Abstract
Despite significant advances in cancer precision medicine, a significant hurdle to its broader adoption remains the multitude of variants of unknown significance identified by clinical tumor sequencing and the lack of biologically validated methods to distinguish between functional and benign variants. Here we used functional data on MAP2K1 and MAP2K2 mutations generated in real-time within a co-clinical trial framework to benchmark the predictive value of a three-part in silico methodology. Our computational approach to variant classification incorporated hotspot analysis, three-dimensional molecular dynamics simulation, and sequence paralogy. In silico prediction accurately distinguished functional from benign MAP2K1 and MAP2K2 mutants, yet drug sensitivity varied widely among activating mutant alleles. These results suggest that multifaceted in silico modeling can inform patient accrual to MEK/ERK inhibitor clinical trials, but computational methods need to be paired with laboratory- and clinic-based efforts designed to unravel variabilities in drug response.
Introduction
Prospective tumor sequencing is increasingly used by clinicians to guide treatment selection in patients with cancer (1). Despite widespread enthusiasm for this approach, only a small number of cancer-associated genes have been clinically validated as predictive biomarkers of drug response. Furthermore, even in well-studied genes, the majority of mutations identified by clinical tumor profiling are of unknown biologic and clinical significance.
Here, we set out to formally evaluate the accuracy and predictive value of a multifaceted in silico approach to distinguish functional from benign mutations within the context of a co-clinical trial paradigm. We used real-time tumor mutation profiles generated as part of an ongoing institution-wide prospective tumor sequencing initiative, to direct preclinical discovery efforts with the goal of informing patient care. To do so, we conducted a computational and biochemical comparative analysis of mutations in the mitogen activated protein kinase (MAPK) kinase genes, MAP2K1 and MAP2K2, which encode the MEK1 and MEK2 kinases, respectively. Upon activation by RAF kinases, MEK1/2 phosphorylate ERK, thereby promoting proliferation and survival. Activation of MAPK signaling is common in human cancer, most often mediated by mutations in the RAS genes, BRAF, NF1, or upstream receptors. Mutations in MAP2K1/2 are less common, and their phenotypic contribution remains poorly understood. A small number of recurrent MEK1 mutations have been shown to be oncogenic (2–4). Additionally, dramatic and durable anti-tumor responses to MEK inhibition have been reported in patients with MEK1-mutant histiocytosis, but only anecdotally in other cancer types (2,5–7). The likelihood that such responses will be observed more broadly remains unknown. We therefore sought to biologically validate an in silico discovery platform that could distinguish between functional and therapeutically actionable versus benign MAP2K1 and MAP2K2 mutations.
Materials and Methods
42K MSK-IMPACT sequencing cohort and hotspot data analysis
We assembled somatic mutational data from 42,434 retrospectively and prospectively sequenced human cancers using an approach analogous to those previously described (8,9). As compared to prior hotspot analyses, all mutational data from the studies associated with The Cancer Genome Atlas were derived from the single multi-center mutation calling in multiple cancers project (MC3)(10). We also included incrementally accrued patients enrolled on our institutional prospective tumor profiling initiative (11), numbering a total of 21,918 patients (51.6% of the full study cohort). For this prospective study, beginning in May 2014, cancer patients seen at Memorial Sloan Kettering Cancer Center were offered matched tumor-germline DNA sequencing at physician discretion on an institutional protocol (ClinicalTrials.gov identifier: NCT01775072). Written informed consent was obtained from all participating patients and the study was conducted in accordance with recognized ethical guidelines. MSK-IMPACT data is broadly available via AACR Genie (12). After excluding studies and samples of insufficient quality and breadth, we performed hotspot analysis of MAP2K1 and MAP2K2 as previously described (8,9). MAP2K1 and MAP2K2 mutational hotspots were considered significant if they exceeded a false discovery rate of 1% (q-value<0.01). Trending hotspots were those with a q-value between 0.01 and 0.25, corresponding to a false discovery rate of 25%.
Cell lines and culture conditions
HEK-293H cells were purchased from ATCC and HEK-293T cells from Invitrogen and maintained in DME-HG with 10% FBS, 2mM L-glutamine and 50units/ml each of penicillin and streptomycin and used within three months of passages after receipt. CRISPR knock-in lines were derived by subcloning guide RNA sequences targeting MEK1 F53L and K57N (gcagcagcgaaagcgccttg and cttctgcttctgggtaagaa) and E102_I103del (tggagatcaaacccgcaatc and tgatctccagatgaattagc) into a px330-U6-Chimeric-CBh-hSpCas9 mammalian expression vector (Addgene 42230) followed by transient transfection into 293T cells along with a 200bp donor vector encoding the missense mutation or deletion of interest plus a mutated PAM site (F53L:tttggaacaggaccaacttggaggccttgcagaagaagctggaggagctagagcttgatgagcagcagcgaaagcgcct tgaagcacttcttacccagaagcagaaggtgggagaactgaaggatgacgactttgagaagatcagtgagctgggggctggcaat ggcggtgtg;K57N:acttggaggccttgcagaagaagctggaggagctagagcttgatgagcagcagcgaaagcgccttgaagc atttcttacccagaatcagaaggtgggagaactgaaggatgacgactttgagaagatcagtgagctgggggctggcaatggcggtgt ggtgttcaaggtct;E102_I103del:1)tatctttcatcccttcctccctctttctttcataaaacctctctttcttccacctttctccagctaatt catctgaaacccgcaatccgaaaccagatcataagggagctgcaggttctgcatgagtgcaactctccgtacatcgtgggc,2)tatct ttcatcccttcctccctctttctttcataaaacctctctttcttccacctttctacagctaattcatctgaaacccgcaatccggaaccagatcat aagggagctgcaggttctgcatgagtgcaactctccgtacatcgtgggc). Cells were then single cell sorted into 96 well plates and single colonies were screened for perfect integration of the mutant basepair by PCR of the affected region of MEK1, followed by Sanger sequencing. As further confirmation select subclones were analyzed by MSK-IMPACT genomic sequencing and PCR/Sanger sequencing to confirm the presence of the mutant and lack of off-target mutations. CRISPR lines were maintained in the same conditions as parental 293T and limited to use within 15 passages. All lines tested mycoplasma negative.
MEK1/2 transfections, drug treatment and western blotting
MAP2K1 mutants were generated from the C-terminal GFP-tagged MEK1 plasmid (pEGPF-N1-MEK1, Addgene #14746) and MAP2K2 mutant constructs were generated off of the N-terminal myc-tagged MEK2 plasmid (pcDNA3.1-Hygro_MEK2, Addgene #40776) using the QuikChange II XL Site-Directed Mutagenesis Kit (Agilent #200522) and verified by Sanger sequencing. HEK-293H or HEK-293T cells were transiently transfected with the wild-type or mutant MEK1-GFP plasmid using Lipofectamine 2000 Transfection Reagent (Invitrogen #11668500). Trametinib was obtained from GlaxoSmithKline and SCH772984 from Merck. All drugs were dissolved in DMSO to yield 10 mmol/L stock and stored at −80°C. At 24hr post-transfection, media was aspirated and refreshed with or without targeted inhibitors for various times (1hr for large mutant panels without drug, or as indicated with drug), at which point cells were washed with cold PBS and harvested on ice and pelleted. Briefly, cells were then lysed in 1% NP-40 buffer with protease and phosphatase inhibitors, protein quantified using the Pierce BCA kit and prepared and ran on western blots as previously reported (13).
Antibodies, ECL and western blot imaging
Rabbit polyclonal antibodies recognizing phosphorylated MEK1/2 (S217/221) (#9121L), MEK1/2 (#9122), phosphorylated ERK1/2 (T202/Y204) (#9101), ERK1/2 (#9102), and phosphorylated p90RSK T356/S363 (#9344) were obtained from Cell Signaling. Rabbit monoclonal antibodies recognizing RSK (#9355), p90RSK S380 (#9341), GFP (D5.1 XP(R)(#2956) and GAPDH (#2118) were obtained from Cell Signaling. After incubation with primary overnight, membranes were probed with horseradish peroxidase-conjugated secondary antibody (Amersham NA934) for 1hr, washed and proteins were detected by chemiluminescence (SuperSignal West Pico Plus, Thermo Scientific, #34577) and visualized using the Fuji LAS-4000 imager (GE Life Sciences).
Molecular dynamics simulation (MD simulation)
Protein preparation: The crystal structure (PDB ID: 3SLS)(14) of wild-type MEK1 was retrieved from the Protein Data Bank (PDB) (www.rcsb.org)(15). Structures of mutant MEK1 mutations were constructed using SWISS-MODEL (16) with the wild-type structure as a template. The ligand (ANP) from the crystal structure was used as the ligand of each MEK1 simulated structures.
Simulation: All of the molecular dynamics simulations were conducted in a periodic boundary box with SPC water model using the Gromacs 5.1.4 package (17) with the Gromos 53a6 force field (18). The parameterization of the ligand was produced by PRODRG2.5 server (19,20). To neutralize the systems, chloride ions and sodium ions were added to a random place in the simulation box (18). The MD simulations were subjected to three steps. First, the main chain of the residues in the protein and heavy atoms (C, O and N) of the ligand were constrained to minimize the energy, and the same process was performed with the unconstrained system. In the second step, each system was slowly heated from 0K to 300K with constrained and unconstrained conditions successively. Finally, the 100ns unconstrained production MD simulation was performed. The hydrogen atoms were constrained using the SHAKE algorithm (21). The particle mesh Ewald summation algorithm was taken to calculate the long-range electrostatic interactions (22). All production simulations were performed under constant (atmospheric) pressure maintained by the Langevin piston method (23) and an optimum temperature (300 K) with 2fs time step. System configurations were updated for every 2.0ps. The protein secondary structure analysis was performed with the plug-in of Gromacs software for the simulation of trajectories and the secondary structure data was extracted for the probability analysis. % probability was based on the time of the existence of the α-helix or β-sheet during the simulation divided by the whole simulation time. Each secondary structure was represented by different letters in the simulation data (e.g. α-Helix corresponds to G, β-sheet corresponds to E). The probability of the secondary structure of each residue in the simulation process was obtained by counting the probability of each letter. First the total number of frames in the simulation (Total) was determined, followed by the total number of frames for each residue in the α-Helix secondary structure (G). Then the % probability of α-Helix for each residue was calculated using G/Total*100. Similarly, the probability of β-Sheet for each residue was determined by E/Total*100.
MEK2 paralogy analysis
Paralogous residue positions were determined between MEK1 and MEK2 using pairwise alignment between their respective peptide sequences (MEK1: ENSP00000302486 and MEK2: ENSP00000262948) based on the BLOSUM62 matrix to assign amino acid similarity using a gap-opening penalty of 11 and a gap extension penalty of 1. For each MEK1 residue, a 30 amino acid window on either side was used as the region of alignment to MEK2, which avoids biasing the alignment at a specific residue based on a region of worse overall alignment. E-values indicate that only the N-terminal sequence was of lower alignment quality and therefore paralogy more difficult to determine.
Results
Pan-cancer landscape of MAP2K1 mutations
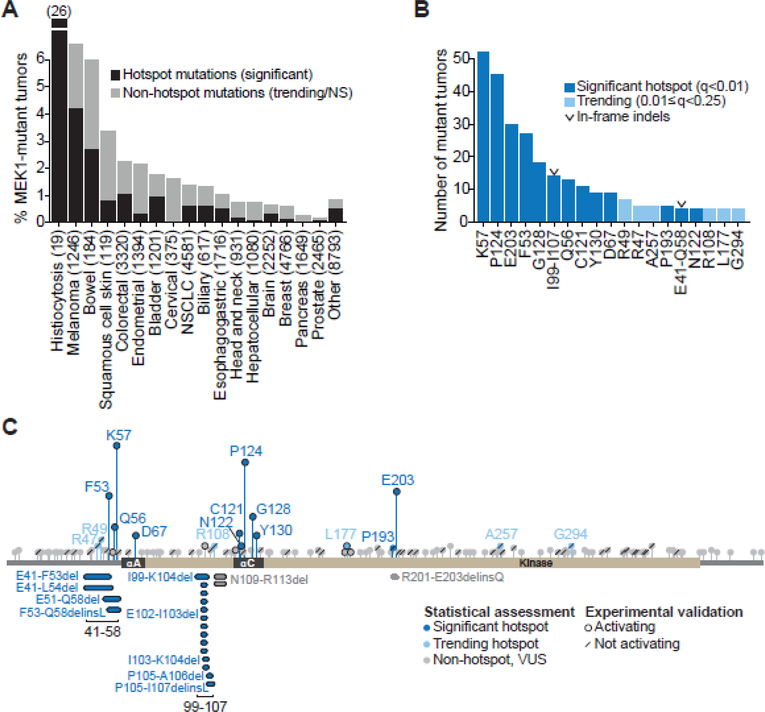
To define the landscape of MAP2K1 alterations pan-cancer, we analyzed sequencing data from 42,434 patient tumors encompassing 32 organ types. 20,516 tumor samples originated from previously published resources (TCGA, individual publications, etc.). The remaining 21,918 tumor/normal pairs were tumors from our ongoing prospective MSK-IMPACT genomic profiling initiative (11). MAP2K1 alterations were identified in 1.1% of all tumors (458 of 42,434 tumors), of which the majority were missense mutations (81%), followed by in-frame deletions (4.3%). The incidence of MAP2K1 mutations ranged from 6.6% in melanoma and 6% in bowel cancers to less than 2% in other tumor types (Fig. 1A). A notable outlier was histiocytosis, where MAP2K1 mutations were identified in 26% of tumors (Fig. 1A). MAP2K1 mutations were distributed throughout the gene, with mutational clusters localized to helix-A (αA) in the negative regulatory domain and helix-C (αC) in the kinase domain (Fig. 1B and C)(3,24,25).
Figure 1. Landscape of MAP2K1 (MEK1) alterations in human cancer.
A, Percentage of tumors with somatic MAP2K1 mutations as a function of cancer type in a cohort of 42,434 sequenced tumor/normal pairs. Mutations were classified as significant hotspots (black) versus trending or not-significant (NS) non-hotspots (gray). B, Number of tumor samples per indicated significant hotspot (dark blue) or trending hotspot site (light blue). Two significant clusters of in-frame deletions are labeled with a “v” symbol. C, MEK1 protein schematic depicting the amino acid position, domain location, and recurrence of the 458 MEK1 mutants. Missense mutants were plotted above, and in-frame deletions were plotted below the domain schematic. Mutants were categorized as significant hotspots (q<0.01, dark blue), trending hotspots (0.01≤q<0.25, light blue) or non-hotspots (q>0.25, gray). Variants shown in subsequent functional studies to increase p-ERK expression are highlighted with a black ring, whereas those that had no effect on p-ERK are noted with a forward slash.
We next performed an in-depth in silico analysis to catalog MAP2K1 mutations as driver versus benign events. First, we employed a statistical framework for mutational hotspot detection which adjusts for gene- and site-specific background mutation rates, among other variables (8,9). Thirteen statistically significant mutational hotspot sites in MAP2K1 were identified, which accounted for 49.3% of the MAP2K1-mutated tumors (Figs. 1A, black, 1B and C, dark blue, and Supplementary Table S1). Eleven were missense mutants, including K57, the most frequently mutated site, and novel variants at N122 and P193. Two were clusters of largely uncharacterized in-frame deletions, one encompassing residues E41-Q58, and a second spanning I99-I107 (Figs. 1B and C). We observed lineage-specific patterns of mutation with K57N most common in lung and bowel cancers and P124S/L in melanoma (Supplementary Fig. S1). The balance of patients harbored one or more of six trending hotspots, defined as those that fell just below the threshold of statistical significance (Figs. 1B and C, light blue), and non-hotspot mutations, defined as those consistent with the background mutation rate (Figs. 1A and C, gray).
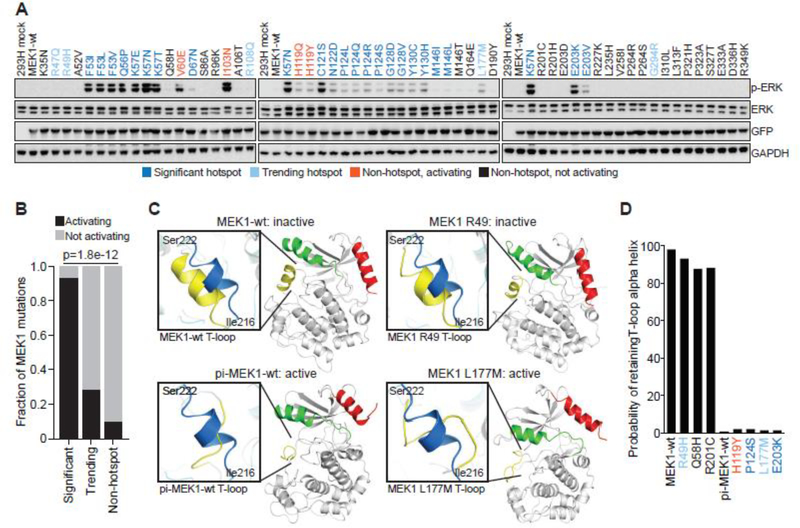
We next interrogated the predictive value of hotspot status with the ability of MEK1 variants to activate ERK. In total, we generated 115 MEK1 mutants, including 92 missense and 23 indel variants that were either identified within our 42K cohort, previously reported, or were mutants not yet found somatically, but generated based on paralogy. Twelve of 13 MAP2K1 hotspot sites (93.6%; 29/31 alleles tested at these 13 sites) validated as biologically active in our screen (Figs. 2A and B). By contrast, only 1/6 trending hotspot sites (28.6%; 2/7 alleles tested at these 6 sites) and 4/45 non-hotspot sites (10.4%; 5/48 alleles tested at these 45 sites) induced ERK phosphorylation (Fig. 2B).
Figure 2. Concordance of biochemical and 3D modeling of MEK1 mutants.
A, GFP-tagged wildtype MEK1 (MEK1-wt) and MEK1 missense mutants were expressed in 293H cells. Expression of phosphorylated ERK (p-ERK), total ERK, GFP (MEK1) and GAPDH levels were assessed by western blot. Mutations were color coded as follows: statistically significant hotspot (dark blue); trending hotspot (light blue); non-hotspot, activating (orange); non-hotspot, not activating (black) B, Quantification of the accuracy of the computational inference of hotspot as compared to functional validation. Bars represent the fraction of either significant, trending, or not-significant mutants that were validated as activating in biochemical assays (black) versus not activating (gray). Statistical significance reflects enrichment of biochemical validation with hotspot prediction, p value = 1.8e−12. C, The structural changes to the MEK1 protein predicted to occur upon mutation of specific residues as assessed by molecular dynamics (MD) simulation. Results were compared to the simulated structure of MEK1-wt and phosphomimetic-MEK1-wt (pi-MEK1-wt). D, Quantitation of the probability that the natural T-loop alpha helix structure (inactive conformation) is retained following MD simulation of select MEK1 mutants.
Among the hotspot sites, we confirmed that MEK1 point mutants predicted to disrupt helix-A in the negative regulatory domain (F53L, Q56P, K57E/N and D67N) or distort the position of helix-C in the kinase domain (C121S, P124S/L, G128D/V, and Y130C) (Fig. 2A) were activating, consistent with the literature (2,3,25–32). We additionally expressed rare mutant alleles at these hotspot sites, including F53I/V, K57T, P124Q/R and Y130H, and found them to be activating as well. MEK1 E203K (n=26), the lone hotspot peak in the distal kinase domain, also validated as ERK-activating, consistent with prior studies. Interestingly, our efforts to assess the impact of all patient-derived variants within a hotspot site revealed that while the less common E203R and E203V mutants (n=2 each) were functional, E203D, a conservative amino acid change present in two patients, did not induce p-ERK levels above wildtype MEK, and E203Q did so only minimally (Fig. 2A and Supplementary Fig. S2). The two remaining missense hotspots were previously uncharacterized variants at N122 and P193. N122D activated p-ERK and would be predicted to disrupt helix-C similarly to C121S (Fig. 2A). P193S was the only hotspot mutation that did not induce p-ERK. (Supplementary Fig. S2).
Among the trending hotspots, only L177M/V induced p-ERK expression (Fig. 2A and Supplementary Fig. S2). Its mechanism of action is unclear as it is not located near other activating mutations in either sequence or structure. The other trending hotspots (R47, R49, R108, A257 and G294) were not activating, even though R49 clusters in three-dimensional proximity to other activating MEK1 mutations (Fig. 2A)(33).
Of the 45 non-hotspot sites tested, only the previously known MEK1 I103N and the novel T55P, H119Q/Y and N109_R113del mutations (see below for indel) were activating, all of which are situated in or near critical domains (Fig. 2A and Supplementary Fig. S2). Notably, MEK1 T55P was previously identified as a germline variant in a patient with a RASopathy syndrome, as have seven of the activating MEK1 somatic hotspots (F53, D67, P124, G128, Y130 and E203), and the non-hotspot E44 mutation (34–36). Despite its previous association with a RASopathy (Noonan syndrome), the E44G/K mutants did not activate ERK, consistent with a thorough review of this patient’s clinical records which noted that the patient’s mother was an asymptomatic carrier of the E44G variant (36). We also confirmed that MEK1 L42F, a germline variant yet to arise somatically, induced p-ERK (Supplementary Fig. S2). Finally, only one of ten mutants N-terminal to F53 was activating (L42F) nor were any of the 25 mutants C-terminal to I204. In sum, the results robustly support the value of hotspot analysis for predicting the oncogenic status of previously uncharacterized genomic variants.
To further refine our ability to computationally predict the functional status of MEK1 mutants, we performed three-dimensional molecular dynamics simulation (MD simulation) to infer the ability of specific amino acid substitutions to impact MEK1 conformational activation. Systematic evaluation of the structural changes that occur following RAF-mediated phosphorylation of wildtype MEK1 demonstrated that disordering of the empiric alpha-helical structure of the activation (T-) loop is critical for kinase activation and could be used to predict MEK1 mutant activation status (29–31,37). MD simulation of the biochemically benign, non-hotspot MEK1 R49H, Q58H, and R201C mutants predicted retention of the alpha-helix structure similar to wildtype MEK1, despite their location within regulatory domains or near known hotspots (Figs. 2C and D). In contrast, MD simulation predicted that the novel activating non-hotspot variants MEK1 H119Y and L177M, as well as previously characterized activating hotspots P124S and E203K, would significantly disrupt the alpha-helix conformation of the T-loop similar to phosphomimetic-MEK1 (Figs. 2C and D). Thus, MD simulation could distinguish activating MEK1 mutants and therefore help prioritize non-recurrent MEK1 mutations for functional evaluation.
Hotspot and MD simulation analysis of two novel clusters of MEK1 in-frame deletions
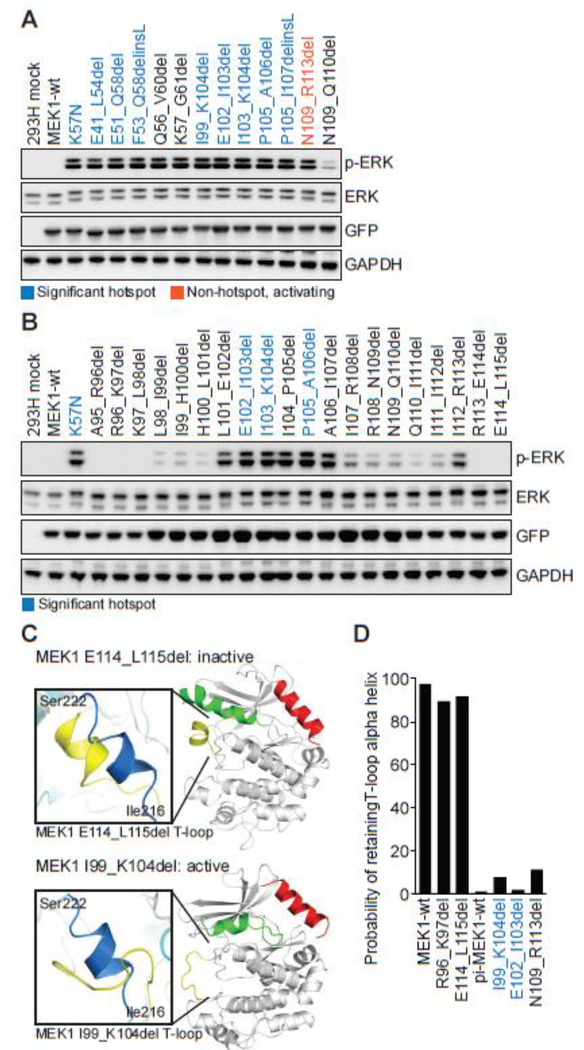
Hotspot analysis identified two clusters of somatic MEK1 in-frame insertions/deletions (indels). Four unique negative regulatory domain mutants were identified in patients with histiocytosis (E41_F53del), lung adenocarcinoma (E41_L54del), melanoma (E51_Q58del) and pheochromocytoma (F53_Q58delinsL). Further, fourteen patients with diverse tumor types harbored five unique helix-C deletions, the most common being E102_I103del, a previously characterized activating deletion (Fig. 1C)(4,25,38,39). All nine of these MEK1 hotspot in-frame indels robustly induced ERK phosphorylation (Fig. 3A and Supplementary Fig. S2).
Figure 3. Functional and molecular dynamics characterization of two clusters of in-frame MEK1 deletions involving the negative regulatory domain and helix C.
A, MEK1 in-frame deletions were expressed in 293H cells and p-ERK expression compared to the K57N mutant. Expression of total ERK, GFP (MEK1) and GAPDH were assessed as controls. Mutations identified in patient tumors from our 42K cohort were color coded as statistically significant hotspots (dark blue), novel non-hotspot activating indels (orange), or other MEK1 in-frame deletions reported in the literature or used for comparison (black). B, Step-wise, two-base pair deletion mutants of MEK1 from A96 to K115 were expressed in 293H cells and p-ERK expression was assessed by western blot. C-D, The conformational changes and probability of retaining the wildtype T-loop alpha helix structure following molecular dynamics simulation for MEK1 in-frame deletion mutants, as in Figs. 2C–D.
As prior large-scale sequencing studies, including in melanoma, failed to report MEK1 indels, we reviewed subsequent MSK-IMPACT analyses, the multi-institutional AACR GENIE dataset (12), and the literature to substantiate our discovery of these clusters. This analysis identified fourteen additional non-recurrent somatic or germline MEK1 indels, only two of which had been previously characterized (Supplementary Table S2)(2,40). Twelve of these 14 MEK1 indels induced p-ERK expression, including the lone insertion mutation (L50_E51insI), as well as all seven negative regulatory deletions and all four helix-C deletions (Fig. 3A and Supplementary Fig. S2). Notably, a germline MEK1 K59del identified in cardio-facio-cutaneous syndrome patients was less activating than other indels in that region (35,41,42). Also of note, MEK1 N109_R113del (n=2), localized within helix C, was activating but fell just below the statistical threshold used to define trending hotspots (Supplementary Table S1). Of the two remaining indels, R201_E203delinsQ was not activating in our assay as it effectively creates a E203Q mutation, which was similarly minimally activating (Supplementary Fig. S2). D152_L155del did not fall within either hotspot cluster nor near any other functional domain and was not activating (Supplementary Fig. S2).
To extend prior work which revealed the activating potential of disrupting helix-A of the negative regulatory domain (43), we sought to define the boundaries within which deletions in helix-C induced MEK1 activity. We therefore designed 6-bp deletions (2 amino acids, paired) walking 5’ to 3’ in the helix-C region from residues A95 to L115 and observed robust activation of ERK in the sub-region from L98 to R113, with the highest activity involving or including deleted residues between L101 to I107 (Fig. 3B). These data suggest that all novel MEK1 indels involving amino acids L98 to R113 should be considered as likely activating.
Consistent with our biochemical and hotspot analyses, MD simulation also predicted that MEK1 I99_K104del, E102_I103del and N109_R113del would induce loss of the alpha-helical conformation of the T-loop and thus induce kinase activation (Figs. 3C and D). Conversely, MD simulation of R96_K97del and E114_L115del, functionally benign MEK1 indels that lie outside of the essential helix-C residues, were predicted to retain the alpha-helical T-loop structure similar to wildtype MEK1 (Figs. 3C and D). Thus, individually and collectively, hotspot, biological and structural analyses revealed the presence of two clusters of activating MEK1 in-frame indels.
Leveraging paralogy between MEK1 and MEK2 to predict the functionality of MEK2 mutants
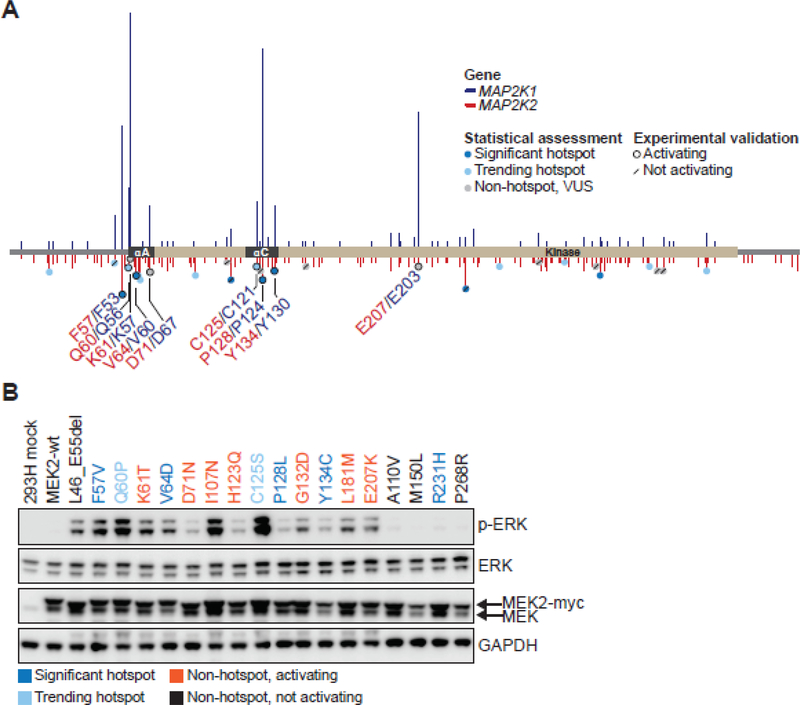
MAP2K2 alterations were present in only 0.7% of tumors (294 of 42,434) in our combined retrospective and prospective cohort, likely less than the requisite cohort size for adequately powered hotspot analysis. Given that MAP2K1 and MAP2K2 are highly homologous genes that share 80% base pair identity, with 90% conservation of their kinase domains, MAP2K2 was an attractive candidate to benchmark paralogy analysis as a method to distinguish between functional and benign variants (44–46). Multiple sequence alignment of the MEK1 and MEK2 protein coding sequences demonstrated that all residues within the negative regulatory and helix-C domains were identical, except for one amino acid (Supplementary Fig. S3). Therefore, we tested whether paralogy to MEK1 could reliably predict the activation status of previously uncharacterized MEK2 mutations (Fig. 4A).
Figure 4. MEK2 residues paralogous to MEK1 hotspots are activating.
A, MEK2 protein schematic depicting the amino acid position, domain location, and recurrence of MEK2 mutants (below x axis, red) aligned with paralogous MEK1 mutants (above x axis, blue). MEK2 mutants were categorized as significant hotspots (q<0.01, dark blue), trending hotspots (0.01≤q<0.25, light blue) or non-hotspots (q>0.25, gray). Experimentally validated calls were outlined with a black ring indicating a mutation that induced p-ERK expression, or a forward slash signifying variants that had no effect on p-ERK expression. Functional MEK2 mutants identified in patient tumors within our 42K cohort are noted in red lettering, along with their paralogous MEK1 residues in navy blue. B, myc-tagged MEK2 mutants were generated, overexpressed in 293H cells and induction of ERK phosphorylation (p-ERK) was assessed by western blot. Expression of total ERK, MEK and GAPDH were assessed as controls. Hotspot MEK2 mutants (dark blue), trending hotspots (light blue), non-hotspot activating (orange), and non-hotspot, not activating (black) mutants are indicated.
All fourteen MEK2 mutations paralogous to activating MEK1 mutants, hotspot and non-hotspot alike, induced ERK phosphorylation. These mutations included four classified as hotspots (MEK2 F57, V64, P128, Y134) and two as trending hotspots (Q60, C125) (Fig. 4B and Supplementary Tables S3–4). As predicted based on paralogy to activating MEK1 negative regulatory domain indels, we also found that MEK2 L46_E55del, a germline RASopathy variant yet to be identified somatically (36), was activating (Fig. 4B). None of twelve MEK2 mutations paralogous to experimentally validated benign MEK1 variants induced ERK phosphorylation (Fig. 4B, Supplementary Fig. S3 and Supplementary Table S3). Overall, assessment of paralogy to MEK1 accurately discriminated activating from benign variants in MEK2.
Differential MEK and ERK inhibitor sensitivity among MEK1/2 mutants
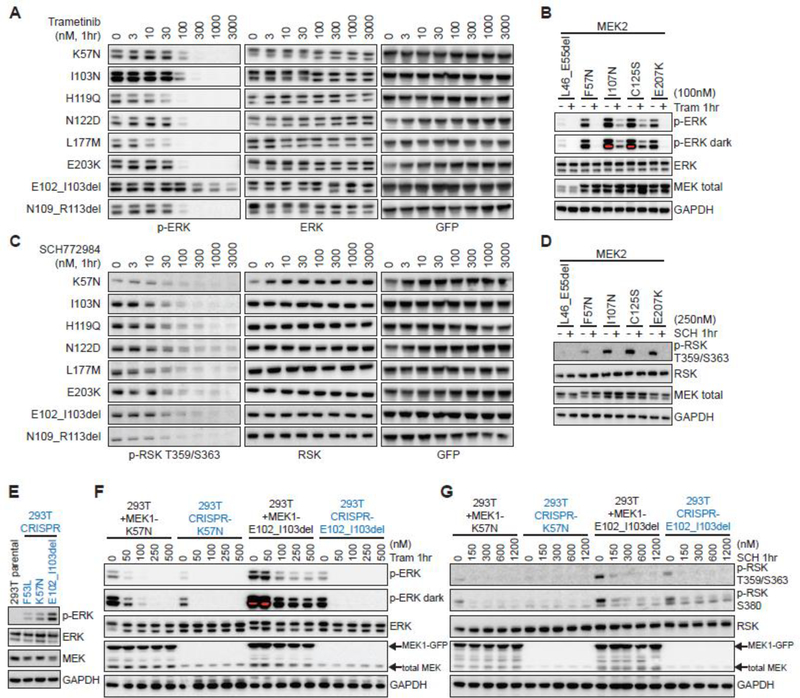
Both durable responses and acquired resistance to MEK inhibition have been reported in MEK1-mutant patients (2,5–7,26,40,47,48). To define the MEK/ERK inhibitor sensitivity of the novel MEK1 and MEK2 variants identified above, we compared the ability of MEK and ERK inhibitors to abrogate ERK pathway output. We observed significant differences among ERK-activating MEK1 and MEK2 mutations in the ability of trametinib, an FDA-approved MEK inhibitor, to inhibit ERK phosphorylation (Fig. 5A and Supplementary Fig. S4). The novel missense alleles L42F, T55P, H119Q/Y, N122D and L177M were sensitive to MEK inhibition, though at different thresholds. Negative regulatory domain indels spanning E41-E62 were also highly sensitive to trametinib, whereas helix-C indels from I99-I107 were less responsive. This is consistent with recent reports demonstrating that cells expressing MEK1 E102_I103del require higher concentrations of trametinib to inhibit ERK than MEK1 point mutants such as K57N (4). The novel N109_R113del, just C-terminal to the helix-C hotspot cluster, proved to be an exception as it was highly sensitive to trametinib.
Figure 5. MEK1/2 mutants are variably sensitive to MEK and ERK inhibition.
A-B, Hotspot and activating MEK1 (A) or MEK2 (B) missense and in-frame deletion mutants were expressed in 293H cells for 24hr, then treated for 1hr with vehicle or (A) the MEK inhibitor trametinib (33000nM) or (B) trametinib at 100nM followed by assessment of p-ERK expression by western blot. C-D, Hotspot and activating MEK1 (C) or MEK2 (D) missense and in-frame deletion mutants were expressed in 293H cells for 24hr, then treated for 1hr with vehicle or (C) the ERK inhibitor SCH772984 (3–3000nM) or (D) SCH772984 at 250nM followed by assessment of p-RSK expression by western blot. E, p-ERK expression in 293T cells in which MEK1 F53L, K57N or MEK E102_I103del mutants were knocked-in using CRISPR/Cas9. F-G, MEK1 K57N or E102_103del constructs were overexpressed in 293T and compared to their respective 293T MEK1 CRISPR knock-in derived cell lines (in blue) for changes in p-ERK (F) or p-RSK (G) at baseline and after 1hr treatment with vehicle or increasing doses of (F) trametinib (50–500nM) or (G) SCH772984 (150–1200nM).
The sensitivity of activating MEK2 mutants to trametinib also varied as a function of mutant allele in a pattern similar to their paralogous MEK1 alleles (Fig. 5B and Supplementary Fig. S4). We next extended this therapeutic analysis to the effects of ERK inhibition using the ATP-competitive ERK1/2 kinase inhibitor SCH772984 (49). In contrast to trametinib, SCH772984 uniformly inhibited downstream pathway activity, as assessed by downregulation of RSK phosphorylation, in MEK1 and MEK2 missense and indel mutants (Figs. 5C and D, and Supplementary Fig. S5).
Transient or stable overexpression models enable faster and broader assessment of mutant allele activity and drug sensitivity than knock-in cell line or mouse models. While time-efficient, oncogene overexpression can confound assessment of drug sensitivity by altering relative levels of mutant and wild-type protein expression or by influencing other protein-protein interactions (50). We therefore used CRISPR/Cas9 to generate isogenic 293T cells in which the F53L, K57N and E102_I103del mutants were knocked-in and expressed under the control of the endogenous MEK1 promoter. As predicted, CRISPR knock-in MEK1 F53L, K57N and E102_I103del cells had increased ERK phosphorylation, with E102_I103del resulting in the greatest induction (Fig. 5E). Basal p-ERK was, however, markedly lower in each of the CRISPR knock-in lines as compared to 293T cells transfected with K57N or E102_I103del (Fig. 5F, 0nM trametinib lanes). As such, trametinib was able to more efficiently suppress p-ERK in CRISPR knock-in cells, especially in the case of MEK1 E102_I103del (Fig. 5F). Moreover, SCH772984 effectively abrogated p-RSK in both models (Fig. 5G).
Discussion
One limitation to the broader adoption of cancer precision medicine is the large number of variants of unknown significance identified during clinical tumor and germline next-generation sequencing. Emerging algorithmic strategies and high-throughput phenotyping screens have attempted to address this challenge, but experimentally validated consensus methodology is lacking. To gauge the utility of in silico methods for the stratification of variants of unknown significance as functional or benign, we leveraged a cohort of 42,434 tumor/normal matched pairs and comprehensive patient- and paralogy-directed functional analysis of the MAP2K1 and MAP2K2 genes. We found that an integrated, multifaceted computational analysis could, with strong predictive value, categorize mutants as biologically active versus likely inert.
It has long been presumed that mutational recurrence within a population is indicative of positive selection and thus likely to be predictive of functionality. More recently, this dogma has been questioned based on data suggesting that a subset of hotspots are non-functional variants arising at inherently mutable sites (51). For instance, APOBEC3A-mediated deamination of cytosine within DNA short hairpin loops can increase passenger mutation rates 200-fold compared to non-stem-loop sites enriched in known drivers (52). The hotspot algorithm used here was designed to account for many of these innate covariates including nucleotide context mutability, gene- and site-specific mutation rates; maximizing sensitivity for hotspot discovery (minimizing false negatives) while seeking to maintain a low false discovery rate of 1%. In support of the robustness of this computational approach, 93.5% of MEK1 hotspots were confirmed to induce ERK activation versus only five of 52 trending and non-hotspot sites. Our review of the postulated 27 optimal APOBEC3A substrate sites in MEK1 and 15 in MEK2 (52), indicate that none were mutated in MEK1 in the >42K sequenced tumors to date, suggesting that APOBEC3A-induced mutagenesis is not a confounding factor in this case.
The differences in inherent mutability of individual sites throughout the human genome does, however, reinforce the importance of careful biological verification of statistical methods as performed here using large, collaborative sequencing datasets to test and train algorithmic models coupled with subsequent laboratory validation. Our experience suggests that no single algorithm is likely to account for all genomic covariates or achieve a balance of sensitivity and specificity that is appropriate for all settings. For example, in a research versus clinical context, an algorithm designed to maximize specificity may fail to flag a subset of rare actionable variants that could be critical to individual active cancer patients in need of novel therapies.
To account for the limitations of the hotspot approach to variant classification and to help identify driver mutations in the longtail of the frequency distribution, we have proposed systematic integration of multiple in silico techniques that add critical dimensions and context, such as protein folding, sequence conservation or association with germline pathogenicity, to help strengthen or refine hotspot predictions. In our study, disordering of the T-loop alpha-helix served as a structural proxy for distinguishing activating from benign mutants. Provided that one can isolate discrete conformational changes which influence protein activation, MD simulation could also be a valuable method for inferring the mechanism by which a particular mutant induces kinase activation when sequence location is uninformative; information that could also influence drug sensitivity. Paralogy was also highly effective for candidate mutation discovery in MAP2K2, a gene for which the mutation rate is sufficiently low as to make hotspot analyses equivocal. For example, paralogy accurately predicted the functionality of the nine trending or non-hotspot MEK2 variants that aligned to activating MEK1 mutants. Moreover, paralogy analysis refined one trending (R53Q) and two hotspot (R112Q and R231H) calls to non-functional, similar to their inert MEK1 counterparts.
Whereas our functional studies were guided by and largely restricted to the spectrum of MAP2K1 and MAP2K2 mutations identified to date in cancer patients, several laboratories have taken the reciprocal approach of systematically screening pooled variants using high-throughput cancer phenotyping assays (53–58). These methods provide a rich array of data, but are time and labor intensive, require conversion of phenotypes into computational endpoints, and work best when a single phenotype (e.g. proliferation) is a robust surrogate of tumorigenic output. There is also the potential for masking of weaker variants in such screens by the confounding effects of protein overexpression or the failure to correctly classify variants that have neomorphic phenotypes. We acknowledge that our use of ERK phosphorylation as a surrogate for MEK1/2 variant activity, while straightforward, may also not account for neomorphic phenotypes, or phenotypes that only manifest in the context of specific co-alterations or lineage specific factors.
As several MEK inhibitors are FDA-approved, one primary motivation for delineating all activating MEK1/2 mutations is to identify the most appropriate candidates for treatment with selective ERK pathway inhibitors. However, while our in silico methods robustly identified activating MEK1/2 mutants, MEK inhibitor sensitivity varied widely among mutants. Neither inferences from domain location nor position in three-dimensional space explained the variable MEK inhibitor responses observed. MEK1/2 mutants were uniformly sensitive to ERK inhibition, suggesting that ERK inhibition may be a more broadly effective treatment approach for patients harboring MAP2K1/2 mutations. These data highlight the critical need to generate and train new robust algorithmic methodology to predict response to treatment to help guide the management of patients with previously uncharacterized variants of unknown significance.
Accrual of MEK-mutant patients to clinical trials of MEK or ERK inhibitors has been limited to date, and review of patient records at our institution revealed few examples of off-label MEK inhibitor use. Still, dramatic and durable response of MEK1-mutant tumors to MEK inhibitors have been observed in histiocytosis and in an exceptional responder with low-grade serous ovarian cancer (2,5–7). However, some of the exact mutants (e.g. Q56P, P124L) that were present in histiocytosis patients that responded to MEK inhibitors have been postulated as a basis for acquired resistance to RAF and/or MEK inhibitor therapy in melanoma and colorectal cancer (26,47,48,59). This clinical result suggests that cellular context including lineage, co-mutation pattern, gene dosage, or allelic imbalance may have equal or even greater influence on patient response than biochemical allele sensitivity. If so, patients with mutant alleles that are less sensitive to trametinib, such as E102_I103del, could still respond if the mutation arose in certain lineage contexts, such as histiocytosis, that are less likely to harbor co-mutations that diminish MEK dependence or that exhibit exquisite lineage dependence on ERK signaling. In such patients, even partial inhibition of MAP kinase signaling for short duration may be sufficient to induce a tumor response. Such tumors would be analogous to BCR-ABL mutant chronic myelogenous leukemia, where even transient exposure to dasatinib can provoke an irreversible apoptotic program and clinical response (60).
In sum, our efforts indicate that multifaceted in silico analysis is an efficient and accurate predictor of mutant allele biologic activity. We find that MD simulation and sequence paralogy are robust tools that can complement hotspot analysis and help prioritize recurrent and non-recurrent variants for in depth biologic, therapeutic and clinical validation.
Supplementary Material
Acknowledgments
This research was supported by the Marie-Josée and Henry R. Kravis Center for Molecular Oncology (CMO), Cycle for Survival, the NIH [R01 CA229624 (D. Solit), R01 CA234361 (D. Solit), U54 OD020355 (D. Solit, B. Taylor), R01 CA207244 (B. Taylor), R01 CA204749 (B. Taylor), R35-GM126985 (D. Xu) and R21-LM012790 (D. Xu), T32 TROT fellowship (B. Sylvester), 1 R01 CA201247(O. Abdel-Wahab)], the NIH/NCI [Cancer Center Support Grant P30 CA008748R01, R01 CA056821 (T. Merghoub)] and the NCI [ITCR grant U24-CA220457-01 (J. Gao)]. B. Taylor was also supported by research grants from the American Cancer Society (RSG-15-067-01-TBG), Prostate Cancer Foundation, Anna Fuller Fund, and the Josie Robertson Foundation. J. Gao was supported by the Fund for Innovation in Cancer Informatics from the Brown Performance Group (the-ici-fund.org). O. Abdel-Wahab was also supported by grants from the Histiocyte Society, the Erdheim-Chester Disease Global Alliance, the Functional Genomics Initiative of Memorial Sloan Kettering Cancer Center, the Histiocytosis Association, the Leukemia & Lymphoma Society and the Frame Fund. T. Merghoub was also funded in part through Swim Across America, Ludwig Institute for Cancer Research, Parker Institute for Cancer Immunotherapy and Virginia B. Squiers Foundation. The authors would like to acknowledge the Integrated Genomics Operation (S. Duygu Selcuklu and Pavitra Rao) and MSKCC Diagnostic Molecular Pathology. We would also like to acknowledge the AACR Project GENIE registry and consortium for their commitment to data sharing. We further acknowledge Rony Seger and Dustin Maly for sharing their MEK1/2 vectors via Addgene.
Conflict of Interest Statement:
D.B. Solit has served as a consultant/advisory board member for Pfizer, Loxo Oncology, Vivideon Therapeutics and Illumina. B.S. Taylor reports advisory board activities for Boehringer Ingelheim and honoria and research funding from Genentech. M.F. Berger has served as a consultant/advisory board member for Roche. O. Abdel-Wahab has served as a consultant for H3B Biomedicine, Foundation Medicine Inc, Merck, and Janssen and has received prior research funding from H3B Biomedicine unrelated to the current manuscript. A.N. Shoushtari has served as a consultant/advisory board member for Bristol-Myers Squibb, Immunocore, and Castle Biosciences and reports institutional research funding from AstraZeneca, Bristol-Myers Squibb, Immunocore, and Xcovery. Z. Yao has served as a consultant/advisory board member for MAPKURE, LLC. T. Merghoub has served as a consultant for Leap Therapeutics, Immunos Therapeutics and Pfizer, and co-founder of Imvaq therapeutics. T. Merghoub has equity in Imvaq therapeutics and reports grants from Bristol Myers Squibb, Surface Oncology, Kyn Therapeutics, Infinity Pharmaceuticals, Peregrine Pharmeceuticals, Adaptive Biotechnologies, Leap Therapeutics and Aprea. T. Merghoub is an inventor on patent applications related to work on oncolytic viral therapy, alphavirus-based vaccines, neo-antigen modeling, CD40, GITR, OX40, PD-1 and CTLA-4. The remaining authors have nothing to disclose.
References
- 1.Cheng ML, Berger MF, Hyman DM, Solit DB. Clinical tumour sequencing for precision oncology: time for a universal strategy. Nature reviews Cancer 2018;18:527–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Diamond EL, Durham BH, Haroche J, Yao Z, Ma J, Parikh SA, et al. Diverse and Targetable Kinase Alterations Drive Histiocytic Neoplasms. Cancer Discov 2016;6:154–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Van Allen EM, Wagle N, Sucker A, Treacy DJ, Johannessen CM, Goetz EM, et al. The genetic landscape of clinical resistance to RAF inhibition in metastatic melanoma. Cancer Discov 2014;4:94–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gao Y, Chang MT, McKay D, Na N, Zhou B, Yaeger R, et al. Allele-Specific Mechanisms of Activation of MEK1 Mutants Determine Their Properties. Cancer Discov 2018;8:648–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gounder MM, Solit DB, Tap WD. Trametinib in Histiocytic Sarcoma with an Activating MAP2K1 (MEK1) Mutation. The New England journal of medicine 2018;378:1945–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Papapanagiotou M, Griewank KG, Hillen U, Schimming TT, Moeller LC, Führer D, et al. Trametinib-Induced Remission of an MEK1-Mutated Langerhans Cell Histiocytosis. JCO Precision Oncology 2017:1–5 [DOI] [PubMed] [Google Scholar]
- 7.Diamond EL, Durham BH, Ulaner GA, Drill E, Buthorn J, Ki M, et al. Efficacy of MEK inhibition in patients with histiocytic neoplasms. Nature 2019;567:521–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang MT, Asthana S, Gao SP, Lee BH, Chapman JS, Kandoth C, et al. Identifying recurrent mutations in cancer reveals widespread lineage diversity and mutational specificity. Nature biotechnology 2016;34:155–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang MT, Bhattarai TS, Schram AM, Bielski CM, Donoghue MTA, Jonsson P, et al. Accelerating Discovery of Functional Mutant Alleles in Cancer. Cancer Discov 2018;8:174–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ellrott K, Bailey MH, Saksena G, Covington KR, Kandoth C, Stewart C, et al. Scalable Open Science Approach for Mutation Calling of Tumor Exomes Using Multiple Genomic Pipelines. Cell Syst 2018;6:271–81 e7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zehir A, Benayed R, Shah RH, Syed A, Middha S, Kim HR, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nature medicine 2017;23:703–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Consortium APG. AACR Project GENIE: Powering Precision Medicine through an International Consortium. Cancer Discov 2017;7:818–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hanrahan AJ, Schultz N, Westfal ML, Sakr RA, Giri DD, Scarperi S, et al. Genomic complexity and AKT dependence in serous ovarian cancer. Cancer Discov 2012;2:56–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meier C, Brookings DC, Ceska TA, Doyle C, Gong H, McMillan D, et al. Engineering human MEK-1 for structural studies: A case study of combinatorial domain hunting. J Struct Biol 2012;177:329–34 [DOI] [PubMed] [Google Scholar]
- 15.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, et al. The Protein Data Bank. Nucleic Acids Res 2000;28:235–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Biasini M, Bienert S, Waterhouse A, Arnold K, Studer G, Schmidt T, et al. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res 2014;42:W252–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hess B, van der Vegt NF. Hydration thermodynamic properties of amino acid analogues: a systematic comparison of biomolecular force fields and water models. J Phys Chem B 2006;110:17616–26 [DOI] [PubMed] [Google Scholar]
- 18.Abraham MJ, Gready JE. Optimization of parameters for molecular dynamics simulation using smooth particle-mesh Ewald in GROMACS 4.5. J Comput Chem 2011;32:2031–40 [DOI] [PubMed] [Google Scholar]
- 19.van Aalten DM, Bywater R, Findlay JB, Hendlich M, Hooft RW, Vriend G. PRODRG, a program for generating molecular topologies and unique molecular descriptors from coordinates of small molecules. J Comput Aided Mol Des 1996;10:255–62 [DOI] [PubMed] [Google Scholar]
- 20.Schuttelkopf AW, van Aalten DM. PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr D Biol Crystallogr 2004;60:1355–63 [DOI] [PubMed] [Google Scholar]
- 21.Kräutler V, van Gunsteren WF, Hünenberger PH. A fast SHAKE algorithm to solve distance constraint equations for small molecules in molecular dynamics simulations. The Journal of Computational Chemistry 2001;22:501–8 [Google Scholar]
- 22.Laino T, Hutter J. Notes on “Ewald summation of electrostatic multipole interactions up to quadrupolar level” [J. Chem. Phys. 119, 7471 (2003)]. J Chem Phys 2008;129:074102. [DOI] [PubMed] [Google Scholar]
- 23.Jakobsen AF. Constant-pressure and constant-surface tension simulations in dissipative particle dynamics. J Chem Phys 2005;122:124901. [DOI] [PubMed] [Google Scholar]
- 24.Nikolaev SI, Rimoldi D, Iseli C, Valsesia A, Robyr D, Gehrig C, et al. Exome sequencing identifies recurrent somatic MAP2K1 and MAP2K2 mutations in melanoma. Nature genetics 2011;44:133–9 [DOI] [PubMed] [Google Scholar]
- 25.Arcila ME, Drilon A, Sylvester BE, Lovly CM, Borsu L, Reva B, et al. MAP2K1 (MEK1) Mutations Define a Distinct Subset of Lung Adenocarcinoma Associated with Smoking. Clinical cancer research : an official journal of the American Association for Cancer Research 2015;21:1935–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Emery CM, Vijayendran KG, Zipser MC, Sawyer AM, Niu L, Kim JJ, et al. MEK1 mutations confer resistance to MEK and B-RAF inhibition. Proceedings of the National Academy of Sciences of the United States of America 2009;106:20411–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marks JL, Gong Y, Chitale D, Golas B, McLellan MD, Kasai Y, et al. Novel MEK1 mutation identified by mutational analysis of epidermal growth factor receptor signaling pathway genes in lung adenocarcinoma. Cancer research 2008;68:5524–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carlino MS, Fung C, Shahheydari H, Todd JR, Boyd SC, Irvine M, et al. Preexisting MEK1P124 mutations diminish response to BRAF inhibitors in metastatic melanoma patients. Clinical cancer research : an official journal of the American Association for Cancer Research 2015;21:98–105 [DOI] [PubMed] [Google Scholar]
- 29.Zheng CF, Guan KL. Activation of MEK family kinases requires phosphorylation of two conserved Ser/Thr residues. The EMBO journal 1994;13:1123–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alessi DR, Saito Y, Campbell DG, Cohen P, Sithanandam G, Rapp U, et al. Identification of the sites in MAP kinase kinase-1 phosphorylated by p74raf-1. The EMBO journal 1994;13:1610–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Caunt CJ, Sale MJ, Smith PD, Cook SJ. MEK1 and MEK2 inhibitors and cancer therapy: the long and winding road. Nature reviews Cancer 2015;15:577–92 [DOI] [PubMed] [Google Scholar]
- 32.Wagle N, Emery C, Berger MF, Davis MJ, Sawyer A, Pochanard P, et al. Dissecting therapeutic resistance to RAF inhibition in melanoma by tumor genomic profiling. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 2011;29:3085–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gao J, Chang MT, Johnsen HC, Gao SP, Sylvester BE, Sumer SO, et al. 3D clusters of somatic mutations in cancer reveal numerous rare mutations as functional targets. Genome Med 2017;9:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rodriguez-Viciana P, Tetsu O, Tidyman WE, Estep AL, Conger BA, Cruz MS, et al. Germline mutations in genes within the MAPK pathway cause cardio-facio-cutaneous syndrome. Science 2006;311:1287–90 [DOI] [PubMed] [Google Scholar]
- 35.Dentici ML, Sarkozy A, Pantaleoni F, Carta C, Lepri F, Ferese R, et al. Spectrum of MEK1 and MEK2 gene mutations in cardio-facio-cutaneous syndrome and genotype-phenotype correlations. Eur J Hum Genet 2009;17:733–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nava C, Hanna N, Michot C, Pereira S, Pouvreau N, Niihori T, et al. Cardio-facio-cutaneous and Noonan syndromes due to mutations in the RAS/MAPK signalling pathway: genotype-phenotype relationships and overlap with Costello syndrome. J Med Genet 2007;44:763–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu Y, Zhu J, Guo X, Huang T, Han J, Gao J, et al. How oncogenic mutations activate human MAP kinase 1 (MEK1): a molecular dynamics simulation study. Journal of Biomolecular Structure and Dynamics 2019:1–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chakraborty R, Hampton OA, Shen X, Simko SJ, Shih A, Abhyankar H, et al. Mutually exclusive recurrent somatic mutations in MAP2K1 and BRAF support a central role for ERK activation in LCH pathogenesis. Blood 2014;124:3007–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yuan J, Ng WH, Tian Z, Yap J, Baccarini M, Chen Z, et al. Activating mutations in MEK1 enhance homodimerization and promote tumorigenesis. Science signaling 2018;11. [DOI] [PubMed] [Google Scholar]
- 40.Grisham RN, Sylvester BE, Won H, McDermott G, DeLair D, Ramirez R, et al. Extreme Outlier Analysis Identifies Occult Mitogen-Activated Protein Kinase Pathway Mutations in Patients With Low-Grade Serous Ovarian Cancer. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 2015;33:4099–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nystrom AM, Ekvall S, Berglund E, Bjorkqvist M, Braathen G, Duchen K, et al. Noonan and cardio-facio-cutaneous syndromes: two clinically and genetically overlapping disorders. J Med Genet 2008;45:500–6 [DOI] [PubMed] [Google Scholar]
- 42.Rodriguez-Viciana P, Rauen KA. Biochemical characterization of novel germline BRAF and MEK mutations in cardio-facio-cutaneous syndrome. Methods Enzymol 2008;438:277–89 [DOI] [PubMed] [Google Scholar]
- 43.Mansour SJ, Matten WT, Hermann AS, Candia JM, Rong S, Fukasawa K, et al. Transformation of mammalian cells by constitutively active MAP kinase kinase. Science 1994;265:966–70 [DOI] [PubMed] [Google Scholar]
- 44.Bromberg-White JL, Andersen NJ, Duesbery NS. MEK genomics in development and disease. Brief Funct Genomics 2012;11:300–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aoidi R, Maltais A, Charron J. Functional redundancy of the kinases MEK1 and MEK2: Rescue of the Mek1 mutant phenotype by Mek2 knock-in reveals a protein threshold effect. Science signaling 2016;9:ra9. [DOI] [PubMed] [Google Scholar]
- 46.Roskoski R, Jr. MEK1/2 dual-specificity protein kinases: structure and regulation. Biochemical and biophysical research communications 2012;417:5–10 [DOI] [PubMed] [Google Scholar]
- 47.Trunzer K, Pavlick AC, Schuchter L, Gonzalez R, McArthur GA, Hutson TE, et al. Pharmacodynamic effects and mechanisms of resistance to vemurafenib in patients with metastatic melanoma. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 2013;31:1767–74 [DOI] [PubMed] [Google Scholar]
- 48.Ahronian LG, Sennott EM, Van Allen EM, Wagle N, Kwak EL, Faris JE, et al. Clinical Acquired Resistance to RAF Inhibitor Combinations in BRAF-Mutant Colorectal Cancer through MAPK Pathway Alterations. Cancer Discov 2015;5:358–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Morris EJ, Jha S, Restaino CR, Dayananth P, Zhu H, Cooper A, et al. Discovery of a novel ERK inhibitor with activity in models of acquired resistance to BRAF and MEK inhibitors. Cancer Discov 2013;3:742–50 [DOI] [PubMed] [Google Scholar]
- 50.Bielski CM, Donoghue MTA, Gadiya M, Hanrahan AJ, Won HH, Chang MT, et al. Widespread Selection for Oncogenic Mutant Allele Imbalance in Cancer. Cancer cell 2018;34:852–62 e4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hess JM, Bernards A, Kim J, Miller M, Taylor-Weiner A, Haradhvala NJ, et al. Passenger Hotspot Mutations in Cancer. Cancer cell 2019;36:288–301 e14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Buisson R, Langenbucher A, Bowen D, Kwan EE, Benes CH, Zou L, et al. Passenger hotspot mutations in cancer driven by APOBEC3A and mesoscale genomic features. Science 2019;364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Giacomelli AO, Yang X, Lintner RE, McFarland JM, Duby M, Kim J, et al. Mutational processes shape the landscape of TP53 mutations in human cancer. Nature genetics 2018;50:1381–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim E, Ilic N, Shrestha Y, Zou L, Kamburov A, Zhu C, et al. Systematic Functional Interrogation of Rare Cancer Variants Identifies Oncogenic Alleles. Cancer Discov 2016;6:714–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Berger AH, Brooks AN, Wu X, Shrestha Y, Chouinard C, Piccioni F, et al. Highthroughput Phenotyping of Lung Cancer Somatic Mutations. Cancer cell 2016;30:214–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brenan L, Andreev A, Cohen O, Pantel S, Kamburov A, Cacchiarelli D, et al. Phenotypic Characterization of a Comprehensive Set of MAPK1/ERK2 Missense Mutants. Cell Rep 2016;17:1171–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kohsaka S, Nagano M, Ueno T, Suehara Y, Hayashi T, Shimada N, et al. A method of high-throughput functional evaluation of EGFR gene variants of unknown significance in cancer. Sci Transl Med 2017;9. [DOI] [PubMed] [Google Scholar]
- 58.Ng PK, Li J, Jeong KJ, Shao S, Chen H, Tsang YH, et al. Systematic Functional Annotation of Somatic Mutations in Cancer. Cancer cell 2018;33:450–62 e10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shi H, Moriceau G, Kong X, Koya RC, Nazarian R, Pupo GM, et al. Preexisting MEK1 exon 3 mutations in V600E/KBRAF melanomas do not confer resistance to BRAF inhibitors. Cancer Discov 2012;2:414–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shah NP, Kasap C, Weier C, Balbas M, Nicoll JM, Bleickardt E, et al. Transient potent BCR-ABL inhibition is sufficient to commit chronic myeloid leukemia cells irreversibly to apoptosis. Cancer cell 2008;14:485–93 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.