Abstract
Compartmental epidemic models have been used extensively to study the historical spread of infectious diseases and to inform strategies for future control. A critical parameter of any such model is the transmission rate. Temporal variation in the transmission rate has a profound influence on disease spread. For this reason, estimation of time-varying transmission rates is an important step in identifying mechanisms that underlie patterns in observed disease incidence and mortality. Here, we present and test fast methods for reconstructing transmission rates from time series of reported incidence. Using simulated data, we quantify the sensitivity of these methods to parameters of the data-generating process and to mis-specification of input parameters by the user. We show that sensitivity to the user’s estimate of the initial number of susceptible individuals—considered to be a major limitation of similar methods—can be eliminated by an efficient, “peak-to-peak” iterative technique, which we propose. The method of transmission rate estimation that we advocate is extremely fast, for even the longest infectious disease time series that exist. It can be used independently or as a fast way to obtain better starting conditions for computationally expensive methods, such as iterated filtering and generalized profiling.
Author summary
Many pathogens cause recurrent epidemics. Patterns of recurrence are strongly affected by seasonality of the transmission rate, which can arise from seasonal changes in weather and host population behaviour (e.g., aggregation of children in schools). To understand and predict recurrent epidemic patterns, it is essential to reconstruct the time-varying transmission rate, which is never observed directly. Existing transmission rate estimation methods tend to fall into one of two categories: accurate but too slow to apply to long time series of reported incidence, or fast but inaccurate unless the number of individuals initially susceptible to infection is known precisely. Here, we introduce and compare fast methods inspired by the algorithm that Fine and Clarkson pioneered in the early 1980s. The method that we suggest accurately reconstructs seasonally varying transmission rates, even with crude information about the initial size of the susceptible population.
1 Introduction
The transmission rate of an infectious disease is a salient quantity in the study of epidemics. Changes in the transmission rate over time greatly influence the spread of infection [1, 2]. Quantifying how it changes over time can elucidate factors governing disease spread (e.g., weather [3], contact patterns [4]), inform epidemic forecasts, and suggest strategies for epidemic control [5].
In practice, we do not observe transmission directly. Instead, we observe the number of cases of infection (disease incidence) or number of deaths from infection (disease mortality) reported over time, and must reconstruct time-varying transmission rates from these data [6–13]. Utilizing historical mortality records, it is possible to identify patterns in transmission dating far back in time. Most notably, the London Bills of Mortality and the Registrar General’s Weekly Returns enable investigation of transmission patterns continuously from the mid-17th century to the present, for a number of infectious diseases including cholera [14] and smallpox [15].
A mechanistic understanding of long infectious disease time series—three centuries of weekly data in the case of smallpox [15]—requires methods of transmission rate estimation that are both accurate and fast, and therefore tractable for long time scales. Simulation-based methods of transmission rate estimation from reported incidence or mortality have been developed using the susceptible-infected-removed (SIR) model for infectious disease dynamics [16]. Markov chain Monte Carlo (MCMC [17, 18]) and sequential Monte Carlo (as in iterated filtering [8, 19, 20]) methods are statistically rigorous, but not tractable for long time scales owing to high computational cost. Generalized profiling [21, 22], which combines trajectory and gradient matching, is faster, but still too slow for convenient exploration of time series spanning hundreds of years. (Several CPU hours were required to apply generalized profiling to just 26 years of weekly data [22].)
In comparison, Finkenstädt and Grenfell’s popular “time series SIR” (tSIR) method [7, 23] is extremely fast, using a simple discretization of a continuous-time SIR model to reduce transmission rate estimation to a local regression problem. However, the tSIR method assumes that the duration of infection is equal to the time step, that natural death of susceptible individuals can be ignored, and that cumulative incidence approximates cumulative births. The latter two assumptions are reasonable for pre-vaccination measles, when most susceptible individuals were eventually infected [6]. However, in many contexts (e.g., with pathogens less transmissible than measles), susceptible mortality over time scales of interest and the difference between incidence and births are non-negligible.
In their unpublished PhD and MSc theses, Krylova (Ch. 4 in [24]) and deJonge [25] separately modified a fast discretization method originally proposed by Fine and Clarkson [6]. Krylova’s approach has been employed to estimate the amplitude of seasonal variation in measles transmission in 20th century New York City [9]. Unlike the tSIR method and unlike Fine and Clarkson, Krylova’s and deJonge’s methods do not place constraints on the infectious period or ignore susceptible mortality.
Here, we present a new algorithm based on deJonge’s method and compare its performance to the methods of Fine and Clarkson and Krylova. Our main investigative approach is to apply each method to simulated reported incidence data with known underlying transmission rate, so that error in transmission rate estimates can be quantified exactly.
Our analysis of the methods reveals a shared sensitivity to process and observation error. We mitigate this issue by introducing a smoothing step. The methods are additionally sensitive to error in the user’s estimate of the initial number of susceptible individuals, which is rarely known with any precision. We propose a fast, iterative technique for estimating this parameter from time series of incidence, births, and natural mortality, eliminating a long-standing barrier to the use of fast methods of transmission rate reconstruction.
2 Methods
In §§2.1 and 2.2 below, we present three fast methods for estimating time-varying transmission rates, based on a mechanistic model of disease spread. In §§2.3–2.7, we outline our systematic analysis of the sensitivity of the methods to parameters of the data-generating process and to error in the user-specified values of input parameters. Finally, in §2.8, we introduce peak-to-peak iteration (PTPI), a technique for estimating the initial number of susceptible individuals. Essential notation is summarized in Table 1.
Table 1. Notation.
Unless otherwise stated, simulations of reported incidence time series use the reference values listed here. If a symbol is to be interpreted differently in relation to disease incidence and disease mortality data, then the correct definition is indicated by (I) and (M), respectively.
| Symbol | Name | Definition | Ref. val. | Unit | Notes |
|---|---|---|---|---|---|
| tk | kth observation time | Time of the kth observation in time series data, for k = 0, …, n. | t0 + kΔt | years | |
| t0 | Transient period | Duration of the transient in system (1) that is ignored in simulations of reported incidence, before observations are recorded. | 2000 | years | System (1) is numerically integrated between t = 0 years and t = t0, and observed starting at t = t0. This is done so that simulations reflect dynamics near the attractor of system (1). |
| Δt | Observation interval | Time between successive observations in time series data. | 1 | weeks | Disease mortality is reported weekly in the London Bills of Mortality. |
| n | Time series length | Time between the initial and final observations in time series data, in units Δt, given by (tn − t0)/Δt. | 1042 | — | If Δt = 1 week, then 1042Δt = ⌊20 × 365/7⌋Δt ≃ 20 years. |
| [·]Δt | Nearest kΔt rounding | For time lengths t, , where [⋅] denotes nearest integer rounding. | — | — | |
| 〈⋅〉 | Long-term averaging | For functions x(t), . For sequences xk, . | — | — | |
| (S(t), I(t), R(t)) | State | Number of (susceptible, infected, removed) individuals in the population at time t. | — | — | “Removed” individuals have either recovered from the disease and gained permanent immunity or died from the disease. |
| N(t) | Population size | S(t) + I(t) + R(t). | — | — | |
| B(t) | Births | Number of births that occur during the time interval [t − Δt, t). | — | — | |
| Q(t) | Cumulative incidence | Number of susceptibles who become infected during the time interval [t0, t). | — | — | |
| Z(t) | Incidence | Number of susceptibles who become infected during the time interval [t − Δt, t). | Q(t) − Q(t − Δt) | — | |
| C(t) | Reported disease (I) incidence or (M) mortality | Number of (I) infections or (M) disease-induced deaths reported during the time interval [t − Δt, t). | Eq (31) | — | C is an abbreviation of “cases”, which are reported as infections or as deaths. |
| (S0, I0, R0) | Initial state (t = t0) | (S(t0), I(t0), R(t0)). | (S*, I*, R*) | — | (S*, I*, R*) denotes the state of system (1) after numerical integration between t = 0 years and t = t0 with seasonally forced transmission rate β(t) (Eq (27)), constant vital rates νc and μc, and initial state (t = 0 years) (Eq (32); see below). |
| N0 | Initial population size (t = t0) | N(t0). | S* + I* + R* | — | |
| Endemic equilibrium | Endemic equilibrium of system (1) with constant transmission rate (β ≡ 〈β〉) and constant vital rates (ν ≡ μ ≡ μc). | Eq (32) | — | ||
| Initial population size (t = 0 years) | N(0). | 106 | — | ||
| xk | Estimation input/output | Within an estimation algorithm (Boxes 1–3), the supplied or estimated value of x(tk) (x = C, B, μ, Z, S, I, β). | — | varies | |
| ν(t) | Birth rate | Number of births per unit time relative to , at time t. | νc | year−1 | In simulations of reported incidence, ν(t) is modeled as a constant νc. In general, estimation of β(t) from data does not require the underlying ν(t) to be constant. |
| νc | Birth rate (constant) | See ν(t) above. | 0.04 | year−1 | |
| μ(t) | Natural mortality rate | Number of natural deaths per unit time per capita, at time t. | μc | year−1 | In simulations of reported incidence, μ(t) is modeled as a constant μc. In general, estimation of β(t) from data does not require the underlying μ(t) to be constant. |
| μc | Natural mortality rate (constant) | See μ(t) above. | 0.04 | year−1 | |
| tgen | Mean generation interval | Mean time between onset of infection (in infector) and subsequent transmission of infection (by infector) [26, 27]. | 13 | days | The reference value is the sum of the observed mean latent and infectious periods, which for measles are 8 days and 5 days, respectively [16]. |
| γ | Removal rate | Number of removals (recoveries or deaths from disease) per unit time per infected. | 1/tgen | day−1 | |
| prep | Case reporting probability | (I) Probability that an infection is reported, or (M) the case fatality ratio times the probability that a death from disease is reported. | 0.25 or 1 | — | If we simulate data with under-reporting, then we use prep = 0.25 as a reference value. Otherwise, we set prep = 1. |
| trep | Mean case reporting delay | Mean time between infection and reporting of (I) infection or (M) disease-induced death. | 2 or 0 | weeks | If we simulate data with reporting delays, then we use trep = 2 weeks as a reference value. Otherwise, we set trep = 0 weeks. |
| β(t) | Transmission rate | Number of infections per unit time per susceptible per infected, at time t. | Eq (27) | year−1 | In simulations of reported incidence, β(t) is modeled by the seasonal forcing function defined in Eq (27). In general, estimation of β(t) from data does not require it vary seasonally or even periodically. |
| 〈β〉 | Mean transmission rate | Continuous-time average of the seasonally forced β(t), equal to . | β* | year−1 | β* ≈ 5.6 × 10−4year−1 is the value of 〈β〉 that satisfies Eq (2) with , νc = μc = 0.04 year−1, tgen = γ−1 = 13 days, and . |
| Basic reproduction number | Number of individuals that a typical infected person is expected to infect in an otherwise completely susceptible population. | Eq (2) | — | For measles in the 20th century, [16]. | |
| α | Seasonal amplitude | Amplitude of the seasonally forced β(t) relative to 〈β〉. | 0.08 | — | For measles, α ≈ 0.08 [2]. We require α ∈ [0, 1] to ensure that the seasonal forcing function defined in Eq (27) is non-negative. |
| ϕ(t; ϵ) | Environmental noise (realized) | Phase shift in the seasonally forced β(t), at time t. | Normal(0, ϵ2) | — | ϕ is a realization of a continuous-time stochastic process defined by a set {Φ(t; ϵ)} of independent and Normal(0, ϵ2)-distributed random variables. |
| ϵ | Standard deviation of environmental noise | See ϕ(t; ϵ) above. | 0.5 | — | |
| q | Loess smoothing parameter | Rough number of nearest neighbours weighted in local regression (i.e., when fitting loess curves to time series), determining the degree of smoothing. | — | — | See §2.2.6 for an exact definition. |
2.1 Model of disease transmission
We assume that the principal mechanisms of disease spread in the focal population are captured by the SIR model [16], formulated with time-varying rates of birth, death, and transmission. Expressing the model as a system of ordinary differential equations, we write
| (1a) |
| (1b) |
| (1c) |
where S, I, and R are the numbers of individuals who are susceptible, infected, and removed, respectively; N = S + I + R is the population size; and is the population size at an initial time, defined to be 0 years for simplicity. (We reserve the notation N0 for N(t0), where t0 > 0 years is the length of a transient; see Table 1.)
The time-varying parameters are
ν(t) birth rate, the number of births per unit time relative to ;
μ(t) natural mortality rate, the number of natural deaths per unit time per capita (i.e., relative to N); and
β(t) transmission rate, the number of infections per unit time per susceptible per infected.
The constant parameter γ is the rate of removal from the infected compartment (due to recovery or death from disease) per infected individual.
In Eq (1a) and Eq (1b), we use mass action incidence β(t)SI rather than standard incidence β(t)SI/N. Mass action incidence is essential for reproducing transitions in epidemic patterns resulting from changes in the birth rate [2, 28]. In Eq (1a), we write the net birth rate as rather than ν(t). This formulation is for convenience: the factor of does not affect dynamics, but ensures that ν(t) and μ(t) have the same scale.
The SIR model (1) assumes that the focal population is homogeneously mixed and subject to the mass action principle, which states that incidence is proportional to the product of the densities of susceptibles and infecteds [16]. The model further assumes that the latent period (time from infection to onset of infectiousness) can be ignored and that the infectious period (time from onset of infectiousness to recovery or death from disease) is exponentially distributed [29]. The distributions of the latent and infectious periods affect disease dynamics [28, 30, 31], but Krylova and Earn [28] showed that the effect on long-term dynamical structure is typically small when the mean generation interval is fixed (see Fig 11 in [28]). For this reason, we assign the mean generation interval implied by the SIR model (1) (tgen = γ−1) the value of the sum of the observed mean latent and infectious periods. This sum is the true mean generation interval if the latent and infectious periods are both exponentially distributed, and is a good estimate of the true mean generation interval more generally [28].
Transmissibility of infection is typically measured by the basic reproduction number , defined as the number of individuals that a typical infected person is expected to infect in an otherwise completely susceptible population [16]. If the birth and death rates are constant (ν ≡ νc and μ ≡ μc), and if the transmission rate has a well-defined average 〈β〉 [32], then the basic reproduction number for the SIR model (1) can be written as [28]
| (2) |
2.2 Estimating β(t) from time series data
Here, we examine three fast methods for estimating time-varying transmission rates β(t). The methods take as input (i) time series of reported disease incidence or disease mortality, (ii) time series of births and natural mortality, and (iii) values for input parameters, such as the mean generation interval tgen. By assumption, the time series data are available at discrete, equally spaced time points
| (3) |
where Δt is the observation interval. The methods return as output a time series estimate of β(t), denoted by or simply βk, which can be averaged (§2.2.5) or smoothed (§2.2.6) to distill temporal patterns of interest.
Missing data must be imputed: the three methods are recursive, so they break down as soon as they encounter a missing value. Imputation can be accomplished most simply via linear interpolation between available data. More sophisticated techniques accounting for uncertainty in missing values are described in [33].
2.2.1 FC method
We review the method first described by Fine and Clarkson [6], referred to here as the “FC method”. Let S(t) and I(t) be the number of susceptibles and infecteds in the population at time t. S decreases when susceptibles become infected or die and increases when susceptibles are born. Let Z(t) and B(t) be the number of infections and births, respectively, that occur during the time interval [t − Δt, t). Assuming that natural mortality was negligible, Fine and Clarkson reconstructed S from Z and B with the recursion
| (4) |
Fine and Clarkson further assumed that the observation interval Δt was equal to the mean generation interval tgen, so that prevalence could be approximated by incidence. That is,
| (5) |
for all t. They derived an expression for Z(t + Δt) via the mass action principle
| (6) |
Rearranging Eq (6), they obtained an estimate of β(t), given by
| (7) |
Fine and Clarkson applied Eqs (4), (5), and (7) to estimate S(tk), I(tk), and β(tk) (for k = 0, …, n), after specifying (i) the initial number of susceptibles S0 = S(t0), and (ii) values of Z(tk) and B(tk) from incidence and birth data, respectively.
A limitation of the FC method is the constraint requiring Δt = tgen. For some diseases, this is a minor issue, because incidence and birth data can be aggregated so that the time between successive aggregates is approximately equal to tgen. For example, the mean generation interval of measles is approximately two weeks, so Fine and Clarkson [6] aggregated pairs of weekly observations. A second, more serious limitation is the assumption, implicit in Eqs (4) and (5), that natural mortality is negligible. We discuss this issue in §3.1.
2.2.2 S method
Krylova (Ch. 4 in [24]) generalized the FC method in order to eliminate the constraint requiring Δt = tgen and account for natural mortality. Her approach is based on the SEIR model, which distinguishes “exposed” individuals in the latent stage of infection from infectious individuals. Here, we adapt Krylova’s approach to the SIR model (1) and refer to our approach as the “S method.”
We define S, I, Z, and B as in the FC method. Let μ(t) be the per capita natural mortality rate at time t, and let Q(t) be the total number of infections occurring between the initial observation time t0 and current time t (i.e., cumulative incidence). The observation interval Δt is no longer constrained to be equal to the mean generation interval tgen.
We reconstruct S recursively by discretizing Eq (1a):
| (8) |
Eq (8) is the result of applying the forward Euler method for numerical integration to Eq (1a), and replacing the incidence and birth terms with Z(t + Δt) and B(t + Δt), respectively. Eq (8) is identical to Eq (4) of the FC method, except that it includes a natural mortality term.
In order to estimate β(t), we note that, by definition, dQ/dt is the rate at which individuals enter the infected compartment. From Eq (1b), this is
| (9) |
If the mean generation interval tgen is short enough that I and μ are roughly constant between times t and t + tgen, then dI/dt ≈ 0 in that interval, and using Eq (1b) we can write
| (10) |
In this case, dQ/dt is also (approximately) the rate at which individuals leave the infected compartment, tgen time after infection:
| (11) |
Note that Eq (11) is also valid if the generation interval is narrowly distributed around its mean tgen (even if tgen is long).
Discretizing Eqs (9) and (11) using forward Euler, we obtain two approximations of Z(t + Δt):
| (12) |
Rearranging Eq (12) yields an estimate of β(t), given by
| (13) |
and an estimate of I(t), given by
| (14) |
Since data are available only at the observation times tk (Eq (3)), the value of Z(t + Δt − tgen) in Eq (14) will be observed only if tgen is an integer multiple of Δt. In general, tgen is not divisible by Δt. Therefore, in practice, we replace tgen in Z(t + Δt − tgen) with the nearest integer multiple of Δt, denoted here by [tgen]Δt:
| (15) |
Thus, the S method is defined by Eq (13), coupled with Eqs (8) and (15) for the reconstruction of S and I. The S method requires users to specify (i) input parameters S0 = S(t0) and tgen = γ−1, and (ii) values of Z(tk), B(tk), and μ(tk) from incidence, birth, and natural mortality data, respectively.
The FC method is a special case of the S method, obtained by setting Δt = tgen and μ(t) ≡ 0.
2.2.3 SI method
DeJonge [25] improved Krylova’s method (Ch. 4 in [24]) by reconstructing I directly from Eq (1b) instead of relying on the approximation in Eq (11). Here, we improve deJonge’s discretization, which employs the forward Euler method, by instead combining forward and backward Euler. One way to do this is to use the trapezoidal method: whereas forward and backward Euler take f ′(t)Δt and f ′(t + Δt)Δt, respectively, to approximate integrals , the trapezoidal method takes the average , which is less prone to error. Our discretization, which we call the “SI method”, is consistently more accurate than deJonge’s and others (see §S9 of S1 Text for a comparison of nine possible algorithms). Numerically integrating Eq (1a) and Eq (1b) using the trapezoidal method, and replacing the incidence and birth terms with Z(t + Δt) and B(t + Δt), respectively, we obtain
| (16) |
and
| (17) |
Eq (17) eliminates an important problem with Eq (15) of the S method, which estimates I(t) ≈ 0 if Z(t + Δt − [tgen]Δt) = 0, leading to division by zero in Eq (13).
Discretizing Eq (9) using forward and backward Euler, we obtain two approximations of Z(t + Δt):
| (18) |
Rearranging Eq (18) yields two estimates of β(t),
| (19) |
whose average supplies a more accurate estimate (see §S9 of S1 Text), given by
| (20) |
Thus, the SI method is defined by Eq (20), coupled with Eqs (16) and (17) for the reconstruction of S and I. Compared to the S method, the SI method, in principle, requires one additional input parameter, namely the initial number of infecteds I0 = I(t0). In §3.6, we show that, in practice, this additional information is not necessary.
2.2.4 Estimating true incidence from reported incidence
Let C(t) be the number of infections reported during the time interval [t − Δt, t). We estimate true incidence Z from reported incidence C via
| (21) |
where prep is the probability that an infection is reported and [trep]Δt is the mean time between infection and reporting, rounded to the nearest integer multiple of the observation interval Δt.
Eq (21) has the limitation that multiplying by does not correct for under-reporting if, by chance, C(t + [trep]Δt) = 0. In this situation, not only is the result Z(t) ≈ 0 incorrect, but we divide by zero in the FC and S methods when we substitute Eqs (5) and (15) in Eqs (7) and (13), respectively. If C(t + [trep]Δt) = C(t + [trep]Δt + Δt) = 0, then the SI method also suffers: Eq (20) gives β(t) ≈ 0. To circumvent these issues, we replace zeros in reported incidence time series by linearly interpolating between nonzero values prior to estimating true incidence using Eq (21). We do not replace leading and trailing zeros.
If what we observe is deaths from disease, rather than infections, then we have the complication that only a fraction of infections end in death. In this situation, we can still use Eq (21) to estimate Z, provided we interpret (i) C as reported disease mortality, (ii) prep as the case fatality ratio times the probability that a death from disease is reported, and (iii) trep as the mean time between infection and reporting of disease-induced death.
A more sophisticated method of inferring true incidence from reported data is described in [34].
2.2.5 Averaging raw estimates of β(t)
Given fixed time series data and input parameters, the FC, S, and SI methods return estimates of β(t) that are entirely determined (not random). In the absence of additional data observed from the same population, it is difficult to assign confidence to the output.
However, if an estimate is approximately periodic (with apparent period T) and contains m complete cycles, and if we assume β(t) is truly periodic, then we can view as containing a sample of m estimates of the true cycle, with some variance, and use its mean as an estimator instead of any one of the m cycles. For such an estimate defined on the interval [t0, t0 + mT), the mean and variance are given by
| (22a) |
| (22b) |
In §3.3, we apply the S and SI methods to simulated data to estimate an underlying, seasonally forced β(t) (Eq (27)), which has a period of 1 year. We linearly interpolate the raw time series estimate βk and compute the average 1-year cycle in the interpolant βint(t) using Eq (22a). Comparing this average to the true 1-year cycle, we are able to assess bias in the two methods.
Note that and s2(t) can be used to obtain a formal, likelihood-based measure of confidence in estimates (see §2.3.4 in [35]).
2.2.6 Smoothing raw estimates of β(t)
Process and observation error introduce random fluctuations in reported incidence on top of longer-term (e.g., seasonal) variation. In §3.2, we show that noise in reported incidence is spuriously amplified in βk, the raw time series estimate of β(t).
To distill temporal patterns of interest from the noise, we fit a smooth loess (short for local regression; see Ch. 8.1 in [36]) curve βloess(t; q) to the points and use βloess(t; q) as our final estimate of β(t). Here, q ∈ {5, …, n + 1} is an integer-valued parameter controlling the degree of smoothing. At times t ∈ [t0, tn], the fitted value βloess(t; q) is obtained as follows:
Order the distances dk = |tk − t| of the time points tk (Eq (3)) from t, letting denote the ith smallest distance (for i = 1, …, n + 1).
- Fit a quadratic polynomial p2(t) to the points . This is done by weighted least squares using tricube weights
Hence only time points tk nearer to t than the qth nearest time point are weighted in the fit.(23) Define βloess(t; q) = p2(t).
Typically, smoother fits are obtained with greater q [36, 37].
The optimal value of q for a given time series βk, denoted by qopt, is that which minimizes error in βloess(t; q) relative to β(t). In §3.4, we estimate β(t) from simulated data, smooth βk using each value of q on a grid, and use our knowledge of β(t) to determine qopt. We show that it is possible for smoothing to eliminate much of the error in βk attributable to process and observation error. Thus, in §2.2.7, we explicitly define the FC, S, and SI methods with loess smoothing as a final step.
In practice, β(t) is not known, so we cannot determine qopt. In this case, qopt can be estimated using statistical methods, such as time series cross-validation [38]. However, reasonable results can be obtained much more quickly by inspecting βloess(t; q) directly and increasing q from 4 until a desirable degree of smoothing is achieved (e.g., until noise on the time scale of weeks is visibly reduced, and patterns on the time scale of months are easier to discern).
2.2.7 Summary
In Boxes 1–3 below, we summarize the three methods derived in §§2.2.1–2.2.6 for estimating time-varying transmission rates β(t) from time series data with observation times tk (Eq (3)). We use the notation xk to refer to the value supplied or computed for x(tk) within the estimation algorithms (x = C, B, μ, Z, S, I, β).
Box 1. FC method (Fine & Clarkson 1982 [6])
| (24a) |
| (24b) |
| (24c) |
| (24d) |
where Δt is assumed to be roughly equal to tgen, and natural mortality is assumed to be negligible. Users must specify:
a time series of reported incidence or reported disease mortality, with zeros replaced via linear interpolation between nonzero values;
a time series of births;
input parameters S0, tgen, prep, and trep.
Box 3. SI method (adapted from deJonge 2014 [25])
| (26a) |
| (26b) |
| (26c) |
| (26d) |
| (26e) |
Users must specify:
a time series of reported incidence or reported disease mortality, with zeros replaced via linear interpolation between nonzero values;
a time series of births;
a time series of the per capita natural mortality rate;
input parameters S0, I0, tgen = γ−1, prep, and trep;
loess smoothing parameter q.
Box 2. S method (adapted from Krylova 2011 [24])
| (25a) |
| (25b) |
| (25c) |
| (25d) |
| (25e) |
Users must specify:
a time series of reported incidence or reported disease mortality, with zeros replaced via linear interpolation between nonzero values;
a time series of births;
a time series of the per capita natural mortality rate;
input parameters S0, tgen = γ−1, prep, and trep;
loess smoothing parameter q.
In Box 4, we provide instructions for input specification based on our analysis of the methods.
Box 4. Instructions for input specification
βk is sensitive to mis-specification of S0, but not I0 (cf. §3.6.1). If the user’s estimate of S0 is uncertain, and if the incidence time series Zk is roughly periodic, then a more accurate estimate of S0 may be obtained via peak-to-peak iteration (PTPI; cf. §3.7).
If Sk is negative for any k, then it is likely that the case reporting probability prep was underestimated or that births were systematically under-reported by Bk. This can be resolved by correcting the estimate of prep or correcting Bk, then restarting the algorithm. Users should apply close to the minimal correction necessary to prevent negative Sk.
q must be tuned to the βk time series. An estimate of qopt can be obtained using statistical methods, such as time series cross-validation [38]. However, q can be tuned quickly through visual inspection of βloess(t; q): one can increase q from 5 until a desirable degree of smoothing is achieved (e.g., until noise on the time scale of weeks is visibly reduced, and patterns on the time scale of months are easier to discern).
2.3 Simulating reported incidence data
In order to compare the performance of the FC, S, and SI methods in estimating β(t), we apply the methods to simulated reported incidence data with known underlying β(t). Here, we outline our methods for simulating these data using the SIR model (1).
2.3.1 Seasonal forcing of β(t) with environmental stochasticity
We reproduce seasonal fluctuation in the transmission rate by modeling β(t) in Eq (1) as a sinusoidal forcing function with period equal to one year:
| (27) |
Here, α ∈ [0, 1] is the amplitude of seasonal forcing relative to the mean 〈β〉. We introduce stochastic fluctuation by adding a randomly generated phase shift:
| (28) |
ϕ is a realization of a continuous-time stochastic process consisting of independent, Normal(0, ϵ2)-distributed random variables. It models environmental stochasticity leading to random noise in the transmission rate. Modeling environmental stochasticity with a random phase shift rather than additive noise conveniently avoids negative βϕ(t): βϕ(t) oscillates between 〈β〉(1 − α) and 〈β〉(1 + α) regardless of the distribution of the noise. In practice, we take the values of ϕ at times tk (Eq (3)) and linearly interpolate to obtain values in between. This helps to make simulations of Eqs (1) and (9) with adaptive time steps (cf. §2.3.2) reproducible.
2.3.2 Generating incidence time series with demographic stochasticity
We supplement Eq (1) with Eq (9), so that trajectories of the resulting system record changes in cumulative incidence Q. In this system, we employ the noisy transmission rate βϕ(t) (Eq (28)) and constant vital rates νc and μc. We then either (i) numerically integrate the differential equations to approximate their solution, or (ii) treat the system more realistically as an event-driven, continuous-time Markov process (with event rates specified by terms in the differential equations) and use the adaptive tau-leaping algorithm for stochastic simulation [39, 40]. The latter approach accounts for demographic stochasticity in disease dynamics. We prevent disease fadeout in simulations with demographic stochasticity by setting the rates of infected recovery and death to zero whenever I = 1.
In both methods of simulation, we record the state of the system at times tk (Eq (3)), choosing initial state
| (29) |
where S0 + I0 + R0 = N0 = N(t0). Finally, we derive incidence Z from Q via first differences:
| (30) |
2.3.3 Introducing observation error
Observation error due to under-reporting (prep < 1) and reporting delays (trep > 0 weeks) creates discrepancies between true incidence Z and reported incidence C. We introduce random observation error to simulated incidence time series with delayed binomial sampling:
| (31) |
For simulations without observation error, we set prep < 1 and trep > 0 weeks.
2.3.4 Parametrization
The simulation method outlined in §§2.3.1–2.3.3 is parametrized by
For most simulations, we assign parameters the reference values listed in Table 1. We consider different values when we investigate the sensitivity of β(t) estimates to data-generating parameters (cf. §2.6.1).
We bypass transient dynamics by choosing t0 = 2000 years and numerically integrating system (1) between 0 years and t0 in order to obtain a point (S*, I*, R*) very near the attractor. For this step, we exclude environmental noise, defining β(t) as in Eq (27), and take the initial state to be the endemic equilibrium of the unforced system (system (1) with β ≡ 〈β〉 and ν ≡ μ ≡ μc):
| (32) |
2.4 Creating mock birth and natural mortality time series
In addition to reported incidence data, the FC, S, and SI methods require time series of births and the per capita natural mortality rate. For simplicity, we create mock time series by (i) choosing constant vital rates and , then (ii) setting and for all k. Note that is the result of integrating the net birth rate in the SIR model (1), given by , between successive observation times using .
We specify and , where νc and μc are the data-generating vital rates (cf. §2.3.4), except when we investigate the sensitivity of β(t) estimates to incorrect vital data (cf. §2.6.2). For example, to model under-reporting of births, we simply set .
2.5 Measuring β(t) estimation error
When we simulate reported incidence data, the underlying transmission rate β(t) is defined beforehand via Eq (27) and known for all t. We use this knowledge to quantify the error in estimates of β(t) obtained from the data. Specifically, given an estimate defined at time points tk (Eq (3)), we compute the relative root mean square error (RRMSE), defined as
| (33) |
where
| (34) |
Note that by “underlying transmission rate” we mean the transmission rate excluding environmental noise. Although we simulate data using the noisy βϕ(t), defined in Eq (28), our aim is to reconstruct the noiseless β(t), defined in Eq (27).
2.6 Sensitivity analysis
Error in β(t) estimation from reported incidence data depends on how the data were generated. The number of cases reported over time is influenced by features of the disease (e.g., the natural history of infection), population (e.g., contact patterns), and case reporting (e.g., the frequency and accuracy of reports). In our simulations of reported incidence, there are 14 data-generating parameters (cf. §2.3.4), whose values are summarized in the vector
| (35) |
Estimation error also depends on how accurately certain data-generating parameters are specified by users of the FC, S, and SI methods. The initial observation time t0, observation interval Δt, and time series length n are always known exactly. Other parameters (〈β〉, α, ϵ, , νc, and μc) influence our simulations of reported incidence, but in practice are not parameters of the FC, S, and SI methods. In practice, users are required to specify only S0, tgen, prep, trep, and (with the SI method) I0. However, when we test the methods here, we do specify vital rates νc and μc in order to create mock (constant) birth and natural mortality time series (cf. §2.4). The specified values of these 7 input parameters are summarized in the vector
| (36) |
First, we investigate the sensitivity of the methods to the data-generating parameter values θ. Then, we examine their sensitivity to error in the user’s specification θ′ of the input parameters. Here, we describe our analysis using the notation to refer to transmission rate estimates constructed with user input θ′, from data generated by parameter values θ.
2.6.1 Sensitivity to data-generating parameters
In §3.5, we consider the ideal situation in which the input θ′ corresponds exactly to the data-generating θ. In this case, how sensitive is error in to θ? For example, is β(t) estimated more accurately for diseases with longer mean generation interval tgen, etc.? To answer these questions, we perform the following steps on a grid of data-generating parameter values θ:
Simulate 1000 reported incidence time series using θ.
Create corresponding mock (constant) birth and natural mortality time series (cf. §2.4), specifying and in the input θ′.
Estimate β(t) from the simulated data, specifying , , , , and in the input θ′.
Compute the median RRMSE in the estimates (1000 estimates corresponding to 1000 simulations).
We repeat this analysis 6 times, corresponding to 2 methods of β(t) estimation (S or SI) and 3 methods of data simulation:
without demographic stochasticity and without observation error (fixing prep = 1, trep = 0 weeks),
with demographic stochasticity but without observation error (fixing prep = 1, trep = 0 weeks), or
with demographic stochasticity and with observation error (fixing prep = 0.25 unless sensitivity to prep is considered, trep = 2 weeks).
Environmental stochasticity (ϵ = 0.5) is included in all simulations.
2.6.2 Sensitivity to mis-specification of input parameters
In §3.6, we fix the data-generating θ and consider the more realistic situation in which components of the input θ′ differ from the corresponding components of θ by a potentially large factor. In this case, how sensitive is error in to error in θ′? For example, how important is having an accurate estimate of tgen, etc.? To answer these questions, we perform the following steps:
Simulate 1000 reported incidence time series using fixed data-generating parameter values θ. (We assign the reference values listed in Table 1.)
-
For each point on a grid of input parameter values θ′:
Create mock (constant) birth and natural mortality time series, taking and from the input θ′.
Estimate β(t) from the simulated data, taking , , , , and from the input θ′.
Compute the median RRMSE in the estimates (1000 estimates corresponding to 1000 simulations).
We repeat this analysis 6 times, as outlined at the end of §2.6.1.
2.7 Asymptotic analysis
Here, we examine analytically the propagation of input error to the output of the SI method. (Similar expressions for propagated errors are obtained by analyzing the S method.) Our analysis here supports numerical results presented in §3.6 concerning the sensitivity of β(t) estimation error to mis-specification of input parameters.
2.7.1 Explicit solutions of the (Sk, Ik) difference equations
The SI method uses Eq (26a) to Eq (26c) to recursively reconstruct S(t) and I(t) from time series of reported incidence, births, and natural mortality. After substitution of Eq (26a), Eq (26b) and Eq (26c) can be written as
| (37a) |
| (37b) |
where r = [trep]Δt/Δt is the mean case reporting delay in units of the observation interval, rounded to the nearest integer. Eq (37) are linear, first order difference equations, whose explicit solutions are obtained using standard algebraic techniques (see Eq 1.2.4 in [41]) and given by
| (38a) |
| (38b) |
with the conventions and if a < b. As we show in §2.7.2, explicit solutions of Eq (37) facilitate asymptotic analysis.
2.7.2 Propagation of input error to (Sk, Ik)
We consider the special case in which the vital rates are constant and set and μk = μc for all k (cf. §2.4). Then Eq (38) simplify to
| (39a) |
| (39b) |
where we have made explicit the dependence of Sk and Ik on input parameters S0, I0, νc, μc, tgen = γ−1, and prep. Using Eq (39), we can derive exact expressions for the error propagated to Sk and Ik in the SI method as a result of assigning an incorrect value to an input parameter.
If the initial number of susceptibles is truly S0, but we specify , where ω > 0, then the error propagated to Sk is
| (40) |
where is the life expectancy in the population. Similarly, specifying for I0 yields an error
| (41) |
in Ik, where tinf = (γ + μc)−1 is the mean time between infection and removal from the infected compartment, accounting for the possibility of natural death during infection. Eqs (40) and (41) show that the errors propagated to Sk and Ik vanish as k → ∞; we exploit this fact to improve susceptible reconstruction (cf. §2.8).
Mis-specifying νc by assigning a value creates an error in Sk that increases in magnitude over time and converges to a limit:
| (42) |
Unlike Eq (42), the exact expression for Err(Sk, μc ← ωμc) is not readily simplified and is difficult to interpret:
| (43) |
However, if Ck has a well-defined long-term average 〈C〉 (this will be true if, for instance, Ck is periodic), then Err(Sk, μc ← ωμc) has a well-defined long-term average 〈Err(Sk, μc ← ωμc)〉 with a simple form. Replacing Ci+1+r in Eq (43) with 〈C〉, simplifying the resulting expression, then taking the limit as k → ∞, we obtain
| (44) |
We can similarly show the following, still assuming that 〈C〉 is well-defined:
| (45) |
| (46) |
| (47) |
| (48) |
Here, is the (incorrect) mean time spent infected that results when ωμc is incorrectly specified for μc (Eq (45)) or ωtgen is incorrectly specified for tgen (Eq (46)).
2.7.3 Propagation of error in (Sk, Ik) to βk
Let βk(Zk, Zk+1, Sk, Ik) be the raw SI method estimate of β(tk), given by the right hand side of Eq (26d). Suppose that, due to propagated error (cf. §2.7.2), the arguments are incorrect by a factor, so that
| (49) |
where ωZ, ωS, ωI > 0. Then the computed βk will have relative error
| (50) |
Hence severe underestimation of Sk or Ik (ωS ≪ 1 or ωI ≪ 1) causes the relative error in βk to blow up.
2.8 Estimating S0 via peak-to-peak iteration
Reconstruction of susceptibles S(t) is a necessary step in the reconstruction of β(t) using the FC, S, and SI methods. In §3.6, we show that susceptible reconstruction requires accurate specification of the initial number of susceptibles S0 = S(t0). However, reliable estimates of S0 have, to this point, been difficult to obtain in practice.
We propose a technique for iteratively improving estimates of S0, requiring only incidence, birth, and natural mortality data at times tk (Eq (3)). Crucially, our technique, which we call “peak-to-peak iteration” (PTPI), enables accurate susceptible reconstruction without direct observation of the susceptible population size at the initial time.
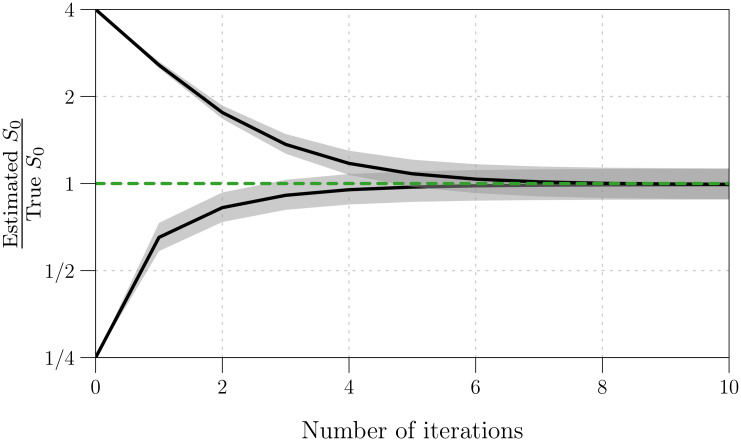
Our approach is motivated by application of the SI method to simulated data. When we incorrectly guessed the value of S0 and attempted to reconstruct S(t) via Eq (26b), the absolute error in the reconstruction decreased monotonically over time (k). (Eq (40) shows that the error propagated from S0 to Sk vanishes as k → ∞.) Consequently, if the underlying dynamics are at least approximately periodic, and if t0 and tn occur at the same phase of the cycle, then Sn is actually a better estimate of S0 than our initial guess. In this situation, instead of reconstructing β(t) directly, we can use Sn as an updated estimate of S0, and reconstruct S(t) more accurately. This procedure can be repeated any number of times, and, with simulated data, we observe rapid convergence to an accurate estimate of S0 (cf. §3.7).
The key point is that the reconstructed final state can be used as an improved estimate of the initial state only if the initial and final states occur at the same phase of the cycle. This will not be true unless the observation period (the time between the first and last observations in time series data) is an integer multiple of the period of the underlying dynamics. We can ensure this by choosing appropriate times at which to start and stop S(t) reconstruction. In noisy periodic data, the points in a cycle that are easiest to identify robustly are the peaks. Consequently, we ignore observations (i) prior to the time ta of the first peak in the incidence time series and (ii) after the time tb of the last peak that occurs near an integer multiple of the apparent period after the first peak. For the truncated time series, the iterations converge to an accurate estimate of S(ta) starting from an initial guess, and we recover the corresponding accurate estimate of S0 by solving Eq (26b) backwards in time, from ta to t0:
| (51) |
The complete PTPI algorithm, which consists of finding ta and tb (truncation step) and estimating S0 (iteration step), is outlined in Boxes 5 and 6 below. In §3.7, we assess the performance of PTPI by applying the technique to simulated data with known underlying S0, starting from an incorrect initial estimate of S0.
Box 5. Peak-to-peak iteration: Truncation step
Goal: Given a roughly periodic time series of incidence, we want to find the time ta of the first peak and the time tb of the last peak occurring at the same phase of the cycle. These times are necessary for the iteration step (Box 6).
Algorithm:
- Smooth the raw incidence time series Zk by applying a (2ℓ1 + 1)-point central moving average, computed via
Choose minimal ℓ1 large enough to remove spurious peaks in Zk caused by noise, while retaining true peaks.(52) Identify the period T of the smoothed incidence time series from its power spectrum, and calculate the number of embedded cycles, given by .
- Construct the set indexing peaks in :
Choose minimal ℓ2 large enough to ensure that indexes true peaks in , but not spurious peaks caused by noise (any that remain after smoothing).(53) Define , the set of times of peaks in , and record the time of the first peak, given by .
For i = 0, …, m, define τi = ta+ iT and find the element of nearest τi, namely . The resulting subset should contain successive time points that are roughly one period apart, i.e., the corresponding peaks in should occur at the same phase of the cycle.
Record the time of the last such peak, given by .
Box 6. Peak-to-peak iteration: Iteration step
Goal: We want to produce an accurate estimate of the initial number of susceptibles S0 = S(t0), given
a roughly periodic time series of incidence,
a time series of births,
a time series of the per capita natural mortality rate,
times ta and tb as defined in the truncation step (Box 5), and
an initial estimate of S0.
Algorithm:
Define an initial estimate of S(ta). (We use the initial estimate of S0.)
Reconstruct S(t) between times ta and tb using Eq (26b), starting with the current estimate of S(ta).
Update the estimate of S(ta) with the estimate of S(tb) obtained in (ii).
Repeat (ii) and (iii) until the sequence of estimates of S(ta) converges (to within a desirable tolerance).
Reconstruct S(t) between times t0 and ta using Eq (51), starting with the final estimate of S(ta) obtained in (iv). The reconstruction is performed backwards in time, from ta to t0.
Record the estimate of S0 = S(t0) computed in (v). This value can be passed back to Eq (26b), allowing for reconstruction of S(t) between times t0 and tn, as usual.
3 Results
In §3.1, we compare the performance of the FC, S, and SI methods in estimating β(t) from an idealized reported incidence time series. In §3.2, we show how process and observation error create spurious noise in estimates of β(t). In §§3.3 and 3.4, we examine averaging and smoothing as ways to distill temporal patterns of interest from noisy estimates of β(t). In §§3.5 and 3.6, we summarize our systematic analysis of the sensitivity of β(t) estimation error to data-generating parameters and to mis-specification of input parameters by the user. In §3.7, addressing apparent sensitivity to mis-specification of the initial number of susceptibles S0, we assess the performance of PTPI as a method of estimating S0. Finally, in §3.8, we report the run times of the S and SI methods and PTPI.
The results reported here are entirely reproducible using the annotated R code available in S1 File.
3.1 Example of β(t) estimation using the FC, S, and SI methods
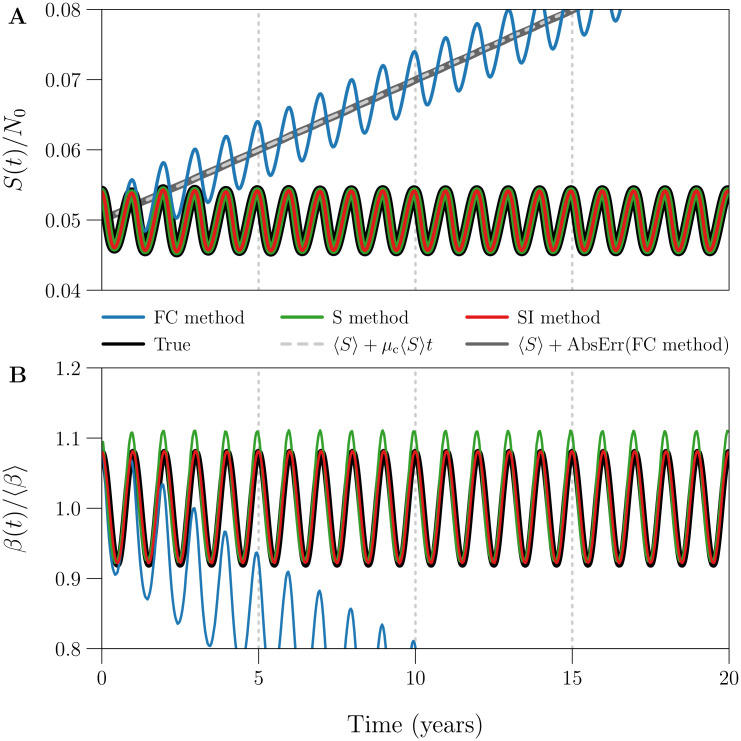
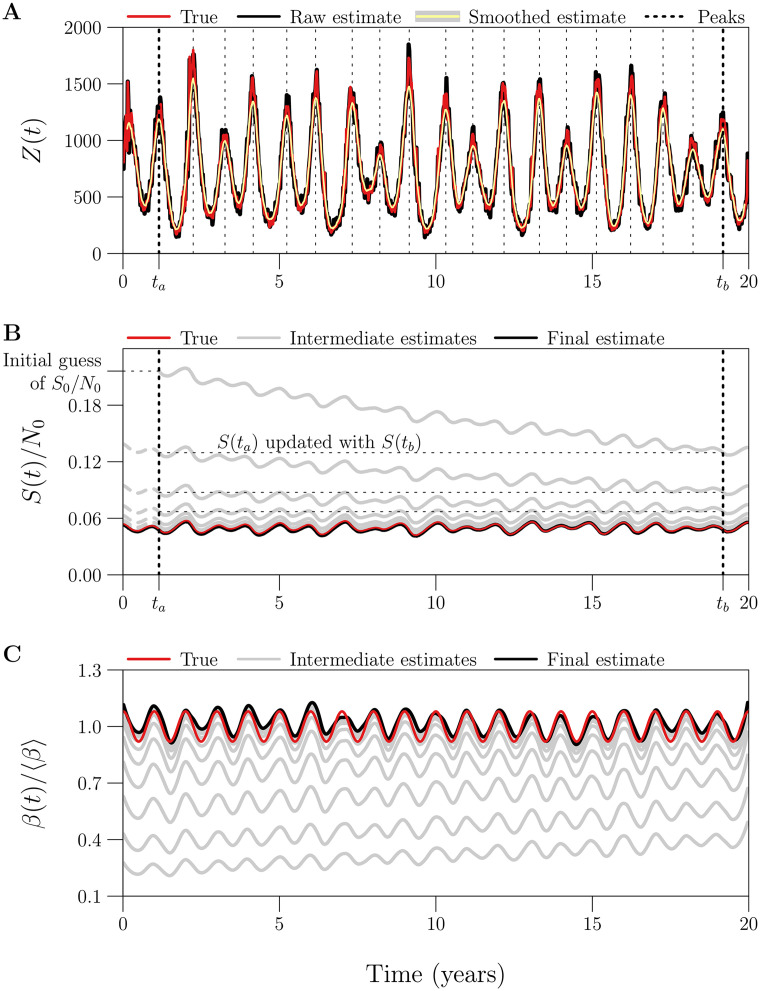
We applied the FC, S, and SI methods without input error to estimate S(t) and β(t) from an idealized reported incidence time series, simulated without process or observation error. The time series estimates Sk and βk are shown in Fig 1. The S and SI methods estimated S(t) and β(t) accurately at every time point, whereas the FC method captured seasonality but failed otherwise. In the FC method, Sk neglects natural mortality (Eq (24b)), so it increases without bound while βk decays to zero due to division by Sk (Eq (24d)).
Fig 1. Example of S(t) and β(t) estimation using the FC, S, and SI methods.
Plotted are the susceptible population size S(t) and seasonally forced transmission rate β(t) (Eq (27)) underlying 20 years of weekly reported incidence, together with time series estimates Sk and βk obtained from the data by the FC [blue], S [green], and SI [red] methods. The reported incidence time series (Δt = 1 week, n = ⌊20 × 365/7⌋ = 1042) was simulated without process or observation error (ϵ = 0, prep = 1), using reference values (Table 1) for all other data-generating parameters. The three estimation methods were applied without input error, i.e., all input parameters were assigned their true (data-generating) values. [Panel A] S(t) scaled by 1/N0, describing the number of susceptibles as a proportion of the initial population size. Grey lines show that the absolute error in the FC method estimate of S(t) increases linearly as μc 〈S〉t, where μc is the constant per capita natural mortality rate and 〈S〉 is the continuous-time average of S(t). [Panel B] β(t) scaled by 1/〈β〉, describing the transmission rate relative to its mean. RRMSE (Eq (33)) in the βk time series generated by the (FC, S, SI) method is roughly (0.3355, 0.0240, 0.0021).
Fig 1A confirms that the absolute error in the FC method estimate of S(t) increases linearly as μc 〈S〉t, where μc is the constant per capita natural mortality rate and 〈S〉 is the continuous-time average of S(t). In practice, the FC method fails whenever natural mortality in the underlying population is non-negligible. Since the S and SI methods address this limitation at effectively no computational cost, we do not present further analysis of the FC method.
In Fig 1B, the SI method estimate of β(t) was very accurate (RRMSE ≈ 0.2%), whereas the S method estimate peaked too early and too high (RRMSE ≈ 2.4%).
3.2 Effects of process and observation error
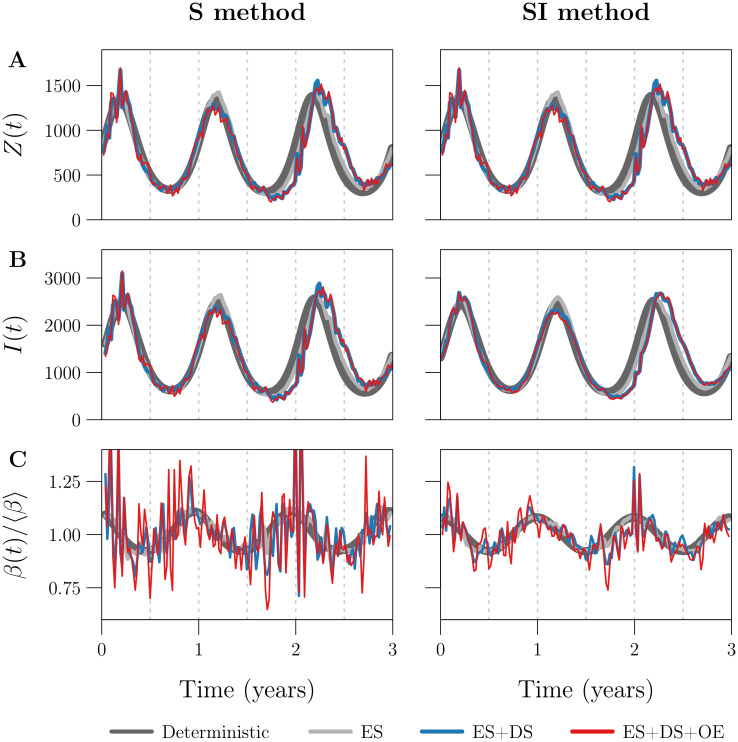
We applied the S and SI methods without input error to four reported incidence time series Ck, simulated using the same parameter values but with different levels of process and observation noise. The first simulation was purely deterministic, while the remaining three included (i) environmental stochasticity [ES], (ii) ES and demographic stochasticity [ES+DS], or (iii) ES, DS, and observation error [ES+DS+OE]. Fig 2 shows the resulting estimates Zk, Ik, and βk of true incidence Z(t), prevalence I(t), and the seasonally forced transmission rate β(t).
Fig 2. Effects of process and observation error on the S and SI methods.
Plotted are the estimates [Row A] Zk, [Row B] Ik, and [Row C] βk of true incidence Z(t), prevalence I(t), and the seasonally forced transmission rate β(t) (Eq (27)) obtained by applying the [Left] S and [Right] SI methods without input error to each of four simulated reported incidence time series (indicated by the legend; Δt = 1 week, n = ⌊3 × 365/7⌋ = 156). The first simulation was purely deterministic [dark grey] (ϵ = 0, prep = 1), while the remaining three accounted for (i) environmental stochasticity [ES, light grey] (ϵ = 0.5, prep = 1), (ii) ES and demographic stochasticity [ES+DS, blue] (ϵ = 0.5, prep = 1), or (iii) ES, DS, and observation error [ES+DS+OE, red] (ϵ = 0.5, prep = 0.25). Reference values (Table 1) were assigned to all other data-generating parameters, in all four simulations. The left and right panels in Row A are identical, because the S and SI methods compute Zk identically (compare Eqs (25a) and (26a)). RRMSE in the βk time series is (0.0239, 0.0375, 0.1126, 0.1432) with the S method and (0.0021, 0.0153, 0.0494, 0.0591) with the SI method (order follows the legend). Note that the underlying β(t) was the same in all simulations; it is not plotted in Row C, but is close to perfectly represented by the dark grey curve in the right panel (RRMSE ≈ 0.2%). Due to process error, the underlying Z(t) and I(t) (also not shown) varied between the deterministic, ES, and ES+DS simulations.
Noise of any type introduces random fluctuations in Ck on top of longer-term (e.g., seasonal) variation. Noise in Ck is propagated to Zk (Fig 2A) and Ik (Fig 2B), because (i) in both the S and SI methods, we scale Ck+r by a constant factor of to compute Zk (Eqs (25a) and (26a)); (ii) in the S method, we scale Ck+1−g+r by a constant factor of [prep(γ + μk)Δt]−1 to compute Ik (Eq (25c) after substitution of Eq (25a)); and (iii) in the SI method, Ik contains a weighted sum of Ci terms (Eq (38b)).
Noise in Zk and Ik is amplified in βk (Fig 2C), distorting the correct temporal pattern, for the following reason. When Z and I are close to zero, small absolute changes in either yield large relative changes in the ratio Z/I and in turn βk, which contains a factor of Zk+1/Ik in the S method (Eq (25d)) and (Zk + Zk+1)/(2Ik) in the SI method (Eq (26d)). Hence low amplitude noise in Zk and Ik appears as spurious, higher amplitude noise in βk. This is an important issue in practice, because the incidence of endemic diseases is typically very small relative to the population size, and periodic fluctuations bringing incidence even closer to zero are common for many diseases [4, 14, 42].
Fig 2 shows that the SI method is much better than the S method at resisting noise propagation. One reason is the effective smoothing of incidence in the SI method, which replaces Zk+1 with (Zk + Zk+1)/2 in the computation of βk (compare Eqs (25d) and (26d)). We expose a second reason in §3.2.1 below by comparing the variance in Ik induced by observation error, between the two methods. (We expect similar results for process error.)
3.2.1 Propagation of noise from Ck to Ik
Consider the S and SI method estimates of prevalence I(tk),
| (54a) |
| (54b) |
Here, g = [tgen]Δt/Δt and r = [trep]Δt/Δt are the mean generation interval and case reporting delay in units of the observation interval, rounded to the nearest integer. These estimates are obtained from Eq (25c) (after substitution of Eqs (25a)) and (38b) when we assume a constant natural mortality rate μc. Following §2.3.3, suppose reported incidence is generated from true incidence Z(tk) via . Then the variance of Ck+r is
| (55) |
It follows from Eqs (54) and (55) and the identity Var(aX) = a2 Var(X) that
| (56a) |
| (56b) |
If Z(t) has a well-defined average 〈Z〉, then replacing instances of Z in Eq (56) with 〈Z〉 and taking the limit as k → ∞, we obtain the average variances
| (57a) |
| (57b) |
Comparing these with 〈Var(Ck)〉 = 〈Z〉prep(1 − prep) using reference parameter values tgen = γ−1 = 13 days, μc = 0.04year−1, and Δt = 1 week, we obtain
| (58a) |
| (58b) |
where tinf = (γ + μc)−1 is the mean time spent infected. Hence, while both the S and SI methods suffer from propagation of noise from reported incidence Ck to estimated prevalence Ik, particularly for prep ≪ 1, the S method tends to be much worse (by a factor of 3.44/0.93 ≈ 3.7 in this example). Comparative resistance to noise propagation is a distinct advantage of the SI method over the S method.
3.3 Averaging the raw estimate of β(t)
Fig 3A displays two raw estimates βk (S and SI methods, applied without input error) of a seasonally forced β(t), each spanning 1000 years (only the first 10 years are shown). The estimates embed 1000 1-year cycles, which are displayed in Fig 3B and 3C together with their 1-year average (cf. §2.2.5).
Fig 3. Bias and variance in 1-year cycles embedded in three estimates of a seasonally forced β(t).
[Panel A] In black, the seasonally forced β(t) (Eq (27)) underlying 1000 years of simulated reported incidence data. In (transparent) colour, raw estimates βk obtained from the data by the S [green] and SI [red] methods, both applied without input error. Only the first 10 of 1000 years are shown. [Panels B and C] In black, the true 1-year cycle in the seasonally forced β(t). In light (transparent) colour, the 1000 1-year cycles embedded in the linear interpolant βint(t) of βk. In dark colour, the average 1-year cycle (Eq (22a)) in βint(t). Results are shown for both the S [Panel B, green] and SI [Panel C, red] methods. [Panel D] Like Panel C, except for a smooth loess curve βloess(t; q) (q = 53) fit to βk, instead of the interpolant βint(t). [Details] A reported incidence time series with 1000 years of weekly observations (Δt = 1 week, n = 52153) was simulated with environmental noise in transmission (ϵ = 0.5), demographic stochasticity, and random under-reporting of cases (prep = 0.25), using reference values (Table 1) for the remaining parameters.
Both estimates suffered from spurious noise distorting the correct seasonal pattern, caused by process and observation error in the data-generating process (cf. §3.2). As in Fig 2C, the variance was markedly smaller with the SI method. Averaging the embedded 1-year cycles recovered the true 1-year cycle from the noise. In the absence of input error, the S method appears to carry a slight bias (peaking early and too high, as in Fig 1), whereas the SI method is nearly unbiased.
While some existing infectious disease time series span several centuries [15], in practice, averaging as in Fig 3B and 3C is sensible only over time intervals during which the underlying seasonal pattern in transmission is roughly stationary.
3.4 Smoothing the raw estimate of β(t)
Regardless of whether averaging is employed, comparison of Fig 3C and 3D shows that it is helpful to smooth the βk time series by fitting a loess curve βloess(t; q) (cf. §2.2.6). An appropriate degree of smoothing (i.e., choice of loess smoothing parameter q) eliminated spurious noise without significantly increasing bias.
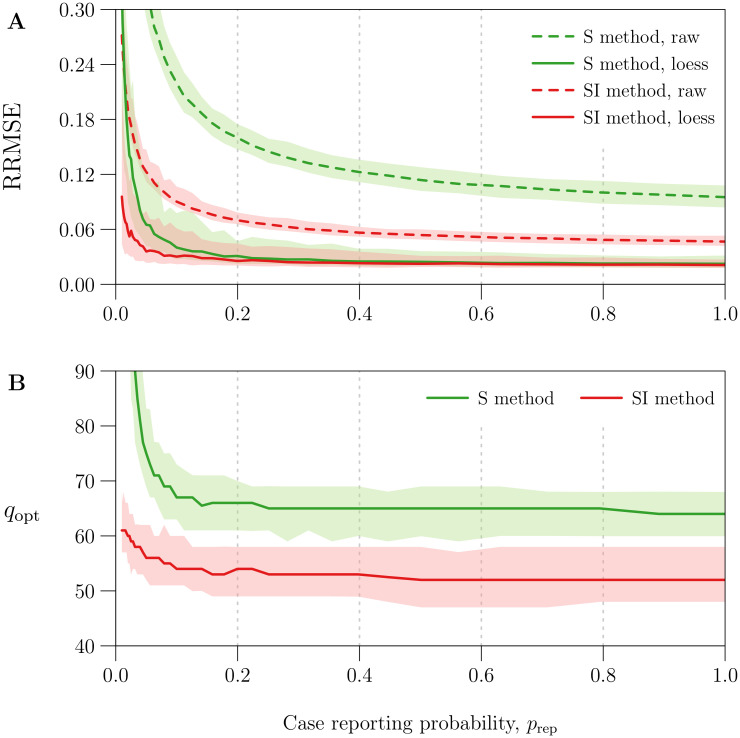
Fig 4A quantifies the effect of smoothing βk using the optimal value qopt for parameter q (cf. §2.2.6). It plots RRMSE before and after smoothing as a function of the amount of noise in the simulated reported incidence data, which was modulated by varying the case reporting probability prep between 0.01 and 1 (more noise for smaller prep; see Eq (31)).
Fig 4. Reduction in β(t) estimation error with optimal loess smoothing.
The horizontal axis measures the case reporting probability prep, for which 41 values equally spaced on a logarithmic scale between 0.01 and 1 were considered. Using each value of prep and reference values (Table 1) for all other parameters, 100 reported incidence time series (Δt = 1 week, n = 1042) were simulated accounting for environmental noise in transmission (ϵ = 0.5), demographic stochasticity, and random under-reporting of cases (measured by prep). The underlying seasonally forced β(t) (Eq (27)) was estimated from reported incidence using the S and SI methods, both applied without input error, yielding two raw estimates βk per simulation. Smooth loess curves βloess(t; q) (q = 10, …, 110; cf. §2.2.6) were fit to each βk time series. The optimal q for a given time series, denoted by qopt, was defined as the value that minimized RRMSE (Eq (33)) in βloess(tk; q). Overall, for each value of prep and each β(t) estimation method (S and SI), 100 values of qopt were obtained corresponding to 100 βk time series. Plotted on the vertical axis as functions of prep are the median and 5th and 95th percentiles of [Panel A] RRMSE in the raw estimates βk [dashed lines] and optimal loess estimates βloess(tk; qopt) [solid lines] and [Panel B] qopt. Lines and bands indicate the median and 5th–95th percentile range, respectively. Results for the S and SI methods are shown in green and red, respectively.
Using the optimal loess estimate βloess(tk; qopt) instead of the raw estimate βk significantly reduced RRMSE—by at least 46% for the S method and 17% for the SI method across all simulations. Although raw estimates generated by the SI method were consistently more accurate (expected in light of Fig 3B and 3C), optimal loess estimates were comparable between the S and SI methods for prep > 0.2 (RRMSE ≈ 3%). For prep < 0.2 (severe under-reporting of cases), optimal smoothing failed to an increasing extent to recover the underlying β(t) from noise in βk. In this setting, the S method was greatly outperformed by the SI method, which is more resilient to noise in reported incidence (cf. §3.2).
Fig 4B shows that median qopt was roughly constant for prep > 0.1, with
| (59) |
More smoothing (greater q) was required to minimize RRMSE for prep > 0.1. More generally, Fig 4 indicates that the S and SI methods should always include a smoothing step. Hence, in the remaining analysis, we always smooth βk.
3.5 Sensitivity to data-generating parameters
Here, we characterize the sensitivity of β(t) estimation error to parameters of the data-generating process. As in §§3.1–3.4, we consider the ideal case in which the user-specified values of all input parameters are equal to the true (data-generating) values. The details of our analysis are outlined in §2.6.1.
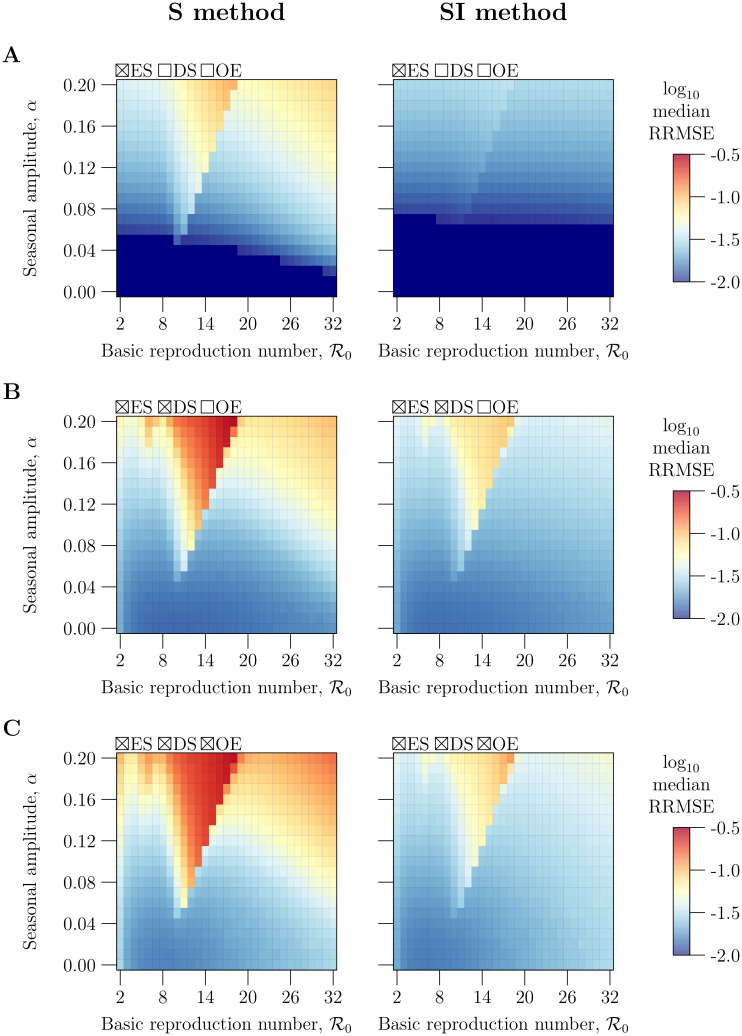
Fig 5 plots the median RRMSE in estimates of a seasonally forced β(t) (Eq (27)) from 1000 realizations of a reported incidence time series, as a bivariate function of the mean 〈β〉 and amplitude α of seasonal forcing. To aid interpretation, the 〈β〉 axis was scaled to measure the basic reproduction number (Eq (2)).
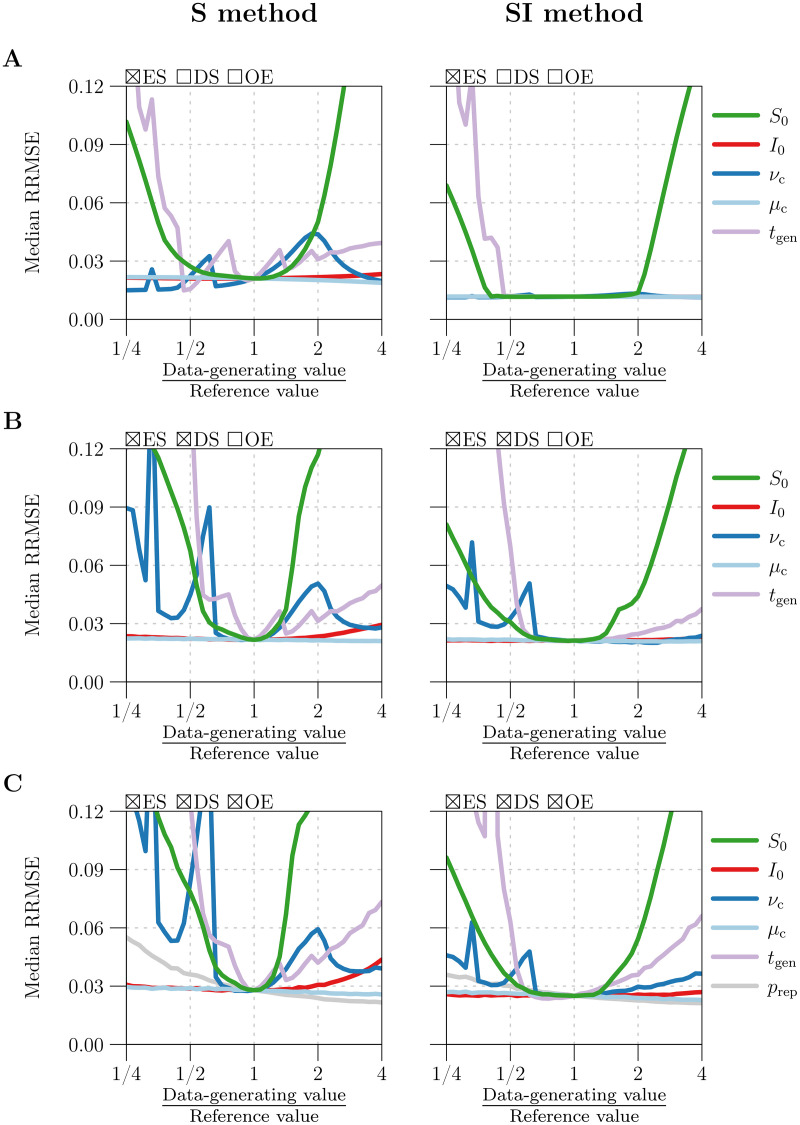
Fig 5. Sensitivity of β(t) estimation error to the mean 〈β〉 and amplitude α of seasonal forcing.
Contained in each panel are heatmaps of median RRMSE (Eq (33)) in estimates of a seasonally forced β(t) (Eq (27)) from simulated reported incidence time series, as a bivariate function of the mean 〈β〉 and amplitude α of seasonal forcing. The 〈β〉 axis has been scaled to measure the basic reproduction number (Eq (2)). When simulating reported incidence, reference values (Table 1) were assigned to all data-generating parameters except 〈β〉 and α. A grid of pairs with levels and α = 0, 0.01, …, 0.2 was considered, with 〈β〉 defined for each value of via Eq (2). For each parametrization, 1000 simulations were performed with environmental stochasticity [ES] (ϵ = 0.5) and with or without demographic stochasticity [DS] and observation error [OE], as indicated by row: [Row A] without DS or OE (prep = 1, trep = 0 weeks), [Row B] with DS but without OE (prep = 1, trep = 0 weeks), [Row C] with DS and OE (prep = 0.25, trep = 2 weeks). Corresponding mock birth and natural mortality time series were created, then β(t) was estimated from the data using [Left] the S method and [Right] the SI method, all without input error. For each set of estimates of β(t) (1000 estimates per parametrization, per simulation method, per estimation method), the median RRMSE was calculated (after smoothing with fixed q; see Eq (59)) and displayed as one point in the appropriate heatmap, coloured according to the logarithmic scale on the right. The darkest blue indicates median RRMSE less than 0.01.
Fig 6 plots median RRMSE as a univariate function of each of 6 additional parameters—the initial states S0 and I0, vital rates νc and μc, mean generation interval tgen, and case reporting probability prep—with the focal parameter assigned values between and 4 times its reference value (Table 1). The horizontal axis measures the ratio of the focal parameter’s data-generating value to its reference value, so that commensurate deviations from the reference case can be compared across the 6 parameters.
Fig 6. Sensitivity of β(t) estimation error to data-generating parameters other than 〈β〉 and α.
Plotted in each panel is the median RRMSE (Eq (33)) in estimates of a seasonally forced β(t) (Eq (27)) from simulated reported incidence time series (Δt = 1 week, n = 1042), as a univariate function of each of 5 or 6 data-generating parameters (indicated by the legend). When simulating reported incidence, reference values (Table 1) were assigned to all but the focal parameter, which was assigned 41 values logarithmically spaced between and 4 times its reference value. The horizontal axis (logarithmic scale) measures the ratio of the focal parameter’s true value to its reference value, so that commensurate deviations from the reference case can be compared across parameters. For each parametrization, 1000 simulations were performed with environmental stochasticity [ES] (ϵ = 0.5) and with or without demographic stochasticity [DS] and observation error [OE], as indicated by row: [Row A] without DS or OE (prep = 1, trep = 0 weeks), [Row B] with DS but without OE (prep = 1, trep = 0 weeks), or [Row C] with DS and OE (prep = 0.25 except when prep is the focal parameter, trep = 2 weeks). Corresponding mock birth and natural mortality time series were created, then β(t) was estimated from the data using [Left] the S method and [Right] the SI method, all without input error. For each set of estimates of β(t) (1000 estimates per parametrization, per simulation method, per estimation method), the median RRMSE was calculated (after smoothing with fixed q; see Eq (59)) and displayed as one point in the appropriate panel and graph.
In order to produce Figs 5 and 6, we assigned reference values (Table 1) to all but the focal data-generating parameter(s) (e.g., all except 〈β〉 and α in Fig 5). We fit loess curves βloess(t; q) to all raw estimates βk of β(t), and recorded the RRMSE in βloess(tk; q). Motivated by Fig 4B and Eq (59), we fixed q = q*, taking q* = 65 with the S method and q* = 53 with the SI method.
A pattern in our interpretation of Figs 5 and 6 below is that error in β(t) estimation is sensitive to a parameter if changes in that parameter (i) cause incidence Z(t) or prevalence I(t) to approach zero more frequently or more closely, or (ii) increase noise in estimated incidence Zk or estimated prevalence Ik. Both outcomes incorrectly increase noise in βk (cf. §3.2).
When the noise in βk is extreme, setting q = q* can undersmooth the time series (q* < qopt). In this case, smaller RRMSE is attainable by determining qopt and setting q = qopt. Nevertheless, we did not find qopt for each of the 5 × 106 time series considered by Figs 5 and 6, which would have increased the total computation time by a factor of 100. Consequently, Figs 5 and 6 may overestimate the sensitivity of β(t) estimation error to data-generating parameters. (In §S5.3 of S1 Text, we show that the quantitative effect of choosing q* over qopt is likely to be small.)
3.5.1 Sensitivity to the basic reproduction number and seasonal amplitude α (Fig 5)
For fixed α, median RRMSE was a non-monotonic function of . The reason is that changes in (effective) are responsible for dynamical transitions that alter the structure of solutions of the SIR model (1) [28, 42, 43]. Specifically, as is increased from 2 to 32, minimum incidence Zmin and minimum prevalence Imin on the attractor varies non-monotonically (see Fig 2 in [28]). Smaller Zmin and Imin yield more noise in βk, and correspondingly greater RRMSE. For fixed , Imin decreases monotonically as α is increased from 0 to 1 (see Fig 11 in [43]), so we expect median RRMSE to increase monotonically with α, as observed in Fig 5.
3.5.2 Sensitivity to the initial state (S0, I0) (Fig 6)
RRMSE is sensitive to the data-generating S0, but not I0. The reference values of S0 and I0 are taken from a point (S*, I*, R*) on the attractor of the SIR model (1) with seasonally forced β(t) and constant vital rates νc and μc (cf. §2.3.4). When S0 is far from S*, the solution of system (1) undergoes extreme fluctuation before relaxing to the attractor, and both Z and I approach zero during the transient, generating spurious noise at the start of the βk time series.
Note that I0 differing from I* has a much smaller effect on dynamics than S0 differing from S* by the same factor. Since I* ≪ S*, the perturbation of (S0, I0, R0) from the attractor is much smaller.
3.5.3 Sensitivity to vital rates νc and μc (Fig 6)
Median RRMSE was a non-monotonic function of the data-generating birth rate νc. This behaviour arises because scaling νc is dynamically equivalent to scaling by the same factor [2, 28], and median RRMSE is a non-monotonic function of (cf. §3.5.1 above).
Changing the data-generating natural mortality rate μc had a negligible effect on RRMSE. This is unsurprising, because natural death is dominated by recovery and disease-induced death in governing the rate of infected decrease. That is, γ ≫ μ(t) in Eq (1b), so changes in μc by up to a factor of 4 have little effect on dynamics.
3.5.4 Sensitivity to the mean generation interval tgen (Fig 6)
Median RRMSE increased rapidly as the data-generating tgen was made smaller than 2−4/5 (roughly 0.57) times its reference value of 13 days. A period-doubling bifurcation occurs near this value of tgen, and the attractor of the SIR model (1) acquires a 2-year cycle with much smaller Zmin and Imin (see §S5.3.1 of S1 Text). Propagation of noise to βk intensifies, resulting in greater RRMSE.
The performance of the S method fluctuates more as a function of tgen than that of the SI method. This occurs because the S method rounds tgen in the numerator of Eq (25c) to the nearest integer multiple of Δt, and the rounding error oscillates as a function of tgen. The SI method does not require rounding, so these fluctuations are not observed.
3.5.5 Sensitivity to the case reporting probability prep (Fig 6)
When the reported incidence data contain observation error (Fig 6C), RRMSE is additionally sensitive to the case reporting probability prep. Decreasing prep increases noise in reported incidence Ck (Eq (31)), which is propagated to estimated incidence Zk, estimated prevalence Ik, and in turn βk (cf. §3.2).
Fig 6 suggests weak sensitivity to prep. However, noise in Zk and Ik is amplified in βk to the extent that Z and I are close to zero (cf. §3.2). Hence, for example, if the data-generating tgen were assigned a value smaller than half its reference value of 13 days, then we would have observed more acute sensitivity to prep as a result of closer approaches to zero by Z and I (cf. §3.5.4 above).
3.5.6 S method versus SI method (Figs 5 and 6)
Both the S and SI methods performed well, estimating β(t) with median RRMSE less than 10% across most parametrizations. However, by resisting noise propagation (cf. §3.2), the SI method was significantly less sensitive to the data-generating parameters and to the addition of demographic stochasticity and observation error.
3.6 Sensitivity to mis-specification of input parameters
In §3.5, we considered the ideal situation in which the user knows the true (data-generating) values of the input parameters. Here, we examine the more realistic situation in which the user specifies input parameters with some error. The effect of mis-specification is particularly important for parameters that are difficult to estimate accurately, such as the case reporting probability prep. The details of our analysis are outlined in §2.6.2.
We restrict our attention to application of the SI method to reported incidence data simulated with process and observation error. Differences in RRMSE between methods of data simulation and β(t) estimation are dominated (by an order of magnitude) by the increase in RRMSE resulting from mis-specified input parameters.
Fig 7A plots the median RRMSE in estimates of β(t) from 1000 realizations of a reported incidence time series, as a univariate function of the factor by which an input parameter—one of the initial states S0 and I0, mean generation interval tgen, vital rates νc and μc, and case reporting parameters prep and trep—was mis-specified. The specified value of the focal parameter was varied between and 4 times its true (data-generating) value, and the remaining parameters were specified without error.
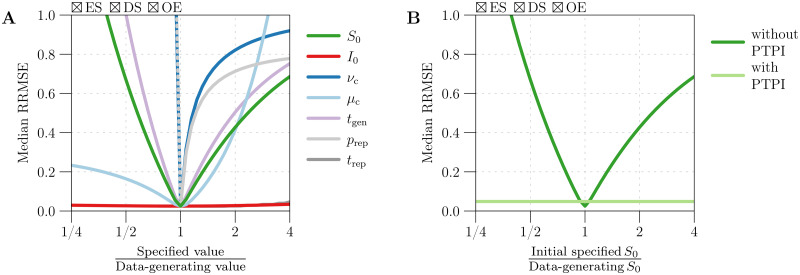
Fig 7. Sensitivity of β(t) estimation error to the user-specified values of input parameters.
[Panel A] Median RRMSE (Eq (33)) in estimates of β(t) from simulated reported incidence time series (Δt = 1 week, n = 1042), as a univariate function of the factor by which an input parameter was mis-specified. One thousand simulations were performed using fixed values (Table 1) for all data-generating parameters. The simulations accounted for environmental stochasticity [ES] (ϵ = 0.5), demographic stochasticity [DS], and observation error [OE] (prep = 0.25, trep = 2 weeks). For each simulation, corresponding mock birth and natural mortality time series were created, and β(t) was estimated from the data using the SI method. True (data-generating) values were specified for all input parameters except the focal parameter (indicated by the legend), for which 41 values logarithmically spaced between and 4 times the true value were specified in turn. Each input parametrization yielded 1000 estimates of β(t), whose median RRMSE was calculated (after smoothing with fixed q; see Eq (59)) and displayed as one point in the appropriate graph. [Panel B] Result of repeating the analysis from Panel A in which S0 was specified with varying amounts of error, but with the initially erroneous value of S0 updated using the method of peak-to-peak iteration (PTPI; 25 iterations) prior to β(t) estimation. The original result, obtained without PTPI, is presented for comparison.
3.6.1 Sensitivity to error in the specified initial state
Fig 7 shows that error in the specified value of S0 is propagated non-negligibly to estimates of β(t), while mis-specification of I0 has practically no effect on β(t) estimation error. Eqs (40) and (41) show that specifying incorrect values and for S0 and I0 creates errors in Sk and Ik that vanish geometrically as k → ∞. However, since tlife ≫ tinf, the decay is significantly slower in Sk. Indeed, with reference values μc = 0.04 year−1, tgen = γ−1 = 13 days, and Δt = 1 week, we find that a factor of 10 reduction in error between times tk and tk+i requires just i = 5 in the infected time series, compared to i = 3002 in the susceptible time series (roughly 58 years with Δt = 1 week). Hence, in practice, accurate reconstruction of S(t), I(t), and in turn β(t) relies on accurate specification of S0, but not I0. We address sensitivity to mis-specification of S0 in §3.7 below.
3.6.2 Sensitivity to error in the specified birth rate and case reporting probability
Mis-specifying νc or prep by a factor of 21/10 (7.2%) yielded median RRMSE greater than 30%. Mis-specifying by a factor of 2−1/10 (−6.7%) led to even worse estimates of β(t), with median RRMSE exceeding 100% (not visible in Fig 7A). Eqs (42) and (47) show that specifying incorrect values and for νc and prep generates absolute errors in Sk that tend to increase over time (k) to a limit. In practice, systematic underestimation of births by the Bk time series (modeled here by ) and overestimation of incidence by the Zk time series () can cause Sk to eventually take negative values. Once this happens, attempts by the S and SI methods to reconstruct β(t) fail completely.
While this failure may seem concerning, it should be viewed as a tool for diagnosing incorrect birth and case reporting rates: if the S or SI method yields negative Sk for any k, then one should speculate that births were underestimated or that incidence was overestimated, and retry the algorithm with a scaled up Bk time series and/or with greater prep (as Zk is computed by scaling reported incidence by a factor of ; see Eqs (25a) and (26a)). Of course, overcorrection is also undesirable (cf. right half of Fig 7A). In our work, we have found that a brief exploration of possible adjustments—factors by which to increase Bk and/or prep—suffices to identify ones that prevent both negative Sk and pronounced transient dynamics at the start of the susceptible time series (indicating under- or overcorrection).
3.7 Solution of the S0 estimation problem using PTPI
In §3.6, we showed that the performance of the S and SI methods is highly sensitive to mis-specification of the initial number of susceptibles S0. Here, we assess PTPI as a way to iteratively improve initially poor estimates of S0 prior to reconstruction of S(t) and β(t).
Fig 8 demonstrates PTPI for an example in which S0 was overestimated by a factor of 4 by a user of the SI method. PTPI yielded increasingly accurate estimates of S0 and correspondingly more accurate reconstructions of S(t) (Fig 8B) and β(t) (Fig 8C). Fig 7B repeats our analysis from §3.6, except using PTPI (25 iterations) to update the incorrect estimate of S0 prior to reconstructing β(t). We see that application of PTPI in conjunction with the SI method enables accurate β(t) reconstruction independently of errors in the initial estimate of S0. This result is unsurprising in light of Fig 9, which shows that PTPI converges rapidly (in fewer than 10 iterations) to an accurate estimate of S0 independently of the initial guess. Due to process error in the underlying dynamics, the relative error in the limiting estimate of S0 varied between the 1000 realizations of reported incidence considered (5th–95th percentile range [−11.9, 12.5]%, median 0.9%). Process error creates variance in the time between peaks in incidence (see Fig 8A), violating the periodicity assumption of PTPI (the theoretical basis of the technique; cf. §2.8). Nevertheless, Figs 7–9 demonstrate that PTPI can significantly improve S(t) and β(t) reconstruction from roughly periodic incidence data.
Fig 8. Example of S(t) and β(t) reconstruction with an overestimate of S0 corrected by peak-to-peak iteration (PTPI).
[Panel A] Truncation step of PTPI (Box 5). Plotted is a reconstruction of true incidence Z(t) from a simulated reported incidence time series, before [Zk, black] and after [, yellow] smoothing with a 13-point central moving average. Vertical lines indicate peaks in . The times of the first peak in and the last peak occurring at the same phase of the cycle (in this case, the last peak) are denoted by ta and tb. [Panel B] Iteration step of PTPI (Box 6), where the initial estimates of both S0 = S(0) and S(ta) were taken to be 4 times the true (data-generating) value of S0. Plotted in grey are successive reconstructions of S(t) between times ta and tb, generated by updating the estimate of S(ta) with the estimate of S(tb) obtained in the previous iteration. Dashed continuations to the left of ta display estimation of S0 backwards in time from estimates of S(ta). Plotted in black is the result of reconstructing S(t) starting from the final estimate of S0, which was obtained after 25 iterations and had a relative error of roughly 1.4% (compared to 300% in the initial estimate). [Panel C] The sequence of reconstructions of β(t) corresponding to the estimates of S0 shown in Panel B. [Details] Twenty years of weekly reported incidence (Δt = 1 week, n = 1042) were simulated with environmental noise in transmission (ϵ = 0.5), demographic stochasticity, and random under-reporting of cases (prep = 0.25), using reference values (Table 1) for the remaining parameters. Z(t), S(t) and β(t) were reconstructed from reported incidence using the SI method without input error (apart from mis-specification of S0).
Fig 9. Convergence of estimates of S0 obtained using peak-to-peak iteration (PTPI).
S0 was estimated by applying PTPI (25 iterations) to 1000 incidence time series (i.e., 1000 realizations of a reported incidence time series, scaled by ). An initial guess for S0 was taken to be or 4 times the true (data-generating) value. For each initial guess, this process generated 1000 sequences of 26 estimates of S0. Plotted are the median [black lines] and 5th–95th percentile range [grey bands] of the estimate of S0 at each iteration, for the first 10 iterations. The vertical axis measures (on a logarithmic scale) the ratio of the estimated and true values of S0, hence convergence close to 1 [dashed green line] represents convergence of the estimates close to the true value. [Details] One thousand reported incidence time series (Δt = 1 week, n = 1042) were simulated with environmental noise in transmission (ϵ = 0.5), demographic stochasticity, and random under-reporting of cases (prep = 0.25), using reference values (Table 1) for the remaining parameters, including S0 (hence S0 was the same in all simulations). True incidence was estimated from reported incidence via Eq (26a) (with reporting parameters prep and trep correctly specified), yielding 1000 time series of estimated incidence. Corresponding mock (constant) birth and natural mortality time series were created (with vital rates νc and μc correctly specified), and these data (estimated incidence, births, natural mortality) were passed to the PTPI algorithm, allowing for iterative re-estimation of S0.
3.8 Run time
We implemented the S and SI methods and PTPI in R and ran them on a MacBook Pro with a 2.4 GHz Quad-Core Intel Core i5 chip. The S and SI methods are both extremely fast, requiring a total of 0.124 and 0.376 seconds, respectively, to generate a reconstruction of β(t) from 1000 years of weekly reported incidence (Δt = 1 week, n = 52142). Application of PTPI in conjunction with either method increases the run time with each iteration, but the total run time remains inconsequential due to the rate of convergence of the iterations to a limiting estimate of S0. For example, when we applied PTPI to the same simulated data, the truncation step (Box 5) added 0.094 seconds to the total run time, while the iteration step (Box 5) added 1.01 seconds per iteration on average.
4 Discussion
We have compared three fast methods of estimating the time-varying transmission rate β(t) from reported incidence time series, all based on discretizations of the SIR model (1). Fine and Clarkson’s method [6], referred to here as the FC method, fails rapidly in practice, because it treats natural mortality in the susceptible population as negligible. Although Krylova’s method [24], adapted here as the S method, corrects this limitation of the FC method and is accurate for certain simulated data, her method suffers from extreme sensitivity to process and observation error. Specifically, noise in reported incidence is spuriously propagated to its estimates of β(t). Our algorithm for transmission rate estimation, referred to here as the SI method and based on deJonge’s method [25], is much more resilient to noise in reported incidence and therefore superior to the S method.
Like its predecessors, the SI method is sensitive to (i) certain input parameters: the initial number of susceptible individuals S0, the case reporting probability prep, and the mean generation interval tgen; as well as (ii) vital data: times series of births and natural mortality without substantial systematic errors.
The requirement of a good estimate of S0 has been a major barrier to use of existing fast methods of β(t) estimation (including those presented in [6, 24, 25]). We have proposed and demonstrated PTPI as a valid and fast technique for obtaining accurate estimates of S0 from poor initial guesses, conditional on periodic dynamics (epidemic recurrence with a fixed period). Use of the SI method in conjunction with PTPI represents a major advance over the existing fast methods.
Estimation of the case reporting probability prep is possible using maximum likelihood approaches, including trajectory matching. However, a fast way to obtain a crude estimate of prep is to divide cumulative reported incidence over the time interval [t0, tn], by the cumulative incidence that is expected from the unforced SIR model (system (1) with β ≡ 〈β〉, ν ≡ νc, and μ ≡ μc) at equilibrium:
| (60) |
This approximation can be made in temporal subintervals to obtain a time-varying reporting rate, which would replace the constant prep in Eq (26a). Sensitivity of the SI method to mis-specification of the mean generation interval (tgen) may be of greater concern, though if the distribution of the incubation period (time from infection to onset of symptoms) is narrow, then tgen will be well approximated by the (observable) mean serial interval [44].
Overall, the SI method, in conjunction with PTPI, represents a highly tractable approach to reconstructing susceptibles and β(t) from infectious disease time series that span decades or centuries. It makes fewer assumptions about the disease and population of interest than the regression-based tSIR method [7, 23] (i.e., it does not require an infectious period equal to the observation interval, ignore susceptible mortality, or assume that cumulative incidence approximates cumulative births). Moreover, it is significantly less complex and much less computationally demanding than simulation-based methods of inference, such as iterated filtering [8, 19, 20] and generalized profiling [21, 22].
Even when the observed infectious disease time series is short enough that simulation-based methods are tractable, the approach to transmission rate reconstruction that we promote here can be usefully employed to provide better starting conditions at negligible computational cost.
Supporting information
A .pdf document containing annotated R code, making the results reported here completely reproducible by the reader.
(PDF)
A .zip archive containing all of the source files needed to compile S1 Text.
(ZIP)
Acknowledgments
We thank Ben Bolker, Jonathan Dushoff, and Sang Woo Park for helpful comments and discussion.
Data Availability
All relevant data are found within the manuscript and Supporting Information files. Specifically, all of the data are simulated and can be generated reproducibly by running the R scripts contained in the Supporting Information files.
Funding Statement
MJ was supported by an Undergraduate Student Research Award from the Natural Sciences and Engineering Research Council of Canada (NSERC) and a Student Fellowship from the M. G. DeGroote Institute for Infectious Disease Research. OK was supported by a Postgraduate Scholarship from NSERC. DJDE was supported by a Discovery Grant from NSERC. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Dietz K. The incidence of infectious diseases under the influence of seasonal fluctuations. In: Mathematical Models in Medicine. vol. 11 of Lecture Notes in Biomathematics. Springer-Verlag Berlin / Hiedelberg; 1976. p. 1–15.
- 2. Earn DJD, Rohani P, Bolker BM, Grenfell BT. A simple model for complex dynamical transitions in epidemics. Science. 2000;287(5453):667–670. 10.1126/science.287.5453.667 [DOI] [PubMed] [Google Scholar]
- 3. Shaman J, Kohn M. Absolute humidity modulates influenza survival, transmission, and seasonality. Proceedings of the National Academy of Sciences. 2009;106(9):3243–3248. 10.1073/pnas.0806852106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. London W, Yorke JA. Recurrent outbreaks of measles, chickenpox and mumps. I. Seasonal variation in contact rates. American Journal of Epidemiology. 1973;98(6):453–468. 10.1093/oxfordjournals.aje.a121575 [DOI] [PubMed] [Google Scholar]
- 5. Hethcote HW. The mathematics of infectious diseases. SIAM Review. 2000;42(4):599–653. 10.1137/S0036144500371907 [DOI] [Google Scholar]
- 6. Fine PEM, Clarkson JA. Measles in England and Wales—I: an analysis of factors underlying seasonal patterns. International Journal of Epidemiology. 1982;11(1):5–14. 10.1093/ije/11.1.5 [DOI] [PubMed] [Google Scholar]
- 7. Finkenstädt B, Grenfell B. Time series modelling of childhood diseases: a dynamical systems approach. Journal of the Royal Statistical Society C (Applied Statistics). 2000;49(2):187–205. 10.1111/1467-9876.00187 [DOI] [Google Scholar]
- 8. He D, Ionides EL, King AA. Plug-and-play inference for disease dynamics: measles in large and small populations as a case study. Journal of the Royal Society Interface. 2010;7:271–283. 10.1098/rsif.2009.0151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hempel K, Earn DJD. A century of transitions in New York City’s measles dynamics. Journal of the Royal Society Interface. 2015;12(106):20150024 10.1098/rsif.2015.0024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pollicott M, Wang H, Weiss H. Extracting the time-dependent transmission rate from infection data via solution of an inverse ODE problem. Journal of Biological Dynamics. 2012;6(2):509–523. 10.1080/17513758.2011.645510 [DOI] [PubMed] [Google Scholar]
- 11. Lange A. Reconstruction of disease transmission rates: applications to measles, dengue, and influenza. Journal of Theoretical Biology. 2016;400:138–153. 10.1016/j.jtbi.2016.04.017 [DOI] [PubMed] [Google Scholar]
- 12. Wallinga J, Teunis P. Different epidemic curves for severe acute respiratory syndrome reveal similar impacts of control measures. American Journal of Epidemiology. 2004;160:509–516. 10.1093/aje/kwh255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Smirnova A, deCamp L, Chowell G. Forecasting epidemics through nonparametric estimation of time-dependent transmission rates using the SEIR model. Bulletin of Mathematical Biology. 2019;81:4343–4365. 10.1007/s11538-017-0284-3 [DOI] [PubMed] [Google Scholar]
- 14. Tien JH, Poinar HN, Fisman DN, Earn DJD. Herald waves of cholera in nineteenth century London. Journal of the Royal Society Interface. 2011;8(58):756–760. 10.1098/rsif.2010.0494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Krylova O, Earn DJD. Patterns of smallpox mortality in London, England, over three centuries. PLoS Biology. 2020. 10.1371/journal.pbio.3000506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Anderson RM, May RM. Infectious Diseases of Humans: Dynamics and Control. Oxford, UK: Oxford University Press; 1991. [Google Scholar]
- 17. Morton A, Finkenstädt B. Discrete time modelling of disease incidence time series by using Markov chain Monte Carlo methods. Journal of the Royal Statistical Society C (Applied Statistics). 2005;54(3):575–594. 10.1111/j.1467-9876.2005.05366.x [DOI] [Google Scholar]
- 18. Cauchemez S, Ferguson NM. Likelihood-based estimation of continuous-time epidemic models from time series data: application to measles transmission in London. Journal of the Royal Society Interface. 2008;5(25):885–897. 10.1098/rsif.2007.1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ionides EL, Breto C, King AA. Inference for nonlinear dynamical systems. Proceedings of the National Academy of Sciences. 2006;103(49):18438–18443. 10.1073/pnas.0603181103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. King AA, Nguyen D, Ionides EL. Statistical inference for partially observed Markov processes via the R package pomp. Journal of Statistical Software. 2009;69(12):1–43. [Google Scholar]
- 21. Ramsay JO, Hooker G, Campbell D, Cao J. Parameter estimation for differential equations: a generalized smoothing approach. Journal of the Royal Statistical Society B (Statistical Methodology). 2007;69(5):741–796. 10.1111/j.1467-9868.2007.00610.x [DOI] [Google Scholar]
- 22. Hooker G, Ellner SP, De Vargas Roditi L, Earn DJD. Parameterizing state-space models for infectious disease dynamics by generalized profiling: measles in Ontario. Journal of the Royal Society Interface. 2011;8(60):961–974. 10.1098/rsif.2010.0412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Becker A, T GB. tsiR: An R package for time series susceptible-infected-recovered models of epidemics. PLoS ONE. 2017;12(9):0185528 10.1371/journal.pone.0185528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krylova O. Predicting epidemiological transitions in infectious disease dynamics. Smallpox in historic London (1664–1930). Hamilton, Ontario, Canada: McMaster University; 2011. Available from: https://macsphere.mcmaster.ca/handle/11375/11231.
- 25.deJonge MS. Fast estimation of time-varying transmission rates. Hamilton, Ontario, Canada: McMaster University; 2014. Available from: https://macsphere.mcmaster.ca/handle/11375/14230.
- 26. Wallinga J, Lipsitch M. How generation intervals shape the relationship between growth rates and reproductive numbers. Proceedings of the Royal Society B (Biological Sciences). 2007;274:599–604. 10.1098/rspb.2006.3754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Champredon D, Dushoff J. Intrinsic and realized generation intervals in infectious-disease transmission. Proceedings of the Royal Society B (Biological Sciences). 2015;282(1821):20152026 10.1098/rspb.2015.2026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Krylova O, Earn DJD. Effects of the infectious period distribution on predicted transitions in childhood disease dynamics. Journal of the Royal Society Interface. 2013;10:20130098 10.1098/rsif.2013.0098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Brauer F, Castillo-Chavez C. Mathematical models in population biology and epidemiology. New York, NY: Springer; 2012. [Google Scholar]
- 30. Lloyd AL. Destabilization of epidemic models with the inclusion of realistic dvistributions of infectious periods. Proceedings of the Royal Society B (Biological Sciences). 2001;268(1470):985–993. 10.1098/rspb.2001.1599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lloyd AL. Realistic distributions of infectious periods in epidemic models: changing patterns of persistence and dynamics. Theoretical Population Biology. 2001;60(1):59–71. 10.1006/tpbi.2001.1525 [DOI] [PubMed] [Google Scholar]
- 32. Ma J, Ma Z. Epidemic threshold conditions for seasonally forced SEIR models. Mathematical Biosciences and Engineering. 2006;3(1):161–172. 10.3934/mbe.2006.3.161 [DOI] [PubMed] [Google Scholar]
- 33. Little RJA, Rubin DB. Statistical Analysis with Missing Data. Hoboken, NJ: John Wiley & Sons; 2019. [Google Scholar]
- 34. Goldstein E, Dushoff J, Ma J, Plotkin JB, Earn DJD, Lipsitch M. Reconstructing influenza incidence by deconvolution of daily mortality time series. Proceedings of the National Academy of Sciences. 2009;106:21825–21829. 10.1073/pnas.0902958106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. He D, Earn DJD. The cohort effect in childhood disease dynamics. Journal of the Royal Society Interface. 2016;13:20160156 10.1098/rsif.2016.0156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cleveland WS, Grosse E, Shyu WM. Local regression models. In: Chambers JM, Hastie TJ, editors. Statistical models in S. London, UK: Chapman & Hall; 1991. p. 309–376.
- 37. Loader C. Local Regression and Likelihood. New York, NY: Springer-Verlag New York; 1999. [Google Scholar]
- 38. Hart JD. Automated kernel smoothing of dependent data by using time series cross-validation. Journal of the Royal Statistical Society B (Statistical Methodology). 1994;56(3):529–542. [Google Scholar]
- 39. Gillespie DT. Stochastic simulation of chemical kinetics. Annual Review of Physical Chemistry. 2007;58:35–55. 10.1146/annurev.physchem.58.032806.104637 [DOI] [PubMed] [Google Scholar]
- 40.Johnson P. adaptivetau: Tau-leaping stochastic simulation; 2016. Available from: https://CRAN.R-project.org/package=adaptivetau.
- 41. Elaydi S. An Introduction to Difference Equations. New York, NY: Springer; 2005. [Google Scholar]
- 42. Bauch CT, Earn DJD. Transients and attractors in epidemics. Proceedings of the Royal Society of London B. 2003;270(1524):1573–1578. 10.1098/rspb.2003.2410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Earn DJD. Mathematical epidemiology of infectious diseases. In: Lewis MA, Chaplain MAJ, Keener JP, Maini PK, editors. Mathematical biology. vol. 14 of IAS Park City Mathematics Series. American Mathematical Society; 2009. p. 151–186.
- 44. Fine PEM. The interval between successive cases of an infectious disease. American Journal of Epidemiology. 2003;158(11):1039–1047. 10.1093/aje/kwg251 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A .pdf document containing annotated R code, making the results reported here completely reproducible by the reader.
(PDF)
A .zip archive containing all of the source files needed to compile S1 Text.
(ZIP)
Data Availability Statement
All relevant data are found within the manuscript and Supporting Information files. Specifically, all of the data are simulated and can be generated reproducibly by running the R scripts contained in the Supporting Information files.