2018; Jia et al., 2007). During cardiovascular development, Hey family genes function in response to Notch signaling, including atrioventricular boundary formation, arterial-venous differentiation and possible trabecular specification (Fischer et al., 2004; Koibuchi and Chin, 2007; Kokubo et al., 2007; Miao et al., 2018; Rutenberg et al., 2006; Tian et al., 2017; Xin et al., 2007; Zhong et al., 2001). In humans, HEY2 variants are associated with Brugada syndrome, a rare disease with high risk of sudden cardiac death (Bezzina et al., 2013). Although Hey proteins play pivotal roles during cardiovascular development and potential human disease, their functions in heart regeneration have not been investigated and remain unknown.
Lysine methylation is a widespread post-translational modification (Biggar and Li, 2015; Black et al., 2012). The Smyd family (Smyd1-Smyd5), containing SET and MYND domains, represents a novel class of lysine methyltransferases (KMTs) with a dual function in histone and non-histone methylation (Spellmon et al., 2015). Smyd2 methylates H3K4 to promote gene transcriptional activation, and its histone methyltransferase activity can be enhanced by interaction with HSP90α (Abu-Farha et al., 2008). By contrast, trimethylation of histone H4K20 by Smyd5 represses gene expression (Stender et al., 2012). Importantly, Smyd2 can methylate non-histone substrates during cell cycle progression, cell survival, apoptosis and other events (Gao et al., 2017; Li et al., 2017; Saddic et al., 2010).
In this study, we reveal cell-autonomous effects of Grl on regulation of myocardial regeneration using loss-of-function and conditional gain-of-function studies. Grl functions as a negative regulator by restraining injury-induced CM dedifferentiation and proliferation. We identify KMT Smyd2 upregulation at the injured myocardium as a direct target of Grl. Smyd2 induction, triggered by injury-induced Grl reduction, is required for CM proliferation during regeneration. Our findings reveal novel roles and mechanisms of the Grl-Smyd2 network in governing vertebrate CM renewal and heart regeneration.
RESULTS
Cardiac injury triggers a grl dynamic expression pattern
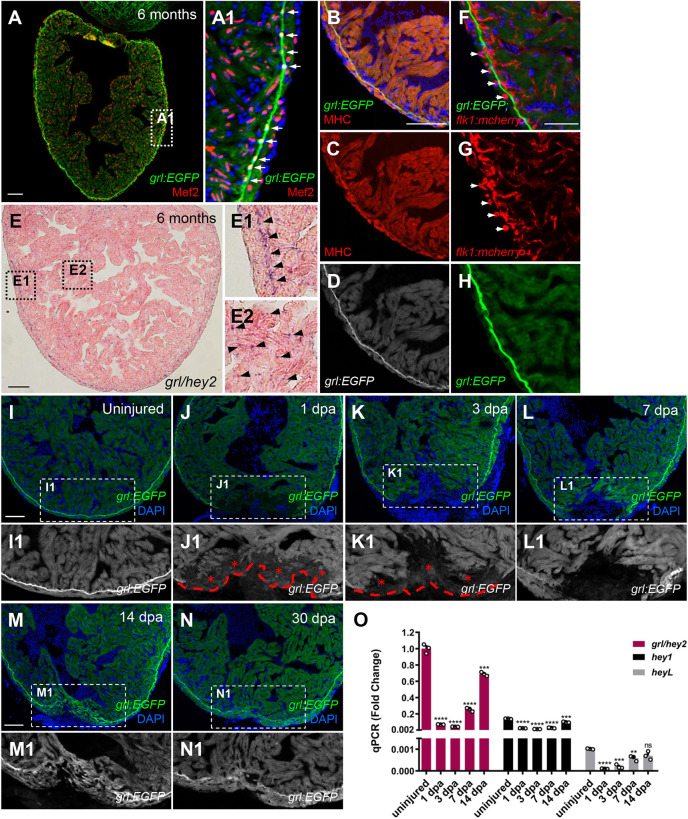
To define the spatiotemporal expression pattern of grl during adult heart regeneration, we generated grl reporter lines Tg(grl:EGFP) and Tg(grl:mCherry) in zebrafish, where EGFP and mCherry were driven by grl promoter-based upstream region. In the adult heart, grl:EGFP expression was detectable throughout the myocardium labeled by myocyte enhance factor 2 (Mef2) and enriched in the primordial myocardial layer (PML) (Fig. 1A,A1; Fig. S1A,B). grl:EGFP was also overlapped with myosin heavy chain (MHC) that marked the CM cytoplasm (Fig. 1B-D). In situ hybridization (ISH) analyses validated grl expression throughout the myocardium, with enrichment in the PML (Fig. 1E). Although grl was expressed in the arterial endothelium of aortae during embryogenesis (Jia et al., 2007; Li et al., 2020; Rowlinson and Gering, 2010; Satow et al., 2001; Zhong et al., 2000), grl:EGFP was absent in the endocardium or coronary vascular endothelium lining with flk1:mCherry expression in the adult heart (Fig. 1F-H). Furthermore, Tg(grl:mCherry) transgenic hearts displayed no colocalization of mCherry with the epicardial marker tcf21:nucEGFP (Fig. S1C). grl:EGFP was also absent in the epicardium marked by injury-induced Raldh2 protein (Fig. S1D). These findings indicate that the myocardium is the primary source of Grl under homeostatic conditions.
Fig. 1.
Reduction of myocardial grl correlates with regenerative responses of the zebrafish heart to injury. (A-D) grl:EGFP (green or white) overlaps with a nuclear CM marker Mef2 (red) (A) and a cytoplasmic CM marker MHC (red) (B-D). (A1) Enlarged image of the dashed box in A. Arrows indicate the grl-enriched primordial layer (PML). (E) ISH for grl displays enriched expression in the PML and its expression throughout the myocardium in adult zebrafish hearts. (E1) Higher-magnification image of the dashed box in E; arrowheads point to the grl-enriched PML. (E2) Enlarged image of the dashed box in E; arrowheads point to grl expression in the myocardium. (F-H) grl:EGFP (green) does not colocalize with endocardial or coronary endothelial cells marked by flk1:mCherry (red) in adult Tg(grl:EGFP;flk1:mCherry) hearts. Arrows indicate the circular coronary vessels. (I-N) Tg(grl:EGFP) adult heart show grl:EGFP (green) expression in uninjured and regenerating ventricles. (I1-N1) Higher-magnification images of the dashed boxes in I-N. Red dashed line indicates approximate plane of resection. Red asterisks mark decreased expression of grl:EGFP in the injury border zone. (O) Expression of grl, hey1, and heyL were examined using qPCR analyses in uninjured and regeneration ventricular samples. Expression levels were normalized to that of β-actin and further normalized to that of grl in uninjured sample (n=3). Data presents as mean±s.e.m. **P<0.01, ***P<0.001, ****P<0.0001, Student's t-test (unpaired, two-tailed). Scale bars: 100 µm (A-E, I-N); 50 µm (F-H).
To explore the injury response of grl during zebrafish heart regeneration, we performed ventricular apex resection using Tg(grl:EGFP) animals and investigated grl temporal expression profiles from injury onset until 30 days post amputation (dpa). We observed that EGFP fluorescence was reduced at the apical edge of the injured myocardium labeled by Mef2 at 1 dpa (Fig. 1J,J1; Fig. S1I,J) and declined to the lowest level at 3 dpa (Fig. 1K,K1), compared with uninjured hearts (Fig. 1I,I1). Thereafter, grl:EGFP increased to some extent at the injured myocardial cell edge at 7 dpa (Fig. 1L,L1;M,M1) and gradually returned to the uninjured level with formation of the PML from 14 dpa to 30 dpa (Fig. 1N,N1). Consistent with grl:EGFP expression, quantitative PCR (qPCR) analyses indicated a great reduction in grl expression at 1 dpa, which reduced to the lowest level at 3 dpa (Fig. 1O). Although grl expression increased to some extent at 7 dpa, it was still reduced by 75% compared with the uninjured level (Fig. 1O). Reduced expression of other Hey family genes, hey1 and heyL, was also detectable by qPCR. Expression of heyL was much lower than that of hey1, despite extremely low expression of both genes (Fig. 1O). We noticed that the increased level of grl:EGFP transgene at 7 dpa seemed to be much higher than that in grl transcripts measured by qPCR (Fig. 1L,O). These differences might be because the 7.8 kb upstream region in the grl:EGFP transgene does not fully reflect the endogenous grl transcripts during regeneration. Indeed, our ISH analyses revealed a discernible increase in grl transcripts at the injury site at 14 dpa rather than 7 dpa (Fig. S1G,H), consistent with qPCR analyses. Collectively, our findings indicate that the reduced expression of myocardial grl correlates with the regenerative responses of the zebrafish heart to injury.
Mutations in grl augment heart regeneration by reducing fibrotic scars and enhancing CM proliferation
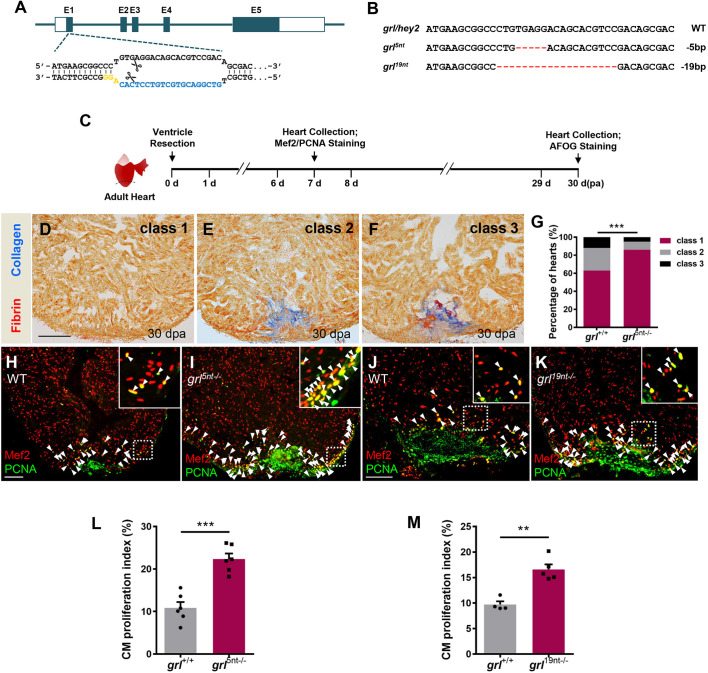
To investigate the effects of grl loss of function on heart regeneration, we generated zebrafish nonsense mutations in grl using the CRISPR/Cas9 technique, as a previous grlm145 mutant caused a point mutation of the terminator codon that is predicted to produce an extended protein with some residue activity (Zhong et al., 2000). Single guide RNA (sgRNA) was designed to target the first exon of grl (Fig. 2A). Two grl deletion mutations, grl5nt−/− mutants with a 5-nuleotide deletion and grl19nt−/− mutants with a 19-nuleotide deletion, were identified (Fig. 2B). These mutants were predicted to produce a premature stop codon and encode a truncated peptide containing 4 amino acids in grl5nt−/− mutants or 29 amino acids in grl19nt−/− mutants, both of which lack the bHLH, Orange and YRPW domains (Fig. S2A). Some mutants survived to adulthood and were fertile. Considering that grl5nt−/− is almost a null mutation, we chose grl5nt−/− mutants to test the effects of grl loss of function on heart regeneration (Fig. S2B). Hearts from 5-month-old grl5nt−/− mutants were subjected to ventricular resection and then assayed for fibrin and fibrotic scar tissue using acid fuchsin-orange G (AFOG) staining at 30 dpa (Fig. 2C). We observed that more grl5nt−/− mutant hearts contained large cardiac myofiber deposits and minimal fibrin or collagen deposits than injured wild-type (WT) sibling hearts in wound regions at 30 dpa (Fig. 2D-G). Therefore, the grl mutant enhances cardiac muscle regeneration and reduces fibrotic scarring.
Fig. 2.
Mutations in Grl lead to enhanced CM proliferation and reduced fibrotic scar tissue following injury. (A) The sgRNA target sequence of the grl allele (blue) and the PAM (yellow) designed in the first exon of grl for mutation generation. (B) Targeted deletion mutations induced by CRISPR/Cas9 technique at the grl genes. The WT sequence is shown at the top. Deletions are shown as red dashes. The mutation deletion is indicated at the right of each sequence. (C) Experimental design for PCNA and Mef2 immunostaining and fibrotic scar (AFOG) analysis. (D-F) Representative AFOG staining images (blue for collagen, red for fibrin) of injured ventricles from WT sibling and grl5nt−/− fish at 30 dpa, scored as ‘class 1’ (complete regeneration) (D), ‘class 2’ (partial regeneration) (E) and ‘class 3’ (blockade in regeneration) (F). (G) Quantification of regenerative status of ventricles from WT sibling fish (n=6, sections=194) and grl5nt−/− fish (n=8, sections=288) at 30 dpa. Heart sections were scored according to the criteria described in Materials and Methods. Histograms show the percentage of heart regeneration represented by each score for each group. ***P<0.001, Chi-square test. (H-K) Section images of injured ventricles from WT (H) and mutant grl5nt−/− fish (I), as well as WT (J) and grl19nt−/− fish (K) at 7 dpa, stained with anti-PCNA (green) and anti-Mef2 (red) antibodies. Insets show higher-magnification images of the dashed boxes. Arrowheads indicate proliferating CMs. (L) Quantification of CM proliferation indices in 7 dpa ventricles of grl+/+ (n=6) and grl5nt−/− fish (n=6). ***P<0.001, Student's t-test (unpaired, two-tailed). (M) Quantification of CM proliferation indices in 7 dpa ventricles of grl+/+ (n=4) and grl19nt−/− fish (n=5). **P<0.01, Student's t-test (unpaired, two-tailed). Data presents as mean±s.e.m. Scale bars: 100 µm.
We next assessed injury-induced CM proliferation in grl5nt−/− mutant hearts by conducting immunostaining of the DNA replication marker proliferating cell nuclear antigen (PCNA) and CM marker Mef2 following ventricular resection (Fig. 2C). We observed that PCNA-positive CMs were markedly increased in grl5nt−/− hearts in comparison to injured WT sibling hearts (Fig. 2H,I). Quantification revealed that the CM proliferation index (number of PCNA+Mef2+ cells/number of Mef2+ cells) was significantly increased by 105% in grl5nt−/− mutant hearts compared with control hearts (Fig. 2L). Similarly, in grl19nt−/− mutants the CM proliferation index was elevated by 70% over control hearts (Fig. 2J,K,M). In contrast, the coronary vasculature was not impaired in grl5nt−/− mutant hearts (Fig. S2C,D). Collectively, these findings demonstrate that mutations in grl enhance injury-induced CM proliferation.
Myocardial-specific grl induction in the adult heart impairs injury-induced CM dedifferentiation and proliferation
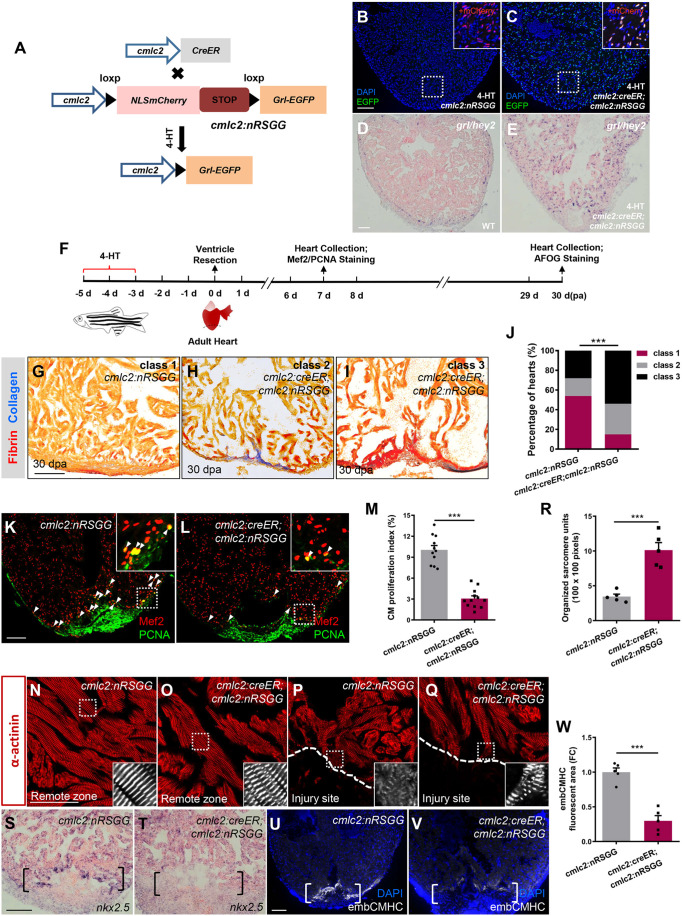
To define the potential contribution of grl gain of function during heart regeneration, we created a transgenic strain of zebrafish that enabled inducible expression of grl in CMs [Tg(cmlc2:loxP-nlsmCherry-STOP-loxP-grl-EGFP), hereafter referred to as Tg(cmlc2:nRSGG)] (Fig. 3A). We crossed Tg(cmlc2:nRSGG) zebrafish with Tg(cmlc2:CreER) animals, in which Cre recombinase fused with estrogen receptor (ER) is under the control of the cmlc2 promoter, permitting Cre-mediated recombination in CMs after 4-hydroxytamoxifen (4-HT) treatment (Kikuchi et al., 2010) (Fig. 3A). To test whether the inducible system works, we administrated 4-HT to Tg(cmlc2:CreER;cmlc2:nRSGG) and Tg(cmlc2:nRSGG) control animals aged 5 months. Following 4-HT treatment, Grl-EGFP signals were detectable in CM nuclei, which overlapped with nucleus-localized mCherry proteins, in Tg(cmlc2:CreER;cmlc2:nRSGG) hearts (Fig. 3C). In contrast, control Tg(cmlc2:nRSGG) hearts exhibited only mCherry nuclear signals in CMs (Fig. 3B). ISH analyses also validated abundant grl transcripts over the whole myocardium in Tg(cmlc2:CreER;cmlc2:nRSGG) hearts compared with controls (Fig. 3D,E).
Fig. 3.
Conditional grl induction in the adult myocardium impairs CM dedifferentiation and proliferation during regeneration. (A) Transgenic zebrafish used for inducible expression of Grl-EGFP in CMs. Tg(cmlc2:nRSGG) zebrafish were crossed with Tg(cmlc2:CreER) animals, which permitted Cre-mediated recombination to induce Grl in CMs after 5 µM 4-HT treatment. (B,C) Grl was induced in CMs of 4-HT-treated Tg(cmlc2:creER;cmlc2:nRSGG) hearts (C) but not Tg(cmlc2:nRSGG) hearts (B), as indicated by EGFP protein (green). Insets show higher-magnification images of the dashed boxes adding mCherry channel (red). (D,E) ISH analyses indicate the endogenous expression of grl in WT hearts (D) and the induced expression of grl in CMs of the whole ventricle in 4-HT-treated Tg(cmlc2:creER;cmlc2:nRSGG) animals (E) at 7 dpa. (F) Experimental design for PCNA and Mef2 immunostaining and fibrotic scar (AFOG) analysis after ventricular resection. 4-HT treatment was 5 µM 4-HT for bath treatment for 2 days. (G-I) Section images of 30 dpa ventricles of 4-HT-treated Tg(cmlc2:nRSGG) control fish (G) and 4-HT-treated Tg(cmlc2:creER;cmlc2:nRSGG) animals (H,I) stained with AFOG (blue for collagen, red for fibrin). Heart sections were scored as ‘class 1’, ‘class 2’ or ‘class 3’ according to the criteria described in Materials and Methods. (J) Quantification of regenerative status of ventricles in 4-HT-treated Tg(cmlc2:creER;cmlc2:nRSGG) (n=6; 129 sections) and Tg(cmlc2:nRSGG) control group (n=4; 84 sections) at 30 dpa. Histograms show the percentage of wounded hearts represented by each score for each group. ***P<0.001, Chi-square test. (K,L) Immunofluorescent section images of injured ventricles from 4-HT-treated Tg(cmlc2:nRSGG) control fish (K) and 4-HT-treated Tg(cmlc2:creER;cmlc2:nRSGG) animals (L) at 7 dpa, stained with anti-PCNA (green) and anti-Mef2 (red) antibodies. Insets show higher-magnification images of the dashed boxes. Arrowheads indicate proliferating CMs. (M) Quantification of CM proliferation indices in 7 dpa ventricles; n=11 (cmlc2:nRSGG), n=12 (cmlc2:creER;cmlc2:nRSGG); ***P<0.001, Student's t-test (unpaired, two-tailed). (N-Q) Confocal images of sections of injured ventricles from 4-HT-treated Tg(cmlc2:nRSGG) control fish (N,P) and 4-HT-treated Tg(cmlc2:creER;cmlc2:nRSGG) animals (O,Q) at 7 dpa, stained with anti-α-actinin antibody, showing regions of the remote zone (N and O) and injury site (P and Q). Insets show higher-magnification images of the dashed boxes. Dashed line indicates the apical edge of the regeneration. (R) Quantification of organized sarcomere units in α-actinin-labeled myocardial tissue (100×100 pixels) in injury border zone of 7 dpa ventricles from 4-HT-treated Tg(cmlc2:nRSGG) control fish and 4-HT-treated Tg(cmlc2:creER;cmlc2:nRSGG) animals (n=5); ***P<0.001, Student's t-test (unpaired, two-tailed). (S,T) ISH analyses of nkx2.5 expression in injured ventricles from 4-HT-treated Tg(cmlc2:nRSGG) control fish (S) and 4-HT-treated Tg(cmlc2:creER;cmlc2:nRSGG) animals (T) at 7 dpa (n=5). Brackets indicate injury site. (U,V) Fluorescent images of injured ventricles from 4-HT-treated Tg(cmlc2:nRSGG) control fish (U) and 4-HT-treated Tg(cmlc2:creER;cmlc2:nRSGG) animals (V) at 7 dpa, stained with embCMHC antibody (white). Brackets indicate injury site. (W) Quantification of embCMHC fluorescent area in injury border zone of 7 dpa ventricles from 4-HT-treated Tg(cmlc2:nRSGG) control fish and 4-HT-treated Tg(cmlc2:creER;cmlc2:nRSGG) animals. Data are relative to area in 4-HT-treated Tg(cmlc2:nRSGG) group (n=5); ***P<0.001, Student's t-test (unpaired, two-tailed). Data presents as mean±s.e.m. Scale bars: 100 µm (B-E,G-I,K,L,S-V); 50 µm (N-Q).
Because grl deficiency enhances cardiac muscle regeneration, we tested whether inducible grl overexpression would have an opposite effect. We treated Tg(cmlc2:CreER;cmlc2:nRSGG) and Tg(cmlc2:nRSGG) control animals with 4-HT, and performed ventricular resections and fibrin and collagen assays at 30 dpa (Fig. 3F). Although the majority of 4-HT-treated control hearts largely contained cardiac muscle with minimal fibrin and fibrotic scar tissue (Fig. 3G,J), myocardial grl induction hearts displayed excessive fibrin and collagen deposits (Fig. 3H-J). Furthermore, CMs in grl-overexpressing hearts appeared to be sparsely distributed (Fig. 3H,I). We next assessed the status of CM proliferation in myocardial grl-overexpressing hearts following injury. Resected hearts were isolated from 4-HT-treated control and Tg(cmlc2:CreER;cmlc2:nRSGG) animals and assayed for PCNA and Mef2 immunostaining at 7 dpa (Fig. 3F). We observed that PCNA-positive CMs were markedly reduced in myocardial grl-overexpressing hearts compared with control hearts (Fig. 3K,L). The CM proliferation index was significantly decreased by 69% in Tg(cmlc2:CreER;cmlc2:nRSGG) hearts in comparison with control hearts (Fig. 3M). Thus, inducible grl overexpression in adult CMs reduces injury-induced CM proliferation.
Regenerating CMs normally undergo dedifferentiation, which is characterized by reactivation of cardiac embryonic or fetal genes as well as less organized sarcomeres in the myocardial injury region (D'Uva et al., 2015; Jopling et al., 2010; Kikuchi et al., 2010). We tested whether CM dedifferentiation might be regulated by Grl. In the region remote from injury sites, myofibrils organize in regular sarcomere units exhibiting cross-striations revealed by α-actinin in both conditional grl-overexpressing and control hearts (Fig. 3N,O). At the wound edge, control Tg(cmlc2:nRSGG) hearts exhibited disassembled cytoplasmic sarcomeres, indicative of CM dedifferentiation (Fig. 3P,R) (Jopling et al., 2010). In contrast, induced grl-overexpressing hearts displayed organized sarcomeres at cardiac wounds, suggesting a reduction in dedifferentiation (Fig. 3Q,R). Notably, expression of the nkx2.5 progenitor marker and embryonic-specific cardiac myosin heavy chains (embCMHC) were markedly reduced at injury sites in myocardial grl-overexpressing hearts compared with control hearts at 7 dpa (Fig. 3S-W). Conversely, expression of nkx2.5 and embCMHC were increased at wound edges in grl mutant hearts compared with control hearts (Fig. S3A-E). Moreover, grl5nt−/− mutant hearts exhibited conspicuously disassembled sarcomeres in comparison with control hearts (Fig. S3F-H). Collectively, these findings indicate that Grl reduces the dedifferentiation tendency of CMs during regeneration.
Grl repressor negatively regulates transcripts encoding lysine methyltransferase Smyd2 in the regenerating myocardium
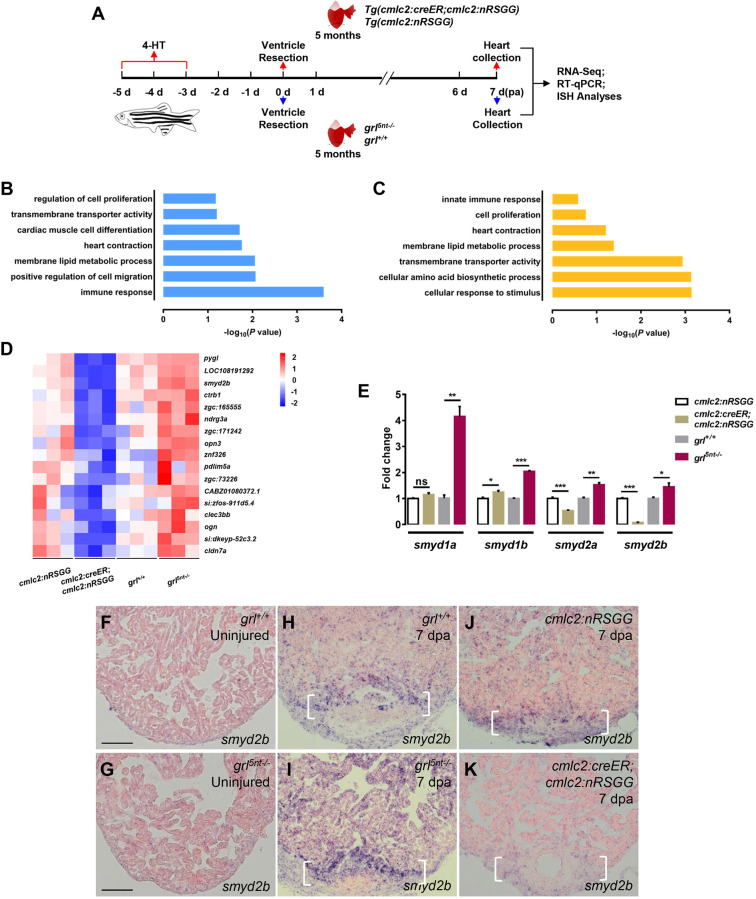
Grl mediates transcriptional repression by binding E-box motifs (Fischer et al., 2005, 2002; Heisig et al., 2012). We reasoned that potential Grl downstream genes should be downregulated in injured hearts overexpressing grl, but simultaneously upregulated in grl-deficient hearts following damage. Therefore, we performed RNA sequencing (RNA-seq) on ventricular wound tissues collected from myocardial grl-overexpressing hearts, grl5nt−/− mutant hearts and their respective control groups. The 4-HT-treated Tg(cmlc2:creER;cmlc2:nRSGG) and Tg(cmlc2:nRSGG) control animals, as well as grl5nt−/− mutant and WT sibling animals, were subjected to ventricle resections. Total RNAs were isolated from injured ventricles at 7 dpa, which corresponds to when CM proliferation actively occurs (Fig. 4A), and sequenced. We generated and analyzed transcriptional profiles in grl-overexpressing hearts, grl5nt−/− mutant hearts and their control groups (Fig. S4A,B, Tables S1 and S2). As expected, gene ontology (GO) analyses identified categories in ‘regulation of cell proliferation’, ‘cardiac muscle cell differentiation’ and ‘heart contraction’ that were enriched in the downregulated gene set in grl-overexpressing RNA profiles and in the increased gene set in grl5nt−/− mutant RNA profiles (Fig. 4B,C). However, we also obtained the enrichment of genes related to metabolic and biosynthetic processes, and to the immune response. We surveyed the list to identify potential Grl target genes that should be upregulated in grl5nt−/− mutant hearts and simultaneously reduced in grl-overexpressing hearts following injury. About 50 genes were identified as fitting the criteria and being present in overlapped RNA profile datasets (Fig. S4C, Table S2). After filtering out genes with low expression levels (FPKM<1), we obtained 17 differential expression genes containing epigenetic regulatory genes (such as smyd2b), metabolism regulatory genes (such as znf326) and other unknown factors (Fig. 4D).
Fig. 4.
Identification of Smyd2 methyltransferase as candidate Grl target in the regenerating heart. (A) Experimental design for RNA-seq, RT-qPCR and ISH analyses. Red arrows represent experimental steps for Tg(cmlc2:creER;cmlc2:nRSGG) animals and Tg(cmlc2:nRSGG) control fish. Blue arrows represent experimental steps for grl5nt−/− mutant fish and WT sibling fish. (B,C) Bar graph showing −log10 P values for GO terms significantly represented in the downregulated gene category of the grl-overexpressing groups (B) and in the upregulated gene category of the grl mutant groups (C). (D) Heatmap indicating genes downregulated in Tg(cmlc2:creER;cmlc2:nRSGG) hearts and upregulated in grl5nt−/− mutant hearts after resection, compared with control wounded hearts. Red, higher expression; blue, lower expression. FC>1.5, P<0.05. (E) qPCR analyses of smyd1a, smyd1b, smyd2a and smyd2b in injured hearts extracted from Tg(cmlc2:creER;cmlc2:nRSGG), Tg(cmlc2:nRSGG), grl5nt−/− and grl+/+ animals. Data presents as mean±s.e.m. (n=3); *P<0.05, **P<0.01, ***P<0.001, Student's t-test (unpaired, two-tailed). (F-K) ISH analyses of smyd2b expression in uninjured hearts of WT sibling fish (F), grl5nt−/− fish (G), and injured ventricles from WT sibling fish (H), grl5nt−/− fish (I), 4-HT-treated Tg(cmlc2:nRSGG) control fish (J) and 4-HT-treated Tg(cmlc2:creER;cmlc2:nRSGG) animals (K) at 7 dpa, respectively (n=5). Brackets indicate injury site. Scale bars: 100 µm.
We next queried these potential target genes for Grl-binding E-box content using the JASPAR database, and found that the KMT smyd2b contains multiple E-boxes in the regulatory upstream region (Fig. S4D). Recent studies report that Smyd family genes (smyd1 and smyd2) play crucial roles in cardiomyogenesis and myofibrillogenesis (Donlin et al., 2012; Gottlieb et al., 2002; Just et al., 2011; Li et al., 2013; Tan et al., 2006; Voelkel et al., 2013). The genes smyd1a and smyd1b, smyd2a and smyd2b are duplicated in the zebrafish genome. Similar to smyd2b, the regulatory upstream region in smyd2a and human SMYD2 contains E-box motifs (Fig. S4E,F). Quantitative PCR analyses indicated that expression of smyd1a and smyd1b, smyd2a and smyd2b were upregulated in wounded grl5nt−/− mutant hearts (Fig. 4E). However, expression of smyd2a and smyd2b but not smyd1a and smyd1b were reduced in 4-HT-treated grl-overexpressing hearts following injury (Fig. 4E), suggesting that expression of smyd2a and smyd2b is transcriptionally repressed by Grl during cardiac regeneration.
To assess whether smyd2 expression is normally upregulated by injury, we analyzed and compared smyd2b expression in uninjured and amputated WT hearts, considering smyd2a expression is very low (data not shown). Using ISH analyses, we observed a marked induction of smyd2b expression at the wound edge and a slight increase in the remote myocardial area in WT hearts at 7 dpa (Fig. 4H), whereas smyd2b transcripts were hardly detectable in uninjured WT hearts (Fig. 4F). Next, we determined whether smyd2 induction was controlled by Grl during regeneration. We noticed that grl5nt−/− mutant hearts following resection displayed much stronger smyd2b upregulation at wound edges and remote areas compared with that in injured WT sibling hearts (Fig. 4I,H). Similar to uninjured WT hearts, smdy2b transcripts were hardly detectable in unresected grl5nt−/− mutant hearts (Fig. 4G), suggesting that grl reduction is not sufficient to induce smyd2b expression in uninjured hearts. Remarkably, myocardial-specific grl overexpression abolished smyd2b induction at the wound region and remote areas, compared with that in injured Tg(cmlc2:nRSGG) control hearts (Fig. 4J,K). Collectively, these findings demonstrate that Grl negatively regulates smyd2b expression throughout the myocardium in response to injury.
Smyd2, acting as a key Grl transcriptional target, is required for heart regeneration by enhancing CM proliferation
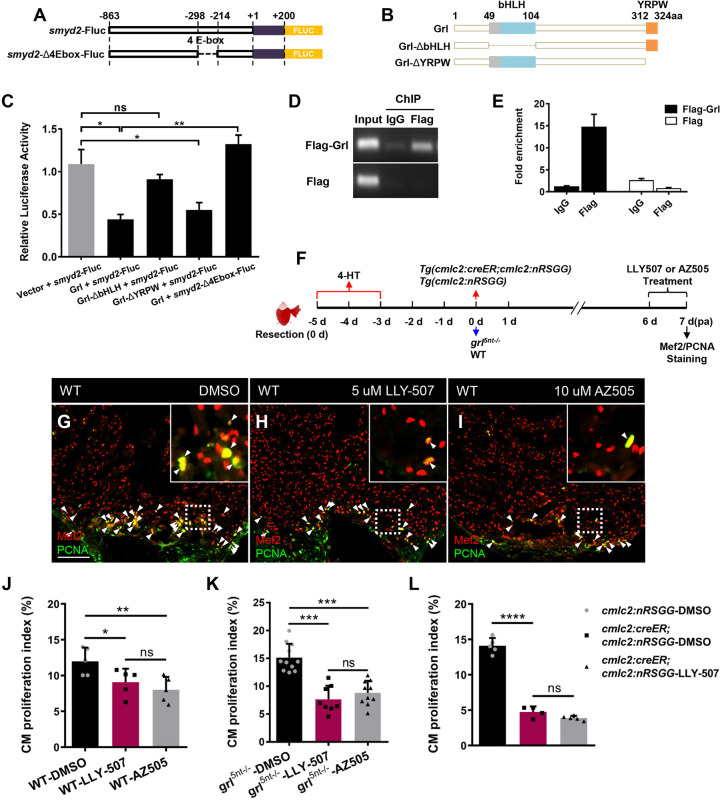
To assess whether Grl directly mediates transcriptional repression of smyd2, we performed luciferase reporter assays in HEK 293T cells. We generated a pGL3-smyd2-Fluc vector that links the smyd2 promoter containing four E-box motifs with firefly luciferase (Fluc) (Fig. 5A), as well as a pGL3-smyd2-Δ4Ebox-Fluc plasmid containing the Fluc-fused smyd2 promoter deleting four E-boxes (Fig. 5A). Grl-truncated mutants harboring deletions of the bHLH domain (Grl-ΔbHLH) and the YRPW motif (Grl-ΔYRPW) were also constructed (Fig. 5B). We co-transfected dual luciferase expression plasmid pGL3-smyd2-Fluc with pcDNA3-Grl, pcDNA3-Grl-ΔbHLH or pcDNA3-Grl-ΔYRPW. Grl transfection efficiently reduced smyd2 luciferase activity by 60% in comparison with pcDNA3 controls (Fig. 5C). By contrast, Grl-ΔbHLH mutant failed to reduce smyd2-luciferase activity, whereas the Grl-ΔYRPW mutant maintained the repression of smyd2-luciferase activity (Fig. 5C), indicating that Grl represses smyd2 transcription via its bHLH domain. Remarkably, deletion of four E-box motifs in the smyd2 promoter abrogated repression responsiveness to Grl when pcDNA3-Grl and pGL3-smyd2-Δ4Ebox-Fluc were co-transfected (Fig. 5C). To assess whether Grl binds the smyd2 promoter region, chromatin-immunoprecipitation (ChIP) was conducted by transfection of pEGFP-Flag-Grl or pEGFP-Flag control plasmids into cells. ChIP-PCR analyses revealed enrichment of the E-box-containing promoter region of smyd2 in the chromatin-immunoprecipitates using anti-Flag antibody, whereas the control IgG antibody did not enrich smyd2 promoter region (Fig. 5D,E). Whole genome ChIP-seq analyses for human HEY2 also identified enrichment of the SMYD2 promoter region (Heisig et al., 2012). Collectively, these results demonstrate that Grl mediates transcriptional repression of smyd2 by binding to the E-box-containing promoter, indicating that smyd2 is a direct transcriptional target of Grl, consistent with RNA profiling analyses in the setting of heart regeneration.
Fig. 5.
Smyd2 acts as a transcriptional target of Grl to promote heart regeneration. (A) E-box deletion in the smyd2 promoter for luciferase assays. (B) Deletion (Δ) fragments of Grl created for luciferase assays. (C) Luciferase activity in cells after co-transfection of pGL3-smyd2-Fluc with control plasmid vector, Grl, Grl-ΔbHLH or Grl-ΔYRPW expression plasmids, as well as transfection of pGL3-smyd2-Δ4Ebox-Fluc with Grl expression plasmids. The relative flyfire luciferase activity was normalized by Renilla luciferase and calculated as the ratio of each experimental group to the control group (n=3). (D,E) ChIP-PCR (D) and ChIP-qPCR (E) analyses of enrichment of Grl at its predicted binding sites in smyd2 promoter in HEK 293T cell. Enrichment levels of smyd2 promoter fragment were examined in pEGFP-Flag-grl and pEGFP-Flag transfected groups immunoprecipitated with anti-IgG or anti-Flag antibody. Genomic DNA isolated before IP was analyzed as the input control. Enrichment levels of qPCR analysis were normalized to that in the pEGFP-Flag-grl transfected group immunoprecipitated with IgG. (F) Experimental design for inhibitor treatment and CM proliferation analyses after ventricular resection in WT, grl mutant hearts, as well as in 4-HT-treated Tg(cmlc2;CreER;cmlc2:nRSGG) hearts and 4-HT-treated Tg(cmlc2;nRSGG) control hearts. Treatment at 6-7 dpa was with 5 µM LLY-507, 10 µM AZ505 or 0.5‰ DMSO. Red arrows represent experimental steps for Tg(cmlc2:creER;cmlc2:nRSGG) animals and Tg(cmlc2:nRSGG) control fish. Blue arrows represent experimental steps for grl5nt−/− mutant fish and WT sibling fish. (G-I) Immunofluorescent section images of injured ventricles from DMSO- (G), LLY-507- (H) and AZ505-treated (I) WT fish at 7 dpa, stained with anti-PCNA (green) and anti-Mef2 (red) antibodies. Insets show higher-magnification images of the dashed boxes. Arrowheads indicate proliferating CMs. (J) Quantification of CM proliferation indices in 7 dpa ventricles derived from DMSO-, LLY-507- or AZ505-treated WT fish (n=5). (K) Quantification of CM proliferation indices in 7 dpa ventricles derived from DMSO- (n=11), LLY-507- (n=8) or AZ505-treated grl5nt−/− fish (n=10). (L) Quantification of CM proliferation indices in 7 dpa ventricles derived from DMSO-treated Tg(cmlc2:nRSGG) control fish (n=5), DMSO-treated Tg(cmlc2:creER;cmlc2:nRSGG) fish (n=4) and LLY-507-treated Tg(cmlc2:creER;cmlc2:nRSGG) fish (n=5). Data presents as mean±s.e.m. *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001, Student's t-test (unpaired, two-tailed). Scale bars: 100 µm.
Because Smyd2 is induced mainly in the myocardium following injury, we tested whether Smyd2 functions as an essential regulator to promote CM proliferation during regeneration. LLY-507 and AZ505 are chemical inhibitors that specifically block the KMT activity of Smyd2 (Gao et al., 2017; Li et al., 2017; Nguyen et al., 2015). We administrated resected WT animals with LLY-507 or AZ505, and performed immunostaining with PCNA and Mef2 (Fig. 5F). Inhibition of Smyd2 KMT activity by LLY-507 treatment caused a marked reduction in CM proliferation, leading to a 25% reduction in the CM proliferation index in comparison with vehicle treatment (Fig. 5G,H,J). Similarly, administration of injured WT hearts with AZ505 inhibitor reduced the CM proliferation index by 34% (Fig. 5G,I,J). These findings indicate that the KMT activity of Smyd2 is required for CM proliferation during regeneration. Next, we assessed whether the elevated CM proliferation in grl mutant hearts might be due to Smyd2 induction following damage. We administrated grl5nt−/− mutant animals with LLY-507 or AZ505 treatment following resections and analyzed the CM proliferation indices (Fig. 5F). Inactivation of Smyd2 activity with LLY-507 or AZ505 treatment resulted a significant reduction in the CM proliferation indices in grl5nt−/− hearts compared with vehicle-treated grl5nt−/− hearts (Fig. 5K; Fig. S5A-C). Considering that Smyd2 is a transcriptional repression target of Grl, we assessed CM proliferation in myocardial grl induction hearts with Smyd2 inactivation (Fig. 5F). LLY-507-treated grl induction hearts displayed almost the same percentage reduction in CM proliferation indices as in vehicle-treated hearts following resection (Fig. 5L; Fig. S5D-F), suggesting that smyd2 functions in the grl pathway during regeneration.
Smyd2 lysine methyltransferase mediates Stat3 activation during heart regeneration
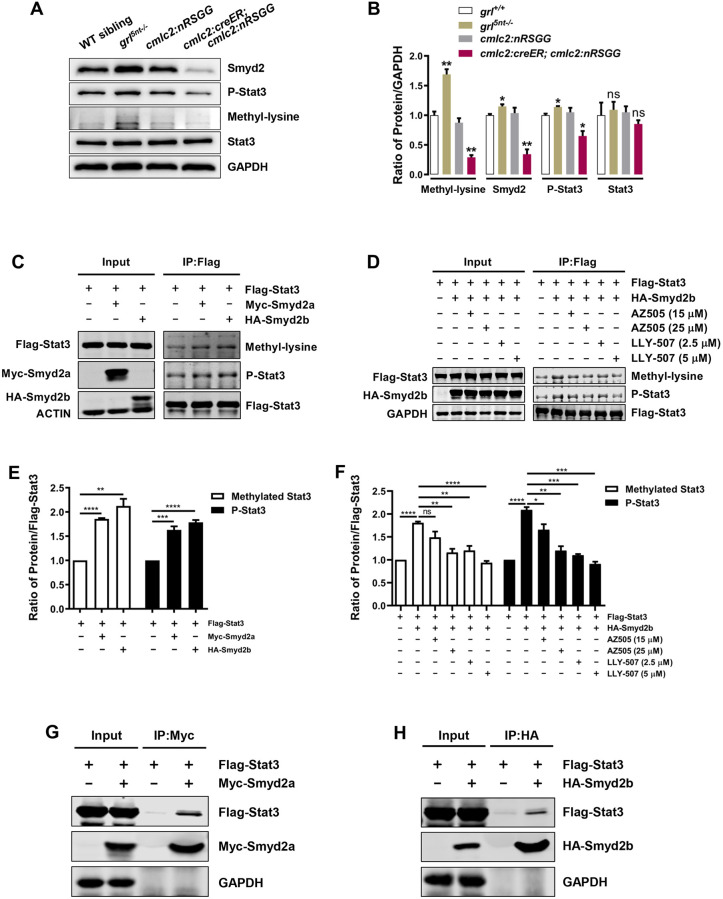
As a KMT, Smyd2 conducts its function by methylation of histones or non-histone substrates, including Stat3, during cell proliferation, survival and other events (Donlin et al., 2012; Gao et al., 2017; Li et al., 2017). We first assessed whether Smyd2 regulates histone methylation during Grl-mediated heart regeneration. Methylation of H3K4 and H3K9 were examined using cardiac wound tissues extracted from grl5nt−/− mutant hearts or grl-overexpressing hearts. We learned that various methylation patterns of H3K4 and H3K9 appeared not to be affected in grl loss- or gain-of-function hearts (Fig. S6). On the contrary, levels of phosphorylated Stat3 (P-Stat3) and Smyd2 were both increased or reduced in grl5nt−/− mutant hearts or grl-overexpressing hearts, respectively, whereas Stat3 levels failed to be affected (Fig. 6A,B), revealing correlation of the phosphorylated Stat3 level with the Smyd2 level during regeneration. Previous studies report that, during renal cystic tissue growth, Stat3 phosphorylation is induced by Smyd2 through methylation of its own protein (Li et al., 2017, 2018a), leading to increased renal epithelial cell proliferation. Phosphorylated active Stat3 is also mitogenic and required for heart regeneration in zebrafish (Fang et al., 2013). We hypothesized that Smyd2 regulates myocardial regeneration by modulating the Stat3 methylation and phosphorylation activity. Anti-methylated lysine (methyl-lysine) antibody, a pan-methyl antibody available in the field, was used to recognize lysine-methylated proteins. We found that methylated proteins corresponding to the size of Stat3 were correlated with the P-Stat3 level or the Smyd2 level in grl5nt−/− mutant hearts or grl-overexpressing hearts, respectively (Fig. 6A,B). We tested whether zebrafish Smyd2a or Smyd2b directly regulates Stat3 methylation and phosphorylation. Plasmids containing pcDNA3-Flag-Stat3 and pcDNA3-Myc-Smyd2a or pcDNA3-HA-Smyd2b were co-transfected into HEK 293T cells. We found that coexpression of Stat3 with Smyd2a or Smyd2b in HEK 293T cells resulted in an increase in levels of both Stat3 methylation and phosphorylation (Fig. 6C,E). In contrast, inactivating Smyd2 KMT activity with inhibitor AZ505 or LLY-507 not only caused a reduction in methylated Stat3, but also led to a decrease in phosphorylated Stat3 (Fig. 6D,F). Furthermore, co-immunoprecipitation (Co-IP) analyses revealed physical interactions between Flag-Stat3 with Myc-Smyd2a or HA-Smyd2b (Fig. 6G,H). Together, these findings indicate that zebrafish Smyd2 interacts with Stat3 to regulate the methylation and phosphorylation of Stat3 in cultured cells, in agreement with previous murine studies (Li et al., 2018a).
Fig. 6.
Smyd2 functions as a KMT and controls Stat3 methylation and phosphorylation. (A) western blot analysis exhibiting Smyd2, Stat3, P-Stat3 or corresponding lysine methylated proteins using total lysates extracted from WT sibling, grl5nt−/−, 4-HT-treated control Tg(cmlc2:nRSGG) hearts or 4-HT-treated Tg(cmlc2:creER;cmlc2:nRSGG) hearts at 7 dpa, respectively. GAPDH was used as a loading control. (B) Quantification of western blots using ImageJ software and normalized to GAPDH (n=3). (C) Methylation and phosphorylation assay showing increased levels of methylated Stat3 and P-Stat3 in Flag-Stat3 and Myc-Smyd2a/HA-Smyd2b co-transfected HEK 293T cells. The cell lysates were used to immunoprecipitate (IP) Stat3 with anti-Flag antibody and then blotted with anti-methyl-lysine or anti-P-Stat3 antibodies. (D) Treatment with AZ505 or LLY-507 diminishes Stat3 methylation and phosphorylation in cells co-transfected with Flag-Stat3 and HA-Smyd2b. The cell lysates were treated with AZ505 (15 µM for 6 h, 25 µM for 6 h) or LLY-507 (2.5 µM for 28 h, 5 µM for 28 h), used to IP Stat3 using anti-Flag antibody and then blotted with anti-methyl-lysine and anti-P-Stat3 antibodies. (E) Quantification of methylated Stat3 and phosphorylated Stat3 levels normalized to Flag-Stat3 (n=3). (F) Quantification of methylated Stat3 and phosphorylated Stat3 levels normalized to Flag-Stat3 in cells treated with AZ505 or LLY-507 (n=3). (G,H) Smyd2a or Smyd2b immunoprecipitation with anti-Myc antibody (G) or anti-HA antibody (H), respectively, detects Stat3 using anti-Flag antibody. HEK 293T cells were co-transfected with constructs of Flag-Stat3 and Myc-Smyd2a (G) or HA-Smyd2b (H). GAPDH was used as a loading control. Data represents mean±s.e.m. *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001, Student's t-test (unpaired, two-tailed).
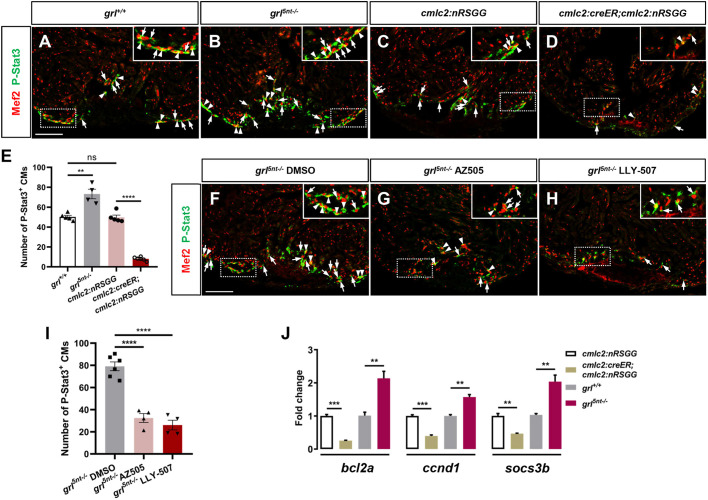
Next, we assessed whether Grl-Smyd2 modulates Stat3 methylation and phosphorylation activity during heart regeneration. Following injury, injured hearts derived from grl mutants and WT siblings, as well as grl-overexpressing and control animals, were subjected to double immunostaining with anti-P-Stat3 and anti-Mef2 antibodies (Fig. 7A-D). Previous studies report that P-Stat3 is primarily detectable in the nucleus, but certain human tissues and cells also display cytosolic and perinuclear P-Stat3, some of which goes to the nuclei afterwards (Chen et al., 2006; Lee et al., 2017; Li et al., 2018b; Zhang et al., 2019). During heart regeneration, we observed that P-Stat3 partially overlapped with Mef2 in CM nuclei in the injury region, whereas some P-Stat3 localized in the cytoplasmic area closely around CM nuclei (Fig. 7A). P-Stat3 was also detected in non-CM cells at the wound region (Fig. 7A). However, P-Stat3 was hardly detectable in CMs or non-CM cells in the remote area of injured hearts (Fig. 7A), suggesting that the murine P-Stat3 antibody recognizes proliferative CMs at the injury region in zebrafish hearts, despite some nonspecific signals. We distinguished and quantified P-Stat3-positive CMs in the injury region, including CMs with nuclear P-Stat3 labeling and perinuclear cytosolic P-Stat3 staining (Fig. 7A-I). Notably, P-Stat3-positive CMs were increased at injury regions in grl5nt−/− mutant hearts compared with WT sibling hearts (Fig. 7A,B,E). Inversely, myocardial grl induction caused a marked reduction in P-Stat3-labeled CMs at wound areas in comparison with Tg(cmlc2:nRSGG) control hearts (Fig. 7C-E), indicating the negative regulation of P-Stat3-labeled CMs by Grl at the cardiac wound. To establish connections between the methyltransferase activity of Smyd2 and Stat3 phosphorylation, we treated injured grl5nt−/− mutant hearts with inhibitors AZ505 and LLY-507. Consistent with our hypothesis, inhibiting Smyd2 KMT activity with AZ505 or LLY507 following injury reduced P-Stat3-labeled CMs in grl5nt−/− mutant hearts compared with vehicle-treated grl5nt−/− hearts (Fig. 7F-I), suggesting the contribution of Smyd2 KMT activity to the increased P-Stat3-labeled CMs in grl mutant hearts. Furthermore, expressions of B-cell lymphoma 2 (bcl2a), cyclin D1 (ccnd1) and suppressor of cytokine signaling 3b (socs3b), target genes in the Stat3 pathway, were reduced in grl-overexpressing hearts and increased in grl mutant hearts (Fig. 7J). Taken together, these findings suggest that the effects of Grl on heart regeneration are mediated at least in part by Smyd2-dependent Stat3 activation.
Fig. 7.
Grl-Smyd2 mediates Stat3 activation during heart regeneration. (A-D) Immunofluorescent images of injured ventricles stained with anti-P-Stat3 antibody (green) and anti-Mef2 antibody (red) from WT siblings (A), grl5nt−/− mutant fish (B), 4-HT-treated Tg(cmlc2:nRSGG) control animals (C) and 4-HT-treated Tg(cmlc2:creER;cmlc2:nRGG) animals (D) at 7 dpa. Insets show higher-magnification images of the dashed boxes. Arrowheads indicate P-Stat3 overlapping with Mef2 in CM nuclei; Arrows indicate perinuclear cytosolic P-Stat3 around Mef2 nuclei. P-Stat3 immunostaining was also detectable in non-CM cells in the injured area. (E) Quantification of number of P-Stat3-positive CMs in the injury border zone of 7 dpa ventricles from WT siblings (n=5), grl5nt−/− mutants (n=4), Tg(cmlc2:nRSGG) control fish (n=5), or Tg(cmlc2:creER;cmlc2:nRGG) fish (n=6). (F-H) Immunofluorescent section images of injured ventricles stained with anti-P-Stat3 antibody (green) and anti-Mef2 antibody (red) from DMSO-treated (F), AZ505-treated (G) and LLY-507-treated grl5nt−/− mutant fish (H) at 7 dpa. Insets show higher-magnification images of the dashed boxes. Arrowheads indicate P-Stat3 overlapping with Mef2; Arrows indicate perinuclear cytosolic P-Stat3 around Mef2 nuclei. (I) Quantification of number of P-Stat3-positive CMs in injury border zones at 7 dpa from DMSO-treated (n=6), AZ505-treated (n=4) and LLY-507-treated grl5nt−/− mutant fish (n=4). (J) qPCR analyses of bcl2a, ccnd1 and socs3b in injured hearts extracted from Tg(cmlc2:creER;cmlc2:nRSGG) and Tg(cmlc2:nRSGG) control fish, as well as WT sibling and grl5nt−/− mutant fish (n=3). Data presents as mean±s.e.m. **P<0.01, ***P<0.001, ****P<0.0001, Student's t-test (unpaired, two-tailed). Scale bars: 100 µm.
DISCUSSION
In this study, we describe the transcriptional control of methylation modulation in governing CM renewal and heart regeneration. We demonstrate that Grl functions as a negative regulator to restrain heart regeneration in zebrafish, and identify KMT Smyd2 as the Grl-repressing target in this context. Following cardiac injury, Grl reduction triggers Smyd2 induction at the injured myocardial cell edge, leading to CM proliferation and heart regeneration. We show that zebrafish Smyd2 functions as a KMT and regulates CM renewal in part by modulating the activity of Stat3. These findings suggest that manipulation of the Grl-Smyd2 network might be instrumental in potential strategies to relieve cardiac barriers and accelerate heart regeneration in mammalian hearts.
The limited regenerative potential of adult mammalian hearts can be attributed to cell-intrinsic barriers that prevent CMs from entering the cell cycle (González-Rosa et al., 2018; Li et al., 2015; Tzahor and Poss, 2017). Recent studies have uncovered a few molecular blocks that impede zebrafish heart regeneration (Dogra et al., 2017; Jopling et al., 2012; Missinato et al., 2018). We demonstrate that Grl acts as a transcriptional repressor to limit injury-induced CM proliferation. This is consistent with its roles in negative regulation of embryonic heart development and growth in zebrafish (Jia et al., 2007). Hey family genes play multiple roles during embryogenesis, with expression in various cell types (Fischer et al., 2002; Nakagawa et al., 1999; Zhong et al., 2000). In the embryonic heart, grl is enriched in the myocardial compact zone, as well as weakly expressed in the trabecular region and ventricular endocardium (Jia et al., 2007; Miao et al., 2018). During murine heart development, hey2 expression in myocardial compact zones and ventricular endocardium is regulated by Notch signaling (Miao et al., 2018), but its functional mechanisms remain unknown. In the adult zebrafish heart, we observed that grl expression is restricted to myocardial cells but not the endocardium or coronary vasculature. Notably, myocardial grl expression in the adult heart is reduced in response to heart damage. By contrast, Notch signaling is upregulated in endocardial and epicardial cells following injury, stimulating CM renewal in zebrafish (Zhao et al., 2019). Thus, the role of myocardial Grl in limiting heart regeneration is independent of endocardial Notch signaling. We observed that grl expression declines to the lowest level from 1 dpa to 3 dpa, increases to some extent at 7 dpa, and gradually restores from 14 dpa to 30 dpa. During regeneration, many factors that stimulate CM proliferation are induced after injury, which can counteract inhibitory factors such as Grl for CM proliferation. We believe that the spatiotemporal regulation of genome-wide networks enables CMs to regenerate after injury. Overall, the temporal expression patterns of grl are consistent with the functional model in which Grl negatively regulates CM proliferation during zebrafish heart regeneration. It is important to test whether murine Hey2 expression is reduced following cardiac injury, and whether conditional Hey2 deletion in mice can promote CM regeneration.
Smyd2 represents a novel class of KMTs and has been reported to methylate histone and non-histone substrates (Donlin et al., 2012; Gao et al., 2017; Li et al., 2017). Our studies identify a crucial role for Smyd2 during heart regeneration. However, Smyd2 is dispensable for cardiac development in mice (Diehl et al., 2010), suggesting that Smyd2 is required for injury-induced heart regeneration or functions in response to other cardiac stresses, consistent with our observations that zebrafish smyd2 expression was not detectable in uninjured adult hearts. In addition to Stat3, Smyd2 methylates other non-histone substrates, including BMP receptor 2 (BMPR2), NF-κB and HSP90 (Donlin et al., 2012; Gao et al., 2017; Li et al., 2017). It is likely that Smyd2 has multiple methylation substrates during cardiac regeneration. It is important to assess whether Smyd2 regulates methylation of NF-κB or BMPR2 following injury. Alternatively, Smyd2 might methylate chaperone Hsp90 to promote interaction of the methyl-Hsp90-sarcomeric protein complex and myofilament organization during heart regeneration (Donlin et al., 2012). Future studies are warranted to investigate various methylation substrates of Smyd2 during cardiac regeneration, and to assess the extent to which methylation modulations play crucial roles during Grl-mediated transcriptional controls. The findings of such studies will reveal novel transcriptional regulations and various methylation controls that direct vertebrate heart regeneration, which will be relevant in developing strategies for regeneration interventions in humans.
MATERIALS AND METHODS
Zebrafish strains
WT zebrafish of the AB strain were used for generating grl mutants and various transgenic lines. Published transgenic lines used in this study included Tg(cmlc2:mCherry) (Palencia-Desai et al., 2011), Tg(flk1:EGFP) (Jin et al., 2005), Tg(flk1:mCherry) (Ren et al., 2013), Tg(tcf21:nucEGFP) (Wang et al., 2011) and Tg(cmlc2:CreER) (Kikuchi et al., 2010). The Institutional Animal Care and Use Committee at East China Normal University advises animal care and research.
Generation of grl mutants
The grl mutants were generated using CRISPR/Cas9-mediated mutagenesis. Cas9 capped mRNA was produced by in vitro transcription from a pGH-T7-zCas9 vector (Liu et al., 2014) using mMESSAGE mMACHINE T7 Transcription Kit (Ambion). The sgRNA target site of grl was designed via the CRISPRscan website. The sgRNA transcription template was PCR amplified and then transcribed using MAXIscript T7 Kit (Ambion). Cas9 capped mRNA and sgRNA were co-injected into zebrafish embryos at the one-cell stage. The final concentrations of Cas9 mRNA and sgRNA were 400 ng/µl and 30 ng/µl, respectively.
Generation of Tg(grl:EGFP), Tg(grl:mCherry) and Tg(cmlc2:loxP-nlsmCherry-STOP-loxP-grl-EGFP) zebrafish
To generate Tg(grl:EGFP) zebrafish, a 7.8 kb fragment containing the grl promoter sequence was amplified from BAC14 (Zhong et al., 2000) and cloned into the pCR-Blunt II-TOPO vector. A 2888 bp tol2 3′end-tol2 5′end-EGFP cassette was amplified from pT2KXIGΔin vector (Urasaki et al., 2006), discarding the original EF1p promoter. After verification of the grl promoter sequence, a 7.6 kb grl promoter fragment flanked by a 24-nucleotide overlap of tol2 3′end-tol2 5′end-EGFP cassette was amplified and assembled with this cassette using Gibson Assembly Master Mix (NEB). DNA construct (40 ng/µl) and Tol2 transposase mRNA (50 ng/µl) prepared from pCS-TP were co-injected into zebrafish embryos at the one-cell stage. To generate the Tg(grl:mCherry) line, the EGFP sequence in pT2KXIGΔin vector was replaced with the mCherry fragment. The methods used were the same as described above.
To generate the Tg(cmlc2:nRSGG) strain, full name as Tg(cmlc2:loxP-nlsmCherry-STOP-loxP-grl-EGFP), the MultiSite Gateway Kit (Invitrogen) was used to simultaneously clone a cmlc2:loxP-nlsmCherry-STOP-loxP cassette, grl CDS fragment and EGFP fragment into the pDESTol2pA2 destination vector. The cmlc2:loxp-nlsmCherry-STOP-loxp cassette was flanked by attB4 and attB1r. The grl CDS fragment was flanked by attB1 and attB2. The EGFP fragment was flanked by attB2r and attB3. Three attP-containing donor vectors (pDONR P4-P1R, pDONR 221 and pDONR P2R-P3) were used in separate BP recombination reactions with the above-mentioned attB-containing PCR fragments to generate three entry clones. The three entry clones and the pDESTol2pA2 destination vector were used together in an LR recombination reaction between attL- and attR-flanked regions to create the cmlc2:loxP-nlsmCherry-STOP-loxP-grl-EGFP construct. The construct was co-injected with Tol2 transposase mRNA into one-cell stage embryos to generate transgenic lines, which were screened for nlsmCherry and then EGFP upon Cre-mediated recombination.
Ventricular apex resection and chemical treatment
Zebrafish at 5-6 months of age were used for ventricular resection surgery as previously described (Poss et al., 2002). About 20% of ventricular muscle was excised from the ventricle apex after anesthesia as previously described (Poss et al., 2002). 4-HT (Sigma) was dissolved in ethanol to give a 20 mM stock solution. To induce expression of grl in adult zebrafish heart, Tg(cmlc2:creER;cmlc2:nRSGG) fish were incubated with 5 µM 4-HT for bath treatment for 2 days. After 24 hours, fresh 4-HT was prepared and added for an additional 24 h treatment. AZ505 ditrifluoroacetate (MCE) was dissolved in DMSO to give a 20 mM stock solution and diluted in aquarium water to 10 µM for bath treatment from 6 to 7 dpa. LLY-507 (MCE) was dissolved in DMSO to give a 10 mM stock solution and diluted in aquarium water to 5 µM for bath treatment from 6 to 7 dpa. DMSO (0.5‰) was used as a vehicle control for AZ505 and LLY-507 treatment.
Fibrin and fibrotic scar analysis, in situ hybridization and immunofluorescence
Zebrafish hearts were fixed in 4% paraformaldehyde (PFA) at room temperature for 1 h. All histological experiments were performed using 10 µm cryosections. Acid fuchsin-orange G (AFOG) staining for fibrin and fibrotic scar analyses was performed as described (Poss et al., 2002). ISH was performed using digoxigenin-labeled RNA antisense probes as described (Lepilina et al., 2006). Probes were used at a concentration of 2 ng/µl diluted with hybridization buffer. The hybridization was performed overnight at 65°C. Detection was done using 1:5000 dilution of anti-digoxigenin-AP (Roche) and visualized by NBT/BCIP (Roche) substrate reacting at 37°C. Immunofluorescence was performed as described (Kikuchi et al., 2011). Cryosections were administrated with antigen retrieval at 98°C for 20 min (for PCNA staining), washed three times in PBS containing 0.1% Tween-20 and incubated in blocking buffer (3% BSA, 5% normal sheep serum, 1% DMSO, 0.1% Triton-X100, 0.1% Tween-20 in PBS) at 37°C for 30 min. The sections were incubated with primary antibody overnight at 4°C. After washing, the sections were incubated with secondary antibody for 2 h at 37°C and then mounted with Vectashield mounting medium (Vector laboratories) for imaging. Primary antibodies used in this study were as follows: anti-mCherry (Invitrogen; M11217; 1:200), anti-Mef2 (Santa Cruz; sc-313; 1:100), anti-MHC (DSHB; MF20; 1:200), anti-PCNA (Sigma; P8825; 1:200), anti-Raldh2 (ABclonal; A7503; 1:200), anti-α-actinin (Abcam; ab9465; 1:100), anti-α-actinin (Abcam; ab68167; 1:100), embCMHC antibody (DSHB; N2.261; 1:100) and anti-P-Stat3 (Santa Cruz; sc-8059; 1:100). Secondary antibodies used in this study were as follows: Alexa Fluor 555 goat anti-rat IgG (H+L) (Invitrogen; A-21434; 1:800), Alexa Fluor 594 goat anti-rabbit IgG (H+L) (Invitrogen; A-11012; 1:800), Alexa Fluor 488 goat anti-mouse IgG (H+L) (Invitrogen; A-11001; 1:800), Alexa Fluor 594 goat anti-mouse IgG (H+L) (Invitrogen; A-11005; 1:800) and Alexa Fluor 647 goat anti-mouse IgG (H+L) (Invitrogen; A-21235; 1:800).
To quantify regenerative status shown by AFOG staining at 30 dpa, we scored heart sections for regeneration, with ‘class 1’ indicating complete regeneration showing contiguous ventricle wall, ‘class 2’ indicating partial regeneration showing some collagen deposition and ‘class 3’ indicating a block in regeneration showing fibrin and collagen deposition. Multiple sections of each heart were scored according to these criteria.
To quantify CM proliferation index, four to six sections showing the largest injury area were selected from each heart, and images were taken using a 10× objective. The number of Mef2+ and Mef2+ PCNA+ cells were manually counted using ImageJ software within a defined region including almost all Mef2+ PCNA+ cells around the injury site (1344×384 pixel region of the 1388×1040 pixel images). The percentages of Mef2+ PCNA+ cells from all sections were averaged to determine a proliferation index for each heart.
qPCR analysis
For cDNA synthesis using PrimeScript II 1st Strand cDNA Synthesis Kit (TaKaRa), 1 µg of total RNA extracted from ventricles was used. RT-qPCR was carried out on biological triplicates with SYBR Premix Ex Taq II (Takara) using a Roche LightCycler 480 II system. The relative expression levels of target genes were normalized to β-actin and quantified by the 2−ΔΔCT method (Livak and Schmittgen, 2001).
RNA-seq and data analysis
Ventricles at 7 dpa were collected from 4-HT-treated Tg(cmlc2:creER;cmlc2:nRSGG) and Tg(cmlc2:nRSGG) animals, as well as grl5nt−/− and WT sibling zebrafish. Six ventricles were pooled per biological replicate. Each group contained three biological replicates. Total RNAs of the ventricles were isolated using TRIzol Reagents (Life Technologies) and checked for a RIN number to inspect RNA integrity using the Agilent 2100 Bioanalyzer system (Agilent Technologies). Qualified total RNAs were further purified by RNAClean XP Kit (Beckman Coulter) and RNase-Free DNase Set (Qiagen). Libraries were constructed using the TruSeq RNA Sample Preparation Kit (Illumina) followed by a quality check with Qubit 2.0 Fluorometer (Invitrogen) and the Agilent 2100 Bioanalyzer system (Agilent Technologies), Sequencing was carried out on the Illumina HiSeq X Ten (Illumina) at Shanghai Biotechnology Corporation.
Sequenced raw reads were preprocessed using Seqtk to filter out rRNA reads, sequencing adapters, short-fragment reads and other low-quality reads to obtain clean reads for data analyses. Hisat2 (version 2.0.4) was used to map the clean reads to the zebrafish GRCz10 reference genome. After genome mapping, the mapped reads of genes were converted to FPKM (fragments per kilobase of exon model per million mapped reads) using Stringtie (version 1.3.0) (Pertea et al., 2016, 2015) for standardization of gene expression. The fold-changes of genes were calculated according to the FPKM in each sample. Genes with differential expression levels between samples were analyzed using edgeR (Robinson et al., 2010), with GO enrichment and KEGG pathway enrichment analyses.
Transfection, ChIP and Luciferase assay
The Flag tag sequence fused with zebrafish grl CDS was ligated into the pEGFP-N1 vector. The plasmids were transfected into HEK 293T cells using Lipo 3000 Transfection Reagent (Invitrogen) as previously described (Ni et al., 2011). At 48 h after transfection, ChIP was performed using EZ-ChIP Kit (Millipore). HEK 293T cells were crosslinked with 1% formaldehyde for 10 min. After incubation with glycine to stop crosslinking, cells were rinsed in PBS and homogenized in SDS lysis buffer. Cell lysates were sonicated with a FS-300 Ultrasonic Processing apparatus. ChIP was then performed by using an antibody against Flag (mouse; Sigma) or a normal mouse IgG antibody (Millipore) as negative control, according to the manufacturer's instructions.
HEK 293T cells were cultured overnight at 8×104 cells per well in 24-well plates. Firefly luciferase pGL3-smyd2-Fluc plasmid vector (0.5 µg) and Renilla luciferase pRL-TK plasmid vector (5 ng, as a normalization control) were co-transfected with different Grl mutant plasmids (0.5 µg) using 1 µl of Lipofectamine 2000 (Life Technologies) according to the manufacturer's protocol. Approximately 24 h later, cell lysates were collected and the firefly luciferase and Renilla luciferase activities measured using a Dual-Luciferase Reporter Assay System (Promega). The experiment was repeated three times.
Western blot analysis for injured heart tissue
Ventricles from 4-HT-treated Tg(cmlc2:creER;cmlc2:nRSGG) and Tg(cmlc2:nRSGG) animals, as well as grl5nt−/− and WT sibling zebrafish at 7 dpa were collected and homogenized with an electric homogenizer in cold RIPA lysis buffer (50 mM Tris pH 7.4, 150 mM NaCl, 1% Triton X-100 and 0.1% SDS) supplemented with proteinase inhibitors (Millipore) and phosphatase inhibitor cocktail (Apex Bio). The lysates were loaded into 12.5% SDS-PAGE after boiling for 5 min in SDS-PAGE sample loading buffer. Proteins were then transferred to PVDF membranes using a Trans-Blot Turbo Transfer System (Bio-Rad). Signals were detected by the Clarity Western ECL Substrate (Bio-Rad) and scanned using a ChemiScope series system (Clinx). Antibodies used for western blot in this study were as follows: anti-Smyd2 (Cell Signaling; #9734; 1:500), anti-P-Stat3 (Santa Cruz; sc-8059; 1:500), anti-Stat3 (Santa Cruz; sc-8019; 1:500), anti-methyl-lysine (Abbkine; ABM0060; 1:1000), anti-GAPDH (Abcam; ab181602; 1:10,000), anti-GAPDH (Abmart; M20006L; 1:4000), anti-actin (Sigma; A5441; 1:4000), anti-Flag (Sigma; 7425/1804; 1:4000), anti-Myc (HuaAn Biotechnology; R1208-1; 1:4000), anti-HA (Santa Cruz; sc-805; 1:4000), HRP-anti-rabbit IgG (Cwbio; CW0103S; 1:5000) and HRP-anti-mouse IgG (Cwbio; CW0102S; 1:5000).
Co-IP assay
HEK 293T cells were transfected with pcDNA3-Flag-Stat3, pcDNA3-Myc-Smyd2a or pcDNA3-HA-Smyd2b plasmids and lysed 48 h later with IP lysis buffer (50 mM Tris-HCl pH7.5, 150 mM NaCl, 1% Triton X-100, 1 mM EDTA, 1× protease inhibitor cocktail and 1 mM DTT). The lysates were cleared by centrifugation at 13,600 g for 20 min at 4°C. The supernatants were diluted with binding buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 8% glycerol, 1 mM EDTA, 1× protease inhibitor cocktail and 1 mM DTT) to obtain a final Triton X-100 concentration of 0.2%, and then directly incubated with anti-Myc/HA-affinity beads for 4 h at 4°C. After centrifugation and washing three times with washing buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 0.1% Triton X-100, 1 mM EDTA, 1× protease inhibitor cocktail and 1 mM DTT), the protein-bead complexes were boiled in 1× SDS loading buffer and analyzed by western blot.
Methylation and phosphorylation assay
For detection of methylation and phosphorylation of Stat3, HEK 293T cells were transfected with pcDNA3-Myc-Smyd2a, pcDNA3-HA-Smyd2b and pcDNA3-Flag-Stat3 as indicated. When inhibitors were used, cells were treated with AZ505 at different concentrations for 6 h before sample collection, whereas LLY-507 treatment was for 28 h. At 48 h post transfection, cells were washed with PBS twice and lysed in denaturing IP lysis buffer (150 mM NaCl, 0.1% NP40 or Triton X-100, 25 mM Tris-HCl, pH 8.0, 5 mM EDTA, 10% glycerol, 1% SDS, 1× protease inhibitor cocktail, 1× phosphatase inhibitor cocktail) and treated at 100°C for 20 min. The lysates were cleared by centrifugation at 13,600 g for 20 min. The supernatant was diluted with denaturing binding buffer without SDS to give a final SDS concentration of 0.1%, and then directly incubated with anti-Flag-M2-affinity beads (Bimake, Houston, TX) for 4 h. After extensive washing with IP washing buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.1% Triton X-100, 1 mM EDTA), the immunoprecipitated proteins were boiled in 1× SDS loading buffer and analyzed by western blot.
Imaging
AFOG and ISH images were taken using a Nikon Eclipse Ni microscope with a Nikon Digital Sight DS-Ri1 camera. Immunostaining images were taken using a Zeiss Axio Observer.Z1 microscope, Zeiss LSM 710 confocal microscope and Andor Dragonfly 500 High Speed confocal microscope.
Statistical analysis
GraphPad software was used to perform statistical analysis. Data were analyzed by the two-tailed Student's t-test and Chi-square tests. For the Student's t-test, data are represented as mean±s.e.m. and considered significant at P<0.05.
Acknowledgements
We thank Jinmin Ma for helping with cell transfection; Yuanyuan Peng, Guozhen Wu and Wenyuan Ma for zebrafish care; Xiaoli Zhu, Xiang Jia for helping with ChIP assays; ECNU Multifunctional Platform for Innovation (004) for image acquisition; and T.P.Z. lab members for comments on the manuscript.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: P.S., T.P.Z.; Methodology: P.S., X.P.; Software: P.S., J.S., B.G.; Validation: P.S., H.Z., B.G., Y.Z., X.Z., X.H.; Formal analysis: P.S., J.S., B.G., Y.Z., X.Z., X.H., K.S.L., J.W., B.Z., L.W.; Investigation: P.S., H.Z., J.S., B.G., Y.Z., X.Z., X.H., K.S.L.; Writing - original draft: P.S., T.P.Z.; Supervision: L.W., T.P.Z.; Project administration: L.W., T.P.Z.; Funding acquisition: T.P.Z.
Funding
This work was supported by grants from Ministry of Science and Technology of the People's Republic of China (2018YFA0800103, 2018YFA0801004) and National Natural Science Foundation of China (NSFC31530044, NSFC31970780). Deposited in PMC for immediate release.
Data availability
The raw RNA-seq data have been deposited in GEO under accession number GSE129499.
Supplementary information
Supplementary information available online at https://dev.biologists.org/lookup/doi/10.1242/dev.190678.supplemental
References
- Abu-Farha M., Lambert J.-P., Al-Madhoun A. S., Elisma F., Skerjanc I. S. and Figeys D. (2008). The tale of two domains: proteomics and genomics analysis of SMYD2, a new histone methyltransferase. Mol Cell. Proteomics 7, 560-572. 10.1074/mcp.M700271-MCP200 [DOI] [PubMed] [Google Scholar]
- Bezzina C. R., Barc J., Mizusawa Y., Remme C. A., Gourraud J.-B., Simonet F., Verkerk A. O., Schwartz P. J., Crotti L., Dagradi F. et al. (2013). Common variants at SCN5A-SCN10A and HEY2 are associated with Brugada syndrome, a rare disease with high risk of sudden cardiac death. Nat. Genet. 45, 1044-1049. 10.1038/ng.2712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggar K. K. and Li S. S.-C. (2015). Non-histone protein methylation as a regulator of cellular signalling and function. Nat. Rev. Mol. Cell Biol. 16, 5-17. 10.1038/nrm3915 [DOI] [PubMed] [Google Scholar]
- Black J. C., Van Rechem C. and Whetstine J. R. (2012). Histone lysine methylation dynamics: establishment, regulation, and biological impact. Mol. Cell 48, 491-507. 10.1016/j.molcel.2012.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.-L., Hsieh F.-C. and Lin J. (2006). Systemic evaluation of total Stat3 and Stat3 tyrosine phosphorylation in normal human tissues. Exp. Mol. Pathol. 80, 295-305. 10.1016/j.yexmp.2005.11.003 [DOI] [PubMed] [Google Scholar]
- D'Uva G., Aharonov A., Lauriola M., Kain D., Yahalom-Ronen Y., Carvalho S., Weisinger K., Bassat E., Rajchman D., Yifa O. et al. (2015). ERBB2 triggers mammalian heart regeneration by promoting cardiomyocyte dedifferentiation and proliferation. Nat. Cell Biol. 17, 627-638. 10.1038/ncb3149 [DOI] [PubMed] [Google Scholar]
- Diehl F., Brown M. A., van Amerongen M. J., Novoyatleva T., Wietelmann A., Harriss J., Ferrazzi F., Bottger T., Harvey R. P., Tucker P. W. et al. (2010). Cardiac deletion of Smyd2 is dispensable for mouse heart development. PLoS ONE 5, e9748 10.1371/journal.pone.0009748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dogra D., Ahuja S., Kim H.-T., Rasouli S. J., Stainier D. Y. R. and Reischauer S. (2017). Opposite effects of Activin type 2 receptor ligands on cardiomyocyte proliferation during development and repair. Nat. Commun. 8, 1902 10.1038/s41467-017-01950-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donlin L. T., Andresen C., Just S., Rudensky E., Pappas C. T., Kruger M., Jacobs E. Y., Unger A., Zieseniss A., Dobenecker M.-W. et al. (2012). Smyd2 controls cytoplasmic lysine methylation of Hsp90 and myofilament organization. Genes Dev. 26, 114-119. 10.1101/gad.177758.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y., Gupta V., Karra R., Holdway J. E., Kikuchi K. and Poss K. D. (2013). Translational profiling of cardiomyocytes identifies an early Jak1/Stat3 injury response required for zebrafish heart regeneration. Proc. Natl. Acad. Sci. USA 110, 13416-13421. 10.1073/pnas.1309810110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer A., Leimeister C., Winkler C., Schumacher N., Klamt B., Elmasri H., Steidl C., Maier M., Knobeloch K. P., Amann K. et al. (2002). Hey bHLH factors in cardiovascular development. Cold Spring Harbor Symp. Quant. Biol. 67, 63-70. 10.1101/sqb.2002.67.63 [DOI] [PubMed] [Google Scholar]
- Fischer A., Schumacher N., Maier M., Sendtner M. and Gessler M. (2004). The Notch target genes Hey1 and Hey2 are required for embryonic vascular development. Genes Dev. 18, 901-911. 10.1101/gad.291004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer A., Klattig J., Kneitz B., Diez H., Maier M., Holtmann B., Englert C. and Gessler M. (2005). Hey basic helix-loop-helix transcription factors are repressors of GATA4 and GATA6 and restrict expression of the GATA target gene ANF in fetal hearts. Mol. Cell. Biol. 25, 8960-8970. 10.1128/MCB.25.20.8960-8970.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao S., Wang Z., Wang W., Hu X., Chen P., Li J., Feng X., Wong J. and Du J. X. (2017). The lysine methyltransferase SMYD2 methylates the kinase domain of type II receptor BMPR2 and stimulates bone morphogenetic protein signaling. J. Biol. Chem. 292, 12702-12712. 10.1074/jbc.M117.776278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibb N., Lazic S., Yuan X., Deshwar A. R., Leslie M., Wilson M. D. and Scott I. C. (2018). Hey2 regulates the size of the cardiac progenitor pool during vertebrate heart development. Development (Cambridge, England) 145, dev167510. [DOI] [PubMed] [Google Scholar]
- González-Rosa J. M., Sharpe M., Field D., Soonpaa M. H., Field L. J., Burns C. E. and Burns C. G. (2018). Myocardial polyploidization creates a barrier to heart regeneration in zebrafish. Dev. Cell 44, 433-446. 10.1016/j.devcel.2018.01.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlieb P. D., Pierce S. A., Sims R. J., Yamagishi H., Weihe E. K., Harriss J. V., Maika S. D., Kuziel W. A., King H. L., Olson E. N. et al. (2002). Bop encodes a muscle-restricted protein containing MYND and SET domains and is essential for cardiac differentiation and morphogenesis. Nat. Genet. 31, 25-32. 10.1038/ng866 [DOI] [PubMed] [Google Scholar]
- Han Y., Chen A., Umansky K.-B., Oonk K. A., Choi W.-Y., Dickson A. L., Ou J., Cigliola V., Yifa O., Cao J. et al. (2019). Vitamin D Stimulates Cardiomyocyte Proliferation and Controls Organ Size and Regeneration in Zebrafish. Dev. Cell 48, 853-863. 10.1016/j.devcel.2019.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison M. R., Feng X., Mo G., Aguayo A., Villafuerte J., Yoshida T., Pearson C. A., Schulte-Merker S. and Lien C. L. (2019). Late developing cardiac lymphatic vasculature supports adult zebrafish heart function and regeneration. eLife 8, e42762 10.7554/eLife.42762.sa2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heisig J., Weber D., Englberger E., Winkler A., Kneitz S., Sung W.-K., Wolf E., Eilers M., Wei C.-L. and Gessler M. (2012). Target gene analysis by microarrays and chromatin immunoprecipitation identifies HEY proteins as highly redundant bHLH repressors. PLoS Genet. 8, e1002728 10.1371/journal.pgen.1002728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia H., King I. N., Chopra S. S., Wan H., Ni T. T., Jiang C., Guan X., Wells S., Srivastava D. and Zhong T. P. (2007). Vertebrate heart growth is regulated by functional antagonism between Gridlock and Gata5. Proc. Natl. Acad. Sci. USA 104, 14008-14013. 10.1073/pnas.0702240104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin S.-W., Beis D., Mitchell T., Chen J. N. and Stainier D. Y. (2005). Cellular and molecular analyses of vascular tube and lumen formation in zebrafish. Development (Cambridge, England) 132, 5199-5209. 10.1242/dev.02087 [DOI] [PubMed] [Google Scholar]
- Jopling C., Sleep E., Raya M., Martí M., Raya A. and Belmonte J. C. I. (2010). Zebrafish heart regeneration occurs by cardiomyocyte dedifferentiation and proliferation. Nature 464, 606-609. 10.1038/nature08899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jopling C., Suñè G., Morera C. and Izpisua Belmonte J. C. (2012). p38α MAPK regulates myocardial regeneration in zebrafish. Cell cycle 11, 1195-1201. 10.4161/cc.11.6.19637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Just S., Meder B., Berger I. M., Etard C., Trano N., Patzel E., Hassel D., Marquart S., Dahme T., Vogel B. et al. (2011). The myosin-interacting protein SMYD1 is essential for sarcomere organization. J. Cell Sci. 124, 3127-3136. 10.1242/jcs.084772 [DOI] [PubMed] [Google Scholar]
- Kathiriya I. S., King I. N., Murakami M., Nakagawa M., Astle J. M., Gardner K. A., Gerard R. D., Olson E. N., Srivastava D. and Nakagawa O. (2004). Hairy-related transcription factors inhibit GATA-dependent cardiac gene expression through a signal-responsive mechanism. J. Biol. Chem. 279, 54937-54943. 10.1074/jbc.M409879200 [DOI] [PubMed] [Google Scholar]
- Kikuchi K., Holdway J. E., Werdich A. A., Anderson R. M., Fang Y., Egnaczyk G. F., Evans T., MacRae C. A., Stainier D. Y. and Poss K. D. (2010). Primary contribution to zebrafish heart regeneration by gata4(+) cardiomyocytes. Nature 464, 601-605. 10.1038/nature08804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi K., Holdway J. E., Major R. J., Blum N., Dahn R. D., Begemann G. and Poss K. D. (2011). Retinoic acid production by endocardium and epicardium is an injury response essential for zebrafish heart regeneration. Dev. Cell 20, 397-404. 10.1016/j.devcel.2011.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koibuchi N. and Chin M. T. (2007). CHF1/Hey2 plays a pivotal role in left ventricular maturation through suppression of ectopic atrial gene expression. Circ. Res. 100, 850-855. 10.1161/01.RES.0000261693.13269.bf [DOI] [PubMed] [Google Scholar]
- Kokubo H., Tomita-Miyagawa S., Hamada Y. and Saga Y. (2007). Hesr1 and Hesr2 regulate atrioventricular boundary formation in the developing heart through the repression of Tbx2. Development (Cambridge, England) 134, 747-755. 10.1242/dev.02777 [DOI] [PubMed] [Google Scholar]
- Lee S. H., Kim E.-K., Kwon J.-E., Lee J.-K., Lee D., Kim S.-Y., Seo H.-B., Na H. S., Jung K., Kwok S.-K. et al. (2017). Ssu72 attenuates autoimmune arthritis via targeting of STAT3 signaling and Th17 activation. Sci. Rep. 7, 5506 10.1038/s41598-017-05421-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepilina A., Coon A. N., Kikuchi K., Holdway J. E., Roberts R. W., Burns C. G. and Poss K. D. (2006). A dynamic epicardial injury response supports progenitor cell activity during zebrafish heart regeneration. Cell 127, 607-619. 10.1016/j.cell.2006.08.052 [DOI] [PubMed] [Google Scholar]
- Li H., Zhong Y., Wang Z., Gao J., Xu J., Chu W., Zhang J., Fang S. and Du S. J. (2013). Smyd1b is required for skeletal and cardiac muscle function in zebrafish. Mol. Biol. Cell 24, 3511-3521. 10.1091/mbc.e13-06-0352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Yang H. and Zhong T. P. (2015). Regeneration across metazoan phylogeny: lessons from model organisms. J. Genet. Genomics 42, 57-70. 10.1016/j.jgg.2014.12.002 [DOI] [PubMed] [Google Scholar]
- Li L. X., Fan L. X., Zhou J. X., Grantham J. J., Calvet J. P., Sage J. and Li X. (2017). Lysine methyltransferase SMYD2 promotes cyst growth in autosomal dominant polycystic kidney disease. J. Clin. Investig. 127, 2751-2764. 10.1172/JCI90921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L. X., Zhou J. X., Calvet J. P., Godwin A. K., Jensen R. A. and Li X. (2018a). Lysine methyltransferase SMYD2 promotes triple negative breast cancer progression. Cell Death Dis. 9, 326 10.1038/s41419-018-0347-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Liu W., Yin J., Chen Y., Guo S., Fan H., Li X., Zhang X., He X. and Duan C. (2018b). TSG-6 attenuates inflammation-induced brain injury via modulation of microglial polarization in SAH rats through the SOCS3/STAT3 pathway. J. Neuroinflammation 15, 231 10.1186/s12974-018-1279-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Li H.-Y., Gu S.-Y., Zi H.-X., Jiang L. and Du J.-L. (2020). One-step generation of zebrafish carrying a conditional knockout-knockin visible switch via CRISPR/Cas9-mediated intron targeting. Sci. China. Life Sci. 63, 59-67. 10.1007/s11427-019-1607-9 [DOI] [PubMed] [Google Scholar]
- Liu D., Wang Z., Xiao A., Zhang Y., Li W., Zu Y., Yao S., Lin S. and Zhang B. (2014). Efficient gene targeting in zebrafish mediated by a zebrafish-codon-optimized cas9 and evaluation of off-targeting effect. J. Genet. Genomics 41, 43-46. 10.1016/j.jgg.2013.11.004 [DOI] [PubMed] [Google Scholar]
- Livak K. J. and Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25, 402-408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Marin-Juez R., Marass M., Gauvrit S., Rossi A., Lai S.-L., Materna S. C., Black B. L. and Stainier D. Y. R. (2016). Fast revascularization of the injured area is essential to support zebrafish heart regeneration. Proc. Natl. Acad. Sci. USA 113, 11237-11242. 10.1073/pnas.1605431113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao L., Li J., Li J., Tian X., Lu Y., Hu S., Shieh D., Kanai R., Zhou B.-Y., Zhou B. et al. (2018). Notch signaling regulates Hey2 expression in a spatiotemporal dependent manner during cardiac morphogenesis and trabecular specification. Sci. Rep. 8, 2678 10.1038/s41598-018-20917-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Missinato M. A., Saydmohammed M., Zuppo D. A., Rao K. S., Opie G. W., Kuhn B. and Tsang M. (2018). Dusp6 attenuates Ras/MAPK signaling to limit zebrafish heart regeneration. Development (Cambridge, England) 145, dev157206 10.1242/dev.157206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamed T. M. A., Ang Y.-S., Radzinsky E., Zhou P., Huang Y., Elfenbein A., Foley A., Magnitsky S. and Srivastava D. (2018). Regulation of cell cycle to stimulate adult cardiomyocyte proliferation and cardiac regeneration. Cell 173, 104-116. 10.1016/j.cell.2018.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monroe T. O., Hill M. C., Morikawa Y., Leach J. P., Heallen T., Cao S., Krijger P. H. L., de Laat W., Wehrens X. H. T., Rodney G. G. et al. (2019). YAP Partially reprograms chromatin accessibility to directly induce adult cardiogenesis In Vivo. Dev. Cell 48, 765-779.e7. 10.1016/j.devcel.2019.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa O., Nakagawa M., Richardson J. A., Olson E. N. and Srivastava D. (1999). HRT1, HRT2, and HRT3: a new subclass of bHLH transcription factors marking specific cardiac, somitic, and pharyngeal arch segments. Dev. Biol. 216, 72-84. 10.1006/dbio.1999.9454 [DOI] [PubMed] [Google Scholar]
- Nguyen H., Allali-Hassani A., Antonysamy S., Chang S., Chen L. H., Curtis C., Emtage S., Fan L., Gheyi T., Li F. et al. (2015). LLY-507, A cell-active, potent, and selective inhibitor of protein-lysine methyltransferase SMYD2. J. Biol. Chem. 290, 13641-13653. 10.1074/jbc.M114.626861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni T. T., Rellinger E. J., Mukherjee A., Xie S., Stephens L., Thorne C. A., Kim K., Hu J., Lee E., Marnett L. et al. (2011). Discovering small molecules that promote cardiomyocyte generation by modulating Wnt signaling. Chem. Biol. 18, 1658-1668. 10.1016/j.chembiol.2011.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palencia-Desai S., Kohli V., Kang J., Chi N. C., Black B. L. and Sumanas S. (2011). Vascular endothelial and endocardial progenitors differentiate as cardiomyocytes in the absence of Etsrp/Etv2 function. Development (Cambridge, England) 138, 4721-4732. 10.1242/dev.064998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea M., Pertea G. M., Antonescu C. M., Chang T.-C., Mendell J. T. and Salzberg S. L. (2015). StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33, 290-295. 10.1038/nbt.3122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea M., Kim D., Pertea G. M., Leek J. T. and Salzberg S. L. (2016). Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 11, 1650-1667. 10.1038/nprot.2016.095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porrello E. R., Mahmoud A. I., Simpson E., Hill J. A., Richardson J. A., Olson E. N. and Sadek H. A. (2011). Transient regenerative potential of the neonatal mouse heart. Science (New York, N.Y.) 331, 1078-1080. 10.1126/science.1200708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poss K. D., Wilson L. G. and Keating M. T. (2002). Heart regeneration in zebrafish. Science (New York, N.Y.) 298, 2188-2190. 10.1126/science.1077857 [DOI] [PubMed] [Google Scholar]
- Ren C.-G., Wang L., Jia X.-E., Liu Y.-J., Dong Z.-W., Jin Y., Chen Y., Deng M., Zhou Y., Zhou Y. et al. (2013). Activated N-Ras signaling regulates arterial-venous specification in zebrafish. J. Hematol. Oncol. 6, 34 10.1186/1756-8722-6-34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M. D., McCarthy D. J. and Smyth G. K. (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139-140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowlinson J. M. and Gering M. (2010). Hey2 acts upstream of Notch in hematopoietic stem cell specification in zebrafish embryos. Blood 116, 2046-2056. 10.1182/blood-2009-11-252635 [DOI] [PubMed] [Google Scholar]
- Rutenberg J. B., Fischer A., Jia H., Gessler M., Zhong T. P. and Mercola M. (2006). Developmental patterning of the cardiac atrioventricular canal by Notch and Hairy-related transcription factors. Development (Cambridge, England) 133, 4381-4390. 10.1242/dev.02607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saddic L. A., West L. E., Aslanian A., Yates J. R. III, Rubin S. M., Gozani O. and Sage J. (2010). Methylation of the retinoblastoma tumor suppressor by SMYD2. J. Biol. Chem. 285, 37733-37740. 10.1074/jbc.M110.137612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto M., Hirata H., Ohtsuka T., Bessho Y. and Kageyama R. (2003). The basic helix-loop-helix genes Hesr1/Hey1 and Hesr2/Hey2 regulate maintenance of neural precursor cells in the brain. J. Biol. Chem. 278, 44808-44815. 10.1074/jbc.M300448200 [DOI] [PubMed] [Google Scholar]
- Satow T., Bae S.-K., Inoue T., Inoue C., Miyoshi G., Tomita K., Bessho Y., Hashimoto N. and Kageyama R. (2001). The basic helix-loop-helix gene hesr2 promotes gliogenesis in mouse retina. J. Neurosci. 21, 1265-1273. 10.1523/JNEUROSCI.21-04-01265.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spellmon N., Holcomb J., Trescott L., Sirinupong N. and Yang Z. (2015). Structure and function of SET and MYND domain-containing proteins. Int. J. Mol. Sci. 16, 1406-1428. 10.3390/ijms16011406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stender J. D., Pascual G., Liu W., Kaikkonen M. U., Do K., Spann N. J., Boutros M., Perrimon N., Rosenfeld M. G. and Glass C. K. (2012). Control of proinflammatory gene programs by regulated trimethylation and demethylation of histone H4K20. Mol. Cell 48, 28-38. 10.1016/j.molcel.2012.07.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan X., Rotllant J., Li H., De Deyne P. and Du S. J. (2006). SmyD1, a histone methyltransferase, is required for myofibril organization and muscle contraction in zebrafish embryos. Proc. Natl. Acad. Sci. USA 103, 2713-2718. 10.1073/pnas.0509503103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian X., Li Y., He L., Zhang H., Huang X., Liu Q., Pu W., Zhang L., Li Y., Zhao H. et al. (2017). Identification of a hybrid myocardial zone in the mammalian heart after birth. Nat. Commun. 8, 87 10.1038/s41467-017-00118-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzahor E. and Poss K. D. (2017). Cardiac regeneration strategies: staying young at heart. Science (New York, N.Y.) 356, 1035-1039. 10.1126/science.aam5894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urasaki A., Morvan G. and Kawakami K. (2006). Functional dissection of the Tol2 transposable element identified the minimal cis-sequence and a highly repetitive sequence in the subterminal region essential for transposition. Genetics 174, 639-649. 10.1534/genetics.106.060244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voelkel T., Andresen C., Unger A., Just S., Rottbauer W. and Linke W. A. (2013). Lysine methyltransferase Smyd2 regulates Hsp90-mediated protection of the sarcomeric titin springs and cardiac function. Biochim. Biophys. Acta 1833, 812-822. 10.1016/j.bbamcr.2012.09.012 [DOI] [PubMed] [Google Scholar]
- Wang J., Panáková D., Kikuchi K., Holdway J. E., Gemberling M., Burris J. S., Singh S. P., Dickson A. L., Lin Y.-F., Sabeh M. K. et al. (2011). The regenerative capacity of zebrafish reverses cardiac failure caused by genetic cardiomyocyte depletion. Development (Cambridge, England) 138, 3421-3430. 10.1242/dev.068601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C.-C., Kruse F., Vasudevarao M. D., Junker J. P., Zebrowski D. C., Fischer K., Noël E. S., Grun D., Berezikov E., Engel F. B. et al. (2016). Spatially resolved genome-wide transcriptional profiling identifies BMP signaling as essential regulator of zebrafish cardiomyocyte regeneration. Dev. Cell 36, 36-49. 10.1016/j.devcel.2015.12.010 [DOI] [PubMed] [Google Scholar]
- Xin M., Small E. M., van Rooij E., Qi X., Richardson J. A., Srivastava D., Nakagawa O. and Olson E. N. (2007). Essential roles of the bHLH transcription factor Hrt2 in repression of atrial gene expression and maintenance of postnatal cardiac function. Proc. Natl. Acad. Sci. USA 104, 7975-7980. 10.1073/pnas.0702447104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin M., Olson E. N. and Bassel-Duby R. (2013). Mending broken hearts: cardiac development as a basis for adult heart regeneration and repair. Nat. Rev. Mol. Cell Biol. 14, 529-541. 10.1038/nrm3619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Liu R.-X., Chan K.-W., Hu J., Zhang J., Wei L., Tan H., Yang X. and Liu H. (2019). Exosomal transfer of p-STAT3 promotes acquired 5-FU resistance in colorectal cancer cells. J. Exp. Clin. Cancer Res. 38, 320 10.1186/s13046-019-1314-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L., Ben-Yair R., Burns C. E. and Burns C. G. (2019). Endocardial notch signaling promotes cardiomyocyte proliferation in the regenerating zebrafish heart through Wnt pathway antagonism. Cell Rep 26, 546-554.e5. 10.1016/j.celrep.2018.12.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong T. P., Rosenberg M., Mohideen M. A., Weinstein B. and Fishman M. C. (2000). gridlock, an HLH gene required for assembly of the aorta in zebrafish. Science (New York, N.Y.) 287, 1820-1824. 10.1126/science.287.5459.1820 [DOI] [PubMed] [Google Scholar]
- Zhong T. P., Childs S., Leu J. P. and Fishman M. C. (2001). Gridlock signalling pathway fashions the first embryonic artery. Nature 414, 216-220. 10.1038/35102599 [DOI] [PubMed] [Google Scholar]