Abstract
Chromatin regulatory landscape plays a critical role in many disease processes and embryo development. Epigenome sequencing technologies such as chromatin immunoprecipitation sequencing (ChIP-seq) and assay for transposase-accessible chromatin with high-throughput sequencing (ATAC-seq) have enabled us to dissect the pan-genomic regulatory landscape of cells and tissues in both time and space dimensions by detecting specific chromatin state and its corresponding transcription factors. Pioneered by the advancement of chromatin immunoprecipitation-chip (ChIP-chip) technology, abundant epigenome profiling technologies have become available such as ChIP-seq, DNase I hypersensitive site sequencing (DNase-seq), ATAC-seq and so on. The advent of single-cell sequencing has revolutionized the next-generation sequencing, applications in single-cell epigenetics are enriched rapidly. Epigenome sequencing technologies have evolved from low-throughput to high-throughput and from bulk sample to the single-cell scope, which unprecedentedly benefits scientists to interpret life from different angles. In this review, after briefly introducing the background knowledge of epigenome biology, we discuss the development of epigenome sequencing technologies, especially ChIP-seq & ATAC-seq and their current applications in scientific research. Finally, we provide insights into future applications and challenges.
Keywords: Chromatin regulatory landscape, Epigenome sequencing, Single cell, Developmental biology
Introduction
The genome is packaged by histone proteins that are decorated with a wide variety of modifications. Histone acetylation is one of the best-characterized chromatin modifications and correlates with the opening of local chromatin structures and transcriptional activation (e.g., H3K27ac correlates with enhancers) [1]. Compared with histone acetylation, histone methylation is more diverse in terms of both functions and forms. Histon methylation includes H3K4me1, H3K4me3, H3K9me3, H3K36, H3K79, etc. Specific methylation of lysines can exist as monomethylation, dimethylation or trimethylation with different functions [2]. Repressive histone methylation, such as H3K9me3, is highly associated with condensed and constitutive heterochromatin [3]. Meanwhile, active histone methylation such as H3K4me3 contributes to active transcription. Several studies even revealed a class of bivalent chromatin with both active and repressive features, which exhibits overlapping patterns of H3K4me3 and H3K27me3 [4]. The discovery of the bivalent signature of such poised genes was unexpected and very important. For instance, it can be a critical landmark in the maternal-to-zygotic transition process, providing the first clues about the “intermediate” state [5]. Also, bivalent chromatin is not specific to embryonic stem cells and has been well documented in other cell types [6, 7]. In more complex cases, elements that behave as promoters in some tissues can act as enhancers in other tissues, called cREDS (cis-regulatory elements with dynamic signatures), and the same regulatory elements can have both promoter and enhancer signatures [8].
More histone modifications also include phosphorylation, ubiquitination, sumoylation and ADP ribosylation. The current exploration of histone modifications still has the following problems:
Modifications on histone surfaces are often dynamic: some modifications can be added and erased in just a few minutes after a cell has been stimulated, so that the histone modifications detected in a population of cells under specific conditions at a given moment are in fact only partially representative of the types of potential modifications.
Antibodies to detect histone modifications are essential for many epigenetic chromatin sequencing techniques: in ChIP-seq, for example, antibody specificity testing is required for both histones and transcription factors [9].
The mechanisms of heterochromatin formation and diffusion, and the “memory” and “fading” of histone modifications, have been studied only to a minimal extent.
The concept of histone code can not be widely used to accurately describe and predict a specific biological phenomenon in many cases [10, 11].
Some histone modifications are active in some genomic regions. In contrast, repressive in others: for example, methylation of the H3K9 locus can be repressive in the promoter region and active in the coding region [12].
In contrast to single histone modifications, the chromatin regulatory landscape is a higher-level annotation on the biological function of chromatin that combines histone modifications, transcription factor binding, and the regulatory function of genomic elements. The renowned Roadmap Epigenomics Consortium has led large-scale human reference epigenomic studies that have provided detailed and accurate descriptions and classifications of the functional states of regulatory elements [7, 13]. In addition, by combining these chromatin epigenomic states with existing genome-wide methylation information and gene expression profiles from RNA sequencing (RNA-seq) [14], scientists can interpret the tissue-specific epigenomic landscape from a multi-dimensional perspective.
Many studies of chromatin dynamics require, besides access to information on histone modifications and genomic regulatory elements, a proper understanding of the interactions between transcription factors and chromatin, which is often essential for understanding development and disease progression [15, 16]. Some transcription factors binding to chromatin regulatory regions require specific histone modifications, while others require the assistance of open chromatin and other activators. Binding of some transcription factors to the regulatory regions facilitates the recruitment of additional transcription factors. It may promote the diffusion of chromatin status, which in turn further influences transcription factor binding. The in-depth research on the interaction mechanism is still limited. Recent research has explored epigenetic regulatory mechanisms by constructing synthetic read-write modules, which will help us better understand the basic principles of epigenetic inheritance [17].
The development of technologies for investigating chromatin dynamics
ChIP-based methods
The earliest technology applied to large-scale epigenetic mapping was chromatin immunoprecipitation (ChIP) followed by microarray hybridization (chip) (ChIP-chip), which allowed scientists to detect DNA-protein interactions on a genome-wide scale [18]. ChIP-chip is based on microarray hybridization, where a large number of probes covering a genome or a specified region are seeded on a high-density chip. However, this method has a few shortcomings: low resolution, ambiguous factors introduced by the probe design, signal bias, difficult in broad application to more species [19].
Compared to Chip-chip, chromatin immunoprecipitation sequencing (ChIP-seq) provides higher resolution, less noise, and greater coverage [20, 21]. With the rapid decline in the cost of second-generation sequencing, ChIP-seq will become one of the indispensable tools for studying gene regulation and epigenetics. In addition to better identification of sequence motifs, ChIP-seq can also be utilized to find key transcription factors, enhancers, and other regulatory elements [22].
Conventional ChIP-seq
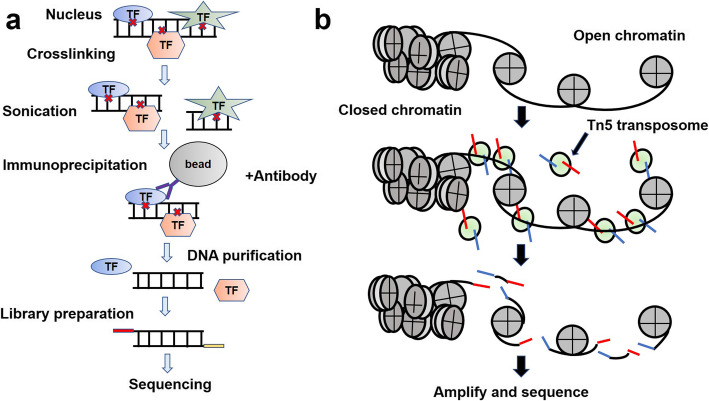
For DNA-binding proteins, ChIP-seq experiments are aimed at enriching DNAs bound to specific proteins. The procedure consists of multiple steps (Fig. 1a): first crosslinking DNA and proteins in situ via formaldehyde, followed by sonication of the DNA into small 200–600 bp fragments, and then immunoprecipitation of the DNA-protein complexes of interest with antibodies. The DNA is then uncross-linked, and the released DNA is subjected to end repair, adapter ligation and other library preparation steps. Finally, sequencing is performed.
Fig. 1.
Workflows of ChIP-seq and ATAC-seq. a In ChIP-seq, chromatin is crosslinked using formaldehyde and sonicated to obtain DNA fragments of 200–600 base pairs. Then the DNA-protein complex of interest can be immunoprecipitated by the antibody. Library preparation steps: end repair, A-tailing and adapters ligation, library sequencing. b ATAC-seq identifies regions of open chromatin using a hyperactive Tn5 transposase, which preferentially inserts into accessible chromatin and tags the sites with sequencing adaptors
However, ChIP-seq has some limitations: the introduction of potential bias by PCR amplification, length limitations of PCR amplification, GC bias in both fragmentation and sequencing processes, the need for 105 ∼ 107 cells due to a massive loss in the immunoprecipitation process, and the potential for epitope masking due to the formaldehyde crosslinking process [23]. Formaldehyde can crosslink transcription factors to DNA, maintaining their binding status in vivo, but it may also mask the antigenic epitopes of the transcription factors, producing false-positive results. This formaldehyde crosslinked ChIP is also known as X-ChIP [20]. Natural ChIP (N-ChIP), on the other hand, does not have the problem of epitope masking and can be used for a much smaller number of cells [24, 25]. This method abandons formaldehyde fixation, using micrococcal nuclease (MNase) for chromatin digestion. MNase can quickly and gently sever chromatin DNA and maximize the preservation of the original chromatin structure and the binding of the target protein, increasing the reliability of the ChIP results. However, this approach may result in the loss of many binding sites when target proteins do not bind strongly to DNA, so it is generally used for ChIP of histones other than transcription factors [26]. For antibody-dependent epigenetic sequencing techniques, the selection of the appropriate antibody is often the most critical step. In terms of choosing between monoclonal or polyclonal antibodies, monoclonal antibodies may result in weak signals due to interference from other protein components. In contrast, polyclonal antibodies may produce unwanted false positives [23].
Lowering cellular input
A major limitation of conventional ChIP-seq is the large number of cells required (105 ∼ 107 cells). Various strategies have been adapted to optimize the protocol to measure low-abundance cells over the past few years. ChIPmentation combines chromatin immunoprecipitation with sequencing library preparation by Tn5 transposase. The fast, low-cost library preparation protocol allows histone ChIP-seq using 10,000 cells [27]. The ultra-low-input MNase-based native ChIP (ULI-NChIP) generates high-quality maps of histone modifications from 103 to 106 embryonic stem cells by adjusting NChIP procedure [28]. Microfluidic oscillatory washing-based ChIP-seq (MOWChIP-seq) can even be applied to genome-wide analysis of histone modifications using as few as 100 cells [29]. While these methods can yield relatively accurate profiles of histone modifications such as H3K4me3 in a small number of cells, they are ineffective for many transcription factors with significantly fewer binding sites across the genome.
Cleavage under targets and release using nuclease (CUT&RUN), like ChIP-seq, is for detecting DNA-protein interactions, and it does not require formaldehyde crosslinking and sonication-based fragmentation, but instead uses MNase fused to Protein A/G to cut and release target DNA fragments in situ, thus significantly increasing the signal-to-noise ratio, and can be applied to as low as 100 ~ 1000 cells [30]. The ultra-low-input CUT&RUN (uliCUT&RUN) further improves this method to the single-cell level. By using this method, researchers found that the binding sites of CTCF in hESC cells were more concentrated, while H3K4me3 occupied a relatively wide region. Also, the binding pattern of SOX2 and Nanog, as well as other essential transcription factors in early embryonic development, were precisely described [31].
Cleavage under targets and tagmentation (CUT&Tag) is another technology that can detect DNA-protein interaction in low cellular input samples or even single cells [32]. Instead of Protein A/G-fused MNase, CUT&Tag applies Protein A/G-fused Tn5 transposase (pA/G-Tn5) to cut DNA, demanding high-quality core enzyme. The core enzyme pA/G-Tn5 has high activity, high sensitivity and high affinity for trace amounts of DNA and can effectively capture limited binding sites in a small number of cells. Another great feature of CUT&Tag is that all the library preparation steps are performed in the same tube after the addition of concanavalin protein A beads. As a result, the sequencing data has a lower background. So compared to CUT&RUN, CUT&Tag without end repair and additional adapter ligation is easier and more efficient. Since pA/G-Tn5 can bind and cut non-specific open chromatin under different conditions, CUT&Tag has the potential to simultaneously access some of the open chromatin and specific transcription factor binding sites by changing buffers composition. However, non-specific DNA cutting is a disturbance to the desired transcription factor binding profile in more scientific contexts, where the known open chromatin data need to be combined to remove these disturbances. Too complicated data cleaning process is generally not recommended, as combining other data to remove background often risks introducing batch effects.
Single-cell ChIP-seq
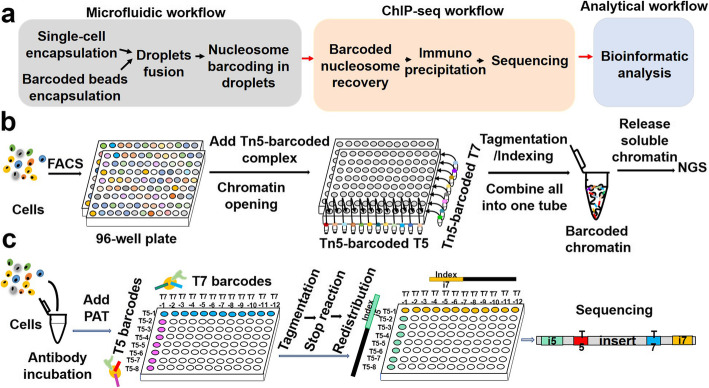
In contrast to bulk ChIP-seq, which is unable to detect chromatin signatures of individual cells, single-cell ChIP-seq (scChIP-seq) helps study genetic diversity in heterogeneous cell populations and understand the evolution of tumor populations. Droplet-based single-cell ChIP-seq (Drop-ChIP) combines a microfluidic device with single-cell DNA barcoding, allowing researchers to obtain a relatively low-coverage map per cell [33]. The basic experimental procedure consists of four steps (Fig. 2a): 1) Droplet formation: each cell encapsulated in a droplet is mixed with lysate and MNase; cells are then lysed in drops and their chromatin is fragmented; the second barcode drop contains DNA barcodes for ligation to the chromatin fragments. The two drops are mixed to form an indexing microreactor. 2) Nucleosome barcoding in droplets. 3) Immunoprecipitation of barcoded nucleosomes. 4) Library construction and sequencing. The downstream data analysis pipeline is similar to that of single-cell RNA-seq [34]. scChIP-seq enables the clustering of cell populations based on the diversity of the chromatin landscape and the identification of chromatin features specific to each population, such as the loss of H3K27me3 markers in some cells may associate with chemoresistance [35]. However, because the data generated by Drop-ChIP of individual cells are too sparse, thousands of cells are required to obtain good results for clustering.
Fig. 2.
Workflows of single-cell ChIP-seq. a Workflow of Drop-ChIP. b Workflow of sc-itChIP-seq. FACS: fluorescence-activated cell sorting. NGS: next-generation sequencing. c Workflow of coBatch. PAT: the fusion of the N-terminal of Tn5 transposase with protein A (pA-Tn5 [PAT])
Epigenomic tools that largely reduce cellular input, including CUT&RUN and CUT&Tag, do not necessarily perform better than ChIP-seq, depending on the situation. Both single-cell CUT&Tag and single-cell ATAC-seq (scATAC-seq) rely on a special device called Takara ICELL8 [36]. The microfluidic-based Drop-ChIP can only generate about 1000 unique reads per cell, the too sparse data coupled with expensive devices limit its application [33]. Therefore, ChIP methods that can be applied more widely, perform more robustly, and cost less on low-abundance cells or even a single cell are urgently needed. Single-cell simultaneous indexing and tagmentation-based ChIP-seq (sc-itChIP-seq) is a ChIP-based technology with high-resolution and its experimental procedure begins with a whole-genome chromatin opening of fixed cells/native cells sorted by FACS (fluorescence activated cell sorting), allowing Tn5 to fragment DNA homogeneously [37]. The released DNA fragments are enriched by immunoprecipitation (Fig. 2b). DNA-protein interaction peaks of individual cells can be obtained by demultiplexing and debarcoding. The library preparation steps are all carried out in the same tube, which dramatically reduces the loss. sc-itChIP-seq is useful for the identification of lineage-specific enhancers and key transcription factors during the differentiation process. Scientists have applied this approach to successfully identify the epigenetic trajectory of naive mouse embryonic stem cells and to reveal the reprogramming events of the lineage-specific enhancers in the fate determination of cardiac progenitor cells [37]. Given that sc-itChIP-seq does not depend on costly equipment, its application in laboratories is more widespread than the previous scChIP-seq (Table 1).
Table 1.
Comparisons between single-cell ChIP-seq methods
| Methods | Strategy | Device for cell sorting | Cell state | Mapping rate |
|---|---|---|---|---|
| scDrop-ChIP | ChIP and microfluidic system for droplet-forming | Costly microfluidic device | Native | 70% [33] |
| sc-itChIP-seq | ChIP and Tn5-barcoding (single round) | FACS | Native, fixed | 94% [37] |
| CUT&Tag | ChIP-free, Tn5-barcoding (for single round) | Costly Takara ICELL8 | Native | 97% [32] |
| CoBATCH | ChIP-free, Tn5-barcoding (for two rounds) | FACS | Native, fixed | 94% [38] |
Combinatorial barcoding and targeted chromatin release (CoBatch) can not only profile the epigenetic landscape of samples with relatively low cellular input, but also be applied to the scale of thousands of single cells in the native or fixed state with a high signal-to-noise ratio [38]. This approach applies an enzyme called PAT, which is the fusion of the N-terminal of Tn5 transposase with protein A. Cells incubated with antibodies are distributed into wells (200 ~ 2000 cells per well), PAT with different barcodes are then added for the first round of indexing. All cells are pooled and redistributed to different wells (20 ~ 25 cells per well), and finally amplified with varying primers of PCR for the second round of indexing (Fig. 2c). Using the single-cell data generated from coBatch, researchers successfully achieved high-throughput identification of cell types such as endothelial cells and mesenchymal cells, revealing the heterogeneity of the endothelial cell population in depth. However, the shortcoming of this method is that it cannot be directly applied to samples with input as low as a dozen cells, such as preimplantation embryos [38].
Nucleosome positioning and chromatin accessibility
One of the most classical technologies for investigating open chromatin regions is DNase I hypersensitive site sequencing (DNase-seq) [39]. DNaseI has endonuclease activity and can be utilized to obtain open chromatin fragments of appropriate length by controlling the cutting efficiency. DNaseI is used to cut DNase-sensitive sites on the genome. Then the digested fragments are amplified, the sequencing data are analyzed for peaks to acquire information on relatively open chromatin regions as well as protein-protected regions, which are usually the sites of transcription factor binding.
MNase-seq is employed to probe nucleosome positioning, and experiments are performed using MNase digestion to fragment chromatin without crosslinking [40]. Unlike DNase, MNase has both exonuclease and endonuclease activity, after binding to open chromatin. It digests DNA right up until it encounters obstacles such as transcription factors and nucleosomes and removes the linker DNA, which make MNase-seq ideal for exploring nucleosome positioning. MNase-seq requires fathoming the appropriate enzymatic conditions, inevitably resulting in difficult to control confounding factors, such as sequence binding preferences and enzymatic activity of the enzyme itself [41]. ChIP-seq, DNase-seq and MNase-seq measure transcription factor mapping, chromatin accessibility and nucleosome positioning, respectively. They all require large amounts of input material and yield ‘averaged’ profiles that are insensitive to cellular heterogeneity, which significantly limits the application of these technologies in some rare and precious samples like early embryos. Moreover, these technologies involve complicated and time-consuming sample preparation and library construction, and cannot directly investigate the interactions between nucleosome positioning, chromatin accessibility, and transcription factor mapping. Specifically, the most considerable limitations are:
High cellular input masks heterogeneity between cell populations; the requirement for input material limits the application of DNase-seq and MNase-seq in specific clinical samples and makes them difficult to achieve personalized epigenomics studies.
To obtain the required amount of cells, cells often undergo an in vitro culture for amplification. However, in vitro culture does not fully mimic in vivo conditions and may further add extraneous factors that may cause alterations in chromatin state, thus increasing the risk of experimental failure.
The development of ATAC-seq
Assay for transposase-accessible chromatin with high-throughput sequencing (ATAC-seq) technology successfully achieves simultaneous identification of open chromatin regions, nucleosome positioning, and regulatory motifs while reducing input material to 500 ~ 5000 cells [42]. Using motif inference, scientists have successfully inferred the binding sites of DNA-binding proteins in the B cell line. Also, ATAC-seq has a relatively simple and efficient ‘two-step’ library-preparation procedure: transposition and PCR (Fig. 1b). In ATAC-seq experiments, the nuclei of cells are collected after nucleus-cytoplasm separation, and the chromatin inside the nucleus is fragmented by Tn5 transposase and ligated to sequencing adapters, which largely simplifies the protocol. Tightly packed chromatin DNA is not accessible for transposome, whereas chromatin DNA in open regions is randomly inserted and fragmented by transposome. Fragmented DNAs are collected for subsequent analysis. The most significant innovation of ATAC-seq is based on the application of Tn5 transposase: wide-type Tn5 transposase has low activity in transposition [43, 44]. For better usage in scientific research, after directed evolution, it can become hyperactive Tn5 transposase with increased affinity for DNA. Tn5 transposase can be assembled with designed adapters in vitro to form the active transposome complex. Although Tn5 transposase unavoidably brings about bias due to sequence-dependent binding, this transposition bias can be corrected by the development of computational tools [45].
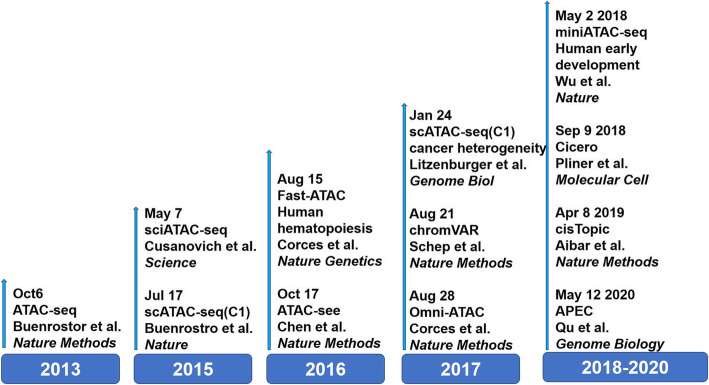
ATAC-seq has the advantage of high efficiency and low cellular input requirement, but its applicability to different types of samples is still limited. ATAC-seq-derived technologies generated for expanding applications include fast-ATAC, Omni-ATAC, and miniATAC-seq (Fig. 3) [46–48]. For example, the fast-ATAC is optimized for blood samples, using a digitonin-contained transposition buffer to combine the two steps of permeabilization and transposition into one step. It not only increases the fragment yield per cell but also dramatically reduces the proportion of mitochondrial reads [46]. Omni-ATAC can be applied to a variety of cell types and long-term preserved frozen samples. Improvements, including a variety of different detergents, washing with Tween-20 after cell lysis, and the use of phosphate buffer saline to enhance the signal-to-noise ratio in the transposition reaction. All those improvements allow Omni-ATAC’s application to a wide range of cell lines, tissue types, and frozen samples stored for long periods while improving data quality [47]. miniATAC-seq has been optimized primarily for DNA purification steps and lysis buffer (the optimized concentration of NP-40 for embryos), and can even be applied in 20 cells (e.g. early embryos) with high-quality sequencing data [48]. Optimization of the ATAC-seq protocol has also been extensively used to cells from neural tissues and bio-banked specimens [49, 50].
Fig. 3.
Development of ATAC-seq and analysis tools
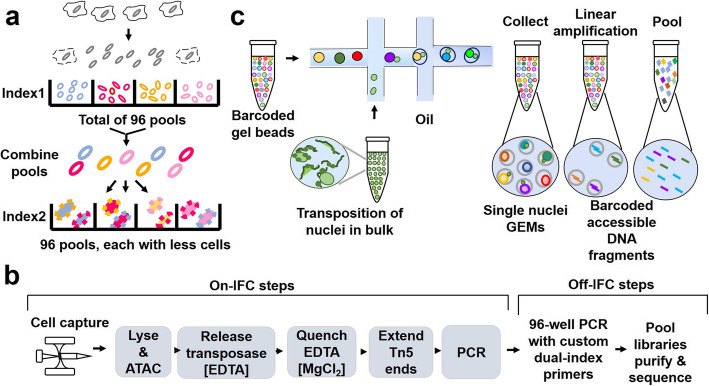
ATAC-seq can measure samples with as low as 500 cells, but it has not been able to decipher cell-to-cell variability at the single-cell level. The advent of single-cell sequencing technology has enabled us to understand life at a finer single-cell level [34, 51]. The information that can be obtained from single-cell transcriptome sequencing is quite limited, and further study of the epigenomic dynamics of regulatory elements at the single-cell level is essential to figure out the more detailed mechanisms of cell differentiation and development. In 2015, the single-cell combinatorial indexing assay for transposase accessible chromatin with sequencing (sci-ATAC-seq) was developed, which enabled the sequencing of a large number of individual cells at the same time [52]. In this strategy, cells were uniquely labeled and sequenced for chromatin accessibility at the single-cell level. After first barcoding all nuclei in the 96-well plate with barcoded Tn5, the nuclei are then pooled and redistributed to a new set of wells. Thus a second barcode could be added by PCR amplification (Fig. 4a). The first scATAC-seq technology based on a microfluidic platform is integrated with Fluidigm single-cell platform C1 (integrated fluidic circuit) (Fig. 4b), it is an automated method for single-cell chromatin accessibility mapping. It can capture more reads per cell than sci-ATAC-seq, opening up the exploring of the diversity of inter-cellular regulators (regulome) [53]. Droplet-based scATAC-seq (10 × ATAC-seq) performed on the Chromium platform (10 × Genomics) achieves a dramatic and unprecedented increase in the throughput of scATAC-seq experiments (Fig. 4c). Single-cell nuclei suspensions are loaded into microfluidics to promote the formation of gel bead in emulsion (GEM). Within each GEM, gel beads oligos were newly designed to consist of a 29-bp sequencing adapter, a 16 bp barcode (to index droplets) and the first 14 bp of read 1 N (primers of the linear amplification reaction), allowing thousands of cells to be measured per experiment. Applying 10 × scATAC-seq, researchers achieved a more excellent mapping of differential open chromatin in immune cells within the tumor microenvironment [54]. To interpret the scATAC-seq data, which are more sparse compared to single-cell RNA-seq data, scientists have developed a variety of computational tools such as chromVAR [55], Cicero [56], cisTopic [57], APEC [58], etc.
Fig. 4.
Workflows of single-cell ATAC-seq. a The combinatorial indexing method of sci-ATAC-seq. The first barcodes are introduced by Tn5 transposase and the second indexing is introduced by amplification using primers containing a second barcode. b scATAC-seq based on the integrated fluidic circuit (IFC). In scATAC-seq using a microfluidics platform (Fluidigm), after transposition and PCR on the IFC, libraries were collected and PCR amplified with cell-identifying barcoded primers. Single-cell libraries were then pooled and sequenced on a high-throughput sequencing instrument. c Workflow of droplet-based scATAC-seq (10 × ATAC-seq) implemented on the Chromium platform (10 × Genomics). GEM: gel bead in emulsion
Interactions between ChIP-seq & ATAC-seq
ChIP-seq alone may be challenging to find meaningful regulatory elements when comparing two or more samples. In this regard, ATAC-seq has the advantage of high resolution and high signal-to-noise ratio, which allows researchers to identify differentially regulated sequences, such as enhancers. Combining ChIP-seq with ATAC-seq makes it easier to determine significant peaks. ChIP-seq methods are still irreplaceable although methods for detecting open chromatin, such as DNase-seq and ATAC-seq can help to infer genomic features. All those techniques are indirect for detecting the binding sites of transcription factors to DNA, which means that the transcription factors mapping inferred from the enriched motifs still needs to be further validated [35, 59]. The probable limitations of using ATAC-seq alone are:
Not all chromatin regulators have a corresponding motif, regulators such as chromatin remodeling proteins do not have a DNA sequence preference. In contrast, the interaction between chromatin remodeling proteins and nucleosomes is often an essential factor in early embryonic development as well as cell fate decisions [60]. Open chromatin information alone cannot be used to infer the binding profiles of such factors.
Some motifs have the potential to be bound by multiple sequence-specific transcription factors [61].
Some of the homologous transcription factors bind in similar motif patterns, in many cases, the direct assignment of open chromatin peaks to specific individual transcription factors is less reliable. Results obtained from direct detection of interactions between specific transcription factors and DNA are often more reliable.
The genome-wide chromatin accessibility profile detected using ATAC-seq represents another level of the chromatin regulatory landscape compared to ChIP-seq, which directly determines specific DNA-protein interactions (Table 2). Therefore, combining ATAC-seq and ChIP-seq will help scientists gain a more comprehensive and in-depth understanding of chromatin regulatory dynamics and their biological significance.
Table 2.
Comparisons between ChIP-seq and ATAC-seq
| Factor | ChIP-seq | ATAC-seq |
|---|---|---|
| Dependence on antibody | Yes, ChIP-seq requires good and specific antibodies | No |
| Throughput for detection | Only for Specific proteins | Global chromatin accessibility |
| Controls | Required, for distinguishing real peak regions from artifacts | Usually not required, but naked DNA controls are useful for characterizing the sequence bias of enzymatically induced cleavage |
| Results | Direct interactions between proteins and DNA | Potential binding sites for regulators |
Applications resetting understanding of the biological question
ChIP-seq & ATAC-seq approaches to refine our understanding of chromatin remodeling in early embryos
Mammalian embryo development involves genome-wide epigenetic changes, including DNA methylation, histone modifications, open chromatin and chromatin conformation [62]. Crucially, many genomic regulatory elements such as promoters, enhancers, insulators, and locus control regions guide embryonic development through interactions with cell type-specific transcription factors [63]. Also, long-range interactions between these regulatory elements are intriguing (Fig. 5a). By innovatively optimizing the protocol of ChIP, it can be available for histone modification reconstruction studies in very low cellular input such as embryos (Fig. 5b). In early mouse embryos, H3K4me3 undergoes extensive reprogramming events, where it disappears in the zygote and is reconstructed again in the offspring, accompanied by zygotic genome activation (ZGA). Thus, the application of ChIP-seq optimization in embryos may help to reveal the detailed process of inheritance of mammalian histone modifications from parents to offspring, i.e., the differences in parental modification patterns before and after fertilization [5, 64, 65]. More importantly, the application of improved ATAC-seq methods in embryonic tissues facilitates the mapping of genome-wide chromatin accessibility at critical periods of embryo development. In preimplantation embryos, for example, ATAC-seq is utilized to reveal the temporal dynamics of high-resolution chromatin changes in ZGA and minor zygotic genome activation (minorZGA) [66]. Additional epigenomic studies have also shown unique chromatin states at different stages of embryo development [67, 68]. These findings provide valuable clues for further interrogating human embryo development and clinical guidance in the future. More questions, such as the key factors and transposons that regulate chromatin state transitions, remain to be discovered.
Fig. 5.

Future applications of single-cell epigenomics. a Long-range interaction of regulatory elements. b Chromatin dynamics in rare cell types (e.g., early embryos). c Cell lineage tracking. d Deconvolution of intercellular heterogeneity
Single-cell approaches to probe developmental processes in organs
Epigenome sequencing technology plays an irreplaceable role for experimental sytem to track the developmental trajectories and explore the cell fate determination mechanisms (Fig. 5c). ChIP-seq and ATAC-seq at the single-cell level help to comprehensively resolve the developmental dynamics of tissues and organs. In 2018, scATAC-seq was applied to cluster cells at different stages of mouse forebrain development and used to identify key regulatory factors inferred from the accessible chromatin peaks [69]. Integrating ChIP-seq, ATAC-seq, and DNase-seq in organoid models [70] with transcriptomic data will help to reveal the developmental dynamics of specific cells, discover key transcriptional regulators, and identify disease susceptible cell populations [71–73]. Multi-omics integration of single-cell transcriptome and ATAC-seq in organ development can provide a robust foundation for the clinical treatment of diseases. For example, by revealing the critical time points and gene regulatory networks of human hippocampus development from a comprehensive perspective, researchers provided information on potential cell groups involved in the pathology of Parkinson’s disease, Alzheimer’s disease and Huntington’s disease [74].
In addition to neurobiology studies, chromatin dynamics analysis has shed light on muscle development [75], mammary gland development [76], and cardiac precursor cell fate determination [77]. Fine-grained studies of single-cell chromatin dynamics will help to create a global map of human organ development in the future, enabling us to trace the embryonic origin of every tissue and organ.
Single-cell approaches to assess the complexity of cancer
There is currently limited understanding of the highly heterogeneous tumor tissues, including differences in the tumor microenvironment, differences between original primary cancer and metastases, and the evolution of tumor subclones. Immune cells in the tumor microenvironment are often critical for the immune escape and infiltration processes of cancer cells [78, 79]. scATAC-seq and its expanded application will help to reveal the heterogeneity of the epigenetic landscape in tumor development and provide potential targets for therapy (Fig. 5d) [80–82]. For example, the application of scATAC-seq has determined the regulatory network of malignant, stromal and immune cells in the tumor microenvironment. The intra-tumor T cell exhaustion in the tumor microenvironment of patients was compared before and after immunotherapy by detecting the dynamics of immune cell development at single cell level [54]. The key regulatory T cell populations that respond to immunotherapy can be identified. Integrated analysis of ChIP-seq, ATAC-seq and DNA mutation profile in the same cell will enable scientists to uncover new subclones of cancer cells, allowing for personalized clinical trials. Thus, understanding the chromatin regulatory landscape at the single-cell level will significantly accelerate biomedical advances in cancer therapy [83].
Extended technologies of ATAC-seq can also provide new insights into tumor heterogeneity. Assay of transposase-accessible chromatin with visualization (ATAC-see) helps to image open chromatin in situ by fluorescently labeling accessible loci [84]. For example, by using ATAC-see and fluorescence in situ hybridization [85], scientists have provided physical evidence of the co-localization of extrachromosomal DNA (ecDNA) [86] and ATAC-see fluorescence signals. Confirmed by ATAC-seq and MNase-seq data, this result suggests that ecDNA is highly accessible, which may explain why oncogenes on ecDNA can be expressed in large quantities [87]. Therefore, the adaptation of ChIP-seq and ATAC-seq technologies can provide new directions for targeted therapies, and supply physical evidence of imaging to the heterogeneity of cancers, making scientific findings more comprehensive and reliable.
Conclusion and future directions
Combination of scRNA-seq and scATAC-seq
To construct a complete regulatory network, RNA-seq, ATAC-seq and ChIP-seq often need to be combined. Although many algorithms have been developed to integrate multi-omics data, it is difficult to assess the performance of these algorithms and whether they fully preserve the biological variance. A growing number of studies in recent years have demonstrated the potential of parallel analysis of multiple modalities in the same cells [88–90]. Single-nucleus chromatin accessibility and mRNA expression sequencing (SNARE-seq) based on micro-droplet platform enables simultaneous sequencing of transcriptome and chromatin accessibility in a single cell by dual-omics capturing, and can appropriately correlate the results of both modalities to obtain detailed information on the regulation of gene expression [91]. sci-CAR is a combinatorial indexing–based method that allows co-assay of chromatin accessibility and mRNA (CAR) in thousands of single cells, combining sci-ATAC-seq and sci-RNA-seq [92]. Paired-seq is an ultra-high-throughput method for parallel analysis of transcriptome and accessible chromatin in millions of individual cells [93]. In this method, a ligation-based combinatorial indexing strategy is adopted to simultaneously tag both the open chromatin fragments generated by the Tn5 transposases and the cDNA molecules produced from reverse transcription of RNA in millions of cells. Compared to SNARE-seq and sci-CAR, Paired-seq has a much higher throughput, which makes it possible for analysis of gene regulatory programs at an organismal scale. A significant advantage of open chromatin mapping over the transcriptome [94] is that accessibility maps provide expression status of genes and the network of interactions between regulatory elements, so by combining ATAC-seq with RNA-seq, more compelling biological questions will be solved. For example, spatiotemporal gene expression patterns are highly correlated with cis-regulatory elements and thus differ between individual cells. Based on this premise, how these heterogeneous cells act in coordination to construct a comprehensive network of cell communication is intriguing.
The promise and limitations of probing chromatin in single cells
Compared to open chromatin data derived from populations of cells, scATAC-seq signals are binary and sparse, so a new analytic framework to account for fundamental differences from bulk data is needed. A feasible approach is to aggregate information from many single cells to identify determinants of cell-to-cell chromatin variation [54]. As the ends of each fragment are indicative of regions of open chromatin, it is possible to analyze the combined signal from these fragments to determine regions of the genome enriched for open chromatin and thus, putatively of regulatory and functional significance. However, a theoretical disadvantage of such a method would be not being able to identify rare peaks appearing only in scarce populations. One major limitation to current scATAC-seq approaches is that they capture only a subset of the open chromatin sites in single cells, a lot of sites may be lost or not detectable during both experimental and computational procedures. It seems unlikely that more comprehensive coverage can be achieved in the near future. Higher per-cell coverage would allow new questions to be answered. For example, it is still confusing how chromatin accessibility differs between the two alleles in a single cell, or how do open chromatin regions correlate in an individual cell [95].
Prospects for spatial epigenomes
Tn5 transposase is widely used for chromatin accessibility sequencing, linear amplification via transposon insertion [96] and even detection of potential pathogens co-infected with coronavirus [97]. The ability of Tn5 to anneal a variety of adapters for personalized applications is an essential aspect of proposing innovative research designs, such as coupling adapters with fluorescence for accessible chromatin imaging [84]. Besides, modifications to Tn5 and the design of protocols would further realize high-throughput and large-scale experiments as undisclosed reagents severely limit the development of novel applications [98]. If scientists could efficiently personalize library preparation procedures, it would allow epigenome sequencing technology to benefit more large-scale projects like The Encyclopedia of DNA Elements (ENCODE) [99, 100]. For example, draw upon the principles of the spatial transcriptome [101], once Tn5 is available in bulk, it may be possible to sequence samples directly without cell dissociation and centrifugation steps, thereby retaining the original state of cells and significantly reducing loss. Also, spatial epigenomes can be realized by mapping the chromatin states of individual cells to their location information. Although many studies have yielded an excellent temporal map of tissue and organ development, the challenge remains to improve further the spatial resolution of developmental studies, such as cell migration trajectories in embryonic organs at different stages.
Not only the spatial location and migration of cells are required, but also the spatial dynamics on the molecular level is crucial. Genome-wide chromatin conformational changes within cells are important mechanisms for regulating cell behavior [102]. The HiChIP technology has successfully resolved the dynamics of chromatin structure with high sensitivity and high resolution by combining ChIP with chromatin conformation capture [103]. Therefore, with the further development and improvement of the technologies involved in analyzing the dynamics of the epigenome, there are broad and attractive prospects to integrate different omics and experimental techniques for multi-dimensional life science research. A more comprehensive map would also provide a better understanding of the interplay of multiple regulatory elements within individual cells.
Acknowledgments
We apologize to colleagues whose work has not been cited due to space limitations and the broad scope of this review.
Consent to participate
Not applicable.
Code availability
Not applicable.
Abbreviations
- ATAC-see
Assay of transposase-accessible chromatin with visualization
- ATAC-seq
Assay for transposase-accessible chromatin with high-throughput sequencing
- ChIP
Chromatin immunoprecipitation
- ChIP-chip
Chromatin immunoprecipitation (ChIP) followed by microarray hybridization (chip)
- ChIP-seq
Chromatin immunoprecipitation sequencing
- CoBatch
Combinatorial barcoding and targeted chromatin release
- CUT&RUN
Cleavage under targets and release using nuclease
- CUT&Tag
Cleavage under targets and tagmentation
- DNase-seq
DNase I hypersensitive site sequencing
- Drop-ChIP
Droplet-based single-cell ChIP-seq
- ecDNA
Extrachromosomal DNA
- FACS
Fluorescence activated cell sorting
- GEM
Gel bead in emulsion
- IFC
Integrated fluidic circuit
- MNase
Micrococcal nuclease
- MNase-seq
MNase digestion with deep sequencing
- pA/G-Tn5
Protein A/G-fused Tn5 transposase
- RNA-seq
RNA sequencing
- scATAC-seq
Single-cell ATAC-seq
- scChIP-seq
Single-cell ChIP-seq
- sci-ATAC-seq
Single-cell combinatorial indexing assay for transposase accessible chromatin with sequencing
- sci-CAR
Single-cell combinatorial indexing-based co-assay of chromatin accessibility and mRNA
- sc-itChIP-seq
Single-cell simultaneous indexing and tagmentation-based ChIP-seq
- SNARE-seq
Single-nucleus chromatin accessibility and mRNA expression sequencing
- ZGA
Zygotic genome activation
Authors’ contributions
Shaoqian Ma provided the idea for the article and conducted literature searching and draft. Yongyou Zhang reviewed and/or edited of the manuscript before submission. All authors read and approved the final manuscript.
Funding
This work was supported by a research grant of the national undergraduate training program for innovation and entrepreneurship. (no. 2019X0670 to Shaoqing Ma).
Availability of data and materials
Not applicable.
Ethics approval
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Shaoqian Ma, Email: shaoqianma@qq.com.
Yongyou Zhang, Email: yongyouzhang@xmu.edu.cn.
References
- 1.Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci U S A. 2010;107(50):21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128(4):693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 3.Hublitz P, Albert M, Peters AHFM. Mechanisms of transcriptional repression by histone lysine methylation. Int J Dev Biol. 2009;53(2–3):335–354. doi: 10.1387/ijdb.082717ph. [DOI] [PubMed] [Google Scholar]
- 4.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125(2):315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 5.Xia W, Xu J, Yu G, Yao G, Xu K, Ma X, et al. Resetting histone modifications during human parental-to-zygotic transition. Science. 2019;365(6451):353–360. doi: 10.1126/science.aaw5118. [DOI] [PubMed] [Google Scholar]
- 6.Allis CD, Jenuwein T. The molecular hallmarks of epigenetic control. Nat Rev Genet. 2016;17(8):487–500. doi: 10.1038/nrg.2016.59. [DOI] [PubMed] [Google Scholar]
- 7.Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A, et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518(7539):317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leung D, Jung I, Rajagopal N, Schmitt A, Selvaraj S, Lee AY, et al. Integrative analysis of haplotype-resolved epigenomes across human tissues. Nature. 2015;518(7539):350–354. doi: 10.1038/nature14217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meyer CA, Liu XS. Identifying and mitigating bias in next-generation sequencing methods for chromatin biology. Nat Rev Genet. 2014;15(11):709–721. doi: 10.1038/nrg3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293(5532):1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 11.Liu CL, Kaplan T, Kim M, Buratowski S, Schreiber SL, Friedman N, et al. Single-nucleosome mapping of histone modifications in S. cerevisiae. PLoS Biol. 2005;3(10):e328. doi: 10.1371/journal.pbio.0030328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vakoc CR, Mandat SA, Olenchock BA, Blobel GA. Histone H3 lysine 9 methylation and HP1gamma are associated with transcription elongation through mammalian chromatin. Mol Cell. 2005;19(3):381–391. doi: 10.1016/j.molcel.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 13.Romanoski CE, Glass CK, Stunnenberg HG, Wilson L, Almouzni G. Epigenomics: roadmap for regulation. Nature. 2015;518(7539):314–316. doi: 10.1038/518314a. [DOI] [PubMed] [Google Scholar]
- 14.Stark R, Grzelak M, Hadfield J. RNA sequencing: the teenage years. Nat Rev Genet. 2019;20(11):631–656. doi: 10.1038/s41576-019-0150-2. [DOI] [PubMed] [Google Scholar]
- 15.Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447(7143):407–412. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- 16.Bernstein BE, Meissner A, Lander ES. The mammalian epigenome. Cell. 2007;128(4):669–681. doi: 10.1016/j.cell.2007.01.033. [DOI] [PubMed] [Google Scholar]
- 17.Park M, Patel N, Keung AJ, Khalil AS. Engineering epigenetic regulation using synthetic read-write modules. Cell. 2019;176(1–2):227–38.e20. doi: 10.1016/j.cell.2018.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ren B, Robert F, Wyrick JJ, Aparicio O, Jennings EG, Simon I, et al. Genome-wide location and function of DNA binding proteins. Science. 2000;290(5500):2306–2309. doi: 10.1126/science.290.5500.2306. [DOI] [PubMed] [Google Scholar]
- 19.Park PJ. ChIP-seq: advantages and challenges of a maturing technology. Nat Rev Genet. 2009;10(10):669–680. doi: 10.1038/nrg2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barski A, Cuddapah S, Cui K, Roh T-Y, Schones DE, Wang Z, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129(4):823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 21.Robertson G, Hirst M, Bainbridge M, Bilenky M, Zhao Y, Zeng T, et al. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat Methods. 2007;4(8):651–657. doi: 10.1038/nmeth1068. [DOI] [PubMed] [Google Scholar]
- 22.Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316(5830):1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- 23.Kidder BL, Hu G, Zhao K. ChIP-Seq: technical considerations for obtaining high-quality data. Nat Immunol. 2011;12(10):918–922. doi: 10.1038/ni.2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O'Neill L. Immunoprecipitation of native chromatin: NChIP. Methods. 2003;31(1):76–82. doi: 10.1016/s1046-2023(03)00090-2. [DOI] [PubMed] [Google Scholar]
- 25.Kasinathan S, Orsi GA, Zentner GE, Ahmad K, Henikoff S. High-resolution mapping of transcription factor binding sites on native chromatin. Nat Methods. 2014;11(2):203–209. doi: 10.1038/nmeth.2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gilfillan GD, Hughes T, Sheng Y, Hjorthaug HS, Straub T, Gervin K, et al. Limitations and possibilities of low cell number ChIP-seq. BMC Genomics. 2012;13:645. doi: 10.1186/1471-2164-13-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schmidl C, Rendeiro AF, Sheffield NC, Bock C. ChIPmentation: fast, robust, low-input ChIP-seq for histones and transcription factors. Nat Methods. 2015;12(10):963–965. doi: 10.1038/nmeth.3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brind'Amour J, Liu S, Hudson M, Chen C, Karimi MM, Lorincz MC. An ultra-low-input native ChIP-seq protocol for genome-wide profiling of rare cell populations. Nat Commun. 2015;6:6033. doi: 10.1038/ncomms7033. [DOI] [PubMed] [Google Scholar]
- 29.Cao Z, Chen C, He B, Tan K, Lu C. A microfluidic device for epigenomic profiling using 100 cells. Nat Methods. 2015;12(10):959–962. doi: 10.1038/nmeth.3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Skene PJ, Henikoff JG, Henikoff S. Targeted in situ genome-wide profiling with high efficiency for low cell numbers. Nat Protoc. 2018;13(5):1006–1019. doi: 10.1038/nprot.2018.015. [DOI] [PubMed] [Google Scholar]
- 31.Hainer SJ, Bošković A, McCannell KN, Rando OJ, Fazzio TG. Profiling of pluripotency factors in single cells and early embryos. Cell. 2019;177(5):1319–29.e11. doi: 10.1016/j.cell.2019.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaya-Okur HS, Wu SJ, Codomo CA, Pledger ES, Bryson TD, Henikoff JG, et al. CUT&tag for efficient epigenomic profiling of small samples and single cells. Nat Commun. 2019;10(1):1930. doi: 10.1038/s41467-019-09982-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rotem A, Ram O, Shoresh N, Sperling RA, Goren A, Weitz DA, et al. Single-cell ChIP-seq reveals cell subpopulations defined by chromatin state. Nat Biotechnol. 2015;33(11):1165–1172. doi: 10.1038/nbt.3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tang F, Barbacioru C, Wang Y, Nordman E, Lee C, Xu N, et al. mRNA-Seq whole-transcriptome analysis of a single cell. Nat Methods. 2009;6(5):377–382. doi: 10.1038/nmeth.1315. [DOI] [PubMed] [Google Scholar]
- 35.Grosselin K, Durand A, Marsolier J, Poitou A, Marangoni E, Nemati F, et al. High-throughput single-cell ChIP-seq identifies heterogeneity of chromatin states in breast cancer. Nat Genet. 2019;51(6):1060–1066. doi: 10.1038/s41588-019-0424-9. [DOI] [PubMed] [Google Scholar]
- 36.Mezger A, Klemm S, Mann I, Brower K, Mir A, Bostick M, et al. High-throughput chromatin accessibility profiling at single-cell resolution. Nat Commun. 2018;9(1):3647. doi: 10.1038/s41467-018-05887-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ai S, Xiong H, Li CC, Luo Y, Shi Q, Liu Y, et al. Profiling chromatin states using single-cell itChIP-seq. Nat Cell Biol. 2019;21(9):1164–1172. doi: 10.1038/s41556-019-0383-5. [DOI] [PubMed] [Google Scholar]
- 38.Wang Q, Xiong H, Ai S, Yu X, Liu Y, Zhang J, et al. CoBATCH for high-throughput single-cell epigenomic profiling. Mol Cell. 2019;76(1):206–16.e7. doi: 10.1016/j.molcel.2019.07.015. [DOI] [PubMed] [Google Scholar]
- 39.Boyle AP, Song L, Lee B-K, London D, Keefe D, Birney E, et al. High-resolution genome-wide in vivo footprinting of diverse transcription factors in human cells. Genome Res. 2011;21(3):456–464. doi: 10.1101/gr.112656.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schones DE, Cui K, Cuddapah S, Roh T-Y, Barski A, Wang Z, et al. Dynamic regulation of nucleosome positioning in the human genome. Cell. 2008;132(5):887–898. doi: 10.1016/j.cell.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gaffney DJ, McVicker G, Pai AA, Fondufe-Mittendorf YN, Lewellen N, Michelini K, et al. Controls of nucleosome positioning in the human genome. PLoS Genet. 2012;8(11):e1003036. doi: 10.1371/journal.pgen.1003036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods. 2013;10(12):1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reznikoff WS. Tn5 as a model for understanding DNA transposition. Mol Microbiol. 2003;47(5):1199–1206. doi: 10.1046/j.1365-2958.2003.03382.x. [DOI] [PubMed] [Google Scholar]
- 44.Goryshin IY, Reznikoff WS. Tn5 in vitro transposition. J Biol Chem. 1998;273(13):7367–7374. doi: 10.1074/jbc.273.13.7367. [DOI] [PubMed] [Google Scholar]
- 45.Viswanadham VV, Mahajan VS, Pillai S. A Bayesian approach for correcting Tn5 transposition bias in ATAC-seq footprinting. bioRxiv. 2019:525808. 10.1101/525808.
- 46.Corces MR, Buenrostro JD, Wu B, Greenside PG, Chan SM, Koenig JL, et al. Lineage-specific and single-cell chromatin accessibility charts human hematopoiesis and leukemia evolution. Nat Genet. 2016;48(10):1193–1203. doi: 10.1038/ng.3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Corces MR, Trevino AE, Hamilton EG, Greenside PG, Sinnott-Armstrong NA, Vesuna S, et al. An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat Methods. 2017;14(10):959–962. doi: 10.1038/nmeth.4396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu J, Xu J, Liu B, Yao G, Wang P, Lin Z, et al. Chromatin analysis in human early development reveals epigenetic transition during ZGA. Nature. 2018;557(7704):256–260. doi: 10.1038/s41586-018-0080-8. [DOI] [PubMed] [Google Scholar]
- 49.Milani P, Escalante-Chong R, Shelley BC, Patel-Murray NL, Xin X, Adam M, et al. Cell freezing protocol suitable for ATAC-Seq on motor neurons derived from human induced pluripotent stem cells. Sci Rep. 2016;6:25474. doi: 10.1038/srep25474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Scharer CD, Blalock EL, Barwick BG, Haines RR, Wei C, Sanz I, et al. ATAC-seq on biobanked specimens defines a unique chromatin accessibility structure in naïve SLE B cells. Sci Rep. 2016;6:27030. doi: 10.1038/srep27030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Picelli S. Single-cell RNA-sequencing: the future of genome biology is now. RNA Biol. 2017;14(5):637–650. doi: 10.1080/15476286.2016.1201618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cusanovich DA, Daza R, Adey A, Pliner HA, Christiansen L, Gunderson KL, et al. Multiplex single cell profiling of chromatin accessibility by combinatorial cellular indexing. Science. 2015;348(6237):910–914. doi: 10.1126/science.aab1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Buenrostro JD, Wu B, Litzenburger UM, Ruff D, Gonzales ML, Snyder MP, et al. Single-cell chromatin accessibility reveals principles of regulatory variation. Nature. 2015;523(7561):486–490. doi: 10.1038/nature14590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Satpathy AT, Granja JM, Yost KE, Qi Y, Meschi F, McDermott GP, et al. Massively parallel single-cell chromatin landscapes of human immune cell development and intratumoral T cell exhaustion. Nat Biotechnol. 2019;37(8):925–936. doi: 10.1038/s41587-019-0206-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schep AN, Wu B, Buenrostro JD, Greenleaf WJ. chromVAR: inferring transcription-factor-associated accessibility from single-cell epigenomic data. Nat Methods. 2017;14(10):975–978. doi: 10.1038/nmeth.4401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pliner HA, Packer JS, McFaline-Figueroa JL, Cusanovich DA, Daza RM, Aghamirzaie D, et al. Cicero predicts cis-regulatory DNA interactions from single-cell chromatin accessibility data. Mol Cell. 2018;71(5):858–71.e8. doi: 10.1016/j.molcel.2018.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bravo González-Blas C, Minnoye L, Papasokrati D, Aibar S, Hulselmans G, Christiaens V, et al. cisTopic: cis-regulatory topic modeling on single-cell ATAC-seq data. Nat Methods. 2019;16(5):397–400. doi: 10.1038/s41592-019-0367-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li B, Li Y, Li K, Zhu L, Yu Q, Cai P, et al. APEC: an accesson-based method for single-cell chromatin accessibility analysis. Genome Biol. 2020;21(1):116. doi: 10.1186/s13059-020-02034-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473(7345):43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zaret KS, Mango SE. Pioneer transcription factors, chromatin dynamics, and cell fate control. Curr Opin Genet Dev. 2016;37:76–81. doi: 10.1016/j.gde.2015.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Long HK, Prescott SL, Wysocka J. Ever-changing landscapes: transcriptional enhancers in development and evolution. Cell. 2016;167(5):1170–1187. doi: 10.1016/j.cell.2016.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hemberger M, Dean W, Reik W. Epigenetic dynamics of stem cells and cell lineage commitment: digging Waddington's canal. Nat Rev Mol Cell Biol. 2009;10(8):526–537. doi: 10.1038/nrm2727. [DOI] [PubMed] [Google Scholar]
- 63.Rivera CM, Ren B. Mapping human epigenomes. Cell. 2013;155(1):39–55. doi: 10.1016/j.cell.2013.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang B, Zheng H, Huang B, Li W, Xiang Y, Peng X, et al. Allelic reprogramming of the histone modification H3K4me3 in early mammalian development. Nature. 2016;537(7621):553–557. doi: 10.1038/nature19361. [DOI] [PubMed] [Google Scholar]
- 65.Zheng H, Huang B, Zhang B, Xiang Y, Du Z, Xu Q, et al. Resetting epigenetic memory by reprogramming of histone modifications in mammals. Mol Cell. 2016;63(6):1066–1079. doi: 10.1016/j.molcel.2016.08.032. [DOI] [PubMed] [Google Scholar]
- 66.Wu J, Huang B, Chen H, Yin Q, Liu Y, Xiang Y, et al. The landscape of accessible chromatin in mammalian preimplantation embryos. Nature. 2016;534(7609):652–657. doi: 10.1038/nature18606. [DOI] [PubMed] [Google Scholar]
- 67.Xiang Y, Zhang Y, Xu Q, Zhou C, Liu B, Du Z, et al. Epigenomic analysis of gastrulation identifies a unique chromatin state for primed pluripotency. Nat Genet. 2019;52(1):95–105. doi: 10.1038/s41588-019-0545-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xie W, Schultz MD, Lister R, Hou Z, Rajagopal N, Ray P, et al. Epigenomic analysis of multilineage differentiation of human embryonic stem cells. Cell. 2013;153(5):1134–1148. doi: 10.1016/j.cell.2013.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Preissl S, Fang R, Huang H, Zhao Y, Raviram R, Gorkin DU, et al. Single-nucleus analysis of accessible chromatin in developing mouse forebrain reveals cell-type-specific transcriptional regulation. Nat Neurosci. 2018;21(3):432–439. doi: 10.1038/s41593-018-0079-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Takebe T, Wells JM. Organoids by design. Science. 2019;364(6444):956–959. doi: 10.1126/science.aaw7567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Trevino AE, Sinnott-Armstrong N, Andersen J, Yoon S-J, Huber N, Pritchard JK, et al. Chromatin accessibility dynamics in a model of human forebrain development. Science. 2020;367(6476). 10.1126/science.aay1645. [DOI] [PMC free article] [PubMed]
- 72.Amiri A, Coppola G, Scuderi S, Wu F, Roychowdhury T, Liu F, et al. Transcriptome and epigenome landscape of human cortical development modeled in organoids. Science. 2018;362(6420). 10.1126/science.aat6720. [DOI] [PMC free article] [PubMed]
- 73.Xie H, Zhang W, Zhang M, Akhtar T, Li Y, Yi W, et al. Chromatin accessibility analysis reveals regulatory dynamics of developing human retina and hiPSC-derived retinal organoids. Sci Adv. 2020;6(6):eaay5247. doi: 10.1126/sciadv.aay5247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhong S, Ding W, Le S, Lu Y, Dong H, Fan X, et al. Decoding the development of the human hippocampus. Nature. 2020;577(7791):531–536. doi: 10.1038/s41586-019-1917-5. [DOI] [PubMed] [Google Scholar]
- 75.Pliner HA, Packer J, McFaline-Figueroa JL, Cusanovich DA, Daza R, Srivatsan S, et al. Chromatin accessibility dynamics of myogenesis at single cell resolution. bioRxiv. 2017:155473. 10.1101/155473.
- 76.Chung C-Y, Ma Z, Dravis C, Preissl S, Poirion O, Luna G, et al. Single-cell chromatin analysis of mammary gland development reveals cell-state transcriptional regulators and lineage relationships. Cell Rep. 2019;29(2):495–510.e6. doi: 10.1016/j.celrep.2019.08.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jia G, Preussner J, Chen X, Guenther S, Yuan X, Yekelchyk M, et al. Single cell RNA-seq and ATAC-seq analysis of cardiac progenitor cell transition states and lineage settlement. Nat Commun. 2018;9(1):4877. doi: 10.1038/s41467-018-07307-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Beatty GL, Gladney WL. Immune escape mechanisms as a guide for cancer immunotherapy. Clin Cancer Res. 2015;21(4):687–692. doi: 10.1158/1078-0432.CCR-14-1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Dunn GP, Old LJ, Schreiber RD. The three Es of cancer immunoediting. Annu Rev Immunol. 2004;22:329–360. doi: 10.1146/annurev.immunol.22.012703.104803. [DOI] [PubMed] [Google Scholar]
- 80.Al-Ali R, Bauer K, Park J-W, Al Abdulla R, Fermi V, Deimling A, et al. Single-nucleus chromatin accessibility reveals intratumoral epigenetic heterogeneity in IDH1 mutant gliomas. Acta Neuropathol Commun. 2019;7(1):201. doi: 10.1186/s40478-019-0851-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mazor T, Pankov A, Song JS, Costello JF. Intratumoral heterogeneity of the epigenome. Cancer Cell. 2016;29(4):440–451. doi: 10.1016/j.ccell.2016.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ramón Y, Cajal S, Sesé M, Capdevila C, Aasen T, Mattos-Arruda L, Diaz-Cano SJ, et al. Clinical implications of intratumor heterogeneity: challenges and opportunities. J Mol Med (Berl) 2020;98(2):161–177. doi: 10.1007/s00109-020-01874-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Clark SJ, Lee HJ, Smallwood SA, Kelsey G, Reik W. Single-cell epigenomics: powerful new methods for understanding gene regulation and cell identity. Genome Biol. 2016;17:72. doi: 10.1186/s13059-016-0944-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chen X, Shen Y, Draper W, Buenrostro JD, Litzenburger U, Cho SW, et al. ATAC-see reveals the accessible genome by transposase-mediated imaging and sequencing. Nat Methods. 2016;13(12):1013–1020. doi: 10.1038/nmeth.4031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Langer-Safer PR, Levine M, Ward DC. Immunological method for mapping genes on Drosophila polytene chromosomes. Proc Natl Acad Sci U S A. 1982;79(14):4381–4385. doi: 10.1073/pnas.79.14.4381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Turner KM, Deshpande V, Beyter D, Koga T, Rusert J, Lee C, et al. Extrachromosomal oncogene amplification drives tumour evolution and genetic heterogeneity. Nature. 2017;543(7643):122–125. doi: 10.1038/nature21356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wu S, Turner KM, Nguyen N, Raviram R, Erb M, Santini J, et al. Circular ecDNA promotes accessible chromatin and high oncogene expression. Nature. 2019;575(7784):699–703. doi: 10.1038/s41586-019-1763-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhu C, Preissl S, Ren B. Single-cell multimodal omics: the power of many. Nat Methods. 2020;17(1):11–14. doi: 10.1038/s41592-019-0691-5. [DOI] [PubMed] [Google Scholar]
- 89.Kamies R, Martinez-Jimenez CP. Advances of single-cell genomics and epigenomics in human disease: where are we now? Mamm Genome. 2020;31(5–6):170–180. doi: 10.1007/s00335-020-09834-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hasin Y, Seldin M, Lusis A. Multi-omics approaches to disease. Genome Biol. 2017;18(1):83. doi: 10.1186/s13059-017-1215-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chen S, Lake BB, Zhang K. High-throughput sequencing of the transcriptome and chromatin accessibility in the same cell. Nat Biotechnol. 2019. 10.1038/s41587-019-0290-0. [DOI] [PMC free article] [PubMed]
- 92.Cao J, Cusanovich DA, Ramani V, Aghamirzaie D, Pliner HA, Hill AJ, et al. Joint profiling of chromatin accessibility and gene expression in thousands of single cells. Science. 2018;361(6409):1380–1385. doi: 10.1126/science.aau0730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhu C, Yu M, Huang H, Juric I, Abnousi A, Hu R, et al. An ultra high-throughput method for single-cell joint analysis of open chromatin and transcriptome. Nat Struct Mol Biol. 2019;26(11):1063–1070. doi: 10.1038/s41594-019-0323-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Svensson V, Vento-Tormo R, Teichmann SA. Exponential scaling of single-cell RNA-seq in the past decade. Nat Protoc. 2018;13(4):599–604. doi: 10.1038/nprot.2017.149. [DOI] [PubMed] [Google Scholar]
- 95.Pott S, Lieb JD. Single-cell ATAC-seq: strength in numbers. Genome Biol. 2015;16:172. doi: 10.1186/s13059-015-0737-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chen C, Xing D, Tan L, Li H, Zhou G, Huang L, et al. Single-cell whole-genome analyses by linear amplification via transposon insertion (LIANTI) Science. 2017;356(6334):189–194. doi: 10.1126/science.aak9787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lu B, Dong L, Yi D, Zhu C, Zhang M, Li X, et al. Transposase assisted tagmentation of RNA/DNA hybrid duplexes. bioRxiv. 2020. 10.1101/2020.01.29.926105. [DOI] [PMC free article] [PubMed]
- 98.Picelli S, Björklund AK, Reinius B, Sagasser S, Winberg G, Sandberg R. Tn5 transposase and tagmentation procedures for massively scaled sequencing projects. Genome Res. 2014;24(12):2033–2040. doi: 10.1101/gr.177881.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Maher B. ENCODE: the human encyclopaedia. Nature. 2012;489(7414):46–48. doi: 10.1038/489046a. [DOI] [PubMed] [Google Scholar]
- 100.Landt SG, Marinov GK, Kundaje A, Kheradpour P, Pauli F, Batzoglou S, et al. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res. 2012;22(9):1813–1831. doi: 10.1101/gr.136184.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ståhl PL, Salmén F, Vickovic S, Lundmark A, Navarro JF, Magnusson J, et al. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science. 2016;353(6294):78–82. doi: 10.1126/science.aaf2403. [DOI] [PubMed] [Google Scholar]
- 102.Eagen KP. Principles of chromosome architecture revealed by hi-C. Trends Biochem Sci. 2018;43(6):469–478. doi: 10.1016/j.tibs.2018.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Mumbach MR, Rubin AJ, Flynn RA, Dai C, Khavari PA, Greenleaf WJ, et al. HiChIP: efficient and sensitive analysis of protein-directed genome architecture. Nat Methods. 2016;13(11):919–922. doi: 10.1038/nmeth.3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.