Abstract
Resistance to cancer therapy is a major barrier to cancer management. Conventional views have proposed that acquisition of resistance may result from genetic mutations. However, accumulating evidence implicates a key role of non-mutational resistance mechanisms underlying drug tolerance, the latter of which is the focus that will be discussed here. Such non-mutational processes are largely driven by tumor cell plasticity, which renders tumor cells insusceptible to the drug-targeted pathway, thereby facilitating the tumor cell survival and growth. The concept of tumor cell plasticity highlights the significance of re-activation of developmental programs that are closely correlated with epithelial–mesenchymal transition, acquisition properties of cancer stem cells, and trans-differentiation potential during drug exposure. From observations in various cancers, this concept provides an opportunity for investigating the nature of anticancer drug resistance. Over the years, our understanding of the emerging role of phenotype switching in modifying therapeutic response has considerably increased. This expanded knowledge of tumor cell plasticity contributes to developing novel therapeutic strategies or combination therapy regimens using available anticancer drugs, which are likely to improve patient outcomes in clinical practice.
Subject terms: Cancer therapy, Cancer therapy
Introduction
The rapid development of novel therapeutic strategies, represented by targeted therapy, has made great contributions to the improvement of clinical outcomes in patients with cancer.1,2 However, such improvements have not been translated into complete remission (CR) due to the inevitable emergence of drug resistance, which is regarded as a major impediment in clinics for achieving complete cures.1,3 For decades, along with the identification of various resistance-conferring mutations, researchers have theorized that this therapeutic failure is mainly attributable to genomic mechanisms, such as the acquisition of mutations that occur on the drug target, thus impairing the drug binding and mutation-induced continuous activation of pro-survival pathways.4,5 This would suggest that reagents designed to selectively repress such bona fide resistance mechanisms hold great promise for the realization of long-term curative effects and the improvement of living quality in patients with cancer. However, drug resistance frequently occurs and remains a clinical challenge.6,7 The development of secondary mutations may also provide a mechanistic explanation for such resistance, and may even present a treatment option for patients (e.g., the so-called “next-generation” tyrosine kinase inhibitor [TKI] for non-small cell lung cancer [NSCLC] patients).8 The observation that clones with resistance-conferring mutations can pre-exist within an individual tumor prior to drug exposure and be further selected during treatment indicates that merely targeting the validated genetic resistance mechanisms is not enough.9–12
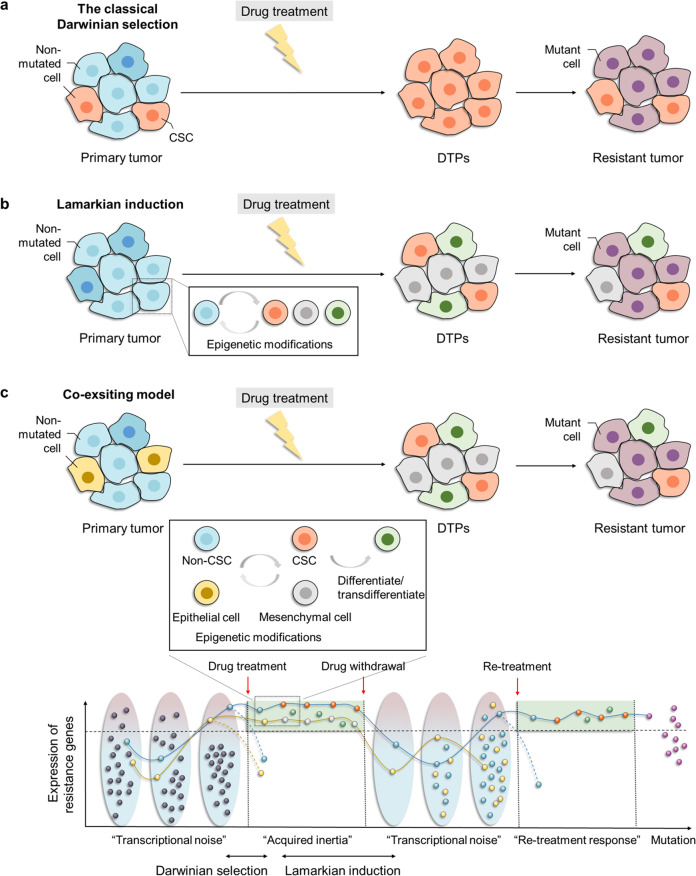
Occurring in parallel are numerable cases that are not related to genomic/genetic alterations, raising the possibility of non-mutational mechanisms involved in maintaining cancer cell survival and growth upon treatment.13–16 For instance, a rare subpopulation of cancer stem cells (CSCs), or poorly differentiated cancer cells equipped with enhanced drug efflux properties and heightened self-renewal potential, is intrinsically more refractory to multiple cancer therapies, suggesting a fundamental role of CSCs as a reservoir for tumor recurrence.17 Indeed, such stem cell-like phenotype-dependent relapses have been previously described in patients with chronic myelogenous leukemia following imatinib mesylate treatment18,19 and have been further documented in various types of solid tumors.20–22 Being regarded as the source of non-mutational resistance, this subpopulation—named drug-tolerant persisters (DTPs), has been widely recognized for its dormant, slow-cycling state and stem-like signature.13 Such a so-called quiescent condition of DTPs allows them to survive for long periods of time (weeks to months) in the time frame between being killed and developing mutations.13 This window of opportunity seems essential for DTPs—or at least parts of DTPs, to acquire mutation-driven resistance mechanisms by which they can evolve into clinically relevant drug-resistant cells.23,24 As such, the tolerance/dormancy/persistence state, which is accepted as an alternative route for acquiring resistance, tends to serve as a “bridge” to link the non-mutational mechanisms with bona fide resistance mechanisms (i.e., to connect phenotype-dependent DTPs with genotype-dependent resistant cells24,25) (Fig. 1).
Fig. 1.
The genesis of DTPs according to natural selection theory (classical Darwinian selection), the Lamarckian induction concept, and the coexisting model. a The natural selection theory shows that the preexisting DTPs, here represented by CSCs, can be selected and enriched upon drug exposure. b The concept of Lamarckian induction attaches importance to the natural aptitude of tumor cells in adapting to pharmacologic interventions through different levels of epigenetic modifications, giving rise to the emergence and coexistence of DTPs in varying tolerant states. c The coexisting model suggests the dynamic transcriptional fluctuation at a single-cell level of resistance-related markers (“transcriptional noise”). A small fraction of tumor cells, whose expression of these resistance-related genes exceeds a certain threshold at the moment of treatment, can survive and be selected (the blue and yellow dot), marking a return to classical Darwinian selection. However, with increasing duration of drug exposure, such a stochastic, transient, fluctuated “survival mode” arrives at drug-refractory state through epigenetic modifications, ultimately resulting in the establishment of a DTP pool. These alterations in the epigenome, which can be summed up as “acquired inertia,” are in agreement with the concept of Lamarckian induction. The solid line represents the changes of resistance-related markers expression with treatment, while the dotted line represents those without treatment (below). CSC cancer stem cell, DTPs drug-tolerant persisters
Despite knowing the significant contributions made by DTPs to both non-mutational and mutational processes during resistance, controversies still exist concerning the genesis of DTPs between the natural selection theory (classical Darwinian selection), Lamarckian induction concept, and the coexisting model, as described below26 (Fig. 1). The natural selection theory is a simple and intuitive principle. Specifically, DTPs, here represented by CSCs in an inconspicuous but preexisting form which are hidden by the overwhelming number of non-CSCs, can be selected and enriched upon drug exposure17,27 (Fig. 1a). This theory, based on phenotypic behavior, can also be interpreted as a process for selecting the pre-existing slow-cycling cells under treatment, for example, pre-existing JARID1B-expressing melanoma cells or ZEB2-expressing colorectal cancer cells.28,29
In contrast to the “passive” mode of Darwinian selection, the concept of Lamarckian induction attaches importance to the natural aptitude of tumor cells in adapting to internal or external stimuli actively, especially in response to pharmacologic interventions, essentially a concept of therapy-triggered “adaptation” (Fig. 1b). This adaptation, rather than the “one mutation, one outcome” dualistic model, is predominantly reflected in the dynamic change of a number of resistance-related genes through epigenetic events during treatment, laying a mechanistic foundation for the emergence and coexistence of DTPs in varying tolerant states26,30 (Fig. 1b). Among the resistance-related markers, the well-characterized drug efflux pump—multidrug resistance protein-1 (MDR1), serves as an example.31 In this case, a quick and robust response to vincristine exposure manifesting as phenotypic switching from a low- to high-efflux state, has been observed, which is proved to be a direct consequence of “active” MDR1 induction via single-cell longitudinal surveillance.31 More importantly, once such an induction is triggered, transcriptomic alterations tend to persist for a relatively long time after drug withdrawal31 termed “epigenetic memory.”32 This is in accordance with the notion that DTPs can transiently evade treatment and maintain the pro-survival phenotype or transcriptome alterations for some time.12,26
In actual fact, the dynamic transcriptional fluctuation of resistance-related markers at a single-cell level is more likely to occur before the addition of drug in a manner similar to the so-called “transcriptional noise,” thus giving rise to an incremental source of transcriptional variability for drug selection16,32–35 (Fig. 1c). As a result, a small fraction of tumor cells, whose expression of these resistance-related genes exceeds a certain threshold at the moment of treatment, can survive or be selected.16 The “internal noise” (e.g., random pattern of transcriptional variability on resistance-related genes) can be viewed as a loaded “weapon” within the “arsenal” of tumor cells to cope with “external noise”36 (e.g., drug exposure), marking a return to classical Darwinian selection (Fig. 1c). However, with increasing duration of drug exposure, such a stochastic, transient, fluctuated “survival mode” develops into an adaptive, stable, dormant, drug-refractory state through epigenetic modifications, ultimately resulting in the establishment of a pool of DTPs.16,34 These alterations in the epigenome (i.e., “adapting to shape change instead of being shaped”) are in agreement with the concept of Lamarckian induction31 (Fig. 1c). Hence, throughout the entire process of the emergence and maintenance of DTPs, these two concepts are not opposite, but rather intertwined and complementary to each other (Fig. 1c).
If one regards the profound transcriptional variability16 as the “innate skill” of tumor cells to pursue greater phenotypic diversity, the epigenome-associated dormant state caused by long-term treatment will be more likely the “acquired inertia” of DTPs due to the assumption that the survival skills, that is, overexpression of resistance-related genes, have been gained from the cells surviving initial therapy. This raises the question of “when treatment is discontinued, will the ‘acquired inertia’ fade away and/or ‘innate ability’ be restored?” Consistent with in vitro laboratory experiments, the so-called ‘re-treatment response’’ phenomenon observed clinically supports the occurrence of a reversible process from acquired drug-refractory to initial drug-susceptible state following drug withdrawal37 (Fig. 1c). Specifically, a significant fraction of patients with NSCLC who have been through a failed treatment with epidermal growth factor receptor (EGFR)-TKI-based therapy (gefitinib) can immediately achieve remarkable tumor regression following re-treatment with gefitinib after a drug-free interval, demonstrating a second response “window” to treatment with TKIs.37,38 Similar re-treatment responses in different cancer types have also been observed with other anticancer agents, including daratumumab,39 trastuzumab,40 radium-223,41 and pembrolizumab.42,43 The prerequisite for such a secondary response is that the timing of re-treatment needs to precede the presence of a novel resistance-conferring mutation in DTPs. This can be interpreted as a process of residual DTPs getting rid of the “acquired inertia” while re-activating the “innate skill” or, put another way, a transition from a slow-cycling, drug-refractory to a fast-cycling, drug-susceptible phenotype (Fig. 1c).
Indeed, this reversible phenotype switching, at first glance, can be attributed to the proactive behavioral “changes” of tumor cells to adapt to environmental “changes” albeit in an uncontrollable manner. This also implies that hijacking the mechanisms underlying these “changes” for therapeutic purposes, transforming such a process from uncontrolled to controlled, could be a promising approach. For this reason, studies revolving around the complicated cellular mechanisms involved in the “war” of “hide (phenotype switching)-and-seek (cancer therapy)44” have gained increasing prominence in recent years.
In terms of phenotype switching, cell plasticity (the fundamental ability of cells to change their properties in a reversible way actively or passively) plays a prominent role in postinjury tissue repair and regeneration, as well as the restoration of disrupted homeostasis.45–47 Besides making contributions to such physiological processes, when activated aberrantly, cell plasticity is involved in the evolution and progression of multiple diseases, particularly cancer.46–49 This sheds new light on the explanation of the intratumoral heterogeneity of phenotypic features of cancer during which tumor cells exhibit varying degrees of phenotypic interconversion between drug-susceptible and drug-refractory states.50 The above general description of phenotype switching in cases of drug exposure or drug withdrawal represents a universally applicable model of tumor cell plasticity, regardless of what types of cancer are treated or what kind of therapies are employed. However, behind this universally plastic behavior, there exist differences in exactly how cancer cells evade therapy including epithelial–mesenchymal transition (EMT), acquiring properties of CSCs or trans-differentiation potential26,47,51–54 (Fig. 1c). Intriguingly, these somewhat functionally overlapping processes are more or less associated with the aberrant (re-)activation of developmental programs, suggesting that similar molecular mechanisms underlying plasticity-driven resistance to therapy may be involved.55,56
In summary, in this review, we present a comprehensive description of tumor cell plasticity in response to treatment of various cancers with respect to targeted therapies, chemotherapy, and immunotherapy, and will highlight the mechanisms involved.
Epithelial–mesenchymal transition (EMT)
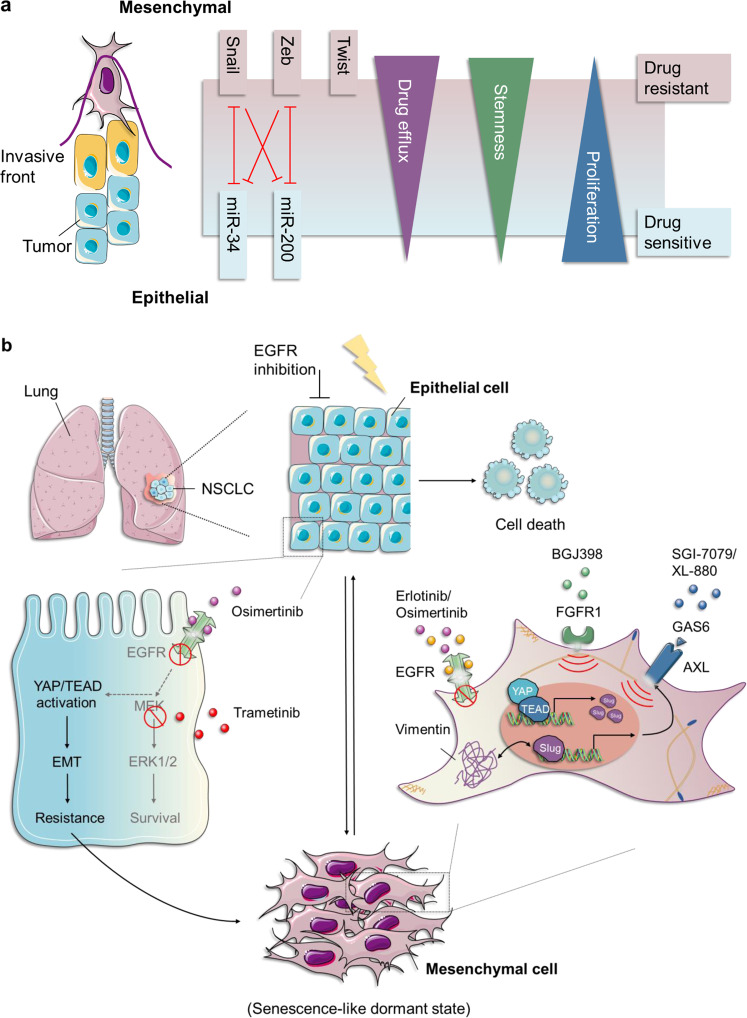
The programs of EMT and its inverse process, mesenchymal-to-epithelial transition (MET), are involved in governing vertebrate embryonic development in a highly dynamic, transitory and reversible manner, representing a prime example of cell plasticity, both in normal and neoplastic cells.55,57,58 At conceptual and morphological levels, cells undergoing EMT are characterized by loss of apical–basal polarity and the disruption of cell–cell contacts, including tight (e.g., ZO-1), adherens (e.g., E-cadherin), and gap junctions (e.g., connexins), while acquiring the front–rear polarity and dramatic remodeling of the cytoskeleton organization. This ultimately results in the morphotype switching from “cobblestone-like” shapes to “fibroblast-like” (e.g., vimentin) forms.55,59–61 Mechanistically, this process is generally performed by several EMT‑inducing transcription factors (EMT-TFs), such as Snail, zinc-finger E-box-binding (Zeb), and basic helix–loop–helix TFs, and noncoding microRNAs (miRNAs), epigenetic, and post-translational regulators, as well as alterative splicing factors, which are further integrated and controlled by multiple signaling pathways, such as the transforming growth factor-β (TGF-β), wingless/integrated (Wnt), Notch, and Ras-mitogen-activated protein kinase (Ras-MAPK) pathways, in response to paracrine and autocrine stimuli62–64 (Fig. 3a). Notably, the EMT-TFs are orchestrated and dynamically regulated themselves by each other and/or other factors in every step of EMT programming, in particular, the two well-established double-negative feedback loops, miR-34/Snail1 and miR-200/Zeb (Fig. 3a). The former regulatory circuit preferentially participates in the initial phase of EMT induction in epithelial cells, while the latter tends to be involved in the development and maintenance of the mesenchymal state.65–71 Functionally, it is generally recognized that the EMT programs not only play an irreplaceable role in multiple physiological processes throughout the whole course of an individual’s life, especially during embryonic development (tissue morphogenesis and organogenesis), wound healing, tissue repair, and the induction of pluripotency, but also contribute to various pathological events, including formation of fibrosis and tumor malignancy—from its genesis to development.59,72–75
Fig. 3.
The role of EMT in EGFR-TKI resistance. a Cancer cells undergoing EMT are characterized by morphotypic switching from “cobblestone-like” shapes to “fibroblast-like” forms. This process can be achieved via several EMT-TFs (Snail, Zeb, and Twist) and miRNAs in response to paracrine and autocrine stimuli, endowing cancer cell with a more aggressive phenotype, including enhanced invasive capacity, therapeutic resistance (enhanced drug efflux and slow cell proliferation), and stemness properties. b In EGFR-mutant NSCLC, upregulation of TEAD-mediated YAP promotes the transcription of Slug, which further induces the upregulation of AXL in NSCLC cells. AXL signaling, whose activation relies on interactions with its specific ligand GAS6, promotes EMT that drives Slug-overexpressing mesenchymal cells to acquire resistance with erlotinib. In addition, the mesenchymal cells display enhanced resistance to EGF816 accompanied by a significant activation of the FGFR1 pathway, implicating the potential of FGFR1 as a drug target for evading resistance to EGF816. A subpopulation of cancer cells can enter a senescence-like state to escape cell death upon administration of EGFRi (osimertinib) in combination with MEKi (tretinamib), resulting in resistance. This change is characterized by YAP/TEAD-mediated activation of EMT programs. The therapeutic strategy of pharmacologically cotargeting YAP/TEAD (by MYF-01-37) and EGFR/MEK leads to synthetic lethality. AXL anexelekto, GAS6 growth arrest-specific protein 6, SGI-7079/XL-880 AXL inhibitor, EGF816 the third-generation EGFR-TKIs, FGFR1 fibroblast growth factor receptor 1, BGJ398: FGFR inhibitor
EMT in carcinoma progression
From embryonic development (physiology) to cancer progression (pathology)
Before discussing the impacts of EMT programs on carcinoma progression, it is necessary to mention the inspirations provided by the considerable amount of theoretical and experimental studies on their physiological roles. To be precise, in case of embryonic development, several sequential cycles of EMT and MET—termed as primary, secondary, and tertiary EMT, are highly organized and carefully orchestrated according to separate biological requirements, resulting in the terminal differentiation of specialized cell types and the organization of the extremely intricate three-dimensional (3D) structure of internal organs.62,74,75 A typical exemplar is the formation of embryonic heart during which all three cycles are shown to be experienced successively.62,74,76 This process is also characterized by the fact that EMT programs take place in well-differentiated epithelia, laying a theoretical and realistic foundation for the occurrence of EMT in other well-differentiated epithelia, including tumor cells. In addition, during the process of wound healing, keratinocytes residing at the wound edge initiate part of the EMT programming autonomously, which leads to the acquisition of an intermediate phenotype—also described as the “metastable” state, along with the capability of migrating towards the middle regions to seal the wound.73,74,77,78 Such a functional conversion from stationary to migratory phenotype of keratinocytes, when mapped to cancer progression, denotes that the influence of EMT on the biological behavior of carcinoma cells may be primarily embodied in their ability to migrate or invade—or, even more evocatively, in tipping the scale of “Go (migration) or Grow (proliferation)” towards the “Go/migration,”79 which perhaps foreshadows a more aggressive phenotype and a higher metastatic potential of tumor cells.
From “complete” to “partial”: the perfect paradigm for tumor cell plasticity
Not surprisingly, the occurrence, performance, and potential roles of EMT in carcinoma cells, as proposed theoretically, have already been determined through compelling experimental evidence in the past two decades, although contradictory opinions exist.80,81 These anomalies have stemmed from a lack of convincing evidence at the surgical pathological level to support the concept that EMT is involved in the onset and development of cancers, resulting in a noticeable mismatch of results between laboratory models and human tissue sections.80,81 Faced with this mismatch, researchers have provided an appropriate explanation that, in the vast majority of cases of cancer, the induction of EMT may not be as straightforward as it seems—that is, not strictly abiding by the binary-based “all or nothing” principle, but rather a complicated multistage process containing one or more intermediate phenotype(s) with a varying degree(s) of EMT—currently known as the “partial EMT(s)” state.82,83 Moreover, a complementary explanation for this mismatch is the indistinguishable expression of EMT markers that results from the coexistence of carcinoma cells with tissue-resident stroma-associated cells inside and around individual tumors, the latter of which can naturally exhibit variable levels of the mesenchymal signature.84 Therefore, it seems neither accurate nor objective to measure the “partial EMT” within clinical cancer tissues by the standards of the “complete” one. More importantly, this manifestation of EMT in human cancers echoes the aforementioned “metastable” state in wound healing and the reversible process in renal fibrosis,85,86 all of which points to the conclusion that the incomplete activation state appears to be an essential trait of EMT programs, not just during carcinoma progression.
Since the introduction and recognition of the “partial EMT” concept, it should be pointed out that understanding the true meaning of “partial” is the key issue to unlocking the secrets of EMT programs in cancer. As its name implies, the “partial EMT” concept in carcinoma cells can be interpreted simply as a hybrid epithelial and mesenchymal phenotype, existing in the form of clusters and even single cells.87–92 Such formats endow cancer cells with the possibility of accessing multifunctional cell clusters and multi-identity single cells so that they can readily cope with the changeable environments.91 It is this strong ability of self-adaptation and self-adjustment in tumor cells that represents a perfect mechanism for plasticity, but an enormous threat to cancer patients. Various attempts have already been made in the field to address this experimentally. The ideal approach, proposed by Weinberg,81 is to trace the dynamic changes of cancer cells at both an individual and multicellular cluster level, from the time they depart the primary tumor until the point at which metastatic colonization is detectable clinically at a distant organ. In contrast to the initiation (primary tumor) or termination (metastatic tumor) phases, effective monitoring for the intermediate phase (e.g., by using circulating tumor cells, CTCs) is of fundamental importance in exploring the exact role of “partial EMT” in human cancer. However, this is technically challenging due to their very low abundance in blood.93
To address this issue, Yu et al.94 have developed a quantifiable, dual-colorimetric RNA–in situ hybridization approach to investigate the contributions made by EMT to primary tumors, tumor-infiltrating lymphocytes (TILs), and CTCs from patients suffering from metastatic breast cancer. Compared to primary tumors where EMT occurs very rarely, a significant fraction of patient-derived CTCs display obvious mesenchymal features, the level and quantity of which are positively related to disease progression during anticancer drug treatment.94 Further evidence has been obtained through long-term longitudinal surveillance of EMT traits in CTCs from an index patient, whose blood samples were collected serially.94 The results from this serial monitoring demonstrate that dynamic changes in the ratio between epithelial and mesenchymal phenotype in CTCs may largely determine the final clinical outcome, that is, response or resistance, both to targeted therapy and chemotherapy.94 Similarly, using single-cell RNA-sequencing (scRNA-seq), a subsequent study found direct evidence that the partial EMT program of head and neck squamous cell carcinoma may serve as a valid, independent predictor for adverse clinicopathologic features and malignant biological behaviors, particularly nodal metastasis, through comparative analyses of primary and metastatic specimens.90 Together, these observations on clinical samples are in agreement with those from cultured cells and animal models that have tightly linked “partial EMT” to cancer progression, in which therapeutic resistance and metastatic potential are shown to be the closely linked, and extremely threatening.
More recently, with the establishment of an effective, rapid, large-scale single-cell resolution 3D (LSR-3D) imaging protocol capable of visualizing the cellular organization of an entire mammary tumor, Rios et al.95 discovered that epithelial and mesenchymal subsets coexist within the same clone in most observable cases of Pten/Trp53 deletion models, offering solid evidence that the induction of partial EMT acts as a ubiquitous adjusting and controlling mode at a clonal level. This finding, from a space perspective, highlights that the induction of partial EMT is not confined to the traditional concept that whether or not carcinoma cells undergo EMT is determined by their localities within an individual tumor, but is more likely an inherent property of most clones wherever they reside. This adds a twist to the traditional view that EMT usually occurs along the invasive front.92,95 From a temporal perspective, there seem to be no specific “timestamps” indicating when the partial EMT state of carcinoma cells first occurs as it is seen throughout the period of LSR-3D imaging, including clones from an Elf5-driven tumor at its early stage.95 This visual evidence ties in closely with data from previous in vivo or in vitro experimental studies, which showed that prior to the development of a malignant phenotype, EMT programs have already started imperceptibly in certain types of human carcinomas, including breast,96–98 pancreatic,99 and prostate cancer (PCa).100 This is in accord with perplexing clinical observations of early metastatic dissemination before the formation of a detectably localized tumor,101–104 and the preresistance state of a minor subpopulation of tumor cells prior to drug exposure.21,22 The temporal mode of EMT may provide a plausible mechanism by which the above paradoxes can be, at least partially, explained.
Taken collectively, the progressive notion arising from both spatial and temporal perspectives has brought the concept of EMT into a new level of complexity and universality. These two properties can be simultaneously embodied in that carcinoma cells with varying degrees of EMT localizing randomly within an individual tumor, display their respective functional attributes of each clone, or even single cell, ranging from atypical hyperplasia to late-stage metastasis and/or therapeutic resistance. All this suggests that EMT programming during the process of cancer progression is a perfect paradigm for investigating the nature of tumor cell plasticity.
From metastasis to resistance
Indeed, from the observations mentioned above, as well as other studies, the proposition that EMT may act as the main driver of metastatic process, drug resistance, maintenance of stemness, and immunosuppression seems justified55,105–107 (Fig. 3a). Unfortunately, quite a few patients have metastatic diseases at initial diagnosis, especially in regions where regular health checks and screening are not routine.108 There is ample evidence to provide support for the major role of EMT in all steps of the “invasion-metastasis cascade.”109 However, the existence of a causal relationship between them remains a long-standing subject of dispute.110
Specifically, the contributions made by EMT to metastasis was initially proposed due to the demonstration that inhibiting the expression of Twist and the resulting EMT could significantly alleviate pulmonary metastasis of highly metastatic mammary carcinoma cells in vivo.111 Subsequently, similar biological impacts on invasion and metastasis induced by other key EMT-TFs, such as Snail1,112,113 Slug,114–116 and ZEB1,117 were extensively documented in different types of carcinomas. In fact, underlying such functional similarities, these EMT-TFs appear to specialize in handling their precise biological subfunctions in a tissue (spatial)- and/or clinical-phase (temporal)-specific manner; that is, they are organized in a way that tends to be coordinated and complementary, but not redundant.63 For example, using a mutant KRAS and p53 driven (KPC) mouse model of pancreatic ductal adenocarcinoma (PDAC), Krebs et al.117 have demonstrated that depletion of the EMT-TF Zeb1, but not Snail or Twist,118 markedly inhibited PDAC progression from its genesis to advanced metastatic disease. By contrast, using the MMTV-PyMT spontaneous breast cancer model that carries wild-type TP53, obvious inhibitory effects on the self-renewal capacity and metastatic potential were observed following Snail1 excision,119 rather than by downregulating Zeb1/2 via forced expression of miR-200.118 These repressions can be explicitly reversed in vivo by transient overexpression of Snail1.120 When connecting these conflicting findings to the aforementioned observations of the contribution made by Twist to breast cancer metastasis,111,121 it can be safely concluded that the expression pattern and regulative mechanism of an individual EMT-TF depend critically on the site where the primary tumor occurs, as demonstrated by the roles of Zeb1 in PDAC,117 Snail1 and Twist1 in breast cancer,111,119–122 and Zeb1/2 in melanoma.123,124
Besides differences in the expression of EMT-TFs among cancer types, there also exist differences within each cancer type, represented by the spatiotemporal, synergistic effects of Snail1/Twist1-mediated EMT programs on the promotion of breast cancer progression, especially during metastasis.125 This parallel-cooperative functioning mode is reflected by the realization that transitory activated Snail1 plays an indispensable role in EMT initiation, while Twist1 is mainly responsible for the maintenance of late-stage EMT programming, echoing their physiological roles during Drosophila mesoderm development.120,125–127 Besides functioning synergistically as described above, biological influences exerted by interactions between EMT-TFs vary from one type of cancer to another, and can even perform antagonistically, as shown by the contrasting behaviors of Zeb1 and Zeb2 during initiation and metastatic progression of melanoma.123,124,128 Based on this, when mapping the motile, invasive, and dedifferentiated traits acquired through EMT programming to the multistep process of “invasion-metastasis cascade” in the context of mammary carcinoma, it can be hypothesized that Snail-induced initial EMT is associated with early dissemination of carcinoma cells, including local invasion and subsequent intravasation into blood circulation, whereas Twist1-triggered late EMT tends to take place during extravasation and the formation of dormant, scattered micrometastases due to the overlapping functions between EMT and CSC.55,125,129 Such a dormant, growth-arrested state, persisting for months or even years, indicates that micrometastatic clusters or single disseminated tumor cells (DTCs) can tilt their functional balance towards stem-like attributes—referred to as “tumor-initiating CSCs,” from a previous migratory phenotype—termed “migratory CSCs’’,129 in order to survive in, and adapt to, a distant, unfamiliar microenvironment, serving as potential initiators of macroscopic metastatic lesions.129,130 Starting from this concept, re-initiation of tumor growth in a foreign tissue would require maintenance of the self-renewal capacity of DTCs through asymmetric division while differentiating into non-CSCs to spawn fast-cycling epithelial progeny, ultimately giving rise to overt metastases (also called colonization).129 Although the mechanism of metastatic colonization shows organ preference,131,132 stemming largely from the organ-specific premetastatic niches (PMNs),133,134 the restoration of epithelial characteristics, induced presumably by a process of reversible EMT–MET, seems to be a common feature shared by multiple cancer types during seeding of a secondary tumor, only by which means can the EMT-induced invasive phenotype be functionally equivalent to metastatic potential.59,129,135–143 With MET programs enabled, macroscopic metastases therefore exhibit the same histopathological trait of epithelial cell predominance as that of their corresponding primary carcinomas, while reconstructing the typical lineage hierarchy between CSCs and non-CSCs lost during EMT induction, as if EMTs had not actually occurred.84,129,144 However, many believe that it is such inherent plasticity that ultimately leads to a lack of persuasive evidence at the pathological level to support the essential role of EMT in metastasis, leading to the controversy.80,81
Taking all this into consideration, although having yielded conflicting evidence on whether the EMT programs contribute to metastases, the spatiotemporal regulation, and pleiotropic, non-redundant functions of EMT-TFs, the dynamic, transient, and reversible EMT–MET operating system, as well as the extended concept of EMT from dualism (a complete form) to pluralism (multilayered, partial forms), goes a long way in explaining why the controversy occurred and how it can be resolved.
The fact that cancer metastasis, therapeutic resistance, and immunosuppression are three complex and poorly understood processes, which often coexist clinically, is of particular note.55,107,109 Although metastasis rather than the primary tumor is the reason for ~90% of cancer-associated deaths,129 drug resistance must also be addressed. More recently, two seminal papers by Fischer et al.118 and Zheng et al.145 have highlighted an irreplaceable role of the EMT in cyclophosphamide and gemcitabine resistance of breast and pancreatic cancer cells, respectively, while challenging the conventional role of EMT in cancer metastasis.
Here, in this review, we focus on the mechanistic inter-relationships between the EMT programs—representative of cancer cell plasticity, and the resistance to cancer therapies, including targeted therapy, chemotherapy, immunotherapy, and radiotherapy.
The relationship between EMT and drug resistance
It is commonly believed that re-activation of developmental programs is one of the principal mechanisms controlling many adult disease processes, including the EMT programs in drug resistance.56,135,146 To better understand the relationship between gene- and protein-expression profiles in tumor tissues of cancer patients and their corresponding clinical responses, multiple studies have been performed. These showed a positive correlation between the expression of mesenchymal-/stroma-related markers and therapeutic resistance, including for chemotherapy, targeted therapy, radiotherapy, and immunotherapy, although at times this has been controversial.147–157 For example, in the context of estrogen receptor (ER)-negative breast cancer, Farmer et al.147 have reported that upregulation of the genes within stromal metagene exhibits a significant predictive effect on the resistance to neoadjuvant chemotherapy with 5-fluorouracil, epirubicin, and cyclophosphamide. This signature seems to, at least in part, depend on the activation of EMT programs within carcinoma cells.147 Analogously, employing integrative analyses of gene expression and proteomic profiling, a robust 76-gene mesenchymal signature was derived and verified to have the potential to predict whether or not the resistance to EGFR-TKIs and phosphatidylinositol 3-kinase/protein kinase B (PI3K/Akt) inhibitors would be acquired in various NSCLC cell lines and clinical samples, highlighting the significant impact of different phenotypic (epithelial and mesenchymal) states on drug responsiveness.148 Furthermore, in melanoma, two markers (PTRF and IGFBP7) related to phenotype switching from melanocytic to mesenchymal state were shown to distinguish MAPK inhibitor-resistant cells from MAPK inhibitor-sensitive melanoma cells by proteomic screening.149 In short, considering all the above, the concept that EMT programs serve as a direct contributor to the acquisition of resistance to both cytotoxic and targeted therapeutic agents in a variety of cancer types is fairly convincing.
Over the years, the rapid development of immune checkpoint blockade (ICB) therapies (e.g., inhibitors of cytotoxic T lymphocyte-associated protein 4 [CTLA-4] and programmed cell death-1/programmed cell death-ligand 1 [PD-1/PD-L1]) has revolutionized the clinical treatment landscape in a wide range of advanced tumor types. Nevertheless, low response rates as well as ensuing immunotherapy resistance and delayed relapse represent a significant challenge in the field of cancer immunotherapy for the treatment of a variety of tumors, including lung adenocarcinoma,158,159 melanoma,160,161 PCa,162 and pancreatic cancers.163 However, the molecular mechanisms involved in immune escape remain elusive,164 and the influence of EMT programs on immunotherapies remains controversial. A number of studies have suggested a positive correlation between EMT-related signatures and T cell infiltration, leading to enhanced sensitivity to ICB.150–154 By contrast, others have indicated that tumors with EMT/stroma-related gene expression are closely connected with lower clinical responses and poorer progression-free and overall survival.156,157
Tumor microenvironment (TME) contributes to resistance via EMT
(1) TGF-β within TME
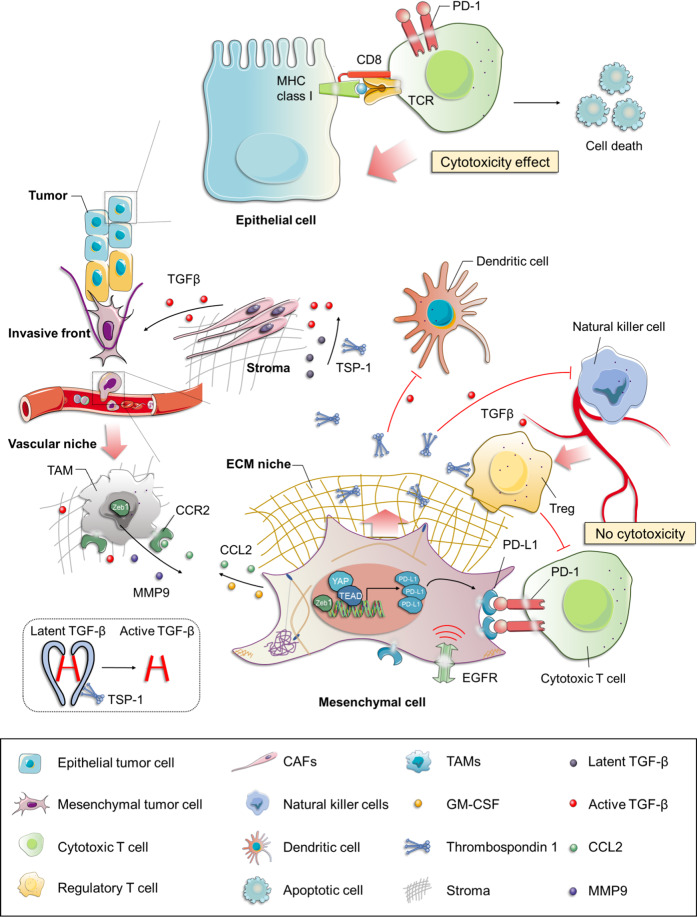
TGF-β, a well-established key promoter and sustainer of mesenchymal and/or CSC state,165,166 contributes to the induction and function of immunosuppressive regulatory T cells (Tregs)167,168 and inhibition of metabolic activity of natural killer (NK) cells,169,170 laying the foundation for molecular mechanisms underlying the significant role of EMT programs in antitumor immune response (Fig. 2). It is known that TGF-β can be activated by removing the N-terminal latency-associated peptide through serine proteases (plasmin171 and cathepsin D172) and matrix metalloproteinases (MMP9 and MMP14).173 Activated TGF-β can bind to a subset of integrins (including αvβ6,174,175 αvβ8,173,176 and αvβ1,177) or bind to the secreted and matricellular protein thrombospondin-1 (TSP-1, which is regarded as the first discovered activator of TGF-β1) under both physiological and pathological conditions in vivo.178–182 An increased level of TSP-1 secreted by mesenchymal cells, especially Snail-overexpressing cells, on the one hand facilitates the further activation of the TGF-β signaling pathway, thus contributing to a positive feedback effect on EMT. On the other hand, it promotes the continual generation of Foxp3+ Tregs from naive T cells, which antagonize the effects of cytotoxic T lymphocytes (CTLs), together with the induction of impaired dendritic cells (DCs) and inhibition of NK cells within the TME, thus ultimately resulting in resistance to immunotherapy, and even chemotherapy183–186 (Fig. 2).
Fig. 2.
The tumor microenvironment (TME) contributes to resistance via EMT. Increased levels of TSP-1 (the activator of TGF-β1) secreted by mesenchymal tumor cells on the one hand facilitates the further activation of the TGF-β signaling pathway, contributing to a positive feedback effect on EMT; on the other hand, it promotes the generation of Foxp3+ Tregs from naive CD4+ CD25− T cells that antagonizes the activity of cytotoxic T cells, together with the induction of impaired DCs and inhibition of NK cells within the TME, thus ultimately resulting in immunotherapy, and even chemotherapy resistance. Tumors arising from the mesenchymal cells express a higher level of PD-L1 and lower level of MHC-I, together with more Tregs within TME in comparison with those formed by the epithelial cells, supporting the immunosuppressive role of EMT programs, which at least in part contributes to the resistance to cancer therapies. The TAMs, known as the most plentiful immune-related stromal components in TME, have been shown to infiltrate mainly at the invasive fronts of tumors. CCL2, synthesized by cancer cells, triggers the recruitment of circulating monocytes with the expression of CCR2 into tumors with the subsequent acquisition of a TAM phenotype. ZEB1-expressing macrophages promote their own polarization toward a stronger protumor phenotype; and meanwhile, upregulate the expression of CCL2 and CD74 in cancer cells through an increased release of MMP9, resulting in a mesenchymal/stem-like state. This forms a CCR2-MMP9-CCL2+ feedback loop between TAMs and the cancer cells. TSP-1 thrombospondin-1, PD-L1 programmed cell death-ligand 1, Tregs regulatory T cells, DCs dendritic cells, MHC-I the class-I major histocompatibility complex, TAMs tumor-associated macrophages, CAFs cancer-associated fibroblasts
To extend these observations further, epithelial and mesenchymal cell lines derived from the transgenic MMTV-PyMT breast cancer model have been established.187 Tumors arising from the more mesenchymal-like cell lines (with high levels of vimentin and EMT-TFs) exhibit the reduction of the class-I major histocompatibility complex together with more Tregs within TME in comparison with those formed by the more epithelial-like cell lines (with significant levels of E-cadherin and epithelial cell adhesion molecule).188 This is consistent with the immunosuppressive role of EMT programs described above. In addition, tumors arising from the more mesenchymal cell lines are frequently accompanied by an obvious infiltration of protumor and anti-inflammatory alternatively activated (M2) macrophages (CD206+ and Arginase1+), instead of antitumor and proinflammatory classically activated (M1) macrophages (inducible nitric oxide synthase, iNOS+ and IL-12+), which occur in tumors arising in the more epithelial cells.188–190 This mirrors the switchable potential between two different polarization states due to the high plasticity of macrophages depending on changes in the local TME—in this case, referring to the induction of EMT programs of cancer cells within the TME.190–192 While the binary system of polarization states seems an attractive hypothesis, accumulating evidence demonstrates that tumor-associated macrophages (TAMs) prefer to share a mixed phenotype and express both M1 (HLA-DR, interleukin-1β [IL-1β], or TNF-α) and M2 (CD163 and IL-10) markers rather than being considered as two cell types completely independent of each other.193,194 This may explain why TAMs exhibiting characteristics of either tumor killing (M1-like) or tumor-promoting (M2-like) macrophages can play an equally important role in the induction of EMT programs in PDAC.195
(2) The role of inflammation in EMT
The concept that there exists a causal link between chronic inflammation and the onset of cancer is well established and widely accepted owing to comprehensive biochemical and clinical evidence.196–198 Indeed, the effects exerted by inflammatory reactions on cancer is not limited to its initial stage, but can also be observed during its progression, including late-stage disease characterized by the acquisition of malignant behaviors, particularly those related to the induction of EMT.198 Before discussing the role of inflammation on EMT programs, it is beneficial to explore how inflammation is involved in other physiological and pathological processes, among which wound healing is highly representative.199,200 Of note, the implementation of wound healing or tissue regeneration, a highly conserved process that largely depends on an EMT-induced migratory phenotype of keratinocytes, tends to be a result of the combined effect of the inflammatory microenvironment and EMT, serving as a perfect paradigm for investigating the interactions between these two events. In this sense, when extending this paradigm to cancer progression, there is every reason to believe that such a crosstalk between inflammation and EMT can be replicated, hijacked, and extended by carcinoma cells, with EMT programs being exploited for the metastatic process. This echoes the previous notion that tumors have characteristics similar to long-term unhealing wounds.201 More importantly, cancer therapy, especially chemotherapy and radiotherapy, are both capable of stimulating inflammatory responses per se, providing the mechanistic foundation for the involvement of inflammation in therapeutic resistance, and even immunosuppressive TME.202,203 In this context, recent advances regarding the roles of inflammation and EMT programming, and their interactions in resistance will be summarized, with a particular focus on the functions of two sources of inflammatory cellular components, namely, macrophages and myeloid-derived suppressor cells (MDSCs).
- Recruitment of macrophages into TME and resistance. From the macroscopic viewpoint, apart from tissue-resident cells, there also exist a high proportion of multiple immune cells recruited to the TME, which have been termed as “partners in crime” in the “sanctuary of the devil.”204,205 Among these, the TAMs, known as the most plentiful immune-related stromal components in TME,206 might account, in part, for the poor prognosis of patients with different types of tumor, such as hepatocellular carcinoma (HCC),207 breast,208–210 gastric,211–214 lung,215–217 pancreatic,218 PCa,219 esophageal220 and thyroid cancers,221 and Hodgkin’s lymphoma.222,223 Therefore, not only the numerical superiority of infiltrating cells within the TME, but also the significant effect on patient prognosis indicates a pivotal role of TAMs in the crosstalk between cancer cells and TME.224 In general, the emergence of a TAM phenotype firstly requires for the recruitment of monocytes into TME triggered though various tumor cell-derived cytokines, chemokines, and growth factors in a collaborative way, which contain granulocyte–macrophage colony-stimulating factor (GM-CSF), chemokine (C-C motif) ligand 2 (CCL2), CSF-1, macrophage-stimulating protein, vascular endothelial growth factor-A, and TGF-β1, and so on. Subsequently, these recruited monocytes can further differentiate into M2 macrophages fueled by IL-4, IL-6, or IL-10.190
-
GM-CSF and IL-6. Similar to cancer-associated fibroblasts (CAFs), TAMs have also been shown to infiltrate mainly at the invasive fronts of tumors, the sites where cancer cells most frequently undergo EMT (Fig. 2). Such spatial overlap between host and tumor indicates that the signals resulting from bidirectional communications may be responsible for the commonly accepted spatial characteristics of EMT induction mentioned previously.225–229Based on that, Bonde et al.230 demonstrated a significant role for intratumoral TAMs in the activation of EMT programs in cancer cells through depletion of macrophages in F9-teratocarcinoma bearing mice, thus broadening and challenging the traditional view that TAMs-mediated EMT is confined to the invasive fronts. Furthermore, TGF-β derived from macrophages has been identified as the major cytokine controlling this highly context-dependent phenotype switching using a candidate-based screen. Supporting this notion, a systematic analysis of a large number of NSCLC tissue samples revealed that overexpression of EMT-associated markers in tumor cells was significantly and positively correlated with intratumoral CD68+ macrophage density and intraepithelial TGF-β levels, together with concomitant higher histologic grade and tumor heterogeneity, all of which contributed to drug resistance and patient relapse.230 In addition, by comparing the effects on human monocytes of coculturing with the medium derived from epithelial-like or mesenchymal-like cell lines, respectively, Su et al.227,231 reported that breast cancer cells with mesenchymal signature could activate macrophages to differentiate into the M2-macrophage phenotype, which was partially attributed to the secretion of GM-CSF from mesenchymal tumor cells (Fig. 2). This immunosuppressive phenotype, in turn, further strengthened EMT programming of tumor cells in various cancer types, including, but not limited to, breast,227,232 gallbladder,233 and pancreatic,234 as well as head and neck cancers,235 via releasing CCL18 from TAMs, forming a positive feedback loop both in vitro and in vivo227,231 (Fig. 2). A recent study that focused on investigating the role of oncoprotein MCT-1/MCTS1 (multiple copies in T cell malignancy 1) in triple-negative breast cancer (TNBC) identified significant enrichment of proinflammatory cytokines/chemokines, such as IL-6, GM-CSF, and monocyte chemotactic protein-1 (also known as CCL2) released from MCT-1-overexpressing cells compared to those observed in control cells.236 Among those cytokines, the relative abundance of IL-6 within TME has been demonstrated to promote the trans-polarization of infiltrating THP-1 monocytic cells into an immunosuppressive M2-like macrophages phenotype (CD163+ and CD206+);237 the activation of EMT processes, together with the maintenance of cancer stemness in TNBC, jointly result in the suppression of antitumor immune responses and tumor recurrences following therapy.236,238,239
-
CCL2. Aside from GM-CSF and IL-6, the chemokine CCL2, synthesized by cancer and stroma cells within the TME, triggers the recruitment of proinflammatory F4/80low circulating monocytes with expression of CCR2 (the receptor for CCL2) into tumors with the subsequent acquisition of a TAM phenotype240,241 is emerging as a prominent regulator of cancer metastasis,242 especially metastatic growth,243,244 and angiogenesis.245 Interestingly, continuous recruitment and enhanced infiltration of macrophages resulting from upregulation of tumor cell-derived CCL2 is observed upon targeting androgen receptor (AR) with short interfering RNAs, which leads to the establishment of an immunosuppressive microenvironment, induction of EMT programming, and a growing population of stem/progenitor cells.246,247 These eventually result in the development of androgen deprivation therapy (ADT) resistance in PCa.246,247 Similar CCL2-mediated monocyte/macrophage trafficking was also observed in the inducible KrasG12D p53-null (iKPC) PDAC mouse model, which largely depends on overexpression of HDAC5.248 Subsequently, TGFβ secreted by these recruited TAMs endows tumor cells with a mesenchymal-like phenotype enabling them to survive in case of oncogenic KRAS (KRAS*) extinction, indicating a significant role of the CCL2-TGFβ/EMT signaling pathway in the resistance to KRAS* targeting therapy.248 While a similar quasi-mesenchymal phenotype has already been validated by previous studies focused on the acquisition of resistance to KRAS* functional suppression both in KRAS*-driven PDAC and lung cancer,249,250 the results reported by Hou et al.248 extend the mechanisms involved in bypassing KRAS* addiction from the tumor cell per se (YAP1 activation) to host-tumor interactions in PDAC. Given the growing emphasis on the role of YAP1 or Hippo pathway in the TME,251–253 it seems worth exploring how CCL2 and YAP1 could be integrated together to promote EMT-related resistance.254Using subcutaneously implanted tumor models in mice with PDAC cell lines derived from spontaneous tumors of KrasLSL-G12D/+, Trp53LSL-R172H/+, and Pdx1-Cre (KPC), Kalbasi et al.255 demonstrated that the accumulation of tumor-derived inflammatory cytokines and chemokines, particularly a sharp increase of CCL2 compared to baseline in response to the stress of ablative radiotherapy, boosts the recruitment of Ly6C+CCR2+ monocytes/macrophages into the TME. Aided by this radiotherapy-induced macrophage trafficking, tumor cells acquire strengthened survival capacity and heightened intratumoral neovascularization, instead of T cell infiltration, ultimately giving rise to radiotherapy resistance in PDAC.255 Furthermore, the blockade of the CCL2–CCR2 axis by a neutralizing anti-CCL2 antibody significantly abrogates the recruitment and infiltration of inflammatory monocytes upon ablative radiotherapy, supporting a novel therapeutic role for targeting tumor-derived CCL2 against resistance to radiotherapy in PDAC.255 Because of the convincing evidence from many studies on various cancers, which highlight the contribution CCL expression has made to the activation of EMT programs,256–259 we put forward the hypothesis that the mechanisms underlying radiotherapy resistance involve the transition towards a mesenchymal phenotype in cancer cells with CCL2 expression. This hypothesis is in line with observed critical role of TAMs infiltration in EMT induction.More recently, based on a transgenic mice model of ovarian carcinoma, Cortés et al.260 demonstrated that the tumor-promoting functions of TAMs, as represented by chemotherapy resistance, requires full Zeb1 expression by TAMs with the release of CCL2 by the cancer cells. It is generally known that expression of ZEB1 (the well-characterized key activator of EMT) by cancer cells endows them a more aggressive phenotype, including enhanced invasive capacities, therapeutic resistance, and stemness properties, resulting in poor clinical outcomes in a variety of human cancer types261–263 (Fig. 2). Rather than simply focusing on tumor cells, it is important to understand which stromal cell types also expresses ZEB1 and how these cells perform their functions within TME.264 Cortés et al. and other researchers showed that in the context of ovarian cancer, ZEB1-expressing macrophages promote their own polarization toward a stronger protumor phenotype (F4/80low, CCR2+),265,266 and meanwhile, upregulate the expression of CCL2 and CD74 in cancer cells through an increased release of MMP9, thus resulting in phenotype switching towards a mesenchymal/stem-like state of carcinoma cells260 (Fig. 2). This forms a CCR2-MMP9-CCL2+ feedback loop between TAMs and the cancer cells, which significantly contributes to resistance to chemotherapeutic drugs (e.g., cisplatin) due to the expression of ZEB1 by both cancer and stroma cells (TAMs) (Fig. 2).260 Targeting ZEB1 in cancer cells is currently being considered in clinical trials. However, the above data suggest that effective inhibition of tumor growth and improved response to chemotherapy would also require targeting of ZEB1 in TAMs.260Similar contributions by TAMs to the resistance of chemotherapeutics (gemcitabine and 5-fluorouracil) via EMT induction have been validated in other cancer types, including pancreatic and colorectal cancers.267,268 For this reason, strategies targeting TAM or involving EMT can be hijacked and exploited for therapeutic purposes by modulating TAM function, infiltration, or activation. Collectively, it follows that phenotypic and functional switching back and forth between epithelial and mesenchymal states plays a crucial role in the resistance to immunotherapy and the establishment of an immunosuppressive TME by its effect on multiple immune cell types, perhaps in a coordinated fashion.189Indeed, given the existing evidence detailed above, it is plausible that EMT-induced resistance to different therapeutic strategies, including targeted therapy, chemotherapy, immunotherapy, and radiotherapy, in certain cases seems to coexist simultaneously despite relying on different molecular mechanisms. This raises the possibility that these distinct biological processes interrelate closely with each other, similar to the role played by TAMs, based on the inflammatory microenvironment.
-
-
Interactions between MDSCs and EMT induce resistance. Together with the positive feedback role that TAMs, growth factors (i.e., TGF-β) and chemokines (i.e., CCL18) play in the EMT programming of carcinoma cells, the recruitment of other inflammatory cells within TME and a surge of tumor-promoting soluble factors associated with inflammation, as well as the activation of key inflammatory signaling pathways, also promote the malignant behaviors of multiple cancers, especially those in relation to the EMT induction—including resistance and metastasis.55,269,270 MDSCs, a heterogeneous, immunosuppressive population of immature myeloid cells, which tends to be accumulated within TME under chronic inflammation271,272 can be used as an exemplar. These heterogeneous MDSCs, characterized by the multiplicity and complexity of their phenotypic markers, have been classified into two main categories: monocytic MDSC (mMDSC) and polymorphonuclear or granulocytic MDSC (PMN-/Gr-MDSC).273 Using RETAAD (the activated RET) transgenic mouse model of melanoma, a comparative analysis of immune infiltrates from primary and metastatic sites noted that CD11b+Gr1hiF4/80− PMN-MDSCs could be selectively recruited to and infiltrate in the primary tumor mass, where inflammatory cells are relatively plentiful, by CXCR2-CXCR2 ligand (i.e., CXCL5) interactions. This contributes to the induction of EMT and its associated tumor dissemination and therapeutic resistance.274 Additionally, in a lethal PCa model triggered by deletion of Pten and Smad4,275 a similar communication between cancer and tumor-associated inflammatory cell, that is, an elevated recruitment of CXCR2-expressing MDSCs attracted by upregulated expression of CXCL5 in the carcinoma cells, has been identified. This largely depends on Hippo-YAP signaling in a non-cell-autonomous manner.276 On the basis of such a dependency, coupled with the well-recognized cell-autonomous role of YAP1 and the involvement of CXCL5 in EMT,250,274,277 it is reasonable to hypothesize that a subpopulation of cancer cells characterized by a mesenchymal signature may be localized at invasive fronts, which would facilitate the establishment of an immunosuppressive TME through selective secretion of chemoattractants like CXCL5, thus resulting in resistance and metastasis.
In addition to the similarities detailed above, different and sometimes opposing effects of PMN-MDSCs involvement in EMT programming in multiple cancer types are worth to be acknowledged, thereby allowing the heterogeneity of phenotypes (cell-surface markers) to connect with that of functions. For example, based on syngeneic mouse models of mammary carcinoma, Ouzounova et al.278 demonstrated that a preferential, regional recruitment of PMN-MDSC to the lung facilitated the establishment of a premetastatic, inflammatory environment, which could induce, to some extent at least, DTCs to regain epithelial characteristics, particularly the fast-growing phenotype, by activating MET programs, ultimately resulting in colonization and overt pulmonary metastases in vivo.279 In contrast, the model also showed that mMDSCs infiltrated and gathered at the invasive fronts of the primary tumor, tending to play a role in the process of tumor dissemination by inducing a motile, drug-refractory, mesenchymal-like phenotype.278 Such an enhanced migration of mMDSCs to the primary tumor has been further validated by positron emission tomography imaging in a PyMT breast cancer model,279 and also observed in mice bearing platinum-resistant bladder tumors,280 implying a potential role for anti-inflammatory therapy (in this case, MDSC-targeted therapy) in increasing the susceptibility of cancer cells to antitumor drugs.
Supporting this notion, the significance and feasibility of the above therapeutic strategy has been enhanced due to the robust curative effects in a chimeric mouse model of metastatic castration-resistant PCa, which were achieved by a combination of immune checkpoint inhibitors with anti-MDSC agents (e.g., cabozantinib [Cabo] and BEZ235 [BEZ]).162 Mechanistically, the success of such synergistic responses largely depends on the Cabo- and/or BEZ-induced diminishment of intratumoral Gr-MDSCs (CD11b+Gr1+Ly6G+Ly6Clow) and reduction in secretion of MDSC-promoting cytokines (e.g., CCL5, CCL12) from carcinoma cells without impairing the function of CTLs.162 Further studies indicated that these recruited Gr-MDSCs, also known as PMN-MDSCs, were predominantly enriched in both human castration-resistant prostate cancer (CRPC) biopsies and castrated mice tumors, which could in turn release IL-23 to promote acquired resistance to androgen-deprived therapies by upregulating AR signaling in PCa cells.281 IL-23 is a heterodimeric and immunomodulatory cytokine that, when activated inappropriately in esophageal cancer (e.g., secreted by MDSCs within the TME) can result in protumor inflammatory responses and immune escape,282 during which the EMT programming is involved.283 These observations provide direct evidence to support the view that MDSC-mediated therapeutic resistance seems to be a consequence of the synergistic action of tumor and recruited host cells, including the induction of EMT in carcinoma cells and MDSC-induced immunosuppressive TME through a bidirectional, chemokine–cytokine crosstalk mechanism. However, from a more macroscopic level, in the context of drug resistance, the elementary question about the causal relationship between EMT and MDSCs, namely whether the mesenchymal signature of cancer cells can cause the recruitment of MDSCs by releasing chemoattractant chemokines, or can be caused by these recruited MDSCs through secretion of cytokines, remains unanswered. Further studies, centered on the functional and mechanistic links between EMT and MDSCs, as well as their relevance to therapeutic response, are therefore needed.
(3) Hypoxia signaling in driving EMT and resistance
The contribution made by hypoxia to cancer progression and therapeutic resistance has long been observed in a wide spectrum of cancers, since it was demonstrated by Wenger and colleagues that inactivation of hypoxia-inducible factor (HIF) could sensitize carcinoma cells to chemotherapeutic agents, including carboplatin and etoposide.284,285 The mechanistic basis for the effects of hypoxia on drug resistance is complex and varies from cancer to cancer. In general, hypoxia can impede drug sensitivity by manipulating drug efflux, cell proliferation and survival signaling pathways, DNA damage repair, metabolic reprogramming, tumor vascularization, stemness maintenance, and modification of stromal cells.286–289 Although the mechanism underlying hypoxia-mediated drug tolerance is not fully understood, the effect of EMT has attracted major attention.
In several incidences of EMT, including cancer and fibrosis, hypoxia is experienced as a dynamic stimulus in the local microenvironment under ischemic conditions.290 Therefore, it is reasonable to infer the existence of a crosstalk between hypoxia and EMT. The influences of low oxygen on cancer cells are orchestrated by HIF.291 HIF-1α is a TF, which can be degraded by prolyl hydroxylases, such as PHD2, under normoxic conditions.292 Notably, lack of oxygen can inactivate PHD, leading to the accumulation and subsequent activation of HIF-1α. Activated HIF-1α can directly bind to the hypoxia-responsive element of the promoter of several EMT-associated genes, such as TWIST1 and TGF-β, to stimulate the induction of EMT.293 Furthermore, HIF-1α can also promote EMT through mediating PI3K/Akt, Wnt, and Notch signaling pathways. For instance, HIF-1α cooperates with N1ICD as a transcriptional complex to be recruited to the Snail promoter, thus promoting SNAIL expression. In addition, Notch can potentiate the recruitment of HIF-1α to the lysyl oxidase (LOX) promoter and enhance the expression of LOX, which can further stabilize the Snail protein and induce the EMT process.294 In addition, hypoxia can also induce EMT by regulating the communication of exosomes derived from bone marrow-derived mesenchymal stem cells (MSCs) and cancer cells.295 Indeed, hypoxia confers cancer cells with cues for preserving a stem-like phenotype and bridges the linkage between EMT and drug resistance.290 For example, hypoxia in the central region of HCC considerably decreases the drug sensitivity of tumor cells through inducing EMT programs. Salidroside can promote the degradation of HIF-1α, thus inhibiting the EMT of HCC cells, leading to enhanced antitumor efficacy of platinum drugs.296 Furthermore, hypoxia can activate EMT by activating the nuclear factor-κB (NF-κB) pathway, evidenced by the observed EMT-like morphology and EMT protein markers in hypoxic lung cancer cells. 20(R)-Ginsenoside (Rg3), known as the ginseng extract, can increase the sensitivity to cisplatin in hypoxic lung cancer cells by inhibiting EMT.297
(4) EMT-induced immunosuppressive TME in drug resistance
In the context of the TME, contributions made by EMT programs to treatment resistance are reflected in two major drivers: one is the secretion of cytokines and/or chemokines derived from non-tumor cells that triggers the phenotype switching of epithelial cancer cells towards a mesenchymal state, directly resulting in drug resistance via EMT itself; occurring in parallel are alteration in the distribution and function of multiple tissue-resident cells and/or recruited immune cells within TME, which is a consequence of the EMT induction of cancer cells either directly or indirectly. This creates an immunosuppressive microenvironment upon drug exposure, ultimately giving rise to immune escape and therapy resistance.298–300 Further studies are required to address the coexistence and dependence of these two aspects, and the underlying interlinkage mechanisms by which EMT programs and immunosuppression function together to evade the lethal effect of antitumor drugs.
-
EMT and PD-L1 expression. The positive correlation between EMT programs and the expression of PD-L1 (also called as B7-H1 or CD274), a ligand binding to the immune receptor PD-1 (also known as PDCD1), widely occurs in healthy tissue cells, antigen-presenting cells and a variety of tumor cells for escaping antitumor immune responses. It serves as a “bridge” to connect EMT programs and immunosuppression.153,301,302 For example, based on the EMT signature in lung cancer previously mentioned,148 an analysis of patient-derived, pan-cancer EMT signature reveals that tumors of mesenchymal status exhibit significant enrichment of multiple immune checkpoints, especially PD-L1, which may act as novel biological targets or therapeutic vulnerabilities in mesenchymal-like tumors.150 Even before this was realized, studies had built a large body of credible evidence for the contributions made by PD-1/PD-L1 inhibitory pathway to immune escape. This was supported by the following facts: inhibition of CD8+ CTL proliferation and function, and enhanced production of Foxp3+ Tregs from CD4+ T cells, resulting in peripheral immune tolerance303–306 of patients in several types of carcinomas, including NSCLC,307,308 lung squamous cell carcinoma,303 liver cancer,309 and myeloproliferative neoplasms.310 This functional connectivity suggests that the expression of PD-L1 in cancer cells, in some sense at least, may play a role as an indicator since the levels of immunosuppression rise during cancer progression. This has led to intensive discussions about the relationship between EMT programs and immunosuppression, with a particular emphasis on PD-L1.
Subsequently, following a succession of recent studies, a similar positive relationship between EMT programs and PD-L1 expression has been further validated in multiple cancer types, including lung,153,301,302 breast,188,311 and head and neck cancers.312 Furthermore, this relationship has the potential to perform in a bidirectional regulating and controlling mode. In general terms, the induction of EMT can cause significant upregulation of PD-L1. In turn, several studies have shown that the activation of PD-L1 signaling is also crucial for maintaining the characteristic manifestation of malignant tumors with aggressive clinical features, in particular EMT programming,311,313,314 immune escape,307,308 and stem cell properties.315,316 Consequently, higher expression of PD-L1 has been connected to worse prognoses in multiple types of carcinomas, including esophageal cancer,317 renal cell carcinoma,318 gastric carcinoma,319 ovarian cancer,320 and melanoma.321
Reinforcing this concept, a raft of studies have suggested that the miR-200/ZEB1 axis, a well-understood double-negative feedback loop that governs the reversible phenotypic transformation of cancer cells between epithelial and mesenchymal states,322–325 participates in multiple processes of auxiliary cellular functions associated with cancer progression, including immune escape,154,260,326,327 drug resistance,260,328,329 stem cell properties,330 and endothelial trans-differentiation.331,332 It is especially interesting that under most conditions, the target genes for these functionally overlapping biologic processes are more likely to function in a non-overlapping mode, which means the EMT-related miR-200/ZEB1 axis is endowed with an incredible potential for regulating pleiotropic downstream targets specialized in handling various cellular functions.154,333–335 With increasing in-depth studies, the list of target genes is constantly being expanded. In line with this, PD-L1 has been identified as a direct downstream target of the miR-200/ZEB1 axis in NSCLC cells by Chen et al.,154 resulting in a diminished antitumorigenic immune response due to CD8+ T cell (PD-1+ TIM-3+) exhaustion and a reduced number of CD8+ TILs.326 Similar mechanisms have been further validated by various studies on breast cancer and NSCLC.327,336 In short, these observations provide direct experimental evidence that EMT induction and PD-L1-mediated immunosuppressive TME are closely associated via miR-200/ZEB.154 In addition, a retrospective study on colon cancer at the histological level has shown that the regulation of miR-200/ZEB axis, manifesting itself as upregulated expression of ZEB and downregulated expression of miR-200, occurs preferentially in regions of tumor budding at the invasive front where EMT is frequently accompanied by significantly elevated expression of PD-L1, rather than at the tumor center.337,338 This spatial overlap can also be interpreted as a consequence of the close interactions between EMT programming and PD-L1 expression in cancer cells. From a macroscopic perspective, it may explain the observations that carcinoma cells at the invasive front, supposedly more vulnerable to autoimmune attack because of direct exposure to immunocytes, exhibit improved survival and a capability of avoiding host immune surveillance.
These findings have gained further support from another study, which indicated the activation of PD-L1 transcription can be driven by Mucin 1-C (MUC1-C)-induced formation of transcription initiation complexes with NF-κB p65 on the PD-L1 promoter.339 These MUC1-C–NF-κB p65 complexes are also recruited to occupy the promoter of other NF-κB target genes, comprising MUC1-C and ZEB1, among which the former results in an auto-inductive circuit, while the latter leads to the repression of miR-200c expression; in turn, the transcriptional activation of ZEB1 in NSCLC cells triggers EMT programming through the negative feedback regulation of the ZEB1/miR-200c axis.65,339–342 Echoing the previous finding that PD-L1 acts as a target of miR-200, it seems quite logical and reasonable to speculate that the MUC1-C functioning in conjunction with NF-κB p65 drives the induction of EMT and upregulates PD-L1 gene expression at both the transcriptional and post-transcriptional levels via a ZEB1/miR-200-dependent mechanism. This raises the possibility that MUC1-C may act as an upstream regulator coordinating these two responses.154,339 As a consequence of this coordination, researchers have hypothesized that patients with mesenchymal tumors are more likely to benefit from immunotherapy, particularly for anti-PD-L1 or PD-1-neutralizing antibodies against PD-L1 overexpression.122 The possible role of PD-L1 as a prognostic factor in patients undergoing PD-1/PD-L1 inhibitor treatment is still under investigation and needs further studies and testing in clinical practice. As far as the current research findings are concerned, it may be concluded that patients whose tumors show high expression of PD-L1 (mesenchymal signature) may experience improved clinical outcomes with anti-PD-1/PD-L1 treatment. However, this is not always the case. Some patients with PD-L1-negative cancer (epithelial signature), across a wide variety of human cancer types, also show robust responses to PD-1/PD-L1 antibodies (e.g., nivolumab), suggesting that PD-L1 overexpression in tumor tissues is neither a sufficient nor necessary condition for guaranteeing improved clinical benefits (survival time) for patients.343–349
A change of approach to predict patient response to immunotherapy may therefore be required. Emerging evidence shows that exosomes secreted by tumor cells have bioactive PD-L1 on their surface, which can suppress the immune response.350–354 Based on the clinical data of patients with melanoma or NSCLC,351,355 exosome-derived PD-L1 in response to treatment with anti-PD-1/PD-L1 antibodies (e.g., nivolumab and pembrolizumab) may have the potential to predict the clinical outcomes of anti-PD-1 therapy, or even to become a novel therapeutic target, in spite of the ambiguity of the relationship between EMT and PD-L1 levels within exosomes.
Besides playing an essential role in immunosuppression and a possible role in resistance to immunotherapy as discussed previously,356 it is interesting to note that the “bidirectional regulation between EMT and PD-L1” is also involved in resistance to targeted therapy357 as well as chemotherapy, linking it to functions beyond its immunoregulation activities. Researchers have demonstrated that upregulation of PD-L1 mediated by YAP at the transcript level leads to the acquired resistance to EGFR-TKIs (e.g., gefitinib) in NSCLC, a process that largely depends on the induction of EMT.357–360 Similar findings have also emerged for malignant pleural mesothelioma, an extremely aggressive cancer originating from membrane covering the lungs and the inner side of the ribs.361–363 Besides making important contributions for the acquired resistance to gefitinib (a first-generation TKI), elevated PD-L1 expression also exerts a positive effect on the acquisition of intrinsic resistance to EGFR-TKIs in EGFR-mutant NSCLC by inducing an EMT phenotype, which might rely, at least partially, on the activation of the TGF-β/Smad canonical signaling pathway.364–367 These observations, together with the aforementioned bidirectional regulation between EMT and PD-L1, suggest that phenotypic switching between epithelial and mesenchymal states in carcinoma cells is usually accompanied by a dynamic transcriptional fluctuation of PD-L1 at the single-cell level.16 This enables PD-L1-expressing cells to survive following exposure to molecularly targeted agents. More importantly, activation of the EGFR pathway in turn induces upregulation of PD-L1 in EGFR-mutant NSCLC cells by alternative mechanisms, including the IL-6/Janus kinase (JAK)/signal transducer and activator of transcription 3 (STAT3)368 and ERK1/2/c-Jun307 signal transduction pathways. Similar instances of EGFR-driven PD-L1 expression have been observed in malignant melanoma cells (A875 and A375)321 as well as salivary adenoid cystic carcinoma (SACC) cells (SACC-83 and HN13) that underwent EMT,369 although the underlying molecular mechanisms vary from cancer to cancer.308,321,368,369 In addition to promoting PD-L1 expression, through a combined investigation of human glioblastoma (GBM) specimens and cell lines, EGFR signaling has also been characterized by its ability to maintain the stability of PD-L1 protein in a COP9 signalosome 6-dependent manner in which EMT programs are presumably involved.370–372 Indeed, the knowledge that activated EGFR plays an important role in adjusting PD-L1, either by promoting the expression or maintaining the stability of PD‐L1 protein, suggests that PD-L1 is less like a “bystander,” but rather a key participant in promoting biological functions of EGFR signaling. This would explain why the upregulated PD-L1 has been shown to more frequently occur in EGFR-TKI-resistant carcinoma cells.365,367,373,374 From the above studies, it can be reasonably assumed that the two therapies—targeted therapy (EGFR-TKIs, BRAF inhibitors, etc.) and immunotherapy (PD-1/PD-L1 or CTLA-4 blockade), when combined, could be a potential approach for improving outcomes for patients with resistance to either therapy alone. However, investigations on the clinical feasibility of this combined strategy remain at the initial stage due to the high frequency of adverse reactions, represented by immune-related adverse events and even interstitial pneumonitis in NSCLC patients,375–378 as well as hepatotoxicity in patients with melanoma.379
Given the +nd on the mutual interplay between EMT programs and PD-L1 in certain types of human cancer.357–363 Such a dependency, consistent with the observations on resistance to targeted therapy, has been further confirmed from the acquisition of resistance to various cytotoxic chemotherapies. For instance, in HCC cells, transcriptional upregulation of PD-L1 mediated by Y-box binding protein-1, a multifunctional transcription/translation factor shown to be important in the regulation of multidrug resistance gene (MDR1) and EMT inducers (Zeb1, Snail1, Twist, etc.) by being recruited and binding to their promoter regions or mRNAs,380–384 results in an immunosuppressive TME (decreased CD8+ T cells and increased Tregs) and MDR1 overexpression, as well as simultaneous EMT induction.385 Because of the extensive functional links among these biological processes, it is most likely that the acquisition of resistance in HCC cells to multiple chemotherapeutic drugs—particularly doxorubicin (DOX), is a consequence of joint, but not isolated, effects of the three aspects mentioned above.385 These results are consistent with previous research reported by Li et al.,386 using a comparative analysis between chemo-resistant and chemo-sensitive breast cancer cell lines and patient samples, demonstrated that high levels of PD-L1, together with low levels of miR-3609, are crucial for a heightened resistance to DOX and poor patient prognosis. In addition, another study that used the CRISPR/Cas9 system to probe the role of PD-L1 in the acquisition of chemoresistance showed that knockout of PD-L1 in osteosarcoma cells can significantly lower their resistance to DOX, and paclitaxel,387 further supporting the aforementioned studies.385,386 In addition to being involved in DOX resistance, the contributions to chemotherapy resistance made by reciprocal and complementary interactions between EMT and PD-L1 have been further validated in cisplatin (DDP) resistance of NSCLC cells. A possible mechanism in this study was attributed to the activation of the JAK/STAT3 pathway in an ataxia telangiectasia mutated-dependent manner,388 largely in agreement with the previous reports.368,389 Collectively, these observations provide strong evidence that the tight interactions between EMT programming and PD-L1 expression contribute critically to the development of resistance to either chemotherapeutics or targeted drugs, with or without the involvement of EMT/PD-L1-related immune escape.
-
EMT and other immune checkpoints. As discussed above, immunotherapy based on immune checkpoint blockers has indeed revolutionized the therapeutic strategies for cancer; however, this treatment is still in its infancy owing to a lack of understanding regarding the interactions between tumor and host cells within TME, especially immune cells, during immunotherapy. Among the complicated dynamic, interactions between cancer cells and immune modulators, particular attention here should be paid to the induction of EMT because of its strong relevance to the expression of immune checkpoints, as well as their regulators or ligands like the aforementioned PD-L1. This can provide insight into the potential value of hijacking EMT signatures for therapeutic purposes, such as for improving response to immunotherapy.390,391 Supporting this concept, it has been widely reported that mesenchymal tumors are more refractory to immunotherapy, indicating that targeting the EMT process in combination with immunotherapy may hold great potential for improving the clinical outcome of immunotherapeutic strategies.52 In parallel, another study showed that Snail-expressing breast cancer cells exhibited reduced susceptibility to CTLs-mediated lysis.392
Indeed, current observations suggest that the biological effects exerted by EMT on different immune checkpoints prefer to coordinate with each other, jointly promoting the establishment of an immunosuppressive TME.391 This EMT-mediated immune escape has been regarded as a major driver of resistance to multiple cancer therapies, including chemotherapy, targeted therapy, and immunotherapy. From this perspective, in order to overcome intratumoral immunosuppression, developing an in-depth knowledge of exactly how other immune checkpoints (i.e., excluding PD-L1) are regulated by EMT is of great importance and urgently required. To address this, Noman et al.393 have found that EMT-associated TFs (i.e., ZEB1 and Snail) play essential roles in regulating the expression of CD47 in human mammary cancer cells. Similar to the influence on PD-L1, Snail1/Zeb1 can directly bind to the E-box motif of the CD47 promoter and thus induce their expression, which is closely linked to poor outcomes for patients with TNBC.327,393,394 In addition to CD47, the expression of CTLA-4, B and T lymphocyte attenuator, B7-H3 (CD276) and T cell immunoglobulin, and mucin protein-3 (TIM-3), along with the ratio of tumor immunosuppressive CD4+Foxp3+ Treg cells, are positively related to EMT phenotype in lung adenocarcinomas.153 In addition, a highly positive correlation has also been identified between EMT and newly emerging immune checkpoint-associated genes, including CD276, TGFB1, and OX40 in kidney cancer.395,396 However, the molecular basis of EMT-mediated dysregulation of these checkpoint genes warrants further elucidation to provide insights for the use of EMT signatures as predictive biomarkers for ICB compounds and other immunotherapies in a broad range of cancers. Despite the prominent links between EMT and immunosuppression, the mechanism underlying EMT-mediated dysregulated immune checkpoint-associated genes still remains largely unknown and further investigations are urgently needed.
The role of EMT in resistance to EGFR-TKIs
In the previous section, the means by which PD-L1 expression on EMT programs contributes to the acquisition of resistance during EGFR-TKIs treatment has been extensively reviewed. We now explore the mechanics of EMT induction during the development of drug resistance for three generations of EGFR-TKIs, focusing on the tumor cells per se.
Initially, based on extensive experimental evidence from studies on EGFR-mutant NSCLCs, the features of EMT represented by the downregulation of E-cadherin, together with overexpression of vimentin and/or EMT-TFs Snail1/2, were observed in the course of overriding cytostatic effects of EGFR inhibition first-/second-generation EGFR-TKIs (such as gefitinib, erlotinib, and afatinib) or antagonistic monoclonal antibodies, prior to the development of genetic resistance mechanisms.47–52 The Hippo pathway, involved in tumor suppressor signaling, plays a critical role in the regulation of development, cell fate (e.g., cell senescence, proliferation, and apoptosis), and organ size, mainly by repressing the oncogenic TFs YAP and its paralog transcription co-activator with PDZ binding motif (TAZ).397–401 Previous studies have delineated the role of YAP/TAZ in the acquisition of drug resistance and the promotion of EMT programming in multiple cancers, including PDAC,249 melanoma,402,403 neuroblastomas,404 PCa,405 HCC,406 and lung cancer (ALK-rearranged,407 KRAS-mutant,250 and EGFR-mutant NSCLC.360) In EGFR-mutant NSCLC, more specifically, upregulation of TEAD-mediated YAP promotes the transcription of the EMT‑TF Snail2 (Slug), but not Snail1, Twist, or Zeb1, which further induces the upregulation of the membrane-protein anexelekto (AXL) receptor tyrosine kinase in NSCLC cells. In this case at least, AXL acts as a downstream target of YAP148,360,408–412 (Fig. 3b). AXL signaling, whose activation relies on interactions with its specific ligand—growth arrest-specific protein 6 (GAS6), has been proven to be inextricably linked to the acquisition of the classic EMT-related signature,413–415 notably vimentin411 and E-cadherin416,417 (Fig. 3b). In this context, activated AXL directly drives Slug-overexpressing mesenchymal cells to acquire resistance with erlotinib in the absence of secondary mutations, such as the EGFR p.Thr790Met (T790M) alteration or MET activation408,418,419 (Fig. 3b). On the other hand, Slug overexpression can also antagonize p53-mediated apoptosis by repressing the transcription of proapoptotic effector PUMA, further promoting mesenchymal cell stemness and resistance to both targeted therapy (e.g., erlotinib) and chemotherapy (e.g., cisplatin).420–422 As expected, using the EGFR inhibitor (EGFRi) erlotinib in combination with the AXL inhibitor (SGI-7079/XL-880) or YAP1 inhibitor verteporfin can significantly diminish the expression of EMT-related markers and successfully restore the sensitivity to erlotinib of NSCLC cells with a mesenchymal signature, both in drug-resistant cell lines as well as in mouse xenograft models.148,408–410
Taken together, these results reveal that blockade of the GAS6/AXL pathway is sufficient to increase erlotinib susceptibility by reversing the EMT process, suggesting potential evidence for the treatment of advanced NSCLCs. Accordingly, the development of AXL inhibitors has already gained substantial interest from both academia and pharmaceutical companies. BGB324, known as a first-in-class AXL-selective small-molecule inhibitor, was developed by BergenBio and entered phase I clinical trials in 2013.423 Subsequently, a phase I/II clinical and pharmacokinetic study of BGB324 confirmed its safety and efficacy in patients with advanced NSCLC, a number of whom had sustained responses for at least 6 months without disease progression.424 A review of current strategies that target AXL, including application of small-molecule inhibitors, anti-AXL monoclonal antibody (20G7-D9),425 and high-affinity AXL decoy-receptor (MYD1-72),426,427 shows the multitarget AXL inhibitors (such as SGI-7079 and sunitinib) as combination partners have achieved the best curative effect with respect to drug resistance according to available preclinical and clinical evidence.148,415,428,429
In addition, treatment with the third-generation, irreversible EGFR-TKIs osimertinib (AZD9291), which has successfully doubled the median progression-free survival compared to that of the first-generation drugs, has been approved for the treatment of late malignant EGFR-mutant NSCLC and recommended as the preferred first-line therapy for those patients.430,431 Importantly, osimertinib, designed to target activated EGFR- and T790M-resistant mutations instead of wild-type EGFR, also remains the preferred second-line therapy for patients with advanced first-/second-generation TKI-resistant NSCLC.432–434 However, despite an initial obvious curative effect initially, patients usually suffer tumor recurrences within 1 to 2 years treatment with osimertinib due to acquired resistance.357 Prior studies based on clinical or preclinical models have revealed several mechanisms of acquired resistance to osimertinib, such as the secondary mutation (EGFR C797S),7,435,436 bypass pathway activation (MET and HER2 amplifications),437,438 and an increased dependence on RAS signaling.439 These resistance mechanisms are mostly caused by genetic alterations, but non-genetic resistance mechanisms also exist, such as EMT induction, which is also necessary and sufficient to develop acquired resistance.440 Interestingly, consistent with the above mechanism, the GAS6/AXL axis plays a vital role both in the development of de novo resistance to osimertinib and also in the initiation and maintenance of a “persistent” state from EMT induction with or without re-activation of HER3 and EGFR.440,441 Therefore, the combination of the AXL inhibitor (Cabo or NPS1034) or the AXL degrader (Yuanhuadine, YD) with osimertinib is expected to be an effective therapeutic strategy for delaying or overcoming resistance in EGFR-mutant NSCLC, as seen in both in vitro and in vivo experimental models440–443 (Fig. 3b). Moreover, by the establishment of gefitinib- and osimertinib (AZD9291)-resistant NSCLC cell lines,7,444 representing first- or third-generation EGFR-TKIs, respectively, a recent study has demonstrated that the EMT induction tends to be a general phenomenon during the formation of acquired resistance in both situations when compared to parental cell lines.445 Although the underlying mechanisms of the above two cases are different, one depending on the activation of Src/Hakai, while the other depends on the upregulation of Zeb1, the mesenchymal, drug-refractory phenotype of gefitinib- and osimertinib-resistant NSCLC cells can be reversed via the simultaneous double inhibition of histone deacetylase (HDAC) and 3-hydroxy-3-methylglutaryl coenzyme A reductase using JMF3086.445 Based on these observations, Raoof et al.446 used mesenchymal cell lines derived from tissue samples of NSCLC patients suffering from TKI-resistant tumor, which simulated EMT-like phenotype switching. Using whole-genome screening, these mesenchymal cells showed increased resistance to the third-generation EGFR-TKIs (EGF816) in vitro accompanied by a significant activation of the fibroblast growth factor receptor 1 (FGFR1) pathway, implicating the potential of FGFR1 as a druggable target for evading resistance to EGF816446 (Fig. 3b). As expected, the novel use of EGF816 in combination with FGFR inhibitor BGJ398 can significantly inhibit the survival of mesenchymal cells and the development of full resistance in EGFR-mutant NSCLC446 (Fig. 3b). Collectively, from the aforementioned observations, as well as previous findings that the tumors undergo drug-induced phenotype switching while maintaining their primary mutations in EGFR, one can conclude that tumor cell plasticity-induced EMT is essential for acquired resistance upon multiple TKIs treatment, during which there are no genetic alterations.
Prevention of EGFR pathway re-activation by pharmacologically inhibiting downstream pathway components, such as RAF, MEK, or ERK, is another common strategy that has been implemented to delay resistance.447,448 Studies have indicated that, in comparison to treatment with the single-agent EGFRi (WZ4002), combination therapy with both WZ4002 and an MEK inhibitor (MEKi) (tretinamib/selumetinib) can significantly slow down resistance in EGFR-mutant NSCLC.447 However, even with this combined therapy, acquired resistance ultimately occurs.447,449 Kurppa et al.450 observed that a subpopulation of cancer cells can enter a senescence-like, dormant state to escape cell death upon the administration of EGFRi (osimertinib) in combination with MEKi (tretinamib), leading to resistance (Fig. 2b). This change is regarded as a highly reversible process characterized by YAP/TEAD-mediated activation of EMT or MET programs.451 As expected, the therapeutic strategy of pharmacologically cotargeting YAP/TEAD (by MYF-01-37) and EGFR/MEK leads to synthetic lethality, the realization of which predominantly depends on the phenotypic switching from the dormant/senescence, mesenchymal-like, EGFRi/MEKi-refractory state to a proliferative, epithelial-like, EGFRi/MEKi-susceptible state of EGFR-mutant NSCLC450,451 (Fig. 3b). Analogously, in the context of resistance to KRAS suppression, Shao et al.250 also found that increased activity of YAP helps bypass loss of KRAS signaling, which at least in part depends on EMT programming. Taken together, this phenotype switching between actively proliferative and senescence-like dormant state may reflect one particular manifestation of EMT programs under drug exposure, which acts in a YAP-dependent manner and serves as an adaptation mechanism against loss of oncogene (EGFR and KRAS) signaling.250,450,451
EMT-targeted compounds in clinical trials
As apparent from the above discussion, targeting the EMT process or cell plasticity holds great potential for circumventing drug resistance. However, only a few compounds specifically designed to inhibit the EMT process are currently in clinical trial. Specifically, AB-16B5 is an antibody which directly against secreted clusterin, a stimulator of EMT programming and subsequent cancer progression.297,452 AB-16B5 is now being evaluated in a phase I clinical trial in advanced solid malignancy. Moreover, AB-16B5 combined with docetaxel is being evaluated in subjects with metastatic NSCLC (Clinicaltrials.gov identifier NCT04364620). Inhibitors targeting Notch, TGF-β, and Wnt signaling pathways are also promising candidates for suppressing EMT and cell plasticity. For example, PF-03446962 and galunisertib are antagonists designed to inhibit the TGF-β receptor and are currently in phase I clinical studies on solid cancers (Clinicaltrials.gov identifier NCT00557856, NCT02423343). Both PF-03446962 and galunisertib can inhibit the EMT programs and thus impede cancer development.453,454 In addition, Wnt inhibitors such as ETC-1922159 and OMP-54F28 have been reported to inhibit the EMT programs and are now in phase I clinical trial in solid cancers (Clinicaltrials.gov identifier NCT02521844, NCT01608867). ETC-1922159 can inhibit the secretion of Wnt ligands, while OMP-54F28 is a recombinant protein that directly binds to Wnt ligands, and thus blocks Wnt signaling effectively and its resulting cancer progression, such as those related to EMT programs.
Transition between non-CSC and CSC states
Definition and characteristics of CSCs
The emergence of CSCs relies on the existence of rare, immature subpopulations of tumorigenic cells within solid tumors or hematological malignancies that display the potential of indefinite proliferation and clonal long-term repopulation, along with the capability of self-renewal and differentiation (the defining features of a CSC), which contributes to tumor initiation and heterogeneity.455–458 The features summarized above are responsible for a number of clinical observations with regard to CSCs, including the frequent tumor recurrence after initially effective therapies, the emergence of a tumor dormancy state, and distant metastasis.458–460 It is known that CSCs share considerable commonalities with adult stem cells, such as identical surface markers (ALDH1, CD133, and CD44), re-activation of development-related pathways, and lack of differentiation.461 It is worth mentioning here that, similar to the normal stem cell that can give rise to a new stem cell with a committed progenitor, CSCs possess the ability of asymmetrical mitosis, a process regulated by multiple intricate mechanisms, yielding one daughter cell that remains as a CSC to sustain its self-renewal potential, and a progenitor or committed cell (transient amplifying [TA] cell), which is equipped with a high proliferative capability committed to differentiating into non-CSCs, constituting the bulk of the tumor.455,461,462 In addition to asymmetric division, symmetric division, which is characterized by the fact that daughter cells derived from a CSC appear to be either two CSCs (symmetric renewal) or two stem-committed cells (symmetric commitment), can also exist simultaneously.463,464 In light of these two modes of division, it is important to note that TA cells generated from asymmetric division, under certain circumstances (e.g., anti-CSCs therapy, or CSC loss), are more inclined towards dedifferentiation rather than differentiation, which allows their re-acquisition of a stem cell-like phenotype, serving to “replenish” the CSC pool on an inexhaustible basis.464,465 Indeed, such a functional plasticity of TA cells between “forming a tumor” and “recharging a CSC pool” is essentially a bidirectional and hierarchical plasticity of CSC regulation endowed by asymmetric division, through which a dynamic equilibrium between CSCs and non-CSCs (i.e., dedifferentiation and differentiation, or dormancy and proliferation) can be maintained.464,465 Intriguingly, two biological events both relevant to cellular plasticity (the EMT programs mentioned above and CSC division) are not in parallel but in tandem; or more specifically, EMT can regulate the process of CSC division in a desired orientation depending on certain scenarios. For example, several lines of evidence show that some EMT-TFs (i.e., Twist2 and Snail) are able to elicit robust regulation of stemness traits in lung CSCs by augmenting symmetric division, yet repressing asymmetric division.466 Conversely, in another recent study, EMT seems to play a role in maintaining and enhancing a stem cell-like phenotype by directing breast CSCs towards asymmetric division.467 Together, these conflicting findings regarding the links between EMT programs and CSC divisional profiling could induce a pattern of plasticity adjustment in specific types of cancer, where CSCs tend towards either symmetric or asymmetric division with EMT-TF involvement. This is a manifestation of cell fate plasticity, leading to alterations on the proportion and composition of different phenotypic subtypes within an individual tumor.
Although it is difficult to distinguish CSCs from non-CSCs, particularly special cancer types with a relative shallow hierarchy, identification of CSCs with various frequencies has been obtained using combinations of multiple cell-surface antigens in a variety of carcinomas, including leukemia,468 breast cancer,469,470 PCa,471,472 colorectal cancer,473,474 melanomas,475 and brain cancer.476 Accumulating reports also reveal that certain cancer cells can exhibit plasticity via a reversible transitioning between the CSC and non-CSC state, which repopulates the CSC pool and enables the cells to survive therapy.49,390,477 Specifically, in comparison to the non-CSC state, CSCs acquire resistance via several alternate mechanisms, including the upregulation of multidrug efflux pump, elevated DNA-repair capacity, improved adaption to reactive oxygen species, maintenance of a slow-cycling state, and higher trans-differentiation potential.478,479 All the above-mentioned molecular mechanisms, except for trans-differentiation, have been elegantly discussed in recent reviews.458,459,480 We will therefore focus on CSC trans-differentiation (another type of cell plasticity) and the latest observations on potential pharmacological intervention.
The role of CSCs trans-differentiation in drug resistance
Lineage plasticity, also known as trans-differentiation or lineage switching, is the process by which cells acquire phenotypic characteristics of another cell lineage, occurring during the process of physiological regeneration of damaged tissue.481,482 In the context of cancer therapy, tumor cells can transdifferentiate from a cell type dependent on the drug target to a specialized cell type that is not.483,484 For example, using varying types of conditioned medium, melanoma spheroid cells (CSCs) can transdifferentiate into multiple cell lineages, such as melanocytes, adipocytes, chondrocytes, or osteocytes.485 It has been reported that a similar trans-differentiation process could be induced by treating melanoma CSCs with unsaturated fatty acids, or by upregulation of peroxisomal proliferator receptor-γ (PPARγ).486,487 In line with these observations for melanoma, PPARγ agonists, as represented by the antidiabetic drug, rosiglitazone,488 have also been found to induce cellular re-differentiation in a variety of malignancies, including myxoid liposarcomas,489,490 GBM,491 breast cancer,492 and chronic myeloid leukemia.493
Interestingly, on the basis of a well-established adipogenesis induction protocol,494,495 the recent research advances achieved by Ishay-Ronen et al. have provided strong evidence, indicating that the plasticity of carcinoma cells, in this case mesenchymal-like breast cancer cells (e.g., MT▵Ecad496 and Py2T-LT cells,496) can be hijacked and exploited for therapeutic purposes by forcing their trans-differentiation process towards postmitotic and functional adipocytes both in vitro and in vivo, rather than by killing directly497–499 (Fig. 4). Of note, this unpredicted trans-differentiation—from cancer to fat, only occurs in cell lines with mesenchymal features rather than those with epithelial features (e.g., MTflECad500 and Py2T cells.496) This was confirmed497 using specific markers or stains for different stages during adipogenesis, such as C/EBPα (CCAAT/enhancer-binding protein α) for preadipocytes,501 and Nile Red for lipid droplets.502 Such findings, to a certain degree, mirror the results from previous studies, which have concluded that EMT-derived cells, akin to MSCs, are equipped with the potential capacity of multilineage trans-differentiation, particularly the three mesodermal lineages: osteoblasts, chondrocytes, and adipocytes.503–506 This supports the notion that carcinoma cells must meet the explicit precondition of achieving a high plasticity level, and then sustaining its superiority, prior to trans-differentiation being implemented endogenously or exogenously. To put this in context, the superiority of stronger plasticity of tumor cells with mesenchymal attributes (e.g., those exhibiting partial/intermediate EMT programs) is not embodied in their increased capability for invading the surrounding stroma, but in their enormous potential for trans- and re-differentiation due to the mechanistic connection and functional overlap between EMT process and the CSC phenotype55,497–499,503,507 (Fig. 4). Further analysis has found that TGF-β—key promoter and sustainer of EMT programming,165,166 which is also known for its negative role in adipocyte development508,509 represses the adipogenic trans-differentiation of EMT-derived breast cancer cells by activating non-canonical MEK/ERK signaling pathways.497–499 As predicted, the combination therapy of an MEKi (trametinib) with an adipogenesis inducer (rosiglitazone), termed adipogenesis therapy or trans-differentiation therapy, strongly promotes the direct lineage conversion of those tumor cells that transdifferentiate from an “aggressive” mesenchymal phenotype to “peaceful” adipocytes in a PDX model497–499 (Fig. 4). This promotion, however, has proven to be strictly limited to cancer cells with strengthened plasticity, more specifically to those at the invasive front of the primary tumor, which is hypothesized to be the region where EMT most frequently occurs77 (Fig. 4). This so-called “limitation” of adipogenesis therapy, manifesting itself as spatial and functional specificity, is also a “benefit” precisely because those adipogenesis therapy-targeted cells are intrinsically more refractory to existing therapeutic approaches due to the closely mechanistic link between EMT, CSCs, and drug resistance (as mentioned earlier in this review and discussed comprehensively by Shibue and Weinberg.55) (Fig. 4). This suggests the possibility of avoiding treatment failures by trans-differentiation therapy alone or in combination with multiple conventional therapies, including chemotherapy, targeted therapy, radiotherapy, or immunotherapy497–499 (Fig. 4). The combination of conventional therapy and trans-differentiation therapy likened to the proverbial “killing two birds with one stone” where the conventional therapies efficiently kill the proliferative cancer cells that form the bulk of the tumor,510,511 while trans-differentiation therapy497 eradicates invasive cells in areas of tumor budding that escape conventional therapies by the development of a dedifferentiated EMT/CSC phenotype497–499 (Fig. 4). However, this promising trans-differentiation-based strategy still needs to be further validated, refined, and extended from current preclinical proof-of-concept trials to a proven clinical demonstration of successful breast cancer treatment.
Fig. 4.
EMT can be hijacked for therapeutic purposes by forcing trans-differentiation. EMT frequently occurs at the invasive front of the individual tumor, which also allows cancer cells to achieve a high plasticity level due to the mechanistic correlation and functional overlap between the EMT process and the CSC phenotype. The mesenchymal characteristics of those tumor cells are embodied in their potential for re-differentiation and possibly even trans-differentiation. Meanwhile, cancer cells also achieve resistance to a variety of conventional therapeutics during the EMT process, commonly resulting in tumor recurrence. However, the plasticity of those cancer cells can be utilized for therapeutic purposes by forcing their trans-differentiation process towards postmitotic and well-differentiated phenotypes rather than by direct killing. The treatment of an MEK inhibitor—trametinib—together with an adipogenesis inducer—rosiglitazone—can strongly promote the direct lineage conversion of those aggressive cancer cells to “peaceful” adipocytes. This provides the potential of preventing treatment failure by combining trans-differentiation therapy with multiple conventional therapies, efficiently killing the proliferative cancer cells that form the bulk of the tumor as well as eradicating invasive cells that escape conventional therapies by the development of an EMT/CSC phenotype. EMT epithelial–mesenchymal transition, CSC cancer stem cell
In the clinical setting, it is worth contemplating all-trans retinoic acid (ATRA)-based regimens for treating acute promyelocytic leukemia (APL), during which accumulating abnormal promyelocytes break the differentiation block by both a change in configuration as well as degradation of the PML-RARα (promyelocytic leukemia-retinoic acid receptor α), the oncogenic fusion protein in APL pathogenesis, leading to mature granulocytes.512–516 This has significant clinical benefits with ~85% CR.512–514 Although by definition there are differences between ATRA-based differentiation therapy and adipogenesis-based trans-differentiation therapy,484,517,518 it is undeniable that APL cells share a great deal in common with EMT-derived carcinoma cells. Specifically, high plasticity and stem cell-like (re-)differentiation capability either empower tumor cells themselves with a spontaneous drug-refractory phenotype or render them vulnerable to a terminal differentiation state through appropriate pharmacological interventions. However, in current practice, progressive resistance to monotherapy with ATRA emerges over a short period, typically within 3–6 months.512–514 Adipogenesis therapy could be the beginning of novel trans-differentiation-based therapies yet to be developed.
The above examples would suggest that the activation of cancer cell plasticity by driving CSCs (trans-)differentiation could be a promising therapeutic approach for overcoming drug resistance. However, “opportunities” frequently come with “risks”. On the one hand, considering the close correlation between adipose tissue and breast cancer, the adipocyte-rich TME resulting from trans-differentiation therapy has an increased risk of further supporting the growth and metastasis of residual cancer cells, functioning as so-called cancer-associated adipocytes.519,520 On the other hand, trans-differentiation, particularly neuroendocrine trans-differentiation, has frequently been linked to disturbing side effects, including aggressiveness and resistance,54,521 warning that hijacking such a process for therapeutic purposes will not be without risk. Some examples are given below.
Neuroendocrine trans-differentiation from CRPC to NEPC
Although trans-differentiation of CSCs may offer a novel therapeutic avenue, drug-induced neuroendocrine trans-differentiation observed in PCa and NSCLC shows evidence to the contrary.54,521 PCa can be taken as a convincing example. PCa, a hormone-dependent cancer, is characterized by the high dependence of AR-related signaling for tumor growth and survival during the early stages.522 Treatment advantage can be taken made of this dependence. Current first-line therapy, based on ADTs and targeting the AR itself, has already been proven to be effective clinically for the prevention of PCa growth523 (Fig. 5a). Unfortunately, this effect is often limited, ranging from months to a few years.524,525 This is mainly due to the potential of PCa cells to adjust to ADTs, reflected in the re-activation of AR-mediated signaling through various mechanisms, including functional residual androgens, genomic amplification of the AR locus, AR ligand-binding domain mutations, and AR splice variants.54,524,525 All these result in the same outcome: a more aggressive form of PCa known as CRPC526 (Fig. 5).
Fig. 5.
Trans-differentiation from castration-sensitive PCa to CRPC to NEPC: involvement of two generations of AR pathway-targeted agent. a ADTs mediate CRPC generation in an AR-independent manner, while ARPIs trigger NEPC formation dependent on AR signaling. ADTs blocking AR-related signaling, which exhibit remarkable activity causing tumor regression, lead to the emergence of aggressive CRPC with tumor recurrence. Likewise, the novel ARPIs by targeting specific ADT resistance contribute to tumor regression, whereas inducing a more aggressive NEPC phenotype in the process of neuroendocrine trans-differentiation promotes later acquisition of therapy resistance. b The characteristics of lineage switching from CRPC to NEPC in terms of clinical histology and molecular levels. Alteration of cellular identity from CRPC to NEPC is mainly characterized by the absence of AR and PSA. NEPC is also different from CRPC due to deletion of TP53 and RB1, enhancement of MYCN or AURKA, and upregulation of EZH2. c Aggressive behavior accompanying functional transformation from CRPC to NEPC. As lineage switching occurs, CRPC is converted to NEPC accompanied by increased invasiveness, intensive drug resistance, and elevated stem-like cell properties. CRPC castration-resistant prostate cancer, NEPC neuroendocrine prostate cancer, AR androgen receptor, ADTs androgen deprivation therapies, ARPIs AR pathway inhibitors, PSA serum prostate-specific antigen
However, even at this stage, there is a dependence on AR signaling.524–526 This has given rise to the emergence of second-generation AR pathway inhibitors (ARPIs), which act by targeting the specific ADT-resistant mechanisms by which AR signaling can be re-activated.525–528 As expected, satisfactory clinical therapeutic effects have been achieved with ARPIs (abiraterone acetate [abiraterone] and enzalutamide) in patients with CRPC529,530 (Fig. 5a). Despite this, new resistance to ARPIs inevitably occurs.54,525 Mechanistically, unlike the process of resistance to ADTs, emerging evidence suggests that resistance to ARPIs tends to be developed in an AR-independent manner by significantly altering the typical course of CRPC, including lineage conversion (neuroendocrine trans-differentiation) and/or the induction of EMT programs26,54,525 (Fig. 5). The former is termed as therapy-induced lineage crisis.531 Histologically, such a cellular identity crisis manifests as lineage switching from CRPC to neuroendocrine prostate cancer (NEPC), which is characterized by the presence of neuroendocrine markers (chromogranin A, synaptophysin, etc.) with the absence of AR and serum prostate-specific antigen532,533 (Fig. 5). At the molecular level, NEPC shares similar features to primary neuroendocrine tumors, such as combined inactivation of TP53 and RB1, as well as amplification of MYCN54,533–535 (Fig. 5b). Functionally, NEPC cells are associated with multiple aggressive behaviors, such as strong metastatic potential, enhanced stem cell-like (re-)differentiation capability, and heightened therapeutic resistance, all of which ultimately result in worse prognosis54,536 (Fig. 5c).
The precise mechanisms by which ARPIs induce lineage crisis in CRPC still remain to be elucidated. Despite that imperfect understanding, a large body of evidence suggests that lineage plasticity of tumor cell may act as a plausible explanation for the emergence of NEPC and ARPIs resistance.26,54 There also exists a possibility that NEPC could originate from a small subpopulation of neuroendocrine cells surrounding the primary tumor without accompanying lineage conversion,537 which needs to be clarified (Fig. 6a). Proceeding from that, from the analysis of clinical data, the frequency of PCa-specific genetic alterations—represented by TMPRSS2-ERG gene rearrangement538 in castration-sensitive prostate cancer (CSPC), has been shown to be similar to that of NEPC (46%, 45% respectively).539,540 This coincidence helps to confirm that late-stage NEPC is derived from early-stage CSPC.533 Further evidence has been gathered from an investigation on divergent clonal evolution, which identified that CRPC and NEPC share essential genetic alterations, not limited to TMPRSS2-ERG fusions, with each other.534 According to the above clinical and laboratory findings, it can be safely concluded that, in most cases, the emergence of NEPC under ARPIs treatment is the consequence of lineage plasticity, also termed as neuroendocrine trans-differentiation. Second, ADT-induced neuroendocrine trans-differentiation initially requires developmental reprogramming to prostate cancer stem cells (PCSCs) of a neural class in AR-dependent cell lines541–543 (Fig. 6b). Such reprogrammed cells lose their features of prostate differentiation becoming neural/neural crest stem cells, resulting in a malignant phenotype resistant to ARPIs in vitro and in vivo543 (Fig. 6b). Further research performed in genetically engineered mouse models (GEMM) showed that the resistance acquired by lineage trans-differentiation from AR-dependent CRPC to AR-independent NEPC is largely reliant on the upregulation of the well-established undifferentiated cell marker SOX2 (SRY-Box Transcription Factor 2) and epigenetic regulator EZH2 (histone methyltransferase enhancer of zeste homolog 2)544,545 (Fig. 6b). Given the reversibility of tumor cell plasticity, such a basal-like NEPC can partially be reverted to its original lineage—luminal epithelial-like CRPC, by the re-establishment of TP53 and RB1 biological functions, re-exposure to androgens or inhibition of SOX2 expression543–545 (Fig. 6b). Notably, TF SOX2 is shown to play an important role in embryonic development and transcription modification, which is indeed implicated in the acquisition and maintenance of stem-like cell properties, especially for the central nervous system.546,547 In addition, a model of MUC1-driven lineage plasticity in PCa shows that MUC1 can upregulate the expression of BRN2 by recruitment of MYC and subsequent binding to the promoter region of BRN2, which further contributes to SOX2 expression.548,549 This series of molecular regulations indicates that MUC1 may act as an upstream effector to regulate SOX2-induced lineage plasticity of neuroendocrine trans-differentiation in PCa.548 Finally, by integrating systemic analyses of GEMM with patient clinical data, Zou et al.550 provided conclusive genetic evidence that drug-induced neuroendocrine trans-differentiation of PCSCs is one of the main reasons behind treatment failure.551 Taken together, findings from different approaches, including clinical data, together with multiple in vitro and in vivo experimental models, strongly demonstrate that tumor cell plasticity-induced lineage switching enables PCa to escape from ARPI treatment.543–545,548–551
Fig. 6.
Two models describing the mechanism of lineage switching from CRPC to NEPC. a Following ARPI treatment, NEPC can originate from a small subpopulation of mutated neuroendocrine cells surrounding the primary tumor in CRPC, or derive from overgrown CSCs in CRPC, which undergo a differentiation process to acquire an AR-independent basal-like phenotype. b AR-dependent luminal epithelial cells initially undergo developmental reprogramming to neurological PCSCs, followed by differentiation into an AR-independent basal-like NEPC by ARPI-induced neuroendocrine trans-differentiation. Due to dynamic reversible cancer cell plasticity, the newly acquired NEPC can be reverted to the luminal epithelial-like CRPC by restoring TP53 and RB1, re-exposure to androgen or inhibition of EZH2 and SOX2 implicated in pluripotency networks. PCSCs prostate cancer stem cells, SOX2 SRY-box transcription factor 2, EZH2 enhancer of zeste homolog 2
Neuroendocrine trans-differentiation from NSCLC to SCLC
Similar observations of neuroendocrine trans-differentiation have been made for lung cancer, in which the transition from EGFR-mutant NSCLC to small cell lung cancer (SCLC) (Fig. 7), the most thoroughly described example of lineage plasticity to date, is closely correlated to resistance to EGFR-TKIs.26,521,552 SCLC is a highly aggressive neuroendocrine tumor with characteristics of neuroendocrine organ-like nest-like structure, a rapid growth rate, and a natural propensity for early metastatic spread, not typically observed in patients with no history of smoking.553–555 The first description of this trans-differentiation was a case report showing that a 45-year-old patient diagnosed with EGFR-mutant lung adenocarcinoma had obtained a partial response to erlotinib treatment, but 18 months later relapsed with metastatic synaptophysin (a typical neuroendocrine marker)-positive SCLC in multiple organs, with no trace of adenocarcinoma at autopsy.556 The contributions made by neuroendocrine trans-differentiation to the resistance to TKI-based therapy in EGFR-mutant NSCLC have been excellently presented in a number of recent reviews.26,521
Fig. 7.
Overview of the molecular basis of re-activation of developmental programs contributing to cancer cell plasticity—major inhibitors of Hh, Wnt, and Notch signaling pathways for targeted therapy. The Hedgehog (Hh), wingless/integrated (Wnt), and Notch signaling pathways play a crucial part in acquisition and expansion of CSC phenotype after being stimulated by internal factors or extrinsic stimuli, for instance, HGF and docetaxel. Re-activation of these developmental programs promotes the corresponding transcription factors entering into the nucleus to regulate expression of downstream effectors that are closely related to CSCs regeneration and maintenance, as well as multiple biological functions. To prevent cancer cell plasticity, particularly phenotypic switching from non-CSC to CSC states induced by Hh, Wnt, and Notch signaling pathways, a range of inhibitors targeting these pathways have been approved for clinical use or are under development, as shown in blue, purple, and green. CAFs cancer-associated fibroblasts, Ihh/Dhh/Shh: Sonic hedgehog/Indian hedgehog/Desert hedgehog, PTCH1 2 patched1 and patched2, SMO smoothened, ATO arsenic trioxide, HDAC6i histone deacetylase 6 inhibitor, GLI glioma-associated oncogene homolog, LRP5/6 lipoprotein receptor-related protein 5/6, HGF hepatocyte growth factor, VA valproic acid, EMT epithelial–mesenchymal transition, NICD Notch intracellular region, ADAM a disintegrin and metalloprotease, DLL1, 3, 4 Delta-like ligand 1, 3, 4, Jagged 1, 2 Serrate-like ligand 1, 2, IR ionizing radiation, NT neoadjuvant therapy, Rova-T rovalpituzumab tesirine, DIM 3,3′-diindolylmethane, DiFiD 3,5-bis (2,4-difluorobenzylidene)-4-piperidone, EGCG epigallocatechin-3-gallate
The mechanism behind re-activation of developmental programs
Although somatic and neoplastic stem cells reside at the top of the lineage hierarchy, extensive studies have shown that, under certain conditions, not all carcinoma types strictly conform to the generally accepted unidirectional hierarchical model of CSC. This results in a phenotypic switching whereby non-CSCs acquire characteristics of CSCs, such as self-renewal capacity and a slow-cycling state.557–559 This reversible plasticity of carcinoma cells, a bidirectional interconversion between CSC and non-CSC states, is central to its role in tumor progression and prognosis of individual cancer patients, given the extensive evidence that CSCs are intrinsically more prone to disseminating to distant organs and are, at the same time, more refractory to existing antitumor therapies. Further in-depth understanding of this switching process is clearly urgently needed.560,561
From a macroscopic point of view, re-activation of transcriptional cascades rewrites cell fate in response to a wide variety of signals derived from different developmental signaling pathways involved in embryonic development, tissue homeostasis, and adult diseases processes (e.g., carcinoma progression, harboring Hedgehog [Hh], Wnt, and Notch pathways).562–564 The (re-)activation of developmental programs can be either beneficial or harmful to organ functions, depending on whether it occurs during early development or in the adult.562–564 By narrowing the focus to carcinoma progression, growing evidence reveals that these evolutionarily conserved developmental pathways are re-activated during tumorigenesis and are crucial to the acquisition and expansion of the CSC phenotype by interacting with each other, or other oncogenic signaling pathways including NF-κB, MAPK, PI3K/AKT/mTOR (mammalian target of rapamycin), and EGF.564,565 Of note, the contributions made by the reciprocity of these complicated pathways to CSC generation and biological function can be traced to Hh/Wnt/Notch-induced fluctuation of expression of their downstream effectors in response to external or internal stimuli. These include various cytokines and growth factors, and markers of apoptosis/antiapoptosis, proliferation, metastasis, and resistance.566
The above suggests that cancer cell plasticity induced by the re-activation of developmental programs to some extent determines the organizational structure and highly heterogeneous nature of individual tumors, arising from cancer cells in varying stages of differentiation. This highlights the concept that CSCs, except for those that are pre-existing, can be generated de novo from bulk non-CSCs at a low but non-negligible rate. More importantly, this provides biological insight into the therapeutic potential of targeting these pathways.567 Undoubtedly, this bidirectional phenotypic transition between non-CSC and CSC states has brought enormous challenge for effective clinical treatment and is driving a marked shift of attention towards the optimization of developmental pathway-targeted therapy.568 Details on the relationship between the acquisition and maintenance of CSC phenotype and CSC-dependent signaling pathways, as well as the development of appropriate Hh/Wnt/Notch signal-targeted drugs for preclinical or clinical trials will be discussed below.
Hh pathway
The Hh signaling pathway is intimately concerned with cell proliferation and differentiation, together with tissue homeostasis and regeneration throughout life. However, aberrant activation of Hh has also made it a force to be reckoned with in oncology due to its involvement in tumorigenesis and progression, from the formation of tumor-initiating cells to angiogenesis as well as tumor immune escape.569,570 It is known that Hh signal transmission is largely managed by two multitransmembrane receptors on the target cell: Patched (PTCH1 and PTCH2) and Smoothened (SMO): the former is characterized by antagonizing effects, while the latter has positive regulatory functions.571 When levels of secreted Hh ligands (Sonic hedgehog [SHH]/Indian hedgehog/Desert hedgehog) are low or non-existent, the transmembrane receptor PTCH is stably situated in the primary cilium (PC), thereby inhibiting SMO activity and preventing further signal transduction.572 However, in the presence of Hh ligands, PTCH can dissociate from the PC and simultaneously relieve its repressive effects on SMO following binding by extracellular Hh ligands. This in turn facilitates signal transmission and consequent activation and nuclear localization of the Hh pathway downstream effector GLI (glioma-associated oncogene homolog, GLI1, GLI2, and GLI3) accompanied by upregulated expression of Hh target genes participating in differentiation, proliferation, and survival.563,565,571,573
Deregulation of the Hh pathway occurs in diverse cancers, including those of the breast, lung, bladder, pancreas, and stomach.574–578 Since there exists closely functional overlap between EMT programs and the CSC phenotype,55 the associated signaling pathways involved in sustaining the mesenchymal (or quasi-mesenchymal) state of carcinoma cells in various tissues has been a major topic of research. In particular, radiotherapy has been found to stimulate re-activation of Hh signaling, which further induces the EMT process by overexpression of EMT-stimulating factors and mesenchymal markers.579
Studies on multiple myeloma have suggested that Hh signaling is involved in interactions between CSCs, differentiated cancer cells, and the microenvironment, whereby blocking signaling can result in CSC differentiation.580 In TNBC, the acquisition of a chemo-resistant and stem-like phenotype benefits from a supportive niche with expression of fibroblast growth factor 5 and production of fibrillar collagen, which is provided by CAFs that are reprogrammed by newly secreted Hh ligands.581 The administration of docetaxel leads to the release of SHH ligand, followed by activation of the Hh pathway. Furthermore, increases in the expression of stemness signature with breast mammosphere formation provide information on the connection between chemotherapy-induced Hh signaling and expansion of breast CSC populations.582 Intriguingly, in kidney cancer, cigarette smoke triggers the activation of the SHH pathway, thus enhancing tumorsphere formation and elevating renal CSC populations. This finding from a series of experiments supports a molecular mechanism of cigarette smoke-elicited stemness by Hh signaling activation.583
Aberrations in Hh cascade contributing to tumorigenesis and tumor progress indicate that the Hh pathway represents a valid target for cancer therapy clinically. In particular, drugs targeting SMO have attracted considerable interest. Cyclopamine, an alkaloid extracted from Veratrum californium, was the first identified Hh inhibitor suppressing CSC proliferation with effective control of Hh-dependent tumors.584,585 As a first-in-class, the cyclopamine-competitive SMO antagonist, vismodegib, is effective in reducing the content and/or viability of breast CSCs. It was licensed for the treatment of metastatic basal cell carcinoma by the US Food and Drug Administration (FDA) in 2012 and the European Medicines Agency in 2013.586,587 There is also considerable interest in the selective SMO antagonist sonidegib due to its success in the treatment of advanced basal cell carcinoma, resulting in its launch in the USA in 2015.568,588 Other selective SMO inhibitors (e.g., glasdegib, saridegib, and taladegib) have entered a number of clinical trials, including metastatic or recurrent head and neck squamous cell carcinoma, and acute myeloid leukemia.589–591 Regrettably, a number of approved SMO inhibitors that serve as single-target agents display a ubiquitous toxic reaction and build up chemotherapy resistance, indicating that further development of Hh signaling pathway inhibitors is required to overcome these common side effects.
SMO-dependent activation of GLI TFs, a late stage of the Hh pathway that regulates the expression of critical developmental genes, is another possible target.592 Arsenic trioxide (ATO), an FDA-approved drug which directly binds to GLI1 and GLI2, is highly effective in inhibiting Hh signaling, further causing induction of differentiation and apoptosis of CSCs.593 This results in higher remission rates and significantly longer survival in APL.593 Analogously, GANT-61 is another type of GLI inhibitor, currently under preclinical study, which is capable of preventing DNA binding to GLI1 and GLI2.594 HDAC6 inhibitor, which can promote differentiation and decrease the stemness of glioma stem cells via inactivation of SHH/GLI1 signaling, is another drug that can overcome stemness by targeting that pathway.595
Wnt pathway
Generally, during embryonic development, extracellular Wnt proteins monitor and modulate a variety of cellular processes, including cell proliferation and differentiation, while in adulthood Wnt signaling participates in the maintenance of somatic stem cell identity and orientation differentiation of MSCs.596,597 In brief, intracellular Wnt signaling functions through an autocrine or paracrine mode, either by the canonical pathway (Wnt/β-catenin pathway) or non-canonical pathways (the planar cell polarity pathway, which involves jun N-terminal kinase and the Wnt/Ca2+ pathway).596,598 The β-catenin-dependent Wnt pathway is highly conserved through evolution and is activated by interactions between Wnt proteins and their respective receptors, the seven-transmembrane receptor Frizzled (FZD) and the single-pass, low-density lipoprotein receptor-related proteins 5 or 6.599,600 In addition, in the case of Wnt signal, β-catenin accumulates in the cytoplasm and then localizes in the nucleus instead of being ubiquitinated and degraded, thus driving the transcription of the stemness-related target genes and inducing a series of cellular reactions.601
Much of the research on CSC characteristics has examined whether Wnt signaling is crucially tethered to EMT with acquisition of stem-like properties.135 It is known that Wnt signaling can stabilize β-catenin proteins along with the typical EMT marker—Snail, in a tandem fashion and generate TCF/LEF (T cytokine/lymphocyte enhancer) transcriptional machinery so as to cooperatively govern EMT, thereby initiating tumor cell dedifferentiation.602,603 It has also been postulated that Wnt5 signals via the FZD2 receptor and FYN (an Src family kinase) activate STAT3 transcription to trigger EMT programming through the previously unrecognized, Wnt5-FZD2 non-canonical pathway. This has been observed in multiple cancer cell lines as well as a mouse xenograft model.604,605 Wnt/β-catenin signaling has been discovered to affect EMT stimulated by ionizing radiation (IR), whereby upregulation of Wnt ligand and nuclear accumulation of β-catenin with elevated β-catenin/T cell factor transcriptional activities can be induced by IR.606–608 IR-induced Wnt/β-catenin signaling expedites activation of EMT by enhancing Snail protein stability.609 In the case of radioresistance, ribosomal S6 protein kinase 4, which has been reported to contribute to therapeutic resistance and poor prognosis, phosphorylates GSK-3β directly at Ser9, activating the Wnt/β-catenin pathway and acquiring CSC properties in esophageal squamous cell carcinoma.610 It is worth mentioning that Wnt signal can be coupled with Notch to induce a liver CSC phenotype; there appears to be a decrease in expression levels of a number of TFs implicated in EMT with a loss of CSC properties like self-renewal and tumorigenicity when Wnt or Notch signaling is blocked.611
Besides intrinsic factors, a dynamic shift from a differentiated to a stemness state of cancer cells can occur in response to extrinsic environmental cues.612 Consistent with this notion, hepatocyte growth factor, a myofibroblast-secreted factor, assists colorectal cancer cells to attain a stemness-like state from a differentiated, mature phenotype by β-catenin-dependent transcription both in vitro and in vivo.612 A study utilizing patient-derived colorectal cancer organoids has demonstrated that clinical use of MEKi (selumetinib, trametinib, and PD318088) unfortunately enhances Wnt activity and enrichment of gene signatures of stemness and relapse, ultimately inducing cancer cell plasticity.613 Likewise, valproic acid, used as an HDAC inhibitor and an anticancer agent in breast cancer clinical trials, has been found to be responsible for the upregulation of Wnt reporter activity, which enlarges the breast CSC pool through dedifferentiation of non-stem-like cells and promotes their capacity to generate tumors.614 Another study in B cell lymphoma concluded that once cancer cells escape from chemotherapy-induced senescence, they are much more likely to re-enter the cell cycle with strongly elevated Wnt-dependent clonogenicity as well as substantial upregulation of stem cell signatures.615 In human GBM and breast cancer, it has been proven that activation of TGF-β associated with Wnt pathways can induce an undifferentiated state to promote stemness under hypoxia.165,616
Review of the literature shows that dysregulation of the Wnt pathways exerts distinct functions in the dedifferentiation of CSCs. Once Wnt/β-catenin signaling is activated, PMP22 (peripheral myelin protein 22), an integral membrane glycoprotein, causes differentiation of gastric CSCs, whose mRNA levels decline dramatically.617 In contrast, tumor necrosis factor receptor-associated protein-1 inhibits the differentiation of CSCs by adjusting ubiquitination or phosphorylation of β-catenin in colorectal carcinoma.618
Although promising advances have been made in the development of inhibitors blocking the Hh pathway in early phase clinical trials, the development of drugs targeting the Wnt pathway still seems to face serious challenges. To date, relatively few agents have successfully reached clinical development, although DKN-01, a humanized monoclonal antibody that binds to and blocks the activity of the Dickkopf-1 protein, modulating Wnt/β-catenin signaling, is undergoing clinical trials in a wide range of cancer types.619 The suppressors of the Wnt signaling pathway, ipafricept and vantictumab (both first-in-class antibodies), are well tolerated in patients and reduce the abundance and frequency of CSCs in patient-derived tumor xenograft models of numerous cancer types.620,621
Published results of clinical trials showed that cirmtuzumab, a monoclonal antibody targets ROR1, which serves as a receptor for Wnt5a in the Wnt-planar cell polarity pathway successfully led to a reduction in dedifferentiation marker expression in chronic lymphocytic leukemia.622 In addition, Foxy-5, a Wnt5a-mimicking peptide in phase I study, causes activation of downstream Wnt5a signaling in colorectal, prostate cancer, and metastatic breast cancer owing to its antimetastatic activity.623–625 IWP2 and LGK974, small-molecule inhibitors, have been shown effective in rodent tumor models by preventing palmitoylation of Wnt ligands and targeting a Wnt-specific acyltransferase, porcupine. On the one hand, these compounds block autocrine signaling, which sustains the stem phenotypes of existing CSCs, while on the other hand, they curb paracrine signaling transmission that triggers formation of regenerative CSCs.626,627 A recent study has shown that Myc decoy oligodeoxynucleotide (ODN) attacks the transcription targets of Wnt/β-catenin signaling, accelerating the differentiation of simulated mouse CSC models. This suggests optimizing the Myc decoy ODN approach as a prospective strategy for differentiation therapy.628
Notch pathway
Dysregulation of the Notch pathway occurs in many cancers, including leukemia, GBM, and cancers of breast, cervix, colon, endometrium, kidney, lung, pancreas, and prostate.629 In mammals, there are five Notch ligands (Delta-like ligand: [DLL] 1, 3, 4 and Serrate-like ligand: Jagged 1, 2), forming a class of transmembrane proteins with conserved molecular structure. There are four Notch receptors (Notch1–4) that consist of an extracellular region, transmembrane region, and intracellular region (NICD/ICN) comprising a highly evolutionarily conserved Notch pathway, together with ligands above. In the absence of Notch signal, DNA-binding protein CSL (collective name of CBF-1, Suppressor of hairless and Lag) is bound to a co-repressor complex, which leads to repression of transcription. Binding of ligands to the extracellular domain of their receptor triggers two consecutive proteolytic cleavages: initially by ADAM (a disintegrin and metalloprotease), followed by γ-secretase, generating a soluble intracellular domain (NICD) that is transferred into the nucleus upon Notch signaling. Thus, when a ligand expressed on one cell specifically binds to a receptor on the adjacent cell, NICD together with CSL protein complex lead to the conversion from the original “synergistic inhibition complex” to a “synergistic activation complex.”630
As has been observed for the Wnt pathway, overwhelming evidence indicates that Notch signaling exerts a major influence on the security of a pool of stem or progenitor cells during embryonic or adult developmental programs.631 In addition, the Notch pathway is a fundamental master pathway closely controlling the fate of CSCs.632 As with the aforementioned signaling pathways, multiple evidence has been generated regarding the preternatural re-activation of Notch signaling causing acceleration of proliferation and restriction of differentiation in various cancers.633 The Notch pathway has also been considered to be involved in mediating resistance to chemoradiotherapy in several human malignancies.634 Similar to the Hh and Wnt pathways, there is considerable evidence to support the functional connection between EMT and Notch signaling by dominating central processes such as stemness generation.635 Moreover, EMT programs in colorectal cancer can be induced by constitutively active Notch1 by retroviral transduction that activates CD44, Slug, and Smad-3 via a cascade of other Notch receptors through induction of Jagged 1 expression.635 In addition, modeling the effect of inflammatory cytokines in the tumor microenvironment suggests that these cytokines are likely to stabilize a hybrid epithelial/mesenchymal phenotype and improve the frequency of CSCs by activating Notch-Jagged signaling.636 Experimental data have implied that Notch signaling can induce EMT programming by upregulation of Snail following irradiation.637 Another representative finding is that irradiation is capable of inducing de novo generation of breast CSCs relying on Notch signaling, which coincides with overexpression of the TFs as well as stem cell markers (Oct4, SOX2, Nanog, and Klf4). This reprogramming can be partially prevented by Notch inhibition.567
Previous research reports showed that an increase in CSC subpopulations, attributed to the activation of Notch signaling together with EMT induction, occurs in breast cancer mouse models after being treated with docetaxel.638 In current clinical practice, to allow optimal surgery and improved prognosis, neoadjuvant therapy (NT) is widely used in patients with locally advanced or inflammatory breast cancer. However, conjunctive chemotherapy-triggered events potentially contribute to the formation of a CSC phenotype, with higher levels of nuclear Notch and stemness markers being detected in primary breast cancers following NT.639 Alternatively, the interplay between Wnt and Notch signaling with other critical pathways like the Hh pathway mentioned earlier specifies the differentiation/stem states of cells.640 Indeed, it has been demonstrated that ectopic activation of Notch is sufficient to prompt dedifferentiation and drive tumorigenic transformation of mature adipocytes in vivo.641
Notably, the subsequent failure of secondary tumor growth upon re-transplantation indicates that loss of Notch results in a 50% reduction of cancer-initiating cell populations in xenograft models of esophageal adenocarcinoma cells.642 Another study has verified that the antitumor effects of Notch blockade assist in guiding the differentiation of liver CSCs into mature hepatocytes. This depends on the inverse process of EMT, namely, MET.643 In addition, enhanced miR-200b-3p reduces Notch signaling followed by a depletion of pancreatic CSC populations due to their tendency for asymmetric division. Coincidentally, the miR-34a-Numb-Notch feedback loop prevents ionizing radiation-induced EMT, blocking transformation from a differentiated state to a stem-like state in NSCLC.644 It should be mentioned that, among the four homologs that act as ligand-activated TFs in Notch signal transduction, in contrast to Notch3 and Notch4, there are trans-activation domains present in Notch1 and Notch2. This increases the functional complexity of Notch1 and Notch2, possibly conferring on them multiple roles in cancer biology to some extent.645 Among such functionalities, the possible relevance of Notch1 and Notch2 to the regulation of EMT course and CSCs has been suggested. One study indicated that there could be a latent interaction network between Notch1, HIF-1α, and GPER (an alternative ER), in which Notch1 responds to distinct microenvironmental cues (e.g., estrogen or hypoxia) in the context of the interplay of HIF-1α and GPER, thus promoting the activation of EMT programming in several cancers.646–648 By extension, elevated expression of ERα and subsequent estrogen effects could activate the Notch pathway through its binding to the promoter region of Notch1, which enhances EMT status together with basal stem-like properties of prostate cancer cells.648 Furthermore, hypoxia has been proved to be conducive to Snail1 transcription by the promotion of a HIF-1α/NICD synergistic interaction, in turn triggering the recruitment of NICD to the Snail1 promoter. In this way, HIF-1α facilitates EMT programming by improving activation of Snail in a Notch-dependent manner in oral squamous cell carcinoma.647 Perhaps, more importantly, in terms of the collaboration between GPER and HIF-1α, estrogen appears to strengthen Notch-mediated EMT by increasing HIF-1α recruitment at the Snail promoter via nuclear GPER.646 With respect to the mechanistic links between Notch2 and CSCs or EMT, in vivo and in vitro investigations on NSCLC have shown that Notch2 plays a central role in miR-181b-mediated stemness, whereas silencing Notch2 causes a striking reduction in tumorsphere formation of NSCLC cells.649 In breast cancer, highly active Notch2 has been regarded as a key mediator and major contributor in fractionated radiation-induced EMT via the IL-6/JAK/STAT3 signaling axis, leading to the loss of E-cadherin and elevated N-cadherin and vimentin levels.637 Likewise, deregulation of miR-195-5p is likely to modulate Notch2 translation and further upregulate Notch2 expression, thereby motivating EMT in colorectal cancer cell lines.650 In comparison to deletion of Notch1, forced overexpression of Notch2 in bladder cancer displays oncogenic effects, including EMT with its effector HES1 targeting the vimentin promoter in a Snail/Slug-dependent manner, and in addition to that, Notch2 facilitates dedifferentiation accompanied by increased CSC production in vitro and in vivo.651
There is accumulating evidence indicating the therapeutic potential of targeting Notch signaling for its roles in the enrichment of colon and breast CSCs.652–654 Clearly, the inhibition of signaling through the Notch receptors reduces the subpopulations of breast CSCs and impairs tumor-initiating capacity, indicating that targeting Notch signaling can be regarded as a potential therapeutic strategy.655 In this respect, two approaches to inhibiting Notch signal have been tried clinically: use of γ-secretase inhibitors (GSIs) as well as antibodies against the Notch receptor or ligand.565
Since γ-Secretase, a multisubunit intramembrane protein complex, plays a pivotal role in Notch signal transduction by exhibiting proteolysis, it is projected to be an effective therapeutic target in cancer.656,657 Based on this, GSIs are the most broadly developed Notch pathway inhibitors to date. In vitro studies have presented ample evidence that GSIs decrease CSC subpopulations and tumorsphere formation, indicating that Notch signal activation is required for CSCs stemness.658 RO4929097, a novel molecular inhibitor of γ-secretase, impairs colony formation in primary melanoma cell lines and affects tumor formation in human primary melanoma xenografts.659 Weekly oral delivery of MRK003, a cyclic sulfamide GSI, exhibits prominent inhibition of tumor growth, decreased expression of stemness markers, and efficient suppression of clonogenicity potency in brain cancer, supporting its further clinical use.660 According to clinical/preclinical data, treatment with other functional GSIs, such as MK-0752 and PF-03084014, can cause tumor regression or induce tumor growth arrest by targeting CSCs in breast and colorectal cancer. In liver cancer, a low dose of PF-03084014 induces tumorsphere differentiation and contributes to chemosensitization, further demonstrating its future clinical potential.634,661,662
The atypical Notch receptor ligand DLL3 may also provide a new practicable target for treatment of neuroendocrine carcinomas. The blockbuster drug, rovalpituzumab tesirine (Rova-T), an antibody–drug conjugate targeting the protein DLL3 on tumor cells, showed good safety and efficacy when given as a monotherapy in a phase I trial on recurrent SCLCs, particularly in individuals with high levels of DLL3.663 It has also been noted that natural agents downregulating Notch signal, including curcumin (from turmeric), 3,3′-diindolylmethane (found in cruciferous vegetables), 3,5-bis (2,4-difluorobenzylidene)-4-piperidone (from turmeric), and epigallocatechin-3-gallate (from tea), have been proposed as alternative strategies for cancer therapy and have successfully undergone clinical trials.664,665
Targeting cell plasticity of non‑CSC and CSC transition
An issue of great concern is that a single approach aimed at merely eradicating CSCs tends to be restrictive and not comprehensive enough due to its efficacy only for low-grade cancers with the acquisition of therapeutic resistance in most cases.464,666 This highlights the possibility that the CSC subpopulation, along with its plasticity influenced by numerous TFs (i.e., Sox gene family), multiple signaling pathways (i.e., Wnt-β-catenin, IL-6-STAT3, and retinoid X receptor signaling pathway), and tumor microenvironment containing secreted factors and extracellular matrix, may impact on clinical trials. In combination with those signaling pathways discussed above, various other pathways interact with each other, for example, Wnt and Notch, uniting in a vast and complicated network. It is unrealistic to try and block all cancer-causing pathways in a therapeutic manner. Rather, there should be a focus on identifying and then abolishing the dominant drivers of plasticity among CSCs and nearby differentiated non-CSCs in the CSC niche in situ to assist CSC-targeted therapy.667 Emerging systems biology data provide a means to make it possible to explore how the various elements interact and influence one another to normalize neoplastic cells. Specific core TFs might contribute to phenotypic switching by triggering alterations in the expression of a battery of genes within the corresponding regulatory network.668 One other point worth emphasizing is that certain epigenetic regulators, such as EZH2 and REST (repressor element-1 silencing transcription factor), involved in differentiation to a neuroendocrine phenotype (the aforementioned trans-differentiation) and resistance to routine therapy in prostate or lung cancer apply genetic or pharmacological means to inhibit their activity, aiming to reverse this phenotypic transformation, and regenerate, or maintain the drug-susceptible state.26 On the basis of these discoveries, there is a trend towards the development of differentiation and normalization therapy (e.g., ATRA, tranylcypromine analogs, rosmantuzumab, and oncostatin M), and combined therapy with regimens designed to target cellular components and/or related pathways within the TME (e.g., NCT01839487, NCT02030860, and NCT01621243 [a series of clinical trials of PDAC]) rather than anti-CSC therapy alone, with the potential to increase the life expectancy of a far wider range of cancer patients.458,669–674
Despite these advances, in order to fully achieve CR in clinical practice, novel rationally designed therapeutic approaches developed on the basis of an in-depth understanding of CSC dynamics are urgently needed. However, it is unavoidable that plasticity-targeted therapy will also be confronted with many challenges, as patients who suffer from the same type of cancer vary considerably in their response to similar treatments, highlighting the need for a personalized/precision medicine approach. The persistence of minimal residual disease (MRD) characterized by drug-tolerant cancer cells, following cancer therapy due to various forms of phenotypic switching, requires an understanding of the intratumoral heterogeneity within individual tumors through a systematic and integrated analysis of potential plasticity-associated factors. Only in this way can an optimal, effective, and personalized therapeutic strategy be formulated.
Conclusions
For cancer cells: better to change than be killed
At first sight, accompanied by the development of emerging therapeutic strategies (e.g., targeted therapy and immunotherapy), coupled with a solid understanding of the genetic mutations involved, advanced or even chemo-/ radiation-resistant cancers seem to be curable clinically. However, the facts suggest otherwise. While initial clinical responses to patients with later-stage carcinomas typically appear encouraging, tumor recurrence inevitably occurs in these patients after a short-lived period of non-progression. This can be evidenced by the development of molecularly targeted therapies, i.e., three generations of EGFR-TKIs, to treat EGFR-mutant NSCLC, the results of which still have not been able to meet clinical expectations due to the acquisition of resistance. What then is the cause of this phenomenon? It could be interpreted as a consequence of de novo mutations, or similar mechanisms, which endow tumor cells with the capability of bypassing inhibition of the targeted pathway under drug exposure. However, these explanations from the perspective of genetic alterations do not fully account for the accumulating clinical and laboratory observations, thus leading to a shift in research priority, at least in part, from mutational mechanisms to those related to non-genetic alterations.
The non-mutational process largely depends on tumor cell plasticity, which is regulated by highly integrated and complex interactions between transcriptional factors, epigenetic modulators as well as a variety of growth factors, cytokines, and chemokines released from non-neoplastic cells within the TME. The impressive ability of tumor cells to switch their identities or phenotypes is more likely a common mechanism by which they can escape treatment. It should be noted that phenotypic “change” is often accompanied by the acquisition of a more aggressive behavior, especially enhanced flexibility, mainly manifested in the processes such as EMT, transition from non-CSC to CSC, or CRPC to NEPC, which will exacerbate the difficulty of clinical treatment. Even more surprising, in most cases, tumor cells can achieve a new phenotype without losing their original properties, suggesting that phenotype switching between two functionally independent states is not strictly adhering to a binary-based “all or nothing” principle, but rather is a complicated multistage dynamic process involving several intermediate phenotypes with varying degrees of maintained biological characteristics. Alternatively, plasticity may have already existed in the “arsenal” of tumor cells prior to drug exposure and thus cancer therapy actually serves as a “trigger” to stimulate “change” to avoid cell death—better to change than be killed. Although tumor plasticity has been proven to play a key role in resistance to cancer therapy, there remain numerous questions to be answered and challenges to face.
For treatment: only “change” can prevent “change”, and make it changeless
Given its malleable nature and consequent poor clinical outcomes, understanding the true meaning of plasticity (“change”) is fundamental to unlocking the secrets of non-mutational resistance mechanisms during cancer therapy. To deal better with the “change” of carcinoma cells, it will be necessary to change both experimental methods and treatment strategies.
Using the example of EMT described earlier in this review, the cognitive evolution of the EMT concept from a “complete” to a “partial” form, to a great extent, could be viewed as a reflection of the development of experimental techniques (i.e., from dual-colorimetric RNA-ISH to scRNA-seq to LSR-3D imaging). This suggests that the ideal approach would monitor the whole dynamic process of cancer development from one phenotype to another, at both an individual and multicellular cluster level. Only when the nature of tumor plasticity is fully understood can complete prevention be truly achieved. This is likely to be based on not only existing strategies, such as intermittent treatment and combination therapy, but also the development of new strategies, such as adipogenesis therapy, which can take advantage of the vulnerability of tumor plasticity.
Finally, knowing that tumor cell plasticity plays an important role in therapeutic resistance, the prevention of this dynamic process seems to be a necessary prerequisite for the improvement of clinical outcomes for cancer patients. This assumes that the “change” of experimental methods is conducive to increasing our understanding of the mechanisms of the phenotypic “change” in cancer cells during treatment, which in turn could accelerate the “change” of therapeutic strategies to prevent tumor cell plasticity. In essence, only “change” can prevent “change,” and make it changeless.
Acknowledgements
This work was supported by project of the State Key Laboratory of Trauma, Burn and Combined Injury, Third Military Medical University (SKLJYJF20).
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
These authors contributed equally: Siyuan Qin, Jingwen Jiang
Contributor Information
Canhua Huang, Email: hcanhua@hotmail.com.
Jian Zhang, Email: zhangjian@sustech.edu.cn.
Weifeng He, Email: whe761211@hotmail.com.
References
- 1.Bedard PL, Hyman DM, Davids MS, Siu LL. Small molecules, big impact: 20 years of targeted therapy in oncology. Lancet. 2020;395:1078–1088. doi: 10.1016/S0140-6736(20)30164-1. [DOI] [PubMed] [Google Scholar]
- 2.Carneiro, B. A. & El-Deiry, W. S. Targeting apoptosis in cancer therapy. Nat. Rev. Clin. Oncol. 17, 395–417 (2020). [DOI] [PMC free article] [PubMed]
- 3.Vasan N, Baselga J, Hyman DM. A view on drug resistance in cancer. Nature. 2019;575:299–309. doi: 10.1038/s41586-019-1730-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Redmond KM, Wilson TR, Johnston PG, Longley DB. Resistance mechanisms to cancer chemotherapy. Front. Biosci. 2008;13:5138–5154. doi: 10.2741/3070. [DOI] [PubMed] [Google Scholar]
- 5.Fojo T. Multiple paths to a drug resistance phenotype: mutations, translocations, deletions and amplification of coding genes or promoter regions, epigenetic changes and microRNAs. Drug Resist. Updat. 2007;10:59–67. doi: 10.1016/j.drup.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 6.Jia Y, et al. Overcoming EGFR(T790M) and EGFR(C797S) resistance with mutant-selective allosteric inhibitors. Nature. 2016;534:129–132. doi: 10.1038/nature17960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thress KS, et al. Acquired EGFR C797S mutation mediates resistance to AZD9291 in non-small cell lung cancer harboring EGFR T790M. Nat. Med. 2015;21:560–562. doi: 10.1038/nm.3854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tan CS, Gilligan D, Pacey S. Treatment approaches for EGFR-inhibitor-resistant patients with non-small-cell lung cancer. Lancet Oncol. 2015;16:e447–e459. doi: 10.1016/S1470-2045(15)00246-6. [DOI] [PubMed] [Google Scholar]
- 9.Bhang HE, et al. Studying clonal dynamics in response to cancer therapy using high-complexity barcoding. Nat. Med. 2015;21:440–448. doi: 10.1038/nm.3841. [DOI] [PubMed] [Google Scholar]
- 10.Turke AB, et al. Preexistence and clonal selection of MET amplification in EGFR mutant NSCLC. Cancer Cell. 2010;17:77–88. doi: 10.1016/j.ccr.2009.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schmitt MW, Loeb LA, Salk JJ. The influence of subclonal resistance mutations on targeted cancer therapy. Nat. Rev. Clin. Oncol. 2016;13:335–347. doi: 10.1038/nrclinonc.2015.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vallette FM, et al. Dormant, quiescent, tolerant and persister cells: four synonyms for the same target in cancer. Biochem. Pharmacol. 2019;162:169–176. doi: 10.1016/j.bcp.2018.11.004. [DOI] [PubMed] [Google Scholar]
- 13.Xue Y, et al. An approach to suppress the evolution of resistance in BRAF(V600E)-mutant cancer. Nat. Med. 2017;23:929–937. doi: 10.1038/nm.4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sharma SV, et al. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell. 2010;141:69–80. doi: 10.1016/j.cell.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Salgia R, Kulkarni P. The genetic/non-genetic duality of drug ‘resistance’ in cancer. Trends Cancer. 2018;4:110–118. doi: 10.1016/j.trecan.2018.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shaffer SM, et al. Rare cell variability and drug-induced reprogramming as a mode of cancer drug resistance. Nature. 2017;546:431–435. doi: 10.1038/nature22794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Trumpp A, Wiestler OD. Mechanisms of Disease: cancer stem cells-targeting the evil twin. Nat. Clin. Pract. Oncol. 2008;5:337–347. doi: 10.1038/ncponc1110. [DOI] [PubMed] [Google Scholar]
- 18.Bhatia R, et al. Persistence of malignant hematopoietic progenitors in chronic myelogenous leukemia patients in complete cytogenetic remission following imatinib mesylate treatment. Blood. 2003;101:4701–4707. doi: 10.1182/blood-2002-09-2780. [DOI] [PubMed] [Google Scholar]
- 19.Cortes J, O’Brien S, Kantarjian H. Discontinuation of imatinib therapy after achieving a molecular response. Blood. 2004;104:2204–2205. doi: 10.1182/blood-2004-04-1335. [DOI] [PubMed] [Google Scholar]
- 20.Yu Y, Ramena G, Elble RC. The role of cancer stem cells in relapse of solid tumors. Front. Biosci. 2012;4:1528–1541. doi: 10.2741/e478. [DOI] [PubMed] [Google Scholar]
- 21.Merlos-Suárez A, et al. The intestinal stem cell signature identifies colorectal cancer stem cells and predicts disease relapse. Cell Stem Cell. 2011;8:511–524. doi: 10.1016/j.stem.2011.02.020. [DOI] [PubMed] [Google Scholar]
- 22.Vanner RJ, et al. Quiescent sox2(+) cells drive hierarchical growth and relapse in sonic hedgehog subgroup medulloblastoma. Cancer Cell. 2014;26:33–47. doi: 10.1016/j.ccr.2014.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hata AN, et al. Tumor cells can follow distinct evolutionary paths to become resistant to epidermal growth factor receptor inhibition. Nat. Med. 2016;22:262–269. doi: 10.1038/nm.4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ramirez M, et al. Diverse drug-resistance mechanisms can emerge from drug-tolerant cancer persister cells. Nat. Commun. 2016;7:10690. doi: 10.1038/ncomms10690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dawson CC, Intapa C, Jabra-Rizk MA. “Persisters”: survival at the cellular level. PLoS Pathog. 2011;7:e1002121. doi: 10.1371/journal.ppat.1002121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boumahdi S, de Sauvage FJ. The great escape: tumour cell plasticity in resistance to targeted therapy. Nat. Rev. Drug Discov. 2020;19:39–56. doi: 10.1038/s41573-019-0044-1. [DOI] [PubMed] [Google Scholar]
- 27.Saha S, et al. Aspirin suppresses the acquisition of chemoresistance in breast cancer by disrupting an NFκB-IL6 signaling axis responsible for the generation of cancer stem cells. Cancer Res. 2016;76:2000–2012. doi: 10.1158/0008-5472.CAN-15-1360. [DOI] [PubMed] [Google Scholar]
- 28.Roesch A, et al. Overcoming intrinsic multidrug resistance in melanoma by blocking the mitochondrial respiratory chain of slow-cycling JARID1B(high) cells. Cancer Cell. 2013;23:811–825. doi: 10.1016/j.ccr.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Francescangeli F, et al. A pre-existing population of ZEB2(+) quiescent cells with stemness and mesenchymal features dictate chemoresistance in colorectal cancer. J. Exp. Clin. Cancer Res. 2020;39:2. doi: 10.1186/s13046-019-1505-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Glasspool RM, Teodoridis JM, Brown R. Epigenetics as a mechanism driving polygenic clinical drug resistance. Br. J. Cancer. 2006;94:1087–1092. doi: 10.1038/sj.bjc.6603024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pisco AO, et al. Non-Darwinian dynamics in therapy-induced cancer drug resistance. Nat. Commun. 2013;4:2467. doi: 10.1038/ncomms3467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arias AM, Hayward P. Filtering transcriptional noise during development: concepts and mechanisms. Nat. Rev. Genet. 2006;7:34–44. doi: 10.1038/nrg1750. [DOI] [PubMed] [Google Scholar]
- 33.Charlebois DA, Abdennur N, Kaern M. Gene expression noise facilitates adaptation and drug resistance independently of mutation. Phys. Rev. Lett. 2011;107:218101. doi: 10.1103/PhysRevLett.107.218101. [DOI] [PubMed] [Google Scholar]
- 34.Chisholm RH, et al. Emergence of drug tolerance in cancer cell populations: an evolutionary outcome of selection, nongenetic instability, and stress-induced adaptation. Cancer Res. 2015;75:930–939. doi: 10.1158/0008-5472.CAN-14-2103. [DOI] [PubMed] [Google Scholar]
- 35.Luria SE, Delbrück M. Mutations of bacteria from virus sensitivity to virus resistance. Genetics. 1943;28:491–511. doi: 10.1093/genetics/28.6.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Singer, E. Inside the Din, Cells Fight Noise With Noise. https://www.quantamagazine.org/biological-noise-in-an-unpredictable-world-20140522 (2014).
- 37.Kurata T, et al. Effect of re-treatment with gefitinib (‘Iressa’, ZD1839) after acquisition of resistance. Ann. Oncol. 2004;15:173–174. doi: 10.1093/annonc/mdh006. [DOI] [PubMed] [Google Scholar]
- 38.Yano S, et al. Retreatment of lung adenocarcinoma patients with gefitinib who had experienced favorable results from their initial treatment with this selective epidermal growth factor receptor inhibitor: a report of three cases. Oncol. Res. 2005;15:107–111. doi: 10.3727/096504005775082020. [DOI] [PubMed] [Google Scholar]
- 39.Nooka AK, et al. Clinical efficacy of daratumumab, pomalidomide, and dexamethasone in patients with relapsed or refractory myeloma: utility of re-treatment with daratumumab among refractory patients. Cancer. 2019;125:2991–3000. doi: 10.1002/cncr.32178. [DOI] [PubMed] [Google Scholar]
- 40.Xu B, et al. Outcomes of re-treatment with first-line trastuzumab plus a taxane in HER2 positive metastatic breast cancer patients after (neo)adjuvant trastuzumab: a prospective multicenter study. Oncotarget. 2016;7:50643–50655. doi: 10.18632/oncotarget.9331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sartor O, et al. Re-treatment with radium-223: first experience from an international, open-label, phase I/II study in patients with castration-resistant prostate cancer and bone metastases. Ann. Oncol. 2017;28:2464–2471. doi: 10.1093/annonc/mdx331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fujita K, et al. Retreatment with pembrolizumab in advanced non-small cell lung cancer patients previously treated with nivolumab: emerging reports of 12 cases. Cancer Chemother. Pharmacol. 2018;81:1105–1109. doi: 10.1007/s00280-018-3585-9. [DOI] [PubMed] [Google Scholar]
- 43.Santini FC, et al. Safety and efficacy of re-treating with immunotherapy after immune-related adverse events in patients with NSCLC. Cancer Immunol. Res. 2018;6:1093–1099. doi: 10.1158/2326-6066.CIR-17-0755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hanahan D. Rethinking the war on cancer. Lancet. 2014;383:558–563. doi: 10.1016/S0140-6736(13)62226-6. [DOI] [PubMed] [Google Scholar]
- 45.Tata PR, Rajagopal J. Cellular plasticity: 1712 to the present day. Curr. Opin. Cell Biol. 2016;43:46–54. doi: 10.1016/j.ceb.2016.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Varga J, Greten FR. Cell plasticity in epithelial homeostasis and tumorigenesis. Nat. Cell Biol. 2017;19:1133–1141. doi: 10.1038/ncb3611. [DOI] [PubMed] [Google Scholar]
- 47.Nieto MA. Epithelial plasticity: a common theme in embryonic and cancer cells. Science. 2013;342:1234850. doi: 10.1126/science.1234850. [DOI] [PubMed] [Google Scholar]
- 48.Gupta PB, et al. Phenotypic plasticity: driver of cancer initiation, progression, and therapy resistance. Cell Stem Cell. 2019;24:65–78. doi: 10.1016/j.stem.2018.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Meacham CE, Morrison SJ. Tumour heterogeneity and cancer cell plasticity. Nature. 2013;501:328–337. doi: 10.1038/nature12624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hammerlindl H, Schaider H. Tumor cell-intrinsic phenotypic plasticity facilitates adaptive cellular reprogramming driving acquired drug resistance. J. Cell Commun. Signal. 2018;12:133–141. doi: 10.1007/s12079-017-0435-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Arozarena I, Wellbrock C. Phenotype plasticity as enabler of melanoma progression and therapy resistance. Nat. Rev. Cancer. 2019;19:377–391. doi: 10.1038/s41568-019-0154-4. [DOI] [PubMed] [Google Scholar]
- 52.Horn LA, Fousek K, Palena C. Tumor plasticity and resistance to immunotherapy. Trends Cancer. 2020;6:432–441. doi: 10.1016/j.trecan.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Doherty, M. R., Smigiel, J. M., Junk, D. J. & Jackson, M. W. Cancer stem cell plasticity drives therapeutic resistance. Cancers8, 8 (2016). [DOI] [PMC free article] [PubMed]
- 54.Davies AH, Beltran H, Zoubeidi A. Cellular plasticity and the neuroendocrine phenotype in prostate cancer. Nat. Rev. Urol. 2018;15:271–286. doi: 10.1038/nrurol.2018.22. [DOI] [PubMed] [Google Scholar]
- 55.Shibue T, Weinberg RA. EMT, CSCs, and drug resistance: the mechanistic link and clinical implications. Nat. Rev. Clin. Oncol. 2017;14:611–629. doi: 10.1038/nrclinonc.2017.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Manzo G. Similarities between embryo development and cancer process suggest new strategies for research and therapy of tumors: a new point of view. Front. Cell Dev. Biol. 2019;7:20. doi: 10.3389/fcell.2019.00020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hay ED. The mesenchymal cell, its role in the embryo, and the remarkable signaling mechanisms that create it. Dev. Dyn. 2005;233:706–720. doi: 10.1002/dvdy.20345. [DOI] [PubMed] [Google Scholar]
- 58.Boyer B, Thiery JP. Epithelium-mesenchyme interconversion as example of epithelial plasticity. APMIS. 1993;101:257–268. doi: 10.1111/j.1699-0463.1993.tb00109.x. [DOI] [PubMed] [Google Scholar]
- 59.Nieto MA, Huang RY, Jackson RA, Thiery JP. EMT: 2016. Cell. 2016;166:21–45. doi: 10.1016/j.cell.2016.06.028. [DOI] [PubMed] [Google Scholar]
- 60.Zhu X, Chen L, Liu L, Niu X. EMT-mediated acquired EGFR-TKI resistance in NSCLC: mechanisms and strategies. Front. Oncol. 2019;9:1044. doi: 10.3389/fonc.2019.01044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Laird DW, Lampe PD. Therapeutic strategies targeting connexins. Nat. Rev. Drug Discov. 2018;17:905–921. doi: 10.1038/nrd.2018.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 63.Stemmler MP, Eccles RL, Brabletz S, Brabletz T. Non-redundant functions of EMT transcription factors. Nat. Cell Biol. 2019;21:102–112. doi: 10.1038/s41556-018-0196-y. [DOI] [PubMed] [Google Scholar]
- 64.Carver EA, et al. The mouse snail gene encodes a key regulator of the epithelial-mesenchymal transition. Mol. Cell. Biol. 2001;21:8184–8188. doi: 10.1128/MCB.21.23.8184-8188.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Burk U, et al. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep. 2008;9:582–589. doi: 10.1038/embor.2008.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gregory PA, et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat. Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 67.Kim NH, et al. A p53/miRNA-34 axis regulates Snail1-dependent cancer cell epithelial-mesenchymal transition. J. Cell Biol. 2011;195:417–433. doi: 10.1083/jcb.201103097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Siemens H, et al. miR-34 and SNAIL form a double-negative feedback loop to regulate epithelial-mesenchymal transitions. Cell Cycle. 2011;10:4256–4271. doi: 10.4161/cc.10.24.18552. [DOI] [PubMed] [Google Scholar]
- 69.Nieto MA. Context-specific roles of EMT programmes in cancer cell dissemination. Nat. Cell Biol. 2017;19:416–418. doi: 10.1038/ncb3520. [DOI] [PubMed] [Google Scholar]
- 70.Tian XJ, Zhang H, Xing J. Coupled reversible and irreversible bistable switches underlying TGFβ-induced epithelial to mesenchymal transition. Biophys. J. 2013;105:1079–1089. doi: 10.1016/j.bpj.2013.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gregory PA, et al. An autocrine TGF-beta/ZEB/miR-200 signaling network regulates establishment and maintenance of epithelial-mesenchymal transition. Mol. Biol. Cell. 2011;22:1686–1698. doi: 10.1091/mbc.e11-02-0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yang J, Weinberg RA. Epithelial-mesenchymal transition: at the crossroads of development and tumor metastasis. Dev. Cell. 2008;14:818–829. doi: 10.1016/j.devcel.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 73.Stone RC, et al. Epithelial-mesenchymal transition in tissue repair and fibrosis. Cell Tissue Res. 2016;365:495–506. doi: 10.1007/s00441-016-2464-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kim, D. H. et al. Epithelial mesenchymal transition in embryonic development, tissue repair and cancer: a comprehensive overview. J. Clin. Med. 7, 1 (2017). [DOI] [PMC free article] [PubMed]
- 75.Yang J, et al. Guidelines and definitions for research on epithelial-mesenchymal transition. Nat. Rev. Mol. Cell. Biol. 2020;21:341–352. doi: 10.1038/s41580-020-0237-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nakajima Y, Yamagishi T, Hokari S, Nakamura H. Mechanisms involved in valvuloseptal endocardial cushion formation in early cardiogenesis: roles of transforming growth factor (TGF)-beta and bone morphogenetic protein (BMP) Anat. Rec. 2000;258:119–127. doi: 10.1002/(SICI)1097-0185(20000201)258:2<119::AID-AR1>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 77.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J. Clin. Invest. 2009;119:1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Savagner P, et al. Developmental transcription factor slug is required for effective re-epithelialization by adult keratinocytes. J. Cell. Physiol. 2005;202:858–866. doi: 10.1002/jcp.20188. [DOI] [PubMed] [Google Scholar]
- 79.Hatzikirou H, et al. ‘Go or grow’: the key to the emergence of invasion in tumour progression? Math. Med. Biol. 2012;29:49–65. doi: 10.1093/imammb/dqq011. [DOI] [PubMed] [Google Scholar]
- 80.Tarin D, Thompson EW, Newgreen DF. The fallacy of epithelial mesenchymal transition in neoplasia. Cancer Res. 2005;65:5996–6000. doi: 10.1158/0008-5472.CAN-05-0699. [DOI] [PubMed] [Google Scholar]
- 81.Ledford H. Cancer theory faces doubts. Nature. 2011;472:273. doi: 10.1038/472273a. [DOI] [PubMed] [Google Scholar]
- 82.Grigore, A. D. et al. Tumor budding: the name is EMT. Partial EMT. J. Clin. Med. 5, 51 (2016). [DOI] [PMC free article] [PubMed]
- 83.Thiery JP, Lim CT. Tumor dissemination: an EMT affair. Cancer Cell. 2013;23:272–273. doi: 10.1016/j.ccr.2013.03.004. [DOI] [PubMed] [Google Scholar]
- 84.Brabletz T. To differentiate or not-routes towards metastasis. Nat. Rev. Cancer. 2012;12:425–436. doi: 10.1038/nrc3265. [DOI] [PubMed] [Google Scholar]
- 85.Grande MT, et al. Snail1-induced partial epithelial-to-mesenchymal transition drives renal fibrosis in mice and can be targeted to reverse established disease. Nat. Med. 2015;21:989–997. doi: 10.1038/nm.3901. [DOI] [PubMed] [Google Scholar]
- 86.Allison SJ, Fibrosis Targeting EMT to reverse renal fibrosis. Nat. Rev. Nephrol. 2015;11:565. doi: 10.1038/nrneph.2015.133. [DOI] [PubMed] [Google Scholar]
- 87.Hong T, et al. An Ovol2-Zeb1 mutual inhibitory circuit governs bidirectional and multi-step transition between epithelial and mesenchymal states. PLoS Comput. Biol. 2015;11:e1004569. doi: 10.1371/journal.pcbi.1004569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lu, M. et al. MicroRNA-based regulation of epithelial-hybrid-mesenchymal fate determination. Proc. Natl. Acad. Sci. USA110, 18144–18149 (2013). [DOI] [PMC free article] [PubMed]
- 89.Steinway SN, et al. Combinatorial interventions inhibit TGFβ-driven epithelial-to-mesenchymal transition and support hybrid cellular phenotypes. NPJ Syst. Biol. Appl. 2015;1:15014. doi: 10.1038/npjsba.2015.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Puram SV, et al. Single-cell transcriptomic analysis of primary and metastatic tumor ecosystems in head and neck cancer. Cell. 2017;171:1611–1624.e1624. doi: 10.1016/j.cell.2017.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Pastushenko I, et al. Identification of the tumour transition states occurring during EMT. Nature. 2018;556:463–468. doi: 10.1038/s41586-018-0040-3. [DOI] [PubMed] [Google Scholar]
- 92.Wong IY, et al. Collective and individual migration following the epithelial-mesenchymal transition. Nat. Mater. 2014;13:1063–1071. doi: 10.1038/nmat4062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yu M, et al. Circulating tumor cells: approaches to isolation and characterization. J. Cell Biol. 2011;192:373–382. doi: 10.1083/jcb.201010021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yu M, et al. Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science. 2013;339:580–584. doi: 10.1126/science.1228522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rios AC, et al. Intraclonal plasticity in mammary tumors revealed through large-scale single-cell resolution 3D imaging. Cancer Cell. 2019;35:618–632.e616. doi: 10.1016/j.ccell.2019.02.010. [DOI] [PubMed] [Google Scholar]
- 96.Hüsemann Y, et al. Systemic spread is an early step in breast cancer. Cancer Cell. 2008;13:58–68. doi: 10.1016/j.ccr.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 97.Ye X, et al. Distinct EMT programs control normal mammary stem cells and tumour-initiating cells. Nature. 2015;525:256–260. doi: 10.1038/nature14897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Harper KL, et al. Mechanism of early dissemination and metastasis in Her2(+) mammary cancer. Nature. 2016;540:588–592. doi: 10.1038/nature20609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Rhim AD, et al. EMT and dissemination precede pancreatic tumor formation. Cell. 2012;148:349–361. doi: 10.1016/j.cell.2011.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Buczek ME, et al. Cytoplasmic PML promotes TGF-β-associated epithelial-mesenchymal transition and invasion in prostate cancer. Oncogene. 2016;35:3465–3475. doi: 10.1038/onc.2015.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Pantel K, Alix-Panabières C, Riethdorf S. Cancer micrometastases. Nat. Rev. Clin. Oncol. 2009;6:339–351. doi: 10.1038/nrclinonc.2009.44. [DOI] [PubMed] [Google Scholar]
- 102.Chemi F, et al. Pulmonary venous circulating tumor cell dissemination before tumor resection and disease relapse. Nat. Med. 2019;25:1534–1539. doi: 10.1038/s41591-019-0593-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Shain AH, et al. The genetic evolution of metastatic uveal melanoma. Nat. Genet. 2019;51:1123–1130. doi: 10.1038/s41588-019-0440-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hu Z, et al. Quantitative evidence for early metastatic seeding in colorectal cancer. Nat. Genet. 2019;51:1113–1122. doi: 10.1038/s41588-019-0423-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.De Craene B, Berx G. Regulatory networks defining EMT during cancer initiation and progression. Nat. Rev. Cancer. 2013;13:97–110. doi: 10.1038/nrc3447. [DOI] [PubMed] [Google Scholar]
- 106.Mani SA, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kudo-Saito C, Shirako H, Takeuchi T, Kawakami Y. Cancer metastasis is accelerated through immunosuppression during Snail-induced EMT of cancer cells. Cancer Cell. 2009;15:195–206. doi: 10.1016/j.ccr.2009.01.023. [DOI] [PubMed] [Google Scholar]
- 108.Siegel RL, Miller KD, Jemal A. Cancer statistics 2020. CA Cancer J. Clin. 2020;70:7–30. doi: 10.3322/caac.21590. [DOI] [PubMed] [Google Scholar]
- 109.Fidler IJ. The pathogenesis of cancer metastasis: the ‘seed and soil’ hypothesis revisited. Nat. Rev. Cancer. 2003;3:453–458. doi: 10.1038/nrc1098. [DOI] [PubMed] [Google Scholar]
- 110.Williams ED, Gao D, Redfern A, Thompson EW. Controversies around epithelial-mesenchymal plasticity in cancer metastasis. Nat. Rev. Cancer. 2019;19:716–732. doi: 10.1038/s41568-019-0213-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Yang J, et al. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117:927–939. doi: 10.1016/j.cell.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 112.Zhang K, et al. The collagen receptor discoidin domain receptor 2 stabilizes SNAIL1 to facilitate breast cancer metastasis. Nat. Cell Biol. 2013;15:677–687. doi: 10.1038/ncb2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Lee JH, et al. A20 promotes metastasis of aggressive basal-like breast cancers through multi-monoubiquitylation of Snail1. Nat. Cell Biol. 2017;19:1260–1273. doi: 10.1038/ncb3609. [DOI] [PubMed] [Google Scholar]
- 114.Guo W, et al. Slug and Sox9 cooperatively determine the mammary stem cell state. Cell. 2012;148:1015–1028. doi: 10.1016/j.cell.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Chakrabarti R, et al. Elf5 inhibits the epithelial-mesenchymal transition in mammary gland development and breast cancer metastasis by transcriptionally repressing Snail2. Nat. Cell Biol. 2012;14:1212–1222. doi: 10.1038/ncb2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lee DG, et al. Loss of NDRG2 promotes epithelial-mesenchymal transition of gallbladder carcinoma cells through MMP-19-mediated Slug expression. J. Hepatol. 2015;63:1429–1439. doi: 10.1016/j.jhep.2015.08.007. [DOI] [PubMed] [Google Scholar]
- 117.Krebs AM, et al. The EMT-activator Zeb1 is a key factor for cell plasticity and promotes metastasis in pancreatic cancer. Nat. Cell Biol. 2017;19:518–529. doi: 10.1038/ncb3513. [DOI] [PubMed] [Google Scholar]
- 118.Fischer KR, et al. Epithelial-to-mesenchymal transition is not required for lung metastasis but contributes to chemoresistance. Nature. 2015;527:472–476. doi: 10.1038/nature15748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ni T, et al. Snail1-dependent p53 repression regulates expansion and activity of tumour-initiating cells in breast cancer. Nat. Cell Biol. 2016;18:1221–1232. doi: 10.1038/ncb3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Tran HD, et al. Transient SNAIL1 expression is necessary for metastatic competence in breast cancer. Cancer Res. 2014;74:6330–6340. doi: 10.1158/0008-5472.CAN-14-0923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Cao J, et al. Twist promotes tumor metastasis in basal-like breast cancer by transcriptionally upregulating ROR1. Theranostics. 2018;8:2739–2751. doi: 10.7150/thno.21477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Xu Y, et al. Twist1 promotes breast cancer invasion and metastasis by silencing Foxa1 expression. Oncogene. 2017;36:1157–1166. doi: 10.1038/onc.2016.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Caramel J, et al. A switch in the expression of embryonic EMT-inducers drives the development of malignant melanoma. Cancer Cell. 2013;24:466–480. doi: 10.1016/j.ccr.2013.08.018. [DOI] [PubMed] [Google Scholar]
- 124.Denecker G, et al. Identification of a ZEB2-MITF-ZEB1 transcriptional network that controls melanogenesis and melanoma progression. Cell Death Differ. 2014;21:1250–1261. doi: 10.1038/cdd.2014.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Tran DD, et al. Temporal and spatial cooperation of Snail1 and Twist1 during epithelial-mesenchymal transition predicts for human breast cancer recurrence. Mol. Cancer Res. 2011;9:1644–1657. doi: 10.1158/1541-7786.MCR-11-0371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Leptin M. twist and snail as positive and negative regulators during Drosophila mesoderm development. Genes Dev. 1991;5:1568–1576. doi: 10.1101/gad.5.9.1568. [DOI] [PubMed] [Google Scholar]
- 127.Zeitlinger J, et al. Whole-genome ChIP-chip analysis of Dorsal, Twist, and Snail suggests integration of diverse patterning processes in the Drosophila embryo. Genes Dev. 2007;21:385–390. doi: 10.1101/gad.1509607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Vandamme N, et al. The EMT transcription factor ZEB2 promotes proliferation of primary and metastatic melanoma while suppressing an invasive, mesenchymal-like phenotype. Cancer Res. 2020;80:2983–2995. doi: 10.1158/0008-5472.CAN-19-2373. [DOI] [PubMed] [Google Scholar]
- 129.Lambert AW, Pattabiraman DR, Weinberg RA. Emerging biological principles of metastasis. Cell. 2017;168:670–691. doi: 10.1016/j.cell.2016.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Luzzi KJ, et al. Multistep nature of metastatic inefficiency: dormancy of solitary cells after successful extravasation and limited survival of early micrometastases. Am. J. Pathol. 1998;153:865–873. doi: 10.1016/S0002-9440(10)65628-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Fidler IJ, Nicolson GL. Organ selectivity for implantation survival and growth of B16 melanoma variant tumor lines. J. Natl Cancer Inst. 1976;57:1199–1202. doi: 10.1093/jnci/57.5.1199. [DOI] [PubMed] [Google Scholar]
- 132.Hart IR, Fidler IJ. Role of organ selectivity in the determination of metastatic patterns of B16 melanoma. Cancer Res. 1980;40:2281–2287. [PubMed] [Google Scholar]
- 133.Peinado H, et al. Pre-metastatic niches: organ-specific homes for metastases. Nat. Rev. Cancer. 2017;17:302–317. doi: 10.1038/nrc.2017.6. [DOI] [PubMed] [Google Scholar]
- 134.Obenauf AC, Massagué J. Surviving at a distance: organ-specific metastasis. Trends Cancer. 2015;1:76–91. doi: 10.1016/j.trecan.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Brabletz T, Kalluri R, Nieto MA, Weinberg RA. EMT in cancer. Nat. Rev. Cancer. 2018;18:128–134. doi: 10.1038/nrc.2017.118. [DOI] [PubMed] [Google Scholar]
- 136.Ocaña OH, et al. Metastatic colonization requires the repression of the epithelial-mesenchymal transition inducer Prrx1. Cancer Cell. 2012;22:709–724. doi: 10.1016/j.ccr.2012.10.012. [DOI] [PubMed] [Google Scholar]
- 137.Esposito M, et al. Bone vascular niche E-selectin induces mesenchymal-epithelial transition and Wnt activation in cancer cells to promote bone metastasis. Nat. Cell Biol. 2019;21:627–639. doi: 10.1038/s41556-019-0309-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Tsai JH, et al. Spatiotemporal regulation of epithelial-mesenchymal transition is essential for squamous cell carcinoma metastasis. Cancer Cell. 2012;22:725–736. doi: 10.1016/j.ccr.2012.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Del Pozo Martin Y, et al. Mesenchymal cancer cell-stroma crosstalk promotes niche activation, epithelial reversion, and metastatic colonization. Cell Rep. 2015;13:2456–2469. doi: 10.1016/j.celrep.2015.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Ruscetti M, et al. Tracking and functional characterization of epithelial-mesenchymal transition and mesenchymal tumor cells during prostate cancer metastasis. Cancer Res. 2015;75:2749–2759. doi: 10.1158/0008-5472.CAN-14-3476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Pastushenko I, Blanpain C. EMT transition states during tumor progression and metastasis. Trends Cell Biol. 2019;29:212–226. doi: 10.1016/j.tcb.2018.12.001. [DOI] [PubMed] [Google Scholar]
- 142.Beerling E, et al. Plasticity between epithelial and mesenchymal states unlinks EMT from metastasis-enhancing stem cell capacity. Cell Rep. 2016;14:2281–2288. doi: 10.1016/j.celrep.2016.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Lee CC, et al. Macrophage-secreted interleukin-35 regulates cancer cell plasticity to facilitate metastatic colonization. Nat. Commun. 2018;9:3763. doi: 10.1038/s41467-018-06268-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Maheswaran S, Haber DA. Cell fate: transition loses its invasive edge. Nature. 2015;527:452–453. doi: 10.1038/nature16313. [DOI] [PubMed] [Google Scholar]
- 145.Zheng X, et al. Epithelial-to-mesenchymal transition is dispensable for metastasis but induces chemoresistance in pancreatic cancer. Nature. 2015;527:525–530. doi: 10.1038/nature16064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Höckel M. Morphogenetic fields of embryonic development in locoregional cancer spread. Lancet Oncol. 2015;16:e148–e151. doi: 10.1016/S1470-2045(14)71028-9. [DOI] [PubMed] [Google Scholar]
- 147.Farmer P, et al. A stroma-related gene signature predicts resistance to neoadjuvant chemotherapy in breast cancer. Nat. Med. 2009;15:68–74. doi: 10.1038/nm.1908. [DOI] [PubMed] [Google Scholar]
- 148.Byers LA, et al. An epithelial-mesenchymal transition gene signature predicts resistance to EGFR and PI3K inhibitors and identifies Axl as a therapeutic target for overcoming EGFR inhibitor resistance. Clin. Cancer Res. 2013;19:279–290. doi: 10.1158/1078-0432.CCR-12-1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Paulitschke V, et al. Proteomic identification of a marker signature for MAPKi resistance in melanoma. EMBO J. 2019;38:e95874. doi: 10.15252/embj.201695874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Mak MP, et al. A patient-derived, pan-cancer EMT signature identifies global molecular alterations and immune target enrichment following epithelial-to-mesenchymal transition. Clin. Cancer Res. 2016;22:609–620. doi: 10.1158/1078-0432.CCR-15-0876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Bu X, Mahoney KM, Freeman GJ. Learning from PD-1 resistance: new combination strategies. Trends Mol. Med. 2016;22:448–451. doi: 10.1016/j.molmed.2016.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Datar I, Schalper KA. Epithelial-mesenchymal transition and immune evasion during lung cancer progression: the chicken or the egg? Clin. Cancer Res. 2016;22:3422–3424. doi: 10.1158/1078-0432.CCR-16-0336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Lou Y, et al. Epithelial-mesenchymal transition is associated with a distinct tumor microenvironment including elevation of inflammatory signals and multiple immune checkpoints in lung adenocarcinoma. Clin. Cancer Res. 2016;22:3630–3642. doi: 10.1158/1078-0432.CCR-15-1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Chen L, et al. Metastasis is regulated via microRNA-200/ZEB1 axis control of tumour cell PD-L1 expression and intratumoral immunosuppression. Nat. Commun. 2014;5:5241. doi: 10.1038/ncomms6241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Zhao L, et al. LncRNA SNHG14/miR-5590-3p/ZEB1 positive feedback loop promoted diffuse large B cell lymphoma progression and immune evasion through regulating PD-1/PD-L1 checkpoint. Cell Death Dis. 2019;10:731. doi: 10.1038/s41419-019-1886-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Hugo W, et al. Genomic and transcriptomic features of response to Anti-PD-1 therapy in metastatic melanoma. Cell. 2016;165:35–44. doi: 10.1016/j.cell.2016.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Wang L, et al. EMT- and stroma-related gene expression and resistance to PD-1 blockade in urothelial cancer. Nat. Commun. 2018;9:3503. doi: 10.1038/s41467-018-05992-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Rizvi NA, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348:124–128. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Skoulidis F, et al. STK11/LKB1 mutations and PD-1 inhibitor resistance in KRAS-mutant lung adenocarcinoma. Cancer Discov. 2018;8:822–835. doi: 10.1158/2159-8290.CD-18-0099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Zaretsky JM, et al. Mutations associated with acquired resistance to PD-1 blockade in melanoma. N. Engl. J. Med. 2016;375:819–829. doi: 10.1056/NEJMoa1604958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Chromatin-remodeling genes promote immunotherapy resistance. Cancer Discov. 8, Of1 (2018). [DOI] [PubMed]
- 162.Lu X, et al. Effective combinatorial immunotherapy for castration-resistant prostate cancer. Nature. 2017;543:728–732. doi: 10.1038/nature21676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Xu JW, et al. Immunotherapy for pancreatic cancer: a long and hopeful journey. Cancer Lett. 2018;425:143–151. doi: 10.1016/j.canlet.2018.03.040. [DOI] [PubMed] [Google Scholar]
- 164.O’Donnell JS, et al. Resistance to PD1/PDL1 checkpoint inhibition. Cancer Treat. Rev. 2017;52:71–81. doi: 10.1016/j.ctrv.2016.11.007. [DOI] [PubMed] [Google Scholar]
- 165.Scheel C, et al. Paracrine and autocrine signals induce and maintain mesenchymal and stem cell states in the breast. Cell. 2011;145:926–940. doi: 10.1016/j.cell.2011.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Hao, Y., Baker, D. & Ten Dijke, P. TGF-β-mediated epithelial-mesenchymal transition and cancer metastasis. Int. J. Mol. Sci. 20, 2767 (2019). [DOI] [PMC free article] [PubMed]
- 167.Liu M, Li S, Li MO. TGF-β control of adaptive immune tolerance: a break from Treg cells. BioEssays. 2018;40:e1800063. doi: 10.1002/bies.201800063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Shahbaz S, et al. CD71+VISTA+ erythroid cells promote the development and function of regulatory T cells through TGF-β. PLoS Biol. 2018;16:e2006649. doi: 10.1371/journal.pbio.2006649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Viel S, et al. TGF-β inhibits the activation and functions of NK cells by repressing the mTOR pathway. Sci. Signal. 2016;9:ra19. doi: 10.1126/scisignal.aad1884. [DOI] [PubMed] [Google Scholar]
- 170.Crane CA, et al. TGF-beta downregulates the activating receptor NKG2D on NK cells and CD8+ T cells in glioma patients. Neuro-Oncology. 2010;12:7–13. doi: 10.1093/neuonc/nop009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Lyons RM, Gentry LE, Purchio AF, Moses HL. Mechanism of activation of latent recombinant transforming growth factor beta 1 by plasmin. J. Cell Biol. 1990;110:1361–1367. doi: 10.1083/jcb.110.4.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Lyons RM, Keski-Oja J, Moses HL. Proteolytic activation of latent transforming growth factor-beta from fibroblast-conditioned medium. J. Cell Biol. 1988;106:1659–1665. doi: 10.1083/jcb.106.5.1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Mu D, et al. The integrin alpha(v)beta8 mediates epithelial homeostasis through MT1-MMP-dependent activation of TGF-beta1. J. Cell Biol. 2002;157:493–507. doi: 10.1083/jcb.200109100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Huang XZ, et al. Inactivation of the integrin beta 6 subunit gene reveals a role of epithelial integrins in regulating inflammation in the lung and skin. J. Cell Biol. 1996;133:921–928. doi: 10.1083/jcb.133.4.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Munger JS, et al. The integrin alpha v beta 6 binds and activates latent TGF beta 1: a mechanism for regulating pulmonary inflammation and fibrosis. Cell. 1999;96:319–328. doi: 10.1016/S0092-8674(00)80545-0. [DOI] [PubMed] [Google Scholar]
- 176.Cambier S, et al. Integrin alpha(v)beta8-mediated activation of transforming growth factor-beta by perivascular astrocytes: an angiogenic control switch. Am. J. Pathol. 2005;166:1883–1894. doi: 10.1016/S0002-9440(10)62497-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Munger JS, Harpel JG, Giancotti FG, Rifkin DB. Interactions between growth factors and integrins: latent forms of transforming growth factor-beta are ligands for the integrin alphavbeta1. Mol. Biol. Cell. 1998;9:2627–2638. doi: 10.1091/mbc.9.9.2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Crawford SE, et al. Thrombospondin-1 is a major activator of TGF-beta1 in vivo. Cell. 1998;93:1159–1170. doi: 10.1016/S0092-8674(00)81460-9. [DOI] [PubMed] [Google Scholar]
- 179.Kumar R, et al. TGF-β activation by bone marrow-derived thrombospondin-1 causes Schistosoma- and hypoxia-induced pulmonary hypertension. Nat. Commun. 2017;8:15494. doi: 10.1038/ncomms15494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Travis MA, Sheppard D. TGF-β activation and function in immunity. Annu. Rev. Immunol. 2014;32:51–82. doi: 10.1146/annurev-immunol-032713-120257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Atanasova VS, et al. Thrombospondin-1 is a major activator of TGF-β signaling in recessive dystrophic epidermolysis bullosa fibroblasts. J. Invest. Dermatol. 2019;139:1497–1505.e1495. doi: 10.1016/j.jid.2019.01.011. [DOI] [PubMed] [Google Scholar]
- 182.Murphy-Ullrich JE, Suto MJ. Thrombospondin-1 regulation of latent TGF-β activation: a therapeutic target for fibrotic disease. Matrix Biol. 2018;68-69:28–43. doi: 10.1016/j.matbio.2017.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Mir FA, Contreras-Ruiz L, Masli S. Thrombospondin-1-dependent immune regulation by transforming growth factor-β2-exposed antigen-presenting cells. Immunology. 2015;146:547–556. doi: 10.1111/imm.12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Dave M. TSP-1-mediated induction of T regulatory cell by adipose-derived mesenchymal stem cells: a mechanism of immunosuppression. Dig. Dis. Sci. 2017;62:1975–1976. doi: 10.1007/s10620-017-4645-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Weng TY, et al. A novel cancer therapeutic using thrombospondin 1 in dendritic cells. Mol. Ther. 2014;22:292–302. doi: 10.1038/mt.2013.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Nath PR, et al. Natural killer cell recruitment and activation are regulated by CD47 expression in the tumor microenvironment. Cancer Immunol. Res. 2019;7:1547–1561. doi: 10.1158/2326-6066.CIR-18-0367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Hosseini H, et al. Early dissemination seeds metastasis in breast cancer. Nature. 2016;540:552–558. doi: 10.1038/nature20785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Dongre A, et al. Epithelial-to-mesenchymal transition contributes to immunosuppression in breast carcinomas. Cancer Res. 2017;77:3982–3989. doi: 10.1158/0008-5472.CAN-16-3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Terry S, et al. New insights into the role of EMT in tumor immune escape. Mol. Oncol. 2017;11:824–846. doi: 10.1002/1878-0261.12093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Shrivastava R, Shukla N. Attributes of alternatively activated (M2) macrophages. Life Sci. 2019;224:222–231. doi: 10.1016/j.lfs.2019.03.062. [DOI] [PubMed] [Google Scholar]
- 191.Rückerl D, Allen JE. Macrophage proliferation, provenance, and plasticity in macroparasite infection. Immunol. Rev. 2014;262:113–133. doi: 10.1111/imr.12221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Zhu J, et al. The role of tumor associated macrophages in the tumor microenvironment: mechanism and functions. Anticancer Agents Med. Chem. 2016;16:1133–1141. doi: 10.2174/1871520616666160520112622. [DOI] [PubMed] [Google Scholar]
- 193.Qian BZ, Pollard JW. Macrophage diversity enhances tumor progression and metastasis. Cell. 2010;141:39–51. doi: 10.1016/j.cell.2010.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Noy R, Pollard JW. Tumor-associated macrophages: from mechanisms to therapy. Immunity. 2014;41:49–61. doi: 10.1016/j.immuni.2014.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Helm O, et al. Tumor-associated macrophages exhibit pro- and anti-inflammatory properties by which they impact on pancreatic tumorigenesis. Int. J. Cancer. 2014;135:843–861. doi: 10.1002/ijc.28736. [DOI] [PubMed] [Google Scholar]
- 196.Coussens LM, Werb Z. Inflammation and cancer. Nature. 2002;420:860–867. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Alderton GK. Immunology: skin inflammation predisposes to cancer. Nat. Rev. Cancer. 2016;16:678. doi: 10.1038/nrc.2016.120. [DOI] [PubMed] [Google Scholar]
- 198.Taniguchi K, Karin M. NF-κB, inflammation, immunity and cancer: coming of age. Nat. Rev. Immunol. 2018;18:309–324. doi: 10.1038/nri.2017.142. [DOI] [PubMed] [Google Scholar]
- 199.Pellicoro A, Ramachandran P, Iredale JP, Fallowfield JA. Liver fibrosis and repair: immune regulation of wound healing in a solid organ. Nat. Rev. Immunol. 2014;14:181–194. doi: 10.1038/nri3623. [DOI] [PubMed] [Google Scholar]
- 200.Karin M, Clevers H. Reparative inflammation takes charge of tissue regeneration. Nature. 2016;529:307–315. doi: 10.1038/nature17039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Dvorak HF. Tumors: wounds that do not heal. Similarities between tumor stroma generation and wound healing. N. Engl. J. Med. 1986;315:1650–1659. doi: 10.1056/NEJM198612253152606. [DOI] [PubMed] [Google Scholar]
- 202.Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell. 2010;140:883–899. doi: 10.1016/j.cell.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Shalapour S, Karin M. Immunity, inflammation, and cancer: an eternal fight between good and evil. J. Clin. Invest. 2015;125:3347–3355. doi: 10.1172/JCI80007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Hanahan D, Coussens LM. Accessories to the crime: functions of cells recruited to the tumor microenvironment. Cancer Cell. 2012;21:309–322. doi: 10.1016/j.ccr.2012.02.022. [DOI] [PubMed] [Google Scholar]
- 205.Hui L, Chen Y. Tumor microenvironment: sanctuary of the devil. Cancer Lett. 2015;368:7–13. doi: 10.1016/j.canlet.2015.07.039. [DOI] [PubMed] [Google Scholar]
- 206.Allavena P, et al. The inflammatory micro-environment in tumor progression: the role of tumor-associated macrophages. Crit. Rev. Oncol. Hematol. 2008;66:1–9. doi: 10.1016/j.critrevonc.2007.07.004. [DOI] [PubMed] [Google Scholar]
- 207.Zhang Q, et al. Landscape and dynamics of single immune cells in hepatocellular carcinoma. Cell. 2019;179:829–845.e820. doi: 10.1016/j.cell.2019.10.003. [DOI] [PubMed] [Google Scholar]
- 208.Yang M, et al. Stromal infiltration of tumor-associated macrophages conferring poor prognosis of patients with basal-like breast carcinoma. J. Cancer. 2018;9:2308–2316. doi: 10.7150/jca.25155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Zhao X, et al. Prognostic significance of tumor-associated macrophages in breast cancer: a meta-analysis of the literature. Oncotarget. 2017;8:30576–30586. doi: 10.18632/oncotarget.15736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Shabo I, et al. Breast cancer expression of CD163, a macrophage scavenger receptor, is related to early distant recurrence and reduced patient survival. Int. J. Cancer. 2008;123:780–786. doi: 10.1002/ijc.23527. [DOI] [PubMed] [Google Scholar]
- 211.Yan Y, et al. High tumor-associated macrophages infiltration is associated with poor prognosis and may contribute to the phenomenon of epithelial-mesenchymal transition in gastric cancer. OncoTargets Ther. 2016;9:3975–3983. doi: 10.2147/OTT.S103112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Xu J, et al. Tumor-associated macrophages induce invasion and poor prognosis in human gastric cancer in a cyclooxygenase-2/MMP9-dependent manner. Am. J. Transl. Res. 2019;11:6040–6054. [PMC free article] [PubMed] [Google Scholar]
- 213.Yamaguchi T, et al. Tumor-associated macrophages of the M2 phenotype contribute to progression in gastric cancer with peritoneal dissemination. Gastric Cancer. 2016;19:1052–1065. doi: 10.1007/s10120-015-0579-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Yin S, et al. The prognostic and clinicopathological significance of tumor-associated macrophages in patients with gastric cancer: a meta-analysis. PLoS ONE. 2017;12:e0170042. doi: 10.1371/journal.pone.0170042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Chen L, et al. Stromal PD-1(+) tumor-associated macrophages predict poor prognosis in lung adenocarcinoma. Hum. Pathol. 2020;97:68–79. doi: 10.1016/j.humpath.2019.12.007. [DOI] [PubMed] [Google Scholar]
- 216.Mei J, et al. Prognostic impact of tumor-associated macrophage infiltration in non-small cell lung cancer: a systemic review and meta-analysis. Oncotarget. 2016;7:34217–34228. doi: 10.18632/oncotarget.9079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Ho CC, et al. TREM-1 expression in tumor-associated macrophages and clinical outcome in lung cancer. Am. J. Respir. Crit. Care Med. 2008;177:763–770. doi: 10.1164/rccm.200704-641OC. [DOI] [PubMed] [Google Scholar]
- 218.Cai L, et al. Role of tumor-associated macrophages in the clinical course of pancreatic neuroendocrine tumors (PanNETs) Clin. Cancer Res. 2019;25:2644–2655. doi: 10.1158/1078-0432.CCR-18-3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Lissbrant IF, et al. Tumor associated macrophages in human prostate cancer: relation to clinicopathological variables and survival. Int. J. Oncol. 2000;17:445–451. doi: 10.3892/ijo.17.3.445. [DOI] [PubMed] [Google Scholar]
- 220.Yagi T, et al. Tumour-associated macrophages are associated with poor prognosis and programmed death ligand 1 expression in oesophageal cancer. Eur. J. Cancer. 2019;111:38–49. doi: 10.1016/j.ejca.2019.01.018. [DOI] [PubMed] [Google Scholar]
- 221.Ryder M, et al. Increased density of tumor-associated macrophages is associated with decreased survival in advanced thyroid cancer. Endocr. Relat. Cancer. 2008;15:1069–1074. doi: 10.1677/ERC-08-0036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Steidl C, et al. Tumor-associated macrophages and survival in classic Hodgkin’s lymphoma. N. Engl. J. Med. 2010;362:875–885. doi: 10.1056/NEJMoa0905680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Guo B, Cen H, Tan X, Ke Q. Meta-analysis of the prognostic and clinical value of tumor-associated macrophages in adult classical Hodgkin lymphoma. BMC Med. 2016;14:159. doi: 10.1186/s12916-016-0711-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Franklin RA, Li MO. Ontogeny of tumor-associated macrophages and its implication in cancer regulation. Trends Cancer. 2016;2:20–34. doi: 10.1016/j.trecan.2015.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Wei C, et al. Crosstalk between cancer cells and tumor associated macrophages is required for mesenchymal circulating tumor cell-mediated colorectal cancer metastasis. Mol. Cancer. 2019;18:64. doi: 10.1186/s12943-019-0976-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Pinto ML, et al. The two faces of tumor-associated macrophages and their clinical significance in colorectal cancer. Front. Immunol. 2019;10:1875. doi: 10.3389/fimmu.2019.01875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Su S, et al. A positive feedback loop between mesenchymal-like cancer cells and macrophages is essential to breast cancer metastasis. Cancer Cell. 2014;25:605–620. doi: 10.1016/j.ccr.2014.03.021. [DOI] [PubMed] [Google Scholar]
- 228.Wu Y, et al. Stabilization of snail by NF-kappaB is required for inflammation-induced cell migration and invasion. Cancer Cell. 2009;15:416–428. doi: 10.1016/j.ccr.2009.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Bates RC, Mercurio AM. Tumor necrosis factor-alpha stimulates the epithelial-to-mesenchymal transition of human colonic organoids. Mol. Biol. Cell. 2003;14:1790–1800. doi: 10.1091/mbc.e02-09-0583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Bonde AK, et al. Intratumoral macrophages contribute to epithelial-mesenchymal transition in solid tumors. BMC Cancer. 2012;12:35. doi: 10.1186/1471-2407-12-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Su S, et al. Breaking the vicious cycle between breast cancer cells and tumor-associated macrophages. Oncoimmunology. 2014;3:e953418. doi: 10.4161/21624011.2014.953418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Lin L, et al. CCL18 from tumor-associated macrophages promotes angiogenesis in breast cancer. Oncotarget. 2015;6:34758–34773. doi: 10.18632/oncotarget.5325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Zhou Z, et al. CCL18 secreted from M2 macrophages promotes migration and invasion via the PI3K/Akt pathway in gallbladder cancer. Cell. Oncol. 2019;42:81–92. doi: 10.1007/s13402-018-0410-8. [DOI] [PubMed] [Google Scholar]
- 234.Meng F, et al. CCL18 promotes epithelial-mesenchymal transition, invasion and migration of pancreatic cancer cells in pancreatic ductal adenocarcinoma. Int. J. Oncol. 2015;46:1109–1120. doi: 10.3892/ijo.2014.2794. [DOI] [PubMed] [Google Scholar]
- 235.She L, et al. Tumor-associated macrophages derived CCL18 promotes metastasis in squamous cell carcinoma of the head and neck. Cancer Cell Int. 2018;18:120. doi: 10.1186/s12935-018-0620-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Weng YS, et al. MCT-1/miR-34a/IL-6/IL-6R signaling axis promotes EMT progression, cancer stemness and M2 macrophage polarization in triple-negative breast cancer. Mol. Cancer. 2019;18:42. doi: 10.1186/s12943-019-0988-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Fu XL, et al. Interleukin 6 induces M2 macrophage differentiation by STAT3 activation that correlates with gastric cancer progression. Cancer Immunol. Immunother. 2017;66:1597–1608. doi: 10.1007/s00262-017-2052-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Mantovani A, et al. Tumour-associated macrophages as treatment targets in oncology. Nat. Rev. Clin. Oncol. 2017;14:399–416. doi: 10.1038/nrclinonc.2016.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Cassetta L, Pollard JW. Targeting macrophages: therapeutic approaches in cancer. Nat. Rev. Drug Discov. 2018;17:887–904. doi: 10.1038/nrd.2018.169. [DOI] [PubMed] [Google Scholar]
- 240.Kolattukudy PE, Niu J. Inflammation, endoplasmic reticulum stress, autophagy, and the monocyte chemoattractant protein-1/CCR2 pathway. Circ. Res. 2012;110:174–189. doi: 10.1161/CIRCRESAHA.111.243212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Ostuni R, Kratochvill F, Murray PJ, Natoli G. Macrophages and cancer: from mechanisms to therapeutic implications. Trends Immunol. 2015;36:229–239. doi: 10.1016/j.it.2015.02.004. [DOI] [PubMed] [Google Scholar]
- 242.Qian BZ, et al. CCL2 recruits inflammatory monocytes to facilitate breast-tumour metastasis. Nature. 2011;475:222–225. doi: 10.1038/nature10138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Zhang L, et al. Microenvironment-induced PTEN loss by exosomal microRNA primes brain metastasis outgrowth. Nature. 2015;527:100–104. doi: 10.1038/nature15376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Zhang J, Lu Y, Pienta KJ. Multiple roles of chemokine (C-C motif) ligand 2 in promoting prostate cancer growth. J. Natl Cancer Inst. 2010;102:522–528. doi: 10.1093/jnci/djq044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Bonapace L, et al. Cessation of CCL2 inhibition accelerates breast cancer metastasis by promoting angiogenesis. Nature. 2014;515:130–133. doi: 10.1038/nature13862. [DOI] [PubMed] [Google Scholar]
- 246.Izumi K, et al. Targeting the androgen receptor with siRNA promotes prostate cancer metastasis through enhanced macrophage recruitment via CCL2/CCR2-induced STAT3 activation. EMBO Mol. Med. 2013;5:1383–1401. doi: 10.1002/emmm.201202367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Zhu P, et al. Macrophage/cancer cell interactions mediate hormone resistance by a nuclear receptor derepression pathway. Cell. 2006;124:615–629. doi: 10.1016/j.cell.2005.12.032. [DOI] [PubMed] [Google Scholar]
- 248.Hou, P. et al. Tumor microenvironment remodeling enables bypass of oncogenic KRAS dependency in pancreatic cancer. Cancer Discov. 10, 1058–1077 (2020). [DOI] [PMC free article] [PubMed]
- 249.Kapoor A, et al. Yap1 activation enables bypass of oncogenic Kras addiction in pancreatic cancer. Cell. 2014;158:185–197. doi: 10.1016/j.cell.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Shao DD, et al. KRAS and YAP1 converge to regulate EMT and tumor survival. Cell. 2014;158:171–184. doi: 10.1016/j.cell.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Kim W, et al. Hepatic Hippo signaling inhibits protumoural microenvironment to suppress hepatocellular carcinoma. Gut. 2018;67:1692–1703. doi: 10.1136/gutjnl-2017-314061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Ni X, et al. YAP is essential for Treg-mediated suppression of antitumor immunity. Cancer Discov. 2018;8:1026–1043. doi: 10.1158/2159-8290.CD-17-1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Moroishi T, et al. The Hippo pathway kinases LATS1/2 suppress cancer immunity. Cell. 2016;167:1525–1539.e1517. doi: 10.1016/j.cell.2016.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Cao MX, et al. Interplay between cancer cells and M2 macrophages is necessary for miR-550a-3-5p down-regulation-mediated HPV-positive OSCC progression. J. Exp. Clin. Cancer Res. 2020;39:102. doi: 10.1186/s13046-020-01602-1. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 255.Kalbasi A, et al. Tumor-derived CCL2 mediates resistance to radiotherapy in pancreatic ductal adenocarcinoma. Clin. Cancer Res. 2017;23:137–148. doi: 10.1158/1078-0432.CCR-16-0870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Xia L, et al. Forkhead box Q1 promotes hepatocellular carcinoma metastasis by transactivating ZEB2 and VersicanV1 expression. Hepatology. 2014;59:958–973. doi: 10.1002/hep.26735. [DOI] [PubMed] [Google Scholar]
- 257.Liu R, et al. FGFR4 promotes stroma-induced epithelial-to-mesenchymal transition in colorectal cancer. Cancer Res. 2013;73:5926–5935. doi: 10.1158/0008-5472.CAN-12-4718. [DOI] [PubMed] [Google Scholar]
- 258.Li S, et al. MCP-1-induced ERK/GSK-3β/Snail signaling facilitates the epithelial-mesenchymal transition and promotes the migration of MCF-7 human breast carcinoma cells. Cell. Mol. Immunol. 2017;14:621–630. doi: 10.1038/cmi.2015.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Shen H, et al. PLEK2 promotes gallbladder cancer invasion and metastasis through EGFR/CCL2 pathway. J. Exp. Clin. Cancer Res. 2019;38:247. doi: 10.1186/s13046-019-1250-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Cortés M, et al. Tumor-associated macrophages (TAMs) depend on ZEB1 for their cancer-promoting roles. EMBO J. 2017;36:3336–3355. doi: 10.15252/embj.201797345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Caramel J, Ligier M, Puisieux A. Pleiotropic roles for ZEB1 in cancer. Cancer Res. 2018;78:30–35. doi: 10.1158/0008-5472.CAN-17-2476. [DOI] [PubMed] [Google Scholar]
- 262.Sánchez-Tilló E, et al. The EMT activator ZEB1 promotes tumor growth and determines differential response to chemotherapy in mantle cell lymphoma. Cell Death Differ. 2014;21:247–257. doi: 10.1038/cdd.2013.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Sánchez-Tilló E, et al. EMT-activating transcription factors in cancer: beyond EMT and tumor invasiveness. Cell. Mol. Life Sci. 2012;69:3429–3456. doi: 10.1007/s00018-012-1122-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Isella C, et al. Stromal contribution to the colorectal cancer transcriptome. Nat. Genet. 2015;47:312–319. doi: 10.1038/ng.3224. [DOI] [PubMed] [Google Scholar]
- 265.Cain DW, et al. Identification of a tissue-specific, C/EBPβ-dependent pathway of differentiation for murine peritoneal macrophages. J. Immunol. 2013;191:4665–4675. doi: 10.4049/jimmunol.1300581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Rei M, et al. Murine CD27(−) Vγ6(+) γδ T cells producing IL-17A promote ovarian cancer growth via mobilization of protumor small peritoneal macrophages. Proc. Natl Acad. Sci. USA. 2014;111:E3562–E3570. doi: 10.1073/pnas.1403424111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Kuwada K, et al. The epithelial-to-mesenchymal transition induced by tumor-associated macrophages confers chemoresistance in peritoneally disseminated pancreatic cancer. J. Exp. Clin. Cancer Res. 2018;37:307. doi: 10.1186/s13046-018-0981-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Wei C, et al. M2 macrophages confer resistance to 5-fluorouracil in colorectal cancer through the activation of CCL22/PI3K/AKT signaling. Onco Targets Ther. 2019;12:3051–3063. doi: 10.2147/OTT.S198126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Greten FR, Grivennikov SI. Inflammation and cancer: triggers, mechanisms, and consequences. Immunity. 2019;51:27–41. doi: 10.1016/j.immuni.2019.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Dominguez C, David JM, Palena C. Epithelial-mesenchymal transition and inflammation at the site of the primary tumor. Semin. Cancer Biol. 2017;47:177–184. doi: 10.1016/j.semcancer.2017.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Veglia F, Perego M, Gabrilovich D. Myeloid-derived suppressor cells coming of age. Nat. Immunol. 2018;19:108–119. doi: 10.1038/s41590-017-0022-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Kumar V, Patel S, Tcyganov E, Gabrilovich DI. The nature of myeloid-derived suppressor cells in the tumor microenvironment. Trends Immunol. 2016;37:208–220. doi: 10.1016/j.it.2016.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Bronte V, et al. Recommendations for myeloid-derived suppressor cell nomenclature and characterization standards. Nat. Commun. 2016;7:12150. doi: 10.1038/ncomms12150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Toh B, et al. Mesenchymal transition and dissemination of cancer cells is driven by myeloid-derived suppressor cells infiltrating the primary tumor. PLoS Biol. 2011;9:e1001162. doi: 10.1371/journal.pbio.1001162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Ding Z, et al. SMAD4-dependent barrier constrains prostate cancer growth and metastatic progression. Nature. 2011;470:269–273. doi: 10.1038/nature09677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Wang G, et al. Targeting YAP-dependent MDSC infiltration impairs tumor progression. Cancer Discov. 2016;6:80–95. doi: 10.1158/2159-8290.CD-15-0224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Zhao J, et al. Tumor-derived CXCL5 promotes human colorectal cancer metastasis through activation of the ERK/Elk-1/Snail and AKT/GSK3β/β-catenin pathways. Mol. Cancer. 2017;16:70. doi: 10.1186/s12943-017-0629-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Ouzounova M, et al. Monocytic and granulocytic myeloid derived suppressor cells differentially regulate spatiotemporal tumour plasticity during metastatic cascade. Nat. Commun. 2017;8:14979. doi: 10.1038/ncomms14979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Eisenblaetter M, et al. Visualization of tumor-immune interaction—target-specific imaging of S100A8/A9 reveals pre-metastatic niche establishment. Theranostics. 2017;7:2392–2401. doi: 10.7150/thno.17138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Takeyama Y, et al. Myeloid-derived suppressor cells are essential partners for immune checkpoint inhibitors in the treatment of cisplatin-resistant bladder cancer. Cancer Lett. 2020;479:89–99. doi: 10.1016/j.canlet.2020.03.013. [DOI] [PubMed] [Google Scholar]
- 281.Calcinotto A, et al. IL-23 secreted by myeloid cells drives castration-resistant prostate cancer. Nature. 2018;559:363–369. doi: 10.1038/s41586-018-0266-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Langowski JL, et al. IL-23 promotes tumour incidence and growth. Nature. 2006;442:461–465. doi: 10.1038/nature04808. [DOI] [PubMed] [Google Scholar]
- 283.Chen D, et al. Interleukin-23 promotes the epithelial-mesenchymal transition of oesophageal carcinoma cells via the Wnt/β-catenin pathway. Sci. Rep. 2015;5:8604. doi: 10.1038/srep08604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Rohwer N, Cramer T. Hypoxia-mediated drug resistance: novel insights on the functional interaction of HIFs and cell death pathways. Drug Resist. Updat. 2011;14:191–201. doi: 10.1016/j.drup.2011.03.001. [DOI] [PubMed] [Google Scholar]
- 285.Unruh A, et al. The hypoxia-inducible factor-1 alpha is a negative factor for tumor therapy. Oncogene. 2003;22:3213–3220. doi: 10.1038/sj.onc.1206385. [DOI] [PubMed] [Google Scholar]
- 286.Calin GA, Pardini B. Mirroring hypoxia in EGFR-TKI tolerance. Nat. Metab. 2019;1:418–419. doi: 10.1038/s42255-019-0057-4. [DOI] [PubMed] [Google Scholar]
- 287.Ye Y, et al. Characterization of hypoxia-associated molecular features to aid hypoxia-targeted therapy. Nat. Metab. 2019;1:431–444. doi: 10.1038/s42255-019-0045-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Hou PC, et al. Hypoxia-induced downregulation of DUSP-2 phosphatase drives colon cancer stemness. Cancer Res. 2017;77:4305–4316. doi: 10.1158/0008-5472.CAN-16-2990. [DOI] [PubMed] [Google Scholar]
- 289.Jing X, et al. Role of hypoxia in cancer therapy by regulating the tumor microenvironment. Mol. Cancer. 2019;18:157. doi: 10.1186/s12943-019-1089-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Marie-Egyptienne DT, Lohse I, Hill RP. Cancer stem cells, the epithelial to mesenchymal transition (EMT) and radioresistance: potential role of hypoxia. Cancer Lett. 2013;341:63–72. doi: 10.1016/j.canlet.2012.11.019. [DOI] [PubMed] [Google Scholar]
- 291.Schito L, Semenza GL. Hypoxia-inducible factors: master regulators of cancer progression. Trends Cancer. 2016;2:758–770. doi: 10.1016/j.trecan.2016.10.016. [DOI] [PubMed] [Google Scholar]
- 292.Tirpe, A. A. et al. Hypoxia: overview on hypoxia-mediated mechanisms with a focus on the role of HIF genes. Int. J. Mol. Sci. 20, 6140 (2019). [DOI] [PMC free article] [PubMed]
- 293.Gonzalez DM, Medici D. Signaling mechanisms of the epithelial-mesenchymal transition. Sci. Signal. 2014;7:re8. doi: 10.1126/scisignal.2005189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Sahlgren C, et al. Notch signaling mediates hypoxia-induced tumor cell migration and invasion. Proc. Natl Acad. Sci. USA. 2008;105:6392–6397. doi: 10.1073/pnas.0802047105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Zhang X, et al. Hypoxic BMSC-derived exosomal miRNAs promote metastasis of lung cancer cells via STAT3-induced EMT. Mol. Cancer. 2019;18:40. doi: 10.1186/s12943-019-0959-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Qin Y, et al. Salidroside improves the hypoxic tumor microenvironment and reverses the drug resistance of platinum drugs via HIF-1α signaling pathway. EBioMedicine. 2018;38:25–36. doi: 10.1016/j.ebiom.2018.10.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Wang J, et al. Ginsenoside Rg3 sensitizes hypoxic lung cancer cells to cisplatin via blocking of NF-κB mediated epithelial-mesenchymal transition and stemness. Cancer Lett. 2018;415:73–85. doi: 10.1016/j.canlet.2017.11.037. [DOI] [PubMed] [Google Scholar]
- 298.Zhan HX, et al. Crosstalk between stromal cells and cancer cells in pancreatic cancer: new insights into stromal biology. Cancer Lett. 2017;392:83–93. doi: 10.1016/j.canlet.2017.01.041. [DOI] [PubMed] [Google Scholar]
- 299.Najafi M, et al. Tumor microenvironment: interactions and therapy. J. Cell. Physiol. 2019;234:5700–5721. doi: 10.1002/jcp.27425. [DOI] [PubMed] [Google Scholar]
- 300.Ma HY, Liu XZ, Liang CM. Inflammatory microenvironment contributes to epithelial-mesenchymal transition in gastric cancer. World J. Gastroenterol. 2016;22:6619–6628. doi: 10.3748/wjg.v22.i29.6619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Mathew M, Safyan RA, Shu CA. PD-L1 as a biomarker in NSCLC: challenges and future directions. Ann. Transl. Med. 2017;5:375. doi: 10.21037/atm.2017.08.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Kim S, et al. PD-L1 expression is associated with epithelial-to-mesenchymal transition in adenocarcinoma of the lung. Hum. Pathol. 2016;58:7–14. doi: 10.1016/j.humpath.2016.07.007. [DOI] [PubMed] [Google Scholar]
- 303.Xu C, et al. Loss of Lkb1 and Pten leads to lung squamous cell carcinoma with elevated PD-L1 expression. Cancer Cell. 2014;25:590–604. doi: 10.1016/j.ccr.2014.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Barber DL, et al. Restoring function in exhausted CD8 T cells during chronic viral infection. Nature. 2006;439:682–687. doi: 10.1038/nature04444. [DOI] [PubMed] [Google Scholar]
- 305.Francisco LM, et al. PD-L1 regulates the development, maintenance, and function of induced regulatory T cells. J. Exp. Med. 2009;206:3015–3029. doi: 10.1084/jem.20090847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Adeegbe DO, Nishikawa H. Natural and induced T regulatory cells in cancer. Front. Immunol. 2013;4:190. doi: 10.3389/fimmu.2013.00190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Chen N, et al. Upregulation of PD-L1 by EGFR activation mediates the immune escape in EGFR-Driven NSCLC: implication for optional immune targeted therapy for NSCLC Patients with EGFR Mutation. J. Thorac. Oncol. 2015;10:910–923. doi: 10.1097/JTO.0000000000000500. [DOI] [PubMed] [Google Scholar]
- 308.Peng S, et al. EGFR-TKI resistance promotes immune escape in lung cancer via increased PD-L1 expression. Mol. Cancer. 2019;18:165. doi: 10.1186/s12943-019-1073-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Xu Y, et al. Translation control of the immune checkpoint in cancer and its therapeutic targeting. Nat. Med. 2019;25:301–311. doi: 10.1038/s41591-018-0321-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Prestipino, A. et al. Oncogenic JAK2(V617F) causes PD-L1 expression, mediating immune escape in myeloproliferative neoplasms. Sci. Transl. Med. 10, eaam7729 (2018). [DOI] [PMC free article] [PubMed]
- 311.Alsuliman A, et al. Bidirectional crosstalk between PD-L1 expression and epithelial to mesenchymal transition: significance in claudin-low breast cancer cells. Mol. Cancer. 2015;14:149. doi: 10.1186/s12943-015-0421-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Ock CY, et al. PD-L1 expression is associated with epithelial-mesenchymal transition in head and neck squamous cell carcinoma. Oncotarget. 2016;7:15901–15914. doi: 10.18632/oncotarget.7431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Wang Y, et al. PD-L1 induces epithelial-to-mesenchymal transition via activating SREBP-1c in renal cell carcinoma. Med. Oncol. 2015;32:212. doi: 10.1007/s12032-015-0655-2. [DOI] [PubMed] [Google Scholar]
- 314.Qiu XY, et al. PD-L1 confers glioblastoma multiforme malignancy via Ras binding and Ras/Erk/EMT activation. Biochim. Biophys. Acta. 2018;1864:1754–1769. doi: 10.1016/j.bbadis.2018.03.002. [DOI] [PubMed] [Google Scholar]
- 315.Almozyan S, et al. PD-L1 promotes OCT4 and Nanog expression in breast cancer stem cells by sustaining PI3K/AKT pathway activation. Int. J. Cancer. 2017;141:1402–1412. doi: 10.1002/ijc.30834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Mansour FA, et al. PD-L1 is overexpressed on breast cancer stem cells through notch3/mTOR axis. Oncoimmunology. 2020;9:1729299. doi: 10.1080/2162402X.2020.1729299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Ohigashi Y, et al. Clinical significance of programmed death-1 ligand-1 and programmed death-1 ligand-2 expression in human esophageal cancer. Clin. Cancer Res. 2005;11:2947–2953. doi: 10.1158/1078-0432.CCR-04-1469. [DOI] [PubMed] [Google Scholar]
- 318.Thompson RH, et al. Tumor B7-H1 is associated with poor prognosis in renal cell carcinoma patients with long-term follow-up. Cancer Res. 2006;66:3381–3385. doi: 10.1158/0008-5472.CAN-05-4303. [DOI] [PubMed] [Google Scholar]
- 319.Wu C, et al. Immunohistochemical localization of programmed death-1 ligand-1 (PD-L1) in gastric carcinoma and its clinical significance. Acta Histochem. 2006;108:19–24. doi: 10.1016/j.acthis.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 320.Hamanishi J, et al. Programmed cell death 1 ligand 1 and tumor-infiltrating CD8+ T lymphocytes are prognostic factors of human ovarian cancer. Proc. Natl Acad. Sci. USA. 2007;104:3360–3365. doi: 10.1073/pnas.0611533104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Li T, et al. IGFBP2 regulates PD-L1 expression by activating the EGFR-STAT3 signaling pathway in malignant melanoma. Cancer Lett. 2020;477:19–30. doi: 10.1016/j.canlet.2020.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Bracken CP, et al. A double-negative feedback loop between ZEB1-SIP1 and the microRNA-200 family regulates epithelial-mesenchymal transition. Cancer Res. 2008;68:7846–7854. doi: 10.1158/0008-5472.CAN-08-1942. [DOI] [PubMed] [Google Scholar]
- 323.Park SM, Gaur AB, Lengyel E, Peter ME. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 2008;22:894–907. doi: 10.1101/gad.1640608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Lamprecht S, et al. PBX3 is part of an EMT regulatory network and indicates poor outcome in colorectal cancer. Clin. Cancer Res. 2018;24:1974–1986. doi: 10.1158/1078-0432.CCR-17-2572. [DOI] [PubMed] [Google Scholar]
- 325.Korpal M, Lee ES, Hu G, Kang Y. The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J. Biol. Chem. 2008;283:14910–14914. doi: 10.1074/jbc.C800074200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Guan T, et al. ZEB1, ZEB2, and the miR-200 family form a counterregulatory network to regulate CD8(+) T cell fates. J. Exp. Med. 2018;215:1153–1168. doi: 10.1084/jem.20171352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Noman MZ, et al. The immune checkpoint ligand PD-L1 is upregulated in EMT-activated human breast cancer cells by a mechanism involving ZEB-1 and miR-200. Oncoimmunology. 2017;6:e1263412. doi: 10.1080/2162402X.2016.1263412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Zhou X, et al. miR-200c inhibits TGF-β-induced-EMT to restore trastuzumab sensitivity by targeting ZEB1 and ZEB2 in gastric cancer. Cancer Gene Ther. 2018;25:68–76. doi: 10.1038/s41417-017-0005-y. [DOI] [PubMed] [Google Scholar]
- 329.Zhou G, et al. miR-200c enhances sensitivity of drug-resistant non-small cell lung cancer to gefitinib by suppression of PI3K/Akt signaling pathway and inhibites cell migration via targeting ZEB1. Biomed. Pharmacother. 2017;85:113–119. doi: 10.1016/j.biopha.2016.11.100. [DOI] [PubMed] [Google Scholar]
- 330.Tsai SC, et al. The miR-200b-ZEB1 circuit regulates diverse stemness of human hepatocellular carcinoma. Mol. Carcinogen. 2017;56:2035–2047. doi: 10.1002/mc.22657. [DOI] [PubMed] [Google Scholar]
- 331.Luo Z, et al. MicroRNA-200C and -150 play an important role in endothelial cell differentiation and vasculogenesis by targeting transcription repressor ZEB1. Stem Cells. 2013;31:1749–1762. doi: 10.1002/stem.1448. [DOI] [PubMed] [Google Scholar]
- 332.Langer EM, et al. ZEB1-repressed microRNAs inhibit autocrine signaling that promotes vascular mimicry of breast cancer cells. Oncogene. 2018;37:1005–1019. doi: 10.1038/onc.2017.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Wellner U, et al. The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nat. Cell Biol. 2009;11:1487–1495. doi: 10.1038/ncb1998. [DOI] [PubMed] [Google Scholar]
- 334.Roybal JD, et al. miR-200 Inhibits lung adenocarcinoma cell invasion and metastasis by targeting Flt1/VEGFR1. Mol. Cancer Res. 2011;9:25–35. doi: 10.1158/1541-7786.MCR-10-0497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Pecot CV, et al. Tumour angiogenesis regulation by the miR-200 family. Nat. Commun. 2013;4:2427. doi: 10.1038/ncomms3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Tuo Z, et al. PD-L1 regulation by SDH5 via β-catenin/ZEB1 signaling. Oncoimmunology. 2019;8:1655361. doi: 10.1080/2162402X.2019.1655361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Martinez-Ciarpaglini C, et al. Low miR200c expression in tumor budding of invasive front predicts worse survival in patients with localized colon cancer and is related to PD-L1 overexpression. Mod. Pathol. 2019;32:306–313. doi: 10.1038/s41379-018-0124-5. [DOI] [PubMed] [Google Scholar]
- 338.Prall F, Hühns M. PD-L1 expression in tumour buds of colorectal carcinoma. Histopathology. 2016;69:158–160. doi: 10.1111/his.12915. [DOI] [PubMed] [Google Scholar]
- 339.Bouillez A, et al. MUC1-C integrates PD-L1 induction with repression of immune effectors in non-small-cell lung cancer. Oncogene. 2017;36:4037–4046. doi: 10.1038/onc.2017.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340.Ahmad R, et al. MUC1-C oncoprotein functions as a direct activator of the nuclear factor-kappaB p65 transcription factor. Cancer Res. 2009;69:7013–7021. doi: 10.1158/0008-5472.CAN-09-0523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Rajabi H, et al. MUC1-C oncoprotein activates the ZEB1/miR-200c regulatory loop and epithelial-mesenchymal transition. Oncogene. 2014;33:1680–1689. doi: 10.1038/onc.2013.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Yang Y, et al. ZEB1 sensitizes lung adenocarcinoma to metastasis suppression by PI3K antagonism. J. Clin. Invest. 2014;124:2696–2708. doi: 10.1172/JCI72171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Henriksen A, Dyhl-Polk A, Chen I, Nielsen D. Checkpoint inhibitors in pancreatic cancer. Cancer Treat. Rev. 2019;78:17–30. doi: 10.1016/j.ctrv.2019.06.005. [DOI] [PubMed] [Google Scholar]
- 344.Zheng, L. PD-L1 expression in pancreatic cancer. J. Natl. Cancer Inst. 109, djw304 (2017). [DOI] [PMC free article] [PubMed]
- 345.Patel SP, Kurzrock R. PD-L1 expression as a predictive biomarker in cancer immunotherapy. Mol. Cancer Ther. 2015;14:847–856. doi: 10.1158/1535-7163.MCT-14-0983. [DOI] [PubMed] [Google Scholar]
- 346.Brahmer J, et al. Nivolumab versus docetaxel in advanced squamous-cell non-small-cell lung cancer. N. Engl. J. Med. 2015;373:123–135. doi: 10.1056/NEJMoa1504627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Borghaei H, et al. Nivolumab versus docetaxel in advanced nonsquamous non-small-cell lung cancer. N. Engl. J. Med. 2015;373:1627–1639. doi: 10.1056/NEJMoa1507643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Fehrenbacher L, et al. Atezolizumab versus docetaxel for patients with previously treated non-small-cell lung cancer (POPLAR): a multicentre, open-label, phase 2 randomised controlled trial. Lancet. 2016;387:1837–1846. doi: 10.1016/S0140-6736(16)00587-0. [DOI] [PubMed] [Google Scholar]
- 349.Baral A, et al. B7-H3 and B7-H1 expression in cerebral spinal fluid and tumor tissue correlates with the malignancy grade of glioma patients. Oncol. Lett. 2014;8:1195–1201. doi: 10.3892/ol.2014.2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Kalluri R. The biology and function of exosomes in cancer. J. Clin. Invest. 2016;126:1208–1215. doi: 10.1172/JCI81135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351.Chen G, et al. Exosomal PD-L1 contributes to immunosuppression and is associated with anti-PD-1 response. Nature. 2018;560:382–386. doi: 10.1038/s41586-018-0392-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Yang Y, et al. Exosomal PD-L1 harbors active defense function to suppress T cell killing of breast cancer cells and promote tumor growth. Cell Res. 2018;28:862–864. doi: 10.1038/s41422-018-0060-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Diaz, A. A. Exosomal PD-L1 induces immunosuppressive non-classical monocytes. Neuro-Oncology22, 901–902 (2020). [DOI] [PMC free article] [PubMed]
- 354.Xie F, et al. The role of exosomal PD-L1 in tumor progression and immunotherapy. Mol. Cancer. 2019;18:146. doi: 10.1186/s12943-019-1074-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355.Del Re,M, et al. PD-L1 mRNA expression in plasma-derived exosomes is associated with response to anti-PD-1 antibodies in melanoma and NSCLC. Br. J. Cancer. 2018;118:820–824. doi: 10.1038/bjc.2018.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.Jiang Y, Zhan H. Communication between EMT and PD-L1 signaling: new insights into tumor immune evasion. Cancer Lett. 2020;468:72–81. doi: 10.1016/j.canlet.2019.10.013. [DOI] [PubMed] [Google Scholar]
- 357.Lee BS, et al. Hippo effector YAP directly regulates the expression of PD-L1 transcripts in EGFR-TKI-resistant lung adenocarcinoma. Biochem. Biophys. Res. Commun. 2017;491:493–499. doi: 10.1016/j.bbrc.2017.07.007. [DOI] [PubMed] [Google Scholar]
- 358.Miao J, et al. YAP regulates PD-L1 expression in human NSCLC cells. Oncotarget. 2017;8:114576–114587. doi: 10.18632/oncotarget.23051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.Hsu, P. C., Jablons, D. M., Yang, C. T. & You, L. Epidermal growth factor receptor (EGFR) pathway, Yes-associated protein (YAP) and the regulation of programmed death-ligand 1 (PD-L1) in non-small cell lung cancer (NSCLC). Int. J. Mol. Sci. 20, 3821 (2019). [DOI] [PMC free article] [PubMed]
- 360.Lee JE, et al. Hippo pathway effector YAP inhibition restores the sensitivity of EGFR-TKI in lung adenocarcinoma having primary or acquired EGFR-TKI resistance. Biochem. Biophys. Res. Commun. 2016;474:154–160. doi: 10.1016/j.bbrc.2016.04.089. [DOI] [PubMed] [Google Scholar]
- 361.Hsu, P. C., Yang, C. T., Jablons, D. M. & You, L. The role of Yes-associated protein (YAP) in regulating programmed death-ligand 1 (PD-L1) in thoracic cancer. Biomedicines6, 114 (2018). [DOI] [PMC free article] [PubMed]
- 362.Hsu PC, et al. Inhibition of yes-associated protein down-regulates PD-L1 (CD274) expression in human malignant pleural mesothelioma. J. Cell. Mol. Med. 2018;22:3139–3148. doi: 10.1111/jcmm.13593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 363.Flores RM, et al. Prognostic factors in the treatment of malignant pleural mesothelioma at a large tertiary referral center. J. Thorac. Oncol. 2007;2:957–965. doi: 10.1097/JTO.0b013e31815608d9. [DOI] [PubMed] [Google Scholar]
- 364.Zhang Y, et al. PD-L1 confers primary resistance to EGFR-TKI in EGFR mutant non-small cell lung cancer via inducing EMT phenotype. Ann. Oncol. 2019;30(Suppl. 2):ii52–ii52. doi: 10.1093/annonc/mdz063.036. [DOI] [Google Scholar]
- 365.Su S, et al. Strong programmed death ligand 1 expression predicts poor response and de novo resistance to EGFR tyrosine kinase inhibitors among NSCLC patients with EGFR mutation. J. Thorac. Oncol. 2018;13:1668–1675. doi: 10.1016/j.jtho.2018.07.016. [DOI] [PubMed] [Google Scholar]
- 366.Zhang Y, et al. The canonical TGF-β/Smad signalling pathway is involved in PD-L1-induced primary resistance to EGFR-TKIs in EGFR-mutant non-small-cell lung cancer. Respir. Res. 2019;20:164. doi: 10.1186/s12931-019-1137-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 367.Hsu KH, et al. High PD-L1 expression correlates with primary resistance to EGFR-TKIs in treatment naïve advanced EGFR-mutant lung adenocarcinoma patients. Lung Cancer. 2019;127:37–43. doi: 10.1016/j.lungcan.2018.11.021. [DOI] [PubMed] [Google Scholar]
- 368.Zhang N, et al. The EGFR pathway is involved in the regulation of PD-L1 expression via the IL-6/JAK/STAT3 signaling pathway in EGFR-mutated non-small cell lung cancer. Int. J. Oncol. 2016;49:1360–1368. doi: 10.3892/ijo.2016.3632. [DOI] [PubMed] [Google Scholar]
- 369.Wang Y, et al. EGFR activation induced Snail-dependent EMT and myc-dependent PD-L1 in human salivary adenoid cystic carcinoma cells. Cell Cycle. 2018;17:1457–1470. doi: 10.1080/15384101.2018.1489177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 370.Su L, et al. EGFR-ERK pathway regulates CSN6 to contribute to PD-L1 expression in glioblastoma. Mol. Carcinogen. 2020;59:520–532. doi: 10.1002/mc.23176. [DOI] [PubMed] [Google Scholar]
- 371.Xu, M. et al. Overexpression of CSN6 promotes the epithelial-mesenchymal transition and predicts poor prognosis in hepatocellular carcinoma. Clin. Res. Hepatol. Gastroenterol. 44, 340–348 (2019). [DOI] [PubMed]
- 372.Wen D, et al. Downregulation of CSN6 attenuates papillary thyroid carcinoma progression by reducing Wnt/β-catenin signaling and sensitizes cancer cells to FH535 therapy. Cancer Med. 2018;7:285–296. doi: 10.1002/cam4.1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373.Jiang L, et al. Continuous targeted kinase inhibitors treatment induces upregulation of PD-L1 in resistant NSCLC. Sci. Rep. 2019;9:3705. doi: 10.1038/s41598-018-38068-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 374.Azuma K, et al. Association of PD-L1 overexpression with activating EGFR mutations in surgically resected nonsmall-cell lung cancer. Ann. Oncol. 2014;25:1935–1940. doi: 10.1093/annonc/mdu242. [DOI] [PubMed] [Google Scholar]
- 375.Schoenfeld AJ, et al. Severe immune-related adverse events are common with sequential PD-(L)1 blockade and osimertinib. Ann. Oncol. 2019;30:839–844. doi: 10.1093/annonc/mdz077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 376.Oshima Y, Tanimoto T, Yuji K, Tojo A. EGFR-TKI-associated interstitial pneumonitis in nivolumab-treated patients with non-small cell lung cancer. JAMA Oncol. 2018;4:1112–1115. doi: 10.1001/jamaoncol.2017.4526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377.Jia Y, et al. Impact of EGFR-TKIs combined with PD-L1 antibody on the lung tissue of EGFR-driven tumor-bearing mice. Lung Cancer. 2019;137:85–93. doi: 10.1016/j.lungcan.2019.09.016. [DOI] [PubMed] [Google Scholar]
- 378.Ahn MJ, et al. EGFR TKI combination with immunotherapy in non-small cell lung cancer. Expert Opin. Drug Saf. 2017;16:465–469. doi: 10.1080/14740338.2017.1300656. [DOI] [PubMed] [Google Scholar]
- 379.Ribas A, et al. Hepatotoxicity with combination of vemurafenib and ipilimumab. N. Engl. J. Med. 2013;368:1365–1366. doi: 10.1056/NEJMc1302338. [DOI] [PubMed] [Google Scholar]
- 380.Maurya PK, et al. Role of Y box protein-1 in cancer: as potential biomarker and novel therapeutic target. J. Cancer. 2017;8:1900–1907. doi: 10.7150/jca.17689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 381.Lyabin DN, Eliseeva IA, Ovchinnikov LP. YB-1 protein: functions and regulation. Wiley Interdiscip. Rev. RNA. 2014;5:95–110. doi: 10.1002/wrna.1200. [DOI] [PubMed] [Google Scholar]
- 382.Deng SJ, et al. Hypoxia-induced LncRNA-BX111 promotes metastasis and progression of pancreatic cancer through regulating ZEB1 transcription. Oncogene. 2018;37:5811–5828. doi: 10.1038/s41388-018-0382-1. [DOI] [PubMed] [Google Scholar]
- 383.Evdokimova V, et al. Translational activation of snail1 and other developmentally regulated transcription factors by YB-1 promotes an epithelial-mesenchymal transition. Cancer Cell. 2009;15:402–415. doi: 10.1016/j.ccr.2009.03.017. [DOI] [PubMed] [Google Scholar]
- 384.Mouneimne G, Brugge JS. YB-1 translational control of epithelial-mesenchyme transition. Cancer Cell. 2009;15:357–359. doi: 10.1016/j.ccr.2009.04.006. [DOI] [PubMed] [Google Scholar]
- 385.Tao Z, et al. Targeting the YB-1/PD-L1 axis to enhance chemotherapy and antitumor immunity. Cancer Immunol. Res. 2019;7:1135–1147. doi: 10.1158/2326-6066.CIR-18-0648. [DOI] [PubMed] [Google Scholar]
- 386.Li D, et al. miR3609 sensitizes breast cancer cells to adriamycin by blocking the programmed death-ligand 1 immune checkpoint. Exp. Cell Res. 2019;380:20–28. doi: 10.1016/j.yexcr.2019.03.025. [DOI] [PubMed] [Google Scholar]
- 387.Liao Y, et al. Targeting programmed cell death ligand 1 by CRISPR/Cas9 in osteosarcoma cells. Oncotarget. 2017;8:30276–30287. doi: 10.18632/oncotarget.16326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 388.Shen M, et al. Inhibition of ATM reverses EMT and decreases metastatic potential of cisplatin-resistant lung cancer cells through JAK/STAT3/PD-L1 pathway. J. Exp. Clin. Cancer Res. 2019;38:149. doi: 10.1186/s13046-019-1161-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 389.Shen MJ, et al. Radiation alters PD-L1/NKG2D ligand levels in lung cancer cells and leads to immune escape from NK cell cytotoxicity via IL-6-MEK/Erk signaling pathway. Oncotarget. 2017;8:80506–80520. doi: 10.18632/oncotarget.19193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 390.Lu W, Kang Y. Epithelial- Dev. Cell. 2019;49:361–374. doi: 10.1016/j.devcel.2019.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 391.De Matteis S, et al. Advances in molecular mechanisms and immunotherapy involving the immune cell-promoted epithelial-to-mesenchymal transition in lung cancer. J. Oncol. 2019;2019:7475364. doi: 10.1155/2019/7475364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 392.Akalay I, et al. Epithelial-to-mesenchymal transition and autophagy induction in breast carcinoma promote escape from T-cell-mediated lysis. Cancer Res. 2013;73:2418–2427. doi: 10.1158/0008-5472.CAN-12-2432. [DOI] [PubMed] [Google Scholar]
- 393.Noman MZ, et al. CD47 is a direct target of SNAI1 and ZEB1 and its blockade activates the phagocytosis of breast cancer cells undergoing EMT. Oncoimmunology. 2018;7:e1345415. doi: 10.1080/2162402X.2017.1345415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 394.Yuan J, et al. High expression of CD47 in triple negative breast cancer is associated with epithelial-mesenchymal transition and poor prognosis. Oncol. Lett. 2019;18:3249–3255. doi: 10.3892/ol.2019.10618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 395.Liang J, et al. The correlation between the immune and epithelial-mesenchymal transition signatures suggests potential therapeutic targets and prognosis prediction approaches in kidney cancer. Sci. Rep. 2018;8:6570. doi: 10.1038/s41598-018-25002-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 396.Lequeux A, et al. Impact of hypoxic tumor microenvironment and tumor cell plasticity on the expression of immune checkpoints. Cancer Lett. 2019;458:13–20. doi: 10.1016/j.canlet.2019.05.021. [DOI] [PubMed] [Google Scholar]
- 397.Harvey KF, Zhang X, Thomas DM. The Hippo pathway and human cancer. Nat. Rev. Cancer. 2013;13:246–257. doi: 10.1038/nrc3458. [DOI] [PubMed] [Google Scholar]
- 398.Moroishi T, Hansen CG, Guan KL. The emerging roles of YAP and TAZ in cancer. Nat. Rev. Cancer. 2015;15:73–79. doi: 10.1038/nrc3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 399.Zanconato F, Cordenonsi M, Piccolo S. YAP/TAZ at the roots of cancer. Cancer Cell. 2016;29:783–803. doi: 10.1016/j.ccell.2016.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 400.Hong AW, Meng Z, Guan KL. The Hippo pathway in intestinal regeneration and disease. Nat. Rev. Gastroenterol. Hepatol. 2016;13:324–337. doi: 10.1038/nrgastro.2016.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 401.Xie Q, et al. YAP/TEAD-mediated transcription controls cellular senescence. Cancer Res. 2013;73:3615–3624. doi: 10.1158/0008-5472.CAN-12-3793. [DOI] [PubMed] [Google Scholar]
- 402.Lin L, et al. The Hippo effector YAP promotes resistance to RAF- and MEK-targeted cancer therapies. Nat. Genet. 2015;47:250–256. doi: 10.1038/ng.3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 403.Keren-Paz A, Emmanuel R, Samuels Y. YAP and the drug resistance highway. Nat. Genet. 2015;47:193–194. doi: 10.1038/ng.3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 404.Coggins GE, et al. YAP1 mediates resistance to MEK1/2 inhibition in neuroblastomas with hyperactivated RAS signaling. Cancer Res. 2019;79:6204–6214. doi: 10.1158/0008-5472.CAN-19-1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 405.Bainbridge A, et al. IKBKE activity enhances AR levels in advanced prostate cancer via modulation of the Hippo pathway. Nucleic Acids Res. 2020;48:5366–5382. doi: 10.1093/nar/gkaa271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 406.Huang Z, et al. PDLIM1 inhibits tumor metastasis through activating Hippo signaling in hepatocellular carcinoma. Hepatology. 2020;71:1643–1659. doi: 10.1002/hep.30930. [DOI] [PubMed] [Google Scholar]
- 407.Yun MR, et al. Targeting YAP to overcome acquired resistance to ALK inhibitors in ALK-rearranged lung cancer. EMBO Mol. Med. 2019;11:e10581. doi: 10.15252/emmm.201910581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 408.Zhang Z, et al. Activation of the AXL kinase causes resistance to EGFR-targeted therapy in lung cancer. Nat. Genet. 2012;44:852–860. doi: 10.1038/ng.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 409.Yu M, et al. YAP1 contributes to NSCLC invasion and migration by promoting Slug transcription via the transcription co-factor TEAD. Cell Death Dis. 2018;9:464. doi: 10.1038/s41419-018-0515-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 410.Ghiso E, et al. YAP-dependent AXL overexpression mediates resistance to EGFR inhibitors in NSCLC. Neoplasia. 2017;19:1012–1021. doi: 10.1016/j.neo.2017.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 411.Vuoriluoto K, et al. Vimentin regulates EMT induction by Slug and oncogenic H-Ras and migration by governing Axl expression in breast cancer. Oncogene. 2011;30:1436–1448. doi: 10.1038/onc.2010.509. [DOI] [PubMed] [Google Scholar]
- 412.Xu MZ, et al. AXL receptor kinase is a mediator of YAP-dependent oncogenic functions in hepatocellular carcinoma. Oncogene. 2011;30:1229–1240. doi: 10.1038/onc.2010.504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 413.Okimoto RA, Bivona TG. AXL receptor tyrosine kinase as a therapeutic target in NSCLC. Lung Cancer. 2015;6:27–34. doi: 10.2147/LCTT.S60438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 414.Gjerdrum C, et al. Axl is an essential epithelial-to-mesenchymal transition-induced regulator of breast cancer metastasis and patient survival. Proc. Natl Acad. Sci. USA. 2010;107:1124–1129. doi: 10.1073/pnas.0909333107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 415.Antony J, Huang RY. AXL-driven EMT state as a targetable conduit in cancer. Cancer Res. 2017;77:3725–3732. doi: 10.1158/0008-5472.CAN-17-0392. [DOI] [PubMed] [Google Scholar]
- 416.Suda K, et al. Epithelial to mesenchymal transition in an epidermal growth factor receptor-mutant lung cancer cell line with acquired resistance to erlotinib. J. Thorac. Oncol. 2011;6:1152–1161. doi: 10.1097/JTO.0b013e318216ee52. [DOI] [PubMed] [Google Scholar]
- 417.Yauch RL, et al. Epithelial versus mesenchymal phenotype determines in vitro sensitivity and predicts clinical activity of erlotinib in lung cancer patients. Clin. Cancer Res. 2005;11:8686–8698. doi: 10.1158/1078-0432.CCR-05-1492. [DOI] [PubMed] [Google Scholar]
- 418.Lee Y, Lee M, Kim S. Gas6 induces cancer cell migration and epithelial-mesenchymal transition through upregulation of MAPK and Slug. Biochem. Biophys. Res. Commun. 2013;434:8–14. doi: 10.1016/j.bbrc.2013.03.082. [DOI] [PubMed] [Google Scholar]
- 419.Chang TH, et al. Slug confers resistance to the epidermal growth factor receptor tyrosine kinase inhibitor. Am. J. Respir. Crit. Care Med. 2011;183:1071–1079. doi: 10.1164/rccm.201009-1440OC. [DOI] [PubMed] [Google Scholar]
- 420.Wu WS, et al. Slug antagonizes p53-mediated apoptosis of hematopoietic progenitors by repressing puma. Cell. 2005;123:641–653. doi: 10.1016/j.cell.2005.09.029. [DOI] [PubMed] [Google Scholar]
- 421.Wu DW, et al. FHIT loss confers cisplatin resistance in lung cancer via the AKT/NF-κB/Slug-mediated PUMA reduction. Oncogene. 2015;34:2505–2515. doi: 10.1038/onc.2014.184. [DOI] [PubMed] [Google Scholar]
- 422.Sun Q, et al. Proapoptotic PUMA targets stem-like breast cancer cells to suppress metastasis. J. Clin. Invest. 2018;128:531–544. doi: 10.1172/JCI93707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 423.Sheridan C. First Axl inhibitor enters clinical trials. Nat. Biotechnol. 2013;31:775–776. doi: 10.1038/nbt0913-775a. [DOI] [PubMed] [Google Scholar]
- 424.Byers L, et al. A phase lilt and pharmacokinetic study of BGB324, a selective AXL inhibitor as monotherapy and in combination with erlotinib in patients with advanced non-small cell lung cancer (NSCLC) Eur. J. Cancer. 2016;69:S18–S19. doi: 10.1016/S0959-8049(16)32636-3. [DOI] [Google Scholar]
- 425.Leconet W, et al. Therapeutic activity of anti-AXL antibody against triple-negative breast cancer patient-derived xenografts and metastasis. Clin. Cancer Res. 2017;23:2806–2816. doi: 10.1158/1078-0432.CCR-16-1316. [DOI] [PubMed] [Google Scholar]
- 426.Kariolis MS, et al. An engineered Axl ‘decoy receptor’ effectively silences the Gas6-Axl signaling axis. Nat. Chem. Biol. 2014;10:977–983. doi: 10.1038/nchembio.1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 427.Kariolis MS, et al. Inhibition of the GAS6/AXL pathway augments the efficacy of chemotherapies. J. Clin. Invest. 2017;127:183–198. doi: 10.1172/JCI85610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 428.Shen Y, et al. Axl inhibitors as novel cancer therapeutic agents. Life Sci. 2018;198:99–111. doi: 10.1016/j.lfs.2018.02.033. [DOI] [PubMed] [Google Scholar]
- 429.Myers SH, Brunton VG, Unciti-Broceta A. AXL inhibitors in cancer: a medicinal chemistry perspective. J. Med. Chem. 2016;59:3593–3608. doi: 10.1021/acs.jmedchem.5b01273. [DOI] [PubMed] [Google Scholar]
- 430.Soria JC, et al. Osimertinib in untreated EGFR-mutated advanced non-small-cell lung cancer. N. Engl. J. Med. 2018;378:113–125. doi: 10.1056/NEJMoa1713137. [DOI] [PubMed] [Google Scholar]
- 431.Ramalingam SS, et al. Overall survival with osimertinib in untreated, EGFR-mutated advanced NSCLC. N. Engl. J. Med. 2020;382:41–50. doi: 10.1056/NEJMoa1913662. [DOI] [PubMed] [Google Scholar]
- 432.Cross DA, et al. AZD9291, an irreversible EGFR TKI, overcomes T790M-mediated resistance to EGFR inhibitors in lung cancer. Cancer Discov. 2014;4:1046–1061. doi: 10.1158/2159-8290.CD-14-0337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 433.Piotrowska Z, et al. Landscape of acquired resistance to osimertinib in EGFR-mutant NSCLC and clinical validation of combined EGFR and RET inhibition with osimertinib and BLU-667 for acquired RET fusion. Cancer Discov. 2018;8:1529–1539. doi: 10.1158/2159-8290.CD-18-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 434.Mok TS, et al. Osimertinib or platinum-pemetrexed in EGFR T790M-positive lung cancer. N. Engl. J. Med. 2017;376:629–640. doi: 10.1056/NEJMoa1612674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 435.Niederst MJ, et al. The allelic context of the C797S mutation acquired upon treatment with third-generation EGFR inhibitors impacts sensitivity to subsequent treatment strategies. Clin. Cancer Res. 2015;21:3924–3933. doi: 10.1158/1078-0432.CCR-15-0560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 436.Yu HA, et al. Acquired resistance of EGFR-mutant lung cancer to a T790M-specific egfr inhibitor: emergence of a third mutation (C797S) in the EGFR tyrosine kinase domain. JAMA Oncol. 2015;1:982–984. doi: 10.1001/jamaoncol.2015.1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 437.Ou SI, Agarwal N, Ali SM. High MET amplification level as a resistance mechanism to osimertinib (AZD9291) in a patient that symptomatically responded to crizotinib treatment post-osimertinib progression. Lung Cancer. 2016;98:59–61. doi: 10.1016/j.lungcan.2016.05.015. [DOI] [PubMed] [Google Scholar]
- 438.Planchard D, et al. EGFR-independent mechanisms of acquired resistance to AZD9291 in EGFR T790M-positive NSCLC patients. Ann. Oncol. 2015;26:2073–2078. doi: 10.1093/annonc/mdv319. [DOI] [PubMed] [Google Scholar]
- 439.Eberlein CA, et al. Acquired resistance to the mutant-selective EGFR inhibitor AZD9291 is associated with increased dependence on RAS signaling in preclinical models. Cancer Res. 2015;75:2489–2500. doi: 10.1158/0008-5472.CAN-14-3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 440.Namba K, et al. Activation of AXL as a preclinical acquired resistance mechanism against osimertinib treatment in EGFR-mutant non-small cell lung cancer cells. Mol. Cancer Res. 2019;17:499–507. doi: 10.1158/1541-7786.MCR-18-0628. [DOI] [PubMed] [Google Scholar]
- 441.Taniguchi H, et al. AXL confers intrinsic resistance to osimertinib and advances the emergence of tolerant cells. Nat. Commun. 2019;10:259. doi: 10.1038/s41467-018-08074-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 442.Kim D, et al. AXL degradation in combination with EGFR-TKI can delay and overcome acquired resistance in human non-small cell lung cancer cells. Cell Death Dis. 2019;10:361. doi: 10.1038/s41419-019-1601-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 443.Okura N, et al. ONO-7475, a novel AXL inhibitor, suppresses the adaptive resistance to initial EGFR-TKI treatment in EGFR-mutated non-small cell lung cancer. Clin. Cancer Res. 2020;26:2244–2256. doi: 10.1158/1078-0432.CCR-19-2321. [DOI] [PubMed] [Google Scholar]
- 444.Lin YC, et al. DUSP1 expression induced by HDAC1 inhibition mediates gefitinib sensitivity in non-small cell lung cancers. Clin. Cancer Res. 2015;21:428–438. doi: 10.1158/1078-0432.CCR-14-1150. [DOI] [PubMed] [Google Scholar]
- 445.Weng CH, et al. Epithelial-mesenchymal transition (EMT) beyond EGFR mutations per se is a common mechanism for acquired resistance to EGFR TKI. Oncogene. 2019;38:455–468. doi: 10.1038/s41388-018-0454-2. [DOI] [PubMed] [Google Scholar]
- 446.Raoof S, et al. Targeting FGFR overcomes EMT-mediated resistance in EGFR mutant non-small cell lung cancer. Oncogene. 2019;38:6399–6413. doi: 10.1038/s41388-019-0887-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 447.Tricker EM, et al. Combined EGFR/MEK inhibition prevents the emergence of resistance in EGFR-mutant lung cancer. Cancer Discov. 2015;5:960–971. doi: 10.1158/2159-8290.CD-15-0063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 448.Vojnic M, et al. Acquired BRAF rearrangements induce secondary resistance to EGFR therapy in EGFR-mutated lung cancers. J. Thorac. Oncol. 2019;14:802–815. doi: 10.1016/j.jtho.2018.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 449.Ramalingam, S. S. et al. Osimertinib plus selumetinib for patients (pts) with EGFR-mutant (EGFRm) NSCLC following disease progression on an EGFR-TKI: results from the Phase Ib TATTON study. Cancer Res. 79 (American Association for Cancer Research Annual Meeting Abstract, 2019).
- 450.Kurppa KJ, et al. Treatment-induced tumor dormancy through YAP-mediated transcriptional reprogramming of the apoptotic pathway. Cancer Cell. 2020;37:104–122.e112. doi: 10.1016/j.ccell.2019.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 451.Bado I, Zhang XH. Senesce to survive: YAP-mediated dormancy escapes EGFR/MEK inhibition. Cancer Cell. 2020;37:1–2. doi: 10.1016/j.ccell.2019.12.008. [DOI] [PubMed] [Google Scholar]
- 452.Wang C, et al. Tumor-derived secretory clusterin induces epithelial-mesenchymal transition and facilitates hepatocellular carcinoma metastasis. Int. J. Biochem. Cell Biol. 2012;44:2308–2320. doi: 10.1016/j.biocel.2012.09.012. [DOI] [PubMed] [Google Scholar]
- 453.Simonelli M, et al. Phase I study of PF-03446962, a fully human monoclonal antibody against activin receptor-like kinase-1, in patients with hepatocellular carcinoma. Ann. Oncol. 2016;27:1782–1787. doi: 10.1093/annonc/mdw240. [DOI] [PubMed] [Google Scholar]
- 454.Clarke JM, et al. A phase Ib study of the combination regorafenib with PF-03446962 in patients with refractory metastatic colorectal cancer (REGAL-1 trial) Cancer Chemother. Pharmacol. 2019;84:909–917. doi: 10.1007/s00280-019-03916-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 455.Pattabiraman DR, Weinberg RA. Tackling the cancer stem cells - what challenges do they pose? Nat. Rev. Drug Discov. 2014;13:497–512. doi: 10.1038/nrd4253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 456.Prasetyanti PR, Medema JP. Intra-tumor heterogeneity from a cancer stem cell perspective. Mol. Cancer. 2017;16:41. doi: 10.1186/s12943-017-0600-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 457.Das M, Law S. Role of tumor microenvironment in cancer stem cell chemoresistance and recurrence. Int. J. Biochem. Cell Biol. 2018;103:115–124. doi: 10.1016/j.biocel.2018.08.011. [DOI] [PubMed] [Google Scholar]
- 458.Batlle E, Clevers H. Cancer stem cells revisited. Nat. Med. 2017;23:1124–1134. doi: 10.1038/nm.4409. [DOI] [PubMed] [Google Scholar]
- 459.Lytle NK, Barber AG, Reya T. Stem cell fate in cancer growth, progression and therapy resistance. Nat. Rev. Cancer. 2018;18:669–680. doi: 10.1038/s41568-018-0056-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 460.Tang DG. Understanding cancer stem cell heterogeneity and plasticity. Cell Res. 2012;22:457–472. doi: 10.1038/cr.2012.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 461.Peiris-Pagès M, et al. Cancer stem cell metabolism. Breast Cancer Res. 2016;18:55. doi: 10.1186/s13058-016-0712-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 462.Santoro A, et al. Molecular mechanisms of asymmetric divisions in mammary stem cells. EMBO Rep. 2016;17:1700–1720. doi: 10.15252/embr.201643021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 463.Wu M, et al. Imaging hematopoietic precursor division in real time. Cell Stem Cell. 2007;1:541–554. doi: 10.1016/j.stem.2007.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 464.Najafi M, Mortezaee K, Ahadi R. Cancer stem cell (a)symmetry & plasticity: tumorigenesis and therapy relevance. Life Sci. 2019;231:116520. doi: 10.1016/j.lfs.2019.05.076. [DOI] [PubMed] [Google Scholar]
- 465.Venkei ZG, Yamashita YM. Emerging mechanisms of asymmetric stem cell division. J. Cell Biol. 2018;217:3785–3795. doi: 10.1083/jcb.201807037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 466.Liu D, et al. Asymmetric division gene Neurl2 mediates Twist2 regulation of self-renewal of mouse Lewis lung cancer stem cells. J. Cancer. 2019;10:3381–3388. doi: 10.7150/jca.31553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 467.Wu MJ, et al. Epithelial-mesenchymal transition directs stem cell polarity via regulation of mitofusin. Cell Metab. 2019;29:993–1002.e1006. doi: 10.1016/j.cmet.2018.11.004. [DOI] [PubMed] [Google Scholar]
- 468.Thomas D, Majeti R. Biology and relevance of human acute myeloid leukemia stem cells. Blood. 2017;129:1577–1585. doi: 10.1182/blood-2016-10-696054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 469.Yang F, Xu J, Tang L, Guan X. Breast cancer stem cell: the roles and therapeutic implications. Cell. Mol. Life Sci. 2017;74:951–966. doi: 10.1007/s00018-016-2334-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 470.Zhang S, et al. Inhibition of chemotherapy resistant breast cancer stem cells by a ROR1 specific antibody. Proc. Natl Acad. Sci. USA. 2019;116:1370–1377. doi: 10.1073/pnas.1816262116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 471.Tsao T, et al. Cancer stem cells in prostate cancer radioresistance. Cancer Lett. 2019;465:94–104. doi: 10.1016/j.canlet.2019.08.020. [DOI] [PubMed] [Google Scholar]
- 472.Mei, W. et al. The contributions of prostate cancer stem cells in prostate cancer initiation and metastasis. Cancers11, 434 (2019). [DOI] [PMC free article] [PubMed]
- 473.Wahab SMR, Islam F, Gopalan V, Lam AK. The identifications and clinical implications of cancer stem cells in colorectal cancer. Clin. Colorectal Cancer. 2017;16:93–102. doi: 10.1016/j.clcc.2017.01.011. [DOI] [PubMed] [Google Scholar]
- 474.Zhou Y, et al. Cancer stem cells in progression of colorectal cancer. Oncotarget. 2018;9:33403–33415. doi: 10.18632/oncotarget.23607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 475.Parmiani, G. Melanoma cancer stem cells: markers and functions. Cancers8, 34 (2016). [DOI] [PMC free article] [PubMed]
- 476.Gimple RC, Bhargava S, Dixit D, Rich JN. Glioblastoma stem cells: lessons from the tumor hierarchy in a lethal cancer. Genes Dev. 2019;33:591–609. doi: 10.1101/gad.324301.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 477.Easwaran H, Tsai HC, Baylin SB. Cancer epigenetics: tumor heterogeneity, plasticity of stem-like states, and drug resistance. Mol. Cell. 2014;54:716–727. doi: 10.1016/j.molcel.2014.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 478.Iyer AK, Singh A, Ganta S, Amiji MM. Role of integrated cancer nanomedicine in overcoming drug resistance. Adv. Drug Deliv. Rev. 2013;65:1784–1802. doi: 10.1016/j.addr.2013.07.012. [DOI] [PubMed] [Google Scholar]
- 479.Semenza GL. Hypoxia-inducible factors: coupling glucose metabolism and redox regulation with induction of the breast cancer stem cell phenotype. EMBO J. 2017;36:252–259. doi: 10.15252/embj.201695204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 480.Vitale I, et al. DNA damage in stem cells. Mol. Cell. 2017;66:306–319. doi: 10.1016/j.molcel.2017.04.006. [DOI] [PubMed] [Google Scholar]
- 481.Merrell AJ, Stanger BZ. Adult cell plasticity in vivo: de-differentiation and transdifferentiation are back in style. Nat. Rev. Mol. Cell. Biol. 2016;17:413–425. doi: 10.1038/nrm.2016.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 482.Tetteh PW, Farin HF, Clevers H. Plasticity within stem cell hierarchies in mammalian epithelia. Trends Cell Biol. 2015;25:100–108. doi: 10.1016/j.tcb.2014.09.003. [DOI] [PubMed] [Google Scholar]
- 483.Beltran H, et al. The role of lineage plasticity in prostate cancer therapy resistance. Clin. Cancer Res. 2019;25:6916–6924. doi: 10.1158/1078-0432.CCR-18-1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 484.de Thé H. Differentiation therapy revisited. Nat. Rev. Cancer. 2018;18:117–127. doi: 10.1038/nrc.2017.103. [DOI] [PubMed] [Google Scholar]
- 485.Fang D, et al. A tumorigenic subpopulation with stem cell properties in melanomas. Cancer Res. 2005;65:9328–9337. doi: 10.1158/0008-5472.CAN-05-1343. [DOI] [PubMed] [Google Scholar]
- 486.Kakizuka A, et al. Chromosomal translocation t(15;17) in human acute promyelocytic leukemia fuses RAR alpha with a novel putative transcription factor, PML. Cell. 1991;66:663–674. doi: 10.1016/0092-8674(91)90112-C. [DOI] [PubMed] [Google Scholar]
- 487.Kersten S, Desvergne B, Wahli W. Roles of PPARs in health and disease. Nature. 2000;405:421–424. doi: 10.1038/35013000. [DOI] [PubMed] [Google Scholar]
- 488.Kahn BB, McGraw TE. Rosiglitazone, PPARγ, and type 2 diabetes. N. Engl. J. Med. 2010;363:2667–2669. doi: 10.1056/NEJMcibr1012075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 489.Frapolli R, et al. Combination of PPARγ agonist pioglitazone and trabectedin induce adipocyte differentiation to overcome trabectedin resistance in myxoid liposarcomas. Clin. Cancer Res. 2019;25:7565–7575. doi: 10.1158/1078-0432.CCR-19-0976. [DOI] [PubMed] [Google Scholar]
- 490.Charytonowicz E, et al. PPARγ agonists enhance ET-743-induced adipogenic differentiation in a transgenic mouse model of myxoid round cell liposarcoma. J. Clin. Invest. 2012;122:886–898. doi: 10.1172/JCI60015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 491.Pestereva E, Kanakasabai S, Bright JJ. PPARγ agonists regulate the expression of stemness and differentiation genes in brain tumour stem cells. Br. J. Cancer. 2012;106:1702–1712. doi: 10.1038/bjc.2012.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 492.Zheng ZH, et al. Mycophenolic acid induces adipocyte-like differentiation and reversal of malignancy of breast cancer cells partly through PPARγ. Eur. J. Pharmacol. 2011;658:1–8. doi: 10.1016/j.ejphar.2011.01.068. [DOI] [PubMed] [Google Scholar]
- 493.Prost S, et al. Erosion of the chronic myeloid leukaemia stem cell pool by PPARγ agonists. Nature. 2015;525:380–383. doi: 10.1038/nature15248. [DOI] [PubMed] [Google Scholar]
- 494.Kopinke D, Roberson EC, Reiter JF. Ciliary Hedgehog signaling restricts injury-induced adipogenesis. Cell. 2017;170:340–351.e312. doi: 10.1016/j.cell.2017.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 495.Wang QA, Scherer PE, Gupta RK. Improved methodologies for the study of adipose biology: insights gained and opportunities ahead. J. Lipid Res. 2014;55:605–624. doi: 10.1194/jlr.R046441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 496.Waldmeier L, Meyer-Schaller N, Diepenbruck M, Christofori G. Py2T murine breast cancer cells, a versatile model of TGFβ-induced EMT in vitro and in vivo. PLoS ONE. 2012;7:e48651. doi: 10.1371/journal.pone.0048651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 497.Ishay-Ronen D, et al. Gain fat-lose metastasis: converting invasive breast cancer cells into adipocytes inhibits cancer metastasis. Cancer Cell. 2019;35:17–32.e16. doi: 10.1016/j.ccell.2018.12.002. [DOI] [PubMed] [Google Scholar]
- 498.Hinz S, LaBarge MA. Hijacking EMT: better fat than dead. Cancer Cell. 2019;35:1–2. doi: 10.1016/j.ccell.2018.12.007. [DOI] [PubMed] [Google Scholar]
- 499.Ishay-Ronen D, Christofori G. Targeting cancer cell metastasis by converting cancer cells into fat. Cancer Res. 2019;79:5471–5475. doi: 10.1158/0008-5472.CAN-19-1242. [DOI] [PubMed] [Google Scholar]
- 500.Fantozzi A, et al. VEGF-mediated angiogenesis links EMT-induced cancer stemness to tumor initiation. Cancer Res. 2014;74:1566–1575. doi: 10.1158/0008-5472.CAN-13-1641. [DOI] [PubMed] [Google Scholar]
- 501.Lefterova MI, et al. PPARgamma and C/EBP factors orchestrate adipocyte biology via adjacent binding on a genome-wide scale. Genes Dev. 2008;22:2941–2952. doi: 10.1101/gad.1709008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 502.Greenspan P, Mayer EP, Fowler SD. Nile red: a selective fluorescent stain for intracellular lipid droplets. J. Cell Biol. 1985;100:965–973. doi: 10.1083/jcb.100.3.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 503.Battula VL, et al. Epithelial-mesenchymal transition-derived cells exhibit multilineage differentiation potential similar to mesenchymal stem cells. Stem Cells. 2010;28:1435–1445. doi: 10.1002/stem.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 504.Sinha S, et al. Pbrm1 steers mesenchymal stromal cell osteolineage differentiation by integrating PBAF-dependent chromatin remodeling and BMP/TGF-β signaling. Cell Rep. 2020;31:107570. doi: 10.1016/j.celrep.2020.107570. [DOI] [PubMed] [Google Scholar]
- 505.Gao W, et al. Melatonin enhances chondrogenic differentiation of human mesenchymal stem cells. J. Pineal Res. 2014;56:62–70. doi: 10.1111/jpi.12098. [DOI] [PubMed] [Google Scholar]
- 506.Liu Y, et al. Intracellular VEGF regulates the balance between osteoblast and adipocyte differentiation. J. Clin. Invest. 2012;122:3101–3113. doi: 10.1172/JCI61209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 507.Wilson, M. M., Weinberg, R. A., Lees, J. A. & Guen, V. J. Emerging mechanisms by which EMT programs control stemness. Trends Cancer6, 775–780 (2020). [DOI] [PubMed]
- 508.Zamani N, Brown CW. Emerging roles for the transforming growth factor-{beta} superfamily in regulating adiposity and energy expenditure. Endocr. Rev. 2011;32:387–403. doi: 10.1210/er.2010-0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 509.Luo H, et al. Growth differentiation factor 11 inhibits adipogenic differentiation by activating TGF-beta/Smad signalling pathway. Cell Prolif. 2019;52:e12631. doi: 10.1111/cpr.12631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 510.Recasens A, Munoz L. Targeting cancer cell dormancy. Trends Pharmacol. Sci. 2019;40:128–141. doi: 10.1016/j.tips.2018.12.004. [DOI] [PubMed] [Google Scholar]
- 511.Searle EJ, et al. Akt inhibition improves long-term tumour control following radiotherapy by altering the microenvironment. EMBO Mol. Med. 2017;9:1646–1659. doi: 10.15252/emmm.201707767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 512.Wang ZY, Chen Z. Acute promyelocytic leukemia: from highly fatal to highly curable. Blood. 2008;111:2505–2515. doi: 10.1182/blood-2007-07-102798. [DOI] [PubMed] [Google Scholar]
- 513.Kenny PA, Bissell MJ. Tumor reversion: correction of malignant behavior by microenvironmental cues. Int. J. Cancer. 2003;107:688–695. doi: 10.1002/ijc.11491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 514.Tallman MS, Nabhan C, Feusner JH, Rowe JM. Acute promyelocytic leukemia: evolving therapeutic strategies. Blood. 2002;99:759–767. doi: 10.1182/blood.V99.3.759. [DOI] [PubMed] [Google Scholar]
- 515.Zhu J, Lallemand-Breitenbach V, de Thé H. Pathways of retinoic acid- or arsenic trioxide-induced PML/RARalpha catabolism, role of oncogene degradation in disease remission. Oncogene. 2001;20:7257–7265. doi: 10.1038/sj.onc.1204852. [DOI] [PubMed] [Google Scholar]
- 516.Segalla S, et al. Retinoic acid receptor alpha fusion to PML affects its transcriptional and chromatin-remodeling properties. Mol. Cell. Biol. 2003;23:8795–8808. doi: 10.1128/MCB.23.23.8795-8808.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 517.Zhang, J. & Liu, F. The de-, re-, and trans-differentiation of β-cells: regulation and function. Semin. Cell Dev. Biol. 103, 68–75 (2020). [DOI] [PubMed]
- 518.Nadkarni RR, Abed S, Draper JS. Stem cells in pulmonary disease and regeneration. Chest. 2018;153:994–1003. doi: 10.1016/j.chest.2017.07.021. [DOI] [PubMed] [Google Scholar]
- 519.Park J, et al. Obesity and cancer—mechanisms underlying tumour progression and recurrence. Nat. Rev. Endocrinol. 2014;10:455–465. doi: 10.1038/nrendo.2014.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 520.Lengyel E, Makowski L, DiGiovanni J, Kolonin MG. Cancer as a matter of fat: the crosstalk between adipose tissue and tumors. Trends Cancer. 2018;4:374–384. doi: 10.1016/j.trecan.2018.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 521.Quintanal-Villalonga Á, et al. Lineage plasticity in cancer: a shared pathway of therapeutic resistance. Nat. Rev. Clin. Oncol. 2020;17:360–371. doi: 10.1038/s41571-020-0340-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 522.Huggins C, Hodges CV. Studies on prostatic cancer. I. The effect of castration, of estrogen and androgen injection on serum phosphatases in metastatic carcinoma of the prostate. CA Cancer J. Clin. 1972;22:232–240. doi: 10.3322/canjclin.22.4.232. [DOI] [PubMed] [Google Scholar]
- 523.Knudsen KE, Scher HI. Starving the addiction: new opportunities for durable suppression of AR signaling in prostate cancer. Clin. Cancer Res. 2009;15:4792–4798. doi: 10.1158/1078-0432.CCR-08-2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 524.Niu Y, et al. ADT with antiandrogens in prostate cancer induces adverse effect of increasing resistance, neuroendocrine differentiation and tumor metastasis. Cancer Lett. 2018;439:47–55. doi: 10.1016/j.canlet.2018.09.020. [DOI] [PubMed] [Google Scholar]
- 525.Watson PA, Arora VK, Sawyers CL. Emerging mechanisms of resistance to androgen receptor inhibitors in prostate cancer. Nat. Rev. Cancer. 2015;15:701–711. doi: 10.1038/nrc4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 526.Mansinho A, Macedo D, Fernandes I, Costa L. Castration-resistant prostate cancer: mechanisms, targets and treatment. Adv. Exp. Med. Biol. 2018;1096:117–133. doi: 10.1007/978-3-319-99286-0_7. [DOI] [PubMed] [Google Scholar]
- 527.Hotte SJ, Saad F. Current management of castrate-resistant prostate cancer. Curr. Oncol. 2010;17(Suppl. 2):S72–S79. doi: 10.3747/co.v17i0.718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 528.Lowrance WT, et al. Castration-resistant prostate cancer: AUA Guideline Amendment 2018. J. Urol. 2018;200:1264–1272. doi: 10.1016/j.juro.2018.07.090. [DOI] [PubMed] [Google Scholar]
- 529.Scher HI, et al. Increased survival with enzalutamide in prostate cancer after chemotherapy. N. Engl. J. Med. 2012;367:1187–1197. doi: 10.1056/NEJMoa1207506. [DOI] [PubMed] [Google Scholar]
- 530.Parker C, Sartor O. Abiraterone and increased survival in metastatic prostate cancer. N. Engl. J. Med. 2011;365:767. doi: 10.1056/NEJMc1107198. [DOI] [PubMed] [Google Scholar]
- 531.Roubaud G, Liaw BC, Oh WK, Mulholland DJ. Strategies to avoid treatment-induced lineage crisis in advanced prostate cancer. Nat. Rev. Clin. Oncol. 2017;14:269–283. doi: 10.1038/nrclinonc.2016.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 532.Epstein JI, et al. Proposed morphologic classification of prostate cancer with neuroendocrine differentiation. Am. J. Surg. Pathol. 2014;38:756–767. doi: 10.1097/PAS.0000000000000208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 533.Beltran H, et al. Molecular characterization of neuroendocrine prostate cancer and identification of new drug targets. Cancer Discov. 2011;1:487–495. doi: 10.1158/2159-8290.CD-11-0130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 534.Beltran H, et al. Divergent clonal evolution of castration-resistant neuroendocrine prostate cancer. Nat. Med. 2016;22:298–305. doi: 10.1038/nm.4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 535.Tan HL, et al. Rb loss is characteristic of prostatic small cell neuroendocrine carcinoma. Clin. Cancer Res. 2014;20:890–903. doi: 10.1158/1078-0432.CCR-13-1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 536.Conteduca V, et al. Clinical features of neuroendocrine prostate cancer. Eur. J. Cancer. 2019;121:7–18. doi: 10.1016/j.ejca.2019.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 537.Palmgren JS, Karavadia SS, Wakefield MR. Unusual and underappreciated: small cell carcinoma of the prostate. Semin. Oncol. 2007;34:22–29. doi: 10.1053/j.seminoncol.2006.10.026. [DOI] [PubMed] [Google Scholar]
- 538.Guo CC, et al. TMPRSS2-ERG gene fusion in small cell carcinoma of the prostate. Hum. Pathol. 2011;42:11–17. doi: 10.1016/j.humpath.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 539.Lotan TL, et al. ERG gene rearrangements are common in prostatic small cell carcinomas. Mod. Pathol. 2011;24:820–828. doi: 10.1038/modpathol.2011.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 540.Mosquera JM, et al. Prevalence of TMPRSS2-ERG fusion prostate cancer among men undergoing prostate biopsy in the United States. Clin. Cancer Res. 2009;15:4706–4711. doi: 10.1158/1078-0432.CCR-08-2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 541.Jia L, et al. Orphan nuclear receptor TLX contributes to androgen insensitivity in castration-resistant prostate cancer via its repression of androgen receptor transcription. Oncogene. 2018;37:3340–3355. doi: 10.1038/s41388-018-0198-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 542.Skvortsov S, Skvortsova II, Tang DG, Dubrovska A. Concise review: prostate cancer stem cells: current understanding. Stem Cells. 2018;36:1457–1474. doi: 10.1002/stem.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 543.Nouri M, et al. Therapy-induced developmental reprogramming of prostate cancer cells and acquired therapy resistance. Oncotarget. 2017;8:18949–18967. doi: 10.18632/oncotarget.14850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 544.Mu P, et al. SOX2 promotes lineage plasticity and antiandrogen resistance in TP53- and RB1-deficient prostate cancer. Science. 2017;355:84–88. doi: 10.1126/science.aah4307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 545.Ku SY, et al. Rb1 and Trp53 cooperate to suppress prostate cancer lineage plasticity, metastasis, and antiandrogen resistance. Science. 2017;355:78–83. doi: 10.1126/science.aah4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 546.Ben-Porath I, et al. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat. Genet. 2008;40:499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 547.Niu W, et al. In vivo reprogramming of astrocytes to neuroblasts in the adult brain. Nat. Cell Biol. 2013;15:1164–1175. doi: 10.1038/ncb2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 548.Yasumizu Y, et al. MUC1-C regulates lineage plasticity driving progression to neuroendocrine prostate cancer. Nat. Commun. 2020;11:338. doi: 10.1038/s41467-019-14219-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 549.Bhagirath D, et al. BRN4 is a novel driver of neuroendocrine differentiation in castration-resistant prostate cancer and is selectively released in extracellular vesicles with BRN2. Clin. Cancer Res. 2019;25:6532–6545. doi: 10.1158/1078-0432.CCR-19-0498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 550.Zou M, et al. Transdifferentiation as a mechanism of treatment resistance in a mouse model of castration-resistant prostate cancer. Cancer Discov. 2017;7:736–749. doi: 10.1158/2159-8290.CD-16-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 551.Sinha S, Nelson PS. The path of most resistance: transdifferentiation underlies exceptional nonresponses to androgen receptor pathway inhibition in prostate cancer. Cancer Discov. 2017;7:673–674. doi: 10.1158/2159-8290.CD-17-0481. [DOI] [PubMed] [Google Scholar]
- 552.Roca E, et al. Outcome of patients with lung adenocarcinoma with transformation to small-cell lung cancer following tyrosine kinase inhibitors treatment: a systematic review and pooled analysis. Cancer Treat. Rev. 2017;59:117–122. doi: 10.1016/j.ctrv.2017.07.007. [DOI] [PubMed] [Google Scholar]
- 553.Gazdar AF, Bunn PA, Minna JD. Small-cell lung cancer: what we know, what we need to know and the path forward. Nat. Rev. Cancer. 2017;17:725–737. doi: 10.1038/nrc.2017.87. [DOI] [PubMed] [Google Scholar]
- 554.Blackhall F, et al. Will liquid biopsies improve outcomes for patients with small-cell lung cancer? Lancet Oncol. 2018;19:e470–e481. doi: 10.1016/S1470-2045(18)30455-8. [DOI] [PubMed] [Google Scholar]
- 555.Pelosof, L. et al. Proportion of never-smoker non-small cell lung cancer patients at three diverse institutions. J. Natl. Cancer Inst. 109 (2017). [DOI] [PMC free article] [PubMed]
- 556.Oser MG, Niederst MJ, Sequist LV, Engelman JA. Transformation from non-small-cell lung cancer to small-cell lung cancer: molecular drivers and cells of origin. Lancet Oncol. 2015;16:e165–e172. doi: 10.1016/S1470-2045(14)71180-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 557.Roesch A, et al. A temporarily distinct subpopulation of slow-cycling melanoma cells is required for continuous tumor growth. Cell. 2010;141:583–594. doi: 10.1016/j.cell.2010.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 558.Chaffer CL, et al. Normal and neoplastic nonstem cells can spontaneously convert to a stem-like state. Proc. Natl Acad. Sci. USA. 2011;108:7950–7955. doi: 10.1073/pnas.1102454108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 559.Gupta PB, et al. Stochastic state transitions give rise to phenotypic equilibrium in populations of cancer cells. Cell. 2011;146:633–644. doi: 10.1016/j.cell.2011.07.026. [DOI] [PubMed] [Google Scholar]
- 560.Dean M, Fojo T, Bates S. Tumour stem cells and drug resistance. Nat. Rev. Cancer. 2005;5:275–284. doi: 10.1038/nrc1590. [DOI] [PubMed] [Google Scholar]
- 561.Malanchi I, et al. Interactions between cancer stem cells and their niche govern metastatic colonization. Nature. 2011;481:85–89. doi: 10.1038/nature10694. [DOI] [PubMed] [Google Scholar]
- 562.Goyal D, Limesand SW, Goyal R. Epigenetic responses and the developmental origins of health and disease. J. Endocrinol. 2019;242:T105–t119. doi: 10.1530/JOE-19-0009. [DOI] [PubMed] [Google Scholar]
- 563.Amakye D, Jagani Z, Dorsch M. Unraveling the therapeutic potential of the Hedgehog pathway in cancer. Nat. Med. 2013;19:1410–1422. doi: 10.1038/nm.3389. [DOI] [PubMed] [Google Scholar]
- 564.Nüsslein-Volhard C, Wieschaus E. Mutations affecting segment number and polarity in Drosophila. Nature. 1980;287:795–801. doi: 10.1038/287795a0. [DOI] [PubMed] [Google Scholar]
- 565.Takebe N, et al. Targeting Notch, Hedgehog, and Wnt pathways in cancer stem cells: clinical update. Nat. Rev. Clin. Oncol. 2015;12:445–464. doi: 10.1038/nrclinonc.2015.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 566.Yang L, et al. Targeting cancer stem cell pathways for cancer therapy. Signal Transduct. Target Ther. 2020;5:8. doi: 10.1038/s41392-020-0110-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 567.Lagadec C, et al. Radiation-induced reprogramming of breast cancer cells. Stem Cells. 2012;30:833–844. doi: 10.1002/stem.1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 568.Du FY, Zhou QF, Sun WJ, Chen GL. Targeting cancer stem cells in drug discovery: current state and future perspectives. World J. Stem Cells. 2019;11:398–420. doi: 10.4252/wjsc.v11.i7.398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 569.Scales SJ, de Sauvage FJ. Mechanisms of Hedgehog pathway activation in cancer and implications for therapy. Trends Pharmacol. Sci. 2009;30:303–312. doi: 10.1016/j.tips.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 570.Beachy PA, et al. Interactions between Hedgehog proteins and their binding partners come into view. Genes Dev. 2010;24:2001–2012. doi: 10.1101/gad.1951710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 571.Infante P, et al. Inhibition of Hedgehog-dependent tumors and cancer stem cells by a newly identified naturally occurring chemotype. Cell Death Dis. 2016;7:e2376. doi: 10.1038/cddis.2016.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 572.Zhou Q, Kalderon D. Hedgehog activates fused through phosphorylation to elicit a full spectrum of pathway responses. Dev. Cell. 2011;20:802–814. doi: 10.1016/j.devcel.2011.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 573.Varjosalo M, Taipale J. Hedgehog: functions and mechanisms. Genes Dev. 2008;22:2454–2472. doi: 10.1101/gad.1693608. [DOI] [PubMed] [Google Scholar]
- 574.Villegas VE, et al. Tamoxifen treatment of breast cancer cells: impact on Hedgehog/GLI1 signaling. Int. J. Mol. Sci. 2016;17:308. doi: 10.3390/ijms17030308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 575.Pietanza MC, et al. A phase I trial of the Hedgehog inhibitor, sonidegib (LDE225), in combination with etoposide and cisplatin for the initial treatment of extensive stage small cell lung cancer. Lung Cancer. 2016;99:23–30. doi: 10.1016/j.lungcan.2016.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 576.Amantini C, et al. Capsaicin triggers autophagic cell survival which drives epithelial mesenchymal transition and chemoresistance in bladder cancer cells in an Hedgehog-dependent manner. Oncotarget. 2016;7:50180–50194. doi: 10.18632/oncotarget.10326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 577.Xu Y, et al. Inhibition of the Hedgehog pathway induces autophagy in pancreatic ductal adenocarcinoma cells. Oncol. Rep. 2014;31:707–712. doi: 10.3892/or.2013.2881. [DOI] [PubMed] [Google Scholar]
- 578.Yoon C, et al. CD44 expression denotes a subpopulation of gastric cancer cells in which Hedgehog signaling promotes chemotherapy resistance. Clin. Cancer Res. 2014;20:3974–3988. doi: 10.1158/1078-0432.CCR-14-0011. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 579.Wang S, et al. Potential role of Hedgehog pathway in liver response to radiation. PLoS ONE. 2013;8:e74141. doi: 10.1371/journal.pone.0074141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 580.Merchant AA, Matsui W. Targeting Hedgehog—a cancer stem cell pathway. Clin. Cancer Res. 2010;16:3130–3140. doi: 10.1158/1078-0432.CCR-09-2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 581.Cazet AS, et al. Targeting stromal remodeling and cancer stem cell plasticity overcomes chemoresistance in triple negative breast cancer. Nat. Commun. 2018;9:2897. doi: 10.1038/s41467-018-05220-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 582.Sims-Mourtada J, et al. Taxane-induced hedgehog signaling is linked to expansion of breast cancer stem-like populations after chemotherapy. Mol. Carcinogen. 2015;54:1480–1493. doi: 10.1002/mc.22225. [DOI] [PubMed] [Google Scholar]
- 583.Qian W, et al. Cigarette smoke stimulates the stemness of renal cancer stem cells via Sonic Hedgehog pathway. Oncogenesis. 2018;7:24. doi: 10.1038/s41389-018-0029-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 584.Chen JK, Taipale J, Cooper MK, Beachy PA. Inhibition of Hedgehog signaling by direct binding of cyclopamine to Smoothened. Genes Dev. 2002;16:2743–2748. doi: 10.1101/gad.1025302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 585.Iovine V, et al. One hundred faces of cyclopamine. Curr. Pharm. Des. 2016;22:1658–1681. doi: 10.2174/1381612822666160112130157. [DOI] [PubMed] [Google Scholar]
- 586.Dirix L. Discovery and exploitation of novel targets by approved drugs. J. Clin. Oncol. 2014;32:720–721. doi: 10.1200/JCO.2013.53.7118. [DOI] [PubMed] [Google Scholar]
- 587.Li W, et al. Signaling pathway inhibitors target breast cancer stem cells in triple-negative breast cancer. Oncol. Rep. 2019;41:437–446. doi: 10.3892/or.2018.6805. [DOI] [PubMed] [Google Scholar]
- 588.Dummer R, et al. The 12-month analysis from basal cell carcinoma outcomes with LDE225 treatment (BOLT): aphase II, randomized, double-blind study of sonidegib in patients with advanced basal cell carcinoma. J. Am. Acad. Dermatol. 2016;75:113–125.e115. doi: 10.1016/j.jaad.2016.02.1226. [DOI] [PubMed] [Google Scholar]
- 589.Bowles DW, et al. A pilot study of cetuximab and the hedgehog inhibitor IPI-926 in recurrent/metastatic head and neck squamous cell carcinoma. Oral Oncol. 2016;53:74–79. doi: 10.1016/j.oraloncology.2015.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 590.Ueno H, et al. A phase I and pharmacokinetic study of taladegib, a Smoothened inhibitor, in Japanese patients with advanced solid tumors. Invest. N. Drugs. 2018;36:647–656. doi: 10.1007/s10637-017-0544-y. [DOI] [PubMed] [Google Scholar]
- 591.Cortes JE, et al. Randomized comparison of low dose cytarabine with or without glasdegib in patients with newly diagnosed acute myeloid leukemia or high-risk myelodysplastic syndrome. Leukemia. 2019;33:379–389. doi: 10.1038/s41375-018-0312-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 592.Wickström M, et al. Targeting the hedgehog signal transduction pathway at the level of GLI inhibits neuroblastoma cell growth in vitro and in vivo. Int. J. Cancer. 2013;132:1516–1524. doi: 10.1002/ijc.27820. [DOI] [PubMed] [Google Scholar]
- 593.Beauchamp EM, et al. Arsenic trioxide inhibits human cancer cell growth and tumor development in mice by blocking Hedgehog/GLI pathway. J. Clin. Invest. 2011;121:148–160. doi: 10.1172/JCI42874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 594.Long B, et al. Targeting GLI1 suppresses cell growth and enhances chemosensitivity in CD34+ enriched acute myeloid leukemia progenitor cells. Cell. Physiol. Biochem. 2016;38:1288–1302. doi: 10.1159/000443075. [DOI] [PubMed] [Google Scholar]
- 595.Yang W, et al. HDAC6 inhibition induces glioma stem cells differentiation and enhances cellular radiation sensitivity through the SHH/Gli1 signaling pathway. Cancer Lett. 2018;415:164–176. doi: 10.1016/j.canlet.2017.12.005. [DOI] [PubMed] [Google Scholar]
- 596.Nusse, R. Wnt signaling. Cold Spring Harb. Perspect. Biol. 4, a011163 (2012). [DOI] [PMC free article] [PubMed]
- 597.Krausova M, Korinek V. Wnt signaling in adult intestinal stem cells and cancer. Cell Signal. 2014;26:570–579. doi: 10.1016/j.cellsig.2013.11.032. [DOI] [PubMed] [Google Scholar]
- 598.Fan J, et al. Noncanonical Wnt signaling plays an important role in modulating canonical Wnt-regulated stemness, proliferation and terminal differentiation of hepatic progenitors. Oncotarget. 2017;8:27105–27119. doi: 10.18632/oncotarget.15637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 599.van Amerongen R, Nusse R. Towards an integrated view of Wnt signaling in development. Development. 2009;136:3205–3214. doi: 10.1242/dev.033910. [DOI] [PubMed] [Google Scholar]
- 600.MacDonald BT, Tamai K, He X. Wnt/beta-catenin signaling: components, mechanisms, and diseases. Dev. Cell. 2009;17:9–26. doi: 10.1016/j.devcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 601.Dodge ME, Lum L. Drugging the cancer stem cell compartment: lessons learned from the hedgehog and Wnt signal transduction pathways. Annu. Rev. Pharmacol. Toxicol. 2011;51:289–310. doi: 10.1146/annurev-pharmtox-010510-100558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 602.Yook JI, et al. Wnt-dependent regulation of the E-cadherin repressor snail. J. Biol. Chem. 2005;280:11740–11748. doi: 10.1074/jbc.M413878200. [DOI] [PubMed] [Google Scholar]
- 603.Yook JI, et al. A Wnt-Axin2-GSK3beta cascade regulates Snail1 activity in breast cancer cells. Nat. Cell Biol. 2006;8:1398–1406. doi: 10.1038/ncb1508. [DOI] [PubMed] [Google Scholar]
- 604.Gujral TS, et al. A noncanonical Frizzled2 pathway regulates epithelial-mesenchymal transition and metastasis. Cell. 2014;159:844–856. doi: 10.1016/j.cell.2014.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 605.Sandsmark E, et al. A novel non-canonical Wnt signature for prostate cancer aggressiveness. Oncotarget. 2017;8:9572–9586. doi: 10.18632/oncotarget.14161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 606.Lento W, et al. Loss of β-catenin triggers oxidative stress and impairs hematopoietic regeneration. Genes Dev. 2014;28:995–1004. doi: 10.1101/gad.231944.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 607.Dong Z, et al. Wnt/β-catenin pathway involvement in ionizing radiation-induced invasion of U87 glioblastoma cells. Strahlenther. Onkol. 2015;191:672–680. doi: 10.1007/s00066-015-0858-7. [DOI] [PubMed] [Google Scholar]
- 608.de Marcondes PG, Morgado-Díaz JA. The role of EphA4 signaling in radiation-induced EMT-like phenotype in colorectal cancer cells. J. Cell. Biochem. 2017;118:442–445. doi: 10.1002/jcb.25738. [DOI] [PubMed] [Google Scholar]
- 609.Bastos LG, et al. Progeny from irradiated colorectal cancer cells acquire an EMT-like phenotype and activate Wnt/β-catenin pathway. J. Cell. Biochem. 2014;115:2175–2187. doi: 10.1002/jcb.24896. [DOI] [PubMed] [Google Scholar]
- 610.Li, M. et al. Ribosomal S6 protein kinase 4 promotes radioresistance in esophageal squamous cell carcinoma. J. Clin. Invest. 130, 4301–4319 (2020). [DOI] [PMC free article] [PubMed]
- 611.Wang R, et al. Notch and Wnt/β-catenin signaling pathway play important roles in activating liver cancer stem cells. Oncotarget. 2016;7:5754–5768. doi: 10.18632/oncotarget.6805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 612.Vermeulen L, et al. Wnt activity defines colon cancer stem cells and is regulated by the microenvironment. Nat. Cell Biol. 2010;12:468–476. doi: 10.1038/ncb2048. [DOI] [PubMed] [Google Scholar]
- 613.Zhan T, et al. MEK inhibitors activate Wnt signalling and induce stem cell plasticity in colorectal cancer. Nat. Commun. 2019;10:2197. doi: 10.1038/s41467-019-09898-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 614.Debeb BG, et al. Histone deacetylase inhibitors stimulate dedifferentiation of human breast cancer cells through WNT/β-catenin signaling. Stem Cells. 2012;30:2366–2377. doi: 10.1002/stem.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 615.Milanovic M, et al. Senescence-associated reprogramming promotes cancer stemness. Nature. 2018;553:96–100. doi: 10.1038/nature25167. [DOI] [PubMed] [Google Scholar]
- 616.Anido J, et al. TGF-β receptor inhibitors target the CD44(high)/Id1(high) glioma-initiating cell population in human glioblastoma. Cancer Cell. 2010;18:655–668. doi: 10.1016/j.ccr.2010.10.023. [DOI] [PubMed] [Google Scholar]
- 617.Cai W, et al. PMP22 regulates self-renewal and chemoresistance of gastric cancer cells. Mol. Cancer Ther. 2017;16:1187–1198. doi: 10.1158/1535-7163.MCT-16-0750. [DOI] [PubMed] [Google Scholar]
- 618.Lettini G, et al. TRAP1 regulates stemness through Wnt/β-catenin pathway in human colorectal carcinoma. Cell Death Differ. 2016;23:1792–1803. doi: 10.1038/cdd.2016.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 619.Kagey MH, He X. Rationale for targeting the Wnt signalling modulator Dickkopf-1 for oncology. Br. J. Pharmacol. 2017;174:4637–4650. doi: 10.1111/bph.13894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 620.Jimeno A, et al. A first-in-human phase I study of the anticancer stem cell agent Ipafricept (OMP-54F28), a decoy receptor for Wnt Ligands, in patients with advanced solid tumors. Clin. Cancer Res. 2017;23:7490–7497. doi: 10.1158/1078-0432.CCR-17-2157. [DOI] [PubMed] [Google Scholar]
- 621.Gurney A, et al. Wnt pathway inhibition via the targeting of Frizzled receptors results in decreased growth and tumorigenicity of human tumors. Proc. Natl Acad. Sci. USA. 2012;109:11717–11722. doi: 10.1073/pnas.1120068109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 622.Choi MY, et al. Phase I trial: cirmtuzumab inhibits ROR1 signaling and stemness signatures in patients with chronic lymphocytic leukemia. Cell Stem Cell. 2018;22:951–959.e953. doi: 10.1016/j.stem.2018.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 623.Mehdawi LM, et al. Non-canonical WNT5A signaling up-regulates the expression of the tumor suppressor 15-PGDH and induces differentiation of colon cancer cells. Mol. Oncol. 2016;10:1415–1429. doi: 10.1016/j.molonc.2016.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 624.Kelsey R. Prostate cancer: Foxy-5 in prostate cancer model. Nat. Rev. Urol. 2017;14:638. doi: 10.1038/nrurol.2017.160. [DOI] [PubMed] [Google Scholar]
- 625.Prasad CP, Manchanda M, Mohapatra P, Andersson T. WNT5A as a therapeutic target in breast cancer. Cancer Metastasis Rev. 2018;37:767–778. doi: 10.1007/s10555-018-9760-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 626.Chen B, et al. Small molecule-mediated disruption of Wnt-dependent signaling in tissue regeneration and cancer. Nat. Chem. Biol. 2009;5:100–107. doi: 10.1038/nchembio.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 627.Liu J, et al. Targeting Wnt-driven cancer through the inhibition of porcupine by LGK974. Proc. Natl Acad. Sci. USA. 2013;110:20224–20229. doi: 10.1073/pnas.1314239110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 628.Johari B, et al. Myc decoy oligodeoxynucleotide inhibits growth and modulates differentiation of mouse embryonic stem cells as a model of cancer stem cells. Anticancer Agents Med. Chem. 2017;17:1786–1795. doi: 10.2174/1871521409666170412142507. [DOI] [PubMed] [Google Scholar]
- 629.Saygin C, et al. Targeting cancer stemness in the clinic: from hype to hope. Cell Stem Cell. 2019;24:25–40. doi: 10.1016/j.stem.2018.11.017. [DOI] [PubMed] [Google Scholar]
- 630.Wang Z, Li Y, Sarkar FH. Notch signaling proteins: legitimate targets for cancer therapy. Curr. Protein Pept. Sci. 2010;11:398–408. doi: 10.2174/138920310791824039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 631.Siebel C, Lendahl U. Notch signaling in development, tissue homeostasis, and disease. Physiol. Rev. 2017;97:1235–1294. doi: 10.1152/physrev.00005.2017. [DOI] [PubMed] [Google Scholar]
- 632.Nwabo Kamdje AH, et al. Developmental pathways associated with cancer metastasis: Notch, Wnt, and Hedgehog. Cancer Biol. Med. 2017;14:109–120. doi: 10.20892/j.issn.2095-3941.2016.0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 633.Rizzo P, et al. Rational targeting of Notch signaling in cancer. Oncogene. 2008;27:5124–5131. doi: 10.1038/onc.2008.226. [DOI] [PubMed] [Google Scholar]
- 634.Schott AF, et al. Preclinical and clinical studies of gamma secretase inhibitors with docetaxel on human breast tumors. Clin. Cancer Res. 2013;19:1512–1524. doi: 10.1158/1078-0432.CCR-11-3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 635.Fender AW, et al. Notch-1 promotes stemness and epithelial to mesenchymal transition in colorectal cancer. J. Cell. Biochem. 2015;116:2517–2527. doi: 10.1002/jcb.25196. [DOI] [PubMed] [Google Scholar]
- 636.Bocci F, et al. Toward understanding cancer stem cell heterogeneity in the tumor microenvironment. Proc. Natl Acad. Sci. USA. 2019;116:148–157. doi: 10.1073/pnas.1815345116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 637.Kim RK, et al. Radiation driven epithelial-mesenchymal transition is mediated by Notch signaling in breast cancer. Oncotarget. 2016;7:53430–53442. doi: 10.18632/oncotarget.10802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 638.Zhang CC, et al. Synergistic effect of the γ-secretase inhibitor PF-03084014 and docetaxel in breast cancer models. Stem Cells Transl. Med. 2013;2:233–242. doi: 10.5966/sctm.2012-0096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 639.Liu L, et al. Chemotherapy induces breast cancer stemness in association with dysregulated monocytosis. Clin. Cancer Res. 2018;24:2370–2382. doi: 10.1158/1078-0432.CCR-17-2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 640.Fessler E, Dijkgraaf FE, De Sousa EMF, Medema JP. Cancer stem cell dynamics in tumor progression and metastasis: is the microenvironment to blame? Cancer Lett. 2013;341:97–104. doi: 10.1016/j.canlet.2012.10.015. [DOI] [PubMed] [Google Scholar]
- 641.Bi P, et al. Notch activation drives adipocyte dedifferentiation and tumorigenic transformation in mice. J. Exp. Med. 2016;213:2019–2037. doi: 10.1084/jem.20160157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 642.Wang Z, et al. Notch signaling drives stemness and tumorigenicity of esophageal adenocarcinoma. Cancer Res. 2014;74:6364–6374. doi: 10.1158/0008-5472.CAN-14-2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 643.Luo H, et al. Differentiation-inducing therapeutic effect of Notch inhibition in reversing malignant transformation of liver normal stem cells via MET. Oncotarget. 2018;9:18885–18895. doi: 10.18632/oncotarget.24421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 644.Kang J, et al. Rhamnetin and cirsiliol induce radiosensitization and inhibition of epithelial-mesenchymal transition (EMT) by miR-34a-mediated suppression of Notch-1 expression in non-small cell lung cancer cell lines. J. Biol. Chem. 2013;288:27343–27357. doi: 10.1074/jbc.M113.490482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 645.Kraman M, McCright B. Functional conservation of Notch1 and Notch2 intracellular domains. FASEB J. 2005;19:1311–1313. doi: 10.1096/fj.04-3407fje. [DOI] [PubMed] [Google Scholar]
- 646.De Francesco, E. M., Maggiolini, M. & Musti, A. M. Crosstalk between Notch, HIF-1α and GPER in breast cancer EMT. Int. J. Mol. Sci. 19, 2011 (2018). [DOI] [PMC free article] [PubMed]
- 647.Ishida T, et al. Notch signaling induces EMT in OSCC cell lines in a hypoxic environment. Oncol. Lett. 2013;6:1201–1206. doi: 10.3892/ol.2013.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 648.Shen Y, et al. Estrogen receptor α-NOTCH1 axis enhances basal stem-like cells and epithelial-mesenchymal transition phenotypes in prostate cancer. Cell Commun. Signal. 2019;17:50. doi: 10.1186/s12964-019-0367-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 649.Wang X, et al. miR-181b/Notch2 overcome chemoresistance by regulating cancer stem cell-like properties in NSCLC. Stem Cell Res. Ther. 2018;9:327. doi: 10.1186/s13287-018-1072-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 650.Lin X, et al. miR-195-5p/NOTCH2-mediated EMT modulates IL-4 secretion in colorectal cancer to affect M2-like TAM polarization. J. Hematol. Oncol. 2019;12:20. doi: 10.1186/s13045-019-0708-7. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 651.Hayashi T, et al. Not all NOTCH is created equal: the oncogenic role of NOTCH2 in bladder cancer and its implications for targeted therapy. Clin. Cancer Res. 2016;22:2981–2992. doi: 10.1158/1078-0432.CCR-15-2360. [DOI] [PubMed] [Google Scholar]
- 652.Shi W, Harris AL. Notch signaling in breast cancer and tumor angiogenesis: cross-talk and therapeutic potentials. J. Mammary Gland Biol. Neoplasia. 2006;11:41–52. doi: 10.1007/s10911-006-9011-7. [DOI] [PubMed] [Google Scholar]
- 653.Bu P, et al. A microRNA miR-34a-regulated bimodal switch targets Notch in colon cancer stem cells. Cell Stem Cell. 2013;12:602–615. doi: 10.1016/j.stem.2013.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 654.D’Angelo RC, et al. Notch reporter activity in breast cancer cell lines identifies a subset of cells with stem cell activity. Mol. Cancer Ther. 2015;14:779–787. doi: 10.1158/1535-7163.MCT-14-0228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 655.Wong NK, et al. Heterogeneity of breast cancer stem cells as evidenced with Notch-dependent and Notch-independent populations. Cancer Med. 2012;1:105–113. doi: 10.1002/cam4.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 656.Andersson ER, Lendahl U. Therapeutic modulation of Notch signalling-are we there yet? Nat. Rev. Drug Discov. 2014;13:357–378. doi: 10.1038/nrd4252. [DOI] [PubMed] [Google Scholar]
- 657.Cook N, et al. A phase I trial of the γ-secretase inhibitor MK-0752 in combination with gemcitabine in patients with pancreatic ductal adenocarcinoma. Br. J. Cancer. 2018;118:793–801. doi: 10.1038/bjc.2017.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 658.So JY, et al. HES1-mediated inhibition of Notch1 signaling by a Gemini vitamin D analog leads to decreased CD44(+)/CD24(-/low) tumor-initiating subpopulation in basal-like breast cancer. J. Steroid Biochem. Mol. Biol. 2015;148:111–121. doi: 10.1016/j.jsbmb.2014.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 659.Huynh C, et al. The novel gamma secretase inhibitor RO4929097 reduces the tumor initiating potential of melanoma. PLoS ONE. 2011;6:e25264. doi: 10.1371/journal.pone.0025264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 660.Chu Q, et al. Prolonged inhibition of glioblastoma xenograft initiation and clonogenic growth following in vivo Notch blockade. Clin. Cancer Res. 2013;19:3224–3233. doi: 10.1158/1078-0432.CCR-12-2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 661.Arcaroli JJ, et al. Tumours with elevated levels of the Notch and Wnt pathways exhibit efficacy to PF-03084014, a γ-secretase inhibitor, in a preclinical colorectal explant model. Br. J. Cancer. 2013;109:667–675. doi: 10.1038/bjc.2013.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 662.Wu CX, et al. Notch inhibitor PF-03084014 inhibits hepatocellular carcinoma growth and metastasis via suppression of cancer stemness due to reduced activation of Notch1-Stat3. Mol. Cancer Ther. 2017;16:1531–1543. doi: 10.1158/1535-7163.MCT-17-0001. [DOI] [PubMed] [Google Scholar]
- 663.Rudin CM, et al. Rovalpituzumab tesirine, a DLL3-targeted antibody-drug conjugate, in recurrent small-cell lung cancer: a first-in-human, first-in-class, open-label, phase 1 study. Lancet Oncol. 2017;18:42–51. doi: 10.1016/S1470-2045(16)30565-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 664.Ahmad A, Sakr WA, Rahman KM. Novel targets for detection of cancer and their modulation by chemopreventive natural compounds. Front Biosci. 2012;4:410–425. doi: 10.2741/e388. [DOI] [PubMed] [Google Scholar]
- 665.Li Y, et al. Curcumin inhibits proliferation and invasion of osteosarcoma cells through inactivation of Notch-1 signaling. FEBS J. 2012;279:2247–2259. doi: 10.1111/j.1742-4658.2012.08607.x. [DOI] [PubMed] [Google Scholar]
- 666.Elshamy WM, Duhé RJ. Overview: cellular plasticity, cancer stem cells and metastasis. Cancer Lett. 2013;341:2–8. doi: 10.1016/j.canlet.2013.06.020. [DOI] [PubMed] [Google Scholar]
- 667.Eun K, Ham SW, Kim H. Cancer stem cell heterogeneity: origin and new perspectives on CSC targeting. BMB Rep. 2017;50:117–125. doi: 10.5483/BMBRep.2017.50.3.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 668.Brock A, Krause S, Ingber DE. Control of cancer formation by intrinsic genetic noise and microenvironmental cues. Nat. Rev. Cancer. 2015;15:499–509. doi: 10.1038/nrc3959. [DOI] [PubMed] [Google Scholar]
- 669.Lewis PW, et al. Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science. 2013;340:857–861. doi: 10.1126/science.1232245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 670.Harris WJ, et al. The histone demethylase KDM1A sustains the oncogenic potential of MLL-AF9 leukemia stem cells. Cancer Cell. 2012;21:473–487. doi: 10.1016/j.ccr.2012.03.014. [DOI] [PubMed] [Google Scholar]
- 671.Pan Y, et al. Therapeutic approaches targeting cancer stem cells. J. Cancer Res. Ther. 2018;14:1469–1475. doi: 10.4103/jcrt.JCRT_976_17. [DOI] [PubMed] [Google Scholar]
- 672.Hingorani SR, et al. HALO 202: Randomized Phase II Study of PEGPH20 Plus Nab-Paclitaxel/Gemcitabine Versus Nab-Paclitaxel/Gemcitabine in Patients With Untreated, Metastatic Pancreatic Ductal Adenocarcinoma. J. Clin. Oncol. 2018;36:359–366. doi: 10.1200/JCO.2017.74.9564. [DOI] [PubMed] [Google Scholar]
- 673.Perera RM, Bardeesy N. Pancreatic cancer metabolism: breaking it down to build it back up. Cancer Discov. 2015;5:1247–1261. doi: 10.1158/2159-8290.CD-15-0671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 674.O’Reilly EM, et al. Randomised phase II trial of gemcitabine and nab-paclitaxel with necuparanib or placebo in untreated metastatic pancreas ductal adenocarcinoma. Eur. J. Cancer. 2020;132:112–121. doi: 10.1016/j.ejca.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]