Abstract
Cilia are plasma membrane protrusions that act as cellular antennae and propellers in eukaryotes. To achieve their sensory and motile functions, cilia maintain protein and lipid compositions that are distinct from those of the cell body. The transition zone (TZ) is a specialized region located at the ciliary base, which functions as a barrier separating the interior and exterior of cilia. The TZ comprises a number of transmembrane and soluble proteins. Meckel syndrome (MKS)1, B9 domain (B9D)1/MKS9, and B9D2/MKS10 are soluble TZ proteins that are encoded by causative genes of MKS and have a B9D in common. We here demonstrate the interaction mode of these B9D proteins to be MKS1–B9D2–B9D1 and demonstrate their interdependent localization to the TZ. Phenotypic analyses of MKS1-knockout (KO) and B9D2-KO cells show that the B9D proteins are involved in, although not essential for, normal cilia biogenesis. Rescue experiments of these KO cells show that formation of the B9D protein complex is crucial for creating a diffusion barrier for ciliary membrane proteins.
INTRODUCTION
Cilia are axonemal microtubule-based appendages that protrude from the surface of various eukaryotic cells and function as cellular antennae and propellers; in particular, cilia play crucial roles in embryonic development and adult tissue organization in vertebrates. To achieve these functions, cilia maintain distinct protein and lipid compositions in the ciliary membrane and cilioplasm from those of the plasma membrane and cytoplasm. The distinct compositions are accomplished by the ciliary gate, which is located at the proximal end of the axoneme and controls the entry and exit of ciliary proteins (Garcia-Gonzalo and Reiter, 2017; Gonçalves and Pelletier, 2017; Jensen and Leroux, 2017). This gate comprises transition fibers (TFs), which are equivalent to the distal appendages of the mother centriole-derived basal body, and the transition zone (TZ). Analyses using superresolution microscopy demonstrated the structural framework of the TZ (Yang et al., 2015; Shi et al., 2017). The importance of these structures has been confirmed by the fact that mutations in the genes for TFs and TZ components cause a broad spectrum of inherited disorders, generally called the ciliopathies (Braun and Hildebrandt, 2017; Reiter and Leroux, 2017), which include Meckel syndrome (MKS), Joubert syndrome, Bardet-Biedl syndrome (BBS), and nephronophthisis (NPHP).
Specific proteins are present on the ciliary membrane, including various G protein-coupled receptors (GPCRs) (Mukhopadhyay and Rohatgi, 2014; Schou et al., 2015; Nachury and Mick, 2019) and lipid-anchored proteins (Jensen and Leroux, 2017). The functions of cilia therefore depend on the selective entry and exit of proteins across the ciliary gate as well as on their proper intraciliary trafficking. Bidirectional trafficking of ciliary proteins is mediated by the intraflagellar transport (IFT) machinery, which is composed of two multisubunit complexes, IFT-A and IFT-B (Taschner and Lorentzen, 2016; Nakayama and Katoh, 2018). The IFT-B complex mediates anterograde trafficking from the base to the tip of cilia driven by kinesin-2, whereas the IFT-A complex mediates retrograde trafficking driven by dynein-2. Besides the IFT complexes, the BBSome complex, composed of eight BBS proteins, connects the IFT machinery to ciliary membrane proteins (Klink et al., 2017; Nachury, 2018; Wingfield et al., 2018; Nakayama and Katoh, 2020). In addition to mediating intraciliary protein trafficking, the IFT machinery and the BBSome regulate the entry and exit of ciliary proteins across the ciliary gate. For example, the BBSome mediates the export of GPCRs across the ciliary gate in mammalian cells (Eguether et al., 2014; Liew et al., 2014; Nozaki et al., 2018; Ye et al., 2018; Nozaki et al., 2019) and the export of phospholipase D in Chlamydomonas flagella (Liu and Lechtreck, 2018). The IFT-A complex and its associated protein TULP3 are involved in the entry of GPCRs into cilia (Mukhopadhyay et al., 2010; Badgandi et al., 2017; Hirano et al., 2017). Furthermore, recent lines of evidence in Caenorhabditis elegans suggest that the IFT-A complex and dynein-2 participate in the integrity of the TZ by yet unknown mechanisms (Jensen et al., 2018; Scheidel and Blacque, 2018).
The TZ is characterized by Y-shaped structures that span from the axoneme to the ciliary membrane. A combination of genetic interaction studies and biochemical analyses of protein–protein interactions has suggested that the TZ mainly comprises three distinct modules (Sang et al., 2011; Williams et al., 2011; Garcia-Gonzalo and Reiter, 2017; Gonçalves and Pelletier, 2017), that is, an MKS module and an NPHP module and a CEP290 module connecting these two modules. Consistent with the phenotypes of individuals with MKS and NPHP, loss-of-function studies of MKS and NPHP components showed that knockout (KO) mice and mutant mice of MKS components are generally embryonic lethal (Weatherbee et al., 2009; Dowdle et al., 2011; Garcia-Gonzalo et al., 2011; Chih et al., 2012; Roberson et al., 2015; Goetz et al., 2017), whereas those of NPHP components are not lethal (Jiang et al., 2008; Won et al., 2011). Despite the drastic phenotypes at the whole-body level, most MKS protein deficiencies have mild effects on ciliogenesis. At the cellular level, however, cells lacking MKS proteins often demonstrate severe defects in the ciliary localization of membrane proteins (Dowdle et al., 2011; Garcia-Gonzalo et al., 2011; Chih et al., 2012; Roberson et al., 2015; Slaats et al., 2016; Goetz et al., 2017), including INPP5E, GPR161, and Smoothened (SMO), the latter two of which are GPCRs involved in Hedgehog signaling.
The MKS module is composed of several transmembrane proteins (TMEM and TCTN proteins) and three nontransmembrane proteins, MKS1, B9D1/MKS9, and B9D2/MKS10, which share a B9 domain (B9D) belonging to the C2 domain superfamily (Bialas et al., 2009; Zhang and Aravind, 2010). Although previous studies suggested the presence of interactions among these B9D proteins (Dowdle et al., 2011; Chih et al., 2012), their modes of interaction have remained unclear. On the other hand, analyses using cells derived from mutant mice and using C. elegans and Drosophila mutants suggested an association among the localization of the three B9D proteins to the TZ (Williams et al., 2008; Bialas et al., 2009; Roberson et al., 2015; Pratt et al., 2016), although these studies did not address localization defects in the context of protein–protein interactions.
In this study, we demonstrate the interaction modes among the three B9D proteins using the visible immunoprecipitation (VIP) assay, which we developed as a convenient and flexible strategy to visually detect protein–protein interactions (Katoh et al., 2015, 2016). We then address the roles of B9D proteins as constituents of the ciliary diffusion barrier by establishing and analyzing CRISPR/Cas9-mediated KO cells and discuss the roles of these B9D proteins in the context of complex formation.
RESULTS
Interactions among the B9D proteins
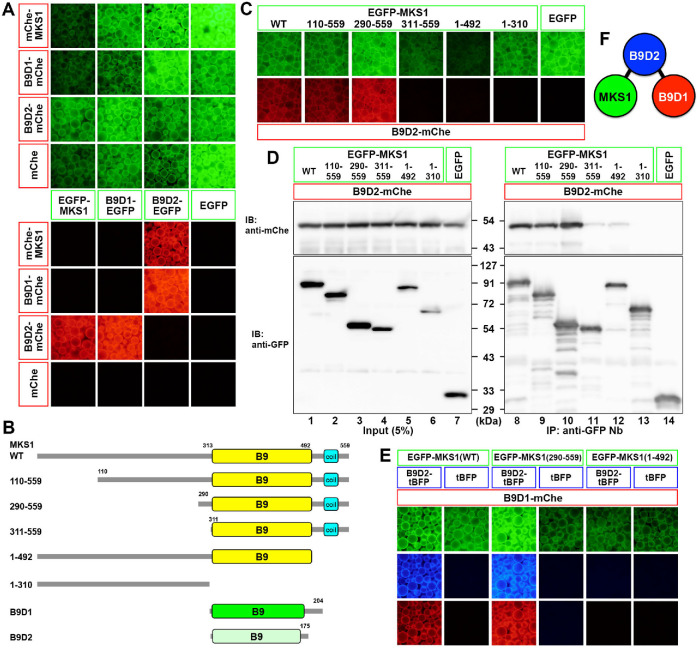
Utilizing the VIP assay, we first determined how the three B9D proteins interact with each other. Lysates were prepared from HEK293T cells coexpressing either EGFP-fused MKS1, B9D1, or B9D2, and any of their mCherry (mChe)-fused constructs in all possible combinations, and then processed for immunoprecipitation with a glutathione S-transferase (GST)-tagged anti-GFP nanobody (Nb) prebound to glutathione–Sepharose 4B beads. Red-fluorescent signals on the precipitated beads indicated that B9D2 interacts directly with MKS1 and B9D1, whereas the latter two proteins do not directly interact with each other (Figure 1A).
FIGURE 1:
Complex formation of B9D proteins. (A) Interactions of B9D proteins. Lysates prepared from HEK293T cells coexpressing the indicated combinations of EGFP-fused and mChe-fused MKS1, B9D1, and B9D2 were processed for the VIP assay using GST–anti-GFP Nb prebound to glutathione–Sepharose beads. EGFP and mChe signals on the precipitated beads were observed under a microscope. (B) Schematic representation of the domain organization of MKS1, B9D1, and B9D2 proteins, as well as the MKS1 deletion constructs used in this study. (C, D) Lysates prepared from HEK293T cells coexpressing B9D2-mChe and the indicated EGFP-MKS1 constructs were processed for the VIP assay (C) or conventional immunoblotting analysis (D). (E) Visible three-hybrid assay to confirm the linear MKS1–B9D2–B9D1 interaction. Lysates prepared from HEK293T cells triply coexpressing B9D1-mChe, either B9D2-tBFP or tBFP, and the indicated EGFP-MKS1 construct were processed for the VIP assay. (F) A model of the tripartite B9D protein complex.
As schematically shown in Figure 1B, the B9D occupies almost the entire length of the B9D1 and B9D2 proteins. On the other hand, in the MKS1 protein, there are approximately 310 and 70 amino acid extensions from the N- and C-terminus, respectively, of the B9D. Therefore, we then determined the region of the MKS1 protein that participates in its interaction with B9D2. As shown in Figure 1, C (VIP assay) and D (immunoblotting), although the N-terminal truncation mutants MKS1(110–559) and MKS1(290–559) retained the ability to interact with B9D2, the MKS1(311–559) construct almost completely lacked this ability. On the other hand, a C-terminal 67-amino acid truncation mutant MKS1(1-492), as well as the MKS1(1–310) construct, which lacks the entire B9D, lacked binding to B9D2. Thus, in addition to the B9D, the short N- and C-terminal extensions from the B9D are also required for the interaction of the MKS1 protein with B9D2.
The linear interaction of MKS1–B9D2–B9D1 was confirmed by the visible three-hybrid assay (Katoh et al., 2015); EGFP-MKS1(WT) and EGFP-MKS1(290–559) coimmunoprecipitated B9D1-mChe in the presence of TagBFP (tBFP)-fused B9D2 but not in the presence of tBFP (Figure 1E). By contrast, EGFP-MKS1(1–492) did not coimmunoprecipitate B9D1-mChe even in the presence of B9D2-tBFP (Figure 1E). Thus, we concluded that the B9D protein complex is composed of the linear interactions of MKS1–B9D2–B9D1 (Figure 1F).
MKS1-KO and B9D2-KO cells are moderately compromised with respect to ciliogenesis but unimpaired with respect to IFT particle trafficking
To analyze the roles of the B9D proteins, we then established KO cell lines of MKS1 and B9D2 from human telomerase reverse transcriptase-immortalized retinal pigment epithelial 1 (hTERT-RPE1) cells using the modified CRISPR/Cas9 system, which we previously reported (Katoh et al., 2017). For both MKS1 and B9D2, we chose two independent KO cell lines (#MKS1-2-2 and #MKS1-3-13 and #B9D2-1-5 and #B9D2-3-6, respectively) established using distinct target sequences; genomic characterization of these KO cell lines are summarized in Supplemental Figure S1.
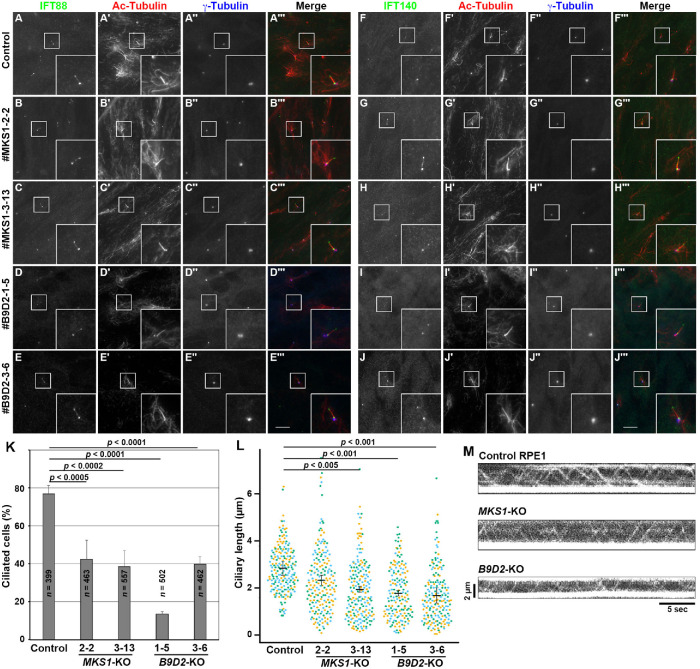
First, the MKS1-KO and B9D2-KO cell lines as well as control RPE1 cells were coimmunostained for acetylated α-tubulin (Ac-tubulin) and γ-tubulin (to visualize the axoneme and basal body, respectively) and either IFT88 (an IFT-B subunit; Figure 2, A–E) or IFT140 (an IFT-A subunit; Figure 2, F–J). Both the MKS1-KO and the B9D2-KO cell lines were moderately compromised with respect to ciliogenesis efficiency (Figure 2K) and the length of the formed cilia (Figure 2L); the moderate ciliogenesis defect was comparable to previous analyses of MEFs from MKS1 and B9D1 mutant mice (Dowdle et al., 2011; Goetz et al., 2017). However, localization of IFT88 and IFT140 in the formed cilia appeared normal. As in control cells, IFT88 was localized mainly to the base of cilia and a minor proportion to the distal tip (Figure 2, A–E), and IFT140 was localized mainly to the ciliary base (Figure 2, F–J) in both MKS1-KO and B9D2-KO cells. Furthermore, total internal reflection fluorescence (TIRF) microscopy of cells stably expressing tRFP-IFT88 showed that the IFT particles positive for tRFP-IFT88 undergo bidirectional movement in both MKS1-KO and B9D2-KO cells, as in control cells (Supplemental Videos S1–S3; also Figure 2M).
FIGURE 2:
Moderate ciliogenesis defects and normal localization of IFT components in the absence of MKS1 or B9D2. (A–J) Control RPE1 cells (A, F), the MKS1-KO cell lines #MKS1-2-2 (B, G) and #MKS1-3-13 (C, H), and the B9D2-KO cell lines #B9D2-1-5 (D, I) and #B9D2-3-6 (E, J) were serum starved for 24 h to induce ciliogenesis and triple immunostained for either IFT88 (A–E) or IFT140 (F–J), Ac-tubulin (A′–J′), and γ-tubulin (A′′–J′′). Insets are 2.5-fold-enlarged images of the boxed regions. Scale bars, 10 µm. (K) Ciliated cells of control cells and MKS1-KO and B9D2-KO cell lines were counted, and percentages of ciliated cells were represented as bar graphs. Values are means ± SD of three independent experiments. (L) Ciliary lengths of individual cells of the control cell line, the MKS1-KO cell lines, and the B9D2-KO cell lines were measured and expressed as scatter plots. Different colored dots represent three independent experiments, horizontal lines are means, and error bars are SD. Statistical significances were calculated using one-way ANOVA followed by the Dunnett multiple comparison test. (M) Kymograms showing the movement of tRFP-IFT88-positive particles in control RPE1 cells, MKS1-KO cells (#MKS1-3-13), and B9D2-KO cells (#B9D2-3-6). See Supplemental Videos S1–S3, respectively.
Movie S1.
TIRF microscopy of RPE1 cells expressing tRFP-IFT88
Movie S2.
TIRF microscopy of #MKS1-3-13 cells expressing tRFP-IFT88
Movie S3.
TIRF microscopy of #B9D2-3-6 cells expressing tRFP-IFT88
The normal localization and movement of IFT88 (an IFT-B subunit) in MKS1-KO and B9D2-KO cells were somewhat unexpected, because a previous study using the yeast two-hybrid system suggested an interaction of B9D2 with IFT70 (Zhao and Malicki, 2011); IFT70 is an IFT-B subunit that directly interacts with the IFT52–IFT88 dimer in the IFT-B complex (Katoh et al., 2016; Takei et al., 2018), and its deficiency results in severe ciliogenesis defects (Takei et al., 2018). However, the physiological relevance of the B9D2–IFT70 interaction was not analyzed in a previous study (Zhao and Malicki, 2011). We therefore attempted to confirm the B9D2–IFT70 interaction using the VIP assay. However, we were unable to show the binary interaction between B9D2 and IFT70, nor a composite interaction between the B9D protein complex and the IFT70–IFT52–IFT88 trimer (Supplemental Figure S2). Thus, it is unlikely that the B9D proteins make a major contribution to the assembly and trafficking of IFT particles. However, in view of the fact that in MKS1-KO and B9D2-KO cells the ciliogenesis efficiency was significantly reduced and the remaining cilia were shorter in length than in control RPE1 cells (Figure 2, K and L), the B9D proteins might participate in the loading to IFT particles of cargo proteins required for ciliary biogenesis, such as the αβ-tubulin dimer.
Defects in ciliary membrane localization of transmembrane and lipidated membrane proteins in MKS1-KO and B9D2-KO cells
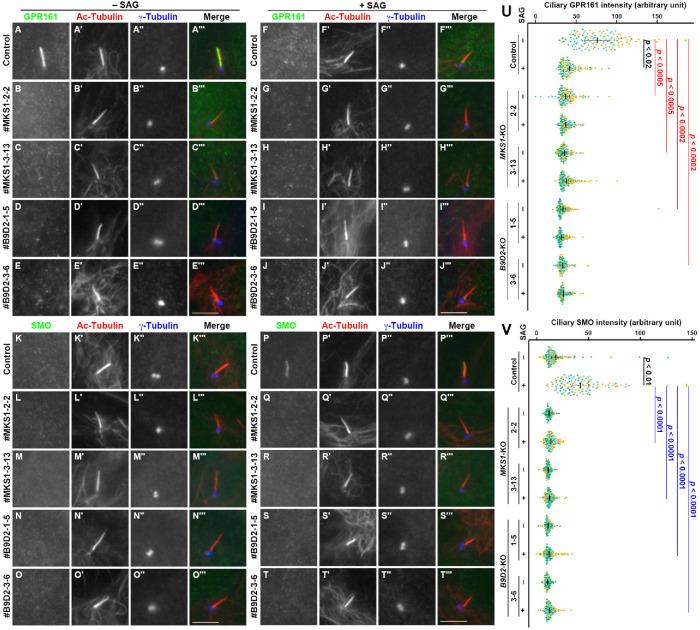
We then analyzed the localization of ciliary GPCRs in MKS1-KO and B9D2-KO cells. In control RPE1 cells, GPR161 localized to the ciliary membrane under basal conditions (Figure 3A), whereas its level in cilia was considerably reduced when cells were stimulated with SMO Agonist (SAG) (Figure 3F). In striking contrast, GPR161 was not detectable on the ciliary membrane, even under basal conditions in MKS1-KO or B9D2-KO cells (Figure 3, B–E; also see Figure 3U). On the other hand, SMO was absent from cilia in nonstimulated control cells (Figure 3K), whereas it entered cilia on stimulation with SAG (Figure 3P). In MKS1-KO and B9D2-KO cells, SMO was not found on the ciliary membrane even under SAG-stimulated conditions (Figure 3, Q–T; also see Figure 3V). These observations suggest that in the absence of MKS1 or B9D2, these ciliary GPCRs are unable to be retained on the ciliary membrane.
FIGURE 3:
Absence of ciliary GPCRs in MKS1-KO and B9D2-KO cells. (A–T) Control RPE1 cells (A, F, K, P), the #MKS1-2-2 cell line (B, G, L, Q), the #MKS1-3-13 cell line (C, H, M, R), the #B9D2-1-5 cell line (D, I, N, S), and the #B9D2-3-6 cell line (E, J, O, T) were serum starved for 24 h and incubated for a further 24 h without (–SAG; A–E, K–O) or with (+SAG; F–J, P–T) 200 nM SAG. The cells were triple immunostained with an antibody against either GPR161 (A–J) or SMO (K–T) and against Ac-tubulin (A′–T′) and γ-tubulin (A′′–T′′). Scale bars, 5 µm. (U, V) Relative ciliary staining intensities of GPR161 and SMO in control, MKS1-KO, and B9D2-KO cells were estimated and expressed as scatter plots. Different colored dots represent three independent experiments, horizontal lines are means, and error bars are SD. Statistical significances among multiple cell lines were calculated using one-way ANOVA followed by the Dunnett multiple comparison test, and those between two groups (–SAG and +SAG) were calculated using the Student t test.
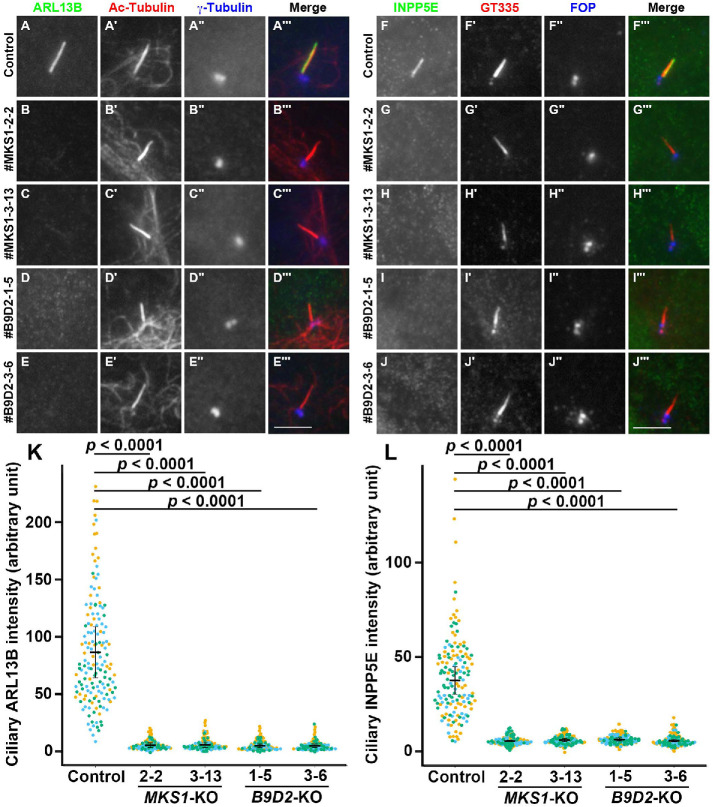
We also analyzed the localization of lipidated membrane proteins in MKS1-KO and B9D2-KO cells; ARL13B is a small GTPase that is anchored on the ciliary membrane via palmitoyl moieties attached to Cys residues located near the N-terminus (Roy et al., 2017), and INPP5E is a phosphoinositide phosphatase with an isoprenylated C-terminus (Humbert et al., 2012). As shown in Figure 4, both ARL13B and INPP5E were located in the cilia of control RPE1 cells (Figure 4, A and F, respectively). In striking contrast, the ciliary localization of these proteins was abolished in MKS1-KO and B9D2-KO cells (Figure 4, B–E and G–J; also see Figure 4, K and L). Thus, in addition to transmembrane proteins, lipidated membrane proteins are also unable to localize to the ciliary membrane in the absence of the B9D proteins.
FIGURE 4:
Delocalization of ciliary lipidated membrane proteins in the absence of MKS1 or B9D2. (A–J) Control RPE1 cells (A, F), the MKS1-KO cell lines #MKS1-2-2 (B, G) and #MKS1-3-13 (C, H), and the B9D2-KO cell lines #B9D2-1-5 (D, I) and #B9D2-3-6 (E, J) were serum starved for 24 h to induce ciliogenesis and triple immunostained with an antibody against either ARL13B (A–E) or INPP5E (F–J) and against either Ac-tubulin (A′–E′) or GT335 (F′–J′), which recognizes polyglutamylated tubulin, and against either γ-tubulin (A′′–E′′) or FOP (F′′–J′′). Scale bars, 5 µm. (K, L) The relative ciliary staining intensities of ARL13B and INPP5E in control, MKS1-KO, and B9D2-KO cells were estimated and expressed as scatter plots. Different colored dots represent three independent experiments, horizontal lines are means, and error bars are SD. Statistical significances among multiple cell lines were calculated using one-way ANOVA followed by the Dunnett multiple comparison test.
The abnormal phenotypes of MKS1-KO and B9D2-KO cell lines were rescued by the exogenous expression of MKS1 (Supplemental Figure S3 and Figure 5A; see below) and B9D2 (Supplemental Figure S3), respectively, excluding the possibility that the mislocalization of these membrane proteins in the KO cell lines resulted from off-target effects.
FIGURE 5:
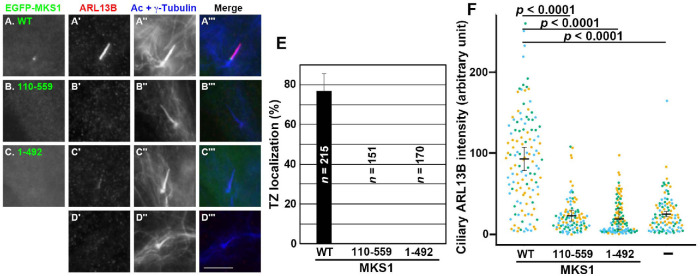
MKS1 deletion mutants lack the ability to localize to the TZ. (A–D) MKS1-KO cells (#MKS1-3-13) or those stably expressing an EGFP-fused MKS1 construct, as indicated, were immunostained for ARL13B, and Ac-tubulin and γ-tubulin. Ciliary staining for ARL13B was restored only by the exogenous expression of EGFP-MKS1(WT) (A′). Scale bar, 5 µm. (E) Cells with TZ localization of the MKS1 constructs shown in (A–C) were counted and expressed as bar graphs. (F) The relative ciliary staining intensities of ARL13B in MKS1-KO cells (–) and in those expressing the MKS1 construct, as indicated, were estimated and expressed as scatter plots. Different colored dots represent three independent experiments, horizontal lines are means, and error bars are SD. Statistical significances among multiple cell lines were calculated using one-way ANOVA followed by the Dunnett multiple comparison test.
Interdependent localization of B9D proteins to the TZ
As shown in Figure 1, MKS1 interacts with B9D2 via its B9D. To address whether the interaction of MKS1 with B9D2 is essential for its function, we expressed MKS1 constructs in MKS1-KO cells and analyzed their localization. As shown in Figure 5A, EGFP-MKS1(WT) expressed in MKS1-KO cells localized to the ciliary base and restored the ciliary localization of ARL13B (compare Figure 5, A′ with D′). By contrast, EGFP-fused MKS1(1–492), which lacks the ability to interact with B9D2 (Figure 1, C and D), did not localize to the TZ or restore ARL13B localization in MKS1-KO cells (Figure 5C). A somewhat unexpected result was that EGFP-fused MKS1(110–559) neither localized to the TZ nor restored ciliary ARL13B localization (Figure 5B), even though this MKS1 construct retained the ability to interact with B9D2 (Figure 1, C and D). These observations indicate that not only the region of MKS1 involved in B9D2 binding but also its N-terminal region with an unknown role is essential for the TZ localization and function of MKS1 (see Discussion).
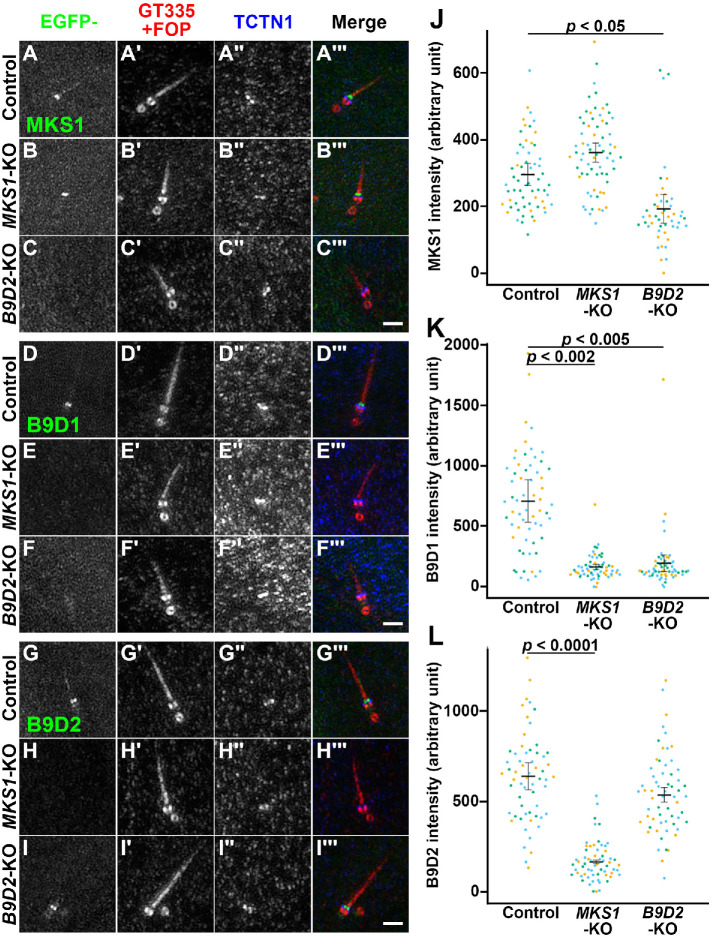
As the above experiments did not clarify whether the binding of MKS1 to B9D2 is essential for its TZ localization and function, we then analyzed the localization of endogenous MKS1 in B9D2-KO cells as well as in MKS1-KO cells. In control RPE1 cells, MKS1 was detected at the TZ (Figure 6A). By contrast, the staining for MKS1 at the TZ was abolished not only in MKS1-KO cells (Figure 6B) but also in B9D2-KO cells (Figure 6C). These observations demonstrate that the TZ localization of MKS1 is dependent on B9D2.
FIGURE 6:
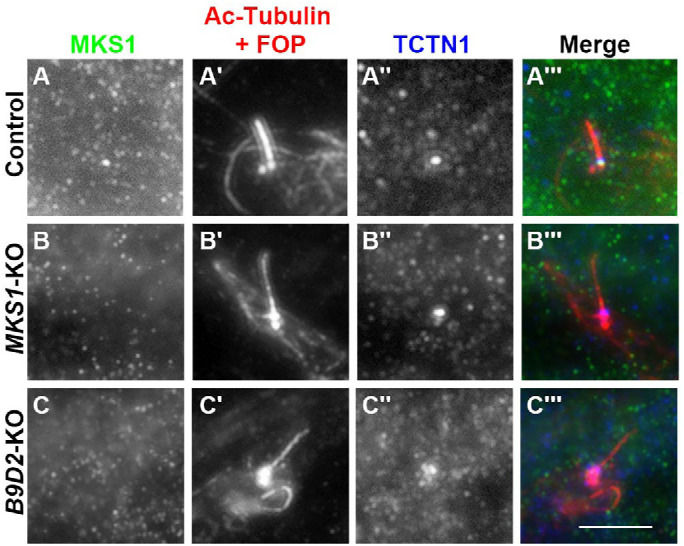
B9D2-dependent TZ localization of MKS1. Control RPE1 cells (A), MKS1-KO cells (#MKS1-3-13) (B), or B9D2-KO cells (#B9D2-3-6) (C) were stained with antibodies against MKS1 (A–C), and Ac-tubulin and FOP (A′–C′), and TCTN1 (A′′–C′′). Scale bar, 5 µm.
However, we could not examine whether B9D1 and B9D2 localize to the TZ in a manner dependent on other B9D proteins, as a commercially available antibody against B9D1 or B9D2 did not work well in immunofluorescence. We therefore examined the localization of EGFP-fused MKS1, B9D1, and B9D2 stably expressed in MKS1-KO and B9D2-KO cells; in these experiments, we observed cells by Airyscan superresolution microscopy to distinguish the localization of expressed proteins with that of TCTN1, which is another TZ protein. When EGFP-MKS1 was expressed, it was localized to the TZ in control RPE1 and MKS1-KO cells, although it was slightly more distal from the basal body marker FOP compared with TCTN1 (Figure 7, A and B, respectively). In striking contrast, EGFP-MKS1 was not localized to the TZ when expressed in B9D2-KO cells (Figure 7C; also see Figure 7J), indicating that the TZ localization of MKS1 depends on B9D2. B9D1-EGFP was localized to the TZ in control RPE1 cells (Figure 7D), but not in MKS1-KO or B9D2-KO cells (Figure 7, E and F; also see Figure 7K), indicating that the TZ localization of B9D1 depends not only on B9D2 but also on MKS1, which does not show a direct interaction with B9D1 (Figure 1, A and E). Finally, B9D2-EGFP was localized to the TZ in control RPE1 and B9D2-KO cells (Figure 7, G and I, respectively), but not in MKS1-KO cells (Figure 7H; also see Figure 7L). These observations demonstrate that the TZ localizations of the three B9D proteins are interdependent; namely, the formation of the MKS1–B9D2–B9D1 complex is necessary for the TZ localization of individual B9D proteins.
FIGURE 7:
Interdependent TZ localization of the B9D proteins. (A–I) Control RPE1 cells (A, D, G), MKS1-KO cells (#MKS1-3-13) (B, E, H), or B9D2-KO cells (#B9D2-3-6) (C, F, I) stably expressing EGFP-fused MKS1 (A–C), B9D1 (D–F), or B9D2 (G–I) were stained with FluoTag-X4 anti-GFP Nb (A–I), antibodies against GT335 and FOP (A′–I′), and against TCTN1 (A′′–I′′). The stained cells were then observed by Airyscan superresolution microscopy. Scale bars, 1 µm. (J–L) Relative TZ staining intensities of EGFP-fused MKS1 (J), B9D1 (K), and B9D2 (L) in the images acquired by confocal laser-scanning microscopy were estimated and expressed as scatter plots. In the scatter plots, different colored dots represent three independent experiments, horizontal lines are means, and error bars are SD. Statistical significances among multiple cell lines were calculated using one-way ANOVA followed by the Dunnett multiple comparison test.
DISCUSSION
In this study, we demonstrated that B9D2 interacts with MKS1 and B9D1 to form a tripartite complex, namely, the linear MKS1–B9D2–B9D1 complex shown in Figure 1F. The B9Ds of these B9D proteins mainly participate in complex formation. MKS1-KO and B9D2-KO cells demonstrated essentially the same phenotypes, in particular, mild defects in ciliogenesis (Figure 2), the absence of integral and lipid-anchored membrane proteins on the ciliary membrane (Figures 3 and 4), and apparently normal localization and trafficking of components of the IFT machinery (Figure 2). Furthermore, the three B9D proteins were found to localize to the TZ in an interdependent manner, as B9D1 and B9D2 did not localize to the TZ in MKS1-KO cells, and MKS1 and B9D1 did not localize to the TZ in B9D2-KO cells (Figures 6 and 7). On the basis of these observations, we conclude that formation of the heterotrimeric B9D complex is essential for the TZ to function as a diffusion barrier for membrane proteins. On the other hand, these B9D proteins appear to be dispensable for formation of the permeability barrier, as the trafficking of the IFT machinery appeared normal. Thus, although the B9D proteins are soluble proteins, they are essential for constituting a barrier for membrane proteins, but are not essential for the barrier for soluble proteins. However, these B9D proteins may make only a minor contribution to the permeability barrier of some soluble proteins, as cilia biogenesis is mildly impaired in MKS1-KO and B9D2-KO cells; tubulins and some other axonemal components within cilia may not be maintained at proper levels in the absence of any of the B9D proteins.
Our data on the interdependent TZ localization of the three B9D proteins are consistent with a previous study on Drosophila spermatocyte cilia, in which the TZ localization of B9D1 and B9D2 was impaired in an mks1 mutant, and the TZ localization of MKS1 and B9D2 was impaired in a b9d1 mutant (Pratt et al., 2016). However, our data are partly incompatible with two previous studies on C. elegans neuronal cilia; compared with the WT worm strain, the TZ localization of B9D1 and B9D2 remained unchanged in mks1 mutant strains (Williams et al., 2008; Bialas et al., 2009). Although we do not know the exact reason for the apparent discrepancy, the complexity of the TZ assembly might be variable among species, as previously proposed (Wiegering et al., 2018). Another possibility is that the roles of B9D proteins might be somewhat different in C. elegans, as the disruption of any of the B9D protein genes in C. elegans had no apparent effects on chemosensation (Williams et al., 2008; Bialas et al., 2009).
For MKS1, its interactions not only with B9D2 to form the B9D complex but also with other TZ proteins are essential for its function as a component of the diffusion barrier, because an N-terminal deletion mutant of MKS1 that retains the ability to interact with B9D2 was unable to rescue the abnormal phenotype of MKS1-KO cells. Some studies based on protein interactions and proximity-based labeling suggested potential interactions of B9D proteins, directly or indirectly, with other TZ proteins and non-TZ proteins (Gupta et al., 2015; Roberson et al., 2015; Boldt et al., 2016; Huttlin et al., 2017). However, as our efforts utilizing the VIP assay to identify potential interacting partners of the B9D proteins other than themselves have been unsuccessful so far, the interactions of B9D proteins with other proteins might involve multipartite and composite complex formation.
As the B9D proteins are soluble proteins, the molecular basis of the B9D complex as a diffusion barrier for membrane proteins remains unclear. Among the TZ components, the MKS module contains transmembrane proteins (TMEM and TCTN proteins) in addition to the B9D proteins, whereas the NPHP and CEP290 modules are constituted only by soluble proteins (Garcia-Gonzalo and Reiter, 2017; Gonçalves and Pelletier, 2017). Previous superresolution imaging studies demonstrated that MKS proteins, including MKS1, are located near the ciliary membrane, whereas NPHP proteins are located near the axoneme (Yang et al., 2015; Shi et al., 2017). Overall, the B9D complex might constitute the outmost portion of the Y-links or the ciliary necklace (Gilula and Satir, 1972) and connect them to the transmembrane proteins of the MKS module (Garcia-Gonzalo and Reiter, 2017).
MATERIALS AND METHODS
Plasmids, antibodies, and reagents
cDNAs for human MKS1 (NM_017777), B9D1 (NM_015681), and B9D2 (NM_030578) were obtained from a cDNA library by PCR amplification. Expression vectors for IFT proteins used in this study are listed in Supplemental Table S1; all of them were constructed in our previous studies (Katoh et al., 2016). Antibodies used in this study are listed in Supplemental Table S2. GST-tagged anti-GFP Nb prebound to glutathione–Sepharose 4B beads were prepared as described previously (Katoh et al., 2015, 2018). SAG was purchased from ENZO Life Sciences.
VIP assay and immunoblotting analysis
The VIP assay and subsequent immunoblotting analyses were performed as described previously (Katoh et al., 2015, 2016) with a minor modification (Nishijima et al., 2017); experimental details of the VIP assay have been described previously (Katoh et al., 2018). Briefly, HEK293T cells on a 6-well plate (approximately 1.6 × 106 cells) were cotransfected with expression vectors for EGFP, mChe/tRFP, and tBFP fusion constructs using Polyethylenimine Max (Polysciences) and cultured for 24 h in DMEM with high glucose (Nacalai Tesque) supplemented with 5% fetal bovine serum (FBS). The transfected cells were lysed in 250 µl of HMDEKN cell-lysis buffer (10 mM HEPES, pH 7.4, 5 mM MgSO4, 1 mM dithiothreitol, 0.5 mM EDTA, 25 mM KCl, 0.05% NP-40) containing protease inhibitor cocktail (Nacalai Tesque). After 15 min on ice, the lysates were centrifuged at 16,100 × g for 15 min. The supernatant (200 µl) was transferred to a 0.2-ml 8-tube strip containing GST-tagged anti-GFP Nb bound to glutathione–Sepharose 4B beads (approximately 5 µl bed volume of the beads), and incubated for 1 h at 4°C with constant rotation of the tube. After centrifugation of the tube at 2,000 × g for 30 s, the precipitated beads were washed three times with lysis buffer (180 µl), transferred to a 96-well glass-bottom plate, and observed using an all-in-one type microscope (BZ-8000, KEYENCE) with a 20× /0.75 NA objective lens under fixed conditions (sensitivity: ISO 400; exposure: 1/30 or 1/15 s for green fluorescence; sensitivity: ISO 800; exposure: 1/10 or 1/15 s for red fluorescence; and sensitivity: ISO 800; exposure: 1/5 s for blue fluorescence), unless otherwise noted.
The beads bearing fluorescent fusion proteins were then processed for conventional immunoblotting analysis. Proteins on the beads were denatured by boiling, separated by SDS–PAGE, and electroblotted onto an Immobilon-P membrane (Merck Millipore). The membrane was blocked in 5% skimmed milk and incubated sequentially with primary antibody and peroxidase–conjugated secondary antibody. Protein bands were detected using a Chemi-Lumi One L kit (Nacalai Tesque).
Immunofluorescence analysis and TIRF microscopy
hTERT-RPE1 cells (CRL-4000, American Type Culture Collection) were grown in DMEM/F-12 (Nacalai Tesque) supplemented with 10% FBS and 0.348% sodium bicarbonate. To induce ciliogenesis, cells were grown on coverslips to 100% confluence and serum starved for 24 h in Opti-MEM containing 0.2% bovine serum albumin.
Immunofluorescence analysis was performed as described previously (Takahashi et al., 2012; Funabashi et al., 2018; Nozaki et al., 2018). For staining of endogenous MKS1, cells were fixed with 10% trichloroacetic acid and permeabilized with 0.1% Triton X-100. The immunostained cells were observed using an AxioObserver microscope (Carl Zeiss) or an A1R-MP confocal laser-scanning microscope (Nikon). Statistical analyses were performed using JMP Pro 14 software (SAS Institute).
Airyscan superresolution imaging was performed using the LSM800 microscope (Carl Zeiss). The images taken in superresolution Airyscan mode were Airy-processed in 3D with a strength value of 7.0–7.5. Image stacks were collected with a z-step size of 0.18 µm and processed with Zen software (Carl Zeiss) to build superresolution and maximum projection images.
TIRF microscopy was performed as described previously (Kubo et al., 2015; Hamada et al., 2018). Control RPE1, MKS1-KO, and B9D2-KO cells expressing tRFP-IFT88 were serum starved for 24 h on a glass-bottom culture dish, placed on a microscope stage prewarmed to 37°C, and observed using a TIRFM ECLIPSE Ti microscope (Nikon) at a video rate using NIS-Elements imaging software. Kymograms were generated with NIS-Elements imaging software.
Establishment of MKS1-KO and B9D2-KO cell lines using the CRISPR/Cas9 system
The CRISPR/Cas9-mediated KO strategy using homology-independent DNA repair (version 2 method) was previously described in detail (Katoh et al., 2017; also see Takahara et al., 2018; Takei et al., 2018; Tsurumi et al., 2019). The single-guide RNA (sgRNA) sequence targeting the human MKS1 or B9D2 gene (see Supplemental Table S3) was designed using CRISPOR (Haeussler et al., 2016). Double-stranded oligonucleotides for the target sequences were inserted into the all-in-one sgRNA expression vector peSpCAS9(1.1)-2 × sgRNA (Addgene 80768). hTERT-RPE1 cells grown on a 12-well plate (approximately 3.0 × 105 cells) were transfected with the sgRNA vector (1 µg) and the donor knock-in vector, pDonor-tBFP-NLS-Neo(universal) (0.25 µg; Addgene 80767), using X-tremeGENETM 9 DNA Transfection Reagent (Roche Applied Science). After the transfected cells were cultured in the presence of G418 (600 µg/ml), cells containing nuclear tBFP signals were isolated under a microscope or using an SH800 Series cell sorter (SONY). Genome DNA extracted from the isolated cells was subjected to PCR using GoTaq Master Mixes (Promega) and three sets of primers (Supplemental Table S3) to distinguish the following three states of integration of the donor knock-in vector: forward integration, reverse integration, and no integration with a small indel (for examples, see Hirano et al., 2017). Disruption of both alleles of the MKS1 or B9D2 gene was confirmed by direct sequencing of the PCR products.
Preparation of cells stably expressing EGFP-fused MKS1, B9D2, and B9D1 constructs
Lentiviral expression vectors for MKS1, B9D2, and B9D1 constructs were prepared in a previously described manner (Takahashi et al., 2012). Briefly, a pRRLsinPPT-based expression vector was transfected into HEK293T cells with packaging vectors (pRSV-REV, pMD2.g, and pMDLg/pRRE; gifts from Peter McPherson, McGill University [ Thomas et al., 2009]). Culture media were replaced 8 h after transfection, and those containing viral particles were collected at 24, 36, and 48 h after transfection. The collected media were passed through a 0.45-µm filter and centrifuged at 32,000 × g at 4°C for 4 h. Precipitated viral particles were resuspended in Opti-MEM (Invitrogen). KO cells expressing an EGFP-fused MKS1, B9D2, or B9D1 construct were prepared by the addition of a lentiviral suspension into the culture medium, followed by a 24-h incubation. These cells were used for immunofluorescence analysis.
Supplementary Material
Acknowledgments
We thank Peter McPherson for providing plasmid vectors for recombinant lentivirus production, Shohei Nozaki and Fumiyoshi Ishidate for technical assistance and advice, and Helena Akiko Popiel for critical reading of the manuscript. This work was supported in part by grants from the Japan Society for the Promotion of Science (JSPS) (grant numbers 15H04370 and 19H00980 to K.N. and 15K07929 and 18H02403 to Y.K.), a grant of JRPs-LEAD with UKRI from JSPS, a grant from the Uehara Memorial Foundation to Y.K., and the Kyoto University internal grant ISHIZUE to K.N. We used Airyscan microscopy at the Research Support Platform in Osaka City University and the SH800 Series cell sorter at the Medical Research Support Center in Graduate School of Medicine, Kyoto University.
Abbreviations used:
- BBS
Bardet-Biedl syndrome
- B9D
B9 domain
- FBS
fetal bovine serum
- GPCR
G protein-coupled receptor
- GST
glutathione S-transferase
- hTERT-RPE1
human telomerase reverse transcriptase-immortalized retinal pigment epithelial 1
- IFT
intraflagellar transport
- KO
knockout
- mChe
mCherry
- MKS
Meckel syndrome
- Nb
nanobody
- NPHP
nephronophthisis
- SAG
Smoothened Agonist
- sgRNA
single-guide RNA
- SMO
Smoothened
- tBFP
TagBFP
- TF
transition fibers
- TIRF
total internal reflection fluorescence
- TZ
transition zone
- VIP
visible immunoprecipitation.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E20-03-0208) on July 29, 2020.
REFERENCES
- Badgandi HB, Hwang S, Shimada IS, Loriot E, Mukhopadhyay S. (2017). Tubby family proteins are adaptors for ciliary trafficking of integral membrane proteins. J Cell Biol , 743–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bialas NJ, Inglis PN, Li C, Robinson JF, Parker JDK, Healey MP, Davis EE, Inglis CD, Cottell DC, Blacque OE, et al. (2009). Functional interactions between the ciliopathy-associated Meckel syndrome 1 (MKS1) protein and two novel MKS1-related (MKSR) proteins. J Cell Sci , 611–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boldt K, van Reeuwijk J, Lu Q, Koutroumpas K, Nguyen TM, Texier Y, van Beersum SEC, Horn N, Willer JR, Mans D, et al. (2016). An organelle-specific protein landscape identifies novel diseases and molecular mechanisms. Nat Commun , 11491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun DA, Hildebrandt F. (2017). Ciliopathies. Cold Spring Harb Perspect Biol , a028191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chih B, Liu P, Chinn Y, Chalouni C, Komuves LG, Hass PE, Sandoval W, Peterson AS. (2012). A ciliopathy complex at the transition zone protects the cilia as a privileged membrane domain. Nat Cell Biol , 61–72. [DOI] [PubMed] [Google Scholar]
- Dowdle WE, Robinson JF, Kneist A, Sirerol-Piquer MS, Frints SG, Corbit KC, Zaghloul NA, van Lijnschoten G, Mulders L, Verver DE, et al. (2011). Disruption of a ciliary B9 protein complex causes Meckel syndrome. Am J Hum Genet , 94–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eguether T, San Agustin JT, Keady BT, Jonassen JA, Liang Y, Francis R, Tobita K, Johnson CA, Abdelhamed ZA, Lo CW, Pazour GJ. (2014). IFT27 links the BBSome to IFT for maintenance of the ciliary signaling compartment. Dev Cell , 279–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funabashi T, Katoh Y, Okazaki M, Sugawa M, Nakayama K. (2018). Interaction of heterotrimeric kinesin-II with IFT-B-connecting tetramer is crucial for ciliogenesis. J Cell Biol , 2867–2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Gonzalo FR, Cotbit KC, Sirerol-Piquer MS, Ramaswami G, Otto EA, Noriega TR, Seol AD, Robinson JF, Bennett CL, Josifova DJ, et al. (2011). A transition zone complex regulates mammalian ciliogenesis and ciliary membrane composition. Nat Genet , 776–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Gonzalo FR, Reiter JF. (2017). Open sesame: how transition fibers and the transition zone control ciliary composition. Cold Spring Harb Perspect Biol , a028134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilula NB, Satir P. (1972). The ciliary necklace. A ciliary membrane specialization. J Cell Biol , 494–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetz SC, Bangs F, Barrington CL, Katsanis N, Anderson KV. (2017). The Meckel syndrome-associated protein MKS1 functionally interacts with components of the BBSome and IFT complexes to mediate ciliary trafficking and hedgehog signaling. PLoS One , e0173399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonçalves J, Pelletier L. (2017). The ciliary transition zone: finding the pieces and assembling the gate. Mol Cells , 243–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta GD, Coyaud É, Gonçalves J, Mojarad BA, Liu Y, Wu Q, Gheiratmand L, Comartin D, Tkach JM, Cheung SW, et al. (2015). A dynamic protein interaction landscape of the human centrosome-cilium interface. Cell , 1484–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeussler M, Schönig K, Eckert H, Eschstruth A, Mianné J, Renaud JB, Schneider-Maunoury S, Shkumatava A, Teboul L, Kent J, et al. (2016). Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR. Genome Biol , 148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada Y, Tsurumi Y, Nozaki S, Katoh Y, Nakayama K. (2018). Interaction of WDR60 intermediate chain with TCTEX1D2 light chain of the dynein-2 complex is crucial for ciliary protein trafficking. Mol Biol Cell , 1628–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T, Katoh Y, Nakayama K. (2017). Intraflagellar transport-A complex mediates ciliary entry as well as retrograde trafficking of ciliary G protein-coupled receptors. Mol Biol Cell , 429–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humbert MC, Weihbrecht K, Searby CC, Li Y, Pope RM, Sheffield VC, Seo S. (2012). ARL13B, PDE6D, and CEP164 form a functional network for INPP5E ciliary targeting. Proc Natl Acad Sci USA , 19691–19696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttlin EL, Bruckner RJ, Paulo JA, Cannon JR, Ting L, Baltier K, Colby G, Gebreab F, Gygi MP, Parzen H, et al. (2017). Architecture of the human interactome defines protein communities and disease networks. Nature , 505–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen VL, Lambacher NJ, Li C, Mohan S, Williams CL, Inglis PN, Yoder BK, Blacque OE, Leroux MR. (2018). Role for intraflagellar transport in building a functional transition zone. EMBO Rep , e45862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen VL, Leroux MR. (2017). Gates for solubule and membrane proteins, and two trafficking systems (IFT and LIFT), establish a dynamic ciliary signaling compartment. Curr Opin Cell Biol , 83–91. [DOI] [PubMed] [Google Scholar]
- Jiang S-T, Chiou Y-Y, Wang E, Lin H-K, Lee S-P, Lu H-Y, Wang C-KL, Tang M-J, Li H. (2008). Targeted disruption of Nphp1 causes male infirtility due to defects in the later steps of sperm morphogenesis in mice. Hum Mol Genet , 3368–3379. [DOI] [PubMed] [Google Scholar]
- Katoh Y, Michisaka S, Nozaki S, Funabashi T, Hirano T, Takei R, Nakayama K. (2017). Practical method for targeted disruption of cilia-related genes by using CRISPR/Cas9-mediated homology-independent knock-in system. Mol Biol Cell , 898–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh Y, Nakamura K, Nakayama K. (2018). Visible immunoprecipitation (VIP) assay: a simple and versatile method for visual detection of protein-protein interactions. Bio-protocol , e2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh Y, Nozaki S, Hartanto D, Miyano R, Nakayama K. (2015). Architectures of multisubunit complexes revealed by a visible immunoprecipitation assay using fluorescent fusion proteins. J Cell Sci , 2351–2362. [DOI] [PubMed] [Google Scholar]
- Katoh Y, Terada M, Nishijima Y, Takei R, Nozaki S, Hamada H, Nakayama K. (2016). Overall architecture of the intraflagellar transport (IFT)-B complex containing Cluap1/IFT38 as an essential component of the IFT-B peripheral subcomplex. J Biol Chem , 10962–10975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klink BU, Zent E, Juneja P, Kuhlee A, Rausner S, Wittinghofer A. (2017). A rocombinant BBSome core complex and how it interacts with ciliary cargo. eLife , e27434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo K, Kobayashi M, Nozaki S, Yagi C, Hatsuzawa K, Katoh Y, Shin H-W, Takahashi S, Nakayama K. (2015). SNAP23/25 and VAMP2 mediate exocytic event of transferrin receptor-containing recycling vesicles. Biol Open , 910–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liew GM, Ye F, Nager AR, Murphy JP, Lee JSH, Aguiar M, Breslow DK, Gygi SP, Nachury MV. (2014). The intraflagellar transport protein IFT27 promotes BBSome exit from cilia through the GTPase ARL6/BBS3. Dev Cell , 265–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Lechtreck KF. (2018). The Bardet-Biedl syndrome protein complex is an adaptor expanding the cargo of range of intraflagellar transport trains for ciliary export. Proc Natl Acad Sci USA , E934–E943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay S, Rohatgi R. (2014). G-protein-coupled receptors, Hedgehog signaling and primary cilia. Sem Cell Dev Biol , 63–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay S, Wen X, Chih B, Nelson CD, Lane WS, Scales SJ, Jackson PK. (2010). TULP3 bridges the IFT-A complex and membrane phosphoinositides to promote trafficking of G protein-coupled receptors into primary cilia. Genes Dev , 2180–2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachury MV. (2018). The molecular machines that traffic signaling receptors into and out of cilia. Curr Opin Cell Biol , 124–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachury MV, Mick DU. (2019). Establishing and regulating the composition of cilia for signal transduction. Nat Rev Mol Cell Biol , 389–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama K, Katoh Y. (2018). Ciliary protein trafficking mediated by IFT and BBSome complexes with the aid of kinesin-2 and dynein-2 motors. J Biochem , 155–164. [DOI] [PubMed] [Google Scholar]
- Nakayama K, Katoh Y. (2020). Architecture of the IFT ciliary trafficking machinery and interplay between its components. Crit Rev Biochem Mol Biol , 179–196. [DOI] [PubMed] [Google Scholar]
- Nishijima Y, Hagiya Y, Kubo T, Takei R, Katoh Y, Nakayama K. (2017). RABL2 interacts with the intraflagellar transport B complex and CEP19 and participates in ciliary assembly. Mol Biol Cell , 1652–1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozaki S, Castro Araya RF, Katoh Y, Nakayama K. (2019). Requirement of IFT-B–BBSome complex interaction in export of GPR161 from cilia. Biol Open , bio043786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozaki S, Katoh Y, Kobayashi T, Nakayama K. (2018). BBS1 is involved in retrograde trafficking of ciliary GPCRs in the context of the BBSome complex. PLoS One , e0195005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt MB, Titlow JS, Davis I, Barker AR, Dawe HR, Raff JW, Roque H. (2016). Drosophila sensory cilia lacking MKS proteins exhibit striking defects in development but only subtle defects in adults. J Cell Sci , 3732–3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter JF, Leroux MR. (2017). Genes and molecular pathways underpinning ciliopathies. Nat Rev Mol Cell Biol , 533–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberson EC, Dowdle WE, Ozanturk A, Garcia-Gonzalo FR, Li C, Halbritter J, Elkhartoufi N, Porath JD, Cope H, Ashley-Koch A, et al. (2015). TMEM231, mutated in orofaciodigital and Meckel syndromes, organizes the ciliary transition zone. J Cell Biol , 129–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy K, Jerman S, Jozsef L, McNamara T, Onyekaba G, Sun Z, Marin EP. (2017). Palmitoylation of the ciliary GTPase ARL13b is necessary for its stability and its role in cilia formation. J Biol Chem , 17703–17717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sang L, Miller AL, Corbit KC, Giles RH, Brauer MJ, Otto EA, Baye LM, Wen X, Scales SJ, Kwong M, et al. (2011). Mapping the NPHP-JBTS-MKS protein network reveals ciliopathy disease genes and pathways. Cell , 513–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheidel N, Blacque O. (2018). Intraflagellar transport complex A genes differentially regulate cilium formation and transition zone gating. Curr Biol , 3279–3287. [DOI] [PubMed] [Google Scholar]
- Schou KB, Pedersen LB, Christensen ST. (2015). Ins and outs of GPCR signaling in primary cilia. EMBO Rep , 1099–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X, Garcia G, III, Van De Weghe JC, McGorty R, Pazour GJ, Doherty D, Huang B, Reiter JF. (2017). Super-resolution microscopy reveals that disruption of ciliary transition-zone architecture causes Joubert syndrome. Nat Cell Biol , 1178–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slaats GG, Isabella CR, Koes HY, Dempsey JC, Gremmels H, Monroe GR, PhelpsIan G, Duran KJ, Adkins J, Kumar SA, et al. (2016). MKS1 regulates ciliary INPP5E levels in Joubert syndrome. J Med Genet , 62–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahara M, Katoh Y, Nakamura K, Hirano T, Sugawa M, Tsurumi Y, Nakayama K. (2018). Ciliopathy-associated mutations of IFT122 impair ciliary protein trafficking but not ciliogenesis. Hum Mol Genet , 516–528. [DOI] [PubMed] [Google Scholar]
- Takahashi S, Kubo K, Waguri S, Yabashi A, Shin H-W, Katoh Y, Nakayama K. (2012). Rab11 regulates exocytosis of recycling vesicles at the plasma membrane. J Cell Sci , 4049–4057. [DOI] [PubMed] [Google Scholar]
- Takei R, Katoh Y, Nakayama K. (2018). Robust interaction of IFT70 with IFT52–IFT88 in the IFT-B complex is required for ciliogenesis. Biol Open , bio033241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taschner M, Lorentzen E. (2016). The intraflagellar transport machinery. Cold Spring Harb Perspect Biol , a028092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas S, Ritter B, Verbich D, Sanson C, Bourbonnière L, McKinney RA, McPherson PS. (2009). Intersectin regulates dendritic spine development and somatodendritic endocytosis but not synaptic vesicle recycling in hippocampal neurons. J Biol Chem , 12410–12419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsurumi Y, Hamada Y, Katoh Y, Nakayama K. (2019). Interactions of the dynein-2 intermediate chain WDR34 with the light chains are required for ciliary retrograde protein trafficking. Mol Biol Cell , 658–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weatherbee SD, Niswander LA, Anderson KV. (2009). A mouse model for Meckel syndrome reveals Mks1 is required for ciliogenesis and Hedgehog signaling. Hum Mol Genet , 4565–4575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiegering A, Dildrop R, Kalfhues L, Spychala A, Kuschel S, Lier JM, Zobel T, Dahmen S, Leu T, Struchtrup A, et al. (2018). Cell type-specific regulation of ciliary transition zone assembly in vertebrates. EMBO J , e97791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams CL, Li C, Kida K, Inglis PN, Mohan S, Semenec L, Bialas NJ, Stupay RM, Chen N, Blacque OE, et al. (2011). MKS and NPHP modules cooperate to establish basal body/transition zone membrane associations and ciliary gate function during ciliogenesis. J Cell Biol , 1023–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams CL, Winkelbauer ME, Schafer JC, Michaud EJ, Yoder BK. (2008). Functional redundancy of the B9 proteins and nephrocystins in Caenorhabditiselegans ciliogenesis. Mol Biol Cell , 2154–2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingfield JL, Lechtreck K-F, Lorentzen E. (2018). Trafficking of ciliary membrane proteins by the intraflagellar transport/BBSome machinery. Essays Biochem , 753–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Won J, de Evsikova CM, Smith RS, Hicks WL, Edwards MM, Longo-Guess C, Li T, Naggert JK, Nishina PM. (2011). NPHP4 is necessary for normal photoreceptor ribbon synapse maintenance and outer segment formation, and for sperm development. Hum Mol Genet , 482–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang TT, Su J, Wang W-J, Craige B, Witman GB, Tsou M-FB, Liao J-C. (2015). Superresolution pattern recognition reveals the architectural map of the ciliary transition zone. Sci Rep , 14096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye F, Nager AR, Nachury MV. (2018). BBSome trains remove activated GPCRs from cilia by enabling passage through the transition zone. J Cell Biol , 1847–1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Aravind L. (2010). Identification of novel families and classification of the C2 domain superfamily elucidate the origin and evolution of membrane targeting activities in eukaryotes. Gene , 18–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Malicki J. (2011). Nephrocyctins and MKS proteins interact with IFT particle and facilitate transport of selected ciliary cargos. EMBO J , 2532–2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.