Abstract
Postpartum depressive disorder (PPD) is a unique subtype of major depressive disorder and a substantial contributor to maternal morbidity and mortality. However, the pathogenesis of PPD has still remained elusive, and it may associate with genetic and environmental factors. Gut microbiota has already been proved to be associated with depression; however, a limited number of studies have concentrated on PPD. The present study aimed to explore the potential correlations between gut microbiota and PPD. In this study, 57 participants were enrolled, in which fecal samples of 28 patients with PPD and 16 healthy controls (HCs) were collected and then analyzed by high-throughput sequencing of the 16S ribosomal RNA (rRNA) gene. The results showed that diversity and composition of gut microbial communities were partly different between PPD patients and HCs. The relative abundance of Firmicutes phyla was lower in PPD patients. The levels of several predominant genera were significantly different between PPD patients and HCs. More importantly, the PPD patients experienced reduced levels of Faecalibacterium, Phascolarctobacterium, Butyricicoccus, and Lachnospiraceae, as well as increased levels of Enterobacteriaceae family. In addition, a correlation was observed between levels of Phascolarctobacterium, Lachnospiraceae, Faecalibacterium, and Tyzzerella.3 and the severity of depressive symptoms. Various kinds of bacteria, such as Lachnospiraceae and Faecalibacterium, were found to be associated with levels of sex hormones. This study indicated the correlation between gut microbiota and PPD, and gut microbiota-based biomarkers may be helpful for the diagnosis and treatment of PPD patients. However, further studies need to be conducted to clarify the cause–effect relationship between PPD patients and gut microbiota and to highlight the suitability of gut microbiome as a biomarker.
Keywords: postpartum depressive disorder, gut microbiota, 16SrRNA gene, gut-brain, sex hormone
Introduction
Postpartum depressive disorder (PPD) is a type of mood disorder associated with childbirth, and it can affect both sexes, associating with the morbidity of 7.1% in the first three postnatal months (Gavin et al., 2005). The symptoms of PPD include extreme sadness, low energy, anxiety, crying episodes, irritability, and changes in sleeping or eating patterns, thereby resembling a major depressive disorder (MDD). PPD severely influences mothers' quality of life and daily activities, and it can also negatively affect the newborn child (Netsi et al., 2018; Weissman, 2018). Despite the fact that multiple antidepressants are recommended for alleviating symptoms of depression, <50% of PPD patients have obvious responses to the existing drug therapies (Hansen et al., 2008). Hormonal abnormalities, neuroendocrine processes, or inflammation all play different roles in depression, although interactions between genetic and environmental factors have significantly attracted scholars' attention. In recent years, mounting evidence has supported the viewpoints that gut microbiome is closely associated with the function and behavior of the brain (Clemente et al., 2012).
The human microbiome, a complex assemblage of the microbes, inhabits, and interacts with human hosts, which are classified into beneficial or pathogenic bacteria. In the human body, great numbers of microbes can be found in the gastrointestinal tract, especially in the colon with the densest and most diverse microbial community (Tremaroli and Backhed, 2012). The gastrointestinal microbiota modulates host function from various aspects: apart from the decomposition of food residues and production of micronutrients, they could produce short-chain fatty acids (SCFAs) (Cummings et al., 1987) and neuroactive substance (Barrett et al., 2012), affect hypothalamic–pituitary–adrenal (HPA) axis (Ait-Belgnaoui et al., 2012) and gut barrier, and promote the balance within the immune system (Clemente et al., 2012).
A number of scholars have supported and characterized “microbiome–gut–brain (MGB) axis” and illuminated a possible role of gut microbiota dysbiosis in a variety of diseases, such as obesity (Torres-Fuentes et al., 2017), inflammatory bowel disease (Lahtinen et al., 2020), and hypertension (Li et al., 2020). Although previous studies have demonstrated that microbial communities may influence our health, further attention needs to be paid to the potential associations between maternal gut microbiome and PPD. However, several studies have suggested that alterations of gut microbiota may influence other depressive disorders. Compared with healthy subjects, the relative abundance of Bacteroidetes, Proteobacteria, and Actinobacteria has markedly increased in MDD patients, and it was unveiled that Faecalibacterium was negatively correlated with the severity of symptoms of depression (Jiang et al., 2015). Another study pointed out that the relative abundance of Firmicutes, Actinobacteria, and Bacteroidetes has remarkably changed in MDD patients, and fecal microbiota transplantation of Germ-free (GF) mice with “depression microbiota” derived from MDD patients resulted in depression-like behaviors (Zheng et al., 2016). In those studies, the gut microbiota related to inflammatory status, oxidative stress, and disease severity could be further identified (Jiang et al., 2015). Additionally, animal studies showed that changes of gut microbiota were associated with depressive-like behaviors (Foster and McVey Neufeld, 2013), and probiotic supplementation could prevent and alleviate anxiety and depression in mice by regulating gut microbiota dysbiosis (Jang et al., 2019). Besides, a number of scholars have reported the characteristics of maternal microbiome during pregnancy or in the postpartum period. A study assessed the relationship between psychosocial stress and fecal microbiota in pregnant women, and their findings revealed a significant association between anxiety in late pregnancy and women's fecal microbiota composition at the genus level. More specifically, the fecal microbiota of mothers with lower anxiety could be characterized by higher abundance of the Eubacterium and Oscillospira compared with mothers with higher prenatal anxiety (Hechler et al., 2019). A randomized controlled trial (RCT) demonstrated that targeted supplementation with probiotics can correct PPD and postpartum anxiety-associated behavioral abnormalities (Slykerman et al., 2017). On the basis of these findings, we speculated that gut microbiota dysbiosis may be involved in the development of PPD.
The present study aimed to investigate whether the gut microbiota could be changed in PPD patients and identify the specific microbiota for PPD via high-throughput sequencing of the 16S ribosomal RNA (rRNA) gene. Additionally, the associations between gut microbiota and clinical patterns were explored.
Materials and Methods
Ethics, Consent, and Permissions
This study was approved by the Ethics Committee of Shenzhen Traditional Chinese Medicine Hospital [Shenzhen, China; Registration No. (2018), 81]. All procedures were designed and conducted in accordance with the Declaration of Helsinki. Eligible participants were informed about all the procedures, benefits, as well as potential risks that they may encounter in this trial, and they could withdraw the study at any time without any specific reason. All the participants signed the written informed consent form prior to commencing the study.
Recruitment of Study Subjects
Healthy participants and patients with PPD were included in this study. All participants were recruited from the Shenzhen Traditional Chinese Medicine Hospital and Shenzhen Maternity & Child Healthcare Hospital (Shenzhen, China). The PPD was diagnosed according to the Fourth Edition of the Diagnostic and Statistical Manual of Mental Disorders (DSM-IV). Participants who gave birth within 1 year were recruited for further evaluation. People were enrolled in this study from June 2019 to October 2019.
The inclusion criteria for patients with PPD were as follows: (1) patients who are aged 20–49 years old; (2) patients who were diagnosed with PPD by a psychiatrist; (3) onset of disease within 12 months after delivery; (4) the scores of 17-item Hamilton depression rating scale (17-HAMD) ranging from 7 to 24; and (5) signing the written informed consent form. Patients with any one of the following items were excluded: (1) bipolar disorder (diagnostic criteria according to DSM-IV) or serious mental disorders, such as schizophrenia; (2) dysnoesia or having difficulty in understanding the content of the questionnaire due to brain diseases or other reasons, or incapable of effective interview; (3) pregnancy; (4) the score of “suicide” item in the 17-HAMD would be more than 2; (5) having committed suicide within 1 year; or (6) having anorexia nervosa.
The inclusion criteria for healthy participants were as follows: (1) participants who are aged 20–49 years old; (2) no obvious discomfort; (3) normal health examination after delivery (routine blood count, liver and kidney functions, and electrocardiogram); (4) 17-HAMD score would be <7; and (5) signing the written informed consent form for voluntarily participation in this study. Participants with any of the following conditions were excluded: (1) with pregnancy; (2) having committed suicide in the last year; (3) the score of “suicide” item in 17-HAMD would be >2; or (4) participants who were involved in other clinical trials.
Evaluation of Clinical Scales
All participants completed the evaluation process of 17-HAMD and the Edinburgh Postnatal Depression Scale (EPDS). The 17-HAMD, a 17-item scale, was designed to measure the frequency and intensity of depressive symptoms in individuals with MDD (Hamilton, 1960). The 17-HAMD scores range from 0 to 52: “7 < scores ≤ 17” indicate clinically mild depression; “17 < scores ≤ 24” are indicative of moderate depression; “scores > 24” denote severe depression. The EPDS, a self-reporting scale, consists of 10 items with acceptable sensitivity, specificity, and positive predictive values (Cox et al., 1987), containing mood, fun, self-accusation, anxiety, fear, insomnia, coping ability, sadness, crying, and self-injury. Each item is divided into four grades: never (0 point), occasionally (1 point), often (2 points), and always (3 points). The total score of EPDS ranges from 0 to 30, and scores higher than 13 are indicative of clinically significant depression.
Measurement of Serum Levels of Sex Hormones
Blood samples were collected immediately after the 17-HAMD and EPDS assessment, then transferred to the laboratory, and stored in −20°C refrigerator for 15 min until preparation for further analysis. The serum levels of sex hormones, including follicle-stimulating hormone (FSH), luteinizing hormone (LH), prolactinemia (PRL), progesterone (PROG), estradiol (E2), and testosterone (TESTO), were detected using commercially available enzyme-linked immunosorbent assay (ELISA) kits (RayBiotech, Norcross, GA, USA).
Analysis of Clinical Characteristics
Demographic and clinical characteristics of healthy controls (HCs) and patients with PPD were compared using SPSS 22.0 software (IBM, Armonk, NY, USA). For continuous data, normally distributed data were analyzed by the Student's t-test and expressed as mean ± standard deviation, while abnormally distributed data were analyzed by the Mann–Whitney U-test and presented as median or interquartile range (IQR). For count data, the chi-square test was used. P < 0.05 was considered statistically significant.
Collection of Fecal Samples
Fecal samples of all participants were collected after being enrolled within 2 days after 17-HAMD and EPDS evaluation, which were put into a sterile plastic cup and immediately stored at −20°C after defecation, and further transported at −80°C storage to the laboratory of Shenzhen Traditional Chinese Medicine Hospital. Additionally, for hospitalized patients, the collected samples were stored in a −80°C refrigerator. The details of fecal sample collection have been previously described (Zhou et al., 2019).
DNA Extraction
DNA extraction was carried out using MOBIO PowerSoil® DNA Isolation kit, and DNA was stored at −80°C in Tris-EDTA buffer solution. To perform amplification of the V4 region of the 16S rRNA gene and add barcode sequences, unique fusion primers were designed based on the universal primer set, 515F (5′-GTGYCAGCMGCCGCGGTAA-3′) and 806R (5′-GGACTACNVGGGTWTCTAAT-3′), along with barcode sequences. PCR mixtures contained 1 μl of each forward and reverse primer (10 μM), 1 μl of template DNA, 4 μl of dNTPs (2.5 mM), 5 μl of 10 × EasyPfu Buffer, 1 μl of Easy Pfu DNA Polymerase (2.5 U/μl), and 1 μl of double-distilled water in a 50-μl reaction volume. Thermal cycling consisted of an initial denaturation step at 95°C for 5 min, followed by 30 cycles of denaturation at 94°C for 30 s, annealing at 60°C for 30 s, and extension at 72°C for 40 s, with a final extension step at 72°C for 4 min. Amplicons were run for each sample on an agarose gel. Expected band size for 515f-806r was ~300–350 bp. Amplicons were quantified with Quant-iT PicoGreen dsDNA Assay Kit (P11496; Thermo Fisher Scientific, Waltham, MA, USA) according to manufacturer's instructions. The amplicon library for high-throughput sequencing on the Illumina MiSeq V3 reagent PE150 (300 cycles) platform was combined to an equal amount and subsequently quantified using KAPA Library Quantification Kit (KK4824; Illumina, Inc., San Diego, CA, USA) according to manufacturer's protocols.
High-Throughput Sequencing of 16S Ribosomal RNA Gene
Using the Quantitative Insights Into Microbial Ecology (QIIME) 2.0, the raw sequences were processed to concatenate reads into tags according to the overlapping relationship; then, reads belonging to each sample were separated with barcodes, and low-quality reads were removed. The processed tags were clustered into the Amplicon Sequence Variants (ASVs) at the commonly used 97% similarity threshold. The ASVs were assigned to taxa by matching to the SILVA database. A phylogenetic tree of representative sequences was constructed. Evenness, observed species, Shannon, and Faith-PD indices were used to estimate the α-diversity. The measurement of β-diversity was undertaken using UniFrac that is a β-diversity measure that uses phylogenetic information, and the principal coordinate analysis (PCoA) was employed to calculate the distance matrixes. To further identify the specific bacteria as biomarkers at the genus level, linear discriminant analysis effect size (LEfSe) was applied through the Huttenhower Lab Galaxy Server (Segata et al., 2011) after taxa summaries were reformatted. In the setting of LEfSe, firstly, the Kruskal–Wallis test (α = 0.05) was employed to detect taxa using differential abundance analysis; secondly, the Wilcoxon rank-sum test was used to investigate the biological consistency among subclasses; finally, the effect size of differentially abundant genera was estimated by linear discriminant analysis (LDA) (Segata et al., 2011), and the threshold on the logarithmic LDA score for discriminative features was 2. All the analyses were conducted using “vegan” package in R 3.4.1 software. Correlations between clinical variables and bacterial taxa were analyzed by using Spearman's Rho test.
Results
Participants' Demographic and Clinical Data
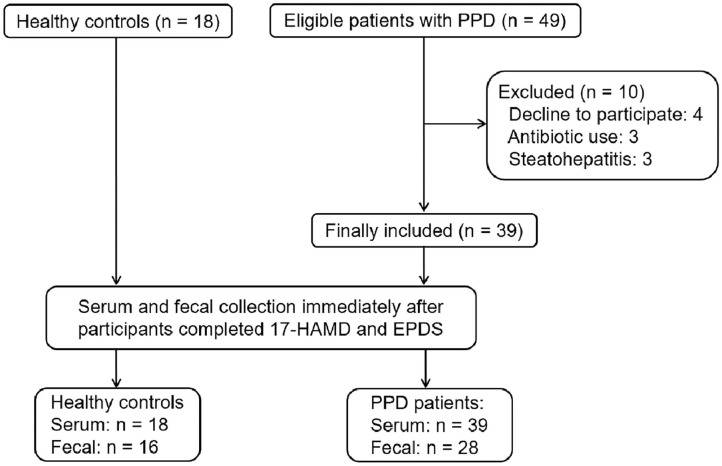
From June 7, 2019 to October 15, 2019, 67 participants were recruited from Shenzhen Traditional Chinese Medicine Hospital and Shenzhen Maternity & Child Healthcare Hospital. Of the 67 participants, 10 patients withdrew from the study. Finally, 18 HCs and 39 patients with PPD were included in the study. All participants completed the 17-HAMD and EPDS evaluation. Among HCs, 18 participants completed the serum sample collection; however, two cases did not complete the fecal sample collection. Among patients with PPD, 39 patients completed the serum sample collection, and only 28 cases completed the fecal sample collection (Figure 1).
Figure 1.
The recruitment of participants and the process of sample collection.
Demographic characteristics, such as age, body mass index (BMI), education background, duration of postpartum, etc., showed no significant difference between the HC group and PPD group (Table 1; P > 0.05). For the EPDS and 17-HAMD, the total score was significantly higher in the PPD group than that in the HC group (Table 1; P < 0.001). Other baseline information was also collected, including special diets, gastrointestinal disorders, metabolic disorders, and antibiotic/probiotic treatments. The surveys showed that all puerpera had no irritable bowel syndrome and bowel cancer; however, some of them had constipation (Table 1). None had hypertension or diabetes after childbirth, while three participants were excluded for steatohepatitis (Figure 1). Additionally, another three patients with antibiotic use were ruled out as well (Figure 1). Furthermore, all the participants were from Shenzhen (China), whose favorable diet was easy-to-digest foods and less consumption of greasy, Atsumi, and spicy food. All the eligible participants had a general appetite and nutritious food with balanced portion of vegetables and meat.
Table 1.
Baseline characteristics and clinical symptoms in patients with PPD and HCs.
| Parameter | HC group | PPD group | P-value |
|---|---|---|---|
| (n = 18) | (n = 39) | ||
| Sociodemographics | |||
| Age, mean (SD), years | 32.57 ± 3.98 | 33.64 ± 4.27 | 0.376 |
| BMI, mean (SD) | 20.90 ± 2.24 | 21.50 ± 2.87 | 0.434 |
| High school or less, No. (%) | 2 (11.11%) | 7 (17.95%) | 0.275 |
| Duration of postpartum, mean | 142.94 ± 129.00 | 112.00 ± 91.84 | 0.372 |
| (SD), days | |||
| Menstruating, No. (%) | 11 (61.1%) | 30 (76.0%) | 0.217 |
| Constipation, No. (%) | 5 (27.8%) | 16 (41.0%) | 0.335 |
| Severity of depressive symptoms | |||
| HAMDs | 3.83 ± 1.98 | 13.46 ± 3.51 | <0.001* |
| EPDS | 5.72 ± 3.72 | 15.33 ± 4.66 | <0.001* |
HC, healthy control; PPD, postpartum depressive disorder; HAMDS, Hamilton's Depression Scale; EPDS, Edinburgh Postnatal Depression Scale. P < 0.05 is marked with *.
The serum levels of sex hormones in the PPD group were compared with those in the HC group (Table 2). No significant differences in the levels of FSH, LH, and PROG were observed between the two groups. However, the levels of E2 and TESTO in the PPD group were lower than those in the HC group (P = 0.036 and 0.012, respectively), and the PRL level in the PPD group was higher than that in the HC group (P = 0.001).
Table 2.
Sex hormone levels in patients with PPD and HCs.
| Parameter | HCs group | PPD group | P-value |
|---|---|---|---|
| (n = 18) | (n = 39) | ||
| FSH, mean (SD), ng/ml | 9.05 ± 5.74 | 8.59 ± 3.01 | 0.757 |
| LH, mean (SD), ng/ml | 14.73.90 ± 28.25 | 6.04 ± 5.26 | 0.212 |
| E2, mean (SD), ng/ml | 240.61 ± 331.17 | 109.42 ± 125.33 | 0.036* |
| PRL, mean (SD), ng/ml | 346.43 ± 265.54 | 872.82 ± 860.03 | 0.001* |
| PROG, mean (SD), ng/ml | 4.86 ± 13.49 | 2.005 ± 4.41 | 0.392 |
| TESTO, mean (SD), ng/ml | 1.27 ± 0.61 | 0.82 ± 0.612 | 0.012* |
HC, healthy control; PPD, postpartum depressive disorder; FSH, follicle-stimulating hormone; LH: luteinizing hormone; E2, estradiol; PRL, prolactinemia; PROG, progesterone; TESTO, testosterone. P < 0.05 is marked with *.
Collection of 16S Ribosomal RNA Sequences
Herein, 44 samples from participants who completed 16S rRNA sequences were collected, in which 1,765,950 qualified sequences from 1,852,840 raw sequences were filtered. Then, a total of 979 qualified ASVs were clustered for downstream analysis. The mean number of ASVs per sample was 92.70, and the standard deviation was 23.86. The sequencing results could be achieved from 28 PPD patients and 16 HCs.
Analysis of Microbial α- and β-Diversity
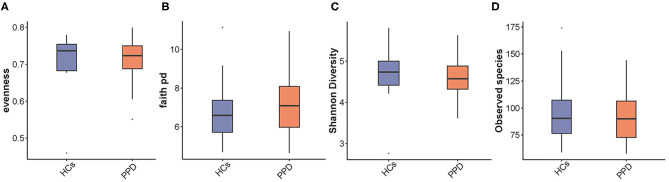
The indices of fecal bacterial α-diversity are shown in Figure 2. There were no significant differences between the PPD group and HC group, including observed species, Evenness, Shannon, and Faith-PD indices (P = 0.669, 0.526, 0.367, and 0.435, respectively) (Figure 2). However, the observed species seemed to be much higher in the HC group than those in the PPD group (Figure 2D).
Figure 2.
An analysis of microbial α-diversity. Evenness index (A); Faith pd index (B); Shannon diversity (C); Observed species (D).
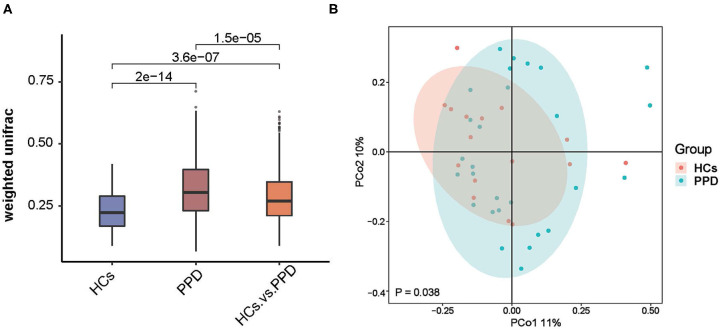
The indices of fecal bacterial β-diversity are illustrated in Figure 3. The Wilcoxon rank-sum test and PCoA were performed to measure differences in β-diversity between the two groups using weighted UniFrac distance metrics (Figures 3A,B, respectively). The sample-based differences in the PPD group were significantly higher than those in the HC group (P = 2e-14) (Figure 3A). Additionally, as displayed in Figure 3B, the result of PCoA unveiled that there was a significant difference in bacterial communities between the PPD group and HC group (P = 0.038). The results disclosed that the indices of fecal bacterial β-diversity in the HC group were more centralized than those in the PPD group.
Figure 3.
An analysis of microbial β-diversity. The Wilcoxon rank-sum test analysis (A) and Principal coordinates analysis plots (B) of the fecal microbiome based on the weighted-UniFrac distance metric.
Composition of Microbial Communities
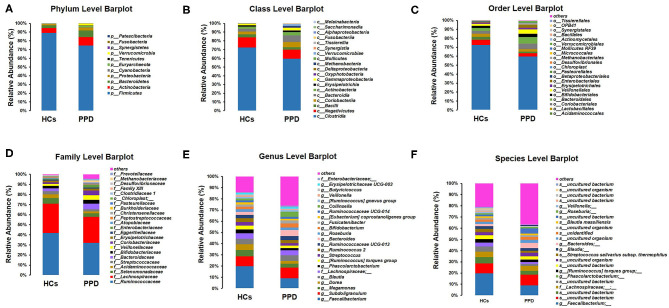
The histograms of species were made for the two groups at the levels of phylum, class, order, family, and genus on the basis of the annotation results. The histograms showing the relative abundance of species uncovered the composition (species and corresponding proportion) of microbial communities in each group with higher relative abundance at different levels (Figures 4A–F). The analysis of the composition of the gut microbiota at the levels of phylum and genus reflected the entire structure of gut microbiota.
Figure 4.
Composition of microbial communities. Phylum level (A); Class level (B); Order level (C); Family level (D); Genus level (E); Species level (F).
On the whole, there were significant differences in the composition of the gut microbiota at the phylum level (Figure 4A). Besides, 10 and 11 phyla were detected in the HC group and PPD group by sequence alignment analysis using the SILVA database, respectively. p_Patescibacteria was only found in the PPD group, although its relative abundance was low. In both groups, the proportion of p_Firmicutes was the highest among all phyla, which was higher in the HC group (88.91%) than that in PPD group (74.57%). p_Actinobacteria and p_Bacteroidetes ranked the second and third, while they were both relatively lower in the HC group than those in the PPD group.
Furthermore, we analyzed the characteristics and alterations in community structure of the gut microbiota in the two groups at the genus level according to the relative abundance (Figure 4E, Table 3). Among all bacteria, g_Faecalibacterium was the most dominant one, accounting for 19.79% in HC group and 9.22% in the PPD group. The prominent changes of community structure of the gut microbiota at the genus level are related to the decrease of g_Faecalibacterium (P = 0.003), g_Phascolarctobacterium (P = 0.022), g_Butyricicoccus (P = 0.024), and g_Megasphaera (P = 0.047) in the PPD group compared to those in the HC group (Table 3).
Table 3.
Relative abundance of gut microbial communities at the genus level.
| Tax_name | HCs (%) | PPD (%) | P-value | |
|---|---|---|---|---|
| 1 | g_Faecalibacterium | 19.7913 | 9.2243 | 0.003 |
| 2 | g_Subdoligranulum | 8.9888 | 9.4907 | 0.778 |
| 3 | g_Megamonas | 5.6413 | 3.4550 | 0.366 |
| 4 | g_Dorea | 5.0988 | 2.3821 | 0.090 |
| 5 | g_Blautia | 4.9138 | 5.1350 | 0.826 |
| 6 | f_Lachnospiraceae;_;_ | 4.7738 | 4.3886 | 0.534 |
| 7 | g_Phascolarctobacterium | 3.4688 | 1.9243 | 0.022 |
| 8 | g_[Ruminococcus] torques group | 3.3713 | 2.3286 | 0.163 |
| 9 | g_Streptococcus | 3.3200 | 2.0829 | 0.227 |
| 10 | g_Ruminococcus 2 | 3.2888 | 2.7879 | 0.864 |
| 11 | g_Ruminococcaceae UCG-013 | 2.9538 | 3.5471 | 0.788 |
| 12 | g_Bacteroides | 2.6638 | 5.5364 | 0.714 |
| 13 | g_Roseburia | 2.4225 | 2.0914 | 0.485 |
| 14 | g_Bifidobacterium | 2.3725 | 3.6564 | 0.288 |
| 15 | g_Fusicatenibacter | 1.9988 | 2.8257 | 0.668 |
| 16 | g_[Eubacterium] coprostanoligenes group | 1.8713 | 1.4979 | 0.837 |
| 17 | g_Ruminococcaceae UCG-014 | 1.7888 | 1.6050 | 0.598 |
| 18 | g_Collinsella | 1.5913 | 4.5729 | 0.225 |
| 19 | g_[Ruminococcus] gnavus group | 1.5125 | 1.4343 | 0.882 |
| 20 | g_Veillonella | 1.1950 | 0.5864 | 0.193 |
| 21 | g_Butyricicoccus | 1.0888 | 0.6300 | 0.024 |
| 22 | g_Erysipelotrichaceae UCG-003 | 1.0800 | 1.2386 | 0.892 |
| 23 | f_Enterobacteriaceae;_ | 0.6763 | 1.5679 | 0.712 |
| 24 | g_Megasphaera | 0.6125 | 0.4636 | 0.047 |
| 25 | g_Anaerostipes | 0.6025 | 0.7200 | 0.345 |
| 26 | g_Tyzzerella 4 | 0.5763 | 0.4671 | 0.516 |
| 27 | g_Adlercreutzia | 0.5600 | 0.2793 | 0.221 |
| 28 | g_Olsenella | 0.5200 | 0.1986 | 0.972 |
| 29 | Others | 11.2575 | 23.8821 |
HC, healthy control; PPD, postpartum depressive disorder.
Comparing Differences in Bacterial Genus Between the Healthy Control Group and Postpartum Depressive Disorder Group
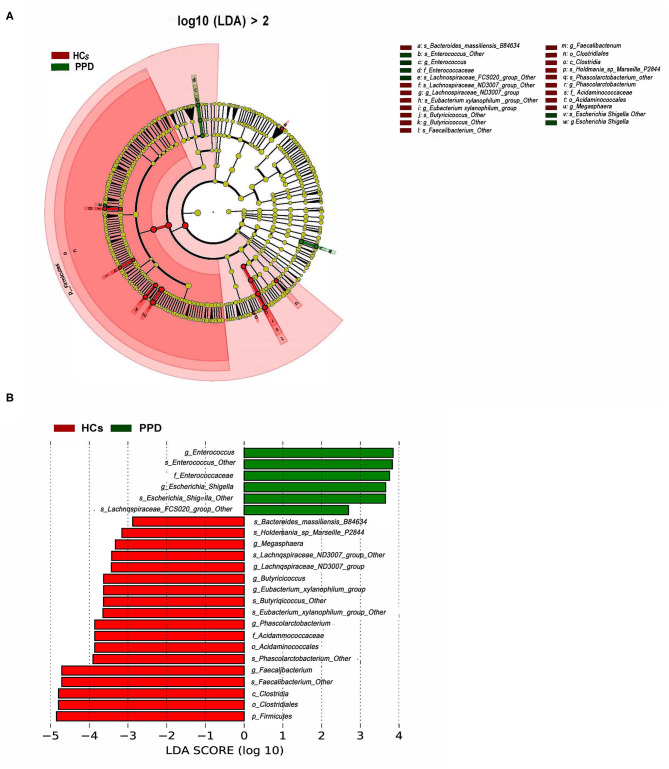
To further investigate the differences in the abundance between the two groups and to explore the specific bacteria associated with PPD, LEfSe analysis was utilized (P < 0.05, LDA value > 2). The most significant difference among the two groups was that Faecalibacterium, Phascolarctobacterium, Butyricicoccus, Lachnospiraceae, Acidaminococcaceae, Eubacterium_xylanophilum, and Megasphaera were mainly enriched in the HC group, and Enterocossus and Escherichia_Shigella were mainly enriched in the PPD group (Figures 5A,B).
Figure 5.
Bacterial taxa differences between healthy control (HC) and postpartum depressive disorder (PPD) patient samples. Cladogram showing the most differentially abundant taxa identified by linear discriminant analysis effect size (LEfSe). Red indicates clades enriched in the HC group, whereas blue indicates clades enriched in the PPD group (A). Comparisons of gut microbiota between the NC and PPD groups (B). Only genera meeting a linear discriminant analysis score threshold >2 are shown.
Associations of Gut Microbiota With Clinical Indicators and Sex Hormones
Associations of Gut Microbiota With Clinical Indicators
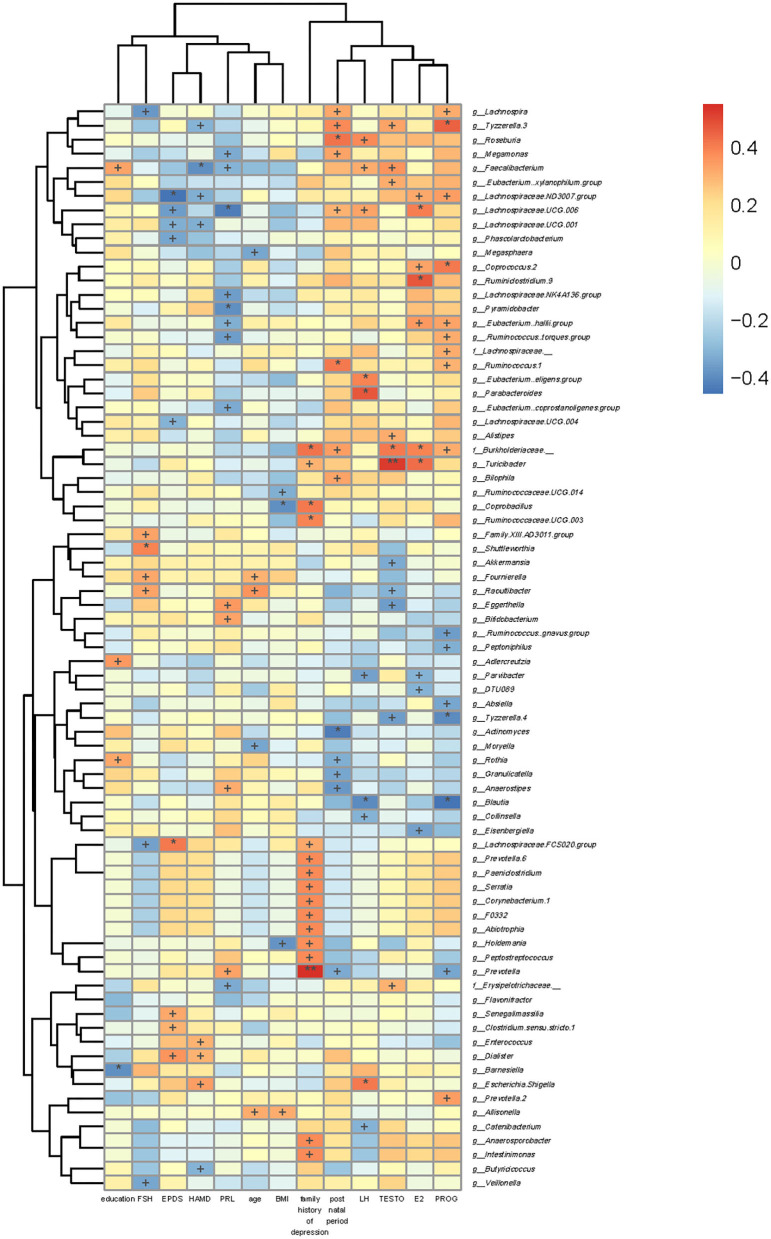
BMI was positively correlated with g_Allisonella but negatively correlated with g_Holdemania, g_Coprobacillus, and g_Ruminococcaceae.UCG.014. Age was positively correlated with g_Allisonella, g_Raoultibacter, and g_Fournierella, while that was negatively correlated with g_Moryella and g_Megasphaera.
EPDS scores were positively correlated with g_Dialister, g_Clostridium.sensu.stricto.1, g_Senegalimassilia, and g_Lachnospiraceae.FCS020.group, while those were negatively correlated with g_Lachnospiraceae.UCG.004, g_Phascolarctobacterium, g_Lachnospiraceae.UCG.001, g_Lachnospiraceae.UCG.006, and g_Lachnospiraceae.ND3007.group. The 17-HAMD scores were positively correlated with g_Escherichia.Shigella, g_Dialister, and g_Enterococcus, while those were negatively correlated with g_Butyricicoccus, g_Lachnospiraceae.UCG.001, g_Lachnospiraceae.ND3007.group, g_Faecalibacterium, and g_Tyzzerella.3 (Figure 6).
Figure 6.
Association of gut microbiota with clinical indicators. The heat map of Spearman's rank correlation coefficients between the gut microbiota and clinical indicators. +P < 0.05; *P < 0.01; **P < 0.001.
Associations of Gut Microbiota With Levels of Sex Hormones
We also analyzed the associations between levels of sex hormones and gut microbiota. FSH level was positively correlated with g_Raoultibacter, g_Fournierella, g_Shuttleworthia, and g_Family.XIII.AD3011.group, while that was negatively correlated with g_Veillonella, g_Lachnospiraceae.FCS020.group, and g_Lachnospira. Moreover, PRL level was negatively correlated with f_Erysipelotrichaceae_,g_Eubacterium..coprostanoligenes.group, g_Ruminococcus..torques.group, g_Lachnospiraceae.NK4A136.group, g_Pyramidobacter, g_Eubacterium..hallii.group, g_Lachnospiraceae.UCG.006, g_Megamonas, and g_Faecalibacterium, while that was positively correlated with g_Prevotella, g_Anaerostipes, g_Bifidobacterium, and g_Eggerthella. PROG level was positively correlated with g_Prevotella.2, f_Burkholderiaceae._, g_.Eubacterium..hallii.group, g_.Ruminococcus..torques.group, f_Lachnospiraceae._, g_Ruminococcus.1, g_Coprococcus.2, g_Lachnospiraceae.ND3007.group, g_Lachnospira, and g_Tyzzerella.3, whereas that was negatively correlated with g_Prevotella, g_Blautia, g_Absiella, g_Tyzzerella.4, g_.Ruminococcus..gnavus.group, and g_Peptoniphilus. TESTO level was positively correlated with f_Erysipelotrichaceae._, g_Alistipes, f_Burkholderiaceae._, g_Turicibacter, g_Faecalibacterium, g_.Eubacterium.xylanophilum.group, and g_Tyzzerella.3, while it was negatively correlated with g_Tyzzerella.4, g_Raoultibacter, g_Eggerthella, and g_Akkermansia. LH level was negatively correlated with g_Catenibacterium, g_Blautia, g_Collinsella, g_Parvibacter, g_Lachnospiraceae.UCG.006, g_Faecalibacterium, and g_Roseburia, while that was positively correlated with g_Escherichia.Shigella, g_.Eubacterium..eligens.group, and g_Parabacteroides. E2 level was g_Eisenbergiella, g_Parvibacter, and g_DTU089, while that was positively correlated with f_Burkholderiaceae._, g_Turicibacter, g_.Eubacterium..hallii.group, g_Coprococcus.2, g_Ruminiclostridium.9, g_Lachnospiraceae.ND3007.group, and g_Lachnospiraceae.UCG.006 (Figure 6).
Discussion
In the current study, we characterized the gut microbiota of PPD and HC patients by high-throughput sequencing of the 16S rRNA gene. Moreover, associations of gut microbiota with clinical indicators and sex hormones were assessed. The results showed that diversity and composition of gut microbial communities were remarkably different between the PPD group and HC group. We found that some butyrate-producing genera were enriched in the HC group, such as Faecalibacterium, Phascolarctobacterium, and Butyricicoccus. Additionally, the overgrowth of Enterococcaceae and Escherichia_Shigella was detected in the PPD group. Moreover, the present study revealed that the amounts of specific genera were correlated with clinical indicators and levels of sex hormones in the PPD group, which indicated that sex hormones might play a significant role in the gut microbiome of patients with PPD.
Additionally, no significant difference in α-diversity was noted between the PPD group and HC group. Similarly, a recent research revealed that there were no significant differences in the Shannon's index and Simpson's index between patients with MDD and non-depressed controls (Sanada et al., 2020). However, β-diversity of gut microbiota, referring to the sample-based differences, was markedly greater in the PPD group than those in the control group. Various factors may influence the diversity of gut microbiota, including age, diet, and health status, and a number of studies suggested that keeping a rich diversity of gut microbiota is highly beneficial for protecting from autoimmune and metabolic diseases (Manichanh et al., 2006; Claesson et al., 2012; Le Chatelier et al., 2013). However, other studies demonstrated that microbial diversity is reduced in patients with MDD (Dantzer et al., 2018; Winter et al., 2018). Hu et al. (2019) found that diversity of gut microbiota was noticeably different between patients with bipolar disorders and HCs. Another one studied the interaction between antidepression treatments and gut microbiota in a mouse model of depression and found an obvious increase of species richness in acupuncture-treated mice, which indicated that the diversity of the gut microbiota may be a factor influencing depression (Fu, 2019). Thus, due to controversial results about bacterial diversity, further research is warranted to explore the differences in bacterial α- and β-diversity between patients with PPD and HCs.
The present study demonstrated that the microbial composition in the PPD group was noticeably different from that in the HC group. Phylum Firmicutes ranked the first in both groups, and it was markedly lower in the PPD group than that in the HC group, which was consistent with findings of a previous research (Hu et al., 2019). Additionally, we found that Escherichia_Shigella was enriched in the PPD group. Escherichia_Shigella, belonging to the Enterobacteriaceae family, is Gram-negative bacteria and observed in normal gut flora. Overgrowth of Enterbacteriaceae could result in gut inflammation and increased permeability of the gut wall, which in turn favors bacterial translocation, promoting systemic inflammation (Winter and Bäumler, 2014). Clinical depression is accompanied by increased pro-inflammatory cytokine interleukin (IL), such as IL-1β and IL-6 (Berk et al., 2013; Wong et al., 2016). Some studies found an anti-inflammatory effect of Faecalibacterium, Bifidobacterium, and Lactobacillus on stress responses and depressive disorders (Jiang et al., 2015; Aizawa et al., 2016). Moreover, previous studies have also found that the increase of Enterobacteriaceae in the gastrointestinal tract can induce behavioral and psychological changes in animals and humans (Goehler et al., 2008; Löwe et al., 2014; Jiang et al., 2015; Borgo et al., 2017). More importantly, various bacteria with decreased abundance were found in the PPD group, including Faecalibacterium, Phascolarctobacterium, and Butyricicoccus. It was in accordance with some studies (Jiang et al., 2015; Zheng et al., 2016) that reported lower abundance of Faecalibacterium in depressive patients compared with non-depressed individuals, while the study by Chen et al. (2018) presented a contrary result. Faecalibacterium is a genus of bacteria, and Faecalibacterium prausnitzii, its only known species, produces butyric acid and other SCFAs (Louis and Flint, 2009). In addition, a recent study demonstrated that the intake of Faecalibacterium prausnitzii improves anxiety-related and depressive-like behaviors in the preclinical setting (Hao et al., 2019), and the study by Jiang et al. (2015) showed a negative correlation between the abundance of Faecalibacterium and the severity of depression symptoms. Besides, Phascolarctobacterium, affiliated with the Acidaminococcaceae at the family level, was significantly higher in the HC group, which is in line with a study that assessed and compared patients with MDD with non-depressed controls (Jiang et al., 2015). However, another study (Jeffery et al., 2012) concentrated on patients with both irritable bowel syndrome and depression and found that Acidaminococcaceae significantly overgrew using pyrosequencing fecal samples. Additionally, we also noted lower abundance of Butyricicoccus at the genus level in the PPD group. Butyricicoccus belongs to the clostridial cluster IV genus of the Firmicutes phylum, which is typically decreased in patients with inflammatory bowel disease (Eeckhaut et al., 2013). The inflammatory bowel conditions have been found to be correlated with high comorbidity with depression and anxiety (Bhandari et al., 2017). Moreover, we found that the abundance of Butyricicoccus was positively associated with EPDS scores. Butyricicoccus pullicaecorum is an anaerobic and butyrate-producing bacterium from the genus Butyricicoccus. Butyrate, one of the main products of colonic microbiota, has been found beneficial for a variety of diseases, such as insulin resistance and ischemic stroke (Canani et al., 2011). Furthermore, butyrate in the central nervous system can influence the function of the hippocampus and promote the expression of brain-derived neurotrophic factor (BDNF), which has been shown to have antidepressant-like effects in animal models (Yamawaki et al., 2012; Wei et al., 2014). Thus, decreased butyrate-producing bacteria in PPD patients may contribute to the disease pathology.
Furtherly, we noted that the gut microbial communities were relevant to confounding factors, including clinical indicators (age, BMI, etc.), disease severity, and levels of sex hormones. Among the PPD patients, BMI was found to be positively correlated with g_Allisonella but negatively correlated with g_Holdemania, g_Coprobacillus, and g_Ruminococcaceae.UCG.014. In another study that involved patients with bipolar disorders, Holdemania at the genus level was also found to be negatively correlated with BMI (Hu et al., 2019). Another study showed that the abundance of Ruminococcaceae increased in obesity patients (Chávez-Carbajal et al., 2019), and Ruminococcaceae was associated with antidepressant effects in rats under chronic mild stress (Tung et al., 2019). Moreover, the increased levels of Ruminococcaceae would decrease after weight reduction (Kang et al., 2017). Ruminococcaceae is a dominant butyrate producer. Butyrate supports the energy for colonic mucosa and makes a different regulatory effect on gene expression and inflammation (Pajak et al., 2007; Hamer et al., 2008; Ivanov and Honda, 2012). These findings indicated that Ruminococcaceae may be related to the development of PPD through metabolic pathways. Besides, Holdemania at the genus level is involved in glucose metabolism and metabolic syndrome (Lippert et al., 2017).
Previous studies have reported some associations between the levels of several genera and depression severity in MDD (Naseribafrouei et al., 2014; Jiang et al., 2015). In the present study, EPDS scores were negatively associated with Phascolarctobacterium and Lachnospiraceae at the genus level, and 17-HAMD scores were positively associated with Escherichia.Shigella, Dialister, and Enterococcus and negatively associated with Butyricicoccus, Lachnospiraceae, Faecalibacterium, and Tyzzerella.3 at the genus level. Hence, gut microbiota may play a pivotal role in the metabolic disturbance in PPD patients.
With respect to the causes of PPD, although it has been poorly understood, a number of human and animal studies supported the role of sex hormones in PPD (Schiller et al., 2015). In the current study, we also found that the serum levels of E2, PROG, and TESTO were noticeably different between the PPD group and HC group. Recently, various studies have reported that changes in the levels of sex hormones could be related to diversity and profiles of gut microbiota (Baker et al., 2017; Shin et al., 2019). The gut microbiota has been shown to be influenced by E2; meanwhile, the gut microbiota also could regulate E2 level through secreting β-glucuronidase (Flores et al., 2012; Huang et al., 2017). In the present research, our results also found that some bacteria genera were associated with serum sex hormone levels in PPD patients, such as Faecalibacterium, Lachnospiraceae, and Megamonas, which were significantly different from those in HCs. A recent study has demonstrated that the regulation of sex hormones–microbiota–inflammation axis could ameliorate polycystic ovary syndrome (PCOS), including Faecalibacterium, Parabacteroides, Bifidobacterium, and so on (Wang et al., 2020). Another study showed that prenatal androgen exposure causes hypertension and gut microbiota dysbiosis (Sherman et al., 2018). Moreover, a study found that small amounts of brain E2 and PROG could improve menopausal symptoms by decreasing serum FSH levels and maintaining the diversity of the gut microbiome in estrogen-deficient rats (Park et al., 2018). Our study used EPDS and 17-HAMD scores to assess symptoms of depression in PPD patients and found that Faecalibacterium and Lachnospiraceae were correlated with disease severity. Hence, the abovementioned findings showed that the interaction of sex hormones and gut microbiota may play a substantial role in PPD.
Conclusions
In summary, our findings may provide further evidence to support a number of previous reports that the gut microbial composition of PPD patients partially differs from that of HCs, and we explored new associations among gut microbiota, disease severity, and sex hormones. We also noted that Faecalibacterium, Phascolarctobacterium, and Butyricicoccus were significantly decreased in patients with PPD, which were all butyrate-producing, as well as Enterobacteriaceae family increased obviously. In addition, we demonstrated that Phascolarctobacterium, Lachnospiraceae, Faecalibacterium, and Tyzzerella.3 were correlated with disease severity; besides, various kinds of bacteria, such as Lachnospiraceae and Faecalibacterium, were found to be associated with the levels of sex hormones. The abovementioned results may assist scholars to further explore the underlying pathogenesis of PPD. Moreover, the identified microbiota in this study could be a potential diagnostic biomarker of PPD. Therefore, our findings may provide significant clues for future researches.
Limitations
Serval limitations of this study should be pointed out. Firstly, this observational study may not contain reliable indicators of causal effects; thus, further longitudinal studies on patients with depression are required to clarify the cause–effect relationship between PPD and gut microbiota. Secondly, due to the small sample size, we could not adjust for multiple testing or ethnicity, and our results still need to be validated by further studies with larger sample sizes. Therefore, future studies may elucidate the temporal and causal relationships between gut microbiota and PPD.
Data Availability Statement
The original contributions presented in the study are publicly available. This data can be found here: https://www.ncbi.nlm.nih.gov/, with accession number PRJNA637228.
Ethics Statement
The studies involving human participants were reviewed and approved by Medical Ethical Review committee of Shenzhen Traditional Chinese Medicine Hospital (Shenzhen, China; Approval No. [2018], 81). The patients/participants provided their written informed consent to participate in this study.
Author Contributions
YZ conceived and planned the experiments and wrote the manuscript. YZ and CC executed the experiments. HY analyzed the data. ZY contributed to revise the final manuscript. All authors approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the Shenzhen Maternity and Child Healthcare Hospital and the Shenzhen Mental Health Center/Shenzhen Kangning Hospital for their expert advice.
Footnotes
Funding. This study was supported by the Natural Science Foundation of Guangdong Province (2019A1515110657), China Postdoctoral Science Foundation (2019M663123), National Key Research and Development Plan of China (2017YFC1703604), and Shenzhen Sanming Project (SZSM201612001).
References
- Ait-Belgnaoui A., Durand H., Cartier C., Chaumaz G., Eutamene H., Ferrier L., et al. (2012). Prevention of gut leakiness by a probiotic treatment leads to attenuated HPA response to an acute psychological stress in rats. Psychoneuroendocrinology 37, 1885–1895. 10.1016/j.psyneuen.2012.03.024 [DOI] [PubMed] [Google Scholar]
- Aizawa E., Tsuji H., Asahara T., Takahashi T., Teraishi T., Yoshida S., et al. (2016). Possible association of Bifidobacterium and Lactobacillus in the gut microbiota of patients with major depressive disorder. J. Affect. Disord. 202, 254–257. 10.1016/j.jad.2016.05.038 [DOI] [PubMed] [Google Scholar]
- Baker J. M., Al-Nakkash L., Herbst-Kralovetz M. M. (2017). Estrogen-gut microbiome axis: physiological and clinical implications. Maturitas 103, 45–53. 10.1016/j.maturitas.2017.06.025 [DOI] [PubMed] [Google Scholar]
- Barrett E., Ross R. P., O'Toole P. W., Fitzgerald G. F., Stanton C. (2012). γ-aminobutyric acid production by culturable bacteria from the human intestine. J. Appl. Microbiol. 113, 411–417. 10.1111/j.1365-2672.2012.05344.x [DOI] [PubMed] [Google Scholar]
- Berk M., Williams L. J., Jacka F. N., O'Neil A., Pasco J. A., Moylan S., et al. (2013). So depression is an inflammatory disease, but where does the inflammation come from? BMC Med. 11:200. 10.1186/1741-7015-11-200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhandari S., Larson M. E., Kumar N., Stein D. (2017). Association of inflammatory bowel disease (IBD) with depressive symptoms in the United States population and independent predictors of depressive symptoms in an IBD population: a NHANES study. Gut Liver 11, 512–519. 10.5009/gnl16347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borgo F., Riva A., Benetti A., Casiraghi M. C., Bertelli S., Garbossa S., et al. (2017). Microbiota in anorexia nervosa: the triangle between bacterial species, metabolites and psychological tests. PLoS ONE 12:e0179739. 10.1371/journal.pone.0179739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canani R. B., Costanzo M. D., Leone L., Pedata M., Meli R., Calignano A. (2011). Potential beneficial effects of butyrate in intestinal and extraintestinal diseases. World J. Gastroenterol. 17, 1519–1528. 10.3748/wjg.v17.i12.1519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chávez-Carbajal A., Nirmalkar K., Pérez-Lizaur A., Hernández-Quiroz F., Ramírez-Del-Alto S., García-Mena J., et al. (2019). Gut microbiota and predicted metabolic pathways in a sample of mexican women affected by obesity and obesity plus metabolic syndrome. Int. J. Mol. Sci. 20:438. 10.3390/ijms20020438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J. J., Zheng P., Liu Y. Y., Zhong X. G., Wang H. Y., Guo Y. J., et al. (2018). Sex differences in gut microbiota in patients with major depressive disorder. Neuropsychiatr. Dis. Treat. 14, 647–655. 10.2147/NDT.S159322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claesson M. J., Jeffery I. B., Conde S., Power S. E., O'Connor E. M., Cusack S., et al. (2012). Gut microbiota composition correlates with diet and health in the elderly. Nature 488, 178–184. 10.1038/nature11319 [DOI] [PubMed] [Google Scholar]
- Clemente J. C., Ursell L. K., Parfrey L. W., Knight R. (2012). The impact of the gut microbiota on human health: an integrative view. Cell 148, 1258–1270. 10.1016/j.cell.2012.01.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J. L., Holden J. M., Sagovsky R. (1987). Detection of postnatal depression. Development of the 10-item Edinburgh postnatal depression scale. Br. J. Psychiatry 150, 782–786. 10.1192/bjp.150.6.782 [DOI] [PubMed] [Google Scholar]
- Cummings J. H., Pomare E. W., Branch W. J., Naylor C. P., Macfarlane G. T. (1987). Short chain fatty acids in human large intestine, portal, hepatic and venous blood. Gut 28, 1221–1227. 10.1136/gut.28.10.1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dantzer R., Cohen S., Russo S. J., Dinan T. G. (2018). Resilience and immunity. Brain Behav. Immun. 74, 28–42. 10.1016/j.bbi.2018.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eeckhaut V., Machiels K., Perrier C., Romero C., Maes S., Flahou B., et al. (2013). Butyricicoccus pullicaecorum in inflammatory bowel disease. Gut 62, 1745–1752. 10.1136/gutjnl-2012-303611 [DOI] [PubMed] [Google Scholar]
- Flores R., Shi J., Fuhrman B., Xu X., Veenstra T. D., Gail M. H., et al. (2012). Fecal microbial determinants of fecal and systemic estrogens and estrogen metabolites: a cross-sectional study. J. Transl. Med. 10:253. 10.1186/1479-5876-10-253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster J. A., McVey Neufeld K. A. (2013). Gut-brain axis: how the microbiome influences anxiety and depression. Trends Neurosci. 36, 305–312. 10.1016/j.tins.2013.01.005 [DOI] [PubMed] [Google Scholar]
- Fu C. (2019). Effects of Acupuncture Treatment on Intestinal Flora in Mice with Chronic Stress Anxiety Model. Beijing: Beijing University of Chinese Medicine. [Google Scholar]
- Gavin N. I., Gaynes B. N., Lohr K. N., Meltzer-Brody S., Gartlehner G., Swinson T. (2005). Perinatal depression: a systematic review of prevalence and incidence. Obstet. Gynecol. 106(5 Pt 1), 1071–1083. 10.1097/01.AOG.0000183597.31630.db [DOI] [PubMed] [Google Scholar]
- Goehler L. E., Park S. M., Opitz N., Lyte M., Gaykema R. P. (2008). Campylobacter jejuni infection increases anxiety-like behavior in the holeboard: possible anatomical substrates for viscerosensory modulation of exploratory behavior. Brain Behav. Immun. 22, 354–366. 10.1016/j.bbi.2007.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamer H. M., Jonkers D., Venema K., vanhoutvin S., Troost F. J., Brummer R. J. (2008). Review article: the role of butyrate on colonic function. Aliment. Pharmacol. Ther. 27, 104–119. 10.1111/j.1365-2036.2007.03562.x [DOI] [PubMed] [Google Scholar]
- Hamilton M. (1960). A rating scale for depression. J. Neurol. Neurosurg. Psychiatr. 23, 56–62. 10.1136/jnnp.23.1.56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen R., Gaynes B., Thieda P., Gartlehner G., Deveaugh-Geiss A., Krebs E., et al. (2008). Meta-analysis of major depressive disorder relapse and recurrence with second-generation antidepressants. Psychiatr. Serv. 59, 1121–1130. 10.1176/ps.2008.59.10.1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Z., Wang W., Guo R., Liu H. (2019). Faecalibacterium prausnitzii (ATCC 27766) has preventive and therapeutic effects on chronic unpredictable mild stress-induced depression-like and anxiety-like behavior in rats. Psychoneuroendocrinology 104, 132–142. 10.1016/j.psyneuen.2019.02.025 [DOI] [PubMed] [Google Scholar]
- Hechler C., Borewicz K., Beijers R., Saccenti E. (2019). Association between psychosocial stress and fecal microbiota in pregnant women. Sci. Rep. 9:4463. 10.1038/s41598-019-40434-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S., Li A., Huang T., Lai J., Li J., Sublette M. E., et al. (2019). Gut microbiota changes in patients with bipolar depression. Adv. Sci. 6:1900752. 10.1002/advs.201900752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang G., Xu J., Lefever D. E., Glenn T. C., Nagy T., Guo T. L. (2017). Genistein prevention of hyperglycemia and improvement of glucose tolerance in adult non-obese diabetic mice are associated with alterations of gut microbiome and immune homeostasis. Toxicol. Appl. Pharmacol. 332, 138–148. 10.1016/j.taap.2017.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov I. I., Honda K. (2012). Intestinal commensal microbes as immune modulators. Cell Host Microbe 12, 496–508. 10.1016/j.chom.2012.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang H. M., Lee K. E., Kim D. H. (2019). The preventive and curative effects of lactobacillus reuteri NK33 and bifidobacterium adolescentis NK98 on immobilization stress-induced anxiety/depression and colitis in mice. Nutrients 11:819. 10.3390/nu11040819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffery I. B., O'Toole P. W., Ohman L., Claesson M. J., Deane J., Quigley E. M., et al. (2012). An irritable bowel syndrome subtype defined by species-specific alterations in faecal microbiota. Gut 61, 997–1006. 10.1136/gutjnl-2011-301501 [DOI] [PubMed] [Google Scholar]
- Jiang H., Ling Z., Zhang Y., Mao H., Ma Z., Yin Y., et al. (2015). Altered fecal microbiota composition in patients with major depressive disorder. Brain Behav. Immun. 48, 186–194. 10.1016/j.bbi.2015.03.016 [DOI] [PubMed] [Google Scholar]
- Kang C., Wang B., Kaliannan K., Wang X., Lang H., Hui S., et al. (2017). Gut microbiota mediates the protective effects of dietary capsaicin against chronic low-grade inflammation and associated obesity induced by high-fat diet. mBio 8:e00470-17. 10.1128/mBio.00470-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahtinen P., Jalanka J., Hartikainen A., Mattila E., Hillilä M., Punkkinen J., et al. (2020). Randomised clinical trial: faecal microbiota transplantation versus autologous placebo administered via colonoscopy in irritable bowel syndrome. Aliment. Pharmacol. Ther. 51, 1321–1331. 10.1111/apt.15740 [DOI] [PubMed] [Google Scholar]
- Le Chatelier E., Nielsen T., Qin J., Prifti E., Hildebrand F., Falony G., et al. (2013). Richness of human gut microbiome correlates with metabolic markers. Nature 500, 541–546. 10.1038/nature12506 [DOI] [PubMed] [Google Scholar]
- Li J., Yang X., Zhou X., Cai J. (2020). The role and mechanism of intestinal flora on blood pressure regulation and hypertension development. Antioxid. Redox Signal. 10.1089/ars.2020.8104 [DOI] [PubMed] [Google Scholar]
- Lippert K., Kedenko L., Antonielli L., Kedenko I., Gemeier C., Leitner M., et al. (2017). Gut microbiota dysbiosis associated with glucose metabolism disorders and the metabolic syndrome in older adults. Benef. Microbes 8, 545–556. 10.3920/BM2016.0184 [DOI] [PubMed] [Google Scholar]
- Louis P., Flint H. J. (2009). Diversity, metabolism and microbial ecology of butyrate-producing bacteria from the human large intestine. FEMS Microbiol. Lett. 294, 1–8. 10.1111/j.1574-6968.2009.01514.x [DOI] [PubMed] [Google Scholar]
- Löwe B., Andresen V., Fraedrich K., Gappmayer K., Wegscheider K., Treszl A., et al. (2014). Psychological outcome, fatigue, and quality of life after infection with shiga toxin-producing Escherichia coli O104. Clin. Gastroenterol. Hepatol. 12, 1848–1855. 10.1016/j.cgh.2014.02.035 [DOI] [PubMed] [Google Scholar]
- Manichanh C., Rigottier-Gois L., Bonnaud E., Gloux K., Pelletier E., Frangeul L., et al. (2006). Reduced diversity of faecal microbiota in Crohn's disease revealed by a metagenomic approach. Gut 55, 205–211. 10.1136/gut.2005.073817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naseribafrouei A., Hestad K., Avershina E., Sekelja M., Linlokken A., Wilson R., et al. (2014). Correlation between the human fecal microbiota and depression. Neurogastroenterol. Motil. 26, 1155–1162. 10.1111/nmo.12378 [DOI] [PubMed] [Google Scholar]
- Netsi E., Pearson R. M., Murray L., Cooper P., Craske M. G., Stein A. (2018). Association of persistent and severe postnatal depression with child outcomes. JAMA Psychiatry 75, 247–253. 10.1001/jamapsychiatry.2017.4363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajak B., Orzechowski A., Gajkowska B. (2007). Molecular basis of sodium butyrate-dependent proapoptotic activity in cancer cells. Adv. Med. Sci. 52, 83–88. [PubMed] [Google Scholar]
- Park S., Kim D. S., Kang E. S., Kim D. B., Kang S. (2018). Low-dose brain estrogen prevents menopausal syndrome while maintaining the diversity of the gut microbiomes in estrogen-deficient rats. Am. J. Physiol. Endocrinol. Metab. 315, E99–E109. 10.1152/ajpendo.00005.2018 [DOI] [PubMed] [Google Scholar]
- Sanada K., Nakajima S., Kurokawa S., Barceló-Soler A., Ikuse D., Hirata A., et al. (2020). Gut microbiota and major depressive disorder: a systematic review and meta-analysis. J. Affect. Disord. 266, 1–13. 10.1016/j.jad.2020.01.102 [DOI] [PubMed] [Google Scholar]
- Schiller C. E., Meltzer-Brody S., Rubinow D. R. (2015). The role of reproductive hormones in postpartum depression. CNS Spectr. 20, 48–59. 10.1017/S1092852914000480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segata N., Izard J., Waldron L., Gevers D., Miropolsky L., Garrett W. S., et al. (2011). Metagenomic biomarker discovery and explanation. Genome Biol. 12:R60. 10.1186/gb-2011-12-6-r60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman S. B., Sarsour N., Salehi M., Schroering A., Mell B., Joe B., et al. (2018). Prenatal androgen exposure causes hypertension and gut microbiota dysbiosis. Gut Microbes 9, 400–421. 10.1080/19490976.2018.1441664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J. H., Park Y. H., Sim M., Kim S. A., Joung H., Shin D. M. (2019). Serum level of sex steroid hormone is associated with diversity and profiles of human gut microbiome. Res. Microbiol. 170, 192–201. 10.1016/j.resmic.2019.03.003 [DOI] [PubMed] [Google Scholar]
- Slykerman R. F., Hood F., Wickens K., Thompson J. M. D., Barthow C., Murphy R., et al. (2017). Effect of Lactobacillus rhamnosus HN001 in pregnancy on postpartum symptoms of depression and anxiety: a randomised double-blind placebo-controlled trial. EBioMedicine 24, 159–165. 10.1016/j.ebiom.2017.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Fuentes C., Schellekens H., Dinan T. G., Cryan J. F. (2017). The microbiota-gut-brain axis in obesity. Lancet Gastroenterol. Hepatol. 2, 747–756. 10.1016/S2468-1253(17)30147-4 [DOI] [PubMed] [Google Scholar]
- Tremaroli V., Backhed F. (2012). Functional interactions between the gut microbiota and host metabolism. Nature 489, 242–249. 10.1038/nature11552 [DOI] [PubMed] [Google Scholar]
- Tung T. H., Tung Y. T., Lin I. H., Shih C. K., Nguyen N. T. K., Shabrina A., et al. (2019). Fish oil, but not olive oil, ameliorates depressive-like behavior and gut microbiota dysbiosis in rats under chronic mild stress. Biomolecules 9:516. 10.3390/biom9100516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T., Sha L., Li Y., Zhu L., Wang Z., Li K., et al. (2020). Dietary α-linolenic acid-rich flaxseed oil exerts beneficial effects on polycystic ovary syndrome through sex steroid hormones-microbiota-inflammation axis in rats. Front. Endocrinol. 11:284. 10.3389/fendo.2020.00284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y., Melas P. A., Wegener G., Mathé A. A., Lavebratt C. (2014). Antidepressant-like effect of sodium butyrate is associated with an increase in TET1 and in 5-hydroxymethylation levels in the Bdnf gene. Int. J. Neuropsychopharmacol. 18:pyu032. 10.1093/ijnp/pyu032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissman M. M. (2018). Postpartum depression and its long-term impact on children: many new questions. JAMA Psychiatry 75, 227–228. 10.1001/jamapsychiatry.2017.4265 [DOI] [PubMed] [Google Scholar]
- Winter G., Hart R. A., Charlesworth R. P. G., Sharpley C. F. (2018). Gut microbiome and depression: what we know and what we need to know. Rev. Neurosci. 29, 629–643. 10.1515/revneuro-2017-0072 [DOI] [PubMed] [Google Scholar]
- Winter S. E., Bäumler A. J. (2014). Dysbiosis in the inflamed intestine: chance favors the prepared microbe. Gut Microbes 5, 71–73. 10.4161/gmic.27129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong M. L., Inserra A., Lewis M. D., Mastronardi C. A., Leong L., Choo J., et al. (2016). Inflammasome signaling affects anxiety- and depressive-like behavior and gut microbiome composition. Mol. Psychiatry 21, 797–805. 10.1038/mp.2016.46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamawaki Y., Fuchikami M., Morinobu S., Segawa M., Matsumoto T., Yamawaki S. (2012). Antidepressant-like effect of sodium butyrate (HDAC inhibitor) and its molecular mechanism of action in the rat hippocampus. World J. Biol. Psychiatry 13, 458–467. 10.3109/15622975.2011.585663 [DOI] [PubMed] [Google Scholar]
- Zheng P., Zeng B., Zhou C., Liu M., Fang Z., Xu X., et al. (2016). Gut microbiome remodeling induces depressive-like behaviors through a pathway mediated by the host's metabolism. Mol. Psychiatry 21, 786–796. 10.1038/mp.2016.44 [DOI] [PubMed] [Google Scholar]
- Zhou Y., Yu H., Guo Y., Chen C., Huang X., Gou Y., et al. (2019). Efficacy of acupuncture versus sham acupuncture for postpartum depression disorder: Study protocol for a randomized controlled trial. Eur. J. Integr. Med. 31:100982. 10.1016/j.eujim.2019.10098230658669 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The original contributions presented in the study are publicly available. This data can be found here: https://www.ncbi.nlm.nih.gov/, with accession number PRJNA637228.