Abstract
The microbiome has been hypothesized to play a role in cancer development. Due to the diversity of published data, an overview of available epidemiological evidence linking the microbiome with cancer is now needed. We conducted a systematic review using a tailored search strategy in Medline and EMBASE databases to identify and summarize the current epidemiologic literature on the relationship between the microbiome and different cancer outcomes published until December 2019. One hundred and twenty-four eligible articles were identified. The large diversity of parameters used to describe microbial composition made it impossible to harmonize the different studies in a way that would allow meta-analysis, therefore only a qualitative description of results could be performed. Fifty studies reported differences in the gut microbiome between colorectal cancer patients and various control groups. The most consistent findings were for Fusobacterium, Porphyromonas and Peptostreptococcus being significantly enriched in fecal and mucosal samples from colorectal cancer patients. For the oral microbiome, significantly increased and decreased abundance was reported for Fusobacterium and Streptococcus, respectively, in oral cancer patients compared to controls. Overall, although there was a large amount of evidence for some of these alterations, most require validation in high quality, preferably prospective, epidemiological studies.
Keywords: Cancer, microbiome, epidemiology, human
Introduction
The human microbiome defines either the microorganisms (bacteria, archaea, lower and higher eukaryotes, and viruses) found in and on the human body or their collective genomes (i.e. genetic material) (1–3). Since the development of high-throughput approaches using next-generation sequencing, our understanding of the human microbiome, even the non-cultivable microorganisms, has increased enormously, shedding light on its complexity (1,4,5). Although the gut microbiome has been most extensively studied, microorganisms also inhabit all of the barrier surfaces of the human body including the skin, the oral cavity, the nasopharynx, the esophagus and stomach, and also the vagina, the urinary tract, the lungs and others (4,6). The composition of the microbiome varies depending on the anatomical site and it also differs between individuals (4). The microbiota, the ensemble of microorganisms living in a specific environment, form a dynamic entity and can change within an individual in response to diet and other external factors such as medication, environment, and lifestyle (1,7,8).
In addition to the role that microbial imbalance (9) may play in infectious and autoimmune conditions; there is a growing appreciation of potential influences on cancer development (3,6). For example, the microbiota and its metabolic products influence the development and maintenance of inflammation, which has been broadly recognised to be one of the hallmarks of cancer (10–12). Inflammation has been demonstrated to promote cancer by promoting genetic instability and, once the tumour has been established, through the creation of a tumour-promoting microenvironment. In addition, the microbiota may also influence the development of cancer-promoting conditions, such as adiposity and insulin resistance (13–15). Due to these effects on host metabolism, cellular proliferation, inflammation, and immunity, the microbiome may influence cancer risk at different levels, including cancer initiation, promotion, dissemination, and response to therapy (6,16). The effects of the microbiota on cancer risk can be local, situated at the interface where the organism interacts with the host tissue in which the cancer originates, or systemic, via the physiological communication of the host organism and the microbiota through intact membranes or following alteration of barrier permeability in pathological situations. This potential link between the microbiome and cancer development might offer new opportunities for cancer prevention by understanding etiologic pathways for screening, diagnosis, and treatment (2,3,17).
A large number of individual studies on the association of the microbiome with cancer have been published in the past decade, and an in-depth review of the literature is now needed. While some published reviews and meta-analyses focusing on microbiome profiles for single cancer sites were recently published (18–20), no systematic overview of the most significant associations between microbiome composition (e.g. taxa and diversity) and different cancer sites has been published to date. Therefore, the objective of this systematic review was not only to summarise the published data, but also to evaluate the strengths and weaknesses of the current evidence on the relationship between the human microbiome and cancer in epidemiologic studies.
Materials and methods
A systematic review was conducted to identify peer-reviewed publications related to the human microbiome and cancer. The review considered the recommendations of the Meta-analysis of Observational Studies in Epidemiology (MOOSE) group (21) and followed the Preferred Reporting Items for Systematic Reviews and Metaanalyses (PRISMA) (22). A protocol (Prospero registration number: CRD42018105860) was prepared following these guidelines (22).
Eligibility (Inclusion and exclusion) criteria considered in this review
Type of studies
Studies that utilized the following designs were considered for inclusion:
Observational studies: case-control, including case-crossover design or cohort studies.
Interventional studies: Randomized Controlled Trials (RCT).
Reviews, case reports and other studies without a comparison group, cross-sectional studies, reports from conferences or annual meetings, editorials, opinions, in vitro studies and studies of animals were excluded from this review. Studies not written in English were also excluded from this review.
Type of participants
This review focused on adult humans with or without any type of cancer. Studies focusing on children (< 18 years) were excluded.
Type of outcome and exposure measurements
Studies investigating the prevalence or incidence of cancer and its association with the human microbiome were considered for inclusion. Our primary interest was cancer development and not cancer survival/mortality. Studies investigating only pre-cancerous lesions/conditions (no malignant conditions) were excluded.
Studies that target specific microbes (e.g. Helicobacter pylori) instead of studying a microbiome (community of microorganisms) were also excluded. Next-generation sequencing approaches, including 16S ribosomal RNA gene amplicons and genome shotgun metagenomics, are the most common methods for investigating communities of microorganisms. However other methods such as aerobic and anaerobic subculturing onto selective agar media were also considered in this review, provided that communities of microorganisms were investigated. Exposures of interest were relative abundancy of taxa and diversity measures (alpha- & beta diversity) in cancer cases versus controls.
Data collection and analysis
Tailored search strings (see Supplementary material Table S1) were used to search relevant articles up from database conception until December 2019 via the PubMed and EMBASE databases. Also, references in the selected publications were checked for further studies.
Selection of studies
Four investigators (A.L. & Z.V. for first search; I.H. & N.M. for 2nd search) independently reviewed titles and abstracts. In case no consensus was reached, the article was maintained for the next selection phase (e.g. selection by full text article). When the disagreement was still present at full-text selection (n=6), the opinion of the remaining co-authors involved in this review was requested.
Data extraction
Data were extracted by three researchers independently (I.H., N.M. and S.Z.). Considering the important differences in outcome measures reported in the different publications, it was decided to report the significant differences in relative abundancy of taxa and/or diversity measures (alpha- & beta diversity) between cancer cases and controls. If multiple analyses were run with different levels of adjustment, then the one with maximal adjustment for confounders was chosen.
Methodological quality and strength of association
Two reviewers (I.H. & N.M.) independently assessed the methodological quality of the included articles via the Newcastle – Ottawa quality assessment scale (23) (See Supplementary Table S2). Each study was evaluated using three broad criteria: 1) appropriate selection of the study population (up to four stars); 2) comparability of the study groups (up to two stars); and 3) ascertainment of the exposure or outcome of interest (up to three stars). The full score was 9, and a high-quality study in our analysis was defined as a study with at least 7 points. The performance of a meta-analysis to calculate adjusted pooled estimates was ruled out because of the large variation in the definition of outcome variables and exposures. We performed instead a qualitative synthesis of results and reported them in categories according to microbiome and study design. An association was considered as “strong” when three or more publications reported a statistically significant association in the same direction and none were in the opposite direction. The association was deemed “suggestive” when two publications reported a statistically significant association in the same direction and none in the opposite direction. All analyses (e.g. counts of cases and controls and articles) were performed in Excel.
Results
Study design and population characteristics of the included studies
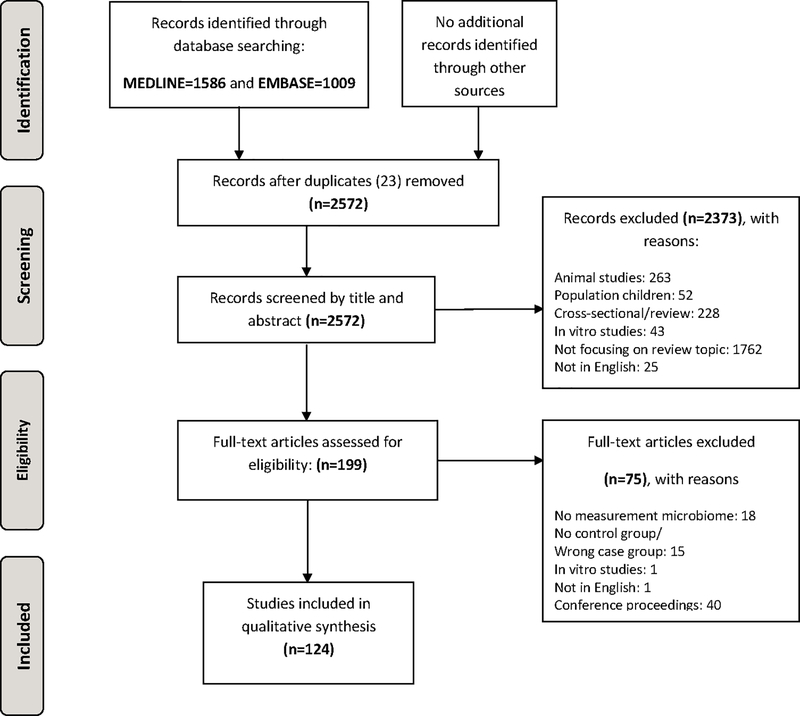
A total of 124 articles (including 15,764 subjects in total; 7,652 cancer cases versus 8,112 controls) were included and used for this review (see Methods and Figure 1: Flowchart). An in depth overview of the different studies and their study design and methods is given in Supplementary table S3. One hundred nineteen studies were case-control studies, including three nested case-control studies (24–26), and five were cohort studies (19,27–30). No RCTs were identified. The gut microbiome was the microbiome most frequently studied (n=50), followed by the oral microbiome (n=16) and then some studies of the bile duct, cervical and intrauterine, oesophagus and gastric, laryngeal, lung, skin, breast, urinary and prostate microbiome (see Table 1).
Figure 1:
PRISMA Flow Diagram
Adapted from: Moher D, Liberati A, Tetzlaff J, Altman DG, The PRISMA Group (2009). Preferred Reporting Items for Systematic Reviews and Meta-Analyses: The PRISMA Statement. PLoS Med 6(7): e1000097. doi:10.1371/journal.pmed1000097.
For more information, visit www.prisma-statement.org.
Table 1.
Number of articles retrieved by microbiome type and the possible links with cancer.
| MICROBIOME | LINKED WITH THE FOLLOWING CANCER(S) | # ARTICLES | # subjects |
||
|---|---|---|---|---|---|
| Total subjects | cases | controls | |||
| Airway and Lung microbiome | Lung cancer | 7 | 309 | 189 | 120 |
| Bile duct microbiome | Cholangiocarcinoma | 2 | 260 | 160 | 100 |
| Breast microbiome | Breast cancer | 2 | 109 | 73 | 36 |
| Cervical microbiome | Cervical cancer | 3 | 289 | 143 | 146 |
| Esophagus microbiome | Esophageal cancer | 1 | 101 | 50 | 51 |
| Gastric microbiome | Gastric cancer | 10 | 999 | 687 | 312 |
| Esophageal cancer | 1 | 91 | 37 | 54 | |
| Gut microbiome | Colorectal cancer | 50 | 5,751 | 2,667 | 3,084 |
| Gastric cancer | 2 | 204 | 116 | 88 | |
| Liver cancer | 1 | 321 | 150 | 171 | |
| Prostate cancer | 1 | 155 | 90 | 65 | |
| Breast cancer | 3 | 325 | 158 | 167 | |
| Thyroid cancer | 1 | 65 | 30 | 35 | |
| Laryngeal microbiome | Laryngeal cancer | 1 | 60 | 29 | 31 |
| Pharyngeal microbiome | Laryngeal cancer | 1 | 96 | 68 | 28 |
| Oral microbiome | Oral cancer | 16 | 1,912 | 724 | 1,188 |
| Head and neck cancer | 4 | 580 | 301 | 279 | |
| Esophageal cancer | 2 | 551 | 193 | 358 | |
| Pancreatic cancer | 5 | 1,729 | 819 | 910 | |
| Gastric cancer | 1 | 51 | 34 | 17 | |
| Colorectal cancer | 3 | 1,019 | 377 | 642 | |
| Colorectal, lung and gastric cancer | 1 | 386 | 286 | 100 | |
| Liver cancer | 1 | 60 | 35 | 25 | |
| Lung cancer | 1 | 66 | 51 | 15 | |
| Ovarian microbiome | Ovarian cancer | 1 | 119 | 99 | 20 |
| Prostate microbiome | Prostate cancer | 1 | 16 | 16 | 0 |
| Skin microbiome | Skin cancer | 1 | 32 | 15 | 17 |
| Urinary microbiome | Bladder cancer | 2 | 78 | 41 | 37 |
| Urinary microbiome | Prostate cancer | 1 | 30 | 14 | 16 |
| All microbiomes | All cancers | 124* | 15,764 | 7,652 | 8,112 |
Thirty-nine of the studies were conducted in North America (n subjects=5,999), five in South America (n subjects=300), fifty-three in Asia (n subjects=6,783), twenty-seven in Europe (n subjects=2,485), two in Africa (n subjects=124) and one in Australia (n=83). The year of publication ranged from 1983 to 2019. The sample size of the studied populations showed large variation. Four articles had a sample size smaller than 10, seventeen articles had a sample size ranging from 10 to 29, twenty-seven articles had a population size from 30 to 49, thirty-two articles included a population size from 50 to 99, and forty-six articles studied over 100 subjects. The largest number of subjects was included for the gut microbiome in relation to colorectal cancer (CRC) risk (n= 5,761), followed by the oral microbiome-oral cancer associations (n= 1,912), and the oral microbiome-pancreatic cancer associations (n= 1,729). All studies included a population with an overall age ranging from 18 to 96 years old. In most of the studies, the cases tended to be slightly older than the healthy controls.
Quality of the studies
The methodological quality assessments are presented in Supplementary table S4. Eighty-eight articles had a good quality score of 7/9 or more of which 51 articles with a total quality score above 8 and two articles reaching the maximum score of 9 stars. For selection, 70 articles received the maximum score, (31), while the most frequent problem was lack of information about the selection of cases and controls and thus their representativeness for the community. For comparability, 82 articles scored the maximum score, while other articles often did not perform any correction for confounders between cases and controls of any kind. This decreases the comparability between the patient group and the healthy controls. For exposure, 20 articles received the maximum score, while low scores were mostly because the studies used another method of ascertainment between cases and controls (e.g. resection for cases while biopsy for controls).
Overall significant findings
As demonstrated in table 1 and supplementary table S4, a large number of samples from different anatomical sites have been investigated in relation to different cancer types. Supplementary table S5 gives an overview of the different bacteria described in these studies. Some of the alterations in the site-specific microbiota were consistent between studies while others were only significant in one study or conflicting between studies (see also overview of strong significant and suggestive findings for each study included in this review in Supplementary Table S4).
In addition, study results were not always comparable due to high heterogeneity in outcome definitions. Many of the articles used different parameters to describe and compare the microbiome including: alpha diversity, beta diversity, relative abundance, absolute abundance, presence/absence of taxa, and some articles clustered microbiota into communities or calculated ratios for the abundance of two microbiota taxonomic groups. Furthermore, the taxonomic level reported differed between articles (mainly the phyla, the families and genera were studied) and some of the articles showed no significant differences in presence and relative abundance of certain phyla while they did show differences at lower taxonomic levels. For example, one study (32) found the same five most abundant phyla within the cases with esophageal cancer or dysplasia and the healthy controls, but different abundances of the orders Clostridiales and Erysipelotrichales. Finally, there are also many different measures and statistical methods used to analyze alpha- and beta- diversity what often limits the comparability of studies reporting these diversity measures.
Due to the large amount of data presented in the different studies included in this review, we have focused on results for which at least two studies reported findings (e.g. diversity measures or taxa) in the same direction in the description below (and visualized in Table 2). A full overview of the results found in the different studies is given in the supplementary Table S4 and the study design of the included studies is presented in supplementary Table S3.
Table 2:
Summary table presenting associations between microbiome composition and colorectal (number of studies=50) and oral cancer risk (n=16)
| Taxa & diversity | Colorectal cancer | N (%) studies for that finding* | Oral cancer | N (%) studies for that finding* | |
|---|---|---|---|---|---|
| PHYLUM | Bacteroidetes | S & T | 3 (19) | ||
| Firmicutes | S | 2 (13) | |||
| Fusobacteria | F & T | 10 (20) | S & T | 2 (13) | |
| FAMILY | Fusobacteriaceae | F & T | 2 (4) | ||
| Lachnospiraceae | F & T | 6 (12) | |||
| Peptostreptococcaceae | F | 2 (4) | |||
| Porphyromonadaceae | F | 2 (4) | |||
| Ruminococcaceae | F &T | 5 (10) | |||
| GENUS | Actinomyces | S | 2 (13) | ||
| Acinetobacter | F | 3 (6) | |||
| Atopobium | F & T | 4 (8) | |||
| Bifidobacterium | F & T | 4 (8) | |||
| Blautia | F & T | 2 (4) | |||
| Capnocytophaga | S & T | 2 (13) | |||
| Desulfovibrio | F | 2 (4) | |||
| Dialister | S & T | 2 (13) | |||
| Fusobacterium | F & T | 16 (32) | S & T | 4 (25) | |
| Gemella | F & T | 3 (6) | |||
| Leptotrichia | F & T & S | 3 (6) | |||
| Odoribacter | F | 2 (4) | |||
| Oscillibacter | F | 2 (4) | |||
| Parvimonas | F | 6 (12) | S & T | 2 (13) | |
| Peptostreptococcus | F & T | 5 (10) | S & T | 2 (13) | |
| Porphyromonas | F & T | 8 (16) | |||
| Prevotella | F | 2 (4) | |||
| Staphylococcus | F | 2 (7) | |||
| Streptococcus | S | 4 (25) | |||
| SPECIES | Bacteroides fragilis | F & T | 4 (8) | ||
| Blautia coccoides | F | 2 (4) | |||
| Escherichia coli | F & T | 2 (4) | |||
| Fusobacterium nucleatum | F & T | 11 (22) | |||
| Parvimonas micra | F | 4 (8) | |||
| Peptostreptococcus stomatis | F | 3 (6) | |||
| Porphyromonas assaccharolytica | F | 2 (4) | |||
| Prevotella intermedia | F & S | 2 (4) | |||
| DIVERSITY | Beta diversity | Sign. in 7 studies | 7 (14) | ||
|
Strong positive association (≥3 publications in same direction; none in opposite direction) |
|||||
|
Suggestive positive association (2 publications in same direction; none in opposite direction) |
|||||
|
Strong negative association (≥3 publications in same direction; none in opposite direction) |
|||||
|
Suggestive negative association (2 publications in same direction; none in opposite direction) |
|||||
Abbreviations: F= faecal samples, S= saliva samples, T= tissue samples (cancer site specific)
Example n (%): 10 and 2 studies for Fusobacteria for CRC & oral cancer respectively, so 20% and 13% of the total number of studies on CRC & oral cancer
Cholangiocarcinoma has been investigated in relation to the bile duct microbiome in two articles (33,34), although one article only focused on the extrahepatic variant. Both studies reported microbiome differences between cases and controls, though not for the same operational taxonomic units (OTU) and a clear separation between cases and controls when analyzing beta diversity.
Two studies investigated the breast tissue microbiome in relation to invasive breast cancer (using benign breast disease as controls), reporting increased relative abundance of certain genera, though these differed between the two studies. Increased abundance was reported in the following low-abundant family and genera in the breast tissue of women with invasive breast cancer: Unclassified Bacteroidetes, Comomonadaceae Enterobacteriaceae, Bacillus and Staphylococcus (35), Fusobacterium, Atopobium, Hydrogenophaga, Gluconacetobacter and Lactobacillus (unadjusted P < 0.05) (36).In both studies, microbial diversity was not considered.
Three studies investigated associations between the cervical microbiome and cervical cancer risk. Conflicting results were found for Lactobacillus iners, however, all studies reported a decrease in abundance of the beneficial Lactobacillus crispatus in cervical cancer cases (37) (38). All three studies also suggested higher alpha-diversity (Shannon index and/or OTU number) among cervical cancer cases compared to controls (37,39) (38).
One study investigated esophageal cancer risk in relation to the esophageal microbiome (40) while another study investigated its association with the gastric microbiome (32). No significant differences were found between the esophageal microbiome of the patients with esophageal cancer and the healthy controls with no evidence of esophageal disease (40). Differences were found in the gastric microbiome composition between controls and cases with esophageal cancer on the order, family, and genus level, though the abundances were different and not always in the same direction in the two studies (32,41). Microbial diversity was considered in one study, in which the number of bacteria between cases and controls was not significant (40).
Ten studies investigated associations between the gastric microbiome and gastric cancer risk (42–51). The gastric microbiome of the cases with gastric cancer showed a significant difference compared to control subjects and in the cancer tissues compared to the adjacent normal tissue in the same cancer patient. Three studies showed an increased abundancy of Fusobacteria among gastric cancer patients or cancer tissue samples (44,47,52) and two studies an increased abundancy of Neisseria among gastric cancer patients when compared to control subjects (46,47). In two of these studies, the analysis of microbial diversity was included, showing a trend to diminish going from non-atrophic gastritis to intestinal type of gastric cancer (42,47).
Seven studies evaluated associations between the airway-lung microbiome and lung cancer risk, using different methodologies (e.g. sputum samples (53,54) versus exhaled breath condensate and bronchial brushing (29,30,55–57). All studies demonstrated clear differences between the microbiome samples of lung cancer patients in comparison with healthy controls, though different taxa were studied (Carpagnano only focused on Aspergillus species). Streptococcus were increased among lung cancer patients in three studies (30,54,56).
Thirty-four studies investigated associations between the oral microbiome and several cancers, of which sixteen considered the association with oral cancer (27,31,58–71) and five with pancreatic cancer (24,72–74). The sixteen studies considering the associations between the oral microbiome and oral cancer risk obtained inconsistent results. Consistent results have been visually presented as strong or suggestive positive/negative associations in Table 2. From the 19 studies that reported results on the phylum Bacteroidetes, three studies confirmed a significant positive association between Bacteroidetes abundance and oral cancer risk. Similarly, four studies confirmed a positive association between the abundance of the genus Fusobacterium and oral cancer risk. Several studies also observed an altered abundance of Streptococcus in oral cancer patients versus controls (27,31,58–60,67,69) with four studies confirming a significant negative association between Streptococcus abundance and oral cancer risk. Furthermore, six studies reported an altered abundance of Prevotella, but results differed in the direction of the association. Two of the five studies investigating associations between the oral microbiome and pancreatic cancer risk reported consistently a lower abundance of Neisseria in the oral microbiome of pancreatic cancer cases compared to controls (72,73). Two studies found a lower abundance of Neisseria and Haemophilus when investigating the tongue coating samples of patients with colorectal, lung and gastric cancer compared to controls (75,76). Among the thirty-four studies analysing the oral microbiome, thirteen studies presented results on the microbial diversity and most of them showed a significant lower bacterial diversity than the healthy controls.
Two studies investigated associations between the urinary microbiome and bladder cancer risk (77,78). Both studies reported a decreased abundancy of Streptococcus and Bucevic reported also increased abundancy of Fusobacterium among bladder cancer patients. Bi et al. also reported a higher alpha diversity among bladder cancer patients.
Fifty studies linked differences in the gut microbiome with CRC, three with breast cancer risk (79–81) and two with gastric cancer risk (82,83). Thirty-three articles used fecal samples as a proxy for the gut microbiome, while twenty-two articles used mucosal biopsies. Three articles investigated both feces and mucosal biopsies (84–86). To compare within consistent specimen types, use of fecal samples or mucosal samples is clearly specified in the description of the results regarding associations between the gut microbiome and CRC risk here below. A summary of the strong and suggestive associations reported for the gut microbiome in relation to CRC risk has been visualized in Table 2.
The phylum Fusobacteria was more often detected in CRC tumor biopsies compared to adjacent normal tissue (25,87–89). Seven studies also found significantly higher levels of the genus Fusobacterium in mucosal samples of CRC cases in comparison with controls (25,84,86,88–92). In five studies, the species Fusobacterium nucleatum was enriched in mucosal samples in CRC cases (85,87,93–95). Thirteen studies also detected a significant enrichment of the genus Fusobacterium in fecal samples of CRC patients (19,52,96–106).
Eight studies found that the genus Porphyromonas was significantly enriched in fecal samples of CRC patients compared with controls (96,99–102,104–107).
Two studies showed that Lactobacillus was significantly more present in fecal samples from CRC cases compared to controls, while the studies of Mira-Pascual et al. and Ohigashi et al. found non-significantly higher abundance of Lactobacillus in controls (85,108–110).
The genus Prevotella was more commonly detected in fecal samples of CRC cases compared to controls in the study of Amiot et al. and two other studies reported the same trend (85,108,109). Peptostreptococcus had significantly higher abundance in fecal samples from CRC cases compared to controls in five studies (52,84,100,101,104,110). Two studies also found a significant increase of Peptostreptococcus in CRC tissue compared to healthy controls (84,88).
Six studies demonstrated an increased abundance of Parvimonas in CRC cases (52,95,97,100,101,105) and three studies a significantly higher presence of Gemella and Leptotrichia in fecal samples of CRC cases compared to controls (25,84,91,100,101).
In the Spanish cohort of Allali et al. Blautia was significantly more abundant in adjacent normal tissue compared to CRC tumor biopsies and Chen et al. also observed higher Blautia levels in healthy controls compared to CRC cases (25,84).
At the family level, Ruminococceae and Lachnospiraceae were significantly more abundant in controls and/or adjacent/’off-tumor’ mucosae compared with CRC tumor samples in four and six studies respectively (84,90,91,95,104,106,111,112).
In summary, the most consistent findings for the gut microbiome were for Fusobacterium (most important species Fusobacterium nucleatum), Parvimonas, Porphyromonas and Peptostreptococcus being significantly enriched in fecal and mucosal samples from CRC patients compared to controls or adjacent normal tissue.
Among the fifty studies investing associations between cancer risk and gut microbiome composition, results on microbial diversity were often reported. While findings on alpha diversity were inconsistent (2,25,52,84,85,87,89,90,92,96,97,100,101,108,110,113,114), five studies on CRC cancer found significant results for beta diversity (84–86,98,100).
Three articles had investigated associations between the gut microbiome and breast cancer risk using a case-control design. All three studies reported microbiome differences between breast cancer cases and controls, though not for the same operational taxonomic units (OTU). The Shannon index was significantly lower in breast cancer cases and the beta-diversity was significantly different between cases and controls (79,80).
Discussion
In total, 124 studies have been included in this systematic review to evaluate the relation between the human microbiome and cancer. The gut microbiome was the microbiome most frequently studied, followed by the oral microbiome and then studies of the bile duct, cervical and intrauterine, oesophagus and gastric, laryngeal, lung, skin, breast, urinary and prostate microbiome. The quality assessment demonstrated rather low quality for the “ascertainment of the exposure of interest” because studies often used different methods of ascertainment for cases and controls e.g. resection for cases while biopsy for controls. The large diversity of parameters used to describe the microbial composition made it impossible to harmonize the different studies in a way that would allow meta-analysis. All articles, apart from one, showed specific differences in microbiome distribution between cases and controls. Some findings were consistent between studies while others were conflicting or only reported in one single study
Concerning the esophageal microbiome, two studies investigated different populations with other sample types, and reported different results, though both found positive associations between Corynebacterium and Peptococcus in the mouth and esophageal cancer risk. Bacteria from the Corynebacterium and Peptococcus genera represent an important component of skin and mucosal membranes, being part of the oral and upper respiratory tract microbiome and exist through commensal interactions with their hosts (115,116). However, some species from the Corynebacterium genus can act as opportunistic pathogens and secrete exotoxins associated with a number of diseases such as diphtheria, pleuropneumonia and sepsis (115). Similarly, Peptococcus, a Gram-positive anaerobic coccus, has been observed to be enriched in different infections such as periodontitis and in wounds such as chronic ulcers (116). Immunocompromised cancer patients might be privileged targets for these opportunistic pathogens. More research and standardized protocols for sample collection are needed to clarify this potential link between the esophageal microbiome and cancer risk.
Many bacteria can be found in the human oral cavity and these are not only associated with oral cancers but also with cancers of the lung, the esophagus, the stomach, the pancreas and the colorectum. While the oral microbiome has been suggested to play a crucial role in carcinogenesis, the oral microbial community is complex and may be impacted by many other environmental and genetic factors and potentially due to this complexity, published research has often shown inconsistent results. However, some potential pathways have been suggested for the putative role between the oral microbiome and cancer risk. For instance, the observed decrease of Streptococcus in pre-cancer and oral cancer cases might reflect early changes of the oral mucosa surface related to tumorigenesis (31,58). Indeed, Streptococci might lose their ability to adhere to mucosa undergoing tumorigenesis while other species, such as Fusobacterium, which are often increased in oral cancer cases (58) might adhere better. Fusobacterium nucleatum is reported to activate the nuclear translocation of NF-κB leading to a pro-inflammatory environment. However, this translocation is inhibited by Streptococcus thus attenuating the pro-inflammatory responses induced by Fusobacterium. This suggests that a decrease of Streptococcus combined with an increase of Fusobacterium might play a role in oral cancers (117). In the gut, F. nucleatum is associated with CRC, potentially because it might increase reactive oxygen species (ROS) and IL-10 production leading to an inhibition of the T-cells and the antitumor immunity (118). Furthermore, some Streptococci can inhibit the oral colonization of certain bacteria such as Aggregatibacter actinomycetemcomitans, Prevotella intermedia and Porphyromonas gingivalis on the oral epithelial surfaces (117). This highlights the importance of interactions between the bacteria themselves and the human mucosa (117).
The gut microbiome was analyzed in both fecal and colorectal mucosal samples. In general, fecal samples had a higher microbial diversity when compared with tissue-associated microbiomes. The results included in this review clearly demonstrate that gut microbiota composition differ between CRC patients and non-cancer patients. The most consistent findings were for Fusobacterium (with most important species Fusobacterium nucleatum), Parvimonas, Porphyromonas and Peptostreptococcus, which were significantly enriched in both fecal and mucosal samples of CRC patients compared to controls across multiple studies. Several of our reviewed articles and a recent meta-analysis (119) reported that Fusobacterium nucleatum -already known as an invasive and pro-inflammatory agent that can cause acute and chronic oral and gastrointestinal infections- was significantly enriched in both fecal and biopsy samples of CRC patients. With its unique FadA adhesin, Fusobacterium nucleatum adheres to, invades and induces inflammatory and oncogenic responses to stimulate growth of CRC cells. (120). Another study suggested that Fusobacterium nucleatum induces nuclear factor-κB (NF-κB) to promote CRC (98). Alternatively, Fusobacterium may act as a “passenger” that multiplies in the more favorable conditions caused by the malignant tumor rather than itself being a causal factor in CRC development (103).
Species of Parvimonas were consistently reported with elevated abundance in CRC tumor biopsies. Parvimonas micra, the only species described at the genus level, is known to cause bacteremia, abdominal abscesses, endocarditis, and other infections (95). Its tumor promoting effect has been suggested to be associated with altered immune responses and enhanced inflammation in the gut (121).
Previously, it has been shown in malignant and primary human oral epithelial cells that Porphyromonas gingivalis and its membrane fraction induces up-regulation of a number of genes involved in inflammation and cell proliferation and control (122), however specific data for CRC are lacking.
A possible explanation for the higher abundance of Peptostreptococcus species in CRC patients could be due specifically to the presence of Peptostreptococcus anaerobius. P. anaerobius has been shown to promote colon dysplasia in a mouse model, specifically interacting with TLR2 and TLR4 on colon cells to increase levels of reactive oxidative species, which promotes cholesterol synthesis and cell proliferation (123).
Overall, many other differences in the abundance of certain bacteria were found between cancer cases and controls, though often not consistent between studies. Differences in results could partly be due to variation in study design and methods used such as tissue sampling (different method or different site), techniques used to characterize the microbiome, or different study populations (e.g. genetic and environmental differences including nutritional intakes and habits, oral hygiene, air pollution etc.), exclusion criteria, or another unrecognized factors. Furthermore, the microbiome may differ depending on tumor stage which could also be a cause of inconsistent results if stage was not considered in the analysis.
Barriers and challenges in microbiome studies
The number of epidemiologic studies that investigate the association between the human microbiome with cancer risk is rising; however, it is challenging to obtain definitive and high quality evidence. First, it remains difficult to establish the causality of the cancer-microbe associations (2,16). When a bacterium is enriched at a tumor site, there are other possible reasons for that association besides a causal relation. For example, the microbe can take advantage of the tumor’s oxygen tension or carbon sources, or find an underused nutritional niche where they can grow freely (16). It is not clear whether the carcinogenic process changes the local environment and creates new niches for microbes, or if an alteration of the microbial composition and its function contribute to carcinogenesis. As such, reverse causation is an important concern (2). Given that many of the previous studies use a case-control design, prospective studies are urgently needed to help understand the temporal nature of the association between the microbiome and cancer development.
Another challenge is that different microbes might contribute to different stages of carcinogenesis (6,16,17). In the initiation stage, the microbiome may promote specific genetic mutations and chronic inflammation, but may also be involved in creating other tumor-promoting environments such as the development of obesity and the metabolic syndrome (6). After the initiation phase, other microbiota could be responsible for tumor growth, angiogenesis and metastasis (6,16). Since cancer development is a process that continues over many years, it is possible that by the time the cancer is diagnosed, the initial causal microbes are no longer present due to the later-stage tumor environment (16,17).
It is also unlikely that many of these cancers are due to an undetected ‘one microbe-one disease’ association (17). It is well established that H. pylori infection causes gastric cancer and there are nine other microorganisms that are designated by the International Agency for Research on Cancer to be carcinogenic to humans (16,17). However, some microbes might have modest and subtle contributions to cancer development, which will be harder to detect. It is also likely that some of those modest contributions depend on the genetic background of the host, sex and age; as well as lifestyle and other environmental exposures (2,17).
In addition, the microbiome composition at one specific anatomical site may be related with cancer development localized elsewhere (e.g. relationship between oral microbiome and pancreatic cancer risk (24,72,73)), making it complex to investigate causal relationships. Indeed, symbiotic bacteria, especially in the gut, are able to influence host metabolism through producing and modifying a plethora of biologically active compounds, including hormones, carbohydrates, and lipids that reach the systemic circulation. For instance, high concentrations of secondary bile acids, derived from host-produced primary bile acids, were associated with increased risk for colorectal, pancreatic, and liver cancer (124–126). Another example is the important role of indole derivatives, produced by gut bacteria from the degradation of tryptophan, such as aryl hydrocarbon receptor ligands, to maintain intestinal homeostasis, limit inflammation and thus, indirectly reduce the risk of colorectal, prostate and breast cancer (127–129). Lastly, short chain fatty acids are produced by the gut microbiome through fermentation of non-digestible fiber and starch and help maintain balance between intestinal immunity and inflammation (130,131). As such microbiota may influence the risk of many cancers, independent of anatomical location due to effects of microbial metabolites on inflammation, an important hallmark of cancer, and other metabolic conditions associated with cancer risk.
Finally, the study design and methods used may limit the validity, generalizability and comparability of results. Since many exogenous and intrinsic host factors influence the human microbiome (>125 factors collectively explain 18.7% of the variation in human gut microbiome composition) (132), a large sample size and an important battery of information on host factors are needed to investigate potential causal relationships between the human microbiome and cancer risk with sufficient statistical power. Further, restrained physical access to certain body sites limits the possibilities for large-scale sample collections. For example, to avoid invasive procedures such as biopsy, other specimen available for studies of prostate cells such as expressed prostate secretions or urine samples containing prostatic cells are often used (133,134). In addition, different sample handling/storage strategies (e.g. room temperature storage versus immediate freezing, different collection media, etc.) (135,136), laboratory methods, sterilization procedures, sequencing strategies (e.g. 16S rRNA gene or shotgun sequencing), and various processing pipelines for the raw sequencing data, can all impact the comparability of different studies (137). Indeed, the techniques used to characterize the microbiome composition may also importantly impact the quality of the results. While bacteriological cultures (in 8 of the 124 included studies) only allow the analysis of certain specific communities of microorganisms as (e.g. <30% of gut microbiota have been cultured to date), most studies were based on sequence analysis of the 16S rRNA gene, which is sufficiently divergent to provide a phylogenetic signal and identification of unknown bacteria. More recent culture-independent techniques such as next-generation sequencing have the advantages that they are high throughput, phylogenetically characterize the microbiota components, and quantify the relative proportions of organisms present (138). In addition, for determining alpha- and beta- diversity, there are many different measures and statistical methods that have been used that may further limit comparability between studies. Gail and colleagues are currently working on a method to have a reference based distance for beta-diversity in order to be able to combine beta-diversity analyses across studies in future (139).
Multi-omics analysis (140) could potentially assist in disentangling complex relationships between the human microbiome and cancer risk by integrating the microbiome data with other omics data of interest for investigating causal factors of cancer development. However, microbiome multi-omics data analysis remains challenging as few tools exist to integrate these data. Current statistical approaches, such as correlation tests between microbiome taxa and specific host metabolites, do not necessarily meet the assumptions of the sparse compositional nature of microbiome data (141,142).
Limitations of our review
A limitation of our review is that tumor stage and grade were not considered due to limited evidence in the studies included in this systematic review. Different stages of the cancer might harbor different microbiomes and this could be a cause of non-consistent results across studies. Tumor severity has been shown to have an impact on microbiome composition (85). On the other hand, another study reported that the gut microbiome did not change as the cancer progressed and that any change might have occurred in the early stage of carcinogenesis (108). Based on these opposing findings, it would be important to further investigate whether tumor stage has an impact on the composition of the microbiome.
Another limitation is that the location of the CRC tumors was not considered although differences in microbiome composition between distal and rectal versus proximal cancers have been shown (86). In addition, microbiome composition in the colorectum can also be influenced by bowel cleansing which is not considered in every study and which was not part of our exclusion criteria.
The microbiome of patients from different countries was also compared with one another (see supplementary Tables S3–S4). The study of Allali et al. showed significantly higher phylogenetic diversity in tumor and adjacent tissues from patients of the US compared with a Spanish cohort (25). So it is not certain whether gut micro-organisms linked to cancer are the same across different countries and populations.
As mentioned above, we compared studies that made use of different techniques to characterize the gut microbiome. These different techniques may lead to different findings. Every study has its own exclusion criteria and although many studies matched their cases and controls for age, sex, body mass index (BMI) and other factors there are some differences in potential confounding variables across reviewed articles which may have impact on our findings (see Supplementary Tables S3–4). Specific study design issues should be considered when conducting longitudinal studies for microbiome research (143). Biological sample collection, processing (DNA extraction, amplification, sequencing, and bioinformatics) and storage as well as quality control standards still require improvement. At the same time, guidelines need to be elaborated to report results and share data to ensure replication and reproducibility of these studies (143).
Finally, a limitation of this review is the lack of quantitative results from meta-analysis due to substantial heterogeneity in the microbiome parameters assessed.
Clinical implications and future perspectives
This review has shown some specific micro-organisms appear to have a different prevalence in cancer cases than in controls. Tumor stage was not considered so it is not clear if these specific micro-organisms are present from the initiation of carcinogenesis. Therefore, more research, in particular via prospective studies, is needed to investigate if the microbiome could be a potential tool for cancer early detection and/or prevention strategies. Few studies already indicated that the gut microbiome, with its specific micro-organisms could potentially be used as an effective tool in early screening for CRC (113).
Knowledge on micro-organisms linked to cancer may also play an important role in cancer therapy. There is limited evidence suggesting that dysbiosis of the gut microbiome can be improved using probiotic and prebiotic supplementations in children with cancer (144). There is limited evidence for cancer prevention, but it is used for many other health outcomes such as improved digestion (17).
More understanding of the specific roles of the microbiome is required before promoting the microbiome as an adjuvant therapy that enhances efficacy or attenuates the toxicity of chemotherapies (6,16). Herein, stronger evidence derived from studies with more optimal study design and methods and from different geographic regions is needed. To detect strong associations between microbiome profiles and various types of cancer, adequate power could be reached by pooling or meta-analysing data across multiple studies. However, heterogeneity of data hinders meta-analysis. Therefore, standardisation across studies is needed. Future prospective studies should coordinate and collect fecal samples using at least one common method in addition to the method of their choice. Recent studies have evaluated multiple fecal and oral sample collection methods and a variety of methods appear to be reproducible, stable, and accurate compared to an immediately frozen fecal sample. (145–147). DNA extraction and sample handling were also reported to contribute to variability in microbiome data (135,136), it is therefore important for future studies to use standardised methods including negative and positive controls at all stages of sample processing and, when possible, use automation when processing samples. In order to have homogeneity of data, data analysis methods need to be transparent, consistently reported and reproducible, which still remains a major challenge in current microbiome reports. Microbiome bioinformatics platforms such as Quantitative Insights Into Microbial Ecology 2 (QIIME 2) (148,149) improve reproducibility of bioinformatics analysis by providing automated, decentralized data provenance tracking helps. Another bioinformatics challenge for future prospective studies is the low access to long-term archives of raw microbiome sequence data with sample and study metadata in common formats (150). Finally, future study designs should focus on the assessment of potential confounding factors by collecting high-quality clinical and nutritional data that are known to influence the microbiome.
Conclusion
Although strong evidence was available for certain taxa of the gut microbiome and CRC risk, and for the oral microbiome and oral cancer risk, for most of the microbiome taxa/indicators the evidence was still too weak to draw firm conclusions in relation to their role in cancer. In addition, based on the state of the field, it is currently not possible to have an overall ascertainment of cancer risk due to the microbiome since the data generated is heterogeneous and merging studies for meta-analysis is extremely difficult. Therefore, future prospective studies with pre-diagnostic specimen collection using standardized methods, consistent laboratory methodology and bioinformatics, and inclusion of quality control samples are required to establish causal links. Also “integromics” analysis (integrating microbiome data with other –omics data e.g. metabolomics, epigenomics) may further assist in disentangling these complex relationships between the human microbiome and cancer risk although these will require significantly increased sample sizes and likely pooling across multiple cohorts.
Supplementary Material
Acknowledgments
Funding
Emily Vogtmann was supported by the Intramural Research Program of the National Cancer Institute at the National Institutes of Health.
Nathalie Michels is financially supported by Research Foundation—Flanders 12H1519N (FWO3EO2015004301).
List of abbreviations
- CRC
colorectal cancer
- PRISMA
Preferred Reporting Items for Systematic reviews and Meta-Analyses
- RCT
Randomized Controlled Trials
Footnotes
Conflict of interest statement: The authors declare no potential conflicts of interest.
Declarations:
Availability of data and material
All data generated or analysed during this study are included in this published article [and its supplementary information files].
IARC disclaimer:
Where authors are identified as personnel of the International Agency for Research on Cancer / World Health Organization, the authors alone are responsible for the views expressed in this article and they do not necessarily represent the decisions, policy or views of the International Agency for Research on Cancer / World Health Organization.
References
- 1.Marchesi JR, Ravel J. The vocabulary of microbiome research: a proposal. Microbiome 2015;3:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vogtmann E, Goedert JJ. Epidemiologic studies of the human microbiome and cancer. British journal of cancer 2016;114:237–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Young VB. The role of the microbiome in human health and disease: an introduction for clinicians. BMJ 2017;356:j831. [DOI] [PubMed] [Google Scholar]
- 4.Blum HE. The human microbiome. Adv Med Sci 2017;62:414–20 [DOI] [PubMed] [Google Scholar]
- 5.Gevers D, Knight R, Petrosino JF, Huang K, McGuire AL, Birren BW, et al. The Human Microbiome Project: a community resource for the healthy human microbiome. PLoS biology 2012;10:e1001377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dzutsev A, Goldszmid RS, Viaud S, Zitvogel L, Trinchieri G. The role of the microbiota in inflammation, carcinogenesis, and cancer therapy. European journal of immunology 2015;45:17–31 [DOI] [PubMed] [Google Scholar]
- 7.Daliri EB, Tango CN, Lee BH, Oh DH. Human microbiome restoration and safety. Int J Med Microbiol 2018;308:487–97 [DOI] [PubMed] [Google Scholar]
- 8.Karkman A, Lehtimaki J, Ruokolainen L. The ecology of human microbiota: dynamics and diversity in health and disease. Ann N Y Acad Sci 2017;1399:78–92 [DOI] [PubMed] [Google Scholar]
- 9.Proal AD, Lindseth IA, Marshall TG. Microbe-microbe and host-microbe interactions drive microbiome dysbiosis and inflammatory processes. Discovery medicine 2017;23:51–60 [PubMed] [Google Scholar]
- 10.Francescone R, Hou V, Grivennikov SI. Microbiome, inflammation, and cancer. Cancer journal (Sudbury, Mass) 2014;20:181–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fouad YA, Aanei C. Revisiting the hallmarks of cancer. American journal of cancer research 2017;7:1016–36 [PMC free article] [PubMed] [Google Scholar]
- 12.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell 2000;100:57–70 [DOI] [PubMed] [Google Scholar]
- 13.Karlsson FH, Tremaroli V, Nookaew I, Bergstrom G, Behre CJ, Fagerberg B, et al. Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature 2013;498:99–103 [DOI] [PubMed] [Google Scholar]
- 14.Pedersen HK, Gudmundsdottir V, Nielsen HB, Hyotylainen T, Nielsen T, Jensen BA, et al. Human gut microbes impact host serum metabolome and insulin sensitivity. Nature 2016;535:376–81 [DOI] [PubMed] [Google Scholar]
- 15.Aron-Wisnewsky J, Prifti E, Belda E, Ichou F, Kayser BD, Dao MC, et al. Major microbiota dysbiosis in severe obesity: fate after bariatric surgery. Gut 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garrett WS. Cancer and the microbiota. Science (New York, NY) 2015;348:80–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bultman SJ. The microbiome and its potential as a cancer preventive intervention. Seminars in oncology 2016;43:97–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wirbel J, Pyl PT, Kartal E, Zych K, Kashani A, Milanese A, et al. Meta-analysis of fecal metagenomes reveals global microbial signatures that are specific for colorectal cancer. Nature medicine 2019;25:679–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yachida S, Mizutani S, Shiroma H, Shiba S, Nakajima T, Sakamoto T, et al. Metagenomic and metabolomic analyses reveal distinct stage-specific phenotypes of the gut microbiota in colorectal cancer. Nature medicine 2019;25:968–76 [DOI] [PubMed] [Google Scholar]
- 20.Weng MT, Chiu YT, Wei PY, Chiang CW, Fang HL, Wei SC. Microbiota and gastrointestinal cancer. Journal of the Formosan Medical Association = Taiwan yi zhi 2019;118 Suppl 1:S32–s41 [DOI] [PubMed] [Google Scholar]
- 21.Stroup DF, Berlin JA, Morton SC, Olkin I, Williamson GD, Rennie D, et al. Meta-analysis of observational studies in epidemiology: a proposal for reporting. Meta-analysis Of Observational Studies in Epidemiology (MOOSE) group. Jama 2000;283:2008–12 [DOI] [PubMed] [Google Scholar]
- 22.Liberati A, Altman DG, Tetzlaff J, Mulrow C, Gøtzsche PC, Ioannidis JPA, et al. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate healthcare interventions: explanation and elaboration. BMJ 2009;339:b2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.The Newcastle - Ottawa quality assessment scale (http://www.ohri.ca/programs/clinical_epidemiology/oxford.asp). Accessed on the fifth of September in 2017.
- 24.Fan X, Alekseyenko AV, Wu J, Peters BA, Jacobs EJ, Gapstur SM, et al. Human oral microbiome and prospective risk for pancreatic cancer: a population-based nested case-control study. Gut 2018;67:120–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Allali I, Delgado S, Marron PI, Astudillo A, Yeh JJ, Ghazal H, et al. Gut microbiome compositional and functional differences between tumor and non-tumor adjacent tissues from cohorts from the US and Spain. Gut microbes 2015;6:161–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hayes RB, Ahn J, Fan X, Peters BA, Ma Y, Yang L, et al. Association of Oral Microbiome With Risk for Incident Head and Neck Squamous Cell Cancer. JAMA oncology 2018;4:358–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guerrero-Preston R, Godoy-Vitorino F, Jedlicka A, Rodríguez-Hilario A, González H, Bondy J, et al. 16S rRNA amplicon sequencing identifies microbiota associated with oral cancer, Human Papilloma Virus infection and surgical treatment. Oncotarget 2016;7:51320–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alanee S, El-Zawahry A, Dynda D, Dabaja A, McVary K, Karr M, et al. A prospective study to examine the association of the urinary and fecal microbiota with prostate cancer diagnosis after transrectal biopsy of the prostate using 16sRNA gene analysis. The Prostate 2019;79:81–7 [DOI] [PubMed] [Google Scholar]
- 29.Lee SH, Sung JY, Yong D, Chun J, Kim SY, Song JH, et al. Characterization of microbiome in bronchoalveolar lavage fluid of patients with lung cancer comparing with benign mass like lesions. Lung cancer (Amsterdam, Netherlands) 2016;102:89–95 [DOI] [PubMed] [Google Scholar]
- 30.Liu HX, Tao LL, Zhang J, Zhu YG, Zheng Y, Liu D, et al. Difference of lower airway microbiome in bilateral protected specimen brush between lung cancer patients with unilateral lobar masses and control subjects. International journal of cancer 2018;142:769–78 [DOI] [PubMed] [Google Scholar]
- 31.Henrich B, Rumming M, Sczyrba A, Velleuer E, Dietrich R, Gerlach W, et al. Mycoplasma salivarium as a dominant coloniser of Fanconi anaemia associated oral carcinoma. PloS one 2014;9:e92297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nasrollahzadeh D, Malekzadeh R, Ploner A, Shakeri R, Sotoudeh M, Fahimi S, et al. Variations of gastric corpus microbiota are associated with early esophageal squamous cell carcinoma and squamous dysplasia. Scientific reports 2015;5:8820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chng KR, Chan SH, Ng AHQ, Li C, Jusakul A, Bertrand D, et al. Tissue Microbiome Profiling Identifies an Enrichment of Specific Enteric Bacteria in Opisthorchis viverrini Associated Cholangiocarcinoma. EBioMedicine 2016;8:195–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aviles-Jimenez F, Guitron A, Segura-Lopez F, Mendez-Tenorio A, Iwai S, Hernandez-Guerrero A, et al. Microbiota studies in the bile duct strongly suggest a role for Helicobacter pylori in extrahepatic cholangiocarcinoma. Clinical microbiology and infection : the official publication of the European Society of Clinical Microbiology and Infectious Diseases 2016;22:178 e11–22 [DOI] [PubMed] [Google Scholar]
- 35.Urbaniak C, Gloor GB, Brackstone M, Scott L, Tangney M, Reid G. The Microbiota of Breast Tissue and Its Association with Breast Cancer. Applied and environmental microbiology 2016;82:5039–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hieken TJ, Chen J, Hoskin TL, Walther-Antonio M, Johnson S, Ramaker S, et al. The Microbiome of Aseptically Collected Human Breast Tissue in Benign and Malignant Disease. Scientific reports 2016;6:30751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Audirac-Chalifour A, Torres-Poveda K, Bahena-Román M, Téllez-Sosa J, Martínez-Barnetche J, Cortina-Ceballos B, et al. Cervical microbiome and cytokine profile at various stages of cervical cancer: A pilot study . PloS one 2016;11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oh HY, Kim BS, Seo SS, Kong JS, Lee JK, Park SY, et al. The association of uterine cervical microbiota with an increased risk for cervical intraepithelial neoplasia in Korea. Clinical microbiology and infection : the official publication of the European Society of Clinical Microbiology and Infectious Diseases 2015;21:674 e1–9 [DOI] [PubMed] [Google Scholar]
- 39.Fang RL, Chen LX, Shu WS, Yao SZ, Wang SW, Chen YQ. Barcoded sequencing reveals diverse intrauterine microbiomes in patients suffering with endometrial polyps. American Journal of Translational Research 2016;8:1581–92 [PMC free article] [PubMed] [Google Scholar]
- 40.Mannell A, Plant M, Frolich J. The microflora of the oesophagus. Ann R Coll Surg Engl 1983;65:152–4 [PMC free article] [PubMed] [Google Scholar]
- 41.Half EE, Amir I, Naftali T, Gophna U, Konikoff FM. Gastric microbiota is altered in esophagitis and barrett’s esophagus and further modified by proton pump inhibitors. Gastroenterology 2012;142:S442. [DOI] [PubMed] [Google Scholar]
- 42.Aviles-Jimenez F, Vazquez-Jimenez F, Medrano-Guzman R, Mantilla A, Torres J. Stomach microbiota composition varies between patients with non-atrophic gastritis and patients with intestinal type of gastric cancer. Scientific reports 2014;4:4202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Seo I, Jha BK, Suh SI, Suh MH, Baek WK. Microbial profile of the stomach: Comparison between normal mucosa and cancer tissue in the same patient. Journal of Bacteriology and Virology 2014;44:162–9 [Google Scholar]
- 44.Castano-Rodriguez N, Goh KL, Fock KM, Mitchell HM, Kaakoush NO. Dysbiosis of the microbiome in gastric carcinogenesis. Scientific reports 2017;7:15957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Coker OO, Dai Z, Nie Y, Zhao G, Cao L, Nakatsu G, et al. Mucosal microbiome dysbiosis in gastric carcinogenesis. Gut 2018;67:1024–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hu YL, Pang W, Huang Y, Zhang Y, Zhang CJ. The Gastric Microbiome Is Perturbed in Advanced Gastric Adenocarcinoma Identified Through Shotgun Metagenomics. Frontiers in cellular and infection microbiology 2018;8:433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li TH, Qin Y, Sham PC, Lau KS, Chu KM, Leung WK. Alterations in Gastric Microbiota After H. Pylori Eradication and in Different Histological Stages of Gastric Carcinogenesis. Scientific reports 2017;7:44935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu X, Shao L, Liu X, Ji F, Mei Y, Cheng Y, et al. Alterations of gastric mucosal microbiota across different stomach microhabitats in a cohort of 276 patients with gastric cancer. EBioMedicine 2019;40:336–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang L, Zhou J, Xin Y, Geng C, Tian Z, Yu X, et al. Bacterial overgrowth and diversification of microbiota in gastric cancer. European journal of gastroenterology & hepatology 2016;28:261–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yu G, Hu N, Wang L, Wang C, Han XY, Humphry M, et al. Gastric microbiota features associated with cancer risk factors and clinical outcomes: A pilot study in gastric cardia cancer patients from Shanxi, China. International journal of cancer 2017;141:45–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yu G, Torres J, Hu N, Medrano-Guzman R, Herrera-Goepfert R, Humphrys MS, et al. Molecular Characterization of the Human Stomach Microbiota in Gastric Cancer Patients. Frontiers in cellular and infection microbiology 2017;7:302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yu J, Feng Q, Wong SH, Zhang D, Liang QY, Qin Y, et al. Metagenomic analysis of faecal microbiome as a tool towards targeted non-invasive biomarkers for colorectal cancer. Gut 2017;66:70–8 [DOI] [PubMed] [Google Scholar]
- 53.Hosgood HD, 3rd, Sapkota AR, Rothman N, Rohan T, Hu W, Xu J, et al. The potential role of lung microbiota in lung cancer attributed to household coal burning exposures. Environ Mol Mutagen 2014;55:643–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cameron SJS, Lewis KE, Huws SA, Hegarty MJ, Lewis PD, Pachebat JA, et al. A pilot study using metagenomic sequencing of the sputum microbiome suggests potential bacterial biomarkers for lung cancer. PloS one 2017;12:e0177062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Carpagnano G Aspergillus spp. colonization in exhaled breath condensate of lung cancer patients from Puglia Region of Italy. European Respiratory Journal 2014;44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tsay JJ, Wu BG, Badri MH, Clemente JC, Shen N, Meyn P, et al. Airway Microbiota Is Associated with Upregulation of the PI3K Pathway in Lung Cancer. American journal of respiratory and critical care medicine 2018;198:1188–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang K, Huang Y, Zhang Z, Liao J, Ding Y, Fang X, et al. A Preliminary Study of Microbiota Diversity in Saliva and Bronchoalveolar Lavage Fluid from Patients with Primary Bronchogenic Carcinoma. Medical science monitor : international medical journal of experimental and clinical research 2019;25:2819–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schmidt BL, Kuczynski J, Bhattacharya A, Huey B, Corby PM, Queiroz ELS, et al. Changes in abundance of oral microbiota associated with oral cancer. PloS one 2014;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Berkovits C, Toth A, Szenzenstein J, Deak T, Urban E, Gacser A, et al. Analysis of oral yeast microflora in patients with oral squamous cell carcinoma. Springerplus 2016;5:1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Homann N, Tillonen J, Meurman JH, Rintamaki H, Lindqvist C, Rautio M, et al. Increased salivary acetaldehyde levels in heavy drinkers and smokers: a microbiological approach to oral cavity cancer. Carcinogenesis 2000;21:663–8 [DOI] [PubMed] [Google Scholar]
- 61.Banerjee S, Tian T, Wei Z, Peck KN, Shih N, Chalian AA, et al. Microbial Signatures Associated with Oropharyngeal and Oral Squamous Cell Carcinomas. Scientific reports 2017;7:4036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bornigen D, Ren B, Pickard R, Li J, Ozer E, Hartmann EM, et al. Alterations in oral bacterial communities are associated with risk factors for oral and oropharyngeal cancer. Scientific reports 2017;7:17686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hsiao JR, Chang CC, Lee WT, Huang CC, Ou CY, Tsai ST, et al. The interplay between oral microbiome, lifestyle factors and genetic polymorphisms in the risk of oral squamous cell carcinoma. Carcinogenesis 2018;39:778–87 [DOI] [PubMed] [Google Scholar]
- 64.Hu X, Zhang Q, Hua H, Chen F. Changes in the salivary microbiota of oral leukoplakia and oral cancer. Oral oncology 2016;56:e6–8 [DOI] [PubMed] [Google Scholar]
- 65.Lee WH, Chen HM, Yang SF, Liang C, Peng CY, Lin FM, et al. Bacterial alterations in salivary microbiota and their association in oral cancer. Scientific reports 2017;7:16540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lim Y, Fukuma N, Totsika M, Kenny L, Morrison M, Punyadeera C. The Performance of an Oral Microbiome Biomarker Panel in Predicting Oral Cavity and Oropharyngeal Cancers. Frontiers in cellular and infection microbiology 2018;8:267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mok SF, Karuthan C, Cheah YK, Ngeow WC, Rosnah Z, Yap SF, et al. The oral microbiome community variations associated with normal, potentially malignant disorders and malignant lesions of the oral cavity. The Malaysian journal of pathology 2017;39:1–15 [PubMed] [Google Scholar]
- 68.Perera M, Al-Hebshi NN, Perera I, Ipe D, Ulett GC, Speicher DJ, et al. Inflammatory Bacteriome and Oral Squamous Cell Carcinoma. Journal of dental research 2018;97:725–32 [DOI] [PubMed] [Google Scholar]
- 69.Wolf A, Moissl-Eichinger C, Perras A, Koskinen K, Tomazic PV, Thurnher D. The salivary microbiome as an indicator of carcinogenesis in patients with oropharyngeal squamous cell carcinoma: A pilot study. Scientific reports 2017;7:5867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yost S, Stashenko P, Choi Y, Kukuruzinska M, Genco CA, Salama A, et al. Increased virulence of the oral microbiome in oral squamous cell carcinoma revealed by metatranscriptome analyses. International journal of oral science 2018;10:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhao H, Chu M, Huang Z, Yang X, Ran S, Hu B, et al. Variations in oral microbiota associated with oral cancer. Scientific reports 2017;7:11773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Farrell JJ, Zhang L, Zhou H, Chia D, Elashoff D, Akin D, et al. Variations of oral microbiota are associated with pancreatic diseases including pancreatic cancer. Gut 2012;61:582–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Torres PJ, Fletcher EM, Gibbons SM, Bouvet M, Doran KS, Kelley ST. Characterization of the salivary microbiome in patients with pancreatic cancer. PeerJ 2015;2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Olson SH, Satagopan J, Xu Y, Ling L, Leong S, Orlow I, et al. The oral microbiota in patients with pancreatic cancer, patients with IPMNs, and controls: a pilot study. Cancer causes & control : CCC 2017;28:959–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Han S, Yang X, Qi Q, Pan Y, Chen Y, Shen J, et al. Potential screening and early diagnosis method for cancer: Tongue diagnosis. International journal of oncology 2016;48:2257–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hu J, Han S, Chen Y, Ji Z. Variations of Tongue Coating Microbiota in Patients with Gastric Cancer. BioMed research international 2015;2015:173729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bi H, Tian Y, Song C, Li J, Liu T, Chen Z, et al. Urinary microbiota - a potential biomarker and therapeutic target for bladder cancer. Journal of medical microbiology 2019;68:1471–8 [DOI] [PubMed] [Google Scholar]
- 78.Bucevic Popovic V, Situm M, Chow CT, Chan LS, Roje B, Terzic J. The urinary microbiome associated with bladder cancer. Scientific reports 2018;8:12157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Goedert JJ, Jones G, Hua X, Xu X, Yu G, Flores R, et al. Investigation of the association between the fecal microbiota and breast cancer in postmenopausal women: a population-based case-control pilot study. Journal of the National Cancer Institute 2015;107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Goedert JJ, Hua X, Bielecka A, Okayasu I, Milne GL, Jones GS, et al. Postmenopausal breast cancer and oestrogen associations with the IgA-coated and IgA-noncoated faecal microbiota. British journal of cancer 2018;118:471–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhu J, Liao M, Yao Z, Liang W, Li Q, Liu J, et al. Breast cancer in postmenopausal women is associated with an altered gut metagenome. Microbiome 2018;6:136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Qi YF, Sun JN, Ren LF, Cao XL, Dong JH, Tao K, et al. Intestinal Microbiota Is Altered in Patients with Gastric Cancer from Shanxi Province, China. Digestive diseases and sciences 2019;64:1193–203 [DOI] [PubMed] [Google Scholar]
- 83.Youssef O, Lahti L, Kokkola A, Karla T, Tikkanen M, Ehsan H, et al. Stool Microbiota Composition Differs in Patients with Stomach, Colon, and Rectal Neoplasms. Digestive diseases and sciences 2018;63:2950–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chen W, Liu F, Ling Z, Tong X, Xiang C. Human intestinal lumen and mucosa-associated microbiota in patients with colorectal cancer. PloS one 2012;7:e39743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mira-Pascual L, Cabrera-Rubio R, Ocon S, Costales P, Parra A, Suarez A, et al. Microbial mucosal colonic shifts associated with the development of colorectal cancer reveal the presence of different bacterial and archaeal biomarkers. Journal of Gastroenterology 2014;50:167–79 [DOI] [PubMed] [Google Scholar]
- 86.Flemer B, Lynch DB, Brown JM, Jeffery IB, Ryan FJ, Claesson MJ, et al. Tumour-associated and non-tumour-associated microbiota in colorectal cancer. Gut 2017;66:633–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Alexander JL, Scott A, Mroz A, Perdones-Montero A, McKenzie J, Rees DN, et al. Mass spectrometry imaging (MSI) of microbiome-metabolome interactions in colorectal cancer. Gastroenterology 2016;150:S23 [Google Scholar]
- 88.Gao Z, Guo B, Gao R, Zhu Q, Wu W, Qin H. Probiotics modify human intestinal mucosa-associated microbiota in patients with colorectal cancer. Mol Med Rep 2015;12:6119–27 [DOI] [PubMed] [Google Scholar]
- 89.McCoy AN, Araújo-Pérez F, Azcárate-Peril A, Yeh JJ, Sandler RS, Keku TO. Fusobacterium Is Associated with Colorectal Adenomas. PloS one 2013;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Burns MB, Lynch J, Starr TK, Knights D, Blekhman R. Virulence genes are a signature of the microbiome in the colorectal tumor microenvironment. Genome medicine 2015;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Warren RL, Freeman DJ, Pleasance S, Watson P, Moore RA, Cochrane K, et al. Co-occurrence of anaerobic bacteria in colorectal carcinomas. Microbiome 2013;1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Marchesi JR, Dutilh BE, Hall N, Peters WHM, Roelofs R, Boleij A, et al. Towards the human colorectal cancer microbiome. PloS one 2011;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tahara T, Yamamoto E, Suzuki H, Maruyama R, Chung W, Garriga J, et al. Fusobacterium in colonic flora and molecular features of colorectal carcinoma. Cancer research 2014;74:1311–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hale VL, Jeraldo P, Chen J, Mundy M, Yao J, Priya S, et al. Distinct microbes, metabolites, and ecologies define the microbiome in deficient and proficient mismatch repair colorectal cancers. Genome medicine 2018;10:78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Shah MS, DeSantis T, Yamal JM, Weir T, Ryan EP, Cope JL, et al. Re-purposing 16S rRNA gene sequence data from within case paired tumor biopsy and tumor-adjacent biopsy or fecal samples to identify microbial markers for colorectal cancer. PloS one 2018;13:e0207002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ahn J, Sinha R, Pei Z, Dominianni C, Wu J, Shi J, et al. Human gut microbiome and risk for colorectal cancer. Journal of the National Cancer Institute 2013;105:1907–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Feng Q, Liang S, Jia H, Stadlmayr A, Tang L, Lan Z, et al. Gut microbiome development along the colorectal adenoma-carcinoma sequence. Nature communications 2015;6:6528. [DOI] [PubMed] [Google Scholar]
- 98.Kasai C, Sugimoto K, Moritani I, Tanaka J, Oya Y, Inoue H, et al. Comparison of human gut microbiota in control subjects and patients with colorectal carcinoma in adenoma: Terminal restriction fragment length polymorphism and next-generation sequencing analyses. Oncology reports 2016;35:325–33 [DOI] [PubMed] [Google Scholar]
- 99.Vogtmann E, Hua X, Zeller G, Sunagawa S, Voigt AY, Hercog R, et al. Colorectal cancer and the human gut microbiome: Reproducibility with whole-genome shotgun sequencing. PloS one 2016;11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wu N, Yang X, Zhang R, Li J, Xiao X, Hu Y, et al. Dysbiosis signature of fecal microbiota in colorectal cancer patients. Microbial ecology 2013;66:462–70 [DOI] [PubMed] [Google Scholar]
- 101.Zeller G, Tap J, Voigt AY, Sunagawa S, Kultima JR, Costea PI, et al. Potential of fecal microbiota for early-stage detection of colorectal cancer. Mol Syst Biol 2014;10:766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Allali I, Boukhatem N, Bouguenouch L, Hardi H, Boudouaya HA, Cadenas MB, et al. Gut microbiome of Moroccan colorectal cancer patients. Medical microbiology and immunology 2018;207:211–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Amitay EL, Werner S, Vital M, Pieper DH, Hofler D, Gierse IJ, et al. Fusobacterium and colorectal cancer: causal factor or passenger? Results from a large colorectal cancer screening study. Carcinogenesis 2017;38:781–8 [DOI] [PubMed] [Google Scholar]
- 104.Baxter NT, Ruffin MTt, Rogers MA, Schloss PD. Microbiota-based model improves the sensitivity of fecal immunochemical test for detecting colonic lesions. Genome medicine 2016;8:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Dai Z, Coker OO, Nakatsu G, Wu WKK, Zhao L, Chen Z, et al. Multi-cohort analysis of colorectal cancer metagenome identified altered bacteria across populations and universal bacterial markers. Microbiome 2018;6:70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sinha R, Ahn J, Sampson JN, Shi J, Yu G, Xiong X, et al. Fecal Microbiota, Fecal Metabolome, and Colorectal Cancer Interrelations. PloS one 2016;11:e0152126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zackular JP, Rogers MA, Ruffin MTt, Schloss PD. The human gut microbiome as a screening tool for colorectal cancer. Cancer prevention research (Philadelphia, Pa) 2014;7:1112–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ohigashi S, Sudo K, Kobayashi D, Takahashi O, Takahashi T, Asahara T, et al. Changes of the intestinal microbiota, short chain fatty acids, and fecal pH in patients with colorectal cancer. Digestive diseases and sciences 2013;58:1717–26 [DOI] [PubMed] [Google Scholar]
- 109.Amiot A, Dona A, Wijeyesekera A, Tournigand C, Lebaleur Y, Sobhani I, et al. 1H HR NMR spectroscopy of fecal extracts enables detection of advanced colorectal neoplasia. United European Gastroenterology Journal 2015;3:A266. [DOI] [PubMed] [Google Scholar]
- 110.Kanazawa K, Konishi F, Mitsuoka T, Terada A, Itoh K, Narushima S, et al. Factors influencing the development of sigmoid colon cancer. Bacteriologic and biochemical studies. Cancer 1996;77:1701–6 [DOI] [PubMed] [Google Scholar]
- 111.Richard ML, Liguori G, Lamas B, Brandi G, da Costa G, Hoffmann TW, et al. Mucosa-associated microbiota dysbiosis in colitis associated cancer. Gut microbes 2018;9:131–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Mori G, Rampelli S, Orena BS, Rengucci C, De Maio G, Barbieri G, et al. Shifts of Faecal Microbiota During Sporadic Colorectal Carcinogenesis. Scientific reports 2018;8:10329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Gao R, Gao Z, Huang L, Qin H. Gut microbiota and colorectal cancer. European journal of clinical microbiology & infectious diseases : official publication of the European Society of Clinical Microbiology 2017;36:757–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Weir TL, Manter DK, Sheflin AM, Barnett BA, Heuberger AL, Ryan EP. Stool microbiome and metabolome differences between colorectal cancer patients and healthy adults. PloS one 2013;8:e70803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Bratcher DF. 131 - Other Corynebacteria In: Long SS, Prober CG, Fischer M, editors. Principles and Practice of Pediatric Infectious Diseases (Fifth Edition): Elsevier; 2018. p 778–81.e2. [Google Scholar]
- 116.Murphy EC, Frick IM. Gram-positive anaerobic cocci--commensals and opportunistic pathogens. FEMS microbiology reviews 2013;37:520–53 [DOI] [PubMed] [Google Scholar]
- 117.Zhang G, Chen R, Rudney JD. Streptococcus cristatus modulates the Fusobacterium nucleatum-induced epithelial interleukin-8 response through the nuclear factor-kappa B pathway. J Periodontal Res 2011;46:558–67 [DOI] [PubMed] [Google Scholar]
- 118.Nosho K, Sukawa Y, Adachi Y, Ito M, Mitsuhashi K, Kurihara H, et al. Association of Fusobacterium nucleatum with immunity and molecular alterations in colorectal cancer. World journal of gastroenterology 2016;22:557–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wang D, Li Y, Zhong H, Ding Q, Lin Y, Tang S, et al. Alterations in the human gut microbiome associated with Helicobacter pylori infection. FEBS open bio 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Rubinstein MR, Wang X, Liu W, Hao Y, Cai G, Han YW. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/beta-catenin signaling via its FadA adhesin. Cell host & microbe 2013;14:195–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Yu J, Zhao L, Zhao R, Long X, Coker OO, Sung JJY. The role of Parvimonas micra in intestinal tumorigenesis in germ-free and conventional APCmin/+ mice. Journal of Clinical Oncology 2019;37:531– [Google Scholar]
- 122.Groeger S, Jarzina F, Domann E, Meyle J. Porphyromonas gingivalis activates NFkappaB and MAPK pathways in human oral epithelial cells. BMC Immunol 2017;18:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Tsoi H, Chu ESH, Zhang X, Sheng J, Nakatsu G, Ng SC, et al. Peptostreptococcus anaerobius Induces Intracellular Cholesterol Biosynthesis in Colon Cells to Induce Proliferation and Causes Dysplasia in Mice. Gastroenterology 2017;152:1419–33.e5 [DOI] [PubMed] [Google Scholar]
- 124.Jia W, Xie G, Jia W. Bile acid-microbiota crosstalk in gastrointestinal inflammation and carcinogenesis. Nature reviews Gastroenterology & hepatology 2018;15:111–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Xie G, Wang X, Huang F, Zhao A, Chen W, Yan J, et al. Dysregulated hepatic bile acids collaboratively promote liver carcinogenesis. International journal of cancer 2016;139:1764–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Li P, Shu Y, Gu Y. The potential role of bacteria in pancreatic cancer: A systematic review. Carcinogenesis 2020 [DOI] [PubMed] [Google Scholar]
- 127.Xie G, Raufman JP. Role of the Aryl Hydrocarbon Receptor in Colon Neoplasia. Cancers 2015;7:1436–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Jin UH, Lee SO, Pfent C, Safe S. The aryl hydrocarbon receptor ligand omeprazole inhibits breast cancer cell invasion and metastasis. BMC cancer 2014;14:498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Fritz WA, Lin TM, Cardiff RD, Peterson RE. The aryl hydrocarbon receptor inhibits prostate carcinogenesis in TRAMP mice. Carcinogenesis 2007;28:497–505 [DOI] [PubMed] [Google Scholar]
- 130.Descamps HC, Herrmann B, Wiredu D, Thaiss CA . The path toward using microbial metabolites as therapies. EBioMedicine 2019;44:747–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Neumann S, Peyroux EM, Woodall MJ, Shields NJ, Young SL, Pattison ST. The Influence of Microbial Metabolites in the Gastrointestinal Microenvironment on Anticancer Immunity. Current Cancer Treatment 2019 [Google Scholar]
- 132.Zhernakova A, Kurilshikov A, Bonder MJ, Tigchelaar EF, Schirmer M, Vatanen T, et al. Population-based metagenomics analysis reveals markers for gut microbiome composition and diversity. Science (New York, NY) 2016;352:565–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Smelov V, Arroyo Muhr LS, Bzhalava D, Brown LJ, Komyakov B, Dillner J. Metagenomic sequencing of expressed prostate secretions. Journal of medical virology 2014;86:2042–8 [DOI] [PubMed] [Google Scholar]
- 134.Smelov V, Bzhalava D, Arroyo Muhr LS, Eklund C, Komyakov B, Gorelov A, et al. Detection of DNA viruses in prostate cancer. Scientific reports 2016;6:25235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Choo JM, Leong LE, Rogers GB. Sample storage conditions significantly influence faecal microbiome profiles. Scientific reports 2015;5:16350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Costea PI, Zeller G, Sunagawa S, Pelletier E, Alberti A, Levenez F, et al. Towards standards for human fecal sample processing in metagenomic studies. Nature biotechnology 2017;35:1069–76 [DOI] [PubMed] [Google Scholar]
- 137.Sinha R, Abu-Ali G, Vogtmann E, Fodor AA, Ren B, Amir A, et al. Assessment of variation in microbial community amplicon sequencing by the Microbiome Quality Control (MBQC) project consortium. Nature biotechnology 2017;35:1077–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Hiergeist A, Glasner J, Reischl U, Gessner A. Analyses of Intestinal Microbiota: Culture versus Sequencing. ILAR journal 2015;56:228–40 [DOI] [PubMed] [Google Scholar]
- 139.Maziarz M, Pfeiffer RM, Wan Y, Gail MH. Using standard microbiome reference groups to simplify beta-diversity analyses and facilitate independent validation. Bioinformatics (Oxford, England) 2018;34:3249–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Le Cao KA, Gonzalez I, Dejean S. integrOmics: an R package to unravel relationships between two omics datasets. Bioinformatics (Oxford, England) 2009;25:2855–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Gloor GB, Macklaim JM, Pawlowsky-Glahn V, Egozcue JJ . Microbiome Datasets Are Compositional: And This Is Not Optional. Front Microbiol 2017;8:2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Filzmoser P, Hron K. Correlation Analysis for Compositional Data. Mathematical Geosciences 2008;41:905 [Google Scholar]
- 143.Sinha R, Ahsan H, Blaser M, Caporaso JG, Carmical JR, Chan AT, et al. Next steps in studying the human microbiome and health in prospective studies, Bethesda, MD, May 16–17, 2017. Microbiome 2018;6:210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Bai J, Behera M, Bruner DW. The gut microbiome, symptoms, and targeted interventions in children with cancer: a systematic review. Supportive care in cancer : official journal of the Multinational Association of Supportive Care in Cancer 2017 [DOI] [PubMed] [Google Scholar]
- 145.Sinha R, Chen J, Amir A, Vogtmann E, Shi J, Inman KS, et al. Collecting Fecal Samples for Microbiome Analyses in Epidemiology Studies. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology 2016;25:407–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Vogtmann E, Chen J, Kibriya MG, Chen Y, Islam T, Eunes M, et al. Comparison of Fecal Collection Methods for Microbiota Studies in Bangladesh. Applied and environmental microbiology 2017;83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Vogtmann E, Chen J, Amir A, Shi J, Abnet CC, Nelson H, et al. Comparison of Collection Methods for Fecal Samples in Microbiome Studies. American journal of epidemiology 2017;185:115–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nature biotechnology 2019;37:852–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Minot SS, Krumm N, Greenfield NB. One Codex: A Sensitive and Accurate Data Platform for Genomic Microbial Identification. bioRxiv 2015:027607 [Google Scholar]
- 150.Overcoming hurdles in sharing microbiome data. Nature microbiology 2017;2:1573– [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.