Abstract
Escherichia coli is a versatile bacterial species that includes both harmless commensal strains and pathogenic strains found in the gastrointestinal tract in humans and warm-blooded animals. The growing amount of DNA sequence information generated in the era of “genomics” has helped to increase our understanding of the factors and mechanisms involved in the diversification of this bacterial species. The pathogenic side of E. coli that is afforded through horizontal transfers of genes encoding virulence factors enables this bacterium to become a highly diverse and adapted pathogen that is responsible for intestinal or extraintestinal diseases in humans and animals. Many of the accessory genes acquired by horizontal transfers form syntenic blocks and are recognized as genomic islands (GIs). These genomic regions contribute to the rapid evolution, diversification and adaptation of E. coli variants because they are frequently subject to rearrangements, excision and transfer, as well as to further acquisition of additional DNA. Here, we review a subgroup of GIs from E. coli termed pathogenicity islands (PAIs), a concept defined in the late 1980s by Jörg Hacker and colleagues in Werner Goebel’s group at the University of Würzburg, Würzburg, Germany. As with other GIs, the PAIs comprise large genomic regions that differ from the rest of the genome by their G + C content, by their typical insertion within transfer RNA genes, and by their harboring of direct repeats (at their ends), integrase determinants, or other mobility loci. The hallmark of PAIs is their contribution to the emergence of virulent bacteria and to the development of intestinal and extraintestinal diseases. This review summarizes the current knowledge on the structure and functional features of PAIs, on PAI-encoded E. coli pathogenicity factors and on the role of PAIs in host–pathogen interactions.
Keywords: Escherichia coli, pathogenicity islands, genomic island, virulence, ExPEC, AIEC, InPEC
Introduction
Escherichia coli is a versatile bacterial species that has an extensive phylogenetic substructure comprising eight phylogroups (A, B1, B2, C, D, E, F, and G) that are roughly linked to the lifestyles of the different strains (Milkman, 1973; Selander et al., 1987; Escobar-Páramo et al., 2004a; Clermont et al., 2019). E. coli strains colonize the gastrointestinal tract of human infants and coexist in good health with the host, with strains providing mutual benefits for decades. These commensal strains rarely cause diseases in healthy hosts, as they lack specialized virulence traits; they typically originate from phylogroup A (Herzer et al., 1990; Escobar-Páramo et al., 2004b). However, other E. coli strains acquire virulence attributes that confer them with the ability to adapt to new niches and cause intestinal and extraintestinal diseases (Le Gall et al., 2007; Tenaillon et al., 2010). Consequently, E. coli strains can be classified into three groups: commensal/probiotic strains, intestinal-pathogenic strains and extraintestinal-pathogenic strains.
Intestinal-pathogenic E. coli (InPEC), which are rarely encountered in the fecal flora of healthy hosts, cause gastroenteritis or colitis. Six well-described pathotypes of diarrheagenic E. coli (DEC) have been identified: enteropathogenic E. coli (EPEC), enterohemorrhagic E. coli (EHEC), enterotoxigenic E. coli (ETEC), enteroaggregative E. coli (EAEC), enteroinvasive E. coli (EIEC) and diffusely adherent E. coli (DAEC) (Nataro and Kaper, 1998; Kaper et al., 2004). The DEC pathotypes can share certain virulence features; however, each pathotype possesses a specific combination of virulence traits that results in a distinctive pathogenic mechanism. The EPEC strains are not specifically classified as a phylogroup, although some studies preferentially associate them with phylogroup B1 (Reid et al., 2000; Wang et al., 2013). The EHEC are distributed preferentially between phylogroups A and B1 (Askari Badouei et al., 2015; Martins et al., 2015). However, the EHEC of serotype O157: H7 belongs to phylogroup E (Girardeau et al., 2005).
Extraintestinal E. coli (ExPEC) are facultative pathogens responsible for 80% of urinary tract infections (UTIs) in outpatients. Infection by ExPEC leads to a large portion of nosocomial UTIs (50%) and is the leading cause of abscesses, accounting for 30% of meningitis in neonates (Gransden et al., 1990; Johnson, 1991; Russo and Johnson, 2000; Foxman, 2003; Nielubowicz and Mobley, 2010). Bacteremia and septic shock can accompany infections at any site. ExPEC typically belong to the phylogroup B2 – an E. coli genetic background that accumulates virulence factors – and occasionally to phylogroups D, F or G (Tourret and Denamur, 2016; Clermont et al., 2019). Group B2 has the greatest diversity among all E. coli phylogroups (Touchon et al., 2009), suggesting that it has subspecies status and includes subgroups correlated with a flexible gene pool (Le Gall et al., 2007; Lescat et al., 2009). This flexible gene pool comprises genes for various combinations of virulence factors, such as adhesins, iron-acquisition systems, host defense-avoidance mechanisms and toxins (Croxen and Finlay, 2010). The link between the evolutionary lineages of E. coli, certain extraintestinal virulence genes and infection sites has led to the concepts of uropathogenic E. coli (UPEC), sepsis-associated E. coli (SEPEC), neonatal meningitis E. coli (NMEC) and avian pathogenic E. coli (APEC; which cause extraintestinal infections in poultry). Finally, a non-diarrheagenic InEC pathotype, called adherent-invasive E. coli (AIEC), has been noted for its association with inflammatory bowel diseases such as Crohn’s disease (Darfeuille-Michaud et al., 2004). AIEC is considered a pathobiont bacterium rather than a bacterium responsible for acute infection, and it frequently belongs to the phylogroup B2, like ExPEC, and shares with ExPEC the most virulence traits.
Whole-genome E. coli phylogeny suggests an evolution of ExPEC from commensal E. coli strains that were originally devoid of virulence factors but became pathogenic because of horizontal gene transfers involving transduction, transformation, and conjugation events (Lo et al., 2015). Phages, plasmids and large parts of the genome, designated as genomic islands (GIs), were transferred from one bacterium to another (Benedek and Schubert, 2007; Schubert et al., 2009; Schneider et al., 2011; Messerer et al., 2017). These types of GIs carrying virulence-associated genes were identified in UPEC in the early 1980s by Hacker et al. (1983, 1990) and were designated as pathogenicity islands (PAIs) (Blum et al., 1994). Since then, PAIs have been described from SEPEC, MNEC and diarrheagenic isolates and in other species, such as Salmonella enterica. The species Escherichia coli and Salmonella enterica share 70% of their genome (McClelland et al., 2001) and have recently diverged (Doolittle et al., 1996). The divergence of both species has been, in large part, due the acquisition of specific PAIs and has resulted in very different lifestyles (Nieto et al., 2016). This review summarizes the current knowledge on the structure and functional features of PAIs, on E. coli PAI-encoded pathogenicity factors and on the role of PAIs in host-pathogen interactions.
Structural Features of Pathogenicity Islands
Pathogenicity islands are a group of large (>10 kb) integrative elements that encode one or more virulence genes that are absent from the genomes of non-pathogenic representative bacteria of the same species or of closely related species (Blum et al., 1994; Hacker and Carniel, 2001; Dobrindt et al., 2004; Schmidt and Hensel, 2004). In contrast to other integrative elements, such as bacteriophages, plasmids or integrative and conjugative elements (ICEs), PAIs are non-replicative and lack the ability to self-mobilize.
Comparison of the genomic region of PAIs and the remaining parts of the host genome shows that PAIs have their own genomic characteristics, which is strong evidence of their foreign origin and horizontal acquisition. The G + C contents (i.e., the percentage of guanine and cytosine bases), the frequency of dinucleotides or high-order oligonucleotides and the codon usage in PAIs often differ from those of the host organisms (Groisman and Ochman, 1996; Karlin, 2001; Dobrindt et al., 2004). For instance, the G + C content of the 536 UPEC core genome is 50%, while the G + C content is 41% in the PAIs I536, II536, IV536, and V536. In the EPEC genome, the G + C content of the LEE (locus of enterocyte effacement) PAI is only 39%, while the G + C content of the E. coli core genome is ∼50%. However, the donor and recipient organisms may possibly have a similar G + C sequence composition, thereby complicating the extraction of PAIs from the core genome. Even for donor and recipient organisms with different sequence compositions, the PAI region can be “ameliorated” throughout evolution, making the sequence composition or codon usage of the PAI region similar to that of the core genome (Lawrence and Ochman, 1997). The divergences in GC content can therefore reflect a recent acquisition and/or an evolutionary mechanism that maintains a divergence in GC content. This divergence can have a functional role, since curved and AT-rich PAIs are preferential targets of the global gene silencer H-NS (Lucchini et al., 2006; Navarre et al., 2006; Oshima et al., 2006). The H-NS-mediated silencing prevents the uncontrolled transcription of genes within PAIs to ensure that bacterial fitness is maintained, and it may also have evolutionary consequences by influencing the acquisition and maintenance of foreign DNA (Lucchini et al., 2006; Navarre et al., 2006; Oshima et al., 2006).
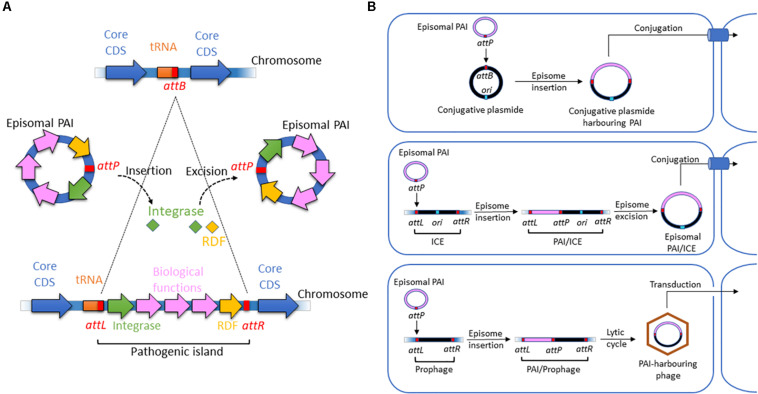
Nonetheless, most GIs contain a recombination module (Figure 1A) that is also observed in other integrative elements, such as phages, integrons, conjugative transposons and ICEs. The module consists of three parts: (i) an integrase of the tyrosine recombinase family; (ii) two flanking attachment sites forming direct repeats resembling the attR and attL sites of prophages; and, in some cases, and (iii) a recombination directionality factor (RDF). This recombination module is characteristic of the integrative elements that insert a circular intermediate into the host genome. However, island-encoded integrases form a separate clade within the tyrosine recombinase family (Boyd et al., 2009). Therefore, PAIs are a distinct class of integrative elements and are not degenerate remnants of other mobile elements (Fogg et al., 2011).
FIGURE 1.
Structure and motility functions of PAIs. (A) Insertion and excision processes of PAIs. PAIs are chromosomal fragments of pathogenic bacteria that encode biological functions involved in virulence. Their insertion in the chromosome is due to the presence of att sites at a chromosomal acceptor site (attB) and in the episomal PAI (attP). They are recognized by integrases, which catalyze a recombination of att sites. It results in the insertion of the episomal element at the attB site and the formation of direct repeated sequences (DRS) also named attL (left DRS) and attR (right DRS) in the ends of the inserted PAI. The excision of the PAI results from recombination between the direct repeats attL and attR. Catalyzed by integrases and recombination directionality factors (RDFs) also called excisionases, it generates an episomal element that contains one of the att sites (attP), while the other att site remains in the chromosome (attB). (B) Horizontal transfer of PAIs via conjugative plasmids, ICEs, and phages harboring att sites. Episomal PAIs can be inserted at att sites in conjugative plasmids, ICEs and phages as described above and then transferred into a bacterial recipient via conjugation for ICE-type and plasmid-type navettes or via transduction for phage-type navettes.
Most island-related integrases are inserted adjacent to tRNA or transfer-messenger RNA (tmRNA) loci (Figure 1A). However, the number of specific tRNA genes used is limited. Within the E. coli genome, ∼87 tRNA genes have been annotated, and integrative genetic elements use only a handful of these sites. Fifteen tDNAs are hotspots of integration, with asnT, aspV, leuX, metV, pheV, and thrW as the most frequently targeted genes (Reiter et al., 1989; Cheetham and Katz, 1995; Williams, 2003). The tDNA pattern targeted by insertions may differ in B2 and A/B1 E. coli strains, potentially influencing their ability to acquire and lose PAIs (Escobar-Páramo et al., 2004a; Germon et al., 2007).
Pathogenicity islands often include mobile elements (or fragments thereof), such as bacteriophages, plasmids and insertion sequences. These mobile elements play an important role in recombination, resulting in genetic rearrangements, insertions, deletions and, therefore, variation of PAIs. The elements contribute to the formation of mosaic-like structures, another hallmark of PAIs that promotes the accretion of traits into islands (Dobrindt et al., 2010).
A number of E. coli PAIs have the PAI features mentioned above; however, some lack one, two or even more features, making the detection of PAIs from sequenced genomes a challenge. The detection methods usually use the following indicative features of the horizontal origin of GIs and PAIs: (i) biased sequence composition, (ii) gene or motif content (i.e., tRNA/tmRNA, direct repeats, integrases, mobility-related genes, high prevalence of hypothetical proteins and virulence/metabolic/antibiotic resistance genes to subclassify the GIs), and (iii) sporadic phylogenetic distribution assessed by the identification of regions only present in a subset of genomes and/or containing genes usually found in PAIs, such as virulence genes, transposase, integrases or genes coding unknown functions (Langille et al., 2010; Lu and Leong, 2016; Bertelli et al., 2019). Composite detection methods, such as IslandViewer (Bertelli et al., 2017) and GIHunter (Han Wang, 2014), are the most sensitive methods (Bertelli et al., 2019). The data with the highest accuracy are provided by tools such as Islander, which can detect tRNA sequence direct repeats (Hudson et al., 2015). Using these in silico approaches, the databases of predicted or curated GIs have been developed from publicly available genome sequences and provide a large sampling of structural variations and gene content in PAIs (Bi et al., 2012; Hudson et al., 2015; Yoon et al., 2015; Bertelli et al., 2017; Li et al., 2018).
Instability and Motiliy of Pathogenicity Islands
Pathogenicity islands in ExPEC and InPEC can undergo deletion at frequencies ranging from 5 × 10–3 to 1 × 10–6 (Turner et al., 2001; Tauschek et al., 2002; Middendorf et al., 2004; Bielaszewska et al., 2011). In many cases, excision and integration seem to be mediated by PAI-encoded integrases (Hochhut et al., 2006). These harbor a highly conserved C-terminal domain involved in recombination and a more divergent N-terminal domain that specifically recognizes the integration site attB (Figure 1A). During the acquisition, the PAI-specific attachment site attP is frequently integrated adjacent to the 3’ end of tDNAs, as observed for phage integration. The recombination results in the direct duplication of the attachment site attB, which forms 16 to 130 bp direct repeats (attL and attR) that flank the PAIs. If multiple isoacceptor tDNAs exist, chromosomal insertion may occur at all available loci, as observed for the high-pathogenicity island (HPI), LEE PAI or PAI IIJ96 (Buchrieser et al., 1998; Tauschek et al., 2002; Bidet et al., 2005). Integrase also excises PAIs from the genome as circular non-replicative intermediates (Figure 1A) using a site-specific recombination process (Rajanna et al., 2003; Hochhut et al., 2006; Wilde et al., 2008). In UPEC strain 536, excision of PAIs I536, II536 and III536 depends on their own integrases. However, PAI V536 undergoes excision even in the absence of its integrase, which can be substituted by the integrase of PAI V536, suggesting crosstalk between PAIs (Hochhut et al., 2006). Enzymes called excisionases or recombination directionality factors (RDFs) can assist integrases (Lewis and Hatfull, 2001; Sakellaris et al., 2004). A bioinformatic analysis showed that each PAI in UPEC 536 contained its own cognate putative RDF (Napolitano et al., 2011). RDFs act as positive or negative integrase transcriptional regulators and offer stability to their integrase protein partners at the excision site (Numrych et al., 1992; Panis et al., 2010). The mobility of PAI can also be independent of the recombination module and involve homologous DNA recombination (Schubert et al., 2009).
Horizontal transfer (HT) is another aspect of PAI mobility (Figure 1B). Because most islands do not contain an origin of replication and are not able to self-mobilize, HT of excised PAIs has been hypothesized to occur with the help of bacteriophages, ICEs or conjugative plasmids (Middendorf et al., 2004). The presence of phage-related sequences on most PAIs suggests that phages have a key role in HT (Boyd et al., 2001; O’Shea and Boyd, 2002). Alternatively, PAIs can be transferred by conjugation in the presence of an attB-presenting helper replicon and accessory transfer genes (Schubert et al., 2009; Schneider et al., 2011). An alternative mechanism, independent of the att site, is homologous DNA recombination, which involves sequences shared by plasmids, a PAI or its environment, and the recipient genome (Schubert et al., 2009). The stabilization of beneficial genetic information localized on mobile genetic elements can then be achieved by the selective loss of transfer or mobilization functions encoded by these elements.
Non-Diarrheagenic Escherichia coli
Physiopathology of ExPEC and AIEC Infections
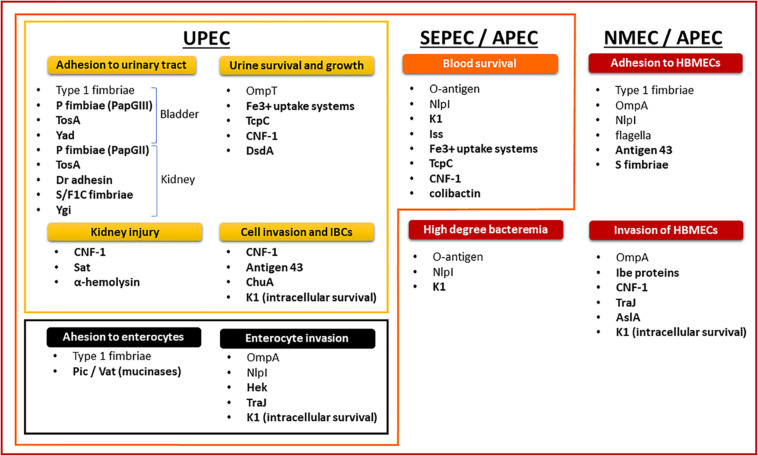
The non-diarrheagenic E. coli are opportunistic pathogens. ExPEC take advantage of host behavior and susceptibility by employing virulence factors (Figure 2) to colonize the digestive tract and then move on to the bladder, where they cause cystitis. The cystitis infection can ascend through the ureters to the kidneys, eventually causing pyelonephritis, and potentially reaching the blood compartment to cause sepsis (Johnson, 1991; Nielubowicz and Mobley, 2010). ExPEC use cell-surface adhesins to adhere to the host’s epithelial cells (Croxen and Finlay, 2010). Adhesin-receptor interactions, invasion factors and bacterial engulfment-enhancing toxins stimulate bacterial internalization into apical uroepithelium cells, leading to invading bacteria that are endocytosed into membrane vesicles (Martinez et al., 2000; Eto et al., 2007). The ExPEC then escape from the endocytic vesicles (Mulvey et al., 1998, 2001; Anderson et al., 2003; Eto et al., 2007; Rosen et al., 2007; Flores-Mireles et al., 2015), proliferate inside the cellular cytoplasm, and form intracellular bacterial communities (IBCs) (Mulvey et al., 1998, 2000; Wright et al., 2007; Flores-Mireles et al., 2015).
FIGURE 2.
Nesting distribution of pathogenicity factors in ExPEC pathotypes and their role in the physiopathology of infections.
Intracellular bacterial communities constitute biofilm-like quiescent reservoirs that preserves ExPEC viability and can lead to recurrent UTIs (Eto et al., 2006). Cytosolic E. coli also induce cell lysis and exfoliation (Mulvey et al., 1998, 2000). The innate immune response initiated by infected uroepithelium cells and exfoliation clears many bacteria from the urinary tract with the flow of urine, but this also leaves the underlying layers of immature bladder epithelial cells exposed and more susceptible to infection (Mulvey et al., 1998, 2000, 2001; Bower et al., 2005; Wiles et al., 2008). In a small proportion of UTIs, ExPEC will decrease the expression of bladder-targeting adhesins and enhance the expression of both kidney-targeting adhesins and bacterial motility (Flores-Mireles et al., 2015). Adhesion and mobility allow ExPEC to ascend through the ureters to the kidneys, where they cause acute pyelonephritis (Svanborg et al., 2001).
Asymptomatic bacteriuria (ABU) occurs in up to 6% of healthy individuals and in 20% of elderly individuals. ABU-associated E. coli persist for months or years without evoking a destructive mucosal host response (Kunin and Halmagyi, 1962; Lindberg, 1975). E. coli strains lacking virulence genes and a defect in TLR4 signaling lead to the asymptomatic carrier state (Andersson et al., 1991; Hull et al., 1999; Frendéus et al., 2001; Svanborg et al., 2001; Bergsten et al., 2005; Fischer et al., 2006; Klemm et al., 2006; Ragnarsdóttir et al., 2008; Grönberg-Hernández et al., 2011).
Bacterial destabilization of the renal epithelium and subsequent inflammation result in renal scarring, which acts as a conduit to the bloodstream. One study showed that 91% of E. coli strains isolated from the blood and urine of the same hospitalized patients were closely related, suggesting that UPEC is the main origin of septicemias and SEPEC (Vollmerhausen et al., 2014). The strains originating from UTIs can also be found in the guts of the patients (Moreno et al., 2008). Accordingly, overgrowing intestinal ExPEC strains can also translocate through the gut epithelium and survive in mesenteric lymph nodes to enter the bloodstream (Bark et al., 1995; MacFie et al., 1999; Ljungdahl et al., 2000; Owrangi et al., 2018). Throughout their progression, ExPEC use numerous capture systems to find the necessary elements, particularly iron, to aid their growth (Braun, 2003). In the bloodstream, SEPEC protect themselves from the immune system by surface structures and effectors which allow them to escape the bactericidal activity of complement and phagocytosis (Cross et al., 1984; Kim et al., 1992; Kim, 2016). This resistance is a factor in the induction of a high degree of bacteremia (Kim, 2016).
Several in vivo studies on NMEC have pointed out a relationship between the magnitude of bacteremia and the development of meningitis (Kim, 2016). E. coli binding and the invasion of human brain microvascular endothelial cells (HBMECs) mediated by E. coli pathogenicity factors is also a prerequisite (Huang et al., 1995, 1999; Prasadarao et al., 1996; Wang et al., 1999; Hoffman et al., 2000; Zhao et al., 2018). The vacuoles containing NMEC that result from HBMEC invasion evolve as endosomes without fusion with lysosomes, thereby allowing E. coli to cross the blood-brain barrier alive (Kim et al., 2003).
Avian pathogenic E. coli strains cause avian colibacillosis, a poultry extraintestinal infection. Extensive genetic similarity has been documented between APEC and ExPEC strains (Rodriguez-Siek et al., 2005; Moulin-Schouleur et al., 2006; Johnson et al., 2007, 2012; Mora et al., 2009, 2012). APEC can cause diseases in mammalian infection models, and these diseases mimic human ExPEC infections (Tivendale et al., 2010). Conversely, human sources of ExPEC can cause diseases in avian species. Human ExPEC and APEC therefore share similar pathogenic potential, and some human-associated ExPEC may have evolved from APEC, and vice versa (Skyberg et al., 2006).
Adherent-invasive E. coli strains have an increased prevalence in inflammatory bowel disease (IBD). AIEC strains harbor genetic similarity with ExPEC in terms of phylogenetic origin and pathogenicity genotype. However, only 6.3% of ExPEC strains exhibit AIEC phenotypic features, suggesting that the AIEC pathotype is disease specific and contributes to the pathophysiology of chronic IBD (Palmela et al., 2018). AIEC bacteria interact with the intestinal mucosa and can (i) cross the mucus layer and resist antimicrobial peptides, (ii) adhere to the surface of the mucosa and increase intestinal permeability, and (iii) penetrate the lamina propria, persist in macrophage vacuoles and chronically stimulate the immune system. Apart from having a high genotypic variability, AIEC belong mainly to phylogroup B2 and resemble ExPEC in terms of virulence factor content. However, AIEC could have selected an original regulatory system and/or pathoadaptive mutations to enable better adaptation to intestinal conditions.
PAIs in ExPEC and AIEC
Experimental and epidemiological investigations of ExPEC pathogenicity factors (PFs) show that no PF is limited to, and no single clone is required for, any particular extraintestinal infection. Statistical analyses of PF content and the resulting lethality in an animal model suggested that the virulence of ExPEC is more likely linked to the number of PFs than to the genetic background (Picard et al., 1999; Johnson and Kuskowski, 2000). In addition, the success of pathogens at various host sites can be due to multiple combinations of PFs, and virulence potential probably exists as a continuum that depends on the number and type of PFs and the inoculum size (Russo and Johnson, 2000). The prevalence of PFs in ExPEC infections is reported in Table 1.
TABLE 1.
Prevalence of virulence factors in ExPEC according to the source (Korhonen et al., 1985; Guyer et al., 2000, 2000; Russo et al., 2001; Bingen-Bidois et al., 2002; Johnson et al., 2002, 2004, 2008; Marrs et al., 2002; Bonacorsi et al., 2003; Russo, 2003; Srinivasan et al., 2003; Buckles et al., 2004; Bonacorsi and Bingen, 2005; Parham et al., 2005a; Rendón et al., 2007; Restieri et al., 2007; Ananias and Yano, 2008; Henderson et al., 2009; Schubert et al., 2010; Ejrnæs et al., 2011; Mahjoub-Messai et al., 2011; Vejborg et al., 2011; Lehti et al., 2012; Qin et al., 2013; Tarchouna et al., 2013; Firoozeh et al., 2014; Basmaci et al., 2015; Nagarjuna et al., 2015; Wijetunge et al., 2015; Beck et al., 2016; Cordeiro et al., 2016; Lee and Lee, 2018; Daga et al., 2019; Nojoomi and Ghasemian, 2019; Raimondi et al., 2019; Sarowska et al., 2019).
| DESIGNATION | ADHESIN | FUNCTION AND ASSOCIATED INFECTIONS | Feces (%) | Cystitis (%) | APN (%) | Blood (%) | LCS (%) | PATHOTYPE |
| Type 1 fimbriae | fimH | Colonization factor and biofilm formation in EIs, especially cystitis and meningitidis, by binding receptor D-Mannose | 68 to 99 | 62 to 100 | 88 to 99 | 90 to 98 | 92 | UPEC, SEPEC, MNEC, APEC |
| ECP fimbriae | Mat | Meningitis associated and temperature regulated fimbriae | 48 | 91 to 98 | 92 | 100 | 96 | NMEC |
| Curli fimbriae | csgAB | Attachment and invasion of host cells, interaction with host proteins, activation of the immune system, bacterial aggregation and biofilm formation | 34 to 36 | 44 to 100 | 24 | 53 to 92 | NF | UPEC, SEPEC, APEC |
| P fimbriae | papGII | Proinflammatory activity and colonization factor in EIs especially pyelonephritis by binding GbO4 and GbO3 | 17 to 23 | 13 | 64 to 67 | 48 to 68 | 20 to 50 | UPEC, SEPEC, MNEC, APEC |
| papGIII | Proinflammatory activity and colonization factor in EIs especially cystitis by binding receptors GbO5 and GloboA | 8 to 9 | 22 | 19 to 35 | 14 to 27 | 1 to 6 | UPEC, SEPEC, APEC | |
| S fimbriae | sfaS | Adhesion to intestinal and urinary tract cells and facilitate penetration into the tissues by binding receptors NeuNAc (α2-3)Gal in UTIs, meningitidis | 7 to 32 | 13 to 34 | 88 to 99 | 39 to 97 | 28 to 59 | UPEC, SEPEC, NMEC |
| F1C fimbriae | focG | Adhesion to epithelial and endothelial cells in the bladder and kidney | 3 to 6 | 14 to 39 | 35 | 21 | 1 to 4 | UPEC, SEPEC |
| G fimbriae | gafD | Binding GlcNAc receptor | 5 | 0 to 4 | 0 to 18 | 0 | 3 | |
| Auf fimbriae | auf | Auf fimbriae | 21 to 27 | 67 | NF | NF | 62 | UPEC, NMEC |
| RTX protein TosA | tosA | Adhesion to host cells derived from the upper urinary tract, enhances survival in disseminated infections and lethality during sepsis | 11 | 16 | 29 | UPEC | ||
| Dr fimbriae | dra | Binding to the receptors DAF of epithelial cells, type IV collagen and CEACAMs. Promotes host cells invasion | 6 | 0 to 4 | 6 to 12 | 5 | NF | UPEC |
| Afimbrial adhesin | afa | Binding to receptor DAF on the epithelial cells and hemagglutination capacity | 2 to 17 | 19 to 81 | 16 | 49 | 9 | UPEC |
| IrgA homolog adhesin | iha | Iron-regulated-gene-homolog adhesin | 14 to 24 | 22 | 41 | 33 to 40 | 19 | UPEC, SEPEC |
| DESIGNATION | INVASIN | FUCNTION AND ASSOCIATED INFECTIONS | Feces (%) | Cystitis (%) | APN (%) | Blood (%) | LCS (%) | PATHOTYPE |
| hemagglutinin | hek/hra | Promote autoaggregation, hemagglutination, epithelial cell invasion and is associated with bacteremia in newborns | 14 to 28 | 28 | 33 to 66 | 45 | 11 | UPEC, SEPEC, NMEC, APEC |
| Endothelial brain invasion | ibeA | Cell invasion into the host tissues such as endothelial cell by binding Vimentine and Caspr1 to promote penetration of the blood–brain barrier | 1 to 15 | 13 to 23 | 29 | 4 to 11 | 32 to 38 | UPEC, NMEC, SEPEC, APEC |
| Arylsulfatase | aslA | Contributes to brain microvascular endothelial cell invasion | 46 | NF | NF | NF | 87 to 89 | NMEC |
| DESIGNATION | TOXINS | FUCNTION AND ASSOCIATED INFECTIONS | Feces (%) | Cystitis (%) | APN (%) | Blood (%) | LCS (%) | PATHOTYPE |
| α-hemolysin | hly | Pore-forming RTX toxin responsible for kidney injury and inflammation | 4 to 15 | 15 to 43 | 38 to 47 | 34 to 77 | 9 to 30 | UPEC, NMEC |
| Cytotoxic necrotizing factor 1 | cnf1 | activation of Rho GTPases, engaging in cell necrosis and promoting resistance to phagocytosis, inflammation, recurrent UTIs and blood-brain barrier penetration | 3 to 13 | 13 to 39 | 28 to 47 | 17 to 49 | 6 to 27 | UPEC, SEPEC, NMEC |
| Cytolethal distending toxin | cdt | DNAse inducing apoptosis and cellular senescence resulting in cell distention | 0 to 1 | 9 to 17 | 12 | 0 to10 | 46 | SEPEC |
| Colibactin | clb | PK-NRP compounds inducing DNA damage resulting preferentially in apoptosis in immune cells and premature cellular senescence in epithelial cells. | 11 to 32 | 33 | 44 | 18 to 58 | 75 | UPEC, SEPEC, NMEC, APEC |
| DESIGNATION | ATs | FUCNTION AND ASSOCIATED INFECTIONS | Feces (%) | Cystitis (%) | APN (%) | Blood (%) | LCS (%) | PATHOTYPE |
| Antigen43 | agn43 | Autotransporter involved in auto-aggregation, adhesion to host cells, biofilm development and persistence | 41 | 29 to 94 | 65 | 69 | NF | UPEC |
| Sap | sap | Autotransporter adhesin | NF | 1,5 | NF | NF | NF | |
| Secreted autotransporter toxin | Sat | Autotransporter having serine protease activity inducing cytotoxic effect and kidney injury | 14 to 24 | 23 to 56 | 55 to 68 | 26 to 39 | 49 | UPEC, NMEC |
| Vacuolating autotransporter toxin | Vat | Autotransporter having serine protease activity affecting mucins, facilitating epithelium colonization and inducing host cell vacuolization | 0 to 49 | 42 to 68 | 44 | 12 to 71 | 51 to 76 | UPEC, APEC, NMEC |
| Temperature-sensitive hemagglutinin | tsh | Autotransporter having temperature-sensitive hemagglutinin activity | 1 to 5 | 4 to 8 | 18 | 20 | 11 | APEC, NMEC |
| Serin protease autotransporter | pic | Autotransporter exhibiting serine protease activity affecting mucins, facilitating epithelium colonization and damaging host cell membrane | 0 to 23 | 21 to 34 | 14 to 40 | 0 to 36 | NF | UPEC |
| Secreted and surface associated lipoprotein | sslE/yghJ | Cytotoxic metalloprotease exihiting mucinase and proinflammatory activity | NF | NF | NF | NF | NF | NMEC,IPEC |
| Contact-dependent growth inibition | cdiAB | Induce the growth inhibition of the bacterial target | NF | NF | NF | NF | NF | UPEC |
| DESIGNATION | IRON UPTAKE | FUNCTION AND ASSOCIATED INFECTIONS | Feces (%) | Cystitis (%) | APN (%) | Blood (%) | LCS (%) | PATHOTYPE |
| Enterobactin | ent, fepA | Acquisition of siderophore Enterobactin-Fe3 + | 100 | 100 | * | * | * | UPEC, SEPEC, MNEC, APEC |
| Aerobactin | aer, iuc, iut | Acquisition of siderophore Aerobactin-Fe ions in the host involved in UTI | 16 to 41 | 24 ot 52 | 38 to 77 | 54 to 80 | 61 to 88 | UPEC, APEC, NMEC |
| Salmochelin | iroN | Acquisition of siderophore Salmochelin-Fe ions in the host involved in UTI and meningitidis | 5 to 26 | 33 to 74 | 76 to 78 | 44 to 63 | 38 to 75 | UPEC, NMEC, SEPEC APEC |
| Yersiniabactin | irp, fuyA | Acquisition of siderophore Yersiniabactin-Fe ions in the host involved in UTI and meningitidis | 23 to 70 | 63 to 96 | 97 | 82 | 90 to 99 | NMEC |
| ChuA, Hma | chu, hma | Acquisition of Fe from hemoglobin in the host system | 53 to 59 | 66 to 80 | 66 to 71 | 48 to 73 | 89 | UPEC, SEPEC |
| Sit | sitDCBA | Transportation of Fe and Mn | 25 | 44 | 56 | 96 | 92 | UPEC, APEC |
| IreA | ireA | Iron-regulated homolog of siderophore receptors involved in UTI | 7 to 19 | 13 | 47 | 32 | NF | UPEC |
| Fec | fecARI | Acquisition of citrate-Fe3 + | NF | NF | NF | NF | NF | |
| Fbp | fbpABCD | Fe acquisition | NF | 58 | 59 | 45 | NF | |
| K1 capsule | kpsMI-neuA | Enable resistance to complement activation and phagocytosis, immunological tolerance and intracellular survival promoting both bacteremia and meningitis | 0 to 27 | 35 | 13 to 47 | 8 to 82 | 75 to 90 | SEPEC, NMEC |
| TcpC | tcpC | Impairs the innate immune response by inhibiting Toll-like receptor and MyD88-specific signaling | 8 | 22 | 42 | 32 to 39 | 0 | UPEC, SEPEC |
| D-serine deaminase | dsdCXA | Promotes bacterial growth by using D-serine as carbon, nitrogen, and energy source, and prevent the bacteriostatic activity of D-serine in the urine and brain. | 0 to 4 | 20 to 22 | NF | 6 | 73 to 87 | UPEC, NMEC |
| TrapT | traT | Inhibition of the classical pathway of complement activity and serum survival | 43 to 88 | 36 to 68 | 49 to 81 | 50 to 81 | NF | UPEC, SEPEC |
| Iss | Resistance to complement and serum survival | 4 to 20 | 13 | 35 | 10 to 34 | 25 | UPEC, SEPEC, NMEC | |
| Outer membrane protein T | OmpT | Resistance to protamine and urine survival | 15 to 68 | 70 | 88 to 95 | 21 to 81 | 96 | UPEC, SEPEC, NMEC |
NF, data not found, ∗Present in almost all Escherichia coli genomes.
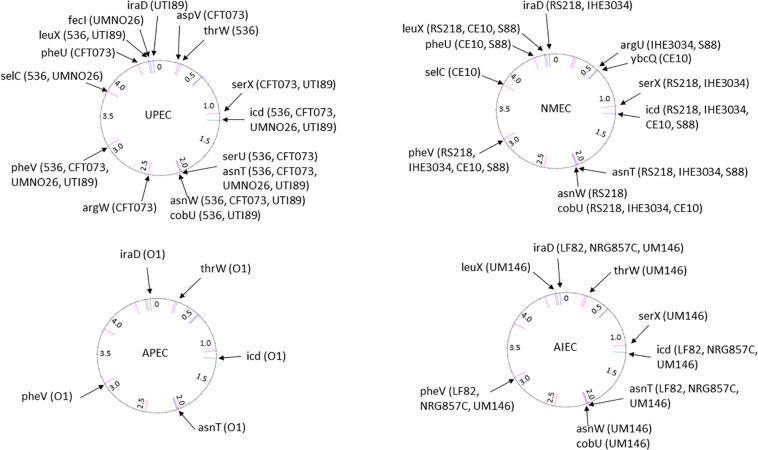
Most ExPEC PFs are encoded by PAIs (Table 2), and the encoded functions are involved in critical steps of the infectious process. The cooperative effect of PAIs has been experimentally confirmed with the prototype strains 536, CFT073 and RS218 (Silver et al., 1980; Hacker et al., 1983; Brzuszkiewicz et al., 2006) by monitoring UTI, septicemia and meningitis mouse models using single-PAI and/or multiple-PAI deletion mutants (Schubert et al., 2002; Xie et al., 2006; Lloyd et al., 2009a; Tourret et al., 2010). These studies showed an additive contribution of PAIs to extraintestinal virulence, but redundancies (i.e., iron uptake) were also evident. The involvement of PAIs in virulence also depends on the genetic background and on the infection model used to test them, suggesting a complex network of pathogenetic possibilities resulting from multiple and independent acquisitions of functions without an unequivocal goal. Given the functional redundancies in PAIs, and to provide an overview of the pathogenicity functions encoded by ExPEC PAIs, their PF content is presented here by PF type and the main structural features of corresponding PAIs is reported in Table 2 and Figure 3 for reference strains.
TABLE 2.
Islands predicted by IslandViewer and/or genomic comparison to E. coli K12 from sequenced strains belonging to pathotypes UPEC (536, CFT073, UMNO26, and UTI89), NMEC (RS218, IHE3034, CE10, and S88), APEC (APECO1) and AIEC (LF82, NRG857C, and UM146) and commensal strains (ED1a and HS).
| PAI name (alternatives) | Chromosomal insertion site | Size (kb) | %GC | Virulence/fitness factors within islands (≥4kb) | Position |
| UPEC strain 536 (Genbank accession number: NC_008253, 4938920-bp) | |||||
| PAI-536-thrW (III536) | thrW | 77 | 47 | S pilus, Salmochelin, Ag43, Vat | 294409.371141 |
| PAI-536-icd | icd | 47 | 50 | Iss, Sit system | 1187870.1235252 |
| PAI-536-serU (VII536) | serU | 22 | 38 | TcpC | 1947667.1969721 |
| PAI-536-asnT (IV536, HPI) | asnT | 30 | 57 | Yersiniabactin | 1971318.2001603 |
| PAI-536-asnW (VI536) | asnW | 54 | 53 | Colibactin | 2015441.2068949 |
| PAI-536-cobU | cobU | 43 | 49 | Hma, Fbp system | 2075300.2116007 |
| PAI-536-pheV (V536) | pheV | 80 | 48 | Pix pilus, Ag43, K15 capsule | 3128357.3208357 |
| PAI-536-selC (I536) | selC | 77 | 46 | α-Hemolysin, PapX, F17-like pilus, CS12-like pilus | 3948173.4025637 |
| PAI-536-leuX (II536) | leuX | 102 | 46 | Hek, P pilus, α-Hemolysin, CdiAB, DsdA | 4735418.4840138 |
| UPEC strain CFT073 (Genbank accession number: NC_004431, 5231428-bp) | |||||
| PAI-CFT073-aspV (IIICFT073) | aspV | 100 | 47 | Pic, Hma, TosA, fbp system | 248751.348573 |
| PAI-CFT073-serX | serX | 113 | 49 | S pilus, Salmochelin, Ag43 | 1127702-1241149 |
| PAI-CFT073-icd | icd | 54 | 50 | Sit system | 1397370.1450849 |
| PAI-CFT073-serU | serU | 22 | 38 | TcpC | 2194512.2216563 |
| PAI-CFT073-asnT (HPI) | asnT | 32 | 57 | Yersiniabactin | 2218159.2250547 |
| PAI-CFT073-asnW | asnW | 54 | 53 | Colibactin | 2262986.2316960 |
| PAI-CFT073-cobU | cobU | 43 | 49 | Hma, Fbp system | 2322324.2365760 |
| PAI-CFT073-argW | argW | 17 | 44 | IpuA, IpuB, DsdA | 2747237.2764564 |
| PAI-CFT073-pheV (ICFT073) | pheV | 123 | 47 | α-Hemolysin, P pilus, Iha, Sat, Aerobactin, Ag43, K2 capsule | 3406498.3529292 |
| PAI-CFT073-pheU (IICFT073) | pheU | 52 | 48 | P pilus, IreA | 4919569.4971387 |
| UPEC strain UMNO26 (Genbank accession number: NC_011751, 5202090-bp) | |||||
| PAI-UMNO26-icd | icd | 46 | 50 | Iss, Sit system | 1411940.1457673 |
| PAI-UMNO26-asnT (HPI) | asnT | 66 | 52 | Yersiniabactin, T4SS | 2277413.2343149 |
| PAI-UMNO26-pheV | pheV | 116 | 47 | P pilus, Iha, Sat, Aerobactin, Ag43, K5 capsule | 3445979.3561885 |
| PAI-UMNO26-selC | selC | 17 | 48 | eaeX | 4301079.4320994 |
| PAI-UMNO26-fecI | fecI | 26 | 50 | Ag43 | 5031002.5057400 |
| UPEC strain UTI89 (Genbank accession number: NC_007946, 5065741-bp) | |||||
| PAI-UTI89-serX | serX | 62 | 48 | S pilus, Salmochelin, Ag43 | 1080842.1142536 |
| PAI-UTI89-icd | icd | 47 | 50 | Iss, Sit system | 1242292.1288830 |
| PAI-UTI89-asnT | asnT | 31 | 57 | Yersiniabactin | 2069935.2101331 |
| PAI-UTI89-asnW | asnW | 54 | 53 | Colibactin | 2113982.2168375 |
| PAI-UTI89-cobU | cobU | 42 | 49 | Hma, Fbp system | 2173987.2216153 |
| PAI-UTI89-pheV | pheV | 18 | 40 | K1 capsule | 3288044. 3306044 |
| PAI-UTI89-leuX | leuX | 122 | 46 | Hek, P pilus, F17-like pilus, CNF1, α-Hemolysin, CdiB, DsdA | 4779711.4900870 |
| PAI-UTI89-iraD | iraD | 20 | 46 | IbeA | 4919657.4940004 |
| NMEC strain RS218 (Genbank accession number: CP007149, 5087638-bp) | |||||
| PAI-RS218-serX | serX | 62 | 48 | S pilus, Salmochelin, Ag43 | 1060280.1123164 |
| PAI-RS218-icd | icd | 49 | 50 | Iss, Sit system | 1219583.1268928 |
| PAI-RS218-asnT (HPI) | asnT | 31 | 57 | Yersiniabactin | 2089060.2120469 |
| PAI-RS218-asnW | asnW | 54 | 53 | Colibactin | 2133298.2134569 |
| PAI-RS218-cobU | cobU | 43 | 49 | Hma, Fbp system | 2191418.2235286 |
| PAI-RS218-pheV | pheV | 18 | 40 | K1 capsule | 3307311.3325311 |
| PAI-RS218-leuX | leuX | 123 | 46 | Hek, P pilus, F17-like pilus, CNF1, α-Hemolysin, CdiB, DsdA | 4841086.4963833 |
| PAI-RS218-iraD (GimA) | iraD | 21 | 46 | IbeA | 4982442.5003432 |
| NMEC strain IHE3034 (Genbank accession number: NC_017628, 5108383-bp) | |||||
| PAI-IHE3034-argU | argU | 50 | 50 | Iss, ompT | 559677.609271 |
| PAI-IHE3034-serX | serX | 62 | 48 | S pilus, Salmochelin, Ag43 | 1123943.1185632 |
| PAI-IHE3034-icd | icd | 48 | 50 | Iss, Sit system | 1285251.1331958 |
| PAI-IHE3034-asnT (HPI) | asnT | 31 | 57 | Yersiniabactin | 2148149.2179443 |
| PAI-IHE3034-cobU | cobU | 43 | 49 | Hma, Fbp system | 2252158.2294325 |
| PAI-IHE3034-pheV | pheV | 18 | 46 | K1 capsule | 3441108.3459812 |
| PAI-IHE3034-iraD (GimA) | iraD | 21 | 46 | IbeA | 4964058 4984404 |
| NMEC strain CE10 (Genbank accession number: NC_017646, 5313531-bp) | |||||
| PAI-CE10-ybcQ | ybcQ | 32 | 52 | Iss | 574012.606099 |
| PAI-CE10-icd | icd | 56 | 52 | Sit system | 1285000.1341381 |
| PAI-CE10-asnT (HPI) | asnT | 31 | 57 | Yersiniabactin | 2259714.2291113 |
| PAI-CE10-cobU | cobU | 27 | 47 | Hma | 2309373.2336831 |
| PAI-CE10-pheV | pheV | 63 | 46 | P pilus, Aerobactin, Sat, Iha, K1 capsule | 3487305.3550576 |
| PAI-CE10-selC | argW | 10 | 42 | IpuA, IpuB (DsdA) | 4303582.4357800 |
| PAI-CE10-pheU | pheU | 20 | 47 | Hek, P pilus | 4961804.4982152 |
| PAI-CE10-leuX | leuX | 42 | 49 | Fec, VirK, MsbB2 | 5127122.5169094 |
| NMEC strain S88 (Genbank accession number: NC_011742, 5032268 bp-bp) | |||||
| PAI-S88-argU | argU | 38 | 52 | ompT | 574349.612629 |
| PAI-S88-icd | icd | 48 | 50 | Iss, Sit system | 1183142.1229738 |
| PAI-S88-asnT (HPI) | asnT | 31 | 57 | Yersiniabactin | 1998347.2029756 |
| PAI-S88-pheV | pheV | 74 | 45 | Hek, P pilus, IreA, K1 capsule | 3219875.3293447 |
| PAI-S88-pheU | pheU | 64 | 48 | PapX | 4621378.4684940 |
| PAI-S88-leuX | leuX | 39 | 50 | Fec, Ag43 | 4821000.4862061 |
| APEC strain APECO1 (Genbank accession number: NC_008563, 5082025-bp) | |||||
| PAI-APECO1-thrW | thrW | 20 | 41 | Tsh | 295124.315041 |
| PAI-APCEO1-icd | icd | 49 | 50 | Iss, Sit system | 1179544.1226126 |
| PAI-APECO1-asnT | asnT | 31 | 57 | Yersiniabactin | 2090794.2122085 |
| PAI-APECO1-pheV | pheV | 73 | 44 | Hek, P pilus, IreA, K1 capsule | 3305091.3378655 |
| PAI-APECO1-iraD (GimA) | iraD | 21 | 46 | IbeA | 4935886.4956233 |
| AIEC strain LF82 (Genbank accession number: NC_011993, 4773108-bp) | |||||
| PAI_LF82-icd | icd | 49 | 50 | Iss, Sit system | 1174302 1223227 |
| PAI-LF82-asnT | asnT | 31 | 57 | Yersiniabactin | 2007750 2039203 |
| PAI-LF82-pheV | pheV | 18 | 42 | K5 capsule | 3110301 3128705 |
| PAI-LF82-iraD (GimA) | iraD | 21 | 46 | IbeA | 4673091 4693405 |
| AIEC strain NRG857C (Genbank accession number: NC_017634, 4747819-bp) | |||||
| PAI-NRG857C-icd | Icd | 61 | 50 | Iss, Sit system | 1171022 1231985 |
| PAI-NRG857C-asnT | asnT | 31 | 57 | Yersiniabactin | 2014814 2046266 |
| PAI-NRG857C-pheV | pheV | 18 | 42 | K5 capsule | 3084523 3102935 |
| PAI-NRG857C-iraD (GimA) | iraD | 21 | 46 | IbeA | 4648143 4668457 |
| AIEC strain UM146 (Genbank accession number: NC_017632, 4993013-bp) | |||||
| PAI-UM146-pheV | pheV | 28 | 46 | K1 capsule | 326687 354390 |
| PAI-UM146-cobU | cobU | 42 | 49 | Hma, Fbp system | 1426242 1468411 |
| PAI-UM146-asnW | asnW | 54 | 53 | Colibactin | 1474024 1529464 |
| PAI-UM146-asnT | asnT | 31 | 57 | Yersiniabactin | 1541958 1573422 |
| PAI-UM146-icd | icd | 29 | 53 | Sit system | 2353562 2382356 |
| PAI-UM146-serX | serX | 62 | 48 | S pilus, Salmochelin, Ag43 | 2499854 2561497 |
| PAI-UM146-thrW | thrW | 20 | 41 | Tsh | 3301638 3321999 |
| PAI-UM146-leuX | leuX | 121 | 46 | Hek, P pilus, F17-like pilus, CNF-1, α-Hemolysin, CdiA, DsdA | 4520979 4642133 |
| PAI-UM146-iraD (GimA) | iraD | 20 | 46 | IbeA | 4660918 4681265 |
| Commensal strain ED1a (Genbank accession number: NC_011745, 5209548-bp) | |||||
| PAI-ED1a-icd | icd | 12 | 49 | Sit system | 1282697.1294467 |
| PAI-ED1a-asnT (HPI) | asnT | 31 | 57 | Yersiniabactin | 2164139.2222325 |
| PAI-ED1a-pheU | pheU | 44 | 48 | Iha, Aerobactin | 4837580 4881090 |
| PAI-ED1a-leuX | leuX | 57 | 50 | MsbB2, VirK, fec, Ag43 | 5065910.5122732 |
| Commensal strain HS (Genbank accession number: NC_009800, 4643538-bp) | |||||
| PAI-HS-hisF | hisI | 39 | 47 | Klebsiella-related LPS and capsule determinants | 2143083.2181967 |
FIGURE 3.
The position of PAI insertion sites in the non-diarrheagenic E. coli pathotypes. The name of genetic features associated with the insertion is indicated with the name of bacterial strains harboring PAI at the corresponding site. The position of the insertion site is indicated in pink for tRNA genes and in blue for coding genes. The numbers reported in the inner ring are the position (megabase) in the genome of the non-pathogenic E. coli strain MG1565 used as a reference.
Adhesins Encoded or Regulated by PAIs
The capacity of E. coli to adhere to host cells plays a fundamental role in the colonization of any site. Adhesion is mediated by proteins called adhesins, which specifically recognize receptors and are often contain antigenic sugars that are present on the surface of host cells. Adhesins are exposed directly to the surface of bacteria or are carried by filamentous structures called “fimbriae” or “pili” (Table 1). Most adhesins are encoded by PAIs, except for conserved fimbriae, such as type 1 fimbriae, curly fibers, outer membrane protein A (OmpA), chitin-binding domain of chiA and new lipoprotein I (NlpI) that are involved in the adhesion of AIEC and NMEC to enterocytes and HBMECs (Kim, 2003, 2008).
Type 1 fimbriae are present in more than 80% of E. coli strains and other Enterobacteriales (Abraham et al., 1988). However, they are major players in ExPEC virulence, and their expression is regulated by PAIs. The type 1 fimbria adhesin FimH binds specifically to α-D-mannose residues attached to membrane glycoproteins exposed at the surface of bladder cells, enterocytes and the endothelial cells of brain capillaries (Keith et al., 1986; Teng et al., 2005). The role of type 1 pili in cystitis has been demonstrated by mutagenesis and in animal models (Keith et al., 1986; Johnson, 1991; Connell et al., 1996). Their adhesin FimH binds to glycosylated uroplakin and α1β3 integrins covering the apical surface of uroepithelium cells, thereby promoting the invasion of host cells and the development of IBCs (Connell et al., 1996; Martinez et al., 2000; Gunther et al., 2002; Mysorekar and Hultgren, 2006; Eto et al., 2007; Flores-Mireles et al., 2015). Notably, in some AIEC, UPEC, MNEC and APEC strains, the protein sequence of FimH adhesin has polymorphisms that confer a greater ability to interact with D-mannose.
The expression of type 1 pili depends on a promoter encoded by the phase-variation invertible element fimS. The orientation of fimS can be reversed by the recombinases FimB and FimE encoded within the operon Fim, and by recombinases FimX, IpuA and IpbA encoded by PAIs (Klemm, 1986; McClain et al., 1991; Bryan et al., 2006; Hannan et al., 2008). IpbA is ubiquitous, as it is found approximately half the time in both commensal fecal and uropathogenic isolates of E. coli (Bryan et al., 2006). FimX-encoded PAI is observed in more than 80% of UPEC and is more prevalent in the UPEC of a lower urinary tract origin (87.5%) than of upper urinary tract origin (74%) or of commensal isolates (36%) (Bryan et al., 2006; Bateman et al., 2013). IpuA-encoding PAIs also tend to occur more frequently among UPEC strains than in commensal fecal isolates (Bateman et al., 2013). This observation is consistent with the hypothesis that FimW and IpuA recombinases play a role in the regulation of E. coli uropathogenesis. FimX, IpuA and IpbA promote inversion of fimS from OFF to ON and probably promote bladder colonization, while FimE inverts the promoter from ON to OFF and could therefore promote the release of bacteria from the bladder epithelium and the infection of the higher tract. The latter might be enhanced by a coordinated upregulation of both bacterial mobility and other adhesins, such as P fimbriae (Klemm, 1986; McClain et al., 1991; Snyder et al., 2005; Bryan et al., 2006; Hannan et al., 2008).
FimX also regulates the predicted LuxR-like response regulator HyxR, which is encoded from the same PAI and is a negative regulator of the nitrosative stress response and intracellular macrophage survival (Bateman and Seed, 2012b,a). IpuA and the associated recombinase IpuB, which lacks activity on fimS, regulate the orientation of a phase-variable invertible element ipuS, which is located in the same PAI, proximal to ipuA and ipuB (Battaglioli et al., 2018). The orientation of ipuS drives the transcription of a two-gene operon containing ipuR (a predicted LuxR-type regulator) and upaE (an autotransporter involved in biofilm formation and extracellular matrix adhesion) (Battaglioli et al., 2018). Consistent with this phenotype, the ipuS orientation ON results in (i) defective swimming motility, (ii) an increase in adhesion to human kidney epithelial cells, and (iii) promotion of kidney colonization in experimental UTI mouse models (Battaglioli et al., 2018). Overall, therefore, PAIs not only encode PFs, but they are also involved in virulence regulation networks.
P fimbriae are encoded by the pap operon (pyelonephritis associated pili), which is associated with different PAIs (Lane and Mobley, 2007). The non-structural gene papX of the pap operon is another example of the PAI gene that is involved in virulence regulation. The protein PapX binds directly to the flhDC promoter, thereby inhibiting the transcription of the master regulator of flagellar biosynthesis, motility and chemotaxis (Li et al., 2001; Simms and Mobley, 2008; Reiss and Mobley, 2011; Reiss et al., 2012). The fimbrial-tip adhesin of the P fimbria PapG (Källenius et al., 1980) binds to glycosphingolipids containing the digalactoside Gal(α1-4β) Gal moieties found in the renal epithelium, P blood-group antigen and gut epithelium (Antão et al., 2009). Three major alleles of the papG locus (GI to III) have been described, with alleles GII and GIII being the most frequently encountered in human disease (Marklund et al., 1992). Allele GII recognizes globotriosylceramide (GbO3) and globotetraosylceramide (GbO4),which are particularly abundant in the kidneys, and it is preferentially found in strains isolated from pyelonephritis because allele GIII recognizes globopentaosylceramide (GbO5) and is mainly found in strains isolated from cystitis (Karr et al., 1989; Lindstedt et al., 1991; Marrs et al., 2002). GbO5 is found in the urothelium of animals but is not present in humans, except in certain individuals expressing an analog (GloboA) and who are particularly susceptible to lower UTIs with PapGIII strains (Lindstedt et al., 1991).
PAI-CFT073-aspV harbors the tos operon, which encodes the repeat-in-toxin (RTX) family member TosA (Lloyd et al., 2009a). The RTX protein family members can be involved in a range of functions, including pore and biofilm formation and adhesion to host cells (Linhartová et al., 2010; Dhakal and Mulvey, 2012). The presence of tosA promotes bladder and kidney colonization in a UTI mouse model and is a marker of UPEC (Vigil et al., 2011b,a). Localized to the bacterial surface, TosA mediates adhesion to host cells derived from the upper urinary tract, increases bacterial survival in disseminated infections and enhances lethality during sepsis (Vigil et al., 2012). The TosCBD proteins mediate the production and export of TosA, while TosE and TosF suppress motility by an unknown regulatory function. TosR is a member of the PapB family of transcriptional regulators, including the fimbria-associated regulators PapB and FocB (Xia et al., 1998; Lindberg et al., 2008; Hultdin et al., 2010). TosR regulates the tos operon (Engstrom and Mobley, 2016) and other genes involved in adhesion (P, F1C, and Auf fimbriae) and biofilm formation (Luterbach et al., 2018).
Other fimbrial gene clusters are found in ExPEC PAIs (Table 2). F1C/S fimbriae promote adhesion to primary human renal proximal-tubular cells by binding to α-sialyl-2-3-β-galactoside (Kreft et al., 1995). The F1C/S fimbriae are frequently found in NMEC and could also be involved in the crossing of the blood-brain barrier, as occurs with type 1 fimbriae (Korhonen et al., 1985; Ott et al., 1986). However, in a meningitis animal model, the deletion of the F1C/S fimbriae operon in E. coli K1 did not significantly affect bacterial binding to and invasion of HBMECs, and the deletion also did not affect bacterial penetration into the central nervous system (Kim, 2008). Ygi fimbriae promote renal tropism, biofilm formation and in vivo fitness in the urine and kidneys (Spurbeck et al., 2011). Yad fimbriae are necessary for adhesion to a bladder epithelial cell line and biofilm formation, and the deletion of these fimbrial genes has been shown to increase motility (Spurbeck et al., 2011). A double deletion strain, Δygi Δyad, showed impaired colonization of the urine, bladder, and kidneys in a mouse model, demonstrating that these fimbriae contribute to uropathogenesis (Spurbeck et al., 2011). ExPEC also produces non-fimbrial adhesin FdeC that is highly prevalent in both commensal strains and InPEC. This adhesin binds epithelial cells and, collagen V and VI, and in vivo experiments suggest that it has a role in UTI infections (Nesta et al., 2012).
Overall, PAIs provide the ability to produce multiple fimbria that confer a selective advantage within particular niches and are involved in crosstalk between regulators of various fimbrial types and in motility.
Invasins Encoded by PAIs
Extraintestinal E. coli PAIs can produce invasins, which are structures that promote bacterial internalization within host cells. This process allows bacteria to escape targeting by the immune system and is involved in the formation of the IBCs observed in UTIs and Crohn’s disease with UPEC and AIEC, respectively. These internalizations also allow bacteria to cross epithelial and endothelial barriers by transcytosis.
The PAI-encoded protein Hek/Tia, which is more frequently found in UPEC than in commensal isolates (Table 1), is responsible for bacterial aggregation on the surface of enterocytes, as is observed for enteroaggregative E. coli, and is involved in the invasion of T84 enterocytic cells (Fagan and Smith, 2007). The mechanism behind the invasion of enterocytes and the bladder epithelium and the requirement for preliminary adhesion are still largely unknown. The invasion of HBMECs by NMEC requires binding to and the invasion of the HBMECs and involves the PAI-encoded microbial factors IbeA and cytotoxic necrotizing factor-1 (CNF-1), as well as the housekeeping factors OmpA, NlpI and arylsulfatase-like gene aslA (Huang et al., 1995, 1999, 2001; Prasadarao et al., 1996; Wang et al., 1999; Hoffman et al., 2000; Khan et al., 2002; Kim, 2003, 2008). Studies have attempted to identify the receptors at the surface of HBMECs for these bacterial factors: CNF-1 interacts with laminin receptors, while IbeA binds to both vimentin and HBMEC contactin-associated protein 1 (CASPR1) (Zou et al., 2006; Zhao et al., 2018). This latter interaction activates focal adhesion and kinase signaling, which causes E. coli internalization (Zhao et al., 2018).
Toxins Encoded by PAIs
Extraintestinal E. coli strains can use PAI-encoded genes to produce several toxins, including α-hemolysin, CNF-1, cytolethal and distending toxin (Cdt), and colibactin (Clb).
E. coli α-hemolysin is a pore-forming toxin belonging to the class of RTX toxins that are encoded by the hly operon. Although most UPEC strains carry one copy of the hemolysin operon, pyelonephritogenic strains 536 and J96 harbor two copies of the hly operon and both loci are required for full virulence (Nagy et al., 2006). The toxin is encoded by the hlyA gene. The gene hlyC encodes an acyl transferase that is required for activation of the toxin, while genes hlyB and hlyD are involved in the energy-dependent secretion of HlyA (Welch, 1991). HlyA is a hemolysin that targets different cell types, including leukocytes (Galmiche and Boquet, 2001). The involvement of α-hemolysin in virulence has been suggested because of its high prevalence in UPEC compared to its prevalence in fecal strains and because of its high expression within IBCs (Johnson, 1991; Reigstad et al., 2007). In upper urinary tract infections, α-hemolysin is thought to play both a direct cytotoxic role in renal cells and a pro-inflammatory role that causes the secretion of cytokines IL-6 and IL-8 and alters Ca2+ membrane flow (Uhlén et al., 2000). The combination of these two actions weakens the renal epithelium and promotes the passage of bacteria into the blood (Bonacorsi et al., 2006). By interfering with NF-kB-mediated proinflammatory signaling pathways and triggering the breakdown of paxillin and other host regulatory proteins, HlyA also dampens the host’s immune response to infection and enhances the exfoliation of bladder epithelial cells (Dhakal and Mulvey, 2012).
CNF-1 induces the formation of giant multinucleated cells, changes actin and tubulin organization, and most likely promotes cell spreading (Fiorentini et al., 1988). Its toxic activity arises because of the post-translational activating mutation of Rho GTPases by deamidation, an essential control factor in the shape, adhesion, mobility, phagocytosis, and oxidative burst in host cells (Flatau et al., 1997). The CNF-1 prevalence is higher in strains isolated from UTIs and prostatitis than in fecal isolates (Caprioli et al., 1987; Bingen-Bidois et al., 2002). An animal model of UTIs has shown an involvement of CNF-1 in resistance to phagocytosis by macrophages and in the induction of deep and persistent infections of the bladder (Rippere-Lampe et al., 2001). The CNF-1-mediated disruption of the Rho GTPase signaling pathways has antiapoptotic abilities in uroepithelium cells and leads to immune dysregulation, while conferring a survival advantage in the presence of neutrophils (Davis et al., 2005). CNF-1 also plays an important role in the rearrangement of the HBMEC cytoskeleton that allows crossing of the blood-brain barrier (Badger et al., 2000; Kim, 2002).
The pks genomic island present in E. coli strains of phylogroup B2 encodes colibactin, a hybrid polyketide/non-ribosomal peptide that causes DNA damage and cell cycle arrest of eukaryotes (Nougayrède et al., 2006). The colibactin-encoding determinant has been detected primarily in extraintestinal pathogenic isolates of E. coli, other Enterobacteriales, and commensal E. coli (Nougayrède et al., 2006; Putze et al., 2009). E. coli persisting in the infant gut microbiota tends more often to carry the pks island than do either intermediate-term colonizers or transient strains, suggesting that the pks island contributes to the gut-colonizing capacity of group B2 strains (Nowrouzian and Oswald, 2012). The frequent detection of the pks island in E. coli isolated from biopsies of patients suffering from colon cancer also suggests involvement in long-term intestinal colonization, raising the question of its role in colorectal cancer (Swidsinski et al., 1998; Martin et al., 2004; Bronowski et al., 2008; Cuevas-Ramos et al., 2010; Arthur et al., 2012; Buc et al., 2013; Cougnoux et al., 2014, 2016; Pleguezuelos-Manzano et al., 2020). The presence of the pks island in ExPEC could also indicate that colibactin contributes to fitness or virulence during extraintestinal infections (Johnson et al., 2008; Krieger et al., 2011; McCarthy et al., 2015). A recent observation indicates that the probiotic effects of the E. coli Nissle 1917 strain to ameliorate colitis severity and to modulate cytokine expression cannot be separated from the strain’s ability to express functional colibactin (Olier et al., 2012). This finding demonstrates that, depending on the niche or context, colibactin-producing PAI can be considered either a virulence factor and/or a probiotic factor.
The TLRs are an important family of innate sensors that recognize diverse microbial products and launch the signaling pathways that ultimately lead to the clearance of the pathogen from the host and the establishment of a memory response in anticipation of any subsequent attack. ExPEC PAI-serU can interfere directly with the TLR signaling pathway by producing an inhibitor homolog of TLR receptors to dampen the NF-kB-induced proinflammatory response (Cirl et al., 2008). This protein, called the TLR domain containing-protein C (TcpC), is secreted by an efflux pump and then internalized into macrophages, where it can disable TLR signaling through direct binding of MyD88, the key TLR signaling adaptor (Cirl et al., 2008). The tcpC gene is present in 30–40% of the E. coli strains isolated from septicemia and in almost half of those causing pyelonephritis but is less common in cystitis (22%), ABU (16%) and E. coli strains of the fecal microbiota (8%) (Table 1). TcpC can increase the severity of UTIs in humans, which is consistent with the important role of TLR4 in host defense in the urinary tract (Samuelsson et al., 2004).
Autotransporters Encoded by PAIs
Extraintestinal E. coli PAIs encode several autotransporters (ATs), which are a family of proteins that mediate their own secretion through the outer membrane of gram-negative bacteria. Also known as the type V secretion system, ATs contain a C-terminal membrane anchor region that forms a pore through which the passenger domain is translocated to the cell surface. The passenger domain of ATs is exposed at the bacterial surface and/or secreted after autoproteolytic cleavage. The AT passenger domains are diverse in function and can act as enzymes (lipases, esterases, or proteases) and/or as adhesins (Tapader et al., 2019).
AT antigen 43 (Ag43) is widely distributed among E. coli strains, including UPEC. Ag43 mediates autoaggregation, biofilm formation and host cell adhesion. It promotes persistent colonization in a UTI mouse model and is expressed within IBCs, suggesting that Ag43 plays a role in the intracellular growth phase of UPEC (Anderson et al., 2003; Kim, 2003, 2008). Ag43 is also involved in the adhesion of NMEC to HBMECs (Ulett et al., 2007). The ATs UpaB and UpaC and their homologs are found in UPEC and in various strains of E. coli (Allsopp et al., 2012). UpaC promotes adhesion to abiotic surfaces and biofilm formation, while UpaB promotes adhesion to components of the extracellular matrix. In vivo experiments have suggested that UpaB, unlike UpaC, is important for bacterial fitness during UTI. Finally, the UpaG autotransporter promotes UPEC biofilm formation on abiotic surfaces and facilitates binding to extracellular matrix proteins, fibronectin, and laminin (Valle et al., 2008).
Extraintestinal E. coli PAIs also produce serine protease ATs that are designated as SPATEs (serine protease autotransporters of Enterobacteriaceae) such as Tsh (temperature sensitive hemagglutinin), Hbp (hemoglobin binding protein or hemoglobin protease), Sat (secreted autotransporter toxin), Vat (vacuolating autotransporter toxin) and Pic (protein involved in colonization, cf. DEC section). Tsh was initially identified in APEC as a mannose-resistant hemagglutinin that is overexpressed at low temperatures (Provence and Curtiss, 1994). It is observed in more than half of APEC isolates and its prevalence increases in high-lethality isolates (Dozois et al., 2000). The association of Tsh with the virulence of APEC isolates was further reinforced by the detection of fewer and less pronounced lesions in the air sacs of chickens infected with a Tsh mutant than with the wild type strain (Dozois et al., 2000). The tsh gene is also observed in UPEC (4.5%) and NMEC (11.5%) (Ewers et al., 2007). Frequently encoded by plasmids, tsh is also observed in PAIs and promotes adhesion to red blood cells, hemoglobin, and the extracellular matrix proteins fibronectin and collagen IV (Kostakioti and Stathopoulos, 2004). Tsh only differs from Hbp by two amino acid residues in the passenger domain and probably shares functional similarities regarding the breakdown of hemoglobin factor V and mucin (Dautin, 2010; Tapader et al., 2019).
The vat gene was also originally identified in an APEC strain as a cytotoxin of chicken embryonic fibroblast cells that contributes to avian cellulitis infection (Parreira and Gyles, 2003). It only shares 78% identity with Tsh and has different proteolytic activities (Parham et al., 2005b). However, Tsh/Hbp, Vat and Pic have mucinolytic activity and enhance epithelium colonization (Dautin, 2010; Gibold et al., 2016; Tapader et al., 2019). Pic and Tsh/Hbp also target the major human leukocyte-adhesion molecules CD43, CD44, CD45, and CD93, thereby deregulating leukocyte migration and inflammation (Ruiz-Perez et al., 2011). The Sat ATs induce vacuolation of renal cells, and the serine-protease activity is required for the cytopathic effects of Sat (Guyer et al., 2002; Maroncle et al., 2006). Mention has been made of the role of Sat in breaking down the glomerular barrier to allow bacteria to pass into the blood (Nataro and Kaper, 1998). Three new SPATEs—Sha (Serine-protease hemagglutinin autotransporter), TagB and TagC (tandem autotransporter genes B and C)—have also been recently reported in an ExPEC O1:K1 strain isolated from turkeys and they induce cytopathic effects on a bladder epithelial cell line (Habouria et al., 2019). However, their functional role in UTIs remains to be investigated.
SslE/YghJ (secreted and surface associated lipoprotein) is secreted by the type II secretion system and probably anchored at the bacterial surface in ExPEC. It is a metalloprotease domain initially observed in NMEC (Tapader et al., 2019). SslE, which is widely found in both commensal and pathogenic E. coli, has greater prevalence in pathogenic isolates than in commensal isolates, which may also be defective for its secretion (Moriel et al., 2010). SsIE targets mucins and has cytotoxic and proinflammatory activities that can promote the virulence of pathogenic E. coli and sepsis in neonates (Nesta et al., 2014).
Iron Uptake Systems Encoded by PAIs
Iron is essential for bacteria because of its involvement in many metabolic functions, such as the transport of oxygen and electrons. Most iron in humans is complexed with the transport molecules transferrin (found in blood) and lactoferrin (found in the digestive tract and in salivary and pulmonary secretions) and with reserve molecules (ferritin) or it is incorporated into the heme of hemoglobin and myoglobin. This complexation results in iron limitation, which in turn serves as one of the innate defenses against the survival of bacteria within hosts. To use iron from the host, bacteria have developed capture systems that target these iron-complexed forms within citrate ions (fec system) or heme (chu system). The bacteria use transport systems (Sit systems) or produce PK-NRP compounds called siderophores that capture iron from host transporters or reserve systems and transport it back to the bacteria via specific receptors (Braun, 2003). The siderophore enterobactin, which is found in almost all E. coli, is encoded by the core genome. Thus, Lipocalin-2, which is capable of sequestering enterobactin, prevents its uptake by the bacteria via the FepA receptor (Fischbach et al., 2006). Therefore, ExPEC acquires PAI-encoding iron-uptake systems to thwart this host defense mechanism. The most frequently encountered siderophores are aerobactin, yersiniabactin and salmochelin (Braun, 2003). Salmochelin is a modified enterobactin formed by glycosylation of one of its derivatives, and this modification makes salmochelin insensitive to inhibition by human Lipocalin-2 (Fischbach et al., 2006). The PAI-encoded siderophores salmochelin and yersiniabactin are both produced at significantly higher levels in ExPEC isolated from urine than in rectal isolates and could play a greater role than enterobactin within the human urinary tract (Henderson et al., 2009). In a newborn rat meningitis model, salmochelin plays an important role in maintaining a high level of bacteremia, which is a necessary step for bacterial crossing of the blood-brain barrier (Nègre et al., 2004).
Receptor-mediated uptake of heme is another iron-uptake mechanism encoded by ExPEC PAIs. Two outer membrane heme receptors, ChuA and Hma, have been characterized, and both were required for bacterial optimal fitness in a UTI mouse model (Hagan and Mobley, 2009). ChuA could also be important during IBC formation (Reigstad et al., 2007). The PAI-encoded Sit system transports manganese and ferrous iron into the ExPEC cytoplasm and enhances resistance to oxidative stress. The sit genes are upregulated during murine UTI and are expressed during human UTIs, suggesting that they contribute to urofitness (Snyder et al., 2004; Hagan et al., 2010).
Protectins Encoded by PAIs
Protectins are the bacterial elements that protect bacteria against host weapons, such as antibacterial factors (i.e., defensins, D-serine), serum complement and immune cells. PAI-encoded capsules (K antigen) confer this type of protection (Whitfield, 2006). The resulting homopolymers exposed at the surface of the bacteria can resemble various glycoconjugates found within vertebrate hosts (e.g., sialic acid), such as K1 or K4 capsules, and can contribute to the bacterium’s immune-evasion strategy. The K1 antigen is associated with E. coli strains responsible for invasive infections (Sarff et al., 1975). K1 antigen is critical for the induction of a high degree of bacteremia, is associated with a high resistance to both phagocytosis and serum and is required for bacterial crossing of the blood-meningeal barrier (Opal et al., 1982; Cross et al., 1984; Kim et al., 1992). Other PAI-encoded protective elements are surface proteins such as the Iss (increased serum survival) protein, which has been implicated in serum resistance (Xu et al., 2018).
D-serine is a bacteriostatic amino acid that is found in urine and brain (Cosloy and McFall, 1973; Kumashiro et al., 1995). UPEC and NMEC strains frequently harbor a PAI-encoded dsdCXA locus (Bloom and McFall, 1975; Moritz and Welch, 2006). The dsdCXA locus has been hypothesized to be important for UPEC/NMEC pathogenesis. Accordingly, isogenic strains defective in dsdA have a growth defect in urine relative to dsdA wild-type strains. However, dsdA neither positively nor negatively affected the ability of UPEC strain CFT073 to colonize urinary tract in a mouse model (Hryckowian et al., 2015).
Diarrheagenic Escherichia coli
As already described above, InPEC essentially regroups (i) E. coli strains responsible for diarrhea (DEC) and (ii) AIEC, which are not involved in diarrhea but are involved in inflammatory bowel diseases. Among the DEC, the EPEC and EHEC have undoubtedly received the most attention regarding PAIs. By contrast, little information is available about PAIs in DAEC, except for the presence PAI pic/set in some strains (Czeczulin et al., 1999). Compared to ExPEC, the information about PAIs in DEC is generally scarce.
Physiopathology of DEC
The six recognized categories of DEC are primarily based on clinical symptoms, including the type of diarrhea, related syndromes or bacterial interaction with intestinal epithelium cells, as revealed by histological observation, as well as associated molecular determinants of virulence and pathogenicity (Nataro and Kaper, 1998; Kaper et al., 2004).
ETEC are mainly associated with traveler’s diarrhea but can also appear in weanling diarrhea among children (Nataro and Kaper, 1998). The diarrhea is generally not bloody, but watery, and is sometimes associated with vomiting that ranges from mild to severe purging, similar to cholera. The pathogenicity of ETEC mainly relies on heat-labile (LT) and/or heat-stable (ST) enterotoxins responsible for net secretion of intestinal fluid (Turner et al., 2006a; Sack, 2011). The LT form belongs to the cholera toxin family and is secreted by a Type II secretion system (T2SS) with the contribution of LeoA (labile enterotoxin output) for its efficient secretion (Tauschek et al., 2002; Jobling and Holmes, 2006). Unlike the cholera toxins that are secreted as free soluble proteins, LT associates with the outer membrane vesicles by binding to LPS (Horstman and Kuehn, 2002). Sequence divergences confer LT with the ability to bind different ganglioside receptors that mediate entry into the host cell (Fukuta et al., 1988; Jobling and Holmes, 2006). LT is an ADP-ribosylating toxin that inactivates the GTPase activity of the G protein complex by ADP-ribosylation. This, in turn, leads to prolonged activation of the adenylate cyclase activity and elevated intracellular levels of cAMP and results in electrolyte release and stimulation of intestinal excretion (Spangler, 1992; Kaper et al., 2004; Jobling and Holmes, 2006).
By contrast, ST can be discriminated into type I (ST-I, also called STa) and type II (ST-II, also called STb), which target different receptors (Peterson and Whipp, 1995; Turner et al., 2006a). ST-I binds and activates a guanylate cyclase C receptor and the subsequent increase in intracellular cGMP levels activates the cystic fibrosis transmembrane conductance regulator (CFTR) chloride channel (Vaandrager, 2002; Turner et al., 2006a). ST-II, which binds the acidic glycosphingolipid sulphatide, activates a pertussis toxin-sensitive GTP-binding regulatory protein that ultimately activates CFTR (Peterson and Whipp, 1995; Fujii et al., 1997; Rousset et al., 1998). Surface colonization is an important in step in the physiopathology of ETEC, which involves a large and diverse repertoire of pili, called colonization factor antigens (CFAs), that bind to different receptors at the surface of the host cells (Turner et al., 2006a; Isidean et al., 2011; Ageorges et al., 2020). Additional virulence factors can be present, including some surface-exposed adhesins like TibA or Tia, the toxins ClyA and EAST1, or the protease EatA; however, this does not appear to be a common feature among ETEC (Turner et al., 2006a,b).
Enteropathogenic E. coli infection manifests as acute diarrhea in infants and is often associated with vomiting (Nataro and Kaper, 1998; Kaper et al., 2004). A hallmark of EPEC is its attaching and effacing (A/E) histopathology. This is accompanied by intimate attachment of bacterial cells expressing cell-surface intimin (Eae) with the cognate translocated intimin receptor (Tir) expressed on the surface of eukaryotic cell following injection into their cytosol by a Type III, subtype a, secretion system (T3aSS) (Nougayrède et al., 2003; Desvaux et al., 2006; Lai et al., 2013). A large repertoire of T3aSS effectors participate in the infection process (Dean et al., 2005; Dean and Kenny, 2009). However, and despite decades of investigations, the exact molecular mechanisms by which EPEC induce diarrhea are not well understood (Ochoa and Contreras, 2011).
The presence or absence of plasmid pEAF (E. coli adhesion factor) encoding bundle-forming pili (BFP), can be used to further categorize EPEC into typical (tEPEC) and atypical EPEC (aEPEC), respectively (Trabulsi et al., 2002; Hernandes et al., 2009; Hu and Torres, 2015). In tEPEC, BFP are type 4 pili (T4P) encoded by the bfp operon and induce a localized adhesion (LA) pattern at the surface of intestinal epithelial cells to form microcolonies (Scaletsky et al., 1984; Ageorges et al., 2020). In aEPEC, no localized LA patterns are observed but instead localized-adhesion-like (LAL) or aggregative adhesion (AA) patterns form loose bacterial cell clusters involving alternative adhesion factors such as EspA filaments, LifA (lymphocyte inhibitory factor) or ECP (E. coli common pili) (Rodrigues et al., 1996; Trabulsi et al., 2002; Ageorges et al., 2020).
EHEC are responsible for pediatric diseases ranging from watery diarrhea to hemorrhagic colitis that can further lead to thrombotic microangiopathies, especially hemolytic and uremic syndrome (HUS) and thrombotic thrombocytopenic purpura (TTP) (Karmali et al., 1983; Ruggenenti and Remuzzi, 1998; Tarr et al., 2005; Tarr, 2009; Wada et al., 2014). EHEC are primarily characterized by the expression of the key virulence shigatoxin (Stx) that binds to glycosphingolipid Gb3 before penetrating the cell and inducing apoptosis of kidney epithelial cells (Melton-Celsa, 2014). Nonetheless, and somewhat surprisingly, the way that Stx crosses the intestinal mucosa is not fully elucidated. While tEHEC possess the locus of enterocyte effacement (LEE) as in any EPEC, the aEHEC do not express the injectisome and instead rely on alternative monomeric and multimeric surface colonization factors, such as adhesins and pili (McWilliams and Torres, 2014; Monteiro et al., 2016; Ageorges et al., 2020). Similar to EPEC, however, the precise molecular mechanisms that induce bloody diarrhea remain elusive (Croxen et al., 2013; Stevens and Frankel, 2014).
Of note, and contrary to EPEC, A/E lesions are never detected on colonic biopsies from EHEC infection (Nataro and Kaper, 1998), although they are observed in vitro using epithelial cell culture or human colonic explants (Golan et al., 2011; Battle et al., 2014; Lewis et al., 2015). Some E. coli isolated from other sources (environment, animals, food and the agri-food chain) can be genotypically characterized as possessing the stx gene and can be regrouped as shigatoxin-encoding E. coli (STEC, or shigatoxin-producing E. coli when Stx production has been experimentally confirmed). The pathogenicity of EHEC has been clinically ascertained, and STEC can have very different virulence levels, up to hyper-virulence; alternatively, they can even be potentially avirulent and are not systematically and rigorously considered as pathogenic (Karmali et al., 2003; Caprioli et al., 2005; Laing et al., 2009; Monteiro et al., 2016). With more than 400 distinct serotypes revealed in STEC, only a handful of EHEC serotypes has been clinically associated with epidemic outbreaks, although they can all be potentially involved in human infections (Karmali et al., 2003; Blanco et al., 2004; Cristancho et al., 2008; Mathusa et al., 2010). The search for genetic markers has been the Grail for decades; nevertheless, the differences in the virulence level is most certainly associated with various compensatory combinations of molecular determinants, including PAIs, together with regulatory networks that can act at various levels and not just regarding the transcriptional aspects of genetic expression (Ageorges et al., 2020).
After ETEC, EAEC is the second most common cause of tourista that causes acute and sometimes persistent diarrhea (Jenkins, 2018). The diarrhea is typically non-bloody and without inflammation or emetic symptoms. Upon adhesion onto intestinal epithelial cells, EAEC exhibit a typical stacked brick-like pattern known as aggregative adhesion (Kaper et al., 2004). EAEC form a loose biofilm at the mucosal surface involving pili and especially AAF (aggregative adhesion fimbriae, encoded by a large plasmid pAA) (Kaper et al., 2004; Ageorges et al., 2020). Additional virulence factors include the pAA-encoded enterotoxins EAST1 and Pet (plasmid-encoded toxin) and the chromosome-encoded ShET1 (Shigella enterotoxin 1) and Pic (protein involved in intestinal colonization) (Chaudhuri et al., 2010; Jenkins, 2018). However, the molecular basis that mediates diarrhea upon infection by EAEC remains to be elucidated.
Extensive and compelling evidence indicates that EIEC and Shigella cannot be discriminated from a taxonomic point of view as they both belong to the same species, E. coli, and basically correspond to the same pathotype (Lan et al., 2004; Peng et al., 2009; Chaudhuri and Henderson, 2012; Pettengill et al., 2015; Michelacci et al., 2016). Differences in the virulence level have sometimes been put forward, with Shigella considered hyper-virulent relative to EIEC, which generally induces less severe disease. Nevertheless, significant exceptions do exist and the intestinal illness is in fact indistinguishable (Parsot, 2005; Michelacci et al., 2016; Belotserkovsky and Sansonetti, 2018; Hendriks et al., 2020). As with EHEC/STEC, the search for genetic markers that could solely explain the difference in the virulence levels might have somehow hindered the potential in the heterogeneity of the regulation of the genetic expression from one EIEC/Shigella strain to another as well as between individual cells in an isogenic population. The hallmarks of EIEC/Shigella are the invasion plasmid pINV, which encodes a T3aSS (like in EPEC and EHEC), and cognate effectors of key importance in host invasion of this intracellular pathogen, which have been extensively investigated (Zagaglia et al., 1991; Lan et al., 2001, 2003; Lan and Reeves, 2002; Schnupf and Sansonetti, 2019). Additional virulence factors include the mucinase Pic (previously called ShMu for Shigella haemagglutin and mucinase), also found in EAEC and contributing to the initial step of intestinal colonization (Henderson et al., 1999; Parham et al., 2004). Another is the protease SigA (Shigella immunoglobulin A protease) that contributes to the watery prodromal phase of the infection (Al-Hasani et al., 2000; Chua et al., 2015), together with the enterotoxins ShET1 and ShET2 (Henderson et al., 1999). The exact function and mode of action of the latter enterotoxins remain to be determined (Mattock and Blocker, 2017; Belotserkovsky and Sansonetti, 2018).
DAEC, rather than DEC, are essentially involved in diarrhea in young children (Servin, 2005, 2014), and some DAEC are ExPEC involved in urinary tract infections. EIEC are known to preferentially colonize the colon, as well as the distal ileum for EHEC, whereas the ETEC and EPEC colonize the small intestine, as well as the colon for EAEC (Clements et al., 2012). By contrast, the primary site of intestinal colonization of DAEC has not been determined (Servin, 2014). DAEC exhibits a scattered adhesion pattern on intestinal epithelial cells, called diffuse adhesion (Kaper et al., 2004; Croxen et al., 2013). DAEC have previously been subdivided into those expressing and those not expressing Afa/Dr (afimbrial adhesin/decay-accelerating factor receptor) adhesins (Servin, 2005), but the latter subclass is now considered to belong to aEPEC, leaving Afa/Dr adhesins as a hallmark of DAEC (Servin, 2014). Previously, daaE and/or daaC were briefly suggested as biomarkers of DAEC, but F1845 pili are rare among DAEC strains (Campos et al., 1999). The group of Afa/Dr adhesins is quite diverse and can induce rearrangement of the cytoskeleton upon binding to the eukaryotic cell and can even promote bacterial internalization (Servin, 2005; Le Bouguénec and Servin, 2006). Additional virulence factors include pks island products (such as colibactin), flagella and the secreted autotransporter toxin (Sat) (Servin, 2014). Notably, while some DAEC strains have been sequenced (Lindsey et al., 2018; Tang et al., 2019), no complete assembled genomes are as yet available. In addition, contrary to EHEC, ETEC, ETEC or EAEC with E. coli strains EDL933, H10407, E2348/69 or O42, respectively (Iguchi et al., 2009; Chaudhuri et al., 2010; Crossman et al., 2010; Latif et al., 2014), no prototypical DAEC strains have been sequenced to date, which limits our understanding of PAIs within this DEC pathotype.
PAI-Encoded Virulence Factors of DEC
While PAIs were originally described in ExPEC, they later appeared to be present in InPEC. To date, 13 PAIs have been characterized, some of which are present in different DEC pathotypes (Figure 4; Table 3).
FIGURE 4.
The position of PAI insertion sites in the diarrheagenic E. coli pathotypes. The name of the genetic features associated with the insertion is indicated with the name of PAIs found at the corresponding site. The position of the insertion site is indicated in pink for tRNA genes and in blue for coding genes. The numbers reported in the inner ring are the positions (megabase) in the genome of the non-pathogenic E. coli strain MG1565 used as a reference.
TABLE 3.
Features of PAIs found in the different DEC pathotypes.
| Name | Alternative names | Insertion site | Size (kb) | Main determinants encoded within the PAIa | DECb |
| LEE PAI | selC, pheV, pheU | 35.6-111 | T3aSS apparatus, Intimin, Tir, Ter system*, LifA* | EPEC, tEHEC | |
| PAI pic/set | SHI-1, She, PAI IICFT073 | pheV | 46.6-71.6 | Pic, ShET1 | EAEC, EIEC, EHEC, EPEC, DAEC |
| LAA PAI | pheV, selC, ileX, serX, thrW | 86.3 | SisA, Hes, IhA, LesP, PagC, SlhA, Ag43 | aEHEC, EPEC, EAEC | |
| PAI iuc/iut | SHI-2, SHI-3 | selC, pheU, pheV | 21-30 | Aerobactin, ShiA*, ShiD* | EIEC |
| PAI tia | selC | 46 | Tia, LeoA | ETEC, EPEC, EHEC | |
| PAI tibA | – | 21.6 | TibA | ETEC | |
| SE PAI | pheV | 8 | SubAB, Tia, ShiA | aEHEC | |
| SHI-O PAI | proAB, thrW, argW | 4.6 | GtrABV | EIEC, EAEC | |
| SRL PAI | serX, serW, orf58 | 66 | StrepR, AmpR, CmR, TetR, FecABCDE-I-R, AspR-DcuA | EIEC, EHEC | |
| PAI espC | orf360, ssrA | 15.2 | EspC, VirA | EPEC, EHEC | |
| PAI sit | aspS, yecN | 10-34 | SitABCD | EIEC, | |
| HPI | asnT, asnV | 40 | Ybt | EAEC, EIEC, EPEC, ETEC, EHEC | |
| PAI IIAL862 | pheV | 61 | Afa/Dr, DeoK | DEC |
aMain virulence or niche factors. ∗Molecular determinants that can be absent from some version of the PAI. bDEC pathotypes where the PAI can be present. EPEC, enteropathogenic E. coli, EHEC, enterohemorrhagic E. coli, tEHEC, typical EHEC (LEE positive); aEHEC, atypical EHEC (LEE negative); ETEC, enterotoxigenic E. coli; EAEC, enteroaggregative E. coli; EIEC, enteroinvasive E. coli (including Shigella); DAEC, diffusely adherent E. coli; DEC, diarrheagenic E. coli (pathotype undefined among the above).
LEE PAI
The LEE is undoubtedly the most investigated PAI in DEC, and readers are referred to recent reviews solely dedicated to this PAI for detailed information (Kirsch et al., 2004; Parham et al., 2005a; Schmidt, 2010; Stevens and Frankel, 2014; Connolly et al., 2015; Franzin and Sircili, 2015; Kendall, 2016; Furniss and Clements, 2018; Turner et al., 2019). The presence of the LEE is the hallmark of EPEC, but it is also found in tEHEC (McDaniel et al., 1995; McDaniel and Kaper, 1997). As first described in prototypical EPEC strain E2348/69 (Perna et al., 1998; Iguchi et al., 2009) and EHEC strain EDL933 (or strain Sakai) (Hayashi et al., 2001; Perna et al., 2001; Latif et al., 2014), the core region of the PAI consists of 41 CDS, organized into (i) 5 operons named LEE-1 to LEE-5 encoding transcriptional regulators, including the master LEE-encoded regulator (Ler), chaperones and structural components of the T3aSS, as well as some effectors, (ii) one grlAB bicistron encoding transcriptional regulators, and (iii) some monocistrons. The T3aSS constitutes a major virulence factor that allows the assembly of a cell-surface needle structure, the injectisome which enables the direct transport of virulence effectors into a host cell (Cornelis, 2006; Desvaux et al., 2006; Brutinel and Yahr, 2008; Mueller et al., 2008; Diepold and Wagner, 2014, 2014; Platenkamp and Mellies, 2018; Lara-Tejero and Galán, 2019; Ageorges et al., 2020). The regulation and function of the T3aSS has been extensively reviewed and readers are invited to consult the above reviews for further details on this vast field of research. Apart from the LEE core region, a certain degree of variability exists between LEEs found in different EPEC or EHEC strains, with estimated sizes ranging from approximately 35 to 110 kb (Sperandio et al., 1998; Jores et al., 2005; Müller et al., 2009). The LEE core region can be completed by prophage genes, as in EHEC ELD933 or Sakai, whereas the fusion with OI-43/48 SpLE1 (Sakai prophage-like element 1) containing a ter operon conferring resistance to tellurite (Tarr et al., 2000; Hayashi et al., 2001). The OI-122 SplE3, which contains a lifA (efa1) region that encodes the virulence factor lymphostatin (Klapproth et al., 2000; Badea et al., 2003; Morabito et al., 2003) or some truncated versions of these islands, can also be found in some variants of the LEE PAI (Gärtner and Schmidt, 2004; Kirsch et al., 2004; Müller et al., 2009). The LEE PAI is inserted at the selC tRNA gene, but the pheV and pheU tRNA genes were also reported as alternative insertion sites for the LEE among these variants (Rumer et al., 2003; Bertin et al., 2004). Phylogenetic analyses based on selected genes indicated that the LEE core region could be discriminated into two main core clusters, one harboring eae-α and -γ and the other one harboring eae-β and -ε independently from the tRNA insertion site (Jores et al., 2004). Nonetheless, the current knowledge on the distribution of the LEE indicates complex events of genetic acquisitions and recombinational rearrangements in E. coli genomes and the evolutionary network remains quite incomplete (Jores et al., 2004; Rasko et al., 2008; Montero et al., 2019).
PAI pic/set
PAI pic/set is inserted into the pheV gene and was identified in EAEC, as well as in EIEC/Shigella (called SHI-1 PAI, previously also known as She) and UPEC (called PAI IICFT073), and in some EHEC and EPEC strains; however, they differ in size, organization, gene composition and genome localization (Al-Hasani et al., 2001a,b; Behrens et al., 2002; Parham et al., 2005a). Island probing revealed this PAI to be quite unstable and to undergo spontaneous excision from the chromosome at a frequency of 10–5–10–6 (Rajakumar et al., 1997). The number of IS-like element flanking the gene (especially IS629 and IS911 also found around other genes encoding SPATEs) suggested that pic (previously called she) is part of a mobile element that could move independently of the PAI (Henderson et al., 1998, 1999; Parham et al., 2005a).
The setB and setA genes, contained within the pic gene but on the complementary strand, encode the oligomeric enterotoxin ShET1 (Fasano et al., 1995; Henderson et al., 1999; Behrens et al., 2002). The transcription of setB was induced in simulated human intestinal microbial systems, although the exact signals promoting gene expression remain to be identified (Behrens et al., 2002). The genetic expression of the pic/set locus is quite unusual and complex. At least three promoters, Ppic1, 2, and 3, could direct the transcription of pic but with different forces and constitutive/inducible expression (Behrens et al., 2002). A PsetA promoter was identified just upstream of setA, whereas a PsetB was identified 1.5 kb upstream of setB with a silencer DRE (downstream regulatory element) region in between that repressed the expression of setBA without any effect on pic transcription (Behrens et al., 2002). Apart from regulation by its own promoter, setA also appears to be expressed by transcription readthrough from setB.
LAA PAI
The locus of adhesion and autoaggregation (LAA) is a composite 86-kb PAI composed of four modules (Montero et al., 2017). In addition to the host immune response suppressor SisA (shiA-like inflammation suppressor genes A) (Lloyd et al., 2009b), module I encodes an autoaggregative factor with Hes (hemagglutinin from STEC), a member of the Hra (heat-resistant agglutinin) family (Montero et al., 2014), together with module IV encoding Ag43, a member of the SAAT (self-associating autotransporter). Module III encodes the adhesin Iha (iron regulated gene homolog adhesin) and the SPATE LesP (LAA encoded SPATE), while module IV actually corresponds to the PAI previously known as PAI ICL3 (Shen et al., 2004).
PAI ICL3 was originally described is a hybrid genomic region that entered B1 E. coli genomes on multiple and independent occasions (Shen et al., 2004). It is composed of segments of the EHEC EDL933 OI-48 at both its extremities and two genomic segments, GS-I and GS-II. GS-I comprises fragments of the Z1640 gene encoding the ShlB transporter family of the T2bSS, whereas GS-II is a composite region encoding a member of the ShlA exoprotein family secreted by the T5bSS. GS-II encodes several transposase genes, genes originating from EHEC EDL933 OI-122, including pagC (phoP-activated gene C), and an additional fragment of Z1640 (Shen et al., 2004; Girardeau et al., 2009). Different variants and often deleted versions of PAI ICL3 were reported as carried by genomic islands, including GIpheV-CRICC168 from Citrobacter rodentium, inserted at pheV, selC or serW tRNA (Girardeau et al., 2009). However, the mosaic structure of LAA PAI was not understood at the time, and these GIs represent complete and incomplete LAA PAI variants (i.e., comprising less or all of the four modules) belonging to two major lineages (Montero et al., 2017). In agreement with PAI ICL3 investigations (Girardeau et al., 2009; Hauser et al., 2013), the LAA PAI appeared to be exclusively present in aEHEC in the first instance and could thus complement the absence of the LEE PAI (Colello et al., 2018, 2019).
PAI iuc/iut
The SHI-2 (Shigella pathogenicity island 2) PAI is inserted at the selC tRNA gene and encodes an integrase almost identical to its ortholog in the LEE PAI. It possesses an abundant number of partial transposases and IS elements, which are remnants of the multiple stepwise assembly of the island (Moss et al., 1999; Vokes et al., 1999). SHI-2 codes the iucABCD operon for synthesis of aerobactin (Lawlor and Payne, 1984), a hydroxamate siderophore involved in iron uptake (Vokes et al., 1999; Braun, 2003), as well as the iutA gene encoding the outer membrane receptor for aerobactin complexed to iron (Crosa, 1989). In Shigella boydi, a 21-kb PAI harboring the iucABCD-iutA operon but inserted at the pheU tRNA gene and possessing LEE prophage genes absent from SHI-2, was called SHI-3 (Purdy and Payne, 2001; Nie et al., 2006). Nonetheless, SHI-3 remains very closely related to the 30-kb SHI-2 in S. flexneri 2a SA100 (Vokes et al., 1999) and 23.8-kb SHI-2 in S. flexneri 5a M90T (Moss et al., 1999), and it belongs to the same family of PAI.
The iut/iuc operon is highly conserved in SHI-2 and SHI-3 and can be inserted in at different loci, especially in paralogs to phe tRNA genes (at least pheV, pheR and pheU) (Al-Hasani et al., 2001a; Nie et al., 2006). The SHI-2/3 PAIs are regrouped under the designation of PAI iuc/iut to rationalize the naming in a way similar to that of PAI pic/set. The PAI iuc/iut would have been acquired horizontally, as suggested by its high degree of conservation with the aerobactin operon harbored on plasmid pColV in some E. coli strains (Moss et al., 1999). SHI-2 also harbors several genes encoding the predicted cytoplasmic proteins, including ShiA (a putative quinone reductase) that has been demonstrated to attenuate inflammation by abrogating the innate T-cell response (Moss et al., 1999; Ingersoll et al., 2003; Ingersoll and Zychlinsky, 2006). SHI-2 also encodes potential integral inner membrane proteins, including ShiF (a putative transporter of the major facilitator superfamily), a dispensable auxiliary factor involved in aerobactin export and/or synthesis (Moss et al., 1999; Genuini et al., 2019). Like pic/set in SHI-1, the shiF and shiG genes overlap and would be transcribed in reverse orientation on a complementary strand, although experimental investigations of genetic regulation and function of ShiG are still required (Moss et al., 1999).
Importantly, the imm (immunity) gene (shiD) appears responsible for immunity to different colicins, including colicin V (ColV) (Vokes et al., 1999). Colicins are proteins produced by E. coli that cause cell lysis of sensitive strains (Cascales et al., 2007). In some strains, PAI iuc/iut is inverted and is thus expressed from the complementary strand, but it is also differently localized and orientated with respect to the oriC (Nie et al., 2006), which could have consequences on the expression of the aerobactin genes (Schmid and Roth, 1987; Eisen et al., 2000; Kothapalli et al., 2005). Among the EIEC/Shigella, the size of PAI iuc/iut clearly differs, but further investigations are needed to determine their diversity and to gain insight into their gene content, organization, phylogeny and distribution in E. coli genomes (Moss et al., 1999; Vokes et al., 1999; Yang et al., 2005).
PAI tia
The prototyptical ETEC strain H10407 contains 25 identified ROD (regions of difference) on the chromosome, two of which are currently viewed as PAIs with the tia (toxigenic invasion locus A) and tibA (toxigenic invasion locus b) loci (Elsinghorst and Weitz, 1994; Crossman et al., 2010). Tia is encoded in a large PAI of approximately 46 kb (corresponding to ROD-20), inserted at selC and flanked by direct repeat sequences (DRSs) identical to those found in PAI-1 in UPEC (Fleckenstein et al., 1996, 2000; Crossman et al., 2010). Tia can act both as an invasin, contributing to the invasion of intestinal epithelium cells (Bondì et al., 2017), and as an adhesin, binding to heparin-sulfate proteoglycans (Mammarappallil and Elsinghorst, 2000; Fleckenstein et al., 2002; Turner et al., 2006b). In addition to int, PAI tia encodes LeoA, an accessory factor for efficient LT secretion, as well as three additional CDS encoding a putative transport system (Fleckenstein et al., 2000). DNA hybridization studies indicate that PAI tia is present in numerous ETECs, as well as in some EPEC and EHEC (Fleckenstein et al., 2000). Nonetheless, the hybridization and multiplex PCR patterns exhibit a considerable level of heterogeneity, suggesting differences in the organization and content of the PAI. The spread of this PAI among DEC and broadly among E. coli strains requires further in-depth investigations (Swenson et al., 1996; Fleckenstein et al., 2000).
PAI tibA
In ETECs, TibA is encoded on a 21.6-kb PAI of low G + C content, corresponding to ROD-13 (Elsinghorst and Weitz, 1994; Crossman et al., 2010). Besides the tibDBCA operon, the PAI tibA codes 14 transposases and IS66, but the insertion site remain elusive. TibA belongs to the SAAT family, is secreted by the T5aSS and is glycosylated by the heptosyltransferase TibC (Masuda et al., 1983; Turner et al., 2006b). Besides cell aggregation and biofilm formation, this glycosylated surface protein is involved in adhesion to intestinal epithelial cells (Lindenthal and Elsinghorst, 1999). The functions of TibD and TibB are yet to be determined. The prevalence and distribution of PAI tibA among E. coli remains to be investigated, though it is not systematically present in all ETEC strains (Elsinghorst and Weitz, 1994; Crossman et al., 2010).
SE PAI
LEE-negative EHEC (i.e., aEHEC) contain an 8-kb PAI coding a subAB2 operon, expressing a subtilase (Michelacci et al., 2013). The subAB1 allelic variant is located on a plasmid, but no additional DNA regions corresponding to the PAI were identified, suggesting that it was not horizontally acquired from this plasmid. SubAB is a well-known cytotoxin inducing apoptosis in LEE-negative EHEC (Wang et al., 2007; Wolfson et al., 2008; Yahiro et al., 2012). This so-called SE (subtilase encoding) PAI also harbors genes encoding an integrase and a sulfate, as well as Tia and ShiA, and is inserted in the pheV tRNA gene. The presence of tia, as in PAI tia, and shiA, as in PAI pic/set, as well as the presence of subAB2 in some ETEC strains (Michelacci et al., 2013), raises questions about the origin and evolution of the SE PAI with respect to these PAIs and demands further phylogenetic analyses. The SE and LEE PAIs have the same insertion site, which led to the hypothesis that mutual exclusion events could have occurred between these PAIs competing for integration at pheV tRNA gene and could further explain the strong association of the subAB2 operon with aEHEC versus tEHEC (Orden et al., 2011; Michelacci et al., 2013). As with PAI iuc/iut, however, insertion at homologous phe tRNA genes cannot be excluded. In general, the prevalence of SE PAI in DEC, and in other E. coli, requires further in-depth investigations.
SHI-O PAI
The SHI-O PAI harbors genes involved in serotype conversion in Shigella by modification of the structure of the O-antigens (Allison and Verma, 2000; Ingersoll et al., 2002; Torres, 2004). SHI-O would appear to be a remnant of an ancient lysogenic bacteriophage that lost its ability to enter into a lytic cycle and underwent excision upon deletions and mutations of sensitive key regions. This PAI codes three genes, gtrA, gtrB and gtrV, that express the glycosyltransferases that allow differential glycosylation of bacterial lipopolysaccharide (LPS) and thus antigenic conversion (Huan et al., 1997; Adams et al., 2001; Sun et al., 2013). SHI-O was also shown to be inverted and to differ in its composition between Shigella strains (Nie et al., 2006) and to have a different insertion in E. coli (Allison and Verma, 2000). Besides the generation of a large variety of shigella serotypes, the modification of O antigens by SHI-O contributes to the modulation of the virulence level through different abilities to evade the host immune response (Robbins et al., 1992; Hong and Payne, 1997; Van den Bosch et al., 1997). The prevalence and distribution of SHI-O in E. coli has not been questioned as yet, although it is present in prototypical EAEC O42 (Chaudhuri et al., 2010).
SRL PAI
The SRL (shigella resistance locus) PAI is a 66-kb element containing at least 59 CDS, especially a 16-kb cluster designated as SLR with genes conferring resistance to streptomycin (aadA1), ampicillin (oxa-1), chloramphenicol (cat) and tetracycline (tetABCD-R) (Rajakumar et al., 1996; Turner et al., 2001, 2003). The SRL PAI is a mobile element flanked by short 14-bp DRSs, which can excise from its insertion site at tRNA gene serX (Turner et al., 2001). The deletion rate was estimated at approximately 10–5 and led to antibiotic-sensitive variants. The int (integrase) gene was required for precision deletion of the island through site-specific recombination. Alternative insertion sites are strongly suspected, such as serW, paralogous to serX, and orf58 (Turner et al., 2003). Besides antibiotic resistance, SRL PAI encodes a functional ferric dicitrate iron transport system (fecABCDE-I-R), which expands the battery of iron acquisition mechanisms in the bacterial strain (Luck et al., 2001). Functional genetic analysis revealed that SRL PAI encoded a functional aspartate racemase converting D- to L-aspartate (AspR) and a cognate transporter (DcuA) enabling catabolism of D-asparate as a sole carbon source (Henríquez et al., 2020). Catabolism of D-amino acids, such as D-aspartate, as a carbon source and/or detoxification of their toxic effects on protein biosynthesis could have an important role in the colonization of the human gastrointestinal tract, an aspect which undoubtedly requires further in-depth investigations. The aspR and dcuA homologs are present in some EHEC O104:H4 isolates (Henríquez et al., 2020), but the broad prevalence of SLR PAI in E. coli needs thorough investigation, although this PAI seems quite prevalent in multiresistant Shigella isolates (Turner et al., 2003).
PAI espC
EspC (E. coli secreted protein C) is a SPATE with enterotoxic activity that induces necrosis and apoptosis in epithelial cells (Mellies et al., 2001; Navarro-Garcia et al., 2014; Serapio-Palacios and Navarro-Garcia, 2016). EspC actually cleaves the cytoskeletal actin-associated protein α-fodrin, the focal adhesion protein paxillin and a focal adhesion kinase (FAK). In EPEC, EspC is encoded in a 15.2-kb PAI inserted between orf360 and tRNA ssrA genes, with respect to the E. coli K12 chromosome (Mellies et al., 2001). Besides five putative transposases, PAI espC encodes an integrase and a homolog to the virulence effector VirA from S. flexneri that is involved in cellular invasion and intracellular spreading (Campbell-Valois et al., 2015; Agaisse, 2016), as well as additional CDS with low sequence similarities that require further analysis to ascertain their function. The low G + C content of this PAI suggests it was acquired horizontally. PAI espC seems present in other DEC, including some EPEC and EHEC, but additional studies are needed to characterize the possible diversity in the genetic organization, prevalence and distribution (Mellies et al., 2001).
PAI sit
Originally described in S. flexneri, the PAI sit primarily harbors a sitABCD locus encoding a ferrous iron and manganese uptake system belonging to the ABC transporter family, homologous to Sit (salmonella iron transport) (Kehres et al., 2002; Runyen-Janecky et al., 2006; Fisher et al., 2009). The PAI sit is found in some commensal and pathogenic E. coli at various chromosomic locations, with insertion sites that fall into four clusters (Fisher et al., 2009). The size and orientation of the island seem to differ considerably, but further studies are required to assess the degree of variation in the gene content of the PAI sit (Chen and Schneider, 2006; Fisher et al., 2009). This PAI has multiple chromosomic insertion sites and a large number of genetic rearrangements and deletion and insertion sequences that would occur through homologous recombination rather than site-specific mechanisms. Therefore, this PAI appears quite unstable (Fisher et al., 2009).
HPI
The HPI (high pathogenicity island) is a PAI originally identified in Yersinia enterocolitica (Schubert et al., 1998; Carniel, 2001). It later appeared widely disseminated in Enterobacteriaceae, including DEC, but essentially in EAEC, as it is rarely present in EIEC/Shigella, EPEC, ETEC or EHEC (Czeczulin et al., 1999; Karch et al., 1999; Schubert et al., 2000). The HPI encodes an iron uptake system based on the siderophore yersioniabactin (Ybt) (Schubert et al., 2004b), which was demonstrated to be functional in several EAEC strains and contributes to the virulence (Carniel, 2001; Hu et al., 2005). While fyuA (also called psn) and ybtPQ (also called irp6-7) encode an outer-membrane receptor and an inner-membrane transporter for iron uptake, respectively, irp1-5 and ybtS (irp9) are involved in the biosynthesis of Ybt (Carniel, 2001). In E. coli, the HPI is approximately 40 kb and is generally inserted at the asnT tRNA gene (Buchrieser et al., 1998; Schubert et al., 2004b). However, it can also be inserted at the asnV tRNA gene harbored in place of IS600, an additional 34.4-kb fragment constituting a functional ICE encoding a T4SS (Schubert et al., 2004a). This element could constitute the progenitor and missing link that would explain the dissemination of HPI among E. coli.
PAI IIAL862
Escherichia coli AL862 was primarily isolated from a septic patient with a cancer, but the PAI containing the afa-8 operon, and further called PAI IIAL862, appeared as a preferential insertion into the pheV gene in several human and bovine pathogenic E. coli strains (Lalioui and Le Bouguénec, 2001; Redford and Welch, 2002). The afa-8 operon, encoding the Afa/Dr adhesin AfaEVIII (Lalioui et al., 1999; Lalioui and Le Bouguénec, 2001), is expressed in some E. coli strains isolated from animals with diarrhea, including calves, pigs, and poultry (Gérardin et al., 2000). PAI IIAL862 can be identified in some human intestinal E. coli isolates but evidence of its implication in diarrhea in humans remains to be demonstrated (Le Bouguénec et al., 2001; Girardeau et al., 2003). This PAI is supposedly present in some DAEC, but investigations are still required (Servin, 2014). Among the four sRNAs encoded in PAI IIAL862, the thermoregulated sRNA AfaR further regulates the expression of the AfaD-VIII subunit. PAI IIAL862 also includes the deoK operon, which confers the ability to use deoxyribose as a carbon source and would increase competitiveness with respect to host infectivity (Lalioui and Le Bouguénec, 2001).
Conclusion
As reviewed here above, PAIs are widespread in E. coli and are major actors in the genome plasticity of this bacterium. The PAIs have shaped the genome of this bacterial species, which further explains the emergence of different pathotypes that result in different intestinal and extraintestinal diseases. The transfer of these GIs in a one-step acquisition of the entire functional determinants, thereby enabling bacterial evolution in quantum leaps, opens the bacterium to new ecological niches. The functional elements encoded by PAIs include pathogenicity effectors, but they also contain regulatory elements involved in crosstalk between PAIs and the genetic background. This crosstalk may be key factors in genome evolution, including PAIs, and probably result in a diversity of expression profiles. This requires more investigations to obtain the best understanding of E. coli virulence and the apparent redundancy of pathogenicity factors in ExPEC.
As exemplified by the LEE and LAA PAIs, these genomic islands are also quite plastic in terms of their insertion or deletion events, resulting in PAIs of different sizes in different strains but with a common evolutionary history. This review has further stressed that, in some cases, further investigations are still required to fill the existing knowledge gaps, for example, about the phylogenetic relationship between SE PA, PAI tia, and PAI pic/set (Michelacci et al., 2013), which may belong to the same family as was later identified for PAI ICL3 and PAI LAA (Montero et al., 2017). However, the PAI nomenclature is quite erratic and requires a more rigorous formulization to avoid different naming of homologous genomic islands, such as SH-2 and SH-3, found in different strains, as was done for HPI. This is necessary to facilitate PAI tracing and our understanding of their phylogeny and evolution.
Additional PAIs are most certainly present in E. coli; examples are the genomic islands OI-57 (Coombes et al., 2008; Imamovic et al., 2010), OI-122 (Karmali et al., 2003; Wickham et al., 2006; Konczy et al., 2008), OI-43/48 (Taylor et al., 2002; Yin et al., 2009; Ju et al., 2013) and GI-selC17584/1 (Schmidt et al., 2001; Saile et al., 2018) in EHEC or some ROD in ETEC (Crossman et al., 2010). However, beyond a correlation of their association with outbreaks or severe diseases, experimental confirmation is still required to demonstrate their involvement and exact role in E. coli virulence and pathogenicity.
The evolution of the pathogenesis of E. coli constitutes a challenge for public health authorities in both developed and developing countries. This is especially evident given the burden of disease and the emergence of new epidemic strains, as dramatically exemplified by the hybrid diarrheagenic EHEC/EAEC O104:H4 strain (Navarro-Garcia, 2014; Prager et al., 2014; Kampmeier et al., 2018). The efficient horizontal gene transfer and high genetic plasticity of the E. coli genome, at both the plasmidic and chromosomic levels, represent potential public health issues that require a full comprehension and determination of the molecular mechanisms at play in the emergence of new E. coli pathotypes and/or E. coli strains with higher virulence levels. The description of numerous E. coli pathotypes with respect to intestinal and extraintestinal pathogenic diseases should not hide the fact that E. coli is primarily a normal and harmless commensal of the human intestinal microbiota, and that all these strains belong to the very same and unique bacterial species, E. coli (including Shigella).
To avoid biases in our understanding of E. coli physiology when solely focusing on anthropocentric medical and pathogenic aspects, we must consider not only pathogenic human and animal isolates but also the commensal strains that occur all along the food chains. Very diverse selection pressures can be encountered in the trophic network and can work together to drive evolution. In the various ecosystems that E. coli calls home (e.g., animal and human GIT, food matrices and soil, water, agri-food environments), the colibiote consists of different commensal and/or pathogenic E. coli strains co-existing and interacting along with the rest of the microbiota of the biocoenoses in the various environmental conditions of the biotopes. E. coli is not intentionally pathogenic, as suggested by its pathoadaptation (Maurelli, 2007; Rahman and Gadjeva, 2014; Pasqua et al., 2017). However, ecocentric and biocentric views allow the perception that these different levels of interactions can induce evolution and selection of genetic traits for specific niche factors for the necessity of fitness and adaptation, which can inadvertently, collaterally, coincidentally and/or randomly result in virulence as a by-product of commensalism (Le Gall et al., 2007; Tenaillon et al., 2010; Hill, 2012; Leimbach et al., 2013). Integrating the physiopathology to the ecophysiology of this species is certainly the next frontier in understanding the forces at play in the E. coli genome plasticity and the emergence of new virulence traits and/or pathotypes.
Author Contributions
MD and RB wrote the first overall draft of the manuscript. RB drew the original pictures. MD, GD, RBe, NB, JD, and RBo wrote sections of the manuscript. RB contributed to conceptualize the overarching aims and had management as well as coordination responsibility for the execution of the work. All authors contributed to the critical revision of the manuscript, read and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abbreviations
- A/E
attaching and effacing
- AA
aggregative adherence
- AAF
aggregative adherence fimbriae
- ABU
asymptomatic bacteriuria
- aEHEC
atypical enterohemorrhagic E. coli
- aEPEC
atypical enteropathogenic E. coli
- Afa
afimbrial adhesin
- Ag43
antigen 43
- AIEC
adherent-invasive E. coli
- APEC
avian pathogenic E. coli
- AT
autotransporter
- CASPR1
contactin-associated protein 1
- CDS
coding sequence
- Cdt
cytolethal and distending toxin
- CFA
colonization factor antigen
- CFTR
cystic fibrosis transmembrane conductance regulator
- CLB
colibactin
- CNF-1
cytotoxic necrotizing factor-1
- DAEC
diffusely adherent E. coli
- DEC
diarrheagenic E. coli
- Dr
decay-accelerating factor receptor
- DRE
downstream regulatory element
- DRS
direct repeat sequences
- EAEC
enteroaggregative E. coli
- ECP
E. coli common pili
- EHEC
enterohemorrhagic E. coli
- EIEC
enteroinvasive E. coli
- EPEC
enteropathogenic E. coli
- EspC
E. coli secreted protein C
- ETEC
enterotoxigenic E. coli
- ExPEC
extraintestinal E. coli
- FAK
focal adhesion kinase
- GbO3
globotriosylceramide
- GbO4
globotetraosylceramide
- GbO5
globopentaosylceramide
- GI
genomic islands
- GS
genome segment
- HBMEC
human brain microvascular endothelial cell
- Hbp
hemoglobin binding protein or hemoglobin protease
- HPI
high-pathogenicity island
- Hra
heat-resistant agglutinin
- HT
horizontal transfer
- HUS
syndrome hemolytic and uremic
- IBC
intracellular bacterial community
- IBD
inflammatory bowel disease
- ICE
integrative and conjugative elements
- Iha
iron regulated gene homolog adhesin
- InPEC
intestinal pathogenic E. coli
- LA
localized adherence
- LAA
Locus of adhesion and autoaggregation
- LAL
localized-adherence like
- LEE
locus of enterocyte effacement
- Ler
LEE-encoded regulator
- LifA
lymphocyte inhibitory factor
- LPS
lipopolysaccharide
- LT
heat-labile enterotoxin
- NlpI
new lipoprotein I
- NMEC
neonatal meningitis E. coli
- OmpA
outer membrane protein A
- PagG
PhoP-activated gene C
- PAI
pathogenicity islands
- Pap
pyelonephritis associated pili
- Pet
plasmid-encoded toxin
- PF
Pathogenicity factors
- Pic
protein involve in colonization
- RDF
recombination directionality factor
- RTX
repeat-in-toxin
- SAAT
self-assocoiating autotransporter
- Sat
secreted autotransporter toxin
- SEPEC
sepsis-associated E. coli
- Sha
serine-protease hemagglutinin autotransporter
- ShET1
shigella enterotoxin 1
- ShET2
shigella enterotoxin 2
- SHI
shigella pathogenicity island
- SigA
Shigella immunoglobulin A protease
- Sit
Salmonella iron transport
- SPATE
serine protease autotransporters of Enterobacteriaceae
- SpLE
sakai prophage-like element
- SslE
secreted and surface associated lipoprotein
- ST
heat-stable enterotoxin
- STEC
shigatoxin-producing E. coli
- Stx
shigatoxin
- T2SS
type II secretion system
- T3aSS
type III, subtype a, secretion system
- T3SS
type III secretion system
- T4SS
type IV secretion system
- Tag
tandem autotransporter gene
- TcpC
TLR domain containing-protein C
- tEHEC
typical enterohemorrhagic E. coli
- tEPEC
typical enteropathogenic E. coli
- Tia
toxigenic invasion locus A
- TLR
toll-like receptor
- Tsh
temperature sensitive hemagglutinin
- TTP
thrombotic thrombocytopenic purpura
- UPEC
uropathogenic E. coli
- UTI
urinary tract infection
- Vat
vacuolating autotransporter toxin
- Ybt
yersioniabactin.
Footnotes
Funding. This study was supported by the Ministère de la Recherche et de la Technologie, Inserm (U1071), and INRAE (USC-2018 and UMR0454).
References
- Abraham S. N., Sun D., Dale J. B., Beachey E. H. (1988). Conservation of the D-mannose-adhesion protein among type 1 fimbriated members of the family Enterobacteriaceae. Nature 336 682–684. 10.1038/336682a0 [DOI] [PubMed] [Google Scholar]
- Adams M. M., Allison G. E., Verma N. K. (2001). Type IV O antigen modification genes in the genome of Shigella flexneri NCTC 8296. Microbiology 147 851–860. 10.1099/00221287-147-4-851 [DOI] [PubMed] [Google Scholar]
- Agaisse H. (2016). Molecular and cellular mechanisms of Shigella flexneri dissemination. Front. Cell. Infect. Microbiol. 6:29. 10.3389/fcimb.2016.00029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ageorges V., Monteiro R., Leroy S., Burgess C. M., Pizza M., Chaucheyras-Durand F., et al. (2020). Molecular Determinants of Surface Colonisation in Diarrhoeagenic Escherichia coli (DEC): from Bacterial Adhesion to Biofilm Formation. FEMS Microbiol. Rev. 44 314–350. 10.1093/femsre/fuaa008 [DOI] [PubMed] [Google Scholar]
- Al-Hasani K., Adler B., Rajakumar K., Sakellaris H. (2001a). Distribution and structural variation of the she pathogenicity island in enteric bacterial pathogens. J. Med. Microbiol. 50 780–786. 10.1099/0022-1317-50-9-780 [DOI] [PubMed] [Google Scholar]
- Al-Hasani K., Henderson I. R., Sakellaris H., Rajakumar K., Grant T., Nataro J. P., et al. (2000). The sigA gene which is borne on the she pathogenicity island of Shigella flexneri 2a encodes an exported cytopathic protease involved in intestinal fluid accumulation. Infect. Immun. 68 2457–2463. 10.1128/iai.68.5.2457-2463.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Hasani K., Rajakumar K., Bulach D., Robins-Browne R., Adler B., Sakellaris H. (2001b). Genetic organization of the she pathogenicity island in Shigella flexneri 2a. Microb. Pathog. 30 1–8. 10.1006/mpat.2000.0404 [DOI] [PubMed] [Google Scholar]
- Allison G. E., Verma N. K. (2000). Serotype-converting bacteriophages and O-antigen modification in Shigella flexneri. Trends Microbiol. 8 17–23. 10.1016/s0966-842x(99)01646-7 [DOI] [PubMed] [Google Scholar]
- Allsopp L. P., Beloin C., Ulett G. C., Valle J., Totsika M., Sherlock O., et al. (2012). Molecular characterization of UpaB and UpaC, two new autotransporter proteins of uropathogenic Escherichia coli CFT073. Infect. Immun. 80 321–332. 10.1128/IAI.05322-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ananias M., Yano T. (2008). Serogroups and virulence genotypes of Escherichia coli isolated from patients with sepsis. Braz. J. Med. Biol. Res. 41 877–883. 10.1590/s0100-879x2008001000008 [DOI] [PubMed] [Google Scholar]
- Anderson G. G., Palermo J. J., Schilling J. D., Roth R., Heuser J., Hultgren S. J. (2003). Intracellular bacterial biofilm-like pods in urinary tract infections. Science 301 105–107. 10.1126/science.1084550 [DOI] [PubMed] [Google Scholar]
- Andersson P., Engberg I., Lidin-Janson G., Lincoln K., Hull R., Hull S., et al. (1991). Persistence of Escherichia coli bacteriuria is not determined by bacterial adherence. Infect. Immun. 59 2915–2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antão E.-M., Wieler L. H., Ewers C. (2009). Adhesive threads of extraintestinal pathogenic Escherichia coli. Gut Pathog. 1:22. 10.1186/1757-4749-1-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arthur J. C., Perez-Chanona E., Mühlbauer M., Tomkovich S., Uronis J. M., Fan T.-J., et al. (2012). Intestinal inflammation targets cancer-inducing activity of the microbiota. Science 338 120–123. 10.1126/science.1224820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Askari Badouei M., Jajarmi M., Mirsalehian A. (2015). Virulence profiling and genetic relatedness of Shiga toxin-producing Escherichia coli isolated from humans and ruminants. Comp. Immunol. Microbiol. Infect. Dis. 38 15–20. 10.1016/j.cimid.2014.11.005 [DOI] [PubMed] [Google Scholar]
- Badea L., Doughty S., Nicholls L., Sloan J., Robins-Browne R. M., Hartland E. L. (2003). Contribution of Efa1/LifA to the adherence of enteropathogenic Escherichia coli to epithelial cells. Microb. Pathog. 34 205–215. 10.1016/s0882-4010(03)00026-3 [DOI] [PubMed] [Google Scholar]
- Badger J. L., Wass C. A., Weissman S. J., Kim K. S. (2000). Application of signature-tagged mutagenesis for identification of Escherichia coli K1 genes that contribute to invasion of human brain microvascular endothelial cells. Infect. Immun. 68 5056–5061. 10.1128/iai.68.9.5056-5061.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bark T., Katouli M., Svenberg T., Ljungqvist O. (1995). Food deprivation increases bacterial translocation after non-lethal haemorrhage in rats. Eur. J. Surg. 161 67–71. [PubMed] [Google Scholar]
- Basmaci R., Bonacorsi S., Bidet P., Biran V., Aujard Y., Bingen E., et al. (2015). Escherichia Coli meningitis features in 325 children from 2001 to 2013 in France. Clin. Infect. Dis. 61 779–786. 10.1093/cid/civ367 [DOI] [PubMed] [Google Scholar]
- Bateman S. L., Seed P. (2012a). Intracellular Macrophage Infections with E. coli under Nitrosative Stress. Bio Protoc. 2:e275. 10.21769/BioProtoc.275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman S. L., Seed P. (2012b). Promoter orientation of prokaryotic phase-variable genes by PCR. Bio Protoc. 2:e274. 10.21769/bioprotoc.274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman S. L., Stapleton A. E., Stamm W. E., Hooton T. M., Seed P. C. (2013). The type 1 pili regulator gene fimX and pathogenicity island PAI-X as molecular markers of uropathogenic Escherichia coli. Microbiology 159 1606–1617. 10.1099/mic.0.066472-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battaglioli E. J., Goh K. G. K., Atruktsang T. S., Schwartz K., Schembri M. A., Welch R. A. (2018). Identification and characterization of a phase-variable element that regulates the autotransporter upae in uropathogenic Escherichia coli. mBio 9:e01360-18. 10.1128/mBio.01360-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battle S. E., Brady M. J., Vanaja S. K., Leong J. M., Hecht G. A. (2014). Actin pedestal formation by enterohemorrhagic Escherichia coli enhances bacterial host cell attachment and concomitant type III translocation. Infect. Immun. 82 3713–3722. 10.1128/IAI.01523-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck C. M., Willett J. L. E., Cunningham D. A., Kim J. J., Low D. A., Hayes C. S. (2016). CdiA effectors from uropathogenic Escherichia coli use heterotrimeric osmoporins as receptors to recognize target bacteria. PLoS Pathog. 12:e1005925. 10.1371/journal.ppat.1005925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens M., Sheikh J., Nataro J. P. (2002). Regulation of the overlapping pic/set locus in Shigella flexneri and enteroaggregative Escherichia coli. Infect. Immun. 70 2915–2925. 10.1128/iai.70.6.2915-2925.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belotserkovsky I., Sansonetti P. J. (2018). Shigella and Enteroinvasive Escherichia Coli. Curr. Top. Microbiol. Immunol. 416 1–26. 10.1007/82_2018_104 [DOI] [PubMed] [Google Scholar]
- Benedek O., Schubert S. (2007). Mobility of the Yersinia High-Pathogenicity Island (HPI): transfer mechanisms of pathogenicity islands (PAIS) revisited (a review). Acta Microbiol. Immunol. Hung. 54 89–105. 10.1556/AMicr.54.2007.2.1 [DOI] [PubMed] [Google Scholar]
- Bergsten G., Wullt B., Svanborg C. (2005). Escherichia coli, fimbriae, bacterial persistence and host response induction in the human urinary tract. Int. J. Med. Microbiol. 295 487–502. 10.1016/j.ijmm.2005.07.008 [DOI] [PubMed] [Google Scholar]
- Bertelli C., Laird M. R., Williams K. P., Simon Fraser University Research Computing Group, Lau B. Y., Hoad G., et al. (2017). IslandViewer 4: expanded prediction of genomic islands for larger-scale datasets. Nucleic Acids Res. 45 W30–W35. 10.1093/nar/gkx343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertelli C., Tilley K. E., Brinkman F. S. L. (2019). Microbial genomic island discovery, visualization and analysis. Brief. Bioinformatics 20 1685–1698. 10.1093/bib/bby042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertin Y., Boukhors K., Livrelli V., Martin C. (2004). Localization of the insertion site and pathotype determination of the locus of enterocyte effacement of shiga toxin-producing Escherichia coli strains. Appl. Environ. Microbiol. 70 61–68. 10.1128/aem.70.1.61-68.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi D., Xu Z., Harrison E. M., Tai C., Wei Y., He X., et al. (2012). ICEberg: a web-based resource for integrative and conjugative elements found in Bacteria. Nucleic Acids Res. 40 D621–D626. 10.1093/nar/gkr846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bidet P., Bonacorsi S., Clermont O., De Montille C., Brahimi N., Bingen E. (2005). Multiple insertional events, restricted by the genetic background, have led to acquisition of pathogenicity island IIJ96-like domains among Escherichia coli strains of different clinical origins. Infect. Immun. 73 4081–4087. 10.1128/IAI.73.7.4081-4087.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielaszewska M., Middendorf B., Tarr P. I., Zhang W., Prager R., Aldick T., et al. (2011). Chromosomal instability in enterohaemorrhagic Escherichia coli O157:H7: impact on adherence, tellurite resistance and colony phenotype. Mol. Microbiol. 79 1024–1044. 10.1111/j.1365-2958.2010.07499.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bingen-Bidois M., Clermont O., Bonacorsi S., Terki M., Brahimi N., Loukil C., et al. (2002). Phylogenetic analysis and prevalence of urosepsis strains of Escherichia coli bearing pathogenicity island-like domains. Infect. Immun. 70 3216–3226. 10.1128/iai.70.6.3216-3226.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco M., Blanco J. E., Mora A., Dahbi G., Alonso M. P., González E. A., et al. (2004). Serotypes, virulence genes, and intimin types of Shiga toxin (verotoxin)-producing Escherichia coli isolates from cattle in Spain and identification of a new intimin variant gene (eae-xi). J. Clin. Microbiol. 42 645–651. 10.1128/jcm.42.2.645-651.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom F. R., McFall E. (1975). Isolation and characterization of D-serine deaminase constitutive mutants by utilization of D-serine as sole carbon or nitrogen source. J. Bacteriol. 121 1078–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum G., Ott M., Lischewski A., Ritter A., Imrich H., Tschäpe H., et al. (1994). Excision of large DNA regions termed pathogenicity islands from tRNA-specific loci in the chromosome of an Escherichia coli wild-type pathogen. Infect. Immun. 62 606–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonacorsi S., Bingen E. (2005). Molecular epidemiology of Escherichia coli causing neonatal meningitis. Int. J. Med. Microbiol. 295 373–381. 10.1016/j.ijmm.2005.07.011 [DOI] [PubMed] [Google Scholar]
- Bonacorsi S., Clermont O., Houdouin V., Cordevant C., Brahimi N., Marecat A., et al. (2003). Molecular analysis and experimental virulence of French and North American Escherichia coli neonatal meningitis isolates: identification of a new virulent clone. J. Infect. Dis. 187 1895–1906. 10.1086/375347 [DOI] [PubMed] [Google Scholar]
- Bonacorsi S., Houdouin V., Mariani-Kurkdjian P., Mahjoub-Messai F., Bingen E. (2006). Comparative prevalence of virulence factors in Escherichia coli causing urinary tract infection in male infants with and without bacteremia. J. Clin. Microbiol. 44 1156–1158. 10.1128/JCM.44.3.1156-1158.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondì R., Chiani P., Michelacci V., Minelli F., Caprioli A., Morabito S. (2017). The Gene tia, harbored by the subtilase-encoding pathogenicity island, is involved in the ability of locus of enterocyte effacement-negative shiga toxin-producing Escherichia coli strains to invade monolayers of epithelial cells. Infect. Immun. 85:e00613-17. 10.1128/IAI.00613-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bower J. M., Eto D. S., Mulvey M. A. (2005). Covert operations of uropathogenic Escherichia coli within the urinary tract. Traffic 6 18–31. 10.1111/j.1600-0854.2004.00251.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd E. F., Almagro-Moreno S., Parent M. A. (2009). Genomic islands are dynamic, ancient integrative elements in bacterial evolution. Trends Microbiol. 17 47–53. 10.1016/j.tim.2008.11.003 [DOI] [PubMed] [Google Scholar]
- Boyd E. F., Davis B. M., Hochhut B. (2001). Bacteriophage-bacteriophage interactions in the evolution of pathogenic bacteria. Trends Microbiol. 9 137–144. 10.1016/s0966-842x(01)01960-6 [DOI] [PubMed] [Google Scholar]
- Braun V. (2003). Iron uptake by Escherichia coli. Front. Biosci. 8 s1409–s1421. 10.2741/1232 [DOI] [PubMed] [Google Scholar]
- Bronowski C., Smith S. L., Yokota K., Corkill J. E., Martin H. M., Campbell B. J., et al. (2008). A subset of mucosa-associated Escherichia coli isolates from patients with colon cancer, but not Crohn’s disease, share pathogenicity islands with urinary pathogenic E. coli. Microbiology 154 571–583. 10.1099/mic.0.2007/013086-0 [DOI] [PubMed] [Google Scholar]
- Brutinel E. D., Yahr T. L. (2008). Control of gene expression by type III secretory activity. Curr. Opin. Microbiol. 11 128–133. 10.1016/j.mib.2008.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan A., Roesch P., Davis L., Moritz R., Pellett S., Welch R. A. (2006). Regulation of type 1 fimbriae by unlinked FimB- and FimE-like recombinases in uropathogenic Escherichia coli strain CFT073. Infect. Immun. 74 1072–1083. 10.1128/IAI.74.2.1072-1083.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzuszkiewicz E., Brüggemann H., Liesegang H., Emmerth M., Olschläger T., Nagy G., et al. (2006). How to become a uropathogen: comparative genomic analysis of extraintestinal pathogenic Escherichia coli strains. Proc. Natl. Acad. Sci. U.S.A. 103 12879–12884. 10.1073/pnas.0603038103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buc E., Dubois D., Sauvanet P., Raisch J., Delmas J., Darfeuille-Michaud A., et al. (2013). High prevalence of mucosa-associated E. coli producing cyclomodulin and genotoxin in colon cancer. PLoS One 8:e56964. 10.1371/journal.pone.0056964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchrieser C., Brosch R., Bach S., Guiyoule A., Carniel E. (1998). The high-pathogenicity island of Yersinia pseudotuberculosis can be inserted into any of the three chromosomal asn tRNA genes. Mol. Microbiol. 30 965–978. 10.1046/j.1365-2958.1998.01124.x [DOI] [PubMed] [Google Scholar]
- Buckles E. L., Bahrani-Mougeot F. K., Molina A., Lockatell C. V., Johnson D. E., Drachenberg C. B., et al. (2004). Identification and characterization of a novel uropathogenic Escherichia coli-associated fimbrial gene cluster. Infect. Immun. 72 3890–3901. 10.1128/IAI.72.7.3890-3901.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell-Valois F.-X., Sachse M., Sansonetti P. J., Parsot C. (2015). Escape of Actively Secreting Shigella flexneri from ATG8/LC3-positive vacuoles formed during cell-to-cell spread is facilitated by IcsB and VirA. mBio 6:e02567-14. 10.1128/mBio.02567-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos L. C., Vieira M. A., Trabulsi L. R., da Silva L. A., Monteiro-Neto V., Gomes T. A. (1999). Diffusely adhering Escherichia coli (DAEC) strains of fecal origin rarely express F1845 adhesin. Microbiol. Immunol. 43 167–170. 10.1111/j.1348-0421.1999.tb02388.x [DOI] [PubMed] [Google Scholar]
- Caprioli A., Falbo V., Ruggeri F. M., Baldassarri L., Bisicchia R., Ippolito G., et al. (1987). Cytotoxic necrotizing factor production by hemolytic strains of Escherichia coli causing extraintestinal infections. J. Clin. Microbiol. 25 146–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caprioli A., Morabito S., Brugère H., Oswald E. (2005). Enterohaemorrhagic Escherichia coli: emerging issues on virulence and modes of transmission. Vet. Res. 36 289–311. 10.1051/vetres:2005002 [DOI] [PubMed] [Google Scholar]
- Carniel E. (2001). The Yersinia high-pathogenicity island: an iron-uptake island. Microbes Infect. 3 561–569. 10.1016/s1286-4579(01)01412-5 [DOI] [PubMed] [Google Scholar]
- Cascales E., Buchanan S. K., Duché D., Kleanthous C., Lloubès R., Postle K., et al. (2007). Colicin biology. Microbiol. Mol. Biol. Rev. 71 158–229. 10.1128/MMBR.00036-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri R. R., Henderson I. R. (2012). The evolution of the Escherichia coli phylogeny. Infect. Genet. Evol. 12 214–226. 10.1016/j.meegid.2012.01.005 [DOI] [PubMed] [Google Scholar]
- Chaudhuri R. R., Sebaihia M., Hobman J. L., Webber M. A., Leyton D. L., Goldberg M. D., et al. (2010). Complete genome sequence and comparative metabolic profiling of the prototypical enteroaggregative Escherichia coli strain 042. PLoS One 5:e8801. 10.1371/journal.pone.0008801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheetham B. F., Katz M. E. (1995). A role for bacteriophages in the evolution and transfer of bacterial virulence determinants. Mol. Microbiol. 18 201–208. 10.1111/j.1365-2958.1995.mmi_18020201.x [DOI] [PubMed] [Google Scholar]
- Chen Z., Schneider T. D. (2006). Comparative analysis of tandem T7-like promoter containing regions in enterobacterial genomes reveals a novel group of genetic islands. Nucleic Acids Res. 34 1133–1147. 10.1093/nar/gkj511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua E. G., Al-Hasani K., Scanlon M., Adler B., Sakellaris H. (2015). Determinants of proteolysis and cell-binding for the Shigella flexneri Cytotoxin, SigA. Curr. Microbiol. 71 613–617. 10.1007/s00284-015-0893-8 [DOI] [PubMed] [Google Scholar]
- Cirl C., Wieser A., Yadav M., Duerr S., Schubert S., Fischer H., et al. (2008). Subversion of Toll-like receptor signaling by a unique family of bacterial Toll/interleukin-1 receptor domain-containing proteins. Nat. Med. 14 399–406. 10.1038/nm1734 [DOI] [PubMed] [Google Scholar]
- Clements A., Young J. C., Constantinou N., Frankel G. (2012). Infection strategies of enteric pathogenic Escherichia coli. Gut Microbes 3 71–87. 10.4161/gmic.19182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clermont O., Dixit O. V. A., Vangchhia B., Condamine B., Dion S., Bridier-Nahmias A., et al. (2019). Characterization and rapid identification of phylogroup G in Escherichia coli, a lineage with high virulence and antibiotic resistance potential. Environ. Microbiol 21 3107–3117. 10.1111/1462-2920.14713 [DOI] [PubMed] [Google Scholar]
- Colello R., Krüger A., Velez M. V., Del Canto F., Etcheverría A. I., Vidal R., et al. (2019). Identification and detection of iha subtypes in LEE-negative Shiga toxin-producing Escherichia coli (STEC) strains isolated from humans, cattle and food. Heliyon 5:e03015. 10.1016/j.heliyon.2019.e03015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colello R., Vélez M. V., González J., Montero D. A., Bustamante A. V., Del Canto F., et al. (2018). First report of the distribution of Locus of Adhesion and Autoaggregation (LAA) pathogenicity island in LEE-negative Shiga toxin-producing Escherichia coli isolates from Argentina. Microb. Pathog. 123 259–263. 10.1016/j.micpath.2018.07.011 [DOI] [PubMed] [Google Scholar]
- Connell I., Agace W., Klemm P., Schembri M., Mãrild S., Svanborg C. (1996). Type 1 fimbrial expression enhances Escherichia coli virulence for the urinary tract. Proc. Natl. Acad. Sci. U.S.A. 93 9827–9832. 10.1073/pnas.93.18.9827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly J. P. R., Finlay B. B., Roe A. J. (2015). From ingestion to colonization: the influence of the host environment on regulation of the LEE encoded type III secretion system in enterohaemorrhagic Escherichia coli. Front. Microbiol. 6:568. 10.3389/fmicb.2015.00568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coombes B. K., Wickham M. E., Mascarenhas M., Gruenheid S., Finlay B. B., Karmali M. A. (2008). Molecular analysis as an aid to assess the public health risk of non-O157 Shiga toxin-producing Escherichia coli strains. Appl. Environ. Microbiol. 74 2153–2160. 10.1128/AEM.02566-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordeiro M. A., Werle C. H., Milanez G. P., Yano T. (2016). Curli fimbria: an Escherichia coli adhesin associated with human cystitis. Braz. J. Microbiol. 47 414–416. 10.1016/j.bjm.2016.01.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornelis G. R. (2006). The type III secretion injectisome. Nat. Rev. Microbiol. 4 811–825. 10.1038/nrmicro1526 [DOI] [PubMed] [Google Scholar]
- Cosloy S. D., McFall E. (1973). Metabolism of D-serine in Escherichia coli K-12: mechanism of growth inhibition. J. Bacteriol. 114 685–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cougnoux A., Dalmasso G., Martinez R., Buc E., Delmas J., Gibold L., et al. (2014). Bacterial genotoxin colibactin promotes colon tumour growth by inducing a senescence-associated secretory phenotype. Gut 63 1932–1942. 10.1136/gutjnl-2013-305257 [DOI] [PubMed] [Google Scholar]
- Cougnoux A., Delmas J., Gibold L., Faïs T., Romagnoli C., Robin F., et al. (2016). Small-molecule inhibitors prevent the genotoxic and protumoural effects induced by colibactin-producing bacteria. Gut 65 278–285. 10.1136/gutjnl-2014-307241 [DOI] [PubMed] [Google Scholar]
- Cristancho L., Johnson R. P., McEwen S. A., Gyles C. L. (2008). Escherichia coli O157:H7 and other Shiga toxin-producing E. coli in white veal calves. Vet. Microbiol. 126 200–209. 10.1016/j.vetmic.2007.06.012 [DOI] [PubMed] [Google Scholar]
- Crosa J. H. (1989). Genetics and molecular biology of siderophore-mediated iron transport in bacteria. Microbiol. Rev. 53 517–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross A. S., Gemski P., Sadoff J. C., Orskov F., Orskov I. (1984). The importance of the K1 capsule in invasive infections caused by Escherichia coli. J. Infect. Dis. 149 184–193. 10.1093/infdis/149.2.184 [DOI] [PubMed] [Google Scholar]
- Crossman L. C., Chaudhuri R. R., Beatson S. A., Wells T. J., Desvaux M., Cunningham A. F., et al. (2010). A commensal gone bad: complete genome sequence of the prototypical enterotoxigenic Escherichia coli strain H10407. J. Bacteriol. 192 5822–5831. 10.1128/JB.00710-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croxen M. A., Finlay B. B. (2010). Molecular mechanisms of Escherichia coli pathogenicity. Nat. Rev. Microbiol. 8 26–38. 10.1038/nrmicro2265 [DOI] [PubMed] [Google Scholar]
- Croxen M. A., Law R. J., Scholz R., Keeney K. M., Wlodarska M., Finlay B. B. (2013). Recent advances in understanding enteric pathogenic Escherichia coli. Clin. Microbiol. Rev. 26 822–880. 10.1128/CMR.00022-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuevas-Ramos G., Petit C. R., Marcq I., Boury M., Oswald E., Nougayrède J.-P. (2010). Escherichia coli induces DNA damage in vivo and triggers genomic instability in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 107 11537–11542. 10.1073/pnas.1001261107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czeczulin J. R., Whittam T. S., Henderson I. R., Navarro-Garcia F., Nataro J. P. (1999). Phylogenetic analysis of enteroaggregative and diffusely adherent Escherichia coli. Infect. Immun. 67 2692–2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daga A. P., Koga V. L., Soncini J. G. M., de Matos C. M., Perugini M. R. E., Pelisson M., et al. (2019). Escherichia coli bloodstream infections in patients at a university hospital: virulence factors and clinical characteristics. Front. Cell. Infect. Microbiol. 9:191. 10.3389/fcimb.2019.00191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darfeuille-Michaud A., Boudeau J., Bulois P., Neut C., Glasser A.-L., Barnich N., et al. (2004). High prevalence of adherent-invasive Escherichia coli associated with ileal mucosa in Crohn’s disease. Gastroenterology 127 412–421. 10.1053/j.gastro.2004.04.061 [DOI] [PubMed] [Google Scholar]
- Dautin N. (2010). Serine protease autotransporters of enterobacteriaceae (SPATEs): biogenesis and function. Toxins 2 1179–1206. 10.3390/toxins2061179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis J. M., Rasmussen S. B., O’Brien A. D. (2005). Cytotoxic necrotizing factor type 1 production by uropathogenic Escherichia coli modulates polymorphonuclear leukocyte function. Infect. Immun. 73 5301–5310. 10.1128/IAI.73.9.5301-5310.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean P., Kenny B. (2009). The effector repertoire of enteropathogenic E. coli: ganging up on the host cell. Curr. Opin. Microbiol. 12 101–109. 10.1016/j.mib.2008.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean P., Maresca M., Kenny B. (2005). EPEC’s weapons of mass subversion. Curr. Opin. Microbiol. 8 28–34. 10.1016/j.mib.2004.12.010 [DOI] [PubMed] [Google Scholar]
- Desvaux M., Hébraud M., Henderson I. R., Pallen M. J. (2006). Type III secretion: What’s in a name? Trends Microbiol. 14 157–160. 10.1016/j.tim.2006.02.009 [DOI] [PubMed] [Google Scholar]
- Dhakal B. K., Mulvey M. A. (2012). The UPEC pore-forming toxin α-hemolysin triggers proteolysis of host proteins to disrupt cell adhesion, inflammatory, and survival pathways. Cell Host Microbe 11 58–69. 10.1016/j.chom.2011.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diepold A., Wagner S. (2014). Assembly of the bacterial type III secretion machinery. FEMS Microbiol. Rev. 38 802–822. 10.1111/1574-6976.12061 [DOI] [PubMed] [Google Scholar]
- Dobrindt U., Hochhut B., Hentschel U., Hacker J. (2004). Genomic islands in pathogenic and environmental microorganisms. Nat. Rev. Microbiol. 2 414–424. 10.1038/nrmicro884 [DOI] [PubMed] [Google Scholar]
- Dobrindt U., Zdziarski J., Salvador E., Hacker J. (2010). Bacterial genome plasticity and its impact on adaptation during persistent infection. Int. J. Med. Microbiol. 300 363–366. 10.1016/j.ijmm.2010.04.010 [DOI] [PubMed] [Google Scholar]
- Doolittle R. F., Feng D. F., Tsang S., Cho G., Little E. (1996). Determining divergence times of the major kingdoms of living organisms with a protein clock. Science 271 470–477. 10.1126/science.271.5248.470 [DOI] [PubMed] [Google Scholar]
- Dozois C. M., Dho-Moulin M., Brée A., Fairbrother J. M., Desautels C., Curtiss R. (2000). Relationship between the Tsh autotransporter and pathogenicity of avian Escherichia coli and localization and analysis of the Tsh genetic region. Infect. Immun. 68 4145–4154. 10.1128/iai.68.7.4145-4154.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen J. A., Heidelberg J. F., White O., Salzberg S. L. (2000). Evidence for symmetric chromosomal inversions around the replication origin in bacteria. Genome Biol. 1:RESEARCH0011. 10.1186/gb-2000-1-6-research0011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ejrnæs K., Stegger M., Reisner A., Ferry S., Monsen T., Holm S. E., et al. (2011). Characteristics of Escherichia coli causing persistence or relapse of urinary tract infections: phylogenetic groups, virulence factors and biofilm formation. Virulence 2 528–537. 10.4161/viru.2.6.18189 [DOI] [PubMed] [Google Scholar]
- Elsinghorst E. A., Weitz J. A. (1994). Epithelial cell invasion and adherence directed by the enterotoxigenic Escherichia coli tib locus is associated with a 104-kilodalton outer membrane protein. Infect. Immun. 62 3463–3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engstrom M. D., Mobley H. L. T. (2016). Regulation of Expression of Uropathogenic Escherichia coli Nonfimbrial Adhesin TosA by PapB Homolog TosR in Conjunction with H-NS and Lrp. Infect. Immun. 84 811–821. 10.1128/IAI.01302-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escobar-Páramo P., Clermont O., Blanc-Potard A.-B., Bui H., Le Bouguénec C., Denamur E. (2004a). A specific genetic background is required for acquisition and expression of virulence factors in Escherichia coli. Mol. Biol. Evol. 21 1085–1094. 10.1093/molbev/msh118 [DOI] [PubMed] [Google Scholar]
- Escobar-Páramo P., Grenet K., Le Menac’h A., Rode L., Salgado E., Amorin C., et al. (2004b). Large-scale population structure of human commensal Escherichia coli isolates. Appl. Environ. Microbiol. 70 5698–5700. 10.1128/AEM.70.9.5698-5700.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eto D. S., Jones T. A., Sundsbak J. L., Mulvey M. A. (2007). Integrin-mediated host cell invasion by type 1-piliated uropathogenic Escherichia coli. PLoS Pathog. 3:e100. 10.1371/journal.ppat.0030100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eto D. S., Sundsbak J. L., Mulvey M. A. (2006). Actin-gated intracellular growth and resurgence of uropathogenic Escherichia coli. Cell. Microbiol. 8 704–717. 10.1111/j.1462-5822.2006.00691.x [DOI] [PubMed] [Google Scholar]
- Ewers C., Li G., Wilking H., Kiessling S., Alt K., Antáo E.-M., et al. (2007). Avian pathogenic, uropathogenic, and newborn meningitis-causing Escherichia coli: how closely related are they? Int. J. Med. Microbiol. 297 163–176. 10.1016/j.ijmm.2007.01.003 [DOI] [PubMed] [Google Scholar]
- Fagan R. P., Smith S. G. J. (2007). The Hek outer membrane protein of Escherichia coli is an auto-aggregating adhesin and invasin. FEMS Microbiol. Lett. 269 248–255. 10.1111/j.1574-6968.2006.00628.x [DOI] [PubMed] [Google Scholar]
- Fasano A., Noriega F. R., Maneval D. R., Chanasongcram S., Russell R., Guandalini S., et al. (1995). Shigella enterotoxin 1: an enterotoxin of Shigella flexneri 2a active in rabbit small intestine in vivo and in vitro. J. Clin. Invest. 95 2853–2861. 10.1172/JCI117991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiorentini C., Arancia G., Caprioli A., Falbo V., Ruggeri F. M., Donelli G. (1988). Cytoskeletal changes induced in HEp-2 cells by the cytotoxic necrotizing factor of Escherichia coli. Toxicon 26 1047–1056. 10.1016/0041-0101(88)90203-6 [DOI] [PubMed] [Google Scholar]
- Firoozeh F., Saffari M., Neamati F., Zibaei M. (2014). Detection of virulence genes in Escherichia coli isolated from patients with cystitis and pyelonephritis. Int. J. Infect. Dis. 29 219–222. 10.1016/j.ijid.2014.03.1393 [DOI] [PubMed] [Google Scholar]
- Fischbach M. A., Lin H., Zhou L., Yu Y., Abergel R. J., Liu D. R., et al. (2006). The pathogen-associated iroA gene cluster mediates bacterial evasion of lipocalin 2. Proc. Natl. Acad. Sci. U.S.A. 103 16502–16507. 10.1073/pnas.0604636103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer H., Yamamoto M., Akira S., Beutler B., Svanborg C. (2006). Mechanism of pathogen-specific TLR4 activation in the mucosa: fimbriae, recognition receptors and adaptor protein selection. Eur. J. Immunol. 36 267–277. 10.1002/eji.200535149 [DOI] [PubMed] [Google Scholar]
- Fisher C. R., Davies N. M. L. L., Wyckoff E. E., Feng Z., Oaks E. V., Payne S. M. (2009). Genetics and virulence association of the Shigella flexneri sit iron transport system. Infect. Immun. 77 1992–1999. 10.1128/IAI.00064-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flatau G., Lemichez E., Gauthier M., Chardin P., Paris S., Fiorentini C., et al. (1997). Toxin-induced activation of the G protein p21 Rho by deamidation of glutamine. Nature 387 729–733. 10.1038/42743 [DOI] [PubMed] [Google Scholar]
- Fleckenstein J. M., Holland J. T., Hasty D. L. (2002). Interaction of an uuter membrane protein of enterotoxigenic Escherichia coli with cell surface heparan sulfate proteoglycans. Infect. Immun. 70 1530–1537. 10.1128/iai.70.3.1530-1537.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleckenstein J. M., Kopecko D. J., Warren R. L., Elsinghorst E. A. (1996). Molecular characterization of the tia invasion locus from enterotoxigenic Escherichia coli. Infect. Immun. 64 2256–2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleckenstein J. M., Lindler L. E., Elsinghorst E. A., Dale J. B. (2000). Identification of a gene within a pathogenicity island of enterotoxigenic Escherichia coli H10407 required for maximal secretion of the heat-labile enterotoxin. Infect. Immun. 68 2766–2774. 10.1128/iai.68.5.2766-2774.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores-Mireles A. L., Walker J. N., Caparon M., Hultgren S. J. (2015). Urinary tract infections: epidemiology, mechanisms of infection and treatment options. Nat. Rev. Microbiol. 13 269–284. 10.1038/nrmicro3432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogg P. C. M., Rigden D. J., Saunders J. R., McCarthy A. J., Allison H. E. (2011). Characterization of the relationship between integrase, excisionase and antirepressor activities associated with a superinfecting Shiga toxin encoding bacteriophage. Nucleic Acids Res. 39 2116–2129. 10.1093/nar/gkq923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foxman B. (2003). Epidemiology of urinary tract infections: incidence, morbidity, and economic costs. Dis. Mon. 49 53–70. 10.1067/mda.2003.7 [DOI] [PubMed] [Google Scholar]
- Franzin F. M., Sircili M. P. (2015). Locus of enterocyte effacement: a pathogenicity island involved in the virulence of enteropathogenic and enterohemorragic Escherichia coli subjected to a complex network of gene regulation. Biomed Res. Int. 2015:534738. 10.1155/2015/534738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frendéus B., Wachtler C., Hedlund M., Fischer H., Samuelsson P., Svensson M., et al. (2001). Escherichia coli P fimbriae utilize the Toll-like receptor 4 pathway for cell activation. Mol. Microbiol. 40 37–51. 10.1046/j.1365-2958.2001.02361.x [DOI] [PubMed] [Google Scholar]
- Fujii Y., Nomura T., Yamanaka H., Okamoto K. (1997). Involvement of Ca(2+)-calmodulin-dependent protein kinase II in the intestinal secretory action of Escherichia coli heat-stable enterotoxin II. Microbiol. Immunol. 41 633–636. 10.1111/j.1348-0421.1997.tb01904.x [DOI] [PubMed] [Google Scholar]
- Fukuta S., Magnani J. L., Twiddy E. M., Holmes R. K., Ginsburg V. (1988). Comparison of the carbohydrate-binding specificities of cholera toxin and Escherichia coli heat-labile enterotoxins LTh-I, LT-IIa, and LT-IIb. Infect. Immun. 56 1748–1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furniss R. C. D., Clements A. (2018). Regulation of the locus of enterocyte effacement in attaching and effacing pathogens. J. Bacteriol. 200:e00336-17. 10.1128/JB.00336-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galmiche A., Boquet P. (2001). Toxines bactériennes?: facteurs de virulence et outils de biologie cellulaire. Med. Sci. 17 691–700. 10.4267/10608/1993 7902565 [DOI] [Google Scholar]
- Gärtner J. F., Schmidt M. A. (2004). Comparative analysis of locus of enterocyte effacement pathogenicity islands of atypical enteropathogenic Escherichia coli. Infect. Immun. 72 6722–6728. 10.1128/IAI.72.11.6722-6728.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genuini M., Bidet P., Benoist J.-F., Schlemmer D., Lemaitre C., Birgy A., et al. (2019). ShiF acts as an auxiliary factor of aerobactin secretion in meningitis Escherichia coli strain S88. BMC Microbiol. 19:298. 10.1186/s12866-019-1677-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gérardin J., Lalioui L., Jacquemin E., Le Bouguénec C., Mainil J. G. (2000). The afa-related gene cluster in necrotoxigenic and other Escherichia coli from animals belongs to the afa-8 variant. Vet. Microbiol. 76 175–184. 10.1016/s0378-1135(00)00234-0 [DOI] [PubMed] [Google Scholar]
- Germon P., Roche D., Melo S., Mignon-Grasteau S., Dobrindt U., Hacker J., et al. (2007). tDNA locus polymorphism and ecto-chromosomal DNA insertion hot-spots are related to the phylogenetic group of Escherichia coli strains. Microbiology 153 826–837. 10.1099/mic.0.2006/001958-0 [DOI] [PubMed] [Google Scholar]
- Gibold L., Garenaux E., Dalmasso G., Gallucci C., Cia D., Mottet-Auselo B., et al. (2016). The Vat-AIEC protease promotes crossing of the intestinal mucus layer by Crohn’s disease-associated Escherichia coli. Cell. Microbiol. 18 617–631. 10.1111/cmi.12539 [DOI] [PubMed] [Google Scholar]
- Girardeau J. P., Bertin Y., Martin C. (2009). Genomic analysis of the PAI ICL3 locus in pathogenic LEE-negative Shiga toxin-producing Escherichia coli and Citrobacter rodentium. Microbiology 155 1016–1027. 10.1099/mic.0.026807-0 [DOI] [PubMed] [Google Scholar]
- Girardeau J. P., Dalmasso A., Bertin Y., Ducrot C., Bord S., Livrelli V., et al. (2005). Association of virulence genotype with phylogenetic background in comparison to different seropathotypes of Shiga toxin-producing Escherichia coli isolates. J. Clin. Microbiol. 43 6098–6107. 10.1128/JCM.43.12.6098-6107.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girardeau J. P., Lalioui L., Said A. M. O., De Champs C., Le Bouguénec C. (2003). Extended virulence genotype of pathogenic Escherichia coli isolates carrying the afa-8 operon: evidence of similarities between isolates from humans and animals with extraintestinal infections. J. Clin. Microbiol. 41 218–226. 10.1128/jcm.41.1.218-226.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golan L., Gonen E., Yagel S., Rosenshine I., Shpigel N. Y. (2011). Enterohemorrhagic Escherichia coli induce attaching and effacing lesions and hemorrhagic colitis in human and bovine intestinal xenograft models. Dis. Model. Mech. 4 86–94. 10.1242/dmm.005777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gransden W. R., Eykyn S. J., Phillips I., Rowe B. (1990). Bacteremia due to Escherichia coli: a study of 861 episodes. Rev. Infect. Dis. 12 1008–1018. 10.1093/clinids/12.6.1008 [DOI] [PubMed] [Google Scholar]
- Groisman E. A., Ochman H. (1996). Pathogenicity islands: bacterial evolution in quantum leaps. Cell 87 791–794. 10.1016/s0092-8674(00)81985-6 [DOI] [PubMed] [Google Scholar]
- Grönberg-Hernández J., Hernández J. G., Sundén F., Connolly J., Svanborg C., Wullt B. (2011). Genetic control of the variable innate immune response to asymptomatic bacteriuria. PLoS One 6:e28289. 10.1371/journal.pone.0028289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunther N. W., Snyder J. A., Lockatell V., Blomfield I., Johnson D. E., Mobley H. L. T. (2002). Assessment of virulence of uropathogenic Escherichia coli type 1 fimbrial mutants in which the invertible element is phase-locked on or off. Infect. Immun. 70 3344–3354. 10.1128/iai.70.7.3344-3354.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guyer D. M., Henderson I. R., Nataro J. P., Mobley H. L. (2000). Identification of sat, an autotransporter toxin produced by uropathogenic Escherichia coli. Mol. Microbiol. 38 53–66. 10.1046/j.1365-2958.2000.02110.x [DOI] [PubMed] [Google Scholar]
- Guyer D. M., Radulovic S., Jones F.-E., Mobley H. L. T. (2002). Sat, the secreted autotransporter toxin of uropathogenic Escherichia coli, is a vacuolating cytotoxin for bladder and kidney epithelial cells. Infect. Immun. 70 4539–4546. 10.1128/iai.70.8.4539-4546.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habouria H., Pokharel P., Maris S., Garénaux A., Bessaiah H., Houle S., et al. (2019). Three new serine-protease autotransporters of Enterobacteriaceae (SPATEs) from extra-intestinal pathogenic Escherichia coli and combined role of SPATEs for cytotoxicity and colonization of the mouse kidney. Virulence 10 568–587. 10.1080/21505594.2019.1624102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacker J., Bender L., Ott M., Wingender J., Lund B., Marre R., et al. (1990). Deletions of chromosomal regions coding for fimbriae and hemolysins occur in vitro and in vivo in various extraintestinal Escherichia coli isolates. Microb. Pathog. 8 213–225. 10.1016/0882-4010(90)90048-u [DOI] [PubMed] [Google Scholar]
- Hacker J., Carniel E. (2001). Ecological fitness, genomic islands and bacterial pathogenicity. A Darwinian view of the evolution of microbes. EMBO Rep. 2 376–381. 10.1093/embo-reports/kve097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacker J., Knapp S., Goebel W. (1983). Spontaneous deletions and flanking regions of the chromosomally inherited hemolysin determinant of an Escherichia coli O6 strain. J. Bacteriol. 154 1145–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagan E. C., Lloyd A. L., Rasko D. A., Faerber G. J., Mobley H. L. T. (2010). Escherichia coli global gene expression in urine from women with urinary tract infection. PLoS Pathog. 6:e1001187. 10.1371/journal.ppat.1001187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagan E. C., Mobley H. L. T. (2009). Haem acquisition is facilitated by a novel receptor Hma and required by uropathogenic Escherichia coli for kidney infection. Mol. Microbiol. 71 79–91. 10.1111/j.1365-2958.2008.06509.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Wang D. C. (2014). An accurate genomic island prediction method for sequenced bacterial and archaeal genomes. J. Proteomics Bioinform. 07 214–221. 10.4172/jpb.1000322 [DOI] [Google Scholar]
- Hannan T. J., Mysorekar I. U., Chen S. L., Walker J. N., Jones J. M., Pinkner J. S., et al. (2008). LeuX tRNA-dependent and -independent mechanisms of Escherichia coli pathogenesis in acute cystitis. Mol. Microbiol. 67 116–128. 10.1111/j.1365-2958.2007.06025.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser E., Mellmann A., Semmler T., Stoeber H., Wieler L. H., Karch H., et al. (2013). Phylogenetic and molecular analysis of food-borne shiga toxin-producing Escherichia coli. Appl. Environ. Microbiol. 79 2731–2740. 10.1128/AEM.03552-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T., Makino K., Ohnishi M., Kurokawa K., Ishii K., Yokoyama K., et al. (2001). Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 8 11–22. 10.1093/dnares/8.1.11 [DOI] [PubMed] [Google Scholar]
- Henderson I. R., Czeczulin J., Eslava C., Noriega F., Nataro J. P. (1999). Characterization of pic, a secreted protease of Shigella flexneri and enteroaggregative Escherichia coli. Infect. Immun. 67 5587– 5596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson I. R., Navarro-Garcia F., Nataro J. P. (1998). The great escape: structure and function of the autotransporter proteins. Trends Microbiol. 6 370–378. 10.1016/s0966-842x(98)01318-3 [DOI] [PubMed] [Google Scholar]
- Henderson J. P., Crowley J. R., Pinkner J. S., Walker J. N., Tsukayama P., Stamm W. E., et al. (2009). Quantitative metabolomics reveals an epigenetic blueprint for iron acquisition in uropathogenic Escherichia coli. PLoS Pathog. 5:e1000305. 10.1371/journal.ppat.1000305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendriks A. C. A., Reubsaet F. A. G., Kooistra-Smid A. M. D. M., Rossen J. W. A., Dutilh B. E., Zomer A. L., et al. (2020). Genome-wide association studies of Shigella spp. and Enteroinvasive Escherichia coli isolates demonstrate an absence of genetic markers for prediction of disease severity. BMC Genomics 21:138. 10.1186/s12864-020-6555-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henríquez T., Salazar J. C., Marvasi M., Shah A., Corsini G., Toro C. S. (2020). SRL pathogenicity island contributes to the metabolism of D-aspartate via an aspartate racemase in Shigella flexneri YSH6000. PLoS One 15:e0228178. 10.1371/journal.pone.0228178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandes R. T., Elias W. P., Vieira M. A. M., Gomes T. A. T. (2009). An overview of atypical enteropathogenic Escherichia coli. FEMS Microbiol. Lett. 297 137–149. 10.1111/j.1574-6968.2009.01664.x [DOI] [PubMed] [Google Scholar]
- Herzer P. J., Inouye S., Inouye M., Whittam T. S. (1990). Phylogenetic distribution of branched RNA-linked multicopy single-stranded DNA among natural isolates of Escherichia coli. J. Bacteriol. 172 6175–6181. 10.1128/jb.172.11.6175-6181.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill C. (2012). Virulence or niche factors: what’s in a name? J. Bacteriol. 194 5725–5727. 10.1128/JB.00980-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochhut B., Wilde C., Balling G., Middendorf B., Dobrindt U., Brzuszkiewicz E., et al. (2006). Role of pathogenicity island-associated integrases in the genome plasticity of uropathogenic Escherichia coli strain 536. Mol. Microbiol. 61 584–595. 10.1111/j.1365-2958.2006.05255.x [DOI] [PubMed] [Google Scholar]
- Hoffman J. A., Badger J. L., Zhang Y., Huang S. H., Kim K. S. (2000). Escherichia coli K1 aslA contributes to invasion of brain microvascular endothelial cells in vitro and in vivo. Infect. Immun. 68 5062–5067. 10.1128/iai.68.9.5062-5067.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong M., Payne S. M. (1997). Effect of mutations in Shigella flexneri chromosomal and plasmid-encoded lipopolysaccharide genes on invasion and serum resistance. Mol. Microbiol. 24 779–791. 10.1046/j.1365-2958.1997.3731744.x [DOI] [PubMed] [Google Scholar]
- Horstman A. L., Kuehn M. J. (2002). Bacterial surface association of heat-labile enterotoxin through lipopolysaccharide after secretion via the general secretory pathway. J. Biol. Chem. 277 32538–32545. 10.1074/jbc.M203740200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hryckowian A. J., Baisa G. A., Schwartz K. J., Welch R. A. (2015). dsdA does not affect colonization of the murine urinary tract by Escherichia coli CFT073. PLoS One 10:e0138121. 10.1371/journal.pone.0138121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J., Kan B., Liu Z.-H., Yu S.-Y. (2005). Enteroaggregative Escherichia coli isolated from Chinese diarrhea patients with high-pathogenicity island of Yersinia is involved in synthesis of siderophore yersiniabactin. World J. Gastroenterol. 11 5816–5820. 10.3748/wjg.v11.i37.5816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J., Torres A. G. (2015). Enteropathogenic Escherichia coli: foe or innocent bystander? Clin. Microbiol. Infect. 21 729–734. 10.1016/j.cmi.2015.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huan P. T., Bastin D. A., Whittle B. L., Lindberg A. A., Verma N. K. (1997). Molecular characterization of the genes involved in O-antigen modification, attachment, integration and excision in Shigella flexneri bacteriophage SfV. Gene 195 217–227. 10.1016/s0378-1119(97)00143-1 [DOI] [PubMed] [Google Scholar]
- Huang S. H., Chen Y. H., Fu Q., Stins M., Wang Y., Wass C., et al. (1999). Identification and characterization of an Escherichia coli invasion gene locus, ibeB, required for penetration of brain microvascular endothelial cells. Infect. Immun. 67 2103–2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S. H., Wan Z. S., Chen Y. H., Jong A. Y., Kim K. S. (2001). Further characterization of Escherichia coli brain microvascular endothelial cell invasion gene ibeA by deletion, complementation, and protein expression. J. Infect. Dis. 183 1071–1078. 10.1086/319290 [DOI] [PubMed] [Google Scholar]
- Huang S. H., Wass C., Fu Q., Prasadarao N. V., Stins M., Kim K. S. (1995). Escherichia coli invasion of brain microvascular endothelial cells in vitro and in vivo: molecular cloning and characterization of invasion gene ibe10. Infect. Immun. 63 4470–4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson C. M., Lau B. Y., Williams K. P. (2015). Islander: a database of precisely mapped genomic islands in tRNA and tmRNA genes. Nucleic Acids Res. 43 D48–D53. 10.1093/nar/gku1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull R. A., Rudy D. C., Donovan W. H., Wieser I. E., Stewart C., Darouiche R. O. (1999). Virulence properties of Escherichia coli 83972, a prototype strain associated with asymptomatic bacteriuria. Infect. Immun. 67 429–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hultdin U. W., Lindberg S., Grundström C., Huang S., Uhlin B. E., Sauer-Eriksson A. E. (2010). Structure of FocB–a member of a family of transcription factors regulating fimbrial adhesin expression in uropathogenic Escherichia coli. FEBS J. 277 3368–3381. 10.1111/j.1742-4658.2010.07742.x [DOI] [PubMed] [Google Scholar]
- Iguchi A., Thomson N. R., Ogura Y., Saunders D., Ooka T., Henderson I. R., et al. (2009). Complete genome sequence and comparative genome analysis of enteropathogenic Escherichia coli O127:H6 strain E2348/69. J. Bacteriol. 191 347–354. 10.1128/JB.01238-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamovic L., Tozzoli R., Michelacci V., Minelli F., Marziano M. L., Caprioli A., et al. (2010). OI-57, a genomic island of Escherichia coli O157, is present in other seropathotypes of Shiga toxin-producing E. coli associated with severe human disease. Infect. Immun. 78 4697–4704. 10.1128/IAI.00512-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingersoll M., Groisman E. A., Zychlinsky A. (2002). Pathogenicity islands of Shigella. Curr. Top. Microbiol. Immunol. 264 49–65. [PubMed] [Google Scholar]
- Ingersoll M. A., Moss J. E., Weinrauch Y., Fisher P. E., Groisman E. A., Zychlinsky A. (2003). The ShiA protein encoded by the Shigella flexneri SHI-2 pathogenicity island attenuates inflammation. Cell. Microbiol. 5 797–807. 10.1046/j.1462-5822.2003.00320.x [DOI] [PubMed] [Google Scholar]
- Ingersoll M. A., Zychlinsky A. (2006). ShiA abrogates the innate T-cell response to Shigella flexneri infection. Infect. Immun. 74 2317–2327. 10.1128/IAI.74.4.2317-2327.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isidean S. D., Riddle M. S., Savarino S. J., Porter C. K. (2011). A systematic review of ETEC epidemiology focusing on colonization factor and toxin expression. Vaccine 29 6167–6178. 10.1016/j.vaccine.2011.06.084 [DOI] [PubMed] [Google Scholar]
- Jenkins C. (2018). Enteroaggregative Escherichia coli. Curr. Top. Microbiol. Immunol. 416 27–50. 10.1007/82_2018_105 [DOI] [PubMed] [Google Scholar]
- Jobling M. G., Holmes R. K. (2006). Heat-Labile Enterotoxins. EcoSal Plus 2 1–28. 10.1128/ecosalplus.8.7.5 [DOI] [PubMed] [Google Scholar]
- Johnson J. R. (1991). Virulence factors in Escherichia coli urinary tract infection. Clin. Microbiol. Rev. 4 80–128. 10.1128/cmr.4.1.80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson J. R., Johnston B., Kuskowski M. A., Nougayrede J.-P., Oswald E. (2008). Molecular epidemiology and phylogenetic distribution of the Escherichia coli pks genomic island. J. Clin. Microbiol. 46 3906–3911. 10.1128/JCM.00949-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson J. R., Kuskowski M. (2000). Clonal origin, virulence factors, and virulence. Infect. Immun. 68 424–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson J. R., Kuskowski M. A., Gajewski A., Sahm D. F., Karlowsky J. A. (2004). Virulence characteristics and phylogenetic background of multidrug-resistant and antimicrobial-susceptible clinical isolates of Escherichia coli from across the United States, 2000-2001. J. Infect. Dis. 190 1739–1744. 10.1086/425018 [DOI] [PubMed] [Google Scholar]
- Johnson J. R., Oswald E., O’Bryan T. T., Kuskowski M. A., Spanjaard L. (2002). Phylogenetic distribution of virulence-associated genes among Escherichia coli isolates associated with neonatal bacterial meningitis in the Netherlands. J. Infect. Dis. 185 774–784. 10.1086/339343 [DOI] [PubMed] [Google Scholar]
- Johnson T. J., Kariyawasam S., Wannemuehler Y., Mangiamele P., Johnson S. J., Doetkott C., et al. (2007). The genome sequence of avian pathogenic Escherichia coli strain O1:K1:H7 shares strong similarities with human extraintestinal pathogenic E. coli genomes. J. Bacteriol. 189 3228–3236. 10.1128/JB.01726-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson T. J., Logue C. M., Johnson J. R., Kuskowski M. A., Sherwood J. S., Barnes H. J., et al. (2012). Associations between multidrug resistance, plasmid content, and virulence potential among extraintestinal pathogenic and commensal Escherichia coli from humans and poultry. Foodborne Pathog. Dis. 9 37–46. 10.1089/fpd.2011.0961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jores J., Rumer L., Wieler L. H. (2004). Impact of the locus of enterocyte effacement pathogenicity island on the evolution of pathogenic Escherichia coli. Int. J. Med. Microbiol. 294 103–113. 10.1016/j.ijmm.2004.06.024 [DOI] [PubMed] [Google Scholar]
- Jores J., Wagner S., Rumer L., Eichberg J., Laturnus C., Kirsch P., et al. (2005). Description of a 111-kb pathogenicity island (PAI) encoding various virulence features in the enterohemorrhagic E. coli (EHEC) strain RW1374 (O103:H2) and detection of a similar PAI in other EHEC strains of serotype 0103:H2. Int. J. Med. Microbiol. 294 417–425. 10.1016/j.ijmm.2004.09.009 [DOI] [PubMed] [Google Scholar]
- Ju W., Shen J., Toro M., Zhao S., Meng J. (2013). Distribution of pathogenicity islands OI-122, OI-43/48, and OI-57 and a high-pathogenicity island in Shiga toxin-producing Escherichia coli. Appl. Environ. Microbiol. 79 3406–3412. 10.1128/AEM.03661-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Källenius G., Möllby R., Svenson S. B., Winberg J., Hultberg H. (1980). Identification of a carbohydrate receptor recognized by uropathogenic Escherichia coli. Infection 8(Suppl. 3), 288–293. 10.1007/bf01639597 [DOI] [PubMed] [Google Scholar]
- Kampmeier S., Berger M., Mellmann A., Karch H., Berger P. (2018). The 2011 German enterohemorrhagic Escherichia Coli O104:H4 outbreak-the danger is still out there. Curr. Top. Microbiol. Immunol. 416 117–148. 10.1007/82_2018_107 [DOI] [PubMed] [Google Scholar]
- Kaper J. B., Nataro J. P., Mobley H. L. T. (2004). Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2 123–140. 10.1038/nrmicro818 [DOI] [PubMed] [Google Scholar]
- Karch H., Schubert S., Zhang D., Zhang W., Schmidt H., Olschläger T., et al. (1999). A genomic island, termed high-pathogenicity island, is present in certain non-O157 Shiga toxin-producing Escherichia coli clonal lineages. Infect. Immun. 67 5994–6001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlin S. (2001). Detecting anomalous gene clusters and pathogenicity islands in diverse bacterial genomes. Trends Microbiol. 9 335–343. 10.1016/s0966-842x(01)02079-0 [DOI] [PubMed] [Google Scholar]
- Karmali M. A., Mascarenhas M., Shen S., Ziebell K., Johnson S., Reid-Smith R., et al. (2003). Association of genomic O island 122 of Escherichia coli EDL 933 with verocytotoxin-producing Escherichia coli seropathotypes that are linked to epidemic and/or serious disease. J. Clin. Microbiol. 41 4930–4940. 10.1128/jcm.41.11.4930-4940.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karmali M. A., Steele B. T., Petric M., Lim C. (1983). Sporadic cases of haemolytic-uraemic syndrome associated with faecal cytotoxin and cytotoxin-producing Escherichia coli in stools. Lancet 1 619–620. 10.1016/s0140-6736(83)91795-6 [DOI] [PubMed] [Google Scholar]
- Karr J. F., Nowicki B., Truong L. D., Hull R. A., Hull S. I. (1989). Purified P fimbriae from two cloned gene clusters of a single pyelonephritogenic strain adhere to unique structures in the human kidney. Infect. Immun. 57 3594–3600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehres D. G., Janakiraman A., Slauch J. M., Maguire M. E. (2002). SitABCD is the alkaline Mn(2+) transporter of Salmonella enterica serovar Typhimurium. J. Bacteriol. 184 3159–3166. 10.1128/jb.184.12.3159-3166.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keith B. R., Maurer L., Spears P. A., Orndorff P. E. (1986). Receptor-binding function of type 1 pili effects bladder colonization by a clinical isolate of Escherichia coli. Infect. Immun. 53 693–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendall M. M. (2016). Interkingdom Chemical Signaling in Enterohemorrhagic Escherichia coli O157:H7. Adv. Exp. Med. Biol. 874 201–213. 10.1007/978-3-319-20215-0_9 [DOI] [PubMed] [Google Scholar]
- Khan N. A., Wang Y., Kim K. J., Chung J. W., Wass C. A., Kim K. S. (2002). Cytotoxic necrotizing factor-1 contributes to Escherichia coli K1 invasion of the central nervous system. J. Biol. Chem. 277 15607–15612. 10.1074/jbc.M112224200 [DOI] [PubMed] [Google Scholar]
- Kim K. J., Elliott S. J., Di Cello F., Stins M. F., Kim K. S. (2003). The K1 capsule modulates trafficking of E. coli-containing vacuoles and enhances intracellular bacterial survival in human brain microvascular endothelial cells. Cell. Microbiol. 5 245–252. 10.1046/j.1462-5822.2003.t01-1-00271.x [DOI] [PubMed] [Google Scholar]
- Kim K. S. (2002). Strategy of Escherichia coli for crossing the blood-brain barrier. J. Infect. Dis. 186(Suppl. 2), S220–S224. 10.1086/344284 [DOI] [PubMed] [Google Scholar]
- Kim K. S. (2003). Pathogenesis of bacterial meningitis: from bacteraemia to neuronal injury. Nat. Rev. Neurosci. 4 376–385. 10.1038/nrn1103 [DOI] [PubMed] [Google Scholar]
- Kim K. S. (2008). Mechanisms of microbial traversal of the blood-brain barrier. Nat. Rev. Microbiol. 6 625–634. 10.1038/nrmicro1952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K. S. (2016). Human meningitis-associated Escherichia coli. EcoSal Plus 7 1–15. 10.1128/ecosalplus.ESP-0015-2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K. S., Itabashi H., Gemski P., Sadoff J., Warren R. L., Cross A. S. (1992). The K1 capsule is the critical determinant in the development of Escherichia coli meningitis in the rat. J. Clin. Invest. 90 897–905. 10.1172/JCI115965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirsch P., Jores J., Wieler L. H. (2004). [Plasticity of bacterial genomes: pathogenicity islands and the locus of enterocyte effacement (LEE)]. Berl. Munch. Tierarztl. Wochenschr. 117 116–129. [PubMed] [Google Scholar]
- Klapproth J. M., Scaletsky I. C., McNamara B. P., Lai L. C., Malstrom C., James S. P., et al. (2000). A large toxin from pathogenic Escherichia coli strains that inhibits lymphocyte activation. Infect. Immun. 68 2148–2155. 10.1128/iai.68.4.2148-2155.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemm P. (1986). Two regulatory fim genes, fimB and fimE, control the phase variation of type 1 fimbriae in Escherichia coli. EMBO J. 5 1389–1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemm P., Roos V., Ulett G. C., Svanborg C., Schembri M. A. (2006). Molecular characterization of the Escherichia coli asymptomatic bacteriuria strain 83972: the taming of a pathogen. Infect. Immun. 74 781–785. 10.1128/IAI.74.1.781-785.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konczy P., Ziebell K., Mascarenhas M., Choi A., Michaud C., Kropinski A. M., et al. (2008). Genomic O island 122, locus for enterocyte effacement, and the evolution of virulent verocytotoxin-producing Escherichia coli. J. Bacteriol. 190 5832–5840. 10.1128/JB.00480-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korhonen T. K., Valtonen M. V., Parkkinen J., Väisänen-Rhen V., Finne J., Orskov F., et al. (1985). Serotypes, hemolysin production, and receptor recognition of Escherichia coli strains associated with neonatal sepsis and meningitis. Infect. Immun. 48 486–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostakioti M., Stathopoulos C. (2004). Functional analysis of the Tsh autotransporter from an avian pathogenic Escherichia coli strain. Infect. Immun. 72 5548–5554. 10.1128/IAI.72.10.5548-5554.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kothapalli S., Nair S., Alokam S., Pang T., Khakhria R., Woodward D., et al. (2005). Diversity of genome structure in Salmonella enterica serovar Typhi populations. J. Bacteriol. 187 2638–2650. 10.1128/JB.187.8.2638-2650.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreft B., Placzek M., Doehn C., Hacker J., Schmidt G., Wasenauer G., et al. (1995). S fimbriae of uropathogenic Escherichia coli bind to primary human renal proximal tubular epithelial cells but do not induce expression of intercellular adhesion molecule 1. Infect. Immun. 63 3235–3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieger J. N., Dobrindt U., Riley D. E., Oswald E. (2011). Acute Escherichia coli prostatitis in previously health young men: bacterial virulence factors, antimicrobial resistance, and clinical outcomes. Urology 77 1420–1425. 10.1016/j.urology.2010.12.059 [DOI] [PubMed] [Google Scholar]
- Kumashiro S., Hashimoto A., Nishikawa T. (1995). Free D-serine in post-mortem brains and spinal cords of individuals with and without neuropsychiatric diseases. Brain Res. 681 117–125. 10.1016/0006-8993(95)00307-c [DOI] [PubMed] [Google Scholar]
- Kunin C. M., Halmagyi N. E. (1962). Urinary-tract infections in schoolchildren. II. Characterization of invading organisms. N. Engl. J. Med. 266 1297–1301. 10.1056/NEJM196206212662502 [DOI] [PubMed] [Google Scholar]
- Lai Y., Rosenshine I., Leong J. M., Frankel G. (2013). Intimate host attachment: enteropathogenic and enterohaemorrhagic Escherichia coli. Cell. Microbiol. 15 1796–1808. 10.1111/cmi.12179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laing C. R., Buchanan C., Taboada E. N., Zhang Y., Karmali M. A., Thomas J. E., et al. (2009). In silico genomic analyses reveal three distinct lineages of Escherichia coli O157:H7, one of which is associated with hyper-virulence. BMC Genomics 10:287. 10.1186/1471-2164-10-287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalioui L., Jouve M., Gounon P., Le Bouguenec C. (1999). Molecular cloning and characterization of the afa-7 and afa-8 gene clusters encoding afimbrial adhesins in Escherichia coli strains associated with diarrhea or septicemia in calves. Infect. Immun. 67 5048–5059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalioui L., Le Bouguénec C. (2001). afa-8 Gene cluster is carried by a pathogenicity island inserted into the tRNA(Phe) of human and bovine pathogenic Escherichia coli isolates. Infect. Immun. 69 937–948. 10.1128/IAI.69.2.937-948.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan R., Alles M. C., Donohoe K., Martinez M. B., Reeves P. R. (2004). Molecular evolutionary relationships of enteroinvasive Escherichia coli and Shigella spp. Infect. Immun. 72 5080–5088. 10.1128/IAI.72.9.5080-5088.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan R., Lumb B., Ryan D., Reeves P. R. (2001). Molecular evolution of large virulence plasmid in Shigella clones and enteroinvasive Escherichia coli. Infect. Immun. 69 6303–6309. 10.1128/IAI.69.10.6303-6309.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan R., Reeves P. R. (2002). Escherichia coli in disguise: molecular origins of Shigella. Microbes Infect. 4 1125–1132. 10.1016/s1286-4579(02)01637-4 [DOI] [PubMed] [Google Scholar]
- Lan R., Stevenson G., Reeves P. R. (2003). Comparison of two major forms of the Shigella virulence plasmid pINV: positive selection is a major force driving the divergence. Infect. Immun. 71 6298–6306. 10.1128/iai.71.11.6298-6306.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane M. C., Mobley H. L. T. (2007). Role of P-fimbrial-mediated adherence in pyelonephritis and persistence of uropathogenic Escherichia coli (UPEC) in the mammalian kidney. Kidney Int. 72 19–25. 10.1038/sj.ki.5002230 [DOI] [PubMed] [Google Scholar]
- Langille M. G. I., Hsiao W. W. L., Brinkman F. S. L. (2010). Detecting genomic islands using bioinformatics approaches. Nat. Rev. Microbiol. 8 373–382. 10.1038/nrmicro2350 [DOI] [PubMed] [Google Scholar]
- Lara-Tejero M., Galán J. E. (2019). The injectisome, a complex nanomachine for protein injection into mammalian cells. EcoSal Plus 8:10.1128/ecosalplus.ESP-0039-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latif H., Li H. J., Charusanti P., Palsson B. Ø., Aziz R. K. (2014). A Gapless, Unambiguous Genome Sequence of the Enterohemorrhagic Escherichia coli O157:H7 Strain EDL933. Genome Announc. 2:e00821-14. 10.1128/genomeA.00821-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawlor K. M., Payne S. M. (1984). Aerobactin genes in Shigella spp. J. Bacteriol. 160 266–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence J. G., Ochman H. (1997). Amelioration of bacterial genomes: rates of change and exchange. J. Mol. Evol. 44 383–397. 10.1007/pl00006158 [DOI] [PubMed] [Google Scholar]
- Le Bouguénec C., Lalioui L., du Merle L., Jouve M., Courcoux P., Bouzari S., et al. (2001). Characterization of AfaE adhesins produced by extraintestinal and intestinal human Escherichia coli isolates: PCR assays for detection of Afa adhesins that do or do not recognize Dr blood group antigens. J. Clin. Microbiol. 39 1738–1745. 10.1128/JCM.39.5.1738-1745.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bouguénec C., Servin A. L. (2006). Diffusely adherent Escherichia coli strains expressing Afa/Dr adhesins (Afa/Dr DAEC): hitherto unrecognized pathogens. FEMS Microbiol. Lett. 256 185–194. 10.1111/j.1574-6968.2006.00144.x [DOI] [PubMed] [Google Scholar]
- Le Gall T., Clermont O., Gouriou S., Picard B., Nassif X., Denamur E., et al. (2007). Extraintestinal virulence is a coincidental by-product of commensalism in B2 phylogenetic group Escherichia coli strains. Mol. Biol. Evol. 24 2373–2384. 10.1093/molbev/msm172 [DOI] [PubMed] [Google Scholar]
- Lee E., Lee Y. (2018). Prevalence of Escherichia coli carrying pks islands in bacteremia patients. Ann. Lab. Med. 38 271–273. 10.3343/alm.2018.38.3.271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehti T. A., Heikkinen J., Korhonen T. K., Westerlund-Wikström B. (2012). The response regulator RcsB activates expression of Mat fimbriae in meningitic Escherichia coli. J. Bacteriol. 194 3475–3485. 10.1128/JB.06596-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leimbach A., Hacker J., Dobrindt U. (2013). E. coli as an all-rounder: the thin line between commensalism and pathogenicity. Curr. Top. Microbiol. Immunol. 358 3–32. 10.1007/82_2012_303 [DOI] [PubMed] [Google Scholar]
- Lescat M., Hoede C., Clermont O., Garry L., Darlu P., Tuffery P., et al. (2009). aes, the gene encoding the esterase B in Escherichia coli, is a powerful phylogenetic marker of the species. BMC Microbiol. 9:273. 10.1186/1471-2180-9-273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis J. A., Hatfull G. F. (2001). Control of directionality in integrase-mediated recombination: examination of recombination directionality factors (RDFs) including Xis and Cox proteins. Nucleic Acids Res. 29 2205–2216. 10.1093/nar/29.11.2205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis S. B., Cook V., Tighe R., Schüller S. (2015). Enterohemorrhagic Escherichia coli colonization of human colonic epithelium in vitro and ex vivo. Infect. Immun. 83 942–949. 10.1128/IAI.02928-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Tai C., Deng Z., Zhong W., He Y., Ou H.-Y. (2018). VRprofile: gene-cluster-detection-based profiling of virulence and antibiotic resistance traits encoded within genome sequences of pathogenic bacteria. Brief. Bioinformatics 19 566–574. 10.1093/bib/bbw141 [DOI] [PubMed] [Google Scholar]
- Li X., Rasko D. A., Lockatell C. V., Johnson D. E., Mobley H. L. (2001). Repression of bacterial motility by a novel fimbrial gene product. EMBO J. 20 4854–4862. 10.1093/emboj/20.17.4854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindberg S., Xia Y., Sondén B., Göransson M., Hacker J., Uhlin B. E. (2008). Regulatory Interactions among adhesin gene systems of uropathogenic Escherichia coli. Infect. Immun. 76 771–780. 10.1128/IAI.01010-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindberg U. (1975). Asymptomatic bacteriuria in school girls. V. The clinical course and response to treatment. Acta Paediatr. Scand. 64 718–724. 10.1111/j.1651-2227.1975.tb03910.x [DOI] [PubMed] [Google Scholar]
- Lindenthal C., Elsinghorst E. A. (1999). Identification of a glycoprotein produced by enterotoxigenic Escherichia coli. Infect. Immun. 67 4084–4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsey R. L., Batra D., Smith P., Patel P. N., Tagg K. A., Garcia-Toledo L., et al. (2018). PacBio genome sequences of Escherichia coli serotype O157:H7, Diffusely Adherent E. coli, and Salmonella enterica Strains, All Carrying Plasmids with an mcr-1 Resistance Gene. Microbiol. Resour. Announc. 7:e01025-18. 10.1128/MRA.01025-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindstedt R., Larson G., Falk P., Jodal U., Leffler H., Svanborg C. (1991). The receptor repertoire defines the host range for attaching Escherichia coli strains that recognize globo-A. Infect. Immun. 59 1086–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linhartová I., Bumba L., Mašín J., Basler M., Osièka R., Kamanová J., et al. (2010). RTX proteins: a highly diverse family secreted by a common mechanism. FEMS Microbiol. Rev. 34 1076–1112. 10.1111/j.1574-6976.2010.00231.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ljungdahl M., Lundholm M., Katouli M., Rasmussen I., Engstrand L., Haglund U. (2000). Bacterial translocation in experimental shock is dependent on the strains in the intestinal flora. Scand. J. Gastroenterol. 35 389–397. 10.1080/003655200750023958 [DOI] [PubMed] [Google Scholar]
- Lloyd A. L., Henderson T. A., Vigil P. D., Mobley H. L. T. (2009a). Genomic islands of uropathogenic Escherichia coli contribute to virulence. J. Bacteriol. 191 3469–3481. 10.1128/JB.01717-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd A. L., Smith S. N., Eaton K. A., Mobley H. L. T. (2009b). Uropathogenic Escherichia coli Suppresses the host inflammatory response via pathogenicity island genes sisA and sisB. Infect. Immun. 77 5322–5333. 10.1128/IAI.00779-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo Y., Zhang L., Foxman B., Zöllner S. (2015). Whole-genome sequencing of uropathogenic Escherichia coli reveals long evolutionary history of diversity and virulence. Infect. Genet. Evol. 34 244–250. 10.1016/j.meegid.2015.06.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu B., Leong H. W. (2016). Computational methods for predicting genomic islands in microbial genomes. Comput. Struct. Biotechnol. J. 14 200–206. 10.1016/j.csbj.2016.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucchini S., Rowley G., Goldberg M. D., Hurd D., Harrison M., Hinton J. C. D. (2006). H-NS mediates the silencing of laterally acquired genes in bacteria. PLoS Pathog. 2:e81. 10.1371/journal.ppat.0020081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luck S. N., Turner S. A., Rajakumar K., Sakellaris H., Adler B. (2001). Ferric dicitrate transport system (Fec) of Shigella flexneri 2a YSH6000 is encoded on a novel pathogenicity island carrying multiple antibiotic resistance genes. Infect. Immun. 69 6012–6021. 10.1128/IAI.69.10.6012-6021.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luterbach C. L., Forsyth V. S., Engstrom M. D., Mobley H. L. T. (2018). TosR-mediated regulation of adhesins and biofilm formation in uropathogenic Escherichia coli. mSphere 3:e00222-18. 10.1128/mSphere.00222-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacFie J., O’Boyle C., Mitchell C. J., Buckley P. M., Johnstone D., Sudworth P. (1999). Gut origin of sepsis: a prospective study investigating associations between bacterial translocation, gastric microflora, and septic morbidity. Gut 45 223–228. 10.1136/gut.45.2.223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahjoub-Messai F., Bidet P., Caro V., Diancourt L., Biran V., Aujard Y., et al. (2011). Escherichia coli isolates causing bacteremia via gut translocation and urinary tract infection in young infants exhibit different virulence genotypes. J. Infect. Dis. 203 1844–1849. 10.1093/infdis/jir189 [DOI] [PubMed] [Google Scholar]
- Mammarappallil J. G., Elsinghorst E. A. (2000). Epithelial cell adherence mediated by the enterotoxigenic Escherichia coli tia protein. Infect. Immun. 68 6595–6601. 10.1128/iai.68.12.6595-6601.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marklund B. I., Tennent J. M., Garcia E., Hamers A., Båga M., Lindberg F., et al. (1992). Horizontal gene transfer of the Escherichia coli pap and prs pili operons as a mechanism for the development of tissue-specific adhesive properties. Mol. Microbiol. 6 2225–2242. 10.1111/j.1365-2958.1992.tb01399.x [DOI] [PubMed] [Google Scholar]
- Maroncle N. M., Sivick K. E., Brady R., Stokes F.-E., Mobley H. L. T. (2006). Protease activity, secretion, cell entry, cytotoxicity, and cellular targets of secreted autotransporter toxin of uropathogenic Escherichia coli. Infect. Immun. 74 6124–6134. 10.1128/IAI.01086-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrs C. F., Zhang L., Tallman P., Manning S. D., Somsel P., Raz P., et al. (2002). Variations in 10 putative uropathogen virulence genes among urinary, faecal and peri-urethral Escherichia coli. J. Med. Microbiol. 51 138–142. 10.1099/0022-1317-51-2-138 [DOI] [PubMed] [Google Scholar]
- Martin H. M., Campbell B. J., Hart C. A., Mpofu C., Nayar M., Singh R., et al. (2004). Enhanced Escherichia coli adherence and invasion in Crohn’s disease and colon cancer. Gastroenterology 127 80–93. [DOI] [PubMed] [Google Scholar]
- Martinez J. J., Mulvey M. A., Schilling J. D., Pinkner J. S., Hultgren S. J. (2000). Type 1 pilus-mediated bacterial invasion of bladder epithelial cells. EMBO J. 19 2803–2812. 10.1093/emboj/19.12.2803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins F. H., Guth B. E. C., Piazza R. M., Leão S. C., Ludovico A., Ludovico M. S., et al. (2015). Diversity of Shiga toxin-producing Escherichia coli in sheep flocks of Paraná State, southern Brazil. Vet. Microbiol. 175 150–156. 10.1016/j.vetmic.2014.11.003 [DOI] [PubMed] [Google Scholar]
- Masuda H., Owaribe K., Hatano S. (1983). Contraction of triton-treated culture cells. A calcium-sensitive contractile model. Exp. Cell Res. 143 79–90. 10.1016/0014-4827(83)90111-8 [DOI] [PubMed] [Google Scholar]
- Mathusa E. C., Chen Y., Enache E., Hontz L. (2010). Non-O157 Shiga toxin-producing Escherichia coli in foods. J. Food Prot. 73 1721–1736. 10.4315/0362-028x-73.9.1721 [DOI] [PubMed] [Google Scholar]
- Mattock E., Blocker A. J. (2017). How do the virulence factors of Shigella work together to cause disease? Front. Cell. Infect. Microbiol. 7:64. 10.3389/fcimb.2017.00064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurelli A. T. (2007). Black holes, antivirulence genes, and gene inactivation in the evolution of bacterial pathogens. FEMS Microbiol. Lett. 267 1–8. 10.1111/j.1574-6968.2006.00526.x [DOI] [PubMed] [Google Scholar]
- McCarthy A. J., Martin P., Cloup E., Stabler R. A., Oswald E., Taylor P. W. (2015). The Genotoxin Colibactin Is a Determinant of Virulence in Escherichia coli K1 Experimental Neonatal Systemic Infection. Infect. Immun. 83 3704–3711. 10.1128/IAI.00716-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClain M. S., Blomfield I. C., Eisenstein B. I. (1991). Roles of fimB and fimE in site-specific DNA inversion associated with phase variation of type 1 fimbriae in Escherichia coli. J. Bacteriol. 173 5308–5314. 10.1128/jb.173.17.5308-5314.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClelland M., Sanderson K. E., Spieth J., Clifton S. W., Latreille P., Courtney L., et al. (2001). Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413 852–856. 10.1038/35101614 [DOI] [PubMed] [Google Scholar]
- McDaniel T. K., Jarvis K. G., Donnenberg M. S., Kaper J. B. (1995). A genetic locus of enterocyte effacement conserved among diverse enterobacterial pathogens. Proc. Natl. Acad. Sci. U.S.A. 92 1664–1668. 10.1073/pnas.92.5.1664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDaniel T. K., Kaper J. B. (1997). A cloned pathogenicity island from enteropathogenic Escherichia coli confers the attaching and effacing phenotype on E. coli K-12. Mol. Microbiol. 23 399–407. 10.1046/j.1365-2958.1997.2311591.x [DOI] [PubMed] [Google Scholar]
- McWilliams B. D., Torres A. G. (2014). Enterohemorrhagic Escherichia coli Adhesins. Microbiol. Spectr. 2:EHEC00032013 10.1128/microbiolspec.EHEC-0003-2013 [DOI] [PubMed] [Google Scholar]
- Mellies J. L., Navarro-Garcia F., Okeke I., Frederickson J., Nataro J. P., Kaper J. B. (2001). espC pathogenicity island of enteropathogenic Escherichia coli encodes an enterotoxin. Infect. Immun. 69 315–324. 10.1128/IAI.69.1.315-324.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melton-Celsa A. R. (2014). Shiga Toxin (Stx) classification, structure, and function. Microbiol. Spectr. 2:EHEC-0024-2013. 10.1128/microbiolspec.EHEC-0024-2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messerer M., Fischer W., Schubert S. (2017). Investigation of horizontal gene transfer of pathogenicity islands in Escherichia coli using next-generation sequencing. PLoS One 12:e0179880. 10.1371/journal.pone.0179880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelacci V., Prosseda G., Maugliani A., Tozzoli R., Sanchez S., Herrera-León S., et al. (2016). Characterization of an emergent clone of enteroinvasive Escherichia coli circulating in Europe. Clin. Microbiol. Infect. 22 287.e11–287.e1. 10.1016/j.cmi.2015.10.025 [DOI] [PubMed] [Google Scholar]
- Michelacci V., Tozzoli R., Caprioli A., Martínez R., Scheutz F., Grande L., et al. (2013). A new pathogenicity island carrying an allelic variant of the Subtilase cytotoxin is common among Shiga toxin producing Escherichia coli of human and ovine origin. Clin. Microbiol. Infect. 19 E149–E156. 10.1111/1469-0691.12122 [DOI] [PubMed] [Google Scholar]
- Middendorf B., Hochhut B., Leipold K., Dobrindt U., Blum-Oehler G., Hacker J. (2004). Instability of pathogenicity islands in uropathogenic Escherichia coli 536. J. Bacteriol. 186 3086–3096. 10.1128/jb.186.10.3086-3096.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milkman R. (1973). Electrophoretic variation in Escherichia coli from natural sources. Science 182 1024–1026. 10.1126/science.182.4116.1024 [DOI] [PubMed] [Google Scholar]
- Monteiro R., Ageorges V., Rojas-Lopez M., Schmidt H., Weiss A., Bertin Y., et al. (2016). A secretome view of colonisation factors in Shiga toxin-encoding Escherichia coli (STEC): from enterohaemorrhagic E. coli (EHEC) to related enteropathotypes. FEMS Microbiol. Lett. 363:fnw179. 10.1093/femsle/fnw179 [DOI] [PubMed] [Google Scholar]
- Montero D., Orellana P., Gutiérrez D., Araya D., Salazar J. C., Prado V., et al. (2014). Immunoproteomic analysis to identify Shiga toxin-producing Escherichia coli outer membrane proteins expressed during human infection. Infect. Immun. 82 4767–4777. 10.1128/IAI.02030-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montero D. A., Canto F. D., Velasco J., Colello R., Padola N. L., Salazar J. C., et al. (2019). Cumulative acquisition of pathogenicity islands has shaped virulence potential and contributed to the emergence of LEE-negative Shiga toxin-producing Escherichia coli strains. Emerg. Microbes Infect. 8 486–502. 10.1080/22221751.2019.1595985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montero D. A., Velasco J., Del Canto F., Puente J. L., Padola N. L., Rasko D. A., et al. (2017). Locus of Adhesion and Autoaggregation (LAA), a pathogenicity island present in emerging Shiga Toxin-producing Escherichia coli strains. Sci. Rep. 7:7011. 10.1038/s41598-017-06999-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mora A., López C., Dabhi G., Blanco M., Blanco J. E., Alonso M. P., et al. (2009). Extraintestinal pathogenic Escherichia coli O1:K1:H7/NM from human and avian origin: detection of clonal groups B2 ST95 and D ST59 with different host distribution. BMC Microbiol. 9:132. 10.1186/1471-2180-9-132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mora A., López C., Herrera A., Viso S., Mamani R., Dhabi G., et al. (2012). Emerging avian pathogenic Escherichia coli strains belonging to clonal groups O111:H4-D-ST2085 and O111:H4-D-ST117 with high virulence-gene content and zoonotic potential. Vet. Microbiol. 156 347–352. 10.1016/j.vetmic.2011.10.033 [DOI] [PubMed] [Google Scholar]
- Morabito S., Tozzoli R., Oswald E., Caprioli A. (2003). A mosaic pathogenicity island made up of the locus of enterocyte effacement and a pathogenicity island of Escherichia coli O157:H7 is frequently present in attaching and effacing E. coli. Infect. Immun. 71 3343–3348. 10.1128/iai.71.6.3343-3348.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno E., Andreu A., Pigrau C., Kuskowski M. A., Johnson J. R., Prats G. (2008). Relationship between Escherichia coli strains causing acute cystitis in women and the fecal E. coli population of the host. J. Clin. Microbiol. 46 2529–2534. 10.1128/JCM.00813-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriel D. G., Bertoldi I., Spagnuolo A., Marchi S., Rosini R., Nesta B., et al. (2010). Identification of protective and broadly conserved vaccine antigens from the genome of extraintestinal pathogenic Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 107 9072–9077. 10.1073/pnas.0915077107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moritz R. L., Welch R. A. (2006). The Escherichia coli argW-dsdCXA genetic island is highly variable, and E. coli K1 strains commonly possess two copies of dsdCXA. J. Clin. Microbiol. 44 4038–4048. 10.1128/JCM.01172-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss J. E., Cardozo T. J., Zychlinsky A., Groisman E. A. (1999). The selC-associated SHI-2 pathogenicity island of Shigella flexneri. Mol. Microbiol. 33 74–83. 10.1046/j.1365-2958.1999.01449.x [DOI] [PubMed] [Google Scholar]
- Moulin-Schouleur M., Schouler C., Tailliez P., Kao M.-R., Brée A., Germon P., et al. (2006). Common virulence factors and genetic relationships between O18:K1:H7 Escherichia coli isolates of human and avian origin. J. Clin. Microbiol. 44 3484–3492. 10.1128/JCM.00548-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller C. A., Broz P., Cornelis G. R. (2008). The type III secretion system tip complex and translocon. Mol. Microbiol. 68 1085–1095. 10.1111/j.1365-2958.2008.06237.x [DOI] [PubMed] [Google Scholar]
- Müller D., Benz I., Liebchen A., Gallitz I., Karch H., Schmidt M. A. (2009). Comparative analysis of the locus of enterocyte effacement and its flanking regions. Infect. Immun. 77 3501–3513. 10.1128/IAI.00090-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulvey M. A., Lopez-Boado Y. S., Wilson C. L., Roth R., Parks W. C., Heuser J., et al. (1998). Induction and evasion of host defenses by type 1-piliated uropathogenic Escherichia coli. Science 282 1494–1497. 10.1126/science.282.5393.1494 [DOI] [PubMed] [Google Scholar]
- Mulvey M. A., Schilling J. D., Hultgren S. J. (2001). Establishment of a persistent Escherichia coli reservoir during the acute phase of a bladder infection. Infect. Immun. 69 4572–4579. 10.1128/IAI.69.7.4572-4579.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulvey M. A., Schilling J. D., Martinez J. J., Hultgren S. J. (2000). Bad bugs and beleaguered bladders: interplay between uropathogenic Escherichia coli and innate host defenses. Proc. Natl. Acad. Sci. U.S.A. 97 8829–8835. 10.1073/pnas.97.16.8829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mysorekar I. U., Hultgren S. J. (2006). Mechanisms of uropathogenic Escherichia coli persistence and eradication from the urinary tract. Proc. Natl. Acad. Sci. U.S.A. 103 14170–14175. 10.1073/pnas.0602136103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagarjuna D., Mittal G., Dhanda R. S., Verma P. K., Gaind R., Yadav M. (2015). Faecal Escherichia coli isolates show potential to cause endogenous infection in patients admitted to the ICU in a tertiary care hospital. New Microbes New Infections 7 57–66. 10.1016/j.nmni.2015.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy G., Altenhoefer A., Knapp O., Maier E., Dobrindt U., Blum-Oehler G., et al. (2006). Both alpha-haemolysin determinants contribute to full virulence of uropathogenic Escherichia coli strain 536. Microbes Infect. 8 2006–2012. 10.1016/j.micinf.2006.02.029 [DOI] [PubMed] [Google Scholar]
- Napolitano M. G., Almagro-Moreno S., Boyd E. F. (2011). Dichotomy in the evolution of pathogenicity island and bacteriophage encoded integrases from pathogenic Escherichia coli strains. Infect. Genet. Evol. 11 423–436. 10.1016/j.meegid.2010.12.003 [DOI] [PubMed] [Google Scholar]
- Nataro J. P., Kaper J. B. (1998). Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11 142–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarre W. W., Porwollik S., Wang Y., McClelland M., Rosen H., Libby S. J., et al. (2006). Selective silencing of foreign DNA with low GC content by the H-NS protein in Salmonella. Science 313 236–238. 10.1126/science.1128794 [DOI] [PubMed] [Google Scholar]
- Navarro-Garcia F. (2014). Escherichia coli O104:H4 pathogenesis: an enteroaggregative E. coli/Shiga Toxin-Producing E. coli explosive cocktail of high virulence. Microbiol. Spectr. 2:EHEC-0008-2013. 10.1128/microbiolspec.EHEC-0008-2013 [DOI] [PubMed] [Google Scholar]
- Navarro-Garcia F., Serapio-Palacios A., Vidal J. E., Salazar M. I., Tapia-Pastrana G. (2014). EspC promotes epithelial cell detachment by enteropathogenic Escherichia coli via sequential cleavages of a cytoskeletal protein and then focal adhesion proteins. Infect. Immun. 82 2255–2265. 10.1128/IAI.01386-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nègre V. L., Bonacorsi S., Schubert S., Bidet P., Nassif X., Bingen E. (2004). The siderophore receptor IroN, but not the high-pathogenicity island or the hemin receptor ChuA, contributes to the bacteremic step of Escherichia coli neonatal meningitis. Infect. Immun. 72 1216–1220. 10.1128/iai.72.2.1216-1220.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesta B., Spraggon G., Alteri C., Moriel D. G., Rosini R., Veggi D., et al. (2012). FdeC, a novel broadly conserved Escherichia coli adhesin eliciting protection against urinary tract infections. mBio 3:e00010-12. 10.1128/mBio.00010-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesta B., Valeri M., Spagnuolo A., Rosini R., Mora M., Donato P., et al. (2014). SslE elicits functional antibodies that impair in vitro mucinase activity and in vivo colonization by both intestinal and extraintestinal Escherichia coli strains. PLoS Pathog. 10:e1004124. 10.1371/journal.ppat.1004124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie H., Yang F., Zhang X., Yang J., Chen L., Wang J., et al. (2006). Complete genome sequence of Shigella flexneri 5b and comparison with Shigella flexneri 2a. BMC Genomics 7:173. 10.1186/1471-2164-7-173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielubowicz G. R., Mobley H. L. T. (2010). Host-pathogen interactions in urinary tract infection. Nat. Rev. Urol. 7 430–441. 10.1038/nrurol.2010.101 [DOI] [PubMed] [Google Scholar]
- Nieto P. A., Pardo-Roa C., Salazar-Echegarai F. J., Tobar H. E., Coronado-Arrázola I., Riedel C. A., et al. (2016). New insights about excisable pathogenicity islands in Salmonella and their contribution to virulence. Microbes Infect. 18 302–309. 10.1016/j.micinf.2016.02.001 [DOI] [PubMed] [Google Scholar]
- Nojoomi F., Ghasemian A. (2019). The relation of phylogroups, serogroups, virulence factors and resistance pattern of Escherichia coli isolated from children with septicemia. New Microbes New Infect 29 100517. 10.1016/j.nmni.2019.100517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nougayrède J.-P., Fernandes P. J., Donnenberg M. S. (2003). Adhesion of enteropathogenic Escherichia coli to host cells. Cell. Microbiol. 5 359–372. 10.1046/j.1462-5822.2003.00281.x [DOI] [PubMed] [Google Scholar]
- Nougayrède J.-P., Homburg S., Taieb F., Boury M., Brzuszkiewicz E., Gottschalk G., et al. (2006). Escherichia coli induces DNA double-strand breaks in eukaryotic cells. Science 313 848–851. 10.1126/science.1127059 [DOI] [PubMed] [Google Scholar]
- Nowrouzian F. L., Oswald E. (2012). Escherichia coli strains with the capacity for long-term persistence in the bowel microbiota carry the potentially genotoxic pks island. Microb. Pathog. 53 180–182. 10.1016/j.micpath.2012.05.011 [DOI] [PubMed] [Google Scholar]
- Numrych T. E., Gumport R. I., Gardner J. F. (1992). Characterization of the bacteriophage lambda excisionase (Xis) protein: the C-terminus is required for Xis-integrase cooperativity but not for DNA binding. EMBO J. 11 3797–3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochoa T. J., Contreras C. A. (2011). Enteropathogenic escherichia coli infection in children. Curr. Opin. Infect. Dis. 24 478–483. 10.1097/QCO.0b013e32834a8b8b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olier M., Marcq I., Salvador-Cartier C., Secher T., Dobrindt U., Boury M., et al. (2012). Genotoxicity of Escherichia coli Nissle 1917 strain cannot be dissociated from its probiotic activity. Gut Microbes 3 501–509. 10.4161/gmic.21737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opal S., Cross A., Gemski P. (1982). K antigen and serum sensitivity of rough Escherichia coli. Infect. Immun. 37 956–960. 10.1128/IAI.37.3.956-960.1982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orden J. A., Horcajo P., de la Fuente R., Ruiz-Santa-Quiteria J. A., Domínguez-Bernal G., Carrión J. (2011). Subtilase cytotoxin-coding genes in verotoxin-producing Escherichia coli strains from sheep and goats differ from those from cattle. Appl. Environ. Microbiol. 77 8259–8264. 10.1128/AEM.05604-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Shea Y. A., Boyd E. F. (2002). Mobilization of the Vibrio pathogenicity island between Vibrio cholerae isolates mediated by CP-T1 generalized transduction. FEMS Microbiol. Lett. 214 153–157. 10.1111/j.1574-6968.2002.tb11339.x [DOI] [PubMed] [Google Scholar]
- Oshima T., Ishikawa S., Kurokawa K., Aiba H., Ogasawara N. (2006). Escherichia coli histone-like protein H-NS preferentially binds to horizontally acquired DNA in association with RNA polymerase. DNA Res. 13 141–153. 10.1093/dnares/dsl009 [DOI] [PubMed] [Google Scholar]
- Ott M., Hacker J., Schmoll T., Jarchau T., Korhonen T. K., Goebel W. (1986). Analysis of the genetic determinants coding for the S-fimbrial adhesin (sfa) in different Escherichia coli strains causing meningitis or urinary tract infections. Infect. Immun. 54 646–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owrangi B., Masters N., Kuballa A., O’Dea C., Vollmerhausen T. L., Katouli M. (2018). Invasion and translocation of uropathogenic Escherichia coli isolated from urosepsis and patients with community-acquired urinary tract infection. Eur. J. Clin. Microbiol. Infect. Dis. 37 833–839. 10.1007/s10096-017-3176-4 [DOI] [PubMed] [Google Scholar]
- Palmela C., Chevarin C., Xu Z., Torres J., Sevrin G., Hirten R., et al. (2018). Adherent-invasive Escherichia coli in inflammatory bowel disease. Gut 67 574–587. 10.1136/gutjnl-2017-314903 [DOI] [PubMed] [Google Scholar]
- Panis G., Duverger Y., Courvoisier-Dezord E., Champ S., Talla E., Ansaldi M. (2010). Tight regulation of the intS gene of the KplE1 prophage: a new paradigm for integrase gene regulation. PLoS Genet. 6:e1001149. 10.1371/journal.pgen.1001149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parham N. J., Pollard S. J., Chaudhuri R. R., Beatson S. A., Desvaux M., Russell M. A., et al. (2005a). Prevalence of pathogenicity island IICFT073 genes among extraintestinal clinical isolates of Escherichia coli. J. Clin. Microbiol. 43 2425–2434. 10.1128/JCM.43.5.2425-2434.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parham N. J., Pollard S. J., Desvaux M., Scott-Tucker A., Liu C., Fivian A., et al. (2005b). Distribution of the serine protease autotransporters of the Enterobacteriaceae among extraintestinal clinical isolates of Escherichia coli. J. Clin. Microbiol. 43 4076–4082. 10.1128/JCM.43.8.4076-4082.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parham N. J., Srinivasan U., Desvaux M., Foxman B., Marrs C. F., Henderson I. R. (2004). PicU, a second serine protease autotransporter of uropathogenic Escherichia coli. FEMS Microbiol. Lett. 230 73–83. 10.1016/S0378-1097(03)00862-0 [DOI] [PubMed] [Google Scholar]
- Parreira V. R., Gyles C. L. (2003). A novel pathogenicity island integrated adjacent to the thrW tRNA gene of avian pathogenic Escherichia coli encodes a vacuolating autotransporter toxin. Infect. Immun. 71 5087–5096. 10.1128/iai.71.9.5087-5096.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsot C. (2005). Shigella spp. and enteroinvasive Escherichia coli pathogenicity factors. FEMS Microbiol. Lett. 252 11–18. 10.1016/j.femsle.2005.08.046 [DOI] [PubMed] [Google Scholar]
- Pasqua M., Michelacci V., Di Martino M. L., Tozzoli R., Grossi M., Colonna B., et al. (2017). the intriguing evolutionary journey of enteroinvasive E. coli (EIEC) toward Pathogenicity. Front. Microbiol. 8:2390. 10.3389/fmicb.2017.02390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J., Yang J., Jin Q. (2009). The molecular evolutionary history of Shigella spp. and enteroinvasive Escherichia coli. Infect. Genet. Evol. 9 147–152. 10.1016/j.meegid.2008.10.003 [DOI] [PubMed] [Google Scholar]
- Perna N. T., Mayhew G. F., Pósfai G., Elliott S., Donnenberg M. S., Kaper J. B., et al. (1998). Molecular evolution of a pathogenicity island from enterohemorrhagic Escherichia coli O157:H7. Infect. Immun. 66 3810–3817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perna N. T., Plunkett G., Burland V., Mau B., Glasner J. D., Rose D. J., et al. (2001). Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409 529–533. 10.1038/35054089 [DOI] [PubMed] [Google Scholar]
- Peterson J. W., Whipp S. C. (1995). Comparison of the mechanisms of action of cholera toxin and the heat-stable enterotoxins of Escherichia coli. Infect. Immun. 63 1452–1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettengill E. A., Pettengill J. B., Binet R. (2015). Phylogenetic analyses of Shigella and enteroinvasive Escherichia coli for the identification of molecular epidemiological markers: whole-genome comparative analysis does not support distinct genera designation. Front. Microbiol. 6:1573. 10.3389/fmicb.2015.01573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard B., Garcia J. S., Gouriou S., Duriez P., Brahimi N., Bingen E., et al. (1999). The link between phylogeny and virulence in Escherichia coli extraintestinal infection. Infect. Immun. 67 546–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platenkamp A., Mellies J. L. (2018). Environment Controls LEE Regulation in Enteropathogenic Escherichia coli. Front. Microbiol. 9:1694. 10.3389/fmicb.2018.01694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleguezuelos-Manzano C., Puschhof J., Rosendahl Huber A., van Hoeck A., Wood H. M., Nomburg J., et al. (2020). Mutational signature in colorectal cancer caused by genotoxic pks+ E. coli. Nature 580 269–273. 10.1038/s41586-020-2080-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prager R., Lang C., Aurass P., Fruth A., Tietze E., Flieger A. (2014). Two novel EHEC/EAEC hybrid strains isolated from human infections. PLoS One 9:e95379. 10.1371/journal.pone.0095379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasadarao N. V., Wass C. A., Weiser J. N., Stins M. F., Huang S. H., Kim K. S. (1996). Outer membrane protein A of Escherichia coli contributes to invasion of brain microvascular endothelial cells. Infect. Immun. 64 146–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provence D. L., Curtiss R. (1994). Isolation and characterization of a gene involved in hemagglutination by an avian pathogenic Escherichia coli strain. Infect. Immun. 62 1369–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purdy G. E., Payne S. M. (2001). The SHI-3 iron transport island of Shigella boydii 0-1392 carries the genes for aerobactin synthesis and transport. J. Bacteriol. 183 4176–4182. 10.1128/JB.183.14.4176-4182.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putze J., Hennequin C., Nougayrède J.-P., Zhang W., Homburg S., Karch H., et al. (2009). Genetic structure and distribution of the colibactin genomic island among members of the family Enterobacteriaceae. Infect. Immun. 77 4696–4703. 10.1128/IAI.00522-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin X., Hu F., Wu S., Ye X., Zhu D., Zhang Y., et al. (2013). Comparison of adhesin genes and antimicrobial susceptibilities between uropathogenic and intestinal commensal Escherichia coli strains. PLoS One 8:e61169. 10.1371/journal.pone.0061169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragnarsdóttir B., Fischer H., Godaly G., Grönberg-Hernandez J., Gustafsson M., Karpman D., et al. (2008). TLR- and CXCR1-dependent innate immunity: insights into the genetics of urinary tract infections. Eur. J. Clin. Invest. 38 12–20. 10.1111/j.1365-2362.2008.02004.x [DOI] [PubMed] [Google Scholar]
- Rahman S., Gadjeva M. (2014). Does NETosis contribute to the bacterial pathoadaptation in cystic fibrosis? Front. Immunol. 5:378. 10.3389/fimmu.2014.00378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raimondi S., Righini L., Candeliere F., Musmeci E., Bonvicini F., Gentilomi G., et al. (2019). Antibiotic resistance, virulence factors, phenotyping, and genotyping of E. coli Isolated from the Feces of Healthy Subjects. Microorganisms 7:251. 10.3390/microorganisms7080251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajakumar K., Sasakawa C., Adler B. (1996). A spontaneous 99-kb chromosomal deletion results in multi-antibiotic susceptibility and an attenuation of contact haemolysis in Shigella flexneri 2a. J. Med. Microbiol. 45 64–75. 10.1099/00222615-45-1-64 [DOI] [PubMed] [Google Scholar]
- Rajakumar K., Sasakawa C., Adler B. (1997). Use of a novel approach, termed island probing, identifies the Shigella flexneri she pathogenicity island which encodes a homolog of the immunoglobulin A protease-like family of proteins. Infect. Immun. 65 4606–4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajanna C., Wang J., Zhang D., Xu Z., Ali A., Hou Y.-M., et al. (2003). The vibrio pathogenicity island of epidemic Vibrio cholerae forms precise extrachromosomal circular excision products. J. Bacteriol. 185 6893–6901. 10.1128/jb.185.23.6893-6901.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasko D. A., Rosovitz M. J., Myers G. S. A., Mongodin E. F., Fricke W. F., Gajer P., et al. (2008). The pangenome structure of Escherichia coli: comparative genomic analysis of E. coli commensal and pathogenic isolates. J. Bacteriol. 190 6881–6893. 10.1128/JB.00619-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redford P., Welch R. A. (2002). Extraintestinal Escherichia coli as a model system for the study of pathogenicity islands. Curr. Top. Microbiol. Immunol. 264 15–30. [PubMed] [Google Scholar]
- Reid S. D., Herbelin C. J., Bumbaugh A. C., Selander R. K., Whittam T. S. (2000). Parallel evolution of virulence in pathogenic Escherichia coli. Nature 406 64–67. 10.1038/35017546 [DOI] [PubMed] [Google Scholar]
- Reigstad C. S., Hultgren S. J., Gordon J. I. (2007). Functional genomic studies of uropathogenic Escherichia coli and host urothelial cells when intracellular bacterial communities are assembled. J. Biol. Chem. 282 21259–21267. 10.1074/jbc.M611502200 [DOI] [PubMed] [Google Scholar]
- Reiss D. J., Howard F. M., Mobley H. L. T. (2012). A novel approach for transcription factor analysis using SELEX with high-throughput sequencing (TFAST). PLoS One 7:e42761. 10.1371/journal.pone.0042761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiss D. J., Mobley H. L. T. (2011). Determination of target sequence bound by PapX, repressor of bacterial motility, in flhD promoter using systematic evolution of ligands by exponential enrichment (SELEX) and high throughput sequencing. J. Biol. Chem. 286 44726–44738. 10.1074/jbc.M111.290684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter W. D., Palm P., Yeats S. (1989). Transfer RNA genes frequently serve as integration sites for prokaryotic genetic elements. Nucleic Acids Res. 17 1907–1914. 10.1093/nar/17.5.1907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rendón M. A., Saldaña Z., Erdem A. L., Monteiro-Neto V., Vázquez A., Kaper J. B., et al. (2007). Commensal and pathogenic Escherichia coli use a common pilus adherence factor for epithelial cell colonization. Proc. Natl. Acad. Sci. U.S.A. 104 10637–10642. 10.1073/pnas.0704104104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Restieri C., Garriss G., Locas M.-C., Dozois C. M. (2007). Autotransporter-encoding sequences are phylogenetically distributed among Escherichia coli clinical isolates and reference strains. Appl. Environ. Microbiol. 73 1553–1562. 10.1128/AEM.01542-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rippere-Lampe K. E., O’Brien A. D., Conran R., Lockman H. A. (2001). Mutation of the gene encoding cytotoxic necrotizing factor type 1 (cnf(1)) attenuates the virulence of uropathogenic Escherichia coli. Infect. Immun. 69 3954–3964. 10.1128/IAI.69.6.3954-3964.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins J. B., Chu C., Schneerson R. (1992). Hypothesis for vaccine development: protective immunity to enteric diseases caused by nontyphoidal Salmonellae and Shigellae may be conferred by serum IgG antibodies to the O-specific polysaccharide of their lipopolysaccharides. Clin. Infect. Dis. 15 346–361. 10.1093/clinids/15.2.346 [DOI] [PubMed] [Google Scholar]
- Rodrigues J., Scaletsky I. C., Campos L. C., Gomes T. A., Whittam T. S., Trabulsi L. R. (1996). Clonal structure and virulence factors in strains of Escherichia coli of the classic serogroup O55. Infect. Immun. 64 2680–2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Siek K. E., Giddings C. W., Doetkott C., Johnson T. J., Fakhr M. K., Nolan L. K. (2005). Comparison of Escherichia coli isolates implicated in human urinary tract infection and avian colibacillosis. Microbiology 151 2097–2110. 10.1099/mic.0.27499-0 [DOI] [PubMed] [Google Scholar]
- Rosen D. A., Hooton T. M., Stamm W. E., Humphrey P. A., Hultgren S. J. (2007). Detection of intracellular bacterial communities in human urinary tract infection. PLoS Med. 4:e329. 10.1371/journal.pmed.0040329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousset E., Harel J., Dubreuil J. D. (1998). Sulfatide from the pig jejunum brush border epithelial cell surface is involved in binding of Escherichia coli enterotoxin b. Infect. Immun. 66 5650–5658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggenenti P., Remuzzi G. (1998). Pathophysiology and management of thrombotic microangiopathies. J. Nephrol. 11 300–310. [PubMed] [Google Scholar]
- Ruiz-Perez F., Wahid R., Faherty C. S., Kolappaswamy K., Rodriguez L., Santiago A., et al. (2011). Serine protease autotransporters from Shigella flexneri and pathogenic Escherichia coli target a broad range of leukocyte glycoproteins. Proc. Natl. Acad. Sci. U.S.A. 108 12881–12886. 10.1073/pnas.1101006108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rumer L., Jores J., Kirsch P., Cavignac Y., Zehmke K., Wieler L. H. (2003). Dissemination of pheU- and pheV-located genomic islands among enteropathogenic (EPEC) and enterohemorrhagic (EHEC) E. coli and their possible role in the horizontal transfer of the locus of enterocyte effacement (LEE). Int. J. Med. Microbiol. 292 463–475. 10.1078/1438-4221-00229 [DOI] [PubMed] [Google Scholar]
- Runyen-Janecky L., Dazenski E., Hawkins S., Warner L. (2006). Role and regulation of the Shigella flexneri sit and MntH systems. Infect. Immun. 74 4666–4672. 10.1128/IAI.00562-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo T. (2003). Medical and economic impact of extraintestinal infections due to Escherichia coli: focus on an increasingly important endemic problem. Microbes Infect. 5 449–456. 10.1016/S1286-4579(03)00049-2 [DOI] [PubMed] [Google Scholar]
- Russo T. A., Carlino U. B., Johnson J. R. (2001). Identification of a new iron-regulated virulence gene, ireA, in an extraintestinal pathogenic isolate of Escherichia coli. Infect. Immun. 69 6209–6216. 10.1128/IAI.69.10.6209-6216.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo T. A., Johnson J. R. (2000). Proposal for a new inclusive designation for extraintestinal pathogenic isolates of Escherichia coli: ExPEC. J. Infect. Dis. 181 1753–1754. 10.1086/315418 [DOI] [PubMed] [Google Scholar]
- Sack R. B. (2011). The discovery of cholera - like enterotoxins produced by Escherichia coli causing secretory diarrhoea in humans. Indian J. Med. Res. 133 171–180. [PMC free article] [PubMed] [Google Scholar]
- Saile N., Schuh E., Semmler T., Eichhorn I., Wieler L. H., Bauwens A., et al. (2018). Determination of virulence and fitness genes associated with the pheU, pheV and selC integration sites of LEE-negative food-borne Shiga toxin-producing Escherichia coli strains. Gut Pathog. 10:43. 10.1186/s13099-018-0271-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakellaris H., Luck S. N., Al-Hasani K., Rajakumar K., Turner S. A., Adler B. (2004). Regulated site-specific recombination of the she pathogenicity island of Shigella flexneri. Mol. Microbiol. 52 1329–1336. 10.1111/j.1365-2958.2004.04048.x [DOI] [PubMed] [Google Scholar]
- Samuelsson P., Hang L., Wullt B., Irjala H., Svanborg C. (2004). Toll-like receptor 4 expression and cytokine responses in the human urinary tract mucosa. Infect. Immun. 72 3179–3186. 10.1128/IAI.72.6.3179-3186.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarff L. D., McCracken G. H., Schiffer M. S., Glode M. P., Robbins J. B., Orskov I., et al. (1975). Epidemiology of Escherichia coli K1 in healthy and diseased newborns. Lancet 1 1099–1104. 10.1016/s0140-6736(75)92496-4 [DOI] [PubMed] [Google Scholar]
- Sarowska J., Futoma-Koloch B., Jama-Kmiecik A., Frej-Madrzak M., Ksiazczyk M., Bugla-Ploskonska G., et al. (2019). Virulence factors, prevalence and potential transmission of extraintestinal pathogenic Escherichia coli isolated from different sources: recent reports. Gut Pathog. 11:10. 10.1186/s13099-019-0290-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaletsky I. C., Silva M. L., Trabulsi L. R. (1984). Distinctive patterns of adherence of enteropathogenic Escherichia coli to HeLa cells. Infect. Immun. 45 534–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M. B., Roth J. R. (1987). Gene location affects expression level in Salmonella typhimurium. J. Bacteriol. 169 2872–2875. 10.1128/jb.169.6.2872-2875.1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt H., Hensel M. (2004). Pathogenicity islands in bacterial pathogenesis. Clin. Microbiol. Rev. 17 14–56. 10.1128/cmr.17.1.14-56.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt H., Zhang W. L., Hemmrich U., Jelacic S., Brunder W., Tarr P. I., et al. (2001). Identification and characterization of a novel genomic island integrated at selC in locus of enterocyte effacement-negative, Shiga toxin-producing Escherichia coli. Infect. Immun. 69 6863–6873. 10.1128/IAI.69.11.6863-6873.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt M. A. (2010). LEEways: tales of EPEC, ATEC and EHEC. Cell. Microbiol. 12 1544–1552. 10.1111/j.1462-5822.2010.01518.x [DOI] [PubMed] [Google Scholar]
- Schneider G., Dobrindt U., Middendorf B., Hochhut B., Szijártó V., Emody L., et al. (2011). Mobilisation and remobilisation of a large archetypal pathogenicity island of uropathogenic Escherichia coli in vitro support the role of conjugation for horizontal transfer of genomic islands. BMC Microbiol. 11:210. 10.1186/1471-2180-11-210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnupf P., Sansonetti P. J. (2019). Shigella pathogenesis: new insights through advanced methodologies. Microbiol. Spectr. 7:BAI-0023-2019. 10.1128/microbiolspec.BAI-0023-2019 [DOI] [PubMed] [Google Scholar]
- Schubert S., Cuenca S., Fischer D., Heesemann J. (2000). High-pathogenicity island of Yersinia pestis in enterobacteriaceae isolated from blood cultures and urine samples: prevalence and functional expression. J. Infect. Dis. 182 1268–1271. 10.1086/315831 [DOI] [PubMed] [Google Scholar]
- Schubert S., Darlu P., Clermont O., Wieser A., Magistro G., Hoffmann C., et al. (2009). Role of intraspecies recombination in the spread of pathogenicity islands within the Escherichia coli species. PLoS Pathog. 5:e1000257. 10.1371/journal.ppat.1000257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert S., Dufke S., Sorsa J., Heesemann J. (2004a). A novel integrative and conjugative element (ICE) of Escherichia coli: the putative progenitor of the Yersinia high-pathogenicity island. Mol. Microbiol. 51 837–848. 10.1046/j.1365-2958.2003.03870.x [DOI] [PubMed] [Google Scholar]
- Schubert S., Nörenberg D., Clermont O., Magistro G., Wieser A., Romann E., et al. (2010). Prevalence and phylogenetic history of the TcpC virulence determinant in Escherichia coli. Int. J. Med. Microbiol. 300 429–434. 10.1016/j.ijmm.2010.02.006 [DOI] [PubMed] [Google Scholar]
- Schubert S., Picard B., Gouriou S., Heesemann J., Denamur E. (2002). Yersinia high-pathogenicity island contributes to virulence in Escherichia coli causing extraintestinal infections. Infect. Immun. 70 5335–5337. 10.1128/iai.70.9.5335-5337.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert S., Rakin A., Heesemann J. (2004b). The Yersinia high-pathogenicity island (HPI): evolutionary and functional aspects. Int. J. Med. Microbiol. 294 83–94. 10.1016/j.ijmm.2004.06.026 [DOI] [PubMed] [Google Scholar]
- Schubert S., Rakin A., Karch H., Carniel E., Heesemann J. (1998). Prevalence of the “high-pathogenicity island” of Yersinia species among Escherichia coli strains that are pathogenic to humans. Infect. Immun. 66 480–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selander R. K., Musser J. M., Caugant D. A., Gilmour M. N., Whittam T. S. (1987). Population genetics of pathogenic bacteria. Microb. Pathog. 3 1–7. 10.1016/0882-4010(87)90032-5 [DOI] [PubMed] [Google Scholar]
- Serapio-Palacios A., Navarro-Garcia F. (2016). EspC, an autotransporter protein secreted by enteropathogenic Escherichia coli, causes apoptosis and necrosis through caspase and calpain activation, including direct procaspase-3 cleavage. mBio 7:e00479-16. 10.1128/mBio.00479-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Servin A. L. (2005). Pathogenesis of Afa/Dr diffusely adhering Escherichia coli. Clin. Microbiol. Rev. 18 264–292. 10.1128/CMR.18.2.264-292.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Servin A. L. (2014). Pathogenesis of human diffusely adhering Escherichia coli expressing Afa/Dr adhesins (Afa/Dr DAEC): current insights and future challenges. Clin. Microbiol. Rev. 27 823–869. 10.1128/CMR.00036-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen S., Mascarenhas M., Rahn K., Kaper J. B., Karmali M. A., Karmal M. A. (2004). Evidence for a hybrid genomic island in verocytotoxin-producing Escherichia coli CL3 (serotype O113:H21) containing segments of EDL933 (serotype O157:H7) O islands 122 and 48. Infect. Immun. 72 1496–1503. 10.1128/iai.72.3.1496-1503.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver R. P., Aaronson W., Sutton A., Schneerson R. (1980). Comparative analysis of plasmids and some metabolic characteristics of Escherichia coli K1 from diseased and healthy individuals. Infect. Immun. 29 200–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simms A. N., Mobley H. L. T. (2008). PapX, a P fimbrial operon-encoded inhibitor of motility in uropathogenic Escherichia coli. Infect. Immun. 76 4833–4841. 10.1128/IAI.00630-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skyberg J. A., Johnson T. J., Johnson J. R., Clabots C., Logue C. M., Nolan L. K. (2006). Acquisition of avian pathogenic Escherichia coli plasmids by a commensal E. coli isolate enhances its abilities to kill chicken embryos, grow in human urine, and colonize the murine kidney. Infect. Immun. 74 6287–6292. 10.1128/IAI.00363-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder J. A., Haugen B. J., Buckles E. L., Lockatell C. V., Johnson D. E., Donnenberg M. S., et al. (2004). Transcriptome of uropathogenic Escherichia coli during urinary tract infection. Infect. Immun. 72 6373–6381. 10.1128/IAI.72.11.6373-6381.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder J. A., Haugen B. J., Lockatell C. V., Maroncle N., Hagan E. C., Johnson D. E., et al. (2005). Coordinate expression of fimbriae in uropathogenic Escherichia coli. Infect. Immun. 73 7588–7596. 10.1128/IAI.73.11.7588-7596.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spangler B. D. (1992). Structure and function of cholera toxin and the related Escherichia coli heat-labile enterotoxin. Microbiol. Rev. 56 622–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperandio V., Kaper J. B., Bortolini M. R., Neves B. C., Keller R., Trabulsi L. R. (1998). Characterization of the locus of enterocyte effacement (LEE) in different enteropathogenic Escherichia coli (EPEC) and Shiga-toxin producing Escherichia coli (STEC) serotypes. FEMS Microbiol. Lett. 164 133–139. 10.1111/j.1574-6968.1998.tb13078.x [DOI] [PubMed] [Google Scholar]
- Spurbeck R. R., Stapleton A. E., Johnson J. R., Walk S. T., Hooton T. M., Mobley H. L. T. (2011). Fimbrial profiles predict virulence of uropathogenic Escherichia coli strains: contribution of ygi and yad fimbriae. Infect. Immun. 79 4753–4763. 10.1128/IAI.05621-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan U., Foxman B., Marrs C. F. (2003). Identification of a gene encoding heat-resistant agglutinin in Escherichia coli as a putative virulence factor in urinary tract infection. J. Clin. Microbiol. 41 285–289. 10.1128/jcm.41.1.285-289.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens M. P., Frankel G. M. (2014). The locus of enterocyte effacement and associated virulence factors of enterohemorrhagic Escherichia coli. Microbiol. Spectr. 2:EHEC-0007-2013. [DOI] [PubMed] [Google Scholar]
- Sun Q., Lan R., Wang Y., Wang J., Wang Y., Li P., et al. (2013). Isolation and genomic characterization of SfI, a serotype-converting bacteriophage of Shigella flexneri. BMC Microbiol. 13:39. 10.1186/1471-2180-13-39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svanborg C., Bergsten G., Fischer H., Frendéus B., Godaly G., Gustafsson E., et al. (2001). The “innate” host response protects and damages the infected urinary tract. Ann. Med. 33 563–570. 10.3109/07853890109002101 [DOI] [PubMed] [Google Scholar]
- Swenson D. L., Bukanov N. O., Berg D. E., Welch R. A. (1996). Two pathogenicity islands in uropathogenic Escherichia coli J96: cosmid cloning and sample sequencing. Infect. Immun. 64 3736–3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swidsinski A., Khilkin M., Kerjaschki D., Schreiber S., Ortner M., Weber J., et al. (1998). Association between intraepithelial Escherichia coli and colorectal cancer. Gastroenterology 115 281–286. [DOI] [PubMed] [Google Scholar]
- Tang F., Wang J., Li D., Gao S., Ren J., Ma L., et al. (2019). Comparative genomic analysis of 127 Escherichia coli strains isolated from domestic animals with diarrhea in China. BMC Genomics 20:212. 10.1186/s12864-019-5588-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapader R., Basu S., Pal A. (2019). Secreted proteases: a new insight in the pathogenesis of extraintestinal pathogenic Escherichia coli. Int. J. Med. Microbiol. 309 159–168. 10.1016/j.ijmm.2019.03.002 [DOI] [PubMed] [Google Scholar]
- Tarchouna M., Ferjani A., Ben-Selma W., Boukadida J. (2013). Distribution of uropathogenic virulence genes in Escherichia coli isolated from patients with urinary tract infection. Int. J. Infect. Dis. 17 e450–e453. 10.1016/j.ijid.2013.01.025 [DOI] [PubMed] [Google Scholar]
- Tarr P. I. (2009). Shiga toxin-associated hemolytic uremic syndrome and thrombotic thrombocytopenic purpura: distinct mechanisms of pathogenesis. Kidney Int. Suppl. S29–S32. 10.1038/ki.2008.615 [DOI] [PubMed] [Google Scholar]
- Tarr P. I., Bilge S. S., Vary J. C., Jelacic S., Habeeb R. L., Ward T. R., et al. (2000). Iha: a novel Escherichia coli O157:H7 adherence-conferring molecule encoded on a recently acquired chromosomal island of conserved structure. Infect. Immun. 68 1400–1407. 10.1128/iai.68.3.1400-1407.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarr P. I., Gordon C. A., Chandler W. L. (2005). Shiga-toxin-producing Escherichia coli and haemolytic uraemic syndrome. Lancet 365 1073–1086. 10.1016/S0140-6736(05)71144-2 [DOI] [PubMed] [Google Scholar]
- Tauschek M., Strugnell R. A., Robins-Browne R. M. (2002). Characterization and evidence of mobilization of the LEE pathogenicity island of rabbit-specific strains of enteropathogenic Escherichia coli. Mol. Microbiol. 44 1533–1550. 10.1046/j.1365-2958.2002.02968.x [DOI] [PubMed] [Google Scholar]
- Taylor D. E., Rooker M., Keelan M., Ng L.-K., Martin I., Perna N. T., et al. (2002). Genomic variability of O islands encoding tellurite resistance in enterohemorrhagic Escherichia coli O157:H7 isolates. J. Bacteriol. 184 4690–4698. 10.1128/jb.184.17.4690-4698.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenaillon O., Skurnik D., Picard B., Denamur E. (2010). The population genetics of commensal Escherichia coli. Nat. Rev. Microbiol. 8 207–217. 10.1038/nrmicro2298 [DOI] [PubMed] [Google Scholar]
- Teng C.-H., Cai M., Shin S., Xie Y., Kim K.-J., Khan N. A., et al. (2005). Escherichia coli K1 RS218 interacts with human brain microvascular endothelial cells via type 1 fimbria bacteria in the fimbriated state. Infect. Immun. 73 2923–2931. 10.1128/IAI.73.5.2923-2931.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tivendale K. A., Logue C. M., Kariyawasam S., Jordan D., Hussein A., Li G., et al. (2010). Avian-pathogenic Escherichia coli strains are similar to neonatal meningitis E. coli strains and are able to cause meningitis in the rat model of human disease. Infect. Immun. 78 3412–3419. 10.1128/IAI.00347-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres A. G. (2004). Current aspects of Shigella pathogenesis. Rev. Latinoam. Microbiol. 46 89–97. [PubMed] [Google Scholar]
- Touchon M., Hoede C., Tenaillon O., Barbe V., Baeriswyl S., Bidet P., et al. (2009). Organised genome dynamics in the Escherichia coli species results in highly diverse adaptive paths. PLoS Genet. 5:e1000344. 10.1371/journal.pgen.1000344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tourret J., Denamur E. (2016). “Population phylogenomics of extraintestinal pathogenic Escherichia coli,” in Urinary Tract Infections: Molecular Pathogenesis and Clinical Management, 2nd Edn, eds Mulvey M., Klumpp D., Stapleton A., (Washington, DC: ASM Press; ), 207–233. 10.1128/microbiolspec.UTI-0010-2012 [DOI] [Google Scholar]
- Tourret J., Diard M., Garry L., Matic I., Denamur E. (2010). Effects of single and multiple pathogenicity island deletions on uropathogenic Escherichia coli strain 536 intrinsic extra-intestinal virulence. Int. J. Med. Microbiol. 300 435–439. 10.1016/j.ijmm.2010.04.013 [DOI] [PubMed] [Google Scholar]
- Trabulsi L. R., Keller R., Tardelli Gomes T. A. (2002). Typical and atypical enteropathogenic Escherichia coli. Emerg. Infect. Dis. 8 508–513. 10.3201/eid0805.010385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner N. C. A., Connolly J. P. R., Roe A. J. (2019). Control freaks-signals and cues governing the regulation of virulence in attaching and effacing pathogens. Biochem. Soc. Trans. 47 229–238. 10.1042/BST20180546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner S. A., Luck S. N., Sakellaris H., Rajakumar K., Adler B. (2001). Nested deletions of the SRL pathogenicity island of Shigella flexneri 2a. J. Bacteriol. 183 5535–5543. 10.1128/JB.183.19.5535-5543.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner S. A., Luck S. N., Sakellaris H., Rajakumar K., Adler B. (2003). Molecular epidemiology of the SRL pathogenicity island. Antimicrob. Agents Chemother. 47 727–734. 10.1128/aac.47.2.727-734.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner S. M., Chaudhuri R. R., Jiang Z.-D., DuPont H., Gyles C., Penn C. W., et al. (2006a). Phylogenetic comparisons reveal multiple acquisitions of the toxin genes by enterotoxigenic Escherichia coli strains of different evolutionary lineages. J. Clin. Microbiol. 44 4528–4536. 10.1128/JCM.01474-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner S. M., Scott-Tucker A., Cooper L. M., Henderson I. R. (2006b). Weapons of mass destruction: virulence factors of the global killer enterotoxigenic Escherichia coli. FEMS Microbiol. Lett. 263 10–20. 10.1111/j.1574-6968.2006.00401.x [DOI] [PubMed] [Google Scholar]
- Uhlén P., Laestadius A., Jahnukainen T., Söderblom T., Bäckhed F., Celsi G., et al. (2000). Alpha-haemolysin of uropathogenic E. coli induces Ca2+ oscillations in renal epithelial cells. Nature 405 694–697. 10.1038/35015091 [DOI] [PubMed] [Google Scholar]
- Ulett G. C., Valle J., Beloin C., Sherlock O., Ghigo J.-M., Schembri M. A. (2007). Functional analysis of antigen 43 in uropathogenic Escherichia coli reveals a role in long-term persistence in the urinary tract. Infect. Immun. 75 3233–3244. 10.1128/IAI.01952-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaandrager A. B. (2002). Structure and function of the heat-stable enterotoxin receptor/guanylyl cyclase C. Mol. Cell. Biochem. 230 73–83. [PubMed] [Google Scholar]
- Valle J., Mabbett A. N., Ulett G. C., Toledo-Arana A., Wecker K., Totsika M., et al. (2008). UpaG, a new member of the trimeric autotransporter family of adhesins in uropathogenic Escherichia coli. J. Bacteriol. 190 4147–4161. 10.1128/JB.00122-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Bosch L., Manning P. A., Morona R. (1997). Regulation of O-antigen chain length is required for Shigella flexneri virulence. Mol. Microbiol. 23 765–775. 10.1046/j.1365-2958.1997.2541625.x [DOI] [PubMed] [Google Scholar]
- Vejborg R. M., Hancock V., Schembri M. A., Klemm P. (2011). Comparative genomics of Escherichia coli strains causing urinary tract infections. Appl. Environ. Microbiol. 77 3268–3278. 10.1128/AEM.02970-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigil P. D., Alteri C. J., Mobley H. L. T. (2011a). Identification of in vivo-induced antigens including an RTX family exoprotein required for uropathogenic Escherichia coli virulence. Infect. Immun. 79 2335–2344. 10.1128/IAI.00110-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigil P. D., Stapleton A. E., Johnson J. R., Hooton T. M., Hodges A. P., He Y., et al. (2011b). Presence of putative repeat-in-toxin gene tosA in Escherichia coli predicts successful colonization of the urinary tract. mBio 2:e00066-11. 10.1128/mBio.00066-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigil P. D., Wiles T. J., Engstrom M. D., Prasov L., Mulvey M. A., Mobley H. L. T. (2012). The repeat-in-toxin family member TosA mediates adherence of uropathogenic Escherichia coli and survival during bacteremia. Infect. Immun. 80 493–505. 10.1128/IAI.05713-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vokes S. A., Reeves S. A., Torres A. G., Payne S. M. (1999). The aerobactin iron transport system genes in Shigella flexneri are present within a pathogenicity island. Mol. Microbiol. 33 63–73. 10.1046/j.1365-2958.1999.01448.x [DOI] [PubMed] [Google Scholar]
- Vollmerhausen T. L., Woods J. L., Faoagali J., Katouli M. (2014). Interactions of uroseptic Escherichia coli with renal (A-498) and gastrointestinal (HT-29) cell lines. J. Med. Microbiol. 63 1575–1583. 10.1099/jmm.0.076562-0 [DOI] [PubMed] [Google Scholar]
- Wada H., Matsumoto T., Yamashita Y. (2014). Natural history of thrombotic thrombocytopenic purpura and hemolytic uremic syndrome. Semin. Thromb. Hemost. 40 866–873. 10.1055/s-0034-1395154 [DOI] [PubMed] [Google Scholar]
- Wang H., Paton J. C., Paton A. W. (2007). Pathologic changes in mice induced by subtilase cytotoxin, a potent new Escherichia coli AB5 toxin that targets the endoplasmic reticulum. J. Infect. Dis. 196 1093–1101. 10.1086/521364 [DOI] [PubMed] [Google Scholar]
- Wang L., Wakushima M., Aota T., Yoshida Y., Kita T., Maehara T., et al. (2013). Specific properties of enteropathogenic Escherichia coli isolates from diarrheal patients and comparison to strains from foods and fecal specimens from cattle, swine, and healthy carriers in Osaka City. Japan. Appl. Environ. Microbiol. 79 1232–1240. 10.1128/AEM.03380-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Huang S. H., Wass C. A., Stins M. F., Kim K. S. (1999). The gene locus yijP contributes to Escherichia coli K1 invasion of brain microvascular endothelial cells. Infect. Immun. 67 4751–4756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch R. A. (1991). Pore-forming cytolysins of Gram-negative bacteria. Mol. Microbiol. 5 521–528. 10.1111/j.1365-2958.1991.tb00723.x [DOI] [PubMed] [Google Scholar]
- Whitfield C. (2006). Biosynthesis and assembly of capsular polysaccharides in Escherichia coli. Annu. Rev. Biochem. 75 39–68. 10.1146/annurev.biochem.75.103004.142545 [DOI] [PubMed] [Google Scholar]
- Wickham M. E., Lupp C., Mascarenhas M., Vazquez A., Coombes B. K., Brown N. F., et al. (2006). Bacterial genetic determinants of non-O157 STEC outbreaks and hemolytic-uremic syndrome after infection. J. Infect. Dis. 194 819–827. 10.1086/506620 [DOI] [PubMed] [Google Scholar]
- Wijetunge D. S. S., Gongati S., DebRoy C., Kim K. S., Couraud P. O., Romero I. A., et al. (2015). Characterizing the pathotype of neonatal meningitis causing Escherichia coli (NMEC). BMC Microbiol. 15:211. 10.1186/s12866-015-0547-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilde C., Mazel D., Hochhut B., Middendorf B., Le Roux F., Carniel E., et al. (2008). Delineation of the recombination sites necessary for integration of pathogenicity islands II and III into the Escherichia coli 536 chromosome. Mol. Microbiol. 68 139–151. 10.1111/j.1365-2958.2008.06145.x [DOI] [PubMed] [Google Scholar]
- Wiles T. J., Kulesus R. R., Mulvey M. A. (2008). Origins and virulence mechanisms of uropathogenic Escherichia coli. Exp. Mol. Pathol. 85 11–19. 10.1016/j.yexmp.2008.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams K. P. (2003). Traffic at the tmRNA gene. J. Bacteriol. 185 1059–1070. 10.1128/jb.185.3.1059-1070.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfson J. J., May K. L., Thorpe C. M., Jandhyala D. M., Paton J. C., Paton A. W. (2008). Subtilase cytotoxin activates PERK, IRE1 and ATF6 endoplasmic reticulum stress-signalling pathways. Cell. Microbiol. 10 1775–1786. 10.1111/j.1462-5822.2008.01164.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright K. J., Seed P. C., Hultgren S. J. (2007). Development of intracellular bacterial communities of uropathogenic Escherichia coli depends on type 1 pili. Cell. Microbiol. 9 2230–2241. 10.1111/j.1462-5822.2007.00952.x [DOI] [PubMed] [Google Scholar]
- Xia Y., Forsman K., Jass J., Uhlin B. E. (1998). Oligomeric interaction of the PapB transcriptional regulator with the upstream activating region of pili adhesin gene promoters in Escherichia coli. Mol. Microbiol. 30 513–523. 10.1046/j.1365-2958.1998.01080.x [DOI] [PubMed] [Google Scholar]
- Xie Y., Kolisnychenko V., Paul-Satyaseela M., Elliott S., Parthasarathy G., Yao Y., et al. (2006). Identification and characterization of Escherichia coli RS218–derived islands in the pathogenesis of E. coli Meningitis. J. Infect. Dis. 194 358–364. 10.1086/505429 [DOI] [PubMed] [Google Scholar]
- Xu W.-Y., Li Y.-J., Fan C. (2018). Different loci and mRNA copy number of the increased serum survival gene of Escherichia coli. Can. J. Microbiol. 64 147–154. 10.1139/cjm-2017-0363 [DOI] [PubMed] [Google Scholar]
- Yahiro K., Tsutsuki H., Ogura K., Nagasawa S., Moss J., Noda M. (2012). Regulation of subtilase cytotoxin-induced cell death by an RNA-dependent protein kinase-like endoplasmic reticulum kinase-dependent proteasome pathway in HeLa cells. Infect. Immun. 80 1803–1814. 10.1128/IAI.06164-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F., Yang J., Zhang X., Chen L., Jiang Y., Yan Y., et al. (2005). Genome dynamics and diversity of Shigella species, the etiologic agents of bacillary dysentery. Nucleic Acids Res. 33 6445–6458. 10.1093/nar/gki954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin X., Wheatcroft R., Chambers J. R., Liu B., Zhu J., Gyles C. L. (2009). Contributions of O island 48 to adherence of enterohemorrhagic Escherichia coli O157:H7 to epithelial cells in vitro and in ligated pig ileal loops. Appl. Environ. Microbiol. 75 5779–5786. 10.1128/AEM.00507-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon S. H., Park Y.-K., Kim J. F. (2015). PAIDB v2.0: exploration and analysis of pathogenicity and resistance islands. Nucleic Acids Res. 43 D624–D630. 10.1093/nar/gku985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zagaglia C., Casalino M., Colonna B., Conti C., Calconi A., Nicoletti M. (1991). Virulence plasmids of enteroinvasive Escherichia coli and Shigella flexneri integrate into a specific site on the host chromosome: integration greatly reduces expression of plasmid-carried virulence genes. Infect. Immun. 59 792–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W.-D., Liu D.-X., Wei J.-Y., Miao Z.-W., Zhang K., Su Z.-K., et al. (2018). Caspr1 is a host receptor for meningitis-causing Escherichia coli. Nat. Commun. 9:2296. 10.1038/s41467-018-04637-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Y., He L., Huang S.-H. (2006). Identification of a surface protein on human brain microvascular endothelial cells as vimentin interacting with Escherichia coli invasion protein IbeA. Biochem. Biophys. Res. Commun. 351 625–630. 10.1016/j.bbrc.2006.10.091 [DOI] [PubMed] [Google Scholar]