Significance
The plasma membrane (PM) acts as the interface between intra- and extracellular environments and is thus important for intercellular communication and extracellular signal perception. The composition and amounts of PM proteins are tightly regulated, by molecular mechanisms that remain largely unknown in plant cells. We identified a pair of ANTH domain-containing proteins functioning as adaptors for the retrieval of VAMP72 members, which are components of the membrane fusion machinery, during clathrin-mediated endocytosis. Our results further indicate that the recycling mechanisms of homologous VAMP7 proteins are different in plants and animals, suggesting a divergence of the endocytosis mechanism between these two kingdoms.
Keywords: PICALM1, ANTH domain, SNARE, clathrin-mediated endocytosis
Abstract
The plasma membrane (PM) acts as the interface between intra- and extracellular environments and exhibits a tightly regulated molecular composition. The composition and amount of PM proteins are regulated by balancing endocytic and exocytic trafficking in a cargo-specific manner, according to the demands of specific cellular states and developmental processes. In plant cells, retrieval of membrane proteins from the PM depends largely on clathrin-mediated endocytosis (CME). However, the mechanisms for sorting PM proteins during CME remain ambiguous. In this study, we identified a homologous pair of ANTH domain-containing proteins, PICALM1a and PICALM1b, as adaptor proteins for CME of the secretory vesicle-associated longin-type R-SNARE VAMP72 group. PICALM1 interacted with the SNARE domain of VAMP72 and clathrin at the PM. The loss of function of PICALM1 resulted in faulty retrieval of VAMP72, whereas general endocytosis was not considerably affected by this mutation. The double mutant of PICALM1 exhibited impaired vegetative development, indicating the requirement of VAMP72 recycling for normal plant growth. In the mammalian system, VAMP7, which is homologous to plant VAMP72, is retrieved from the PM via the interaction with a clathrin adaptor HIV Rev-binding protein in the longin domain during CME, which is not functional in the plant system, whereas retrieval of brevin-type R-SNARE members is dependent on a PICALM1 homolog. These results indicate that ANTH domain-containing proteins have evolved to be recruited distinctly for recycling R-SNARE proteins and are critical to eukaryote physiology.
The plasma membrane (PM) acts as the limiting membrane in eukaryotic cells, functioning as the interface between intra- and extracellular environments. Various solutes and water are transported across the PM by transporters and channels, which is fundamental for the maintenance of cellular homeostasis. Various receptor proteins are also included in the PM, whose amounts, localization, and activities are tightly regulated for accurate perception of extracellular signals and intercellular communications, and this regulation depends largely on balanced exo- and endocytic transport to and from the PM.
In the last step of exocytic trafficking, secretory vesicles are tethered to the PM, which is generally mediated by the exocyst tethering complex and low-molecular-weight GTPases belonging to the RAB GTPase family. These GTPases act as a molecular switch to promote the assembly of tethering factors (1). Once tethered properly to the PM, secretory vesicles fuse with the PM, which is mediated by soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) proteins (2). SNARE proteins can be classified into two groups, i.e., Q-SNARE and R-SNARE, based on the amino acid in the center of the coiled-coil SNARE domain. The Q-SNARE domains are further subdivided into Qa-, Qb-, and Qc-SNARE domains, based on their amino acid sequence similarity. In general, Q-SNAREs localize to the target membrane, whereas R-SNAREs localize to transport intermediates, such as membrane vesicles, including secretory vesicles. The two membranes bearing Q- or R-SNAREs are fused to each other by each of the four SNARE domains (Qa, Qb, Qc, and R) that assemble into a tight complex. After membrane fusion, the SNARE complex is disassembled, which is mediated by N-ethylmaleimide-sensitive factor (NSF) and soluble NSF attachment protein alpha (α-SNAP) (3). For sustainable membrane fusion, disassembled R-SNAREs must be retrieved from the target compartment and transported to new transport vesicles for the subsequent round of membrane fusion. However, mechanisms of R-SNARE recycling in the plant membrane trafficking system remain unclear.
Animal R-SNAREs are further divided into two subgroups: longins and brevins. Both these R-SNARE groups consist of the R-SNARE domain followed by a transmembrane domain at the C terminal, in addition to which longins harbor a profilin-like globular-folded domain, called the longin domain, at the N terminal (4). Conversely, brevins harbor a short and variable N-terminal sequence instead of the longin domain. Although brevins play important roles in membrane trafficking events, including neurotransmission in animals (e.g., synaptobrevin, which mediates membrane fusion between the synaptic vesicle and the presynaptic membrane), and similar brevin-type R-SNAREs have been identified in other opisthokonts, such as yeasts, other eukaryotic lineages do not possess R-SNAREs with a similar N-terminal structure (5), suggesting that brevin was acquired in an ancestor of opisthokonts. Conversely, longins are broadly conserved in eukaryotic lineages and play important roles in membrane trafficking in several organisms. Longins are also distributed throughout the plant lineages, with notable expansion of a subgroup, i.e., the VAMP7 group, during land plant evolution (6, 7). While animals possess only one or few VAMP7 members, Arabidopsis harbors 11 VAMP7 members, which are further classified into two subgroups, namely, VAMP71 and VAMP72. These VAMPs participate in distinct membrane fusion events. VAMP71 mediates homotypic fusion of the vacuolar membrane in Arabidopsis (8), and remarkable functional differentiation has been reported in the VAMP72 group. Of the seven VAMP72 members in Arabidopsis, VAMP727 is localized mainly to the multivesicular bodies (MVBs), mediating the fusion of MVBs with the vacuolar and plasma membranes (9, 10), whereas the other VAMP72 members (VAMP721 to 726) localize to the trans-Golgi network (TGN), secretory vesicles, forming cell plates, and PM, reflecting their major role in the secretory pathway (11–14). Animal VAMP7 also mediates membrane fusion at several locations, including the PM and autophagosomes (15, 16). Thus, VAMP7 underwent functional diversification/specialization in a lineage-specific manner during evolution, and this underpins various cellular functions in diverse organisms.
VAMP7 members mediate membrane fusion at the PM in both animals and plants. After membrane fusion, VAMP7 must be retrieved from the PM, through the endocytic pathway, for subsequent fusion events. In animal cells, VAMP7 is sorted into clathrin-coated vesicles, through the interaction of its longin domain with a clathrin adaptor, known as HIV Rev-binding protein (Hrb or AGFG1) (17). However, the mechanism for the retrieval of VAMP72 members from the plant PM remains unknown. An Hrb homolog with a similar domain composition is not encoded in the Arabidopsis genome; therefore, VAMP72 must be loaded into endocytic vesicles in a distinct manner from the animal system. Brevin-type secretory R-SNAREs should also be retrieved from the PM via endocytic transport. In animals, the retrieval of brevin-type R-SNAREs, including VAMP8, VAMP3, and VAMP2, from the PM is mediated by another class of clathrin adaptor protein containing the AP180 N-terminal Homology (ANTH) domain, that is, CALM/PICALM (18, 19). In plants, the PICALM family has also expanded during evolution (20), and some of its members are required for normal pollen tube growth (21–23). However, it is still not known if any of the PICALM members are involved in the recycling of SNARE proteins in plants.
In the present study, we attempted to identify a clathrin adaptor responsible for recycling VAMP72 from the PM in Arabidopsis. We further revealed divergent retrieval mechanisms of secretory longin-type SNAREs in plants and animals.
Results
ANTH-Domain Containing Proteins Interact with the SNARE Domain of VAMP721.
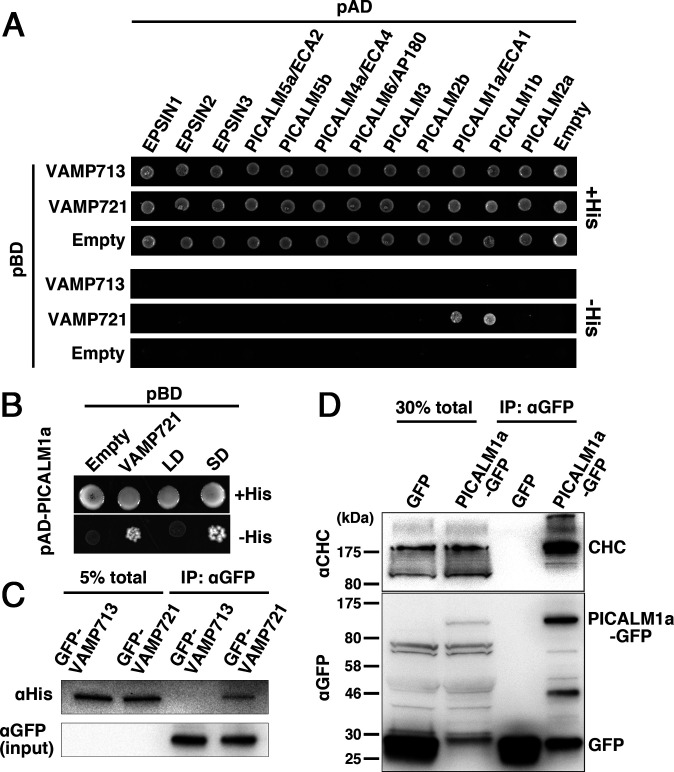
To determine the molecules interacting with VAMP72 proteins in Arabidopsis, we performed yeast two-hybrid screening using the cytoplasmic domain of VAMP727 as bait, which allowed us to identify several candidate interacting partners. We hypothesized that the adaptor protein mediating the retrieval of VAMP72 members from the PM would also interact with other VAMP72 members; therefore, we performed a second screening using the cytoplasmic domain of VAMP721 as bait, and an ANTH domain-containing protein, PICALM1a/ECA1, was identified to interact with the VAMP72 members (Fig. 1A and SI Appendix, Fig. S1). This interaction should be specific to the VAMP72 group, because we did not detect an interaction between PICALM1a and a vacuolar membrane-residing VAMP7 member, VAMP713, in the yeast two-hybrid assay (Fig. 1A). The PICALM family expanded during land plant evolution. Eighteen PICALM proteins are encoded in the Arabidopsis genome (20), in addition to putative clathrin adaptor proteins with the Epsin N-terminal Homology (ENTH) domain, which has a similar structure to the ANTH domain. We further tested whether VAMP72 could interact with other E/ANTH-containing proteins or whether its interaction was specific to PICALM1. As shown in Fig. 1A, VAMP721 was found to interact with PICALM1b, a PICALM1a paralog (20), but not with other E/ANTH members in the yeast two-hybrid assay. We then investigated whether PICALM1a interacted with the longin or the SNARE domain of the VAMP72 members by splitting the cytoplasmic part of VAMP721 and VAMP727 into each domain. In the yeast two-hybrid assay, PICALM1a interacted with the SNARE domain of VAMP721 and VAMP727, but not with their longin domains (Fig. 1B and SI Appendix, Fig. S1).
Fig. 1.
PICALM1 interacts with the SNARE domain of VAMP721 and clathrin. (A) Interaction between VAMP721 and A/ENTH domain-containing proteins was tested by yeast two-hybrid assays. The cytoplasmic domain of VAMP721 or VAMP713 was expressed as a fusion with the DNA-binding domain (pBD), and A/ENTH domain-containing proteins were expressed as fusions with a transcriptional activation domain (pAD). Interactions between two proteins were tested using the HIS3 reporter gene. (B) The cytoplasmic domain of VAMP721 was split into the longin (LD) and SNARE (SD) domains and expressed as fusion proteins with the DNA-binding domain together with the fusion protein of PICALM1a and the transcriptional activation domain (pAD-PICALM1a). (C) Coimmunoprecipitation analysis for interaction between PICALM1a and VAMP721. Lysates prepared from Arabidopsis plants expressing GFP-tagged VAMP721 or VAMP713 and His6-tagged PICALM1a were subjected to immunoprecipitation using an anti-GFP antibody. Coprecipitated PICALM1a was detected using an anti-His antibody. (D) Coimmunoprecipitation analysis for interaction between PICALM1a and clathrin heavy chain. Lysates prepared from Arabidopsis plants expressing GFP-tagged PICALM1a or GFP were subjected to immunoprecipitation using an anti-GFP antibody. The precipitates were then subjected to immunoblotting using an anti-clathrin heavy chain antibody or anti-GFP antibody.
We further investigated the in planta interaction between VAMP721 and PICALM1a. We constructed transgenic plants expressing His6-tagged PICALM1a and GFP-tagged VAMP721 or VAMP713, which were subjected to immunoprecipitation using the anti-GFP antibody. Although comparable amounts of GFP-VAMP713 and GFP-VAMP721 were precipitated, PICALM1a-His6 was coprecipitated with only VAMP721 (Fig. 1C), indicating the specificity of interaction between PICALM1 and VAMP72 in plants. Because ANTH domain-containing proteins are assumed to act as clathrin adaptors, we examined the interaction between PICALM1a and clathrin in plants. We generated transgenic plants expressing GFP-tagged PICALM1a, which was found to retain its function (as described below) and performed an immunoprecipitation analysis using the anti-GFP antibody. As shown in Fig. 1D, PICALM1a-GFP was coprecipitated with the clathrin heavy chain (CHC), whereas GFP alone was not. This result indicates that PICALM1 interacts with clathrin, probably during clathrin-mediated vesicle formation in Arabidopsis.
PICALM1 Acts in CME.
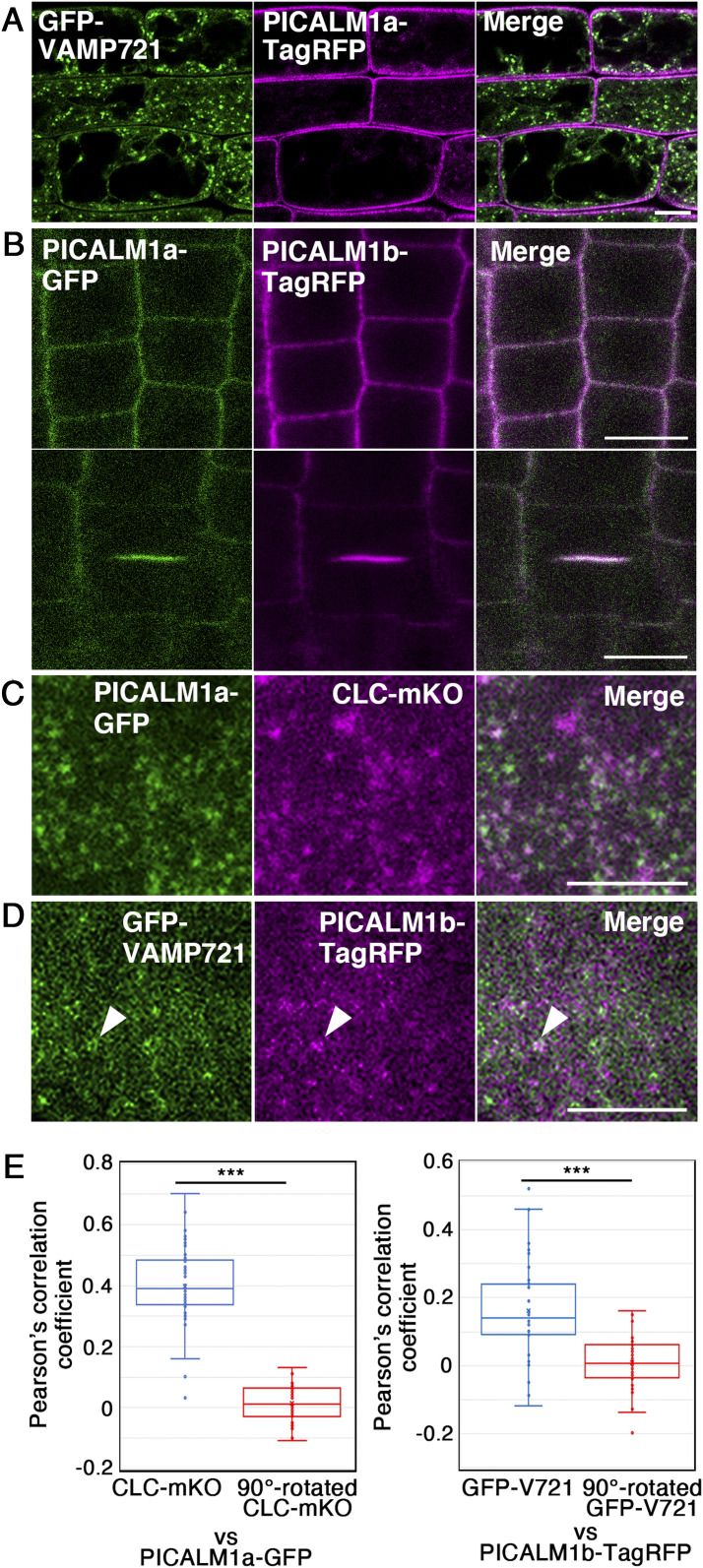
Clathrin acts at multiple locations in the post-Golgi trafficking network, including the PM and TGN in plant cells (24), and VAMP721 also localizes to the PM and TGN (12, 14). Thus, we examined the location in the plant cell where PICALM1a cooperates with clathrin. On observing transgenic plants expressing GFP-VAMP721 and PICALM1a-TagRFP by confocal microscopy, we found that GFP-VAMP721 was localized primarily to the punctate compartments, most likely the TGN, and also to the PM to a smaller extent, as previously reported (Fig. 2A). Conversely, PICALM1a-TagRFP was almost exclusively localized to the PM in nondividing cells, suggesting its function at the PM (Fig. 2A). We then compared the localization of PICALM1a and PICALM1b by expressing these proteins after tagging with distinct fluorescent proteins and found that both proteins targeted to the PM in nondividing cells (Fig. 2B). In dividing cells, PICALM1a accumulated at the forming cell plate, as reported previously (25), similarly to PICALM1b (Fig. 2B). As PICALM1a and PICALM1b exhibited similar subcellular localization, we used either PICALM1a or PICALM1b for further analyses.
Fig. 2.
Subcellular localization of PICALM1. (A) Subcellular localization of GFP-VAMP721 and PICALM1a-TagRFP in root epidermal cells observed by confocal microscopy. (B) Subcellular localization of PICALM1a-GFP and PICALM1b-TagRFP in root epidermal cells observed by confocal microscopy. (C) Localization of PICALM1a-GFP and CLATHRIN LIGHT CHAIN 2 tagged with mKO (CLC-mKO) near the PM observed by VIAFM. (D) Localization of GFP-VAMP721 and PICALM1b-TagRFP near the PM observed by VIAFM. The arrowheads indicate a dot bearing both GFP and TagRFP signals. (E) Quantification of colocalization between PICALM1a-GFP and CLC-mKO (Left) and GFP-VAMP721 and PICALM1b-TagRFP (Right) observed by VIAFM. The 90°-rotated CLC-mKO or PICALM1b-TagRFP images were used to test whether the detected colocalization was random or not. Wilcoxon signed rank test was used for statistical analyses. ***P < 0.001. n = 38 images for PICALM1a-GFP and CLC-mKO and 40 images for GFP-VAMP721 and PICALM1b-TagRFP. (Scale bars: 10 μm in A and B; 5 μm in C and D.)
We compared the localization of PICALM1a-GFP and CLATHRIN LIGHT CHAIN 2 tagged with monomeric Kusabira Orange (CLC-mKO) near the PM in root epidermal cells of transgenic plants expressing these fluorescently tagged proteins, by variable incidence angle fluorescent microscopy (VIAFM or VAEM) (26, 27). CLC-mKO was localized to the PM in foci, as previously reported (28, 29), where significant colocalization with PICALM1a-GFP was observed (Fig. 2 C and E). Significant colocalization was also detected between PICALM1b and VAMP721 when observed by VIAFM, further supporting the role of PICALM1 in the CME of VAMP721 (Fig. 2 D and E). However, the colocalization frequency was lower than that between PICALM1 and clathrin. This is probably due to the multiple localization of VAMP721 near the PM (i.e., secretory vesicles, sites of secretion, and clathrin-coated pits), although we do not rule out the possibility that PICALM1 could also be involved in CME of other cargo molecules, besides the VAMP72 proteins. Noticeable localization of fluorescently tagged PICALM1a was not detected at the vacuolar membrane visualized with GFP-VAMP713, on punctate compartments bearing CLC-mKO in the cytoplasm, on the TGN labeled with VHAa1-mRFP, or on MVBs visualized with mRFP-ARA7/RABF2b (SI Appendix, Fig. S2). The subcellular localization of PICALM1a at the PM, together with its interaction with clathrin and VAMP721, strongly suggested that PICALM1 acts as an adaptor for sorting VAMP721 to clathrin-coated vesicles at the PM.
PICALM1 Is Required for Normal Vegetative Development.
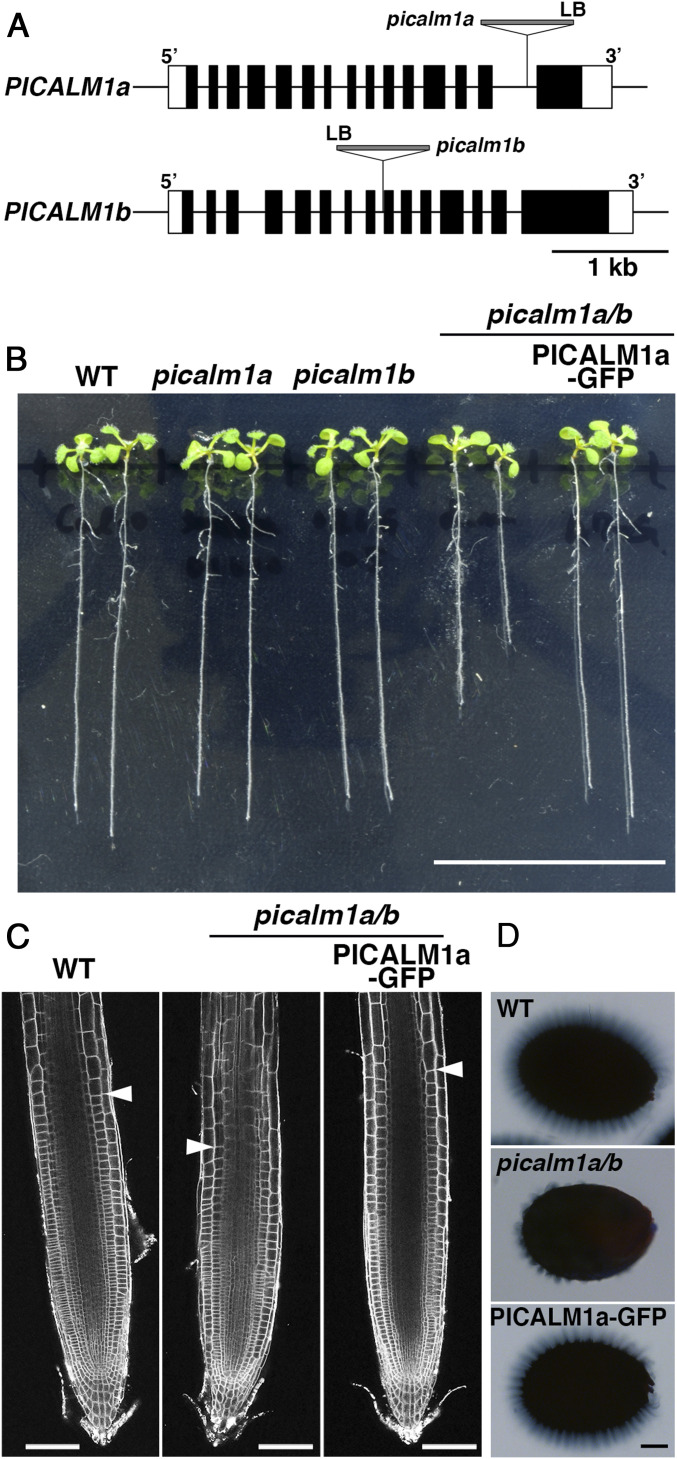
To determine the function of PICALM1 in plants, we analyzed the effects of loss-of-function mutations in the PICALM1a/b genes. Single mutants of PICALM1a (SALK_043625) and PICALM1b (GABI_026G05) did not exhibit macroscopic phenotypic abnormalities, although full-length transcripts were not detected by RT-PCR in these mutants (Fig. 3 A and B and SI Appendix, Figs. S3A and S4A). We therefore obtained the double mutant by crossing the single mutants. The picalm1a/b double mutant exhibited semidwarfism throughout its vegetative growth (Fig. 3B and SI Appendix, Figs. S3 A and B and S4 B–D), indicating important and redundant functions of PICALM1a and PICALM1b in the vegetative growth of Arabidopsis. The growth defect was rescued by expressing PICALM1a-GFP under the regulation of PICALM1a regulatory elements (promoter and introns) (Fig. 3B and SI Appendix, Fig. S3 A and B), indicating that the semidwarfism of the picalm1a/b mutant is due to mutations in the PICALM1 genes, further demonstrating that GFP-tagged PICALM1a retains its original function. The double mutant was fertile and produced seeds, although the silique was shorter than that of the wild type, indicating that PICALM1 does not play a critical role in reproductive processes, distinguishing it from PICALM5 members that are required for sustainable pollen tube growth (23). The size and cortical cell number in the root meristematic region were reduced in the picalm1a/b double mutant, whereas the final cell size was comparable with that in wild-type plants (Fig. 3C and SI Appendix, Fig. S4 E–G), suggesting that the semidwarfism caused by the picalm1a/b mutation is mainly due to a compromised meristematic activity, resulting in a reduced cell number, rather than a reduced cell size.
Fig. 3.
Effects of picalm1 mutations on plant development. (A) Schematic presentation of gene models for PICALM1a and PICALM1b and T-DNA insertion sites in picalm1a and picalm1b mutants used in this study. (B) Ten-day-old Arabidopsis seedlings of wild-type (WT), picalm1a, and picalm1b single mutants, picalm1a picalm1b (picalm1a/b) double mutant, and the double mutant transformed with PICALM1a-GFP. (C) Confocal images of root meristematic zones of the WT, picalm1a picalm1b double mutant (picalm1a/b), and double mutant transformed with PICALM1a-GFP and stained with FM4-64. The arrowheads indicate the boundary between the meristematic and elongation zones. (D) Seed coat mucilage after the imbibition of the WT, picalm1a picalm1b double mutant (picalm1a/b), and double mutant transformed with PICALM1a-GFP (PICALM1a-GFP) stained with ruthenium red. (Scale bars: 3 cm in B; 100 μm in C and D.)
A volcano-shaped secondary cell wall structure of mucilage secretory cells in the seed coat was rarely observed in the picalm1a/b mutant (SI Appendix, Fig. S3C), whereas the seed coat of the mutant complemented with PICALM1a-GFP harbored the volcano-shaped structure, similar to wild-type plants. We then examined the extrusion of mucilage after the imbibition of seeds by ruthenium red staining, which demonstrated that the amount of mucilage was substantially reduced in the picalm1a/b mutant (Fig. 3D and SI Appendix, Fig. S4H). Given that defective mucilage deposition and/or extrusion may also occur due to the impairment of secretion-related proteins such as ECHIDNA and subunits of the exocyst complex (30, 31), the data indicate that the secretory activity is compromised by the picalm1a/b mutation. This is also consistent with the notion that PICALM1 acts in the recycling of VAMP72 members acting in the secretory pathway.
PICALM1 Is Required for Loading VAMP721 to Clathrin-Coated Endocytic Vesicles.
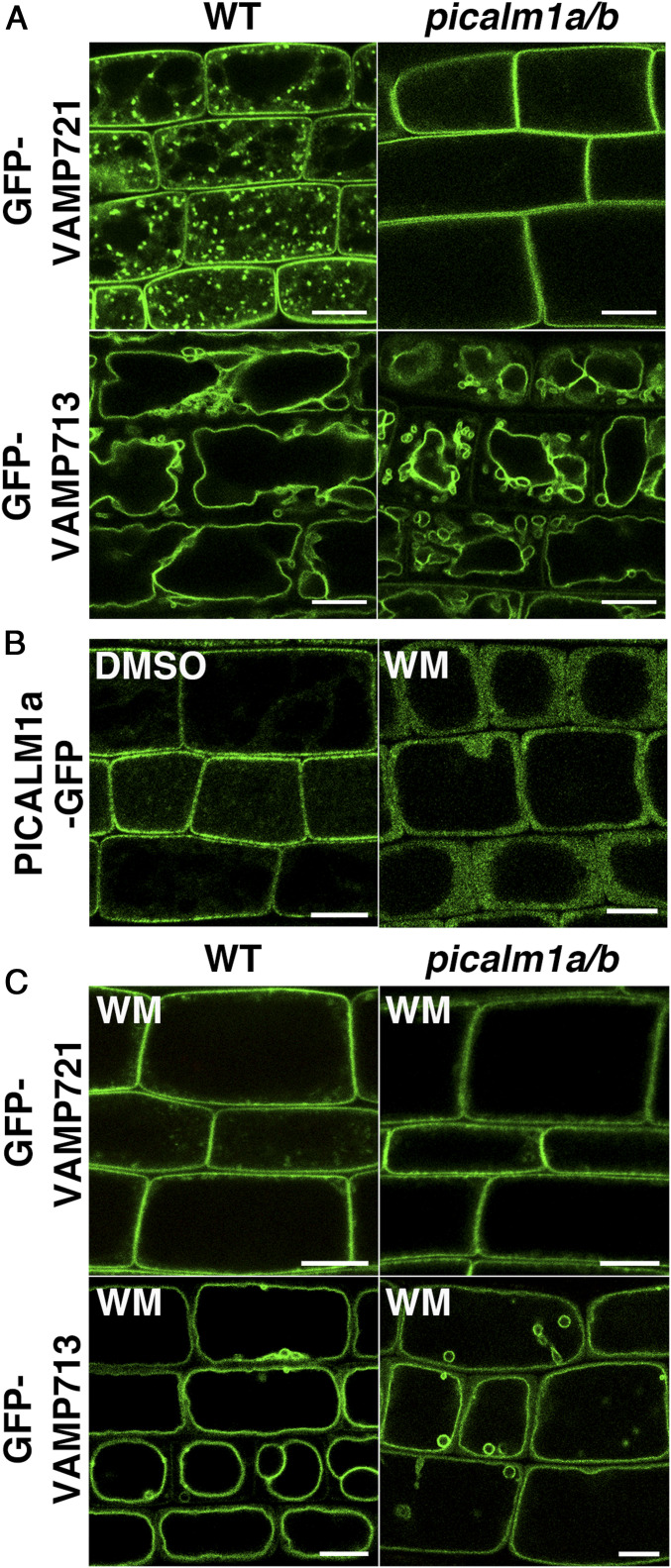
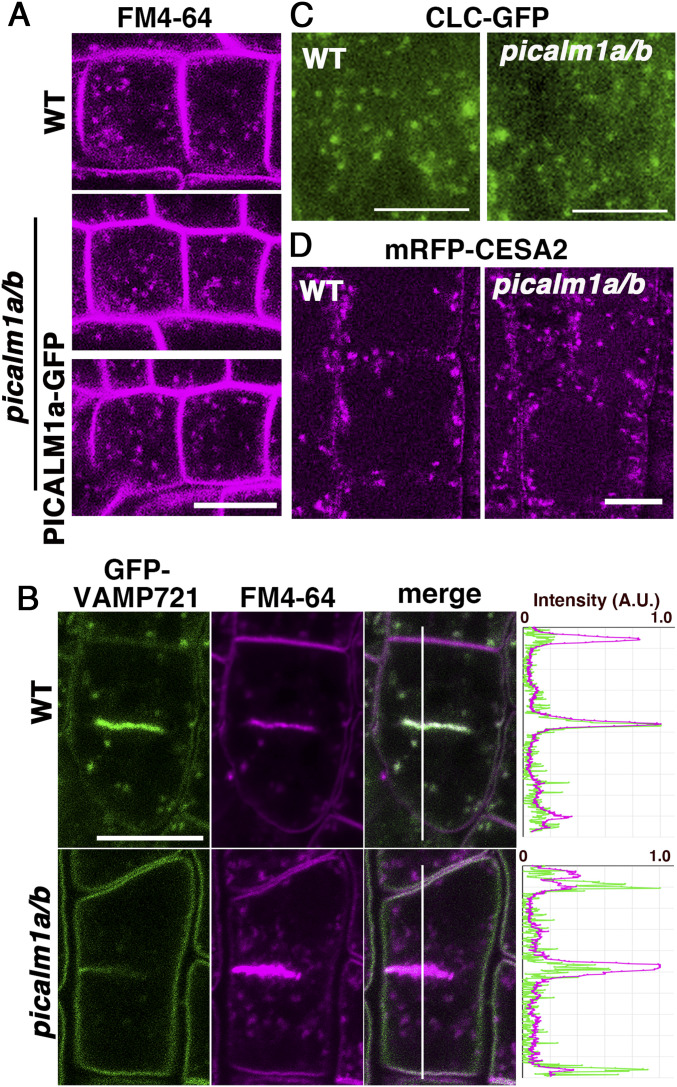
If PICALM1 is an adaptor mediating VAMP72 loading to clathrin-coated vesicles, VAMP72 would accumulate at the PM in the picalm1a/b double mutant because of defective retrieval from the PM. To verify this, we expressed GFP-VAMP721 in picalm1a/b and observed its localization in root epidermal cells. We observed that GFP-VAMP721 was localized almost exclusively to the PM in the double mutant, contrary to the punctate cytoplasmic localization with minor localization at the PM in wild-type plants (Fig. 4A). Thus, PICALM1 is a major factor for retrieving the longin-type SNARE VAMP721 in Arabidopsis, which strongly supports the notion that PICALM1 is the adaptor protein bridging the clathrin cage to the VAMP72 proteins during CME.
Fig. 4.
PICALM1 is required for the retrieval of VAMP721 from the PM. (A) Confocal images of root epidermal cells of 7-d-old WT or picalm1a/b double mutant plants expressing GFP-VAMP721 or GFP-VAMP713. (B) Confocal images of root epidermal cells of 7-d-old Arabidopsis plants expressing PICALM1a-GFP treated with DMSO or WM. (C) Confocal images of root epidermal cells of 7-d-old WT or picalm1a/b plants expressing GFP-VAMP721 or GFP-VAMP713 treated with WM. (Scale bars: 10 μm.)
The ANTH domain is also known to bind phosphatidylinositol 4,5-bisphosphate [PtdIns(4,5)P2], which recruits ANTH domain proteins to the membrane domain containing PtdIns(4,5)P2 in animal cells (32). This would also be the case for Arabidopsis PICALM1, because treatment with wortmannin (WM) or phenylarsine oxide (PAO), which act as inhibitors of phosphatidylinositol 3- and 4-kinases or phosphatidylinositol 4-kinase in plant cells, respectively, caused PICALM1a-GFP to dissociate from the PM and disperse in the cytosol (Fig. 4B and SI Appendix, Fig. S5). We further investigated whether the WM treatment hampered the retrieval of VAMP721 from the PM. If PICALM1 indeed acts as an adaptor in CME of VAMP721, WM treatment would hamper this process by inhibiting the association of PICALM1 with the PM. As expected, there was a substantial reduction in the amount of GFP-VAMP721 on punctate compartments in the cytoplasm of root epidermal cells treated with WM, and GFP-VAMP721 accumulated almost exclusively at the PM (Fig. 4C). Conversely, WM did not affect the localization of GFP-VAMP721 to the PM in the picalm1a/b double mutant. The vacuolar membrane localization of VAMP713 was not affected by WM in either wild-type or the double mutant plants. These results further support that PICALM1 is an adaptor protein required for the retrieval of VAMP72 from the PM in Arabidopsis.
PICALM1 Is Not Required for General Endocytosis.
We further investigated whether the picalm1a/b mutation hampered general endocytosis, by monitoring the internalization of the lipophilic endocytic tracer dye FM4-64 in root epidermal cells. In nondividing cells, internalization of FM4-64 was not markedly affected by the double mutation (Fig. 5A and SI Appendix, Fig. S6A), indicating that endocytic flow from the PM to endosomes was not severely affected by this mutation. Consistent with this notion, FM4-64 accumulated rapidly at the forming cell plate in wild-type and double mutant plants, as previously reported (33), suggesting that endocytic transport to the forming cell plate was also not severely affected by the picalm1a/b mutation (Fig. 5B). However, the picalm1a/b mutation affected the localization of VAMP721 at the forming cell plate, which presented lower density of GFP-VAMP721 in the picalm1a/b mutant than in the wild-type plant expressing GFP-VAMP721 (Fig. 5B). This effect of the double mutation reflects the contribution of VAMP721 recycled from the PM via PICALM1-dependent retrieval in membrane fusion at the forming cell plate in wild-type plants. Whereas, VAMP721 synthesized de novo could somehow accomplish cytokinesis in the picalm1a/b mutant, considering that the double mutant did not exhibit a severe cytokinesis defect like the vamp721 vamp722 double mutant, which is defective in cell plate formation (14).
Fig. 5.
General endocytosis is not markedly affected in the picalm1a/b mutant. (A) Internalization of FM4-64 into root epidermal cells in 6-d-old WT (Top) and picalm1a/b mutant plants without (Middle) or with the PICALM1a-GFP transgene (Bottom). (B) Dividing root epidermal cells in 9-d-old WT (Top) and picalm1a/b (Bottom) mutant plants expressing GFP-VAMP721 and stained with FM4-64. Graphs on the Right indicate relative intensities of fluorescence from GFP and FM4-64 along the white line in merged images. A.U., arbitrary unit. (C) Localization of CLC-GFP near the PM observed by VIAFM in root epidermal cells of 7-d-old WT (Left) and picalm1a/b mutant (Right) plants. (D) Subcellular localization of mRFP-CESA2 in root epidermal cells of 8-d-old WT (Top) and picalm1a/b (Bottom) mutant plants. (Scale bars: 10 μm in A, B, and D; 5 μm in C.)
To examine whether the loss of function of PICALM1 affected the formation and/or distribution of clathrin-coated pits at the PM, we observed CLC-GFP in wild-type and picalm1a/b mutant plants by VIAFM and found no significant abnormality in the mutant (Fig. 5C and SI Appendix, Fig. S6B). This result suggested that PICALM1 is not required for the assembly of and vesicle formation by clathrin at the PM, consistent with unaffected general endocytosis visualized with FM4-64. To test whether PICALM1 is generally involved in the retrieval of membrane proteins from the PM, we also examined whether a component of the cellulose synthase complex involved in primary cell wall synthesis, CESA2, which is cycled between the PM and intracellular compartments through CME (34–36), was affected in the picalm1a/b mutant. We found that mRFP-tagged CESA2 exhibited similar localization in root epidermal cells of both wild-type and double mutant plants (Fig. 5D). These results indicated that PICALM1 is required for the CME of the VAMP72 proteins, acting as the adaptor protein during the sorting of VAMP72 into clathrin-coated pits/vesicles at the PM, although they are not required for general endocytosis.
Discussion
In this study, we demonstrated that a pair of paralogous ANTH domain-containing proteins, PICALM1a and PICALM1b, act as adaptors for loading the longin-type R-SNARE VAMP72 to clathrin-coated vesicles at the PM in Arabidopsis. In mammalian cells, ANTH domain-containing CALM/PICALM mediates the sorting of R-SNARE proteins into clathrin-coated vesicles at the PM by recognizing the SNARE domain (18, 19). However, only brevin-type R-SNAREs, such as VAMP8, VAMP3, and VAMP2, which are the short versions of R-SNAREs, are recycled by CALM. Mammalian VAMP7, which is a longin-type R-SNARE orthologous to Arabidopsis VAMP72, is recycled distinctly from brevins; the clathrin adaptor Hrb binds to the longin domain, but not to the SNARE domain, of VAMP7, to load it to clathrin-coated vesicles formed at the PM. Thus, the mechanisms of retrieving VAMP7 are different between animals and plants. This is surprising, considering that the distribution of brevins is restricted in eukaryotic lineages, and longin is widely conserved and is believed to have an ancient origin (5). In Dictyostelium discoideum, an ANTH-domain protein has been reported to interact with VAMP7B, further regulating its localization at contractile vacuoles (37). This might imply that the ANTH-domain protein was originally involved in recycling VAMP7 proteins, and the Hrb-dependent system in mammals could have been acquired in restricted lineages during evolution. However, it remains unknown whether the PM-resident VAMP7 is retrieved by ANTH-domain proteins in D. discoideum. Investigation of VAMP7 recycling mechanisms in other organisms is required to reconstitute the diversification of longin-SNARE retrieval systems during evolution.
Our findings provide interesting insights into the functional differentiation of ANTH domain-containing proteins during plant evolution. Four ANTH-domain proteins are encoded in the human genome (38), and the genome of the basal land plant, the liverwort Marchantia polymorpha, harbors three genes encoding ANTH proteins (39). Surprisingly, the Arabidopsis genome encodes 18 ANTH proteins, suggesting divergent functions of this protein family in Arabidopsis. A comparative genomic analysis further suggested that this group expanded during land plant evolution (20). However, although distinct expression patterns and subcellular localizations have been reported for these proteins (21, 22, 25, 40), their molecular functions remain largely ambiguous in plants. Recently, we found that one subgroup of the Arabidopsis PICALM family, PICALM5, is responsible for CME of ANXUR receptor kinases, which is required for sustained pollen tube growth and therefore, normal fertility (23). The present study revealed the molecular function of another subgroup of ANTH proteins, PICALM1, which was found to mediate CME of VAMP72 and was required for normal vegetative development. Together with distinctive expression patterns and subcellular localizations of related members (21, 22, 25, 40), our results indicated that PICALM functions have diverged for the retrieval of distinct cargo proteins via CME during land plant evolution and are involved in various physiological events during both vegetative and reproductive developmental processes in angiosperms.
PICALM1 is required for the retrieval of VAMP721 from the PM, and the loss of this function resulted in the accumulation of VAMP721 at the PM. Because PICALM1 also interacted with VAMP727 in yeast two-hybrid analysis, it may mediate recycling of other VAMP72 members, in addition to VAMP721, in Arabidopsis. VAMP72 members are involved in fundamental cellular activities, in Arabidopsis, including cytokinesis, and the double mutation in VAMP721 and VAMP722 results in seedling lethality (11, 14). However, the picalm1a/b mutant completed its life cycle, although it did exhibit semidwarfism. This mitigative effect could be attributed to the VAMP72 members synthesized de novo, which could partially perform their functions, whereas reuse of this R-SNARE is required for complete vegetative growth in Arabidopsis. Because secretory trafficking is also critical to reproductive processes, such as the pollen tube growth, and is strictly regulated (41), VAMP72 members may also play pivotal roles in reproductive processes. Expression data deposited in the public database also indicate that VAMP725 and VAMP726 are highly expressed in mature pollen (bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi). An absence of any detectable defect in the reproduction of picalm1a/b mutants might reflect the involvement of other PICALM members in the recycling of these pollen-dominant VAMP72 members, and this warrants further investigation.
Our study demonstrated that regulatory mechanisms of the membrane trafficking system diverged between the plant and animal systems, and the clathrin adaptor PICALM family underwent neofunctionalization and/or subfunctionalization during the evolution of plants. However, complete visualization of PICALM functions in Arabidopsis would require the identification of molecular functions and cargo proteins of subgroups other than PICALM1 and PICALM5, which would further highlight the significance of CME in plant physiology. Functional analyses of ANTH-domain proteins in basal land plants, such as liverworts and mosses, can effectively reconstitute the functional diversification of this protein family during land plant evolution. PICALM members have also been identified as components of the TPLATE/TSET complex, which is a clathrin adaptor complex conserved only in plants and some limited clades of eukaryotes (42); and mammalian ANTH domain proteins are known to act with the AP-2 clathrin adaptor complex (43–45). Similar to the case of PICALM1, Arabidopsis TPLATE and AP-2 are reported to dissociate from the PM upon WM treatment (46, 47). It could also be possible that these adaptor complexes cooperate with PICALM1 in sorting the VAMP72 members into clathrin-coated pits/vesicles during CME. Functional coordination of plant ANTH proteins with other clathrin adaptors is important to understand the unique regulatory mechanisms of CME in plant cells.
Materials and Methods
Plant Materials, Growth Condition, and Plasmids.
Arabidopsis thaliana plants from the Col-0 accession were used in this study. Arabidopsis seedlings were grown on half-strength Murashige and Skoog (1/2 MS) agar medium containing 1% sucrose, at 23 °C under continuous light. Fourteen-day-old plants were transplanted to soil and grown at 23 °C under a long-day light/dark cycle (16 h light and 8 h dark). Arabidopsis picalm1a (SALK_043625) and picalm1b (GABI_026G05) mutants were obtained from the Arabidopsis Biological Resource Center (ABRC) and German plant genomics research program-Kölner Arabidopsis T-DNA lines (GABI-Kat) (48, 49), respectively. The mutants were backcrossed to wild-type Arabidopsis (Col-0) at least three times. To construct transgenic plants expressing fluorescently or His-tagged PICALM1a and PICALM1b under the regulation of their own regulatory elements, GFP cDNA, TagRFP, or 6xHis-tag was inserted in front of the stop codon of the 4.5-kb genomic fragment of PICALM1a or the 5.7-kb genomic fragment of PICALM1b. These genomic fragments contained the coding regions, introns, and 1.1 kb and 2.1 kb of 5′-flanking sequences of PICALM1a and PICALM1b, respectively. The chimeric genomic fragments of PICALM1a and PICALM1b were subcloned into binary vectors, i.e., pB7FWG, pGWB4, pGWB559, pGWB659, or pGWB7 (28, 50–52). To construct transgenic plants expressing fluorescently tagged VAMP713, VAMP721, ARA7, and CLATHRIN LIGHT CHAIN 2 (CLC) under their native promoters, previously described chimeric genomic fragments (9, 53, 54), containing GFP, mRFP, or monomeric Kusabira Orange (mKO) inserts in front of the start or stop codon of each gene, were subcloned into pBGW or pGWB1 binary vectors (50, 51). The binary vector used for constructing transgenic plants expressing mRFP-tagged VHAa1 (VHAa1-mRFP) was provided by K. Schumacher, University of Heidelberg, Heidelberg, Germany (33). Wild-type or picalm1a picalm1b mutant Arabidopsis plants were transformed or cotransformed with resultant binary vectors by the floral dipping method using Agrobacterium tumefaciens (strain GV3101::pMP90) (55). Transgenic plants expressing GFP were provided by S. Mano, National Institute for Basic Biology (NIBB), Okazaki, Japan (56).
Yeast Two-Hybrid Assays.
Yeast two-hybrid screening was performed using the Matchmaker Two-Hybrid Library Construction and Screening Kit (Clontech) and the normalized Arabidopsis cDNA library (Mate & Plate Library, Universal Arabidopsis, Clontech), containing cDNA sequences derived from seedlings, flowers, buds, pollen, leaves, siliques, and stems, introduced into the yeast strain Y187. The cDNA fragment for the cytoplasmic domain of VAMP727 was subcloned into pGBKT7, which was then transformed into the yeast strain Y2H Gold. After mating between the Y2H Gold strain expressing VAMP72 and the Y187 strain with the expression library, resultant zygotes were selected by culturing on SD/−Leu/−Trp/+X-α-Gal/+Aureobasidin A plate for 5 d at 25 °C. Plasmids collected from positive clones were then sequenced using the T7 primer.
To construct GAL4 AD fusion-expressing vectors, open reading frames (ORFs) for ANTH/ENTH-domain proteins (PICALM1a/ECA1/AT2G01600, PICALM1b/AT1G14910, PICALM2a/AT5G57200, PICALM2b/AT4G25940, PICALM3/AT5G35200, PICALM4a/ECA4/AT2G25430, PICALM5a/ECA2/AT1G03050, PICALM5b/AT4G02650, PICALM6/AP180/AT1G05020, EPSIN1/AT5G11710, EPSIN2/AT2G43160, and EPSIN3/AT3G59290) were subcloned into pAD-GAL4-GWRFC. These members were selected according to the review of Holstein and Oliviusson (57); in addition, an ANTH protein close to the PICALM1 group, PICALM2a (20), was included. To construct GAL4 BD fusion-expressing vectors, ORFs for the cytoplasmic domains of VAMP7 proteins (VAMP713, VAMP727, and VAMP721) and the longin or SNARE domains of VAMP72 proteins (VAMP727 and VAMP721) were subcloned into pBD-GAL4-GWRFC. pAD-GAL4-GWRFC and pBD-GAL4-GWRFC were provided by T. Demura, Nara Institute of Science and Technology (NAIST), Ikoma, Japan. Empty vectors, i.e., pAD-GAL4-2.1 and pBD-GAL4 Cam, were used as negative controls. Each pair of the pAD-GAL4-GWRFC- and pBD-GAL4-GWRFC-derived vectors was introduced into Saccharomyces cerevisiae strain AH109 (Clontech). The AH109 transformants were selected on SD/−Leu/−Trp plates, cultured in SD/−Leu/−Trp liquid medium for 24 h at 28 °C, and then diluted with water to achieve an OD600 of 5.0. Then, 5 μL of each culture was spotted on SD/−Leu/−Trp and SD/−Leu/−Trp/−His and incubated for 72 h at 28 °C. Transformation was performed independently at least twice, and at least two colonies per transformation were checked for interaction.
Primers.
Primers used for genotyping, cloning, plasmid construction, and RT-PCR are listed in SI Appendix, Table S1.
RT-PCR.
Total RNA was extracted from 10-d-old seedlings with genotypes presented in SI Appendix, Fig. S3A, using RNeasy Plant Mini Kit (Qiagen) and TURBO DNA-free Kit (Ambion, Thermo Fisher Scientific), according to the manufacturer’s instructions. The extracted RNA (400 ng) was then reverse transcribed using SuperScript III Reverse Transcriptase (Invitrogen), and ORFs of the indicated genes were amplified using PrimeSTAR Max DNA Polymerase (Takara Bio) with gene-specific primers listed in SI Appendix, Table S1.
Confocal Microscopy.
Root epidermal or lateral root cap cells of seedlings expressing fluorescently tagged PICALM1a, PICALM1b, VAMP713, VAMP721, ARA7, VHAa1, CESA2, and/or CLC were observed using the LSM780 confocal microscope (Carl Zeiss) with a 63× oil immersion lens (N.A. = 1.40), operated using ZEN software (Carl Zeiss). GFP and mRFP/TagRFP/FM4-64 were excited with 488 nm Ar/Kr and 561 nm diode lasers, respectively. For WM treatment, seedlings were soaked in liquid 1/2 MS medium with dimethyl sulfoxide (DMSO, 0.1%, vol/vol; Wako) or WM (33 µM; Sigma) dissolved in DMSO for 120 min before microscopic observation. For PAO treatment, the seedlings were soaked in liquid 1/2 MS medium with DMSO (0.1%, vol/vol; Wako) or PAO (10 µM; Sigma) dissolved in DMSO for 90 min before microscopic observation. For FM4-64 staining, seedlings were soaked in liquid 1/2 MS medium with FM4-64 dissolved in DMSO (2 µM; Thermo Fisher Scientific) for 5 min (Figs. 3C and 5B) or 30 min (Fig. 5A) and washed twice before microscopic observation. The root meristematic zone of 6-d-old seedlings stained with FM4-64 was observed under an LSM780 confocal microscope (Carl Zeiss) with a 40× oil immersion lens (N.A. = 1.40) in the tile scan mode. The acquired images were processed using ZEN software (Carl Zeiss) and Photoshop CC (Adobe Systems).
Observation of Mucilage Secretory Cells and Ruthenium Red Staining of Mucilage.
Mucilage secretory cells in the seed coat of dry seeds were observed using Miniscope TM-1000 (Hitachi). To visualize the mucilage released from the seed coat, dry seeds were soaked in distilled water for 20 min and stained with 1% ruthenium red (Wako) for 1 h. The stained seeds were observed using a BX60 (Olympus) equipped with a UPlanApo 10× lens (N.A. = 0.40). Images were acquired using a DP73 digital camera (Olympus) operated using cellSens standard software (Olympus).
VIAFM Observation.
Root epidermal cells of 7-d-old seedlings expressing fluorescently tagged PICALM1 and VAMP721 or CLC were observed using an IX-71 (Olympus) equipped with a UAPON 100× O TIRF lens (Olympus) for VIAFM. The roots were placed on a glass slide (76 mm × 26 mm, Matsunami) and covered with a 0.12- to 0.17-mm-thick coverslip (24 mm × 60 mm, Matsunami). For VIAFM observations, GFP and mKO or TagRFP were simultaneously excited using 473- and 561-nm lasers, respectively. A FF01-523/610-625 filter (Semrock) was used to remove autofluorescence of samples. The fluorescence emission spectra were separated using a FF560-FDi01-25 × 36 LP dichroic mirror (Semrock) and filtered through a FF01-523/35 filter (Semrock) for GFP and a ET620/60M filter (Chroma) for mKO and TagRFP, using W-View GEMINI (Hamamatsu Photonics). VIAFM images were acquired using the iXon X3 EMCCD camera (Andor Technology) operated with Metamorph software (Molecular Devices). Each frame was exposed for 200 ms. The acquired images were analyzed using ImageJ and Photoshop CC (Adobe Systems).
Antibodies.
Anti-GFP and anti-His-tag antibodies for the coimmunoprecipitation analysis of PICALM1a and VAMP721 interaction were purchased from MBL (598 and PM032, respectively). The anti-GFP antibody used for the coimmunoprecipitation analysis of PICALM1a and CHC was prepared as described previously (8). The anti-CHC antibody was purchased from Agrisera (AS10 690). Antibodies were diluted for immunoblotting as follows: anti-GFP, ×1,000; anti-His-tag, ×1,000; anti-CHC, ×2,000.
Immunoprecipitation.
For immunoprecipitation followed by Western blotting (Fig. 1B), 0.8 g of 14-d-old transgenic seedlings expressing PICALM1a-His6 and GFP-VAMP713 or GFP-VAMP721 was soaked in cross-linking buffer (20 mM Hepes pH 7.5, 1% sucrose, and 1 mM dithiobis [succinimidyl propionate] [DSP] [Thermo Fisher Scientific]) for 1 h at room temperature (RT). Tris⋅HCl (20 mM, pH 7.5) was then added, and the mixture was incubated for 30 min at RT. Samples were homogenized in grinding buffer (400 mM sucrose, 50 mM Hepes pH 7.5, 50 mM ethylenediamine-N,N,N',N'-tetraacetic acid (EDTA) pH 8.0, and one tablet of cOmplete EDTA-free [Roche]/50 mL). Lysates were centrifuged at 1,000 × g for 10 min and 8,000 × g for 15 min at 4 °C to remove any debris. Supernatants were incubated for 30 min with 1% Triton X-100 at 4 °C for solubilization and then centrifuged at 20,000 × g for 30 min at 4 °C. For analyzing the interaction between PICALM1a and VAMP721 (Fig. 1C), 0.8 g of 14-d-old transgenic seedlings expressing GFP or PICALM1a-GFP was homogenized in the aforementioned grinding buffer, and the lysates were treated as previously described. Supernatants were incubated for 30 min with 1% Triton X-100 and 1 mM dithiothreitol (DTT) at 4 °C for solubilization and centrifuged at 20,000 × g for 30 min at 4 °C. Immunoprecipitation with detergent extracts was performed using the micro-MACS GFP-tagged protein isolation kit (Miltenyi Biotec), according to the manufacturer’s instructions. Immunoprecipitates were then subjected to immunoblotting.
Statistical Analysis.
The Wilcoxon signed rank test (Fig. 2E) and Wilcoxon rank-sum test (SI Appendix, Fig. S6B) were used for statistical analyses between the two groups. For statistical analysis among three or more groups, Tukey’s honestly significant difference post hoc test was used for samples with normal distribution (SI Appendix, Fig. S4B) and the Steel–Dwass test was used for nonparametric sample (others) after one-way ANOVA. Normality of data were tested using the Jarque–Bera test.
Supplementary Material
Acknowledgments
We thank Dr. S. Mano (NIBB), Dr. T. Demura (NAIST), and Dr. T. Nakagawa (Shimane University) for sharing materials for the experiments, as well as the ABRC and the Salk Institute for providing Arabidopsis mutants. Technical support was provided by the Model Plant Research Facility, NIBB BioResource Center. This work was financially supported by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (T.U., 18H02470, 19H05670, and 19H05675) (N.T., 24248001) (K.E., 18K06303 and 19H04872) and (K.N., 18H03941); Grant-in-Aid for Japan Society for the Promotion of Science Fellows (M.F., 10J08869); the Mitsubishi Foundation; and Yamada Science Foundation.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2011152117/-/DCSupplemental.
Data Availability.
All study data are included in the article and SI Appendix.
References
- 1.Grosshans B. L., Ortiz D., Novick P., Rabs and their effectors: Achieving specificity in membrane traffic. Proc. Natl. Acad. Sci. U.S.A. 103, 11821–11827 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Saito C., Ueda T., “Functions of RAB and SNARE proteins in plant life” in International Review of Cell and Molecular Biology, Jeon K. W., Ed. (Academic Press, 2009), pp. 183–233. [DOI] [PubMed] [Google Scholar]
- 3.Ryu J. K., Jahn R., Yoon T. Y., Review: Progresses in understanding N-ethylmaleimide sensitive factor (NSF) mediated disassembly of SNARE complexes. Biopolymers 105, 518–531 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Daste F., Galli T., Tareste D., Structure and function of longin SNAREs. J. Cell Sci. 128, 4263–4272 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Venkatesh D. et al., Evolution of the endomembrane systems of trypanosomatids-conservation and specialisation. J. Cell Sci. 130, 1421–1434 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kanazawa T. et al., SNARE molecules in Marchantia polymorpha: Unique and conserved features of the membrane fusion machinery. Plant Cell Physiol. 57, 307–324 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Sanderfoot A., Increases in the number of SNARE genes parallels the rise of multicellularity among the green plants. Plant Physiol. 144, 6–17 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takemoto K. et al., Distinct sets of tethering complexes, SNARE complexes, and Rab GTPases mediate membrane fusion at the vacuole in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 115, E2457–E2466 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ebine K. et al., A membrane trafficking pathway regulated by the plant-specific RAB GTPase ARA6. Nat. Cell Biol. 13, 853–859 (2011). [DOI] [PubMed] [Google Scholar]
- 10.Ebine K. et al., A SNARE complex unique to seed plants is required for protein storage vacuole biogenesis and seed development of Arabidopsis thaliana. Plant Cell 20, 3006–3021 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.El Kasmi F. et al., SNARE complexes of different composition jointly mediate membrane fusion in Arabidopsis cytokinesis. Mol. Biol. Cell 24, 1593–1601 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Uemura T. et al., A Golgi-released subpopulation of the trans-Golgi network mediates protein secretion in Arabidopsis. Plant Physiol. 179, 519–532 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Uemura T. et al., Systematic analysis of SNARE molecules in Arabidopsis: Dissection of the post-Golgi network in plant cells. Cell Struct. Funct. 29, 49–65 (2004). [DOI] [PubMed] [Google Scholar]
- 14.Zhang L. et al., Arabidopsis R-SNARE proteins VAMP721 and VAMP722 are required for cell plate formation. PLoS One 6, e26129 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chaineau M., Danglot L., Galli T., Multiple roles of the vesicular-SNARE TI-VAMP in post-Golgi and endosomal trafficking. FEBS Lett. 583, 3817–3826 (2009). [DOI] [PubMed] [Google Scholar]
- 16.Takáts S. et al., Autophagosomal Syntaxin17-dependent lysosomal degradation maintains neuronal function in Drosophila. J. Cell Biol. 201, 531–539 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pryor P. R. et al., Molecular basis for the sorting of the SNARE VAMP7 into endocytic clathrin-coated vesicles by the ArfGAP Hrb. Cell 134, 817–827 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koo S. J. et al., SNARE motif-mediated sorting of synaptobrevin by the endocytic adaptors clathrin assembly lymphoid myeloid leukemia (CALM) and AP180 at synapses. Proc. Natl. Acad. Sci. U.S.A. 108, 13540–13545 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller S. E. et al., The molecular basis for the endocytosis of small R-SNAREs by the clathrin adaptor CALM. Cell 147, 1118–1131 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zouhar J., Sauer M., Helping hands for budding prospects: ENTH/ANTH/VHS accessory proteins in endocytosis, vacuolar transport, and secretion. Plant Cell 26, 4232–4244 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaneda M. et al., Plant AP180 N-terminal homolog proteins are involved in clathrin-dependent endocytosis during pollen tube growth in Arabidopsis thaliana. Plant Cell Physiol. 60, 1316–1330 (2019). [DOI] [PubMed] [Google Scholar]
- 22.Li H. et al., The REN4 rheostat dynamically coordinates the apical and lateral domains of Arabidopsis pollen tubes. Nat. Commun. 9, 2573 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Muro K. et al., ANTH domain-containing proteins are required for the pollen tube plasma membrane integrity via recycling ANXUR kinases. Commun. Biol. 1, 152 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ito E. et al., Dynamic behavior of clathrin in Arabidopsis thaliana unveiled by live imaging. Plant J. 69, 204–216 (2012). [DOI] [PubMed] [Google Scholar]
- 25.Song K. et al., An A/ENTH domain-containing protein functions as an adaptor for clathrin-coated vesicles on the growing cell plate in Arabidopsis root cells. Plant Physiol. 159, 1013–1025 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fujimoto M., Arimura S., Nakazono M., Tsutsumi N., Imaging of plant dynamin-related proteins and clathrin around the plasma membrane by variable incidence angle fluorescence microscopy. Plant Biotechnol. 24, 449–455 (2007). [Google Scholar]
- 27.Konopka C. A., Bednarek S. Y., Variable-angle epifluorescence microscopy: A new way to look at protein dynamics in the plant cell cortex. Plant J. 53, 186–196 (2008). [DOI] [PubMed] [Google Scholar]
- 28.Fujimoto M. et al., Arabidopsis dynamin-related proteins DRP2B and DRP1A participate together in clathrin-coated vesicle formation during endocytosis. Proc. Natl. Acad. Sci. U.S.A. 107, 6094–6099 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Konopka C. A., Bednarek S. Y., Comparison of the dynamics and functional redundancy of the Arabidopsis dynamin-related isoforms DRP1A and DRP1C during plant development. Plant Physiol. 147, 1590–1602 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gendre D. et al., Trans-Golgi network localized ECHIDNA/Ypt interacting protein complex is required for the secretion of cell wall polysaccharides in Arabidopsis. Plant Cell 25, 2633–2646 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kulich I. et al., Arabidopsis exocyst subunits SEC8 and EXO70A1 and exocyst interactor ROH1 are involved in the localized deposition of seed coat pectin. New Phytol. 188, 615–625 (2010). [DOI] [PubMed] [Google Scholar]
- 32.Ford M. G. et al., Simultaneous binding of PtdIns(4,5)P2 and clathrin by AP180 in the nucleation of clathrin lattices on membranes. Science 291, 1051–1055 (2001). [DOI] [PubMed] [Google Scholar]
- 33.Dettmer J., Hong-Hermesdorf A., Stierhof Y. D., Schumacher K., Vacuolar H+-ATPase activity is required for endocytic and secretory trafficking in Arabidopsis. Plant Cell 18, 715–730 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bashline L., Li S., Anderson C. T., Lei L., Gu Y., The endocytosis of cellulose synthase in Arabidopsis is dependent on μ2, a clathrin-mediated endocytosis adaptin. Plant Physiol. 163, 150–160 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bashline L., Li S., Zhu X., Gu Y., The TWD40-2 protein and the AP2 complex cooperate in the clathrin-mediated endocytosis of cellulose synthase to regulate cellulose biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 112, 12870–12875 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fujimoto M., Suda Y., Vernhettes S., Nakano A., Ueda T., Phosphatidylinositol 3-kinase and 4-kinase have distinct roles in intracellular trafficking of cellulose synthase complexes in Arabidopsis thaliana. Plant Cell Physiol. 56, 287–298 (2015). [DOI] [PubMed] [Google Scholar]
- 37.Wen Y. et al., AP180-mediated trafficking of Vamp7B limits homotypic fusion of Dictyostelium contractile vacuoles. Mol. Biol. Cell 20, 4278–4288 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.De Craene J. O. et al., Evolutionary analysis of the ENTH/ANTH/VHS protein superfamily reveals a coevolution between membrane trafficking and metabolism. BMC Genomics 13, 297 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bowman J. L. et al., Insights into land plant evolution garnered from the Marchantia polymorpha genome. Cell 171, 287–304.e15 (2017). [DOI] [PubMed] [Google Scholar]
- 40.Zhao Y. et al., Phosphoinositides regulate clathrin-dependent endocytosis at the tip of pollen tubes in Arabidopsis and tobacco. Plant Cell 22, 4031–4044 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grebnev G., Ntefidou M., Kost B., Secretion and endocytosis in pollen tubes: Models of tip growth in the spot light. Front. Plant Sci. 8, 154 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gadeyne A. et al., The TPLATE adaptor complex drives clathrin-mediated endocytosis in plants. Cell 156, 691–704 (2014). [DOI] [PubMed] [Google Scholar]
- 43.Hao W., Luo Z., Zheng L., Prasad K., Lafer E. M., AP180 and AP-2 interact directly in a complex that cooperatively assembles clathrin. J. Biol. Chem. 274, 22785–22794 (1999). [DOI] [PubMed] [Google Scholar]
- 44.Miller S. E. et al., CALM regulates clathrin-coated vesicle size and maturation by directly sensing and driving membrane curvature. Dev. Cell 33, 163–175 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Traub L. M., Regarding the amazing choreography of clathrin coats. PLoS Biol. 9, e1001037 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Di Rubbo S. et al., The clathrin adaptor complex AP-2 mediates endocytosis of brassinosteroid insensitive1 in Arabidopsis. Plant Cell 25, 2986–2997 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Van Damme D. et al., Adaptin-like protein TPLATE and clathrin recruitment during plant somatic cytokinesis occurs via two distinct pathways. Proc. Natl. Acad. Sci. U.S.A. 108, 615–620 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kleinboelting N., Huep G., Kloetgen A., Viehoever P., Weisshaar B., GABI-Kat SimpleSearch: New features of the Arabidopsis thaliana T-DNA mutant database. Nucleic Acids Res. 40, D1211–D1215 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rosso M. G. et al., An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol. Biol. 53, 247–259 (2003). [DOI] [PubMed] [Google Scholar]
- 50.Karimi M., Inzé D., Depicker A., GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7, 193–195 (2002). [DOI] [PubMed] [Google Scholar]
- 51.Nakagawa T. et al., Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J. Biosci. Bioeng. 104, 34–41 (2007). [DOI] [PubMed] [Google Scholar]
- 52.Nakamura S. et al., Gateway binary vectors with the bialaphos resistance gene, bar, as a selection marker for plant transformation. Biosci. Biotechnol. Biochem. 74, 1315–1319 (2010). [DOI] [PubMed] [Google Scholar]
- 53.Ebine K. et al., Plant vacuolar trafficking occurs through distinctly regulated pathways. Curr. Biol. 24, 1375–1382 (2014). [DOI] [PubMed] [Google Scholar]
- 54.Huang J. et al., Arabidopsis dynamin-related proteins, DRP2A and DRP2B, function coordinately in post-Golgi trafficking. Biochem. Biophys. Res. Commun. 456, 238–244 (2015). [DOI] [PubMed] [Google Scholar]
- 55.Clough S. J., Bent A. F., Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743 (1998). [DOI] [PubMed] [Google Scholar]
- 56.Mano S., Hayashi M., Nishimura M., Light regulates alternative splicing of hydroxypyruvate reductase in pumpkin. Plant J. 17, 309–320 (1999). [DOI] [PubMed] [Google Scholar]
- 57.Holstein S. E., Oliviusson P., Sequence analysis of Arabidopsis thaliana E/ANTH-domain-containing proteins: Membrane tethers of the clathrin-dependent vesicle budding machinery. Protoplasma 226, 13–21 (2005). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and SI Appendix.