The folding and insertion of β-barrel outer membrane proteins (OMPs) are conserved processes in mitochondria, chloroplasts, and Gram-negative bacteria. In Gram-negative bacteria, OMPs are assembled into the outer membrane (OM) by the heteropentomeric β-barrel assembly machine (BAM complex). In this study, we probe the function of the individual BAM proteins and how they coordinate assembly of a diverse family of OMPs. Furthermore, we identify a gain-of-function bamA mutant capable of assembling OMPs independently of all four other BAM proteins. This work advances our understanding of OMP assembly and sheds light on how this process is distinct in Gram-negative bacteria.
KEYWORDS: BAM complex, Escherichia coli, Gram-negative bacteria, outer membrane, outer membrane biogenesis
ABSTRACT
The heteropentomeric β-barrel assembly machine (BAM complex) is responsible for folding and inserting a diverse array of β-barrel outer membrane proteins (OMPs) into the outer membrane (OM) of Gram-negative bacteria. The BAM complex contains two essential proteins, the β-barrel OMP BamA and a lipoprotein BamD, whereas the auxiliary lipoproteins BamBCE are individually nonessential. Here, we identify and characterize three bamA mutations, the E-to-K change at position 470 (bamAE470K), the A-to-P change at position 496 (bamAA496P), and the A-to-S change at position 499 (bamAA499S), that suppress the otherwise lethal ΔbamD, ΔbamB ΔbamC ΔbamE, and ΔbamC ΔbamD ΔbamE mutations. The viability of cells lacking different combinations of BAM complex lipoproteins provides the opportunity to examine the role of the individual proteins in OMP assembly. Results show that, in wild-type cells, BamBCE share a redundant function; at least one of these lipoproteins must be present to allow BamD to coordinate productively with BamA. Besides BamA regulation, BamD shares an additional essential function that is redundant with a second function of BamB. Remarkably, bamAE470K suppresses both, allowing the construction of a BAM complex composed solely of BamAE470K that is able to assemble OMPs in the absence of BamBCDE. This work demonstrates that the BAM complex lipoproteins do not participate in the catalytic folding of OMP substrates but rather function to increase the efficiency of the assembly process by coordinating and regulating the assembly of diverse OMP substrates.
IMPORTANCE The folding and insertion of β-barrel outer membrane proteins (OMPs) are conserved processes in mitochondria, chloroplasts, and Gram-negative bacteria. In Gram-negative bacteria, OMPs are assembled into the outer membrane (OM) by the heteropentomeric β-barrel assembly machine (BAM complex). In this study, we probe the function of the individual BAM proteins and how they coordinate assembly of a diverse family of OMPs. Furthermore, we identify a gain-of-function bamA mutant capable of assembling OMPs independently of all four other BAM proteins. This work advances our understanding of OMP assembly and sheds light on how this process is distinct in Gram-negative bacteria.
INTRODUCTION
The distinctive asymmetric structure of the outer membrane (OM) contributes to its essential function and its distinctive permeability properties. Large and hydrophobic molecules, including many antibiotics, are excluded from entering the cell due to the strong lateral interactions between lipopolysaccharide (LPS) molecules that make up the outer leaflet of the OM. Conversely, small and hydrophilic nutrients and signaling molecules are able to cross through β-barrel outer membrane proteins (OMPs) that function as generalized or specialized porins (1, 2). OMPs further contribute to the selective permeability of the OM by maintaining envelope integrity and participating in toxic molecule efflux (2, 3).
While all β-barrel OMPs share the common feature of a β-barrel domain, which is an antiparallel β-sheet folded into a cylinder, the size and complexity of OMPs differ significantly (3, 4). The dimensions of the β-barrel domain can vary from 8 strands to 26 strands (5, 6) in Escherichia coli, and recently, a 36-strand β-barrel was described in the Gram-negative microbe Flavobacterium johnsoniae (7). OMPs can have additional functional domains (3), including passenger domains (8, 9), protein-protein interaction motifs (6, 10, 11), and extracellular modules (12). Furthermore, OMPs are found as monomers (5), dimers (13), trimers (14–16), and in higher oligomeric states (17, 18), and several are folded around lipoprotein plugs (3, 19, 20).
The β-barrel assembly machine (BAM complex) is a heteropentomeric complex responsible for the assembly of OMPs into the OM. BAM is composed of four lipoproteins, BamBCDE, and one OMP, BamA. Only BamA and BamD are essential for viability in wild-type cells, while BamBCE are individually nonessential (21–23). The BAM complex lipoproteins scaffold around the periplasmic domain of BamA to form a top hat in the OM (24–29). The vestibule formed by the BAM complex lipoproteins and the periplasmic domain of BamA protects OMP substrates during the initial stages of folding that occur at the periplasmic face of the OM (30–33).
The current model of OMP assembly suggests that BamD regulates OMP engagement with the complex, and the bulk of the folding process relies on BamA. Substrate recognition by BamD represents an assembly checkpoint; abnormal binding results in substrates being rejected from the BAM machinery while proper binding allows for assembly to proceed (31, 34–36). Once the quality of the OMP substrate has been confirmed, BamD communicates to BamA to engage with the OMP substrate (34, 35, 37, 38). BamA then catalyzes OMP folding and membrane insertion, though the mechanism through which this occurs is under current investigation (21, 30, 32, 33, 39, 40).
Despite the diversity of OMP substrates, all are assembled by the BAM complex through utilization of distinct assembly pathways. For example, the assembly of the LPS insertase LptD around its lipoprotein plug LptE (19, 41, 42) is orders of magnitude slower than the assembly of simple, smaller multimers like LamB (14, 43), and assembly of these two substrates relies on different BAM lipoproteins (44). The mechanism by which the BAM complex prioritizes substrate selection and regulates the assembly of the entire OMP profile of the cell remains unknown but likely requires one or more of the BAM lipoproteins.
Mutants that lack one of the nonessential lipoproteins display only minor defects in OMP assembly and OM integrity (21, 23). Double mutants of the accessory lipoproteins exhibit varying phenotypes ranging from the modest temperature sensitivity of a ΔbamB ΔbamC mutant to the more pronounced conditional lethality of the ΔbamB ΔbamE mutant (23, 45, 46). A ΔbamB ΔbamC ΔbamE triple mutant is lethal. Thus, BAM complex function correlates with the number of accessory lipoproteins present, indicating that BamBCE likely share a redundant function(s) that becomes increasingly essential only when more than one has been removed. The functional redundancy of the BAM lipoproteins complicates studies of their individual importance.
Here, we identify suppressor mutations that allow viability of otherwise lethal lipoprotein deletions (ΔbamB ΔbamC ΔbamE, ΔbamD, and ΔbamC ΔbamD ΔbamE). In these suppressed strains, the function(s) of the remaining BAM lipoprotein(s) becomes increasingly important. We utilize genetic analysis to probe these functions by comparing and contrasting how suppressors identified in one deletion background behave in another. Strikingly, one of the bamA suppressor mutations allows cells to survive without any of the BAM lipoproteins. Thus, none of the redundant roles of the lipoproteins are essential for BAM complex catalytic activity, and all must function to increase the efficiency of OMP assembly via regulatory mechanisms. The suppressor mutations described here identify key residues in BamA that help define the active site of this remarkable protein-folding factor.
RESULTS
Suppression of ΔbamB ΔbamC ΔbamE.
Previous studies have shown that the stress-sensing lipoprotein RcsF is a substrate of the BAM complex (20, 47). During assembly, RcsF is threaded through the lumen of an abundant OMP to anchor the lipidated amino terminus of the protein in the outer leaflet of the OM (20). This complex, interlocked structure represents a challenging substrate for the BAM complex to assemble. Indeed, several BAM complex mutations, notably the ΔbamB ΔbamE double mutant, can be suppressed by simply removing RcsF (48, 49).
The discrepancy in viability between ΔbamB ΔbamE mutant and ΔbamB ΔbamC ΔbamE mutant cells led us to test for suppression of the ΔbamB ΔbamC ΔbamE mutant by removal of RcsF. We used linkage disruption to quantitate the ability to inherit bamB::kan in a ΔbamC ΔbamE mutant background either in the presence or absence of RcsF. As expected, kanamycin-resistant colonies were not recovered in ΔbamC ΔbamE mutant transductants, and deletion of rcsF did not increase the frequency of bamB::kan integration (see Table S1 in the supplemental material). Thus, the lethality of the ΔbamB ΔbamC ΔbamE triple mutant is not due to stalled assembly of RcsF.
We conclude that in a strain lacking BamBE (with or without RcsF), BamC function becomes critical. This is noteworthy because, in a wild-type strain, taking away BamC has no obvious consequence (21). In this triple mutant, we can now use suppressor analysis to probe the function of BamC.
We isolated spontaneous suppressor mutations that allowed for survival of ΔbamC ΔbamE mutant cells under conditions of BamB depletion in the absence of RcsF. Targeted sequencing of bamA identified these suppressor mutations as the A-to-P change at position 496 (bamAA496P) and the A-to-S change at position 499 (bamAA499S), both of which alter residues located on extracellular loop 3 of the BamA β-barrel domain (24).
The E-to-K change at position 470 (bamAE470K) confers resistance to the BamA inhibitor MRL-494 (50), and other changes at codon E470 have also been identified as suppressors of additional assembly-defective mutations (30, 51). Previously, we showed that bamD can be deleted in a bamAE470K mutant background, demonstrating that BamAE470K bypasses the requirement for BamD in OMP assembly (52). The potent suppression demonstrated by bamAE470K led us to investigate if this mutation was able to suppress other toxic BAM complex deletions.
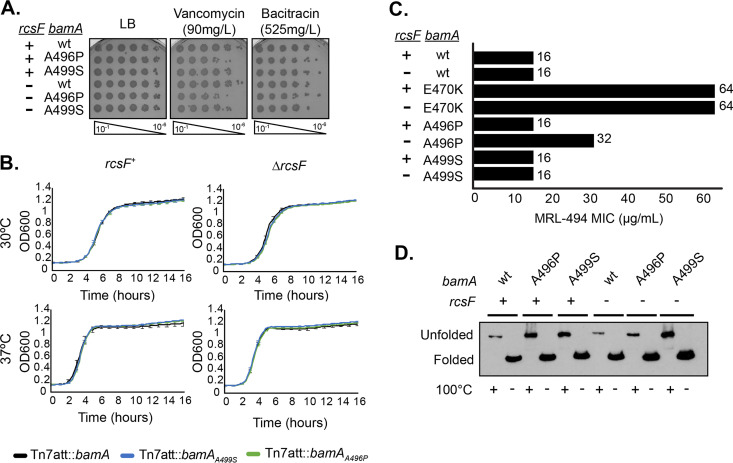
We monitored both the growth and OM permeability barrier in the bamAE470K, bamAA496P, and bamAA499S strains both in the presence and absence of RcsF (Table 1; Fig. 1A and B). Cells expressing these suppressor alleles showed similar barrier function, as measured by ability to grow on medium containing antibiotics and growth, as cells expressing wild-type bamA. In contrast to the bamAE470K strain, the bamAA496P and bamAA499S strains are not resistant to MRL-494 and exhibit a stable β-barrel domain (Fig. 1C and D).
TABLE 1.
Summary of bamA suppressor phenotypes
| bamA allele | Phenotype |
||||
|---|---|---|---|---|---|
| OM barrier | Growth | β-barrel stability | MRL-494 resistance | Basal Rcs activation | |
| bamAE470K | + | + | − | + | + |
| bamAA496P | + | + | + | − | + |
| bamAA499S | + | + | + | − | − |
FIG 1.
Phenotypic characterization of bamAA496P and bamAA499S mutations. (A) The indicated strains were normalized by OD600, serially diluted, and spotted onto medium containing vancomycin, bacitracin, or erythromycin. Plates were grown overnight at 37°C. (B) The indicated stains were normalized by OD600, inoculated into LB, and grown overnight at 30°C or 37°C. Error bars represent standard error of the mean (SEM). (C) MRL-494 resistance of the bamA suppressor alleles was evaluated using the MIC protocol. Data represents the average of two biological replicates. (D) Heat modifiability of BamA mutants. Exponentially growing cultures were normalized by OD600, lysed, and either boiled (denatured) or incubated at room temperature (nondenatured) for 10 min. Samples were electrophoresed at 4°C and probed for BamA.
To test for suppression of the ΔbamB ΔbamC ΔbamE strain by bamAE470K, bamAA496P, and bamAA499S, we again used linkage disruption. Kanamycin-resistant bamB::kan transductants were isolated from ΔbamC ΔbamE bamAE470K mutant cells in the presence or absence of RcsF (Table S1). bamB::kan was inherited by the ΔbamC ΔbamE bamAA496P and ΔbamC ΔbamE bamAA499S mutant cell only in the absence of RcsF. The inability of BamAA496P and BamAA499S to promote viability of the ΔbamB ΔbamC ΔbamE mutant cells in the presence of RcsF suggests that these mutants struggle to assemble RcsF/OMP complexes when the nonessential lipoproteins are absent. Thus, bamB can be deleted in cells expressing all three of the BamA suppressors in the absence of BamC and BamE. However, for two of the suppressors, bamAA496P and bamAA499S, rcsF must be deleted (Table 2).
TABLE 2.
Summary of suppression phenotypes
| bamA allele | BAM mutant |
|||
|---|---|---|---|---|
| ΔbamD | ΔbamC ΔbamD ΔbamE | ΔbamB ΔbamC ΔbamE | ΔbamB ΔbamC ΔbamE BamD depletion | |
| bamAE470K | + | + | + | + |
| ΔrcsF bamAE470K | + | + | + | − |
| bamAA496P | + | + | − | − |
| ΔrcsF bamAA496P | + | + | + | − |
| bamAA499S | − | − | − | − |
| ΔrcsF bamAA499S | + | + | + | − |
To conclusively demonstrate that the BamA suppressors support viability in the absence of all three accessory lipoproteins, we built clean deletion strains in which bamB, bamC, and bamE are directly deleted or disrupted. PCR and immunoblot analysis (Fig. 2A and B) confirm that the gene and protein products are absent from these strains.
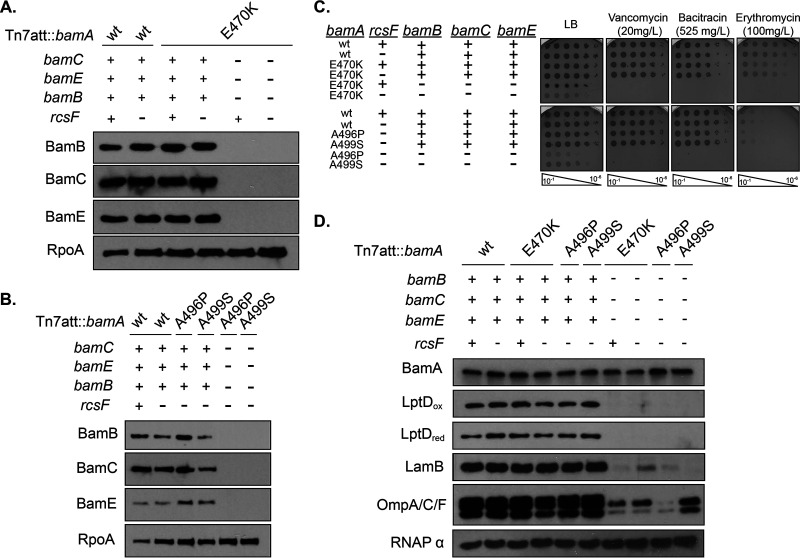
FIG 2.
bamAE470K, bamAA496P, and bamAA499S mutations promote viability of ΔbamB ΔbamC ΔbamE mutants. (A and B) Immunoblot analysis of stationary phase suppressed ΔbamB ΔbamC ΔbamE mutants expressing bamAE470K (A) or bamAA496P or bamAA499S (B) probing for BamB, BamC, and BamE. RpoA served as a loading control. (C) The indicated strains were normalized by OD600, serially diluted, and spotted onto medium containing vancomycin, bacitracin, or erythromycin. Plates were incubated overnight at 30°C. (D) Immunoblot analysis of stationary-phase cultures. Samples were electrophoresed and probed for a variety of β-barrel OMPs. RpoA served as a loading control.
Phenotypic characterization of the ΔbamB ΔbamC ΔbamE null strains revealed that all strains exhibit growth defects (Table 3), permeability (Fig. 2C), and OMP assembly defects (Fig. 2D), suggesting that the OM barrier is impaired. Notably, the levels of all OMPs tested are reduced in this background rather than defective assembly of only a subset of OMP substrates (Fig. 2D).
TABLE 3.
Viability assessment of ΔbamB ΔbamC ΔbamE, ΔbamD, and ΔbamC ΔbamD ΔbamE mutants in stationary-phase overnight cultures
| Genotype | CFU/ml ± SD (×108) (n = 3) |
|---|---|
| Tn7att::bamA | 11.5 ± 1.82 |
| Tn7att::bamAE470K | 11.7 ± 3.09 |
| Tn7att::bamAA496P | 12.4 ± 0.55 |
| Tn7att::bamAA499S | 13.9 ± 1.10 |
| ΔrcsF Tn7att::bamA | 11.0 ± 1.31 |
| ΔrcsF Tn7att::bamAE470K | 11.6 ± 2.32 |
| ΔrcsF Tn7att::bamAA496P | 12.7 ± 0.84 |
| ΔrcsF Tn7att::bamAA499S | 12.5 ± 3.98 |
| ΔbamB ΔbamC ΔbamE Tn7att::bamAE470K | 1.08 ± 0.19 |
| ΔbamB ΔbamC ΔbamE ΔrcsF Tn7att::bamAE470K | 0.32 ± 0.33 |
| ΔbamB ΔbamC ΔbamE ΔrcsF Tn7att::bamAA496P | 0.32 ± 0.18 |
| ΔbamB ΔbamC ΔbamE ΔrcsF Tn7att::bamAA499S | 0.463 ± 0.68 |
| bamD::kan Tn7att::bamAA496P | 0.241 ± 0.08 |
| bamD::kan ΔrcsF Tn7att::bamAA496P | 0.587 ± 0.23 |
| bamD::kan ΔrcsF Tn7att::bamAA499S | 0.611 ± 0.13 |
| ΔbamC ΔbamD ΔbamE Tn7att::bamAE470K | 1.16 ± 0.49 |
| ΔbamC ΔbamD ΔbamE ΔrcsF Tn7att::bamAE470K | 1.35 ± 0.33 |
| ΔbamC ΔbamD ΔbamE Tn7att::bamAA496P | 0.385 ± 0.06 |
| ΔbamC ΔbamD ΔbamE ΔrcsF Tn7att::bamAA496P | 0.7.32 ± 0.02 |
| ΔbamC ΔbamD ΔbamE ΔrcsF Tn7att::bamAA499S | 0.298 ± 0.06 |
We conclude that the critical function of BamC that becomes apparent in strains lacking BamBE and RcsF can be suppressed by bamAE470K, bamAA496P, and bamAA499S.
Suppression of ΔbamD.
As noted above, bamAE470K bypasses the requirement for BamD (52). Since bamAE470K, bamAA496P, and bamAA499S can all suppress the ΔbamB ΔbamC ΔbamE mutant in strains lacking RcsF, we tested whether the new suppressor mutations would also be able to bypass the function of BamD.
The ability to delete bamD in cells expressing bamAA496P and bamAA499S using linkage disruption with a P1 lysate carrying a bamD::kan disruption allele linked to a nearby nadB::Tn10 marker. Cells expressing bamD in diploid were able to inherit the bamD::kan allele along with nadB::Tn10. Conversely, bamA+ strains (in the presence or absence of rcsF [±rcsF]) were unable to inherit bamD::kan, demonstrating the essentiality of BamD. Cells expressing bamAA496P were able to inherit bamD::kan in both the presence and absence of rcsF, whereas cells expressing bamAA499S were able to receive bamD::kan only in a ΔrcsF background (see Table S2 in the supplemental material). The requirement of rcsF to be deleted in order for BamAA499S to allow viability of cells lacking BamD highlights the difficult nature of RcsF/OMP complex assembly. We show that this effect is specific to RcsF and not Rcs signaling, as deletion of the response regulator of the Rcs stress response, rcsB, which disrupts the signal transduction pathway downstream of RcsF, does not allow bamD to be disrupted in a bamAA499S mutant background (Table S2).
We have observed that the levels of the Rcs stress response are noticeably higher in the bamAE470K and bamAA496P strains than in bamAA499S strains (see Fig. S1 in the supplemental material). Since unassembled RcsF could accumulate in a cellular location where it might activate the stress response, this may reflect the fact that assembly of RcsF/OMP complexes is more efficient in bamAA499S strains and is instead rejected from the BAM machine by BamAE470K and BamAA496P. If so, then this would explain why the presence of RcsF is more of a problem in the bamAA499S strain than in the other two suppressors.
Based on these results, we constructed clean deletion strains in which bamD is directly disrupted in bamAA496P (±rcsF) and ΔrcsF bamAA499S mutant backgrounds (Table 2). PCR and immunoblot analysis (Fig. 3) confirmed that BamD protein was absent in these strains.
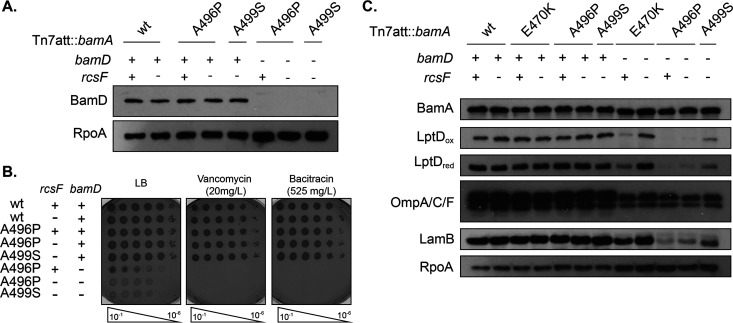
FIG 3.
bamAA496P and bamAA499S mutations bypass the essentiality of BamD. (A) Immunoblot analysis of stationary-phase bamD null strains expressing bamAA496P or bamAA499S. RpoA was used as a loading control. (B) The indicated strains were normalized by OD600, serially diluted, and spotted onto medium containing vancomycin or bacitracin. Plates were grown overnight at 30°C. (C) Stationary-phase cultures were normalized by OD600, electrophoresed, and probed for the indicated proteins. RpoA served as a loading control.
We characterized the suppressed bamD mutants expressing bamAA496P and bamAA499S and observed similar defects to those in bamD::kan bamAE470K strains (52). Cells exhibited growth (Table 3), OM permeability (Fig. 3B), and OMP assembly defects (Fig. 3C).
We conclude that bamAE470K, bamAA496P, and bamAA499S suppressors can all bypass the essential requirement for BamD.
Suppression of ΔbamC ΔbamD ΔbamE.
Previous studies have demonstrated that the BAM complex can be functionally dissected into two subcomplexes of BamAB and BamCDE. Structural studies have identified contacts between BamAD, BamAC, and BamAE (24); however, only the interaction between the BamAD proteins is essential (37, 38), leading to the hypothesis that the interaction of BamC and BamE to BamA is largely mediated through BamD. Since the essential function of BamD is bypassed by bamAE470K (±rcsF) (52), bamAA496P (±rcsF), and bamAA499S ΔrcsF, we predicted that bamC and bamE could be deleted in these suppressor backgrounds as well.
We assayed our ability to delete bamD using linkage disruption experiments with the P1 phage carrying a bamD::kan allele linked to a nadB::Tn10 marker described earlier. The bamD::kan deletion allele could not be inherited by haploid bamD cells lacking BamC and BamE (±rcsF), reflecting the essentiality of BamD in cells expressing bamA+. Coinheritance of bamD::kan with the nadB::Tn10 marker was also observed in ΔbamC ΔbamE mutant cells expressing bamAE470K and bamAA496P, regardless of the presence of RcsF. The inheritance of the marker was observed only in ΔbamC ΔbamE bamAA499S mutant cells that lacked RcsF (Table S2). Based on these results, clean disruption strains were constructed, and PCR and immunoblot analyses (Fig. 4A and B) confirmed that these strains lacked BamC, BamD, and BamE.
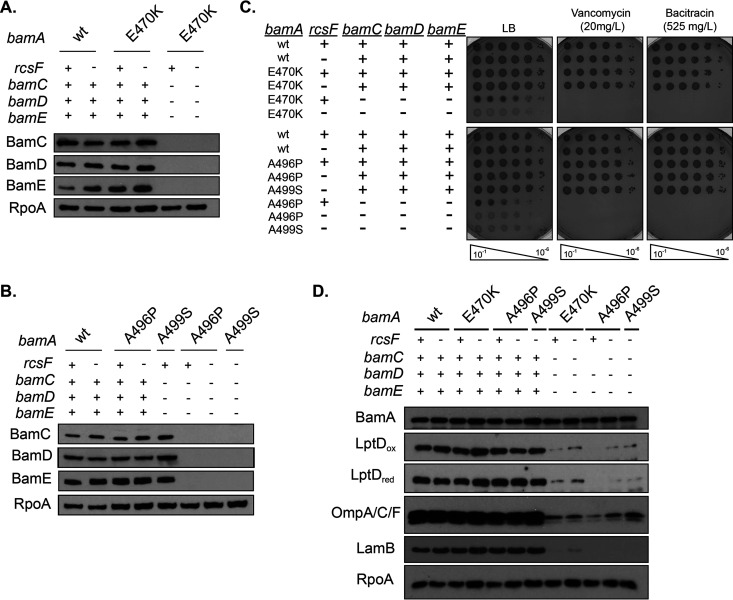
FIG 4.
bamA alleles promote viability of ΔbamC ΔbamD ΔbamE mutants. (A and B) Immunoblot analysis probing for BamC, BamD, and BamE in stationary-phase ΔbamC ΔbamD ΔbamE mutants expressing bamAE470K (A) or bamAA496P or bamAA499S (B). RpoA served as a loading control. (C) Cells were normalized by OD600, serially diluted, and spotted onto medium containing the indicated antibiotics. Plates were incubated overnight at 30°C. (D) Stationary-phase cultures were analyzed by immunoblot analysis probing for OMPs. RpoA was used as a loading control.
We characterized the phenotypes of the suppressed ΔbamC ΔbamD ΔbamE mutant strains. Similar to suppressed ΔbamD mutants, cells lacking BamC, BamD, and BamE have growth defects (Table 3), OM barrier (Fig. 4C), and OMP assembly defects (Fig. 4D).
We conclude that in bamB+ ΔbamD strains carrying the bamA suppressors, BamC and BamE have little, if any, effect on OMP assembly.
A minimal BAM complex.
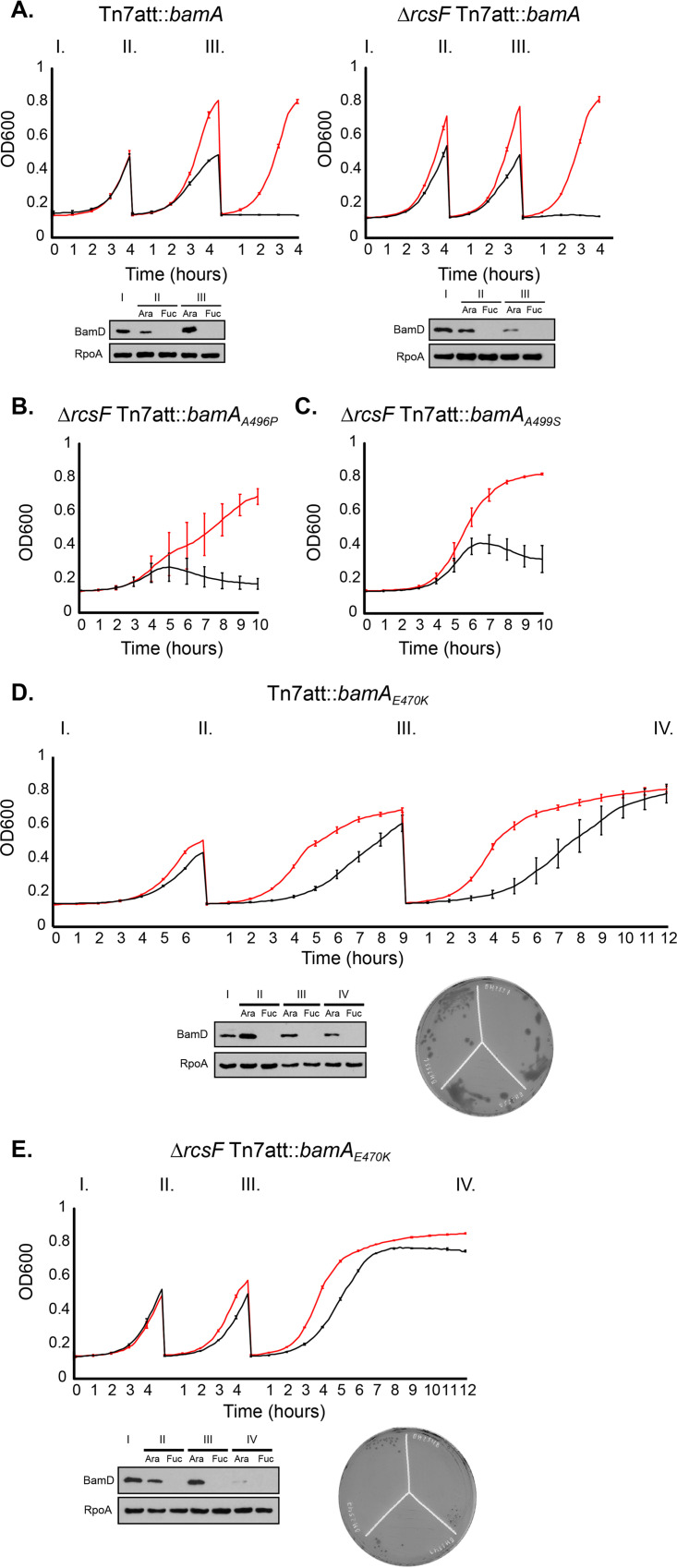
The identification of suppressor alleles that promote viability of ΔbamB ΔbamC ΔbamE and ΔbamC ΔbamD ΔbamE mutant cells led us to wonder if we could construct a viable strain with a minimal BAM complex lacking all of the lipoproteins, BamBCDE. Our strategy was to deplete BamD from the background that had the most stringent requirement for suppression, the ΔbamB ΔbamC ΔbamE mutant strain (Table 2). Again, we utilized an arabinose-inducible ectopic copy of bamD in a genetic background expressing the bamA suppressor alleles in which the native loci of bamD, bamC, bamE, and bamB were deleted. In cells expressing bamAA496P or bamAA499S, rcsF was also deleted, as these alleles cannot suppress the ΔbamB ΔbamC ΔbamE mutant when RcsF is present. Depletion of bamD by growing the strains on the anti-inducer fucose showed that the bamAE470K (±rcsF) mutant supported growth, albeit weakly, whereas ΔrcsF bamAA496P and ΔrcsF bamAA499S mutants did not support growth in the absence of the BAM complex lipoproteins (see Fig. S2 in the supplemental material).
To monitor cell viability over the course of BamD depletion, we passaged exponentially growing cells expressing the above-mentioned depletion constructs in arabinose or fucose. In control strains expressing wild-type bamA, cells inoculated into fucose survived two subcultures, or about 12 doublings, before the cells died (Fig. 5A). The depletion of the BamD protein can also be monitored by immunoblot analysis. In the ΔbamB ΔbamC ΔbamE ΔrcsF bamAA496P and ΔbamB ΔbamC ΔbamE ΔrcsF bamAA499S mutant backgrounds, the cells grown in arabinose survived due to the viability of the ΔbamB ΔbamC ΔbamE mutant backgrounds (Fig. 5B and C, red; Table 2). Cells grown in fucose lysed before reaching the mid-log-phase back dilution benchmark, suggesting that this genetic background is hypersensitive to BamD depletion (Fig. 5B and C, black; Table 2).
FIG 5.
BamAE470K allows for cell viability in the absence of BamB, BamC, BamD, and BamE. Overnight cultures of wild-type and ΔrcsF strain controls (A) or ΔbamB ΔbamC ΔbamE mutants expressing an arabinose-inducible copy of bamD and the labeled bamA alleles (±rcsF) (B to E) were grown overnight at 30°C in medium supplemented with arabinose. Cells were normalized by OD600 and inoculated into medium containing arabinose or fucose. Cultures were grown to an OD600 of ∼0.5 and back-diluted to cycle cells during logarithmic growth. Growth was monitored by OD600, and BamD depletion was confirmed by immunoblot analysis probing for BamD and RpoA as a loading control. The red line indicates growth in medium supplemented with arabinose, and the black line indicates growth in medium supplemented with fucose. Each depletion curve represents the average of three biological replicates and error bars indicate SEM.
In contrast, ΔbamB ΔbamC ΔbamE bamAE470K mutant cells (±rcsF) were able to survive depletion of BamD and grow past the point at which wild-type cells die due to a lack of BamD (Fig. 5D and E). Removal of RcsF accelerated the growth of these strains (Fig. 5E). The BamD-depleted ΔbamB ΔbamC ΔbamE ΔrcsF bamAE470K mutant cells, however, could not be successfully isolated on agar plates (Fig. 5E). BamD-depleted ΔbamB ΔbamC ΔbamE bamAE470K mutant cells that express RcsF could be successfully isolated on agar plates, but viability of these cells is severely compromised (Fig. 5D; see also Fig. S2). The absence of BamBCDE in the depleted strain was confirmed by immunoblot analysis (see Fig. S3 in the supplemental material).
The seemingly paradoxical behavior of RcsF in the bamAE470K suppressor strain merits further comment. As noted above, we suggest that the assembly of the RcsF/OMP complexes is defective in bamAE470K strains (Fig. S1). Accordingly, the presence of RcsF does not further compromise BAM function significantly in this mutant, but the mislocalized protein does activate the Rcs stress response. We suspect that removal of RcsF in the ΔbamB ΔbamC ΔbamE bamAE470K mutant background accelerates growth because the mutant cells do not waste energy producing capsule. However, if RcsF is present, the capsule produced strengthens the cell envelope enough to allow individual colonies to be isolated on agar plates (53, 54).
We monitored OMP assembly in the depleted ΔbamB ΔbamC ΔbamD ΔbamE bamAE470K mutant cells (Fig. S3). OMP levels were strongly reduced in this strain and were decreased in comparison to the isogenic ΔbamB ΔbamC ΔbamE bamAE470K strain that was grown in the presence of arabinose to induce expression of BamD. BamAE470K, though, was able to successfully assemble detectable levels of OMPs in the absence of BamBCDE.
We conclude that in the absence of all four of the BAM lipoproteins, BamAE470K can assemble OMPs well enough to support cell viability.
BamB and BamD have a redundant function in OMP assembly.
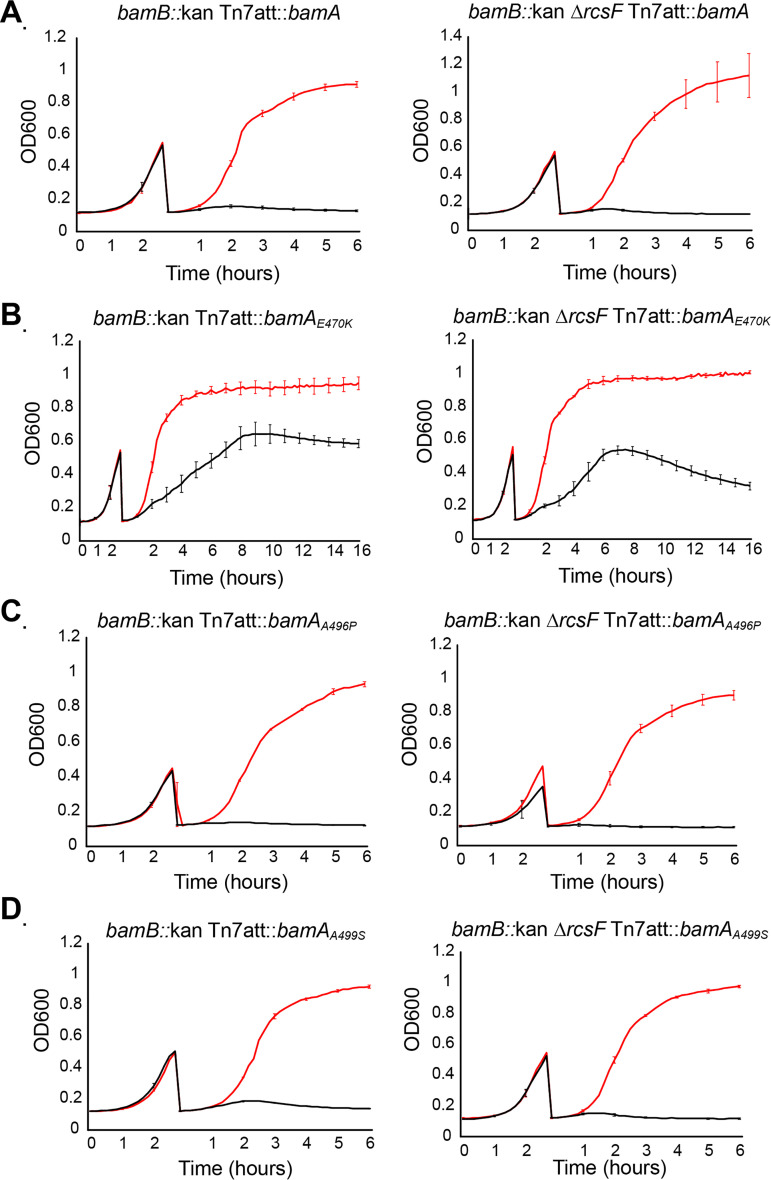
It is clear from the analysis of the phenotypes of the ΔbamD, ΔbamC ΔbamD ΔbamE, ΔbamB ΔbamC ΔbamE, and ΔbamB ΔbamC ΔbamD ΔbamE strains carrying the various bamA suppressors that bamAA496P and bamAA499S cannot tolerate the simultaneous deletion of bamB and bamD (Table 2). These results suggest that BamB and BamD share a redundant function in OMP assembly.
We depleted BamD from bamB mutants in strains expressing the bamA suppressor alleles (±rcsF) (Fig. 6). Cells carrying an ectopic copy of bamD expressed from an arabinose-inducible promoter were inoculated into medium containing arabinose or fucose and cycled in exponential phase to deplete BamD. Cells expressing bamA+ were unable to survive BamD depletion in either bamB+ (see Fig. S4 in the supplemental material) or bamB null backgrounds (Fig. 6A) due to the essentiality of BamD. The bamAA496P (±rcsF) and bamAA499S ΔrcsF strains were able to survive BamD depletion, as previously demonstrated (Fig. S4), but were unable to survive BamD depletion in bamB null backgrounds (Fig. 6C and D). Thus, in bamAA496P and bamAA499S mutant backgrounds, BamB and BamD have a redundant function.
FIG 6.
BamB and BamD have redundant functions. Depletion of BamD from an arabinose-inducible PBAD promoter in bamB null backgrounds expressing the wild-type bamA or the bamA suppressor alleles in the presence or absence of RcsF. The red line indicates growth in medium supplemented with arabinose, and the black line indicates growth in medium supplemented with fucose. bamA+ (A), bamAE470K (B), bamAA496P (C), or bamAA499S (D) strains in the presence or absence of rcsF. Cells were inoculated into medium supplemented with arabinose (inducer) or fucose (repressor). Cultures were back-diluted at an OD600 of ∼0.5. Depletions were performed at 37°C.
In contrast, bamAE470K promoted survival in the absence of BamB and BamD (Fig. 6B). Intriguingly, bamB::kan ΔrcsF bamAE470K mutant cells survived longer than the wild-type control during BamD depletion; however, these cells eventually lysed, suggesting that BamAE470K only promotes survival in the absence of BamB and BamD when RcsF was present. This phenotype mimics the ability of ΔbamB ΔbamC ΔbamE ΔrcsF bamAE470K mutant cells to grow during BamD depletion; however, individual cells were not able to survive on agar plates. As described in more detail in Discussion, this demonstrates that BamD has two essential functions, one that is redundant with BamB and another that is unique.
DISCUSSION
In this study, we have used three bamA suppressor mutations, bamAE470K, bamAA496P, and bamAA499S, to provide insight into the functions performed by the BAM complex lipoproteins during OMP assembly. These studies were facilitated by the knowledge that the interlocked RcsF/OMP complex poses a significant assembly challenge for the BAM complex. The lipidated amino terminus of RcsF is exposed on the cell surface by being threaded through the lumen of an OMP (20, 47). Efficient formation of the complex requires BamE (47), and the conditional lethal phenotype of a ΔbamB ΔbamE double mutant can be suppressed by simply removing RcsF (48, 49). Phenotypes conferred by one of these bamA suppressor alleles, bamAA499S, are apparent only in strains in which rcsF is deleted and suppression by the bamAA496P suppressor allele sometimes requires the absence of RcsF as well.
In a ΔrcsF mutant background, all three bamA mutations bypass the normally essential requirement for BamD. This reinforces the conclusion that BamD does not perform a critical role in OMP folding and membrane insertion but rather functions as an assembly checkpoint to prevent BamA from engaging defective substrates (35, 36, 52). Indeed, bamAE470K and bamAA496P, but not bamAA499S, can suppress the ΔbamD mutant even in cells that produce RcsF (Table 2). Therefore, bamAE470K, bamAA496P, and bamAA499S suppress the ΔbamD mutant by bypassing the assembly checkpoint imposed by BamD on BamA.
We have shown that the suppression phenotypes conferred by the bamA suppressor alleles to ΔbamC ΔbamD ΔbamE strains are identical to those conferred to the ΔbamD strain (Table 2). This correlation confirms that the deletion of ΔbamD likely dissociates most of the BamC and BamE proteins from the BamAB subcomplex, explaining the ability of the same bamA suppressor mutations to suppress both conditions. In wild-type cells, we propose that BamD mediates interaction of BamC and BamE with the BAM holocomplex. In the absence of BamD, BamC and BamE are not required for OMP assembly, as they can be removed with little consequence to cell viability in the suppressor strains.
Our results demonstrate that the nonessential BAM lipoproteins, BamBCE, all share a redundant function. Indeed, in an otherwise wild-type strain lacking RcsF, this function can be performed by any one of the three, even the enigmatic BamC. It is also clear that this function can be bypassed by suppressor mutations in bamA that bypass the essential function of BamD. Thus, the redundant function shared by these three lipoproteins is to facilitate proper cooperation between the essential proteins BamA and BamD. In the absence of all three lipoproteins, BamAD coordination is lethally disrupted, resulting in cell death.
In addition to this shared redundant function, the nonessential lipoproteins also have unique functions. BamE is required to assemble the surface-exposed RcsF/OMP interlocked complex (47). Also, in the absence of BamE, BamB prevents the lethal jamming of the BAM complex by RcsF (48, 49). The absence of BamB also reduces the assembly of the abundant porins but does not affect the assembly of lower volume substrates, such as the LptD/E complex (44). Indeed, for reasons discussed below, we think it is likely that BamB has an additional function(s).
The synthetic lethality of bamB and bamD deletion in the bamAA496P and bamAA499S suppressor backgrounds led us to identify a functional redundancy between BamB and BamD (Table 2). Thus, despite the fact that BamAA496P and BamAA499S do not require BamD for function, in its absence, BamB becomes essential. Alternatively stated, BamD has two essential functions as follows: (i) regulation of BamA activity (35, 37, 38, 52, 55) and (ii) a function shared specifically with BamB. In the absence of bamB, the second function of BamD becomes essential even in genetic backgrounds that have bypassed the first function. This demonstrates that BamB has two functions, one that is redundant with BamC and BamE and one that is redundant with BamD.
Previous in vitro studies have demonstrated that BamB and BamD are individually sufficient to assemble substrate BamA (55). Additionally, BamA folding is impaired in bamB mutants under conditions of BamD depletion (56). If the redundant function of BamB and BamD affects only the assembly of substrate BamA, then it is possible that this function is catalytic. If this is so, then the bamAE470K suppressor mutation must affect not only the catalytic function of BamA but also its properties as an assembly substrate. We would predict that bamAE470K mutant cells are able to survive in the absence of both BamB and BamD because BamAE470K self-assembles without the aid of any of the BAM lipoproteins. Alternatively, if the redundant function of BamB and BamD extends to other OMP substrates beyond BamA, then this function may be regulatory as well, rather than catalytic.
The potency of the bamA suppressor alleles prompted us to test if they would support cell viability in the absence of all four BAM lipoproteins. Cells expressing BamAA496P and BamAA499S could not survive in the absence of BamB, BamC, BamD, and BamE. Incredibly, we observed that strains expressing bamAE470K were able to survive in the absence of BamBCDE; however, cells require RcsF to be present in order to grow on agar medium. We propose that the capsule produced in the ΔbamB ΔbamC ΔbamD ΔbamE bamAE470K mutant cells offers mechanical support to the weakened OM, whereas deletion of rcsF prevents production of the protective capsule (53, 54).
The synthetic phenotype of bamB and bamD deletion in bamAA496P and bamAA499S mutants was predictive of the inability of these cells to survive in the absence of all four BAM lipoproteins.
Thus, we have identified a genetic background that permits cell survival despite removal of all of the BAM complex lipoproteins, suggesting that BamA alone is able to catalyze the folding and insertion of OMPs into the OM. The BAM complex lipoproteins, then, may have evolved to improve the accuracy and efficiency of OMP assembly and aid with assembly of different OMP substrates, such as RcsF/OMP complexes. This result is consistent with in vitro work demonstrating that BamA can catalyze OMP insertion on its own (55, 57, 58). However, the in vitro work is done under optimized conditions with a single OMP substrate. Cell survival is a much more demanding test that requires a potent suppressor mutation.
We have shown that the bamAE470K mutant bypasses the requirement for BamD, the redundant function that BamD shares with BamB, and the redundant function shared by the three nonessential lipoproteins BamBCE. However, in striking contrast to the observation that the phenotypes conferred by this bamA suppressor to ΔbamC ΔbamD ΔbamE strains are identical to that conferred to the ΔbamD strain(Table 2), the bamAE470K suppressed ΔbamC ΔbamD ΔbamE mutant grows far better than the suppressed ΔbamB ΔbamC ΔbamD ΔbamE mutant. Thus, there must be an additional function for BamB that we have not yet identified.
All of the BAM lipoproteins are located in the periplasm. Residue E470 is located on the fourth β-strand with its side chain pointing into the lumen, and residues A496P and A499S are located on extracellular loop 3 (24). None of these locations are near the periplasm, but they clearly alter BamA activity in a striking manner. Recent work shows that considerable barrel folding occurs within the BamA barrel (33), and residue E470 has been demonstrated to lie close to a stalled substrate (30). Extracellular loop 3 is near the seam of BamA as a part of the exit pore, which must open to allow substrates into the OM (40). We suggest that these residues are an important part of the active site of this remarkable protein-folding catalyst.
Accepted evolutionary hypotheses posit that mitochondria and chloroplasts are descended from diderm bacteria (59, 60). Both organelles have an OM containing β-barrel proteins that are inserted by an Omp85 family protein, Sam50 in mitochondria and OEP80 in chloroplasts (61–64). However, additional accessory lipoproteins involved in organelle OMP assembly do not share sequence similarity to the BAM complex lipoproteins (65). Mitochondria have a number of accessory proteins that aid OMP assembly by Sam50, but they are located on the cytoplasmic side of the mitochondrial OM, not the intramembrane space, which would be the equivalent of the bacterial periplasm (66–68). Accessory proteins of the chloroplast β-barrel assembly complex have not been characterized (65). Both mitochondria and chloroplasts are located in a constant, never-changing environment and are responsible for assembling only a small set of substrates (69–71). The bacterial lipoproteins evolved to broaden the substrate specificity and fortify BAM to allow function in many different, often hostile environments. Indeed, BamA and BamD are proposed to be the ancestral components of the BAM complex in proteobacteria, and many of the nonessential BAM lipoproteins are highly conserved as well (72).
MATERIALS AND METHODS
Bacterial strains and growth conditions.
All strains, plasmids, and oligonucleotides are listed in Table S3 in the supplemental material of this study. Strains were constructed using standard microbiological techniques as previously described (73). Strains were grown in LB medium supplemented with 0.2% arabinose, 0.1% fucose, 50 mg/liter kanamycin, 25 mg/liter tetracycline (high), 10 mg/liter tetracycline (low), 20 mg/liter chloramphenicol (high), 10 mg/liter chloramphenicol (low), 525 mg/liter bacitracin, and 20 mg/liter vancomycin when appropriate. Unless otherwise noted, all strains were grown at 30°C. bamD and bamA deletion alleles originated from previous studies (21, 22). Construction of the λatt bamD and bamA depletion alleles is described elsewhere (21–23). Other deletion alleles originated from the Keio collection, Carol Gross Tn10 collection, or Blattner collection (74–76). FLP recombination target (FRT)-flanked resistance cassettes were removed by use of the Flp recombinase system as previously described (77).
Efficiency of plating assay.
Stationary-phase cultures were normalized by optical density at 600 nm (OD600), serially diluted, and spotted onto the indicated medium. Plates were incubated at 30°C overnight. In the case of depletions, cells were grown overnight in medium containing 0.2% arabinose and diluted in LB medium.
Immunoblot analysis.
Stationary-phase cultures were normalized by OD600 and lysed in a mixture of BugBuster (Millipore), Benzonase (1:100; Sigma-Aldrich), 1 M MgCl2 (1:100), and protease inhibitor cocktail (1:100; Sigma-Aldrich) for 10 min at room temperature. Samples were diluted 1:2 in 4× Laemmli sample buffer (Bio-Rad) supplemented with β-mercaptoethanol for reduced samples. Samples were boiled for 10 min and electrophoresed on a 10% (BamA, OmpA/C/F, LamB, RNA polymerase [RNAP] α, BamD, and BamC), 8% (LptD), or 15% (BamE) SDS-PAGE gel. Proteins were transferred to a nitrocellulose membrane and immunoblotted with rabbit polyclonal primary antibody probing for anti-BamA (1:25,000), anti-LamB (1:11,500), anti-LptD (1:10,000), anti-BamC (1:60,000), anti-BamE (1:5,000), anti-BamD (1:10,000), anti-OmpA/C/F (1:25,000), or anti-RpoA (1:50,000). Donkey anti-rabbit IgG–horseradish peroxidase secondary antibody (GE Healthcare) was used at a dilution of 1:10,000.
Heat modifiability assay.
Samples were normalized by OD600 and lysed in BugBuster (Millipore), Benzonase (1:100; Sigma-Aldrich), 1 M MgCl2 (1:100), and protease inhibitor cocktail (1:100; Sigma-Aldrich) at room temperature for 10 min. Samples were diluted 1:2 in 4× Laemmli sample buffer (Bio-Rad) supplemented with β-mercaptoethanol and divided in two. One aliquot was boiled for 10 min (denatured/unfolded protein), and the other was incubated at room temperature for 10 min (nondenatured/folded protein). Samples were electrophoresed on a 10% SDS-PAGE gel at 4°C, and immunoblotting was carried out as described above.
MIC assay.
Evaluation of MIC was performed as previously described (50, 78). Briefly, 2-fold serial dilutions of MRL-494 were made in dimethyl sulfoxide (DMSO) to a concentration 50-fold higher than the target concentration to account for dilution upon treatment. The tested MRL-494 concentrations ranged from 64 μg/ml to 0.03125 μg/ml. Overnight cultures of the indicated strains were first diluted 1:50 and then diluted 1:1,000 in fresh medium. A total of 98 μl of the cell dilution was inoculated into a 96-well plate, and 2 μl of the appropriate MRL-494 concentration was added. The plate was incubated overnight at 37°C, and the OD600 was measured at 16- and 24-h time points in a BioTek Synergy H1 plate reader.
Cell viability measurements.
The number of viable cells in an overnight culture reflects the growth rate and viability of the strain. Viability was assessed by measuring CFU (CFU per milliliter). Stationary-phase cultures were diluted, plated onto LB medium, and grown overnight at 30°C. The number of colonies was used to determine CFU per milliliter. Measurements were performed in biological triplicate.
Linkage disruption assay.
P1 phage carrying bamD::kan nadB::Tn10 or bamB::kan xyz::cam was used to transduce the indicated strains at 30°C. Transductants were selected for integration of the marker allele (i.e., tetracycline for nadB::Tn10 and chloramphenicol for xyz::cam), and colonies were patched to kanamycin to check for integration of the allele of interest. The number of kanamycin colonies over total number of transductants was used to calculate linkage frequency. Calculations represent the average of at least three transductions. In the case of linkage disruption to ascertain viability of ΔbamC ΔbamD ΔbamE strains, cells carried a bamE::cam allele and were cross-patched to chloramphenicol to ensure that integration of bamD::kan nadB::Tn10 did not transduce wild-type bamE. Similarly, in linkage disruption experiments testing viability of ΔbamB ΔbamC ΔbamE mutant cells, bamC was amplified by colony PCR from kanamycin-resistant transductants to ensure that bamB::kan xyz::cam integration did not transduce wild-type bamC.
BamD liquid depletion.
Cells were grown overnight at 30°C in LB medium supplemented with 0.2% arabinose. Cultures were normalized to an OD600 of 0.01 and inoculated into LB medium containing either arabinose or fucose in 2-ml volumes in a 24-well plate (Sarstedt). Cultures were grown shaking at the indicated temperature until cells reached an OD600 of 0.5 in a BioTek Synergy H1 plate reader. Cells were then diluted 1:50 into fresh medium, and growth was repeated. Back dilutions were performed until cell survival of wild-type control ceased. At each back dilution, samples were collected for immunoblot analysis as described above. Growth measurements were performed in biological triplicate, and immunoblot analysis with anti-BamD to ensure depletion of BamD was performed on one biological replicate.
Supplementary Material
ACKNOWLEDGMENTS
We thank members of the Silhavy laboratory for productive discussion.
Research performed in this publication was supported by the National Institute of General Medical Sciences of the National Institutes of Health under grant number R35-GM118024 (to T.J.S.) and R01-GM034821 (to T.J.S.).
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
E.M.H. and T.J.S. designed the research. E.M.H. performed the research. E.M.H. and T.J.S. wrote the manuscript.
We declare no competing interests.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Silhavy TJ, Kahne D, Walker S. 2010. The bacterial cell envelope. Cold Spring Harb Perspect Biol 2:a000414. doi: 10.1101/cshperspect.a000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nikaido H. 2003. Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol Biol Rev 67:593–656. doi: 10.1128/mmbr.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wimley WC. 2003. The versatile β-barrel membrane protein. Curr Opin Struct Biol 13:404–411. doi: 10.1016/s0959-440x(03)00099-x. [DOI] [PubMed] [Google Scholar]
- 4.Koebnik R, Locher KP, Van Gelder P. 2000. Structure and function of bacterial outer membrane proteins: barrels in a nutshell. Mol Microbiol 37:239–253. doi: 10.1046/j.1365-2958.2000.01983.x. [DOI] [PubMed] [Google Scholar]
- 5.Vogt J, Schulz GE. 1999. The structure of the outer membrane protein OmpX from Escherichia coli reveals possible mechanisms of virulence. 7:1301–1309. doi: 10.1016/s0969-2126(00)80063-5. [DOI] [PubMed] [Google Scholar]
- 6.Qiao S, Luo Q, Zhao Y, Zhang XC, Huang Y. 2014. Structural basis for lipopolysaccharide insertion in the bacterial outer membrane. Nature 511:108–111. doi: 10.1038/nature13484. [DOI] [PubMed] [Google Scholar]
- 7.Lauber F, Deme JC, Lea SM, Berks BC. 2018. Type 9 secretion system structures reveal a new protein transport mechanism. Nature 564:77–82. doi: 10.1038/s41586-018-0693-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oomen CJ, van Ulsen P, Van Gelder P, Feijen M, Tommassen J, Gros P. 2004. Structure of the translocator domain of a bacterial autotransporter. EMBO J 23:1257–1266. doi: 10.1038/sj.emboj.7600148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dautin N, Bernstein HD. 2007. Protein secretion in Gram-negative bacteria via the autotransporter pathway. Annu Rev Microbiol 61:89–112. doi: 10.1146/annurev.micro.61.080706.093233. [DOI] [PubMed] [Google Scholar]
- 10.Noinaj N, Guillier M, Barnard TJ, Buchanan SK. 2010. TonB-dependent transporters: regulation, structure, and function. Annu Rev Microbiol 64:43–60. doi: 10.1146/annurev.micro.112408.134247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gatzeva-Topalova PZ, Walton TA, Sousa MC. 2008. Crystal structure of YaeT: conformational flexibility and substrate recognition. Structure 16:1873–1881. doi: 10.1016/j.str.2008.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leonard-Rivera M, Misra R. 2012. Conserved residues of the putative L6 loop of Escherichia coli BamA play a critical role in the assembly of β-barrel outer membrane proteins, including that of BamA itself. J Bacteriol 194:4662–4668. doi: 10.1128/JB.00825-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stenberg F, Chovanec P, Maslen SL, Robinson CV, Ilag LL, Heijne von G, Daley DO. 2005. Protein complexes of the Escherichia coli cell envelope. J Biol Chem 280:34409–34419. doi: 10.1074/jbc.M506479200. [DOI] [PubMed] [Google Scholar]
- 14.Schirmer T, Keller TA, Wang YF, Rosenbusch JP. 1995. Structural basis for sugar translocation through maltoporin channels at 3.1A resolution. Science 267:512–514. doi: 10.1126/science.7824948. [DOI] [PubMed] [Google Scholar]
- 15.Cowan SW, Schirmer T, Rummel G, Steiert M, Ghosh R, Pauptit RA, Jansonius JN, Rosenbusch JP. 1992. Crystal structures explain functional properties of two E. coli porins. Nature 358:727–733. doi: 10.1038/358727a0. [DOI] [PubMed] [Google Scholar]
- 16.Baslé A, Rummel G, Storici P, Rosenbusch JP, Schirmer T. 2006. Crystal structure of osmoporin OmpC from E. coli at 2.0 Å. J Mol Biol 362:933–942. doi: 10.1016/j.jmb.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 17.Chami M, Guilvout I, Gregorini M, Rémigy HW, Müller SA, Valerio M, Engel A, Pugsley AP, Bayan N. 2005. Structural insights into the secretin PulD and its trypsin-resistant core. J Biol Chem 280:37732–37741. doi: 10.1074/jbc.M504463200. [DOI] [PubMed] [Google Scholar]
- 18.Meng G, Fronzes R, Chandran V, Remaut H, Waksman G. 2009. Protein oligomerization in the bacterial outer membrane (review). Mol Membr Biol 26:136–145. doi: 10.1080/09687680802712422. [DOI] [PubMed] [Google Scholar]
- 19.Wu T, McCandlish AC, Gronenberg LS, Chng S-S, Silhavy TJ, Kahne D, Kahne D. 2006. Identification of a protein complex that assembles lipopolysaccharide in the outer membrane of Escherichia coli. Proc Natl Acad Sci U S A 103:11754–11759. doi: 10.1073/pnas.0604744103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Konovalova A, Perlman DH, Cowles CE, Silhavy TJ. 2014. Transmembrane domain of surface-exposed outer membrane lipoprotein RcsF is threaded through the lumen of β-barrel proteins. Proc Natl Acad Sci U S A 111:E4350–E4358. doi: 10.1073/pnas.1417138111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu T, Malinverni J, Ruiz N, Kim S, Silhavy TJ, Kahne D. 2005. Identification of a multicomponent complex required for outer membrane biogenesis in Escherichia coli. Cell 121:235–245. doi: 10.1016/j.cell.2005.02.015. [DOI] [PubMed] [Google Scholar]
- 22.Malinverni JC, Werner J, Kim S, Sklar JG, Kahne D, Misra R, Silhavy TJ. 2006. YfiO stabilizes the YaeT complex and is essential for outer membrane protein assembly in Escherichia coli. Mol Microbiol 61:151–164. doi: 10.1111/j.1365-2958.2006.05211.x. [DOI] [PubMed] [Google Scholar]
- 23.Sklar JG, Wu T, Gronenberg LG, Malinverni JC, Malinverni J, Kahne D, Silhavy TJ. 2007. Lipoprotein SmpA is a component of the YaeT complex that assembles outer membrane proteins in Escherichia coli. Proc Natl Acad Sci U S A 104:6400–6405. doi: 10.1073/pnas.0701579104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gu Y, Li H, Dong H, Zeng Y, Zhang Z, Paterson NG, Stansfeld PJ, Wang Z, Zhang Y, Wang W, Dong C. 2016. Structural basis of outer membrane protein insertion by the BAM complex. Nature 531:64–69. doi: 10.1038/nature17199. [DOI] [PubMed] [Google Scholar]
- 25.Iadanza MG, Higgins AJ, Schiffrin B, Calabrese AN, Brockwell DJ, Ashcroft AE, Radford SE, Ranson NA. 2016. Lateral opening in the intact β-barrel assembly machinery captured by cryo-EM. Nat Commun 7:12865. doi: 10.1038/ncomms12865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Noinaj N, Kuszak AJ, Gumbart JC, Lukacik P, Chang H, Easley NC, Lithgow T, Buchanan SK. 2013. Structural insight into the biogenesis of β-barrel membrane proteins. Nature 501:385–390. doi: 10.1038/nature12521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bakelar J, Buchanan SK, Noinaj N. 2016. The structure of the β-barrel assembly machinery complex. Science 351:180–186. doi: 10.1126/science.aad3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Han L, Zheng J, Wang Y, Yang X, Liu Y, Sun C, Cao B, Zhou H, Ni D, Lou J, Zhao Y, Huang Y. 2016. Structure of the BAM complex and its implications for biogenesis of outer-membrane proteins. Nat Struct Mol Biol 23:192–196. doi: 10.1038/nsmb.3181. [DOI] [PubMed] [Google Scholar]
- 29.Albrecht R, Zeth K. 2011. Structural basis of outer membrane protein biogenesis in bacteria. J Biol Chem 286:27792–27803. doi: 10.1074/jbc.M111.238931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee J, Tomasek D, Santos TM, May MD, Meuskens I, Kahne D. 2019. Formation of a β-barrel membrane protein is catalyzed by the interior surface of the assembly machine protein BamA. eLIFE 8:e49787. doi: 10.7554/eLife.49787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee J, Xue M, Wzorek JS, Wu T, Grabowicz M, Gronenberg LS, Sutterlin HA, Davis RM, Ruiz N, Silhavy TJ, Kahne D. 2016. Characterization of a stalled complex on the β-barrel assembly machine. Proc Natl Acad Sci U S A 113:8717–8722. doi: 10.1073/pnas.1604100113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Doyle MT, Bernstein HD. 2019. Bacterial outer membrane proteins assemble via asymmetric interactions with the BamA β-barrel. Nat Commun 10:3358. doi: 10.1038/s41467-019-11230-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tomasek D, Rawson S, Lee J, Wzorek JS, Harrison SC, Li Z, Kahne D. 2020. Structure of a nascent membrane protein as it folds on the BAM complex. Nature 583:473–478. doi: 10.1038/s41586-020-2370-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pavlova O, Peterson JH, Ieva R, Bernstein HD. 2013. Mechanistic link between β-barrel assembly and the initiation of autotransporter secretion. Proc Natl Acad Sci U S A 110:E938–E947. doi: 10.1073/pnas.1219076110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee J, Sutterlin HA, Wzorek JS, Mandler MD, Hagan CL, Grabowicz M, Tomasek D, May MD, Hart EM, Silhavy TJ, Kahne D. 2018. Substrate binding to BamD triggers a conformational change in BamA to control membrane insertion. Proc Natl Acad Sci U S A 115:2359–2364. doi: 10.1073/pnas.1711727115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hagan CL, Wzorek JS, Kahne D. 2015. Inhibition of the β-barrel assembly machine by a peptide that binds BamD. Proc Natl Acad Sci U S A 112:2011–2016. doi: 10.1073/pnas.1415955112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ricci DP, Hagan CL, Kahne D, Silhavy TJ. 2012. Activation of the Escherichia coli β-barrel assembly machine (Bam) is required for essential components to interact properly with substrate. Proc Natl Acad Sci U S A 109:3487–3491. doi: 10.1073/pnas.1201362109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McCabe AL, Ricci D, Adetunji M, Silhavy TJ. 2017. Conformational changes that coordinate the activity of BamA and BamD allowing β-barrel assembly. J Bacteriol 199:e00373-17. doi: 10.1128/JB.00373-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hagan CL, Kim S, Kahne D. 2010. Reconstitution of outer membrane protein assembly from purified components. Science 328:890–892. doi: 10.1126/science.1188919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Noinaj N, Kuszak AJ, Balusek C, Gumbart JC, Buchanan SK. 2014. Lateral opening and exit pore formation are required for BamA function. Structure 22:1055–1062. doi: 10.1016/j.str.2014.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chng SS, Ruiz N, Chimalakonda G, Silhavy TJ, Kahne D. 2010. Characterization of the two-protein complex in Escherichia coli responsible for lipopolysaccharide assembly at the outer membrane. Proc Natl Acad Sci U S A 107:5363–5368. doi: 10.1073/pnas.0912872107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chimalakonda G, Ruiz N, Chng S-S, Garner RA, Kahne D, Silhavy TJ. 2011. Lipoprotein LptE is required for the assembly of LptD by the β-barrel assembly machine in the outer membrane of Escherichia coli. Proc Natl Acad Sci U S A 108:2492–2497. doi: 10.1073/pnas.1019089108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ureta AR, Endres RG, Wingreen NS, Silhavy TJ. 2007. Kinetic analysis of the assembly of the outer membrane protein LamB in Escherichia coli mutants each lacking a secretion or targeting factor in a different cellular compartment. J Bacteriol 189:446–454. doi: 10.1128/JB.01103-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Charlson ES, Werner JN, Misra R. 2006. Differential effects of yfgL mutation on Escherichia coli outer membrane proteins and lipopolysaccharide. J Bacteriol 188:7186–7194. doi: 10.1128/JB.00571-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rigel NW, Schwalm J, Ricci DP, Silhavy TJ. 2012. BamE modulates the Escherichia coli β-barrel assembly machine component BamA. J Bacteriol 194:1002–1008. doi: 10.1128/JB.06426-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tellez R, Misra R. 2012. Substitutions in the BamA β-barrel domain overcome the conditional lethal phenotype of a ΔbamB ΔbamE strain of Escherichia coli. J Bacteriol 194:317–324. doi: 10.1128/JB.06192-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Konovalova A, Mitchell AM, Silhavy TJ. 2016. A lipoprotein/β-barrel complex monitors lipopolysaccharide integrity transducing information across the outer membrane. eLIFE 5:e15276. doi: 10.7554/eLife.15276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hart EM, Gupta M, Wühr M, Silhavy TJ. 2019. The synthetic phenotype of ΔbamB ΔbamE double mutants results from a lethal jamming of the bam complex by the lipoprotein RcsF. mBio 10:e00662-19. doi: 10.1128/mBio.00662-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tata M, Konovalova A. 2019. Improper coordination of BamA and BamD results in Bam complex jamming by a lipoprotein substrate. mBio 10:e00660-19. doi: 10.1128/mBio.00660-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hart EM, Mitchell AM, Konovalova A, Grabowicz M, Sheng J, Han X, Rodriguez-Rivera FP, Schwaid AG, Malinverni JC, Balibar CJ, Bodea S, Si Q, Wang H, Homsher MF, Painter RE, Ogawa AK, Sutterlin H, Roemer T, Black TA, Rothman DM, Walker SS, Silhavy TJ. 2019. A small-molecule inhibitor of BamA impervious to efflux and the outer membrane permeability barrier. Proc Natl Acad Sci U S A 116:21748–21757. doi: 10.1073/pnas.1912345116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Browning DF, Bavro VN, Mason JL, Sevastsyanovich YR, Rossiter AE, Jeeves M, Wells TJ, Knowles TJ, Cunningham AF, Donald JW, Palmer T, Overduin M, Henderson IR. 2015. Cross-species chimeras reveal BamA POTRA and β-barrel domains must be fine-tuned for efficient OMP insertion. Mol Microbiol 97:646–659. doi: 10.1111/mmi.13052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hart EM, Gupta M, Wühr M, Silhavy TJ. 2020. The gain-of-function allele bamAE470K bypasses the essential requirement for BamD in β-barrel outer membrane protein assembly. Proc Natl Acad Sci U S A 117:18737–18743. doi: 10.1073/pnas.2007696117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sledjeski DD, Gottesman S. 1996. Osmotic shock induction of capsule synthesis in Escherichia coli K-12. J Bacteriol 178:1204–1206. doi: 10.1128/jb.178.4.1204-1206.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Meredith TC, Mamat U, Kaczynski Z, Lindner B, Holst O, Woodard RW. 2007. Modification of lipopolysaccharide with colanic acid (M-antigen) repeats in Escherichia coli. J Biol Chem 282:7790–7798. doi: 10.1074/jbc.M611034200. [DOI] [PubMed] [Google Scholar]
- 55.Hagan CL, Westwood DB, Kahne D. 2013. Bam lipoproteins assemble BamA in vitro. Biochemistry 52:6108–6113. doi: 10.1021/bi400865z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Misra R, Stikeleather R, Gabriele R. 2015. In vivo roles of BamA, BamB and BamD in the biogenesis of BamA, a core protein of the β-barrel assembly machine of Escherichia coli. J Mol Biol 427:1061–1074. doi: 10.1016/j.jmb.2014.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schiffrin B, Calabrese AN, Higgins AJ, Humes JR, Ashcroft AE, Kalli AC, Brockwell DJ, Radford SE. 2017. Effects of periplasmic chaperones and membrane thickness on BamA-catalyzed outer-membrane protein folding. J Mol Biol 429:3776–3792. doi: 10.1016/j.jmb.2017.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gessmann D, Chung YH, Danoff EJ, Plummer AM, Sandlin CW, Zaccai NR, Fleming KG. 2014. Outer membrane β-barrel protein folding is physically controlled by periplasmic lipid head groups and BamA. Proc Natl Acad Sci U S A 111:5878–5883. doi: 10.1073/pnas.1322473111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tommassen J. 2010. Assembly of outer-membrane proteins in bacteria and mitochondria. Microbiology 156:2587–2596. doi: 10.1099/mic.0.042689-0. [DOI] [PubMed] [Google Scholar]
- 60.Sagan L. 1967. On the origin of mitosing cells. J Theoretical Biology 14:225–274. doi: 10.1016/0022-5193(67)90079-3. [DOI] [PubMed] [Google Scholar]
- 61.Gentle I, Gabriel K, Beech P, Waller R, Lithgow T. 2004. The Omp85 family of proteins is essential for outer membrane biogenesis in mitochondria and bacteria. J Cell Biol 164:19–24. doi: 10.1083/jcb.200310092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kozjak V, Wiedemann N, Milenkovic D, Lohaus C, Meyer HE, Guiard B, Meisinger C, Pfanner N. 2003. An essential role of Sam50 in the protein sorting and assembly machinery of the mitochondrial outer membrane. J Biol Chem 278:48520–48523. doi: 10.1074/jbc.C300442200. [DOI] [PubMed] [Google Scholar]
- 63.Paschen SA, Waizenegger T, Stan T, Preuss M, Cyrklaff M, Hell K, Rapaport D, Neupert W. 2003. Evolutionary conservation of biogenesis of β-barrel membrane proteins. Nature 426:862–866. doi: 10.1038/nature02208. [DOI] [PubMed] [Google Scholar]
- 64.Sommer MS, Daum B, Gross LE, Weis BLM, Mirus O, Abram L, Maier U-G, Kühlbrandt W, Schleiff E. 2011. Chloroplast Omp85 proteins change orientation during evolution. Proc Natl Acad Sci U S A 108:13841–13846. doi: 10.1073/pnas.1108626108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Misra R. 2012. Assembly of the β-barrel outer membrane proteins in Gram-negative bacteria, mitochondria, and chloroplasts. ISRN Mol Biol 2012:708203. doi: 10.5402/2012/708203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Milenkovic D, Kozjak V, Wiedemann N, Lohaus C, Meyer HE, Guiard B, Pfanner N, Meisinger C. 2004. Sam35 of the mitochondrial protein sorting and assembly machinery is a peripheral outer membrane protein essential for cell viability. J Biol Chem 279:22781–22785. doi: 10.1074/jbc.C400120200. [DOI] [PubMed] [Google Scholar]
- 67.Waizenegger T, Habib SJ, Lech M, Mokranjac D, Paschen SA, Hell K, Neupert W, Rapaport D. 2004. Tob38, a novel essential component in the biogenesis of β-barrel proteins in mitochondria. EMBO Rep 5:704–709. doi: 10.1038/sj.embor.7400183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ishikawa D, Yamamoto H, Tamura Y, Moritoh K, Endo T. 2004. Two novel proteins in the mitochondrial outer membrane mediate β-barrel protein assembly. J Cell Biol 166:621–627. doi: 10.1083/jcb.200405138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zeth K. 2010. Structure and evolution of mitochondrial outer membrane proteins of β-barrel topology. Biochim Biophys Acta 1797:1292–1299. doi: 10.1016/j.bbabio.2010.04.019. [DOI] [PubMed] [Google Scholar]
- 70.Ulrich T, Gross LE, Sommer MS, Schleiff E, Rapaport D. 2012. Chloroplast β-barrel proteins are assembled into the mitochondrial outer membrane in a process that depends on the TOM and TOB complexes. J Biol Chem 287:27467–27479. doi: 10.1074/jbc.M112.382093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schleiff E, Eichacker LA, Eckart K, Becker T, Mirus O, Stahl T, Soll J. 2003. Prediction of the plant β-barrel proteome: a case study of the chloroplast outer envelope. Protein Sci 12:748–759. doi: 10.1110/ps.0237503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Anwari K, Webb CT, Poggio S, Perry AJ, Belousoff M, Celik N, Ramm G, Lovering A, Sockett RE, Smit J, Jacobs-Wagner C, Lithgow T. 2012. The evolution of new lipoprotein subunits of the bacterial outer membrane BAM complex. Mol Microbiol 84:832–844. doi: 10.1111/j.1365-2958.2012.08059.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Silhavy TJ, Berman ML, Enquist LW. 1984. Experiments with gene fusions. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [Google Scholar]
- 74.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2:2006.0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Singer M, Baker TA, Schnitzler G, Deischel SM, Goel M, Dove W, Jaacks KJ, Grossman AD, Erickson JW, Gross CA. 1989. A collection of strains containing genetically linked alternating antibiotic resistance elements for genetic mapping of Escherichia coli. Microbiol Rev 53:1–24. doi: 10.1128/MMBR.53.1.1-24.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kang Y, Durfee T, Glasner JD, Qiu Y, Frisch D, Winterberg KM, Blattner FR. 2004. Systematic mutagenesis of the Escherichia coli genome. J Bacteriol 186:4921–4930. doi: 10.1128/JB.186.15.4921-4930.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Konovalova A, Grabowicz M, Balibar CJ, Malinverni JC, Painter RE, Riley D, Mann PA, Wang H, Garlisi CG, Sherborne B, Rigel NW, Ricci DP, Black TA, Roemer T, Silhavy TJ, Walker SS. 2018. Inhibitor of intramembrane protease RseP blocks the σE response causing lethal accumulation of unfolded outer membrane proteins. Proc Natl Acad Sci U S A 115:E6614–E6621. doi: 10.1073/pnas.1806107115. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.