Abstract
The ability to synthesize simple aromatic compounds is well known from bacteria, fungi and plants, which all share an exclusive biosynthetic route—the shikimic acid pathway. Some of these organisms further evolved the polyketide pathway to form core benzenoids via a head-to-tail condensation of polyketide precursors. Arthropods supposedly lack the ability to synthesize aromatics and instead rely on aromatic amino acids acquired from food, or from symbiotic microorganisms. The few studies purportedly showing de novo biosynthesis via the polyketide synthase (PKS) pathway failed to exclude endosymbiotic bacteria, so their results are inconclusive. We investigated the biosynthesis of aromatic compounds in defence secretions of the oribatid mite Archegozetes longisetosus. Exposing the mites to a diet containing high concentrations of antibiotics removed potential microbial partners but did not affect the production of defensive benzenoids. To gain insights into benzenoid biosynthesis, we fed mites with stable-isotope labelled precursors and monitored incorporation with mass spectrometry. Glucose, malonic acid and acetate, but not phenylalanine, were incorporated into the benzenoids, further evidencing autogenous biosynthesis. Whole-transcriptome sequencing with hidden Markov model profile search of protein domain families and subsequent phylogenetic analysis revealed a putative PKS domain similar to an actinobacterial PKS, possibly indicating a horizontal gene transfer.
Keywords: biosynthetic pathways, Benzenoids, chemical ecology, Oribatid mites, Chemical defence
1. Introduction
Simple aromatic compounds (i.e. chemicals containing a benzene ring) are important products in chemical science and industry, but also in nature [1,2]. Overall, about 550 different simple and many more complex aromatics have been described from bacteria, fungi, plants and animals [3,4]. The unique electronic structure of the benzene ring—a delocalized π-electron system with a six-ringed carbon skeleton—provides aromatics with key structural motifs that shape interactions on both molecular and organismal levels [5,6]. In the primary metabolism of arthropods, aromatic amino acids are important building blocks of proteins, but also are used in cuticle formation, sclerotization and melanization [7,8]. Among secondary metabolites, arthropods use simple benzenoids in pheromonal communication and as defensive compounds, while the more complex polyketide aromatics also function as potent antibiotics [3,9,10].
Bacteria, fungi and plants evolved two biosynthetic routes to form benzene rings: the shikimic acid pathway [1,10] and the polyketide pathway [3]. In the latter benzene rings form via a head-to-tail condensation of poly-β-carbonyl intermediates followed by an intramolecular condensation forming the aromatic system [11,12]. Sponges, sea urchins and some vertebrates also can synthesize simple aromatics via the polyketide pathway [3,13,14]. Among arthropods, aromatics are important for chemical interactions in insects, arachnids and myriapods [9], yet it is still unclear whether these benzenoids are acquired from diet or from endosymbiotic bacteria, or are synthesized de novo by the animals [3,10,15]. It appears that especially the complex, large ringed polyketide aromatics are mostly produced by bacteria [16–20], while simple aromatics may originate from a potential arthropod polyketide pathway [21–24]. Pankewitz & Hilker [15] reviewed available studies on polyketides and found no unequivocal evidence for de novo biosynthesis of aromatics in arthropods, since potential endosymbiont contributions were not excluded. A decade later, this still holds true [10,25,26]. Only the studies by Bestmann et al. [21] on aromatic trail pheromones of formicine ants and Pankewitz et al. [27] on anthraquinones isolated from leaf beetle eggs indicated that bacteria may not be involved in the biosynthesis of these chemicals, yet in both studies the success of antibiotic treatment was not confirmed. Also, leaf beetle anthraquinones are formed via a eukaryotic polyketide folding mode, rendering a direct bacterial involvement unlikely [28].
The oribatid mite Archegozetes longisetosus Aoki (figure 1i) produces defensive secretions containing two simple aromatics, 2-hydroxy-6-methyl-benzaldehyde (2,6-HMBD) and 3-hydroxybenzene-1,2-dicarbaldehyde (γ-acaridial), in a pair of opisthonotal exocrine glands [26]. We reared animals in a controlled, sterile environment to investigate their ability to synthesize these benzene-ringed compounds de novo. We performed stable isotope labelling experiments with different precursors under intensive antibiotic treatment to maintain symbiont-free animals and to uncover the biosynthetic pathway of these aromatics. We demonstrate that 2,6-HMBD and γ-acaridial are both synthesized de novo, probably from poly-β-carbonyl (e.g. acetyl-CoA and malonyl-CoA). Furthermore, whole-transcriptome sequencing (RNAseq) uncovered putative PKS domains similar to those of bacteria, indicating a potential horizontal gene transfer.
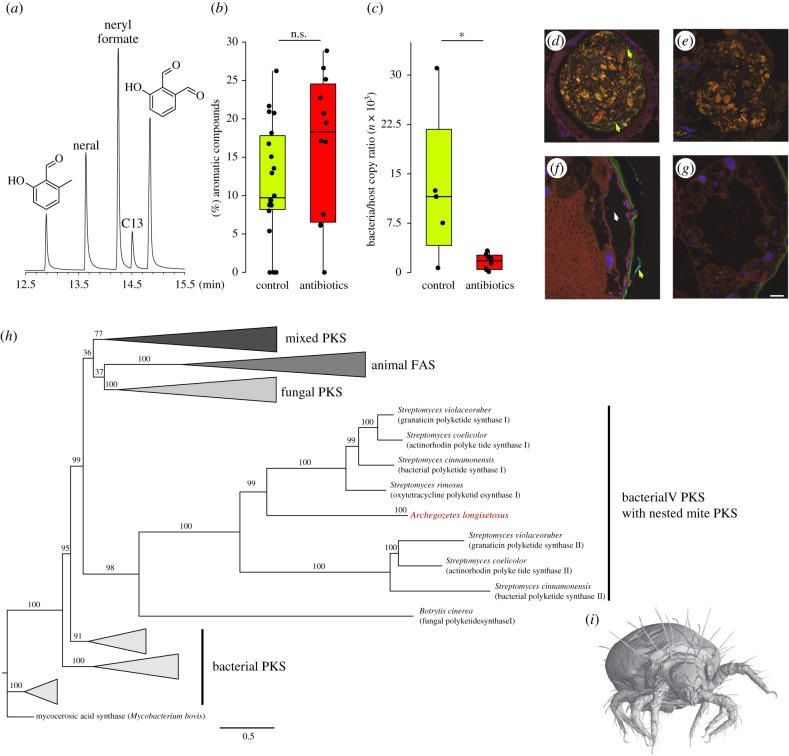
Figure 1.
Mites (Archegozetes longisetosus) contain two simple aromatic compounds in their defensive secretions, and reduction of bacteria did not affect their production. (a) Representative GC trace of mite gland extracts; in order of retention time: 2-hydroxy-6-methyl-benzaldehyde (2,6-HMBD), neral, neryl formate, tridecane, 3-hydroxybenzene-1,2-dicarbaldehyde (γ-acaridial). Pentadec-7-ene, pentadecane, heptadeca-6,9-diene, heptadec-8-ene, heptadecane are not shown. (b) Supplementing a wheat grass diet (control) with a mixture of antibiotics (10% w/w; combined amoxicillin, streptomycin and tetracycline; ‘antibiotics’) did not affect the relative amount of aromatic compounds but resulted in significantly lower bacterial load in the mite (c). Fluorescence in situ hybridization (FISH) revealed a high bacterial prevalence in the food bolus of control group mites (d), while no detectable bacteria (e) were found in similar bolus in antibiotic-treated mites. Even in the control group, no bacteria were detected in the gland (f); the white arrowhead marks the centre of the gland) or in the caeca (i.e. pairwise sac-like organ that are located close to the glandular tissue; (g). Maximum-likelihood tree (h) based on an alignment of the KS-domains of fatty-acid and PKSs from animals (including those of the mite—depicted in dark red), fungi and bacteria. Bootstrap values (based on 1000 replicates) are indicated along branches and the scale bar below the tree denotes substitutions per site. The tree was rooted by the outgroup mycocerosic acid synthase from Mycobacterium bovis. Frontal view (i) of a µCT-reconstruction of A. longisetosus. In the FISH images, bacteria are stained in green with the general bacterial probe EUB338-Cy5; the green arrow heads mark bacterial signals detected in the food bolus (d) or on the cuticle (f). The scale bar in FISH images is 20 µm. U-test: n.s. = not significant, p > 0.05; *= significant, p < 0.05. (Online version in colour.)
2. Results and discussion
The defensive chemicals of the oribatid mite A. longisetosus consist of a blend of ten compounds (figure 1a,i) including two terpenes (approx. 45%), six hydrocarbons (approx. 15%) and two aromatic compounds (approx. 40%) [29,30]. While the hydrocarbons probably serve as solvents, the terpenes and aromatics are bioactive compounds used as alarm pheromones and predator repellents [31].
In a first feeding experiment, we tested if elimination of potentially symbiotic gut bacteria influences the biosynthesis of 2,6-HMBD and γ-acaridial by feeding the mites with food containing 10% antibiotics (a mixture of amoxicillin, streptomycin and tetracycline; see methods below). We found that production of 2,6-HMBD and γ-acaridial was unaffected by treatments with individual antibiotics or with all three in concert (figure 1b; U-test: z = − 1.3, p = 0.19; n = 32; electronic supplementary material, figure S1). The combined treatment, which was used in further incorporation experiments with labelled precursors, effectively eliminated bacteria in the mites (figure 1c; U-test: z = − 2.1, p = 0.027; n = 13). These results also were supported by fluorescence in situ hybridization (FISH), revealing that all detectable bacteria found on the food and in the alimentary tract (figure 1d) were eliminated in the antibiotic-treated mites (figure 1e). We detected no bacteria in the glands (figure 1f), and no or only single bacteria in the digestive caeca (figure 1g), irrespective of antibiotic treatment, further supporting the general absence of endosymbiotic bacteria. By contrast, bacteria on the outer cuticle of the mites remained mostly unaffected by antibiotic treatment and served as an internal control for FISH (electronic supplementary material, figure S2). Hence, the gland does not house bacteria involved in the production of defensive compounds or precursors, as described for defensive symbioses in other arthropods [16,18,32]. The paired digestive caeca, which are directly connected with the glandular tissue via a plasma mass [33], do not appear to be brood chambers for bacteria or yeasts (electronic supplementary material, figure S2 andS3). Also, in none of the numerous histological and ultrastructural studies on the ovarial ultrastructure, egg development, vitellogenesis and cleavage was there any indication of vertically transmitted prokaryotic or eukaryotic symbionts [34–38].
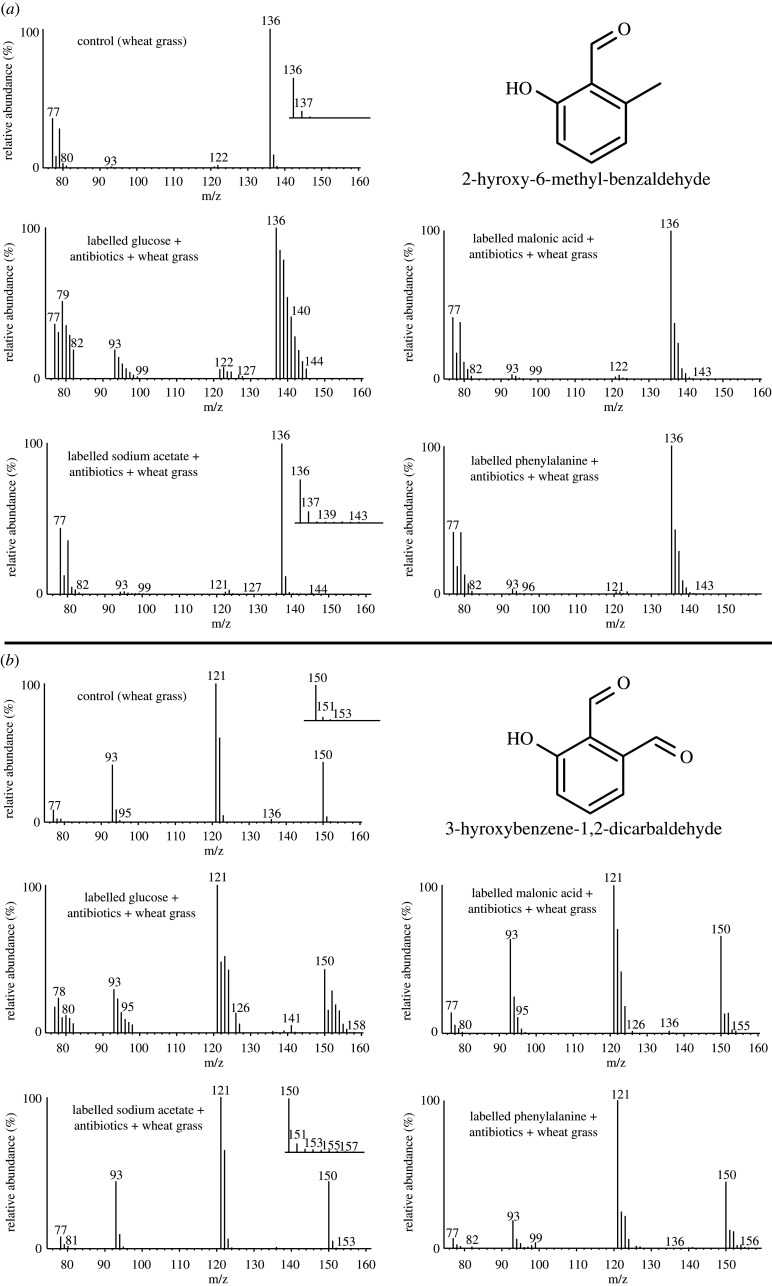
In a second experiment, the diet of A. longisetosus was supplemented with food containing 25% of a stable isotope-labelled precursor—[13C6, d7] D-glucose, [13C3] malonic acid, sodium [13C1] acetate or [13C6] phenylalanine—and 10% antibiotics. To examine the incorporation and possible biosynthesis of 2,6-HMBD (figure 2a) and γ-acaridial (figure 2b), we compared selected fragment ions using mass spectrometry and calculated an enrichment factor based on relative intensities. Both aromatics showed consistently enriched [M+1]+ to [M+8]+-ion series for [13C6, d7] D-glucose, [13C3] malonic acid and sodium [13C1] acetate. Enrichment was strongest for the most general source, the heavily labelled D-glucose, while the more specific β-carbonyl precursors showed less enrichment (electronic supplementary material, table S1) for both compounds (see figure 2a for 2,6-HMBD and figure 2b for γ-acaridial). This stepwise enrichment pattern indicates that short β-carbonyls joined to form complex molecules via multiple head-to-tail condensation reactions, which is indicative of a PKS-like mechanism [22–24]. When feeding [13C6] phenylalanine, we found no enrichment of the [M+6]+-ion (electronic supplementary material, table S1; figure 2), which would be expected for the incorporation of the completely 13C-labelled benzene ring [39]. Instead, we again found a stepwise enrichment in the [M+1]+ to [M+8]+-ion series (electronic supplementary material, table S1; figure 2), indicating that only fragments of the benzene ring of [13C6] phenylalanine had been incorporated into 2,6-HMBD and γ-acaridial via a PKS-like reaction after the mites had catabolized the amino acids to poly-β-carbonyl units [40,41]. As we can exclude a direct influence of bacteria (antibiotics treatment, qPCR, microscopy; figure 1; electronic supplementary material figure S2), fungi (microscopy; electronic supplementary material figure S3) and protists (microscopy) [34–38], this strongly supports a de novo biosynthesis of both aromatic compounds.
Figure 2.
Representative mass spectra of 2-hydroxy-6-methyl-benzaldehyde (a) and 3-hydroxybenzene-1,2-dicarbaldehyde (b) extracted from defensive glands of mites fed with unlabelled wheatgrass (control), or wheatgrass infused with 13C/d-labelled glucose, 13C-malonic acid, 13C-acetate and 13C-phenylalanine recorded in single-ion mode. Inserts show the [M + 1]+-ion series in the control and 13C-acetate. Mites fed with wheatgrass infused with a mixture of labelled precursors and antibiotics show enriched ions.
Given this mass spectrometric evidence, the mite must ultimately possess the necessary enzymes to produce 2,6-HMBD and γ-acaridial via a PKS-like pathway. To explore whether A. longisetosus expresses the respective genes encoding for such an enzyme and to trace down its potential evolutionary origin, we performed whole-body RNAseq of adult mites, assembled the transcriptome and predicted the longest open reading frame (ORFs) of the transcripts. Subsequent searching of the predicted protein sequences for ketoacyl-synthase (KS) domains—one module of both fatty acid (FAS) and polyketide synthases (PKSs) commonly used for phylogenetic analyses of these enzyme families [13,14]—revealed 10 candidate KS-domains in A. longisetosus (see Material and methods; electronic supplementary material, table S2). A phylogenetic analysis of the mite's KS-domains using a maximum-likelihood (ML) approach—including a pre-existing dataset of 139 KS-domains [13,14] of bacterial, fungal and animal PKS as well as animal FAS—revealed that eight of the mite's KS-domains were well nested within the animal FAS-clade (see electronic supplementary material, figure S4 for full ML tree), while two KS-domains were nested with a clade of bacterial PKS, specifically PKS from the actinobacterial genus Streptomyces (figure 1h). As expected, a SPARCLE (=Subfamily Protein Architecture Labelling Engine) [42] search identified both protein sequences as beta-ketoacyl-synthases known from both FAS and PKS, yet based on their phylogenetic placement with bacterial KS-domains (figure 1h), we refer to them as putative PKS domains. Both putative PKS domains had a GC content of approximately 37%, which is slightly higher than the usual GC content of known oribatid mite genomes (approx. 30%) [43], but much lower than in Streptomyces genomes (approx. 70%) [44]. Furthermore, the putative PKS domains have an amino acid composition more similar to those of other mites than to those of Streptomyces (electronic supplementary material, figure S5). Thus, the putative PKS domain genes are most likely integrated into the mite's genome and not due to Streptomyces contamination. The nested position within a Streptomyces clade (figure 1h) suggests an ancient horizontal gene transfer (HGT) of PKS-encoding genes from bacteria to A. longisetosus, as previously discussed for arthropods by Pankewitz & Hilker [15].
We were not able to isolate the entire biosynthetic PKS gene cluster for A. longisetosus, if such clusters indeed exist in animals. This might be due to the many short transcripts in the transcriptome and a lack of confirmed, contiguous gene models, but also to the diffuse genomic organization of pathway loci within the genomes of animals. Unlike in prokaryotes, where components of a biosynthetic cluster are conveniently organized as operons [45], identifying all components of biosynthetic pathways is much more challenging in animals [46]. Enzymes (or domains) are seldom clustered as tandems, but instead are scattered across the genome, and regulatory elements controlling expression are cryptic and sometimes even very distant from the open reading frame [47]. Furthermore, since there is no functional arthropod PKS-cluster for reference, it remains unknown whether PKS gene clusters of arthropods—in case they exist—are similar to those of bacteria, fungi or other animals. Our study provides evidence that HGT—a common mechanisms of acquiring new genes in mites [48] as well as other arthropods [49,50]—could explain the presence of putative PKS-domains and the mode of benzenoid synthesis via head-to-tail condensation in A. longisetosus (figure 3).
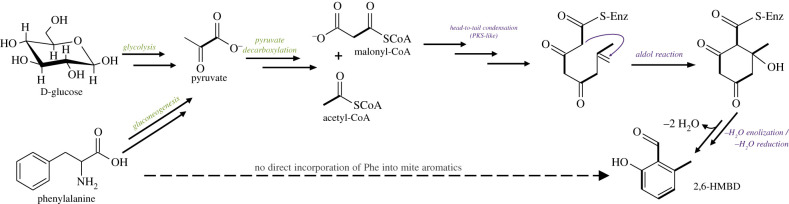
Figure 3.
Proposed biosynthetic scenario leading to 2-hydroxy-6-methyl-benzaldehyde (2,6-HMBD) in the oribatid mite Archegozetes longisetosus starting with D-glucose and phenylalanine as primary substrates. Common metabolic pathways are labelled in green, pathway-specific reactions are depicted in purple. Multiple arrows indicate multiple reaction steps. (Online version in colour.)
Since all experimental data (mass spectrometry of stable isotopes and RNAseq) point to a polyketide-like mechanism, we propose the following as the most likely biochemical pathway (figure 3): the mites harness (poly)-β-carbonyls like malonyl-CoA and/or acetyl-CoA produced via glycolysis and gluconeogenesis from sugars and amino acids, respectively, to form a C8-polyketid-like intermediate via a head-to-tail condensation. Subsequently, a ring closing cyclization via an aldol-reaction yields a second intermediate, which eventually undergoes several –H2O enolization and –H2O reduction reactions [22], inducing aromatization of the ring and the formation of the final product 2,6-HMBD. The second aromatic compound (3-hydroxybenzene-1,2-dicarbaldehyde) may be biosynthesized from 2,6-HMBD by an enzymatic oxidation of the methyl group to the corresponding aldehyde or via a different head-to-tail condensation.
This biosynthetic scenario accords well with studies on benzenoid ant pheromones [21,23,24] and benzoquinoid defensive chemicals of harvestmen [22] that indicated a polyketide origin of these compounds, but did not exclude or specifically test for an involvement of symbiotic microorganisms [51–53]. A more closely related arachnid, the storage mite Chortoglyphus arcuatus Troupeau, produces an aliphatic polyketide-derived aggregation pheromone—(4R,6R,8R)-4,6,8-trimethyldecan-2-one—synthesized from one acetate and four propionates [54], further supporting the ability of mites to produce polyketides de novo, yet potential bacterial participation was not excluded, as well.
Overall, the mass spectrometric analysis of labelled precursors under controlled conditions, as well as the molecular evolutionary assessment, indicates that this oribatid mite produces small aromatic compounds using a horizontally acquired putative PKS. With more examples, we may find that ancient horizontal gene transfer had a more general role in the evolution of aromatic compound synthesis across different arthropod groups.
3. Material and methods
(a). Mites
The lineage ‘ran’ [55] of the pantropical, parthenogenetic oribatid mite Archegozetes longisetosus was used in this study. Experimental cultures were established from an already existing line fed on wheat grass (Triticum sp.) powder from Naturya (Bath, UK) as follows: first, we collected eggs and surface-washed them with sodium hypochlorite solution (3% w/v), ethanol (70% v/v) and sterilized water for 5 s, 15 s and 30 s, respectively. Eggs were then transferred to sterile Petri dishes (diameter 45 mm) lined with 1 cm sterilized plaster of Paris. Sterile cultures were maintained in a laminar-flow closet at 28°C and 90% relative humidity. Sterilized water and 3–5 mg wheat grass or different food mixtures (see below) were provided three times each week.
(b). Antibiotics feeding experiment
In the first experiment, we fed four different mixtures of antibiotic-laden wheat grass (10% antibiotics, w/w) and pure wheat grass as a control to different groups of 150 mites for one generation (approx. 50 days). We prepared 10% (w/w) mixtures of sterilized wheat grass powder with amoxicillin, streptomycin and tetracycline as well as a mixture of all three antibiotics (3.3% w/w for each). One week after the adult eclosion, the defensive glands were extracted in hexane and chemically analysed (see below).
(c). Feeding experiments with labelled precursors
For the second experiment, we used only the 10% (w/w) mixture which contained all three antibiotics. Additionally, we added 25% (w/w) stable isotope-labelled precursors. We prepared four different mixtures with [13C6, d7] D-glucose, [13C3] malonic acid, sodium [13C1] acetate and [13C6] phenylalanine (all greater than 99% enrichment, Sigma-Aldrich, St Louis, USA) as well as a control with untreated wheat grass. Again, cultures were maintained for one generation and glands of adult specimens were extracted one week after eclosion using hexane (see below). Furthermore, a subset of these mites was used for FISH and qPCR experiments (see below).
(d). Fluorescence in situ hybridization
For the control as well as the [13C6, d7] D-glucose and [13C6] phenylalanine treatments (both with 10% antibiotics, see above) three entire specimens of A. longisetosus were fixated in 4% paraformaldehyde in PBS, and FISH was performed on semi-thin sections as described previously [56,57]. The fixated samples were dehydrated in a graded ethanol series and then embedded in cold-polymerizing resin (Technovit 8100; Heraeus Kulzer, Hanau, Germany) according to manufacturer's instructions. Sections of 7 µm thickness were obtained with a steel knife on a HM355S microtome (Leica, Germany) and mounted on microscope slides coated with poly-l-lysine (Kindler, Freiburg, Germany). FISH was done with the general eubacterial probes EUB388-Cy5 (5′-GCTGCCTCCCGTAGGAGT-3′) [58] and EUB784-Cy3 (5′-TGGACTACCAGGGTATCTAATCC-3′) [59] (two individuals each), or a combination of EUB338-Cy3 and the general yeast probe PF2-Cy5 (5′-CTCTGGCTTCACCCTATTC-3′) [60] (one individual each). Samples were incubated for 90 min at 60°C in 100 µl hybridization buffer (0.9 M NaCl, 0.02 M Tris/HCl pH 8.0, 0.01% SDS) containing 5 µl of each probe (500 nM) as well as DAPI (4′,6-diamidino-2-phenylindole) for counterstaining of host cell nuclei. Two wash steps with pre-warmed washing buffer (0.1M NaCl, 0.02M Tris/HCl pH8.0, 0.01% SDS, 5 mM EDTA), the second for 20 min at 60°C, as well as rinsing with dH2O, served to remove residual probes. After drying at room temperature, slides were covered with VectaShield and inspected on an AxioImager.Z1 fluorescence microscope (Zeiss, Jena, Germany).
(e). Quantitative PCR
To assess the effect of antibiotic treatment on absolute numbers of bacteria associated with the mites, bacterial 16S rRNA copy numbers were determined by quantitative PCR (qPCR). Since FISH experiments had revealed the presence of bacteria on the surface, the mites were surface washed in 5% (v/v) sodium dodecyl sulfate solution before bacterial quantification. For the control and the [13C6, d7] D-glucose + 10% antibiotics treatment, DNA was extracted from eight replicates of 15–25 mites each, using the MasterPure DNA purification kit (Epicentre Technologies) according to manufacturer's instructions. As a quality control of the DNA extracts and for later standardization of bacterial titres, the DNA extracts were subjected to a qPCR with primers targeting the host 28S rRNA gene (D3A_F: 5'-GACCCGTCTTGAAACACGGA-3′; and D3B_R: 5'-TCGGAAGGAACCAGCTACTA-3′) [61]. Subsequently, samples that showed amplification for the host 28S (five of the control replicates and all eight of the antibiotic treatment) were subjected to a qPCR with general eubacterial 16S rRNA gene primers (Univ16SRT-F: 5'-ACTCCTACGGGAGGCAGCAGT-3′; Univ16SRT-R: 5'-TATTACCGCGGCTGCTGGC-3′) [62]. qPCRs were done on a RotorGene-Q cycler (Qiagen, Hilden, Germany) in final reaction volumes of 25 µl, including the following components: 1 µl of DNA template, 2.5 µl of each primer (10 µM), 6.5 µl of autoclaved distilled H2O, and 12.5 µl of SYBR Green Mix (Qiagen, Hilden, Germany). PCR conditions included 95°C for 5 min, followed by 40 cycles of 95°C for 10 s, 70°C for 15 s, and 72°C for 10 s. A melting curve analysis was performed by increasing the temperature from 60°C to 95°C within 20 min. Standard curves were established for the host 28S and bacterial 16S assays by using 103–1010 copies of PCR product as templates. A Qubit fluorometer (Thermo Fisher Scientific) was used to measure DNA concentrations for the templates of the standard curve. The ratio between absolute copy numbers of bacterial 16S and host 28S (= bacterial/host copy ratio) was used as a standardized measure of bacterial abundance per mite sample.
(f). Chemical analysis
Gland exudates, containing the two studied aromatics, were extracted from living mites by submersing individuals (antibiotics feeding experiment) or groups of 15 (labelling experiment) in 50 µl hexane for 3 min, which is a well-established method to obtain oil gland compounds from mites [29,30]. Crude hexane extracts (2–5 µl) were analysed with a GCMS-QP2010 Ultra gas chromatography—mass spectrometry (GCMS) system from Shimadzu (Kyōto, Japan) equipped with a ZB-5MS capillary column (0.25 mm × 30 m, 0.25 µm film thickness) from Phenomenex (Torrance, USA). Hydrogen was used a carrier gas with a flow rate of 3.00 ml min−1, with splitless injection and a temperature ramp was set to increase from 50°C (5 min) to 210°C at a rate of 6°C min−1, followed by 35°C min−1 up to 320°C (for 5 min). Electron ionization mass spectra were recorded at 70 eV and characteristic fragment ions were monitored in single ion mode.
(g). Data analysis
For the antibiotic feeding experiment, we quantified the ion abundance and calculated the relative composition of aromatics (2,6-HMBD and γ-acaridial combined) compared to ion abundance of the other compounds on an individual base. Then we compared the (%) aromatic compounds among groups or treatments with a Kruskal–Wallis test or Mann–Whitney U-tests, respectively. The bacterial/host copy ratio was analysed with a Mann–Whitney U-test as well. Statistics were performed in PAST 3.17 [63]. For stable isotope enrichment, we compared the four treatment groups with the control and calculated the enrichment factors (EF) as EF = (rtreatment − rcontrol)/rcontrol, where rtreatment is the relative abundance (% relative to M+) of a respective ion in the treatment GCMS analyses and rcontrol is the relative intensity (%) of the same ion in the control group.
(h). DNA/RNA extraction, genome/RNA sequencing
For both extractions, about 200 adult mites were taken from the non-treated stock culture, starved for 24 h to avoid possible contamination from food in the gut, subsequently washed with 1% SDS for 10 s. Finally, DNA or RNA was extracted from living specimens using the ‘Quick-DNA Miniprep Plus Kit’ or the ‘Quick-RNA MiniPrep Kit’ from Zymo Research (Irvine, CA, USA) according to the manufacturer's protocols, respectively. Amounts and quality of DNA/RNA were accessed using a Qubit fluorometer and NanoDrop One (Thermo Fisher Scientific), respectively. Extracted DNA/ RNA was shipped to Omega Bioservices (Norcross, GA, USA) on dry ice for library preparation and sequencing. DNA library preparation followed the KAPA HyperPrep Kit protocol, RNA was used for poly-A selection, cDNA synthesis and library preparation followed the Illumina TruSeq mRNA Stranded Kit protocol. Both libraries were 150 bp paired-end sequenced on a HighSeq4000 platform.
(i). Genome and transcriptome assembly
Quality of reads was assessed using FastQC v. 0.11.8, Illumina adapters were trimmed with Cutadapt v. 1.18 [64]. Illumina short reads for the genome were assembled using the Platanus Genome Assembler pipeline v. 1.2.4 [65], yielding an assembly with a total length of 172.0 Mbp, an N50 = 25 196 bp and a BUSCO score [66] of C:94.4%[S:92.6%, D:1.8%], F:1.8%, M:3.8%. For the genome-guided assembly of the transcriptome, a bam-file was created from the genome fasta-file using STAR [67], while RNAseq reads were in silico normalized and subsequently used together with the bam-file to assemble the transcripts using Trinity v. 2.8.4 [68], yielding an assembly with a total length of 162.8 Mbp, an N50 = 2994 bp and a BUSCO score of C:96.3%[S:36.5%, D:59.8%], F:1.3%, M:2.4%. TransDecoder (v. 5.5.0) [69] was used to identify putative candidate coding regions in the transcriptome data and translate them into protein sequences.
(j). HMMR search, alignment and phylogenetic analysis
One way of identifying putative PKS domains is a phylogenetic analysis of the KS-domain. Therefore, we isolated potential KS-domain sequences of fatty acid and PKSs, by searching the transdecoded protein sequences with profile hidden Markov models as implemented in HMMER v3 [70] using the pfam domains ‘PF16197’, ‘PF00109’ and ‘PF02801’ as queries. Subsequently, protein sequences of fatty acid and PKS s previously used for phylogenetic analyses [13,14], as well as sequences from the acariform mite species Tetranychus urticae [48] were downloaded and the same HMMER searches were performed to extract the KS-domain sequences from the respective fasta-files. All putative KS-domain protein sequences were concatenated and sequences shorter than 300 amino acids were removed. All remaining sequences were aligned using MUSCLE [71]; ends were manually inspected and trimmed. The resulting final protein sequence alignment (electronic supplementary material, table S2; GenBank MT849380) was used to construct a maximum-likelihood (ML) phylogenetic tree for the KS domains using the iqtree pipeline [72], including automated model selection. The ML tree was constructed under the LG + R7 model using 1000 bootstrap runs and rooted by the outgroup sequence for mycocerosic acid synthase from Mycobacterium bovis. The similarity of amino acid frequency of the mite and Streptomyces KS-domains was assessed using kmer counting and principal component analysis in R [73].
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Acknowledgements
We thank Roy A. Norton for improving the manuscript and Benjamin Weiss, Dagmar Klebsch and Maximilian Maschler for their technical assistance. We are further grateful to Julian Wagner who helped with bioinformatics.
Ethics
There are no legal restrictions on working with mites.
Data accessibility
The datasets supporting this article have been uploaded as part of the electronic supplementary material.
Authors' contributions
A.B. designed research, performed chemical analysis and transcriptomic work, analysed the data and took lead in drafting the manuscript; M.K. performed molecular and microscopic research, analysed the data and helped drafting the manuscript; M.H. designed research, performed chemical analysis, analysed the data and drafted the manuscript. All authors gave final approval for publication and agree to be held accountable for the work performed therein.
Competing interests
The authors declare no conflict of interest.
Funding
A.B. is a Simons Fellow of the Life Sciences Research Foundation and was previously supported by a PhD scholarship from the German National Academic Foundation. This study was supported by the German Research Foundation (DFG; HE 4593/5-1) to MH, a pilot grant of Caltech's Center for Environmental Microbial Interactions (CEMI; CEMI-19-028) to A.B. and a Consolidator Grant of the European Research Council (ERC CoG 819585 ‘SYMBeetle’) to M.K.
References
- 1.Walsh C, Tang Y. 2017. Natural product biosynthesis: chemical logic and enzymatic machinery. London, UK: Royal Society of Chemistry. [Google Scholar]
- 2.Rocke AJ. 2015. It began with a daydream: the 150th anniversary of the Kekule benzene structure. Angew. Chem. Int. Ed. Engl. 54, 46–50. ( 10.1002/anie.201408034) [DOI] [PubMed] [Google Scholar]
- 3.Hertweck C. 2009. The biosynthetic logic of polyketide diversity. Angew. Chem. Int. Ed. Engl. 48, 4688–4716. ( 10.1002/anie.200806121) [DOI] [PubMed] [Google Scholar]
- 4.El-Sayed AM. 2020. The Pherobase: database of pheromones and semiochemicals. See http://www.pherobase.com.
- 5.Meyer EA, Castellano RK, Diederich F. 2003. Interactions with aromatic rings in chemical and biological recognition. Angew. Chem. Int. Ed. Engl. 42, 1210–1250. ( 10.1002/anie.200390319) [DOI] [PubMed] [Google Scholar]
- 6.Riley KE, Hobza P. 2013. On the importance and origin of aromatic interactions in chemistry and biodisciplines. Acc. Chem. Res. 46, 927–936. ( 10.1021/ar300083h) [DOI] [PubMed] [Google Scholar]
- 7.Brunet P. 1980. The metabolism of the aromatic amino acids concerned in the cross-linking of insect cuticle. Insect Biochem. 10, 467–500. ( 10.1016/0020-1790(80)90082-7) [DOI] [Google Scholar]
- 8.Chen P. 1966. Amino acid and protein metabolism in insect development. In Advances in insect physiology (eds Beament JWL, Treherne JE, Wigglesworth VB), pp. 53–132. Amsterdam, The Netherlands: Elsevier. [Google Scholar]
- 9.Eisner T, Eisner M, Siegler M. 2005. Secret weapons. Cambridge, MA: Harvard University Press. [Google Scholar]
- 10.Morgan ED. 2010. Biosynthesis in insects. Cambridge, UK: RSC Publishing. [Google Scholar]
- 11.Birch AJ, Donovan FW. 1953. Studies in relation to biosynthesis. I. Some possible routes to derivatives of orcinol and phloroglucinol. Aust. J. Chem. 6, 360–368. ( 10.1071/CH9530360) [DOI] [Google Scholar]
- 12.Harris TM, Harris CM. 1977. Synthesis of polyketide-type aromatic natural products by biogenetically modeled routes. Tetrahedron 33, 2159–2185. ( 10.1016/0040-4020(77)80001-X) [DOI] [Google Scholar]
- 13.Castoe TA, Stephens T, Noonan BP, Calestani C. 2007. A novel group of type I polyketide synthases (PKS) in animals and the complex phylogenomics of PKSs. Gene 392, 47–58. ( 10.1016/j.gene.2006.11.005) [DOI] [PubMed] [Google Scholar]
- 14.Cooke TF, et al. 2017. Genetic mapping and biochemical basis of yellow feather pigmentation in budgerigars. Cell 171, 427–439.e421. ( 10.1016/j.cell.2017.08.016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pankewitz F, Hilker M. 2008. Polyketides in insects: ecological role of these widespread chemicals and evolutionary aspects of their biogenesis. Biol. Rev. 83, 209–226. ( 10.1111/j.1469-185X.2008.00040.x) [DOI] [PubMed] [Google Scholar]
- 16.Kellner RL. 1999. What is the basis of pederin polymorphism in Paederus riparius rove beetles? The endosymbiotic hypothesis. Entomo Exp Appl 93, 41–49. ( 10.1046/j.1570-7458.1999.00560.x) [DOI] [Google Scholar]
- 17.Kellner RL. 2001. Suppression of pederin biosynthesis through antibiotic elimination of endosymbionts in Paederus sabaeus. J. Insect. Physiol. 47, 475–483. ( 10.1016/S0022-1910(00)00140-2) [DOI] [PubMed] [Google Scholar]
- 18.Piel J. 2009. Metabolites from symbiotic bacteria. Nat. Prod. Rep. 26, 338–362. ( 10.1039/b703499g) [DOI] [PubMed] [Google Scholar]
- 19.Piel J, Butzke D, Fusetani N, Hui D, Platzer M, Wen G, Matsunaga S. 2005. Exploring the chemistry of uncultivated bacterial symbionts: antitumor polyketides of the pederin family. J. Nat. Prod. 68, 472–479. ( 10.1021/np049612d) [DOI] [PubMed] [Google Scholar]
- 20.Piel J, Hui D, Wen G, Butzke D, Platzer M, Fusetani N, Matsunaga S. 2004. Antitumor polyketide biosynthesis by an uncultivated bacterial symbiont of the marine sponge Theonella swinhoei. Proc. Natl Acad. Sci. USA 101, 16 222–16 227. ( 10.1073/pnas.0405976101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bestmann HJ, Übler E, Hölldobler B. 1997. First biosynthetic studies on trail pheromones in ants. Angew. Chem. Int. Ed. Engl. 36, 395–397. ( 10.1002/anie.199703951) [DOI] [Google Scholar]
- 22.Rocha DF, Wouters FC, Zampieri DS, Brocksom TJ, Machado G, Marsaioli AJ. 2013. Harvestman phenols and benzoquinones: characterisation and biosynthetic pathway. Molecules 18, 11 429–11 451. ( 10.3390/molecules180911429) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun C-M, Toia RF. 1993. Biosynthetic studies on ant metabolites of m-mellein and 2,4-dihydroxyacetophenone from 1,2-13C2 acetate. J. Nat. Prod. 56, 953–956. ( 10.1021/np50096a024) [DOI] [Google Scholar]
- 24.Tecle B, Briophy JJ, Toia RF. 1986. Biosynthesis of 2-hydroxy-6-methylacetophenone in an Australian ponerine ant, Rhytidoponera aciculata (Smith). Insect Biochem. 16, 333–336. ( 10.1016/0020-1790(86)90044-2) [DOI] [Google Scholar]
- 25.Nazareth TM, Sudatti DB, Machado G. 2016. Chemical defense as a condition-dependent trait in harvestmen. J. Chem. Ecol. 42, 1047–1051. ( 10.1007/s10886-016-0749-0) [DOI] [PubMed] [Google Scholar]
- 26.Raspotnig G, Schaider M, Föttinger P, Schönhofer A. 2017. A model for phylogenetic chemosystematics: evolutionary history of quinones in the scent gland secretions of harvestmen. Front. Ecol. Evol. 5, 139 ( 10.3389/fevo.2017.00139) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pankewitz F, Zöllmer A, Gräser Y, Hilker M. 2007. Anthraquinones as defensive compounds in eggs of Galerucini leaf beetles: biosynthesis by the beetles? Arch. Insect. Biochem. Physiol. 66, 98–108. ( 10.1002/arch.20215) [DOI] [PubMed] [Google Scholar]
- 28.Bringmann G, et al. 2006. Different polyketide folding modes converge to an identical molecular architecture. Nat. Chem. Biol. 2, 429 ( 10.1038/nchembio805) [DOI] [PubMed] [Google Scholar]
- 29.Sakata T, Norton RA. 2003. Opisthonotal gland chemistry of a middle-derivative oribatid mite, Archegozetes longisetosus (Acari: Trhypochthoniidae). Int. J. Acarol. 29, 345–350. ( 10.1080/01647950308684351) [DOI] [Google Scholar]
- 30.Brückner A, Heethoff M. 2017. The ontogeny of oil gland chemistry in the oribatid mite Archegozetes longisetosus Aoki (Oribatida, Trhypochthoniidae). Int. J. Acarol. 43, 337–342. ( 10.1080/01647954.2017.1321042) [DOI] [Google Scholar]
- 31.Raspotnig G. 2006. Chemical alarm and defence in the oribatid mite Collohmannia gigantea (Acari: Oribatida). Exp. Appl. Acarol 39, 177–194. ( 10.1007/s10493-006-9015-4) [DOI] [PubMed] [Google Scholar]
- 32.Florez LV, Biedermann PHW, Engl T, Kaltenpoth M. 2015. Defensive symbioses of animals with prokaryotic and eukaryotic microorganisms. Nat. Prod. Rep. 32, 904–936. ( 10.1039/c5np00010f) [DOI] [PubMed] [Google Scholar]
- 33.Raspotnig G, Schuster R, Krisper G. 2003. Functional anatomy of oil glands in Collohmannia gigantea (Acari, Oribatida). Zoomorphology 122, 105–112. ( 10.1007/s00435-003-0075-2) [DOI] [Google Scholar]
- 34.Bergmann P, Heethoff M. 2012. The oviduct is a brood chamber for facultative egg retention in the parthenogenetic oribatid mite Archegozetes longisetosus Aoki (Acari, Oribatida). Tissue Cell 44, 342–350. ( 10.1016/j.tice.2012.05.002) [DOI] [PubMed] [Google Scholar]
- 35.Bergmann P, Heethoff M. 2012. Development of the internal reproductive organs in early nymphal stages of Archegozetes longisetosus Aoki (Acari, Oribatida, Trhypochthoniidae) as obtained by synchrotron X-ray microtomography (SR-μCT) and transmission electron microscopy (TEM). Soil Org. 84, 459–470. ( 10.1016/j.tice.2012.05.002) [DOI] [Google Scholar]
- 36.Bergmann P, Laumann M, Cloetens P, Heethoff M. 2008. Morphology of the internal reproductive organs of Archegozetes longisetosus Aoki (Acari, Oribatida). Soil Org. 80, 171–195. ( 10.1051/acarologia/20132108) [DOI] [Google Scholar]
- 37.Bergmann P, Laumann M, Heethoff M. 2010. Ultrastructural aspects of vitellogenesis in Archegozetes longisetosus Aoki, 1965 (Acari, Oribatida, Trhypochthoniidae). Soil Org. 82, 193–208. ( 10.1051/acarologia/20132108) [DOI] [Google Scholar]
- 38.Laumann M, Bergmann P, Norton RA, Heethoff M. 2010. First cleavages, preblastula and blastula in the parthenogenetic mite Archegozetes longisetosus (Acari, Oribatida) indicate holoblastic rather than superficial cleavage. Arthr. Struc. Dev. 39, 276–286. ( 10.1016/j.asd.2010.02.003) [DOI] [PubMed] [Google Scholar]
- 39.Babst BA, Harding SA, Tsai CJ. 2010. Biosynthesis of phenolic glycosides from phenylpropanoid and benzenoid precursors in populus. J. Chem. Ecol. 36, 286–297. ( 10.1007/s10886-010-9757-7) [DOI] [PubMed] [Google Scholar]
- 40.Bender DA. 2012. Amino acid metabolism. Hoboken, NJ: John Wiley & Sons, Ltd. [Google Scholar]
- 41.Mazelis M. 1980. Amino acid catabolism. In Amino acids and derivatives: a comprehensive treatise (ed. Miflin BJ.), pp. 541–567, Cambridge, MA: Academic Press. [Google Scholar]
- 42.Marchler-Bauer A, et al. 2016. CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucl. Acids Res. 45, D200–D203. ( 10.1093/nar/gkw1129) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bast J, Schaefer I, Schwander T, Maraun M, Scheu S, Kraaijeveld K. 2016. No accumulation of transposable elements in asexual arthropods. Mol. Biol. Evol. 33, 697–706. ( 10.1093/molbev/msv261) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tian X, et al. 2016. Comparative genomics analysis of Streptomyces species reveals their adaptation to the marine environment and their diversity at the genomic level. Front. Microbiol. 7, 998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ermolaeva MD, White O, Salzberg SL. 2001. Prediction of operons in microbial genomes. Nucl. Acid Res. 29, 1216–1221. ( 10.1093/nar/29.5.1216) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brückner A, Parker J. 2020. Molecular evolution of gland cell types and chemical interactions in animals. J. Exp. Biol. 223 ( 10.1242/jeb.211938) [DOI] [PubMed] [Google Scholar]
- 47.Shlyueva D, Stampfel G, Stark A. 2014. Transcriptional enhancers: from properties to genome-wide predictions. Nat Rev Gen 15, 272–286. ( 10.1038/nrg3682) [DOI] [PubMed] [Google Scholar]
- 48.Grbić M, et al. 2011. The genome of Tetranychus urticae reveals herbivorous pest adaptations. Nature 479, 487 ( 10.1038/nature10640) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Husnik F, et al. 2013. Horizontal gene transfer from diverse bacteria to an insect genome enables a tripartite nested mealybug symbiosis. Cell 153, 1567–1578. ( 10.1016/j.cell.2013.05.040) [DOI] [PubMed] [Google Scholar]
- 50.Wybouw N, Dermauw W, Tirry L, Stevens C, Grbić M, Feyereisen R, Van Leeuwen T. 2014. A gene horizontally transferred from bacteria protects arthropods from host plant cyanide poisoning. Elife 3, e02365 ( 10.7554/eLife.02365) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Feldhaar H. 2011. Bacterial symbionts as mediators of ecologically important traits of insect hosts. Physiol Entomo 36, 533–543. ( 10.1111/j.1365-2311.2011.01318.x) [DOI] [Google Scholar]
- 52.Ferrari J, Vavre F. 2011. Bacterial symbionts in insects or the story of communities affecting communities. Philos. Trans. R Soc. Lond. B Biol. Sci. 366, 1389–1400. ( 10.1098/rstb.2010.0226) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gibson CM, Hunter MS. 2010. Extraordinarily widespread and fantastically complex: comparative biology of endosymbiotic bacterial and fungal mutualists of insects. Ecol. Lett. 13, 223–234. ( 10.1111/j.1461-0248.2009.01416.x) [DOI] [PubMed] [Google Scholar]
- 54.Schulz S, Fuhlendorff J, Steidle JL, Collatz J, Franz JT. 2004. Identification and biosynthesis of an aggregation pheromone of the storage mite Chortoglyphus arcuatus. Chembiochem 5, 1500–1507. ( 10.1002/cbic.200400110) [DOI] [PubMed] [Google Scholar]
- 55.Heethoff M, Bergmann P, Laumann M, Norton RA. 2013. The 20th anniversary of a model mite: A review of current knowledge about Archegozetes longisetosus (Acari, Oribatida). Acarologia 53, 353–368. ( 10.1051/acarologia/20132108) [DOI] [Google Scholar]
- 56.Weiss B, Kaltenpoth M. 2016. Bacteriome-localized intracellular symbionts in pollen-feeding beetles of the genus Dasytes (Coleoptera, Dasytidae). Front Microbiol 7, 1486 ( 10.3389/fmicb.2016.01486) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kaltenpoth M, Yildirim E, Gürbüz MF, Herzner G, Strohm E. 2012. Refining the roots of the beewolf-Streptomyces symbiosis: Antennal symbionts in the rare genus Philanthinus (Hymenoptera, Crabronidae). Appl Environ Microbiol 78, 822–827. ( 10.1128/AEM.06809-11) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Amann RI, Binder BJ, Olson RJ, Chisholm SW, Devereux R, Stahl DA. 1990. Combination of 16S ribosomal RNA targeted oligonucleotide probes with flow-cytometry for analyzing mixed microbial populations. App. Environ Microbiol 56, 1919–1925. ( 10.1128/AEM.56.6.1919-1925.1990) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pakwan C, Kaltenpoth M, Weiss B, Chantawannakul P, Jun G, Disayathanoowat T. 2017. Bacterial communities associated with the ectoparasitic mites Varroa destructor and Tropilaelaps mercedesae of the honey bee (Apis mellifera). FEMS Microbiol. Ecol. 94. [DOI] [PubMed] [Google Scholar]
- 60.Kempf VAJ, Trebesius K, Autenrieth IB. 2000. Fluorescent in situ hybridization allows rapid identification of microorganisms in blood cultures. J. Clin. Microbiol. 38, 830–838. ( 10.1128/JCM.38.2.830-838.2000) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Laumann M, Norton RA, Weigmann G, Scheu S, Maraun M, Heethoff M. 2007. Speciation in the parthenogenetic oribatid mite genus Tectocepheus (Acari, Oribatida) as indicated by molecular phylogeny. Pedobiologia 51, 111–122. ( 10.1016/j.pedobi.2007.02.001) [DOI] [Google Scholar]
- 62.Clifford RJ, Milillo M, Prestwood J, Quintero R, Zurawski DV, Kwak YI, Waterman PE, Lesho EP, Mc Gann P. 2012. Detection of bacterial 16S rRNA and identification of four clinically important bacteria by real-time PCR. PLoS ONE 7, e48558 ( 10.1371/journal.pone.0048558) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hammer Ø, Harper DAT, Ryan PD. 2001. PAST: Paleontological statistics software package for education and data analysis. Palaeo Electr. 4, 9. [Google Scholar]
- 64.Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, 10–12. ( 10.14806/ej.17.1.200) [DOI] [Google Scholar]
- 65.Kajitani R, et al. 2014. Efficient de novo assembly of highly heterozygous genomes from whole-genome shotgun short reads. Gen. Res. 24, 1384–1395. ( 10.1101/gr.170720.113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinform 31, 3210–3212. ( 10.1093/bioinformatics/btv351) [DOI] [PubMed] [Google Scholar]
- 67.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinform 29, 15–21. ( 10.1093/bioinformatics/bts635) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Grabherr MG, et al. 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotech. 29, 644 ( 10.1038/nbt.1883) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Haas BJ, et al. 2013. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 8, 1494 ( 10.1038/nprot.2013.084) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Eddy SR. 2011. Accelerated profile HMM searches. PLoS Comp. Biol. 7, e1002195 ( 10.1371/journal.pcbi.1002195) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucl. Acid Res. 32, 1792–1797. ( 10.1093/nar/gkh340) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. 2014. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. ( 10.1093/molbev/msu300) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.R Core Development Team. 2020. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; See http://www.R-project.org. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets supporting this article have been uploaded as part of the electronic supplementary material.