Key Points
Question
How does an artificial intelligence (AI) algorithm compare with radiology residents in full-fledged preliminary reads of anteroposterior (AP) frontal chest radiographs?
Findings
This diagnostic study was conducted among 5 third-year radiology residents and an AI algorithm using a study data set of 1998 AP frontal chest radiographs assembled through a triple consensus with adjudication ground truth process covering more than 72 chest radiograph findings. There was no statistically significant difference in sensitivity between the AI algorithm and the radiology residents, but the specificity and positive predictive value were statistically higher for AI algorithm.
Meaning
These findings suggest that well-trained AI algorithms can reach performance levels similar to radiology residents in covering the breadth of findings in AP frontal chest radiographs, which suggests there is the potential for the use of AI algorithms for preliminary interpretations of chest radiographs in radiology workflows to expedite radiology reads, address resource scarcity, improve overall accuracy, and reduce the cost of care.
This diagnostic study examines the area under the curve, sensitivity, specificity, and positive predictive value of chest radiograph readings performed by an artificial intelligence algorithm vs by radiology residents.
Abstract
Importance
Chest radiography is the most common diagnostic imaging examination performed in emergency departments (EDs). Augmenting clinicians with automated preliminary read assistants could help expedite their workflows, improve accuracy, and reduce the cost of care.
Objective
To assess the performance of artificial intelligence (AI) algorithms in realistic radiology workflows by performing an objective comparative evaluation of the preliminary reads of anteroposterior (AP) frontal chest radiographs performed by an AI algorithm and radiology residents.
Design, Setting, and Participants
This diagnostic study included a set of 72 findings assembled by clinical experts to constitute a full-fledged preliminary read of AP frontal chest radiographs. A novel deep learning architecture was designed for an AI algorithm to estimate the findings per image. The AI algorithm was trained using a multihospital training data set of 342 126 frontal chest radiographs captured in ED and urgent care settings. The training data were labeled from their associated reports. Image-based F1 score was chosen to optimize the operating point on the receiver operating characteristics (ROC) curve so as to minimize the number of missed findings and overcalls per image read. The performance of the model was compared with that of 5 radiology residents recruited from multiple institutions in the US in an objective study in which a separate data set of 1998 AP frontal chest radiographs was drawn from a hospital source representative of realistic preliminary reads in inpatient and ED settings. A triple consensus with adjudication process was used to derive the ground truth labels for the study data set. The performance of AI algorithm and radiology residents was assessed by comparing their reads with ground truth findings. All studies were conducted through a web-based clinical study application system. The triple consensus data set was collected between February and October 2018. The comparison study was preformed between January and October 2019. Data were analyzed from October to February 2020. After the first round of reviews, further analysis of the data was performed from March to July 2020.
Main Outcomes and Measures
The learning performance of the AI algorithm was judged using the conventional ROC curve and the area under the curve (AUC) during training and field testing on the study data set. For the AI algorithm and radiology residents, the individual finding label performance was measured using the conventional measures of label-based sensitivity, specificity, and positive predictive value (PPV). In addition, the agreement with the ground truth on the assignment of findings to images was measured using the pooled κ statistic. The preliminary read performance was recorded for AI algorithm and radiology residents using new measures of mean image-based sensitivity, specificity, and PPV designed for recording the fraction of misses and overcalls on a per image basis. The 1-sided analysis of variance test was used to compare the means of each group (AI algorithm vs radiology residents) using the F distribution, and the null hypothesis was that the groups would have similar means.
Results
The trained AI algorithm achieved a mean AUC across labels of 0.807 (weighted mean AUC, 0.841) after training. On the study data set, which had a different prevalence distribution, the mean AUC achieved was 0.772 (weighted mean AUC, 0.865). The interrater agreement with ground truth finding labels for AI algorithm predictions had pooled κ value of 0.544, and the pooled κ for radiology residents was 0.585. For the preliminary read performance, the analysis of variance test was used to compare the distributions of AI algorithm and radiology residents’ mean image-based sensitivity, PPV, and specificity. The mean image-based sensitivity for AI algorithm was 0.716 (95% CI, 0.704-0.729) and for radiology residents was 0.720 (95% CI, 0.709-0.732) (P = .66), while the PPV was 0.730 (95% CI, 0.718-0.742) for the AI algorithm and 0.682 (95% CI, 0.670-0.694) for the radiology residents (P < .001), and specificity was 0.980 (95% CI, 0.980-0.981) for the AI algorithm and 0.973 (95% CI, 0.971-0.974) for the radiology residents (P < .001).
Conclusions and Relevance
These findings suggest that it is possible to build AI algorithms that reach and exceed the mean level of performance of third-year radiology residents for full-fledged preliminary read of AP frontal chest radiographs. This diagnostic study also found that while the more complex findings would still benefit from expert overreads, the performance of AI algorithms was associated with the amount of data available for training rather than the level of difficulty of interpretation of the finding. Integrating such AI systems in radiology workflows for preliminary interpretations has the potential to expedite existing radiology workflows and address resource scarcity while improving overall accuracy and reducing the cost of care.
Introduction
The increase in imaging orders and the availability of high-resolution scanners have led to large workloads for radiologists.1,2 With advances in artificial intelligence (AI), there is now potential for radiologists to be aided in clinical workflows by machine assistants in a manner similar to the duties performed by radiology residents,3 through preliminary interpretations that can be later corrected or approved by attending radiologists. This can help expedite workflows, improve accuracy, and ultimately reduce overall costs.3
Recent work has focused on chest radiography, the most common imaging examination conducted in emergency departments (EDs) and urgent care settings. Several studies have reported machine learning models achieving radiologist-level performance for different chest radiograph findings.4,5,6,7 However, large-scale adoption of these models is still lacking, owing to 3 main factors: limited findings, limited generalizability across data sets, and lack of rigorous comparative assessment studies vis-à-vis radiologists against criterion standard data sets. There has been no systematic effort to catalog the number of findings that constitute sufficient coverage of the anomalies seen in chest radiographs.8 The generalizability of the models across data sets even for limited target findings has been difficult owing to lack of sufficient variety in training data and the choice of thresholds used for estimation that are not necessarily optimized on relevant metrics. Typically, models are trained and tested on a single hospital’s data, which are not necessarily representative of prevalence distributions seen in other hospitals. Lastly, to our knowledge, there has been no systematic study that objectively compares the performance of machine learning models vs radiologists for a comprehensive preliminary read on a sufficiently large test data set of chest radiographs. Existing studies report comparisons of model performance on a limited number of labels, with ground truth obtained by majority vote from board-certified radiologists.9,10,11,12 For machines to serve as cognitive assistants, they must prove themselves through the comprehensiveness of coverage of findings based on meaningful metrics that match the realistic use scenarios and objective evaluation through rigorous comparative studies.
In this diagnostic study, we present an AI algorithm that was developed keeping the aforementioned requirements in mind. Since our target use case was for emergency settings, and since other viewpoints would need a broader cataloging effort or integration of information from lateral views, we focused on anteroposterior (AP) frontal chest radiographs for cataloging and field testing. Specifically, we used a systematic clinician-guided multistep approach to first catalog the space of possible findings in AP frontal chest radiographs. We then acquired a large multihospital data set and developed a text analysis algorithm to label the radiographs from their associated reports. Next, we designed a novel deep learning architecture for the simultaneous recognition of a large number of findings in chest radiographs and optimized its prediction using F1 score–based thresholds.14 Finally, we assessed the readiness of the AI algorithm for preliminary reads of AP frontal chest radiographs through an objective comparison study with reads by radiology residents on a triple consensus with adjudication ground truth data of 1998 AP frontal chest radiographs drawn from a source that was representative of realistic inpatient and ED settings.
Methods
All data used in this diagnostic study were deidentified and covered under the secondary use of health data, per Safran et al.13 Informed patient consent was waived by the National Institutes of Health (NIH) institutional review board for the NIH data set.4 The data in the MIMIC dataset has been previously deidentified, and the institutional review boards of the Massachusetts Institute of Technology and Beth Israel Deaconess Medical Center both approved the use of the database for research. This study is reported following the Transparent Reporting of a Multivariable Prediction Model for Individual Prognosis or Diagnosis (TRIPOD) reporting guideline.
Characterizing the Space of Findings Seen in AP Chest Radiographs
To assemble a comprehensive list of radiology findings in chest radiographs, a team of 4 clinicians (3 radiologists [A. Sharma, M. Morris, and B.S.] and 1 internal medicine physician [J.T.W.]) used a process shown in eFigure 1 in the Supplement. Specifically, they iteratively searched through the best practice literature, including Fleishner’s glossary,15 consulted several radiologists for a raw list of everyday use terms, and arrived at a list of initial core findings. They were found to be in 6 major categories: anatomical findings, tubes and lines, placements of tubes and lines, external devices, viewpoint-related issues, and diseases associated with findings. These were used as starting vocabulary to accumulate related terms from more than 200 000 radiology reports obtained from the MIMIC-III16 intensive care unit database, which includes other reports relating to admission. Specifically, we used a tool called the Domain Learning Assistant17 (DLA) to assemble related terms in 3 iterative steps. In the preprocessing step, the text corpus of reports was analyzed for n-grams, and a 2-layer word2Vec18 neural net model was built for learning term embeddings from raw text. In the explore phase, the starting vocabulary phrases were projected using this model to find the nearest n-dimensional vectors and hence their corresponding original terms from textual reports. These were then presented in the user interface for a clinician to validate and select (eFigure 2 in the Supplement). In the exploit phase, the given terms were modified by substitution and expanded terms to include abbreviations, synonyms, and ontologically related and visually similar concepts, which were further adjudicated in the tool by the radiologists. Finally, the created entries in the lexicon were reviewed by 2 radiologists (H.A. and A. Syed) for semantic consistency. Currently, the lexicon consists of more than 11 000 unique terms covering the space of 72 core findings, as listed in Table 1, and represents the largest set of finding labels assembled for chest radiographs, to our knowledge.
Table 1. Core Finding Labels Derived From the Chest Radiograph Lexicon .
| Finding | Samples in modeling data set, No. | AUC of AI algorithm | |
|---|---|---|---|
| Type | Label | ||
| Anatomical | Not otherwise specified opacity (eg, pleural or parenchymal opacity) | 81 013 | 0.736 |
| Anatomical | Linear or patchy atelectasis | 79 218 | 0.776 |
| Anatomical | Pleural effusion or thickening | 76 954 | 0.887 |
| Anatomical | No anomalies | 55 894 | 0.847 |
| Anatomical | Enlarged cardiac silhouette | 49 444 | 0.846 |
| Anatomical | Pulmonary edema or hazy opacity | 40 208 | 0.861 |
| Anatomical | Consolidation | 29 986 | 0.79 |
| Anatomical | Not otherwise specified calcification | 14 333 | 0.82 |
| Anatomical | Pneumothorax | 11 686 | 0.877 |
| Anatomical | Lobar or segmental collapse | 10 868 | 0.814 |
| Anatomical | Fracture | 9738 | 0.758 |
| Anatomical | Mass or nodule (not otherwise specified) | 8588 | 0.742 |
| Anatomical | Hyperaeration | 8197 | 0.905 |
| Anatomical | Degenerative changes | 7747 | 0.83 |
| Anatomical | Vascular calcification | 4481 | 0.873 |
| Anatomical | Tortuous aorta | 3947 | 0.814 |
| Anatomical | Multiple masses or nodules | 3453 | 0.754 |
| Anatomical | Vascular redistribution | 3436 | 0.705 |
| Anatomical | Enlarged hilum | 3106 | 0.734 |
| Anatomical | Scoliosis | 2968 | 0.815 |
| Anatomical | Bone lesion | 2879 | 0.762 |
| Anatomical | Hernia | 2792 | 0.828 |
| Anatomical | Postsurgical changes | 2526 | 0.834 |
| Anatomical | Mediastinal displacement | 1868 | 0.907 |
| Anatomical | Increased reticular markings or ILD pattern | 1828 | 0.891 |
| Anatomical | Old fractures | 1760 | 0.762 |
| Anatomical | Subcutaneous air | 1664 | 0.913 |
| Anatomical | Elevated hemidiaphragm | 1439 | 0.775 |
| Anatomical | Superior mediastinal mass or enlargement | 1345 | 0.709 |
| Anatomical | Subdiaphragmatic air | 1258 | 0.75 |
| Anatomical | Pneumomediastinum | 915 | 0.807 |
| Anatomical | Cyst or Bullae | 778 | 0.76 |
| Anatomical | Hydropneumothorax | 630 | 0.935 |
| Anatomical | Spinal degenerative changes | 454 | 0.818 |
| Anatomical | Calcified nodule | 439 | 0.736 |
| Anatomical | Lymph node calcification | 346 | 0.603 |
| Anatomical | Bullet or foreign bodies | 339 | 0.715 |
| Anatomical | Other soft tissue abnormalities | 334 | 0.652 |
| Anatomical | Diffuse osseous irregularity | 322 | 0.89 |
| Anatomical | Dislocation | 180 | 0.728 |
| Anatomical | Dilated bowel | 92 | 0.805 |
| Anatomical | Osteotomy changes | 76 | 0.942 |
| Anatomical | New fractures | 70 | 0.696 |
| Anatomical | Shoulder osteoarthritis | 70 | 0.698 |
| Anatomical | Elevated humeral head | 69 | 0.731 |
| Anatomical | Azygous fissure (benign) | 47 | 0.652 |
| Anatomical | Contrast in the GI or GU tract | 17 | 0.724 |
| Device | Other internal postsurgical material | 26 191 | 0.831 |
| Device | Sternotomy wires | 12 262 | 0.972 |
| Device | Cardiac pacer and wires | 12 109 | 0.985 |
| Device | Musculoskeletal or spinal hardware | 5481 | 0.848 |
| Technical | Low lung volumes | 25 546 | 0.877 |
| Technical | Rotated | 3809 | 0.803 |
| Technical | Lungs otherwise not fully included | 1440 | 0.717 |
| Technical | Lungs obscured by overlying object or structure | 653 | 0.68 |
| Technical | Apical lordotic | 620 | 0.716 |
| Technical | Apical kyphotic | 566 | 0.872 |
| Technical | Nondiagnostic radiograph | 316 | 0.858 |
| Technical | Limited by motion | 290 | 0.628 |
| Technical | Limited by exposure or penetration | 187 | 0.834 |
| Technical | Apices not included | 175 | 0.822 |
| Technical | Costophrenic angle not included | 62 | 0.807 |
| Tubes and lines | Central intravascular lines | 57 868 | 0.891 |
| Tubes and lines | Tubes in the airway | 32 718 | 0.96 |
| Tubes and lines | Enteric tubes | 27 998 | 0.939 |
| Tubes and lines | Incorrect placement | 11 619 | 0.827 |
| Tubes and lines | Central intravascular lines: incorrectly positioned | 4434 | 0.769 |
| Tubes and lines | Enteric tubes: incorrectly positioned | 4372 | 0.931 |
| Tubes and lines | Coiled, kinked, or fractured | 4325 | 0.857 |
| Tubes and lines | Tubes in the airway: incorrectly positioned | 1962 | 0.919 |
Abbreviations: AI, artificial intelligence; AUC, area under the curve; GI, gastrointestinal; GU, genitourinary; ILD, interstitial lung disease.
Assembling the Data Sets for Model Training
The data set for model training (hereafter, the modeling data set) was assembled from 2 hospital sources: MIMIC-4,19 and the NIH4 depicting high-quality Digital Imaging and Communications in Medicine imagery (ie, 1024 × 1024 pixels). The MIMIC-4 was collected from 2011 to 2016, while the NIH data set was collected during 1992 to 2015. They showed a wide range of clinical settings, including intensive care units, urgent care, inpatient care, and EDs.
The MIMIC-419 data set had associated radiology reports, while the NIH data set4 came with few labels, requiring recreation of reports through fresh reads. The sampling procedure, as well as the resulting training data set created, are shown in Figure 1. Both AP and posteroanterior (PA) images were used for training the model to get good coverage of findings of AP chest radiographs that are also seen in PA images. While all AP (240 764 images) and PA (101 362 images) images in the MIMIC-4 data set were used owing to available reports, the NIH data set was randomly sampled for report generation, from which only a subset of 11 692 images could be manually reread.
Figure 1. Sampling of Data Distributions for Artificial Intelligence Algorithm Training and Evaluation.
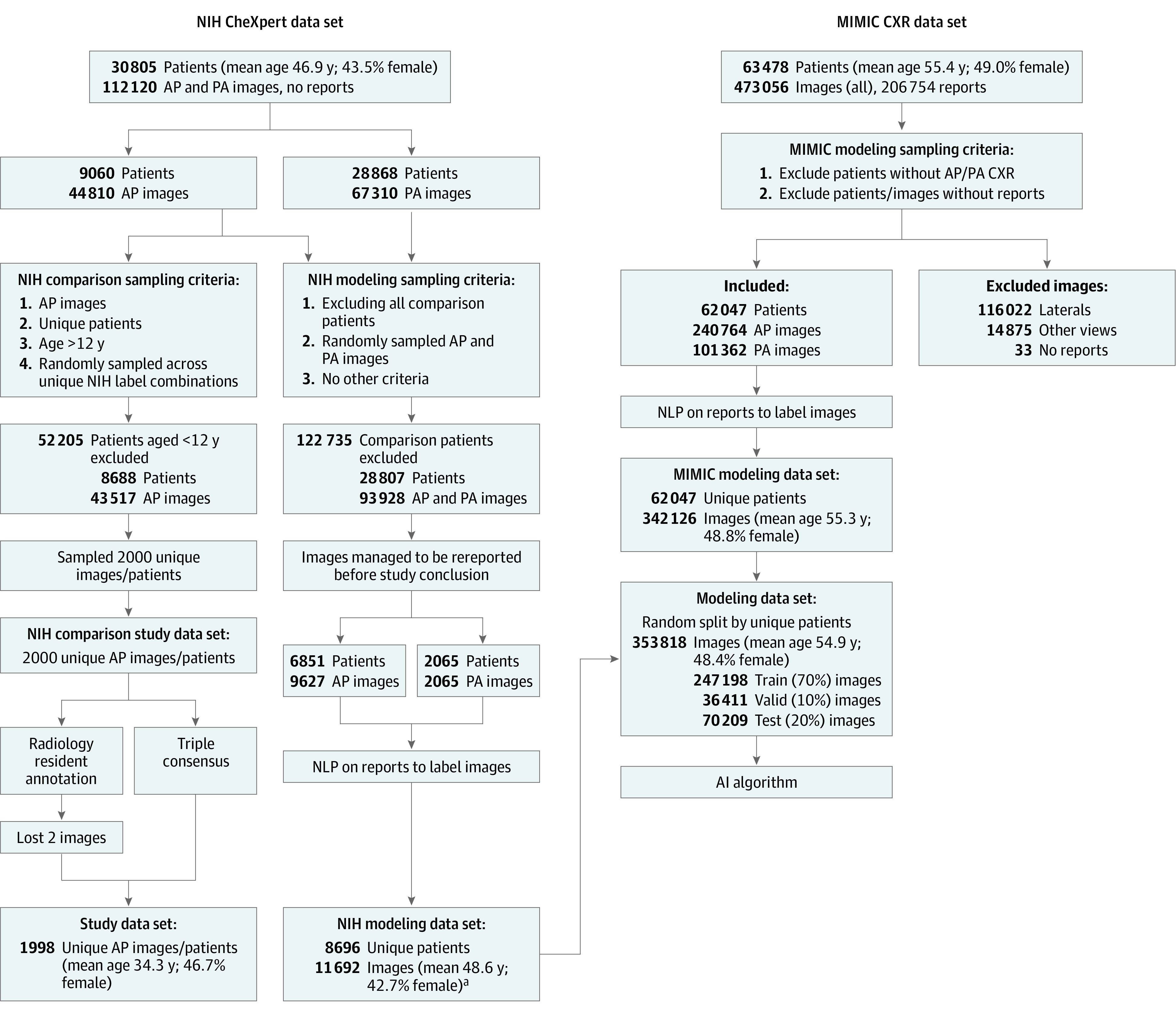
Two images were excluded from the comparison study data set owing to radiology resident annotations missing. The prevalence distribution of training and study data sets are different owing to the difference in the sampling process.
Extracting Finding Labels From Reports
To derive labels for images from their associated radiological reports, we extracted sentences from relevant sections of the report (those labeled findings or overall impression). Using the generated lexicon, we systematically checked for each finding term and its variants in the sentences using stemming and word-form normalization for robust localization. To reduce false positives in labeling, we detected negation terms (occurring prior to and after the target finding label), unchanged statuses of a mentioned label, hypothetically mentioned label, and optionally, the associated anatomical context. Examples of radiology reports and the automatically extracted findings from these reports are shown in eTable 1 in the Supplement.
On a sample collection of 2771 radiological reports and using 2 radiologists (A. Sharma and A. Syed) to manually validate the resulting 10 842 findings flagged by the algorithm, we found 84 semantically inaccurate detections and an overall precision of 99.2% and recall of 92.6%. Since the misses mainly corresponded to missing lexicon vocabulary, this was found to be sufficient for our image labeling purposes.
Designing a Neural Network Architecture
A deep neural network architecture was designed (Figure 2). It combines the advantages of pretrained features with a multiresolution image analysis through the feature pyramid network20 for fine grained classification. Specifically VGGNet21 (16 layers) and ResNet22 (50 layers) were used as the initial feature extractors, which were trained on several million images from ImageNet.23 Dilated blocks composed of multiscale features24 and skip connections25 were used to improve convergence while spatial dropout was used to reduce overfitting. Group normalization26 (16 groups) was used, along with Rectified Linear Unit27 as activation function. Dilated blocks with different feature channels were cascaded with maxpooling to learn more abstract features. Bilinear pooling was used for effective fine-grained classification.28
Figure 2. Deep Learning Network Architecture for Anteroposterior Chest Radiographs.
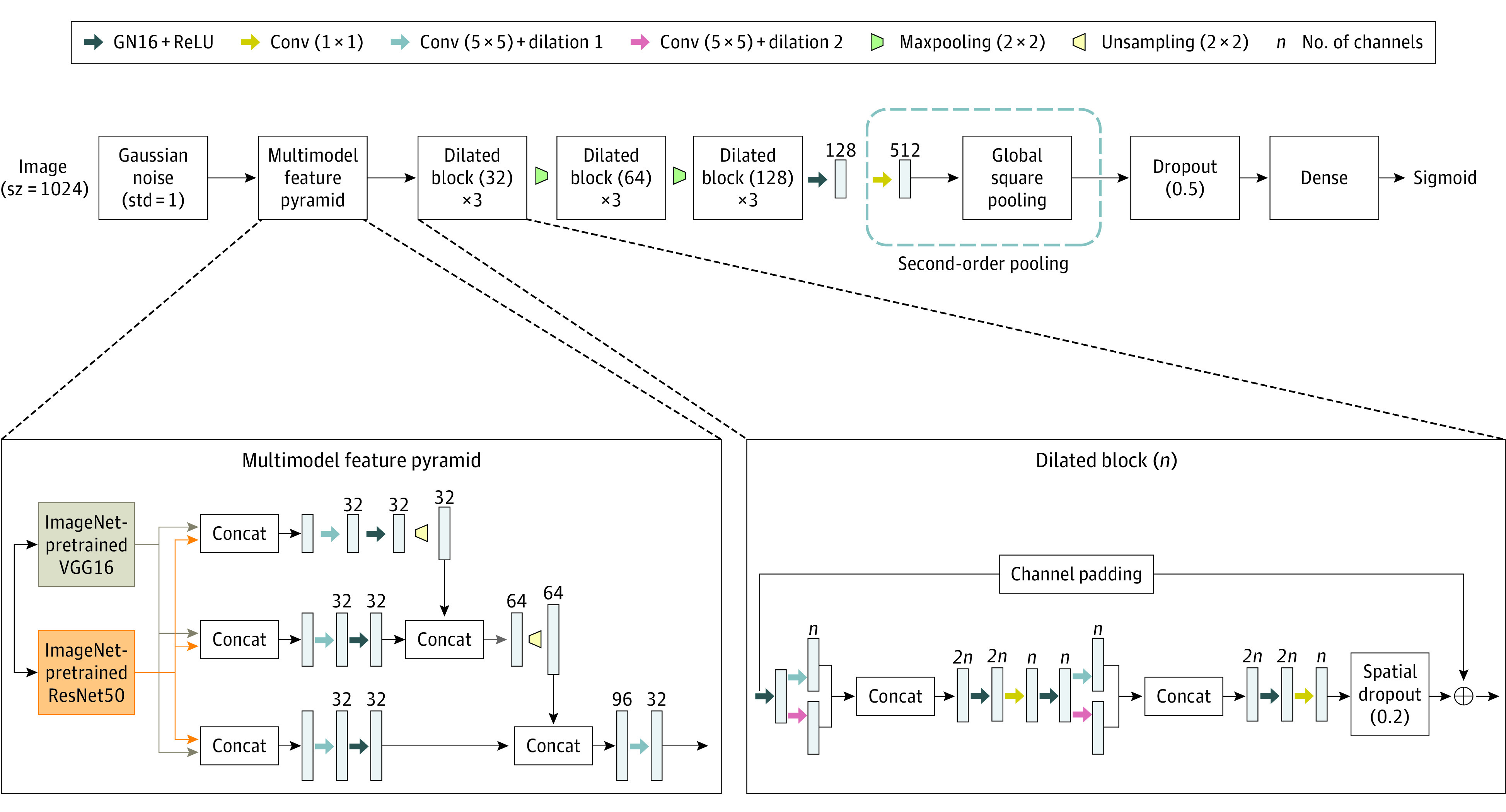
The architecture can be seen as a combination of ResNet5022 and VGG1621 and was selected after experimenting with many alternatives. For example, a baseline VGG1621 model was also developed using ImageNet pretraining,23 with all layers up to the final convolutional layer followed by global average pooling and a fully connected layer for classification. It was abandoned after it yielded worse performance in terms of mean area under the curve (AUC).
Algorithm Training
To train the deep learning model, the modeling data set was split into 3 partitions for training, validation, and testing. Since existing methods of random splitting29 cannot ensure adequate number of images for low-incidence–label training, our splitting algorithm sorted the labels by their frequencies of occurrences. It then iteratively assigned the images from distinct patients to the 3 partitions in the ratio of 70% for training, 10% for validation, and 20% for testing. Once the number of patients in each split was determined per label, the assignment of the patients and images was still random. Thus the algorithm ensured that the prevalence distributions were similar for training, validation, and testing partitions while minimizing the selection bias through random sampling of images. eAppendix 1 in the Supplement details the algorithm and eFigure 3 in the Supplement shows the resulting similarity in the prevalence distribution in the 3 partitions of the modeling data set of Figure 1.
The deep learning model was trained on all 72 finding labels. As the images were of high resolution (ie, 1024 × 1024 pixels), training took approximately 10 days. We used the Nadam optimizer for fast convergence, with the learning rate as 2 × 10−6. Two NVIDIA Tesla V100 graphics processing units with 16 GB memory were used for multi–graphics processing unit training with a batch size of 12 and 30 epochs.
The performance of the trained model on the testing partition of the modeling data set for all the 72 labels is shown through the area under the respective receiver operating characteristics (ROC) curves in Table 1. The ROC curve shows the tradeoff between sensitivity and specificity as a function of thresholds on the estimated values. The mean AUC across all the labels was 0.807 (weighted mean AUC, 0.841). A comparable VGG-16 based implementation gave a mean AUC of 0.604. Although individual label accuracies could still be improved with more training data, this network covers the largest number of chest radiograph findings to date, to our knowledge.
Algorithm Prediction
While the ROC curve shows the algorithm performance as a function of thresholds, a reasonable threshold had to be chosen per label for use in estimation on unseen data sets. Since our objective was to maximize the number of correct detections per image while minimizing the false positives, we selected the thresholds by maximizing the mean image-based F1 score,14 a well-known measure of accuracy that is the harmonic mean of positive predictive value (PPV) and sensitivity. The validation partition of the modeling data set was used for this optimization. Further details are available in eAppendix 2 in the Supplement.
Comparative Study on Preliminary Reads
To assess the readiness of the trained AI algorithm for realistic inpatient settings, we performed a comparative study with radiology residents involving full-fledged preliminary read of frontal AP chest radiographs. The data set was drawn from the same NIH hospital data source,4 but this time focusing on single AP frontal chest radiographs from patients older than 12 years, based on the target use case (Figure 1). A random sampling across the initial label combinations provided in the NIH data set4 was used to create the data set of 1998 AP frontal chest radiographs (hereafter, study data set). Since this was performed prior to creating the ground truth labels, the breadth of label coverage was uncertain. Furthermore, the resulting prevalence distribution was not expected to match the modeling data set, thus serving as a good case to test the AI algorithm’s generalization ability.
Ground Truth Labeling for Study Data Set
For the purpose of the comparison study, we created criterion standard labels through a process in which 3 board-certified radiologists (A. Sharma, M. Morris, and B.S.) with 3 to 5 years of experience each independently labeled all the images first. The annotation platform showed full-resolution images (ie, 1024 × 1024 pixels) with zoom, inversion, windowing, and contrast adjustment to recreate a typical radiological read setting. The discrepancies in labels were recorded per image and were shown back to these radiologists in the interface as shown in eFigure 5 in the Supplement. Consensus was achieved through an in-person video adjudication discussion. The criterion-standard label creation process took more than 400 hours during a period of 9 months. Coverage was achieved for at least 68 of 72 finding labels. The resulting prevalence distribution of labels in the study data set (eFigure 4 in the Supplement) was different from the one used for training (eFigure 3 in the Supplement).
Recording Reads of Radiology Residents
Five radiology residents were selected from academic medical centers around the US after they passed a reading adequacy test on 5 unrelated chest radiographs. Each of the residents evaluated approximately 400 nonoverlapping set of images and were unaware of AI algorithm estimates. A web-based structured form interface was used to show the images and collect their discrete read on the 72 possible findings (eFigure 6 in the Supplement). This objective capture avoided the accidental miss typical of free-form reporting. Furthermore, the label names were selected to be similar to those in routine reporting. Prior training on finding definitions was provided to residents so that the selection of anomalies through the list shown in the user interface did not artificially constrain the radiologists.
Statistical Analysis
The learning performance of the AI algorithm was judged using the conventional ROC curve and the area under the curve (AUC) during training and field testing on the study data set. The individual finding label performance was measured using the conventional measures of label-based sensitivity, specificity, and positive predictive value (PPV). The agreement with the ground truth on the assignment of findings to images was measured using the pooled κ statistic. The preliminary read performance was recorded using mean image-based sensitivity, specificity, and PPV designed for recording the fraction of misses and overcalls on a per image basis. Analysis of variance (ANOVA) and 95% CIs were calculated using the Python scipy.stats module version 1.5.2 (Python Software Foundation. P values reported for our ANOVA test are from 1-way ANOVA tests. We considered P < .05 as statistically significant and report the P values for each test.
Results
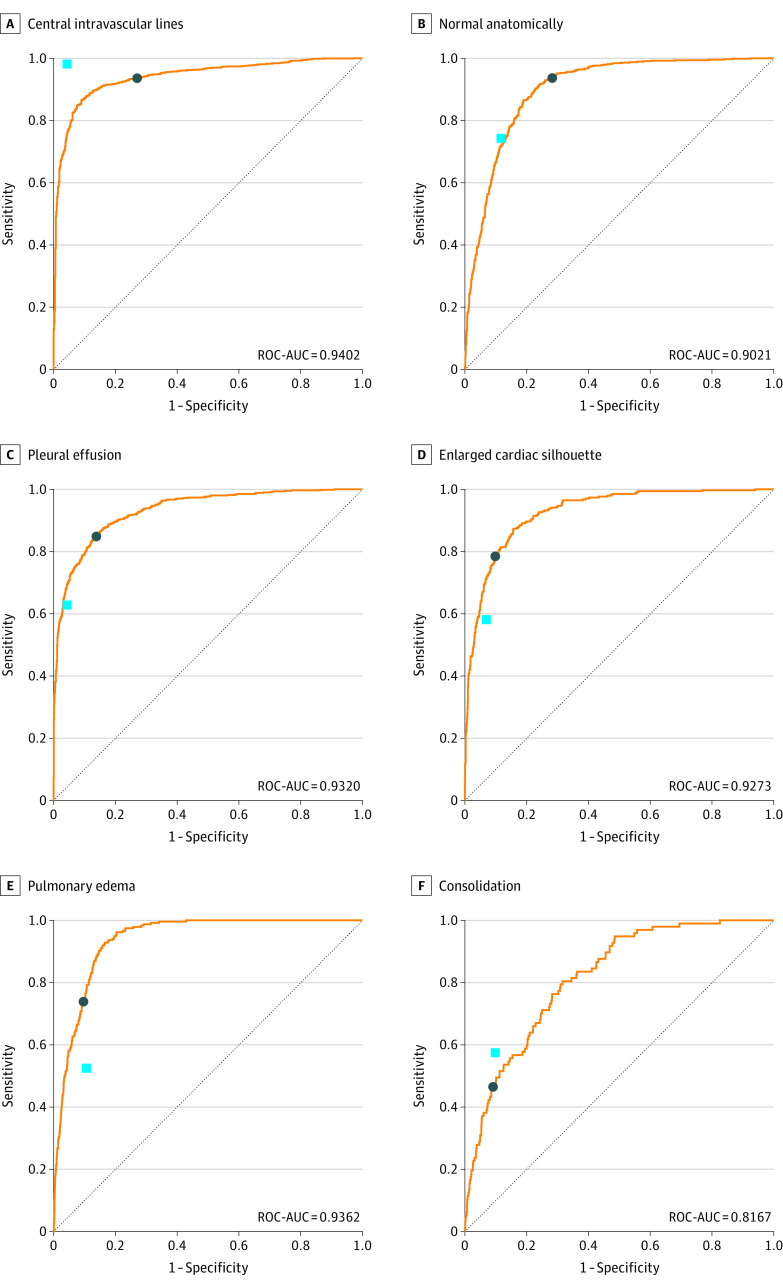
The AI algorithm generalized well to the study data set despite it having a different prevalence distribution, particularly for prevalent findings. Figure 3 shows the ROC curves for 6 of the most prevalent labels on the study data set. Comparing the AUCs for all the prevalent finding labels (ie, with at least 50 images) (eTable 2 in the Supplement) with the corresponding data in the training data set (Table 1), we observed that generalization was more a function of label prevalence than interpretation difficulty (eg, pneumothorax). The mean AUC of the AI algorithm for all 72 finding labels on the study data set was 0.772 (weighted mean, 0.865).
Figure 3. Receiver Operating Characteristic Curves of Artifical Intelligence Algorithm on Study Data Set and Relative Performance.
The findings selected were from the most prevalent ones in the modeling data set. The light blue square indicates mean sensitivity and 1 − specificity of the radiology residents on the comparison study data set; dark blue circle, operating point of the artificial intelligence algorithm based on the F1 score–based threshold derived from training data.
Comparison of Per-Finding Performance
Using the F1 score–based thresholds selected during training, we retained the estimated labels from the AI algorithm. Similarly, the radiology residents’ discrete label choices for each image were recorded. The per-finding label performances for both AI algorithm and radiology residents were each measured using conventional label-based sensitivity, specificity, and PPV30 measures using the ground truth as reference. eTable 3 in the Supplement lists the performance for all prevalent findings labels within the study data set. Figure 3 illustrates the relative performance of AI algorithm and radiology residents for the 6 most prevalent findings. Of the 9 most prevalent findings, we observed that the residents’ operating points were on or very near the ROC curve for 4 findings (ie, no anomalies, opacities, pleural effusion, and airway tubes), below for 2 findings (ie, pulmonary edema and cardiomegaly), and above for 3 findings (ie, atelectasis, central vascular lines, and consolidation).
The relative agreement of the estimations across all estimated findings was measured through the pooled κ scores31 as 0.543 for the AI algorithm and 0.585 for the radiology residents. The overall distribution of κ scores is shown in eFigure 7 in the Supplement. Overall, the AI algorithm performed similarly to residents for tubes and lines and nonanomalous reads, and generally outperformed for high-prevalence labels, such as cardiomegaly, pulmonary edema, subcutaneous air, and hyperaeration. Conversely, the AI algorithm generally performed worse for lower-prevalence findings that also had a higher level of difficulty of interpretation, such as masses or nodules and enlarged hilum. eTable 3 in the Supplement summarizes the overall agreement results.
Comparison of the Preliminary Read Performance of the AI Algorithm vs Radiology Residents
Since the read quality is assessed in clinical workflows by the number of overcalls or misses per image, we measured the preliminary read performance using image-based sensitivity, PPV, and specificity measures that record the ratios of true positives, false positives, and true negatives on a per-image basis, as outlined in eAppendix 3 in the Supplement. We calculated the means of these measures across images for the AI algorithm and radiology residents. Since multiple radiology residents were involved, these measures were further meaned across residents. We then performed a 1-sided analysis of variance test to compare the means of 2 groups using the F distribution. The null hypothesis of the test was that the groups would have similar means. The results are shown in Table 2. The mean image-based sensitivity was 0.716 (95% CI, 0.704-0.729) for the AI algorithm and 0.720 (95% CI, 0.709-0.732) for the radiology residents , (P = 0.66), while the PPV was 0.730 (95% CI, 0.718-0.742) for the AI algorithm and 0.682 (95% CI, 0.670-0.694) for the radiology residents (P < .001), and specificity was 0.980 (95% CI, 0.980-0.981)for the AI algorithm and 0.973 (95% CI, 0.971-0.974) for the radiology residents (P < .001). Thus, while no statistically significant difference was found in mean image-based sensitivity between groups, the specificity and PPV were statistically higher for the AI algorithms compared with radiology residents. eFigure 8 in the Supplement shows the box plots for the preliminary read performance differences of radiology residents and our AI algorithm, reinforcing the same conclusion.
Table 2. Preliminary Read Performance Differences Between Radiology Residents and AI Algorithm.
| Method | No. | Image-based measure, mean (95% CI) | |||
|---|---|---|---|---|---|
| Images | Findings | PPV | Sensitivity | Specificity | |
| All radiology residents | 1998 | 72 | 0.682 (0.670-0.694) | 0.720 (0.709-0.732) | 0.973 (0.971-0.974) |
| AI algorithm | 1998 | 72 | 0.730 (0.718-0.742) | 0.716 (0.704-0.729) | 0.980 (0.979-0.981) |
| AI vs radiology residents, P value | NA | NA | .001 | .66 | <.001 |
Abbreviations: AI, artificial intelligence; NA, not applicable; PPV, positive predictive value.
Discussion
This diagnostic study found that the variation in per-finding performance of both the residents and AI algorithm was primarily a function of the level of interpretation difficulty for an anomaly, the number of training images provided, and the generalizability across the varieties in anomaly appearance across data sets. In general, residents performed better for more subtle anomalies, such as masses and nodules, misplaced lines and tubes, and various forms of consolidation, while the AI algorithm was better at detecting nonanomalous findings, the presence of tubes and lines, and clearly visible anomalies, such as cardiomegaly, pleural effusion, and pulmonary edema.
We also note that threshold choices made to maximize the preliminary read performance of the AI algorithm could imply a suboptimal choice of threshold for the specific finding itself (eg, consolidation), which may lead to some labels never being called. For the target use cases involving expedited workflow, this was still appropriate, as the most common findings could be caught in the AI-driven preliminary read while the more complex findings would benefit from expert overreads. Finally, as shown in eTable 4 in the Supplement, there is also considerable variation across radiology residents themselves, pointing to the variations in training received in respective schools.
Limitations
This study has some limitations. First, since the study data set was drawn before labeling, the ground truth data after labeling did not have sufficient representation from the less prevalent findings for adequate testing of the AI algorithm, although it was already trained for these findings. If the MIMIC-4 data set had been available earlier, the study data set could have been preselected using automatic labeling from the associated reports. This could increase the likelihood of adequate coverage of low-prevalence labels after triple consensus ground truth labeling. Second, the comparison with radiology residents was done using data obtained from only 5 radiology residents. A more comprehensive study of the performance of a larger pool of radiology residents could strengthen the conclusions.
Conclusions
This diagnostic study is significant to the AI and radiology communities in several ways. First, it has a large clinician involvement in the design and execution of the study, indicating the integral role of radiologists and clinical experts in the development of AI algorithms. In our study, we have used clinical expertise in many phases, including the cataloguing of possible findings in chest radiographs, formation of a chest radiograph lexicon with synonyms and term variants curated from reports, creation of triple-consensus ground truth, and objective recording of comparable radiology read performances. Second, with the wide spectrum of findings labeled in a large training data set acquired from multiple hospital sources, confounding factors due to hidden stratification within anomalies was more fully covered than any other existing efforts. Third, we have shown that it is possible to build a single neural network to capture a wide variety of fine-grained findings and optimizing their prediction by selecting operating points based on the F1 score. Fourth, using a systematic comparative study, we have shown that it is possible to build AI algorithms that reach the typical level of performance of third-year radiology residents for a large set of findings. Thus this study established a practical benchmark for making AI algorithms clinically usable in future.
Overall, this study points to the potential use AI systems in future radiology workflows for preliminary interpretations that target the most prevalent findings, leaving the final reads performed by the attending physician to still catch any potential misses from the less-prevalent fine-grained findings. Having attending physicians quickly correct the automatically produced reads, we can expect to significantly expedite current dictation-driven radiology workflows, improve accuracy, and ultimately reduce the overall cost of care.
eFigure 1. Curation Process for Generating the List of Possible Findings in AP Chest Radiographs
eFigure 2. Vocabulary Expansion Process Used for the Chest Radiograph Lexicon Construction
eFigure 3. Splitting Algorithm for Producing the Partitions for Training, Validation, and Testing in the Modeling Data Set
eFigure 4. Prevalence Distribution of the Labels in the Comparison Study Data Set
eFigure 5. User Interface Used by Radiologists for Building Consensus After Independent Read Discrepancies Were Catalogued
eFigure 6. Web-Based User Interface Used for Collecting the Reads from Radiology Residents on the Comparative Study Data Set
eFigure 7. Extent of Agreement With the Ground Truth for AI Algorithm and Radiology Residents on Labels in the Comparison Study Data Set With at Least 2.5% Prevalence
eFigure 8. Preliminary Read Performance Differences of Radiology Residents and the AI Algorithm
eTable 1. Finding Label Extraction From Reports Through Text Analytics
eTable 2. Performance of AI Algorithm vs Radiology Residents Across Labels With at Least 2.5% Prevalence in the Comparison Study Data Set
eTable 3. Comparative Finding Label Recognition Performance Between Radiologists and AI Algorithm
eTable 4. Variation in Read Performance Across Radiology Residents
eAppendix 1. Splitting Algorithm for Model Training
eAppendix 2. Method of Threshold Selection for Finding Labels
eAppendix 3. Measuring Deep Learning Model Performances for Multilabel Reads
References
- 1.Zha N, Patlas MNDR, Duszak R Jr. Radiologist burnout is not just isolated to the United States: perspectives from Canada. J Am Coll Radiol. 2019;16(1):121-123. doi: 10.1016/j.jacr.2018.07.010 [DOI] [PubMed] [Google Scholar]
- 2.Kane L. Medscape national physician burnout, depression and suicide report 2019. Medscape. January 16, 2019. Accessed September 11, 2020. https://www.medscape.com/slideshow/2019-lifestyle-burnout-depression-6011056
- 3.Do HM, Spear LG, Nikpanah M, et al. Augmented radiologist workflow improves report value and saves time: a potential model for implementation of artificial intelligence. Acad Radiol. 2020;27(1):96-105. doi: 10.1016/j.acra.2019.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang X, Peng Y, Lu L, Lu Z, Bagheri M SR. ChestX-ray8: hospital-scale chest x-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases. Paper presented at: 2017 IEEE Conference on Computer Vision and Pattern Recognition; July 21-26, 2017; Honolulu, HI. Accessed September 11, 2020. doi: 10.1109/CVPR.2017.369 [DOI] [Google Scholar]
- 5.Taylor AG, Mielke C, Mongan J. Automated detection of moderate and large pneumothorax on frontal chest X-rays using deep convolutional neural networks: a retrospective study. PLoS Med. 2018;15(11):e1002697. doi: 10.1371/journal.pmed.1002697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rajpurkar P, Irvin J, Zhu Ket al. CheXNet: radiologist-level pneumonia detection on chest x-rays with deep learning. arXiv. Preprint posted online December 25, 2017. Accessed September 11, 2020. https://arxiv.org/abs/1711.05225 [Google Scholar]
- 7.Pan I, Cadrin-Chênevert A, Cheng PM. Tackling the Radiological Society of North America Pneumonia Detection Challenge. AJR Am J Roentgenol. 2019;213(3):568-574. doi: 10.2214/AJR.19.21512 [DOI] [PubMed] [Google Scholar]
- 8.Morris MA, Saboury B, Burkett B, Gao J, Siegel EL. Reinventing radiology: big data and the future of medical imaging. J Thorac Imaging. 2018;33(1):4-16. doi: 10.1097/RTI.0000000000000311 [DOI] [PubMed] [Google Scholar]
- 9.Irvin J, Rajpurkar P, Ko Met al. CheXpert: a large chest radiograph dataset with uncertainty labels and expert comparison. Paper presented at: AAAI-19 Thirty-Third AAAI Conference on Artificial Intelligence. January 27-February 1, 2019; Honolulu, HI. Accessed September 11, 2020. doi: 10.1609/aaai.v33i01.3301590 [DOI] [Google Scholar]
- 10.Majkowska A, Mittal S, Steiner DF, et al. Chest radiograph interpretation with deep learning models: assessment with radiologist-adjudicated reference standards and population-adjusted evaluation. Radiology. 2020;294(2):421-431. doi: 10.1148/radiol.2019191293 [DOI] [PubMed] [Google Scholar]
- 11.Annarumma M, Withey SJ, Bakewell RJ, Pesce E, Goh V, Montana G. Automated triaging of adult chest radiographs with deep artificial neural networks. Radiology. 2019;291(1):196-202. doi: 10.1148/radiol.2018180921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wong KCL, Moradi M, Wu J. Identifying disease-free chest x-ray images with deep transfer learning. Deep AI. April 2, 2019. Accessed September 11, 2020. https://deepai.org/publication/identifying-disease-free-chest-x-ray-images-with-deep-transfer-learning
- 13.Safran C, Bloomrosen M, Hammond WE, et al. ; Expert Panel . Toward a national framework for the secondary use of health data: an American Medical Informatics Association white paper. J Am Med Inform Assoc. 2007;14(1):1-9. doi: 10.1197/jamia.M2273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brownlee J. Classification accuracy is not enough: more performance measures you can use. Accessed June 26, 2020. https://machinelearningmastery.com/classification-accuracy-is-not-enough-more-performance-measures-you-can-use/
- 15.Hansell DM, Bankier AA, MacMahon H, McLoud TC, Müller NLRJ, Remy J. Fleischner Society: glossary of terms for thoracic imaging. Radiology. 2008;246(3):697-722. doi: 10.1148/radiol.2462070712 [DOI] [PubMed] [Google Scholar]
- 16.Johnson AEW, Pollard TJ, Shen L, et al. MIMIC-III, a freely accessible critical care database. Sci Data. 2016;3(1):160035. doi: 10.1038/sdata.2016.35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Coden A, Gruhl D, Lewis N, Tanenblatt M TJ. Spot the drug: an unsupervised pattern matching method to extract drug names from very large clinical corpora. Paper presented at IEEE Second International Conference on Healthcare Informatics, Imaging and Systems Biology; September 28-29, 2012; San Diego, CA. Accessed September 11, 2020. doi: 10.1109/HISB.2012.16 [DOI] [Google Scholar]
- 18.Mikolov T, Chen K, Corrado G, Dean J. Efficient estimation of word representations in vector space. Paper presented at: 1st International Conference on Learning Representations, ICLR 2013. May 2-4, 2013; Scottsdale, AZ. Accessed September 11, 2020. https://arxiv.org/abs/1301.3781 [Google Scholar]
- 19.Johnson AEW, Pollard TJ, Berkowitz SJ, et al. MIMIC-CXR, a de-identified publicly available database of chest radiographs with free-text reports. Sci Data. 2019;6(1):317. doi: 10.1038/s41597-019-0322-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin T-Y, Dollár P, Girshick R, He K, Hariharan B BS. Feature pyramid networks for object detection. Paper presented at: 2017 IEEE Conference on Computer Vision and Pattern Recognition. July 21-26, 2017; Honolulu, HI. Accessed September 11, 2020. https://arxiv.org/abs/1612.03144 [Google Scholar]
- 21.Simonyan K, Zisserman A. Very deep convolutional networks for large-scale image recognition. arXiv. Preprint posted online April 10, 2015. Accessed September 11, 2020. https://arxiv.org/abs/1409.1556 [Google Scholar]
- 22.He K, Zhang X, Ren S, Sun J. Deep residual learning for image recognition. Paper presented at: 2016 IEEE Conference on Computer Vision and Pattern Recognition. June 27-30, 2016; Las Vegas, NV. Accessed September 11, 2020. doi: 10.1109/CVPR.2016.90 [DOI] [Google Scholar]
- 23.Deng J, Dong W, Socher R, Li L-J, Li K, Fei-Fei L. Imagenet: a large-scale hierarchical image database. Paper presented at: 2009 IEEE Conference on Computer Vision and Pattern Recognition. June 20-25, 2009; Miami, FL. Accessed September 11, 2020. doi: 10.1109/CVPR.2009.5206848 [DOI] [Google Scholar]
- 24.Yu F, Koltun V. Multi-scale context aggregation by dilated convolutions. arXiv. Preprint posted online April 20, 2016. https://arxiv.org/abs/1511.07122 [Google Scholar]
- 25.He K, Zhang X, Ren S, Sun J. Identity mappings in deep residual networks. Poster presented at: 14th European Conference on Computer Vision. October 8-16, 2016; Amsterdam, the Netherlands. Accessed September 11, 2020. http://www.eccv2016.org/files/posters/S-3A-07.pdf [Google Scholar]
- 26.Wu Y, He K. Group normalization. Paper Presented at: 16th European Conference on Computer Vision. September 8-14, 2018; Munich, Germany. Accessed September 11, 2020. https://arxiv.org/abs/1803.08494 [Google Scholar]
- 27.Brownlee J. A gentle introduction to the rectified linear unit (ReLU). Accessed June 26, 2020. https://machinelearningmastery.com/rectified-linear-activation-function-for-deep-learning-neural-networks/
- 28.Lin T-Y, Maji S. Improved bilinear pooling with CNNs. arXiv Preprinted posted online July 21, 2017. Accessed September 11, 2020. https://arxiv.org/abs/1707.06772 [Google Scholar]
- 29.Kumar S. Data splitting technique to fit any machine learning model. Accessed September 11, 2020. https://towardsdatascience.com/data-splitting-technique-to-fit-any-machine-learning-model-c0d7f3f1c790
- 30.Parikh R, Mathai A, Parikh S, Chandra Sekhar G, Thomas R. Understanding and using sensitivity, specificity and predictive values. Indian J Ophthalmol. 2008;56(1):45-50. doi: 10.4103/0301-4738.37595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.De Vries H, Elliott MN, Kanouse DE, Teleki SS. Using pooled kappa to summarize interrater agreement across many items. Field Methods. 2008;20(3):272-282. doi: 10.1177/1525822X08317166 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
eFigure 1. Curation Process for Generating the List of Possible Findings in AP Chest Radiographs
eFigure 2. Vocabulary Expansion Process Used for the Chest Radiograph Lexicon Construction
eFigure 3. Splitting Algorithm for Producing the Partitions for Training, Validation, and Testing in the Modeling Data Set
eFigure 4. Prevalence Distribution of the Labels in the Comparison Study Data Set
eFigure 5. User Interface Used by Radiologists for Building Consensus After Independent Read Discrepancies Were Catalogued
eFigure 6. Web-Based User Interface Used for Collecting the Reads from Radiology Residents on the Comparative Study Data Set
eFigure 7. Extent of Agreement With the Ground Truth for AI Algorithm and Radiology Residents on Labels in the Comparison Study Data Set With at Least 2.5% Prevalence
eFigure 8. Preliminary Read Performance Differences of Radiology Residents and the AI Algorithm
eTable 1. Finding Label Extraction From Reports Through Text Analytics
eTable 2. Performance of AI Algorithm vs Radiology Residents Across Labels With at Least 2.5% Prevalence in the Comparison Study Data Set
eTable 3. Comparative Finding Label Recognition Performance Between Radiologists and AI Algorithm
eTable 4. Variation in Read Performance Across Radiology Residents
eAppendix 1. Splitting Algorithm for Model Training
eAppendix 2. Method of Threshold Selection for Finding Labels
eAppendix 3. Measuring Deep Learning Model Performances for Multilabel Reads