Abstract
Purpose
Artemisia nilagirica (AN), which is known to have antimicrobial, antioxidant, antiulcer, and anti-asthmatic properties, has been recently shown to have anti-cancer activity. However, the mechanism responsible for the anti-cancer property and its effect on cellular properties and functions are not known.
Material and methods
We have characterized the biochemical and biomechanical properties of MDA-MB-231 cells treated with the methanolic extract from AN.
Results
We show that AN-treatment decreases cell-eccentricity, increases expression of actin and microtubules, and do not affect cell-area. Increased expression of cytoskeletal proteins is known to change the mechanical properties of the cells, which was confirmed using micropipette aspiration and Atomic Force Microscopy. We identified the upregulation of the tumorigenic pathway (TGF-β) leading to activation of Rho-A as the molecular mechanism responsible for actin upregulation. Since the initial stages of TGF-β upregulation are known to suppress tumor growth by activating apoptosis, we hypothesized that the mechanism of cell death due to AN-treatment is through TGF-β activation. We have validated this hypothesis by partially recuing cell death through inhibition of TGF-β using Alk-5.
Conclusion
In summary, our study reveals the mechanism of action of Artemisia nilagirica using a synergy between biochemical and biomechanical techniques.
Keywords: Artemisia nilagirica, MDA-MB-231 breast cancer cells, Anti-tumor effects, TGF-β, Cell stiffness, Engineering, Chemistry, Agricultural science, Biological sciences, Health sciences, Pharmaceutical science, Pharmaceutical chemistry, Natural product chemistry, Cell biology
Artemisia nilagirica; MDA-MB-231 breast cancer cells; Anti-tumor effects; TGF-β; Cell stiffness; Engineering; Chemistry; Agricultural science; Biological sciences; Health sciences; Pharmaceutical science; Pharmaceutical chemistry; Natural product chemistry; Cell biology
1. Introduction
Breast cancer is the second leading cause of cancer deaths in women worldwide [1]. Approximately 81% breast cancers are invasive although considered to be a single disease, it consists of four molecular subtypes and 12 distinct histological subtypes, where each type vary in terms of risk factors, treatments and outcomes [2]. Currently, a combination of various techniques such as radiation therapy, chemotherapy, immunotherapy, and surgery are used. Although proven effective, most of these strategies and combinations adversely affect the patient's quality of life through severe side effects [3]. In comparison, plant-extracts are known to have lower side effects and hence can be a better option against cancer. Previous studies have shown anti-cancer properties for curcumin from turmeric, gingiberin from ginger, piperine from black pepper, apigenin from parsley, carvacrol from oregano, crocetin from saffron, and resveratrol from grapes, moringa, yarrow, garlic, and Berberis libanotica Ehrenb (BLE) [4, 5, 6, 7]. These plant extracts induce changes in cell cycle, expression of oncogenes, metastasis, apoptosis, etc [8]. Plant-derived products are globally gaining interest for various treatments as they are evolving as effective drugs with reduced side effects even at high dosages [9]. Approximately 40–50% of cancer patients intake plant parts and its derivatives in their diet due to their anti-cancer properties [10].
Artemisia Nilagirica (Clarke) is a plant known for its antiseptic, anthelmintic, antifungal, antimicrobial, antibacterial, insecticidal, antioxidant, antiulcer, and anti-asthmatic activity. This extract has been previously characterized chemically and has been used to treat asthma, cough, and leprosy [11, 12]. Its methanolic-extract has also been shown to have anti-cancer properties [13, 14]. These studies hypothesized that the anti-cancer properties may be due to the presence of flavonoids and sesquiterpene lactones, which have free radical quenching properties. However, the mode of action of this extract and its effect on the physical properties of the cell are unexplored.
We have first performed biochemical and biomechanical characterization of breast cancer cells treated with AN with a motivation to identify the mechanism responsible for its anti-cancer properties. Since metastasis is the leading cause of mortality in breast cancers, we have chosen MDA-MB-231 cells, as our model system for breast cancer. Another extensively characterized plant extract with anti-cancer properties, Piper nigrum Linn. (PN) which is an alkaloid extracted from black pepper, was used as positive control. PN is known to arrest the cell cycle at G1 phase and cause cell death due to apoptosis [15, 16]. Untreated MDA-MB-231 cells were used as control. We first characterized the physical properties of cells such as morphology, migration and stiffness because of the close relationship between cytoskeleton and cancer. Atomic Force Microscopy and Micropipette Aspiration were used to characterize cell stiffness. To characterize the changes in biochemical properties, we have quantified the DNA content, analyzed changes in cell-cycle and the differential regulation of genes related to the cytoskeleton, tumor growth, cell adhesion and Epithelial to Mesenchymal Transition (EMT).
Our results show that the activation of the TGF-β pathway by AN-treatment contributes to cell-death and thereby to its anti-cancer property. Another interesting observation is that the changes in biomechanical properties due to the treatment manifest before we could detect changes in the biochemical markers.
2. Materials and methods
2.1. Cell culture
MDA-MB-231 cells were cultured in Dulbecco's modified Eagle's medium-high glucose (DMEM, Sigma–Aldrich) supplemented with 10% heat-inactivated fetal bovine serum (FBS) (Sigma-Aldrich, North American origin) and antibiotics penicillin streptomycin powder (Hi Media) at a concentration of 0.4 μg/mL. The cells were cultured at 37 °C, 95% relative humidity and 5% CO2. Cells from an 80% confluent dish were taken for experiments.
2.2. Plant extract and determination of IC50 values
Twenty-five gram of Piper nigrum (PN) matured (green inflorescence) and Artemisia nilagirica (AN) leaves were ground in ice cold HPLC grade methanol using pestle and mortar. The ground plant-material was transferred to a 250 mL standard measuring flask. The volume was made up with ice-cold methanol and extracted overnight. We further filtered the extract using Whatman No. 1 filter paper. The solvent from the extract was removed by air-drying. The residues were collected and used for the experiment.
MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide] assay [17] was performed to calculate the IC50 value of PN and AN extract for MDA-MB-231 cells. Cells were trypsinized from an 80% confluent flask. Approximately 5,000 cells/well were seeded in 96-well plates and was kept for 24 h of incubation. After incubation the cells were treated with various concentrations of AN and PN for 24 h. The medium with extract was aspirated out from the wells and MTT reagent was added to each well and incubate for 4 h. MTT reagent was aspirated out and DMSO (Dimethyl sulfoxide) was added to dissolve the formazan (purple crystals) formed in live cells. The absorbance was measured using a Micro-Plate reader at an excitation wavelength of 570 nm. Average of three individual biological experiments were analyzed. The IC50 value for PN and AN, for MDA-MB-231 cells was expressed as percentage of cell inhibition. It was calculated using equation given below. Further experiments were carried out with these concentrations.
| (1) |
The mean IC50 value, calculated from three independent MTT assays, was used for further experiments. We obtained IC50 values of PN and AN as 6.6 μg/mL and 13.5 μg/mL respectively.
2.3. Immunostaining and fluorescence microscopy
Cells were fixed with 4% paraformaldehyde for 10 min at room temperature followed by permeabilization with 0.5% TritonX-100 for 10 min at 37 °C. Microtubules were stained with Alpa-tubulin antibody FITC conjugate (Invitrogen) for 20 min at 37 °C, actin filaments were stained with Rhodamin-Phalloidin (Molecular probes) for 20 min at 37 °C, and Nucleus were stained with DAPI (4,6-diamidino-2- phenylindole, dihydrochloride) (Molecular probes) for 10 min at 37 °C. Images were acquired with either a Leica AF6000 inverted wide-field fluorescence microscope or with a TCS SP5 II confocal microscope. The confocal images were analyzed using ImageJ software to determine the cell area, cell roundness. The total wide-field fluorescence intensity per cell was used to estimate the difference in the amount of actin and microtubules. In brief confocal images are opened in Image J software and converted to 8bit image. Scale was set by calculating the pixel to micron ratio for each image. Each cell boundary is marked manually and correspondingly the cell area, cell roundness was calculated. Average of 30–60 cells were taken to determine to cell area, cell roundness and intensity measurements. The results of each experiment are expressed as the mean values ± standard deviation of the mean (SD). The significance of the data was analyzed using a Student's t-test, and differences between two means with p < 0.05 (∗).
2.4. Cell cluster expansion rate for treated and non-treated cells
The rate of expansion for the cell clusters was determined after treatment with AN and PN. The expansion was compared with untreated cells to confirm the effect of these extracts. Cells from an 80% confluent culture dish were trypsinized and approximately 100,000 cells were taken and placed as a drop at the center of three separate Perti-plates. After one hour of incubation, cells adhered and appeared like a cluster of cells. Culture medium was then added to the Petri-plate. After 24 h of incubation, bright - field images of these cell clusters were taken; this was considered time, t = 0. AN and PN extracts where added in two separate Perti-plate with MDA-MB-231 cell clusters and the third one is untreated where no extract was added. These Petri-plate were incubated for 24 h; after incubation brigh t -field images were taken. This was considered time, t = 24 h. The cell cluster expansion rate was calculated using the increase in area of the cell cluster on petriplate from t = 0 to 2 4 h. The cell area was obtained using a custom-built code written in MATLAB (www.mathworks.com). The results of three biological repeats were analyzed using a student's t-test, and a p < 0.05 was considered to be significant.
2.5. Flow cytometry for cell-cycle and apoptosis analysis
Approximately 50,000 cells were seeded in a 35mm Petri dish. After 24 h of incubation, cells were treated with PN and AN extract. Cells were incubated for two time points 24 h and 48 h in case of cell cycle study and for apoptosis one time point 48 h. After the incubation, the cells were harvested from the Petri-dish using cell scraper. Cells were transferred to an eppendorf tube and centrifuged at 1500 rpm for 3 min. The supernatant was discarded, and the pellet resuspended in 1 mL of complete medium. The numbers of cells were counted using a Haemocytometer. After cell counting, the cells were again centrifuged, and the supernatant was carefully removed in order to obtain the cell pellet. For cell cycle analysis, to the cell pellet, 100 μL of hypotonic solution containing propidium iodide, at a concentration of 4 μl/mL, was added for every 1,00,000 cells. By using this cell suspension, cell-cycle analysis was done using Flow Cytometry (Accuri C6) as described in [18].
For cell apoptosis, to the cell pellet 100 μL of PBS containing propidium iodide, at a concentration of 5 μL/ml was added and incubated for 3 min at room temperature, followed by centrifugation. The supernatant was discarded, and cells were resuspended on 100 μL of annexin-binding buffer with 2.5 μL of annexin 5 conjugate, incubate for 10 min at room temperature, followed by centrifugation. The supernatant was discarded, and the cells were suspended in 100 μL PBS. This cell suspension was used for apoptosis analysis using Flow Cytometry (Accuri C6). Data was analyzed using FlowJo 10 software. Cell debris and clumps were excluded from the analysis in all samples. The results of three biological repeats were analyzed using a student's t-test, and a p < 0.05 was considered to be significant.
2.6. DNA quantification
The proliferation of cells in untreated and treated conditions was evaluated at day 1, 3 and 5. The cellular DNA content was measured using the Picogreen ds-DNA quantification kit (Invitrogen), as described in [19]. Briefly, cells were lysed using lysis buffer (0.02% SDS with Proteinase K 0.2 mg/ml). The lysate was mixed with the Picogreen dye and the DNA content was determined by measuring the fluorescence intensity in a well-plate reader at 485 nm excitation and 528 nm emission. The results of three biological repeats were analyzed.
2.7. RNA isolation and quantitative real time-polymerase chain reaction (qRT-PCR)
Cells were cultured on a 60 mm Petri-plate for up to 80% confluence. Extracts were added in two separate Petri-plate PN and AN respectively, after incubation period of 24 h and 48 h, two time points for each, RNA was isolated using the RNase Mini Kit (Quigen) as per the manufacturer's instruction. A total of 0.5–2 μg of RNA was used for cDNA synthesis using a high-capacity cDNA reverse transcription kit (Applied Biosystems) as per the manufacturer's instruction.
Quantitative real-time PCR (qRT-PCR) was carried out using a Power up SYBR Green master mix (Thermo Scientific) with 10 ng of the cDNA as the template. Gene expression was normalized to glyceraldehyde 3-phosphate dehydrogenase (GAPDH). Fold change was calculated using 2-ΔΔct. EMT markers Epithelial-cadherin (E-cad), Neural-cadherin (N- cad), Vimentin, Snail, Slug, Twist, Paxillin, along with Matrix metalloproteinase-2 (MMP2), Ras homology gene-member A(RhoA), Transforming growth factor Beta 1 (TGF-β), Activin Receptor-Like Kinase (ALK-5) primers were used for gene expression study. Gene expression across three individual biological experiments was analyzed. A paired t-test was performed, and p < 0.05 was taken as statistically significant.
2.8. Mechanical characterization
We have performed two mechanical characterization techniques viz., micropipette aspiration [20] and AFM [21]. Experiments were completed within 40 min after trypsinization of cells. To confirm that cells are alive during the mechanical testing, Trypan blue test (stains the dead cells blue) was used.
2.9. Micropipette aspiration
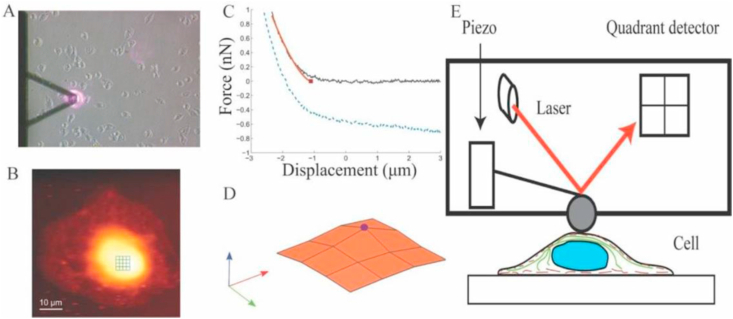
In this technique, cells are aspirated into a micropipette by exerting a negative pressure. Borosilicate glass pipette (OD 1 mm, ID 0.5 mm, and length 10 cm) was pulled to micrometer dimensions using a micropipette puller (Flaming/brown micropipette puller, Sutter Instrument Co, model P-97). The tip of the micropipette was then cut to the desired diameter (d) using a microforge (MF-900, NARISHIGE). The pipette was then fixed to a pipette holder mounted on a XYZ positioner (Figure 1). shows the setup used for aspiration, which includes an inverted microscope (MP-285), micro injector, XYZ positioners, controller, controller box, CCD camera, micropipette holder and a computer. Trypsinized cells were suspended in cell-culture medium and transferred to a glass coverslip for aspiration. To identify live cells during the experiments, Trypan blue (stains the dead cells blue) was added to the cell suspension. The micropipette was brought adjacent to a cell using the XYZ positioner. Following this, the pressure is gradually reduced using a controller until the cell sits firmly at the tip of the micropipette. The pipette was raised to ensure that the cell is not dislodged at this initial pressure. Subsequently, the pressure was decreased in unit steps until the cell enters the pipette. This reading is taken as the final pressure [20]. The difference between the initial and final pressures was used to calculate the force required to aspirate the cell as shown in Eq. (2).
| (2) |
Figure 1.
Experimental set-up for Micropipette Aspiration with Controller Box, CCD camera, Micropipette holder, Micro-injector, XYZ positioner, and Controller.
Experiments with each sample were completed within 40 min of trypsinization. The results of experiment are expressed as the mean values ± standard deviation of the mean (SD). The significance of test of the data was analyzed using a Student's t-test, and differences between two means with p < 0.05 (∗).
2.10. Atomic Force Microscopy
Cell elasticity measurements were performed using Park Systems XE-Bio Atomic Force Microscope (AFM). App Nano Hydra 6V-200NG-TL cantilever with a 5.2 μm Silicon di-oxide spherical bead was used for cell indentations. The stiffness of the cantilever was measured using a thermal tuning method and was found to be 0.041 N/m. The A-B sensitivity, which represents the relation between the difference in the voltage on the photodiode and the cantilever deformation, was calibrated every time the laser position was adjusted. The topography of the cell was first obtained by contact imaging using a set point of ~ 0.7 nN. Multiple force–displacement curves, on a 4 × 4 grid, were obtained on a small area of 5 μm × 5 μm above the nucleus as shown in (Figure 2). The modulus of elasticity and the point of contact of the cantilever with the cell were obtained for each of these curves using Hertzian contact model. The highest contact point was designated as the apex of the nucleus; the modulus corresponding to this curve was used as the modulus of the elasticity of the cell. The results of experiment are expressed as the mean values ± standard deviation of the mean (SD). The significance of the difference in means was analyzed using a Student's t-test and p-value < 0.05, represented by ∗, was considered significant.
Figure 2.
Measuring cell stiffness using Atomic Force Microscope (A) Bright-field image of AFM scanning on a cell, (B) its corresponding topography, (C) Typical force-displacement curve during indentation, (D) Surface profile obtained from scan profile used to determine highest point of the cell, (E) Schematic representation of AFM experiment.
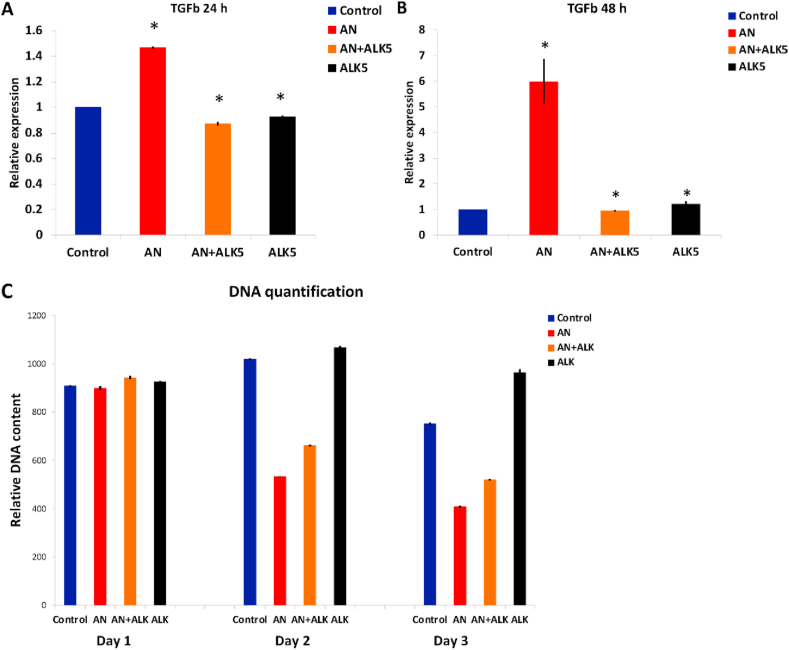
For inhibiting over-expression of TGF-β AN-treated cells were incubated with 10 μM Activin Receptor-Like Kinase (ALK-5, TGF-βR1 inhibitor, SB 431542, Sigma Aldrich U.S.A) [22] for 24 h and 48 h. Inhibition of TGF-β upon treating with ALK5 was confirmed using qRT-PCR. Total DNA content of AN-treated cells incubated with ALK5 was measured to ascertain if TGF-β upregulation was the mechanism of cell-death by AN. The results of experiment (performed in triplicates) are expressed as the mean values ± standard deviation of the mean (SD). The significance of the difference in means was analyzed using a Student's t-test, and a p-value < 0.05 (∗) was considered significant.
2.11. Statistical analysis
The results of each experiment (performed in triplicates) are expressed as the mean values ± standard deviation of the mean (SD). Control and the treated cells were compared using ANOVA with Bonifferoni correction in case of cell area expansion. The significance of the difference in means was analyzed using a Student's t-test, and a p-value < 0.05 (∗) was considered significant.
3. Results
3.1. Cytoskeleton and cell morphology
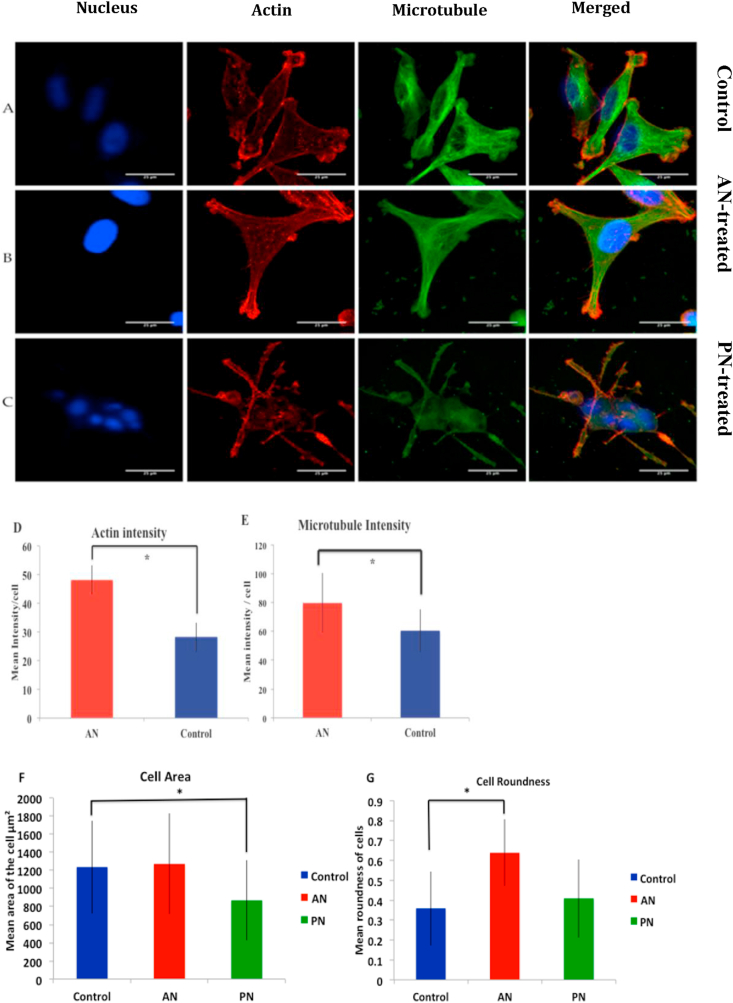
Cells treated with PN appeared stressed whereas those treated with AN spread well (Figure 3 A–C). Expression of actin and microtubule in AN-treated cells were significantly higher than in control cells (Figure 3 D and E). Since PN-treated cells were peculiar, which was confirmed using DNA quantification, we did not quantify the expression of actin and microtubule in those cells. The area of PN-treated cells (869 ± 442 μm2), was significantly lower than that of control cells (1236 ± 514μm2). AN-treatment (1271 ± 555 μm2) did not lead to any significant change in cell area (Figure 3 F). In contrast, AN-treatment affects the roundness of cells whereas PN-treatment does not. The roundness of AN-treated cells (0.64 ± 0.167) was significantly higher than that of control cells (0.36 ± 0.185) and PN treated cells were (0.41 ± 0.195) (Figure 3 G).
Figure 3.
Perturbation in the cytoskeleton and cellular morphology due to AN-treatment. Confocal images of MDA-MB-231 cells stained for (from left to right) nucleus, actin, microtubule. Length of the scale bar is 25 μm. From top to bottom (A) Control, (B) AN-treated cells and (C) PN-treated cells. Quantification of (D)Actin intensity, (E) Microtubule intensity, (F) Cell area and (G) Cell roundness. Statistical analysis was performed using ANOVA with ∗ representing p < 0.05.
3.2. Migration
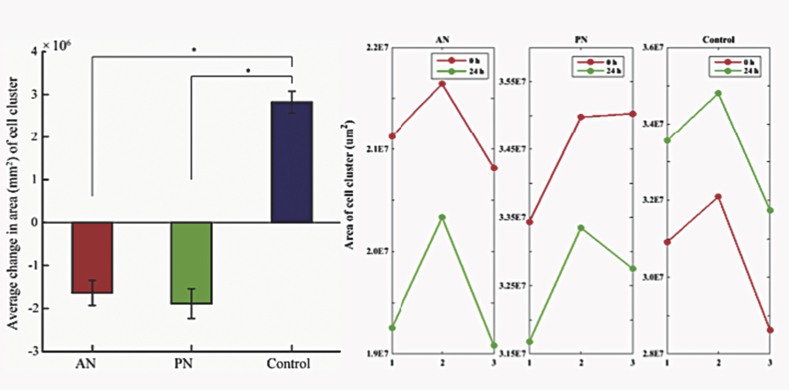
To establish the functional effect of cytoskeletal perturbations, we studied migration of these cells. For quantifying migration, we estimated the changes in size of cell cluster with time. We observed that cells treated with both these extracts showed significantly lower migration than control cells. Interestingly, both these treatments reduced the size of the cell cluster suggesting contraction of cells or cell death. Since AN-treatment did not decrease cell size (Figure 4), we suspected that the decrease in migration would be due to cytoskeletal perturbations or cell death.
Figure 4.
Cell cluster area of control cells and treated cells, its significance percentage change in cell cluster area. Statistical analysis was performed using ANOVA with ∗ representing p < 0.05.
3.3. Cell proliferation, cell cycle and apoptosis
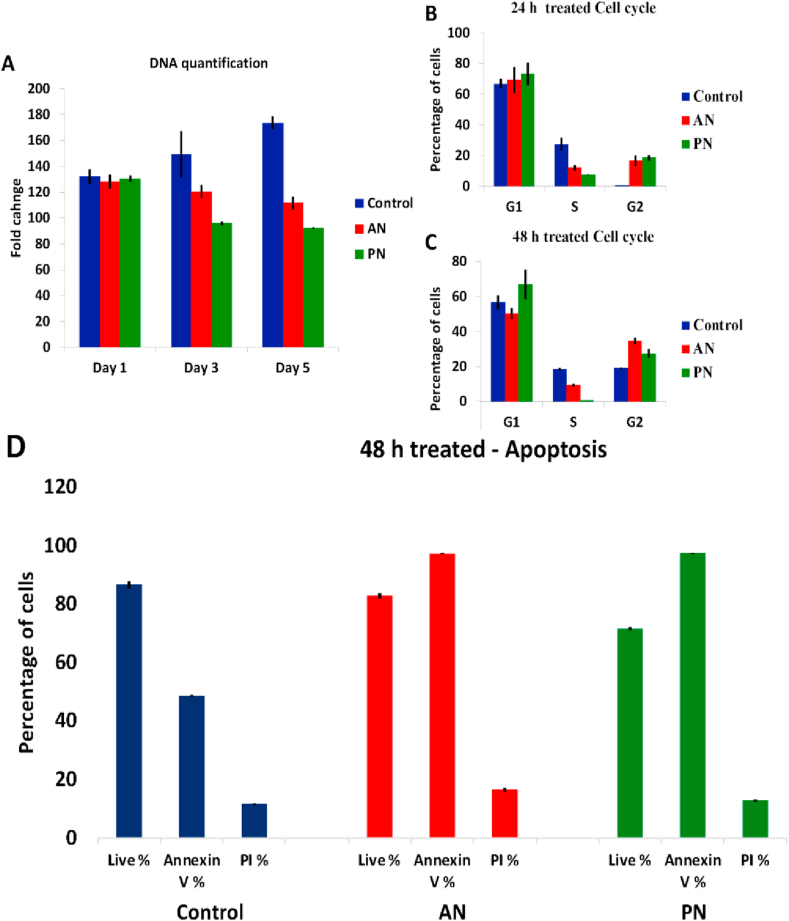
Both AN and PN treatments lead to decrease in DNA content (Figure 5 A). In comparison to AN- treatment, PN-treatment results in lower DNA content suggesting enhanced cell death. Moreover, cells treated with PN showed faster decrease in DNA content in comparison to AN-treated cells, which showed a gradual decrease. We further studied the effect of these plant extracts on cell-cycle. We observed that PN-treatment led to a larger number of cells in the G1 phase (67%) in comparison with control (50.2%). This agrees with previous studies that showed cell-cycle arrest at G1 phase by PN [16]. Cells treated with AN did not show any difference in cell cycle when compared to control (Figure 5 B, C). Previous studies have established that the anti-cancer property of PN is due to activation of apoptotic pathway upon G1 phase arrest. Both AN and PN treated cells were checked for apoptosis. Our results suggest that the loss of cell viability by AN-treatment is not due to such a cell-cycle dependent mechanism, but the cells showed increased percentage of death due to apoptosis (Figure 5 D).
Figure 5.
DNA quantification (A), cell cycle analysis after drug-treatment for 24 h (B) and 48 h (C) and Apoptosis after drug-treatment for 48 h (D).
3.4. Quantitative real time- polymerase chain reaction (qRT-PCR)
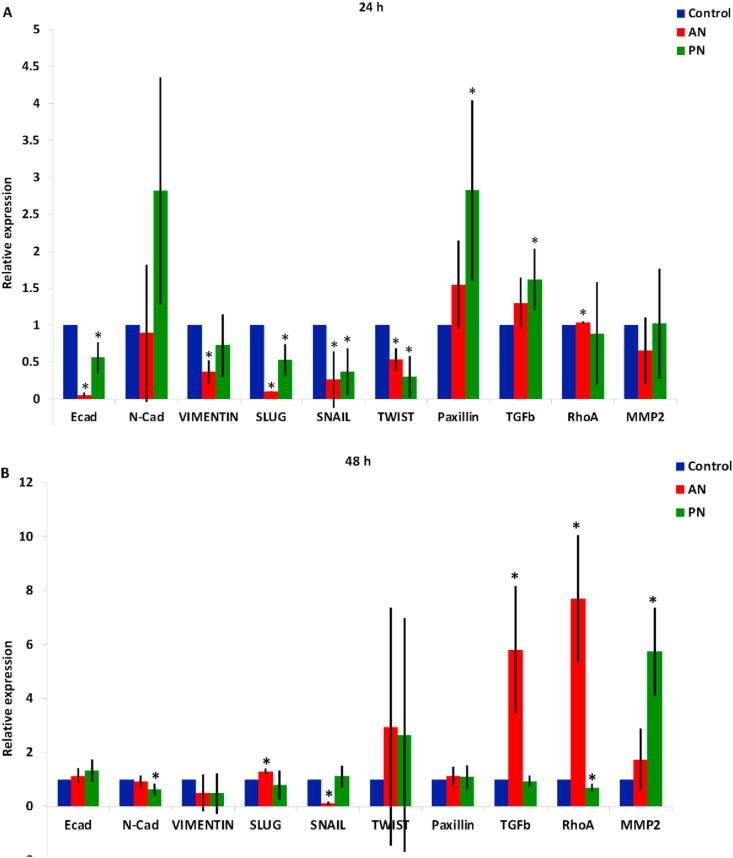
To study the anti-cancer properties of these compounds, we studied their effect on Epithelial to Mesenchymal Transition (EMT). For this, we estimated the expression of several well-characterized EMT markers such as E-cadherin, N-cadherin and Vimentin, and associated transcription factors such as Snail, Slug and Twist(). We also looked at other markers related to cell adhesion (Paxillin), tumor progression (TGF-β), cytoskeleton (RhoA) and migration (MMP2).
AN - At 24 h, we observed that there was a reduction in the expression of E-cadherin suggesting a mesenchymal transition (Figure 6). However, the down-regulation of Vimentin, Slug, Snail and Twist indicates transition to an epithelial phenotype. Even at 48 h, we do not see any conclusive pattern suggesting a switch to the epithelial or mesenchymal cell phenotype [23, 24, 25, 26]. There was no change in the expression of MMP2 ruling out any change in migration potential due to AN-treatment. For both 24 and 48 h, there was no significant change in the expression of Paxillin negating any changes in cell adhesion. Interestingly, we observed a consistent increase in the expression of TGF-β at 24 and 48 h. TGF-β is known to suppress tumor in the early stages of tumorigenesis by activating apoptosis [27, 28]. Hence, cell-death due to this compound, observed by DNA quantification (Figure 5 A), could be due to the activation of the TGF-β pathway leading to apoptosis. We confirmed this using inhibitor experiments. By treating cells with ALK5, which inhibits TGF-βR1, we were able to partially rescue cell-death (Figure 7). We further observed an increase in the expression of RhoA indicating enhanced contractility in the actin cytoskeleton [29]. This is consistent with the increased expression of TGF−β since TGF-β is known to upregulate RhoA [27].
Figure 6.
Change in the relative expression of genes due to AN and PN-treatment for (A) 24 h and (B) 48 h measure using q RT_PCR. Statistical analysis was performed using ANOVA with ∗ representing p < 0.05.
Figure 7.
Rescue of cell-death upon inhibiting TGF- β. Inhibition of TGF- β, measured using qRT_PCR after (A) 24 h and (B) 48 h of treatment with ALK5 and (C) Change in total DNA content with time. Day 2 and 3 represents 24 H and 48 H of treatment. Statistical analysis was performed using ANOVA with ∗representing p < 0.05.
PN – PN-treated cells show downregulation of EMT markers and upregulation of adhesion marker (Paxilin) and tumor progression marker (TGF-β) at 24 h. However, by 48 h we do not see any significant change in any of these markers except for MMP2. Since our DNA quantification study showed that by 48 h PN-treated cells are in the death phase, we did not further probe these observations.
3.5. Mechanical characterization
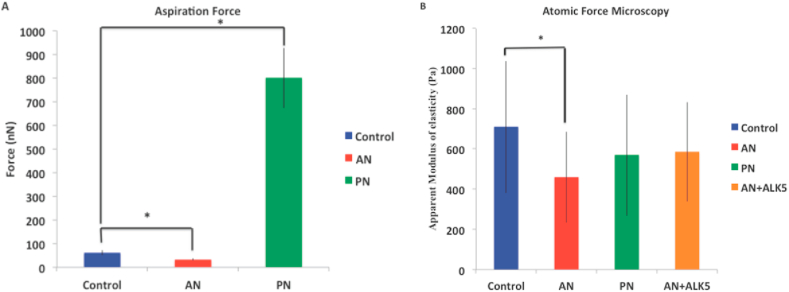
We have used two popular techniques, micropipette aspiration and AFM, for mechanical characterization of cells treated with these compounds (Figure 8). In micropipette aspiration, we observed that both AN and PN have significantly altered the mechanical properties of the cells. While AN-treatment has reduced the force required for aspirating cells into the micropipette, PN-treatment has increased the aspiration force in comparison to control (Figure 8A). The increase in aspiration force could be because PN-treatment arrests cells in G1 phase and initiates cell death (Figure 5B, C). Similarly, AFM measurements show significant reduction in cell stiffness due to AN-treatment. The mean stiffness of cells treated with PN is also lower than that of control. By inhibiting TGF-β using ALK5, the mean cell stiffness was partially rescued. Mean and standard deviation of the mechanical parameters estimated using aspiration and AFM are listed in Table 1.
Figure 8.
Biomechanical characterization using Micropipette aspiration (A) and Atomic Force Microscopy (B). PN-treated cells are arrested in G1 phase and initiates the cell death hence in case of aspiration the force required to aspirate the cell is larger in comparison to control and AN-treated cells. Statistical analysis was performed using ANOVA with ∗ representing p < 0.05.
Table 1.
Comparison of mechanical properties measured using micropipette aspiration and AFM.
| Micropipette aspiration |
Atomic Force Microscopy |
|
|---|---|---|
| Aspiration force (nN) | Apparent modulus of elasticity (Pa) | |
| Control | 61.3 ± 11.9 | 709 ± 334 |
| AN-treated | 32.4 ± 5.6 | 459 ± 233 |
| PN-treated | 801 ± 127.6 | 569 ± 337 |
4. Discussion
We have performed both biochemical and biomechanical characterization of breast cancer cells (MDAMB-231) treated with AN for identifying the mechanism responsible for its anti-cancer properties. Since metastasis is the leading cause of mortality in breast cancers, we have chosen MDAMB-231 cells, as our model system for breast cancer. Another extensively characterized plant extract with anti-cancer properties, Piper nigrum Linn. (PN) which is an alkaloid extracted from black pepper, was used as positive control. PN is known to arrest the cell cycle at G1 phase and cause cell death through apoptosis [15, 16].
Cell death due to AN was inferred from the reduction in DNA content. Cell death by the reduction in DNA and RNA content through the induction of apoptotic pathways have been previously reported for many other plants extracts [30, 31, 32, 33, 34]. Many of these cytotoxic agents attach to DNA through covalent or non-polar binding [35]. They inhibit cell survival in cancer cells by arresting cell cycle and inducing apoptosis [36].
Cells treated with PN had reduced cell area in comparison to control whereas there was no significant change in the case of AN-treatment [36]. Interestingly, AN-treatment enhanced the roundness of cells while there was no significant change due to PN-treatment. By using immunofluorescence assay, we observed that cells treated with PN showed granular nuclei, irregular cell boundaries and abnormal cytoskeleton probably due to cell death [32, 33, 36, 37]. In contrast, AN-treated cells showed well-spread cellular and nuclear morphology, and enhanced expression of actin and microtubule [33]. The cells are much more stretched and spindle-shaped, resulting in a smaller cell area. The formation of the actin fibers as well as the distribution of cortical actin did not change, substantially. This increase in expression of cytoskeletal proteins did not lead to increased migration potential when treated with AN-extract. Migration of PN- treated cells was also lower than that of control cells. AN-treated cancer cells successfully repressed the migration of cells. Since the actin cytoskeleton is stabilized, cells cannot move and divide, thereby arresting the cell cycle to G2/M [33, 34, 38].
One of the hallmarks of cancer is the transformation of cells from epithelial to mesenchymal (EMT). This is a reversible process since cells can transform from mesenchymal to epithelial (MET). Switching between these phenotypes and thereby between occurrence and recurrence is a fundamental characteristic of cancer [39]. EMT is majorly influenced by change in several genes expression, in which TGFβ plays a major role by inducing metabolic reprogramming. Our gene-expression studies showed that TGF-β and its downstream target RhoA are both up-regulated when treated with AN extract [40, 41, 42]. TGF-β signaling is known to reduce cell migration by upregulating expression of Snail and Slug [43]. A physical manifestation of TGF-β and RhoA upregulation was increased actin and microtubule expression, which in turn enhanced the mechanical properties. These mechanical changes were characterized using AFM and micropipette aspiration. The apparent modulus of elasticity measured using AFM increased whereas the aspiration force decreased for AN-treated cells in comparison to the control. In comparison, the aspiration force for PN-treated cells was almost 10 times higher than control probably because the cells are dying. From previous studies, we know that PN-treatment leads to cell cycle arrest thereby leading to apoptosis. However, the mechanism responsible for loss of cell viability due to AN-treatment was unknown.
Our results suggest that one of the mechanisms responsible for cell-death in AN-treated cells is TGF-β upregulation. TGF-β is a multifunctional cytokine that regulates EMT and several other cellular processes such as cell cycle, differentiation, morphogenesis, and apoptosis [44]. Interestingly, TGF-β is known to suppress the tumor in the early stages of tumorigenesis [45]. Hence, upregulation of TGF-β could be the mechanism of cell-death due to AN-treatment [46, 47]. This was further confirmed by incubating AN-treated cells with ALK5, which inhibits TGF-β. AN-treated cells treated with ALK5 showed higher cell viability compared to AN-treated cells [40, 41, 42].
We have also observed the known downstream effects of TGF-β pathway upregulation such as enhanced expression of RhoA [48] and actin [49], and reduction in cell stiffness [50]. We observed a significant difference in the mechanical properties by 24 h of AN-treatment (Figure 8) even though biochemical signals such as the expression of TGF-β and RhoA were significantly different only by 48 h (Figure 6). A similar observation was made in a previous study, wherein the authors showed that cancerous cells obtained from pleural effusion displayed altered mechanical properties before changes in cell morphology were discernible [51]. These observations are in line with a growing body of evidence, which show that mechanical properties can be used as biomarkers for early detection of cancer. In summary, we show that the molecular mechanism of cell death due to AN extract is by upregulation of TGF-β expression. The anti-cancer property of this extract makes it an attractive option for cancer therapy.
5. Conclusion
We have characterized the changes in the biochemical and biomechanical properties of MDAMB-231 cell upon treatment with the methanolic-extract from Artemesia nilagirica. Previous studies have showed the anti-cancer property of AN extract, but the exact mode of action was unexplored. Our results show that the reduction in cell viability is due to activation of TGF-β, which is known to activate apoptotic pathway in the early stages of tumor-growth. Some of the known consequences of TGF-β upregulation such as upregulation of RhoA pathway and enhanced expression of actin were also observed. The changes in actin led to alterations in the mechanical properties of cells. Interestingly, the changes in mechanical properties were detectable earlier than the changes in TGF-β and its downstream targets. Our study demonstrates the mode of action of AN on MDA-MB-231 breast cancer cells to prove its anti-cancer property and change in the mechanical property of the cells can be used as biomarkers for early detection of cancer.
Declarations
Author contribution statement
S.R. Raju, S. Balakrishnan and S. Kollimada: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
K.N. Chandrashekara: Conceived and designed the experiments; Contributed reagents, materials, analysis tools or data; Wrote the paper.
A. Jampani: Conceived and designed the experiments; Wrote the paper.
Funding statement
This work was supported by the Indian Council of Medical Research, New Delhi, India.
Competing interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Acknowledgements
The authors would like to thank Ms. Monisha Mohandas from BioSystems Science and Engineering, Indian Institute of Science for help with the AFM study.
References
- 1.Momenimovahed Zohre, Hamid Salehiniya. Epidemiological characteristics of and risk factors for breast cancer in the world. Breast Cancer. 2019;11:151–164. doi: 10.2147/BCTT.S176070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.American Cancer Society . American Cancer Society, Inc; Atlanta: 2019. Breast Cancer Facts & Figures 2019–2020. [Google Scholar]
- 3.Qiu K., He C., Feng W., Wang W., Zhou X., Yin Z., Chen L., Wang H., Mo X. Doxorubicin- loaded electrospun poly(l-lactic acid)/mesoporous silica nanoparticles composite nanofibers for potential postsurgical cancer treatment. J. Mater. Chem. B. 2013;1:4601–4611. doi: 10.1039/c3tb20636j. [DOI] [PubMed] [Google Scholar]
- 4.Abdullaev F.I., Espinosa-Aguirre J.J. Biomedical properties of saffron and its potential use in canccancer er therapy and chemoprevention trials. Cancer Detect. Prev. 2004 doi: 10.1016/j.cdp.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 5.Habib S.H.M., Makpol S., Hamid N.A.A., Das S., Ngah W.Z.W., Yusof Y.A.M. Ginger extract (Zingiber officinale) has anti-cancer and anti-inflammatory effects on ethionine-induced hepatoma rats. Clinics. 2008;63:2008. doi: 10.1590/S1807-59322008000600017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kuttan Ramadasan, Bhanumathy P., Nirmala K., George M.C. Potential anticancer activity of turmeric (Curcuma longa) Cancer Lett. 1985;29:197–202. doi: 10.1016/0304-3835(85)90159-4. [DOI] [PubMed] [Google Scholar]
- 7.Tinhofer I., Bernhard D., Senfter M., Anether G., Loeffler M., Kroemer G., Kofler R., Csordas A., Greil R. Resveratrol, a tumor-suppressive compound from grapes, induces apoptosis via a novel mitochondrial pathway controlled by Bcl-2. Faseb. J. 2001;15:1613–1615. doi: 10.1096/fj.00-0675fje. [DOI] [PubMed] [Google Scholar]
- 8.Karade Preeti Gajendra, Jadhav Namdeo Ramhari. In vitro studies of the anticancer action of Tectaria cicutaria in human cancer cell lines: G0/G1 p53-associated cell cycle arrest-Part I. J. Tradit. Compl. Med. 2018;8:459–464. doi: 10.1016/j.jtcme.2017.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang H., Oo Khor T., Shu L., Su Z.-Y., Fuentes F., Lee J.-H., Tony Kong A.-N. Anti-Cancer Agents in Medicinal Chemistry. 2012. Plants vs. Cancer: a review on natural phytochemicals in preventing and treating cancers and their druggability; pp. 1281–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Donaldson M.S. Nutrition and cancer: a review of the evidence for an anti-cancer diet. Nutrition. 2004;3:19. doi: 10.1186/1475-2891-3-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ahameethunisa A.R., Hopper W. Antibacterial activity of Artemisia nilagirica leaf extracts against clinical and phytopathogenic bacteria. BMC Compl. Alternative Med. 2010;10 doi: 10.1186/1472-6882-10-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kirtikar K.R., Basu S.D. Vol. 8. 1945. Indian Medicinal Plants, 1975.Vol. III, Period. Expert. Delhi, India; pp. 2310–2311. [Google Scholar]
- 13.Devmurari V.P., Jivani N.P. Anticancer evaluation of artemisia nilagirica. 2010;2:1603–1608. [Google Scholar]
- 14.Mohanty B., Puri S., Kesavan V. A review on therapeutic potential of Artemisia nilagirica. J. Plant Biochem. Physiol. 2018;6 [Google Scholar]
- 15.De Souza Grinevicius V.M.A., Kviecinski M.R., Santos Mota N.S.R., Ourique F., Porfirio Will Castro L.S.E., Andreguetti R.R., Gomes Correia J.F., Filho D.W., Pich C.T., Pedrosa R.C. Piper nigrum ethanolic extract rich in piperamides causes ROS overproduction, oxidative damage in DNA leading to cell cycle arrest and apoptosis in cancer cells. J. Ethnopharmacol. 2016;189:139–147. doi: 10.1016/j.jep.2016.05.020. [DOI] [PubMed] [Google Scholar]
- 16.Fofaria N.M., Kim S.H., Srivastava S.K. Piperine causes G1 phase cell cycle arrest and apoptosis in melanoma cells through checkpoint kinase-1 activation. PloS One. 2014;9 doi: 10.1371/journal.pone.0094298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Senthilraja P., Kathiresan K. In vitro cytotoxicity MTT assay in vero, HepG2 and MCF-7 cell lines study of marine yeast. J. Appl. Pharm. Sci. 2015;5 [Google Scholar]
- 18.Kandala P.K., Srivastava S.K. Activation of checkpoint kinase 2 by 3,3’-diindolylmethane is required for causing G2/M cell cycle arrest in human ovarian cancer cells. Mol. Pharmacol. 2010;78:297–309. doi: 10.1124/mol.110.063750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kumar S., Raj S., Sarkar K., Chatterjee K. Engineering a multi-biofunctional composite using poly(ethylenimine) decorated graphene oxide for bone tissue regeneration. Nanoscale. 2016;8:6820–6836. doi: 10.1039/c5nr06906h. [DOI] [PubMed] [Google Scholar]
- 20.Sliogeryte K., Thorpe S.D., Wang Z., Thompson C.L., Gavara N., Knight M.M. Differential effects of LifeAct-GFP and actin-GFP on cell mechanics assessed using micropipette aspiration. J. Biomech. 2016;49:310–317. doi: 10.1016/j.jbiomech.2015.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guck J., Schinkinger S., Lincoln B., Wottawah F., Ebert S., Romeyke M., Lenz D., Erickson H.M., Ananthakrishnan R., Mitchell D., Kas J., Ulvick S., Bilby C. Optical deformability as an inherent cell marker for testing malignant transformation and metastatic competence. Biophys. J. 2005;88:3689–3698. doi: 10.1529/biophysj.104.045476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gareth J. Inman, Francisco J. Nicolás, James F. Callahan, John D. Harling, Laramie M. Gaster, Alastair D. Reith, N.J. Laping and C.S. Hill, n.d. SB-431542 is a potent and specific inhibitor of transforming growth factor-β superfamily type I Activin receptor-like kinase (ALK) receptors ALK4, ALK5, and ALK7. Mol. Pharmacol., 62, 65–74. [DOI] [PubMed]
- 23.Moyret-Lalle C. Epithelial-mesenchymal transition transcription factors and miRNAs: “Plastic surgeons” of breast cancer. World J. Clin. Oncol. 2014;5:311. doi: 10.5306/wjco.v5.i3.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sánchez-Tilló E., Liu Y., De Barrios O., Siles L., Fanlo L., Cuatrecasas M., Darling D.S., Dean D.C., Castells A., Postigo A. EMT-activating transcription factors in cancer: beyond EMT and tumor invasiveness. Cell. Mol. Life Sci. 2012 doi: 10.1007/s00018-012-1122-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu 1 Y.-N., Yin J.J., Abou-Kheir W., Hynes P.G., Casey O.M., Fang L., Yi M., Stephens R.M., Seng V., Sheppard-Tillman H., Martin P., Kelly K. MiR-1 and miR-200 inhibit EMT via Slug-dependent and tumorigenesis via Slug-independent mechanisms. Oncogene. 2013;32(3):296–306. doi: 10.1038/onc.2012.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu Yen-Nien, Abou-Kheir Wassim, Yin Juan Juan, Fang Lei, Hynes Paul, Casey Orla, Hu Dong, Wan Yong, Victoria Seng, Sheppard-Tillman Heather, Martin Philip, Kelly Kathleen. Critical and reciprocal regulation of KLF4 and SLUG in transforming growth factor β-initiated prostate cancer epithelial-mesenchymal transition. Mol Cell Biol. 2012;32(5):941–953. doi: 10.1128/MCB.06306-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fleming Y.M., Ferguson G.J., Spender L.C., Larsson J., Karlsson S., Ozanne B.W., Grosse R., Inman G.J. TGF-β-mediated activation of RhoA signalling is required for efficient V12HaRas and V600EBRAF transformation. Oncogene. 2009;28:983–993. doi: 10.1038/onc.2008.449. [DOI] [PubMed] [Google Scholar]
- 28.Leight J.L., Wozniak M.A., Chen S., Lynch M.L., Chen C.S. Matrix rigidity regulates a switch between TGF- 1-induced apoptosis and epithelial-mesenchymal transition. Mol. Biol. Cell. 2012;23:781–791. doi: 10.1091/mbc.E11-06-0537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roovers K., Assoian R.K. Effects of Rho kinase and actin stress fibers on sustained extracellular signal-regulated kinase activity and activation of G 1 phase cyclin-dependent kinases. Mol. Cell Biol. 2003;23:4283–4294. doi: 10.1128/MCB.23.12.4283-4294.2003. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 30.Lahlou M. Te success of natural products in drug discovery. Pharmacol. Pharm. 2013;4:17. [Google Scholar]
- 31.Beutler J.A. Natural products as a foundation for drug discovery. Curr. Protoc. Pharmacol. 2009;46:9. doi: 10.1002/0471141755.ph0911s46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Srivastava S. Quercetin, a natural favonoid interacts with DNA, arrests cell cycle and causes tumor regression by activating mitochondrial pathway of apoptosis. Sci. Rep. 2016;6:24049. doi: 10.1038/srep24049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Engel N., Ali I., Adamus A. Antitumor evaluation of two selected Pakistani plant extracts on human bone and breast cancer cell lines. BMC Compl. Alternative Med. 2016;16:244. doi: 10.1186/s12906-016-1215-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gothai S., Muniandy K., Mohd Esa N., Subbiah S.K., Arulselvan P. Anticancer potential of Alternanthera sessilis extract on HT-29 human colon cancer cells. Asian Pac. J. Trop. Biomed. 2018;8:394–402. [Google Scholar]
- 35.Palchaudhuri R., Hergenrother P.J. DNA as a target for anticancer compounds: methods to determine the mode of binding and the mechanism of action. Curr. Opin. Biotechnol. 2007;18:497. doi: 10.1016/j.copbio.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 36.Greenwell M., Rahman P.K.S.M. Medicinal plants: their use in anticancer treatment. Int. J. Pharm. Sci. Res. 2015;6(10):4103–4112. doi: 10.13040/IJPSR.0975-8232.6(10).4103-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Solowey E., Lichtenstein M., Sallo S., Paavilainen H., Solowet E., Lorberboum-Galski H. Evaluating medicinal plants for anticancer activity. Sci. World J. 2014;2014:1–12. doi: 10.1155/2014/721402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hosseini A., Ghorbani A. Cancer therapy with phytochemicals: evidence from clinical studies. Avicenna J. Phytomed. 2015;5(2):84–97. [PMC free article] [PubMed] [Google Scholar]
- 39.Cheaito Katia A., Bahmad Hisham F., Hadadeh Ola, Saleh Eman, Dagher Christelle, Hammoud Miza Salim, Shahait Mohammad, Mrad Zaki Abou, Nassif Samer, Tawil Ayman, Bulbul Muhammad, Khauli Raja, Wazzan Wassim, Nasr Rami, Shamseddine Ali, Temraz Sally, El-Sabban Marwan E., El-Hajj Albert, Mukherji Deborah, Abou-Kheir Wassim. EMT markers in locally-advanced prostate cancer: predicting recurrence? Front. Oncol. 2019;9:131. doi: 10.3389/fonc.2019.00131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yeh H.W., Hsu E.C., Lee S.S., Lang Y.D., Lin Y.C., Chang C.Y., Lee S.Y., Gu D.L., Shih J.H., Ho C.M., Chen C.F., Chen C.T., Tu P.H., Cheng C.F., Chen R.H., Yang R.B., Jou Y.S. PSPC1 mediates TGF-β1 autocrine signalling and Smad2/3 target switching to promote EMT, stemness and metastasis. Nat. Cell Biol. 2018;20(4):479–491. doi: 10.1038/s41556-018-0062-y. [DOI] [PubMed] [Google Scholar]
- 41.Hua W., ten Dijke P., Kostidis S. TGFβ-induced metabolic reprogramming during epithelial-to-mesenchymal transition in cancer. Cell. Mol. Life Sci. 2019 doi: 10.1007/s00018-019-03398-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Caballero-Díaz D., Bertran E., Peñuelas-Haro I., Moreno-Càceres J., Malfettone A., López-Luque J., Addante A., Herrera B. Clathrin switches Transforming Growth Factor-β role to pro-tumorigenic in liver cancer. J. Hepatol. 2019;72 doi: 10.1016/j.jhep.2019.09.012. [DOI] [PubMed] [Google Scholar]
- 43.Dhasarathy Archana, Phadke Dhiral, Mav Deepak, Shah Ruchir R., Wade Paul A. The transcription factors Snail and Slug activate the transforming growth factor-beta signaling pathway in breast cancer. PloS One. 2011;6(10) doi: 10.1371/journal.pone.0026514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee J., Choi J.-H., Joo C.-K. TGF-β1 regulates cell fate during epithelial–mesenchymal transition by upregulating surviving. Cell Death Dis. 2013;4(7):e714. doi: 10.1038/cddis.2013.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu Shuang, Chen Shuang, Zeng Jun. TGF-β signaling: a complex role in tumorigenesis. Mol. Med. Rep. 2018;17:699–704. doi: 10.3892/mmr.2017.7970. (Review) [DOI] [PubMed] [Google Scholar]
- 46.Syed Viqar. TGF-β signaling in cancer. J. Cell. Biochem. 2016;117(6):1279–1287. doi: 10.1002/jcb.25496. Epub 2016 Feb 11. [DOI] [PubMed] [Google Scholar]
- 47.Gu Shuchen, Feng Xin-Hua. TGF-β signaling in cancer. Acta Biochim. Biophys. 2018 Oct 1;50(10):941–949. doi: 10.1093/abbs/gmy092. [DOI] [PubMed] [Google Scholar]
- 48.Chen W., Delongchamps N.B., Mao K. High RhoA expression at the tumor front in clinically localized prostate cancer and association with poor tumor differentiation. Oncol. Lett. 2016;11(2):1375–1381. doi: 10.3892/ol.2015.4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Giehl K., Keller C., Muehlich S., Goppelt-Struebe M. Actin-Mediated gene expression depends on RhoA and Rac1 signaling in proximal tubular epithelial cells. PloS One. 2015;10(3) doi: 10.1371/journal.pone.0121589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kulkarni A.H., Chatterjee A., Kondaiah P., Gundiah N. TGF-β induces changes in breast cancer cell deformability. Phys. Biol. 2018;15 doi: 10.1088/1478-3975/aac3ba. [DOI] [PubMed] [Google Scholar]
- 51.Cross S.E., Jin Y.S., Tondre J., Wong R., Rao J.Y., Gimzewski J.K. AFM-based analysis of human metastatic cancer cells. Nanotechnology. 2008;19 doi: 10.1088/0957-4484/19/38/384003. [DOI] [PubMed] [Google Scholar]