Abstract
The outbreak of the recent coronavirus (SARS-CoV-2), which causes a severe pneumonia infection, first identified in Wuhan, China, imposes significant risks to public health. Around the world, researchers are continuously trying to identify small molecule inhibitors or vaccine candidates by targeting different drug targets. The SARs-CoV-2 macrodomain-I, which helps in viral replication and hijacking the host immune system, is also a potential drug target. Hence, this study targeted viral macrodomain-I by using drug similarity, virtual screening, docking and re-docking approaches. A total of 64,043 compounds were screened, and potential hits were identified based on the docking score and interactions with the key residues. The top six hits were subjected to molecular dynamics simulation and Free energy calculations and repeated three times each. The per-residue energy decomposition analysis reported that these compounds significantly interact with Asp22, Ala38, Asn40, Val44, Phe144, Gly46, Gly47, Leu127, Ser128, Gly130, Ile131, Phe132 and Ala155 which are the critical active site residues. Here, we also used ADPr as a positive control to compare our results. Our results suggest that our identified hits by using such a complicated computational pipeline could inhibit the SARs-CoV-2 by targeting the macrodomain-1. We strongly recommend the experimental testing of these compounds, which could rescue the host immune system and could help to contain the disease caused by SARs-CoV-2.
Communicated by Ramaswamy H. Sarma
Keywords: Macrodomain-I, immune system, SARs-CoV-2, drug similarity, free energy
Introduction
At the end of 2019, the emergence of the new coronavirus infection in Wuhan, China, has rapidly spread across the world and has affected many countries very severely. This virus causes a disease named as COVID-19 (Coronavirus Disease), and the virology nomenclature committee has named it SARs-CoV-2 (Wu et al., 2020). The virus belongs to the beta coronaviruses and is believed to be transferred from bat through pangolin. This virus has been reported to highly contagious and can transfer from human-to-human through different mediums. The SARS-CoV-2 virus-like SARS-CoV and MERS-CoV, a beta-coronavirus and all of them evolved from bats, are considered to be capable of causing unprecedented health, economic and societal repercussions (Lu et al., 2020). The world health organization (WHO) declared this virus a global pandemic and constituted a Public Health Emergency of International Concern. The epidemic has dramatically evolved, with more than 180 countries worldwide reporting laboratory-confirmed SARS-CoV-2 cases (Raoult et al., 2020).
Like the other coronaviruses, specifically the beta coronaviruses, this SARs-CoV-2 also possess similar cellular machinery. The SARs-CoV-2 owns a single stand + RNA with a total of sixteen nonstructural proteins (nsp) (Chan et al., 2020). These proteins are responsible for different cellular functions, from self-replication to infection and host immune invasion. Among the total 16 nonstructural proteins, nsp3 is one among those with an important role such as viral life cycle, interaction with host proteins and involved in anchoring the coronavirus replication/transcription complex (RTC) to modified membranous structures originating from the endoplasmic reticulum (ER) (Angeletti et al., 2020). The sixteen different domains of nsp3 perform these various functions. Among the 16 different domains, macrodomain-1 or SARS-unique domain is an essential domain with host immune evasion function (Fehr et al., 2018). The macrodomain-I or X-domain has been reported to be indispensable for SARs coronaviruses RNA replication (Angeletti et al., 2020; Kusov et al., 2015). The ADP-ribose polymerases (PARPs) are primarily attached to this domain to perform its function. PARPs regulate important cellular processes such as cell-cycle progression, cell division, host–virus interaction, DNA damage repair, genome maintenance, transcription regulation, protein degradation, ageing and cell death (Putics et al., 2005). In SARs-CoVs, the binding of ADPR to the macrodomain of nsp3 is important (Fehr & Perlman, 2015). This binding initiates the signaling of RNA replication and virulence. Interferon (IFN) regulates the anti-viral function of PARPs, and thus, evolutionary this domain (macrodomain) neutralizes the host immune response upon infection (Eriksson et al., 2008; Fehr et al., 2016). Accordingly, macrodomain by removing ADP-ribosylation from modified host proteins, prevent host responses to viral infection (Egloff et al., 2006).
Conversely, in the presence of free ADPRs in the cell directed by the PARP activity, activates the TRPM2 receptor. TRPM2, which is Transient Receptor Potential 2 receptor, contributes to different cellular functions such as the production of cytokines, regulation of calcium signaling, oxidative stress response, insulin production, cell mortality and death (Sumoza-Toledo & Penner, 2011). The Severe inflammatory response, cytokine storms and necrosis in the patient's lungs with COVID-19 can explain the activation of the TRPM2 signaling pathway, which can result from the action of the macrodomain or stress conditions that lead to accumulation of ADPR (Zheng et al., 2020). Macrodomain has been reported as one of the potential drug targets in coronaviruses, Chikungunya viruses and also in cancer (Shimizu et al., 2020; Vijayasri & Hopper, 2017).
The macrodomain possesses a sandwich like α/β/α architecture. This domain possesses seven beta-sheets and six helices. Previous studies reported that the binding of ADPr mainly involves five different stretches, which could vary from specie to specie (Piotrowski et al., 2009). The Asp21 residue, which corresponds to a different position in different species, is conserved and fixes the one end of ADP. Conversely, Asn39 fixes the other end of the ADPr (Chatterjee et al., 2009). These residues are reported to be essential in some SARs and MERs viruses for the important activity of the ADPr. Recently, structure-based drug designing has dramatically helped to design drugs against different disease and coronaviruses (Khan, Ali, et al., 2020; M. T. Khan et al., 2020; Quimque et al., 2020).
The crystallographic structure of SARs-CoV-2 has recently been resolved in complex with ADPr. In the macrodomain of SARs-CoV-2, the two important residues which facilitate the binding of ADPr correspond to Asp22 and Asn40. The over-structure possesses similar architecture to that of SARs and MERs macrodomain. Hence, this study uses structure-based drug designing approaches to design a potential inhibitor to target the SARs-CoV-2 macrodomain-I. Drug similarity and virtual screening approaches were combined with molecular dynamics simulation, and free energy calculation methods identify high binding inhibitors to target macrodomain. This study will ease a way of designing potential inhibitors to fight against the COVID-19.
Material & methods
Protein structure retrieval and preparation
The crystallographic structure of SARs-CoV-2 macrodomain-I recently reported was retrieved using the PDB ID: 6YWL and processed to remove extra chains and was analyzed for missing residues and other structural defects. The structure was minimized by using YASARA and MolProbity (Chen et al., 2010; Land & Humble, 2018). Hydrogens were added by using H++ webserver (Anandakrishnan et al., 2012). Water molecules and other atoms were removed from the structure. The structure was visualized in PyMOL (DeLano, 2002).
Drug similarity search & databases retrieval
The binding of ADPr to the macrodomain reveals important information regarding the interacting residues and the ADPr structural scaffold. Here, the SMILES of ADPr was used as input and search against ZINC, PDB and Chembl databases was performed using SwissSimilarity (Zoete et al., 2016). All the compounds were retrieved and were converted mdb format. Furthermore, two different databases NANPDB and TCM (Traditional Chinese Medicine Database) (Chen, 2011) were also screened against the macrodomain-I. NANPDB database retrieved from http://african-compounds.org/nanpdb/ was screened against the Mac-I structure (Ntie-Kang et al., 2017). Resource from 617 species (146 families of plants, animals, bacteria and fungi) comprised of 6482 compounds are included in this database. Conversely, the TCM database possesses 57,320 compounds from Chinese herbs.
Ligands preparation and virtual screening protocol
The retrieved compounds were subjected to charges correction, washing and minimization using MMFF94x force field. The database was converted to .mdb format to be used as input for Molecular Operating Environment (MOE v2016) (Vilar et al., 2008). Previously, it has been reported that five stretches of amino acids 20–22, 37–39, 43–49, 45–47 and 124–131 are reported to be involved in the binding of ADPr (Lei et al., 2018). So, using the selected residues option, an active site was defined. With ten conformations, each ligand was screened against the active site using a triangle matcher as a placement while London dG as a scoring method. Docking scores and visual interactions were used as a criterion for selecting the best hits. ADPr was used as positive control.
Induced-fit docking
After the screening of the two databases and the identified similar compounds, a composite database of the top 80 compounds was generated. This database contains ligands with high docking scores and interactions with the key residues. IFD produces better accuracy. Thus, using IFD docking protocol, the top 80 hits were re-docked to validate the results. The compounds were further lessened to a reasonable figure using the IFD protocol, which could then be assessed and docked individually against the Mac-I active site.
Re-docking of top hits using genetic algorithm
For the re-docking, we used AutoDock Vina software, which is based on a Genetic Algorithm (GA) (Morris et al., 2009; Trott & Olson, 2010). AutoDock software was used to define the grid dimension and box based on the defined residues. The protein structure was converted to .pdbqt format while using the ligands preparation criteria such as root detection, charges, hydrogen and aromaticity criteria were used for ligands preparation. Each ligand molecule was prepared individually and converted to .pdbqt file. To achieve high accuracy, we set exhaustiveness to 64. Thus here, multisteps docking and re-docking approaches were utilized to identify the most potential hits that could probably inhibit the SARs-CoV-2.
ADMET and bioactivity prediction
To predict the bioactivity of each top ligand Molinspiration Cheminformatics tool was used while for ADMET analysis, SwissADME was utilized (Daina et al., 2017). Molinspiration has been used by approximately 4500 studies to predict the bioactivity results.
Molecular dynamics simulation of the top hits
To understand the dynamics and interacting behaviour, the top hits identified complexed with Mac-I were subjected to molecular dynamics simulation. To obtain better and more accurate simulation results, all of the complexes were submitted to Propka 3.1, which is an online web server, for protonation state correction. Amber 18 package with PMEMD.CUDA implementation was used to perform the simulations (Case et al., 2005; Pearlman et al., 1995). For protein AMBER ff14SB while for drug molecules GAFF2 (generalized Amber force field) (Vassetti et al., 2019). Before protein–ligands topology generation, antechamber and frcmod were used to obtain the ligand topologies file for simulation (Wang et al., 2001). Systems were solvated with TIP3P water box with 8.0 Å distance each side and were neutralized by adding Na + ions. Using the 300 K and a pressure of 1.0 bar controlled by Langevin thermostat and Berendsen Barostat was used (Davidchack et al., 2009; Lin et al., 2017). For the hydrogen bonding SHAKE algorithm, while for long-range interactions, particle mesh Ewald summation (PME) algorithm was used (Kräutler et al., 2001; Toukmaji et al., 2000). The nonbonded cut-off was fixed at 10.0 Å. Each system was minimized by using two-step minimization approach. A time step was set as 2.0 fs. Followed by heating and equilibration, the production simulation was carried for 100 ns at the NPT ensemble, and the Cartesian coordinates were stored at every 10 ps. Overall, 5000 frames were obtained from each production simulation. Each simulation was repeated three times.
Post-simulation analysis and visualization
Post-simulation analyses such as root mean square deviation (RMSD) was carried out on the trajectories obtained from each system to estimate the stability of each system while root mean square fluctuation (RMSF) to access flexibility at residues level. For compactness of the structure, we calculated the Rg (radius of gyration) as criteria for determining the structural compactness during the simulation time using CPPTRAJ and PTRAJ (Roe & Cheatham, 2013).
Binding free energy calculations
The binding of each ligand molecule was measured using the Poisson–Boltzmann surface area molecular mechanics (MMGBSA) method. (Junaid et al., 2019; Khan, Junaid, et al., 2018, 2020; M. T. Khan et al., 2020; Sun et al., 2014). The most extensively used MMPBSA.py script was utilized as an input, which contains all the protocols for calculating free energy. For each system, 5000 structural frames were used to calculate the free energy using the following equation.
The total binding energy is represented ΔGbind, while the others demonstrate the binding energy of complex, protein and ligand. The whole energy can be divided into specific energy term, which contributes to the total binding free energy. To calculate the contribution of particular energy term, the following equation was used:
The above equation contains representation for each energy term, such as vdW and electrostatic. In addition, both polar and nonpolar interaction energy terms are given. This method of calculating the total binding free energy is widely accepted and used by colossal studies (Khan, Ali, et al., 2020; Khan, Rehman, et al., 2020; Khan, Saleem, et al., 2018; Khan et al., 2019; M. T. Khan et al., 2020; Wang et al., 2019).
Per-Residue energy decomposition analysis
Free energy decomposition for each residue was conducted using the MM-GBSA integrated script to obtain a comprehensive view of the protein–ligand interaction and classify the main residues responsible for the interaction. This study of energy decomposition is carried out on the same snapshots that were used in the above analysis.
Hydrogen bonding analysis
Hydrogen bonding occupancy analysis was used to monitor the bonding lifetime during the simulation. Both Intra and inter molecular hydrogen bonding analysis were performed.
Results
Drug similarity search
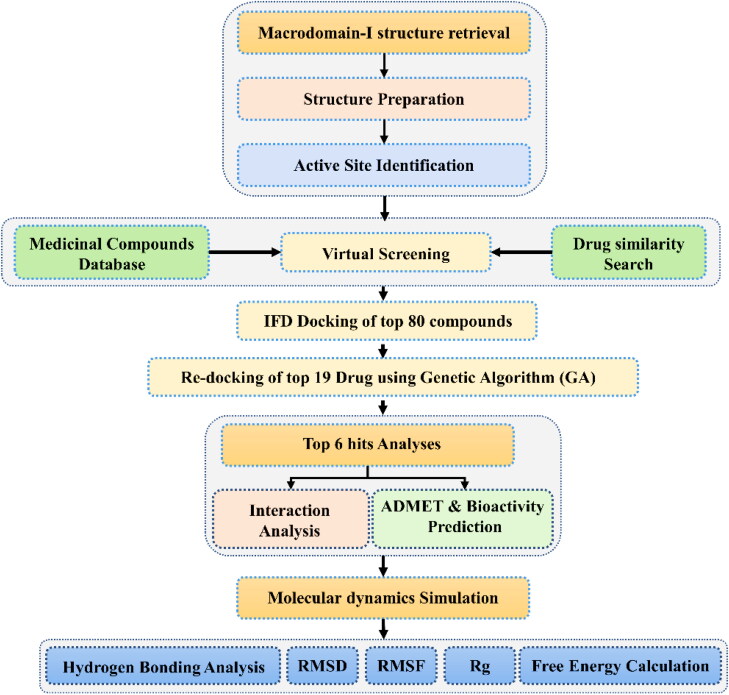
SwissSimilarity webserver was utilized to identify similar compounds like ADPr, which could bind to Mac-I. The server follows a criterion of Tanimoto, also known as Jaccard score to classify compounds similarity from 0 to 1. Compounds with a greater score (near to 1) are considered as the most similar while the low score shows diversity. Hence, using SwissSimilarity server, a total of 400 similar compounds were predicted from the RCSB databank. For compounds docking, we set a criterion of 0.8. Compounds with score >0.8 will be considered for screening while the rest will be discarded. Using this criterion, only 131 compounds were found to have a score higher than 0.8. In contrast, the rest of 269 compounds were reported to have a score less than 0.8. In the same way, only 11 compounds were predicted from Chembl to have a score higher than 0.8. These 11 compounds were also from a total of 400 predicted compounds. However, using the same criteria, no molecule Zinc Drug like a molecule database was found to have a similar scaffold. Thus, a total of 142 compounds were obtained for screening from a similarity search. Results from this protocol were analyzed, and the top six hits were subjected to molecular dynamics simulation and post-simulation analyses. The overall flow of the work is given in Figure 1.
Figure 1.
The overall flow of the work. This methodology contains multisteps such as docking and re-docking and bioactivity prediction, molecular dynamics simulation and post-simulation analyses are given.
Virtual screening and molecular docking
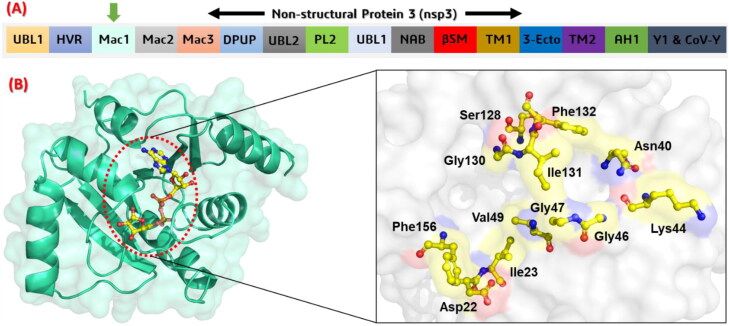
A computational approach, virtual screening, is an ultrafast paradigm to develop novel potential drugs against the latest coronavirus (SARs-CoV-2). Computational virtual screening is a time saving and reliable approach to find novel hits. Herein, we used VS approach to screen the screen 141 compounds from Drug similarity search result, 6482 compounds were screened from NANPDB database while 57,320 compounds were screened from the TCM database. The active site was carefully selected based on the ADPr. The active site residues include the two conserved residues Asp22 and Asn40 which are indispensable for the function of macrodomain-I. Among the active site residues Asp22, Ile23, Asn40, Lys44, Gly46, Gly47, Val49, Ser128, Gly130, Ile131, Phe132 and Phe156 makes the active site cavity. The overall information regarding the nps3 domains, the crystal structure of Macrodomain-I and its active site residues are given in Figure 2.
Figure 2.
(A) showing the structural organization of SARs-CoV-2 nsp3. The sixteen different domains area given in panel A. The macrodomain-I used in this study is shown with a green arrow. (B) showing the ADPr (yellow stick) bound structure of macrodomain-I from SARs-CoV-2. In panel B, the active site residues which are involved in interactions with Mac-I are given and labelled. The active site residues are shown in yellow stick colour.
First screening
In the first round of screening a total of 64,043 compounds were tested. The docking score for these 64,043 compounds ranges from −11.02 to −7.5 kcal/mol. Among these only compounds with docking score greater than −9.0 kcal/mol were selected for interaction analysis. Using this criterion, only 80 compounds were found to have a score higher than −9.0 kcal/mol. Among these, the top-scoring compounds 73 compounds were from similar drug compounds, while three compounds from NANPDB and TCM were reported to have scores higher than −9.0 kcal/mol. Compounds were analyzed for interactions, and only those which form interactions with Asp22 and Asn40 were selected. Furthermore, these compounds were also analyzed for hydrogen bonding vdW and electrostatic interactions with the key residues.
Second screening
In the second round of screening to the top 80 compounds were subjected to induced-fit docking. The docking scores for these compounds range from −11.0 to −9.50 kcal/mol. To select the best compounds from all these, a criterion based on docking score and multiple interactions with the defined active site residues was used to filter the top hits. This screening resulted in 36 best compounds satisfying the specified criteria. Each conformation was manually visualized for this purpose. The obtained 36 compounds were then subjected to ADMET analysis, which excluded 17 compounds while the remaining were the best fit. Hence, from these 80 compounds, only 19 compounds were found to form the best interactions with active site residues and to have a good binding affinity.
Third screening
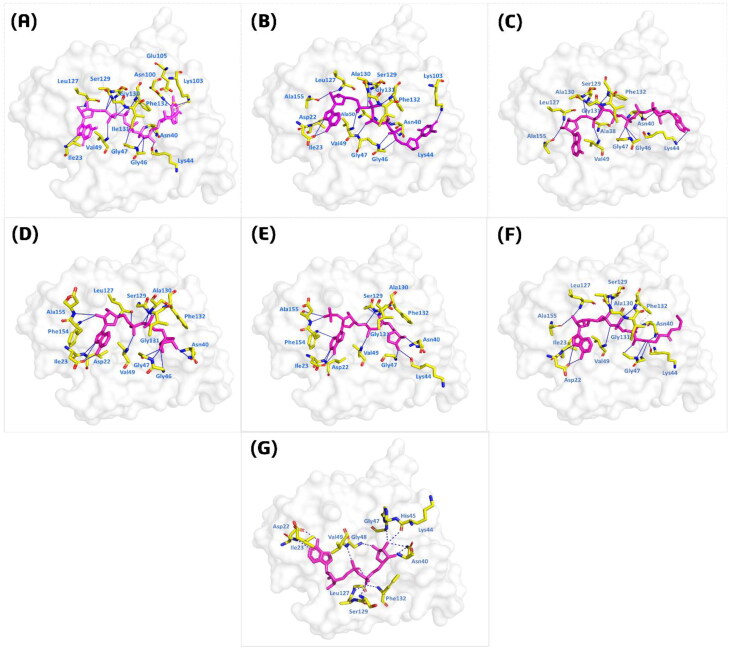
In the third round of screening GA implemented in AutoDock Vina was used with improved parameters to increase the accuracy. A total of 19 compounds with 64 exhaustiveness and Num_mode set to 25 was used. The top-scoring compounds were analyzed for final selection to be subjected to molecular dynamics simulation. The docking scores for these top compounds were predicted from −12.152 to −10.007 kcal/mol. The top six complexes were analyzed for interaction analysis and given in Figure 3. Compound P1,P5-Di(guanosine-5′) pentaphosphate ammonium salt (ChemSpider ID: 4484327) with the docking score −12.152 was found to form 9 hydrogen bonds with the key active site residues. Among these interacting residues, Asn40, which is essential for the function of this domain also form multiple interactions with the compound. A single pie-cation and salt bridge with Ala102 and Ala104 were formed. To understand the mechanism of how this compounds interacts with the receptor molecule an atomic level pattern was explored. It was observed that the [N51] of the ligand formed hydrogen bond with the Ile123 [N] by accepting a hydrogen. The same way [O7] formed interaction with [CA] of the Gln130 and [N] of Ile131. O [16] and O [19] from the ligand interacted with the [ND2] of Asn40 and [N] of gly47. Furthermore, O [43] formed a H-pi interaction by interacting with the 6-ring of the Phe130. Conversely, compound (R)-RETRO-THIORPHAN (ChemSpider ID: 394363) formed only 11 hydrogen bonds, but no other interactions were reported. The docking score for compounds (R)-RETRO-THIORPHAN was reported to be −12.27 kcal/mol. Among the 11 hydrogen bonds formed by (R)-RETRO-THIORPHAN, useful interactions with the two key residues Asp22 and Asn40 were also formed. Herein, the N [34] of the second ligand formed a hydrogen bond by interacting with the [N41] of Ile23. While in case of Asp22 [OD1] was reported to be involved in the interaction. Likewise, O [31], O [44], O [10], O [19] and O [52] interact with the [O] of Ala154, Ala138 while the [N] of Gly147, Ser128, Leu126, Ala50 and the [NZ] of Lys102 are actively involved in the interactions. Based on the docking score, the third-ranked compound significant interactions with Ala38, Asn40, Gly46, Gly47, Val49 and many other residues formed hydrogen bonds with the compound P(1),P(6)-bis(5′-adenosyl)hexaphosphate. The docking score for compound P(1),P(6)-bis(5′-adenosyl)hexaphosphate (ChemSpider ID: 110267) was reported to be −11.19 kcal/mol. No other interactions such as pie-cation, hydrophobic and salt bridges were reported. To further give an insight into the mechanism we have explored its atomic level interaction pattern. It was observed that O [45]-[O]Ala38, O [51]-[O]Ala154, O [7]-[CA]Gly30, O [7]-[N]Ile131, O [10]-[N]Gly130 and Ph2132, O [16] and [O19]- [ND2] Asn40 while the 6-ring of the ligand form a pi-hydrogen bond with the [CG] of Lys44. In addition, compound (Rib5)ppA (ChemSpider ID: 30975) with the docking score −11.03 kcal/mol also formed almost 10 hydrogen bonds the key residues, including significant interactions with Asp22 and Asn40. O [22]-[O]Leu126 acting as H-donor formed hydrogen interaction. N [36] and N [29] formed hydrogen bonds with [OD1] of Asp22 and [N] of Ile23. O [7]-[N]Val49, O [10]-[N]Gly130 and Phe132, O [20]-[ND2] Asn40, O [21]-[N]Ile131 and O [26]-[N]Phe156 were involved in hydrogen bonding. Compound known as NA7 (ChemSpider ID: 393480) formed multiple hydrogen interactions with Asp22 and Asn40. Besides, Lys44, Val49, Ser128, Gly30 and many other residues also formed multiple interactions. The docking score for compound NA7 was reported to be −11.03 kcal/mol. Furthermore, compound dephospho-CoA (ChemSpider ID: 24785028) formed two hydrophobic interactions with Ile131 and Phe132. Multiple hydrogen bonds with other active site residues including Asp22 and Asn40 were also reported. N [39] with [OD1] of Asp22, O [7]-[N]Val49, O [10]-[N]Gly130 and Phe132, O [18]-[ND2] Asn40, O [19]-[N]Gly46, O [20]-[N]Ser128, O [20]-[N]Val49, O [29]-[N]Asp157 and N [37]-[N]Ile23 act as H-acceptor formed significant interactions. The docking score for compound dephospho-CoA was found to be −10.94 kcal/mol. In this case the N [39] with [OD1] of Asp22 formed hydrogen bond, N [32] with Ile12, O [44] Gly47 [O], O [10]-[N]Gly130, O [41] and O [17]-[N]Ile131 while O [29]-[O]Ala154 formed hydrogen bonds. The positive control ADPr also formed interactions with the key residues and the docking was reported to be −9.76 kcal/mol. Hence, these results suggest that these compounds possess high affinity towards the macrodomain-I and its interactions with the key active site residues suggest that these compound possess strong inhibitory properties. Detail information regarding the interactions with the key residues such as hydrogen interaction, hydrophobic interactions, pie-cation interactions, salt-bridges and the docking scores are given in Table 1.
Figure 3.
Interaction pattern of the top 6 compounds. The light magenta colour shows the specific ligand while the yellow colour sticks are the interacting residues. (A) P1, P5-Di(guanosine-5') pentaphosphate ammonium salt (ChemSpider ID: 4484327), (B) (R)-RETRO-THIORPHAN (ChemSpider ID: 394363), (C) P(1), P(6)-bis(5'-adenosyl)hexaphosphate (ChemSpider ID: 110267), (D) (Rib5)ppA (ChemSpider ID: 30975), (E) NA7 (ChemSpider ID: 393480) (F) dephospho-CoA (ChemSpider ID: 24785028), (G) ADPr native ligand.
Table 1.
Detail information regarding the interactions with the key residues such as hydrogen interaction, hydrophobic interactions, pie-cation interactions, salt-bridges and the docking scores are given kcal/mol.
| ChemSpider Compound ID |
Interactions |
Docking score | Bioactivity | ||
|---|---|---|---|---|---|
| Hydrogen bonding | Hydrophobic bonds | Other bonds | |||
P1,P5-Di(guanosine-5′) pentaphosphate ammonium salt (ChemSpider ID: 4484327)
|
Ala38, Asn40, Gly46, Gly47, Val49, Ser128, Gly130, Ile131,Phe132 | – | Ala102, Ala104 | –12.15 | 0.74 |
|
(R)-RETRO-THIORPHAN (ChemSpider ID: 394363) 
|
Asp22, Ile23, Asn40, Gly46, Gly47, Val49, Ser128, Gly130, Ile131,Phe132, | – | – | –12.27 | 0.82 |
P(1),P(6)-bis(5′-adenosyl) hexaphosphate (ChemSpider ID: 110267)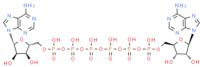
|
Asn40, Ly44, Gly46, Val49, Gly47, Leu127, Ser128, Gly130, Ile131,Phe132, Ala155 | – | – | –11.19 | 0.75 |
(Rib5)ppA (ChemSpider ID: 30975)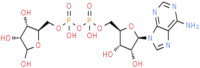
|
Asp22, Ile23, Asn40, Gly46, Gly47, Val49, Ser128, Gly130, Ile131,Phe132, | – | – | –11.03 | 0.58 |
NA7 (ChemSpider ID: 393480)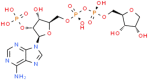
|
Asn40, Ly44, Val49, Gly47, Leu127, Ser128, Gly130, Ile131,Phe132, Ala155 | – | – | –11.03 | 0.52 |
dephospho-CoA (ChemSpider ID: 24785028)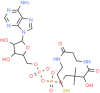
|
Asp22, Asn40, Ly44, Val49, Gly47, Leu127, Ser128, Gly130, Ile131,Phe132, Ala155 | Ile131,Phe132 | – | –10.94 | 0.64 |
ADPr (ChemSpider ID:24785028)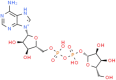
|
Asp22, Asn40, Ly44, Gly46, Gly47, Ser128, Gly130, Ala155 | Als50, Ile131, Phe132 | – | –9.76 | 0.59 |
Furthermore, results obtained from the server shows that all these shortlisted compounds are active against the enzyme targets. From the scores, it can be seen that compound 1 with score 0.74 possesses inhibitory effects against the enzymes while the others the reported score for compound 2 (0.82), compound 3 (0.75), compound 4 (0.58), compound 5 (0.52) while compound 6 possess (0.64) bioactivity score against the macrodomain-I. Conversely, the bioactivity score predicted for the ADPr positive control was 0.59. Thus, these results strongly suggest that the shortlisted compounds could efficiently bind with the macrodomain-I in the experimental setup and should be tested for clinical trials as all the compounds are obeying the ADMET rules expect few.
Stability of the protein–ligand complexes
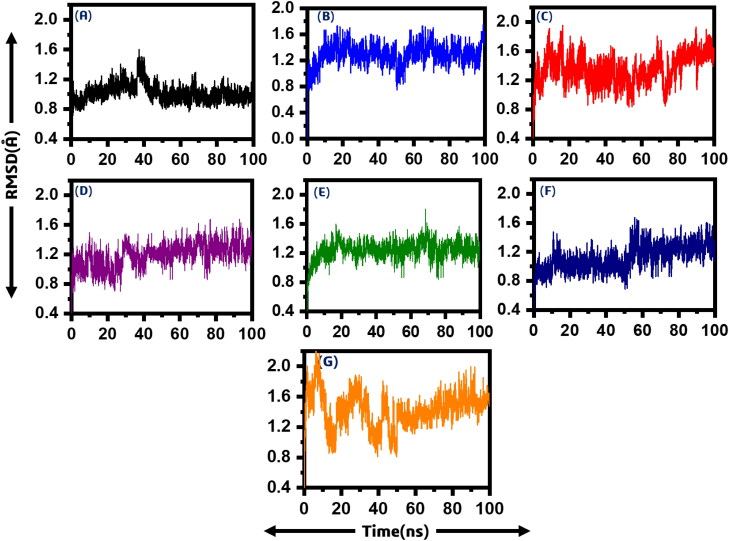
After the screening of all the drugs and re-scoring, the best hits were subjected to molecular dynamics simulation to understand its dynamic behaviour. To calculate the dynamic stability of all the systems root mean square deviation (RMSD) was calculated as a function of time. In the case of compound (ChemSpider ID: 4484327) it can be seen that the average RMSD value is 1.0 Å. Soon, the system entered the production state. No significant convergence was observed; however, at 38 ns, a little convergence was observed. However, after 38 ns the system, RMSD value again fell to the normal. On the hand, the average RMSD value for compound (ChemSpider ID: 394363) was found to be relatively high. It was reported to be 1.4 Å. A major convergence between 50 and 58 ns was observed, which is due to the relatively large size of the ligand. In addition, the third system (ChemSpider ID: 110267) relatively showed unstable behaviour till 25 ns then soon the system entered the equilibrium state, and the average RMSD was found to 1.4 Å. It can be seen that the RMSD converged between 60 and 80 ns but again the system gained the stability and remained stable.
Furthermore, compound (ChemSpider ID: 30975) showed convergence between 30 and 35 ns with an average RMSD 1.0 Å. A little convergence at different intervals was also observed. On the other hand, the systems compound (ChemSpider ID: 393480) and compound 6 relatively remained more stable, and no major convergence was observed. A small fluctuation between 65 and 75 ns was reported. It can be seen that the average RMSD for compound 5 was reported to be 1.0 Å while 1.2 Å for ChemSpider ID: 24785028. A major convergence between 55 and 60 ns was observed but the system remained stable until 100 ns. Thus, these results strongly indicate that these compounds, when bound to the macrodomain-I, remained stable. Furthermore, ADPr was used as positive control to compare the results. It can be seen that the ADPr complex is relatively more unstable during simulation until 50 ns but afterward the system gain the stability. Figure 4 shows the RMSD graphs of all the six systems. The x-axis is showing the simulation time in nanoseconds while the y-axis is showing the RMSD in Angstrom. The RMSDs for the replicate 2 and replicate 3 are given in Supporting Information Figures S1 and S2.
Figure 4.
Showing the RMSD of all the seven systems. The x-axis is showing the time in nanoseconds while the y-axis is showing RMSD in Angstrom. (A) P1, P5-Di(guanosine-5') pentaphosphate ammonium salt (ChemSpider ID: 4484327), (B) (R)-RETRO-THIORPHAN (ChemSpider ID: 394363), (C) P(1), P(6)-bis(5'-adenosyl)hexaphosphate (ChemSpider ID: 110267), (D) (Rib5)ppA (ChemSpider ID: 30975), (E) NA7 (ChemSpider ID: 393480) (F) dephospho-CoA (ChemSpider ID: 24785028), (G) ADPr native ligand.
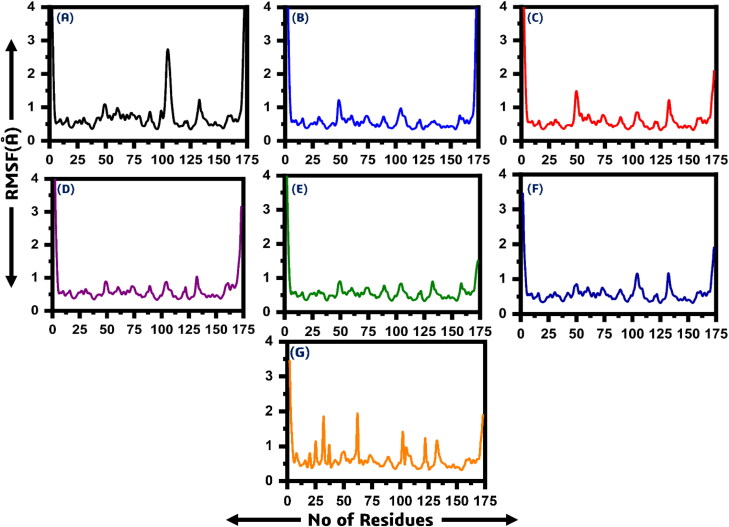
Residual flexibility of all the systems
The flexibility of each system was accessed by using Root mean square fluctuation (RMSF). It can be seen that all the systems showed a more similar pattern of residual fluctuations. In the case of compound 1 a little different pattern of residual flexibility can be observed at residue position 50–55 and 100–105. It is due to the binding of the ligand with these residues and thus decreases the flexibility between 50 and 55 residues while the flexibility was increased between 100 and 105. The average RMSF value for all the systems was observed to be 1.0 Å. Figure 5 shows the RMSFs graphs of all the six systems. The x-axis is showing the number of residues while the y-axis is showing the RMSF in Angstrom. Using ADPr as positive control the results shows that ADPr complex possess higher residual fluctuations at different regions.
Figure 5.
Showing the RMSF of all the seven systems. The x-axis is showing the time in number of residues while the y-axis is showing RMSF in Angstrom. (A) P1, P5-Di(guanosine-5') pentaphosphate ammonium salt (ChemSpider ID: 4484327), (B) (R)-RETRO-THIORPHAN (ChemSpider ID: 394363), (C) P(1), P(6)-bis(5'-adenosyl)hexaphosphate (ChemSpider ID: 110267), (D) (Rib5)ppA (ChemSpider ID: 30975), (E) NA7 (ChemSpider ID: 393480) (F) dephospho-CoA (ChemSpider ID: 24785028), (G) ADPr native ligand.
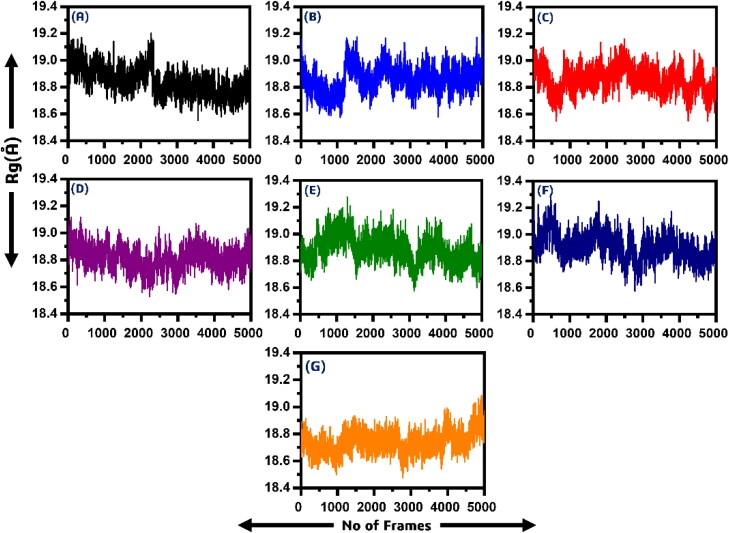
Structural compactness through Rg
In order to calculate the compactness of all the ligand-bound systems, Rg (radius of gyration) was calculated. The stability of the complexes formed also depended on the compactness between the ligand and the target. From Figure 6, it can be easily observed that the average Rg value for all the systems is between 18.2–18.6 Å. In the case of P1, P5-Di(guanosine-5′) pentaphosphate ammonium salt (ChemSpider ID: 4484327) the Rg value remained high till 10 ns. Soon after reaching 40 ns a little convergence was observed until 42 ns. However, the compactness remained 18.8 Å till the end of the simulation. In the case of (R)-RETRO-THIORPHAN (ChemSpider ID: 394363) the system remained relatively more compact than the first one. The average Rg value was reported to be 19 Å with significant fluctuation during the simulation time.
Figure 6.
Showing the Rg of all the six systems. The x-axis is showing the number of frames while the y-axis is showing the Rg in Angstrom. (A) P1, P5-Di(guanosine-5') pentaphosphate ammonium salt (ChemSpider ID: 4484327), (B) (R)-RETRO-THIORPHAN (ChemSpider ID: 394363), (C) P(1), P(6)-bis(5'-adenosyl)hexaphosphate (ChemSpider ID: 110267), (D) (Rib5)ppA (ChemSpider ID: 30975), (E) NA7 (ChemSpider ID: 393480) (F) dephospho-CoA (ChemSpider ID: 24785028), (G) ADPr native ligand.
Furthermore, P(1), P(6)-bis(5′-adenosyl)hexaphosphate (ChemSpider ID: 110267) showed no significant divergence of Rg. Initially, the Rg value remained low till 20 ns, but soon the Rg value fluctuated and the value remained between 18.8 and 19.00 Å. After reaching 20 ns, the system converged its compactness again, and the Rg value remained 18.8 Å until the end of the simulation. The average Rg value for (Rib5)ppA (ChemSpider ID: 30975) was 18.6–18.8 Å with no major fluctuation was observed. Thus, this system also showed relatively more uniform compactness than the others. In addition, the similar pattern of Rg was also observed for NA7 (ChemSpider ID: 393480), but the average Rg was seen to be higher than (Rib5)ppA. The Rg value for compound 5 was reported to be 18.8 Å. Significant convergence at different intervals was observed. In the case of dephospho-CoA (ChemSpider ID: 24785028) the Rg value remained a little higher in the start, but major fluctuation at 15 ns and 60 ns can be observed while the Rg value for rest of simulation remained to 18.8 Å. Thus, these results also imply that the ligands bound to the macrodomain-I are dynamically stable and the binding is also dynamically favourable. It can be also seen that the ADPr complex also possess more compactness. Figure 6 shows the Rg graphs of all the six systems. The x-axis is showing the number of frames while the y-axis is showing the Rg in Angstrom. The Rg of replicate 2 and replicate 3 are given in Supporting Information Figures S3 and S4.
Binding free energy
A common technique, MM-GBSA, has been used to estimate the binding free energy of all six systems. The free energy binding specifies the binding interaction between the ligands and the target protein or receptor. Table 2 provides each energy expression, like van der Waals energy, electrostatic energy, polar solvent energy, solvent-accessible surface energy and total binding free energy of all the systems. It can be seen that the P1,P5-Di(guanosine-5′) pentaphosphate ammonium salt (ChemSpider ID: 4484327) system possesses the highest total binding energy of −71.996 kcal/mol. The total binding energy for (R)-RETRO-THIORPHAN (ChemSpider ID: 394363) was reported to be −70.114 kcal/mol. Conversely, the total binding energies for P(1),P(6)-bis(5′-adenosyl)hexaphosphate (ChemSpider ID: 110267), (Rib5)ppA (ChemSpider ID: 30975), NA7 (ChemSpider ID: 393480) and dephospho-CoA (ChemSpider ID: 24785028) was reported to be −66.201 kcal/mol, −66.949 kcal/mol, −70.268 kcal/mol and −66.965 kcal/mol respectively. We used ADPr the native ligand as positive control to compare our results. The total binding energy for ADPr was reported to be −67.123 kcal/mol. Thus it confirms that these compound possess significant binding free energy against the Macrodomain-I. While the other energy terms such as van der Waals energy (ΔvdW), electrostatic energy (Δelec), polar solvent energy (Δps), solvent-accessible surface area (ΔSASA) are listed in Table 2, these findings strongly indicate that these shortlisted compounds must be experimentally tested against SARs-CoV-2 as soon as possible. The MMGBSA results of replicate 2 and replicate 3 are given in Supporting Information Tables S1 and S2.
Table 2.
The table listed different energy terms calculated for the six complexes subjected to MM-GBSA analysis.
| Complexes | MM-GBSA (kcal/mol) |
||||
|---|---|---|---|---|---|
| ΔvdW | Δelec | Δps | ΔSASA | ΔGTotal | |
| P1,P5-Di(guanosine-5′) pentaphosphate ammonium salt (ChemSpider ID: 4484327) | –82.663 | –14.465 | 30.700 | –5.471 | –71.996 |
| (R)-RETRO-THIORPHAN(ChemSpider ID: 394363) | –78.452 | –22.563 | 32.785 | –4.125 | –70.114 |
| P(1),P(6)-bis(5′-adenosyl)hexaphosphate (ChemSpider ID: 110267) | –66.357 | –20.241 | 32.354 | –3.478 | –66.201 |
| (Rib5)ppA (ChemSpider ID: 30975) | –65.187 | –13.175 | 205.14 | –3.247 | –66.949 |
| NA7 (ChemSpider ID: 393480) | –66.257 | –12.785 | 23.478 | –3.658 | –70.268 |
| dephospho-CoA (ChemSpider ID: 24785028) | –72.613 | –19.223 | 21.365 | –3.007 | –66.965 |
| ADPr | –64.136 | –17.985 | 23.278 | –4.356 | –67.123 |
All the energies are given in kcal/mol.
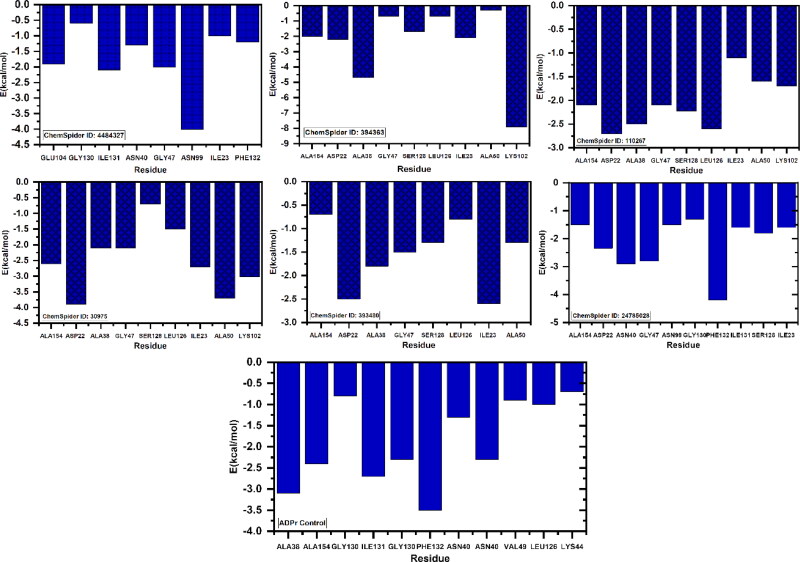
Per-residue energy decomposition
To understand the impact of each residue and its contribution to the total energy, per-residue energy decomposition analysis was carried out on the trajectory file obtained from 100 ns simulation. The per-residue contribution of each system was calculated and given in Figure 7. As given it can be seen that Asp22, Ala38, Asn40, Val44, Phe144 significantly contributed to the total energy while the other residues such as Gly46, Gly47, Leu127, Ser128, Gly130, Ile131, Phe132 and Ala155 also contributed to the total energy.
Figure 7.
The figure is showing the per-residue energy decomposition of the residues contributing to the total binding energy. All the energies are given in kcal/mol. (A) P1, P5-Di(guanosine-5') pentaphosphate ammonium salt (ChemSpider ID: 4484327), (B) (R)-RETRO-THIORPHAN (ChemSpider ID: 394363), (C) P(1), P(6)-bis(5'-adenosyl)hexaphosphate (ChemSpider ID: 110267), (D) (Rib5)ppA (ChemSpider ID: 30975), (E) NA7 (ChemSpider ID: 393480) (F) dephospho-CoA (ChemSpider ID: 24785028), (G) ADPr native ligand.
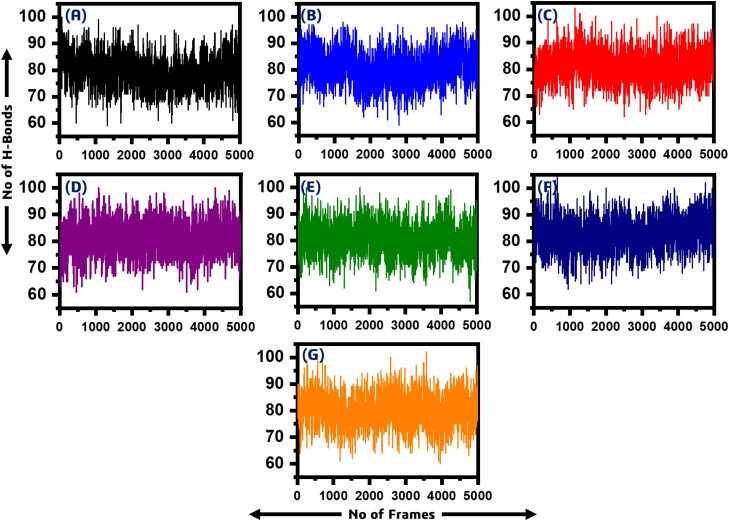
Hydrogen bonding analysis
In order to determine the half-life of each interaction with the key residues we used hydrogen bonding occupancy approach. A 50 ns trajectory was used for each system to monitor the hydrogen bonding during the course of simulation. It can be seen that most of the ligands formed significant hydrogen bonds for longer time. In case of all the six ligands it can be observed that the bonding with Asp22 and Asn40 remained for longer time than the others. All the results are calculated in percentage trajectories (0.79 mean 79% of the trajectories) and given in Table 3.
Table 3.
The hydrogen bonding occupancy of the key residues during the course of simulation.
| Complex | Life time (hydrogen bonding) |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Asp22 | Asn40 | Val44 | Gly46 | Gly47 | Ser128 | Gly130 | Ile131 | Ala155 | |
| P1,P5-Di(guanosine-5′) pentaphosphate ammonium salt (ChemSpider ID: 4484327) | 0.73 | 0.72 | 0.47 | 0.52 | 0.68 | 0.31 | 0.55 | 0.42 | 0.52 |
|
(R)-RETRO-THIORPHAN (ChemSpider ID: 394363) |
0.69 | 0.65 | 0.57 | 0.52 | 0.52 | 0.44 | 0.59 | 0.36 | 0.55 |
| P(1),P(6)-bis(5′-adenosyl)hexaphosphate (ChemSpider ID: 110267) | 0.58 | 0.56 | 0.50 | 0.43 | 0.52 | 0.48 | 0.49 | 0.36 | 0.51 |
| (Rib5)ppA (ChemSpider ID: 30975) | 0.67 | 0.63 | 0.42 | 0.41 | 0.29 | 0.51 | 0.37 | 0.39 | 0.48 |
| NA7 (ChemSpider ID: 393480) | 0.58 | 0.66 | 0.51 | 0.48 | 0.39 | 0.20 | 0.47 | 0.12 | 0.42 |
| dephospho-CoA (ChemSpider ID: 24785028) | 0.42 | 0.56 | 0.25 | 0.62 | 0.52 | 0.34 | 0.41 | 0.28 | 0.42 |
| ADPr | 0.61 | 0.63 | 0.14 | 0.32 | 0.45 | 0.39 | 0.57 | 0.45 | 0.37 |
Each residue bonding is given in percentage trajectories.
Furthermore, we also calculated intra-molecular H-bonds which revealed that the wild type has 79.40 (average H-Bonds), P1, P5-Di(guanosine-5′) pentaphosphate ammonium salt has 80.12, (R)-RETRO-THIORPHAN has 80.64, P(1),P(6)-bis(5′-adenosyl)hexaphosphate has 81.24, (Rib5)ppA has 81.10, NA7 has 80.79 while dephospho-CoA formed 82.82 hydrogen bonds in average. The intra-molecule H-bonds are given in Figure 8.
Figure 8.
A total number of intramolecular H-bond in all the systems. (A) wild type (79.40), (B) P1, P5-Di(guanosine-5') pentaphosphate ammonium salt (80.12) (C) (R)-RETRO-THIORPHAN (80.64), (D) P(1),P(6)-bis(5'-adenosyl)hexaphosphate (81.24), (E) (Rib5)ppA (81.10), (F) NA7 (80.79) (G) while dephospho-CoA formed (82.82).
Discussion
The outbreak of SAR-CoV-2 has now affected almost every corner of the world. Increasing fatalities and mortalities has created a devastating scenario. With the health issue, the world has suffered from economic loss too. This pandemic has forced the world scientists, economists and leaders to find a solution to cope with this disease (A. Khan, M. T. Khan, et al., 2020). This virus belongs to beta-coronaviruses, and previously some viruses such as SARs and MERs are already reported to infect humans through bat and camel. However, the recent SARs-CoV-2 seems to be more devastating because it can be transmitted from human-to-human and reinfections are also reported.
To cope with this infection, scientists and researchers are focusing on many aspects of this virus and host factors. Scientists are targeting every gene/protein of this virus to get some breakthrough and control this pandemic finally. Many studies reported targeting the RdRp protein, PLpro, 3CLpro while others focus on host proteins such as TRPM2. The role of these genes/proteins in the transmission, pathogenesis and spread is indispensable. Besides, sixteen domains long (∼1945) amino acids protein nsp3 is also a major nonstructural protein with its precise role in transcription/translation, interaction with the host proteins and hijacking the host immune system. These diverse functions are domain-specific. Among the total sixteen domains, macrodomain-I, also known as SARs-unique domain or X-domain, play an essential role in the binding of G-quadruplex and hijacking the host immune response. The binding of ADPr to this domain is required for essential processes. Thus, targeting this domain with high binding affinity drugs could help to reduce the burden and contain this pandemic. Therefore, using different approaches such as rational drug designing, conventional computer-aided drug designing approach or the most recent deep learning and artificial intelligence-based approaches could help to find a drug that could target this domain and rescue the host immune system.
Meanwhile, the best approach is to repurpose the old drugs such as Chloroquine, Remdesivir, Dexamethasone and other anti-viral drugs. These drugs are reported to work in some cases, but the use of these drugs is limited by the associated problems. With the practice of repurposing the old drugs, the quest for finding a new drug is still on. In this regard, the use of structure-based drug screening methods is the most useful approach to find a new drug with acceptable pharmacokinetics and pharmacodynamics properties. Likewise, here, we also used CADD approaches to search for a potent molecule that could target the macrodomain-I. In addition to the drug similarity search, we also screened two medicinal compounds databases including south African northern medicinal compounds database and traditional Chinese medicines database (TCM). Promising hits were identified, and their validity was checked again and again. The top hits were confirmed by performing IFD, which further shortlisted the top hits list very precisely. Using another round of docking with different algorithm exempted further hits from the list and shortlisted the top hits. The use of molecular dynamics simulation technique and free energy calculations is the most widely practiced approaches while studying the protein–ligand interaction. Integrating this pipeline further increased the reliability of the quest to test our top hits experimentally because of its promising results. Thus, this study comprised of a complicated and multiple validations stresses on the experimental assays of the top hits to help to contain the recent outbreak.
Based on the findings of the bioinformatics study, we targeted Macrodomain-I from SARS-COV-2 utilizing virtual drug screening and drug similarity frameworks to shortlist the most active compounds. The results of the entire article emphasize the prospective inhibiting effect of the top hits. We have not yet performed more anti-viral studies in vivo and in vitro, as we want to share our findings in anti-SARS-CoV-2 work with scientists as early as possible. This research will assist repurpose the drug design, conduct in vivo and in vitro evaluations of candidate drugs obtained in this study, and prepare for applications for clinical trials.
Conclusion
In conclusion, a thorough investigation of different drugs against the macrodomain-I was performed. A total of 64,043 drugs were screened, and the most potential hits were re-docked using the IFD algorithm. The final best hits were validated by using molecular dynamics simulation and free energy calculations. These results suggest that the top six hits identified efficiently bind to the receptor. Our analysis is based on computational pipeline and demands the experimental test of these top hits, which could help to contain the disease caused by SARs-CoV-2.
Supplementary Material
Disclosure statement
No potential conflict of interest was reported by the authors.
Authors contribution
All the authors were involved in the analysis, writing and revision of the manuscript.
References
- Anandakrishnan, R., Aguilar, B., & Onufriev, A. V. (2012). H++ 3.0: Automating pK prediction and the preparation of biomolecular structures for atomistic molecular modeling and simulations. Nucleic Acids Research, 40, W537–W541. 10.1093/nar/gks375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angeletti, S., Benvenuto, D., Bianchi, M., Giovanetti, M., Pascarella, S., & Ciccozzi, M. (2020). COVID-2019: The role of the nsp2 and nsp3 in its pathogenesis. Journal of Medical Virology, 92(6), 584–588. 10.1002/jmv.25719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case, D. A., Cheatham, T. E., Darden, T., Gohlke, H., Luo, R., Merz, K. M., Onufriev, A., Simmerling, C., Wang, B., & Woods, R. J. (2005). The Amber biomolecular simulation programs. Journal of Computational Chemistry, 26(16), 1668–1688. 10.1002/jcc.20290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, J. F.-W., Kok, K.-H., Zhu, Z., Chu, H., To, K. K.-W., Yuan, S., & Yuen, K.-Y. (2020). Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerging Microbes & Infections, 9(1), 221–236. 10.1080/22221751.2020.1719902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee, A., Johnson, M. A., Serrano, P., Pedrini, B., Joseph, J. S., Neuman, B. W., Saikatendu, K., Buchmeier, M. J., Kuhn, P., & Wüthrich, K. (2009). Nuclear magnetic resonance structure shows that the severe acute respiratory syndrome coronavirus-unique domain contains a macrodomain fold. Journal of Virology, 83(4), 1823–1836. 10.1128/JVI.01781-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, C. Y.-C. (2011). TCM Database@Taiwan: The world's largest traditional Chinese medicine database for drug screening in silico. PLoS One, 6(1), e15939. 10.1371/journal.pone.0015939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, V. B., Arendall, W. B., Headd, J. J., Keedy, D. A., Immormino, R. M., Kapral, G. J., Murray, L. W., Richardson, J. S., & Richardson, D. C. (2010). MolProbity: All-atom structure validation for macromolecular crystallography . Acta Crystallographica Section D Biological Crystallography, 66(Pt 1), 12–21. 10.1107/S0907444909042073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daina, A., Michielin, O., & Zoete, V. (2017). SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Scientific Reports, 7, 42717. 10.1038/srep42717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidchack, R. L., Handel, R., & Tretyakov, M. (2009). Langevin thermostat for rigid body dynamics. The Journal of Chemical Physics, 130(23), 234101. 10.1063/1.3149788 [DOI] [PubMed] [Google Scholar]
- DeLano, W. L. (2002). Pymol: An open-source molecular graphics tool. CCP4 Newsletter on Protein Crystallography, 40(1), 82–92. [Google Scholar]
- Egloff, M.-P., Malet, H., Putics, A., Heinonen, M., Dutartre, H., Frangeul, A., Gruez, A., Campanacci, V., Cambillau, C., Ziebuhr, J., Ahola, T., & Canard, B. (2006). Structural and functional basis for ADP-ribose and poly(ADP-ribose) binding by viral macro domains. Journal of Virology, 80(17), 8493–8502. 10.1128/JVI.00713-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson, K. K., Cervantes-Barragán, L., Ludewig, B., & Thiel, V. (2008). Mouse hepatitis virus liver pathology is dependent on ADP-ribose-1''-phosphatase, a viral function conserved in the alpha-like supergroup. Journal of Virology, 82(24), 12325–12334. 10.1128/JVI.02082-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehr, A. R., Channappanavar, R., Jankevicius, G., Fett, C., Zhao, J., Athmer, J., Meyerholz, D. K., Ahel, I., & Perlman, S. (2016). The conserved coronavirus macrodomain promotes virulence and suppresses the innate immune response during severe acute respiratory syndrome coronavirus infection. mBio, 7(6), 598-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehr, A. R., Jankevicius, G., Ahel, I., & Perlman, S. (2018). Viral macrodomains: Unique mediators of viral replication and pathogenesis. Trends in Microbiology, 26(7), 598–610. 10.1016/j.tim.2017.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehr, A. R., & Perlman, S. (2015). Coronaviruses: An overview of their replication and pathogenesis Coronaviruses (pp. 1–23). Springer. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junaid, M., Shah, M., Khan, A., Li, C.-D., Khan, M. T., Kaushik, A. C., Ali, A., Mehmood, A., Nangraj, A. S., Choi, S., & Wei, D.-Q. (2019). Structural-dynamic insights into the H. pylori cytotoxin-associated gene A (CagA) and its abrogation to interact with the tumor suppressor protein ASPP2 using decoy peptides. Journal of Biomolecular Structure & Dynamics, 37(15), 4035–4050. 10.1080/07391102.2018.1537895 [DOI] [PubMed] [Google Scholar]
- Khan, A., Ali, S. S., Khan, M. T., Saleem, S., Ali, A., Suleman, M., Babar, Z., Shafiq, A., Khan, M., & Wei, D.Q. (2020). Combined drug repurposing and virtual screening strategies with molecular dynamics simulation identified potent inhibitors for SARS-CoV-2 main protease (3CLpro). Journal of Biomolecular Structure and Dynamics, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan, A., Junaid, M., Kaushik, A. C., Ali, A., Ali, S. S., Mehmood, A., & Wei, D.-Q. (2018). Computational identification, characterization and validation of potential antigenic peptide vaccines from hrHPVs E6 proteins using immunoinformatics and computational systems biology approaches. PLoS One, 13(5), e0196484. 10.1371/journal.pone.0196484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan, A., Rehman, A-U., Junaid, M., Li, C.-D., Saleem, S., Humayun, F., Shamas, S., Ali, S. S., Babar, Z., & Wei, D.-Q. (2020). Dynamics insights into the gain of flexibility by Helix-12 in ESR1 as a mechanism of resistance to drugs in breast cancer cell lines. Frontiers in Molecular Biosciences, 6, 159. 10.3389/fmolb.2019.00159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan, A., Kaushik, A. C., Ali, S. S., Ahmad, N., & Wei, D.-Q. (2019). Deep-learning-based target screening and similarity search for the predicted inhibitors of the pathways in Parkinson's disease. RSC Advances, 9(18), 10326–10339. 10.1039/C9RA01007F [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan, A., Saleem, S., Idrees, M., Ali, S. S., Junaid, M., Kaushik, A. C., & Wei, D.-Q. (2018). Allosteric ligands for the pharmacologically important Flavivirus target (NS5) from ZINC database based on pharmacophoric points, free energy calculations and dynamics correlation. Journal of Molecular Graphics & Modelling, 82, 37–47. 10.1016/j.jmgm.2018.03.004 [DOI] [PubMed] [Google Scholar]
- Khan, A., Khan, M. T., Saleem, S., Junaid, M., Ali, A., Shujait Ali, S., Khan, M., & Wei, D.Q. (2020). Structural Insights into the mechanism of RNA recognition by the N-terminal RNA-binding domain of the SARS-CoV-2 nucleocapsid phosphoprotein. Computational and Structural Biotechnology Journal. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan, M. T., Ali, A., Wang, Q., Irfan, M., Khan, A., Zeb, M. T., Zhang, Y.J., Chinnasamy, S., & Wei, D.Q. (2020). Marine natural compounds as potents inhibitors against the main protease of SARS-CoV-2. A molecular dynamic study. Journal of Biomolecular Structure and Dynamics, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kräutler, V., van Gunsteren, W. F., & Hünenberger, P. H. (2001). A fast SHAKE algorithm to solve distance constraint equations for small molecules in molecular dynamics simulations. Journal of Computational Chemistry, 22(5), 501–508. [DOI] [Google Scholar]
- Kusov, Y., Tan, J., Alvarez, E., Enjuanes, L., & Hilgenfeld, R. (2015). A G-quadruplex-binding macrodomain within the “SARS-unique domain” is essential for the activity of the SARS-coronavirus replication-transcription complex. Virology, 484, 313–322. 10.1016/j.virol.2015.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Land, H., & Humble, M. S. (2018). YASARA: A tool to obtain structural guidance in biocatalytic investigations protein engineering (pp. 43–67). Springer. [DOI] [PubMed] [Google Scholar]
- Lei, J., Kusov, Y., & Hilgenfeld, R. (2018). Nsp3 of coronaviruses: Structures and functions of a large multi-domain protein. Antiviral Research, 149, 58–74. 10.1016/j.antiviral.2017.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, Y., Pan, D., Li, J., Zhang, L., & Shao, X. (2017). Application of Berendsen barostat in dissipative particle dynamics for nonequilibrium dynamic simulation. The Journal of Chemical Physics, 146(12), 124108. 10.1063/1.4978807 [DOI] [PubMed] [Google Scholar]
- Lu, R., Zhao, X., Li, J., Niu, P., Yang, B., Wu, H., Wang, W., Song, H., Huang, B., Zhu, N., Bi, Y., Ma, X., Zhan, F., Wang, L., Hu, T., Zhou, H., Hu, Z., Zhou, W., Zhao, L., … Tan, W. (2020). Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. The Lancet, 395(10224), 565–574. 10.1016/S0140-6736(20)30251-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris, G. M., Huey, R., Lindstrom, W., Sanner, M. F., Belew, R. K., Goodsell, D. S., & Olson, A. J. (2009). AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. Journal of Computational Chemistry, 30(16), 2785–2791. 10.1002/jcc.21256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ntie-Kang, F., Telukunta, K. K., Döring, K., Simoben, C. V., A Moumbock, A. F., Malange, Y. I., Njume, L. E., Yong, J. N., Sippl, W., & Günther, S. (2017). NANPDB: A resource for natural products from Northern African sources. Journal of Natural Products, 80(7), 2067–2076. 10.1021/acs.jnatprod.7b00283 [DOI] [PubMed] [Google Scholar]
- Pearlman, D. A., Case, D. A., Caldwell, J. W., Ross, W. S., Cheatham, T. E., DeBolt, S., Ferguson, D., Seibel, G., & Kollman, P. (1995). AMBER, a package of computer programs for applying molecular mechanics, normal mode analysis, molecular dynamics and free energy calculations to simulate the structural and energetic properties of molecules. Computer Physics Communications, 91(1–3), 1–41. 10.1016/0010-4655(95)00041-D [DOI] [Google Scholar]
- Piotrowski, Y., Hansen, G., Boomaars, van der Zanden, A. L., Snijder, E. J., Gorbalenya, A. E., & Hilgenfeld, R. (2009). Crystal structures of the X-domains of a Group-1 and a Group-3 coronavirus reveal that ADP-ribose-binding may not be a conserved property. Protein Science, 18(1), 6–16. 10.1002/pro.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putics, A., Filipowicz, W., Hall, J., Gorbalenya, A. E., & Ziebuhr, J. (2005). ADP-ribose-1"-monophosphatase: A conserved coronavirus enzyme that is dispensable for viral replication in tissue culture. Journal of Virology, 79(20), 12721–12731. 10.1128/JVI.79.20.12721-12731.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quimque, M. T. J., Notarte, K. I. R., Fernandez, R. A. T., Mendoza, M. A. O., Liman, R. A. D., Lim, J. A. K., Pilapil, L. A. E., Ong, J. K. H., Pastrana, A. M., Khan, A., Wei, D.Q., & Macabeo, A. P. G. (2020). Virtual screening-driven drug discovery of SARS-CoV2 enzyme inhibitors targeting viral attachment, replication, post-translational modification and host immunity evasion infection mechanisms. Journal of Biomolecular Structure and Dynamics, 1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raoult, D., Zumla, A., Locatelli, F., Ippolito, G., & Kroemer, G. (2020). Coronavirus infections: Epidemiological, clinical and immunological features and hypotheses. Cell Stress, 4(4), 66–75. 10.15698/cst2020.04.216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roe, D. R., & Cheatham, I. I. I., T. E. (2013). PTRAJ and CPPTRAJ: Software for processing and analysis of molecular dynamics trajectory data. Journal of Chemical Theory and Computation, 9(7), 3084–3095. 10.1021/ct400341p [DOI] [PubMed] [Google Scholar]
- Shimizu, J. F., Martins, D. O. S., McPhillie, M. J., Roberts, G. C., Zothner, C., Merits, A., Harris, M., & Jardim, A. C. G. (2020). Is the ADP ribose site of the Chikungunya virus NSP3 Macro domain a target for antiviral approaches? Acta Tropica, 207, 105490. 10.1016/j.actatropica.2020.105490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumoza-Toledo, A., & Penner, R. (2011). TRPM2: A multifunctional ion channel for calcium signalling. The Journal of Physiology, 589(Pt 7), 1515–1525. 10.1113/jphysiol.2010.201855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, H., Li, Y., Tian, S., Xu, L., & Hou, T. (2014). Assessing the performance of MM/PBSA and MM/GBSA methods. 4. Accuracies of MM/PBSA and MM/GBSA methodologies evaluated by various simulation protocols using PDBbind data set. Physical Chemistry Chemical Physics, 16(31), 16719–16729. 10.1039/c4cp01388c [DOI] [PubMed] [Google Scholar]
- Toukmaji, A., Sagui, C., Board, J., & Darden, T. (2000). Efficient particle-mesh Ewald based approach to fixed and induced dipolar interactions. The Journal of Chemical Physics, 113(24), 10913–10927. 10.1063/1.1324708 [DOI] [Google Scholar]
- Trott, O., & Olson, A. J. (2010). AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. Journal of Computational Chemistry, 31(2), 455–461. 10.1002/jcc.21334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassetti, D., Pagliai, M., & Procacci, P. (2019). Assessment of GAFF2 and OPLS-AA general force fields in combination with the water models TIP3P, SPCE, and OPC3 for the solvation free energy of druglike organic molecules. Journal of Chemical Theory and Computation, 15(3), 1983–1995. 10.1021/acs.jctc.8b01039 [DOI] [PubMed] [Google Scholar]
- Vijayasri, S., & Hopper, W. (2017). Towards the identification of novel phytochemical leads as macrodomain inhibitors of chikungunya virus using molecular docking approach. Journal of Applied Pharmaceutical Science, 7, 74–82. [Google Scholar]
- Vilar, S., Cozza, G., & Moro, S. (2008). Medicinal chemistry and the molecular operating environment (MOE): Application of QSAR and molecular docking to drug discovery. Current Topics in Medicinal Chemistry, 8(18), 1555–1572. 10.2174/156802608786786624 [DOI] [PubMed] [Google Scholar]
- Wang, J., Wang, W., Kollman, P. A., & Case, D. A. (2001). Antechamber: An accessory software package for molecular mechanical calculations. Journal of the American Chemical Society, 222U, 403. [Google Scholar]
- Wang, Y., Khan, A., Chandra Kaushik, A., Junaid, M., Zhang, X., & Wei, D.-Q. (2019). The systematic modeling studies and free energy calculations of the phenazine compounds as anti-tuberculosis agents. Journal of Biomolecular Structure & Dynamics, 37(15), 4051–4069. 10.1080/07391102.2018.1537896 [DOI] [PubMed] [Google Scholar]
- Wu, Y., Ho, W., Huang, Y., Jin, D.-Y., Li, S., Liu, S.-L., Liu, X., Qiu, J., Sang, Y., Wang, Q., Yuen, K.-Y., & Zheng, Z.-M. (2020). SARS-CoV-2 is an appropriate name for the new coronavirus. The Lancet, 395(10228), 949–950. 10.1016/S0140-6736(20)30557-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, M., Gao, Y., Wang, G., Song, G., Liu, S., Sun, D., Xu, Y., & Tian, Z. (2020). Functional exhaustion of antiviral lymphocytes in COVID-19 patients. Cellular & Molecular Immunology, 17(5), 533–535. 10.1038/s41423-020-0402-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoete, V., Daina, A., Bovigny, C., & Michielin, O. (2016). SwissSimilarity: A web tool for low to ultra high throughput ligand-based virtual screening. ACS Publications. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.