Abstract
Fate determination in self-renewal and differentiation of hematopoietic stem and progenitor cells (HSCs and HPCs) is ultimately controlled by gene expression, which is profoundly influenced by the global and local chromatin state. Cellular metabolism directly influences the chromatin state through the dynamic regulation of the enzymatic activities that modify DNA and histones, but also generates genotoxic metabolites that can damage DNA and thus pose threat to the genome integrity. On the other hand, mechanisms modulating the chromatin state impact metabolism by regulating the expression and activities of key metabolic enzymes. Moreover, through regulating either DNA damage response directly or expression of genes involved in this process, chromatin modulators play active and crucial roles in guarding the genome integrity, breaching of which results in defective HSPC function. Therefore, HSPC function is regulated by the dynamic and two-way interactions between metabolism and chromatin. Here, we review recent advances that provide a chromatin perspective on the major impacts the metabolic and genotoxic factors can have on HSPC function and fate determination.
Keywords: Chromatin, Histone modifications, Demethylation, Hematopoietic stem cells, Metabolism, Genome integrity, DNA damage response
Introduction
The mission of somatic stem cells is to maintain tissue homeostasis throughout the lifespan of an organism. To accomplish this mission, stem cells have two defining properties, self-renewal to replenish themselves and differentiation to give arise to cells of all lineages in the specific tissue. Hematopoietic stem cells (HSCs) provide a continuous supply of blood cells by strictly regulating self-renewal and balanced lineage differentiation through generating hematopoietic progenitor cells (HPCs) and further downstream cells. Blood disease can arise as a result of dysregulated activities of HSCs and/or HPCs (HSPCs), as seen in bone marrow failure or hematologic malignancy [1]. Fate determination is thus probably the most pertinent question in stem cell biology. Chromatin is at the forefront of stem cell biology for two main reasons: (1) chromatin is the physiological substrate of transcription reaction and profoundly regulates gene expression including transcription and RNA splicing, and it is the gene expression profile of a cell that ultimately determines the cell to be still a stem cell or a daughter cell of a particular lineage. (2) Chromatin constitutes the physical carrier of genetic information, whose integrity is influenced by chromatin state and directly linked to how stem cells decide to choose the path forward.
Chromatin in eukaryotic cells consists of nucleosomes with genomic DNA wrapping around histone proteins, and can be packaged at different levels. Two copies each of H2A, H2B, H3, and H4 form the histone octamer in the core nucleosome particle, and histone H1 sits at the base of the nucleosome and binds to the linker DNA. In general, chromatin represents a barrier to all DNA-based processes including transcription, replication, and DNA repair. However, chromatin provides an important regulatory platform for these processes by integrating signals from many aspects of cellular state, through epigenetic mechanisms to regulate how compact and accessible the local chromatin is [2]. These epigenetic mechanisms include histone and DNA modifications, chromatin remodeling, and high-order chromatin structure formation. Histones are subject to a wide range of post-translational modifications including acetylation, methylation, phosphorylation, ubiquitylation, succinylation, formylation, hydroxylation, O-GlcNAcylation, and ADP ribosylation, among many others [2, 3]. DNA is also subject to chemical modifications, mainly cytosine methylation and subsequent oxidation including 5-hydroxymethylcytosine (5hmC) [4]. These modifications are dynamically controlled by a balanced act of “writers” (enzymes that add these modifications) and “erasers” (enzymes that remove these modifications) [3]. The functions of these modifications can be through a direct physical impact on the compact level of chromatin, through an impact on charge, or, more commonly, mediated by “readers” that selectively bind to the modified chromatin [2, 3]. Aberrant chromatin modifications and remodeling are frequently associated with human diseases including cancer, and drugs targeting chromatin regulators are being developed to treat cancers including blood cancer [2, 3, 5–7].
Adult HSCs reside in a hypoxic bone marrow environment with limited nutrients. To adapt to this condition, HSCs remain in a quiescent state and mainly utilize glycolysis for energy production, but switch to mitochondrial oxidative phosphorylation upon activation, such as acute blood loss or viral infection [8–10]. Mutations of metabolic enzymes and dysregulation of metabolic pathways are commonly associated with human disease [11], including disorders and cancer in the blood system [12–14]. Like all cellular activities, chromatin-based processes depend on and are regulated by metabolism. Metabolism provides the basic building blocks of nucleic acids and histones, as well as energy that is required for the chromatin-based processes. Moreover, many small molecules used or produced in metabolism are essential co-substrates or co-factors in reactions that add or remove modifications of histones and DNA. In addition, certain metabolites can influence chromatin activity via covalently or allosterically modulating the activities of the proteins that modify histones and DNA [15–17]. Metabolism is also reciprocally regulated by these chromatin-based processes, mostly through control of the expression level and activities of key enzymes involved in metabolic reactions (Fig. 1a). Certain core metabolic enzymes can translocate to nucleus and modulate gene expression either by modifying chromatin or chromatin modulators or by forming complexes with transcription and chromatin modulators [17], but this is not strict “metabolic regulation” of chromatin and thus not reviewed here.
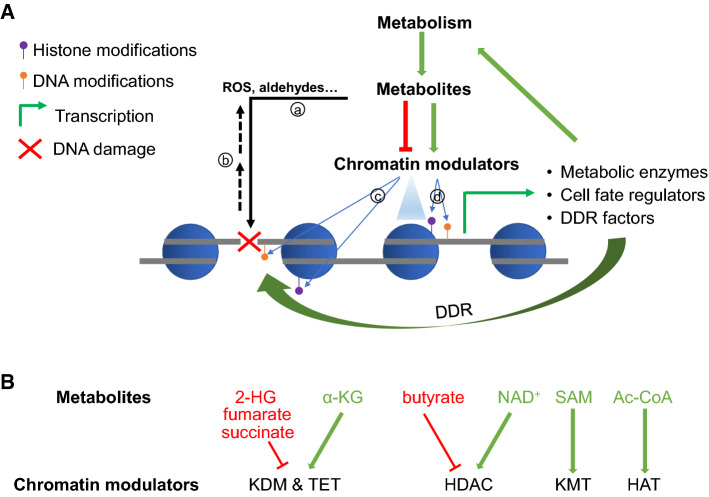
Fig. 1.
Cross-regulation of metabolism and chromatin that control HSPC function. a Metabolic reactions use precursors and produce metabolites, some of which are (a) genotoxic and damage DNA, and some of which can directly regulate (enhance or inhibit) the activities of chromatin modulators in modifying DNA and/or histones and remodeling nucleosomes. b DNA damage is also suggested to cause increased ROS. c Changes in the chromatin state at the damaged DNA sites can directly influence the sensing and repair of the damage. d Chromatin changes also activate or repress transcription and expression of genes encoding key metabolic enzymes, cell fate regulators, and DDR factors that participate in DDR, which together influence the function and fate determination of cells in different lineages, including HSPCs. b Examples of metabolites that inhibit (in red) or promote (in green) the corresponding chromatin enzymes
All cells constantly face genotoxic insults induced by both environmental agents and cellular processes, which can cause various types of DNA damage and catastrophic consequences to the function of cells and organism. Many metabolic pathways produce these genotoxic molecules, including reactive oxygen species (ROS) and aldehydes. Due to the special role of stem cells in tissue homeostasis, maintaining genetic stability is particularly crucial for them to ensure the propagation of correct genetic information throughout the daughter cells in the tissue [18]. Following the detection of DNA lesions, the intricate network of DNA damage response (DDR) orchestrates a signal transduction pathway that activates multiple layers of decisions, including cell cycle arrest, apoptosis, senescence, and DNA repair [19]. DDR profoundly influences both normal hematopoiesis and hematologic malignancies [20], as demonstrated by human blood disease such as Fanconi anemia that results from mutations in DNA repair genes [21], and HSC defects in mice deficient of active components in either recognition (such as ATM [22]) or repair [23, 24] of DNA lesions in the DDR pathways. As DDR occurs in the context of chromatin, it is governed by epigenetic mechanisms regulating chromatin structure and accessibility [25, 26].
Many excellent reviews have covered the biochemical relationships between metabolism and chromatin [15–17, 27–29], and between DNA damage and chromatin [25, 26, 30]. Others have covered how HSCs are regulated by epigenetics [31, 32], metabolism [10, 33–35], and DNA damage [18, 20, 36]. It is possible that most metabolic impacts on stem cells fate determination ultimately cause changes in global or local chromatin states and transcription, but many of these changes are probably indirect. Here, we focus on our current understanding of how metabolism and chromatin modifications mutually and directly regulate each other and impact genome integrity, to fulfill their roles in normal as well as malignant hematopoietic cells (Fig. 1). Many biochemical studies and studies from other cellular systems are formulating conceptual frameworks that have not yet been tested or shown in HSPCs. Therefore, in addition to reviewing published works that provide evidence of these cellular processes in HSPCs, we also try to briefly include these findings here to prompt interest in testing them in hematopoiesis and thus lead to better understanding of normal and diseased hematopoiesis.
Metabolic regulation of chromatin in HSPCs
Regulation of chromatin enzyme activities by metabolic molecules
It is increasingly evident that metabolism is dynamically regulated during cellular differentiation, and alteration of the metabolites direct impacts chromatin modifications, thereby influencing fate determination of embryonic stem cells and somatic cells in different lineages, including muscle and immune cell lineages [15, 16, 37]. Histone modifications have been reported to be affected by metabolites such as s-adenosylmethionine (SAM), alpha-ketoglutarate (α-KG), 2-hydroxyglutarate (2-HG), acetyl-coA, butyrate, and NAD+ [15, 16, 37] (some examples are shown in Fig. 1b). Moreover, through direct impact on chromatin modifications, metabolites can influence spatial interactions of chromatin regions and higher order chromatin structure, thereby influencing gene expression for T-cell fate determination [38] and cancer [39]. Below, we will mainly review the direct metabolism–chromatin connections in HSPCs, focusing on the metabolic regulation of α-ketoglutarate (α-KG)-dependent dioxygenases, a paradigm for metabolic factors regulating chromatin and cell fate. A large variety of enzymes in the α-KG-dependent dioxygenases family directly or indirectly regulate chromatin, employ Fe(II), oxygen, and α-KG as co-factors or co-substrates, and are stimulated by ascorbate (vitamin C, VC) [40]. These small molecules are all intrinsically involved in metabolism, thus offering rich connections between chromatin and the cellular metabolic states (Fig. 2).
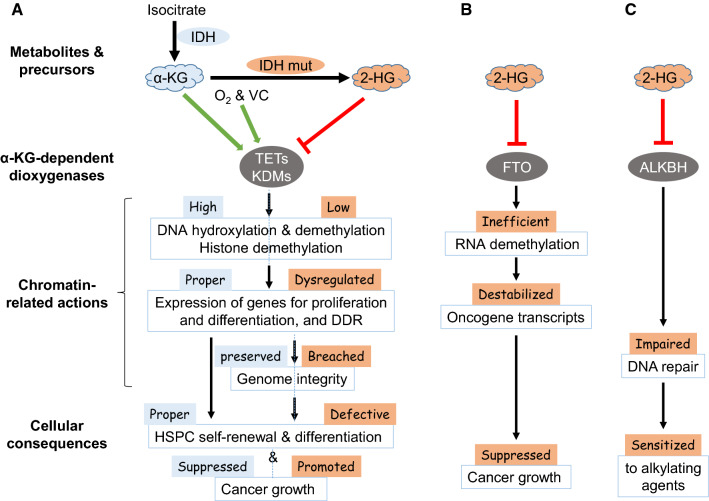
Fig. 2.
Different chromatin regulation of cell functions by the metabolites from isocitrate. α-KG is generated from isocitrate by IDH, and serves as an essential cofactor for a large number of α-KG-dependent dioxygenases including those shown here. These enzymes exert different molecular activities on chromatin (a), RNA modifications (b), and DNA repair proteins (c), which affect gene expression and DDR to ensure proper gene expression and preserve genome integrity for normal hematopoietic cell fate determination and function. In contrast, 2-HG is generated from α-KG by IDH mutants frequently found in cancer, and inhibits these α-KG-dependent dioxygenases and affect the different activities on chromatin (a), RNA (b), and DNA repair (c), and eventually lead to aberrant hematopoietic fate determination and different outcome of cancer growth
Oxygen
Oxygen has profound impact on cell fate determination through both indirect and direct effects on chromatin and gene expression. The indirect effects are mainly through the intricate network around the HIF (hypoxia-inducible factor) transcription factors, which induce target genes for hypoxia adaptation. Oxygen sensing is accomplished by the oxygen- and α-KG-dependent dioxygenase family members that prolyl hydroxylate HIFα, causing its ubiquitination and degradation under normoxic conditions [41]. The HIF network, partly by promoting the expression of glycolytic enzymes and key enzymes that suppress tricarboxylic acid cycle, plays an important role in keeping HSCs in a quiescent state mainly engaged in anaerobic glycolysis instead of the TCA cycle and mitochondrial respiration in the hypoxic bone marrow niche [10].
A more direct regulation of chromatin by oxygen is mediated by Jmjc domain-containing histone lysine demethylases (KDMs) and the 10–11 translocation (TET) proteins, which are α-KG-dependent dioxygenase family members that carry out demethylation of histones and DNA, respectively [40]. The intracellular oxygen level can impact activities of some of these enzymes that have oxygen affinity commensurate with physiological oxygen concentration [42]. The activity of KDM5A as a H3K4 demethylase is inhibited by low oxygen level, and is important for chromatin and transcriptional response to hypoxia in cells [43, 44]. KDM6A, a histone H3K27 demethylase, has the lowest oxygen affinity of all tested KDMs and is thus highly sensitive to oxygen levels. Hypoxia promotes the persistence of H3K27me3 by directly inactivating KDM6A and inhibits transcriptional reprogramming and muscle cell fate transition [45]. Profound hypoxia found in tumors can inhibit TETs and lead to DNA hyper-methylation, which contributes to tumor progression under hypoxic conditions [46]. It remains to be tested if the hypoxic condition in bone marrow contributes to the maintenance of HSCs in an undifferentiated state partly by suppressing the activity of these oxygen-sensitive chromatin enzymes and thus keeping differentiation genes from aberrant activation. On the other hand, it is possible that as HPCs move into microenvironments with higher oxygen levels, the increased activities of these enzymes may promote lineage differentiation by removal of suppressive marks from the lineage-specific genes. Obviously, different KDMs have different impact on the chromatin and transcription states, and their activities need to be coordinated to respond to oxygen levels to govern stem cell fate.
Vitamin C
Recent studies have shown a crucial role of VC, an essential vitamin for human, in stem cells and leukemogenesis through directly influencing chromatin modifications. VC can enhance the activity of JmjC domain-containing KDMs, TET enzymes, and various other FeII- and α-ketoglutarate-dependent dioxygenases [47]. VC treatment of mouse embryonic stem (ES) cells induces a blastocyst-like state through promoting TET-mediated DNA demethylation [48], and maternal VC is important for the development of female fetal germ cells likely via regulating TET-dependent DNA demethylation [49]. Moreover, VC can enhance the reprograming efficiency of mouse embryonic fibroblasts to a pluripotent state [50], through the activation of KDMs for H3K36 and H3K9 [51, 52], and also by elevating DNA 5hmC levels in a TET-dependent manner [53].
VC is enriched in human and mouse HSCs and multipotent progenitor cells (MPPs) compared to the hematopoietic progenitors and downstream committed blood cells [54]. Such a high VC level limits the frequency and function of HSCs, and also inhibits leukemogenesis, in part through promoting the activities of TET2 in DNA demethylation [54]. Depletion or loss-of-function mutations of TET2 are frequently found in hematologic malignancies, and its deficiency leads to skewed differentiation and enhanced repopulating capacity of HSCs [55–59]. Treatment of TET2-mutated HSCs with VC blocks their aberrant self-renewal activity, mimicking the effect of TET2 restoration [56]. Incubation of VC with human leukemia cells impairs their colony formation, and administration of high dose of VC in vivo increases 5hmC level and inhibits disease progression in leukemia xenograft models [56], suggesting a therapeutic potential of VC in leukemia treatment.
VC is also required for HPC generation from human pluripotent stem cell differentiation in vitro and also during zebrafish development [60]. In the absence of VC, endothelial cells fail to undergo endothelial-to-hematopoietic transition to generate functional hemogenic endothelial cells, likely because of failure to open up chromatin regions important for hematopoiesis. Deletion of VC-dependent H3K27 demethylases, KDM6A or KDM6B, or DNA demethylase TET1 impaired HPC generation in the presence of VC. These results demonstrate a critical role of VC in HPC generation through regulation of key histone and DNA demethylase activities [60].
α-KG
Intracellular α-KG is mainly generated by catalyzing the oxidative decarboxylation of isocitrate by isocitrate dehydrogenase 1/2 (IDH1/2), which are frequently mutated in human cancers including acute myeloid leukemia. IDH1/2 mutants convert α-KG to 2-hydroxyglutarate (2-HG), which competitively inhibit a wide range of α-KG-dependent dioxygenases including TETs and KDMs [61]. However, the effects of IDH mutations on cancer phenotypes may differ greatly for different stages and types of cancer, due to the different enzymes that are affected (Fig. 2). In contrast to the oncogenic role of 2-HG in IDH mutant cancers largely through inhibiting DNA and histone demethylation (Fig. 2a), 2-HG also has anti-leukemic activity by inhibiting FTO, an α-KG-dependent dioxygenase that functions as an RNA N6-methyladenosine (m6A) demethylase, and the subsequent destabilization of MYC transcript [62] (Fig. 2b). IDH mutations also impact DNA repair either indirectly through influencing histone methylation state and expression of DDR gene [63] (Fig. 2a) or directly through inhibiting ALKBH proteins that are α-KG-dependent dioxygenases responsible for repairing DNA alkylation lesions [64] (Fig. 2c). The latter mechanism sensitizes IDH-mutated cancer cells to alkylating agents [64]. 2-HG also inhibits prolyl hydroxylases that control the stability of HIF-1α, but the role of this regulation in cancer and cell fate determination remains controversial [61].
NAD+
Mammalian sirtuins are NAD+-dependent deacetylases for both histones and numerous non-histone proteins, and regulate a large number of cellular pathways [65]. As different sirtuins have different subcellular localization, probably only the nuclear SIRTs (SIRT1, 6, and 7) can impact stem cell functions by directly regulating chromatin. SIRT1 is the best studied sirtuin in HSC function [66]. SIRT1-deficient HSPCs show aberrant expansion under stress [67], and myeloid bias in differentiation and aging-like phenotype [68]. SIRT1 binds to the Hoxa9 gene, erases its promoter H4K16 acetylation, and helps to maintain the repressive H3K27me3 mark [67]. In line with such a direct role of SIRT1 in repressing HSC maintenance genes, another study shows that the deacetylation activity of SIRT1 counteracts the non-catalytic activity of MLL1 in recruiting histone acetyltransferase to HSC maintenance genes [69]. SIRT1 can also indirectly impact chromatin by directly deacetylating lysines of TET2 in its catalytic domain and enhancing TET2 activity, which is known to restrict HSPC transformation. By doing so, SIRT1 activation can inhibit the aberrant HSPC functions in Myelodysplastic syndrome, a type of hematologic malignancy [70]. SIRT1 and other sirtuins also regulate normal hematopoietic and leukemic stem cells through mechanisms with indirect or unclear involvement of chromatin [65, 71]. The NAD requirement indicates an intrinsic link of metabolism and sirtuins’ activities. Indeed, muscle stem cells undergo a metabolic shift leading to reduction of NAD+ and SIRT1 activity, with increase in H4K16 acetylation and altered gene expression during activation [72]. However, it remains to be demonstrated that NAD-related metabolism directly affects HSPC function via impacting the sirtuins’ activities.
The mitochondria–chromatin connections
As a central player in metabolism, mitochondria are reviewed here on their connections with chromatin in HSPC function through their impacts on metabolites. Mitochondria are responsible for major energy production, and also play many important roles including the generation of redox molecules and metabolites, regulation of cell signaling, cell death, and biosynthetic metabolism [73]. Relative inactive in quiescent HSCs, mitochondria are rapidly activated to fulfill the metabolic demand upon differentiation [10]. Perturbations to increase [74] or disrupt [75] mitochondrial oxidative phosphorylation aberrantly activate HSCs or block their differentiation, respectively.
The mitochondrial activity can have major impact on cell fate determination through affecting chromatin. Short-term opening of the mitochondrial permeability transition pore enhances reprogramming of mouse embryonic fibroblasts, as the transient release of mitochondrial ROS upregulates a KDM to remove the H3K9 and H3K27 methylation marks from pluripotency gene promoters [76]. Loss of the mitochondrial complex III subunit Rieske iron–sulfur protein (RISP) in mouse fetal HSCs impairs their differentiation with little effect on proliferation. RISP-deficient adult HSCs also have multiple defects, including impaired differentiation, loss of quiescence, and increased DNA damage and apoptosis [77]. RISP-deficient HPCs have altered global gene expression and histone modifications, including increase in methylation of H3K4, H3K9, and H3K79. As the authors also detected relative increases in L(S)-2-HG, succinate, and fumarate, all known inhibitors of DNA and histone demethylases, the altered gene expression, and histone modifications are attributed to direct impact of the metabolic changes following mitochondrial inactivation [77]. Mitochondrial inactivation by depletion of mitochondrial transcription factor A in erythroid progenitors causes defective erythropoiesis, partly due to histone hyperacetylation and dysregulated gene expression following increased levels of β-hydroxybutyrate, a histone deacetylase inhibitor [77]. These studies are thus consistent with the notion that mitochondrial activities and metabolites directly control chromatin activity to govern HSPC fate and function.
While the aforementioned studies show the importance of sufficient mitochondrial activity in maintaining HSPC function through epigenetic mechanisms, another study reveals the other side of the coin for a balanced mitochondrial activity in chromatin control for HSC stemness. In this study, autophagy is shown to be important in maintaining HSC quiescence by clearing metabolically active mitochondria [78]. Genetic disruption of autophagy in the hematopoietic system results in premature blood aging phenotypes associated with metabolic activation, mitochondria accumulation, and accelerated myeloid differentiation of HSCs. Autophagy-deficient HSCs exhibit altered (mainly reduced) DNA methylation and an increasing trend in α-KG level. Moreover, treatment of differentiating HSCs in vitro with metabolites (SAM and α-KG) or metabolism-affecting agent impacts HSC differentiation. This study thus suggests that the uncleared active mitochondria alter HSC fate determination by directly affecting chromatin modifications [78]. On a similar line, treatment of an NAD+-boosting agent in mice potently enhances hematopoiesis and HSC functionality through promoting mitochondrial clearance and asymmetric HSC division, although it is unclear if chromatin is directly affected by the NAD+-mediated metabolic changes [79]. Therefore, these studies underscore the tight control of mitochondrial activity in maintaining the normal function of HSCs.
Metabolic impacts on genome integrity
While metabolic reactions provide energy and molecules for chromatin and chromatin-based processes, they also generate molecules that can seriously damage DNA. In addition, stress associated with DNA replication in proliferative cells also causes DNA lesions. Although the hypoxic condition and low metabolic and replicative state in quiescent HSCs help reduce intracellular oxidative and genotoxic stress, quiescent HSCs also have limited choice (error-prone non-homologous end joining) [80] and capacity in DNA repair [81]. Consequently, quiescent HSCs actually accumulate DNA damage that can be repaired upon entry into cell cycle when cells upgrade their capacity and mode (high-fidelity homologous recombination) to repair DNA [81]. In response to physiologic stress (infection or chronic blood loss), however, activation out of the quiescent state expose HSCs to greater endogenous DNA damage from the increased metabolic and replicative activities, and contributes to HSC attrition and aging phenotypes over time [82].
ROS
ROS are the best-known metabolites that can impair genome integrity. Intracellular ROS are generated mainly from metabolic reactions, such as oxidative phosphorylation in mitochondria and long-chain fatty acid oxidation in peroxisomes [83], and can also be induced by exposure to environmental toxins [80, 84]. HSC cells in the hypoxic niche mainly utilize glycolysis to produce energy, consequently making less ROS [10]. HIF proteins play a crucial role in controlling ROS production [10]. Keeping ROS levels low is important for maintaining genomic stability and self-renewal capacity of HSCs [85]. Metabolic switch to oxidative phosphorylation upon HSC activation results in increase in ROS, which further promote differentiation and maturation [10].
Low or moderate levels of ROS appear to play an important role in stem cell proliferation, differentiation, mobilization, and maintenance [10, 86]. Physiological levels of ROS are required to activate DNA repair pathway and maintain genomic stability in stem cells [87]. However, excessive amount of ROS in HSCs leads to oxidative DNA damage and, consequently, bone marrow failure. FOXO family transcription factors induce the production of many ROS-detoxifying proteins, and their deficiency results in high ROS levels and loss of self-renewal of HSCs [88]. In vitro or in vivo addition of anti-oxidative agent such as N-acetyl-l-cysteine (NAC) and catalase often greatly restores the HSC functions, suggesting a functional role of ROS in bone marrow dysfunction [22, 88, 89]. In contrast to the bulk of the highly proliferative cancer cells, the majority of leukemia stem cells (LSCs) have relatively low ROS levels associated with a quiescent cell cycle status and a low rate of energy metabolism, which may contribute to their resistance to the commonly used chemotherapy strategies that involve the induction of oxidative stress [90].
Aldehydes
Aldehydes, a common class of highly reactive metabolites, constitute a serious threat to genomic integrity that is counteracted by aldehyde-catabolizing enzymes and the Fanconi anemia (FA) DNA repair pathway [91–94]. FA is a rare disorder with congenital abnormalities including bone marrow failure and increased cancer risks. FA arises from mutations in a group of genes that regulate response to genotoxic stresses, especially DNA interstrand crosslinks. Mice double-deficient in aldehyde catabolism and FA pathway show developmental defects, predisposition to acute leukemia, and increased bone marrow failure upon consumption of ethanol, which is metabolized to acetaldehyde in cells [91, 92]. Endogenous and alcohol-derived aldehydes in these mice result in persistent DNA damage in HSPCs that lead to dramatic loss of functional HSC pool [92–94]. FA human patients with mutations in ALDH2 (acetaldehyde dehydrogenase 2) show accelerated bone marrow failure [95]. Therefore, HSCs have a coordinated pathway in response to genotoxic stresses induced by reactive aldehydes. The aldehyde-detoxifying enzymes provide the primary protection, and if this protection is compromised (by genetic mutations) or saturated (e.g., over-drinking), the potential genotoxic damage is counteracted by DDR including the FA pathway, non-homologous end joining, and homologous recombination [94]. These findings have important public health implications linking genetic mutations in metabolite-detoxifying genes and DNA repair genes, alcohol consumption, and blood diseases.
Replicative stress
Replicative stress refers to problems in DNA replication that causes the replication fork to stall, and is an important endogenous source of genome instability and a feature of cancer cells [96]. HSPCs are particularly sensitive to replicative stress as shown by impaired functionality of HSCs and erythrocyte precursors with limited replication origin licensing, a mechanism relieving replication stress [97]. While HSCs expand during aging, their functional activity declines over time. Persistence of replication stress in old HSCs is associated with cell cycle defects and chromosome breaks, and is an important driver of functional decline in aging HSCs [98].
Chromatin regulation of metabolic enzymes in hematopoietic cells
Chromatin does not just get passively impacted by metabolism and DNA damage, but plays an active role in regulating metabolism and response to DNA damage (Fig. 1) [29]. Metabolism is controlled by the expression levels and activities of numerous enzymes, especially those catalyzing the rate-limiting reactions for the entire pathway. Similarly, expression of proteins in DDR is also subject to chromatin regulation. Therefore, chromatin regulation of expression of genes in metabolism and DDR plays an important role in stem cell fate determination.
DPY30, a common core subunit of the SET1/MLL complexes that catalyze H3K4 methylation, is critical in sustaining long-term HSC self-renewal and also in enabling their differentiation [99, 100]. Loss of DPY30 in mouse results in defective HSC function associated with impaired energy metabolism in both glycolytic and mitochondrial activities. DPY30 directly regulates the expression of several genes encoding rate-limiting enzymes in glycolysis [101]. Overexpression of one of these enzymes, PKLR, could transiently and modestly rescue HSC differentiation [101], suggesting that DPY30 supports HSPC function partially through regulation of glycolysis. As DPY30-deficient cells are in an aberrant quiescent state of cell cycle, DPY30 also functions to enable HSC activation by fueling metabolic reprogramming at the chromatin level [101].
KDM4C/JMJD2C removes the repressive H3K9 methyl mark at target genes including those encoding key enzymes in the serine metabolic pathway and transcription factors in amino acid deprivation response, thereby activating amino acid biosynthesis and transportation and promoting cancer cell proliferation [102]. Loss of KDM3C/JMJD1C, another histone H3K9 demethylase, greatly impairs the self-renewal of LSC, but has minor effect on normal hematopoiesis [103, 104]. JMJD1C overexpression promotes LSC proliferation partially by reprogramming energy metabolism-related gene expression, including upregulation of genes that promote glycolytic and oxidative pathways, with consequent increase in energy production in LSCs [105]. Interestingly, the thyroid-hormone receptor-binding domain, but not the demethylase-related domains, of JMJD1C is required for this regulation [105]. Another study used CRISPR/Cas-mediated domain screening approach to show that endogenous JMJD1C requires its demethylase activity and zinc-finger domain for supporting MLL-rearranged leukemia [104]. The apparent discrepancy in domain requirement between these studies may arise from the different approaches of overexpression [105] versus deletion of endogenous gene domains [104], the former needing cautious interpretation in the absence of evidence for JMJD1C overexpression in leukemia. This is an important question as a requirement of the demethylation activity would support a rationale in targeting this enzymatic region. Further studies on identification of the direct target genes and deeper biochemical dissection of JMJD1C will help to tease out the specific contribution of different molecular activities of this chromatin modulator in connecting metabolism and LSC activity.
Role of chromatin modulators in guarding HSPC genome integrity
Much like its impact on transcription, chromatin organization also represents a major barrier to DDR, as both the sensing and repair factors need to gain access to the damaged DNA that is associated with the packed chromatin. Therefore, the cooperation of DDR factors and chromatin modulators is crucial for efficient recognition and repair of DNA lesions [106, 107]. In addition to the direct involvement in DDR, chromatin modulators also indirectly control DDR by regulating expression of DDR genes (Fig. 1). Consequently, aberrant chromatin modifications can result in impaired DDR, increased genomic instability, and defective HSPC function, as demonstrated by a number of studies targeting various chromatin modulators in hematopoietic cells (Table 1).
Table 1.
Targeting chromatin modulators showing a role in maintaining genome integrity and function of hematopoietic cells
| Protein | Molecular function | DNA damage | DDR, repair | ROS | Possible mechanism | Refs | |
|---|---|---|---|---|---|---|---|
| Direct | Indirect | ||||||
| TET2&3 | DNA hydroxylation & demethylation |
DDR↑ Repair ↓ in mature cells |
Accumulate at damage sites | Regulating repair gene expression | [111] | ||
| KMT2A/MLL1 | H3K4 methylation and more | S-phase checkpoint execution | [121] | ||||
| KMT2D/MLL4 | H3K4 methylation (and more?) | ↑ | DDR↑ | ↑ | Relieving transcription stress; Promoting replication fork degradation | Regulating redox gene expression and ROS | [126, 129, 135] |
| KMT2E/MLL5 | Organizing chromatin | ↑ |
DDR↑ Repair ↓ |
↑ | [134] | ||
| KMT2F/SET1A | H3K4 methylation and more | ↑ |
DDR↑ Repair ↓ |
↓ | Protecting replication fork degradation | Regulating repair gene expression | [136–138] |
| DPY30 | H3K4 methylation (and more?) | ↑ |
DDR↑ Repair ↓ |
↓ | Regulating repair gene expression | [101] | |
| BMI1 | ↑ | DDR↑ | ↑ | Metabolically increased ROS | [89] | ||
| MYSM1 | H2A deubiquitination | DDR↑ | ↑ | [145] | |||
| SETD2 | H3K36 methylation | ↑ |
DDR↑ Repair ↓ |
Facilitating repair | [149, 150] | ||
| SIRT1 | Deacetylation of histones and other proteins | ↑ | DDR↑ | ↑ | Regulating redox gene expression | [67, 68] | |
DNA modifications
DNA methylation is thought to have an important role in maintaining genome integrity against the extensive retrotransposons in our genome [108, 109]. The dynamic removal of DNA methylation is also important for genome integrity. TET1 deficiency in mouse results in hematologic malignancy mainly in the B-cell lineage, increase in DNA damage, and downregulation of DNA repair genes in early progenitors and pro-B cells [110]. Loss of both TET2 and TET3 results in aberrant HSPC differentiation with myeloid bias at the expense of erythropoiesis and lymphopoiesis, and induces the development of myeloid leukemia. The double-deficient bone marrow cells, mainly the late HPCs and mature myeloid cells, show accumulation of phospho-H2AX, impaired DNA repair after irradiation, and altered expression of DNA repair genes [111]. These studies support a role of the TET enzymes in guarding genome integrity through regulating DDR genes.
A direct role of 5hmC in DDR is suggested by its enriched localization to sites of DNA damage and repair [112]. Local 5hmC deposition is increased by DNA damaging agents and requires TET enzymes (especially TET2), and loss of TET compromises genome integrity in mouse ES cells during replication stress. As BRCA2, a key DNA repair gene, is downregulated by TET deficiency, the contribution from regulation of DDR gene expression cannot be excluded, but the overall data suggest a direct role of TET enzymes in DDR [112]. Another study shows that, in response to DNA damage, TET3 protein is upregulated and activated (via ATR-dependent phosphorylation), and plays an important role in DNA damage signaling and DNA repair [113]. Therefore, it is possible that DNA modifications can impact DDR through both direct (DDR per se) and indirect (expression of DDR genes) mechanisms to ultimately impact the activities of stem cells including HSPCs.
On the other hand, overexpression of catalytically active, but not inactive, TET2 in mouse pro-B-cell line results in increased level of 5hmC and chromosomal instabilities, and can lead to increased mutagenesis if the subsequent modifications cannot be efficiently eliminated in the context of deficiency of the base excision repair pathway [114]. When taken together with the role of TETs and 5hmC in guarding genome integrity shown in the loss-of-function studies, these findings suggest that the balanced activity of TET2 and the proper levels of different DNA modifications are important for genome integrity.
Histone modifications/chromatin remodeling
KMT2 family proteins and complexes
The KMT2 family proteins include KMT2A/MLL1/MLL, KMT2B/MLL2 (also known as MLL4), KMT2C/MLL3, KMT2D/MLL4 (also known as MLL2), KMT2E/MLL5, KMT2F/SET1A, and KMT2G/SET1B. With the exception of MLL5, they are the major H3K4 methyltransferases in mammals by individually acting as the catalytic subunit of the SET1/MLL complexes, which also contain all of WDR5, RBBP5, ASH2L, and DPY30 as the common core subunits that are important for the catalytic activity of the complexes [115, 116]. In general, SET1A is responsible for the bulk H3K4me3 at promoters, and MLL1 and MLL2 catalyze promoter H3K4me3 at regulated developmental stages, whereas MLL3 and MLL4 mainly catalyze H3K4me1 at gene enhancers [117]. It is worth mentioning that these proteins have physiologically relevant activities independent of H3K4 methylation [116]. Many of these complex components are known to play important roles in HSPC function, and are extensively associated with human cancers [116, 118].
KMT2A/MLL1/MLL
MLL1 plays a critical role in normal hematopoiesis and hematologic malignancy [119, 120]. Chromosomal fusion of MLL1 with a large number of partners drives mixed lineage leukemia. MLL1 is essential for hematopoiesis and self-renewal of HSCs. MLL1 participates in multiple cellular activities, including transcriptional regulation of key targets (e.g., HOXA9) through H3K4 methylation-dependent and -independent mechanisms [119], and integration of cell cycle control and DDR [121]. In response to genotoxic stress in S phase of the cell cycle, MLL1 is phosphorylated by ATR and, consequently, escapes Skp2-mediated degradation, and the accumulated MLL1 methylates H3K4 at late replication and delays DNA replication. MLL1 is thus directly involved in DDR and acts as a key effector of S-phase checkpoint. It is likely that the unrestrained HSC proliferation in MLL1 deficiency [120] is mediated by its roles in executing S-phase checkpoint and in transcriptional control of target genes involved in cell proliferation [122]. Importantly, MLL fusions act as dominant negative mutants that interfere with the ATR-regulated stabilization of MLL1 [121]. These studies suggest that dysregulation of the cell cycle control and DDR incurred by MLL translocations may contribute to leukemogenesis [121].
KMT2C/MLL3 and KMT2D/MLL4
MLL3 and MLL4 are highly mutated in a wide range of human cancers including blood cancer, and have established roles as important tumor suppressors [118, 123]. The detailed tumor suppression mechanisms are not very clear [123], and may be in part related to their emerging role in maintaining genome stability. MLL3 suppression impairs HSPC function in differentiation [124]. Depletion of MLL3 in bladder cancer cells results in the downregulation of genes encoding key components of DDR (ATM and ATR) and the homologous recombination-mediated DNA repair pathway, thereby substantially increasing DNA damage and genomic instability in cells [125].
Loss of MLL4 results in increased DDR, genome instability, and transcription stress (referring to RNA polymerase stalling, pausing, arrest, and/or backtracking), especially at the early replicating fragile sites. MLL4-mediated co-transcriptional H3K4 methylation is proposed to help resolve the transcription stress and/or impact replication [126]. The latter is supported by a very recent work in yeast, showing that H3K4 methylation relieves transcription–replication conflicts by acting as speed bumps to slow down replication machinery [127]. The feasibility of directly mutating histone residue in yeast [127] (not feasible in mammalian cells) greatly helps to establish the involvement of the histone modification in these processes. MLL4-mediated H3K4 methylation and PTIP (an MLL3/4 complex subunit) are also important for degradation of stalled and compromised replication fork (in the absence of BRCA2) by facilitating recruitment of the MRE11 nuclease [128]. These studies together support the important role of MLL4 and its H3K4 methylation activity in the maintenance of genome integrity facing the challenge of transcription and replication stresses.
In another report, MLL4 is shown to be required for normal HSC function and MLL-rearranged leukemia as its loss leads to defects in HSC function and induces myeloid differentiation of leukemic blasts [129]. MLL4 deficiency causes increase in ROS and DNA damage and dysregulation of transcriptional programs involved in the antioxidant response [129]. Treatment of ROS scavengers mitigates DNA damage and keeps the MLL4-deficient leukemia from differentiation, and artificial induction of DNA damage by homing endonuclease induces leukemia differentiation without affecting ROS levels, indicating a causal role of ROS and DNA damage in leukemia differentiation and probably disruption of HSC function. Therefore, MLL4 both directly participates in pathways regulating genome integrity and also transcriptionally controls genes that influence genome stability, consistent with its very frequent mutations in cancers.
KMT2E/MLL5
MLL5, a distinct KMT2 protein [130], is essential for hematopoiesis and normal HSPC function [131–133]. MLL5 deficiency in HSPCs impairs DNA repair and leads to accumulation of DNA damage, which triggers type 1 interferon response and mitochondrial accumulation of BID protein followed by increase in intracellular ROS levels. Interestingly, interferon inactivation or ROS quenching markedly restores HSPC function without alleviating DNA damage or improving DNA repair. The findings in this report suggest that (1) there is a two-way impact between DNA damage and ROS levels (Fig. 1a), as also seen by ROS increase upon loss of ATM [22], and (2) increased DNA damage itself, but not ROS, is compatible with normal HSPC function [134].
What is far unclear is how MLL5 deficiency impairs DNA repair and increases DNA damage. MLL5 lacks methyltransferase activity, but an MLL5 isoform can reorganize chromatin structure and affect local chromatin openness and accessibility (biased toward a more compact state), and promote tumor cell self-renewal via repressing H3 variant H3.3 expression and reducing global H3K4me3 level [135]. The altered chromatin state may directly impair the accessibility to DNA repair factors. Moreover, MLL5 helps to maintain genomic stability by regulating chromosome spatial organization and spindle bipolarity [130]. MLL5 may thus represent a good example of chromatin state influencing HSPC function by directly impacting DNA damage and ROS levels.
KMT2F/SET1A
SET1A deficiency in mouse HSCs disrupts hematopoiesis and impairs HSC proliferation and repopulation [136]. While ROS levels are reduced, SET1A-deficient HSCs accumulate DNA damage as a result of impaired DDR gene expression and DNA repair capacity. SET1A protects DNA damage-induced HSC attrition following inflammation-induced activation. As energy metabolism and oxidative homeostasis genes are significantly enriched in genes upregulated in SET1A-deficient HSPCs, SET1A may also regulate metabolism [136]. SET1A supports the survival of MLL-rearranged leukemia cells by promoting expression of DNA repair genes (including FANCD2) through interaction with Cyclin K, rather than its H3K4 methylation activity [137]. H3K4 methylation by SET1A protects stalled replication forks from deleterious resection in replication stress by enhancing FANCD2-dependent histone chaperone activity, thereby maintaining genome stability [138]. The seemingly inconsistent effects of SET1A- [138] and MLL4- [128] mediated H3K4 methylation at replication fork actually suggest the coordinated activities of these KMT2s in the divergent cellular and/or genomic context, e.g., SET1A has a constitutive role in protecting fork degradation, whereas MLL4 acts to remove the fork whose stability is already compromised (in the absence of BRCA2). The close corporation of SET1A and DDR is also reflected by its regulation of FANCD2 at both transcription [137] and protein levels [138]. A direct involvement of SET1A in DDR is also suggested by its physical and functional association with DNA repair protein RAD18, apparently independent of the core subunits of the SET1A complex, thus independent of its H3K4 methyltransferase activity [139].
DPY30
The DPY30 and WDR5 core subunits of SET1/MLL complexes have also been shown to play a role in maintaining the genome integrity [116]. In addition to the impaired energy metabolism mentioned above, DPY30-deficient HSPCs show increase in DDR, DNA damage, and impaired DNA repair ability. The colony formation ability of DPY30-deficient HSPCs can be rescued by treatment of DDR inhibitors, and the hematopoietic functionality in vivo can be partially rescued by inactivation of CDK inhibitor p21 [101], suggesting that DPY30 regulates hematopoiesis partially through protecting genome integrity. Moreover, it seems that response to DNA damage, rather than the damage per se, has a major impact in HSPC function, echoing conclusions from the other studies [89, 134]. However, it is likely that the preserved HSPCs with unrepaired genome will have higher potential to give rise to malignancy later on.
Therefore, the common increase in DNA damage upon loss of the SET1/MLL complex subunits suggests an important role of H3K4 methylation in regulating genome integrity in HSPCs, while also compatible with the important contribution from the non-catalytic chromatin-regulatory activities of at least SET1A and MLL5. The SET1/MLL complexes regulate the expression of DNA repair genes [101, 136, 137] or genes in redox homeostasis [129]. Moreover, SET1/MLL complex-mediated H3K4 methylation may directly influence the generation of DNA damage in transcription and replication stress, and also regulate DNA repair at the damaged sites including stressed replication sites. The genome-protection role of H3K4 methylation is conserved throughout evolution as evident in yeast [140], where the H3K4 methylation enzyme and the mark itself are found at newly created DNA breaks and C. elegans [141]. However, the direct influence of H3K4 methylation on chromatin integrity is likely to be complicated, as it may facilitate DNA repair by promoting the accessibility of DNA repair proteins, but also predispose chromatin for DNA double-stranded breaks through decondensing local chromatin [142].
Other histone modifications and chromatin remodelers
Studies of IDH mutations show a great example of how metabolic dysregulation results in HSC defects through epigenetic impact on DDR pathway. A mouse model-bearing endogenous IDH1 mutation shows HSC attrition and impaired self-renewal capacity [63]. The IDH1 mutation causes downregulation of Atm, a key DDR gene, and accumulation of DNA damage and impaired DNA repair in HSCs. The authors show that this regulation is independent of TET2, but likely via direct inhibition of KDM4 that demethylates H3K9, as they detected increase in H3K9 methylation globally and also at Atm promoter (Fig. 2a) [63].
BMI1, a chromatin modulator in the Polycomb family, is essential for the maintenance of HSCs, in part through repressing p16Ink4a and p19Arf [143]. BMI1-deficient HSCs have impaired mitochondrial oxidative respiration capacity, elevated ROS levels, and increased DNA damage and DDR [89]. The functional importance of ROS and DDR is shown by the mitigation of many hematopoietic phenotypes of the BMI1-deficient mice by either treatment of antioxidant or genetic inactivation of Chk2, a cell cycle checkpoint in response to DNA damage. However, the long-term repopulation defect cannot be rescued by DDR inactivation. Regarding the mechanisms underlying the increase in ROS levels, the authors show that it is a result of the impaired electron transport chain function. In addition, the dysregulation of many redox regulatory genes and the derepression of p16Ink4a and p19Arf [134] in BMI1-deficient cells may all contribute to ROS-level alteration. These results demonstrate a crucial role of BMI1 in maintaining stem cell function by coordinating energy metabolism, redox homeostasis, and protection of genome integrity, likely through chromatin regulation of key genes for mitochondrial function and redox homeostasis [89].
Depletion of DMAP1, a core component of the NuA4/Tip60 histone acetyltransferase complex, induces DNA damage and severely impairs proliferation in vitro and long-term repopulating of mouse HSCs [144]. Loss of SIRT1, an NAD+-dependent deacetylase, in adult HSPCs results in aberrant HSPC fate determination, associated with increase in ROS, DNA damage, and dysregulation of antioxidant gene expression [67, 68]. These studies suggest the importance of the dynamic regulation of histone acetylation in protecting genome integrity for HSPC function.
Ubiquitination of histones H2A and H2B is prominent chromatin modifications enriched at damaged genomic sites, and plays an important role in efficient repair of DNA double-strand breaks [30]. Loss of Mysm1, one of the H2A-deubiquitinase, results in defective differentiation of hematopoietic cell lineages and impaired HSC function [145]. Mysm1-deficient HSPCs have increased DNA damage and ROS, indicating an important role of Mysm1 in maintaining genome integrity and oxidative stress. However, the molecular mechanisms underlying such regulation are unclear, and may include the regulation of expression of DDR genes and direct regulation of DDR through H2A ubiquitination control at the sites of DNA damage [145].
SETD2-mediated H3K36 tri-methylation is a chromatin modification associated with active gene expression, and also plays a direct role in DNA repair including mismatch repair [146] and homologous recombination repair [147, 148] by facilitating the recruitment of repair factors to DNA damage sites. SETD2 is recurrently mutated in a number of human cancers including blood cancer. Perturbation of SETD2 in vitro or in an animal model of leukemia leads to increased resistance to DNA-damaging agents as a result of impaired local DDR including inefficient DNA damage repair and increased survival, and pharmacologic inhibition of H3K9/36 demethylase can restore the sensitivity to the chemotherapy agents [149]. However, severe depletion of SETD2 in MLL-rearranged leukemia leads to growth arrest and myeloid differentiation, accompanied by increased DNA damage, DDR, and DNA damage-associated gene expression [150]. This is in line with leukemia differentiation upon experimentally induced DNA damage [129].
The role of many chromatin modulators in HSPC function is usually explained based on their impact on transcription. Given the fundamental role of chromatin conformation in DDR, it is likely that the HSPC phenotypes following perturbation of many chromatin modulators arise from dysregulated DDR. For instance, the requirement of PRC2 proteins (including EZH2) for HSC function is largely considered via epigenetic repression of target expression [151, 152]. It is likely that some of the phenotypes are mediated by the direct role of EZH2 in DDR [153, 154]. In particular, EZH2-mediated H3K27 methylation at stalled replication forks helps to recruit MUS81 nuclease to promote fork degradation [155]. This regulation thus joins the MLL4–H3K4 methylation–MRE11 [128] and the SET1A–H3K4 methylation–FANCD2 [138] pathways in a direct and coordinated control of fork stability by histone modifications that have been well established for transcription regulation.
Conclusions and perspectives
Chromatin represents the central stage where signals and impacts from different cellular processes, including metabolism, are integrated and relayed to the DNA-based activities, which ultimately influence cell function. As underscored in this review, chromatin does not merely take the impacts from metabolic and genotoxic molecules, but actively and crucially regulates pathways in metabolism and DDR. These cellular processes constitute a major target of chromatin regulation to maintain the normal function and fate determination of HSPCs, as in part reflected by a profound control of HSPC fate determination by both the Polycomb (BMI1 [89]) and Trithorax (DPY30 [101]) families of chromatin modulators [156] via regulating energy metabolism and genome integrity. Considering the important roles played by many chromatin modulators in hematologic malignancies [6, 7], it will be of great interest to study if and how their roles in blood cancer are at least partially linked to regulation of cellular metabolism and genome integrity. These studies will likely provide combinatorial strategies for cancer treatment, given the high druggability of many enzymes involved in metabolism, chromatin modifications, and DDR.
Many challenges remain. Although many reports show concomitant alterations of metabolism, chromatin state, and HSPC functions, it is difficult to establish a precise causality for a particular molecular process in generating a biological outcome, due to the ubiquitous influence of both metabolism and chromatin on cellular physiology. For instance, dysregulated mitochondrial activities are often accompanied by altered expression of oxidative homeostasis genes, making it difficult to discern their causal versus a compensatory role in ROS-level regulation. Moreover, the functional interplay among the metabolic regulation, ROS levels, DNA damage, DDR, and the chromatin state in controlling HSPC activities clearly requires further studies. Apparently inconsistent or paradoxical conclusions are not uncommon regarding which process(es) lead to the others. For instance, while high levels of ROS are widely believed to cause (thus upstream of) DNA damage, the reverse also seems true [22, 134]. Despite their close connections, ROS and DNA damage are separable and can impact HSPC function independently of each other [129, 134]. Many studies suggest that active DDR protects, and accrued DNA damage disrupts, HSPC functionality, whereas others suggest that ongoing DDR, rather than the damage itself, results in defective HSPCs [89, 101]. While these observations are not contradictory to each other and probably reflect the interconnectivity of these processes, they clearly indicate a need of further work to tease out the cause and consequence of each molecular event.
Another challenging question is whether and how metabolism specifically affects gene expression through regulating the chromatin state, considering the universal usage of certain metabolites as co-factors for many chromatin modifications that can have very different impacts on the chromatin-associated processes. For example, the dynamic regulation of chromatin methylation by methyl writers and erasers is directly influenced by levels of SAM and α-KG (and related metabolites), respectively [17]. Perturbations to affect these metabolite levels, such as methionine restriction [157] or mitochondrial inactivation [77], alter several histone methyl marks including those on H3K4 and H3K9, as well as gene expression. As H3K4 and H3K9 methylation have opposite effects on gene expression, the extent of effect on these marks can lead to different outcome in gene expression. The biochemical basis for a selective impact is the different kinetic properties of the chromatin enzymes in relationship with the metabolite concentrations [16]. However, it may not be easy to accurately determine these enzymatic properties in cells. For instance, while the catalytic domains of some KMT2 family H3K4 methyltransferases have been kinetically characterized in vitro [158], the properties of the full-length enzymes in complexes assembled at higher order in cells [116] and, thus, their responsiveness to the SAM level may differ significantly. Moreover, we do not know well how the metabolites directly affect chromatin modifications at specific regions given the rapidly diffusive nature of metabolites. One mechanism is through the local supply of metabolites provided by the chromatin-bound metabolic enzymes, typically through interaction with transcription factors [17, 159, 160]. Finally, we know very little about the specific gene regulation by metabolism in the context of HSPC functions.
Aberrant chromatin modifications are commonly associated with genomic instability which is represented as increase in DDR. Many studies merely use gamma-H2AX as a readout of increased DDR, which can be a result of increased DNA damage, increased susceptibility or response to the same amount of damage, and/or impaired DNA repair. Chromatin modifications can influence DDR either indirectly by affecting expression of DNA repair genes, or directly by regulating DNA damage sensing and repair at the site of modifications. More studies are needed to better distinguish these mechanisms in cells, including identification of the precise genomic locations of DNA damage and the chromatin modulators and modifications, and biochemical characterizations of how the chromatin modulators and modifications directly link to DDR.
Looking forward, it is highly likely that new connections between metabolism and chromatin will be discovered to regulate HSPC functions. New connections may come from newly identified chromatin modifications. For instance, the newly identified lactate-derived histone lactylation responds to metabolic lactate level and directly stimulates chromatin transcription [161], but its physiological role in HSCs has not been studied. HSCs are more reliant on anaerobic glycolysis and thus produce more lactate from glucose normalized to cellular ATP, but the lactate level is lower than in the other hematopoietic cells due to the overall low level of metabolism in HSCs [8].
New connections may also come from some other fundamentally new chromatin-regulatory mechanisms. As a biopolymer, chromatin’s activity is subject to regulation by factors that affect many physical properties of a polymer. Chromatin and its modulators have been recently shown to adopt the form of phase-separated biomolecular condensates [162–169], which are now recognized as a fundamental principle in organizing cellular space and biochemistry [170–172]. These condensates are driven by weak and multivalent interactions and display a continuum of material properties and molecular dynamics, which profoundly impact the activities of the molecules in the condensates. Importantly, their properties are critically affected by intrinsic factors including histone modifications and extrinsic factors including ionic strength, pH, hydrotropes, Mg2+ concentration, temperature, etc.[173], many of which are influenced by metabolic activities in cells. Of particular relevance, ATP at physiological concentrations (~ mM levels) acts as a biological hydrotrope and has widespread influence on protein solubility, phase separation, and stability [174, 175]. This offers a completely different mechanism by which metabolites may affect chromatin activities, since most ATPases only require ATP concentrations roughly thousands of times lower than the cellular levels and are thus insensitive to metabolic changes of ATP levels [16]. More studies will likely provide unprecedented understanding of how chromatin is influenced by and responds to metabolic pathways for HSPC function.
Acknowledgements
Zhenhua Yang is supported by startup fund from Huazhong University of Science and Technology, and Hao Jiang is supported by a grant (R01DK105531) from the National Institute of Health, American Society of Hematology Scholar Award, American Cancer Society Research Scholar Award (128609-RSG-15-166-01-DMC), and the Leukemia & Lymphoma Society Scholar Award.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Zhenhua Yang, Email: zhenhua@hust.edu.cn.
Hao Jiang, Email: hj8d@virginia.edu.
References
- 1.Kondo M, et al. Biology of hematopoietic stem cells and progenitors: implications for clinical application. Annu Rev Immunol. 2003;21:759–806. doi: 10.1146/annurev.immunol.21.120601.141007. [DOI] [PubMed] [Google Scholar]
- 2.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128(4):693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 3.Allis CD, Jenuwein T. The molecular hallmarks of epigenetic control. Nat Rev Genet. 2016;17(8):487–500. doi: 10.1038/nrg.2016.59. [DOI] [PubMed] [Google Scholar]
- 4.Breiling A, Lyko F. Epigenetic regulatory functions of DNA modifications: 5-methylcytosine and beyond. Epigenetics Chromatin. 2015;8:24. doi: 10.1186/s13072-015-0016-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Audia JE, Campbell RM. Histone modifications and cancer. Cold Spring Harb Perspect Biol. 2016;8(4):a019521. doi: 10.1101/cshperspect.a019521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dawson MA, Kouzarides T. Cancer epigenetics: from mechanism to therapy. Cell. 2012;150(1):12–27. doi: 10.1016/j.cell.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 7.Brien GL, Valerio DG, Armstrong SA. Exploiting the epigenome to control cancer-promoting gene-expression programs. Cancer Cell. 2016;29(4):464–476. doi: 10.1016/j.ccell.2016.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Simsek T, et al. The distinct metabolic profile of hematopoietic stem cells reflects their location in a hypoxic niche. Cell Stem Cell. 2010;7(3):380–390. doi: 10.1016/j.stem.2010.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Suda T, Takubo K, Semenza GL. Metabolic regulation of hematopoietic stem cells in the hypoxic niche. Cell Stem Cell. 2011;9(4):298–310. doi: 10.1016/j.stem.2011.09.010. [DOI] [PubMed] [Google Scholar]
- 10.Ito K, Suda T. Metabolic requirements for the maintenance of self-renewing stem cells. Nat Rev Mol Cell Biol. 2014;15(4):243–256. doi: 10.1038/nrm3772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DeBerardinis RJ, Thompson CB. Cellular metabolism and disease: what do metabolic outliers teach us? Cell. 2012;148(6):1132–1144. doi: 10.1016/j.cell.2012.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mason PJ, Bautista JM, Gilsanz F. G6PD deficiency: the genotype-phenotype association. Blood Rev. 2007;21(5):267–283. doi: 10.1016/j.blre.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 13.Medeiros BC, et al. Isocitrate dehydrogenase mutations in myeloid malignancies. Leukemia. 2017;31(2):272–281. doi: 10.1038/leu.2016.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang F, et al. Targeted inhibition of mutant IDH2 in leukemia cells induces cellular differentiation. Science. 2013;340(6132):622–626. doi: 10.1126/science.1234769. [DOI] [PubMed] [Google Scholar]
- 15.Janke R, Dodson AE, Rine J. Metabolism and epigenetics. Annu Rev Cell Dev Biol. 2015;31:473–496. doi: 10.1146/annurev-cellbio-100814-125544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reid MA, Dai Z, Locasale JW. The impact of cellular metabolism on chromatin dynamics and epigenetics. Nat Cell Biol. 2017;19(11):1298–1306. doi: 10.1038/ncb3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li X, et al. Regulation of chromatin and gene expression by metabolic enzymes and metabolites. Nat Rev Mol Cell Biol. 2018;19(9):563–578. doi: 10.1038/s41580-018-0029-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blanpain C, et al. DNA-damage response in tissue-specific and cancer stem cells. Cell Stem Cell. 2011;8(1):16–29. doi: 10.1016/j.stem.2010.12.012. [DOI] [PubMed] [Google Scholar]
- 19.Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol Cell. 2010;40(2):179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Delia D, Mizutani S. The DNA damage response pathway in normal hematopoiesis and malignancies. Int J Hematol. 2017;106(3):328–334. doi: 10.1007/s12185-017-2300-7. [DOI] [PubMed] [Google Scholar]
- 21.Ceccaldi R, Sarangi P, D'Andrea AD. The Fanconi anaemia pathway: new players and new functions. Nat Rev Mol Cell Biol. 2016;17(6):337–349. doi: 10.1038/nrm.2016.48. [DOI] [PubMed] [Google Scholar]
- 22.Ito K, et al. Regulation of oxidative stress by ATM is required for self-renewal of haematopoietic stem cells. Nature. 2004;431(7011):997–1002. doi: 10.1038/nature02989. [DOI] [PubMed] [Google Scholar]
- 23.Nijnik A, et al. DNA repair is limiting for haematopoietic stem cells during ageing. Nature. 2007;447(7145):686–690. doi: 10.1038/nature05875. [DOI] [PubMed] [Google Scholar]
- 24.Rossi DJ, et al. Deficiencies in DNA damage repair limit the function of haematopoietic stem cells with age. Nature. 2007;447(7145):725–729. doi: 10.1038/nature05862. [DOI] [PubMed] [Google Scholar]
- 25.Lukas J, Lukas C, Bartek J. More than just a focus: The chromatin response to DNA damage and its role in genome integrity maintenance. Nat Cell Biol. 2011;13(10):1161–1169. doi: 10.1038/ncb2344. [DOI] [PubMed] [Google Scholar]
- 26.Price BD, D'Andrea AD. Chromatin remodeling at DNA double-strand breaks. Cell. 2013;152(6):1344–1354. doi: 10.1016/j.cell.2013.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gut P, Verdin E. The nexus of chromatin regulation and intermediary metabolism. Nature. 2013;502(7472):489–498. doi: 10.1038/nature12752. [DOI] [PubMed] [Google Scholar]
- 28.Suganuma T, Workman JL. Chromatin and metabolism. Annu Rev Biochem. 2018;87:27–49. doi: 10.1146/annurev-biochem-062917-012634. [DOI] [PubMed] [Google Scholar]
- 29.Crispo F, et al. Metabolic dysregulations and epigenetics: a bidirectional interplay that drives tumor progression. Cells. 2019;8(8):E798. doi: 10.3390/cells8080798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burgess RC, Misteli T, Oberdoerffer P. DNA damage, chromatin, and transcription: the trinity of aging. Curr Opin Cell Biol. 2012;24(6):724–730. doi: 10.1016/j.ceb.2012.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sashida G, Iwama A. Epigenetic regulation of hematopoiesis. Int J Hematol. 2012;96(4):405–412. doi: 10.1007/s12185-012-1183-x. [DOI] [PubMed] [Google Scholar]
- 32.Cullen SM, et al. Hematopoietic stem cell development: an epigenetic journey. Curr Top Dev Biol. 2014;107:39–75. doi: 10.1016/B978-0-12-416022-4.00002-0. [DOI] [PubMed] [Google Scholar]
- 33.Ito K, Ito K. Metabolism and the control of cell fate decisions and stem cell renewal. Annu Rev Cell Dev Biol. 2016;32:399–409. doi: 10.1146/annurev-cellbio-111315-125134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Folmes CD, et al. Metabolic plasticity in stem cell homeostasis and differentiation. Cell Stem Cell. 2012;11(5):596–606. doi: 10.1016/j.stem.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chandel NS, et al. Metabolic regulation of stem cell function in tissue homeostasis and organismal ageing. Nat Cell Biol. 2016;18(8):823–832. doi: 10.1038/ncb3385. [DOI] [PubMed] [Google Scholar]
- 36.Weiss CN, Ito K. DNA damage response, redox status and hematopoiesis. Blood Cells Mol Dis. 2014;52(1):12–18. doi: 10.1016/j.bcmd.2013.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chisolm DA, Weinmann AS. Connections between metabolism and epigenetics in programming cellular differentiation. Annu Rev Immunol. 2018;36:221–246. doi: 10.1146/annurev-immunol-042617-053127. [DOI] [PubMed] [Google Scholar]
- 38.Chisolm DA, et al. CCCTC-binding factor translates interleukin 2- and alpha-ketoglutarate-sensitive metabolic changes in T cells into context-dependent gene programs. Immunity. 2017;47(2):251–267.e7. doi: 10.1016/j.immuni.2017.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Flavahan WA, et al. Insulator dysfunction and oncogene activation in IDH mutant gliomas. Nature. 2016;529(7584):110–114. doi: 10.1038/nature16490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Islam MS, et al. 2-oxoglutarate-dependent oxygenases. Annu Rev Biochem. 2018;87:585–620. doi: 10.1146/annurev-biochem-061516-044724. [DOI] [PubMed] [Google Scholar]
- 41.Semenza GL. Oxygen homeostasis. Wiley Interdiscip Rev Syst Biol Med. 2010;2(3):336–361. doi: 10.1002/wsbm.69. [DOI] [PubMed] [Google Scholar]
- 42.Batie M, Del Peso L, Rocha S. Hypoxia and chromatin: a focus on transcriptional repression mechanisms. Biomedicines. 2018;6(2):E47. doi: 10.3390/biomedicines6020047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou X, et al. Hypoxia induces trimethylated H3 lysine 4 by inhibition of JARID1A demethylase. Cancer Res. 2010;70(10):4214–4221. doi: 10.1158/0008-5472.CAN-09-2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Batie M, et al. Hypoxia induces rapid changes to histone methylation and reprograms chromatin. Science. 2019;363(6432):1222–1226. doi: 10.1126/science.aau5870. [DOI] [PubMed] [Google Scholar]
- 45.Chakraborty AA, et al. Histone demethylase KDM6A directly senses oxygen to control chromatin and cell fate. Science. 2019;363(6432):1217–1222. doi: 10.1126/science.aaw1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thienpont B, et al. Tumour hypoxia causes DNA hypermethylation by reducing TET activity. Nature. 2016;537(7618):63–68. doi: 10.1038/nature19081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Monfort A, Wutz A. Breathing-in epigenetic change with vitamin C. EMBO Rep. 2013;14(4):337–346. doi: 10.1038/embor.2013.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Blaschke K, et al. Vitamin C induces Tet-dependent DNA demethylation and a blastocyst-like state in ES cells. Nature. 2013;500(7461):222–226. doi: 10.1038/nature12362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.DiTroia SP, et al. Maternal vitamin C regulates reprogramming of DNA methylation and germline development. Nature. 2019;573(7773):271–275. doi: 10.1038/s41586-019-1536-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Esteban MA, et al. Vitamin C enhances the generation of mouse and human induced pluripotent stem cells. Cell Stem Cell. 2010;6(1):71–79. doi: 10.1016/j.stem.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 51.Wang T, et al. The histone demethylases Jhdm1a/1b enhance somatic cell reprogramming in a vitamin-C-dependent manner. Cell Stem Cell. 2011;9(6):575–587. doi: 10.1016/j.stem.2011.10.005. [DOI] [PubMed] [Google Scholar]
- 52.Chen J, et al. H3K9 methylation is a barrier during somatic cell reprogramming into iPSCs. Nat Genet. 2013;45(1):34–42. doi: 10.1038/ng.2491. [DOI] [PubMed] [Google Scholar]
- 53.Chen J, et al. Vitamin C modulates TET1 function during somatic cell reprogramming. Nat Genet. 2013;45(12):1504–1509. doi: 10.1038/ng.2807. [DOI] [PubMed] [Google Scholar]
- 54.Agathocleous M, et al. Ascorbate regulates haematopoietic stem cell function and leukaemogenesis. Nature. 2017;549(7673):476–481. doi: 10.1038/nature23876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Moran-Crusio K, et al. Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer Cell. 2011;20(1):11–24. doi: 10.1016/j.ccr.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cimmino L, et al. Restoration of TET2 function blocks aberrant self-renewal and leukemia progression. Cell. 2017;170(6):1079–1095.e20. doi: 10.1016/j.cell.2017.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Delhommeau F, et al. Mutation in TET2 in myeloid cancers. N Engl J Med. 2009;360(22):2289–2301. doi: 10.1056/NEJMoa0810069. [DOI] [PubMed] [Google Scholar]
- 58.Tefferi A, Lim KH, Levine R. Mutation in TET2 in myeloid cancers. N Engl J Med. 2009;361(11):1117–1118. doi: 10.1056/NEJMc091348. [DOI] [PubMed] [Google Scholar]
- 59.Tan L, Shi YG. Tet family proteins and 5-hydroxymethylcytosine in development and disease. Development. 2012;139(11):1895–1902. doi: 10.1242/dev.070771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang T, et al. Vitamin C-dependent lysine demethylase 6 (KDM6)-mediated demethylation promotes a chromatin state that supports the endothelial-to-hematopoietic transition. J Biol Chem. 2019;294(37):13657–13670. doi: 10.1074/jbc.RA119.009757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dang L, Su SM. Isocitrate dehydrogenase mutation and (r)-2-hydroxyglutarate: from basic discovery to therapeutics development. Annu Rev Biochem. 2017;86:305–331. doi: 10.1146/annurev-biochem-061516-044732. [DOI] [PubMed] [Google Scholar]
- 62.Su R, et al. R-2HG exhibits anti-tumor activity by targeting FTO/m(6)A/MYC/CEBPA signaling. Cell. 2018;172(1–2):90–105.e23. doi: 10.1016/j.cell.2017.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Inoue S, et al. Mutant IDH1 downregulates ATM and alters DNA repair and sensitivity to DNA damage independent of TET2. Cancer Cell. 2016;30(2):337–348. doi: 10.1016/j.ccell.2016.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang P, et al. Oncometabolite D-2-hydroxyglutarate inhibits ALKBH DNA repair enzymes and sensitizes IDH mutant cells to alkylating agents. Cell Rep. 2015;13(11):2353–2361. doi: 10.1016/j.celrep.2015.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.O'Callaghan C, Vassilopoulos A. Sirtuins at the crossroads of stemness, aging, and cancer. Aging Cell. 2017;16(6):1208–1218. doi: 10.1111/acel.12685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li L, Bhatia R. Role of SIRT1 in the growth and regulation of normal hematopoietic and leukemia stem cells. Curr Opin Hematol. 2015;22(4):324–329. doi: 10.1097/MOH.0000000000000152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Singh SK, et al. Sirt1 ablation promotes stress-induced loss of epigenetic and genomic hematopoietic stem and progenitor cell maintenance. J Exp Med. 2013;210(5):987–1001. doi: 10.1084/jem.20121608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rimmele P, et al. Aging-like phenotype and defective lineage specification in SIRT1-deleted hematopoietic stem and progenitor cells. Stem Cell Rep. 2014;3(1):44–59. doi: 10.1016/j.stemcr.2014.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mishra BP, et al. The histone methyltransferase activity of MLL1 is dispensable for hematopoiesis and leukemogenesis. Cell Rep. 2014;7(4):1239–1247. doi: 10.1016/j.celrep.2014.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sun J, et al. SIRT1 activation disrupts maintenance of myelodysplastic syndrome stem and progenitor cells by restoring TET2 function. Cell Stem Cell. 2018;23(3):355–369.e9. doi: 10.1016/j.stem.2018.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Abraham A, et al. SIRT1 regulates metabolism and leukemogenic potential in CML stem cells. J Clin Invest. 2019;129(7):2685–2701. doi: 10.1172/JCI127080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ryall JG, et al. The NAD(+)-dependent SIRT1 deacetylase translates a metabolic switch into regulatory epigenetics in skeletal muscle stem cells. Cell Stem Cell. 2015;16(2):171–183. doi: 10.1016/j.stem.2014.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Vyas S, Zaganjor E, Haigis MC. Mitochondria and cancer. Cell. 2016;166(3):555–566. doi: 10.1016/j.cell.2016.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Maryanovich M, et al. An MTCH2 pathway repressing mitochondria metabolism regulates haematopoietic stem cell fate. Nat Commun. 2015;6:7901. doi: 10.1038/ncomms8901. [DOI] [PubMed] [Google Scholar]
- 75.Yu WM, et al. Metabolic regulation by the mitochondrial phosphatase PTPMT1 is required for hematopoietic stem cell differentiation. Cell Stem Cell. 2013;12(1):62–74. doi: 10.1016/j.stem.2012.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ying Z, et al. Short-term mitochondrial permeability transition pore opening modulates histone lysine methylation at the early phase of somatic cell reprogramming. Cell Metab. 2018;28(6):935–945.e5. doi: 10.1016/j.cmet.2018.08.001. [DOI] [PubMed] [Google Scholar]
- 77.Anso E, et al. The mitochondrial respiratory chain is essential for haematopoietic stem cell function. Nat Cell Biol. 2017;19(6):614–625. doi: 10.1038/ncb3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ho TT, et al. Autophagy maintains the metabolism and function of young and old stem cells. Nature. 2017;543(7644):205–210. doi: 10.1038/nature21388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Vannini N, et al. The NAD-booster nicotinamide riboside potently stimulates hematopoiesis through increased mitochondrial clearance. Cell Stem Cell. 2019;24(3):405–418.e7. doi: 10.1016/j.stem.2019.02.012. [DOI] [PubMed] [Google Scholar]
- 80.Mohrin M, et al. Hematopoietic stem cell quiescence promotes error-prone DNA repair and mutagenesis. Cell Stem Cell. 2010;7(2):174–185. doi: 10.1016/j.stem.2010.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Beerman I, et al. Quiescent hematopoietic stem cells accumulate DNA damage during aging that is repaired upon entry into cell cycle. Cell Stem Cell. 2014;15(1):37–50. doi: 10.1016/j.stem.2014.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Walter D, et al. Exit from dormancy provokes DNA-damage-induced attrition in haematopoietic stem cells. Nature. 2015;520(7548):549–552. doi: 10.1038/nature14131. [DOI] [PubMed] [Google Scholar]
- 83.Testa U, et al. Oxidative stress and hypoxia in normal and leukemic stem cells. Exp Hematol. 2016;44(7):540–560. doi: 10.1016/j.exphem.2016.04.012. [DOI] [PubMed] [Google Scholar]
- 84.Bigarella CL, Liang R, Ghaffari S. Stem cells and the impact of ROS signaling. Development. 2014;141(22):4206–4218. doi: 10.1242/dev.107086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ito K, et al. Reactive oxygen species act through p38 MAPK to limit the lifespan of hematopoietic stem cells. Nat Med. 2006;12(4):446–451. doi: 10.1038/nm1388. [DOI] [PubMed] [Google Scholar]
- 86.Chaudhari P, Ye Z, Jang YY. Roles of reactive oxygen species in the fate of stem cells. Antioxid Redox Signal. 2014;20(12):1881–1890. doi: 10.1089/ars.2012.4963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Li TS, Marban E. Physiological levels of reactive oxygen species are required to maintain genomic stability in stem cells. Stem Cells. 2010;28(7):1178–1185. doi: 10.1002/stem.438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tothova Z, et al. FoxOs are critical mediators of hematopoietic stem cell resistance to physiologic oxidative stress. Cell. 2007;128(2):325–339. doi: 10.1016/j.cell.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 89.Liu J, et al. Bmi1 regulates mitochondrial function and the DNA damage response pathway. Nature. 2009;459(7245):387–392. doi: 10.1038/nature08040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lagadinou ED, et al. BCL-2 inhibition targets oxidative phosphorylation and selectively eradicates quiescent human leukemia stem cells. Cell Stem Cell. 2013;12(3):329–341. doi: 10.1016/j.stem.2012.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Langevin F, et al. Fancd2 counteracts the toxic effects of naturally produced aldehydes in mice. Nature. 2011;475(7354):53–58. doi: 10.1038/nature10192. [DOI] [PubMed] [Google Scholar]
- 92.Pontel LB, et al. Endogenous formaldehyde is a hematopoietic stem cell genotoxin and metabolic carcinogen. Mol Cell. 2015;60(1):177–188. doi: 10.1016/j.molcel.2015.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Garaycoechea JI, et al. Genotoxic consequences of endogenous aldehydes on mouse haematopoietic stem cell function. Nature. 2012;489(7417):571–575. doi: 10.1038/nature11368. [DOI] [PubMed] [Google Scholar]
- 94.Garaycoechea JI, et al. Alcohol and endogenous aldehydes damage chromosomes and mutate stem cells. Nature. 2018;553(7687):171–177. doi: 10.1038/nature25154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hira A, et al. Variant ALDH2 is associated with accelerated progression of bone marrow failure in Japanese Fanconi anemia patients. Blood. 2013;122(18):3206–3209. doi: 10.1182/blood-2013-06-507962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Berti M, Vindigni A. Replication stress: getting back on track. Nat Struct Mol Biol. 2016;23(2):103–109. doi: 10.1038/nsmb.3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Alvarez S, et al. Replication stress caused by low MCM expression limits fetal erythropoiesis and hematopoietic stem cell functionality. Nat Commun. 2015;6:8548. doi: 10.1038/ncomms9548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Flach J, et al. Replication stress is a potent driver of functional decline in ageing haematopoietic stem cells. Nature. 2014;512(7513):198–202. doi: 10.1038/nature13619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yang Z, et al. Dpy30 is critical for maintaining the identity and function of adult hematopoietic stem cells. J Exp Med. 2016;213(11):2349–2364. doi: 10.1084/jem.20160185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yang Z, et al. The DPY30 subunit in SET1/MLL complexes regulates the proliferation and differentiation of hematopoietic progenitor cells. Blood. 2014;124(13):2025–2033. doi: 10.1182/blood-2014-01-549220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yang Z, et al. Control of hematopoietic stem and progenitor cell function through epigenetic regulation of energy metabolism and genome integrity. Stem Cell Reports. 2019;13(1):61–75. doi: 10.1016/j.stemcr.2019.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zhao E, et al. KDM4C and ATF4 cooperate in transcriptional control of amino acid metabolism. Cell Rep. 2016;14(3):506–519. doi: 10.1016/j.celrep.2015.12.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zhu N, et al. MLL-AF9- and HOXA9-mediated acute myeloid leukemia stem cell self-renewal requires JMJD1C. J Clin Invest. 2016;126(3):997–1011. doi: 10.1172/JCI82978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Izaguirre-Carbonell J, et al. Critical role of Jumonji domain of JMJD1C in MLL-rearranged leukemia. Blood Adv. 2019;3(9):1499–1511. doi: 10.1182/bloodadvances.2018026054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lynch JR, et al. JMJD1C-mediated metabolic dysregulation contributes to HOXA9-dependent leukemogenesis. Leukemia. 2019;33(6):1400–1410. doi: 10.1038/s41375-018-0354-z. [DOI] [PubMed] [Google Scholar]
- 106.Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461(7267):1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Soria G, Polo SE, Almouzni G. Prime, repair, restore: the active role of chromatin in the DNA damage response. Mol Cell. 2012;46(6):722–734. doi: 10.1016/j.molcel.2012.06.002. [DOI] [PubMed] [Google Scholar]
- 108.Robertson KD, Wolffe AP. DNA methylation in health and disease. Nat Rev Genet. 2000;1(1):11–19. doi: 10.1038/35049533. [DOI] [PubMed] [Google Scholar]
- 109.Yoder JA, Walsh CP, Bestor TH. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 1997;13(8):335–340. doi: 10.1016/s0168-9525(97)01181-5. [DOI] [PubMed] [Google Scholar]
- 110.Cimmino L, et al. TET1 is a tumor suppressor of hematopoietic malignancy. Nat Immunol. 2015;16(6):653–662. doi: 10.1038/ni.3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.An J, et al. Acute loss of TET function results in aggressive myeloid cancer in mice. Nat Commun. 2015;6:10071. doi: 10.1038/ncomms10071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kafer GR, et al. 5-Hydroxymethylcytosine marks sites of DNA damage and promotes genome stability. Cell Rep. 2016;14(6):1283–1292. doi: 10.1016/j.celrep.2016.01.035. [DOI] [PubMed] [Google Scholar]
- 113.Jiang D, et al. TET3-mediated DNA oxidation promotes ATR-dependent DNA damage response. EMBO Rep. 2017;18(5):781–796. doi: 10.15252/embr.201643179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Mahfoudhi E, et al. TET2-mediated 5-hydroxymethylcytosine induces genetic instability and mutagenesis. DNA Repair (Amst) 2016;43:78–88. doi: 10.1016/j.dnarep.2016.05.031. [DOI] [PubMed] [Google Scholar]
- 115.Shilatifard A. The COMPASS family of histone H3K4 methylases: mechanisms of regulation in development and disease pathogenesis. Annu Rev Biochem. 2012;81:65–95. doi: 10.1146/annurev-biochem-051710-134100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Jiang H (2020) The complex activities of the SET1/MLL complex core subunits in development and disease. Biochim Biophys Acta Gene Regul Mech 1863(7):194560 [DOI] [PMC free article] [PubMed]
- 117.Crump NT, Milne TA. Why are so many MLL lysine methyltransferases required for normal mammalian development? Cell Mol Life Sci. 2019;76(15):2885–2898. doi: 10.1007/s00018-019-03143-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Rao RC, Dou Y. Hijacked in cancer: the KMT2 (MLL) family of methyltransferases. Nat Rev Cancer. 2015;15(6):334–346. doi: 10.1038/nrc3929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Li BE, Ernst P. Two decades of leukemia oncoprotein epistasis: the MLL1 paradigm for epigenetic deregulation in leukemia. Exp Hematol. 2014;42(12):995–1012. doi: 10.1016/j.exphem.2014.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Jude CD, et al. Unique and independent roles for MLL in adult hematopoietic stem cells and progenitors. Cell Stem Cell. 2007;1(3):324–337. doi: 10.1016/j.stem.2007.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Liu H, et al. Phosphorylation of MLL by ATR is required for execution of mammalian S-phase checkpoint. Nature. 2010;467(7313):343–346. doi: 10.1038/nature09350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Artinger EL, Ernst P. Cell context in the control of self-renewal and proliferation regulated by MLL1. Cell Cycle. 2013;12(18):2969–2972. doi: 10.4161/cc.26032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Sze CC, Shilatifard A. MLL3/MLL4/COMPASS family on epigenetic regulation of enhancer function and cancer. Cold Spring Harb Perspect Med. 2016;6(11):a026427. doi: 10.1101/cshperspect.a026427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Chen C, et al. MLL3 is a haploinsufficient 7q tumor suppressor in acute myeloid leukemia. Cancer Cell. 2014;25(5):652–665. doi: 10.1016/j.ccr.2014.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Rampias T, et al. The lysine-specific methyltransferase KMT2C/MLL3 regulates DNA repair components in cancer. EMBO Rep. 2019;20(3):e46821. doi: 10.15252/embr.201846821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Kantidakis T, et al. Mutation of cancer driver MLL2 results in transcription stress and genome instability. Genes Dev. 2016;30(4):408–420. doi: 10.1101/gad.275453.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Chong SY, et al. H3K4 methylation at active genes mitigates transcription-replication conflicts during replication stress. Nat Commun. 2020;11(1):809. doi: 10.1038/s41467-020-14595-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Ray Chaudhuri A, et al. Replication fork stability confers chemoresistance in BRCA-deficient cells. Nature. 2016;535(7612):382–387. doi: 10.1038/nature18325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Santos MA, et al. DNA-damage-induced differentiation of leukaemic cells as an anti-cancer barrier. Nature. 2014;514(7520):107–111. doi: 10.1038/nature13483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Zhang X, et al. MLL5 (KMT2E): structure, function, and clinical relevance. Cell Mol Life Sci. 2017;74(13):2333–2344. doi: 10.1007/s00018-017-2470-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Heuser M, et al. Loss of MLL5 results in pleiotropic hematopoietic defects, reduced neutrophil immune function, and extreme sensitivity to DNA demethylation. Blood. 2009;113(7):1432–1443. doi: 10.1182/blood-2008-06-162263. [DOI] [PubMed] [Google Scholar]
- 132.Madan V, et al. Impaired function of primitive hematopoietic cells in mice lacking the Mixed-Lineage-Leukemia homolog MLL5. Blood. 2009;113(7):1444–1454. doi: 10.1182/blood-2008-02-142638. [DOI] [PubMed] [Google Scholar]
- 133.Emerling BM, et al. MLL5, a homolog of Drosophila trithorax located within a segment of chromosome band 7q22 implicated in myeloid leukemia. Oncogene. 2002;21(31):4849–4854. doi: 10.1038/sj.onc.1205615. [DOI] [PubMed] [Google Scholar]
- 134.Tasdogan A, et al. DNA damage-induced HSPC malfunction depends on ROS accumulation downstream of IFN-1 signaling and bid mobilization. Cell Stem Cell. 2016;19(6):752–767. doi: 10.1016/j.stem.2016.08.007. [DOI] [PubMed] [Google Scholar]
- 135.Gallo M, et al. MLL5 orchestrates a cancer self-renewal state by repressing the histone variant H33 and globally reorganizing chromatin. Cancer Cell. 2015;28(6):715–729. doi: 10.1016/j.ccell.2015.10.005. [DOI] [PubMed] [Google Scholar]
- 136.Arndt K, et al. SETD1A protects HSCs from activation-induced functional decline in vivo. Blood. 2018;131(12):1311–1324. doi: 10.1182/blood-2017-09-806844. [DOI] [PubMed] [Google Scholar]
- 137.Hoshii T, et al. A non-catalytic function of SETD1A regulates cyclin K and the DNA damage response. Cell. 2018;172(5):1007–1021.e17. doi: 10.1016/j.cell.2018.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Higgs MR, et al. Histone methylation by SETD1A protects nascent DNA through the nucleosome chaperone activity of FANCD2. Mol Cell. 2018;71(1):25–41.e6. doi: 10.1016/j.molcel.2018.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Alsulami M, et al. SETD1A Methyltransferase is physically and functionally linked to the DNA damage repair protein RAD18. Mol Cell Proteomics. 2019;18(7):1428–1436. doi: 10.1074/mcp.RA119.001518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Faucher D, Wellinger RJ. Methylated H3K4, a transcription-associated histone modification, is involved in the DNA damage response pathway. PLoS Genet. 2010;6(8):e1001082. doi: 10.1371/journal.pgen.1001082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Herbette M, et al. The C. elegans SET-2/SET1 histone H3 Lys4 (H3K4) methyltransferase preserves genome stability in the germline. DNA Repair (Amst) 2017;57:139–150. doi: 10.1016/j.dnarep.2017.07.007. [DOI] [PubMed] [Google Scholar]
- 142.Burman B, et al. Histone modifications predispose genome regions to breakage and translocation. Genes Dev. 2015;29(13):1393–1402. doi: 10.1101/gad.262170.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Park IK, et al. Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature. 2003;423(6937):302–305. doi: 10.1038/nature01587. [DOI] [PubMed] [Google Scholar]
- 144.Koizumi T, et al. Depletion of Dnmt1-associated protein 1 triggers DNA damage and compromises the proliferative capacity of hematopoietic stem cells. Int J Hematol. 2010;91(4):611–619. doi: 10.1007/s12185-010-0563-3. [DOI] [PubMed] [Google Scholar]
- 145.Nijnik A, et al. The critical role of histone H2A-deubiquitinase Mysm1 in hematopoiesis and lymphocyte differentiation. Blood. 2012;119(6):1370–1379. doi: 10.1182/blood-2011-05-352666. [DOI] [PubMed] [Google Scholar]
- 146.Li F, et al. The histone mark H3K36me3 regulates human DNA mismatch repair through its interaction with MutSalpha. Cell. 2013;153(3):590–600. doi: 10.1016/j.cell.2013.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Pfister SX, et al. SETD2-dependent histone H3K36 trimethylation is required for homologous recombination repair and genome stability. Cell Rep. 2014;7(6):2006–2018. doi: 10.1016/j.celrep.2014.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Carvalho S, et al. SETD2 is required for DNA double-strand break repair and activation of the p53-mediated checkpoint. Elife. 2014;3:e02482. doi: 10.7554/eLife.02482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Mar BG, et al. SETD2 alterations impair DNA damage recognition and lead to resistance to chemotherapy in leukemia. Blood. 2017;130(24):2631–2641. doi: 10.1182/blood-2017-03-775569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Skucha A, et al. MLL-fusion-driven leukemia requires SETD2 to safeguard genomic integrity. Nat Commun. 2018;9(1):1983. doi: 10.1038/s41467-018-04329-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Mochizuki-Kashio M, et al. Dependency on the polycomb gene Ezh2 distinguishes fetal from adult hematopoietic stem cells. Blood. 2011;118(25):6553–6561. doi: 10.1182/blood-2011-03-340554. [DOI] [PubMed] [Google Scholar]
- 152.Xie H, et al. Polycomb repressive complex 2 regulates normal hematopoietic stem cell function in a developmental-stage-specific manner. Cell Stem Cell. 2014;14(1):68–80. doi: 10.1016/j.stem.2013.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Chou DM, et al. A chromatin localization screen reveals poly (ADP ribose)-regulated recruitment of the repressive polycomb and NuRD complexes to sites of DNA damage. Proc Natl Acad Sci U S A. 2010;107(43):18475–18480. doi: 10.1073/pnas.1012946107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Campbell S, et al. Polycomb repressive complex 2 contributes to DNA double-strand break repair. Cell Cycle. 2013;12(16):2675–2683. doi: 10.4161/cc.25795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Rondinelli B, et al. EZH2 promotes degradation of stalled replication forks by recruiting MUS81 through histone H3 trimethylation. Nat Cell Biol. 2017;19(11):1371–1378. doi: 10.1038/ncb3626. [DOI] [PubMed] [Google Scholar]
- 156.Schuettengruber B, et al. Genome regulation by polycomb and trithorax proteins. Cell. 2007;128(4):735–745. doi: 10.1016/j.cell.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 157.Mentch SJ, et al. Histone methylation dynamics and gene regulation occur through the sensing of one-carbon metabolism. Cell Metab. 2015;22(5):861–873. doi: 10.1016/j.cmet.2015.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Vedadi M, et al. Targeting human SET1/MLL family of proteins. Protein Sci. 2017;26(4):662–676. doi: 10.1002/pro.3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Katoh Y, et al. Methionine adenosyltransferase II serves as a transcriptional corepressor of Maf oncoprotein. Mol Cell. 2011;41(5):554–566. doi: 10.1016/j.molcel.2011.02.018. [DOI] [PubMed] [Google Scholar]
- 160.Li X, et al. Nucleus-translocated ACSS2 promotes gene transcription for lysosomal biogenesis and autophagy. Mol Cell. 2017;66(5):684–697.e9. doi: 10.1016/j.molcel.2017.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Zhang D, et al. Metabolic regulation of gene expression by histone lactylation. Nature. 2019;574(7779):575–580. doi: 10.1038/s41586-019-1678-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Gibson BA, et al. Organization of chromatin by intrinsic and regulated phase separation. Cell. 2019;179(2):470–484.e21. doi: 10.1016/j.cell.2019.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Zhang Y, Kutateladze TG (2019) Liquid-liquid phase separation is an intrinsic physicochemical property of chromatin. Nat Struct Mol Biol [DOI] [PMC free article] [PubMed]
- 164.Sanulli S, et al. HP1 reshapes nucleosome core to promote heterochromatin phase separation. Nature. 2019;575(7782):390–394. doi: 10.1038/s41586-019-1669-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Larson AG, et al. Liquid droplet formation by HP1alpha suggests a role for phase separation in heterochromatin. Nature. 2017;547(7662):236–240. doi: 10.1038/nature22822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Strom AR, et al. Phase separation drives heterochromatin domain formation. Nature. 2017;547(7662):241–245. doi: 10.1038/nature22989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Wang L, et al. Histone modifications regulate chromatin compartmentalization by contributing to a phase separation mechanism. Mol Cell. 2019;76(4):646–659. doi: 10.1016/j.molcel.2019.08.019. [DOI] [PubMed] [Google Scholar]
- 168.Hnisz D, et al. A phase separation model for transcriptional control. Cell. 2017;169(1):13–23. doi: 10.1016/j.cell.2017.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Sabari BR, et al. Coactivator condensation at super-enhancers links phase separation and gene control. Science. 2018;361(6400):eaar3958. doi: 10.1126/science.aar3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Hyman AA, Weber CA, Julicher F. Liquid-liquid phase separation in biology. Annu Rev Cell Dev Biol. 2014;30:39–58. doi: 10.1146/annurev-cellbio-100913-013325. [DOI] [PubMed] [Google Scholar]
- 171.Shin Y, Brangwynne CP. Liquid phase condensation in cell physiology and disease. Science. 2017;357(6357):eaaf4382. doi: 10.1126/science.aaf4382. [DOI] [PubMed] [Google Scholar]
- 172.Banani SF, et al. Biomolecular condensates: organizers of cellular biochemistry. Nat Rev Mol Cell Biol. 2017;18(5):285–298. doi: 10.1038/nrm.2017.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Wright RHG, Le Dily F, Beato M. ATP, Mg(2+), nuclear phase separation, and genome accessibility. Trends Biochem Sci. 2019;44(7):565–574. doi: 10.1016/j.tibs.2019.03.001. [DOI] [PubMed] [Google Scholar]
- 174.Patel A, et al. ATP as a biological hydrotrope. Science. 2017;356(6339):753–756. doi: 10.1126/science.aaf6846. [DOI] [PubMed] [Google Scholar]
- 175.Sridharan S, et al. Proteome-wide solubility and thermal stability profiling reveals distinct regulatory roles for ATP. Nat Commun. 2019;10(1):1155. doi: 10.1038/s41467-019-09107-y. [DOI] [PMC free article] [PubMed] [Google Scholar]