Abstract
Background
Using latent class analysis (LCA), in five randomized control trial (RCT) cohorts, two distinct phenotypes of ARDS have been identified (hypo-inflammatory and hyper-inflammatory). The phenotypes are associated with differential outcomes and treatment response. The objective of this project was to develop parsimonious classifier models for phenotype identification that could be accurate and feasible to use in the clinical setting.
Methods
In this retrospective study, three ARDS network RCT cohorts (ARMA, ALVEOLI, and FACTT) were used as the derivation dataset (N=2022), and a fourth (SAILS) was used as the validation dataset (N=715). LCA-derived phenotypes in all of these cohorts served as the reference standard. Machine-learning algorithms were used to select important classifier variables, which were then used to develop nested logistic regression models. The best logistic regression models based on parsimony and predictive accuracy were then evaluated in the validation dataset. Finally, the models’ prognostic validity was tested in two external ARDS clinical trial datasets (START and HARP-2).
Findings
The six most important classifier variables were IL-8, IL-6, protein C, soluble TNF-receptor-1, bicarbonate, and vasopressor-use. From the nested models, 3-variable (IL-8, bicarbonate, and protein C) and 4-variable models (3-variable plus vasopressor use) were adjudicated to be the best performing. In the validation cohort, both models showed good accuracy (AUC 0·94; 95% CI: 0·92–0·95 and 0·95; 95% CI: 0·93–0·96 respectively). In the external datasets, 3-variable models developed in the derivation dataset identified two phenotypes with distinct clinical features and outcomes consistent with prior findings, including differential survival with simvastatin in HARP-2.
Interpretation
ARDS phenotypes can be accurately identified with simple classifier models using 3–4 variables. Pending the development of real-time testing for key biomarkers and prospective validation, these models could facilitate identification of ARDS phenotypes to enable their application in clinical trials and practice.
Introduction
Despite over 50 years of research, disappointingly few clinical trials in the acute respiratory distress syndrome (ARDS) have resulted in positive findings. A few trials that have succeeded include low tidal volume ventilation, prone-positioning and fluid-conservative strategies.1–3 Tellingly, all these interventions were designed to improve supportive care. No clinical trials testing pharmacological interventions in ARDS have identified a benefit. The broad clinical definition of ARDS and the ensuing heterogeneity in aetiology and pathophysiology coalesced under this definition are increasingly implicated as one of the reasons for these “negative” trials.4 To address the issue of heterogeneity, researchers have recently used latent class analysis (LCA) in ARDS. Two phenotypes, termed hyper-inflammatory and hypo-inflammatory, have been consistently identified in five randomized controlled trials (RCTs) cohorts of ARDS.5–8 Mortality and other clinical outcomes are worse in the hyper-inflammatory phenotype.
Whilst other studies have used clinical data to identify phenotypes in ARDS that may be useful for prognostic enrichment,9,10 LCA-derived ARDS phenotypes also offer the potential for predictive enrichment. In secondary analyses of two RCTs, the LCA-derived phenotypes responded differently to positive-end expiratory pressure (PEEP)5 and fluid therapy6. Recently, in a secondary analysis of the completed Hydroxymethylglutaryl-CoA reductase inhibition with simvastatin in Acute lung injury to Reduce Pulmonary dysfunction (HARP-2) trial,11 a survival benefit was observed in the hyper-inflammatory phenotype in patients randomized to simvastatin compared to placebo.7 No treatment effects were observed in the original RCTs. These findings suggest a potential route for prognostic and predictive enrichment in ARDS trials.
Although these results are promising, key barriers limit the identification of these ARDS phenotypes in clinical practice. Most notably, the complexity of the described LCA models, which can consist of up to 40 predictor variables, renders them impractical for prospective clinical use. The main hypothesis of this study was that a simpler model consisting of a maximum of six variables could accurately classify ARDS phenotypes.
In a prior study, a 3-variable model was shown to identify these phenotypes with good accuracy.6 The model, however, had several limitations. Most pertinently, the model used z-scaled values of the classifier variables, rendering them unsuitable for prospective use because prior knowledge of the variables’ population distribution would be necessary. Additionally, the model was derived using a single RCT cohort and was variably accurate in independent cohorts, suggesting suboptimal stability.6 Further, differential treatment effects observed with the original LCA-models were not observed when patients were classified using this model.
The primary objective of this study was to develop and validate parsimonious models that could ultimately be used prospectively to identify ARDS phenotypes. To improve model performance and increase their generalizability, a combined cohort of three RCTs was used to develop the models. Next, three contemporaneous ARDS RCTs, SAILS (Statins for Acutely injured lungs from Sepsis), HARP-2, and START (Stem cells for ARDS treatment) were used to evaluate model performance. The final objective was to test whether, as with LCA-derived phenotypes, a differential treatment effect with simvastatin was observed in phenotypes determined by these parsimonious models in HARP-2.
Methods
Study Population
Two datasets were generated. The ‘derivation’ dataset was used for variable selection and model development. This dataset combined three NHLBI ARDS Network’s RCTs, namely, ARMA (high versus low tidal volume)1, ALVEOLI (high versus low positive end-expiratory pressure)12, and FACCT (conservative versus liberal fluid management).3 For the primary analysis, the ‘validation dataset’ was used to evaluate the accuracy of two of the ‘best’ performing models from the derivation dataset. The validation dataset was derived from the most contemporaneous NHLBI ARDS Network RCT testing rosuvastatin versus placebo in sepsisrelated ARDS.13 Selected trial and population baseline characteristics are presented in Table S1. Additional details on trial protocols and study populations can be found in the original studies.
Data Synthesis and Analysis
Overview of primary analysis
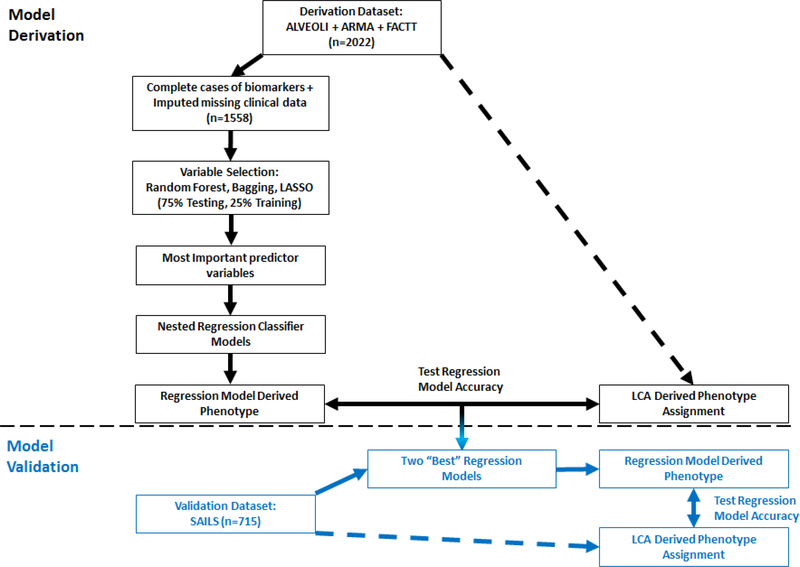
An overview of the primary analysis plan devised a priori is outlined in Figure 1. Briefly, LCA was performed on the derivation dataset (N = 2022), and the resultant phenotypes served as both the dependent variable for machine learning models that were developed for variable selection and as the reference standard to test model performance. The most important variables were, in turn, used to develop nested logistic regression classifier models. Of these, the two ‘best’ models were used for out-of-sample testing in the validation dataset. LCA-derived phenotype assignment in the validation dataset (N = 715) was generated in a prior study and served as the reference standard to test model accuracy.8
Figure 1: Overview of the analysis plan designed a priori for the primary analysis.
The portion of the plan above the dotted line were performed in the derivation dataset (black font) and the portion below the dotted line in the represents the portion of the analysis performed on the validation dataset (blue font).
Latent Class Analysis
We used LCA in the derivation dataset to identify the optimal number of classes that best fit the population. In line with our previous work, we used a combination of demographic, clinical, standard laboratory, and protein biomarkers, all from at or before the time of randomization, as class-defining variables in the models (Table S2).5,6 No clinical outcome variables or severity scores were used in the modeling procedures. Four separate models consisting of one, two, three, and four classes were built. Optimal model selection for the population was judged using the Bayesian information criteria (BIC), Vuong-Lo-Mendell-Rubin (VLMR) likelihood ratio test, the number of observations in the smallest class (classes containing small numbers were not considered clinically meaningful), and entropy. Further details on LCA procedures can be found in the supplementary material.
Selecting Predictor Variables
Two recursive-partitioning machine learning algorithms, classification tree with bootstrapped AGGregatING (bagging) and random forest, were used to identify the most important classifier variables in the derivation dataset. For variable selection, both techniques are known to penalize categorical variables, particularly those with the fewest categories.14 Therefore, a third method, least absolute shrinkage and selection operator (LASSO), was also used to identify important classifier variables. To limit the complexity of the parsimonious models, a priori, a decision was made to limit the maximum number of variables to six for the final modelling.
Prior work indicated that protein biomarkers were likely to be essential components of parsimonious classifier models.6 Only cases with complete biomarker data in the derivation dataset (n=1558) were, therefore, used for variable selection. Multiple imputation with chained equations (MICE) was used to impute missing clinical data in the derivation dataset (see supplementary material for details).
To select the most important variables, a goodness to split score was used for the BAGGING model and the Gini impurity index for the random forest model (see supplementary material for details). For the LASSO modeling, tuning parameter (λ) was sequentially altered such that there were less than eight variables in the final model. The six most important classifier variables common to all three machine learning algorithms were then used to generate nested logistic regression models.
Logistic Regression Models in the Derivation Dataset
The top six variables identified by the machine learning models were used in a forward stepwise regression. Nested logistic regression models of increasing complexity were generated by sequential addition of the variables. The order in which variables were entered into the nested models was determined by findings of stepwise regression analysis.
Model performance was assessed by generating receiver operating characteristic (ROC) curves and calculating area under the curve (AUC) for each model. Akaike information criteria (AIC) and the Youden Index were also generated for each model. Likelihood ratio tests were used to compare nested model performance. In both datasets, data that were not normally distributed were log-transformed (loge) for regression modelling. To test for interaction between outcome and predictor variables, the analysis was repeated by introducing first-order interaction terms to the models.
Model Performance in the Validation Dataset
A priori, a decision was made to take forward two nested logistic regression models and their coefficients from the derivation dataset to test in the validation dataset. The two ‘best’ models were determined by a combination of accuracy in the derivation dataset and model parsimony. These models were used to generate probabilities for phenotype assignment in the validation dataset. For each model, hyper-inflammatory phenotype was assigned using a probability cut-off of either (1) ≥ 0·5 or (2) ≥ Youden Index generated in the derivation dataset. These phenotype assignments were used to calculate sensitivity, specificity, and accuracy of the models. DeLong’s test was used to compare ROC curves and chi-squared test was used to compare model performance. As a sensitivity analysis, the accuracy of alternative classifier models using permutations of the six best predictor variables were also tested in the validation dataset, with each model composed of 3–4 variables.
Model Performance in External Datasets
To test the validity of the models in identifying phenotypes in non-ARDS network RCTs, model performance was evaluated in two recently completed trial datasets. The first RCT used to test model performance was HARP-2. HARP-2 tested the efficacy of simvastatin (80 mg once daily) versus placebo in ARDS.11 The second RCT, the START study, was a phase 2a trial that tested the safety of intravenous human bone marrow-derived mesenchymal stromal cells for moderate to severe ARDS.15 Briefly, START was a double-blind, RCT conducted in five U.S. academic medical centres between 2014–2017. Randomization was based on a 2:1 assignment in favour of the treatment arm. In both studies, clinical and biological data at enrollment were used to assign phenotype using the classifier model developed as above, and outcome data (mortality at day 28, 60, 90 and ventilator-free days to day 28) were used to assess the prognostic validity of phenotype classification.
Assay procedures for plasma biomarker quantification can be found in the original studies.5,7,8 In HARP-2, phenotypes identified by the parsimonious model were evaluated against prior LCAassignment.7 Additionally, randomization data in HARP-2 was used to evaluate treatment interaction with parsimonious model-derived phenotypes and simvastatin. In START, LCA was not performed due to insufficient sample size. The characterization and appropriateness of the identified phenotypes were evaluated using clinical data and outcomes. Details of study protocols and patient populations can be found in the original studies.11
Spearman’s correlation coefficients were calculated to index agreement of class probabilities generated by LCA and parsimonious models. Between-group differences were tested using 2-sample t-test and Mann-Whitney-U-test depending on the distribution of the variable. Difference in outcome in phenotypes were tested using Pearson’s chi-square test. For testing differential response to treatment by class for survival (time to death), time-to-event Kaplan-Meier curves were compared using Wilcoxon test. LCA was performed using Mplus software v8·2. All other analyses were performed using R Studio version 3·3·0.
Role of the Funding Source
Funding sources for this study had no role in study design, data collection, data analysis, interpretation of the data or writing of report. The corresponding author had full access to all of the data and the final responsibility to submit for publication. CSC and KLD also had access to all of the raw data. CMO, DFM and MAM had access to part of the raw data.
Results
Latent Class Analysis
The derivation dataset was comprised of 2022 patients. A 2-class model best fit this dataset. The 2-class model was a significantly improved fit compared to the 1-class model (p<0·0001). Improvement in model fit was not observed when going to a 3-class (p=0·35) or a 4-class model (p=0·13). Good class separation was observed in the 2-class model (entropy=0·83). There were 1431 patients (70·8%) classified as the hypo-inflammatory and 591 (29·2%) as the hyper-inflammatory phenotype. Mean probabilities for class membership were 0.96 (± 0·1) for the hypo-inflammatory class and 0.93 (± 0·1) for the hyper-inflammatory class. The hyper-inflammatory phenotype was associated with higher mortality at day 90 (45% vs 22%, p<0·0001) and with fewer ventilatory-free days (median 3 days, IQR 0 – 19 days vs 20 days, IQR 1 – 24 days; p<0·0001). Key characteristic differences between phenotypes are summarized in Table S2 and are in keeping with prior studies.
Variable Selection (Derivation Dataset)
The most important classifier variables from BAGGING model and Random Forest model are presented in Table S3 and Figure S1 respectively. Using LASSO with a lambda (λ) of 0·1, the seven predictor variables included in the final model were bicarbonate, interleukin-6 (IL-6), interleukin-8 (IL-8), plasminogen-activator inhibitor-1 (PAI-1), protein C, soluble tumournecrosis factor-1 (sTNFR-1), and vasopressors. Bicarbonate, IL-6, IL-8, protein C, sTNFR-1, and vasopressor use were common to all three models and were therefore selected as the six best classifier variables for the parsimonious models.
Multivariate Logistic Regression Models in the Derivation Dataset
Forward stepwise regression failed to eliminate any of the six variables. The six nested models using these variables and their performance in the derivation dataset are summarized in Table 1. No significant interactions were observed when first-order interaction terms were introduced in the models. Increasing model complexity with sequential addition of predictors led to significantly improved model performance (p<0·0001). There was, however, a relative plateauing of AUC and AIC in the 4-variable, 5-variable, and 6-variable models. The 3-variable (IL-8, bicarbonate, and protein C) and 4-variable (IL-8, bicarbonate, protein C, and vasopressor) models were, therefore, considered most optimal in terms of balancing classifying accuracy and model simplicity. The Youden Index generated from the derivation dataset was 0·295 for the 3-variable model and 0·301 for the 4-variable model.
Table 1.
Nested model composition and accuracy in the derivation dataset. IL-8 = interleukin8, sTNFR1= Soluble tumour necrosis factor receptor-1, IL-6 = interleukin-6, AUC = Area under the receiver operator characteristic curve, AIC = Akaike Information Criteria.
| IL-8 | Bicarbonate | Protein C | Vasopressor use | sTNFR1 | IL-6 | AUC | AIC | |
|---|---|---|---|---|---|---|---|---|
| Model 1 | + | - | - | - | - | - | 1268 | |
| Model 2 | + | + | - | - | - | - | 0·92 (0.90–0.93) | 1005 |
| Model 3 | + | + | + | - | - | - | 0·95 (0.93–0.96) | 835 |
| Model 4 | + | + | + | + | - | - | 0·96 (0.95–0.97) | 719 |
| Model 5 | + | + | + | + | + | - | 0·97 (0.96–0.98) | 638 |
| Model 6 | + | + | + | + | + | + | 0·97 (0.97–0.98) | 585 |
CI: Confidence Interval
Model Performance in the Validation Dataset
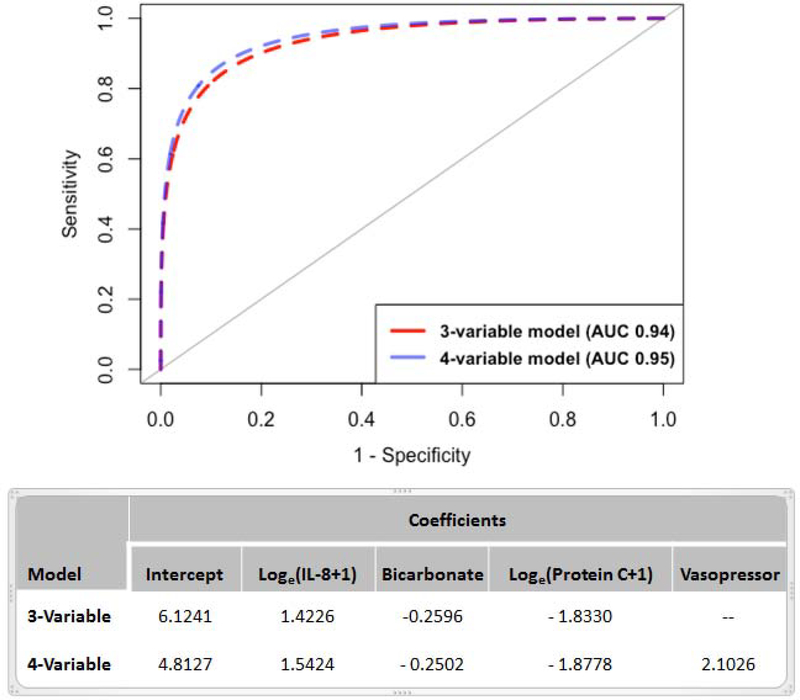
Differences in the baseline characteristics between the derivation and validation datasets are summarized in Table 2. In the validation dataset, AUC for the 3-variable and 4-variable model were 0·94 (95% CI: 0·92–0·95) and 0·95 (95% CI: 0·93–0·96) respectively (Figure 2). Sensitivities and specificities of the models are presented in Table 3. Setting the Youden Index as the probability cut-off to assign phenotype, the 3-variable model had higher specificity compared to the 4-variable model; however, the sensitivity of the 4-variable model was higher. With the probability cut-off set at 0·5, specificity increased in both models to greater than 0·9, with the 3-variable model having higher specificity (0·95).
Table 2.
Comparison of the derivation (ARMA + ALVEOLI + FACTT) and validation (SAILS) datasets in the primary analysis.
| Derivation (complete cases) | Validation (complete cases) | P-Value | ||
|---|---|---|---|---|
| Number of patients | 1558 | 715 | ||
| Sex (Female) | 709 (46%) | 363 (51%) | 0·78b | |
| Race | White | 1107 (71%) | 564 (78·9%) | 0·0001b |
| Non-white | 451 (29%) | 151 (21·1% | ||
| Body mass index kg/m2 | 28·1 ± 7·3 | 30·7 ± 10·1 | <0·0001 | |
| Age (years) | 50·3 ± 16·4 | 54·1 ± 16·3 | <0·0001 | |
| Temperature (°C) | 38·4 ± 1·0 | 38·1 ± 1·0 | <0·0001 | |
| Systolic Blood Pressure (mmHg) | 89 ± 17 | 85 ± 16 | <0·0001 | |
| Heart rate (bpm) | 125 ± 22 | 119 ± 23 | <0·0001 | |
| PaO2/FiO2 ratio | 131 ± 61 | 139 ± 64 | 0·007 | |
| Tidal Volume (mL) | 507 ± 132 | 414 ± 88 | <0·0001 | |
| Minute Ventilation (mL/min) | 12·5 ± 4 | 10·8 ± 3 | <0·0001 | |
| PEEP (cm H2O) | 10 (5–12) | 10 (5 – 10) | 0·90a | |
| PaCO2 (mmHg) | 39 ± 10 | 40 ± 11 | 0·01 | |
| Respiratory rate (breath/min−1) | 33 (26–40) | 32 (27 – 38) | 0·58a | |
| Urine Output (L/24 hr) | 2·2 ± 1·7 | 1·6 ± 1·2 | <0·0001 | |
| Haematocrit (%) | 30 ± 6 | 30 ± 6 | 0·73 | |
| White Blood Cell (103/μL) | 14·4 ± 11 | 15·7 ± 12 | 0·02 | |
| Platelets (103/μL) | 182 ± 121 | 185 ± 121 | 0·58 | |
| Sodium (mmol/L) | 137 ± 6 | 138 ± 5 | 0·006 | |
| Creatinine (mg/dL) | 1·5 ± 1·4 | 1·5 ± 1·2 | 0·98 | |
| Glucose (mg/dL) | 129 ± 61 | 125 ± 46 | 0·06 | |
| Albumin (g/dL) | 2·2 ± 0·6 | 2·2 ± 0·6 | 0·41 | |
| Bilirubin (mg/dL) | 1·6 ± 2·8 | 1·3 ± 1·8 | 0·006 | |
| Bicarbonate (mmol/L) | 21·3 ± 6 | 21·8 ± 6 | 0·08 | |
| Protein C (% control) | 84·8 ± 54 | 80·4 ± 42 | 0·036 | |
| PAI-1 (ng/mL)* | 66 (40 – 110) | 4 (2 – 9) | <0·0001a | |
| Interleukin-6 (pg/mL) | 179 (64 – 567) | 448 (174 –1531) | <0·0001a | |
| Interleukin -8 (pg/mL) | 33 (17 – 83) | 53 (25 – 137) | <0·0001 a | |
| sTNFR1 (pg/mL) | 3964 (2500 – 7176) | 5355 (3090 – 8876) | <0·0001 a | |
| ICAM-1 (ng/mL) | 1074 (632 – 1751) | 360 (236 – 511) | <0·0001 a | |
| APACHE-3 Score | 91·2 (± 30·9) | 93·4 (± 28·2) | 0·10 | |
| ARDS Risk Factors | Trauma | 133 (9%) | 6 (1%) | <0·0001b |
| Sepsis | 369 (24%) | 140 (20%) | ||
| Aspiration | 231 (15%) | 44 (6%) | ||
| Pneumonia | 662 (42%) | 510 (71%) | ||
| Other | 163 (10%) | 15 (2%) | ||
| Vasopressor Use on day of Enrolment(n) | 471 (30·2%) | 394 (55·1%) | <0·0001b | |
| Ventilator Free Days | 17 (0 – 23) | 20 (0 – 25) | 0·0009 a | |
| Mortality at 90 days | 437 (28%) | 199 (27·8%) | 0·96b | |
PAI-1 = Plasminogen activator inhibitor-1, sTNFR1 = soluble tumour necrosis factor-1, ICAM-1 = intercellular adhesion molecule-1.
Observed differences in values in PAI-1 may be due to different assays used for quantification. P-values represent t-test unless annotated (
= Mann-Whitney U test
= chi-squared test)
Figure 2: Receiver operator characteristics (ROC) two best performing regression models in the validation dataset and the respective model coefficients.
3-variable model: IL-8, bicarbonate, and Protein C; 4-variable model: IL-8, bicarbonate, Protein C, and Vasopressor use. AUC = Area under the curve. (Log = logarithm, e = 2·718281, IL-8 = interleukin-8).
Table 3.
Percentage of patients classified as hyperinflammatory subphenotype, sensitivity and specificity of the two ‘best’ models in the validation (SAILS) dataset. A. Probability cut-off set as Youden Index (3-variable: 0·295, 4-variable: 0·301) B. Probability cut off set as 0·5.
| Models | A. Youden Index |
B. Probability ≥ 0·5 |
||||
|---|---|---|---|---|---|---|
| Hyper-Inflammatory | Sensitivity | Specificity | Hyper-Inflammatory | Sensitivity | Specificity | |
|
3-Variable:
IL-8, Bicarbonate, Protein C |
40% | 0·84 | 0·87 | 31% | 0·74 | 0·95 |
|
4-Variable:
IL-8, Bicarbonate, Protein C, Vasopressors |
44% | 0·91 | 0·83 | 36% | 0·82 | 0·91 |
The median probabilities (IQR) generated by the models for belonging to the hyperinflammatory phenotype (cut-off > 0·5) were as follows: 3-variable model 0·85 (IQR 0·68–0·97) and 4-variable model 0·93 (IQR 0·79–0·99). The distribution of probabilities was sparse in the range of 0·3 – 0·7 (Figure S2), suggesting good phenotype discriminatory properties of both the 3 and 4 variable models. The probabilities for phenotype assignment generated by the LCA model showed strong positive correlation with those generated by the parsimonious models (3-variable model r=0·85, 4-variable model r=0·87; p<0·0001 for both).
For the 3-variable model with 0·5 as the probability cut-off, the mean LCA-derived probability was lower for the misclassified subjects compared to the correctly classified subjects in both the hyper-inflammatory (0·88 vs 0·98) and hypo-inflammatory phenotype (0·89 vs 0·96). This finding would suggest that assignment of LCA-derived phenotypes was less certain in subjects misclassified by the parsimonious models. Compared to the hypo-inflammatory phenotype, the hyper-inflammatory phenotype was associated with higher mortality at day 90 (39% vs 23%; p < 0·0001) and fewer ventilator free days (14 days, IQR 0 – 22 days vs 22 days, IQR 0 – 25 days; p < 0·0001). These differences in clinical outcomes were consistent when the four-variable model was used to assign phenotype (data not shown) and when the Youden index was used to assign class in both models (data not shown). Overall, differences in clinical outcomes were similar to the original LCA-derived phenotypes.
Sensitivity Analysis: Ancillary Models
Details of the procedures for the ancillary model development and testing can be found in the supplementary material. The performance of the ancillary models using permutations of the six variables in the validation dataset are presented in Table S4. Most of the ancillary models showed good accuracy in classifying phenotypes, with an AUC ≥ 0·9 in the validation dataset for all models. Replacing Protein C with sTNFR1 resulted in similar AUCs in both the 3-variable and 4-variable models. Replacing IL-8 with IL-6 generally increased model sensitivity, but specificity was lower.
External Validation
HARP-2 Study
In HARP-2, IL-8 and Protein C were not available; therefore, we used an ancillary 3-variable model (Table S4) comprised of IL-6, sTNFR1, and vasopressor use to classify phenotypes in this dataset. 510 of 540 patients had complete data available to estimate classification probabilities. AUC for the model was 0·92 (95% CI: 0·89–0·94). Using the Youden Index from the derivation dataset as the probability cut-off to assign the hyper-inflammatory phenotype (≥ 0·276) resulted in a sensitivity of 0·93 and specificity of 0·62 (Table S5). Using a probability cut-off ≥ 0·5 to assign the hyper-inflammatory phenotype led to a sensitivity of 0·88 and specificity of 0·77.
When a probability cut-off of 0·5 was used for phenotype assignment, 180 (35%) patients were classified as hyper-inflammatory and 328 (65%) patients as hypo-inflammatory. These proportions were similar in comparison to the original LCA-derived phenotypes (Table S5).7 Mortality at day 28 (37% vs 18%; p<0·0001) and at hospital discharge (41% vs 22%; p<0·0001) were significantly higher in the hyper-inflammatory phenotype. The hyperinflammatory phenotype was also associated with fewer ventilator-free days (4 days, IQR 0 – 19 days, vs 17 days, IQR 0 – 23 days; p<0·0001).
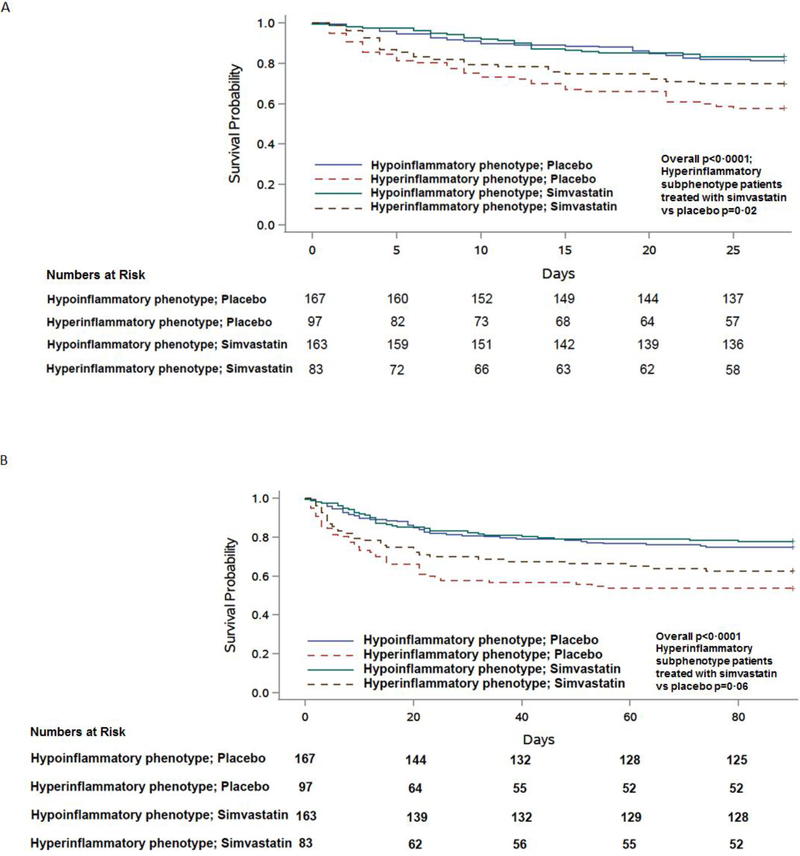
Significantly different survival curves were observed across patients stratified by parsimonious model derived phenotype and treatment (Figure 3, p<0·0001). In the hyper-inflammatory phenotype, compared to placebo, treatment with simvastatin was associated with significantly higher survival at 28 days (p = 0·02). This pattern for survival was also similar at day 90 although the higher observed survival with simvastatin failed to reach statistical significance (overall p<0·0001; p=0·06 for simvastatin compared to placebo in the hyper-inflammatory phenotype). These treatment effects were not observed in the hypo-inflammatory phenotype. Overall, these findings were similar to our prior analysis using LCA-derived phenotypes.7
Figure 3. Kaplan-Meier survival curve in HARP-2 stratified by phenotypes assigned using a 3-variable ancillary parsimonious model (interleukin-6, soluble tumour necrosis factor receptor-1, and vasopressor-use) and treatment (simvastatin or placebo).
Class was assigned using a probability cut-off of ≥ 0·5 to assign phenotype. The number of patients censored at the analysis end-point for each phenotype and treatment level are presented in brackets. A. Censored at 28 days; B. Censored at 90 days.
START Trial
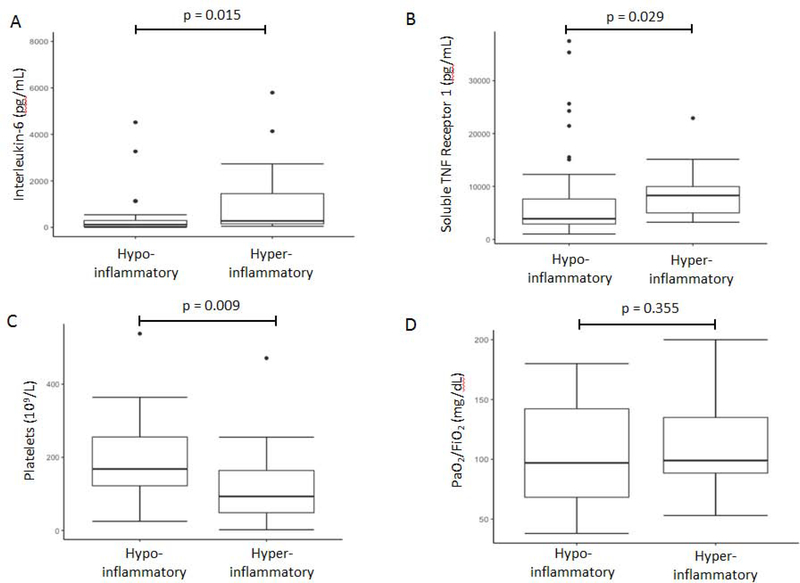
Biomarker data was available for 58 of the 60 patients. The 3-variable and 4-variable models both identified the phenotypes with similar prevalence as in our prior studies (approximately 70% in hypo-inflammatory, 30% in hyper-inflammatory; Table S6). Mortality at day 60 was significantly higher in the hyper-inflammatory group regardless of the model (Table S6). Likewise, as illustrated by the 3-variable model using 0·5 as a cut-off, other metrics of clinical outcome, such as mortality at day 28 (60% vs 14%; p=0·0015) and ventilator-free days (0 days, IQR 0 – 2 days, vs 13 days, IQR 0 – 24 days; p = 0·0040), were also significantly worse in the hyper-inflammatory phenotype. Significant differences in mortality were not observed when patients were stratified by APACHE III score (p=0·13). Aside from clinical outcomes, plasma levels of several inflammatory biomarkers were higher in the hyper-inflammatory phenotype compared to the hypo-inflammatory phenotype (Figure 4A–4B), and platelets were lower in the hyper-inflammatory phenotype (Figure 4C). As with LCA-derived phenotypes, there was no significant difference in PaO2/FiO2 between the identified phenotypes in START (Figure 4D).
Figure 4. Box-Whisker plot depicting difference in key variables between the hyperinflammatory and hypo-inflammatory phenotypes in START using the 3-variable model (Interleukin-8, bicarbonate, and protein C) with a probability cut-off of ≥ 0·5 to assign phenotype.
A. Interleukin-6 (one value not shown in hypo-inflammatory class due to y-axis censoring for visual interpretation) B. Soluble tumour necrosis factor receptor-1 C. Platelet count D. PaO2/FiO2 (P-values are representative of Man-Whitney-U test).
Discussion
Latent class analysis has consistently identified two ARDS phenotypes that show differential outcomes and response to treatment, but the complexity of latent class models has to date rendered ARDS phenotypes inaccessible in the clinical setting. In these analyses, simple classifier models are presented that can accurately identify ARDS phenotypes. The ability to identify phenotypes using a limited set of variables is a critical step towards their clinical application and has important implications for the feasibility of future phenotype-guided clinical trials.
Elevated levels of pro-inflammatory cytokines, such as IL-8, IL-6, and sTNFR1, are known individually to be associated with worse outcomes in ARDS and unsurprisingly emerged as the most important phenotype-defining variables. Protein C, a zymogen with anti-coagulant and anti-inflammatory properties, was also an important variable, and lower levels have been independently associated with increased mortality and adverse outcomes in ARDS.16 Lower levels of bicarbonate in the setting of acute inflammation act as a surrogate for worsening metabolic acidosis, which in turn may reflect tissue hypoxia and dysregulated inflammation. Both Protein C and bicarbonate, therefore, had negative coefficients in the models predicting the hyper-inflammatory phenotype. In comparison to prior studies that have used these variables in isolation to predict outcomes, the presented models developed and validated in this study have the additional benefit of using a composite of these variables and their values relative to each other.
Although the two ‘best’ models both performed with high accuracy, the 3-variable model (IL-8, Bicarbonate, and Protein C) offers some obvious practical advantages for prospective clinical use. The added complexity of the 4-variable model was insufficiently offset by additional accuracy. Moreover, the fourth variable, vasopressor use, is an ambiguous predictor variable. First, it does not factor in dose, thereby providing little insight into severity of shock. Second, the threshold to commence vasopressors in shock varies considerably and is often dictated by institutional, if not individual, discretion.17 Therefore, the 3-variable model which does not incorporate vasopressor use might be preferred. At the same time, given that the 4-variable model itself, and vasopressor use independently, both identify patients with higher mortality 18, it may be potentially valuable in certain ARDS trials.
A priori, a decision was made to compare two probability cut-offs to assign phenotype: the Youden Index from the derivation dataset and 0·5. In all models, the Youden Index cut-off was lower than the 0·5 cut-off and therefore unsurprisingly led to higher sensitivity but lower specificity (Table 3). The proportion of patients with LCA-derived hyper-inflammatory phenotype in the validation dataset and HARP-2 was approximately 35%; this value was more closely matched when using 0·5 as the cut-off in all models. Calculating the Youden Index from the derivation (in-sample) dataset may have led to an over-estimation of model accuracy. In practical terms, the purpose of identifying phenotypes and the potential risk/benefit ratio of the proposed treatment strategy may ultimately dictate the best cut-off. For example, in a trial of a low risk intervention, it may be reasonable to accept lower specificity in order to enhance sensitivity, whereas when studying a higher risk intervention, it might be more important to maximize specificity. Prospectively conducted studies are needed to further test optimal probability cut-offs.
In addition to need for prospective validation, immediate implementation of these models is limited by the lack of a real-time test for biomarker quantification. To our knowledge, there are no commercially available point-of-care or real-time quantifiable assays for IL-8, protein C, or sTNFR1. The current study adds to the increasing weight of evidence which suggests that rapid measurement of plasma protein biomarkers may be crucial in delivering precision-based care in critical illness.19 Recently, the NHLBI convened a multidisciplinary working group to discuss the development of rapid biomarker testing in cardiovascular medicine.20 Similar initiatives in critical care would be timely and essential to shift from the current over-reliance on a ‘one-size fits all’ approach to treating syndromes such as sepsis and ARDS.
Keeping this limitation in mind, we adopted a pragmatic view on model development and sought to develop and evaluate ancillary models using permutations of the six most important variables (Table S4). Most of these models were sufficiently accurate; however, those based on IL-6 were more sensitive and less specific for identifying the hyper-inflammatory phenotype compared to IL-8 based models. One of the ancillary models afforded the opportunity to test its accuracy in the HARP-2 trial, where only a limited set of variables were available for phenotyping. The ability of one of the least accurate ancillary models to not only identify phenotypes in HARP-2 but also to detect the disparate treatment effect in this dataset supports the robustness of the findings and their potential validity in trial cohorts beyond the ARDS network. The performance of the 3-variable model in the START trial adds face-validity to this argument. In START, albeit in a small cohort, the models identified phenotypes that were distinct from each other and also had vastly divergent clinical outcomes (Table S6). More importantly, when stratified by APACHE III score, the same differences in mortality were not observed in the phenotypes, suggesting that the severity of illness identified by phenotypes cannot be extracted from standard measures of severity. Pending rapid biomarker quantification, these models offer a simple and unique method for prognostic, and potentially, predictive enrichment.
This study has several strengths. First, we used four large RCT cohorts, where, in order to avoid overfitting, the validation cohort was kept completely naïve to model development. Additionally, the derivation dataset was the largest in which we have applied LCA. The finding that in this population the two-phenotype model was the optimal fit suggests that ARDS phenotypes are consistent despite changing practice over two decades and across diverse populations. Second, the validation dataset was a contemporary trial of infection-associated ARDS and had a higher incidence of hyper-inflammatory phenotype and significantly different levels of biomarkers and vasopressor-use (Table 2). Despite these key differences in the derivation and validation datasets, the parsimonious models performed with high accuracy in the latter. Taken together with the model performance in the START and HARP-2 trials, the results suggest that the models are likely to be generalizable to other clinical trial populations in ARDS and robust to changes in assays and clinical practice over time, though prospective validation will still be required.
The study also has several limitations. All the presented data are secondary analyses of previously conducted RCTs. Interpretation of the performance of parsimonious models must, therefore, be limited to trial populations. These models must be evaluated in observational cohorts and prospectively before they can be generalized to the ARDS population and used in the clinical setting.
A further limitation is that all the studies used for this analysis except for the START trial were conducted prior to 2014. Since then, prone-positioning has been shown to be beneficial in select populations of ARDS and is now in widespread use for some severe ARDS patients.2 We were unable to test the impact of prone-positioning on phenotype allocation due to lack of data on this therapy. Additionally, the SAILS cohort represents a specific subset of ARDS patients with infection/sepsis, albeit a subset that makes up the majority of ARDS patients.
In addition, the time from ARDS diagnosis to enrollment was different among cohorts (TABLE S1). This variability may have resulted in clinical management strategies playing an important, yet undetermined, role in patient phenotype. A prior study by our group has reported that phenotypes remained stable over a period of 72 hours, suggesting limited impact of management strategy on patient phenotype in this time frame.21 Due to the retrospective study design, however, it not feasible to ascertain the extent to which ventilatory and other management strategies leading up to enrollment altered the inflammatory response in these patients.
The limited sets of variables available in HARP-2 meant that the accuracy of the two primary classifier models was not tested in this dataset. A further limitation of this study is that heterogeneous treatment interaction with phenotype assignment using the parsimonious model was only tested in HARP-2. Differential treatment responses in FACTT and ALVEOLI were not evaluated because both studies served as the derivation dataset, and positive results would be subject to bias and data circularity.
What are some of the key knowledge gaps in the field going forward? Currently, identification of ARDS phenotypes using LCA has been limited to patients enrolled in RCTs, so it is unknown whether these phenotypes are generalizable to broader ARDS populations. Further, it is also not known whether these phenotypes may be identifiable in critical care clinical syndromes beyond ARDS. In particular, given that SAILS was a sepsis-associated cohort, these phenotypes may be applicable to sepsis. In order to fully realise the potential of these phenotypes to deliver precision-based care in ARDS, a better understanding is needed of the underlying biology of the phenotypes; this objective will require more experimental research. Additionally, a better understanding of the longitudinal kinetics of the phenotypes and their response to interventions is needed. For example, the diagnosis of ARDS itself is known to be volatile to standard ventilatory practice in the first 24-hours;22 whether these changes are specific to either phenotype or impact phenotype assignment themselves is unknown.
In summary, this study provides evidence for accurate parsimonious classifier models for ARDS phenotypes. These simple models may facilitate the study of phenotypes in the prospective setting and improve selection of patients for clinical trials.
Supplementary Material
Research in context.
Evidence before the study
Using latent class analysis (LCA), previous studies have consistently identified two phenotypes across five randomised controlled trial (RCT) cohorts of acute respiratory distress syndrome (ARDS). The phenotypes have distinct biological and clinical characteristics with divergent clinical outcomes and differential responses to therapy in secondary analyses of randomized controlled trials. The complexity of the LCA models that identify the phenotypes is a major impediment to their application in the clinical setting. Whether parsimonious models using a selection of key variables could be used to identify the two ARDS phenotypes remains unknown.
Added value of this study
Using an array of machine learning algorithms, the presented study identifies parsimonious models comprised of three to four variables that can accurately classify ARDS phenotypes in two validation cohorts. The phenotypes identified using these parsimonious models shared similar characteristics and outcomes to phenotypes identified using LCA. The survival benefit observed with simvastatin in a prior analysis was also observed in the hyper-inflammatory phenotype identified using the parsimonious model. In a recent trial testing the efficacy of mesenchymal stem cells in ARDS, the hyper-inflammatory phenotype identified by parsimonious models was associated with significantly higher mortality at day 60.
Implication of all the available evidence
Heterogeneity in ARDS is increasingly being recognized as a potential contributing factor to failed clinical trials. LCA-identified phenotypes offer researchers more biologically and clinically uniform subgroups to test hypotheses and interventions. With the simpler models described in this study, identification of the phenotypes may become more feasible and may herald a new era of prospective, phenotype-specific trials in ARDS.
Acknowledgements
“We thank all patients who participated in the trial and their legal representatives, all research nurses and pharmacists in the participating centres, and medical and nursing staff in participating centres who cared for patients and collected data. For the HARP-2 trial we thank the staff of the Northern Ireland Clinical Trials Unit for their support in conducting the trial, the staff from the Health Research Board Galway Clinical Research Facility, Galway, Ireland, for help in conducting the study in Ireland, the staff of the Northern Ireland Clinical Research Network and the NIHR Clinical Research Network for help with patient recruitment and data acquisition, and the UK Intensive Care Society. Work on the HARP-2 study was supported by the UK EME Programme, an MRC and NIHR partnership (08/99/08 and 16/33/01), by a Health Research Award (HRA_POR-2010-131) from the Health Research Board, Dublin; and by additional funding from the HSC Research and Development Division, Public Health Agency for Northern Ireland, the Intensive Care Society of Ireland, and REVIVE. The EME Programme is funded by the MRC and NIHR, with contributions from the Chief Scientist Office in Scotland, the National Institute for Social Care and Health Research in Wales, and the Health and Social Care (HSC) Research and Development Division, Public Health Agency for Northern Ireland. The views expressed in this article are those of the authors and not necessarily those of the Medical Research Council (MRC), National Health Service, NIHR, or Department of Health.”
Funding: HL131621, HL133390, HL140026 (CSC), GM008440-21 (PS)
Declaration of Conflicts
PS was supported by the National Institute of Health during the period that the study was conducted. CSC reports grants from NIH, during the conduct of the study; grants from GlaxoSmithKline, grants and personal fees from Bayer, personal fees from Prometic, personal fees from Roche/Genentech, personal fees from CSL Behring, personal fees from Quark, outside the submitted work. CMO reports grants from NIHR EME during the conduct of the study. In addition, she has received grant funding from NIHR, Wellcome Trust, NI HSC R&D and other funders for ARDS studies. CMO reports her spouse has received consultancy fees from GlaxoSmithKline, Bayer and Boehringer Ingelheim outside the submitted work. DFM reports grants from NIHR EME, HRB, Northern Ireland Public Health Agency Research and Development, ICSI and REVIVE for the conduct of the HARP-2 study. Outside the submitted work, DFM reports personal fees from consultancy for GlaxoSmithKline, Boehringer Ingelheim and Bayer. In addition, his institution has received funds from grants from the UK NIHR, Wellcome Trust, Innovate UK and others. In addition, DFM has a patent issued to his institution for a treatment for ARDS. DFM is a Director of Research for the Intensive Care Society and NIHR EME Programme Director. MAM reports grants from NIH/NHLB I, grants from Department of Defense, grants from Bayer Pharmaceuticals, grants from GlaxoSmithKline and personal fees from Cerus Therapeutics, outside the submitted work. KLD has nothing to disclose.
Footnotes
This article has an online data supplement.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference
- 1.Acute Respiratory Distress Syndrome N, Brower RG, Matthay MA, et al. Ventilation with lower tidal volumes as compared with traditional tidal volumes for acute lung injury and the acute respiratory distress syndrome. N Engl J Med 2000; 342(18): 1301–8. [DOI] [PubMed] [Google Scholar]
- 2.Guerin C, Reignier J, Richard JC, et al. Prone positioning in severe acute respiratory distress syndrome. N Engl J Med 2013; 368(23): 2159–68. [DOI] [PubMed] [Google Scholar]
- 3.National Heart L, Blood Institute Acute Respiratory Distress Syndrome Clinical Trials N, Wiedemann HP, et al. Comparison of two fluid-management strategies in acute lung injury. N Engl J Med 2006; 354(24): 2564–75. [DOI] [PubMed] [Google Scholar]
- 4.Matthay MA, McAuley DF, Ware LB. Clinical trials in acute respiratory distress syndrome: challenges and opportunities. Lancet Respir Med 2017; 5(6): 524–34. [DOI] [PubMed] [Google Scholar]
- 5.Calfee CS, Delucchi K, Parsons PE, et al. Subphenotypes in acute respiratory distress syndrome: latent class analysis of data from two randomised controlled trials. Lancet Respir Med 2014; 2(8): 611–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Famous KR, Delucchi K, Ware LB, et al. Acute Respiratory Distress Syndrome Subphenotypes Respond Differently to Randomized Fluid Management Strategy. Am J Respir Crit Care Med 2017; 195(3): 331–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Calfee CS, Delucchi KL, Sinha P, et al. Acute respiratory distress syndrome subphenotypes and differential response to simvastatin: secondary analysis of a randomised controlled trial. Lancet Respir Med 2018; 6(9): 691–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sinha P, Delucchi KL, Thompson BT, et al. Latent class analysis of ARDS subphenotypes: a secondary analysis of the statins for acutely injured lungs from sepsis (SAILS) study. Intensive Care Med 2018; 44(11): 1859–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Villar J, Ambros A, Mosteiro F, et al. A Prognostic Enrichment Strategy for Selection of Patients With Acute Respiratory Distress Syndrome in Clinical Trials. Crit Care Med 2019; 47(3): 377–85. [DOI] [PubMed] [Google Scholar]
- 10.Sinha P, Calfee CS, Beitler JR, et al. Physiologic Analysis and Clinical Performance of the Ventilatory Ratio in Acute Respiratory Distress Syndrome. Am J Respir Crit Care Med 2019; 199(3): 333–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McAuley DF, Laffey JG, O’Kane CM, et al. Simvastatin in the acute respiratory distress syndrome. N Engl J Med 2014; 371(18): 1695–703. [DOI] [PubMed] [Google Scholar]
- 12.Brower RG, Lanken PN, MacIntyre N, et al. Higher versus lower positive end-expiratory pressures in patients with the acute respiratory distress syndrome. N Engl J Med 2004; 351(4): 327–36. [DOI] [PubMed] [Google Scholar]
- 13.National Heart L, Blood Institute ACTN, Truwit JD, et al. Rosuvastatin for sepsis-associated acute respiratory distress syndrome. N Engl J Med 2014; 370(23): 2191–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Altmann A, Tolosi L, Sander O, Lengauer T. Permutation importance: a corrected feature importance measure. Bioinformatics 2010; 26(10): 1340–7. [DOI] [PubMed] [Google Scholar]
- 15.Matthay MA, Calfee CS, Zhuo H, et al. Treatment with allogeneic mesenchymal stromal cells for moderate to severe acute respiratory distress syndrome (START study): a randomised phase 2a safety trial. Lancet Respir Med 2019; 7(2): 154–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Terpstra ML, Aman J, van Nieuw Amerongen GP, Groeneveld AB. Plasma biomarkers for acute respiratory distress syndrome: a systematic review and meta-analysis*. Crit Care Med 2014; 42(3): 691–700. [DOI] [PubMed] [Google Scholar]
- 17.Bangash MN, Kong ML, Pearse RM. Use of inotropes and vasopressor agents in critically ill patients. Br J Pharmacol 2012; 165(7): 2015–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.National Heart L, Blood Institute Acute Respiratory Distress Syndrome Clinical Trials N, Matthay MA, et al. Randomized, placebo-controlled clinical trial of an aerosolized beta(2)-agonist for treatment of acute lung injury. Am J Respir Crit Care Med 2011; 184(5): 561–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garcia-Laorden MI, Lorente JA, Flores C, Slutsky AS, Villar J. Biomarkers for the acute respiratory distress syndrome: how to make the diagnosis more precise. Ann Transl Med 2017; 5(14): 283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.King K, Grazette LP, Paltoo DN, et al. Point-of-Care Technologies for Precision Cardiovascular Care and Clinical Research: National Heart, Lung, and Blood Institute Working Group. JACC Basic Transl Sci 2016; 1(1–2): 73–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Delucchi K, Famous KR, Ware LB, et al. Stability of ARDS subphenotypes over time in two randomised controlled trials. Thorax 2018; 73(5): 439–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Villar J, Perez-Mendez L, Lopez J, et al. An early PEEP/FIO2 trial identifies different degrees of lung injury in patients with acute respiratory distress syndrome. Am J Respir Crit Care Med 2007; 176(8): 795–804. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.