Abstract
S-adenosyl-l-methionine dependent methyltransferases catalyze methyl transfers onto a wide variety of target molecules, including DNA and RNA. We discuss a family of methyltransferases, those that act on the amino groups of adenine or cytosine in DNA, have conserved motifs in a particular order in their amino acid sequence, and are referred to as class beta MTases. Members of this class include M.EcoGII and M.EcoP15I from Escherichia coli, Caulobacter crescentus cell cycle–regulated DNA methyltransferase (CcrM), the MTA1-MTA9 complex from the ciliate Oxytricha, and the mammalian MettL3-MettL14 complex. These methyltransferases all generate N6-methyladenine in DNA, with some members having activity on single-stranded DNA as well as RNA. The beta class of methyltransferases has a unique multimeric feature, forming either homo- or hetero-dimers, allowing the enzyme to use division of labor between two subunits in terms of substrate recognition and methylation. We suggest that M.EcoGII may represent an ancestral form of these enzymes, as its activity is independent of the nucleic acid type (RNA or DNA), its strandedness (single or double), and its sequence (aside from the target adenine).
INTRODUCTION
MTase families defined by amino acid sequences
Early analysis, of the amino acid sequences of 13 bacterial DNA methyltransferases (MTases) generating 5-methylcytosine (5mC), revealed a set of ten conserved blocks of amino acid residues (1). These conserved motifs, numbered I to X from amino to carboxyl end, were found to have a constant linear order, simplifying their identification in protein sequences (particularly for the shorter or less-conserved motifs), though one alternative permutation of 5mC MTase motif order was later found (2,3). This aided in the discovery of mammalian 5mC-generating DNA methyltransferases Dnmt1 (4) and Dnmt3 (5). The structural analysis of HhaI MTase, from the bacterium Haemophilus haemolyticus, allowed the functions of conserved sequence motifs to be inferred, particularly those responsible for SAM binding (motif I consensus: FxGxG) and for the chemistry of methyltransfer reaction on an inert carbon (motif IV consensus: PC), along with a varied target recognition domain (TRD) for specific binding of the substrate DNA sequence (6,7).
Not surprisingly, the same set of sequence motifs also occur in mammalian Dnmt2 (8–10), a tRNA 5mC MTase (11). The conservation in Dnmt sequence and structure reflects the conserved nature of SAM binding, which occurs so as to optimize the catalysis of methyl transfer onto cytosine-C5 in nucleic acids. Before and after the discovery that Dnmt2 homologs are actually tRNA MTases, a number of studies on the activity of Dnmt2 on DNA yielded kinetic rate constants varying between zero and very low, though the human enzyme can methylate cytidine in a short segment of single-stranded DNA ligated into a tRNA molecule (12).
In addition to cytosine-C5, methylated bases on DNA include N4-methylcytosine (N4mC) and N6-methyladenine (N6mA), both of which are modified on their exocyclic primary amines. Bacteriophages, particularly lytic ones, modify their DNA in many additional ways, including other methylations (13), but our focus here is on methylations that are consistent with normal cell physiology. Recognition of the base to be methylated, and access to it by catalytic residues, occurs after the base is swung out of the DNA duplex into a typical concave binding pocket, in a process known as ‘base flipping’ (14–16). Interestingly, the DNA MTases that generate N4mC and N6mA do not group separately from one another on the basis of sequence (17). This highlights the constraints of the chemistry of amino methylation as being evolutionarily dominant, instead of recognition of cytosine versus adenine in the active-site binding pocket. In fact, there is evidence of independent derivations of N4mC MTases from N6mA MTases (18), and at least one MTase that generates N4mC can generate N6mA on a DNA substrate in which a C-to-A substitution is made at the target base (19).
A multiple sequence alignment of 42 known DNA amino MTases revealed the existence of (at least) three classes of amino MTases, differing from one another by circular permutation in their order of motifs important for three essential functions: binding the methyl donor SAM, binding substrate DNA and catalyzing the chemical reaction between the donor and substrate (20) (Figure 1). Briefly, class α is arranged in the order (N-to-C termini): motif I (SAM-binding), TRD (substrate recognition and binding), and motif IV (methylation catalysis). Class β is arranged in the order: motif IV-TRD-motif I. Class γ is arranged in the order: motif I-motif IV-TRD. The class γ amino MTases, exemplified by M.TaqI (21,22), have the motif order comparable to that of the 5mC MTases (23). The residues of motif IV vary with the target atom—the PC (proline-cysteine) motif in 5mC MTases is responsible for ring carbon-C5 methylation of cytosine, with the Cys sulfhydryl acting as a nucleophile to attack the cytosine ring and initiate the reaction (24–27). In contrast, motif IV for MTases catalyzing exocyclic amino methylation to generate N6mA or N4mC have the four-residue consensus (D/N/S)-P-P-(Y/F/W) [(Asp/Asn/Ser)-Pro-Pro-(Tyr/Phe/Trp)]; where the last bulky hydrophobic side chain stacks against the target base, while the first one abstracts a proton from the target amino group (22,28).
Figure 1.
Classes of amino-MTases. (A) A duplication model of SAM-binding motif I, with either of the duplicated regions diverging prior to introduction of the target recognition domain (TRD) and serving as the ancestor for either the α and γ or the β classes. (B) Schematic of three classes of MTases with altered orders of motifs responsible for SAM binding (motif I), methylation substrate binding (TRD) and catalysis (motif IV). The regions containing motifs I and IV are folded into a seven-stranded sheet. This linear representation does not reflect the fact that, in the three-dimensional structure of just class β MTases, the TRD is oriented oppositely to motifs I/IV. (C) Relative numbers of each class of MTases were obtained from the REBASE database as of spring 2020 (44).
As was illustrated by Dnmt2, the apparent sequence and structural similarities do not reveal with certainty whether an enzyme acts on a particular methylation substrate (DNA, RNA or protein). Other examples include Escherichia coli HemK and human HemK2, which were thought to be DNA N6mA MTases (29,30), yet were found to be protein MTases active on glutamine (31–36). The human HemK2 is also active as a histone lysine MTase (37). The common feature of the potential substrates is the amino group (NH2) of protein glutamine or lysine (in the deprotonated state) or DNA adenine, at which all three methylation reactions are catalyzed by a NPPY motif (a subset of the motif IV possibilities).
While the MTase families differ in motif order, their structures are remarkably similar, comprising a seven-stranded β sheet (1-to-7) with a central topological switch-point between strands β1 and β4, and a characteristic reversed β hairpin (β6 and β7) at one end of the sheet next to strand β5 (Figure 2A and B). This topology allows for circular permutation, where the same structure simply has the break between its amino beginning and carboxyl end at different points (38) and, in fact, it is possible to circularly permute a MTase in the laboratory with retention of function (3). The two most significant motifs – motif I (FxGxG) for binding SAM and motif IV (DPPY) for binding substrate Ade – juxtapose the target N6 atom of Ade in line with the methyl group and sulfur atom of SAM for catalysis, and are positioned at the carboxyl ends of the two parallel neighboring strands β1 and β4 (Figure 2C). Based on the chemical similarity of the SAM–adenosyl and DNA–adenosyl moieties, it makes sense that the two structural elements responsible for the binding (β1–β2 and β4–β5) do, in fact, have striking similarity (20). It was suggested that the original MTases arose after tandem gene duplication (39) converted a SAM-binding domain into a protein that bound two molecules of SAM (20), and there is some evidence to support that hypothesis (40).
Figure 2.
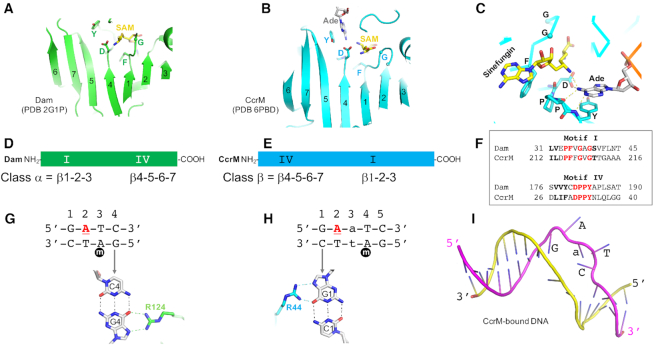
Comparison of Dam (class α) and CcrM (class β). (A, B) The catalytic domain of the seven-stranded β structure in Dam (panel A) and CcrM (panel B). (C) Interactions with the SAM–adenosyl and DNA–adenosyl moieties in the active-site of CcrM. The phenylalanine of motif I provides an edge-to-face interaction to the face of SAM-adenosyl ring. The DPPY motif interacts with DNA adenine. (D–F) The locations of motif I and motif IV are reversed in the amino acid sequences of Dam and CcrM. (G) Dam interacts with guanine G4 of the non-target strand. (H) CcrM interacts with guanine G1 of the target strand. The underlined letter A in red is the methylation target. (I) Two strand separation in the CcrM-bound DNA.
Based on the proposal that the ancestral tandem duplication had two copies of motif I (represented as I–I), one evolutionary model is that the two halves diverged with one SAM-binding motif I changing to an adenine-binding motif IV (Figure 1A). If this change involved the amino-proximal motif I, it would yield the ancestor to Class β MTases (IV–I), while conversion of the carboxyl-proximal SAM binding site would yield the ancestor to Class α and γ MTases (I–IV). An alternative would be that just one of the two changed, with the other motif order occurring subsequently via a true circular permutation event (38,41) (not shown). Presumably, the ancestral MTase(s) as described here would methylate free adenine or a free adenine nucleotide, perhaps contributing to pre-protein metabolism (42,43).
This ancestral nucleotide MTase could then have gained the ability to modify DNA or RNA adenines following an additional fusion of the target (nucleic acid substrate) recognition domain (TRD, Figure 1B). The three classes are roughly the same size in number of known members (Figure 1C). While the majority of 5mC and N4mC DNA MTases each fall into single classes defined by motif order, the N6mA MTases are fairly evenly distributed among the three classes (α, β and γ) (20). This could also be explained by the tandem duplication model in which the ancestral nucleic acid MTase(s) generated N6mA. In addition to considering MTase evolution, we also note here that the β arrangement of motifs has intrinsic structural consequences that, we suggest, impose a unique requirement for this family of MTases to dimerize in order to function. We begin by comparing two MTases that play similar biochemical and physiological roles, but one of them is a β MTase and the other is not.
Escherichia coli Dam versus Caulobacter crescentus CcrM (monomer vs. dimer)
The great majority of bacterial and archaeal DNA MTases are associated with restriction–modification (RM) systems, where the MTase protects a cell's own DNA from digestion by the paired (cognate) restriction endonuclease (44,45). RM systems are important for defense against bacteriophage predation (46,47), although they play other roles as well (48,49). Bacterial ‘orphan’ MTases are so named as, unlike most bacterial DNA MTases, they are not paired with a restriction endonuclease as part of a RM system (50). Orphan MTases are sometimes involved in chromosome replication, DNA repair, and epigenetic gene regulation (51). Examples of such regulatory orphan MTases include the DNA adenine MTase (Dam) in Escherichia coli (Gammaproteobacteria) and cell cycle–regulated DNA MTase (CcrM) in Caulobacter crescentus (Alphaproteobacteria) which are, respectively, responsible for maintenance of daughter strand adenine methylation at GATC or GAnTC sequences (n = any nucleotide), immediately after their replication (52,53). We will first compare these two ‘orphan’ enzymes, and then consider β MTases associated with RM systems, and finally examine the mammalian β MTases.
Although the substrate sequences of Dam (GATC) and CcrM (GAnTC) are both palindromic, both enzymes act on hemi-methylated DNA substrates (Figure 2G-H) – that is, sequences that already contain N6mA in one strand. Both enzymes can act on unmethylated sites, but most often encounter hemimethylated sites resulting from replication of fully-methylated sites, and both enzymes show a preference for hemimethylated over unmethylated sites (54,55).
Besides the aforementioned difference in the order of motifs (Dam is a member of class α and CcrM is a class β MTase), there are three major differences between the two enzymes. First, though both enzymes use an arginine to interact with a 5′ guanine (Figure 2G and H) and both Arg-Gua interactions are sequence-discriminatory contacts, these contacts are made to different strands relative to the substrate Ade. In Dam, the recognized Gua is on the opposite strand, and 2-base pairs away, from the target Ade; whereas in CcrM, the recognized Gua is on the same strand as, and adjacent to, the target Ade. Second, except for the flipped-out target adenine, the Dam-bound DNA conformation has intact intra-helical paired bases, whereas CcrM pulls the two DNA strands apart, creating a bubble comprising four enzyme-recognized, unpaired bases (Figure 2I). These two features of CcrM, strand separation and base recognition on the same strand that contains the target Ade, allow CcrM (but not Dam) to be active on both double-stranded (ds) and single-stranded (ss) DNA, as well as on mismatches within or immediately outside of the recognition sequence (56). Neither enzyme is active on ssRNA. Lastly, Dam is a monomer under most conditions (57,58), while CcrM is a homodimer (Figure 3A) (59), reflecting a broader phenomenon that we will discuss next.
Figure 3.
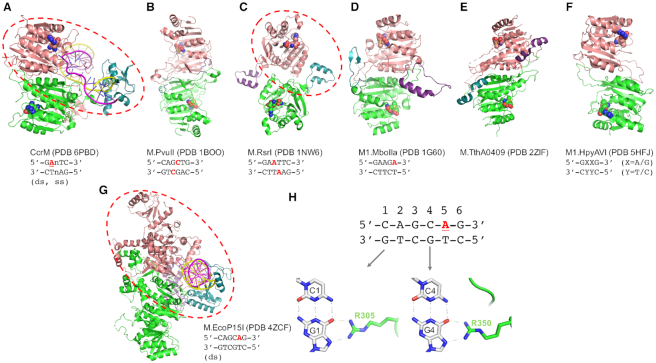
Dimeric structures of class β MTases. (A) CcrM-DNA complex. (B) M.PvuII. (C) M.RsrI, (D) M1.MboII. (E) M.TthA0409 from Thermus thermophilus HB8. (F) M1.HpyAVI from Helicobacter pylori. (G) M.EcoP15I-DNA complex. (H) M.EcoP15I interacts with guanines G1 and G4 of the non-target strand. The dashed red ovals in panels A, C and G indicate the DNA duplex binding region.
Are class-β MTases required to form homodimers for activity?
The homodimeric subunit structure of CcrM has been observed previously in other MTases of structurally-characterized class β MTases. M.PvuII of Proteus vulgaris is (at this writing) the only structurally-characterized N4mC MTase (60), and yielded the first structure for a MTase in the β family. It methylates the central cytosine in its symmetric recognition sequence 5′-CAGCTG-3′. The M.PvuII structure, consistent with the circular permutation model, shares a common fold with MTases of other families, while having the major functional regions (particularly motifs I and IV) permuted into distinct linear order. The major unexpected finding was that M.PvuII forms a homodimer (Figure 3B). This dimeric feature was subsequently observed by others in other β MTase structures. M.RsrI, from Rhodobacter sphaeroides (61,62) (Figure 3C), methylates the internal adenine of the palindromic DNA sequence GAATTC, and its dimeric structure was consistent with earlier biochemical evidence for this MTase (63,64). M1.MboII, from Moraxella bovis (Figure 3D), methylates the 3′ adenine of an asymmetric sequence 5′-GAAGA-3′, and the homodimeric structure again supported by biochemical results (65). Dimer formation was also observed in structures for THA0409 from Thermus thermophilus HB8 (Figure 3E) (66) and M1.HpyAVI from Helicobacter pylori (Figure 3F) (67). Furthermore, even where structures do not yet exist, there is biochemical evidence for dimerization in several other class β MTases (M.BamHI (68), M.LiaCI (69), M.KpnI (70,71), M.HpyAXVII (72)). However, none of these class β MTases were structurally characterized in complex with DNA, and their postulated TRDs were disordered in the absence of bound DNA, so the significance of the dimeric character was not grasped until the structure determination of M.EcoP15I.
‘Division of labor’ between two dimeric subunits of M.EcoP15I homodimers
M.EcoP15I is an archetype of the Type III RM family, and consists of two methylation (M) and 0–2 restriction (R) subunits (73), resulting in M2, M2R1 or M2R2 complexes. The M.EcoP15I M-subunit functions as a dimer (74,75) but recognizes an asymmetric sequence, 5′-CAGCAG-3′ and methylates the 3′ internal adenine of the A-containing strand of the dsDNA (the complementary strand is 5′-CTGCTG-3′). Other Type III MTases have also been shown biochemically to form dimers in solution (76,77).
The M.EcoP15I structure is the first one for a class-β amino MTase bound to its substrate DNA, and it suggests a division of labor between two M subunits in terms of DNA recognition (one M subunit) and catalysis of methyltransfer (the other M subunit) (78) (Figure 3G). The DNA-recognizing subunit of the homodimer provides side chains, such as Arg305 and Arg350, for recognition of guanines on the non-target (non-A-containing) strand (Figure 3H). These Arg-Gua interactions with the non-target strand are reminiscent of Escherichia coli Dam (Figure 2G), and (like Dam) limit M.EcoP15I to methylating dsDNA and not ssDNA. In contrast, while differing roles for each subunit are also seen for CcrM, each protomer contacts primarily a different DNA strand. Specifically, one CcrM subunit binds the target strand, recognizes the target sequence and catalyzes methyl transfer, while the second molecule simply binds the non-target strand (in the case of a dsDNA substrate) (Figure 3A) (59). This division of labor allows CcrM to methylate both ds and ssDNA.
Looking at these dimeric class β MTase structures together, whether in the presence or absence of DNA, they share a striking feature—the catalytic domain of one subunit and the DNA binding domain of the second subunit are arranged so as to face each other, forming one integral binding surface appropriate for one DNA duplex (dashed circles in Figure 3A, C and G). In contrast, the catalytic site and DNA binding of the same subunit face away from one another, such that they cannot cooperate to accomplish the DNA recognition and methylation in one binding event. This is distinct from the association of MTases with other proteins in order to modulate their activity (see Discussion); and also implies that—while there may be kinetic or other reasons for which dimerization may be advantageous to other MTases (examples include mammalian Dnm3L-3a-3a-3L tetramer (79–81))—for the β MTases dimerization is essential for activity.
A sequence non-specific class β MTase: M.EcoGII
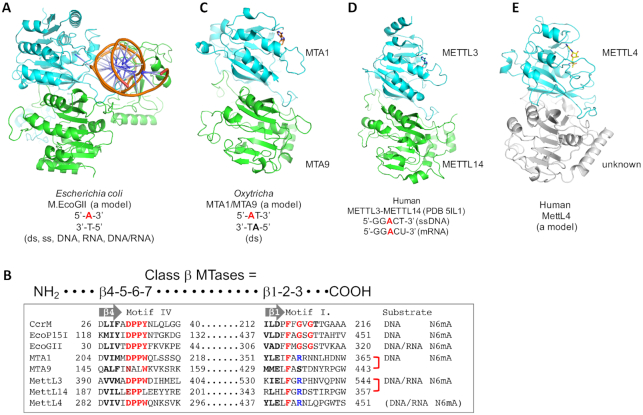
M.EcoGII is encoded in the genome of the pathogenic strain E. coli O104:H4 C227–11 (82), which was responsible for a severe outbreak of hemorrhagic uremia in Europe (83). The gene appears to reside within an integrated prophage genome, but is not expressed during normal bacterial growth. When the gene encoding M.EcoGII is expressed in vivo - using a high copy plasmid vector and a methylation–deficient E. coli host—extensive in vivo adenine methylation activity is revealed. It appears that M.EcoGII methylates adenine residues in any sequence context, in DNA and RNA, double-stranded and single stranded, and even adenine bases in either strand of a DNA/RNA-hybrid oligonucleotide duplex (84). There is not yet a M.EcoGII-DNA complex structure, though modeling based on a homolog (using threading with the M.EcoP15I-DNA structure) predicts a dimeric M.EcoGII complexed with one duplex (Figure 4A). Strikingly, from Figure 4A it appears to be impossible for M.EcoGII to function as a monomer. If it does function as a dimer, which seems likely, it is surprising that there appears to be no quadratic rate dependence of activity on the enzyme concentration [see figure 4 in (84)]. In contrast, such rate dependence is clear for two other β MTases, M.KpnI and M.RsrI [see figure 4 in (64) and (70)]. One possible explanation of this apparent paradox is that M.EcoGII has a particularly high dimerization constant. In fact, the known homodimer CcrM also shows a linear rate dependence on the enzyme concentration [see figure 7 in (85)].
Figure 4.
Homology models of class β MTases. (A) A model of the M.EcoGII-DNA complex based on the actual M.EcoP15I–DNA complex structure. (B) Sequence alignment of motif IV and motif I of class β MTases analyzed in this study. (C) A model of ciliate MTA1 and MTA9 heterodimer based on MettL3-MettL14. (D) A structure of human MettL3-MettL14 heterodimer complex (Structural Genomics Consortium). (E) A dimer model of human MettL4 based on MettL3–MettL14.
Even if we assume that M.EcoGII functions as a dimer, however, it is unclear how the 'division of labor' between the two M.EcoGII subunits is accomplished, given its lack of sequence specificity, and the fact that recognition and methylation occur to the same nucleotide. Considering the aforementioned gene duplication model of a tandemly-duplicated SAM-binding protein having one SAM pocket diverge to bind adenine, M.EcoGII might be an example of such a primordial adenine MTase, lacking a well-defined and functional TRD.
However, while it is difficult to identify TRDs from sequence analysis alone (86–88), there is no evidence that the region in which the TRD is expected to occur in M.EcoGII and its orthologs is unusual (e.g., smaller than in other MTases; not shown). Thus it is also possible that M.EcoGII began as a standard N6mA MTase and lost specificity, under selection for its role in the physiology of the phage that carries it. In this regard, it is noteworthy that some class-β RM MTases tested under certain solvent conditions can in fact methylate the DNA strand of a DNA/RNA hybrid duplex (89).
A heterodimeric class β MTase: MTA1-MTA9 in the ciliate Oxytricha
In contrast to metazoa, DNA N6mA is abundant in various unicellular eukaryotes, including ciliates, the green alga Chlamydomonas, and early-diverging fungi (90–92). A recent study identified four ciliate proteins—two class-β-like MTases (MTA1 and MTA9) and two homeobox-like DNA binding proteins—as being necessary for deposition of N6mA in the Oxytricha genome (93). Instead of a gene-targeting approach, the authors used classic biochemistry by identifying candidate proteins, in nuclear extracts, that co-purified with DNA MTase activity. A conserved catalytic motif IV (DPPW) is preserved in MTA1, but not MTA9 (which has an eroded motif NALW) (Figure 4B). The four-subunit ciliate MTase complex preferentially methylates ApT dinucleotides in dsDNA – in agreement with earlier observations that in the unicellular eukaryotes Saccharomyces cerevesiae, Chlamydomonas reinhardii and Tetrahymena thermophila, N6mA is enriched in ApT dinucleotides (92) within nucleosome linker regions near promoters (94). This ApT sequence is symmetrical, like the CpG methylation target in mammals, suggesting a similar mechanism for maintaining methylation following DNA replication. Indeed, the MTase activity was even higher on hemimethylated than on unmethylated dsDNA, though it showed no activity on ssDNA or RNA (93). Homology modeling suggests a heterodimeric association of MTA1 and MTA9 (Figure 4C), in analogy to mammalian MettL3-MettL14, which we will discuss next.
Heterodimeric complex of mammalian MettL3–MettL14
Detection of N6mA in mammalian DNA was reported only recently, but the role of MettL3 and MettL14 in generating N6mA in RNA has been known for some time. In mammalian (HeLa) cells, the mRNA N6mA MTase activity on the degenerate consensus sequence RRACH (R = purine, H is not a G) (95) requires at least two separate subunits, MT-A and MT-B (96). MT-A is itself a multimeric protein, that contains a 70-kDa MT-A70 subunit (now known as MettL3, containing 580-residues and with a predicted molecular weight of 65 kDa). The amino acid sequence of MT-A70/MettL3 was noted to include conserved motifs (e.g. motif IV: DPPW) of the bacterial DNA MTases but not of known RNA MTases (97). MettL3 and MettL14 form a heterodimeric complex (98), and have been studied widely for their role in generating N6mA in RNA ((99,100) and references therein). Like the ciliate MTA1 and MTA9 (preceding section), mammalian MettL3 is the catalytically active subunit, while MettL14 has an eroded motif IV (EPPL) (Figure 4B). An earlier study suggested that MettL14 catalyzed RNA methylation (98). Unlike the ciliate complex, which requires two additional DNA binding subunits, MettL3 itself contains two tandem CCCH-type zinc fingers within its polypeptide, located N-terminal to its MTase domain, that are necessary for RNA binding and thus enzymatic activity (101). Four research groups have independently determined the structures of the heterodimeric complex of the MTase domains of MettL3 and MettL14 ((102–104) and Structural Genomics Consortium) (Figure 4D). However, these partial complexes, lacking the CCCH-zinc fingers, possess no enzymatic activity and there is no RNA substrate-bound complex structure currently available for MettL3–MettL14.
Low levels of N6mA have been reported in DNA from mouse (105,106), human (30,107), and human malignant brain tumor glioblastoma (108), though other studies have failed to detect N6mA in mammalian genomes (109–111). Two reports present evidence that N6mA in mammalian DNA can result from incorporation of RNA-derived ribo-N6mA that was converted to deoxy-N6mA via the nucleotide salvage pathway (112,113). Interestingly, the three class β MTases MettL3, MettL4, and MettL14, from mouse and human, were recently assessed for the ability to generate N6mA on DNA (114,115). These three proteins were previously considered to be analogs of the MT-A70 subunit of the human mRNA N6mA MTase (116).
Prompted by the observations noted above, revealing that characterized class β MTases are active for DNA N6mA methylation, with some members having activity on ssDNA (CcrM) as well as ssRNA (M.EcoGII), we investigated whether the MettL3-MettL14 heterodimer also possesses methyl transfer activity onto DNA adenine. With synthetic substrate oligonucleotides, MettL3-MettL14 shows >10-fold stronger catalytic efficiency of methylation on ssDNA than the corresponding ssRNA under the same conditions (114). Furthermore, MettL3-MettL14 is active on an unpaired region in overall context of dsDNA. This appears to be consistent with the requirement of MettL3 methylation activity for DNA repair (117).
Mammalian MettL4
The third member of MT-A70 class β MTase in human and mouse is MettL4 (116). The C. elegans homolog was the first of the MettL4 family members to be considered as a candidate DNA N6mA MTase, but there was no enzymatic evidence demonstrating that C. elegans MettL4 has intrinsic DNA N6mA activity (118). Murine MettL4 was reported to be responsible for N6mA deposition in genic elements associated with transcriptional silencing (106). Curiously, recombinant human MettL4 expressed in HEK293T (human embryonic kidney) cells has in vitro enzymatic activity on mitochondrial DNA (115), whereas recombinant human MettL4 purified from E. coli has RNA MTase activity (119). The former study showed that MettL4 localizes within mitochondria in tested tissues (115), and examination of 23 high-confidence N6mA-enriched sites of mitochondrial DNA revealed an ApT containing consensus sequence—CTTATC (in the main text) or CT(C/A)ATC (in the figure S2E of (115))—agrees at least partially with an earlier study that N6mA sites across the mitochondrial genome were generally at an ApT dinucleotide (107). In contrast, the latter study found mainly nuclear localization of MettL4 expressed from a lentiviral vector, and failed to identify appreciable levels of N6mA in mitochondrial DNA, but did find N6mA in a small nuclear RNA (the spliceosome-associated U2 snRNA) (119). The differences among these various studies illustrates the complex nature of DNA versus RNA adenine methylation in mammalian genomes (genomic vs. mitochondrial). The accumulation of N6mA in DNA and/or RNA might also reflect diverse cellular and mitochondrial stress responses in different cell lines under different laboratory conditions. Homology modeling suggested a dimeric MettL4, but was indeterminate as to whether it is a homodimer or heterodimer (the latter of which would require a yet-to-be-identified binding partner) (Figure 4E). A proper biochemical approach, such as the one used to identify the ciliate complex (93), will be needed to identify necessary components of the catalytically active MettL4 MTase complex.
DISCUSSION
Here, we build on earlier suggestions that an ancestral protein that bound a single molecule of SAM evolved into an adenine-binding protein; undergoing first tandem gene duplication, and then divergence of the two SAM-binding pockets (Figure 1A). M.EcoGII might reflect properties of the ancestral enzymes, as it acts on any form of nucleic acid (ss or ds, RNA or DNA), in any sequence context, though many nucleic acids-modifying enzymes are still able to modify both DNA and RNA ((120) and references therein). To become a sequence-specific DNA/RNA adenine MTase, an additional fusion (or selection acting on an exposed surface loop) would have brought in a target recognition domain (TRD) (Figure 1B). Distinguishing between gain of specificity from a promiscuous ancestral MTase, and loss of specificity from a more recent sequence-specific MTase, is not straightforward. This question has been addressed in some cases, for example among metabolic enzymes (121), where increased specificity is seen in essential or higher-flux enzymes, or via reconstruction of ancestral enzymes from phylogenetic analyses, e.g. (122–126). It might be revealing to carry out such analysis with M.EcoGII, or even to determine if it can methylate various adenine derivatives.
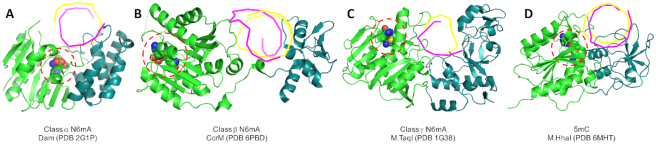
The unique dimeric feature of class β MTases, either in homo- or hetero-form, allows the enzyme to use ‘division of labor’ between two subunits in terms of DNA recognition and methylation. This division could well have resulted from the development or importation of a TRD in an orientation opposite to that of the methylation domain of the same polypeptide, thus requiring the two domains from each subunit to face each other to form one integral functional surface. In contrast, the monomeric enzymes in class α (E. coli Dam), class γ (M.TaqI) and 5mC class (M.HhaI) have TRDs in the same orientation as the methylation domain (Figure 5). The opposing TRD orientation in the class β MTases may help explain why they are primarily nucleic acid enzymes. Examining ∼30 human methyltransferase-like proteins (MettL1 to 27), all—except the three class β MTases discussed here—have the seven-stranded catalytic domain arising from motif order of I-to-IV, and many of them evolved to be non-nucleic acid enzymes (127). We wish to clarify a distinction between the ‘division of labor’ in homodimeric MTases, and non-β MTases that participate in protein–protein interactions for other purposes. For example, Trm112, named for its role in tRNA methylation, is a relatively small protein conserved in all three domains of life (128). In S. cerevisiae, Trm112 interacts with and activates at least four MTases (Bud23, Trm9, Trm11 and Mtq2—all are non-β MTases) that target different components of the translation machinery (rRNA, tRNA and release factors) (129).
Figure 5.
The monomeric structures of (A) E. coli Dam, (B) CcrM monomer taken from a dimer structure, (C) M.TaqI and (D) M.HhaI. The DNA substrates are displayed as ribbons (target A-strand in magenta and non-target strand in yellow) with cofactors as spheres. Note that the active site of CcrM monomer (in red dashed circle in panel B) points away from the DNA.
To summarize, the class β MTases, defined by their unique order of conserved motifs, may have diverged from an ancestral tandemly-duplicated protein that bound two molecules of SAM, differing from the other MTase classes either by which SAM-binding pocket changed to binding adenine, or via circular permutation, with M.EcoGII possibly representing an ancestral β MTase. While these elements are somewhat speculative, there are clear consequences of the β MTase structure on their function—specifically, that the β MTases are likely to be catalytically active only as dimers, while other MTase classes may or may not form dimers, and that this is an intrinsic property distinct from other protein-protein interactions in which MTases may engage.
ACKNOWLEDGEMENTS
We thank members of the Cheng laboratory for discussion.
Authors contributions: The thoughts discussed here in this survey were accumulated over the last two decades during the studies of M.PvuII, Dam, M.EcoGII, CcrM, Trm112-HemK2 and MettL3-14. R.M.B. and X.C. prepared the manuscript during COVID-19 shelter-in-place requirements. All were involved in discussion.
Contributor Information
Clayton B Woodcock, Department of Epigenetics and Molecular Carcinogenesis, University of Texas MD Anderson Cancer Center, Houston, TX 77030, USA.
John R Horton, Department of Epigenetics and Molecular Carcinogenesis, University of Texas MD Anderson Cancer Center, Houston, TX 77030, USA.
Xing Zhang, Department of Epigenetics and Molecular Carcinogenesis, University of Texas MD Anderson Cancer Center, Houston, TX 77030, USA.
Robert M Blumenthal, Department of Medical Microbiology and Immunology, and Program in Bioinformatics, The University of Toledo College of Medicine and Life Sciences, Toledo, OH 43614, USA.
Xiaodong Cheng, Department of Epigenetics and Molecular Carcinogenesis, University of Texas MD Anderson Cancer Center, Houston, TX 77030, USA.
FUNDING
U.S. National Institutes of Health [R35GM134744]; Cancer Prevention and Research Institute of Texas [RR160029]; X.C. is a CPRIT Scholar in Cancer Research. Funding for open access charge: NIH.
Conflict of interest statement. None declared.
REFERENCES
- 1. Posfai J., Bhagwat A.S., Posfai G., Roberts R.J.. Predictive motifs derived from cytosine methyltransferases. Nucleic Acids Res. 1989; 17:2421–2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Xu S., Xiao J., Posfai J., Maunus R., Benner J. 2nd. Cloning of the BssHII restriction-modification system in Escherichia coli: BssHII methyltransferase contains circularly permuted cytosine-5 methyltransferase motifs. Nucleic Acids Res. 1997; 25:3991–3994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Albert P., Varga B., Zsibrita N., Kiss A.. Circularly permuted variants of two CG-specific prokaryotic DNA methyltransferases. PLoS One. 2018; 13:e0197232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bestor T., Laudano A., Mattaliano R., Ingram V.. Cloning and sequencing of a cDNA encoding DNA methyltransferase of mouse cells. The carboxyl-terminal domain of the mammalian enzymes is related to bacterial restriction methyltransferases. J. Mol. Biol. 1988; 203:971–983. [DOI] [PubMed] [Google Scholar]
- 5. Okano M., Xie S., Li E.. Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nat. Genet. 1998; 19:219–220. [DOI] [PubMed] [Google Scholar]
- 6. Cheng X., Kumar S., Posfai J., Pflugrath J.W., Roberts R.J.. Crystal structure of the HhaI DNA methyltransferase complexed with S-adenosyl-L-methionine. Cell. 1993; 74:299–307. [DOI] [PubMed] [Google Scholar]
- 7. Klimasauskas S., Kumar S., Roberts R.J., Cheng X.. HhaI methyltransferase flips its target base out of the DNA helix. Cell. 1994; 76:357–369. [DOI] [PubMed] [Google Scholar]
- 8. Yoder J.A., Bestor T.H.. A candidate mammalian DNA methyltransferase related to pmt1p of fission yeast. Hum. Mol. Genet. 1998; 7:279–284. [DOI] [PubMed] [Google Scholar]
- 9. Okano M., Xie S., Li E.. Dnmt2 is not required for de novo and maintenance methylation of viral DNA in embryonic stem cells. Nucleic Acids Res. 1998; 26:2536–2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dong A., Yoder J.A., Zhang X., Zhou L., Bestor T.H., Cheng X.. Structure of human DNMT2, an enigmatic DNA methyltransferase homolog that displays denaturant-resistant binding to DNA. Nucleic Acids Res. 2001; 29:439–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Goll M.G., Kirpekar F., Maggert K.A., Yoder J.A., Hsieh C.L., Zhang X., Golic K.G., Jacobsen S.E., Bestor T.H.. Methylation of tRNAAsp by the DNA methyltransferase homolog Dnmt2. Science. 2006; 311:395–398. [DOI] [PubMed] [Google Scholar]
- 12. Kaiser S., Jurkowski T.P., Kellner S., Schneider D., Jeltsch A., Helm M.. The RNA methyltransferase Dnmt2 methylates DNA in the structural context of a tRNA. RNA Biol. 2017; 14:1241–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Weigele P., Raleigh E.A.. Biosynthesis and function of modified bases in bacteria and their viruses. Chem. Rev. 2016; 116:12655–12687. [DOI] [PubMed] [Google Scholar]
- 14. Roberts R.J., Cheng X.. Base flipping. Annu. Rev. Biochem. 1998; 67:181–198. [DOI] [PubMed] [Google Scholar]
- 15. Cheng X., Roberts R.J.. AdoMet-dependent methylation, DNA methyltransferases and base flipping. Nucleic Acids Res. 2001; 29:3784–3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hong S., Cheng X.. DNA base Flipping: A general mechanism for writing, reading, and erasing DNA modifications. Adv. Exp. Med. Biol. 2016; 945:321–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bujnicki J.M. Comparison of protein structures reveals monophyletic origin of the AdoMet-dependent methyltransferase family and mechanistic convergence rather than recent differentiation of N4-cytosine and N6-adenine DNA methylation. In Silico Biol. 1999; 1:175–182. [PubMed] [Google Scholar]
- 18. Bujnicki J.M., Radlinska M.. Molecular evolution of DNA-(cytosine-N4) methyltransferases: evidence for their polyphyletic origin. Nucleic Acids Res. 1999; 27:4501–4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jeltsch A. The cytosine N4-methyltransferase M.PvuII also modifies adenine residues. Biol. Chem. 2001; 382:707–710. [DOI] [PubMed] [Google Scholar]
- 20. Malone T., Blumenthal R.M., Cheng X.. Structure-guided analysis reveals nine sequence motifs conserved among DNA amino-methyltransferases, and suggests a catalytic mechanism for these enzymes. J. Mol. Biol. 1995; 253:618–632. [DOI] [PubMed] [Google Scholar]
- 21. Labahn J., Granzin J., Schluckebier G., Robinson D.P., Jack W.E., Schildkraut I., Saenger W.. Three-dimensional structure of the adenine-specific DNA methyltransferase M.Taq I in complex with the cofactor S-adenosylmethionine. Proc. Natl. Acad. Sci. U.S.A. 1994; 91:10957–10961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Goedecke K., Pignot M., Goody R.S., Scheidig A.J., Weinhold E.. Structure of the N6-adenine DNA methyltransferase M.TaqI in complex with DNA and a cofactor analog. Nat. Struct. Biol. 2001; 8:121–125. [DOI] [PubMed] [Google Scholar]
- 23. Schluckebier G., O’Gara M., Saenger W., Cheng X.. Universal catalytic domain structure of AdoMet-dependent methyltransferases. J. Mol. Biol. 1995; 247:16–20. [DOI] [PubMed] [Google Scholar]
- 24. Wu J.C., Santi D.V.. Kinetic and catalytic mechanism of HhaI methyltransferase. J. Biol. Chem. 1987; 262:4778–4786. [PubMed] [Google Scholar]
- 25. Chen L., MacMillan A.M., Chang W., Ezaz-Nikpay K., Lane W.S., Verdine G.L.. Direct identification of the active-site nucleophile in a DNA (cytosine-5)-methyltransferase. Biochemistry. 1991; 30:11018–11025. [DOI] [PubMed] [Google Scholar]
- 26. Zangi R., Arrieta A., Cossio F.P.. Mechanism of DNA methylation: the double role of DNA as a substrate and as a cofactor. J. Mol. Biol. 2010; 400:632–644. [DOI] [PubMed] [Google Scholar]
- 27. Du Q., Wang Z., Schramm V.L.. Human DNMT1 transition state structure. Proc. Natl Acad. Sci. U.S.A. 2016; 113:2916–2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Aranda J., Roca M., Lopez-Canut V., Tunon I.. Theoretical study of the catalytic mechanism of DNA-(N4-cytosine)-methyltransferase from the bacterium Proteus vulgaris. J. Phys. Chem. B. 2010; 114:8467–8473. [DOI] [PubMed] [Google Scholar]
- 29. Bujnicki J.M., Radlinska M.. Is the HemK family of putative S-adenosylmethionine-dependent methyltransferases a ‘missing’ zeta subfamily of adenine methyltransferases? A hypothesis. IUBMB Life. 1999; 48:247–249. [DOI] [PubMed] [Google Scholar]
- 30. Xiao C.L., Zhu S., He M., Chen D, Zhang Q., Chen Y., Yu G., Liu J., Xie S.Q., Luo F. et al.. N(6)-methyladenine DNA modification in the human genome. Mol. Cell. 2018; 71:306–318. [DOI] [PubMed] [Google Scholar]
- 31. Nakahigashi K., Kubo N., Narita S., Shimaoka T., Goto S., Oshima T., Mori H., Maeda M., Wada C., Inokuchi H.. HemK, a class of protein methyl transferase with similarity to DNA methyl transferases, methylates polypeptide chain release factors, and hemK knockout induces defects in translational termination. Proc. Natl Acad. Sci. U.S.A. 2002; 99:1473–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Heurgue-Hamard V., Champ S., Engstrom A., Ehrenberg M., Buckingham R.H.. The hemK gene in Escherichia coli encodes the N(5)-glutamine methyltransferase that modifies peptide release factors. EMBO J. 2002; 21:769–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Graille M., Heurgue-Hamard V., Champ S., Mora L., Scrima N., Ulryck N., van Tilbeurgh H., Buckingham R.H.. Molecular basis for bacterial class I release factor methylation by PrmC. Mol. Cell. 2005; 20:917–927. [DOI] [PubMed] [Google Scholar]
- 34. Figaro S., Scrima N., Buckingham R.H., Heurgue-Hamard V.. HemK2 protein, encoded on human chromosome 21, methylates translation termination factor eRF1. FEBS Lett. 2008; 582:2352–2356. [DOI] [PubMed] [Google Scholar]
- 35. Woodcock C.B., Yu D., Zhang X., Cheng X.. Human HemK2/KMT9/N6AMT1 is an active protein methyltransferase, but does not act on DNA in vitro, in the presence of Trm112. Cell Discov. 2019; 5:50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Li W., Shi Y., Zhang T., Ye J., Ding J.. Structural insight into human N6AMT1-Trm112 complex functioning as a protein methyltransferase. Cell Discov. 2019; 5:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Metzger E., Wang S., Urban S., Willmann D., Schmidt A., Offermann A., Allen A., Sum M., Obier N., Cottard F. et al.. KMT9 monomethylates histone H4 lysine 12 and controls proliferation of prostate cancer cells. Nat. Struct. Mol. Biol. 2019; 26:361–371. [DOI] [PubMed] [Google Scholar]
- 38. Jeltsch A. Circular permutations in the molecular evolution of DNA methyltransferases. J. Mol. Evol. 1999; 49:161–164. [DOI] [PubMed] [Google Scholar]
- 39. Lauster R. Evolution of type II DNA methyltransferases. A gene duplication model. J. Mol. Biol. 1989; 206:313–321. [DOI] [PubMed] [Google Scholar]
- 40. Adams G.M., Blumenthal R.M.. The PvuII DNA (cytosine-N4)-methyltransferase comprises two trypsin-defined domains, each of which binds a molecule of S-adenosyl-L-methionine. Biochemistry. 1997; 36:8284–8292. [DOI] [PubMed] [Google Scholar]
- 41. Bujnicki J.M. Sequence permutations in the molecular evolution of DNA methyltransferases. BMC Evol. Biol. 2002; 2:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. White H.B., 3rd Coenzymes as fossils of an earlier metabolic state. J. Mol. Evol. 1976; 7:101–104. [DOI] [PubMed] [Google Scholar]
- 43. Wolk S.K., Mayfield W.S., Gelinas A.D., Astling D., Guillot J., Brody E.N., Janjic N., Gold L.. Modified nucleotides may have enhanced early RNA catalysis. Proc. Natl Acad. Sci. U.S.A. 2020; 117:8236–8242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Roberts R.J., Vincze T., Posfai J., Macelis D.. REBASE–a database for DNA restriction and modification: enzymes, genes and genomes. Nucleic Acids Res. 2015; 43:D298–D299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Casadesus J. Bacterial DNA methylation and methylomes. Adv. Exp. Med. Biol. 2016; 945:35–61. [DOI] [PubMed] [Google Scholar]
- 46. Loenen W.A., Dryden D.T., Raleigh E.A., Wilson G.G., Murray N.E.. Highlights of the DNA cutters: a short history of the restriction enzymes. Nucleic Acids Res. 2014; 42:3–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Pleska M., Guet C.C.. Effects of mutations in phage restriction sites during escape from restriction-modification. Biol. Lett. 2017; 13:20170646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Vasu K., Nagaraja V.. Diverse functions of restriction-modification systems in addition to cellular defense. Microbiol. Mol. Biol. Rev. 2013; 77:53–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Oliveira P.H., Touchon M., Rocha E.P.. Regulation of genetic flux between bacteria by restriction-modification systems. Proc. Natl Acad. Sci. U.S.A. 2016; 113:5658–5663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Pingoud A., Wilson G.G., Wende W.. Type II restriction endonucleases–a historical perspective and more. Nucleic Acids Res. 2014; 42:7489–7527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Marinus M.G., Casadesus J.. Roles of DNA adenine methylation in host-pathogen interactions: mismatch repair, transcriptional regulation, and more. FEMS Microbiol. Rev. 2009; 33:488–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Messer W., Noyer-Weidner M.. Timing and targeting: the biological functions of Dam methylation in E. coli. Cell. 1988; 54:735–737. [DOI] [PubMed] [Google Scholar]
- 53. Berdis A.J., Lee I., Coward J.K., Stephens C., Wright R., Shapiro L., Benkovic S.J.. A cell cycle-regulated adenine DNA methyltransferase from Caulobacter crescentus processively methylates GANTC sites on hemimethylated DNA. Proc. Natl Acad. Sci. U.S.A. 1998; 95:2874–2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Barel I., Naughton B., Reich N.O., Brown F.L.H.. Specificity versus Processivity in the Sequential Modification of DNA: A Study of DNA Adenine Methyltransferase. J. Phys. Chem. B. 2018; 122:1112–1120. [DOI] [PubMed] [Google Scholar]
- 55. Albu R.F., Jurkowski T.P., Jeltsch A.. The Caulobacter crescentus DNA-(adenine-N6)-methyltransferase CcrM methylates DNA in a distributive manner. Nucleic Acids Res. 2012; 40:1708–1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Woodcock C.B., Yakubov A.B., Reich N.O.. Caulobacter crescentus cell cycle-regulated DNA methyltransferase uses a novel mechanism for substrate recognition. Biochemistry. 2017; 56:3913–3922. [DOI] [PubMed] [Google Scholar]
- 57. Horton J.R., Liebert K., Hattman S., Jeltsch A., Cheng X.. Transition from nonspecific to specific DNA interactions along the substrate-recognition pathway of dam methyltransferase. Cell. 2005; 121:349–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Horton J.R., Liebert K., Bekes M., Jeltsch A., Cheng X.. Structure and substrate recognition of the Escherichia coli DNA adenine methyltransferase. J. Mol. Biol. 2006; 358:559–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Horton J.R., Woodcock C.B., Opot S.B., Reich N.O., Zhang X., Cheng X.. The cell cycle-regulated DNA adenine methyltransferase CcrM opens a bubble at its DNA recognition site. Nat. Commun. 2019; 10:4600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Gong W., O’Gara M., Blumenthal R.M., Cheng X.. Structure of PvuII DNA-(cytosine N4) methyltransferase, an example of domain permutation and protein fold assignment. Nucleic Acids Res. 1997; 25:2702–2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Scavetta R.D., Thomas C.B., Walsh M.A., Szegedi S., Joachimiak A., Gumport R.I., Churchill M.E.. Structure of RsrI methyltransferase, a member of the N6-adenine beta class of DNA methyltransferases. Nucleic Acids Res. 2000; 28:3950–3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Thomas C.B., Scavetta R.D., Gumport R.I., Churchill M.E.. Structures of liganded and unliganded RsrI N6-adenine DNA methyltransferase: a distinct orientation for active cofactor binding. J. Biol. Chem. 2003; 278:26094–26101. [DOI] [PubMed] [Google Scholar]
- 63. Kaszubska W., Webb H.K., Gumport R.I.. Purification and characterization of the M.RsrI DNA methyltransferase from Escherichia coli. Gene. 1992; 118:5–11. [DOI] [PubMed] [Google Scholar]
- 64. Thomas C.B., Gumport R.I.. Dimerization of the bacterial RsrI N6-adenine DNA methyltransferase. Nucleic Acids Res. 2006; 34:806–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Osipiuk J., Walsh M.A., Joachimiak A.. Crystal structure of MboIIA methyltransferase. Nucleic Acids Res. 2003; 31:5440–5448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Morita R., Ishikawa H., Nakagawa N., Kuramitsu S., Masui R.. Crystal structure of a putative DNA methylase TTHA0409 from Thermus thermophilus HB8. Proteins. 2008; 73:259–264. [DOI] [PubMed] [Google Scholar]
- 67. Ma B., Ma J., Liu D., Guo L., Chen H., Ding J., Liu W., Zhang H.. Biochemical and structural characterization of a DNA N6-adenine methyltransferase from Helicobacter pylori. Oncotarget. 2016; 7:40965–40977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Malygin E.G., Ovechkina L.G., Zinov’ev V.V., Lindstrem U.M., Reich N.O.. [DNA-(N4-cytosine)-methyltransferase from Bacillus amyloliquefaciens: kinetic and substrate binding properties]. Mol. Biol. (Mosk). 2001; 35:42–51. [PubMed] [Google Scholar]
- 69. Mruk I., Cichowicz M., Kaczorowski T.. Characterization of the LlaCI methyltransferase from Lactococcus lactis subsp. cremoris W15 provides new insights into the biology of type II restriction-modification systems. Microbiology. 2003; 149:3331–3341. [DOI] [PubMed] [Google Scholar]
- 70. Bheemanaik S., Chandrashekaran S., Nagaraja V., Rao D.N.. Kinetic and catalytic properties of dimeric KpnI DNA methyltransferase. J. Biol. Chem. 2003; 278:7863–7874. [DOI] [PubMed] [Google Scholar]
- 71. Bheemanaik S., Bujnicki J.M., Nagaraja V., Rao D.N.. Functional analysis of amino acid residues at the dimerisation interface of KpnI DNA methyltransferase. Biol. Chem. 2006; 387:515–523. [DOI] [PubMed] [Google Scholar]
- 72. Prasad Y., Kumar R., Chaudhary A.K., Dhanaraju R., Majumdar S., Rao D.N.. Kinetic and catalytic properties of M.HpyAXVII, a phase-variable DNA methyltransferase from Helicobacter pylori. J. Biol. Chem. 2019; 294:1019–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Rao D.N., Dryden D.T., Bheemanaik S.. Type III restriction-modification enzymes: a historical perspective. Nucleic Acids Res. 2014; 42:45–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Ahmad I., Krishnamurthy V., Rao D.N.. DNA recognition by the EcoP15I and EcoPI modification methyltransferases. Gene. 1995; 157:143–147. [DOI] [PubMed] [Google Scholar]
- 75. Urulangodi M., Dhanaraju R., Gupta K., Roy R.P., Bujnicki J.M., Rao D.N.. Asymmetric DNA methylation by dimeric EcoP15I DNA methyltransferase. Biochimie. 2016; 128–129:70–82. [DOI] [PubMed] [Google Scholar]
- 76. Banerjee A., Rao D.N.. Functional analysis of an acid adaptive DNA adenine methyltransferase from Helicobacter pylori 26695. PLoS One. 2011; 6:e16810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Butterer A., Pernstich C., Smith R.M., Sobott F., Szczelkun M.D., Toth J.. Type III restriction endonucleases are heterotrimeric: comprising one helicase-nuclease subunit and a dimeric methyltransferase that binds only one specific DNA. Nucleic Acids Res. 2014; 42:5139–5150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Gupta Y.K., Chan S.H., Xu S.Y., Aggarwal A.K.. Structural basis of asymmetric DNA methylation and ATP-triggered long-range diffusion by EcoP15I. Nat. Commun. 2015; 6:7363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Jia D., Jurkowska R.Z., Zhang X., Jeltsch A., Cheng X.. Structure of Dnmt3a bound to Dnmt3L suggests a model for de novo DNA methylation. Nature. 2007; 449:248–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Veland N., Lu Y., Hardikar S., Gaddis S., Zeng Y., Liu B., Estecio M.R., Takata Y., Lin K., Tomida M.W et al.. DNMT3L facilitates DNA methylation partly by maintaining DNMT3A stability in mouse embryonic stem cells. Nucleic Acids Res. 2019; 47:152–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Gao L., Anteneh H., Song J.. Dissect the DNMT3A- and DNMT3B-mediated DNA Co-methylation through a covalent complex approach. J. Mol. Biol. 2020; 432:569–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Fang G., Munera D., Friedman D.I., Mandlik A., Chao M.C., Banerjee O., Feng Z., Losic B., Mahajan M.C., Jabado O.J. et al.. Genome-wide mapping of methylated adenine residues in pathogenic Escherichia coli using single-molecule real-time sequencing. Nat. Biotechnol. 2012; 30:1232–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Grad Y.H., Lipsitch M., Feldgarden M., Arachchi H.M., Cerqueira G.C., Fitzgerald M., Godfrey P., Haas B.J., Murphy C.I., Russ C. et al.. Genomic epidemiology of the Escherichia coli O104:H4 outbreaks in Europe, 2011. Proc. Natl Acad. Sci. U.S.A. 2012; 109:3065–3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Murray I.A., Morgan R.D., Luyten Y., Fomenkov A., Correa I.R. Jr, Dai N., Allaw M.B., Zhang X., Cheng X., Roberts R.J.. The non-specific adenine DNA methyltransferase M.EcoGII. Nucleic Acids Res. 2018; 46:840–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Shier V.K., Hancey C.J., Benkovic S.J.. Identification of the active oligomeric state of an essential adenine DNA methyltransferase from Caulobacter crescentus. J. Biol. Chem. 2001; 276:14744–14751. [DOI] [PubMed] [Google Scholar]
- 86. Trautner T.A., Pawlek B., Behrens B., Willert J.. Exact size and organization of DNA target-recognizing domains of multispecific DNA-(cytosine-C5)-methyltransferases. EMBO J. 1996; 15:1434–1442. [PMC free article] [PubMed] [Google Scholar]
- 87. Walter J., Trautner T.A., Noyer-Weidner M.. High plasticity of multispecific DNA methyltransferases in the region carrying DNA target recognizing enzyme modules. EMBO J. 1992; 11:4445–4450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Szegedi S.S., Gumport R.I.. DNA binding properties in vivo and target recognition domain sequence alignment analyses of wild-type and mutant RsrI [N6-adenine] DNA methyltransferases. Nucleic Acids Res. 2000; 28:3972–3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Wons E., Mruk I., Kaczorowski T.. Relaxed specificity of prokaryotic DNA methyltransferases results in DNA site-specific modification of RNA/DNA heteroduplexes. J. Appl. Genet. 2015; 56:539–546. [DOI] [PubMed] [Google Scholar]
- 90. Rae P.M., Spear B.B.. Macronuclear DNA of the hypotrichous ciliate Oxytricha fallax. Proc. Natl Acad. Sci. U.S.A. 1978; 75:4992–4996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Hattman S., Kenny C., Berger L., Pratt K.. Comparative study of DNA methylation in three unicellular eucaryotes. J. Bacteriol. 1978; 135:1156–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Mondo S.J., Dannebaum R.O., Kuo R.C., Louie K.B., Bewick A.J., LaButti K., Haridas S., Kuo A., Salamov A., Ahrendt S.R. et al.. Widespread adenine N6-methylation of active genes in fungi. Nat. Genet. 2017; 49:964–968. [DOI] [PubMed] [Google Scholar]
- 93. Beh L.Y., Debelouchina G.T., Clay D.M., Thompson R.E., Lindblad K.A., Hutton E.R., Bracht J.R., Sebra R.P., Muir T.W., Landweber L.F.. Identification of a DNA N6-Adenine methyltransferase complex and its impact on chromatin organization. Cell. 2019; 177:1781–1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Pratt K., Hattman S.. Deoxyribonucleic acid methylation and chromatin organization in Tetrahymena thermophila. Mol. Cell. Biol. 1981; 1:600–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Schibler U., Kelley D.E., Perry R.P.. Comparison of methylated sequences in messenger RNA and heterogeneous nuclear RNA from mouse L cells. J. Mol. Biol. 1977; 115:695–714. [DOI] [PubMed] [Google Scholar]
- 96. Bokar J.A., Rath-Shambaugh M.E., Ludwiczak R., Narayan P., Rottman F.. Characterization and partial purification of mRNA N6-adenosine methyltransferase from HeLa cell nuclei. Internal mRNA methylation requires a multisubunit complex. J. Biol. Chem. 1994; 269:17697–17704. [PubMed] [Google Scholar]
- 97. Bokar J.A., Shambaugh M.E., Polayes D., Matera A.G., Rottman F.M.. Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA (N6-adenosine)-methyltransferase. RNA. 1997; 3:1233–1247. [PMC free article] [PubMed] [Google Scholar]
- 98. Liu J., Yue Y., Han D., Wang X., Fu Y., Zhang L., Jia G., Yu M., Lu Z., Deng X. et al.. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 2014; 10:93–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Frye M., Harada B.T., Behm M., He C.. RNA modifications modulate gene expression during development. Science. 2018; 361:1346–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Liu J., Dou X., Chen C., Chen C., Liu C., Xu M.M., Zhao S., Shen B., Gao Y., Han D. et al.. N (6)-methyladenosine of chromosome-associated regulatory RNA regulates chromatin state and transcription. Science. 2020; 367:580–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Huang J., Dong X., Gong Z., Qin L.Y., Yang S., Zhu Y.L., Wang X., Zhang D., Zou T., Yin P. et al.. Solution structure of the RNA recognition domain of METTL3-METTL14 N(6)-methyladenosine methyltransferase. Protein Cell. 2019; 10:272–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Wang X., Feng J., Xue Y., Guan Z., Zhang D., Liu Z., Gong Z., Wang Q., Huang J., Tang C. et al.. Structural basis of N(6)-adenosine methylation by the METTL3-METTL14 complex. Nature. 2016; 534:575–578. [DOI] [PubMed] [Google Scholar]
- 103. Wang P., Doxtader K.A., Nam Y.. Structural basis for cooperative function of Mettl3 and Mettl14 methyltransferases. Mol. Cell. 2016; 63:306–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Sledz P., Jinek M.. Structural insights into the molecular mechanism of the m(6)A writer complex. eLife. 2016; 5:e18434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Wu T.P., Wang T., Seetin M.G., Lai Y., Zhu S., Lin K., Liu Y., Byrum S.D., Mackintosh S.G., Zhong M. et al.. DNA methylation on N(6)-adenine in mammalian embryonic stem cells. Nature. 2016; 532:329–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Kweon S.M., Chen Y., Moon E., Kvederaviciute K., Klimasauskas S., Feldman D.E.. An adversarial DNA N(6)-Methyladenine-Sensor network preserves polycomb silencing. Mol. Cell. 2019; 74:1138–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Koh C.W.Q., Goh Y.T., Toh J.D.W., Neo S.P., Ng S.B., Gunaratne J., Gao Y.G., Quake S.R., Burkholder W.F., Goh W.S.S.. Single-nucleotide-resolution sequencing of human N6-methyldeoxyadenosine reveals strand-asymmetric clusters associated with SSBP1 on the mitochondrial genome. Nucleic Acids Res. 2018; 46:11659–11670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Xie Q., Wu T.P., Gimple R.C., Li Z., Prager B.C., Wu Q., Yu Y., Wang P., Wang Y., Gorkin D.U. et al.. N(6)-methyladenine DNA Modification in Glioblastoma. Cell. 2018; 175:1228–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Ratel D., Ravanat J.L., Charles M.P., Platet N., Breuillaud L., Lunardi J., Berger F., Wion D.. Undetectable levels of N6-methyl adenine in mouse DNA: Cloning and analysis of PRED28, a gene coding for a putative mammalian DNA adenine methyltransferase. FEBS Lett. 2006; 580:3179–3184. [DOI] [PubMed] [Google Scholar]
- 110. Schiffers S., Ebert C., Rahimoff R., Kosmatchev O., Steinbacher J., Bohne A.V., Spada F., Michalakis S., Nickelsen J., Muller M. et al.. Quantitative LC-MS provides no evidence for m(6) dA or m(4) dC in the genome of mouse embryonic stem cells and tissues. Angew. Chem. 2017; 56:11268–11271. [DOI] [PubMed] [Google Scholar]
- 111. Liu B., Liu X., Lai W., Wang H.. Metabolically generated stable Isotope-Labeled deoxynucleoside code for tracing DNA N(6)-methyladenine in human cells. Anal. Chem. 2017; 89:6202–6209. [DOI] [PubMed] [Google Scholar]
- 112. Musheev M.U., Baumgartner A., Krebs L., Niehrs C.. The origin of genomic N(6)-methyl-deoxyadenosine in mammalian cells. Nat. Chem. Biol. 2020; doi:10.1038/s41589-020-0504-2. [DOI] [PubMed] [Google Scholar]
- 113. Liu X., Lai W., Li Y., Chen S., Liu B., Zhang N., Mo J., Lyu C., Zheng J., Du Y.R. et al.. N(6)-methyladenine is incorporated into mammalian genome by DNA polymerase. Cell Res. 2020; doi:10.1038/s41422-020-0317-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Woodcock C.B., Yu D., Hajian T., Li J., Huang Y., Dai N., Correa I.R. Jr, Wu T., Vedadi M., Zhang X. et al.. Human MettL3-MettL14 complex is a sequence-specific DNA adenine methyltransferase active on single-strand and unpaired DNA in vitro. Cell Discov. 2019; 5:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Hao Z., Wu T., Cui X., Zhu P., Tan C., Dou X., Hsu K.W., Lin Y.T., Peng P.H., Zhang L.S. et al.. N(6)-deoxyadenosine methylation in mammalian mitochondrial DNA. Mol. Cell. 2020; 78:382–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Bujnicki J.M., Feder M., Radlinska M., Blumenthal R.M.. Structure prediction and phylogenetic analysis of a functionally diverse family of proteins homologous to the MT-A70 subunit of the human mRNA:m(6)A methyltransferase. J. Mol. Evol. 2002; 55:431–444. [DOI] [PubMed] [Google Scholar]
- 117. Xiang Y., Laurent B., Hsu C.H., Nachtergaele S., Lu Z., Sheng W., Xu C., Chen H., Ouyang J., Wang S. et al.. RNA m(6)A methylation regulates the ultraviolet-induced DNA damage response. Nature. 2017; 543:573–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Greer E.L., Blanco M.A., Gu L., Sendinc E., Liu J., Aristizabal-Corrales D., Hsu C.H., Aravind L., He C., Shi Y.. DNA methylation on N(6)-Adenine in C. elegans. Cell. 2015; 161:868–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Chen H., Gu L., Orellana E.A., Wang Y., Guo J., Liu Q., Wang L., Shen Z., Wu H., Gregory R.I. et al.. METTL4 is an snRNA m(6)Am methyltransferase that regulates RNA splicing. Cell Res. 2020; doi:10.1038/s41422-019-0270-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Forterre P., Grosjean H.. Grosjean H. The Interplay between RNA and DNA modifications. DNA and RNA Modification Enzymes: Structure, Mechanism, Function and Evolution. 2009; Landes Bioscience; 259–274. [Google Scholar]
- 121. Nam H., Lewis N.E., Lerman J.A., Lee D.H., Chang R.L., Kim D., Palsson B.O.. Network context and selection in the evolution to enzyme specificity. Science. 2012; 337:1101–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Grinshpon R.D., Shrestha S., Titus-McQuillan J., Hamilton P.T., Swartz P.D., Clark A.C.. Resurrection of ancestral effector caspases identifies novel networks for evolution of substrate specificity. Biochem. J. 2019; 476:3475–3492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Marey-Sarwan I., Roer-Strier D., Otto H.. Contextualizing risk and protection: perceptions of Bedouin mothers from unrecognized villages in the Naqab. Am. J. Orthopsychiatry. 2018; 88:306–315. [DOI] [PubMed] [Google Scholar]
- 124. Castro-Fernandez V., Herrera-Morande A., Zamora R., Merino F., Gonzalez-Ordenes F., Padilla-Salinas F., Pereira H.M., Brandao-Neto J., Garratt R.C., Guixe V.. Reconstructed ancestral enzymes reveal that negative selection drove the evolution of substrate specificity in ADP-dependent kinases. J. Biol. Chem. 2017; 292:21218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Clifton B.E., Jackson C.J.. Ancestral protein reconstruction yields insights into adaptive evolution of binding specificity in Solute-Binding proteins. Cell Chem Biol. 2016; 23:236–245. [DOI] [PubMed] [Google Scholar]
- 126. Eick G.N., Colucci J.K., Harms M.J., Ortlund E.A., Thornton J.W.. Evolution of minimal specificity and promiscuity in steroid hormone receptors. PLos Genet. 2012; 8:e1003072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Falnes P.O., Jakobsson M.E., Davydova E., Ho A., Malecki J.. Protein lysine methylation by seven-beta-strand methyltransferases. Biochem. J. 2016; 473:1995–2009. [DOI] [PubMed] [Google Scholar]
- 128. van Tran N., Muller L., Ross R.L., Lestini R., Letoquart J., Ulryck N., Limbach P.A., de Crecy-Lagard V., Cianferani S., Graille M.. Evolutionary insights into Trm112-methyltransferase holoenzymes involved in translation between archaea and eukaryotes. Nucleic Acids Res. 2018; 46:8483–8499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Bourgeois G., Letoquart J., van Tran N., Graille M.. Trm112, a protein activator of methyltransferases modifying actors of the eukaryotic translational apparatus. Biomolecules. 2017; 7:7. [DOI] [PMC free article] [PubMed] [Google Scholar]