Abstract
Nicotinamide adenine dinucleotide (NAD+) is an important natural molecule involved in fundamental biological processes, including the TCA cycle, OXPHOS, β-oxidation, and is a co-factor for proteins promoting healthy longevity. NAD+ depletion is associated with the hallmarks of ageing and may contribute to a wide range of agerelated diseases including metabolic disorders, cancer, and neurodegenerative diseases. One of the central pathways by which NAD+ promotes healthy ageing is through regulation of mitochondrial homeostasis via mitochondrial biogenesis and the clearance of damaged mitochondria via mitophagy. Here, we highlight the contribution of the NAD+-mitophagy axis to ageing and age-related diseases, and evaluate how boosting NAD+ levels may emerge as a promising therapeutic strategy to counter ageing as well as neurodegenerative diseases including Alzheimer’s disease. The potential use of artificial intelligence to understand the roles and molecular mechanisms of the NAD+-mitophagy axis in ageing is discussed, including possible applications in drug target identification and validation, compound screening and lead compound discovery, biomarker development, as well as efficacy and safety assessment. Advances in our understanding of the molecular and cellular roles of NAD+ in mitophagy will lead to novel approaches for facilitating healthy mitochondrial homoeostasis that may serve as a promising therapeutic strategy to counter ageing-associated pathologies and/or accelerated ageing.
Keywords: NAD+, Mitophagy, Artificial intelligence, Ageing, Age-related diseases, Alzheimer’s disease
1. Introduction
Ageing is an inevitable biological process but over the years there has been a dramatic increase in human life expectancy. A recent United Nations report estimated that over 700 million people (representing 9% of the world population) were over 65 years old in 2019, a figure that is expected to double by 2050 (United Nations, 2019). A rise in human longevity has led to an increased global burden of age-related diseases, such as cardiovascular diseases, cancer, and neurodegenerative diseases (especially Alzheimer’s disease/AD) (Fang et al., 2015; Partridge et al., 2018). These age-related conditions not only result in reduced quality of life of the elderly population, but also present as a formidable healthcare and socio-economic challenge. Interestingly, there is a subpopulation of elderly individuals that experience little ill-health. One aim of molecular gerontology is to define the features of such superaged individuals in order to be extended to the general population. In this pursuit, it is necessary to elucidate the intricate molecular mechanisms underlying the human ageing process, which may provide therapeutic interventions to promote “healthy ageing” and possibly halt the progression of age-related pathological conditions (Campisi et al., 2019; Fang et al., 2019, 2016c; Kennedy et al., 2014; Partridge et al., 2018).
Mitochondrial dysfunction and impairment of mitochondrial-autophagy, termed mitophagy, have emerged as important components of the age-related cellular processes that contribute to disease (Kerr et al., 2017b; Rubinsztein et al., 2011). In particular, impairment of the lysosome-targeting and recycling mechanisms have been highlighted as causative for the accumulation of damaged mitochondria, which consequently leads to cellular dysfunction and/or death (Lautrup et al., 2019b; Lou et al., 2019). Evidence stemming from animal models and human cell lines implicate a novel role of NAD+ in autophagy and mitophagy (Fang et al., 2014; Lee et al., 2008; Vannini et al., 2019). An increasing body of evidence over the past decades demonstrates that decline in levels of NAD+ contribute to the ten hallmarks of cellular ageing and age-related diseases (Fivenson et al., 2017; Lautrup et al., 2019a; Nikiforov et al., 2015; Verdin, 2015).
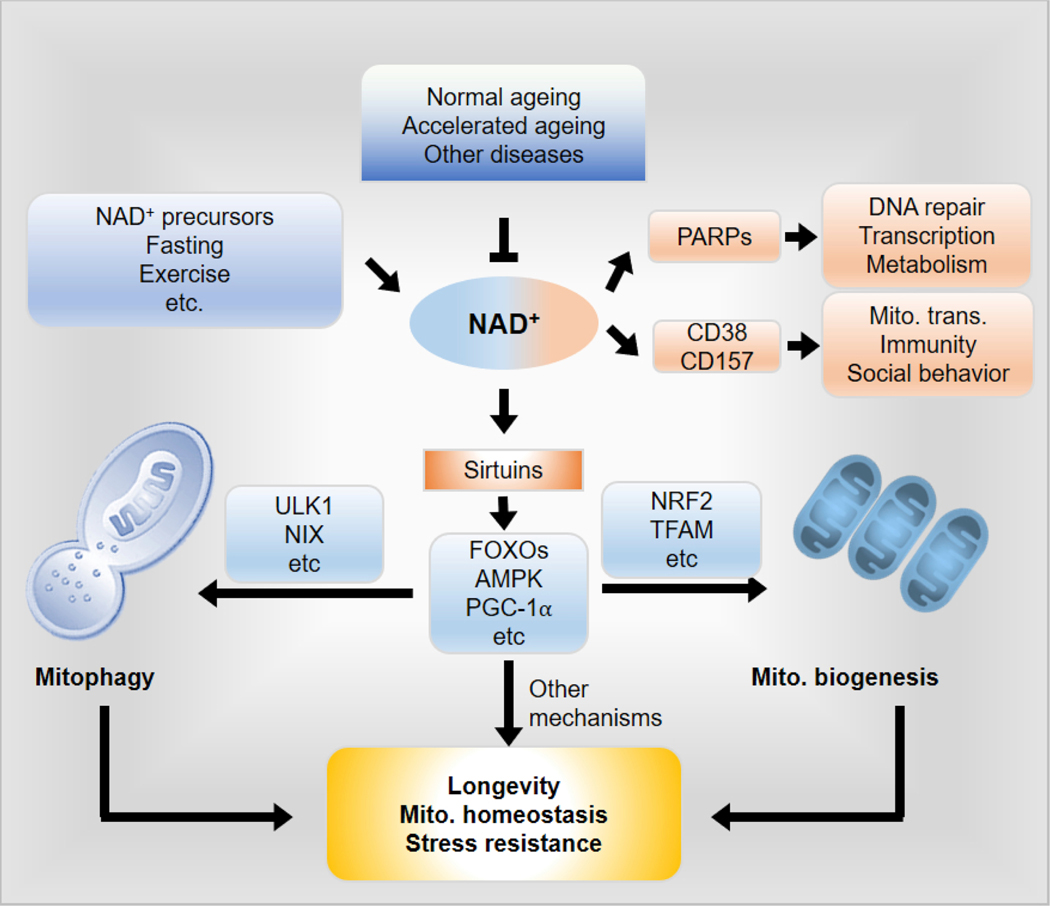
Here, we summarise the roles and molecular mechanisms of NAD+ in ageing, mitochondrial homeostasis, and mitophagy. Specifically, we highlight the contribution of NAD+ depletion to mammalian ageing and evaluate how boosting endogenous NAD+ levels may be a promising therapeutic strategy to counter age-associated pathologies and/or accelerated ageing via promotion of mitochondrial homeostasis and mitophagy (Fig. 1). In addition, we discuss the potential use of artificial intelligence to understand the roles and molecular mechanisms of the NAD+-mitophagy axis in ageing and disease.
Fig. 1. Schematic representation of NAD+ mediated coupling of mitophagy and mitochondrial biogenesis for maintenance of homeostasis and longevity.
Cellular levels of NAD+ decline in an age-dependent manner. NAD+ is a cofactor for sirtuins (i.e. SIRT1) as well as other NAD+ consuming enzymes including: PARPs, CD38, and CD157. The NAD+/sirtuins pathways regulate both mitochondrial biogenesis and induction of mitophagy through designated pathways as detailed in the text. E.g., mitochondrial biogenesis can be triggered by SIRT1 mediated deacetylation and thus activates peroxisome proliferator-activated receptorgamma coactivator 1-alpha (PGC-1α), which in turn translocates to nucleus and interacts with AMP-activated protein kinase (AMPK) to trigger the expression of nuclear factor-erythroid 2-related factor 2 (NRF-2) that in turn orchestrates numerous mitochondrial, stress resistance and longevity genes. Induction of mitophagy is triggered by the NAD+/SIRT1 pathway via multiple pathways, including AMPK-dependent phosphorylation of ULK1, and NIX upregulation. A synergy between mitochondrial biogenesis and mitophagy, as well as the involvement of other known and unknown mechanisms, maintains mitochondrial homeostasis, stress resistance, and finally results in healthy longevity in animals. To note, there may be many other NAD+-dependent but sirtuin-independent pathways that contribute to longevity, mitochondrial homeostasis and stress resistance. Abbreviations: Mito., mitochondrial; trans., transfer.
2. NAD+ metabolic pathways and NAD+ precursors
NAD+ is an essential small molecule that regulates fundamental biological processes such as metabolic redox reactions, cell signalling, gene expression, and DNA repair (Canto et al., 2015; Magni et al., 2004; Nikiforov et al., 2015; Stromland et al., 2019; Verdin, 2015; Yaku et al., 2018). Harden and Young first discovered NAD+ as a molecular fraction (“cozymase”) which accelerated fermentation in yeast extracts. NAD+ has been identified as a necessary cofactor for many cellular metabolism pathways, transporting high energy electrons to support oxidative phosphorylation by reversibly oxidising or reducing NAD+, and serving as a substrate for NAD+-dependent enzymes that link cellular metabolism with epigenetic regulation and DNA damage repair. In particular, an age-dependent decline in NAD+ levels have consistently been reported and is associated with the 10 hallmarks of ageing, including the 10 hallmarks of brain ageing (detailed in our recent review) (Lautrup et al., 2019b), as well as several age-related diseases, including metabolic disorders, cancer and neurodegenerative diseases (Fang et al., 2016c; Fivenson et al., 2017; Lautrup et al., 2019a; Verdin, 2015; Yoshino et al., 2018). Replenishment of NAD+ levels via administration of its precursors has been demonstrated to have beneficial effects against ageing and age-related diseases. Importantly, boosting NAD+ levels have been shown to extend lifespan and or the improvement of healthspan of various laboratory animal models including worms, flies, and rodents (Fivenson et al., 2017; Frederick et al., 2016; Katsyuba and Auwerx, 2017; Rajman et al., 2018; Yaku et al., 2018; Yoshino et al., 2018). Thus, the importance of NAD+ has expanded from a key element in intermediate metabolism to a critical regulator of multiple cell signalling pathways and is now considered a major player contributing to ageing and age-related diseases.
In mammals, NAD+ is abundant within the cell (Canto et al., 2015; Rajman et al., 2018). It is synthesized from a variety of dietary sources, including NAD+ itself; as well as from one or more of its major precursors that include: tryptophan (Trp), nicotinic acid (NA), nicotinamide riboside (NR), nicotinamide mononucleotide (NMN), and nicotinamide (NAM). Based upon the bioavailability of its precursors, there are three pathways for the synthesis of NAD+ in cells: (i) from Trp by the de novo biosynthesis pathway or kynurenine pathway; (ii) from NA in the Preiss–Handler pathway; and (iii) from NAM, NR, and NMN in the salvage pathway (Bogan and Brenner, 2008; Lautrup et al., 2019b). However, the molecular mechanisms that dictate NAD+ synthesis pathway choice remain largely elusive. More studies are needed to further investigate those molecular mechanisms and artificial intelligence (AI) can be a one promising method to accelerate the research process (detailed below).
3. NAD+ regulates mitochondrial homeostasis via mitochondrial biogenesis and mitophagy
Mitochondria are dynamic structures considered as the ‘powerhouses’ of all nucleated cells and play a critical role in developmental and adult neuroplasticity as well as neuronal survival (Lautrup et al., 2019b; Lin and Beal, 2006; Mattson et al., 2008; McWilliams et al., 2018). Thus, they are indispensable for healthy ageing in most eukaryotic organisms (Fang et al., 2016c; Lane and Martin, 2010; Scheibye-Knudsen et al., 2015; Wallace, 1999). Mitochondria are the source the cellular energy currency, namely adenosine triphosphate (ATP). NAD+ has a fundamental role in the production of ATP in the mitochondria. The reduced form of NAD+ (NADH) is utilised as a primary hydride donor in the production of ATP via anaerobic glycolysis and mitochondrial oxidative phosphorylation (OXPHOS) (Fivenson et al., 2017; Wallace, 2012). In recent years it has become apparent that mitochondria coordinate multiple central processes; and mitochondrial homeostasis and maintenance is key to health (Lou et al., 2019); regulation of calcium ions (Ca2+), redox signalling, mitochondria-nuclear communications, and the intricate regulation of cell survival/stress resistance and death (Fang et al., 2016c; Lou et al., 2019; Mattson et al., 2008; Palikaras et al., 2018). Mitochondrial dysfunction is considered a hallmark of ageing and implicated in a spectrum of diseases. While mutations of nuclear- or mitochondria-encoded mitochondrial proteins cause rare mitochondrial disorders (Scheibye-Knudsen et al., 2015), mitochondria-mediated ATP deprivation, oxidative stress and impaired cell signalling have been linked to the pathogenesis of prominent metabolic (e.g., diabetes and cancer) and neurodegenerative diseases (e.g., AD and Parkinson’s disease, PD) (Fang et al., 2019; Kerr et al., 2017b; Wallace, 2012).
Mitochondrial homeostasis can be divided into multiple steps, including mitochondrial biogenesis, mitochondrial fusion and fission, mitochondrial quality control along with repair and the removal of damaged mitochondria via mitophagy. Although, mitochondria are semi- autonomous organelles, the majority of their proteins are encoded by the nuclear genome and have to be imported into the mitochondrion (Calvo and Mootha, 2010). Hence, mitochondrial biogenesis requires a sophisticated communication between the mitochondrion and the nucleus, recruitment of recently synthesized lipids and proteins, mitochondrial import and assembly of both nuclear and mitochondria-derived proteins (Zhu et al., 2013). Mitochondrial homeostasis is necessary for the maintenance of neuronal function and neuronal survival. Mitochondrial quality is tightly regulated by mitophagy, in which defective/superfluous mitochondria are degraded and recycled. Mitophagy in neurons is necessary to prevent neuronal cell death and pathogenic brain ageing, which are at least partially caused by accumulating dysfunctional mitochondria (Fang et al., 2016c; Fivenson et al., 2017; Lin et al., 2017). Several different mitophagy pathways are known, and many are conserved from C. elegans to humans (summarized in Table 1) (Egan et al., 2011; Fivenson et al., 2017; Kerr et al., 2017a).
Table 1.
NAD+ and the mitophagy machinery.
| Protein | Function in mitophagy | Relation to NAD + | Refs |
|---|---|---|---|
| PINK1 | Ser/Thr kinase; initiates mitophagy by phosphorylating components of mitophagy machinery including Ubiquitin and Parkin among others. | NAD+ → ↑PINK1-SARM1-TRAF1 complex→ ↑mitophagy NAD+↑ → ↑SIRT1/SIRT3→ PINK1/PARKIN |
(Das et al., 2014; Lazarou et al., 2015; Murata et al., 2013) |
| PARKIN | E3 ubiquitin ligase: initiates mitophagy by ubiquitinating proteins on the OMM. PTEN-L inhibits PINKl/Parkin dependent mitophagy via dephosphorylation of PINK1- mediated pSer65Ub. | NAD+ → ↑PINK1-SARM1-TRAF1 complex→ ↑ mitophagy | (Murata et al., 2013; Wang et al., 2018) |
| FISI | Protein of the OMM and component of the mitochondrial fission machinery at the mitochondrial surface. Mitochondrial fission and fusion cycles enable a cell to segregate damaged mitochondria from its network. | ↑NAD+→ ↑SIRT1/PGC-1a —FIS1↑ → ↑mitochondrial function and mitochondrial size↓ | (Kang and Hwang, 2009; Stojanovski et al., 2004) |
| OPA1 | Dynamin-related GTPase involved in promoting mitochondrial fusion, interacts with the IMM, structures the cristae & cristae junctions, and contributes to network fusion & mitochondrial respiration. Mitochondrial fission and fusion cycles enable a cell to segregate damaged mitochondria from its network. | ↑NAD+→ ↑SIRT3 ÔPA1 activation | (Lenaers et al., 2009; Samant et al., 2014) |
| DRP1 | DRP1 is located on the OMM and controls distribution and fission of mitochondria. Mitochondrial fission and fusion cycles enable a cell to segregate damaged mitochondria from its network. | NMN→ ↓DRP1→ ↓Fission in AD mice | (Labrousse et al., 1999; Long et al., 2015; Smirnova et al., 1998) |
| MFN1/2 | Mitofusin 1/2 are essential for mitochondrial fusion by tethering mitochondrial membranes. Mitochondrial fission and fusion cycles enable a cell to segregate damaged mitochondria from its network. | NR→ ↑NAD + → ↑SIRT activity → ↑MFN2 | (Canto et al., 2012; Ishihara et al., 2004) |
| LC3 | Essential for autophagosome biogenesis/maturation, mediates the fusion with lysosomes and in addition functions as an adaptor protein for selective autophagy | NAD+ ↑SIRT1 deacetylation of LC3 & ↑expression of LC3 deacetylation and acetylation of FOXO1 and FOXO3 | (Hariharan et al., 2010; Huang et al., 2015; Kume et al., 2010; Lee et al., 2008; Lee and Lee, 2016; Sengupta et al., 2009) |
| ATG5, ATG12 | Essential for autophagosome formation; ATG5 mediates membrane binding whereas ATG12 inhibits binding | NAD + → ↑SIRT1 deacetylation of ATG5 & ↑ expression of ATG12 | (Lee et al., 2008; Romanov et al., 2012; Sengupta et al., 2009) |
| LRPPRC | LRPPRC suppress Parkin mediated mitophagy and acts as a checkpoint protein that prevents mitochondria from autophagy degradation | NAD + → ↑SIRT3 → deacetylation of LRPPRC -→LRPPRC-POLMAT interactions with mtDNA | (Cui et al., 2019; Zou et al., 2014) |
| BNIP3 | Involved in hypoxia-induced mitophagy; regulates OPA1 disassembling, DRP1-mediated fission and recruitment of parkin to mitochondria to facilitate mitophagy; stabilizes PINK1. | NAD+ →↑SIRT1 → deacetylation FOXO3 → ↑BNIP3 | (Hariharan et al., 2010; Landes et al., 2010; Lee et al., 2011; Zhang et al., 2016c) |
| FUNDC1 | OMM protein: initiates hypoxia-induced mitophagy, where phosphorylation of FUNDC1 by CK2 and Src is inhibited and dephosphorylation by PAGM5 is stimulated, hereby allowing FUNDC1 to bind to LC3 and recruit DRP1/DNM1L. | ↑NAD+→ ↑SIRT3-PGAM5-FUNDC1→ ↑FUNDC1- dependent mitophagy | (Cheng et al., 2014; Liu et al., 2012; Yoshino et al., 2018) |
| Cardiolipin | IMM phospholipid: involved in cargo labelling, via interaction with LC3 following mitochondrial membrane rupture. | NAD + → CL binds SIRT5 ↑respiratory chain function | (Chu et al., 2013; Zhang et al., 2017) |
| AMPK | Integrator of mitochondrial homeostasis by controlling mitochondrial life cycle, biogenesis & mitophagy | AMPK→ ↑NAD + → ↑SIRT1 → ↑mitophagy | (Canto et al., 2009; Herzig and Shaw, 2018) |
| ULK1 | Ser/Thr kinase: Activation of AMPK or inhibition of mTOR induces composition of ULK1 initiation complex. | NAD+ →SIRT1 →↓mTOR →↑ULK1 → ↑mitophagy | (Dunlop and Tee, 2014; Egan et al., 2011; Lee et al., 2008) |
| p62 (SQSTM1) | Autophagy/mitophagy receptor: involved in cargo sequestration; degrades sperm-derived mitochondria in fruit flies and mammals. | NAD+ ↑SIRT1 ↑p62- transcription | (Cetrullo et al., 2016; Pankiv et al., 2007; Politi et al., 2014; Song et al., 2016) |
| Beclin 1 | Promotes formation of PI3KC3-C1 and regulates the lipid kinase. | NAD+ → ↑SIRT1 → ↑Beclin1 | (Ou et al., 2014; Wang et al., 2015) |
Abbreviations: PINK1PTEN induced kinase 1; FIS1fission, mitochondrial 1; OPA1OPA1 mitochondrial dynamin like GTPase; DRP1, dynamin-related protein; MFNmitofusin; LC3microtubule-associated protein 1 light chain 3; ATGautophagy-related; LRPPRCleucine rich pentatricopeptide repeat containing; BNIP3BCL2 interacting protein 3; FUNDC1FUN14 domain containing 1; AMPKAMP-activated protein kinase: OMM: outer mitochondrial membrane.
NAD+ does not only play a key-role in cellular energy metabolism and mitochondrial function, it is also involved as a co-factor for a broad array of signalling pathways (O’Bryant et al., 2015). NAD+ acts as a coenzyme for important proteins such as the sirtuins (SIRT1 to SIRT7), CD38, sterile alpha and TIR motif containing 1 (SARM1), and poly [ADP-ribose] polymerase (PARPs). SIRT1 is involved in the regulation of autophagy/mitophagy via multiple pathways namely: (1) deacetylating the autophagy proteins ATG5, ATG7, and LC3 (Huang et al., 2015; Lee et al., 2008); (2) via increase in levels of BECN1 expression, which initiates the class III phosphatidylinositol 3-kinase nucleation complex autophagy (Ou et al., 2014); (3) by enabling the de-acetylation and translocation of LC3 from nucleus to cytoplasm (Huang et al., 2015); (4) by acetylation and deacetylation of FOXO1 and FOXO3 via increased expression of autophagic proteins LC3, Atg12, BNIP3, and Rab7 (Hariharan et al., 2010; Kume et al., 2010; Sengupta et al., 2009); and by upregulation of the expression of proteins involved in autophagy/mitophagy, such as PINK1, Parkin, NIX, etc (Fang et al., 2019, 2016a; Fang et al., 2014). The small GTPase Rab7 interacts with the UVRAG-Vps 16 complex to facilitate autophagosome and endosome maturation (Zhan et al., 2017). Moreover, RAB7A phosphorylation by TBK1 has been shown to promote mitophagy via the PINK1-Parkin pathway (Heo et al., 2018). Furthermore, the NAD+-SIRT1 pathway may stimulate the autophosphorylation and activation of ataxia telangiectasia mutated (ATM), which induces mitophagy by a STK11/LKB1-AMPK-TSC2 pathway (Alexander et al., 2010; Dobbin et al., 2013; Fang, 2019; Zhang et al., 2016b). SIRT2 is a cytoplasmic and mitochondrial-localised NAD-dependent protein, which is a key regulator of mitochondrial function and initiation of mitophagy, in particular via deacetylation of E3 ubiquitin ligase, ATG5 (Liu et al., 2017). The mitochondrial SIRT3 induces mitophagy via the mitochondrial SIRT3PGAM5-FUNDC1-pathway (Fang, 2019; Ma et al., 2018; Onyango et al., 2002). SIRT4 is involved in the mitochondrial morphology/quality control in conjunction with OPA1 as well as regulation of mitophagy in a parkin-independent manner (Fang, 2019; Lang et al., 2017). The function of SIRT5 regulates, among other things, ammonia induced mitophagy (Fang, 2019; Polletta et al., 2015). SIRT6 and SIRT7 are nuclear proteins and they inhibit mTOR, thereby promoting autophagy (Fang, 2019; Takasaka et al., 2014; Xu et al., 2018). Besides sirtuins, CD38, another NAD+-consuming enzyme, plays a crucial role in autophagic fusion with lysosomes (Xiong et al., 2013). Members of the PARPs family, which are mainly involved in DNA repair and cell death through parthanatos, can lead, in conjunction with the cyclic ADP-ribose synthases like CD38, to NAD+ depletion and consequently limiting the activity of SIRT1, therefore decreasing mitophagy (Fang et al., 2014, 2016c). Collectively, such data suggest that NAD+ functions in a multifaceted manner but how the different pathways interact with each other have to be determined.
4. NAD+ in ageing and neurodegenerative diseases
4.1. NAD+ in healthy ageing
An age-related decline in NAD+ levels has been reported across species, ranging from yeast to human, and within multiple types of tissues (Fang et al., 2017, 2014; Mouchiroud et al., 2013). Thus, it can be postulated that replenishment of NAD+ levels may serve as an anti-ageing agent that promotes longevity. Indeed, studies utilising NAD+ precursors, NMN, NR, and NAM in laboratory animal model systems have proved to be effective in promoting healthspan and/or longevity (Canto et al., 2015; Frederick et al., 2016; Lautrup et al., 2019b; Mitchell et al., 2018; Rajman et al., 2018; Yoshino et al., 2018). In a yeast model, NR supplementation increased the replicative lifespan of wild-type yeast by more than ten generations (Belenky et al., 2007). The Nrk1 and the Urh1/Pnp1/Meu1 pathways were identified to induce NR-mediated extension of lifespan (Belenky et al., 2007). In C. elegans, NR extended the average lifespan of wild-type worms (N2) via SIR-2.1 (ortholog to mammalian SIRT1) (Fang et al., 2016b; Mouchiroud et al., 2013). In mice, administration of NR in two-year old mice resulted in extension of lifespan (Zhang et al., 2016a). NR-dependent boosting of mitochondrial function via the mitochondrial unfolded protein response (UPRmt) was identified to a contributor to longevity (Zhang et al., 2016a). NAM, another precursor for NAD+ and a key molecule involved in energy metabolism, was shown to increase lifespan in yeast and C. elegans, but not in WT mice. Higher doses of NAM have, however, been associated with reduced lifespan via inhibition of Sir2 activity (Bitterman et al., 2002; Gallo et al., 2004; Mitchell et al., 2018; Mouchiroud et al., 2013; Schmeisser et al., 2013; van der Horst et al., 2007). NAD+ augmentation does not only promote longevity, but it also has been shown to improve healthspan and prevent age-associated frailty in multiple laboratory models (Canto et al., 2015; Frederick et al., 2016; Lautrup et al., 2019b; Mills et al., 2016; Mitchell et al., 2018; Rajman et al., 2018; Yoshino et al., 2018). In particular, NR supplementation and aerobic exercise, both increase NAD+ levels, improve muscle function and aerobic performance (Crisol et al., 2019). Elevated NAD+ levels trigger SIRT1-mediated deacetylation of peroxisome proliferator-activated receptor-gamma coactivator 1-alpha (PGC-1α), which is associated with skeletal muscle mitochondrial biogenesis (Crisol et al., 2019). NMN improved mitochondrial health, immune function, lipid metabolism, muscle strength, motor and cardiac function (de Picciotto et al., 2016; Fang et al., 2016a; Gomes et al., 2013; Kerr et al., 2017b; Mills et al., 2016; Stein and Imai, 2014). Thus, these studies collectively converge to suggest that NAD+ replenishment promotes healthy longevity in laboratory animal models.
Notably, improvement of mitochondrial health and the NAD+ consuming sirtuins have been identified as common factors coinciding with extension of lifespan and improvement of healthspan. NAD+ has been shown to bolster mitochondrial function via mitochondrial biogenesis as well as degradation of damaged and/or dysfunctional mitochondria through the process of mitophagy. The sirtuins have been shown to promote mitochondrial biogenesis in mouse skeletal muscle tissues, in response to increase in NAD+ levels, via the SIRT1/PGC-1α pathway (Crisol et al., 2019). The NAD+ precursors increase NAD+ levels that is utilised by SIRT1 to deacetylate and thus activate PGC-1α. Deacetylated PGC-1α is translocated to the nucleus where phosphorylation by AMPK triggers the expression of NRF-2 and TFAM, which in turn upregulate mitochondrial biogenesis (Yuan et al., 2016). In addition, the NAD+/SIRT1 pathway may also trigger autophagy/mitophagy via multiple pathways (Fivenson et al., 2017): (1) activation of AMPK, that in turn phosphorylates ULK1, the human autophagy protein 1 (hATG1) (Egan et al., 2011; Kim et al., 2011); (2) activation of AMPK which in turn phosphorylates Tuberous Sclerosis Complex 2 (Tsc2) at Ser1387, and thereby inhibiting the autophagy inhibitor mTOR (Inoki et al., 2003); (3) deacetylation of Foxo1, which induces upregulation of the autophagic protein Rab7 that mediates late autophagosome–lysosome fusion (Hariharan et al., 2010). In fact, AMPK activation and dietary restriction, which also increases NAD+ levels, has been shown to increase lifespan in C. elegans via maintenance of mitochondrial network homeostasis (Mitchell et al., 2016; Weir et al., 2017). Altogether, these findings indicate a key role of NAD+ in longevity by regulating the balance between mitochondrial biogenesis and removal of damaged mitochondria via mitophagy in the effort to maintain healthy and functional mitochondria.
4.2. Premature ageing
NAD+ depletion is not a phenomenon solely associated with normal ageing but is also a common feature underlying various premature ageing conditions. As a result, laboratory models of premature ageing have also provided great insight into the potential of NAD+ in promoting longevity. In the C. elegans model of Xeroderma pigmentosum group A (XPA, a nucleotide excision DNA repair (NER) disorder with severe neurodegeneration), and Ataxia telangiectasia (A–T, due to mutation of ATM which encodes a master regulator of DNA damage response), replenishment of NAD+, using its precursors, was able to extend lifespan and improve healthspan (Fang and Bohr, 2017; Fang et al., 2016b, 2014). In addition, motor deficits in the Atm−/− mice were rescued and lifespan was shown to be extended following NR supplementation (Fang et al., 2016b). NAD+ augmentation was able to restore mitochondrial function, resulting in enhanced neuronal survival and improved motor function in models of premature ageing. In particular, activities of SIRT1 and PGC-1α were elevated along with enhanced DNA repair, which were identified as the underlying mechanisms upon NAD+ supplementation. Similar phenotypes were reported in models of increased replication stress suggesting an improvement in nuclear-to-mitochondria communication as a possible pathway to healthy longevity in the animal models (Fang et al., 2016c). Collectively, these findings highlight mitochondrial dysfunction and impaired mitophagy as a fundamental underlying cause of accelerated ageing, which can be rescued upon NAD+ replenishment.
4.3. Neurodegenerative diseases
Recent findings have highlighted the depletion of NAD+ and compromised mitophagy as contributors to neurodegenerative diseases such as AD, Parkinson’s disease (PD), amyotrophic lateral sclerosis (ALS), and Huntington’s disease (HD) (Fang et al., 2019; Lautrup et al., 2019b; Lloret and Beal, 2019). AD is the most common form of dementia affecting over 45 million people worldwide, a figure estimated to triple by 2050, representing a major healthcare and socio-economic challenge (Prince et al., 2016). It is clinically characterized by an insidious onset and a gradual and progressive deterioration of cognitive abilities ranging from loss of memory to impairment of judgement and reasoning (Canter et al., 2016; Kerr et al., 2017b). The neuropathological hallmarks of AD are extracellular amyloid plaques composed of β-amyloid (Aβ) aggregates and intracellular neurofibrillary tangles (NFT) containing aberrantly hyper-phosphorylated microtubule associated protein tau (MAPT). These changes are accompanied by neuronal loss, synaptic dysfunction, and neuroinflammation (Hou et al., 2017).
Emerging data have shown NAD+ depletion and metabolic dysfunction in mouse models of early-onset familial AD as well as in rat cortical neurons challenged with Aβ (Dong and Brewer, 2019; Fang et al., 2019; Hou et al., 2018; Liu et al., 2013). In fact, NAD+ augmentation inhibited the AD-associated pathology and cognitive decline in multiple AD models, namely, 3xTgAD mice treated with NAM, 3xTgAD/Polβ+/− mice treated with NR, and in Aβ1–42 and the neuronal Tau (pro-aggregating F3ΔK280 tau fragment) transgenic C. elegans models (Fang et al., 2019; Hou et al., 2018; Liu et al., 2013; Sorrentino et al., 2017). In addition, NAD+ augmentation restored mitochondrial function and ameliorated inflammation, synaptic loss as well as protected against neuronal cell death (Gong et al., 2013; Hou et al., 2018; Long et al., 2015; Schondorf et al., 2018; Sorrentino et al., 2017; Wang et al., 2016; Yao et al., 2017). Thus, implicating the contribution of NAD+ depletion in the development and progression of AD in animal models.
In the amyloidogenic models of AD, NAD+ augmentation resulted in rescue of cognitive decline and reduction of Aβ pathology. The underlying mechanisms indicated for reduced Aβ pathology upon NAD+ augmentation were via PGC-1α mediated modulation of β-secretase 1 (BACE-1) activity and/or increased phagocytosis of Aβ by microglia (Fang, 2019; Fang et al., 2019; Lautrup et al., 2019a; Sorrentino et al., 2017). In addition, improvement of mitochondrial proteostasis and mitophagy had the major benefit of NAD+ augmentation (Fang, 2019; Fang et al., 2019; Lautrup et al., 2019a; Sorrentino et al., 2017). In the APP/PS1 mouse model of AD, CD38 depletion resulted in elevation of NAD+ levels that were associated with decrease in Aβ pathology and neuroinflammation accompanied by improvement of spatial learning behaviour (Blacher et al., 2015). CD38 is an NAD+ hydrolase, which increases with the process of ageing, and contributes significantly to age-related decline in NAD+ levels and mitochondrial dysfunction via inhibition of NAD+/SIRT3 activity (Camacho-Pereira et al., 2016). Collectively, NAD+ augmentation improves memory in different APP/PS1 CE mouse models, possibly through reducing Aβ pathology and promoting mitochondrial health and function.
Further evidence stemming from the 3xTgAD mouse studies display reduced phosphorylated tau and rescue of cognitive decline upon NAD+ augmentation (Fang et al., 2019; Hou et al., 2018; Misiak et al., 2017). NAD+ augmentation contributes to neurogenesis and inhibition of AD-associated pathology, neuroinflammation and mitochondrial dysfunction via mitophagy and possibly DNA repair enhancement (through SIRT3 and SIRT6) (Fang et al., 2019; Hou et al., 2018). Thus, NAD+ induced mitophagy is implicated as a key underlying mechanism in rescuing the AD phenotype.
Mitophagy deficits and accumulation of mitochondria were in fact detected in AD human post-mortem tissue as evidenced by reduced phosphorylation of mitophagy initiation proteins, namely ULK1 and TBK1 (Fang et al., 2019). Both ULK1 and TBK1 play an important role induction of mitophagy via recognition and sequestering of damaged mitochondria (Wang et al., 2019). Further evidence from C. elegans, murine and human cell lines overexpressing WT and/or mutant tau implicate impaired mitophagy, which leads to accumulation of damaged mitochondria and functionally to cognitive deficits (Cummins et al., 2019; Fang et al., 2019). Overexpression of WT htau in human cell lines revealed reduced levels of Parkin in the mitochondrial fraction accompanied by localisation of tau in the outer membrane of the mitochondria (Hu et al., 2016). Moreover, upon upregulation of Parkin, the mitophagy deficits were rescued. Thus, suggesting that the insertion of tau in the outer membrane may reduce the interaction between Parkin and the mitochondria (Hu et al., 2016; Perez et al., 2018). However, a recent study highlights an alternative mechanism whereby tau specifically impairs clustering and recruitment of Parkin to the defective mitochondria via sequestering it in the cytosol. The sequestering was indicated to be resulting from an aberrant interaction between the projection domain of tau and Parkin. As a consequence, inhibiting the recognition of damaged mitochondria by the mitophagy machinery; thereby inhibiting the degradation of damaged mitochondria (Cummins et al., 2019). Collectively, these studies suggest that an overexpression of tau impairs the recognition of damaged mitochondria due to impaired Parkin-mitochondria interaction, resulting in accumulation of dysfunctional mitochondria.
Moreover, reduced basal mitophagy as well as impaired induction of mitophagy in response to mitochondrial stress has been shown in C. elegans harbouring pan-neuronal overexpression of human WT and/or mutant tau displayed (Cummins et al., 2019; Fang et al., 2019). Although, administration of mitophagy inducers, small compounds urolithin A (UA) and actinonin (AC), rescued the memory deficits in C. elegans harbouring pan-neuronal expression of tau as well as in the 3xTgAD mouse model of AD (Fang et al., 2019). Furthermore, UA and AC inhibited multiple tau phosphorylation sites in the hippocampi of 3xTgAD mice as well as in human cell lines overexpressing WT htau. Interestingly, inhibition of tau phosphorylation in human cell lines and rescue of memory loss in C. elegans were demonstrated to be strictly mitophagy dependent as specific knockdown of mitophagy initiation genes (cells: ULK1, PINK1; C. elegans: pink1, parkin/pdr-1, nix/dct-1) failed to show the benefits of mitophagy inducing small compounds, like NMN (Fang et al., 2019). How does NAD+ and mitophagy reduce total Tau and p-Tau? One possibility could be autophagic clearance of Tau tangles by NAD+ augmentation (Fang et al., 2019). In line with our study, a recent study shows that p-Tau aggregates were observed localised to mitochondria and within mitophagic vesicles, a possible mode of clearance of tau aggregates (Grassi et al., 2019). Furthermore, we also show that NAD+-induced mitophagy inhibited NLRP3 inflammasome activation in the 3xTgAD mice (Fang et al., 2019). Since NLRP3 inflammasome activation is a driver of Tau pathology, thus NAD+ (including NAD+-induced mitophagy and possibly other NAD+-dependent but mitophagy-independent pathways) may also inhibit Tau pathology through elimination of NLRP3 inflammasome activation. However, whether a possibility of kinases or phosphates directly regulated by NAD+ or mitophagy that could reduce tau phosphorylation needs to be further determined.
Although these data implicate the beneficial effect of boosting mitophagy in AD, partially NAD+ mediated, it remains to be considered as a viable therapeutic strategy. Moreover, studies revealing inhibition of tau phosphorylation and reduction in Aβ pathology following NAD+ augmentation; however, whether NAD+ is inhibiting the development and/or contributing to degradation of these pathological protein deposits remains elusive. For the purpose of translation to humans, the latter is vital as by the time the clinical features of AD are presented by patients, the pathology is well established. Thus, if NAD+ augmentation has the ability to degrade established pathological deposits and promote neurogenesis, it would be a good candidate for AD prevention and/or slowing down the progression of disease.
5. Clinical translation of NAD+ precursors and precautions
5.1. Clinical trials
Encouraging evidence from preclinical models has led to initiation of several clinical trials using NAD+ precursors. NR is orally bioavailable, and it is well-tolerated and elevates NAD+ in healthy middle aged and older adults as well as in old diabetic males. In particular, Trammell et al. reported that oral administration of NR (1000 mg/day for seven days) led to an increase in blood concentration of NAD+ by 2.7-fold and 45.5-fold increase in nicotinic acid adenine dinucleotide (NAAD), an NAD+ biosynthesis intermediate (Trammell et al., 2016). Further evidence arising from an eight-week randomized, double-blind, placebo-controlled study in 120 healthy adults (60–80 years old) revealed that NR (250 mg and 500 mg) induced dose-dependent increase of blood NAD+ levels that became apparent after 4-weeks and was sustained till the end of the study, without any serious adverse effects (Dellinger et al., 2017). In addition, a 2 × 6-week randomized, double-blind, placebo-controlled crossover clinical trial conducted in 55–79 years old individuals showed NR (oral 500 mg/time, twice a day) to be well tolerated and was not only limited to effectively elevating NAD+ levels in healthy adults, but also improve cardiovascular function (Martens et al., 2018). Dollerup et al. showed that 12 weeks of NR supplementation in doses of 2 g/day appears to be safe in diabetic aged men (Dollerup et al., 2018). A more recent study from aged men found good tolerance of NR (1000 mg/day) over a period of 21 days, which resulted in downregulation of energy metabolism and mitochondria pathways as well as an anti-inflammatory effect in skeletal muscle (Elhassan et al., 2019). Collectively, these findings indicate that NR supplementation (up to 2 g/day for up to 3 months) is orally bioavailable and safe. Thus, clinical trials of NAD+ precursors on age-related diseases, such as diabetes, cardiovascular disease, premature ageing diseases, and neurodegenerative diseases are in progress (Lautrup et al., 2019b; Rajman et al., 2018; Yoshino et al., 2018).
5.2. NAD+ supplementation - precaution
Whilst encouraging findings suggesting NAD+ supplementation to be orally bioavailable and safe in healthy individuals; there may be some concerns with regards to administration in certain conditions such as cancer. The expression of major NAD+ metabolic enzymes, namely, iNAMPT, eNAMPT, and/or NMNAT2, are upregulated in a various cancers including: breast, pancreatic, prostate and colorectal cancers (Demarest et al., 2019). Preclinical studies report that elevated levels of NAD+, via NAD+ synthetic enzymes or NAD+ precursors supplementation, promotes cancer cell proliferation, migration, and resistance to anticancer chemo-/radiotherapy (Demarest et al., 2019). Thus, strategies to deplete cellular NAD+ and the inhibition of NAD+ synthetic enzymes can trigger cancer cell death (Buonvicino et al., 2018). Specifically, in pancreatic cancer, cell growth has been shown to be dependent on NAD+ salvage pathway (Chini et al., 2014). Therefore, inhibition of NAD+ synthesis (via Nampt inhibition) and/or promotion of its consumption (via CD38 NADase) inhibits some forms of cancer cell growth (Chini et al., 2014; Ju et al., 2016). A recent study shows that high NAD+ levels may lead to cancer promotion via changes in the secretory activity of senescent cells to release pro-inflammatory cytokines (Nacarelli et al., 2019). Therefore, these studies collectively indicate that there should be a thorough evaluation before the use of and supplementation as this may be a contributor rather than a countermechanism in certain conditions.
6. The application of artificial intelligence to facilitate drug development targeting on the NAD+-mitophagy axis
Development of artificial intelligence (AI), in particular, recent advancement in deep learning techniques, has returned dividend in superior performance in solving computer vision, natural language processing and robotics problems (Schmidhuber, 2015). More recently, AI, including conventional machine learning and newly developed deep learning techniques, have been successfully used in the drug discovery and development pipeline with certain requirements of the data characteristics, e.g., data quality and quantity (Vamathevan et al., 2019). Traditionally, the drug discovery and development process is costly, long, and complicated including drug target identification and validation, compound screening and lead compound discovery (hit to lead generation and lead optimization) within several preclinical and clinical study cycles (Vohora and Singh, 2018). According to DiMasi et al.’s latest research (DiMasi et al., 2016), the mean pre-tax cost of developing a novel prescription drug is around 2.6 billion USD and a complete traditional workflow can take about 10–15 years (Turner, 2010). Although there is huge investment in drug discovery and development, the failure rate is still very high (Takebe et al., 2018).
Essentially, the successful deployment of machine learning and deep learning models is based on reliable data quality and sufficient amount of data, appropriate design of computational algorithms, and prevalence of powerful computing resources such as graphical processing units (GPUs) and tensor processing units (TPUs) for accelerated parallel processing. In general, the models are trained using labelled data (supervised learning) or they can directly learn the characteristics from the unlabelled data (unsupervised learning). Although currently there is no AI algorithm proposed for NAD+ specific drug discovery and mitophagy in ageing, we can envisage such development in near future using the experience gained from previously proposed AI based drug discovery and biomarker identification methods. Here we briefly review AI technologies in drug discovery and biomarker development process and provide a prospective view on AI technologies for understanding roles and molecular mechanisms of NAD+ and mitophagy in ageing.
6.1. Drug target identification and validation
Most diseases (i.e., AD) are caused by protein dysfunctions, and the target identification aims to establish a causal association between the target and the disease. The structure-based drug design methods can be used to discover active small molecules towards the protein targets, but the procedure could be slow. AI algorithms can be used to make predictions of the 3D structure of a protein. Costa et al. proposed a decision tree based method to predict genes associated with morbidity and druggability (Costa et al., 2010). Jeon et al. used supervised support vector machine (SVM) for drug and non-drug targets classification for various cancers (Jeon et al., 2014). Recently, Wang et al. developed a convolutional neural network (CNN) based method for predicting the residue contacts of proteins, which outperformed traditional approaches (Wang et al., 2017). Du et al. applied deep learning for sequence features extraction to predict protein-protein interaction (Du et al., 2016). In addition, natural language processing based techniques have been advanced to identify drug–disease, gene–disease and target–drug associations in the medical literature (Bravo et al., 2015; Kim et al., 2017). In ageing-related studies, deep learning has been used to identify compounds with potential pro-longevity properties, and features used by ageing clocks can potentially be learnt by machines to find new targets (Aliper et al., 2016). Several machine learning based methods have also been proposed to evaluate blood markers, imaging, omics, and epigenetic biomarkers, for age prediction (Bobrov et al., 2018; Mamoshina et al., 2018; Moskalev and Vaiserman, 2018; Putin et al., 2016). Thus, the use of AI may not only provide grounds for orchestration of novel hypothesis in pursuit to unveiling the regulation of NAD+ dependent signalling networks in both health and disease; but it may also possibly pave path for drug discovery and biomarkers development.
6.2. Compound screening and lead compound discovery
A crucial step in drug discovery is the selection of drug candidates that can block or activate the target protein of interest and possess expected characteristics, e.g., bioavailability, bioactivity, and toxicity. These will involve comprehensive virtual and experimental high-throughput screening of large compound libraries (Vamathevan et al., 2019). SVM-based machine learning methods and deep learning methods (e.g., deep neural networks [DNN] and 3D graph CNN) have been used for virtual screening and activity scoring that have reproducible results with superior performance compared to the methods without using AI (Carpenter et al., 2018; Leelananda and Lindert, 2016; Pereira et al., 2016). In addition, drug repurposing (also known as drug repositioning) is a commonly applied alternative strategy to find new targets or novel indications of the approved drugs that can accelerate the drug discovery procedure and reduce the risk of drug development. Advanced AI based approaches have been applied for computational drug repurposing (Luo et al., 2017; Vanhaelen, 2018; Vanhaelen et al., 2017; Yella et al., 2018). Recently, deep learning based methods have been considered in this context, e.g., Aliper et al. designed a DNN based system to classify drugs into different therapeutic categories based on the transcriptomic data alone (Aliper et al., 2016).
6.3. Development of biomarkers
In view of the strong relationship between NAD+ depletion and ageing as well as diseases (i.e., AD); and the potent therapeutic benefits of NAD+ augmentation on such conditions (at least in animal studies as mentioned above), we are working on a possibility to develop the use of blood NAD+ levels, and possibly NAD+-related metabolite(s), as biomarker(s) for ageing and age-related diseases, such as AD. Such biomarkers could be potentially used for the early detection of disease. Furthermore, it could be utilised for measuring rate of change and response to NAD+ related therapeutic intervention, namely, precision medicine. Precision medicine is an emerging approach for disease treatment and prevention that takes into account individual variability. As NAD+ signalling has been suggested as therapeutic intervention in neurodegenerative disease, a compound marker indicating a specific response to NAD+ might help to select individuals who are more likely to respond to such intervention.
The development of biomarkers requires two critical steps. Firstly, is the availability of high dimensionality platform, which could measure NAD+ and its downstream metabolic or proteomic signature. Due to the inherent heterogeneity of the disease, a single biomarker is unable to achieve high accuracy, multimodal biomarkers (e.g. combining both metabolites or proteins) may have greater sensitivity and specificity as a biomarker; and may collectively better reflect and define the disease and its pathology. Secondly, the management and analysis of high-dimensional data. AI methods, such as Support Vector Machine (SVM), a supervised machine learning algorithm, are commonly used in biomarker discovery (Dragomir et al., 2019; Eke et al., 2018). With the rapid evolvement of modern AI technologies, more advanced machine learning algorithms such as lightGBM and Deep Learning based algorithms are developed and widely used in recent years. These methods ought to be employed into the development of NAD+ related biomarkers in ageing and AD to provide better performance and efficiency.
6.4. Efficacy and safety assessment
It is important to perform efficacy and safety assessment for the treatments and drugs that have been developed. Longitudinal studies on patients’ feedback can provide telling information to improve the drug development by optimising the drug discovery procedure. However, in AD and ageing related studies, the actual effects may take years to be revealed and the validation for the effects of anti-ageing treatment might not be straightforward (Zhavoronkov et al., 2019). There is still on-going debate on how to validate AI based algorithms fast and reliably in general healthcare settings, and inputs from domain experts should absolutely be helpful.
Currently, the revolution in AI that has been witnessed by the successful applications in computer vision is speedily disseminating in healthcare and drug discovery and development research. In particular, in ageing related studies, deep learning has been widely used in early diagnosis using multimodal data acquired from various imaging modalities, clinical records, patient’s medical history and etc. There are ongoing research studies on biomarker development using deep learning, which are very promising. How we can utilise deep learning to understand roles and molecular mechanisms of NAD+ in ageing, mitochondrial homeostasis, and mitophagy is still an open question. Regardless how fast the technology can advance, we should perform comprehensive testing on the developed AI methods and deploy them with discretion.
7. Outstanding questions and future perspectives
The inevitable process of ageing is a major cause of various human diseases. Recent findings in preclinical animal models have improved our understanding of NAD+ and NAD+-dependent enzymes in mitochondrial homeostasis, mitophagy and neurodegenerative disorders. Emerging evidence suggest impairment of the NAD+-mitophagy axis as a common feature underlying ageing that is exacerbated in neurodegenerative diseases such as AD. NAD+ augmentation promoted mitophagy and mitochondrial function as well as inhibited AD-associated pathology and cognitive deficits. In particular, the rescue of behavioural phenotype upon NAD+ augmentation was identified to be mitophagy dependent. However, the intricate interaction within the NAD+-mitophagy axis in health and disease is still not fully understood. For this purpose, it is important to initially characterise of NAD+ signalling networks in mitophagy using preclinical models systems (animals and stem cells). Secondly, it is of great significance to uncouple and define how this NAD+-mitophagy axis is altered during the process of ageing and disease. Subsequently, AI can be utilised for: 1) understanding the intricacies of NAD+ signalling networks in mitophagy and novel hypothesis building (Fig. 1); 2) drug discovery; and 3) development of novel biomarkers of ageing and disease. Thus, the combination of traditional drug development with the involvement of AI and machine learning will accelerate the development of biomarkers and drugs for ageing and age-related diseases.
Acknowledgements
The authors acknowledge the valuable work of the many investigators whose published articles they were unable to cite owing to space limitations. The authors thank Dr. Cheonglong Xie and Dr. Guofeng Lou for reading of the manuscript. E.F.F. is supported by the HELSE SØR-ØST (#2017056), the Research Council of Norway (#262175and#277813), the National Natural Science Foundation of China (#81971327), an Akershus University Hospital Strategic grant (#269901), and a Rosa Sløyfe grant (#207819) by the Norwegian Cancer Society. L.J.R. is supported by Nordea-fonden and Novo Nordisk Fonden Challenge Programme: Harnessing the Power of Big Data to Address the Societal Challenge of Aging NNF17OC0027812, and the Olav Thon Foundation. Some of the work described has been supported by funds from the Intramural Program of the National Institute on Aging, NIH, USA (V.A.B.). H.N. was supported by grants from HELSE SØR-ØST (#2019098) and Bergesenstiftelsen.
Footnotes
Declaration of Competing Interest
E.F.F., H.N., V.A.B. have CRADA arrangements with ChromaDex. E.F.F., H.N., L.H.B., J.S.M., L.J.R. are consultants to Aladdin Healthcare Technologies. E.F.F. is consultant to Vancouver Dementia Prevention Centre.
References
- Alexander A, Cai SL, Kim J, Nanez A, Sahin M, MacLean KH, Inoki K, Guan KL, Shen J, Person MD, et al. , 2010. ATM signals to TSC2 in the cytoplasm to regulate mTORC1 in response to ROS. Proc. Natl. Acad. Sci. U. S. A. 107, 4153–4158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aliper A, Plis S, Artemov A, Ulloa A, Mamoshina P, Zhavoronkov A, 2016. Deep learning applications for predicting pharmacological properties of drugs and drug repurposing using transcriptomic data. Mol. Pharm. 13, 2524–2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belenky P, Racette FG, Bogan KL, McClure JM, Smith JS, Brenner C, 2007. Nicotinamide riboside promotes Sir2 silencing and extends lifespan via Nrk and Urh1/Pnp1/Meu1 pathways to NAD+. Cell 129, 473–484. [DOI] [PubMed] [Google Scholar]
- Bitterman KJ, Anderson RM, Cohen HY, Latorre-Esteves M, Sinclair DA, 2002. Inhibition of silencing and accelerated aging by nicotinamide, a putative negative regulator of yeast sir2 and human SIRT1. J. Biol. Chem. 277, 45099–45107. [DOI] [PubMed] [Google Scholar]
- Blacher E, Dadali T, Bespalko A, Haupenthal VJ, Grimm MO, Hartmann T, Lund FE, Stein R, Levy A, 2015. Alzheimer’s disease pathology is attenuated in a CD38-deficient mouse model. Ann. Neurol. 78, 88–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobrov E, Georgievskaya A, Kiselev K, Sevastopolsky A, Zhavoronkov A, Gurov S, Rudakov K, Del Pilar Bonilla, Tobar M, Jaspers S, Clemann S, 2018. PhotoAgeClock: deep learning algorithms for development of non-invasive visual biomarkers of aging. Aging (Albany NY) 10, 3249–3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogan KL, Brenner C, 2008. Nicotinic acid, nicotinamide, and nicotinamide riboside: a molecular evaluation of NAD+ precursor vitamins in human nutrition. Annu. Rev. Nutr. 28, 115–130. [DOI] [PubMed] [Google Scholar]
- Bravo A, Pinero J, Queralt-Rosinach N, Rautschka M, Furlong LI, 2015. Extraction of relations between genes and diseases from text and large-scale data analysis: implications for translational research. BMC Bioinformatics 16, 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buonvicino D, Mazzola F, Zamporlini F, Resta F, Ranieri G, Camaioni E, Muzzi M, Zecchi R, Pieraccini G, Dolle C, et al. , 2018. Identification of the nicotinamide salvage pathway as a new toxification route for antimetabolites. Cell Chem. Biol. 25 (471–482), e477. [DOI] [PubMed] [Google Scholar]
- Calvo SE, Mootha VK, 2010. The mitochondrial proteome and human disease. Annu. Rev. Genomics Hum. Genet. 11, 25–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho-Pereira J, Tarrago MG, Chini CCS, Nin V, Escande C, Warner GM, Puranik AS, Schoon RA, Reid JM, Galina A, et al. , 2016. CD38 dictates agerelated NAD decline and mitochondrial dysfunction through an SIRT3-Dependent mechanism. Cell Metab. 23, 1127–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campisi J, Kapahi P, Lithgow GJ, Melov S, Newman JC, Verdin E, 2019. From discoveries in ageing research to therapeutics for healthy ageing. Nature 571, 183–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canter RG, Penney J, Tsai LH, 2016. The road to restoring neural circuits for the treatment of Alzheimer’s disease. Nature 539, 187–196. [DOI] [PubMed] [Google Scholar]
- Canto C, Gerhart-Hines Z, Feige JN, Lagouge M, Noriega L, Milne JC, Elliott PJ, Puigserver P, Auwerx J, 2009. AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature 458, 1056–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canto C, Houtkooper RH, Pirinen E, Youn DY, Oosterveer MH, Cen Y, Fernandez-Marcos PJ, Yamamoto H, Andreux PA, Cettour-Rose P, et al. , 2012. The NAD(+) precursor nicotinamide riboside enhances oxidative metabolism and protects against high-fat diet-induced obesity. Cell Metab. 15, 838–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canto C, Menzies KJ, Auwerx J, 2015. NAD(+) metabolism and the control of energy homeostasis: a balancing act between mitochondria and the nucleus. Cell Metab. 22, 31–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter KA, Cohen DS, Jarrell JT, Huang X, 2018. Deep learning and virtual drug screening. Future Med. Chem. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cetrullo S, D’Adamo S, Guidotti S, Borzi RM, Flamigni F, 2016. Hydroxytyrosol prevents chondrocyte death under oxidative stress by inducing autophagy through sirtuin 1-dependent and -independent mechanisms. Biochim. Biophys. Acta 1860, 1181–1191. [DOI] [PubMed] [Google Scholar]
- Cheng L, Sharples RA, Scicluna BJ, Hill AF, 2014. Exosomes provide a protective and enriched source of miRNA for biomarker profiling compared to intracellular and cell-free blood. J. Extracell. Vesicles 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chini CC, Guerrico AM, Nin V, Camacho-Pereira J, Escande C, Barbosa MT, Chini EN, 2014. Targeting of NAD metabolism in pancreatic cancer cells: potential novel therapy for pancreatic tumors. Clin. Cancer Res. 20, 120–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu CT, Ji J, Dagda RK, Jiang JF, Tyurina YY, Kapralov AA, Tyurin VA, Yanamala N, Shrivastava IH, Mohammadyani D, et al. , 2013. Cardiolipin externalization to the outer mitochondrial membrane acts as an elimination signal for mitophagy in neuronal cells. Nat. Cell Biol. 15, 1197–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa PR, Acencio ML, Lemke N, 2010. A machine learning approach for genomewide prediction of morbid and druggable human genes based on systems-level data. BMC Genomics 11 (Suppl 5), S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crisol BM, Veiga CB, Braga RR, Lenhare L, Baptista IL, Gaspar RC, Munoz VR, Cordeiro AV, da Silva ASR, Cintra DE, et al. , 2019. NAD(+) precursor increases aerobic performance in mice. Eur. J. Nutr. [DOI] [PubMed] [Google Scholar]
- Cui J, Wang L, Ren X, Zhang Y, Zhang H, 2019. LRPPRC: a multifunctional protein involved in energy metabolism and human disease. Front. Physiol. 10, 595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummins N, Tweedie A, Zuryn S, Bertran-Gonzalez J, Gotz J, 2019. Disease-associated tau impairs mitophagy by inhibiting Parkin translocation to mitochondria. EMBO J. 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S, Mitrovsky G, Vasanthi HR, Das DK, 2014. Antiaging properties of a grape-derived antioxidant are regulated by mitochondrial balance of fusion and fission leading to mitophagy triggered by a signaling network of Sirt1-Sirt3-Foxo3-PINK1PARKIN. Oxid. Med. Cell. Longev. 2014, 345105. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- de Picciotto NE, Gano LB, Johnson LC, Martens CR, Sindler AL, Mills KF, Imai S, Seals DR, 2016. Nicotinamide mononucleotide supplementation reverses vascular dysfunction and oxidative stress with aging in mice. Aging Cell 15, 522–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellinger RW, Santos SR, Morris M, Evans M, Alminana D, Guarente L, Marcotulli E, 2017. Repeat dose NRPT (nicotinamide riboside and pterostilbene) increases NAD(+) levels in humans safely and sustainably: a randomized, double-blind, placebo-controlled study. NPJ Aging Mech. Dis. 3, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demarest TG, Truong GTD, Lovett J, Mohanty JG, Mattison JA, Mattson MP, Ferrucci L, Bohr VA, Moaddel R, 2019. Assessment of NAD(+)metabolism in human cell cultures, erythrocytes, cerebrospinal fluid and primate skeletal muscle. Anal. Biochem. 572, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiMasi JA, Grabowski HG, Hansen RW, 2016. Innovation in the pharmaceutical industry: new estimates of R&D costs. J. Health Econ. 47, 20–33. [DOI] [PubMed] [Google Scholar]
- Dobbin MM, Madabhushi R, Pan L, Chen Y, Kim D, Gao J, Ahanonu B, Pao PC, Qiu Y, Zhao Y, et al. , 2013. SIRT1 collaborates with ATM and HDAC1 to maintain genomic stability in neurons. Nat. Neurosci. 16, 1008–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dollerup OL, Christensen B, Svart M, Schmidt MS, Sulek K, Ringgaard S, Stodkilde-Jorgensen H, Moller N, Brenner C, Treebak JT, et al. , 2018. A randomized placebo-controlled clinical trial of nicotinamide riboside in obese men: safety, insulin-sensitivity, and lipid-mobilizing effects. Am. J. Clin. Nutr. 108, 343–353. [DOI] [PubMed] [Google Scholar]
- Dong Y, Brewer GJ, 2019. Global metabolic shifts in age and alzheimer’s disease mouse brains pivot at NAD +/NADH redox sites. J. Alzheimers Dis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragomir A, Vrahatis AG, Bezerianos A, 2019. A network-based perspective in alzheimer’s disease: current state and an integrative framework. IEEE J. Biomed. Health Inform. 23, 14–25. [DOI] [PubMed] [Google Scholar]
- Du T, Liao L, Wu CH, Sun B, 2016. Prediction of residue-residue contact matrix for protein-protein interaction with Fisher score features and deep learning. Methods 110, 97–105. [DOI] [PubMed] [Google Scholar]
- Dunlop EA, Tee AR, 2014. mTOR and autophagy: a dynamic relationship governed by nutrients and energy. Semin. Cell Dev. Biol. 36, 121–129. [DOI] [PubMed] [Google Scholar]
- Egan DF, Shackelford DB, Mihaylova MM, Gelino S, Kohnz RA, Mair W, Vasquez DS, Joshi A, Gwinn DM, Taylor R, et al. , 2011. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science 331, 456–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eke CS, Jammeh E, Li X, Carroll C, Pearson S, Ifeachor E, 2018. Identification of optimum panel of blood-based biomarkers for alzheimer’s disease diagnosis using machine learning. Conf. Proc. IEEE Eng. Med. Biol. Soc. 2018, 3991–3994. [DOI] [PubMed] [Google Scholar]
- Elhassan YS, Kluckova K, Fletcher RS, Schmidt MS, Garten A, Doig CL, Cartwright DM, Oakey L, Burley CV, Jenkinson N, et al. , 2019. Nicotinamide riboside augments the aged human skeletal muscle NAD(+) metabolome and induces transcriptomic and anti-inflammatory signatures. Cell Rep. 28, 1717–1728 e1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang EF, 2019. Mitophagy and NAD(+) inhibit alzheimer disease. Autophagy 15, 1112–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang EF, Bohr VA, 2017. NAD+: the convergence of DNA repair and mitophagy. Autophagy 13, 442–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang EF, Hou Y, Palikaras K, Adriaanse BA, Kerr JS, Yang B, Lautrup S, Hasan-Olive MM, Caponio D, Dan X, et al. , 2019. Mitophagy inhibits amyloid-beta and tau pathology and reverses cognitive deficits in models of Alzheimer’s disease. Nat. Neurosci. 22, 401–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang EF, Kassahun H, Croteau DL, Scheibye-Knudsen M, Marosi K, Lu H, Shamanna RA, Kalyanasundaram S, Bollineni RC, Wilson MA, et al. , 2016a. NAD(+) replenishment improves lifespan and Healthspan in Ataxia telangiectasia models via Mitophagy and DNA repair. Cell Metab. 24, 566–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang EF, Kassahun H, Croteau DL, Scheibye-Knudsen M, Marosi K, Lu H, Shamanna RA, Kalyanasundaram S, Bollineni RC, Wilson MA, et al. , 2016b. NAD+ replenishment improves lifespan and Healthspan in Ataxia telangiectasia models via Mitophagy and DNA repair. Cell Metab. 24, 566–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang EF, Lautrup S, Hou Y, Demarest TG, Croteau DL, Mattson MP, Bohr VA, 2017. NAD(+) in aging: molecular mechanisms and translational implications. Trends Mol. Med. 23, 899–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang EF, Scheibye-Knudsen M, Brace LE, Kassahun H, SenGupta T, Nilsen H, Mitchell JR, Croteau DL, Bohr VA, 2014. Defective mitophagy in XPA via PARP-1 hyperactivation and NAD(+)/SIRT1 reduction. Cell 157, 882–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang EF, Scheibye-Knudsen M, Chua KF, Mattson MP, Croteau DL, Bohr VA, 2016c. Nuclear DNA damage signalling to mitochondria in ageing. Nat. Rev. Mol. Cell Biol. 17, 308–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang EF, Scheibye-Knudsen M, Jahn HJ, Li J, Ling L, Guo H, Zhu X, Preedy V, Lu H, Bohr VA, et al. , 2015. A research agenda for aging in China in the 21st century. Ageing Res. Rev. 24, 197–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fivenson EM, Lautrup S, Sun N, Scheibye-Knudsen M, Stevnsner T, Nilsen H, Bohr VA, Fang EF, 2017. Mitophagy in neurodegeneration and aging. Neurochem. Int. 109, 202–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frederick DW, Loro E, Liu L, Davila A Jr., Chellappa K, Silverman IM, Quinn WJ 3rd, Gosai SJ, Tichy ED, Davis JG, et al. , 2016. Loss of NAD homeostasis leads to progressive and reversible degeneration of skeletal muscle. Cell Metab. 24, 269–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallo CM, Smith DL Jr., Smith JS, 2004. Nicotinamide clearance by Pnc1 directly regulates Sir2-mediated silencing and longevity. Mol. Cell. Biol. 24, 1301–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes AP, Price NL, Ling AJ, Moslehi JJ, Montgomery MK, Rajman L, White JP, Teodoro JS, Wrann CD, Hubbard BP, et al. , 2013. Declining NAD(+) induces a pseudohypoxic state disrupting nuclear-mitochondrial communication during aging. Cell 155, 1624–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong B, Pan Y, Vempati P, Zhao W, Knable L, Ho L, Wang J, Sastre M, Ono K, Sauve AA, et al. , 2013. Nicotinamide riboside restores cognition through an upregulation of proliferator-activated receptor-gamma coactivator 1alpha regulated beta-secretase 1 degradation and mitochondrial gene expression in Alzheimer’s mouse models. Neurobiol. Aging 34, 1581–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grassi D, Diaz-Perez N, Volpicelli-Daley LA, Lasmezas CI, 2019. Palpha-syn* mitotoxicity is linked to MAPK activation and involves tau phosphorylation and aggregation at the mitochondria. Neurobiol. Dis. 124, 248–262. [DOI] [PubMed] [Google Scholar]
- Hariharan N, Maejima Y, Nakae J, Paik J, Depinho RA, Sadoshima J, 2010. Deacetylation of FoxO by Sirt1 plays an essential role in mediating starvation-induced autophagy in cardiac myocytes. Circ. Res. 107, 1470–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo JM, Ordureau A, Swarup S, Paulo JA, Shen K, Sabatini DM, Harper JW, 2018. RAB7A phosphorylation by TBK1 promotes mitophagy via the PINK-PARKIN pathway. Sci. Adv. 4 eaav0443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzig S, Shaw RJ, 2018. AMPK: guardian of metabolism and mitochondrial homeostasis. Nat. Rev. Mol. Cell Biol. 19, 121–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y, Lautrup S, Cordonnier S, Wang Y, Croteau DL, Zavala E, Zhang Y, Moritoh K, O’Connell JF, Baptiste BA, et al. , 2018. NAD(+) supplementation normalizes key Alzheimer’s features and DNA damage responses in a new AD mouse model with introduced DNA repair deficiency. Proc. Natl. Acad. Sci. U. S. A. 115, E1876–E1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y, Song H, Croteau DL, Akbari M, Bohr VA, 2017. Genome instability in Alzheimer disease. Mech. Ageing Dev. 161, 83–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Li XC, Wang ZH, Luo Y, Zhang X, Liu XP, Feng Q, Wang Q, Yue Z, Chen Z, et al. , 2016. Tau accumulation impairs mitophagy via increasing mitochondrial membrane potential and reducing mitochondrial Parkin. Oncotarget 7, 17356–17368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang R, Xu Y, Wan W, Shou X, Qian J, You Z, Liu B, Chang C, Zhou T, Lippincott-Schwartz J, et al. , 2015. Deacetylation of nuclear LC3 drives autophagy initiation under starvation. Mol. Cell 57, 456–466. [DOI] [PubMed] [Google Scholar]
- Inoki K, Zhu T, Guan KL, 2003. TSC2 mediates cellular energy response to control cell growth and survival. Cell 115, 577–590. [DOI] [PubMed] [Google Scholar]
- Ishihara N, Eura Y, Mihara K, 2004. Mitofusin 1 and 2 play distinct roles in mitochondrial fusion reactions via GTPase activity. J. Cell. Sci. 117, 6535–6546. [DOI] [PubMed] [Google Scholar]
- Jeon J, Nim S, Teyra J, Datti A, Wrana JL, Sidhu SS, Moffat J, Kim PM, 2014. A systematic approach to identify novel cancer drug targets using machine learning, inhibitor design and high-throughput screening. Genome Med. 6, 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju HQ, Zhuang ZN, Li H, Tian T, Lu YX, Fan XQ, Zhou HJ, Mo HY, Sheng H, Chiao PJ, et al. , 2016. Regulation of the Nampt-mediated NAD salvage pathway and its therapeutic implications in pancreatic cancer. Cancer Lett. 379, 1–11. [DOI] [PubMed] [Google Scholar]
- Kang HT, Hwang ES, 2009. Nicotinamide enhances mitochondria quality through autophagy activation in human cells. Aging Cell 8, 426–438. [DOI] [PubMed] [Google Scholar]
- Katsyuba E, Auwerx J, 2017. Modulating NAD(+) metabolism, from bench to bedside. EMBO J. 36, 2670–2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy BK, Berger SL, Brunet A, Campisi J, Cuervo AM, Epel ES, Franceschi C, Lithgow GJ, Morimoto RI, Pessin JE, et al. , 2014. Geroscience: linking aging to chronic disease. Cell 159, 709–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr JS, Adriaanse BA, Greig NH, Mattson MP, Cader MZ, Bohr VA, Fang EF, 2017a. Mitophagy and alzheimer’s disease: cellular and molecular mechanisms. Trends Neurosci. 40, 151–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr JS, Adriaanse BA, Greig NH, Mattson MP, Cader MZ, Bohr VA, Fang EF, 2017b. Mitophagy and alzheimer’s disease: cellular and molecular mechanisms. Trends Neurosci. 40, 151–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Kim JJ, Lee H, 2017. An analysis of disease-gene relationship from Medline abstracts by DigSee. Sci. Rep. 7, 40154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Kundu M, Viollet B, Guan KL, 2011. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat. Cell Biol. 13, 132–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kume S, Uzu T, Horiike K, Chin-Kanasaki M, Isshiki K, Araki S, Sugimoto T, Haneda M, Kashiwagi A, Koya D, 2010. Calorie restriction enhances cell adaptation to hypoxia through Sirt1-dependent mitochondrial autophagy in mouse aged kidney. J. Clin. Invest. 120, 1043–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labrousse AM, Zappaterra MD, Rube DA, van der Bliek AM, 1999. C. Elegans dynamin-related protein DRP-1 controls severing of the mitochondrial outer membrane. Mol. Cell 4, 815–826. [DOI] [PubMed] [Google Scholar]
- Landes T, Emorine LJ, Courilleau D, Rojo M, Belenguer P, Arnaune-Pelloquin L, 2010. The BH3-only Bnip3 binds to the dynamin Opa1 to promote mitochondrial fragmentation and apoptosis by distinct mechanisms. EMBO Rep. 11, 459–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane N, Martin W, 2010. The energetics of genome complexity. Nature 467, 929–934. [DOI] [PubMed] [Google Scholar]
- Lang A, Anand R, Altinoluk-Hambuchen S, Ezzahoini H, Stefanski A, Iram A, Bergmann L, Urbach J, Bohler P, Hansel J, et al. , 2017. SIRT4 interacts with OPA1 and regulates mitochondrial quality control and mitophagy. Aging (Albany NY) 9, 2163–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lautrup S, Lou G, Aman Y, Nilsen H, Tao J, Fang EF, 2019a. Microglial mitophagy mitigates neuroinflammation in Alzheimer’s disease. Neurochem. Int. 129, 104469. [DOI] [PubMed] [Google Scholar]
- Lautrup S, Sinclair DA, Mattson MP, Fang EF, 2019b. NAD+ in brain ageing and neurodegenerative disorders. Cell Metab. 30, 630–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarou M, Sliter DA, Kane LA, Sarraf SA, Wang C, Burman JL, Sideris DP, Fogel AI, Youle RJ, 2015. The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature 524, 309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee IH, Cao L, Mostoslavsky R, Lombard DB, Liu J, Bruns NE, Tsokos M, Alt FW, Finkel T, 2008. A role for the NAD-dependent deacetylase Sirt1 in the regulation of autophagy. Proc. Natl. Acad. Sci. U. S. A. 105, 3374–3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Lee HY, Hanna RA, Gustafsson AB, 2011. Mitochondrial autophagy by Bnip3 involves Drp1-mediated mitochondrial fission and recruitment of Parkin in cardiac myocytes. Am. J. Physiol. Heart Circ. Physiol. 301, H1924–1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YK, Lee JA, 2016. Role of the mammalian ATG8/LC3 family in autophagy: differential and compensatory roles in the spatiotemporal regulation of autophagy. BMB Rep. 49, 424–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leelananda SP, Lindert S, 2016. Computational methods in drug discovery. Beilstein J. Org. Chem. 12, 2694–2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenaers G, Reynier P, Elachouri G, Soukkarieh C, Olichon A, Belenguer P, Baricault L, Ducommun B, Hamel C, Delettre C, 2009. OPA1 functions in mitochondria and dysfunctions in optic nerve. Int. J. Biochem. Cell Biol. 41, 1866–1874. [DOI] [PubMed] [Google Scholar]
- Lin MT, Beal MF, 2006. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature 443, 787–795. [DOI] [PubMed] [Google Scholar]
- Lin MY, Cheng XT, Tammineni P, Xie Y, Zhou B, Cai Q, Sheng ZH, 2017. Releasing syntaphilin removes stressed mitochondria from axons independent of mitophagy under pathophysiological conditions. Neuron 94, 595–610 e596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Pitta M, Jiang H, Lee JH, Zhang G, Chen X, Kawamoto EM, Mattson MP, 2013. Nicotinamide forestalls pathology and cognitive decline in Alzheimer mice: evidence for improved neuronal bioenergetics and autophagy procession. Neurobiol. Aging 34, 1564–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Park SH, Imbesi M, Nathan WJ, Zou X, Zhu Y, Jiang H, Parisiadou L, Gius D, 2017. Loss of NAD-Dependent protein deacetylase Sirtuin-2 alters mitochondrial protein acetylation and dysregulates mitophagy. Antioxid. Redox Signal. 26, 849–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Feng D, Chen G, Chen M, Zheng Q, Song P, Ma Q, Zhu C, Wang R, Qi W, et al. , 2012. Mitochondrial outer-membrane protein FUNDC1 mediates hypoxia-induced mitophagy in mammalian cells. Nat. Cell Biol. 14, 177–185. [DOI] [PubMed] [Google Scholar]
- Lloret A, Beal MF, 2019. PGC-1alpha, sirtuins and PARPs in Huntington’s disease and other neurodegenerative conditions: NAD+ to rule them all. Neurochem. Res. 44, 2423–2434. [DOI] [PubMed] [Google Scholar]
- Long AN, Owens K, Schlappal AE, Kristian T, Fishman PS, Schuh RA, 2015. Effect of nicotinamide mononucleotide on brain mitochondrial respiratory deficits in an Alzheimer’s disease-relevant murine model. BMC Neurol. 15, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou G, Palikaras K, Lautrup S, Scheibye-Knudsen M, Tavernarakis N, Fang EF, 2019. Mitophagy and neuroprotection. Trends Mol. Med. [DOI] [PubMed] [Google Scholar]
- Luo Y, Zhao X, Zhou J, Yang J, Zhang Y, Kuang W, Peng J, Chen L, Zeng J, 2017. A network integration approach for drug-target interaction prediction and computational drug repositioning from heterogeneous information. Nat. Commun. 8, 573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma S, Cao F, Ren J, 2018. NAD+ precursor nicotinamide riboside alleviates alcoholic cardiomyopathy via mitophagy induction: a novel SIRT3-PGAM5-FUNDC1 Axis. Circulation 136. [Google Scholar]
- Magni G, Amici A, Emanuelli M, Orsomando G, Raffaelli N, Ruggieri S, 2004. Enzymology of NAD+ homeostasis in man. Cell. Mol. Life Sci. 61, 19–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamoshina P, Volosnikova M, Ozerov IV, Putin E, Skibina E, Cortese F, Zhavoronkov A, 2018. Machine learning on human muscle transcriptomic data for biomarker discovery and tissue-specific drug target identification. Front. Genet. 9, 242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens CR, Denman BA, Mazzo MR, Armstrong ML, Reisdorph N, McQueen MB, Chonchol M, Seals DR, 2018. Chronic nicotinamide riboside supplementation is well-tolerated and elevates NAD(+) in healthy middle-aged and older adults. Nat. Commun. 9, 1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattson MP, Gleichmann M, Cheng A, 2008. Mitochondria in neuroplasticity and neurological disorders. Neuron 60, 748–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McWilliams TG, Prescott AR, Montava-Garriga L, Ball G, Singh F, Barini E, Muqit MMK, Brooks SP, Ganley IG, 2018. Basal mitophagy occurs independently of PINK1 in mouse tissues of high metabolic demand. Cell Metab. 27, 439–449 e435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills KF, Yoshida S, Stein LR, Grozio A, Kubota S, Sasaki Y, Redpath P, Migaud ME, Apte RS, Uchida K, et al. , 2016. Long-term administration of nicotinamide mononucleotide mitigates age-associated physiological decline in mice. Cell Metab. 24, 795–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misiak M, Vergara Greeno R, Baptiste BA, Sykora P, Liu D, Cordonnier S, Fang EF, Croteau DL, Mattson MP, Bohr VA, 2017. DNA polymerase beta decrement triggers death of olfactory bulb cells and impairs olfaction in a mouse model of Alzheimer’s disease. Aging Cell 16, 162–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell SJ, Bernier M, Aon MA, Cortassa S, Kim EY, Fang EF, Palacios HH, Ali A, Navas-Enamorado I, Di Francesco A, et al. , 2018. Nicotinamide improves aspects of Healthspan, but not lifespan, in mice. Cell Metab. 27, 667–676 e664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell SJ, Madrigal-Matute J, Scheibye-Knudsen M, Fang E, Aon M, Gonzalez-Reyes JA, Cortassa S, Kaushik S, Gonzalez-Freire M, Patel B, et al. , 2016. Effects of sex, strain, and energy intake on hallmarks of aging in mice. Cell Metab. 23, 1093–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moskalev AA, Vaiserman A, 2018. Epigenetics of Aging and Longevity. Elsevier : Academic Press, London ; San Diego ; Cambridge, MA ; Kidlington, Oxford. [Google Scholar]
- Mouchiroud L, Houtkooper RH, Moullan N, Katsyuba E, Ryu D, Canto C, Mottis A, Jo YS, Viswanathan M, Schoonjans K, et al. , 2013. The NAD(+)/Sirtuin pathway modulates longevity through activation of mitochondrial UPR and FOXO signaling. Cell 154, 430–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata H, Sakaguchi M, Kataoka K, Huh NH, 2013. SARM1 and TRAF6 bind to and stabilize PINK1 on depolarized mitochondria. Mol. Biol. Cell 24, 2772–2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nacarelli T, Lau L, Fukumoto T, Zundell J, Fatkhutdinov N, Wu S, Aird KM, Iwasaki O, Kossenkov AV, Schultz D, et al. , 2019. NAD(+) metabolism governs the proinflammatory senescence-associated secretome. Nat. Cell Biol. 21, 397–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikiforov A, Kulikova V, Ziegler M, 2015. The human NAD metabolome: functions, metabolism and compartmentalization. Crit. Rev. Biochem. Mol. Biol. 50, 284–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Bryant SE, Gupta V, Henriksen K, Edwards M, Jeromin A, Lista S, Bazenet C, Soares H, Lovestone S, Hampel H, et al. , 2015. Guidelines for the standardization of preanalytic variables for blood-based biomarker studies in Alzheimer’s disease research. Alzheimers Dement. 11, 549–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onyango P, Celic I, McCaffery JM, Boeke JD, Feinberg AP, 2002. SIRT3, a human SIR2 homologue, is an NAD-dependent deacetylase localized to mitochondria. Proc. Natl. Acad. Sci. U. S. A. 99, 13653–13658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou X, Lee MR, Huang X, Messina-Graham S, Broxmeyer HE, 2014. SIRT1 positively regulates autophagy and mitochondria function in embryonic stem cells under oxidative stress. Stem Cells 32, 1183–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palikaras K, Lionaki E, Tavernarakis N, 2018. Mechanisms of mitophagy in cellular homeostasis, physiology and pathology. Nat. Cell Biol. 20, 1013–1022. [DOI] [PubMed] [Google Scholar]
- Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, Overvatn A, Bjorkoy G, Johansen T, 2007. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 282, 24131–24145. [DOI] [PubMed] [Google Scholar]
- Partridge L, Deelen J, Slagboom PE, 2018. Facing up to the global challenges of ageing. Nature 561, 45–56. [DOI] [PubMed] [Google Scholar]
- Pereira JC, Caffarena ER, Dos Santos CN, 2016. Boosting docking-based virtual screening with deep learning. J. Chem. Inf. Model. 56, 2495–2506. [DOI] [PubMed] [Google Scholar]
- Perez MJ, Jara C, Quintanilla RA, 2018. Contribution of tau pathology to mitochondrial impairment in neurodegeneration. Front. Neurosci. 12, 441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Politi Y, Gal L, Kalifa Y, Ravid L, Elazar Z, Arama E, 2014. Paternal mitochondrial destruction after fertilization is mediated by a common endocytic and autophagic pathway in Drosophila. Dev. Cell 29, 305–320. [DOI] [PubMed] [Google Scholar]
- Polletta L, Vernucci E, Carnevale I, Arcangeli T, Rotili D, Palmerio S, Steegborn C, Nowak T, Schutkowski M, Pellegrini L, et al. , 2015. SIRT5 regulation of ammonia-induced autophagy and mitophagy. Autophagy 11, 253–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prince M, Comas-Herrera A, Knapp M, Guerchet M, Karagiannidou M, 2016. World alzheimer report 2016. Improving Healthcare for People Living With Dementia; (London: ). pp. 140. [Google Scholar]
- Putin E, Mamoshina P, Aliper A, Korzinkin M, Moskalev A, Kolosov A, Ostrovskiy A, Cantor C, Vijg J, Zhavoronkov A, 2016. Deep biomarkers of human aging: application of deep neural networks to biomarker development. Aging (Albany NY) 8, 1021–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajman L, Chwalek K, Sinclair DA, 2018. Therapeutic potential of NAD-Boosting molecules: the in vivo evidence. Cell Metab. 27, 529–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanov J, Walczak M, Ibiricu I, Schuchner S, Ogris E, Kraft C, Martens S, 2012. Mechanism and functions of membrane binding by the Atg5-Atg12/Atg16 complex during autophagosome formation. EMBO J. 31, 4304–4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinsztein DC, Marino G, Kroemer G, 2011. Autophagy and aging. Cell 146, 682–695. [DOI] [PubMed] [Google Scholar]
- Samant SA, Zhang HJ, Hong Z, Pillai VB, Sundaresan NR, Wolfgeher D, Archer SL, Chan DC, Gupta MP, 2014. SIRT3 deacetylates and activates OPA1 to regulate mitochondrial dynamics during stress. Mol. Cell. Biol. 34, 807–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheibye-Knudsen M, Fang EF, Croteau DL, Wilson DM 3rd, Bohr VA, 2015. Protecting the mitochondrial powerhouse. Trends Cell Biol. 25, 158–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmeisser K, Mansfeld J, Kuhlow D, Weimer S, Priebe S, Heiland I, Birringer M, Groth M, Segref A, Kanfi Y, et al. , 2013. Role of sirtuins in lifespan regulation is linked to methylation of nicotinamide. Nat. Chem. Biol. 9, 693–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidhuber J, 2015. Deep learning in neural networks: an overview. Neural Netw. 61, 85–117. [DOI] [PubMed] [Google Scholar]
- Schondorf DC, Ivanyuk D, Baden P, Sanchez-Martinez A, De Cicco S, Yu C, Giunta I, Schwarz LK, Di Napoli G, Panagiotakopoulou V, et al. , 2018. The NAD + precursor nicotinamide riboside rescues mitochondrial defects and neuronal loss in iPSC and fly models of parkinson’s disease. Cell Rep. 23, 2976–2988. [DOI] [PubMed] [Google Scholar]
- Sengupta A, Molkentin JD, Yutzey KE, 2009. FoxO transcription factors promote autophagy in cardiomyocytes. J. Biol. Chem. 284, 28319–28331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnova E, Shurland DL, Ryazantsev SN, van der Bliek AM, 1998. A human dynamin-related protein controls the distribution of mitochondria. J. Cell Biol. 143, 351–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song WH, Yi YJ, Sutovsky M, Meyers S, Sutovsky P, 2016. Autophagy and ubiquitin-proteasome system contribute to sperm mitophagy after mammalian fertilization. Proc. Natl. Acad. Sci. U. S. A. 113, E5261–5270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorrentino V, Romani M, Mouchiroud L, Beck JS, Zhang H, D’Amico D, Moullan N, Potenza F, Schmid AW, Rietsch S, et al. , 2017. Enhancing mitochondrial proteostasis reduces amyloid-beta proteotoxicity. Nature 552, 187–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein LR, Imai S, 2014. Specific ablation of Nampt in adult neural stem cells recapitulates their functional defects during aging. EMBO J. 33, 1321–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stojanovski D, Koutsopoulos OS, Okamoto K, Ryan MT, 2004. Levels of human Fis1 at the mitochondrial outer membrane regulate mitochondrial morphology. J. Cell. Sci. 117, 1201–1210. [DOI] [PubMed] [Google Scholar]
- Stromland O, Niere M, Nikiforov AA, VanLinden MR, Heiland I, Ziegler M, 2019. Keeping the balance in NAD metabolism. Biochem. Soc. Trans. 47, 119–130. [DOI] [PubMed] [Google Scholar]
- Takasaka N, Araya J, Hara H, Ito S, Kobayashi K, Kurita Y, Wakui H, Yoshii Y, Yumino Y, Fujii S, et al. , 2014. Autophagy induction by SIRT6 through attenuation of insulin-like growth factor signaling is involved in the regulation of human bronchial epithelial cell senescence. J. Immunol. 192, 958–968 [DOI] [PubMed] [Google Scholar]
- Takebe T, Imai R, Ono S, 2018. The current status of drug discovery and development as originated in United States academia: the influence of industrial and academic collaboration on drug discovery and development. Clin. Transl. Sci. 11, 597–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trammell SA, Schmidt MS, Weidemann BJ, Redpath P, Jaksch F, Dellinger RW, Li Z, Abel ED, Migaud ME, Brenner C, 2016. Nicotinamide riboside is uniquely and orally bioavailable in mice and humans. Nat. Commun. 7, 12948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner JR, 2010. New Drug Development: An Introduction to Clinical Trials, 2nd edn Springer, New York. [Google Scholar]
- United Nations D.o.E.a.S.A., Population Division, 2019. World population prospects 2019: highlights. World Population Prospects 2019. Highlights, United States: United Nations. [Google Scholar]
- Vamathevan J, Clark D, Czodrowski P, Dunham I, Ferran E, Lee G, Li B, Madabhushi A, Shah P, Spitzer M, et al. , 2019. Applications of machine learning in drug discovery and development. Nat. Rev. Drug Discov. 18, 463–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Horst A, Schavemaker JM, Pellis-van Berkel W, Burgering BM, 2007. The Caenorhabditis elegans nicotinamidase PNC-1 enhances survival. Mech. Ageing Dev. 128, 346–349. [DOI] [PubMed] [Google Scholar]
- Vanhaelen Q, 2018. Computational Methods for Drug Repurposing. Springer Science +Business Media, New York, NY. [Google Scholar]
- Vanhaelen Q, Mamoshina P, Aliper AM, Artemov A, Lezhnina K, Ozerov I, Labat I, Zhavoronkov A, 2017. Design of efficient computational workflows for in silico drug repurposing. Drug Discov. Today 22, 210–222. [DOI] [PubMed] [Google Scholar]
- Vannini N, Campos V, Girotra M, Trachsel V, Rojas-Sutterlin S, Tratwal J, Ragusa S, Stefanidis E, Ryu D, Rainer PY, et al. , 2019. The NAD-Booster nicotinamide riboside potently stimulates hematopoiesis through increased mitochondrial clearance. Cell Stem Cell 24, 405–418 e407. [DOI] [PubMed] [Google Scholar]
- Verdin E, 2015. NAD(+) in aging, metabolism, and neurodegeneration. Science 350, 1208–1213. [DOI] [PubMed] [Google Scholar]
- Vohora D, Singh G, 2018. Pharmaceutical Medicine and Translational Clinical Research. Elsevier/Academic Press, London. [Google Scholar]
- Wallace DC, 1999. Mitochondrial diseases in man and mouse. Science 283, 1482–1488. [DOI] [PubMed] [Google Scholar]
- Wallace DC, 2012. Mitochondria and cancer. Nat. Rev. Cancer 12, 685–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JD, Cao YL, Li Q, Yang YP, Jin M, Chen D, Wang F, Wang GH, Qin ZH, Hu LF, et al. , 2015. A pivotal role of FOS-mediated BECN1/Beclin 1 upregulation in dopamine D2 and D3 receptor agonist-induced autophagy activation. Autophagy 11, 2057–2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Cho YL, Tang Y, Wang J, Park JE, Wu Y, Wang C, Tong Y, Chawla R, Zhang J, et al. , 2018. PTEN-L is a novel protein phosphatase for ubiquitin dephosphorylation to inhibit PINK1-Parkin-mediated mitophagy. Cell Res. 28, 787–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Qi H, Tang Y, Shen HM, 2019. Post-translational modifications of key machinery in the control of mitophagy. Trends Biochem. Sci. [DOI] [PubMed] [Google Scholar]
- Wang S, Sun S, Li Z, Zhang R, Xu J, 2017. Accurate de novo prediction of protein contact map by ultra-deep learning model. PLoS Comput. Biol. 13, e1005324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Hu X, Yang Y, Takata T, Sakurai T, 2016. Nicotinamide mononucleotide protects against beta-amyloid oligomer-induced cognitive impairment and neuronal death. Brain Res. 1643, 1–9. [DOI] [PubMed] [Google Scholar]
- Weir HJ, Yao P, Huynh FK, Escoubas CC, Goncalves RL, Burkewitz K, Laboy R, Hirschey MD, Mair WB, 2017. Dietary restriction and AMPK increase lifespan via mitochondrial network and peroxisome remodeling. Cell Metab. 26, 884–896 e885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong J, Xia M, Xu M, Zhang Y, Abais JM, Li G, Riebling CR, Ritter JK, Boini KM, Li PL, 2013. Autophagy maturation associated with CD38-mediated regulation of lysosome function in mouse glomerular podocytes. J. Cell. Mol. Med. 17, 1598–1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu DH, Chi GN, Zhao CH, Li DY, 2018. Long noncoding RNA MEG3 inhibits proliferation and migration but induces autophagy by regulation of Sirt7 and PI3K/AKT/mTOR pathway in glioma cells. J. Cell. Biochem. [DOI] [PubMed] [Google Scholar]
- Yaku K, Okabe K, Nakagawa T, 2018. NAD metabolism: implications in aging and longevity. Ageing Res. Rev. 47, 1–17. [DOI] [PubMed] [Google Scholar]
- Yao Z, Yang W, Gao Z, Jia P, 2017. Nicotinamide mononucleotide inhibits JNK activation to reverse Alzheimer disease. Neurosci. Lett. 647, 133–140. [DOI] [PubMed] [Google Scholar]
- Yella JK, Yaddanapudi S, Wang Y, Jegga AG, 2018. Changing trends in computational drug repositioning. Pharmaceuticals Basel (Basel) 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshino J, Baur JA, Imai SI, 2018. NAD(+) intermediates: the biology and therapeutic potential of NMN and NR. Cell Metab. 27, 513–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y, Cruzat VF, Newsholme P, Cheng J, Chen Y, Lu Y, 2016. Regulation of SIRT1 in aging: roles in mitochondrial function and biogenesis. Mech. Ageing Dev. 155, 10–21. [DOI] [PubMed] [Google Scholar]
- Zhan L, Chen S, Li K, Liang D, Zhu X, Liu L, Lu Z, Sun W, Xu E, 2017. Autophagosome maturation mediated by Rab7 contributes to neuroprotection of hypoxic preconditioning against global cerebral ischemia in rats. Cell Death Dis. 8, e2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Ryu D, Wu Y, Gariani K, Wang X, Luan P, D’Amico D, Ropelle ER, Lutolf MP, Aebersold R, et al. , 2016a. NAD(+) repletion improves mitochondrial and stem cell function and enhances life span in mice. Science 352, 1436–1443. [DOI] [PubMed] [Google Scholar]
- Zhang L, Wu WK, Gallo RL, Fang EF, Hu W, Ling TK, Shen J, Chan RL, Lu L, Luo XM, et al. , 2016b. Critical role of antimicrobial peptide cathelicidin for controlling Helicobacter pylori survival and infection. J. Immunol. 196, 1799–1809. [DOI] [PubMed] [Google Scholar]
- Zhang T, Xue L, Li L, Tang C, Wan Z, Wang R, Tan J, Tan Y, Han H, Tian R, et al. , 2016c. BNIP3 protein suppresses PINK1 kinase proteolytic cleavage to promote mitophagy. J. Biol. Chem. 291, 21616–21629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Bharathi SS, Rardin MJ, Lu J, Maringer KV, Sims-Lucas S, Prochownik EV, Gibson BW, Goetzman ES, 2017. Lysine desuccinylase SIRT5 binds to cardiolipin and regulates the electron transport chain. J. Biol. Chem. 292, 10239–10249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhavoronkov A, Mamoshina P, Vanhaelen Q, Scheibye-Knudsen M, Moskalev A, Aliper A, 2019. Artificial intelligence for aging and longevity research: recent advances and perspectives. Ageing Res. Rev. 49, 49–66. [DOI] [PubMed] [Google Scholar]
- Zhu J, Wang KZ, Chu CT, 2013. After the banquet: mitochondrial biogenesis, mitophagy, and cell survival. Autophagy 9, 1663–1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J, Yue F, Li W, Song K, Jiang X, Yi J, Liu L, 2014. Autophagy inhibitor LRPPRC suppresses mitophagy through interaction with mitophagy initiator Parkin. PLoS One 9, e94903. [DOI] [PMC free article] [PubMed] [Google Scholar]