Abstract
After three decades of amazing progress made in molecular studies of plant-microbe interactions (MPMI), we start to ask ourselves “what are the major questions still remaining?” as if the puzzle has only a few pieces missing. Such an exercise ultimately leads to the realization that we still have many more questions than answers. Therefore, it would be an impossible task for us to project a coherent “big picture” of the MPMI field in a single review. Instead, we provide our collection of opinions on where we would like to go in our own research (Figure 1) as an invitation to the community to join us in this exploration of new frontiers of MPMI.
Keywords: Extracellular immunity, pattern-triggered immunity (PTI), effector-triggered immunity (ETI), heterogeneity in immune responses, translational regulation, circadian clock
Extracellular immunity at the battlefront of plant defense
Plants encounter a great number of microbes with different lifestyles during their lifetime. The extracellular microbes, from beneficial to pathogenic, are the first challenge faced by the plant immune system. Plants could launch immunity to restrict the colonization and entry of these microbes (Melotto et al., 2006; Boller and He, 2009; Hawes et al., 2012). At the same time, some of these beneficial microbes and successful pathogens have evolved sophisticated strategies to either evade or actively disrupt these surveillance mechanisms (Liang et al., 2013; Melotto et al., 2006; 2017), thereby achieving successful colonization and entry into plants.
At this battlefront, stomatal immunity against leaf-associated microbes and rhizospheric immunity against root-associated microbes are essential layers of the plant’s extracellular defense system. Even after entry into plants, many plant pathogens proliferate in the apoplastic space outside of plant cells. Therefore, immune responses that occur in the apoplast can also be considered extracellular immunity. Significant progress has been made in understanding the immune mechanisms in aerial tissues (Melotto et al., 2006; Boller and Felix, 2009; Zeng and He, 2010; Zeng et al., 2011; Dubiella et al., 2013; Gao et al., 2013; Kadota et al., 2014; Li et al., 2014; Liang et al., 2018; Sun and Zhang, 2020; Zhou and Zhang, 2020). However, in roots, with constant exposure to soil microbes, how plants differentiate beneficial microbes from pathogenic ones and establish rhizospheric immunity remain mostly unknown.
Stomatal immunity
Stomata are natural pores surrounded by specialized guard cells that control gas exchange and water loss in plants. They are typically found in the leaf epidermis and play a critical role in plant growth and development. However, they also provide natural openings for entry of some filamentous and bacterial pathogens (Melotto et al., 2006; Zeng and He, 2010; Zeng et al., 2011). Upon contact, conserved molecular signatures derived from microbes, named microbe-associated molecular patterns (MAMPs) (Ausubel, 2005; Boller and Felix, 2009) are recognized by plant surface pattern-recognition receptors (PRRs). The perception of MAMPs induces stomatal closure, which is important for restricting the entry of microbes lacking a penetration apparatus (Melotto et al., 2006; Thor et al., 2020; Ye et al., 2020). MAMP perception also triggers complicated crosstalk of multiple plant hormone signaling pathways. It has been well-documented that the plant hormone abscisic acid (ABA) can induce stomatal closure (Sirichandra et al., 2009; Joshi-Saha et al., 2011), whereas a derivative of jasmonate (JA), (+)-7-iso-jasmonoyl-L-Ile (JA-Ile), induces stomatal opening (Okada et al., 2009). In addition, salicylic acid (SA)-deficient mutants exhibit compromised stomatal defense mechanisms, indicating a positive role of SA in stomatal defense (Melotto et al., 2006; Zeng and He, 2010; Zeng et al., 2011).
Besides MAMP perception, stomatal development mediated by the ERECTA (ER)-YODA (YDA) signaling pathway also indirectly contributes to plant immunity (Nadeau, 2009; Shpak et al., 2005; Meng et al., 2015). Moreover, this signaling pathway can act in parallel with the canonical PRR signaling pathway to confer broad-spectrum immunity with the ER serving to also recognize yet-to-be-identified MAMPs (Godiard et al., 2003; Llorente et al., 2005; Adie et al., 2007; Shpak, 2013; Häffner et al., 2014; Sopeña-Torres et al., 2018). These findings indicate dual roles of ER-YDA signaling in stomatal development and immunity, and suggest that the key regulators of stomatal development might be co-opted for stomatal immunity during evolution to resolve the dilemma of stomata allowing gas exchange as well as pathogen entry.
To counteract plant immune responses, successful foliar pathogens and some beneficial microbes have evolved strategies to interfere with stomatal immunity. Several pathovars of Pseudomonas syringae produce the phytotoxin coronatine (COR) to induce an interaction between CORONATINE INSENSITIVE1 (COI1) and JASMONATE ZIM-DOMAIN proteins (JAZs). This induced interaction triggers the degradation of JAZs and the activation of NO APICAL MERISTEM [NAM], ARABIDOPSIS TRANSCRIPTION ACTIVATION FACTOR [ATAF], AND CUP-SHAPED COTYLEDON [CUC] (NAC) transcription factors (TFs), thereby inhibiting the accumulation of SA and promoting stomatal opening (Chini et al., 2007; Thines et al., 2007; Yan et al., 2007, 2009; Katsir et al., 2008; Melotto et al., 2008; Sheard et al., 2010; Zheng et al., 2012; Gimenez-Ibanez et al., 2017; Melotto et al., 2017). COR also targets the Arabidopsis RPM1-INTERACTING4 (RIN4) and plasma membrane H+-ATPases, AUTOINHIBITED H(+)-ATPASE ISOFORM 1 (AHA1) and AHA2, to induce stomatal opening (Zhang et al., 2008a; Liu et al., 2009).
Bacterial pathogens secrete effectors to prevent stomatal closure triggered by the perception of MAMPs. The P. syringae effectors HopZ1, AvrB, and HopX1 all target COI1-JAZ complexes to promote stomatal opening (Jiang et al., 2013; Gimenez-Ibanez et al., 2014; Lee et al., 2015; Ma et al., 2015; Zhou et al., 2015b). HopF2 has been reported to target the MITOGEN-ACTIVATED PROTEIN KINASE KINASE 5 (MAPKK5), BRI1-ASSOCIATED RECEPTOR KINASE 1 (BAK1), and RIN4 to suppress MAMP-induced stomatal closure (Wang et al., 2010; Wilton et al., 2010; Hurley et al., 2014; Zhou et al., 2014). The Xanthomonas oryzae effector XopR targets the Arabidopsis RESPIRATORY BURST OXIDASE HOMOLOG D (RBOHD) and interferes with stomatal defense (Wang et al., 2016). The examples of pathogen effector-mediated evasion of stomatal immunity reinforce its important function in plant defense.
Developmentally related to stomata, hydathodes found in leaves of many plants direct the colonization of phytopathogens into the xylem, including Xathomonas campestris (Cerutti et al., 2017). Although hydathode pores do not respond to flagellin treatment, it remains unclear whether hydathodes contribute to sensing and restriction of microbial entry.
Rhizospheric immunity
Although less understood than leaf defense, it is conceivable that the plant rhizosphere is equipped with sophisticated immune mechanisms to differentiate commensal and beneficial microbes from pathogens to allow normal interactions with microbes that are important for plant health.
Extensins are cell wall glycoproteins that are predominantly expressed in plant roots and play an essential role in plant defense (Esquerré-Tugayé et al., 1979; Merkouropoulos and Shirsat, 2003). The composition and structure of extensins in the roots of infected plants undergo significant alterations upon the perception of pathogenic microbes (Xie et al., 2011; Plancot et al., 2013; Wu et al., 2017b), resulting in the strengthening of the plant cell wall to restrict infection. In addition to playing a direct role in modulating the structure and rigidity of the cell wall, extensins and extensin-like molecules are secreted into the rhizosphere and extracellular rhizospheric mucilage (Koroney et al., 2016).
Besides molecules like extensins, border cells are detached cell populations released from the root tip into the rhizosphere (Hamamoto et al., 2006; Hawes et al., 2012). Pathogenic infection can cause an increase in the release of border cells from the root tip (Zhao et al., 2000; Cannesan et al., 2011; Hawes et al., 2012). Accumulating evidence indicates a potential role of border cells in attracting or repelling certain microbes (Gunawardena et al., 2005; Curlango-Rivera and Hawes, 2011; Driouich et al., 2013). Border and border-like cells secrete mucilage containing antimicrobial molecules, polysaccharides, and extracellular DNAs (exDNAs). Both treatment of roots with DNase I and mutating the pathogen’s exDNase-encoding genes reduce root tip immunity, suggesting a positive role of exDNAs in root immunity (Curlango-Rivera et al., 2013; Hawes et al., 2016; Tran et al., 2016). What are the functional mechanisms of exDNAs in the process? Do attraction and repulsion of microbes confer specificity? How is specificity achieved? These are major questions that remain to be addressed.
Extensins and extensin-like molecules, together with root border and border-like cells, have been proposed to form the “root extracellular trap” that functions as a defense network (Brinkmann et al., 2004; Driouich et al., 2013). Additional components that contribute to the formation of root extracellular trap await further investigation. This protective network has potential roles in eliminating pathogenic microbes and facilitating the attachment of beneficial microbes (Brinkmann et al., 2004; Driouich et al., 2013). Secreted extensins and extensin-like molecules can interact with pectins and arabinogalactan proteins to reinforce the mucilage network, thus weakening the attachment of pathogenic microbes to plant roots (Oosterveld et al., 2002; Immerzeel et al., 2006; Cannesan et al., 2012). During a symbiotic interaction, the accumulation of extensins in infected nodules suggests their putative roles in facilitating the attachment of beneficial microbes (Sujkowska-Rybkowska and Borucki, 2014). It remains unknown whether secreted extensins, and extensin-like molecules could serve as signals to modulate microbial behavior. The underlying mechanisms of the root extracellular trap in mediating the interactions with soil microbes await further investigation.
Apoplastic immunity
The apoplast is an enclosed active battle field between plants and microbes and is one of the first environments encountered after pathogen invasion inside the plant body. The plant cell wall is a formidable physical barrier that restricts pathogen colonization. Pathogen attack usually triggers modifications in the plant cell wall and alterations in cell wall integrity (Malinovsky et al., 2014; Bacete et al., 2018; De Lorenzo et al., 2019). The deposition of callose at infection sites is a typical defense response, strengthening the plant cell wall (Stone and Clarke, 1992). Pathogens secrete cell wall-degrading enzymes to modify or hydrolyze cell wall components. The impairment of integrity and degradation of cell walls produces damage or pattern signals that are sensed by plants to activate immune signaling, indicating an active role of the plant cell wall in immunity, rather than a passive role as a barrier only (Malinovsky et al., 2014; Bacete et al., 2018; De Lorenzo et al., 2019).
In the addition to the cell wall, plants also secrete pathogenesis-related (PR) proteins, lytic enzymes and antimicrobial secondary metabolites into the apoplastic space upon pathogen challenge to restrict the colonization of both foliar and rhizospheric microbes (van Loon et al., 2006; Shabab et al., 2008; van Esse et al., 2008). Some MAMPs buried within supramolecular microbial surface structures or larger proteins need to be released by the host lytic enzymes for perception to occur (Liu et al., 2014; Buscaill et al., 2019). Therefore, these enzymes not only serve to destruct microbial structures, but also release immunogenic patterns to trigger plant defenses (Doehlemann and Hemetsberger, 2013; Liu et al., 2014; Jashni et al., 2015; Buscaill et al., 2019).
Roots are also capable of sensing MAMPs in the apoplastic space and mount immune responses (Millet et al., 2010; Beck et al., 2014; Wyrsch et al., 2015; Stringlis et al., 2018). However, perception of the immunogenic peptides flg22, nlp20, and the medium-chain 3-hydroxy fatty acid are spatially restricted in Arabidopsis roots, with the differentiated zone exhibiting very low to no response (Zhou et al., 2020). It has also been observed that there is a higher expression of PRR genes in the inner cellular layers than in the outer cellular layers of Arabidopsis roots (Beck et al., 2014). The lack of flagellin responsiveness correlates with the lack of FLAGELLIN SENSING 2 (FLS2) expression in the differentiated zone. Laser-induced cell ablation of small clusters of cells was shown to be sufficient to induce FLS2 expression and confer MAMP responsiveness in neighboring cells (Zhou et al., 2020). These observations suggest a spatial differentiation of immune responses in root cells that provides a strategy to reduce their responsiveness to beneficial and free-living microbes, while enabling the initiation of specific and strong immune responses to pathogenic microbes through damage associated with the infection. The results highlight the interplay between stresses and suggest a “damage-gated” mechanism for differential response to beneficial and pathogenic microbes.
Microbes employ diverse strategies to evade or disrupt host recognition in the apoplast. Some of them evade host recognition via the divergence of MAMPs. For instance, the nitrogen-fixing symbiont Sinorhizobium meliloti possesses variations in the core sequences of flagellin, evading recognition by the flagellin receptor FLS2 in Arabidopsis (Felix et al., 1999). Another common strategy to suppress apoplastic immunity is through disruption of the recognition of MAMPs by their cognate receptors. Foliar pathogens can secrete effector proteins into the apoplast to achieve this goal. For example, some P. syringae and P. aeruginosa strains secrete the alkaline protease AprA that is capable of degrading flagellin monomers, thereby inhibiting flagellin perception (Bardoel et al., 2011; Pel et al., 2014). P. syringae can also secrete inhibitory molecules to suppress the activity of the plant β-galactosidase 1, which functions to release immunogenic peptides from glycosylated flagellin (Buscaill et al., 2019). Moreover, P. syringae strains possess glycan polymorphisms on flagellin that can evade hydrolysis by plants (Buscaill et al., 2019). The fungal pathogen Cladosporium fulvum secretes the chitin-binding effector proteins Avr4 and Ecp6 to prevent chitin hydrolysis or its recognition by the lysin motif domain-containing receptors (Van den Burg et al., 2006; de Jonge et al., 2010). Although less documented for their specific functions, AprA homologs and Ecp6-like proteins are present in a wide range of rhizospheric beneficial bacterial (Pel et al., 2014) and fungal species (Bolton et al., 2008). The soil-borne pathogen Verticillium dahliae secretes the polysaccharide deacetylase, VdPDA1, to deacetylate chitin oligomers and disrupt chitin perception by plants, providing evidence for the prevention of MAMP recognition by root-infecting fungal pathogens (Gao et al., 2019). In addition, the plant growth-promoting rhizobacterium Piriformospora indica produces the β-glucan-binding lectin, FGB1, to disrupt host perception of chitin (Wawra et al., 2016).
Besides proteins, microbes can also produce other molecules to suppress MAMP-activated immune response. Pseudomonas spp. uses organic acids to lower the environmental pH and inhibit flagellin-activated immune responses (Yu et al., 2019a) while Bradyrhizobium japonicum produces nodulation (Nod) factors to suppress MAMP-induced immune responses in soybean and Arabidopsis (Liang et al., 2013). Lipopolysaccharide of S. meliloti has been shown to suppress MAMP-induced reactive oxygen species (ROS) burst in Medicago truncatula (Scheidle et al., 2005; Tellstrom et al., 2007).
The above evidence demonstrates that the apoplast is a place where the battle between the pathogen and the host intensifies. Despite the progress, the mechanisms that prevent overstimulation of immune responses against beneficial and free-living microbes remain a challenging area for future research.
Sensing immunogenic signals by immune receptors on the cell surface
PRRs are also referred to as cell surface immune receptors that include a large number of receptor kinases (RKs) and receptor-like proteins (RLPs) (Tang et al., 2017; Zipfel and Oldroyd, 2017). In addition to MAMPs, PRRs also sense host-derived, damage-associated molecular patterns (DAMPs), such as structures that are released from plant cell walls, extracellular ATP and NAD (Choi et al., 2014; Wang et al., 2017a; Bacete et al., 2018). These immunogenic patterns are characteristic of a pathological state caused by pathogen invasion (Cook et al., 2015). In addition to MAMPs and DAMPs, plants also produce immune-modulating peptides called phytocytokines whose receptors either directly stimulate immune responses or interact with other PRRs to modulate immune responses (Gust et al., 2017; Segonzac and Monaghan, 2019). Perception of immunogenic patterns by PRRs activates pattern-triggered immunity (PTI) which plays a key role in fending off potential pathogens.
Detailed analyses of several Arabidopsis PRRs, particularly FLS2, EF-TU RECEPTOR (EFR), and LysM-CONTAINING RECEPTOR-LIKE KINASE 5 (LYK5), have uncovered molecular mechanisms underlying the activation of early signaling events downstream of PRRs. Associate with PRRs, receptor-like cytoplasmic kinases (RLCKs) including Botrytis-Induced Kinase 1 (BIK1), PBS1-like kinases (PBLs), and BR-Signaling Kinase 1 (BSK1) directly connect PRRs to downstream components (Lu et al., 2010; Zhang et al., 2010; Shi et al., 2013; Rao et al., 2018; Majhi et al., 2019). The phosphorylation state and/or stability of BIK1is regulated by MAP 4 kinase, PP2C, CPK28 and PLANT U-BOX PROTEIN 25 (PUB25) and PUB26, allowing fine-tuning of responsiveness and degree of immune output (Monaghan et al., 2014; Couto et al., 2016; Wang et al., 2018a; Zhang et al., 2018). A recent study shows that BIK1 is also mono-ubiquitinated by E3 ubiquitin ligases RING-H2 FINGER A3A (RHA3A) and RHA3B and that this modification is necessary for BIK1 to dissociate from PRRs and activate downstream signaling (Ma et al., 2020). Do different PRRs connect to the same RLCKs? A systematic analysis of RLCK higher order mutants showed that while some RLCKs are redundantly required for multiple PRRs, others are specifically required for particular PRRs (Rao et al., 2018). Unlike most other RLCKs that positively regulate PRR-dependent immunity, PBL13 negatively regulates FLS2 function (Lin et al., 2015). Moreover, BIK1 was also found to negatively regulate signaling mediated by RLP23, a receptor for the epitope nlp20 derived from fungal, oomycete and bacterial NLP proteins, through an unknown mechanism (Wan et al., 2019b).
Upon activation by PRRs, RLCKs phosphorylate downstream components, initiating an array of early signaling events including rapid influx of calcium ion, production of ROS, activation of heterotrimeric G proteins, and activation of MAP kinase cascades. BIK1 and its paralog in rice can phosphorylate several cyclic nucleotide-gated channels (CNGCs) to positively regulate calcium influx, although these findings only partially explain MAMP-triggered calcium burst (Tian et al., 2019; Wang et al., 2019b). The increased calcium influx is profoundly important in immune activation, as a number of key immune regulatory proteins are directly regulated by calcium. These include calcium-dependent kinases (CDKs), RBOHs, TFs such as CALMODULIN BINDING PROTEIN 60-LIKE g and CAMODULIN-BINDING TRANSCRIPTION ACTIVATOR 3 (Wang et al., 2009; Huang et al., 2020), CALCIUM-DEPENDENT PROTEIN KINASES (CPKs; Romeis et al., 2001) and calcium-dependent metacaspases (Hander et al., 2019; Shen et al., 2019). BIK1 also directly phosphorylates the N-terminus of RBOHD to promote the rapid production of ROS which serve as a secondary signal for immune activation (Kadota et al., 2014; Li et al., 2014). In contrast, PBL13 phosphorylates the C-terminus of RBOHD to promote its degradation and dampen its activity (Lee et al., 2020). BIK1 additionally phosphorylates components of heterotrimeric G protein complex including regulator of G protein signaling 1 and EXTRA LARGE G protein 2 (XLG2) to activate heterotrimeric G proteins (Liang et al., 2016; 2018). Furthermore, BIK1, multiple PBLs, and BSK1 phosphorylate specific residues in the C-terminus or N-terminus of MAP kinase kinase kinases to activate MAP kinase cascades to induce defense (Yamada et al., 2016; Bi et al., 2018; Yan et al., 2018). Despite of these advances, we still know little about how these early signaling events are linked to downstream defense. For instance, what are the target proteins regulated by ROS and G proteins? How do these proteins regulate defense outputs?
Sensing microbial effector proteins inside the host cell
In addition to PRRs, plants also possess intracellular immune receptors that are nucleotide-binding leucine-rich repeat receptors (NLRs; Jones et al., 2016). NLRs recognize pathogen effectors delivered into the host cell which are initially evolved as virulence factors. This layer of immunity has been referred to as ETI (effector-triggered immunity). The recognition occurs either directly via a physical interaction between an NLR and an effector or indirectly via an interaction between an NLR and an effector-target protein in the host cell. NLR proteins can be broadly divided into three major classes based on the N-terminal domains: coiled-coil, TOLL-INTERLUKIN 1 RECEPTOR, or RPW8 and are thus called CNL, TNL, and RNL (Shao et al., 2016). Recent advances show that these NLRs play different roles in the perception of effectors and immune signaling. Some TNLs and CNLs act as singletons in effector perception and signaling, while others as physically interacting pairs, with one acting as an effector sensor, while the other serving as a helper for signaling (Adachi et al., 2019b). RNLs are a small class of highly conserved NLRs divided into the N. benthamiana N requirement gene1 (NRG1) and Activated Disease Resistance 1 (ADR1) clades (Peart et al., 2005; Bonardi et al., 2011; Collier et al., 2011; Wang et al., 2011) and act downstream of all studied TNLs and some CNLs as “executors” of the hypersensitive response (HR) or salicylate (SA) production (Wang et al., 2011; Qi et al., 2018; Gantner et al., 2019; Lapin et al., 2019). Solanaceous plants carry an additional class of slow-evolving CNLs called NLR-REQUIRED FOR CELL DEATH (NRCs) that function as a network required for the function of different sensor NLRs (Wu et al., 2017a).
Recent breakthroughs are starting to unveil mechanisms by which TNLs and CNLs activate immune signaling upon effector perception. Two reports show that the TIR domain of TNLs possesses the NADase activity upon dimerization, and an unidentified metabolite(s) generated in the reaction is proposed to act as a signal that functions through ENHANCED DISEASE SUSCEPTIBILITY 1 (EDS1) before activating NRGs and ADRs through unknown mechanisms (Horsefield et al., 2019; Wan et al., 2019a). Another study on ZAR1, a singleton NLR with a canonical coiled-coil domain, showed that, upon perception of effector activity, it oligomerizes to form a pentameric complex (Wang et al., 2019a, 2019c). In this complex, which was named the resistosome, the N-terminal α1 helix of ZAR1 forms a funnel-shaped structure, and the resulting pore on the plasma membrane has been found to be required for cell death and antibacterial immunity. The fact that the structural requirements for in vitro assembly of the resistosome are also required for effector-induced oligomerization observed in protoplasts supports the formation of the resistosome during immune activation. How does this pore-forming activity trigger cell death and immune responses? Does it simply cause cell lysis or create an ion channel? Is it also localized to the membranes of organelles? These are urgent questions to be addressed. The mechanism underlying ZAR1 resistosome-mediated immune signaling appears to be shared by a group of CNLs, including the N. benthamiana NRC proteins and some canonical CNLs, which carry a MADA-motif in the N-terminus, as this motif possess similar signaling activity as the ZAR1 α1 helix (Adachi et al., 2019a). The coiled-coil domain of RNLs, referred to as CCR in recent literatures, appear to have a comparable fold as ZAR1’s CC and may also form a pore on membranes, a possibility remains to be tested. This raises an interesting possibility that TNLs and CNLs may use analogous mechanisms for immune execution.
Cross-talk among different receptors and immune activation
It is increasingly clear that integration of different immunogenic signals is critical for proper plant immune output. Two very recent studies showed that activation of NLRs in the absence of PRR activation results in mild immune responses (Ngou et al., 2020; Yuan et al., 2020). When both PRRs and NLRs are stimulated, the plant mounts strong defenses characterized by a much greater ROS production, defense gene transcription, the HR, and fully induced disease resistance. It appears that activation of the two classes of immune receptors can mutually potentiate each other in defense outputs. A study showed that activations of PRRs and NLRs give rise to overlapping phosphoproteomes, with some key signaling components phosphorylated at similar sites, suggesting a very early cross talk of PRR and NLR signaling pathways (Kadota et al., 2019). It is interesting to note that the BIK1-dependent phosphorylation is required but not sufficient for activating CNGCs, RBOHD, heterotrimeric G proteins, and MAP kinase kinase kinases (Kadota et al., 2014; Li et al., 2014; Liang et al., 2016; Bi et al., 2018; Tian et al., 2019). It is possible that multiple inputs at the post translational level are required for the activation of these key signaling components. Indeed, activation of RBOHD also requires phosphorylation by CPKs and likely EF-hand-dependent calcium-binding in the N-terminus of this protein (Dubiella et al., 2013). Do PTI and ETI employ common signaling pathways? While RBOHD and MAP kinases are known to be required for both PTI and ETI (Katota et al., 2019; Su et al., 2018), whether additional PTI components are also required for ETI remains an outstanding question. In addition, it is unknown whether different signal inputs are integrated via direct receptor-receptor interactions or at the level of post-translational modification of signaling components.
Under field conditions, plants are frequently subjected to simultaneous infection with different pathogen classes. Cross talk among immune receptors/co-receptors also plays an important role in cross-protection during complex infections. A recent study shows that CERK1, a co-receptor for chitin, is associated with BAK1, which is a co-receptor for multiple proteinaceous PAMPs including flagellin (Gong et al., 2019). Infection of Arabidopsis plants with bacterial pathogens leads to a BAK1-dependent phosphorylation of CERK1 in the jaxtamembrane domain which potentiates chitin perception and antifungal immunity. Another receptor-like kinase, nuclear shuttle protein-interacting kinase1 (NIK1), is a positive player in anti-virus immunity. NIK1 interacts with FLS2, and the activation of FLS2 can lead to NIK1 phosphorylation to potentiate anti-viral immunity (Li et al., 2019a).
The observation of “damage-gated” MAMP perception in the differentiated zone of Arabidopsis roots represents another example of how different signals are integrated to activate defenses only in response to pathogen infection (Zhou et al., 2020). This unique control of immunity may be particularly important for root microbiota, as constant stimulation of immune responses is not desirable for the interaction with commensal or beneficial microbes. It is currently unknown, however, how cell damage leads to PRR gene expression in the neighboring cells, as application of known DAMPs failed to induce PRR expression in differentiated zone.
Defense mediated by noncanonical resistance genes
In contrast to PRRs and NLRs, other resistance proteins belong to diverse families and often confer broad-spectrum disease resistance. Durable resistance to soybean cyst nematode conferred by Rhg1 is linked to copy number variation in a gene cluster encoding an α-soluble N-ethylmaleimide-sensitive factor attachment protein, a wound-inducible domain protein 12, and an amino acid transporter (Cook et al., 2012). Rhg1 exhibits strong epistatic interaction with Rhg4, a serine hydroxy-methyltransferase, to confer full nematode resistance in the Peking background (Meksem et al., 2001; Liu et al., 2012). Broad-spectrum disease resistance can also be conferred by mutations in susceptibility genes (van Schie and Takken, 2014). For example, loss of function of rice Bsr-k, encoding a tetratricopeptide repeats protein, induces overaccumulation of defense related OsPAL mRNAs, resulting in resistance to Magnaporthe oryzae and Xanthomonas oryzae pv. oryzae (Zhou et al., 2018). Naturally occurring transcription factor alleles, such as the rice Broad-Spectrum Resistance-Dingu 1 (Bsr-d1), can confer broad-spectrum resistance (Li et al, 2017). The rice Ideal Plant Architecture 1 (IPA1) gene encodes a SQUAMOSA promoter binding protein-like (SPL) transcription factor (Jiao et al., 2010; Miura et al., 2010). The ipa1–1D allele is resistant to targeting by miR156 and miR5269, resulting in higher IPA1 transcript and protein accumulation (Jiao et al., 2010; Wang et al., 2015b). The presence of ipa1–1D is able to increase both yield and disease resistance to the fungal pathogen Magnaporthe oryzae (Wang et al., 2018b). Phosphorylation changes this transcription factor’s DNA binding specificity; unphosphorylated IPA1 binds the DEP1 promoter to enhance growth and yield, whereas phosphorylated IPA1 binds the WRKY45 promoter to enhance defense gene expression (Wang et al., 2018b). This elegant example illustrates how a mechanistic understanding of broad-spectrum resistance can be used to enhance crop production. In wheat, WHEAT KINASE START1 (WKS1) encodes a kinase fused to a START lipid-binding domain and renders partial resistance to stripe rust caused by Puccinia striiformis f. sp. tritici (Fu et al., 2009). WKS1 localizes to chloroplasts where it phosphorylates a thylakoid-associated ascorbate peroxidase, reduces the ability of the cells to detoxify ROS and contributes to death of infected cells (Gou et al., 2015). Another noncanonical resistance gene Yr19/Lr34/Sr57/Pm38 encodes an abscisic acid ABC transporter and controls partial resistance against powdery mildew, leaf rust, stripe rust, and stem rust (Krattinger et al., 2019). Yr46/Lr67/Sr55/Pm39 encodes a nonfunctional hexose transporter that confers partial resistance to powdery mildew and all three wheat rusts (Moore et al., 2015). Fhb7 encodes a glutathione S-transferase that confers broad resistance to Fusarium species by detoxification of trichothecenes, such as the deoxynivalenol (DON) toxin in Fusarium graminearum (Wang et al., 2020). It seems that each of these noncanonical resistance genes mediate unique defense mechanisms. Additional studies are required to elucidate which directly perceive the presence of the pathogen and how they activate disease resistance.
An emerging class of broad-spectrum disease resistance genes encode tandem kinase proteins (TKPs), which represent a novel protein family that is widely present in the plant kingdom (Klymiuk et al., 2018). TKPs evolved by either duplication or fusion of two kinase domains, with the most common functional domains belonging to WALL ASSOCIATED KINASEs (WAK) and cysteine-rich kinases. Wheat tandem kinase 1 (WTK1) possesses a kinase-pseudokinase domain architecture and is likely derived from a fusion of WAK and RLCK subfamily VIII domains. WTK1 confers broad-spectrum resistance against more than 3,000 genetically diverse Puccinia striiformis f. sp. tritici isolates (Klymiuk et al., 2018). WTK3 also possesses a kinase-pseudokinase domain architecture and confers resistance to powdery mildew caused by Blumeria graminis f. sp. tritici (Lu et al., 2020). WTK2 and barley RPG1 possess tandem kinase domains and confer resistance to wheat and barley stem rusts, respectively (Brueggeman et al., 2002; Chen et al., 2020). Many outstanding questions remain about this novel class of resistance genes. How are effector(s) or pathogen components recognized? Do pseudokinase domains serve as decoys for pathogen effectors? What downstream signaling events are initiated by diverse TKPs? Does TKP function extend beyond immunity?
Immune responses in vascular tissue
Not only can diverse pathogens of all classes infect plants, but also their distribution and disease phenotypes are variable within a tissue. Various plant pathogens are capable of colonizing specific host tissues and some of them can invade into distinct vascular cell types. Phloem-limited pathogens comprise vector-borne viruses and bacteria whose distribution is influenced by climate change and interconnected with production practices (Juroszek et al., 2020). Bacterial and fungal pathogens such as Xylella, Ralstonia, Clavibacter, Fusarium, and Verticillium employ diverse strategies, including vector transmission, root invasion, and seed/wound entry, to gain entry into and colonize xylem vessels (Michielse and Rep, 2009; Bae et al., 2015).
Fundamental questions remain about mechanisms regulating plant immune responses to pathogens that colonize vascular tissues. Multiple NLR immune receptors have been identified that recognize effectors from xylem-colonizing pathogens. They include RPS4/RRS1-R and ZAR1 that recognize the bacterial pathogens R. solanacearum and Xanthomonas campestris, respectively (Narusaka et al., 2009; Wang et al., 2015a). Using a split-GFP system to visualize effector delivery early during infection, the recognized R. solanacearum effector PopP2 accumulated in the nuclei of host cells surrounding the site of lateral root emergence, which correlates with sites of invasion before xylem entry (Henry et al., 2017). The soilborne fungus Fusarium oxysporum f. sp. lycopersici (Fol) is recognized in tomato by two surface localized immune receptors (the receptor like protein I and the receptor like kinase I-3) as well as one intracellular NLR (I-2) (Takken and Rep, 2010). Tomato lines with the surface localized immune receptors I and I-3 were more effective at restricting fungal spread than the NLR I-2, but all three only inhibited colonization after Fusarium reached xylem tissues (van der Does et al., 2019). These intriguing results expose multiple outstanding questions: What plant cell types are responsible for immune perception? Are specific cell types targeted for effector delivery?
The phloem environment contains carbohydrates, proteins, amino acids, and sugars, representing a specialized niche for pathogens. Some of the most devastating plant pathogens and insects reside and feed, respectively, in phloem sieve elements including the bacterium Ca. Liberibacter asiaticus that has severely impacted the citrus industry worldwide, causing billions of dollars in economic losses (Wang et al., 2017b). To fight infection, the phloem transports mobile defense signals required for systemic acquired resistance (SAR) and systemic wound responses. Recent studies on glutamate, which acts a DAMP, provided striking images of the speed and systemic spread of calcium after glutamate perception (Toyota et al., 2018). Waves of Ca2+, electrical signals, and ROS form a network to promote rapid systemic signaling in plants (Choi et al., 2016). Phloem-specific forisomes and P proteins are also activated by Ca2+, rapidly expand in size, and have been hypothesized to occlude sieve pores (Knoblauch et al., 2014). However, detailed investigation of SEOR (sieve element occlusion-related) P proteins did not reveal alterations in phloem translocation and future studies will be required to understand their physiological roles (Knoblauch et al., 2014). Callose deposition at sieve plates and plasmodesmata is a common defense in response to pathogens and insect feeding which blocks viral spread, but not movement of Ca. Liberibacter (Hao et al., 2008; Achor et al., 2020). The distribution of the bacterial pathogens, phytoplasmas, and Ca. Liberibacters in the vascular tissue cannot be completely explained by source-sink relationships either. Future research is required to understand how both pathogens and the host alter tissue tropism during an infection.
It remains unclear if phloem sieve elements or companion cells are capable of direct pathogen recognition through PRRs or NLRs. There is evidence that phloem-feeding insects can trigger plant immunity while possessing effectors that suppress defense responses. Hemipteran insects use their stylets to occasionally pierce host cells to sample plant material, feed from the phloem, and secrete watery saliva (Tjallingii, 2006; Will et al., 2009). Watery saliva not only contains MAMPs from bacterial symbionts and pathogens that can be perceived by the host, but also contains insect effectors that manipulate host physiology and suppress defense (Chaudhary et al., 2014; Elzinga et al., 2014). For plant defense, a cluster of three lectin receptor kinases were found to confer resistance to both the brown and the white back planthopper insect pests (Liu et al., 2015). Future research identifying additional RLKs or RLPs involved in active perception of piercing-sucking insects and studying their role in controlling plant disease would be a significant advance in our understanding of immune responses.
Investigating heterogeneity of immune responses within tissues and cells
Variations in cellular responses and tissue types determine the outcome of plant-pathogen interactions. Heterogeneity in pathogen distribution and thus uneven targeting of host cells during infection must influence plant responses. For example, microscopy analyses with fluorescently tagged P. syringae reveal preferential colonization of particular sites (substomatal cavity, epidermal cell junctions) (Boureau et al., 2002; Rufián et al., 2018). The fungal pathogen Ustilago maydis induces tumor formation in aerial maize organs by deploying distinct effectors for colonization and virulence in specific organ types (Hemetsberger et al., 2012; Schilling et al., 2014).
One of the most striking demonstrations of the spatial and temporal dynamic of a plant defense response occurs during a typical ETI, where effector-triggered cell death is sharply restricted to the site of infection. The cell death and survival decision is modulated through the spatial and temporal expression of SA and JA at the infection site and in surrounding cells (Enyedi et al., 1992; Liu et al., 2016; Betsuyaku et al., 2018) through the interplay of NONEXPRESSOR of PR (NPR) proteins (Fu et al., 2012; Liu et al., 2016; Zavaliev et al., 2020). In the Zavaliev et al., (2020), the authors found that many NLRs as well as downstream signaling components, such as EDS1, are sequestered in the SA-induced NPR1 condensates (SINCs) in the cytoplasm of cells surrounding the ETI-induced cell death zone. How SA triggers the dynamic formation of SINCs will require future research using technologies with higher resolutions.
Recent advances in single cell RNA-seq, coupled with live cell imaging of reporter lines could facilitate an understanding of how plants dynamically respond to infection (Denyer et al., 2019) to address the questions: How many distinct response clusters occur within a leaf? What is the gradient in PRR and NLR responses in cells directly targeted by pathogen effectors, and in cells proximal and distal to pathogens?
Response to pathogen infection is not uniform even within a cell. For example, the average diameter of P. syringae cells are 1.2 μm, compared with 15–35 μm for the spongy mesophyll, 40–80 μm for the palisade mesophyll, and 10–200 μm for an epidermal pavement cell in fully expanded Arabidopsis leaves (Melaragno et al., 1993; Monier and Lindow, 2003; Wuyts et al., 2010). Visualization of P. syringae effector delivery using a split-GFP system revealed that membrane localized effectors are delivered in small stretches surrounding the plasma membrane and rarely exhibit uniform distribution (Henry et al., 2017; Park et al., 2017). Multiple filamentous pathogens deliver effectors at the haustorial interface (Bozkurt and Kamoun, 2020). Plant organelles cluster at this interface, which may facilitate localized defense (Bozkurt and Kamoun, 2020). Selective autophagy is also deployed at the haustorial interface to counteract pathogen invasion (Dagdas et al., 2018; Bozkurt and Kamoun, 2020). As pathogen-induced modification of individual host cells is not uniform, the corresponding host response may also be compartmentalized.
There is accumulating evidence for localized plant responses. Chitin responses in Arabidopsis plasmodesmata require the RLKs, LYK4 and LYK5, as well as LYSM-CONTAINING GPI-ANCHORED PROTEIN 2 (LYM2) (Faulkner et al., 2013; Cheval et al., 2020). Chitin-induced plasmodesmatal closure results in dynamic changes in the association, mobility and localization of all three proteins, but only LYM2 and LYK4 are found in the plasmodesmatal plasma membrane (Cheval et al., 2020). In response to osmotic stress, the receptor like kinases Qiān Shŏu kinase (QSK1), inflorescence meristem kinase 2, and CYSTEINE RICH RECEPTOR LIKE KINASE 2 (CRK2) rapidly relocate and cluster to plasmodesmata pores (Grison et al., 2019; Hunter et al., 2019). The flagellin receptor FLS2 and the brassinosteroid receptor BRASSENOSTERIOD INSENSITIVE 1 (BRI1) localize to distinct membrane microdomains, despite these two receptors requiring the same co-receptor BAK1 (Bücherl et al., 2017). The differential organization of FLS2 and BRI1 in the plasma membrane may facilitate differentiation between pathogen perception and steroid-mediated growth. Taken together, these data highlight that the reorganization of receptor kinases to distinct membrane domains and subcellular compartments are important layers regulating the specificity and response to the perception of pathogens and abiotic stresses. Future research investigating spatial and temporal responses in plant immune signaling in the context of pathogen localization will shed light onto how plants are capable of integrating rapid, robust, and specific responses to different stimuli.
Translational regulation is a critical layer of control in plant immune induction
Besides the knowledge gaps outlined above, another major blind spot in the plant immune signaling network is translational regulation. This might be due to the intrinsic complexity of the translational process as well as technical limitations in examining the multitude of players involved. However, recent applications of sequencing technology to studies of ribosomal footprinting on mRNAs (Ribo-seq) (Ingolia et.al, 2009), tRNA levels (Zheng et al, 2015a), mRNA modifications (e.g., m6A) (Dominissini et al., 2012; Meyer et al., 2012), as well as in vivo RNA secondary structures (Ding et al., 2014) have made genome-wide investigation of how plants regulate protein production during defense a distinct possibility.
During an infection, the host and the pathogen may compete for access to the host translational machinery. The extreme case on the pathogen side are viruses which rely on the host ribosomes for viral protein synthesis. Various RNA viruses carry internal ribosome entry site (IRES) elements in their genomes that recruit host ribosomes to translate viral RNA despite the absence of a 5’ cap (Martinez-Salas et.al., 2017) required for normal translation initiation in eukaryotes. Perturbation of host metabolism by the pathogen (Scheideler et al., 2002; Ward et al., 2010; Aliferis et al., 2014; Misra et al., 2016; Schwachtje et al., 2018; Yoo et al., 2020) may be an early signal for mounting defense by selectively translating preexisting mRNAs encoding defense proteins. How these defense mRNAs are chosen, whether their translation is at the expense of those involved in growth-related activities, and if there is a correlation between transcriptional and translational regulation need to be addressed.
While examining proteomic changes during a pathogen infection would be the most direct way to tackle the aforementioned questions, this approach currently has significant technical limitations. Jones et al. (2006) took the challenge and performed two-dimensional gel electrophoresis on Arabidopsis inoculated with the virulent bacterial pathogen PstDC3000, the mutant version of PstDC3000, hrpA, which exclusively activates PTI, or PstDC3000 carrying the effector AvrRpm1 which induces ETI. The study detected changes in proteins enriched in defense-related antioxidants and metabolic enzymes and discovered that changes in these proteins occurred prior to significant transcriptional reprogramming, indicating the presence of translational regulation in early immune responses.
Before such a proteomic analysis becomes more accessible, Ribo-seq has been used to estimate translational activity of mRNAs through their association with ribosomes (Ingolia et.al, 2009). Essentially, the amount of mRNA pulled down with the ribosome is compared with the total mRNA level to calculate the translation efficiency (TE). Xu et al. (2017a) performed Ribo-seq on Arabidopsis after treatment with the MAMP signal, elf18. This study was inspired by the earlier finding of Pajerowska-Mukhtar et al. (2012) that translation of the key immune transcription factor, TBF1, is tightly controlled by the 5’-leader sequence (5’-LS) of its mRNA. Consistent with the Jones et al. (2006) findings, Xu et al. (2017a) found no significant correlation (r = 0.41) between the elf18-mediated translational changes and transcriptional dynamics, indicating that translational reprogramming is a distinct regulatory step in PTI induction.
What is the regulatory mechanism of this translational reprogramming during PTI? In yeast, mammals, and plants, stresses such as amino acid starvation could lead to an overall decrease in translational activity, but an increase in translation of stress-responsive proteins such as GCN4 in yeast and ATF4 in mammals (Hinnebusch, 2005; Lageix et al., 2008; Kilberg et al., 2009). This integrated stress response is conserved across eukaryotes and is carried out by GCN2 (or other kinases in mammals)-mediated phosphorylation and the consequent inactivation of the translation initiation factor eIF2α (P-eIF2α), allowing the ribosome to bypass the inhibition of upstream open reading frames (uORFs) and reach the downstream main ORFs (mORFs) (Hinnebusch, 2005; Lageix et al., 2008; Zhang et al., 2008b; Kilberg et al., 2009). Surprisingly, several plant studies found that blocking eIF2α phosphorylation in the gcn2 knockout mutant did not affect immunity-induced translational reprogramming (Luna et al., 2014; Xu et al., 2017a; Izquierdo et al., 2018). For example, elf18-induced TBF1 translation, which is controlled by uORFs in the 5’-LS, and pathogen resistance were appropriately induced in the gcn2 background (Xu et al., 2017a). Moreover, elf18 treatment did not significantly alter the overall translational capacity. Though Liu et al. (2019) found that GCN2 plays a role in stomatal immunity through ABA, the majority of the current data suggest a novel mechanism in reprogramming translatome during plant immune responses.
To elucidate this translational regulatory mechanism, Xu et al. (2017a) searched for sequence features, namely consensus sequences and uORFs, that are associated with mRNAs whose translation is induced during PTI. This search led to the discovery of a significantly enriched consensus sequence consisting of mostly purines (“R-motif”) in the 5’-LSs of those mRNAs and found that the R-motif is not only necessary, but also sufficient for PTI-induced translation. One mRNA feature that Xu et al. (2017a) did not attempt to search for was conserved secondary structure because the prediction algorithms at that time could not accurately reflect the in vivo RNA structural dynamics during stress responses (Ding et al., 2014). Recent developments of in vivo RNA structure probing technologies, such as SHAPE-MaP (Smola and Weeks, 2018), will ultimately enable visualization of RNA structural dynamics inside living cells, which has been shown to be critical in explaining how uORF mediates translation of the mORF (Gunisova and Valasek, 2014; Corley et al., 2017).
The identification of the R-motif in the 5’-LSs of mRNAs with enhanced TE during PTI suggests that its associated protein(s) may play a role in selecting these mRNAs for translation. Poly(A)-binding proteins (PABPs) can bind to the R-motif and affect basal as well as elf18-induced translation (Xu et al., 2017a). Since PABPs link the poly(A) tail of a mRNA to its 5’ cap through interaction with eIF4G and eIF3 to facilitate translation initiation (Sonenberg and Hinnebusch, 2009; Jackson et al., 2010), it is plausible that the 5’ R-motif serves as a cap-independent translation enhancer (CITE) (Shatsky et al., 2018). Indeed, in yeast, cap-independent translation of mRNAs involved in invasive growth induced by nutrient deprivation was found to require recruitment of the poly(A)-binding protein 1 (Pab1) by an A-rich element in the 5’ untranslated regions of these mRNAs (Gilbert et al., 2007).
Besides specialized mRNA features that might serve as CITEs, different components of the translational machinery may be used specifically for translating stress responsive proteins. Through a genetic study performed in Arabidopsis, Izquierdo et al.(2018)found a yeast GCN1 homolog named NOXY7 to act through GCN20, instead of GCN2-P-eIF2α, in mediating translation in response to mitochondrial dysfunction, high boron concentration, and infection by PstDC3000. Future biochemical, proteomic, and genetic studies will lead to identification of more such stress-specific components of the translational machinery.
Recruitment of stress-specific components and/or modifications of the translational machinery is also likely to control translation during ETI that occurs when a pathogen effector is detected by an NLR. Since ETI often leads to programmed cell death, it was surprising that no significant global changes in polysome profiles were observed by either Meteignier et al. (2017) who examined Arabidopsis 2 hr after dexamethasone-induced expression of the bacterial effector AvrRpm1 gene or by Yoo et al. (2020) who performed the experiment 8 hr after inoculation with Ps pv. maculicola ES4326 carrying avrRpt2 (Psm ES4326/avrRpt2). Moreover, while Meteignier et al. (2017) observed a moderate level of overlap between transcriptionally and translationally induced genes (77%), Yoo et al. (2020) found a significant correlation (r = 0.92) between the two activities. It is intuitive to conclude that during ETI, transcriptional induction is the major driving force that defines the ETI proteome. The counter argument may be that since transcription and translation occur in distinct cellular compartments, a good correlation suggests an intricate interplay between the two separately regulated processes. How plants reprogram the proteome without significantly altering the overall translational capacity will be a fascinating question which may be addressed through identification of ETI-specific translation regulators.
What kinds of genes are translationally regulated during PTI and ETI? For those found during PTI, gene ontology (GO) term analyses did not provide a clear answer, probably because the current GO terms were mostly defined by transcriptional dynamics. However, these “first responders” to pathogen challenge are most likely translated from preexisting mRNAs without the involvement of transcriptional regulation. Though they do not have the “defense” GO term yet, many of them (e.g., TFs and enzymes) were found to be critical for PTI (Xu et al., 2017a). In contrast to the PTI translatome analysis, identification of translationally regulated genes during ETI was more difficult due to the strong correlation between transcriptional and translational activities. Perhaps for this very reason, “defense” GO terms turned out to be more helpful in narrowing down the candidate gene list in ETI. Genes involved in aromatic amino acid and phenylpropanoid metabolism and those encoding NLR immune receptors were determined to be translationally regulated by Meteignier et al. (2017) and Yoo et al. (2020). Since NLR expression is regulated at many stages, from mRNA nuclear export to proteasome-mediated degradation (Johnson et al., 2012; Wu et al., 2020; Zavaliev et al., 2020), it was not surprising that some immune receptors are translationally regulated. However, it was somewhat unexpected that knocking out these NLR genes partially compromised ETI triggered by RPS2, a sensor NLR that has been studied extensively, demonstrating the effectiveness of translational profiling in identifying novel players in plant defense.
There are still many unexplored posttranscriptional processes impacting the defense proteome. mRNAs and tRNAs are known to be modified at multiple positions of different nucleotides, such as methylation at N6-adenosine (m6A), which can affect mRNA transport, stability, and translational efficiency (Roundtree et al., 2017). The writers, readers and erasers of m6A have been identified and characterized in plants (Ruzicka et al., 2017; Arribas-Hernandez and Brodersen, 2020), but it is still challenging to quantitatively measure the dynamic changes in m6A to identify defense-specific modifications and distinguish a direct impact of the modification from a pleiotropic effect on plant immunity. Besides RNA modifications, there have been several studies focusing on the various RNA degradation processes during plant immune responses (Gloggnitzer et al., 2014; Maldonado-Bonilla et al., 2014; Roux et al., 2015; Meteignier et al., 2016; Makinen et al., 2017; Li and Wang, 2018; Yu et al., 2019b). For example, Gloggnitzer et al. (2014) found that the nonsense-mediated mRNA decay pathway (NMD) controls the turnover of a large number of NLR mRNAs. During bacterial infection, plants turn down NMD to enhance resistance. Moreover, formation of RNA processing bodies upon different immune responses was found by Yu et al. (2019b) to destabilize PTI-down regulated transcripts and by Meteignier et al. (2016) to inhibit viral RNA translation during ETI. Besides mRNA, tRNA concentrations and charging can also be affected by pathogen challenge (Pajerowska-Mukhtar et al., 2012; Luna et al., 2014; Soprano et al., 2018), which may link pathogen-induced metabolic changes directly to translation of mRNAs through codon-usage, instead of indirectly through activation of GCN2 (Hinnebusch, 2005).
Understanding the regulatory mechanisms controlling translation of plant defense proteins is not only critical for basic science, but also has broader impact on applications in agriculture and beyond (Xu et al., 2017b). After all, most of biological activities are carried out by proteins. Transcripts encoding key regulatory proteins, such as the mRNA of TBF1, carry sequences, such as R-motifs and uORFs, for translation at the proper time, in the specific tissue and of the right amount. Even though the mechanisms by which these sequences control translation still need to be elucidated, they are ready-to-use because they have been optimized through evolution.
The circadian clock is a key mechanism that breaks the disease triangle in plants
So far, most of our studies have been focused on defense against an established infection. However, the battle between the host and the pathogen begins at an even earlier stage. It is known that a successful infection requires at least three factors: a virulent pathogen, a susceptible host, and a favorable environment known as “the disease triangle” (Francl, 2001). Below we will examine how the circadian clock affects each of these factors.
A role of the circadian clock in controlling plant defense against pathogen infection is not as immediately obvious as in regulating plant growth and reproduction, because plants need to be able to respond to infection whenever pathogen invades. Reinforcing this idea, a study by Nagano et al. of transcriptome data of rice plants grown in a paddy field with the corresponding meteorological data found that defense genes had higher stochastic variances than those involved in basic metabolic functions (Nagano et al., 2012). However, the serendipitous discovery that a large number of defense genes against Hyaloperonospora arabidoposidis (Hpa) were directly regulated by the morning circadian clock component, CCA1, unveiled a role for the circadian clock in controlling defense against at least certain pathogens (Wang et al., 2011). The fact that the life cycles of both the plant hosts and some of their pathogens are dictated by the diurnal cycle of the earth suggests that when infection is a predictable event, the host can anticipate it through the function of the circadian clock (Slusarenko and Schlaich, 2003; Bhardwaj et al., 2011; Goodspeed et al., 2012; Zhang et al., 2013; Korneli et al., 2014; Hevia et al., 2015; Ingle et al., 2015; Schumacher, 2017; Lei et al., 2019). For example, for the obligate biotrophic pathogen Hpa to complete its life cycle, darkness is required for the formation of spores, while spore dissemination is triggered by drying of the leaf surface in the morning (Slusarenko and Schlaich, 2003). Meanwhile, the virulence of the fungal pathogen Botrytis cinerea (Bc) on Arabidopsis has been shown to be controlled by the fungal circadian clock regulator, BcFRQ1, resulting in formation of bigger lesion on leaves if the pathogen is inoculated at dusk (Hevia et al., 2015). However, there are still numerous questions to be answered. Is there a general role for the circadian clock in regulating resistance against other pathogens whose life cycles are less predictable than Hpa and Bc? Besides helping plants anticipate infection, does the clock play a role in modulating defense response during an infection? How do changes in environmental conditions, such as humidity, affect plant defense through the circadian clock?
A general role for the circadian clock in plant defense is supported by the Goodspeed et al. study (2012) showing circadian oscillations in basal levels of the plant defense hormones SA and JA. Since SA and JA have antagonistic effects in mediating resistance against biotrophic pathogens and necrotrophs/insects, respectively (Spoel and Dong, 2008), the opposite phasing of SA (peaking at subjective night) and JA (peaking during the subjective day) observed by Goodspeed et al. (2012) suggests that the circadian clock may help plants avoid such a conflict. A direct link between the clock and SA synthesis was made in the Zheng et al. (2015b) study, in which the clock component, CHE, was shown to bind to the promoter of the SA synthesis gene, ICS1, to regulate its transcription. In addition to rhythmic expression of basal SA, CHE was also found to be required in SA synthesis in systemic tissue and disease resistance. However, the biological significance of having a clock component regulating SAR is still unknown. The study by Li et al. (2018) may shed some light on this question. The authors observed that treating a single leaf of two-week-old Arabidopsis seedlings with PstDC3000 or SA dampened the circadian clock in systemic tissues. They hypothesized that this might allow plants to uncouple SA synthesis from the normal clock control and have continuous production of SA for the establishment of SAR. Since CHE has been shown to be a repressor of the core morning clock gene CCA1 (Pruneda-Paz et al., 2009), it is plausible that CHE promotes SA synthesis in systemic tissue by activating ICS1 while dampening the clock.
However, the relationship between SA and the clock appears to be “very complicated”, to use the words of Dr. Steven Kay in describing phenomena associated with the circadian clock. Zhou et al. (2015a) found that spraying mature Arabidopsis with SA, instead of soaking seedlings grown on plates in SA solution, increased the amplitude of the circadian clock genes, such as LHY and TOC1, in contrast to what was observed by Li et al. (2018). This increase was facilitated by the central SA signaling component NPR1 through its associated TGA transcription factors. Moreover, the npr1 mutant was not only insensitive to this SA-mediated reinforcement of the clock, but also had lower-than-WT amplitude of the clock gene expression.
Because the phase and the period of the clock have traditionally been the main focus of chronobiology research, it is easy to overlook the importance of the clock amplitude. The clock amplitude might also play a critical role for the integration of the environmental signals into the regulation of physiological responses. For example, Nagano et al.’s field grown rice transcriptome data showed that the daily amplitudes of core circadian clock genes were affected by environmental stimuli such as temperature and solar radiation, while their phases and periods remained stable (Nagano et al., 2012). Furthermore, Mwimba et al. (2018) found that, in addition to light and temperature, daily humidity oscillation is a novel environmental cue that entrains the plant circadian clock (i.e., a zeitgeber). Interestingly, adding humidity oscillation to the light/dark cycle as it occurs in nature increased amplitude of the circadian clock leading to better plant performance such as higher total biomass and seed weight.
This ability of the plant circadian clock to sense humidity may be critical for defense against pathogen infection because high humidity in the environment is often associated with outbreaks of plant diseases (Thurston et al., 1958; Brown and Ogle, 1997; Xin et al., 2016). High humidity has been shown to induce expression of the HrpL transcription factor in PstDC3000, which activates the type III secretion system for delivery of effectors into the host (Mwimba et al., 2018). High humidity is also required for the pathogen to cause disease by maintaining the aqueous environment in the apoplastic space where the bacterium proliferates (Xin et al., 2016). Therefore, there is “a war over water” between the pathogen and its plant host (Beattie, 2016). The SA-mediated repression of water transport genes found by Zhou et al. suggests that inhibiting genes such as aquaporins may be one of the defense mechanisms of this defense hormone (Zhou et al., 2015a).
Since both SA treatment and daily humidity oscillation could increase the amplitude of the circadian clock, a reasonable question to ask is how this increase affects plant defense. Since the morning clock components CCA1/LHY are known positive regulators (Wang et al., 2011; Zhang et al., 2013) whereas evening clock component TOC1 is a negative regulator of defense genes (Zhou et al., 2015a), an increase in the clock amplitude would result in a higher SA-induced defense gene expression in the morning than in the evening. Indeed, a striking difference in this time-of-the-day sensitivity was observed in the microarray analysis performed by Zhou et al. on plants after treating them with SA in the subjective morning and subjective evening (Zhou et al., 2015a). Zhou et al. found that repression of water transport genes by treating plants with SA in darkness led to significant loss of fresh weight and ultimately death (Zhou et al., 2015a). Therefore, besides its anticipatory function, the circadian clock also serves to gate immune responses upon infection to minimize conflict with other physiological functions, in this case, water transport. As in circadian medicine, which aims to administer drugs at the right time of the day to increase drug efficacy and reduce side effects, the consequence of plants taking “aspirin” at the wrong time of the day (i.e., at night) might be as severe as death by desiccation.
Despite the important role that the circadian clock plays in regulating plant defense, research in this area is technically challenging and financially prohibiting because of the requirement for time series data collection and analysis. The use of clock-driven luciferase reporters is limited to organisms like Arabidopsis which can be readily transformed. To remedy the reliance on time-course experiment in circadian clock studies, the ingenious “molecular timetable” method has been developed first in mice (Ueda et al., 2004) and adapted to plants (Kerwin et al., 2011; Higashi et al., 2016; Li et al., 2019b), which aims to extract circadian information from data taken at a single timepoint. Recently, Li et al. applied this methodology to map abiotic stress inputs into the soybean circadian clock (Li et al., 2019b). These findings were then experimentally confirmed using RASL-seq, a multiplexed targeted RNA sequencing technology (Li et al., 2012). Using this two-module discovery pipeline, one can make good use of all the available plant-microbe interaction transcriptome data regardless whether they are generated from time-course or single-time-point experiments to study the impact that biotic stresses have on the circadian clock.
Future Perspectives
Research discoveries in plant sciences have the potential to guide sustainable increases in agricultural production to feed, shelter, and clothe the world’s population. Crop losses from plant pathogens and pests directly impact human health and range from 17 to 30% average globally in major food crops (Gregory et al., 2009; Savary et al., 2019). The identification and deployment of diverse plant resistance genes using a variety of technologies have clear potential for crop improvement. In contrast to disease-specific resistance, broad-spectrum resistance involving multiple defense mechanisms may have the advantage of increased durability and decreased guesswork involved in the deployment if it can be tightly regulated to minimize potential yield penalty. Therefore, basic research focused on understanding how plants naturally manage their immune responses is the foundation on which applied research can be built. The utility and relevance of fundamental discoveries may not be immediately noticeable, but some of the most impactful discoveries, including genome editing, occurred as the result of curiosity-driven investigations. We believe that investigating novel disease resistance genes, their activation mechanisms, and signaling networks will provide a more holistic understanding of plant defense and can be harnessed for controlling some of the most devastating plant diseases.
Figure 1. Plant immune responses.
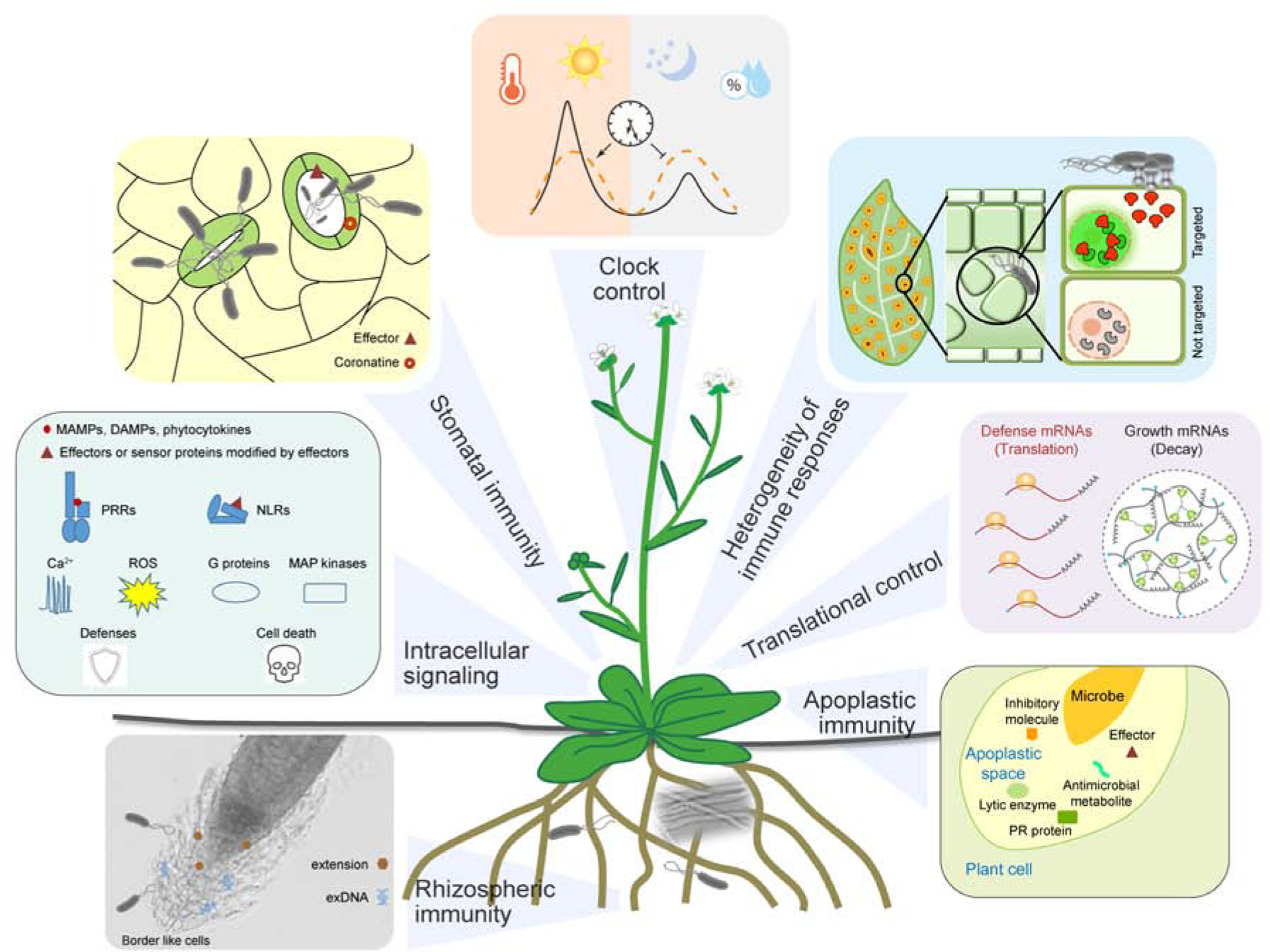
The front line of plant defense occurs in the stomata, rhizosphere and apoplast. Recognition of microbe-associated molecular patterns (MAMPs) can trigger stomatal closure to restrict entry of microbes into plants. To counteract, microbes produce effectors and coronatine to promote stomatal opening. In roots, extensins, border and border-like cells, and extracellular DNAs (exDNAs) contribute to rhizospheric immunity that functions in attracting beneficial microbes while repelling pathogens. In the apoplastic space, plants produce lytic enzymes to release immune-inducing MAMPs and antimicrobial pathogenesis-related (PR) proteins and metabolites to restrict the proliferation of invading pathogens. In the same space, microbes can use effectors and inhibitory molecules to counteract plants’ apoplastic immunity. Recognition of MAMPs and pathogen effectors activate pattern-recognition receptors (PRRs) on the cell surface and nucleotide-binding leucine-rich repeat immune receptors (NLRs) inside the cell to trigger pattern-triggered immunity (PTI) and effector-triggered immunity (ETI), respectively. These immune receptors signal through Ca2+, reactive oxygen species (ROS), G proteins, and MAP kinase cascades to confer resistance which in ETI is often associated with cell death. Activation of both PTI and ETI involve reprogramming of plant proteome through decay of house-keeping mRNAs (perhaps inside stress granules; circle) and activation of translation of defense proteins. Heterogeneity exists in plant responses to infection due to differential pathogen distribution and host immune sensitivity. Plant immune responses are also regulated by the circadian clock which not only helps plants anticipate infection, but also gates immune responses when infection occurs to minimize effects on plant physiology and fitness.
Here we summarize recent progress and share our perspective in the areas of extracellular immunity, sensing of immunogenic signals by cell surface and intracellular immune receptors, heterogeneity of immune responses within tissues and cells, translational regulation as a critical layer of control in plant immune induction and the role of the circadian clock in breaking the disease triangle in plants.
ACKNOWLEDGEMENTS
We thank Tzion Fahima, the Dong lab, and Coaker lab members for their critical reading of the review and Yezi Xiang for helping with the figure. No conflicts of interest declared.
FUNDING
This work was supported by grants from the National Institutes of Health (NIH) (NIH 1R35GM118036), and by the Howard Hughes Medical Institute to X.D., grants from the NIH (NIH 1R35GM136402), National Science Foundation (NSF) (NSF 1937855-0), and United States Department of Agriculture (USDA) (USDA 2019-70016-2979) to G.C., a grant from National Natural Science Foundation of China (31830019) to J.M.Z., and a grant from the Chinese Natural Science Foundation (31922075) and Youth Innovation Promotion Association of the Chinese Academy of Sciences to J.Z.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Achor D, Welker S, Ben-Mahmoud S, Wang C, Folimonova SY, Dutt M, Gowda S, and Levy AJ (2020). Dynamics of Candidatus Liberibacter asiaticus movement and sieve-pore plugging in citrus sink cells. Plant Physiol.182:882–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adachi H, Contreras M, Harant A, Wu CH, Derevnina L, Sakai T, Duggan C, Moratto E, Bozkurt T,O, Maqbool A, et al. (2019a). An N-terminal motif in NLR immune receptors is functionally conserved across distantly related plant species. eLife 8:e49956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adachi H, Derevnina L, and Kamoun S (2019b). NLR singletons, pairs, and networks: evolution, assembly, and regulation of the intracellular immunoreceptor circuitry of plants. Curr. Opin. Plant Biol 50:121–131. [DOI] [PubMed] [Google Scholar]
- Adie BA, Pérez-Pérez J, Pérez-Pérez MM, Godoy M, Sánchez-Serrano JJ, Schmelz EA, and Solano R (2007). ABA is an essential signal for plant resistance to pathogens affecting JA biosynthesis and the activation of defenses in Arabidopsis. Plant Cell 19:1665–1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aliferis KA, Faubert D, and Jabaji S (2014). A metabolic profiling strategy for the dissection of plant defense against fungal pathogens. PLoS One 9:e111930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arribas-Hernandez L, and Brodersen P (2020). Occurrence and functions of m6A and other covalent modifications in plant mRNA. Plant Physiol. 182:79–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel FM (2005). Are innate immune signaling pathways in plants and animals conserved? Nat. Immunol 6:973–979. [DOI] [PubMed] [Google Scholar]
- Bacete L, Mélida H, Miedes E, and Molina A (2018). Plant cell wall-mediated immunity: cell wall changes trigger disease resistance responses. Plant J. 93:614–636. [DOI] [PubMed] [Google Scholar]
- Bae C, Han SW, Song YR, Kim BY, Lee HJ, Lee JM, Yeam I, Heu S, and Oh CS (2015). Infection processes of xylem-colonizing pathogenic bacteria: possible explanations for the scarcity of qualitative disease resistance genes against them in crops. Theor. Appl. Genet 128:1219–1229. [DOI] [PubMed] [Google Scholar]
- Bardoel BW, Van der Ent S, Pel MJ, Tommassen J, Pieterse CM, Van Kessel KP, and Van Strijp JA (2011). Pseudomonas evades immune recognition of flagellin in both mammals and plants. PLoS Pathog. 7:e1002206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beattie GA (2016). A war over water when bacteria invade leaves. Nature 539:506–507. [DOI] [PubMed] [Google Scholar]
- Beck M, Wyrsch I, Strutt J, Wimalasekera R, Webb A, Boller T, and Robatzek S (2014). Expression patterns of FLAGELLIN SENSING 2 map to bacterial entry sites in plant shoots and roots. J. Exp. Bot 65:6487–6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betsuyaku S, Katou S, Takebayashi Y, Sakakibara H, Nomura N, and Fukuda H (2018). Salicylic acid and jasmonic acid pathways are activated in spatially different domains around the infection site during effector-triggered immunity in Arabidopsis thaliana. Plant Cell Physiol. 59: 8–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhardwaj V, Meier S, Petersen LN, Ingle RA, and Roden LC (2011). Defence responses of Arabidopsis thaliana to infection by Pseudomonas syringae are regulated by the circadian clock. PLoS One 6:e26968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi G, Zhou Z, Wang W, Li L, Rao S, Wu Y, Zhang X, Menke FL, Chen S, and Zhou JM (2018). Receptor-like cytoplasmic kinases directly link diverse pattern recognition receptors to the activation of mitogen-activated protein kinase cascades in Arabidopsis. Plant Cell 30:1543–1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boller T, and Felix G (2009). A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol 60:379–406. [DOI] [PubMed] [Google Scholar]
- Boller T, and He SY (2009). Innate immunity in plants: an arms race between pattern recognition receptors in plants and effectors in microbial pathogens. Science 324:742–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolton MD, van Esse HP, Vossen JH, de Jonge R, Stergiopoulos I, Stulemeijer IJ, van den Berg GC, Borrás-Hidalgo O, Dekker HL, de Koster CG, et al. (2008). The novel Cladosporium fulvum lysin motif effector Ecp6 is a virulence factor with orthologues in other fungal species. Mol. Microbiol 69:119–136. [DOI] [PubMed] [Google Scholar]
- Bonardi V, Tang S, Stallmann A, Roberts M, Cherkis K, and Dangl JL (2011). Expanded functions for a family of plant intracellular immune receptors beyond specific recognition of pathogen effectors. Proc. Natl. Acad. Sci. USA 108:16463–16468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boureau T, Routtu J, Roine E, Taira S, and Romantschuk M (2002). Localization of hrpA-induced Pseudomonas syringae pv. tomato DC3000 in infected tomato leaves. Mol. Plant Pathol 3:451–460. [DOI] [PubMed] [Google Scholar]
- Bozkurt TO and Kamoun S (2020). The plant-pathogen haustorial interface at a glance. J. Cell Sci 133:jcs237958.doi: 10.1242/jcs.237958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkmann V, Reichard U, Goosmann C, Fauler B, Uhlemann Y, Weiss DS, Weinrauch Y, and Zychlinsky A (2004). Neutrophil extracellulartraps kill bacteria. Science 303:1532–1535. [DOI] [PubMed] [Google Scholar]
- Brown JF and Ogle HJ (1997). Plant pathogens and plant diseases. Rockvale Publications, Armidale. [Google Scholar]
- Brueggeman R, Rostoks N, Kudrna D, Kilian A, Han F, Chen J, Druka A, Steffenson B, and Kleinhofs A (2002). The barley stem rust-resistance gene Rpg1 is a novel disease-resistance gene with homology to receptor kinases. Proc. Natl. Acad. Sci. USA 99:9328–9333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bücherl CA, Jarsch IK, Schudoma C, Segonzac C, Mbengue M, Robatzek S, MacLean D, Ott T, and Zipfel C (2017). Plant immune and growth receptors share common signalling components but localise to distinct plasma membrane nanodomains. eLife 6: e25114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buscaill P, Chandrasekar B, Sanguankiattichai N, Kourelis J, Kaschani F, Thomas EL, Morimoto K, Kaiser M, Preston GM, Ichinose Y, et al. (2019). Glycosidase and glycan polymorphism control hydrolytic release of immunogenic flagellin peptides. Science 364:eaav0748. [DOI] [PubMed] [Google Scholar]
- Cannesan MA, Durand C, Burel C, Gangneux C, Lerouge P, Ishii T, Laval K, Follet-Gueye ML, Driouich A, and Vicré-Gibouin M (2012). Effect of arabinogalactan proteins from the root caps of pea and Brassica napus on Aphanomyces euteiches zoospore chemotaxis and germination. Plant Physiol. 159:1658–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannesan MA, Gangneux C, Lanoue A, Giron D, Laval K, Hawes MC, Driouich A, and Vicré-Gibouin M (2011). Association between border cell responses and localized root infection by pathogenic Aphanomyces euteiches. Ann. Bot 108:459–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerutti A, Jauneau A, Auriac MC, Lauber E, Martinez Y, Chiarenza S, Leonhardt N, Berthomé R, and Noël LD (2017). Immunity at cauliflower hydathodes controls systemic infection by Xanthomonas campestris pv campestris. Plant Physiol. 174:700–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhary R, Atamian HS, Shen Z, Briggs SP, and Kaloshian I (2014). GroEL from the endosymbiont Buchnera aphidicola betrays the aphid by triggering plant defense. Proc. Natl. Acad. Sci. USA 111:8919–8924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Rouse MN, Zhang W, Zhang X, Guo Y, Briggs J, and Dubcovsky J (2020). Wheat gene Sr60 encodes a protein with two putative kinase domains that confers resistance to stem rust. New Phytol. 225:948–959. [DOI] [PubMed] [Google Scholar]
- Cheval C, Samwald S, Johnston MG, de Keijzer J, Breakspear A, Liu X, Bellandi A, Kadota Y, Zipfel C, and Faulkner C (2020). Chitin perception in plasmodesmata characterizes submembrane immune-signaling specificity in plants. Proc. Natl. Acad. Sci. USA 117: 9621–9629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chini A, Fonseca S, Fernández G, Adie B, Chico JM, Lorenzo O, García-Casado G, López-Vidriero I, Lozano FM, Ponce MR, et al. (2007).The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448:666–671. [DOI] [PubMed] [Google Scholar]
- Choi J, Tanaka K, Cao Y, Qi Y, Qiu J, Liang Y, Lee SY, and Stacey G (2014). Identification of a plant receptor for extracellular ATP. Science 343:290–294. [DOI] [PubMed] [Google Scholar]
- Choi WG, Hilleary R, Swanson SJ, Kim SH, and Gilroy S (2016). Rapid, long-distance electrical and calcium signaling in plants. Annu. Rev. Plant Biol 67:287–307. [DOI] [PubMed] [Google Scholar]
- Collier SM, Hamel LP, and Moffett P (2011). Cell death mediated by the N-terminal domains of a unique and highly conserved class of NB-LRR protein. Mol. Plant Microbe Interact 24: 918–931. [DOI] [PubMed] [Google Scholar]
- Cook DE, Lee TG, Guo X, Melito S, Wang K, Bayless AM, Wang J, Hughes TJ, Willis DK, Clemente TE, et al. (2012). Copy number variation of multiple genes at Rhg1 mediates nematode resistance in soybean. Science 338:1206–1209. [DOI] [PubMed] [Google Scholar]
- Cook DE, Mesarich CH, and Thomma BP (2015). Understanding plant immunity as a surveillace system to detect invasion. Annu. Rev. Phytopathol 53:541–563. [DOI] [PubMed] [Google Scholar]
- Corley M, Solem A, Phillips G, Lackey L, Ziehr B, Vincent HA, Mustoe AM, Ramos SB, Weeks KM, Moorman NJ, et al. (2017). An RNA structure-mediated, posttranscriptional model of human alpha-1-antitrypsin expression. Proc. Natl. Acad. Sci. USA 114:E10244–E10253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couto D, Niebergall R, Liang X, Bücherl CA, Sklenar J, Macho AP, Ntoukakis V, Derbyshire P, Altenbach D, Maclean D, et al. (2016). The Arabidopsis protein phosphatase PP2C38 negatively regulates the central immune kinase BIK1. PLoS Pathog. 12: e1005811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curlango-Rivera G, and Hawes MC (2011). Root tips moving through soil–an intrinsic vulnerability. Plant Signal. Behav 6:1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curlango-Rivera G, Huskey DA, Mostafa A, Kessler JO, Xiong Z, and Hawes MC (2013). Intraspecies variation in cotton border cell production: rhizosphere microbiome implications. Am. J. Bot 100:9–15. [DOI] [PubMed] [Google Scholar]
- Dagdas YF, Pandey P, Tumtas Y, Sanguankiattichai N, Belhaj K, Duggan C, Leary AY, Segretin ME, Contreras MP, Savage ZJ et al. (2018). Host autophagy machinery is diverted to the pathogen interface to mediate focal defense responses against the Irish potato famine pathogen. eLife 7:e37476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jonge R, van Esse HP, Kombrink A, Shinya T, Desaki Y, Bours R,Krol S, Shibuya N, Joosten MH, and Thomma BP (2010). Conserved fungal LysM effector Ecp6 prevents chitin-triggered immunity in plants. Science 329: 953955. [DOI] [PubMed] [Google Scholar]
- De Lorenzo G, Ferrari S, Giovannoni M, Mattei B, and Cervone F (2019). Cell wall traits that influence plant development, immunity, and bioconversion. Plant J. 97:134–147. [DOI] [PubMed] [Google Scholar]
- Denyer T, Ma X, Klesen S, Scacchi E, Nieselt K, and Timmermans MC (2019). Spatiotemporal developmental trajectories in the Arabidopsis root revealed using high-throughput single-cell RNA sequencing. Dev.Cell 48:840–852. [DOI] [PubMed] [Google Scholar]
- Ding Y, Tang Y, Kwok CK, Zhang Y, Bevilacqua PC, Sarah M, and Assmann SM (2014). In vivo genome-wide profiling of RNA secondary structure reveals novel regulatory features. Nature 505:696–700. [DOI] [PubMed] [Google Scholar]
- Doehlemann G, and Hemetsberger C (2013). Apoplastic immunity and its suppression by filamentous plant pathogens. New phytol. 198:1001–1016. [DOI] [PubMed] [Google Scholar]
- Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K, Jacob-Hirsch J, Amariglio N, Kupiec M, et al. (2012). Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485:201–206. [DOI] [PubMed] [Google Scholar]
- Driouich A, Follet-Gueye ML, Vicré-Gibouin M, and Hawes MC (2013). Root border cells and secretions as critical elements in plant host defense. Curr. Opin. Plant Biol 16:489–495. [DOI] [PubMed] [Google Scholar]
- Dubiella U, Seybold H, Durian G, Komander E, Lassig R, Witte CP, Schulze WX, and Romeis T (2013). Calcium-dependent protein kinase/NADPH oxidase activation circuit is required for rapid defense signal propagation. Proc. Natl. Acad. Sci. USA 110: 8744–8749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elzinga DA, De Vos M, and Jander G (2014). Suppression of plant defenses by a Myzus persicae (green peach aphid) salivary effector protein. Mol. Plant Microbe Interact 27:747–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enyedi AJ, Yalpani N, Silverman P, and Raskin I (1992). Localization, conjugation, and function of salicylic acid in tobacco during the hypersensitive reaction to tobacco mosaic virus. Proc. Natl. Acad. Sci. USA 89:2480–2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esquerré-Tugayé MT, Lafitte C, Mazau D, Toppan A, and Touzé A (1979). Cell surfaces in plant-microorganism interactions. II. Evidence for the accumulation of hydroxyproline-rich glycoproteins in the cell wall of diseased plants as a defense mechanism. Plant Physiol. 64:320–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulkner C, Petutschnig E, Benitez-Alfonso Y, Beck M, Robatzek S, Lipka V, and Maule AJ (2013). LYM2-dependent chitin perception limits molecular flux via plasmodesmata. Proc. Natl. Acad. Sci. USA 110:9166–9170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felix G, Duran JD, Volko S, and Boller T (1999). Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J. 18:265–276. [DOI] [PubMed] [Google Scholar]
- Francl LJ (2001). The disease triangle: A plant pathological paradigm revisited. The plant health instructor. [Google Scholar]
- Fu D, Uauy C, Distelfeld A, Blechl A, Epstein L, Chen X, Sela H, Fahima T, and Dubcovsky J (2009). A kinase-START gene confers temperature-dependent resistance to wheat stripe rust. Science 323:1357–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu ZQ, Yan S, Saleh A, Wang W, Ruble J, Oka N, Mohan R, Spoel SH, Tada Y, Zheng N, et al. (2012). NPR3 and NPR4 are receptors for the immune signal salicylic acid in plants. Nature 486:228–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gantner J, Ordon J, Kretschmer C, Guerois R, and Stuttmann J (2019). An EDS1-SAG101 complex is essential for TNL-mediated immunity in Nicotiana benthamiana. Plant Cell 31: 2456–2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao F, Zhang BS, Zhao JH, Huang JF, Jia PS, Wang S, Zhang J, Zhou JM, and Guo HS (2019). Deacetylation of chitin oligomers increases virulence in soil-borne fungal pathogens. Nat. Plants 5:1167–1176. [DOI] [PubMed] [Google Scholar]
- Gao X, Chen X, Lin W, Chen S, Lu D, Niu Y, Li L, Cheng C, Mc, Cormack M, Shxeen J, et al. (2013). Bifurcation of Arabidopsis NLR immune signaling via Ca2+-dependent protein kinases. PLoS Pathog. 9:e1003127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert WV, Zhou K, Butler TK, and Doudna JA (2007). Cap-independent translation is required for starvation-induced differentiation in yeast. Science 317:1224–1227. [DOI] [PubMed] [Google Scholar]
- Gimenez-Ibanez S, Boter M, Fernández-Barbero G, Chini A, Rathjen JP, and Solano R (2014). The bacterial effector HopX1 targets JAZ transcriptional repressors to activate jasmonate signaling and promote infection in Arabidopsis. PLoS. Biol 12:e1001792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimenez-Ibanez S, Boter M, Ortigosa A, García-Casado G, Chini A, Lewsey MG, Ecker JR, Ntoukakis V, and Solano R (2017). JAZ2 controls stomata dynamics during bacterial invasion. New Phytol. 213:1378–1392. [DOI] [PubMed] [Google Scholar]
- Gloggnitzer J, Akimcheva S, Srinivasan A, Kusenda B, Riehs N, Stampfl H, Bautor J, Dekrout B, Jonak C, Jiménez-Gómez JM, et al. (2014). Nonsense-mediated mRNA decay modulates immune receptor levels to regulate plant antibacterial defense. Cell Host Microbe 16:376–390. [DOI] [PubMed] [Google Scholar]
- Godiard L, Sauviac L, Torii KU, Grenon O, Mangin B, Grimsley NH, and Marco Y (2003). ERECTA, an LRR receptor-like kinase protein controlling development pleiotropically affects resistance to bacterial wilt. Plant J. 36:353–365. [DOI] [PubMed] [Google Scholar]
- Gong BQ, Guo J, Zhang N, Yao X, Wang H, and Li JF (2019). Cross-microbial protection via priming a conserved immune co-receptor through juxtamembrane phosphorylation in plants. Cell Host Microbe 26:810–822. [DOI] [PubMed] [Google Scholar]
- Goodspeed D, Chehab EW, Min-Venditti A, Braam J and Covington MF (2012). Arabidopsis synchronizes jasmonate-mediated defense with insect circadian behavior. Proc. Natl. Acad. Sci. USA 109:4674–4677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gou JY, Li K, Wu K, Wang X, Lin H, Cantu D, Uauy C, Dobon-Alonso A, Midorikawa T, and Inoue K (2015). Wheat stripe rust resistance protein WKS1 reduces the ability of the thylakoid-associated ascorbate peroxidase to detoxify reactive oxygen species. Plant Cell 27:1755–1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory PJ, Johnson SN, Newton AC, and Ingram JS (2009). Integrating pests and pathogens into the climate change/food security debate. J. Exp. Bot 60:2827–2838. [DOI] [PubMed] [Google Scholar]
- Grison MS, Kirk P, Brault ML, Wu XN, Schulze WX, Benitez-Alfonso Y, Immel F, and Bayer EM (2019). Plasma membrane-associated receptor-like kinases relocalize to plasmodesmata in response to osmotic stress. Plant Physiol. 181:142–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunawardena U, Rodriguez M, Straney D, Romeo JT, VanEtten HD, and Hawes MC (2005). Tissue specific localization of root infection by Nectria haematococca: mechanisms and consequences. Plant Physiol. 137:1363–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunisova S, and Valasek LS (2014). Fail-safe mechanism of GCN4 translational control--uORF2 promotes reinitiation by analogous mechanism to uORF1 and thus secures its key role in GCN4 expression. Nucleic Acids Res. 42:5880–5893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gust AA, Pruitt R, and Nürnberger T (2017). Sensing danger: key to activating plant immunity. Trends Plant Sci. 22:779–791. [DOI] [PubMed] [Google Scholar]
- Häffner E, Karlovsky P, Splivallo R, Traczewska A, and Diederichsen E (2014). ERECTA, salicylic acid, abscisic acid, and jasmonic acid modulate quantitative disease resistance of Arabidopsis thaliana to Verticillium longisporum. BMC Plant Biol. 14:85.doi: 10.1186/1471-2229-14-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamamoto L, Hawes MC, and Rost TL (2006). The production and release of living root cap border cells is a function of root apical meristem type in dicotyledonous angiosperm plants. Ann. Bot 97:917–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hander T, Fernández-Fernández ÁD, Kumpf RP, Willems P, Schatowitz H, Rombaut D, Staes A, Nolf J, Pottie R, Yao P, Goncalves A, Pavie B, et al. (2019). Damage on plants activates Ca2+-dependent metacaspases for release of immunomodulatory peptides. Science 363: eaar7486. [DOI] [PubMed] [Google Scholar]
- Hao P, Liu C, Wang Y, Chen R, Tang M, Du B, Zhu L, and He G (2008). Herbivore-induced callose deposition on the sieve plates of rice: an important mechanism for host resistance. Plant Physiol. 146:1810–1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawes MC, Allen C, Turgeon BG, Curlango-Rivera G, Minh TT, Huskey DA, and Xiong Z (2016). Root border cells and their role in plant defense. Annu. Rev. Phytopathol 54:143–61. [DOI] [PubMed] [Google Scholar]
- Hawes MC, Curlango-Rivera G, Xiong Z, and Kessler JO (2012). Roles of root border cells in plant defense and regulation of rhizosphere microbial populations by extracellular DNA ‘trapping’. Plant Soil. 355:1–15. [Google Scholar]
- Hemetsberger C, Herrberger C, Zechmann B, Hillmer M, and Doehlemann G (2012). The Ustilago maydis effector Pep1 suppresses plant immunity by inhibition of host peroxidase activity. PLoS Pathog. 8:e1002684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry E, Toruño TY, Jauneau A, Deslandes L, and Coaker G (2017). Direct and indirect visualization of bacterial effector delivery into diverse plant cell types during infection. Plant Cell 29:1555–1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hevia MA, Canessa P, Muller-Esparza H, and Larrondo LF (2015). A circadian oscillator in the fungus Botrytis cinerea regulates virulence when infecting Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 112:8744–8749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashi T, Tanigaki Y, Takayama K, Nagano AJ, Honjo MN, and Fukuda H (2016). Detection of diurnal variation of tomato transcriptome through the molecular timetable method in a sunlight-type plant factory. Front. Plant Sci 7:87. doi: 10.3389/fpls.2016.00087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch AG (2005). Translational regulation of GCN4 and the general amino acid control of yeast. Annu. Rev. Microbiol 59:407–450. [DOI] [PubMed] [Google Scholar]
- Horsefield S, Burdett H, Zhang X, Manik MK, Shi Y, Chen J, Qi T, Gilley J, Lai JS, Rank MX, et al. (2019). NAD+ cleavage activity by animal and plant TIR domains in cell death pathways. Science 365: 793–799. [DOI] [PubMed] [Google Scholar]
- Huang W, Wang Y, Li X, and Zhang Y (2020). Biosynthesis and regulation of salicylic acid and N-hydroxypipecolic acid in plant immunity. Mol. Plant 13:31–41. [DOI] [PubMed] [Google Scholar]
- Hunter K, Kimura S, Rokka A, Tran HC, Toyota M, Kukkonen JP, and Wrzaczek M (2019). CRK2 enhances salt tolerance by regulating callose deposition in connection with PLDα1. Plant Physiol. 180:2004–2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley B, Lee D, Mott A, Wilton M, Liu J, Liu YC, Angers S, Coaker G, Guttman DS, and Desveaux D (2014). The Pseudomonas syringae type III effector HopF2 suppresses Arabidopsis stomatal immunity. PLoS One. 9:e114921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Immerzeel P, Eppink MM, de Vries SC, Schols HA, and Voragen AG (2006). Carrot arabinogalactan proteins are interlinked with pectins. Physiologia Plantarum 128:18–28. [Google Scholar]
- Ingle RA, Stoker C, Stone W, Adams N, Smith R, Grant M, Carré I, Roden LC, and Denby KJ (2015). Jasmonate signalling drives time-of-day differences in susceptibility of Arabidopsis to the fungal pathogen Botrytis cinerea. Plant J. 84:937–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JR, and Weissman JS (2009). Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324:218–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izquierdo Y, Kulasekaran S, Benito P, López B, Marcos R, Cascón T, Hamberg M, and Castresana C (2018). Arabidopsis nonresponding to oxylipins locus NOXY7 encodes a yeast GCN1 homolog that mediates noncanonical translation regulation and stress adaptation. Plant Cell Environ. 41:1438–1452. [DOI] [PubMed] [Google Scholar]
- Jackson RJ, Hellen CU, and Pestova TV (2010). The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol 11:113–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jashni MK, Mehrabi R, Collemare J, Mesarich CH, and de Wit PJ (2015). The battle in the apoplast: further insights into the roles of proteases and their inhibitors in plant-pathogen interactions. Front. Plant Sci 6:584. doi: 10.3389/fpls.2015.00584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang S, Yao J, Ma KW, Zhou H, Song J, He SY, and Ma W (2013). Bacterial effector activates jasmonate signaling by directly targeting JAZ transcriptional repressors. PLoS. Pathog 9:e1003715. doi: 10.1371/journal.ppat.1003715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, Wang Y, Xue D, Wang J, Yan M, Liu G, Dong G, Zeng D, Lu Z, Zhu X, et al. (2010). Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat. Genet 42:541–544. [DOI] [PubMed] [Google Scholar]
- Johnson KC, Dong OX, Huang Y and Li X (2012). A rolling stone gathers no moss, but resistant plants must gather their moses. Cold Spring Harb. Symp. Quant. Biol 77:259–268. [DOI] [PubMed] [Google Scholar]
- Jones AM, Thomas V, Bennett MH, Mansfield J and Grant M (2006). Modifications to the Arabidopsis defense proteome occur prior to significant transcriptional change in response to inoculation with Pseudomonas syringae. Plant Physiol. 142:1603–1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JD, Vance RE, and Dangl JL (2016). Intracellular innate immune surveillance devices in plants and animals. Science 354:aaf6395. [DOI] [PubMed] [Google Scholar]
- Joshi-Saha A, Valon C, and Leung J (2011). Abscisic acid signal off the STARting block. Mol. Plant 4:562–80. [DOI] [PubMed] [Google Scholar]
- Juroszek P, Racca P, Link S, Farhumand J, and Kleinhenz B (2020). Overview on the review articles published during the past 30 years relating to the potential climate change effects on plant pathogens and crop disease risks. Plant Pathol. 69:179–193. [Google Scholar]
- Kadota Y, Liebrand TW, Goto Y, Sklenar J, Derbyshire P, Menke FL, Torres MA, Molina A, Zipfel C, Coaker G, et al. (2019). Quantitative phosphoproteomic analysis reveals common regulatory mechanisms between effector- and PAMP-triggered immunity in plants. New Phytol. 221:2160–2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadota Y, Sklenar J, Derbyshire P, Stransfeld L, Asai S, Ntoukakis V, Jones JD, Shirasu K, Menke F, Jones A, et al. (2014). Direct regulation of the NADPH oxidase RBOHD by the PRR-associated kinase BIK1 during plant immunity. Mol. Cell 54:43–55. [DOI] [PubMed] [Google Scholar]
- Katsir L, Schilmiller AL, Staswick PE, He SY, and Howe GA (2008). COI1 is a critical component of a receptor for jasmonate and the bacterial virulence factor coronatine. Proc. Natl. Acad. Sci. USA 105:7100–7105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerwin RE, Jimenez-Gomez JM, Fulop D, Harmer SL, Maloof JN, and Kliebenstein DJ (2011). Network quantitative trait loci mapping of circadian clock outputs identifies metabolic pathway-to-clock linkages in Arabidopsis. Plant Cell 23:471–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilberg MS, Shan J, and Su N (2009). ATF4-dependent transcription mediates signaling of amino acid limitation. Trends Endocrinol. Metab 20:436–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klymiuk V, Yaniv E, Huang L, Raats D, Fatiukha A, Chen S, Feng L, Frenkel Z, Krugman T, Lidzbarsky G, et al. (2018). Cloning of the wheat Yr15 resistance gene sheds light on the plant tandem kinase-pseudokinase family. Nat. Commun 9:3735–3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoblauch M, Froelich DR, Pickard WF, and Peters WS (2014). SEORious business: structural proteins in sieve tubes and their involvement in sieve element occlusion. J. Exp. Bot 65:1879–1893. [DOI] [PubMed] [Google Scholar]
- Korneli C, Danisman S, and Staiger D (2014). Differential control of pre-invasive and post-invasive antibacterial defense by the Arabidopsis circadian clock. Plant Cell Physiol. 55:1613–1622. [DOI] [PubMed] [Google Scholar]
- Koroney AS, Plasson C, Pawlak B, Sidikou R, Driouich A, Menu-Bouaouiche L, and Vicré-Gibouin M (2016). Root exudate of Solanum tuberosum is enriched in galactose-containing molecules and impacts the growth of Pectobacterium atrosepticum. Ann. of Bot 118:797–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krattinger SG, Kang J, Bräunlich S, Boni R, Chauhan H, Selter LL, Robinson MD, Schmid MW, Wiederhold E, Hensel G, et al. (2019). Abscisic acid is a substrate of the ABC transporter encoded by the durable wheat disease resistance gene Lr34. New Phytol. 223:853–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lageix S, Lanet E, Pouch-Pélissier MN,, Espagnol MC, Robaglia C, Deragon JM, and Pélissier T (2008). Arabidopsis eIF2alpha kinase GCN2 is essential for growth in stress conditions and is activated by wounding. BMC Plant Biol. 8:134. doi: 10.1186/1471-2229-8-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapin D, Kovacova V, Sun X, Dongus JA, Bhandari D, von Born P, Bautor J, Guarneri N, Rzemieniewski J, Stuttmann J, et al. (2019). A coevolved EDS1-SAG101-NRG1 module mediates cell death signaling by TIR-domain immune receptors. Plant Cell 31:2430–2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DH, Bourdais G, Yu G, Robatzek S, and Coaker G (2015). Phosphorylation of the plant immune regulator RPM1-INTERACTING PROTEIN4 enhances plant plasma membrane H+-ATPase activity and inhibits flagellin-triggered immune responses in Arabidopsis. Plant Cell 27:2042–2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DH, Lal NK, Lin ZJD, Ma S, Liu J, Castro B, Toruño T, Dinesh-Kumar SP, and Coaker G (2020). Regulation of reactive oxygen species during plant immunity through phosphorylation and ubiquitination of RBOHD. Nat. Commun 11: 1838.doi: 10.1038/s41467-020-15601-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei J, Jayaprakasha GK, Singh J, Uckoo R, Borrego EJ, Finlayson S, Kolomiets M, Patil BS, Braam J, and Zhu-Salzman K (2019). CIRCADIAN CLOCK-ASSOCIATED1 controls resistance to aphids by altering indole glucosinolate production. Plant Physiol. 181:1344–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Ferreira MA, Huang M, Camargos LF, Yu X, Teixeira RM, Carpinetti PA, Mendes GC, Gouveia-Mageste BC, Liu C, et al. (2019a). The receptor-like kinase NIK1 targets FLS2/BAK1 immune complex and inversely modulates antiviral and antibacterial immunity. Nat. Commun 10:4996.doi: 10.1038/s41467-019-12847-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li FF, and Wang AM (2018). RNA decay is an antiviral defense in plants that is counteracted by viral RNA silencing suppressors. PLoS Pathog. 14:e1007228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Qiu J, and Fu XD (2012). RASL-seq for massively parallel and quantitative analysis of gene expression. Curr. Protoc. Mol. Biol. Chapter 4, Unit 4 13:11–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Li M, Yu L, Zhou Z, Liang X, Liu Z, Cai G, Gao L, Zhang X, Wang Y, et al. (2014). The FLS2-associated kinase BIK1 directly phosphorylates the NADPH oxidase RbohD to control plant immunity. Cell Host Microbe 15:329–338. [DOI] [PubMed] [Google Scholar]
- Li M, Cao L, Mwimba M, Yan Zhou Y, Li L, Zhou M, Schnable PS, O’Rourke JA, Dong X, and Wang W (2019b). Comprehensive mapping of abiotic stress inputs into the soybean circadian clock. Proc. Natl. Acad. Sci. USA 116:23840–23849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Zhu Z, Chern M, Yin J, Yang C, Ran L, Cheng M, He M, Wang K, Wang J, et al. (2017). A natural allele of a transcription factor in rice confers broad-spectrum blast resistance. Cell 170:114–126. [DOI] [PubMed] [Google Scholar]
- Li Z, Bonaldi K, Uribe F and Pruneda-Paz JL (2018). A localized Pseudomonas syringae infection triggers systemic clock responses in Arabidopsis. Curr. Biol 28:630–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X, Ding P, Lian K, Wang J, Ma M, Li L, Li L, Li M, Zhang X, Chen S, et al. (2016). Arabidopsis heterotrimeric G proteins regulate immunity by directly coupling to the FLS2 receptor. eLife 5:e13568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X, Ma M, Zhou Z, Wang J, Yang X, Rao S, Bi G, Li L, Zhang X, Chai J, et al. (2018). Ligand-triggered de-repression of Arabidopsis heterotrimeric G proteins coupled to immune receptor kinases. Cell Res. 28: 529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y, Cao Y, Tanaka K, Thibivilliers S, Wan J, Choi J, ho Kang C, Qiu J, and Stacey G (2013). Nonlegumes respond to rhizobial Nod factors by suppressing the innate immune response. Science 341:1384–1387. [DOI] [PubMed] [Google Scholar]
- Lin ZJD, Liebrand TW, Yadeta KA, and Coaker G (2015). PBL13 is a serine/threonine protein kinase that negatively regulates Arabidopsis immune responses. Plant Physiol. 169:2950–2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Elmore J, Fuglsang A, Palmgren M, Staskawicz B, and Coaker G (2009). RIN4 functions with plasma membrane H+-ATPases to regulate stomatal apertures during pathogen attack. PLoS. Biol 7:e1000139. doi: 10.1371/journal.pbio.1000139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Sonbol FM, Huot B, Gu Y, Withers J, Mwimba M, Yao J, He SY, and Dong X (2016). Salicylic acid receptors activate jasmonic acid signalling through a noncanonical pathway to promote effector-triggered immunity. Nat. Commun 7:13099. doi: 10.1038/ncomms13099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Kandoth PK, Warren SD, Yeckel G, Heinz R, Alden J, Yang C, Jamai A, El-Mellouki T, Juvale PS, et al. (2012). A soybean cyst nematode resistance gene points to a new mechanism of plant resistance to pathogens. Nature 492:256–260. [DOI] [PubMed] [Google Scholar]
- Liu X, Afrin T, and Pajerowska-Mukhtar KM (2019). Arabidopsis GCN2 kinase contributes to ABA homeostasis and stomatal immunity. Commun. Biol 2:302. doi: 10.1038/s42003-019-0544-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XK, Grabherr HM, Willmann R, Kolb D, Brunner F, Bertsche U, Kühner D, Franz-Wachtel M, Amin B, Felix G, et al. (2014). Host-induced bacterial cell wall decomposition mediates pattern-triggered immunity in Arabidopsis. eLife:e01990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YQ, Wu H, Chen H, Liu YL, He J, Kang HY, Sun ZG, Pan G, Wang Q, Hu JL, et al. (2015). A gene cluster encoding lectin receptor kinases confers broad-spectrum and durable insect resistance in rice. Nat. Biotechnol 33:301–305. [DOI] [PubMed] [Google Scholar]
- Llorente F, Alonso-Blanco C, Sanchez-Rodriguez C, Jorda L, and Molina A (2005). ERECTA receptor-like kinase and heterotrimeric G protein from Arabidopsis are required for resistance to the necrotrophic fungus Plectosphaerella cucumerina. Plant J. 43:165–180. [DOI] [PubMed] [Google Scholar]
- Lu D, Wu S, Gao X, Zhang Y, Shan L, and He P (2010). A receptor-like cytoplasmic kinase, BIK1, associates with a flagellin receptor complex to initiate plant innate immunity. Proc. Natl. Acad. Sci. USA 107: 496–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu P, Guo L, Wang Z, Li B, Li J, Li Y, Qiu D, Shi W, Yang L, Wang N, et al. (2020). A rare gain of function mutation in a wheat tandem kinase confers resistance to powdery mildew. Nat. Commun 11: 680. doi: 10.1038/s41467-020-14294-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luna E, van Hulten M, Zhang Y, Berkowitz O, López A, Pétriacq P, Sellwood MA, Chen B, Burrell M, van de Meene A, et al. (2014). Plant perception of beta-aminobutyric acid is mediated by an aspartyl-tRNA synthetase. Nat. Chem. Biol 10:450–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma KW, Jiang S, Hawara E, Lee D, Pan S, Coaker G, Song J, and Ma W (2015). Two serine residues in Pseudomonas syringae effector HopZ1a are required for acetyltransferase activity and association with the host co-factor. New Phytol. 208:1157–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, Claus LA, Leslie ME, Tao K, Wu Z, Liu J, Yu X, Li B, Zhou J, Savatin DV, et al. (2020). Ligand-induced monoubiquitination of BIK1 regulates plant immunity. Nature 581:199–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majhi BB, Sreeramulu S, and Sessa G (2019). BRASSINOSTEROID-SIGNALING KINASE5 associates with immune receptors and is required for immune responses. Plant Physiol. 180:1166–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makinen K, Lohmus A and Pollari M (2017). Plant RNA regulatory network and RNA granules in virus infection. Front. Plant Sci 8:2093. doi: 10.3389/fpls.2017.02093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maldonado-Bonilla LD, Eschen-Lippold L, Gago-Zachert S, Tabassum N, Bauer N, Scheel D, and Lee J (2014). The Arabidopsis tandem zinc finger 9 protein binds RNA and mediates pathogen-associated molecular pattern-triggered immune responses. Plant Cell Physiol. 55:412–425. [DOI] [PubMed] [Google Scholar]
- Malinovsky FG, Fangel JU, and Willats WGT (2014). The role of the cell wall in plant immunity. Front. Plant Sci 5:178. doi: 10.3389/fpls.2014.00178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Salas E, Francisco-Velilla R, Fernandez-Chamorro J, and Embarek AM (2017). Insights into structural and mechanistic features of viral IRES elements. Front. Microbiol 8:2629. doi: 10.3389/fmicb.2017.02629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meksem K, Pantazopoulos P, Njiti VN, Hyten LD, Arelli PR, and Lightfoot DA (2001). ‘Forrest’ resistance to the soybean cyst nematode is bigenic: saturation mapping of the Rhg1 and Rhg4 loci. Theor. Appl. Genet 103:710–717. [Google Scholar]
- Melaragno JE, Mehrotra B, and Coleman AW (1993). Relationship between endopolyploidy and cell size in epidermal tissue of Arabidopsis. Plant Cell 5:1661–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melotto M, Mecey C, Niu Y, Chung HS, Katsir L, Yao J, Zeng W, Thines B, Staswick P, Browse J, et al. (2008). A critical role of two positively charged amino acids in the Jas motif of Arabidopsis JAZ proteins in mediating coronatine- and jasmonoyl isoleucine-dependent interactions with the COI1 F-box protein. Plant J. 55:979–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melotto M, Underwood W, Koczan J, Nomura K, and He SY (2006). Plant stomata function in innate immunity against bacterial invasion. Cell 126:969–980. [DOI] [PubMed] [Google Scholar]
- Melotto M, Zhang L, Oblessuc PR, and He YS (2017). Stomatal defense a decade later. Plant Physiol. 174:561–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X, Chen X, Mang H, Liu C, Yu X, Gao X, Torii K, He P, and Shan L (2015). Differential function of Arabidopsis SERK family receptor-like kinases in stomatal patterning. Curr. Biol 25:2361–2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkouropoulos G, and Shirsat AH (2003). The unusual Arabidopsis extension gene atExt1 is expressed throughout plant development and is induced by a variety of biotic and abiotic stresses. Planta. 217:356–366. [DOI] [PubMed] [Google Scholar]
- Meteignier LV, Oirdi ME, Cohen M, Barff T, Matteau D, Lucier JF, Rodrigue S, Jacques PE, Yoshioka K, and Moffett P (2017). Translatome analysis of an NB-LRR immune response identifies important contributors to plant immunity in Arabidopsis. J. Exp. Bot 68:2333–2344. [DOI] [PubMed] [Google Scholar]
- Meteignier LV, Zhou J, Cohen M, Bhattacharjee S, Brosseau C, Chan MG, Robatzek S, and Moffett P (2016). NB-LRR signaling induces translational repression of viral transcripts and the formation of RNA processing bodies through mechanisms differing from those activated by UV stress and RNAi. J. Exp. Bot 67:2353–2366. [DOI] [PubMed] [Google Scholar]
- Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, and Jaffrey SR (2012). Comprehensive analysis of mRNA methylation reveals enrichment in 3’ UTRs and near stop codons. Cell 149:1635–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michielse CB and Rep M (2009). Pathogen profile update: Fusarium oxysporum. Mol. Plant Pathol 10:311–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millet YA, Danna CH, Clay NK, Songnuan W, Simon MD, Werck-Reichhart D, and Ausubel FM (2010). Innate immune responses activated in Arabidopsis roots by microbe-associated molecular patterns. Plant Cell 22:973–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra BB, de Armas E and Chen S (2016). Differential metabolomic responses of PAMP-triggered immunity and effector-triggered immunity in Arabidopsis suspension cells. Metabolomics 12:doi: 10.1007/s11306-016-0984-y. [DOI] [Google Scholar]
- Miura K, Ikeda M, Matsubara A, Song X, Ito M, Asano K, Matsuoka M, Kitano H, and Ashikari M (2010). OsSPL14 promotes panicle branching and higher grain productivity in rice. Nat. Genet 42:545–549. [DOI] [PubMed] [Google Scholar]
- Monaghan J, Matschi S, Shorinola O, Rovenich H, Matei A, Segonzac C, Malinovsky FG, Rathjen JP, MacLean D, Romeis T, et al. (2014). The calcium-dependent protein kinase CPK28 buffers plant immunity and regulates BIK1 turnover. Cell Host Microbe 16:605–615. [DOI] [PubMed] [Google Scholar]
- Monier JM and Lindow S (2003). Pseudomonas syringae responds to the environment on leaves by cell size reduction. Phytopathology 93:1209–1216. [DOI] [PubMed] [Google Scholar]
- Moore JW, Herrera-Foessel S, Lan C, Schnippenkoetter W, Ayliffe M, Huerta-Espino J, Lillemo M, Viccars L, Milne R, Periyannan S, et al. (2015). A recently evolved hexose transporter variant confers resistance to multiple pathogens in wheat. Nat. Genet 47:1494–1498. [DOI] [PubMed] [Google Scholar]
- Mwimba M, Karapetyan S, Liu L, Marqués J, McGinnis EM, Buchler NE, and Dong X (2018). Daily humidity oscillation regulates the circadian clock to influence plant physiology. Nat. Commun 9:4290. doi: 10.1038/s41467-018-06692-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadeau JA (2009). Stomatal development: new signals and fate determinants. Curr. Opin. Plant Biol 12:29–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano AJ, Sato Y, Mihara M, Antonio BA, Motoyama R, Itoh H, Nagamura Y, and Izawa T (2012). Deciphering and prediction of transcriptome dynamics under fluctuating field conditions. Cell 151:1358–1369. [DOI] [PubMed] [Google Scholar]
- Narusaka M, Shirasu K, Noutoshi Y, Kubo Y, Shiraishi T, Iwabuchi M, and Narusaka Y (2009). RRS1 and RPS4 provide a dual Resistance-gene system against fungal and bacterial pathogens. Plant J. 60:218–226. [DOI] [PubMed] [Google Scholar]
- Ngou BPM, Ahn H-K, Ding P, and Jones JD (2020). Mutual potentiation of plant immunity by cell-surface and intracellular receptors. bioRxiv. doi: 10.1101/2020.04.10.034173. [DOI] [PubMed] [Google Scholar]
- Okada M, Ito S, Marsubara A, Iwakura I, Egoshi S, and Ueda M (2009). Total syntheses of coronatines by exo-selective Diels-Alder reaction and their biological activities on stomatal opening. Org. Biomol. Chem 7:3065–3073. [Google Scholar]
- Oosterveld A, Voragen AG, and Schols HA (2002). Characterization of hop pectins shows the presence of an arabinogalactan-protein. Carbohydrate Polymers. 49:407–413. [Google Scholar]
- Pajerowska-Mukhtar KM, Wang W, Tada Y, Oka N, Tucker CL, Fonseca JP, and Dong X (2012). The HSF-like transcription factor TBF1 is a major molecular switch for plant growth-to-defense transition. Curr. Biol 22:103–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park E, Lee HY, Woo J, Choi D, and Dinesh-Kumar SP (2017). Spatiotemporal monitoring of Pseudomonas syringae effectors via type III secretion using split fluorescent protein fragments. Plant Cell 29:1571–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peart JR, Mestre P, Lu R, Malcuit I, and Baulcombe DC (2005). NRG1, a CC-NBLRR protein, together with N, a TIR-NB-LRR protein, mediates resistance against tobacco mosaic virus. Curr. Biol 15:968–973. [DOI] [PubMed] [Google Scholar]
- Pel MJ, van Dijken AJ, Bardoel BW, Seidl MF, van der Ent S, van Strijp JA, and Pieterse CM (2014). Pseudomonas syringae evades host immunity by degrading flagellin monomers with alkaline protease AprA. Mol. Plant Microbe Interact 27:603–610. [DOI] [PubMed] [Google Scholar]
- Plancot B, Santaella C, Jaber R, Kiefer-Meyer MC, Follet-Gueye ML, Leprince J, Gattin I, Souc C, Driouich A, and Vicré-Gibouin M (2013). Deciphering the responses of root border-like cells of Arabidopsis and flax to pathogen-derived elicitors. Plant Physiol. 163:1584–1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruneda-Paz JL, Breton G, Para A, and Kay SA (2009). A functional genomics approach reveals CHE as a component of the Arabidopsis circadian clock. Science 323:1481–1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi T, Seong K, Thomazella DPT, Kim JR, Pham J, Seo E, Cho MJ, Schultink A, and Staskawicz BJ (2018). NRG1 functions downstream of EDS1 to regulate TIR-NLR-mediated plant immunity in Nicotiana benthamiana. Proc. Natl. Acad. Sci. USA 115: E10979–E10987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao S, Zhou Z, Miao P, Bi G, Hu M, Wu Y, Feng F, Zhang X, and Zhou JM (2018). Roles of receptor-like cytoplasmic kinase VII members in pattern-triggered immune signaling. Plant Physiol. 177:1679–1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romeis T, Ludwig AA, Martin R, and Jones JD (2001). Calcium-dependent protein kinases play an essential role in a plant defense response. EMBO J. 20:5556–5567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roundtree IA, Evans ME, Pan T, and He C (2017). Dynamic RNA modifications in gene expression regulation. Cell 169:1187–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux ME, Rasmussen MW, Palma K, Lolle S, Regué AM, Bethke G, Glazebrook J, Zhang W, Sieburth L, Larsen MR, et al. (2015). The mRNA decay factor PAT1 functions in a pathway including MAP kinase 4 and immune receptor SUMM2. EMBO J. 34:593–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rufián JS, Macho AP, Corry DS, Mansfield JW, Ruiz-Albert J, Arnold DL, and Beuzón CR (2018). Confocal microscopy reveals in planta dynamic interactions between pathogenic, avirulent and non-pathogenic Pseudomonas syringae strains. Mol. Plant Pathol 19:537–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruzicka K, Zhang M, Campilho A, Bodi Z, Kashif M, Saleh M, Eeckhout D, El-Showk S, Li H, Zhong S, et al. (2017). Identification of factors required for m(6) A mRNA methylation in Arabidopsis reveals a role for the conserved E3 ubiquitin ligase HAKAI. New Phytol. 215:157–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savary S, Willocquet L, Pethybridge SJ, Esker P, McRoberts N, and Nelson A (2019). The global burden of pathogens and pests on major food crops. Nat. Ecol. Evol 3:430–439. [DOI] [PubMed] [Google Scholar]
- Scheideler M, Schlaich NL, Fellenberg K, Beissbarth T, Hauser NC, Vingron M, Slusarenko AJ, and Hoheisel JD (2002). Monitoring the switch from housekeeping to pathogen defense metabolism in Arabidopsis thaliana using cDNA arrays. J. Biol. Chem 277:10555–10561. [DOI] [PubMed] [Google Scholar]
- Scheidle H, Gross A, and Niehaus K (2005). The lipid A substructure of the Sinorhizobium meliloti lipopolysaccharides is sufficient to suppress the oxidative burst in host plants. New Phytol. 165:559–565. [DOI] [PubMed] [Google Scholar]
- Schilling L, Matei A, Redkar A, Walbot V, and Doehlemann G (2014). Virulence of the maize smut Ustilago maydis is shaped by organ-specific effectors. Mol. Plant Pathol 15:780–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher J (2017). How light affects the life of Botrytis. Fungal. Genet. Biol 106:26–41. [DOI] [PubMed] [Google Scholar]
- Schwachtje J, Fischer A, Erban A and Kopka J (2018). Primed primary metabolism in systemic leaves: a functional systems analysis. Sci. Rep 8:216. doi: 10.1038/s41598-017-18397-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segonzac C, and Monaghan J (2019). Modulation of plant innate immune signaling by small peptides. Curr. Opin. Plant Biol 51:22–28. [DOI] [PubMed] [Google Scholar]
- Shabab M, Shindo T, Gu C, Kaschani F, Pansuriya T, Chintha R, Harzen A, Colby T, Kamoun S, and van der Hoorn RA (2008). Fungal effector protein Avr2 targets diversifying defense-related cys proteases of tomato. Plant Cell 20:1169–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao ZQ, Xue JY, Wu P, Zhang YM, Wu Y, Hang YY, Wang B, and Chen JQ (2016). Large-scale analyses of angiosperm nucleotide-binding site-leucine-rich repeat genes reveal three anciently diverged classes with distinct evolutionary patterns. Plant Physiol. 170:2095–2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shatsky IN, Terenin IM, Smirnova VV, and Andreev DE (2018). Cap-independent translation: What’s in a name? Trends Biochem. Sci 43: 882–895. [DOI] [PubMed] [Google Scholar]
- Sheard LB, Tan X, Mao H, Withers J, Ben-Nissan G, Hinds TR, Kobayashi Y, Hsu FF, Sharon M, Browse J, et al. (2010). Jasmonate perception by inositolphosphatepotentiated COI1-JAZ co-receptor. Nature 468:400–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen W, Liu J, and Li J (2019). Type-II metacaspases mediate the processing of plant elicitor peptides in Arabidopsis. Mol. Plant 12:1524–1533. [DOI] [PubMed] [Google Scholar]
- Shi H, Shen Q, Qi Y, Yan H, Nie H, Chen Y, Zhao T, Katagiri F, and Tang D (2013). BR-SIGNALING KINASE1 physically associates with FLAGELLIN SENSING2 and regulates plant innate immunity in Arabidopsis. Plant Cell 25:1143–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpak ED (2013). Diverse roles of ERECTA family genes in plant development. J. Integr. Plant Biol 55:1238–1250. [DOI] [PubMed] [Google Scholar]
- Shpak ED, McAbee JM, Pillitteri LJ, and Torii KU (2005). Stomatal patterning and differentiation by synergistic interactions of receptor kinases. Science 309:209–293. [DOI] [PubMed] [Google Scholar]
- Sirichandra C, Wasilewska A, Vlad F, Valon C, and Leung J (2009). The guard cell as a single-cell model towards understanding drought tolerance and abscisic acid action. J. Exp. Bot 60:1439–1463. [DOI] [PubMed] [Google Scholar]
- Slusarenko AJ, and Schlaich NL (2003). Downy mildew of Arabidopsis thaliana caused by Hyaloperonospora parasitica (formerly Peronospora parasitica). Mol. Plant Pathol 4:159–170. [DOI] [PubMed] [Google Scholar]
- Smola MJ, and Weeks KM (2018). In-cell RNA structure probing with SHAPE-MaP. Nat. Protoc 13:1181–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenberg N, and Hinnebusch AG (2009). Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 136:731–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sopeña-Torres S, Jordá L, Sánchez-Rodríguez C, Eva Miedes E, Escudero V, Swami S, López G, Piślewska-Bednarek M, Lassowskat I, and Lee J (2018). YODA MAP3K kinase regulates plant immune responses conferring broad-spectrum disease resistance. New Phytol. 218:661–680. [DOI] [PubMed] [Google Scholar]
- Soprano AS, Smetana JH, and Benedetti CE (2018). Regulation of tRNA biogenesis in plants and its link to plant growth and response to pathogens. Biochem. Biophys. Acta. Gene Regul. Mech 1861:344–353. [DOI] [PubMed] [Google Scholar]
- Spoel SH, and Dong X (2008). Making sense of hormone crosstalk during plant immune responses. Cell Host Microbe 3:348–351. [DOI] [PubMed] [Google Scholar]
- Stone B, and Clarke A (1992). Chemistry and biology of (1→3)-β-glucans. Victoria, Australia: La Trobe University Press. [Google Scholar]
- Stringlis IA, Proietti S, Hickman R, Van Verk MC, Zamioudis C, and Pieterse CMJ (2018). Root transcriptional dynamics induced by beneficial rhizobacteria and microbial immune elicitors reveal signatures of adaptation to mutualists. Plant J. 93:166–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su J, Yang L, Zhu Q, Wu H, He Y, Liu Y, Xu J, Jiang D, and Zhang S (2018). Active photosynthetic inhibition mediated by MPK3/MPK6 is critical to effector-triggered immunity. PLoS Biol. 16, e2004122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sujkowska-Rybkowska M, and Borucki W (2014). Accumulation and localization of extensin protein in apoplast of pea root nodule under aluminum stress. Micron. 67:10–19. [DOI] [PubMed] [Google Scholar]
- Sun L, and Zhang J (2020). Regulatory role of receptor-like cytoplasmic kinases in early immune signaling events in plants. FEMS. Microbiol. Rev doi: 10.1093/femsre/fuaa035. [DOI] [PubMed] [Google Scholar]
- Takken F, and Rep M (2010). The arms race between tomato and Fusarium oxysporum. Mol. Plant Pathol 11:309–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D, Wang G, and Zhou JM (2017). Receptor kinases in plant-pathogen interactions: more than pattern recognition. Plant Cell 29:618–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tellstrom V, Usadel B, Thimm O, Stitt M, Kuster H, and Niehaus K (2007). The lipopolysaccharide of Sinorhizobium meliloti suppresses defense-associated gene expression in cell cultures of the host plant Medicago truncatula. Plant Physiol. 143:825–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thines B, Katsir L, Melotto M, Niu Y, Mandaokar A, Liu G, Nomura K, He SY, Howe GA, and Browse J (2007). JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 448:661–665. [DOI] [PubMed] [Google Scholar]
- Thor K, Jiang S, Michard E, George J, Scherzer S, Huang S, Dindas J, Derbyshire P, Leitão N, DeFalco TA, et al. (2020). The calcium-permeable channel OSCA1.3 regulates plant stomatal immunity. Nature doi: 10.1038/s41586-020-2702-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurston HD, Turston KW, and Eide CJ (1958). The relation of late blight development on potato foliage to temperature and humidity. Ame. Potato J 35:397–406. [Google Scholar]
- Tian W, Hou C, Ren Z, Wang C, Zhao F, Dahlbeck D, Hu S, Zhang L, Niu Q, Li L, et al. (2019). A calmodulin-gated calcium channel links pathogen patterns to plant immunity. Nature 572:131–135. [DOI] [PubMed] [Google Scholar]
- Tjallingii WF (2006). Salivary secretions by aphids interacting with proteins of phloem wound responses. J. Exp. Bot 57:739–745. [DOI] [PubMed] [Google Scholar]
- Toyota M, Spencer D, Sawai-Toyota S, Jiaqi W, Zhang T, Koo AJ, Howe GA, and Gilroy S (2018). Glutamate triggers long-distance, calcium-based plant defense signaling. Science 361:1112–1115. [DOI] [PubMed] [Google Scholar]
- Tran TM, MacIntyre AM, Hawes MC, and Allen C (2016). Escaping underground nets: extracellular DNases degrade plant extracellular traps and contribute to virulence of the plant pathogenic bacterium Ralstonia solanacearum. PLoS Pathog. 12:e1005686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda HR, Chen W, Minami Y, Honma S, Honma K, Iino M, and Hashimoto S (2004). Molecular-timetable methods for detection of body time and rhythm disorders from single-time-point genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 101:11227–11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Burg HA, Harrison SJ, Joosten MH, Vervoort J, and de Wit PJ (2006). Cladosporium fulvum Avr4 protects fungal cell walls against hydrolysis by plant chitinases accumulating during infection. Mol. Plant Microbe Interact 19:1420–1430. [DOI] [PubMed] [Google Scholar]
- van der Does HC, Constantin ME, Houterman PM, Takken FLW, Cornelissen BJC, Haring MA, van den Burg HA, and Rep M (2019). Fusarium oxysporum colonizes the stem of resistant tomato plants, the extent varying with the R-gene present. Eur. J. Plant Pathol 154:55–65. [Google Scholar]
- van Esse HP,Vant Klooster JW, Bolton MD, Yadeta KA, van Baarlen P, Boeren S, Vervoort J, de Wit PJ, and Thomma BP (2008). The Cladosporium fulvum virulence protein Avr2 inhibits host proteases required for basal defense. Plant Cell. 20:1948–1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Loon LC, Rep M, and Pieterse CMJ (2006). Significance of inducible defense-related proteins in infected plants. Annu. Rev. Phytopathol 44:135–162. [DOI] [PubMed] [Google Scholar]
- van Schie CC, and Takken FL (2014). Susceptibility genes 101: how to be a good host. Annu. Rev. Phytopathol 52:551–581. [DOI] [PubMed] [Google Scholar]
- Wan L, Essuman K, Anderson RG, Sasaki Y, Monteiro F, Chung EH, Nishimura EO, DiAntonio A, Milbrandt J, Dangl JL, et al. (2019a). TIR domains of plant immune receptors are NAD+-cleaving enzymes that promote cell death. Science 365:799–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan WL, Zhang L, Pruitt R, Zaidem M, Brugman R, Ma X, Krol E, Perraki A, Kilian J, Grossmann G, et al. (2019b). Comparing Arabidopsis receptor kinase and receptor protein-mediated immune signaling reveals BIK1-dependent differences. New Phytol. 221:2080–2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Zhou M, Zhang X, Yao J, Zhang Y, and Mou Z (2017a). A lectin receptor kinase as a potential sensor for extracellular nicotinamide adenine dinucleotide in Arabidopsis thaliana. eLife 6:e25474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Roux B, Feng F, Guy E, Li L, Li N, Zhang X, Lautier M, Jardinaud MF, Chabannes M, et al. (2015a). The decoy substrate of a pathogen effector and a pseudokinase specify pathogen-induced modified-self recognition and immunity in plants. Cell Host Microbe 18:285–295. [DOI] [PubMed] [Google Scholar]
- Wang HW, Sun SL, Ge WY, Zhao LF, Huo BQ, Wang K, Lyu ZF, Chen LY, Xu SS, Guo J, et al. (2020). Horizontal gene transfer of Fhb7 from fungus underlies Fusarium head blight resistance in wheat. Science 368:doi: 10.1126/science.aba5435. [DOI] [PubMed] [Google Scholar]
- Wang J, Grubb LE, Wang J, Liang X, Li L, Gao C, Ma M, Feng F, Li M, Li L, et al. (2018a). A regulatory module controlling homeostasis of a plant immune kinase. Mol. Cell 69:493–504. [DOI] [PubMed] [Google Scholar]
- Wang J, Hu M, Wang J, Qi J, Han Z, Wang G, Qi Y, Wang HW, Zhou JM, and Chai J (2019a). Reconstitution and structure of a plant NLR resistosome conferring immunity. Science 364:eaav5870. [DOI] [PubMed] [Google Scholar]
- Wang J, Liu X, Zhang A, Ren Y, Wu F, Wang G, Xu Y, Lei C, Zhu S, Pan T, et al. (2019b). A cyclic nucleotide-gated channel mediates cytoplasmic calcium elevation and disease resistance in rice. Cell Res. 29:820–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Wang J, Hu M, Wu S, Qi J, Wang G, Han Z, Qi Y, Gao N, Wang HW, et al. (2019c). Ligand-triggered allosteric ADP release primes a plant NLR complex. Science 364:eaav5868. [DOI] [PubMed] [Google Scholar]
- Wang J, Zhou L, Shi H, Chern M, Yu H, Yi H, He M, Yin J, Zhu X, Li Y, et al. (2018b). A single transcription factor promotes both yield and immunity in rice. Science 361:1026–1028. [DOI] [PubMed] [Google Scholar]
- Wang L, Sun S, Jin. J, Fu D, Yang X, Weng X, Xu C, Li X, Xiao J, and Zhang Q (2015b). Coordinated regulation of vegetative and reproductive branching in rice. Proc. Natl. Acad. Sci. U.S.A 112:15504–15509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Tsuda K, Sato M, Cohen JD, Katagiri F, and Glazebrook J (2009). Arabidopsis CaM binding protein CBP60g contributes to MAMP-induced SA accumulation and is involved in disease resistance against Pseudomonas syringae. PLoS Pathog. 5:e1000301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N, Pierson EA, Setubal JC, Xu J, Levy JG, Zhang Y, Li J, Rangel LT, and Martins J (2017b). The Candidatus Liberibacter–host interface: insights into pathogenesis mechanisms and disease control. Annu. Rev. Phytopathol 55:451–482. [DOI] [PubMed] [Google Scholar]
- Wang S, Sun J, Fan F, Tan Z, Zou Y, and Lu D (2016). A Xanthomonas oryzae pv. oryzae effector, XopR, associates with receptor-like cytoplasmic kinases and suppresses PAMP-triggered stomatal closure. Sci. China Life Sci 59:897–905. [DOI] [PubMed] [Google Scholar]
- Wang W, Barnaby JY, Tada Y, Li H, Tör M, Caldelari D, Lee D, Fu XD, and Dong X (2011). Timing of plant immune responses by a central circadian regulator. Nature 470:110–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Li J, Hou S, Wang X, Li Y, Ren D, Chen S, Tang X, and Zhou JM (2010). A Pseudomonas syringae ADP-ribosyltransferase inhibits Arabidopsis mitogen activated protein kinase kinases. Plant Cell 22:2033–2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward JL, Forcat S, Beckmann M, Bennett M, Miller SJ, Baker JM, Hawkins ND, Vermeer CP, Lu C, Lin W, et al. (2010). The metabolic transition during disease following infection of Arabidopsis thaliana by Pseudomonas syringae pv. tomato. Plant J. 63:443–457. [DOI] [PubMed] [Google Scholar]
- Wawra S, Fesel P, Widmer H, Timm M, Seibel J, Leson L, Nostadt R, Hilbert M, Langen G, and Zuccaro A (2016). The fungal-specific beta-glucan-binding lectin FGB1 alters cell-wall composition and suppresses glucan-triggered immunity in plants. Nat. Commun 7:13188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will T, Kornemann SR, Furch AC, Tjallingii WF, and van Bel AJ (2009). Aphid watery saliva counteracts sieve-tube occlusion: a universal phenomenon? J. Exp. Bot 212:3305–3312. [DOI] [PubMed] [Google Scholar]
- Wilton M, Subramaniam R, Elmore J, Felsensteiner C, Coaker G, and Desveaux D (2010). The type III effector HopF2Pto targets Arabidopsis RIN4 protein to promote Pseudomonas syringae virulence. Proc. Natl. Acad. Sci. USA 107:2349–2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CH, Abd-El-Haliem A, Bozkurt TO, Belhaj K, Terauchi R, Vossen JH, and Kamoun S (2017a). NLR network mediates immunity to diverse plant pathogens. Proc. Natl. Acad. Sci. USA 114:8113–8118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Fan W, Li X, Chen H, Takáč T, Šamajová O, Fabrice MR, Xie L, Ma J, Šamaj J, et al. (2017b). Expression and distribution of extensins and AGPs in susceptible and resistant banana cultivars in response to wounding and Fusarium oxysporum. Sci. Rep 7:42400. doi: 10.1038/srep42400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Tong M, Tian L, Zhu C, Liu X, Zhang Y, and Li X (2020). Plant E3 ligases SNIPER1 and SNIPER2 broadly regulate the homeostasis of sensor NLR immune receptors. EMBO J. 39:e104915 10.15252/embj.2020104915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuyts N, Palauqui JC, Conejero G, Verdeil JL, Granier C, and Massonnet C (2010). High-contrast three-dimensional imaging of the Arabidopsis leaf enables the analysis of cell dimensions in the epidermis and mesophyll. Plant Methods 6:17. doi: 10.1186/1746-4811-6-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyrsch I, Dominguez-Ferreras A, Geldner N, and Boller T (2015). Tissue-specific FLAGELLIN-SENSING 2 (FLS2) expression in roots restores immune responses in Arabidopsis fls2 mutants. New Phytol. 206:774–784. [DOI] [PubMed] [Google Scholar]
- Xie D, Ma L, Samaj J, and Xu C (2011). Immunohistochemical analysis of cell wall hydroxyproline-rich glycoproteins in the roots of resistant and susceptible wax gourd cultivars in response to Fusarium oxysporum f. sp. Benincasae infection and fusaric acid treatment. Plant Cell Rep. 30:1555–1569. [DOI] [PubMed] [Google Scholar]
- Xin XF, Nomura K, Aung K, Velásquez AC, Yao J, Boutrot F, Chang JH, Zipfel C, and He SY (2016). Bacteria establish an aqueous living space in plants crucial for virulence. Nature 539:524–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G, Greene GH, Yoo H, Liu L, Marqués J, Motley J, Dong X (2017a). Global translational reprogramming is a fundamental layer of immune regulation in plants. Nature 545:487–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G, Yuan M, Ai C, Liu L, Zhuang E, Karapetyan S, Wang S, and Dong X (2017b). uORF-mediated translation allows engineered plant disease resistance without fitness costs. Nature 545:491–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K, Yamaguchi K, Shirakawa T, Nakagami H, Mine A, Ishikawa K, Fujiwara M, Narusaka M, Narusaka Y, Ichimura K, et al. (2016). The Arabidopsis CERK1-associated kinase PBL27 connects chitin perception to MAPK activation. EMBO J. 35:2468–2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H, Zhao Y, Shi H, Li J, Wang Y, and Tang D (2018). BRASSINOSTEROID-SIGNALING KINASE1 phosphorylates MAPKKK5 to regulate immunity in Arabidopsis. Plant Physiol. 176:2991–3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Zhang C, Gu M, Bai Z, Zhang W, Qi T, Cheng Z, Peng W, Luo H, Nan F, et al. (2009). The Arabidopsis CORONATINE INSENSITIVE1protein is a jasmonate receptor. Plant Cell 21:2220–2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y, Stolz S, Chételat A, Reymond P, Pagni M, Dubugnon L, and Farmer EE (2007). A downstream mediator in the growth repression limb of the jasmonate pathway. Plant Cell 19: 2470–2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye W, Munemasa S, Shinya T, Wu W, Ma T, Lu J, Kinoshita T, Kaku H, Shibuya N, and Murata Y (2020). Stomatal immunity against fungal invasion comprises not only chitin-induced stomatal closure but also chitosan-induced guard cell death. Proc. Natl. Acad. Sci. USA 117:20932–20942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo H, Greene GH, Yuan M, Xu G, Burton D, Liu L, Marqués J, and Dong X (2020). Translational regulation of metabolic dynamics during effector-triggered immunity. Mol. Plant 13:88–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu K, Tichelaar R, Liu Y, Savant N, Lagendijk E, van Kuijk SJ, Stringlis IA, van Dijken AJH, Pieterse CMJ, Bakker PAHM, et al. (2019a). Rhizosphere-associated Pseudomonas suppress local root immune responses by gluconic acid-mediated lowering of environmental pH. Curr. Biol 29:3913–3920. [DOI] [PubMed] [Google Scholar]
- Yu X, Li B, Jang GJ, Jiang S, Jiang D, Jang JC, Wu SH, Shan L, and He P (2019b). Orchestration of processing body dynamics and mRNA decay in Arabidopsis immunity. Cell Rep. 28:2194–2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M, Jiang Z, Bi. G, Nomura K, Liu M, He SY, Zhou JM, and Xin XF (2020). Pattern-recognition receptors are required for NLR-mediated plant immunity. bioRxiv. doi: 10.1101/2020.04.10.031294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavaliev R, Mohan R, Chen T, and Dong X (2020). Formation of NPR1 condensates promotes cell survival during the plant immune response. Cell 82:1093–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng W, and He SY (2010). A prominent role of the flagellin receptor FLAGELLIN-SENSING2 in mediating stomatal response to Pseudomonas syringae pv. tomato DC3000 in Arabidopsis. Plant Physiol. 153: 1188–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng W, Brutus A, Kremer JM, Withers JC, Gao X, Jones AD, and He SY (2011). A genetic screen reveals Arabidopsis stomatal and/or apoplastic defenses against Pseudomonas syringae pv. tomato DC3000. PLoS Pathog. 7:e1002291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Xie Q, Anderson RG, Ng G, Seitz NC, Peterson T, McClung CR, McDowell JM, Kong D, Kwak JM, et al. (2013). Crosstalk between the circadian clock and innate immunity in Arabidopsis. PLoS Pathog. 9:e1003370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Li W, Xiang T, Liu Z, Laluk K, Ding X, Zou Y, Gao M, Zhang X, Chen S, et al. (2010). Receptor-like cytoplasmic kinases integrate signaling from multiple plant immune receptors and are targeted by a Pseudomonas syringae effector. Cell Host Microbe 7:290–301. [DOI] [PubMed] [Google Scholar]
- Zhang M, Chiang YH, Toruño TY, Lee D, Ma M, Liang X, Lal NK, Lemos M, Lu YJ, Ma S, et al. (2018). The MAP4 kinase SIK1 ensures robust extracellular ROS burst and antibacterial immunity in plants. Cell Host Microbe 24:379–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, He SY, and Assmann SM (2008a). The plant innate immunity response in stomatal guard cells invokes G-protein-dependention channel regulation. Plant J. 56:984–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wang Y, Kanyuka K, Parry MA, Powers SJ, and Halford NG (2008b). GCN2-dependent phosphorylation of eukaryotic translation initiation factor-2alpha in Arabidopsis. J. Exp. Bot 59:3131–3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Misaghi IJ, and Hawes MC (2000). Stimulation of border cell production in response to increased carbon dioxide levels. Plant Physiol. 122:181–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng G, Qin Y, Clark WC, Dai Q, Yi C, He C, Lambowitz AM, and Pan T (2015a). Efficient and quantitative high-throughput tRNA sequencing. Nat. Methods 12:835–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng XY, Zhou M, Yoo H, Pruneda-Paz JL, Spivey NW, Kay SA, and Dong X (2015b). Spatial and temporal regulation of biosynthesis of the plant immune signal salicylic acid. Proc. Natl. Acad. Sci. USA 112:9166–9173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng XY, Spivey NW, Zeng W, Liu PP, Fu ZQ, Klessig DF, He SY, and Dong X (2012). Coronatine promotes Pseudomonas syringae virulence in plants by activating a signaling cascade that inhibits salicylic acid accumulation. Cell Host Microbe 11:587–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F, Emonet A, Tendon VD, Marhavy P, Wu D, Lahaye T, and Geldner N (2020). Co-incidence of damage and microbial patterns controls localized immune responses in roots. Cell 180:e18 440–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Wu S, Chen X, Liu C, Sheen J, Shan L, and He P (2014). The Pseudomonas syringae effector HopF2 suppresses Arabidopsis immunity by targeting BAK1. Plant J. 77:235–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou JM, and Zhang Y (2020). Plant immunity: danger perception and signaling. Cell 181: 978–989. [DOI] [PubMed] [Google Scholar]
- Zhou M, Wang W, Karapetyan S, Mwimba M, Marqués J, Buchler NE, and Dong X (2015a). Redox rhythm reinforces the circadian clock to gate immune response. Nature 523:472–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Liao H, Chern M, Yin J, Chen Y, Wang J, Zhu X, Chen Z, Yuan C, Zhao W, et al. (2018). Loss of function of a rice TPR-domain RNA-binding protein confers broad-spectrum disease resistance. Proc. Natl. Acad. Sci. USA 115:3174–3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou ZY, Wu YJ, Yang YQ, Du M, Zhang XJ, Guo Y, Li C, and Zhou JM (2015b). An Arabidopsis plasma membrane proton ATPase modulates JA signaling and is exploited by the Pseudomonas syringae effector protein AvrB for stomatal invasion. Plant Cell 27:2032–2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel C, and Oldroyd GE (2017). Plant signalling in symbiosis and immunity. Nature 543:328–336. [DOI] [PubMed] [Google Scholar]


